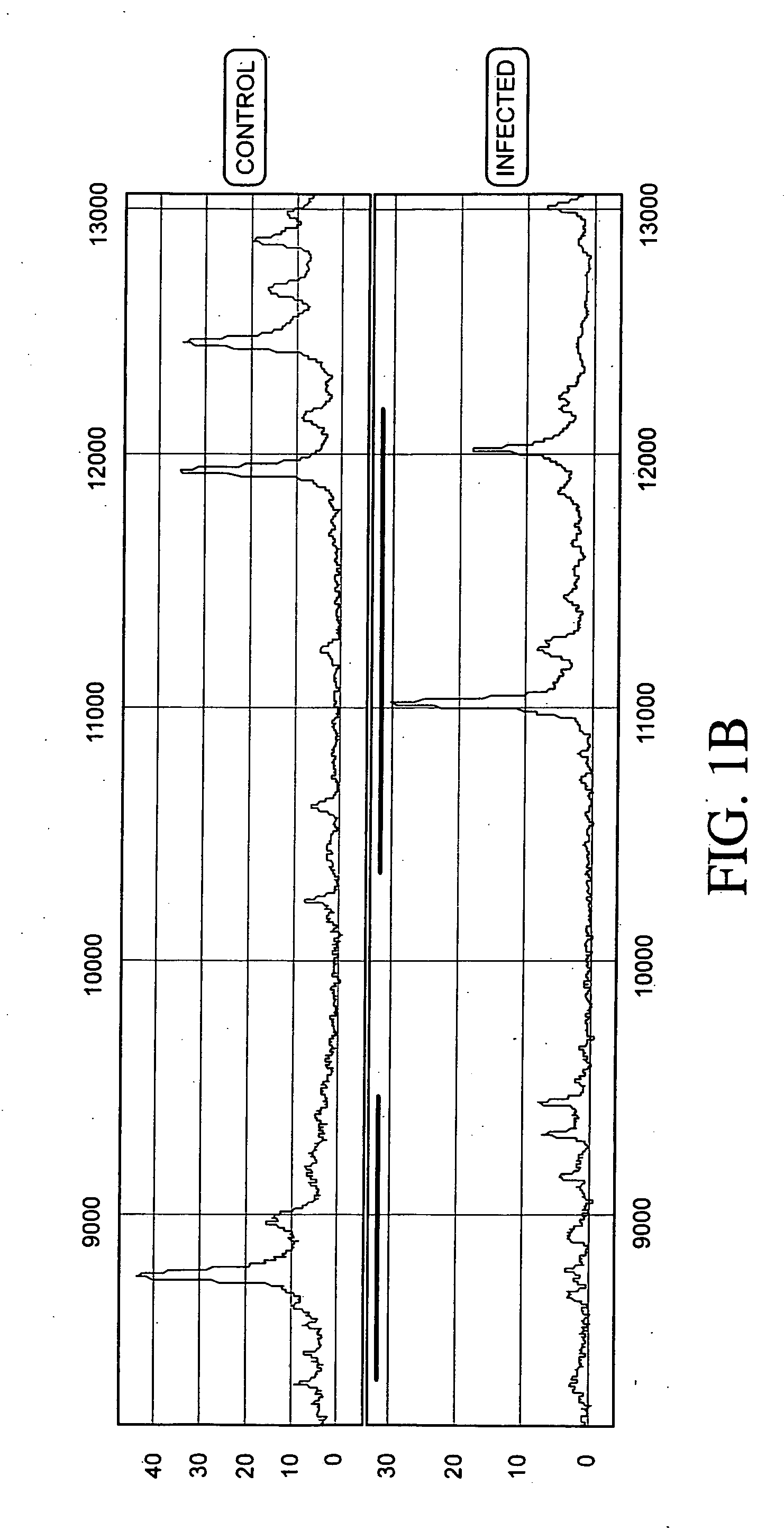
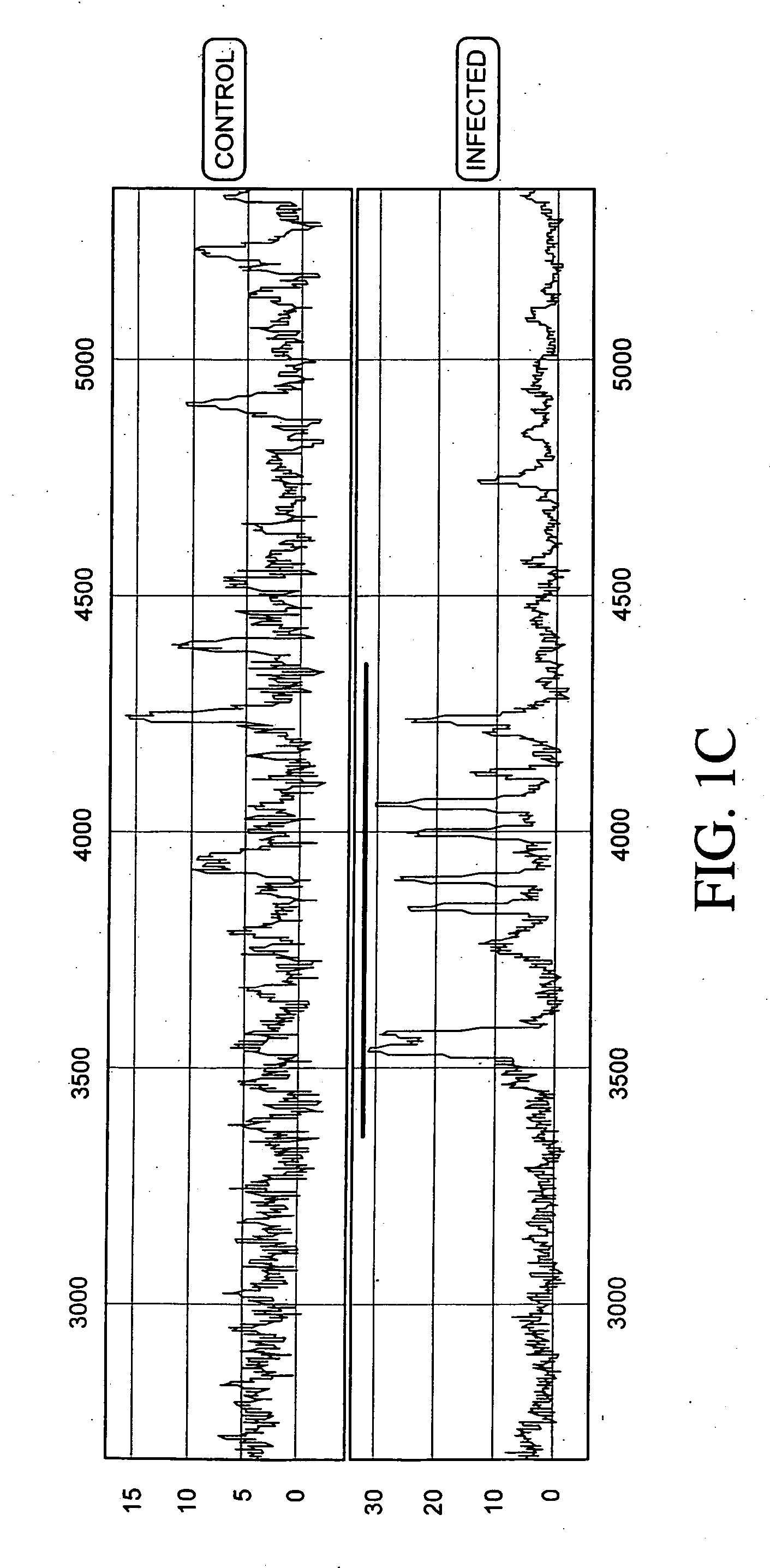
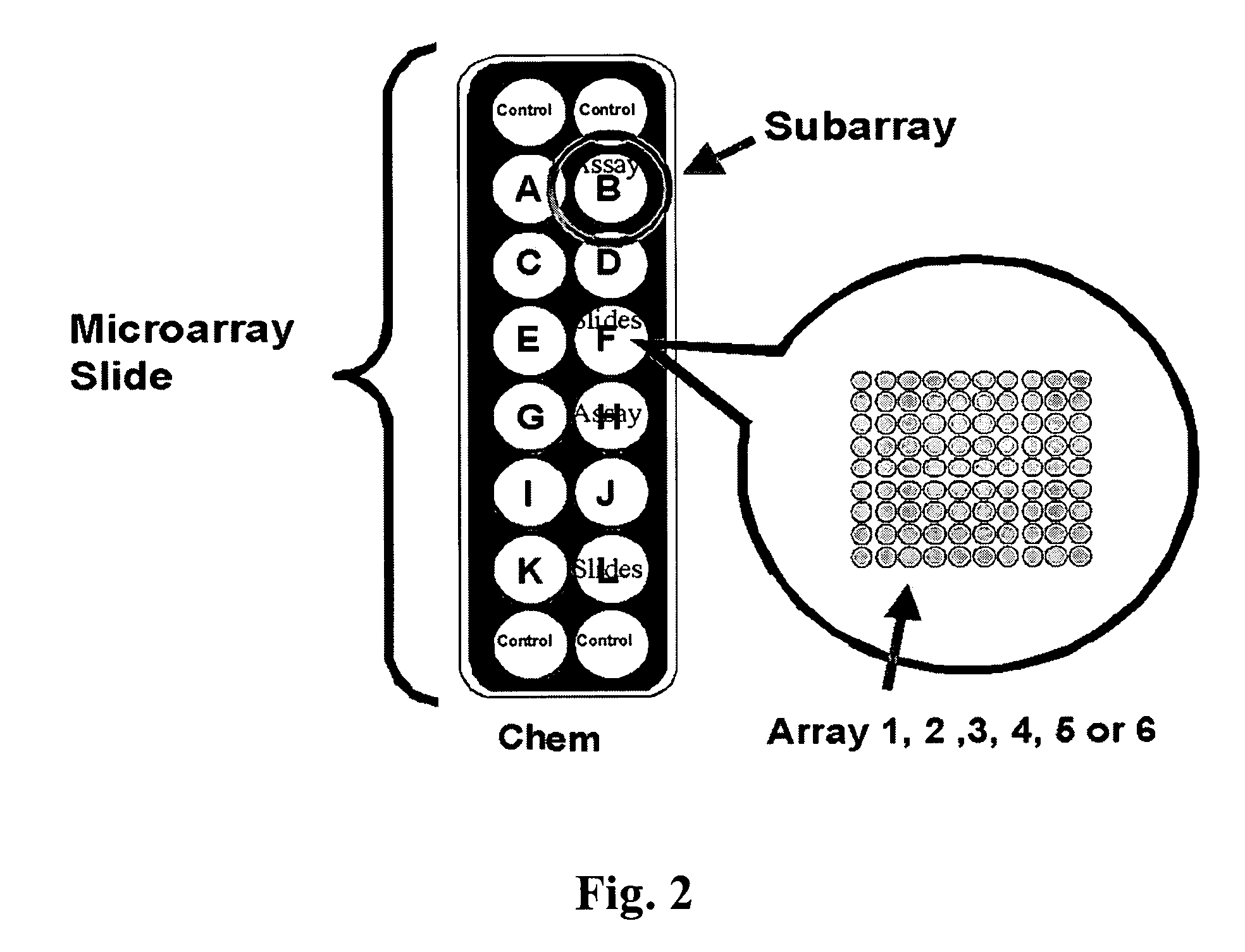
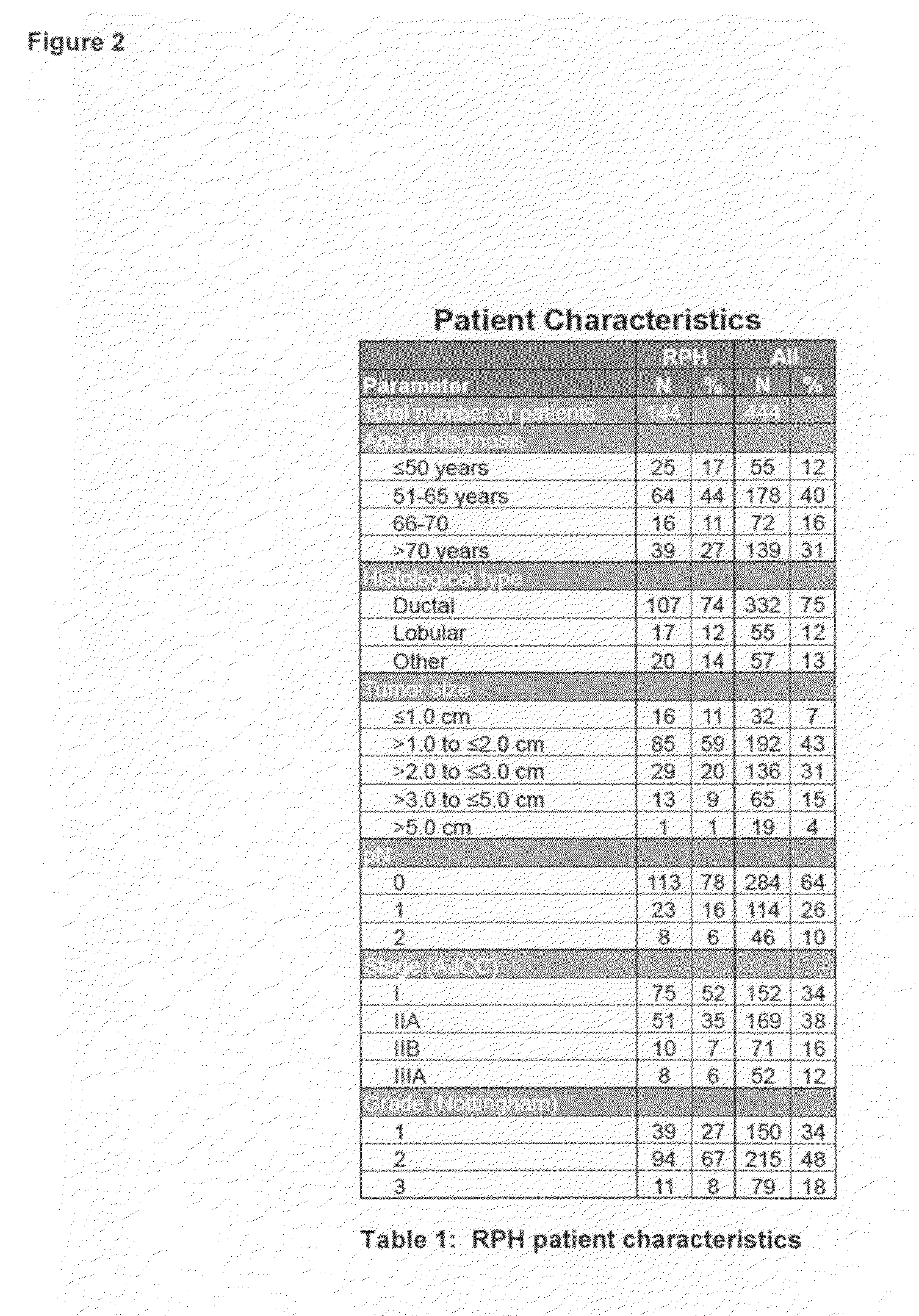
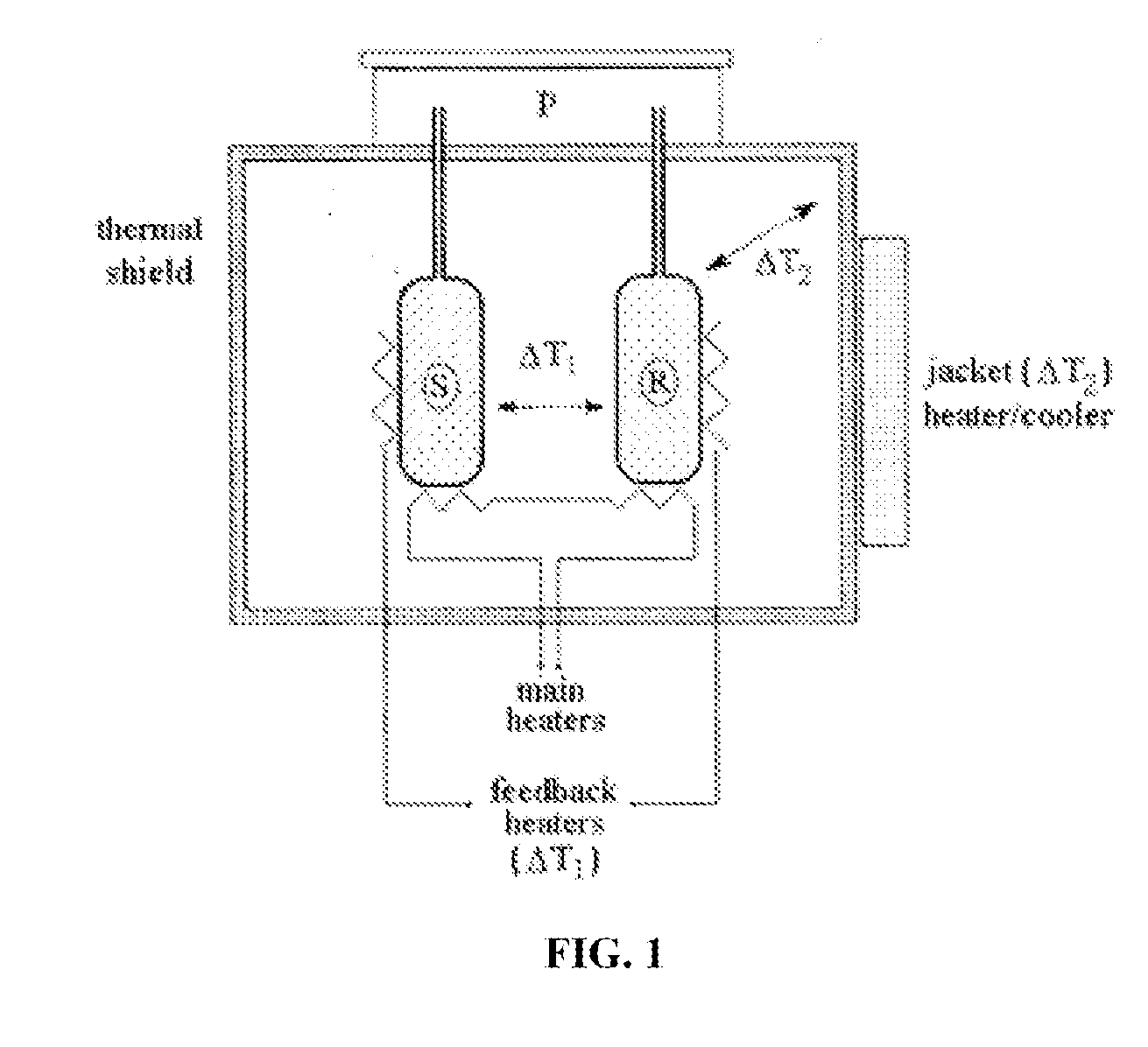
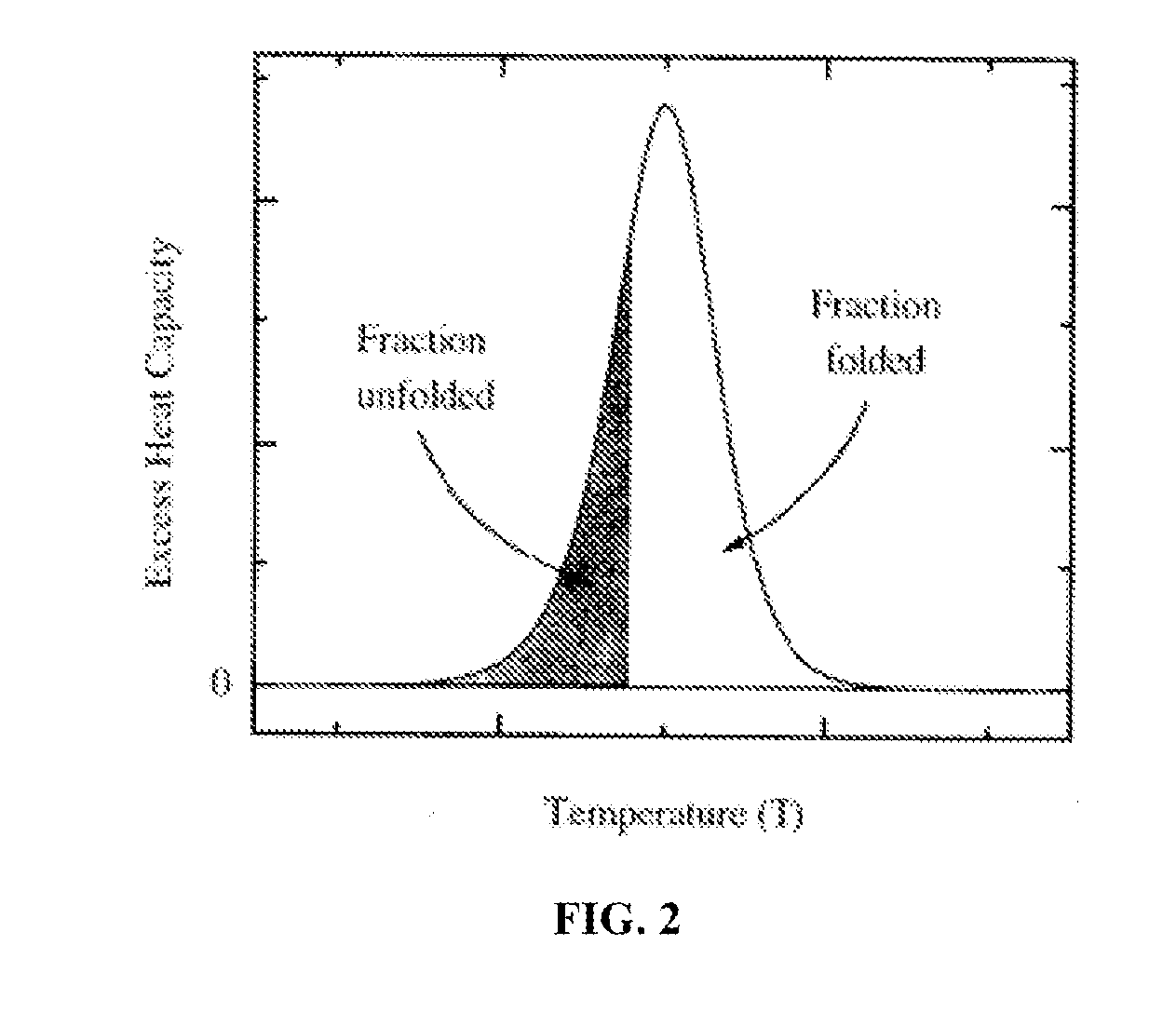
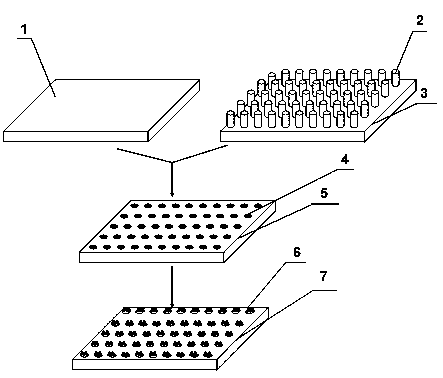
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
140 results about "Proteomic Profiling" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A laboratory procedure that determines both the identity and levels of expression of the proteins in a biological sample or specimen.
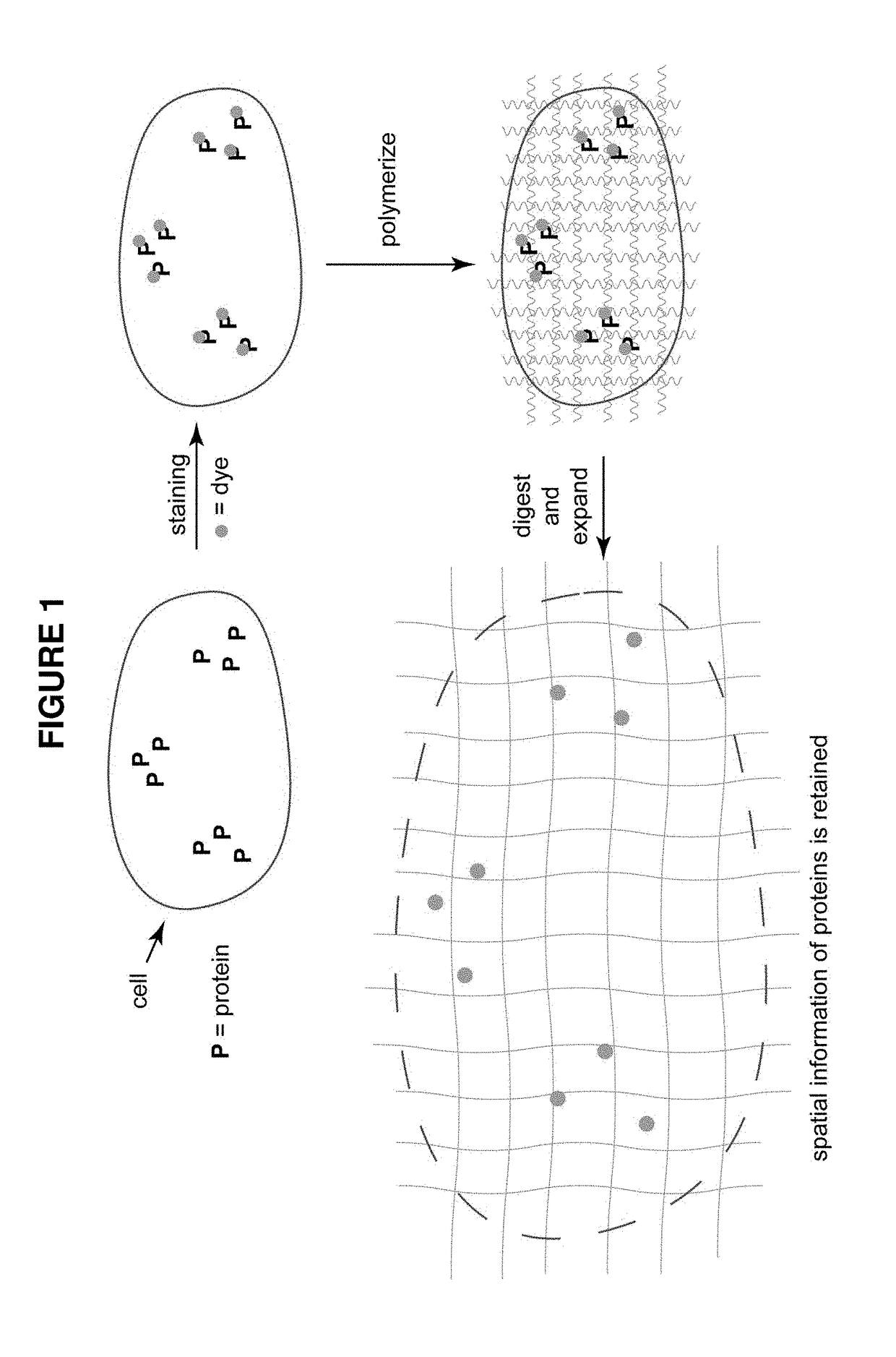
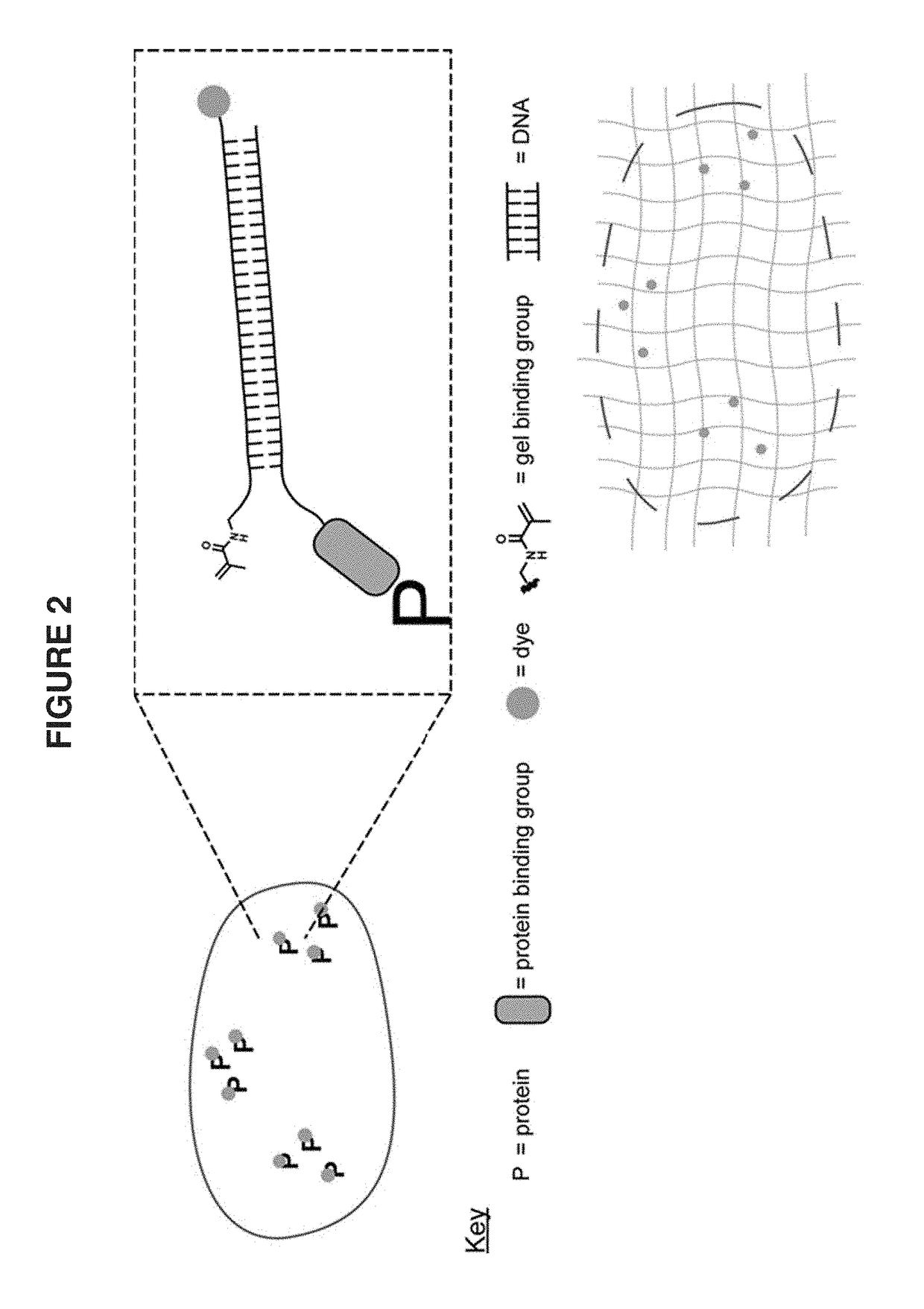
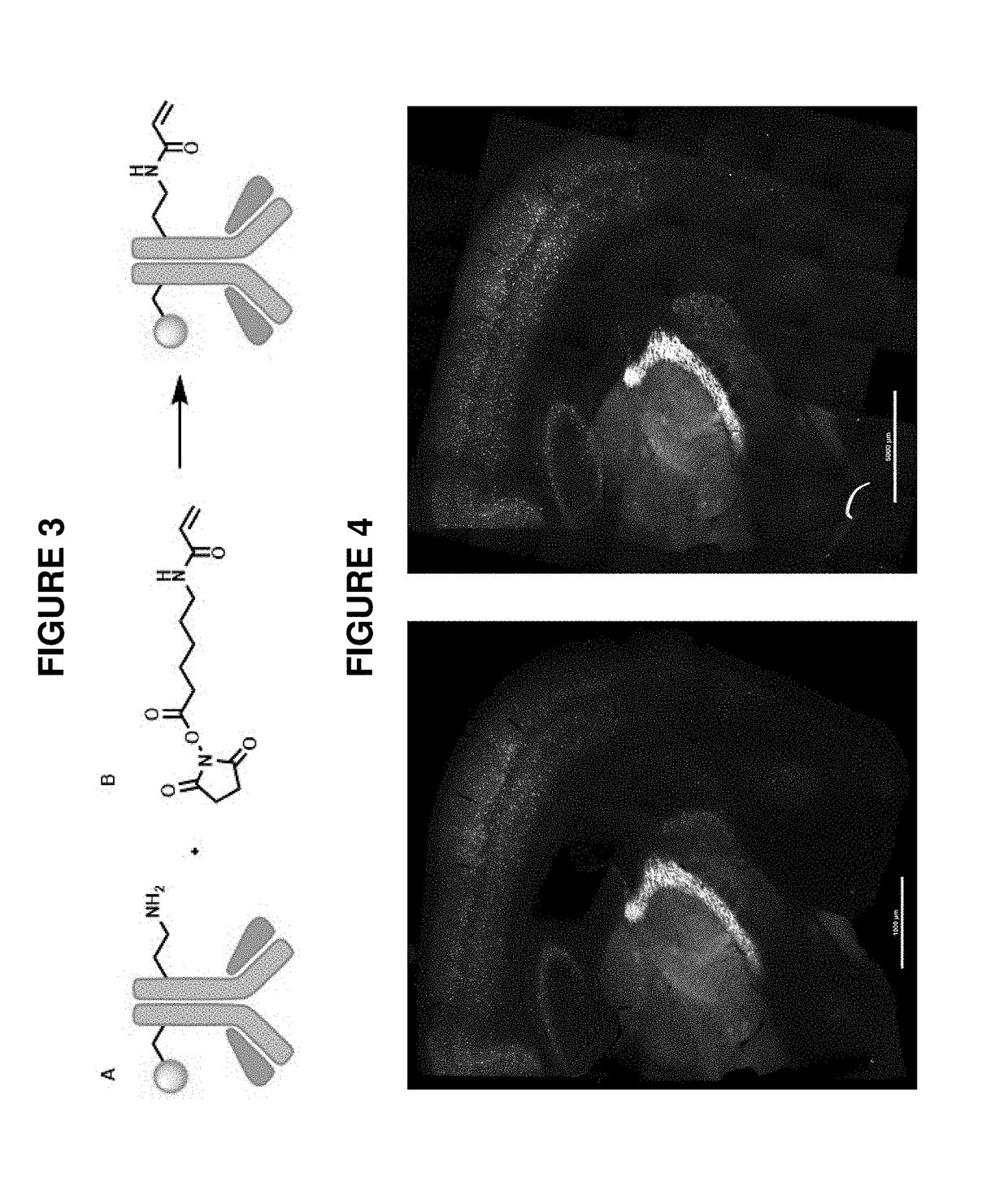
Combining modified antibodies with expansion microscopy for in-situ, spatially-resolved proteomics
InactiveUS20180052081A1Preparing sample for investigationBiological testingSpatially resolvedAntibody
This invention relates to imaging, such as by expansion microscopy, labelling, and analyzing biological samples, such as cells and tissues, as well as reagents and kits for doing so.
Owner:EXPANSION TECH
Proteomic analysis of biological fluids
ActiveUS20070161125A1Eliminate needMicrobiological testing/measurementAnalogue computers for chemical processesDiseaseGynecology
The invention concerns the identification of proteomes of biological fluids and their use in determining the state of maternal / fetal conditions, including maternal conditions of fetal origin, chromosomal aneuploidies, and fetal diseases associated with fetal growth and maturation. In particular, the invention concerns a comprehensive proteomic analysis of human amniotic fluid (AF) and cervical vaginal fluid (CVF), and the correlation of characteristic changes in the normal proteome with various pathologic maternal / fetal conditions, such as intra-amniotic infection, pre-term labor, and / or chromosomal defects. The invention further concerns the identification of biomarkers and groups of biomarkers that can be used for non-invasive diagnosis of various pregnancy-related disorders, and diagnostic assays using such biomarkers.
Owner:HOLOGIC INC
Identification of cancer protein biomarkers using proteomic techniques
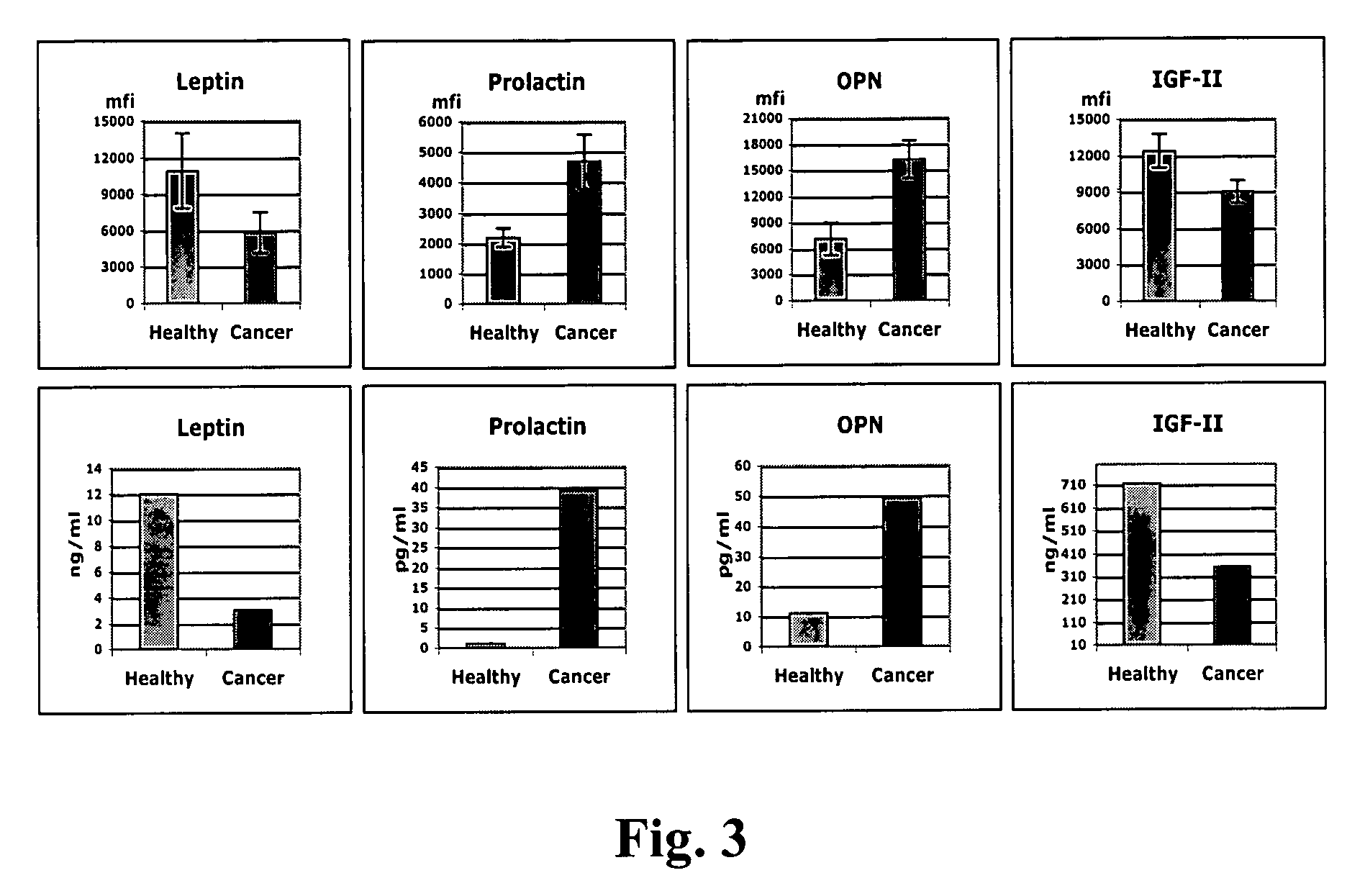
The claimed invention describes methods to diagnose or aid in the diagnosis of cancer. The claimed methods are based on the identification of biomarkers which are particularly well suited to discriminate between cancer subjects and healthy subjects. These biomarkers were identified using a unique and novel screening method described herein. The biomarkers identified herein can also be used in the prognosis and monitoring of cancer. The invention comprises the use of leptin, prolactin, OPN and IGF-II for diagnosing, prognosis and monitoring of ovarian cancer.
Owner:YALE UNIV
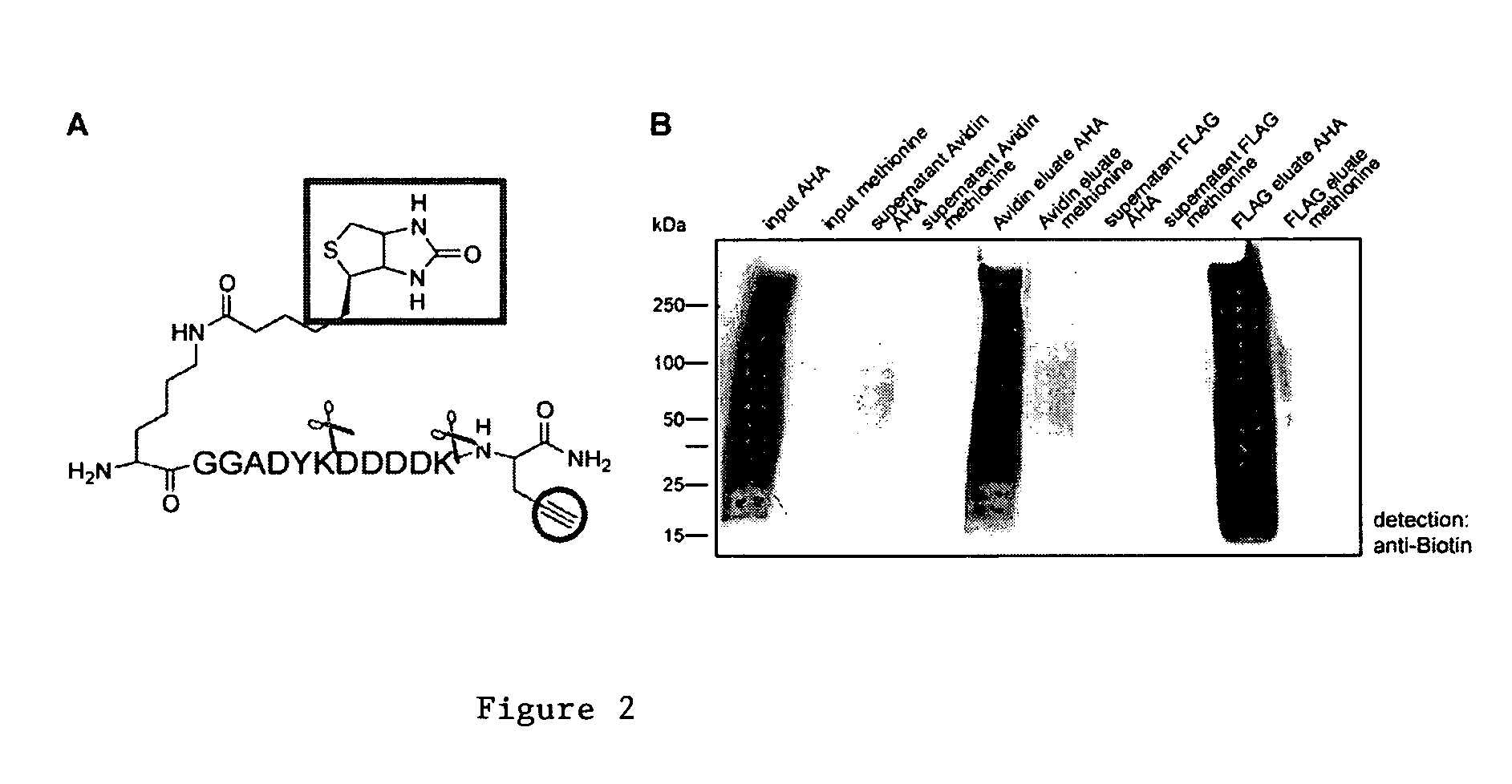
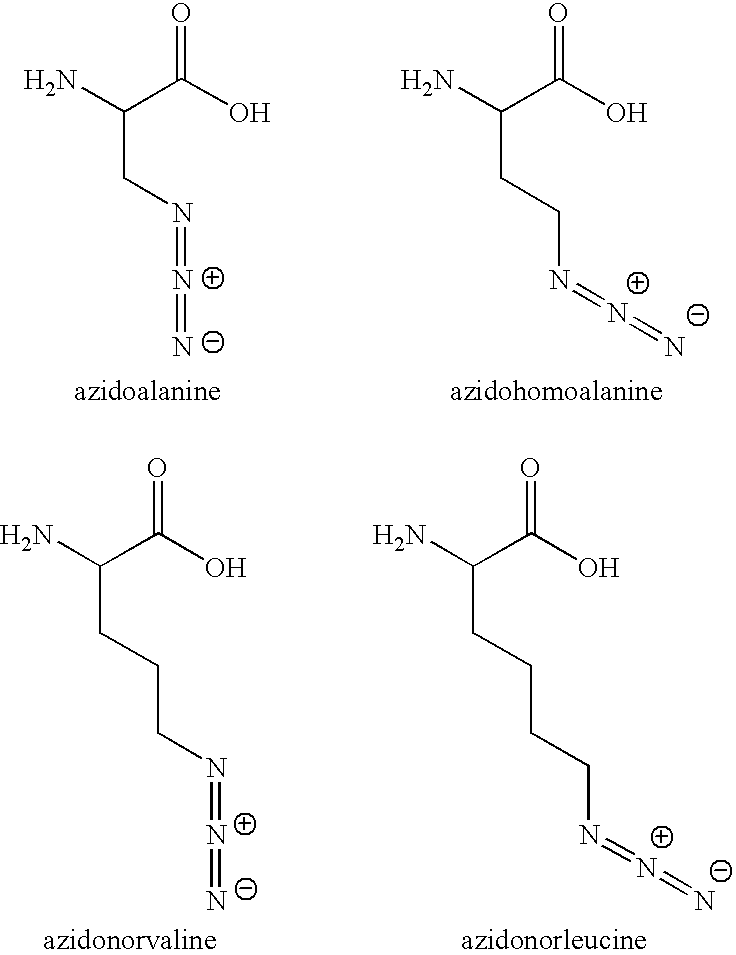
Methods for proteomic profiling using non-natural amino acids
The invention provides methods, reagents and systems for incorporating non-natural amino acids into proteins, preferably in vivo, using the endogenous protein synthesis machinery of an organism. The incorporated non-natural amino acids contain reactive groups for further chemical reagents, which may serve as a “handle” to enrich the proteins or fragments thereof in a number of uses, such as proteomic analysis, imaging of diseased tissues / cells, etc.
Owner:CALIFORNIA INST OF TECH
Artificial neural network proteomic tumor classification
Here the inventors describe a tumor classifier based on protein expression. Also disclosed is the use of proteomics to construct a highly accurate artificial neural network (ANN)-based classifier for the detection of an individual tumor type, as well as distinguishing between six common tumor types in an unknown primary diagnosis setting. Discriminating sets of proteins are also identified and are used as biomarkers for six carcinomas. A leave-one-out cross validation (LOOCV) method was used to test the ability of the constructed network to predict the single held out sample from each iteration with a maximum predictive accuracy of 87% and an average predictive accuracy of 82% over the range of proteins chosen for its construction.
Owner:UNIV OF SOUTH FLORIDA +1
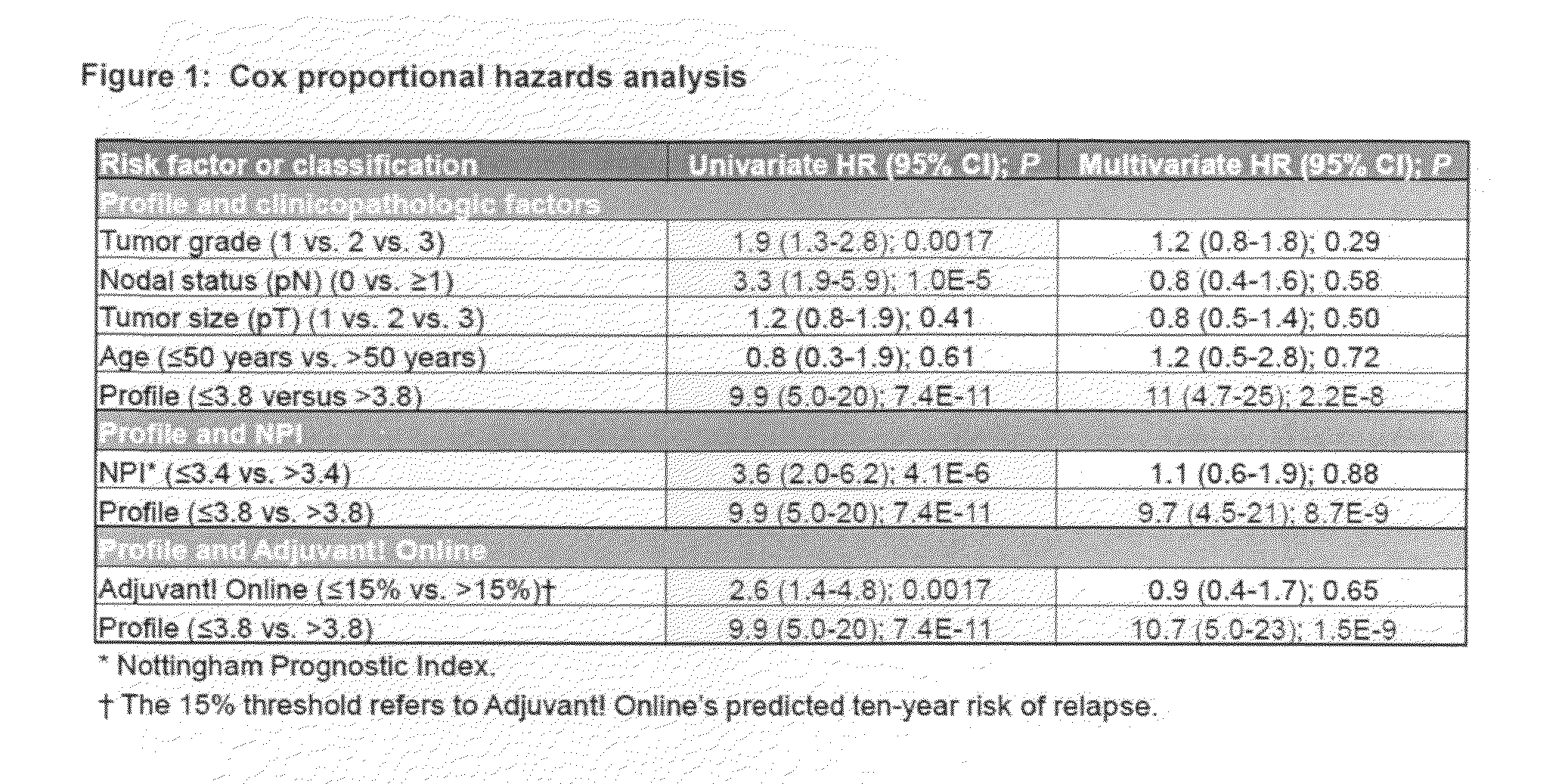
Molecular markers predicting response to adjuvant therapy, or disease progression, in breast cancer
Predicting response to adjuvant therapy or predicting disease progression in breast cancer is realized by (1) first obtaining a breast cancer test sample from a subject; (2) second obtaining clinicopathological data from said breast cancer test sample; (3) analyzing the obtained breast cancer test sample for presence or amount of (a) one or more molecular markers of hormone receptor status, one or more growth factor receptor markers, (b) one or more tumor suppression / apoptosis molecular markers; and (c) one or more additional molecular markers both proteomic and non-proteomic that are indicative of breast cancer disease processes; and then (4) correlating (a) the presence or amount of said molecular markers and, with (b) clinicopathological data from said tissue sample other than the molecular markers of breast cancer disease processes. A kit of (1) a panel of antibodies; (2) one or more gene amplification assays; (3) first reagents to assist said antibodies with binding to tumor samples; (4) second reagents to assist in determining gene amplification; permits, when applied to a breast cancer patient's tumor tissue sample, (A) permits observation, and determination, of a numerical level of expression of each individual antibody, and gene amplification; whereupon (B) a computer algorithm, residing on a computer can calculate a prediction of treatment outcome for a specific treatment for breast cancer, or future risk of breast cancer progression.
Owner:LINKE STEVEN +2
Proteomic profiling method useful for condition diagnosis and monitoring, composition screening, and therapeutic monitoring
InactiveUS20080172184A1Low costLower levelCompound screeningApoptosis detectionTherapy monitoringProtein composition
A method of diagnosing or monitoring a condition of interest in a subject includes comparing thermograms generated using differential scanning calorimetery. A signature thermogram contains a protein composition pattern for a sample obtained from the subject. The signature thermogram is compared to a standard thermogram. Standard thermograms can include a negative standard thermogram containing a protein composition pattern associated with an absence of the condition of interest, and a positive standard thermogram containing a protein composition pattern associated with a presence of the condition of interest.
Owner:UNIV OF LOUISVILLE RES FOUND INC
Cell micro-array chip and preparation method thereof
InactiveCN103060175ABioreactor/fermenter combinationsBiological substance pretreatmentsHigh-Throughput Screening MethodsEngineering
The invention provides a cell micro-array chip and a preparation method thereof and belongs to the technical field of cell chips. The cell micro-array chip is characterized by being prepared by using alginate gels; and the preparation method comprises the following steps: 1, coating a layer of sodium alginate solution on a glass plate, rapidly immerging the glass plate in a calcium chloride solution and soaking the glass plate for 5-30 minutes to form a gel template; 2, preparing a dot matrix template on which a plurality of cylinders are arranged, soaking the top ends of the cylinders in a chitosan or polylysine solution and taking out the cylinders after 5-30 minutes; 3, embossing the top ends of the cylinders on the alginate gel template and forming a complex dot matrix template through electrostatic complexation reaction; and 4, soaking the complex dot matrix template in a culture solution with cell concentration of 1*10<5>-1*10<7> and forming the alginate gel cell micro-array chip when the complex dot matrix template is cultured for 4-16 hours. The cell micro-array chip provided by the invention has the advantage of simplicity and convenience in operation and can be used for cell tissue like culturing, high throughput screening of compounds or medicines, discovery of new genes, proteomic studies and the like.
Owner:TAIYUAN UNIV OF TECH
Highly sensitive proteomic analysis methods, and kits and systems for practicing the same
Methods of determining whether a sample includes one or more analytes, particularly proteinaceous analytes, of interest are provided. In the subject methods, an array of binding agents, where each binding agent includes an epitope binding domain of an antibody, is contacted with the sample. In many embodiments, contact occurs in the presence of a metal ion chelating polysaccharide, e.g., a pectin. Following contact, the presence of binding complexes on the array surface are detected and the resultant data is employed to determine whether the sample includes the one or more analytes of interest. Also provided are kits, systems and other compositions of matter for practicing the subject methods. The subject methods and compositions find use in a variety of applications, including proteomic applications such as protein expression analysis, e.g., differential protein expression profiling.
Owner:CLONTECH LAB
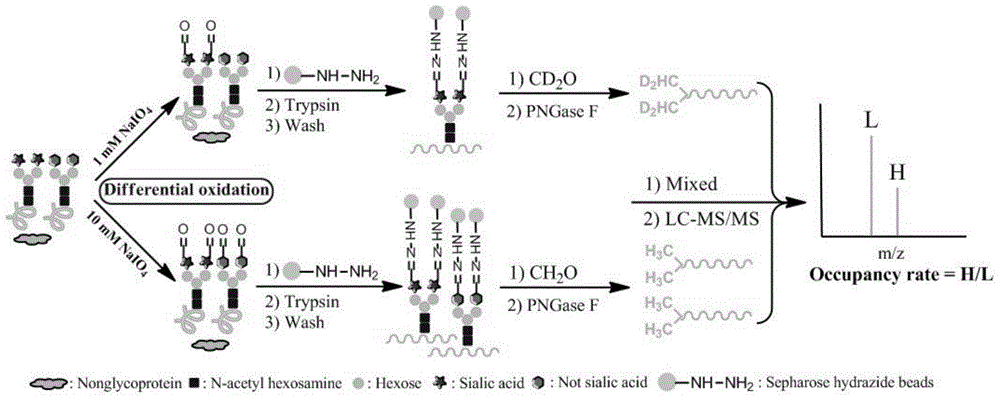
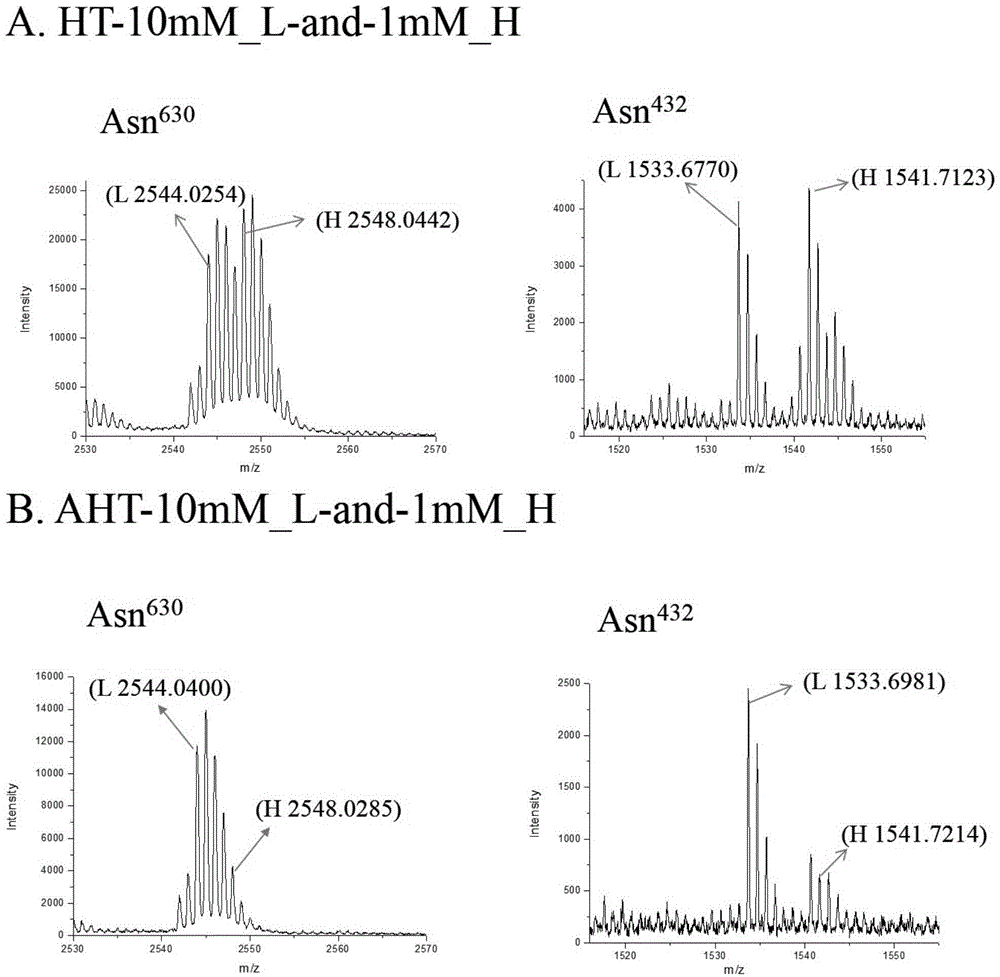
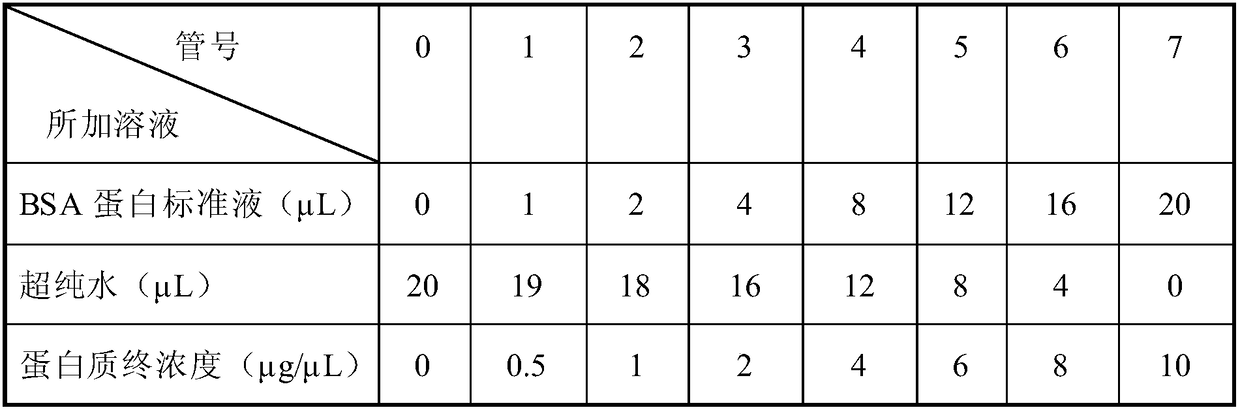
Quantitative method for occupancy of N-bond sialylated sugar chains on glycoprotein and application of quantitative method in hepatoma marker screening
InactiveCN105987945ASimple shareEfficient occupancyMaterial analysis by electric/magnetic meansMicrosphereGlycopeptide
The invention relates to a quantitative method for occupancy of N-bond sialylated sugar chains on glycoprotein and an application of the quantitative method in hepatoma marker screening. The method comprises the following steps: two biological samples containing glycoprotein under a same state are taken, then a sodium periodate solution is used for respectively processing the two biological samples for performing differential oxidation, then a hydrazide chemical method on protein is used for enriching glycoprotein in the samples after differential oxidation, then differences of different stable isotopes can be used for labeling the glycopeptide on hydrazide microspheres, and peptide-N-glycosidase (PNGase) is used for processing, and finally mass spectrometry is carried out on mixing difference-labeled N-glycopeptide so as to quantify the occupancy of N-bond sialylated sugar chains. The method can be used for glycosylation-modified proteomics analysis, can simultaneously obtain the corresponding glycoprotein, glycopeptide and glycosylation bit identification results, and especially can be used for screening of a latent biological marker for hepatic cellular cancer(HCC). The method has the advantages of simpleness and high efficiency.
Owner:DALIAN INST OF CHEM PHYSICS CHINESE ACAD OF SCI
Wild ginseng protein extracting method suitable for dielectrophoresis
InactiveCN101906130AImprove solubilityReduce Denaturing PrecipitationPeptide preparation methodsElectrophoresisCentrifugation
The invention discloses a wild ginseng protein extracting method suitable for dielectrophoresis. The method comprises the following steps of: taking wild ginseng as an experimental material, fully grinding the wild ginseng in liquid nitrogen and transferring the obtained product to a centrifuge tube; adding acetone solution precooled at the temperature of -20 DEG C into the centrifuge tube, washing sample powder for 3 times, collecting sediments by centrifugation and drying the obtained product at room temperature for 20min to obtain the crude extract of the wild ginseng protein; dissolving the crude extract of the wild ginseng protein into lysis buffer, and performing ultrasonic cell disintegration and extracting the supernate by centrifugation to obtain the high-quality solution of the wild ginseng protein. The wild ginseng protein extracting method for the dielectrophoresis is simple and fast, can obtain an electrophoresis pattern having a high repeatability and resolution, is favorable for subsequent protein identification by mass spectrographic analysis and provides a technical support for the proteomic research of the wild ginseng.
Owner:BEIHUA UNIV
Polypeptide chip and application thereof in virus detection
The invention provides a polypeptide chip, which comprises a substrate and n polypeptides distributed on the substrate in an array, each polypeptide sequence has 10-20 amino acids, a group consistingof sequences of first to nth polypeptides covers at least 95% of a viral protein sequence, the adjacent polypeptides have an overlap of 3-8 amino acids, and n is 810-1370. According to the method, proteomics and system biology strategies are adopted, all coded protein sequences of COVID-19 are extracted from NCBI data, an SARS-CoV-2 virus proteome polypeptide chip is designed and prepared, and panoramic scanning of all SARS-CoV-2 virus antibodies in blood of a novel pneumonia virus infected patient is achieved.
Owner:ACADEMY OF MILITARY MEDICAL SCI
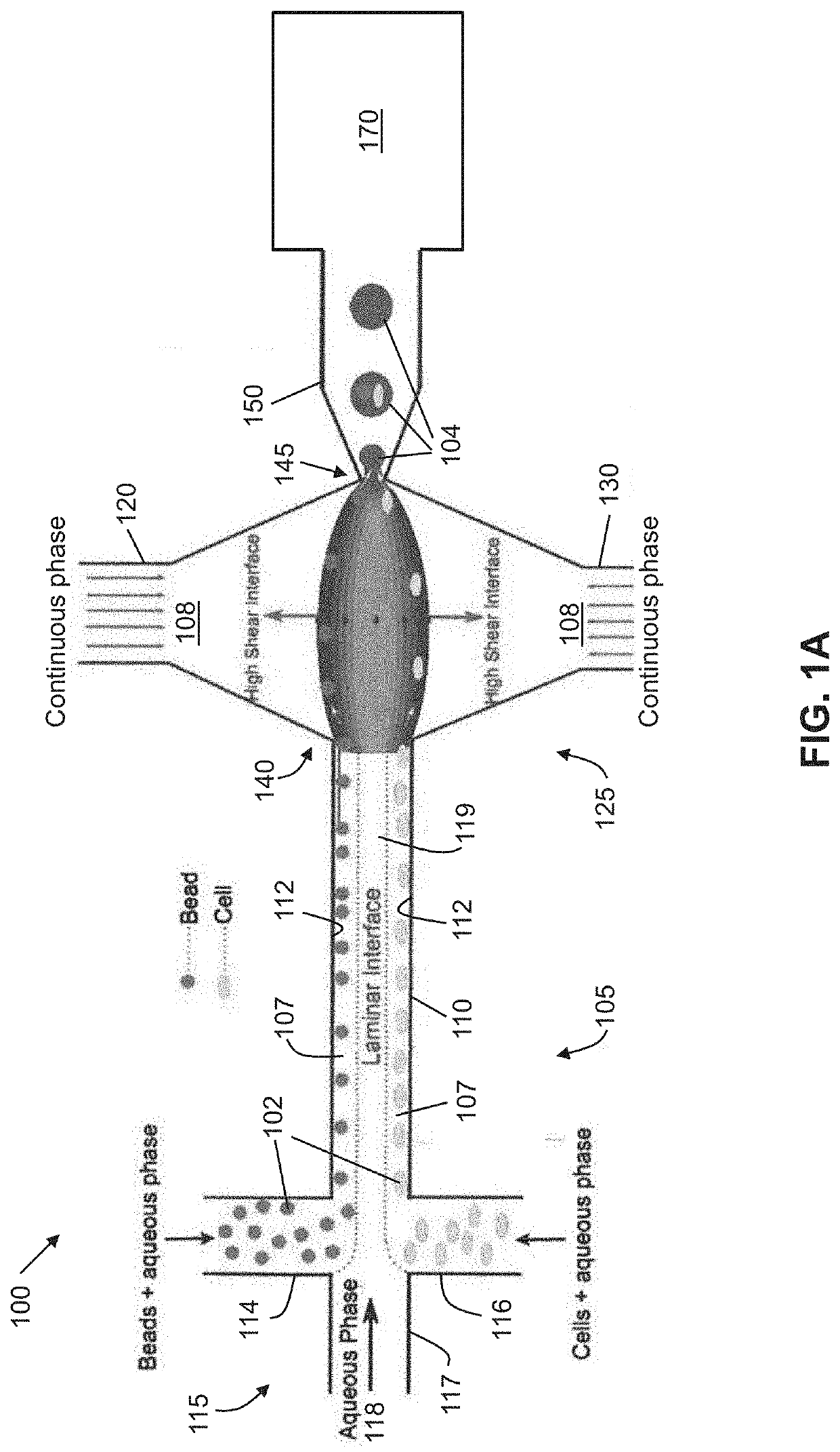
High-efficiency particle encapsulation in droplets with particle spacing and downstream droplet sorting
ActiveUS20200108393A1High throughput experimentationReduce material wasteImmobilised enzymesLaboratory glasswaresGenomicsChemical physics
A passive, hydrodynamic technique implemented using a microfluidic device to perform co-encapsulation of samples in droplets and sorting of said droplets is described herein. The hydrodynamic technique utilizes laminar flows and high shear liquid-liquid interfaces at a microfluidic junction to encapsulate samples in the droplets. A sorting mechanism is implemented to separate sample droplets from empty droplets. This technique can achieve a one-one-one encapsulation efficiency of about 80% and can significantly improve the droplet sequencing and related applications in single cell genomics and proteomics.
Owner:RGT UNIV OF CALIFORNIA
Pressure-assisted molecular recovery (PAMR) of biomolecules, pressure-assisted antigen retrieval (PAAR), and pressure-assisted tissue histology (PATH)
ActiveUS20100136613A1Facilitate re-hydrationEasy to usePreparing sample for investigationDead animal preservationCross-linkHigh pressure
A method is disclosed for reversing fixation-induced cross-linking in tissue specimens that have been preserved for histological examination. The method involves placing the fixed tissue in a liquid under elevated temperature and pressure conditions that are sufficient to reverse the fixation-induced cross-linking, restore antigenicity to proteins, and permit improved molecular and proteomic analysis of the preserved tissue specimen. Methods are also disclosed for processing tissues for histological examination under elevated pressure conditions that enhance the perfusion of liquid reagents into the tissue and reduce overall processing times.
Owner:AMERICA REGISTRY OF PATHOLOGY +2
Methods and vectors for generating antibodies in avian species and uses therefor
The present invention relates to processes for producing polyclonal and monoclonal antibodies to an antigen in an avian species, preferably in a chicken, using polynucleotide vaccination. The present invention also relates to processes for determining the proteomics profile of a set of pre-selected DNA sequences isolated from a bio-sample, preferably the proteomics profile of a human cDNA library. The present invention additionally relates to processes for identifying physiologically distinguishable markers associated with a physiologically abnormal bio-sample. The present invention further relates to antibody arrays, integrated databases for identification of genes and proteins, multi-functional gene expression vectors, and methods of producing and using such antibody arrays, integrated databases and multi-functional gene expression vectors.
Owner:GENWAY BIOTECH
Tumor individualized specific therapeutic vaccine and preparation method thereof
PendingCN111481662ASimple and fast operationShort treatment cycleCancer antigen ingredientsAntineoplastic agentsDendritic cellOncology
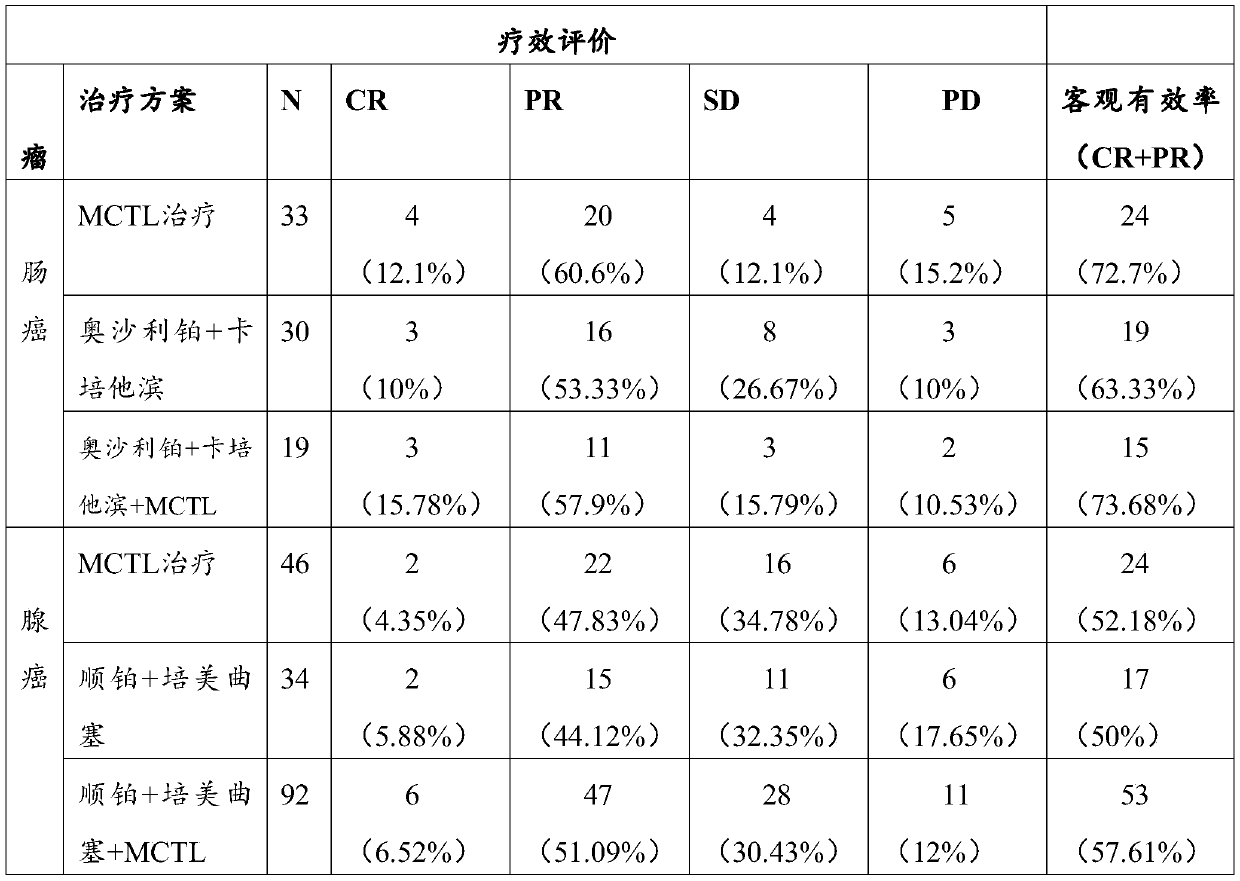
The invention relates to the field of immunology, and discloses a tumor individualized specific therapeutic vaccine and a preparation method thereof. Through MCTL proteomics dendritic cell cluster target detection, a tumor specific molecular target in a subject is analyzed, a therapeutic target is synthesized in vitro and co-cultured with autoimmune cells of the cultured subject, and the therapeutic vaccine is prepared. After the therapeutic vaccine is transfused back to a subject, tumor cells are specifically killed, immune balance is rebuilt, the purpose of effectively treating tumors is achieved, and the therapeutic vaccine has wide clinical application prospects.
Owner:河北博海生物工程开发有限公司
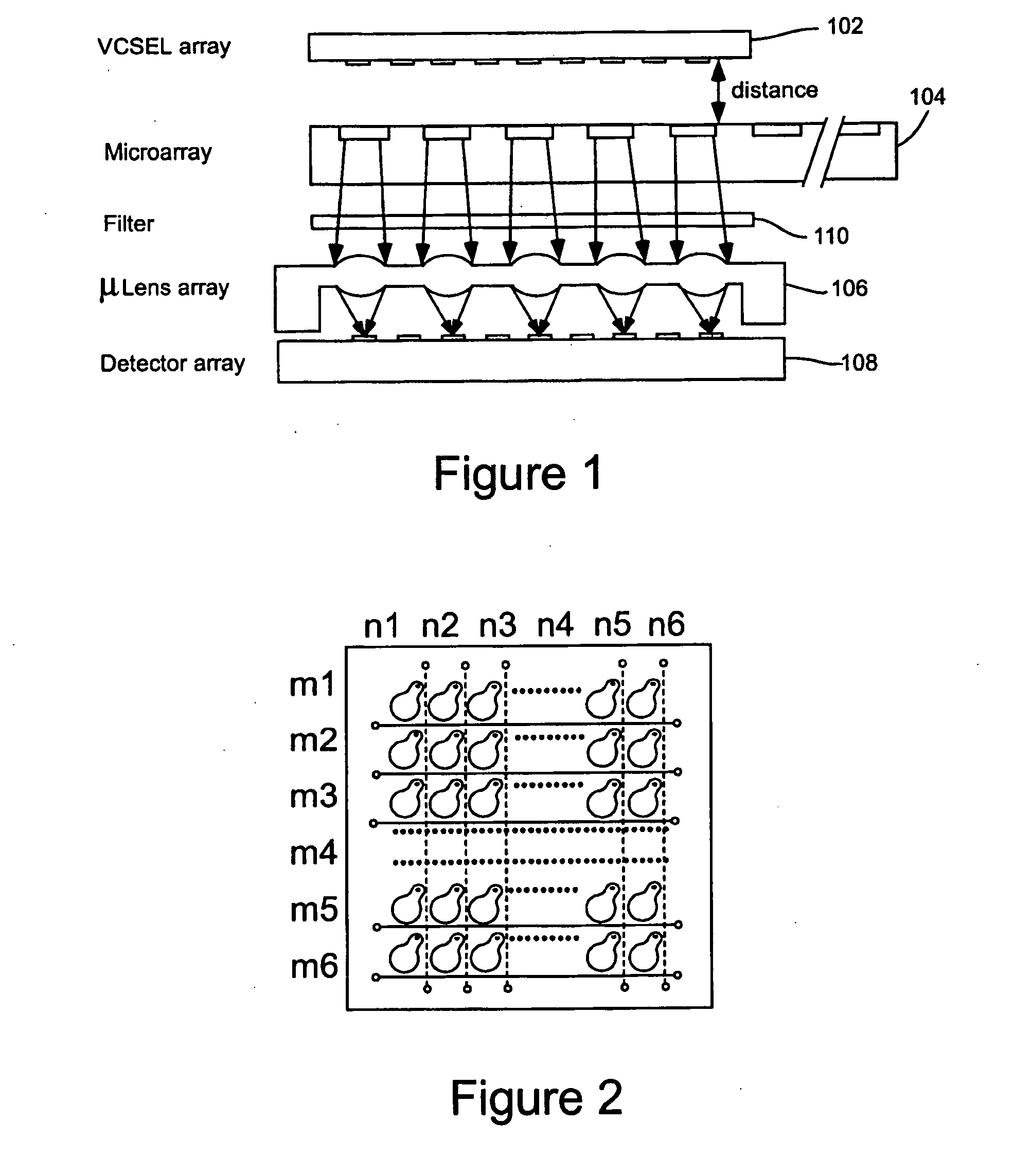
High throughput optical micro-array reader capable of variable pitch and spot size array processing for genomics and proteomics
An optical micro-array reader system includes microchip, VCSEL, microlens and detector arrays. The microchip array includes multiple sample spots to be separately analyzed. The VCSEL array is disposed to simultaneously illuminate more than one of the multiple spots. The microlens array focuses fluorescences or other optical emissions from the sample spots onto the detector array.
Owner:INTEL CORP
A functional enviromics method for cell culture media engineering
InactiveCN103459584AOptimize metabolismImprove productivityFungiMicrobiological testing/measurementBiotechnologyIntracellular
This invention refers to a new method for optimizing the composition of cell culture media. This new method comprises two main stages. In the first stage, a functional enviromics map is built through the joint screening of cell functions and medium factors by the execution of a specific cell culture protocol and exometabolome assays protocol. The functional enviromics map consists of a data array of intensity values of elementary cellular functions against medium factors. In the second stage, optimized cell culture medium formulations are developed that either enhance or repress target elementary cellular functions from columns of the functional enviromics map. The main advantage of this method lies in enabling metabolic engineering through the culture media composition manipulation, wherein an arbitrarily high number of cell functions are optimized through manipulation of medium factors, as opposed to previous methods, which are eminently empirical, are not cell function oriented, and require a much higher number of experiments. Furthermore, this new method is based on cost-effective exometabolome assays and does not require costly intracellular genomic or proteomic assays.
Owner:FACULDADE DE CIENCIAS E TECHA DA UNIV NOVA DE LISBOA
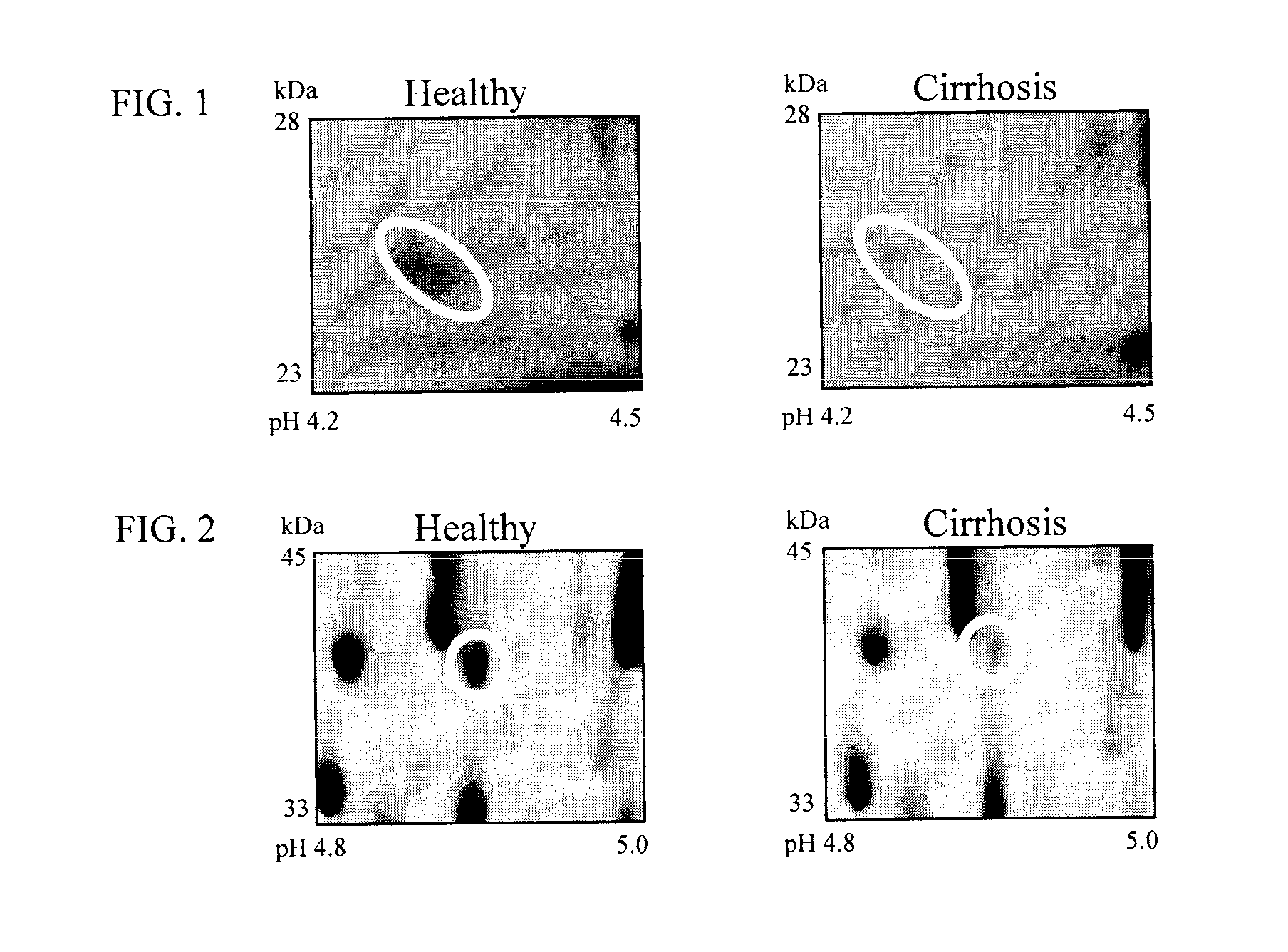
Clinical diagnosis of hepatic fibrosis using a novel panel of low abundant human plasma protein biomarkers
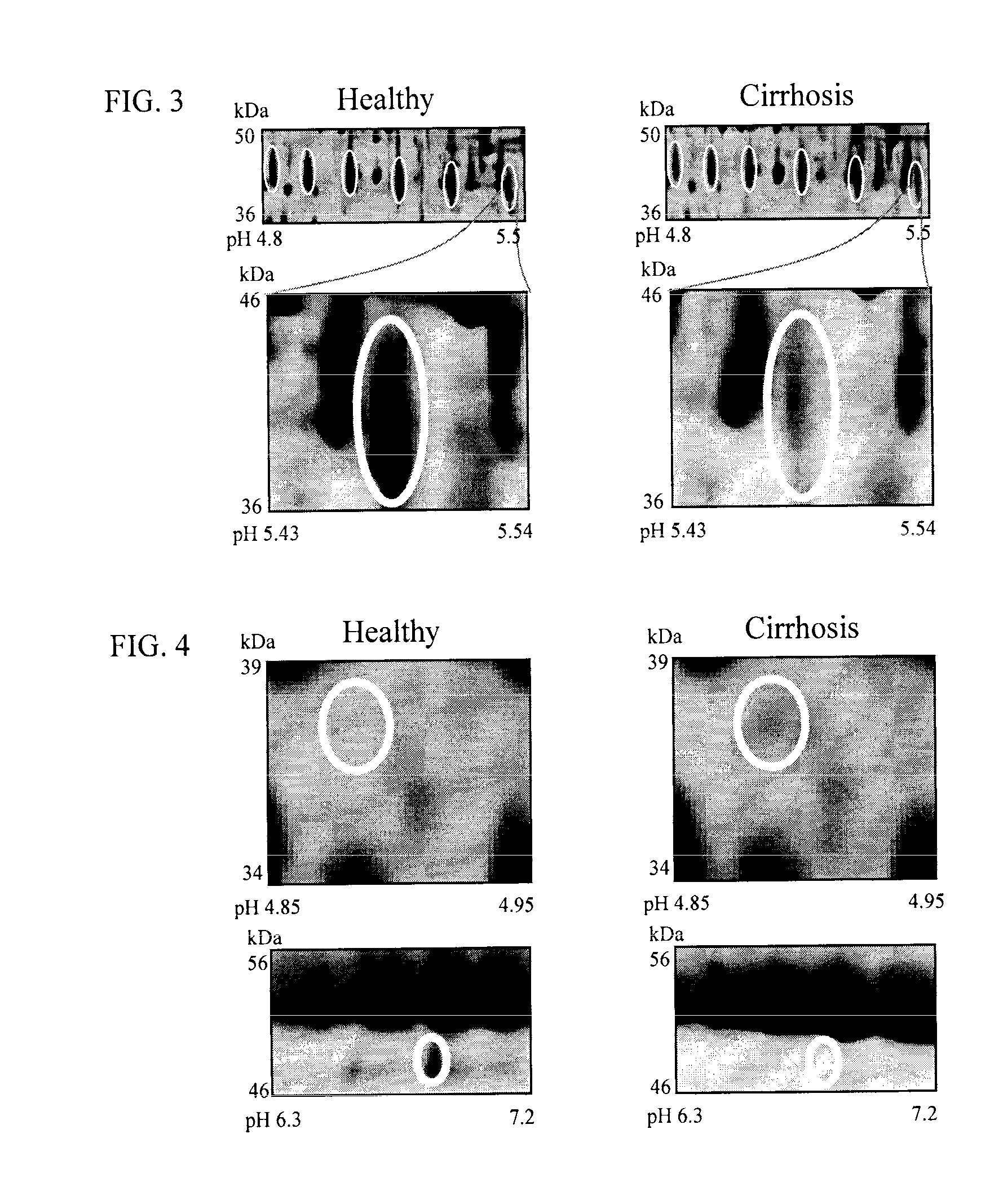
The inventors have proposed a novel panel of human plasma protein biomarkers for diagnosing hepatic fibrosis and cirrhosis. Presently there is no reliable non-invasive way of assessing liver fibrosis. A 2D-PAGE based proteomics study was used to identify potential fibrosis biomarkers. Plasma from patients with hepatic cirrhosis induced by infection with the hepatitis C virus (HCV) were analysed. Several proteins associated with liver scarring and potentially also related to viral infection were identified. These proteins include 14-3-3 protein zeta / delta, adiponectin, afamin, alpha-1-antitrypsin, alpha-2-HS-glycoprotein, apolipoprotein C-III, apolipoprotein E, C4b-binding protein beta chain, intact / cleaved complement C3dg, corticosteroid-binding globulin, fibrinogen gamma chain, beta haptoglobin at pH 5.46-5.49, haptoglobin-related protein, hemopexin, immunoglobulin J chain, leucine-rich alpha-2-glycoprotein, lipid transfer inhibitor protein, retinol-binding protein 4, serum paraoxonase / arylesterase 1, sex hormone-binding globulin and zinc-alpha-2-glycoprotein. These biomarkers can be used in conjunction with polypeptides in WO / 2008 / 031051. The concentrations of these novel biomarkers can be determined using an immunoassay where the concentrations would reflect the extent of fibrosis. A fibrosis scoring scale for each of the novel biomarkers is proposed. The additive result from the scores of all the novel biomarkers would give a more reliable indication of the degree of fibrosis rather than examining individual biomarkers.
Owner:UNIV OF OXFORD
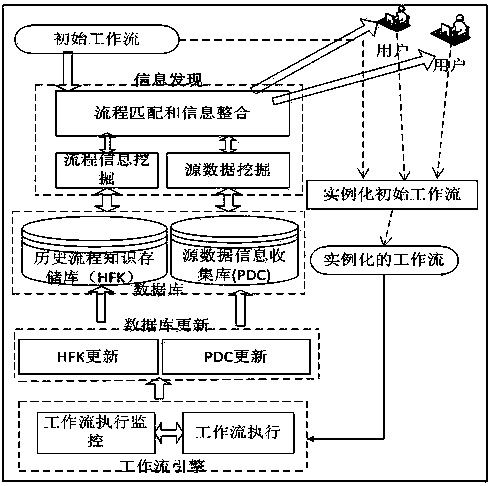
Workflow matching and finding system, based on provenance, facing proteomic data analysis
The invention belongs to the technical field of scientific workflows supported by computers and particularly discloses a workflow matching and finding system, based on provenance, facing proteomic data analysis. When a proteomic data analysis process is set up, the system sufficiently utilizes collected historical process information and provenance information relevant to the historical process information, realizes reuse of knowledge, reduces time consumption and energy consumption at process setting-up time, and quickens the progress of data analysis experiments. The processing process inside the system can be divided into three steps: using scientific workflows based on tasks to describe experiment tasks; utilizing a process matching and provenance information discovery algorithm mechanism to extract the instantiated process and the provenance information of the instantiated process; integrating the extracted process information and showing the information to researchers, and requiring the researchers to reuse or modify the historical process configuration information.
Owner:FUDAN UNIV
Screening method of non-invasive gastric cancer saliva biomarker
InactiveCN108152508ANon-invasive detection and evaluation meansSimple detection and evaluation meansBiological testingLymphatic SpreadFreeze-drying
The invention discloses a screening method of a non-invasive gastric cancer saliva biomarker. The screening method specifically comprises the following steps: protein sample preparation, protein quantification, proteolysis and freeze-drying, labeling a sample by iTRAQ, C18 desalting, liquid-phase separation and mass spectrum identification, and data analysis; the GO function enrichment including biology progress, the molecule function and the cell component is performed on the screened differential protein by using the Panther Classification System in the data analysis; and then the protein mutual effect analysis is performed on the differential protein by using the STRING analysis software; and then the KEGG signal path analysis is performed on the differential protein. Through the screening method disclosed by the invention, the specificity protein marker in the gastric cancer can be discovered through a fast and reliable proteomics technical program, thereby establishing a non-invasive, simple and convenient, fast and practical detection evaluation measure for the early diagnosis, the postoperative recurrence, metastasis, and prognosis observation of the gastric cancer.
Owner:SHENZHEN ELDERLY MEDICAL RES INST +1
Identification and use of glycopeptides as biomarkers for diagnosis and treatment monitoring
PendingCN111148844AMicrobiological testing/measurementMass spectrometric analysisAutoimmune conditionAutoimmune disease
Provided herein are methods for identifying new biomarkers for various diseases using proteomics, peptidomics, metabolics, proteoglycomics, glvcomics, mass spectrometry and machine learning. The present disclosure also provides glycopeptides as biomarkers for various diseases such as cancer and autoimmune diseases.
Owner:VENN BIOSCIENCES CORP
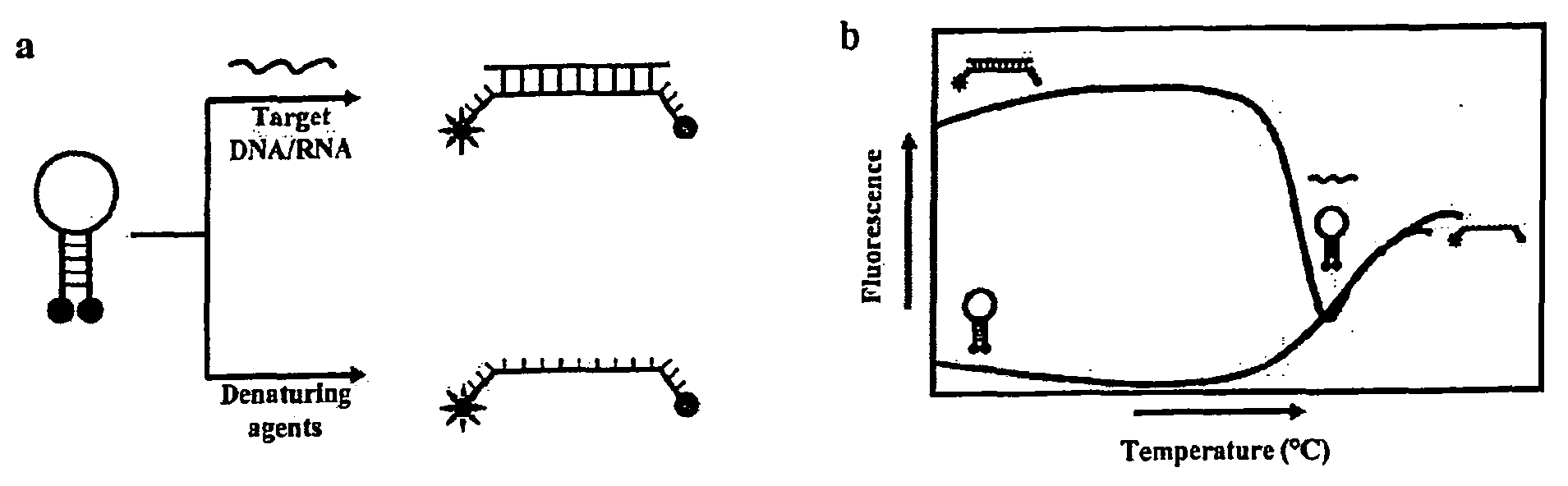
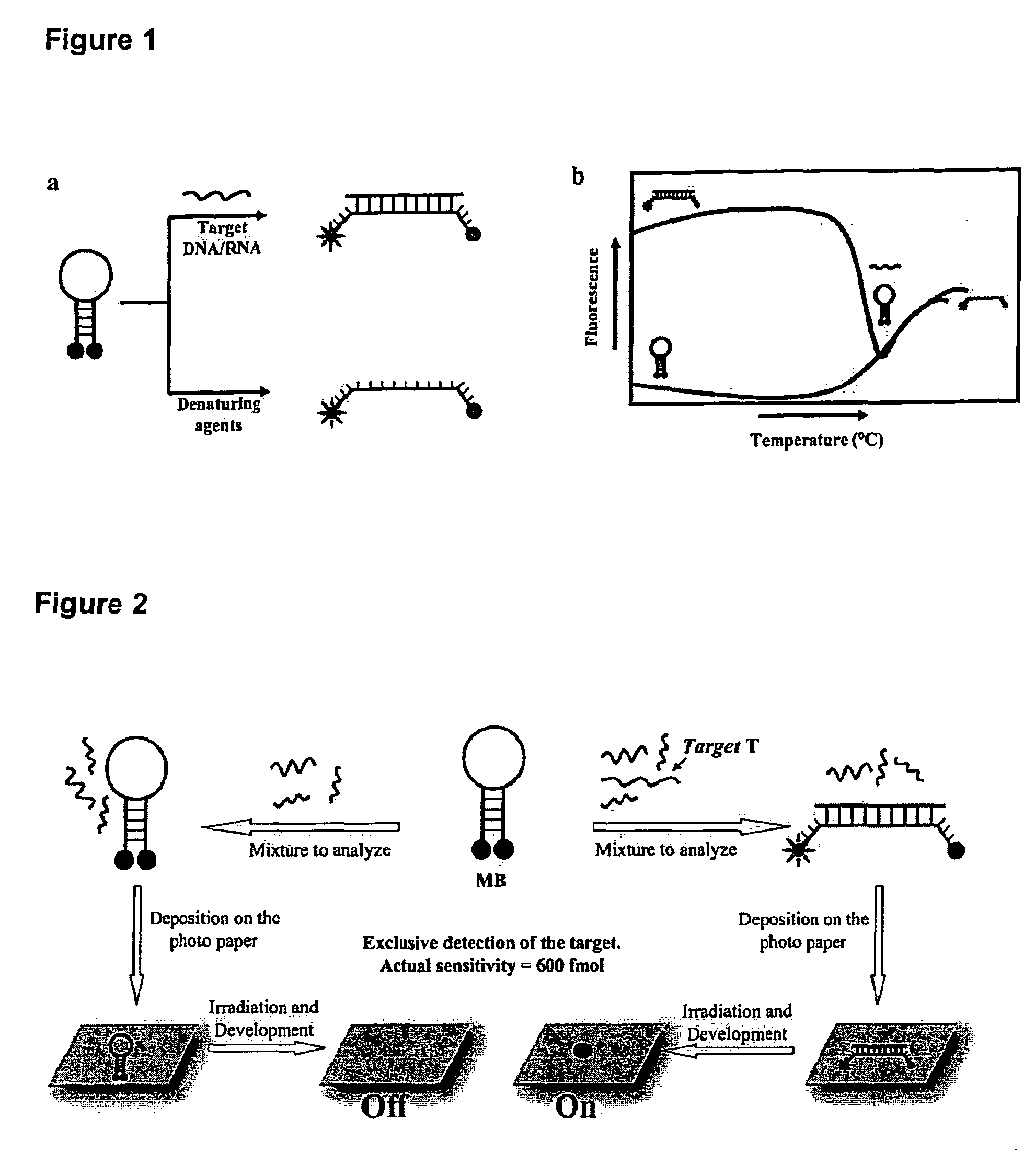
Molecular Beacons for DNA-Photography
InactiveUS20090087857A1High sensitivityQuick checkMicrobiological testing/measurementAnalysis by subjecting material to chemical reactionAnalyteProteomic Profile
The present invention refers to a detection method for analytes using the principle of black-and-white photography and to reagent kits for performing the method, furthermore applied this new technology to detect a biologically relevant sequence in the nanomolar range (femtomoles) in an application circumventing the necessity of a PCR. There are still numerous ways to optimize this methodology that is suitable for a large variety of applications in the genomic diagnostics and proteomics areas.
Owner:BASECLICK
Kit for detecting anti-moesin antibody
ActiveCN101929999AIncreased sensitivityImprove featuresDisease diagnosisColor/spectral properties measurementsEarly predictionMoesin
The invention discloses a kit for detecting an anti-moesin antibody. The kit comprises a solid-phase carrier and a moesin antigenic protein, wherein the moesin antigenic protein is a full-length human moesin. The kit can be applied to a method for early prediction and severity evaluation of connective tissue disease (CTD)-related lung involvement and evaluation is performed by mainly sampling a biological sample of a subject and detecting the quantity of the anti-moesin antibodies in the biological sample. The moesin is prepared by proteomics technology and gene recombination technology and an enzyme-linked immunosorbent assay (ELISA) detection kit for detecting the anti-moesin antibody is provided. The detectable rate of the kit for the CTD-related lung involvement is up to 51.7 percent.
Owner:SHANGHAI KEXIN BIOTECH
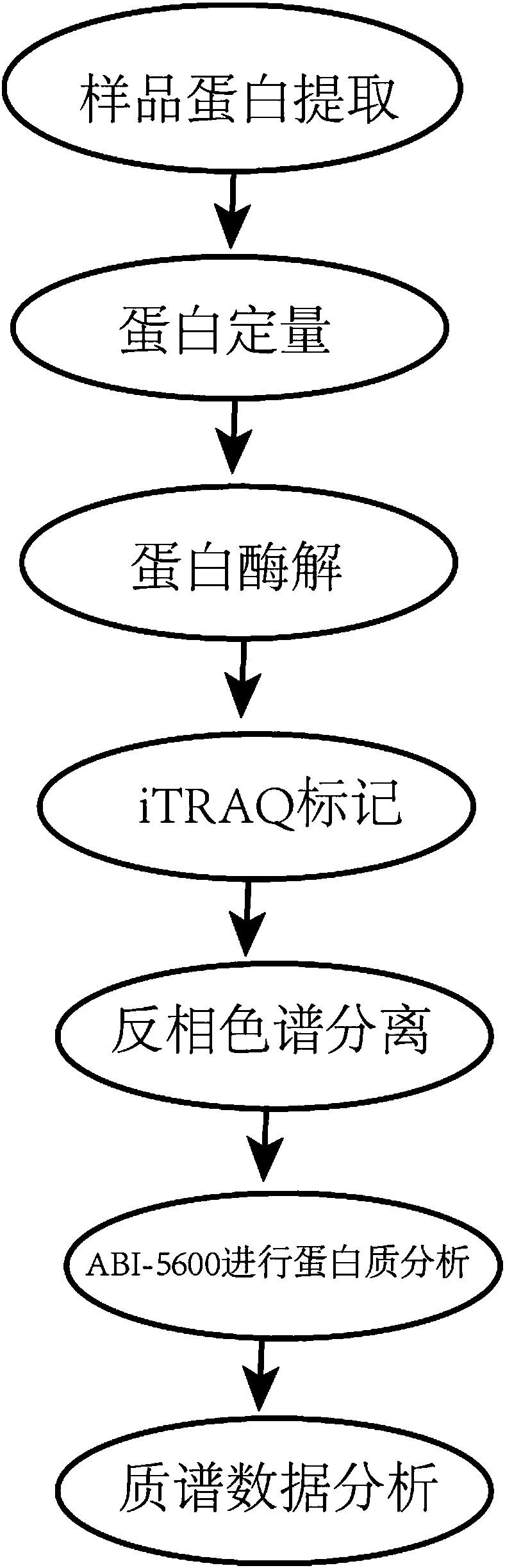
Method for detecting chronic nephritis biomarkers based on salivary proteomics
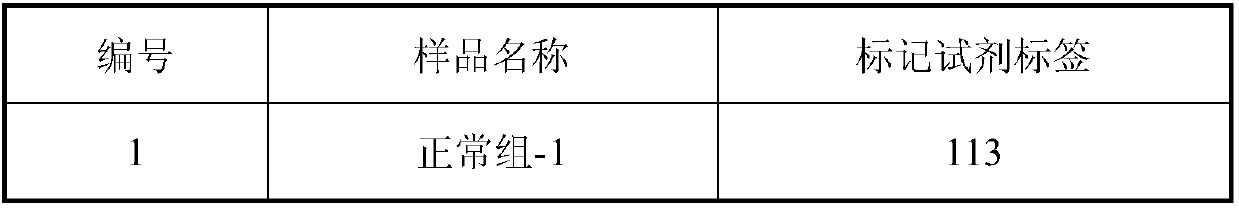
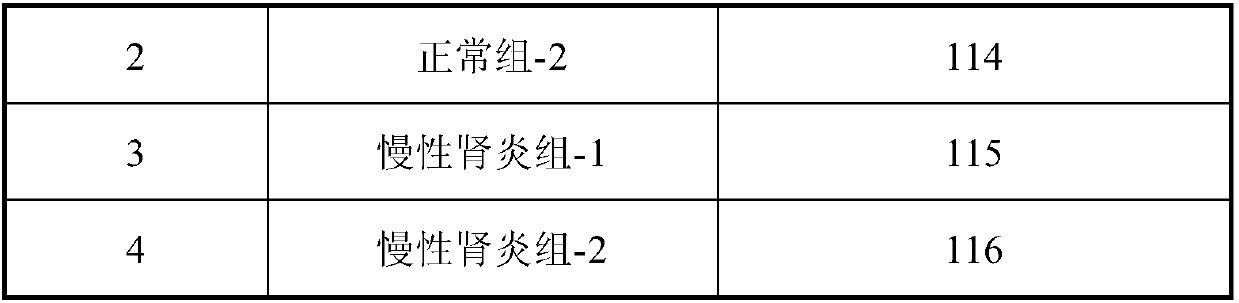
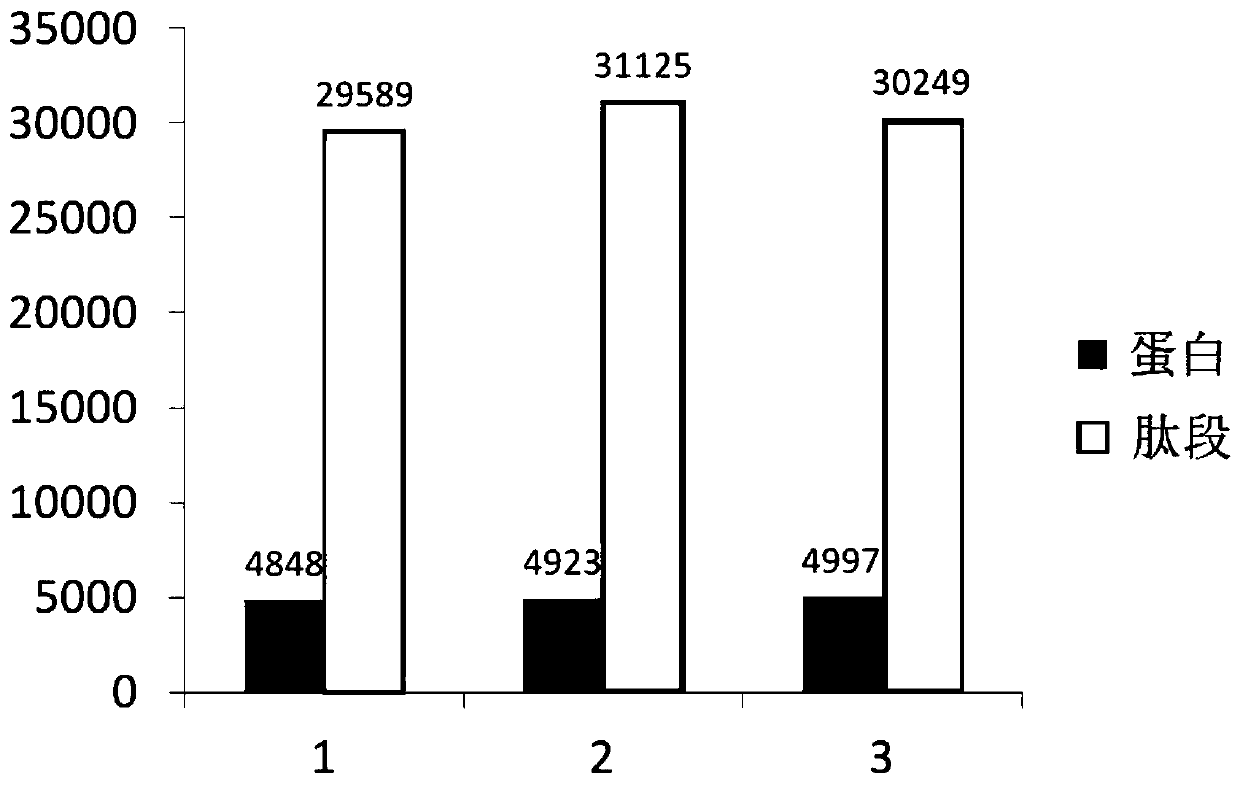
InactiveCN107870245ANon-invasive detection and evaluation meansSimple detection and evaluation meansDisease diagnosisBiological testingLymphatic SpreadFreeze-drying
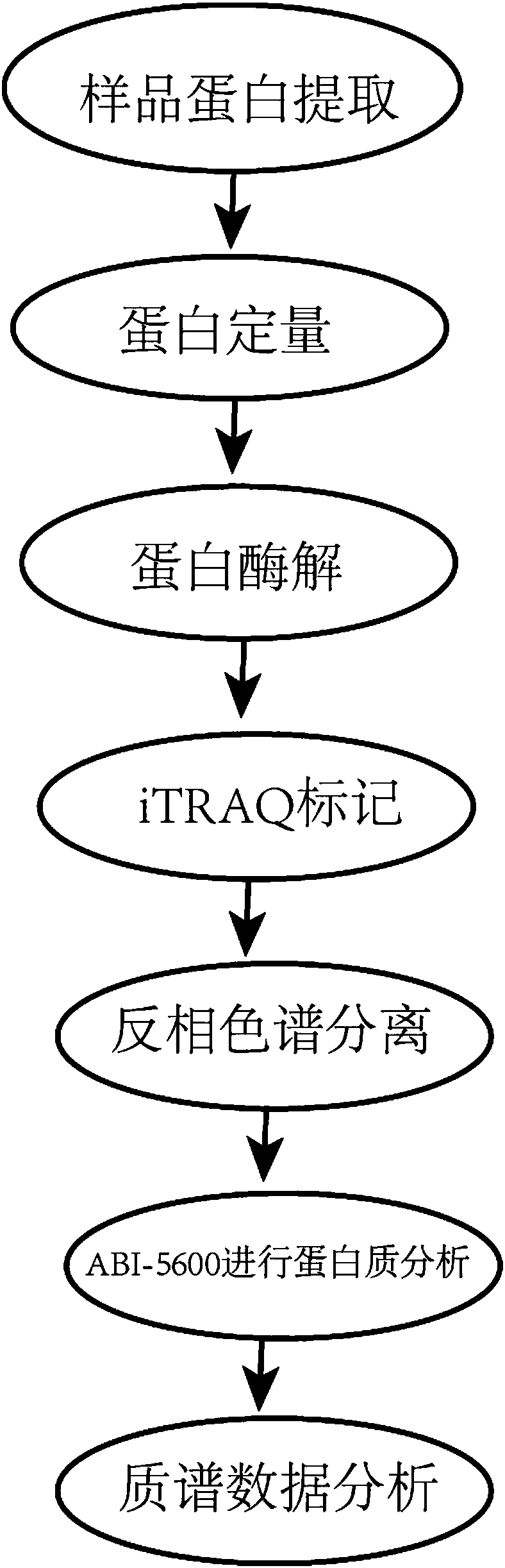
The invention discloses a method for detecting chronic nephritis biomarkers based on salivary proteomics. The method comprises the following specific steps: sample protein extraction; determination ofthe concentration of extracted protein by using a Brad ford method; proteolysis; iTRAQ marking; vacuum refrigerated centrifugation drying; reversed phase chromatography under a high pH condition; protein analysis by using ABI-5600; mass spectral data analysis, namely when selecting a database, in case of an sequenced organism, a database for this species is directly selected, and in case of a non-sequenced organism, a database for a large-range proteome most relevant to a sample to be tested is selected; iTRAQ mass spectrometric analysis by using an ABI-5600 mass spectrometer. The method canbe used for finding out specific protein markers in chronic nephritis by using quick and reliable proteomics technology scheme, so as to establish a non-invasive, easy, convenient, fast, and practicaldetection assessment for early diagnosis and treatment, postoperative recurrence, metastasis and prognosis observation of the chronic nephritis.
Owner:SHENZHEN ELDERLY MEDICAL RES INST +1
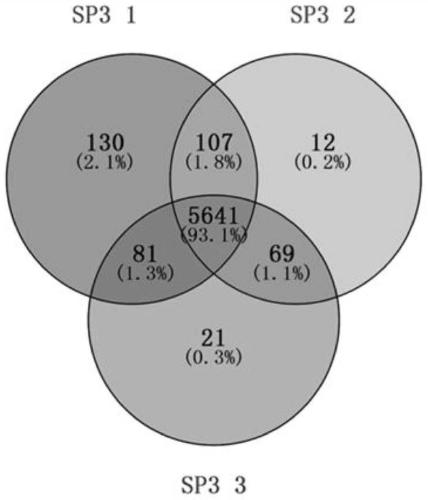
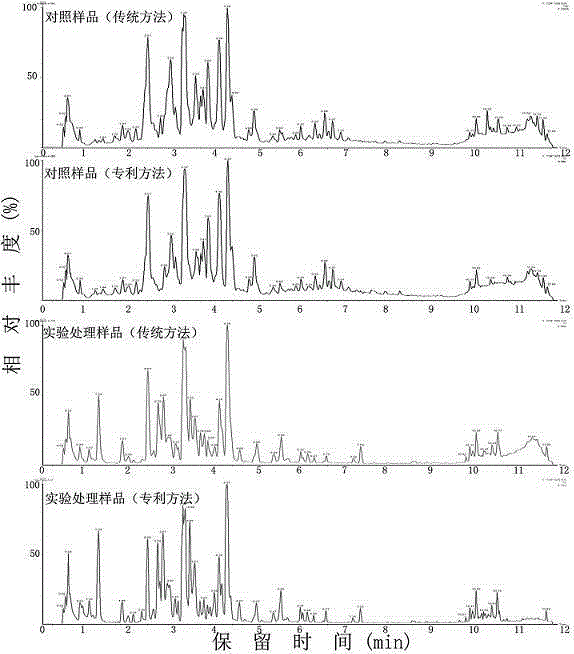
Proteomics analysis method based on SP3 enzymolysis
PendingCN111308097AFully automatedImprove throughputBiological testingFermentationPhenyl EthersMagnetic bead
The invention aims to provide a proteomics analysis method based on SP3 enzymolysis. The method comprises the following steps: adding lysate into cells of a sample to be detected for denaturation; wherein the component of the lysis solution is 50mM of 4-hydroxyethylpiperazine ethanesulfonic acid; 1% lauryl sodium sulfate, 1% of polyethylene glycol octyl phenyl ether; 1% Tween 20, 1% of ethyl phenyl polyethylene glycol; 5 mM of ethylenediamine tetraacetic acid; 50mM sodium chloride, 1% of glycerol; a 1 * protease inhibitor; and 5 mM dithiothreitol, adding a magnetic bead replacement lysate intothe denatured sample; removing a denaturant influencing trypsin activity; after the replacement is finished, carrying out lysine protease enzymolysis and trypsin enzymolysis on the denatured sample;the method has the advantages that automation can be achieved, the repeatability of experiments of different batches is improved, and meanwhile the flux is improved, and meanwhile the method is not inferior to a traditional protein enzymolysis method in terms of evaluation indexes such as the peptide fragment recovery rate and the sample repeatability.
Owner:上海中科新生命生物科技有限公司
Sample pretreatment method for plant metabolome analysis
ActiveCN105289773AReduce direct contactAvoid direct contactPreparing sample for investigationLaboratory glasswaresBiotechnologyPretreatment method
The invention discloses a sample pretreatment method for plant metabolome analysis. The method comprises the following steps: sampling and inactivating, sample homogenizing and grinding, centrifugal filtration and the like; various tissue parts are separately sampled, are frozen and inactivated quickly by adopting liquid nitrogen, and are homogenized and ground by high-speed oscillation in a liquid nitrogen environment; a mixed sample is extracted by adopting ultrasonic in an assistant manner after being obtained; finally, protein is removed through high-speed centrifugation. According to the sample pretreatment method, the defects of temperature change, easiness in pollution, serious sample waste, relatively poor repeatability, uneven sample mixing, relatively low work efficiency, high operating danger coefficient, high requirements on the proficiency and work experience of operating personnel and the like are overcome. The sample pretreatment method can be well applied to hard tissue sampling, homogenizing, grinding and extracting of woody plant samples of tea trees and the like, and is a rapid, nondestructive and pollution-free sample pretreatment technology for performing a metabonomics analysis test. The sample pretreatment method can also be applied to a quality component analysis, RNA (Ribonucleic Acid) extraction, a proteomics analysis and the like.
Owner:TEA RES INST CHINESE ACAD OF AGRI SCI
Method of indentifying an eventual modification of at least one biological parameter implementing young and aged living cells
InactiveUS20040096816A1Evaluate effectCosmetic preparationsToilet preparationsBiochemistryProteomics
An object of the invention is a method of identifying an eventual modification of at least one biological parameter. The present invention relates essentially to a method of identifying an eventual modification of at least one biological parameter, comprising the compared proteomic and / or compared transcriptomic and / or compared genomic analysis: a) of living young living cells, b) of living aged living cells, c) at least one of these two classes of cells being used in a three-dimensional tissue model, enabling eventually identifying at least one biological parameter which is modified further to cell ageing. The invention comprises the use of this process for the screening of active principles.
Owner:BASF BEAUTY CARE SOLUTIONS FRANCE SAS
Photo-induced cell covalent labeled fluorescent molecules as well as preparation method and application thereof
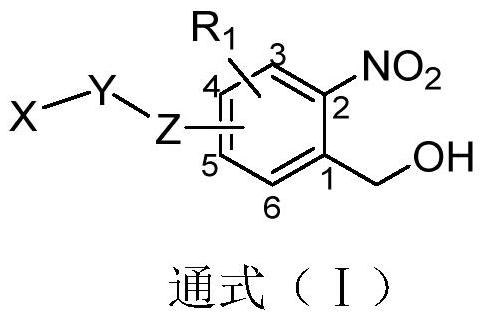
The invention relates to a photoinduced cell covalent labeled fluorescent molecule, and a preparation method and application thereof, in particular to a compound as shown in the following formula (I), or a tautomer, an enantiomer, an enantiomer, a diastereomer, a racemate, a precursor compound, an isotope compound and various forms of salts or hydrates thereof. The compounds can be used for preparing light-induced covalent labeled fluorescent probes and are used for positioning and imaging subcellular organelles in cell biology. The light-induced covalent labeled fluorescent probe has important application potential and outstanding practical value in cell biology research, proteomics research and dynamic change research of biomacromolecules.
Owner:SHANGHAI INST OF MATERIA MEDICA CHINESE ACAD OF SCI
Nano-magnetic microsphere with amino having fixed protease at surface, preparing method and application thereof
InactiveCN1975429AMicrobiological testing/measurementBiological testingProteinase activityMicrosphere
The invention relates to an nm magnetic micro ball with the amino which the surface is fixed with the protease and the preparation method, also it is applied in the protein enzymolysis. The micro ball is composed by the one-step hydro-thermal method and with the particle size of 15-200nm. The glutaraldehyde is as the cross linker and the protease is fixed on the surface of the nm magnetic micro ball by which the protein is solution, slug, target decomposed. The method of the invention is simple, so it is proper for separation and identification of the protein in complex biological sample and it has the good future in the protein study.
Owner:FUDAN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com