A functional enviromics method for cell culture media engineering
A culture medium and cell technology, applied in biochemical equipment and methods, microbial measurement/testing, microorganisms, etc., can solve the problems of difficult, expensive and difficult systematic collection of proteome data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0017] Hereinafter, the best mode for carrying out the present invention is described in detail.
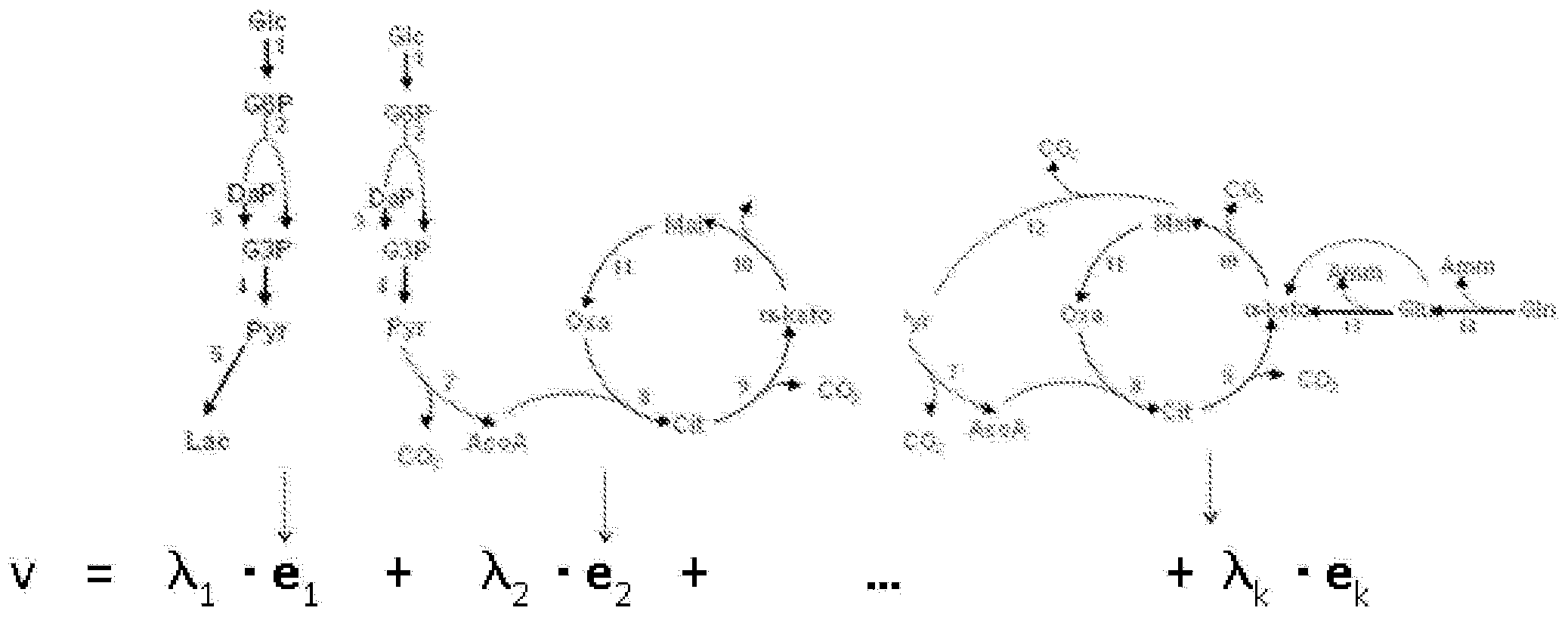
[0018] A unique feature of the method of the present invention is the joint screening of mediator factors and basic cellular functions through a specific protocol to extract data which is subsequently processed into the form of a functional environmental map. The biological configuration into which the medium will be designed (herein referred to as the "target biological configuration") must be clearly articulated before proceeding with the experimental protocol. It can be a whole cell, an organelle, or a coordinated set of biochemical reactions representing a given cellular function. The target biological structure is represented by q biochemical reactions and associated genes and proteins if they are known. Once the structure is clearly elucidated, a working group of N factors and K basic cellular functions is established as the basis.
[0019] The medium formulation is defin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com