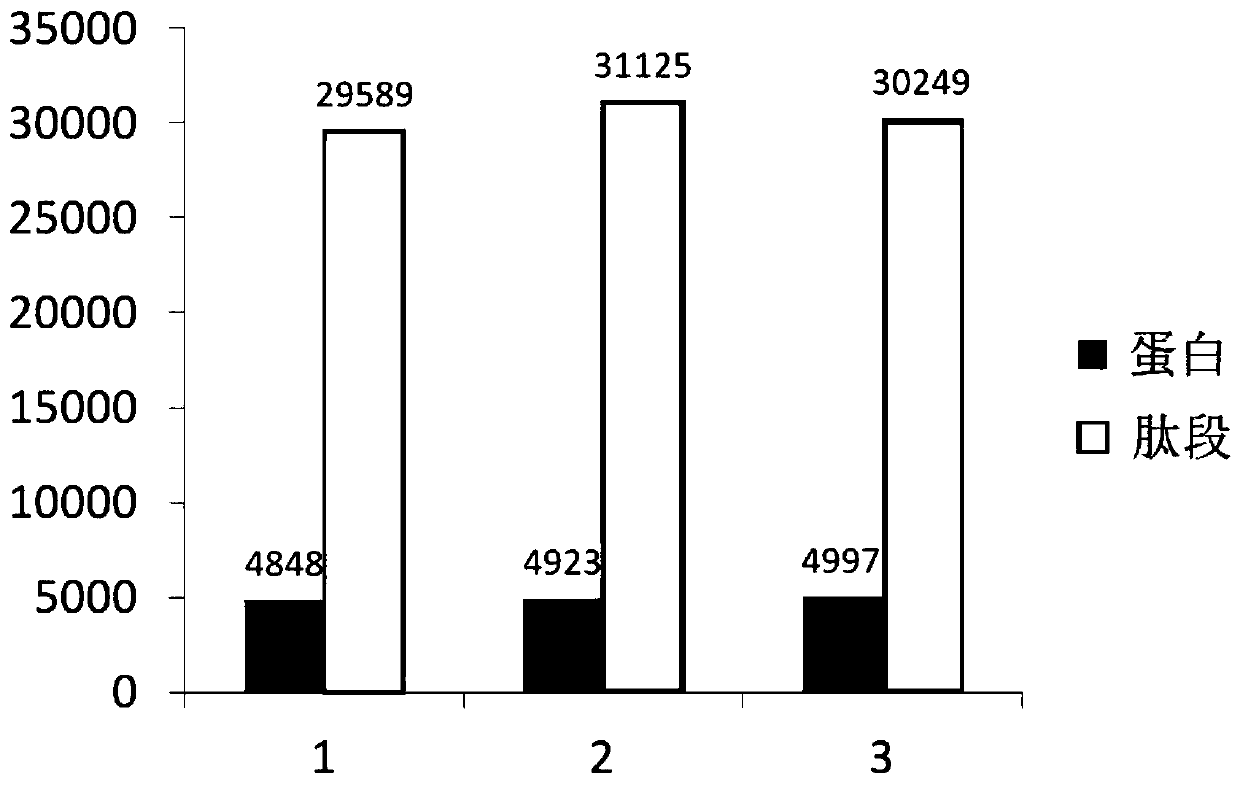
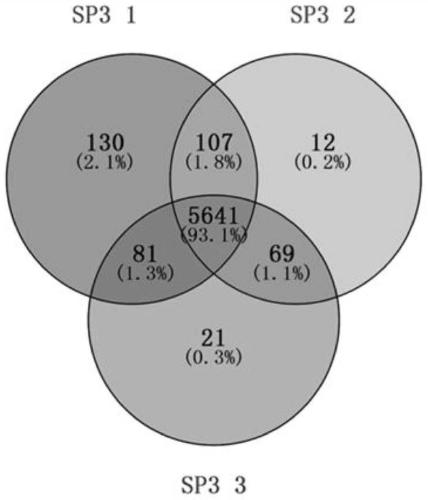
Proteomics analysis method based on SP3 enzymolysis
A technology of proteomics and analysis methods, applied in the field of biological detection, can solve the problems of large sample loss, time-consuming, affecting the effect of enzymatic hydrolysis, etc., and achieve the effect of realizing automation and improving throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0023] The technical solutions of the present invention will be described below in conjunction with the accompanying drawings and embodiments.
[0024] A proteomics analysis method based on SP3 enzymatic hydrolysis. First, add lysate to the cells of the sample to be tested for denaturation, and then add magnetic beads to the denatured sample to replace the lysate. solution, and finally extract the enzymolysis sample for chromatography-tandem mass spectrometry analysis, including the following detailed steps:
[0025] Step S1 For denatured cell samples, take a plate of 293T cells overgrown in a 10cm culture dish, wash the cells with 10mM phosphate buffered saline for 3 times, then add 500μl lysate to the cleaned cell samples, and use a homogenizer After homogenization, a contact ultrasonic instrument was used for assisted denaturation. The ultrasonic power was 200W. The ultrasonic treatment method was 8S, followed by a 5S interval, followed by the second treatment, and the cycl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com