Identification and use of glycopeptides as biomarkers for diagnosis and treatment monitoring
A technology of biomarkers and biological samples, applied in biological testing, disease diagnosis, bioinformatics, etc., can solve problems such as the inability to trace back to the initial protein site of glycan
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
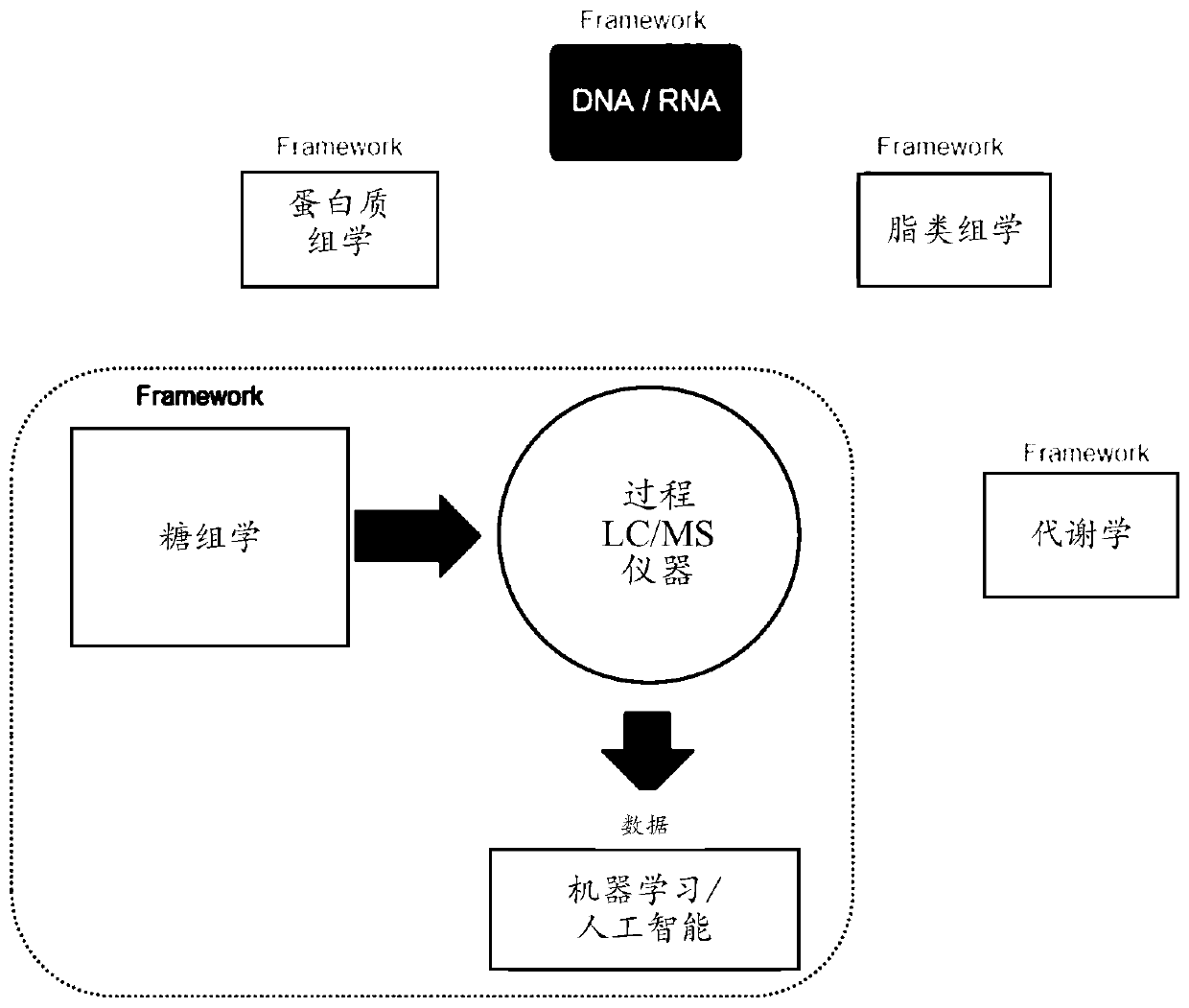
[0097] General approach to biomarker discovery
[0098] In a targeted approach, the target glycoprotein is first identified in a biological sample, and then the peptide fragments are identified and quantified using LC / MS to analyze the site of modification, the nature of the modification, the nature of the modification, and the relative abundance of each modification. The method uses a triple quadrupole (QQQ) mass spectrometer to quantify glycosylated peptide fragments and then analyze their relationship to the subject classification.
[0099] In a non-targeted approach, the glycosylation patterns of all peptide fragments (known and unknown) are analyzed for information on changes in glycosylation patterns in various subjects. Specifically, the up- or down-regulation of glycoproteins is monitored for the classification of subjects. For example, glycoprotein fragments are monitored separately for subjects with a disease or condition versus subjects without the disease or condi...
example 2
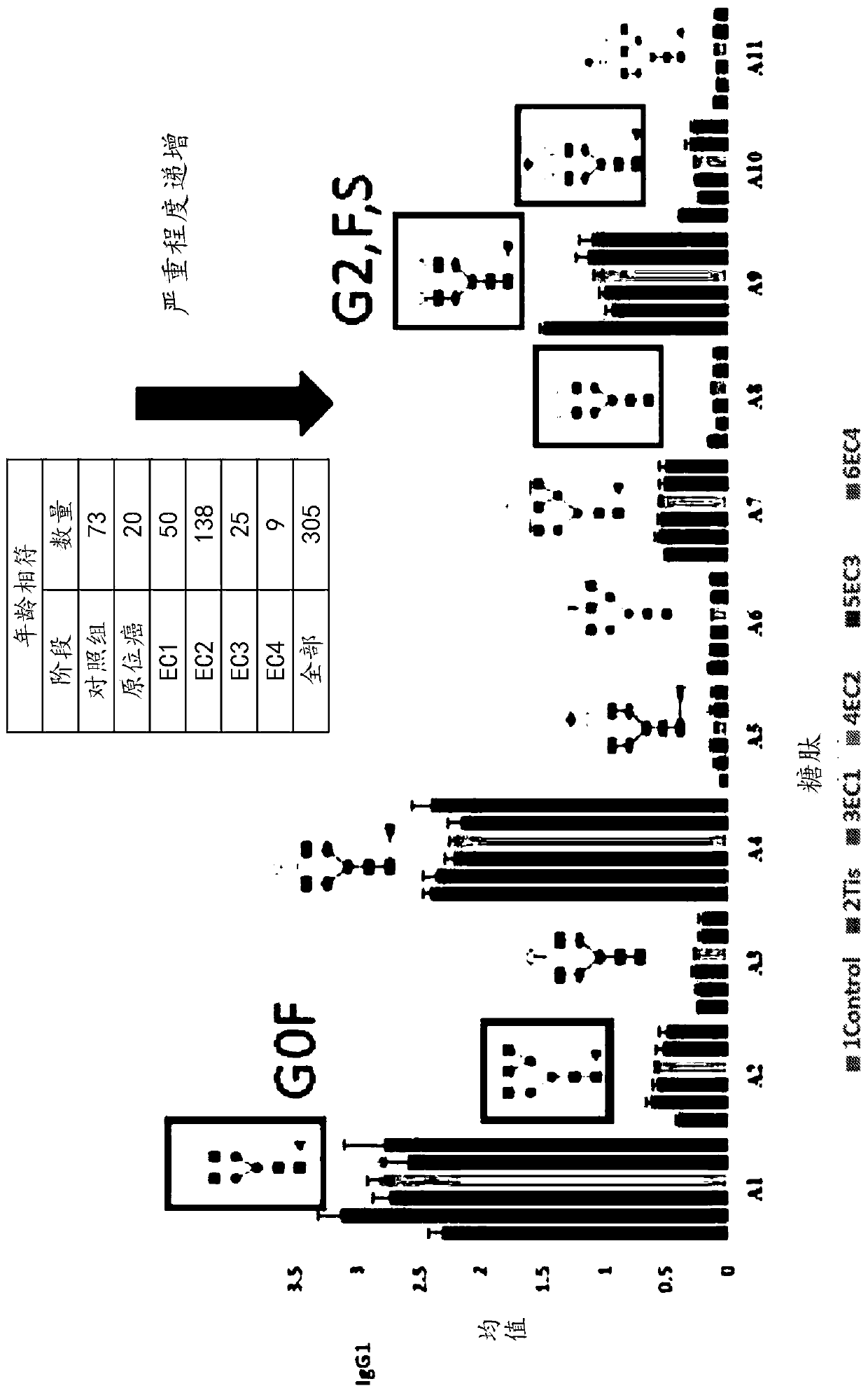
[0101] Quantitative analysis of IgG glycopeptides as potential biomarkers for breast cancer
[0102] Plasma samples of breast cancer patients at different stages and a control group corresponding to the age of said patients were analyzed for IgG1, IgG0 and IgG2 glycopeptides, and their ratio changes were compared. Specifically, 20 samples at the stage of carcinoma in situ, 50 samples at the EC1 stage, 138 samples at the EC2 stage, 25 samples at the EC3 stage, 9 samples at the EC4 stage and 73 A control sample matched to their age was used for MRM quantification. Quantitative results such as figure 2 As shown, in the various stages of breast cancer studied in this experiment, the levels of certain IgG1 glycopeptides were increased and the levels of certain IgG1 glycopeptides were decreased compared with the control group. For example, in the various stages of breast cancer studied in this experiment, the IgG1 glycopeptide Al-A11 was monitored; compared with the control group...
example 3
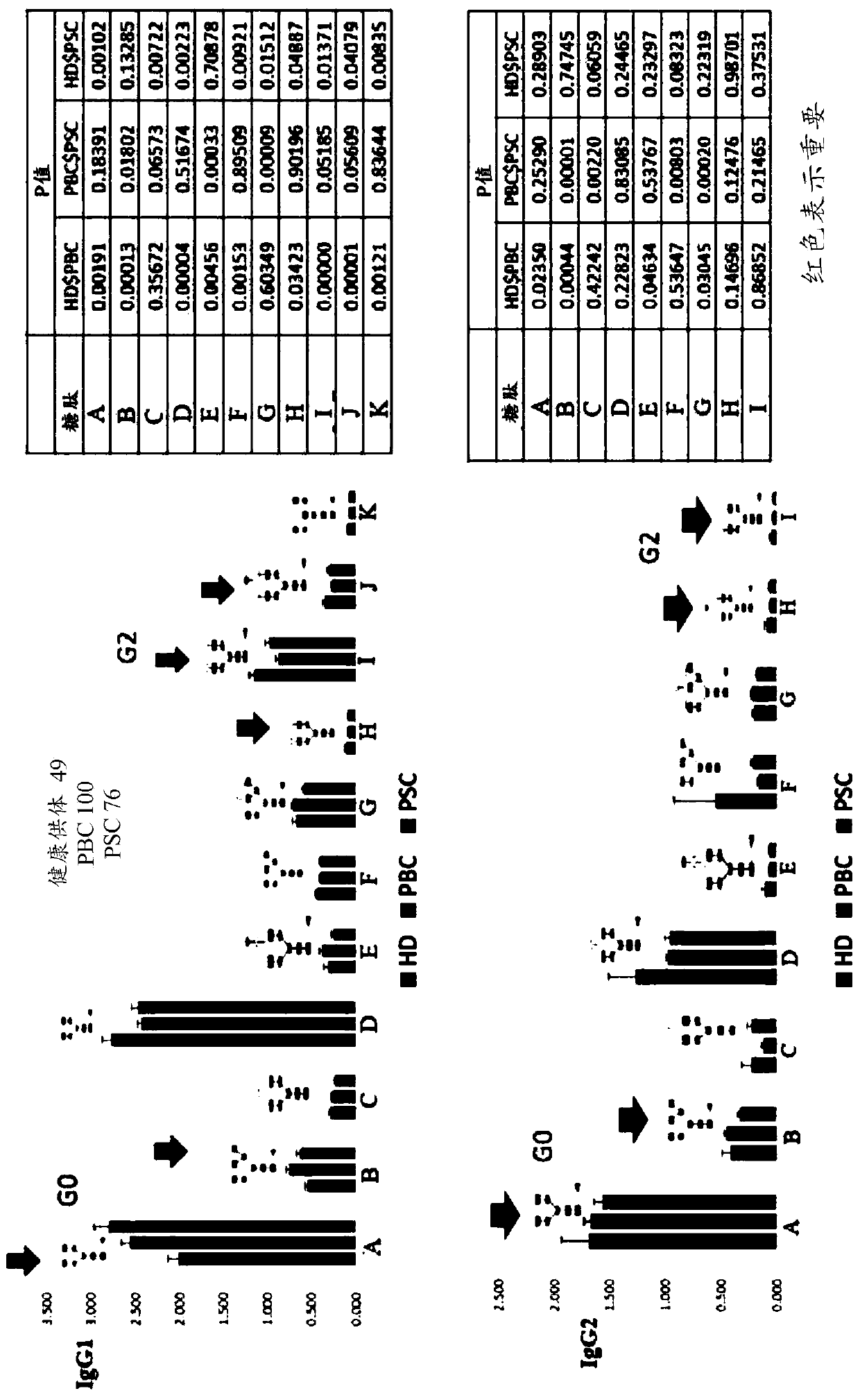
[0104] Quantitative analysis of IgG glycopeptides as potential biomarkers for PSC and PBC
[0105] Analysis of IgG1 and IgG2 glycopeptides in plasma samples from patients with primary sclerosing cholangitis (PSC), patients with primary biliary cirrhosis (PBC), and plasma samples from healthy donors and comparing their glycopeptide ratios The change. Specifically, 100 PBC plasma samples, 76 PSC plasma samples, and 49 healthy donor plasma samples were subjected to MRM quantitative analysis on a QQQ mass spectrometer. from image 3 It can be seen from the quantitative results that some IgG1 glycopeptides were elevated and some IgG1 glycopeptides were decreased in the plasma samples of PBC and PSC patients compared with healthy donors. For example, glycopeptide A was elevated and glycopeptides H, I, and J were decreased in PBC and PSC patients compared with healthy donors. Therefore, glycopeptides A, H, I and J are potential biomarkers for PBC and PSC.
[0106] Similar analyze...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com