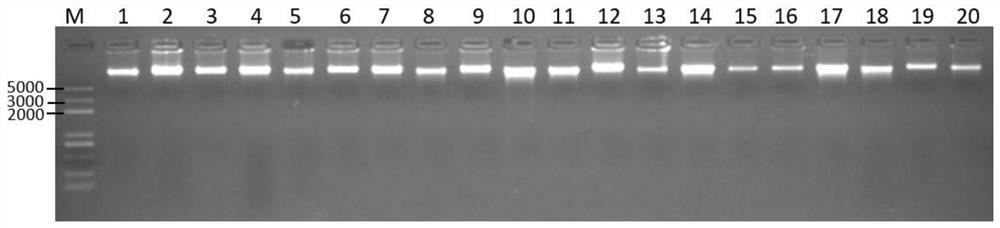
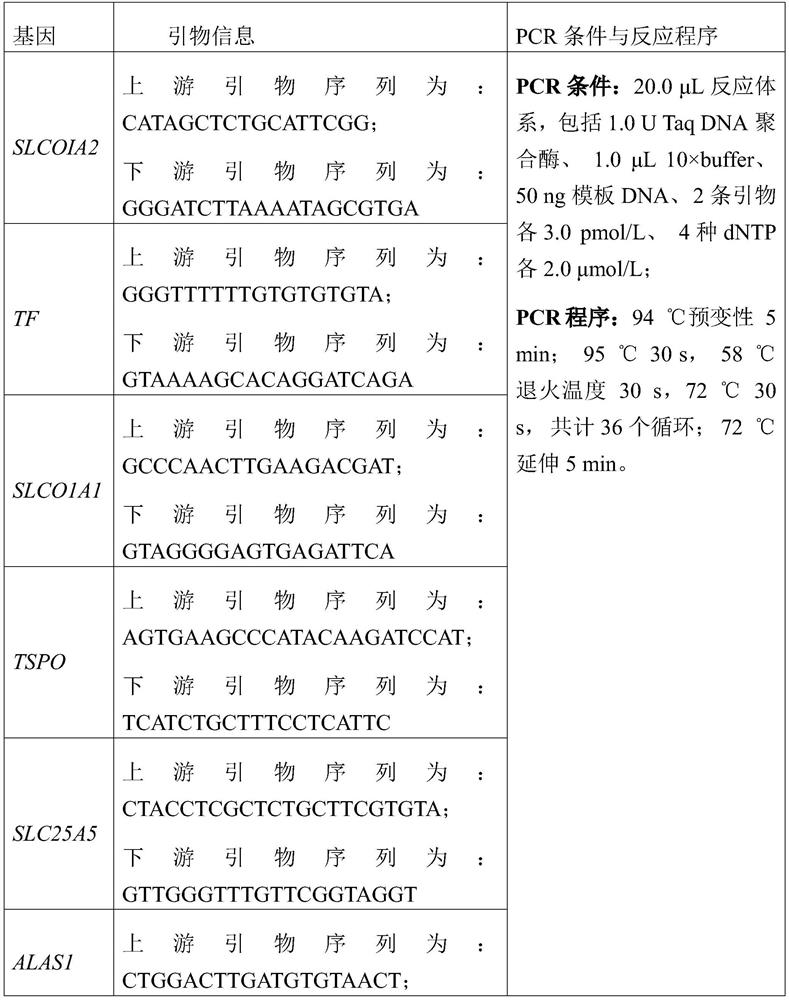
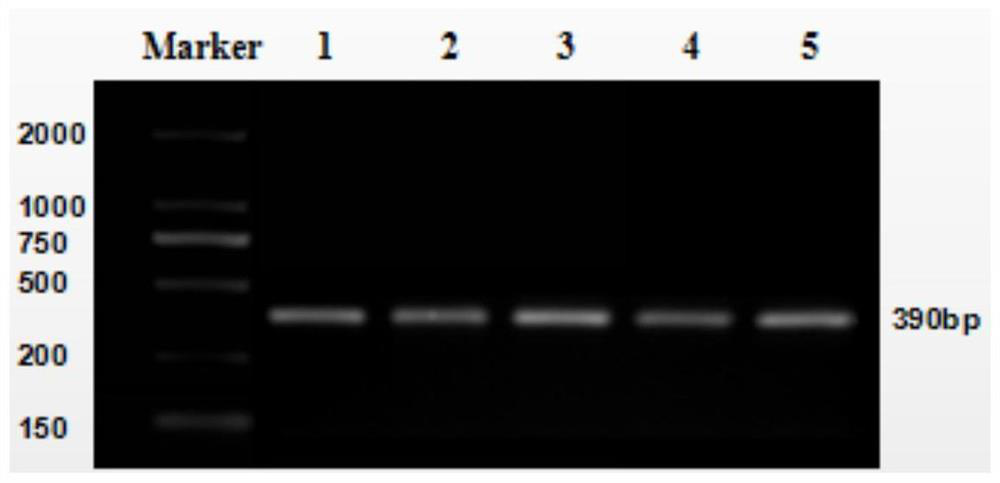
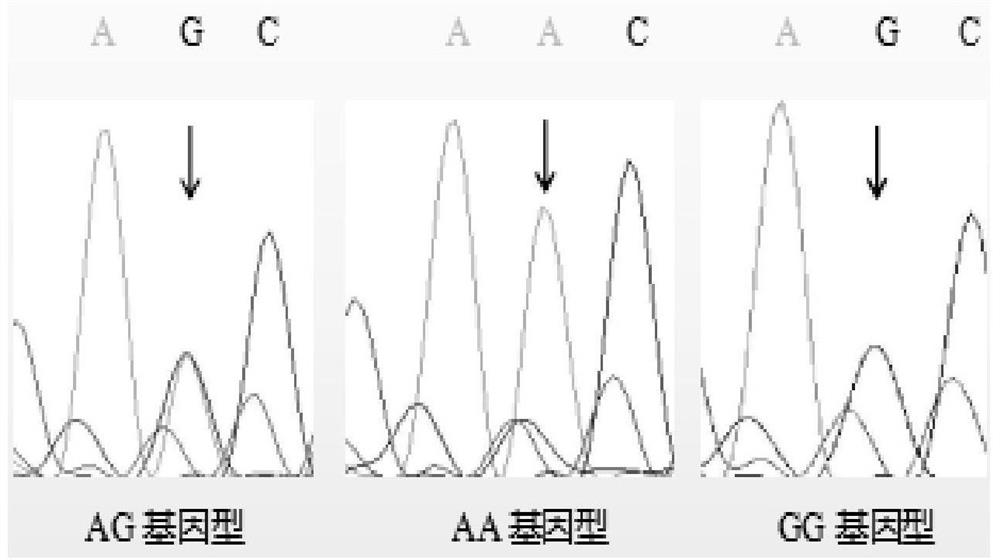
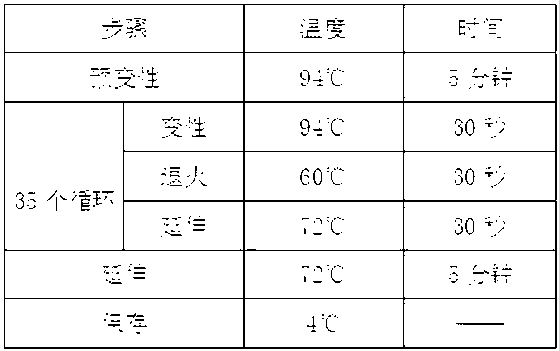
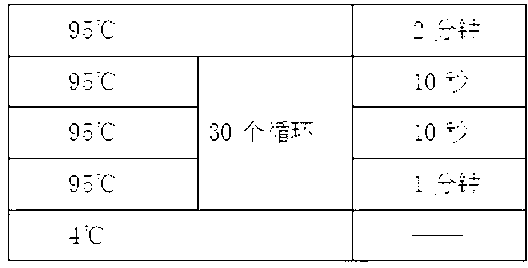
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
74 results about "Pcr sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
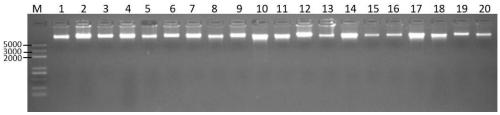
PCR and DNA sequencing are two important techniques in Molecular Biology. Polymerase Chain Reaction (PCR) is the process which creates a large number of copies of a DNA fragment. DNA sequencing is the technique which results in the precise order of the nucleotides of a given DNA fragment.
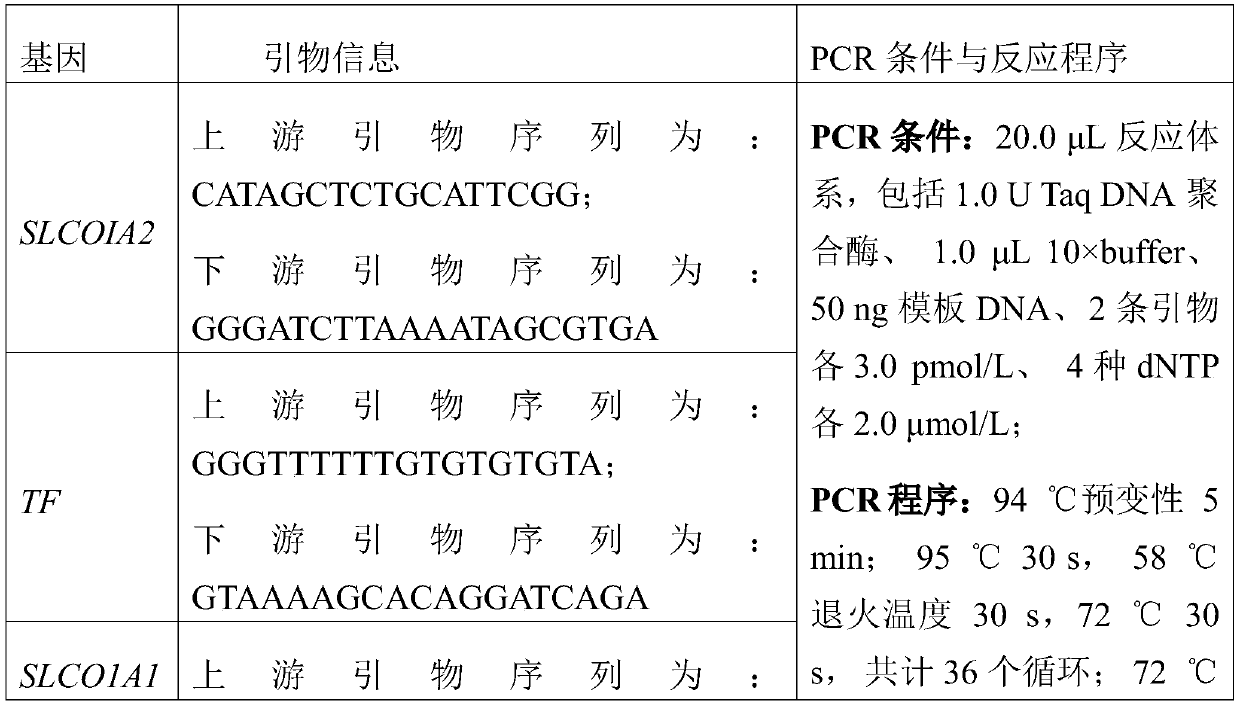
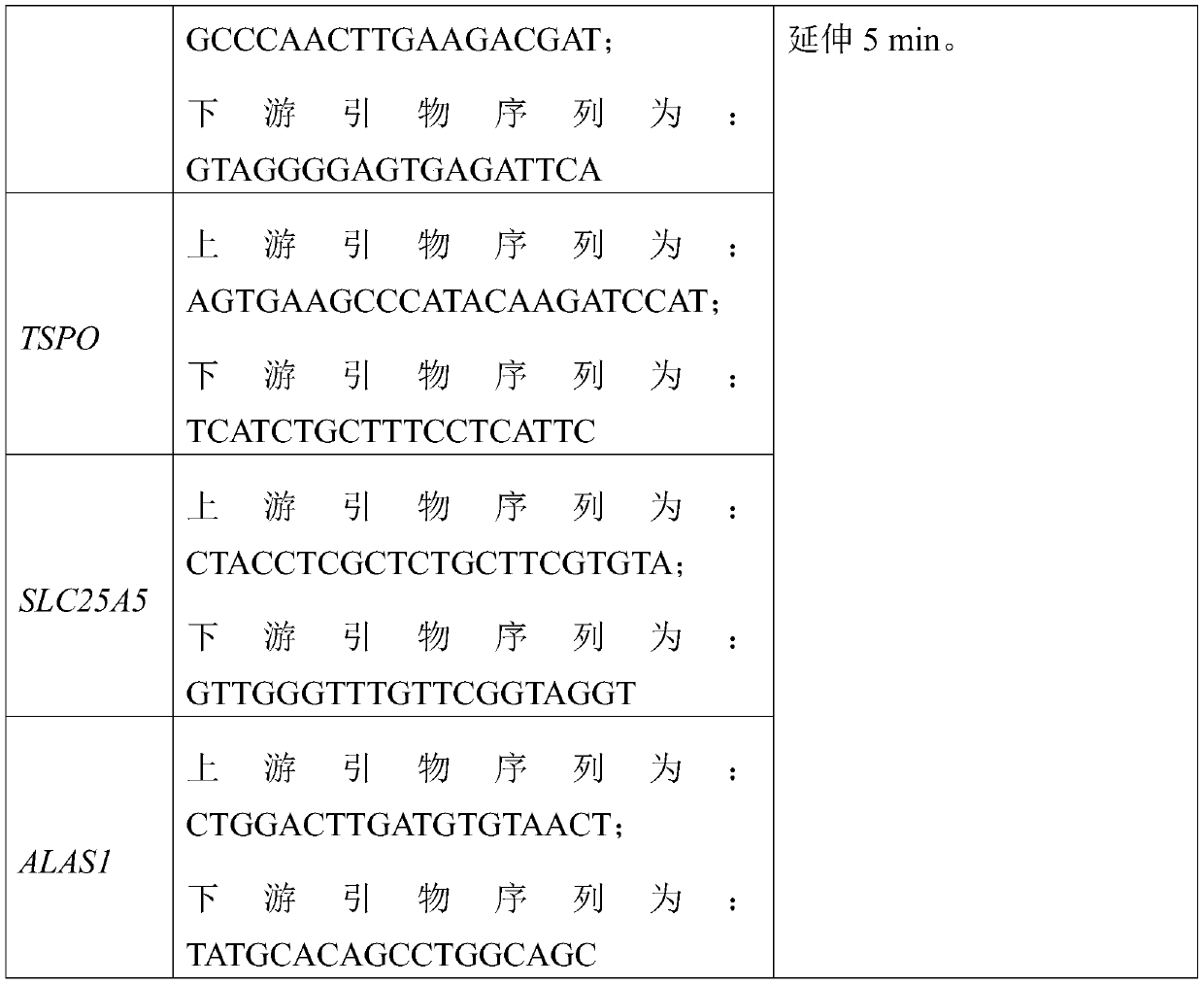
DNA molecular label technology and DNA incomplete interrupt policy-based PCR sequencing method
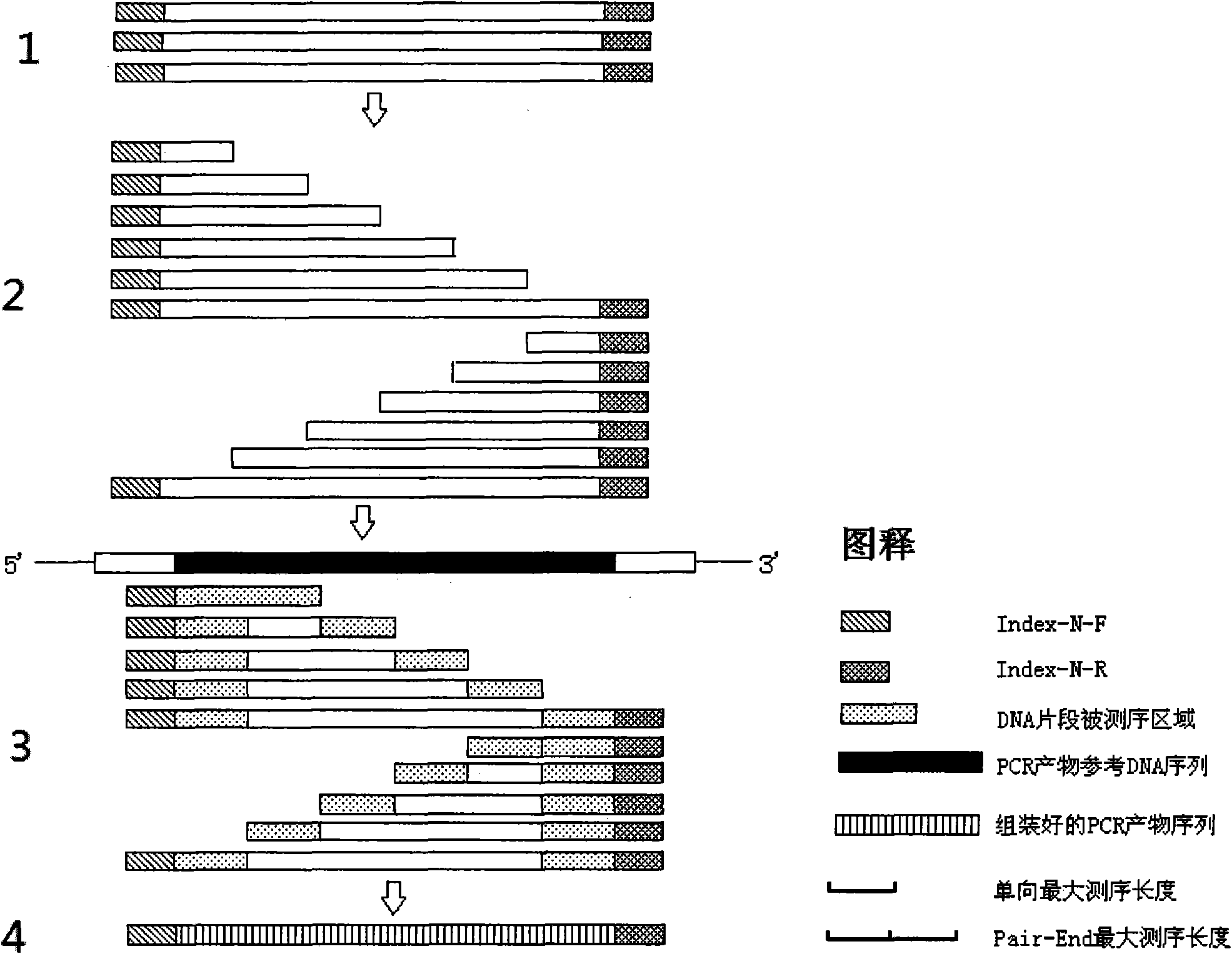
The invention provides a PCR sequencing method. The length of a PCR product which can be completely measured by a sequencer exceeds the measuring length of the sequencer through the combination of a primer label, a DNA incomplete interrupt policy and second-generation sequencing technology (Pair-End sequencing technology) and by taking full advantage of the characteristics of high flux and low cost of a second-generation sequencer at the same time, so that the application range of the method is greatly widened. At the same time, the invention also provides the primer label for the PCR sequencing method and the application of the method to gene typing, particularly to HLA analysis.
Owner:BGI GENOMICS CO LTD
Endogenous protein marking method used for Chip-seq genome-wide binding spectrum
InactiveCN106399311AMeet the packaging ratioHigh infection efficiencyMicrobiological testing/measurementBiological testingMaterial resourcesStable cell line
Owner:TONGJI UNIV
Whole-genome methylation non-bisulfite sequencing library, construction and applications thereof
InactiveCN110820050ASolve the defect of low usage rateReduce usageNucleotide librariesMicrobiological testing/measurementDihydrouracilPhosphoric acid
The invention relates to the technical field of bioinformatics, particularly to a whole-genome methylation non-bisulfite sequencing library, a construction and applications thereof, wherein the sequencing library comprises a TET enzyme reaction solution, the TET enzyme reaction solution comprises the following independently packaged components: a TET enzyme oxidation buffer solution, and the TET enzyme oxidation buffer solution comprises, by micromole, (20-167)*10<3> parts of HEPES or Tris-Cl, (100-333)*10<3> parts of NaCl, 3.3*10<3> parts of alpha-KG or 2-oxoglutarate, 6.67*10<3> parts of ascorbic acid and 4*10<3> parts of adenosine triphosphate. According to the invention, by combining the kit, Fe(NH4)2(SO4)2 and TET enzyme, 5mc can be oxidized into 5cac, the 5cac is reduced into dihydrouracil under the action of a reducing agent, and T is identified through PCR sequencing, so that the the DNA methylation C-to-T conversion under the non-bisulfite condition is achieved, the defects ofbase imbalance and low sequencing data use rate of the existing methylation sequencing library constructed based on the bisulfite conversion are solved; and the formula further has effects of simplecomponents, extremely low TET enzyme use amount and significant cost reducing.
Owner:BEIJING GENEPLUS TECH +1
Kit for detecting polymorphism of VKORC1 and CYP2C9 genes
ActiveCN102329885AHigh detection sensitivityImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationWarfarinVKORC1
The invention provides a kit for quickly detecting polymorphism of VKORC1 and CYP2C9 genes. The kit is used for guiding the dosage of clinical warfarin. The kit comprises three pairs of primers shown as SEQ ID No.1-6, and can also further comprise one or more of the following reagents: polymerase chain reaction (PCR) reaction liquid, negative control, positive control, PCR sequencing reaction liquid and human peripheral blood genome extraction reagent. The kit is used for detecting the polymorphism of the VKORC1 and CYP2C9 genes by adopting a gene sequencing method with relatively high sensitivity and specificity, the specificity and the sensitivity of the detection result of the kit are remarkably improved, and the kit provides a brand-new, quick, simple and convenient gene diagnosis technology for clinically improving the safety and the validity of warfarin application.
Owner:李艳 +1
Kit used for staphylococcus aureus detection
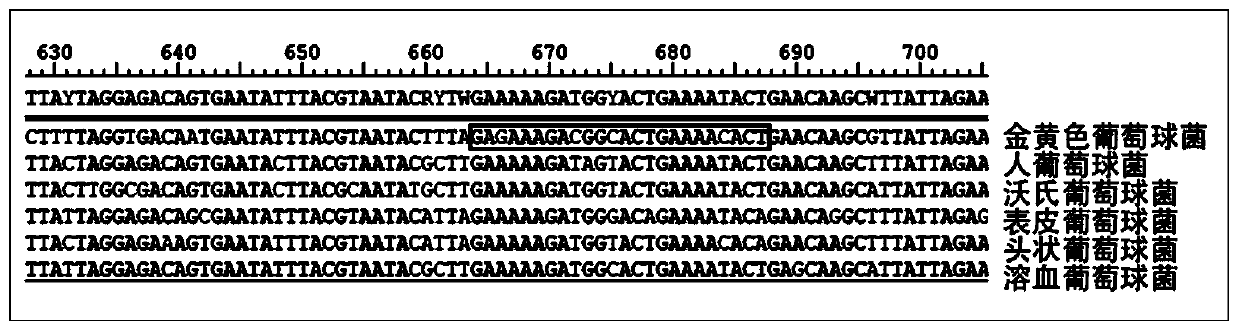
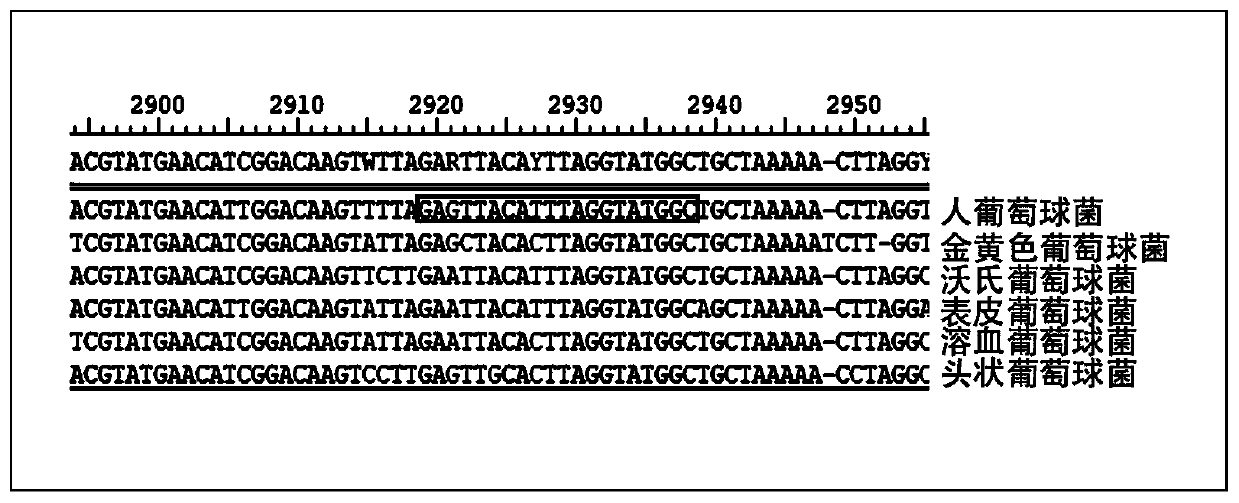
InactiveCN110079586ALower requirementStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationStaphylococcus cohniiStaphylococcus haemolyticus
The invention provides a kit used for staphylococcus aureus detection. The kit used for staphylococcus aureus detection contains guide RNA of a specific targeted staphylococcus aureus rpoB gene, an amplification primer pair, hydrated TwistAmp basic kit reaction drying balls, LbCas12a protein, a single-stranded DNA probe (ssDNA), Ribonuclease Inhibitor and buffer liquid. By using the kit for staphylococcus aureus detection, the detection specificity is high, and staphylococcus aureus and other staphylococcus, including staphylococcus epidermidis, staphylococcus hominis, staphylococcus warneri,staphylococcus capitis and staphylococcus haemolyticus, can be separated. Meanwhile, by using a RNA sequence for detection, elapsed time is about 1 hour, no PCR instruments are used, the equipment requirements are low, and are significantly lower than that of a traditional bacterial culture method or PCR sequencing method, and the clinical application valve is large.
Owner:ZHEJIANG UNIV
SNP molecular marker associated with pig fat deposition, and applications thereof
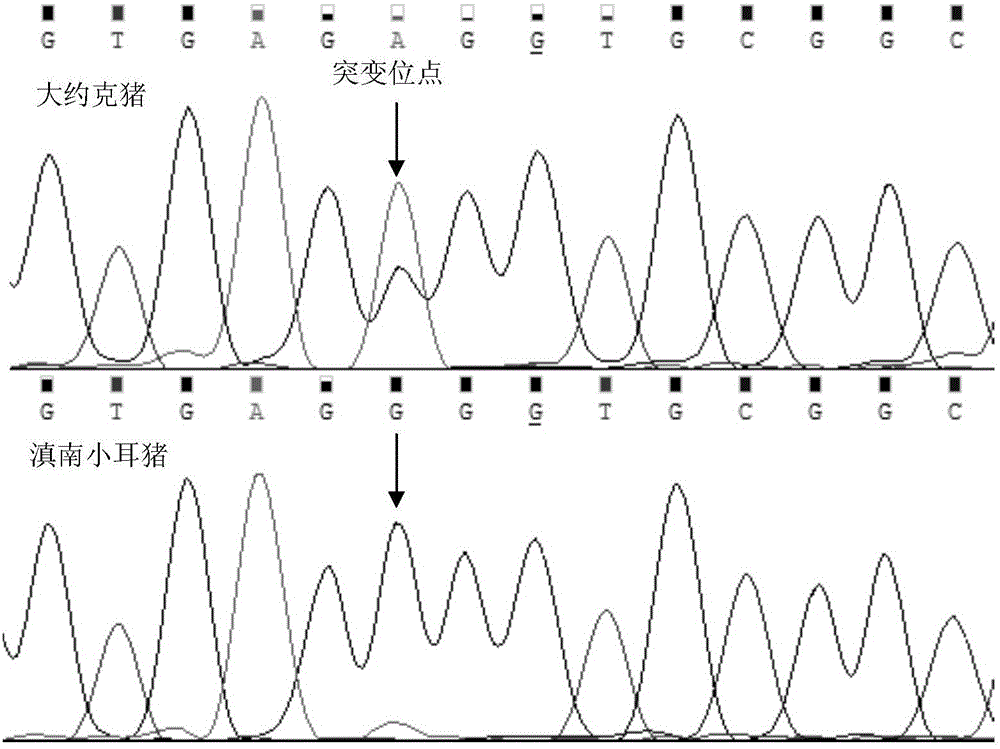
The present invention relates to a SNP molecular marker associated with pig fat deposition, wherein the SNP molecular marker is located at 931bp on the upstream of the initiation codon of the pig FNDC5 gene 5' flanking region, and is recorded as A-931G. According to the present invention, the FNDC5 gene is adopted as the candidate gene affecting the pig fat deposition trait, the different SNP sites among different types of pig populations are obtained by using the PCR sequencing technology, the rapid detection method is established, the relevance between the SNP site and the pig fat deposition capability among the varieties and in the species is identified, and the SNP site affecting the pig fat deposition trait and the genotype are obtained.
Owner:CHINA AGRI UNIV
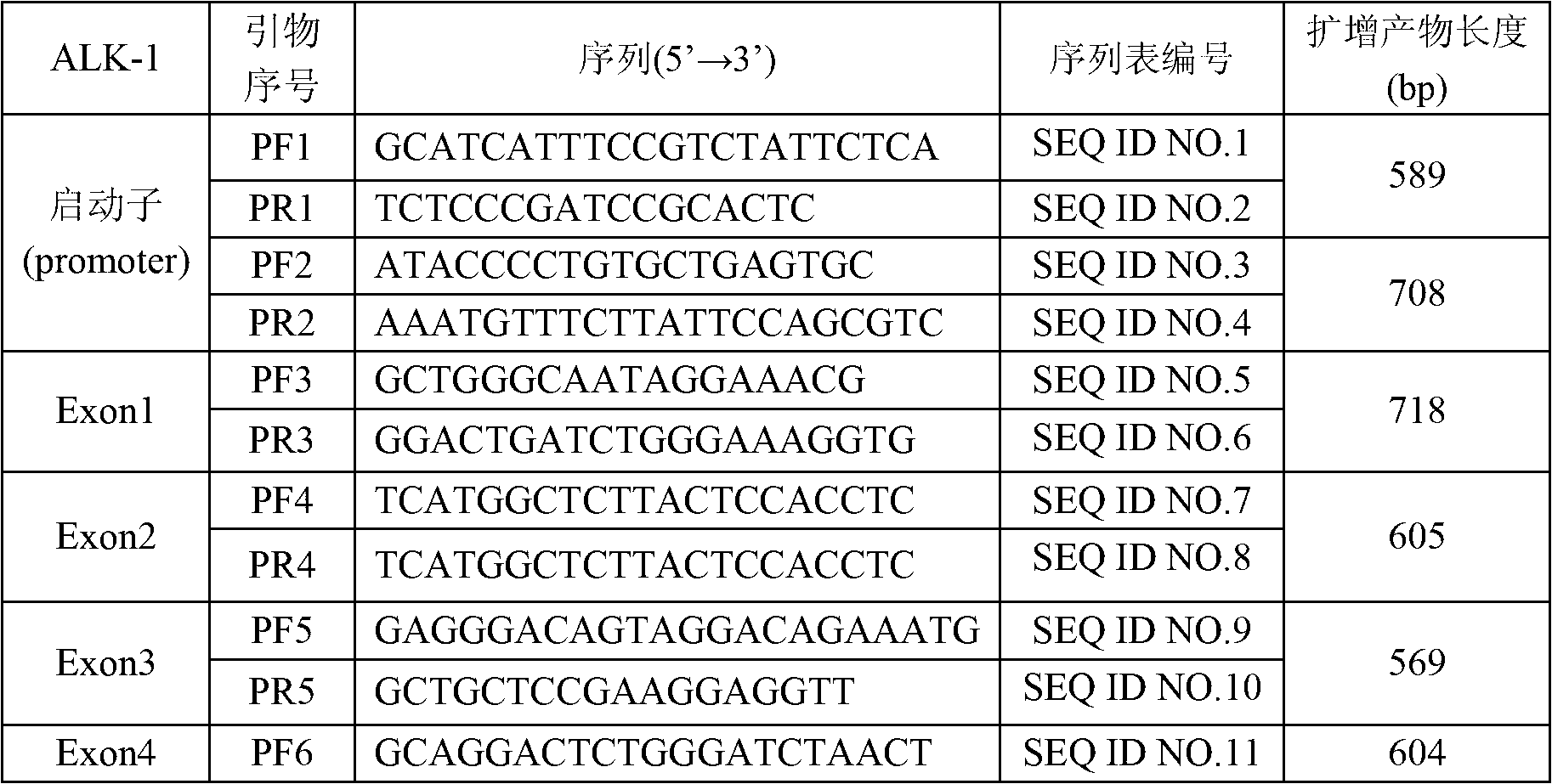
Primer group for detecting ALK-1 (activin receptor type II-like kinase-1) gene mutation, and application thereof as well as kit comprising primer group
ActiveCN102994628AGuaranteed accuracyEnsure uniquenessMicrobiological testing/measurementDNA/RNA fragmentationActivin Receptors Type IIKinase
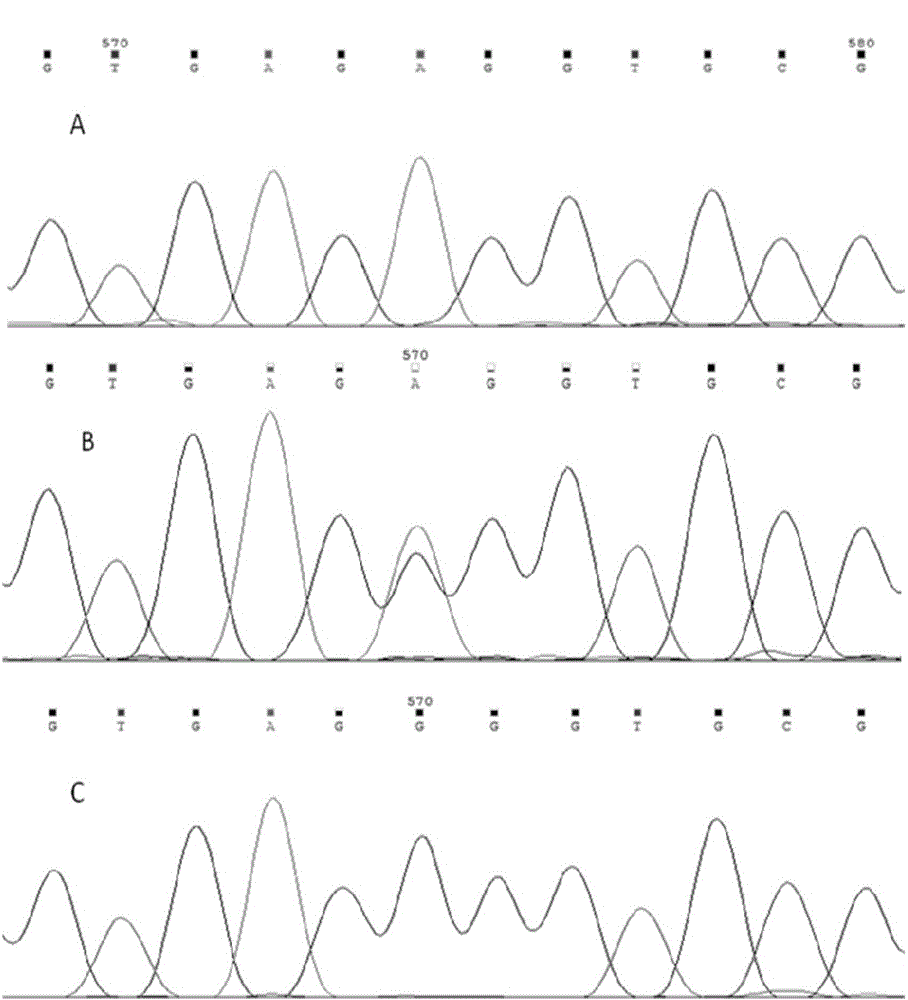
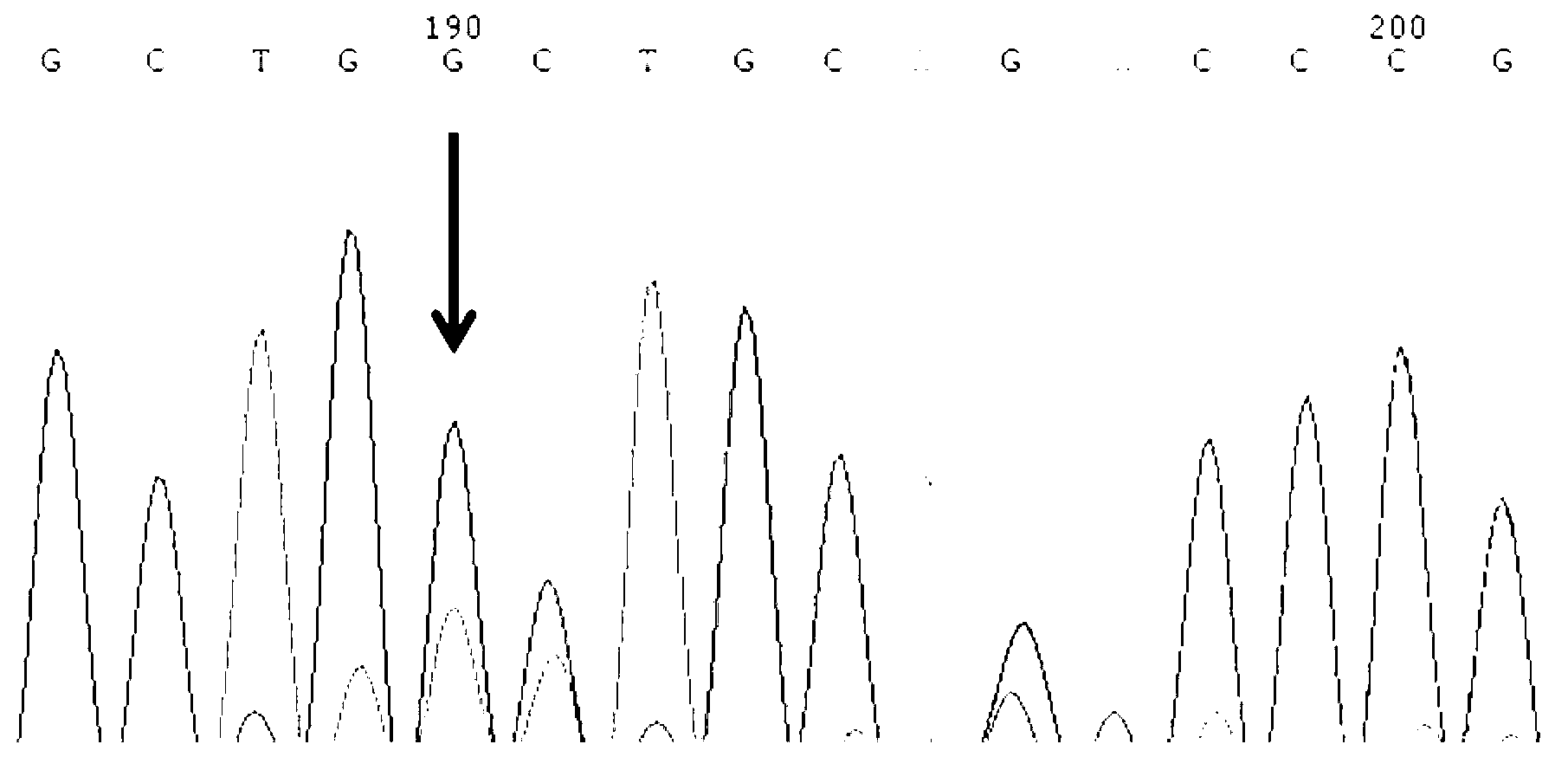
The invention discloses a primer group for detecting ALK-1 (activin receptor type II-like kinase-1) gene mutation, at least comprising ALK-1 gene first to tenth exon regions, and a PCR (Polymerase Chain Reaction) amplification primer group and a PCR sequencing primer group which correspond to any region of a 3' UTR (Untranslated Region) and a promoter. The invention further discloses an application of the primer group, a kit comprising the primer group, and a method for carrying out PCR amplification on an ALK-1 gene by the primer group. The primer group can be used for specially identifying whether the ALK-1 gene is mutated or not so as to ensure the accuracy and the uniqueness of a detected result.
Owner:广州瑞能精准医学科技有限公司
Screening method and application of SNP molecular marker associated with Nandan Yao egg shell color
ActiveCN111705139AImprove color uniformityEasy to filterMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyWhole Genome Association Analysis
The invention discloses a screening method of an SNP molecular marker associated with Nandan Yao egg shell color. The screening method comprises the following steps: (1) finding out genes related to egg quality from known literatures, designing primers, taking Nandan Yao chicken, collecting blood, extracting genomic DNA, and performing sequencing in a mixed pool; (2) taking Nandan Yao hens as testchickens, measuring eggshell color and shape indexes of eggs from 24 weeks old and to 60 weeks old; (3) collecting blood of the 60-week-old test chicken, extracting genome DNA, and performing PCR sequencing on detected SNP sites; and (4) sorting sequencing data and eggshell color data, screening out candidate SNP loci significantly associated with eggshell colors, and providing an SNP molecular marker theoretical basis for eggshell color breeding. The method can be applied to rapid and simple cultivation for improving the eggshell color of Nandan Yao chicken eggs, simplifies whole genome association analysis, directly performs SNP marker screening on trait-related genes, and is suitable for breeding of quantitative traits with high heredity.
Owner:GUANGXI UNIV
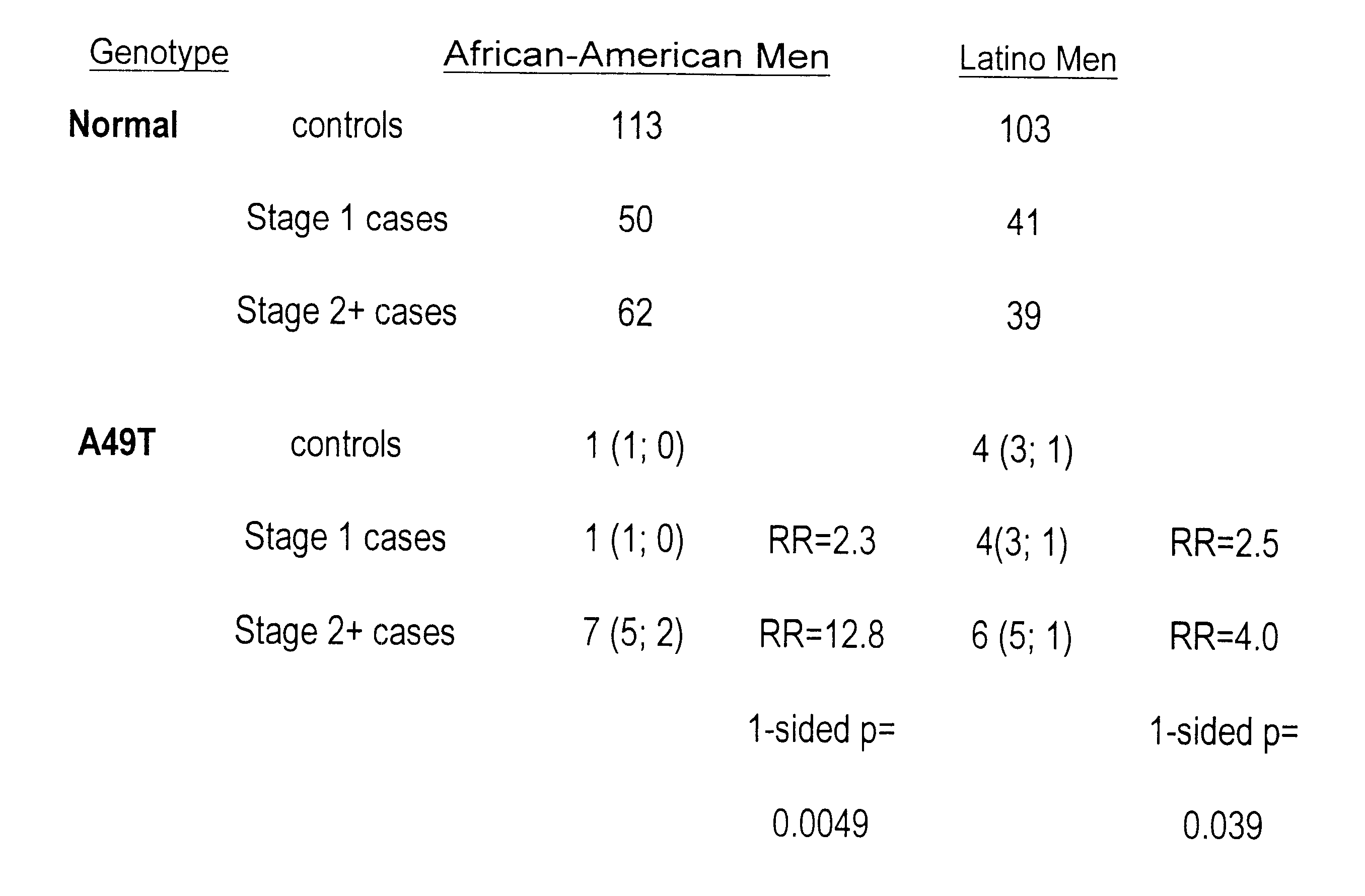
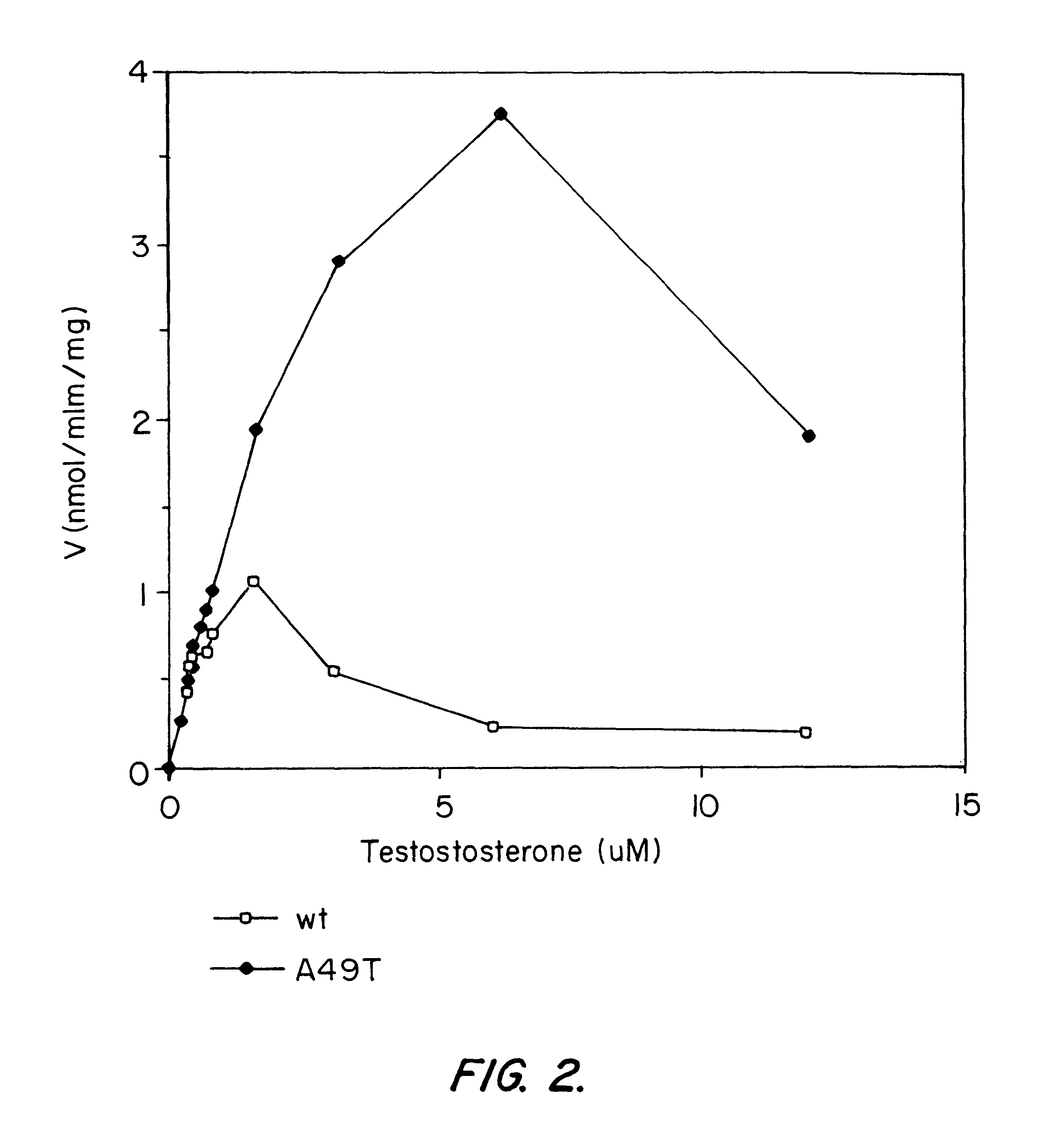
Androgen-metabolic gene mutations and prostate cancer risk
This present invention identifies mutations in several androgen-metabolic genes (SRD5A2, CYP17, HSD3B2, and HSD17B3) and methods of using such mutations in the diagnosis and treatment of inheritable prostate cancer susceptibility. Isolation of genomic DNA of various racial / ethnic populations followed by SSCP scanning and direct PCR sequencing of the aberrant SSCP (single-strand conformation dependent DNA polymorphism) patterns allows for identification of the disclosed polymorphisms. Screening for the disclosed mutations establishes a differential distribution among various racial / ethnic groups as well as altered in vivo enzyme activity that parallels prostate cancer risk.
Owner:UNIV OF SOUTHERN CALIFORNIA
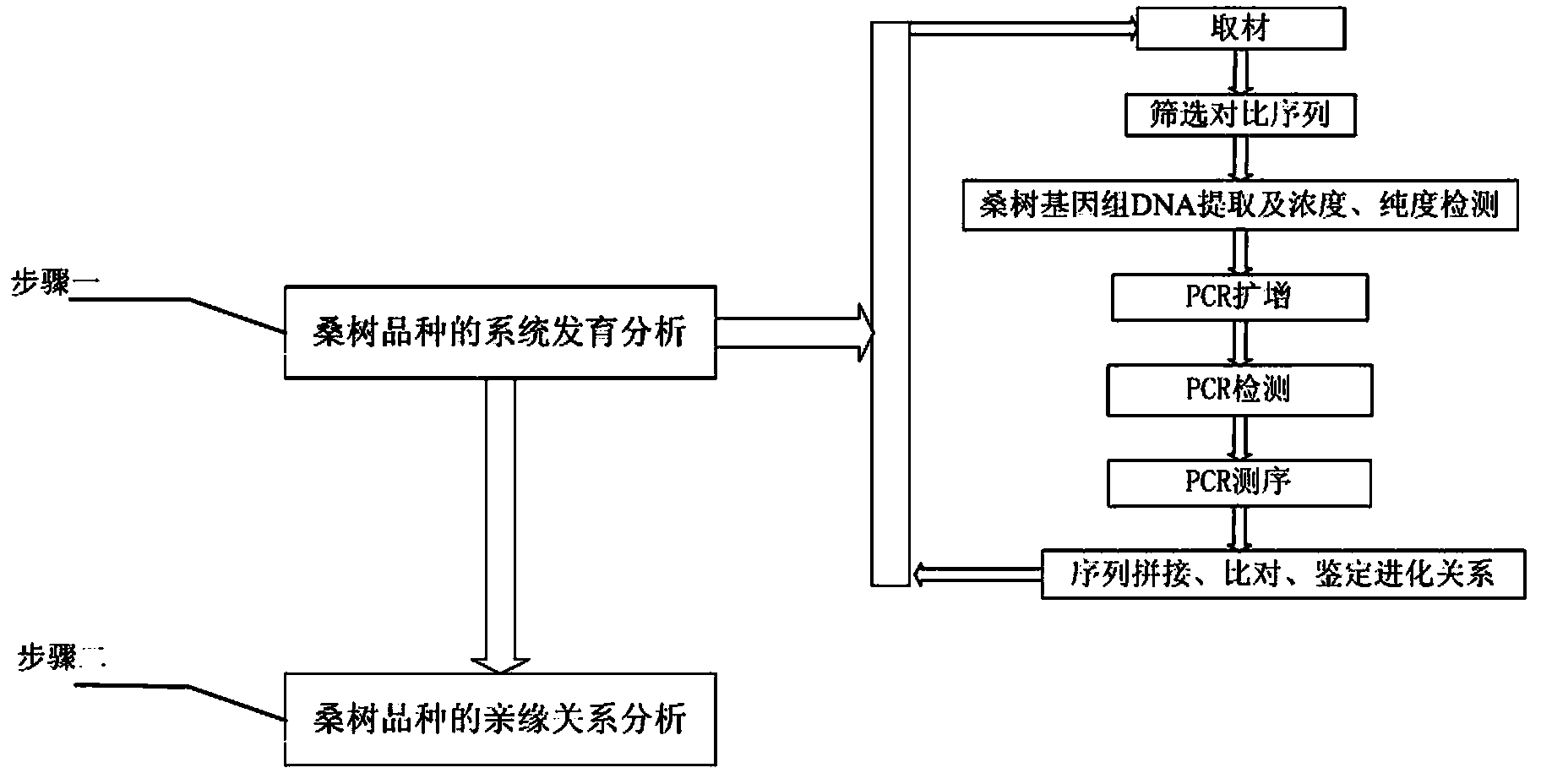
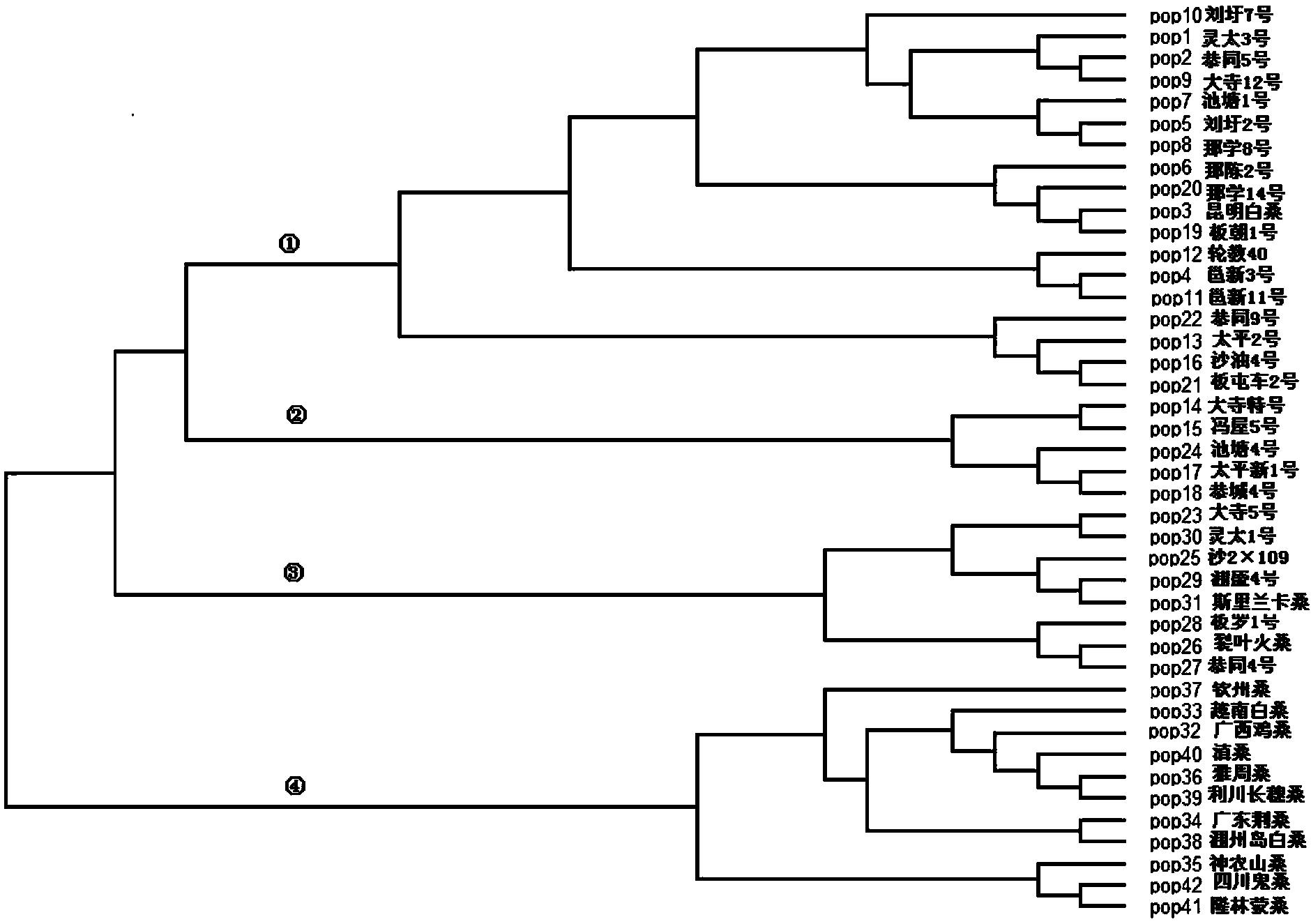
Method for evaluating genetic relationship DNA (deoxyribonucleic acid) molecules of mulberry variety
ActiveCN103898216AImprove accuracyAccurate identificationMicrobiological testing/measurementBiotechnologyGenetic diversity
The invention discloses a method for evaluating genetic relationship DNA (deoxyribonucleic acid) molecules of a mulberry variety. The method comprises the steps of (1) system development analysis of the mulberry variety: 1) material selection; 2) screening of a contrast sequence; 3) extraction of mulberry genome DNA and detection of purity and concentration; 4) PCR (polymerase chain reaction) amplification; 5) PCR detection; 6) PCR sequencing; 7) sequence splicing and comparison, and identification of an evolutionary relationship; (2) genetic relationship analysis of the mulberry variety. According to the method disclosed by the invention, the system development analysis is firstly performed, materials are reasonably selected from worldwide mulberry varieties and variants, and then genetic diversity analysis of mulberry variety populations is performed to further determine the genetic relationship, so that the accuracy and repeatability of evaluating the genetic relationship are significantly improved by using the two-step analysis method, and a great promotion effect is achieved for protection of mulberry variety resources, mulberry variety fingerprint identification, molecular aided breeding and variety resource development and utilization.
Owner:SOUTHWEST UNIV +2
A kit for genotyping hepatitis C virus
ActiveCN102286644AHigh detection sensitivityImprove accuracyMicrobiological testing/measurementMicroorganism based processesForward primerHCV Genotyping
The invention relates to a kit for genotyping hepatitis C virus (HCV), which comprises HCVNS5B gene amplification primers, HCVNCR conserved region gene amplification primers, an HCVRNA extraction reagent, a negative control, a positive control, a cDNA (complementary deoxyribonucleic acid) synthetic reagent, a PCR (polymerase chain reaction) solution and a PCR sequencing reagent, wherein the sequence of the HCVNS5B gene forward primer is disclosed as SEQ ID NO.1, the sequence of the HCVNS5B gene reverse primer is disclosed as SEQ ID NO.2, the sequence of the HCVNCR conserved region gene forward primer is disclosed as SEQ ID NO.3, and the sequence of the HCVNCR conserved region gene forward primer is disclosed as SEQ ID NO.4. The kit provided by the invention has the advantages of high detection sensitivity and good specificity, and can be used for detecting all the reported HCV genotypes (subtypes) at high speed (within 12-14 hours).
Owner:HUAXIN SCI & TECH PANYU CITY
Optimized separation method for human intestine probiotics
ActiveCN105176822AOptimal separation method is practicalOptimized separation method is simpleBacteriaMicrobiological testing/measurementLiquid mediumCulture mediums
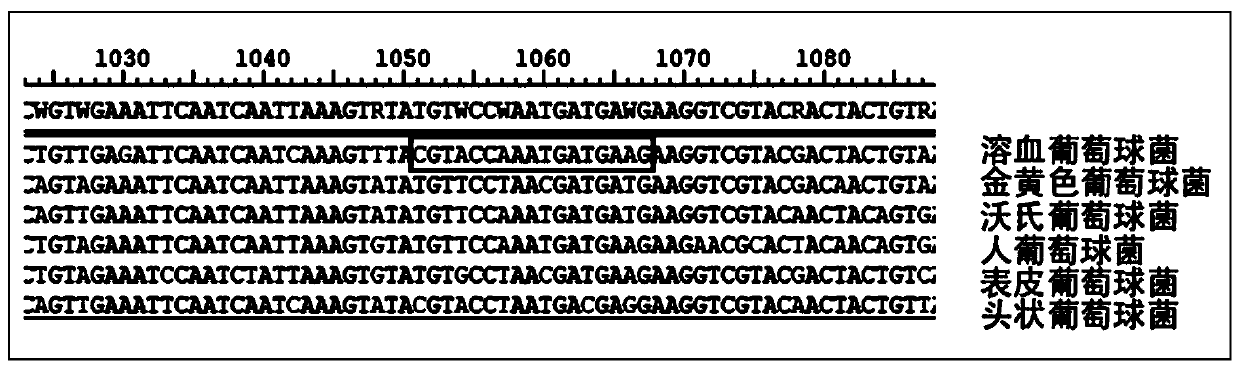
The invention provides an optimized separation method for human intestine probiotics. The optimized separation method comprises the following steps that 1 basic medium preparing, wherein a mucoprotein liquid medium and a mucroprotein solid medium are prepared; 2 screening, wherein a feces specimen is collected and inoculated in the mucroprotein liquid medium for anaerobic culturing to obtain bacterium liquid, the bacterium liquid is taken as a template, a PCR experiment is performed, the bacterium liquid in a PCR positive reaction tube is selected to be inoculated in the mucroprotein solid medium for anaerobic culturing to obtain a bacterium colony, the bacterium colony on the mucroprotein solid medium is inoculated onto a chocolate plate for anaerobic culturing to obtain a bacterium colony, the bacterium colony on the chocolate plate is inoculated onto a Columbia blood plate for purification culturing to obtain a bacterium colony, and PCR sequencing verification is performed on the bacterium colony on the Columbia blood plate through a 16sRNA universal primer to separate the human intestine probiotics. The optimized separation method for the human intestine probiotics is practical, simple, visualized and convenient.
Owner:GUANGZHOU KANGZE MEDICAL TECH CO LTD +1
Kit for detecting staphylococcus hominis
InactiveCN110157821ALower requirementStrong specificityMicrobiological testing/measurementMicroorganism based processesStaphylococcus haemolyticusRpoB
The invention provides a kit for detecting staphylococcus hominis. The kit comprises a guide RNA specifically targeting a staphylococcus hominis rpoB gene, amplification primer pairs, hydrated TwistAmp basic kit reaction drying balls, an LbCas12a protein, a single-stranded DNA probe (ssDNA), a Ribonuclease Inhibitor and a buffer solution. The kit is used for detecting the staphylococcus hominis, the detection specificity is high, and the staphylococcus hominis can be distinguished from other staphylococcocci including staphylococcus aureus, staphylococcus epidermidis, staphylococcus warneri, staphylococcus capitis and staphylococcus haemolyticus. The RNA sequence is used for detection, the consumed time is about 1 hour, no PCR instrument is needed, the requirements for equipment are low and significantly lower than those of a traditional bacterial culture method or PCR sequencing method, and the clinical practical value is high.
Owner:ZHEJIANG UNIV
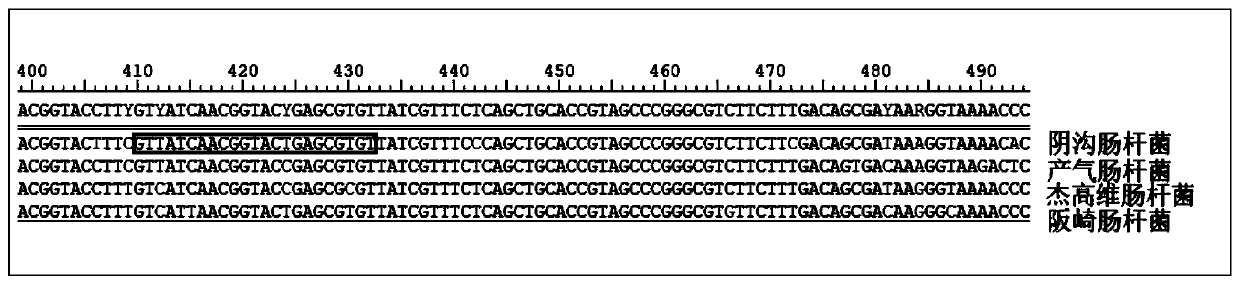
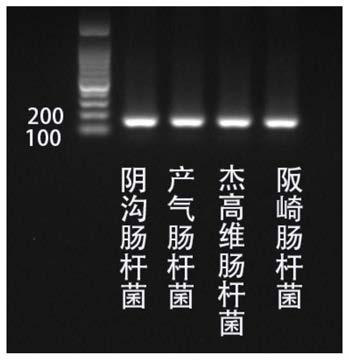
Kit for detection of Enterobacter cloacae
InactiveCN110241239AStrong specificityLow equipment requirementsMicrobiological testing/measurementAgainst vector-borne diseasesRNA SequenceSingle strand dna
The invention provides a kit for the detection of Enterobacter cloacae, including guide RNAs specifically targeting the rpoB gene of the Enterobacter cloacae, a primer pair for amplification, hydrated TwistAmp basic kit reaction drying balls, LbCas12a proteins, a single-stranded DNA probe (ssDNA), Ribonuclease Inhibitor and buffer. The kit can be used to detect the Enterobacter cloacae with high specificity, and can distinguish the Enterobacter cloacae from other Enterobacter bacteria, including Enterobacter aerogenes, Enterobacter gergoviae and Enterobacter sakazakii. At the same time, the method using the RNA sequence described in the invention for detection takes about 1 hour, does not need any PCR instruments, and has low requirements for equipment; and time consumption of the method is significantly lower than that of the conventional bacterial culture method or the PCR sequencing method, with great clinical practical value.
Owner:ZHEJIANG UNIV
Kit for detecting staphylococcus haemolyticus
InactiveCN110129462ALower requirementStrong specificityMicrobiological testing/measurementMicroorganism based processesStaphylococcus haemolyticusRpoB
The invention provides a kit for detecting staphylococcus haemolyticus. The kit comprises guide RNA specifically targeting staphylococcus haemolyticus rpoB gene, an amplification primer pair, hydration TwistAmp basic kit reaction drying balls, LbCas12a protein, a single-chain DNA probe (ssDNA), a Ribonuclease Inhibitor and a buffer solution. When the kit is used for detecting the staphylococcus haemolyticus, high detection specificity is achieved, and the staphylococcus haemolyticus can be distinguished from other staphylococcus such as staphylococcus aureus, staphylococcus hominis, staphylococcus warneri, staphylococcus capitis and staphylococcus epidermidis. Meanwhile, when the RNA sequence is used to perform detection, detection time is about 1 hour, a PCR instrument is not needed, equipment requirements are evidently lower than those of a traditional bacterial culture method or a PCR sequencing method, and a high clinical practical value is achieved.
Owner:ZHEJIANG UNIV
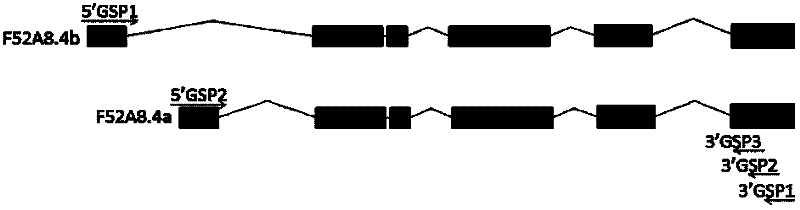
A method and application for mining low-abundance expressed genes and systematically identifying splice variants using nested PCR combined with gene-specific primers for reverse transcription
InactiveCN102260670AImprove digging efficiencyEfficient enrichmentMicrobiological testing/measurementFermentationSequence analysisAgricultural science
The invention discloses a method for excavating a low-abundance expression gene and system identification splicing variants by using a reverse transcription technique combining nested PCR (Polymerase Chain Reaction) with gene-specific primers (GSP), and application thereof. The method is based on reverse transcription PCR of 3'GSP1, gradient PCR is utilized to search for the best annealing temperature of each gene for each primer pair combination, and whether it is a new splicing variant is judged on the basis of GSP nested PCR sequencing and sequence analysis. The method is simple and easy to implement, has the advantages of favorable repetitiveness, high stability and strong reliability, greatly saves the experimental cost, can be applied to excavation of eucaryote low-abundance expression gene under different physiopathological states, and identification and detection of high / low-abundance expression gene splicing variants, and can be used for excavating low-abundance expression gene and identifying and detecting splicing variants of high expression gene and low-abundance expression gene. The method can be used for detecting both known splicing variants of targeted genes and unknown splicing variants of genes, and has wide application prospects.
Owner:INST OF RADIATION MEDICINE ACAD OF MILITARY MEDICAL SCI OF THE PLA
SNP marker related to beef traits of beef cattle and application thereof
The invention belongs to the technical field of biology, and particularly relates to an SNP marker related to beef traits of beef cattle and application thereof. According to the SNP marker related to the beef traits of the beef cattle and the application thereof, SNP loci of all exons of ORL1 genes in a grassland red cattle group are detected by using a PCR sequencing method, and the result discovers that only exons 4 of OLR1 genes in the grassland red cattle group have A / G mutation (at Chr5: 99812807bp); chi-square test proves that the SNP loci are in a Hardy-Weinberg equilibrium state in the grassland red cattle group; and genetic analysis discovers that the genetic homozygosity of the genes is 0.672, the genetic heterozygosity is 0.328, the effective allelic factor is 1.489, the polymorphic information content is 0.274, and the genes belong to moderate polymorphism. The correlation analysis result shows that the genotype of the exons 4 of the OLR1 genes has significant difference from the beef traits such as the eye muscle area of the grassland red cattle.
Owner:JILIN ACAD OF AGRI SCI
Phenylketonuria (PKU) screening kit and application thereof to prenatal screening
InactiveCN103276077AShorten screening timeScreening is quick and easyMicrobiological testing/measurementGeneticsPrenatal screening
The invention discloses a phenylketonuria (PKU) screening kit and application thereof to prenatal screening, and belongs to the technical field of PKU screening. The application method comprises the following steps of: (1) extracting free fetal DNA (Deoxyribonucleic Acid) from peripheral blood of pregnant women; (2) taking the fetal DNA as a template to carry out polymerase chain reaction (PCR) amplification; (3) detecting a PCR product through agarose gel electrophoresis and purifying the PCR product through enzyme reaction; (4) carrying out sequencing reaction on the purified PCR product; (5) purifying a PCR sequencing reaction product; (6) sequencing the purified PCR sequencing reaction product; and (7) comparing the sequencing result with positive and negative control samples. The kit has the beneficial effects that the kit can achieve automatic sequencing, thus shortening the screening time and enabling PKU screening to be simpler, more convenient and faster; the technical gap of prenatal PKU screening is filled; the birth rate of sick children is reduced; and the kit causes no trauma to the fetuses and is safe and reliable.
Owner:邯郸市康业生物科技有限公司
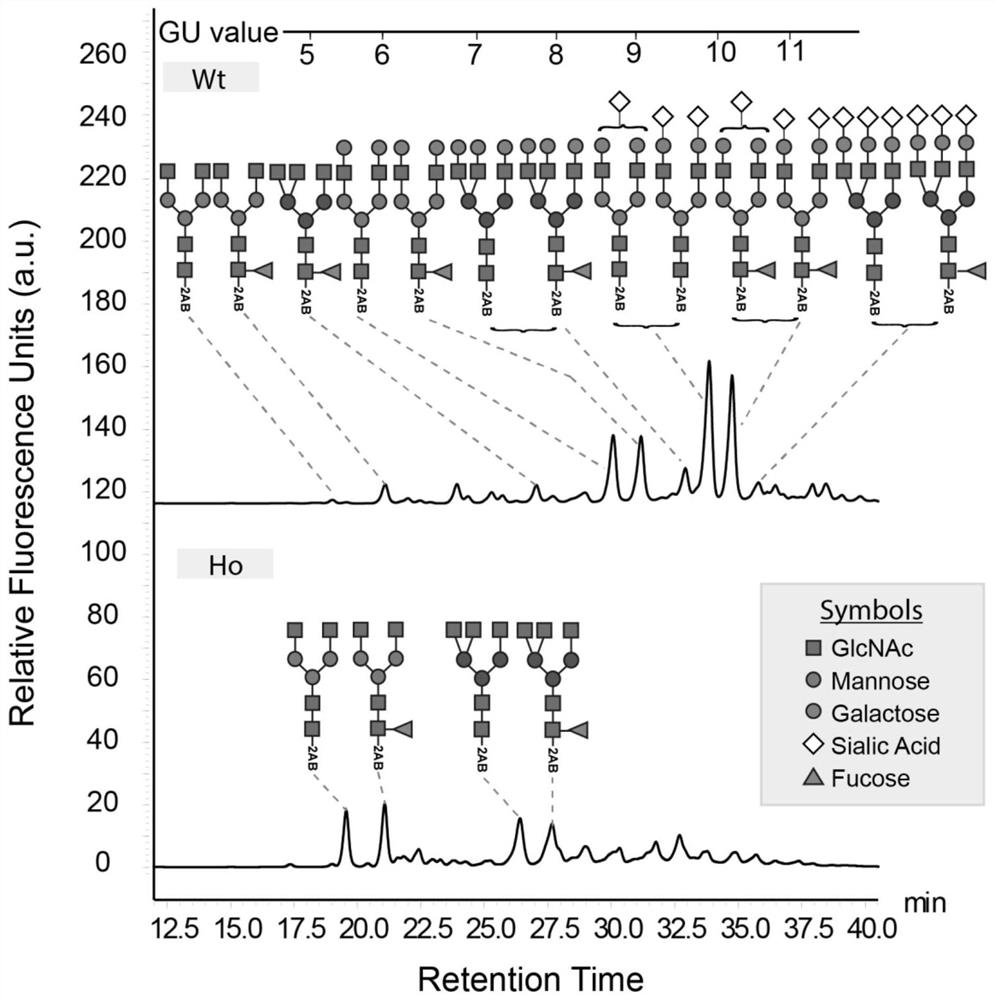
Cell strain capable of expressing glucocerebrosidase with high mannose content as well as preparation method and applications of cell strain
PendingCN108588127AIncrease enzyme activityHigh mannose glycoform contentPeptide/protein ingredientsMetabolism disorderHigh mannoseEukaryotic plasmids
The invention discloses a preparation method of a cell strain capable of expressing glucocerebrosidase with high mannose content. The preparation method comprises the following steps: (1) carrying outlipofection transfection, namely, aiming at the sequence of a GNT1 gene, designing a sgRNA sequence guiding cutting of endonuclease, recombining the sgRNA sequence into a knock-out vector, thus obtaining synthetic plasmids with coexpression of sgRNA and endonuclease, and transfecting a cell pool stably expressing glucocerebrosidase by adopting the synthetic plasmids through a liposome transfection method; (2) carrying out cloning by adopting a limiting dilution method, namely, screening out monoclonal cells, and carrying out enlarged culturing; (3) carrying out mutation cloning screening on the target gene, namely, acquiring monoclone with the GNT1 gene knocked out through cell PCR sequencing, and thus the cell strain capable of expressing glucocerebrosidase with high mannose content is obtained. The cell strain can stably express glucocerebrosidase, meanwhile, the content of the mannose is obviously improved, the binding capacity with mannose receptor is effectively improved, the enzyme activity capacity is improved, and the purpose of increasing the efficacies is achieved.
Owner:WUXI BIOLOGICS IRELAND LTD
Reagent kit for detecting enterobacter aerogenes
InactiveCN110241237AStrong specificityLow equipment requirementsMicrobiological testing/measurementAgainst vector-borne diseasesRpoBSingle strand dna
The invention provides a reagent kit for detecting enterobacter aerogenes. The reagent kit contains a guide RNA specifically targeted to an enterobacter aerogenes rpoB gene, an amplification primer pair, a hydration TwistAmp basic kit reaction drying ball, LbCas12a protein, a single-stranded DNA probe (ssDNA), a Ribonuclease inhibitor and a buffer solution. When the reagent kit is used for detecting the enterobacter aerogenes, the detection specificity is high, and the enterobacter aerogenes can be distinguished from other enterobacter bacteria including enterobacter cloacae, enterobacter gergoviae and enterobacter sakazakii. Besides, when the RNA sequence is used for detection, time consumption is about 1 hour, a PCR instrument is not needed, requirements for equipment are low and are notably lower than those by a traditional bacteria culture method or a PCR sequencing method, and the clinical use value is high.
Owner:ZHEJIANG UNIV
Screening method and application of SNP molecular markers associated with egg shell thickness, eggshell strength and egg shape index of Nandanyao chickens
ActiveCN111518920AImprove resistance to breakageEasy to transportMicrobiological testing/measurementProteomicsGenomic sequencingAnimal science
The invention discloses a screening method of SNP molecular markers associated with egg shell thickness, eggshell strength and egg shape index of Nandanyao chickens. The method comprises the followingsteps: (1) taking Nandanyao chickens, sampling blood to extract genome DNA, performing sequencing, and detecting genes with mutation SNP sites on the Nandanyao chickens; (2) taking Nandanyao hens astest chickens, and measuring eggshell thickness, eggshell strength and egg shape index of eggs; (3) extracting genome DNA, carrying out multiple PCR sequencing on the SNP sites detected in the step (1), and collecting genotype information of the test chickens at the SNP sites; and (4) sorting sequencing data and eggshell thickness, eggshell strength and egg shape index data, and screening out candidate SNP molecular markers which are extremely and significantly associated with eggshell thickness, eggshell strength and egg shape index, wherein the Pr>F value is less than 0.01. The operation steps are simplified, the method is suitable for breeding of high-heredity quantitative characters, the breakage resistance of eggs is improved after screening, and transportation is convenient.
Owner:GUANGXI UNIV
Multi-nucleic acid co-labeling support, preparation method therefor, and application thereof
PendingCN114096678ANucleotide librariesMicrobiological testing/measurementMultiplexChemical reaction
Provided are a variety of nucleic acid co-labeling supports, a preparation method therefor, and an application thereof. The supports comprise a support body, and a variety of nucleic acid labels located on the surface and / or inside of the support body. The nucleic acid labeled on a single support at least comprises: one or more first nucleic acid labels having a function that at least comprises of trapping a specific compound in a reaction system onto the surface of the support; and one or more second nucleic acid labels having a function that at least comprises participating in a specified biochemical reaction process of the specific compound trapped onto the surface of the support. The foregoing variety of nucleic acid co-labeled supports can be used for 5'-terminus single-cell RNA expression profile analysis, construction of a 5'-terminus single-cell VDJ library for a microwell array platform, construction of a 3'-terminus single-cell RNA library, construction of a single-cell transcriptome library, single-cell multi-omics research, multiplex PCR and / or construction of a multiplex PCR sequencing library, etc.
Owner:BEIJING SEEKGENE BIOSCIENCES CO LTD
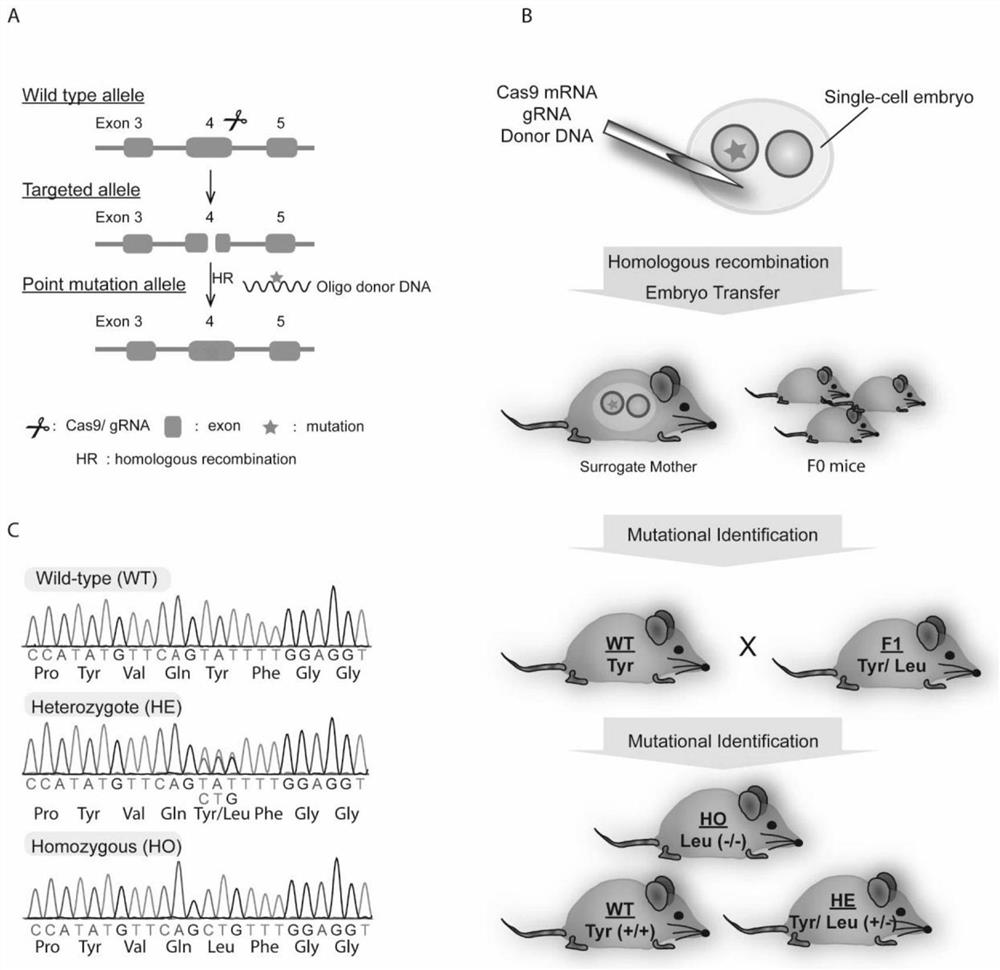
Construction method and application of galactosyltransferase GalT gene point mutation mouse model
The invention provides a construction method and application of a galactosyltransferase GalT gene point mutation mouse model. The construction method comprises the following steps: (1) target construction: searching a CDS region of a galactosyltransferase B4GalT gene, defining an exon part, and determining a gene mutation site; (2) obtaining of fertilized eggs and microinjection: obtaining cas9 mRNA and gRNA in an in vitro transcription mode, obtaining oligo donor DNA through synthesis, and carrying out microinjection on cas9 mRNA, gRNA and oligo donor DNA into fertilized eggs of a C57BL / 6J mouse to obtain an F0-generation mouse; (3) conducting mating propagation on the positive F0-generation mouse and C57BL / 6J to obtain a positive F1-generation heterozygote with B4GalT gene site-specificmutagenesis; and (4) after mating of the positive heterozygote mice, performing PCR sequencing confirmation to obtain positive homozygotes, indicating that the mouse model is successfully constructed.The model is simple to manufacture and good in repeatability, can be used for researching medicines for treating abnormal glycosylation diseases of human beings, and has important practical significance for exploring a treatment method.
Owner:NANJING AGRICULTURAL UNIVERSITY
Kit for detecting staphylococcus epidermidis
InactiveCN110157822ALower requirementStrong specificityMicrobiological testing/measurementMicroorganism based processesStaphylococcus haemolyticusRpoB
The invention provides a kit for detecting staphylococcus epidermidis. The kit comprises a guide RNA specifically targeting a staphylococcus epidermidis rpoB gene, amplification primer pairs, hydratedTwistAmp basic kit reaction drying balls, an LbCas12a protein, a single-stranded DNA probe (ssDNA), a Ribonuclease Inhibitor and a buffer solution. The kit is used for detecting the staphylococcus epidermidis, the detection specificity is high, and the staphylococcus epidermidis can be distinguished from other staphylococcocci including staphylococcus aureus, staphylococcus hominis, staphylococcus warneri, staphylococcus capitis and staphylococcus haemolyticus. The RNA sequence is used for detection, the consumed time is about 1 hour, no PCR instrument is needed, the requirements for equipment are low and significantly lower than those of a traditional bacterial culture method or PCR sequencing method, and the clinical practical value is high.
Owner:ZHEJIANG UNIV
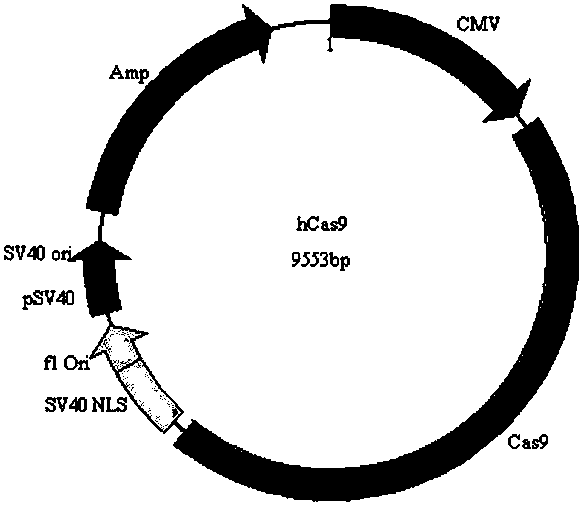
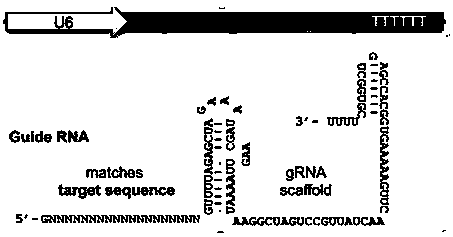
CRISPR-Cas9 system-mediated knockout method for mouse FGF5 (Fibroblast Growth Factor 5) gene
InactiveCN109811008AStable passageImprove securityMicroinjection basedStable introduction of DNAMutantGene mutation
The invention discloses a CRISPR-Cas9 system-mediated knockout method for a mouse FGF5 (Fibroblast Growth Factor 5) gene. The method comprises the following steps: constructing an sgRNA expression vector based on a CRISPR-Cas9 system according to a mouse FGF5 gene sequence; injecting an sgRNA-Cas9 plasmid mixture into a germ cell pronucleus; performing embryo transplantation and strain gene identification to obtain a genetically mutated mouse; performing PCR (Polymerase Chain Reaction) sequencing and confirmation to obtain a hereditary mouse mutant model finally. A built CRISPR-Cas9-mediated targeting vector provides a simple, convenient, rapid and safe way for the knockout of the mouse FGF5 gene. The method realizes site-directed integration of exogenous gene cell lines without any screening markers, greatly improves the safety of transgenic animals, and has important value in genetic breeding of mice.
Owner:INNER MONGOLIA UNIVERSITY
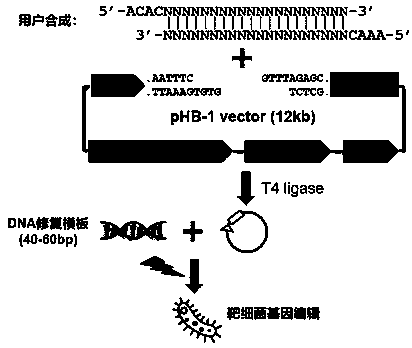
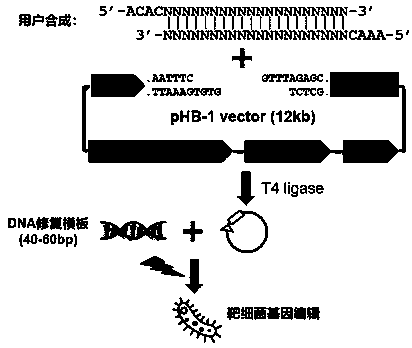
Linearized DNA vector pHB-1 plasmid and test kit prepared from same and used for editing bacterial genomes
PendingCN111139258AEasy to controlFirmly connectedStable introduction of DNANucleic acid vectorEscherichia coliA-DNA
The invention discloses a linearized DNA vector pHB-1 plasmid and a test kit prepared from the same. The genetic sequence of the linearized DNA vector pHB-1 plasmid is shown as SEQ ID NO: 1; the testkit comprises a linearized DNA vector pHB-1 plasmid, an efficient T4DNA ligase, a PCR sequencing primer tL3-seq and an experimental control substance. Wherein the experimental control comprises a specific gRNA sequence primer for editing an escherichia coli MlaC gene, a DNA repair template for introducing MlaC L11R point mutation and a PCR sequencing primer for confirming the MlaC L11R point mutation; the test kit disclosed by the invention can be used for carrying out gene editing experiments on various bacteria, has repeatability and stability, and can be used for editing one or more targetgenes, and the editing efficiency can reach 90-100%.
Owner:义乌市颂健生物科技有限公司
Kit for detecting staphylococcus warneri
InactiveCN110144416ALower requirementStrong specificityMicrobiological testing/measurementMicroorganism based processesStaphylococcus haemolyticusRpoB
The present invention provides a kit for detecting staphylococcus warneri. The kit comprises a guide RNA specifically targeting a staphylococcus warneri rpoB gene, an amplification primer pair, hydration TwistAmp basic kit reaction drying spheres, LbCas12a protein, a single-stranded DNA probe (ssDNA), an ribonuclease inhibitor and a buffer solution. The kit is used for detecting the staphylococcuswarneri and high in detection specificity, and can separate the staphylococcus warneri from other staphylococci, including staphylococcus aureus, staphylococcus hominis, staphylococcus epidermidis, staphylococcus capitis and staphylococcus haemolyticus. At the same time, the use of the RNA sequence for the detection takes about 1 hour, does not require a PCR instrument, has low requirements on equipment, and is significantly lower than a conventional bacterial culture method or a PCR sequencing method and large in clinical practical value.
Owner:ZHEJIANG UNIV
Fast separation method of wild edible fungi
InactiveCN107739719AEasy to collectStrong representativeFungiMicroorganism based processesBiotechnologyBearded tooth
The invention discloses a fast separation method of wild edible fungi. The selected raw materials (including pleurotus djamor, agrocybe aegerita, hericium erinaceus, hypsizygus marmoreus and glossy ganoderma) are widely planted in China; the collection is convenient; the plants belong to different species and have relatively high representativeness. The selected raw materials have very high edibleand medicinal values and also have higher activity in the existing research, so that the raw materials have the further development and utilization potential. An efficient wild fungi tissue isolationmethod is built; a conventional tissue isolation method can easily infect bacteria; the time and the labor are wasted; the PCR (polymerase chain reaction) sequencing result is authenticated in a morphology manner; the equal quantity of triadimefon with the concentration being 1mg / L and gentamicin with the concentration being 40mg / L are respectively added on the first day and the second day of fermentation; the edible fungi with high yield and highest purity can be obtained.
Owner:SICHUAN AGRI UNIV
A kit for detecting drug-resistant mutations of hepatitis B virus
InactiveCN102286643AHigh detection sensitivityImprove accuracyMicrobiological testing/measurementHepatitis B immunizationHepatitis B virus
The invention relates to a test kit for detecting drug-resistant mutations of hepatitis B virus, comprising HBVDNAPol gene region amplification primers, HBVDNA extraction reagents, negative controls and positive controls, PCR reaction solution and PCR sequencing reagents, wherein the upstream primer sequence of the HBVDNAPol gene is As shown in SEQIDNo.1, the downstream primer sequence of HBVDNAPol gene is shown in SEQIDNo.2. The detection kit of the invention has high detection sensitivity, good specificity and high speed, and can be completed within 12 to 14 hours.
Owner:李艳 +3
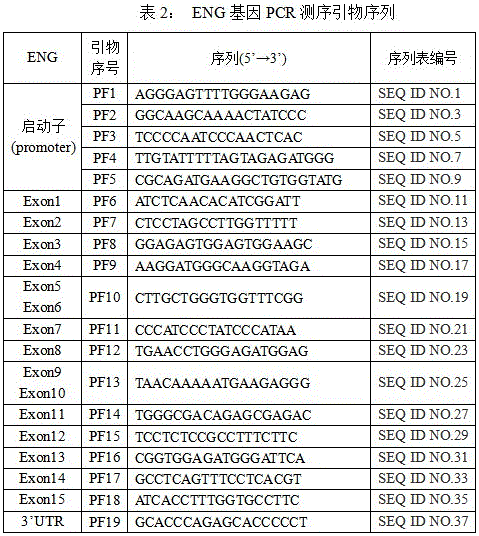
Primer group for detecting ENG gene mutation, purpose of primer group and kit containing primer group
InactiveCN105886634AGuaranteed accuracyEnsure uniquenessMicrobiological testing/measurementDNA/RNA fragmentationMicrobiologyExon
The invention discloses a primer group for detecting ENG gene mutation. The primer group comprises an ENG gene first to fifteenth exon regions, a PCR (polymerase chain reaction) amplification primer group corresponding to 3'UTR and a starter and a PCR sequence testing primer. The invention also discloses a purpose of the primer group and a kit containing the primer group. The primer group can be used for specifically identifying whether the ENG gene mutation occurs or not; the accuracy and the uniqueness of the detection result are ensured.
Owner:THE FIRST AFFILIATED HOSPITAL OF GUANGZHOU MEDICAL UNIV (GUANGZHOU RESPIRATORY CENT)
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com