A kit for detecting drug-resistant mutations of hepatitis B virus
A hepatitis B virus and drug-resistant mutation technology, applied in the field of medical devices, to achieve good specificity, low false positives, and high degree of automation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Preparation of the kit of the invention
[0033] The composition of the kit of the present invention is as follows:
[0034] (1) HBV DNA extraction reagent: 10mmol / L Tris-HCl (pH8.0), 5mmol / L EDTA, 0.5% SDS, 150μg / ml proteinase K
[0035] (2) Primer:
[0036] The sequence of the upstream primer of HBV DNA Pol gene is: 5’-CTGCTGGTGGCTCCAGTT-3’
[0037] The downstream primer sequence of HBV DNA Pol gene is: 5’-GAGTTCCGCAGTATGGATCG-3’
[0038] The above primer sequence was synthesized by Shanghai Life Technology Company.
[0039] (3) Negative control and positive control: deionized water is used as a negative control, and samples containing HBV DNA are used as a positive control.
[0040] (4) PCR reaction solution: 10×PCR Premix (mixed solution), 0.25 pmol / μL primer, 2.5-4.0 mM MgCl 2 , 2U Taq enzyme, 0.2~0.4 mM dNTPs, 0.3~0.6 mM dUTP, usually 1~2 μL template.
[0041] (5) The setting of PCR amplification program: on the ABI9700 instrument, it is usually 95℃ for 5 min, then 9...
Embodiment 2
[0046] Example 2 Using the kit prepared in Example 1 to detect HBV nucleoside analog resistance gene mutation
[0047] To detect HBV nucleoside analog resistance gene mutations in peripheral blood samples of 20 hepatitis B patients.
[0048] Detection process:
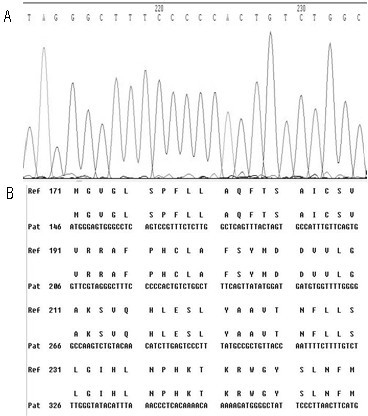
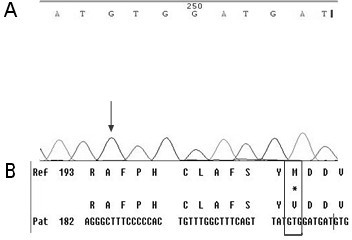
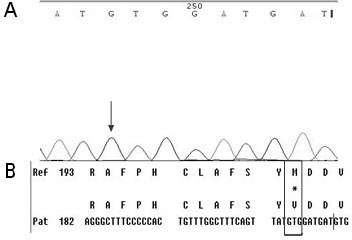
[0049] First, design specific primers based on the HBV RT region gene sequence provided by the HBV nucleic acid database. Obtain peripheral blood samples of clinical hepatitis B patients and quickly extract HBV DNA; configure PCR reaction solution for PCR amplification, then recover PCR amplification products, perform sequencing reactions, and then sequence reaction products after purification, and finally in the NCBI nucleic acid database Perform nucleic acid sequence comparison to find the gene variants of 202, 204, 173, 180, 181, 169, 236, 184, 202, 250 in the reverse transcriptase region.
[0050] Specific steps are as follows:
[0051] (1) Extraction of serum HBV DNA: Extract HBV DNA from peripheral blood of hepatitis B ...
Embodiment 3
[0059] Example 3 Evaluation of the detection capability of the kit
[0060] The detection kit (gene sequencing method) of Example 1 was used to detect HBV resistance in the serum of 30 patients with hepatitis B detected by the probe hybridization method. Experiments show that the sensitivity, specificity and sensitivity of this kit are better than those of probes. The hybrid method is more precise and fully meets the current practical requirements of clinical diagnosis and treatment:
[0061] Comparison of two methods for detecting HBV resistance mutation
[0062]
[0063] among them:
[0064] (1) Specificity: 100%;
[0065] (2) Sensitivity: 95%;
[0066] (3) Positive predictive value: the positive predictive value reaches 100%;
[0067] (4) Negative predictive value: the negative predictive value reaches 91%;
[0068] (5) Repeatability: The results of repeated experiments are consistent;
[0069] (6) Time-consuming: The testing time for a clinical specimen is about 12-14 hours, which is...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com