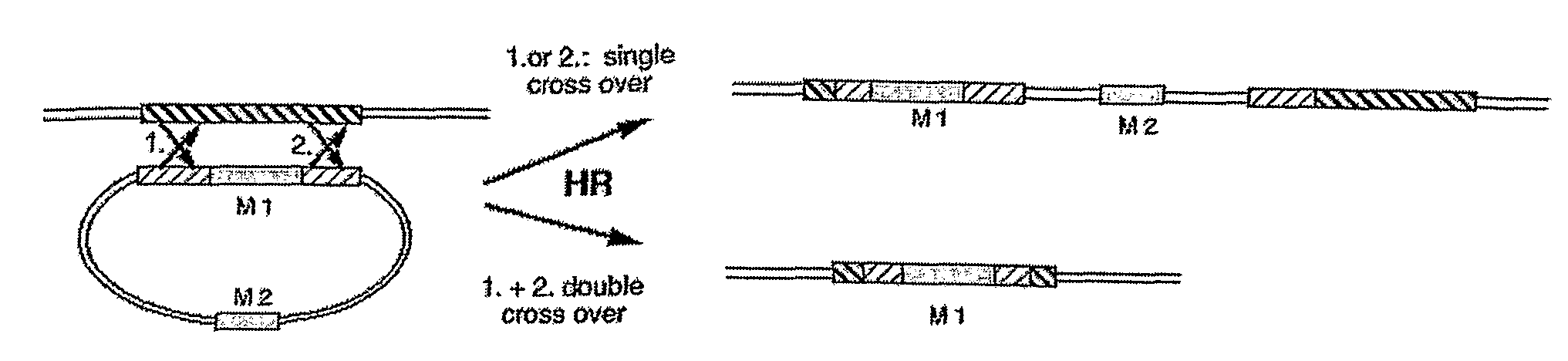
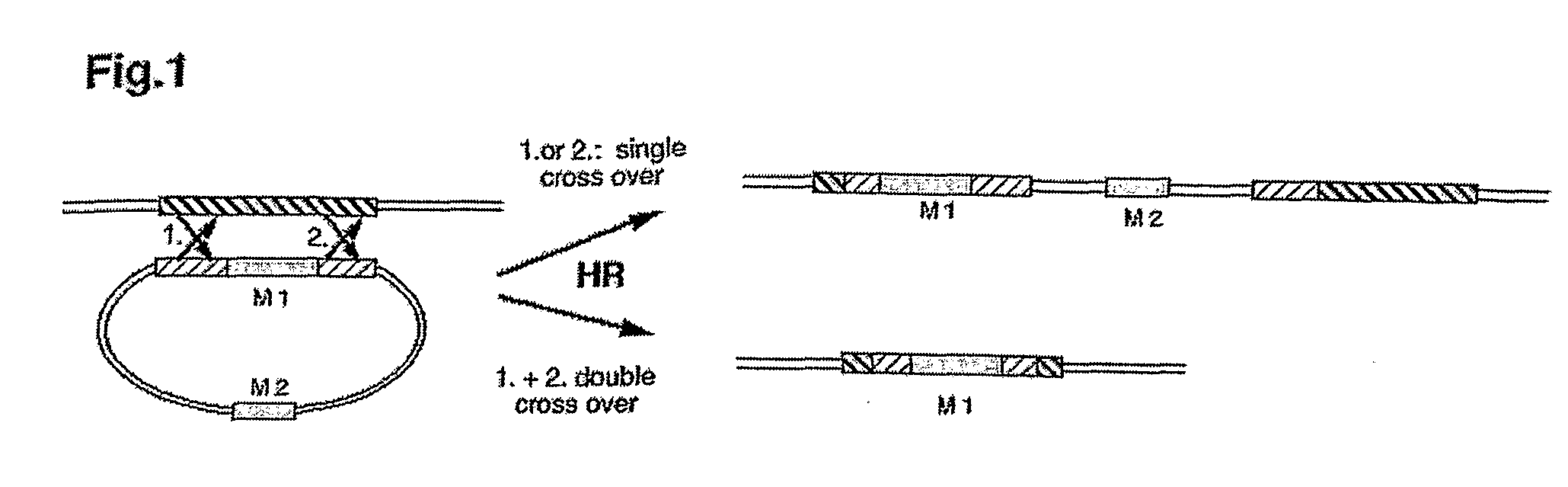
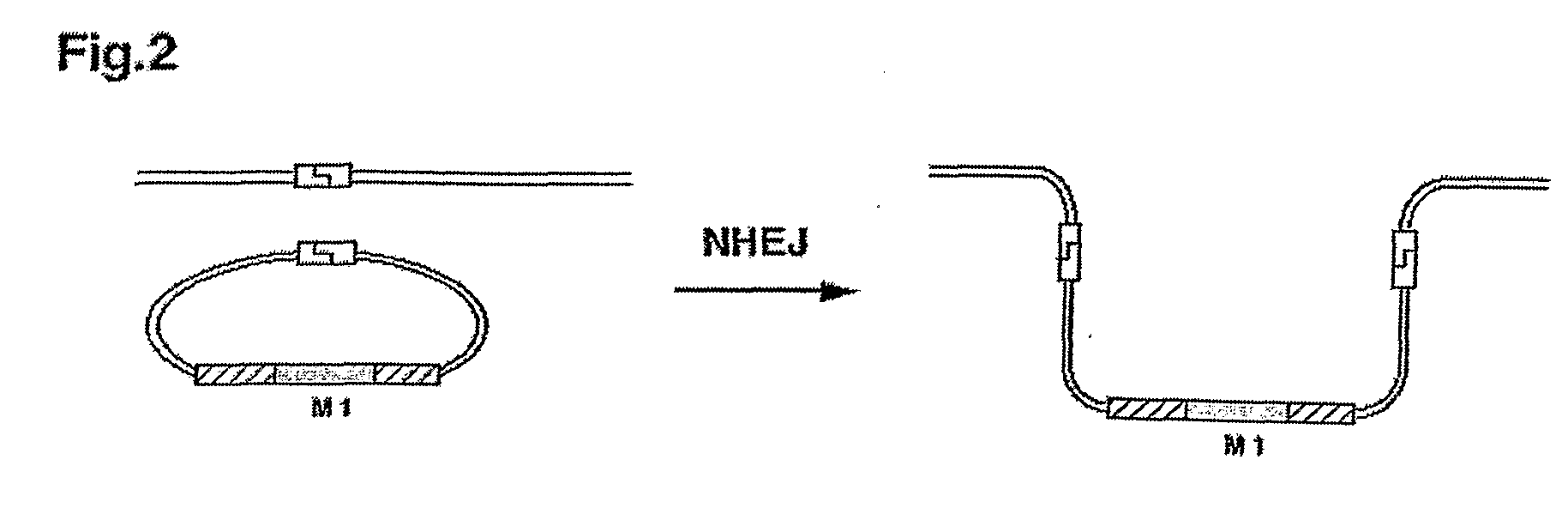
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
1357 results about "Single strand dna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
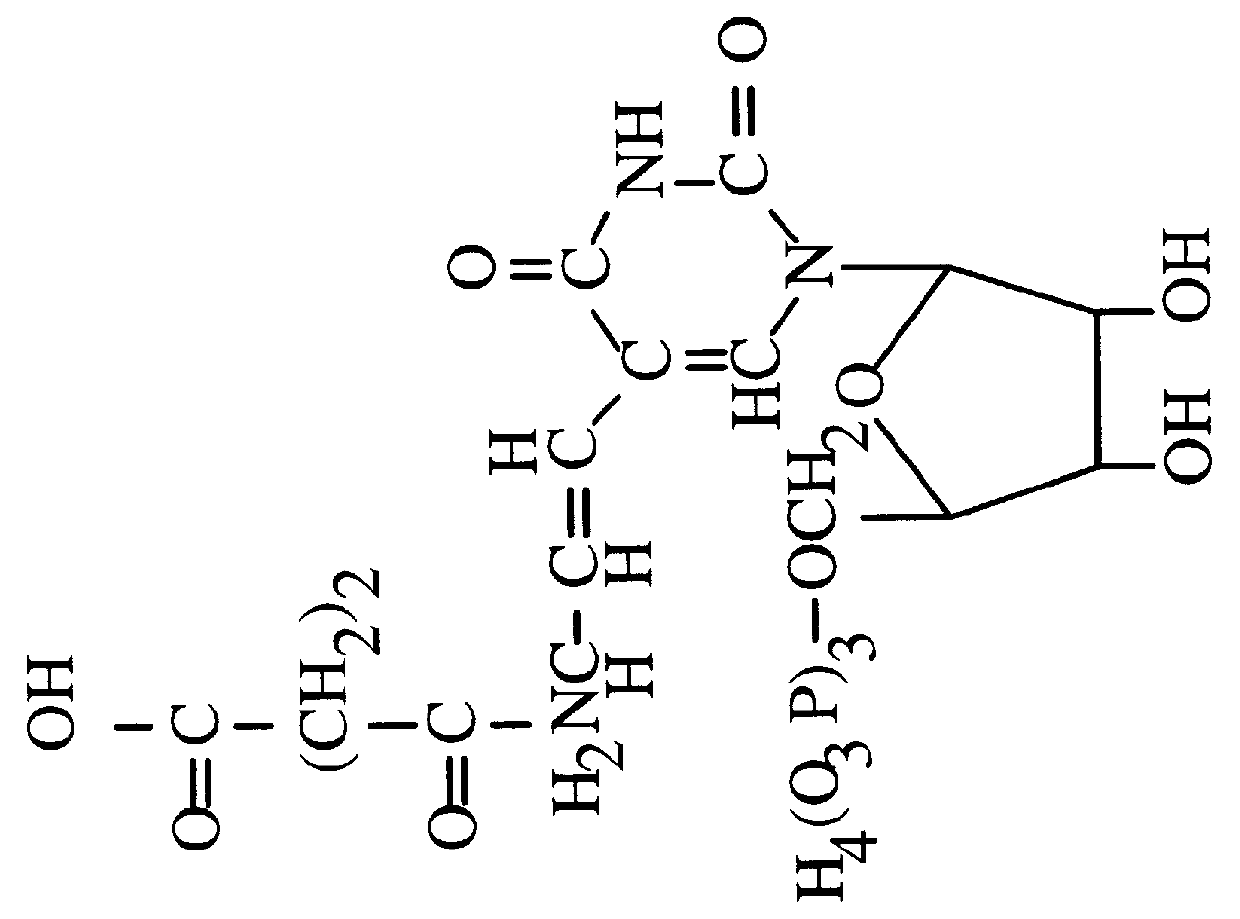
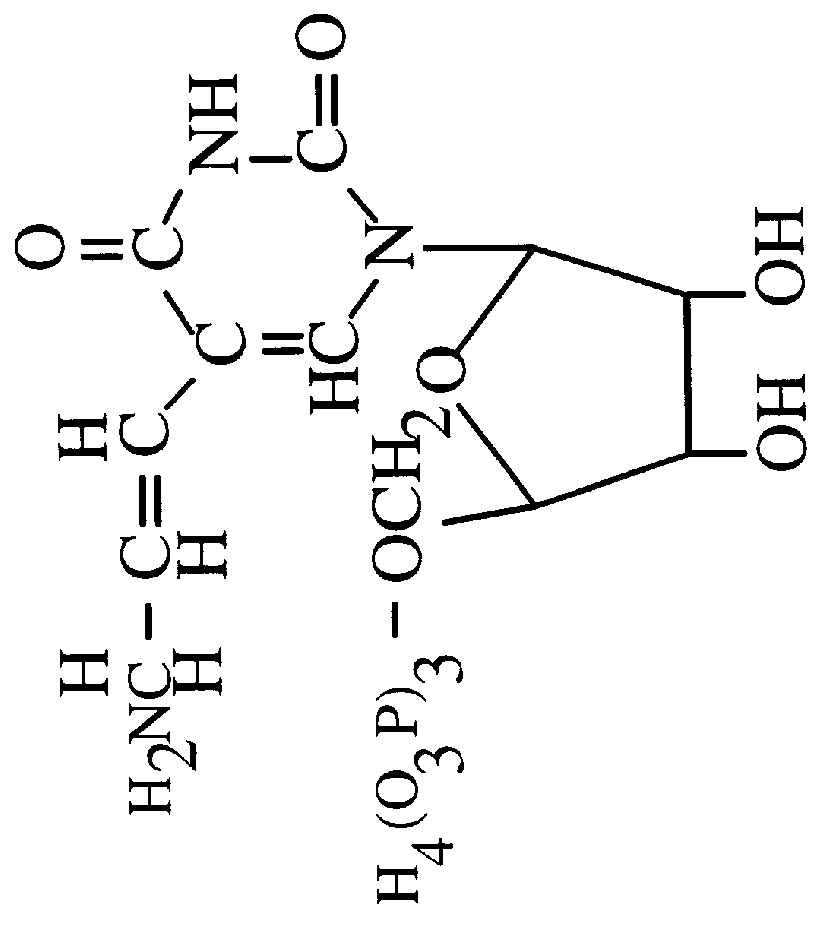
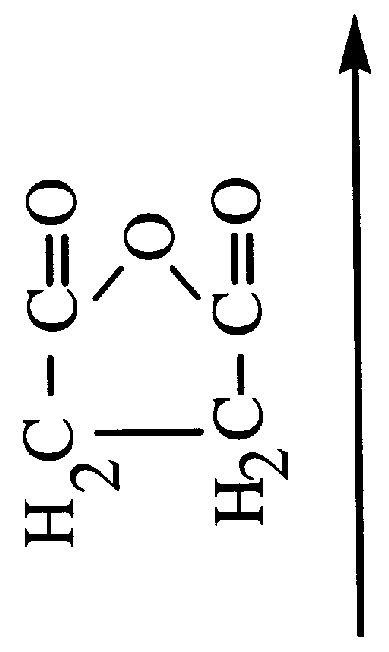
Single stranded dna. A DNA molecule consisting of only a single strand contrary to the typical two strands of nucleotides in helical form. In nature, single stranded DNA genome can be found in Parvoviridae (class II viruses). Single stranded DNA can also be produced artificially by rapidly cooling a heat-denatured DNA.
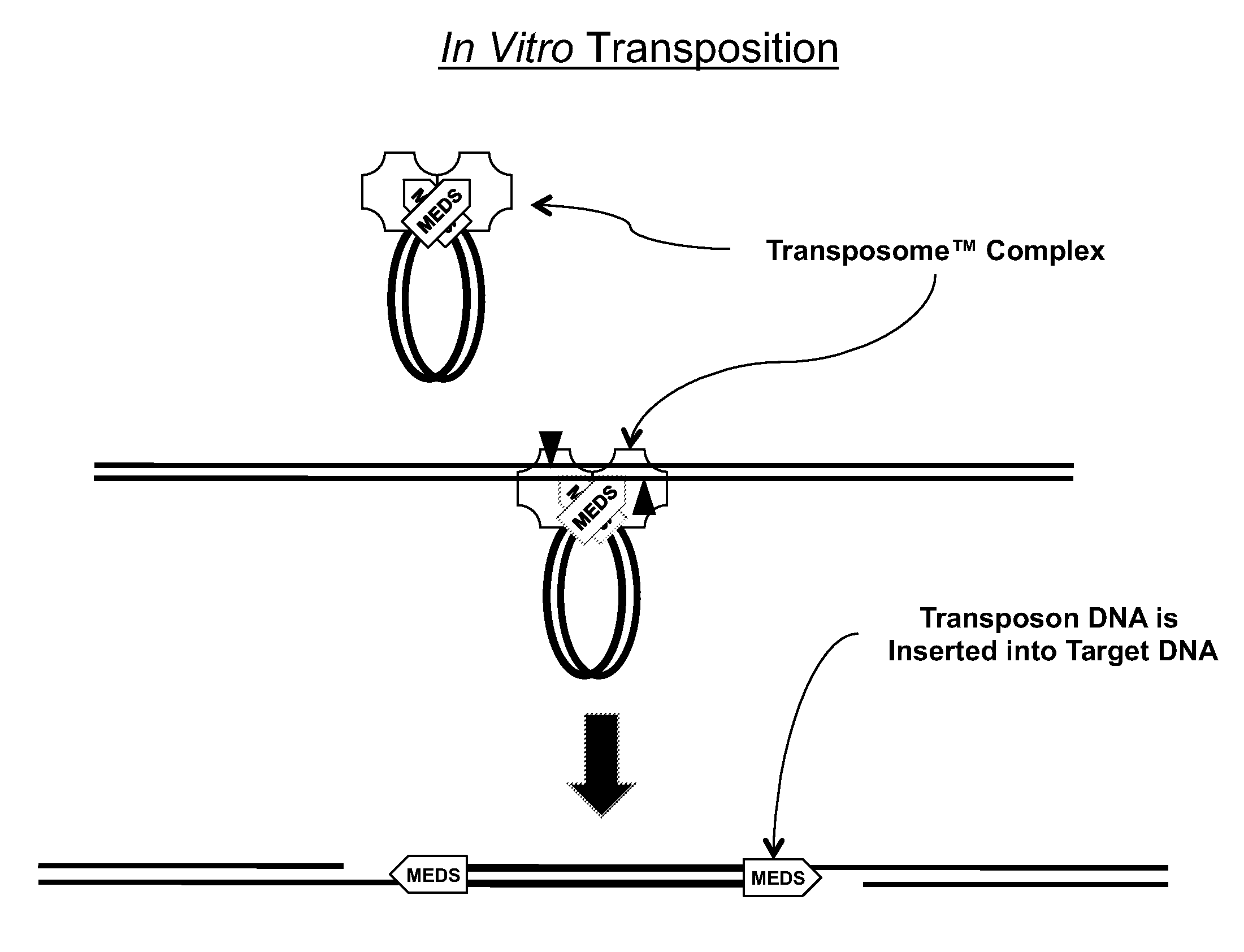
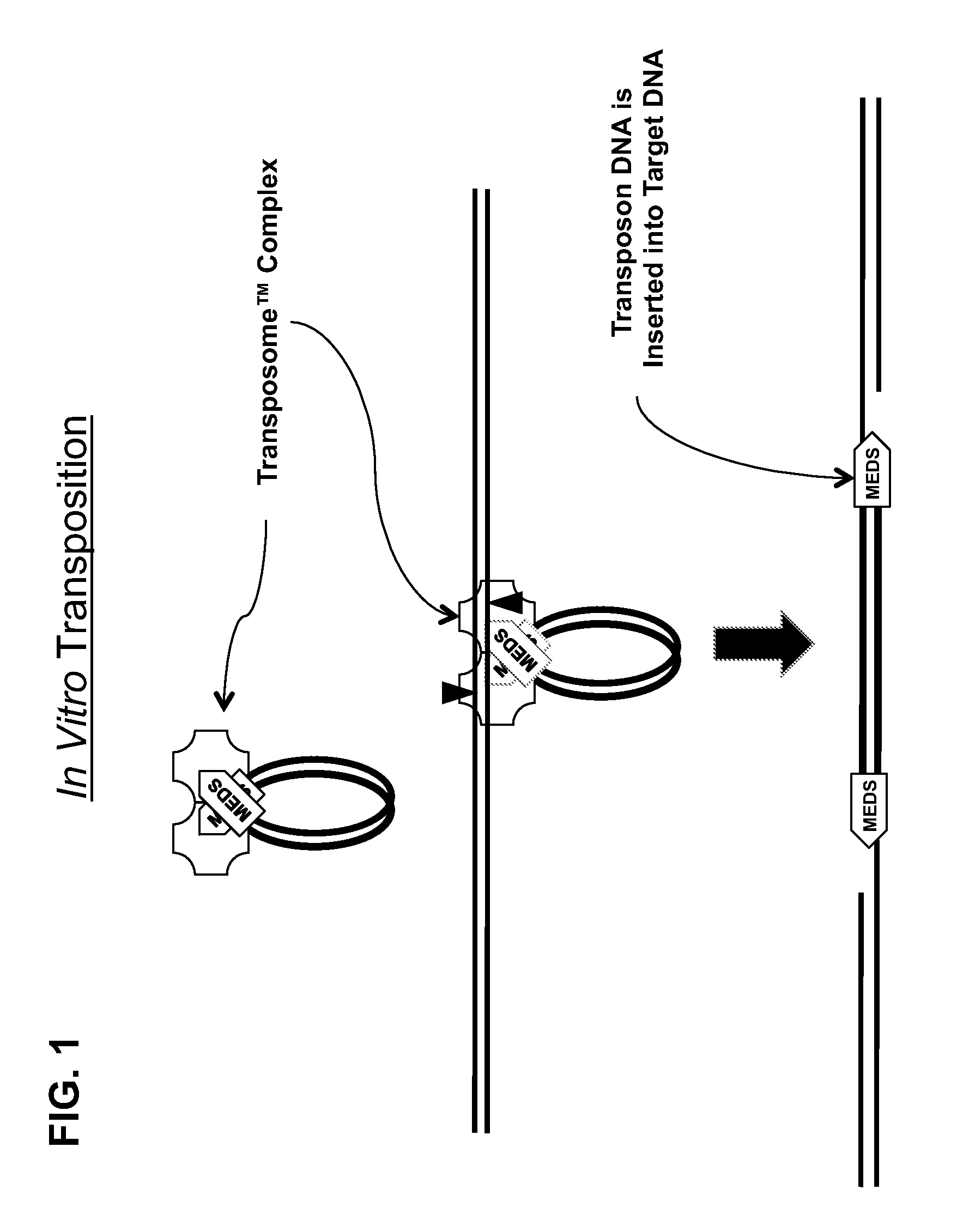
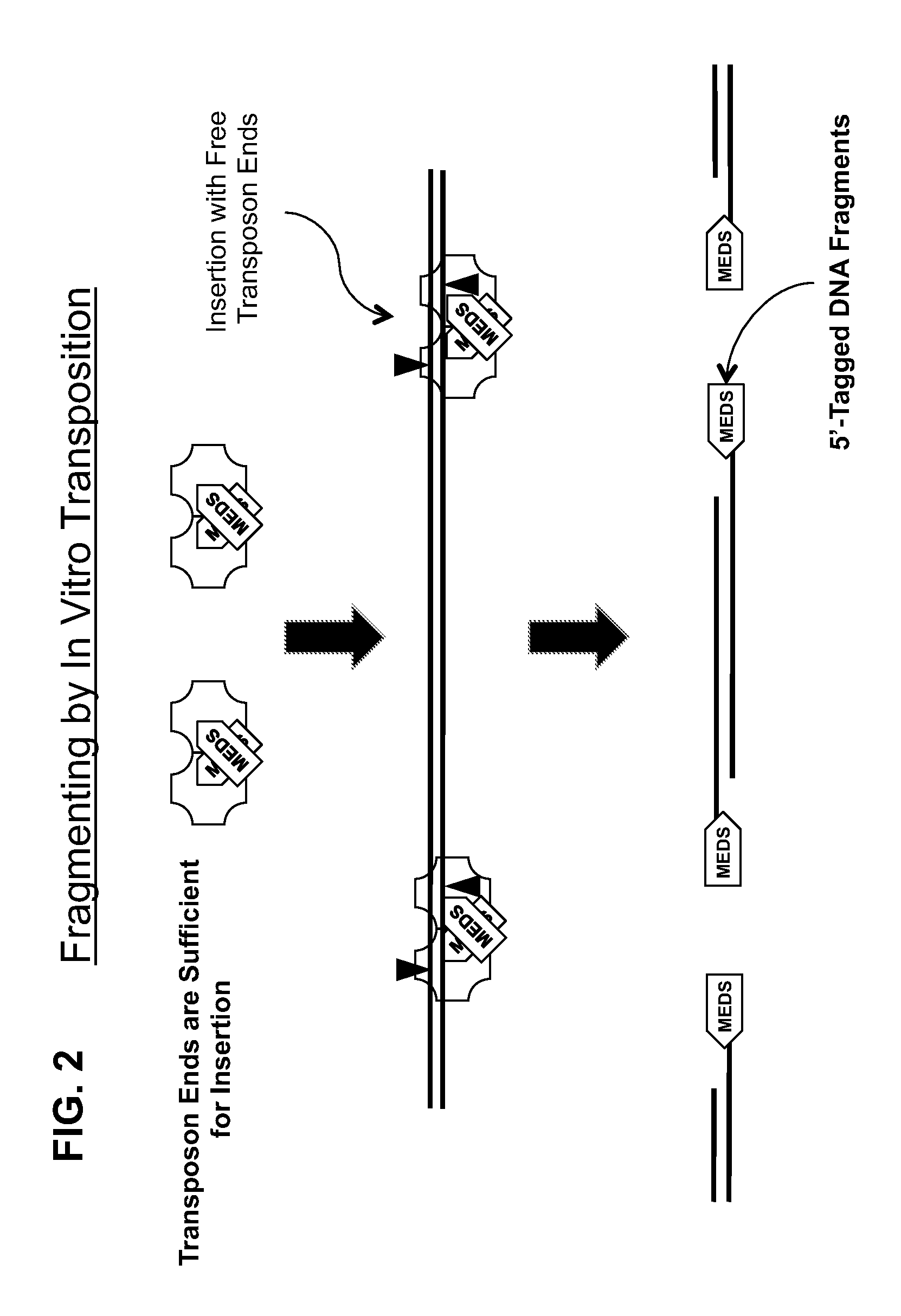
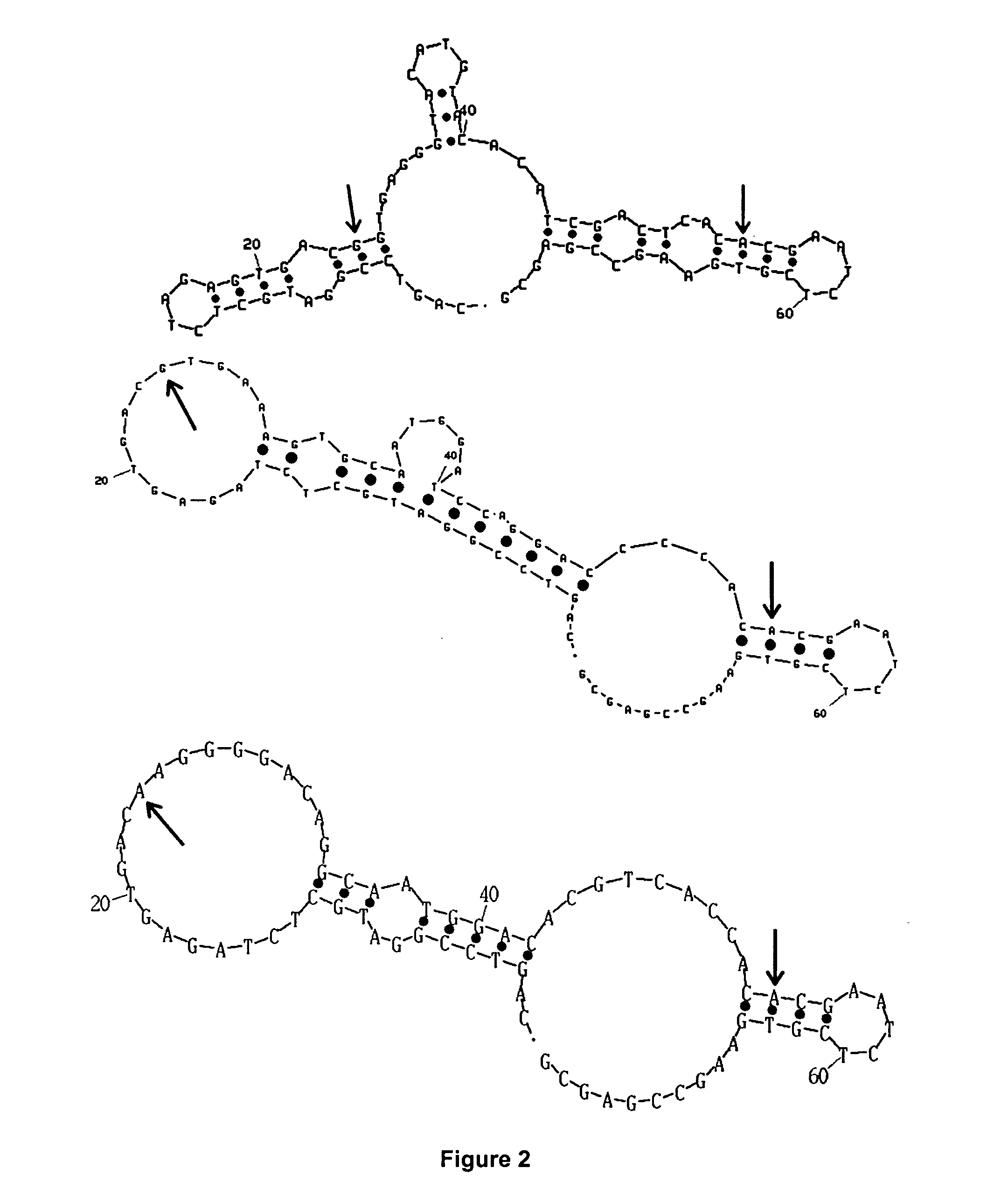
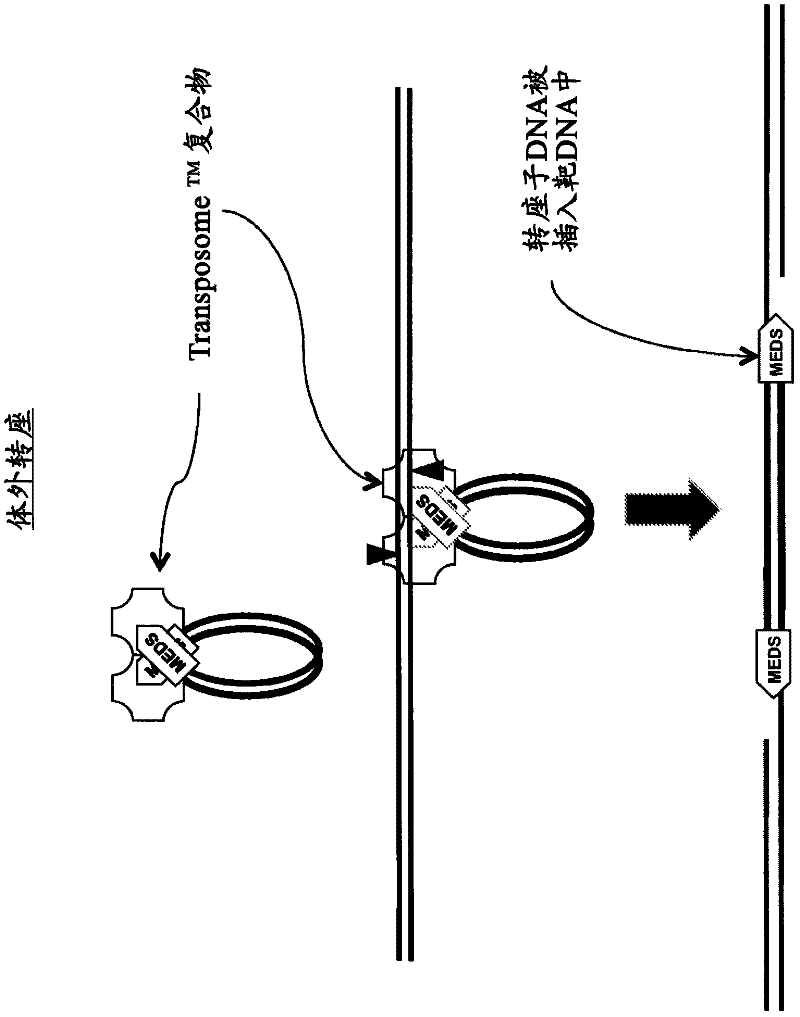
Transposon end compositions and methods for modifying nucleic acids
ActiveUS20100120098A1Sugar derivativesMicrobiological testing/measurementGenomic sequencingPolymerase L
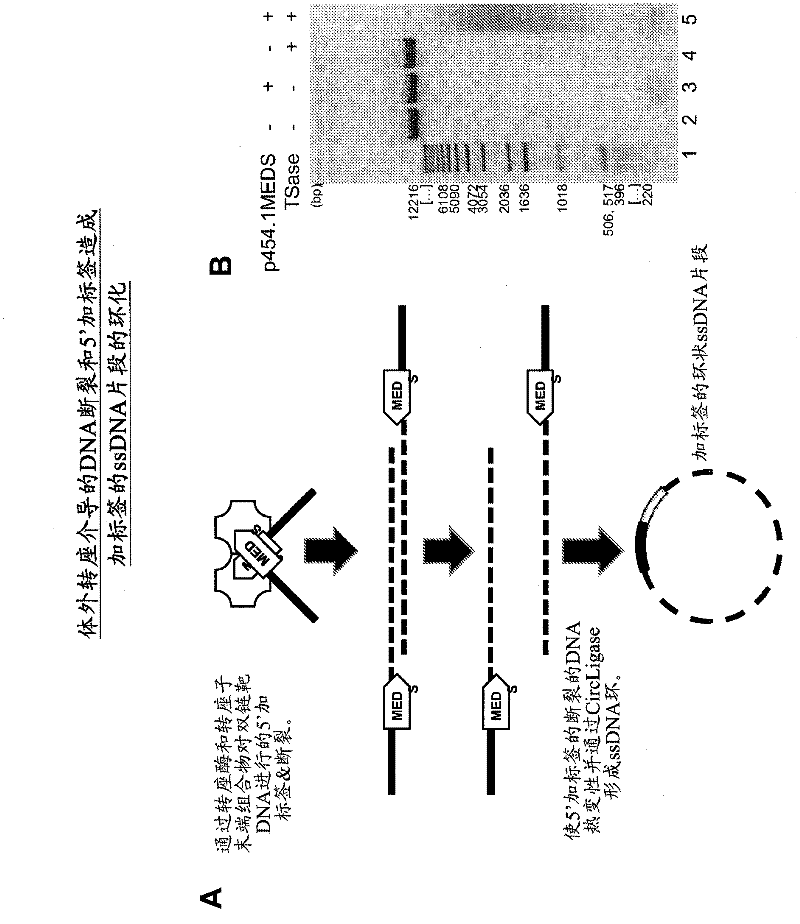
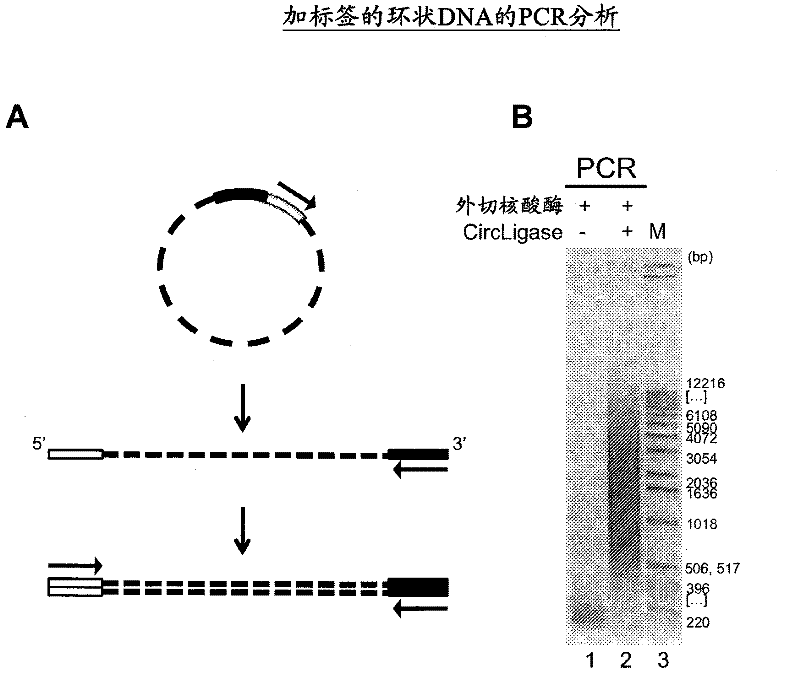
The present invention provides methods, compositions and kits for using a transposase and a transposon end for generating extensive fragmentation and 5′-tagging of double-stranded target DNA in vitro, then using a DNA polymerase for generating 5′- and 3′-tagged single-stranded DNA fragments without performing a PCR amplification reaction, wherein the first tag on the 5′-ends exhibits the sequence of the transferred transposon end and optionally, an additional arbitrary sequence, and the second tag on the 3′-ends exhibits a different sequence from the sequence exhibited by the first tag. The method is useful for generating 5′- and 3′-tagged DNA fragments for use in a variety of processes, including processes for metagenomic analysis of DNA in environmental samples, copy number variation (CNV) analysis of DNA, and comparative genomic sequencing (CGS), including massively parallel DNA sequencing (so-called “next-generation sequencing.)
Owner:ILLUMINA INC
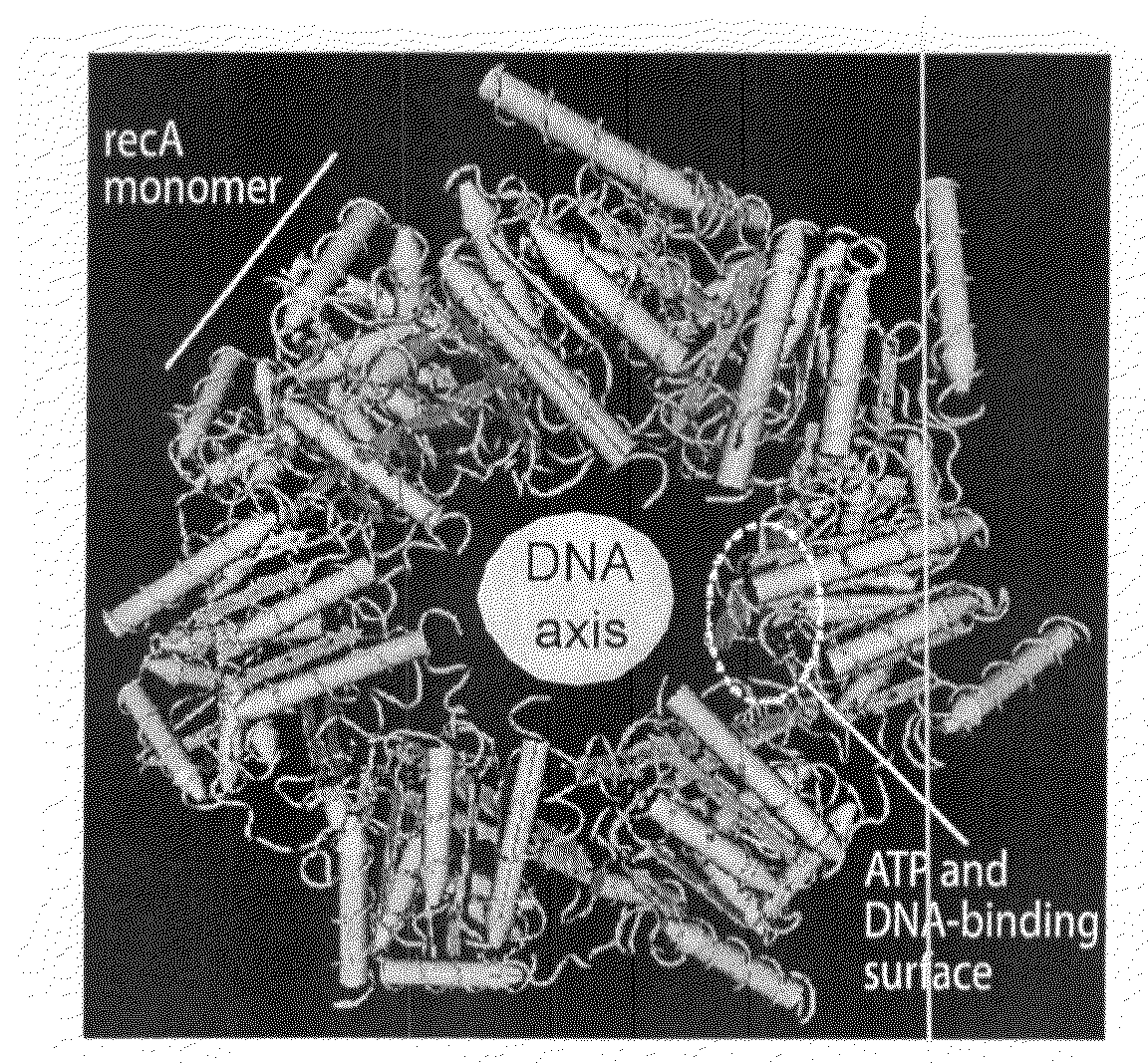
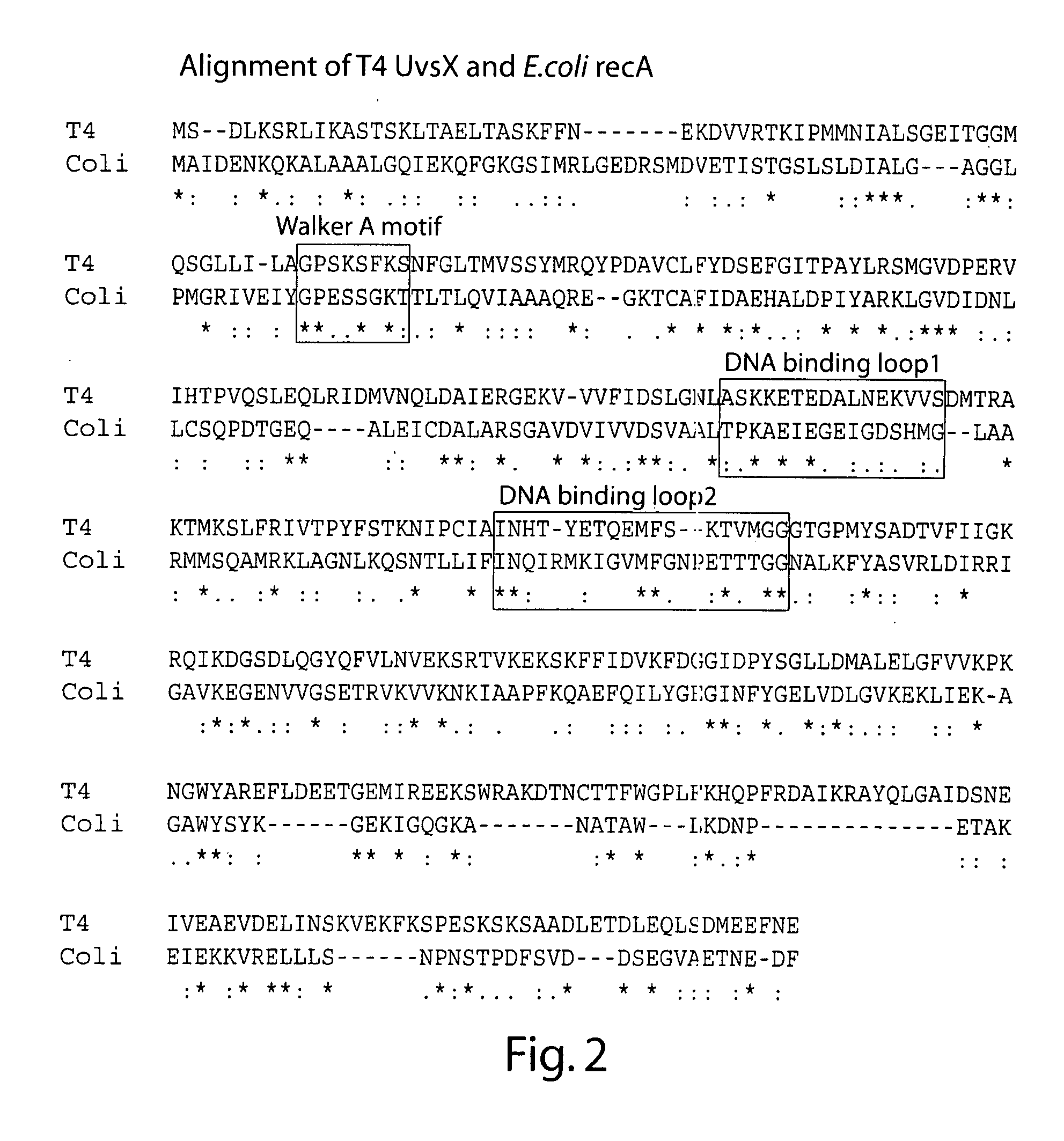
Recombinase polymerase amplification
ActiveUS8071308B2Lower Level RequirementsImprove abilitiesSugar derivativesMicrobiological testing/measurementLoading factorRecombinase Polymerase Amplification
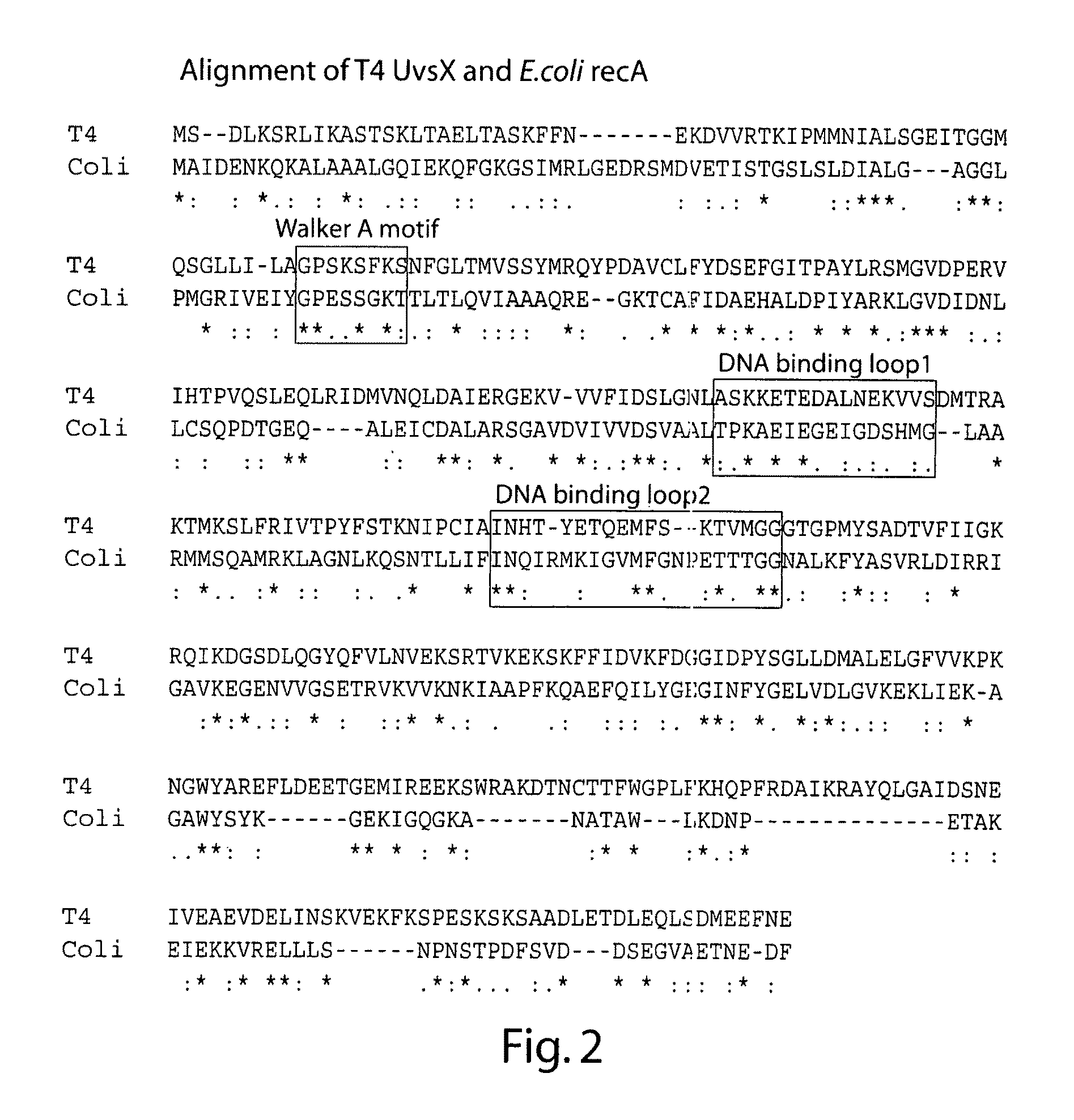
The present invention features novel, diverse, hybrid and engineered recombinase enzymes, and the utility of such proteins with associated recombination factors for carrying out DNA amplification assays. The present invention also features different recombinase ‘systems’ having distinct biochemical activities in DNA amplification assays, and differing requirements for loading factors, single-stranded DNA binding proteins (SSBs), and the quantity of crowding agent employed.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
Nucleic acid archiving
InactiveUS6291166B1Overcomes drawbackEasy to cleanBioreactor/fermenter combinationsBiological substance pretreatmentsRepeat analysisBlood plasma
This invention is directed to a process for irreversibly binding nucleic acid to solid phase and corresponding processes for the utilization thereof. Nucleic acid is bound to solid phase matrixes exhibiting sufficient hydrophilicity and electropositivity to irreversibly bind the nucleic acids from a sample. These processes include nucleic acid (double or single stranded DNA and RNA) capture from high volume:low concentration specimens, buffer changes, washes, and volume reductions, and enable the interface of solid phase bound nucleic acid with enzyme, hybridization or amplification strategies. The invention, solid phase irreversibly bound nucleic acid, may be used, for example, in repeated analyses to confirm results or test additional genes in both research and commercial applications. Further, a method is described for virus extraction, purification, and solid phase amplification from large volume plasma specimens.
Owner:APPL BIOSYSTEMS INC
Detection of chromosomal disorders
InactiveUS20050250111A1Improve accuracyFast and accurate and simple and inexpensive detectionMicrobiological testing/measurementFermentationDiseaseGenomic DNA
Methods for detecting in a single assay any one of multiple chromosomal disorders that result from aneuploidy or certain mutations, particularly microdeletions, and kits for use therein. A polymerase chain reaction (PCR) is carried out to amplify eukaryotic genomic DNA using a plurality of primer oligonucleotide pairs wherein one primer of each pair has a detectable label attached 5′ thereto. A plurality of the primer pairs are targeted to DNA segments of different chromosomes of interest which are indicative of potential chromosomal disorders, and one pair is targeted for a control gene. The amplified PCR products are purified, and single-stranded DNA having the detectable labels is obtained therefrom and hybridized with spots on a microarray that each contain DNA oligonucleotide probes having nucleotide sequences complementary to a nucleotide sequence of one strand of each segment. The microarray is imaged for presence of labels on its respective spots, and the absence or presence of chromosomal disorders as indicated by one or more of the targeted DNA segments of interest is diagnosed by first comparing the imaging results to the imaging of spots specific to the control gene and then to results obtained from imaging normal DNA.
Owner:NOVARTIS AG
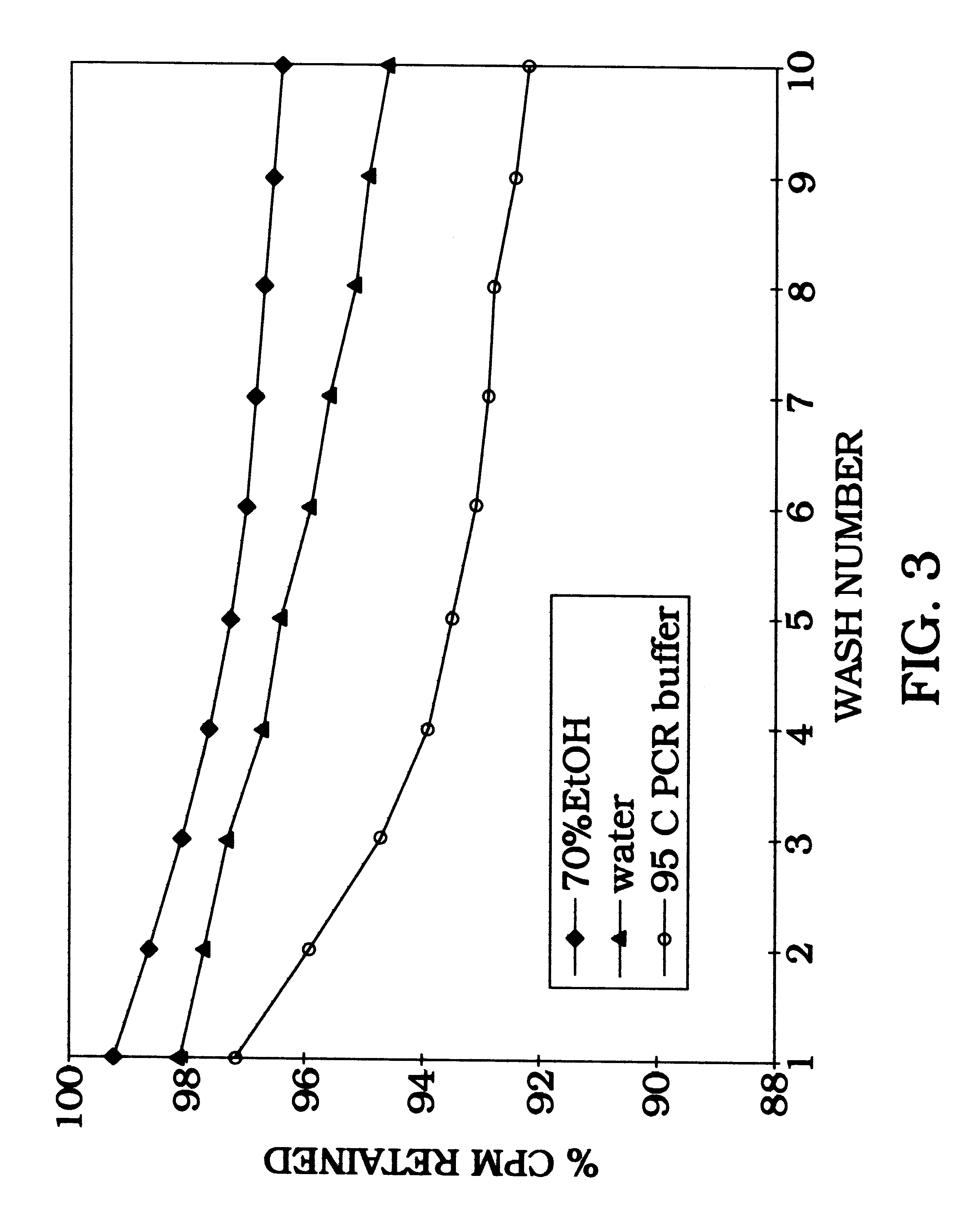
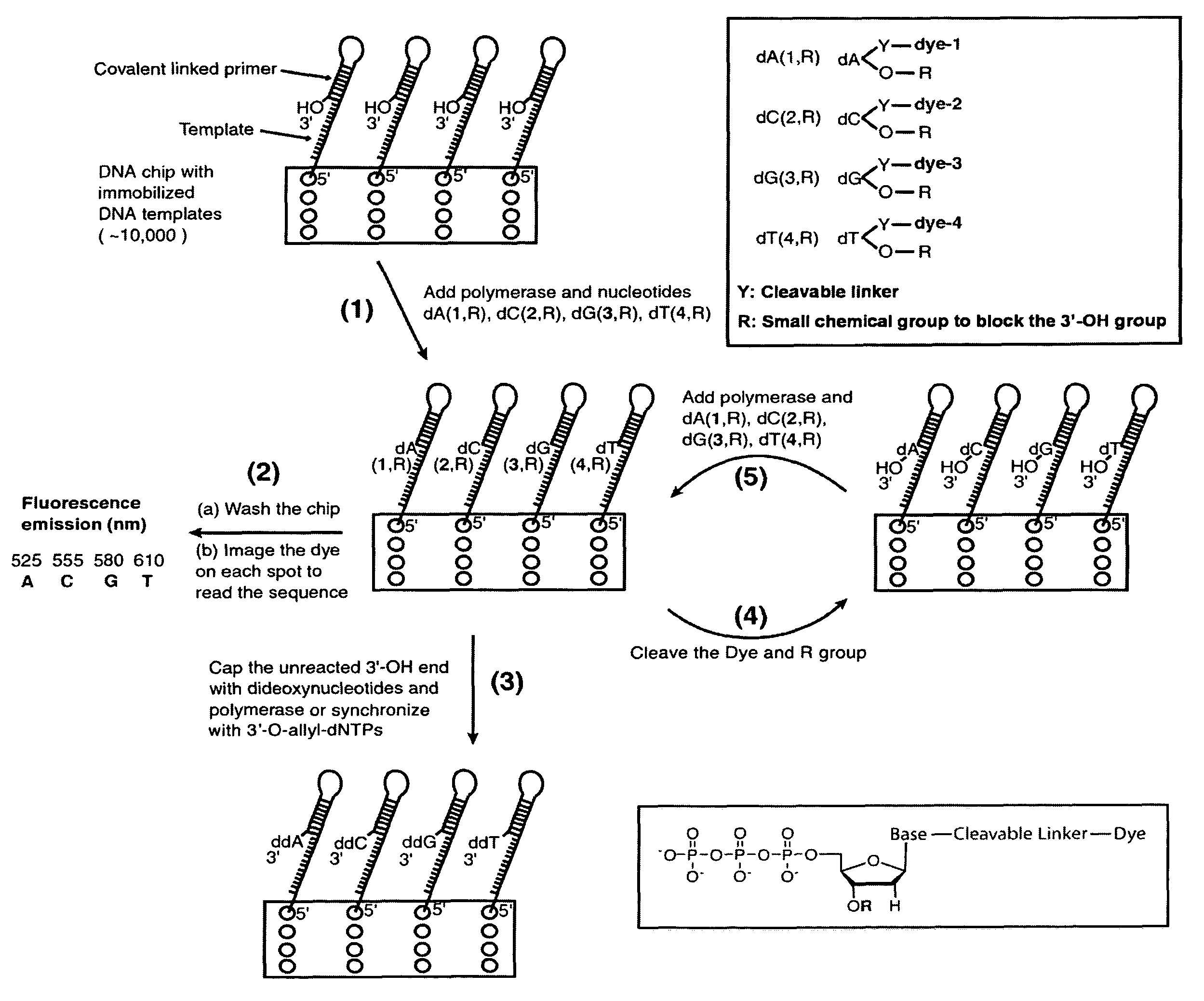
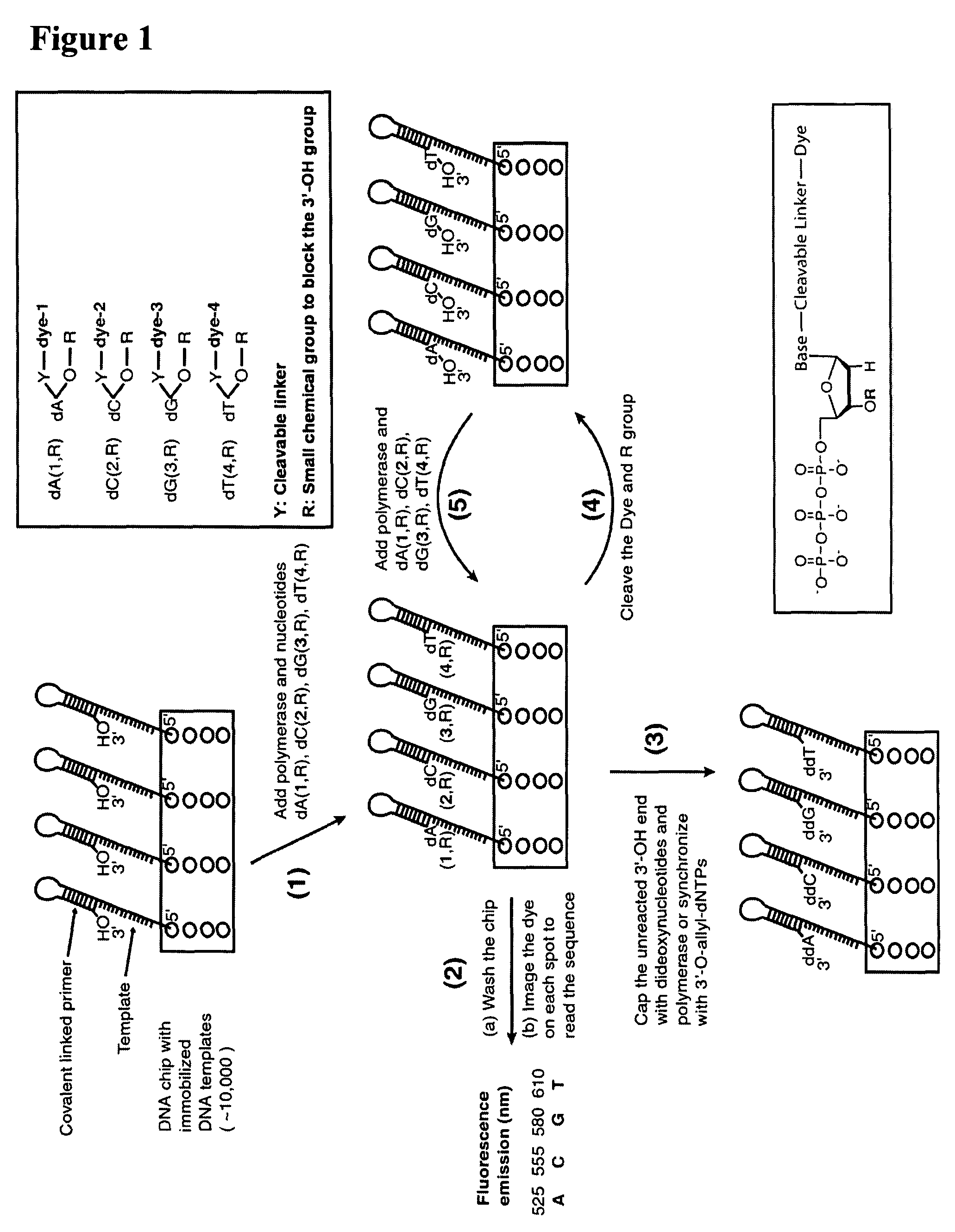
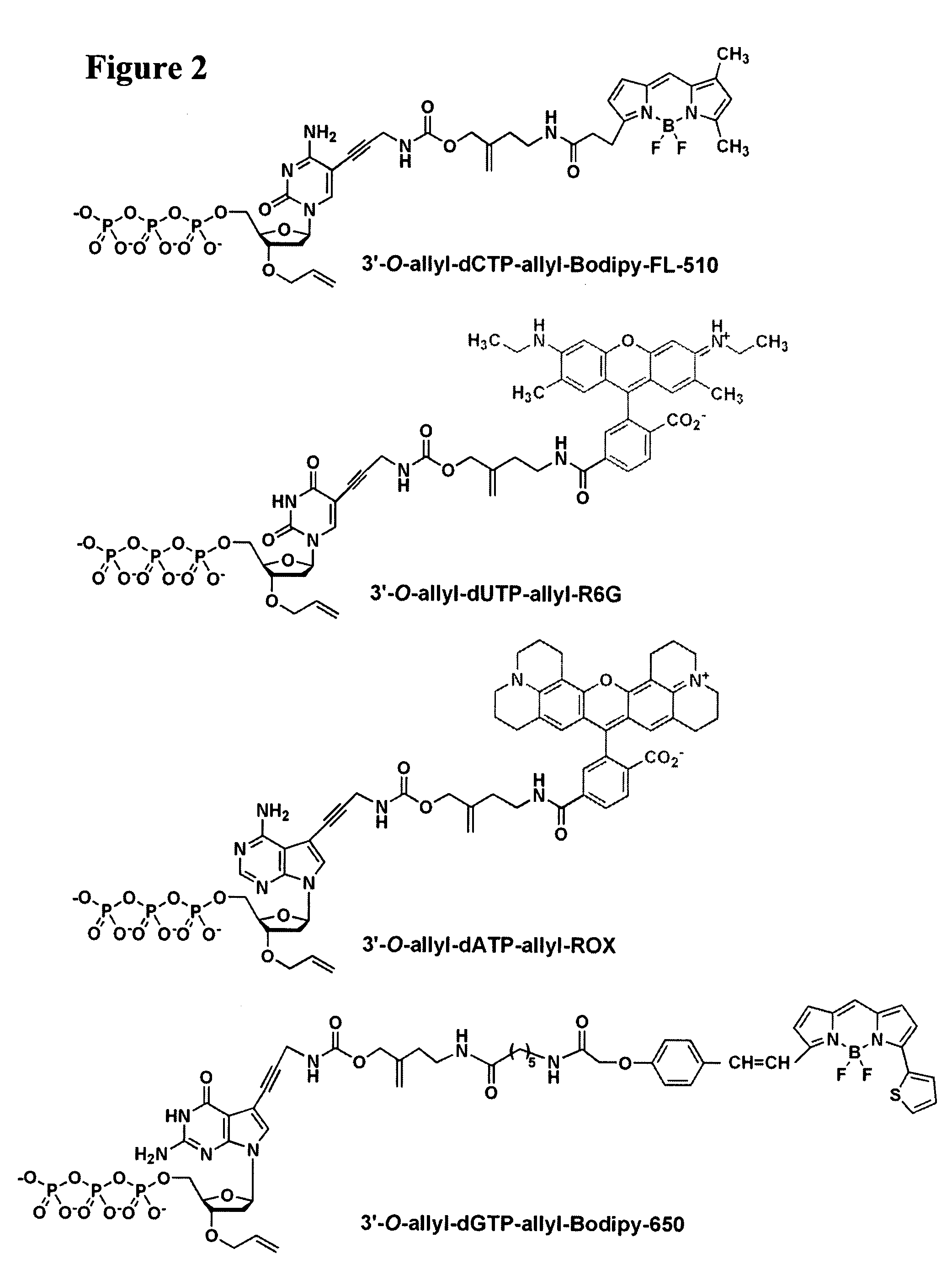
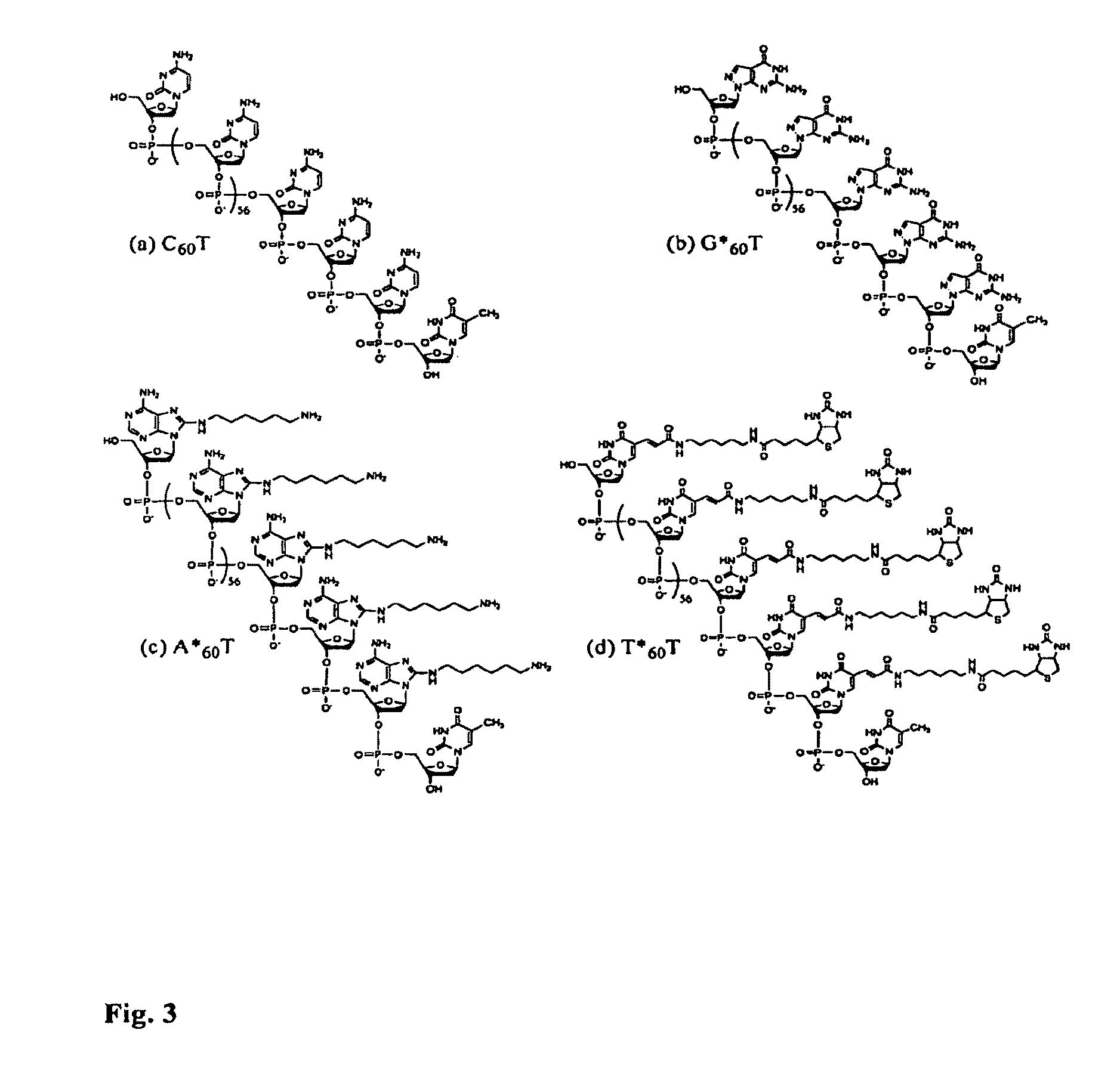
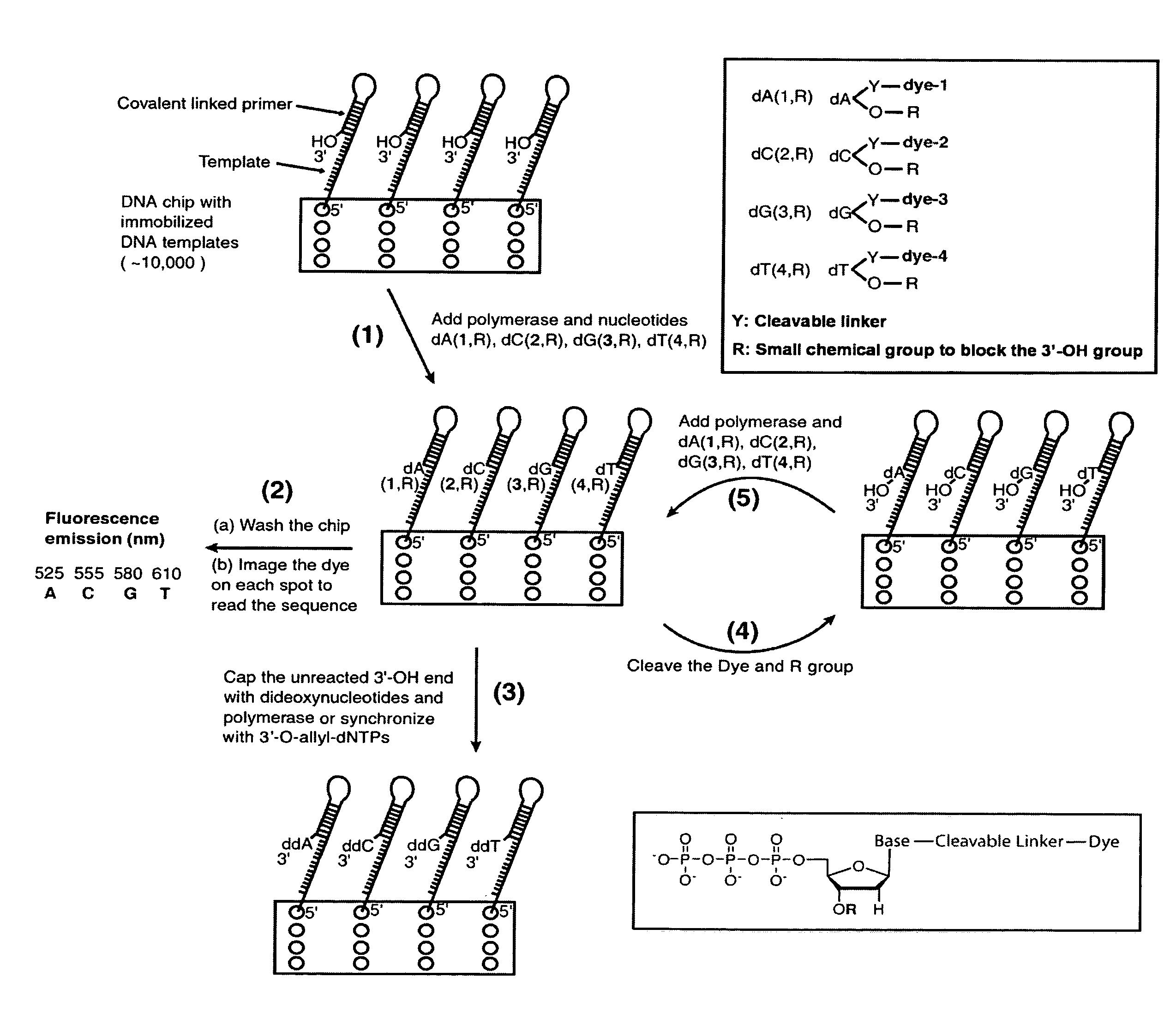
Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
Systematic evolution of ligands exponential enrichment: blended selex
InactiveUS6083696AHigh affinityDesired effectSugar derivativesMicrobiological testing/measurementAla-Ala-Pro-ValKetone
A method is described for generating blended nucleic acid ligands containing non-nucleic acid functional units. Specifically, a SELEX identified RNA ligand to the integrin gpIIbIIIa is conjugated to the peptide Gly-Arg-Gly-Asp-Thr-Pro (SEQ ID NO:1). This blended RNA ligand inhibits the biological activity of gpIIbIIIa with high specificity. Also described is a single-stranded DNA ligand to elastase coupled to N-methoxysuccinyl-Ala-Ala-Pro-Val-chloromethyl (SEQ ID NO:3) ketone. This elastase blended nucleic acid ligand inhibits the biological activity of elastase.
Owner:GILEAD SCI INC
Combinatorial selection of phosphorothioate single-stranded DNA aptamers for TGF-beta-1 protein
InactiveUS20050239134A1Increased proliferationPrevent proliferationPeptide/protein ingredientsAntiviralsDNA AptamersTgf β signalling
The present invention includes the selection and isolation of thioaptamers that target the signaling protein TGF-β1, compositions of such thioaptamers and the use of such thioaptamers to either block or enhance signal transduction of the TGF-β1 protein and thus function as, e.g., immunomodulatory agents. Thioaptamers may also be targeted alone or in combination with other thioaptamers against the ligand, the receptors, the ligand trap protein(s) and / or the co-receptors to modulate TGF-β signaling pathway.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
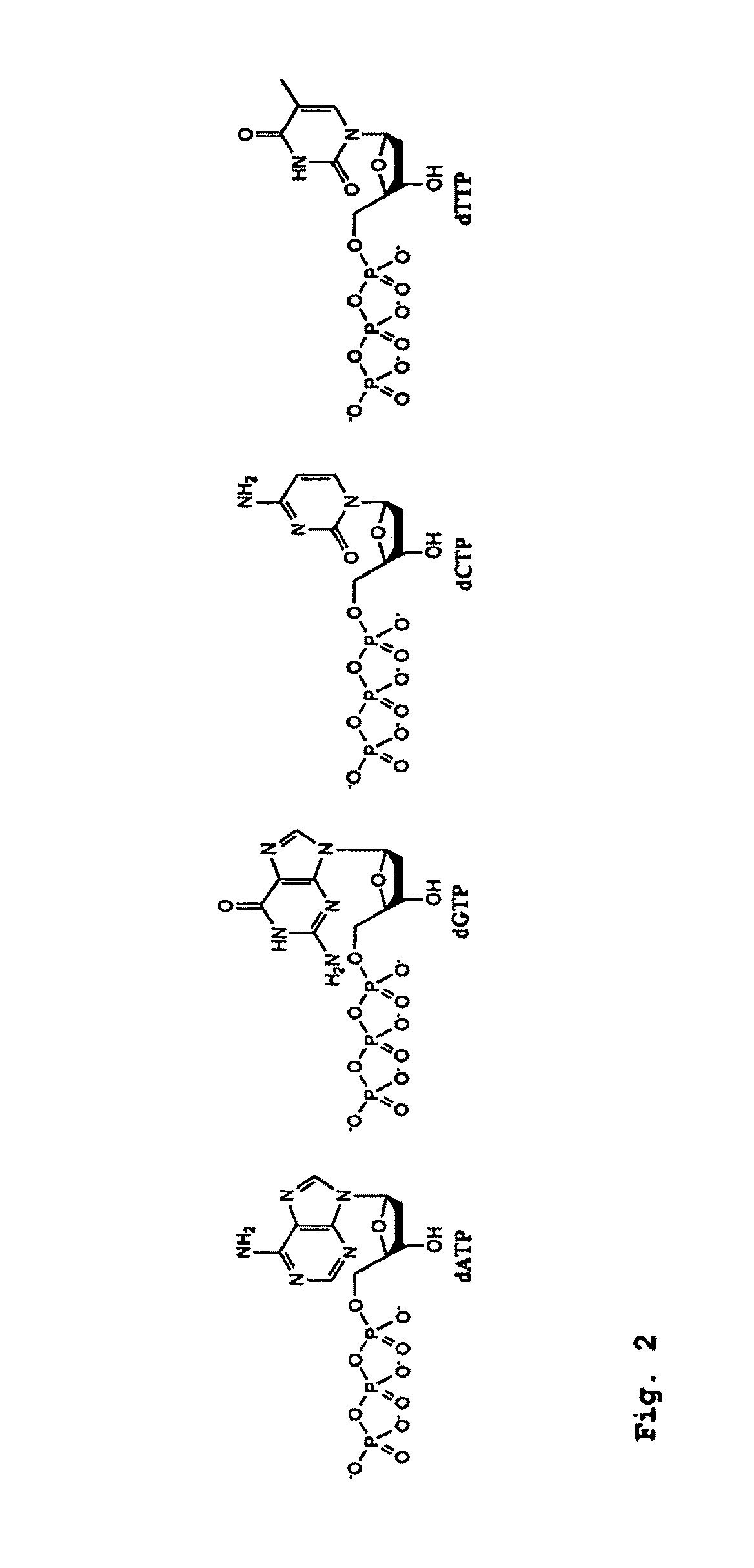
DNA Sequencing by Nanopore Using Modified Nucleotides
ActiveUS20090298072A1Sugar derivativesMicrobiological testing/measurementNucleotideSingle strand dna
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
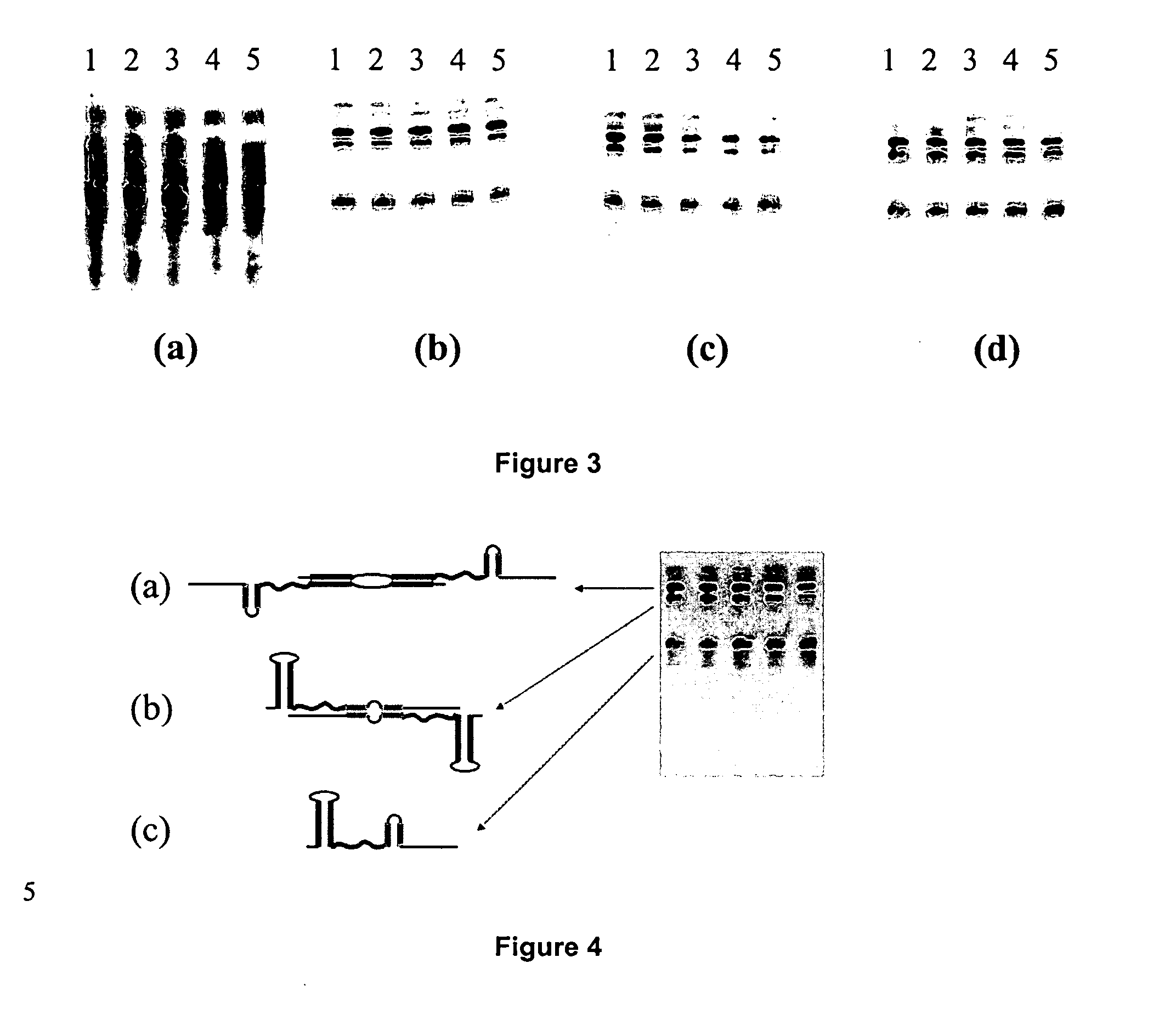
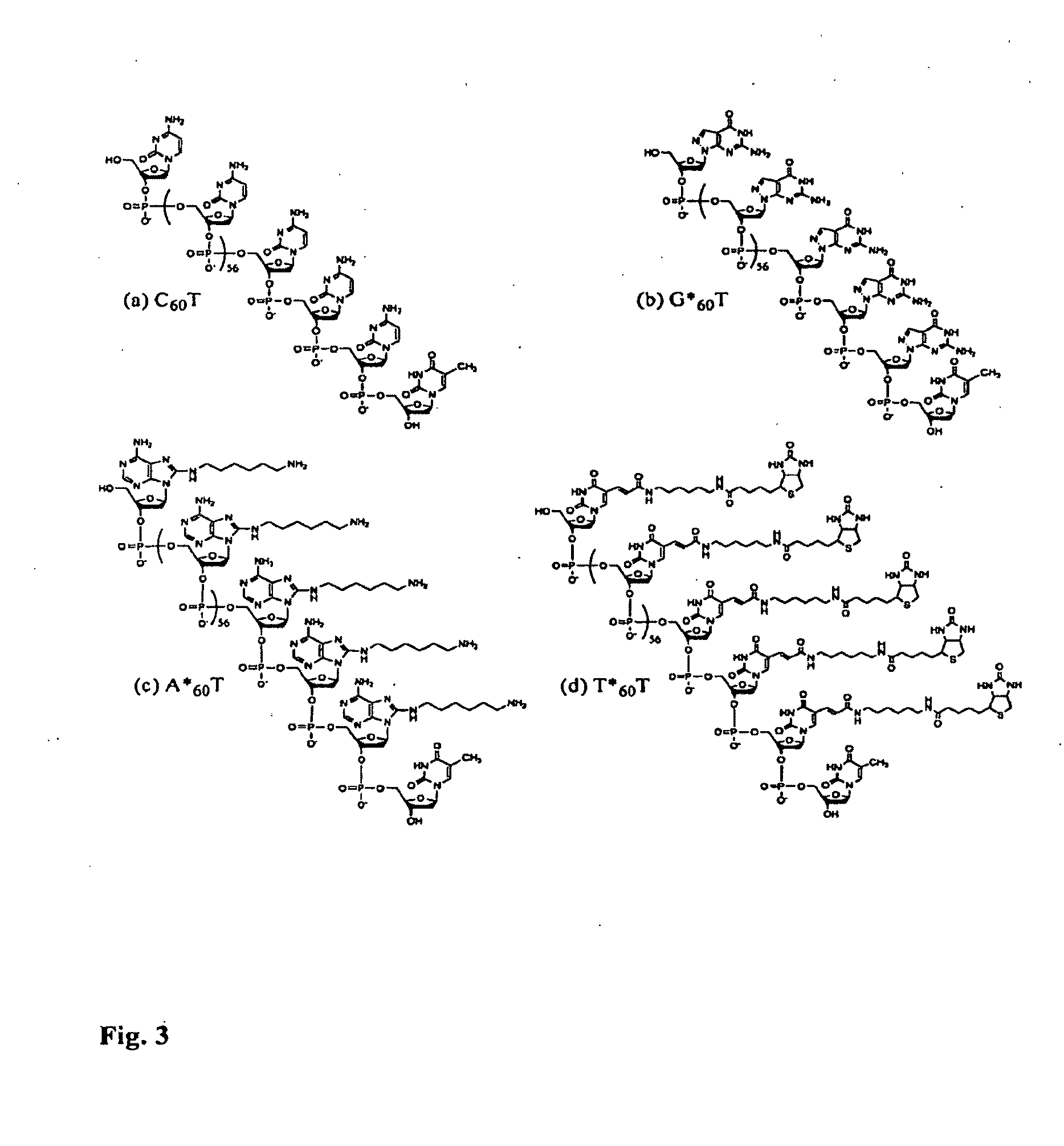
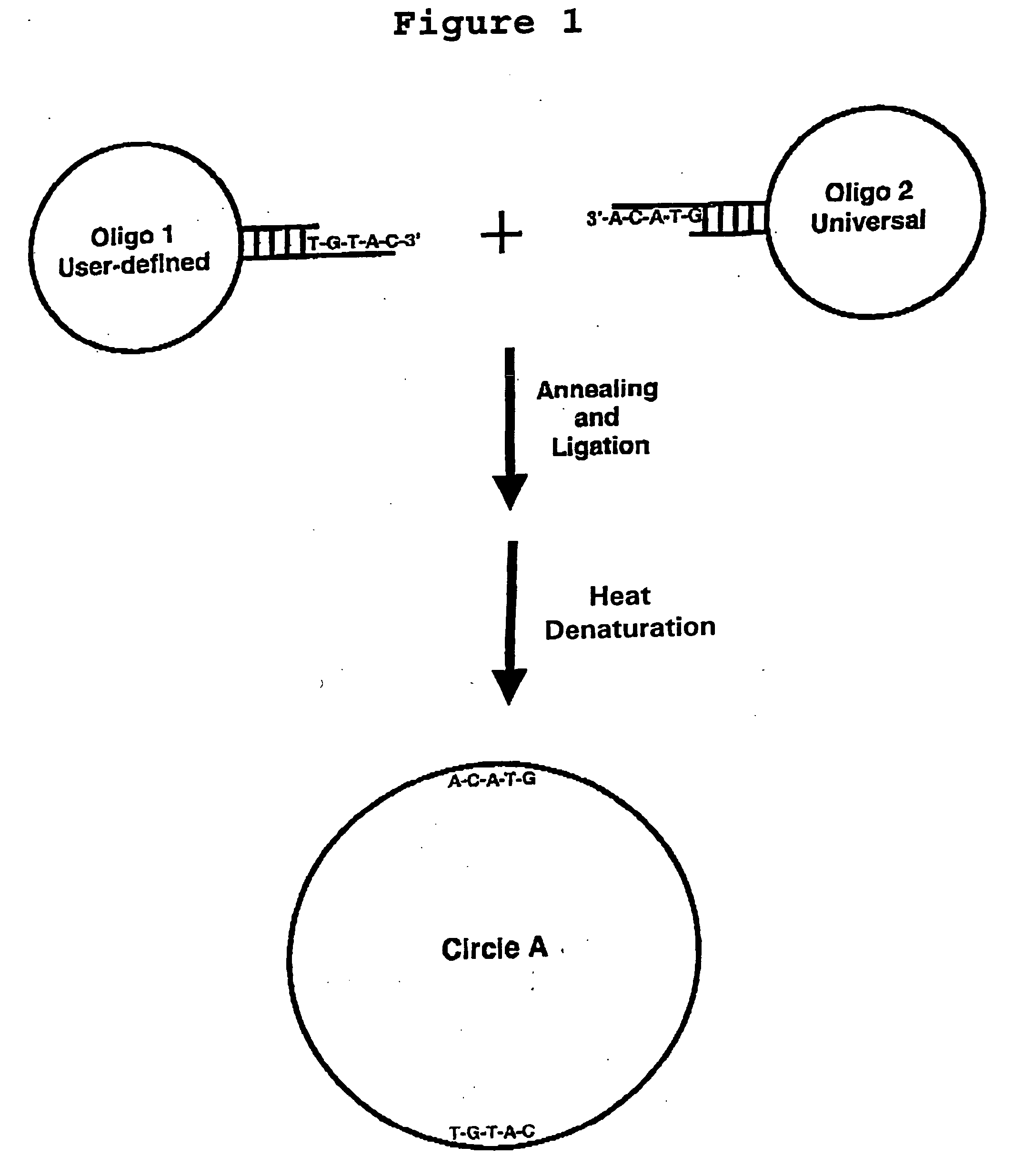
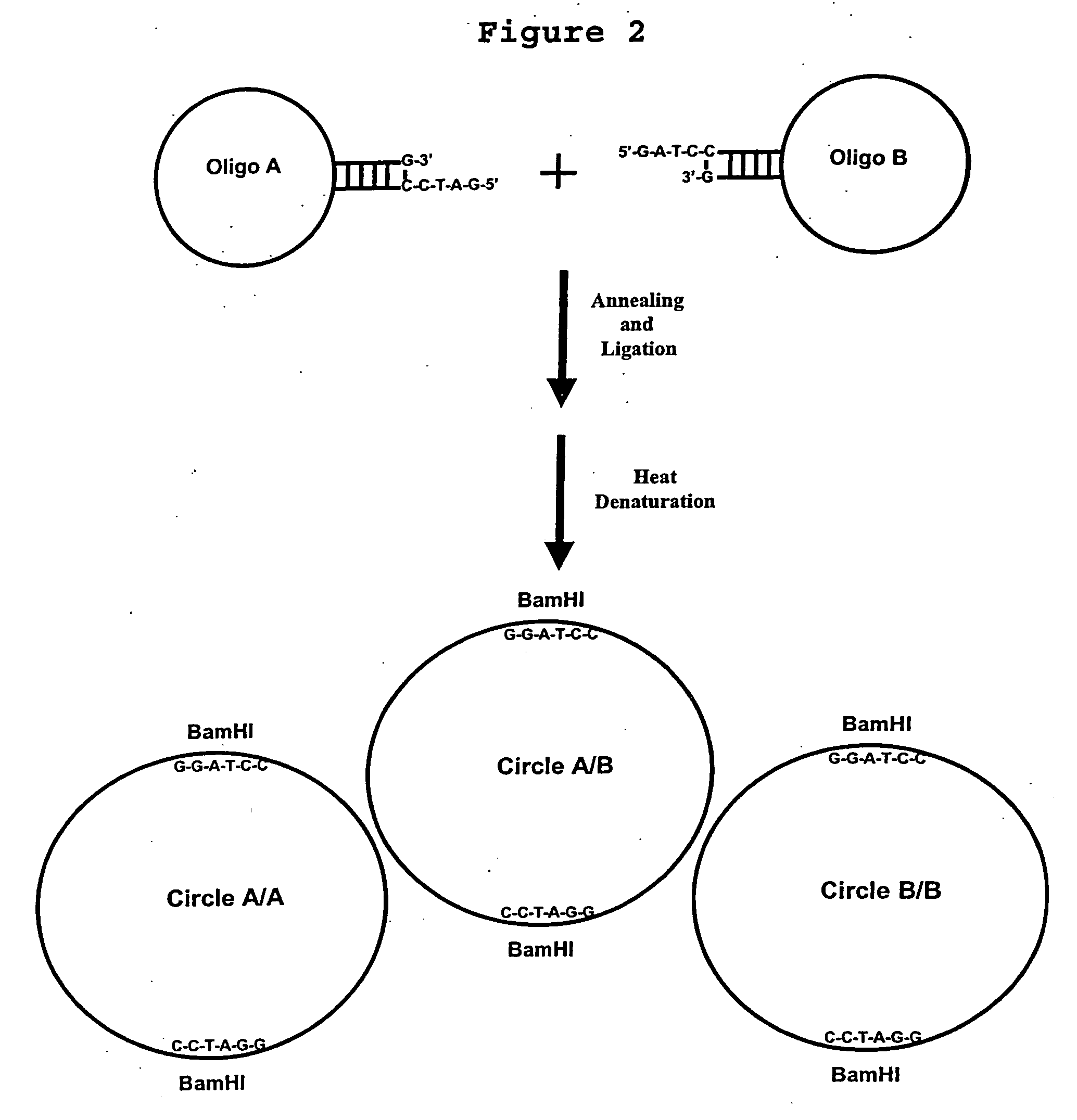
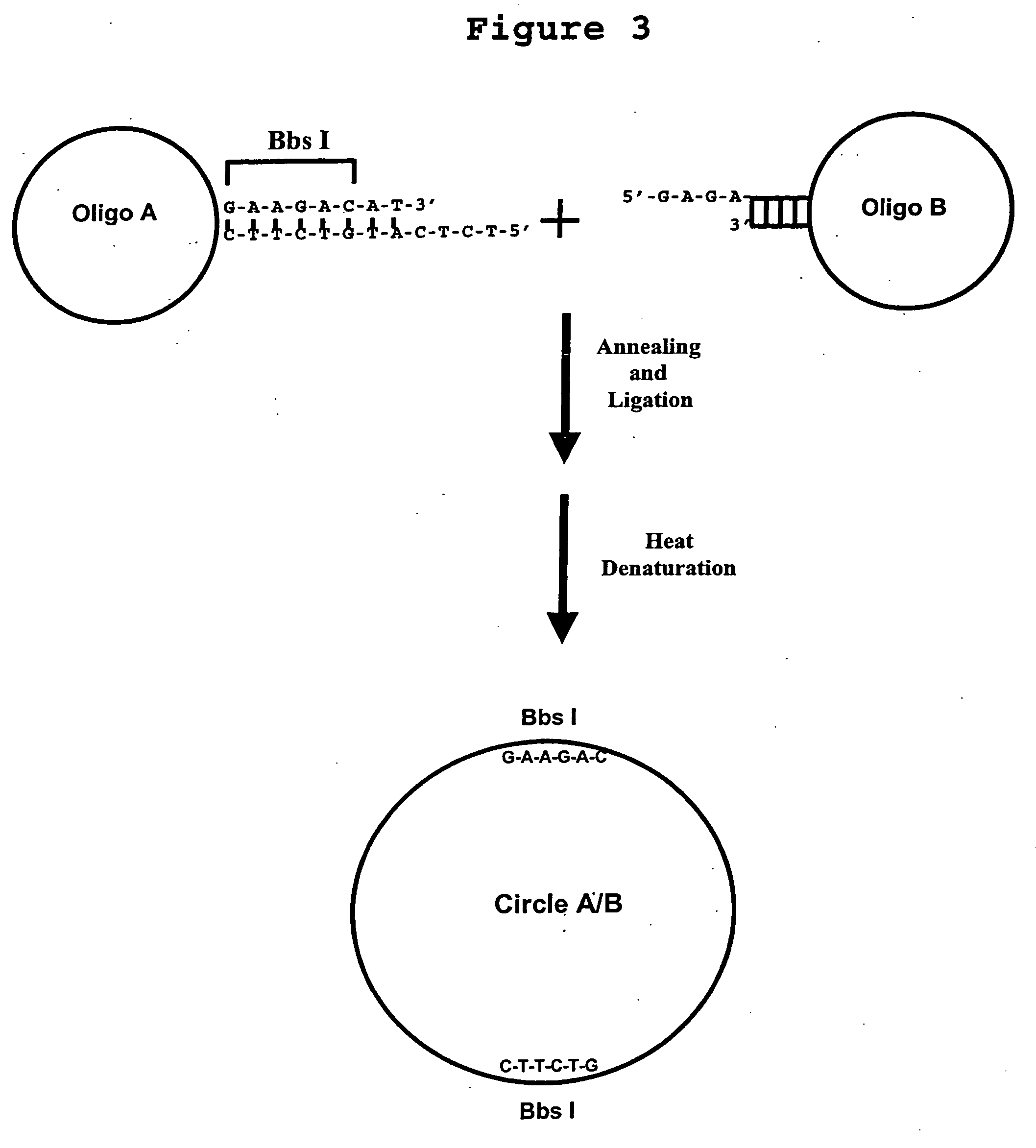
Generation of single-strand circular DNA from linear self-annealing segments
InactiveUS20070015182A1Convenient ligationEasy to copyMicrobiological testing/measurementFermentationNucleotideSingle strand
The present invention provides a method for the rapid simultaneous production of a plurality of single-stranded DNA circles having a predetermined size and nucleotide sequence using pre-designed hairpin oligonucleotides containing complementary sequences for directing ligation to form dumbbell-shaped monomers followed by heat denaturation to yield single-stranded DNA circles.
Owner:ABARZUA PATRICIO
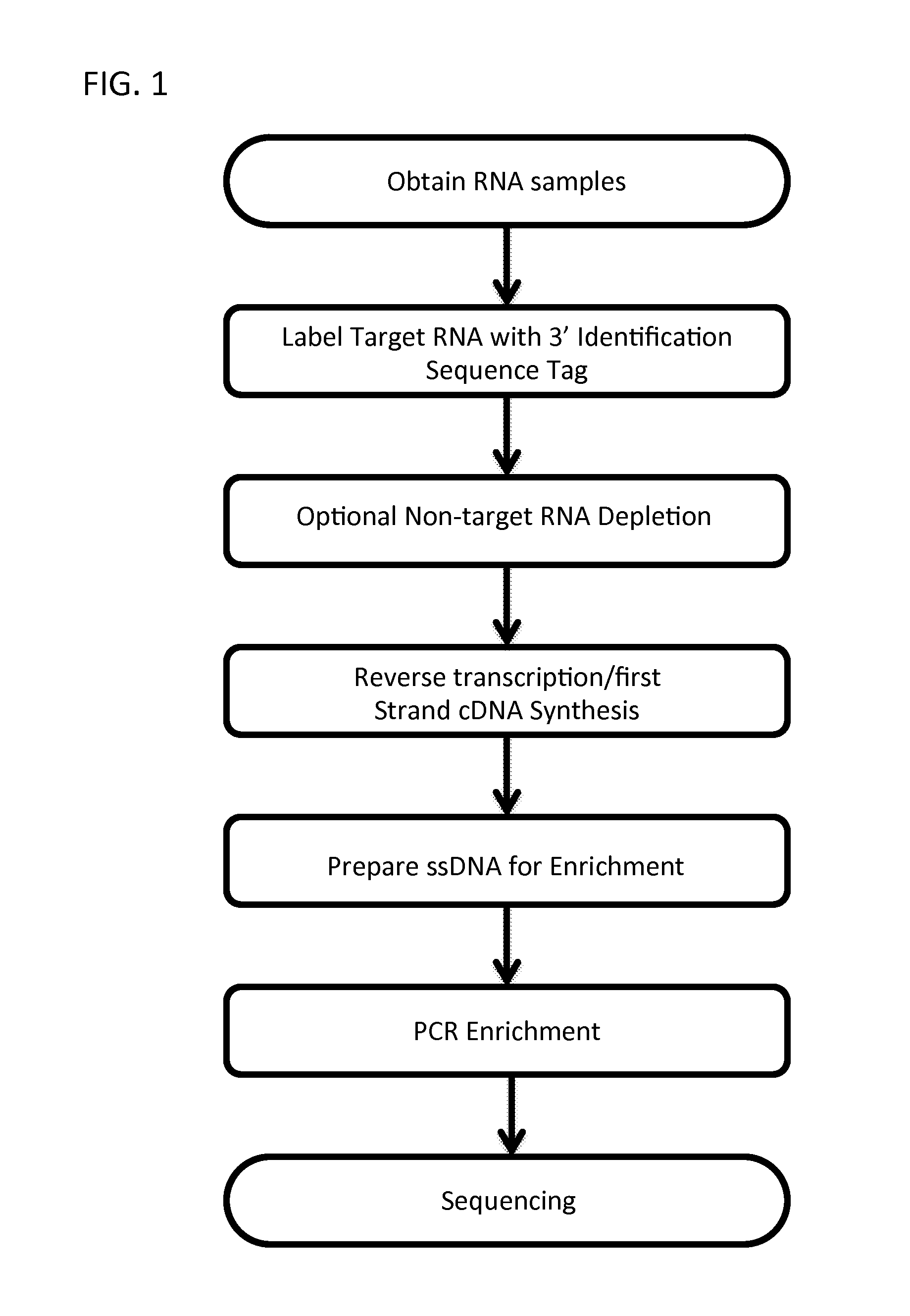
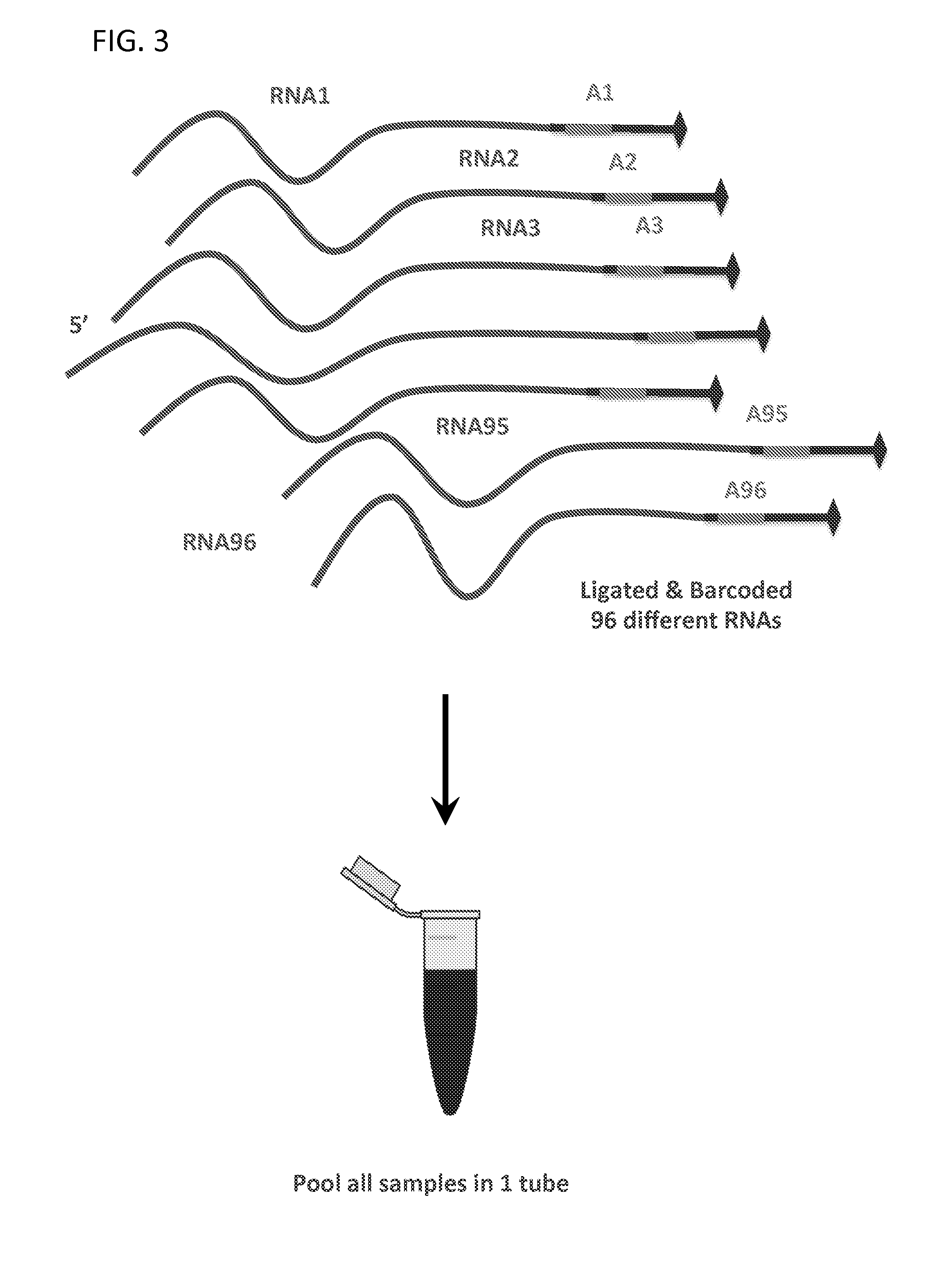
Massively Multiplexed RNA Sequencing
ActiveUS20160024572A1Low costShorten the timeNucleotide librariesMicrobiological testing/measurementRNA SequenceNucleic acid sequencing
A method for parallel sequencing target RNA from samples from multiple sources while maintaining source identification is provided. The method includes providing samples of RNA comprising target RNA from two or more sources; labeling, at the 3′ end, the RNA from the two or more sources with a first nucleic acid adaptor that comprises a nucleic acid sequence that differentiates between the RNA from the two or more sources; reverse transcribing the two or more sources to create a single stranded DNA comprising the nucleic acid sequence that differentiates between the RNA from the two or more sources; amplifying the single stranded DNA to create DNA amplification products that comprise the nucleic acid sequence that differentiates between the RNA from the two or more sources; sequencing the DNA amplification products thereby parallel sequencing target RNA from samples from multiple sources while maintaining source identification.
Owner:THE BROAD INST INC +1
Recombinase polymerase amplification
ActiveUS20090029421A1Lower Level RequirementsImprove abilitiesMicrobiological testing/measurementTransferasesLoading factorRecombinase Polymerase Amplification
The present invention features novel, diverse, hybrid and engineered recombinase enzymes, and the utility of such proteins with associated recombination factors for carrying out DNA amplification assays. The present invention also features different recombinase ‘systems’ having distinct biochemical activities in DNA amplification assays, and differing requirements for loading factors, single-stranded DNA binding proteins (SSBs), and the quantity of crowding agent employed.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
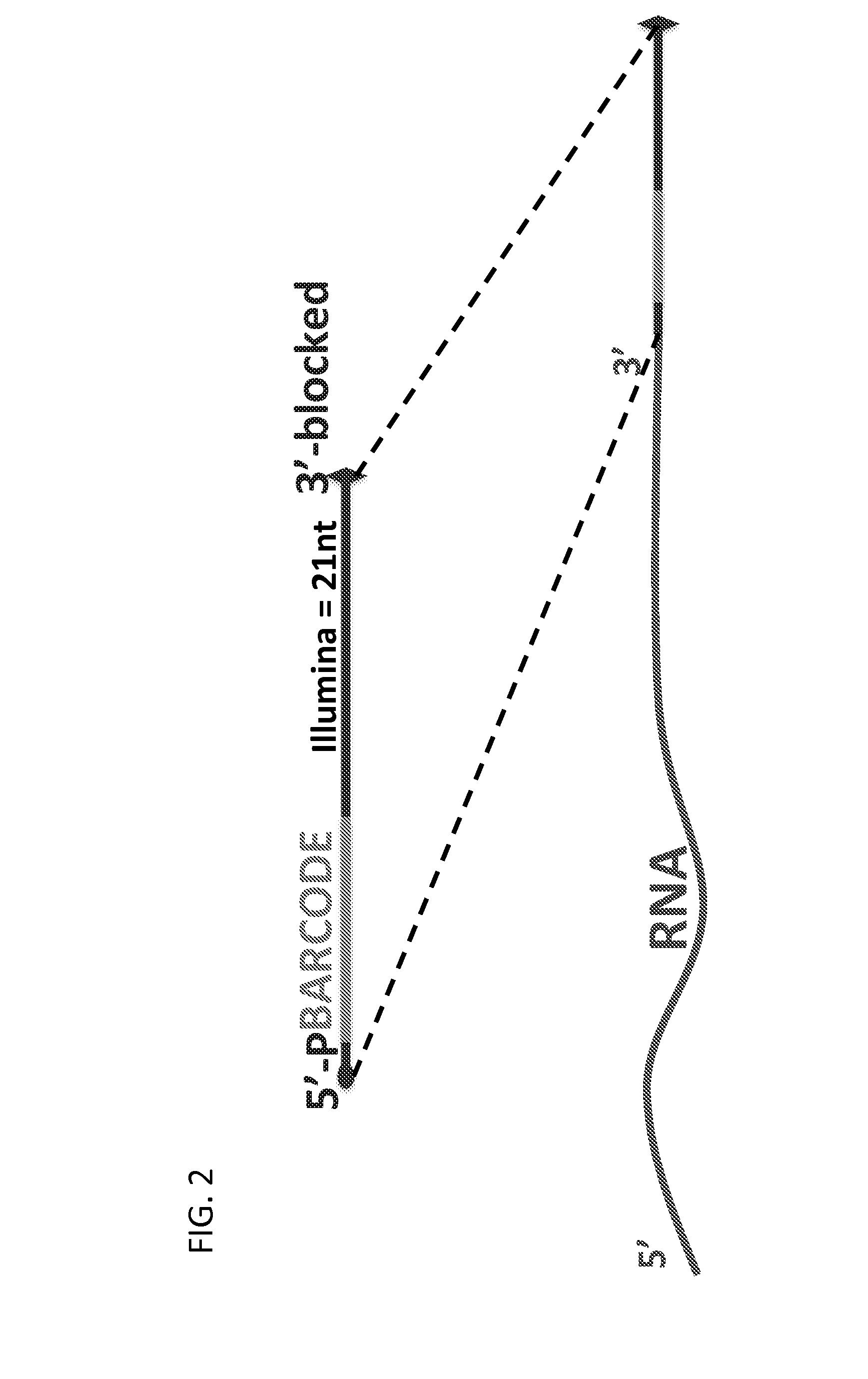
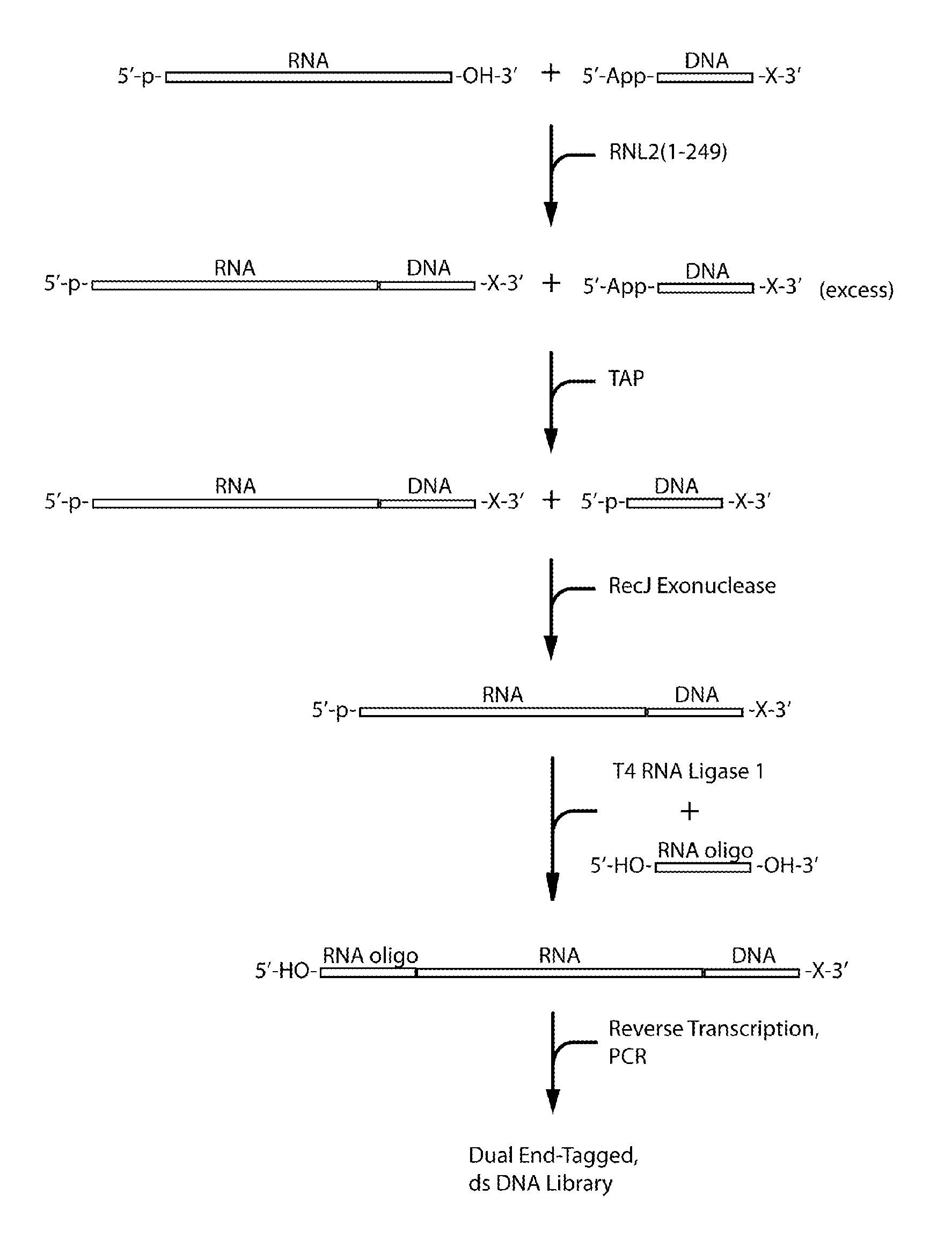
Methods and kits for 3'-end-tagging of RNA
ActiveUS20110104785A1High detection sensitivityRapid and efficient enzymaticHydrolasesMicrobiological testing/measurementRNA SequenceSingle strand dna
The present innovation provides methods and kits that enable rapid and efficient dual end-tagging of RNA to prepare libraries for analysis by applications such as next-generation RNA sequencing, qPCR, microarray analysis, or cloning. The methods do not require time-consuming and inefficient gel-purification steps that are common to methods known in the art. In addition, the present invention provides methods and kits for rapid, high-throughput enzymatic preparation of 5′-activated, 3′-blocked DNA oligonucleotides from standard, single-stranded DNA oligonucleotides.
Owner:ILLUMINA INC
Therapeutic methods for benzodiazepine derivatives
This invention provides an antibody with high affinity for single-stranded DNA, low or no affinity for double-stranded DNA, and capable of specifically binding a DNA hairpin and the hybridoma cell lines which produces these monoclonal antibodies. A chimeric mouse comprising these hybridoma cell lines and a histocompatible mouse is further provided. A method for screening for an agent which will inhibit anti-DNA antibody.DNA binding. One such agent is a benzodiazepine derivative. This invention therefore provides a method of inhibiting the binding of an anti-DNA antibody to its DNA ligand in a sample by contacting the sample with an effective amount of a benzodiazepine derivative.
Owner:RGT UNIV OF MICHIGAN
DNA sequencing by nanopore using modified nucleotides
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
Nanopore electrical sensor
InactiveUS20120037919A1Improve electrical couplingEasy to detectMicrobiological testing/measurementSolid-state devicesImage resolutionEngineering
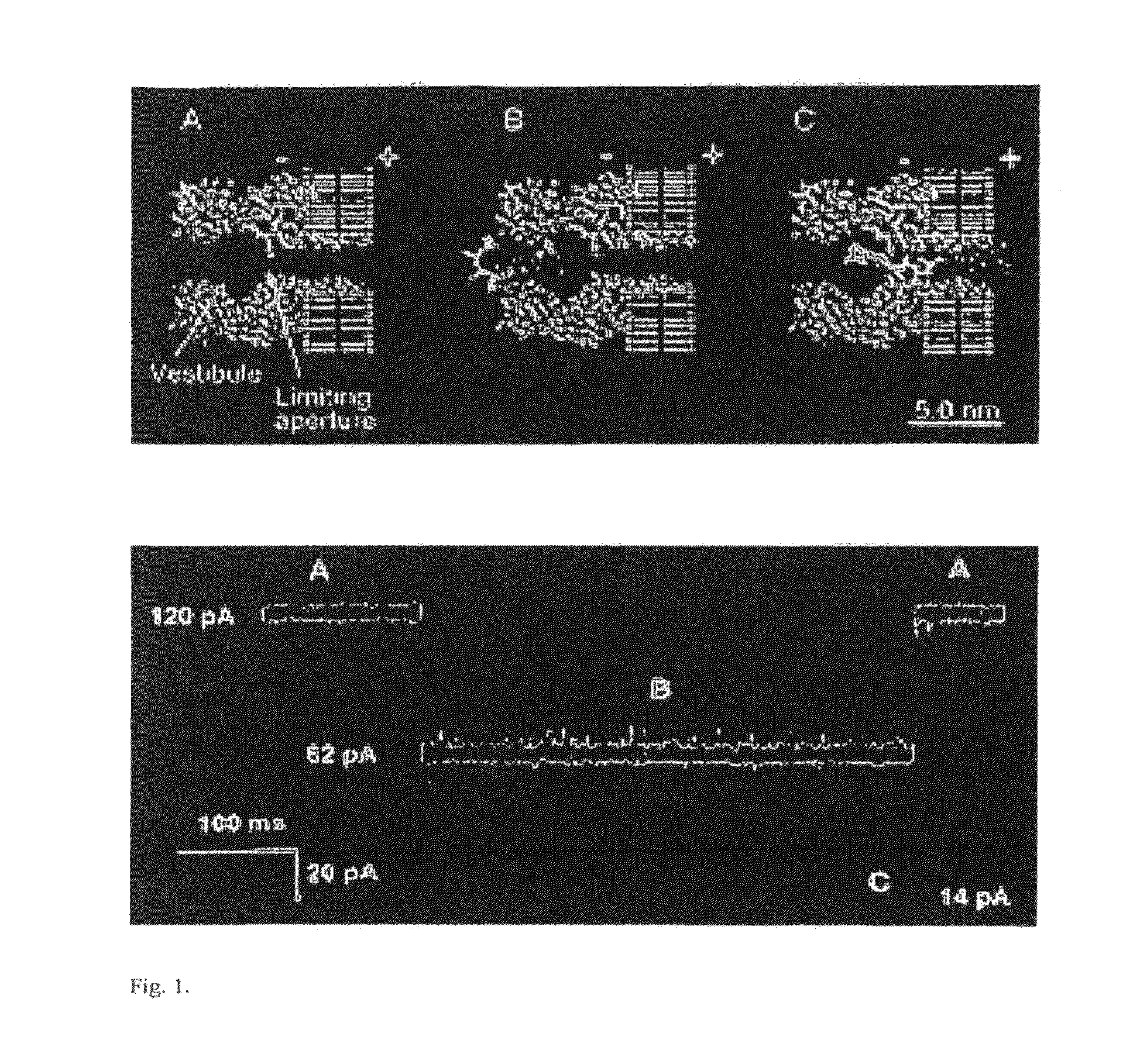
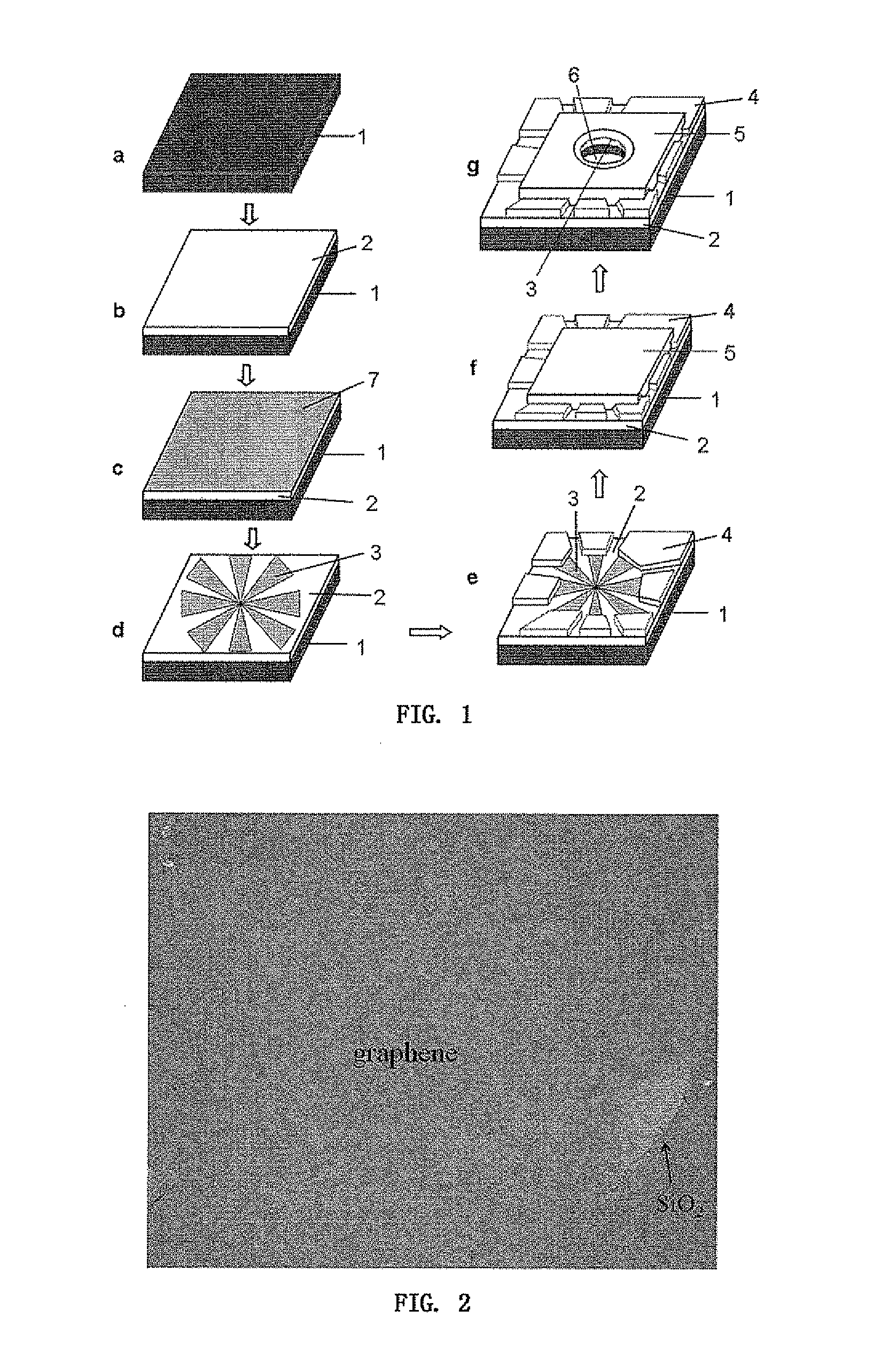
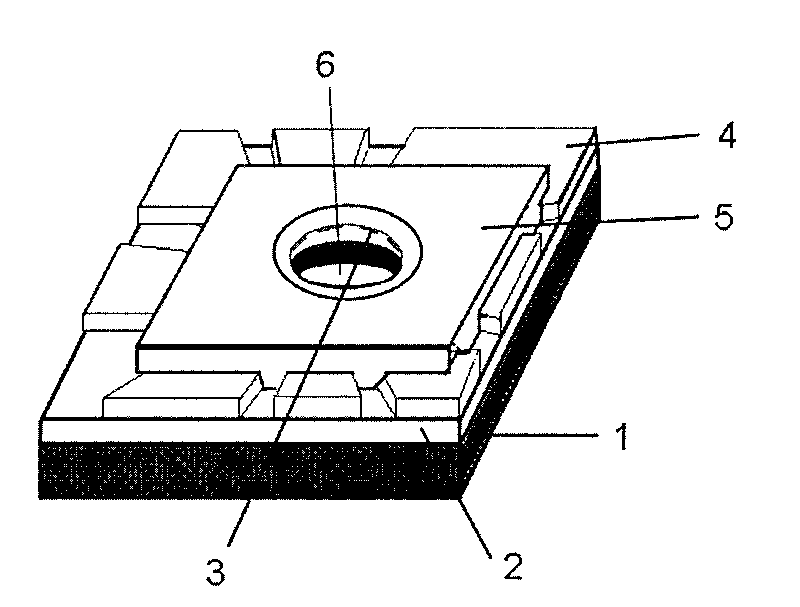
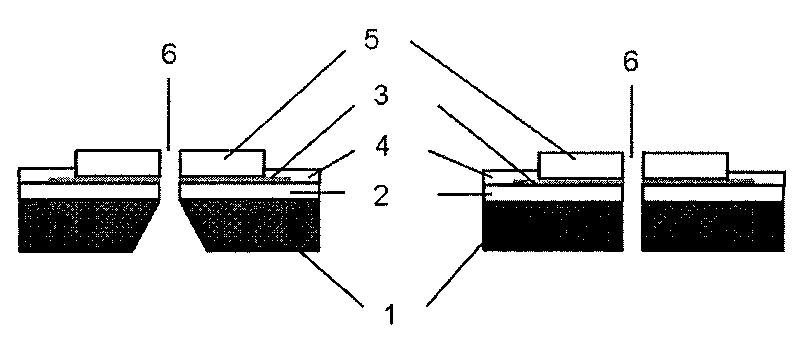
A nanopore electrical sensor is provided. The sensor has layered structure, including a substrate (1), the first insulating layer (2), a symmetrical electrode (3) and the second insulating layer (5) from bottom to top in turn. A nanopore (6) is provided in the center of the substrate (1), the first insulating layer (2), the symmetrical electrode (3) and the second insulating layer (5). The thickness of the symmetrical electrode can be controlled between 0.3 nm and 0.7 nm so as to meet the resolution requirements for detecting a single base in a single-stranded DNA. Thus the sensor is suitable for gene sequencing. The present invention overcomes current technical insufficiency to integrate a nanoelectrode with a nanopore and the method to prepare the nanoelectrode is simple.
Owner:ZHEJIANG UNIV
Synthesis and screening method and kit for lead compound
ActiveCN103882532AEasy to operateSimple and fast operationSequential/parallel process reactionsLibrary tagsCompound (substance)Screening method
Owner:HITGEN INC
In vitro recombination method
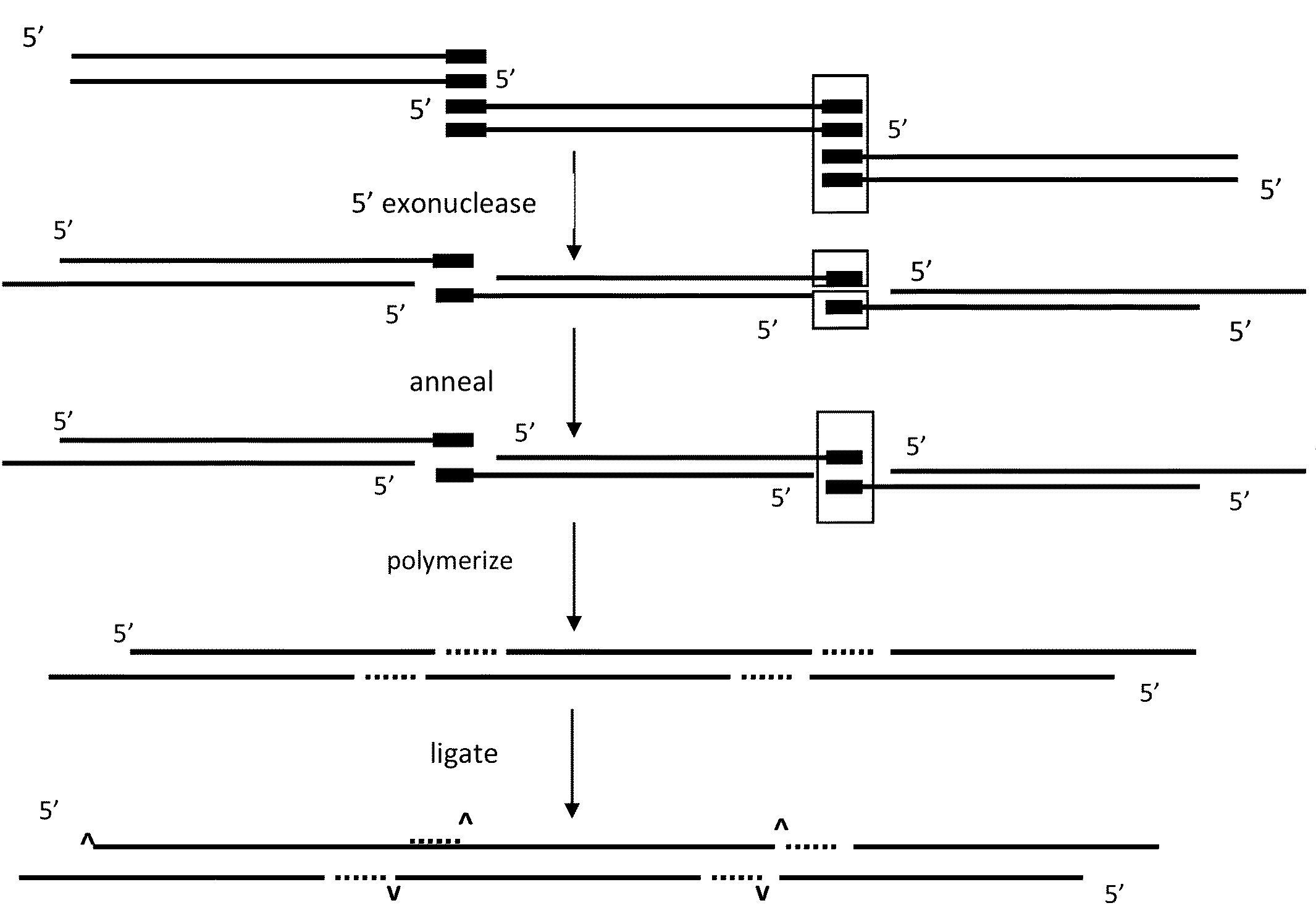
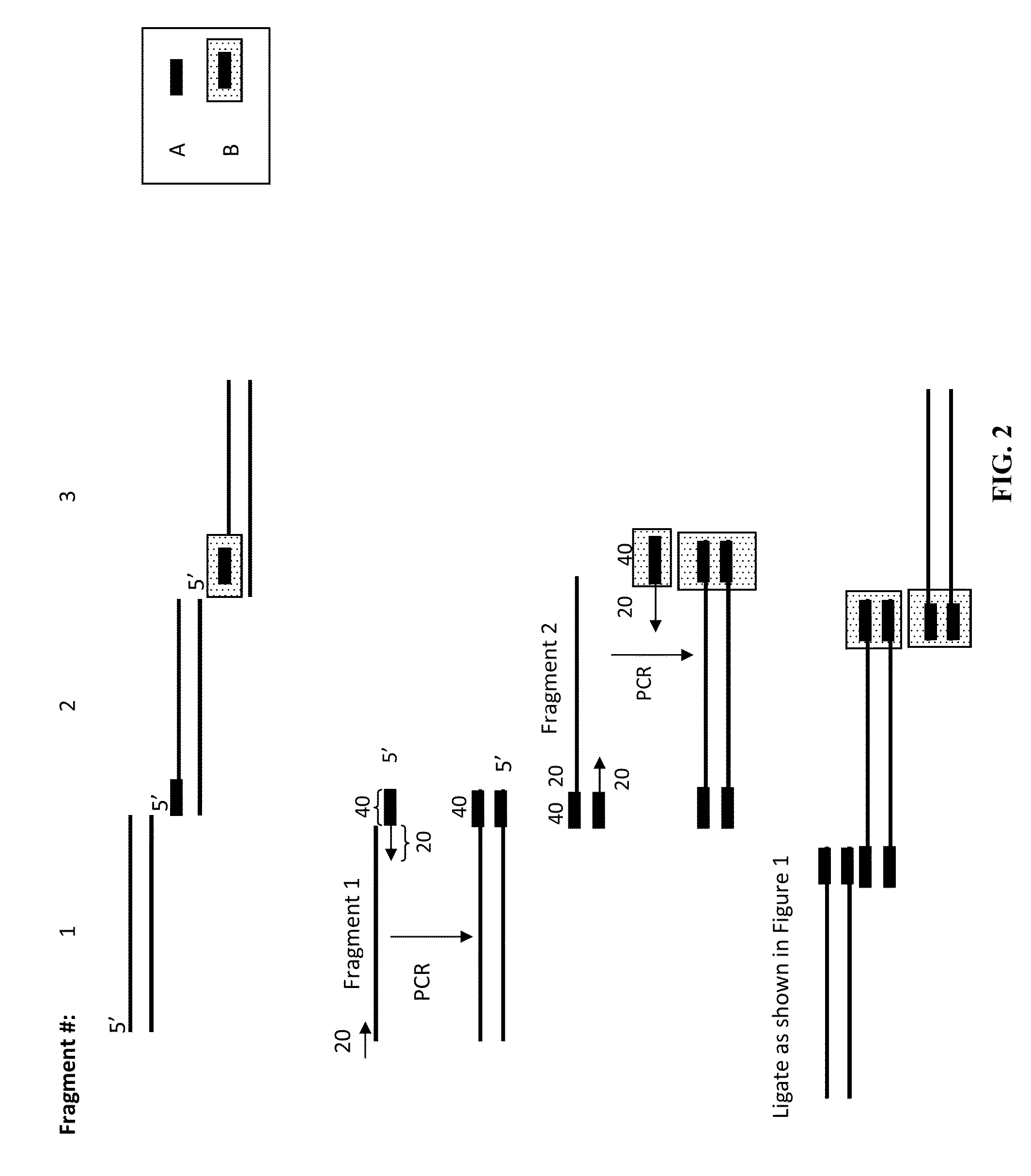
The present invention relates, e.g., to in vitro method, using isolated protein reagents, for joining two double stranded (ds) DNA molecules of interest, wherein the distal region of the first DNA molecule and the proximal region of the second DNA molecule share a region of sequence identity, comprising contacting the two DNA molecules in a reaction mixture with (a) a non-processive 5′ exonculease; (b) a single stranded DNA binding protein (SSB) which accelerates nucleic acid annealing; (c) a non strand-displacing DNA polymerase; and (d) a ligase, under conditions effective to join the two DNA molecules to form an intact double stranded DNA molecule, in which a single copy of the region of sequence identity is retained. The method allows the joining of a number of DNA fragments, in a predetermined order and orientation, without the use of restriction enzymes.
Owner:TELESIS BIO INC
Nanotube sensor devices for DNA detection
InactiveUS20070178477A1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA SolutionsSingle strand dna
A nanotube device is configured as an electronic sensor for a target DNA sequence. A film of nanotubes is deposited over electrodes on a substrate. A solution of single-strand DNA is prepared so as to be complementary to a target DNA sequence. The DNA solution is deposited over the electrodes, dried, and removed from the substrate except in a region between the electrodes. The resulting structure includes strands of the desired DNA sequence in direct contact with nanotubes between opposing electrodes, to form a sensor that is electrically responsive to the presence of target DNA strands. Alternative assay embodiments are described which employ linker groups to attach ssDNA probes to the nanotube sensor device.
Owner:NANOMIX
Transposon end compositions and methods for modifying nucleic acids
The present invention provides the use of transposases and transposon ends to generate extensive fragmentation and 5′-tagging of double-stranded target DNA in vitro, followed by the use of DNA polymerases to generate 5′- and 3′-tagged single DNA without PCR amplification reactions. Methods, compositions and kits for stranded DNA fragments, wherein the first label on the 5' end shows the sequence of the transferred transposon end and optionally an additional arbitrary sequence, and the second label on the 3' end shows the same The first tab shows a sequence different from the sequence. The method can be used to generate 5' and 3' tagged DNA fragments for use in a variety of processes including metagenomic analysis of DNA in environmental samples, copy number variation (CNV) analysis of DNA, and including massively parallel DNA sequencing (so-called "next generation sequencing") involves the process of comparative genome sequencing (CGS).
Owner:EPICENT TECH CORP
Field-effect device for the detection of small quantities of electric charge, such as those generated in bio-molecular process, bound in the vicinity of the surface
InactiveUS7535232B2Resistance/reactance/impedenceMicrobiological testing/measurementSingle strandEngineering
The device (1) allows the detection of small quantities of electrical charge (Qs) utilised for recognition of the gybridisation process of a single strand of DNA. It comprises a chip (2) in which is integrated an MOS device having a floating gate (7) a first portion (7a) of which extends in facing relation to a recess (8) formed in a surface of the chip (2) and accessible from outside the chip (2) and operable to retain an electrical charge (Qs) to be measured bound to it. A second portion (7b) of the gate (7) of the MOS device is coupled to a control electrode, (10) of the chip (2) by means of a capacitor (12) of predetermined value within the chip (2).
Owner:CONVERSANT INTPROP MANAGEMENT INC
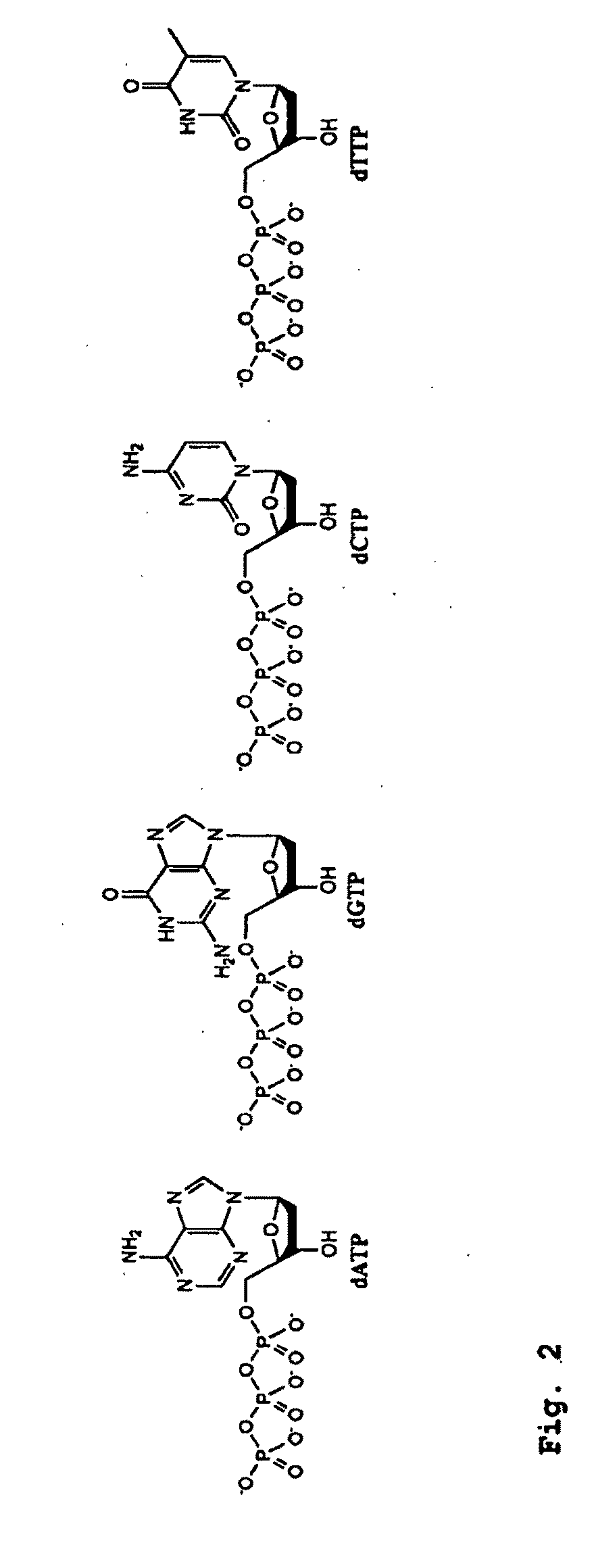
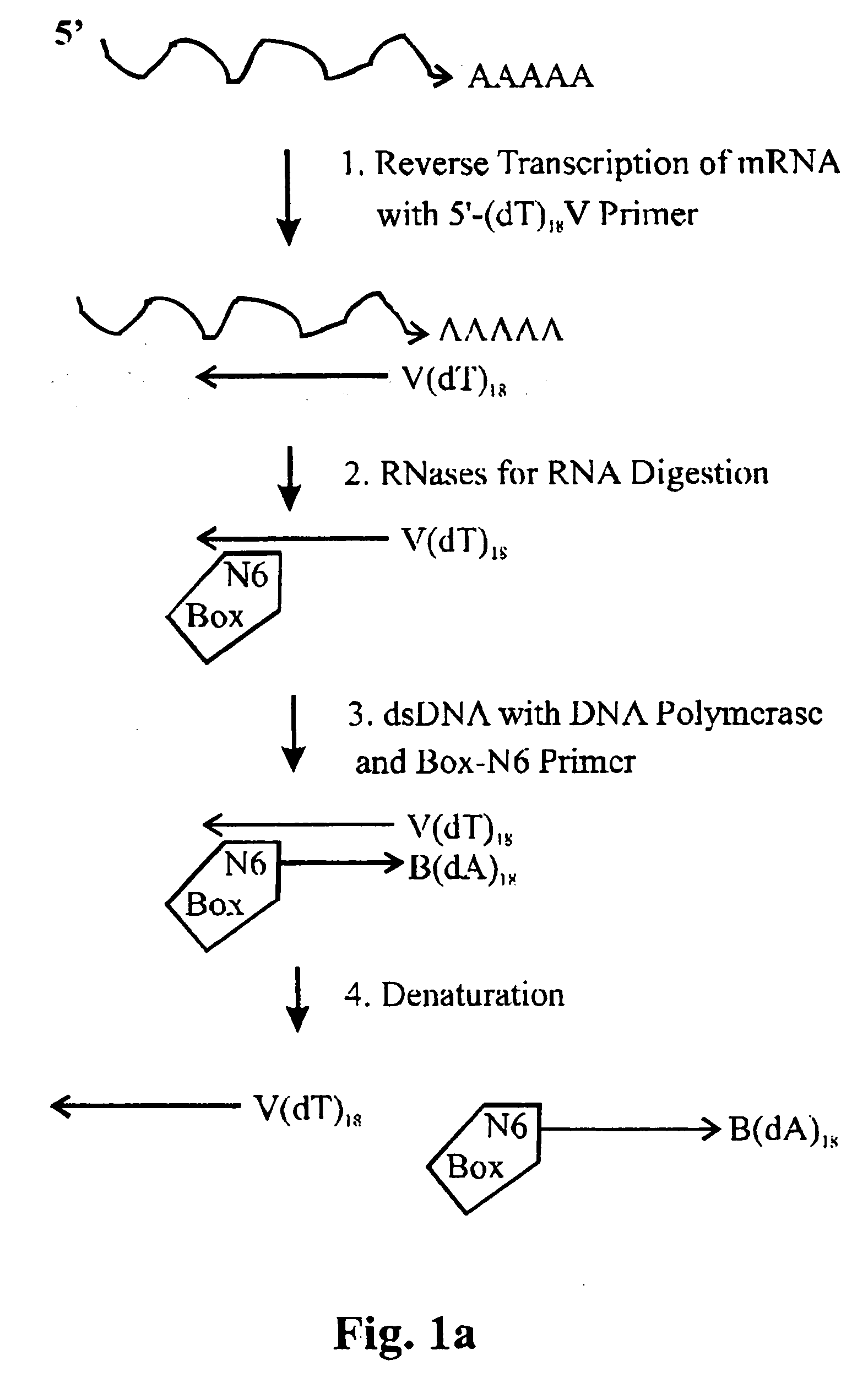
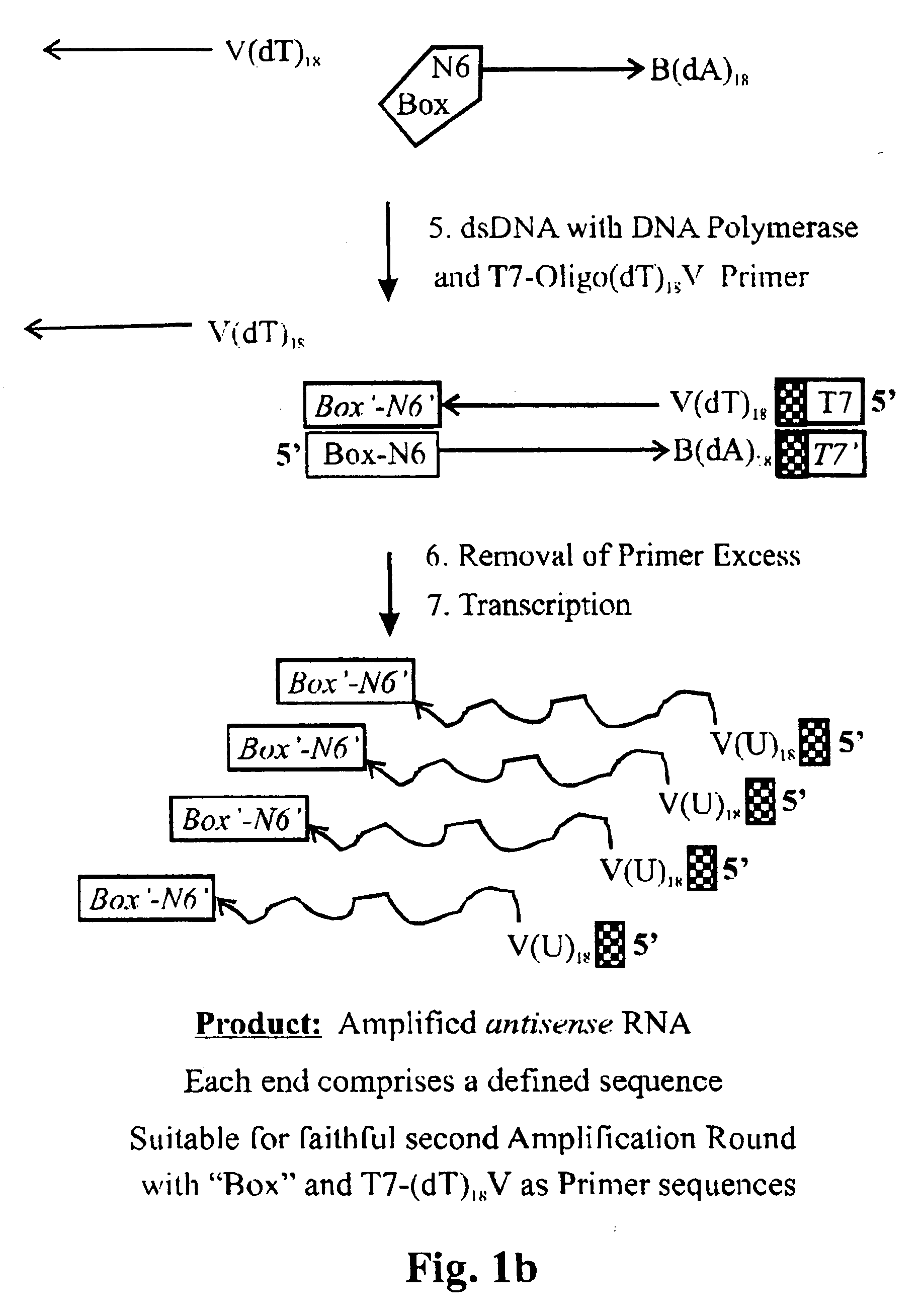
Amplification of ribonucleic acids
The invention relates to methods for the amplification of ribonucleic acids, comprising the following steps: (a) a single stranded DNA is produced from an RNA by means of reverse transcription, using a single-stranded primer having a defined sequence, an RNA-dependent DNA polymerase and deoxyribonucleoside triphosphates; (b) the template RNA is removed; (c) a DNA duplex is produced by means of a single-stranded primer comprising a box sequence, a DNA polymerase and deoxyribonucleoside triphosphates; (d) the duplex is separated into single-stranded DNAs; (e) DNA duplexes are produced from one of the single-stranded DNAs obtained in step (d) by means of a single-stranded primer comprising a promoter sequence at its 5′end and the same defined sequence as the primer used in step (a) at its 3′end, a DNA polymerase and deoxyribonucleoside triphosphates; (f) a plurality of RNA single strands, both ends of which comprise defined sequences, are produced by means of an RNA polymerase and ribonucleoside triphosphates. The invention also relates to kits for amplifying ribonucleic acids according to one of said methods, said kits comprising the following components: (a) at least at least one single-stranded primer, which contains a promoter sequence; (b) at least one single-stranded primer comprising a box sequence; (c) an RNA-dependent DNA polymerase; (d) deoxyribonucleoside triphosphates; (e) a DNA-dependent DNA polymerase; (f) an RNA polymerase; and (g) ribonucleoside triphosphates.
Owner:AMPTEC
Detection And Quantification Of Biomarkers Via A Piezoelectric Cantilever Sensor
InactiveUS20090078023A1High sensitivityShort timeMaterial analysis using sonic/ultrasonic/infrasonic wavesMicrobiological testing/measurementEscherichia coliMultiple sensor
Quantification of a target analyte is performed using a single sample to which amounts of the target analyte are added. Calibration is performed as part of quantification on the same sample. The target analyte is detectable and quantifiable using label free reagents and requiring no sample preparation. Target analytes include biomarkers such as cancer biomarkers, pathogenic Escherichia coli, single stranded DNA, and staphylococcal enterotoxin. The quantification process includes determining a sensor response of a sensor exposed to the sample and configured to detect the target analyte. Sensor responses are determined after sequential additions of the target analyte to the sample. The amount of target analyte detected by the sensor when first exposed to the sample is determined in accordance with the multiple sensor responses.
Owner:DREXEL UNIV
Method for Increasing the Ratio of Homologous to Non-Homologous Recombination
InactiveUS20080194029A1FermentationVector-based foreign material introductionNucleotideSingle-Stranded DNA Binding Proteins
Gene targeting allows the deletion (knock out), the repair (rescuing) and the modification (gene mutation) of a selected gene and the functional analysis of any gene of interest. Targeting of nuclear genes has been a very inefficient process in most eukaryotes including plants and animals due to the dominance of illegitimate integration of the applied DNA into non-homologous regions of the genome. The present invention provides a method for increasing the ratio of homologous to non-homologous recombination of a polynucleotide into a host cell's DNA by suppressing non-homologous recombination. Surprisingly, the number of non-homologous recombination events can be reduced if the polynucleotide is applied as a purified single-stranded DNA, preferably coated with a single strand binding protein.
Owner:HEGEMANN PETER +1
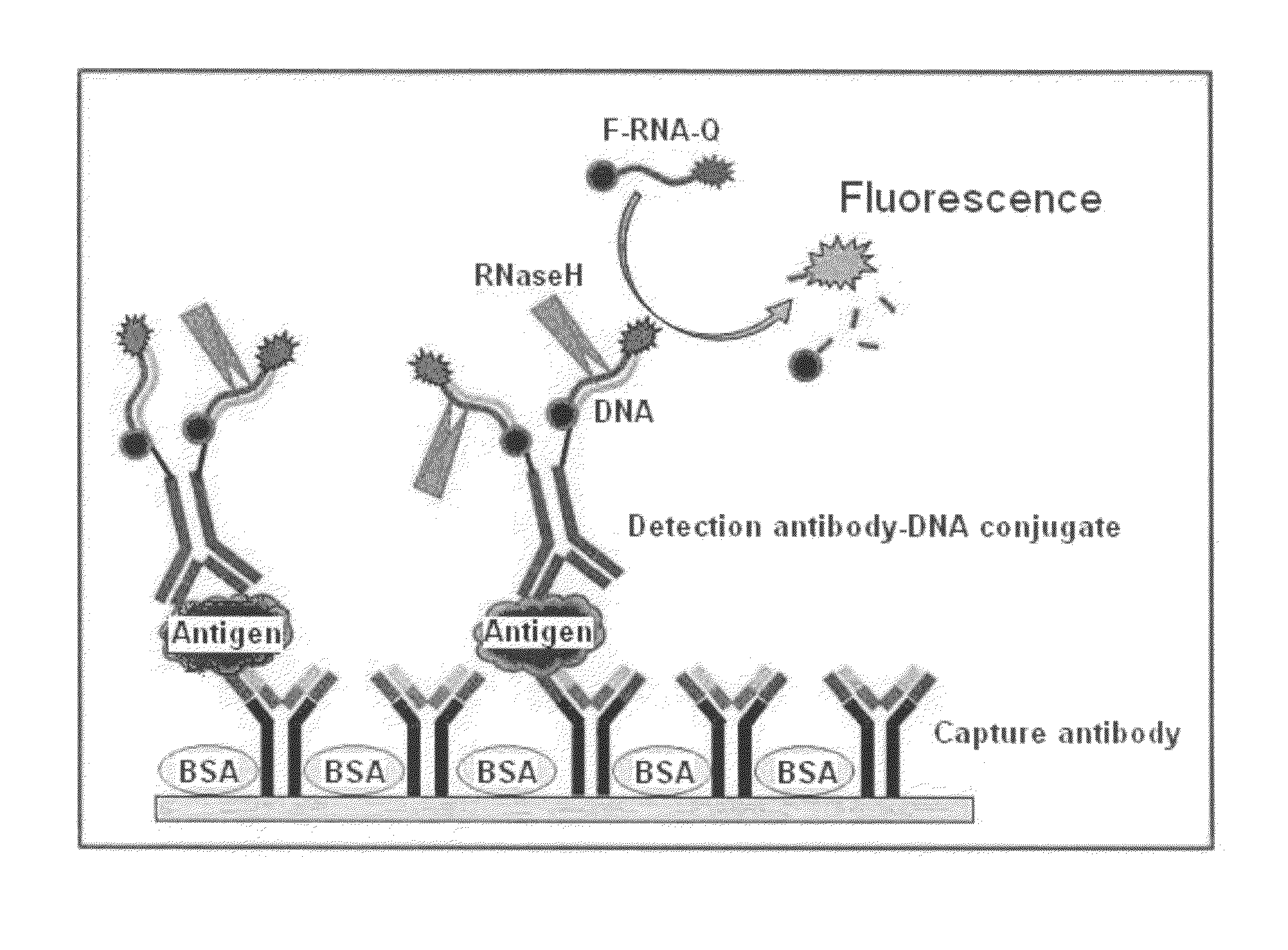
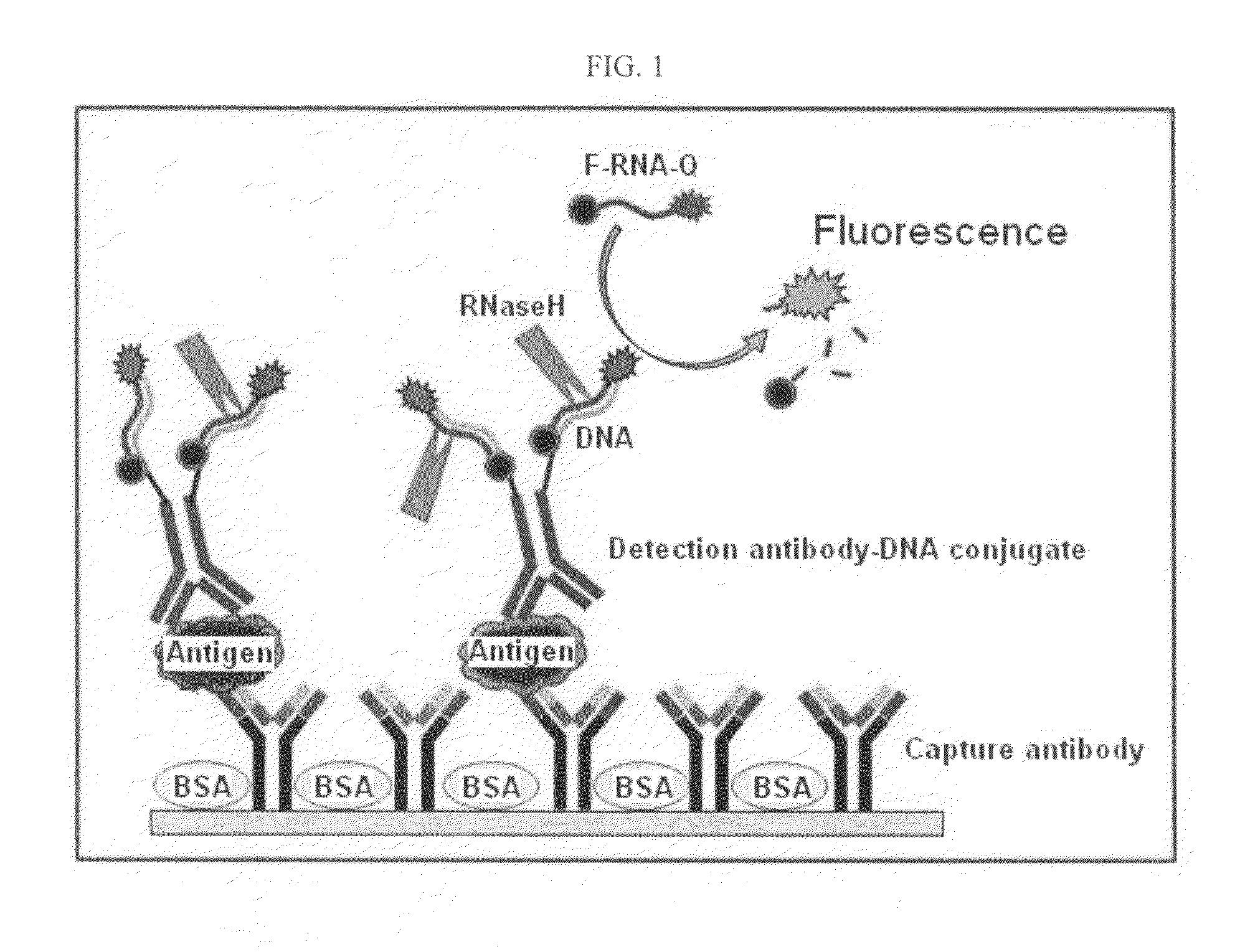
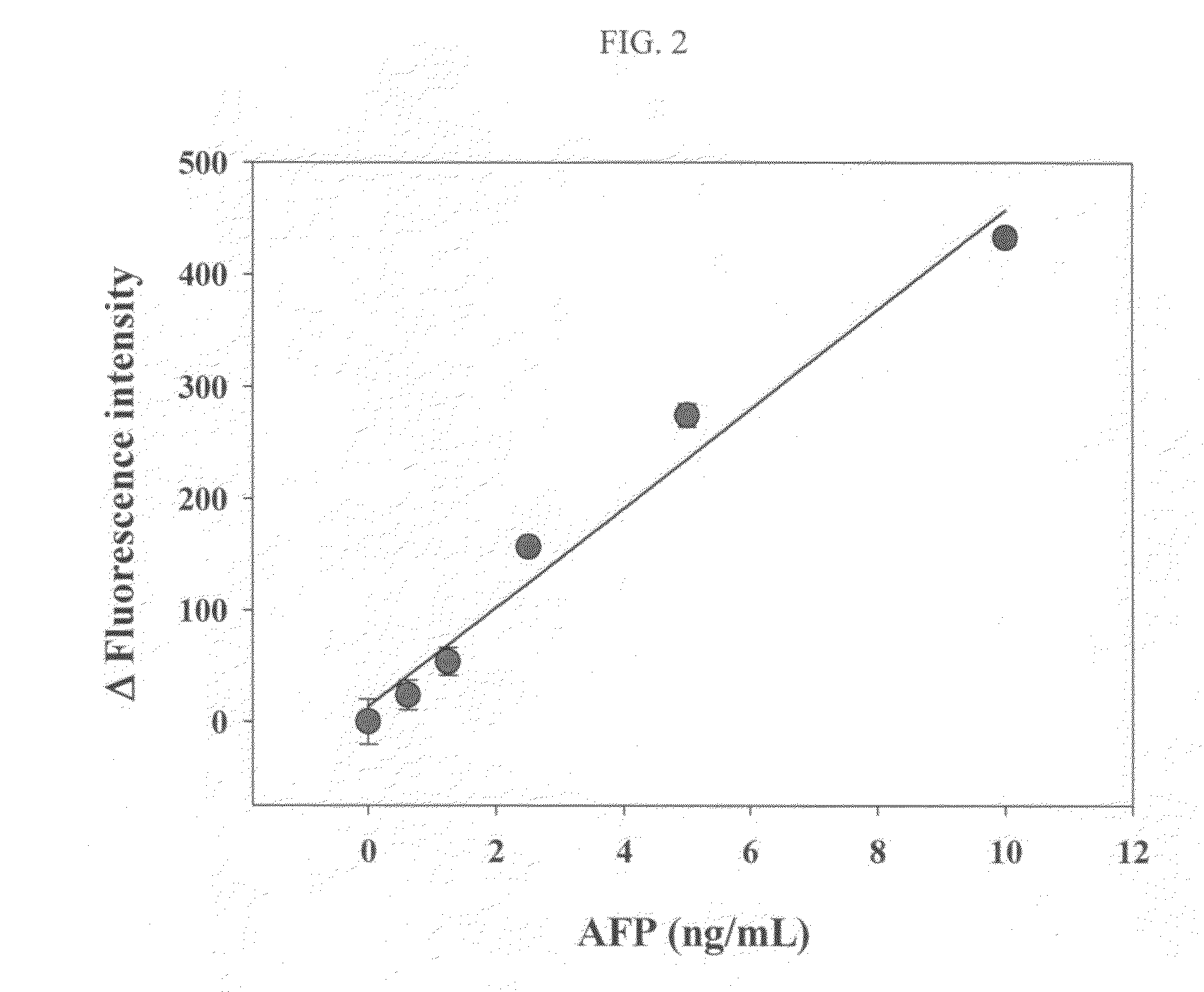
Antigen Detection Kit and Method
InactiveUS20110027772A1Bioreactor/fermenter combinationsBiological substance pretreatmentsSingle-Stranded RNAAntigen
An antigen detection kit and an antigen detection method using the same are provided. The antigen detection kit comprises a capture antibody, a detection antibody bound to a single stranded DNA oligonucleotide, a single stranded RNA oligonucleotide complementary sequence to the DNA oligonucleotide, and an RNase.
Owner:KOREA INST OF SCI & TECH
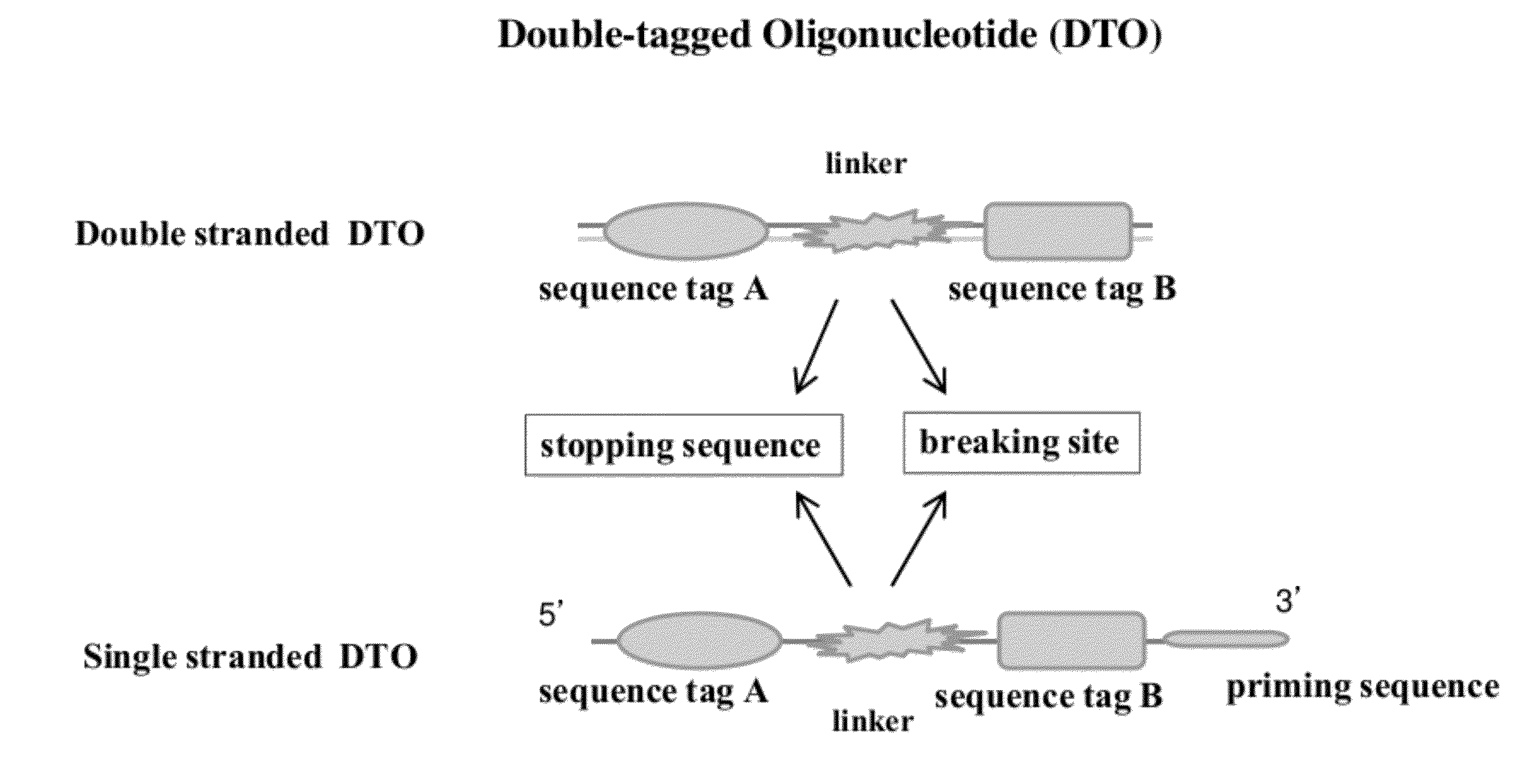
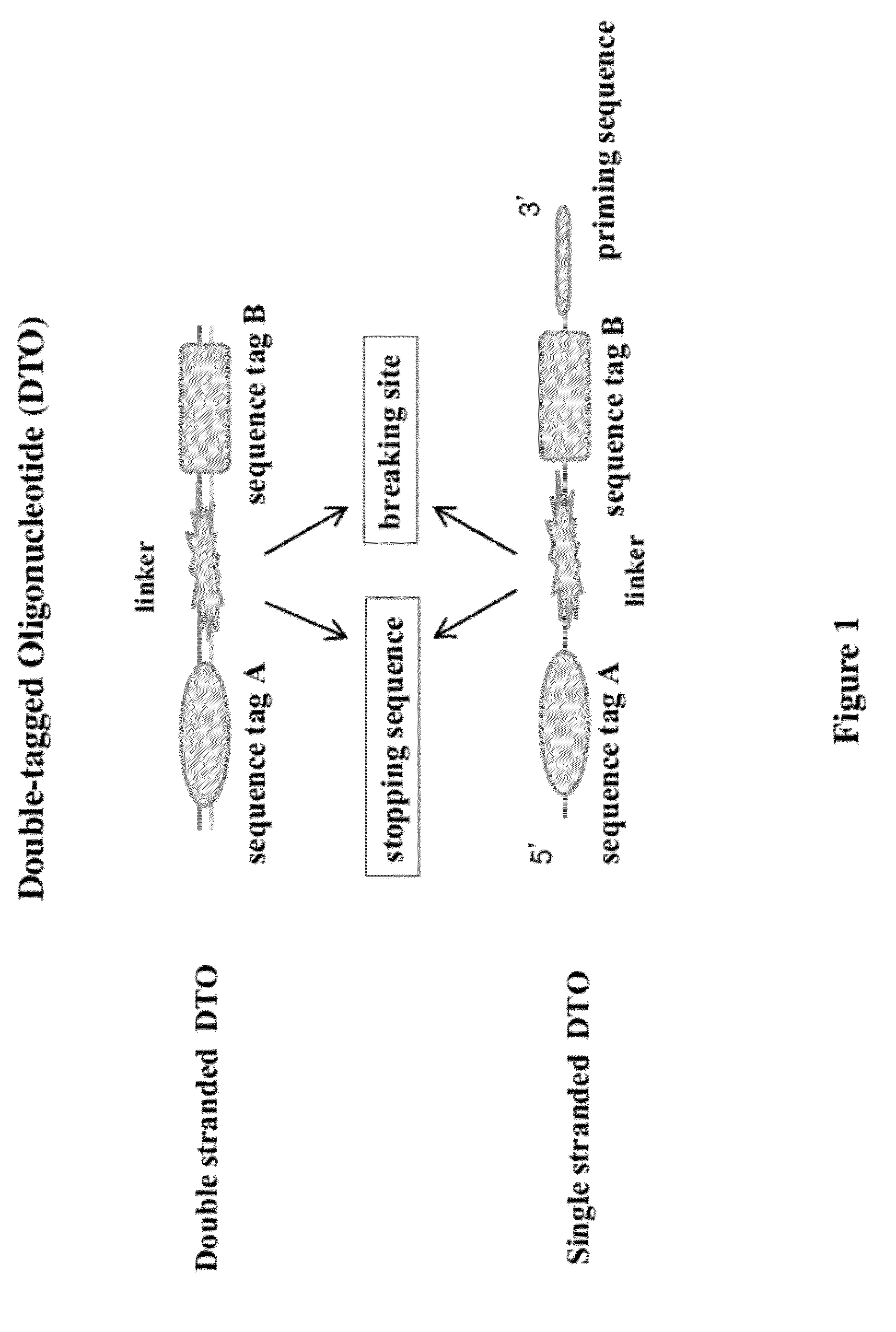
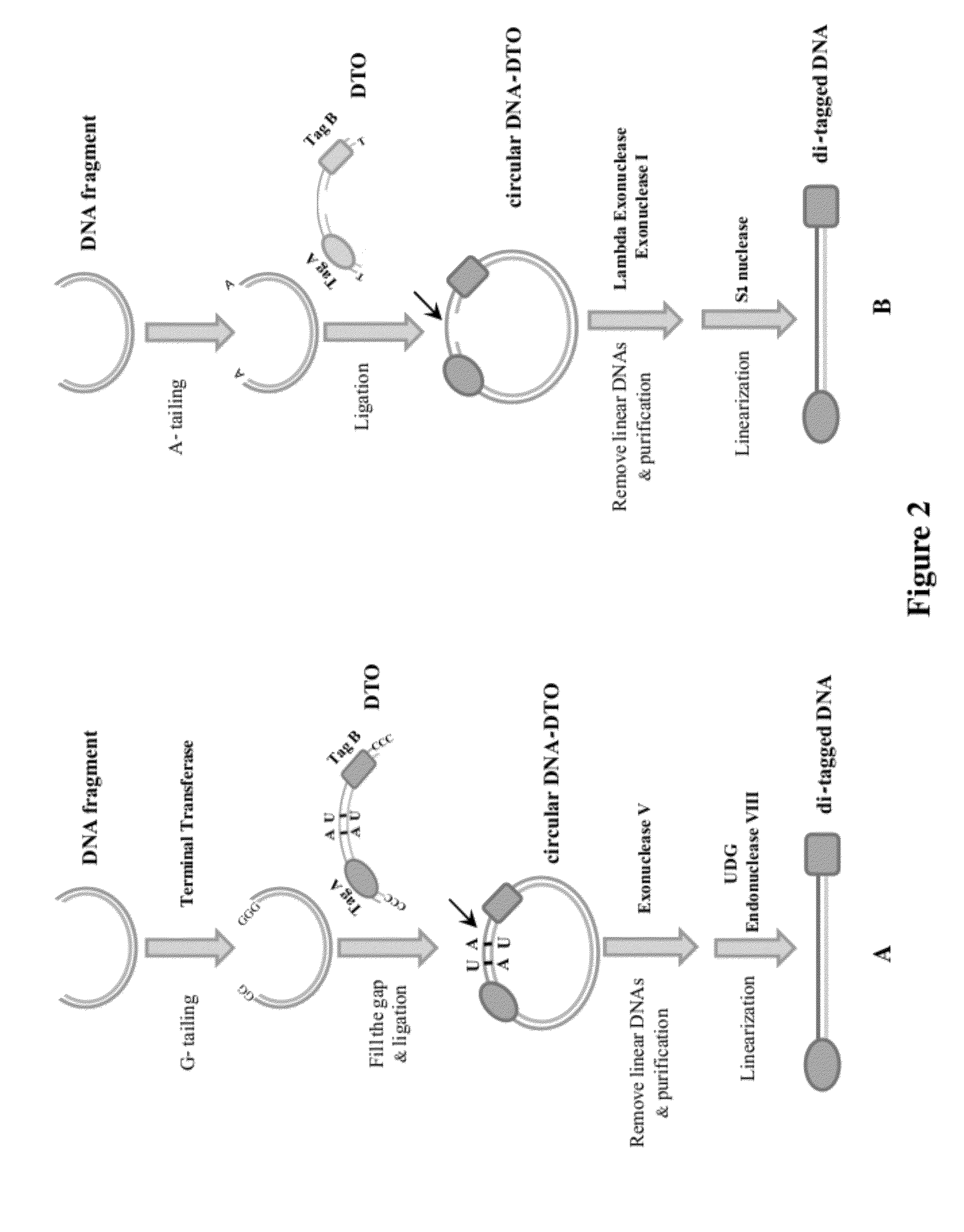
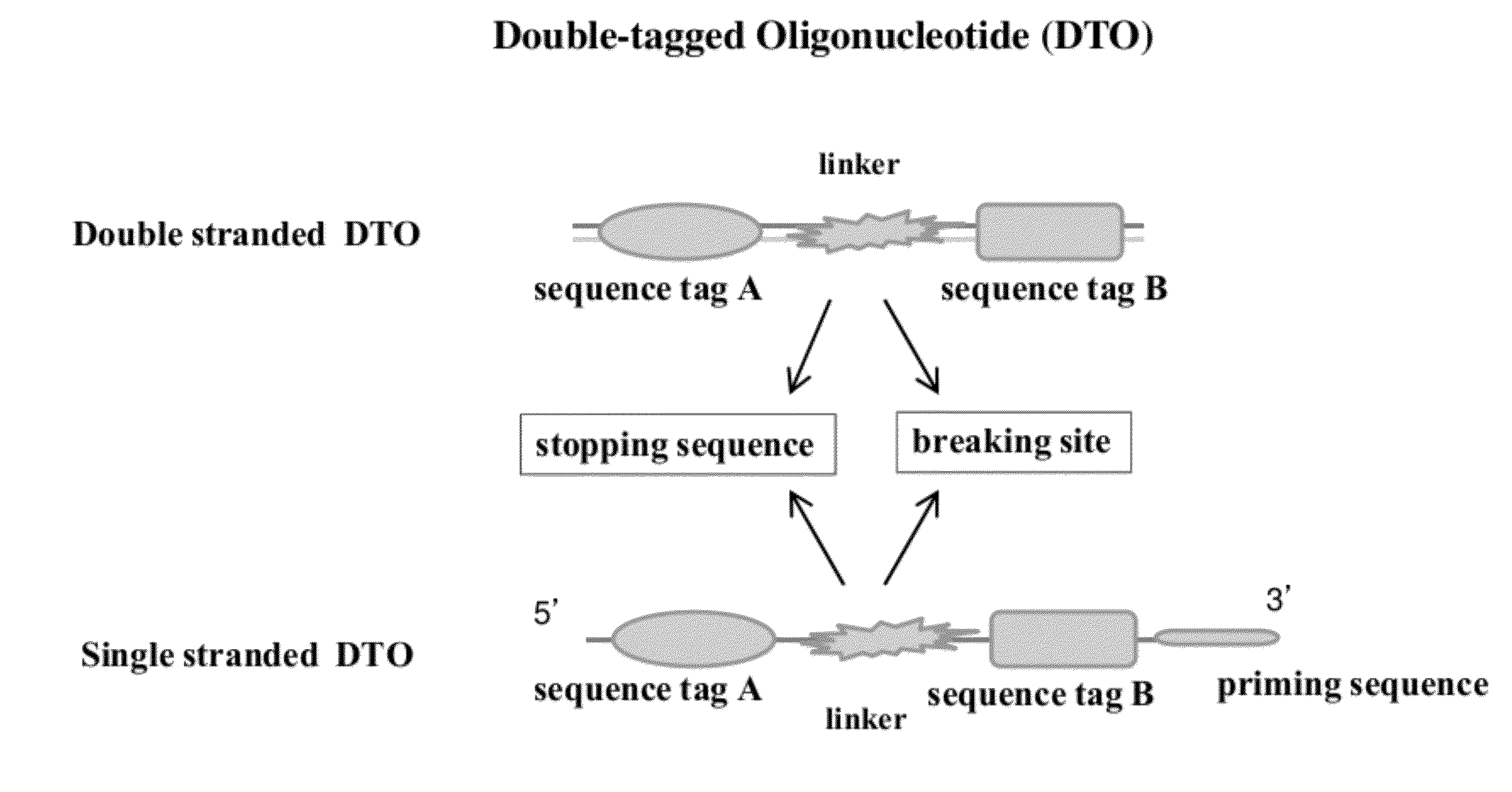
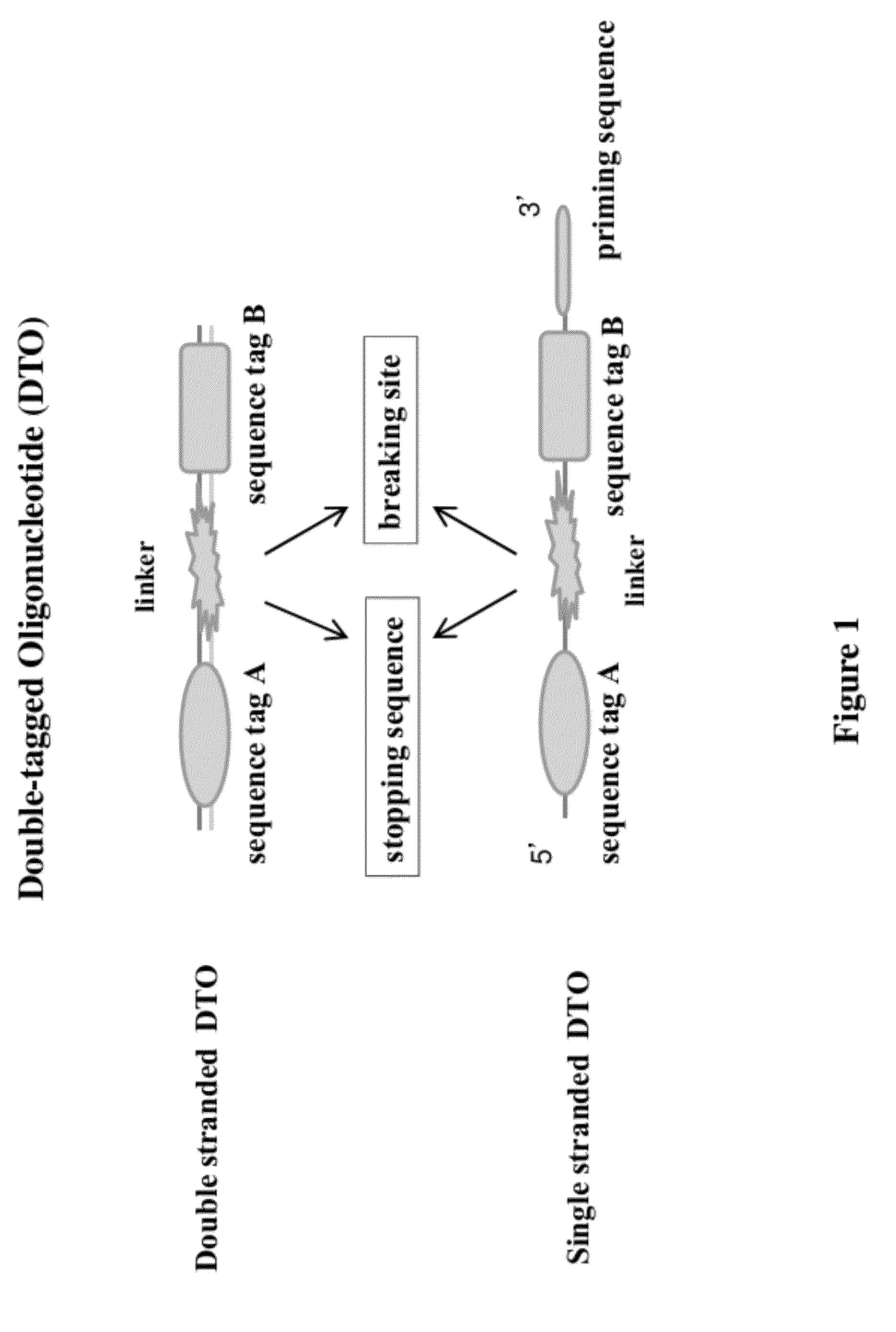
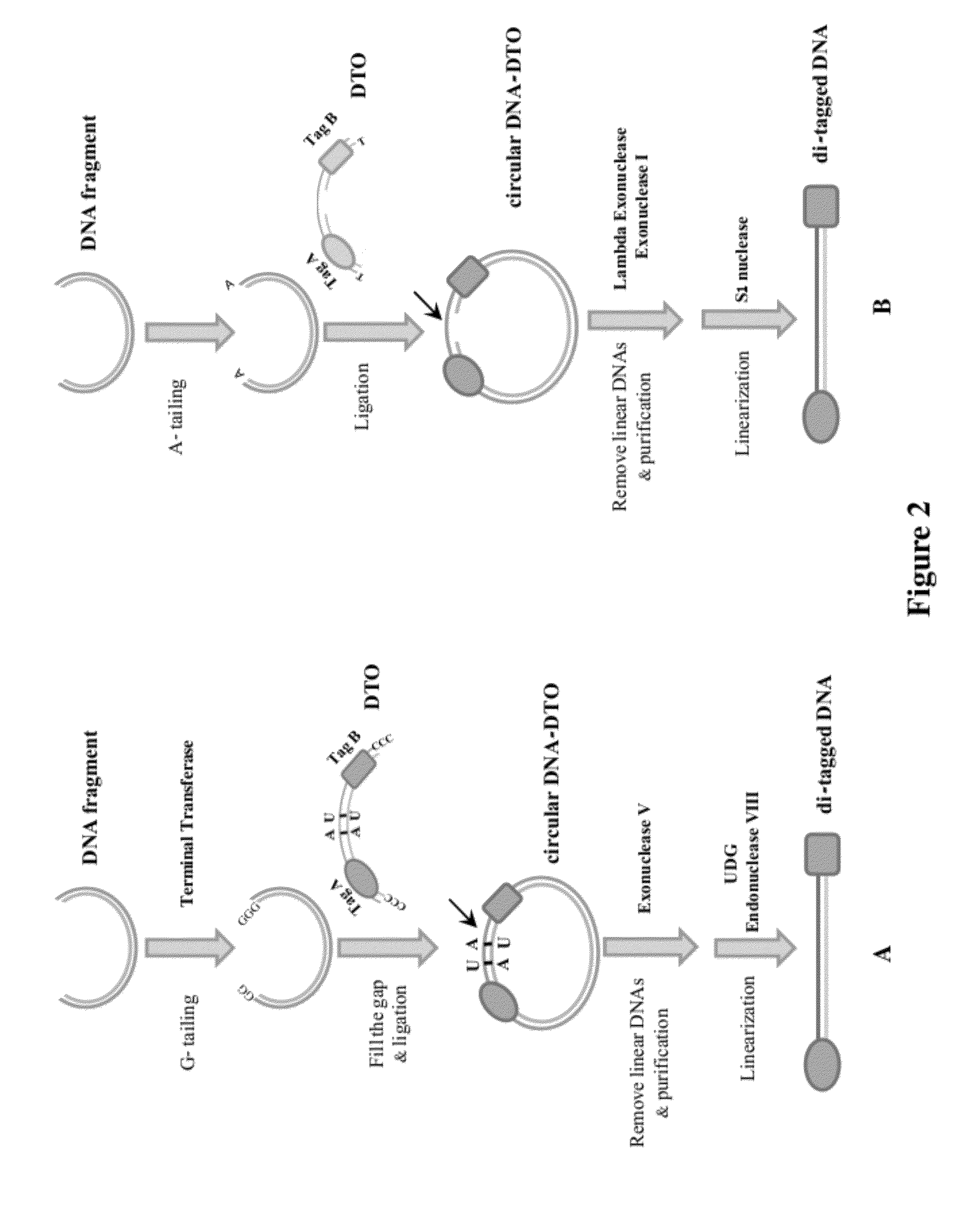
Methods of making di-tagged DNA libraries from DNA or RNA using double-tagged oligonucleotides
ActiveUS8722585B2Easy to separateSimple and highly efficientMicrobiological testing/measurementFermentationNucleotideSingle strand
Disclosed are methods, compositions and kits related to making double-tagged DNA libraries from RNA / DNA samples. A double-tagged oligonucleotide (DTO) is employed to efficiently add two different tags to ends of DNAs to make a double-tagged DNA libraries. Also disclosed are methods to make mate pair libraries using the double-tagged oligonucleotide, and methods to make double-tagged single stranded DNA. The double-tagged DNA libraries of the invention are ready to be used on next generation sequencing machines.
Owner:GNOMEGEN
Nano-pore electric sensor
ActiveCN101694474ASolve the technical difficulties of integrating in nanoporesSimple methodMicrobiological testing/measurementMaterial analysis by electric/magnetic meansImage resolutionSingle strand dna
The invention discloses a nano-pore electric sensor. The nano-pore electric sensor comprises a baseplate, first insulating layers, symmetrical electrodes, electric contact layers, second insulating layers and nano-pores. The first insulating layers and the symmetrical electrodes are sequentially arranged on the baseplate; the electric contact layers are arranged on the first insulating layers and the edges of the symmetrical electrodes; the second insulating layers are arranged on the symmetrical electrodes; and the nano-pores are arranged in the centers of the baseplate, the first insulating layers, the symmetrical electrodes and the second insulating layers. The thickness of each nano-electrode can be controlled within the range from 0.35 to 0.7nm to meet the requirement on resolution for detecting the electric character of a single base group in single-chain DNA, thereby being suitable for the low-cost and fast electronic gene sequence test. The nano-pore electric sensor solves the technical problem for integrating the nano-electrodes in the nano-pores, and the method for preparing the nano-electrodes is simple.
Owner:ZHEJIANG UNIV
Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
Methods of Making Di-Tagged DNA Libraries from DNA or RNA Using Double-tagged Oligonucleotides
ActiveUS20120283145A1Avoid it happening againEasy to separateMicrobiological testing/measurementFermentationCDNA librarySingle strand dna
Disclosed are methods, compositions and kits related to making double-tagged DNA libraries from RNA / DNA samples. A double-tagged oligonucleotide (DTO) is employed to efficiently add two different tags to ends of DNAs to make a double-tagged DNA libraries. Also disclosed are methods to make mate pair libraries using the double-tagged oligonucleotide, and methods to make double-tagged single stranded DNA. The double-tagged DNA libraries of the invention are ready to be used on next generation sequencing machines.
Owner:GNOMEGEN
Mobile rapid test system for nucleic acid analysis
InactiveUS20110039261A1Easy to operateRapid diagnostic informationBioreactor/fermenter combinationsBiological substance pretreatmentsSide chainRechargeable cell
A mobile rapid test system for nucleic acid analysis. A method comprising the steps of amplification of the nucleic acids by means of rapid-PCR technology, conversion of a double-stranded amplification product into a single-stranded DNA fragment, hybridization with a labeled probe and detection of the nucleic acids on a lateral-flow test strip. A device comprising a reaction cavity which preferably consists of a thin film, inlet and outlet openings for the reaction cavity, one or more heatable sample blocks which are connected to miniaturized cooling bodies and a window for reading off the result. The lateral-flow test strip is a component of the mobile rapid test system. Operation of the instrument system requires no external power source, but only batteries or a rechargeable battery.
Owner:AJ INNUSCREEN GMBH
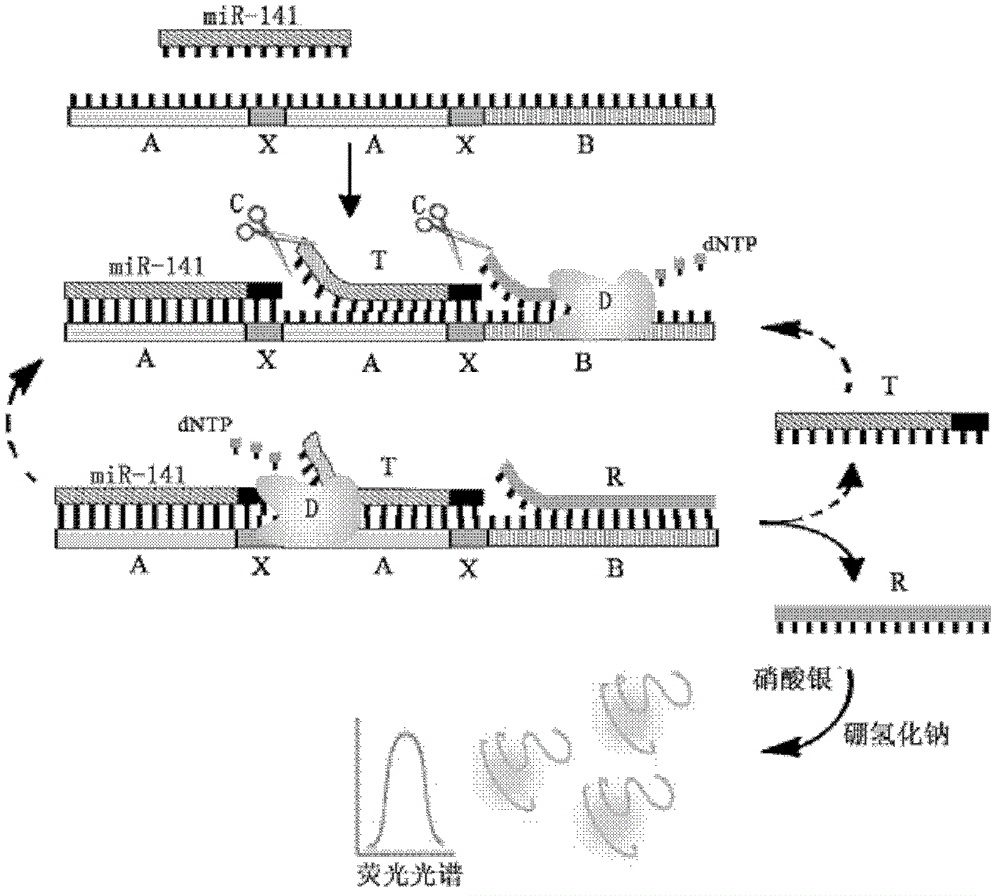
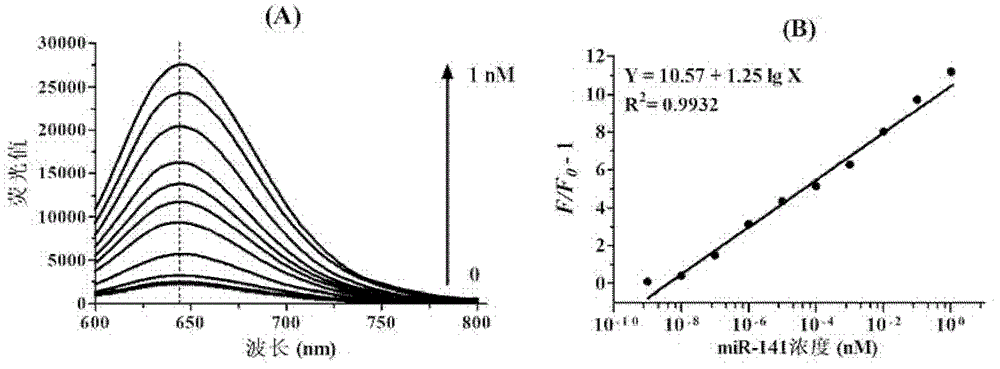
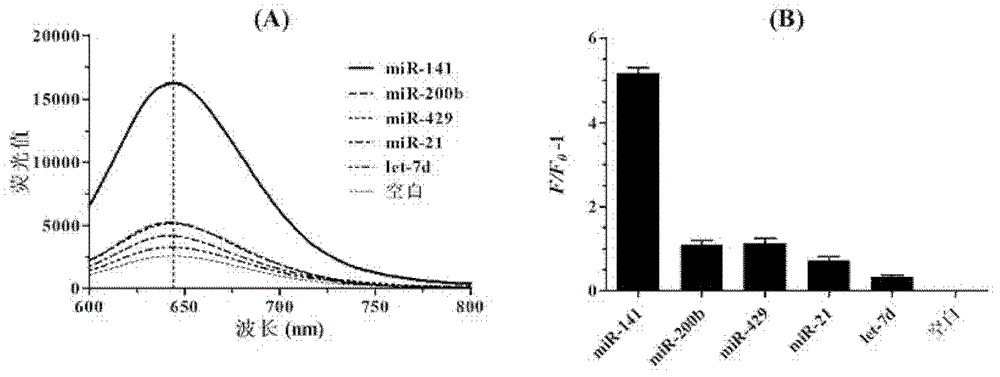
MicroRNA quantitative detection analytic method by utilizing isothermal amplification to synthesize fluorescent nano silver cluster probe
The invention provides a microRNA quantitative detection analytic method by utilizing isothermal amplification to synthesize a fluorescent nano silver cluster probe. The method is as below: a DNA amplification template containing three kinds of functional sequences is designed: a sequence binding with a target microRNA, a nicking endonuclease enzyme sequence and a DNA complementary sequence for synthesis of fluorescent nano cluster; when the target microRNA and the DNA amplification template are combined, isothermal amplification reaction and specific enzyme reaction of nicking endonuclease are induced to obtain a large number of single-stranded DNA products; the DNA sequence for synthesis of fluorescent nano silver cluster and a silver nitrate solution are employed to prepare the fluorescent nano silver cluster probe under the reduction of sodium borohydride; fluorescence signal of the reaction system are determined, and a fluorescence change value is calculated; the value is compared with a standard working curve to calculate the concentration of the target microRNA. The method has the advantages of high sensitivity, strong specificity, wide linear detection range, low background signal and simple operation, and can be widely applied to microRNA detection of biological samples such as tissue, blood or cells.
Owner:EAST CHINA UNIV OF SCI & TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com