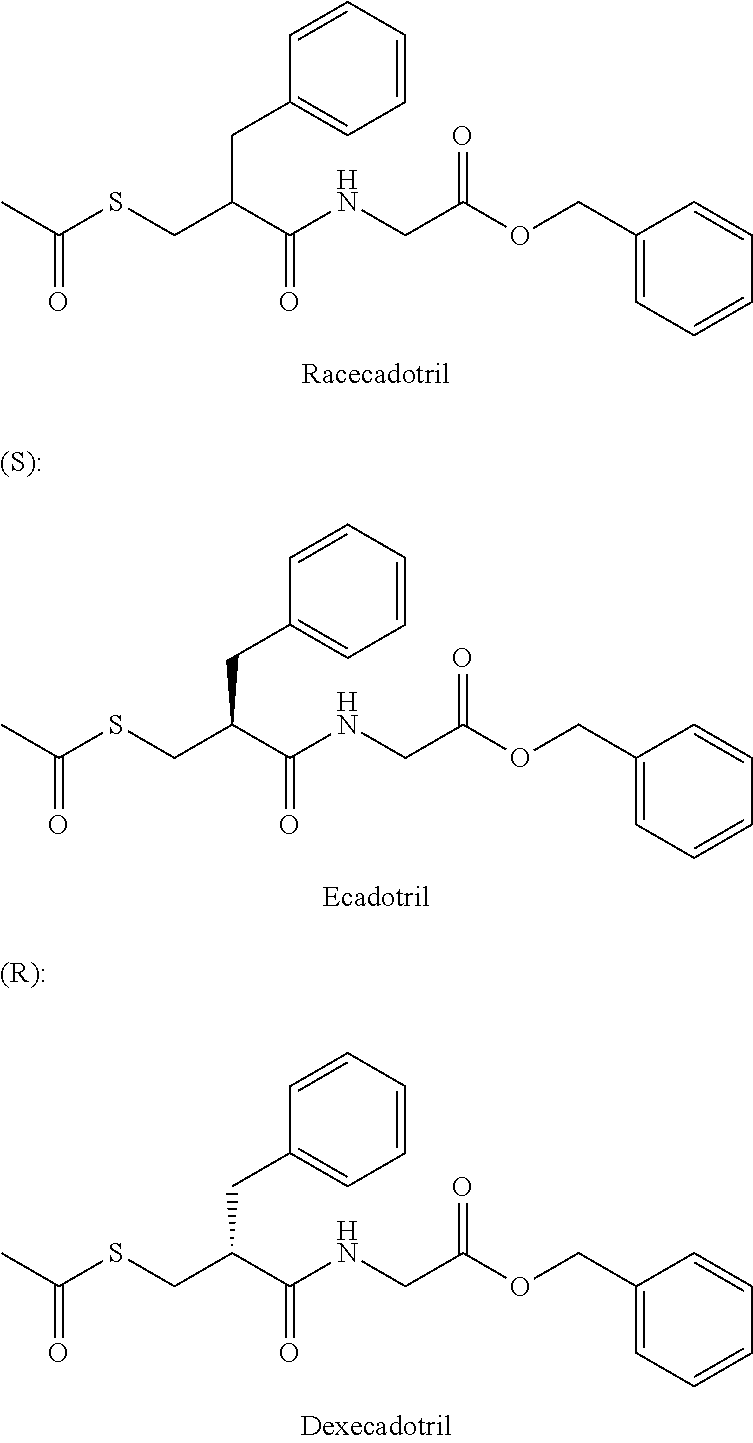
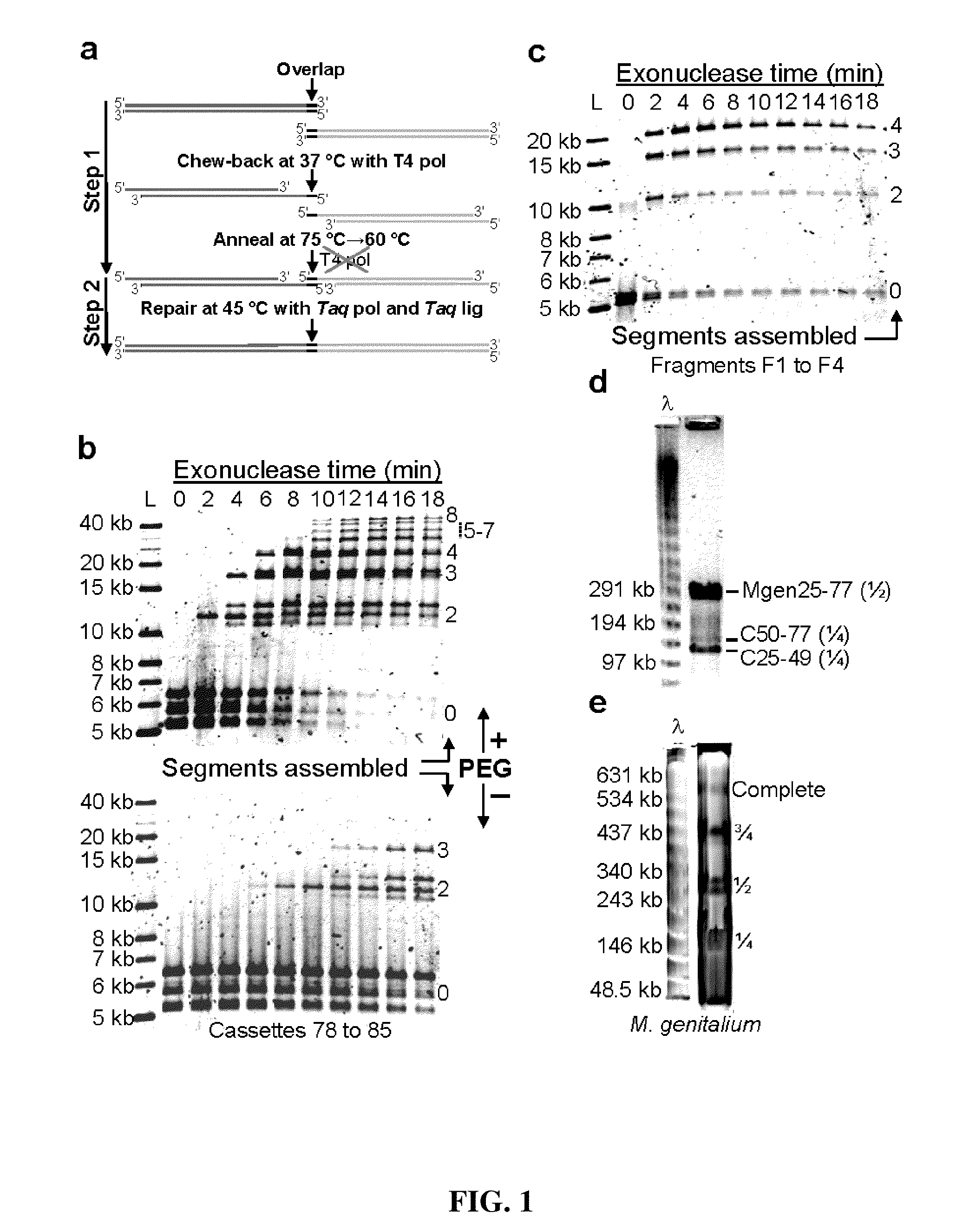
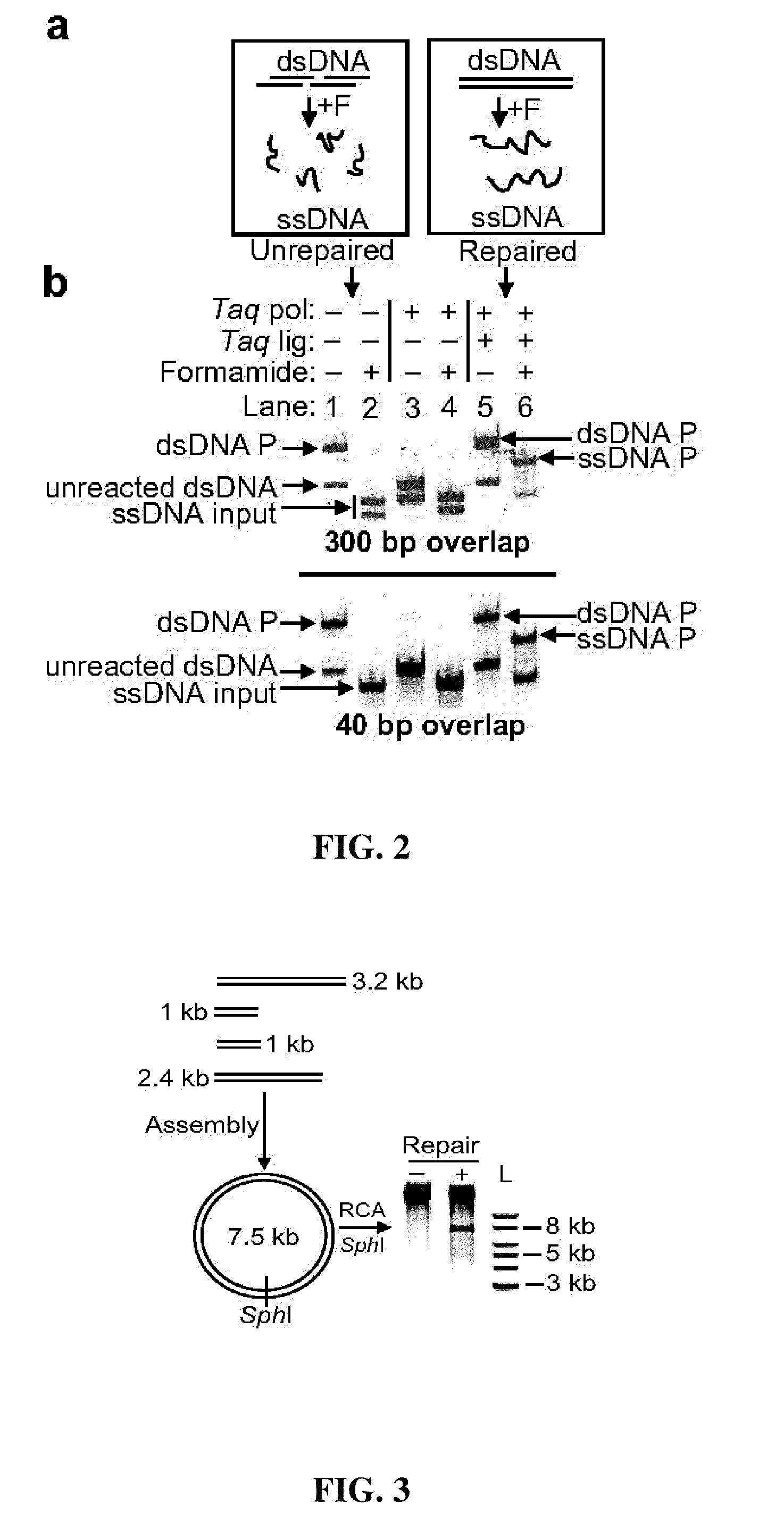
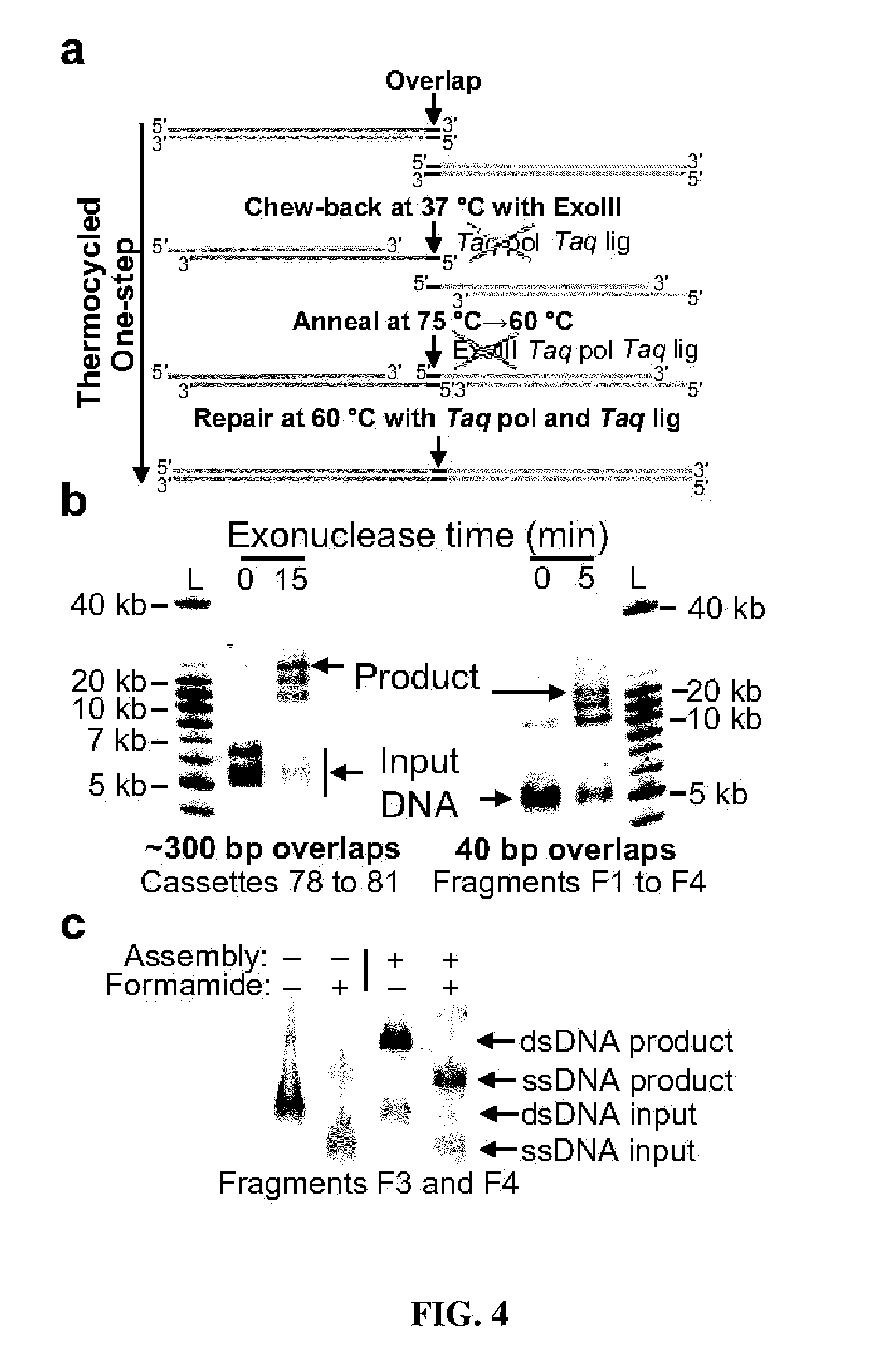
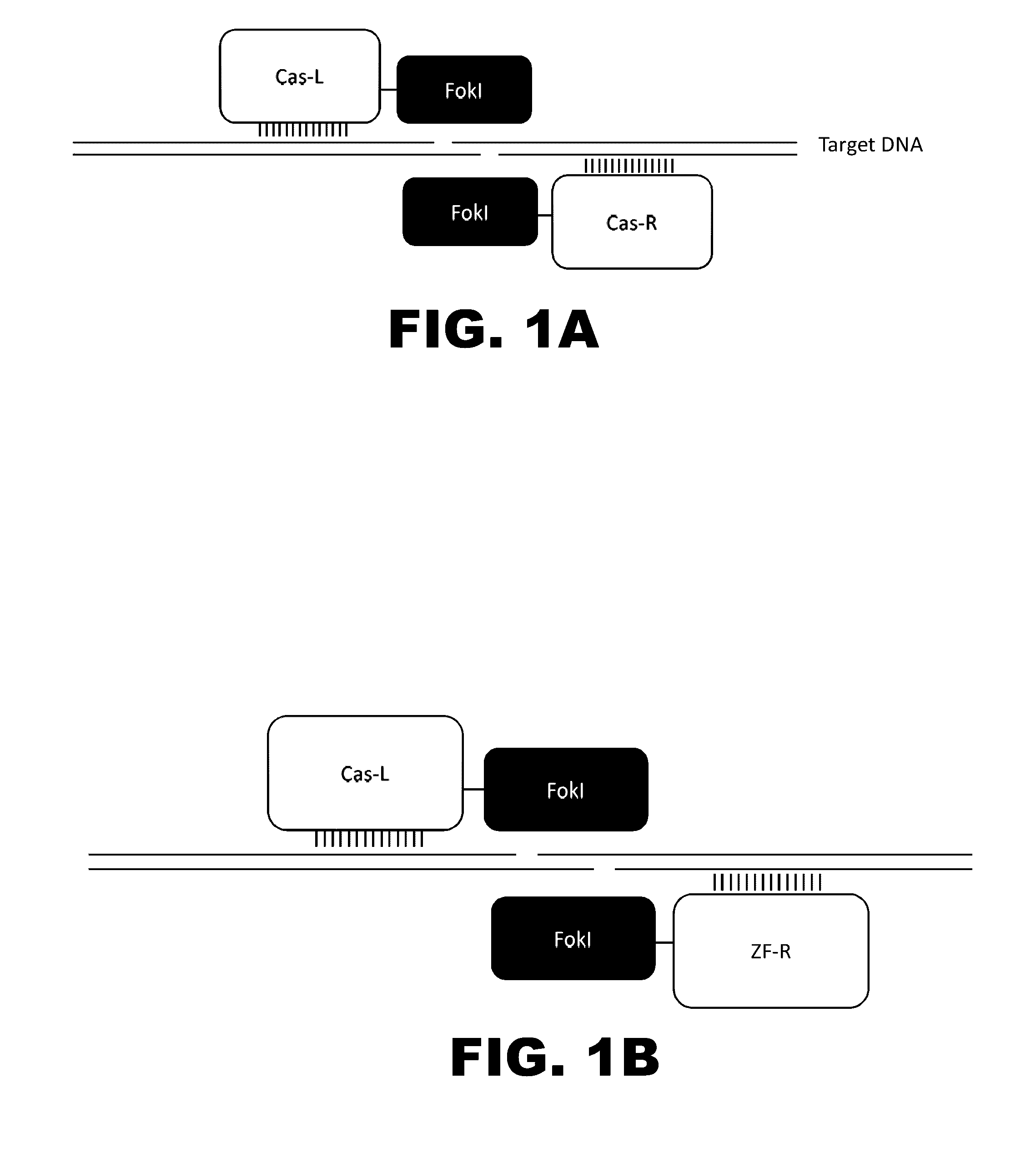
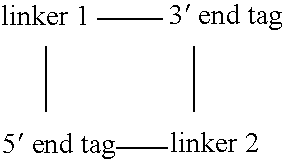Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
1376 results about "Endonuclease" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
Methods for producing a paired tag from a nucleic acid sequence and methods of use thereof
InactiveUS20060024681A1Microbiological testing/measurementFermentationSmaI restriction endonucleaseEcoRI
Methods for producing a paired tag from a nucleic acid sequence are provided in which the paired tag comprises the 5′ end tag and 3′ end tag of the nucleic acid sequence. In one embodiment, the nucleic acid sequence comprises two restriction endonuclease recognition sites specific for a restriction endonuclease that cleaves the nucleic acid sequence distally to the restriction endonuclease recognition sites. In another embodiment, the nucleic acid sequence further comprises restriction endonuclease recognition sites specific for a rare cutting restriction endonuclease. Methods of using paired tags are also provided. In one embodiment, paired tags are used to characterize a nucleic acid sequence. In a particular embodiment, the nucleic acid sequence is a genome. In one embodiment, the characterization of a nucleic acid sequence is karyotyping. Alternatively, in another embodiment, the characterization of a nucleic acid sequence is mapping of the sequence. In a further embodiment, a method is provided for identifying nucleic acid sequences that encode at least two interacting proteins.
Owner:APPL BIOSYSTEMS INC
Modified nucleotide compounds
ActiveUS7495088B1Increased nuclease resistanceSugar derivativesMicrobiological testing/measurementBiologyModified nucleosides
Disclosed is a nuclease resistant nucleotide compound capable of hybridizing with a complementary RNA in a manner which inhibits the function thereof, which modified nucleotide compound includes at least one component selected from the group consisting of MN3M, B(N)xM and M(N)xB wherein N is a phosphodiester-linked modified 2′-deoxynucleoside moiety; M is a moiety that confers endonuclease resistance on said component and that contains at least one modified or unmodified nucleic acid base; B is a moiety that confers exonuclease resistance to the terminus to which it is attached; and x is an integer of at least 2.
Owner:ENZO BIOCHEM
Method for producing complex DNA methylation fingerprints
InactiveUS6214556B1Accurately determineSugar derivativesMicrobiological testing/measurementFingerprintGenomic DNA
Method for characterizing, classifying and differentiating tissues and cell types, for predicting the behavior of tissues and groups of cells, and for identifying genes with changed expression. The method involves obtaining genomic DNA from a tissue sample, the genomic DNA subsequently being subjected to shearing, cleaved by means of a restriction endonuclease or not treated by either one of these methods. The base cytosine, but not 5-methylcytosine, from the thus-obtained genomic DNA is then converted into uracil by treatment with a bisulfite solution. Fractions of the thus-treated genomic DNA are then amplified using either very short or degenerated oligonucleotides or oligonuclcotides which are complementary to adaptor oligonucleotides that have been ligated to the ends of the cleaved DNA. The quantity of the remaining cytosines on the guanine-rich DNA strand and / or the quantity of guanines on the cytosine-rich DNA strand from the amplified fractions are then detected by hybridization or polymerase reaction, which quantities are such that the data generated thereby and automatically applied to a processing algorithm allow the drawing of conclusions concerning the phenotype of the sample material.
Owner:EPIGENOMICS AG
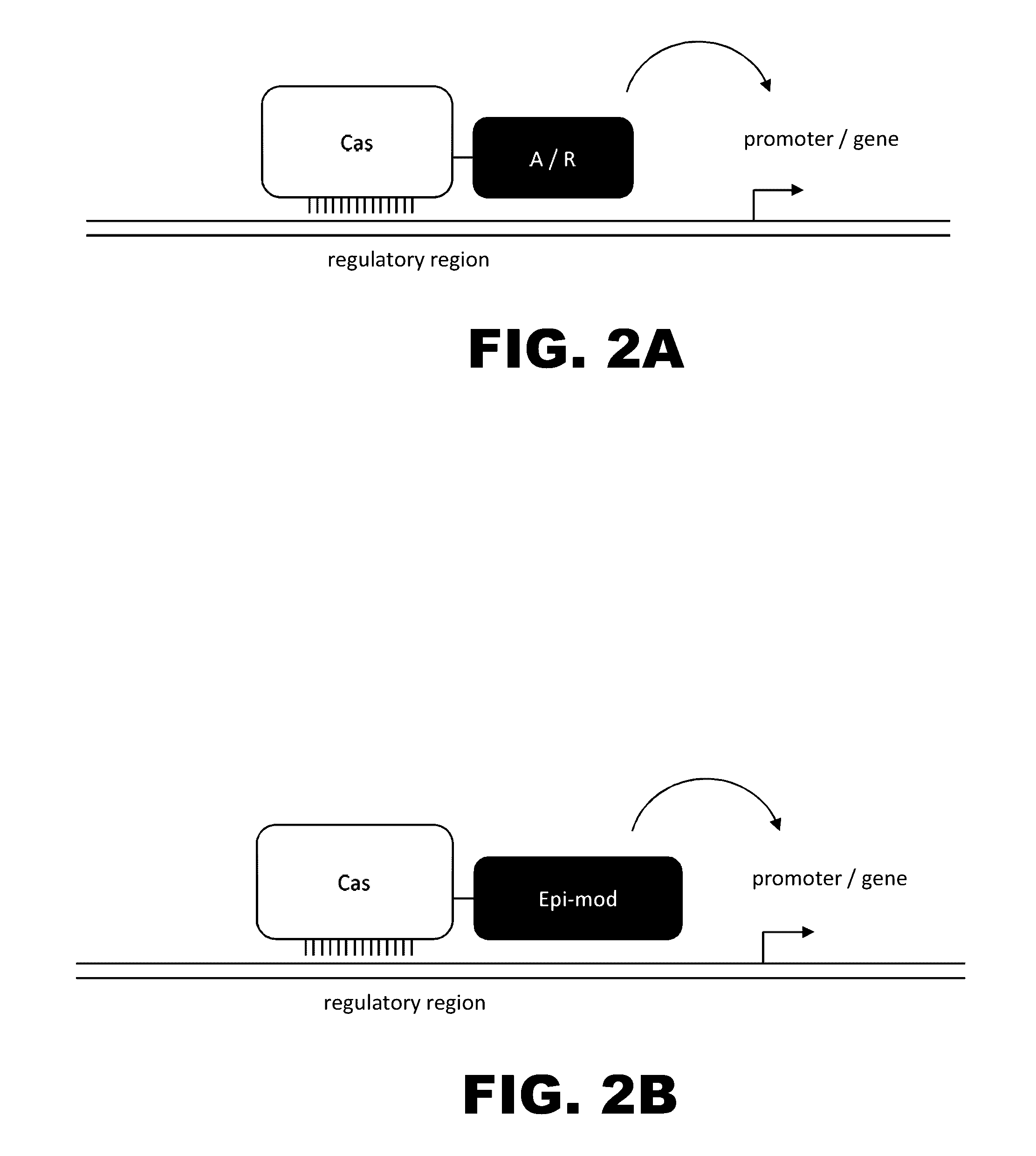
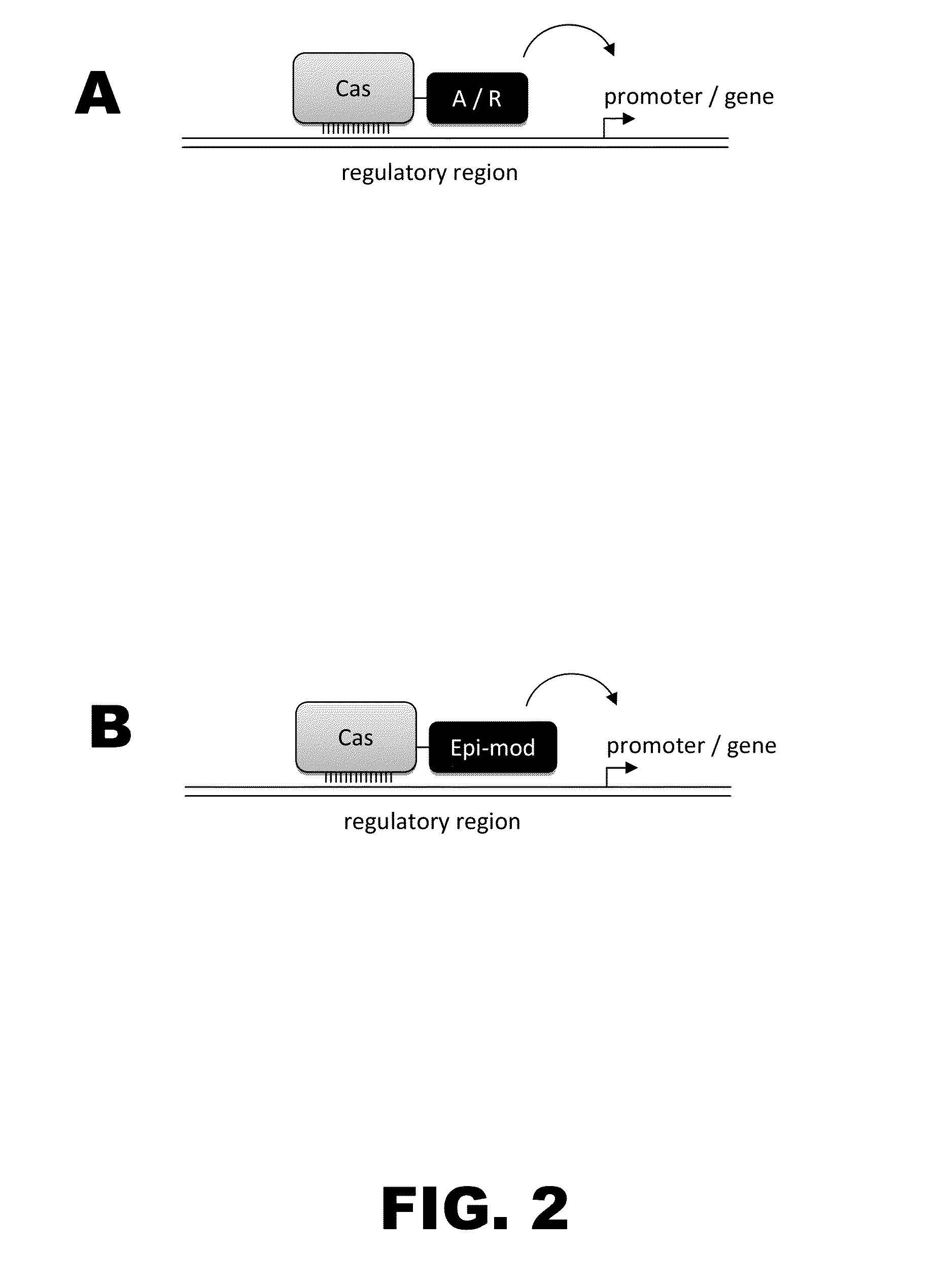
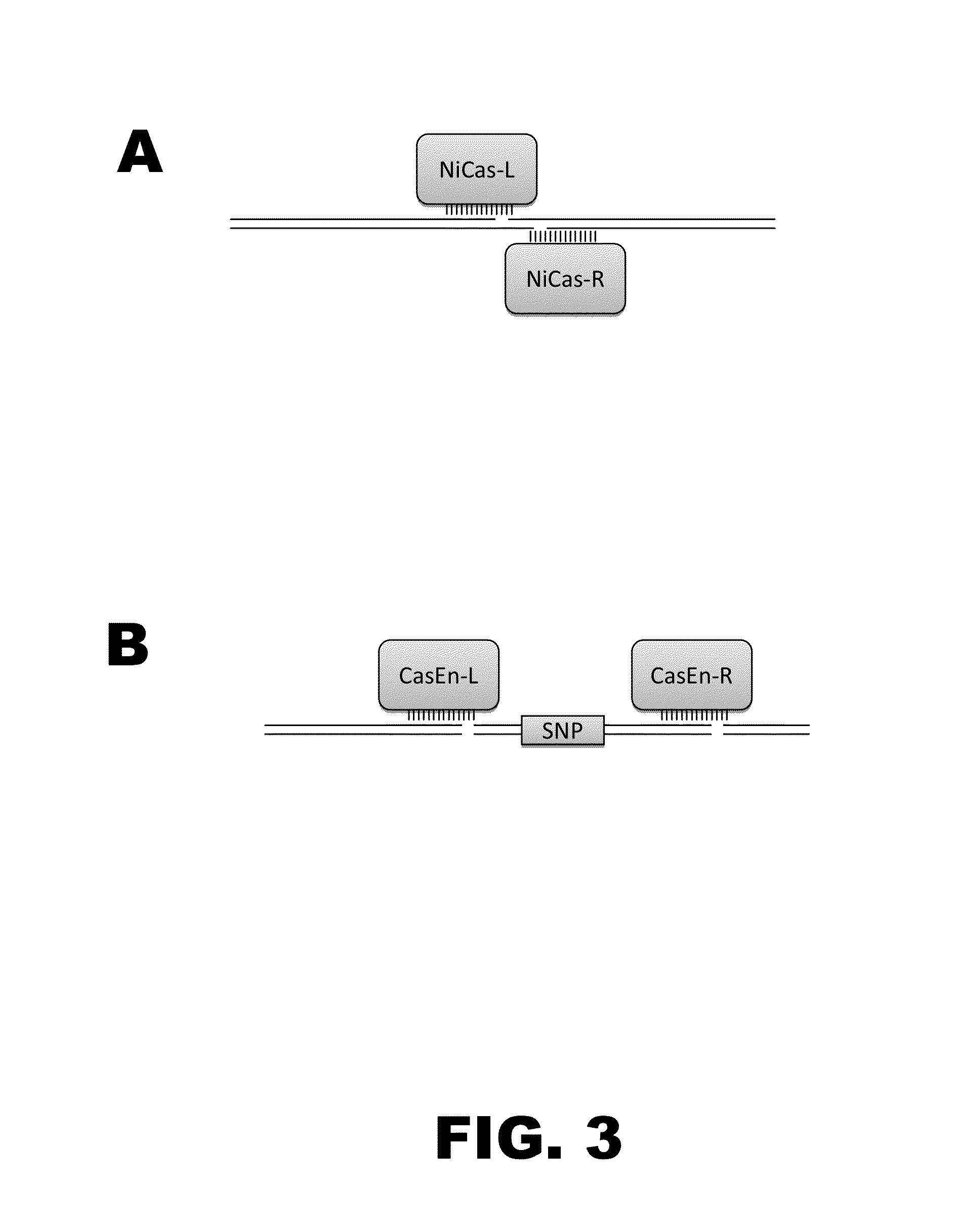
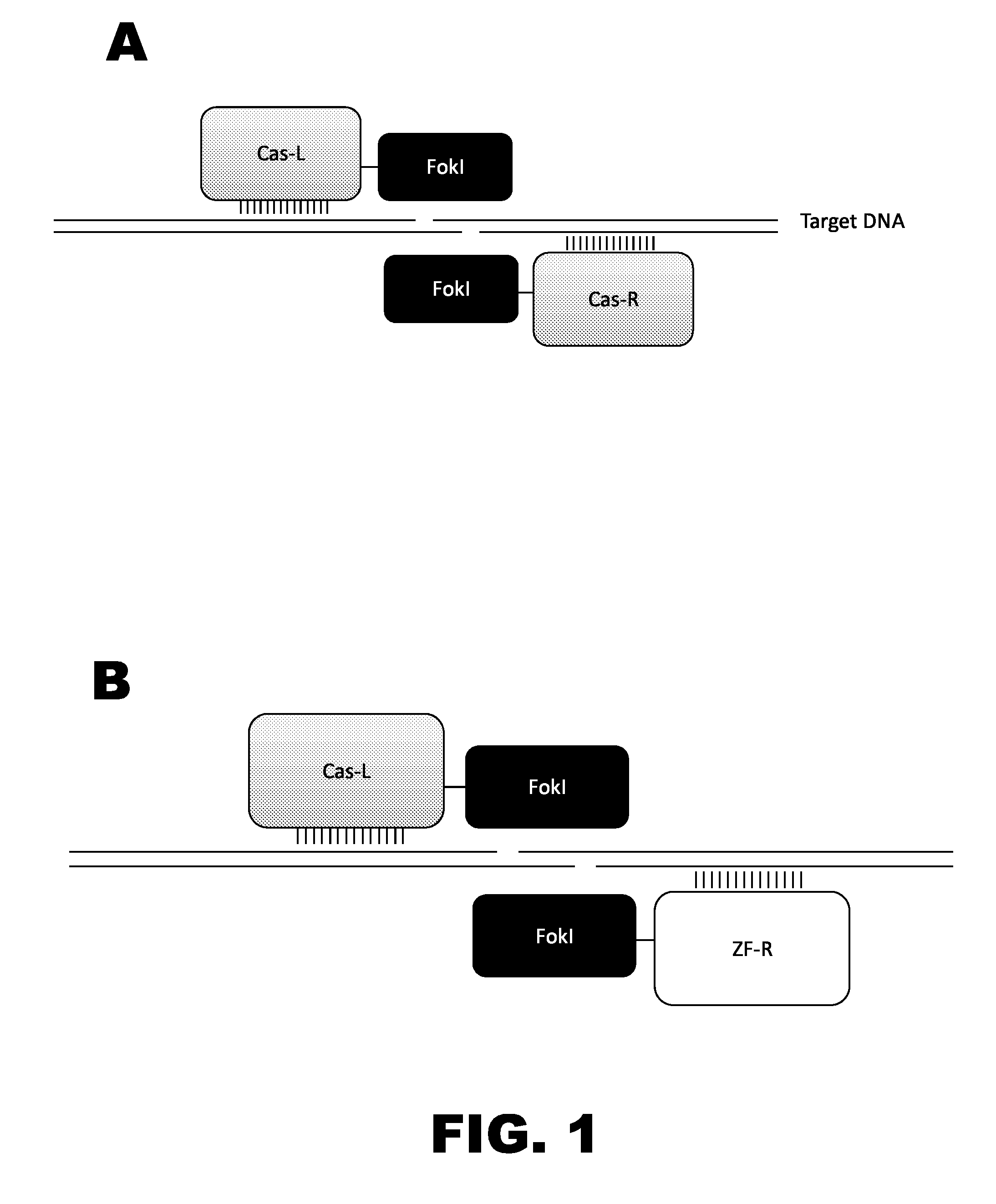
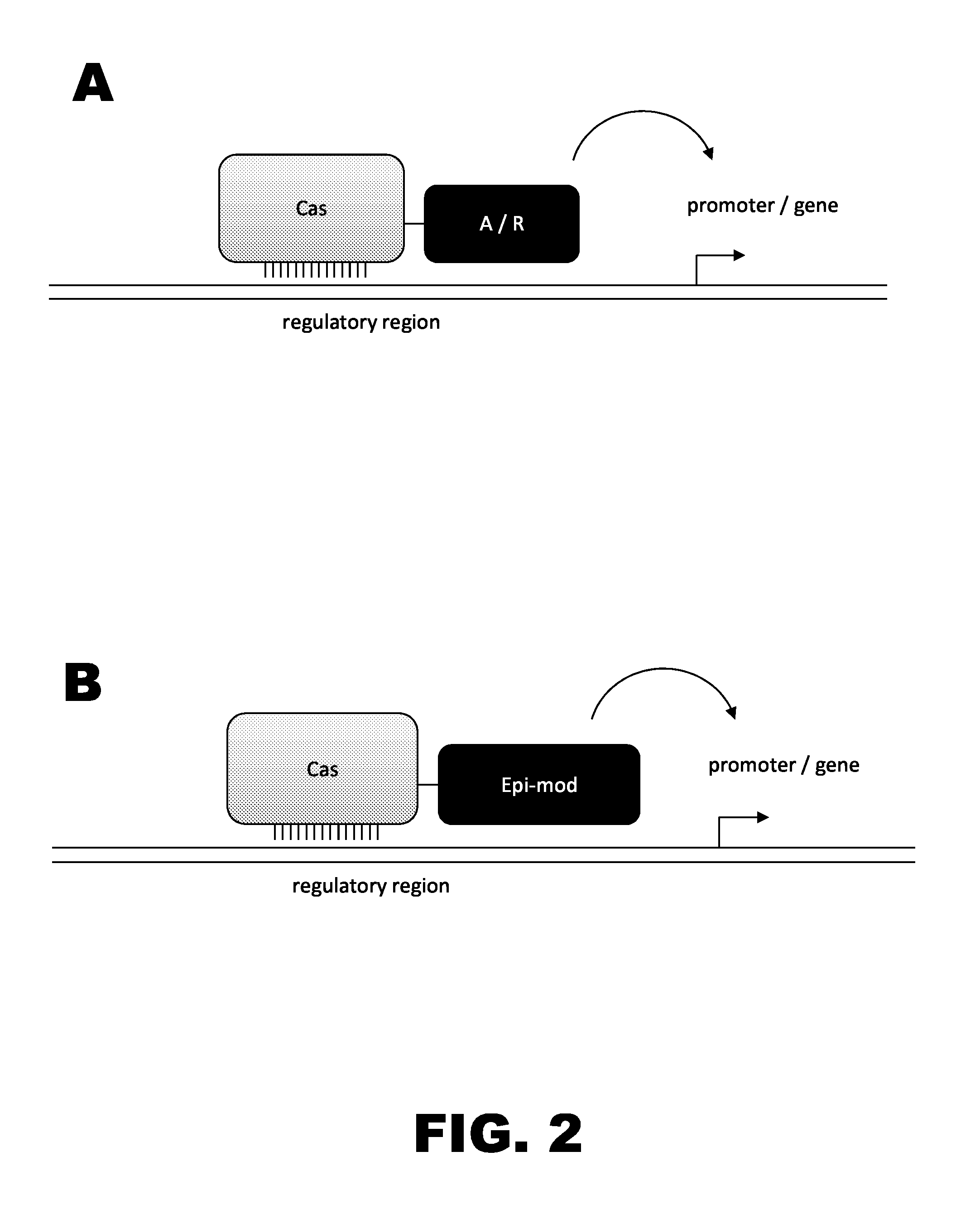
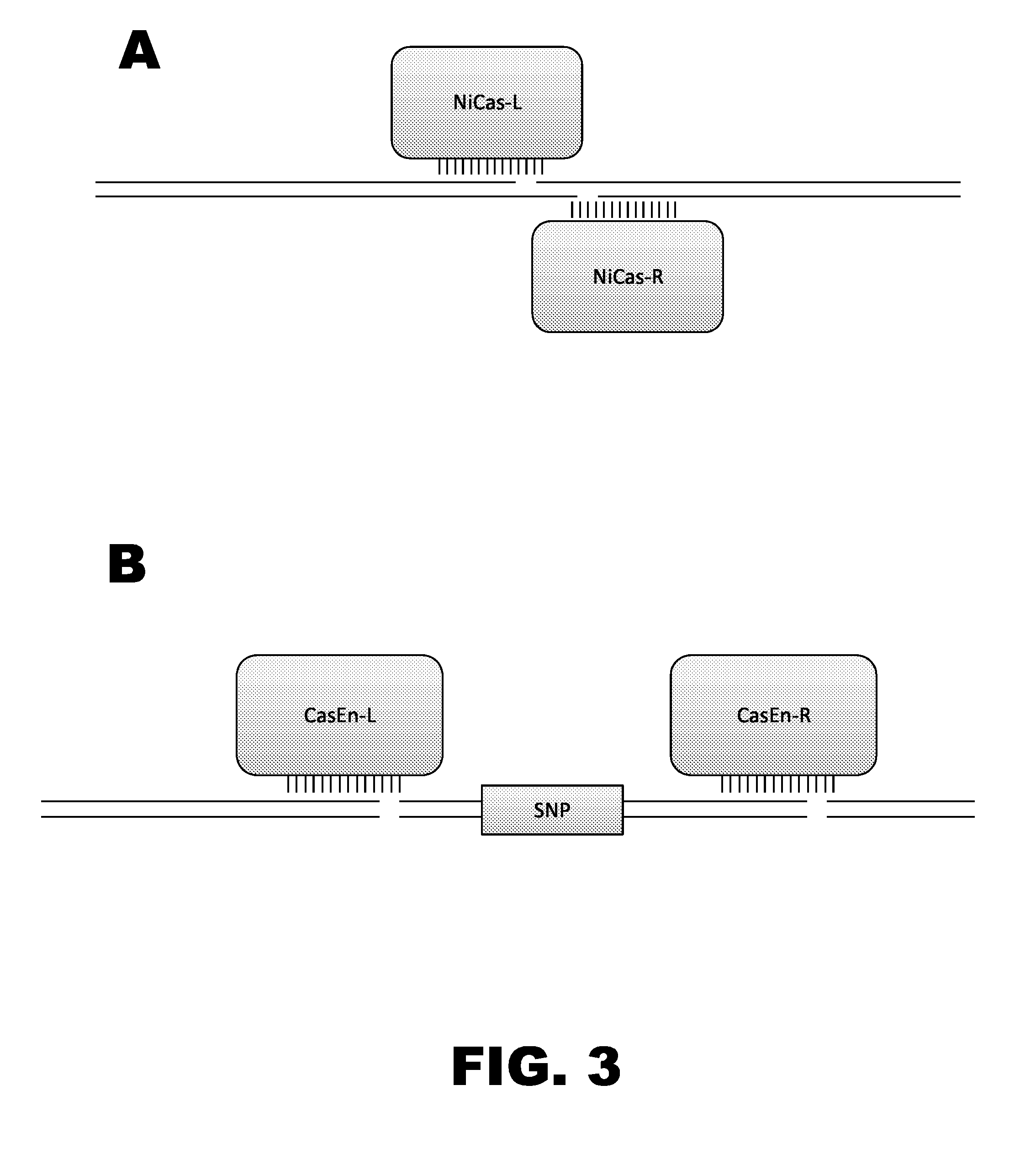
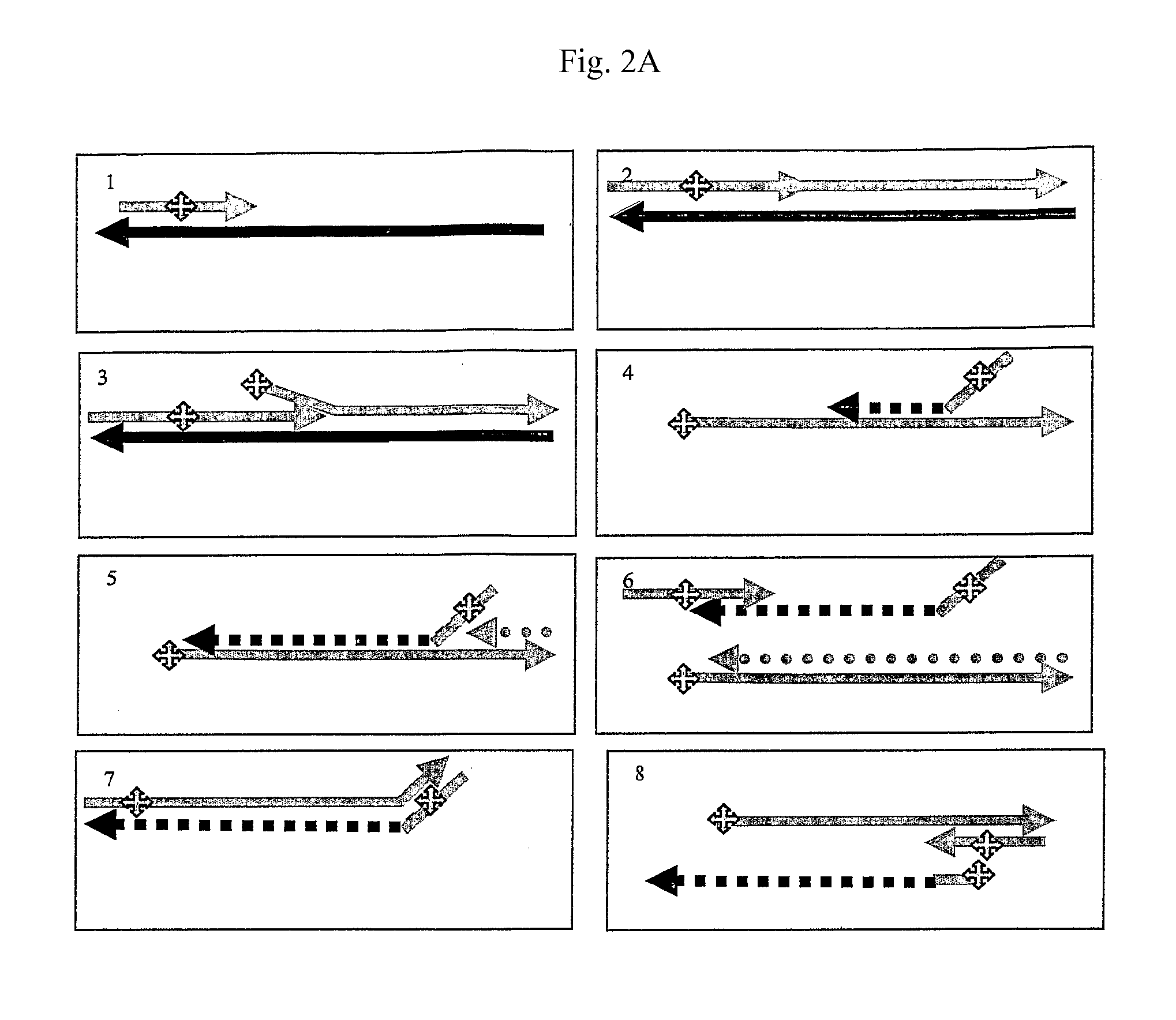
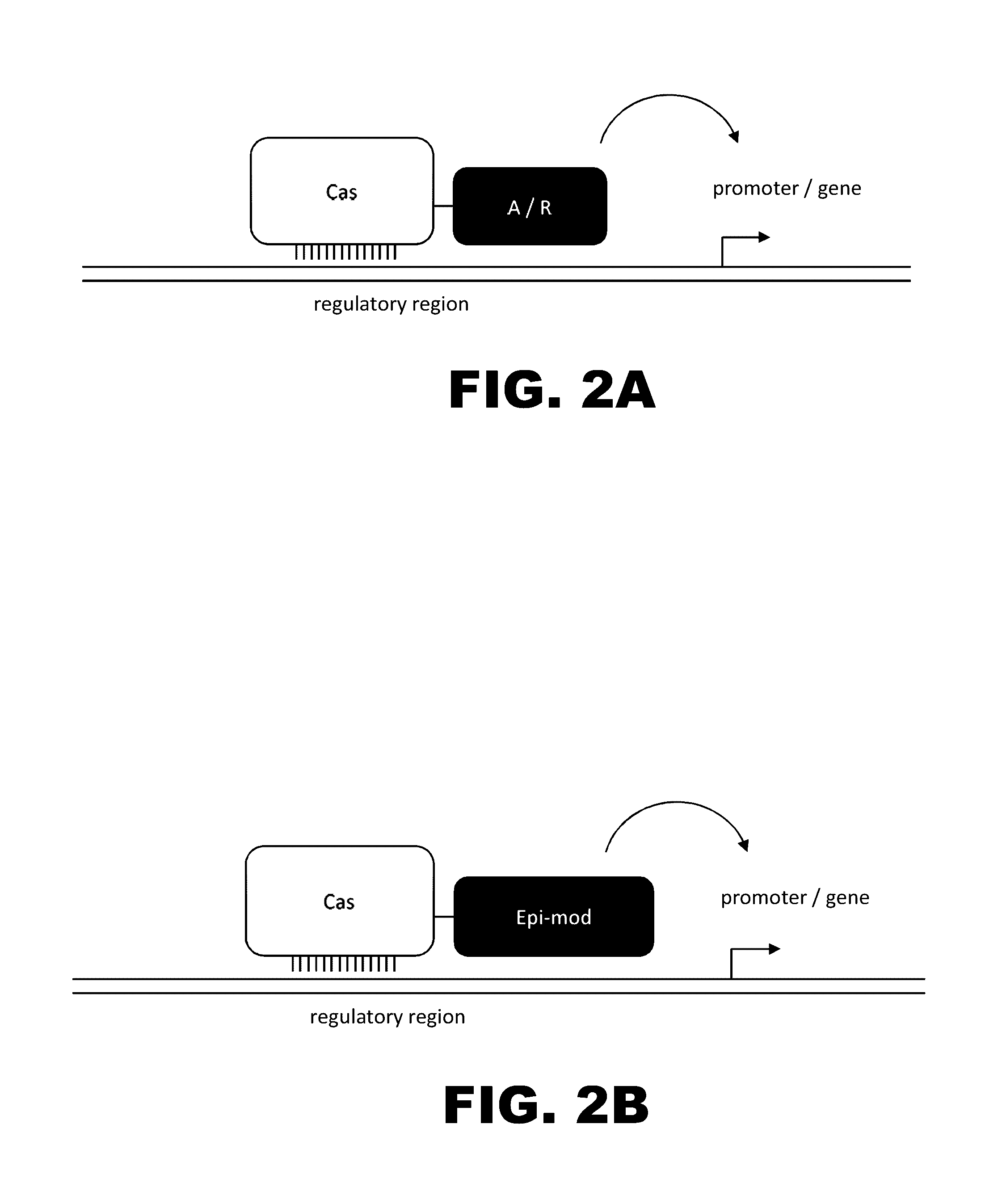
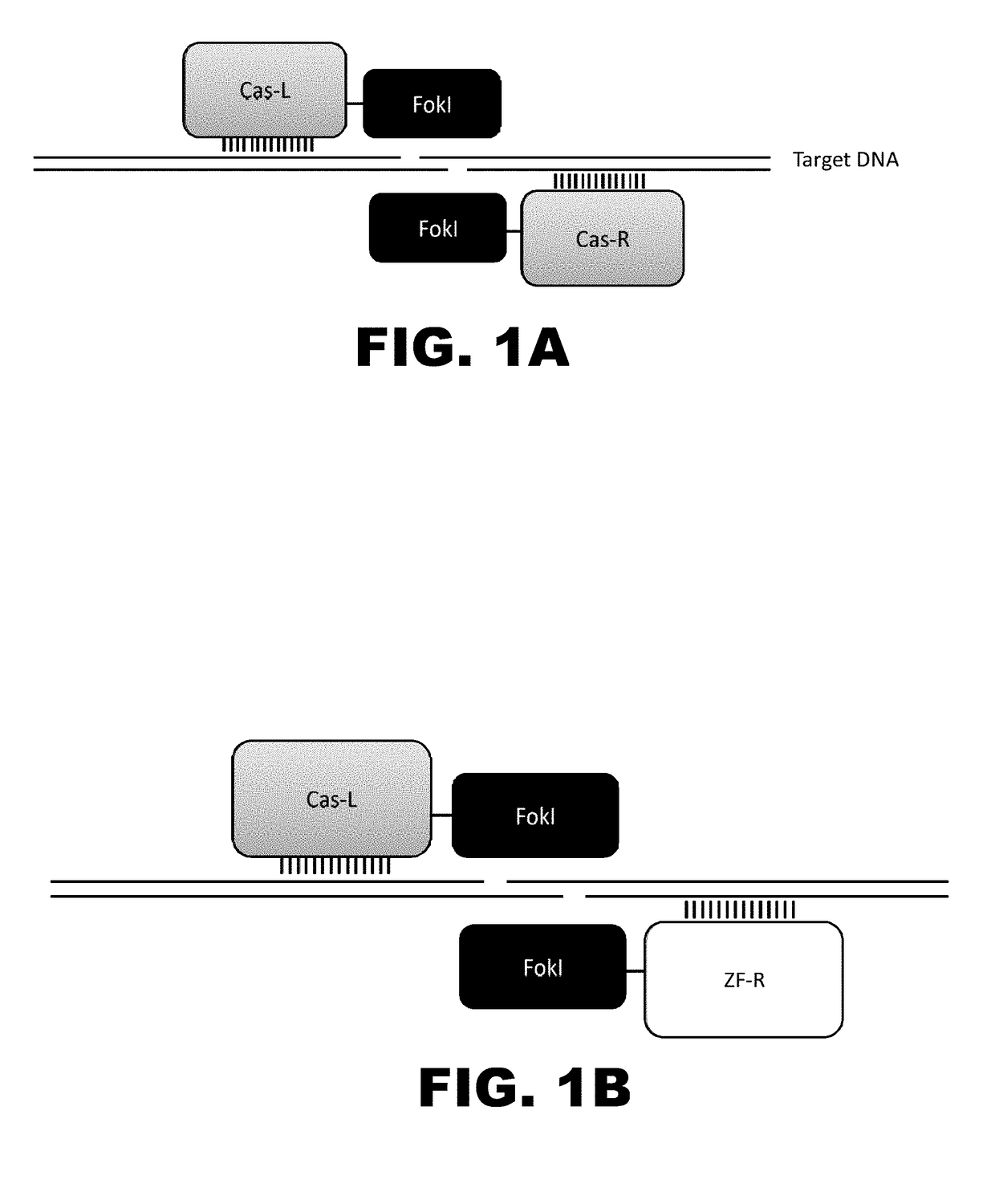
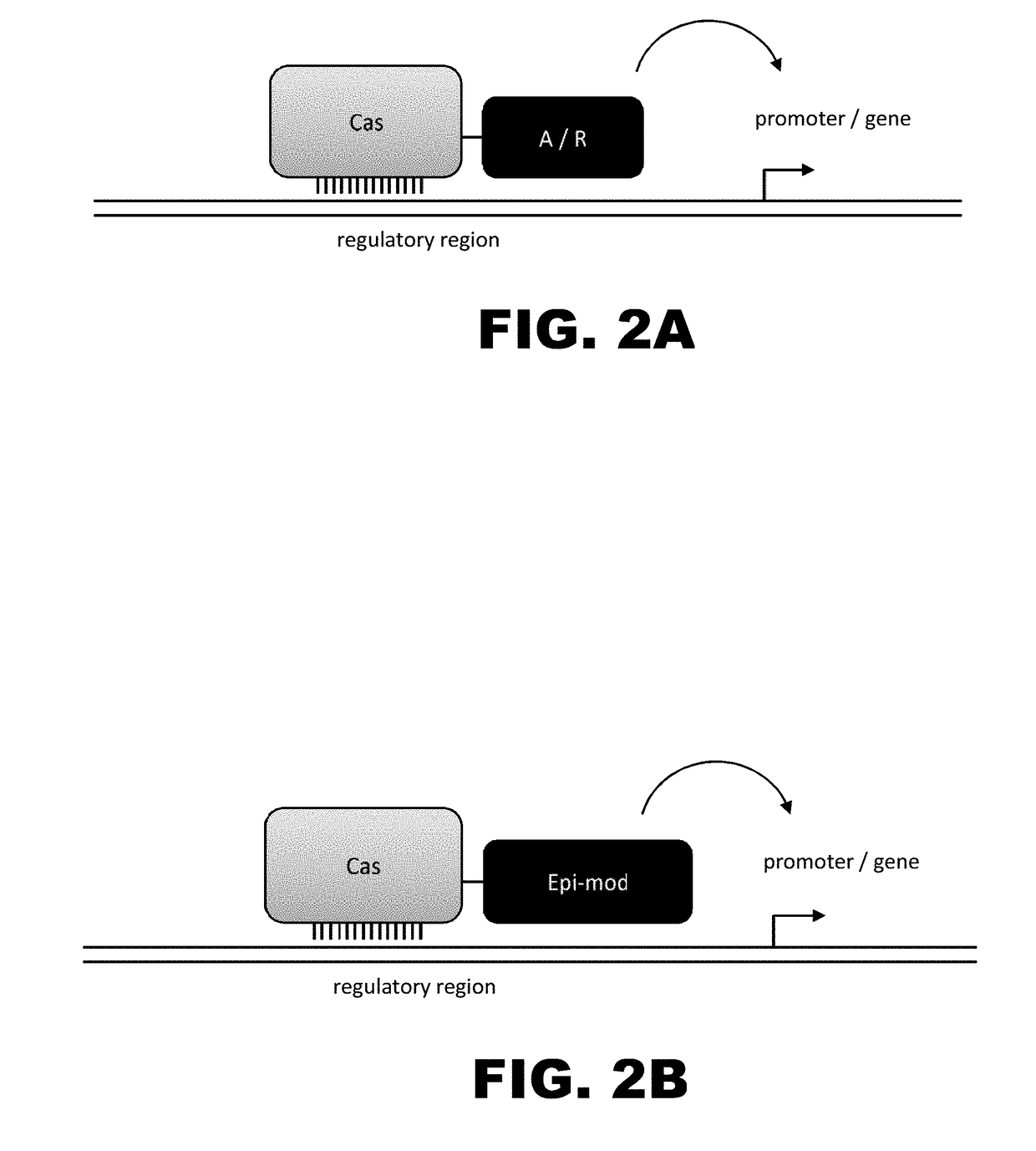
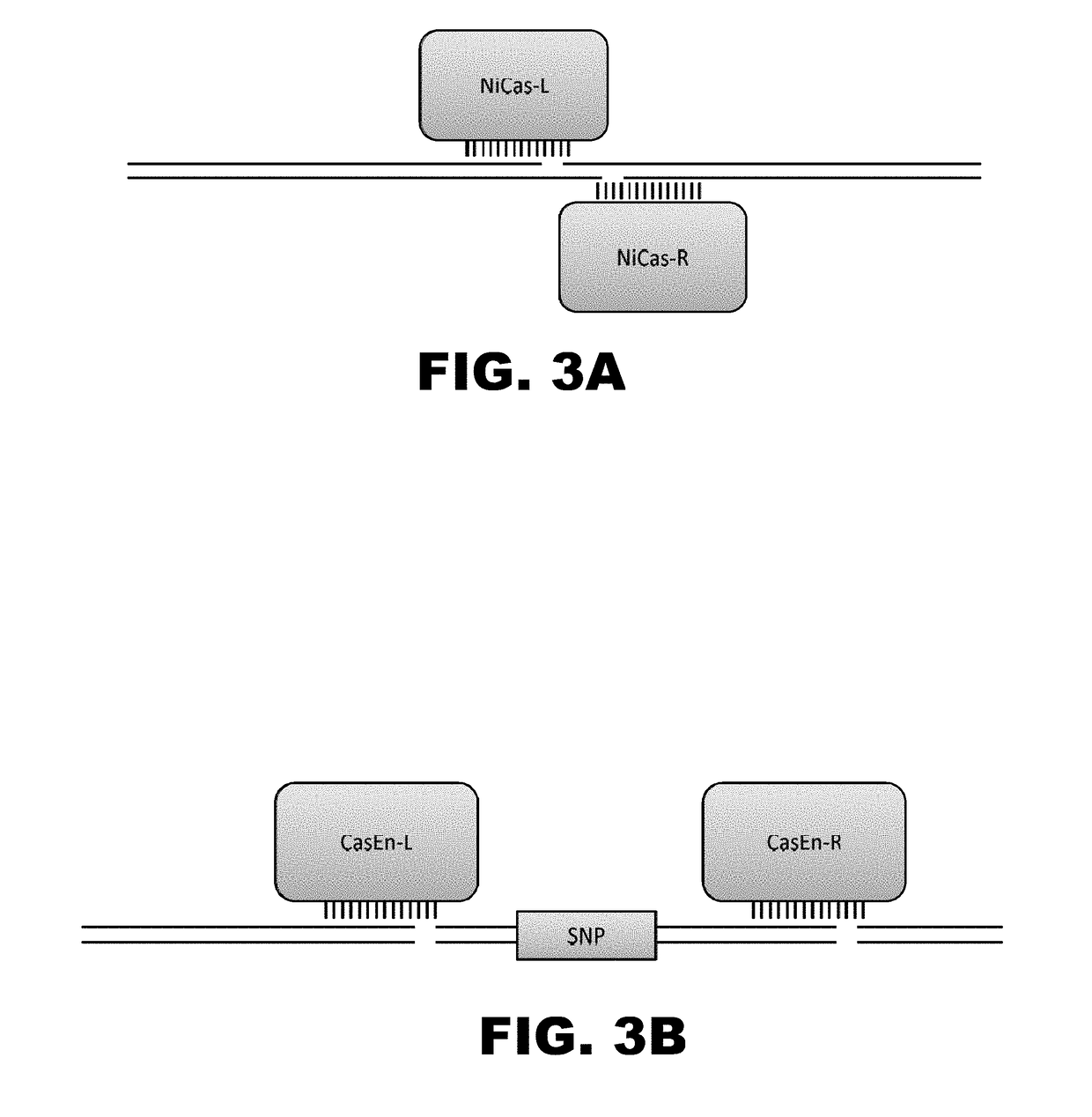
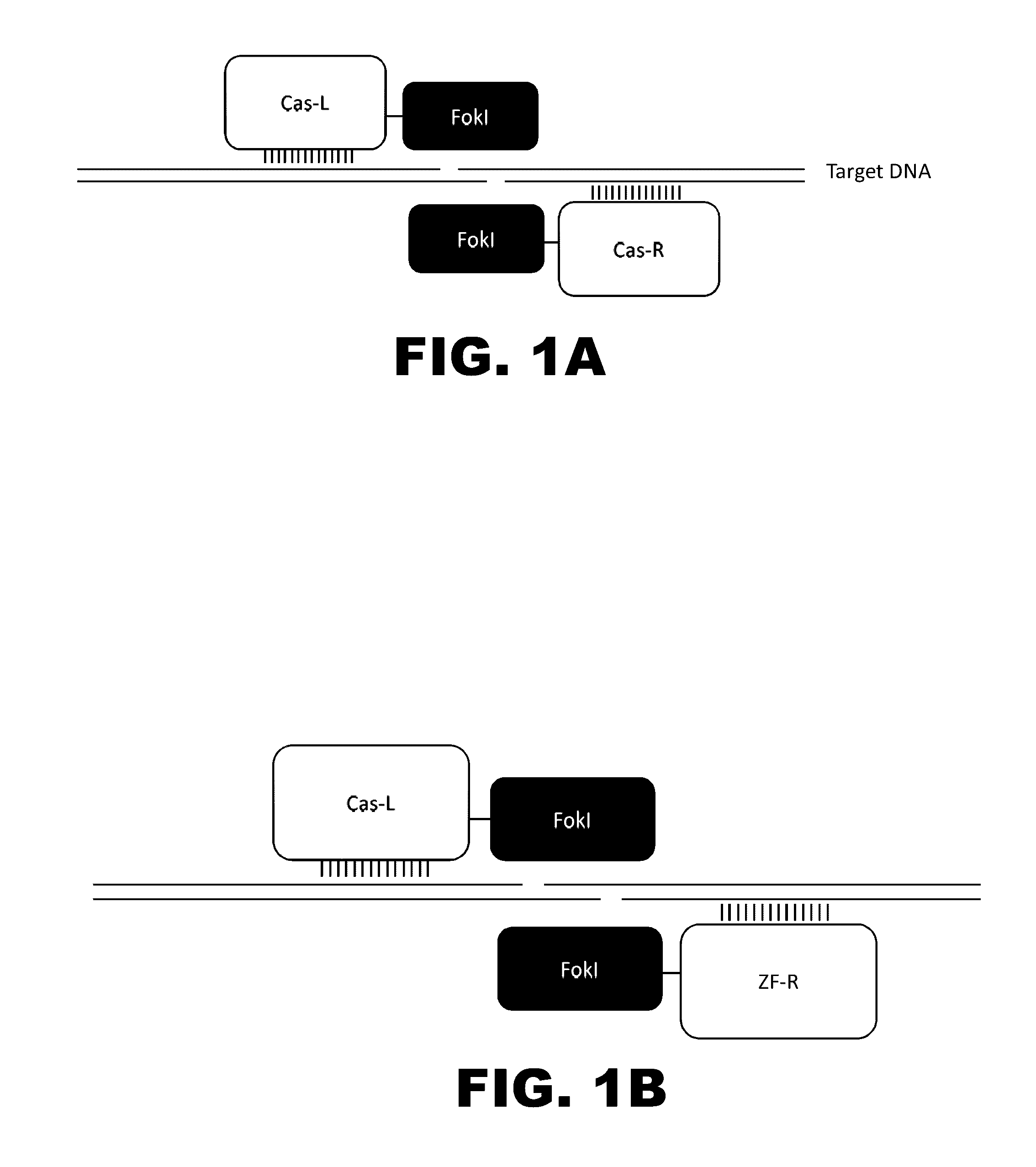
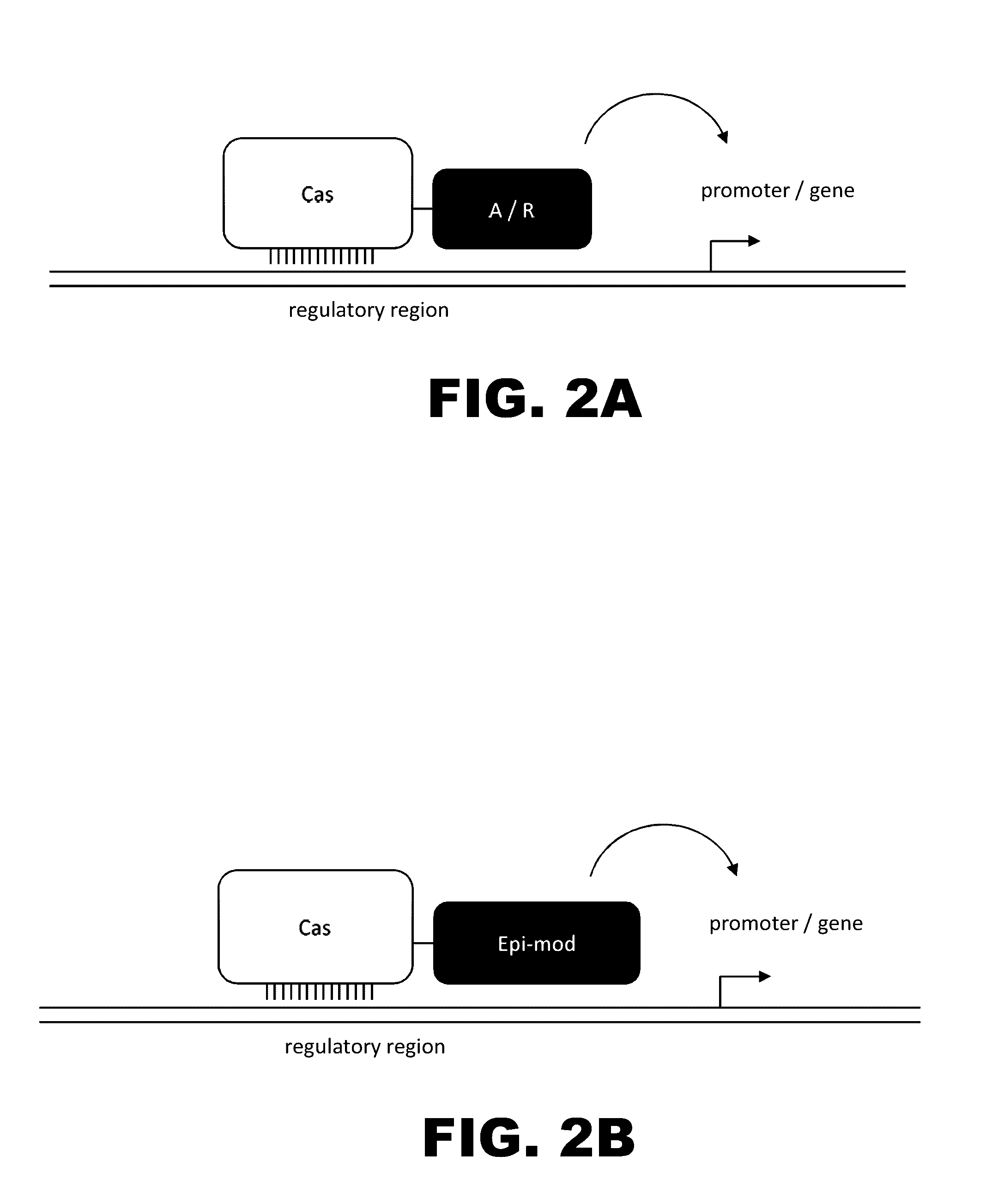
Crispr-based genome modification and regulation
Owner:SIGMA ALDRICH CO LLC
Crispr-based genome modification and regulation
InactiveUS20160017366A1Fusion with DNA-binding domainPeptide/protein ingredientsTranscription RepressionEukaryotic cell
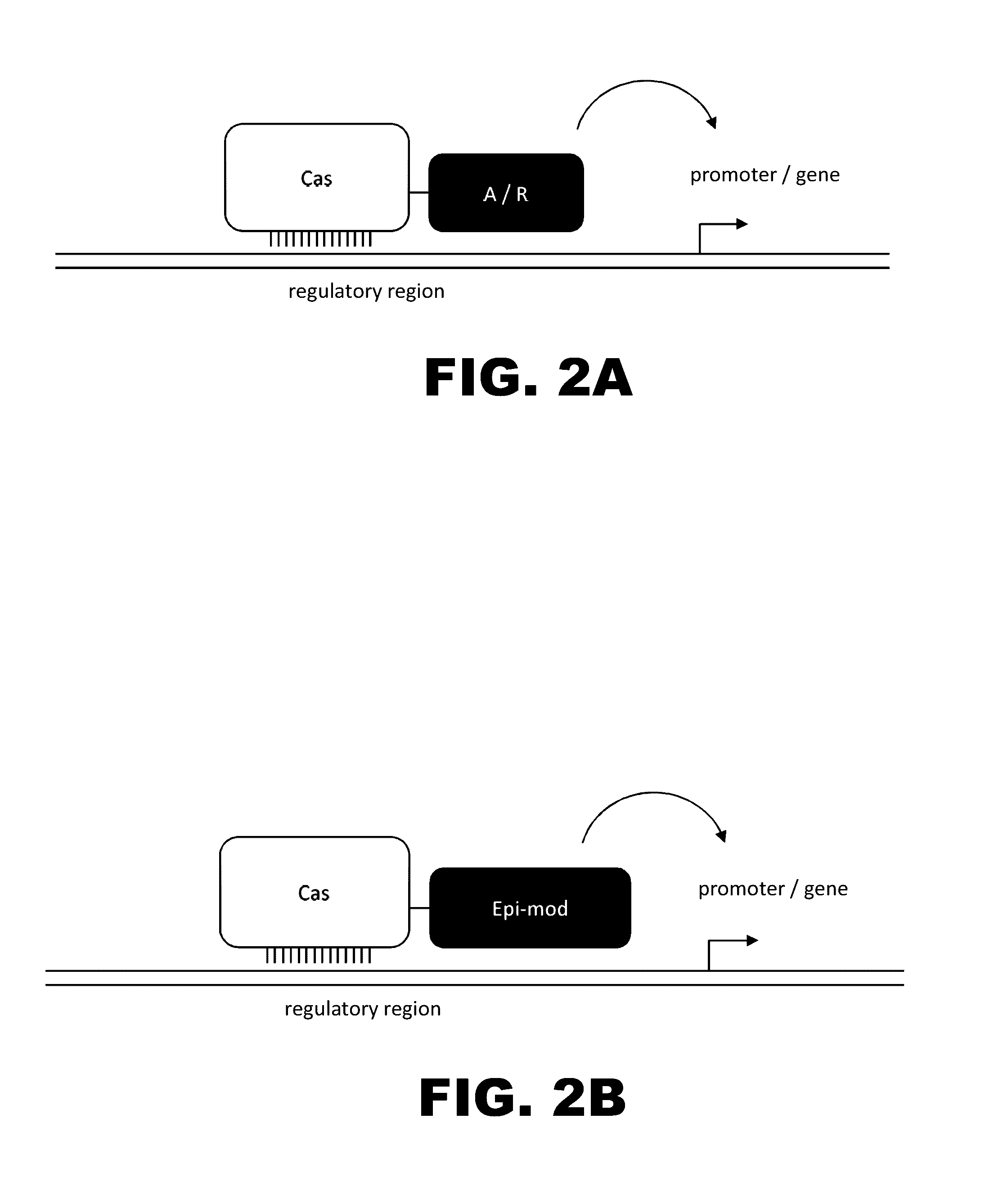
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
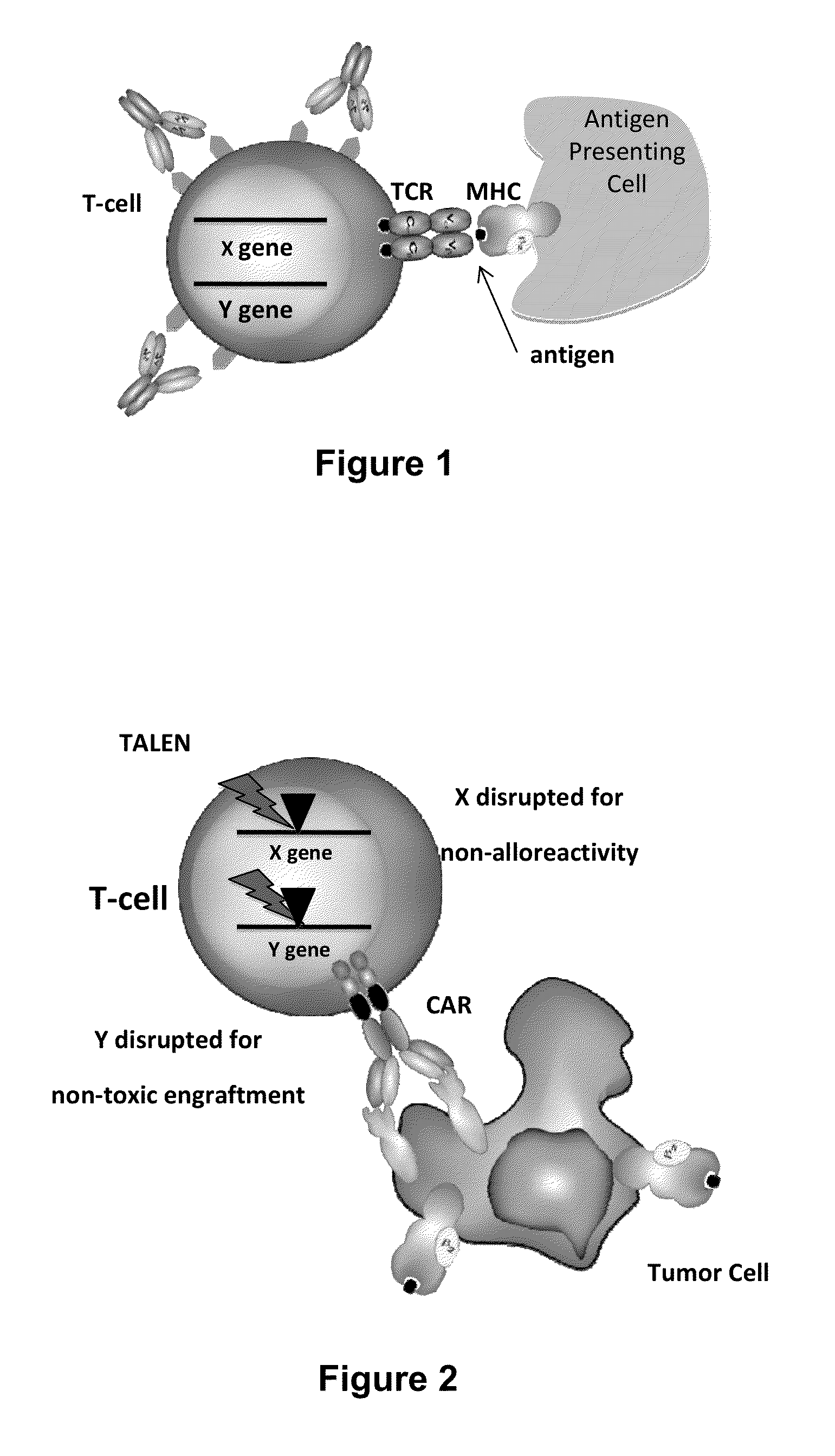
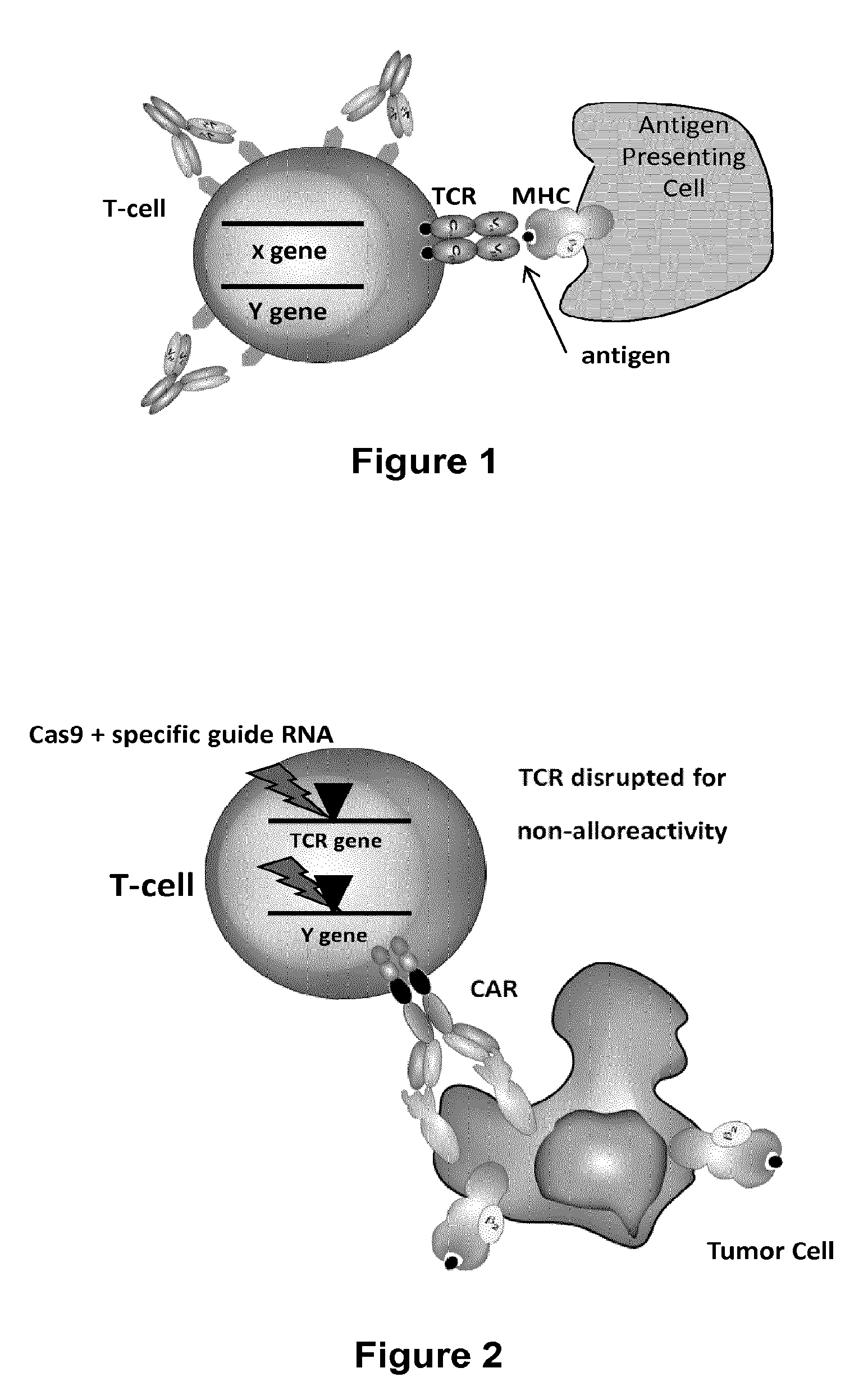
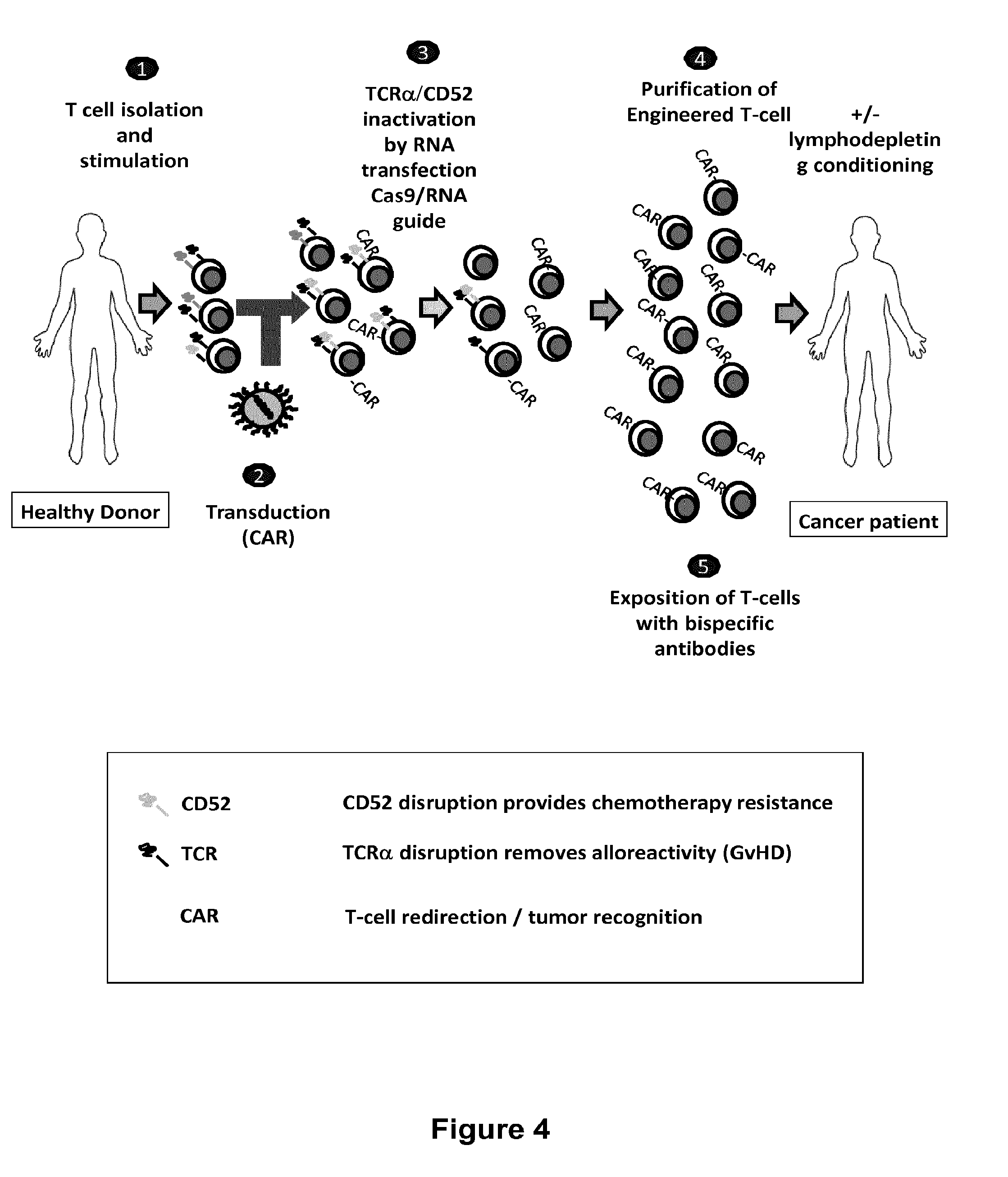
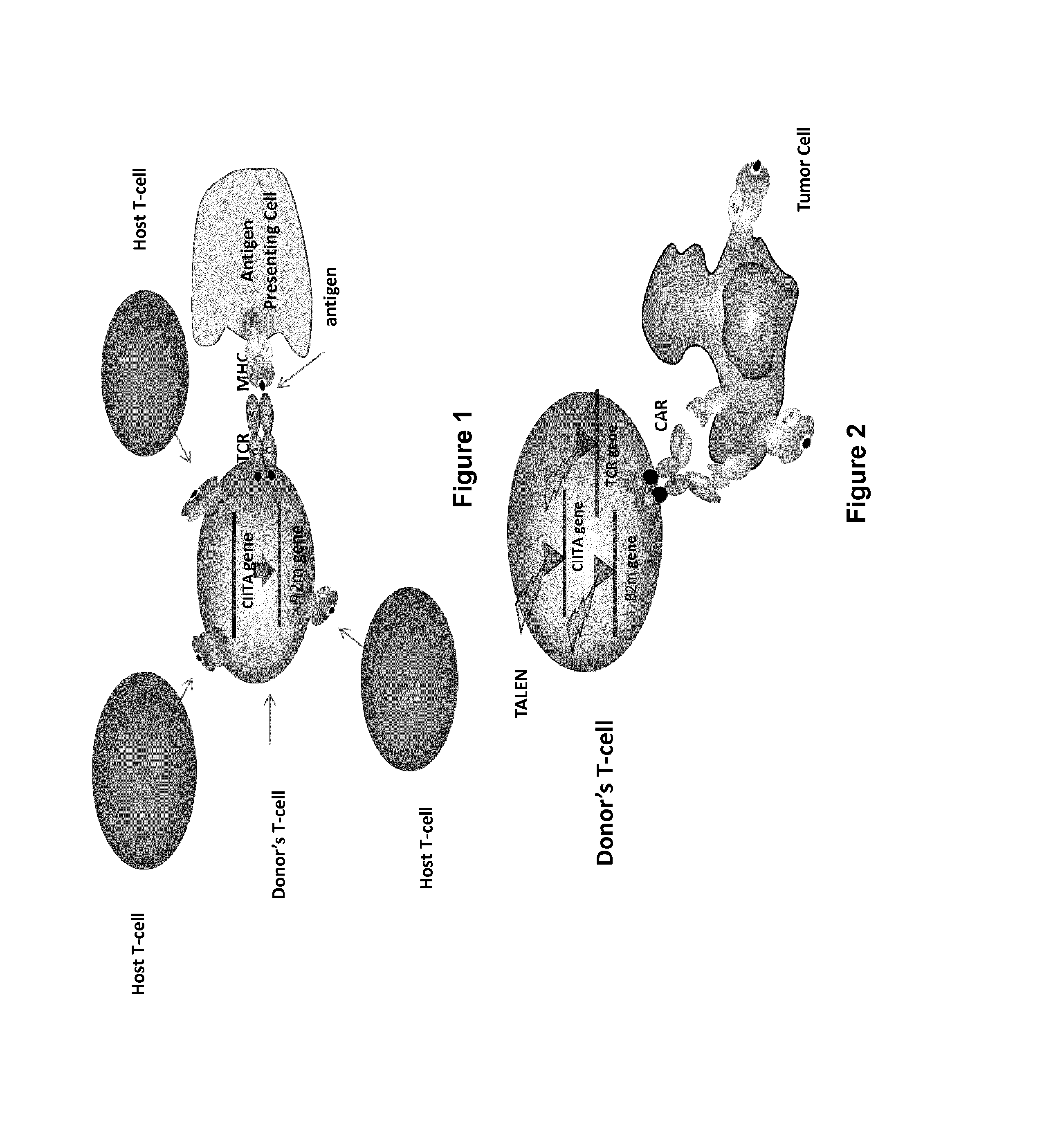
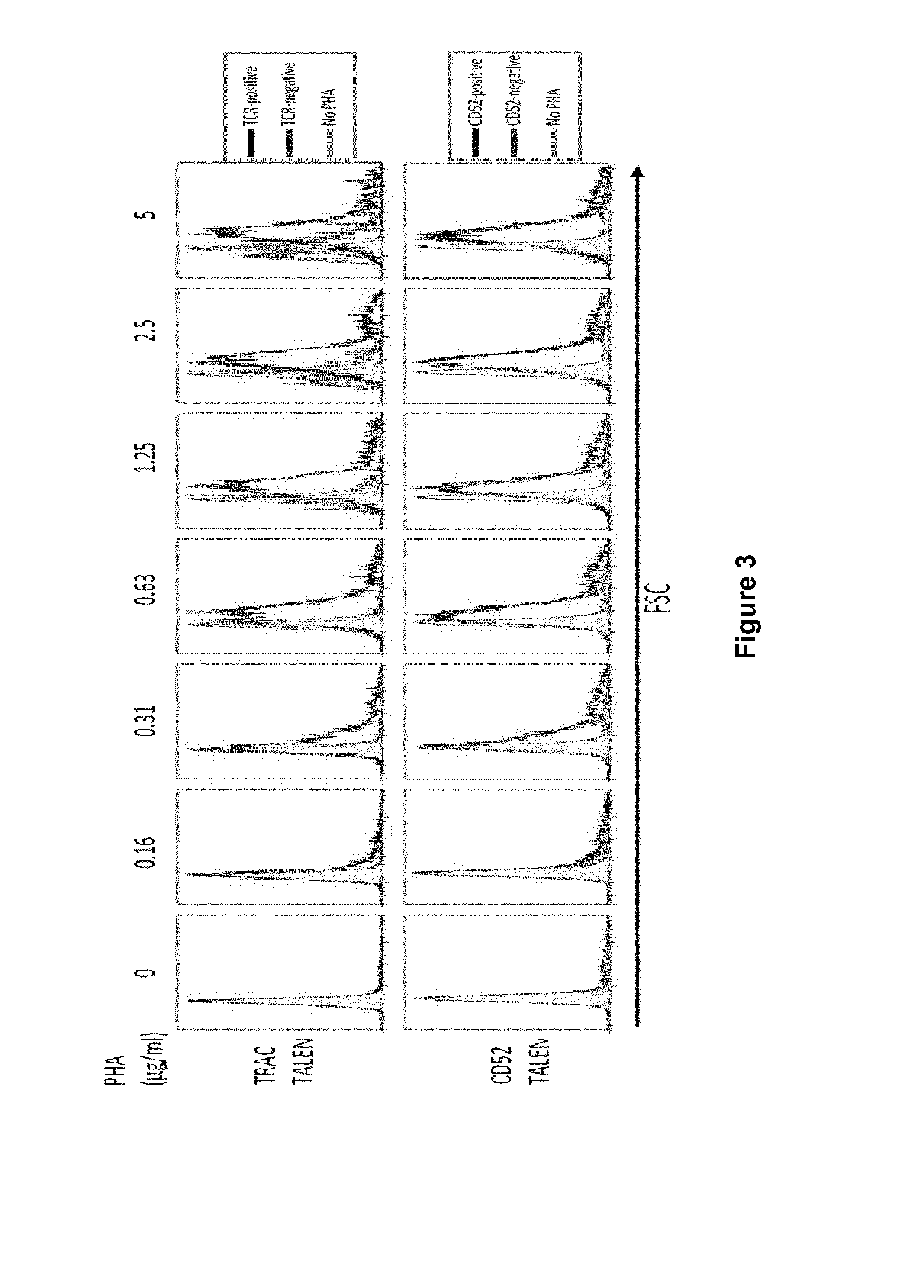
Methods for engineering allogeneic and immunosuppressive resistant t cell for immunotherapy
ActiveUS20130315884A1Precise positioningPeptide/protein ingredientsAntibody mimetics/scaffoldsImmunosuppressive drugPrimary cell
Methods for developing engineered T-cells for immunotherapy that are both non-alloreactive and resistant to immunosuppressive drugs. The present invention relates to methods for modifying T-cells by inactivating both genes encoding target for an immunosuppressive agent and T-cell receptor, in particular genes encoding CD52 and TCR. This method involves the use of specific rare cutting endonucleases, in particular TALE-nucleases (TAL effector endonuclease) and polynucleotides encoding such polypeptides, to precisely target a selection of key genes in T-cells, which are available from donors or from culture of primary cells. The invention opens the way to standard and affordable adoptive immunotherapy strategies for treating cancer and viral infections.
Owner:CELLECTIS SA
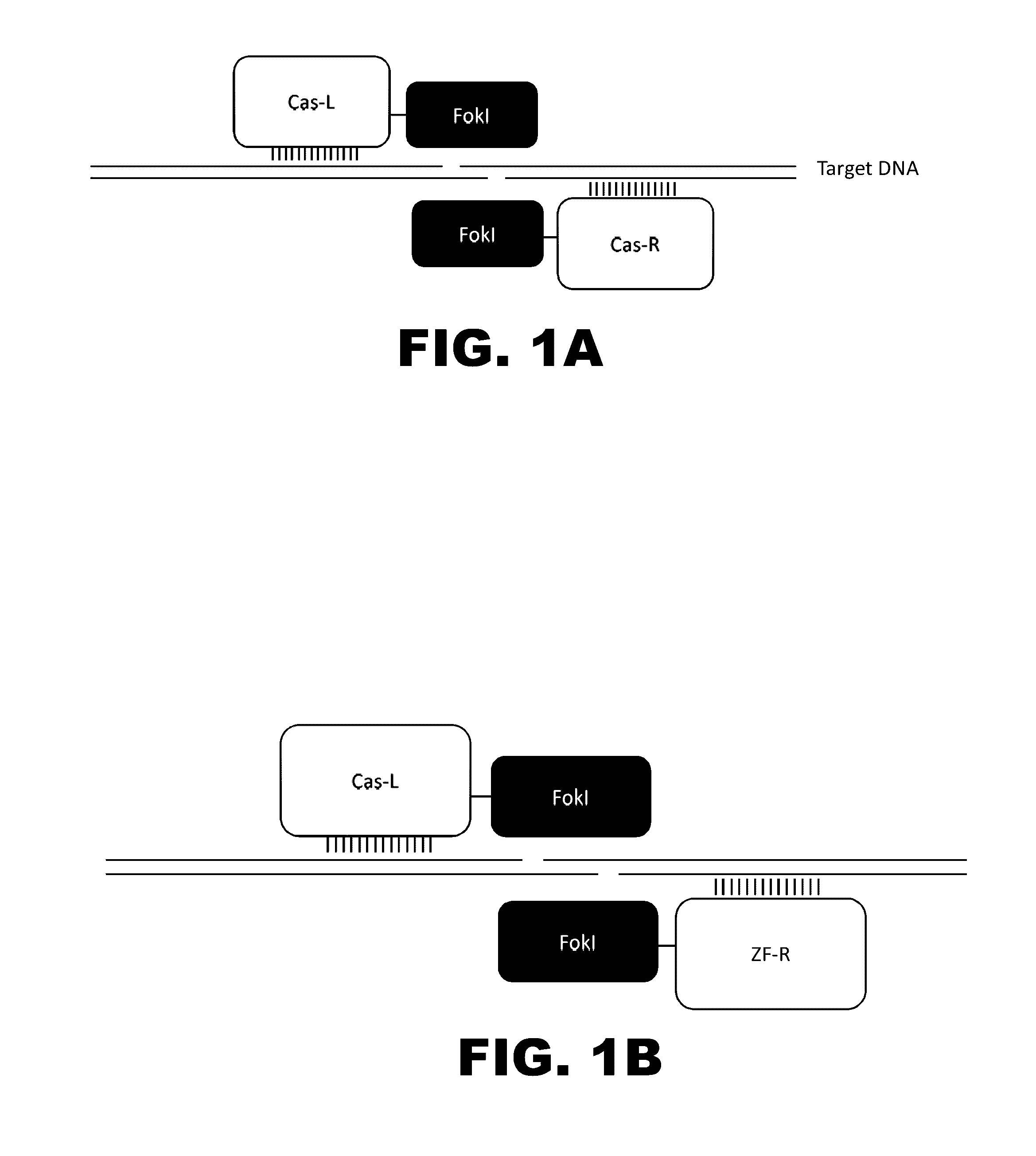
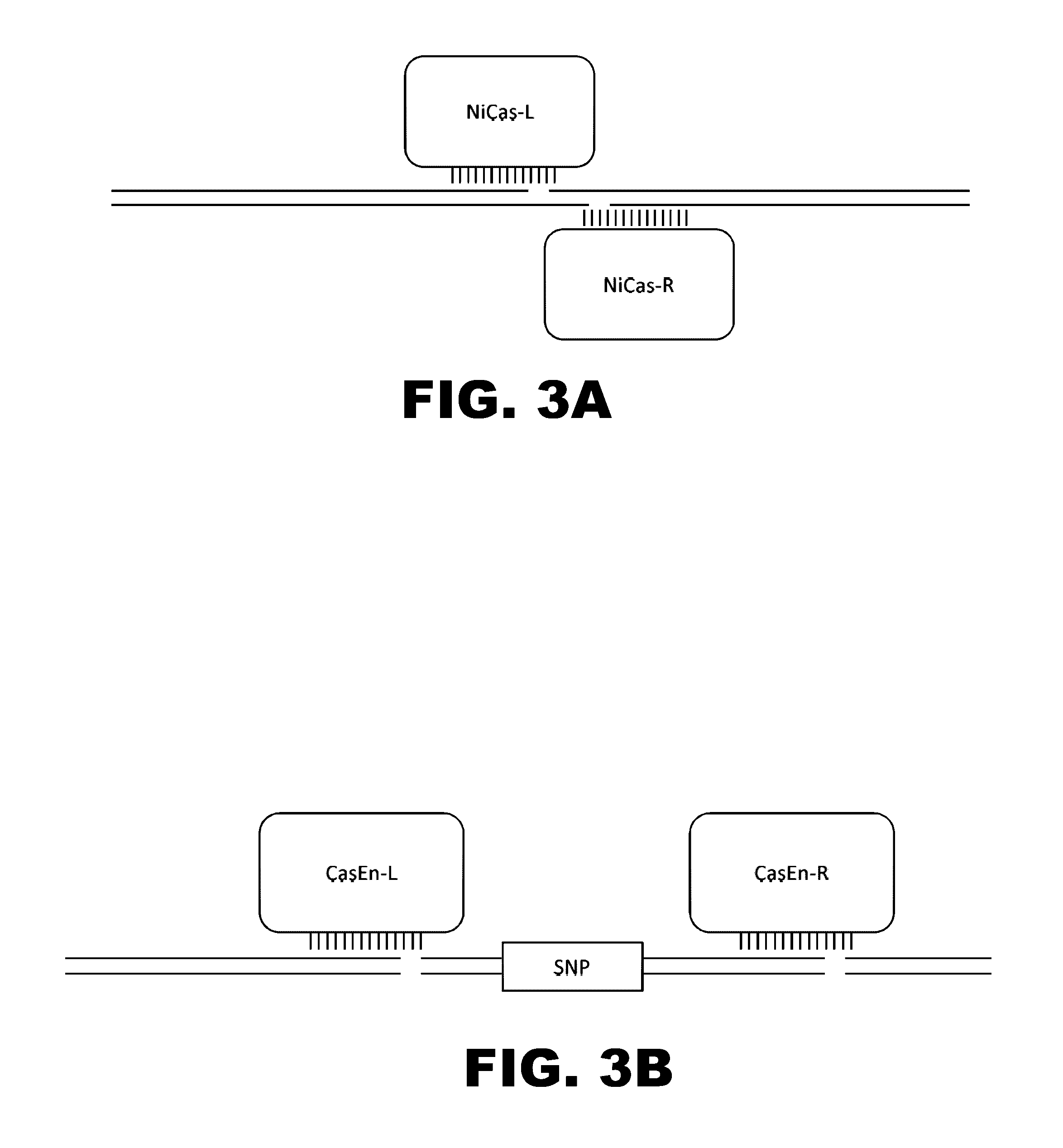
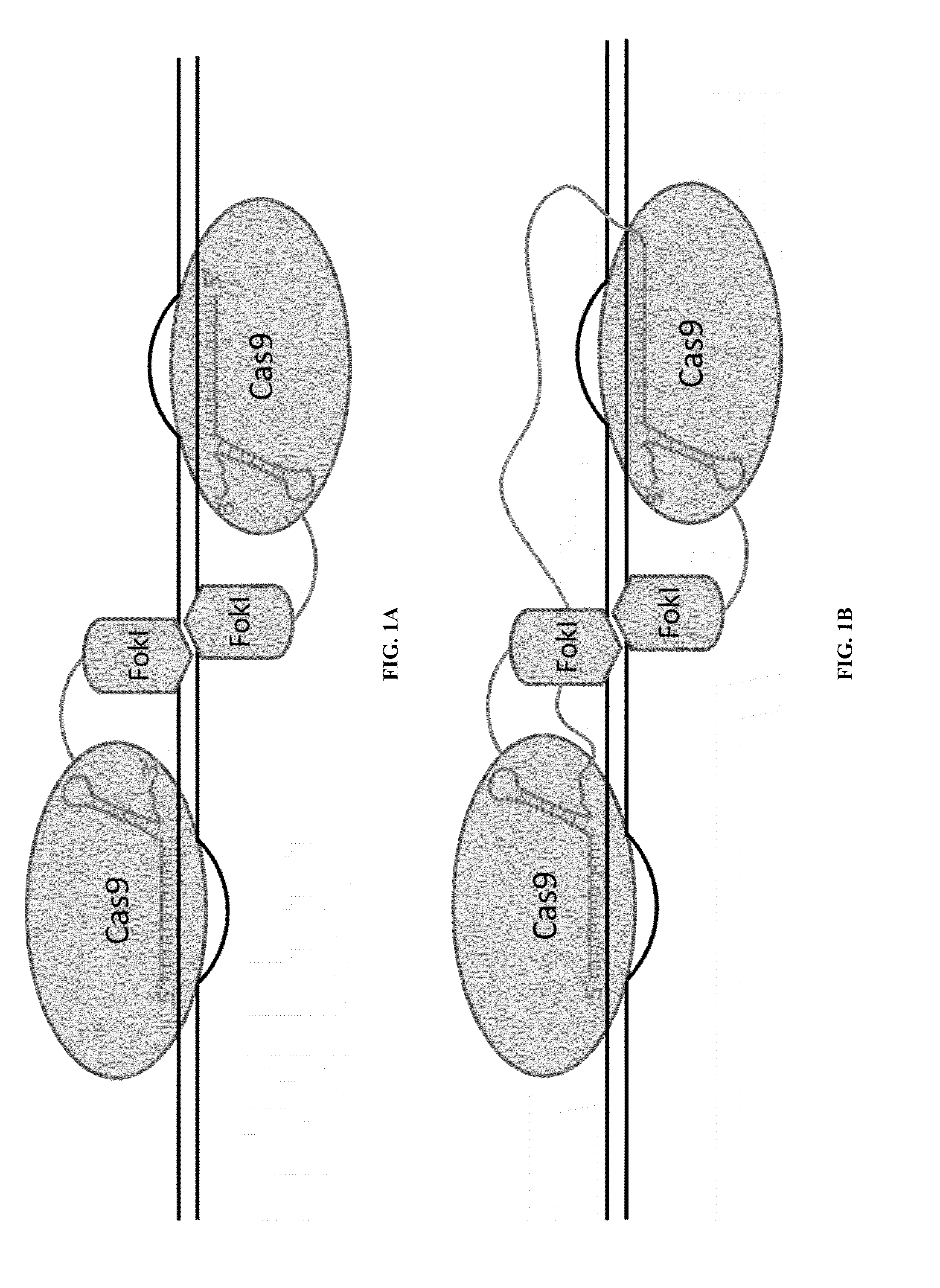
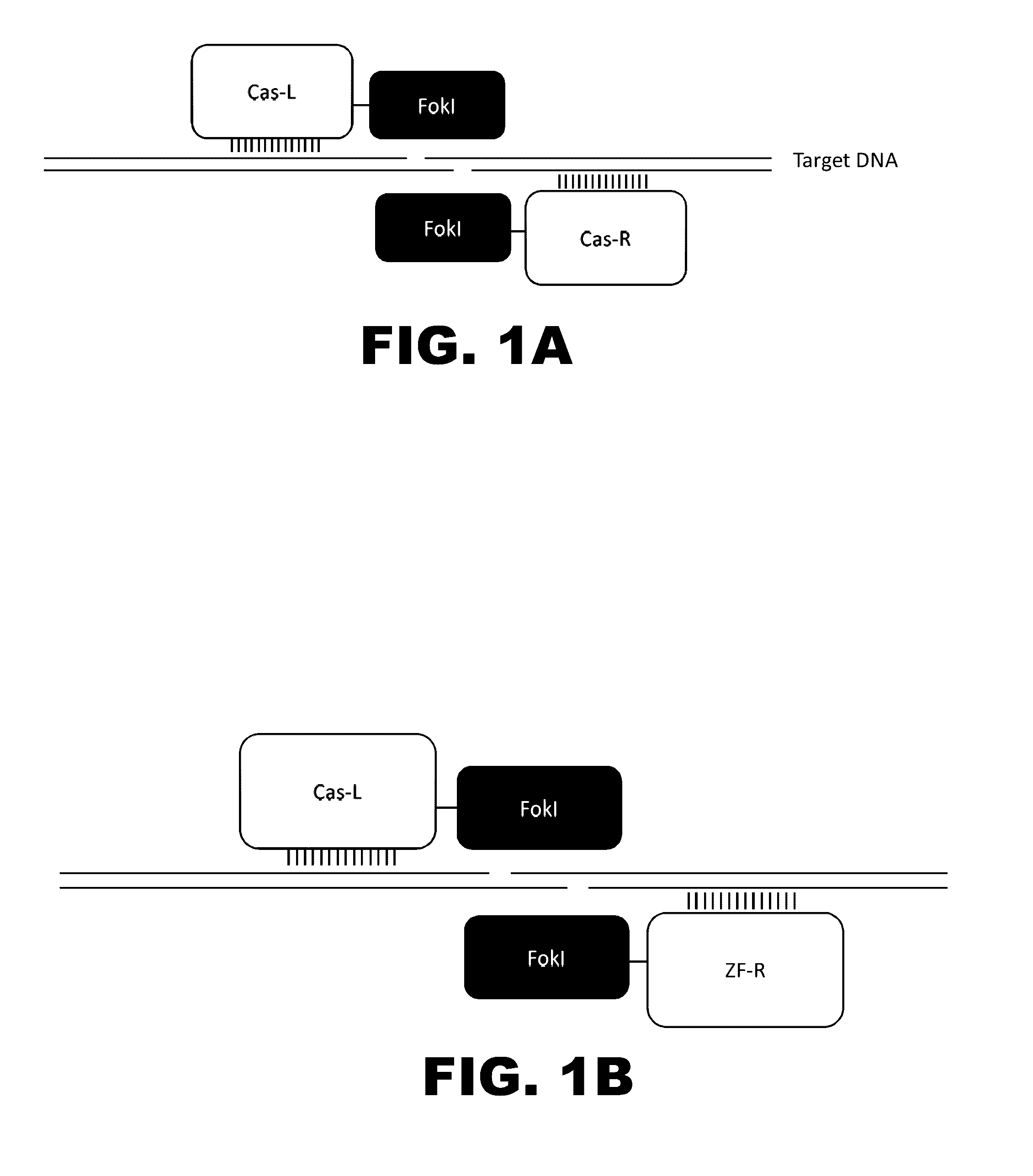
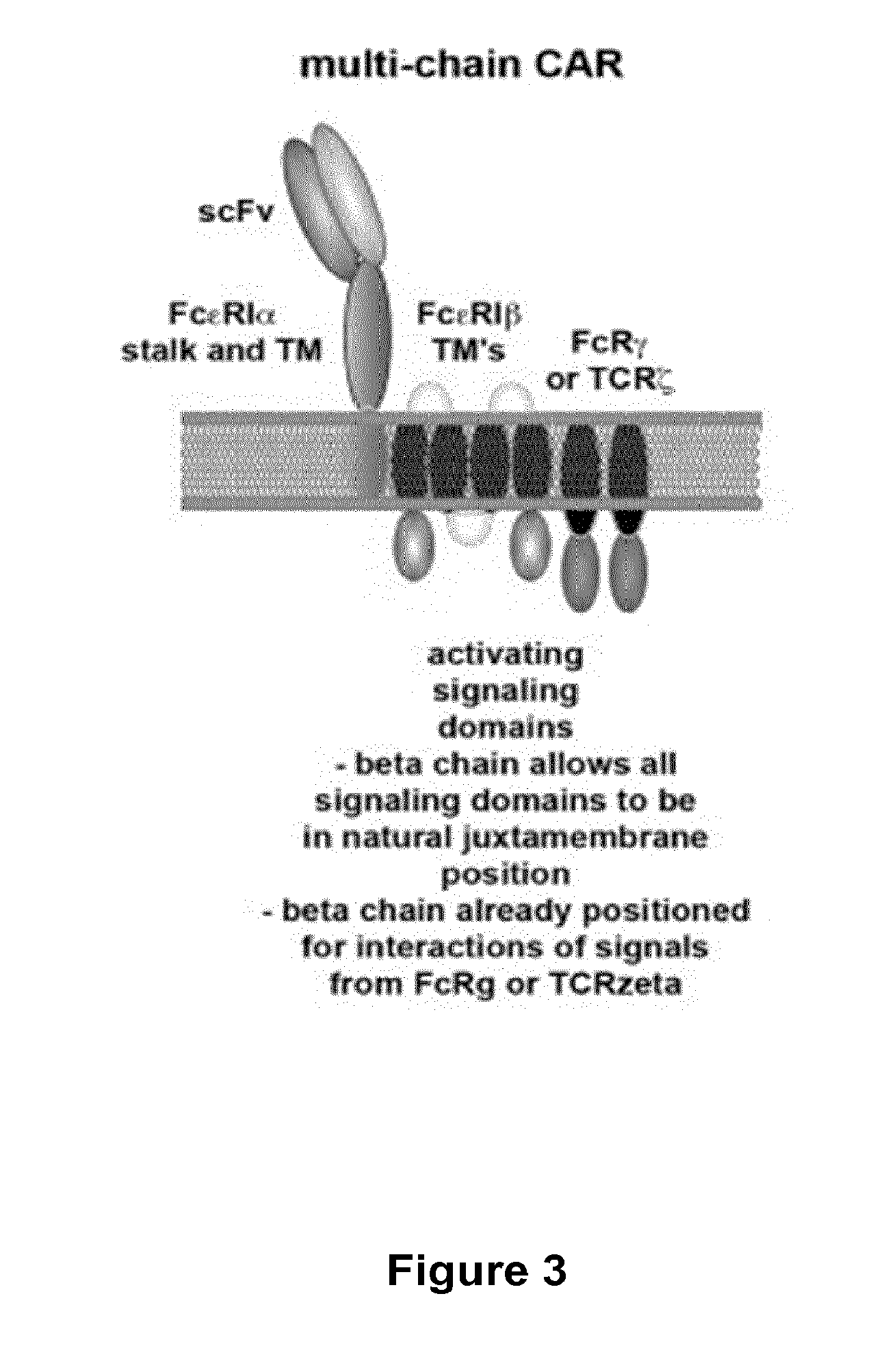
Cas9-recombinase fusion proteins and uses thereof
ActiveUS20150071898A1Strong specificityReduce the possibilityFusion with DNA-binding domainBacteriaSite-specific recombinationResearch setting
Some aspects of this disclosure provide compositions, methods, and kits for improving the specificity of RNA-programmable endonucleases, such as Cas9. Also provided are variants of Cas9, e.g., Cas9 dimers and fusion proteins, engineered to have improved specificity for cleaving nucleic acid targets. Also provided are compositions, methods, and kits for site-specific recombination, using Cas9 fusion proteins (e.g., nuclease-inactivated Cas9 fused to a recombinase catalytic domain). Such Cas9 variants are useful in clinical and research settings involving site-specific modification of DNA, for example, genomic modifications.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Isothermal amplification of nucleic acids
InactiveUS7824890B2Fluorescence enhancementEnhanced signalSugar derivativesMicrobiological testing/measurementStrand invasionEndonuclease
A process of amplifying a nucleic acid template dependent on partial destruction of primer molecules which have extended onto the template molecule followed by strand invasion of the partially destroyed primer template by a replacement primer. The destruction of the primer molecule may be performed by either endonuclease or exonuclease digestion. A signal generation from the amplified products may be obtained by the use of adaptors capable of binding probe molecules as well as the amplified product.
Owner:AVACTA GROUP
Method for making linear, covalently closed DNA constructs
InactiveUS6451563B1Bulking digestionHigh processivitySugar derivativesHydrolasesDNA constructGenomic DNA
A process to obtain linear double-stranded covalently closed DNA "dumbbell" constructs from plasmids by restriction digest, subsequent ligation with hairpin oligodesoxyribonucleotides, optionally in the presence of restriction enzyme, and a final digestion with endo- and exonucleolytic enzymes that degrade all contaminating polymeric DNA molecules but the desired construct. The invention also provides a process to obtain said dumbbell constructs employing endonuclease class II enzymes. Furthermore, the invention provides a process to obtain linear, covalently closed DNA molecules, such as plasmids, free from contamination by genomic DNA, by submitting the DNA preparation to a facultative endonucleolytic degradation step and an obligatory exonucleolytic degradation step.
Owner:MOLOGEN AG +1
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
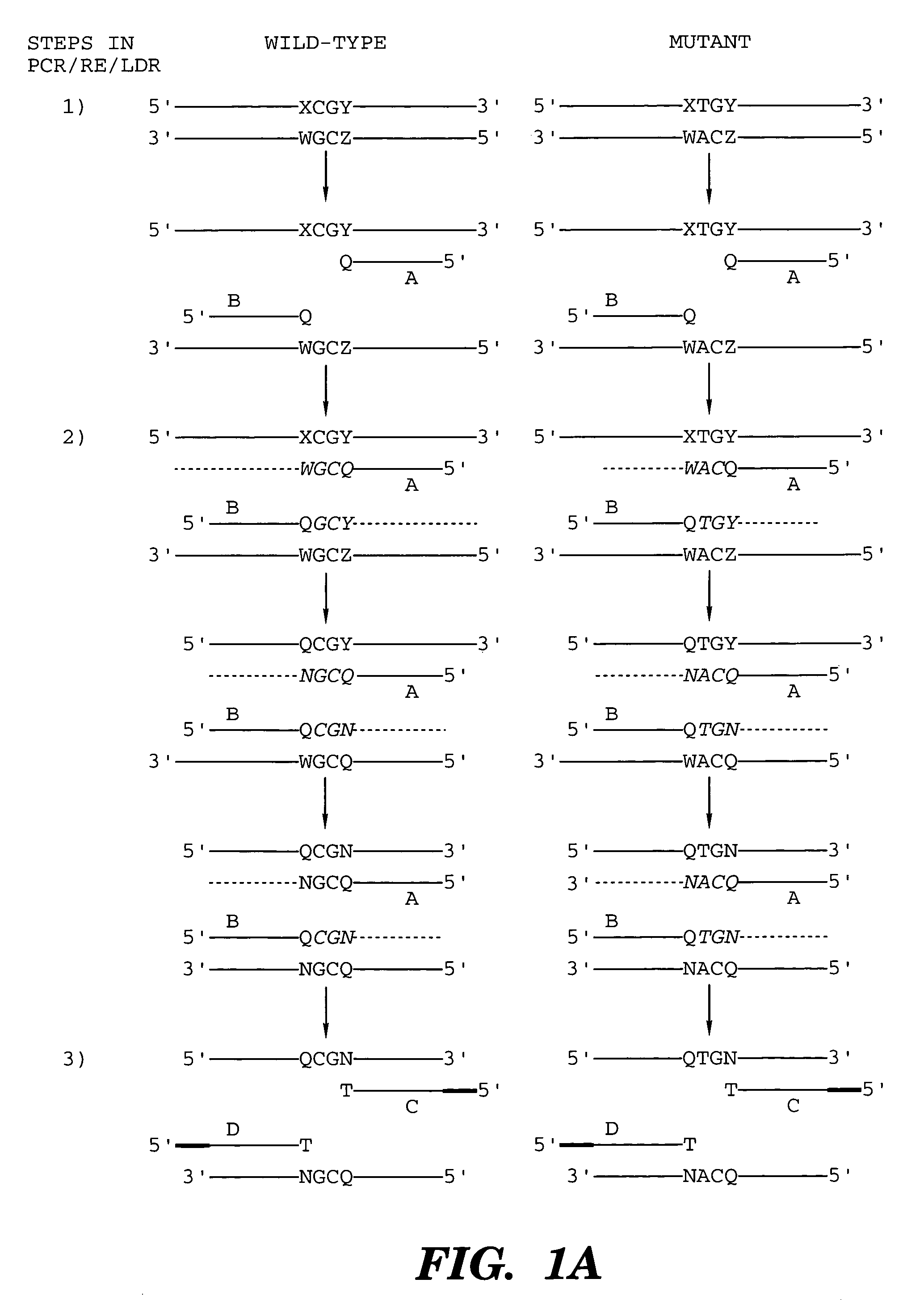
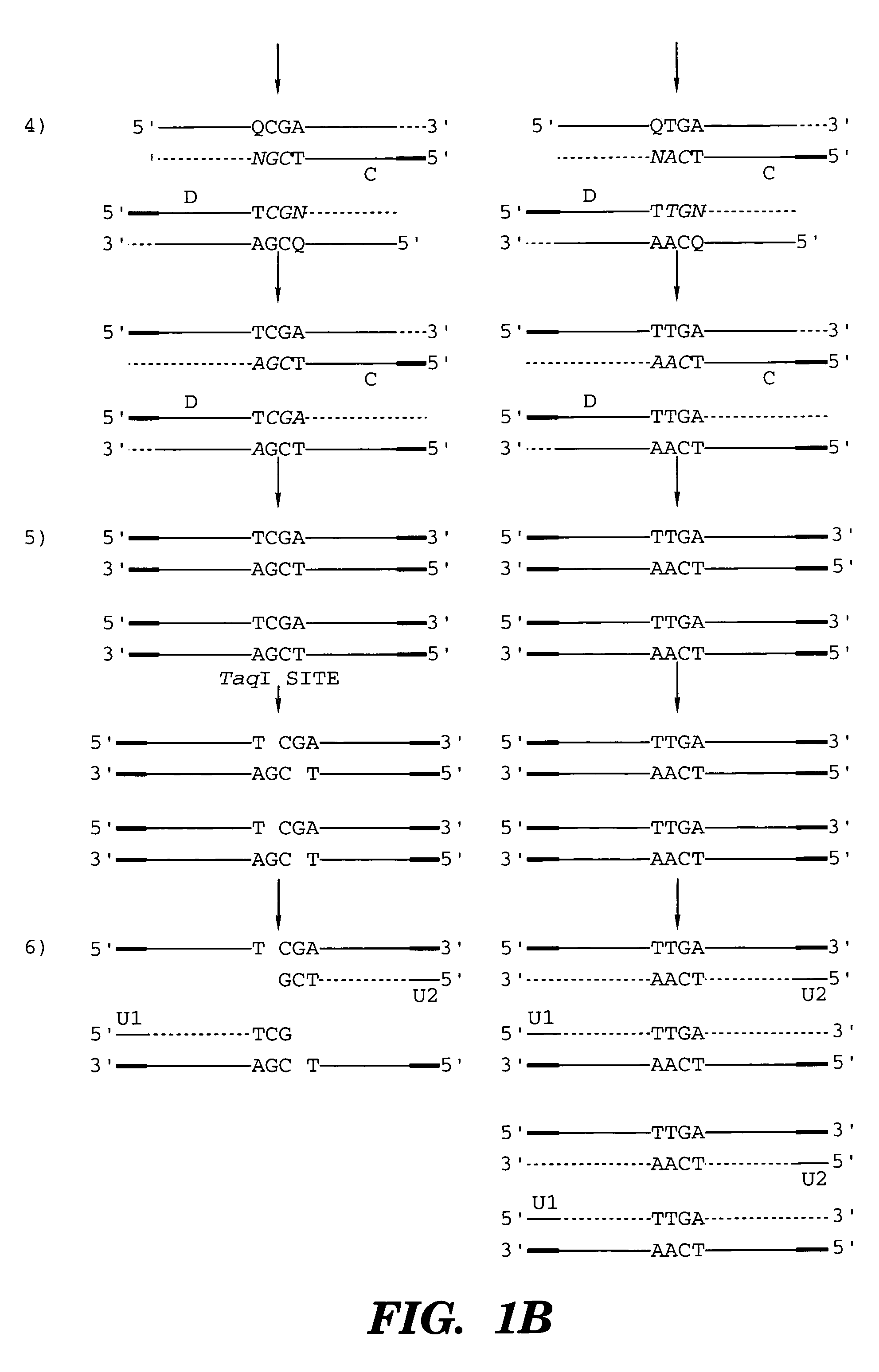
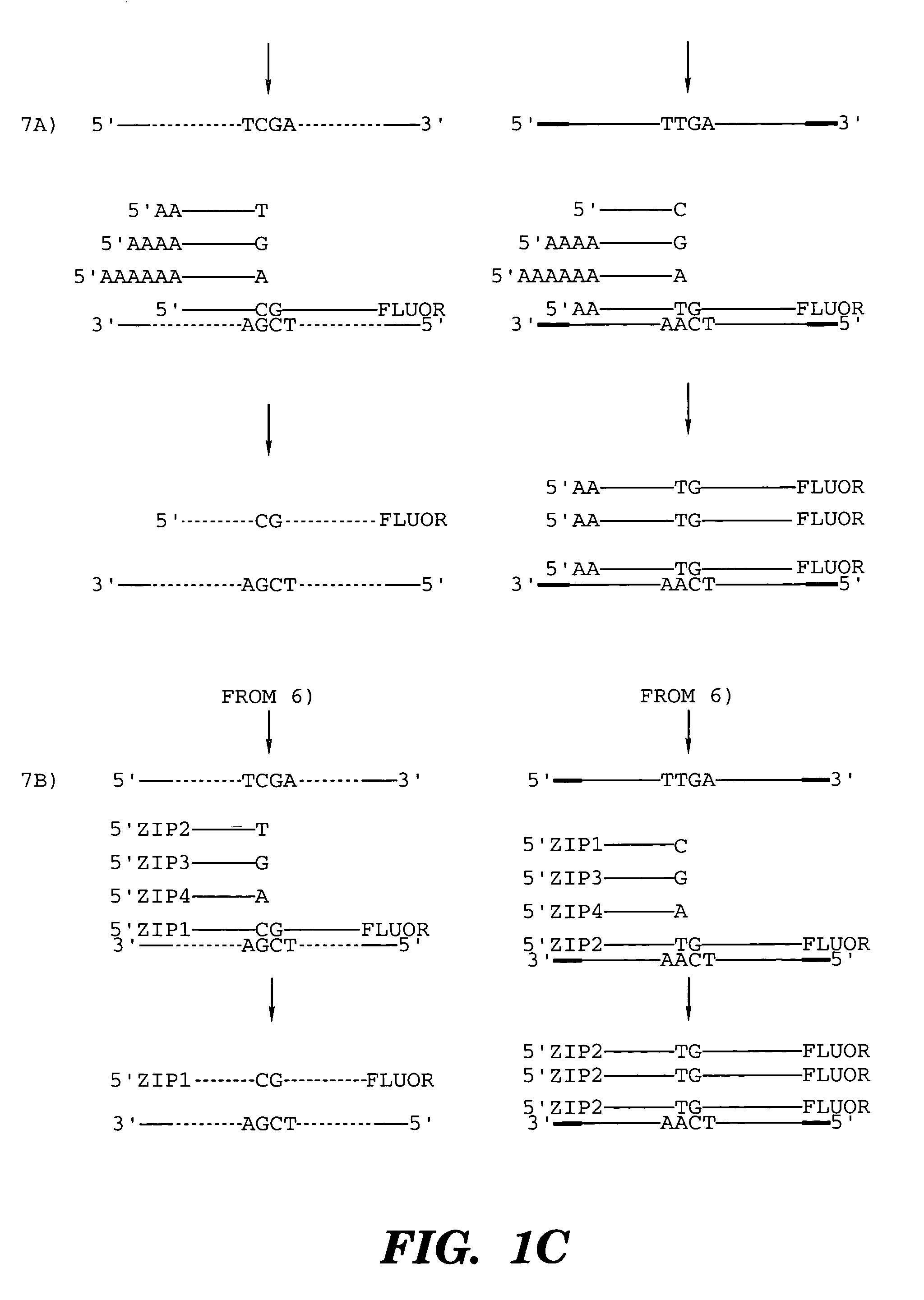
Coupled polymerase chain reaction-restriction-endonuclease digestion-ligase detection reaction process
InactiveUS7014994B1Sensitive highOptimizationSugar derivativesMicrobiological testing/measurementNucleotideWild type
The present invention provides a method for identifying one or more low abundance sequences differing by one or more single-base changes, insertions, or deletions, from a high abundance sequence in a plurality of target nucleotide sequences. The high abundance wild-type sequence is selectively removed using high fidelity polymerase chain reaction analog conversion, facilitated by optimal buffer conditions, to create a restriction endonuclease site in the high abundance wild-type gene, but not in the low abundance mutant gene. This allows for digestion of the high abundance DNA. Subsequently the low abundant mutant DNA is amplified and detected by the ligase detection reaction assay. The present invention also relates to a kit for carrying out this procedure.
Owner:LOUISIANA STATE UNIV +1
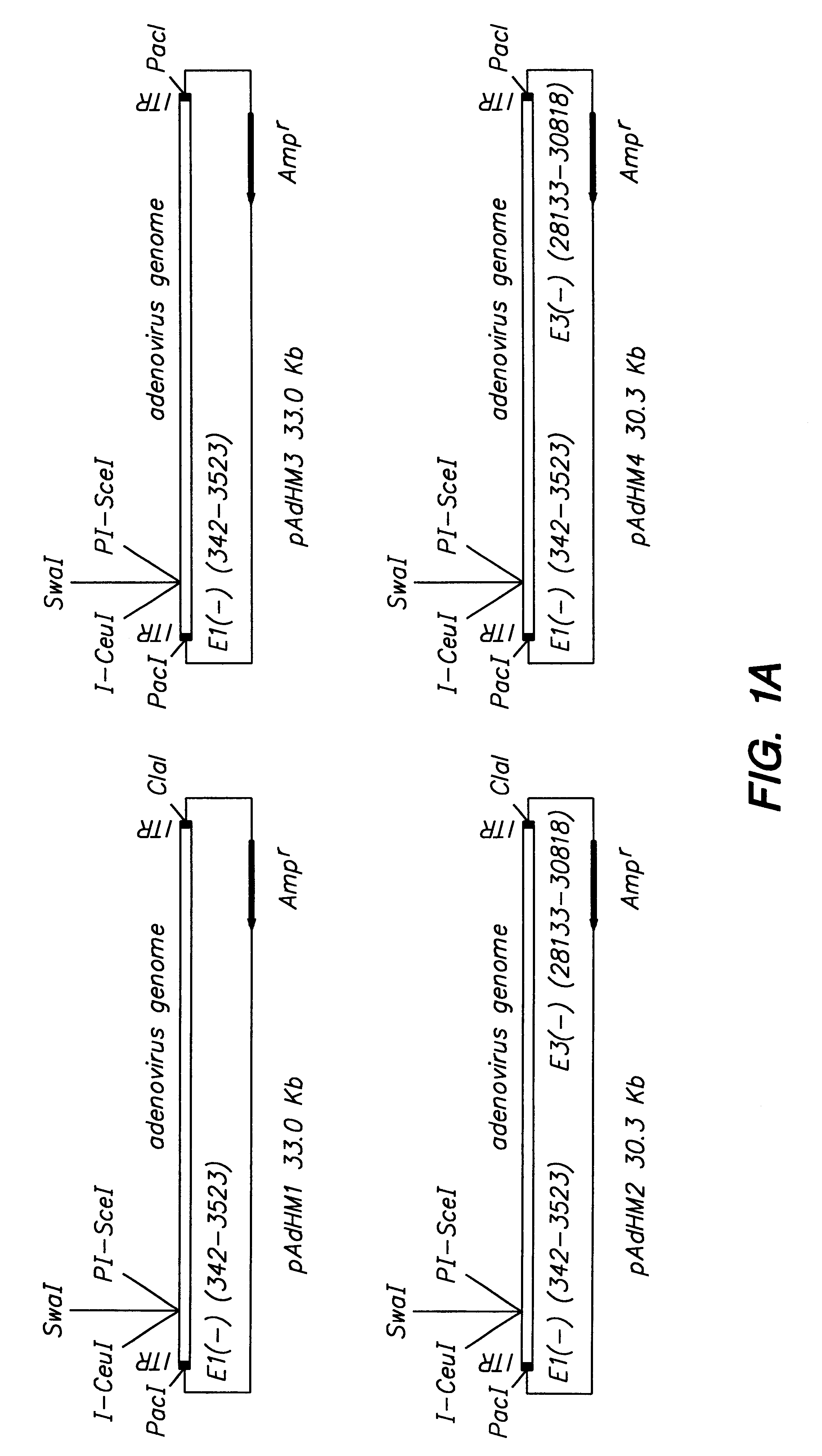
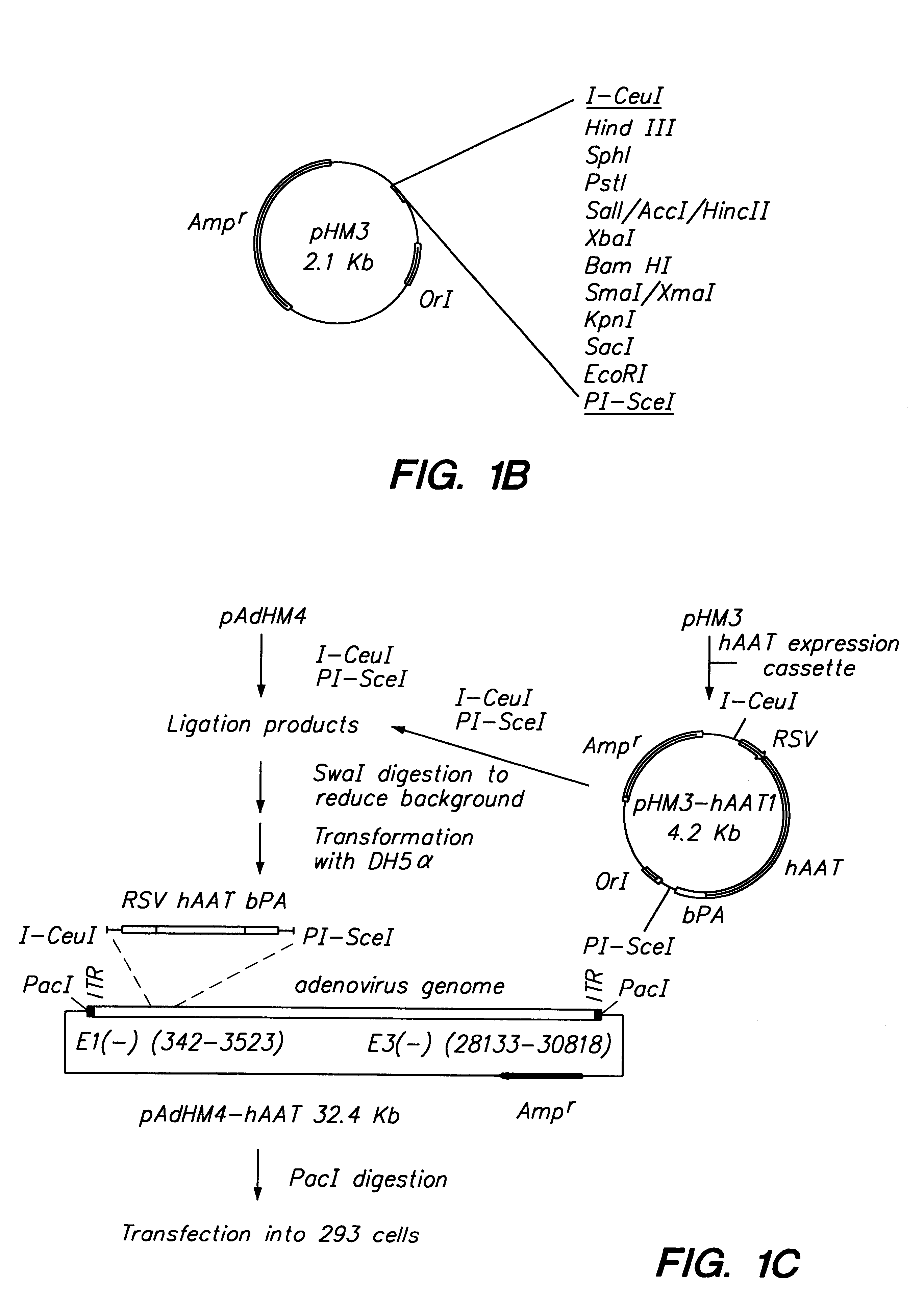
Adenoviral vector and methods for making and using the same
In vitro methods for making a recombinant adenoviral genome, as well as kits for practicing the same and the recombinant adenovirus vectors produced thereby, are provided. In the subject methods, the subject genomes are prepared from first and second vectors. The first vector includes an adenoviral genome having an E region deletion and three different, non-adenoviral restriction endonuclease sites located in the E region. The second vector is a shuttle vector and includes an insertion nucleic acid flanked by two of the three different non-adenoviral restriction endonucleases sites present in the first vector. Cleavage products are prepared from the first and second vectors using the appropriate restriction endonucleases. The resultant cleavage products are then ligated to produce the subject recombinant adenovirus genome. The subject adenoviral genomes find use in a variety of application, including as vectors for use in a variety of applications, including gene therapy.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV +1
In vitro recombination method
ActiveUS20070037197A1HydrolasesMicrobiological testing/measurementSingle-strand DNA-binding proteinSequence identity
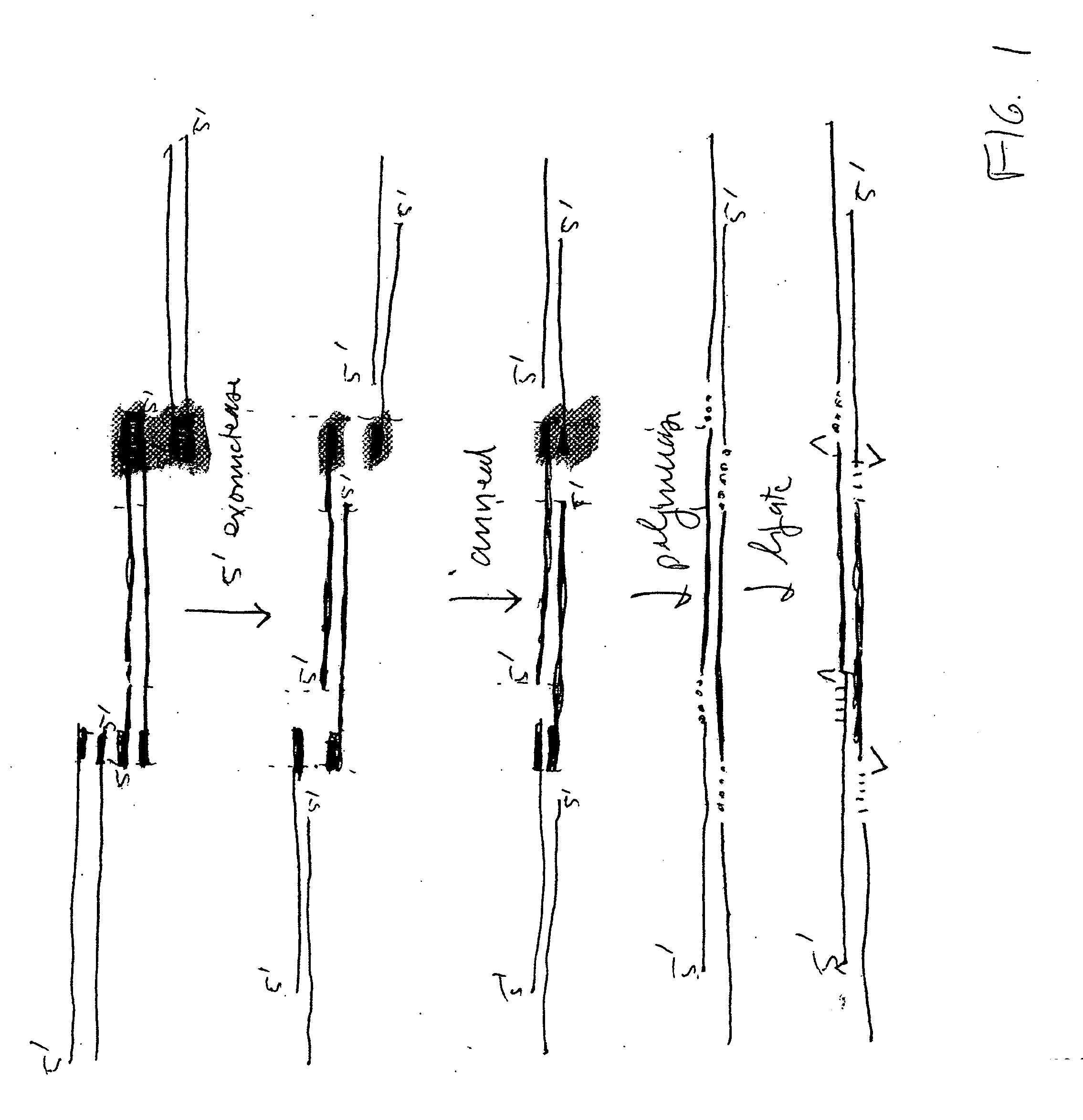
The present invention relates, e.g., to in vitro method, using isolated protein reagents, for joining two double stranded (ds) DNA molecules of interest, wherein the distal region of the first DNA molecule and the proximal region of the second DNA molecule share a region of sequence identity, comprising contacting the two DNA molecules in a reaction mixture with (a) a non-processive 5′ exonculease; (b) a single stranded DNA binding protein (SSB) which accelerates nucleic acid annealing; (c) a non strand-displacing DNA polymerase; and (d) a ligase, under conditions effective to join the two DNA molecules to form an intact double stranded DNA molecule, in which a single copy of the region of sequence identity is retained. The method allows the joining of a number of DNA fragments, in a predetermined order and orientation, without the use of restriction enzymes.
Owner:TELESIS BIO INC
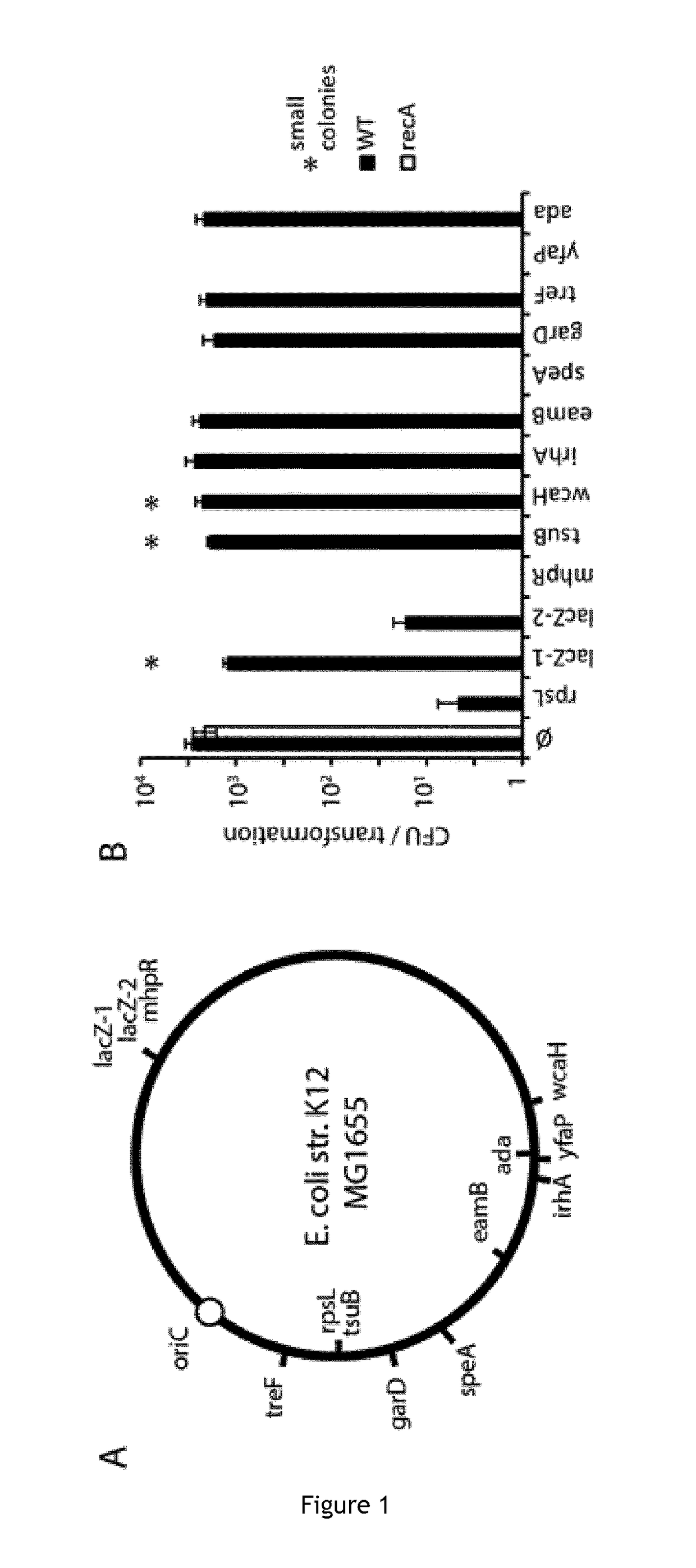
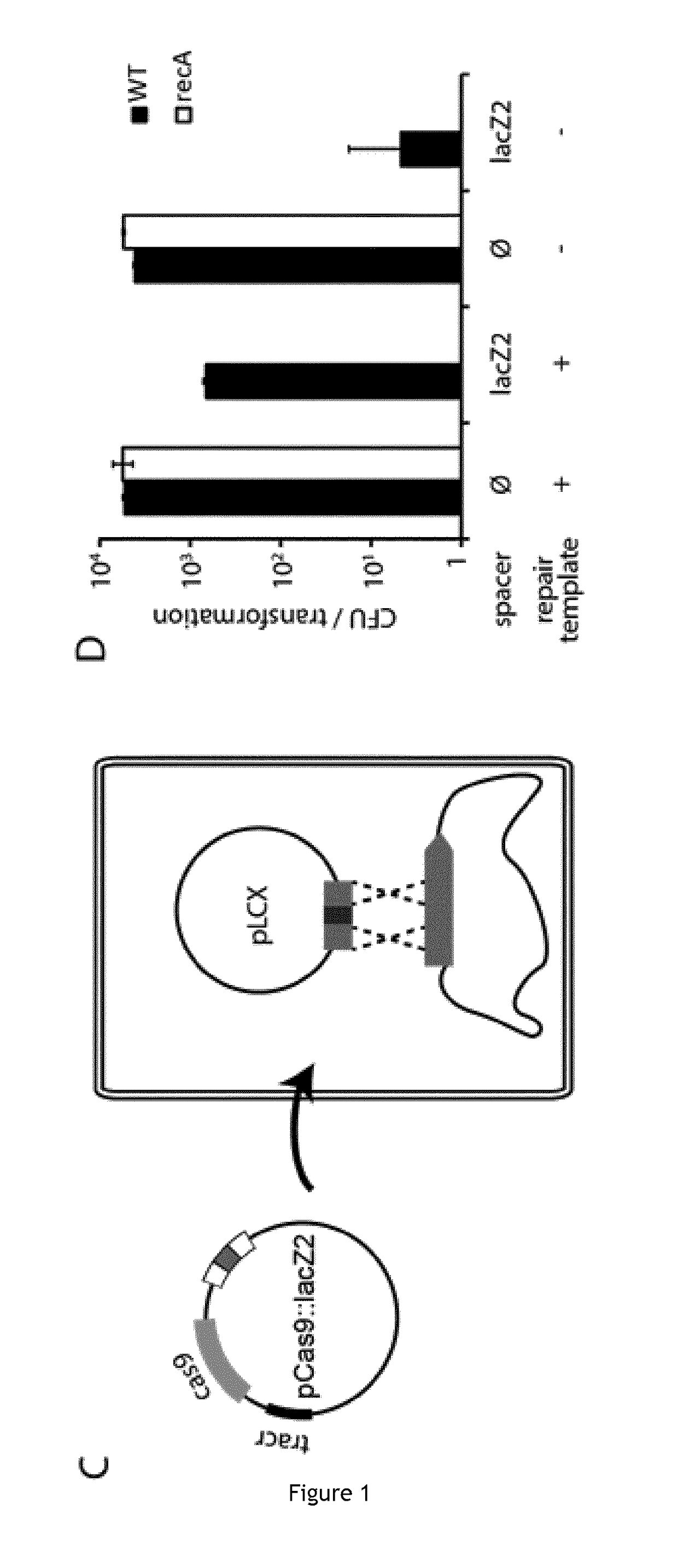
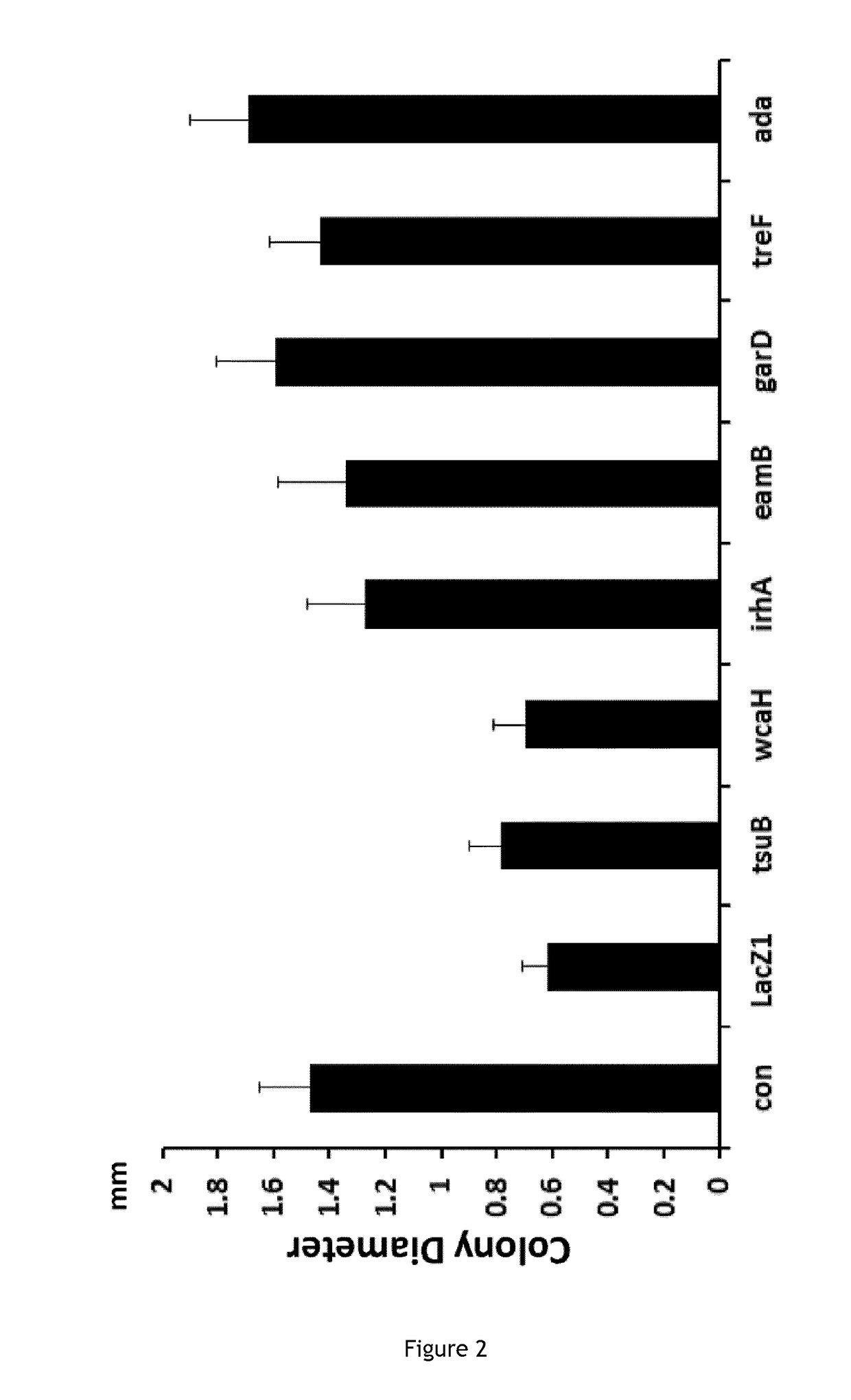
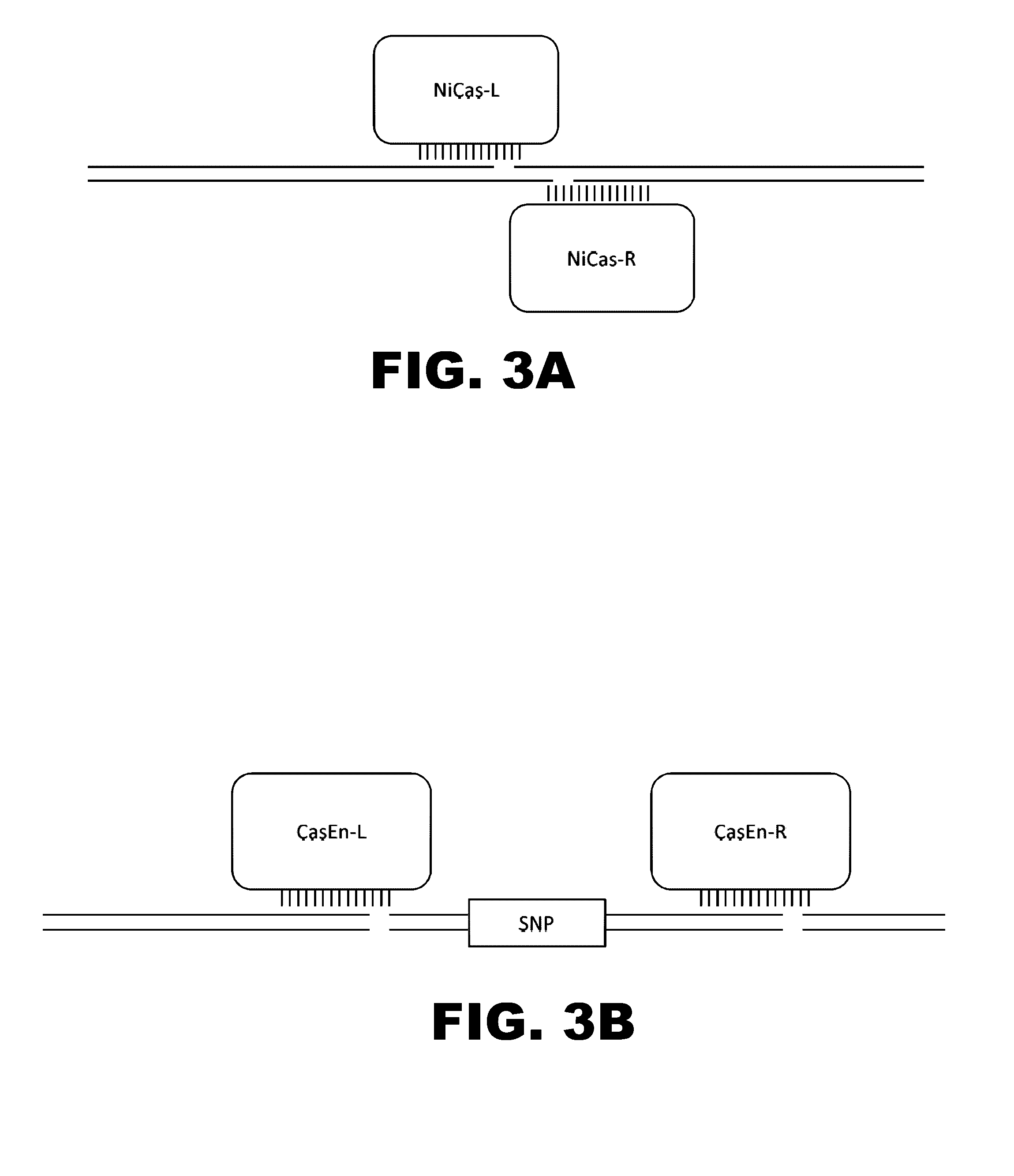
Improving sequence-specific antimicrobials by blocking DNA repair
The invention relates to the improvement of endonuclease-based antimicrobials by blocking DNA repair of double-strand break(s) (DSB(s)) in prokaryotic cells. In this respect, the invention especially concerns a method involving blocking DNA repair after a nucleic acid has been submitted to DSB, in particular by a Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) associated programmable double-strand endonuclease. The invention particularly relates to the use of an exogenous molecule that inhibits DNA repair, preferably a protein that binds to the ends of the double-stranded break to block DSB repair. The invention also relates to vectors, particularly phagemids and plasmids, comprising nucleic acids encoding nucleases and Gam proteins, and a pharmaceutical composition and a product containing these vectors and their application.
Owner:INST PASTEUR +1
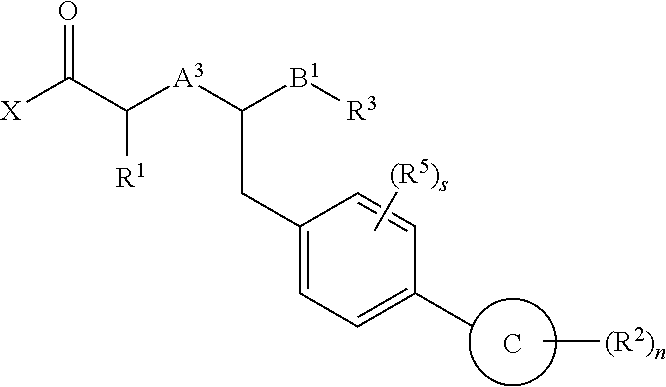
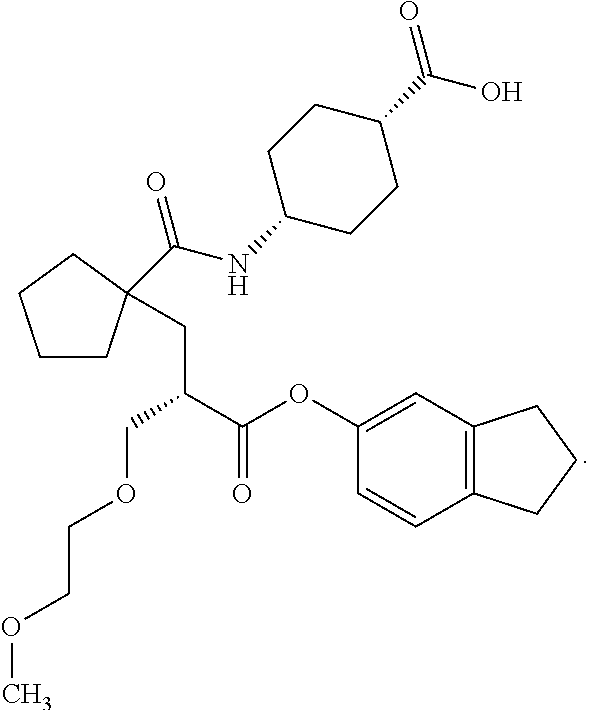
Method of treating contrast-induced nephropathy
InactiveUS20120122844A1Treating and preventing and ameliorating contrast-induced nephropathyImprove the level ofBiocideOrganic chemistryNephrosisActive agent
The present invention provides the use of a neutral endopeptidase inhibitor, in the manufacture of a medicament for the treatment, amelioration and / or prevention of contrast-induced nephropathy. The invention also relates to the use of a compound of Formula I:wherein R1, R2, R3, R5, X, A3, B1, s and n are defined herein, for the treatment, amelioration and / or prevention of contrast-induced nephropathy. The present invention further provides a combination of pharmacologically active agents for use in the treatment, amelioration and / or prevention of contrast-induced nephropathy.
Owner:NOVARTIS AG
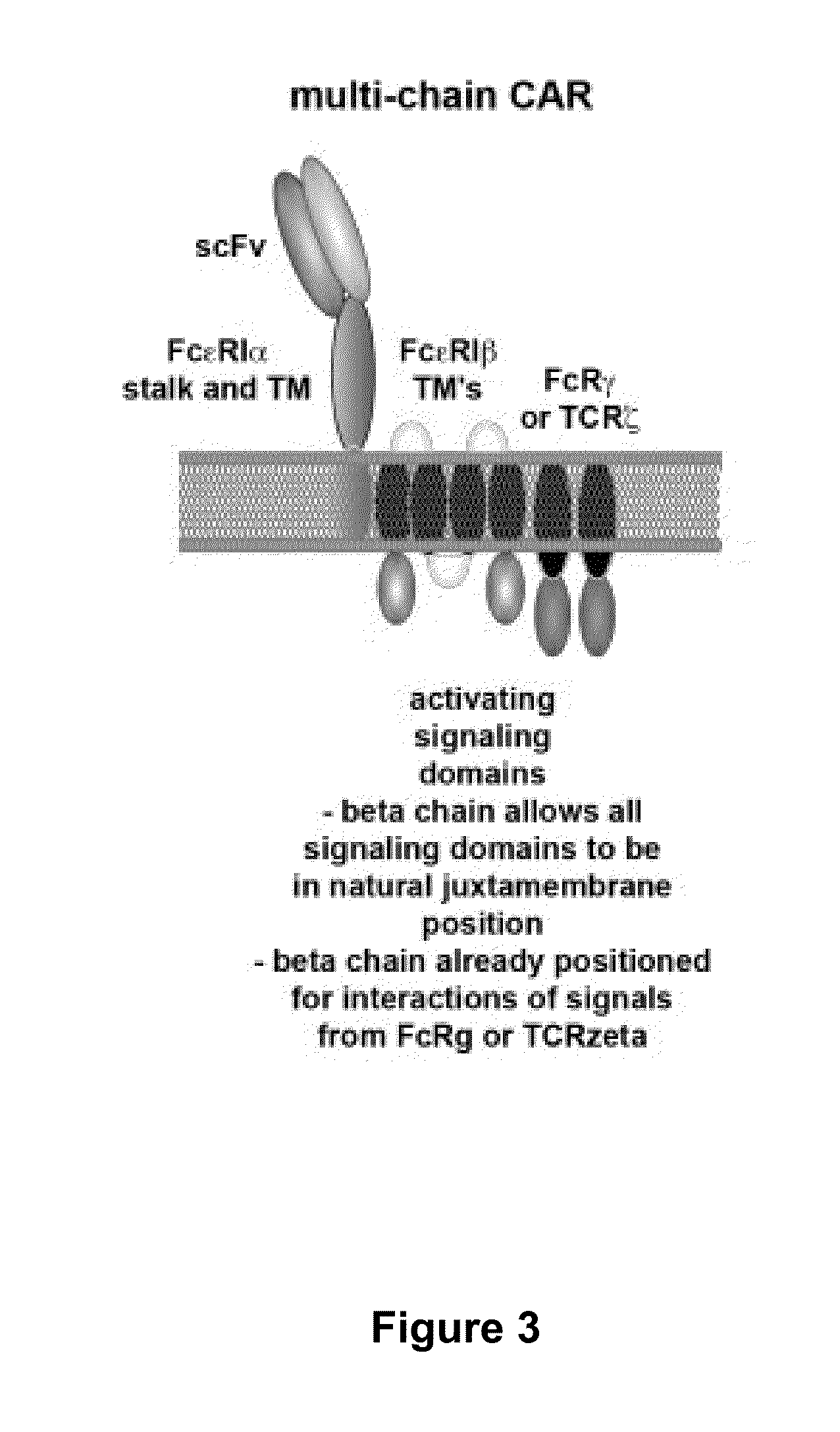
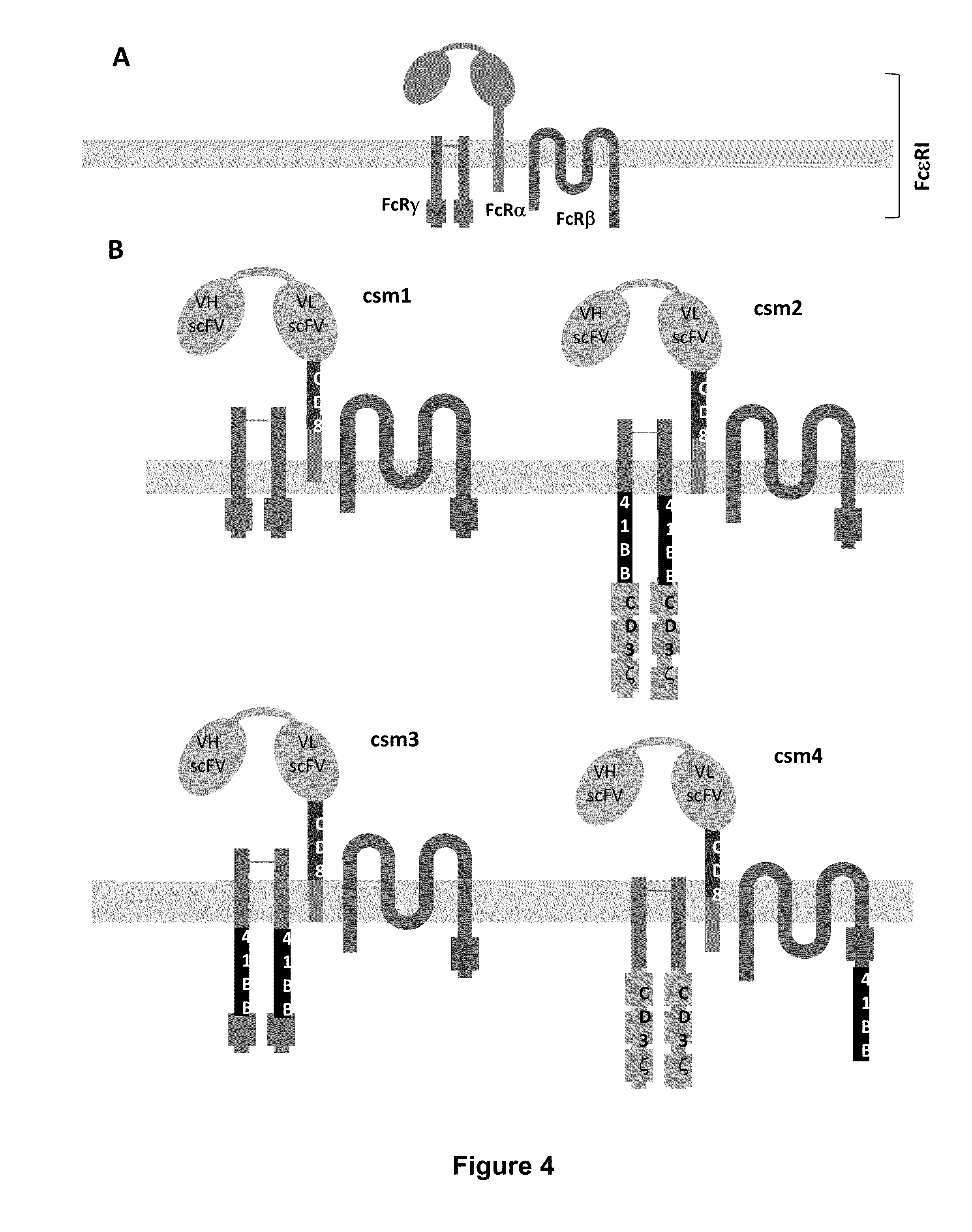
Methods for engineering t cells for immunotherapy by using rna-guided cas nuclease system
ActiveUS20160184362A1Accurate modificationHydrolasesGenetically modified cellsInfected cellAntigen receptors
The present invention relates to methods of developing genetically engineered, preferably non-alloreactive T-cells for immunotherapy. This method involves the use of RNA-guided endonucleases, in particular Cas9 / CRISPR system, to specifically target a selection of key genes in T-cells. The engineered T-cells are also intended to express chimeric antigen receptors (CAR) to redirect their immune activity towards malignant or infected cells. The invention opens the way to standard and affordable adoptive immunotherapy strategies using T-Cells for treating cancer and viral infections.
Owner:CELLECTIS SA
Methods for in vitro joining and combinatorial assembly of nucleic acid molecules
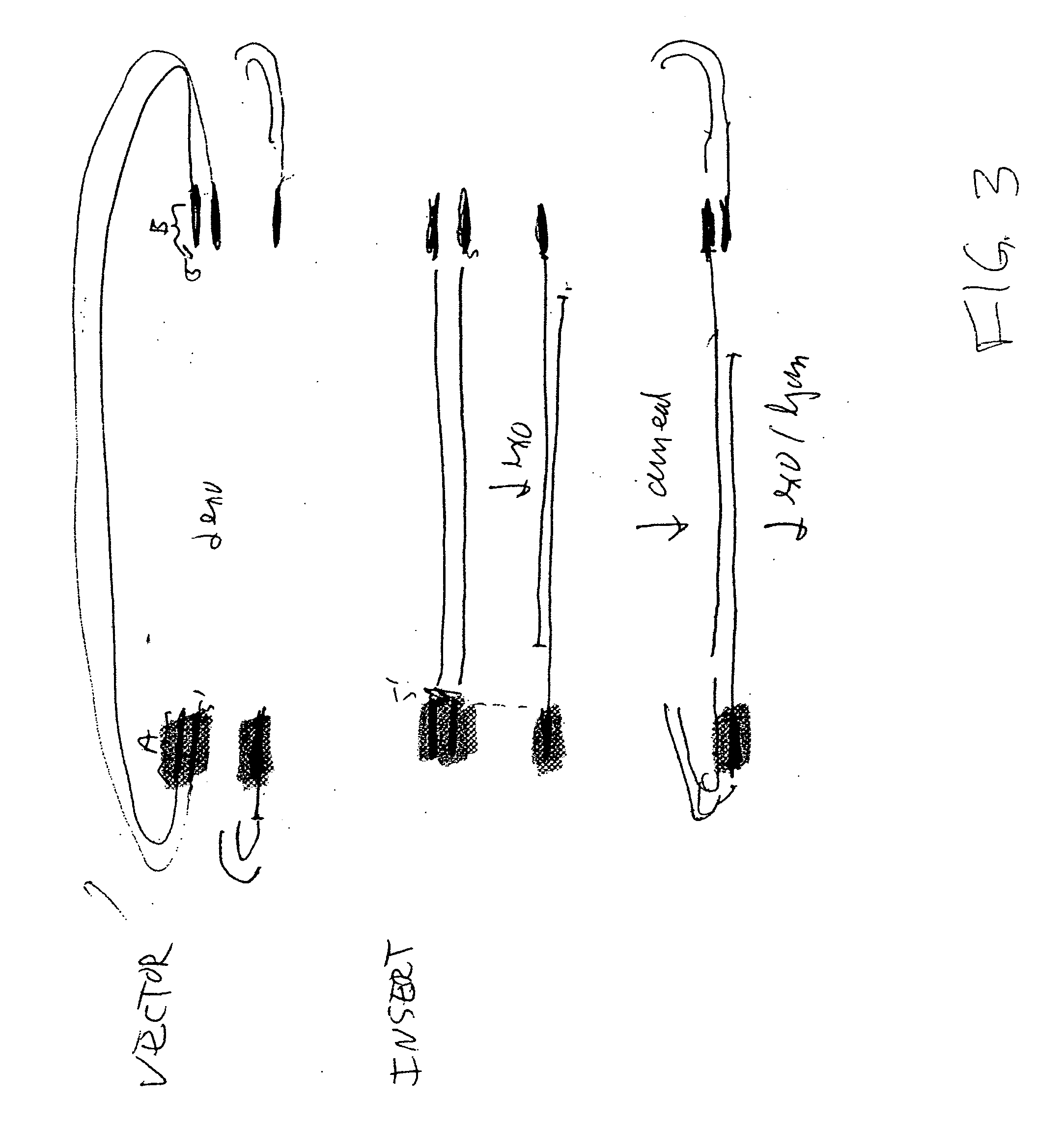
The present invention relates to methods of joining two or more double-stranded (ds) or single-stranded (ss) DNA molecules of interest in vitro, wherein the distal region of the first DNA molecule and the proximal region of the second DNA molecule of each pair share a region of sequence identity. The method allows the joining of a large number of DNA fragments, in a predetermined order and orientation, without the use of restriction enzymes. It can be used, e.g., to join synthetically produced sub-fragments of a gene or genome of interest. Kits for performing the method are also disclosed. The methods of joining DNA molecules may be used to generate combinatorial libraries useful to generate, for example, optimal protein expression through codon optimization, gene optimization, and pathway optimization.
Owner:TELESIS BIO INC
FEN endonucleases
InactiveUS7122364B1Improve performanceA large amountHydrolasesMicrobiological testing/measurementPolymerase LNucleic acid sequencing
The present invention provides novel cleavage agents and polymerases for the cleavage and modification of nucleic acid. The cleavage agents and polymerases find use, for example, for the detection and characterization of nucleic acid sequences and variations in nucleic acid sequences. In some embodiments, the 5′ nuclease activity of a variety of enzymes is used to cleave a target-dependent cleavage structure, thereby indicating the presence of specific nucleic acid sequences or specific variations thereof.
Owner:GEN PROBE INC
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Method and apparatus for identifying classifying or quantifying DNA sequences in a sample without sequencing
InactiveUS6141657ARapid and economical and quantitative and precise determination and classificationSufficient discrimination and resolutionData processing applicationsDigital data processing detailsSequence databaseDNA Sequence Databases
This invention provides methods by which biologically derived DNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information.
Owner:CURAGEN CORP
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
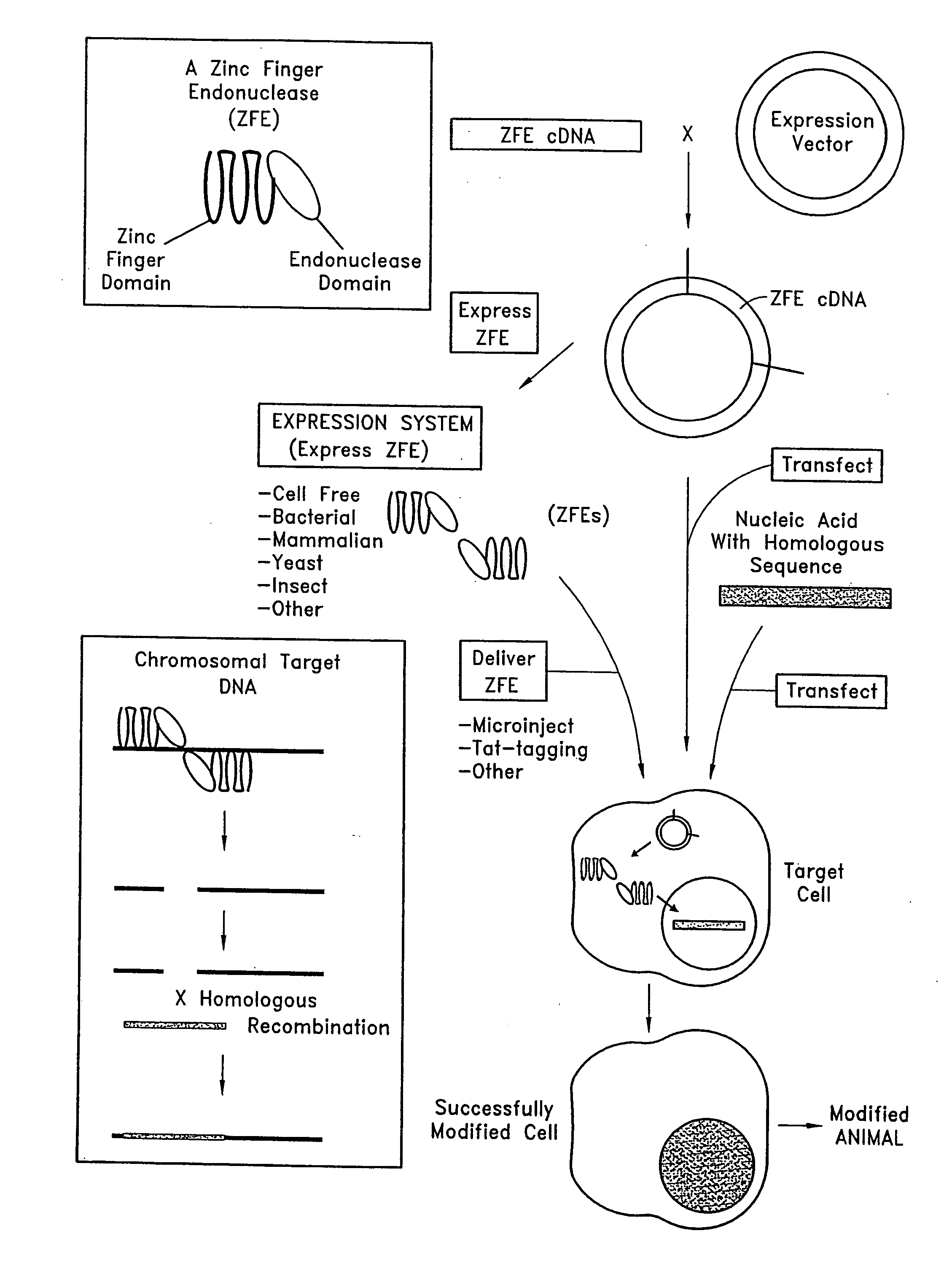
Methods and compositions for using zinc finger endonucleases to enhance homologous recombination
Owner:SANGAMO BIOSCIENCES INC
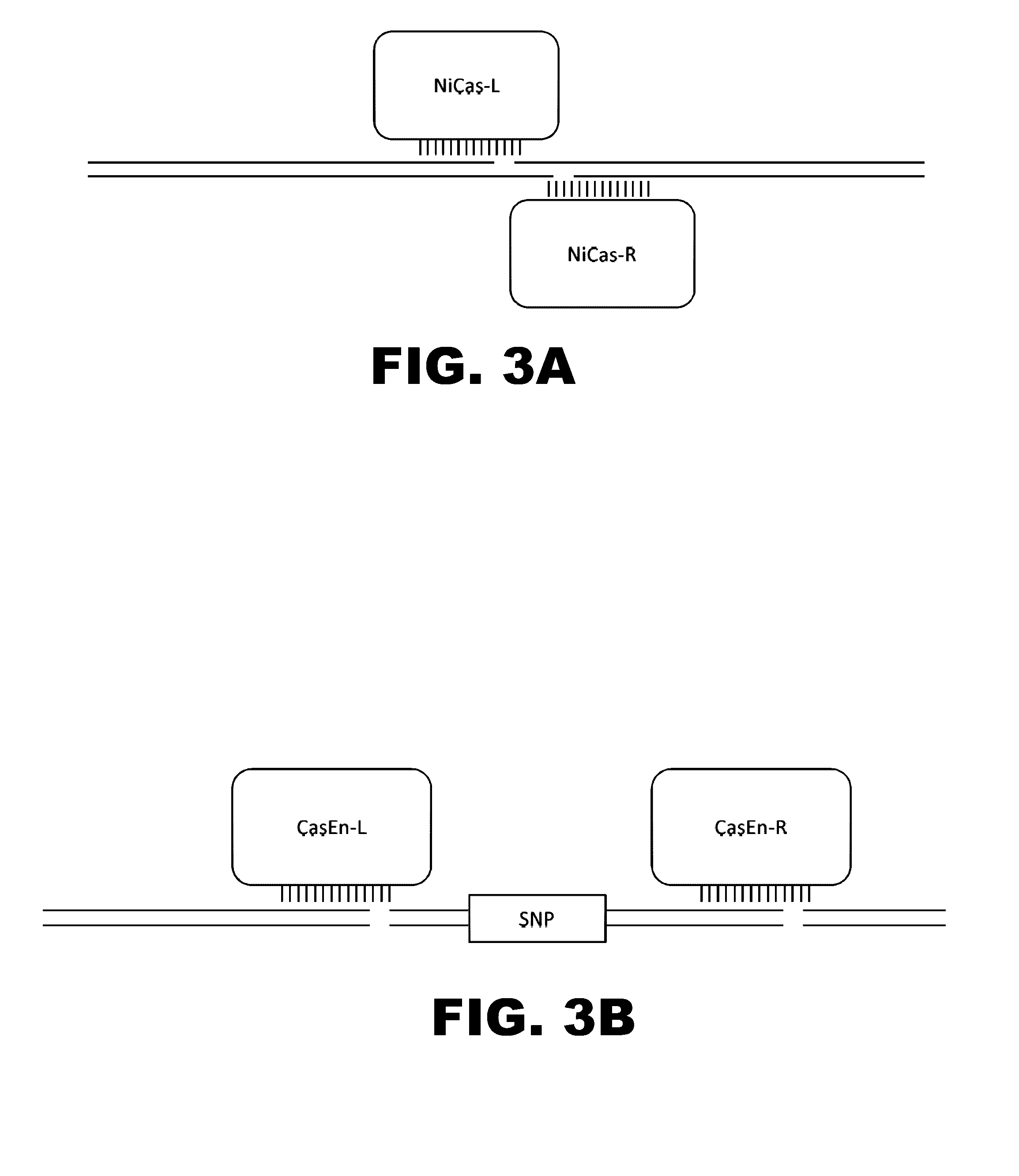
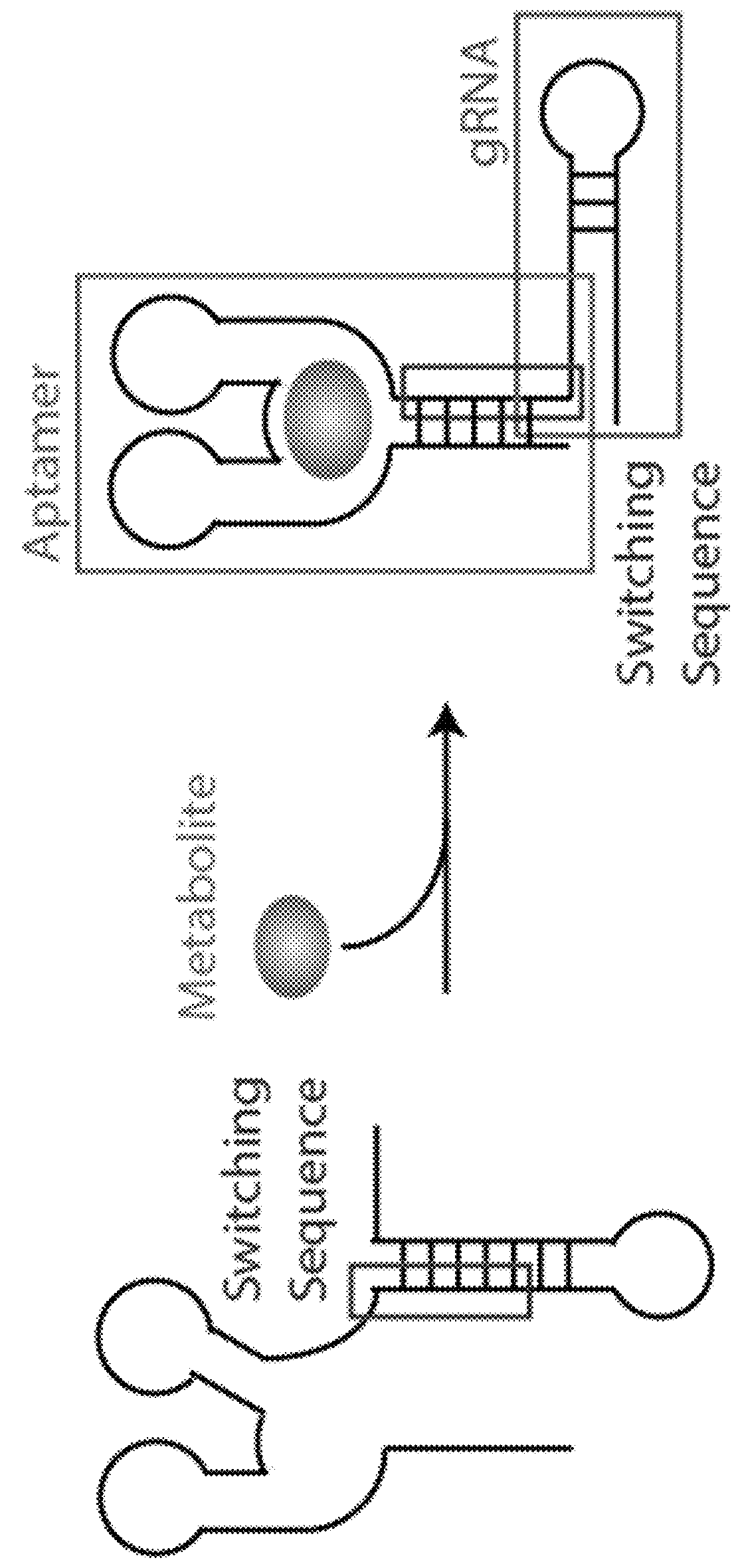
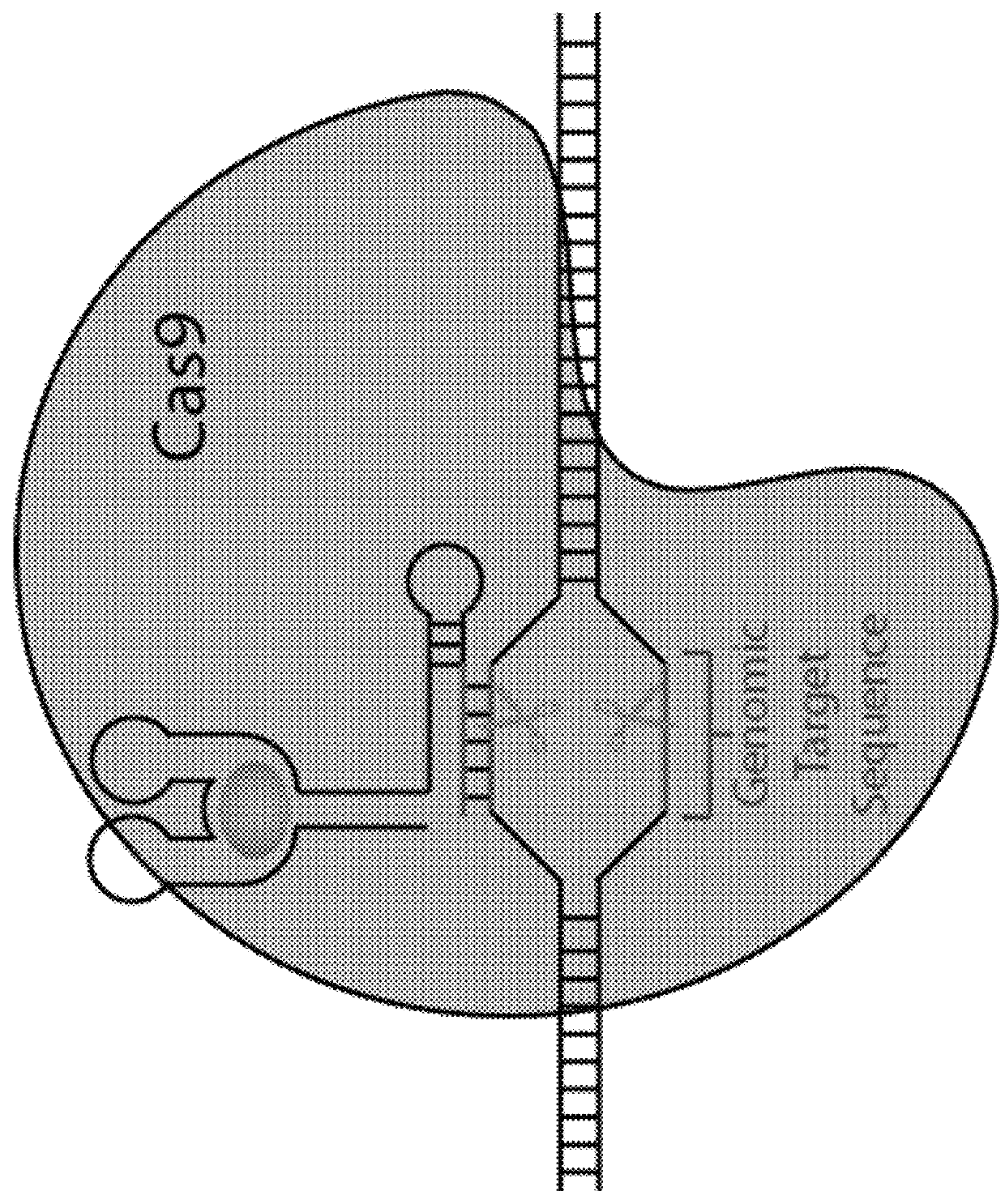
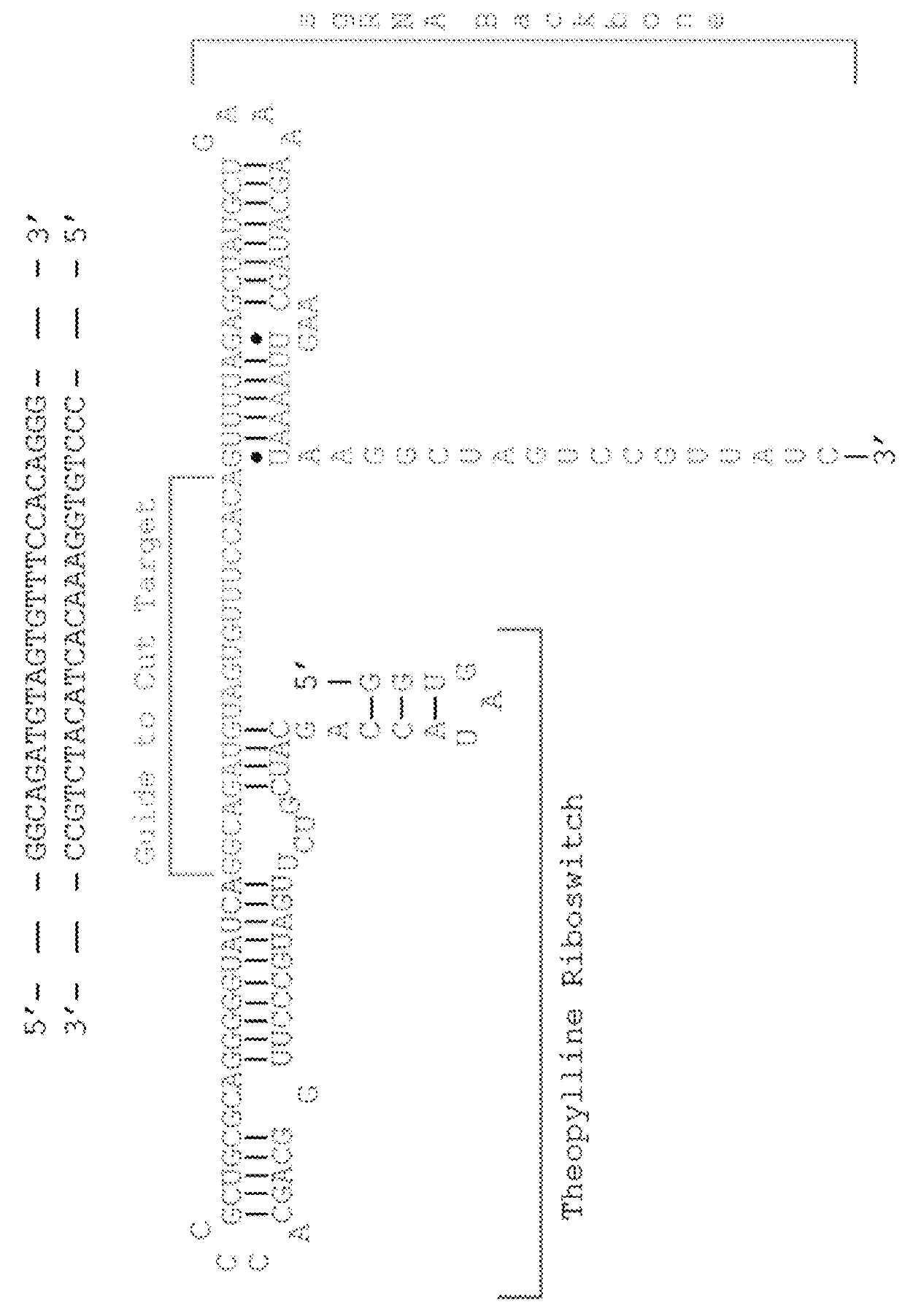
Switchable grnas comprising aptamers
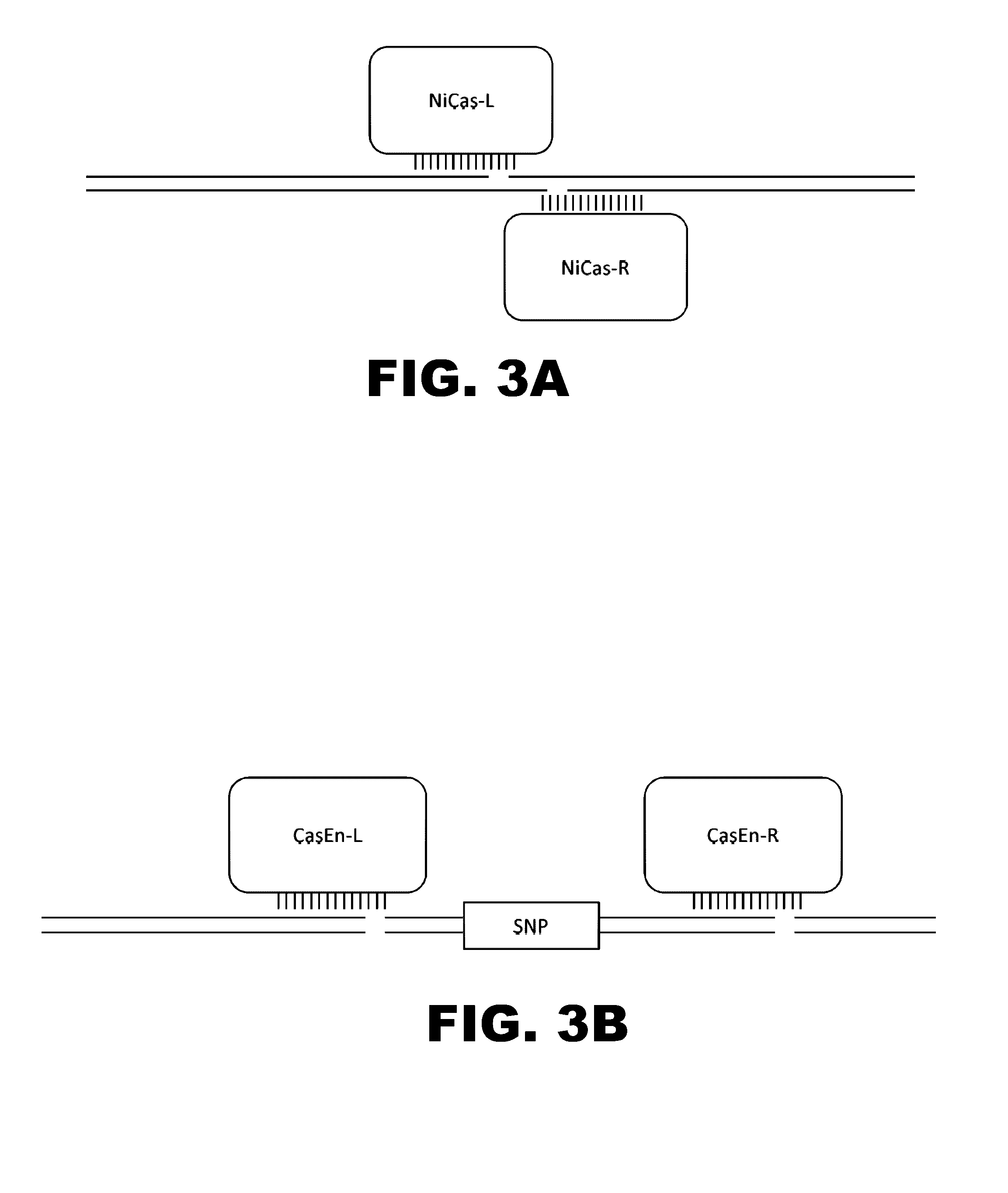
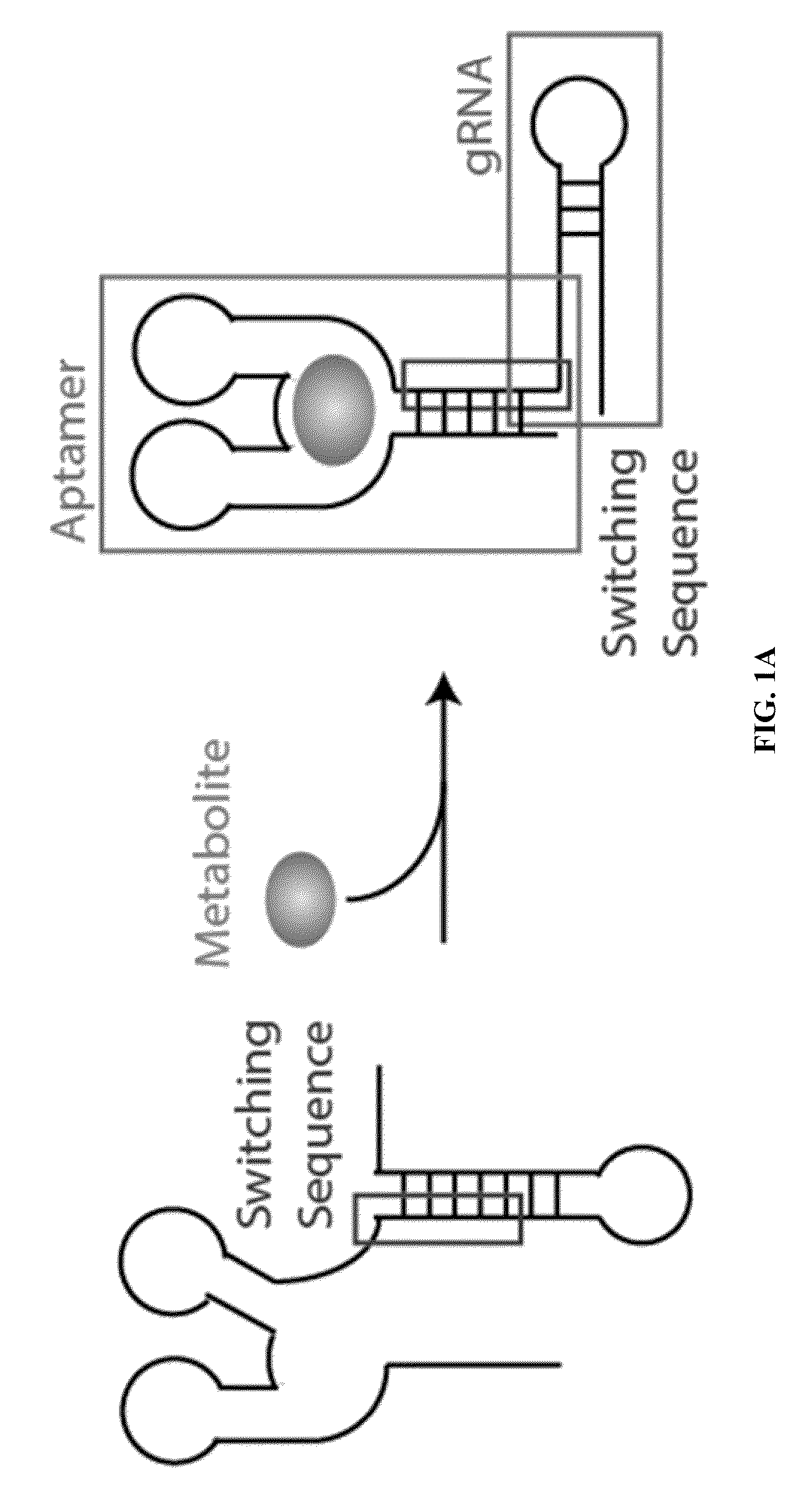
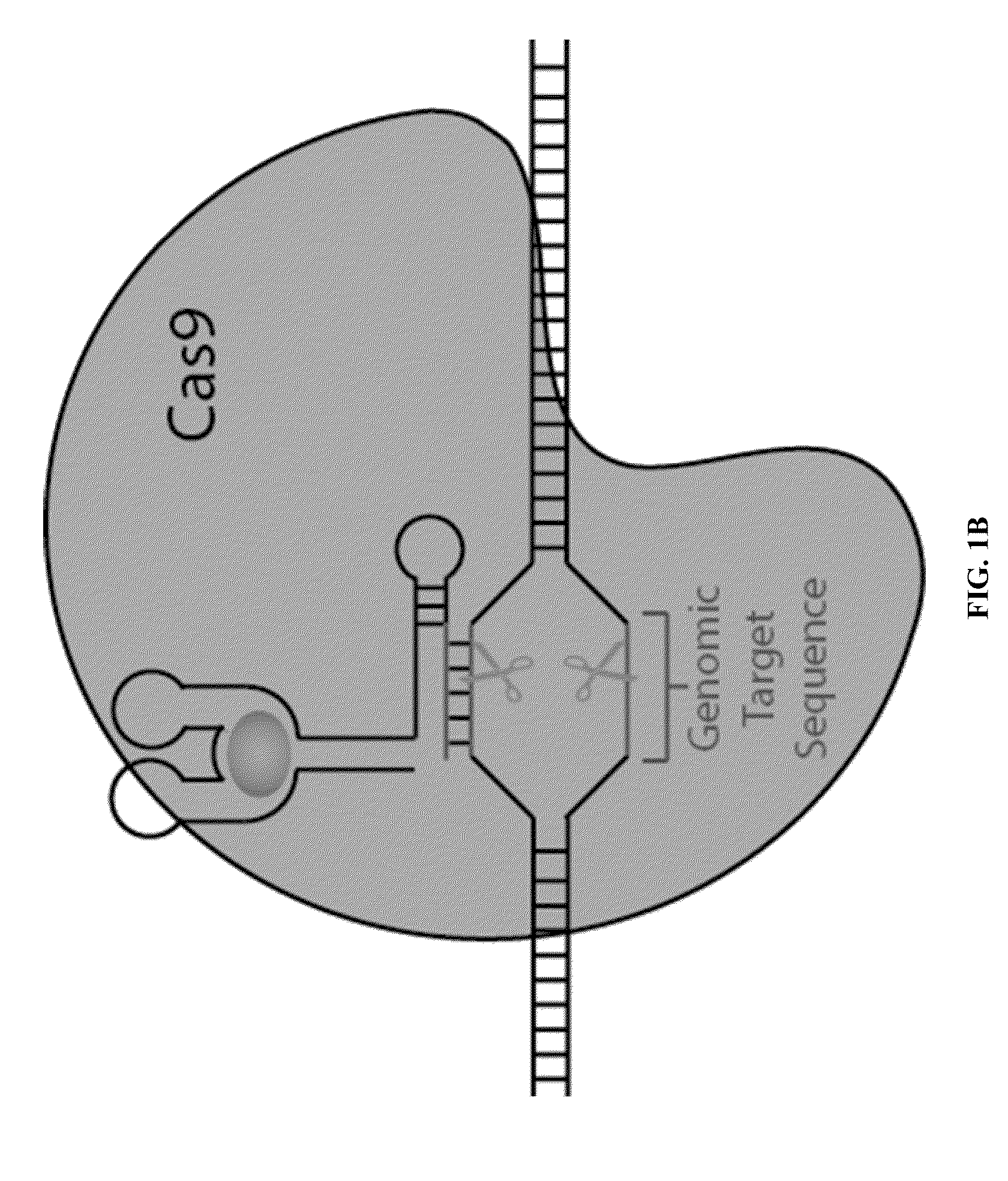
ActiveUS20150071900A1Reduce the possibilityHydrolasesPeptide/protein ingredientsBiologyDNA Endonuclease
Some aspects of this disclosure provide compositions, methods, systems, and kits for controlling the activity and / or improving the specificity of RNA-programmable endonucleases, such as Cas9. For example, provided are guide RNAs (gRNAs) that are engineered to exist in an “on” or “off” state, which control the binding and hence cleavage activity of RNA-programmable endonucleases.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
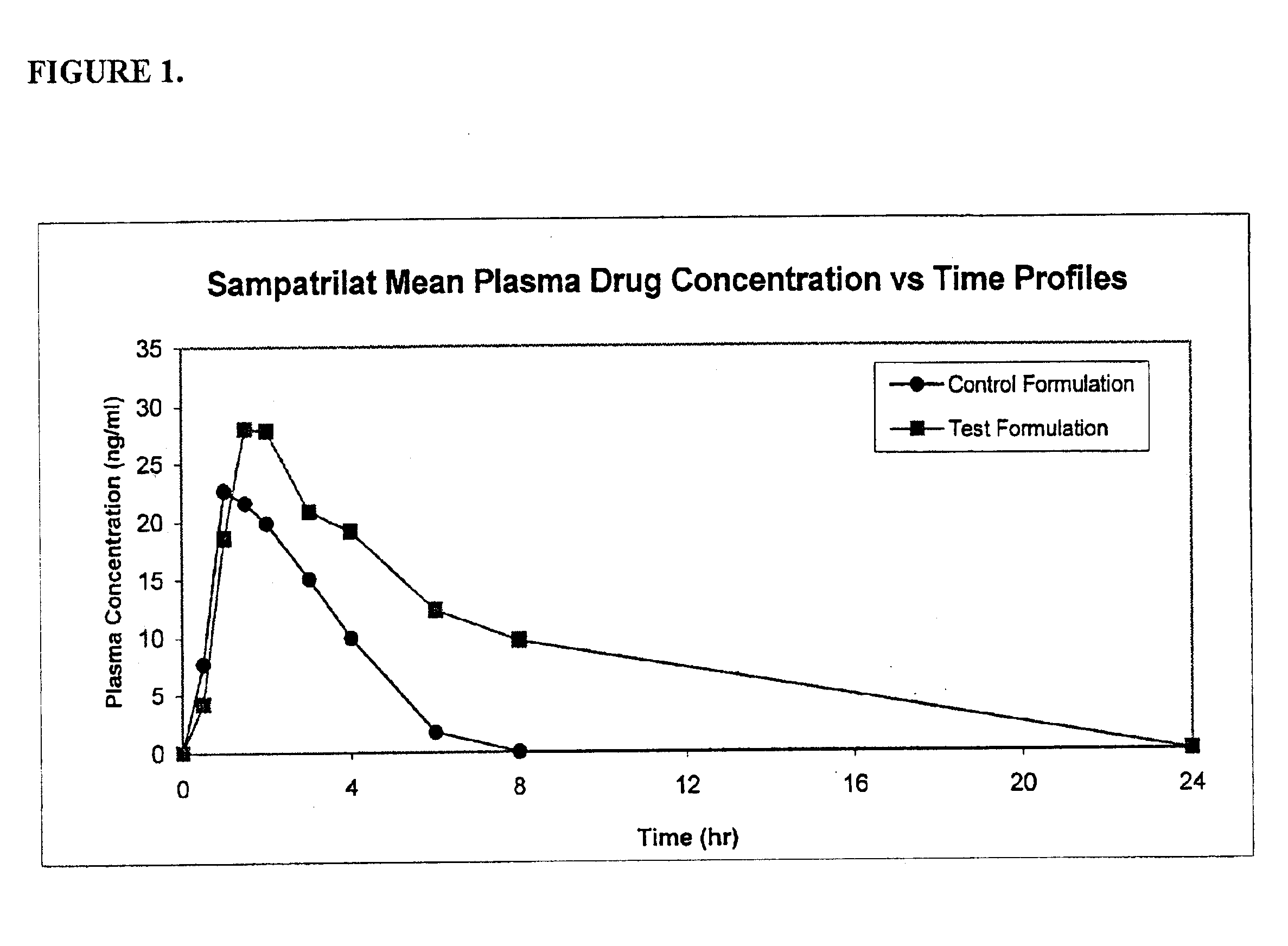
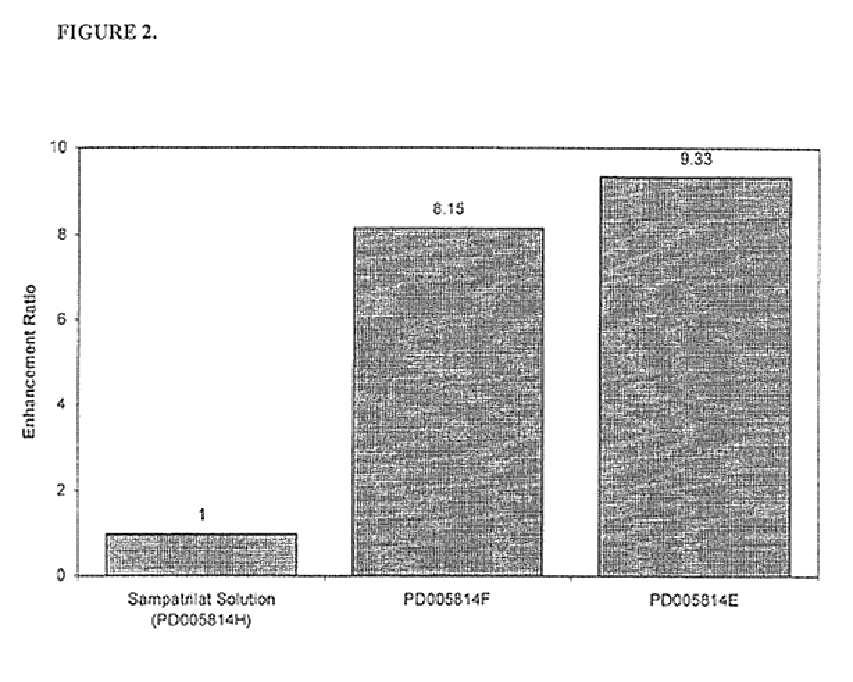
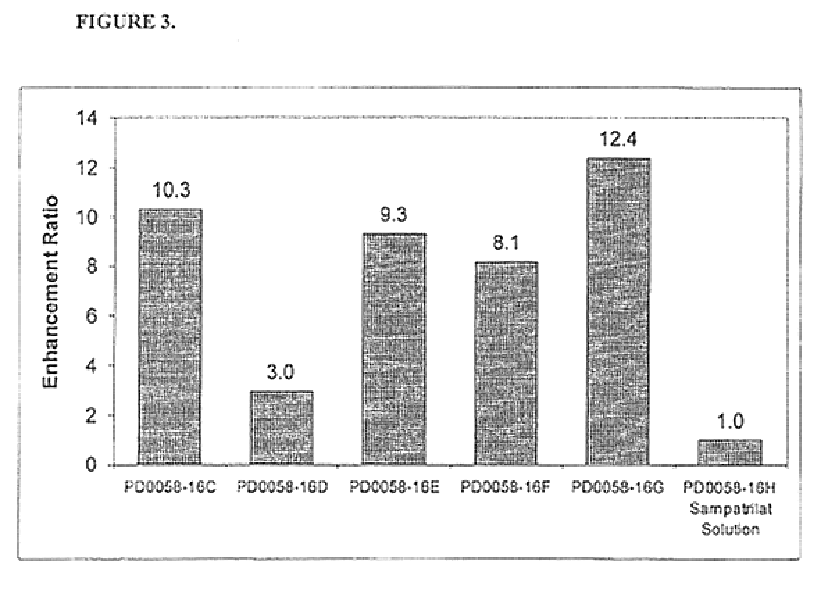
Pharmaceutical compositions including ACE/NEP inhibitors and bioavailability enhancers
InactiveUS6890918B2Enhance the oral bioabsorption of saidBiocideAngiotensinsAngiotensin-converting enzymeOrganic acid
A pharmaceutical composition comprising an inhibitor of angiotensin converting enzyme and neutral endopeptidase, such as sampatrilat, and at least one bioavailability enhancer such as an organic acid, e.g., ascorbic acid. Such a composition has improved systemic bioavailability.
Owner:SUPERNUS PHARM INC
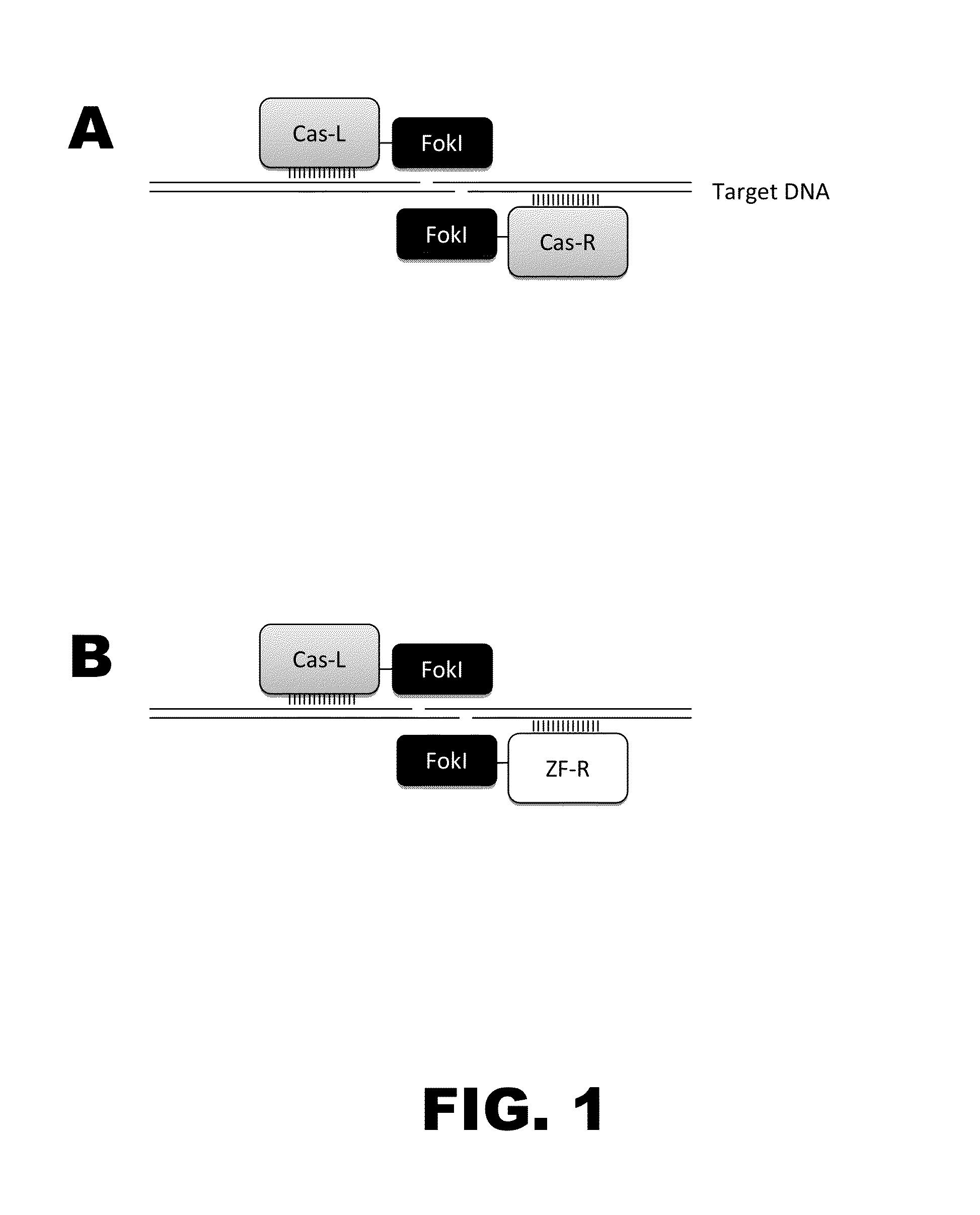
Chimeric Endonucleases and Uses Thereof
InactiveUS20120324603A1Reduce contentFacilitate homologous recombinationFungiFusion with DNA-binding domainBiotechnologyHeterologous
The invention relates to chimeric endonucleases, comprising an endonuclease and a heterologous DNA binding domain, as well as methods of targeted integration, targeted deletion or targeted mutation of polynucleotides using chimeric endonucleases.
Owner:BASF PLANT SCI GMBH
Switchable gRNAs comprising aptamers
Some aspects of this disclosure provide compositions, methods, systems, and kits for controlling the activity and / or improving the specificity of RNA-programmable endonucleases, such as Cas9. For example, provided are guide RNAs (gRNAs) that are engineered to exist in an “on” or “off” state, which control the binding and hence cleavage activity of RNA-programmable endonucleases.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
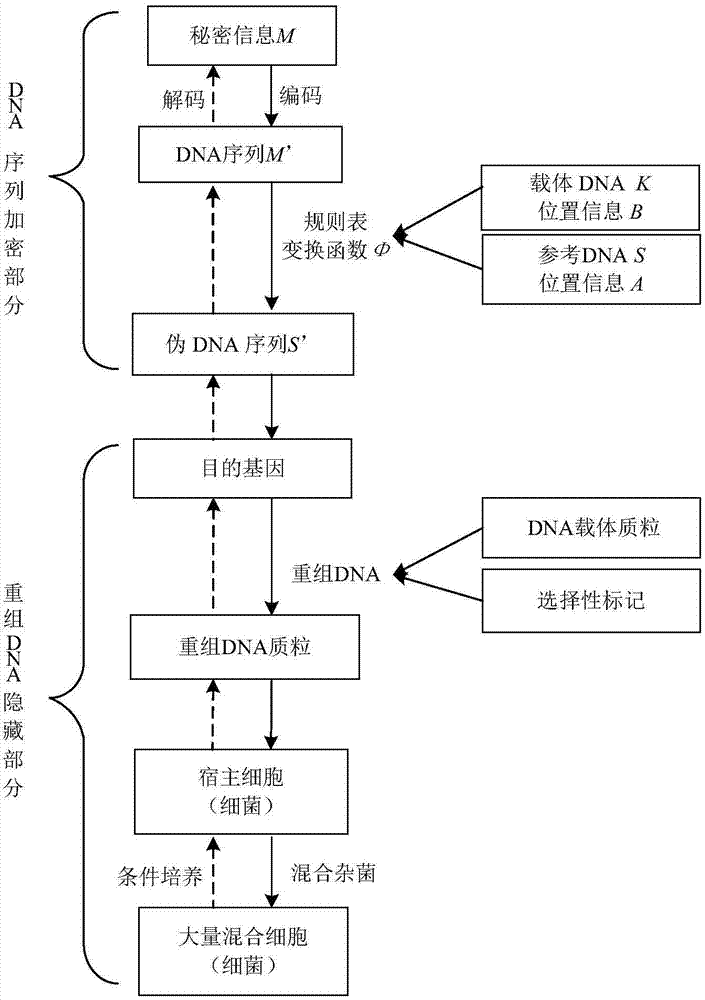
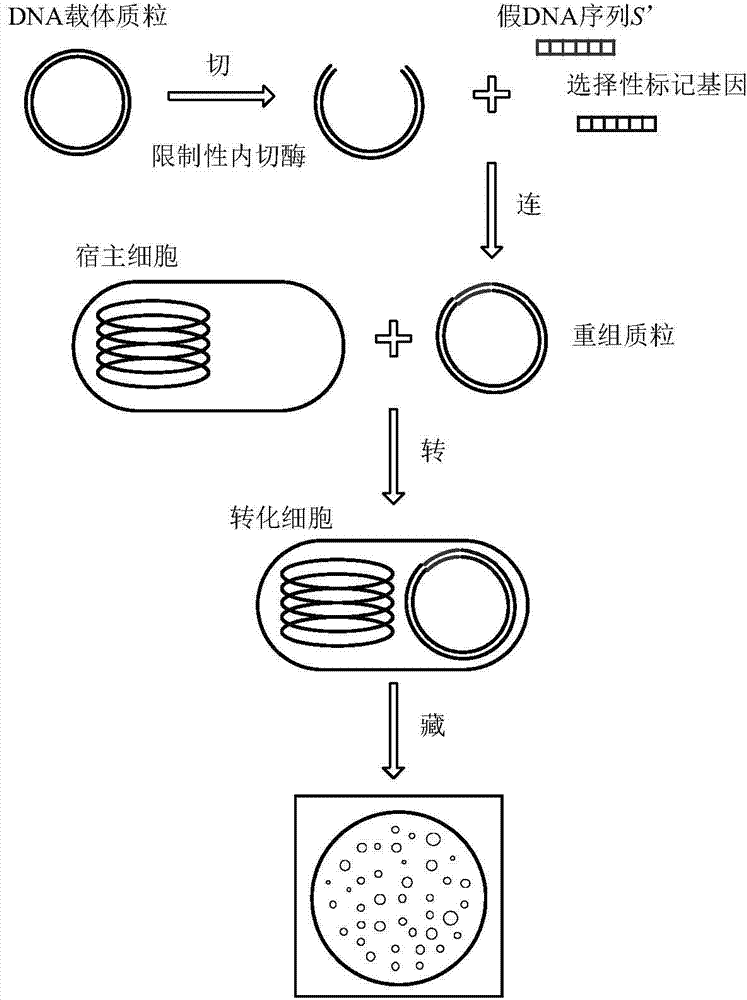
Recombinant DNA technology based information encrypting and hiding method and application
InactiveCN104734848AMeet safety requirementsIncrease the difficulty of crackingVector-based foreign material introductionSecuring communicationPlasmid VectorDNA fragmentation
The invention provides a recombinant DNA technology based information encrypting and hiding method and an application. Firstly, a plain text is coded into a DNA sequence, preprocessing is carried out on the DNA sequence by adopting the encryption algorithm, and a pseudo-DNA segment is generated; and then the pseudo-DNA segment and a marker gene segment are connected to a DNA plasmid vector by using the restriction enzyme and the ligase, and an obtained recombinant plasmid is further hided into a germ body; finally, a cell which contains confidential information is hided into a large number of irrelevant pseudo-cells, and the hided cells can be screened out by the selective cultivation. The encrypting and hiding method is applied to the authentication and signature technology. Encrypting, hiding and transmitting of the confidential information are achieved, and the cracking difficulty of a system is enhanced.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
Method for generating t-cells compatible for allogenic transplantation
InactiveUS20170016025A1Polypeptide with localisation/targeting motifImmunoglobulin superfamilyCIITAAuto immune disease
The present invention pertains to engineered T-cells, method for their preparation and their use as medicament, particularly for immunotherapy. The engineered T-cells of the invention are characterized in that the expression of beta 2-microglobulin (B2M) and / or class II major histocompatibility complex transactivator (CIITA) is inhibited, e.g., by using rare-cutting endonucleases able to selectively inactivating by DNA cleavage the gene encoding B2M and / or CIITA, or by using nucleic acid molecules which inhibit the expression of B2M and / or CIITA. In order to further render the T-cell non-alloreactive, at least one gene encoding a component of the T-cell receptor is inactivated, e.g., by using a rare-cutting endonucleases able to selectively inactivating by DNA cleavage the gene encoding said TCR component. In addition, expression of immunosuppressive polypeptide can be performed on those modified T-cells in order to prolong the survival of these modified T cells in host organism. Such modified T-cell is particularly suitable for allogeneic transplantations, especially because it reduces both the risk of rejection by the host's immune system and the risk of developing graft versus host disease. The invention opens the way to standard and affordable adoptive immunotherapy strategies using T-Cells for treating cancer, infections and auto-immune diseases.
Owner:CELLECTIS SA
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com