Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
59 results about "Transcriptional Repressor" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Crispr-based genome modification and regulation
Owner:SIGMA ALDRICH CO LLC
Crispr-based genome modification and regulation
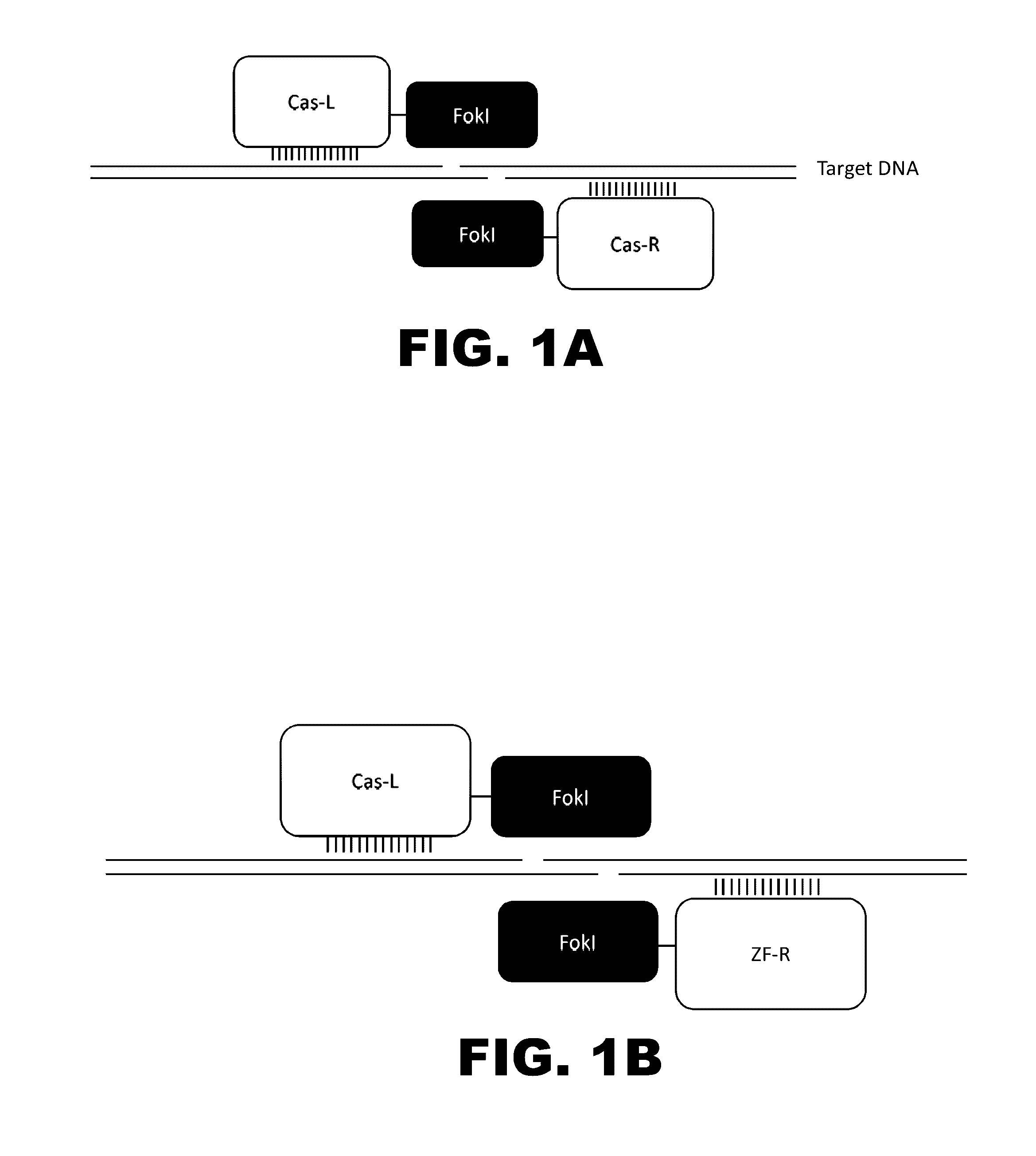
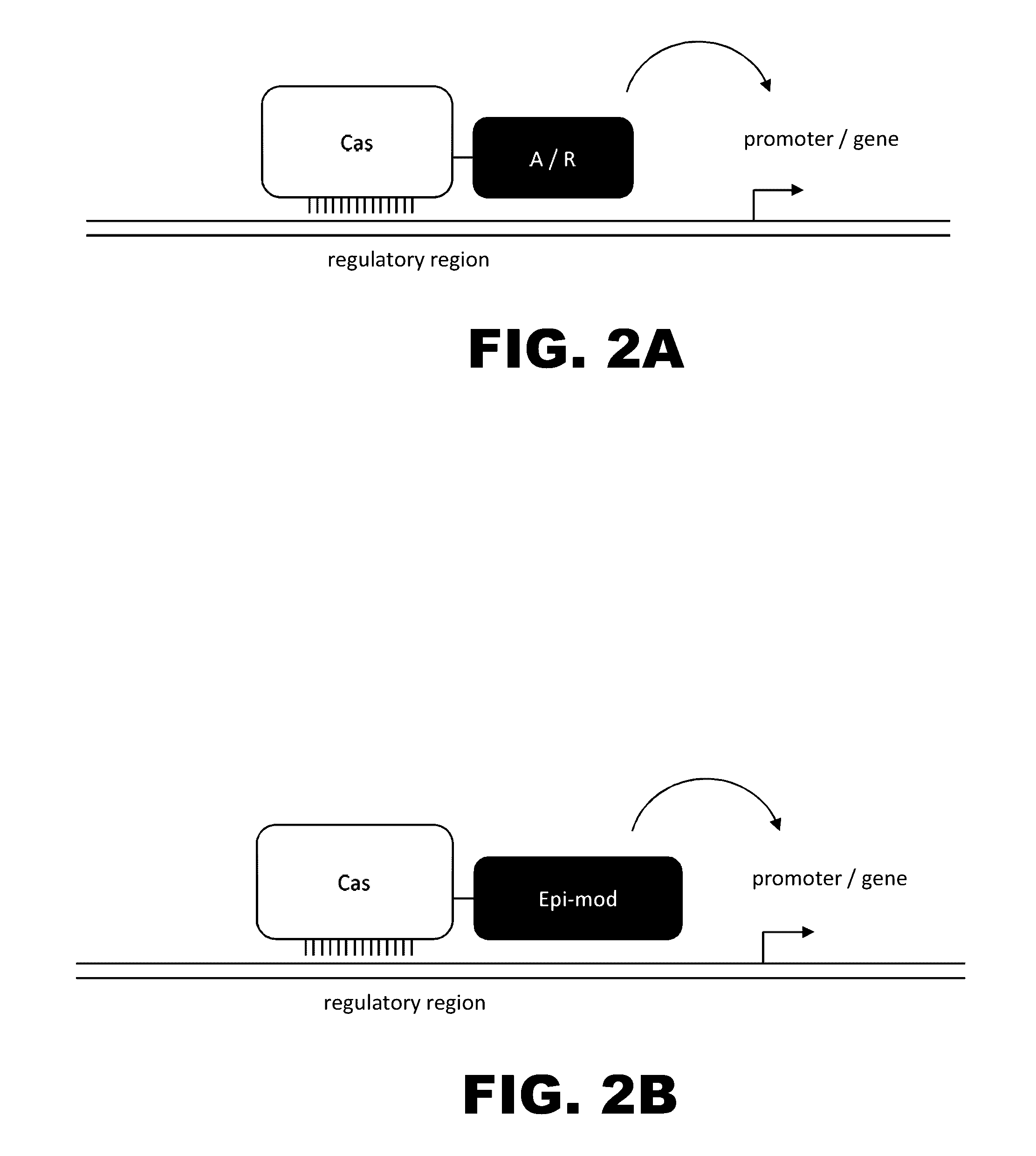
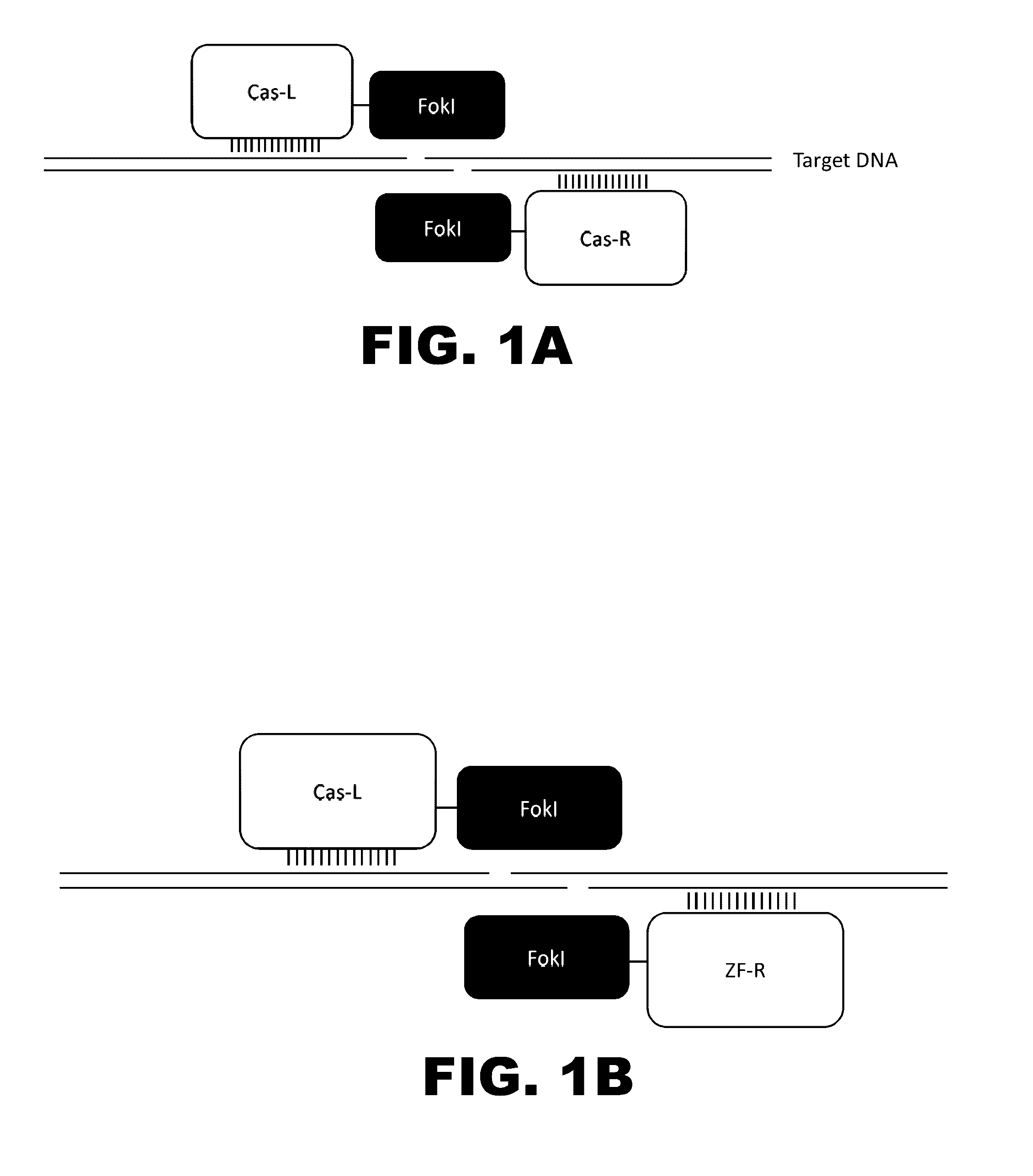
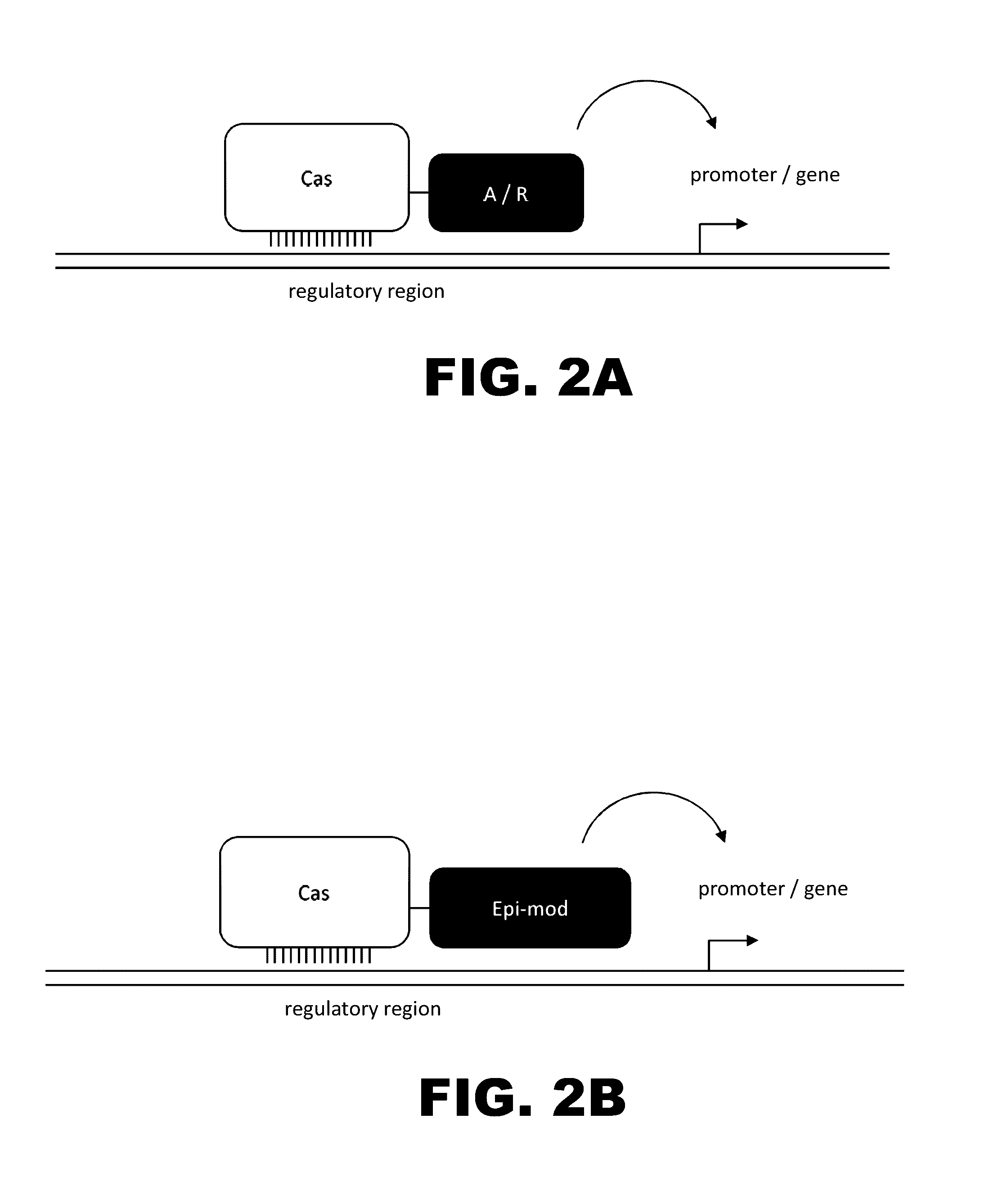
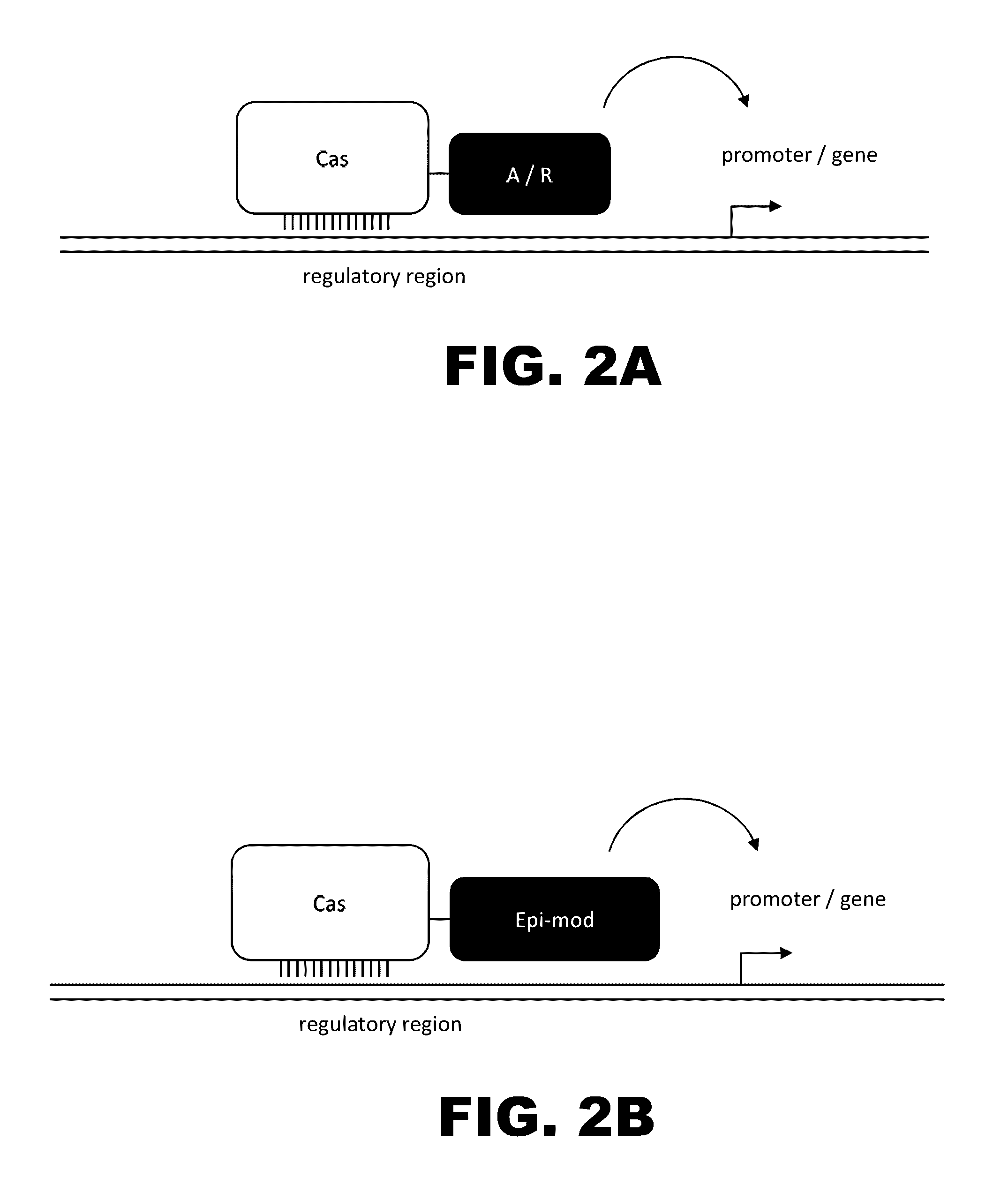
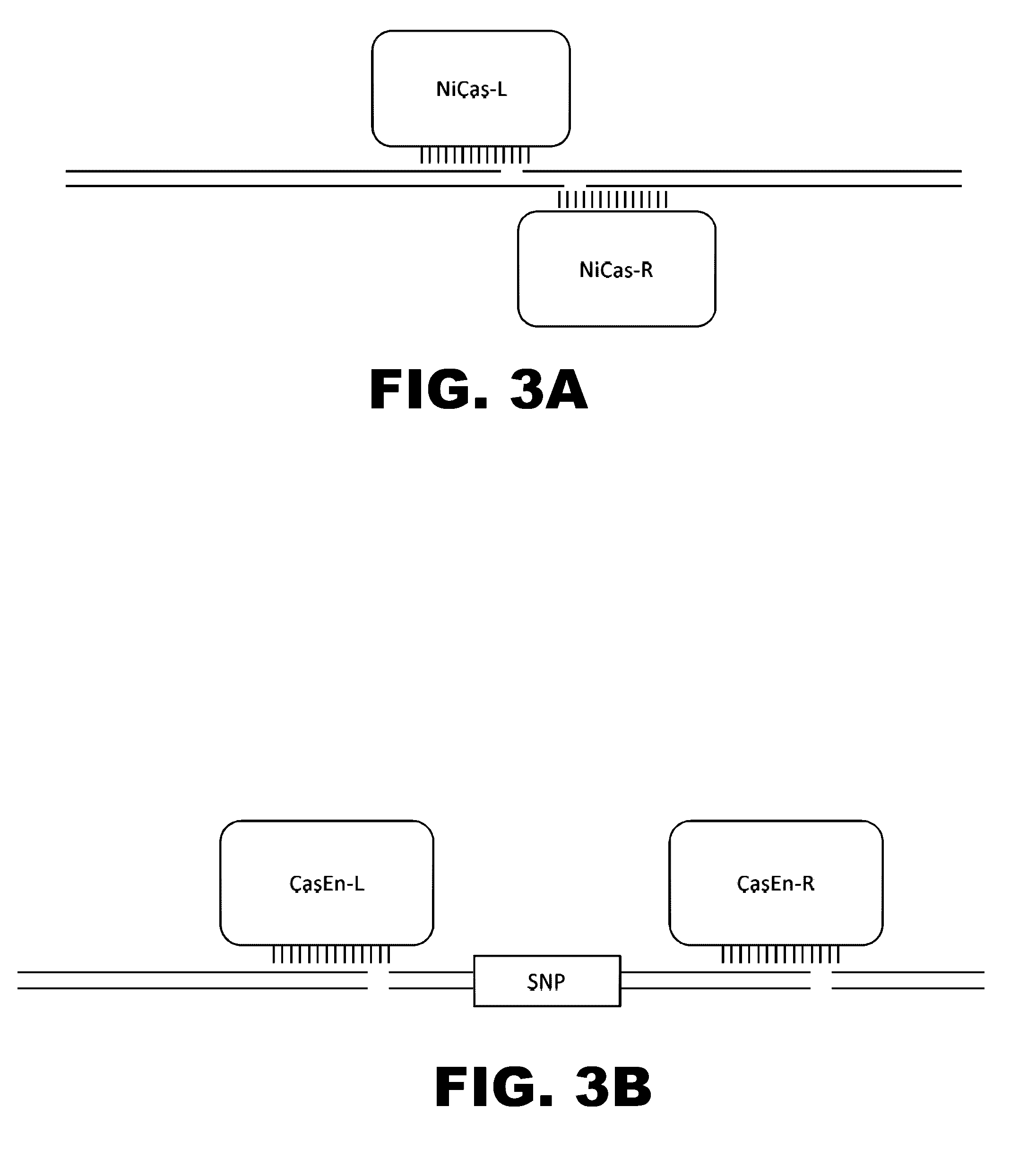
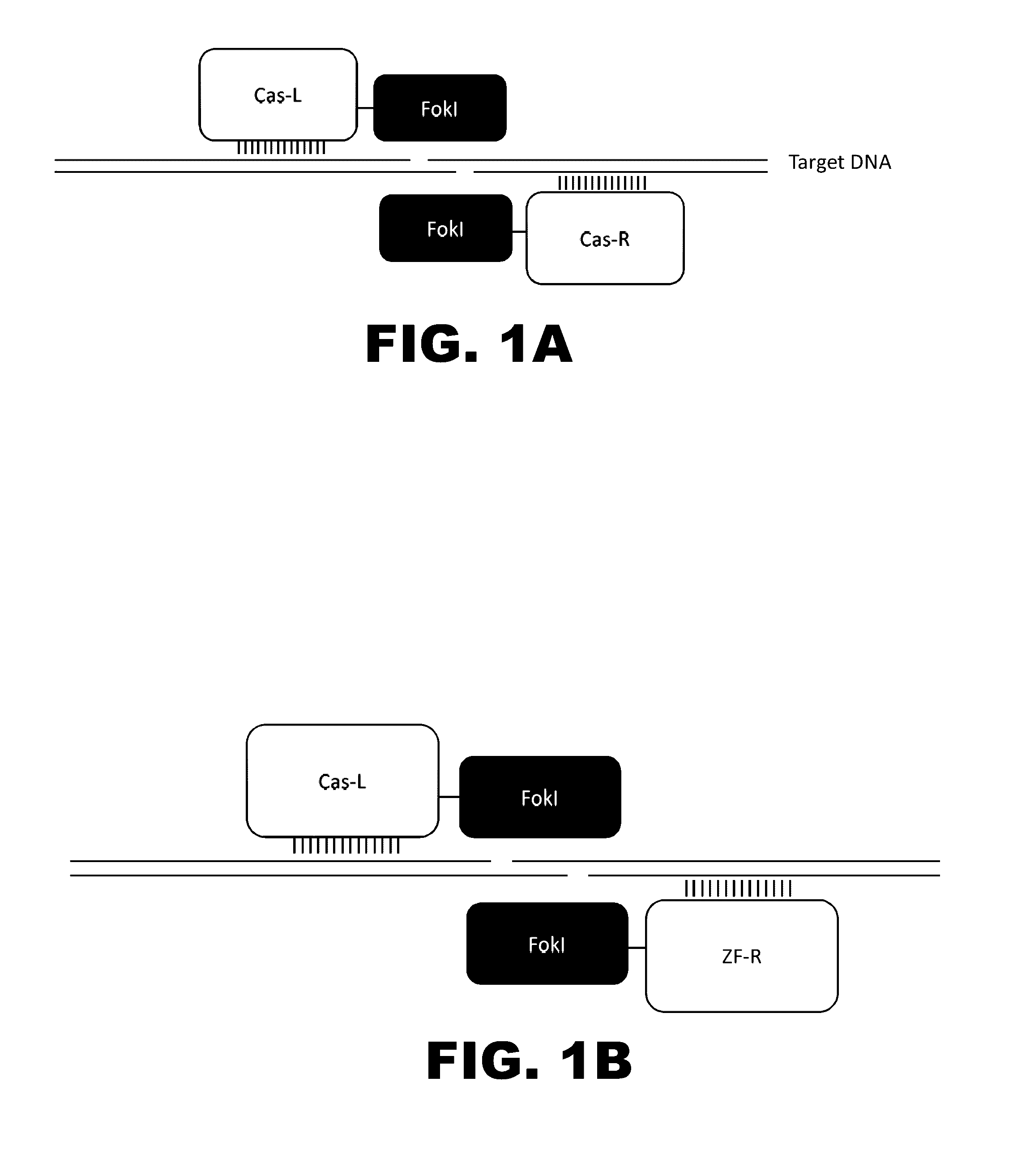
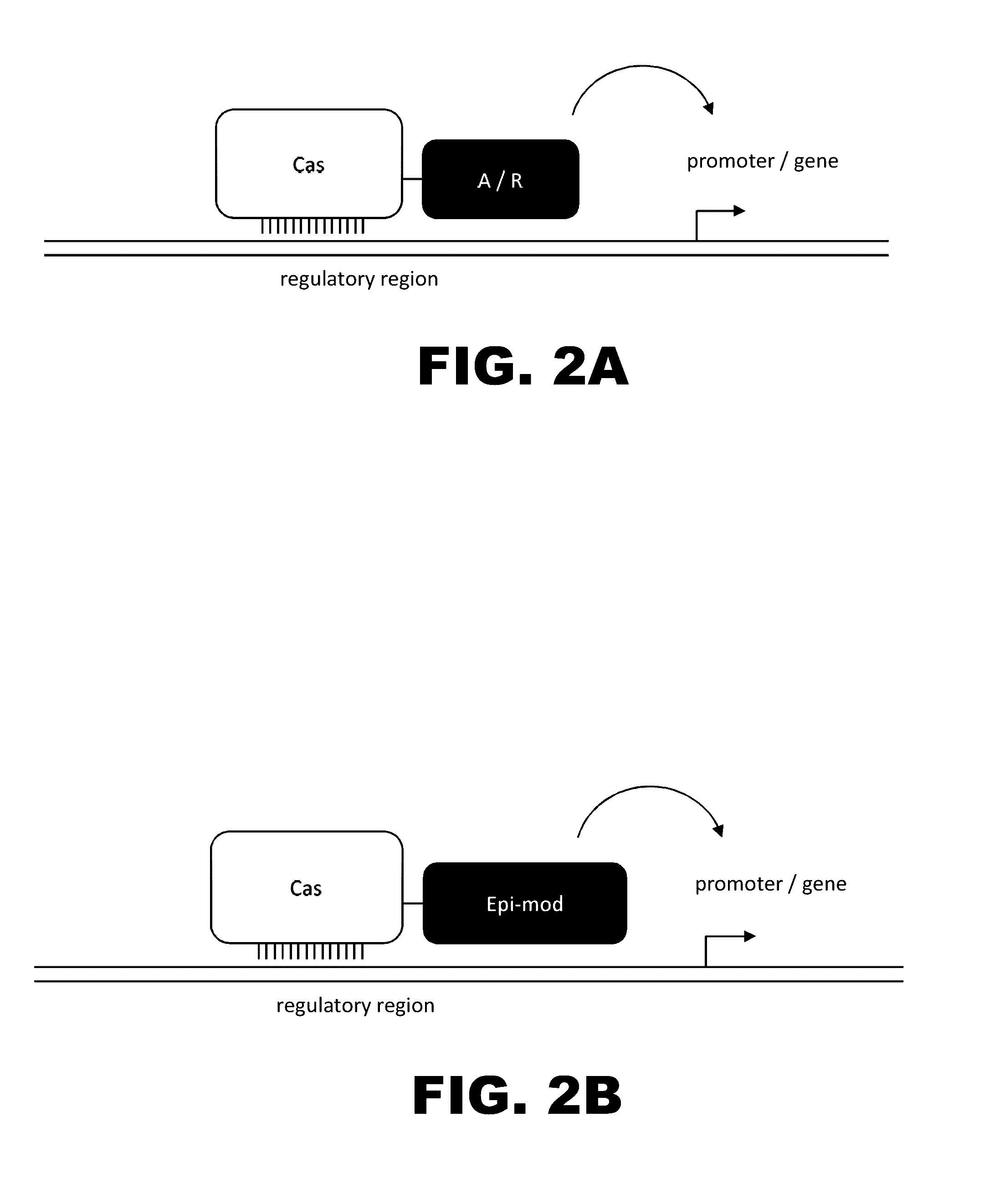
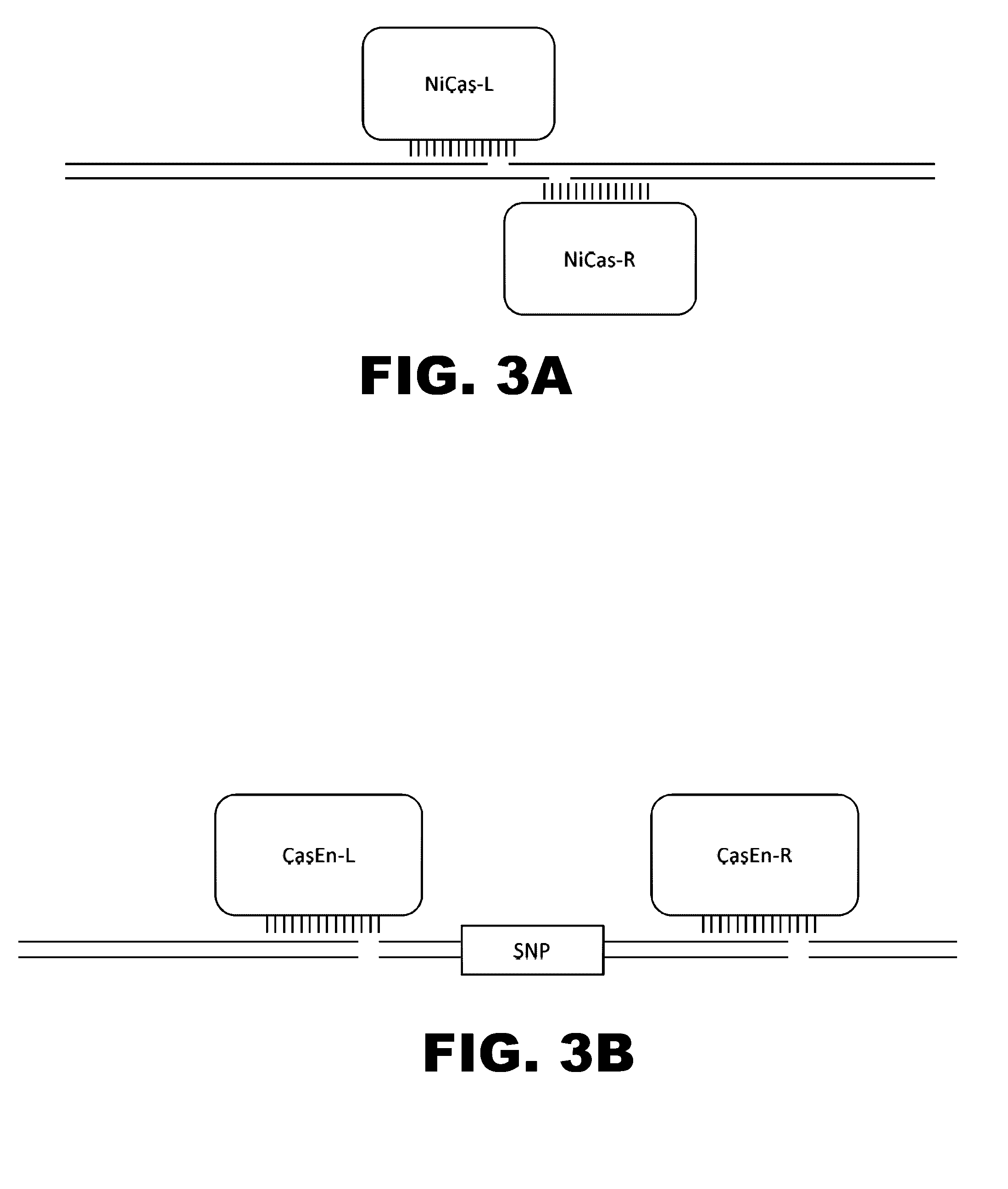
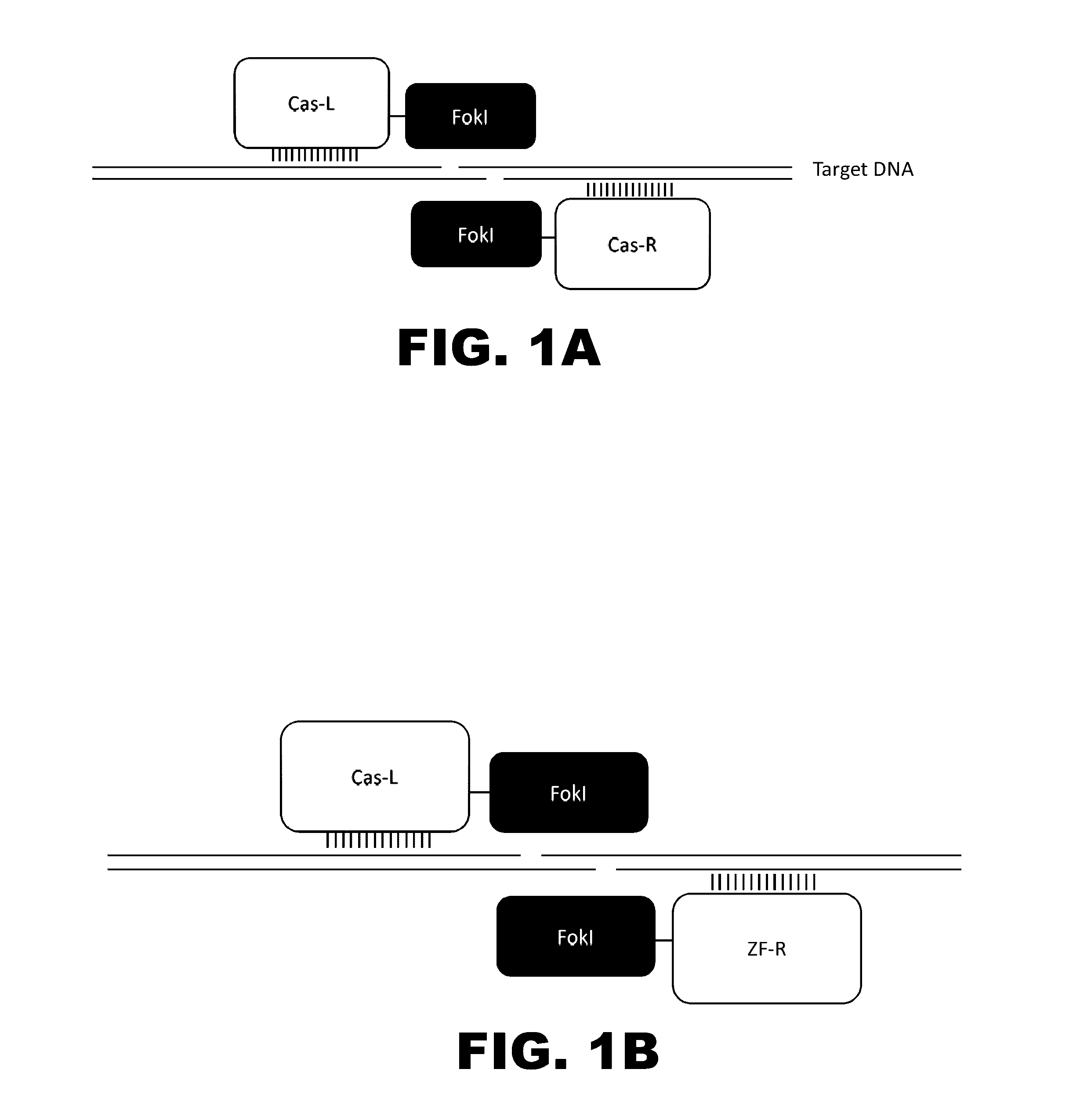
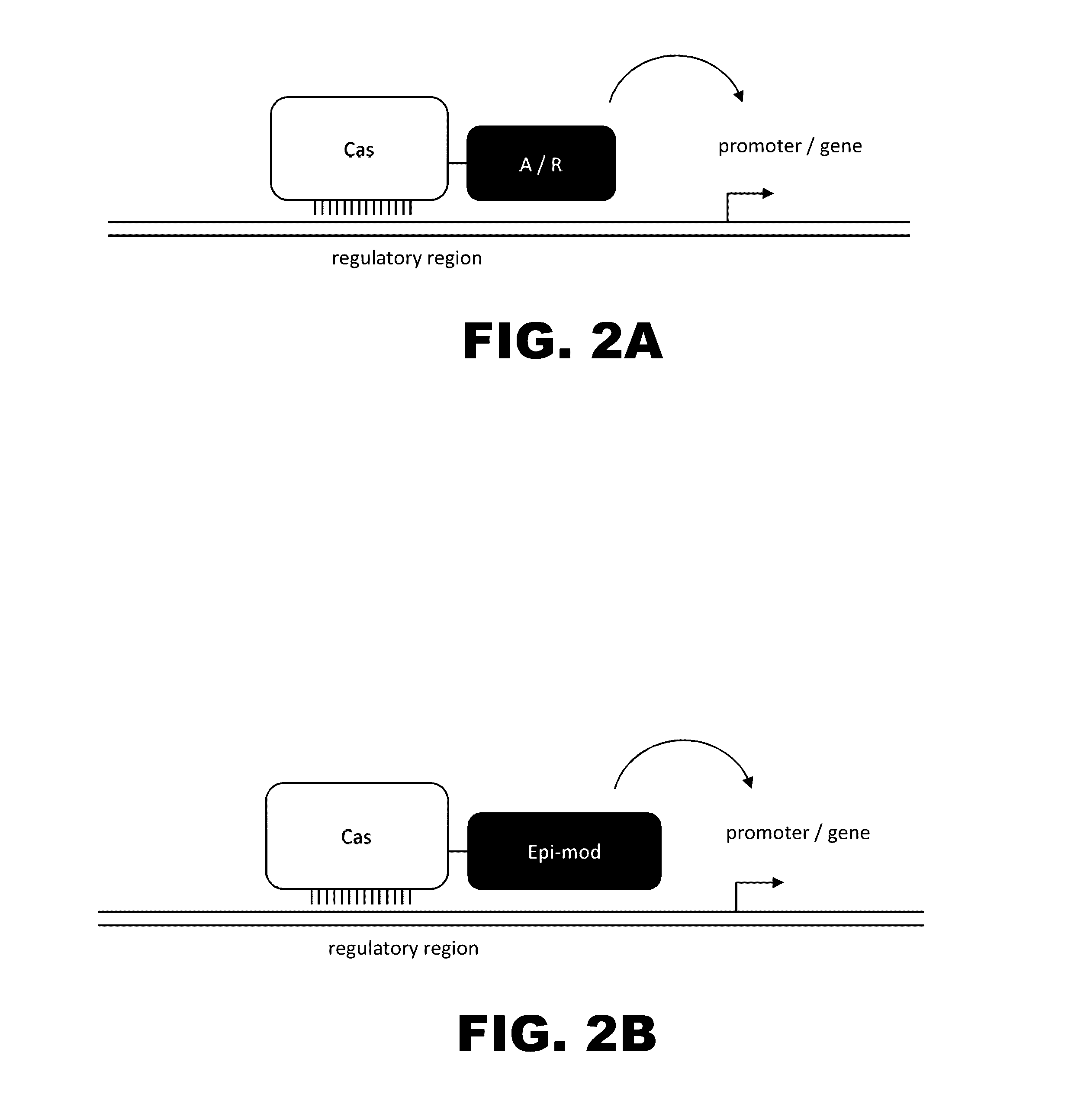
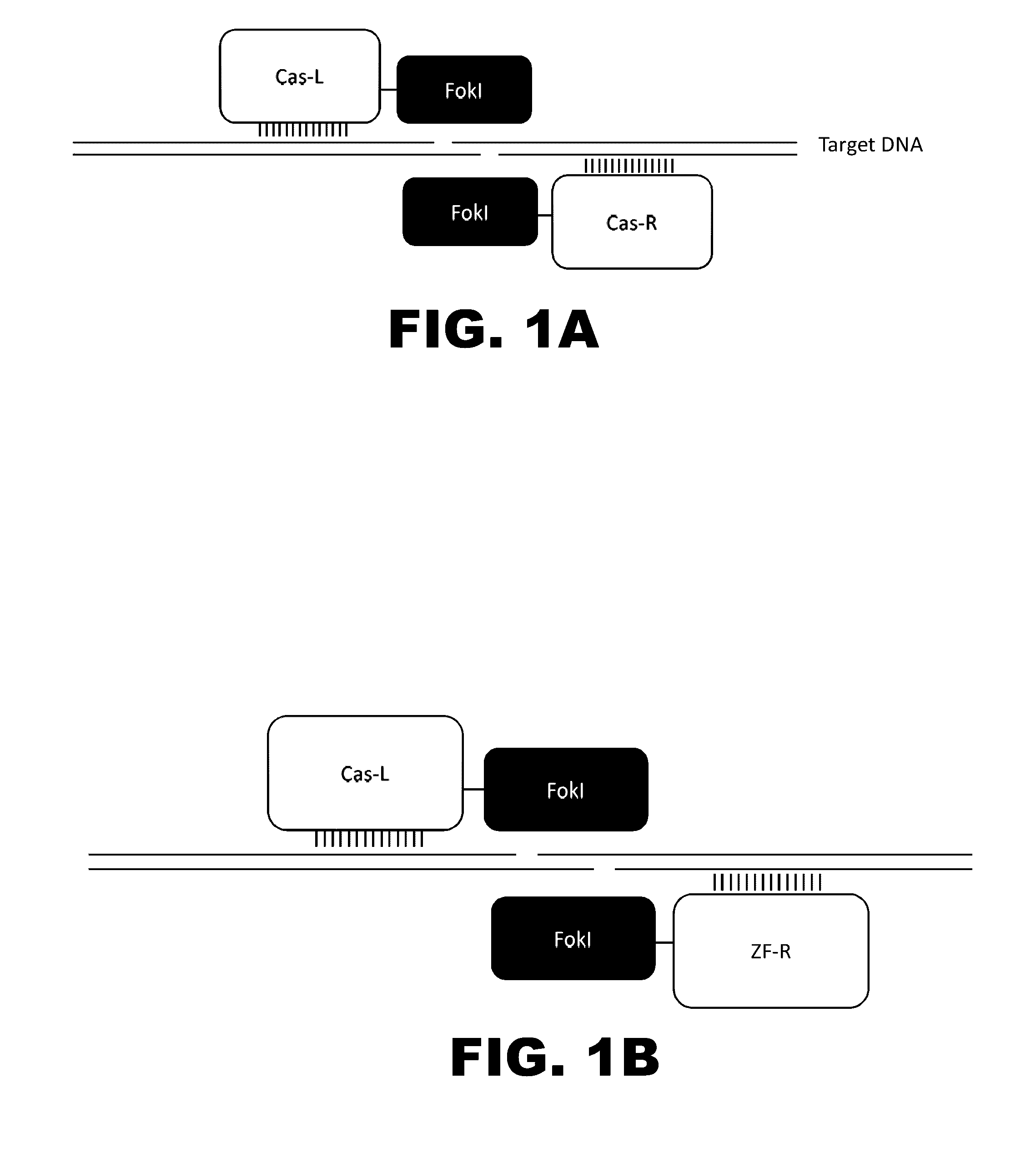
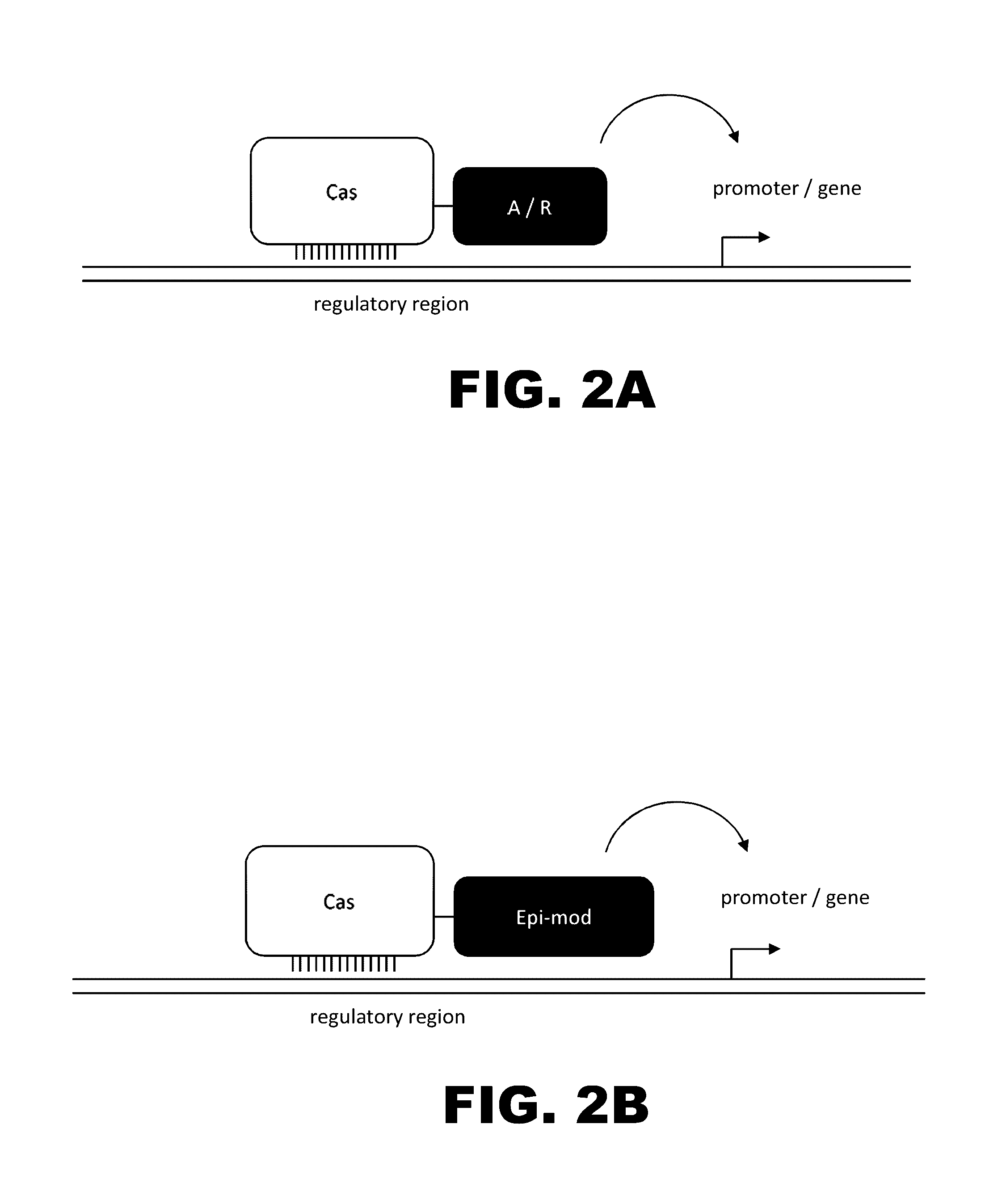
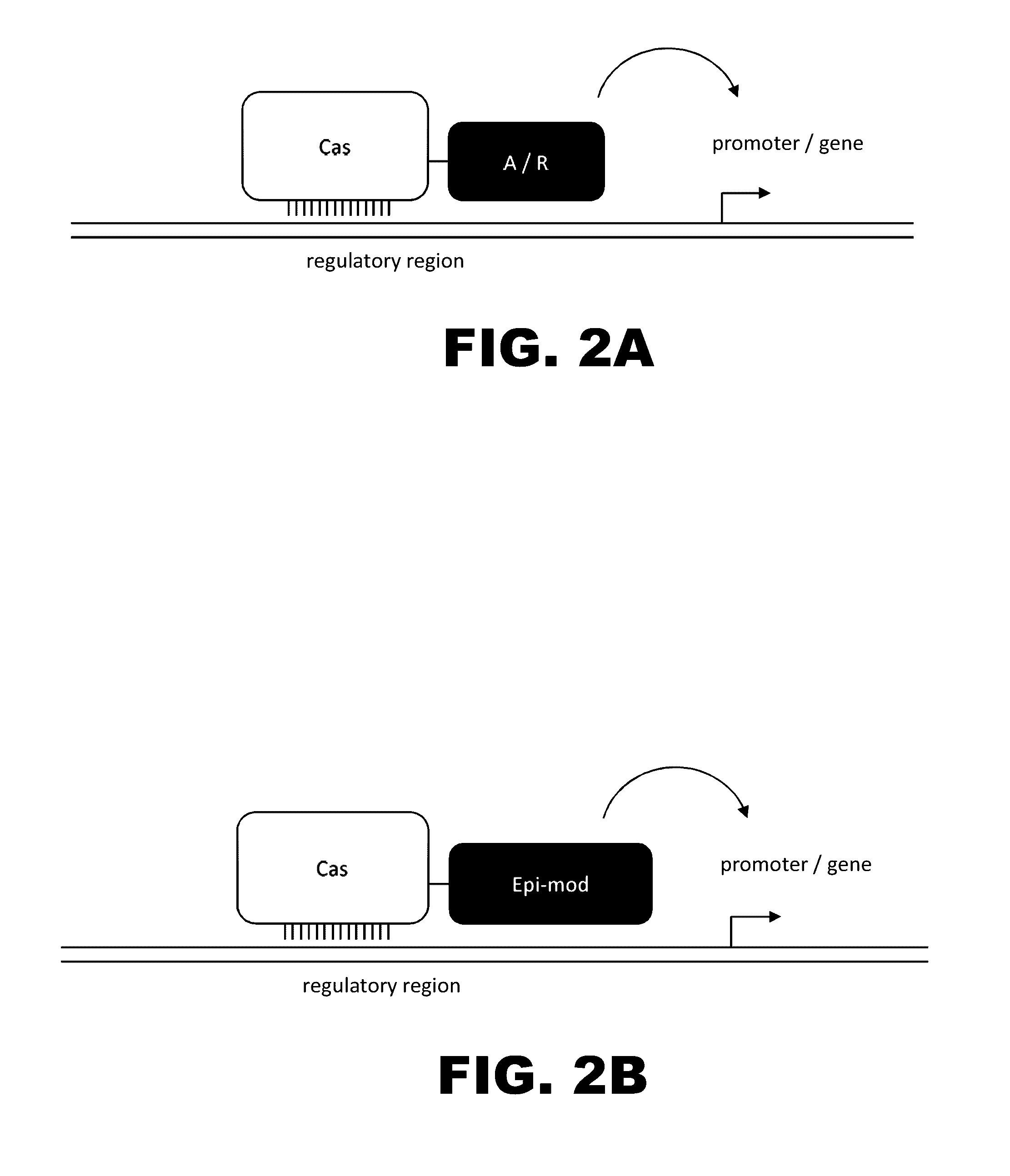
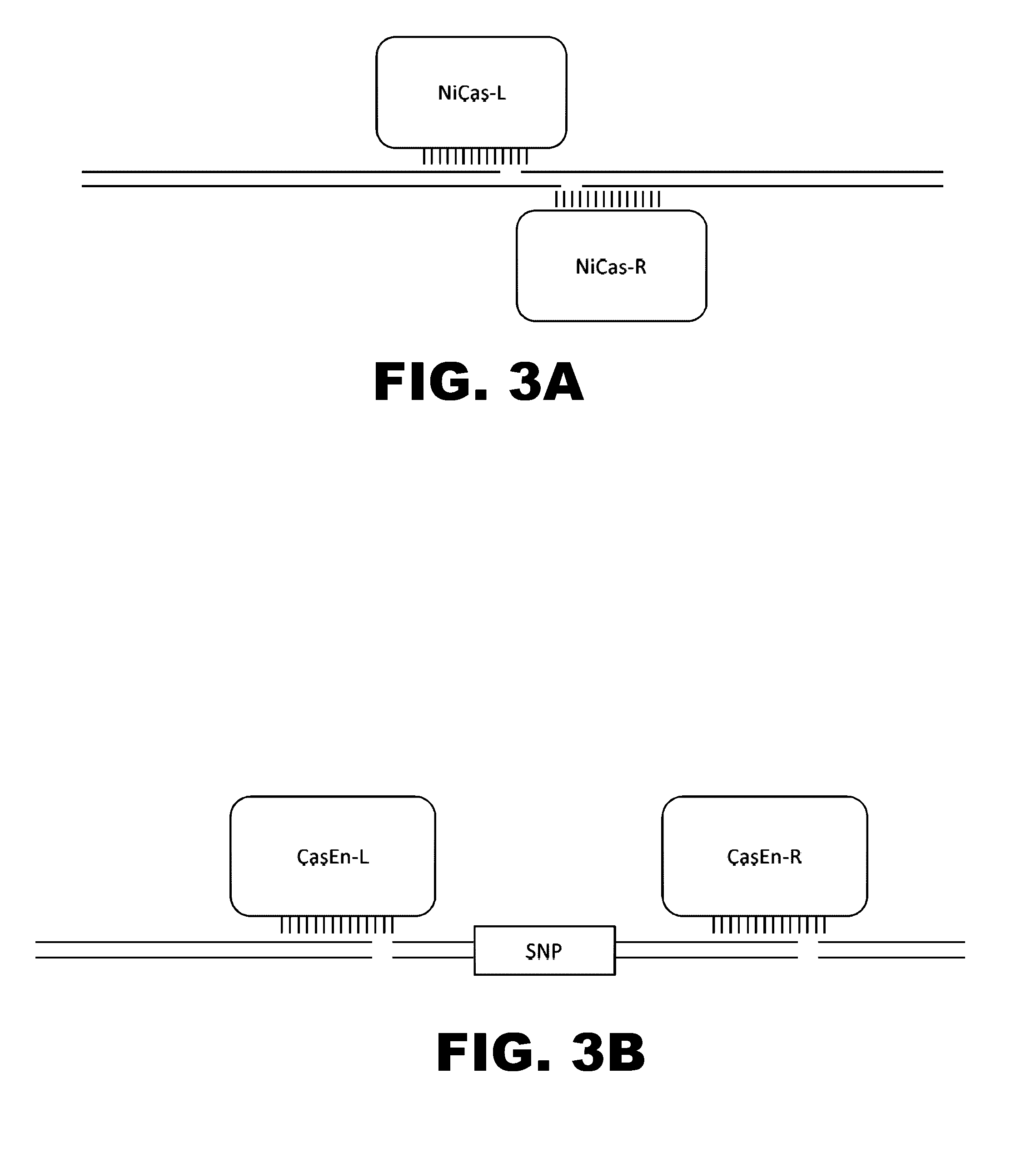
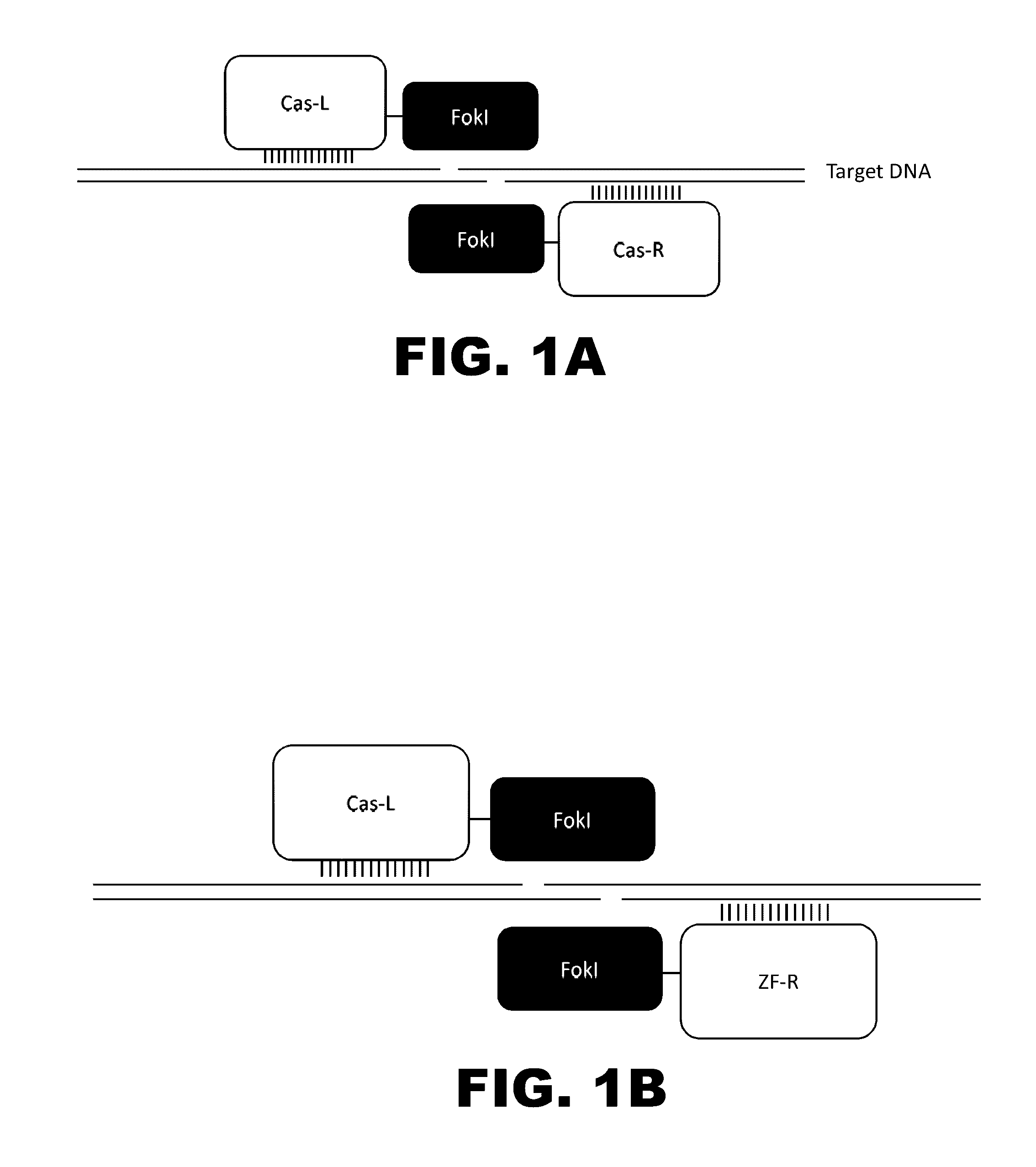
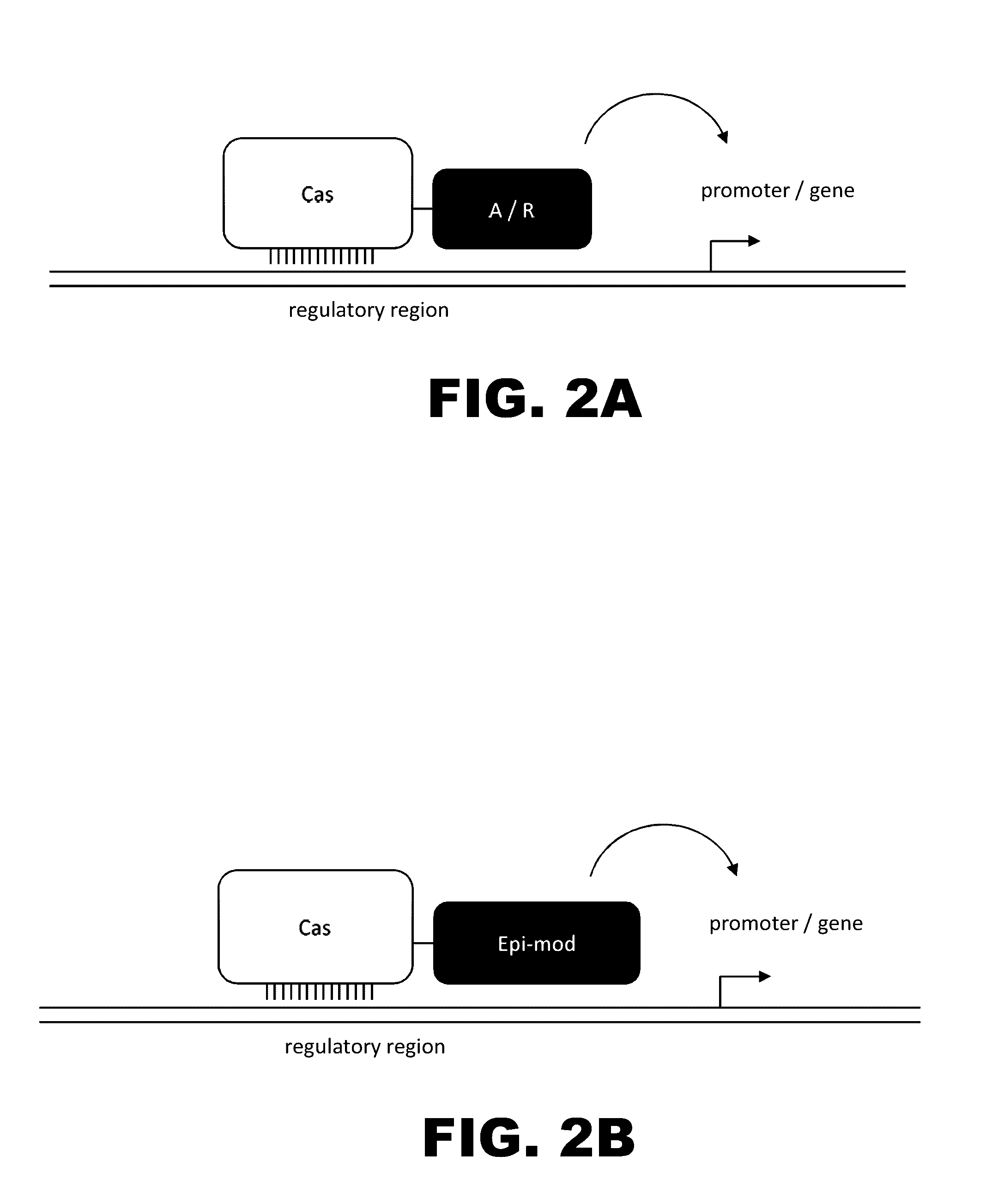
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Crispr-based genome modification and regulation
Owner:SIGMA ALDRICH CO LLC
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Engineered transcription activator-like effector (TALE) domains and uses thereof
ActiveUS20150056177A1Increased on-target cleavage efficiencyReducing unspecific bindingFusion with DNA-binding domainPeptide/protein ingredientsNucleaseGenome
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is their tendency to bind and cleave off-target sequence, which hampers their clinical application and renders applications requiring high-fidelity binding unfeasible. This disclosure provides engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with improved specificity and methods for generating and using such TALEs.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Crispr-based genome modification and regulation
Owner:SIGMA ALDRICH CO LLC
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Engineered transcription activator-like effector (TALE) domains and uses thereof
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is their tendency to bind and cleave off-target sequence, which hampers their clinical application and renders applications requiring high-fidelity binding unfeasible. This disclosure provides engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with improved specificity and methods for generating and using such TALEs.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
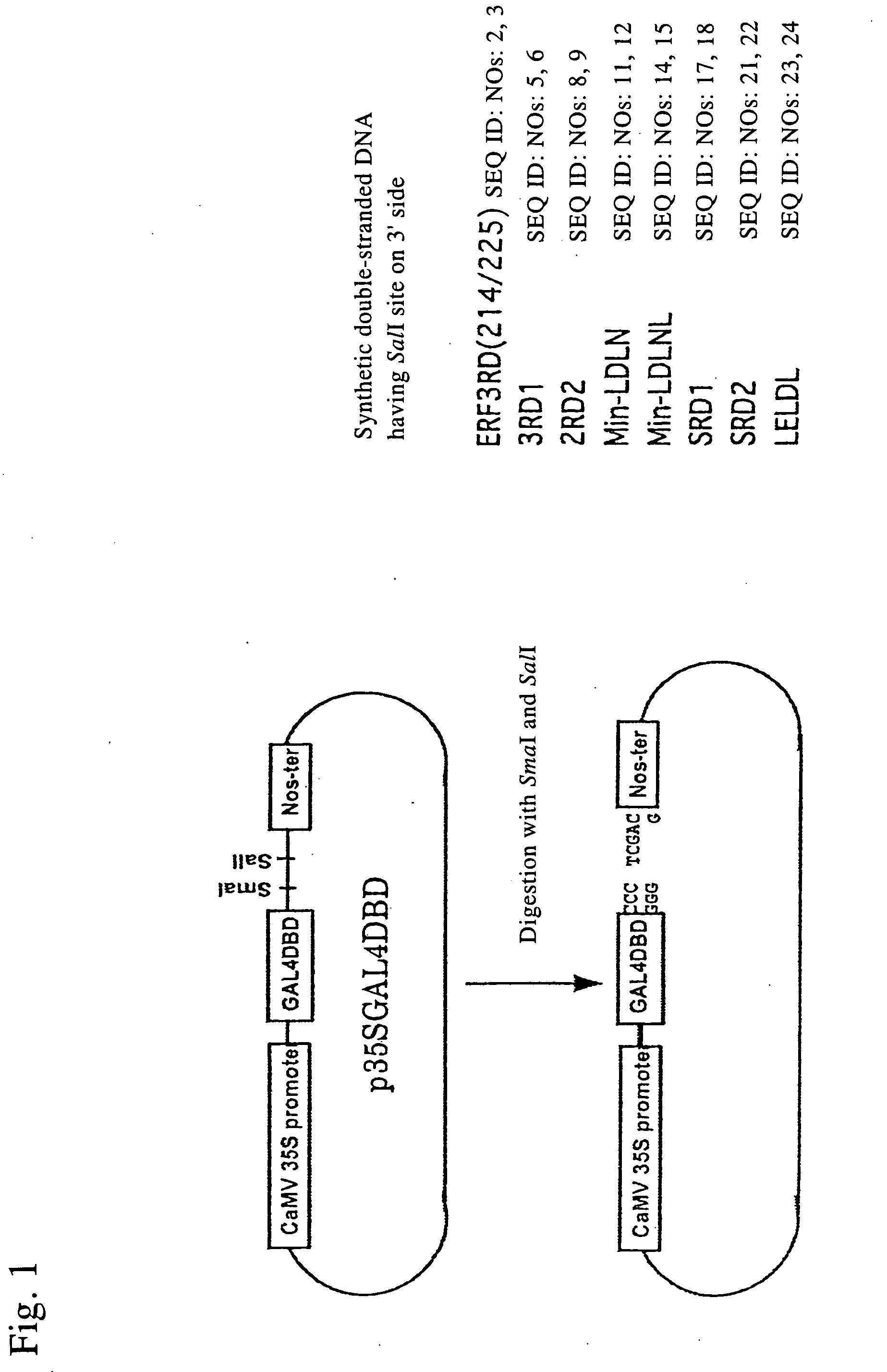
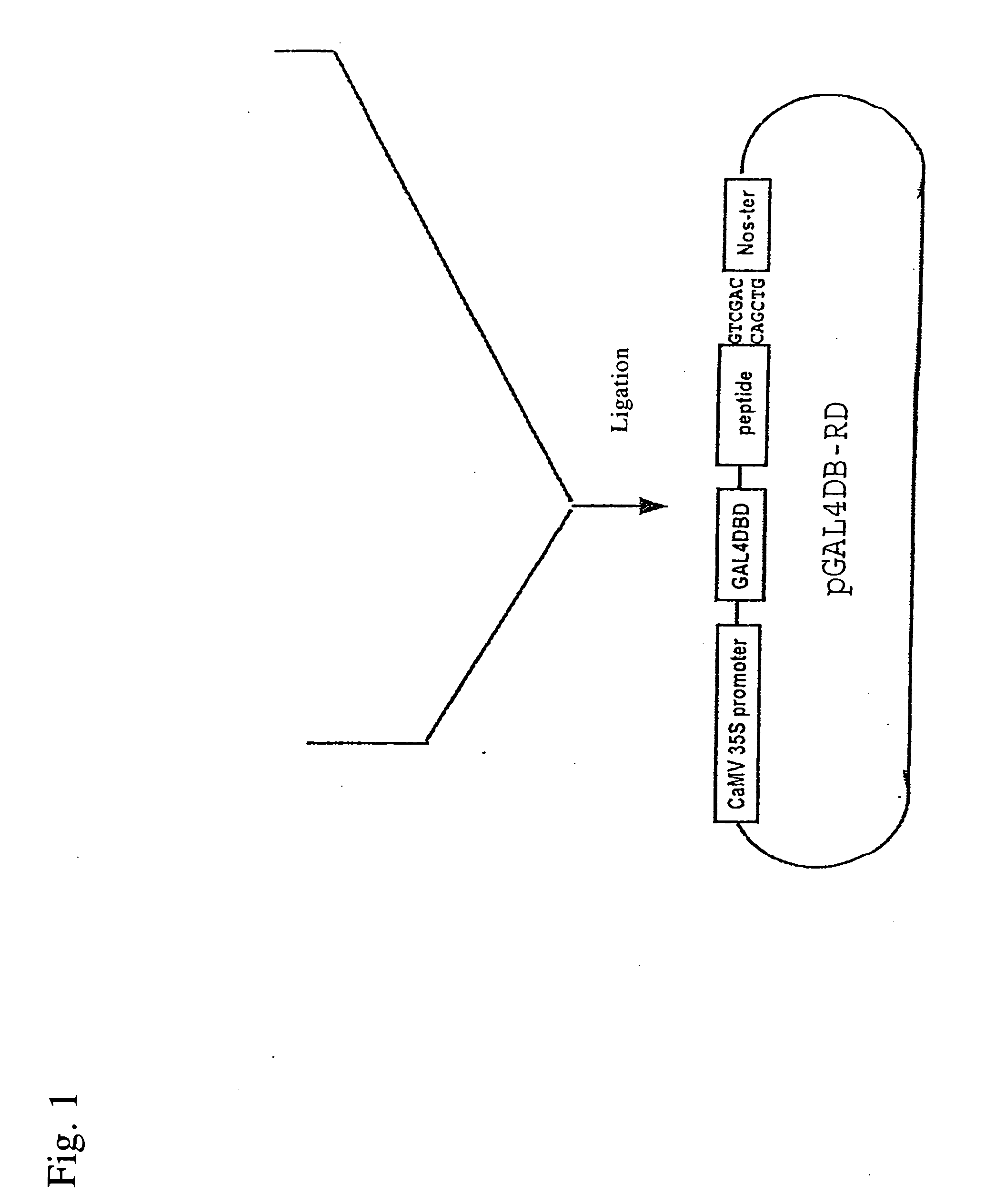
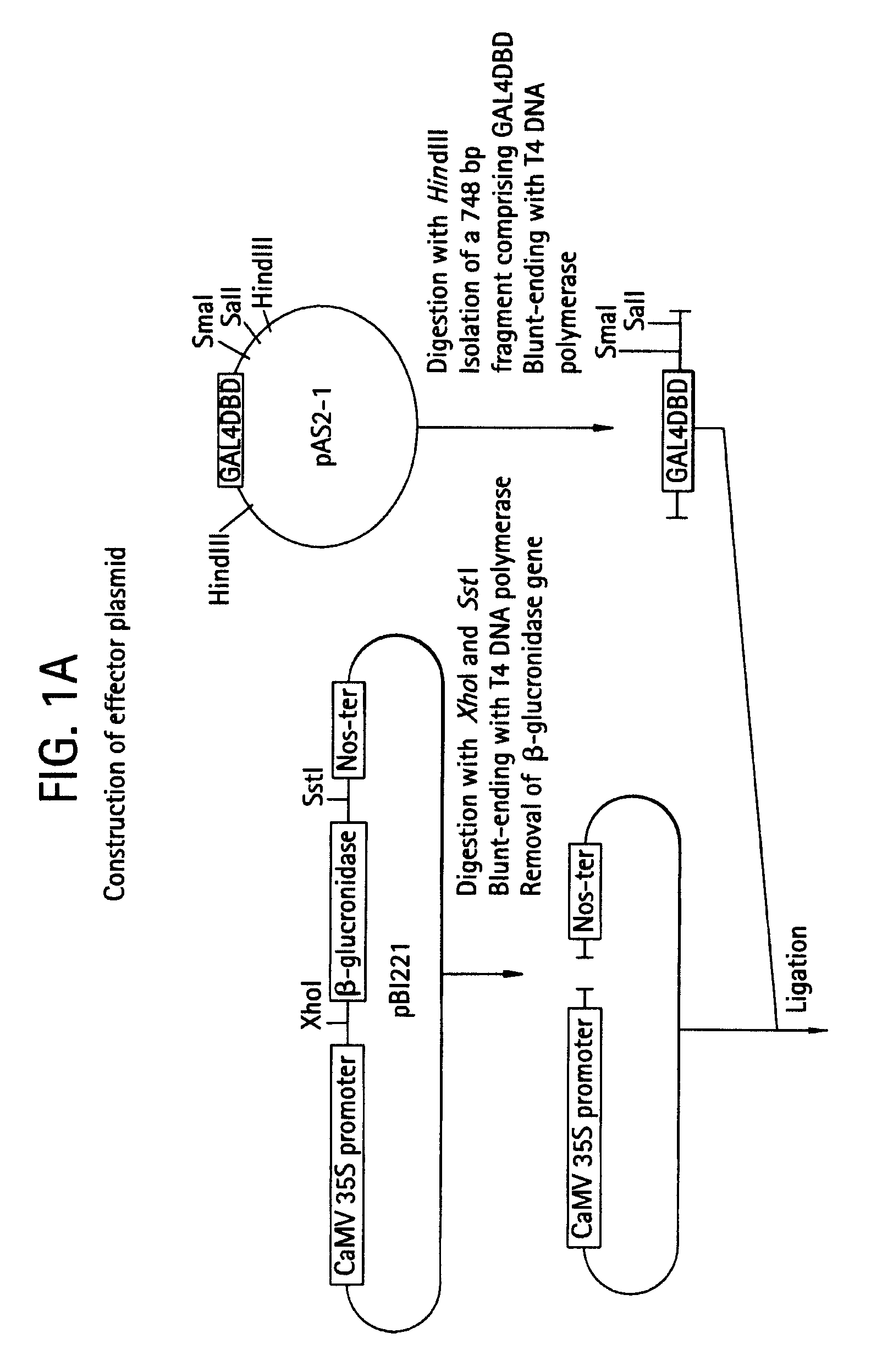
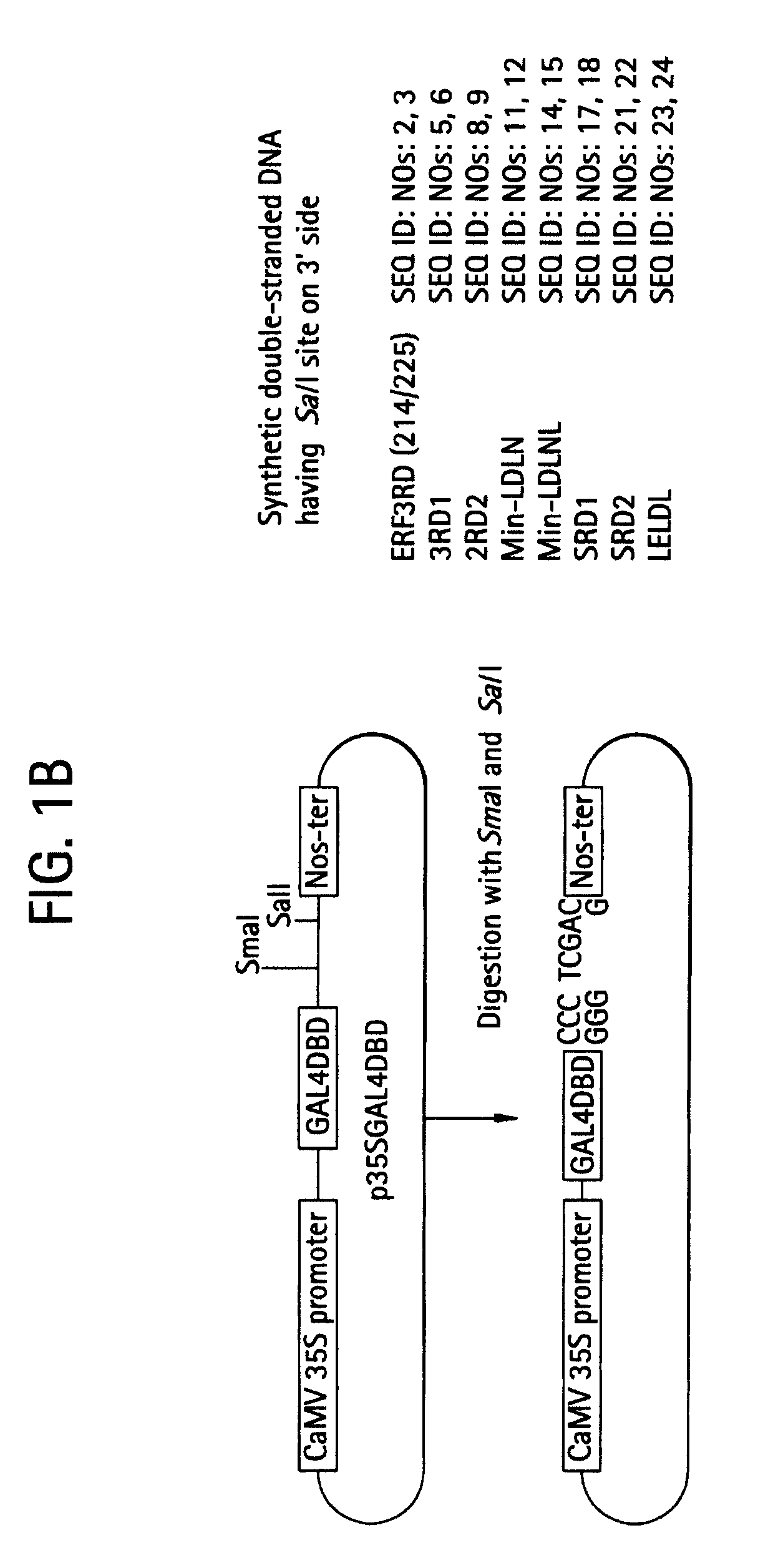
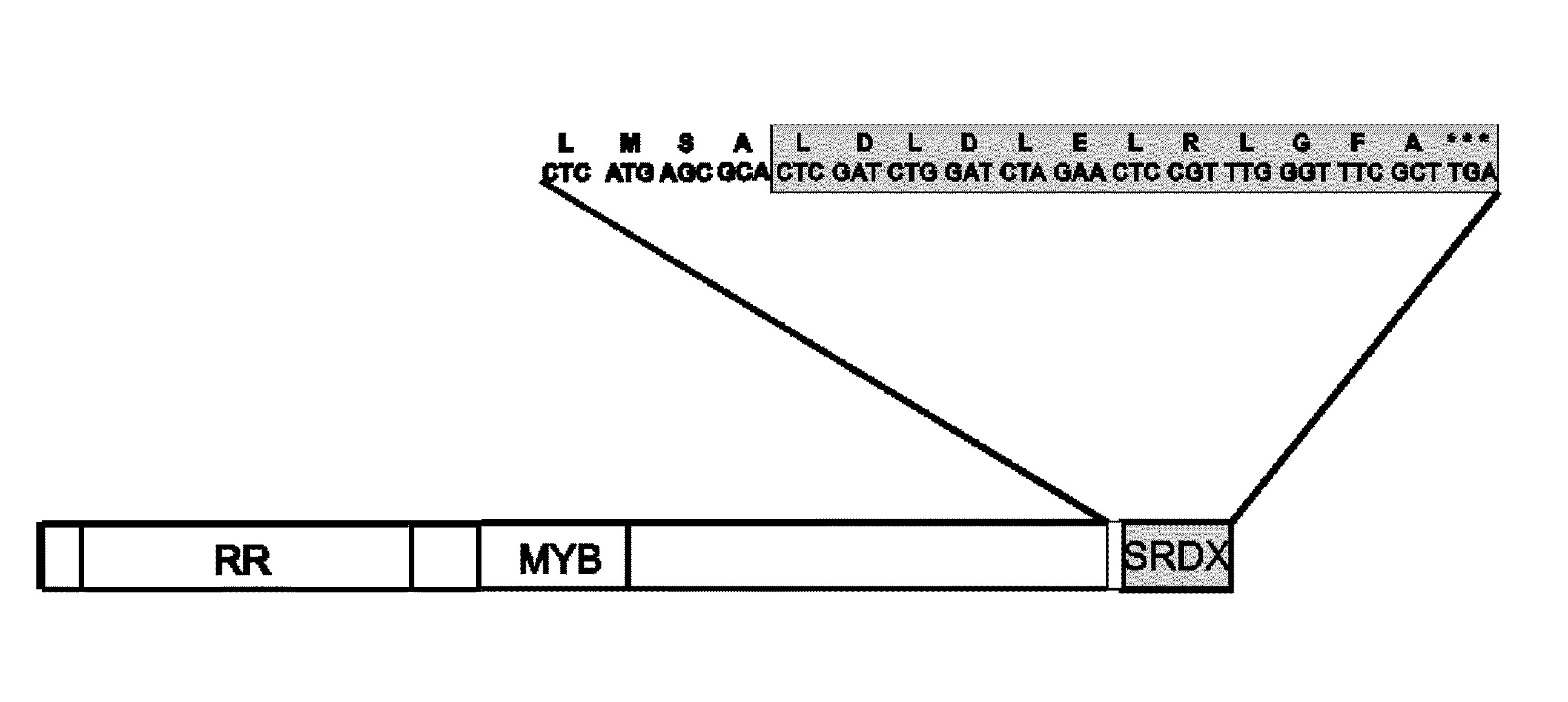
Transcription regulatory gene and peptide
ActiveUS20050183169A1Simple structurePotent capacity for repressing transcription of genePeptide/protein ingredientsImmunoglobulinsTranscription Regulation GeneMetabolic enzymes
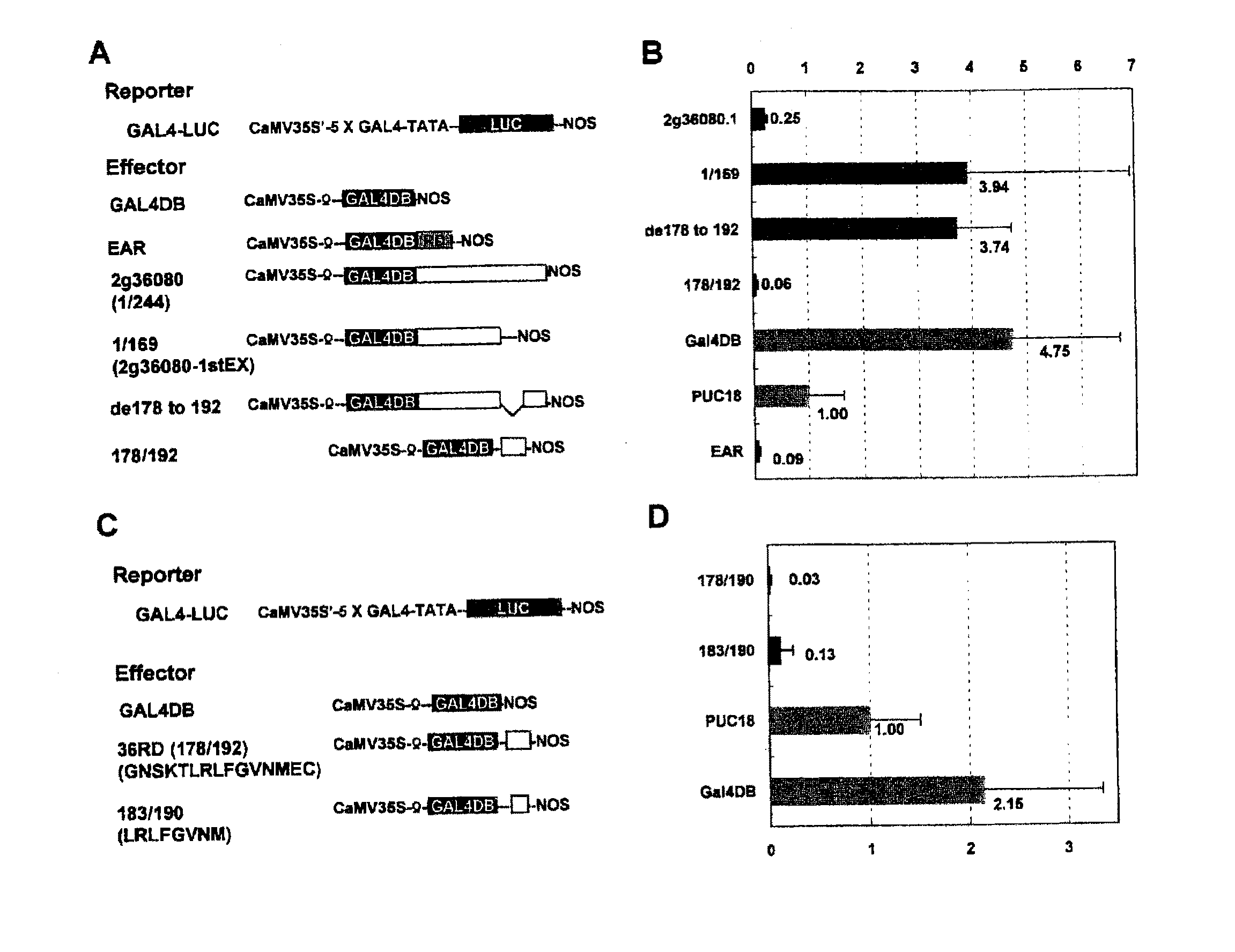
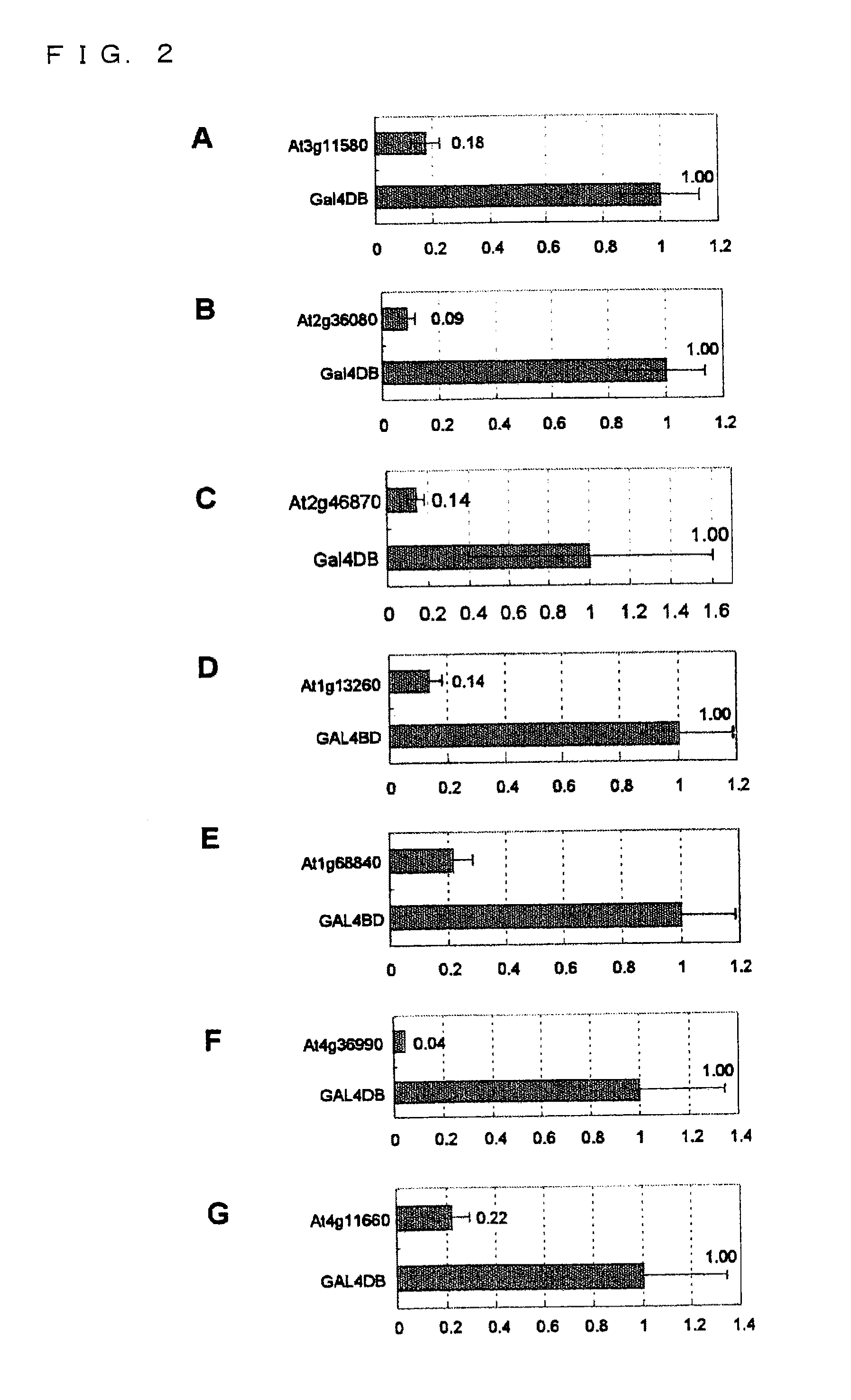
This invention relates to a peptide or protein capable of converting a transcription factor into a transcriptional repressor, a gene encoding such peptide or protein, a chimeric protein in which the aforementioned peptide or protein is fused to a transcription factor, a chimeric gene in which the gene encoding a peptide or protein is fused to a gene encoding a transcription factor, a recombinant vector having such chimeric gene, and a transformant comprising such recombinant vector. The peptide of the invention that is capable of converting a transcription factor into a transcriptional repressor is very short. Thus, it can be very easily synthesized, and it can effectively and selectively repress the transcription of a specific gene. Accordingly, such gene is applicable to and useful in a wide variety of fields, such as repression of the expression of cancerous genes and regulation of the expression of genes encoding pigment-metabolic enzymes.
Owner:GREENSOGNA +1
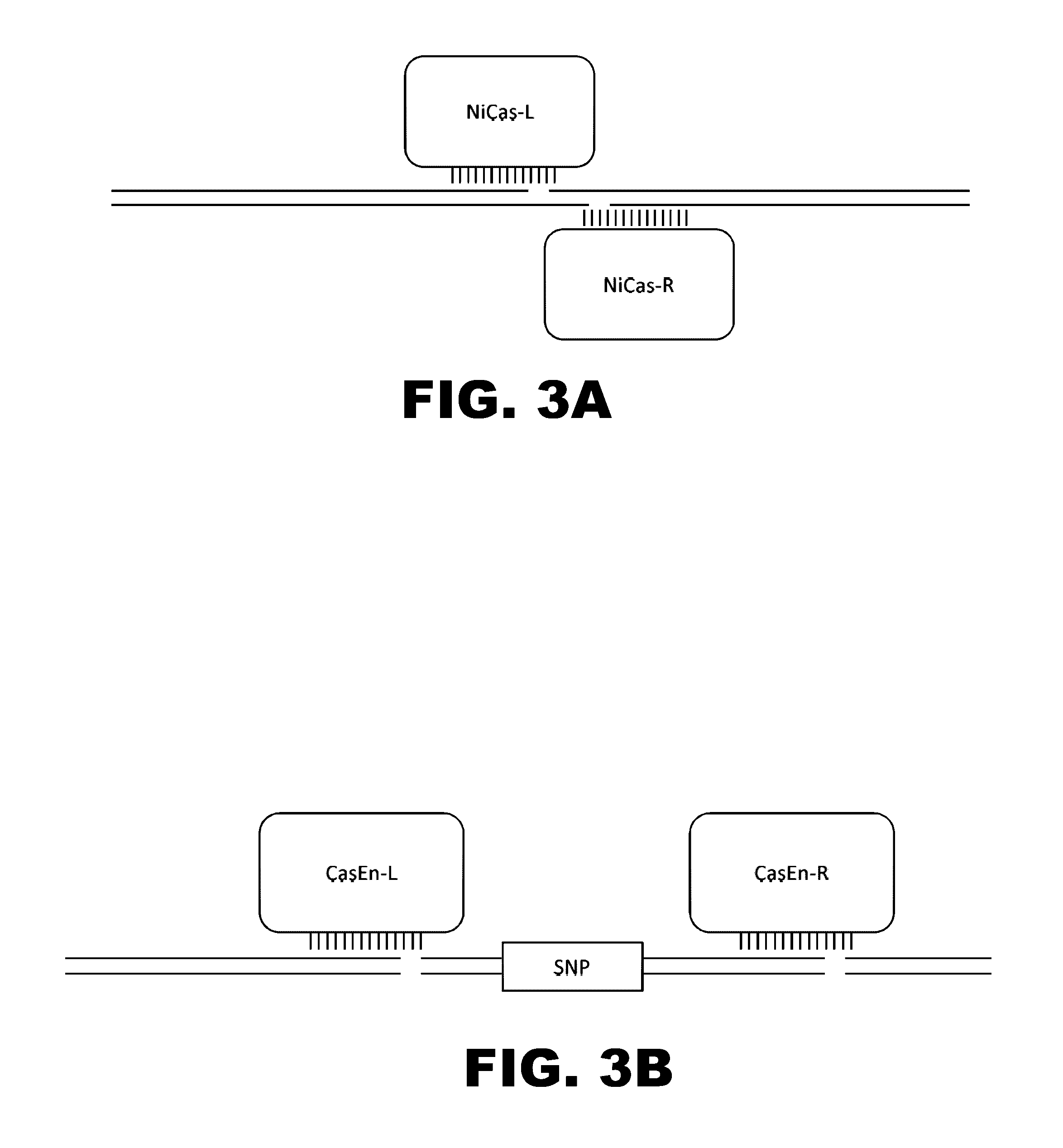
Evolution of talens
ActiveUS20200277587A1Strong specificityHydrolasesGenetic therapy composition manufactureNucleotideEpigenetic Profile
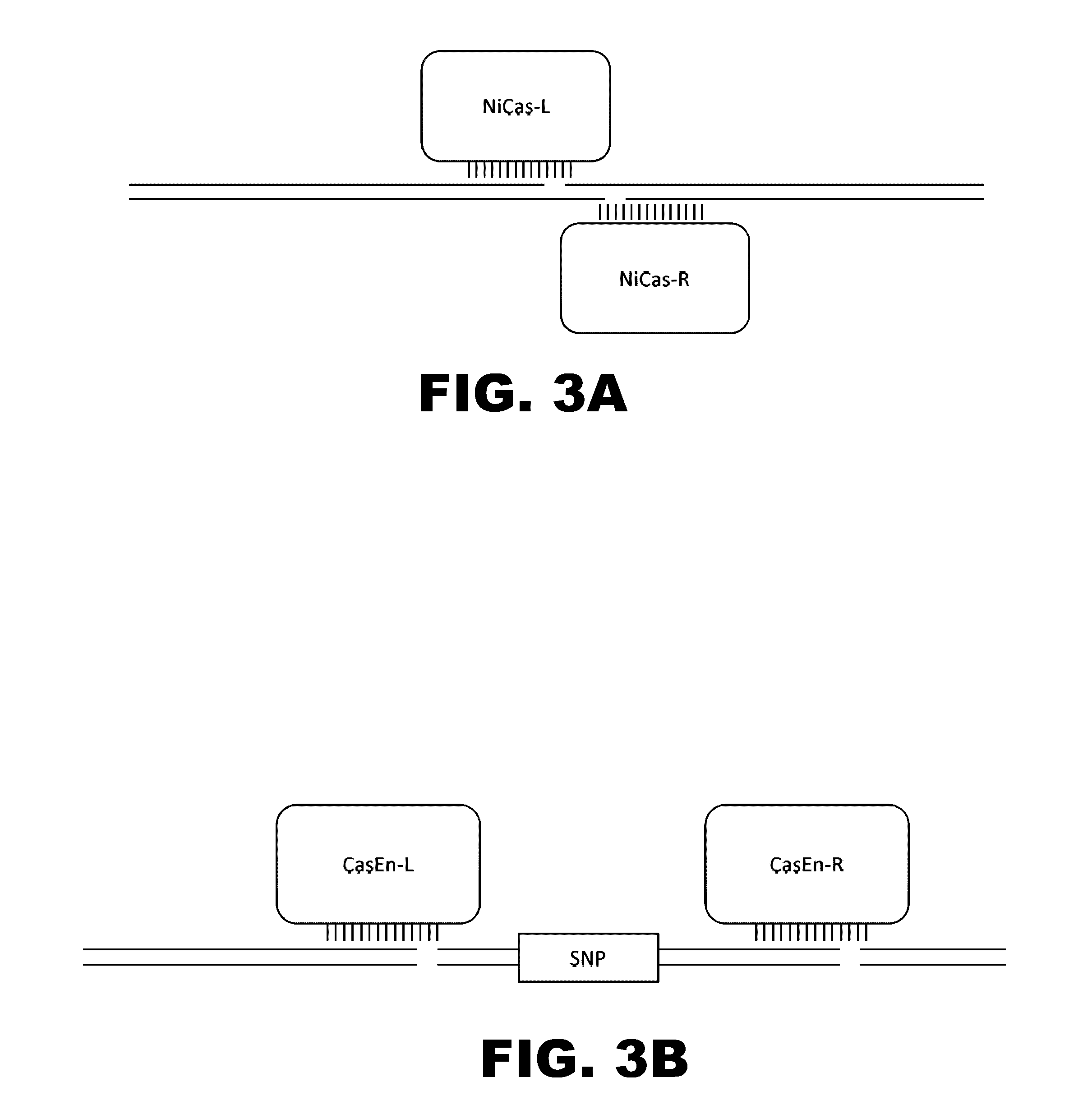
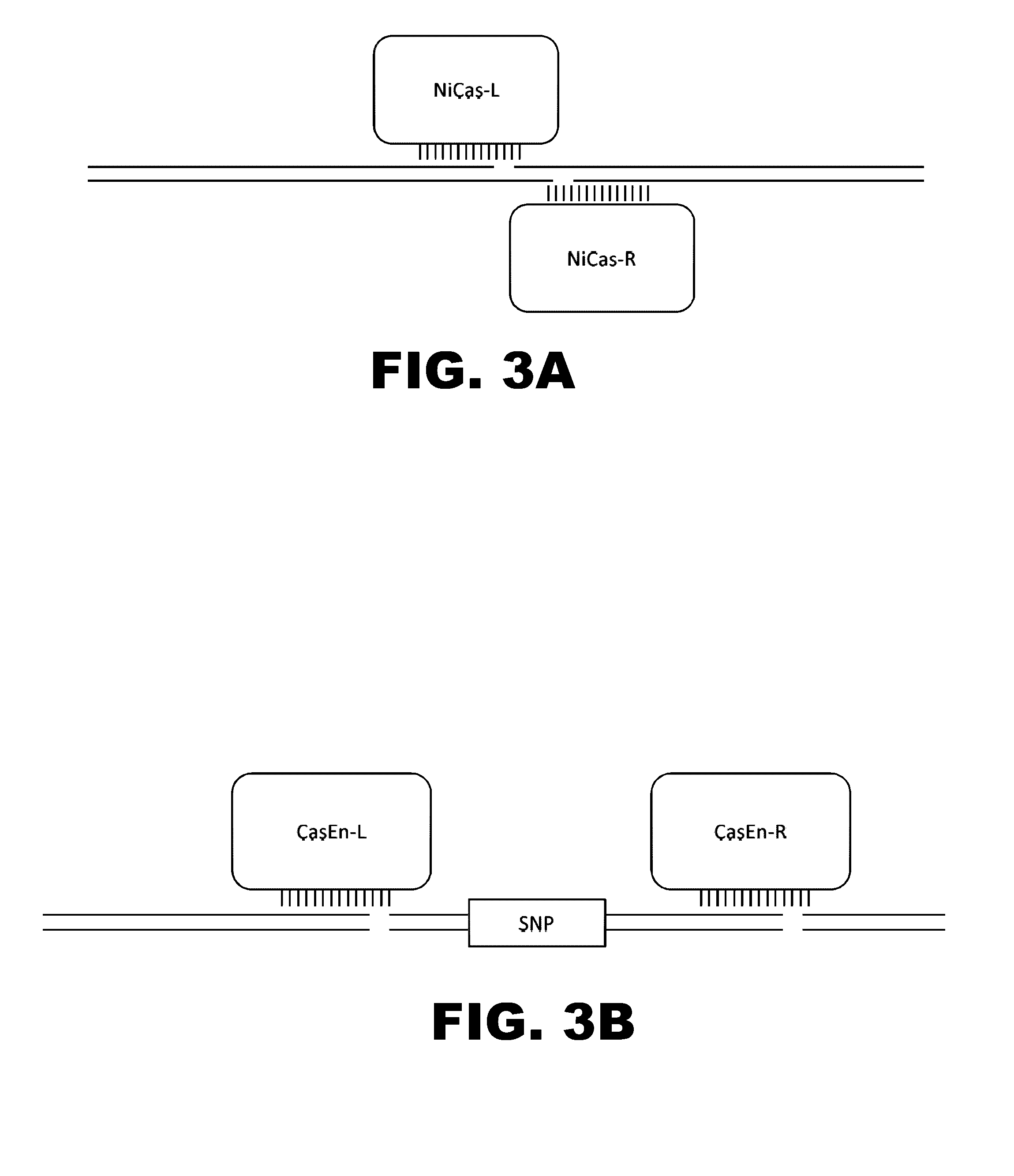
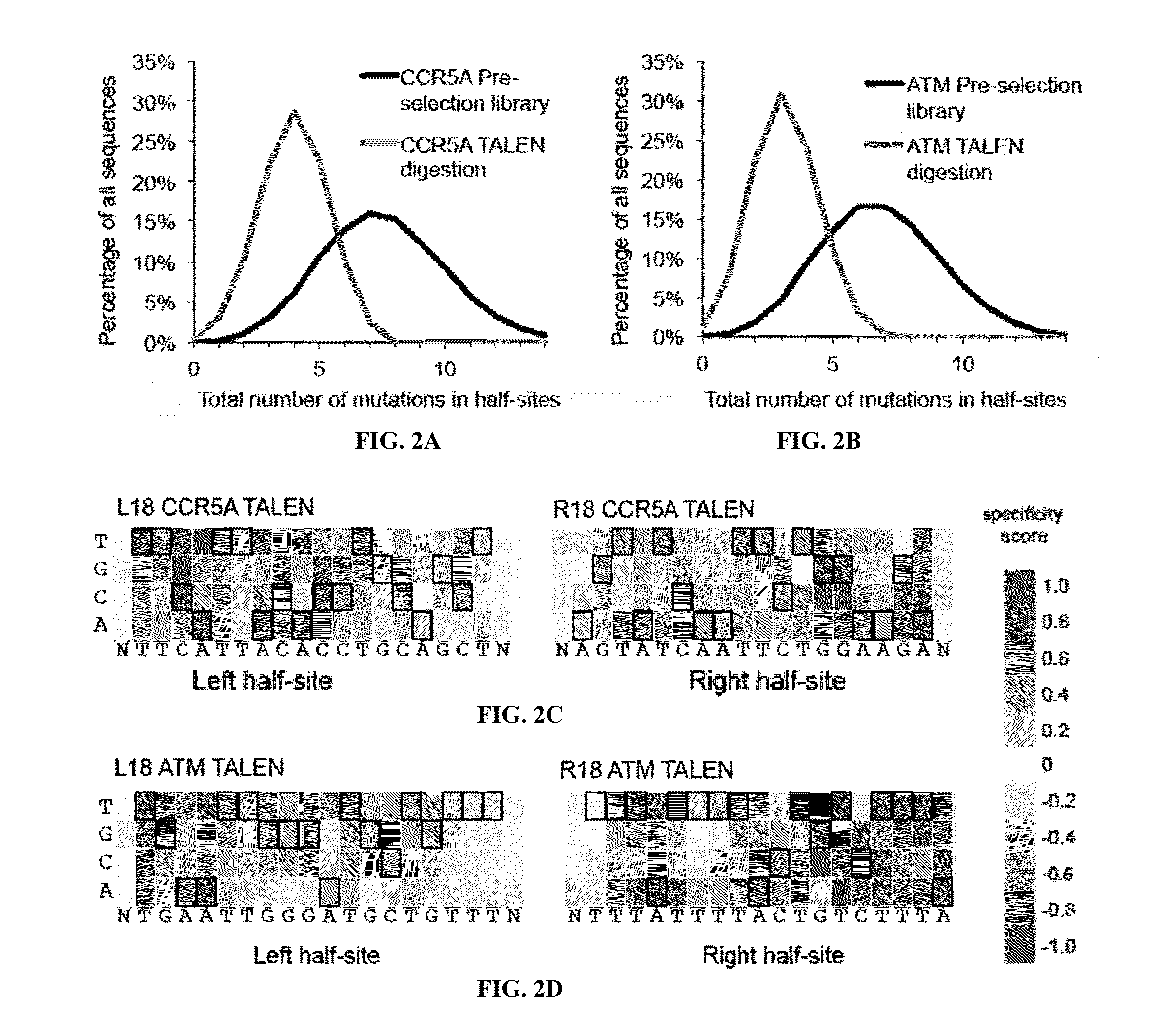
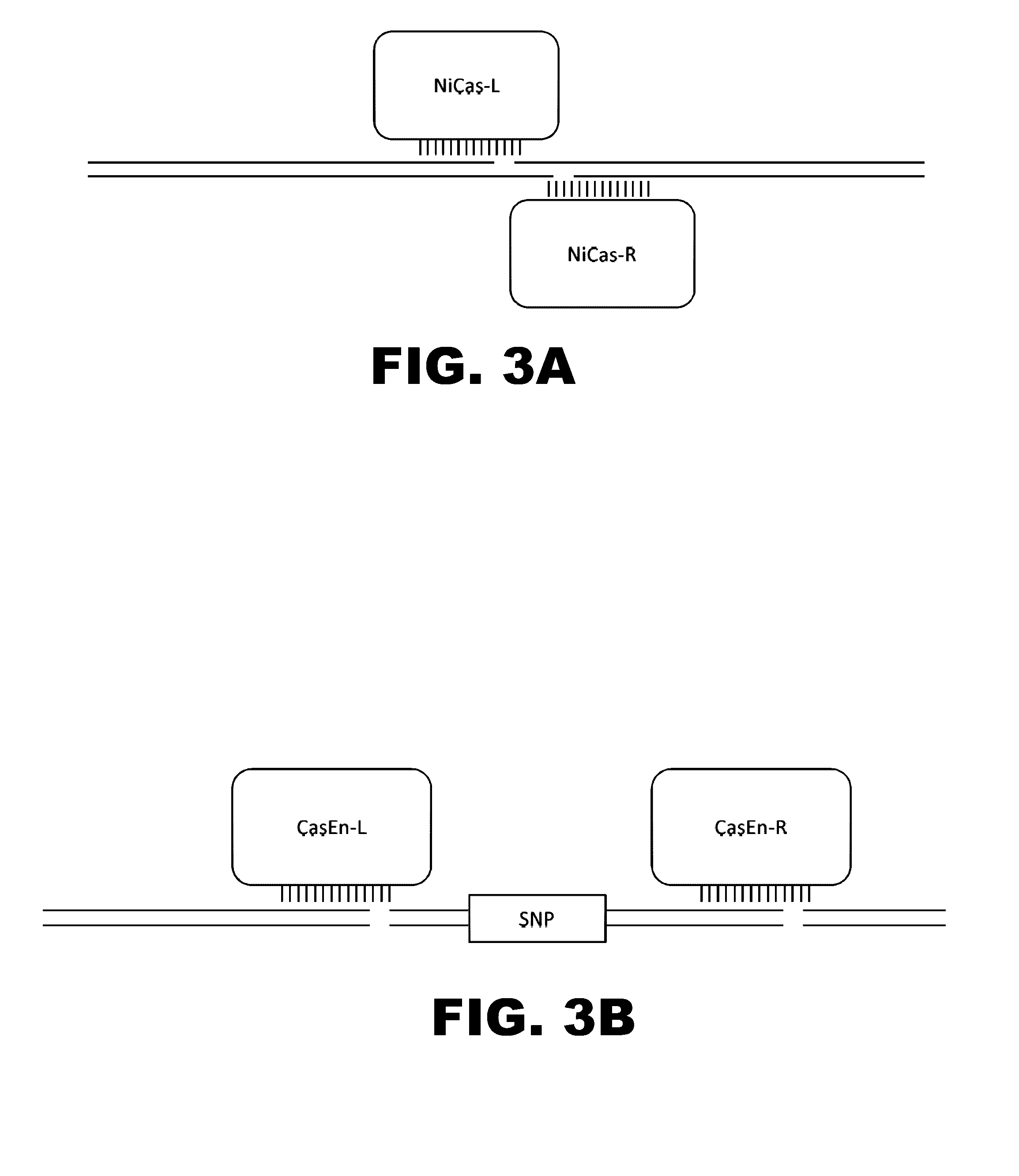
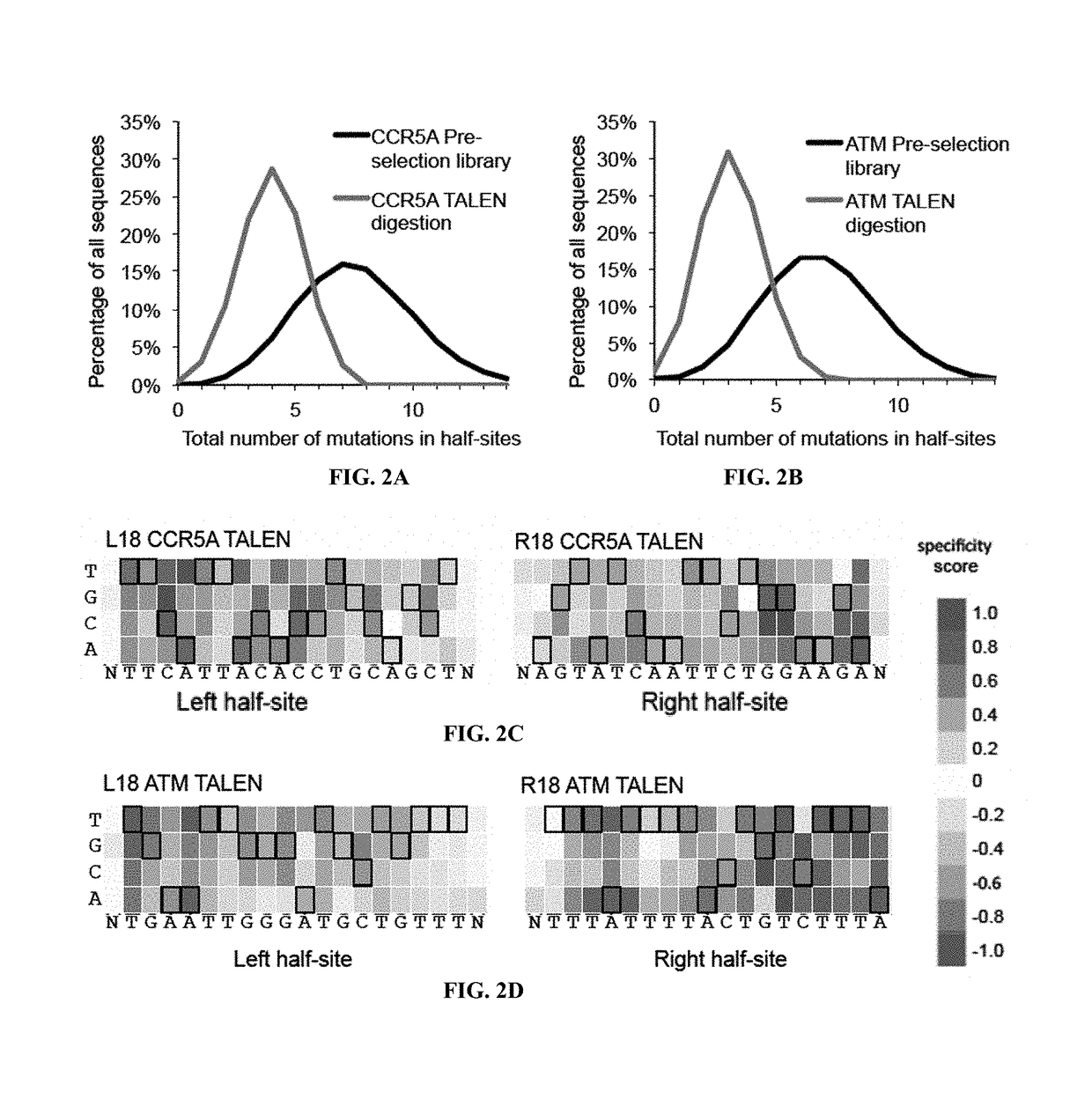
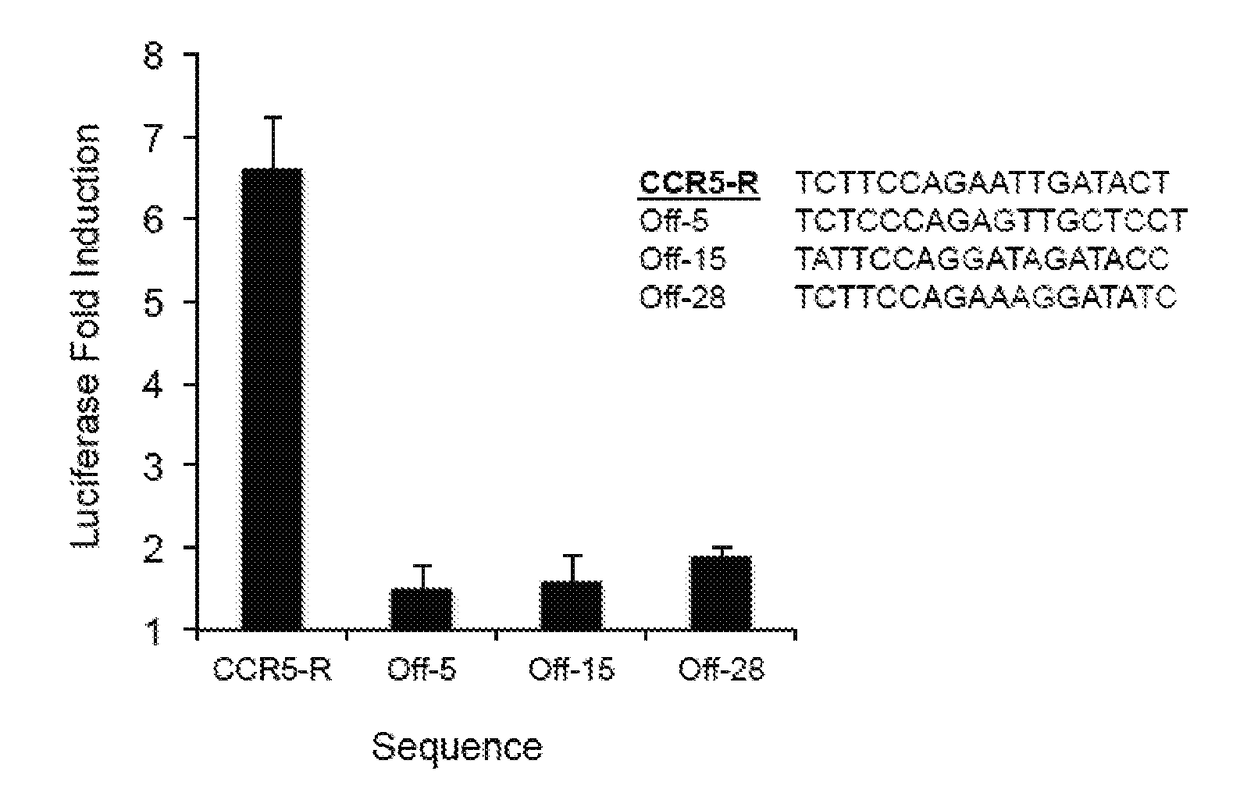
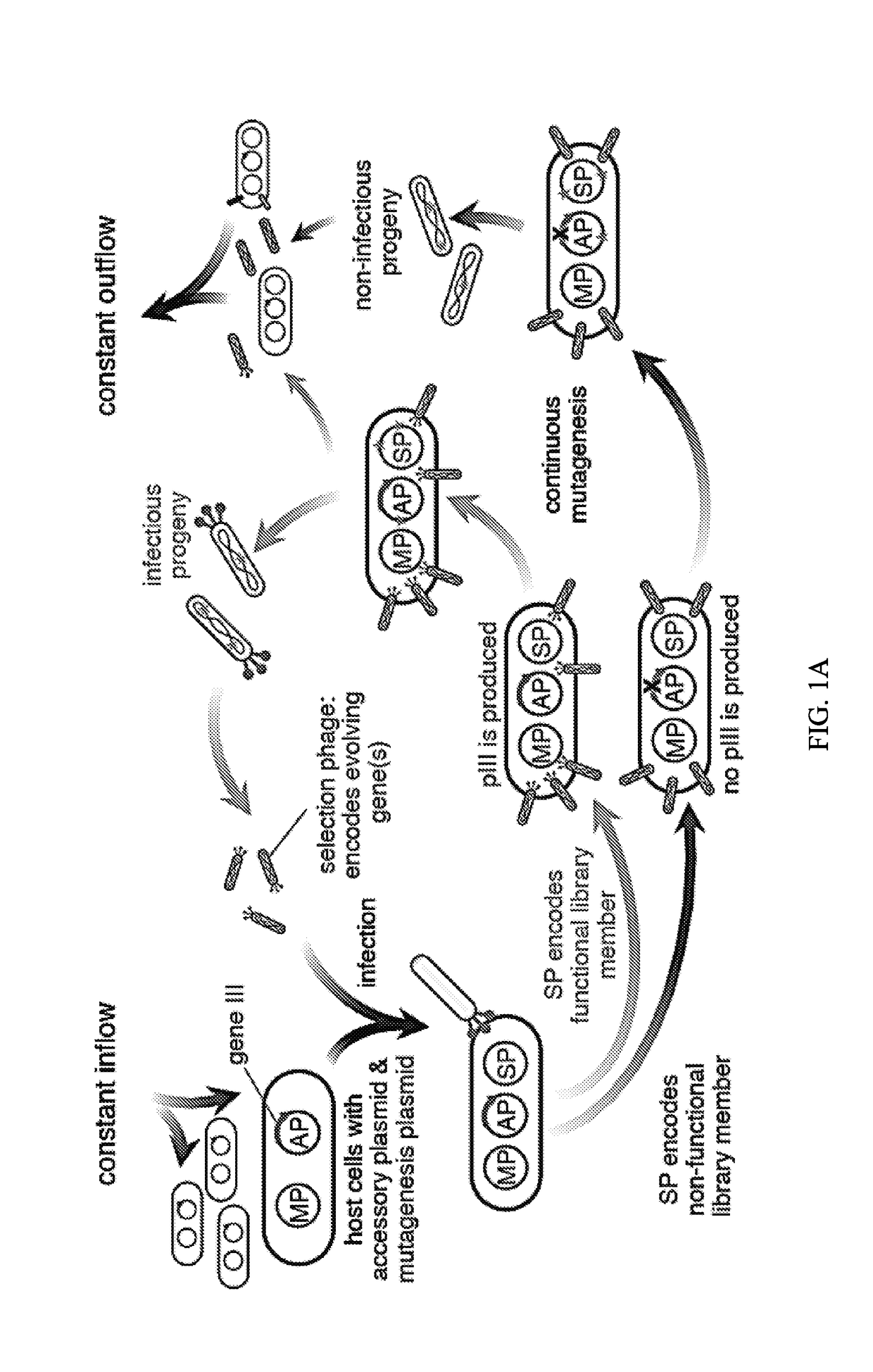
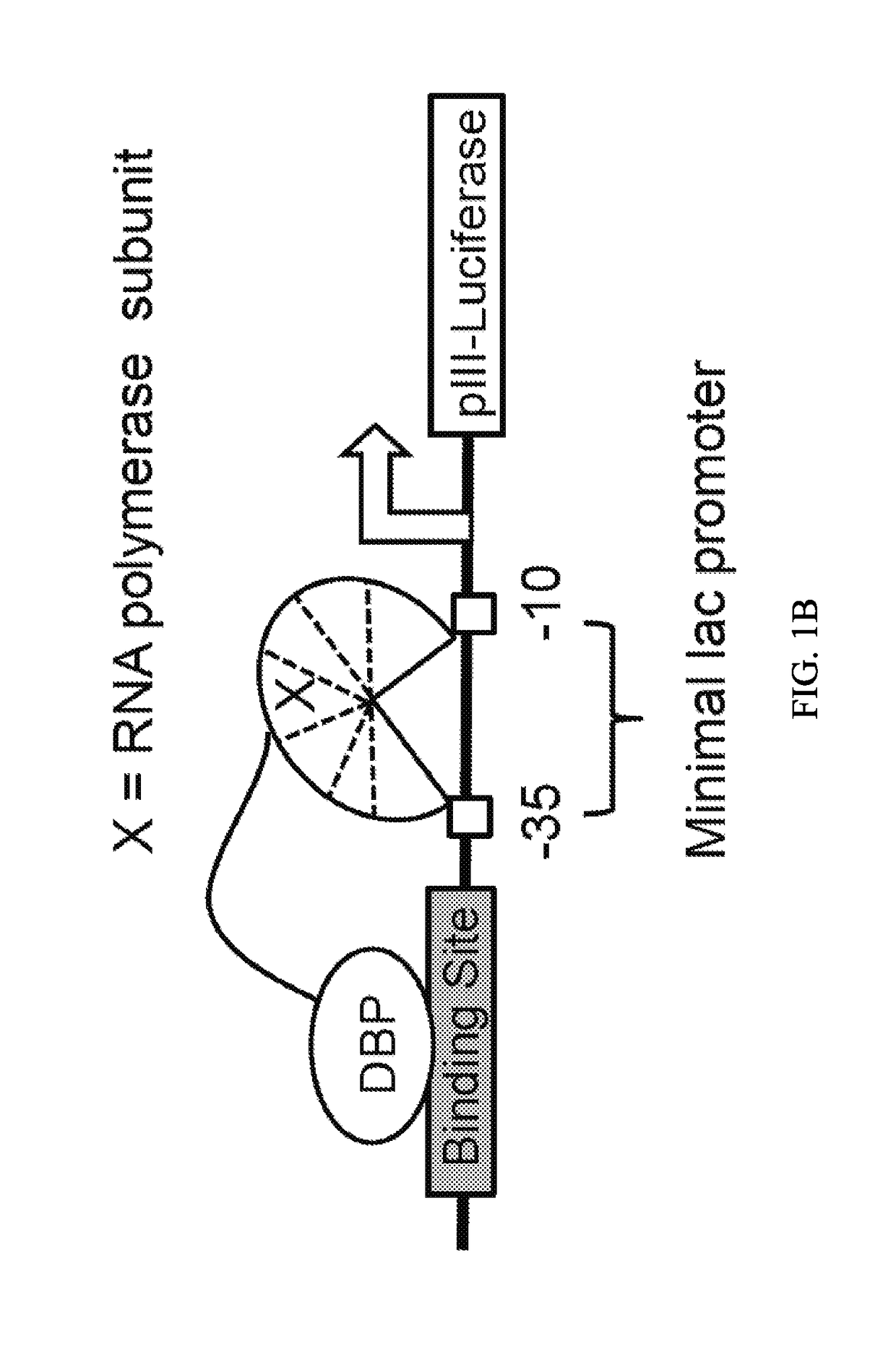
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is that the 5′ nucleotide of the target is specific for thymine (T). TALE domains with alternative 5′ nucleotide specificities could expand the scope of DNA target sequences that can be bound by TALEs. Another drawback of TALEs is their tendency to bind and cleave off-target sequence, which hampers their clinical application and renders applications requiring high-fidelity binding unfeasible. This disclosure provides methods and strategies for the continuous evolution of proteins comprising DNA-binding domains, e.g., TALE domains. In some aspects, this disclosure provides methods and strategies for evolving such proteins under positive selection for a desired DNA-binding activity and / or under negative selection against one or more undesired (e.g., off-target) DNA-binding activities. Some aspects of this disclosure provide engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with altered 5′ nucleotide specificities of target sequences. Engineered TALEs that target ATM with greater specificity are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Gene and peptide for transcriptional repressor
Owner:GREENSOGNA +1
Evolution of TALENs
ActiveUS10612011B2Strong specificityHydrolasesGenetic therapy composition manufactureNucleotideEpigenetic Profile
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is that the 5′ nucleotide of the target is specific for thymine (T). TALE domains with alternative 5′ nucleotide specificities could expand the scope of DNA target sequences that can be bound by TALEs. This disclosure provides methods and strategies for the continuous evolution of proteins comprising DNA-binding domains, e.g., TALE domains. In some aspects, this disclosure provides methods and strategies for evolving such proteins under positive selection for a desired DNA-binding activity and / or under negative selection against one or more undesired (e.g., off-target) DNA-binding activities. Some aspects of this disclosure provide engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with altered 5′ nucleotide specificities of target sequences. Engineered TALEs that target ATM with greater specificity are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Gene capable of increasing seed protein content and method of use thereof
ActiveUS20120159666A1Seed protein content is improvedAchieve mass productionClimate change adaptationOther foreign material introduction processesBiotechnologyProtein composition
According to the present invention, a gene having a novel function that can cause an increase or decrease in seed protein content is searched for. A chimeric protein obtained by fusing a transcription factor consisting of a protein comprising an amino acid sequence shown in any of the even-numbered SEQ ID NOS: 1 to 76 and a functional peptide capable of converting an arbitrary transcription factor into a transcriptional repressor or a transcription factor consisting of a protein comprising an amino acid sequence shown in any of the even-numbered SEQ ID NOS: 77 to 84 is expressed in a plant.
Owner:TOYOTA JIDOSHA KK
Evolution of talens
ActiveUS20180237758A1Strong specificityHydrolasesGenetic therapy composition manufactureDNA-binding domainOff targets
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is that the 5′ nucleotide of the target is specific for thymine (T). TALE domains with alternative 5′ nucleotide specificities could expand the scope of DNA target sequences that can be bound by TALEs. This disclosure provides methods and strategies for the continuous evolution of proteins comprising DNA-binding domains, e.g., TALE domains. In some aspects, this disclosure provides methods and strategies for evolving such proteins under positive selection for a desired DNA-binding activity and / or under negative selection against one or more undesired (e.g., off-target) DNA-binding activities. Some aspects of this disclosure provide engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with altered 5′ nucleotide specificities of target sequences. Engineered TALEs that target ATM with greater specificity are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Gene capable of improving material productivity in seed and method for use thereof
ActiveUS20120144522A1Improve productivityImprove fat contentOther foreign material introduction processesPlant peptidesProduction rateBiotechnology
An object of the present invention is to search for a gene having a novel function that can cause an increase or decrease in material productivity, and particularly, fat and oil content. In the present invention, a chimeric protein obtained by fusing a transcription factor consisting of a protein comprising an amino acid sequence shown in any of the even-numbered SEQ ID NOS: 1 to 158 and a functional peptide capable of converting an arbitrary transcription factor into a transcriptional repressor is expressed in a plant.
Owner:TOYOTA JIDOSHA KK
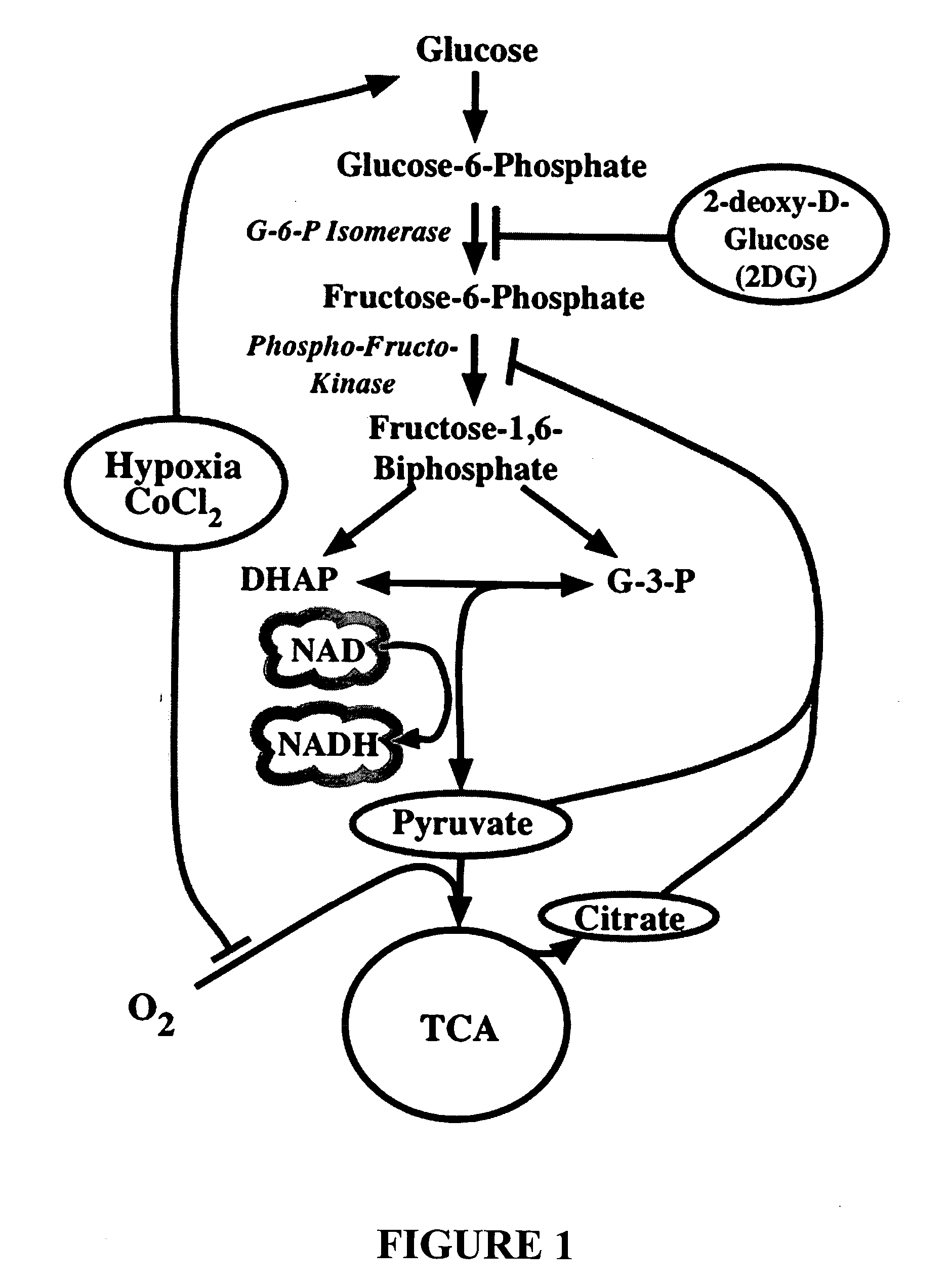
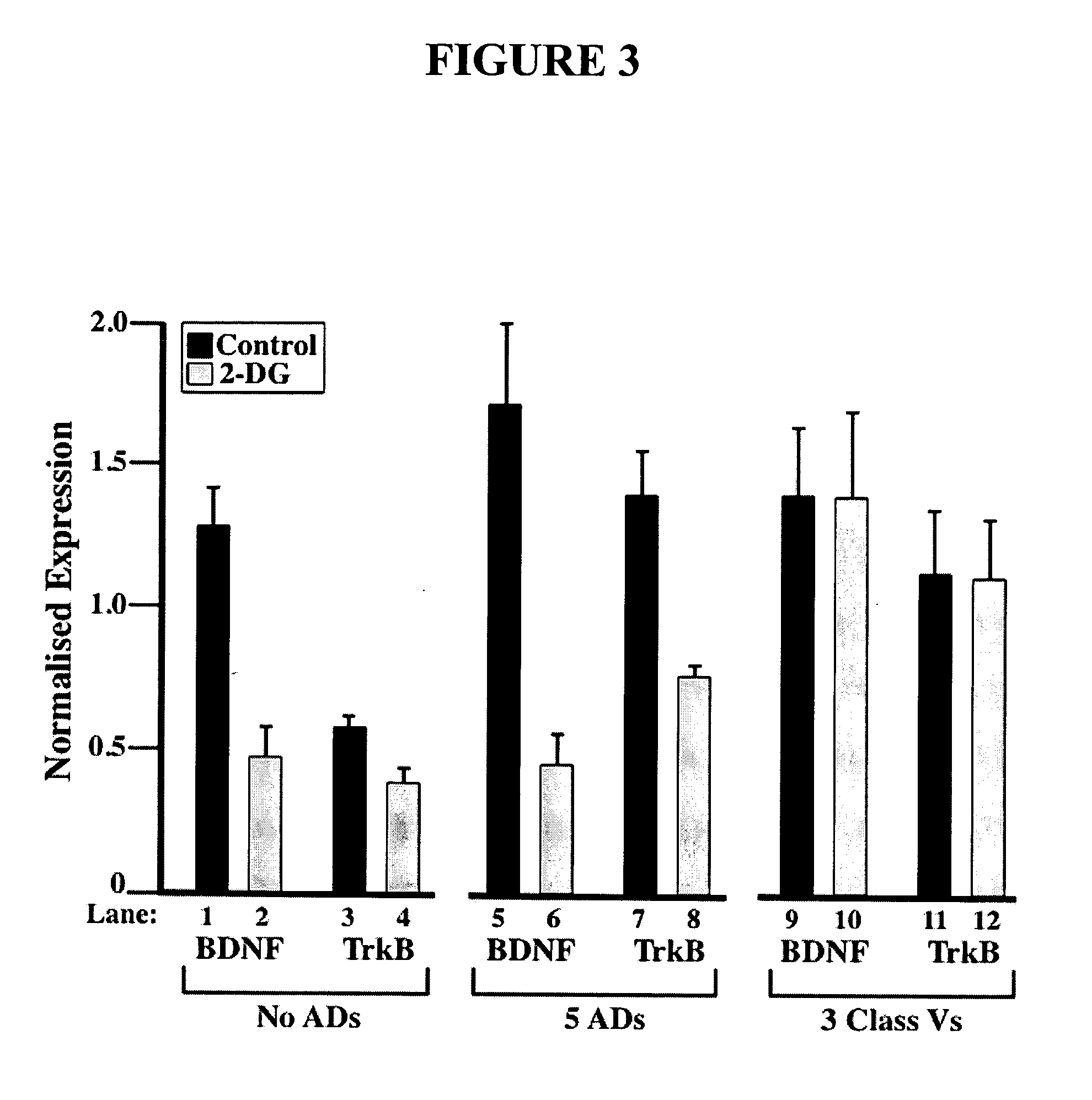
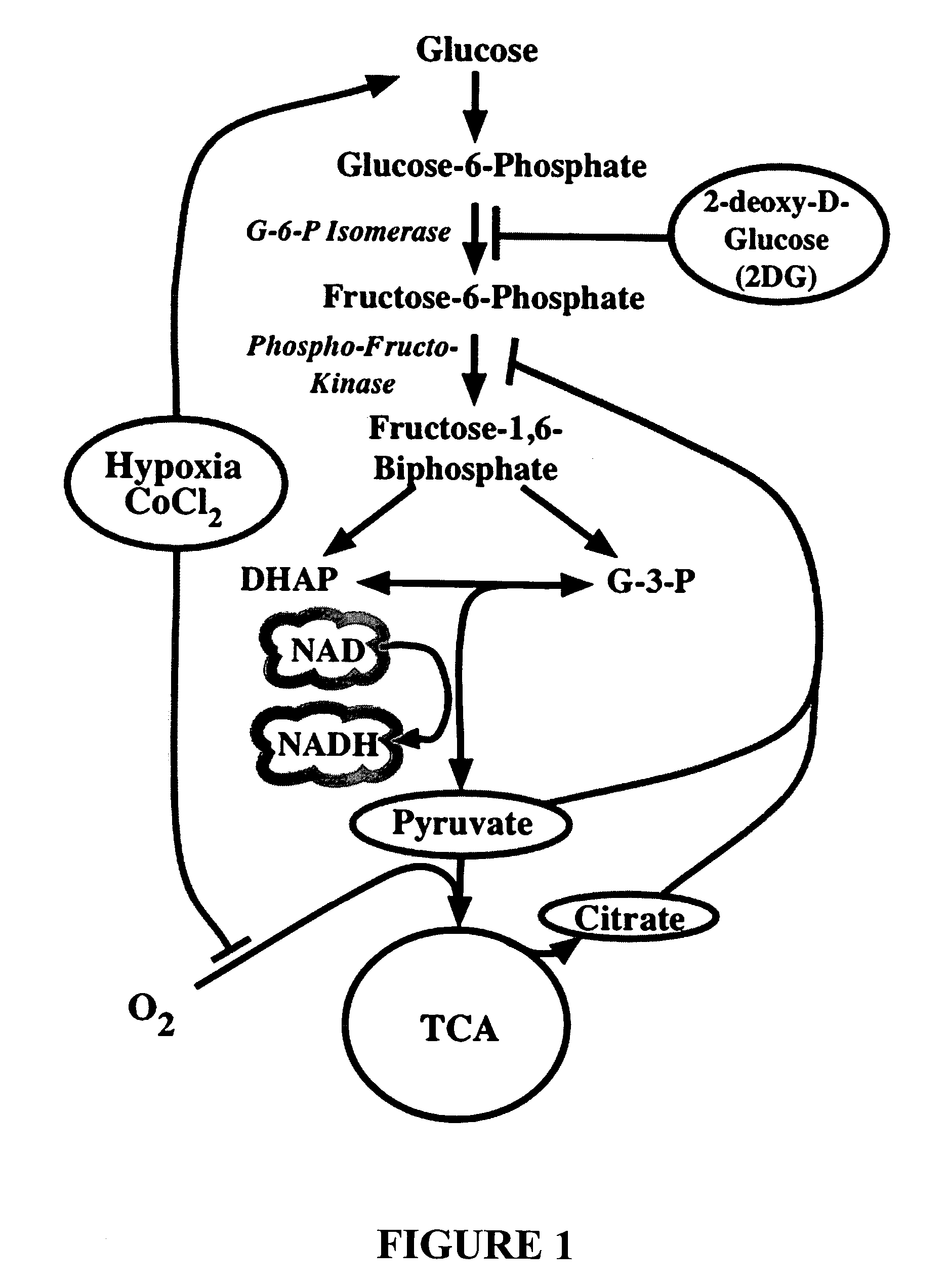
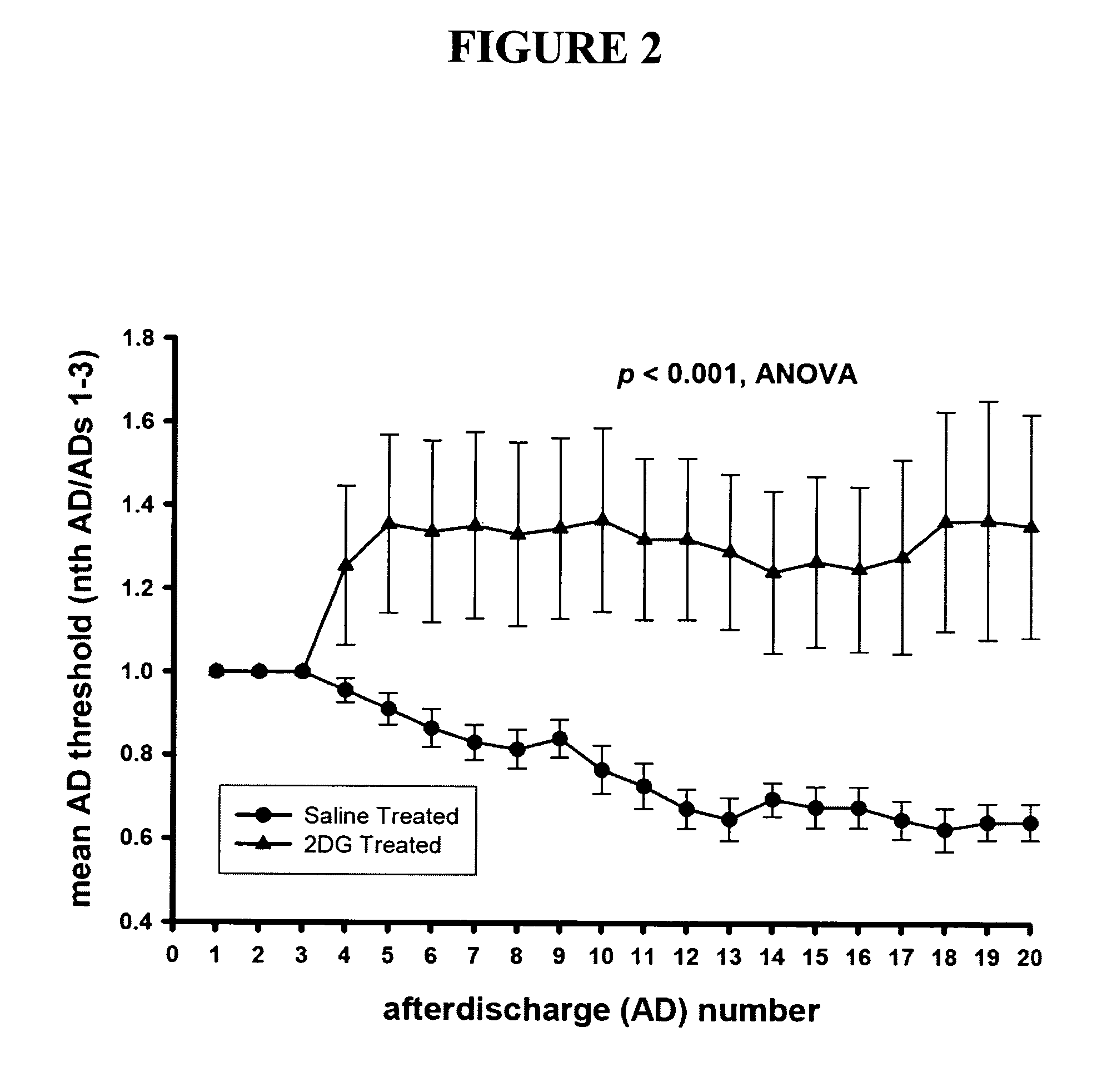
Metabolic-based methods for modulating gene expression
This invention provides methods for modulating cellular gene expression for genes operably linked to an NRSE element that is recognized by an NRSF transcriptional repressor, by changing the concentration of reduced nicotinamide adenine dinucleotide (NADH) in the cell.
Owner:WISCONSIN ALUMNI RES FOUND
Transcriptional Repressors of Cytokinin Signaling and Their Use
ActiveUS20100115666A1Shorten germination timeReduce time lossFusion with DNA-binding domainImmunoglobulinsPolynucleotideTransgene
The invention relates to fusion proteins capable of acting as transcriptional repressors of cytokinin signaling, to polynucleotides encoding these fusion proteins, to vectors and cells comprising these polynucleotides, and to transgenic plants and parts thereof comprising these polynucleotides, vectors, and cells.The invention further relates to a process for making these transgenic plants and to the use of these transgenic plants for producing seeds of enhanced size, with enhanced seed filling, with reduced seed loss and / or with more rapid germination, and / or for producing a live root system with increased root mass, root length and / or root branching. The invention also relates to a method for enhancing the seed size, for enhancing seed filling, for reducing seed loss, and / or for reducing germination time and / or reproduction time, and / or for enhancing the root mass, root length and / or root branching of a plant and to seeds obtainable by the methods of the present invention.
Owner:SCHMULLING THOMAS +2
Isopropyl alcohol-producing bacterium having improved productivity by gntr destruction
ActiveUS20130211170A1Efficient productionImprove efficiencyBacteriaMolecular sieve catalystEscherichia coliCoprecipitation
An isopropyl alcohol-producing Escherichia coli includes an isopropyl alcohol production system, wherein an activity of transcriptional repressor GntR is inactivated, and the isopropyl alcohol-producing Escherichia coli preferably further includes a group of auxiliary enzymes having an enzyme activity expression pattern with which isopropyl alcohol production capacity achieved by the inactivation of the GntR activity is maintained or enhanced. A method of producing isopropyl alcohol includes producing isopropyl alcohol from a plant-derived raw material using the isopropyl alcohol-producing Escherichia coli. A method of producing acetone includes contacting the isopropyl alcohol obtained by the isopropyl alcohol production method with a complex oxide that includes zinc oxide and at least one oxide containing a Group 4 element, and that is prepared by coprecipitation. A method of producing propylene includes contacting isopropyl alcohol and acetone obtained by the production method with a solid acidic substance and a Cu-containing hydrogenation catalyst as catalysts.
Owner:MITSUI CHEM INC
Modification of transcriptional repressor binding site in NF-YC4 promoter for increased protein content and resistance to stress
Method of increasing protein content in a eukaryotic cell comprising an NF-YC4 gene comprising modifying the transcriptional repressor binding site; method of producing a plant with increased proteincontent comprising crossing and selecting for increased protein content; method of increasing resistance to a pathogen or a pest in a plant comprising an NF-YC4 gene comprising modifying the transcriptional repressor binding site, alone or in further combination with expressing QQS in the plant; method for producing a plant with increased resistance to a pathogen or a pest comprising crossing andselecting for increased resistance to the pathogen or the pest; a cell, collection of cells, tissue, organ, or organism in which the NF-YC4 gene comprises a promoter comprising a transcriptional repressor binding site that has been modified so that the transcriptional repressor cannot prevent transcription of the NF-YC4; hybrid plants; and seeds.
Owner:IOWA STATE UNIV RES FOUND
EXO1 Promotor Polymorphism Associated with Exceptional Life Expectancy in Humans
InactiveUS20110027783A1Improve life expectancyPrevents DNA damageSugar derivativesMicrobiological testing/measurementMedicinePromoter polymorphism
The invention relates to an exonuclease 1 (EXO1) promoter polymorphism associated with exceptional life expectancy in humans. More specifically, the invention relates to the promoter region of EXO1 where the gene has been found to be mutated. Moreover, the invention relates to a gene regulation mechanism involving transcription factor E47 as a transcriptional repressor of EXO1.
Owner:UNIVSKLINIKUM SCHLESWIG HOLSTEIN
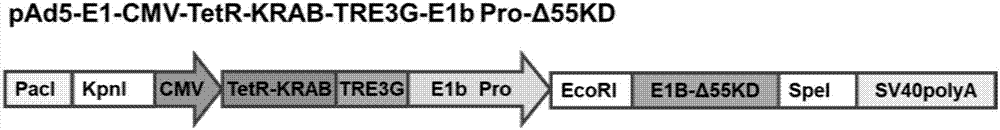
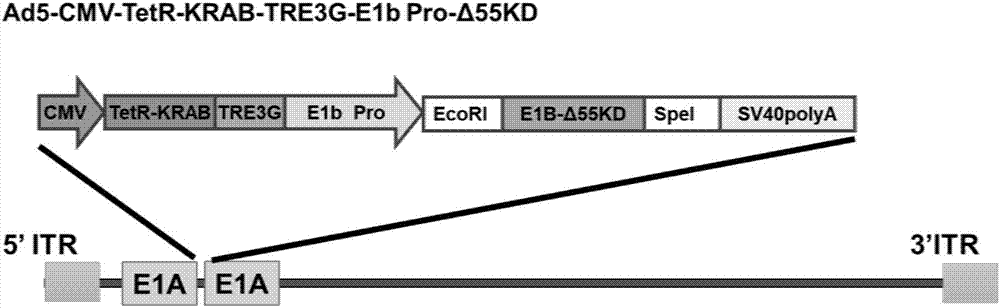
Conditionally replicating adenovirus vector for viral replication regulated by transcription inhibition type Tet-On system and application
The invention discloses a conditionally replicating adenovirus vector for virus replication regulated by a TetR-KRAB-mediated transcription inhibition type Tet-On system, adenovirus E1 region comprises the following components in the following connection sequence: 5'-CMV promoter, TetR-KRAB gene, promoter TRE3G-E1b Pro containing a tetracycline responsive element, EcoR I enzyme cutting site, E1B Delta 55KD protein-deleted adenovirus E1A-E1B19KD gene sequence, enzyme cutting site Spe I and mRNA transcription termination signal SV40polyA-3'; the CMV promoter expresses transcription inhibitor TetR-KRAB; a gene sequence inserted into the two enzyme cutting sites of the EcoR I and the Spe I is a gene fragment from adenovirus E1A translation initiation site to adenovirus E1B19KD protein termination codon, the promoter TRE3G-E1b Pro containing the tetracycline responsive element expresses adenovirus E1A gene, adenovirus E1B19KD gene is expressed by the own promoter of the adenovirus E1B19KD, and the packed adenovirus is named as Ad5-CMV-TetR-KRAB-TRE3G-E1bPro-Delta 55KD. Tests show that the virus can be loaded into mesenchymal stem cells for application in study of tumor therapy.
Owner:SHAANXI NORMAL UNIV
Transcriptional repressor peptides and genes for the same
ActiveUS20110035846A1Effective transcriptionReliably inhibit transcription and expressionPeptide/protein ingredientsImmunoglobulinsAmino acid compositionTranscriptional Repressor
Owner:NAT INST OF ADVANCED IND SCI & TECH
Metabolic-based methods for modulating gene expression
This invention provides methods for modulating cellular gene expression for genes operably linked to an NRSE element that is recognized by an NRSF transcriptional repressor, by changing the concentration of reduced nicotinamide adenine dinucleotide (NADH) in the cell.
Owner:WISCONSIN ALUMNI RES FOUND
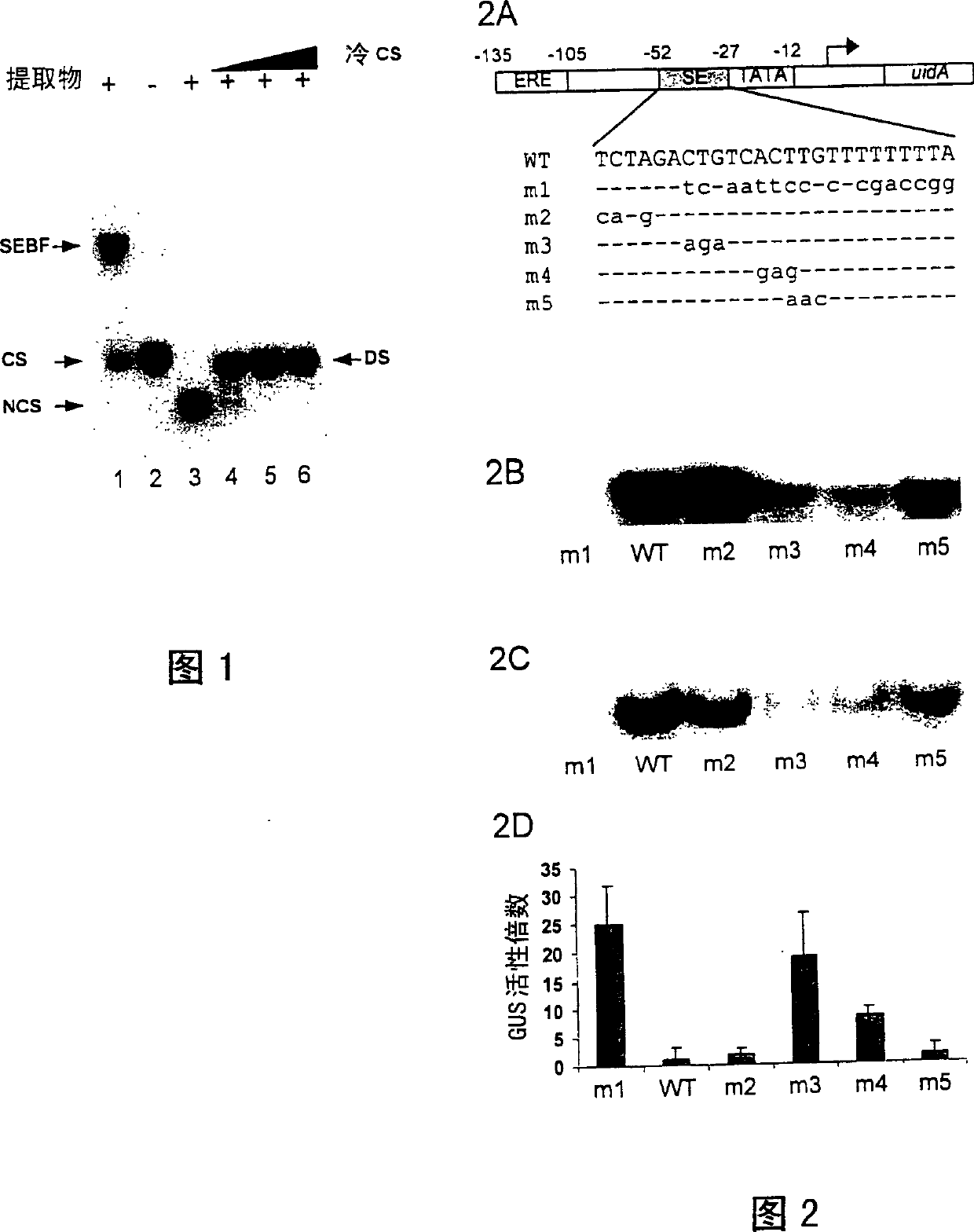
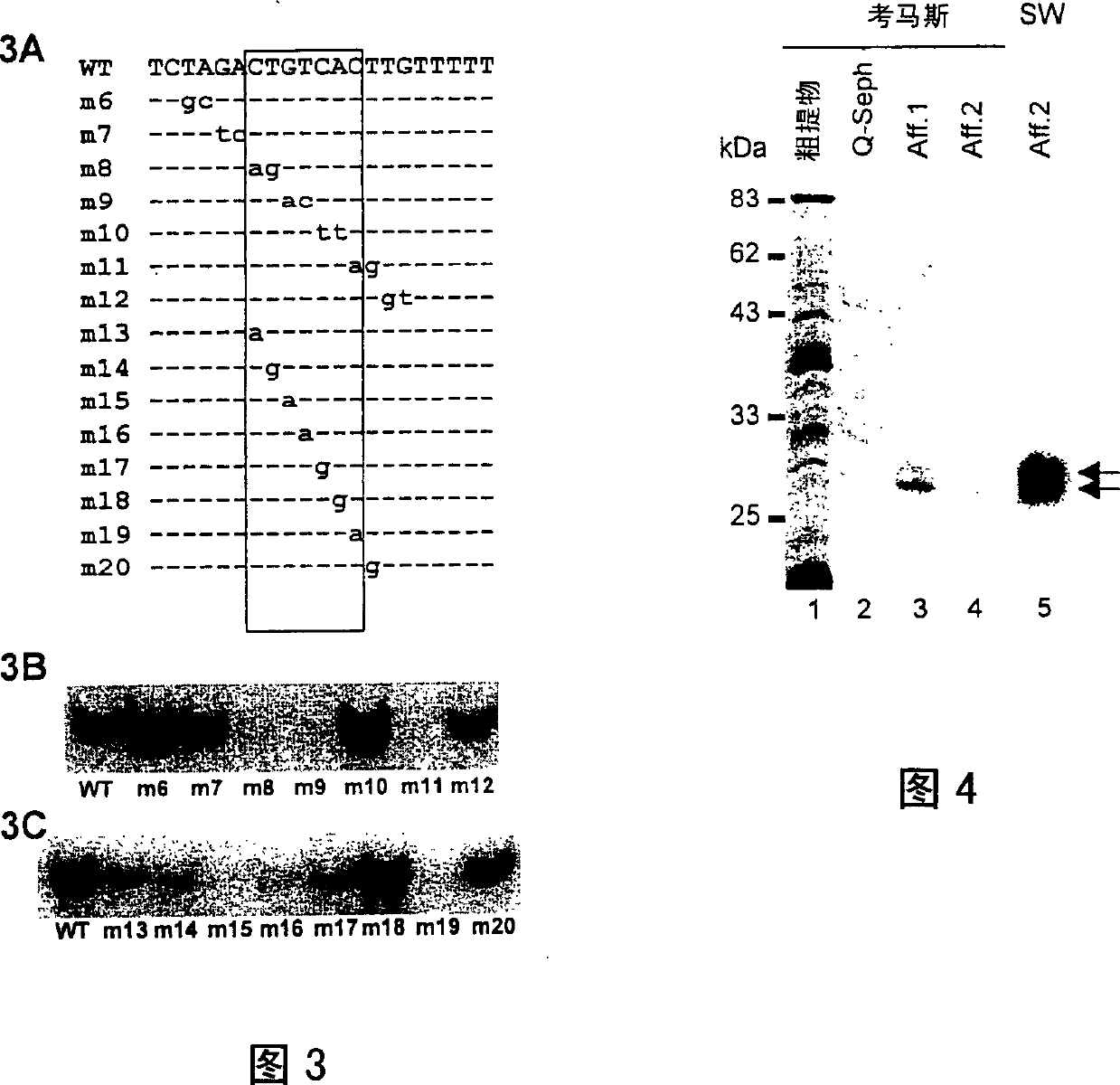
Plant transcriptional repressor, proteic nuclear factors binding thereto, and uses thereof
The present application describes a plant gene regulatory element and the uses thereof. More particularly a silencing element for modulating plant responses to pathogens, auxin and ethylene is described. The invention also describes transcriptional repressors which specifically binds onto the silencing element of the invention. Nucleotide and amino acid sequences of a novel transcriptional repressor referred as 'SEBF' are given.
Owner:造值研究有限公司
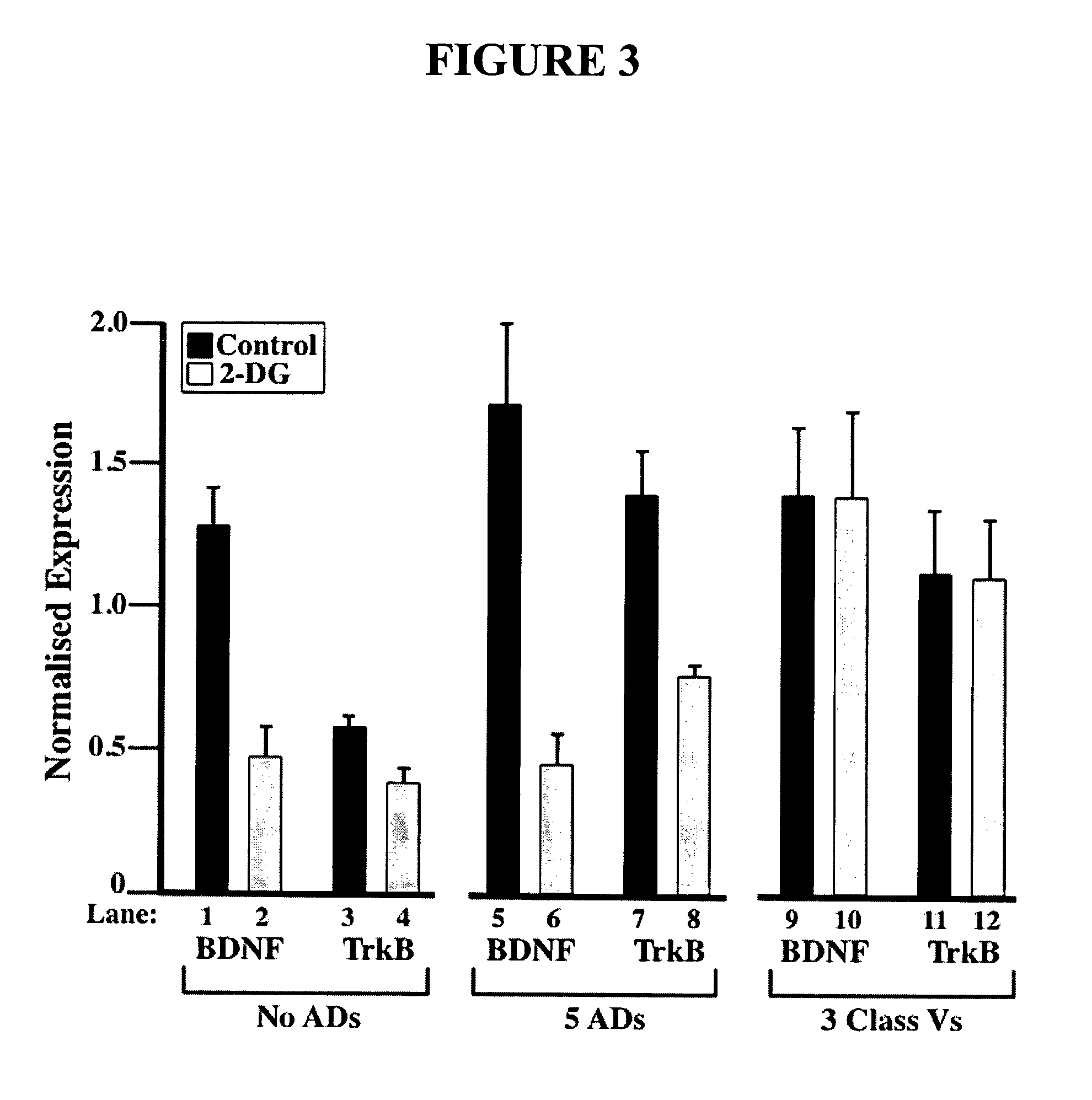
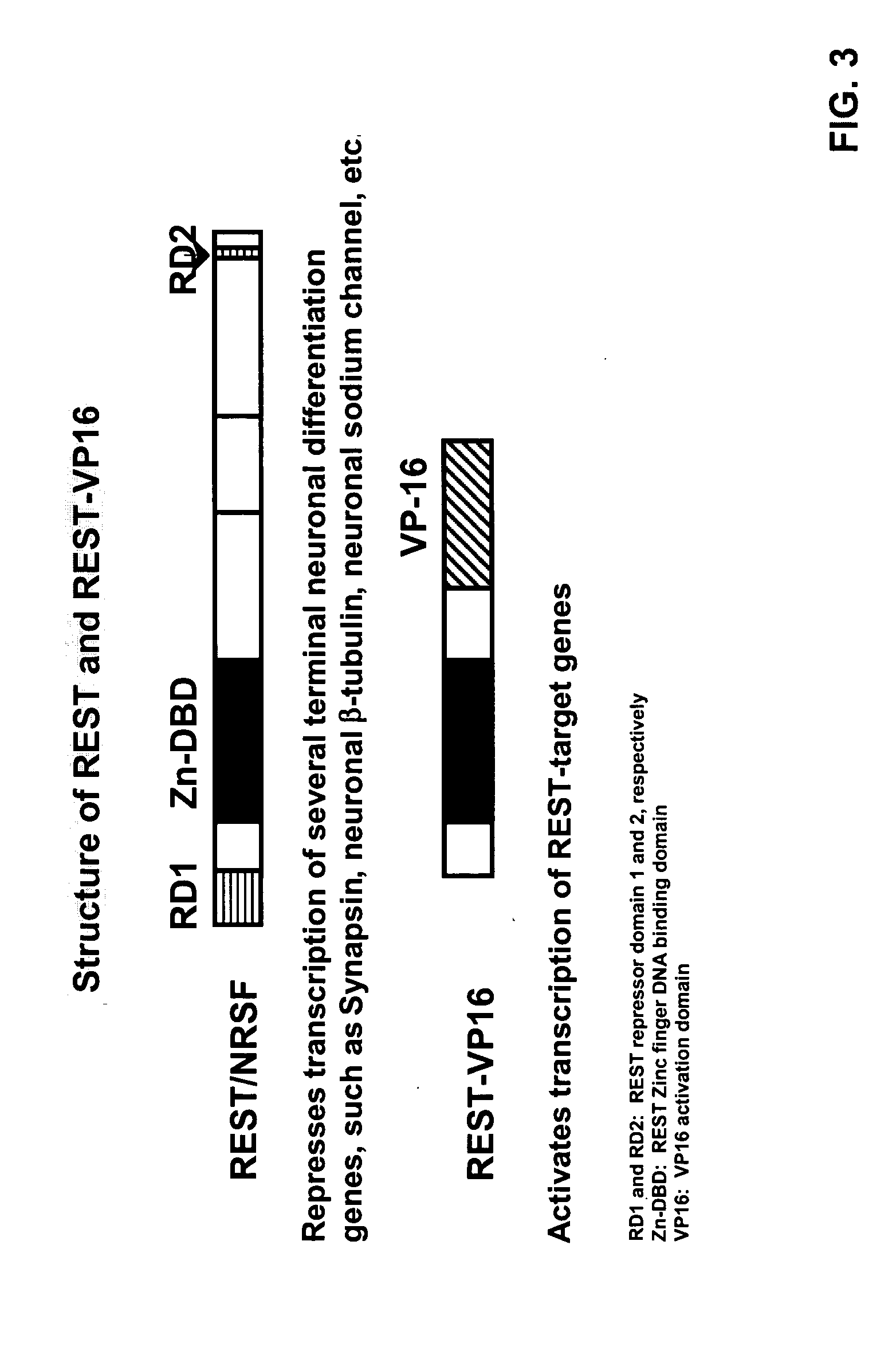
Methods and compositions related to neuronal differentiation
Compositions and methods of the invention use a novel transactivator methodology for manipulation of the molecular mechanisms of cell determination for the production of a cell with a neuronal phentoype. A recombinant transcription factor or transactivator that binds the RE1 promoter element, REST-transactivator, was constructed by replacing the repressor domains of the transcriptional repressor REST with a transcriptional activation domain. The RE1 binding transactivator was designed to induce or manipulate the neuronal differentiation process.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
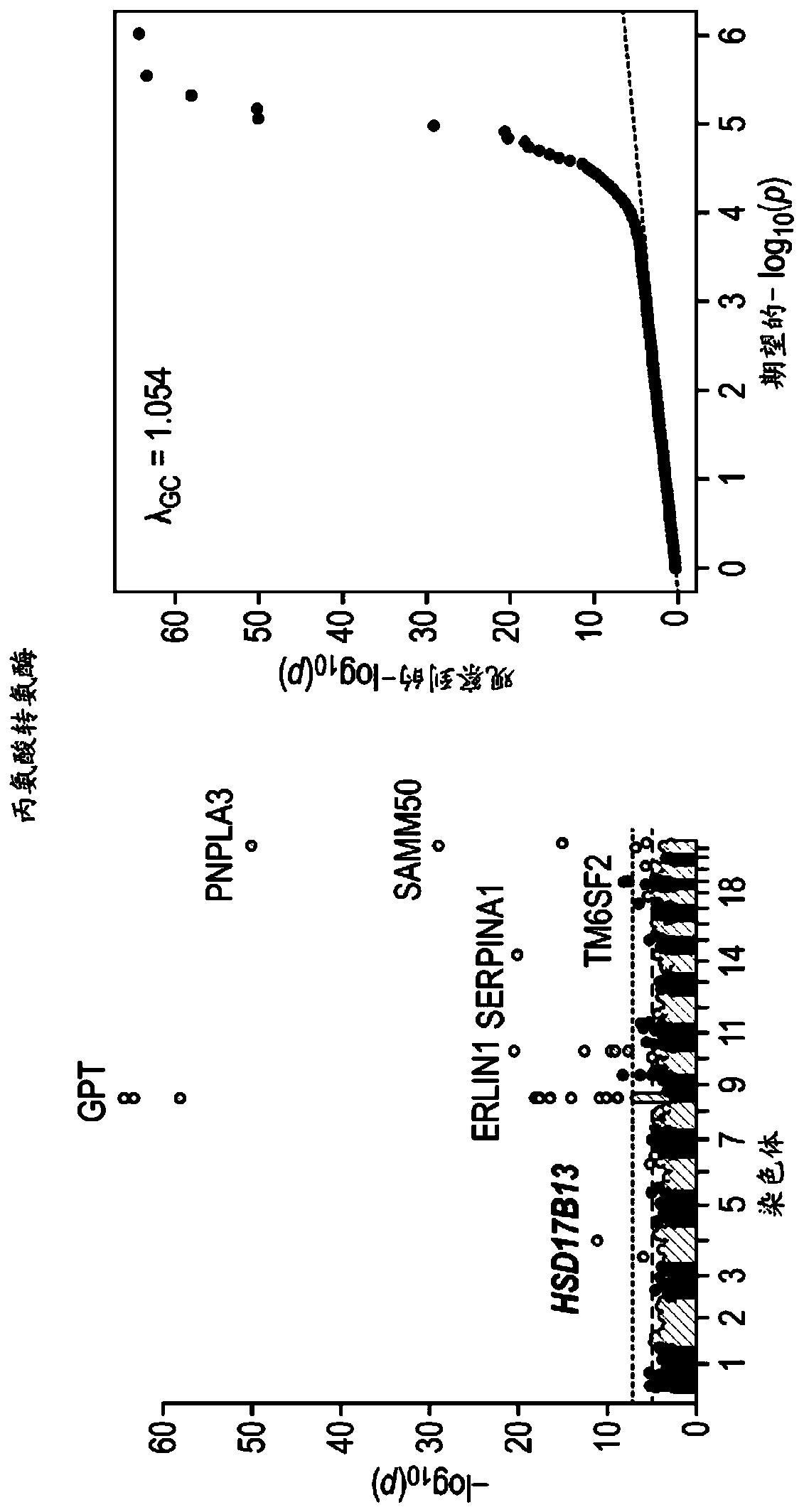
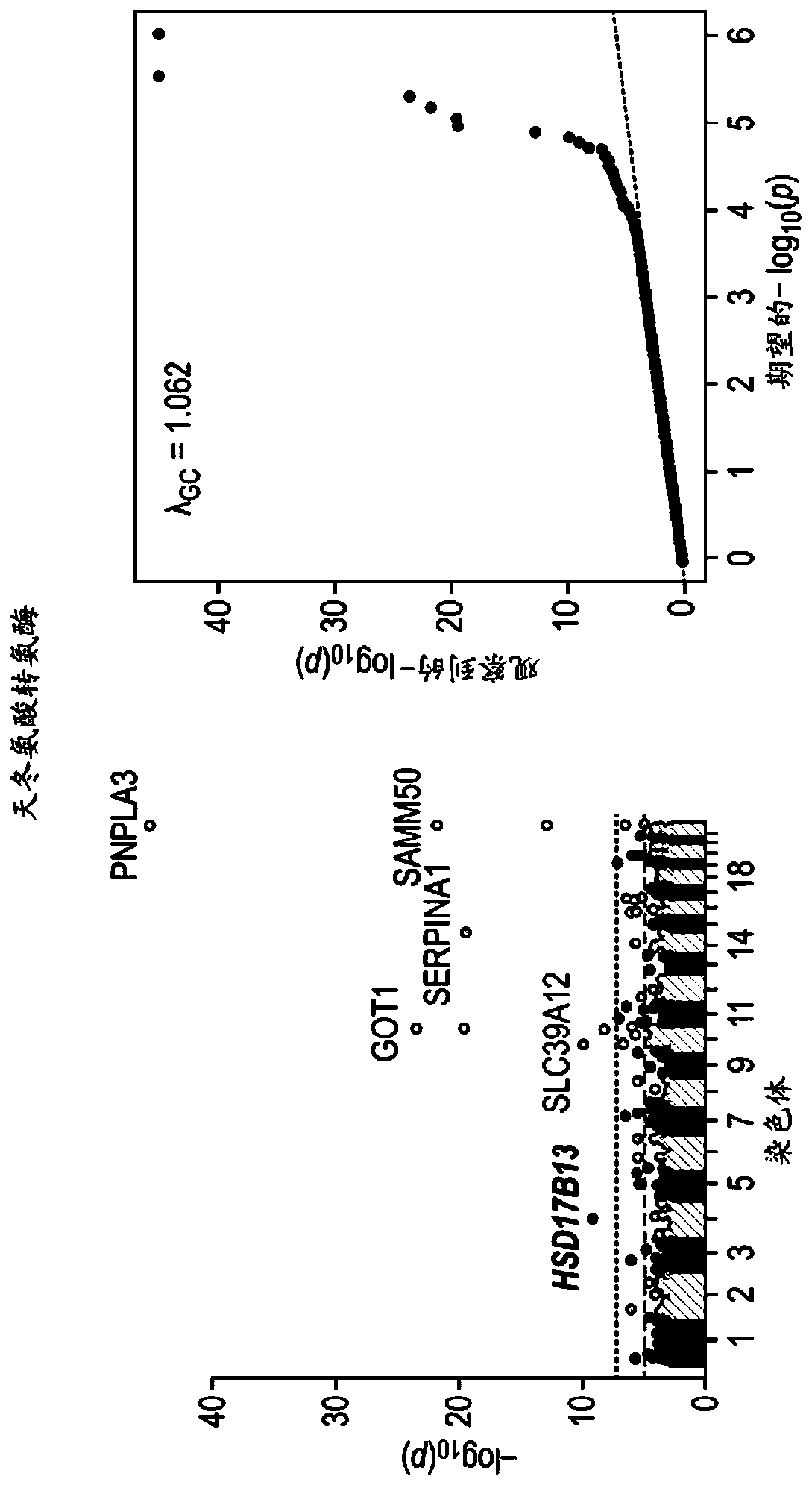
Hsd17b13 variants and uses thereof
Provided are compositions related to HSD17B13 variants, including isolated nucleic acids and proteins related to variants of HSD17B13, and cells comprising those nucleic acids and proteins. Also provided are methods related to HSD17B13 variants. Such methods include methods for modifying a cell through use of any combination of nuclease agents, exogenous donor sequences, transcriptional activators, transcriptional repressors, and expression vectors for expressing a recombinant HSD17B13 gene or a nucleic acid encoding an HSD17B13 protein. Also provided are therapeutic and prophylactic methods for treating a subject having or at risk of developing chronic liver disease.
Owner:REGENERON PHARM INC
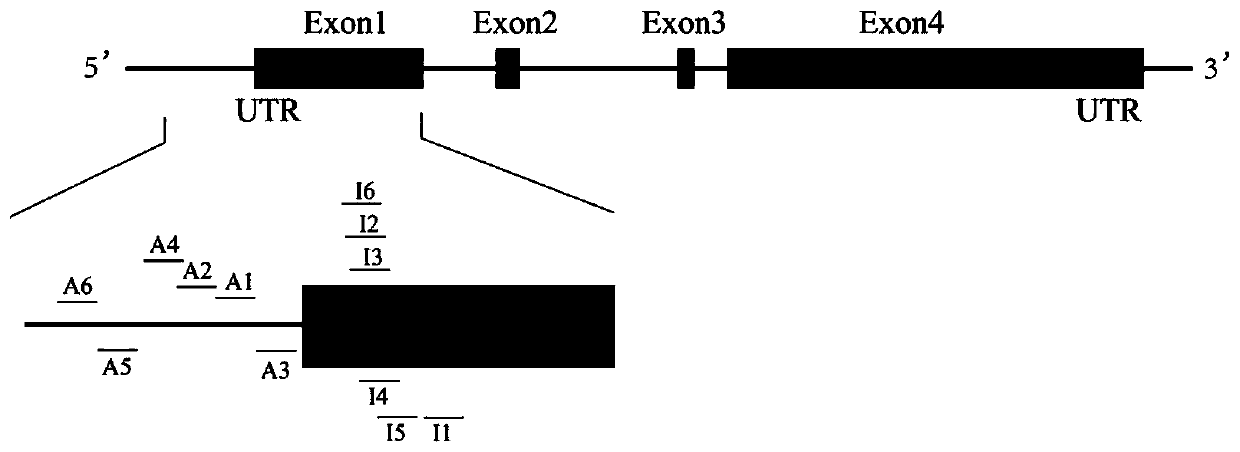
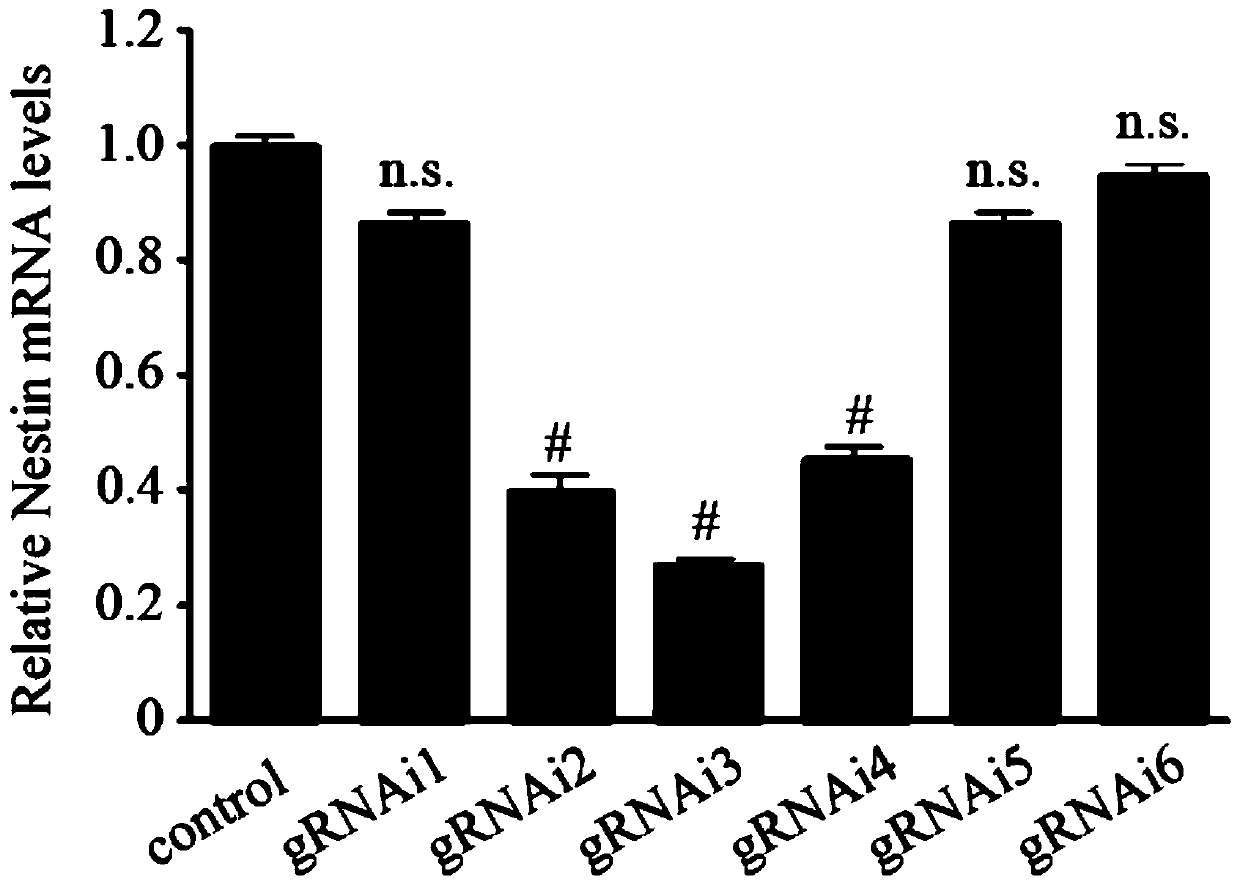
Method for adjusting and controlling expression of nestin gene in neural stem cell and method of targeting cell
ActiveCN110066803AOvercome the disadvantages of constructing stable cell linesOvercoming the disadvantages of stable cell linesStable introduction of DNAAnimals/human peptidesTranscription initiationCell strain
The invention discloses a method for adjusting and controlling expression of a nestin gene in a neural stem cell and a method of targeting a cell, and relates to the technical field of adjustment andcontrol over gene expression. The method for adjusting and controlling the expression of the nestin gene in the neural stem cell comprises the following successively-conducted steps that firstly, gRNAof the nestin gene is designed nearby transcription initiation of the nestin gene, then a carrier is constructed, and the gRNA of the nestin gene can be any one of nestin-i gRNA for guiding a transcriptional repressor and nestin-a gRNA for guiding a transcriptional activator. The method for adjusting and controlling the expression of the nestin gene in the neural stem cell is simple, low in off-target rate and capable of achieving stable expression in cell strains. The defects generated from construction of stable cell strains by a traditional method are overcome, and the stable cell strainscan be obtained in a short time.
Owner:珠海乐维再生医学科技有限公司
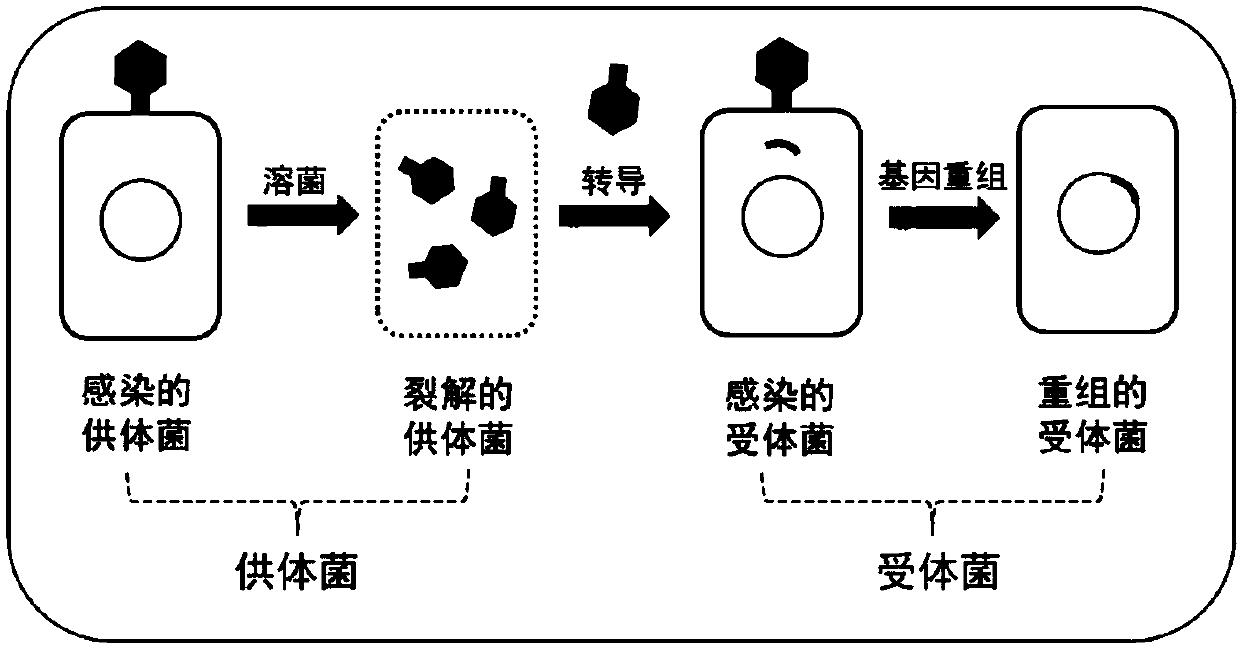
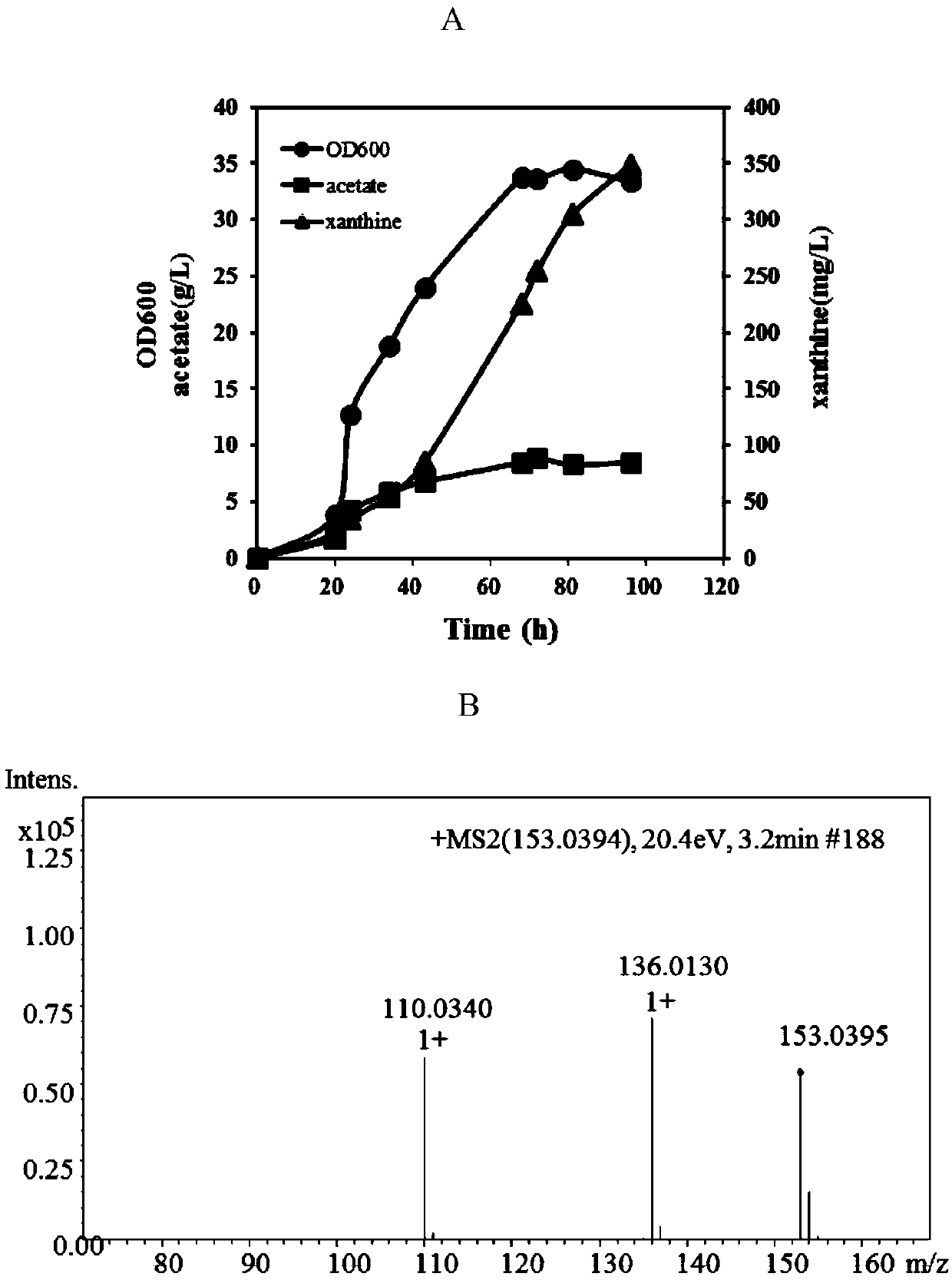
Recombinant bacteria producing xanthine and construction method and application thereof
ActiveCN110656073ARealize accumulationEfficient biosynthesis technologyBacteriaTransferasesEscherichia coliEnzyme Gene
The invention discloses recombinant bacteria producing xanthine and a construction method and application thereof, and belongs to the technical field of genetic engineering. The recombinant bacteria use escherichia coli as original strains, the transcriptional repressor protein purR, phosphoglucose isomerase gene pgi, glucogluconate dehydratase gene edd, adenylosuccinate synthetase gene purA and GMP synthase gene guaA on escherichia coli genome are knocked out, and D128A mutated PRPP synthase gene prs and K326Q and P410W double mutated PRPP amidotrasferase purF are subjected to overexpression.Meanwhile, the invention further provides a method for preparing the recombinant bacteria and a method for producing xanthine by using the recombinant bacteria. For the first time, the high-efficiency biosynthesis of xanthine in recombinant bacteria is realized. The recombinant bacteria are suitable for producing xanthine by fermentation.
Owner:QINGDAO INST OF BIOENERGY & BIOPROCESS TECH CHINESE ACADEMY OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com