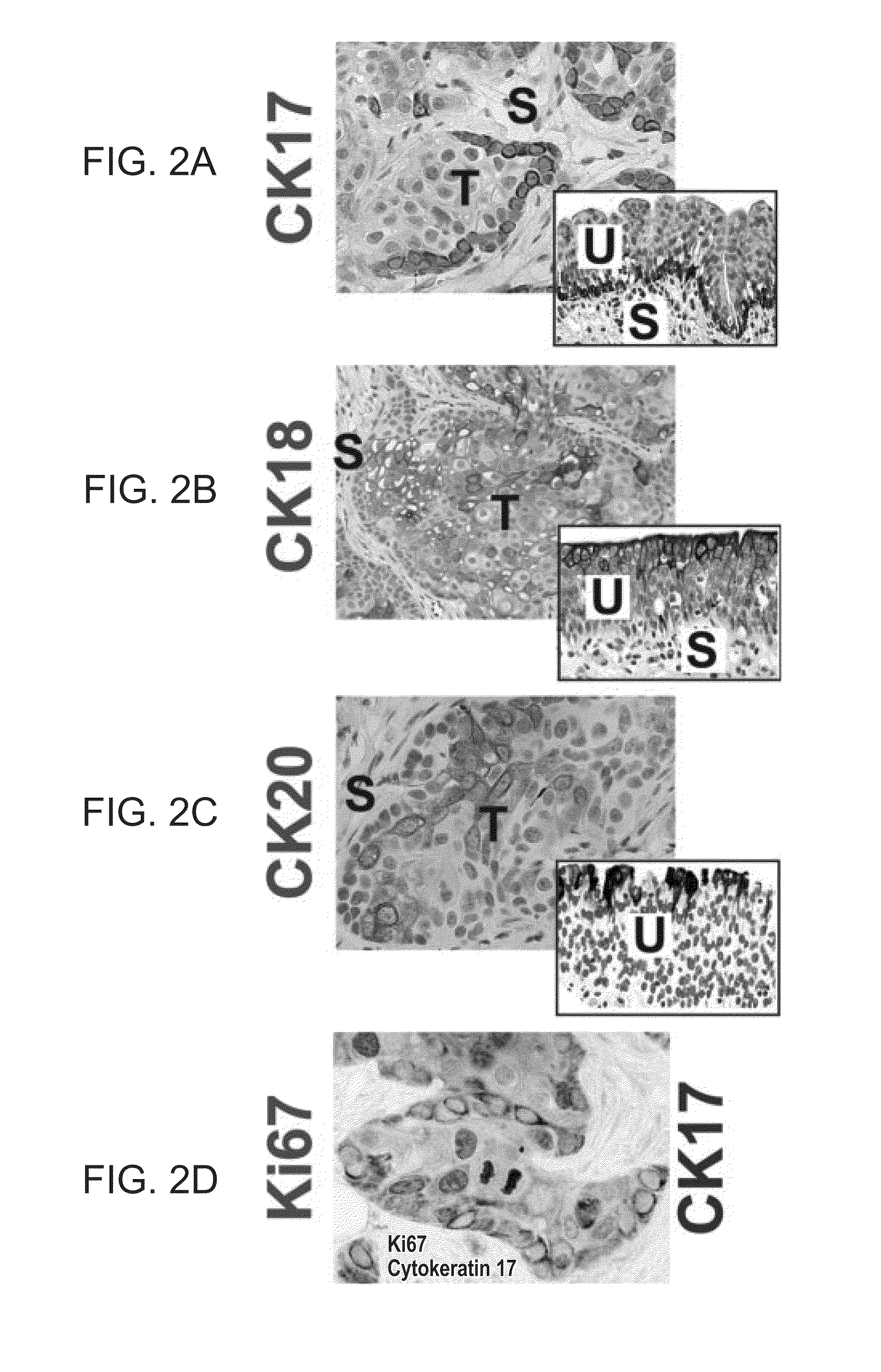
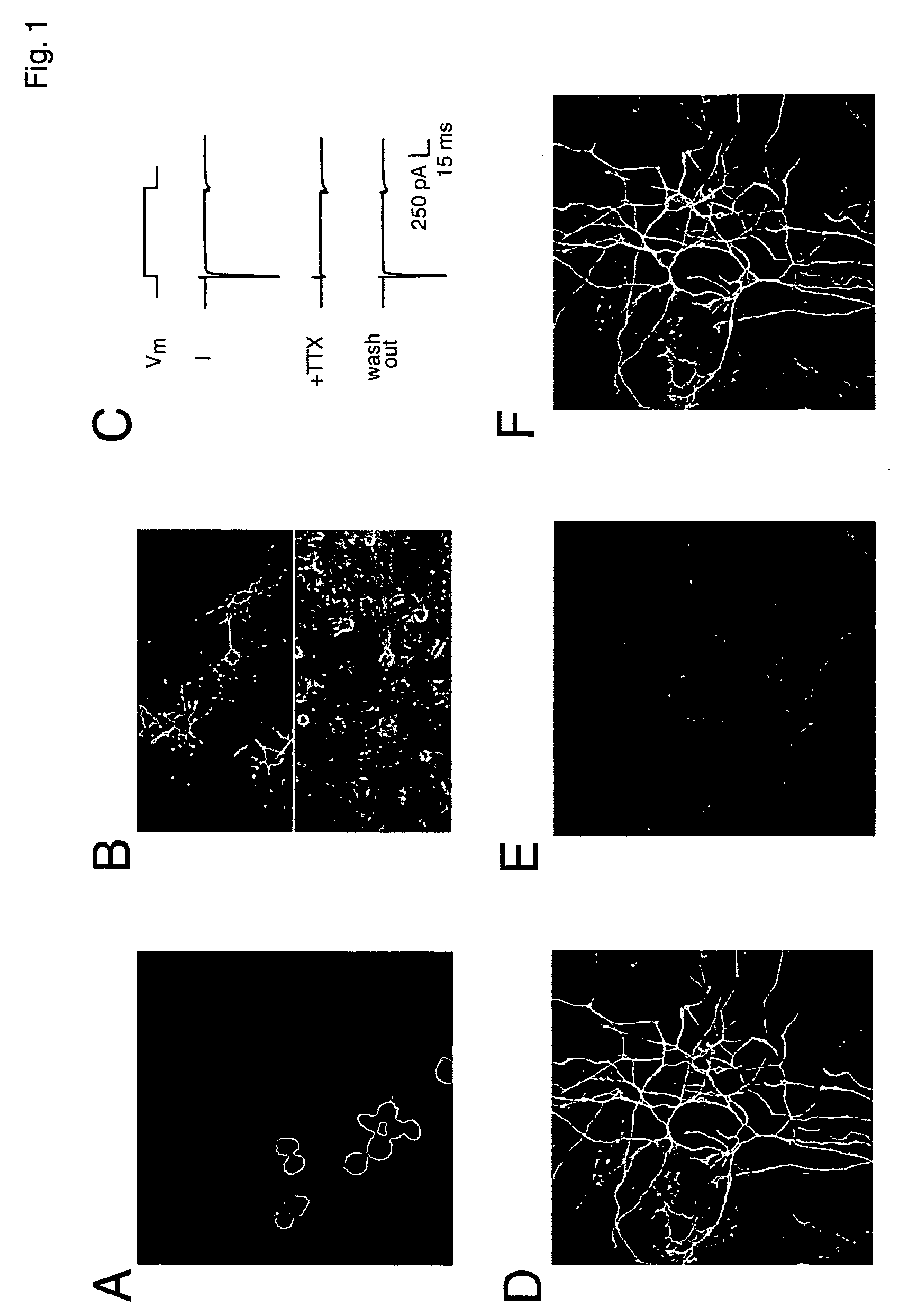
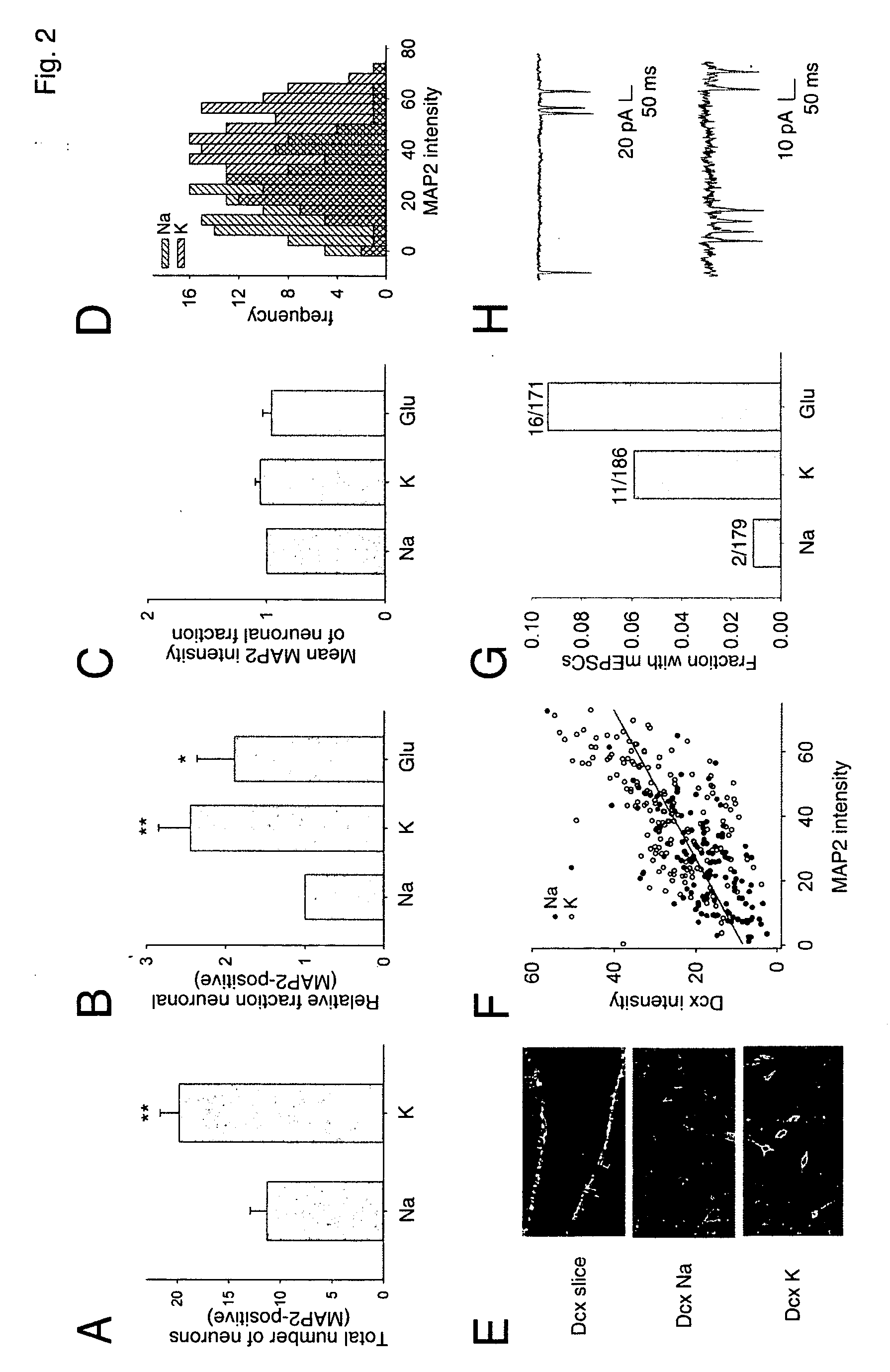
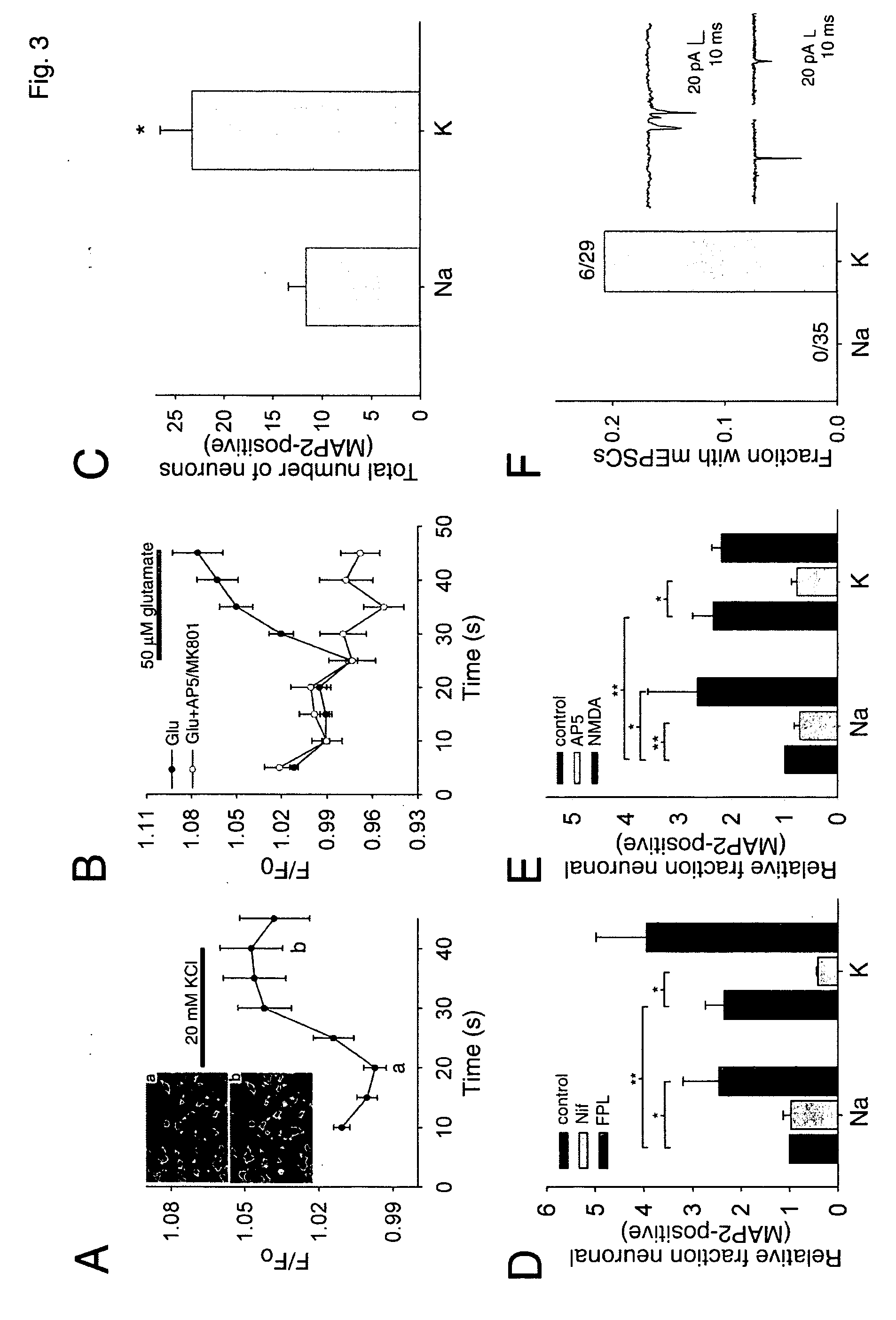
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
306 results about "Expression pattern" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Regular Expression Language - Quick Reference. A regular expression is a pattern that the regular expression engine attempts to match in input text. A pattern consists of one or more character literals, operators, or constructs.
Combinational array for nucleic acid analysis
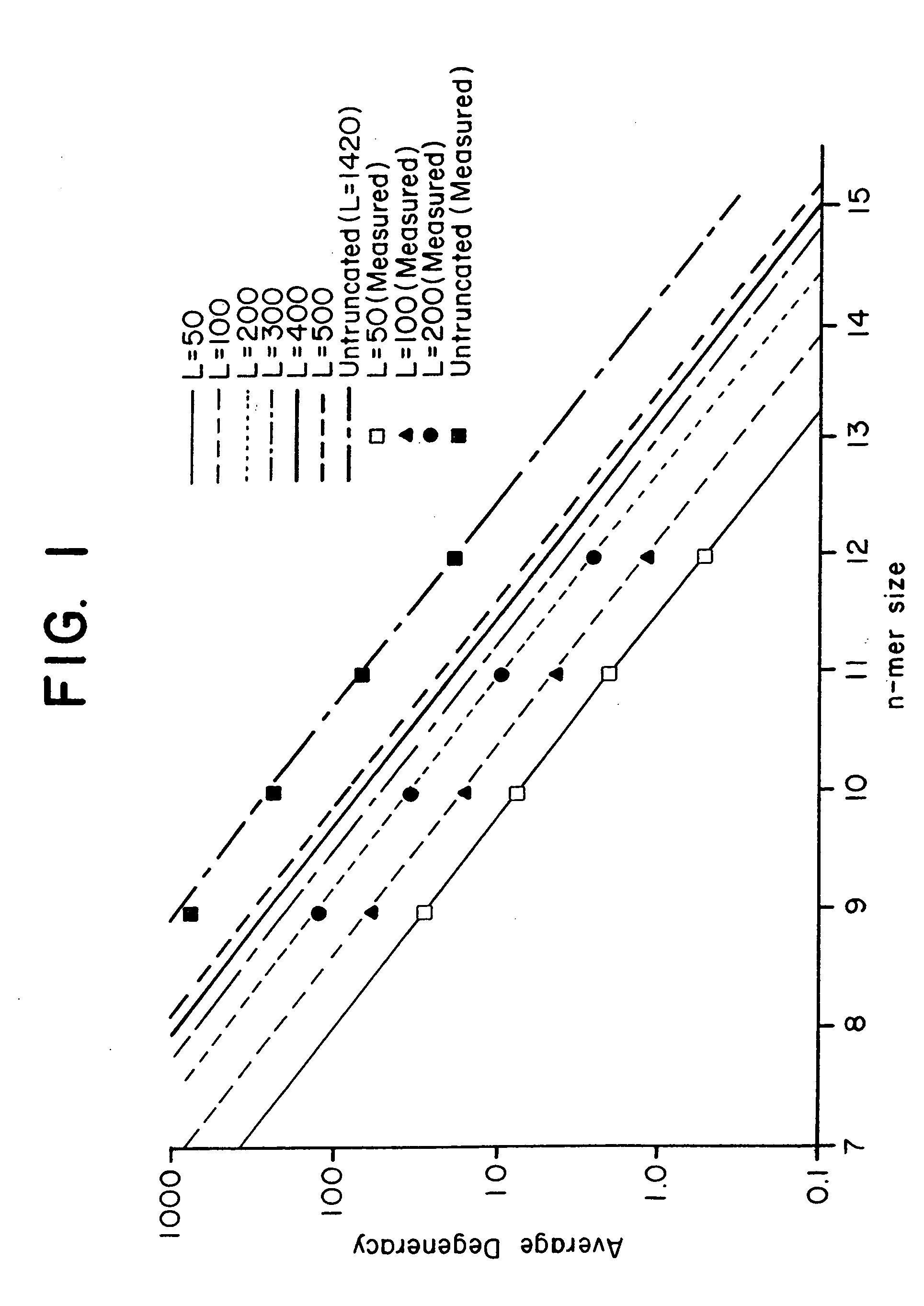
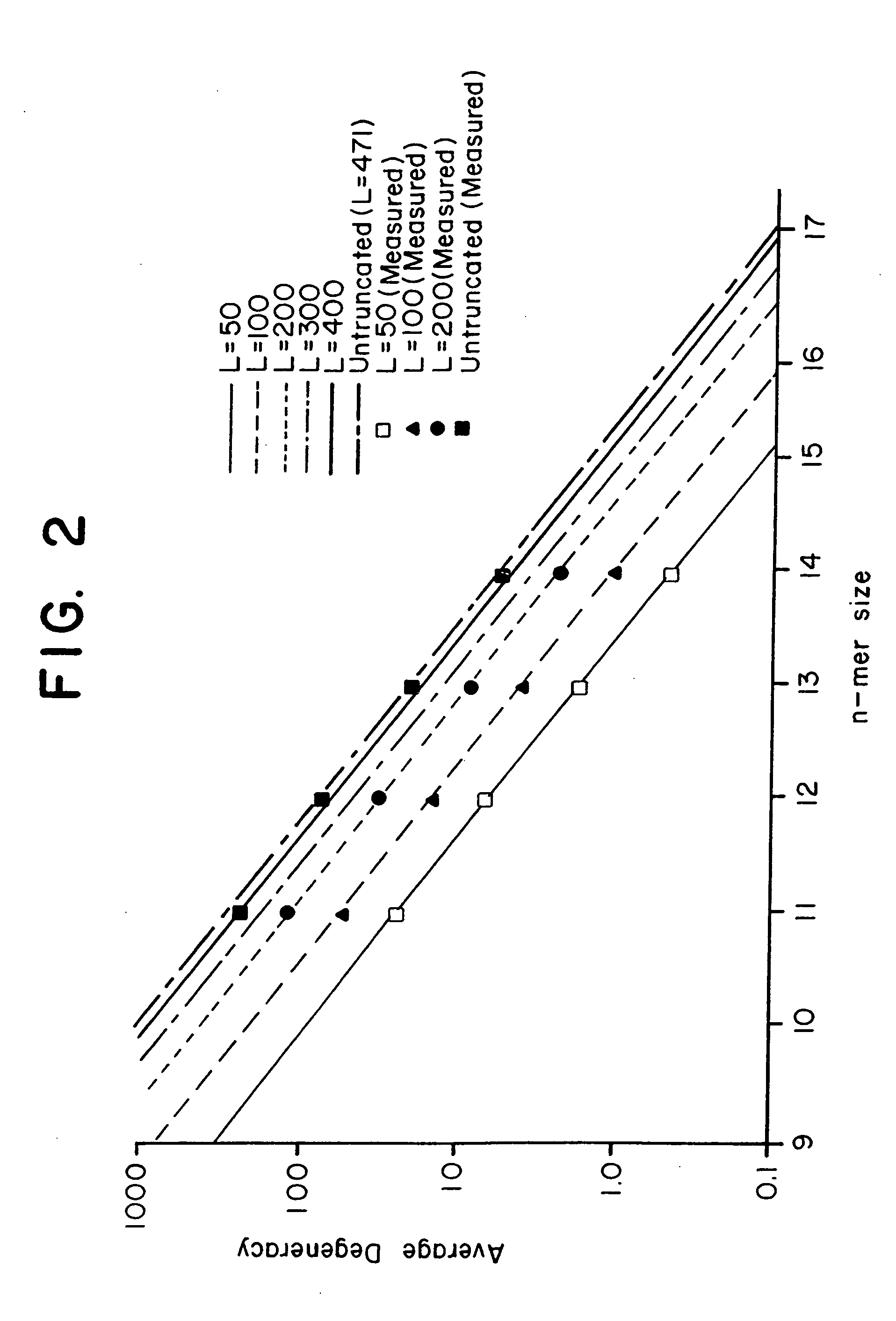
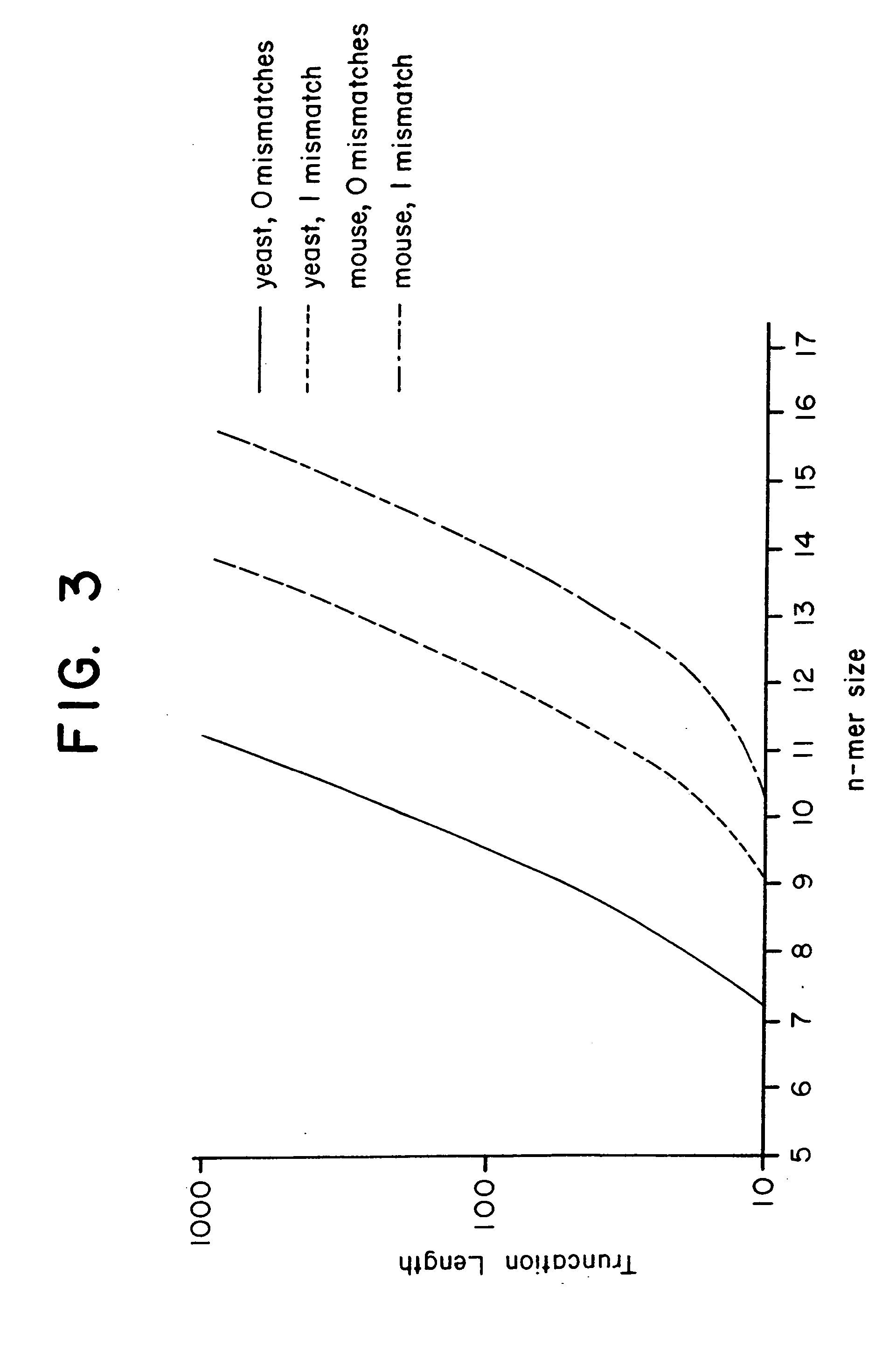
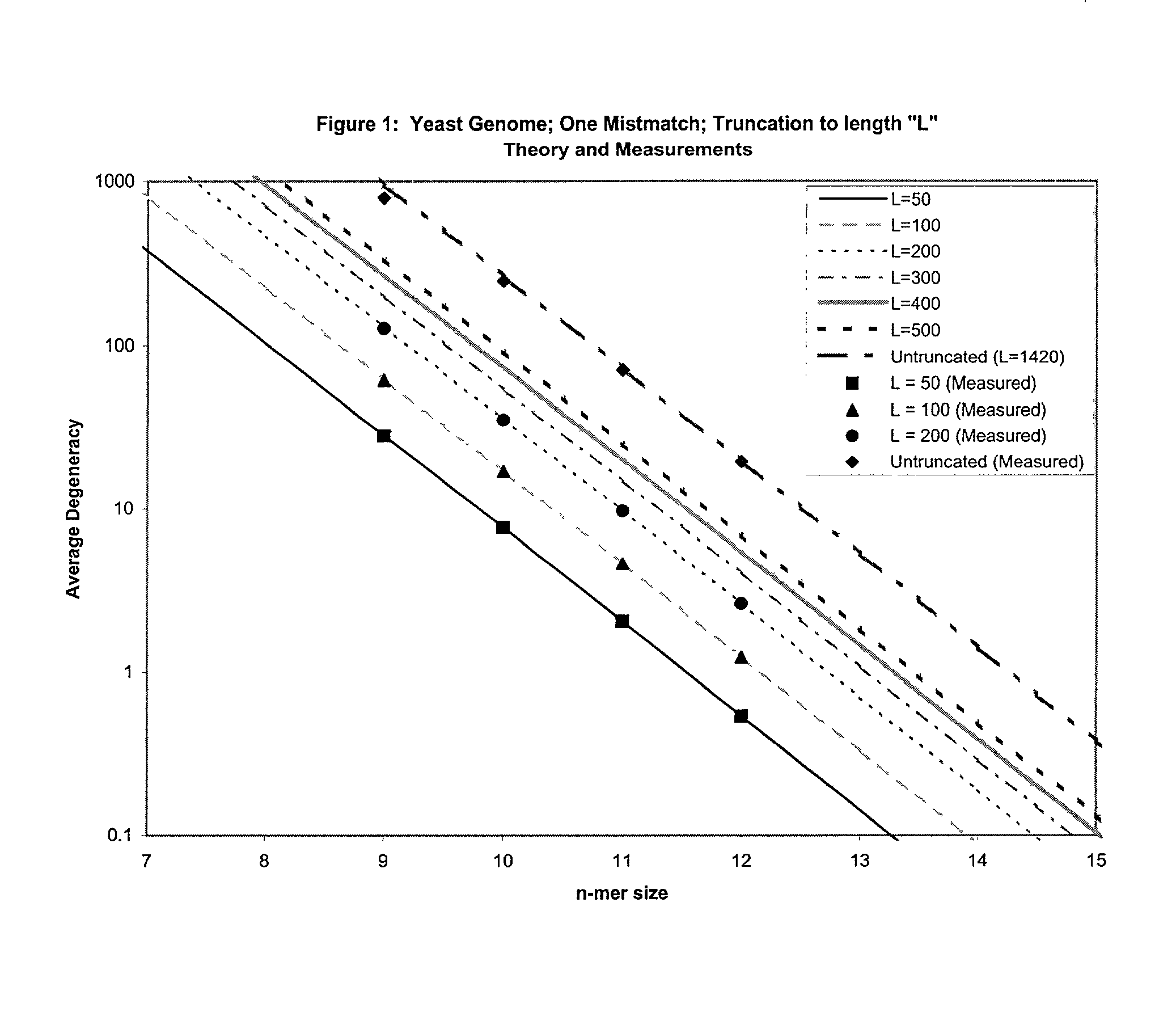
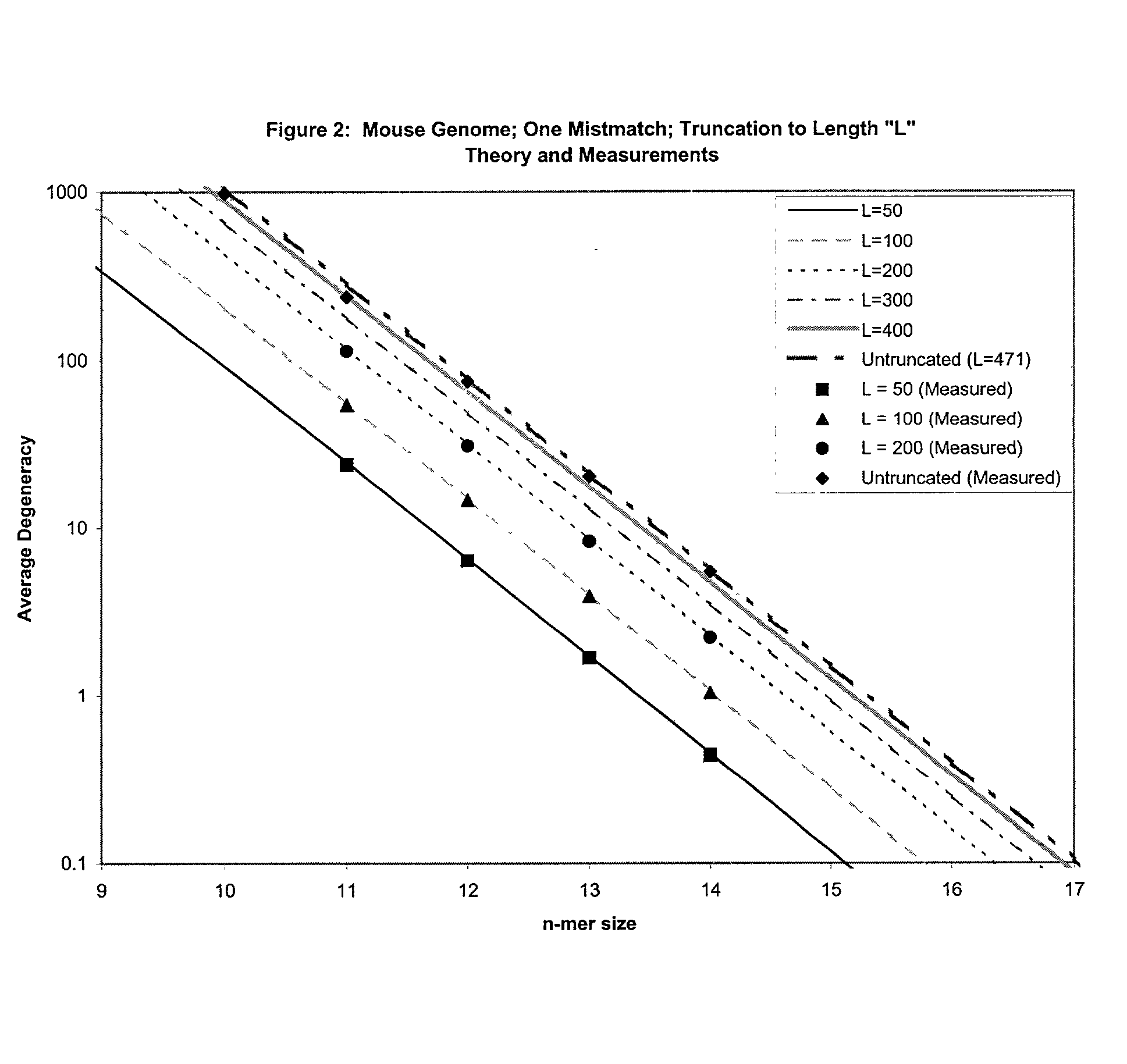
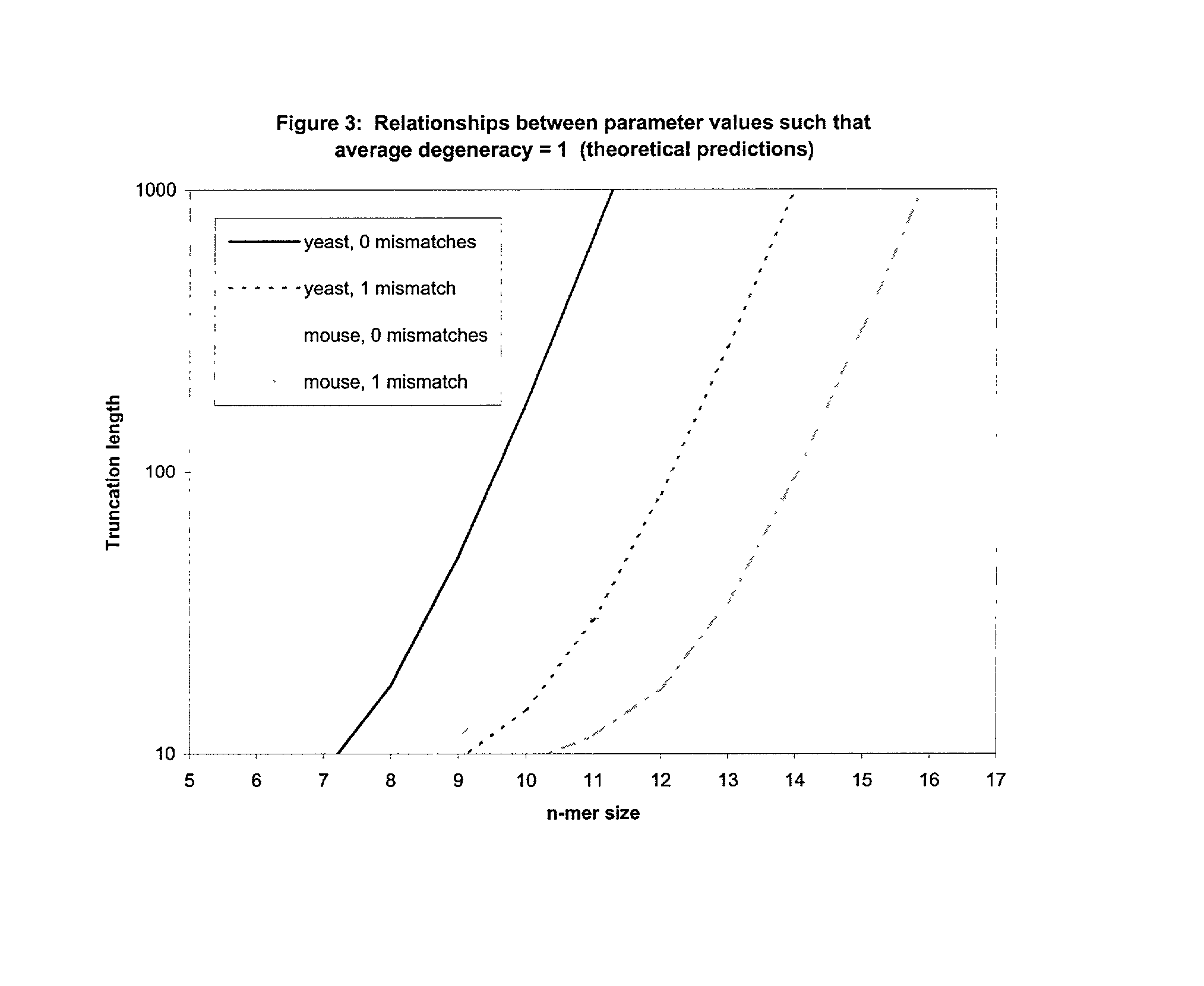
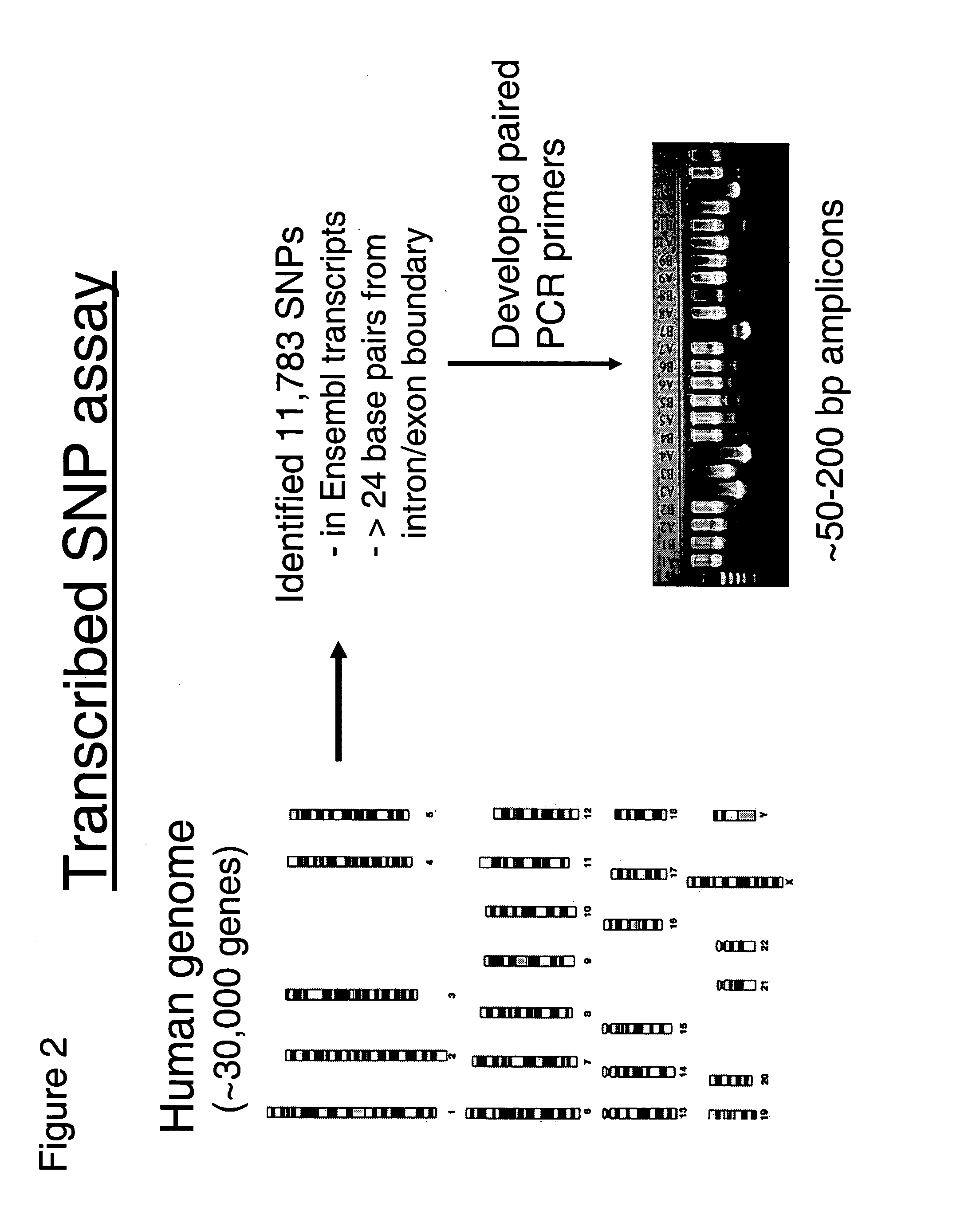
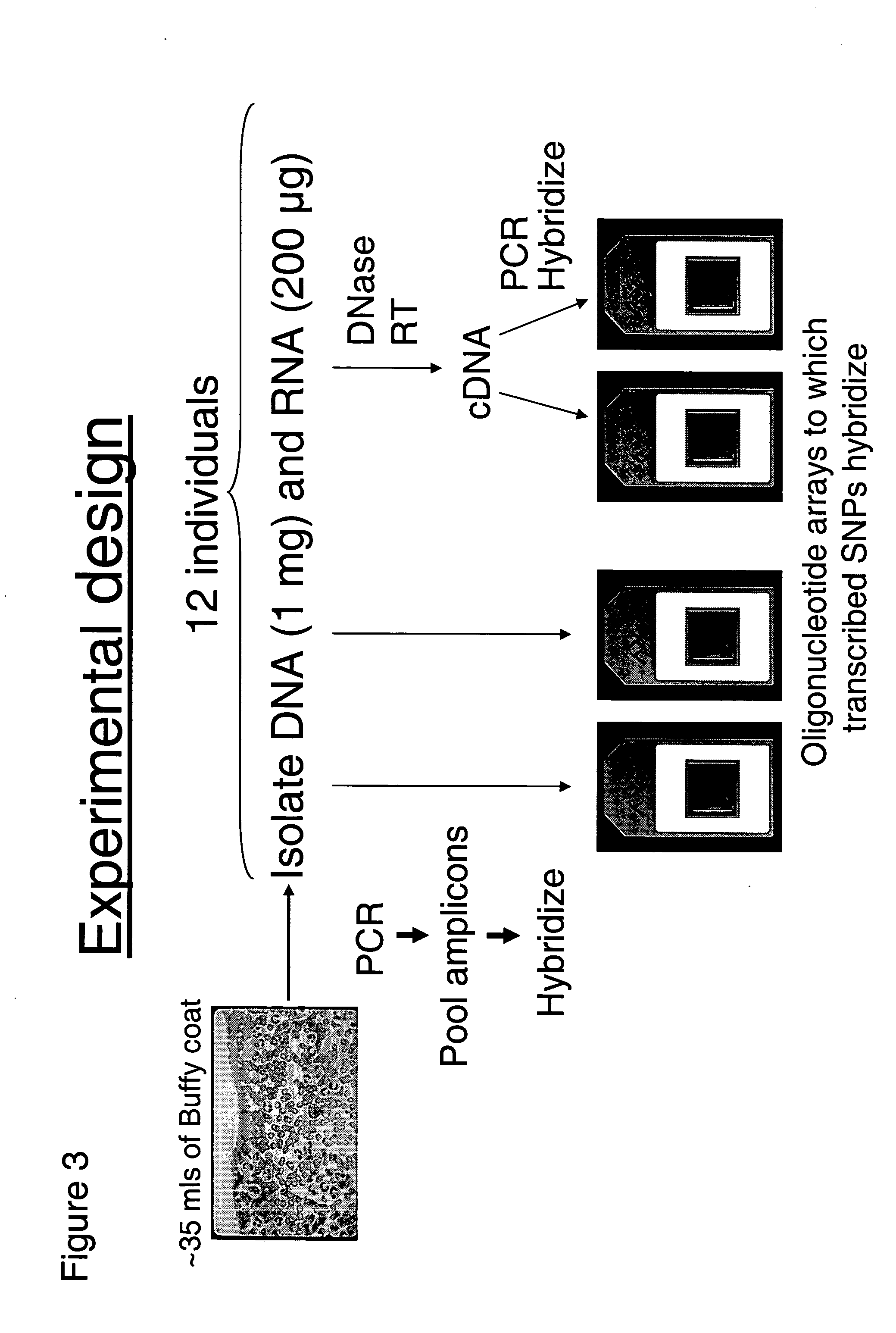
This invention relates to an array, including a universal micro-array, for the analysis of nucleic acids, such as DNA. The devices and methods of the invention can be used for identifying gene expression patterns in any organism. More specifically, all possible oligonucleotides (n-mers) necessary for the identification of gene expression patterns are synthesized. According to the invention, n is large enough to give the specificity to uniquely identify the expression pattern of each gene in an organism of interest, and is small enough that the method and device can be easily and efficiently practiced and made. The invention provides a method of analyzing molecules, such as polynucleotides (e.g., DNA), by measuring the signal of an optically-detectable (e.g., fluorescent, ultraviolet, radioactive or color change) reporter associated with the molecules. In a polynucleotide analysis device according to the invention, levels of gene expression are correlated to a signal from an optically-detectable (e.g. fluorescent) reporter associated with a hybridized polynucleotide. The invention includes an algorithm and method to interpret data derived from a micro-array or other device, including techniques to decode or deconvolve potentially ambiguous signals into unambiguous or reliable gene expression data.
Owner:CALIFORNIA INST OF TECH
Combinatorial array for nucleic acid analysis
InactiveUS20020012926A1Microbiological testing/measurementBiological testingBiological bodyFluorescence
This invention relates to an array, including a universal micro-array, for the analysis of nucleic acids, such as DNA. The devices and methods of the invention can be used for identifying gene expression patterns in any organism. More specifically, all possible oligonucleotides (n-mers) necessary for the identification of gene expression patterns are synthesized. According to the invention, n is large enough to give the specificity to uniquely identify the expression pattern of each gene in an organism of interest, and is small enough that the method and device can be easily and efficiently practiced and made. The invention provides a method of analyzing molecules, such as polynucleotides (e.g., DNA), by measuring the signal of an optically-detectable (e.g., fluorescent, ultraviolet, radioactive or color change) reporter associated with the molecules. In a polynucleotide analysis device according to the invention, levels of gene expression are correlated to a signal from an optically-detectable (e.g. fluorescent) reporter associated with a hybridized polynucleotide. The invention includes an algorithm and method to interpret data derived from a micro-array or other device, including techniques to decode or deconvolve potentially ambiguous signals into unambiguous or reliable gene expression data.
Owner:CALIFORNIA INST OF TECH
Cancer stem cell expression patterns and compounds to target cancer stem cells
InactiveUS20110020221A1Inhibit progressPrevent worseningIn-vivo radioactive preparationsImmunoglobulins against cell receptors/antigens/surface-determinantsCancer targetingMolecular Targeted Therapies
Described herein are therapeutic targets expressed in cancer stem cells and methods for treating and diagnosing cancer by targeting such cells with antibodies, compounds, nucleic acid, or other therapeutic agent. In one embodiment described herein, therapeutic agents for the treatment of cancer are provided based on the identification of cancer stem cell targets. The present invention also includes therapeutic targets for cancer therapy and cancer stem cell-targeted therapy. The invention includes the treatment of cancer by the administration of compounds or agents that target cancer stem cells.
Owner:THE JOHNS HOPKINA UNIV
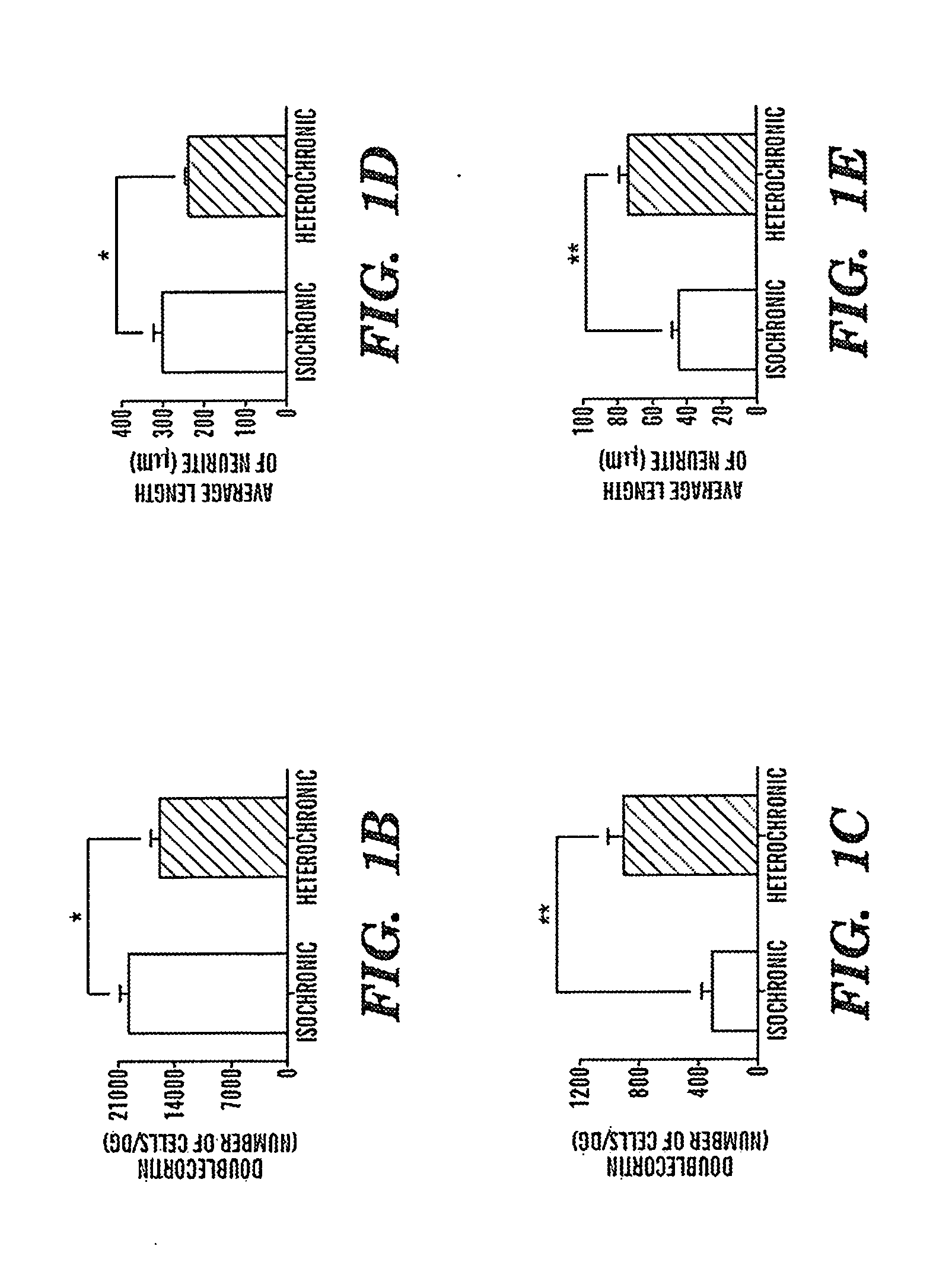
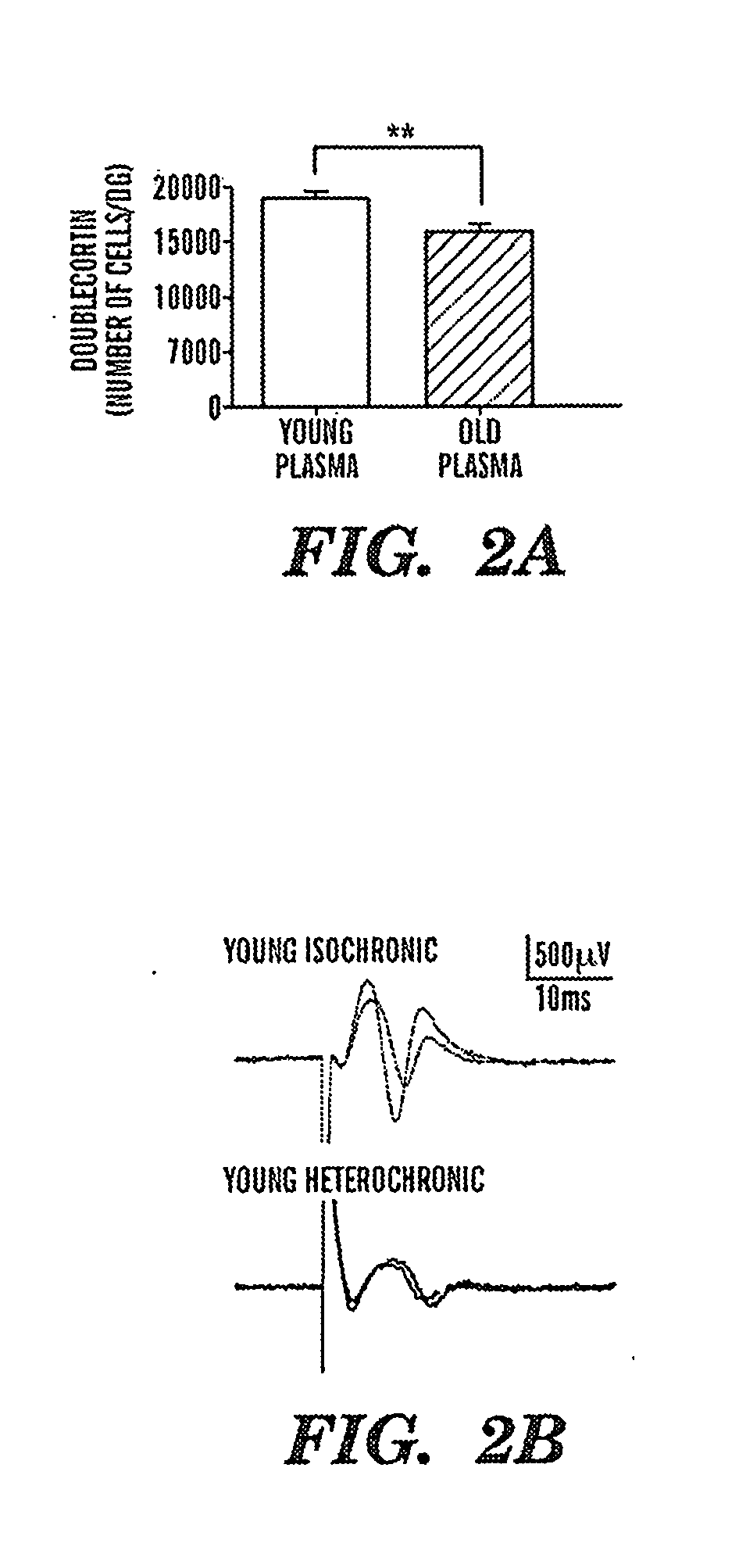
Coupling of excitation and neurogenesis in neural stem/progenitor cells
ActiveUS20050267011A1Increase neuronal cellPromoting neurogenesisElectrotherapyNervous disorderProgenitorNR1 NMDA receptor
Coupling of excitation to neurogenesis in proliferating post-natal NPCs is demonstrated in vitro and in vivo. Neurogenesis is potently enhanced by excitatory stimuli, and involves Cav1.2 / 1.3 channels and NMDA receptors. These Ca2+ influx pathways are located on the proliferating NPCs, allowing them to directly sense and process excitatory stimuli. Excitation increases the fraction of NPC progeny that are neurons, and increases total neuron number. Signaling in this pathway leads to rapid induction of a proneural gene expression pattern involving the bHLH genes HES1, Id2, and NeuroD, and the resulting cells become fully functional neurons defined by neuronal morphology, expression of neuronal structural proteins, expression of neuronal TTX-sensitive voltage gated Na+ channels, and synaptic incorporation into active neural circuits.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Brain endothelial cell expression patterns
InactiveUS20060127902A1Auxiliary diagnosisCompound screeningNervous disorderAbnormal tissue growthBrain tumor
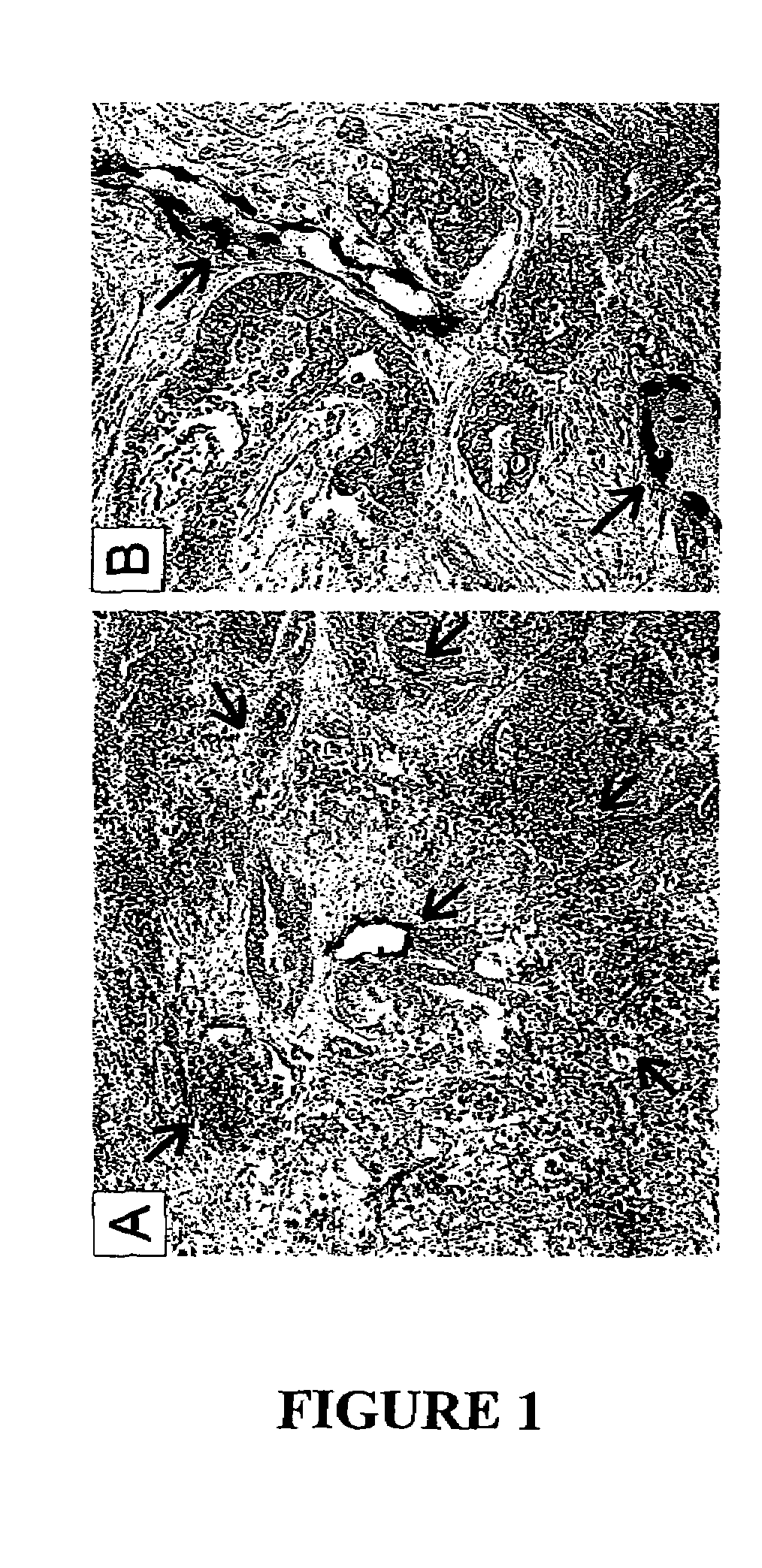
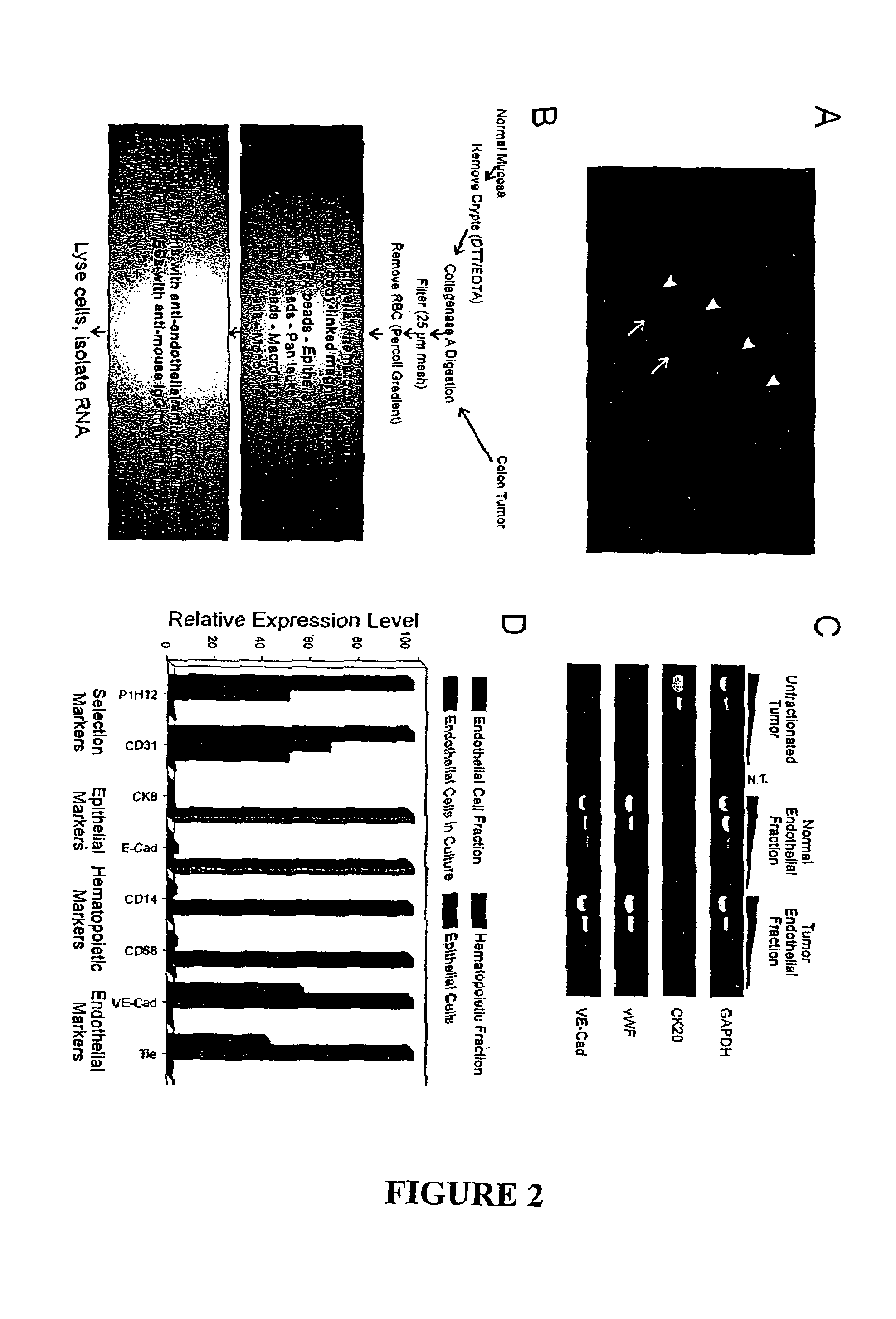
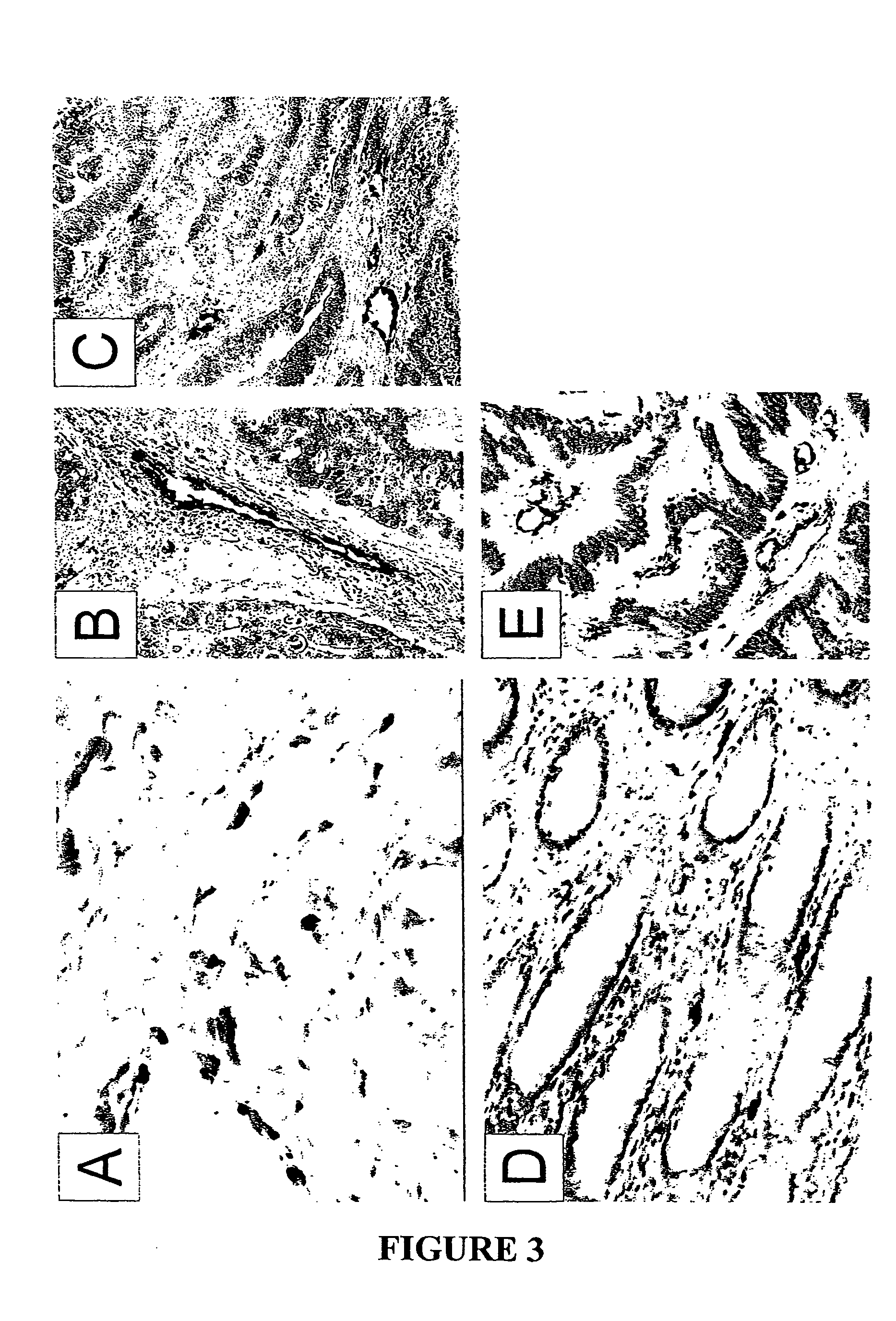
To gain a better understanding of brain tumor angiogenesis, new techniques for isolating brain endothelial cells (ECs) and evaluating gene expression patterns were developed. When transcripts from brain ECs derived from normal and malignant colorectal tissues were compared with transcripts from non-endothelial cells, genes predominantly expressed in the endothelium were identified. Comparison between normal- and tumor-derived endothelium revealed genes that were specifically elevated in tumor-associated brain endothelium. These results confirm that neoplastic and normal endothelium in human brains are distinct at the molecular level, and have significant implications for the development of anti-angiogenic therapies in the future.
Owner:GENZYME CORP +1
Markers for cancer prognosis and therapy and methods of use
InactiveUS20130042333A1Adverse side effectLow costBiocideMicrobiological testing/measurementAdjuvantTreatment field
The invention relates generally to the field of cancer prognosis and treatment. More particularly, the present invention relates to methods and compositions that utilize a particular panel of gene products (“biomarkers”) and their differential expression patterns (“expression signatures”), wherein the expression patterns correlate with responsiveness, or lack thereof, to chemotherapy treatment. The invention is based on the identification of a specific set of biomarkers that are differentially expressed in chemotherapy-treated tumors and which are useful in predicting the likelihood of a therapeutic response, including residual disease persistence and subsequent tumor recurrence in cancer patients receiving chemotherapy. The gene panel is also useful in designing specific adjuvant modalities with improved therapeutic efficiency. Also disclosed are methods for characterizing tumors according to expression of the biomarkers described herein.
Owner:GEN TECH
Rule-based natural language processing
ActiveUS20190102378A1Easy to operateIncrease the amount of calculationSemantic analysisSpeech recognitionData miningMachine learning
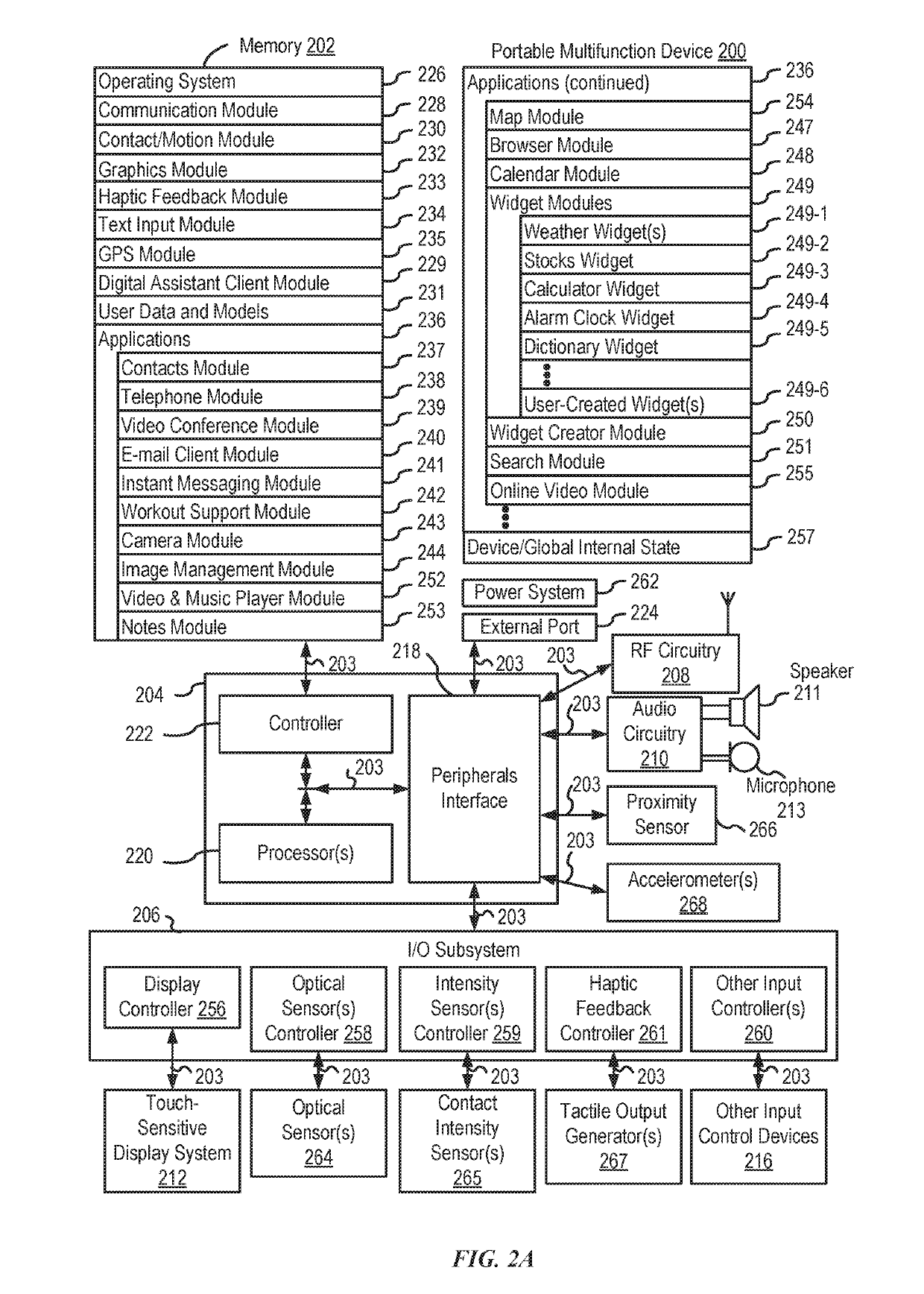
Systems and processes for rule-based natural language processing are provided. In accordance with one example, a method includes, at an electronic device with one or more processors, receiving a natural-language input; determining, based on the natural-language input, an input expression pattern; determining whether the input expression pattern matches a respective expression pattern of each of a plurality of intent definitions; and in accordance with a determination that the input expression pattern matches an expression pattern of an intent definition of the plurality of intent definitions: selecting an intent definition of the plurality of intent definitions having an expression pattern matching the input expression pattern; performing a task associated with the selected intent definition; and outputting an output indicating whether the task was performed.
Owner:APPLE INC
Flax (Linum usitatissimum I.) seed-specific promoters
InactiveUS20070192902A1Sugar derivativesOther foreign material introduction processesLipid formationProtein composition
The present invention is directed to promoters of flax conlinin and ω-3 desaturase genes. The promoters guide high levels of the expression exclusively in flax developing seeds. This specific expression pattern concomitant with the biosynthesis of storage lipids and proteins make these promoters particularly useful for seed-specific modification of fatty acid and protein compositions in plant seeds.
Owner:BIORIGINAL FOOD & SCI
DNA array sequence selection
InactiveUS6706867B1Sugar derivativesMicrobiological testing/measurementBiotechnologyCDNA Microarrays
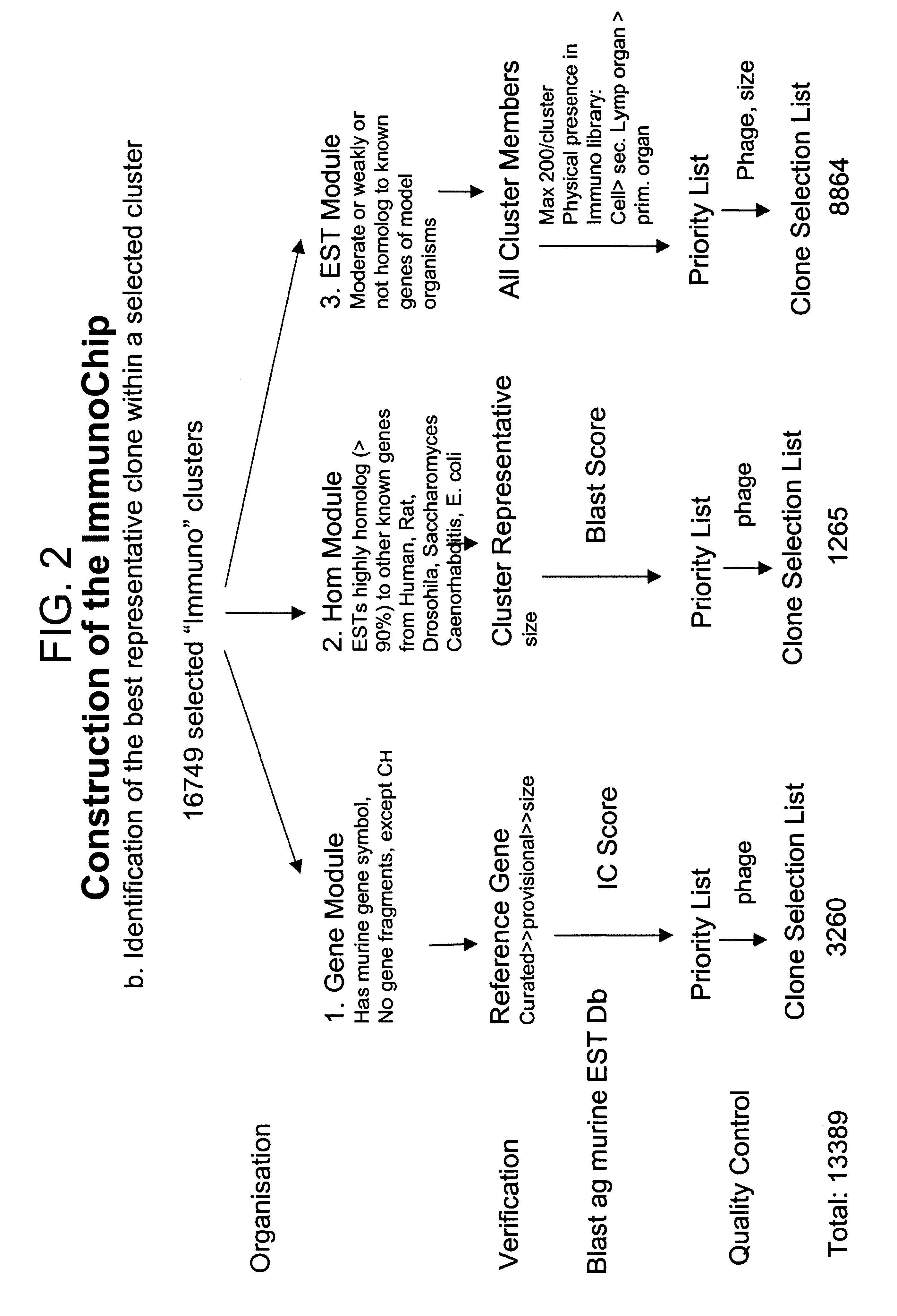
The present invention provides methods and compositions for the construction of custom cDNA microarrays. In particular, the methods involve the selection of relevant clusters based on knowledge and expression patterns using public database information and the identification of the best representative cDNA clones within the selected cluster. The methods facilitate the construction of custom microarrays suitable for use in any biotechnological art. In preferred embodiments, the present invention provides the the ImmunoChip.
Owner:UNITED STATES OF AMERICA
Method for the early detection of pancreatic cancer and other gastrointestinal disease conditions
InactiveUS20080248484A1Early diagnosisSpecific and focused early diagnosisMicrobiological testing/measurementOrgan systemNeoplasm
The present invention uses peripheral blood monocyte-lymphocyte for the early diagnosis of pancreatic cancer, as well as other conditions of the pancreas and other organs. The peripheral blood lymphocytes recognize the new neoplasm in the pancreas, as well as disease processes in other organ systems. The evaluation of this specific recognition of the disease process by the peripheral blood monocyte-lymphocyte through gene microarray expression patterns constitute a successful method for the early detection of pancreatic cancer and other organ disease processes. This document describes the process used in this method of early diagnosis.
Owner:BAUER A ROBERT
Methods, Systems, and Apparatuses for Quantitative Analysis of Heterogeneous Biomarker Distribution
Methods, systems, and apparatuses for detecting and describing heterogeneity in a cell sample are disclosed herein. A plurality of fields of view (FOV) are generated for one or more areas of interest (AOI) within an image of the cell sample are generated. Hyperspectral or multispectral data from each FOV is organized into an image stack containing one or more z-layers, with each z-layer containing intensity data for a single marker at each pixel in the FOV. A cluster analysis is applied to the image stacks, wherein the clustering algorithm groups pixels having a similar ratio of detectable marker intensity across layers of the z-axis, thereby generating a plurality of clusters having similar expression patterns.
Owner:OREGON HEALTH & SCI UNIV OF PORTLAND OREGON +1
Genetic trait breeding method
InactiveUS6946586B1Improve traitsStable introduction of DNAFermentationGenetic traitsPhenotypic trait
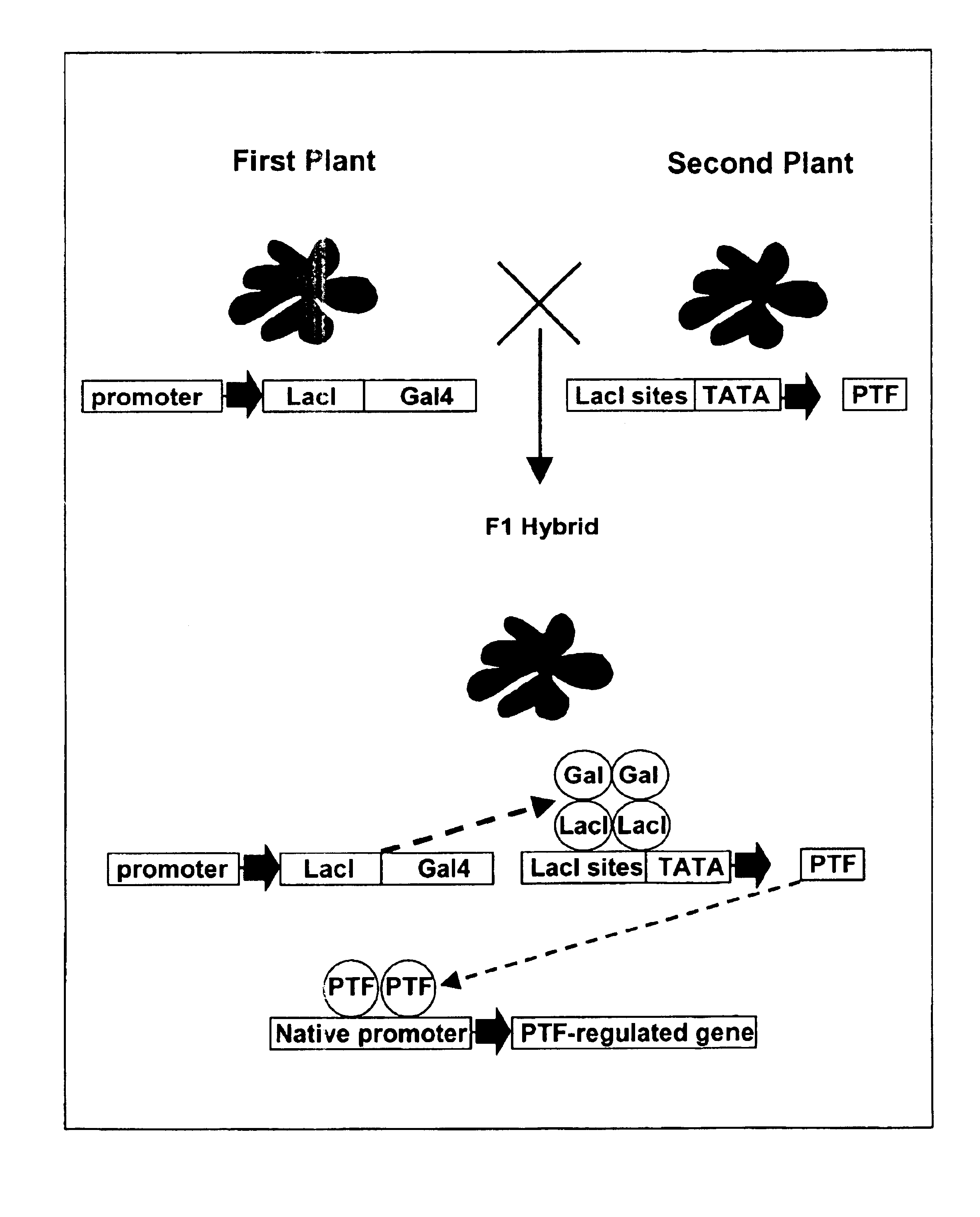
A method for screening for a trait associated with the altered expression of a gene of interest in plants is provided. The method is a combinatorial approach which uses traditional plant breeding techniques for modifying the patterns of expression of a gene of interest. Kits for use in the method and transgenic plants generated by the method are also provided.
Owner:MENDEL BIOTECHNOLOGY INC
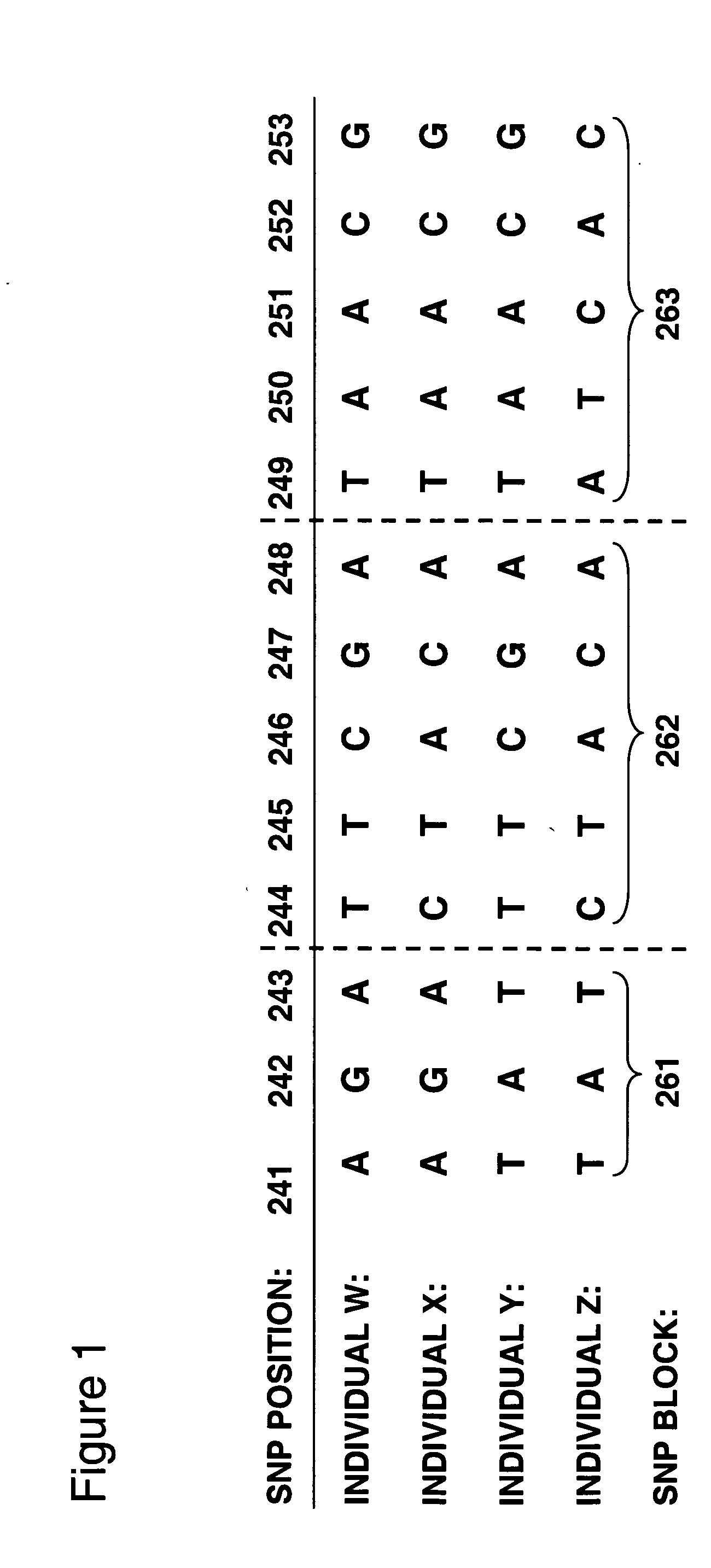
Allele-specific expression patterns
The invention provides methods of analyzing genes for differential relative allelic expression patterns. Haplotype blocks throughout the genomes of individuals are analyzed to identify haplotype patterns that are associated with specific differential relative allelic expression patterns. Haplotype blocks that contain associated haplotype patterns may be further investigated to identify genes or variants of genes involved in differential relative allelic expression patterns.
Owner:PERLEGEN SCIENCES INC
Gene expression signature for classification of cancers
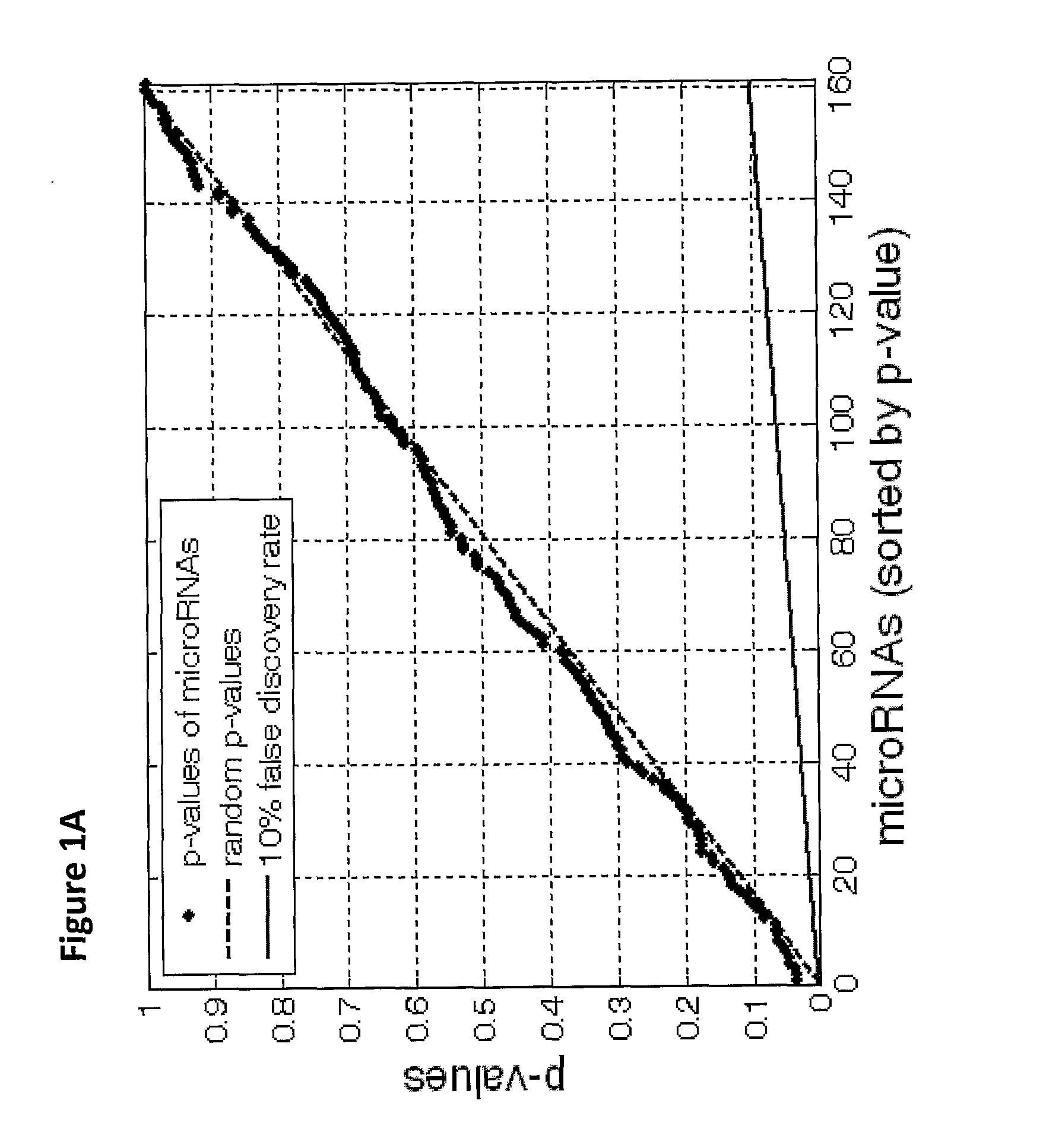
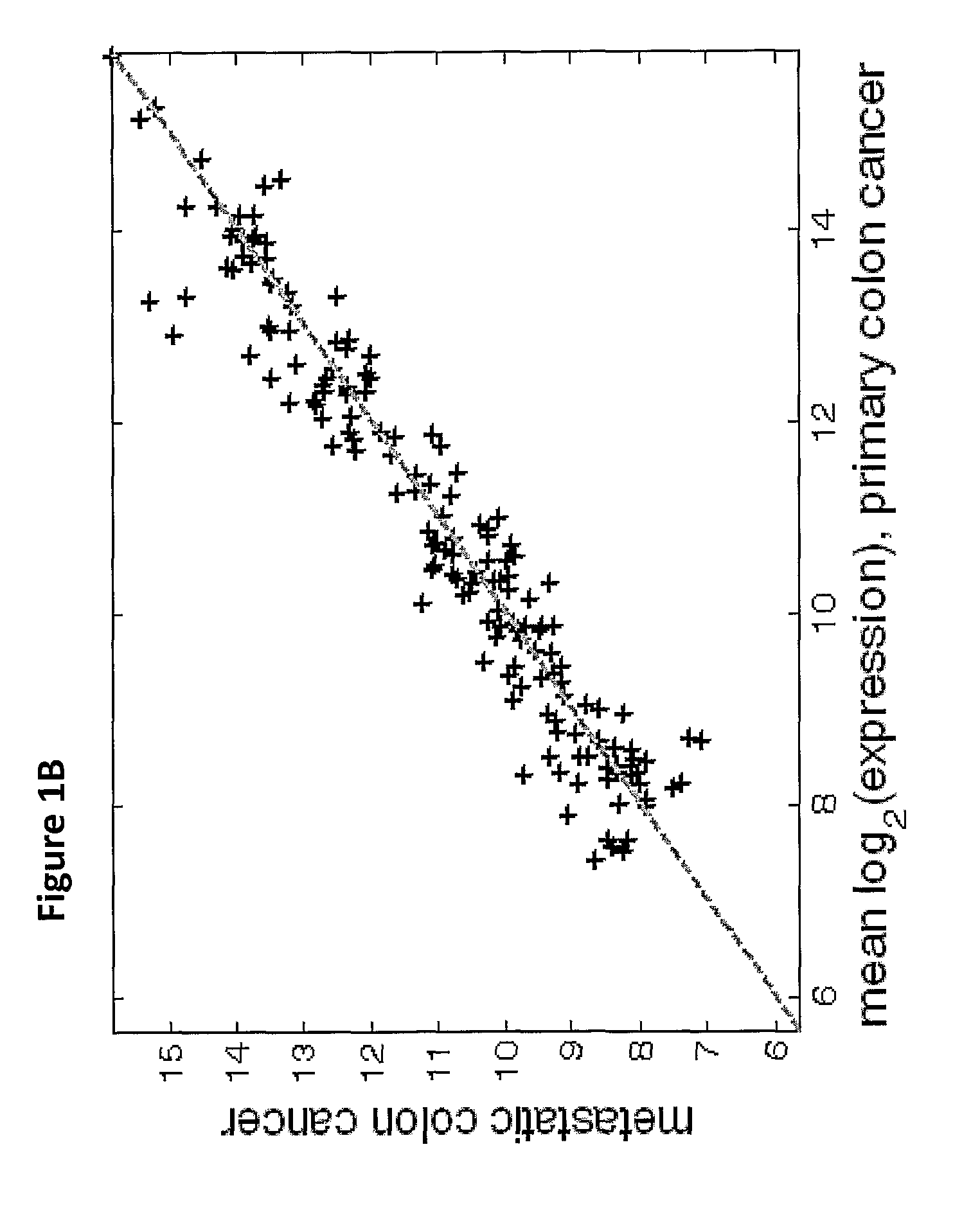
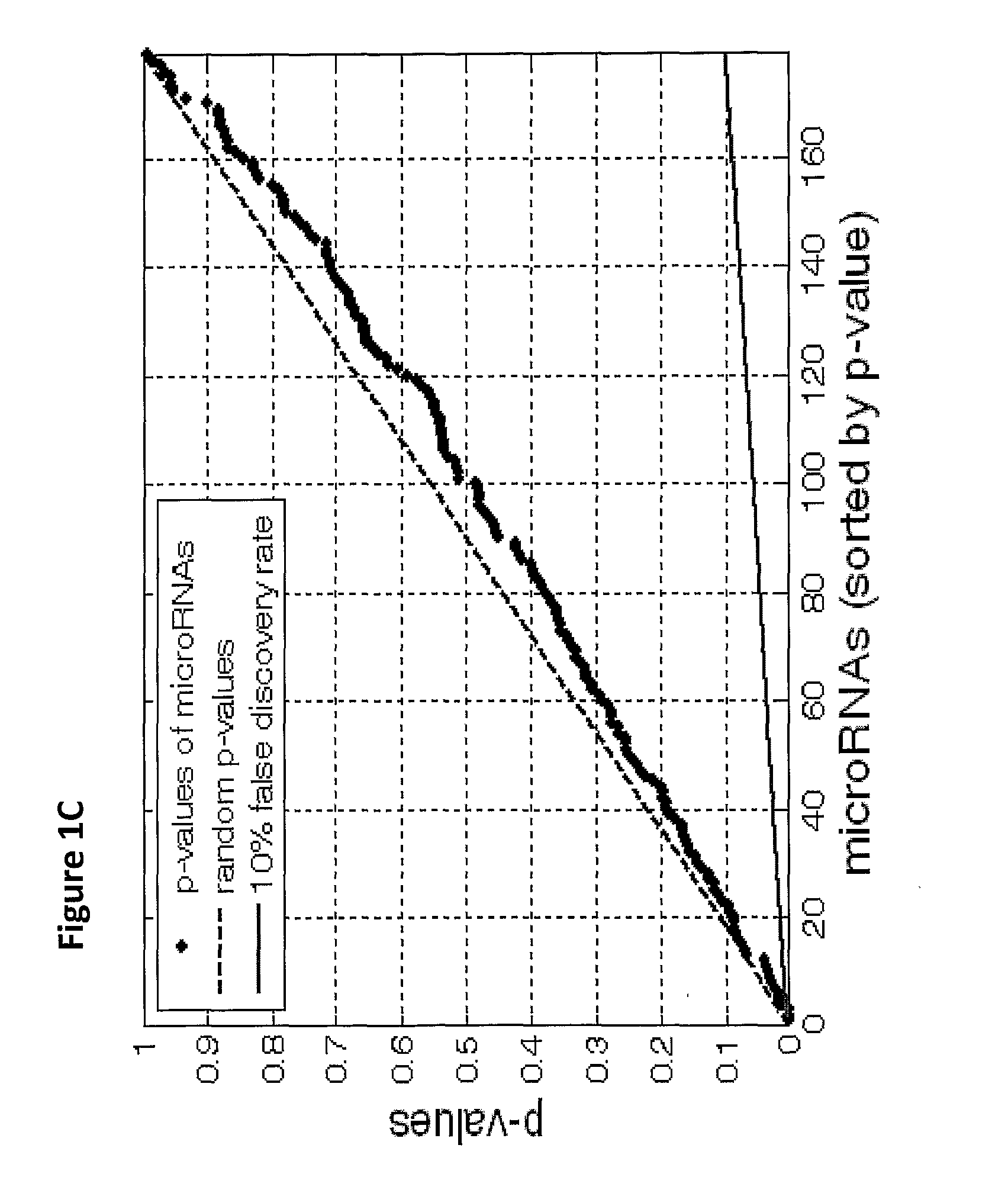
The present invention provides a process for classification of cancers and tissues of origin through the analysis of the expression patterns of specific microRNAs and nucleic acid molecules relating thereto. Classification according to a microRNA tree-based expression framework allows optimization of treatment, and determination of specific therapy.
Owner:TEL HASHOMER MEDICAL RES INFRASTRUCTURE & SERVICES +1
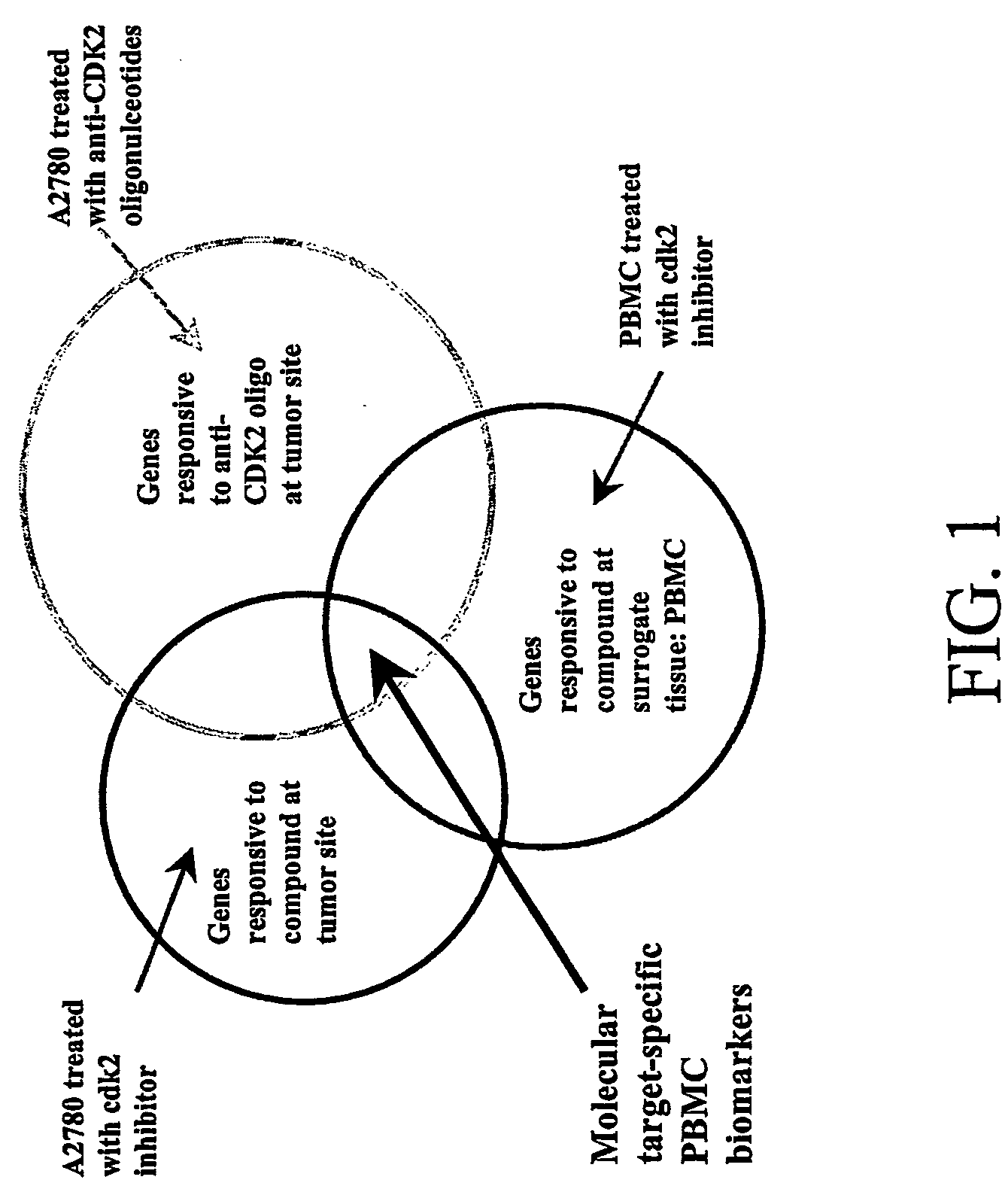
Biomarkers of cyclin-dependent kinase modulation
InactiveUS20070105114A1Modulate cdk activityMicrobiological testing/measurementBiological material analysisMammalCancer research
Biomarkers having expression patterns that correlate with a response of cells to treatment with one or more cdk modulating agents, and uses thereof. Also provided are methods for testing or predicting whether a mammal will respond therapeutically to a method of treating cancer that comprises administering an agent that modulates cdk activity.
Owner:BRISTOL MYERS SQUIBB CO
Biomarkers of aging for detection and treatment of disorders
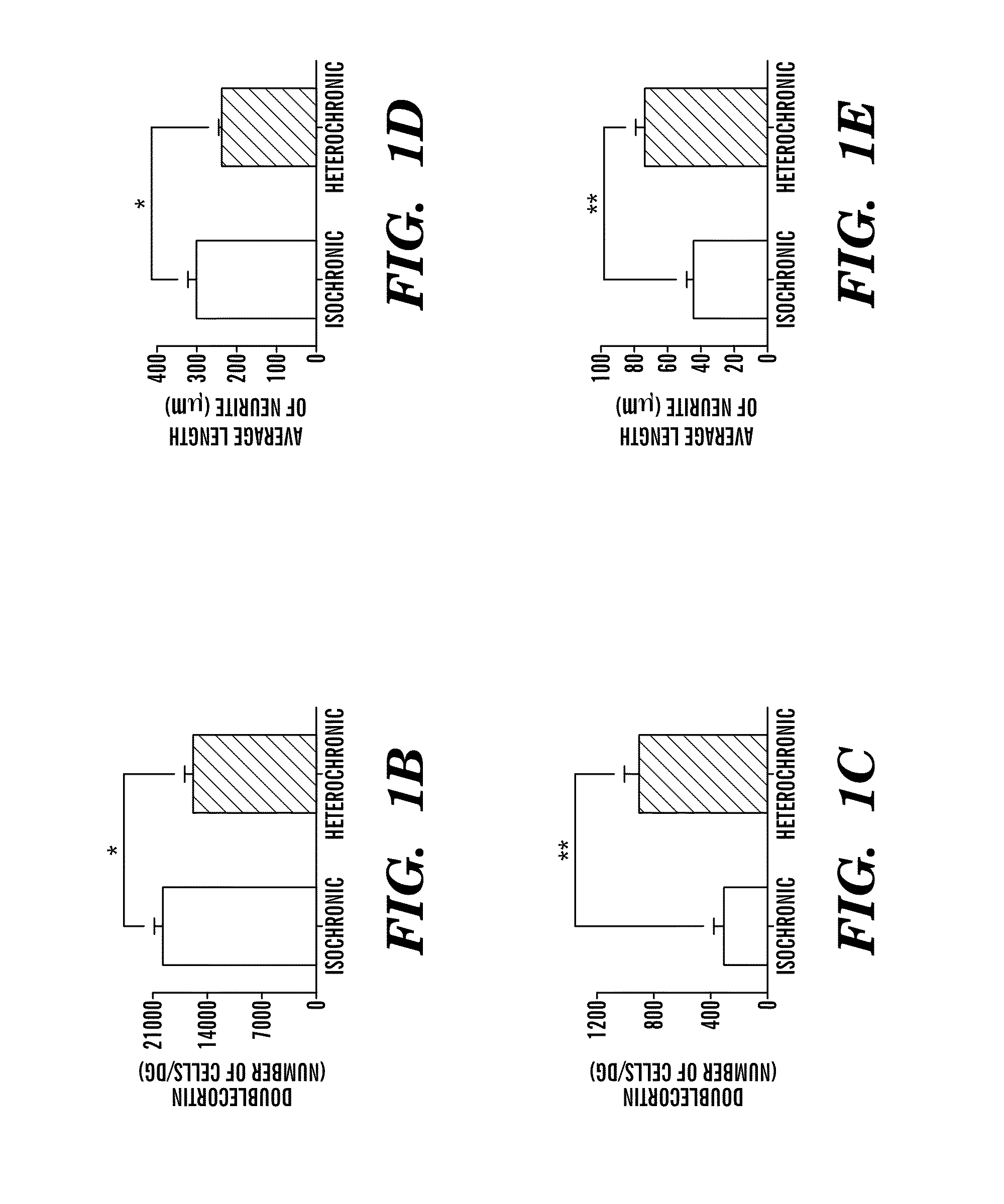
InactiveUS20130040844A1Modulate activityVirusesMicrobiological testing/measurementDiseaseNeural cell
Provided are methods of diagnosis, prognosis, and monitoring of aging using biomarkers that have been discovered to be linked to biological aging process. Methods for increasing neural cell regeneration and cognitive function are also provided. The methods are, at least in part, based on a discovery that altered expression patterns of certain biological markers are associated with biological aging processes. These markers comprise at least Eotaxin / CCL11, 2-microglobulin, MCP-1 and Hap-toglobulin, increased expression of which has been shown to be associated with increase in biological aging process.
Owner:DEPT OF VETERANS AFFAIRS VA +1
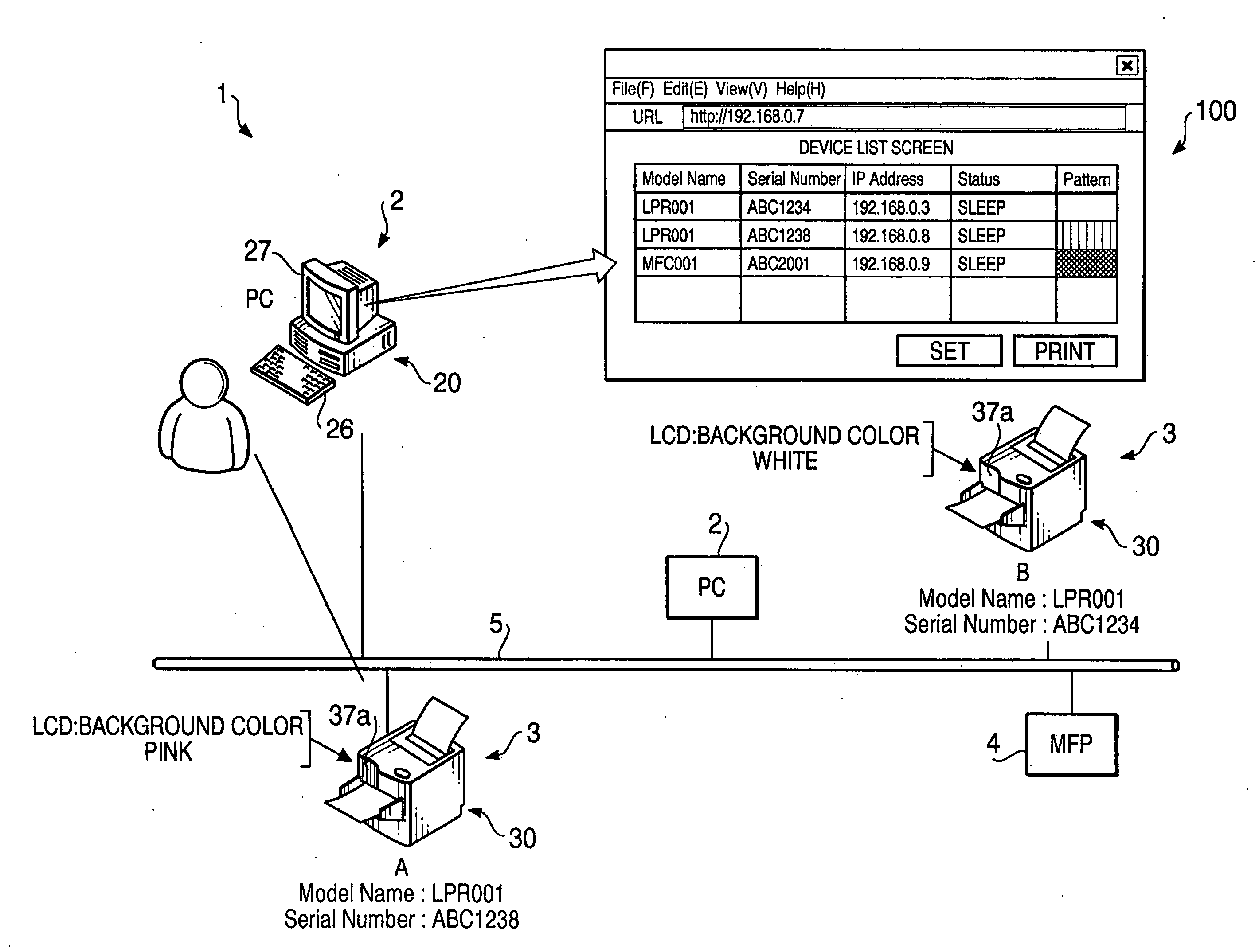
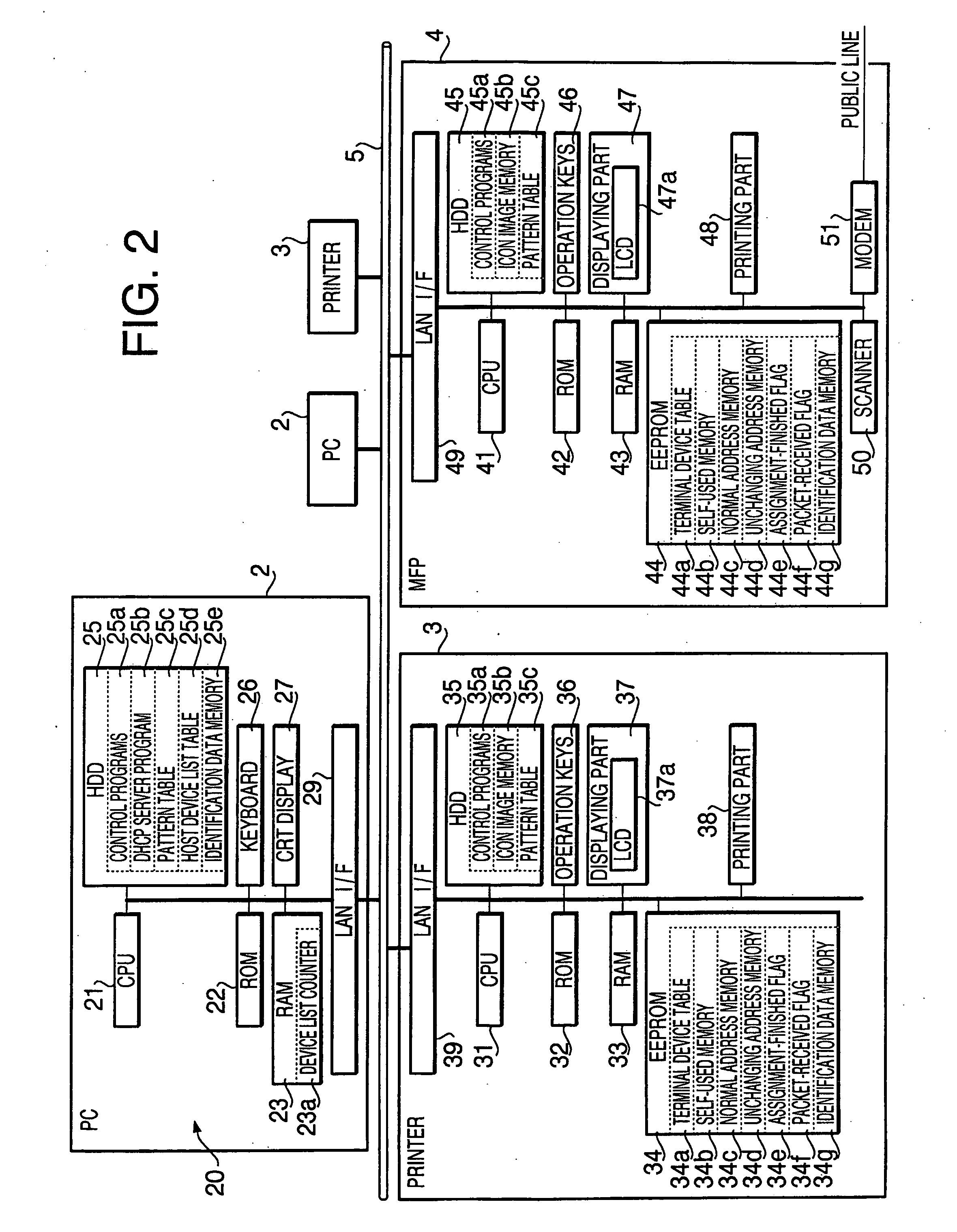
Terminal device
InactiveUS20060221863A1Digital computer detailsData switching by path configurationTerminal equipmentOutput device
A terminal device, which is to be connected to the network together with other terminal devices and a host device to operate the terminal device, includes an output device that outputs perceptually recognized information, a self-pattern storing system that stores information on an expression pattern for the terminal device, a configuring system that configures the information into the self-pattern storing system, an output executing system that makes the output device output the expression pattern, based upon the written information, an obtaining system that obtains information on expression patterns for the other terminal devices, and a setting system that defines an expression pattern different from any of the expression patterns for the other terminal devices as the expression pattern for the terminal device, based upon the obtained information, and sets information on the defined expression pattern as the information to be written.
Owner:BROTHER KOGYO KK
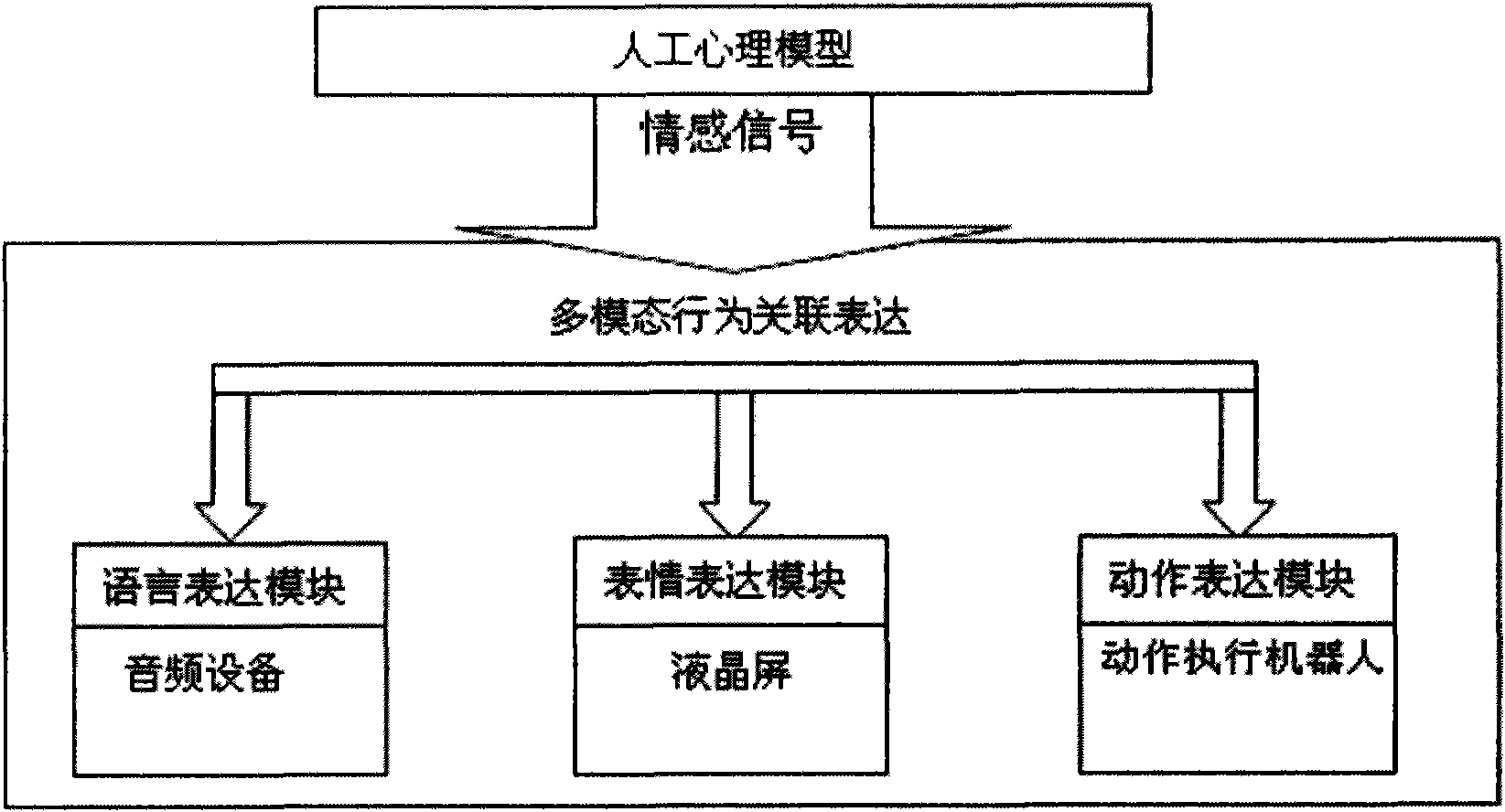
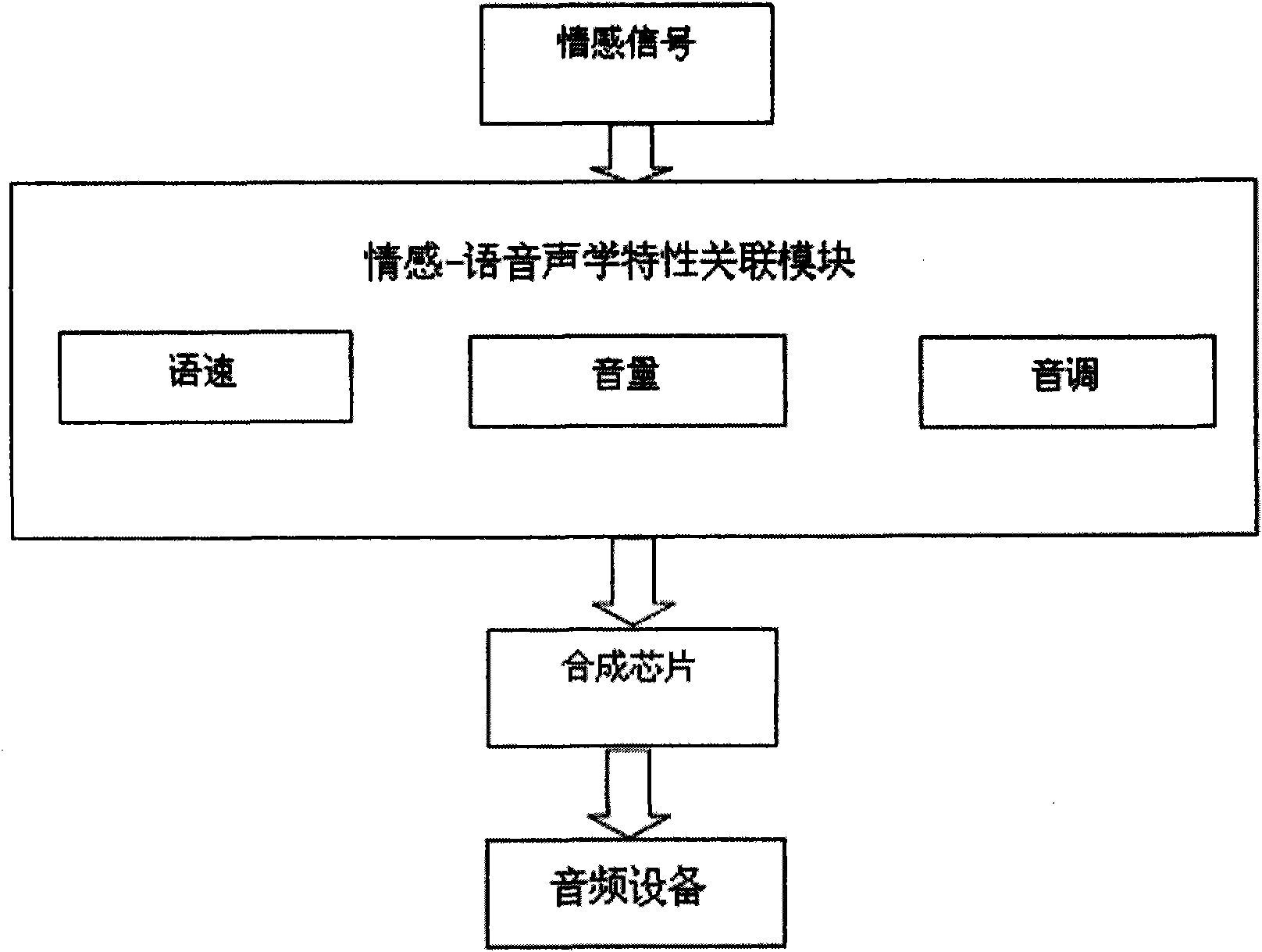
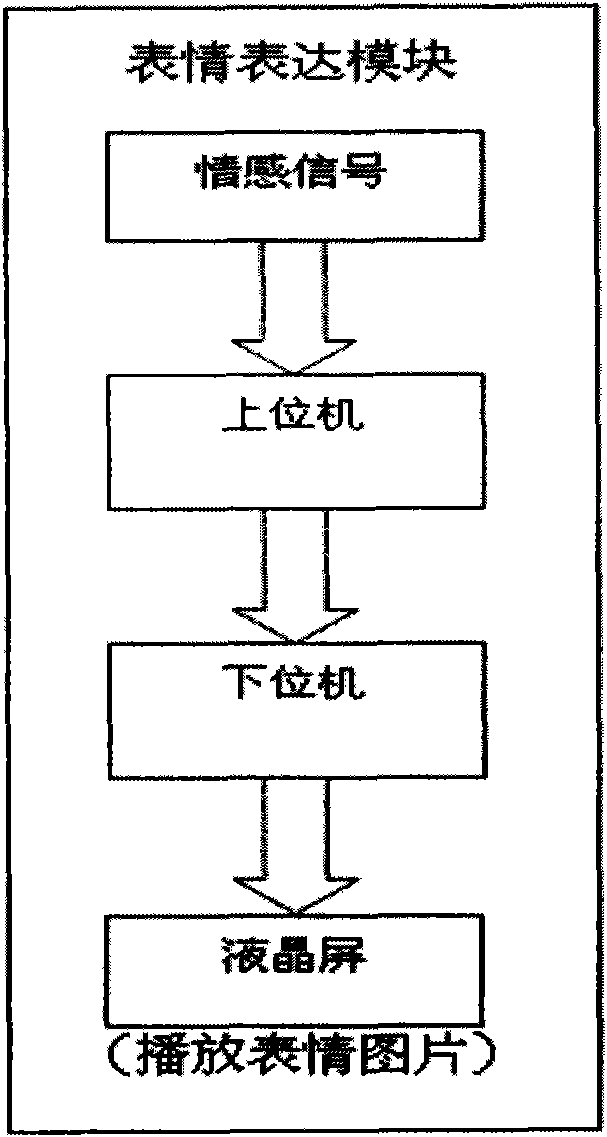
Intelligent emotional robot multi-modal behavioral associative expression system
ActiveCN101661569AConsistentBiological modelsTotal factory controlFacial expressionHuman–computer interaction
On the basis of an artificial mental model, the invention researches a behavioral expression mode and a method of a robot from the view of psychology, and provides an intelligent emotional robot multi-modal behavioral associative expression system. The system accepts the unified driving of emotional signal output by the artificial mental model and is provided with a unified behavioral driving mechanism; therefore, the behavioral expressions are more harmonious. The intelligent emotional robot multi-modal behavioral associative expression system comprises three modules, namely a language expression module, a facial expression module and an action expression module; the three modules accept the unified driving of an emotional signal output by the artificial mental model; and to the expression of the language, face and action of the intelligent emotional robe, the expression of language, face and action are executed synchronously on time sequence, and the unified driving of the emotionalsignal is accepted logically, thereby having consistency.
Owner:UNIV OF SCI & TECH BEIJING
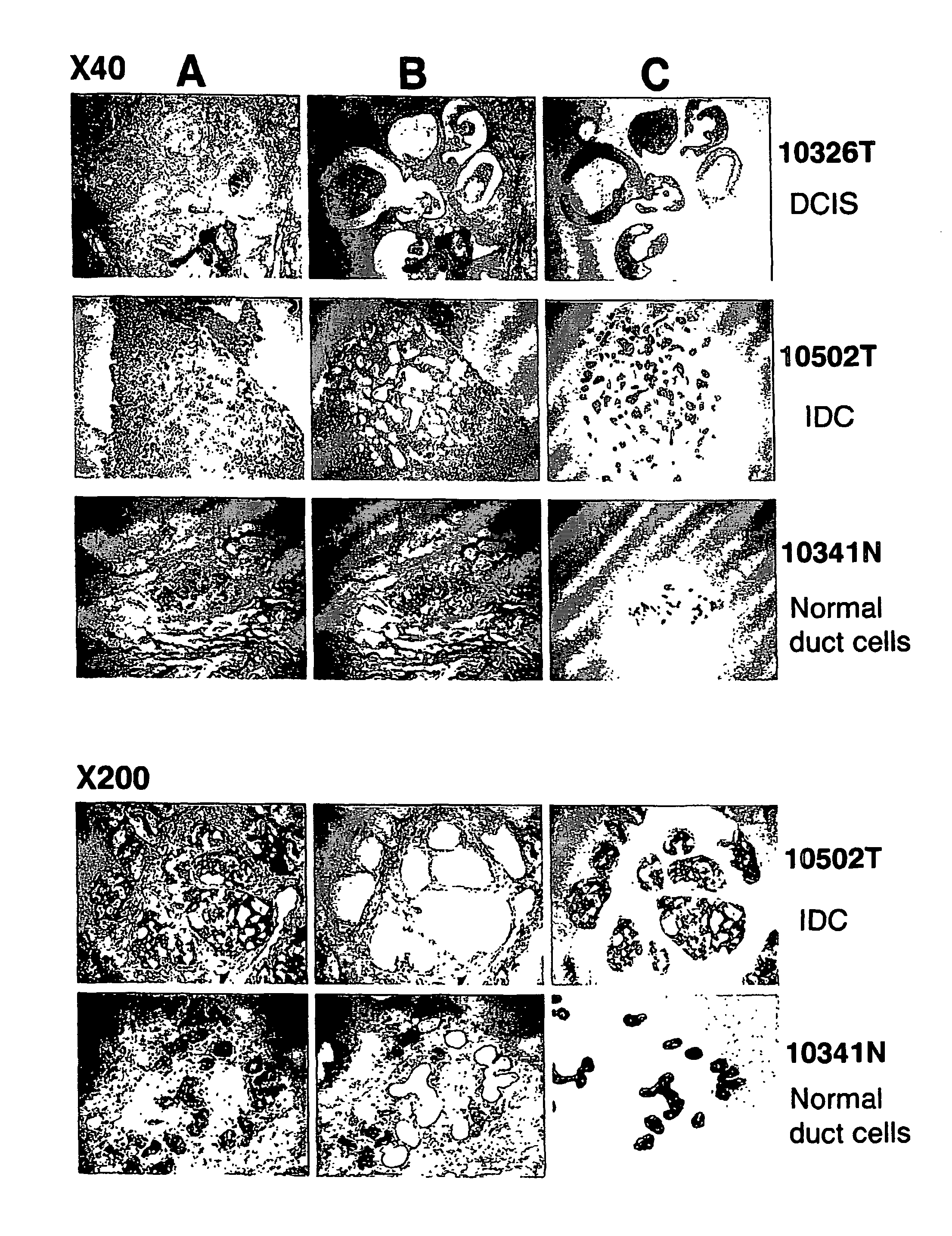
Method of Diagnosing Breast Cancer
InactiveUS20070269432A1Preventing metastasis of breast cancerMetastasis of breast cancer can be treated or preventedOrganic active ingredientsCompound screeningBreast cancer metastasisNormal cell
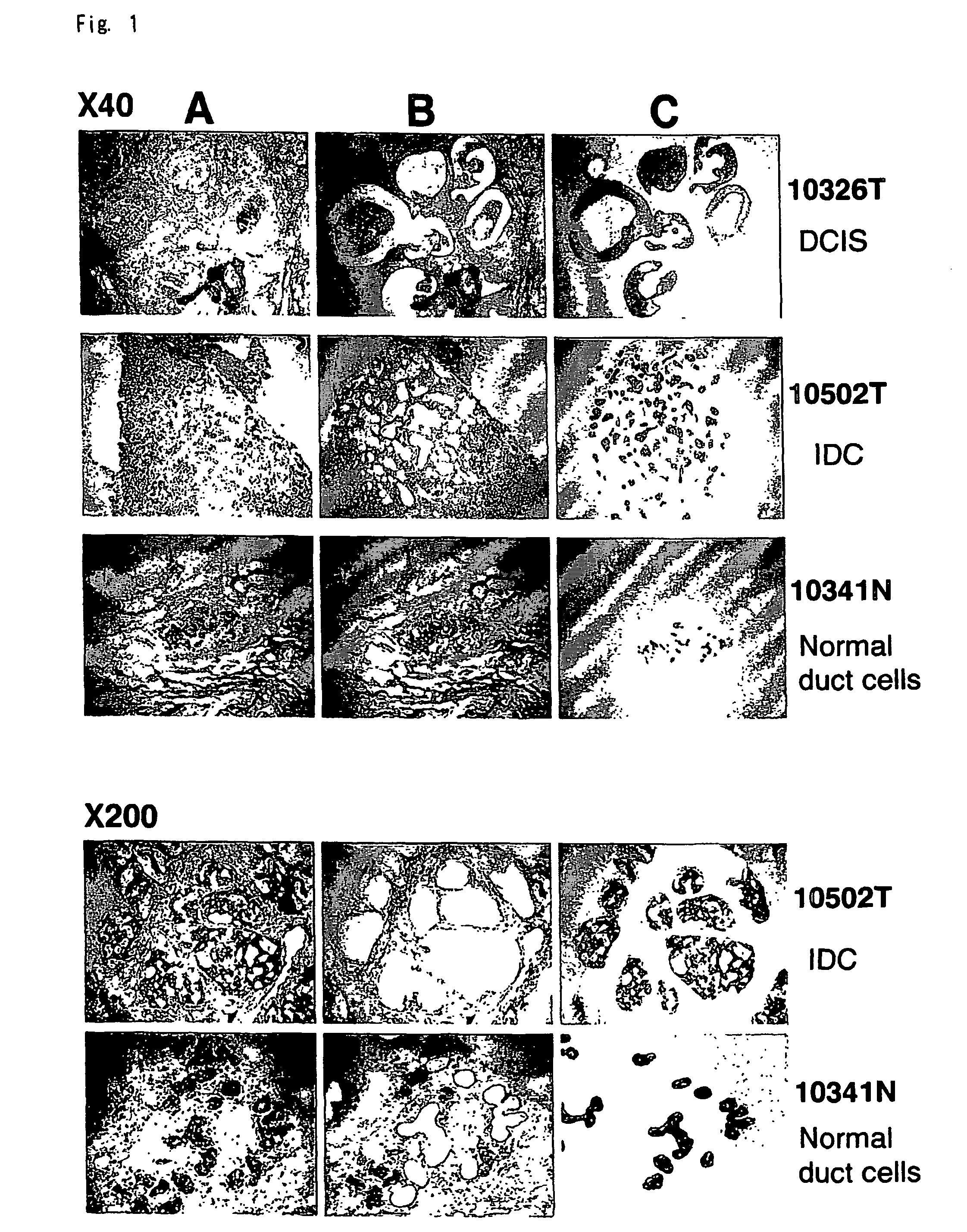
Objective methods for detecting and diagnosing breast cancer (BRC) are described herein. In one embodiment, the diagnostic method involves determining the expression level of a BRC-associated gene that discriminates between BRC cells and normal cells. In another embodiment, the diagnostic method involves determining the expression level of a BRC-associated gene that discriminates among BRC cells, between DCIS and IDC cells. The present invention further provides means for predicting and preventing breast cancer metastasis using BRC-associated genes having unique altered expression patterns in breast cancer cells with lymph-node metastasis. Finally, the present invention provides methods of screening for therapeutic agents useful in the treatment of breast cancer, methods of treating breast cancer and method for vaccinating a subject against breast cancer.
Owner:ONCOTHERAPY SCI INC
Brimonidine compositions and methods for retinal degeneration
InactiveUS20010049369A1Patient compliance is goodReduce incidenceBiocideAnimal repellantsAdjuvantStress marker
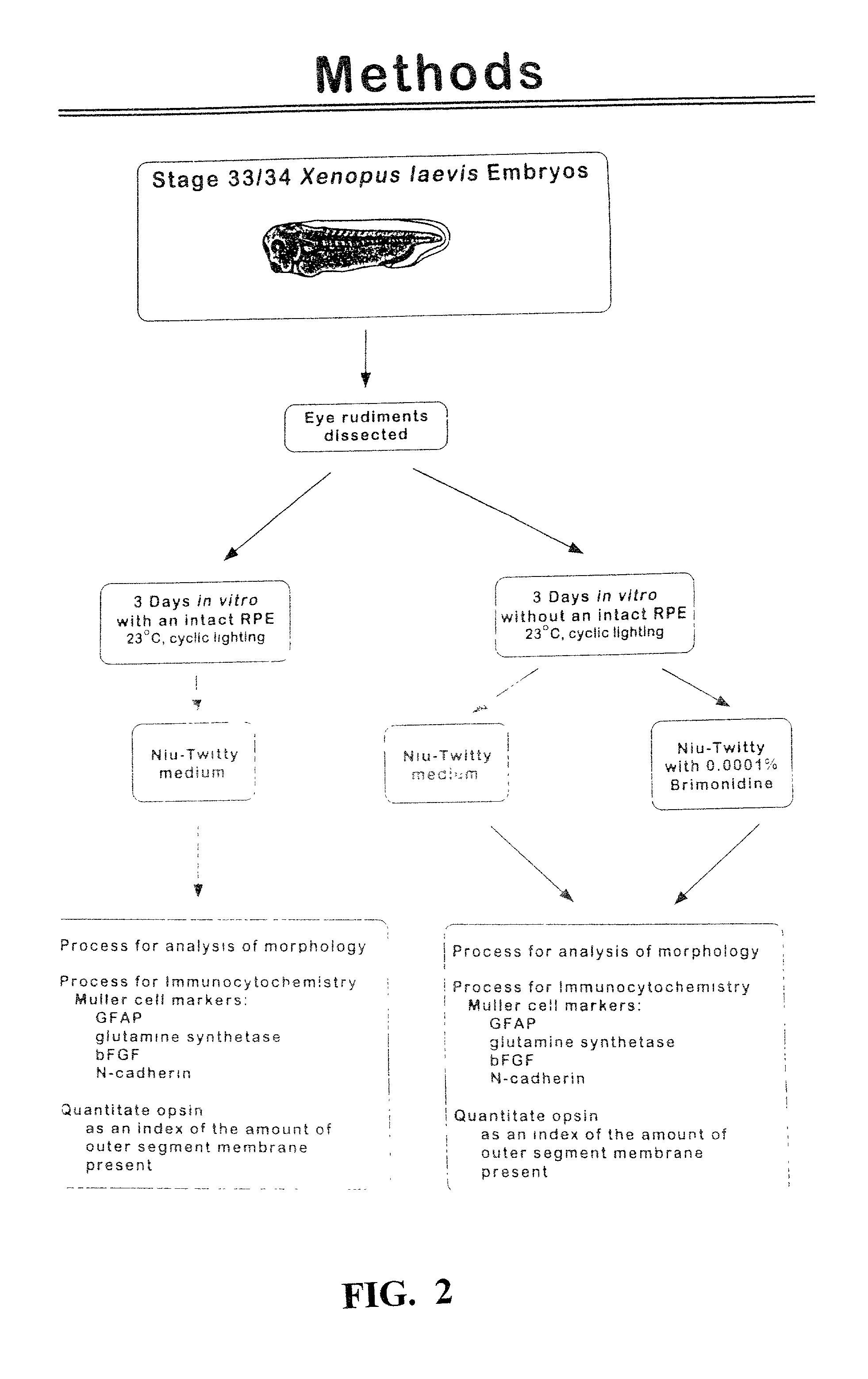
The present study demonstrates that brimonidine tartrate, an alpha-2 adrenergic receptor agonist, can prevent photoreceptor cell degeneration and the associated Muller cell degenerative signs in an in vitro model of retinal degeneration and retinal detachment (separation of the neuroretina from the retinal pigment epithelium). Similar to control conditions, brimonidine allowed for the formation of highly structured photoreceptor outer segments, prevented the expression of stress markers in Müller cells and preserved the expression patterns of Muller cell markers of proper cell-cell contact and differentiation. Ultrastructural studies also indicated that brimonidine favored the formation of cell-cell junctions between photoreceptor cells and Müller cells, indicating that this phenomenon is associated with the exertion of the neuroprotective effect. The results suggest that brimonidine compounds may be utilized as an effective therapeutic agent for early and late onset retinal degenerations caused by defects in photoreceptor cells, Müller cells or both, and as an adjuvant to therapeutic success in retinal detachment surgery or macular translocation surgery for age-related macular degeneration.
Owner:UNIV OF TENNESSEE RES FOUND
Gene expression profiles to predict relapse of prostate cancer
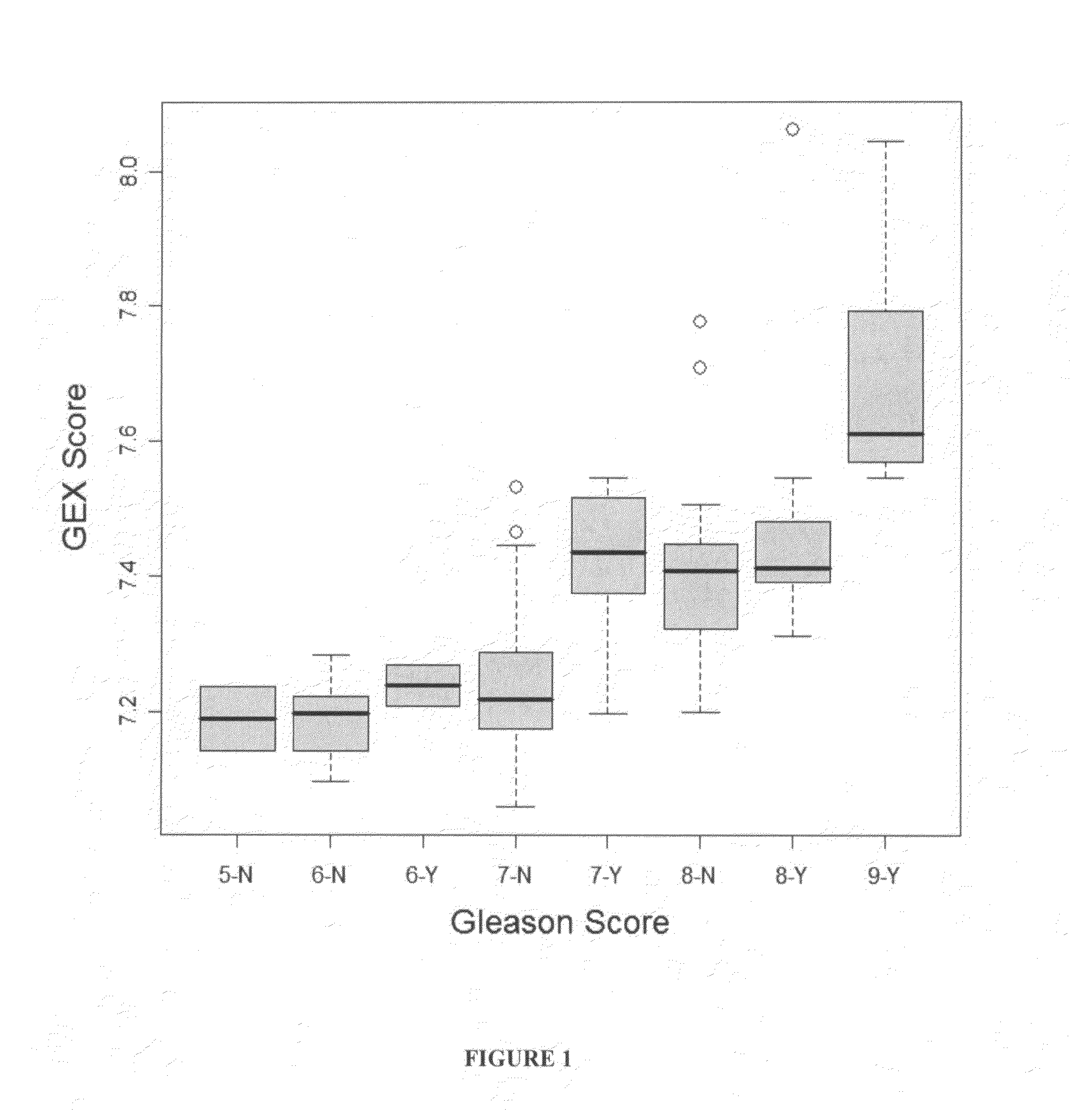
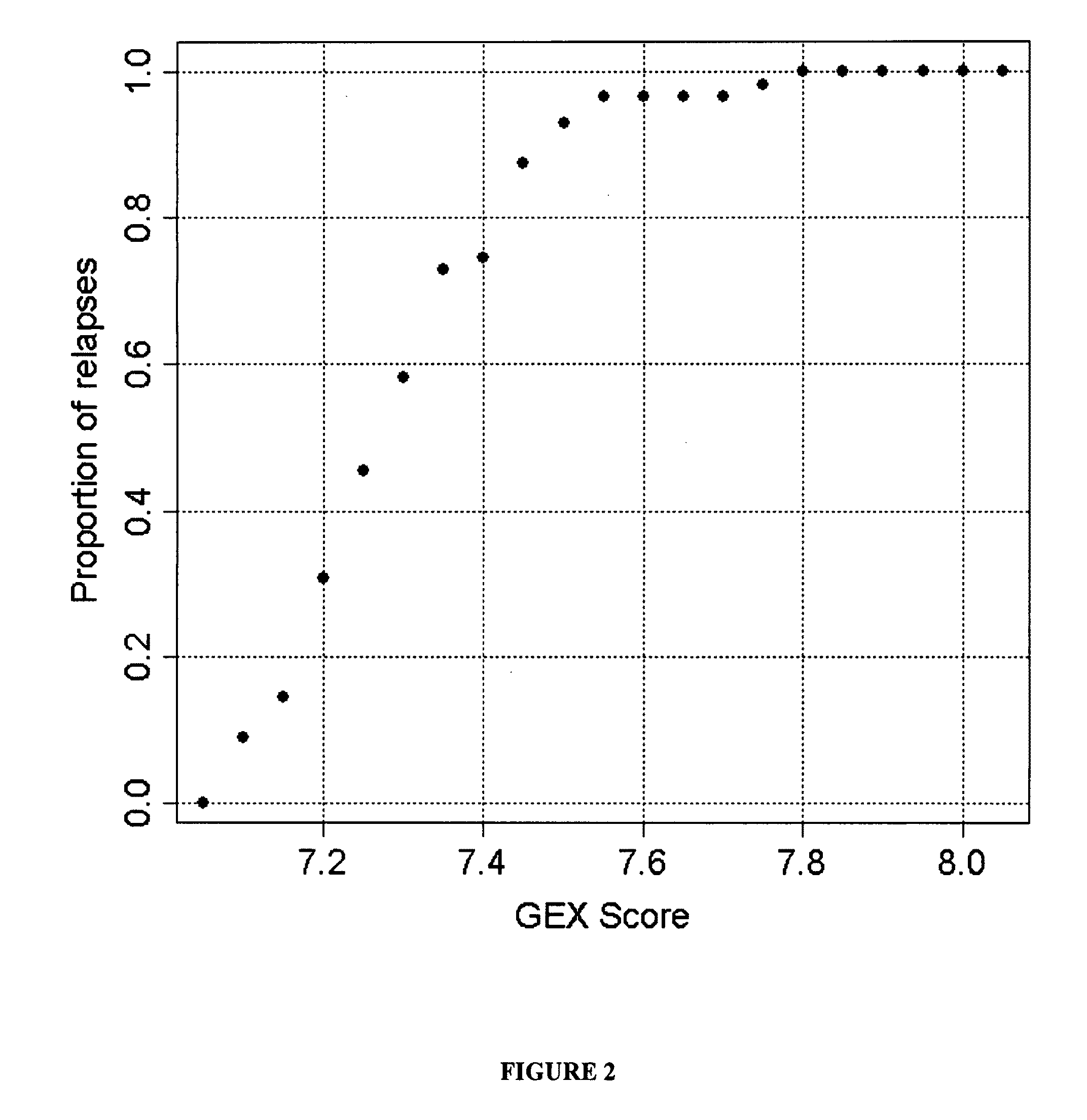
The present invention provides a method for preparing a reference model for cancer relapse prediction that provides higher resolution grading than Gleason score alone. The method encompasses obtaining from different individuals a plurality of prostate carcinoma tissue samples of known clinical outcome representing different Gleason scores; selecting a set of signature genes having an expression pattern that correlates positively or negatively in a statistically significant manner with the Gleason scores; independently deriving a prediction score that correlates gene expression of each individual signature gene with Gleason score for each signature gene in said plurality of prostate carcinoma tissue samples; deriving a prostate cancer gene expression (GEX) score that correlates gene expression of said set of signature genes with the Gleason score based on the combination of independently derived prediction scores in the plurality of prostate cancer tissue samples; and correlating said GEX score with the clinical outcome for each prostate carcinoma tissue sample. A set of signature genes is provided that encompasses all or a sub-combination of GI_2094528, KIP2, NRG1, NBL1, Prostein, CCNE2, CDC6, FBP1, HOXC6, MKI67, MYBL2, PTTG1, RAMP, UBE2C, Wnt5A, MEMD, AZGP1, CCK, MLCK, PPAP2B, and PROK1. Also provided a methods for predicting the probability of relapse of cancer in an individual and methods for deriving a prostate cancer gene expression (GEX) score for a prostate carcinoma tissue sample obtained from an individual.
Owner:ILLUMINA INC
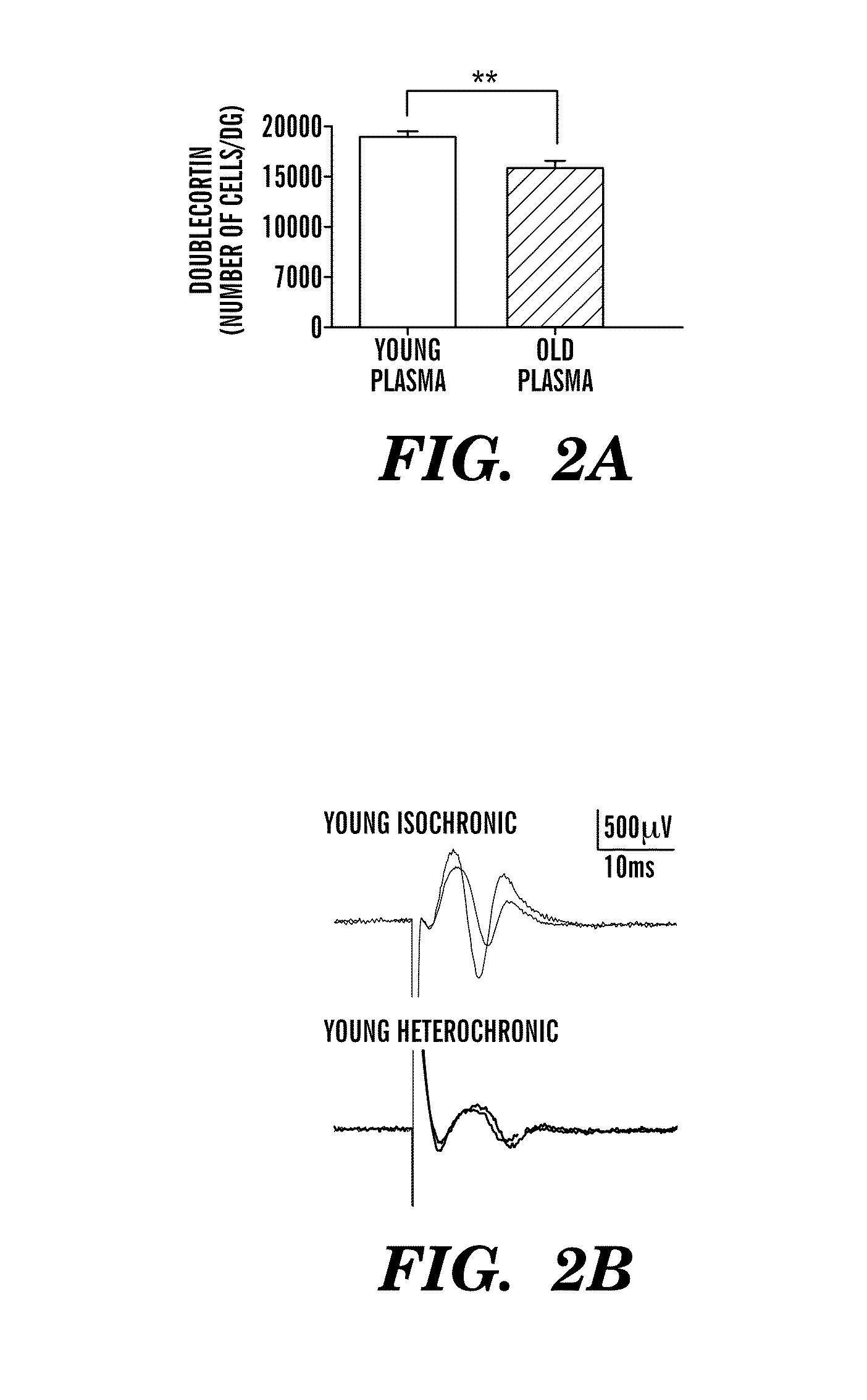
Biomarkers of aging for detection and treatment of disorders
InactiveUS20140255424A1Modulate activityVirusesMicrobiological testing/measurementDiseaseNeural cell
Provided are methods of diagnosis, prognosis, and monitoring of aging using biomarkers that have been discovered to be linked to biological aging process. Methods for increasing neural cell regeneration and cognitive function are also provided. The methods are, at least in part, based on a discovery that altered expression patterns of certain biological markers are associated with biological aging processes. These markers comprise at least Eotaxin / CCL11, β2-microglobulin, MCP-1 and Haptoglobulin, increased expression of which has been shown to be associated with increase in biological aging process.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV +1
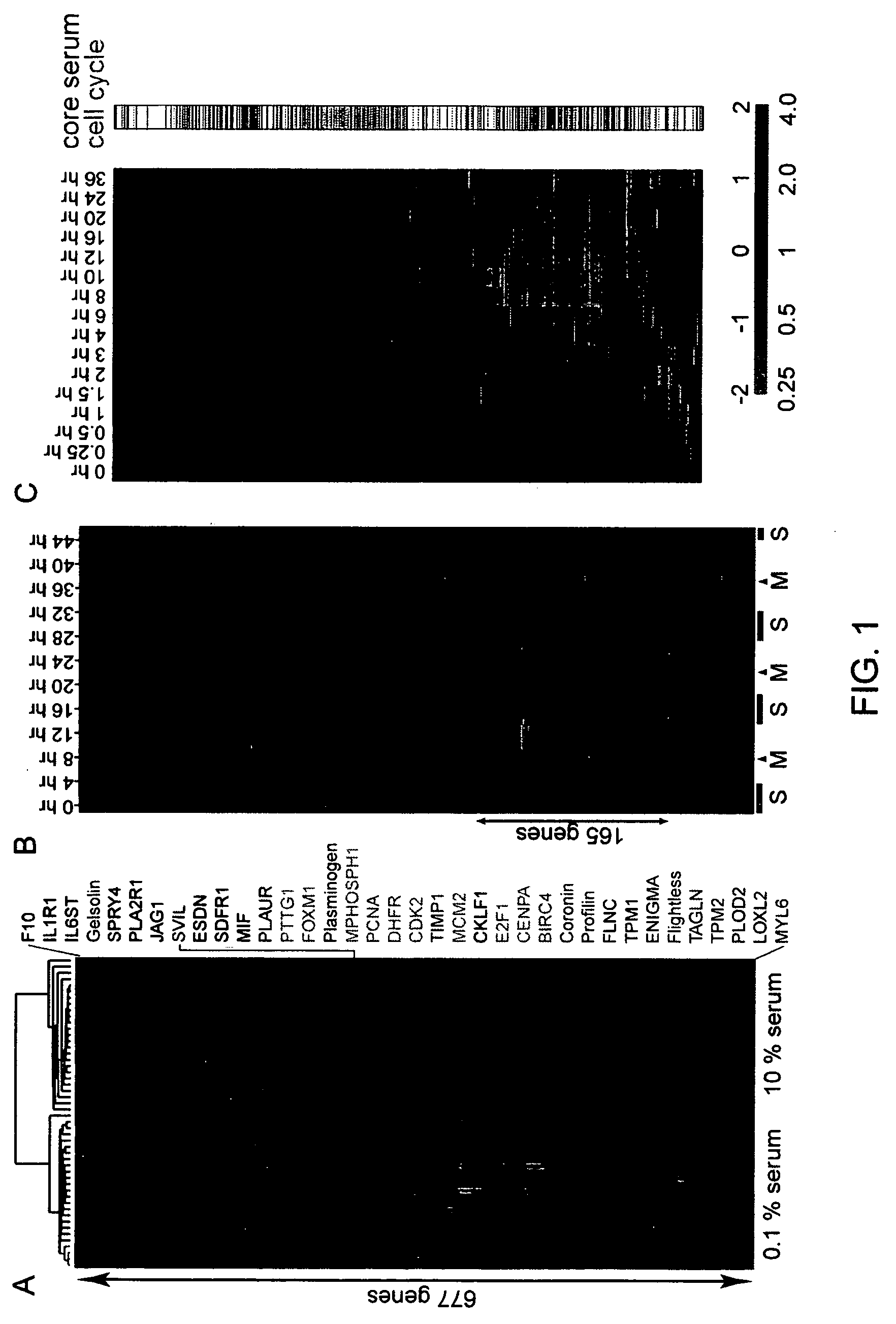
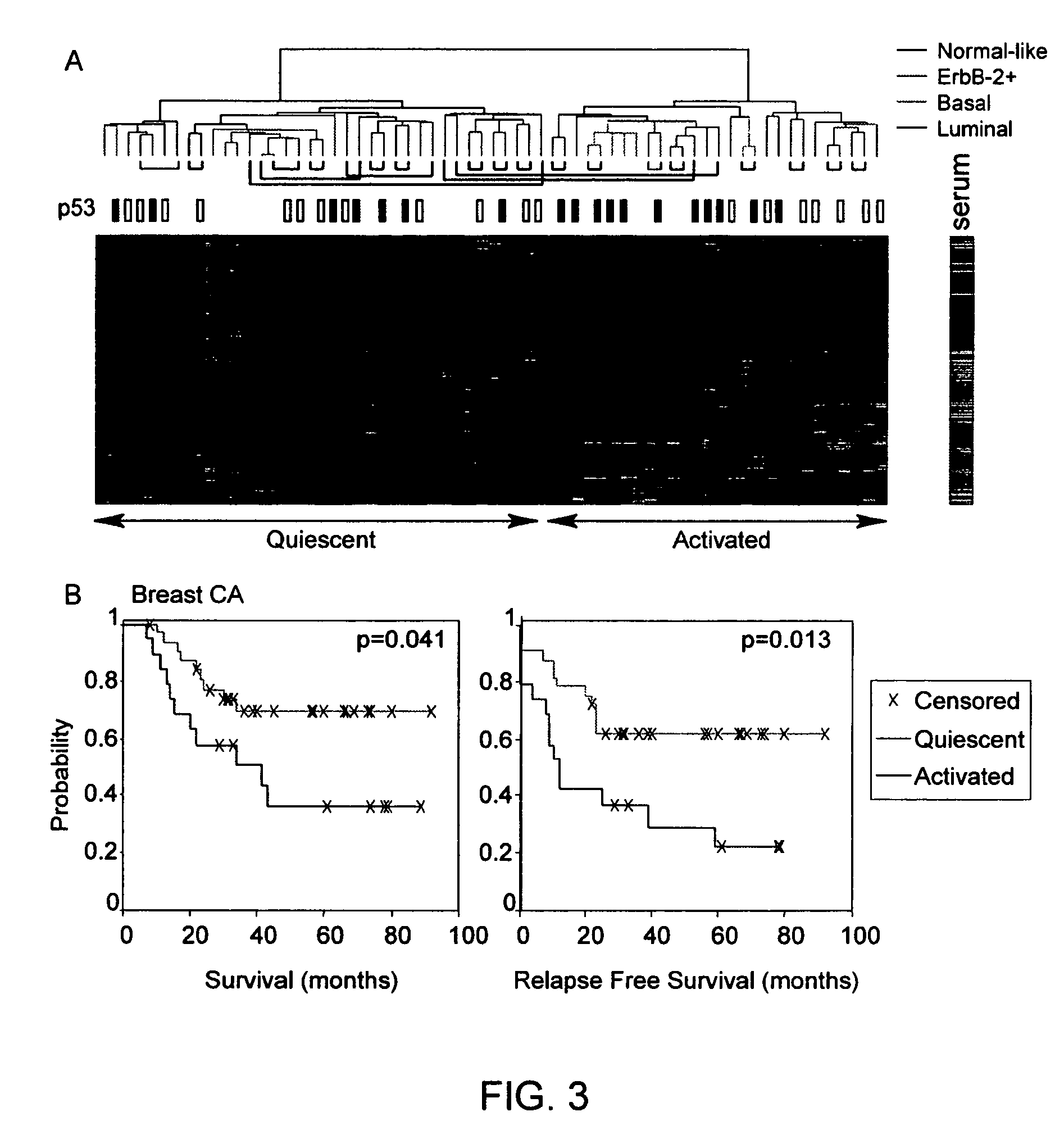
Gene expression signature for prediction of human cancer progression
InactiveUS20060183141A1Increase probabilityOptimize choiceSugar derivativesMicrobiological testing/measurementHuman cancerLymphatic Spread
Methods are provided for classification of cancers by the expression of a set of genes referred to as the core serum response (CSR), or a subset thereof. The expression pattern of the CSR in normal tissues correlates with that seen in quiescent fibroblasts cultured in the absence of serum, while cancer tissues can be classified as having a quiescent or induced CSR signature. Patients with the induced CSR signature have a higher probability of metastasis. Classification according to CSR signature allows optimization of treatment, and determination of whether on whether to proceed with a specific therapy, and how to optimize dose, choice of treatment, and the like.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
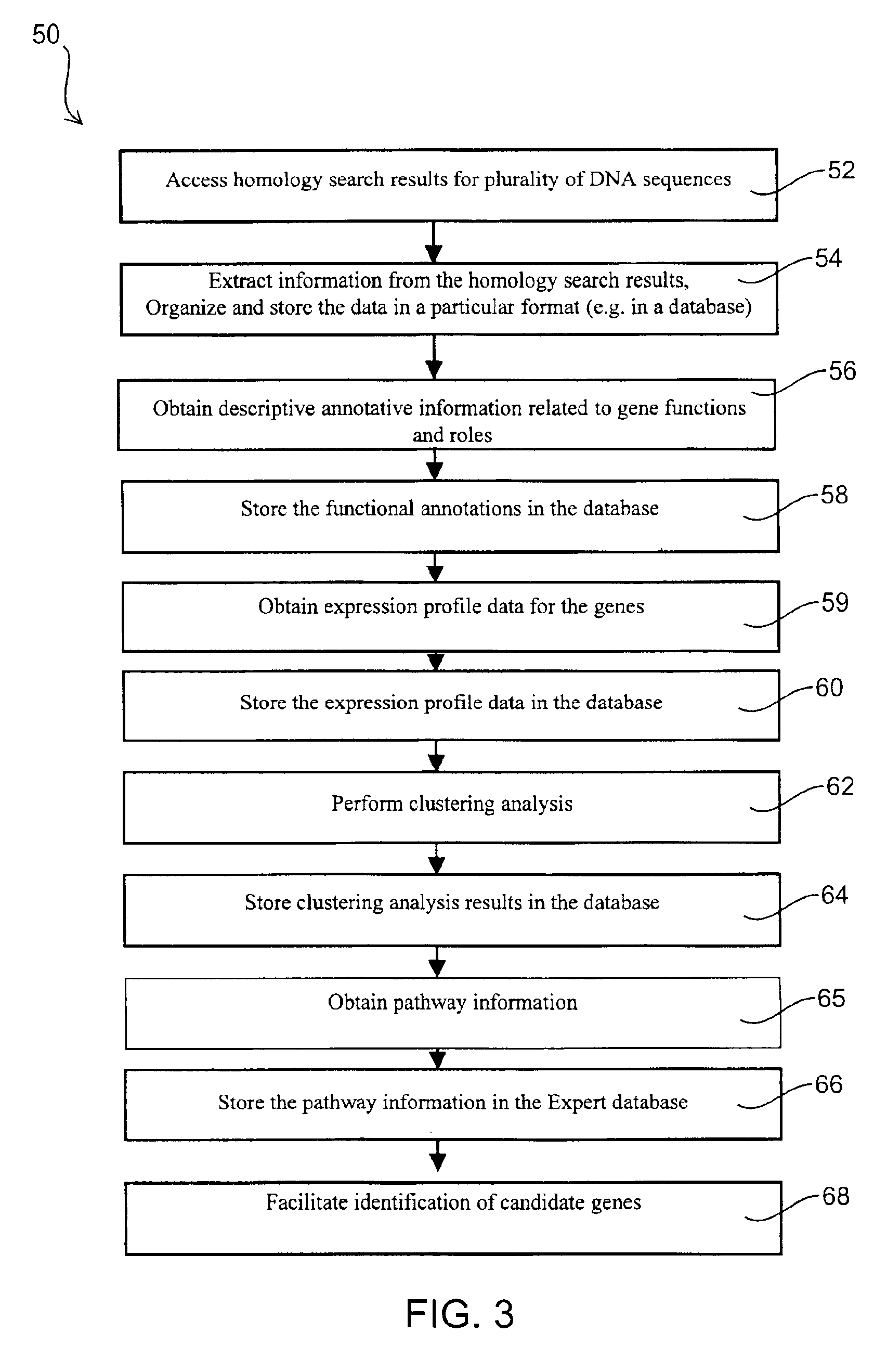
Automated pathway recognition system
InactiveUS6876930B2Easy to identifyQuick analysisData processing applicationsBiostatisticsCandidate Gene Association StudyCandidate gene
There is a pressing need for computer-implemented tools that can summarize and present the enormous amounts of public literature to facilitate analysis of gene expression data. The present invention provides techniques and systems for efficiently integrating public literature regarding gene function with data from gene expression profiling experiments. Information from literature databases relating to a particular set of DNA sequences of known expression pattern is retrieved, processed, cross-referenced and viewed to provide further information about a particular DNA sequence to facilitate its identification as a candidate gene.
Owner:AGY THERAPEUTICS
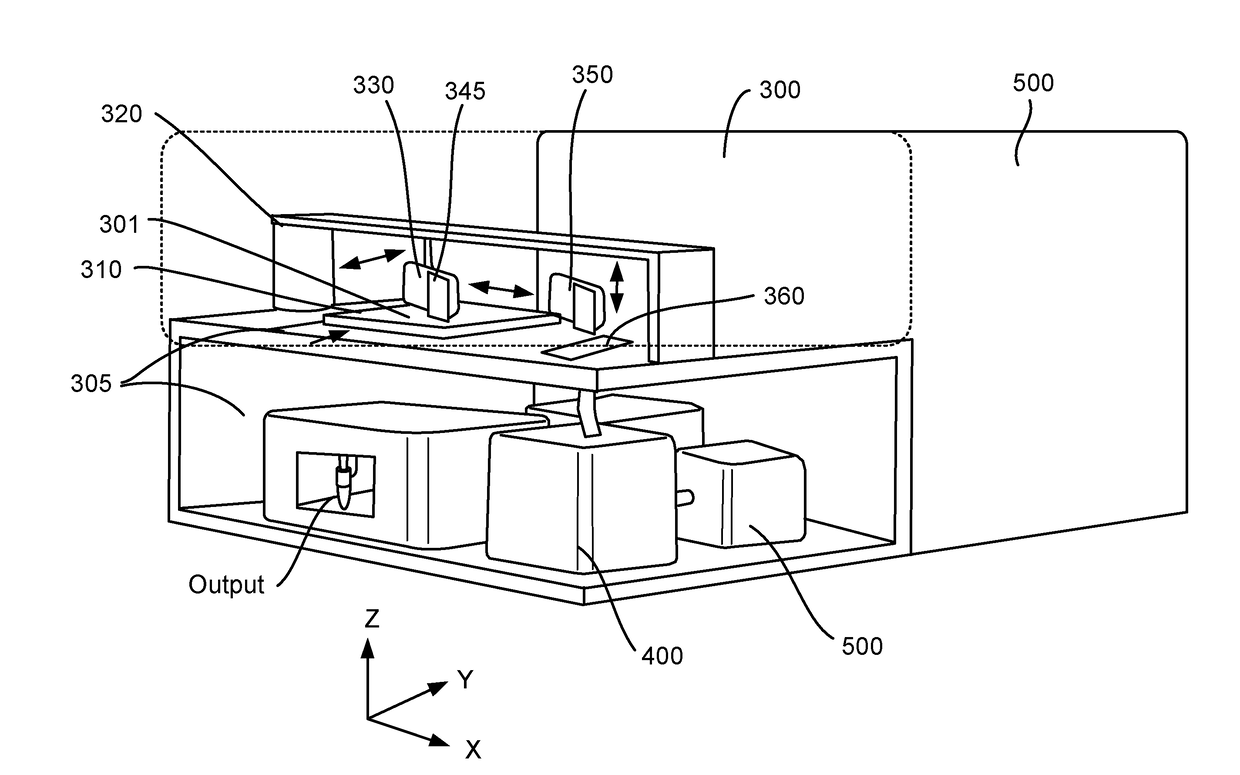
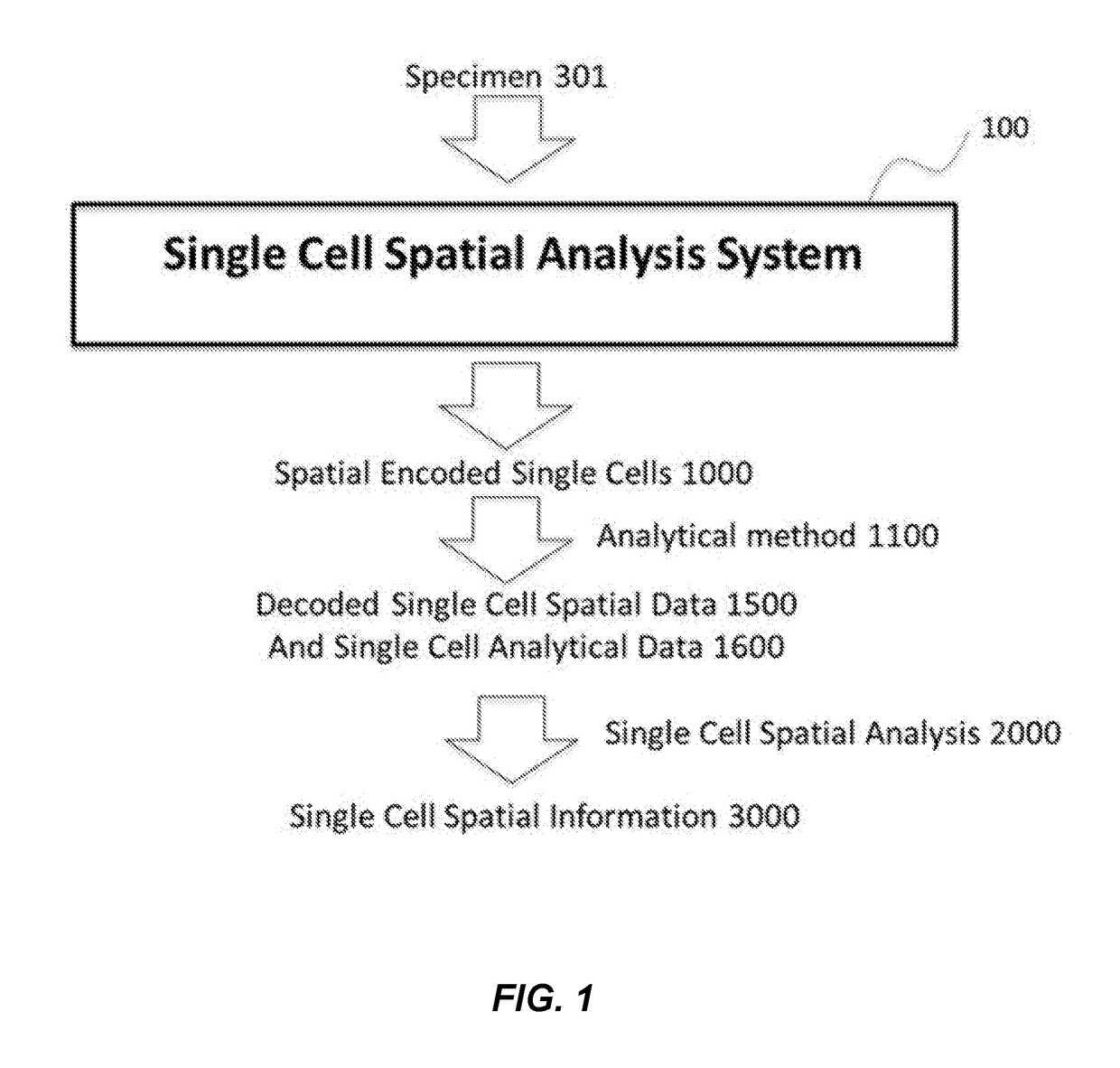
Method and apparatus for encoding cellular spatial position information
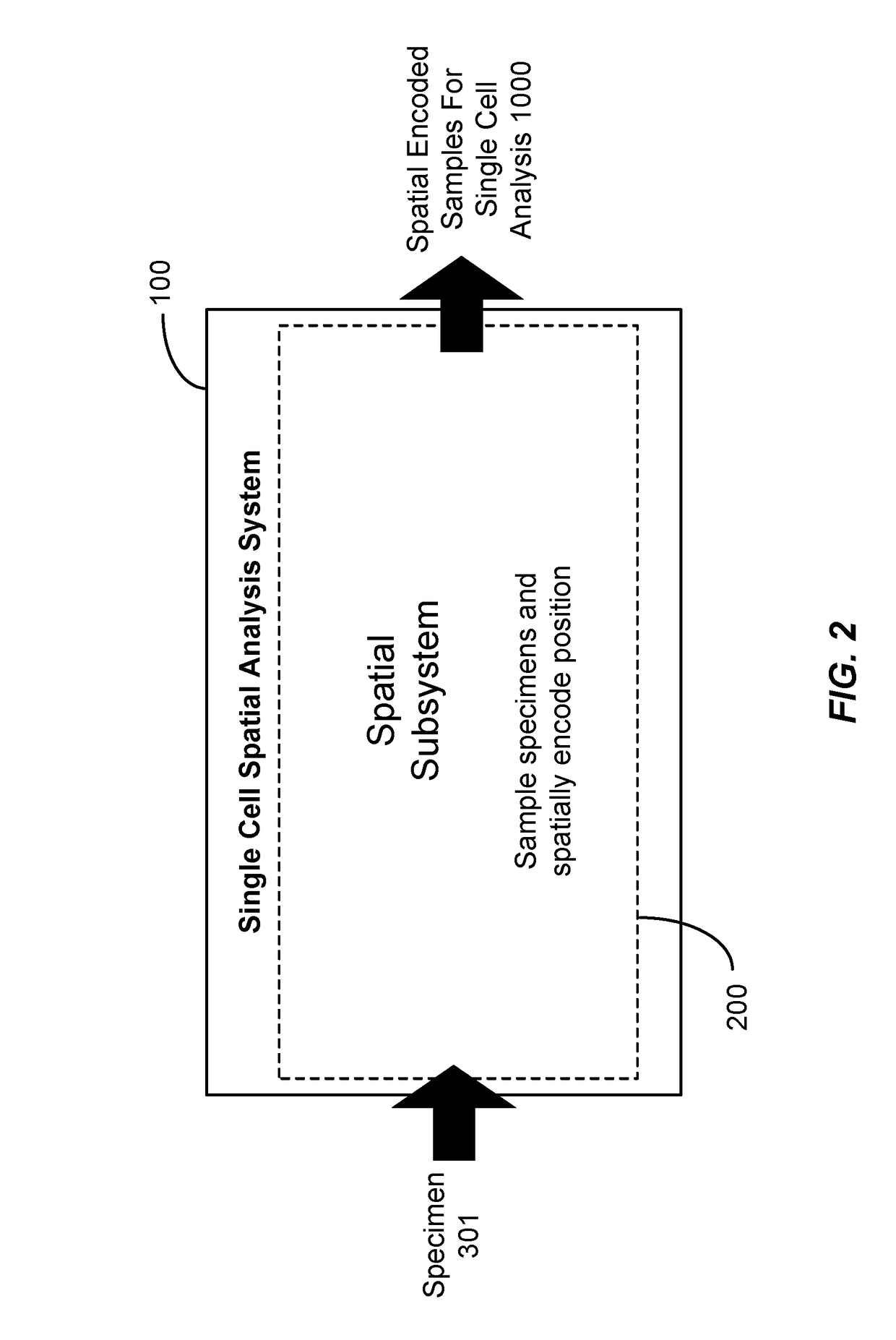
A system, methods, and apparatus are described to collect and prepare single cells and groups of cells from microsamples of specimens and encode spatial information of the physical position of the cells in the specimen. In some embodiment, beads or surfaces with oligonucleotides containing spatial barcodes are used to analyze DNA or RNA. The spatial barcodes allow the position of the cell to be defined and the nucleic acid sequencing information, such as target sequencing, whole genome, gene expression, used to analyze the cells in a microsample for cell type, expression pattern, DNA sequence, and other information, in the context of the cell's physical position in the specimen. In other embodiment, markers such as isotopes are added to a microsample to encode spatial position with mass spectoscopy or other analysis. The spatial encoded information is then readout by analysis such as DNA sequencing, mass spectrometry, fluorescence, or other methods.
Owner:SILICON VALLEY SCI
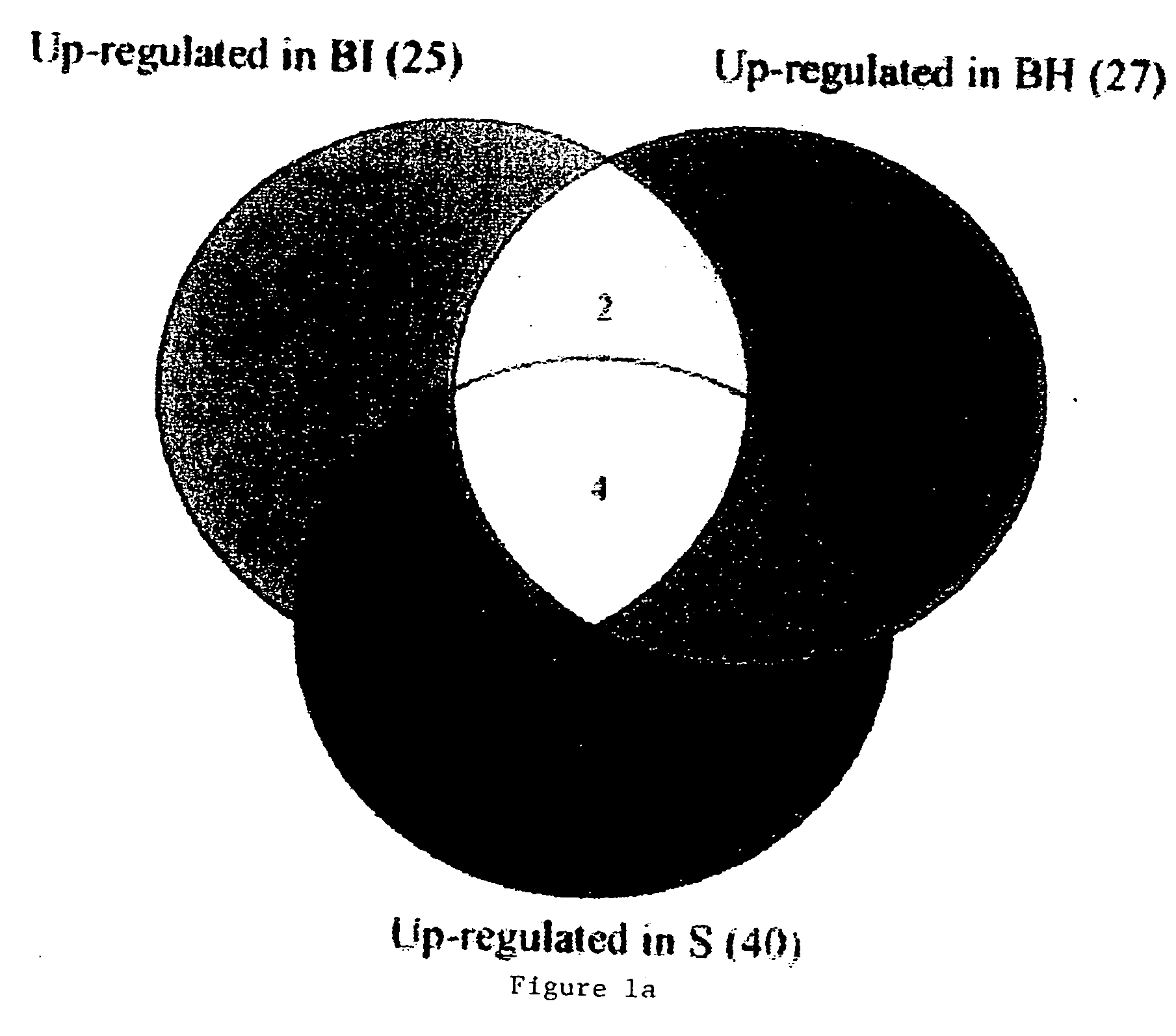
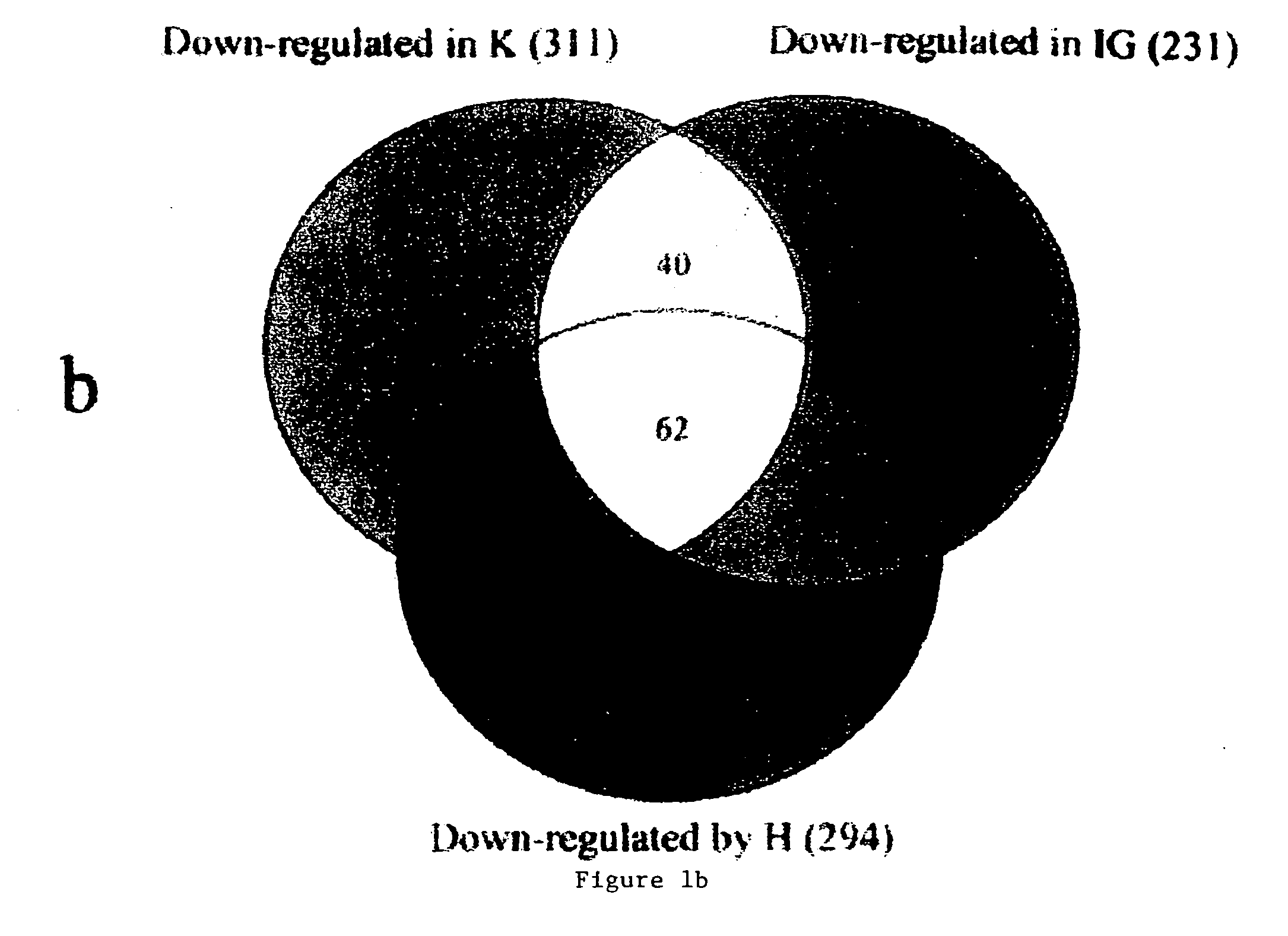
Endothelial cell expression patterns
To gain a better understanding of tumor angiogenesis, new techniques for isolating endothelial cells (ECs) and evaluating gene expression patterns were developed. When transcripts from ECs derived from normal and malignant colorectal tissues were compared with transcripts from non-endothelial cells, over 170 genes predominantly expressed in the endothelium were identified. Comparison between normal- and tumor-derived endothelium revealed 79 differentially expressed genes, including 46 that were specifically elevated in tumor-associated endothelium. Experiments with representative genes from this group demonstrated that most were similarly expressed in the endothelium of primary lung, breast, brain, and pancreatic cancers as well as in metastatic lesions of the liver. These results demonstrate that neoplastic and normal endothelium in humans are distinct at the molecular level, and have significant implications for the development of anti-angiogenic therapies in the future.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
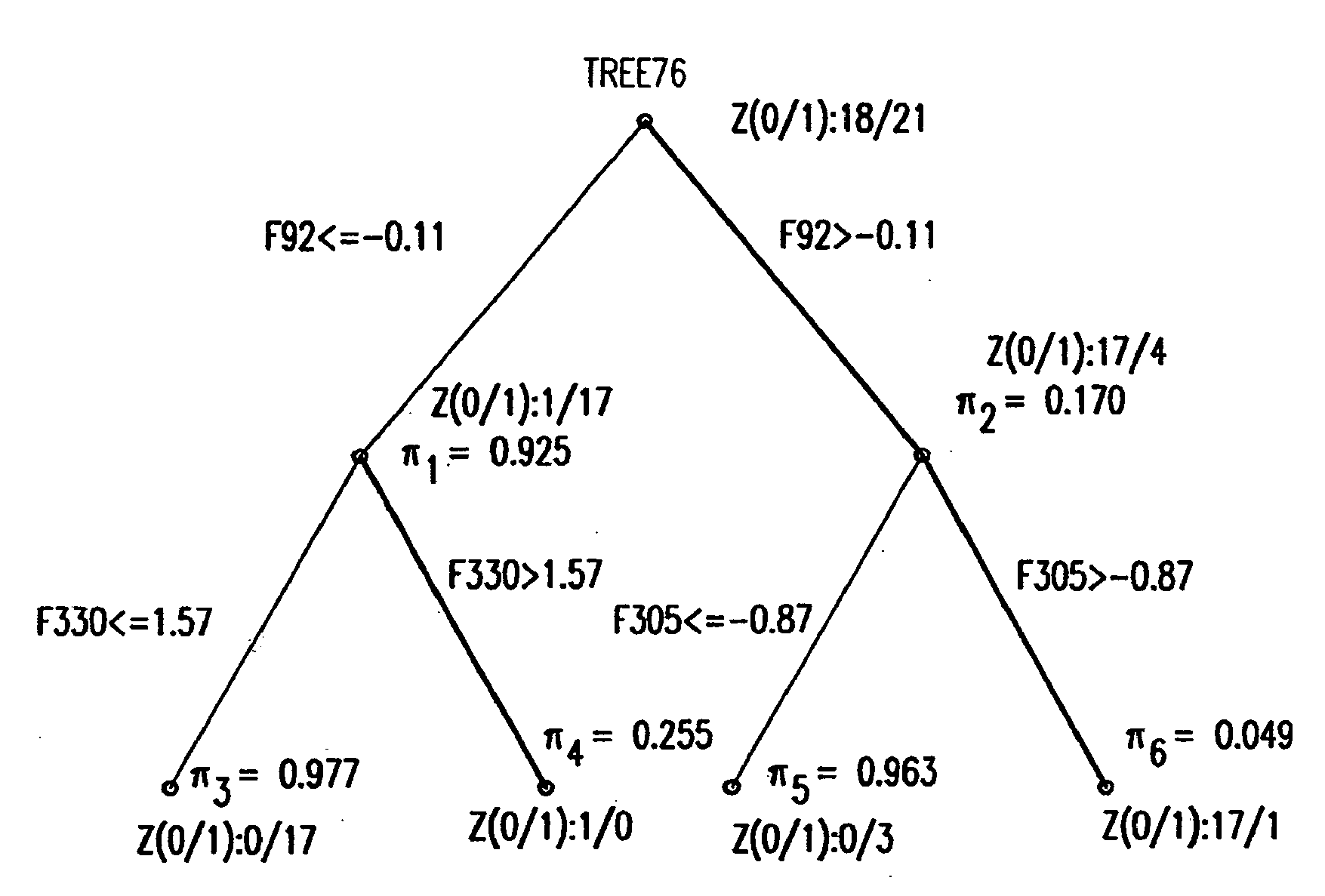
Binary prediction tree modeling with many predictors and its uses in clinical and genomic applications
InactiveUS20090319244A1Mathematical modelsAnalogue computers for chemical processesSingular value decompositionData set
The statistical analysis described and claimed is a predictive statistical tree model that overcomes several problems observed in prior statistical models and regression analyses, while ensuring greater accuracy and predictive capabilities. Although the claimed use of the predictive statistical tree model described herein is directed to the prediction of a disease in individuals, the claimed model can be used for a variety of applications including the prediction of disease states, susceptibility of disease states or any other biological state of interest, as well as other applicable non-biological states of interest. This model first screens genes to reduce noise, applies k-means correlation-based clustering targeting a large number of clusters, and then uses singular value decompositions (SVD) to extract the single dominant factor (principal component) from each cluster. This generates a statistically significant number of cluster-derived singular factors, that we refer to as metagenes, that characterize multiple patterns of expression of the genes across samples. The strategy aims to extract multiple such patterns while reducing dimension and smoothing out gene-specific noise through the aggregation within clusters. Formal predictive analysis then uses these metagenes in a Bayesian classification tree analysis. This generates multiple recursive partitions of the sample into subgroups (the “leaves” of the classification tree), and associates Bayesian predictive probabilities of outcomes with each subgroup. Overall predictions for an individual sample are then generated by averaging predictions, with appropriate weights, across many such tree models. The model includes the use of iterative out-of-sample, cross-validation predictions leaving each sample out of the data set one at a time, refitting the model from the remaining samples and using it to predict the hold-out case. This rigorously tests the predictive value of a model and mirrors the real-world prognostic context where prediction of new cases as they arise is the major goal.
Owner:DUKE UNIV
Biomarkers for neurodegenerative disorders
InactiveUS20070099203A1Medical data miningMicrobiological testing/measurementDementia with Lewy bodiesTreatment effect
The present invention provides methods for diagnosing neurodegenerative disease, such as Alzheimer's Disease, Parkinson's Disease, and dementia with Lewy body disease by detecting a pattern of gene product expression in a cerebrospinal fluid sample and comparing the pattern of gene product expression from the sample to a library of gene product expression pattern known to be indicative of the presence or absence of a neurodegenerative disease. The methods also provide for monitoring neurodegenerative disease progression and assessing the effects of therapeutic treatment. Also provided are kits, systems and devices for practicing the subject methods.
Owner:UNIV OF WASHINGTON
Blood assessment of injury
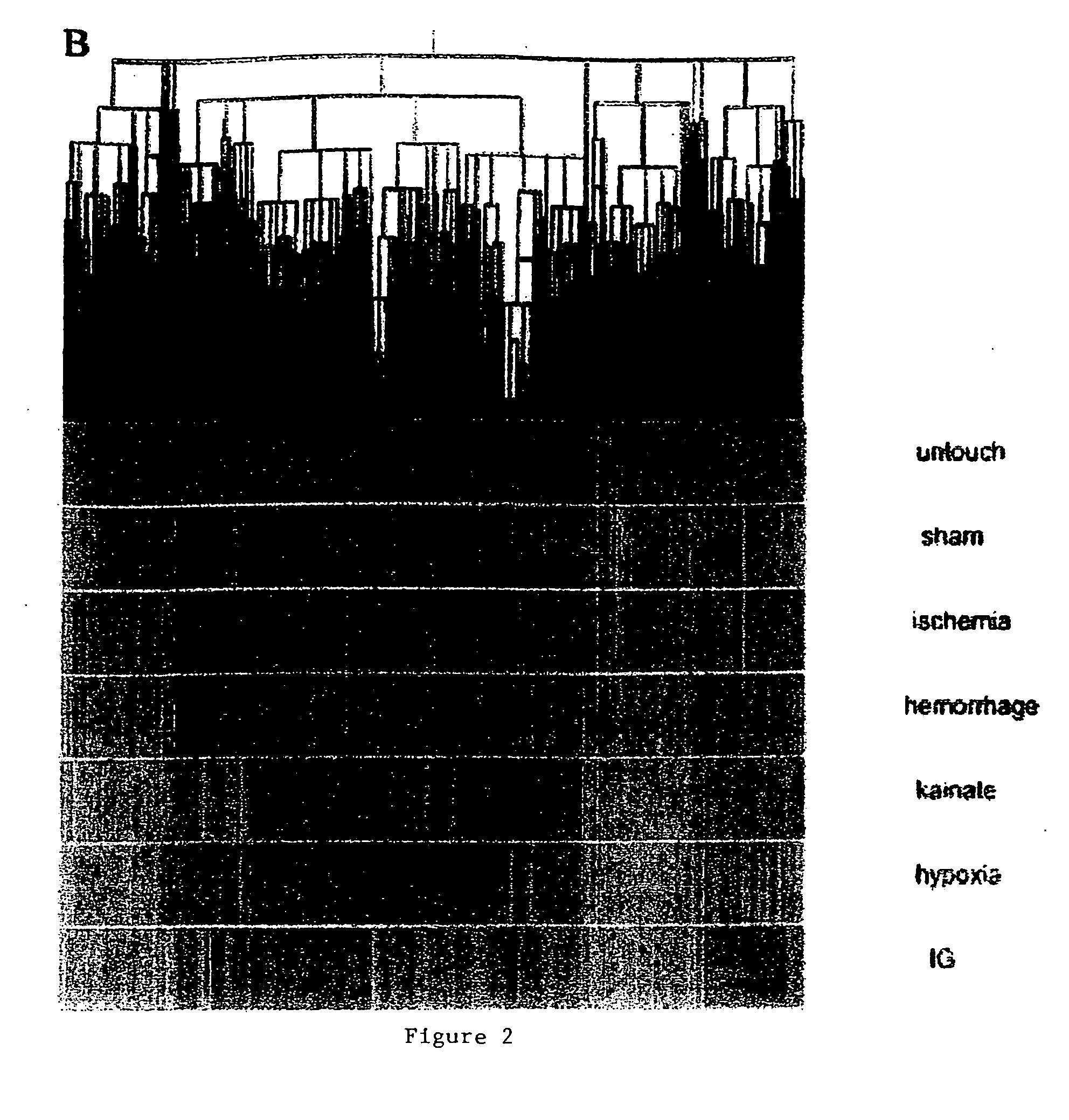
Methods of injury assessment in an individual include the steps of determining a pattern of expression exhibited by blood cells obtained from an individual and comparing the pattern of expression exhibited by the obtained blood cells to an injury database to assess the injury.
Owner:SHARP FR R +2
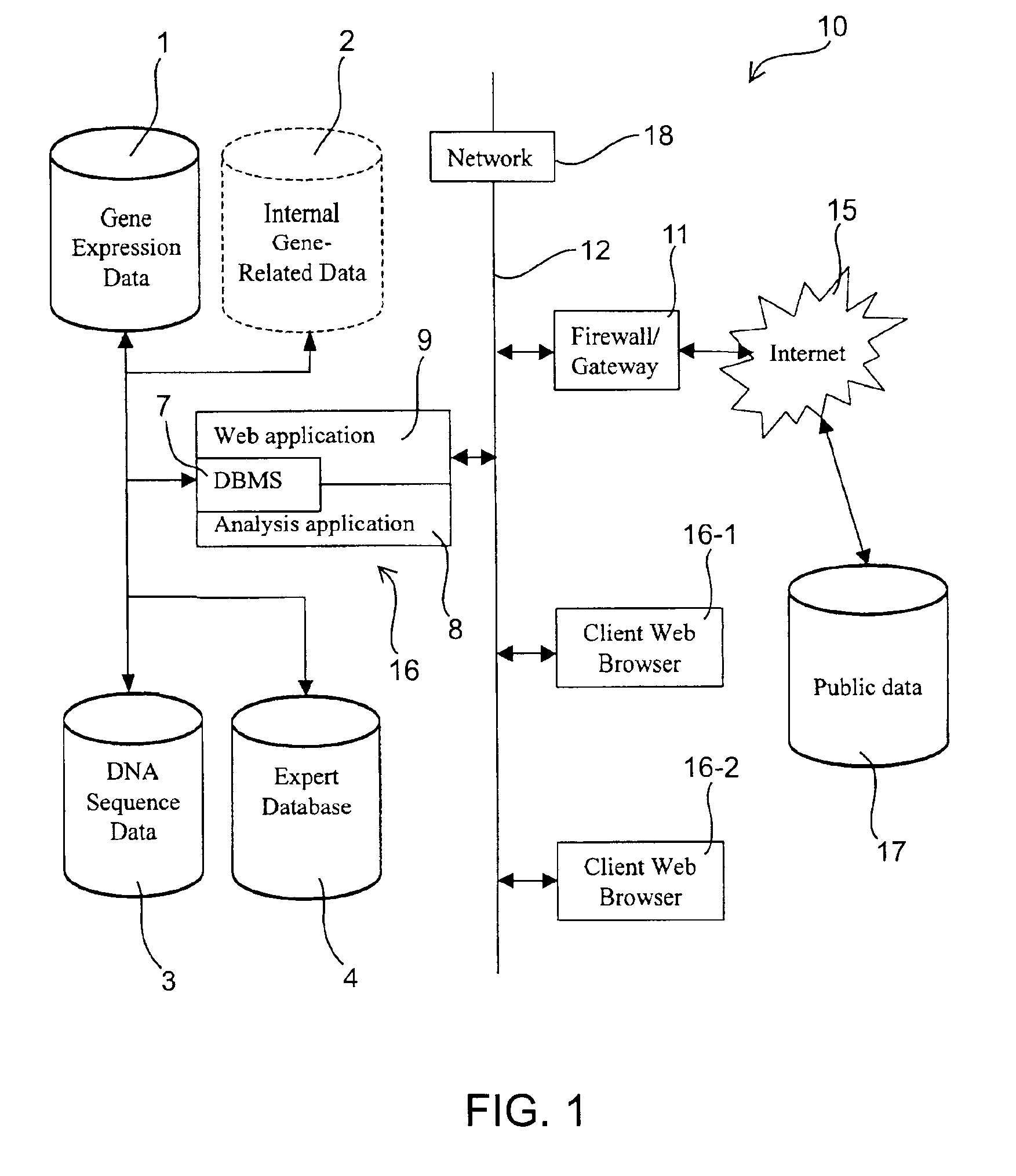
Methods and systems for efficient comparison, identification, processing, and importing of gene expression data
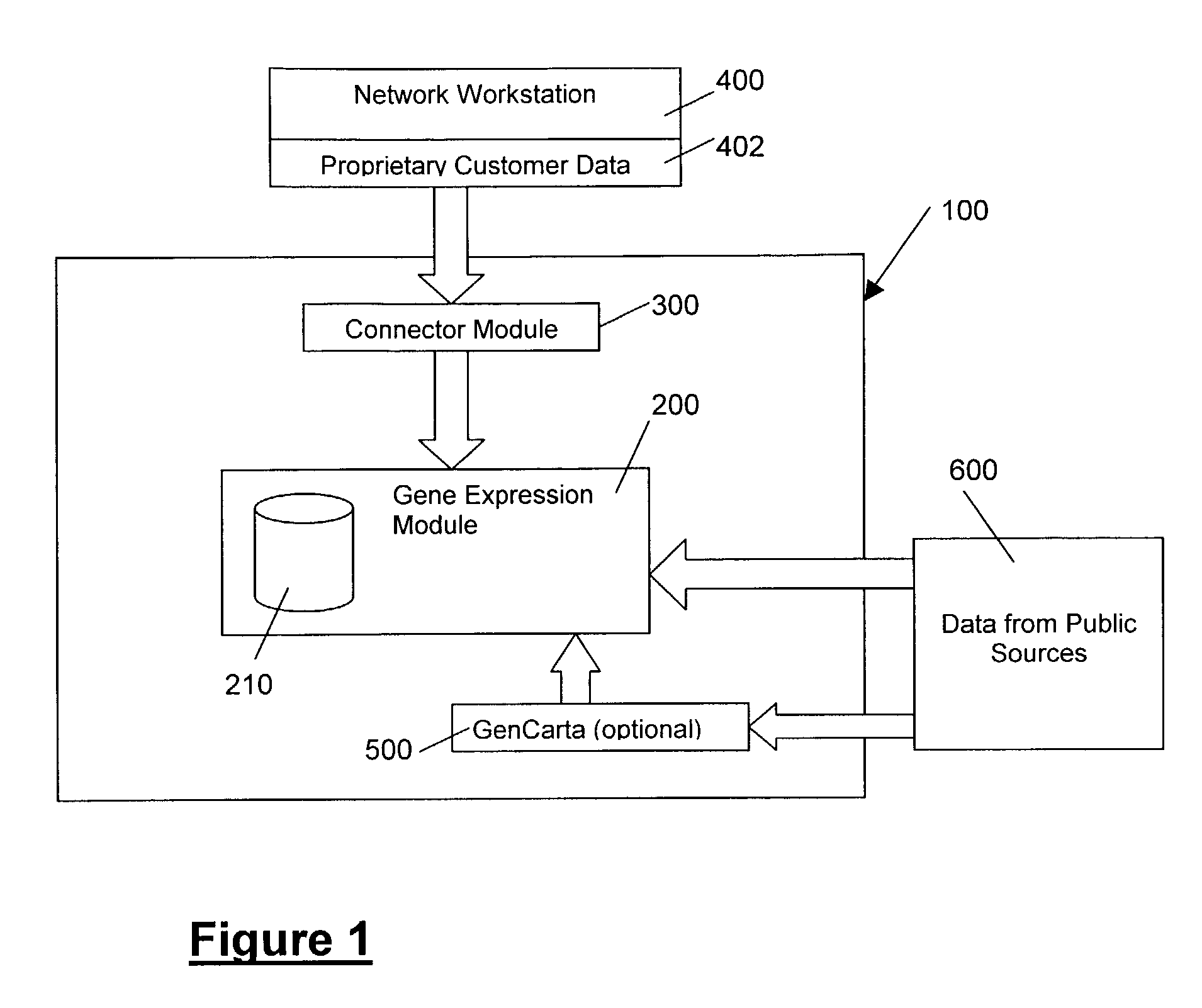
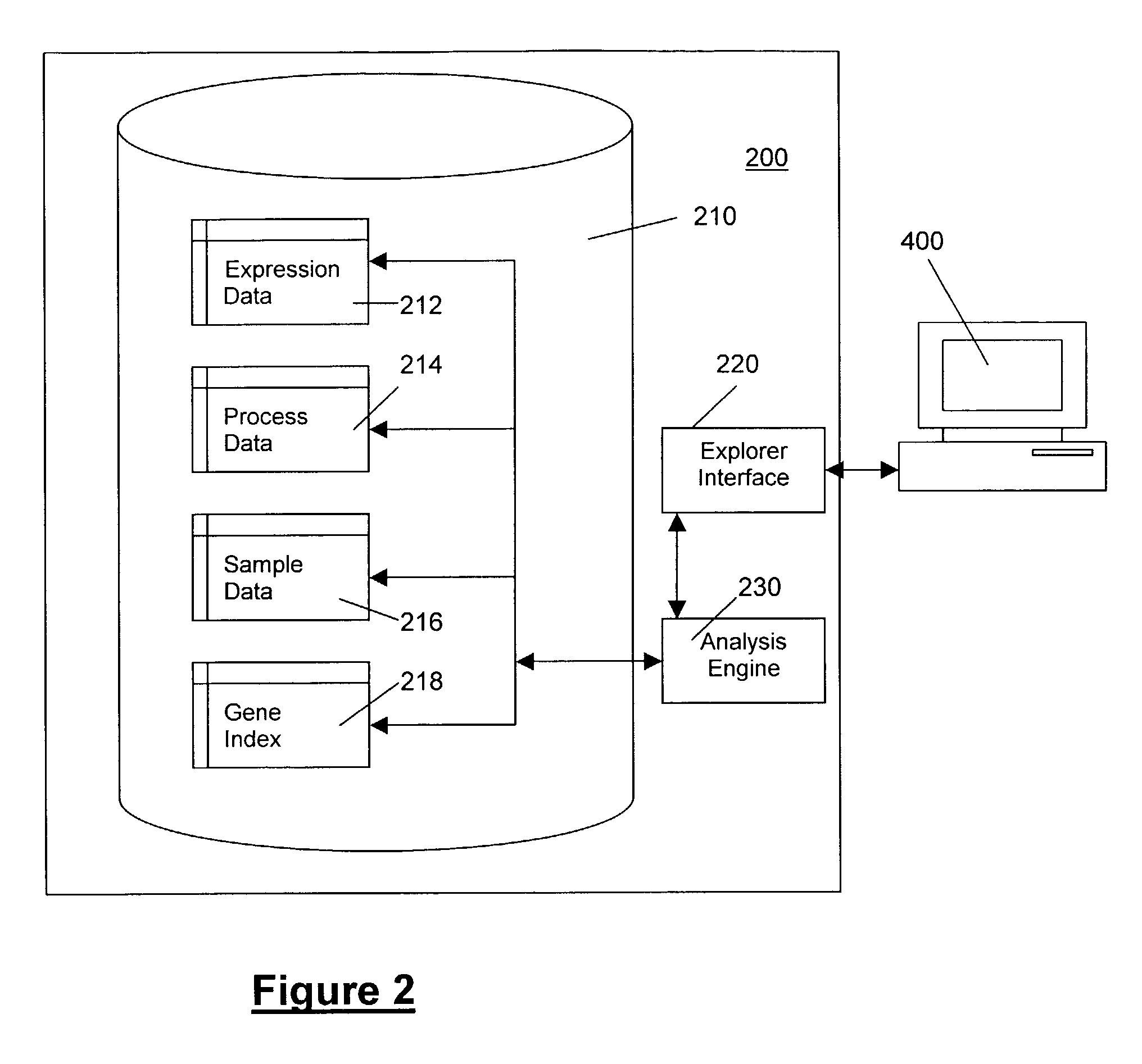
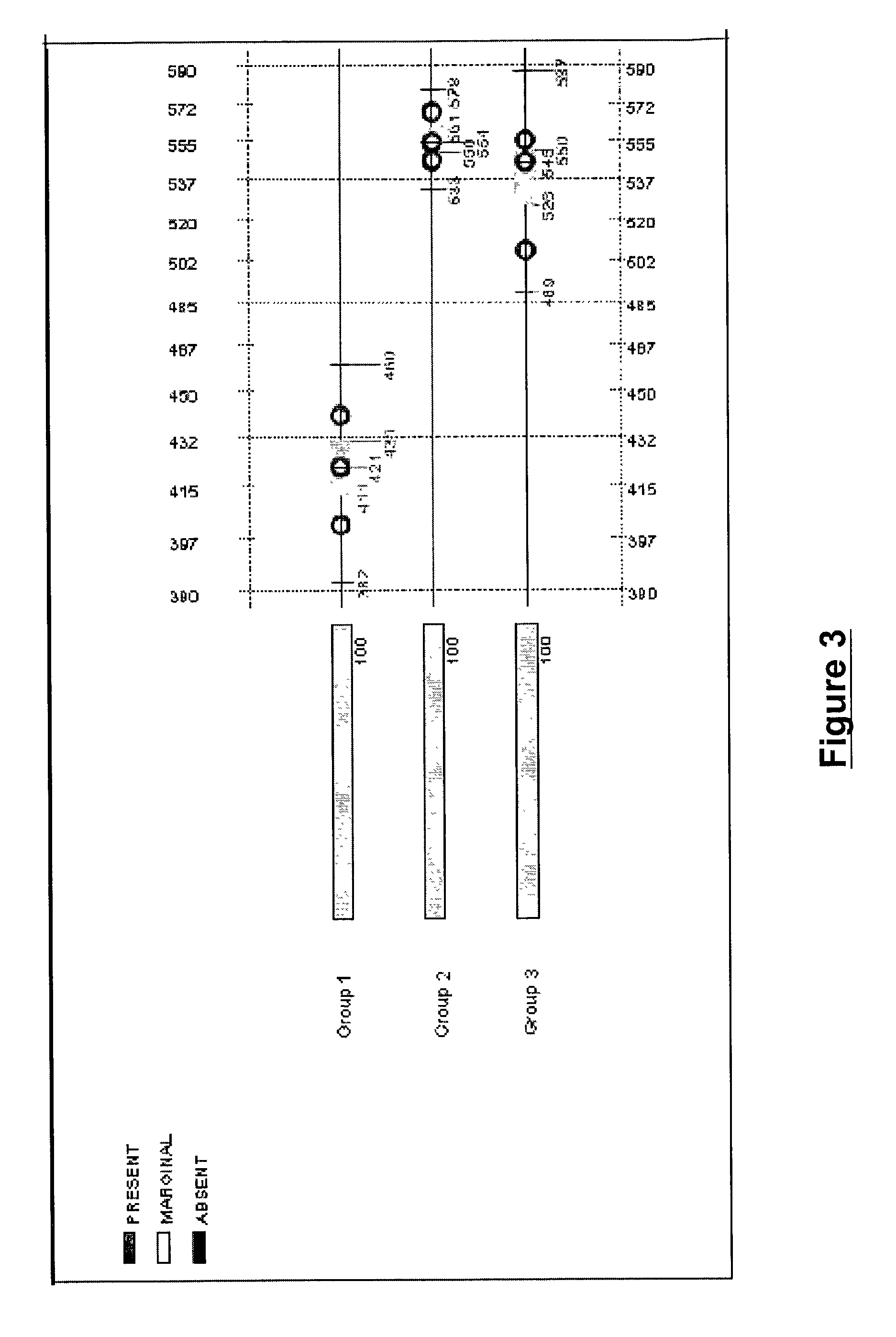
The present invention provides systems and methods for analyzing gene expression, gene annotation, and sample information in a relational format supporting efficient exploration and analysis, including comparative differential expression analysis, expression pattern matching, and mapping of external attributes or identifiers to internal representations of gene fragments or samples.
Owner:OCIMUM BIO SOLUTIONS
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com