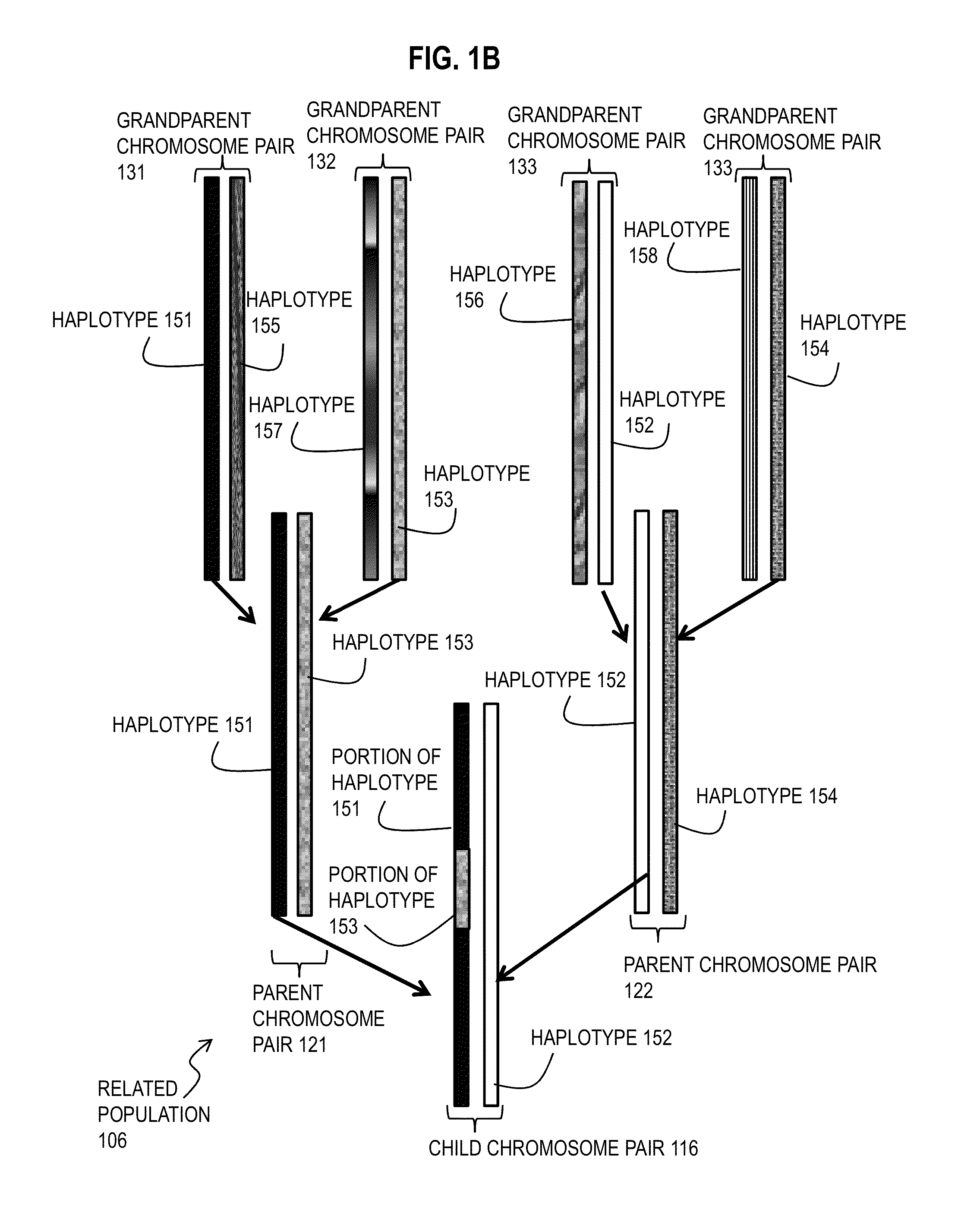
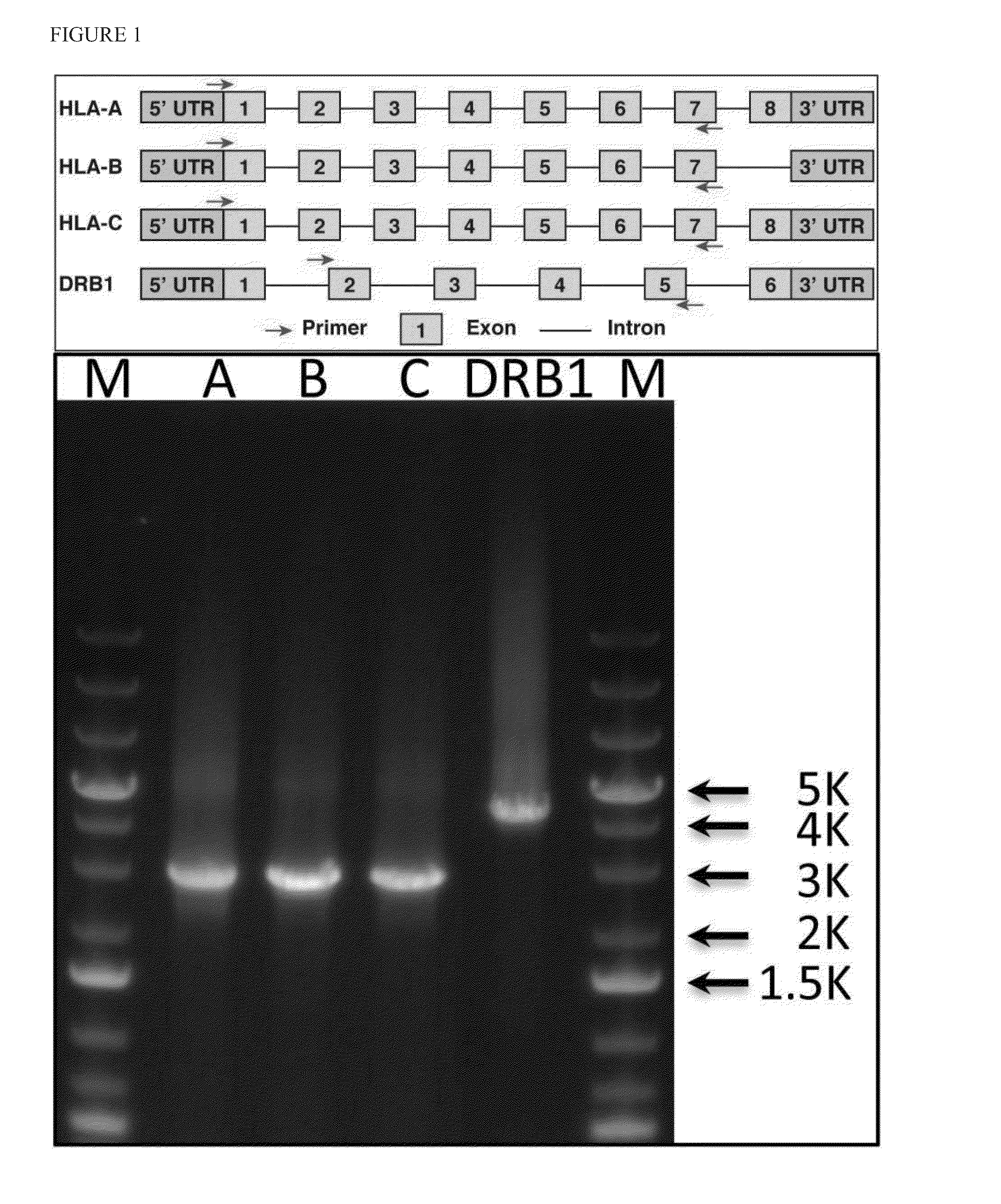
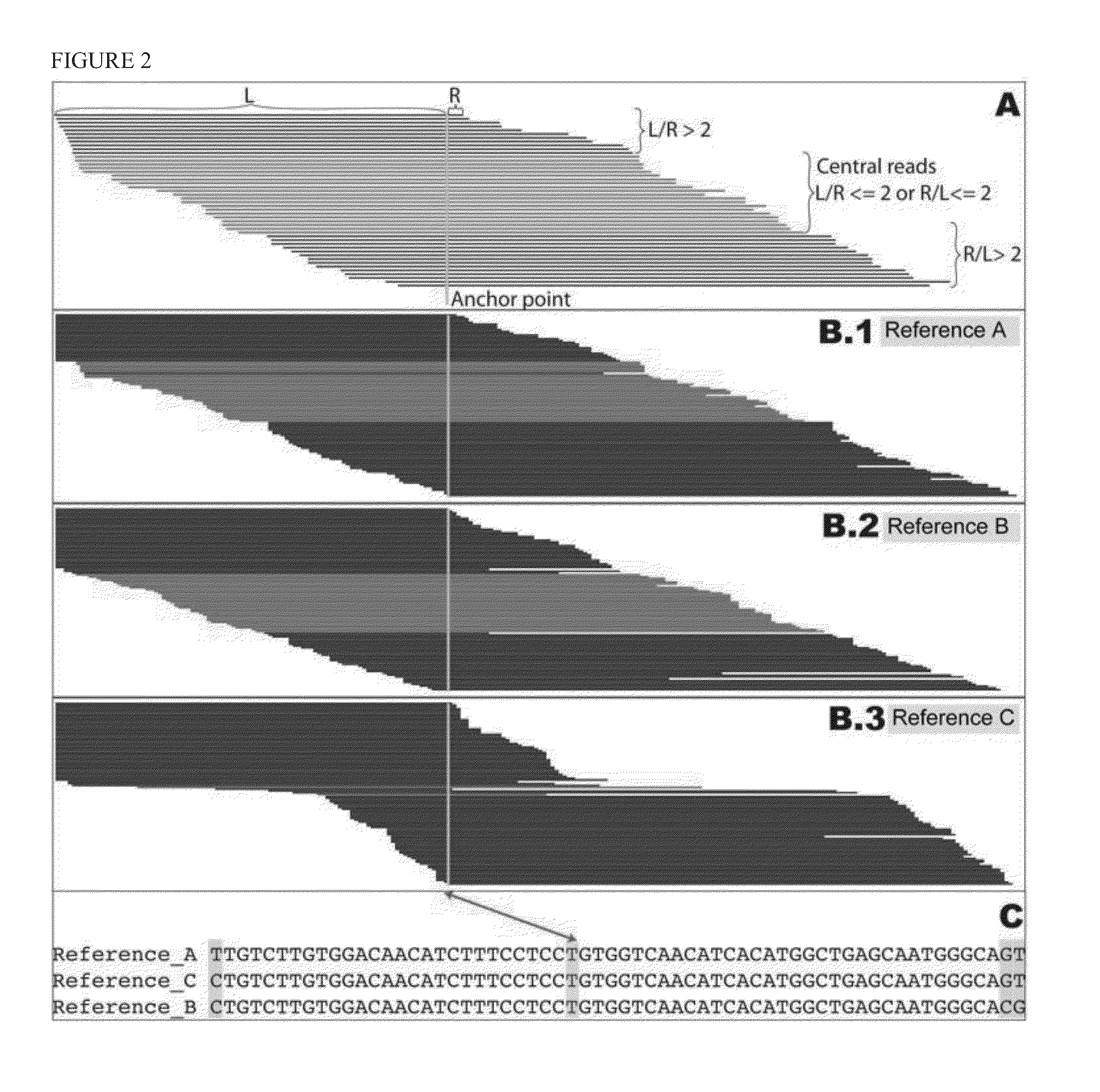
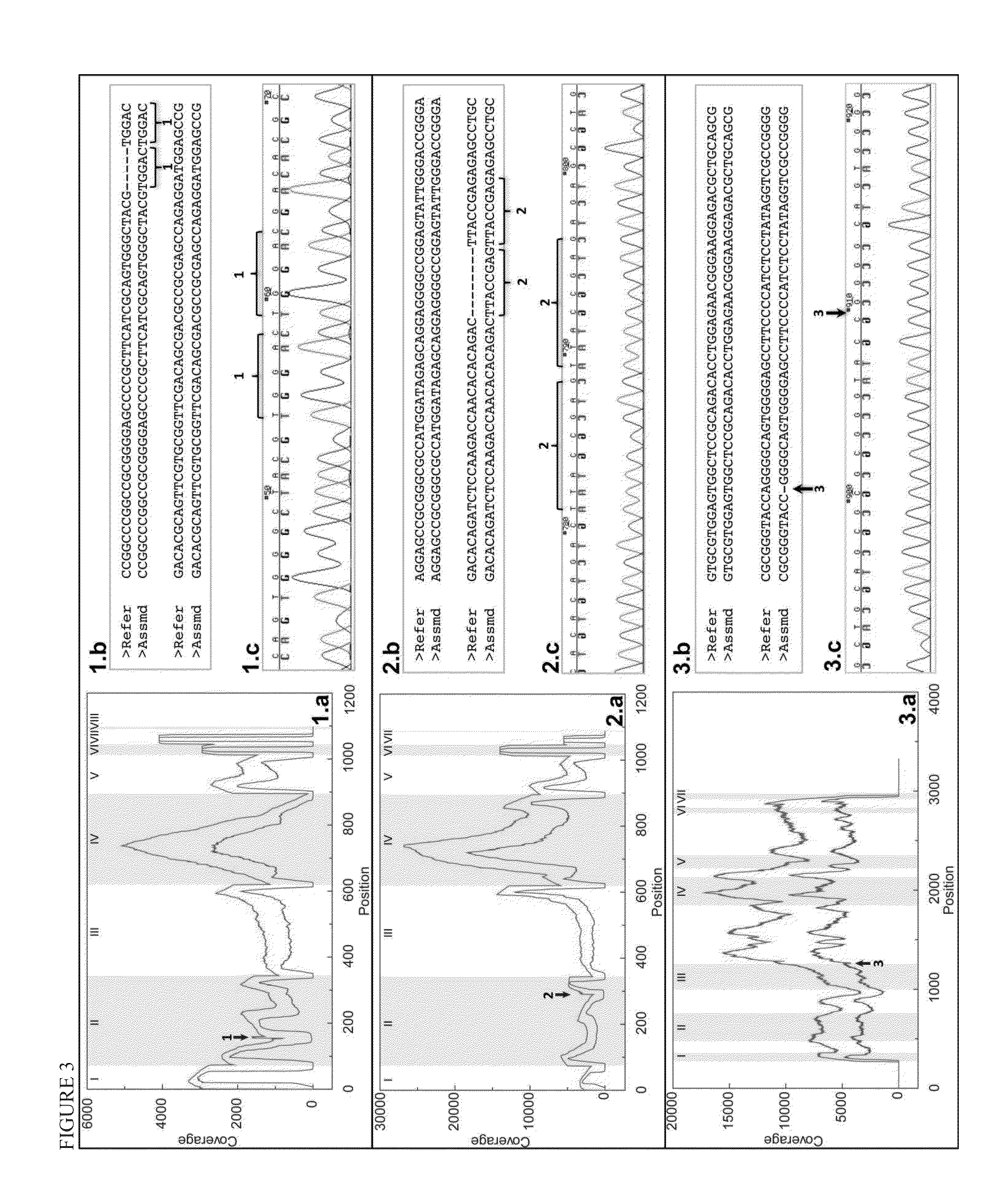
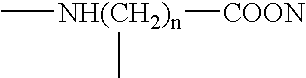Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
806 results about "Haplotype" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
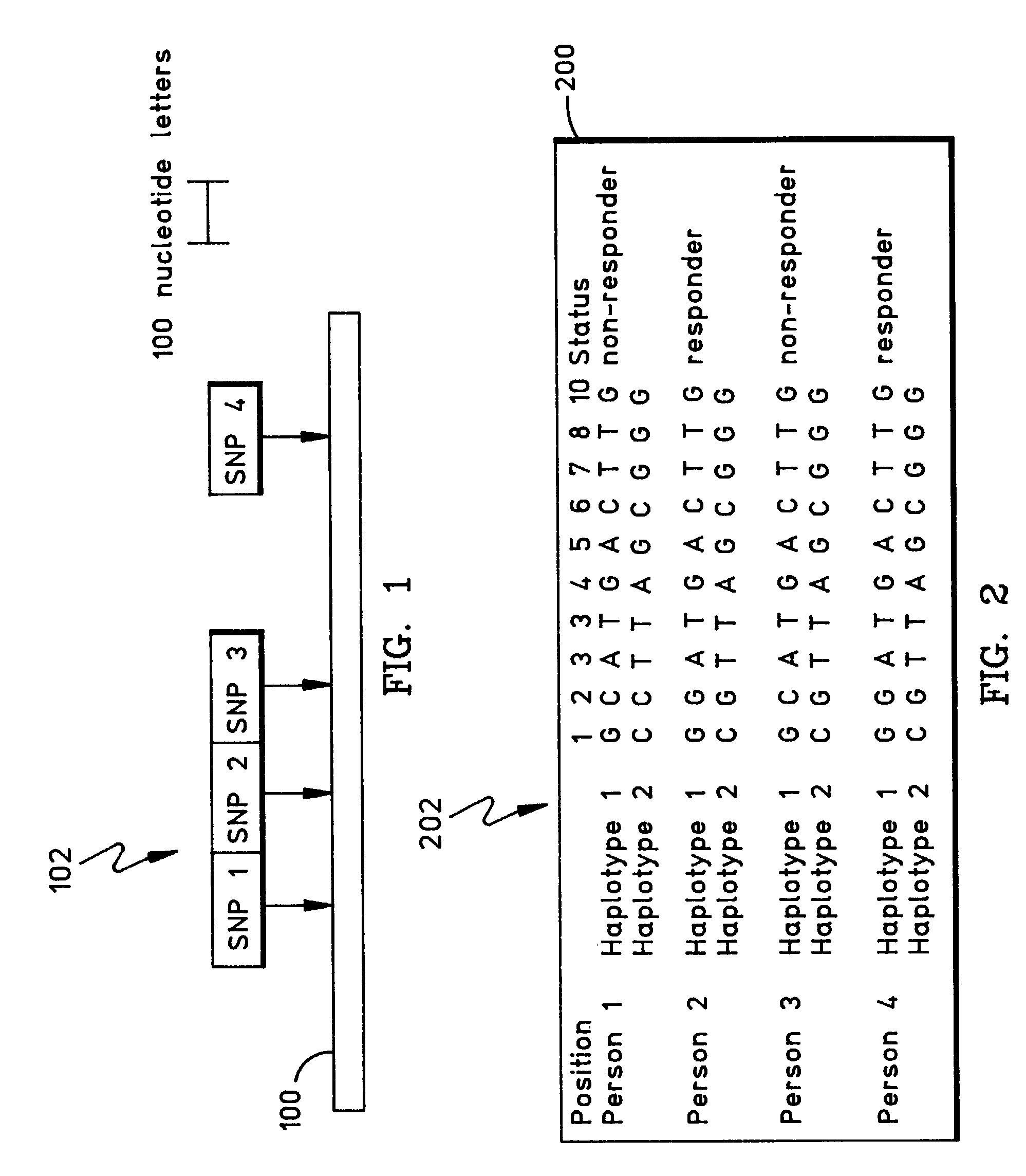
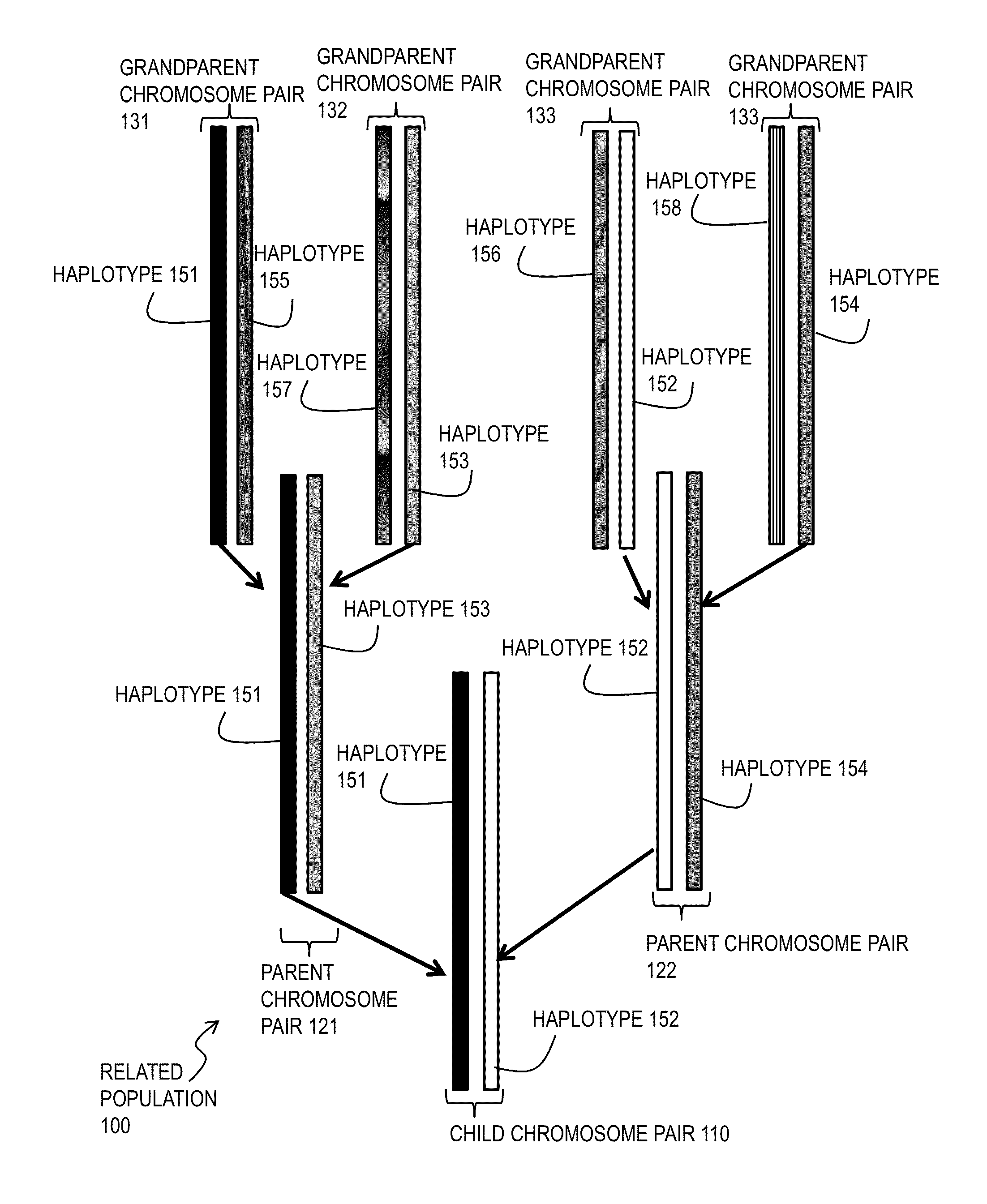
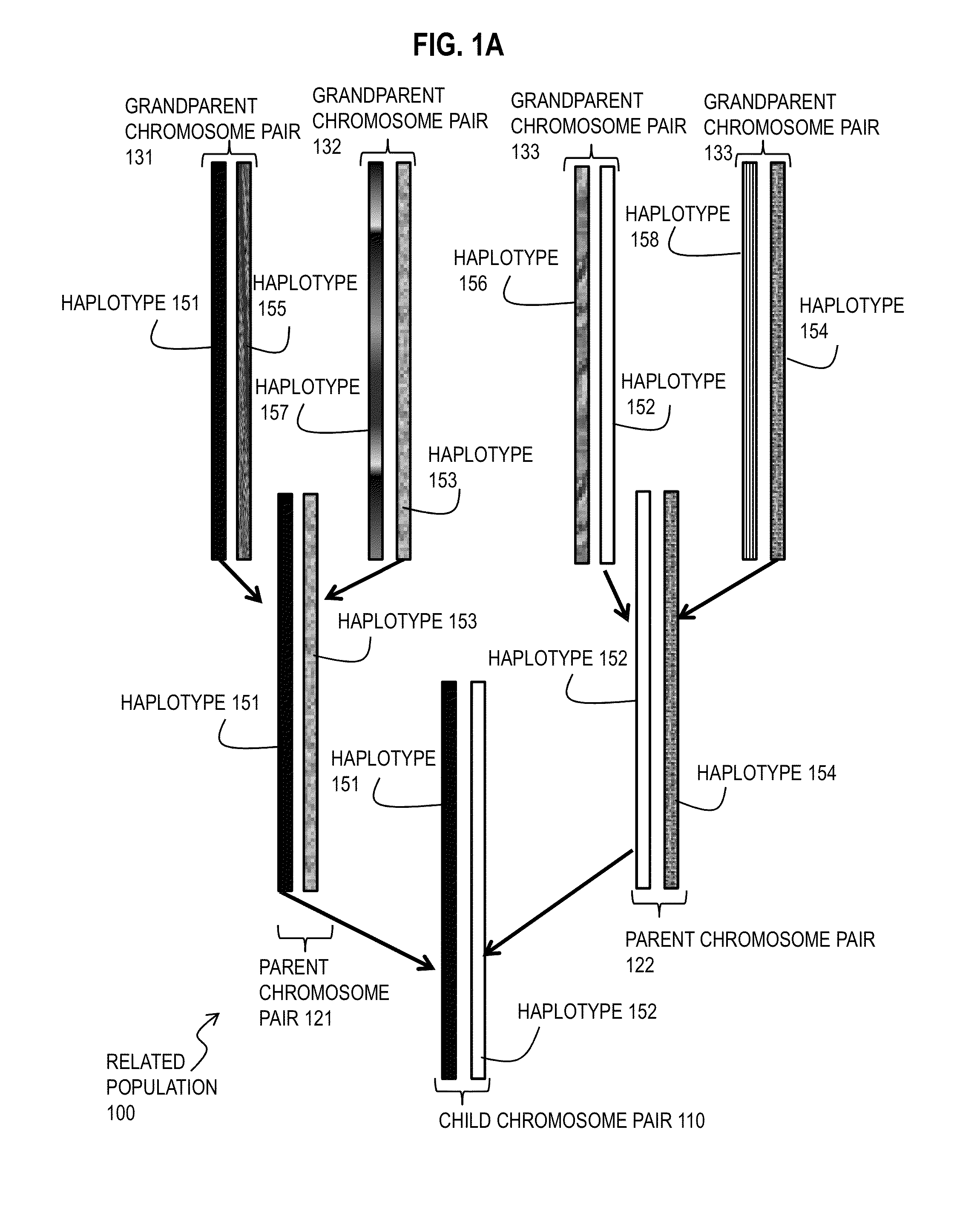
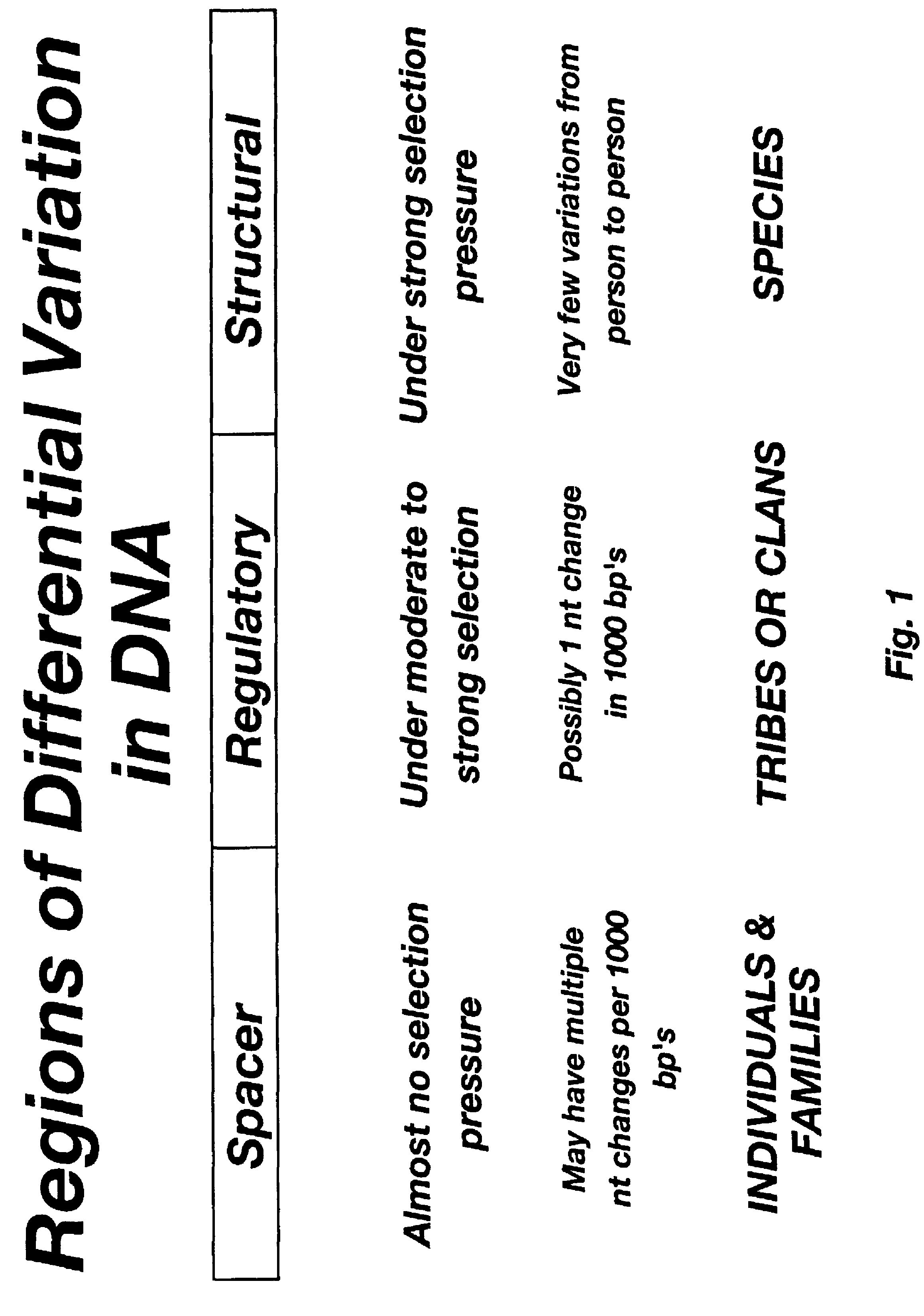
A haplotype (haploid genotype) is a group of alleles in an organism that are inherited together from a single parent. However, there are other uses of this term. First, it is used to mean a collection of specific alleles (that is, specific DNA sequences) in a cluster of tightly linked genes on a chromosome that are likely to be inherited together—that is, they are likely to be conserved as a sequence that survives the descent of many generations of reproduction. A second use is to mean a set of linked single-nucleotide polymorphism (SNP) alleles that tend to always occur together (i.e., that are associated statistically). It is thought that identifying these statistical associations and few alleles of a specific haplotype sequence can facilitate identifying all other such polymorphic sites that are nearby on the chromosome. Such information is critical for investigating the genetics of common diseases; which in fact have been investigated in humans by the International HapMap Project. Thirdly, many human genetic testing companies use the term in a third way: to refer to an individual collection of specific mutations within a given genetic segment; (see short tandem repeat mutation).
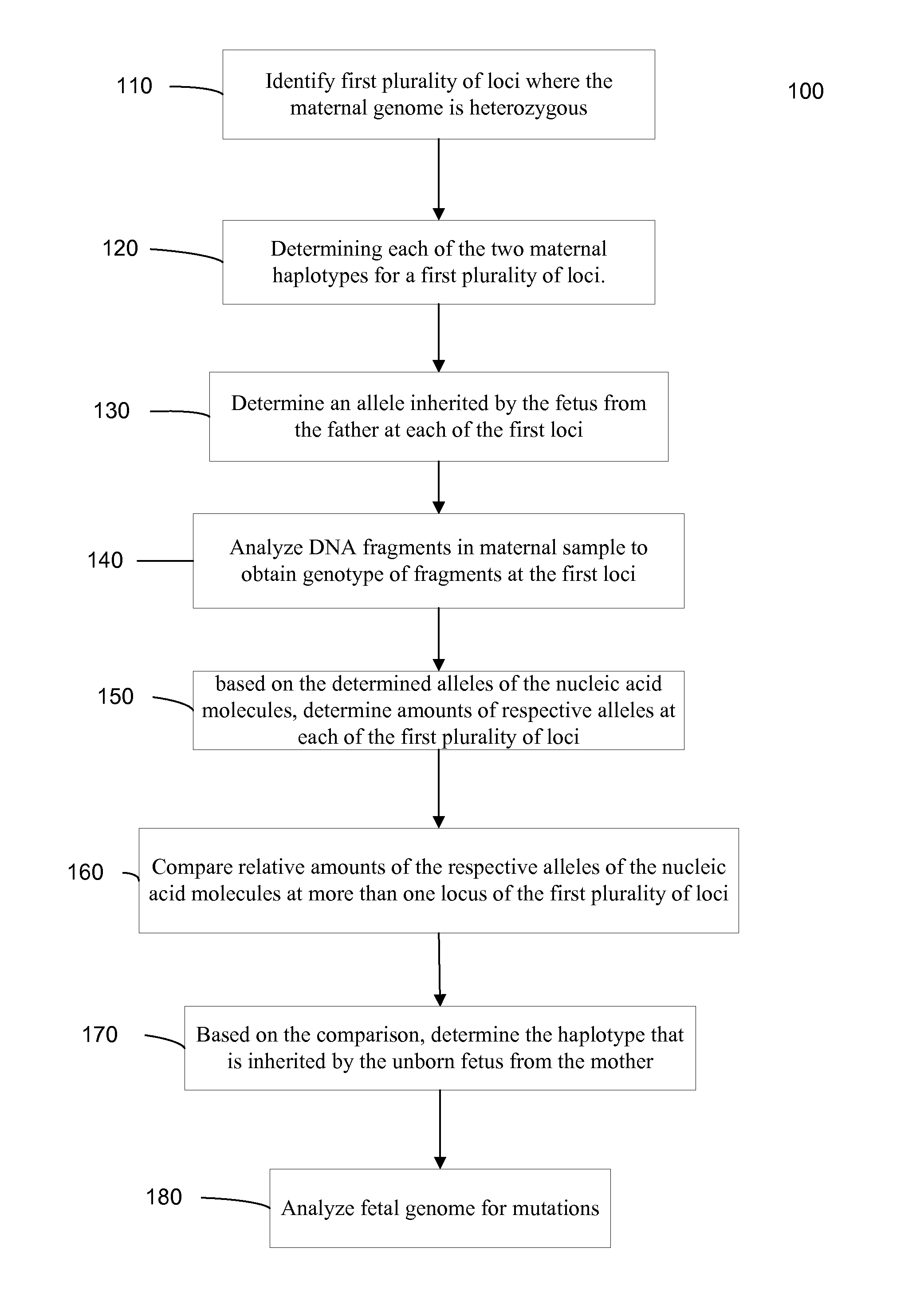
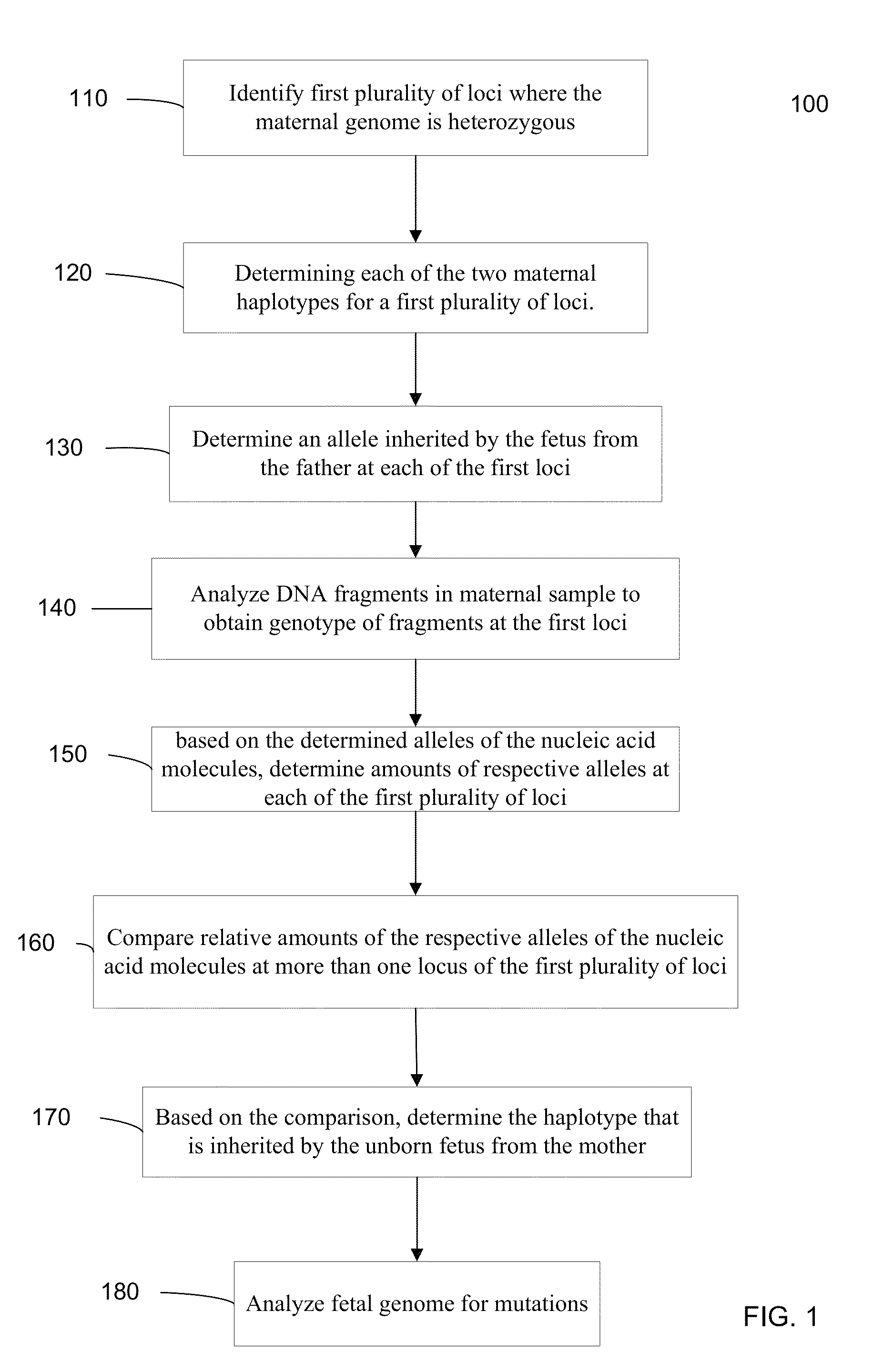
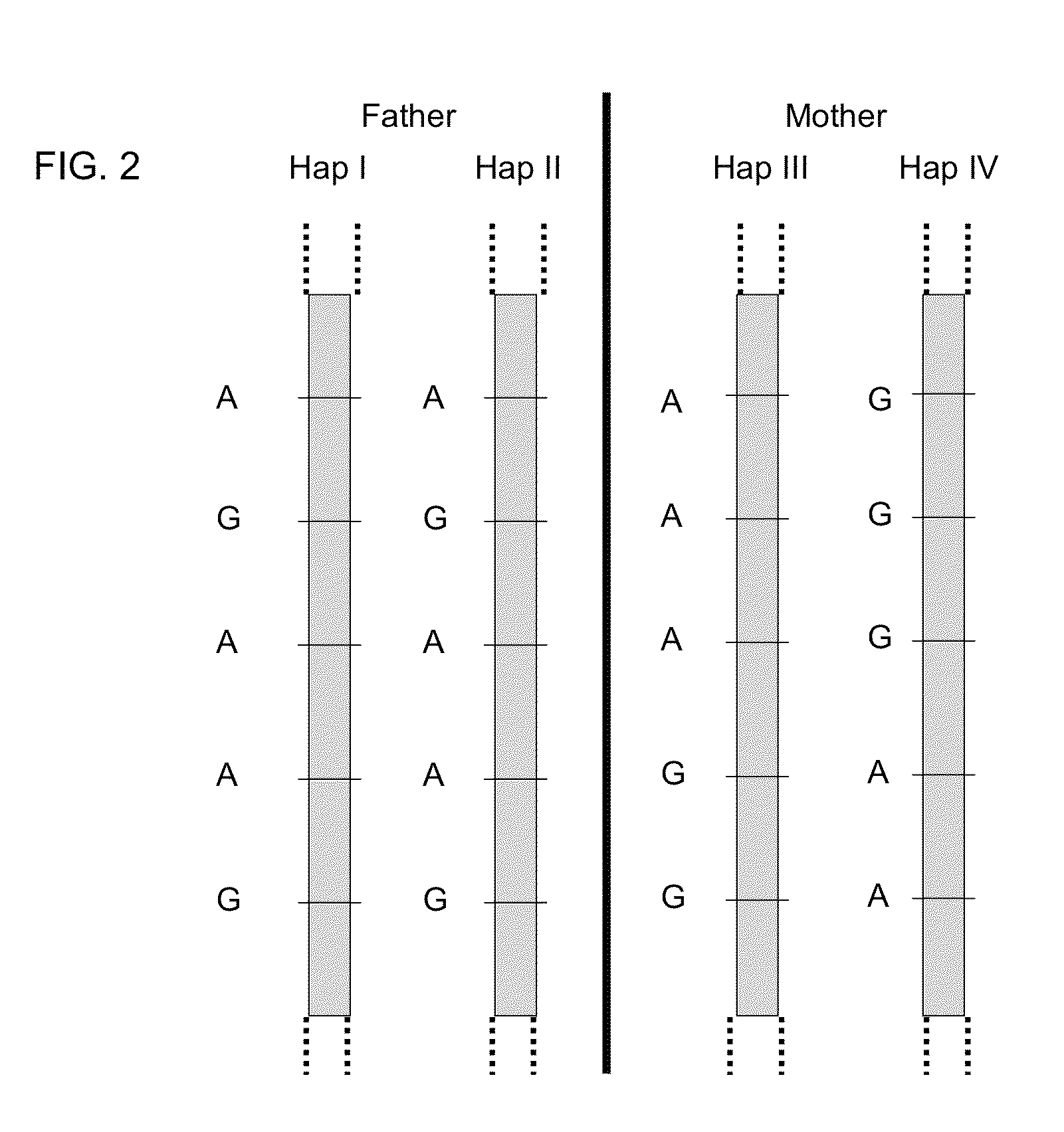
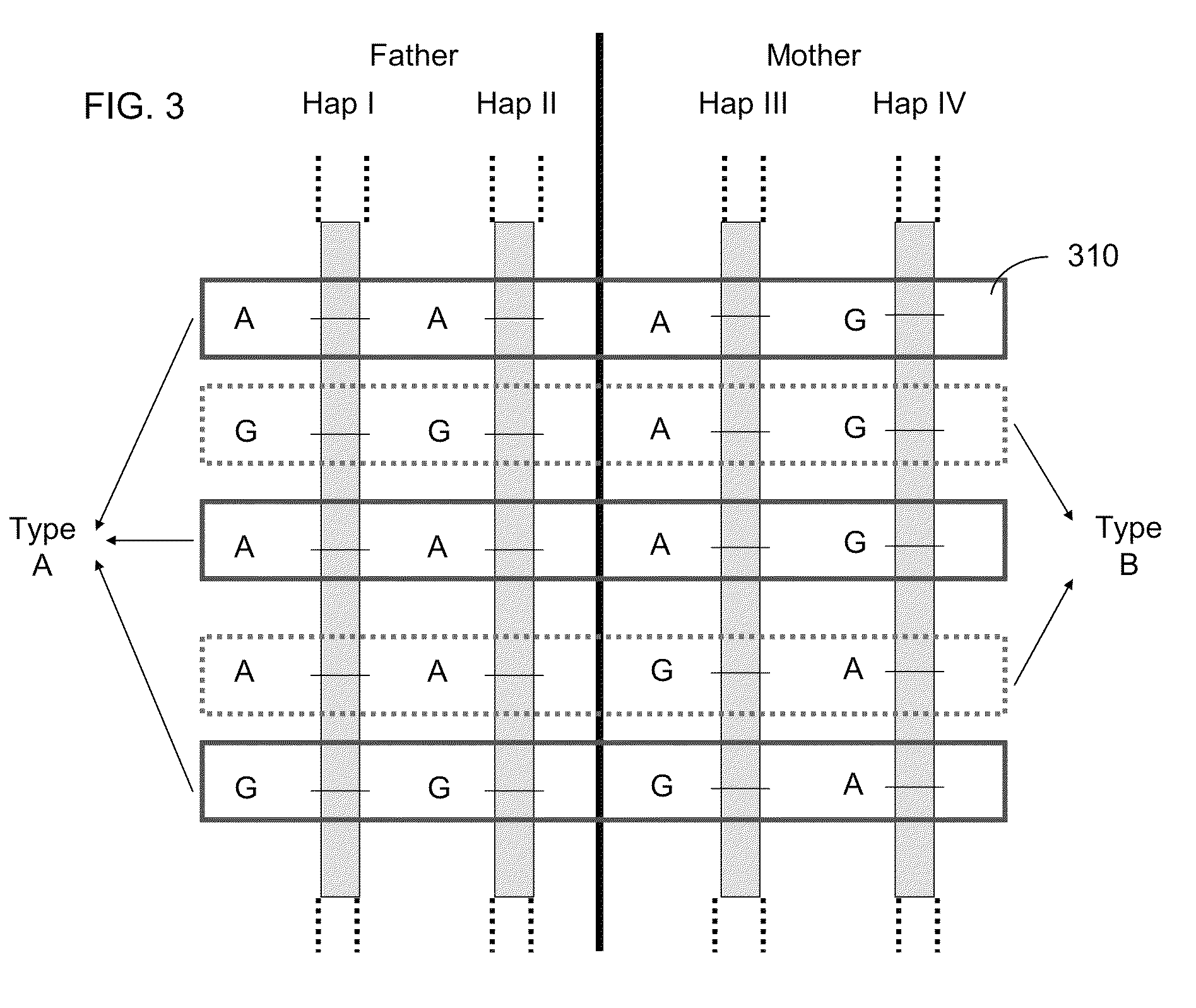
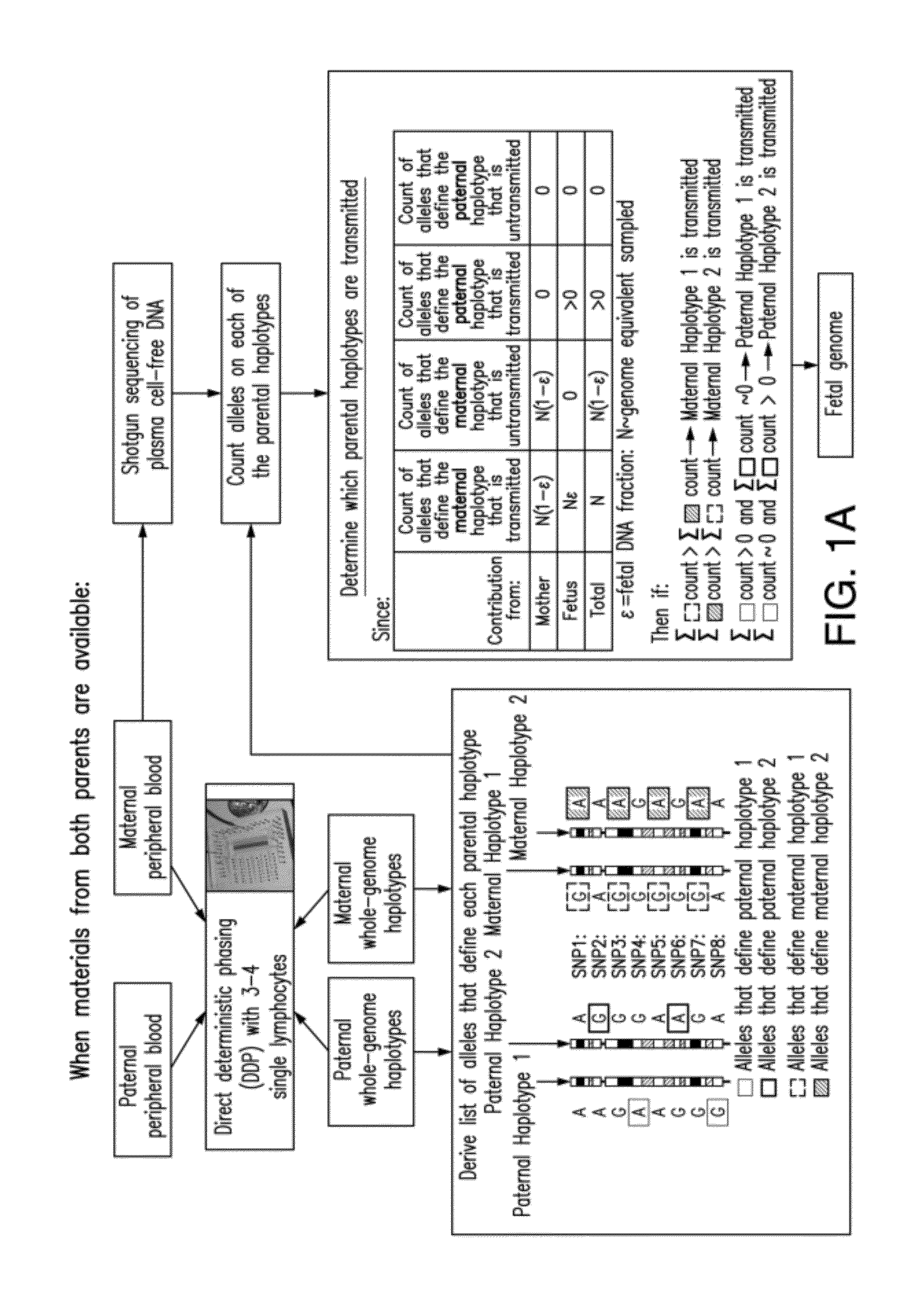
Fetal Genomic Analysis From A Maternal Biological Sample
ActiveUS20110105353A1Microbiological testing/measurementLibrary screeningDna concentrationPopulation
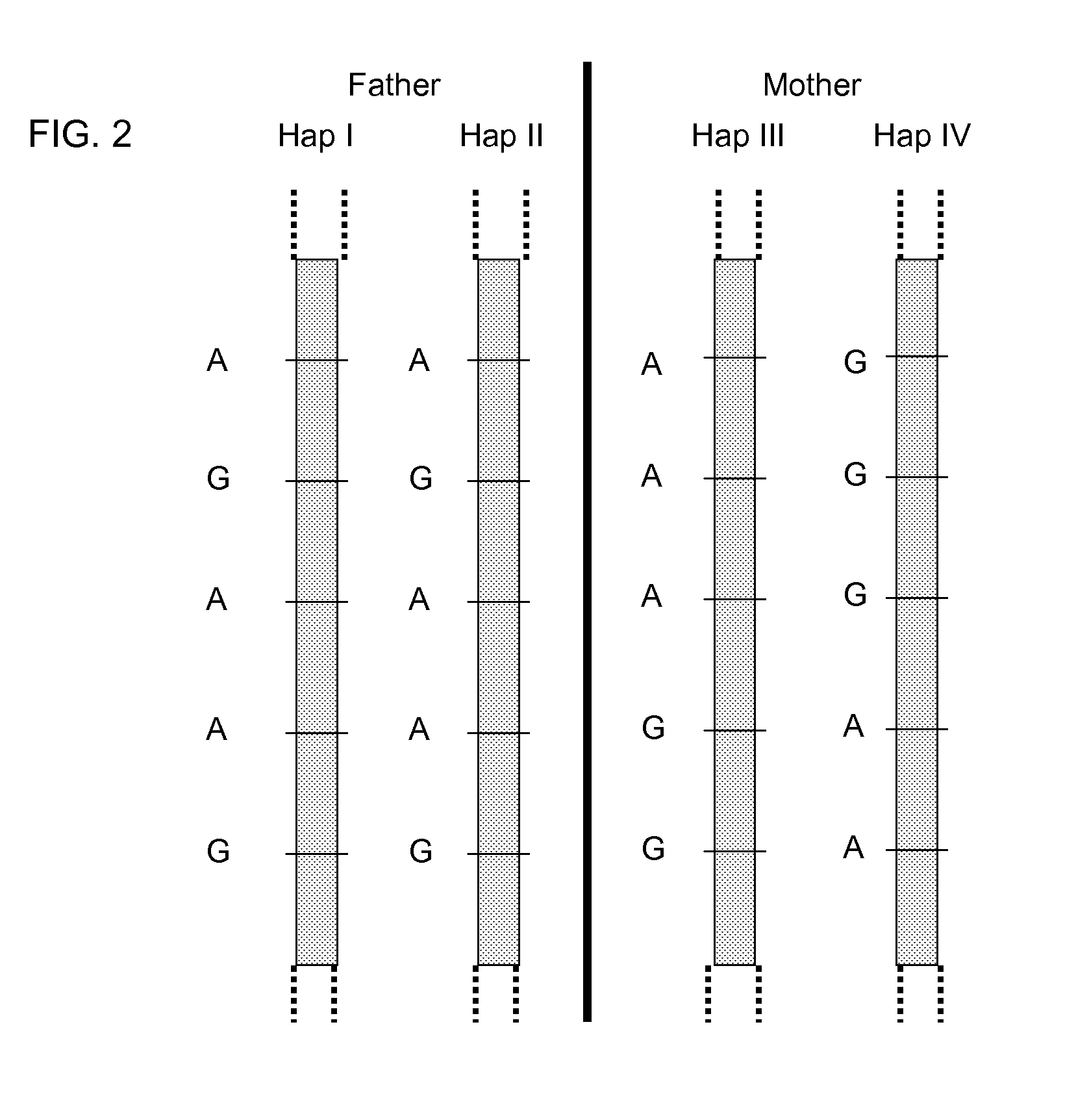
Systems, methods, and apparatus for determining at least a portion of fetal genome are provided. DNA fragments from a maternal sample (maternal and fetal DNA) can be analyzed to identify alleles at certain loci. The amounts of DNA fragments of the respective alleles at these loci can be analyzed together to determine relative amounts of the haplotypes for these loci and determine which haplotypes have been inherited from the parental genomes. Loci where the parents are a specific combination of homozygous and heterozygous can be analyzed to determine regions of the fetal genome. Reference haplotypes common in the population can be used along with the analysis of the DNA fragments of the maternal sample to determine the maternal and paternal genomes. Determination of mutations, a fractional fetal DNA concentration in a maternal sample, and a proportion of coverage of a sequencing of the maternal sample can also be provided.
Owner:SEQUENOM INC +1
Haplotype analysis
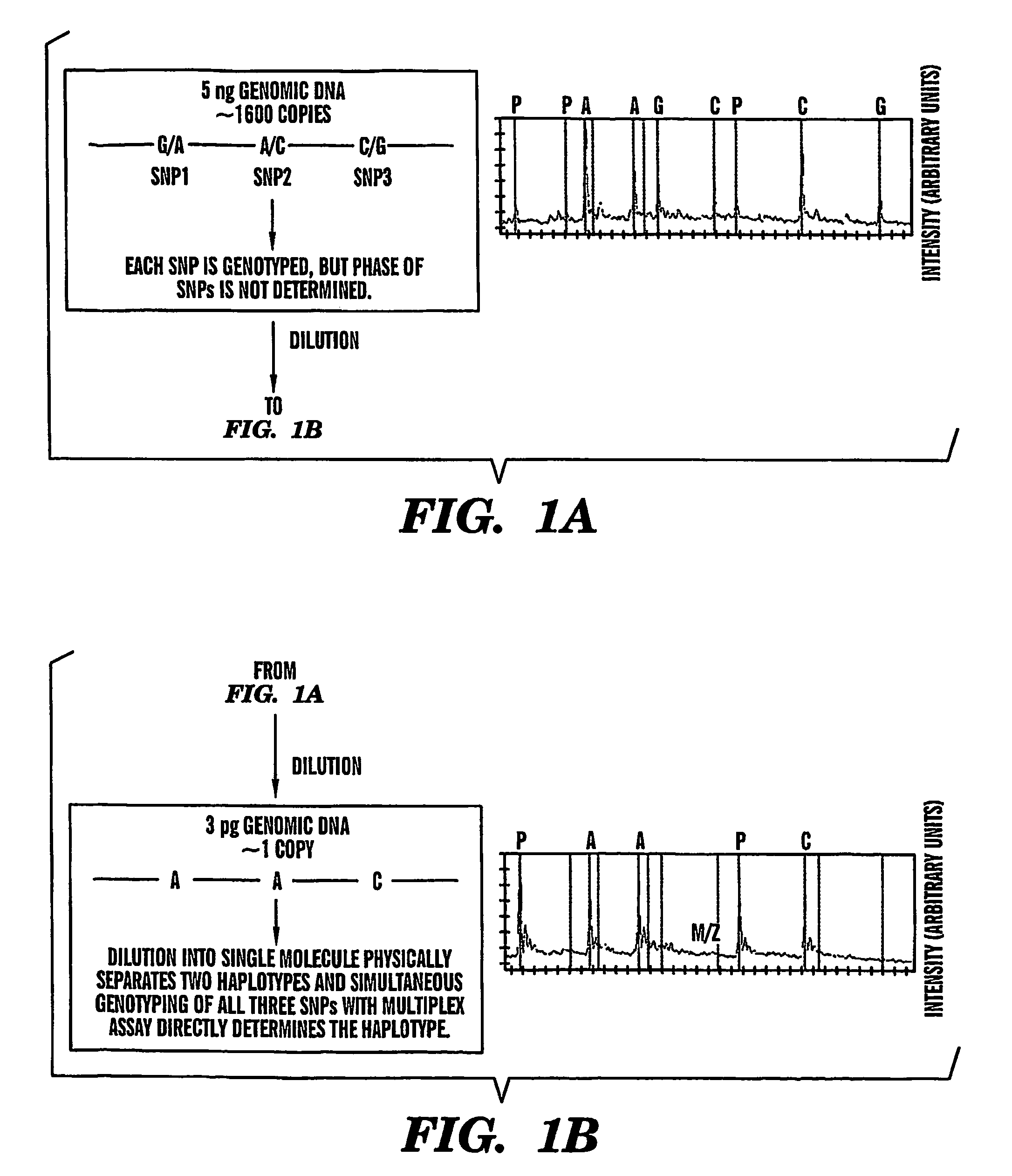
InactiveUS7700325B2High analysisReliable determinationSugar derivativesMicrobiological testing/measurementStatistical analysisHaplotype
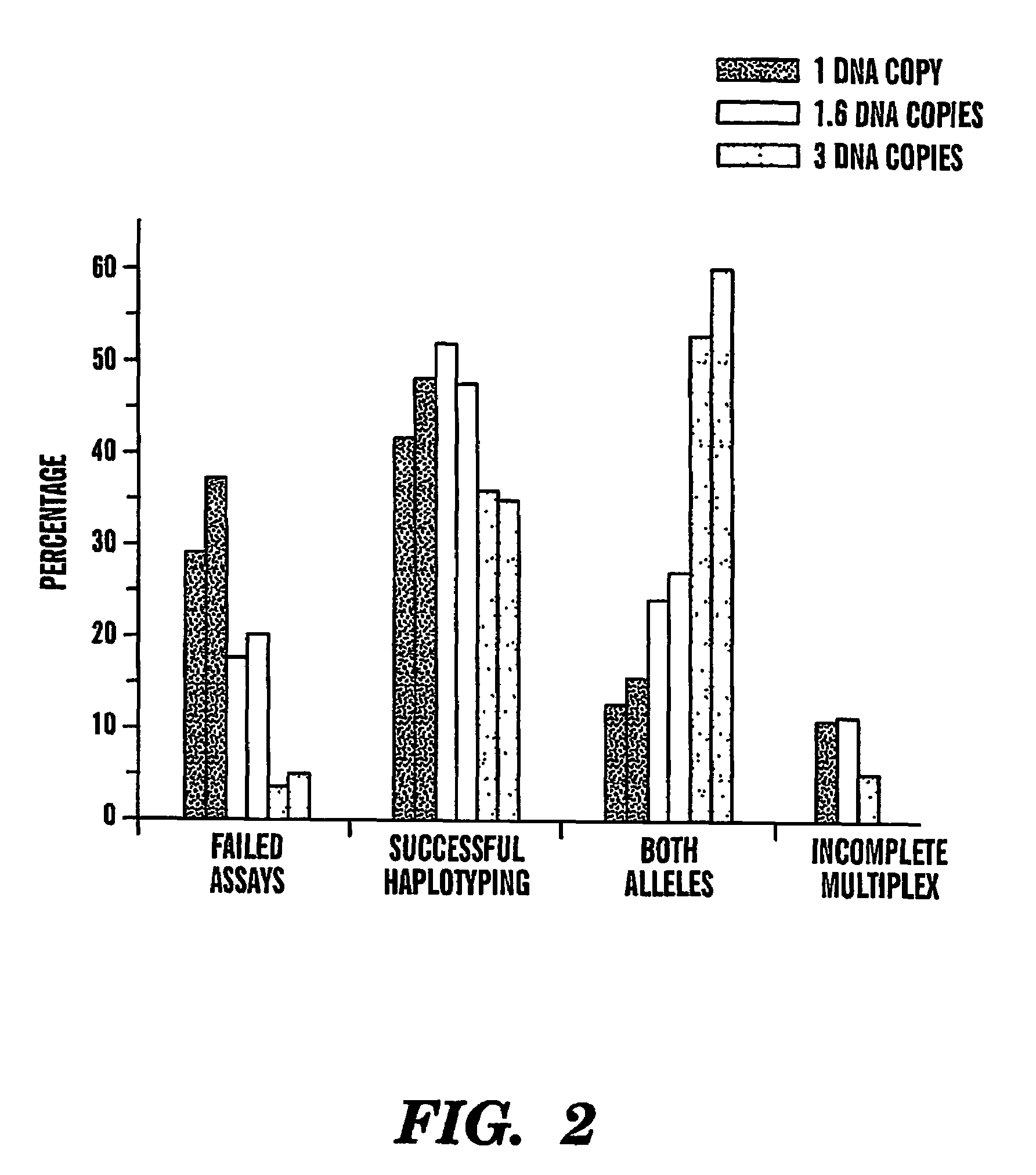
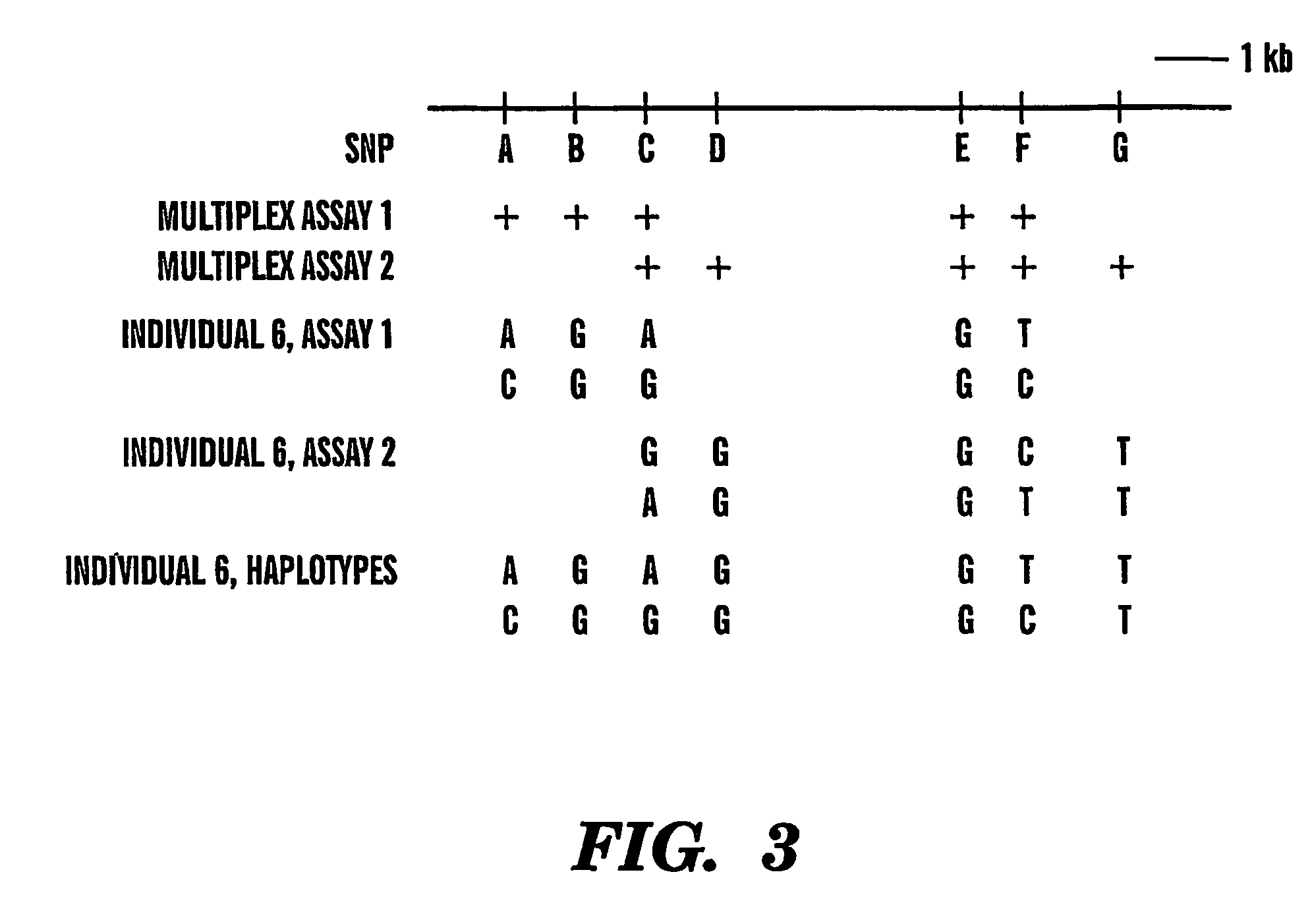
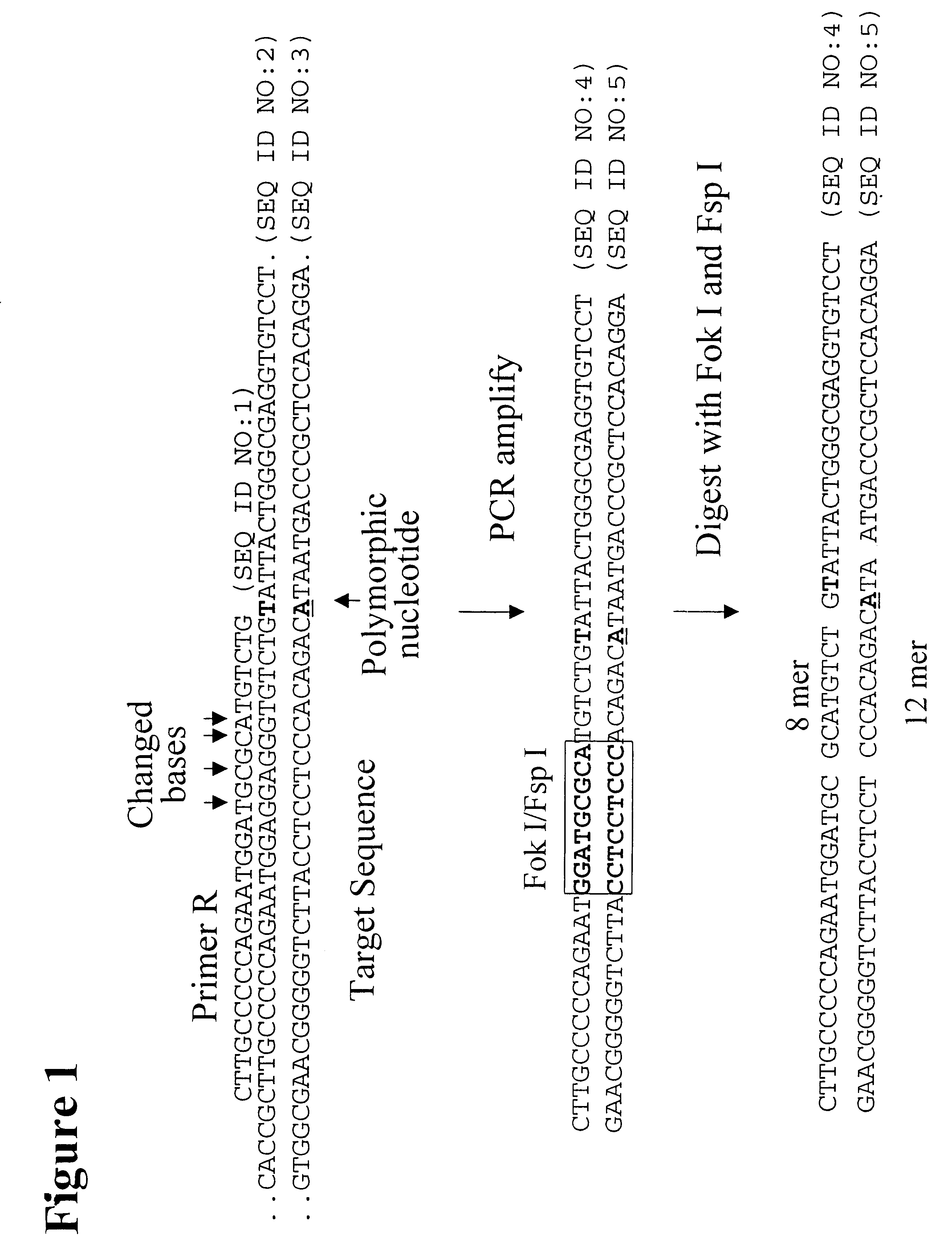
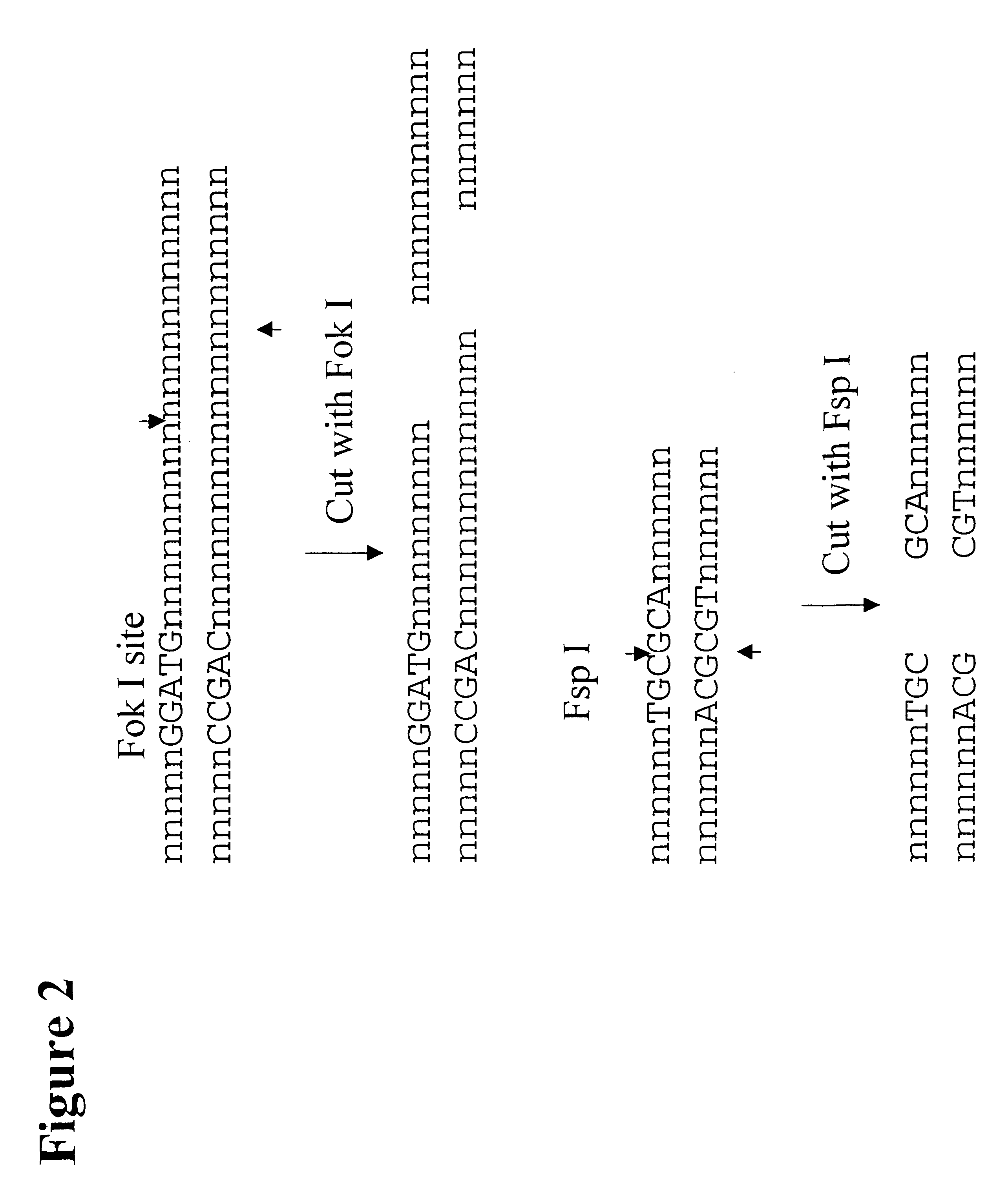
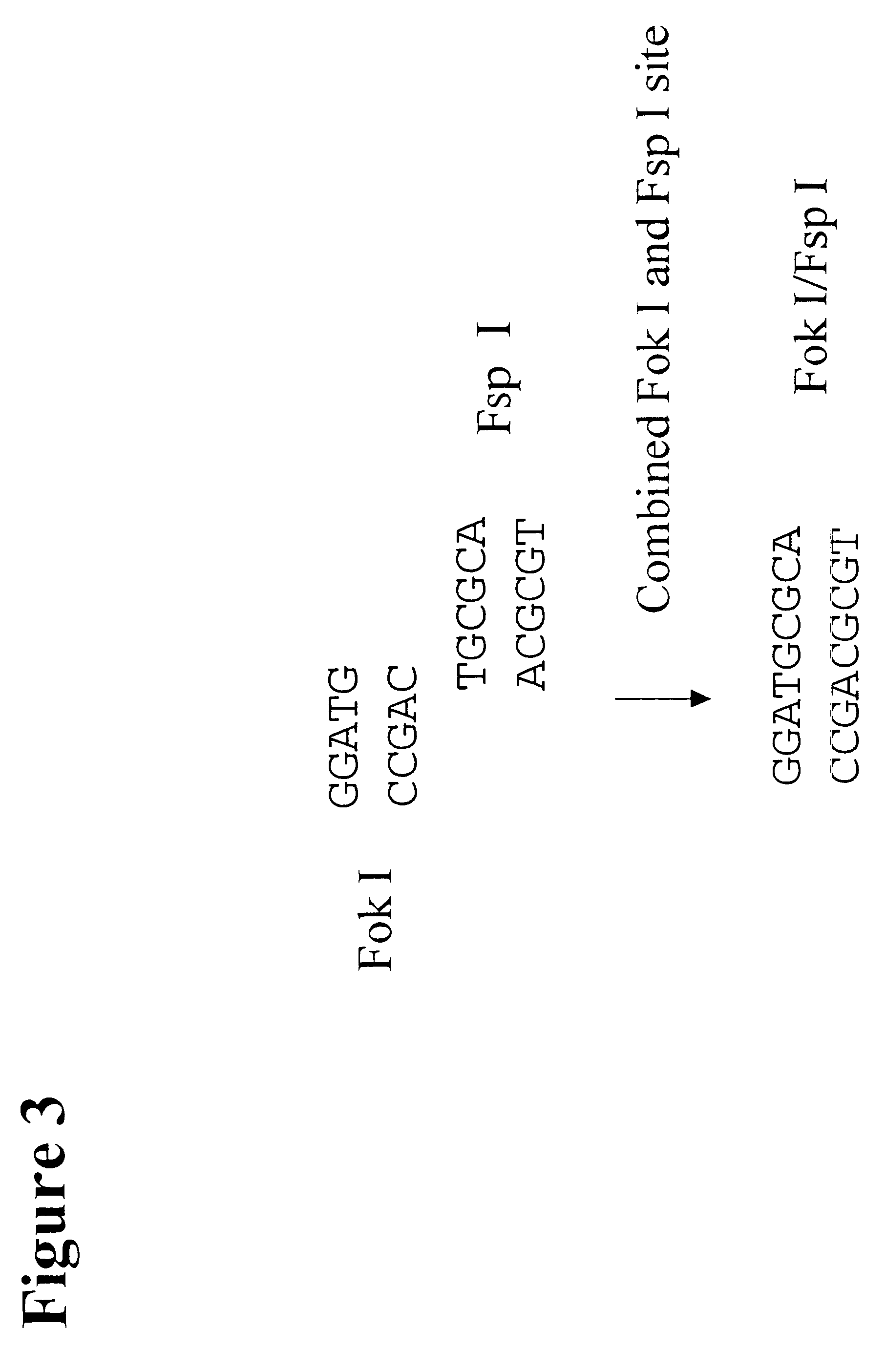
The present invention provides an efficient way for high throughput haplotype analysis. Several polymorphic nucleic acid markers, such as SNPs, can be simultaneously and reliably determined through multiplex PCR of single nucleic acid molecules in several parallel single molecule dilutions and the consequent statistical analysis of the results from these parallel single molecule multiplex PCR reactions results in reliable determination of haplotypes present in the subject. The nucleic acid markers can be of any distance to each other on the chromosome. In addition, an approach wherein overlapping DNA markers are analyzed can be used to link smaller haplotypes into larger haplotypes. Consequently, the invention provides a powerful new tool for diagnostic haplotyping and identifying novel haplotypes.
Owner:TRUSTEES OF BOSTON UNIV
Massively parallel contiguity mapping
ActiveUS20130203605A1Reducing sequenceLimited to sequenceLibrary member identificationChemical recyclingHuman DNA sequencingMammalian genome
Contiguity information is important to achieving high-quality de novo assembly of mammalian genomes and the haplotype-resolved resequencing of human genomes. The methods described herein pursue cost-effective, massively parallel capture of contiguity information at different scales.
Owner:UNIV OF WASHINGTON CENT FOR COMMERICIALIZATION
Fetal genomic analysis from a maternal biological sample
Systems, methods, and apparatus for determining at least a portion of fetal genome are provided. DNA fragments from a maternal sample (maternal and fetal DNA) can be analyzed to identify alleles at certain loci. The amounts of DNA fragments of the respective alleles at these loci can be analyzed together to determine relative amounts of the haplotypes for these loci and determine which haplotypes have been inherited from the parental genomes. Loci where the parents are a specific combination of homozygous and heterozygous can be analyzed to determine regions of the fetal genome. Reference haplotypes common in the population can be used along with the analysis of the DNA fragments of the maternal sample to determine the maternal and paternal genomes. Determination of mutations, a fractional fetal DNA concentration in a maternal sample, and a proportion of coverage of a sequencing of the maternal sample can also be provided.
Owner:SEQUENOM INC +1
Methods for genetic analysis of DNA using biased amplification of polymorphic sites
InactiveUS6475736B1Efficient amplificationEasily and robustly analyzedSugar derivativesMicrobiological testing/measurementHaplotypeDNA
Methods for determining genotypes and haplotypes of genes are described. Also described are single nucleotide polymorphisms and haplotypes in the ApoE gene and methods of using that information.
Owner:AGENA BIOSCI
Long-Range Barcode Labeling-Sequencing
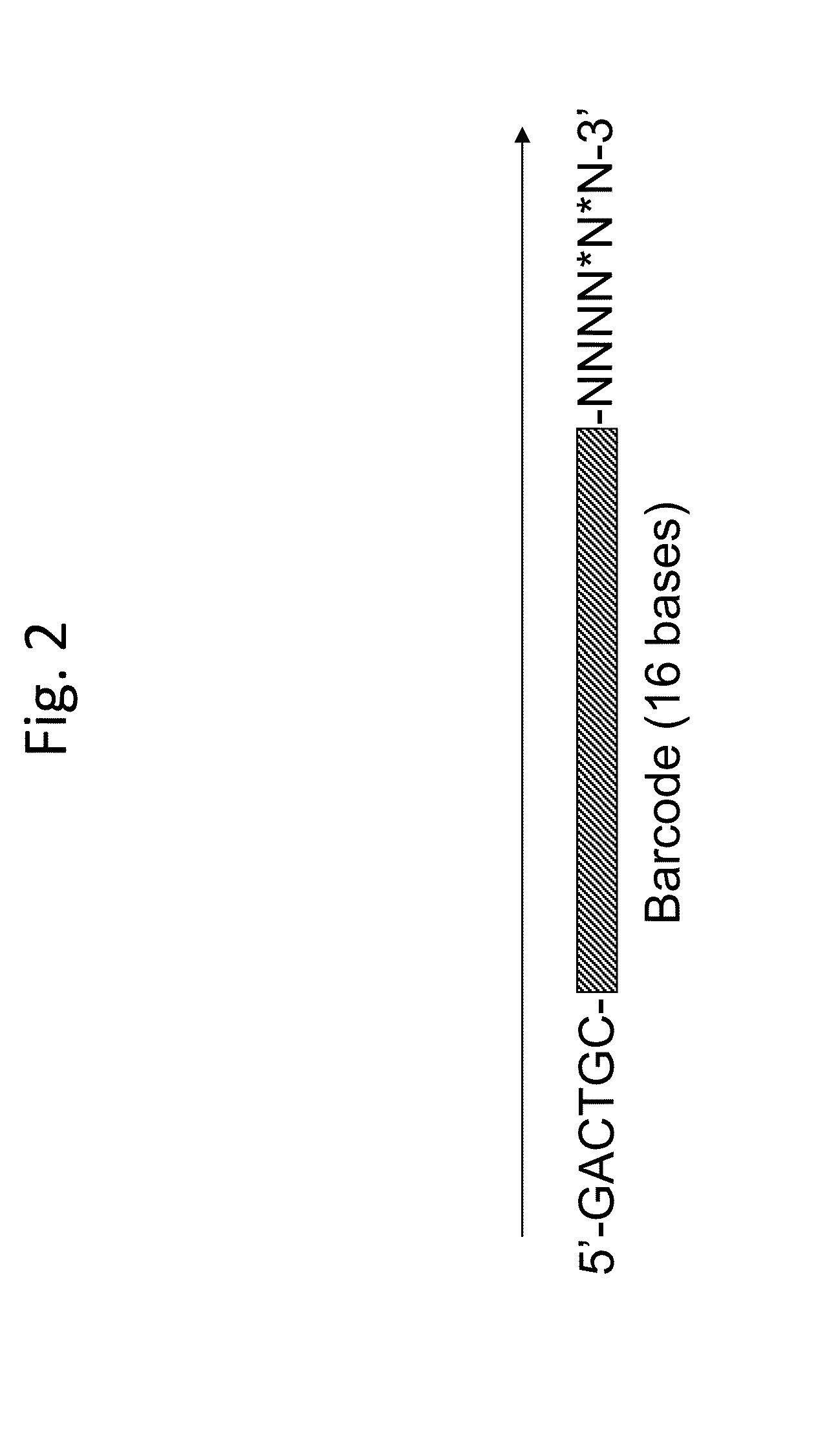
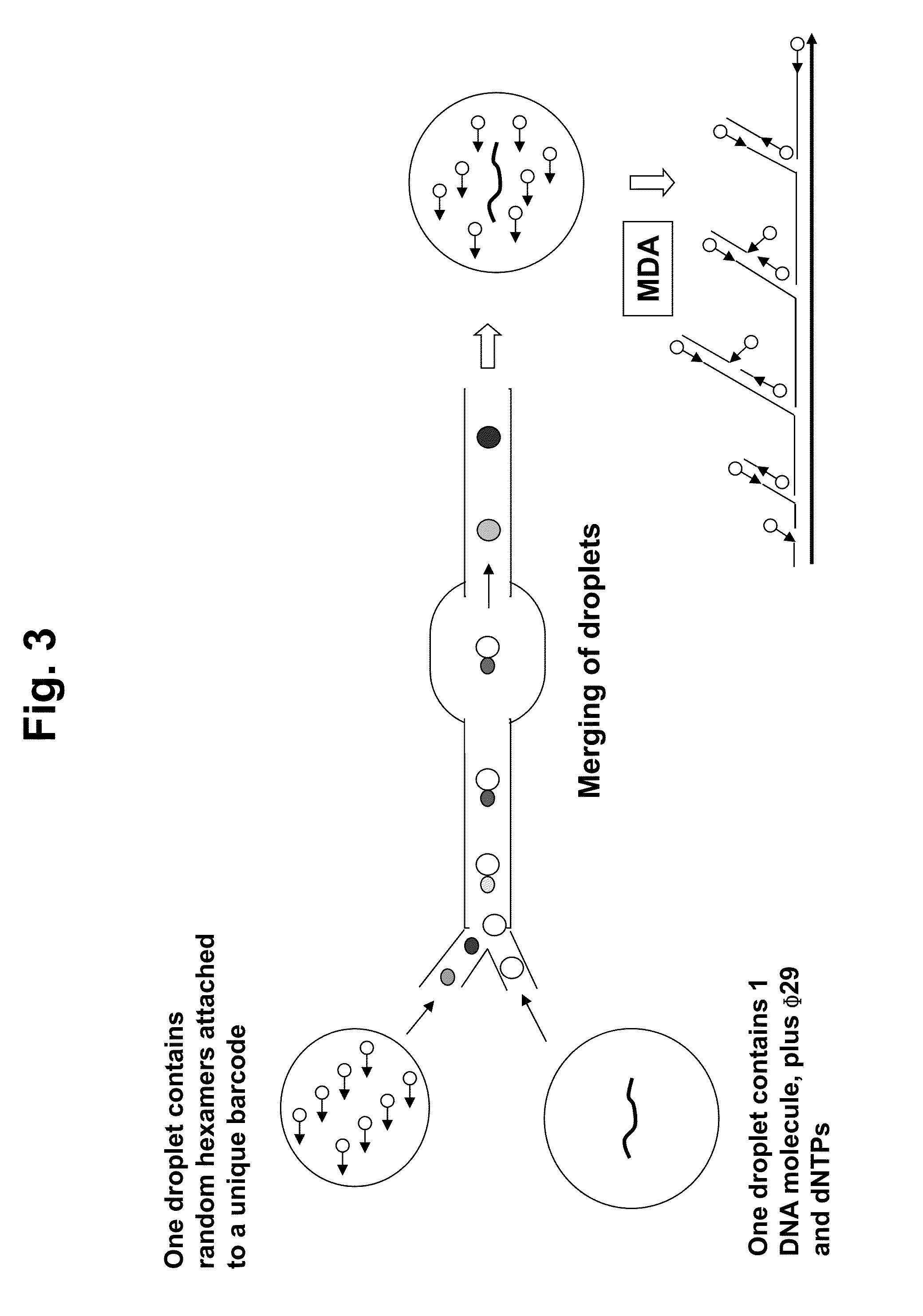
ActiveUS20130130919A1Reduce complexityImprove efficiencyMicrobiological testing/measurementLibrary member identificationBarcodeHaplotype
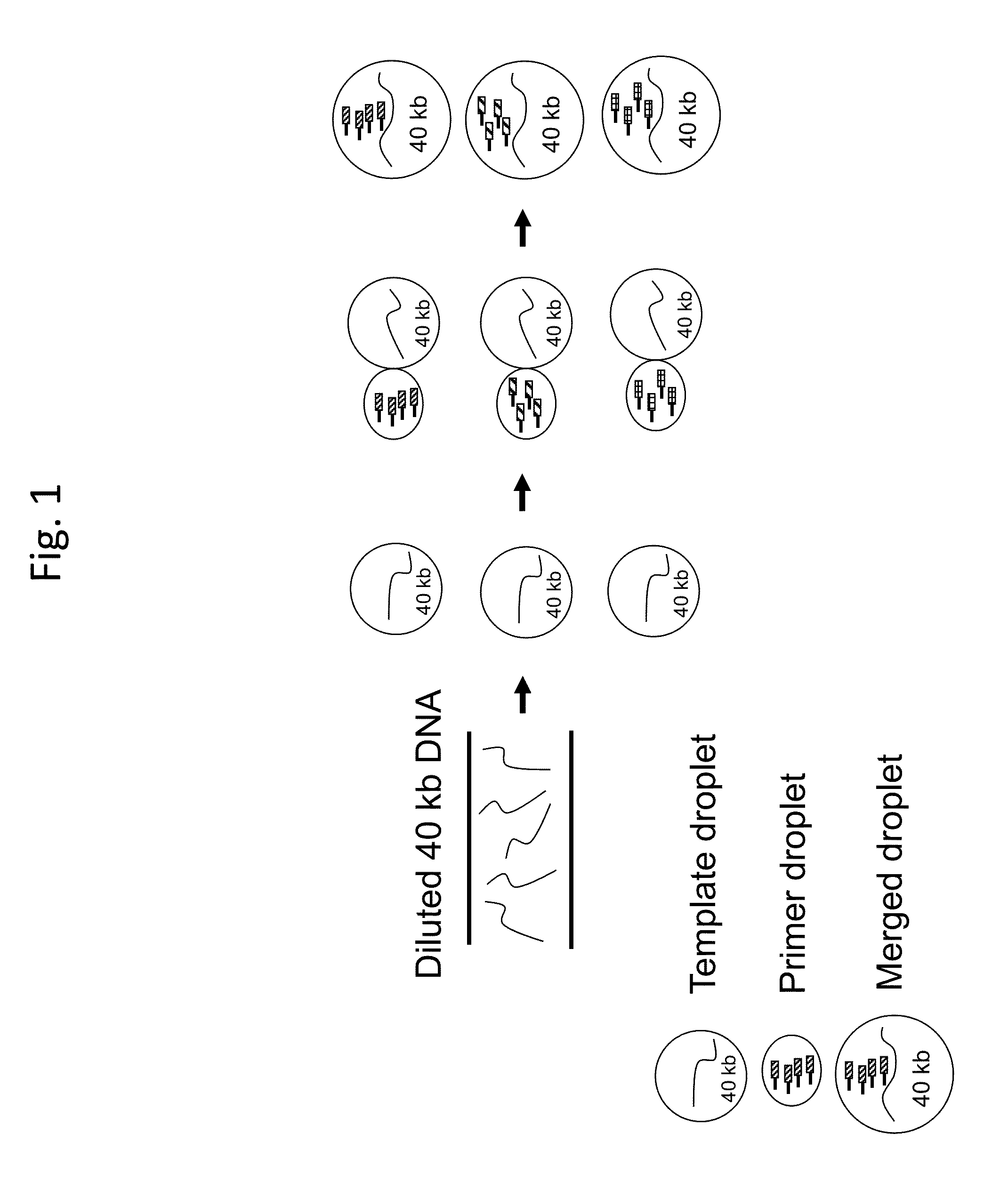
Methods for sequencing single large DNA molecules by clonal multiple displacement amplification using barcoded primers. Sequences are binned based on barcode sequences and sequenced using a microdroplet-based method for sequencing large polynucleotide templates to enable assembly of haplotype-resolved complex genomes and metagenomes.
Owner:RGT UNIV OF CALIFORNIA
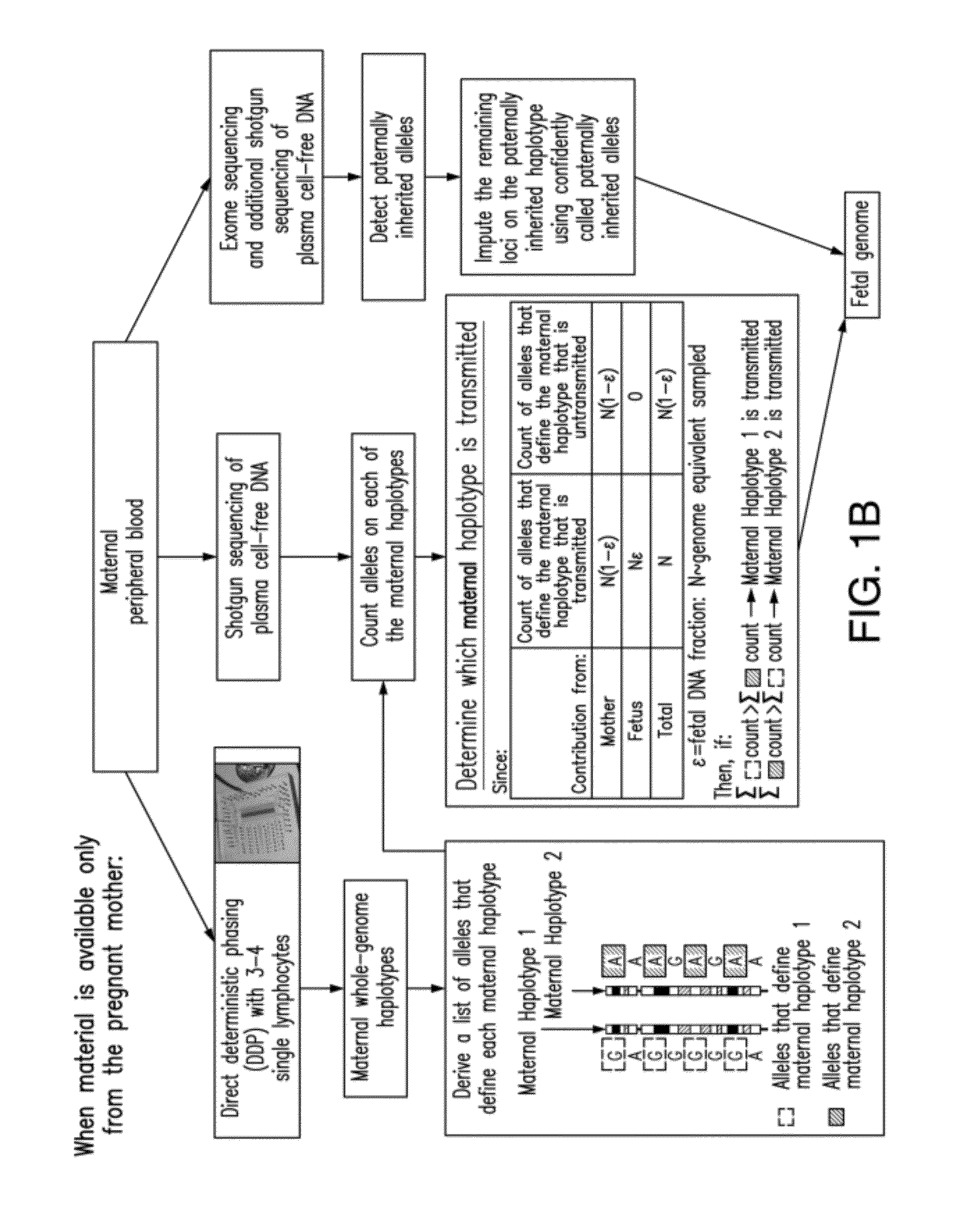
Non-invasive determination of fetal inheritance of parental haplotypes at the genome-wide scale
ActiveUS20120196754A1Reduce riskNon-invasively determiningBioreactor/fermenter combinationsBiological substance pretreatmentsSequence analysisSerum samples
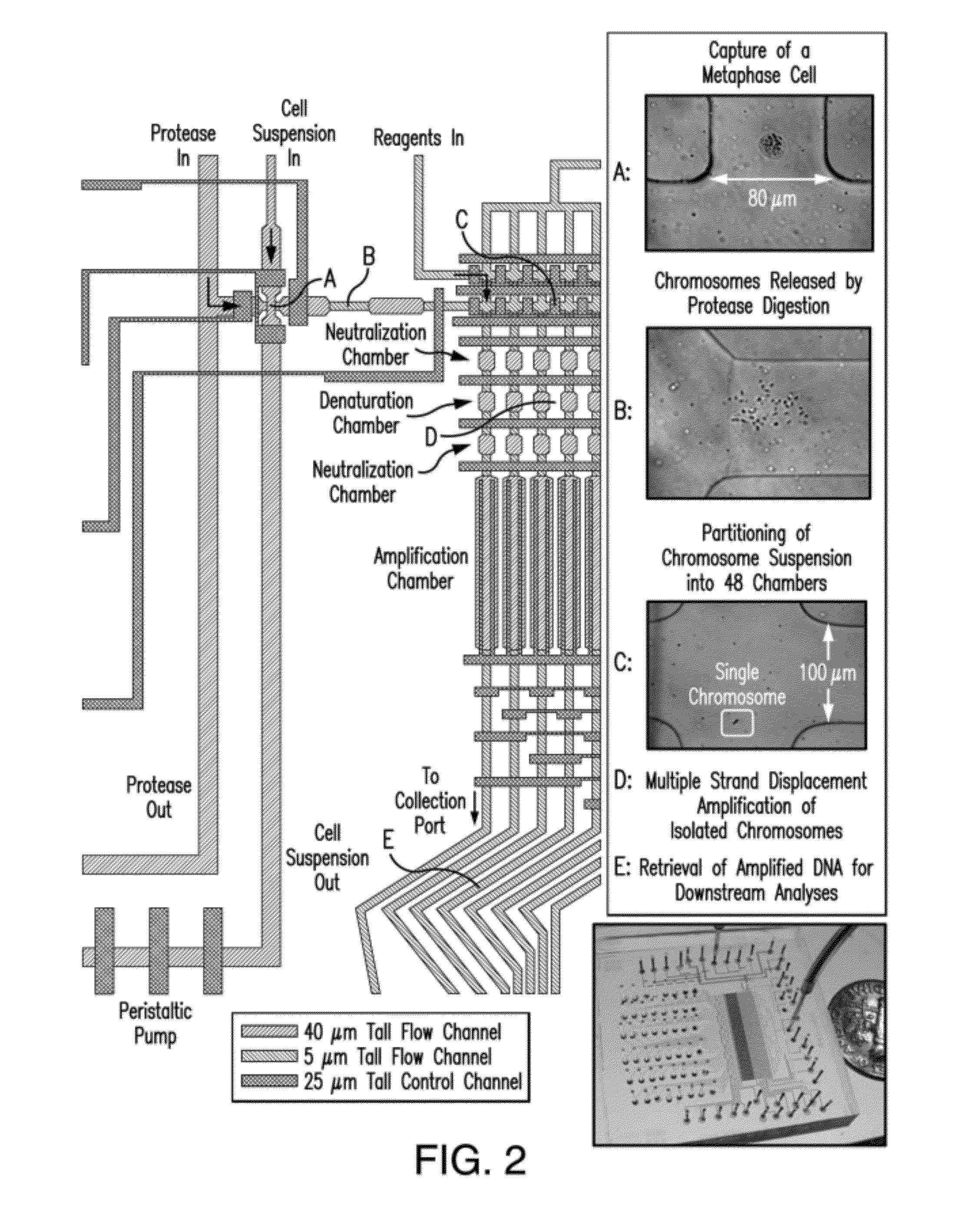
The present invention provides a method, device and a computer program for haplotyping single cells, such that a sample taken from a pregnant female, without directly sampling the fetus, provides the ability to non-invasively determine the fetal genome. The method can be performed by determining the parental and inherited haplotypes, or can be performed merely on the basis of the mother's genetic information, obtained preferably in a blood or serum sample. The novel device allows for sequence analysis of single chromosomes from a single cell, preferably by partitioning single chromosomes from a metaphase cell into long, thin channels where a sequence analysis can be performed.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
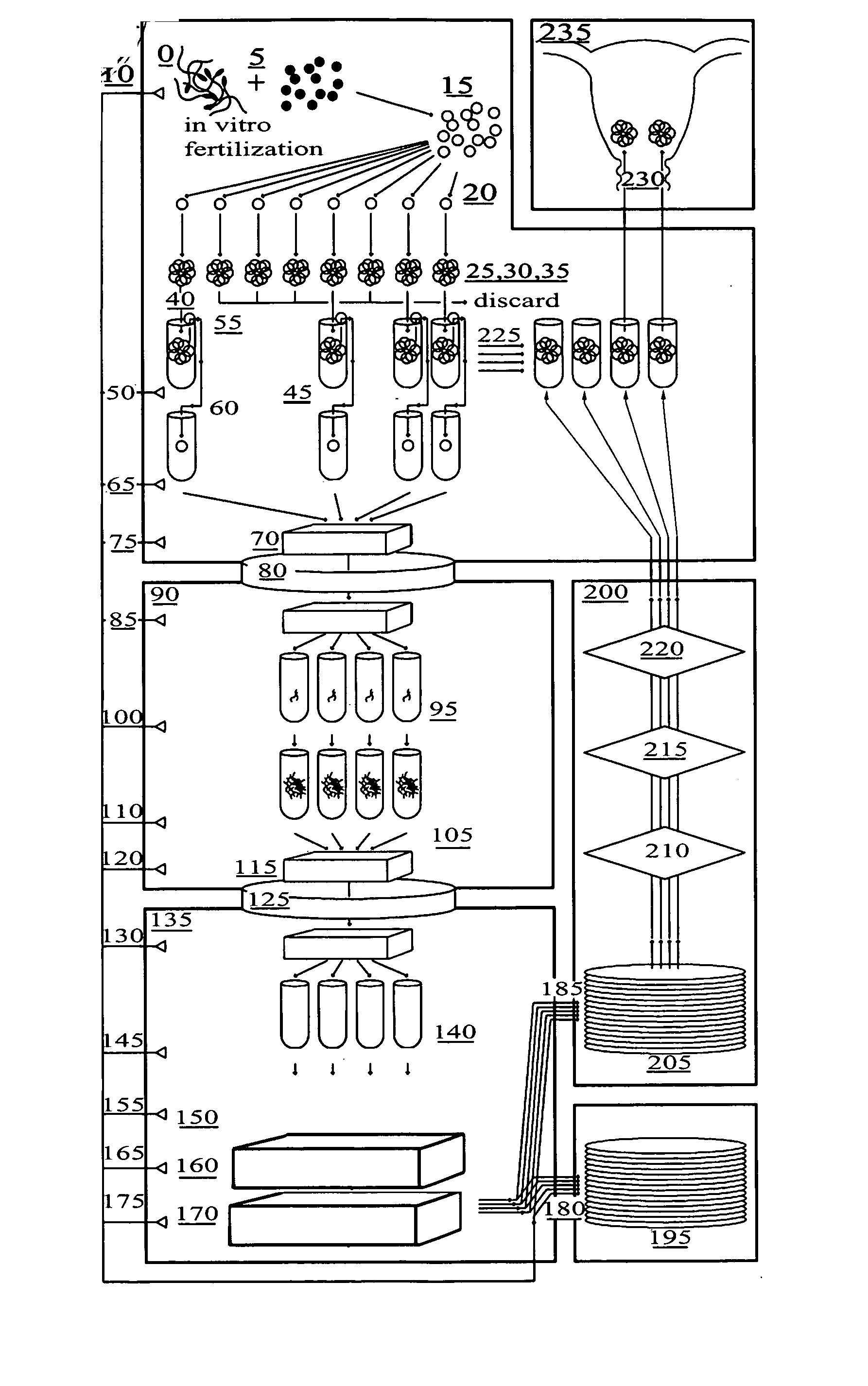
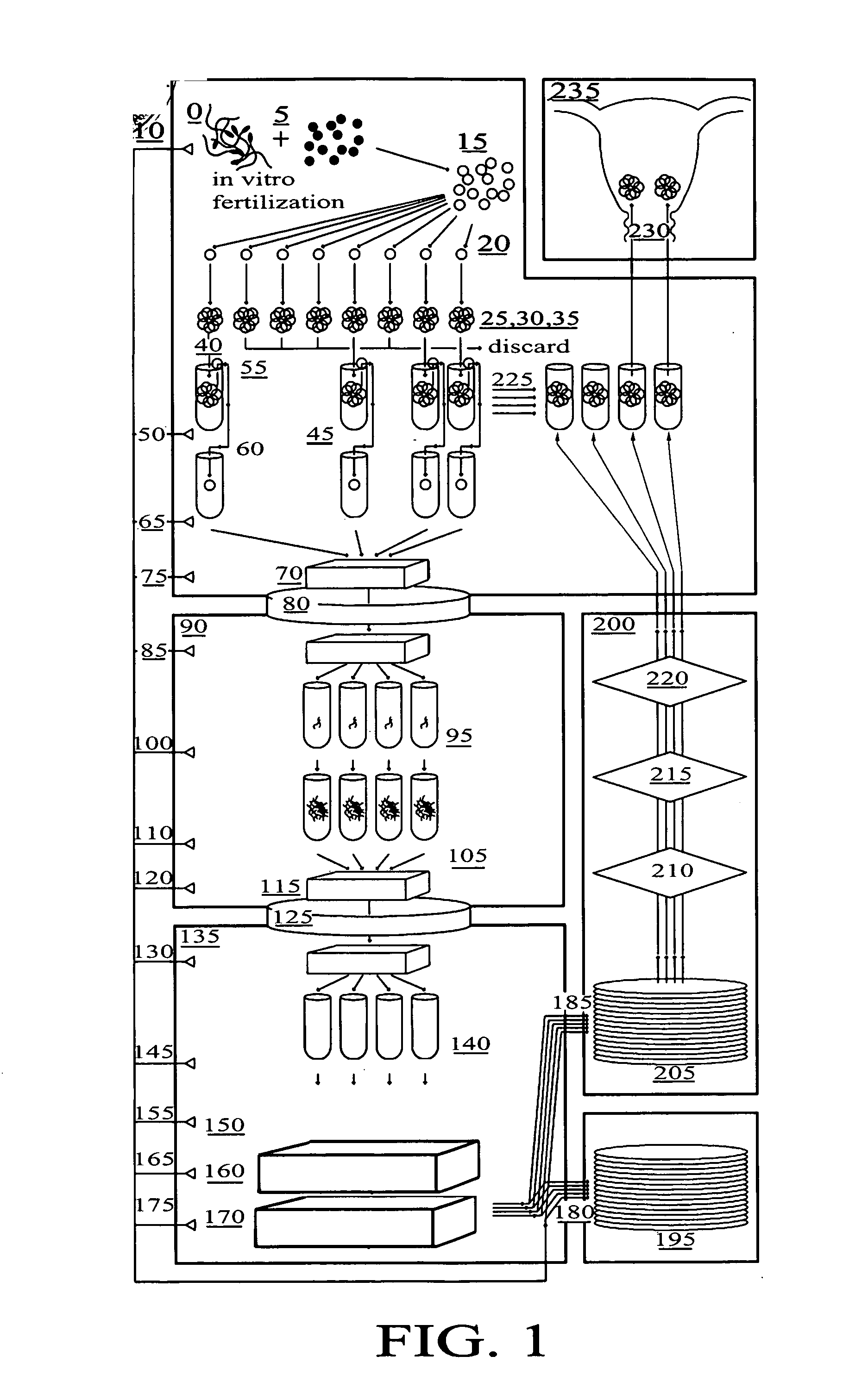
Method for genetic testing of human embryos for chromosome abnormalities, segregating genetic disorders with or without a known mutation and mitochondrial disorders following in vitro fertilization (IVF), embryo culture and embryo biopsy
InactiveUS20080085836A1Reduce significant riskImprove the level ofLibrary screeningLibrary member identificationLess invasiveContamination
We describe a method for interrogating the content and primary structure of DNA by microarray analyses and to provide comprehensive genetic screening and diagnostics prior to embryo transfer within an IVF setting. We will accomplish this by the following claims: 1) an optimized embryo grading system, 2) a less invasive embryo biopsy with reduced cellular contamination, 3) an optimized DNA amplification protocol for single cells, 4) identify aneuploidy and structural chromosome abnormalities using microarrays, 5) identifying sub-telomeric chromosome rearrangements, 6) a modified DNA fingerprinting protocol, 7) determine imprinting and epigenetic changes in developing embryos, 8) performing genome-wide scans to clarify / diagnose multi-factorial genetic disease and to determine genotype / haplotype patterns that may predict future disease, 9) determining single gene disorders with or without a known DNA mutation, 10) determining mtDNA mutations and / or the combination of mtDNA and genomic (nuclear) DNA aberrations that cause genetic disease.
Owner:KEARNS WILLIAM G +1
Methods for obtaining and using haplotype data
InactiveUS6931326B1Accurate predictionReduce costs and risksMicrobiological testing/measurementProteomicsHaplotypeS genotyping
Methods, computer program(s) and database(s) to analyze and make use of gene haplotype information. These include methods, program, and database to find and measure the frequency of haplotypes in the general population; methods, program, and database to find correlation's between an individual's haplotypes or genotypes and a clinical outcome; methods, program, and database to predict an individual's haplotypes from the individual's genotype for a gene; and methods, program, and database to predict an individual's clinical response to a treatment based on the individual's genotype or haplotype.
Owner:PGXHEALTH
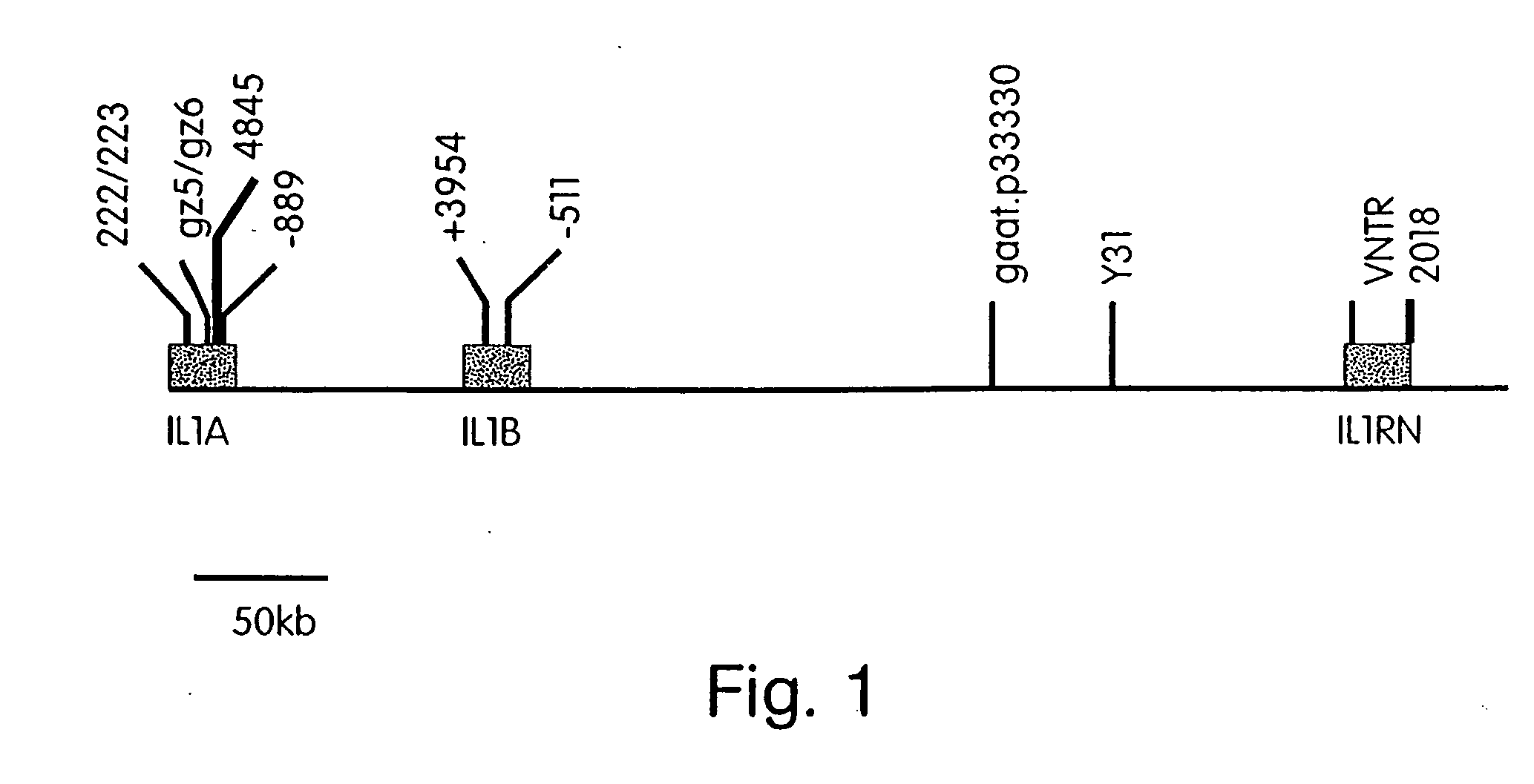
Diagnostics and therapeutics for diseases associated with an IL-1 inflammatory haplotype
Methods and kits for determining whether a subject has or is predisposed to developing a disease which is associated with IL-1 polymorphisms and assays for identifying therapeutics for treating and / or preventing the development of these diseases are provided.
Owner:INTERLEUKIN GENETICS
Sequence analysis using decorated nucleic acids
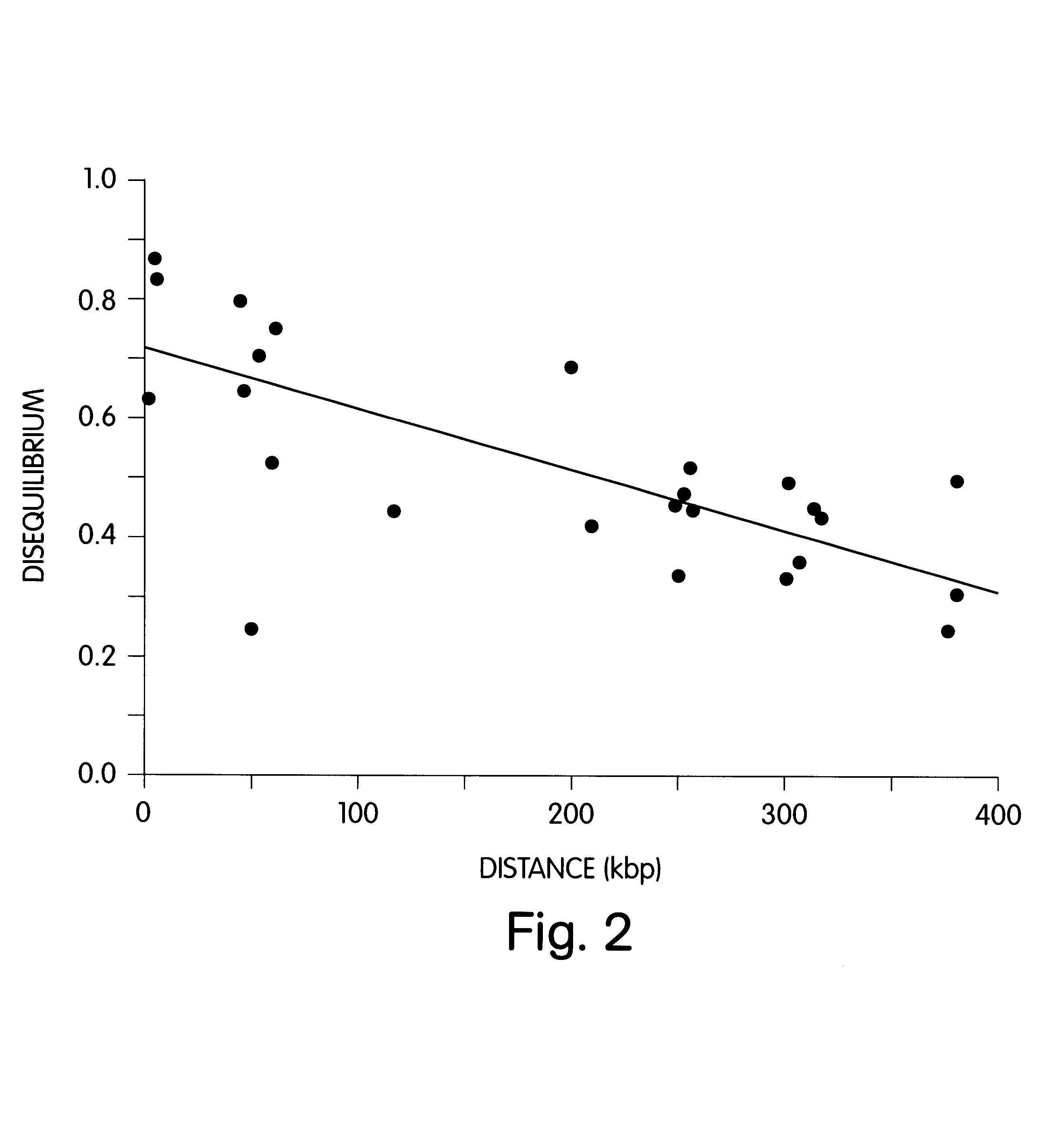
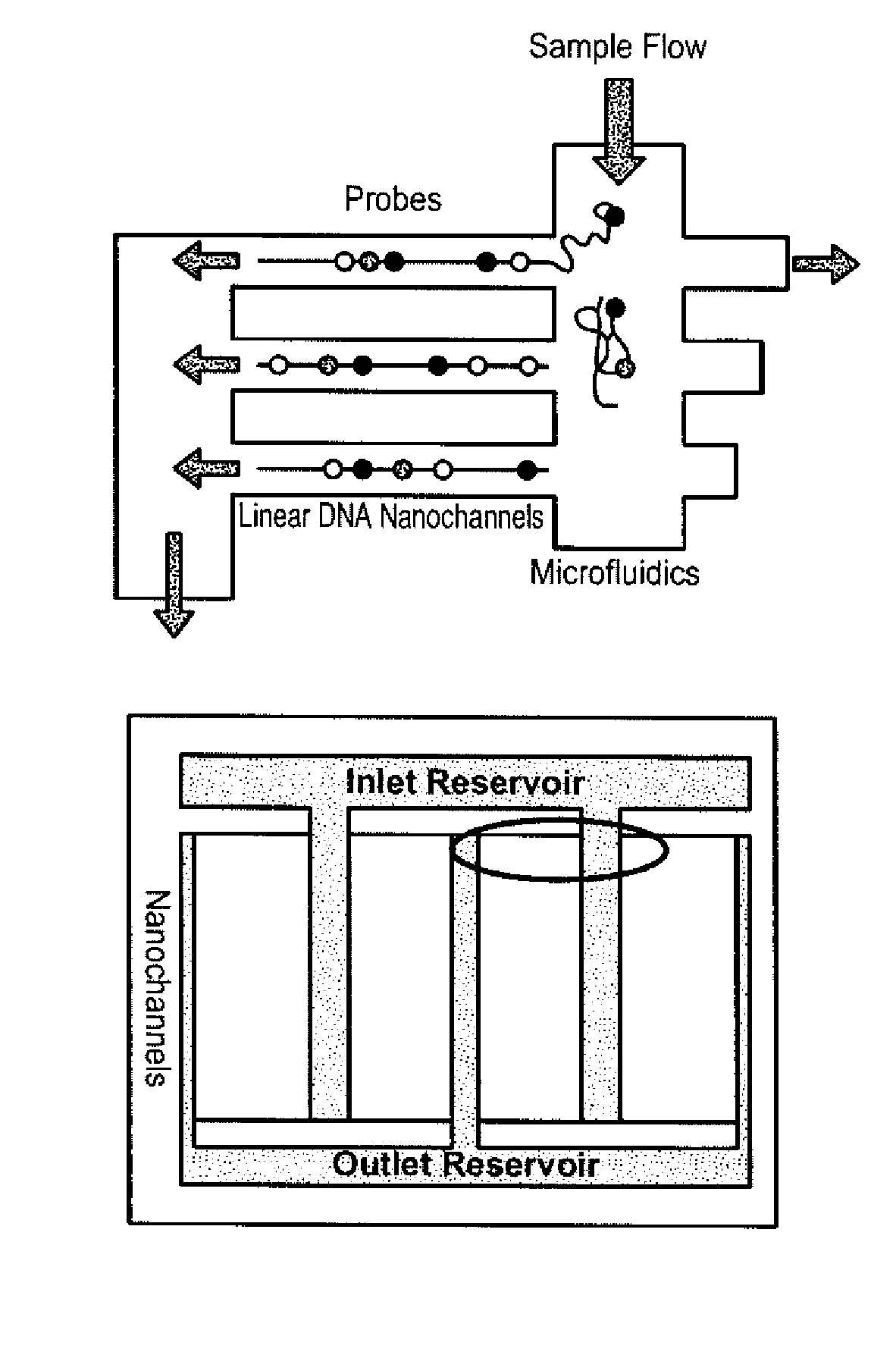
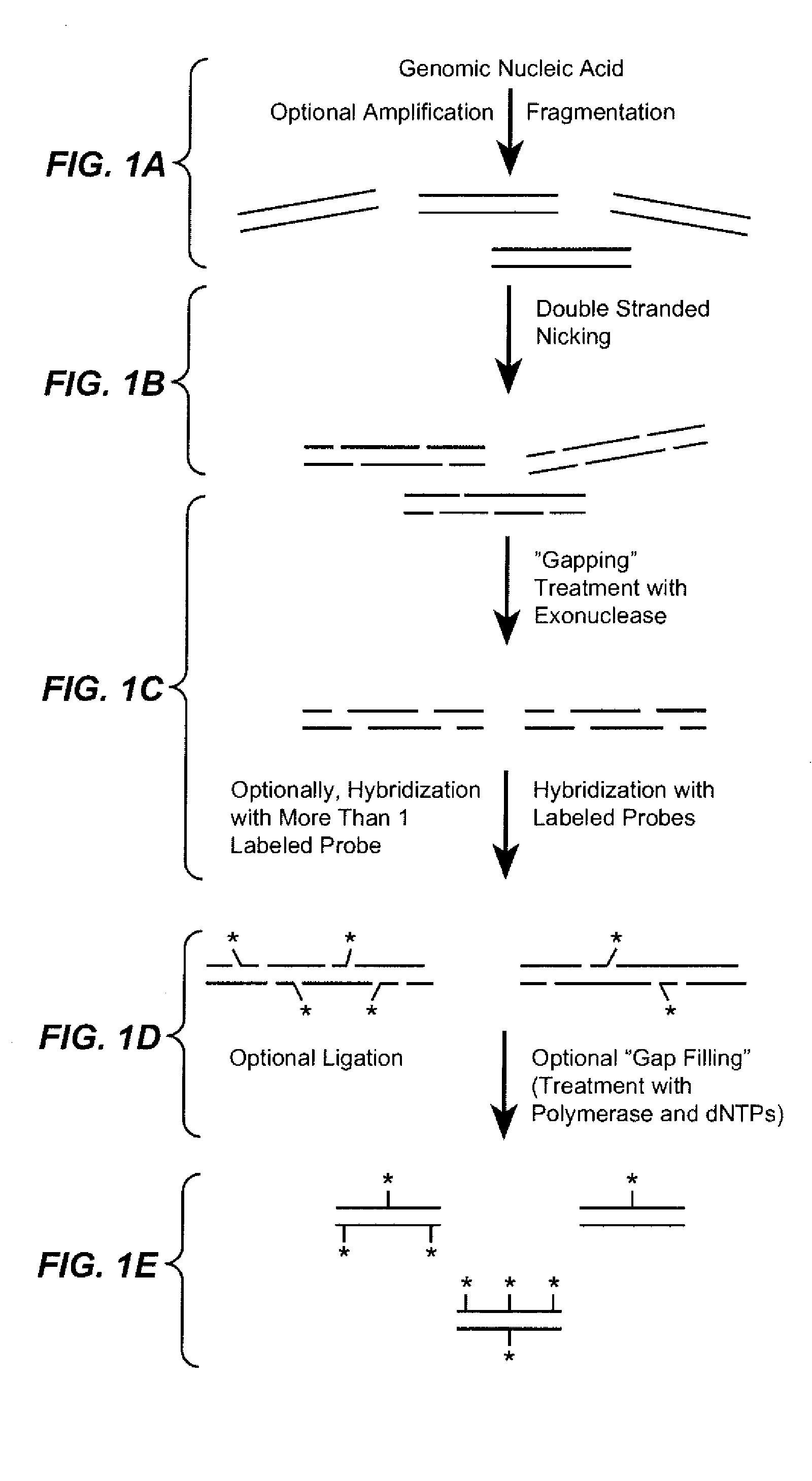
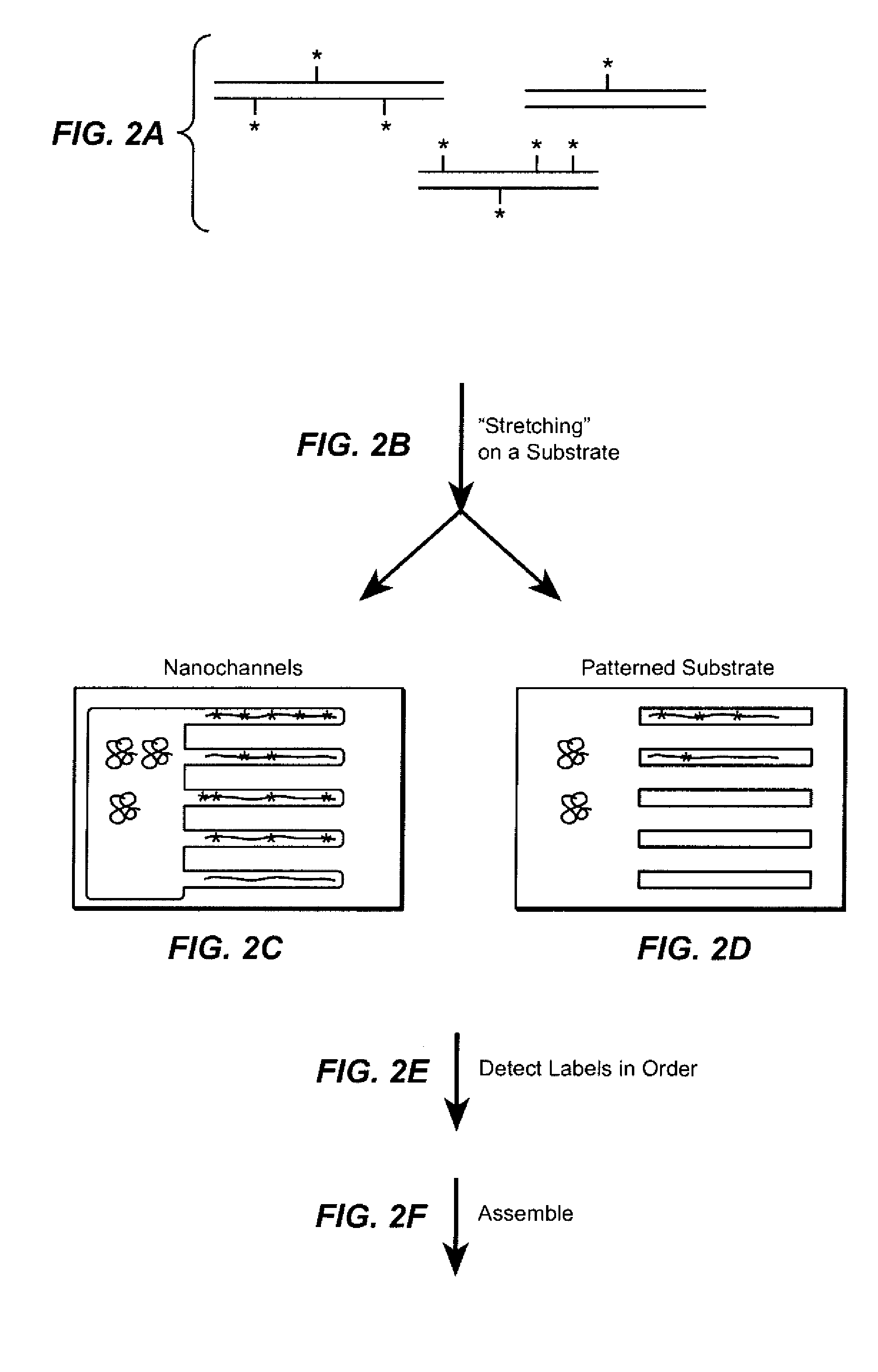
The present invention provides a sequence interrogation chemistry that combines the accuracy and haplotype integrity of long-read sequencing with improved methods of preparing genomic nucleic acids and analyzing sequence information generated from those nucleic acids. The present invention encompasses compositions comprising decorated nucleic acids stretched on substrates. The present invention further encompasses methods of making stretched decorated nucleic acids and methods of using decorated nucleic acids to obtain sequence information.
Owner:COMPLETE GENOMICS INC
Methods for the identification of genetic features for complex genetics classifiers
InactiveUS7107155B2Easy to measureReduce in quantityComputer controlMicrobiological testing/measurementGenetic traitsStatistical analysis
A candidate single nucleotide polymorphism (SNP) combination is selected from a plurality of candidate SNP combinations for a gene associated with a genetic trait. Haplotype data associated with this candidate SNP combination are read for a plurality of individuals and grouped into a positive-responding group and a negative-responding group based on whether a predetermined trait criteria for an individual is met. A statistical analysis on the grouped haplotype data is performed to obtain a statistical measurement. The acts of selecting, reading, grouping, and performing are repeated as necessary to identity the candidate SNP combination having the optimal statistical measurement. In one approach, a directed search based on results of previous statistical analysis of SNP combinations is performed until the optimal statistical measurement is obtained. In addition, the number of SNP combinations selected and analyzed may be reduced based on a simultaneous testing procedure.
Owner:DNAPRINT GENOMICS +1
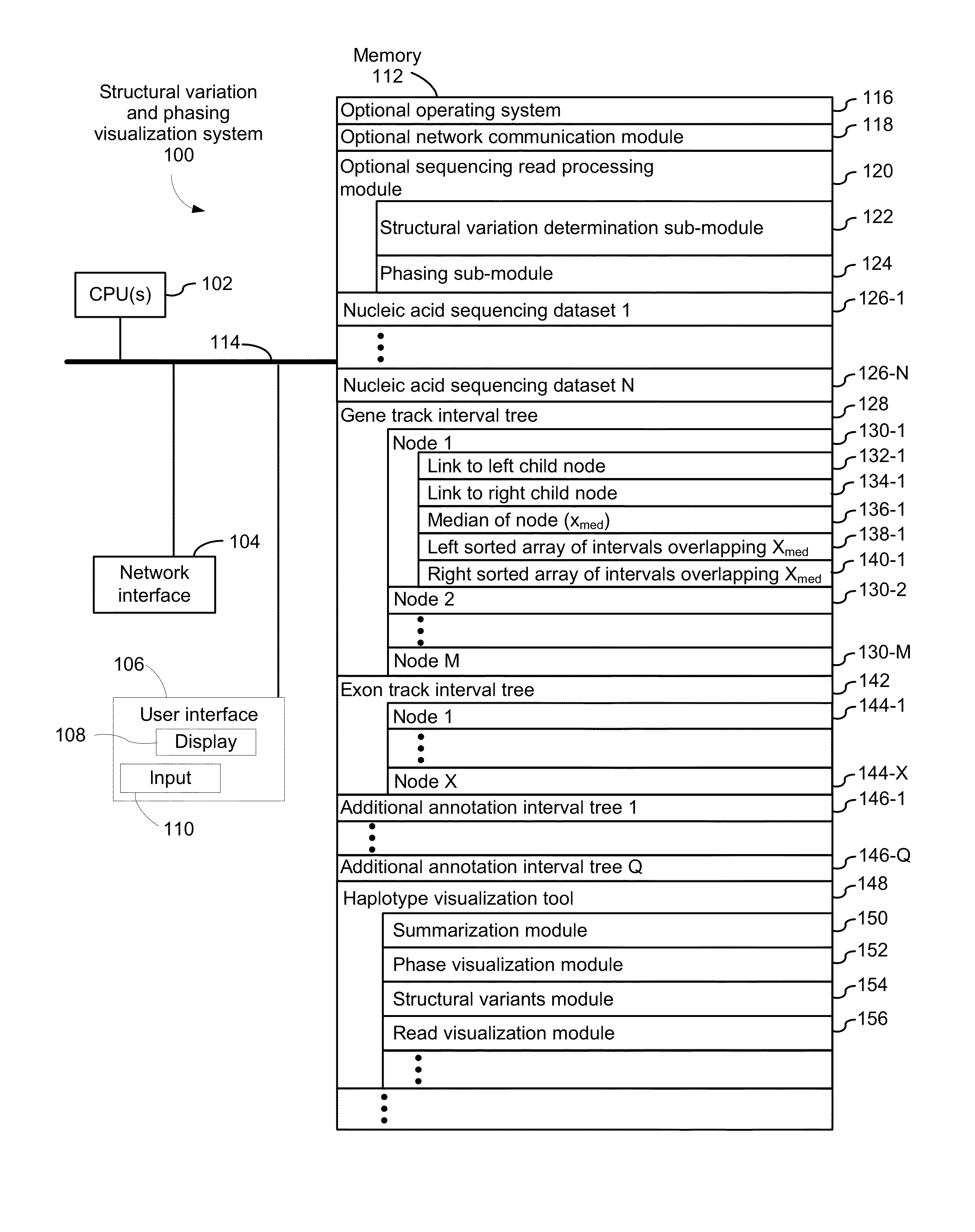
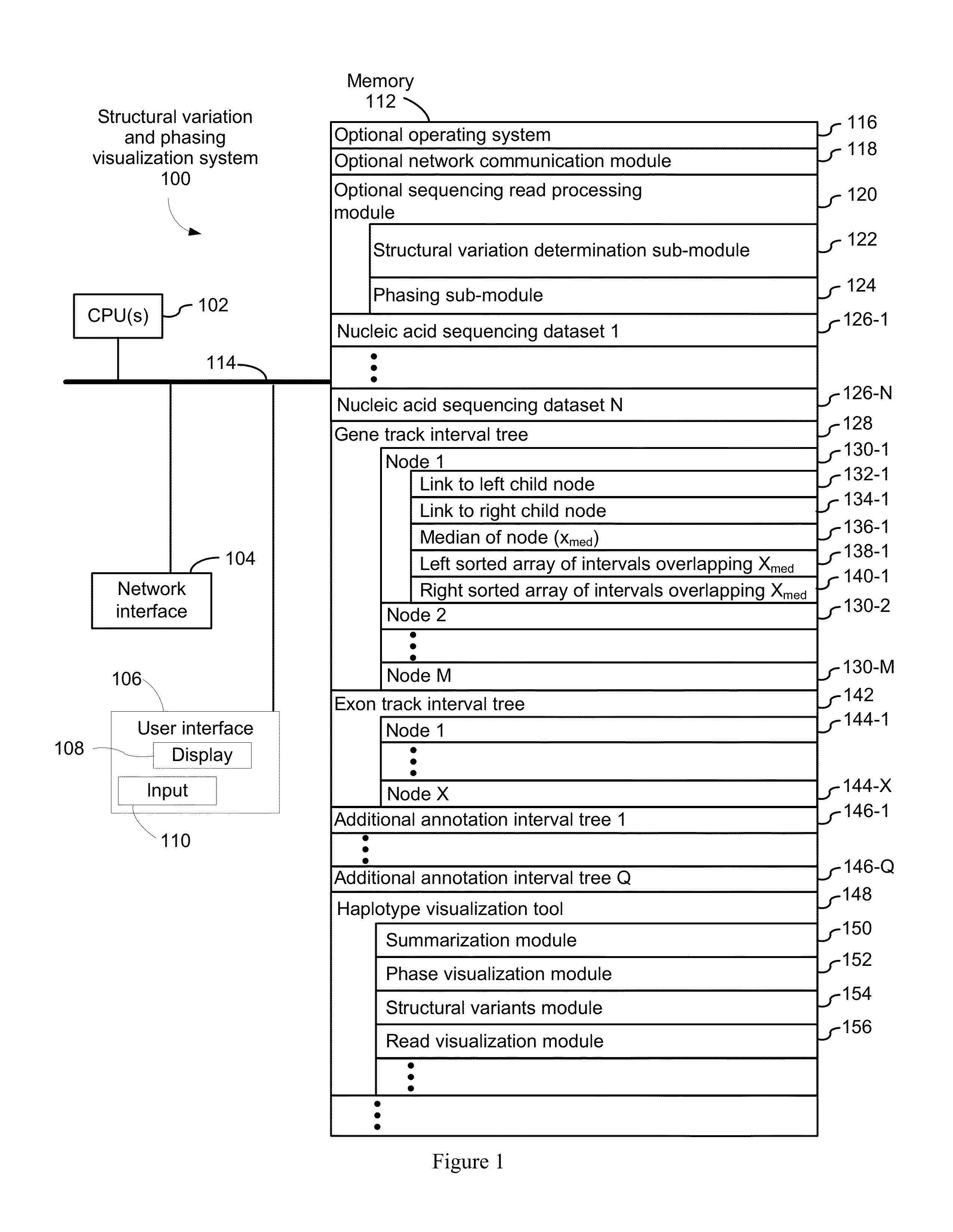
Systems and methods for visualizing structural variation and phasing information
ActiveUS20160203196A1Digital data information retrievalDigital data processing detailsData setNucleic acid sequencing
A system for providing structural variation or phasing information is provided. The system accesses a nucleic acid sequence dataset corresponding to a target nucleic acid in a sample. The dataset comprises a header, synopsis, and data section. The data section comprises a plurality of sequencing reads. Each sequencing read comprises a first portion corresponding to a subset of the target nucleic acid and a second portion that encodes an identifier for the sequencing read from a plurality of identifiers. One or more programs in the memory of the system use a microprocessor of the system to provide a haplotype visualization tool that receives a request for structural variation or phasing information from the dataset. The request is evaluated against the synopsis thereby identifying portions of the data section. Structural variation or phasing information is formatted for display in the haplotype visualization tool using the identified portions of the data section.
Owner:10X GENOMICS
Process, software arrangement and computer-accessible medium for obtaining information associated with a haplotype
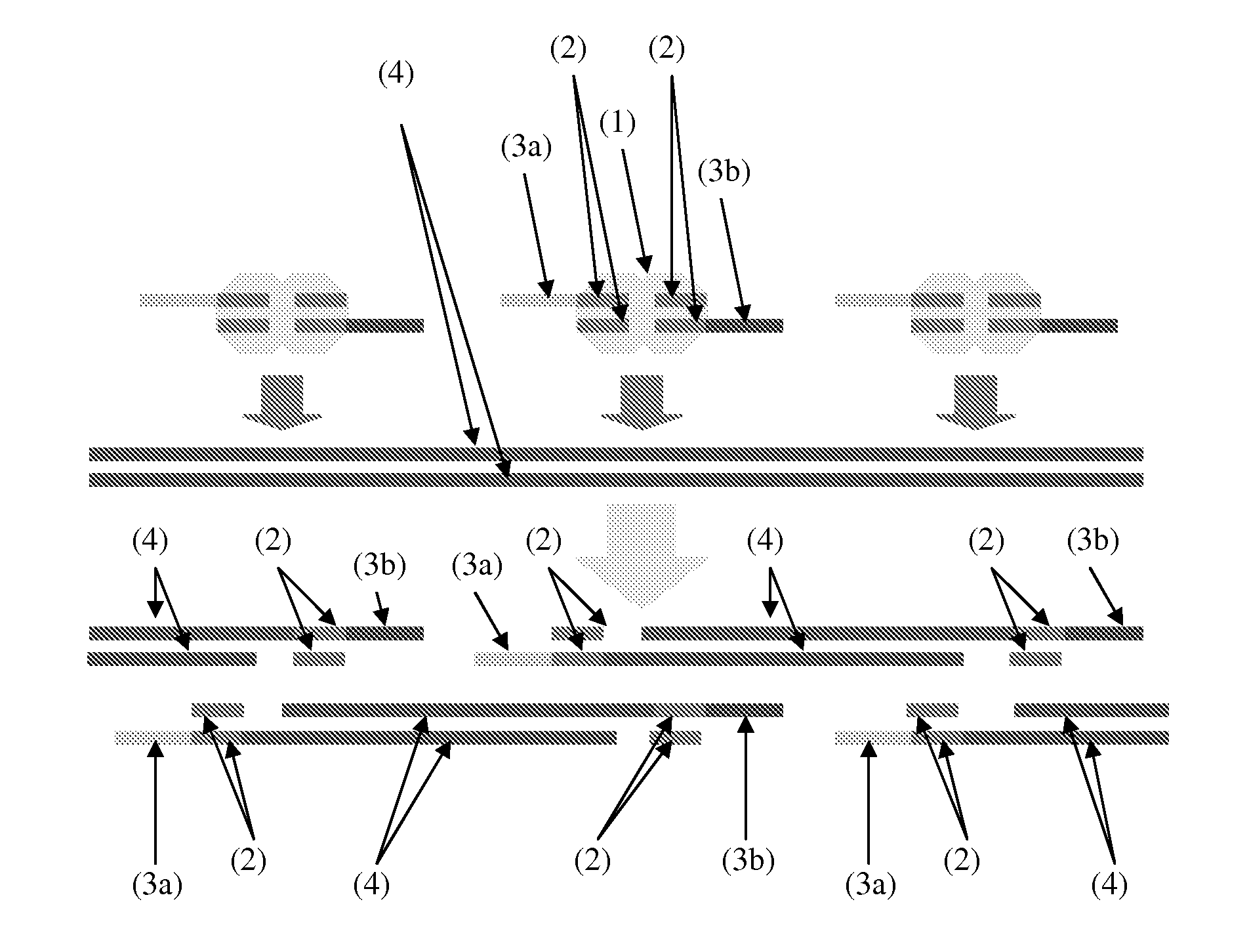
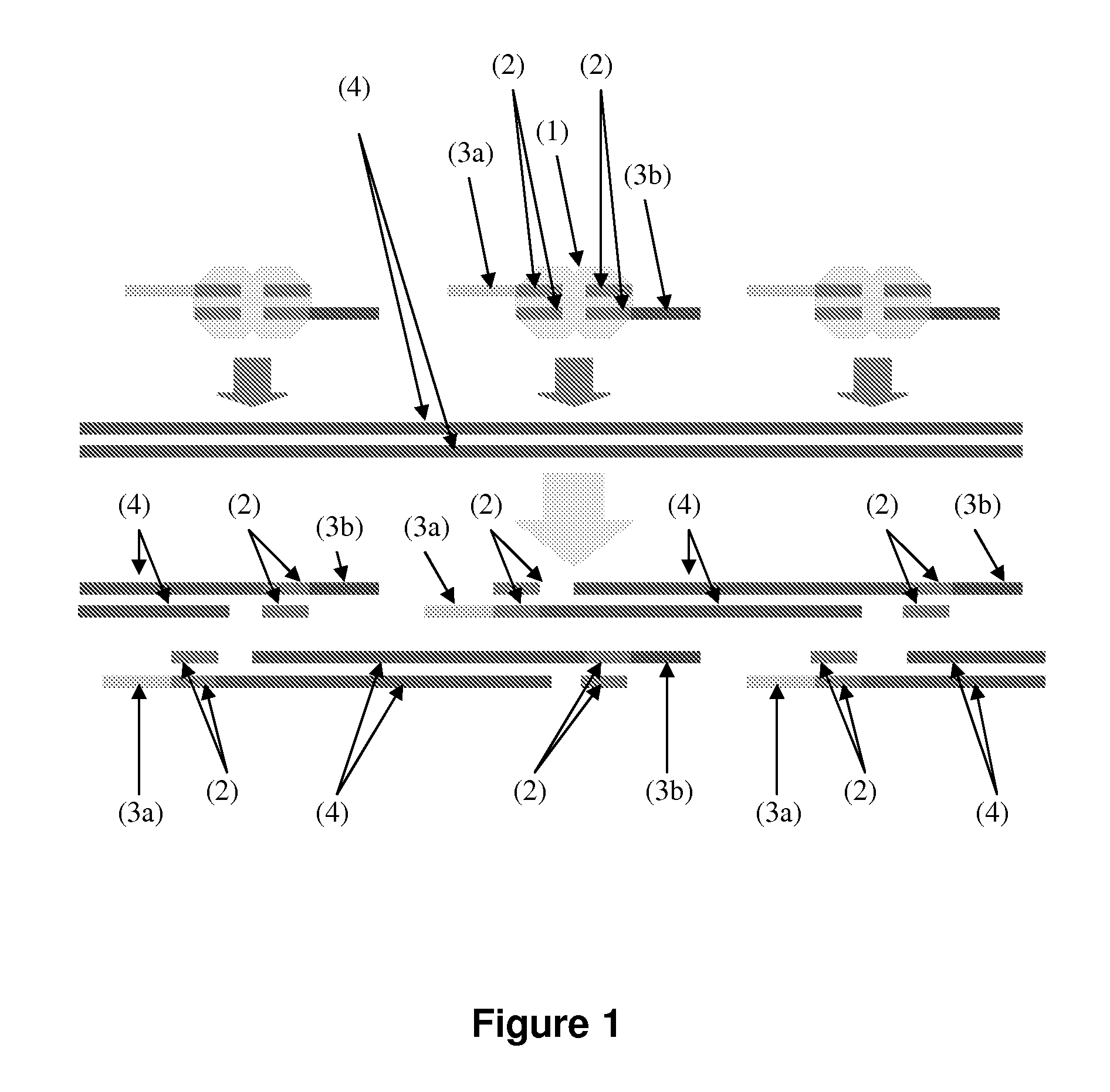
ActiveUS7805282B2Determine an individual's haplotypeMicrobiological testing/measurementComputation using non-denominational number representationHaplotypeComputer science
The present invention relates to a method, system and software arrangement for determining the co-associations of allele types across consecutive loci and hence for reconstructing two haplotypes of a diploid individual from genotype data generated by mapping experiments with single molecules, families or populations. The haplotype reconstruction system, method and software arrangement of the present invention can utilize a procedure that is nearly linear in the number of polymorphic markers examined, and is therefore quicker, more accurate, and more efficient than other population-based approaches. The system, method, and software arrangement of the present invention may be useful to assist with the diagnosis and treatment of any disease, which has a genetic component.
Owner:NEW YORK UNIV +1
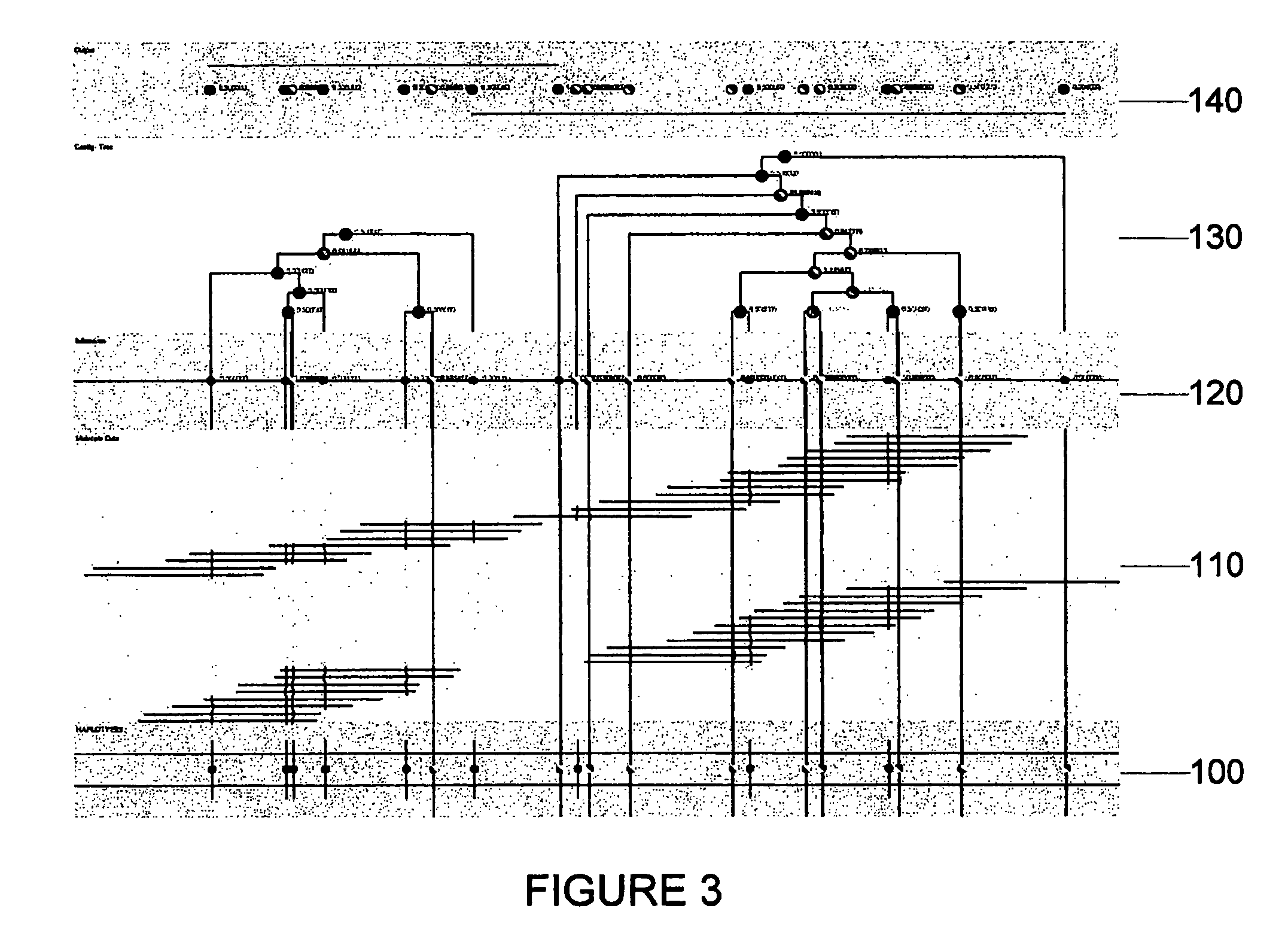
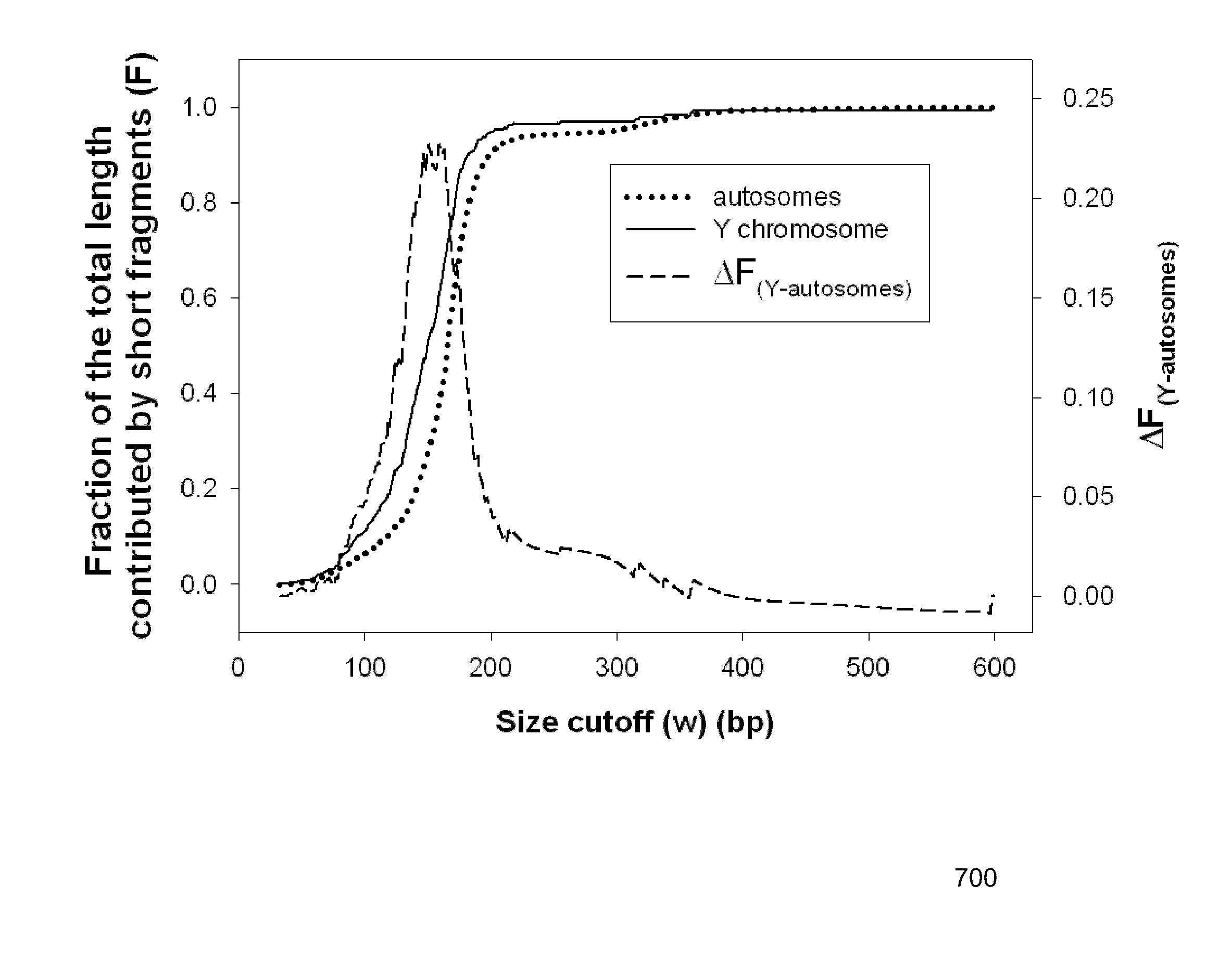
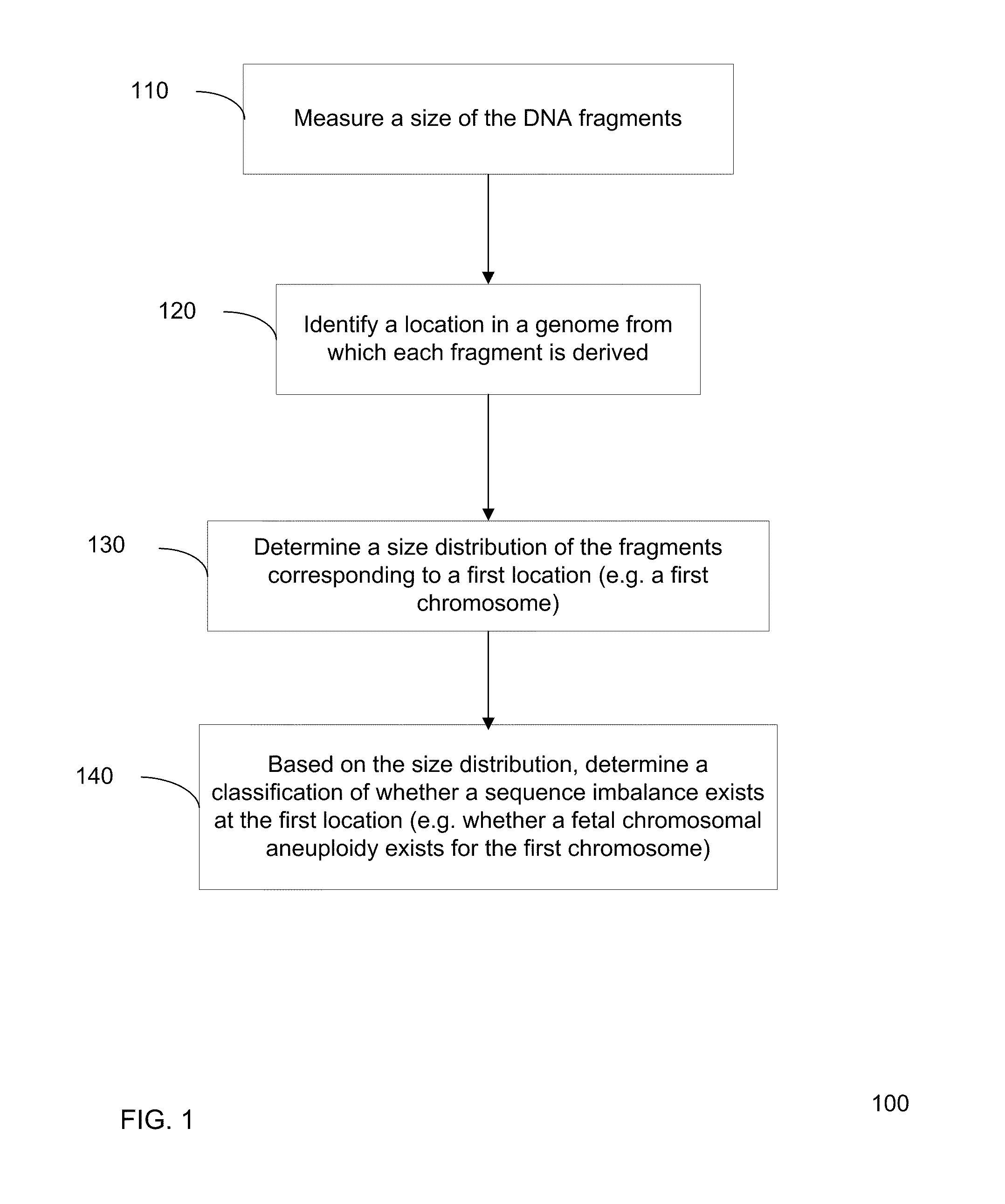
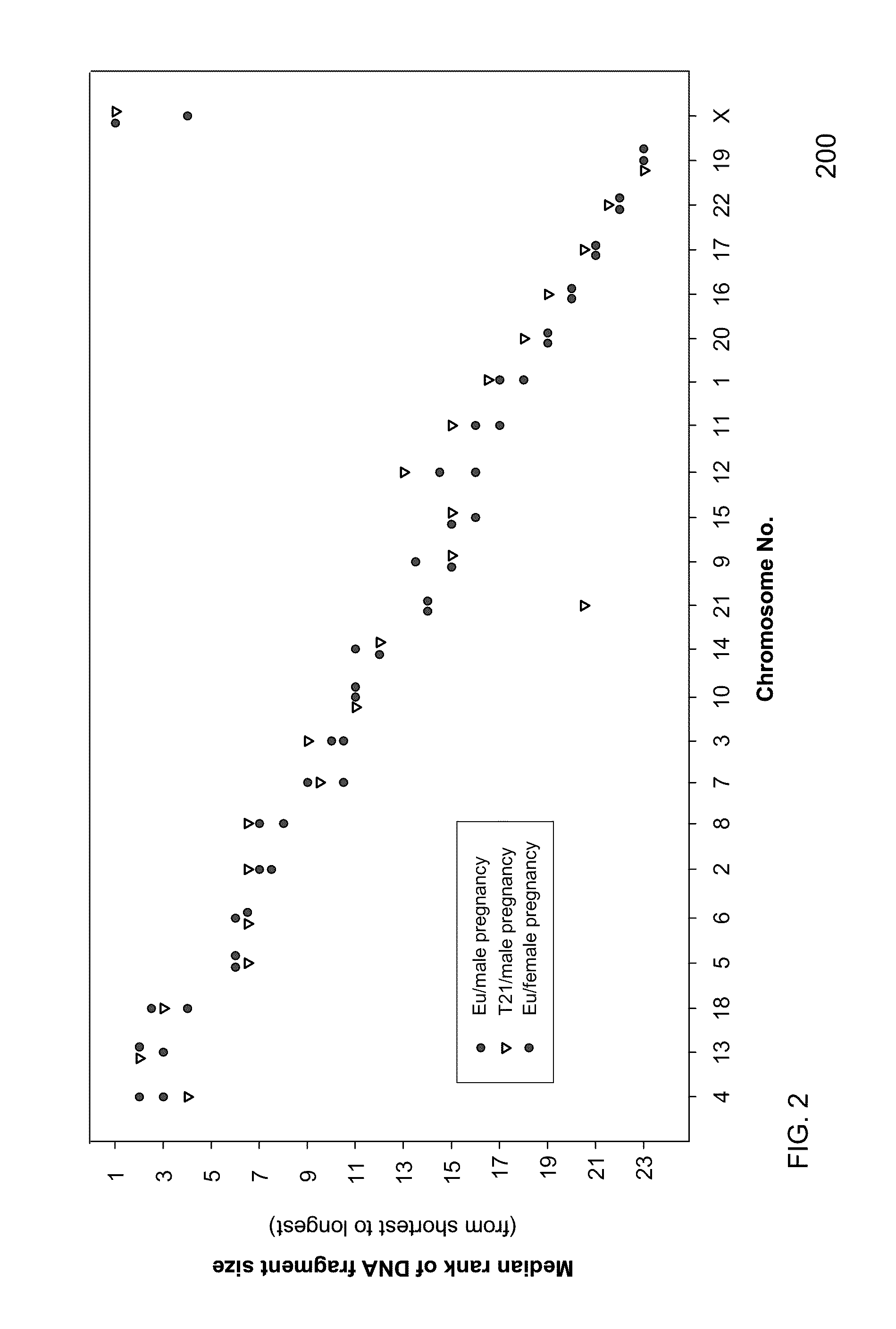
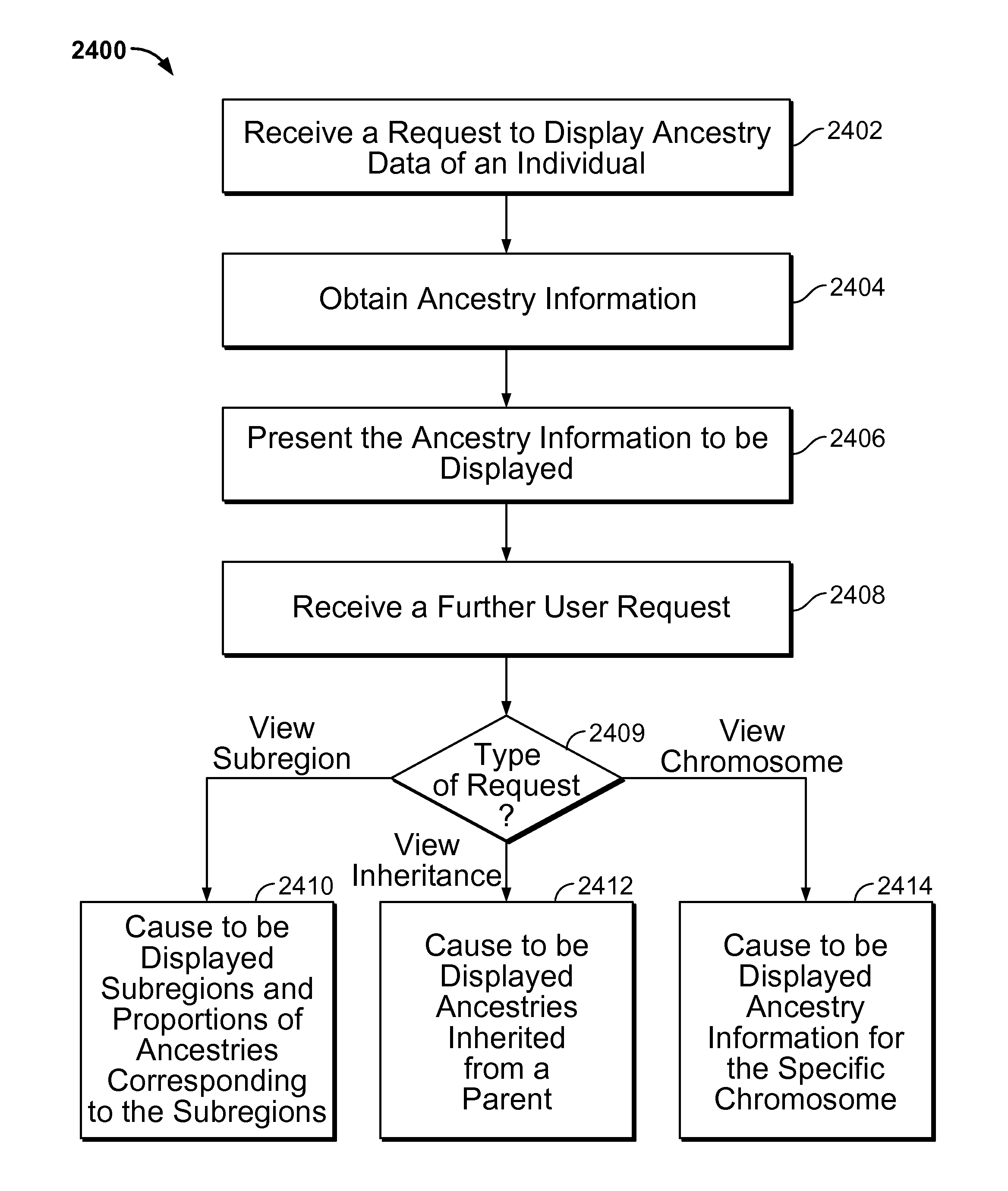
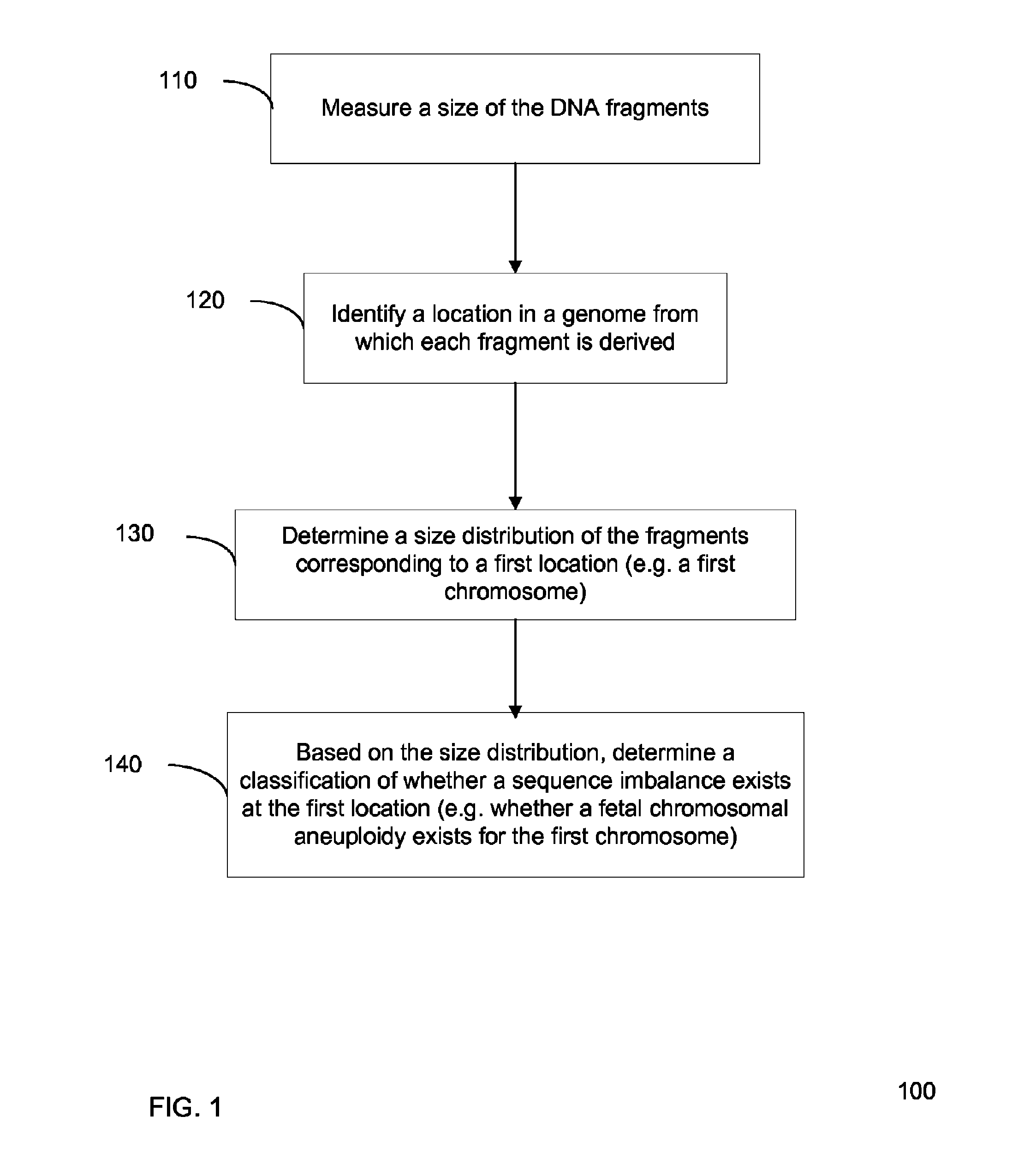
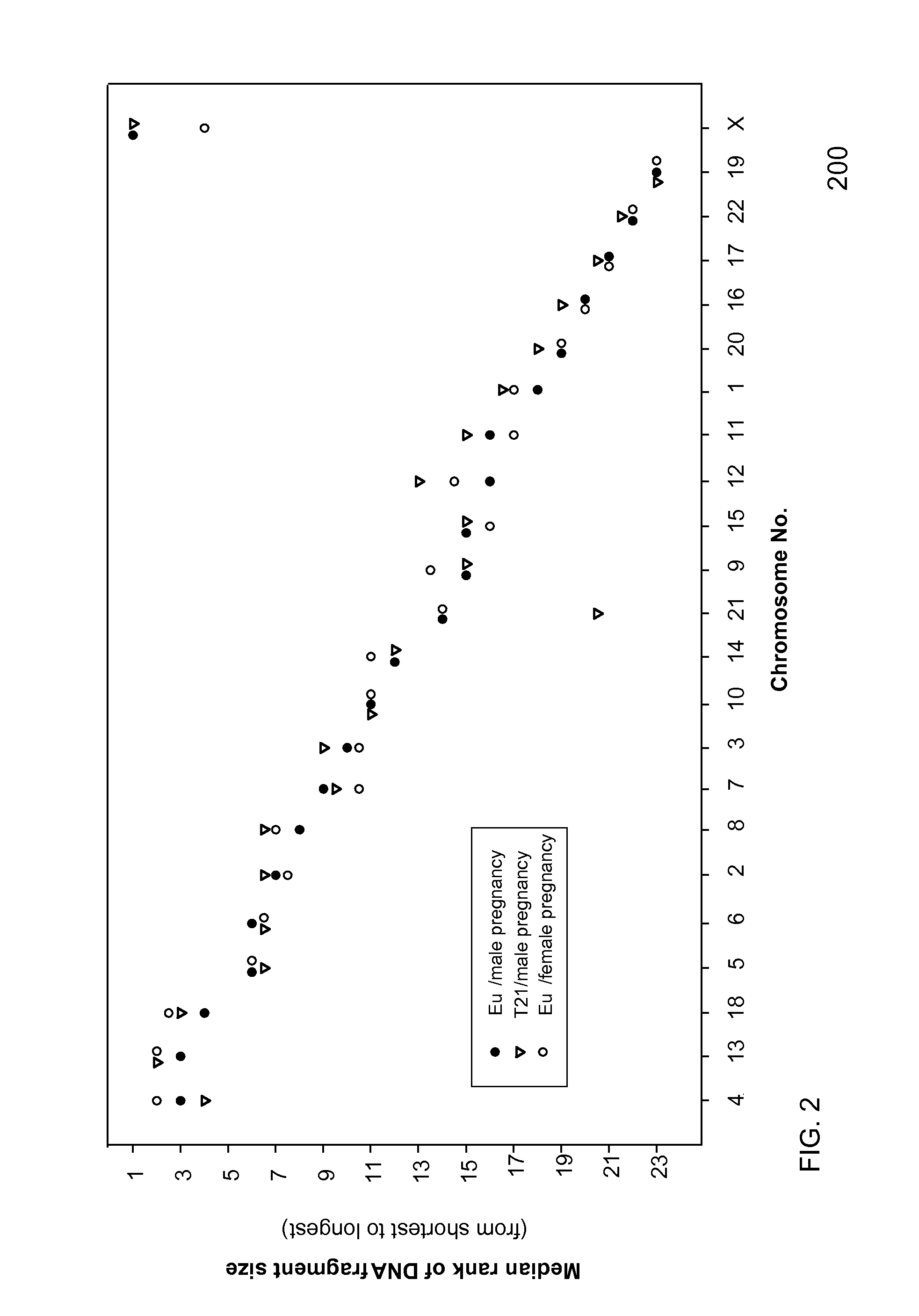
Size-based genomic analysis
Systems, methods, and apparatuses for performing a prenatal diagnosis of a sequence imbalance are provided. A shift (e.g. to a smaller size distribution) can signify an imbalance in certain circumstances. For example, a size distribution of fragments of nucleic acids from an at-risk chromosome can be used to determine a fetal chromosomal aneuploidy. A size ranking of different chromosomes can be used to determine changes of a rank of an at-risk chromosome from an expected ranking. Also, a difference between a statistical size value for one chromosome can be compared to a statistical size value of another chromosome to identify a significant shift in size. A genotype and haplotype of the fetus may also be determined using a size distribution to determine whether a sequence imbalance occurs in a maternal sample relative to a genotypes or haplotype of the mother, thereby providing a genotype or haplotype of the fetus.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
System, method and software arrangement for bi-allele haplotype phasing
ActiveUS20050255508A1Determine an individual's haplotypeMicrobiological testing/measurementProteomicsHaplotypeSystems approaches
The present invention relates to a method, system and software arrangement for determining the co-associations of allele types across consecutive loci and hence for reconstructing two haplotypes of a diploid individual from genotype data generated by mapping experiments with single molecules, families or populations. The haplotype reconstruction system, method and software arrangement of the present invention can utilize a procedure that is nearly linear in the number of polymorphic markers examined, and is therefore quicker, more accurate, and more efficient than other population-based approaches. The system, method, and software arrangement of the present invention may be useful to assist with the diagnosis and treatment of any disease, which has a genetic component.
Owner:NEW YORK UNIV +1
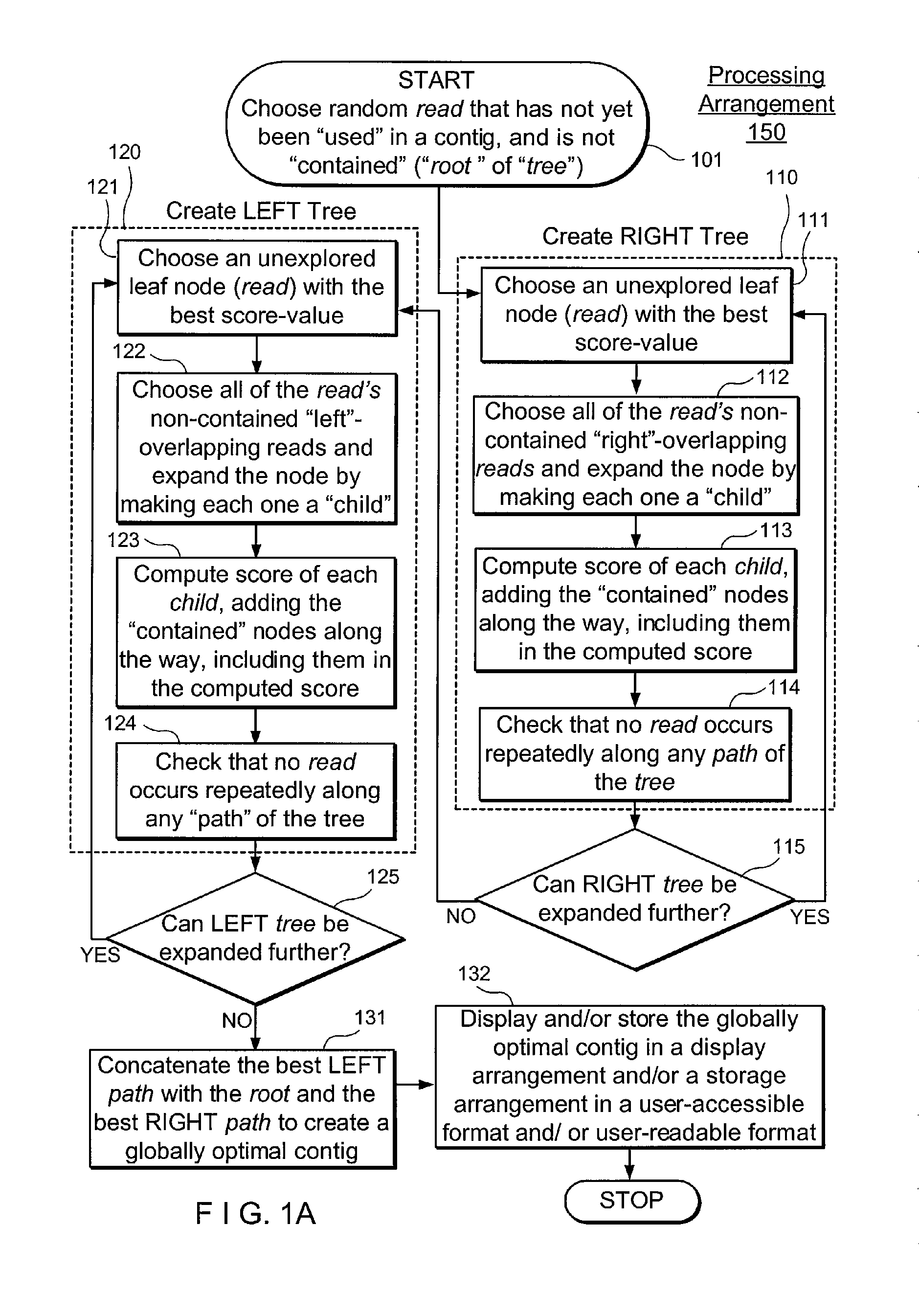
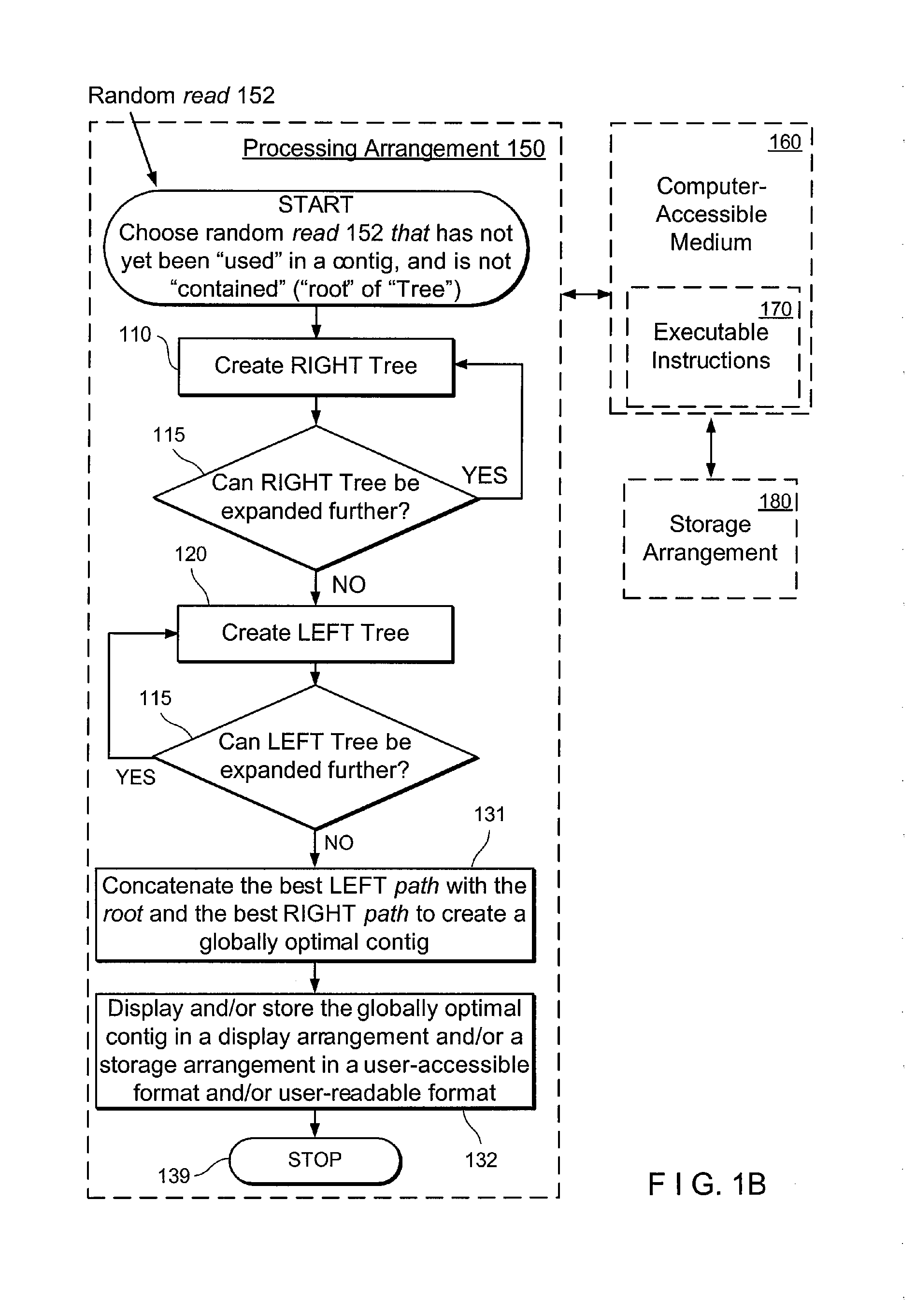
Method, computer-accessible medium and systems for score-driven whole-genome shotgun sequence assemble
ActiveUS20120041727A1Computation using non-denominational number representationSequence analysisHaplotypeData mining
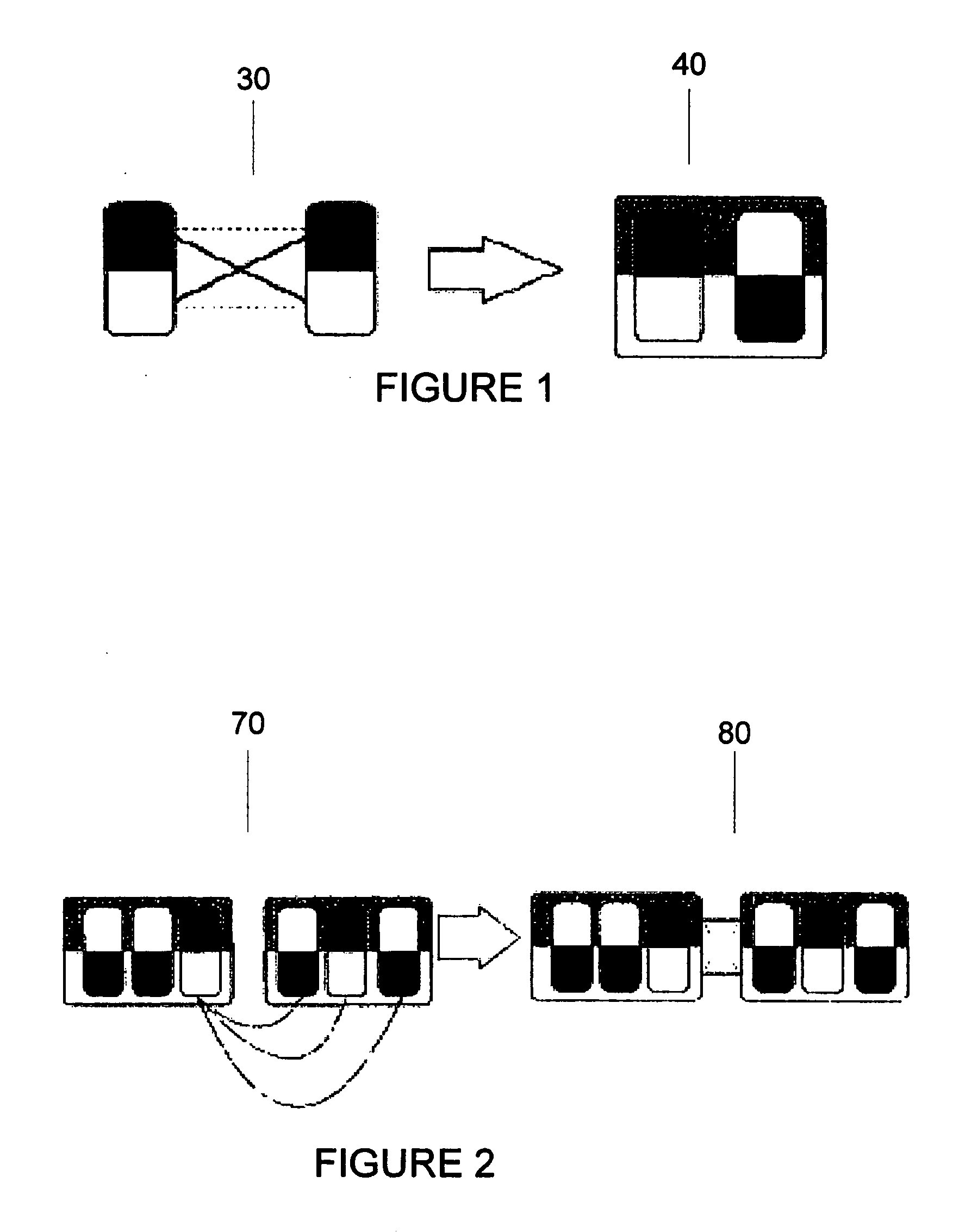
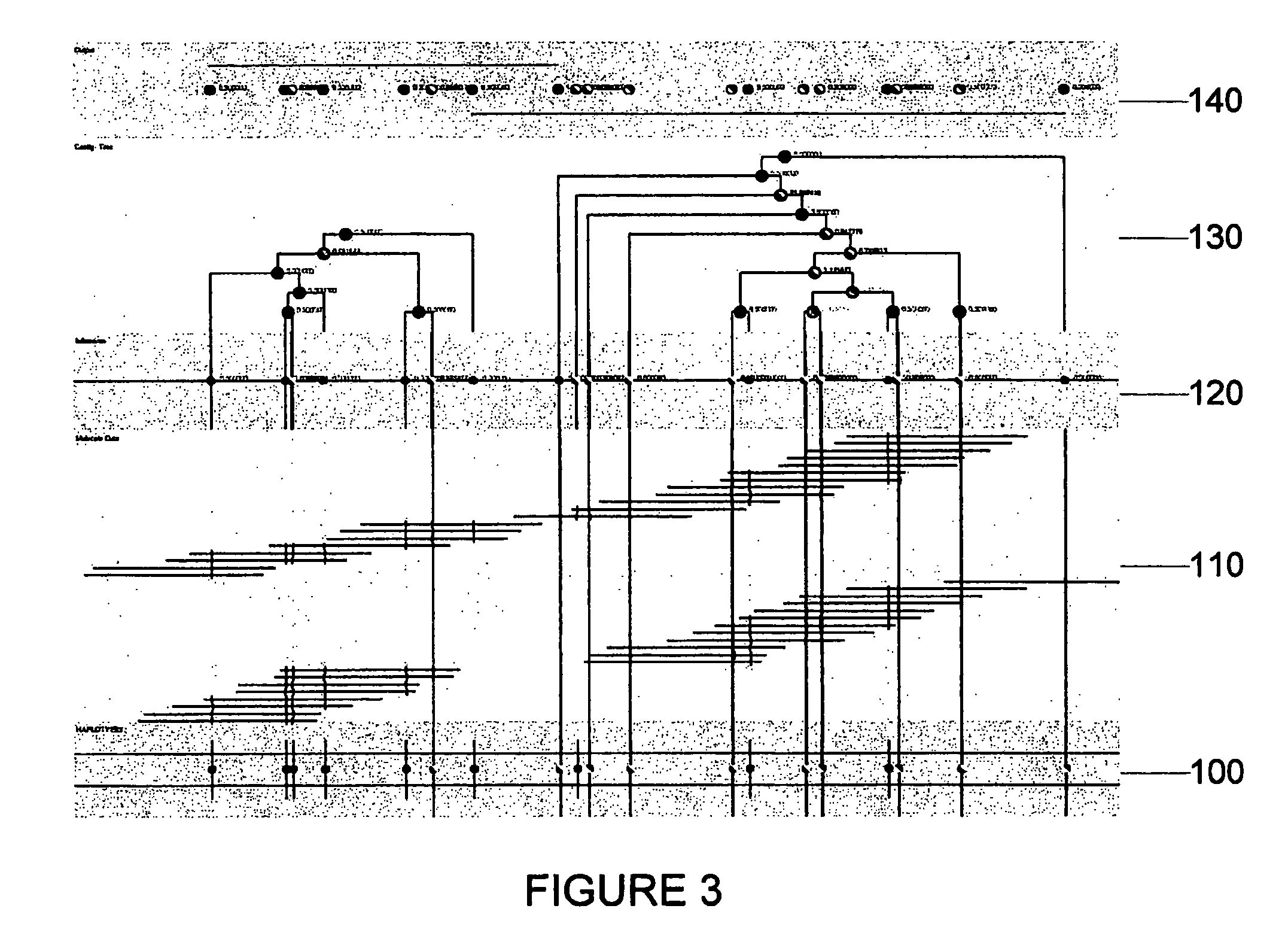
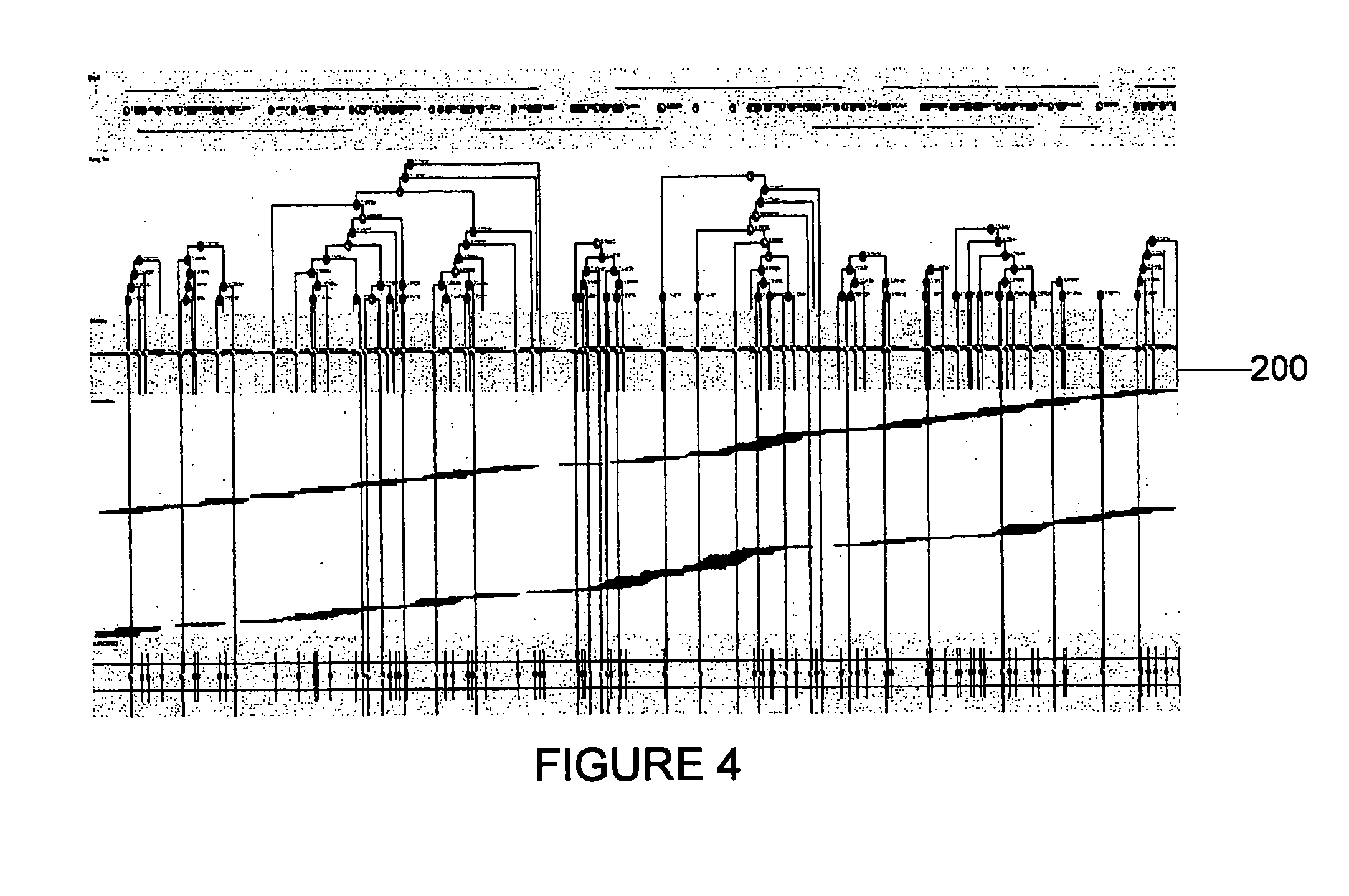
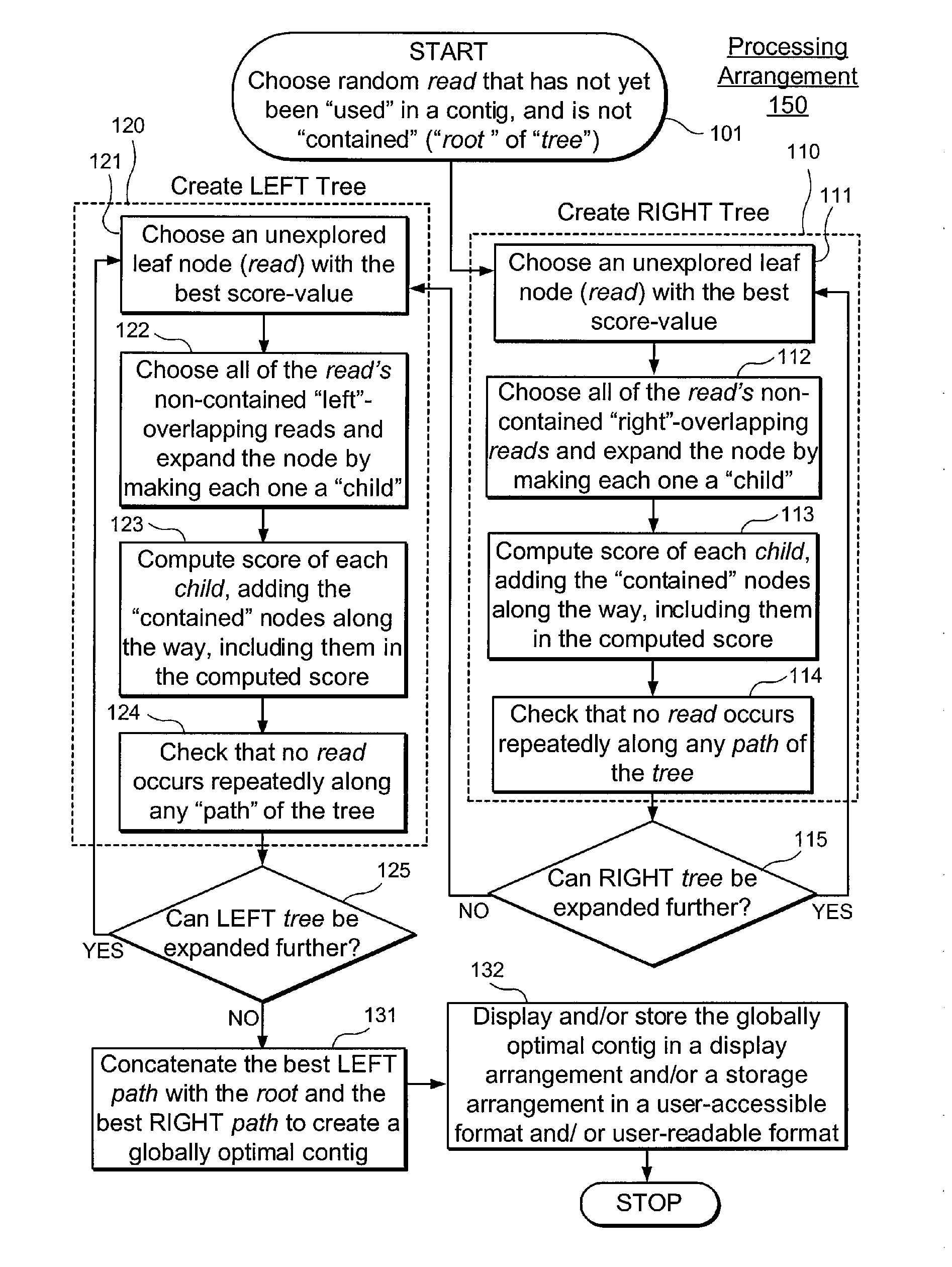
Exemplary embodiments of the present disclosure relate generally to methods, computer-accessible medium and systems for assembling haplotype and / or genotype sequences of at least one genome, which can be based upon, e.g., consistent layouts of short sequence reads and long-range genome related data. For example, a processing arrangement can be configured to perform a procedure including, e.g., obtaining randomly located short sequence reads, using at least one score function in combination with constraints based on, e.g., the long range data, generating a layout of randomly located short sequence reads such that the layout is globally optimal with respect to the score function, obtained through searching coupled with score and constraint dependent pruning to determine the globally optimal layout substantially satisfying the constraints, generating a whole and / or a part of a genome wide haplotype sequence and / or genotype sequence, and converting a globally optimal layout into one or more consensus sequences.
Owner:NEW YORK UNIV
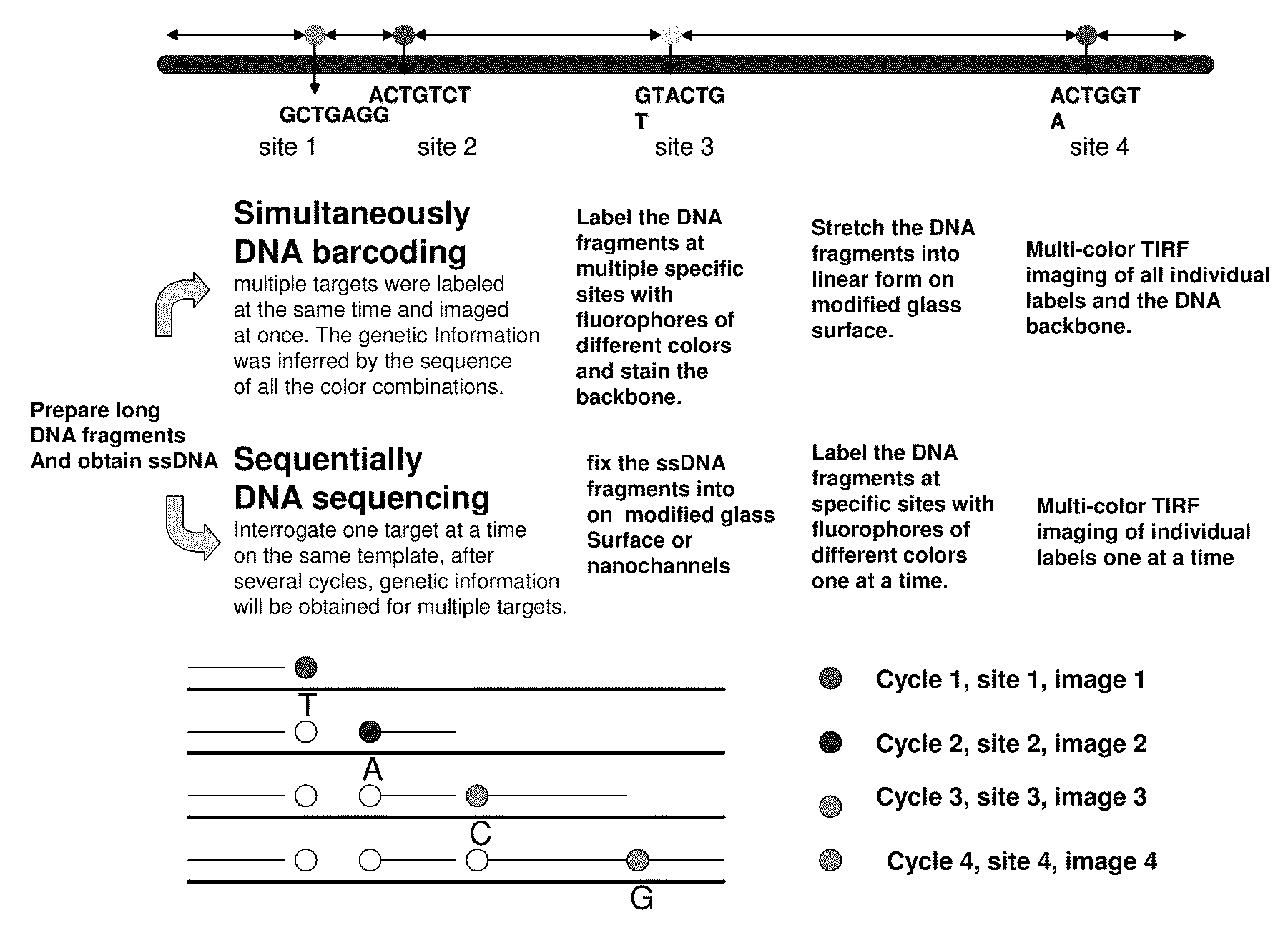
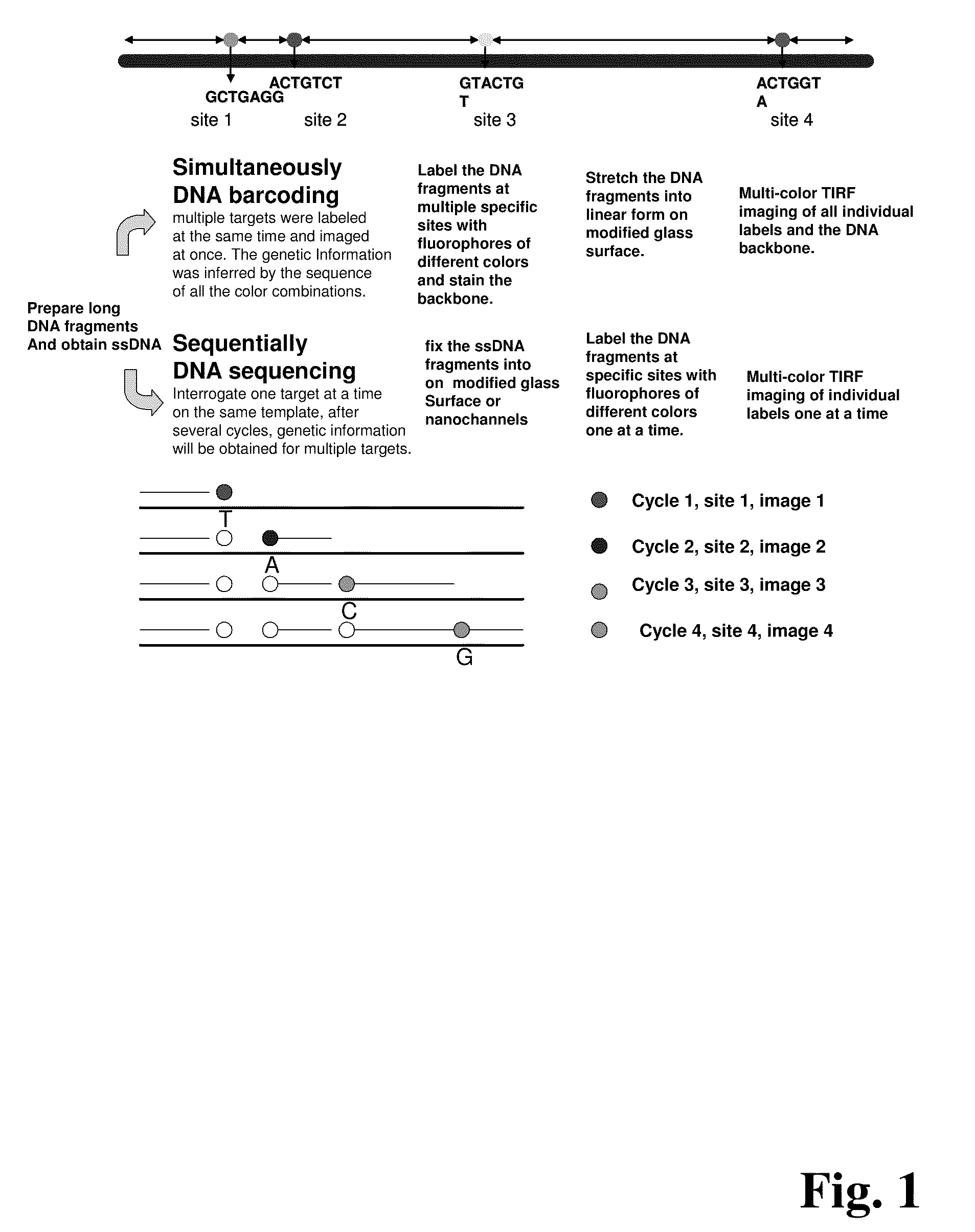
Methods for determining genetic haplotypes and DNA mapping
ActiveUS20090155780A1Improve labeling efficiencyStable labelingMicrobiological testing/measurementFermentationDirect imagingDNA barcoding
Improved methods of genetic haplotyping and DNA sequencing and mapping, including methods for making amplified ssDNA, methods for allele determination, and a DNA barcoding strategy based on direct imaging of individual DNA molecules and localization of multiple sequence motifs or polymorphic sites on a single DNA molecule.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS +1
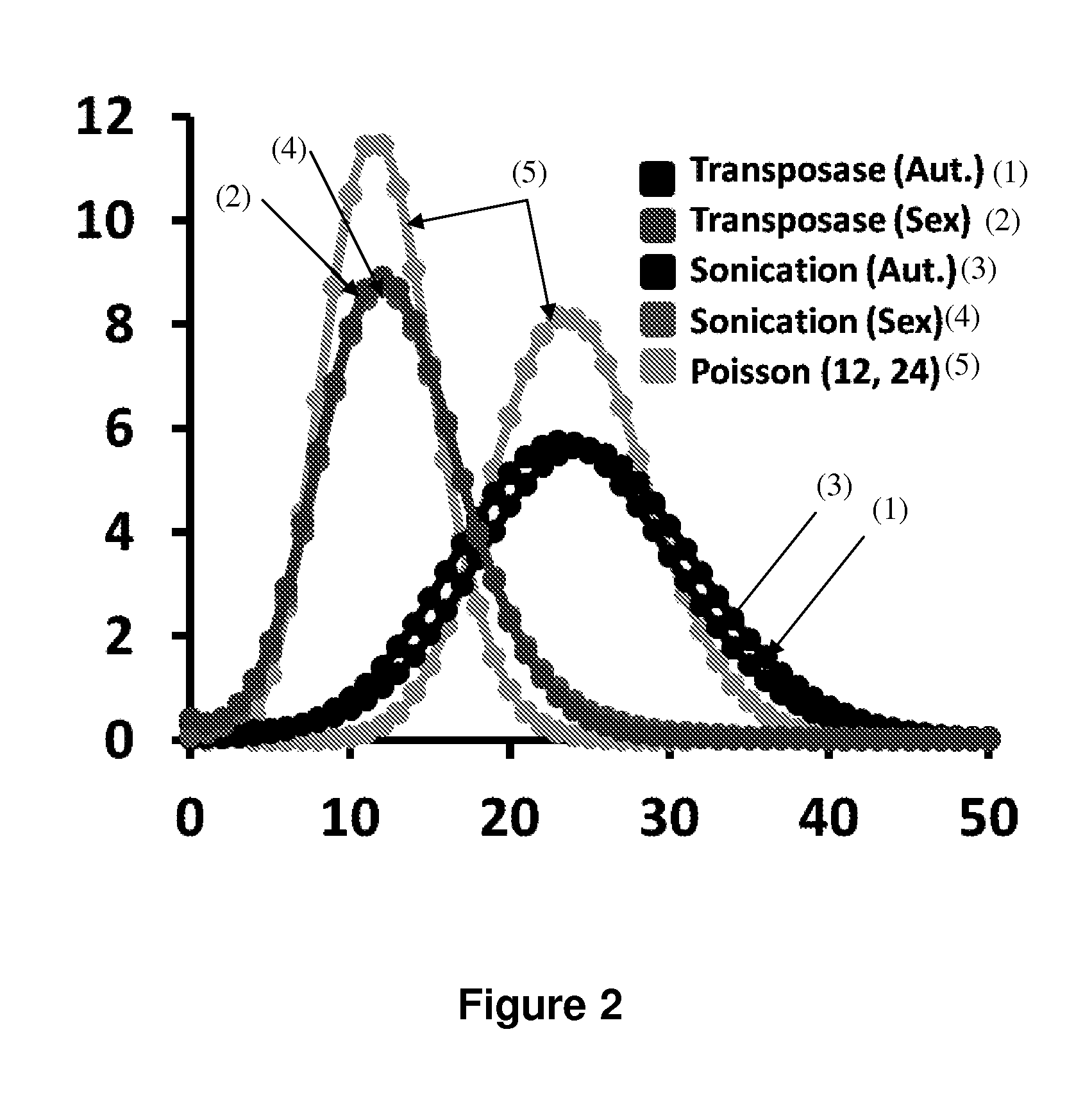
Haploidome determination by digitized transposons
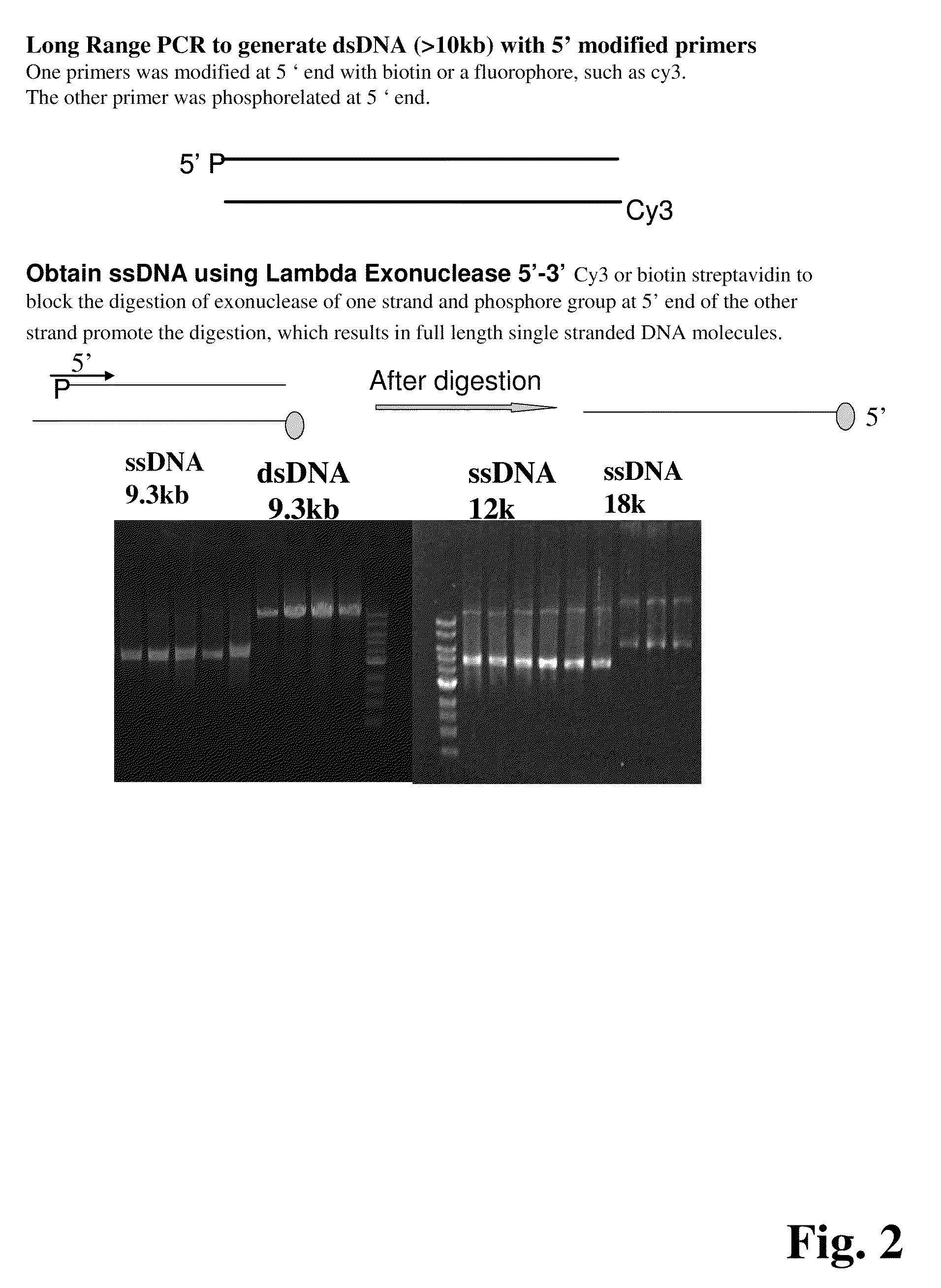
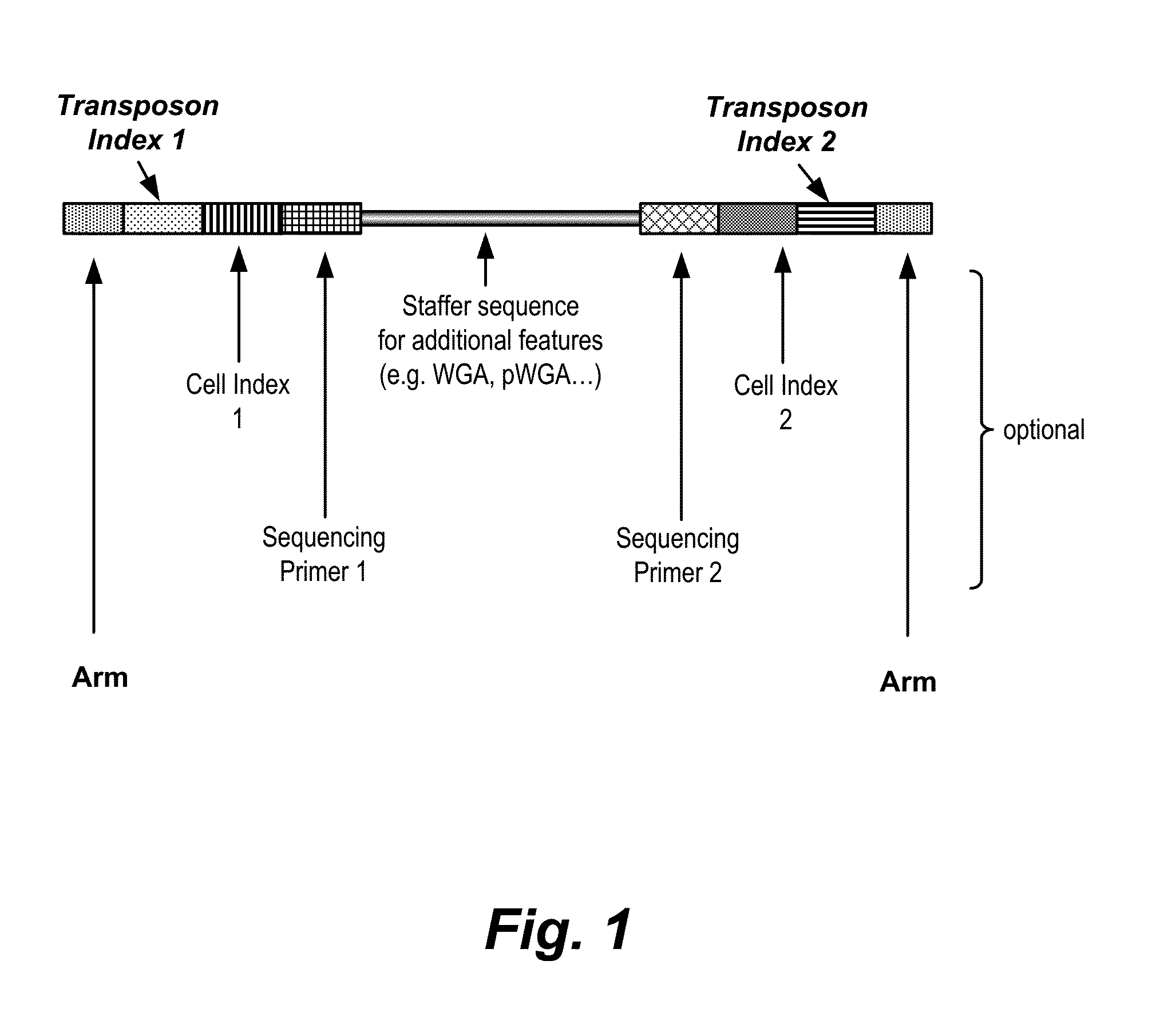
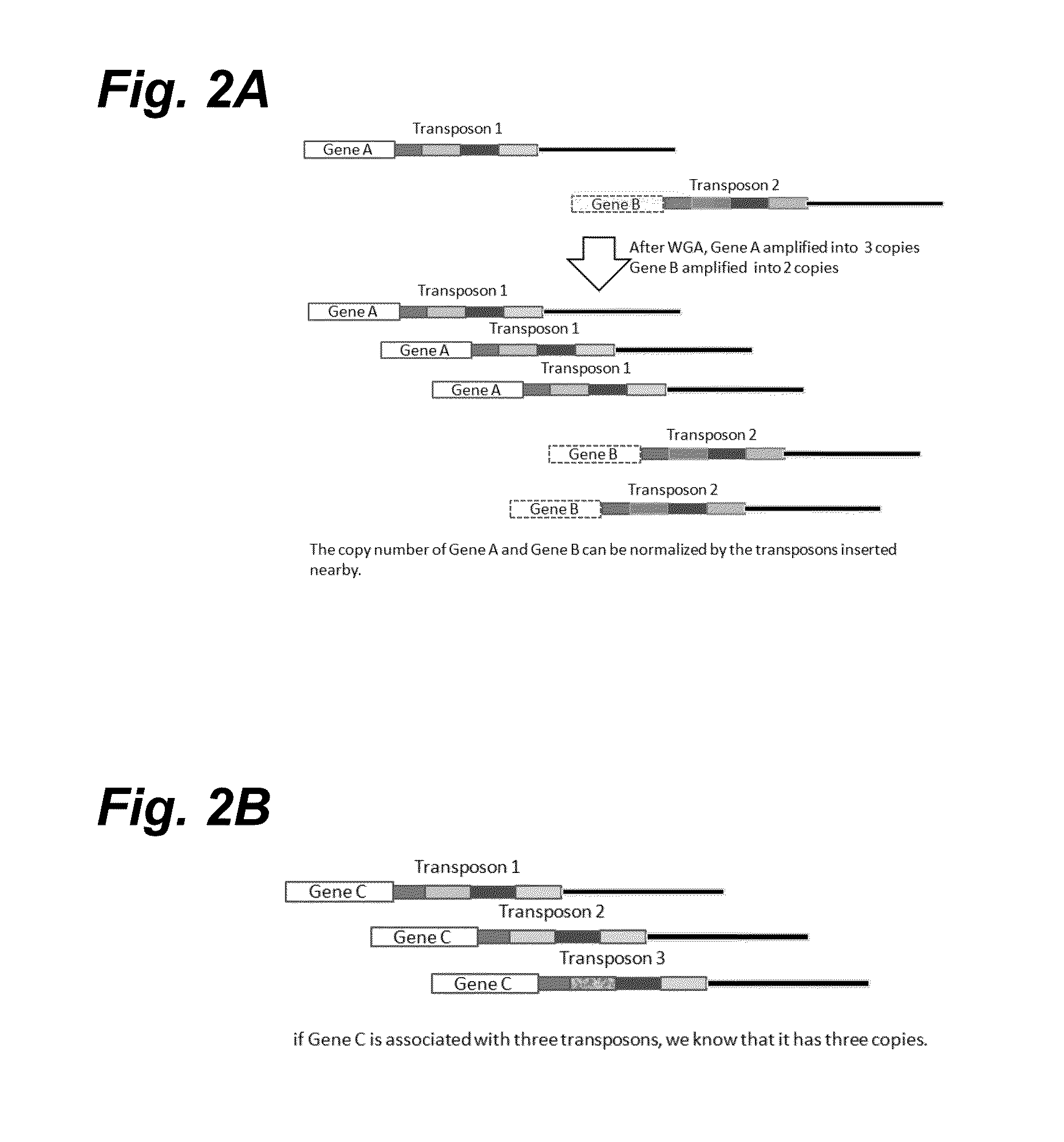
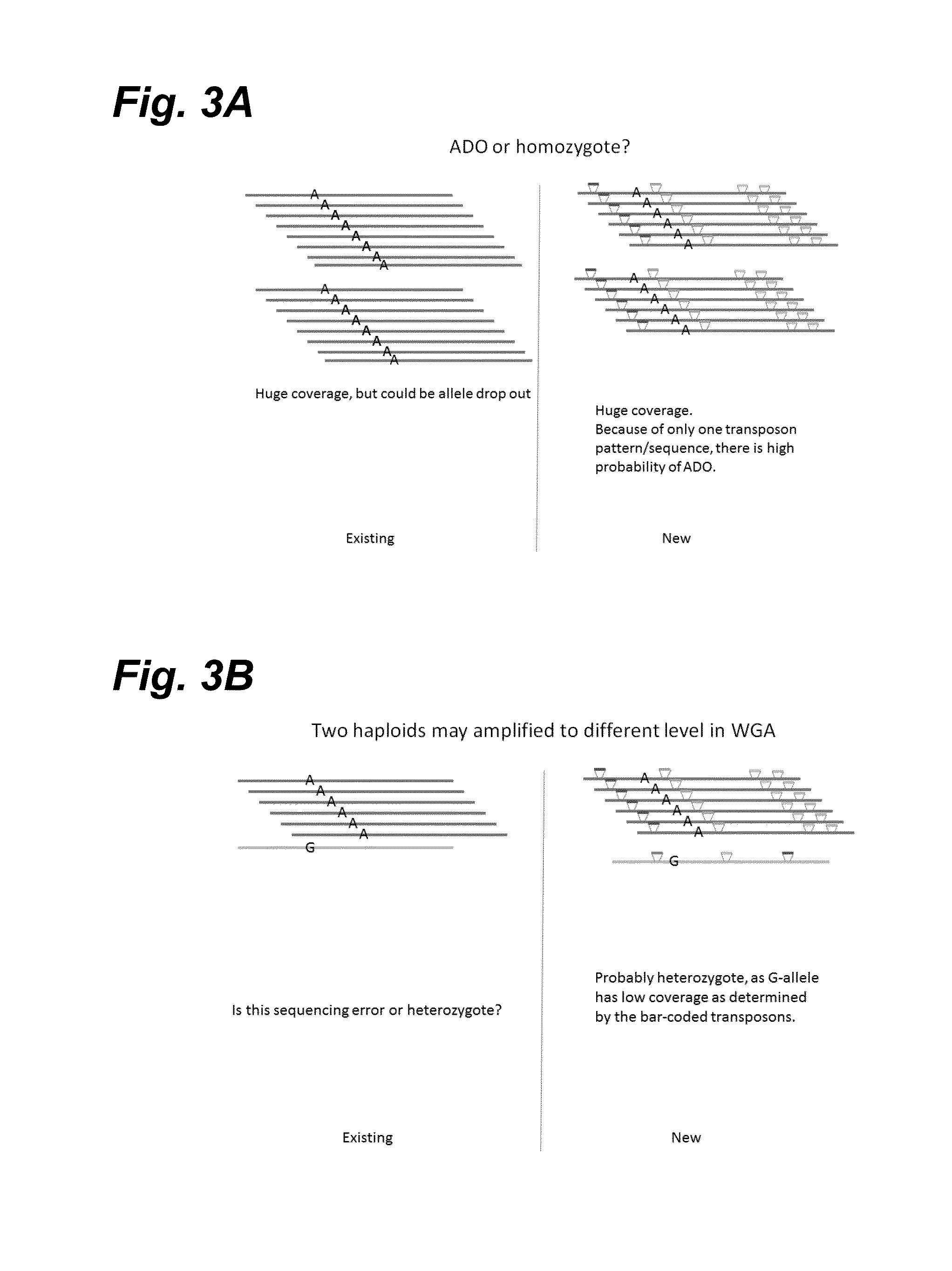
In certain embodiments, the present invention provides a way of “digitally” marking different the alleles of different chromosomes by using a transposase to insert differently barcoded transposons into genomic DNA before further analysis. According to this method, each allele becomes marked with a unique pattern of transposon barcodes. Because each unique pattern of transposon barcodes identifies a particular allele, the method facilitates determinations of ploidy and copy number variation, improves the ability to discriminate among homozygotes, heterozygotes, and patterns arising from sequencing errors, and allows loci separated by uninformative stretches of DNA to be identified as linked loci, thereby facilitating haplotype determinations. Also provided is a novel artificial transposon end that includes a barcode sequence in two or more positions that are not essential for transposition.
Owner:DIGENOMIX CORP
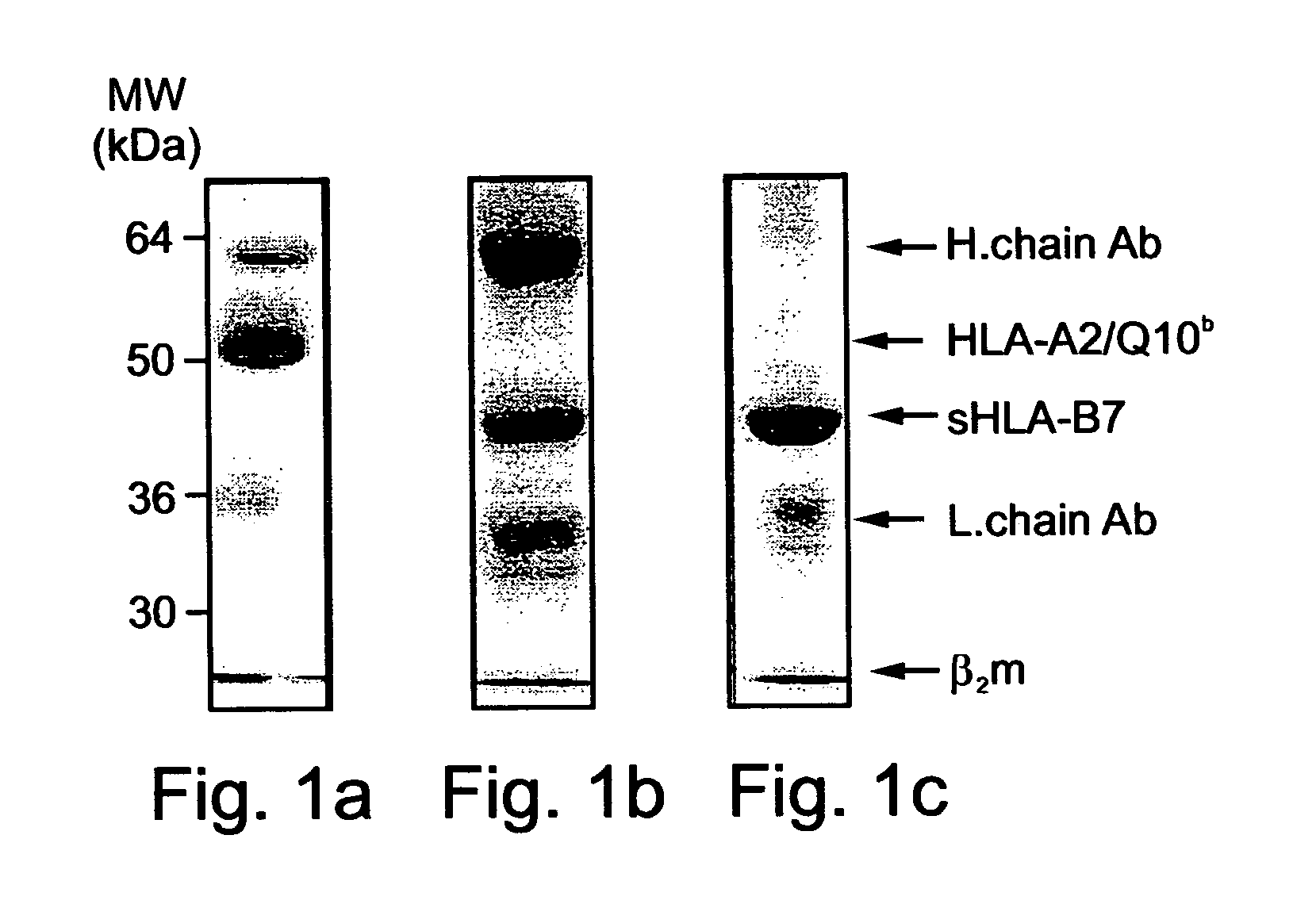
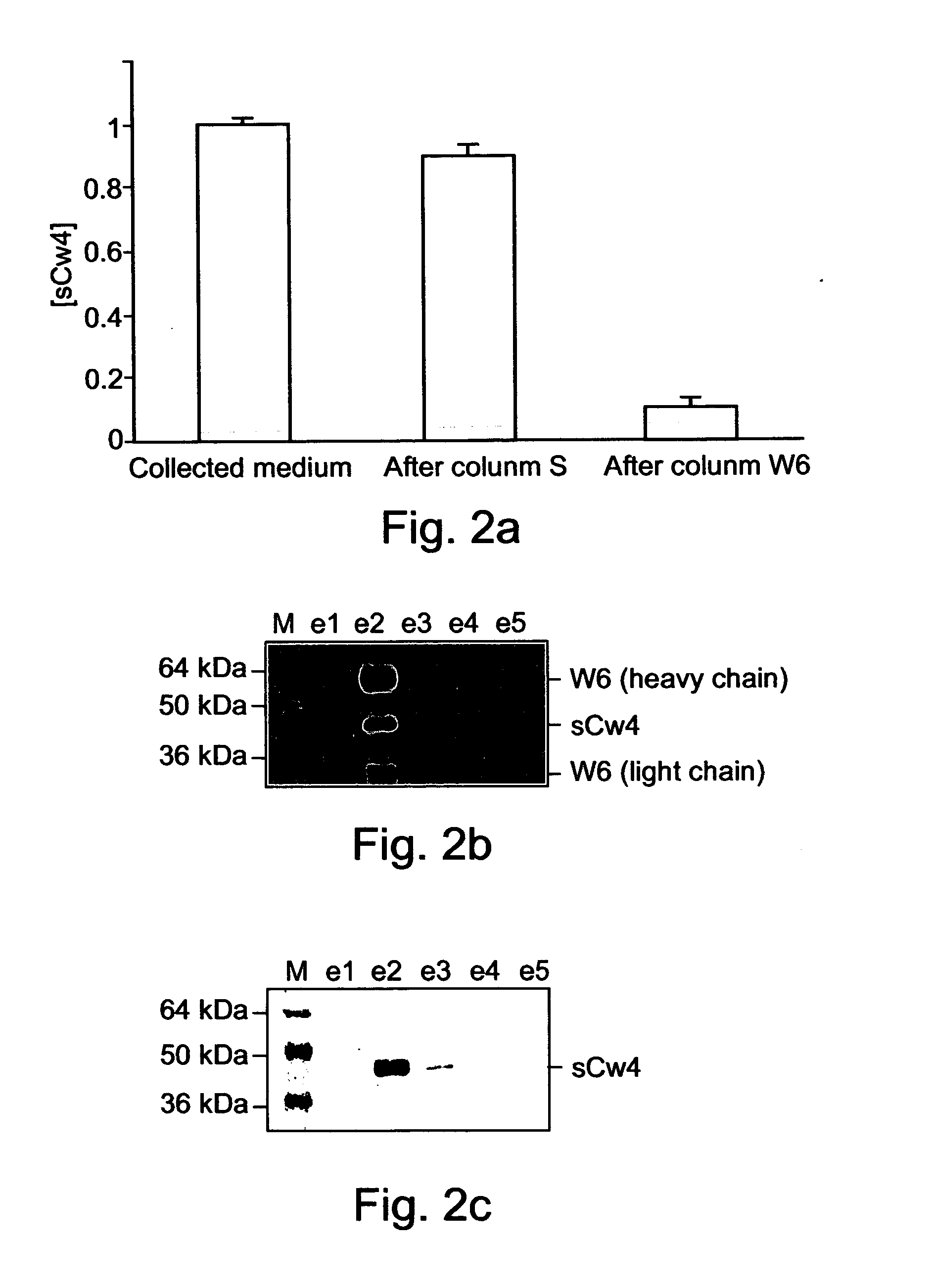
Method of identifying peptides capable of binding to MHC molecules, peptides identified thereby and their uses
A method of identifying peptides originating from a particular cell type and being capable of binding to MHC molecules of a particular haplotype is disclosed. The method comprises obtaining a cell type expressing a soluble and secreted form of the MHC molecules of the particular haplotype; collecting the soluble and secreted form of the MHC molecules of the particular haplotype; and analyzing peptides bound to the soluble and secreted form of the MHC molecules of the particular haplotype, thereby identifying the peptides originating from the particular cell type and being capable of binding to MHC molecules of the particular haplotype.
Owner:TECHNION RES & DEV FOUND LTD
Methods for obtaining and using haplotype data
InactiveUS20050191731A1Accurate predictionReduce costs and risksData visualisationBiostatisticsHaplotypeCrowds
Methods, computer program(s) and database(s) to analyze and make use of gene haplotype information. These include methods, program, and database to find and measure the frequency of haplotypes in the general population; methods, program, and database to find correlation's between an individual's haplotypes or genotypes and a clinical outcome; methods, program, and database to predict an individual's haplotypes from the individual's gen type for a gene; and methods, program, and database to predict an individual's clinical response to a treatment based on the individual's genotype or haplotype.
Owner:JUDSON RICHARD S +3
Methods and compositions to enhance plant breeding
InactiveUS20060282911A1Enhanced agronomicEnhanced transgenic traitBiocideDead animal preservationGermplasmHaplotype
Owner:MONSANTO TECH LLC
Techniques for Determining Haplotype by Population Genotype and Sequence Data
ActiveUS20140045705A1Better phasingImprove accuracyLibrary member identificationProteomicsHaplotypeSequencing data
A novel phasing algorithm harnesses sequencing read information from next generation sequencing technologies to guide and improve local haplotype reconstruction from genotypes. Techniques include determining correlated occurrences of single nucleotide polymorphisms (SNPs) in genes of a population of individuals. A plurality of sequences of nucleotide bases in one or more individuals from the populations of individuals is determined based on ultra-high throughput sequencing of a sample from the one or more individuals. Haplotypes included in the population of individuals are determined based on both the correlated occurrences and the plurality of sequences. The inclusion of paired end read data is especially advantageous for the phasing of rare variants, including singletons.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Haplotying of HLA loci with ultra-deep shotgun sequencing
ActiveUS20140206547A1Minimize biasEfficient reconstructionMicrobiological testing/measurementLibrary member identificationHaploidisationHaplotype
Methods are provided to determine the entire genomic region of a particular HLA locus including both intron and exons. The resultant consensus sequences provides linkage information between different exons, and produces the unique sequence from each of the two genes from the individual sample being typed. The sequence information in intron regions along with the exon sequences provides an accurate HLA haplotype.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
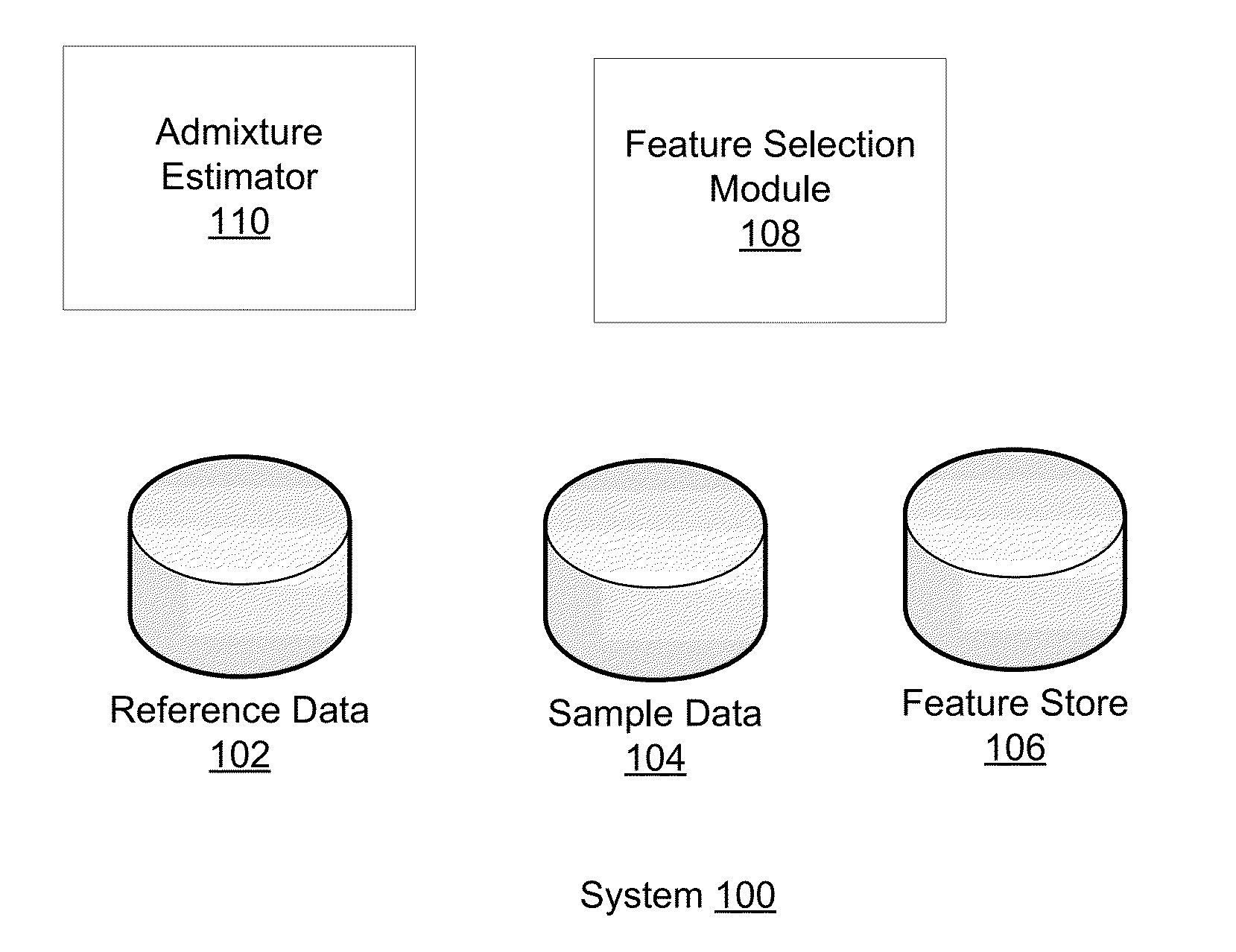
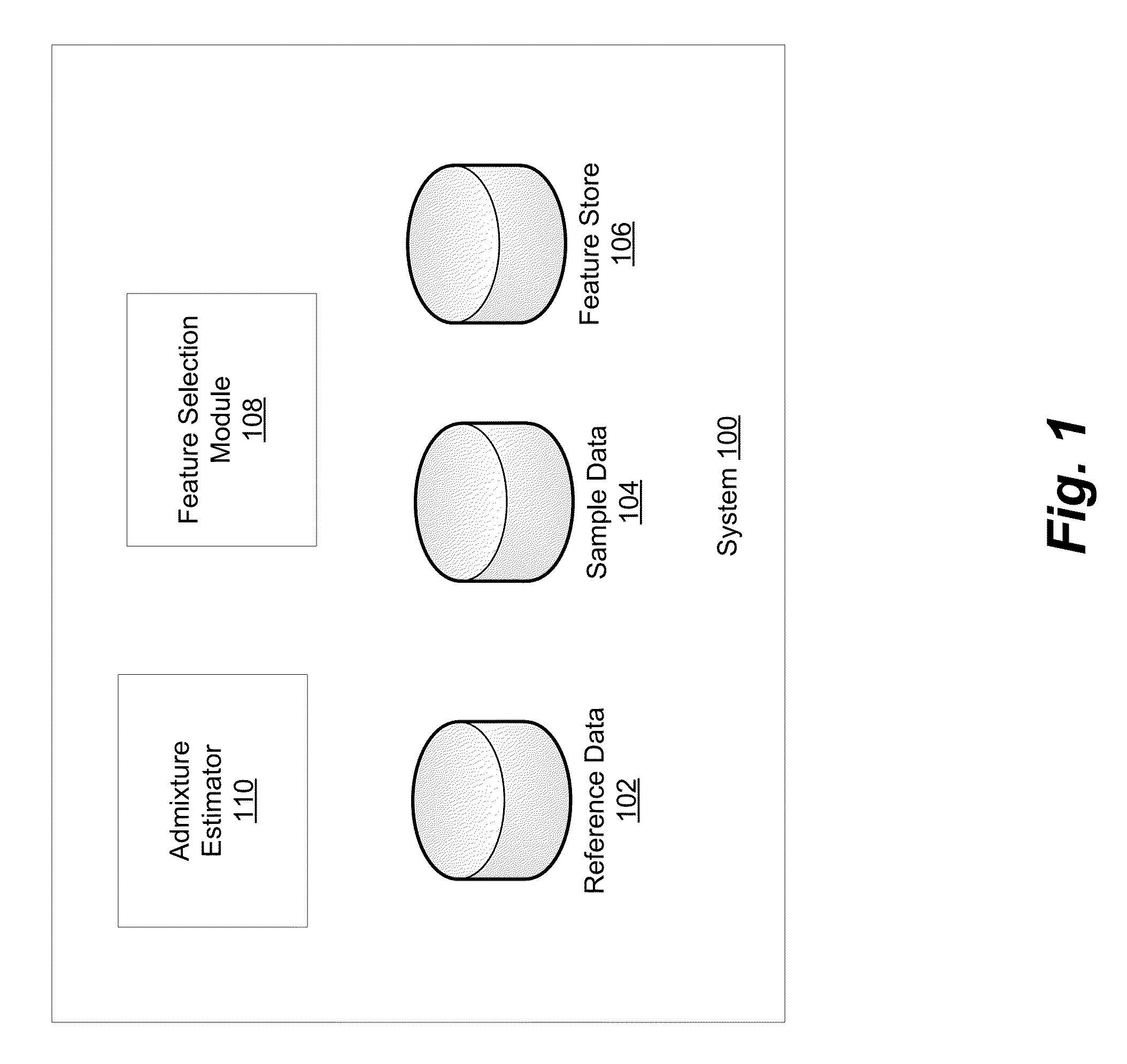
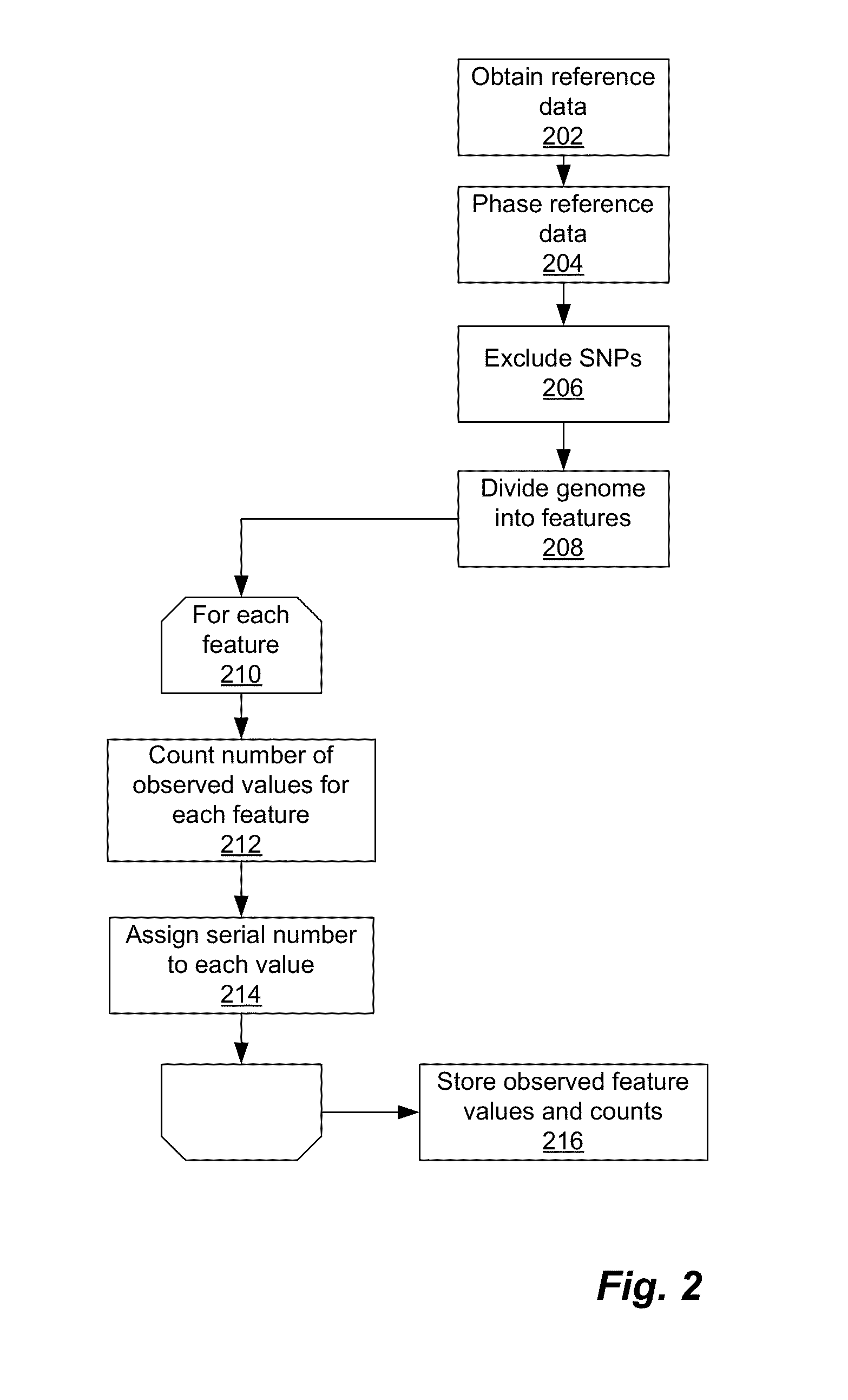
Using Haplotypes to Infer Ancestral Origins for Recently Admixed Individuals
InactiveUS20140067355A1Analogue computers for chemical processesProteomicsPattern recognitionProgenitor
Phased haplotype features are used to infer an individual's ancestry. Reference genomic data is obtained for individuals of known ancestral origin. Haplotype features are identified based on consecutive SNPs from each individual. Sample genomic data is obtained for an individual of unknown ancestral origin. The data is phased and divided into features analogous to the features in the reference data. An admixture estimator then performs an admixture estimation based on the observed feature values in the sample data and the reference data. The estimation indicates a contribution of each of the known populations to the genome of the sample individual.
Owner:ANCESTRY COM DNA
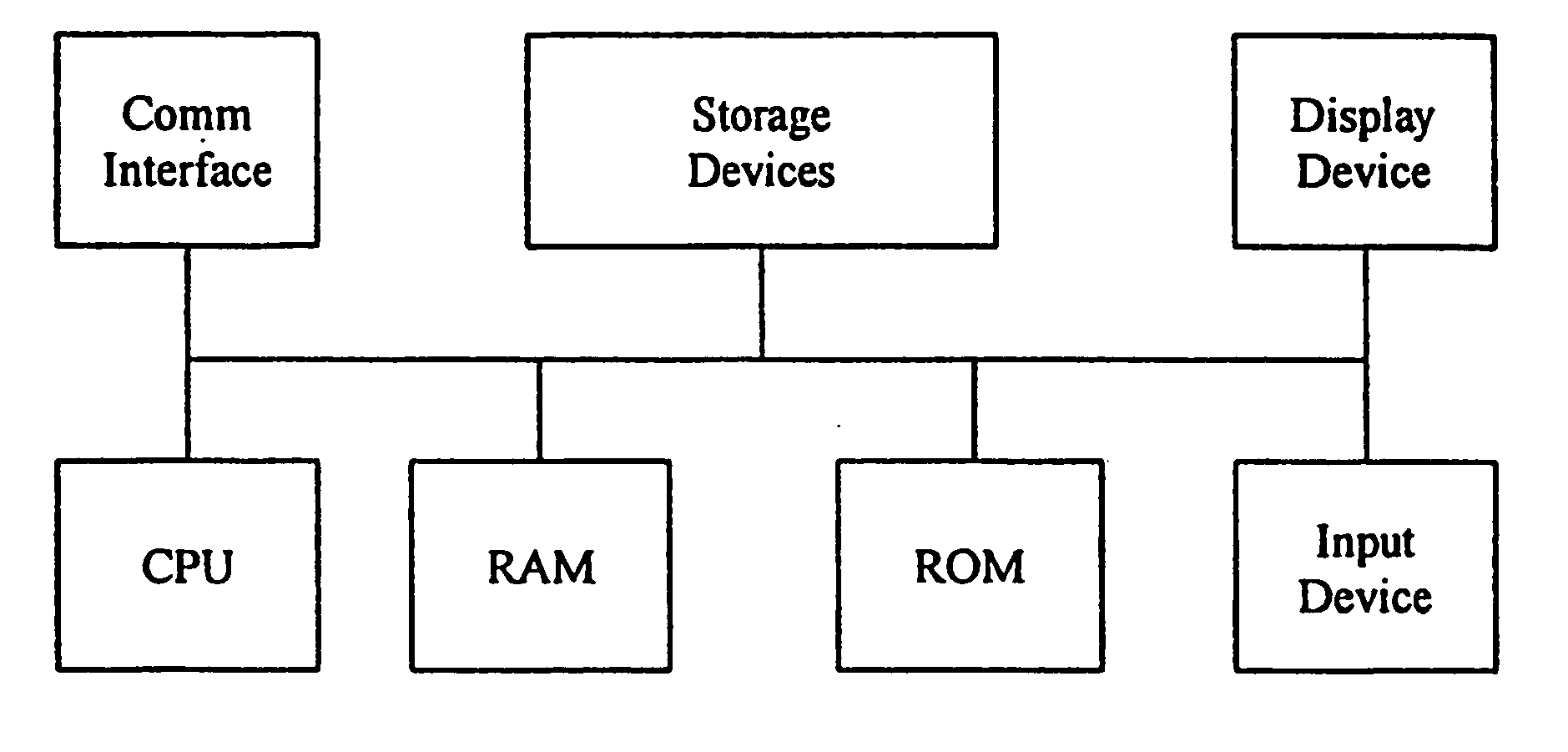
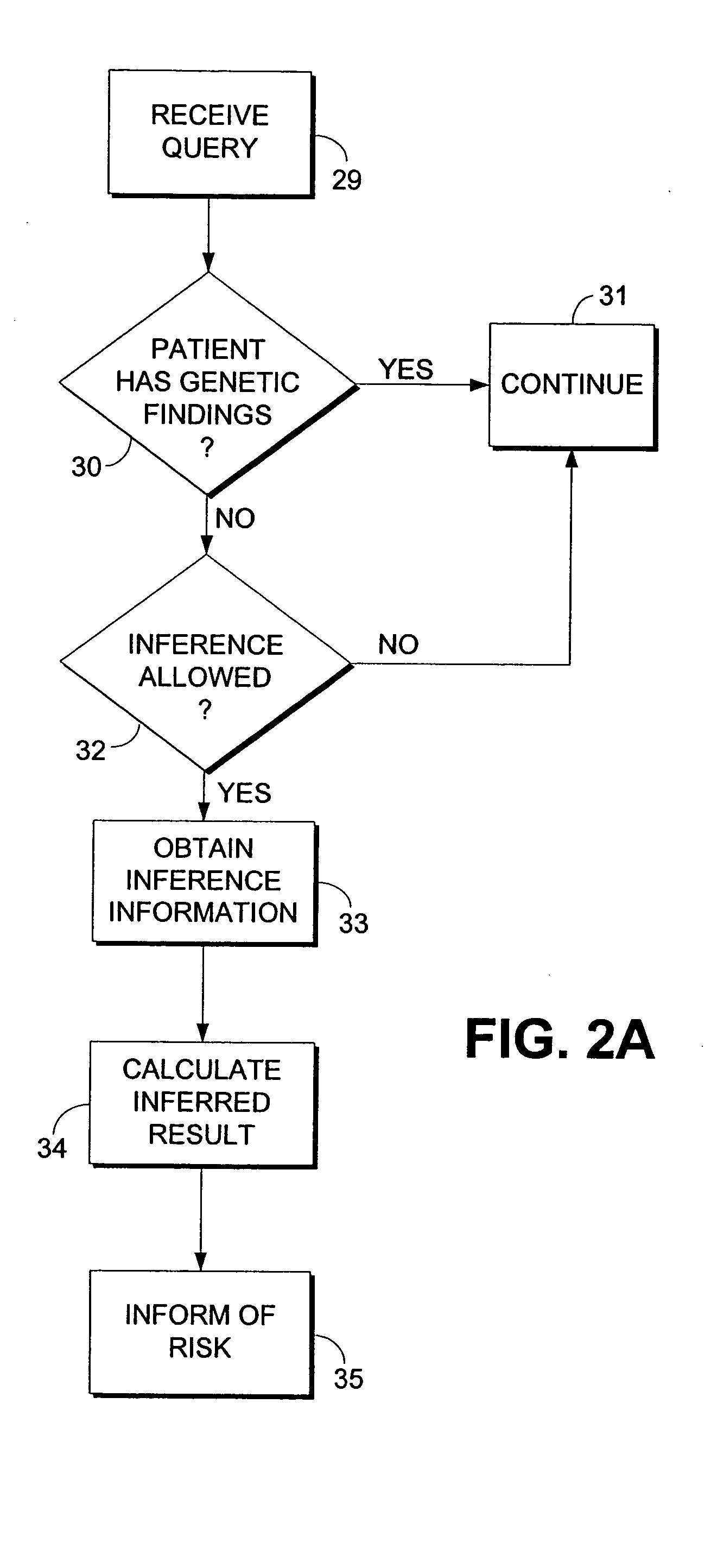
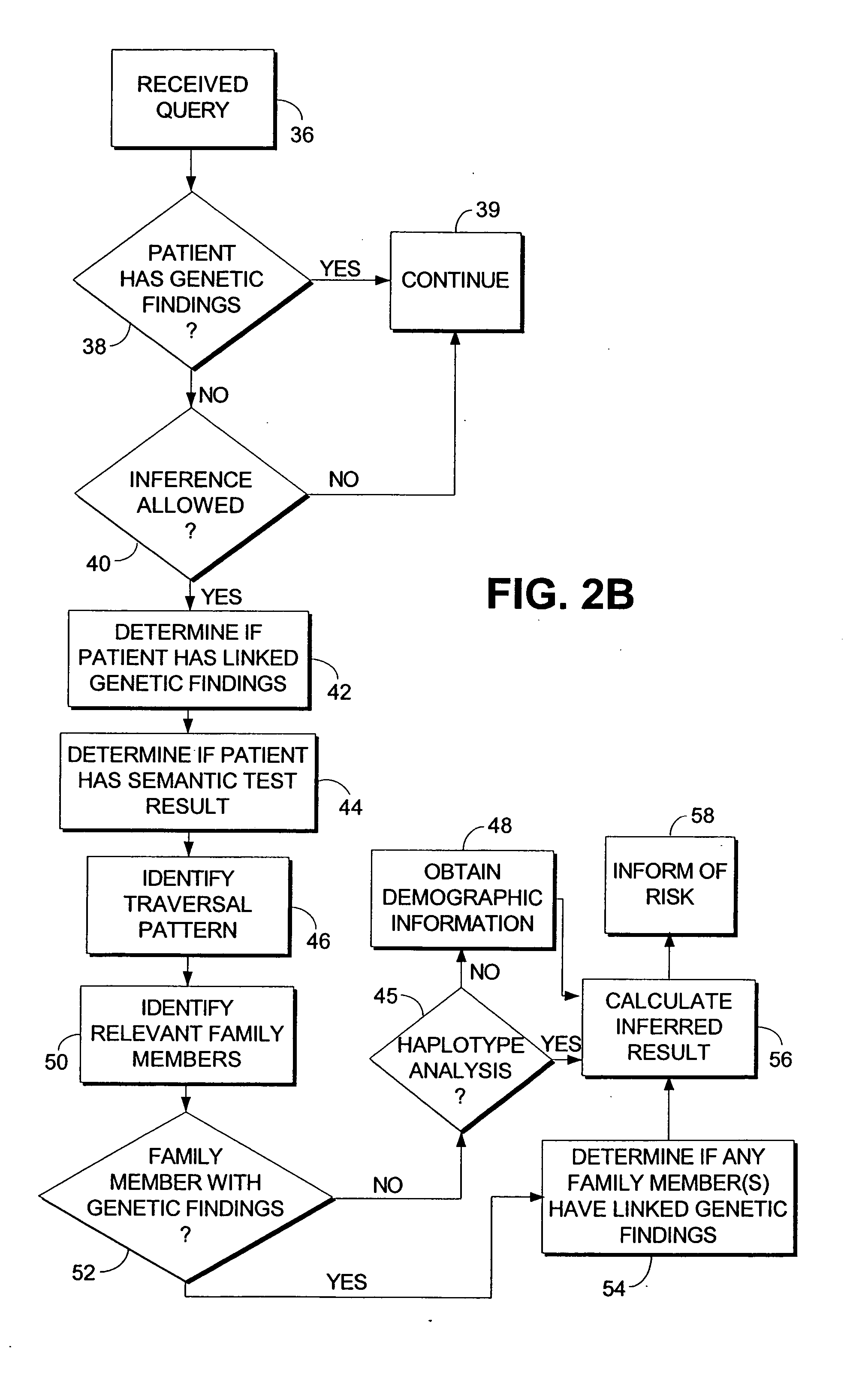
Computerized method and system for inferring genetic findings for a patient
A method and system in a computing environment for inferring genetic findings for a patient is provided. The method includes receiving a request for genetic findings for a person from another application or a user. The method further includes inquiring as to whether the person has the genetic findings. If not, the method automatically provides inferred genetic findings for the person. The inferred genetic findings are calculated using genetic findings for family members of the patient, linkage analysis, haplotype analysis, semantic test results for the person and / or population genetics information.
Owner:CERNER INNOVATION
Diagnostics and therapeutics for diseases associated with an IL-1 inflammatory haplotype
Methods and kits for determining whether a subject has or is predisposed to developing a disease which is associated with IL-1 polymorphisms and assays for identifying therapeutics for treating and / or preventing the development of these diseases are provided.
Owner:UNIV OF SHEFFIELD +1
Trio-based phasing using a dynamic Bayesian network
Performing trio-based phasing includes: obtaining a set of preliminary phased haplotype data of an individual; establishing a dynamic Bayesian network based at least in part on the set of preliminary phased haplotype data of the individual and phased haplotype data of at least one parent of the individual; and determining, based on the dynamic Bayesian network, a set of refined haplotype data of the individual.
Owner:23ANDME
Size-based genomic analysis
Systems, methods, and apparatuses for performing a prenatal diagnosis of a sequence imbalance are provided. A shift (e.g. to a smaller size distribution) can signify an imbalance in certain circumstances. For example, a size distribution of fragments of nucleic acids from an at-risk chromosome can be used to determine a fetal chromosomal aneuploidy. A size ranking of different chromosomes can be used to determine changes of a rank of an at-risk chromosome from an expected ranking. Also, a difference between a statistical size value for one chromosome can be compared to a statistical size value of another chromosome to identify a significant shift in size. A genotype and haplotype of the fetus may also be determined using a size distribution to determine whether a sequence imbalance occurs in a maternal sample relative to a genotypes or haplotype of the mother, thereby providing a genotype or haplotype of the fetus.
Owner:THE CHINESE UNIVERSITY OF HONG KONG
Method for molecular genealogical research
A genealogical research and record keeping system and method for identifying commonalities in haplotypes and other genetic characteristics of two or more individual members of a biological sample. Chromosomal fragments identical by descent identify family ties between siblings, parents and children and ancestors and progeny across many generations. It is particularly useful in corroborating and improving the accuracy of genealogical data, and identifying previously unknown genetic relationships.
Owner:ANCESTRY COM DNA
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com