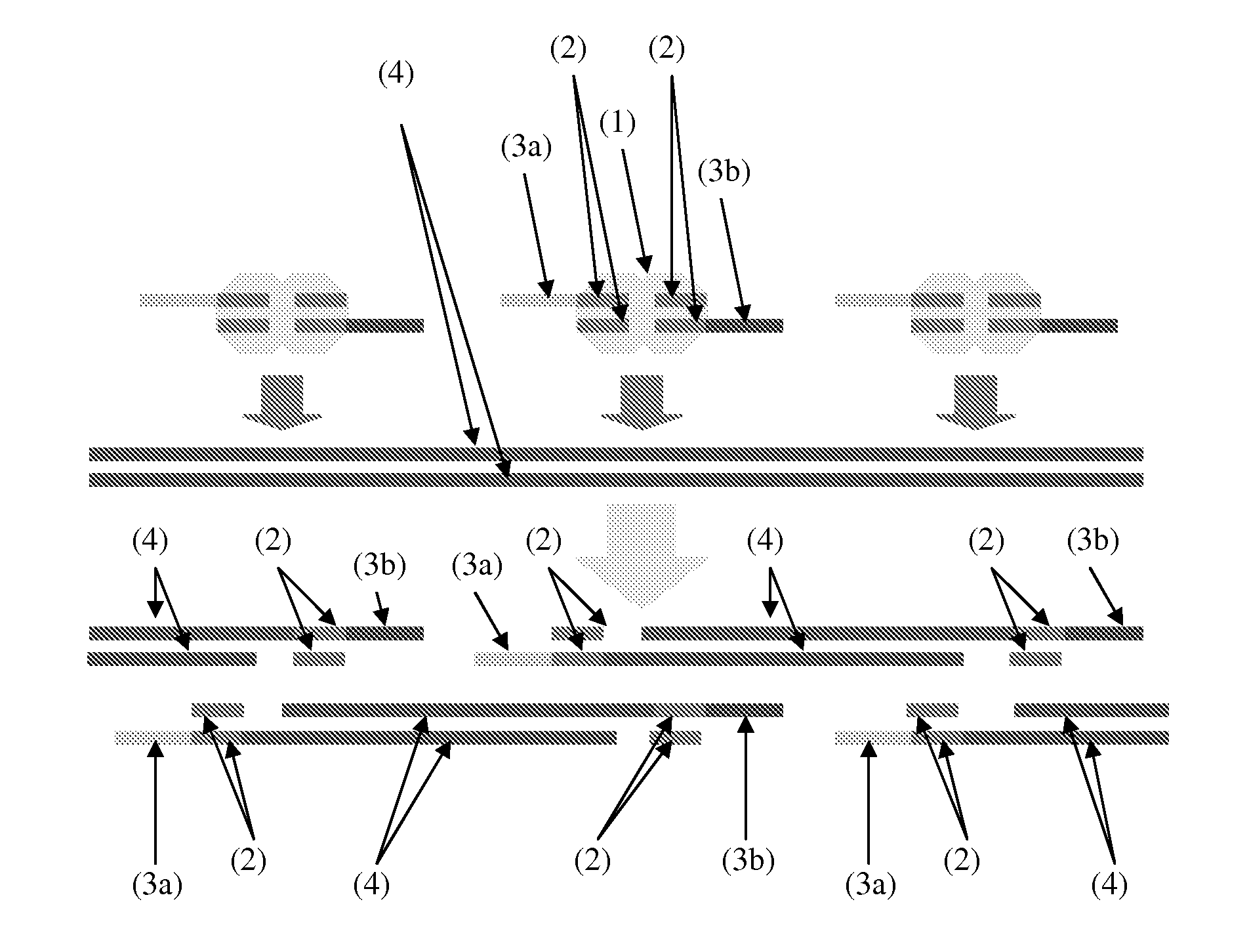
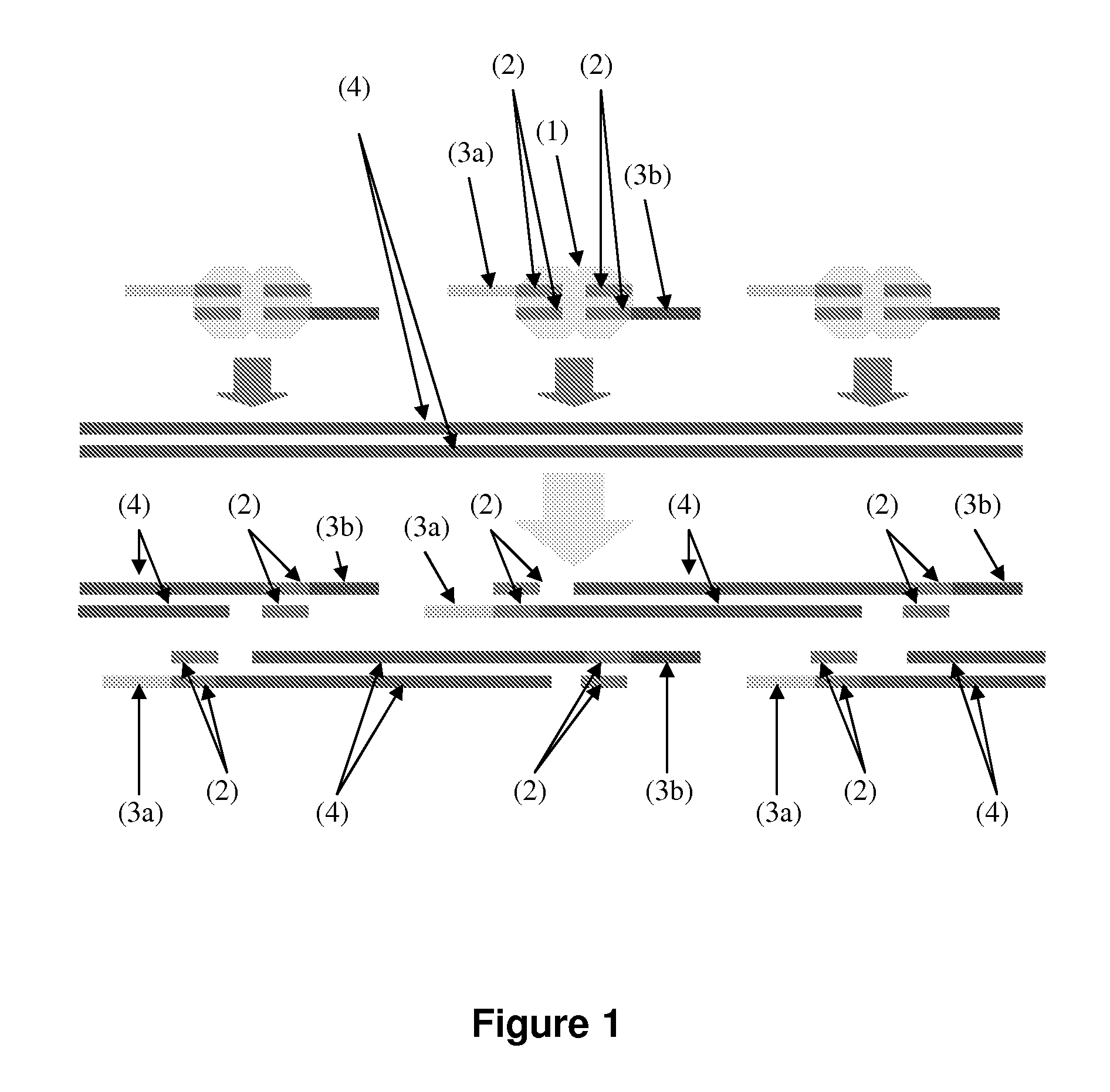
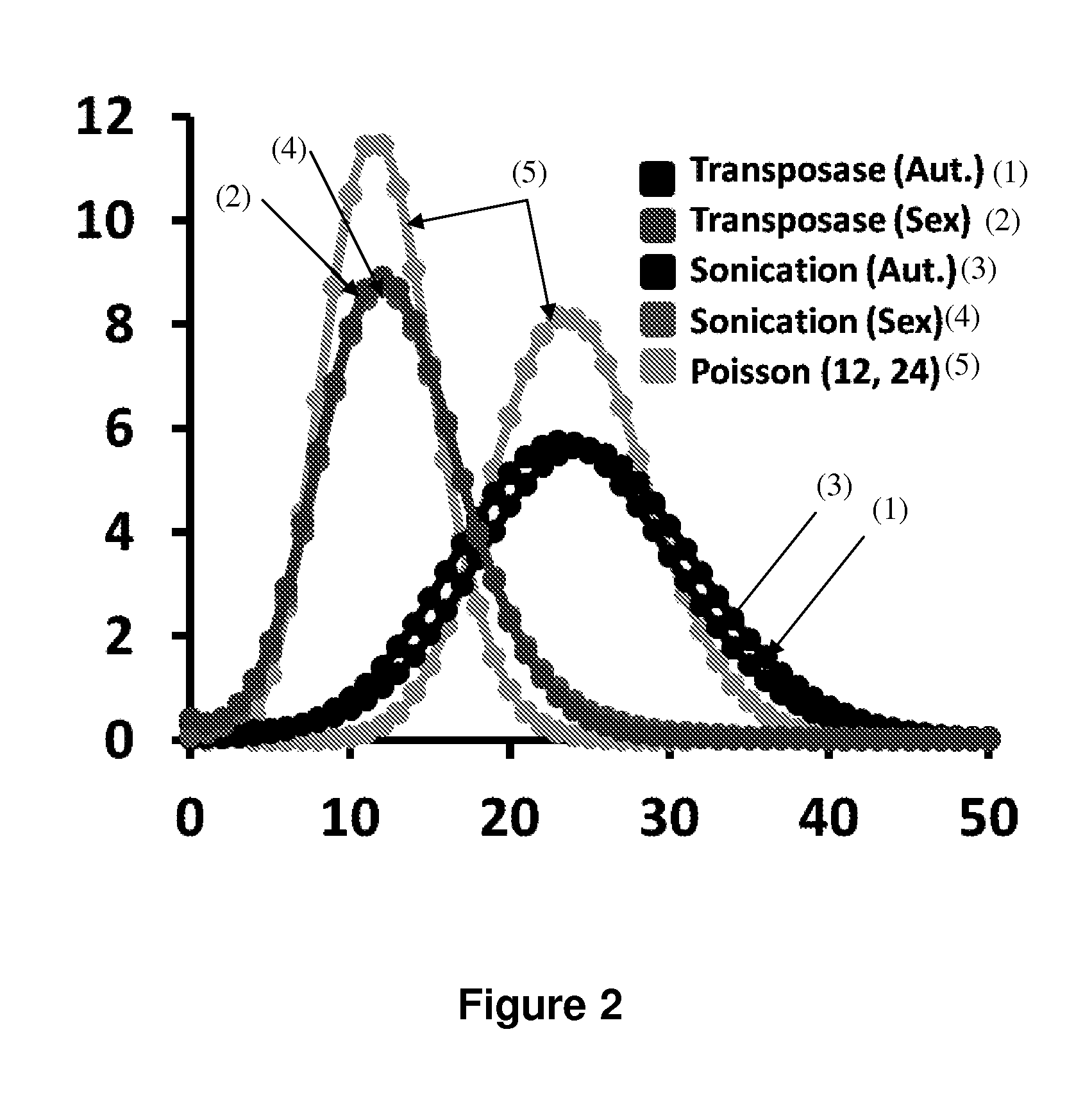
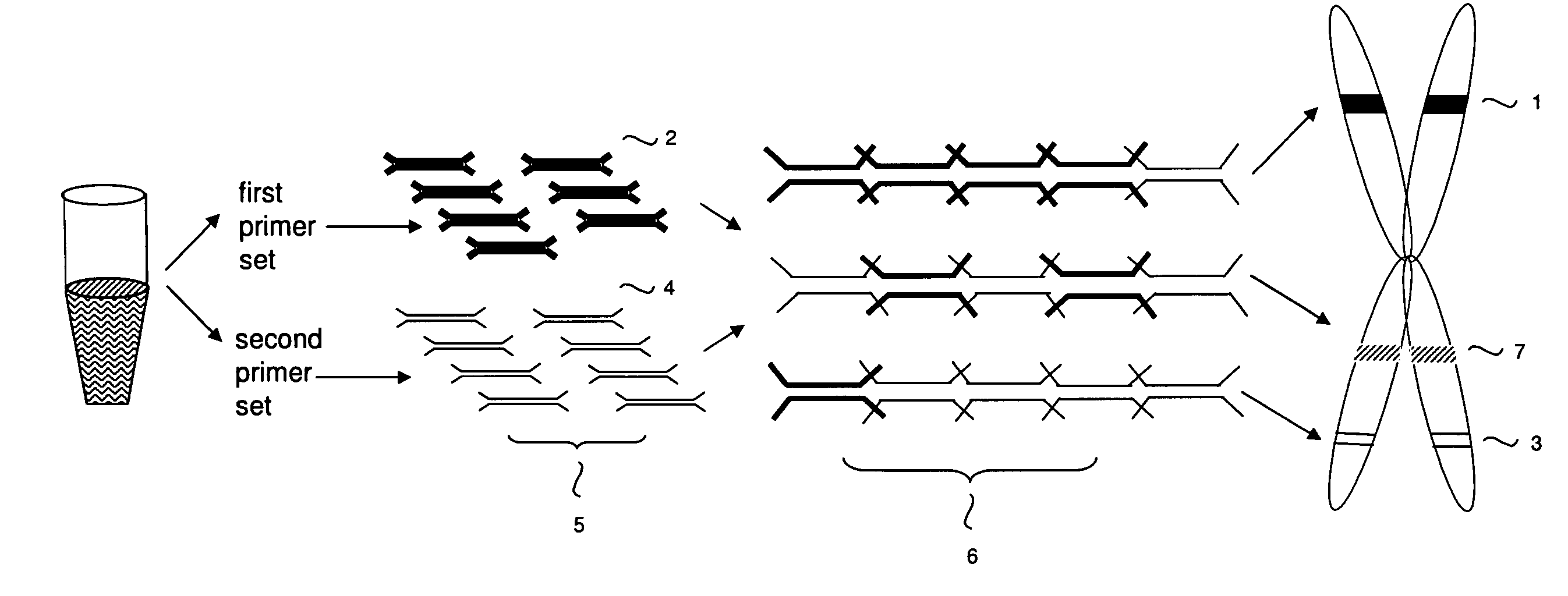
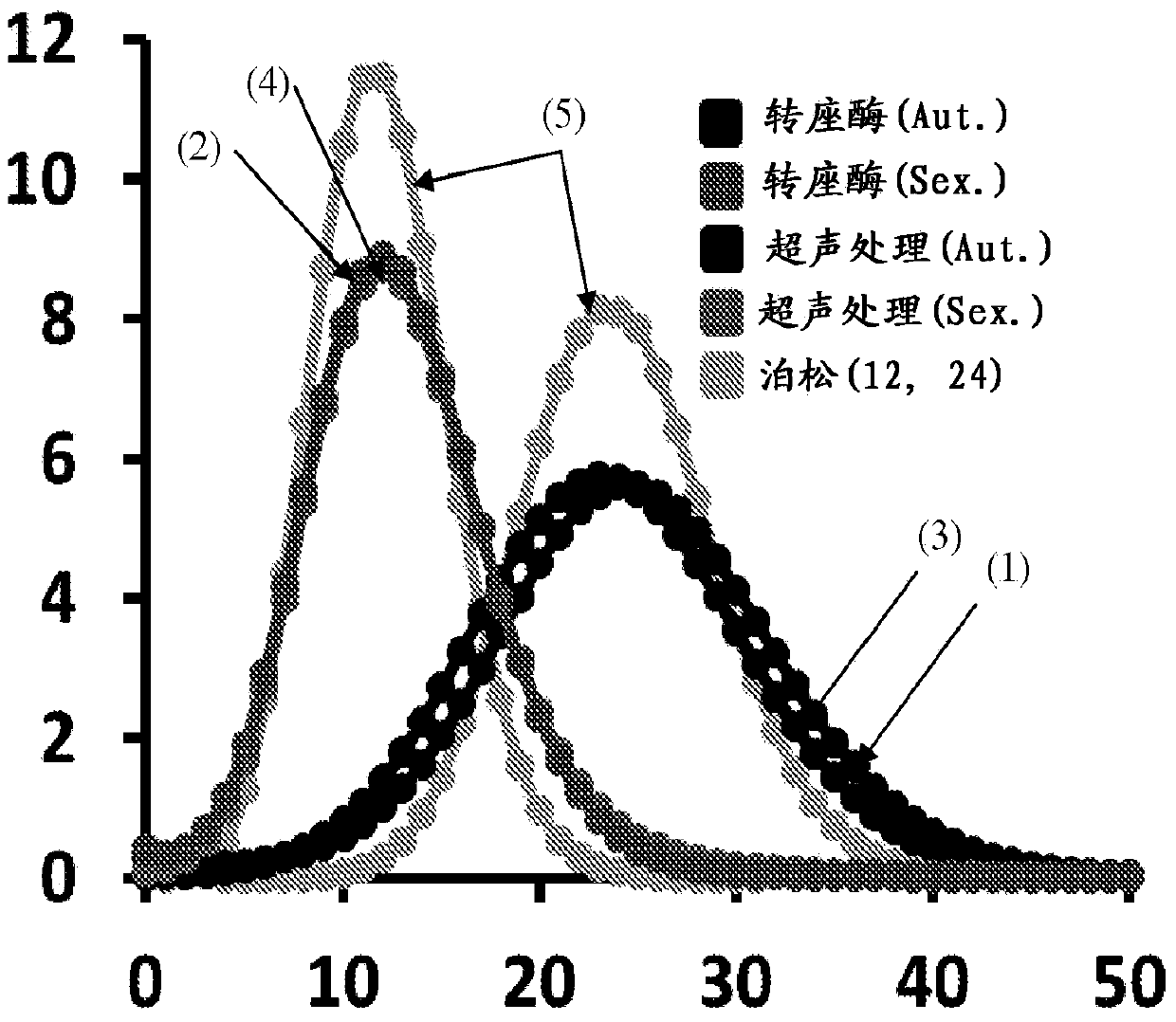
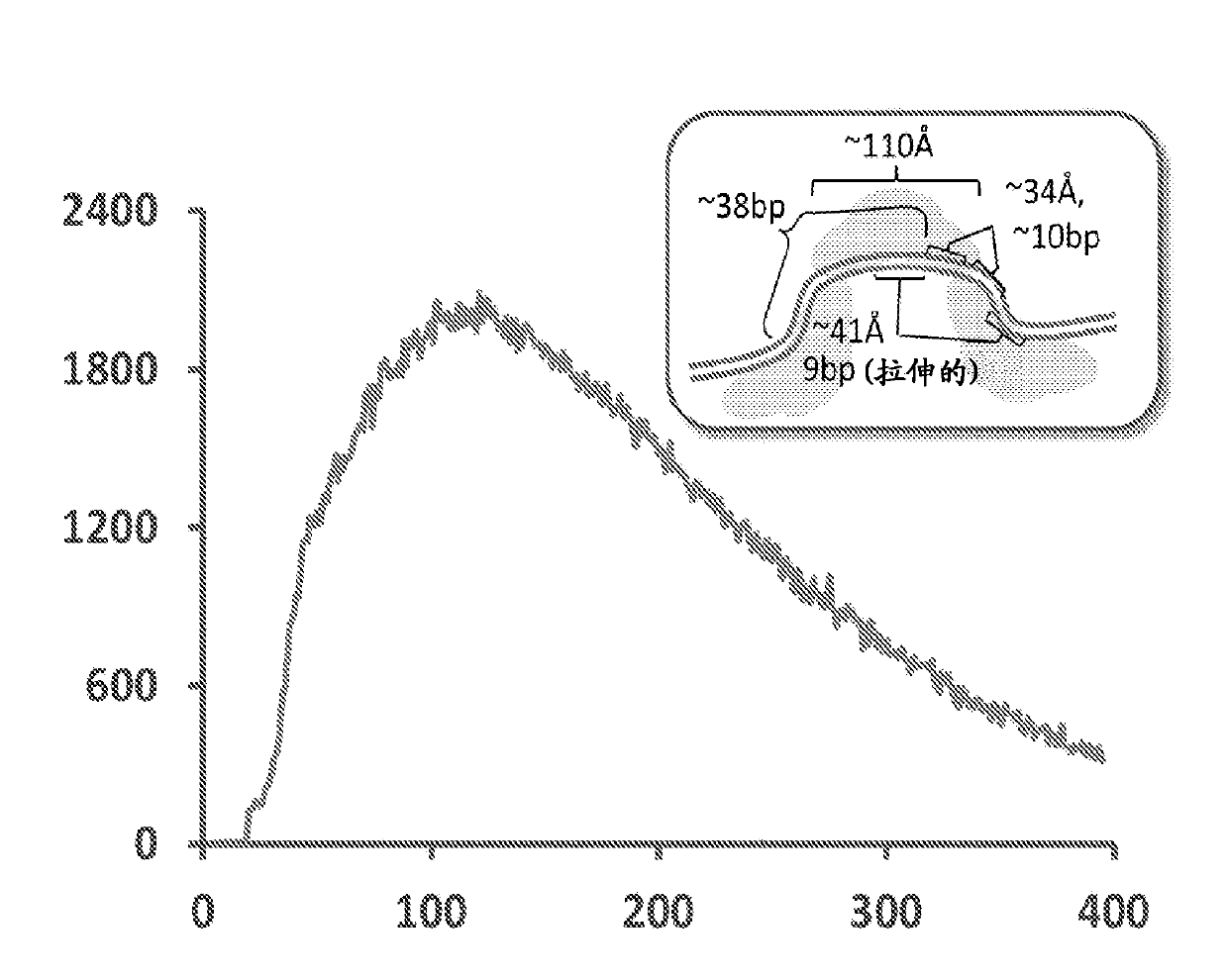
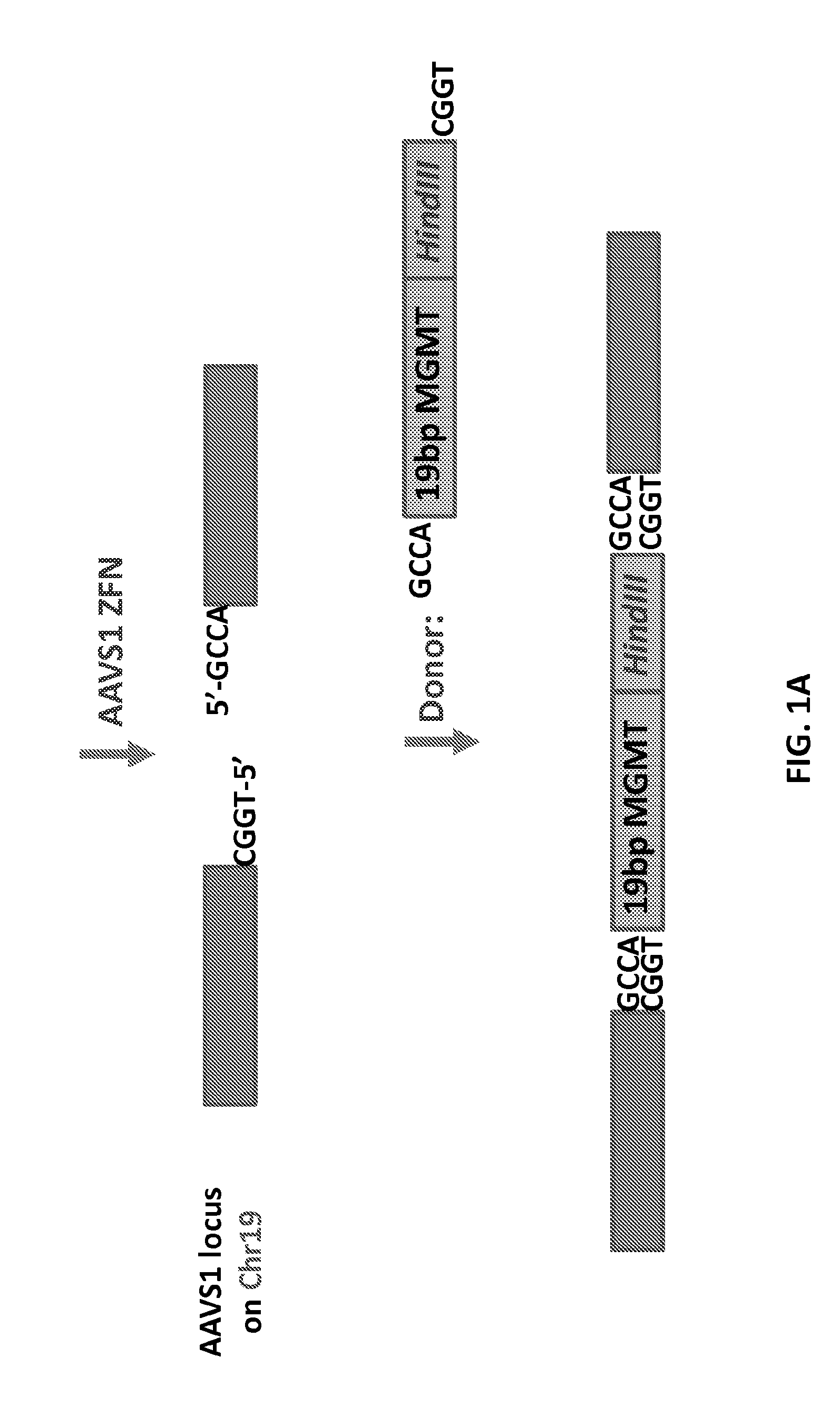
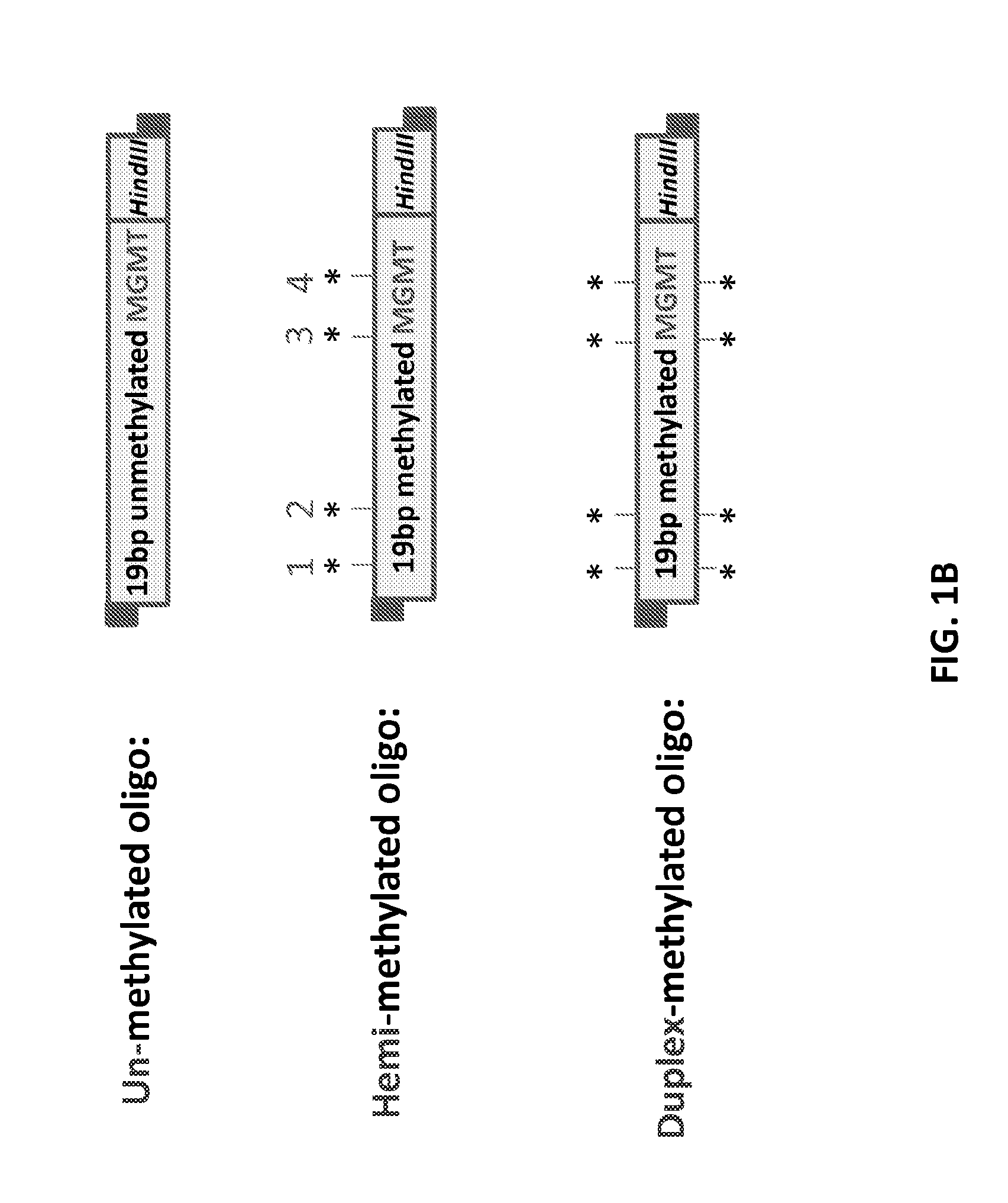
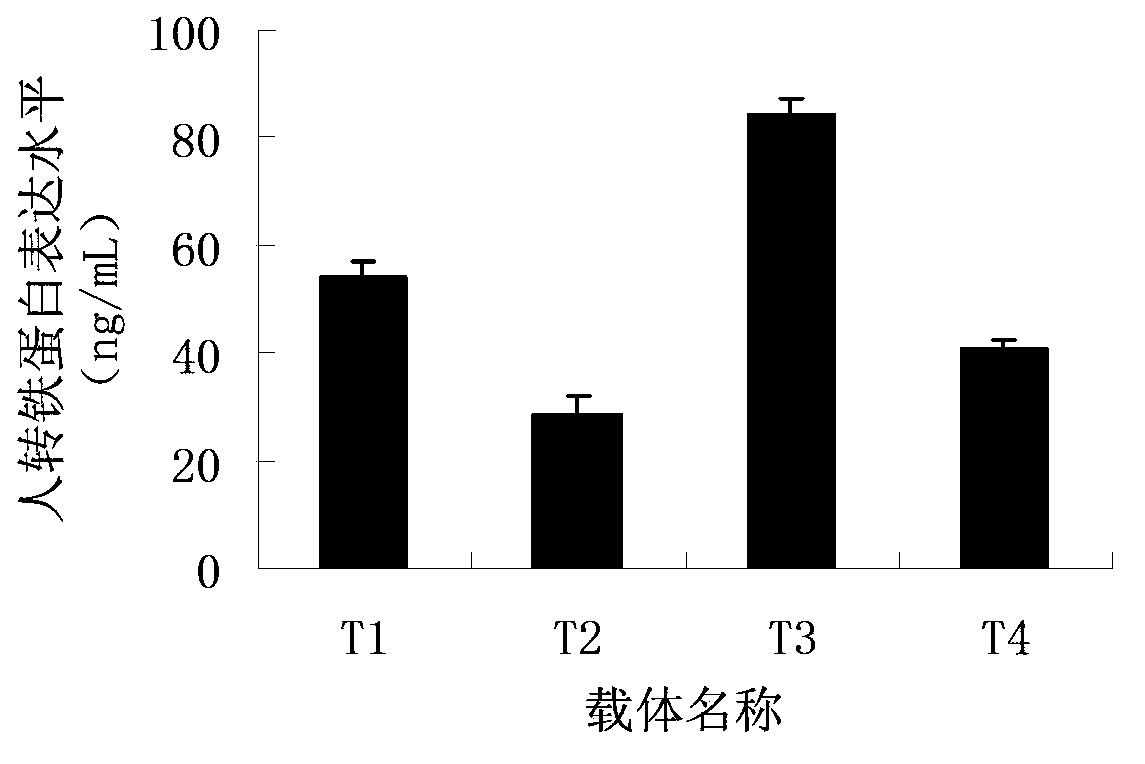
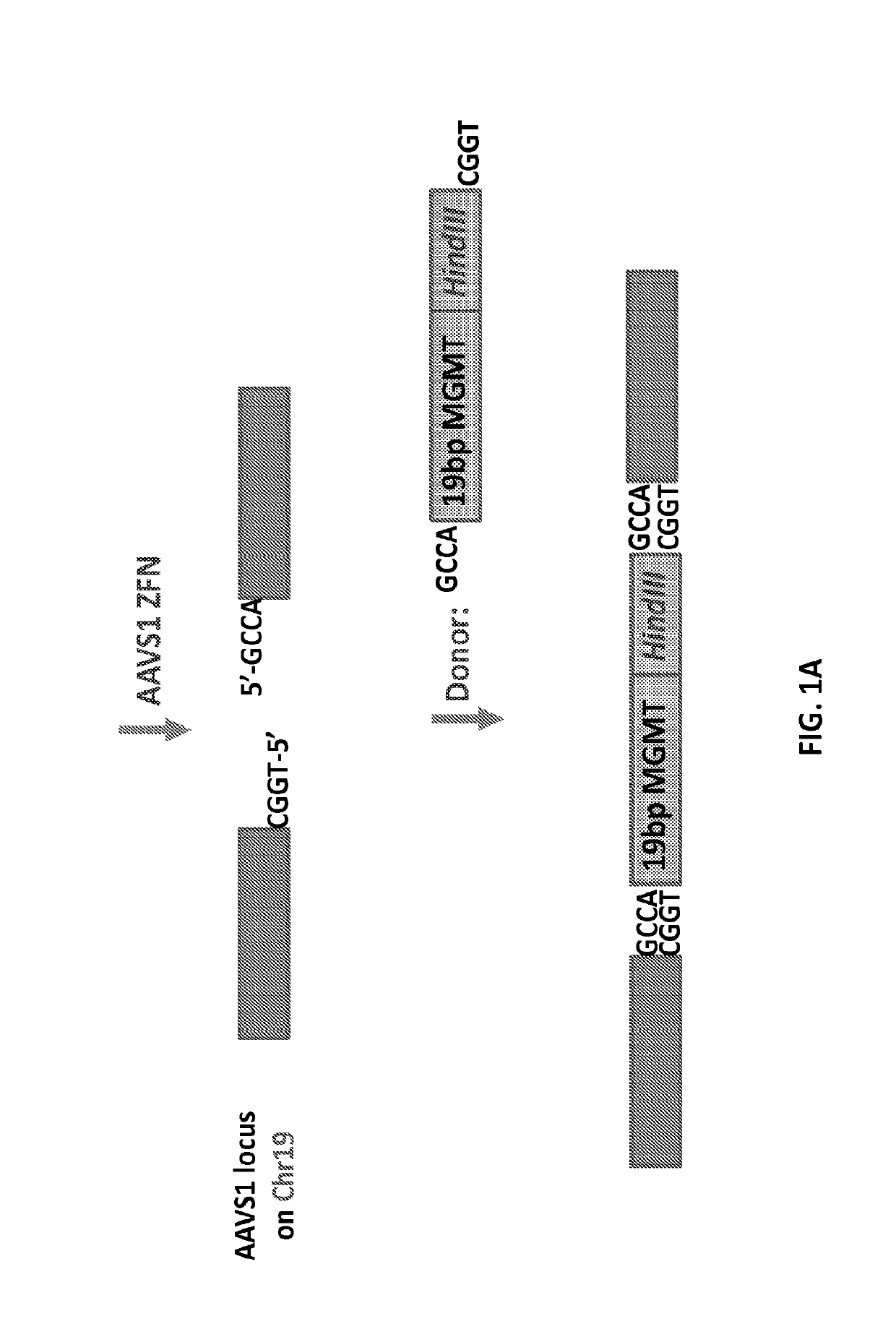
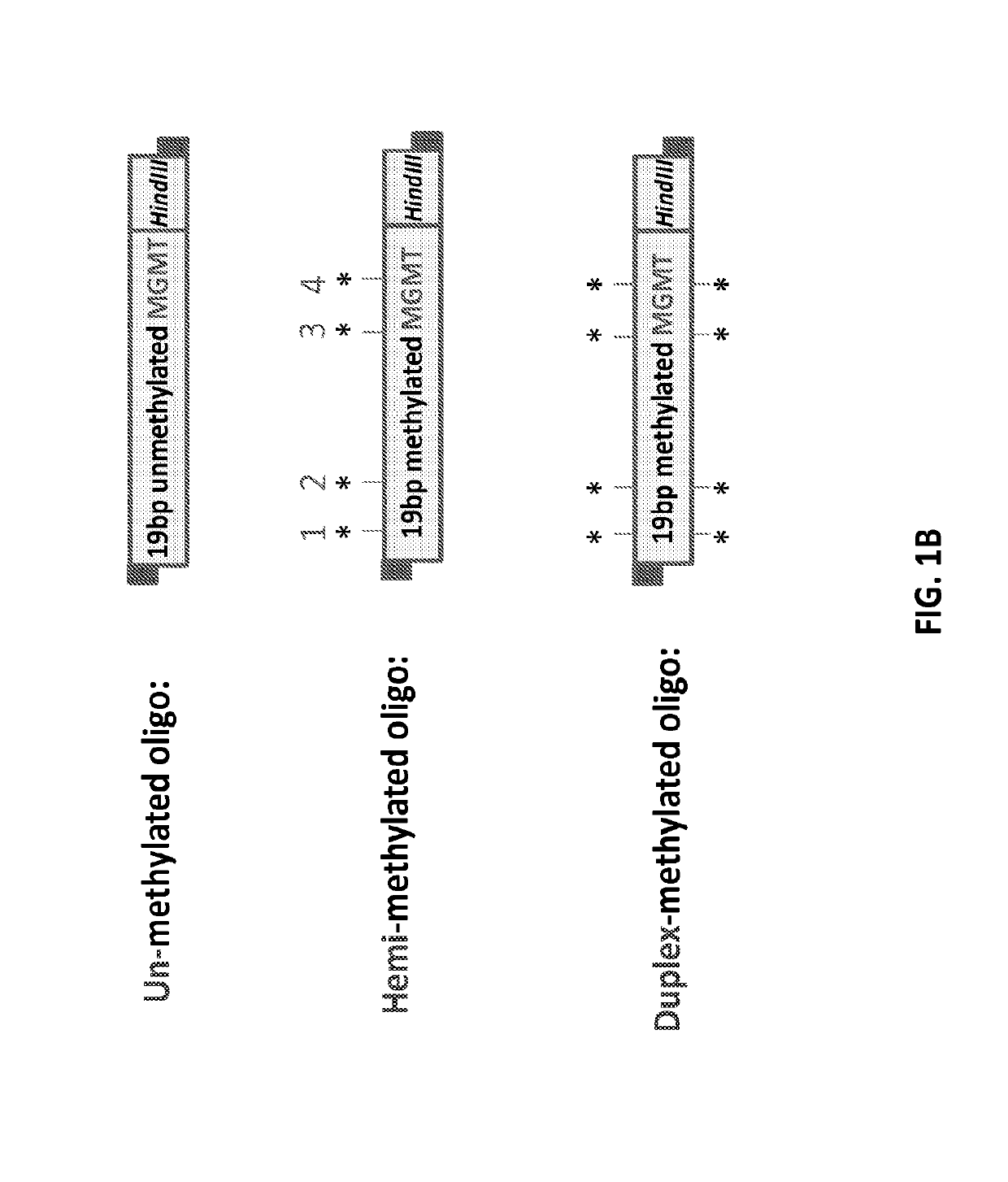
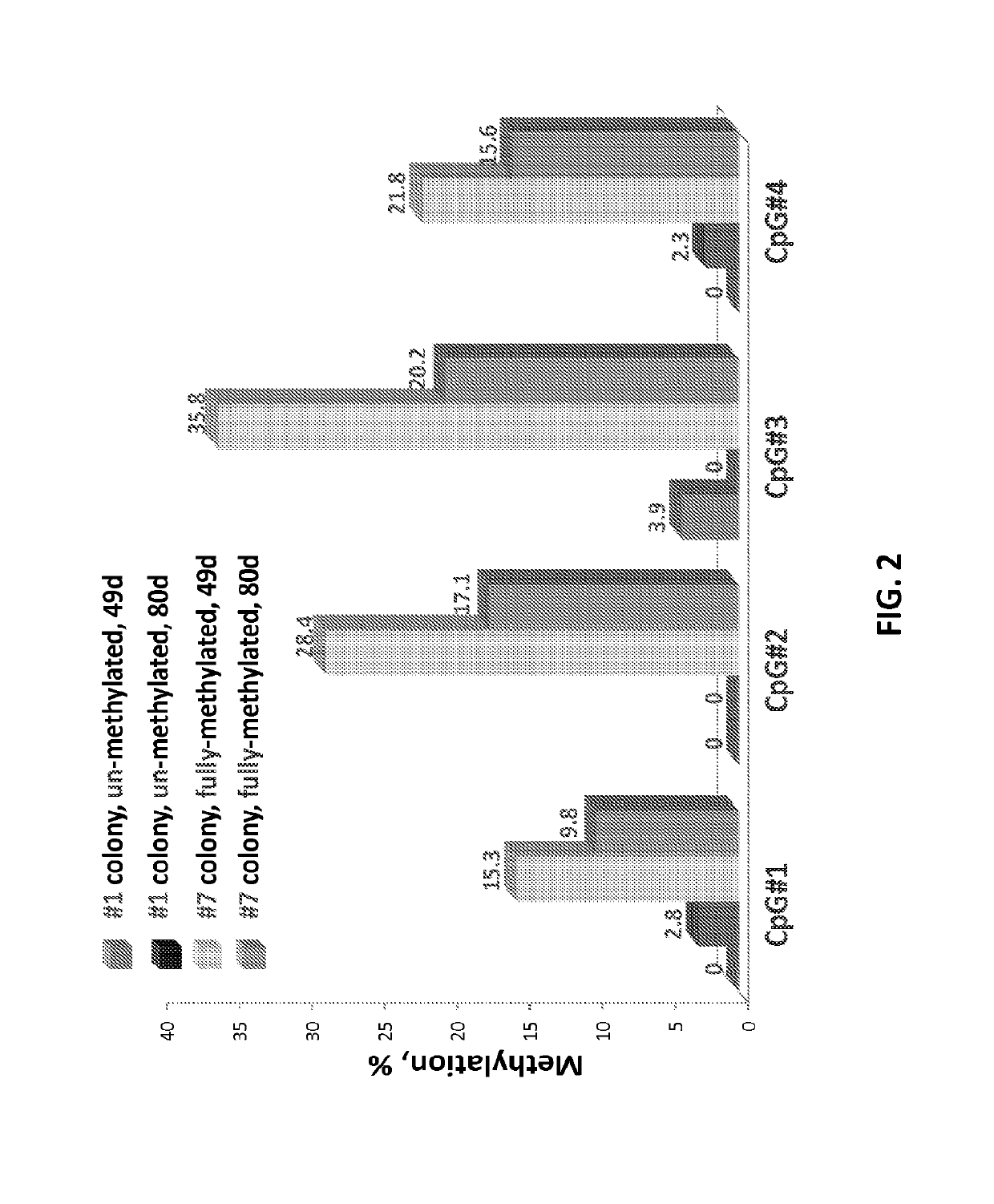
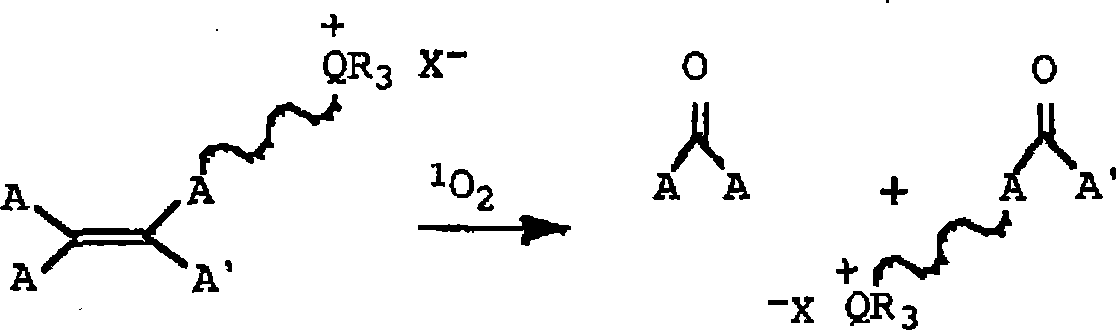Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
45 results about "Mammalian genome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
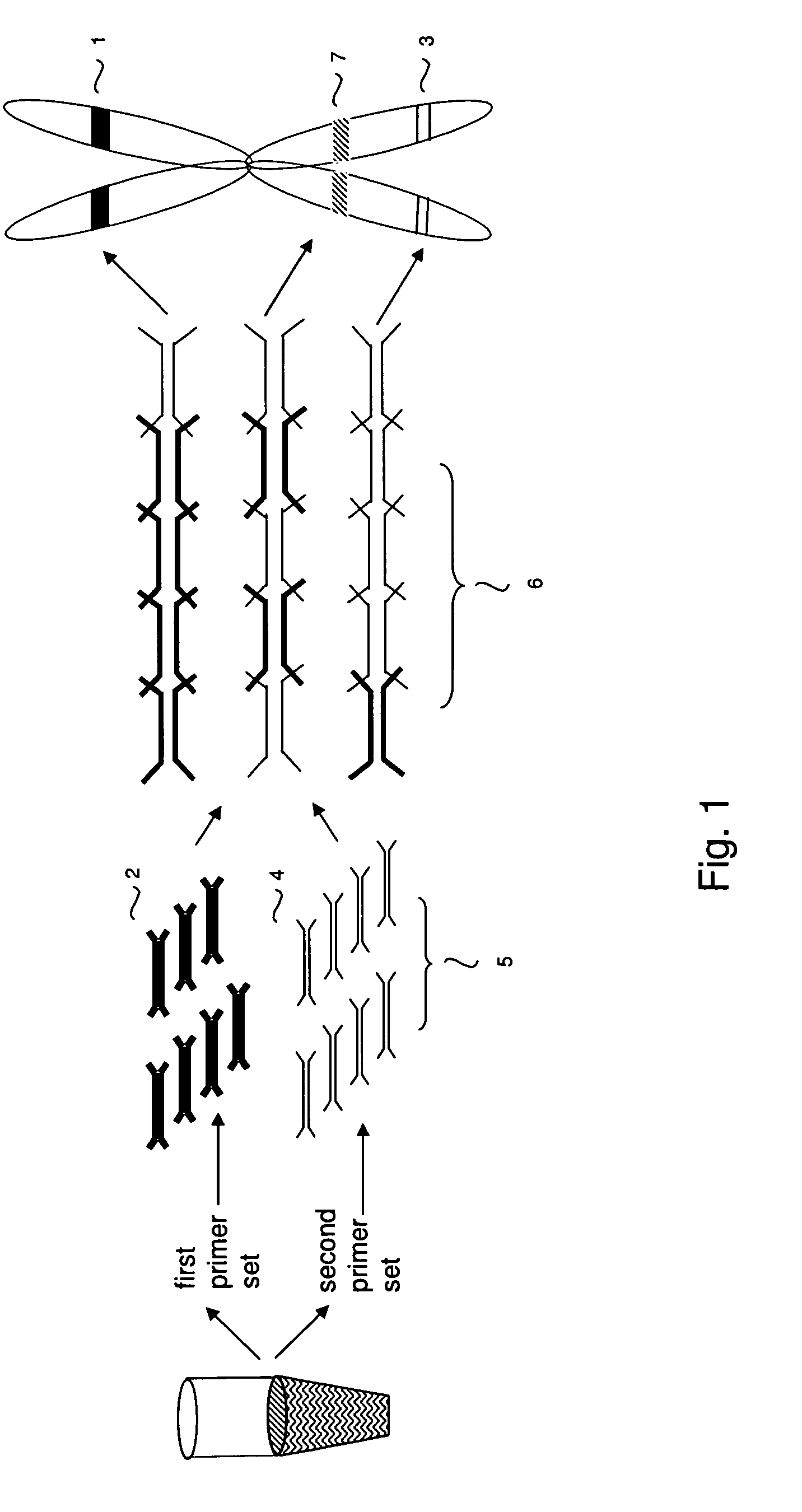
Method of sequencing genomes by hybridization of oligonucleotide probes
InactiveUS6018041ASugar derivativesMicrobiological testing/measurementMammalian genomeHomologous sequence
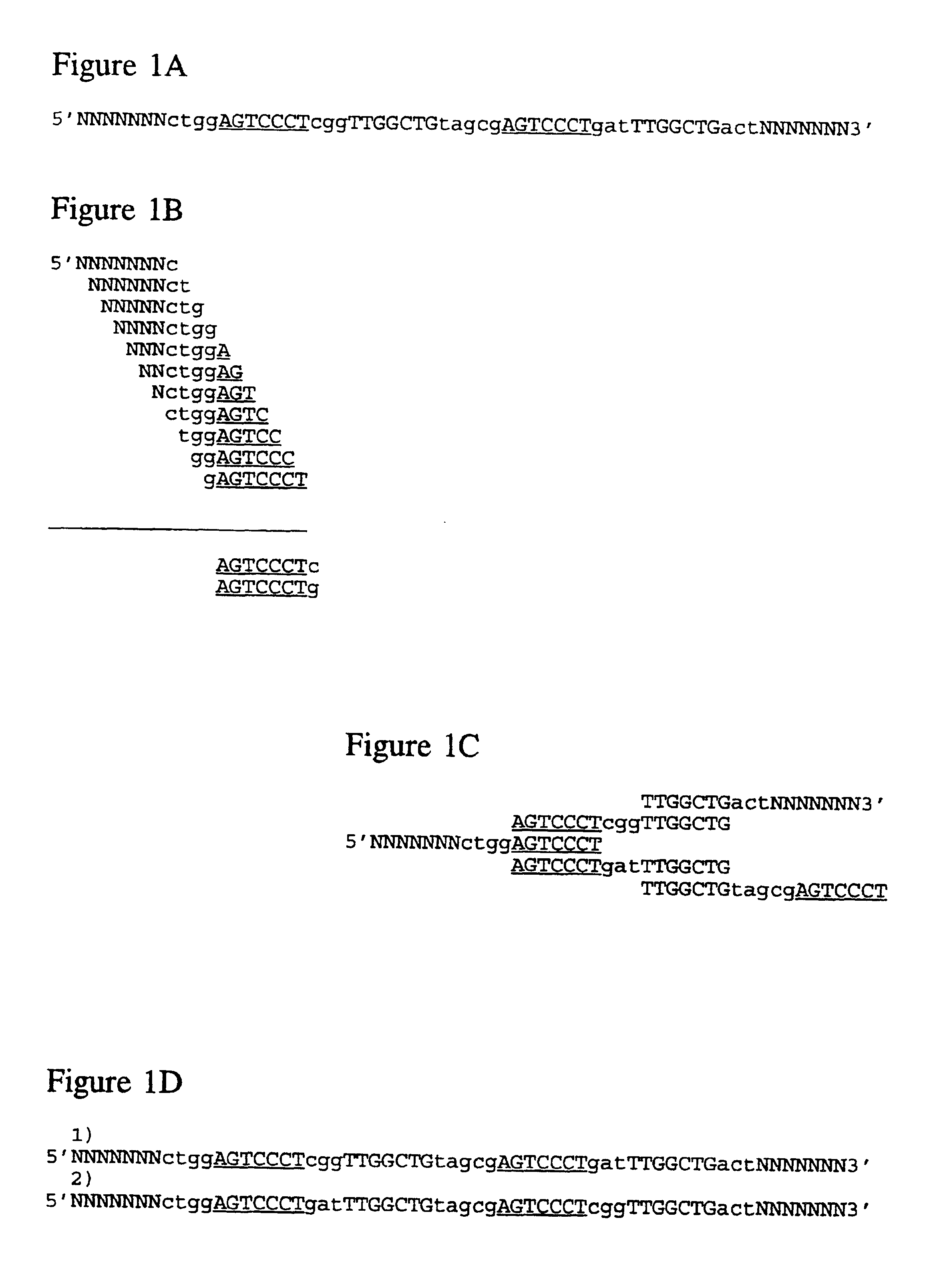
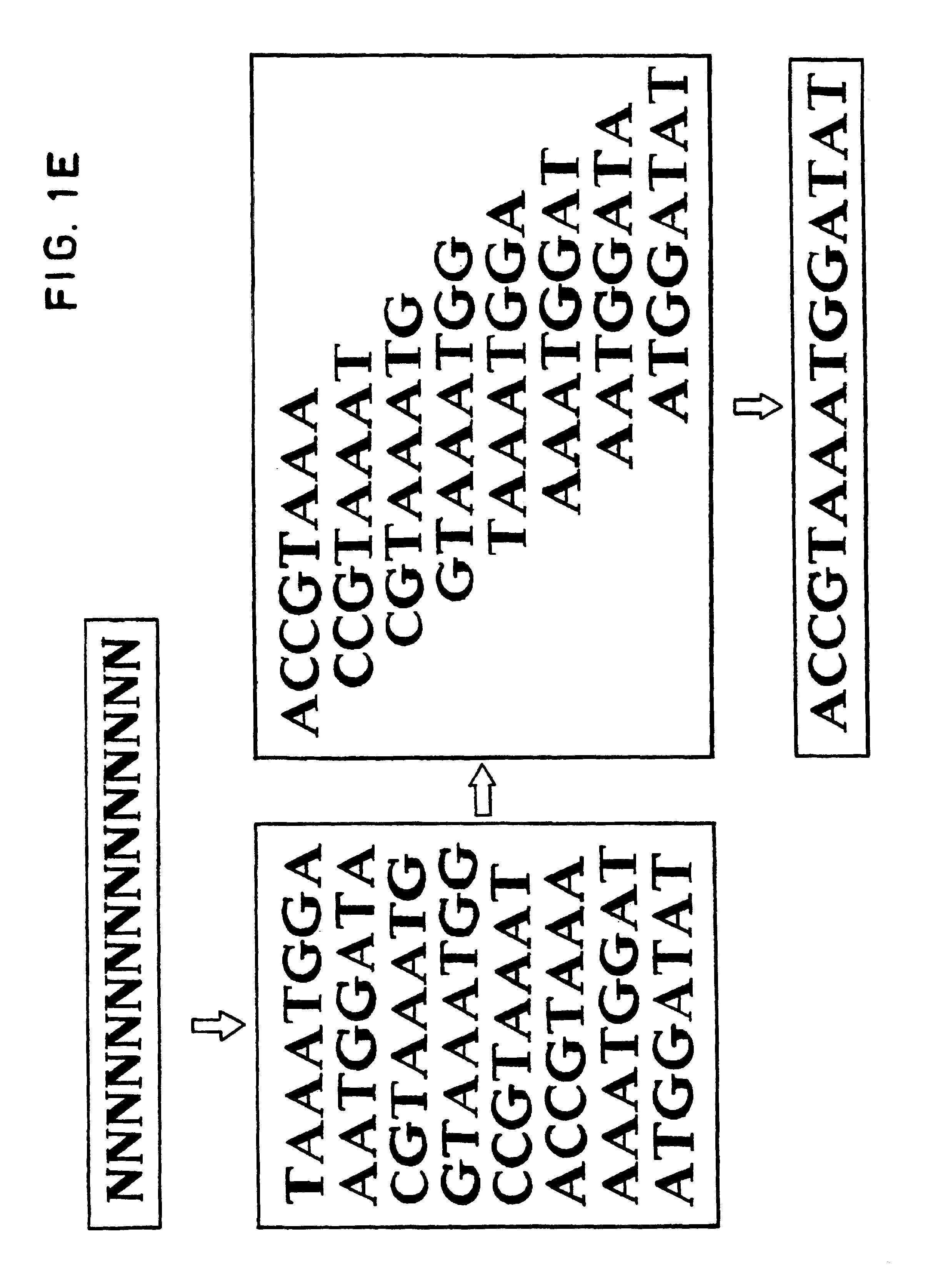
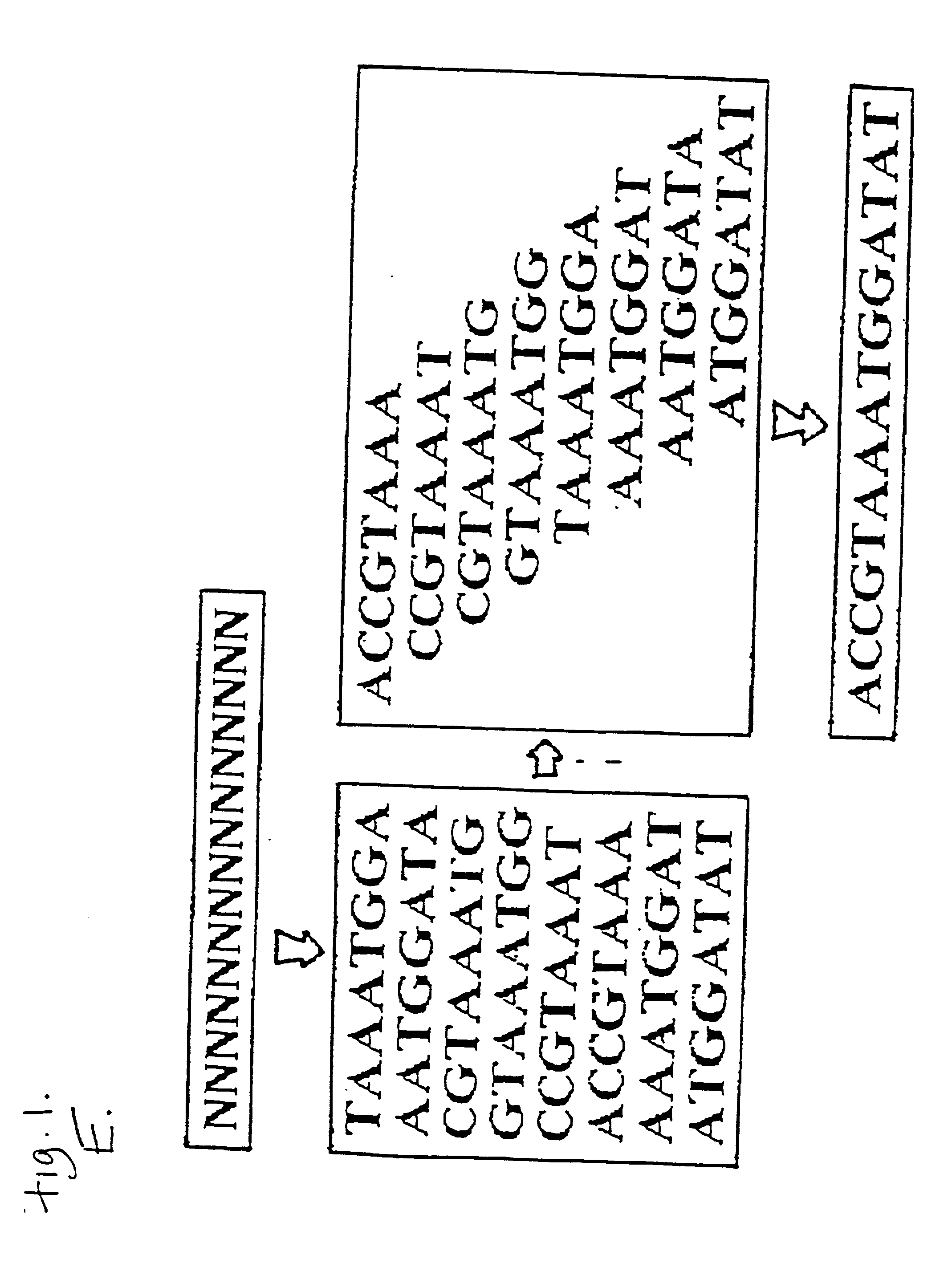
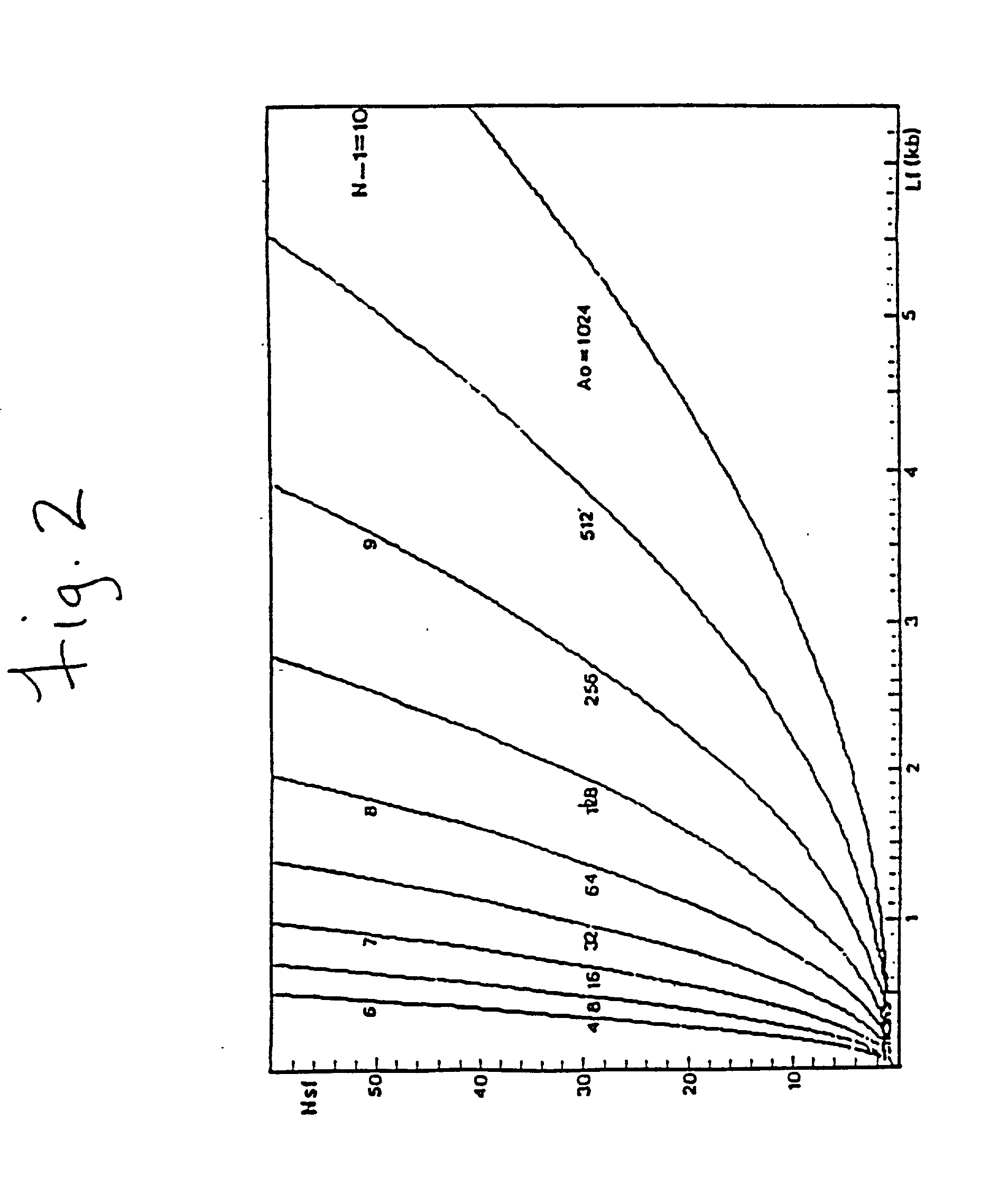
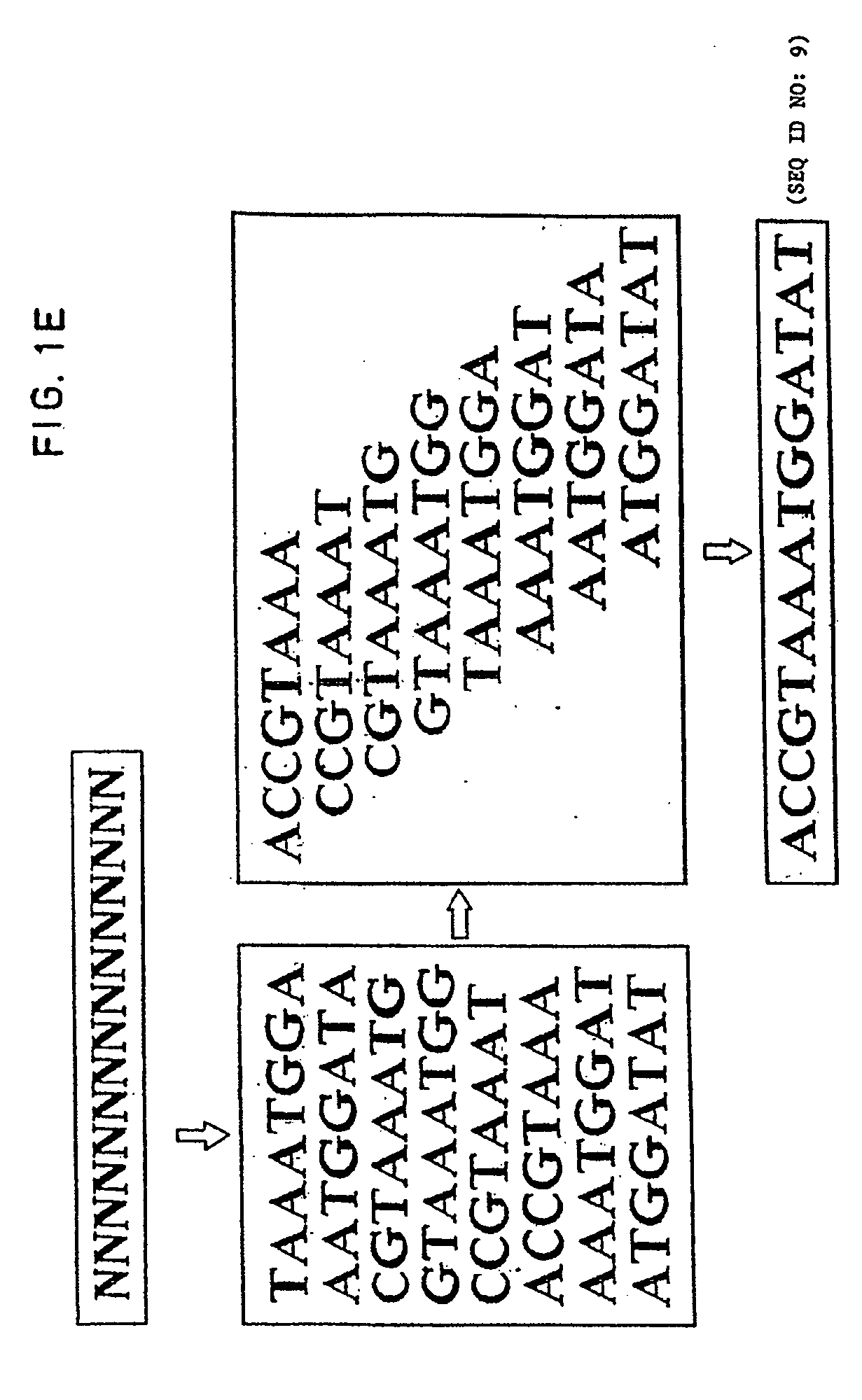
The conditions under which oligonucleotides hybridize only with entirely homologous sequences are recognized. The sequence of a given DNA fragment is read by the hybridization and assembly of positively hybridizing probes through overlapping portions. By simultaneous hybridization of DNA molecules applied as dots and bound onto a filter, representing single-stranded phage vector with the cloned insert, with about 50,000 to 100,000 groups of probes, the main type of which is (A,T,C,G)(A,T,C,G)N8(A,T,C,G), information for computer determination of a sequence of DNA having the complexity of a mammalian genome are obtained in one step. To obtain a maximally completed sequence, three libraries are cloned into the phage vector, M13, bacteriophage are used: with the 0.5 kb and 7 kbp insert consisting of two sequences, with the average distance in genomic DNA of 100 kbp. For a million bp of genomic DNA, 25,000 subclones of the 0.5 kbp are required as well as 700 subclones 7 kb long and 170 jumping subclones. Subclones of 0.5 kb are applied on a filter in groups of 20 each, so that the total number of samples is 2,120 per million bp. The process can be easily and entirely robotized for factory reading of complex genomic fragments or DNA molecules.
Owner:HYSEQ
Massively parallel contiguity mapping
ActiveUS20130203605A1Reducing sequenceLimited to sequenceLibrary member identificationChemical recyclingHuman DNA sequencingMammalian genome
Contiguity information is important to achieving high-quality de novo assembly of mammalian genomes and the haplotype-resolved resequencing of human genomes. The methods described herein pursue cost-effective, massively parallel capture of contiguity information at different scales.
Owner:UNIV OF WASHINGTON CENT FOR COMMERICIALIZATION
Primer-directed chromosome painting
An oligonucleotide composition is provided. The subject composition comprises: a mixture of at least 10 of sets of oligonucleotides, wherein each of the sets of oligonucleotides comprises at least 100 different oligonucleotides of the following formula: X1—V—X2, wherein: X1 and X2 provide binding sites for a pair of PCR primers and V is a variable region that has a variable nucleotide sequence that is complementary to one or more discrete regions of a mammalian genome; the nucleotide sequences of X1 and X2 are the same for each oligonucleotide of a set and different for oligonucleotides of different sets; and the variable regions of each set are complementary to different discrete regions of said mammalian genome. Methods of using the composition and kits containing the composition are also provided.
Owner:AGILENT TECH INC
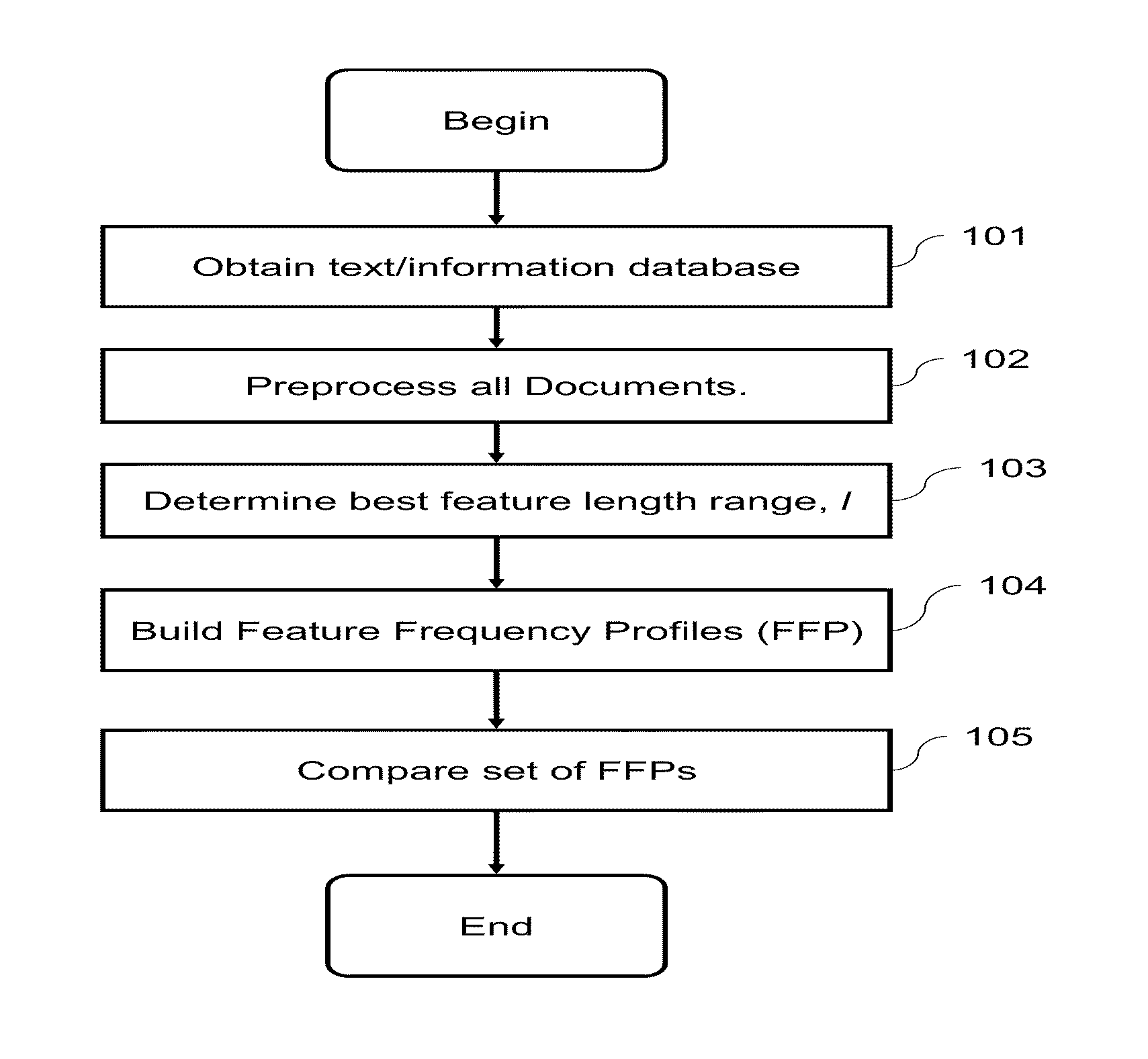
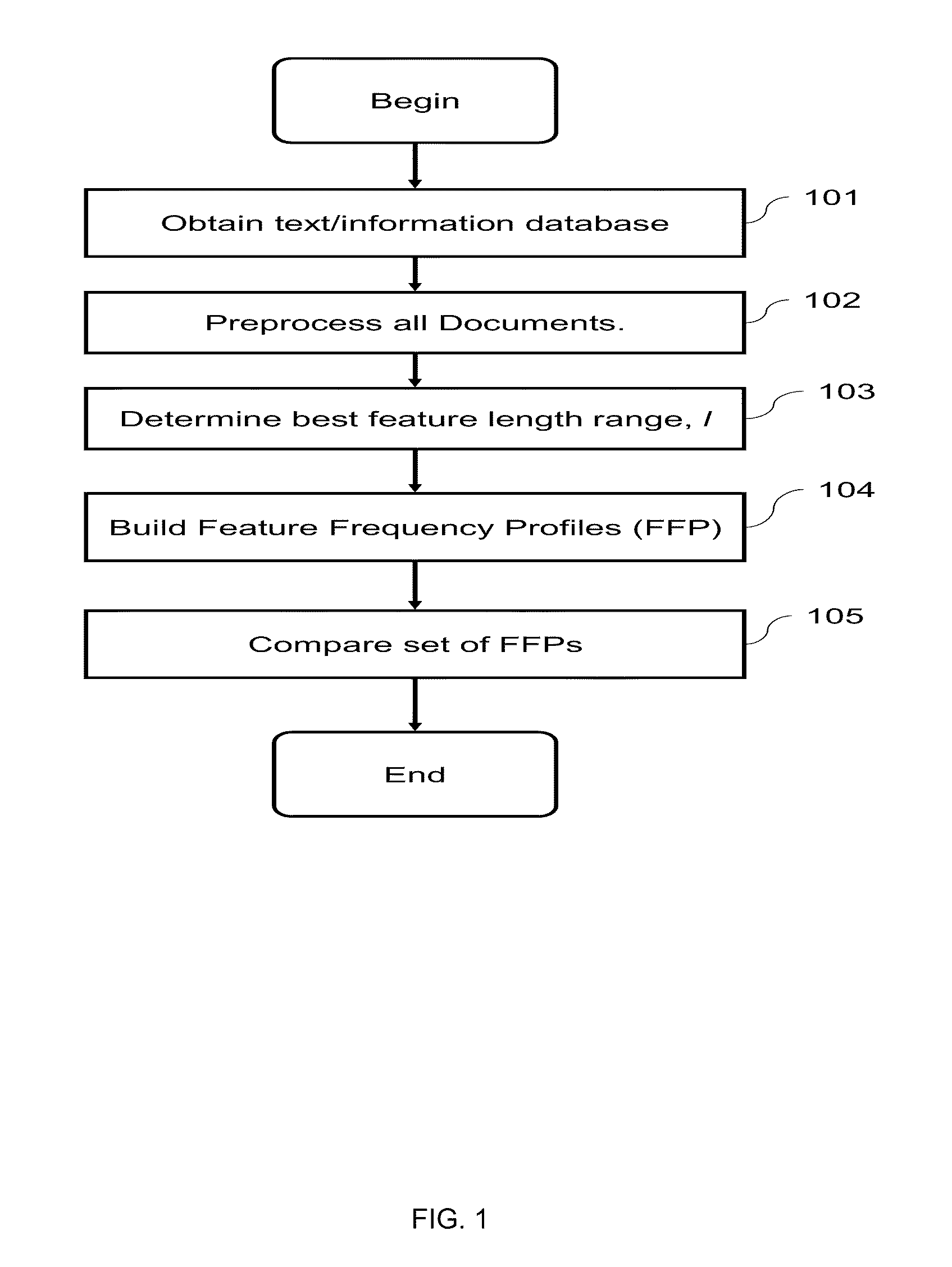
Computational Method for Comparing, Classifying, Indexing, and Cataloging of Electronically Stored Linear Information
InactiveUS20110196872A1Reduce complexityHigh frequencyDigital data information retrievalDigital data processing detailsMammalian genomeAnalysis data
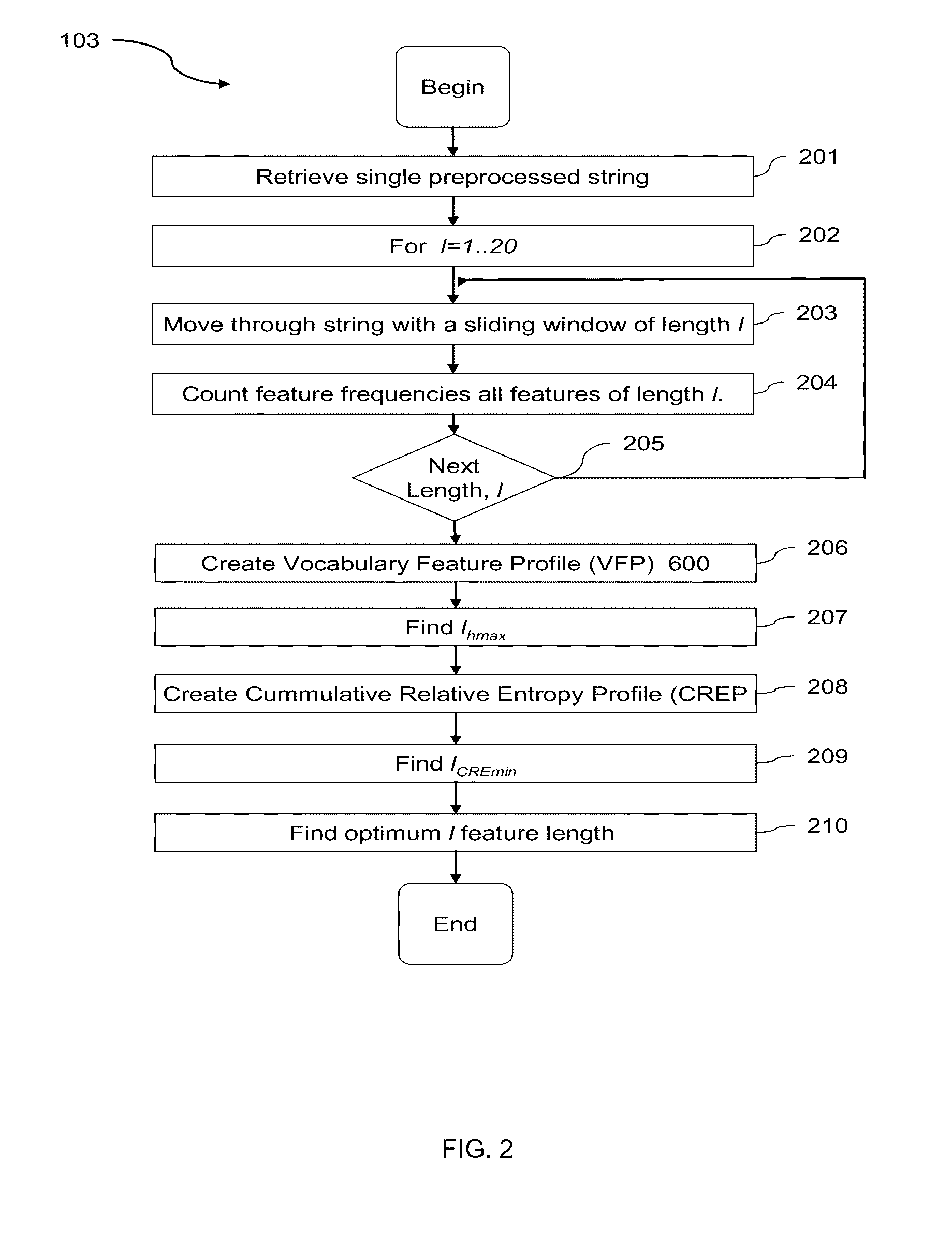
A computational method and system for the comparison and analysis of different objects of information within a database or collection. All objects are compared in a pair-wise fashion so the relative similarity between each object to every other object in the collection is known. A generalized alignment-free method is described for comparing whole genome (coding and non-coding) DNA sequences is used to investigate the relationship among placental mammalian genomes. Differences in word feature frequency profiles (FFP) are used to derive distance and infer evolutionary relationships.
Owner:RGT UNIV OF CALIFORNIA
Method of sequencing of genomes by hybridization of oligonucleotide probes
InactiveUS6451996B1Sugar derivativesMicrobiological testing/measurementMammalian genomeHomologous sequence
The conditions under which oligonucleotides hybridize only with entirely homologous sequences are recognized. The sequence of a given DNA fragment is read by the hybridization and assembly of positively hybridizing probes through overlapping portions. By simultaneous hybridization of DNA molecules applied as dots and bound onto a filter, representing single-stranded phage vector with the cloned insert, with about 50,000 to 100,000 groups of probes, the main type of which is (A,T,C,G)(A,T,C,G)N8(A,T,C,G), information for computer determination of a sequence of DNA having the complexity of a mammalian genome are obtained in one step. To obtain a maximally completed sequence, three libraries are cloned into the phage vector, M13, bacteriophage are used: with the 0.5 kb and 7 kbp insert consisting of two sequences, with the average distance in genomic DNA of 100 kbp. For a million bp of genomic DNA, 25,000 subclones of the 0.5 kbp are required as well as 700 subclones 7 kb long and 170 jumping subclones. Subclones of 0.5 kb are applied on a filter in groups of 20 each, so that the total number of samples is 2,120 per million bp. The process can be easily and entirely robotized for factory reading of complex genomic fragments or DNA molecules.
Owner:COMPLETE GENOMICS INC
ARF-P19, a novel regulator of the mammalian cell cycle
InactiveUS6407062B1Peptide/protein ingredientsAntibody mimetics/scaffoldsAbnormal tissue growthCancer cell
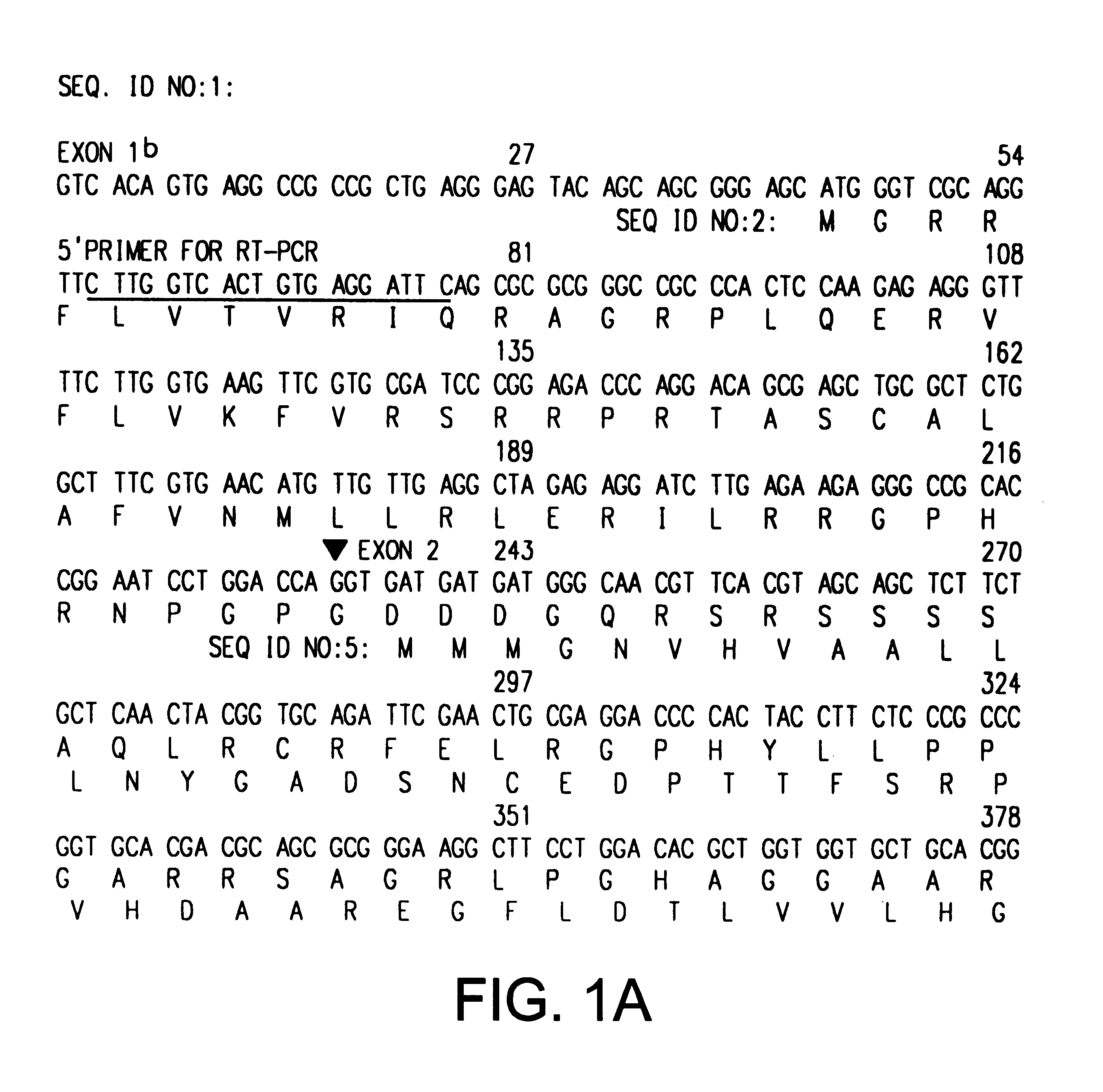
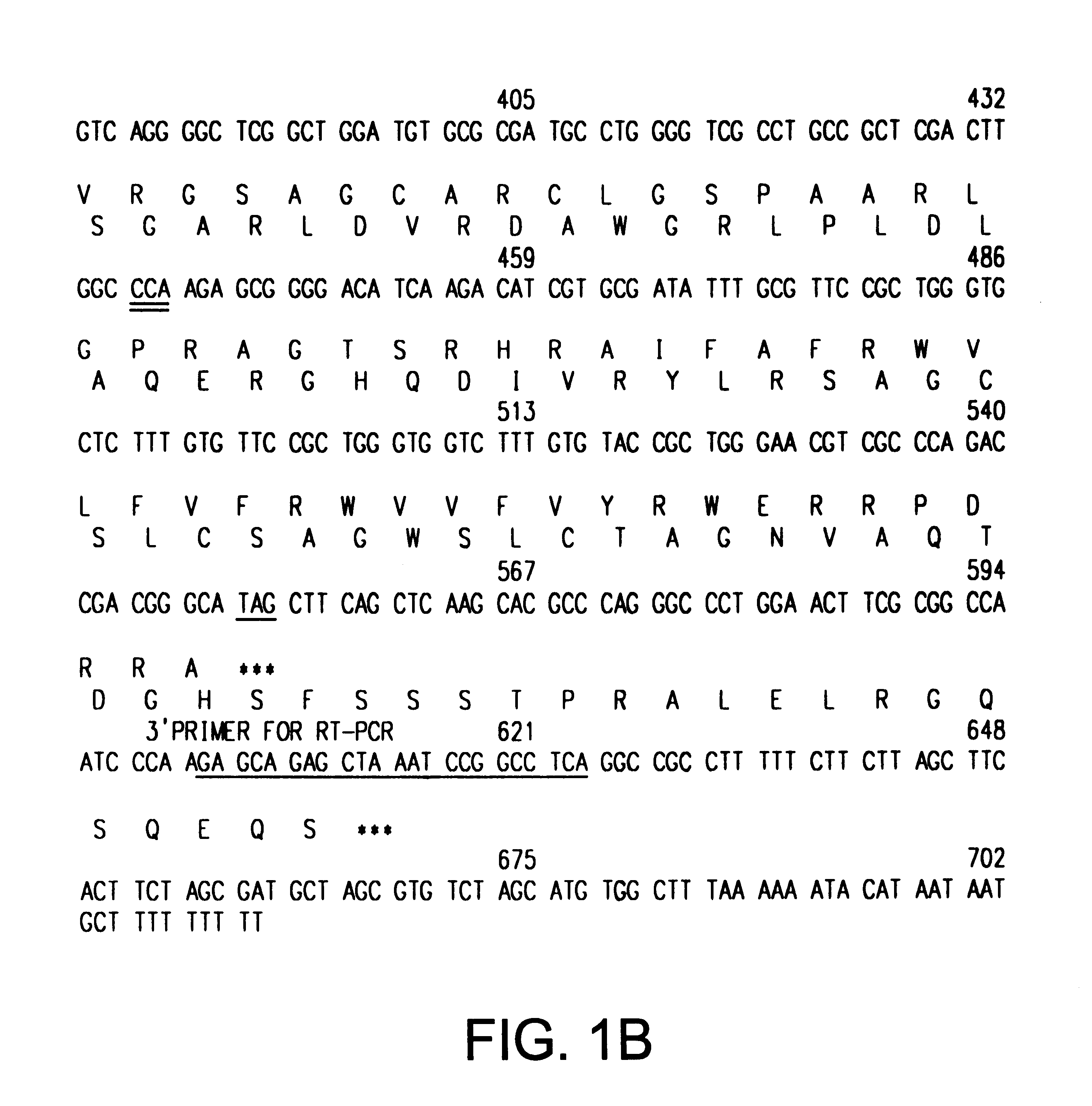
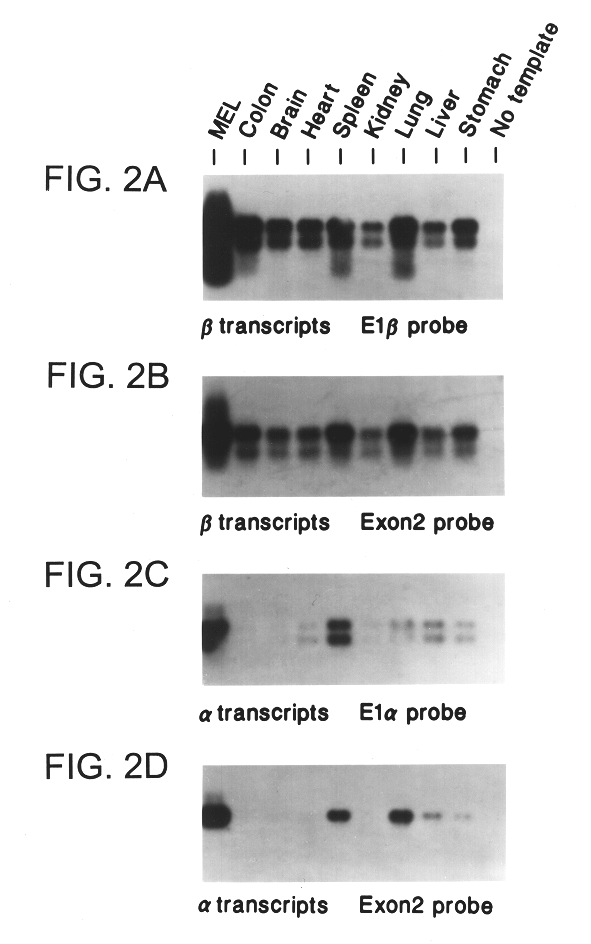
The INK4A (MTS1, CDKN2) gene encodes a specific inhibitor (InK4a-p16) of the cyclin D-dependent kinases CDK4 and CDK6. InK4a-p16 can block these kinase from phosphorylating the retinoblastoma protein (pRb), preventing exit from the G1 phase of the cell cycle. Deletions and mutations involving the gene encoding InK4a-p16, INK4A, occur frequently in cancer cells, implying that INK4a-p16, like pRb, suppresses tumor formulation. However, a completely unrelated protein (ARF-p19) arises in major part from an alternative reading frame of the mouse INK4A gene. Expression of an ARF-p19 cDNA (SEQ ID NO:1) in rodent fibroblasts induces both G1 and G2 phase arrest. Economical reutilization of protein coding sequences in this manner is without precedent in mammalian genomes, and the unitary inheritance of INK4a-p16 and ARF-p19 may reflect a dual requirement for both proteins in cell cycle control.
Owner:ST JUDE CHILDRENS RES HOSPITAL INC
Computer-aided analysis system for sequencing by hybridization
InactiveUS6316191B1Elimination of intensiveLabor intensiveMicrobiological testing/measurementSequence analysisSingle processMammalian genome
The conditions under which oligonucleotide probes hybridize preferentially with entirely complementary and homologous nucleic acid targets are described. Using these hybridization conditions, overlapping oligonucleotide probes associate with a target nucleic acid. Following washes, positive hybridization signals are used to assemble the sequence of a given nucleic acid fragment. Representative target nucleic acids are applied as dots. Up to 100,000 probes of the type (A,T,C,G)(A,T,C,G)N8(A,T,C,G) are used to determine sequence information by simultaneous hybridization with nucleic acid molecules bound to a filter. Additional hybridization conditions are provided that allow stringent hybridization of 6-10 nucleotide long oligomers which extends the utility of the invention. A computer process determines the information sequence of the target nucleic acid which can include targets with the complexity of mammalian genomes. Sequence generation can be obtained for a large complex mammalian genome in a single process.
Owner:COMPLETE GENOMICS INC
Method of sequencing by hybridization of oligonucleotide probes
InactiveUS20020106673A1Resulting in effectivenessRapidity resultingMicrobiological testing/measurementBiological testingSingle processOligomer
The conditions under which oligonucleotide probes hybridize preferentially with entirely complementary and homologous nucleic acid targets are described. Using these hybridization conditions, overlapping oligonucleotide probes associate with a target nucleic acid. Following washes, positive hybridization signals are used to assemble the sequence of a given nucleic acid fragment. Representative target nucleic acids are applied as dots. Up to to 100,000 probes of the type (A,T,C,G) (A,T,C,G)N8(A,T,C,G) are used to determine sequence information by simultaneous hybridization with nucleic acid molecules bound to a filter. Additional hybridization conditions are provided that allow stringent hybridization of 6-10 nucleotide long oligomers which extends the utility of the invention. A computer process determines the information sequence of the target nucleic acid which can include targets with the complexity of mammalian genomes. Sequence generation can be obtained for a large complex mammalian genome in a single process.
Owner:HYSEQ
Pig ROSA26 specific integration site and application thereof
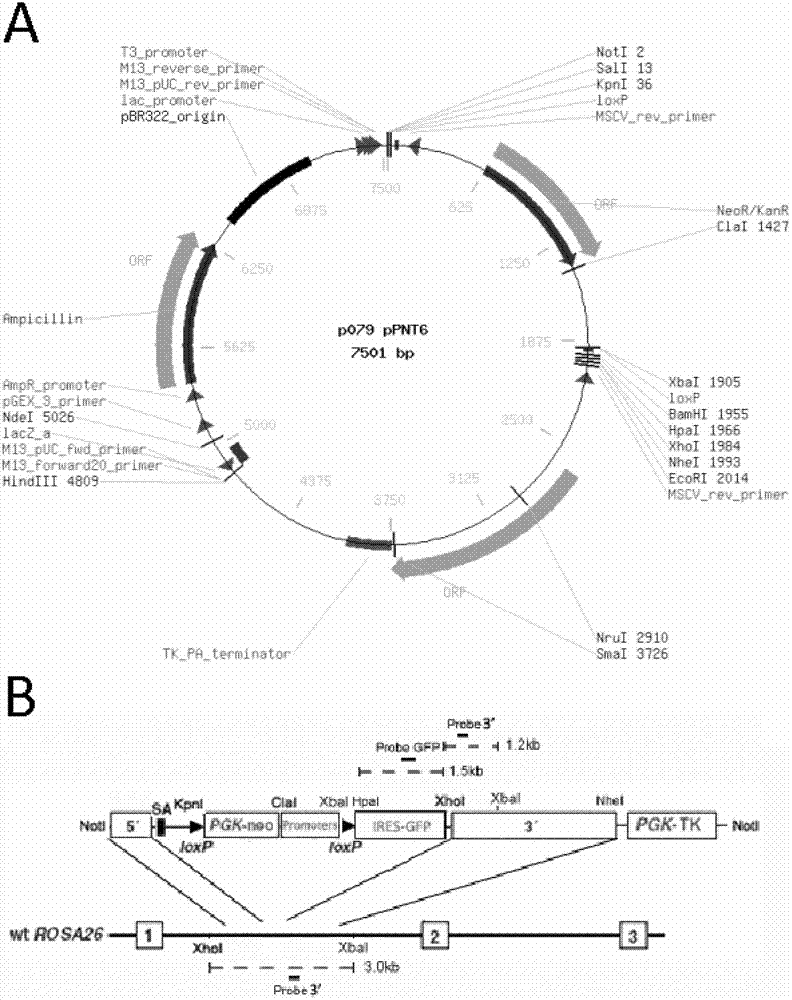
InactiveCN102851279AEfficient and specific targetingEfficient and stable expressionSugar derivativesVector-based foreign material introductionBiotechnologyMammalian genome
The invention discloses a pig ROSA26 specific integration site and an application thereof. The nucleotide sequence of the site is represented by SEQ ID NO.2. A targeting vector derived from the site is represented by SEQ ID NO.1. The ROSA26 specific site adopted by the invention is not subjected to immune-mediated exclusion and epigenetic modification in genomes of pigs or higher mammals. The site can mediate stable and efficient expression of exogenous genes. With the ROSA26 specific site targeting vector, targeting can be realized with high efficiency.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Massively parallel continguity mapping
Contiguity information is important to achieving high-quality de novo assembly of mammalian genomes and the haplotype-resolved resequencing of human genomes. The methods described herein pursue cost-effective, massively parallel capture of contiguity information at different scales.
Owner:UNIV OF WASHINGTON CENT FOR COMMERICIALIZATION
Epigenetic modification of mammalian genomes using targeted endonucleases
The present disclosure provides genetically engineered cell lines comprising chromosomally integrated synthetic sequences having predetermined epigenetic modifications, wherein a predetermined epigenetic modification is correlated with a known diagnosis, prognosis or level of sensitivity to a disease treatment. Also provided are kits comprising said epigenetically modified synthetic nucleic acids or cells comprising said epigenetically modified synthetic nucleic acids that can be used as reference standards for predicting responsiveness to therapeutic treatments, diagnosing diseases, or predicting disease prognosis.
Owner:SIGMA ALDRICH CO LLC
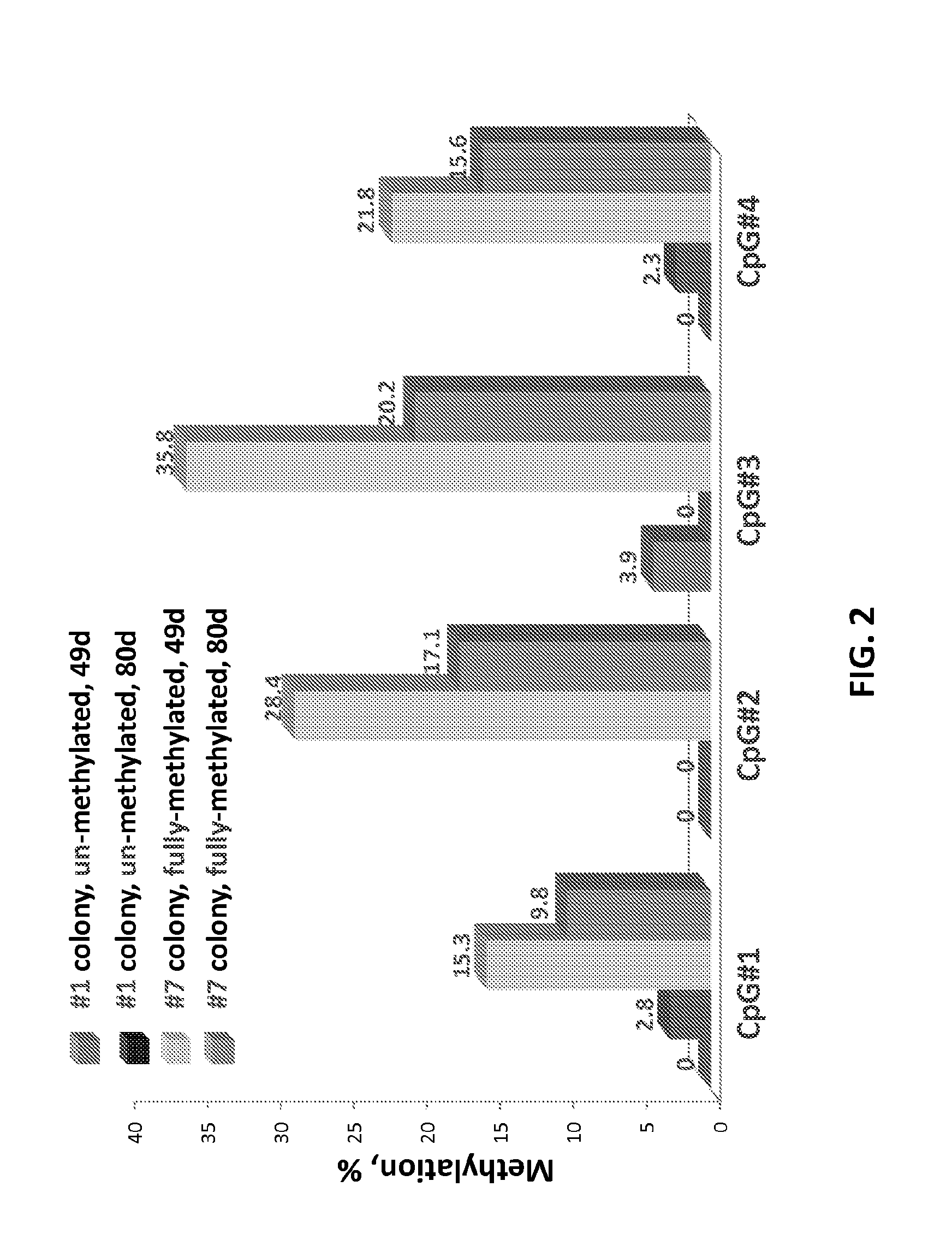
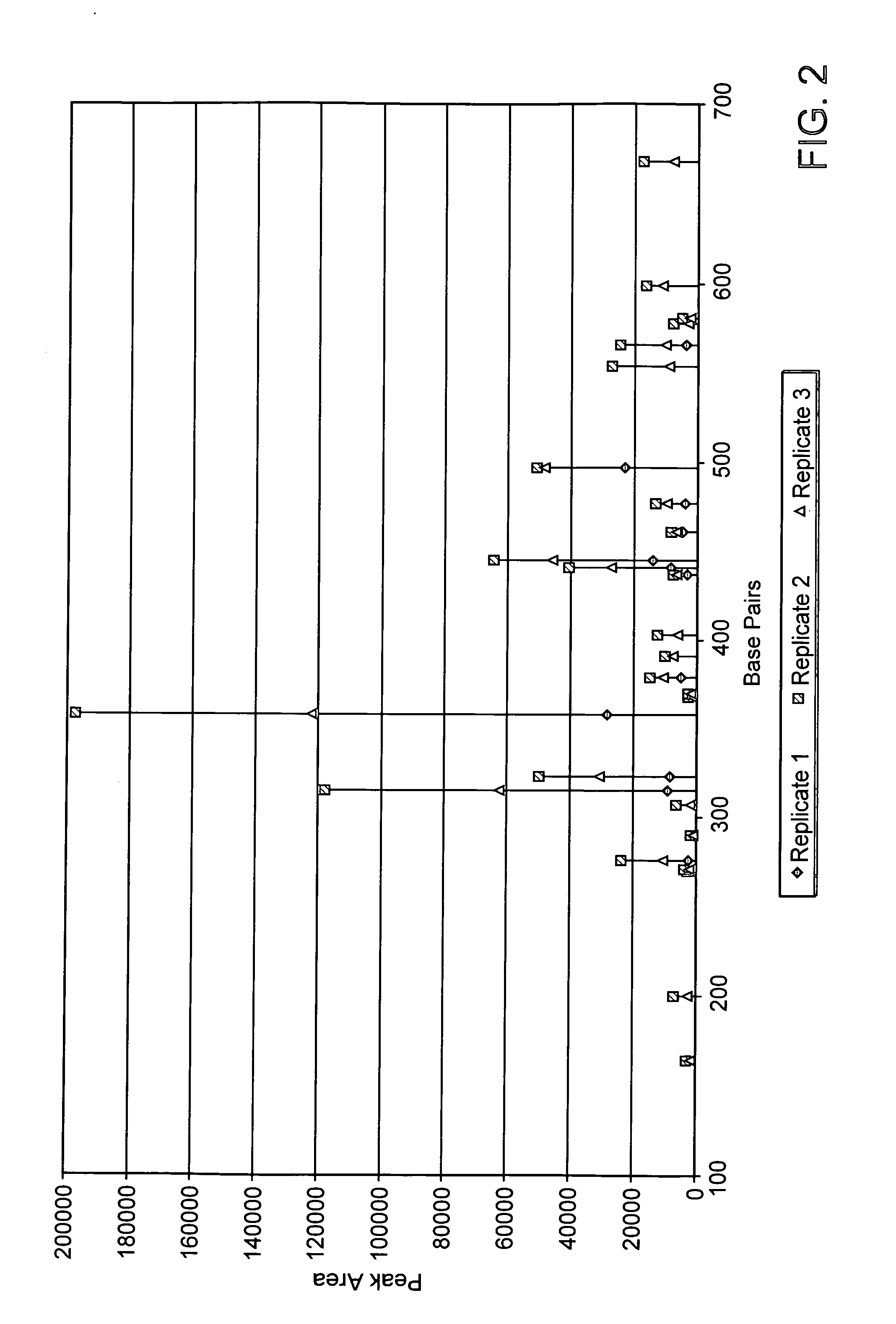
Rapid methods for detecting methylation of a nucleic acid molecule
InactiveUS20050260630A1Rapid and inexpensiveAccurate and easily reproducible resultMicrobiological testing/measurementFermentationMammalian genomeCapillary electrophoresis
Disclosed are methods for determining the methylation status of a target double-stranded nucleic acid molecule using PCR amplification and capillary electrophoresis. The methods are generally useful in measuring the methylation status of a nucleic acid sample, including a mammalian genomic DNA sample, and may be further specifically applied to detecting changes in methylation status of a nucleic acid that are associated with exposure to a toxic compound or treatment, or that are associate with a disease or disorder.
Owner:MICHIGAN STATE UNIV
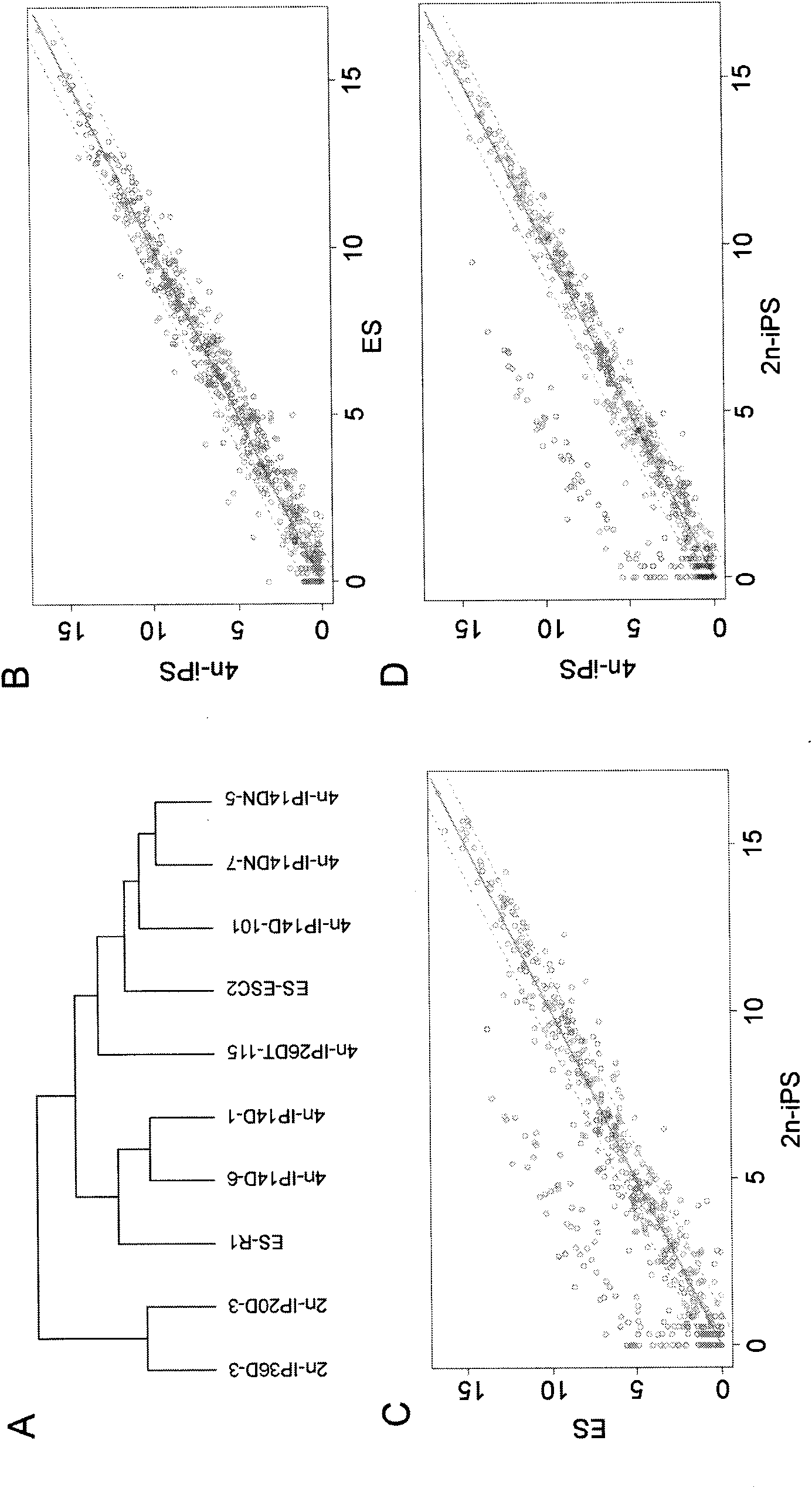
Key genes, microRNAs and other non-coding RNAs or combination thereof used for identifying or regulating cell pluripotency
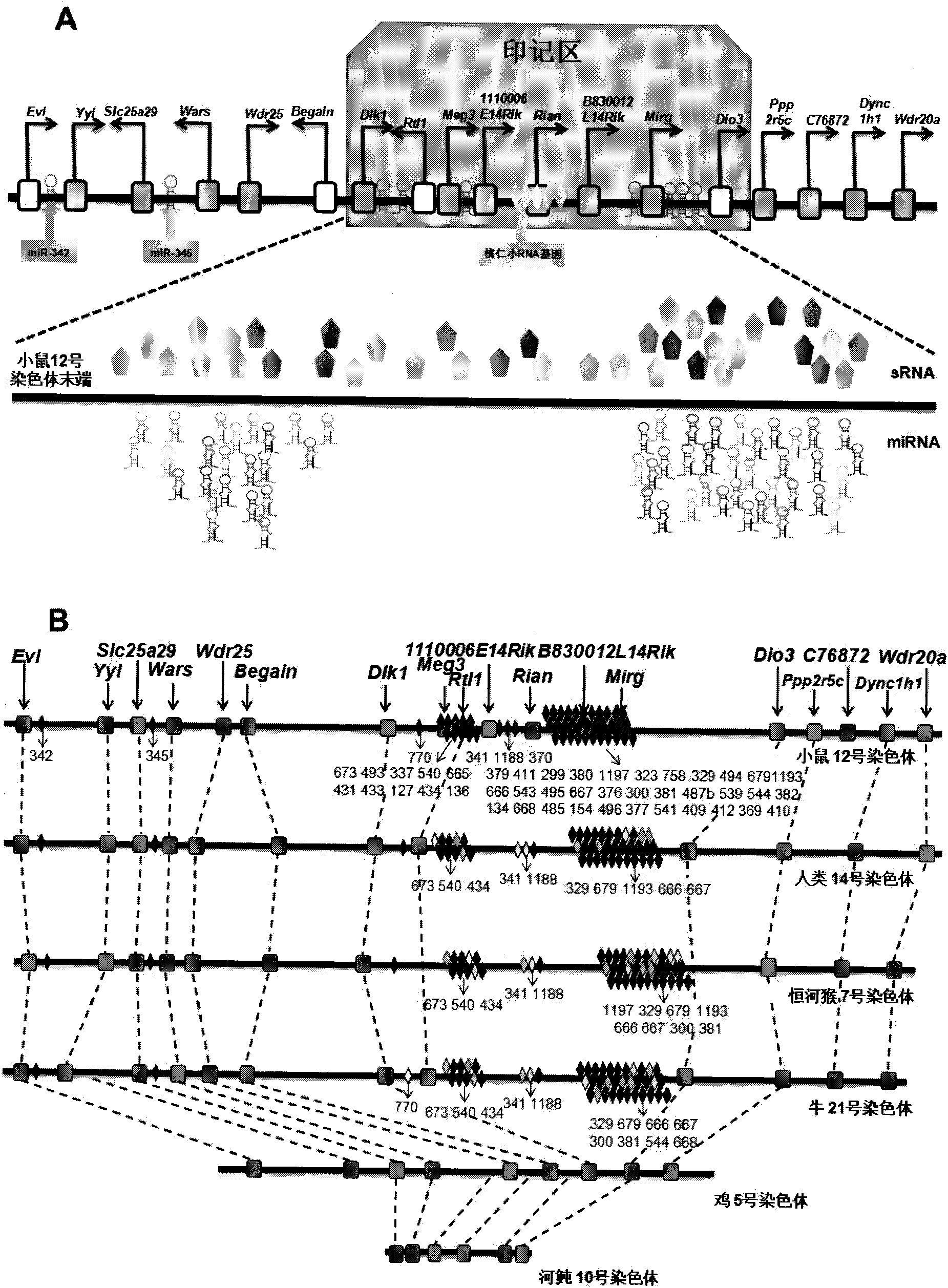
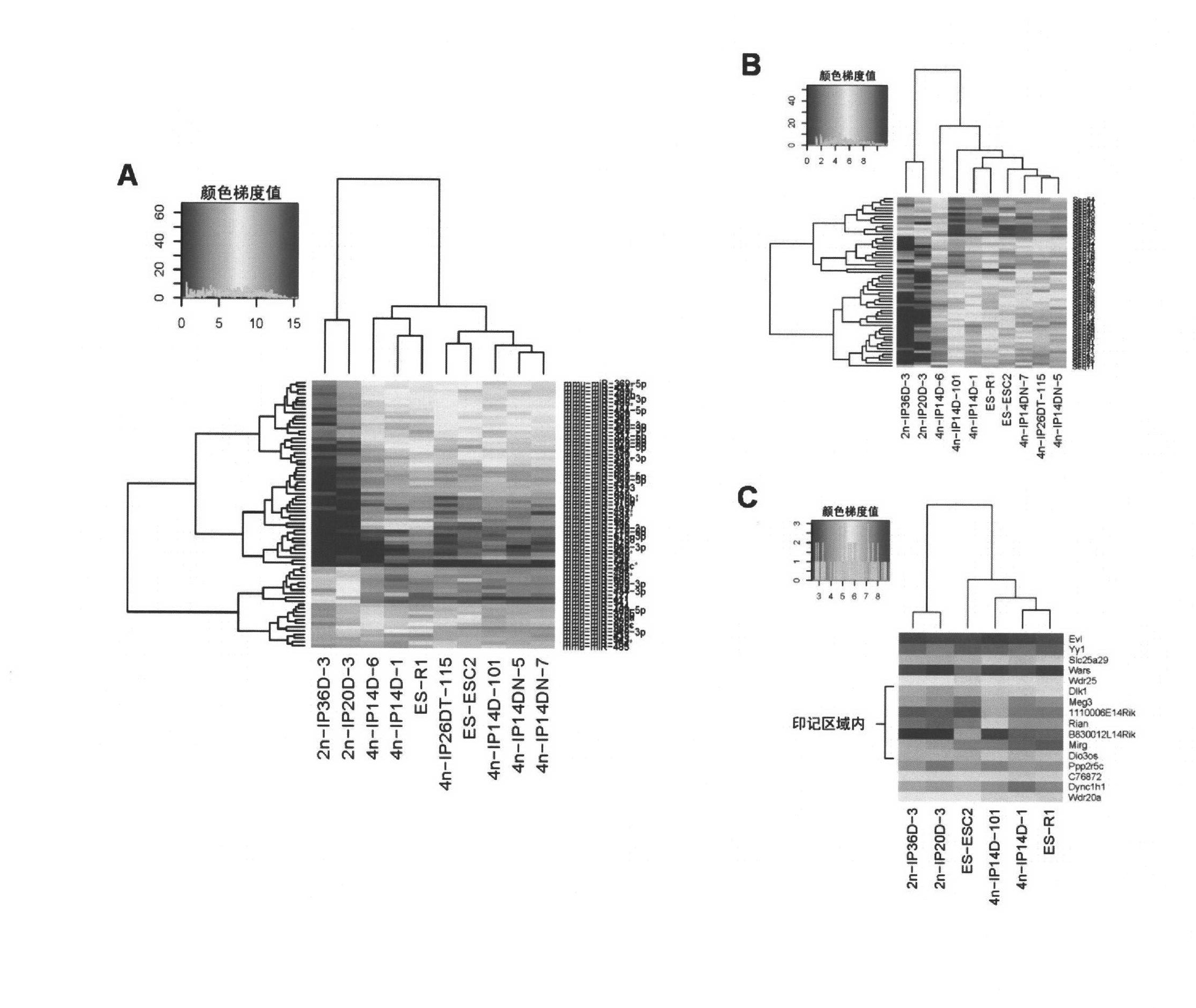
The invention relates to key genes, microRNAs and other non-coding RNAs, or a combination thereof used for identifying or regulating cell pluripotency. The invention is characterized in that the key genes, microRNAs, other non-coding RNAs or a combination is highly expressed in the stem cell with complete pluripotency and the expression is obviously repressed or silent in the stem cell without complete pluripotency. The genes, microRNAs and other non-coding RNAs are the genes, microRNAs and other non-coding RNAs positioned in the chromosome imprinting region named as Dlk1-Dio3 on the long arm of mouse chromosome 12 and the genes, microRNAs and other non-coding RNAs in the genome collinearity regions of other mammals, which have 70%-100% of homology. The invention also relates to applications of the genes, microRNAs and other non-coding RNAs, or the combination thereof used for identifying the pluripotency of the stem cell and regulating cell pluripotency, applications in stem cell typing, applications for regulating the cell pluripotency and the pluripotency state and level of the cell, applications in disease treatment, and applications in the drug target development of tumor treatment or the development of antitumor drugs.
Owner:INST OF ZOOLOGY CHINESE ACAD OF SCI +1
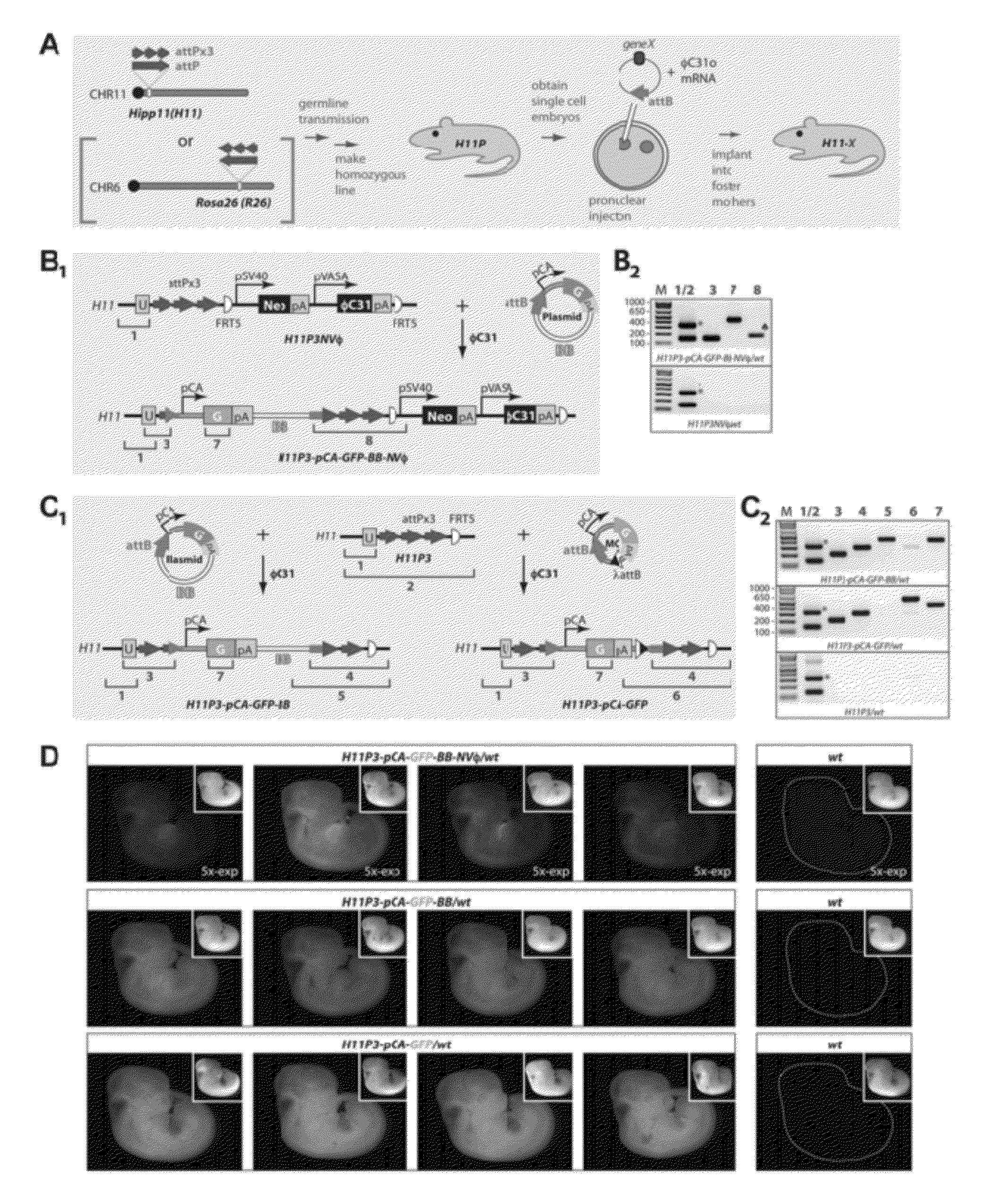
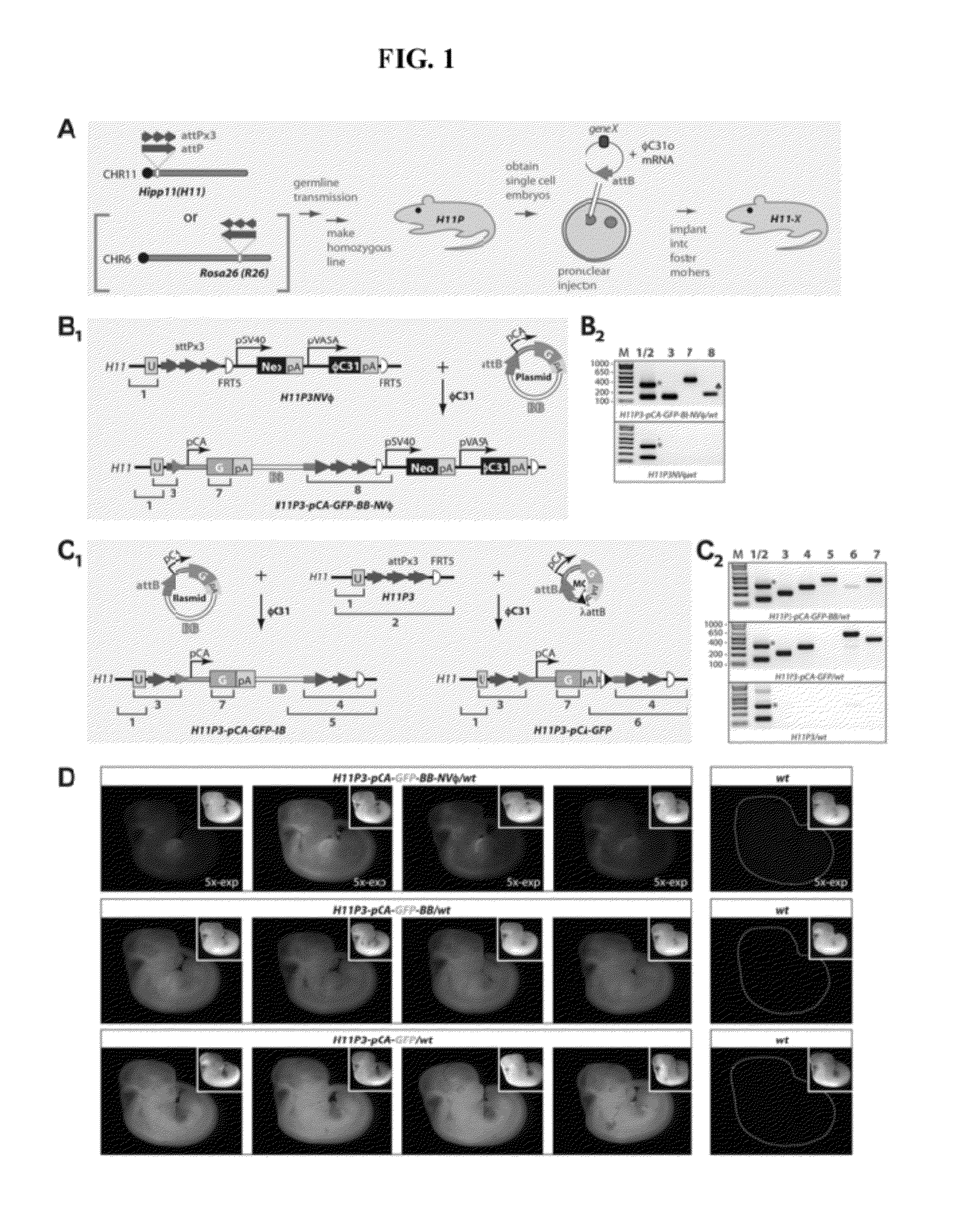
Site-Directed Integration of Transgenes in Mammals
ActiveUS20120124686A1Easy to understandNucleic acid vectorOther foreign material introduction processesMammalian genomeSite-specific recombination
The present disclosure provides a method of making a mammal (e.g., a rodent, such as a mouse) by integrating an intact polynucleotide sequence into a specific genomic locus of the mammal to result in a transgenic mammal. A transgenic mammal made by the methods of the present disclosure would contain a known copy number (e.g., one) of the inserted polynucleotide sequence at a predetermined location. The method involves introducing a site-specific recombinase and a targeting construct, containing a first recombination site and the polynucleotide sequence of interest, into the mammalian cell. The genome of the cell contains a second recombination site and recombination between the first and second recombination sites is facilitated by the site-specific, uni-directional recombinase. The result of the recombination is site-specific integration of the polynucleotide sequence of interest in the genome of the mammal. This inserted sequence is then also transmitted to the progeny of the mammal.
Owner:UNIVERSITY OF OREGON +1
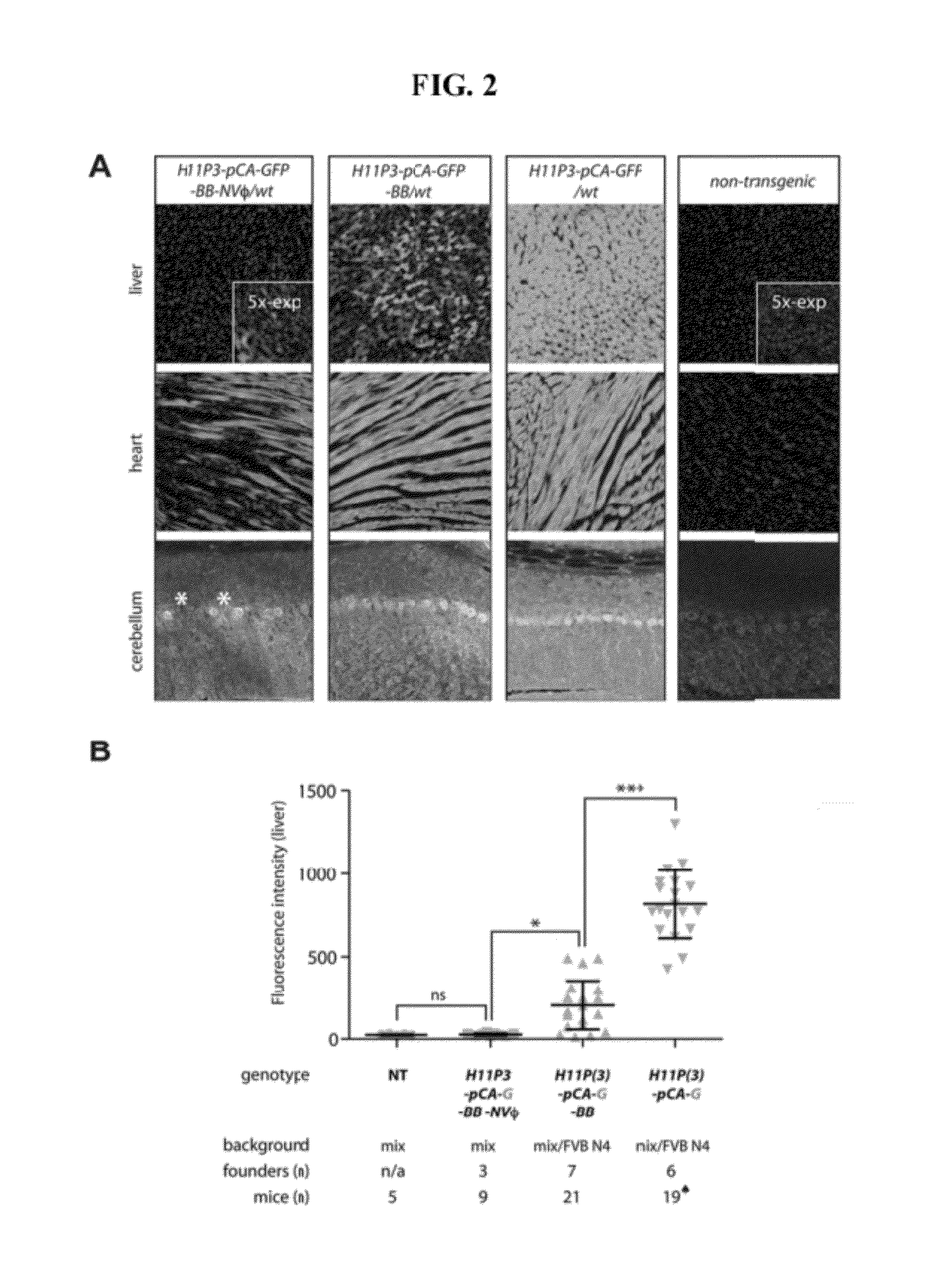
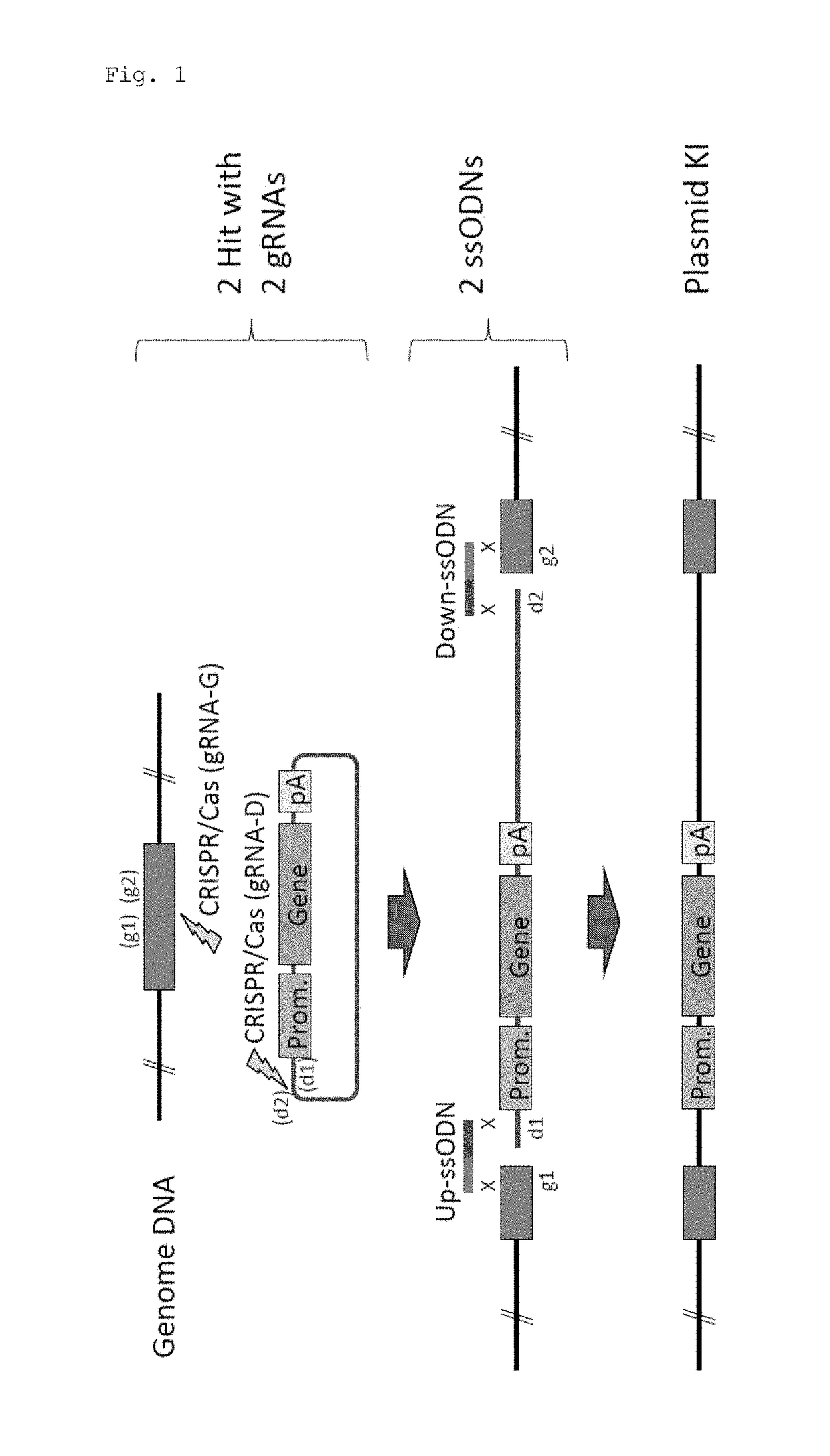
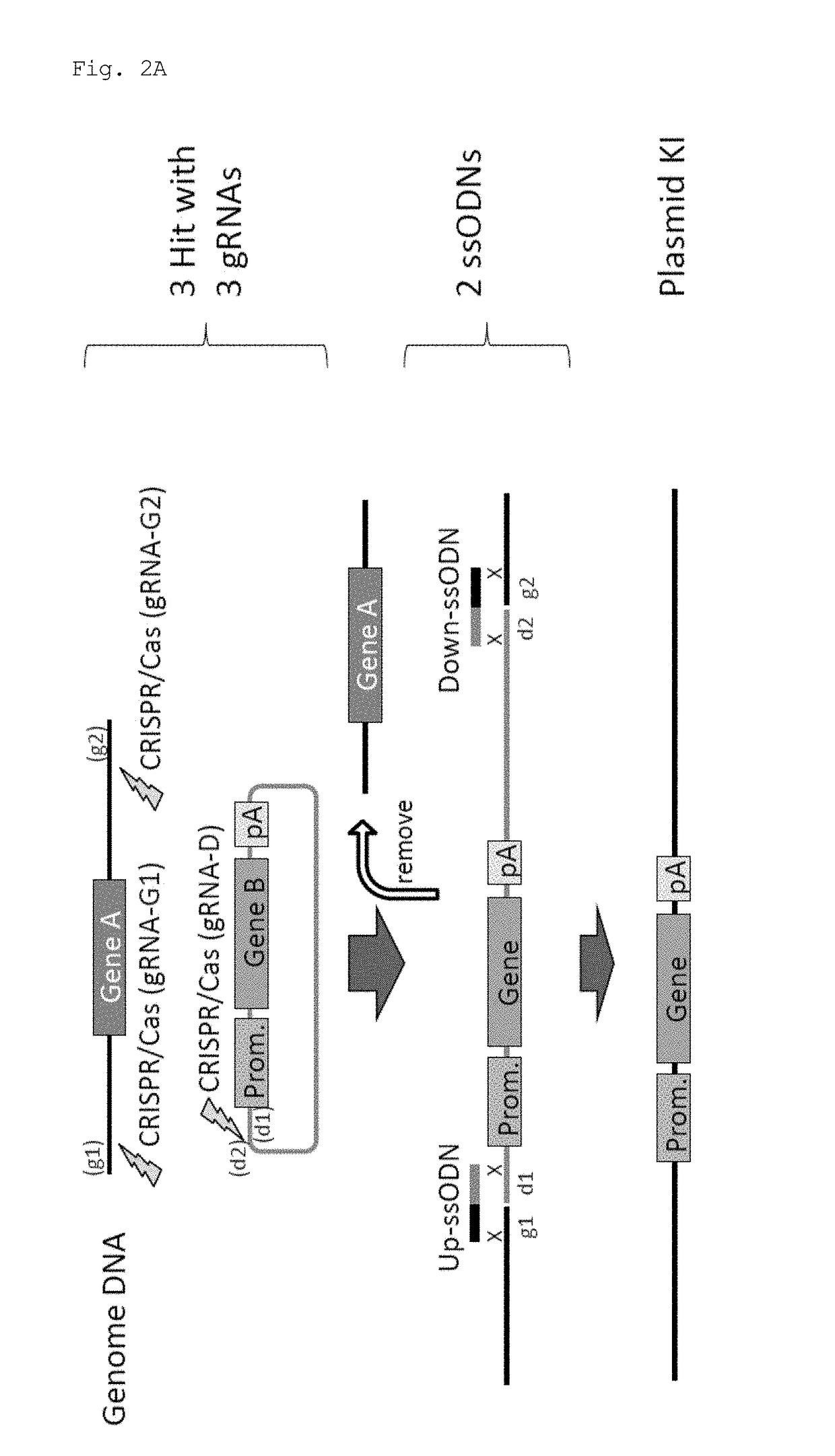
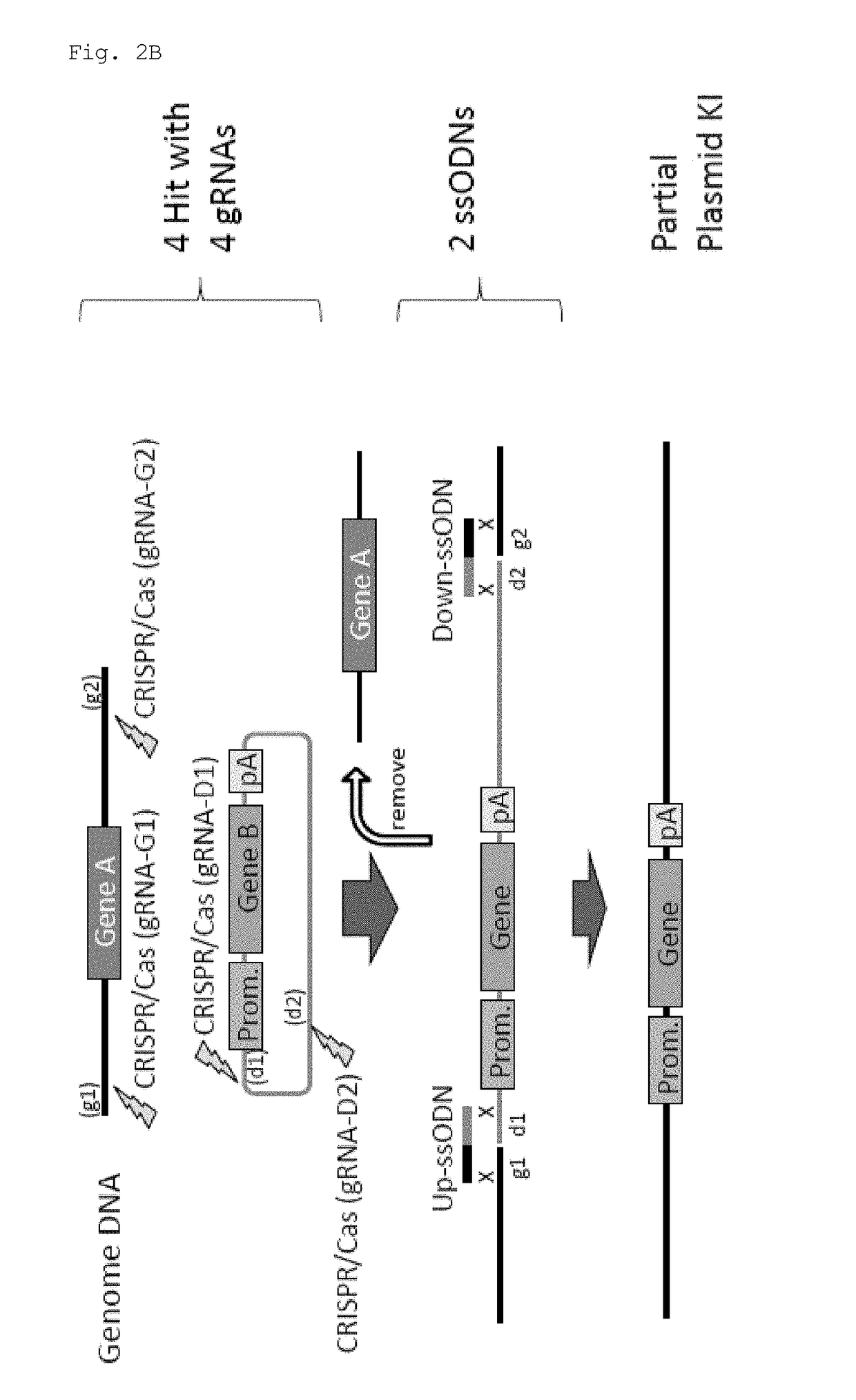
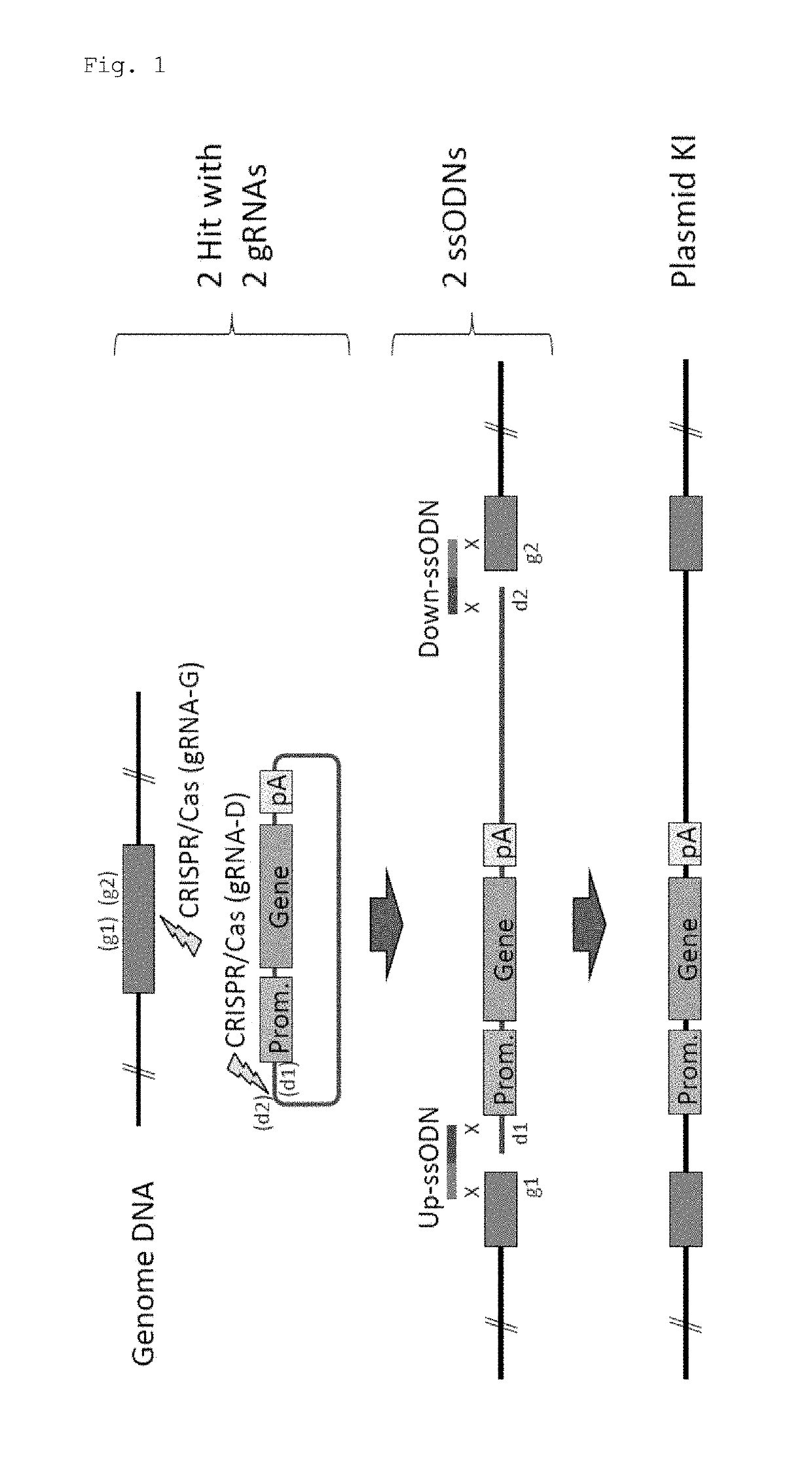
Method for knock-in of DNA into target region of mammalian genome, and cell
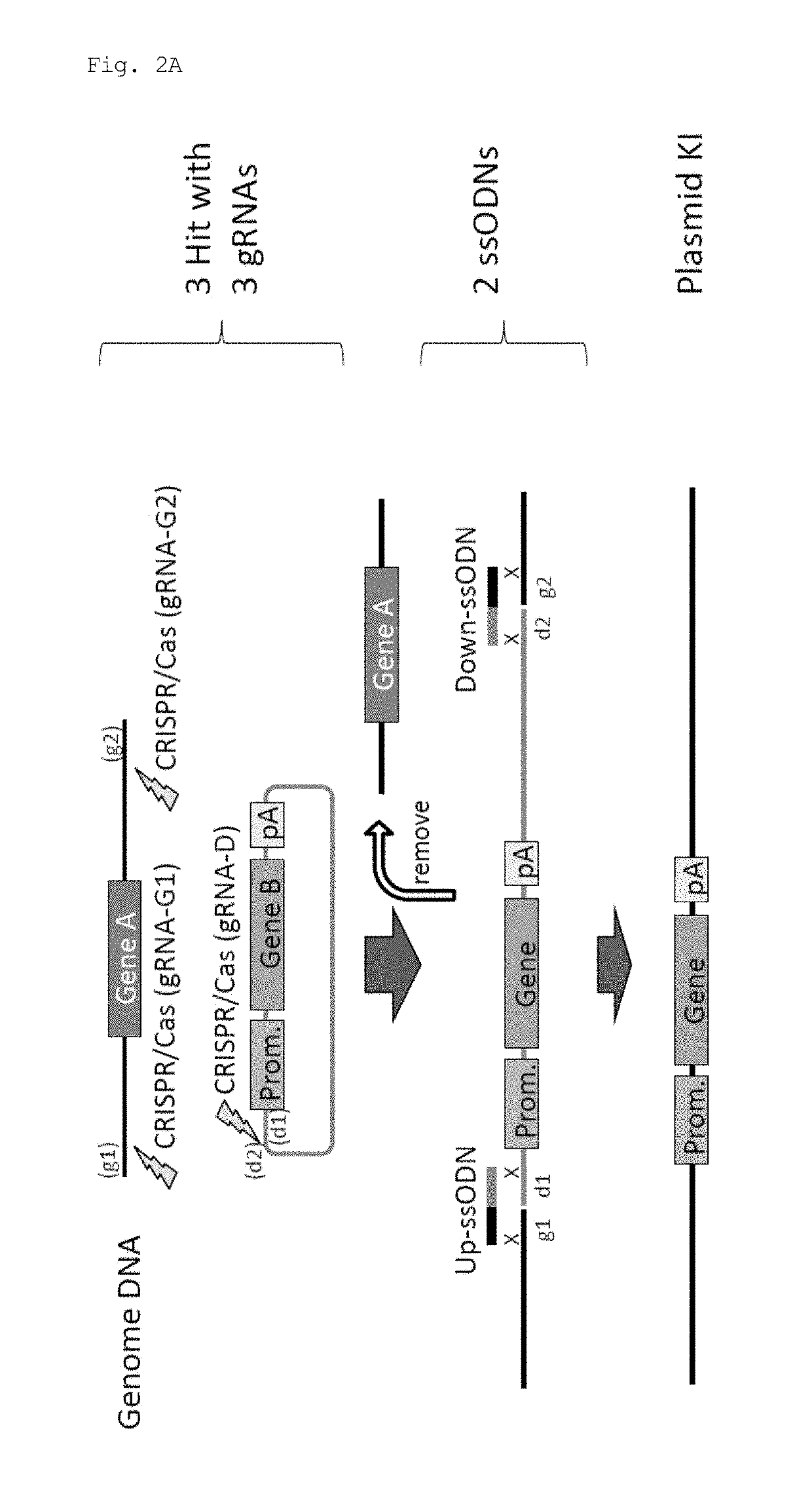
ActiveUS20170251647A1Efficient and accurateGenerate efficientlySugar derivativesStable introduction of DNAMammalian genomeNuclease
This invention provides a method for knock-in of a donor DNA into the genome of a cell, comprising introducing at least one artificial nuclease system capable of cleaving target sequence(s) of the cell genome, the donor DNA, and two single-stranded oligonucleotides (ssODNs) into the cell, the artificial nuclease system cleaving the target sequence(s) on the cell genome, the two ssODNs each complementary to one of the ends generated by the target sequence cleavage in the cell genome and to one of the introduction ends of the donor DNA, the donor DNA being knocked-in at the cleavage site via the two ssODNs.
Owner:KYOTO UNIV
ARF-P19, a novel regulator of the mammalian cell cycle
The INK4A (MTS1, CDKN2) gene encodes a specific inhibitor (InK4a-p16) of the cyclin D-dependent kinases CDK4 and CDK6. InK4a-p16 can block these kinase from phosphorylating the retinoblastoma protein (pRb), preventing exit from the G1 phase of the cell cycle. Deletions and mutations involving the gene encoding InK4a-p16, INK4A, occur frequently in cancer cells, implying that INK4a-p16, like pRb, suppresses tumor formulation. However, a completely unrelated protein (ARF-p19) arises in major part from an alternative reading frame of the mouse INK4A gene. Expression of an ARF-p19 cDNA (SEQ ID NO:1) in rodent fibroblasts induces both G1 and G2 phase arrest. Economical reutilization of protein coding sequences in this manner is without precedent in mammalian genomes, and the unitary inheritance of INK4a-p16 and ARF-p19 may reflect a dual requirement for both proteins in cell cycle control.
Owner:ST JUDE CHILDRENS RES HOSPITAL INC
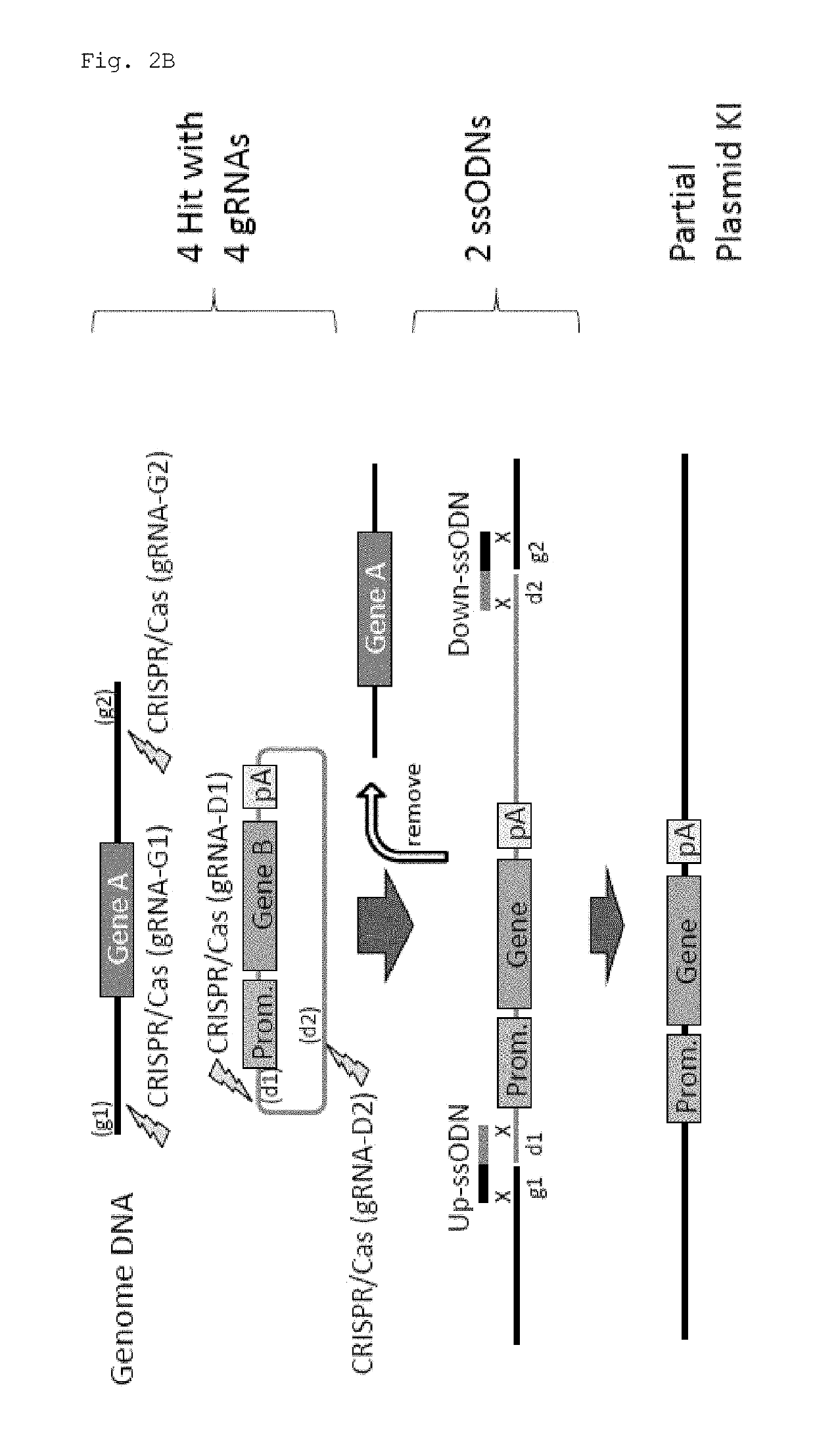
Method for knock-in of DNA into target region of mammalian genome, and cell
ActiveUS10362771B2Accurate and efficientGenerate knock-in animals several timesSugar derivativesStable introduction of DNAMammalian genomeMammal
This invention provides a method for knock-in of a donor DNA into the genome of a cell, comprising introducing at least one artificial nuclease system capable of cleaving target sequence(s) of the cell genome, the donor DNA, and two single-stranded oligonucleotides (ssODNs) into the cell,the artificial nuclease system cleaving the target sequence(s) on the cell genome, the two ssODNs each complementary to one of the ends generated by the target sequence cleavage in the cell genome and to one of the introduction ends of the donor DNA, the donor DNA being knocked-in at the cleavage site via the two ssODNs.
Owner:KYOTO UNIV
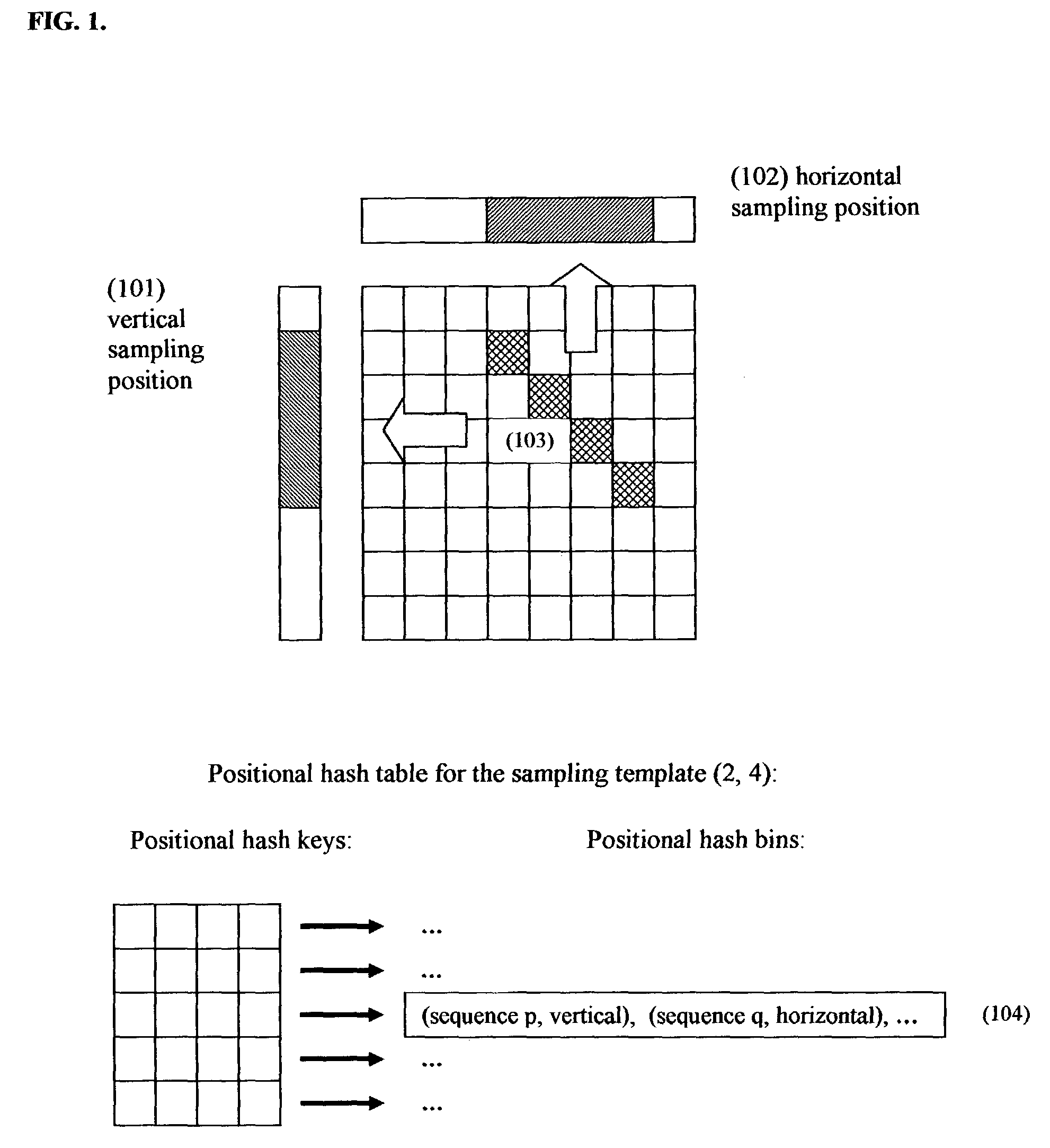
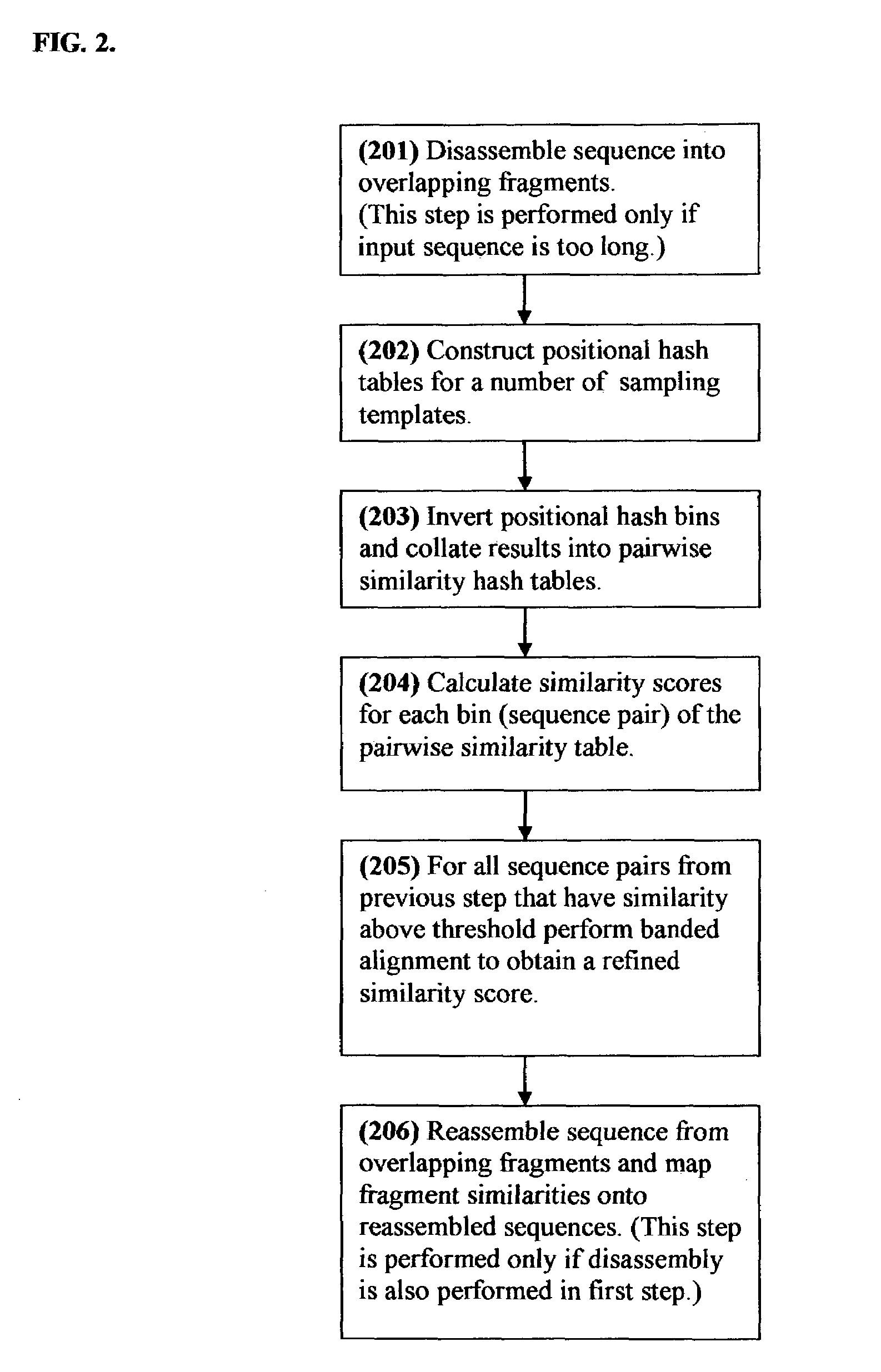
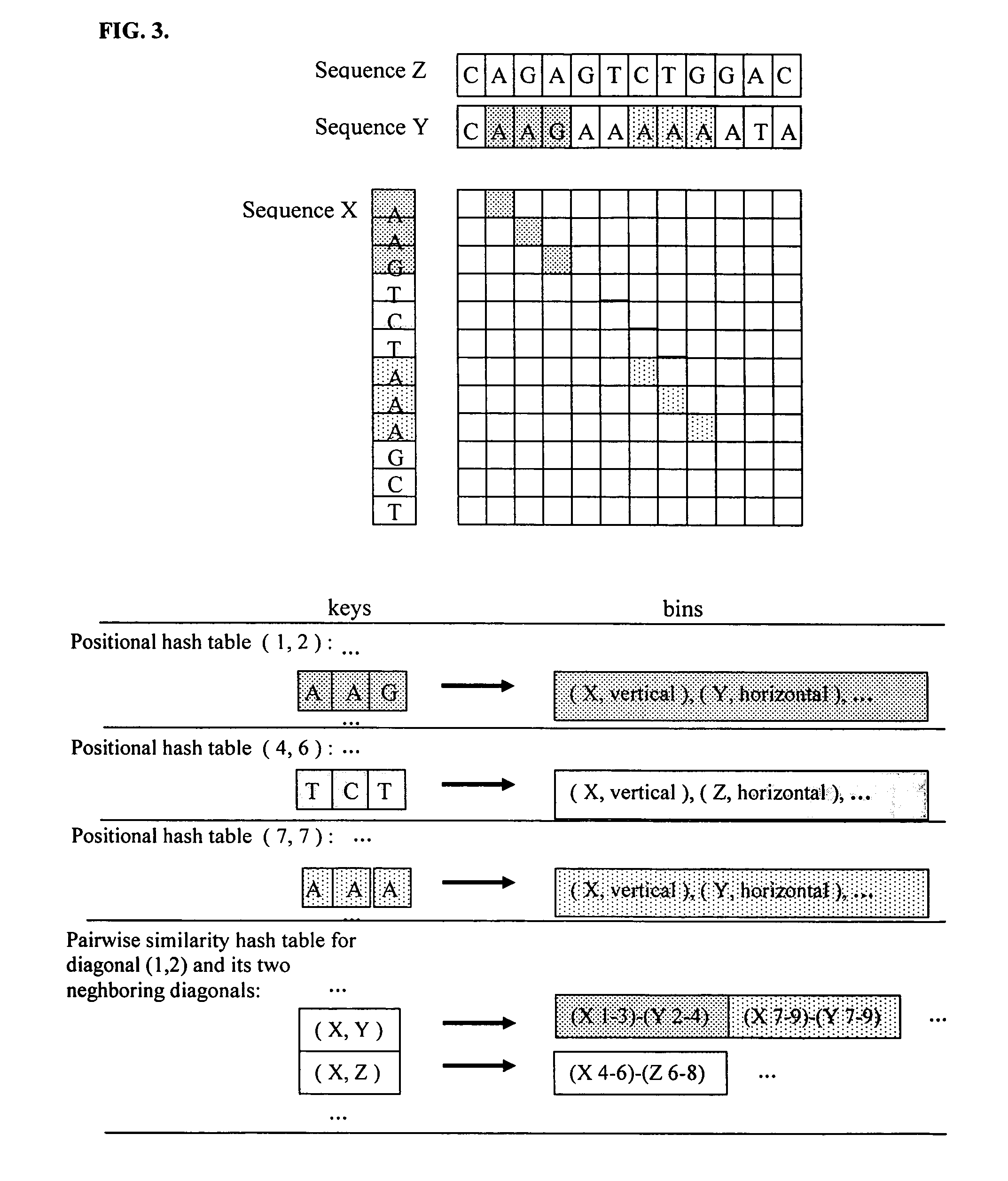
Positional hashing method for performing DNA sequence similarity search
Positional Hashing, a novel method for detecting similarities between texts such as DNA sequences, amino acid sequences, and texts in natural language is disclosed. The method is particularly well suited for large-scale comparisons such as that of mutual comparisons of millions of sequence fragments that result from mammalian-scale sequencing projects and for whole-genome comparisons of multiple mammalian genomes. Positional Hashing is carried out by breaking the sequence comparison problem along its natural structure, solving the subproblems independently, and then collating the solutions into an overall result. The decomposition of the problem into subproblems enables parallelization, whereby a large number of nodes in a computer cluster or a computer farm are concurrently employed on solving the problem without incurring the quadratic time performance penalty characteristic of prior art. Positional Hashing may be used by itself or as a filtering step in conjunction with a variety of sequence comparison algorithms.
Owner:IP GENESIS
Method for producing human serum albumin by using animal mammary gland and expression vector thereof
ActiveCN103898157AIncrease net profitIncrease profitSerum albuminFermentationMammalian genomeGenetics
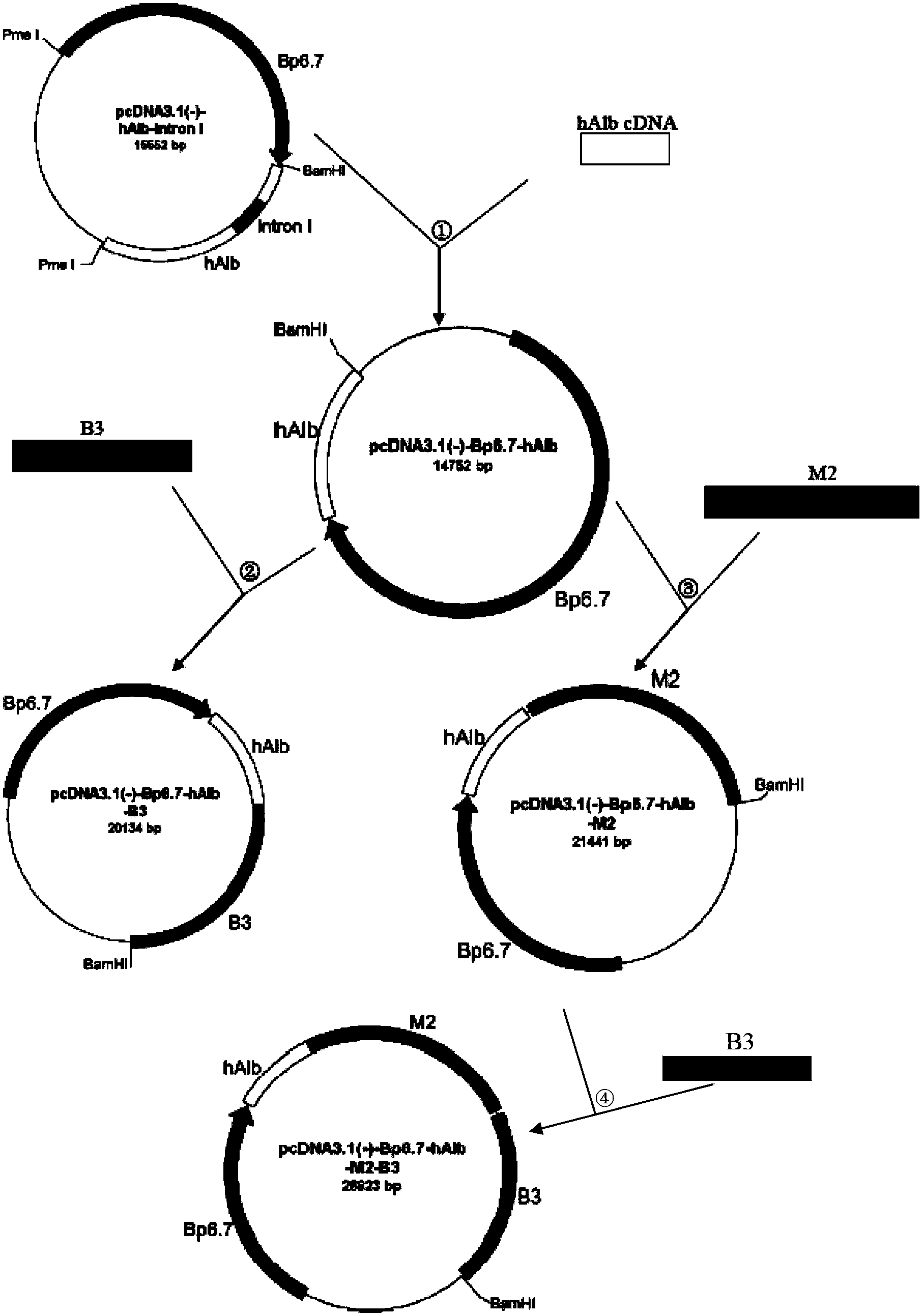
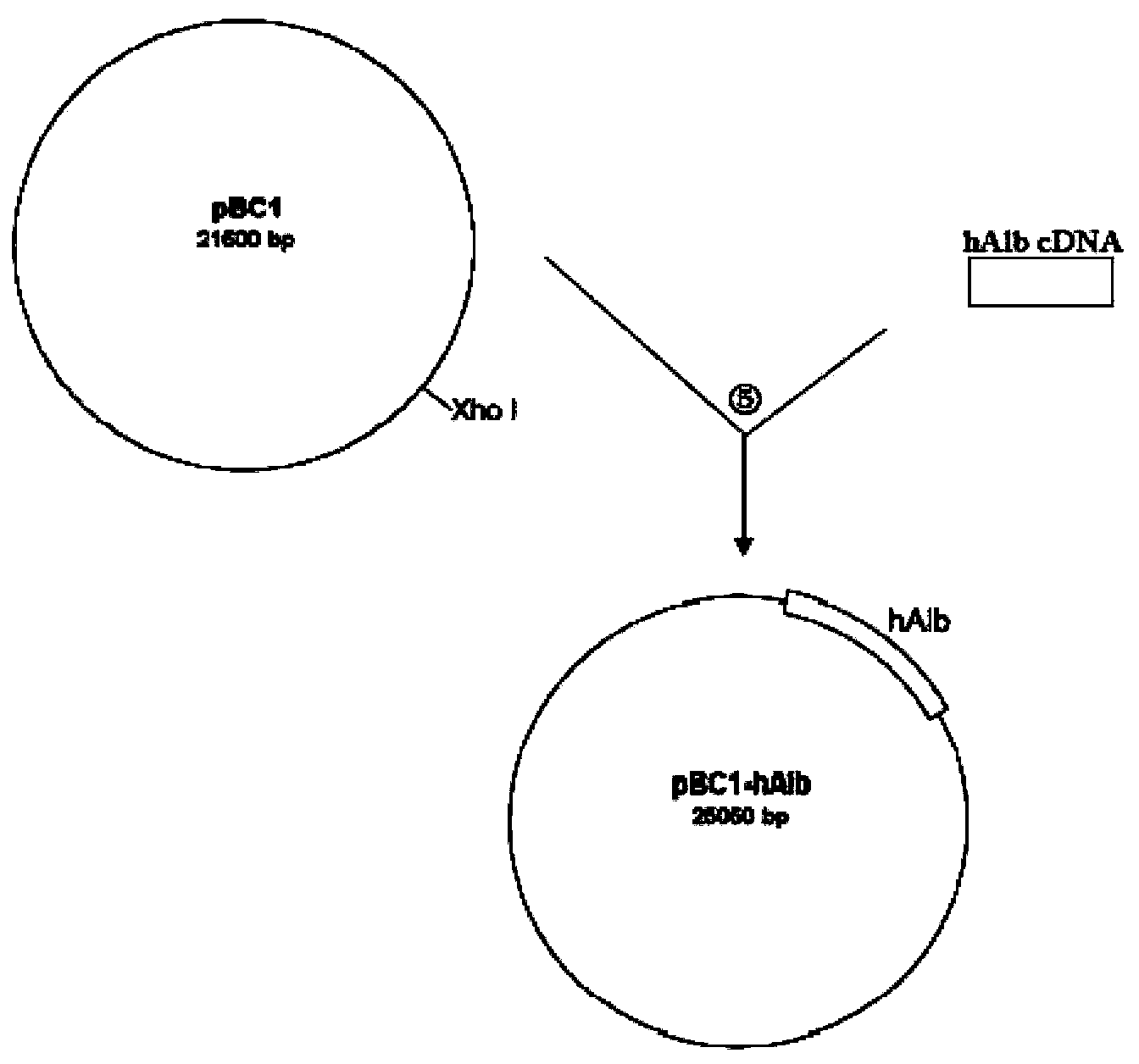
The invention discloses a method for producing human serum albumin by using mammary glands of transgenic animals and an expression vector thereof. The method provided by the invention comprises the steps of: using a vector to transfer a human serum albumin gene expression cassette to a genome of a mammal, so as to obtain a transgenic animal; and acquiring human serum albumin from the milk of the transgenic animal. The human serum albumin gene expression cassette from 5' to 3' includes a promoter, a human serum albumin gene and a terminator in order, also includes one or two selected from a 3' flanking sequence M2 of the human serum albumin gene and a 3' flanking sequence B3 of a beta-casein gene, wherein M2 or B3 are located after the terminator. According to the method provided by the invention, the concentration of human serum albumin in animal milk can at least reach 4.6-6.7 g / L, which can fully meet the commercial utilization level. The method isolates human serum albumin from milk of transgenic cow, so as to provide a low-cost and high-yield method for human serum albumin production.
Owner:SHANGHAI INST OF MEDICAL GENETICS SHANGHAI CHILDRENS HOSPITAL
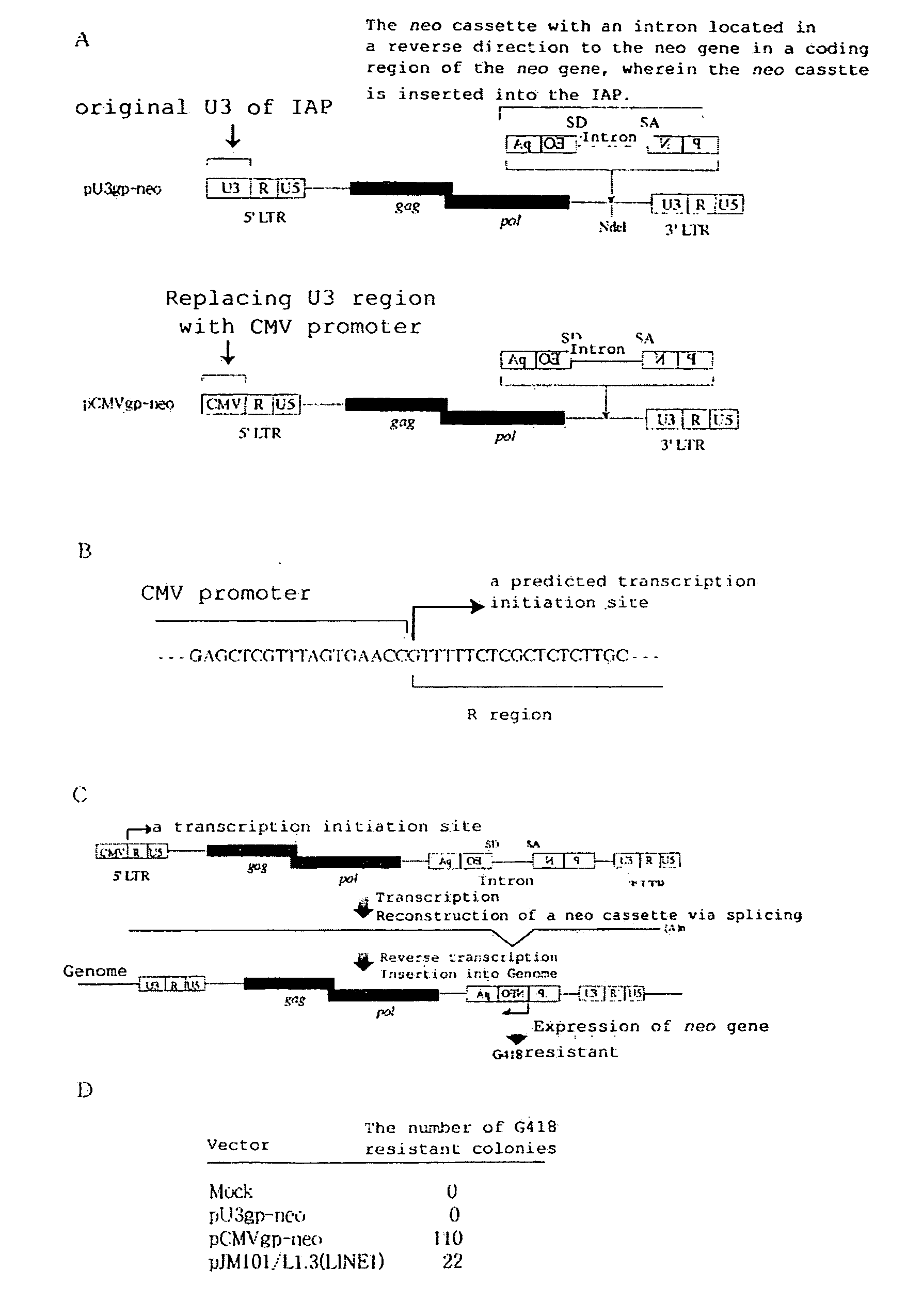
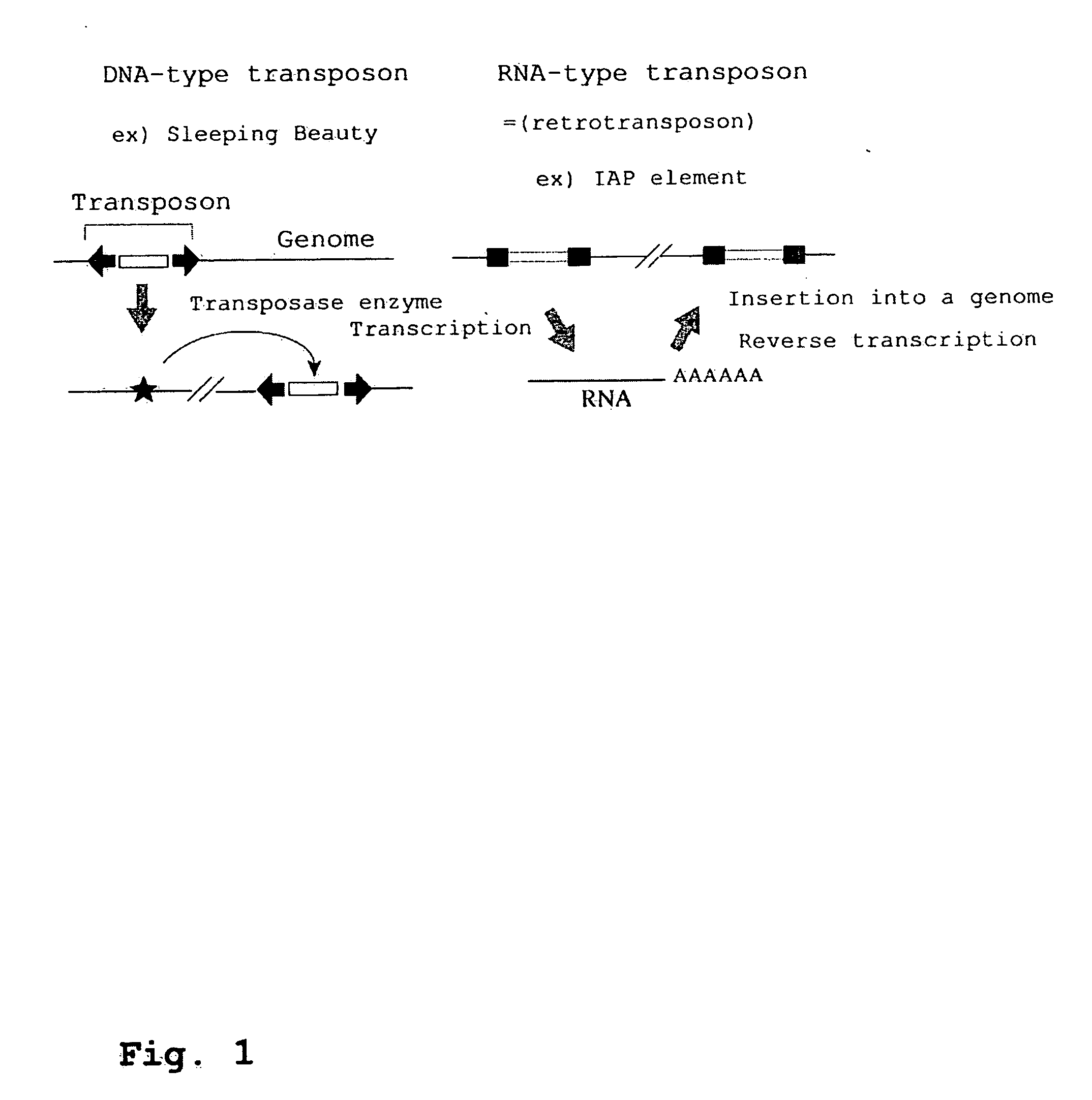
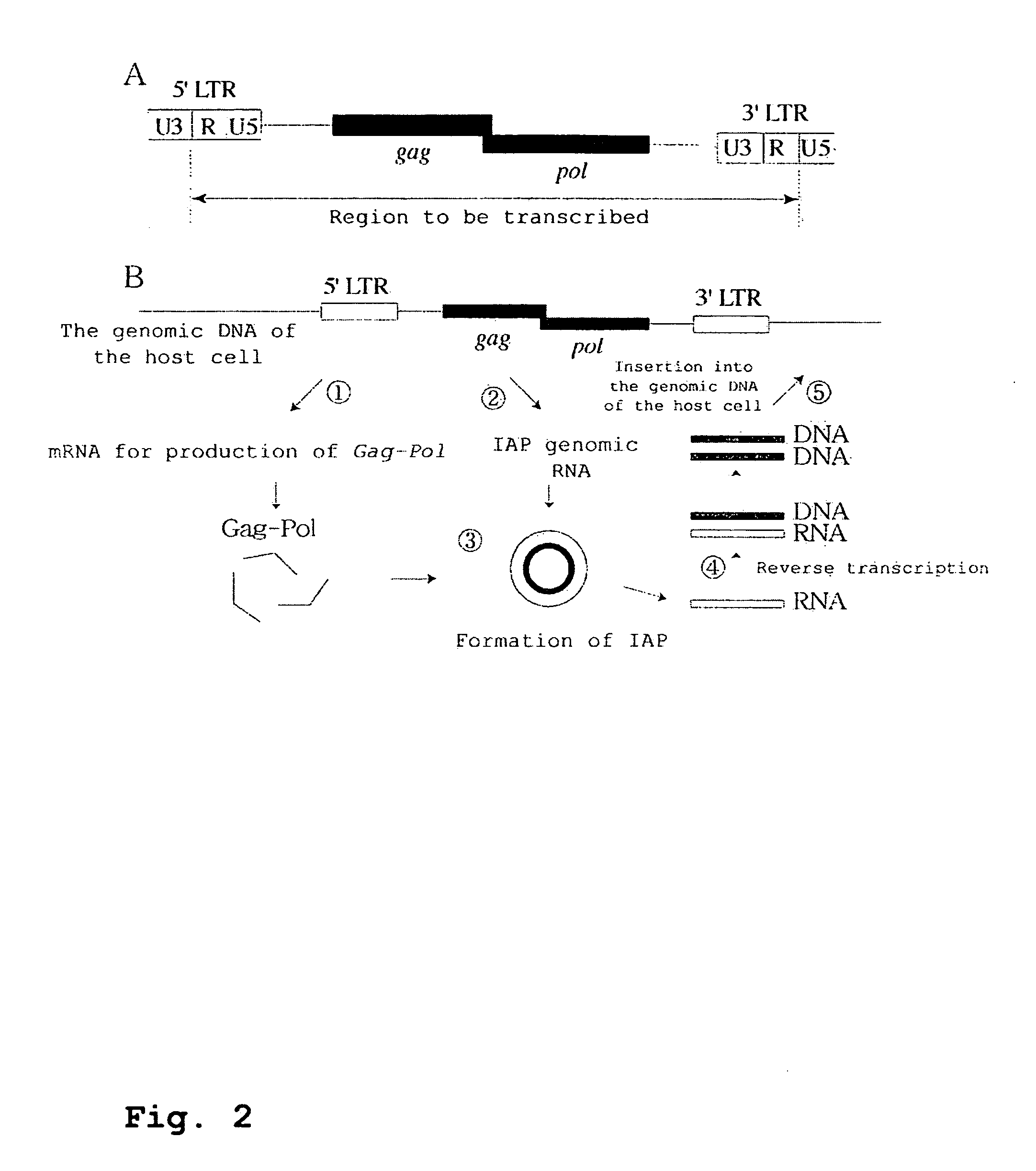
Development of Mammalian Genome Modification Technique Using Retrotransposon
InactiveUS20080104723A1Promoting transpositionEasy to identifySugar derivativesMicrobiological testing/measurementMammalian genomeMammal
To comprehensively modify genome, it is intended to develop a transposition system of the copy and paste type which has an improved efficiency. This object has been achieved by the finding that an LTR retrotransposon is partly usable in a transposition system. Namely, a technique of efficiently transferring a foreign gene into a cell by using a transposon. More specifically speaking, a complete IPA element and a functional promoter sequence are found out. It is clarified that, without a combination of them, a retrotransposon cannot exert its function.
Owner:OSAKA INDAL PROMOTION ORG +1
Identification of multigene biomarkers
Methods for identifying multigene biomarkers for predicting sensitivity or resistance to an anti-cancer drug of interest, or multigene cancer prognostic biomarkers are disclosed. The disclosed methods are based on the classification of the mammalian genome into 51 transcription clusters, i.e., non-overlapping, functionally relevant groups of genes whose intra- group transcript levels are highly correlated. Also disclosed are specific multigene biomarkers for predicting sensitivity or resistance to tivozanib, or rapamycin, and a specific multigene biomarker for determining breast cancer prognosis, all of which were identified using the methods disclosed herein.
Owner:AVEO PHARM INC
Methods of extracting rna
Methods and materials are disclosed for rapid and simple extraction and isolation of RNA from a biological sample involving the use of an acidic solution and a solid phase binding material that has the ability to liberate nucleic acids from biological samples, including whole blood, without first performing any preliminary lysis to disrupt cells or viruses. No detergents or chaotropic substances for lysing cells or viruses are needed or used. Viral, bacterial and mammalian genomic RNA can be isolated using the method of the invention. RNA isolated by the present method is suitable for use in downstream processes such as RT-PCR.
Owner:鲁米金公司
Enhanced recombination of genomic loci
PendingUS20180245091A1Reduce in quantityIncrease the number ofMicrobiological testing/measurementTransferasesMammalian genomeGene cluster
The present disclosure provides methods to accelerate recombination at selected genomic loci, allowing recombination to occur, and selecting events with molecular variation within the selected loci. The accelerated recombination generates novel variations in gene clusters that are present in the plant or mammalian genomes.
Owner:MONSANTO TECH LLC
Methods of extracting nucleic acids
InactiveUS20070190526A1Rapid captureRapid and simple extraction and isolationSugar derivativesMicrobiological testing/measurementSolid phasesLysis
Methods and materials are disclosed for rapid and simple extraction and isolation of nucleic acids, particularly RNA, from a biological sample involving the use of an alkaline reagent followed by an acidic solution and a solid phase binding material that has the ability to liberate nucleic acids from biological samples, including whole blood, without first performing any preliminary lysis to disrupt cells or viruses. No detergents or chaotropic substances for lysing cells or viruses are needed or used. Viral, bacterial and mammalian genomic RNA can be obtained using the method of the invention. RNA obtained by the present method is suitable for use in downstream processes such as RT-PCR.
Owner:NEXGEN DIAGNOSTICS LLC
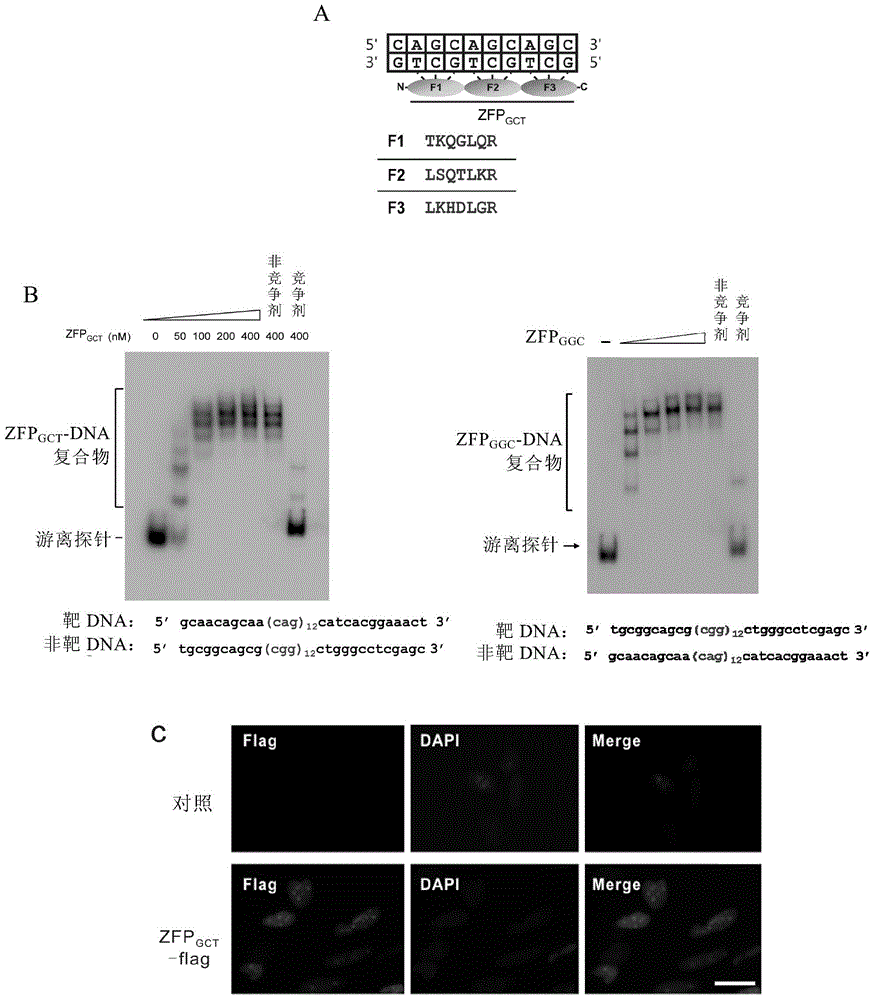
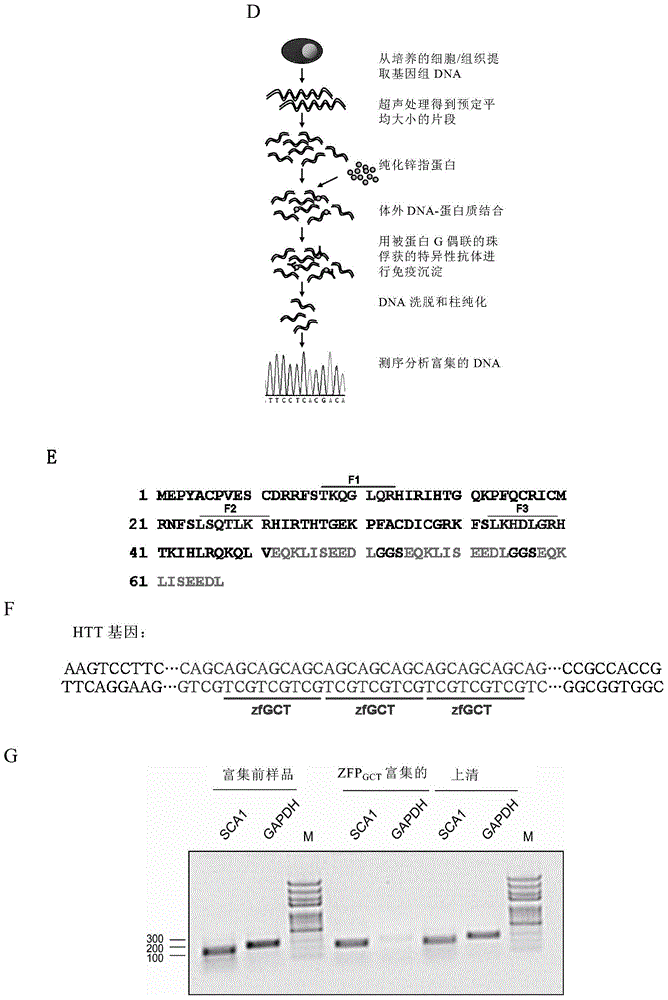
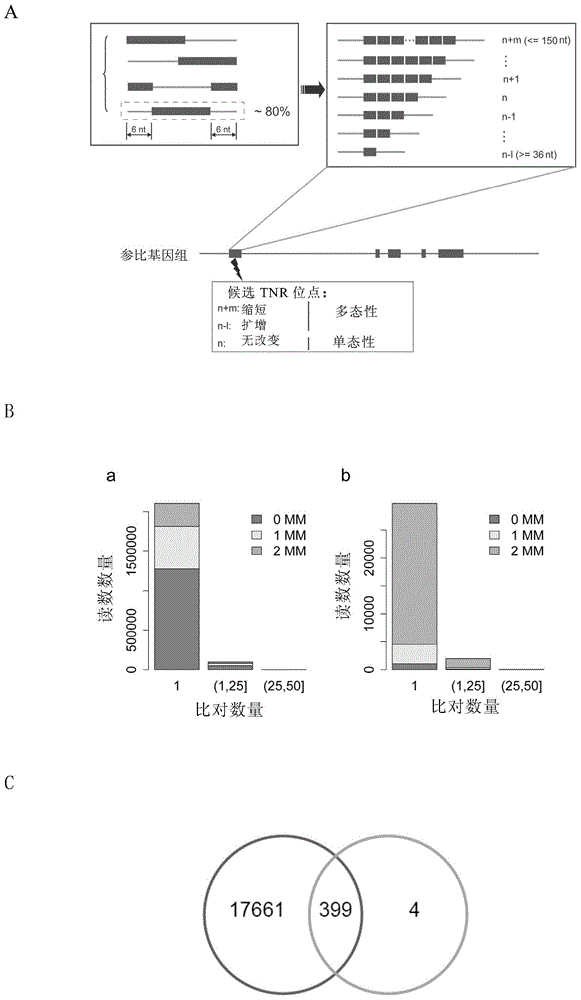
Method for detecting trinucleotide repeated sequence of mammal genome and application thereof
The invention relates to a method for detecting a trinucleotide repeated sequence of a mammal genome and an application thereof, and specifically, the invention relates to a method for detecting the trinucleotide repeated sequence of the mammal genome by using C2H2 zinc finger protein. The invention also relates to the specific C2H2 zinc finger protein, a coding sequence of the specific C2H2 zinc finger protein, polynucleotide constructs containing the coding sequence of the specific C2H2 zinc finger protein, cells, and an application of the C2H2 zinc finger protein, the coding sequence, the polynucleotide constructs and the cells.
Owner:CENT FOR EXCELLENCE IN MOLECULAR CELL SCI CHINESE ACAD OF SCI
Construction and application of mammary specific expression vector for human transferrin
ActiveCN103911393AImprove expression levelIncrease net profitTransferrinsFermentationTransferrin GeneMammal
The invention discloses a method for producing human transferrin by utilizing mammary gland of transgenic animals and an expression vector of human transferrin. The method comprises: utilizing the vector containing a human transferrin gene expression cassette to transfer the human transferrin gene expression cassette into a mammal genome to obtain a transgenic animal and further to obtain human transferrin from milk of the transgenic animal, wherein the expression cassette successively comprises a promoter, human transferrin gene and cattle growth hormone gene terminator from 5' to 3', the promoter is goat beta-casein gene with the sequence of SEQ ID NO. 1, the sequence of human transferrin is shown by SEQ ID NO. 2, and the vector is a eukaryotic expression carrier pcDNA3.1, and the mammals do not contain human. According to the method, the concentration of human transferrin in animal milk is at least 5 g / L and is improved by 160 times or more compared with the human transferrin concentration by employing a conventional method, and therefore the method has great business utilization value.
Owner:SHANGHAI TAO TAO ENG +1
Epigenetic modification of mammalian genomes using targeted endonucleases
The present disclosure provides genetically engineered cell lines comprising chromosomally integrated synthetic sequences having predetermined epigenetic modifications, wherein a predetermined epigenetic modification is correlated with a known diagnosis, prognosis or level of sensitivity to a disease treatment. Also provided are kits comprising said epigenetically modified synthetic nucleic acids or cells comprising said epigenetically modified synthetic nucleic acids that can be used as reference standards for predicting responsiveness to therapeutic treatments, diagnosing diseases, or predicting disease prognosis.
Owner:SIGMA ALDRICH CO LLC
Method and arrangement for determining traits of a mammal
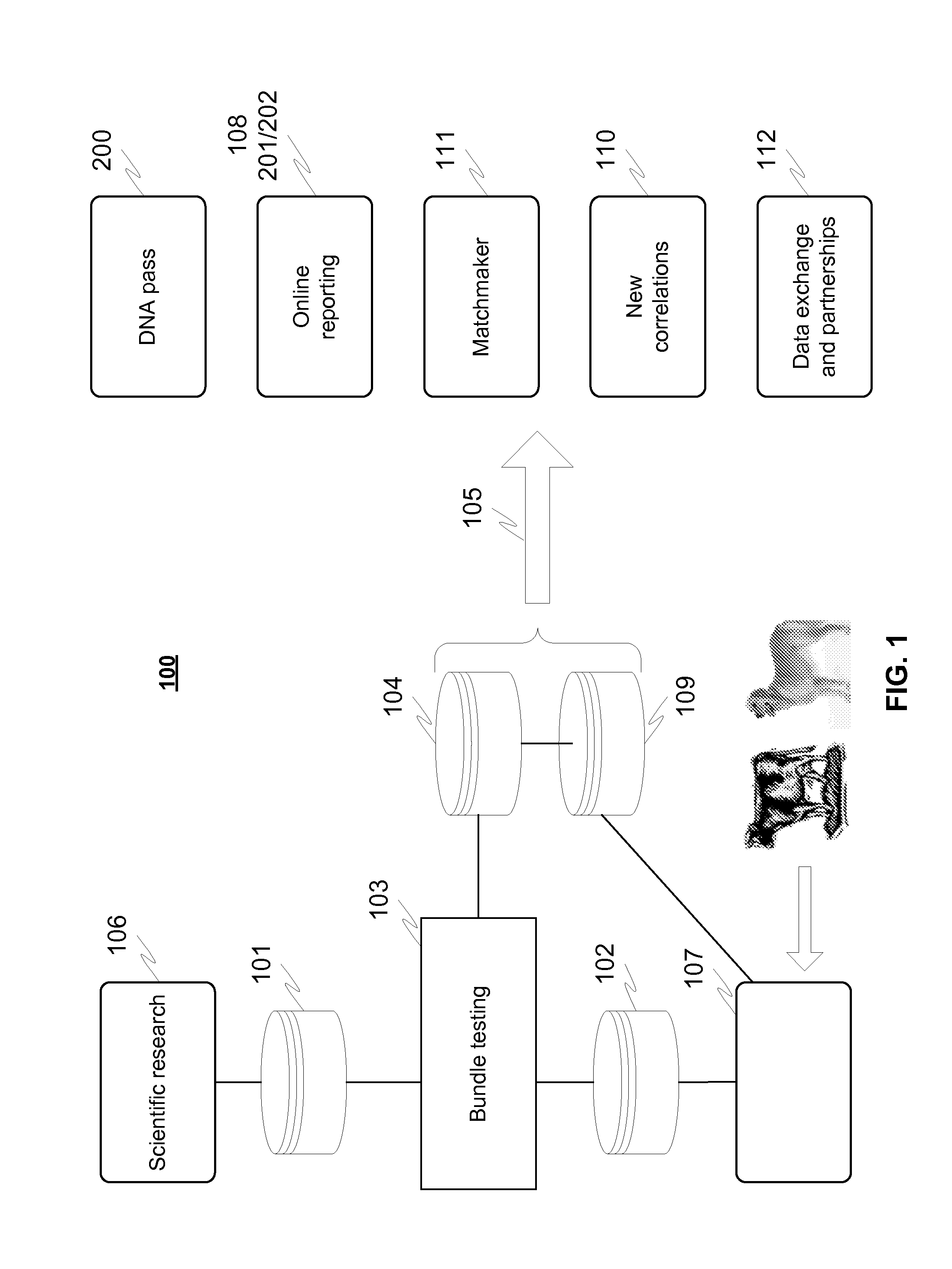
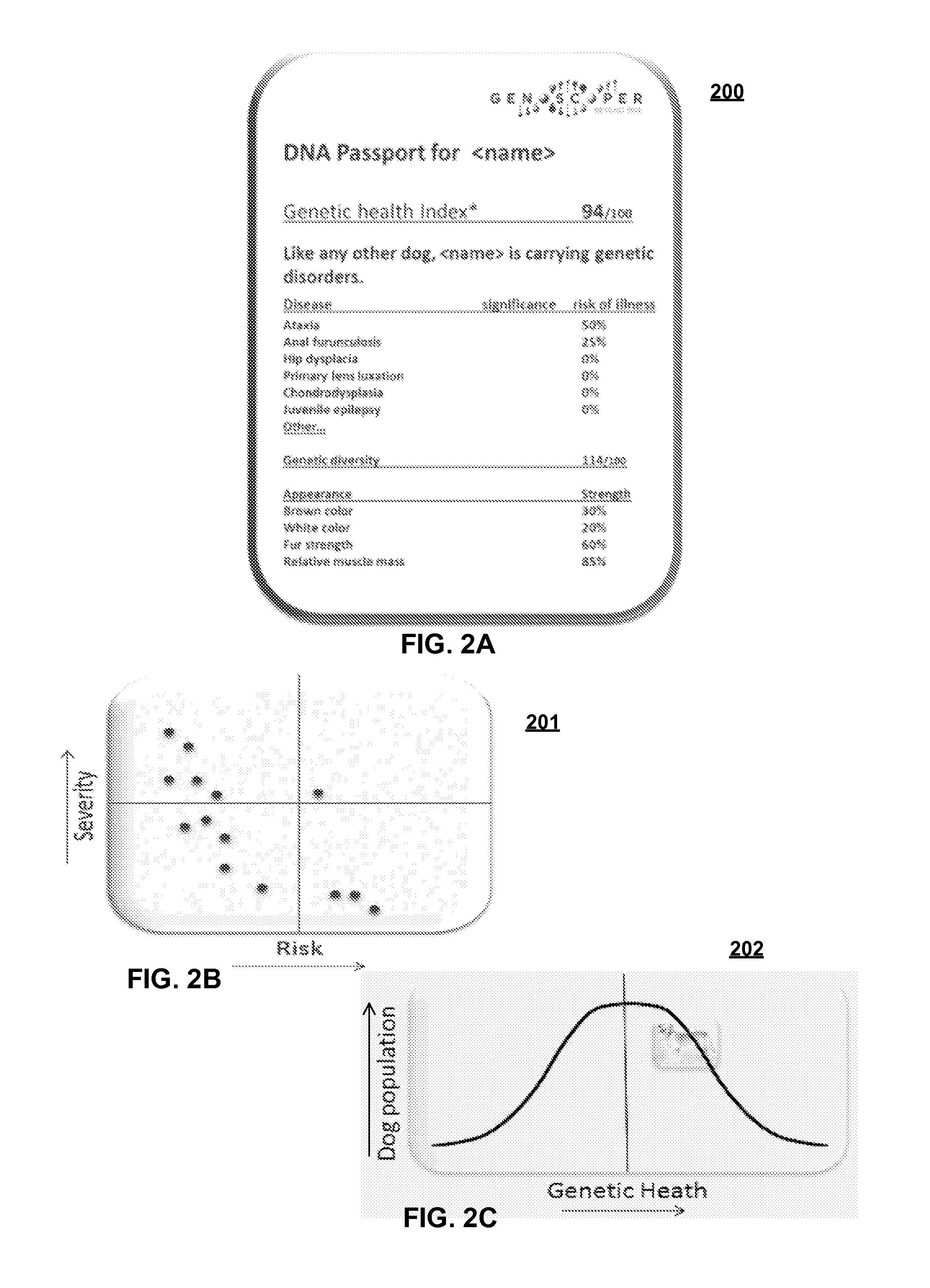
InactiveUS20150286774A1Improve fidelityEasy to produceHealth-index calculationMicrobiological testing/measurementDisease riskMammalian genome
A system for determining traits for an individual mammal includes a first database with genomic data of known traits as well as identified correlations including details of the trait, mutation, disease risk and / or affected breeds of a mammal species, and a second database based on a gene test made for the individual mammal, the database including genotyping data of the individual mammal to be analyzed. A plurality of markers is determined for regions in the mammal's genome so that at least first portion of the markers relates to correlations in various known gene based traits, and at least second portion of the markers used in determining differs from the first portion of the markers. Then the genotyping data of the individual mammal's genome corresponding to the first portion of the markers is compared in order to make correlations and thereby determine a probability or risk or severity of the traits.
Owner:MARS INC
Method of sequencing by hybridization of oligonucleotide probes
The conditions under which oligonucleotide probes hybridize preferentially with entirely complementary and homologous nucleic acid targets are described. Using these hybridization conditions, overlapping oligonucleotide probes associate with a target nucleic acid. Following washes, positive hybridization signals are used to assemble the sequence of a given nucleic acid fragment. Representative target nucleic acids are applied as dots. Up to to 100,000 probes of the type (A,T,C,G)(A,T,C,G)NB(A,T,C,G) are used to determine sequence information by simultaneous hybridization with nucleic acid molecules bound to a filter. Additional hybridization conditions are provided that allow stringent hybridization of 6-10 nucleotide long oligomers which extends the utility of the invention. A computer process determines the information sequence of the target nucleic acid which can include targets with the complexity of mammalian genomes. Sequence generation can be obtained for a large complex mammalian genome in a single process.
Owner:COMPLETE GENOMICS INC
Modification method of mammal genome
InactiveCN106520831AStrong specificityNucleic acid vectorOther foreign material introduction processesSingle strand dnaGenetic engineering
The invention provides a modification method of the mammal genome, belonging to the technical field of genetic engineering. According to the modification method, the target gene DNA sequence of the specific area on the mammal genome is selected according to the research purpose, a pair of single-stranded DNA oligonucleotides for identifying the target gene DNA sequence is designed, the pair of single-stranded DNA oligonucleotides and a plasmid vector pcDNA-DN constructed by adopting the method provided by the invention are used for cotransfection of the mammalian cells at the mass ratio of (1-2): (1-2): (5-20), after 48h, the genome DNA is extracted, the PCR amplification is carried out, the clone sequencing is carried out, and the mutation ratio of the target region sequence is detected. Compared with the prior art, the modification method has the advantages that by synthesizing a pair of short-chain guide DNA, nuclease X is guided to cut the specific gene of the mammal in the cell or individual level, so that the purpose of knocking out or modifying one certain gene is achieved, thus the functions of the genes are analyzed, and a gene mutation pool is constructed. The modification method has the beneficial effects that the total length of the guide DNA is 40nt or longer, and the specificity of genome modification is greatly increased.
Owner:青岛市畜牧兽医研究所
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com