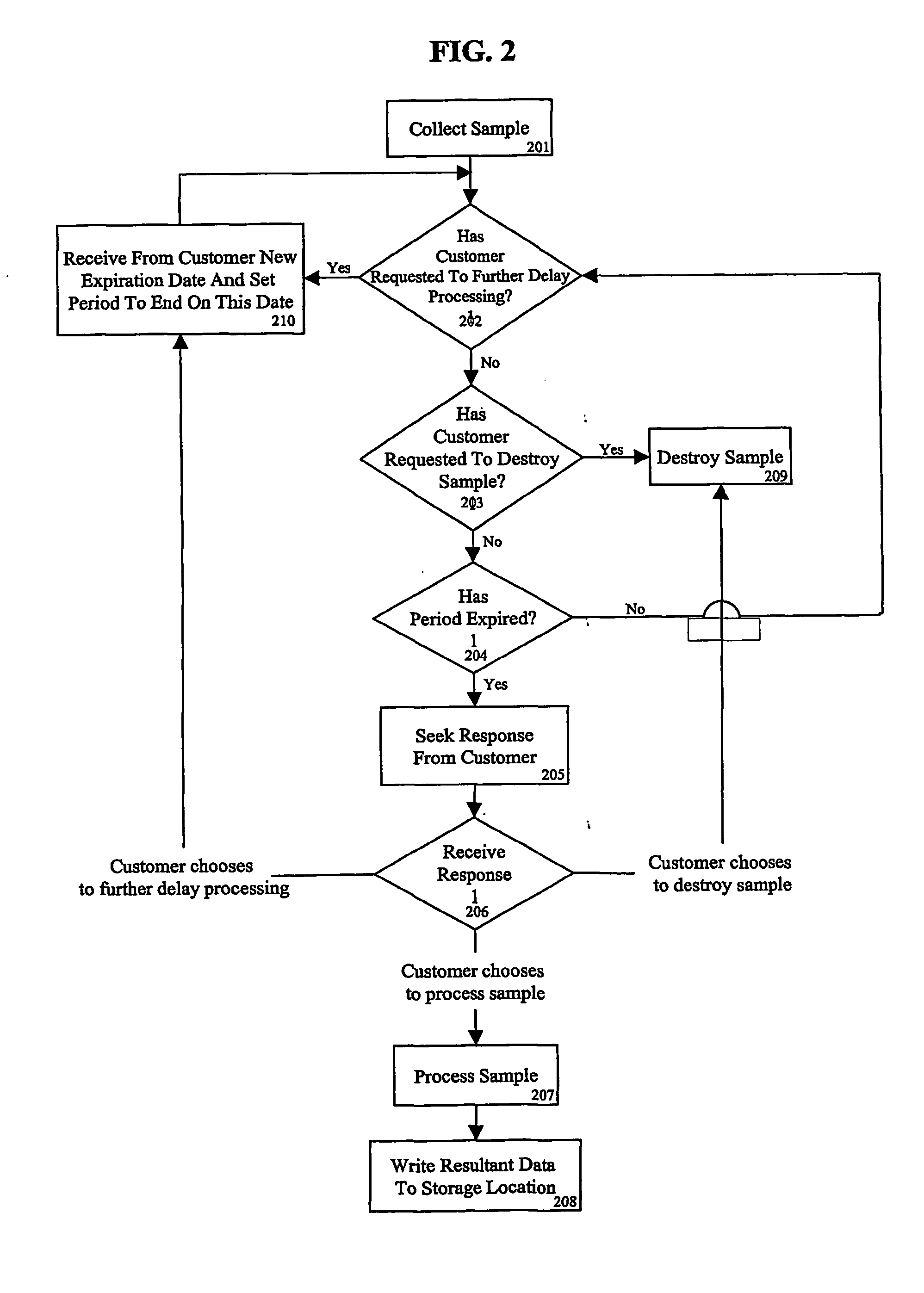
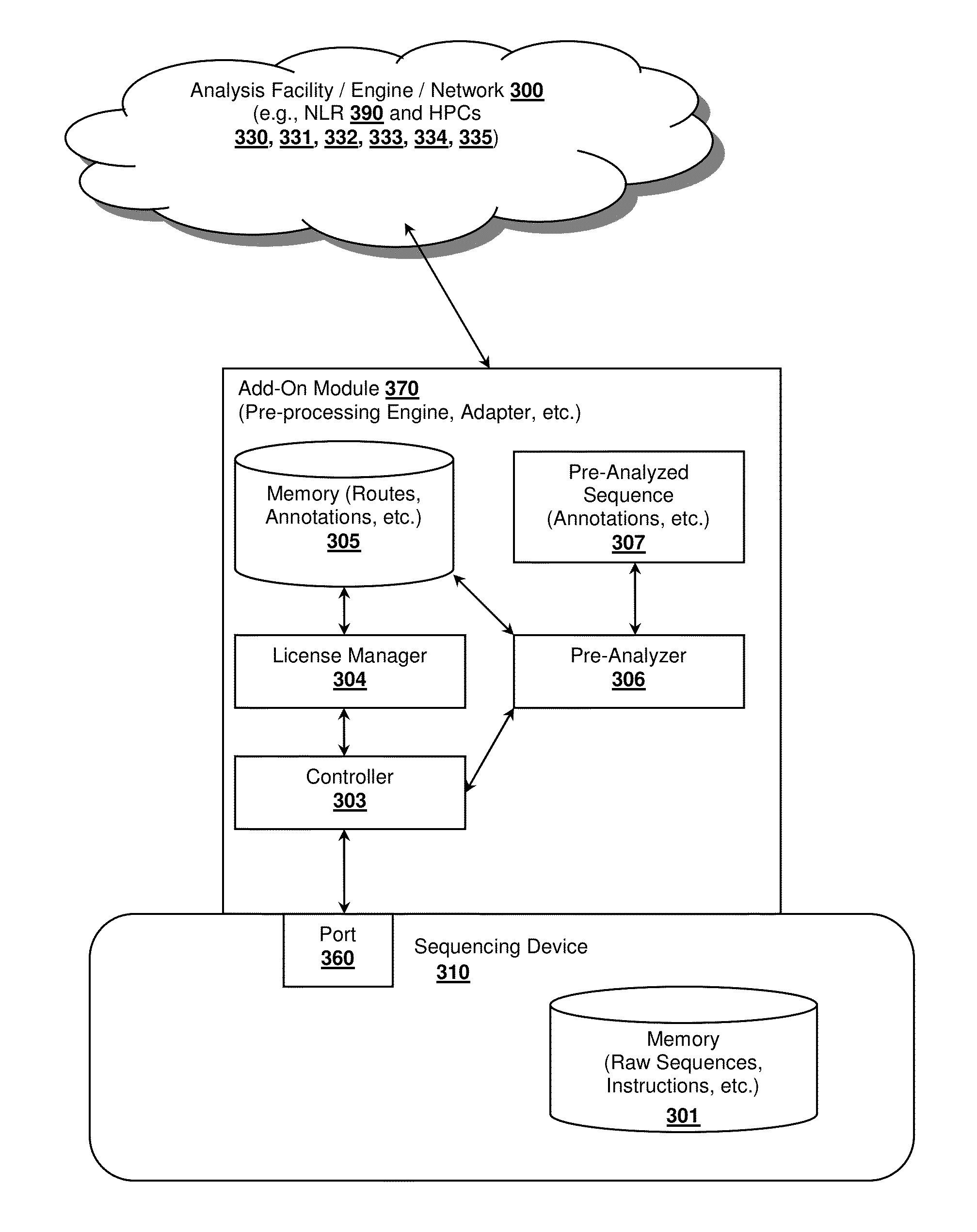
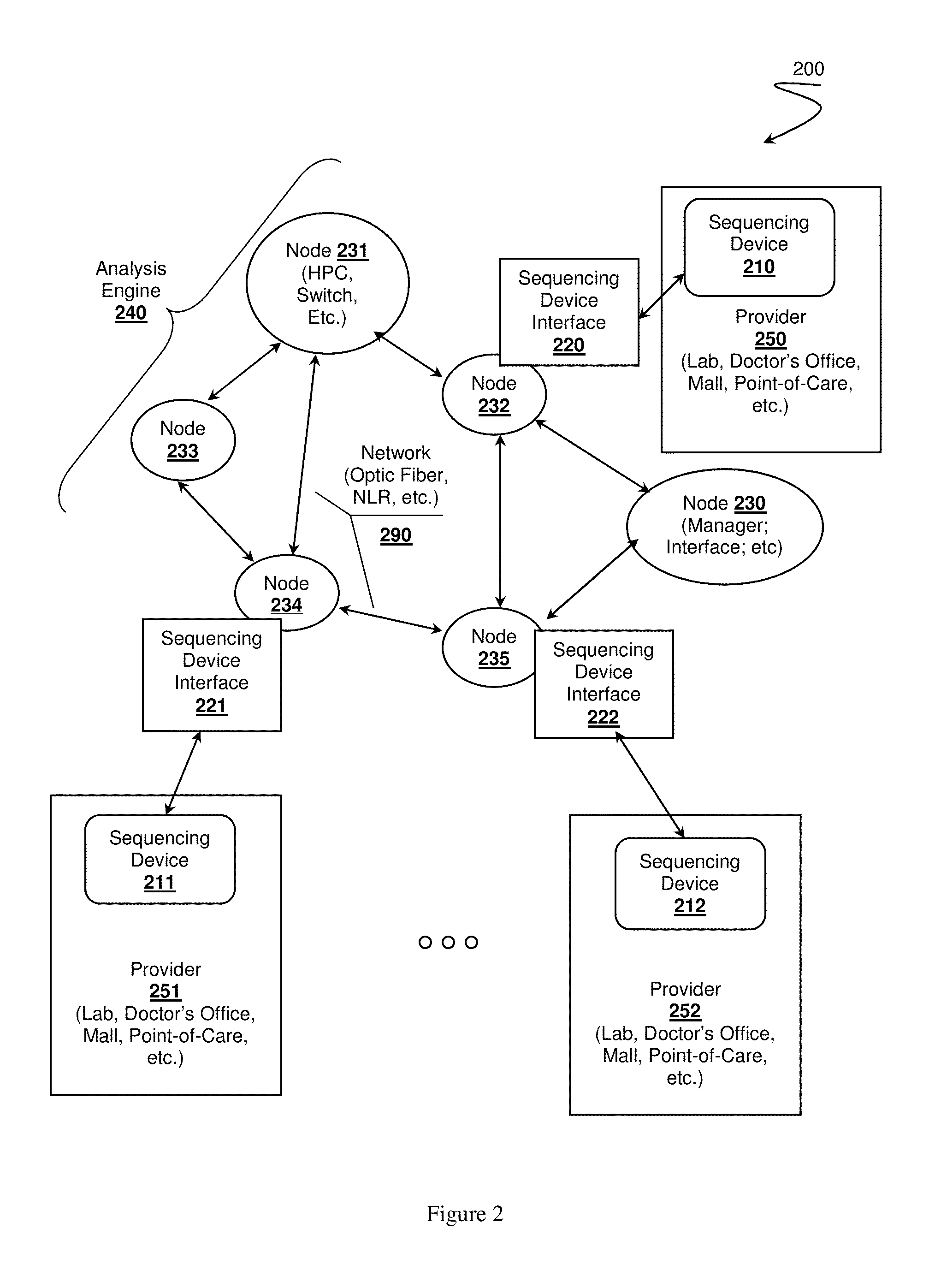
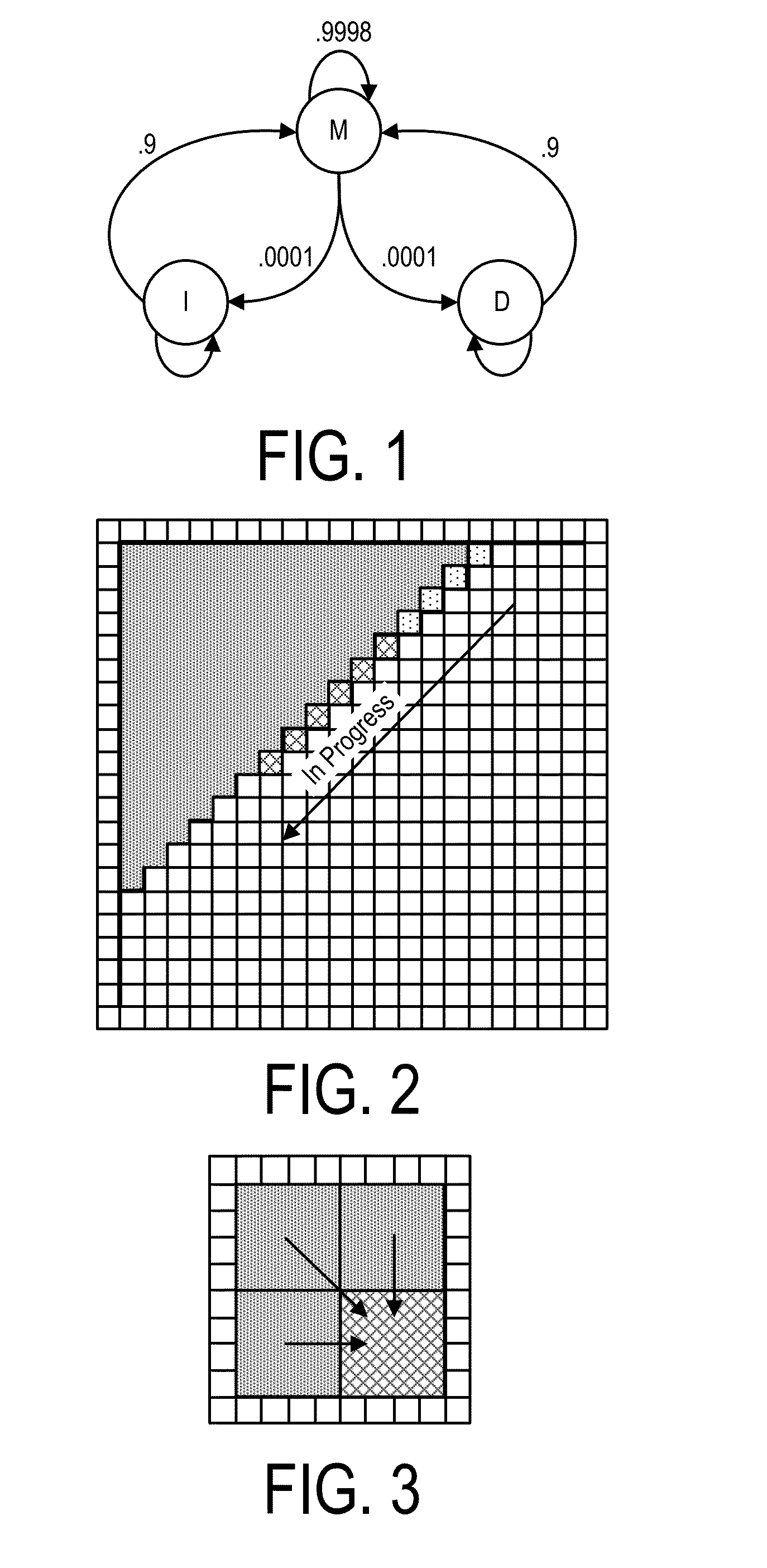
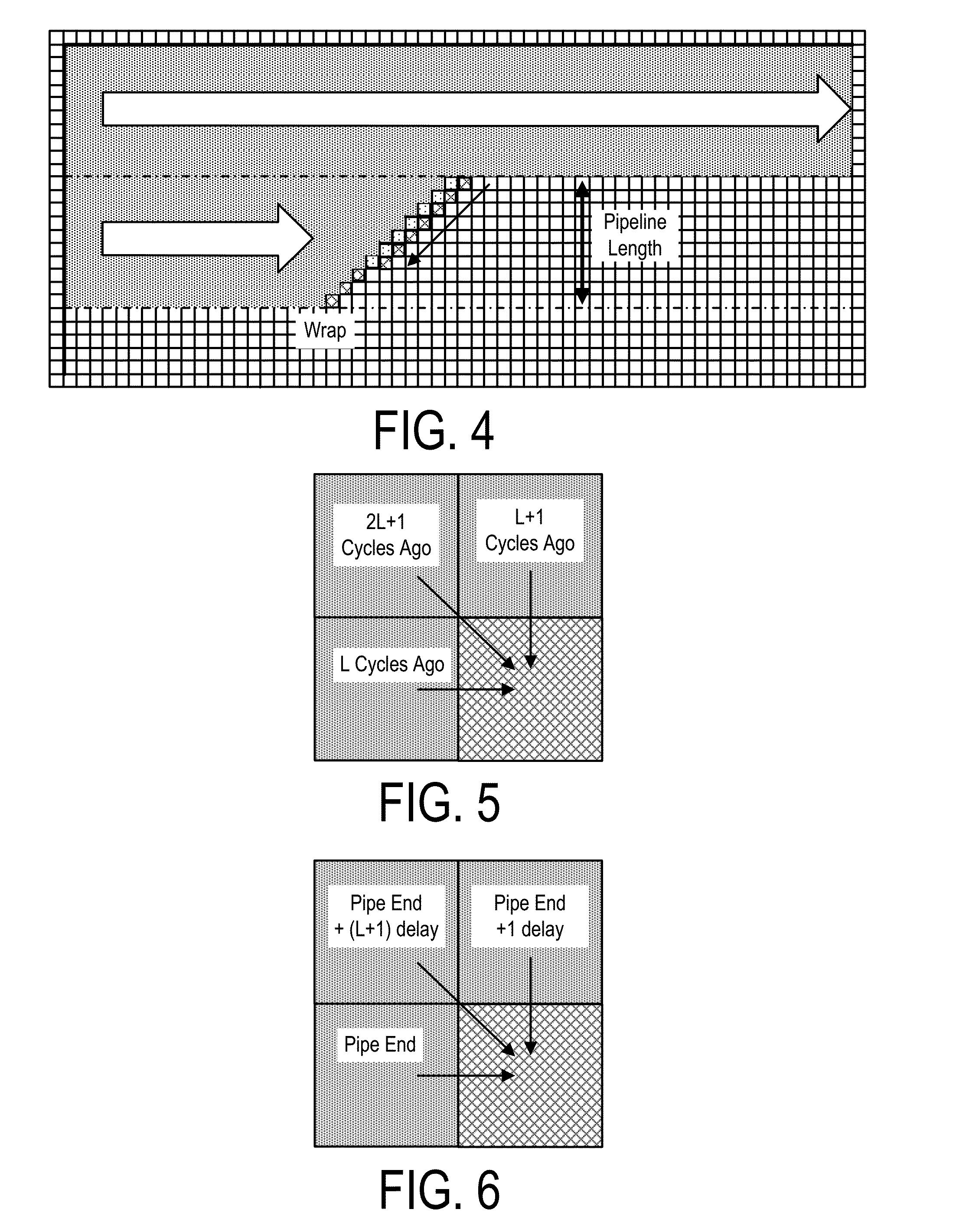
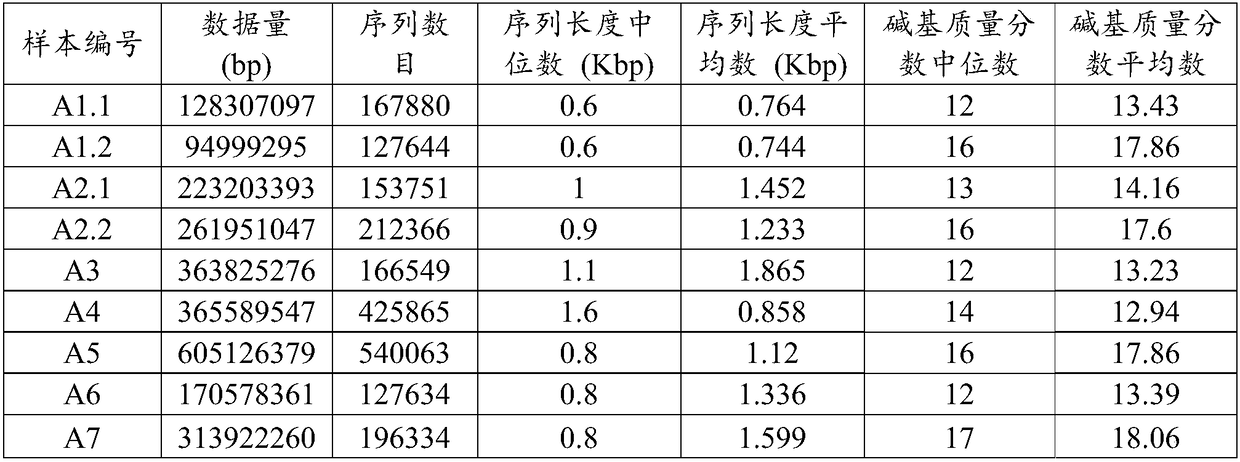
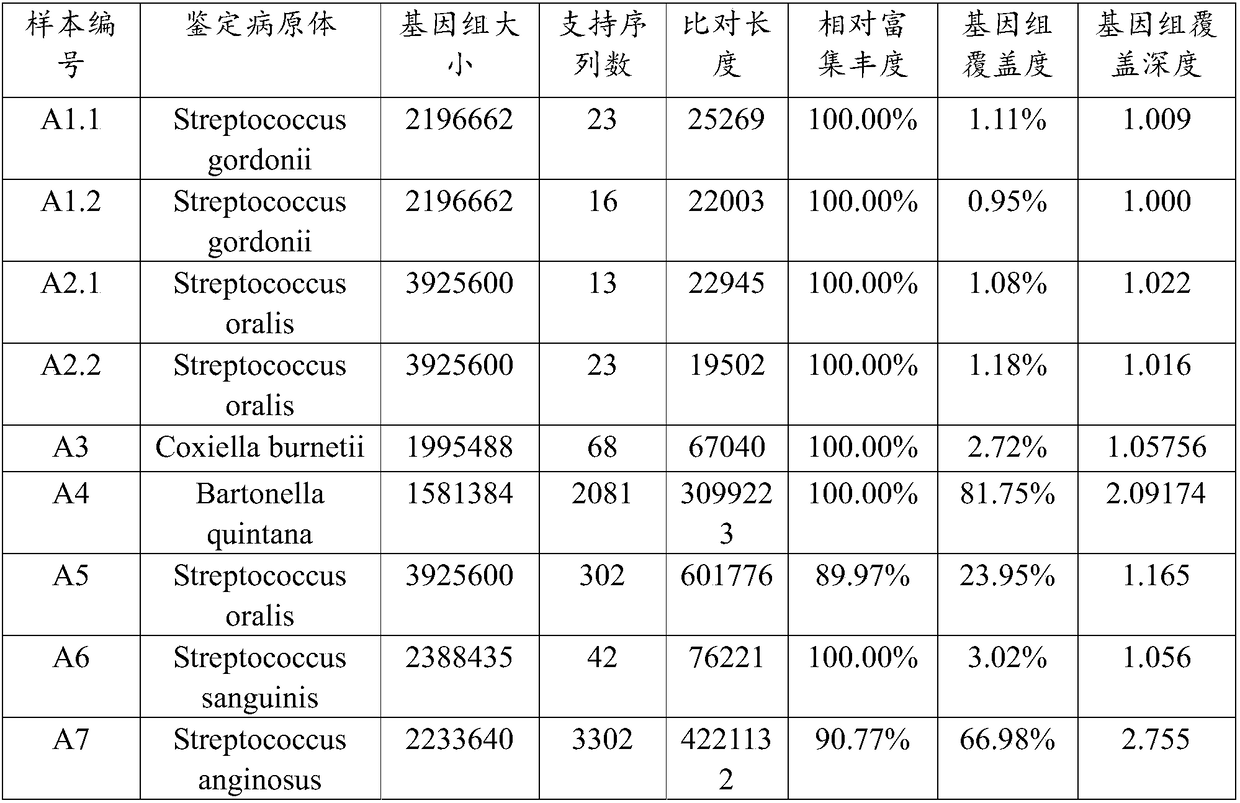
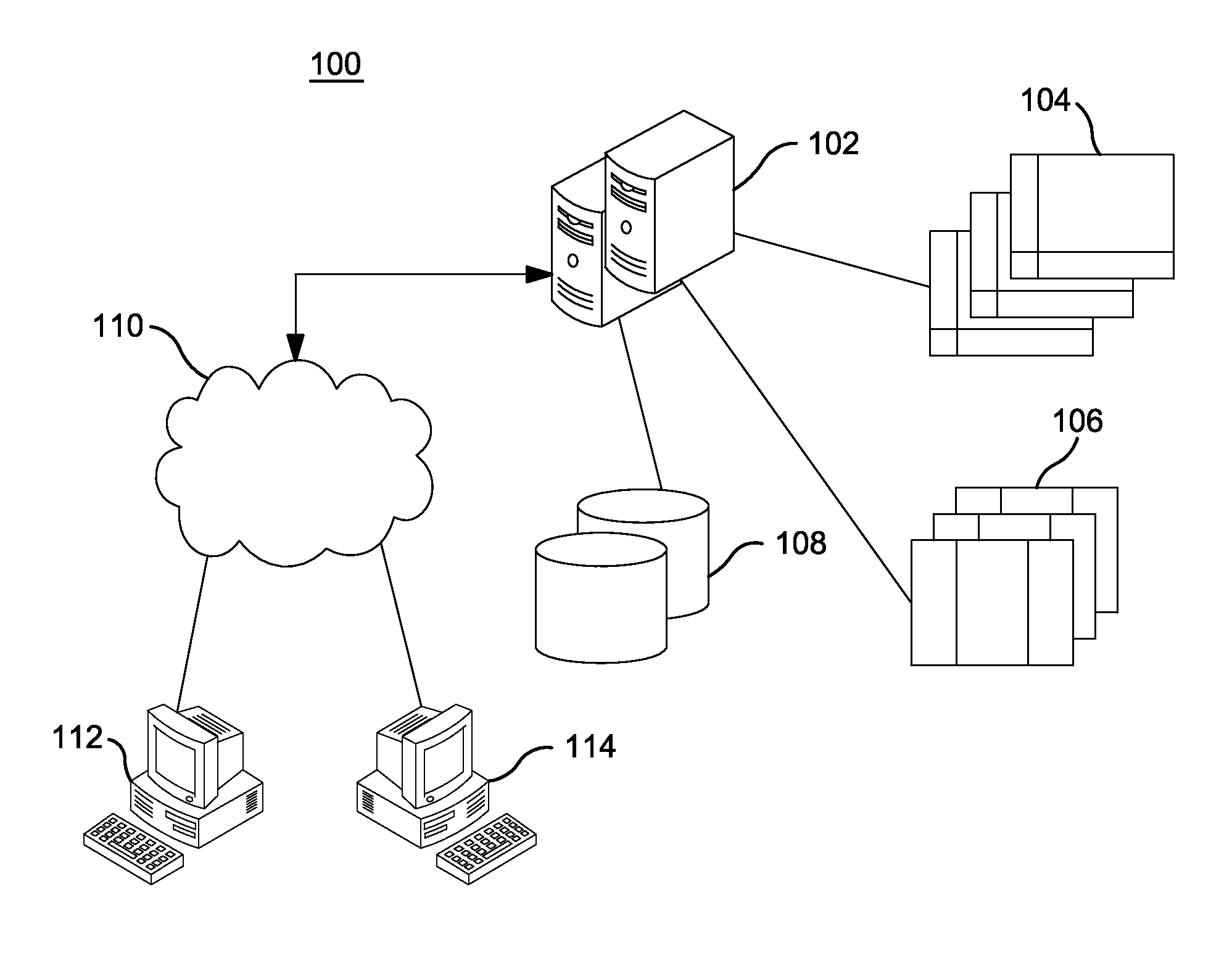
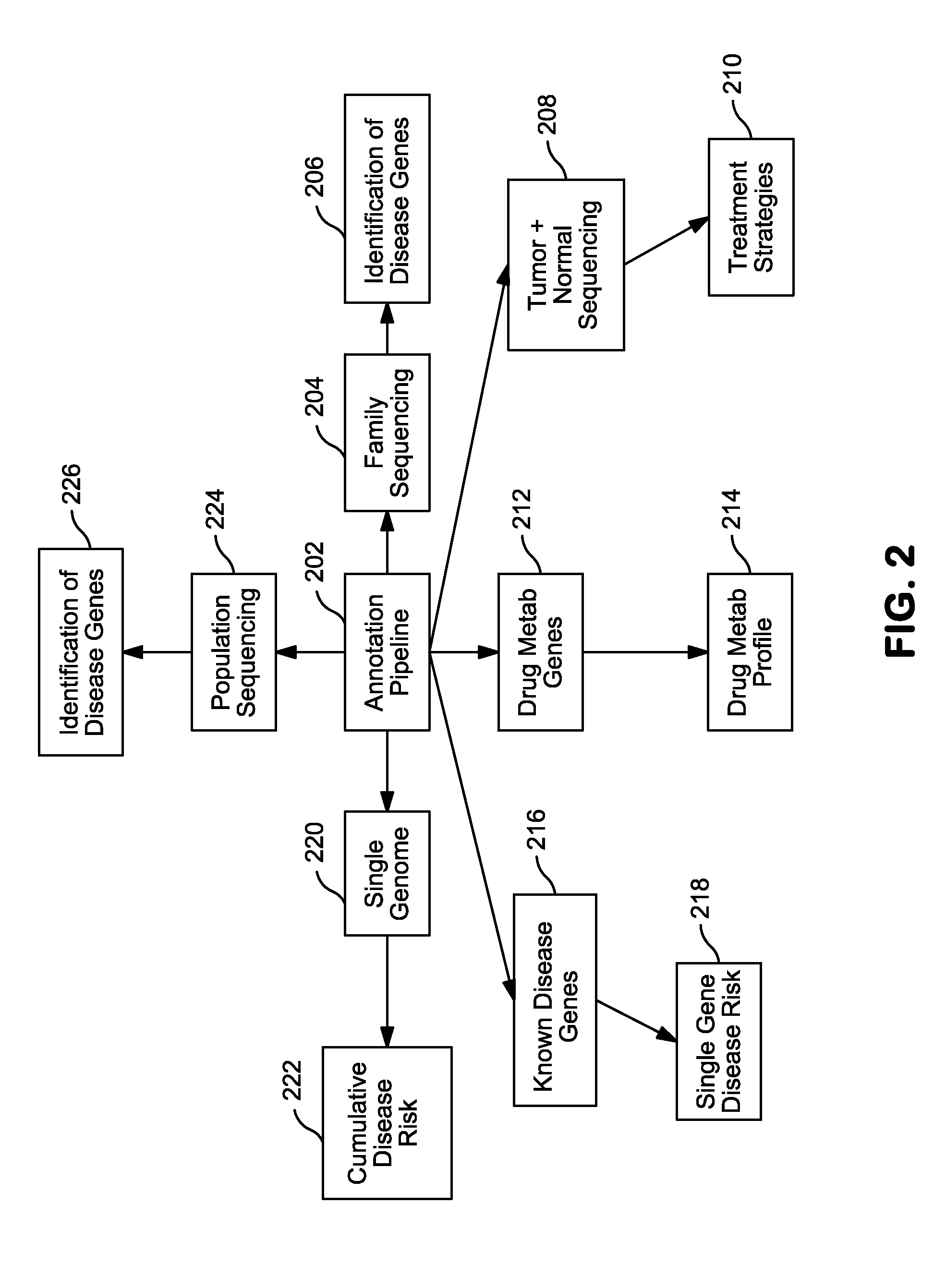
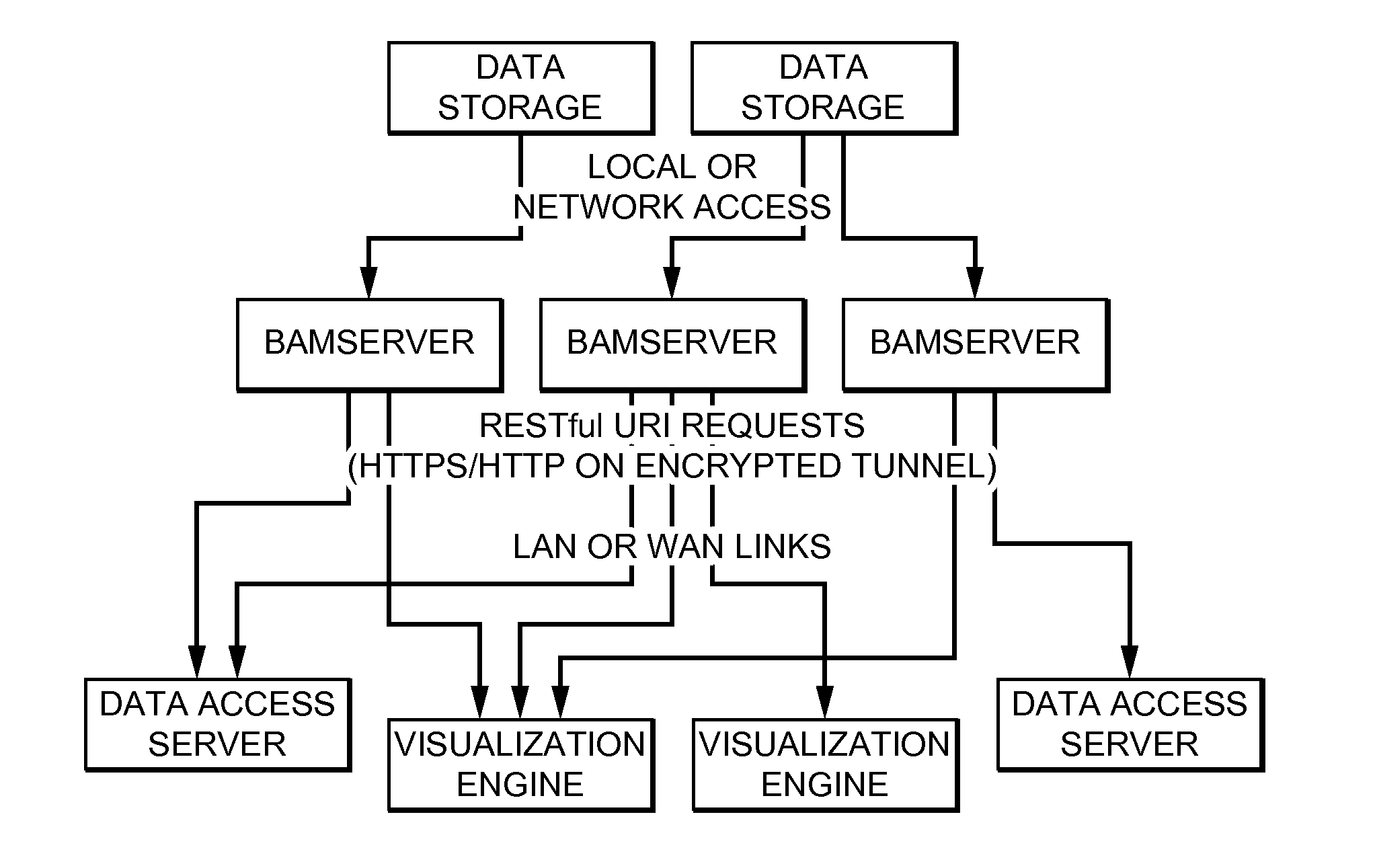
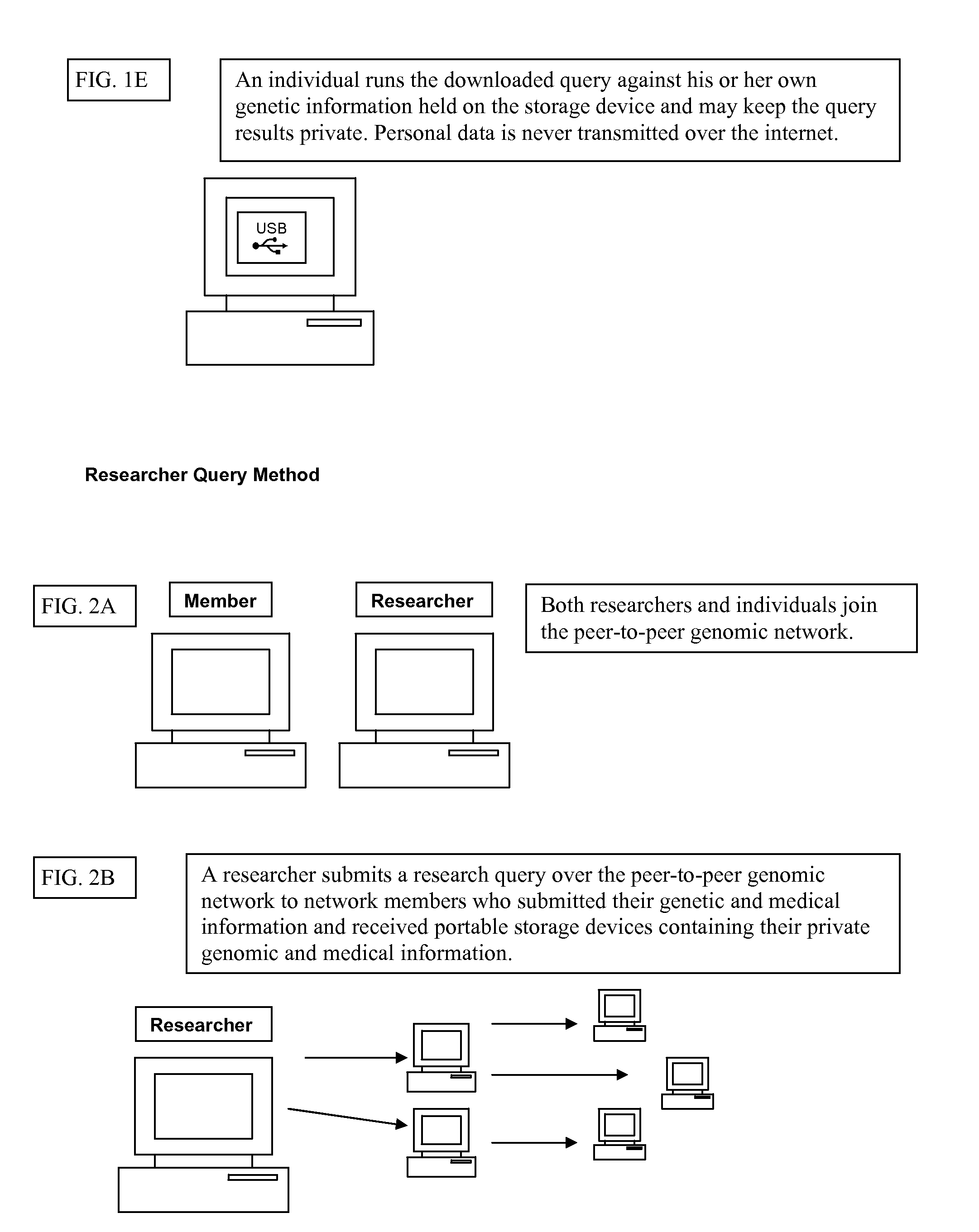
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
341 results about "Genomic data" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Genomic Data. Definition - What does Genomic Data mean? Genomic data refers to the genome and DNA data of an organism. They are used in bioinformatics for collecting, storing and processing the genomes of living things. Genomic data generally require a large amount of storage and purpose-built software to analyze.
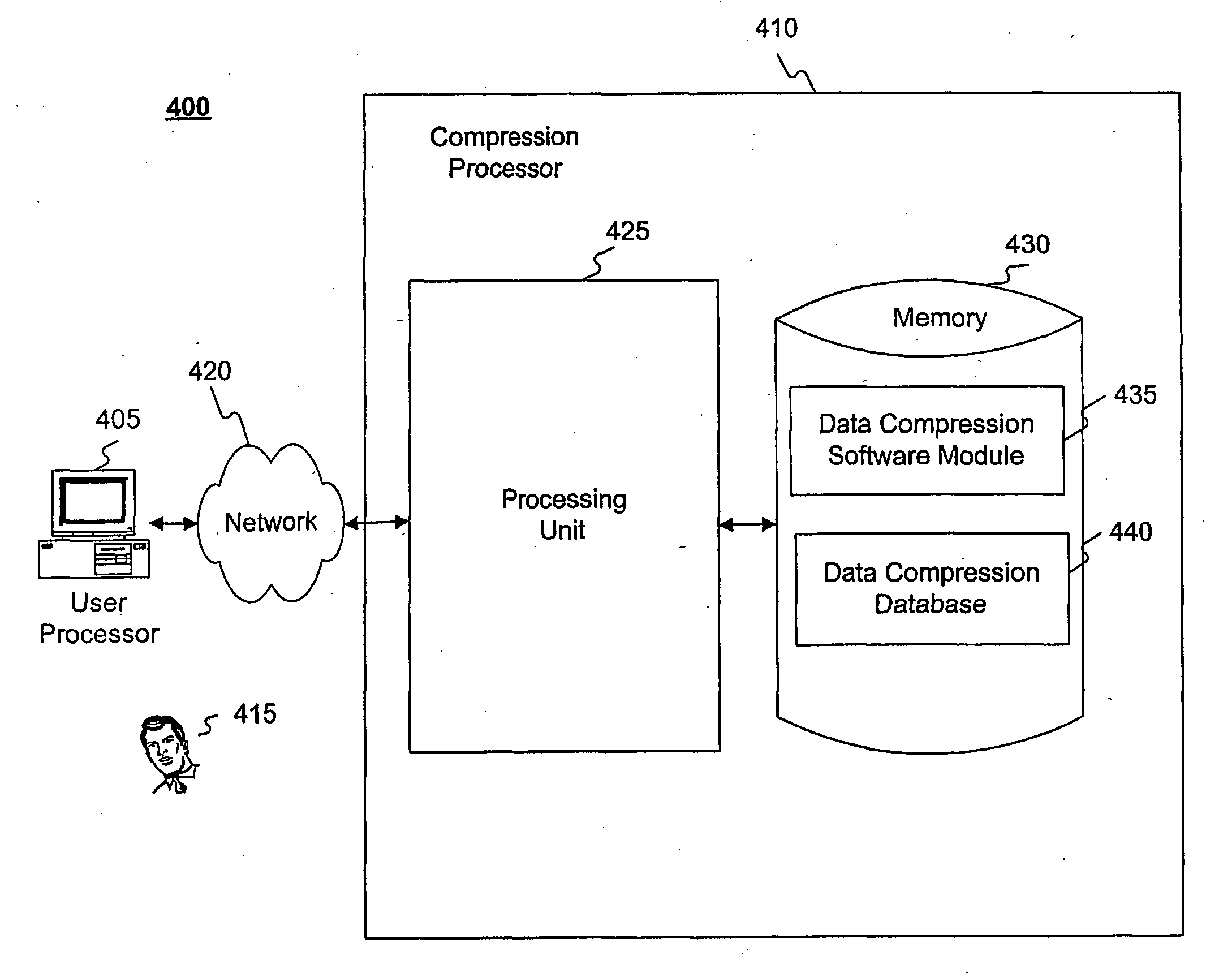
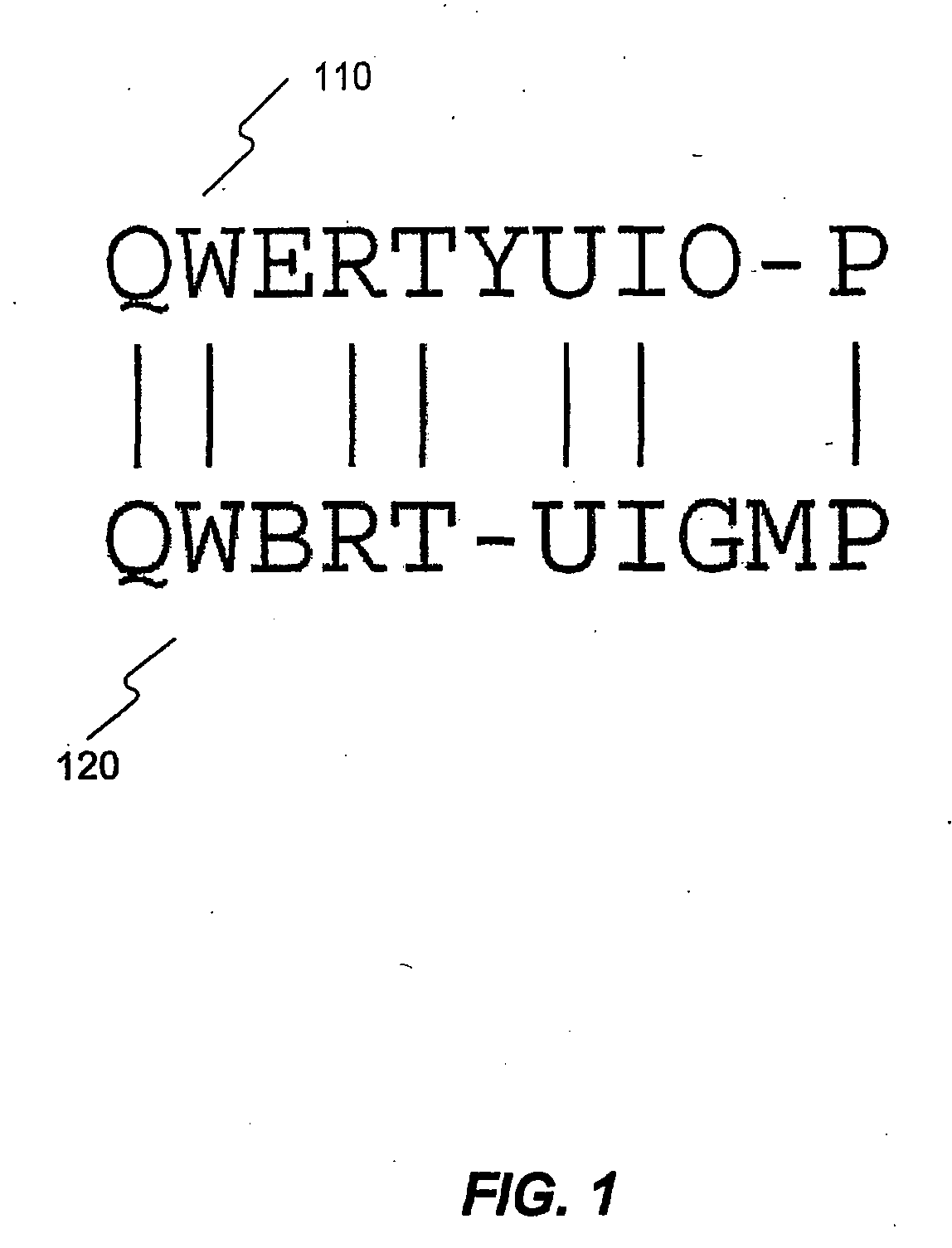
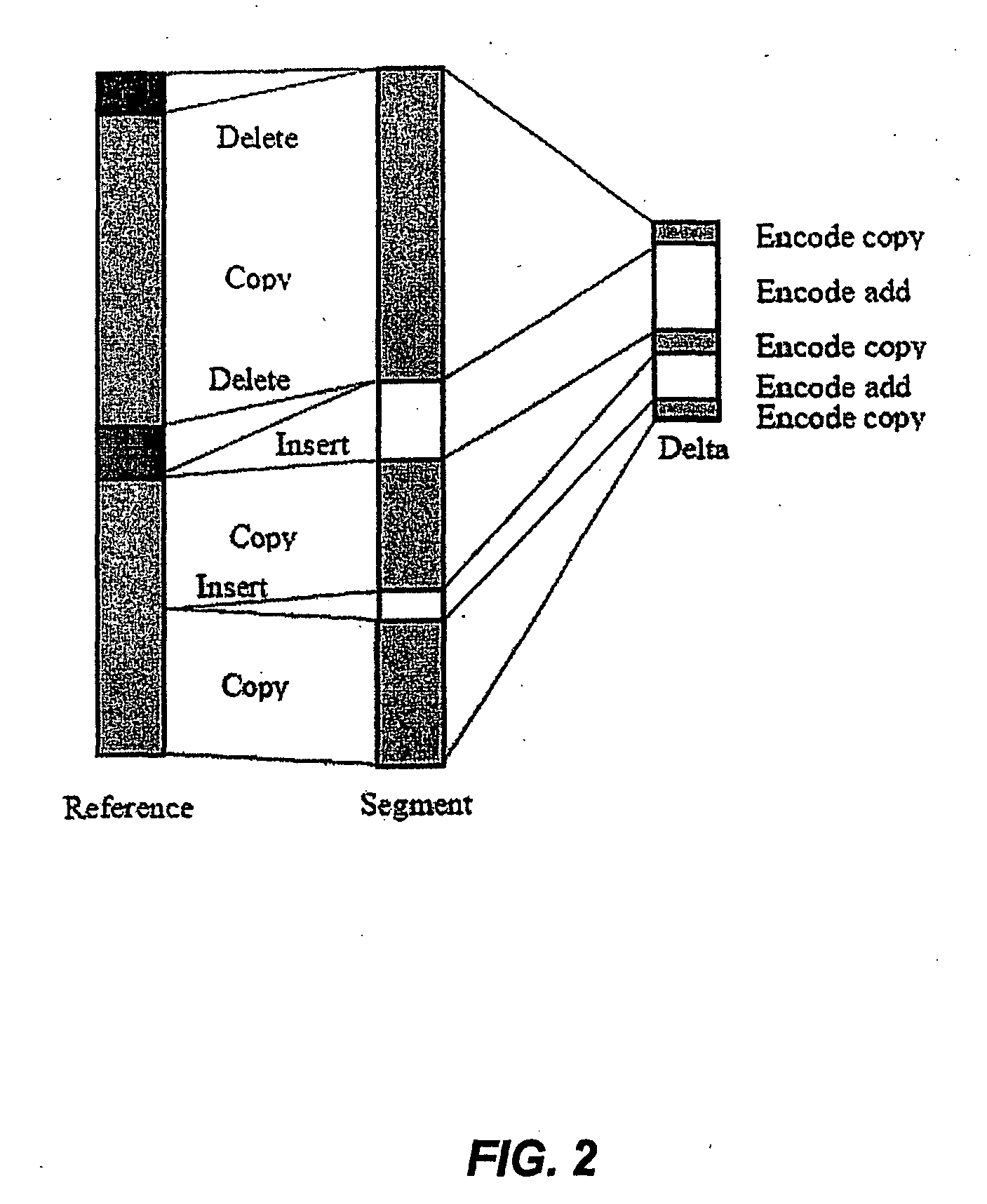
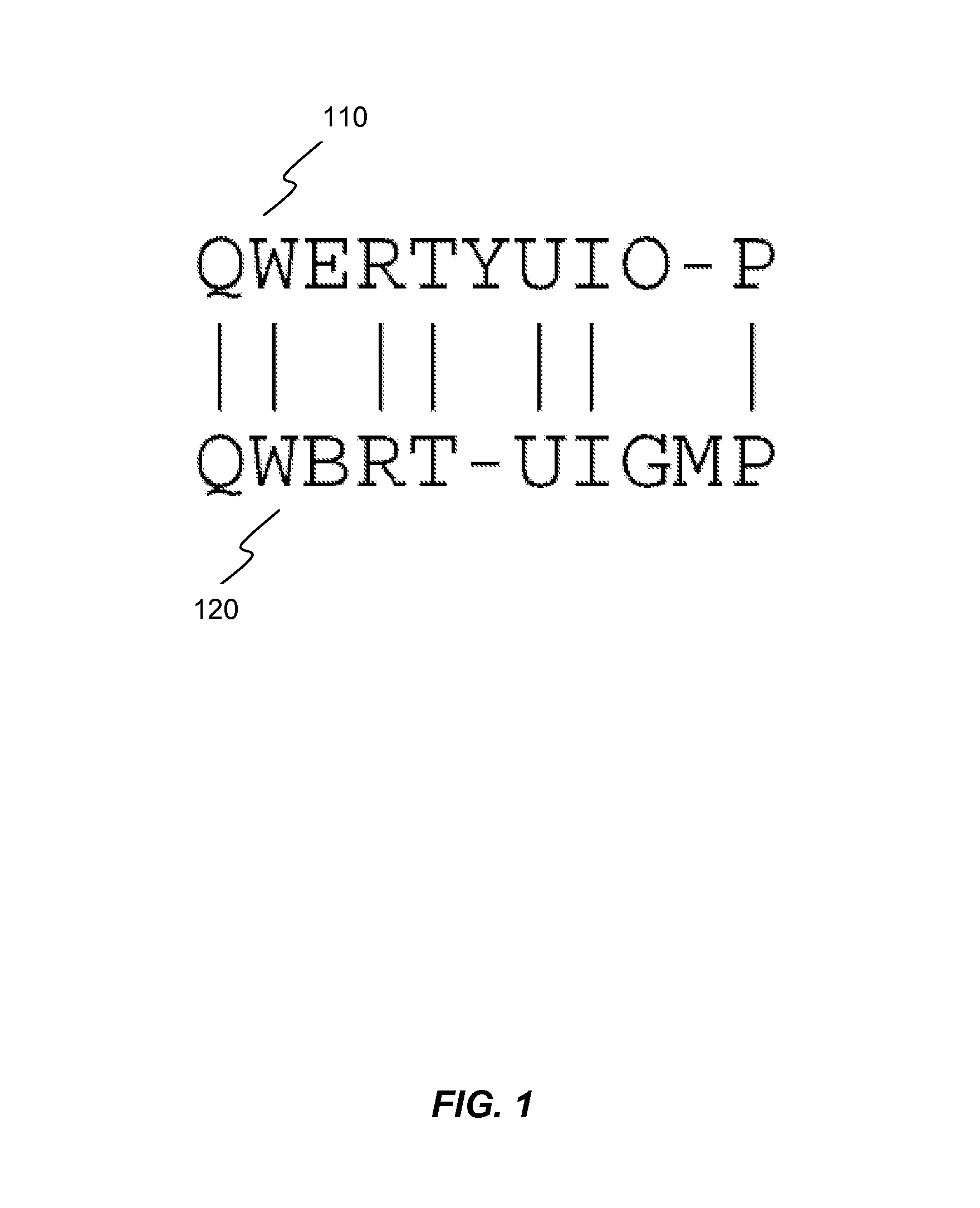
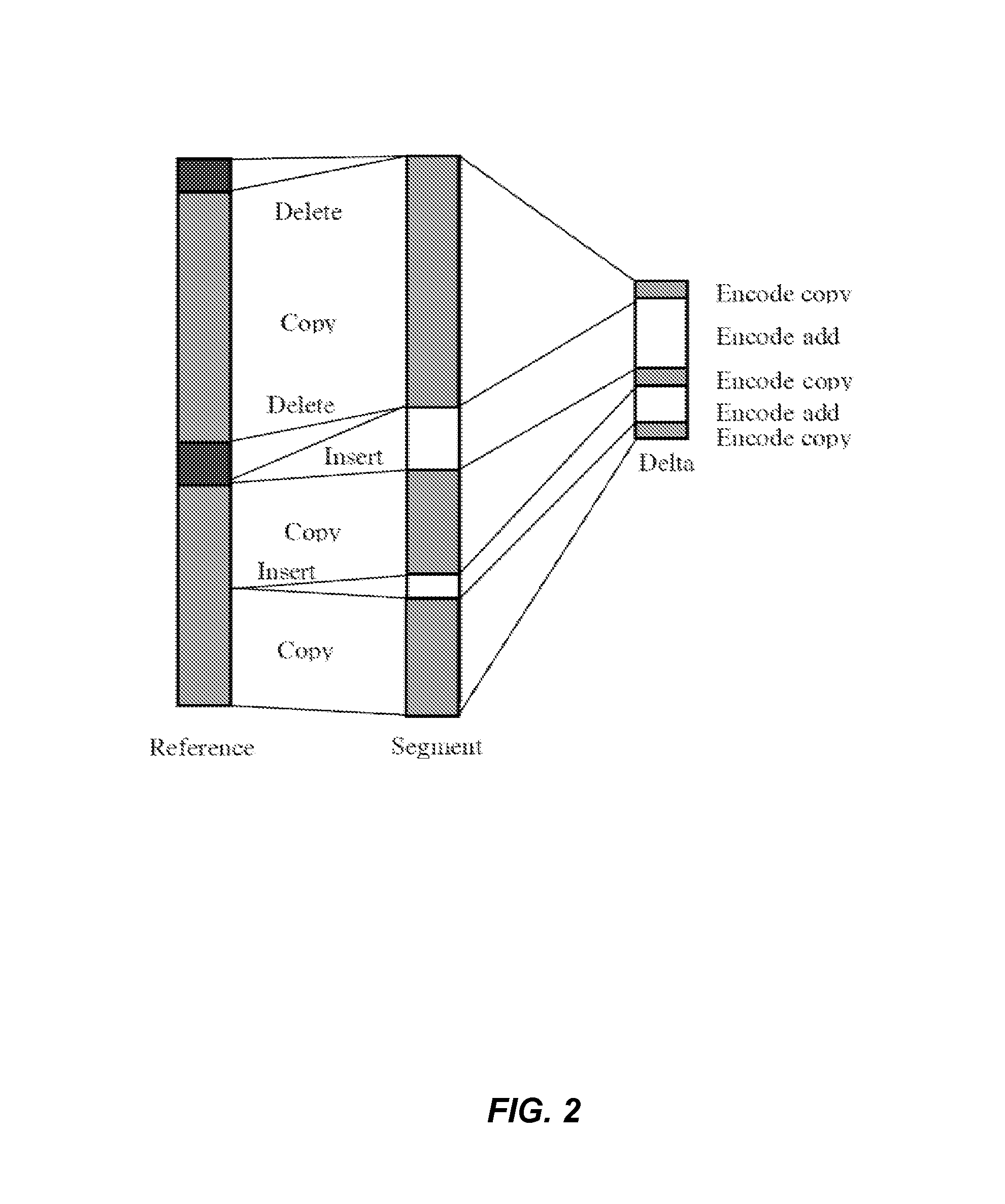
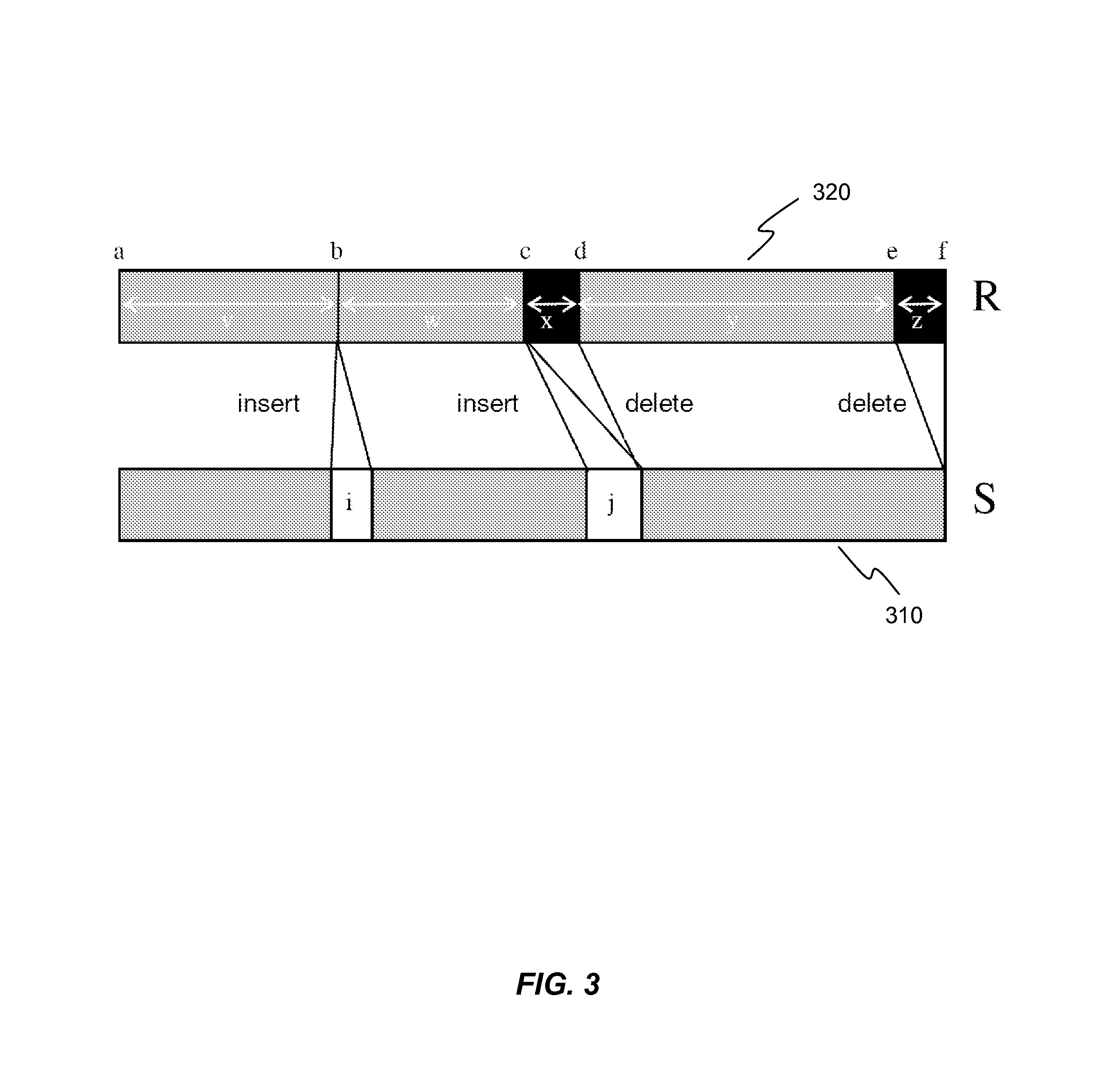
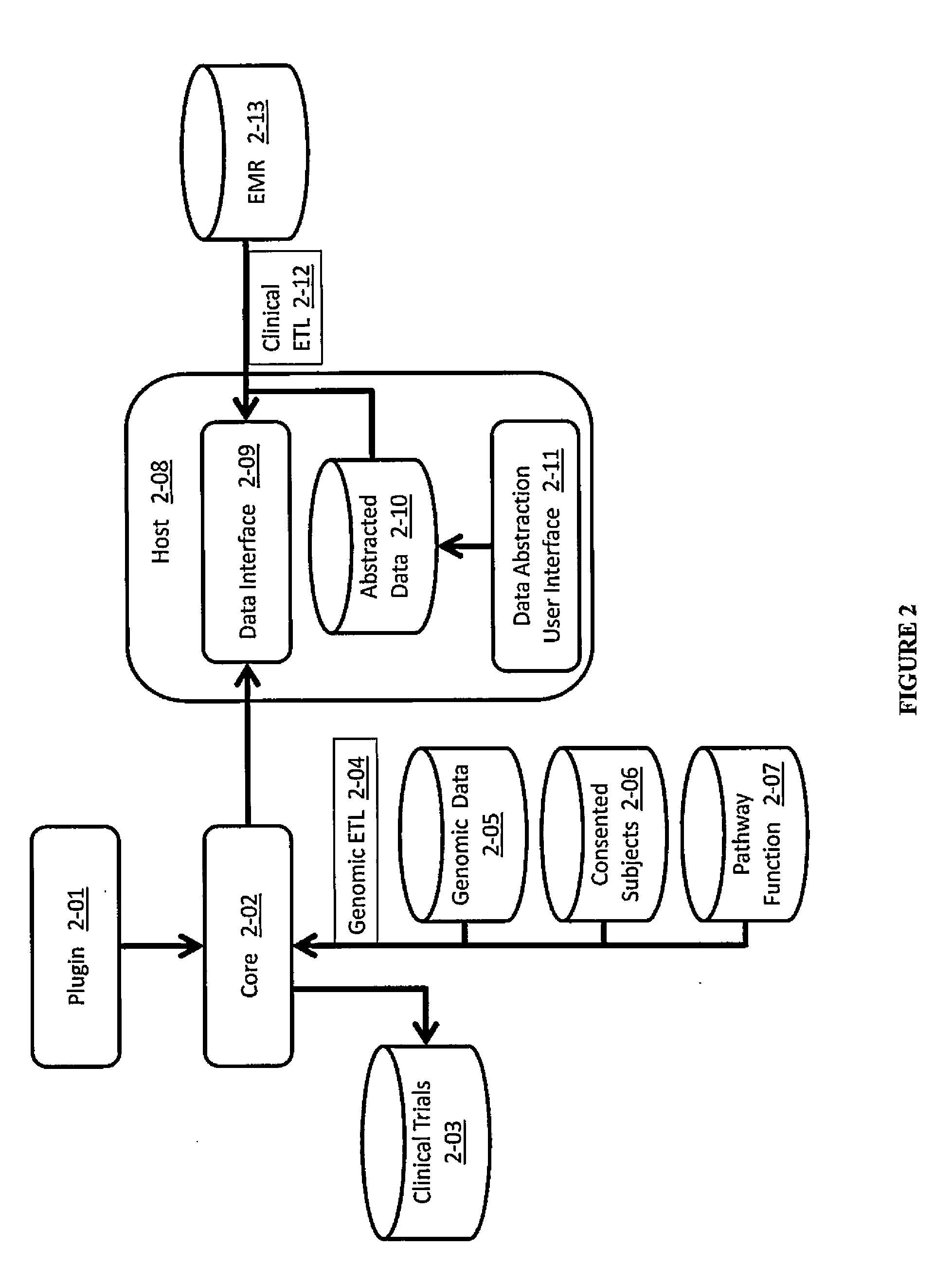
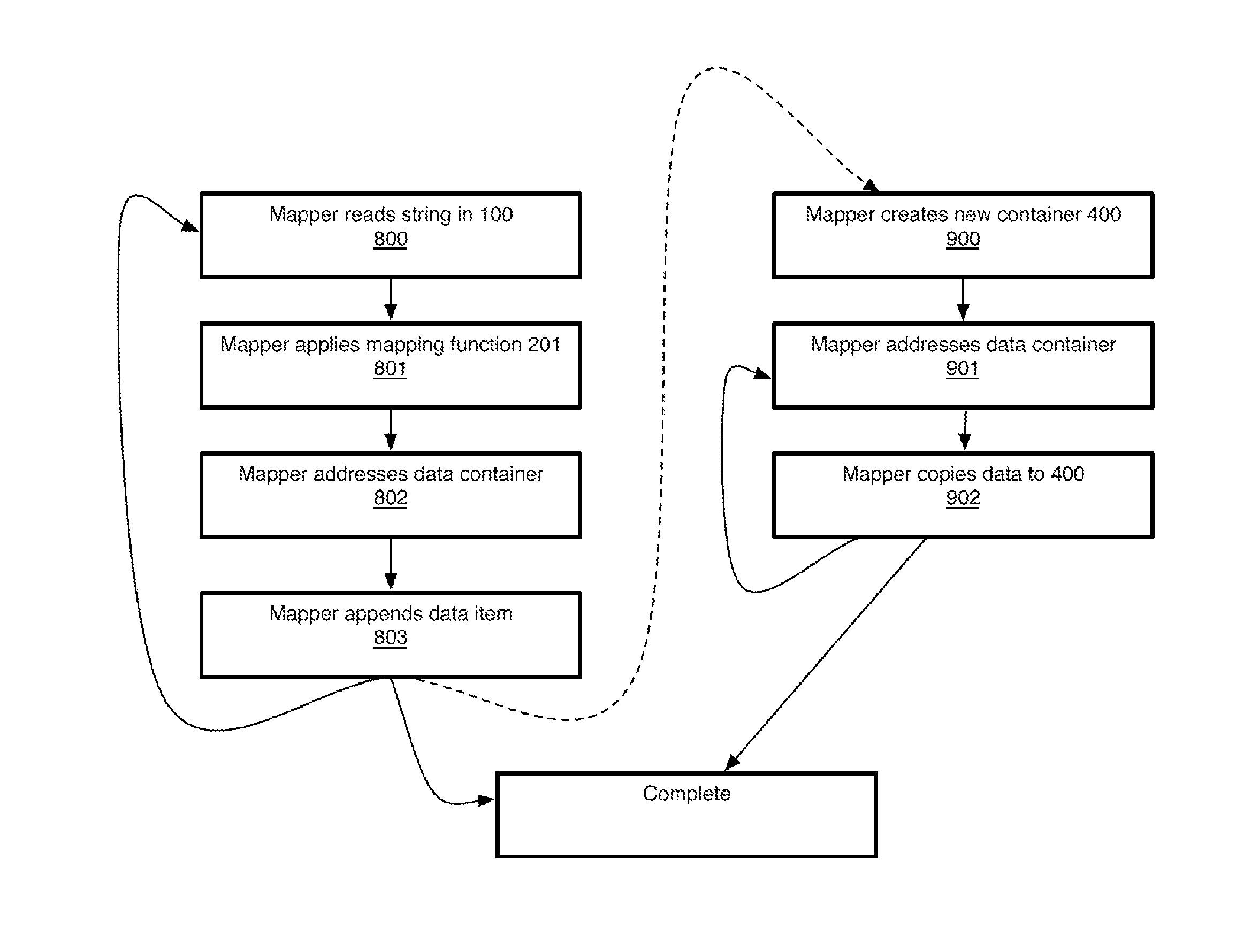
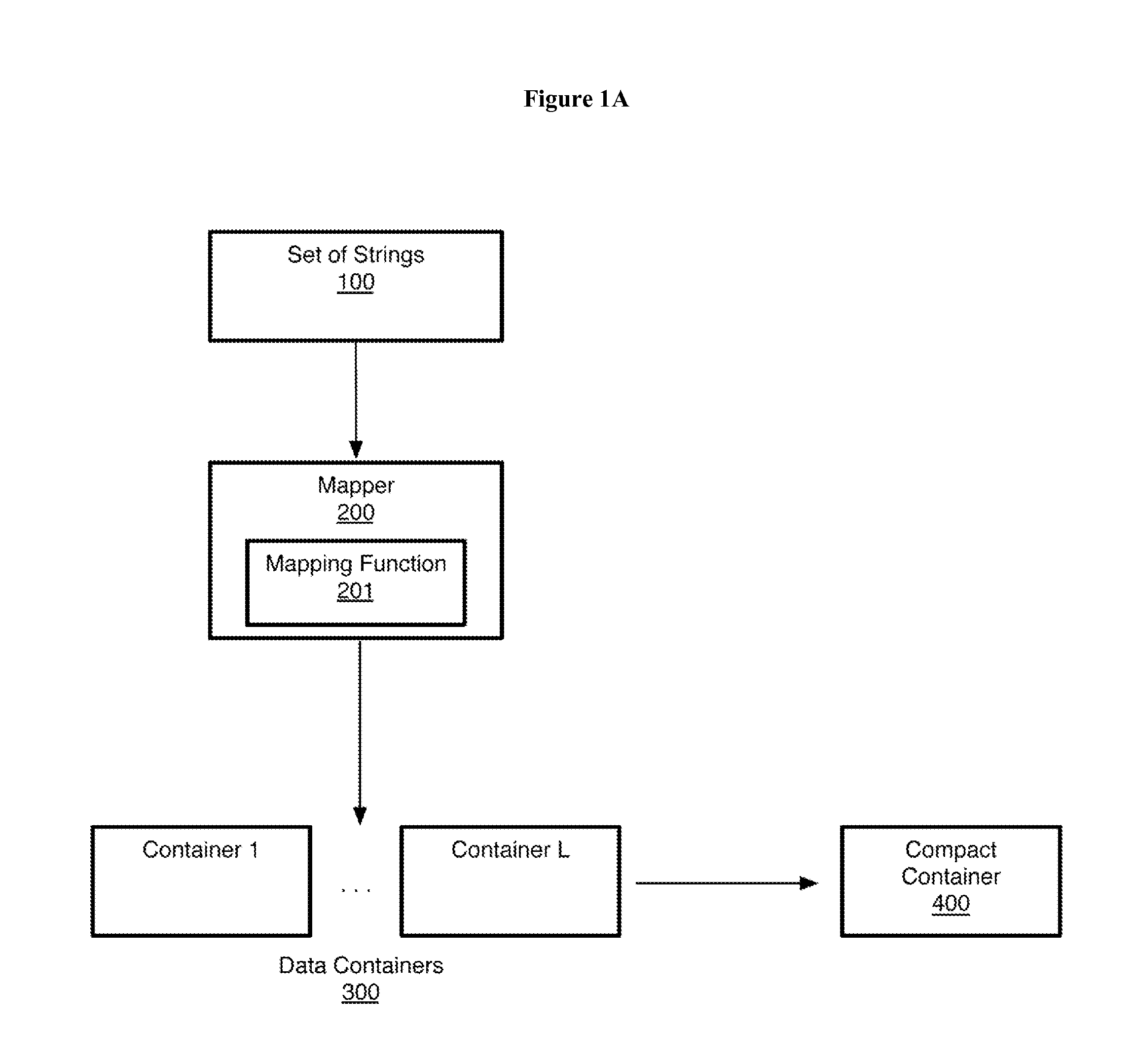
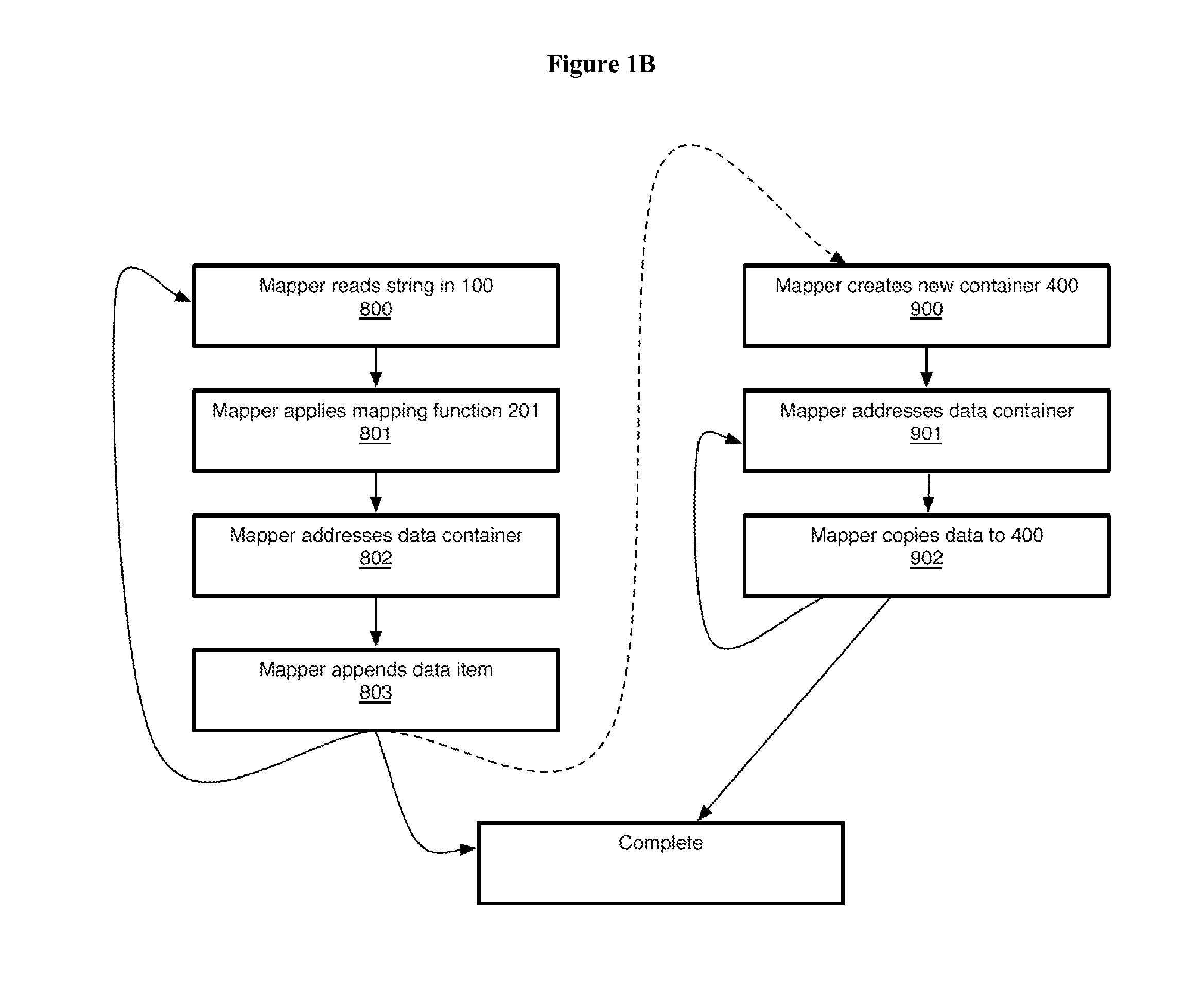
Methods and Systems for Compressing and Comparing Genomic Data
Systems and methods are disclosed for compressing and comparing data such as genomic data. The disclosed systems and methods may include selecting a segment, creating a delta representation of the segment, the delta representation comprising a script, and storing the script. Furthermore, the disclosed systems and methods may include receiving a first script comprising a compressed version of a first segment and receiving a second script comprising a compressed version of a second segment. The disclosed systems and methods may further include comparing the first script to the second script and determining if the first segment matches the second segment based upon the comparison of the first script to the second script.
Owner:MITOTECHNOLOGY LLC
Methods and systems for compressing and comparing genomic data
ActiveUS8340914B2Digital data processing detailsAnalogue computers for chemical processesData systemGenomic data
Systems and methods are disclosed for compressing and comparing data such as genomic data. The disclosed systems and methods may include selecting a segment, creating a delta representation of the segment, the delta representation comprising a script, and storing the script. Furthermore, the disclosed systems and methods may include receiving a first script comprising a compressed version of a first segment and receiving a second script comprising a compressed version of a second segment. The disclosed systems and methods may further include comparing the first script to the second script and determining if the first segment matches the second segment based upon the comparison of the first script to the second script.
Owner:MITOTECHNOLOGY LLC
System and method for the management of genomic data
InactiveUS20050026117A1Low priceSecure transmissionDrug referencesBiological testingGeographic regionsFinancial transaction
A system and method is disclosed for managing users' genomic data, including providing and offering access to genomic-based services, routing genomic data to providers of genomic-based services, brokering financial transactions related to the management of genomic data, securing for users best prices for genomic-based services, allowing users to earn money for the use of their genomic and other data, and using genomic data for marketing and developing products in particular geographic regions or for particular populations.
Owner:GENAISSANCE PHARMA INC
Healthcare analysis stream management
ActiveUS20140012843A1Digital data processing detailsHealthcare managementStream managementGenomic data
Apparatus, systems and methods for pre-processing, analyzing, and storing genomic data through a scalable, distributed analysis system across a network is presented.
Owner:NANT HLDG IP LLC
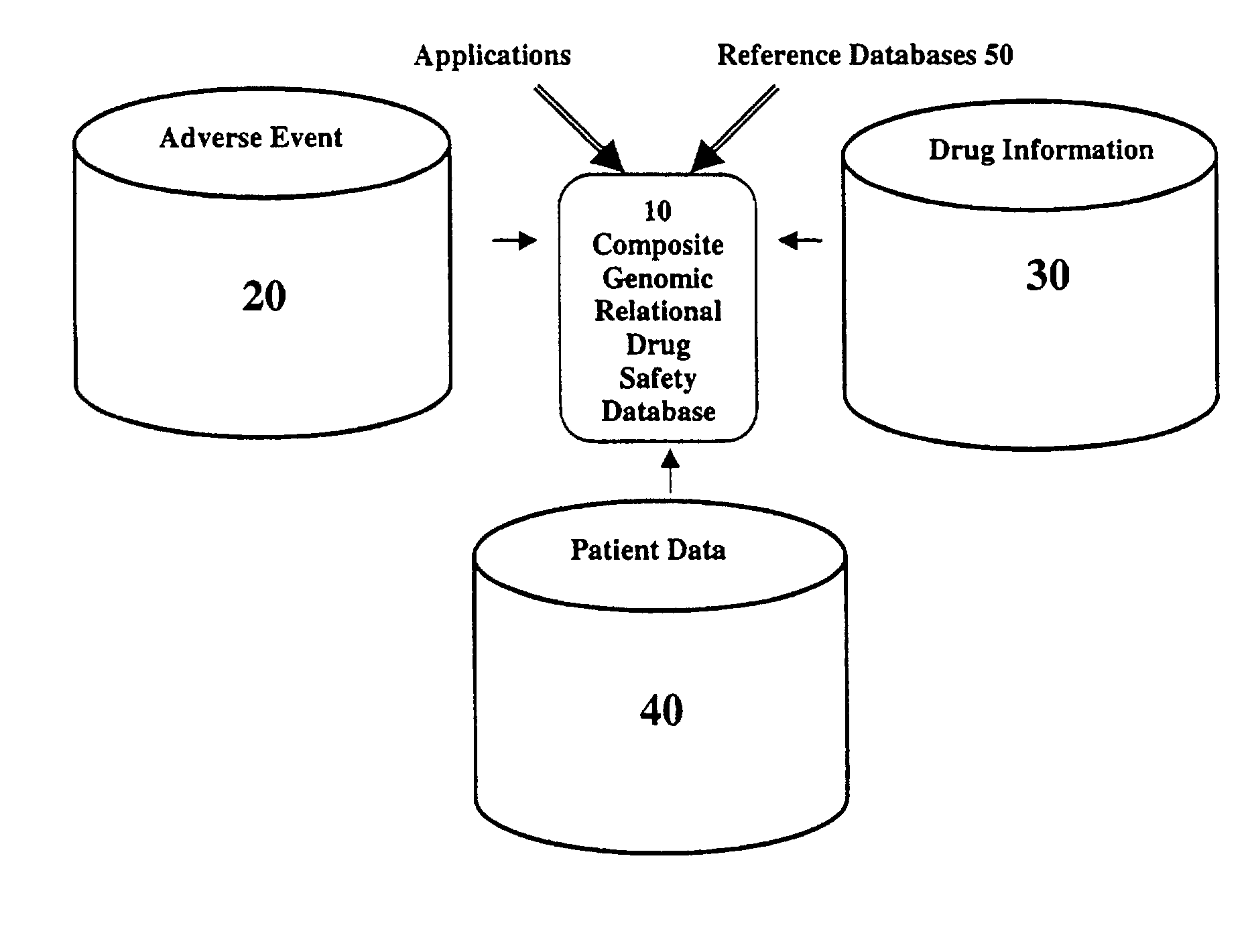
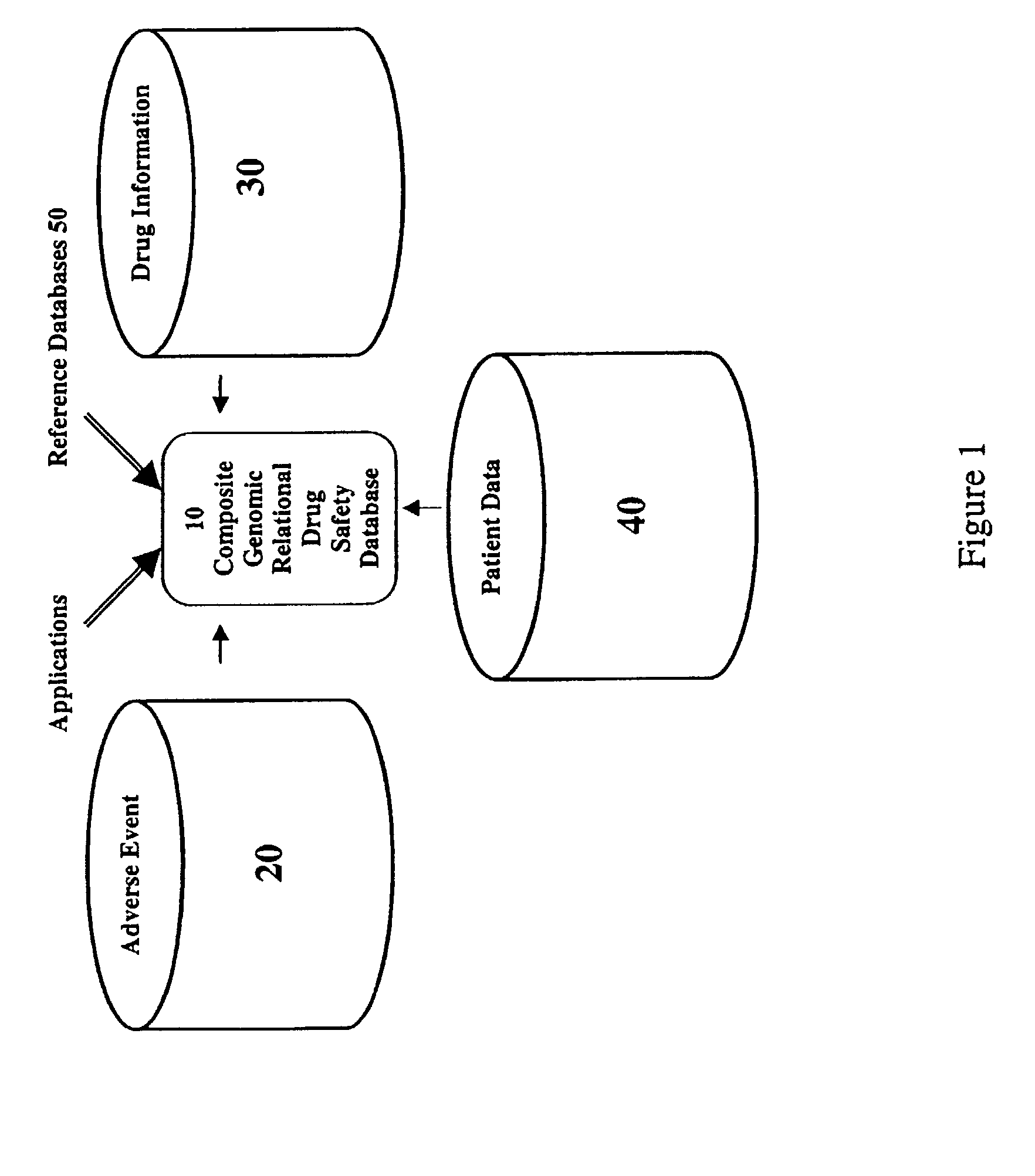
Method and system for the analysis and association of patient-specific and population-based genomic data with drug safety adverse event data
ActiveUS7461006B2Risk of adverse drug reactionIncreased and decreased chanceBiostatisticsComputer-assisted medical data acquisitionPatient characteristicsGenomic DNA
A method for assessing and analyzing one or more drugs, adverse effects and associated risks, and patient characteristics resulting from the use of at least drug of interest is disclosed. The method comprises the steps of selecting one or more cases for analysis, said cases describing the behavior between at least one drug of interest and a patient genotype; profiling statistically derived values from multiple cases related to the safety of the at least one drug, wherein at least one filter is employed for deriving said values; at least one data mining engine; and an output device for displaying the analytic results from the data mining engine. A system for performing the method is likewise disclosed.
Owner:DRUGLOGIC
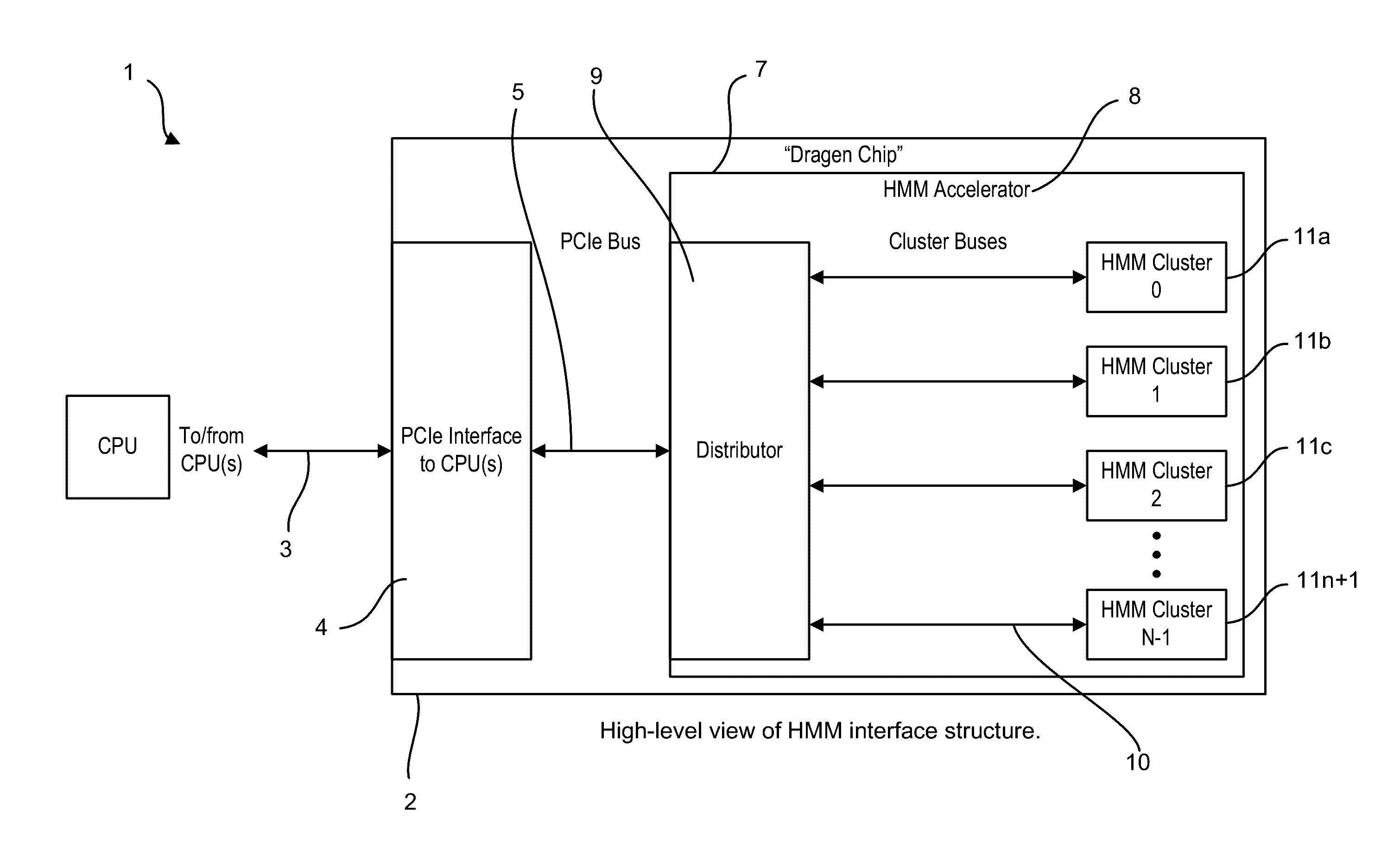
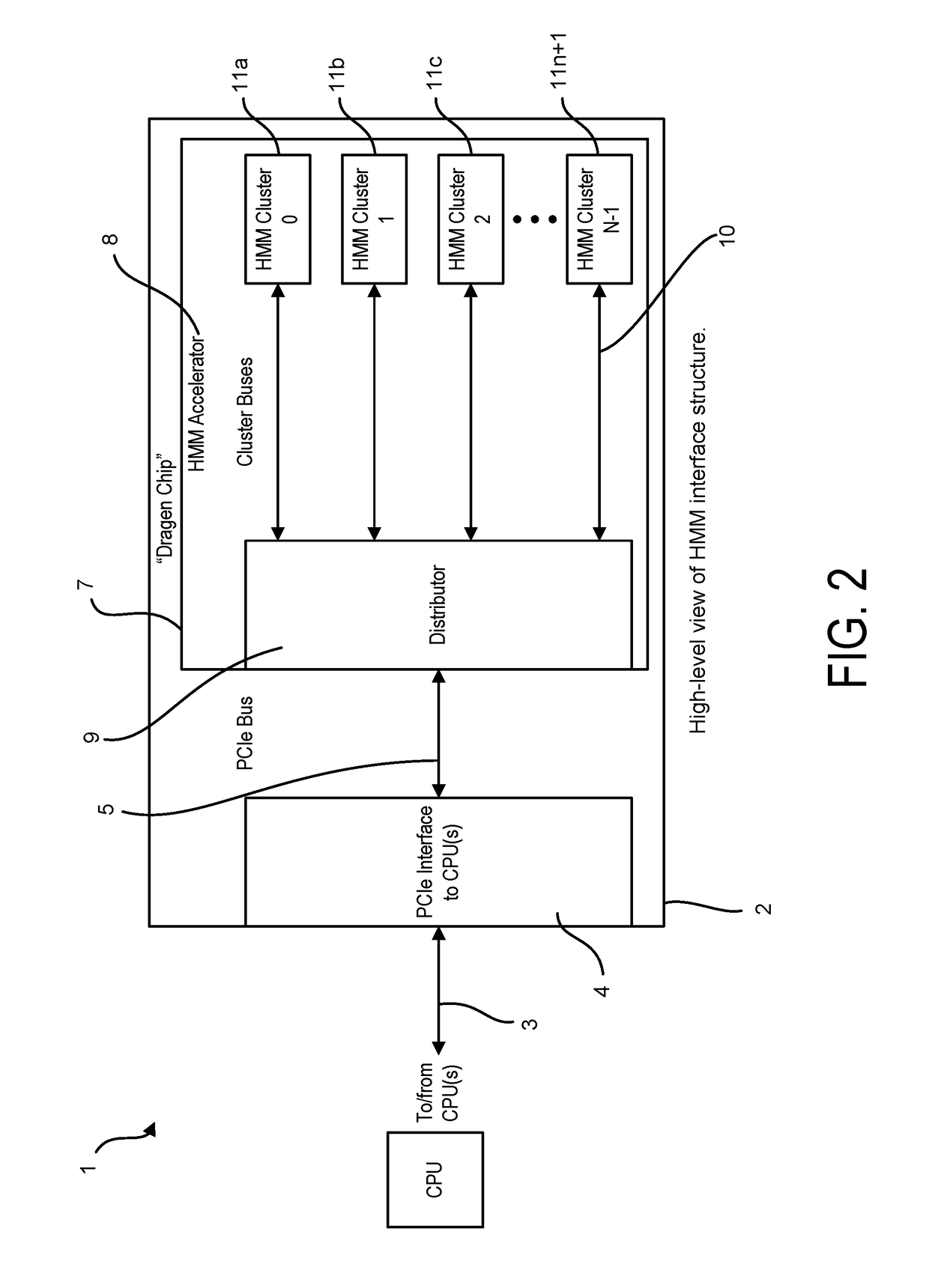
Bioinformatics systems, apparatuses, and methods executed on an integrated circuit processing platform
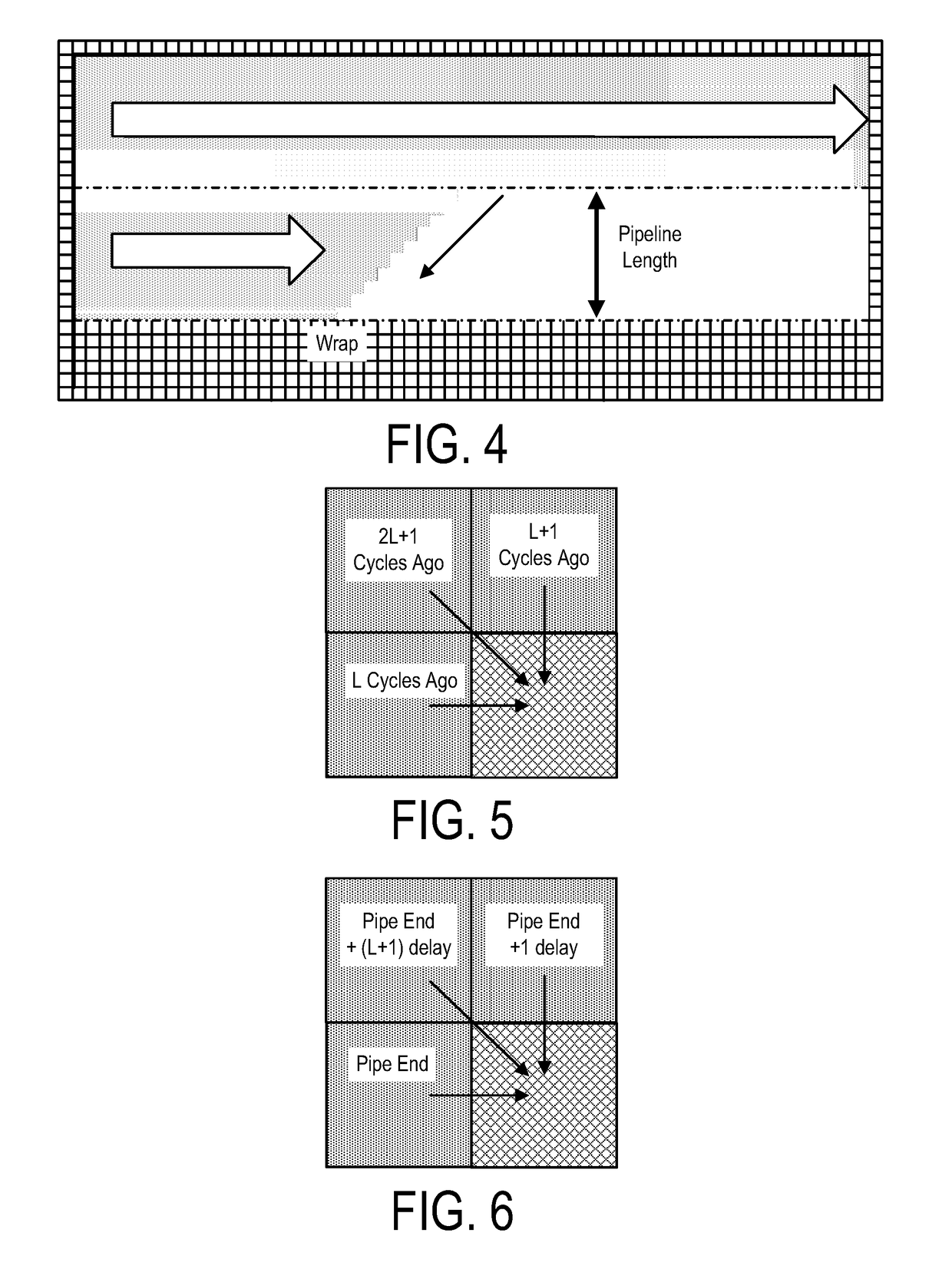
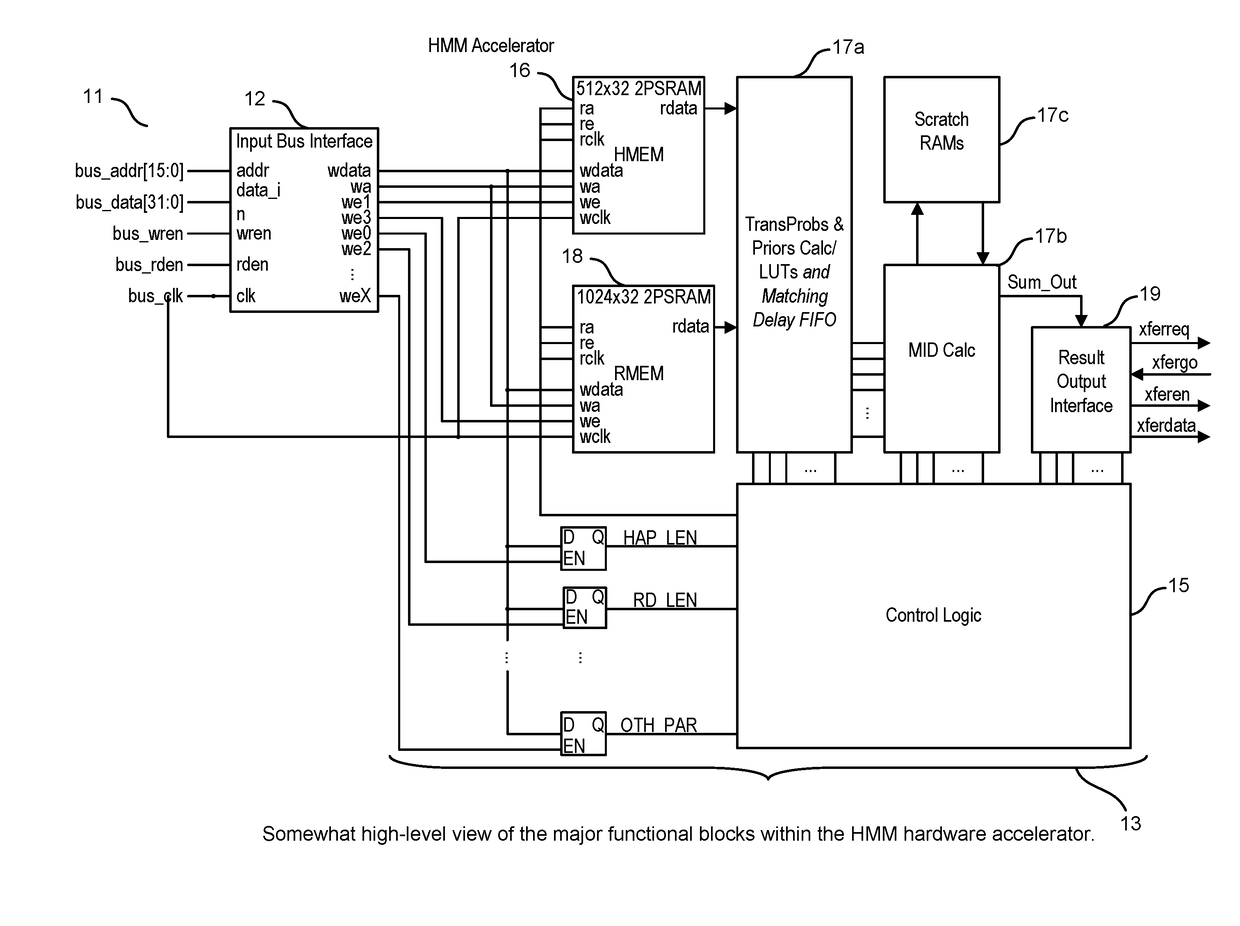
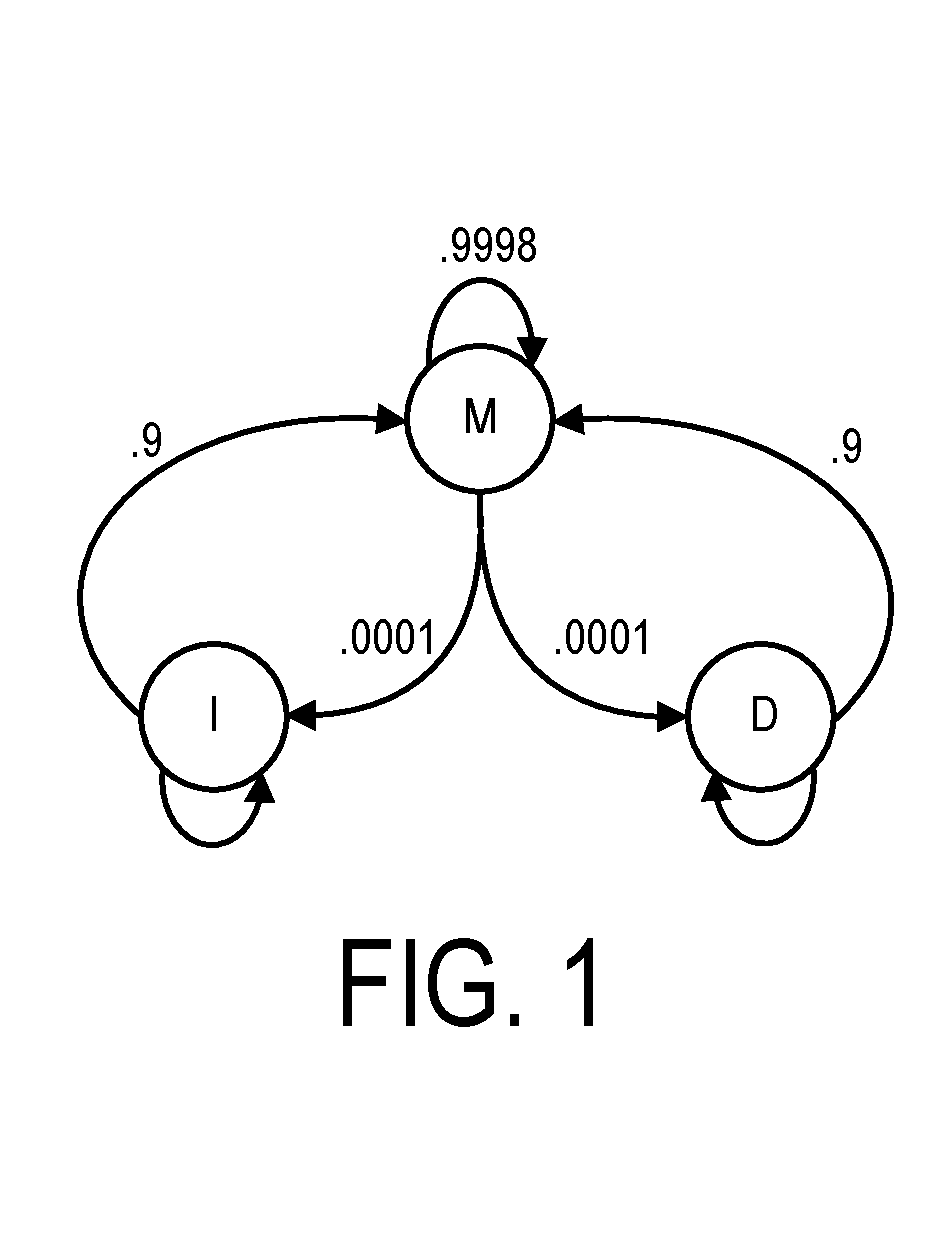
ActiveUS20160306922A1High sensitivityImprove accuracyHealth-index calculationTelemedicineElectricityData source
A system, method and apparatus for executing a bioinformatics analysis on genetic sequence data includes an integrated circuit formed of a set of hardwired digital logic circuits that are interconnected by physical electrical interconnects. One of the physical electrical interconnects forms an input to the integrated circuit that may be connected with an electronic data source for receiving reads of genomic data. The hardwired digital logic circuits may be arranged as a set of processing engines, each processing engine being formed of a subset of the hardwired digital logic circuits to perform one or more steps in the bioinformatics analysis on the reads of genomic data. Each subset of the hardwired digital logic circuits may be formed in a wired configuration to perform the one or more steps in the bioinformatics analysis.
Owner:EDICO GENOME
Bioinformatics systems, apparatuses, and methods executed on an integrated circuit processing platform
ActiveUS20160306923A1Processing speedProcessed result accuracyMemory architecture accessing/allocationMathematical modelsData sourceGenomic data
A system, method and apparatus for executing a bioinformatics analysis on genetic sequence data includes an integrated circuit formed of a set of hardwired digital logic circuits that are interconnected by physical electrical interconnects. One of the physical electrical interconnects forms an input to the integrated circuit that may be connected with an electronic data source for receiving reads of genomic data. The hardwired digital logic circuits may be arranged as a set of processing engines, each processing engine being formed of a subset of the hardwired digital logic circuits to perform one or more steps in the bioinformatics analysis on the reads of genomic data. Each subset of the hardwired digital logic circuits may be formed in a wired configuration to perform the one or more steps in the bioinformatics analysis.
Owner:EDICO GENOME
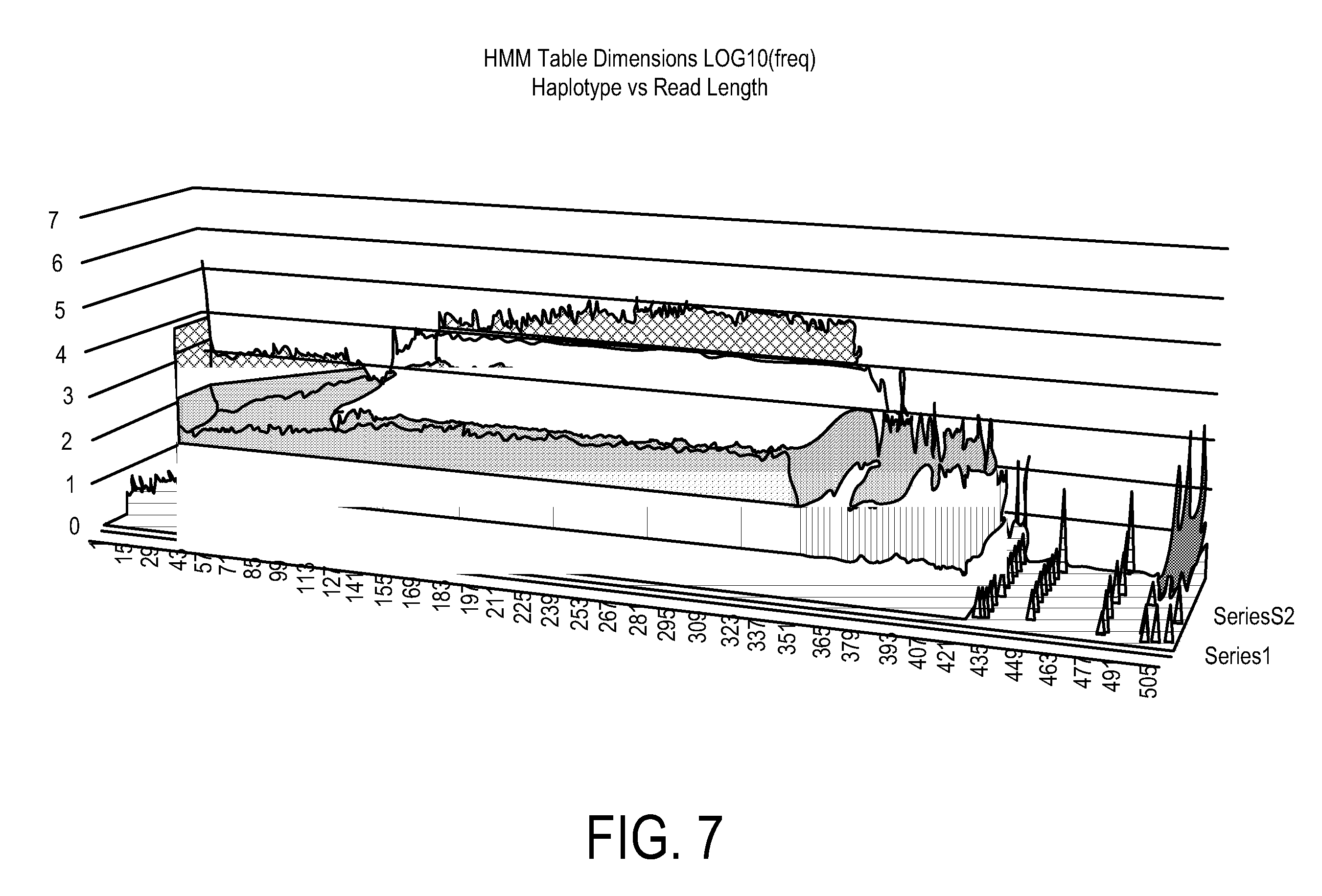
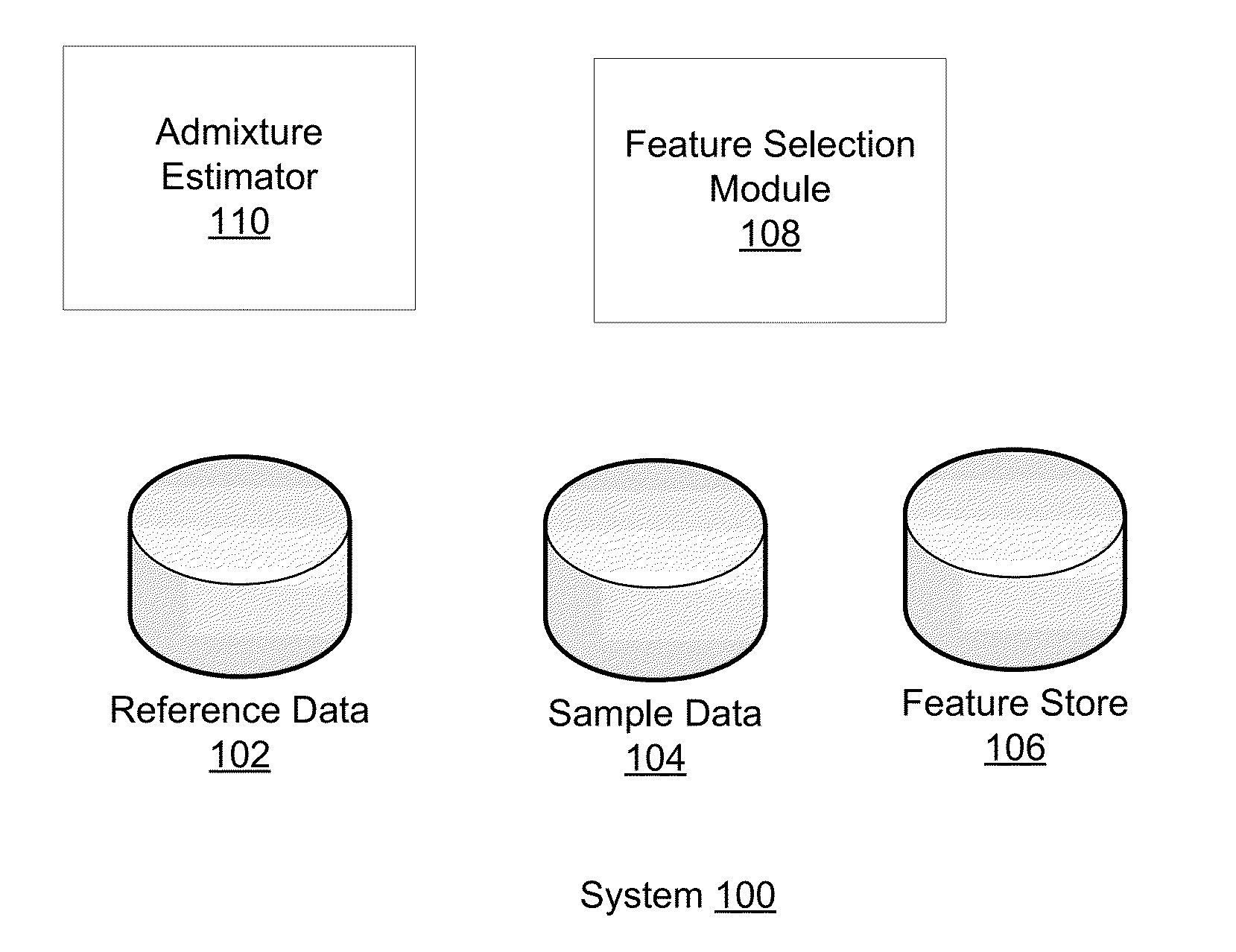
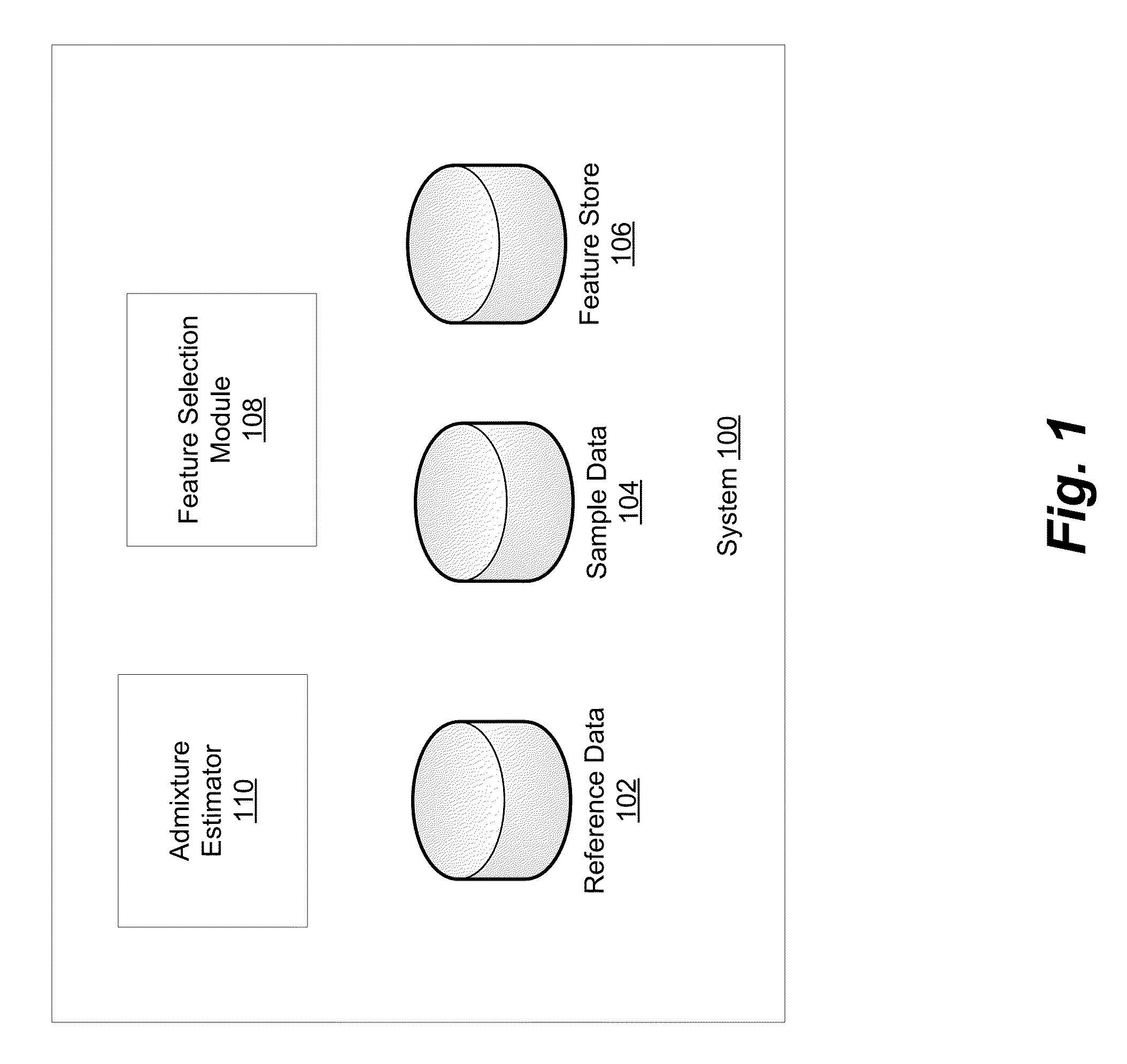
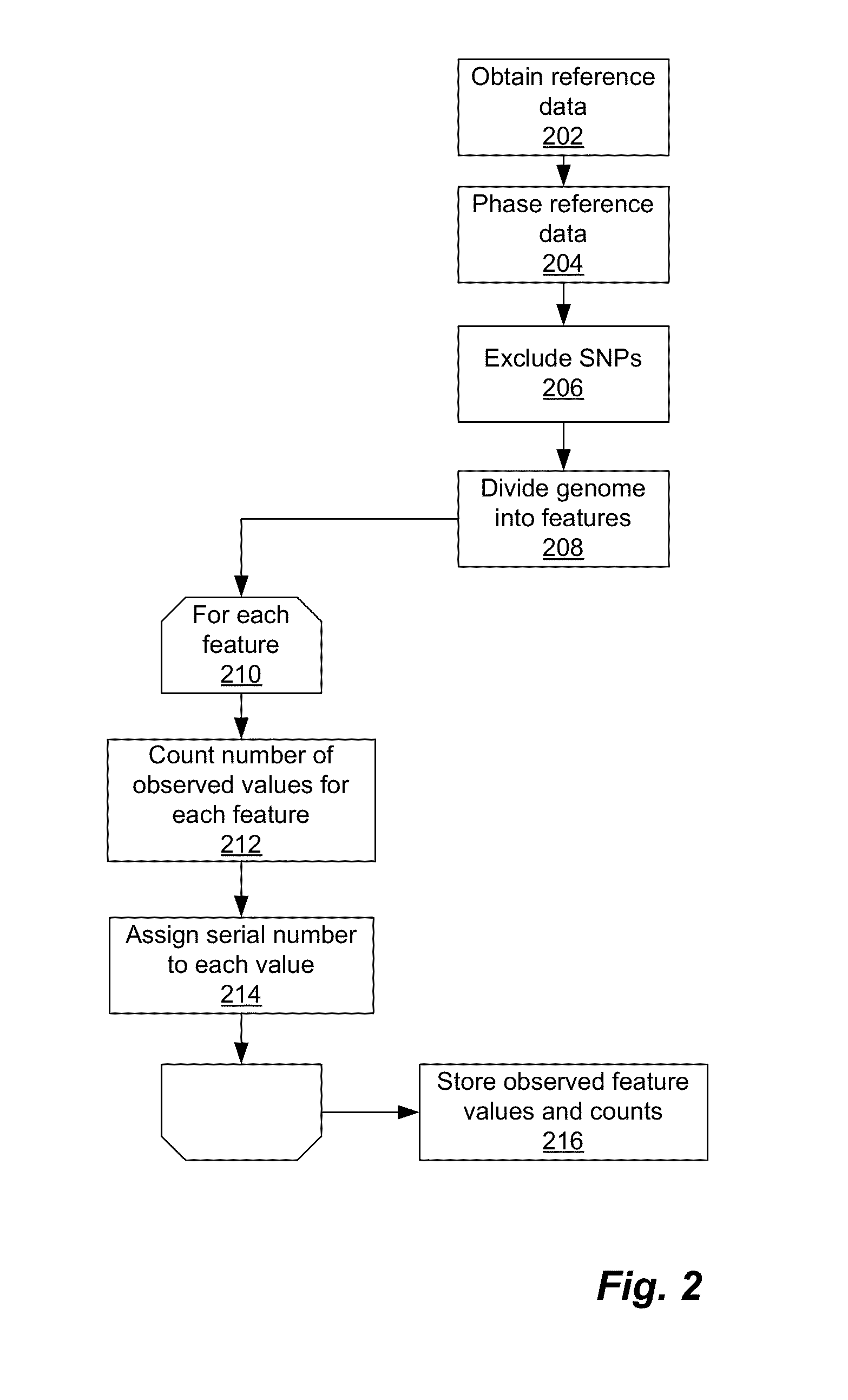
Using Haplotypes to Infer Ancestral Origins for Recently Admixed Individuals
InactiveUS20140067355A1Analogue computers for chemical processesProteomicsPattern recognitionProgenitor
Phased haplotype features are used to infer an individual's ancestry. Reference genomic data is obtained for individuals of known ancestral origin. Haplotype features are identified based on consecutive SNPs from each individual. Sample genomic data is obtained for an individual of unknown ancestral origin. The data is phased and divided into features analogous to the features in the reference data. An admixture estimator then performs an admixture estimation based on the observed feature values in the sample data and the reference data. The estimation indicates a contribution of each of the known populations to the genome of the sample individual.
Owner:ANCESTRY COM DNA
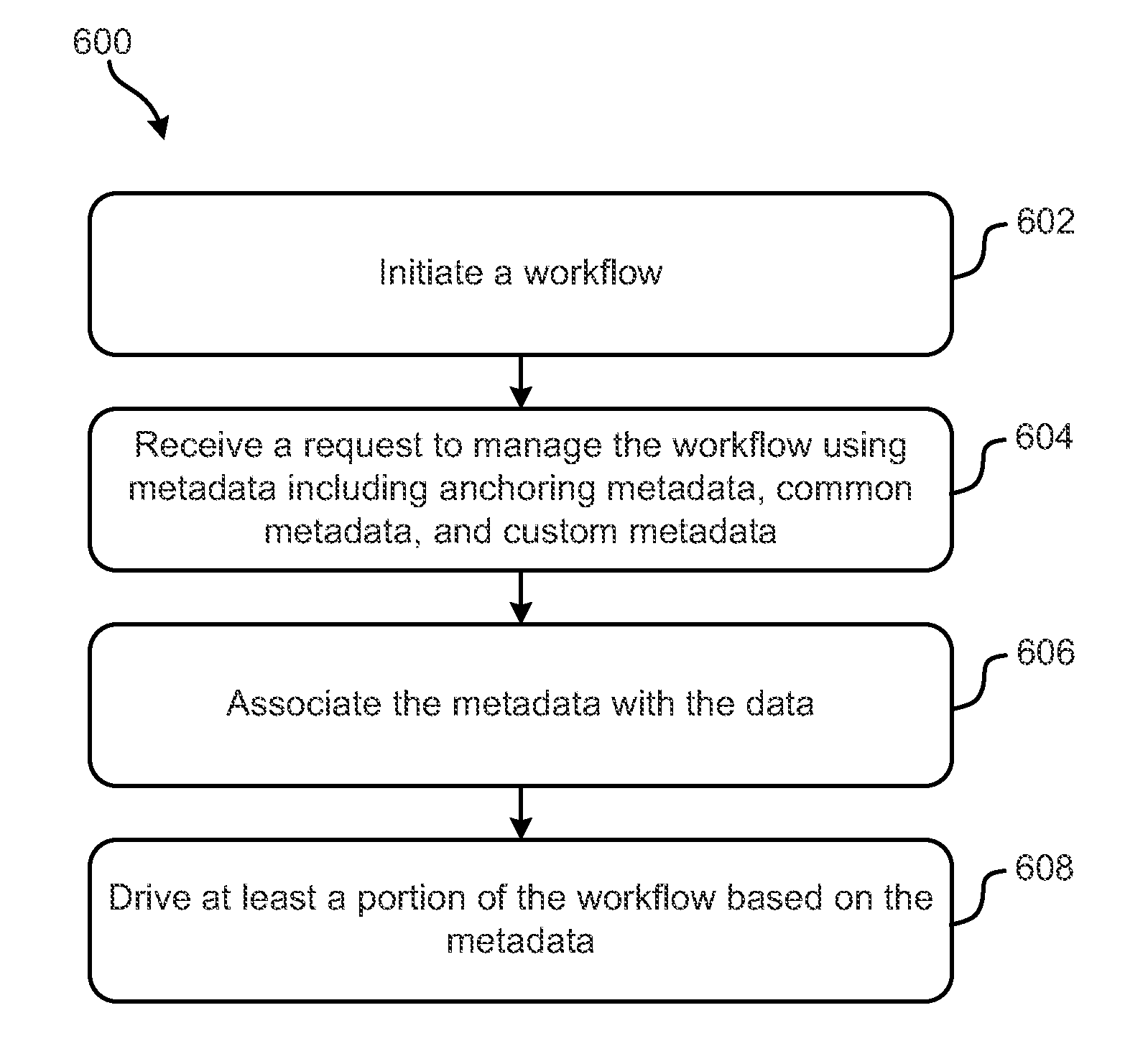
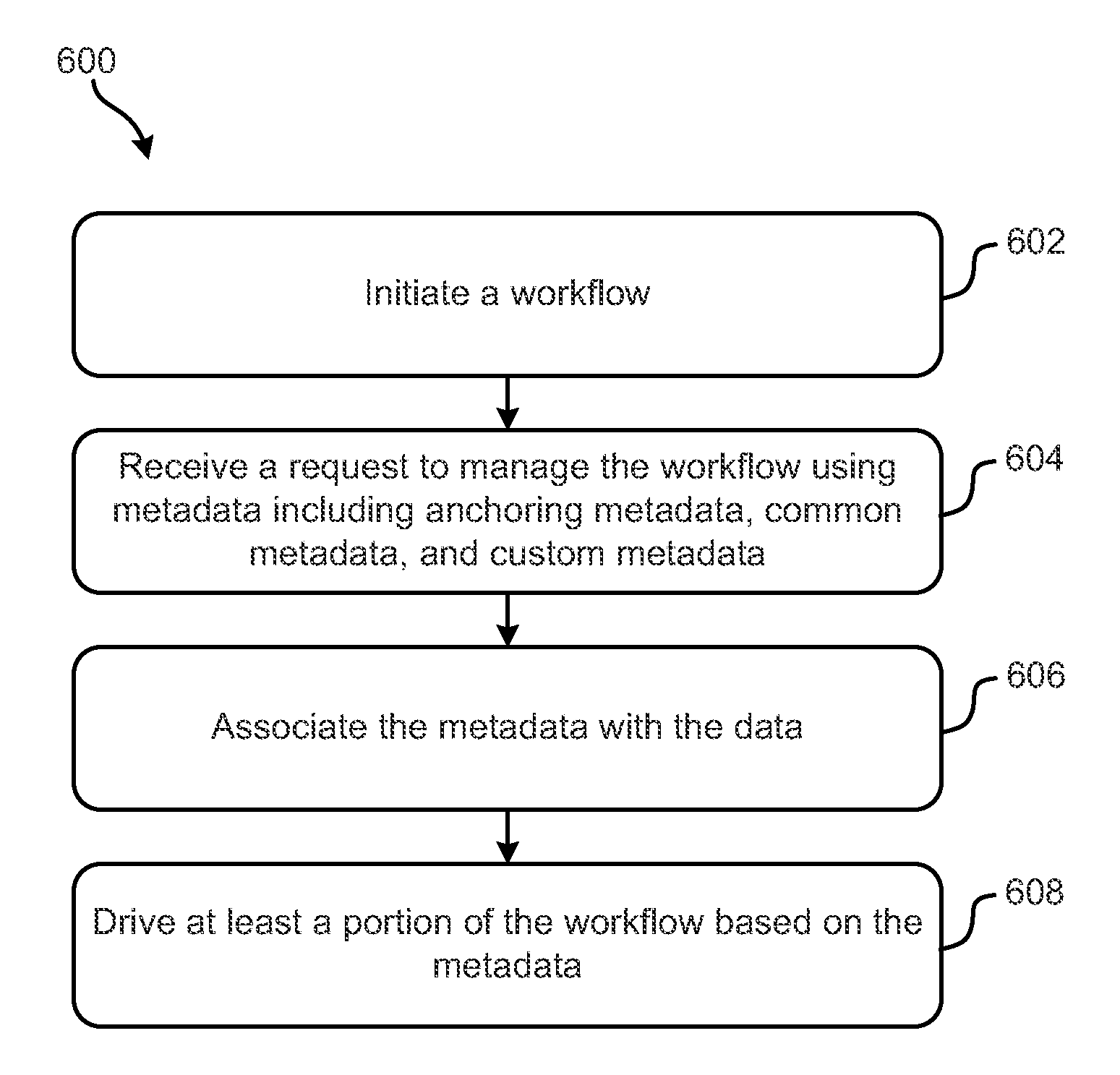
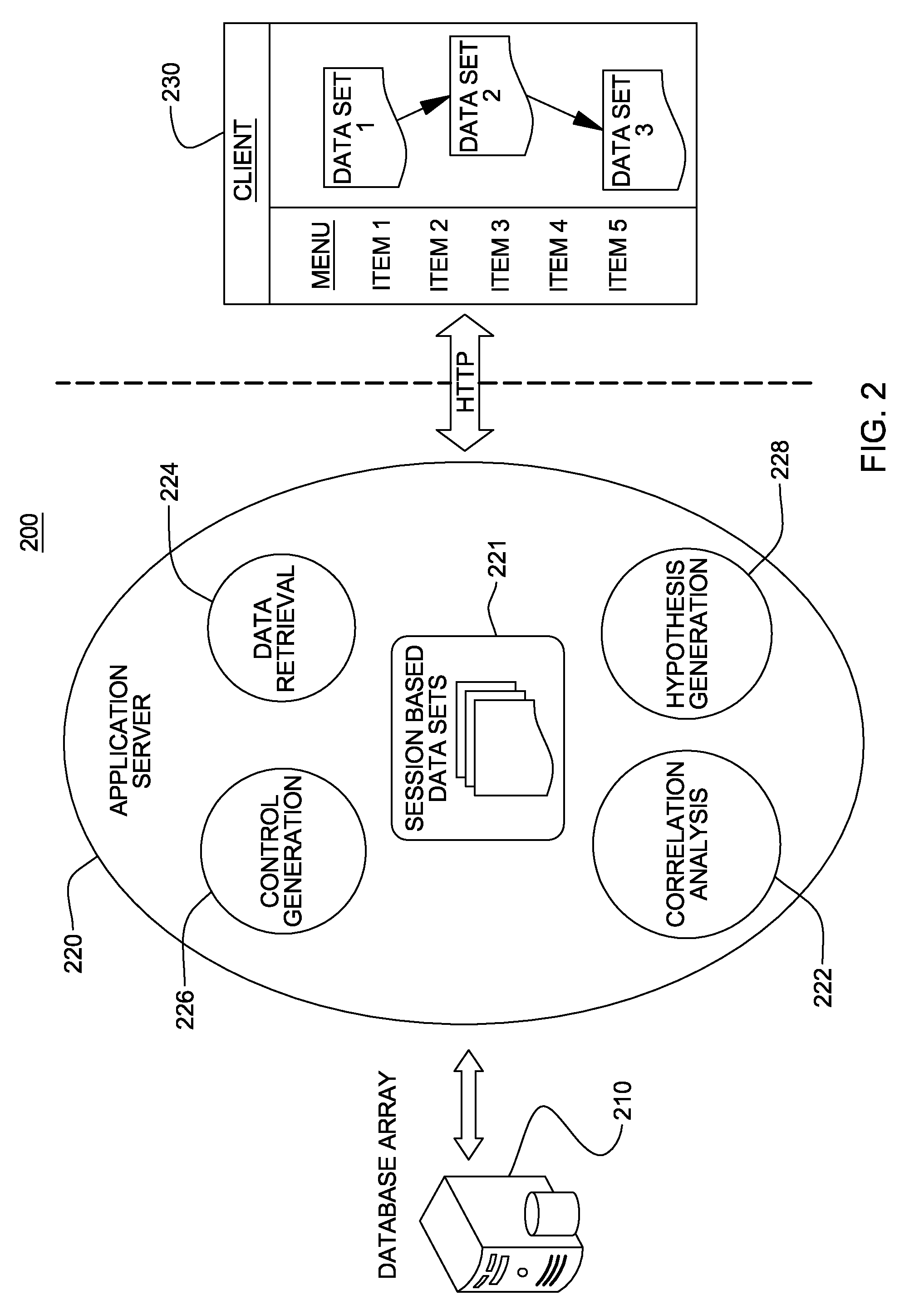
Metadata-driven workflows and integration with genomic data processing systems and techniques
ActiveUS20150286495A1Metadata text retrievalMultimedia data retrievalData processing systemProgram instruction
Systems, methods and computer program products configured to provide and perform metadata-based workflow management are disclosed. The inventive subject matter includes a computer readable storage medium having computer readable program instructions embodied therewith. The computer readable program instructions are configured to: initiate a workflow configured to process data; associate the data with metadata; and drive at least a portion of the workflow based on at least some of the metadata. The metadata include anchoring metadata; common metadata; and custom metadata. Inventive subject matter also encompasses a method for managing genomic data processing workflows using metadata includes: initiating a workflow; receiving a request to manage the workflow using metadata comprising: anchoring metadata, common metadata, and custom metadata, associating the metadata with the data; and driving at least a portion of the workflow based on the metadata. The workflow involves genomic analyses.
Owner:IBM CORP
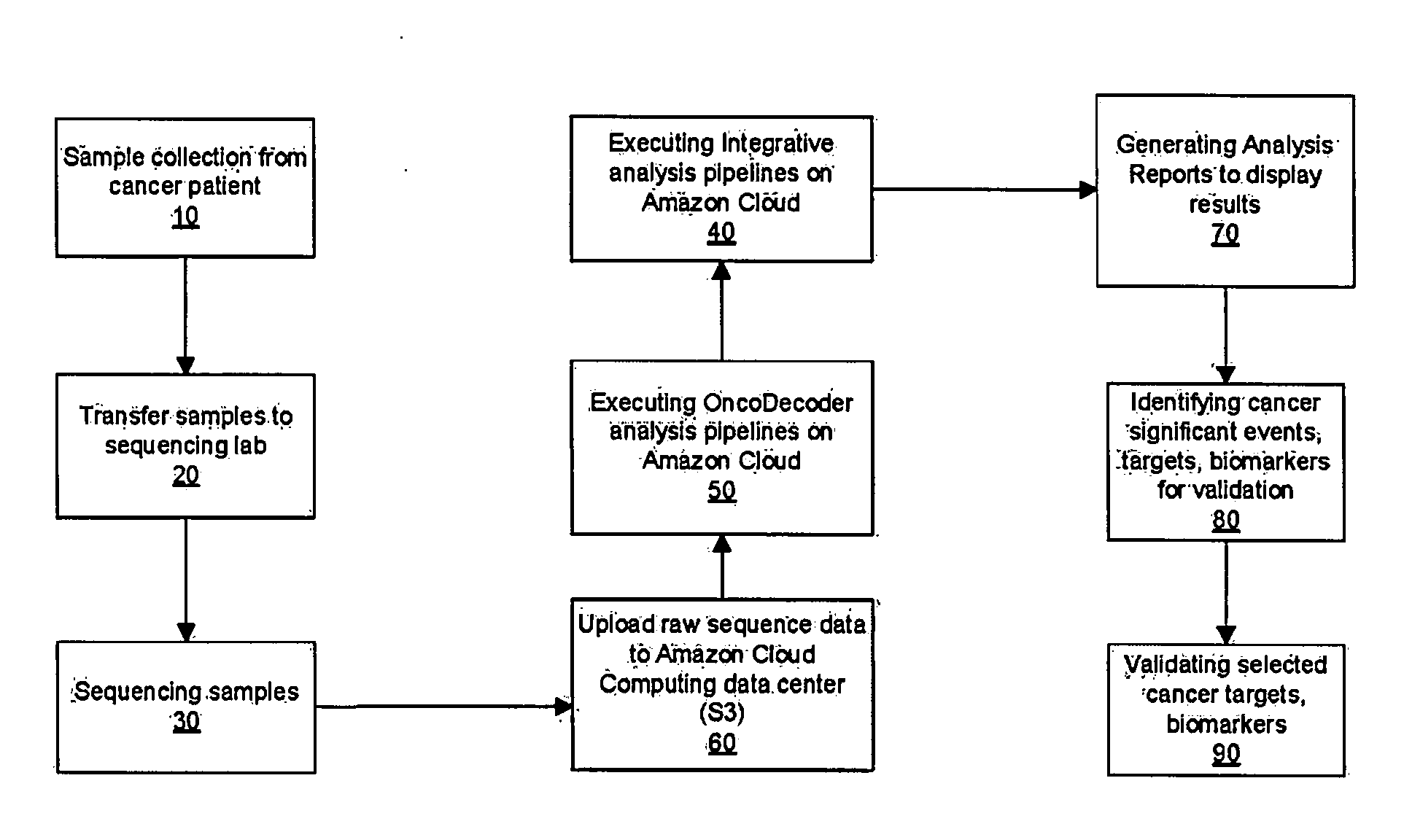
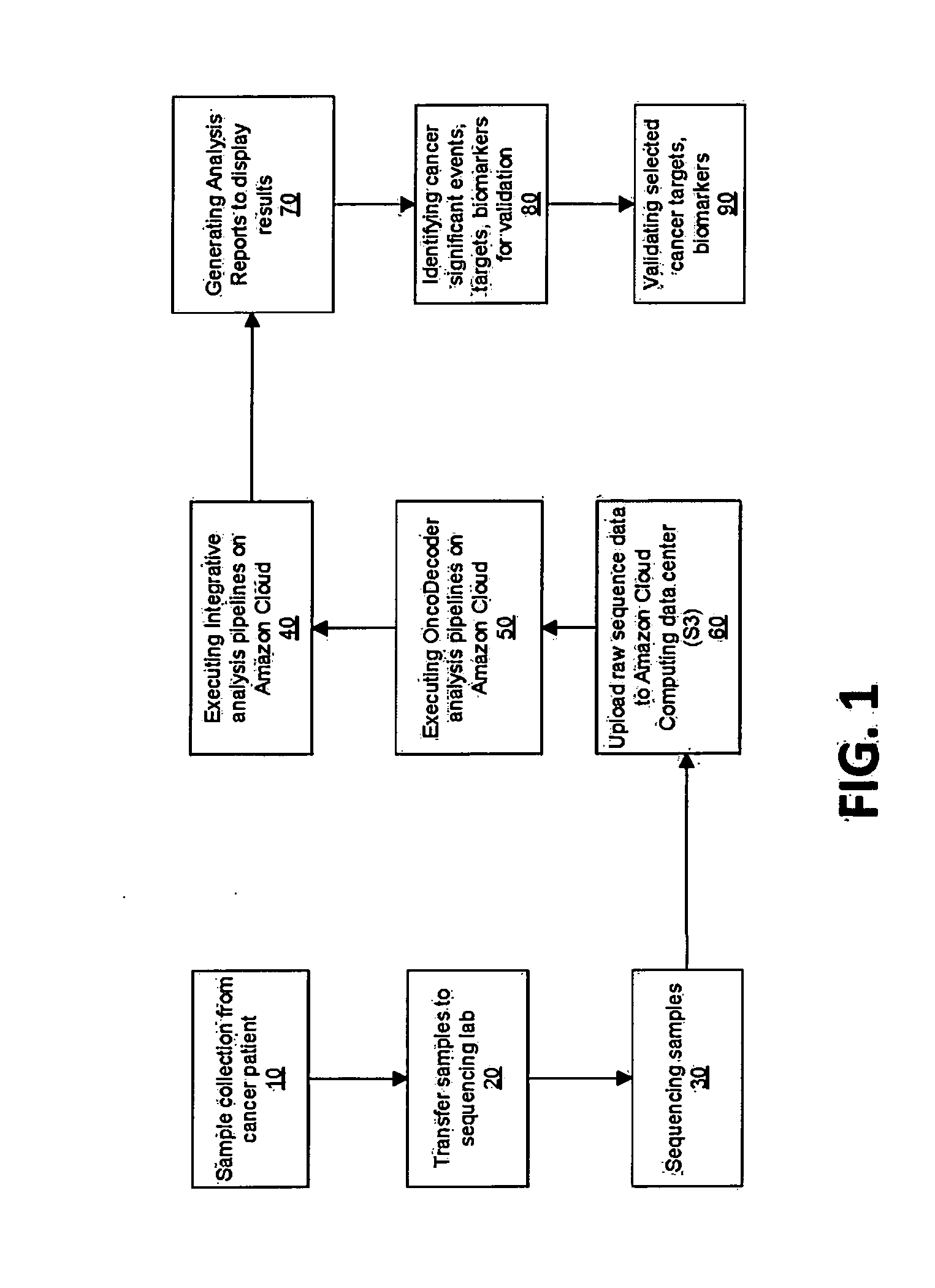
Systems and methods for cancer-specific drug targets and biomarkers discovery
InactiveUS20130184999A1Improve throughputMinimal hardware requirementProteomicsGenomicsHuman cancerBiomarker discovery
The present invention provides users with cloud-based high throughput computing system for integrative analyses of next generation sequencing genomic data, such that human cancer biomarkers and drug targets can be accurately and quickly identified. Advantageously, the present invention harness a comprehensive systematic analysis pipelines for all types of next generation sequencing genomic data, advanced genomic variants calling algorithms and modeling, variant data correlation and integration, and identification of cancer specific biomarkers and therapeutic targets. Thus, the present invention will aid users so that less of their time and efforts are required in order to obtain precisely the desired information for which they are analyzing.
Owner:DING YAN
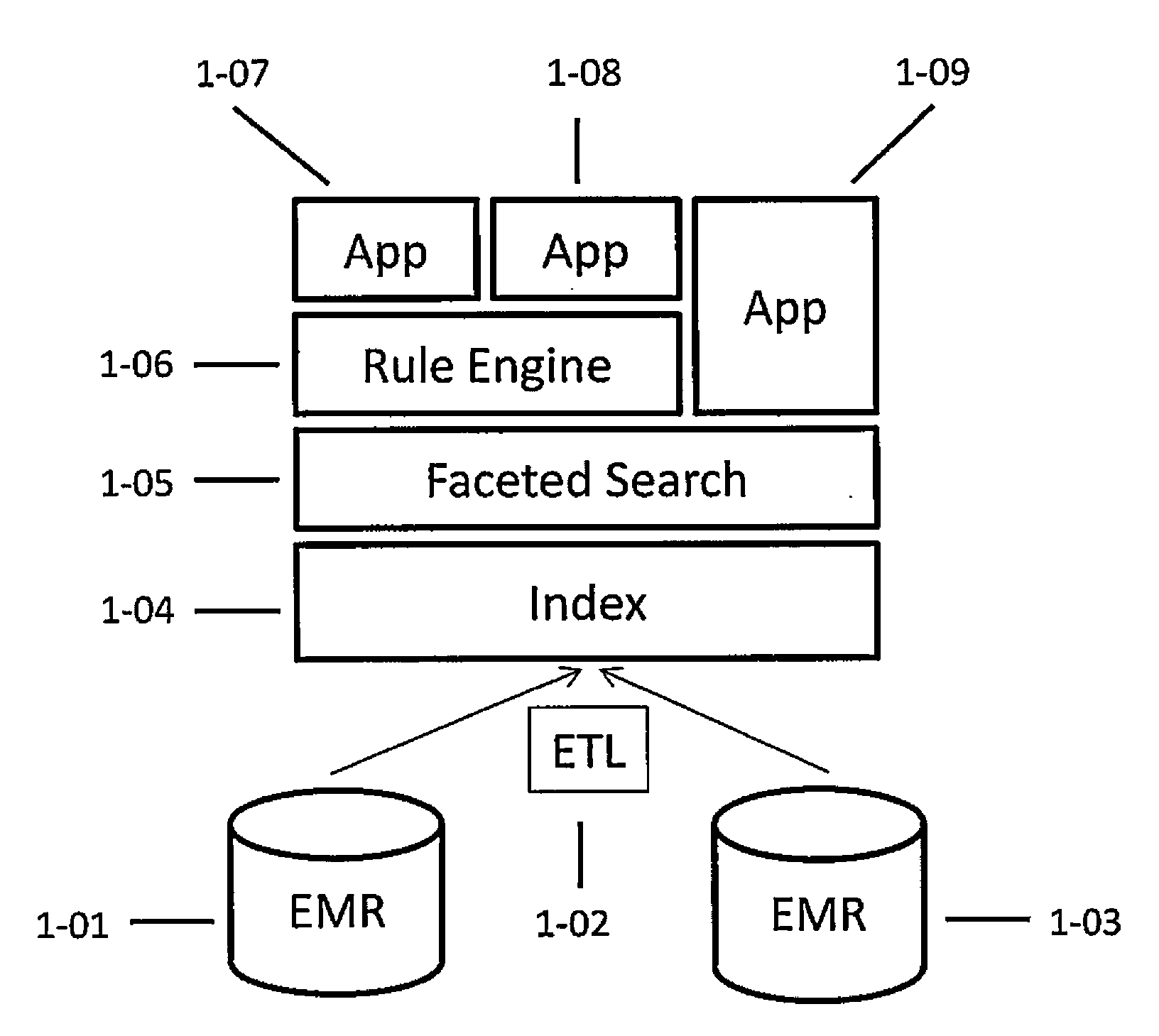
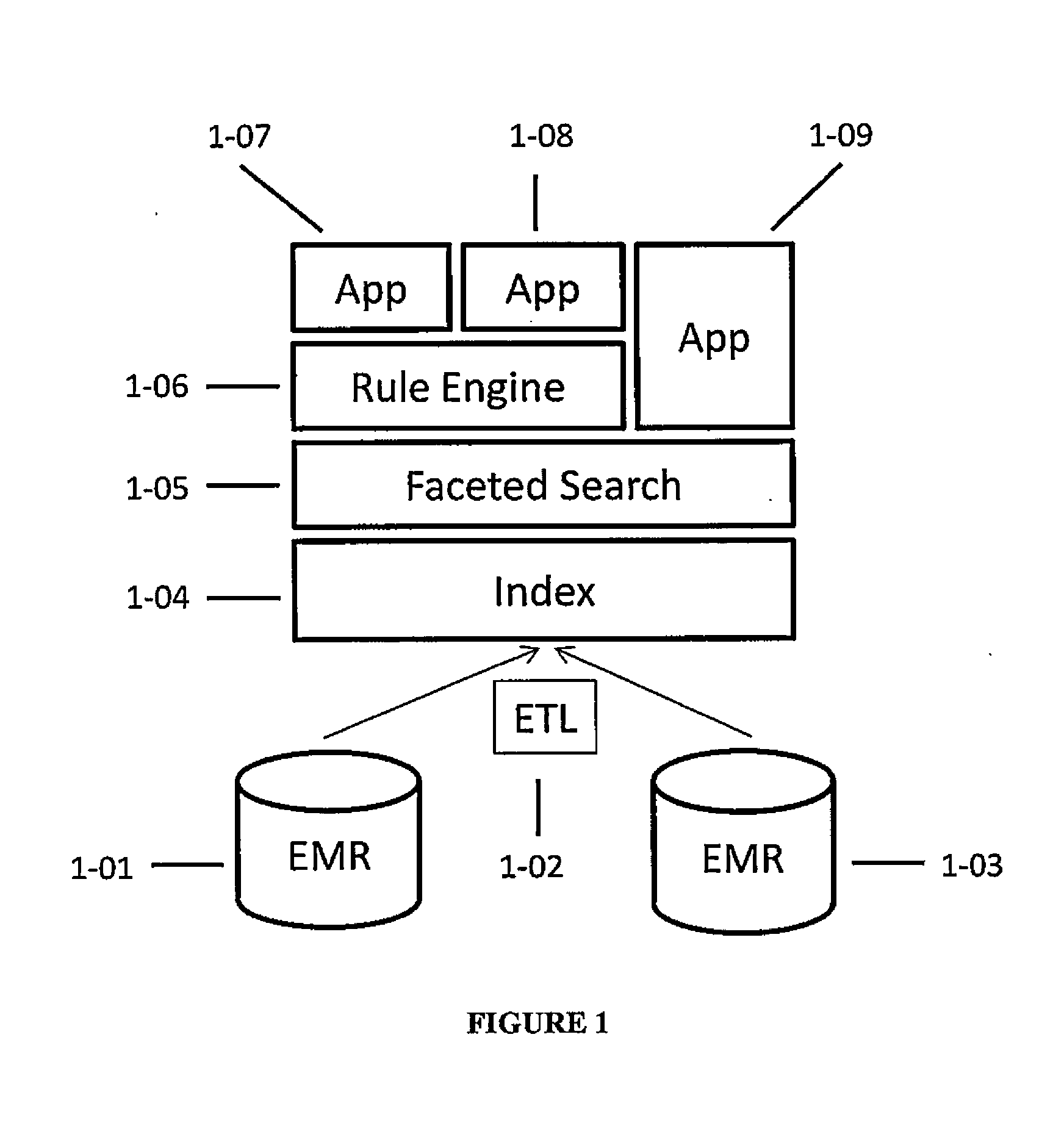
Systems and methods for searching genomic databases
InactiveUS20140046926A1Digital data processing detailsComputer-assisted medical data acquisitionClinical valueData source
The invention described herein solves the challenges encountered in searching for clinical and genomic information from multiple data sources. Systems, methods, and devices of the invention allow a user to search a number of dissimilar information sources simultaneously, and view, process, and perform correlations on the information. The invention uses faceted search to process clinical values, genomic data, subject characteristics, and population characteristics, thereby providing a user with an array of information useful for monitoring or improving the state of health of a subject or a subject population. The invention allows a user to evaluate clinical and research information in a subject-centric way, and analyze information at either the individual or the population level.
Owner:MYCARE
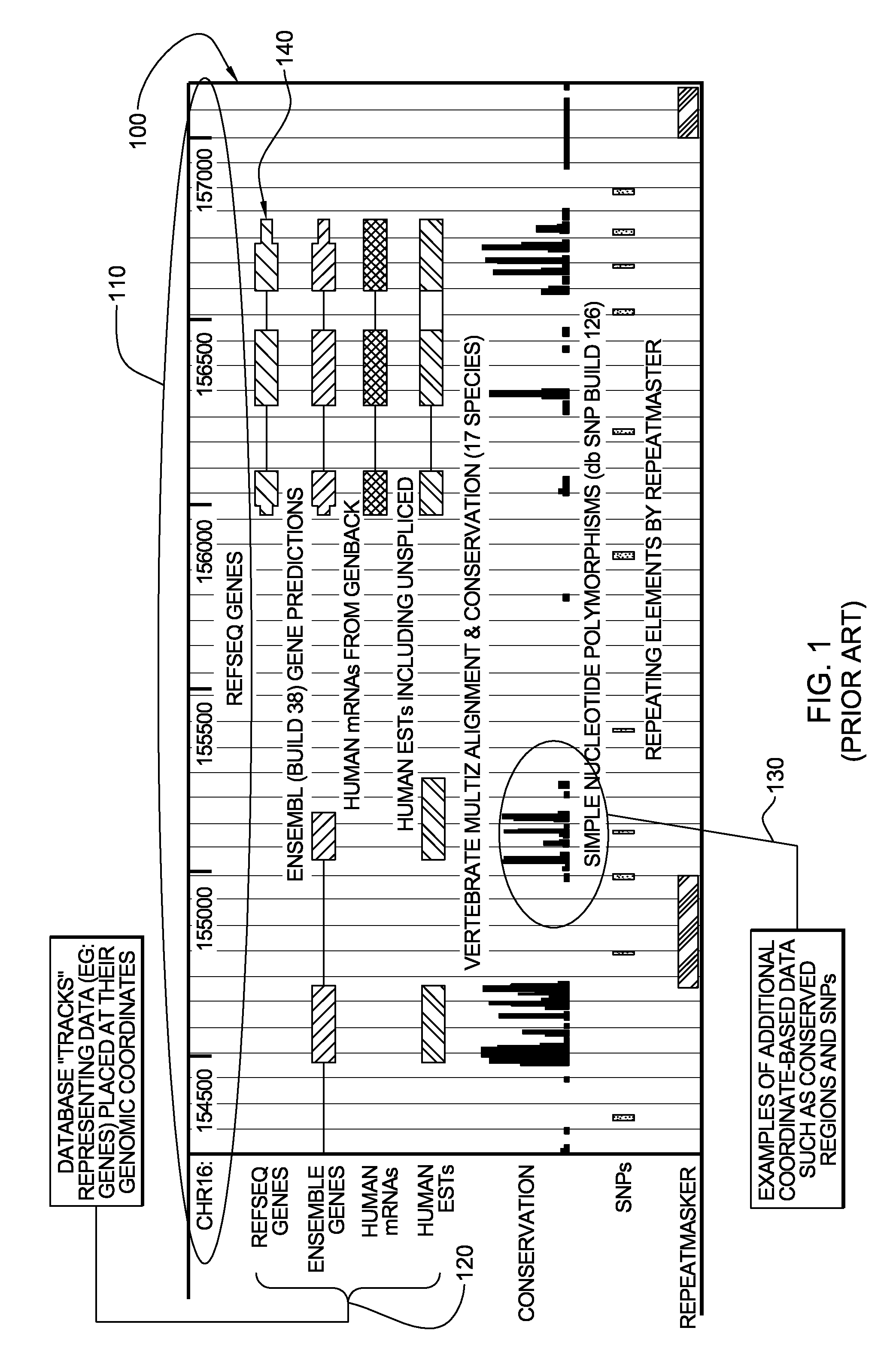
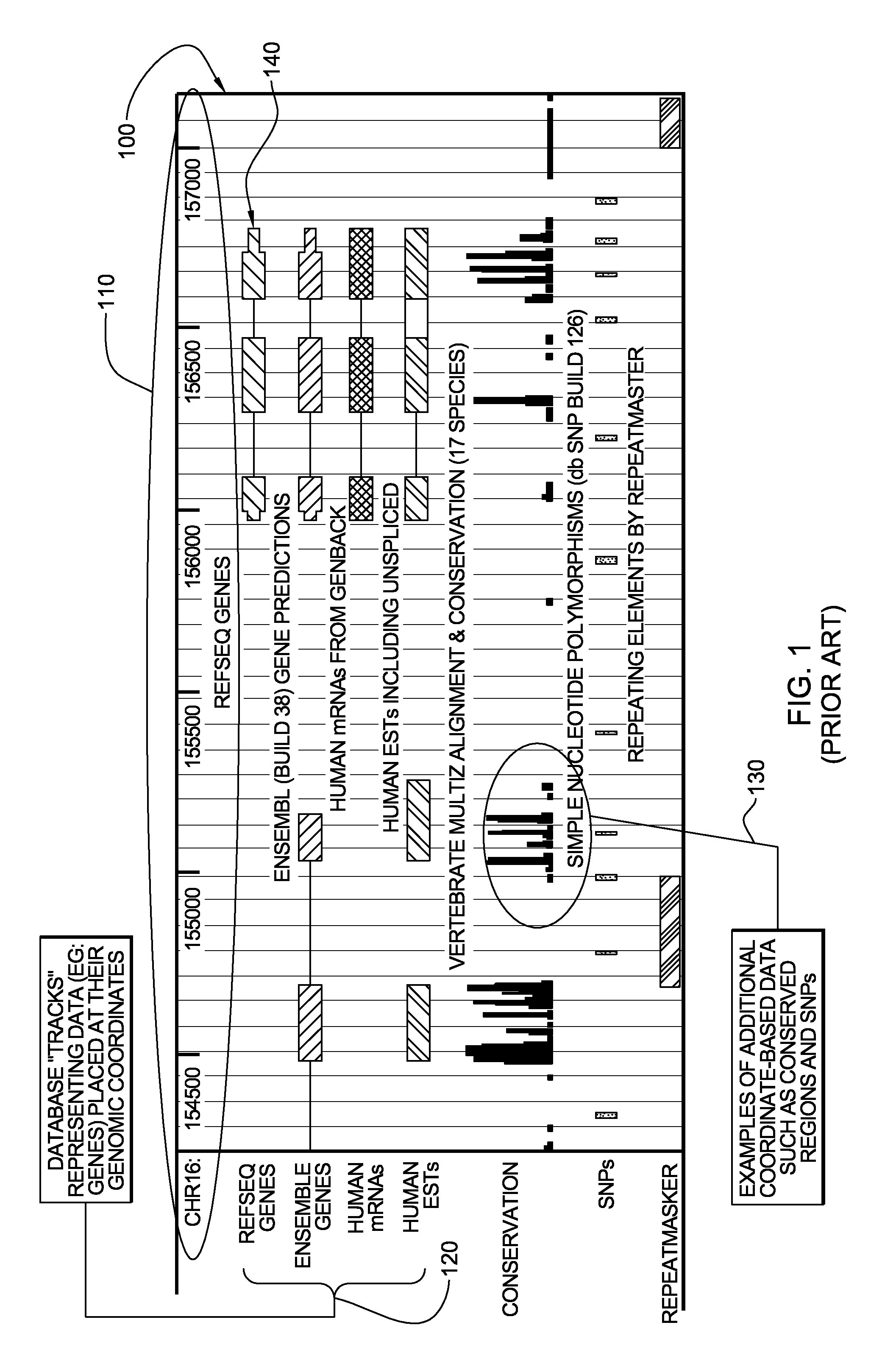
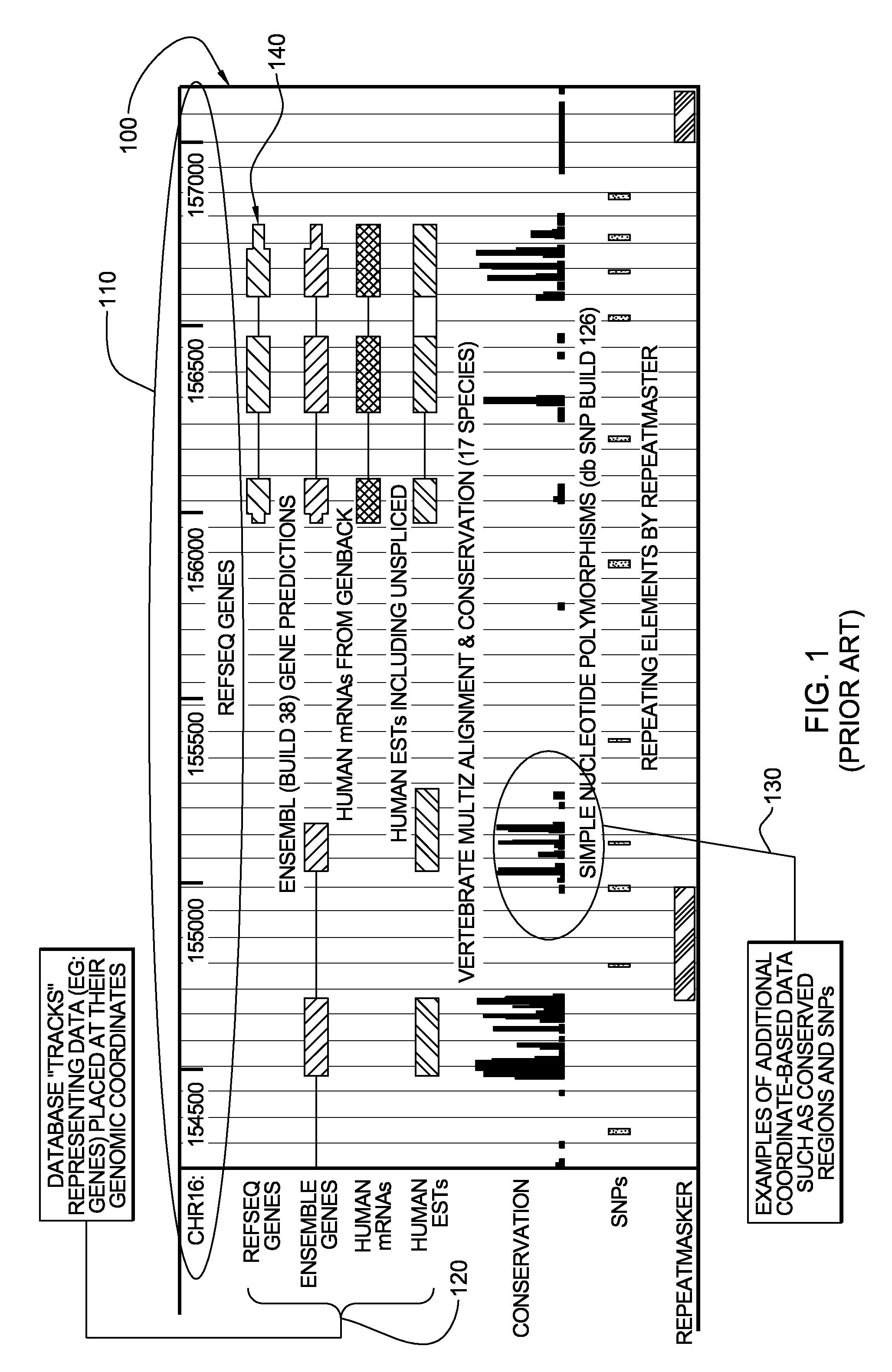
System, Method and computer program product for integrated analysis and visualization of genomic data
Described is a system for analysis and visualization of genomic data. The system allows a user to select at least one individual sample. The sample has chromosomal data representing a genome with a chromosome and also includes chromosomal measurements of at least one event at a particular location on the chromosome. A frequency of event is generated based on the selected sample. The frequency of event is a frequency of occurrence of the event in the selected sample. At least one annotation can be selected that includes chromosomal region specific information as related to the chromosome. Finally, the chromosomal data, the annotation, and the frequency of event on a display can all be simultaneously displayed, thereby allowing a user to view chromosomal region specific information with respect to a particular chromosomal event.
Owner:BIODISCOVERY
Methods and apparatus for sorting data
ActiveUS20150220532A1Digital data information retrievalDigital data processing detailsGenomic dataData sorting
A computer implemented system for genomic data sorting, comprising alignment and position mapping. The system maps each read to a position on the reference genome with which the read is associated, followed by sorting these reads by their mapped positions.
Owner:10X GENOMICS
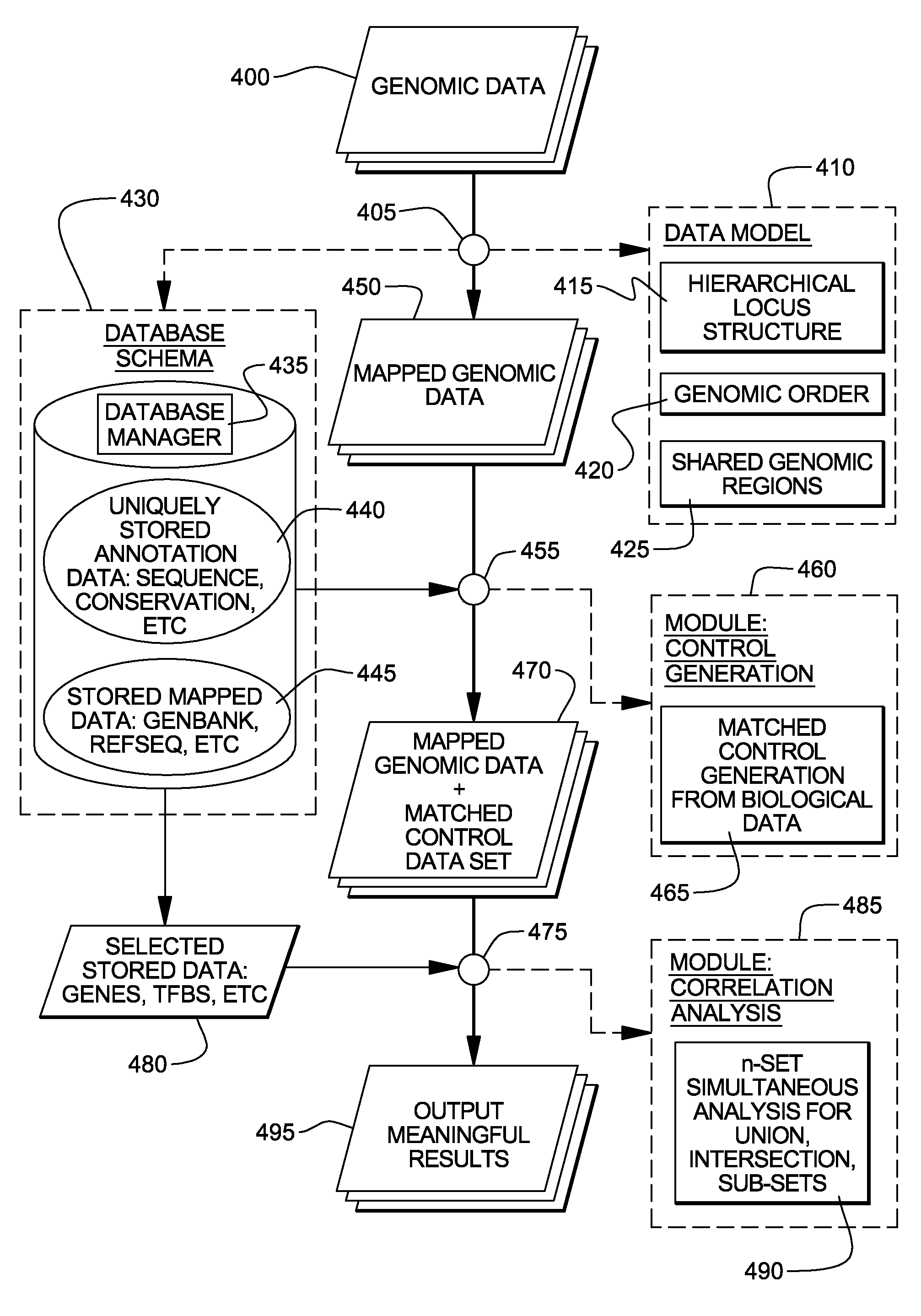
Non-random control data set generation for facilitating genomic data processing
Processing of genomic data is facilitated by providing a control data set generation system wherein a control generator tool or process creates matched data sets for facilitating informatics analysis. These matched data sets may include genomic loci or genomic sequences, or both. The data is taken from a database of actual genomic data, including sequence and annotation data, as opposed to ad-hoc generation, sequence scrambling or the like. This produces biologically relevant and accurate results which allow for stronger controls. The controls are matched against a user-provided data set via a number of parameters.
Owner:THE RES FOUND OF STATE UNIV OF NEW YORK
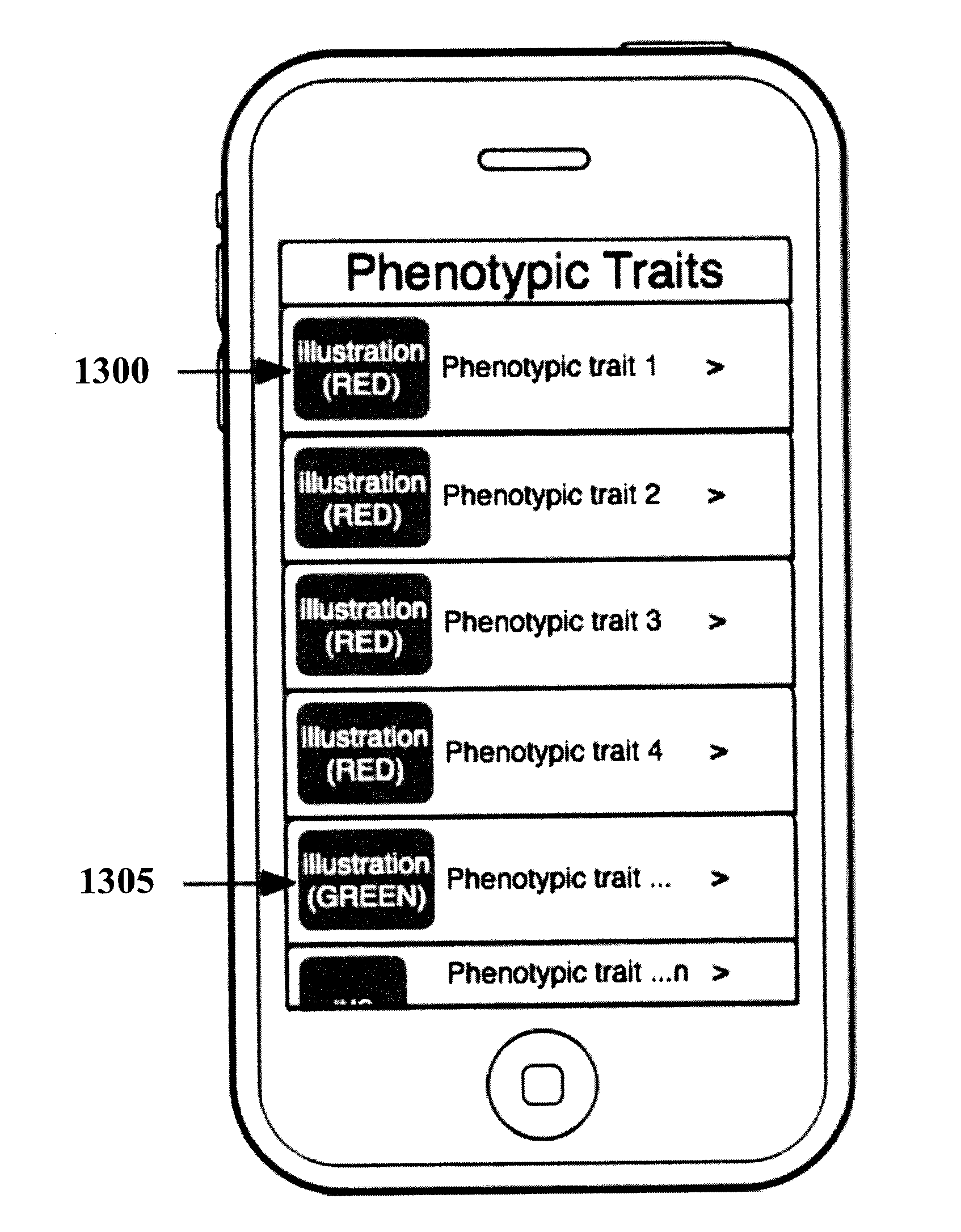
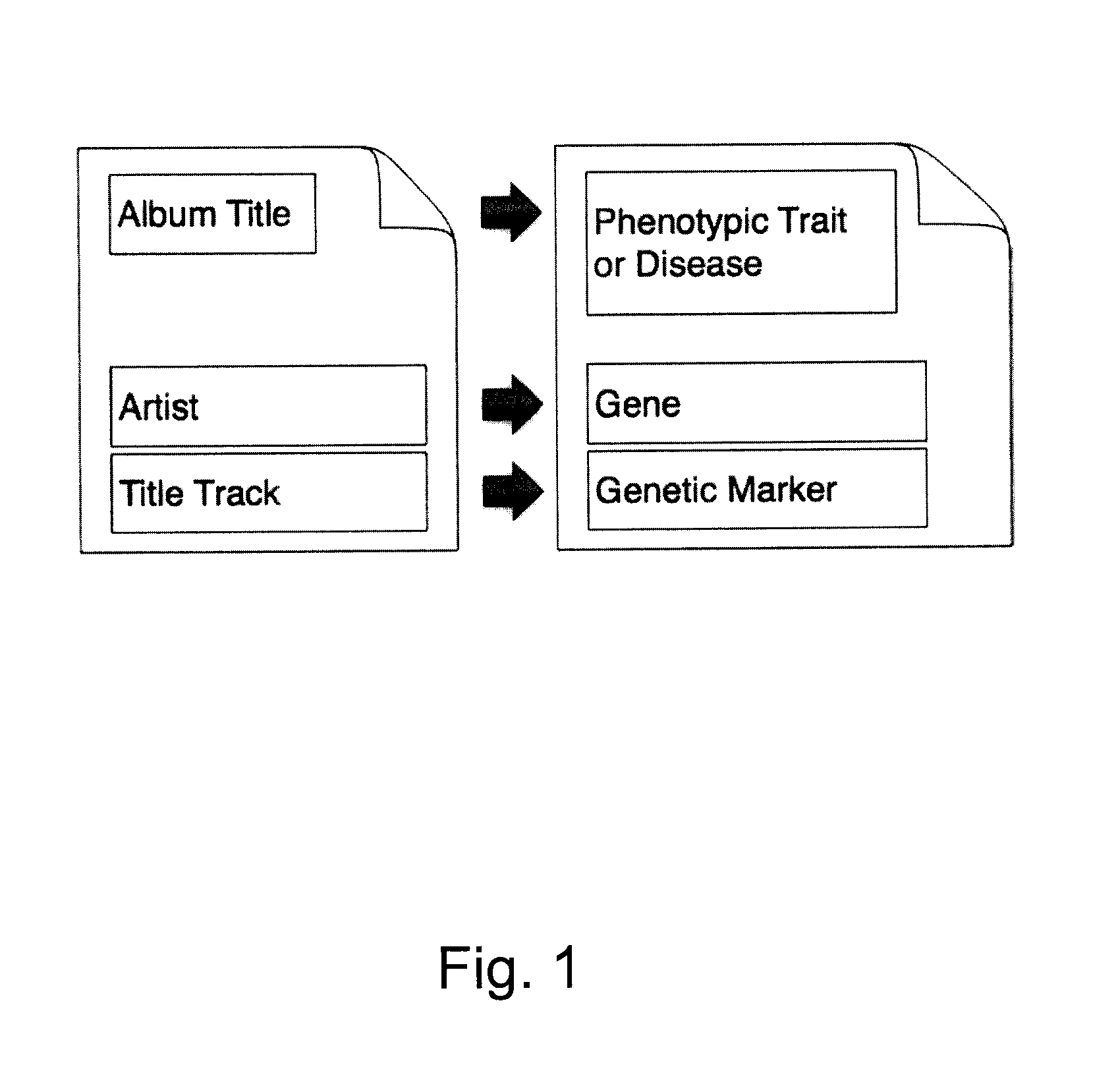
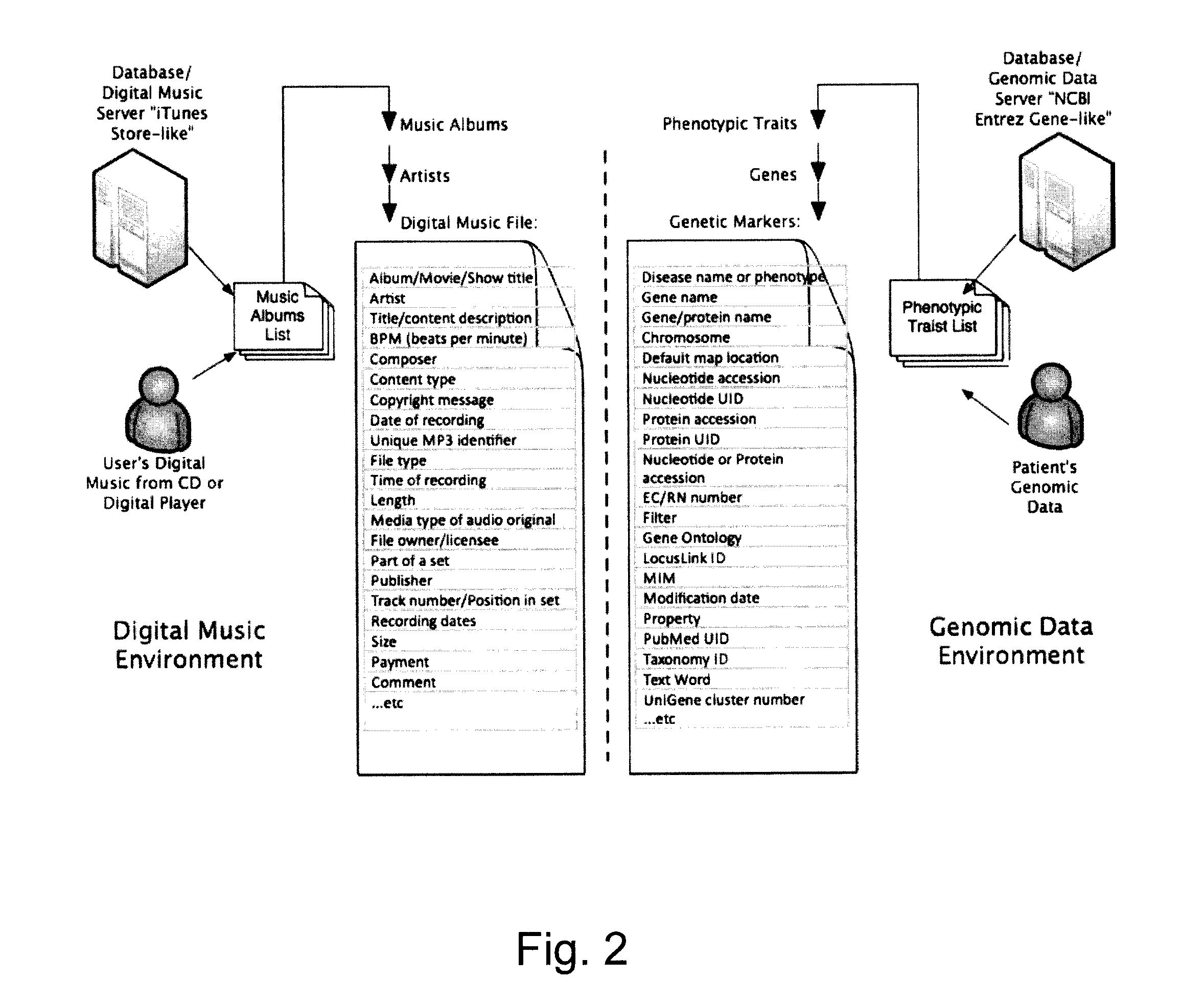
Organization, visualization and utilization of genomic data on electronic devices
InactiveUS20140033125A1Simple organization and visualization and useData visualisationSpecial data processing applicationsHuman DNA sequencingPersonalization
Described herein are methods, devices and systems for simple organization, visualization and use of genome data (e.g. human genome data) on electronic devices (e.g. portable devices). In some embodiments, the data are organized and / or visualized according to phenotype traits, genes, and / or markers in a similar manner to the organization and / or visualization of digital music contents. This concept allows a new procedure for genomic data organization and facilitates the development of genomic data visualization tools. The methods described herein can be implemented with consumer-oriented software on electronic devices, computers, and portable devices, for the use of genomic related data in the field of personalized medicine for predictive, preventive and participative wireless healthcare.
Owner:PORTABLE GENOMICS
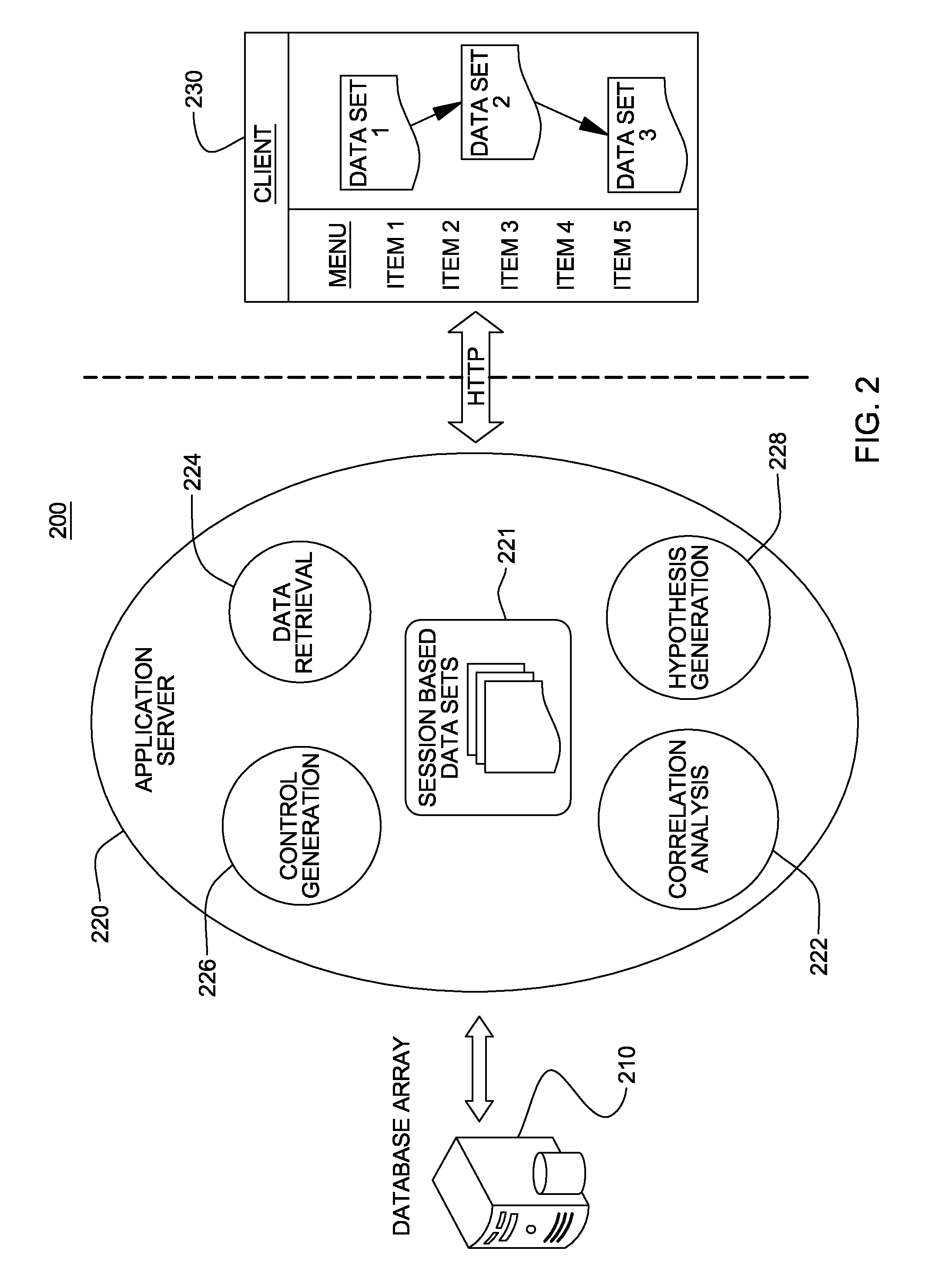
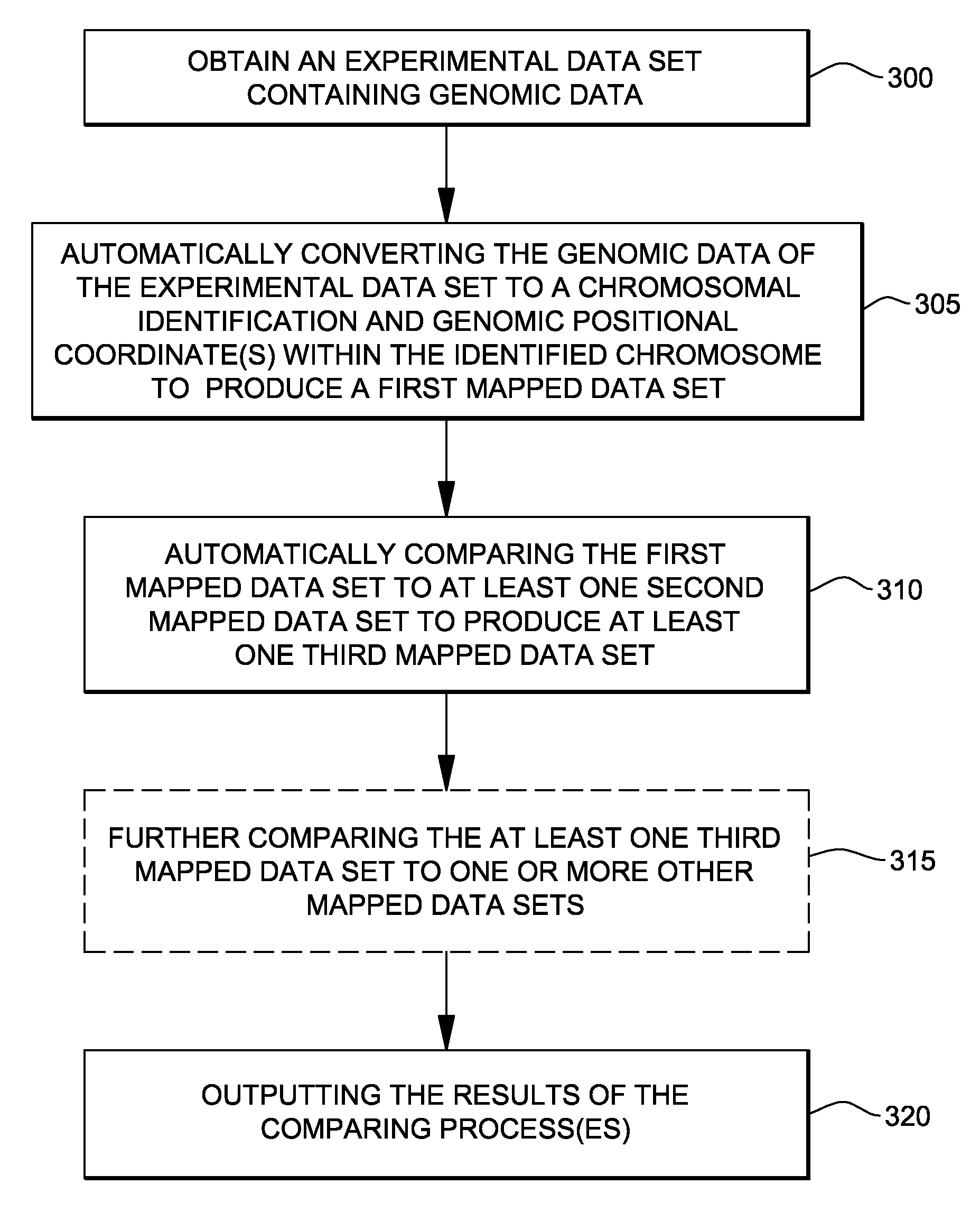
Genomic data processing utilizing correlation analysis of nucleotide loci of multiple data sets
Processing of genomic data is facilitated utilizing correlation analysis of mapped data sets, each data set including genomic data mapped and ordered relative to a genomic coordinate system. Correlation analysis identifies at a nucleotide level nucleotide positions wherein at least one nucleotide locus of each data set correlate. The analysis includes for each data set, selecting a nucleotide locus thereof closest to one end of the coordinate system, comparing the selected nucleotide loci for correlation, and if so, outputting results of the comparing, and updating the selected nucleotide loci by identifying the data set having a next nucleotide locus closest to the one end of the coordinate system, and inserting that next locus into the group of selected loci, and repeating the comparing for the newly selected loci. The process is repeated until nucleotide loci of the mapped data sets are compared and results of the comparison are output.
Owner:THE RES FOUND OF STATE UNIV OF NEW YORK
Metadata-driven workflows and integration with genomic data processing systems and techniques
ActiveUS9354922B2Metadata text retrievalDigital data processing detailsData processing systemProgram instruction
Systems, methods and computer program products configured to provide and perform metadata-based workflow management are disclosed. The inventive subject matter includes a computer readable storage medium having computer readable program instructions embodied therewith. The computer readable program instructions are configured to: initiate a workflow configured to process data; associate the data with metadata; and drive at least a portion of the workflow based on at least some of the metadata. The metadata include anchoring metadata; common metadata; and custom metadata. Inventive subject matter also encompasses a method for managing genomic data processing workflows using metadata includes: initiating a workflow; receiving a request to manage the workflow using metadata comprising: anchoring metadata, common metadata, and custom metadata, associating the metadata with the data; and driving at least a portion of the workflow based on the metadata. The workflow involves genomic analyzes.
Owner:INT BUSINESS MASCH CORP
Bioinformatics Systems, Apparatuses, And Methods Executed On An Integrated Circuit Processing Platform
PendingUS20160171153A1Processing speedProcessed result accuracyBiostatisticsBiological testingData sourceGenomic data
A system, method and apparatus for executing an HMM analysis on genetic sequence data includes an integrated circuit formed of a set of hardwired digital logic circuits that are interconnected by physical electrical interconnects. One of the physical electrical interconnects forms an input to the integrated circuit that may be connected with an electronic data source for receiving reads of genomic data. The hardwired digital logic circuits may be arranged as a set of processing engines, each processing engine being formed of a subset of the hardwired digital logic circuits to perform one or more steps in the HMM analysis on the reads of genomic data. Each subset of the hardwired digital logic circuits may be formed in a wired configuration to perform the one or more steps in the HMM analysis.
Owner:EDICO GENOME
Analysis method and system of metagenome data
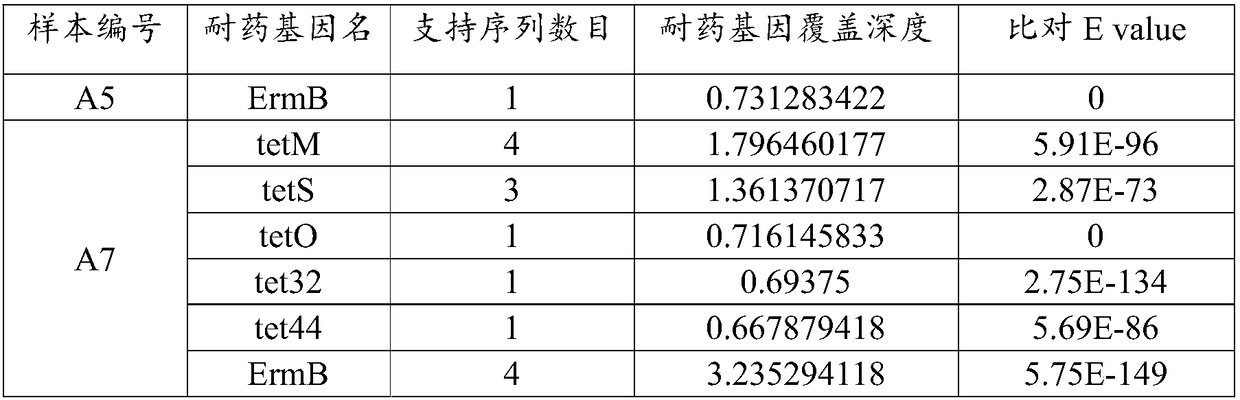
ActiveCN108334750AImprove accuracyReduce processingSequence analysisSpecial data processing applicationsEndocarditisResistant genes
The invention relates to an analysis method and a system of metagenome data. According to the invention, a preliminary species identification result of a sample is obtained on the basis of a k-Mer algorithm, a part or all of supporting sequences are extracted on the basis of the preliminary species identification result, and the preliminary species identification result is verified by using a blast algorithm to judge whether the preliminary species identification result is a reported detected species or not. The method and system disclosed by the invention can lower false positivity, quickly and accurately obtain the reported detected species of the sample in a short time, and are compatible with various mainstream sequencing platforms, thereby being suitable for second-generation sequencing technologies and third sequencing technologies; the method and system of the invention can also accurately identify drug-resistant genes and drug-resistant mutation sites of the sample and map thedrug-resistant genes and the drug-resistant mutation sites of the sample to the reported detected species. Furthermore, the system disclosed by the invention can be used for identifying pathogenic microorganisms, especially endocarditis pathogens to overcome the defect that the endocarditis pathogens are difficultly cultured.
Owner:SIMCERE DIAGNOSTICS CO LTD +2
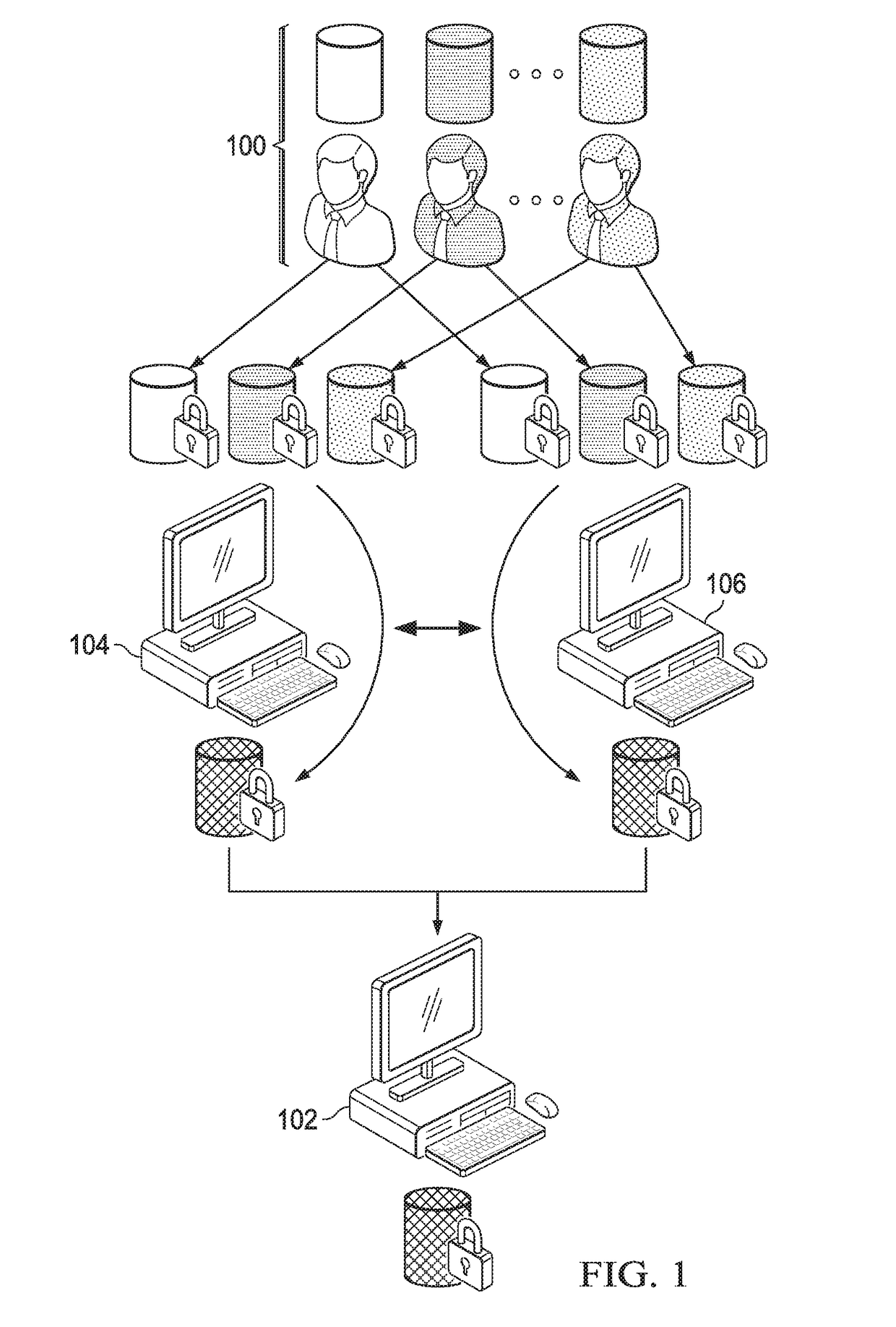
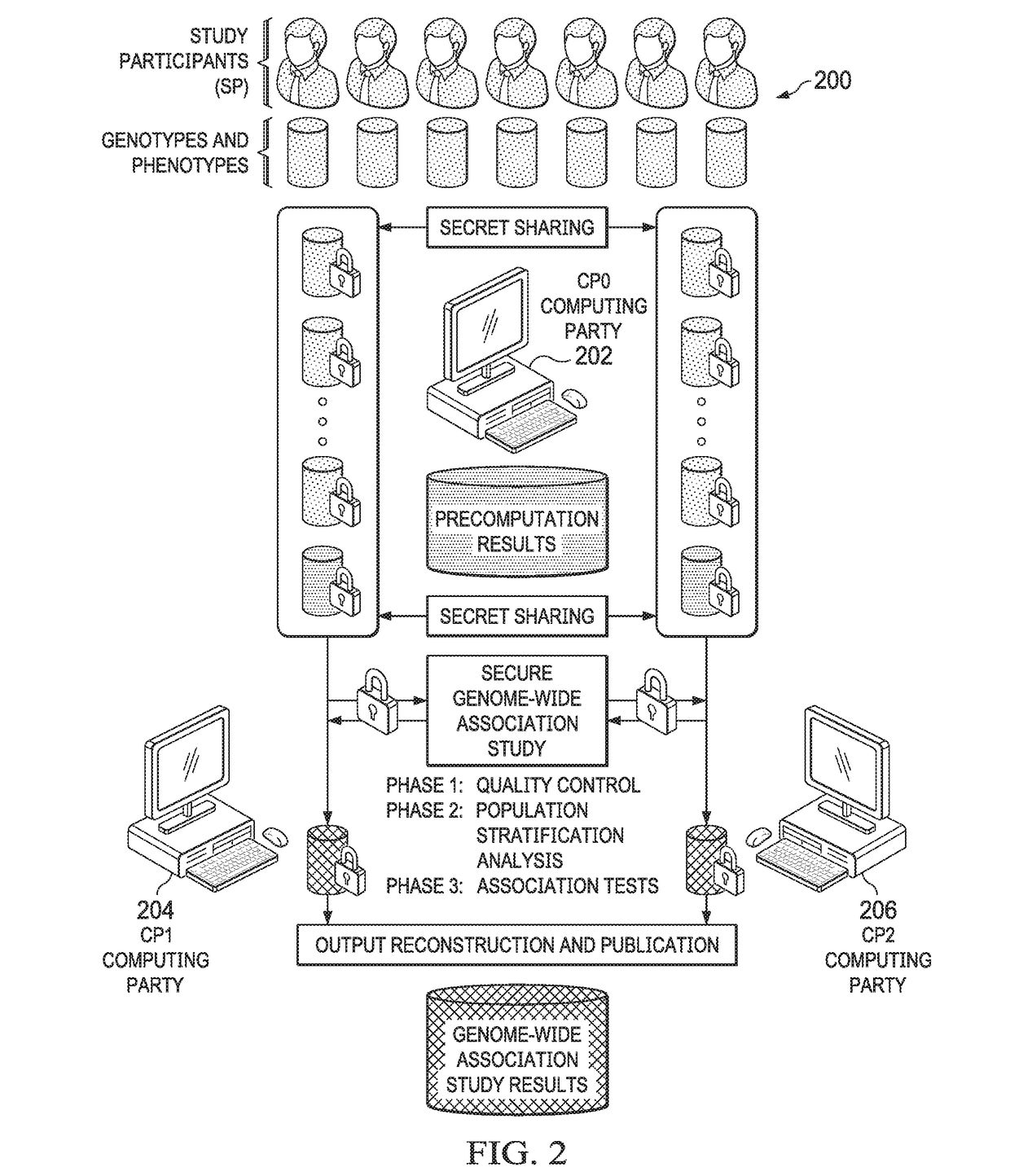
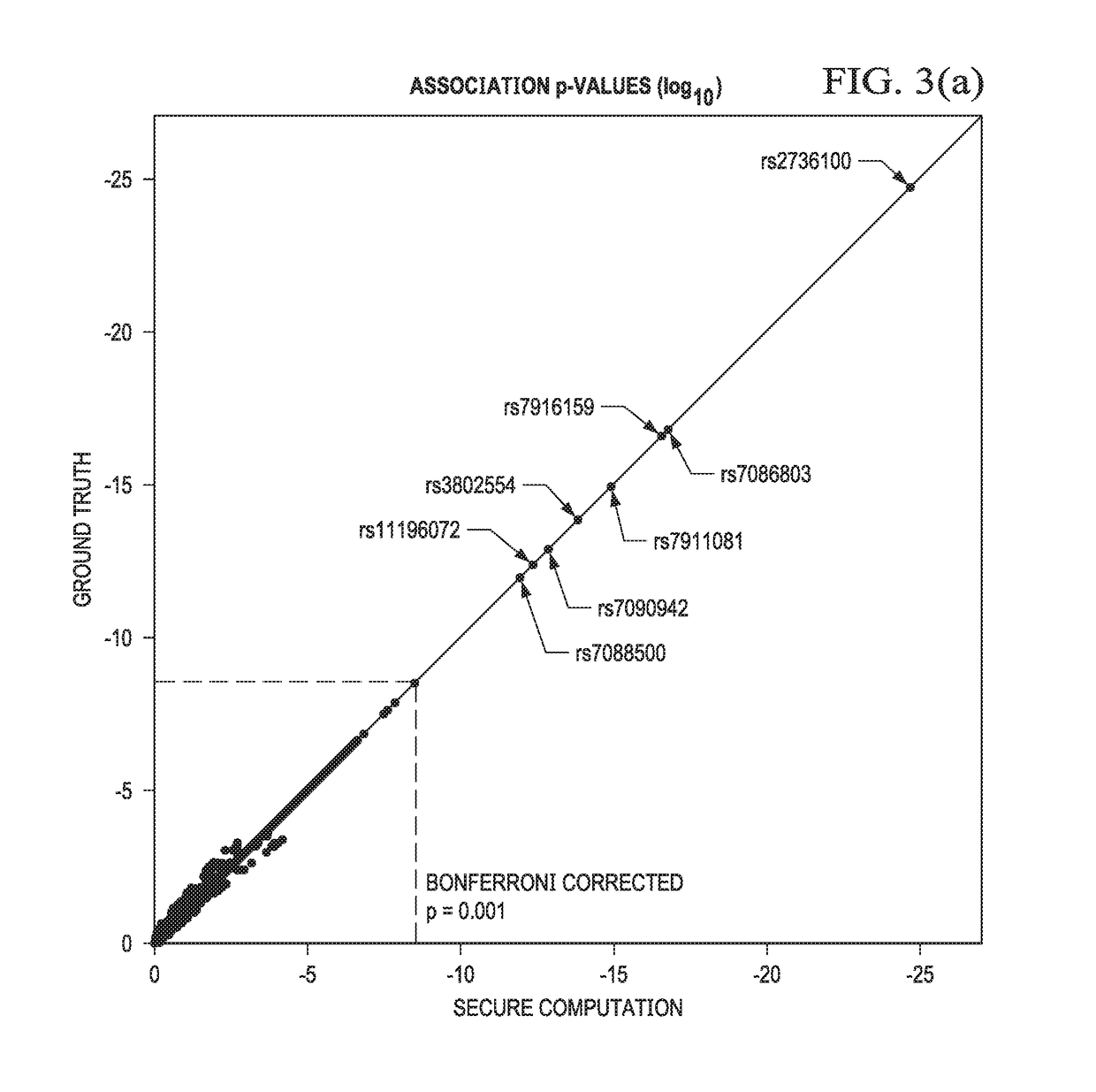
Secure genome crowdsourcing for large-scale association studies
ActiveUS20180373834A1Secure crowdsourcingPrivacy protectionProteomicsGenomicsData setIndividual study
Computationally-efficient techniques facilitate secure crowdsourcing of genomic and phenotypic data, e.g., for large-scale association studies. In one embodiment, a method begins by receiving, via a secret sharing protocol, genomic and phenotypic data of individual study participants. Another data set, comprising results of pre-computation over random number data, e.g., mutually independent and uniformly-distributed random numbers and results of calculations over those random numbers, is also received via secret sharing. A secure computation then is executed against the secretly-shared genomic and phenotypic data, using the secretly-shared results of the pre-computation over random number data, to generate a set of genome-wide association study (GWAS) statistics. For increased computational efficiency, at least a part of the computation is executed over dimensionality-reduced genomic data. The resulting GWAS statistics are then used to identify genetic variants that are statistically-correlated with a phenotype of interest.
Owner:CHO HYUNGHOON +2
Systems and Methods for Genomic Annotation and Distributed Variant Interpretation
InactiveUS20140304270A1Digital data processing detailsRelational databasesGenomic dataGenomic annotation
A computer-based genomic annotation system, including a database configured to store genomic data, non-transitory memory configured to store instructions, and at least one processor coupled with the memory, the processor configured to implement the instructions in order to implement an annotation pipeline and at least one module filtering or analysis of the genomic data.
Owner:THE SCRIPPS RES INST
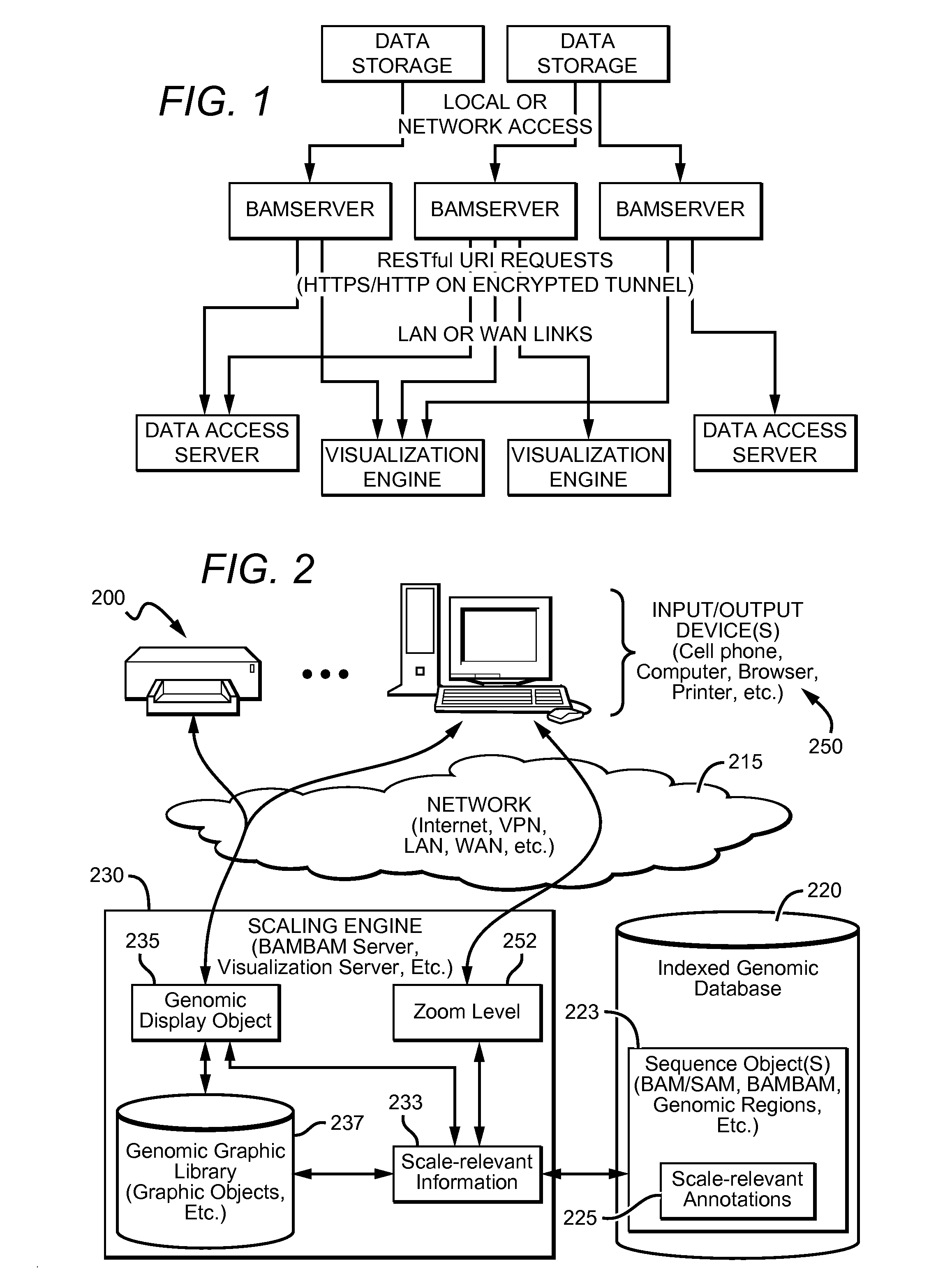
Distributed System Providing Dynamic Indexing And Visualization Of Genomic Data
ActiveUS20140368550A1Reduced data analysisReduce transferGeometric image transformationData visualisationGenomic dataData system
Systems and methods for dynamic visualization of genomic data are provided in which a genomic visualization system adapts presentation of information content according to scale-relevant annotations within a sequence object.
Owner:FIVE3 GENOMICS
Personally controlled storage and testing of personal genomic information
InactiveUS20100121872A1Easy to shareControl over personal dataDigital data processing detailsMultiple digital computer combinationsDiseaseGenomic data
Owner:KNOME
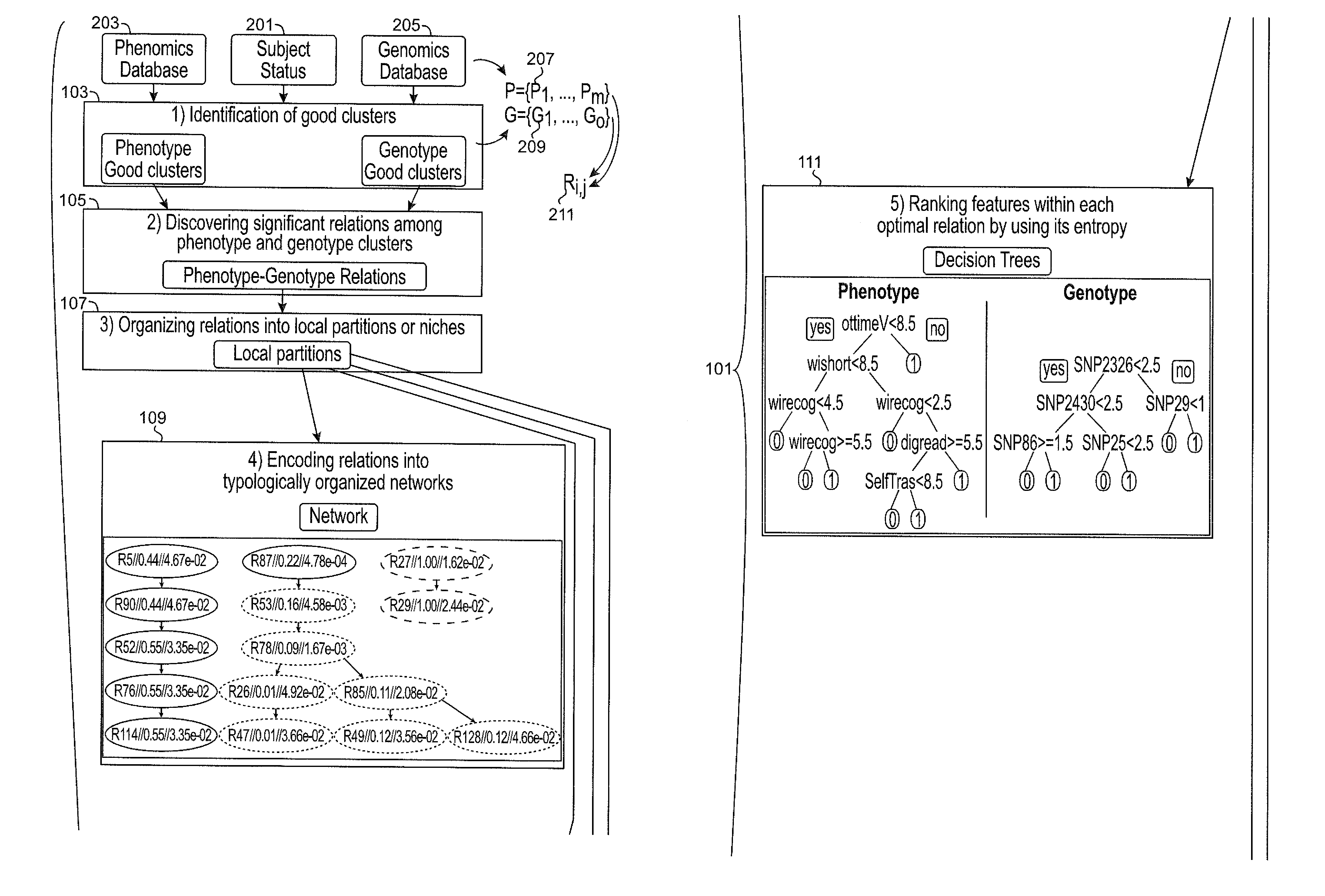
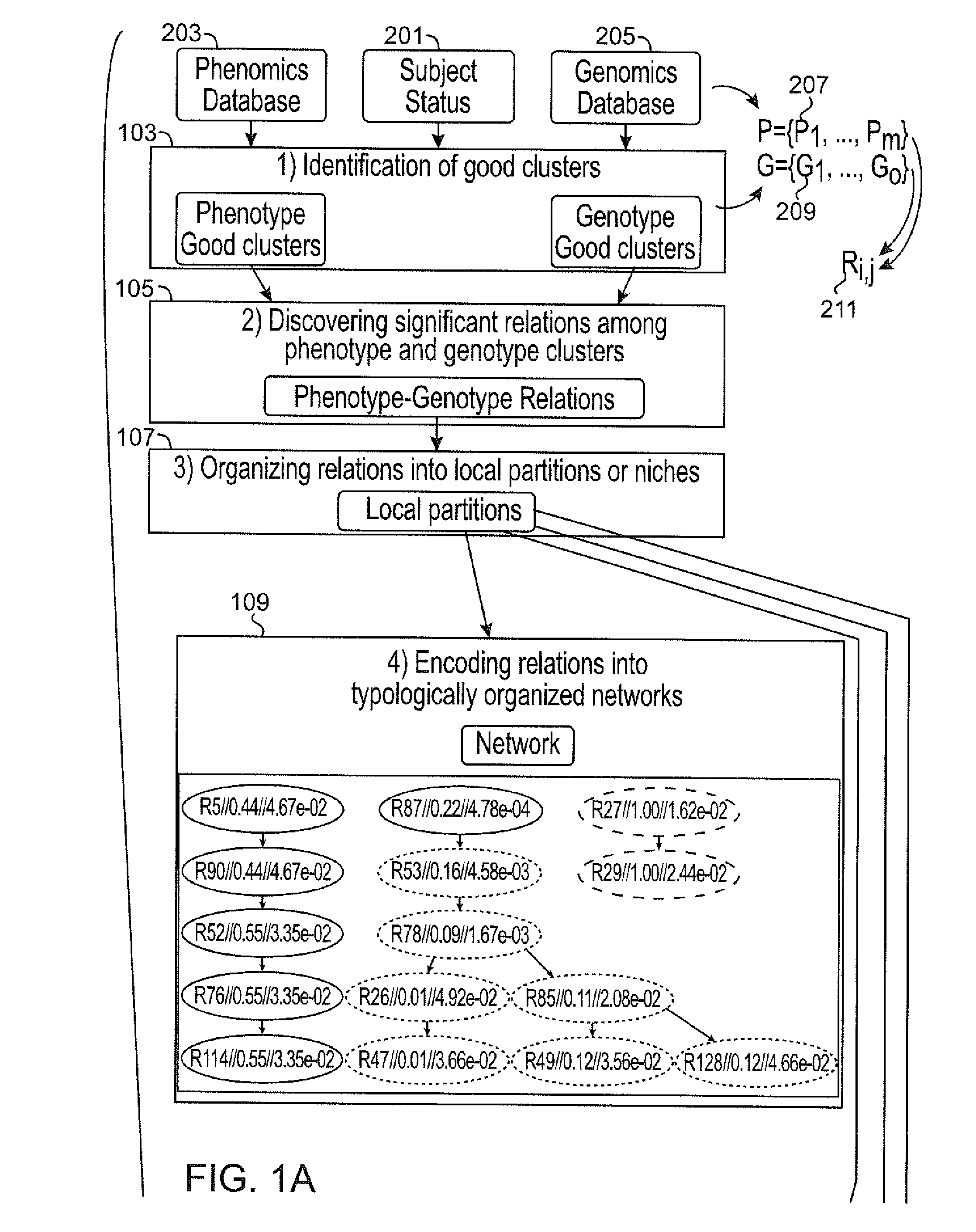
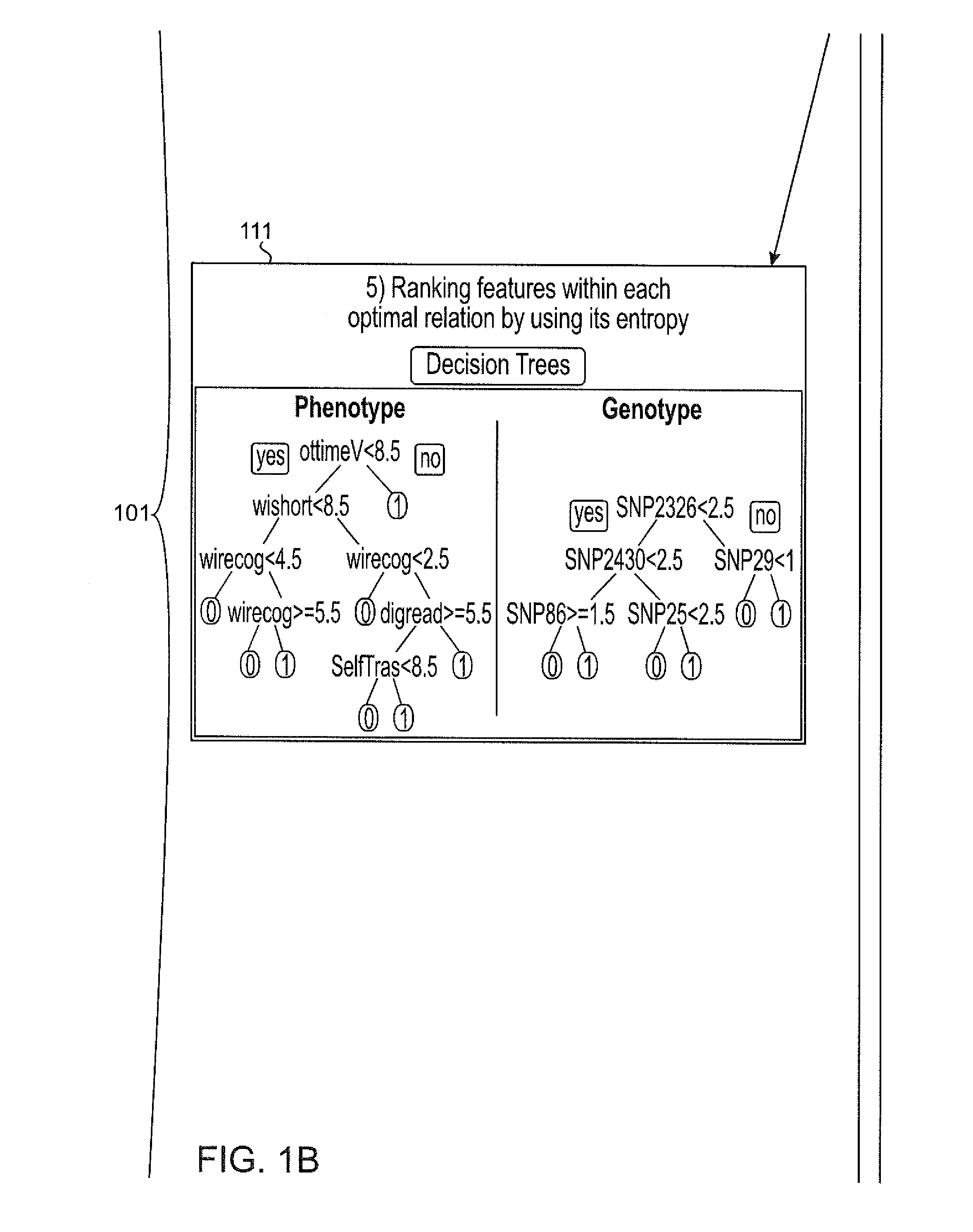
Systems and methods for scalable unsupervised multisource analysis
Systems and methods for identifying genetic variations in a disease and diagnosing a patient with a mental illness, or a generic variant of same. The systems and methods use genomics and phenomics in a computer-implemented methods to identify biclusters in phenomic and genomic data, discover relationships among the biclusters, organize the relations into partitions, rank the predictive utility of features, and map the disease risk function. This can in turn be used to diagnose a patient in a person-centered fashion.
Owner:ZWIR JORGE S
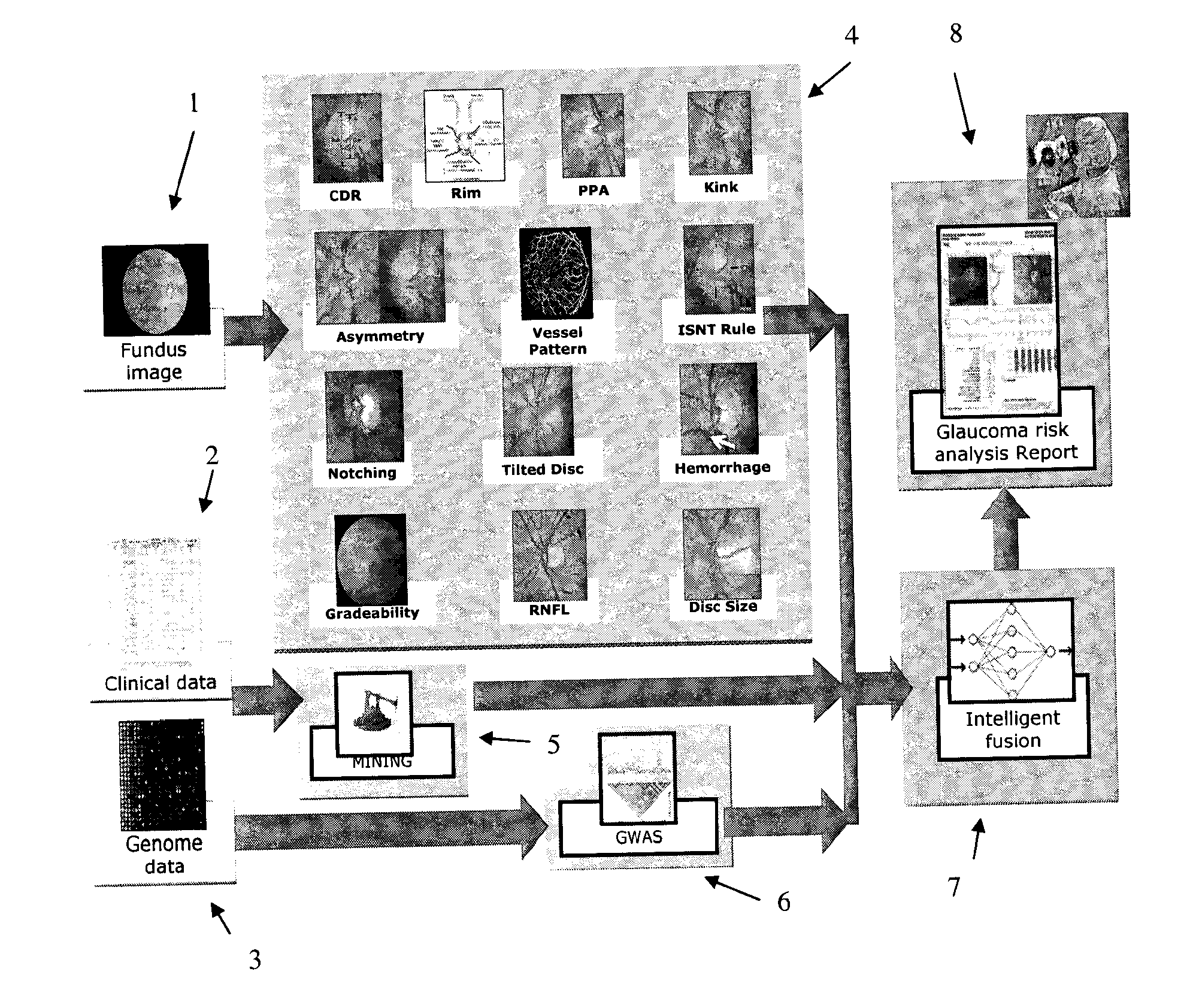
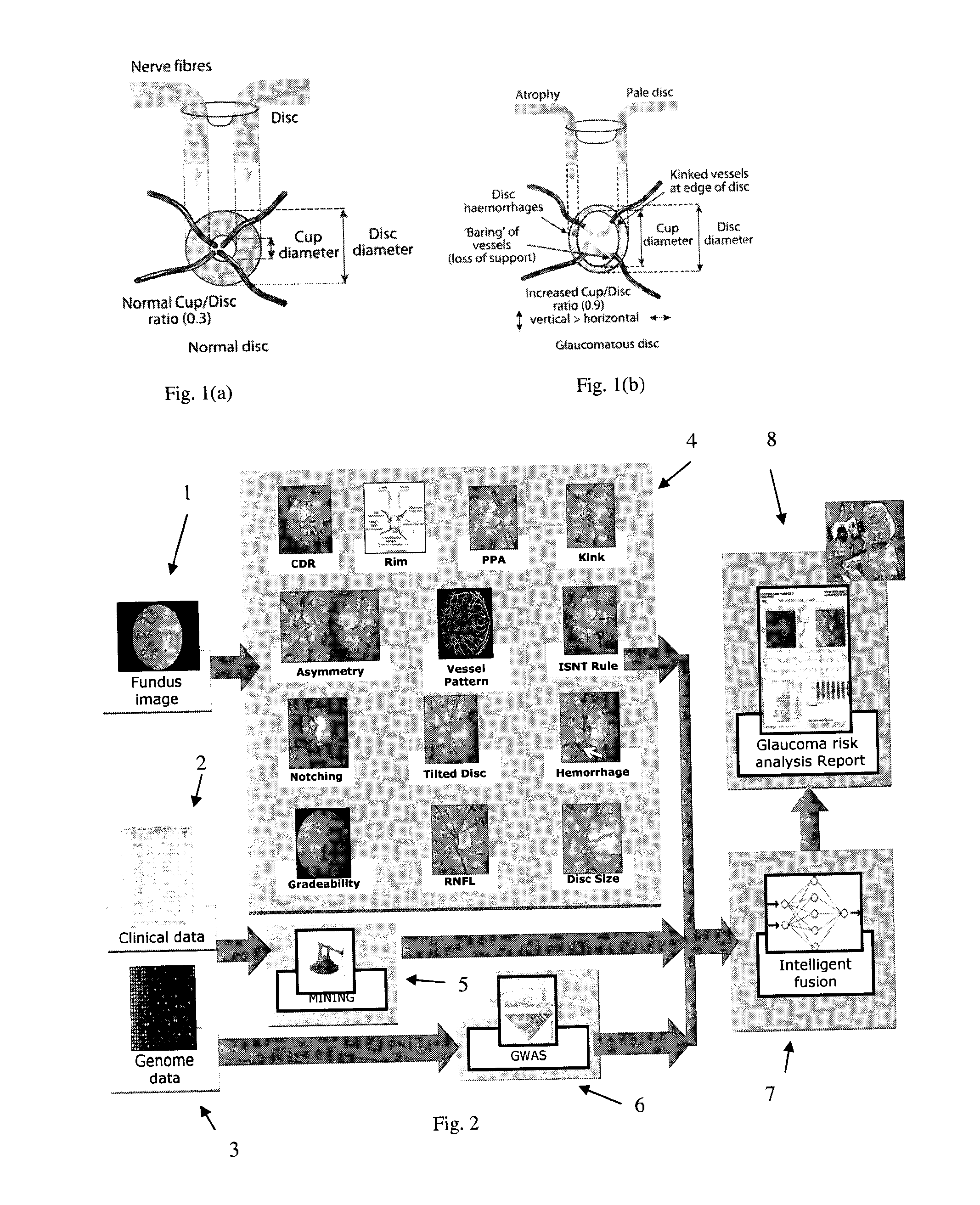
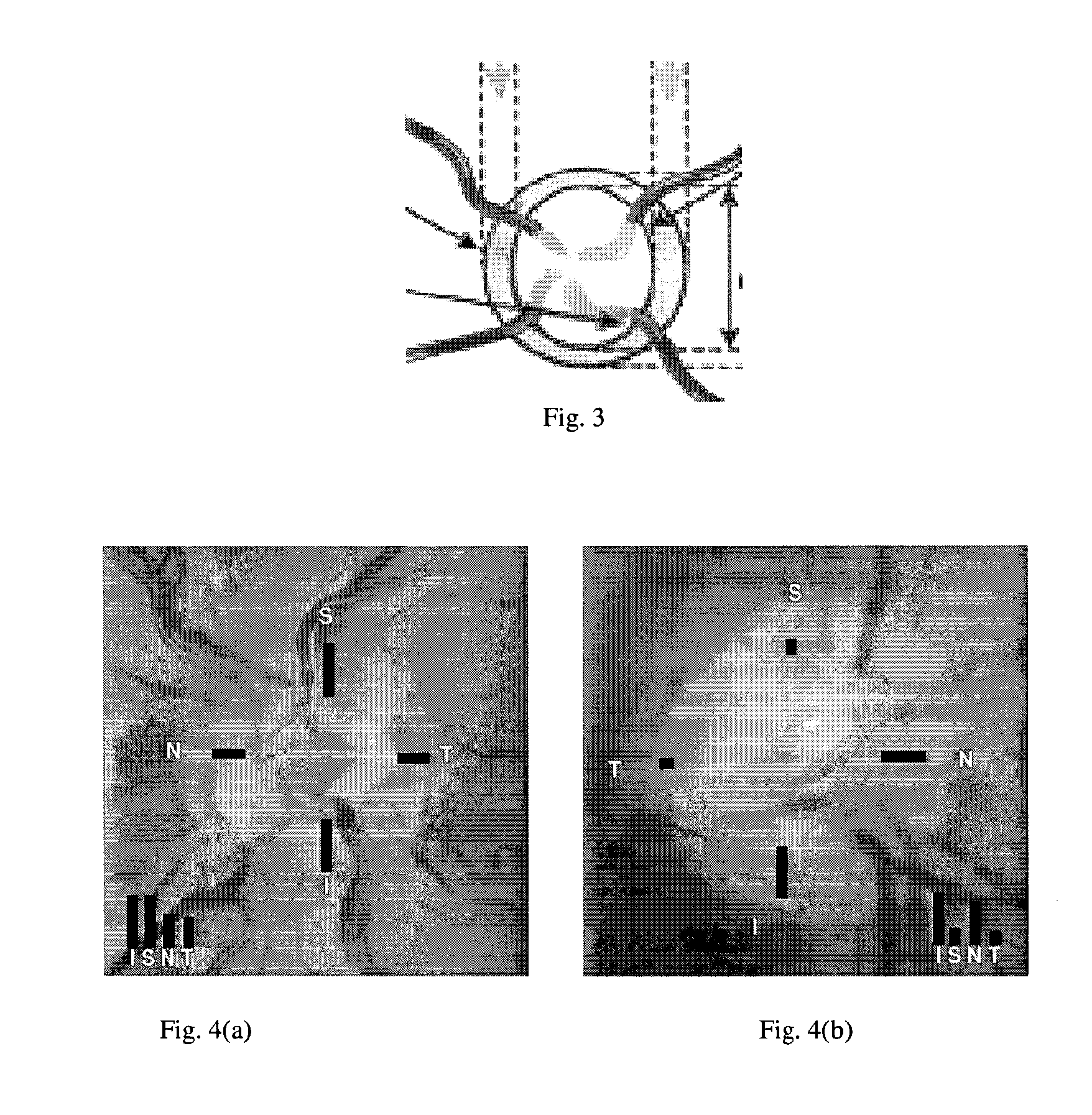
Obtaining data for automatic glaucoma screening, and screening and diagnostic techniques and systems using the data
A non-stereo fundus image is used to obtain a plurality of glaucoma indicators. Additionally, genome data for the subject is used to obtain genetic marker data relating to one or more genes and / or SNPs associated with glaucoma. The glaucoma indicators indicators and genetic marker data are input into an adaptive model operative to generate an output indicative of a risk of glaucoma in the subject. In combination, the genetic indicators and genome data are more informative about the risk of glaucoma than either of the two in isolation. The adaptive model may be a two-stage model, having a first stage in which individual genetic indicators are combined with respective portions of the genome data by first adaptive model modules to form respective first outputs, and a second stage in which the first outputs are combined by a second adaptive mode. Texture analysis is performed on the fundus images to classify them based on their quality, and only images which are determined to meet a quality criterion are subjected to an analysis to determine if they exhibit glaucoma indicators. Also, the images are put into a standard format. The system may include estimating the position of the optic cup by combining results from multiple optic cup segmentation techniques. The system may include estimating the position of the optic disc by applying edge detection to the funds image, excluding edge points that are unlikely to be optic disc boundary points, and estimating the position of an optic disc by fitting an ellipse to the remaining edge points.
Owner:SINGAPORE HEALTH SERVICES PTE +1
Bioinformatics systems, apparatuses, and methods executed on an integrated circuit processing platform
ActiveUS9792405B2High sensitivityImprove accuracyMemory architecture accessing/allocationMathematical modelsElectricityData source
A system, method and apparatus for executing a bioinformatics analysis on genetic sequence data includes an integrated circuit formed of a set of hardwired digital logic circuits that are interconnected by physical electrical interconnects. One of the physical electrical interconnects forms an input to the integrated circuit that may be connected with an electronic data source for receiving reads of genomic data. The hardwired digital logic circuits may be arranged as a set of processing engines, each processing engine being formed of a subset of the hardwired digital logic circuits to perform one or more steps in the bioinformatics analysis on the reads of genomic data. Each subset of the hardwired digital logic circuits may be formed in a wired configuration to perform the one or more steps in the bioinformatics analysis.
Owner:EDICO GENOME
Bioinformatics systems, apparatuses, and methods executed on a quantum processing platform
ActiveUS20180240032A1High sensitivityImprove accuracyQuantum computersDigital data information retrievalDevice formGenomic data
A system, method and apparatus for executing a bioinformatics analysis on genetic sequence data includes a quantum computing device formed of a set of hardwired quantum logic circuits interconnected by a plurality of superconducting connections to process information represented as a quantum state that is configured as a set of one or more qubits. The hardwired quantum logic circuits may be arranged as a set of processing engines, each processing engine being formed of a subset of the hardwired quantum logic circuits to perform one or more steps in the bioinformatics analysis on the reads of genomic data. Each subset of the hardwired quantum logic circuits may be formed in a wired configuration to perform the one or more steps in the bioinformatics analysis.
Owner:ILLUMINA INC
Segmented storage and retrieval of nucleotide sequence information
Processing of genomic data is facilitated by providing a storage device with a database having a segmented sequence table. The table has a plurality of data subsets of common nucleotide sequence size n, wherein≧2, and each data subset of common nucleotide sequence n is separately indexed within the table. A database manager associated with the database retrieves a selected nucleotide sequence locus from the table. The selected nucleotide sequence locus is sized differently from the common nucleotide sequence size n, and the retrieving includes identifying each data subset of the segmented sequence table containing at least a portion of the selected nucleotide sequence locus, and retrieving the identified data subsets. The database manager processes the retrieved, identified data subsets to remove genomic data mapped to the nucleotide positions outside the selected nucleotide sequence locus, and outputs the selected nucleotide sequence locus.
Owner:THE RES FOUND OF STATE UNIV OF NEW YORK
Method for processing genomic data
Owner:KONINKLIJKE PHILIPS ELECTRONICS NV
Peanut oligonucleotide probes and their design method and use method
ActiveCN106987590AImprove general performanceMicrobiological testing/measurementDNA/RNA fragmentationNucleotideMicrosatellite
The invention discloses peanut oligonucleotide probes and their design method and use method. The peanut oligonucleotide probes comprise eight probes having nucleotide sequences shown in the formulas of SEQ ID NO. 1 to NO. 8. The microsatellite and telomere sequences are used for developing the oligonucleotide probes, the novel oligonucleotide probes are combined into a probe set and oligonucleotide types of the peanut cultivar and wild peanut are constructed, an economic, efficient and high universality peanut cytological marker design technique and chromosome recognition technique are built, peanut chromosome markers are enriched, genomes and chromosomes of the peanut cultivar and wild peanut are identified, and chromosomal structural variation of wild peanut species is identified. Through use of a high-throughput small data simplified sequencing (for only measuring the amount of genomic 4Gb data) and bioinformatics analysis, peanut oligonucleotide probe markers are successfully developed. The invention provides a novel effective method for low-cost and high-efficiency development of peanut cytological markers.
Owner:HENAN ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com