Peanut oligonucleotide probes and their design method and use method
An oligonucleotide probe and oligonucleotide technology are applied in the field of peanut oligonucleotide probes and their design, which can solve the problems of difficult development and identification of peanut cytology markers, and achieve economical efficiency, versatility, The effect of good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Embodiment 1, a kind of design method of peanut oligonucleotide probe
[0061] The design method of peanut oligonucleotide probe of the present invention comprises following two modes:
[0062] (a) Development of oligonucleotide probes using microsatellite (SSR) and telomeric sequences
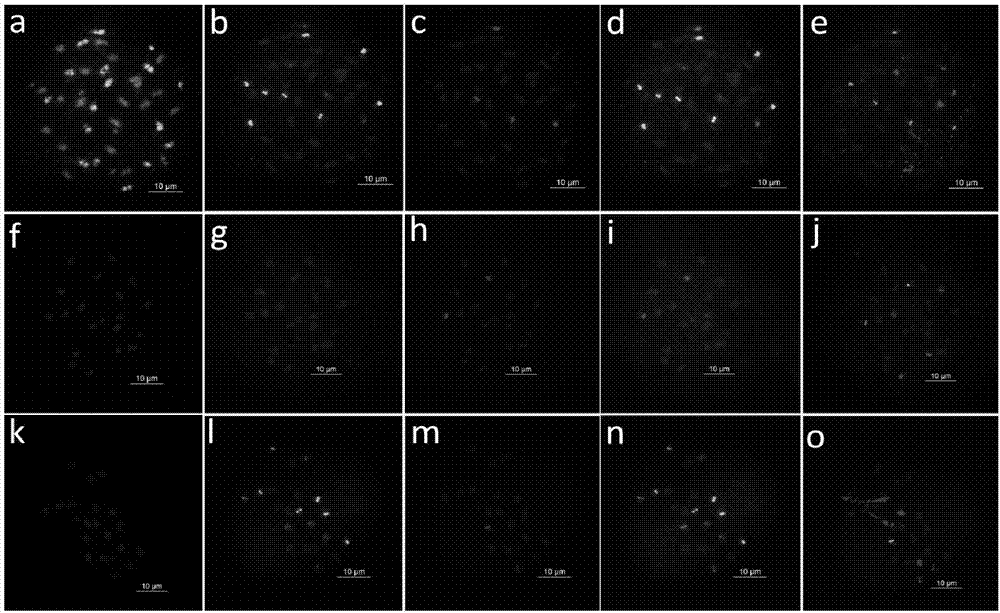
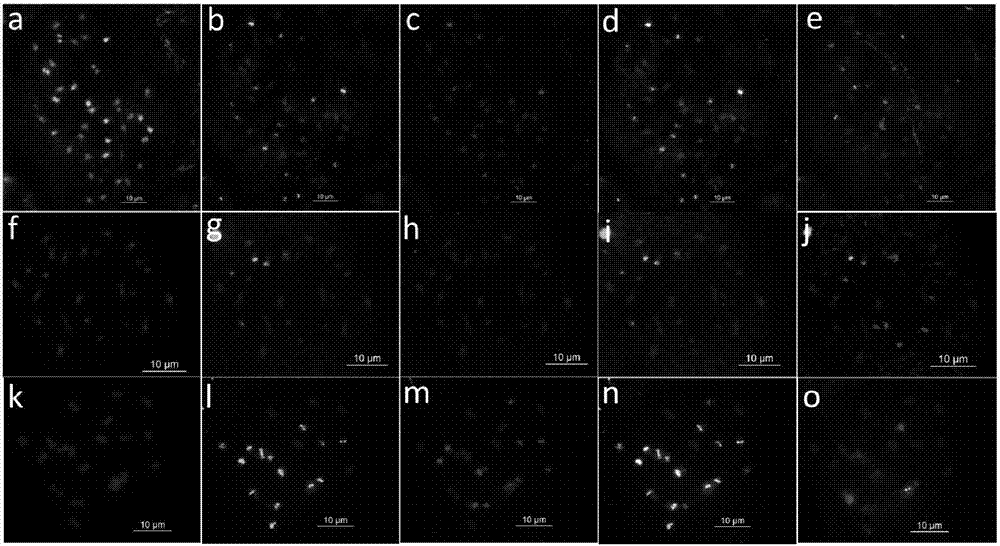
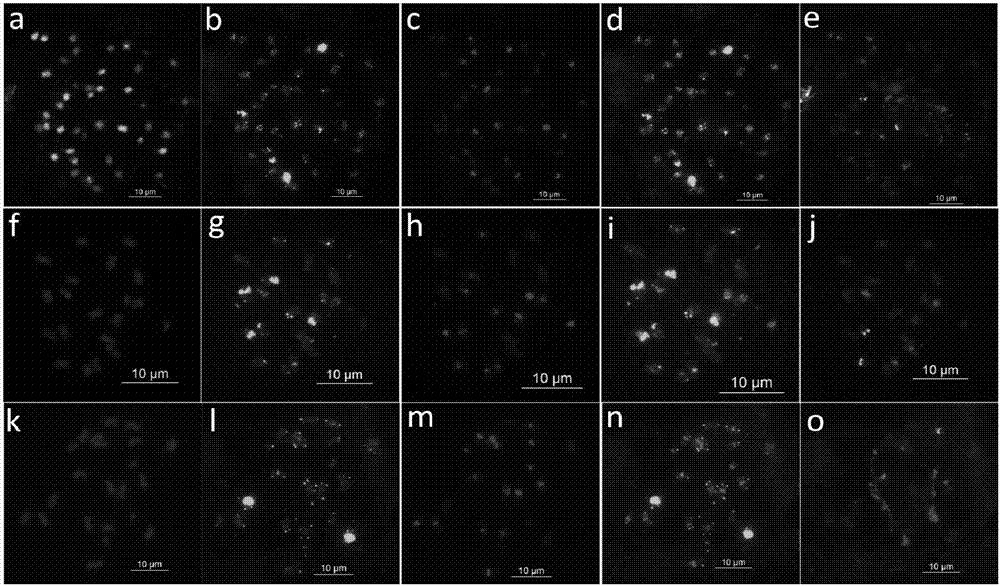
[0063] All ten forms of three-base SSR oligonucleotides (AAC)10, (AAG)10, (AGG)10, (ACT)10, (CAT)10, (CAC)10, (ACG)10, (CAG)10, (ATT)10 and (GCC)10, and then using fluorescence in situ hybridization to screen the root tip metaphase chromosomes of the peanut cultivar "Sizihong", it was found that the probes Oligo DP-2 and Oligo DP-4 were in the four There is a signal on the red chromosome, further use 45S rDNA, 5S rDNA, A.duranensis and Genomic DNA sequence FISH analysis of newly developed oligonucleotide probes OligoDP-2 and Oligo DP-4 in A. duranensis and A. duranensis location on the chromosome. OligoDP-2 and Oligo DP-4 were found in four grains of red, A.duranensis and Stable ...
Embodiment 2
[0076] Embodiment 2, the using method of peanut oligonucleotide probe
[0077] 1. Design of probe cover
[0078] The present invention utilizes Oligo DP-1~Oligo DP-7 to assemble two groups of probe sets, respectively Oligo#1 (FAM marked Oligo DP-2, Oligo DP-5 and Oligo DP-7) and Oligo#2 ( TAMARA-labeled Oligo DP-1, OligoDP-3, Oligo DP-4 and Oligo DP-6), the probe concentration is shown in Table 1.
[0079] 2. Test materials
[0080] The peanut cultivar Sizihong and 8 diploid wild species were preserved by the Economic Crops Research Institute of Henan Academy of Agricultural Sciences (Table 2).
[0081] Table 1: Peanut Oligonucleotide Probe Sets
[0082]
[0083] Table 2: Blocks and codes of Sizihong and 8 wild peanut species
[0084]
[0085] 3. Fluorescence in situ hybridization mixed staining analysis
[0086] The probe set was used to analyze the mixed staining of peanut cultivar Sizihong and 8 diploid wild species, including the following steps:
[0087] (1) P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com