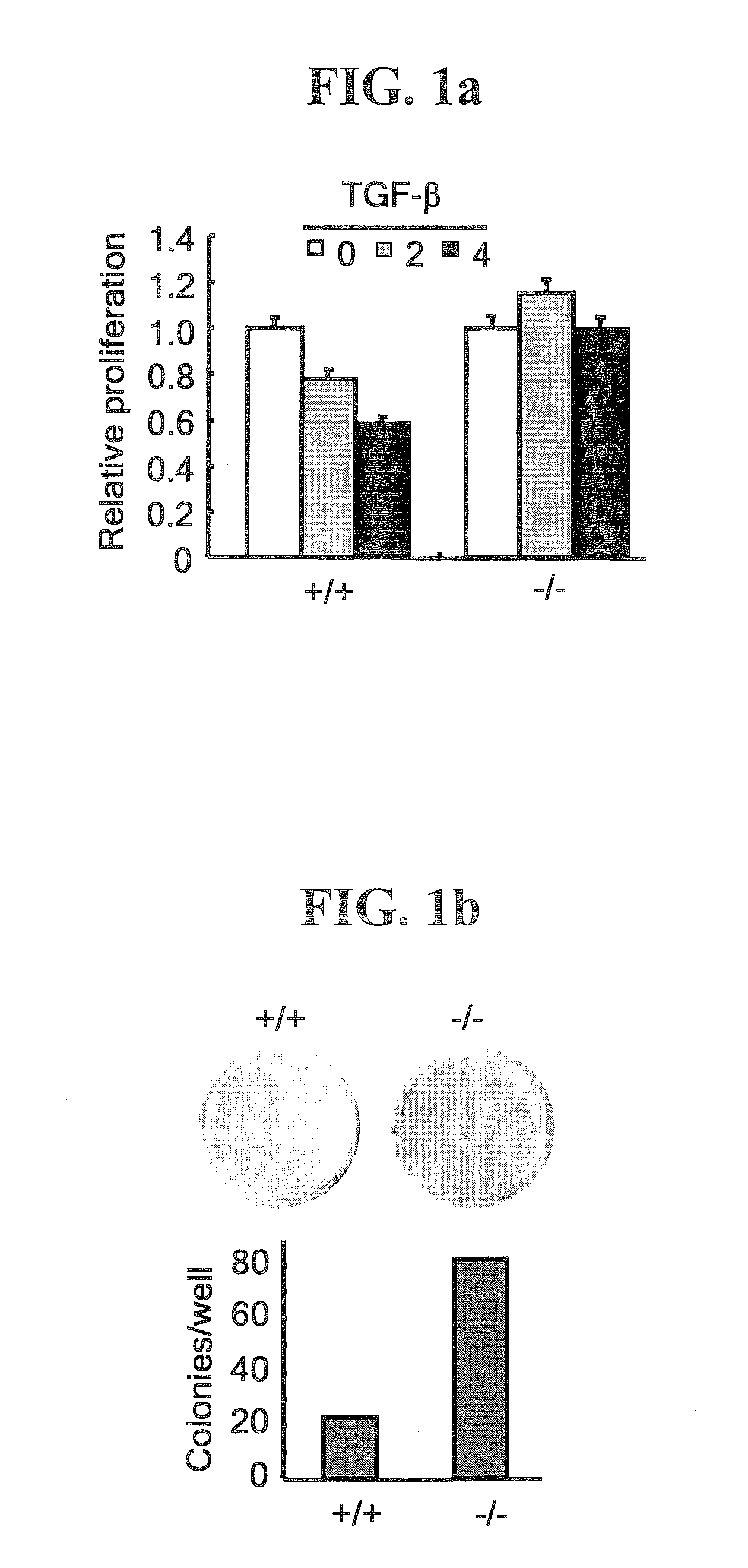
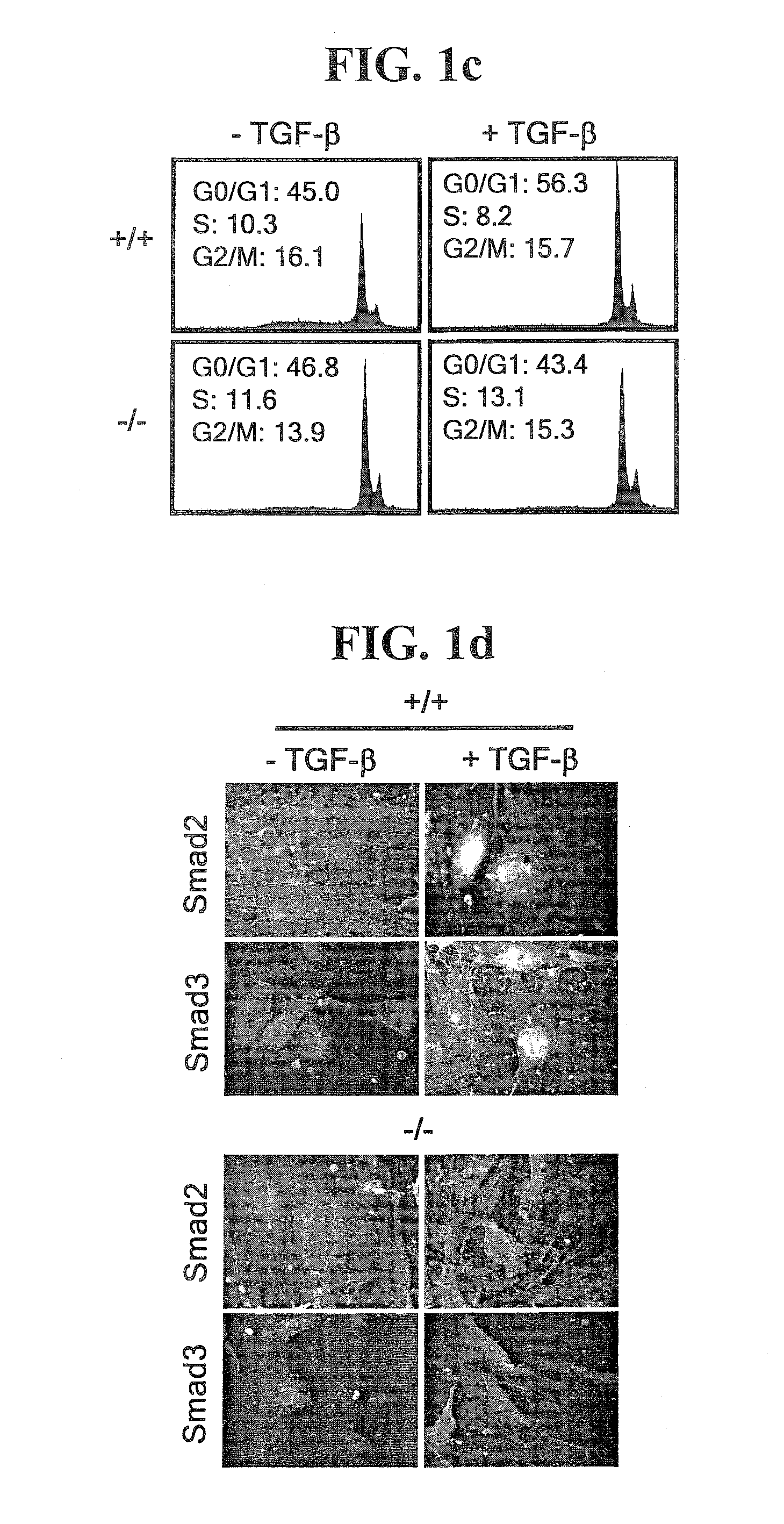
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
2025 results about "Exon" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
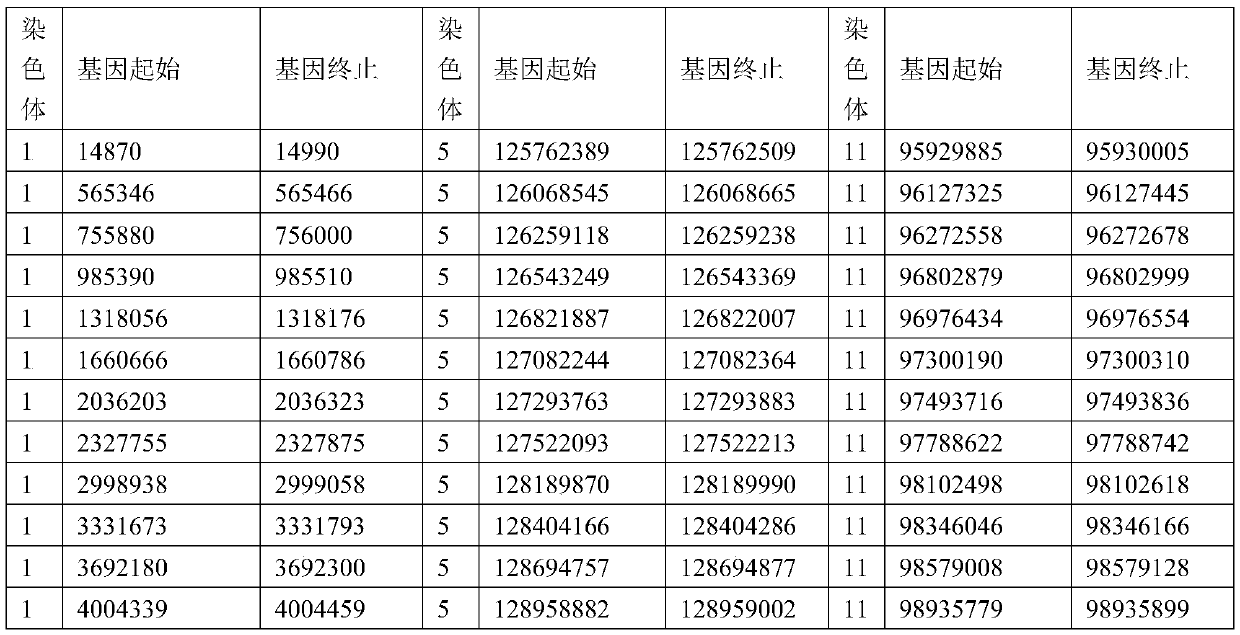
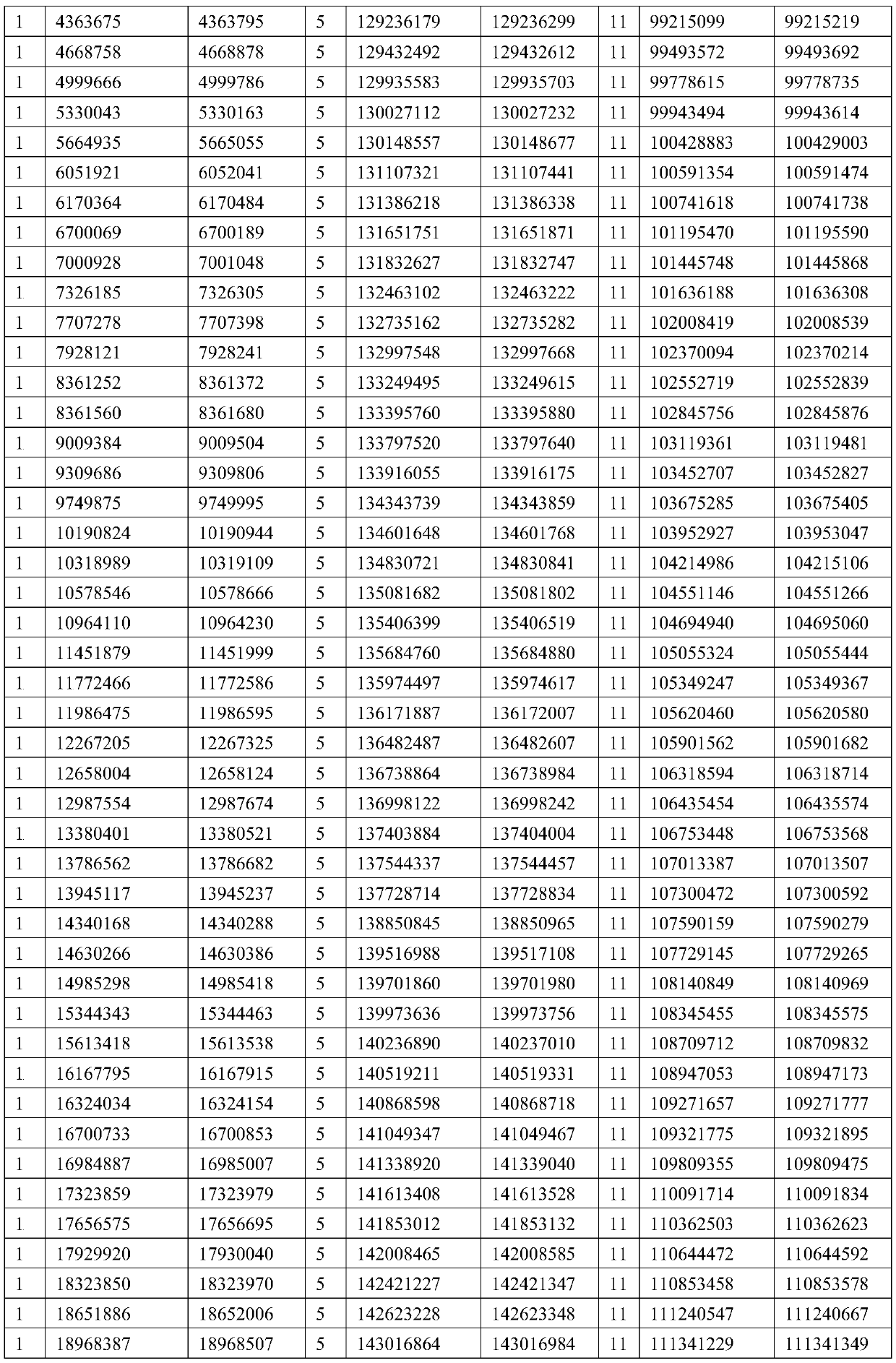
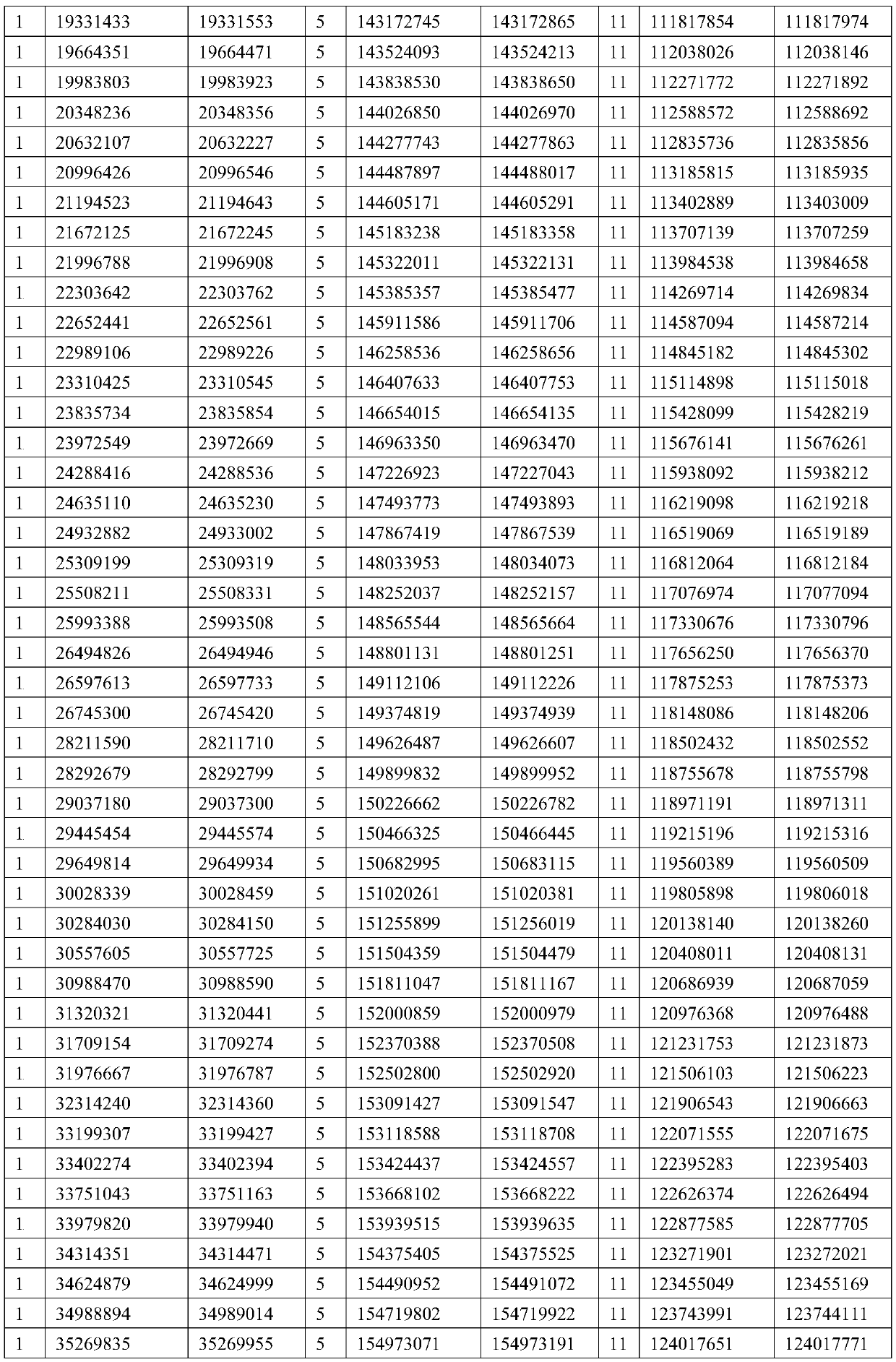
An exon is any part of a gene that will encode a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature messenger RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.
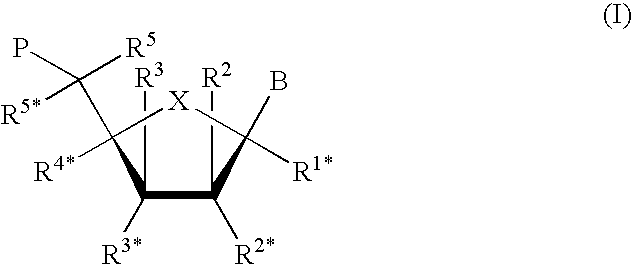
Full-length plant cDNA and uses thereof
Owner:NAT INST OF AGROBIOLOGICAL SCI +1
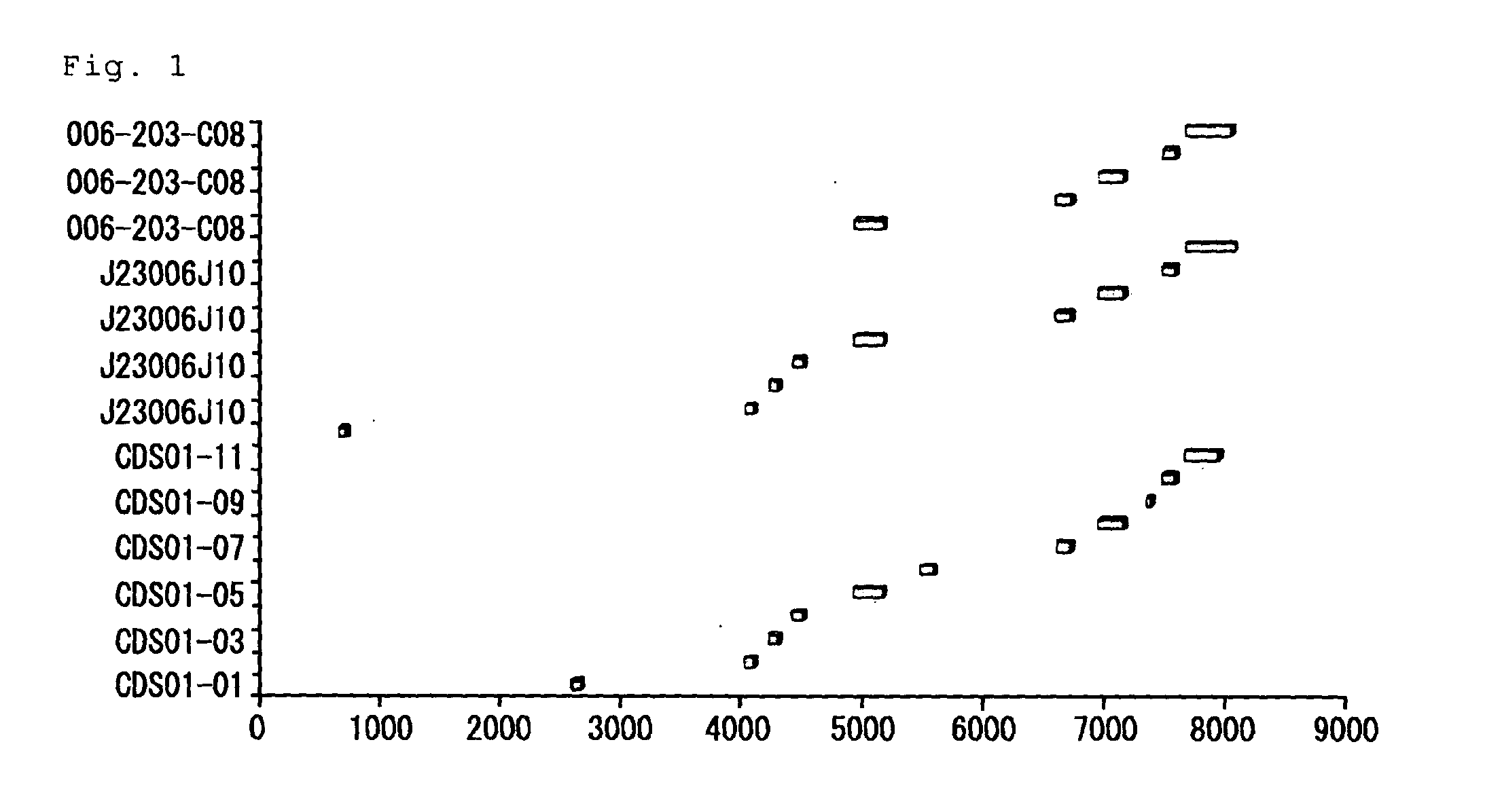
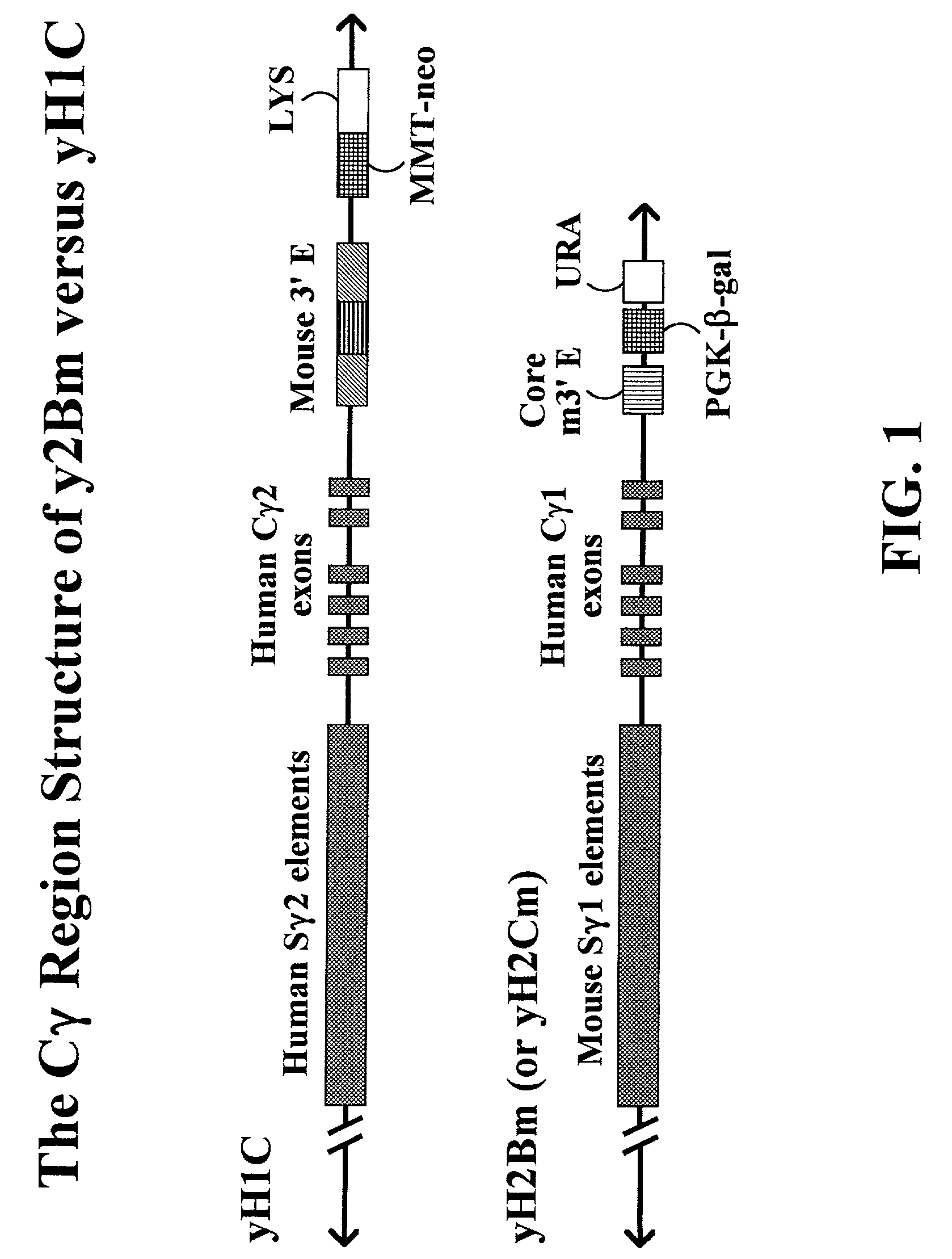
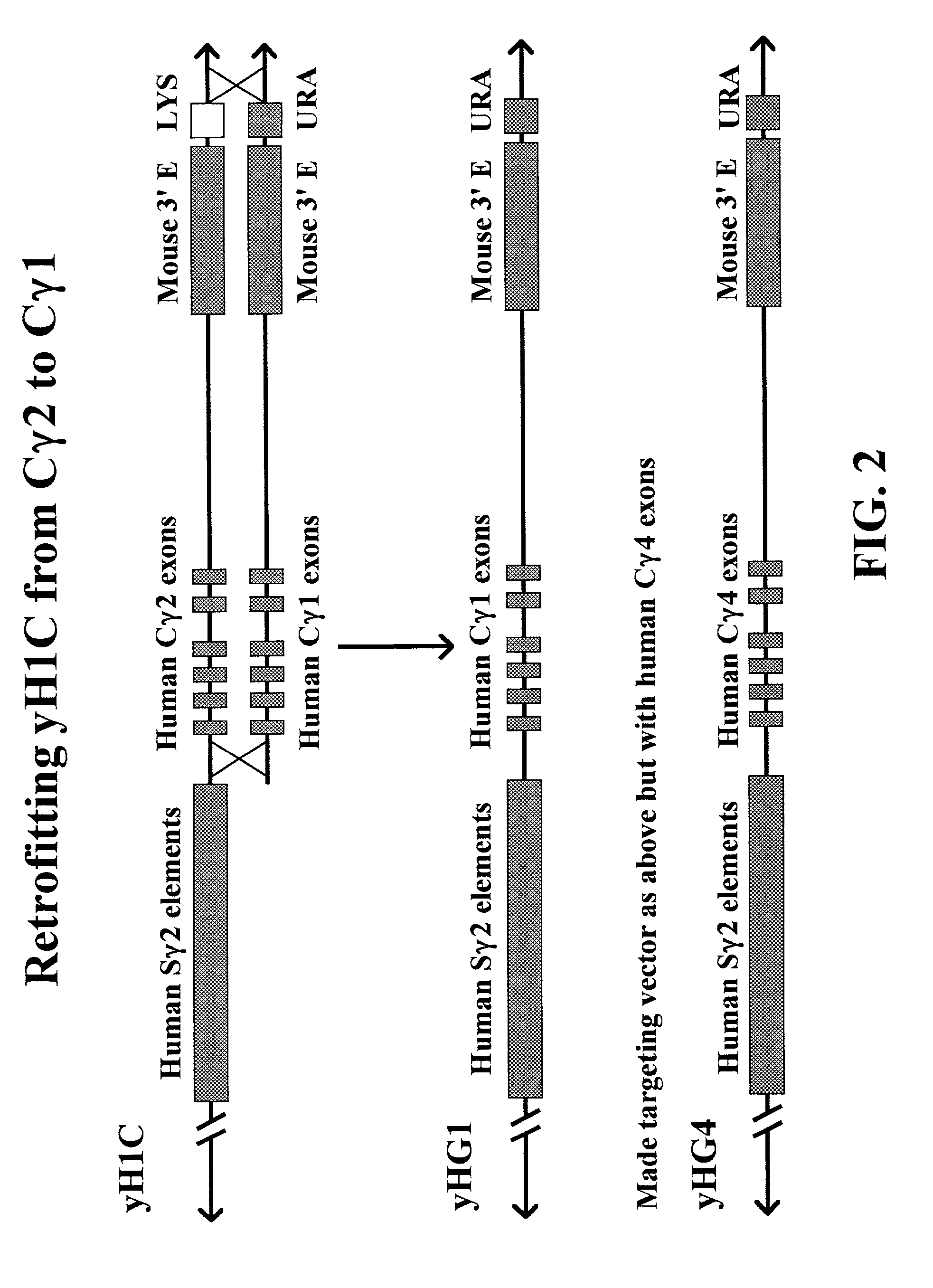
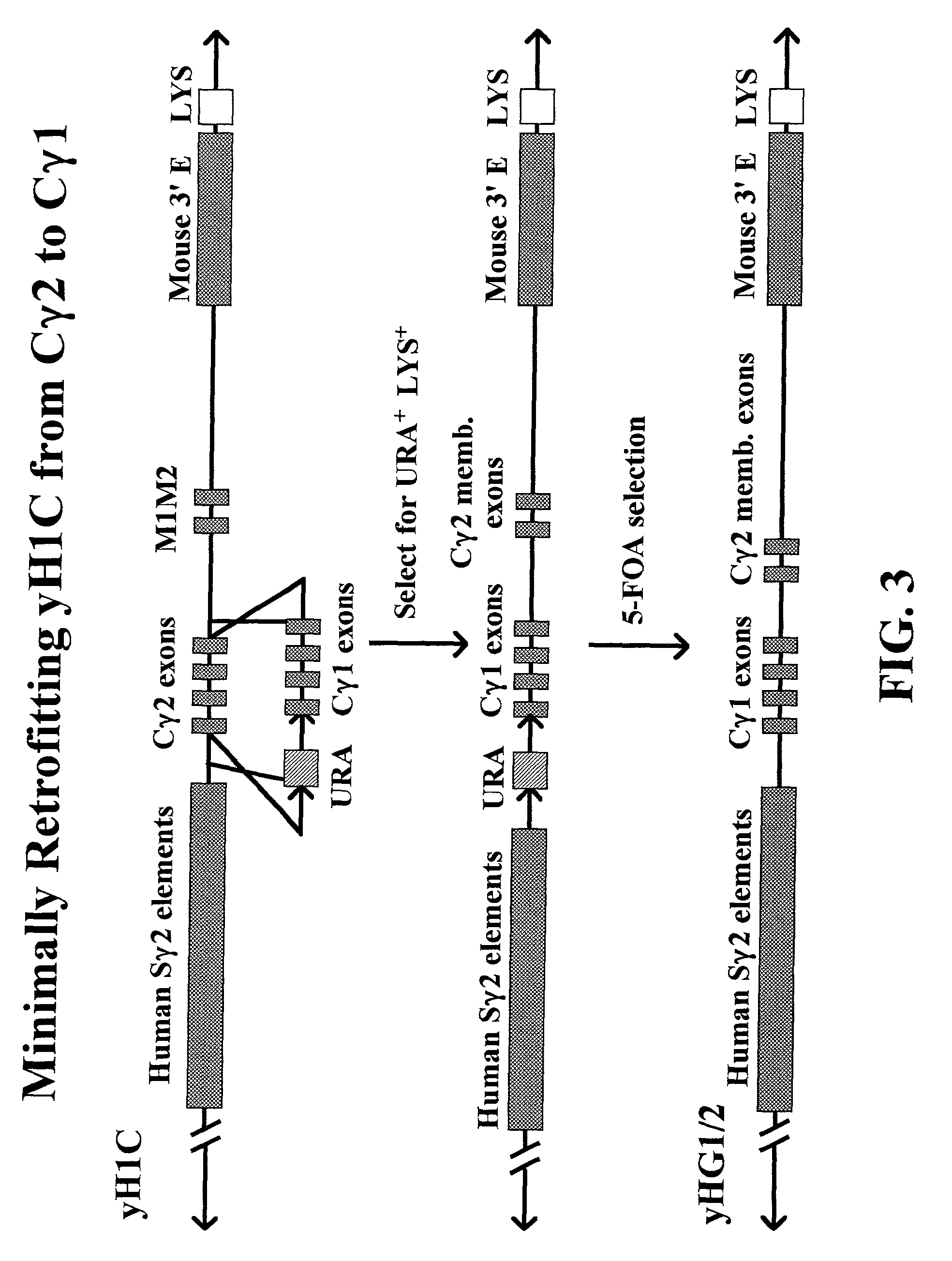
Transgenic animals for producing specific isotypes of human antibodies via non-cognate switch regions
InactiveUS7049426B2Immunoglobulins against cytokines/lymphokines/interferonsImmunoglobulins against cell receptors/antigens/surface-determinantsAntigenExon
The present invention provides fully human antibodies in a transgenic animal of a desired isotype in response to immunization with any virtually any desired antigen. The human immunoglobulin heavy chain transgene in the foregoing animals comprises a human constant region gene segment comprising exons encoding the desired heavy chain isotype, operably linked to switch segments from a constant region of a different heavy chain isotype, i.e., a non-cognate switch region. Said additional constant region segment comprises a switch region and human constant region coding segment, wherein the constant region coding segment is operably linked to a switch region that it is not normally associated with, i.e., a non-cognate switch region. In the transgenes of the invention, the non-cognate switch region may be a switch region from a different species than the constant region coding segment. The switch region and membrane exons of the invention may comprise a human gamma-2 constant region and the secreted constant region exons are from a human gamma-1 or a human gamma-4 constant region.
Owner:ABQENIX INC
Probe set and kit for detecting whole exons of extended genetic diseases and application of probe set
InactiveCN110499364AComprehensive diagnostic extended whole exome testingIncrease positive rateMicrobiological testing/measurementLibrary creationFresh TissueExon
The invention discloses a probe set for detecting whole exons of extended genetic diseases. The probe set for detecting the whole exons of the extended genetic diseases comprises a standard whole exonprobe set, a whole genome copy number variation probe, and a mitochondrial loop full-length probe, and the genetic diseases comprise 6161 genetic diseases; the standard whole exon probe set can detect the genetic diseases caused by whole exon mutation, the genetic diseases comprise nervous system diseases, metabolic system diseases, endocrine system diseases, digestive system diseases, skeletal system diseases, urinary system diseases, immune system diseases, cardiovascular system diseases, blood system diseases, integument system diseases, ophthalmic system diseases, ear system diseases, respiratory system diseases, and genital system diseases; and the density of the mitochondrial probe is 6X; test samples comprise blood, fresh tissue, FFPE samples, and saliva. The invention discloses using method and kit and application of the probe set for detecting the whole exons of the extended genetic diseases.
Owner:北京凯昂医学诊断技术有限公司
Novel methods for quantification of microRNAs and small interfering RNAs
ActiveUS20050272075A1Highly sensitive and specific hybridizationHighly sensitive and specific and ligationSugar derivativesMicrobiological testing/measurementMicroRNAAllele
The invention relates to ribonucleic acids, probes and methods for detection, quantification as well as monitoring the expression of mature microRNAs and small interfering RNAs (siRNAs). The invention furthermore relates to methods for monitoring the expression of other non-coding RNAs, mRNA splice variants, as well as detecting and quantifying RNA editing, allelic variants of single transcripts, mutations, deletions, or duplications of particular exons in transcripts, e.g., alterations associated with human disease such as cancer. The invention furthermore relates to methods for detection, quantification as well as monitoring the expression of deoxy nucleic acids.
Owner:QIAGEN GMBH
Methods and compositions of molecular profiling for disease diagnostics
The present invention relates to compositions, kits, and methods for molecular profiling and cancer diagnostics, including but not limited to gene expression product markers, alternative exon usage markers, and DNA polymorphisms associated with cancer. In particular, the present invention provides molecular profiles associated with thyroid cancer, methods of determining molecular profiles, and methods of analyzing results to provide a diagnosis.
Owner:VERACYTE INC
Novel oligonucleotide compositions and probe sequences useful for detection and analysis of microRNAS and their target mRNAS
InactiveUS20070065840A1Strong specificityHigh sensitivitySugar derivativesMicrobiological testing/measurementTarget mrnaAllele
The invention relates to ribonucleic acids and oligonucleotide probes useful for detection and analysis of microRNAs and their target mRNAs, as well as small interfering RNAs (siRNAs). The invention furthermore relates to oligonucleotide probes for detection and analysis of other non-coding RNAs, mRNAs, mRNA splice variants, allelic variants of single transcripts, mutations, deletions, or duplications of particular exons in transcripts, e.g. alterations associated with human disease, such as cancer.
Owner:EXIQON AS
Breeding method for prolongation of rice fertility stage
ActiveCN104004782AVector-based foreign material introductionAngiosperms/flowering plantsSelf-healingBiotechnology
The invention relates to a breeding method for prolongation of rice fertility stage, and the breeding method comprises the following steps: selecting a target fragment in a rice fertility stage determining gene Ehd3 exon region and constructing a plant CRISPR / Cas9 targeting recombinant vector, introducing into rice cells for regeneration to sprout; using an expression frame in the vector to realize shearing of Ehd3 exon region DNA in the rice cells so as to cause self healing of the rice cells; sequencing regeneration strain genome target fragments to obtain an afunction-mutational strain carrying two equipotential Ehd3 genes, and confirming the prolongation of the regeneration plant fertility stage by phenotypic identification. Experiments show that the method can quickly obtain fertility stage prolonged rice materials.
Owner:RICE RES ISTITUTE ANHUI ACAD OF AGRI SCI
Methods for treating spinal muscular atrophy
InactiveUS20110086833A1Increase inclusivenessImprove the level ofBiocideNervous disorderExonNucleic acid
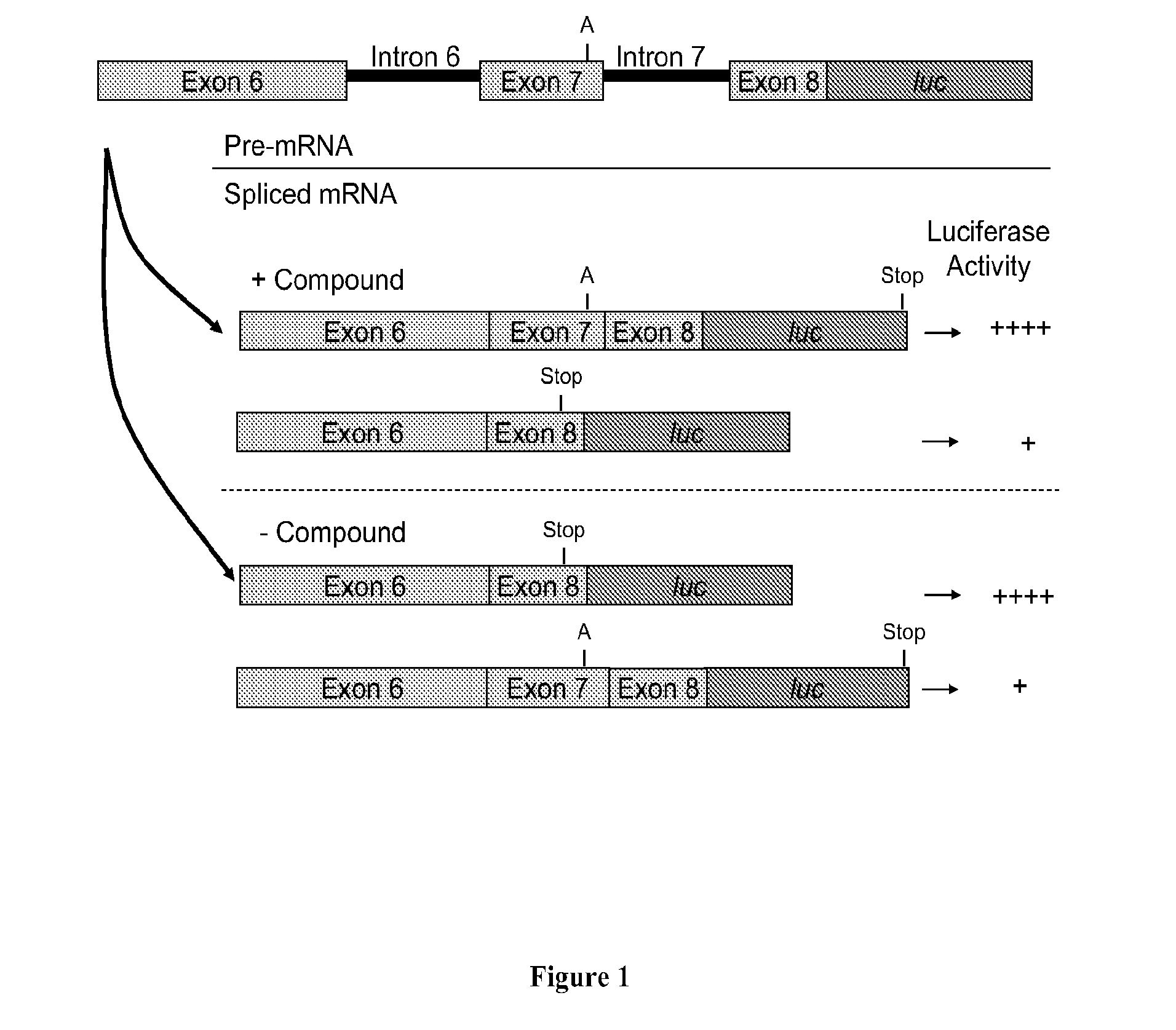
The present invention provides nucleic acid constructs, methods for identifying and validating compounds that increase the inclusion of exon 7 of SMN2 into mRNA transcribed from the SMN2 gene, compounds and pharmaceutical compositions that increase levels of SMN protein produced from the SMN2 gene, and methods for use thereof in treating of SMA.
Owner:PTC THERAPEUTICS INC
Method of cloning reproductive and respiratory syndrome resisting pig
Owner:CHINA AGRI UNIV
Method for knocking off animal FGF5 gene by using CRISPR-Cas9 system
ActiveCN104531704AThe identification rules are simpleEasy to operateMicroinjection basedVector-based foreign material introductionA-DNAExon
The invention provides a method for knocking off an animal FGF5 gene by using a CRISPR-Cas9 system. The method comprises the following steps: firstly, acquiring a DNA sequence aiming at an sgRNA recognition area of a second FGF5 exon, wherein the base sequence of the DNA sequence is as shown in SEQ ID NO.1; secondly, establishing an sgRNA expression structure of the second FGF5 exon, inserting a T7 starter before an sgRNA transcriptional start site, establishing an in-vitro transcription carrier of Cas9 protein, and regulating and controlling by using the T7 starter. Cas9 mRNA and sgRNA are obtained through the in-vitro transcription carrier of Cas9 and sgRNA, and the method can be used for knocking off the animal FGF5 gene.
Owner:CHINA AGRI UNIV
RUNX3 gene showing anti-tumor activity and use thereof
InactiveUS7368255B2Reduce selection requirementsPeptide/protein ingredientsGenetic material ingredientsAbnormal tissue growthCancers diagnosis
The present invention relates to a RUNX3 gene showing anti-tumor activity which is essentially involved in TGF-β dependent-programmed cell death (apoptosis) and use thereof. In addition, the present invention finds that the RUNX3 gene expression is suppressed in the various gastric cancer and lung cancer cell lines. The suppression of the RUNX3 gene expression is due to hyper-methylation of CpG island located around RUNX3 exon (1). The RUNX3 gene and its gene product of the present invention can be used effectively for the development of anti-cancer agents. CpG island around RUNX3 exon (1) could also be used not only for the development of anti-cancer agents which regulate the abnormal DNA methylation and there by induce RUNX3 expression but also for the development of methods for cancer diagnosis by measuring the abnormal DNA methylation.
Owner:NAT UNIV OF SINGAPORE
Methods for assessing disease risk
InactiveUS20120220478A1High copy numberNucleotide librariesMicrobiological testing/measurementDisease riskAutoimmune disease
The invention relates to methods and biomarkers for assessing a subject's risk for a disease, such as cancer, an autoimmune disease or a neurological disease. In particular, the invention provides methods and biomarkers for creating exon copy number variation (ECNV) profiles, and determining disease risk according to the subject's ECNV profiles.
Owner:BAR HARBOR BIOTECH
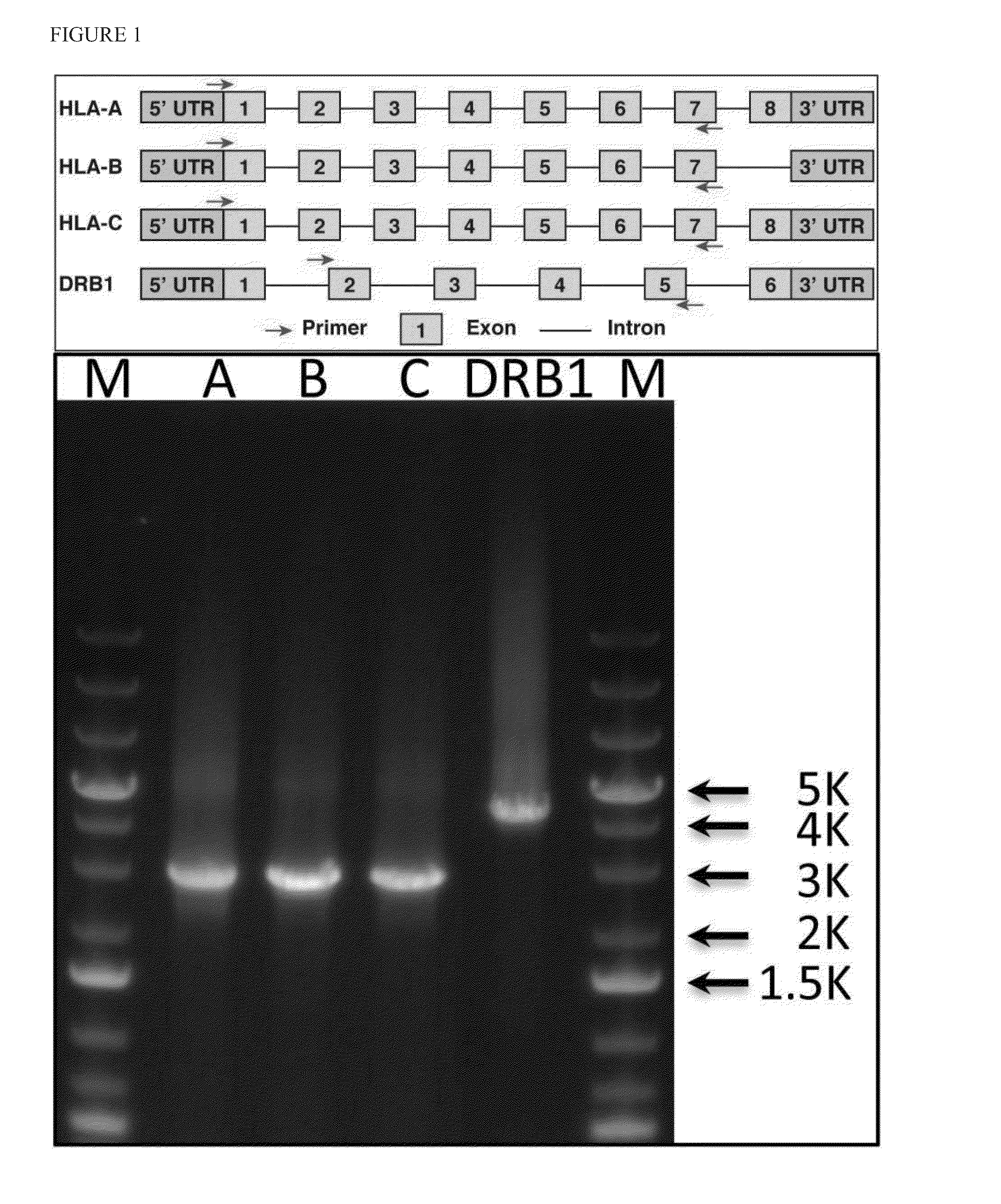
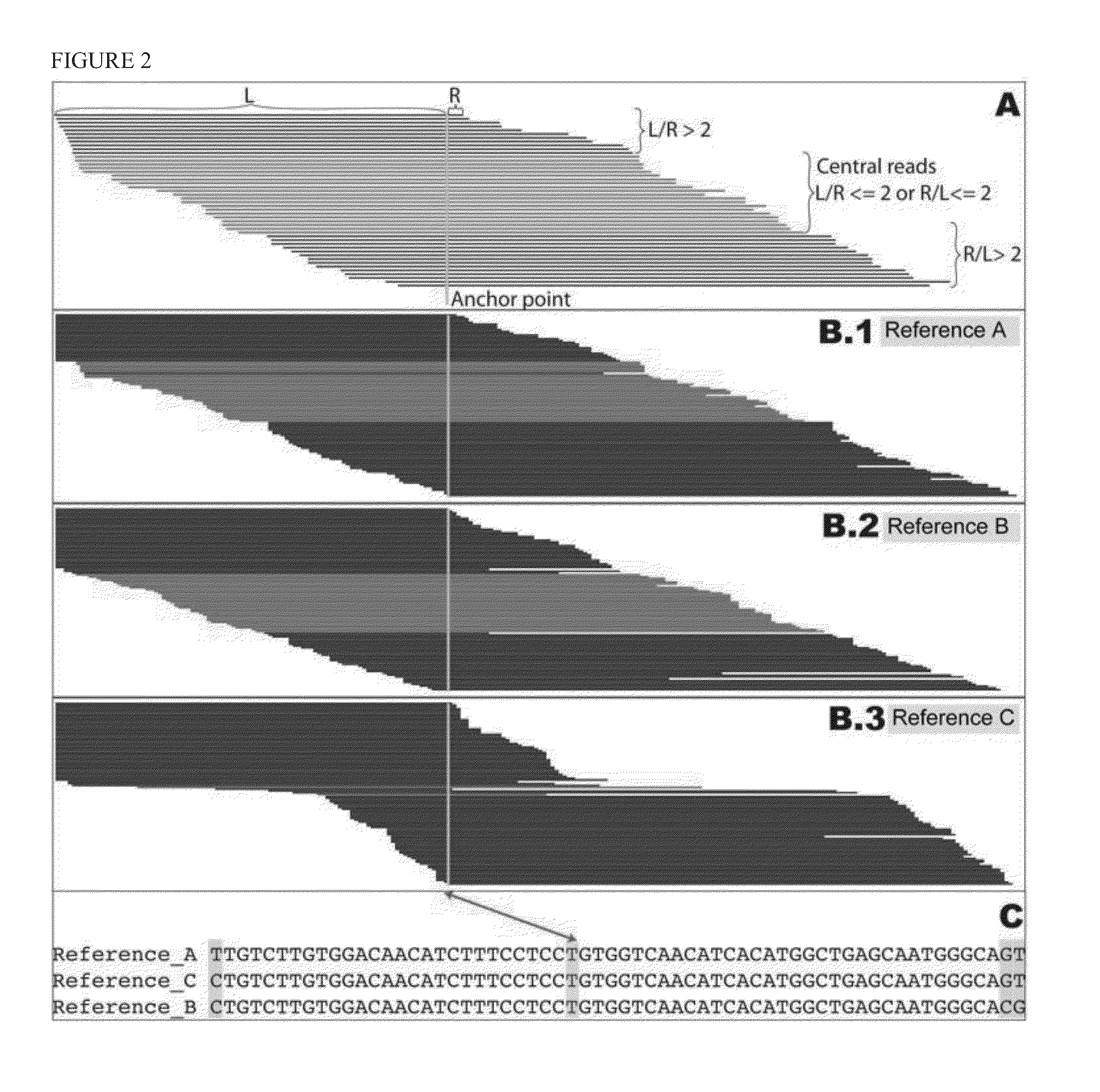
Haplotying of HLA loci with ultra-deep shotgun sequencing
ActiveUS20140206547A1Minimize biasEfficient reconstructionMicrobiological testing/measurementLibrary member identificationHaploidisationHaplotype
Methods are provided to determine the entire genomic region of a particular HLA locus including both intron and exons. The resultant consensus sequences provides linkage information between different exons, and produces the unique sequence from each of the two genes from the individual sample being typed. The sequence information in intron regions along with the exon sequences provides an accurate HLA haplotype.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Method for knocking off animal myostatin gene by using CRISPR-Cas9 system
InactiveCN104531705AThe identification rules are simpleEasy to operateMicroinjection basedVector-based foreign material introductionBiotechnologyTranscription initiation site
The invention provides a method for knocking off an animal myostatin gene by using a CRISPR-Cas9 system. The method comprises the following steps: firstly, acquiring a DNA sequence aiming at an sgRNA recognition area of a second myostatin exon, wherein the base sequence of the DNA sequence is as shown in SEQ ID NO.1; secondly, establishing an sgRNA expression structure of the second myostatin exon, inserting a T7 starter before an sgRNA transcriptional start site, establishing an in-vitro transcription carrier of Cas9 protein, and regulating and controlling by using the T7 starter. Cas9 mRNA and sgRNA are obtained through the in-vitro transcription carrier of Cas9 and sgRNA, and the method can be used for knocking off the animal myostatin gene.
Owner:CHINA AGRI UNIV
Soluble TNF receptors and their use in treatment of disease
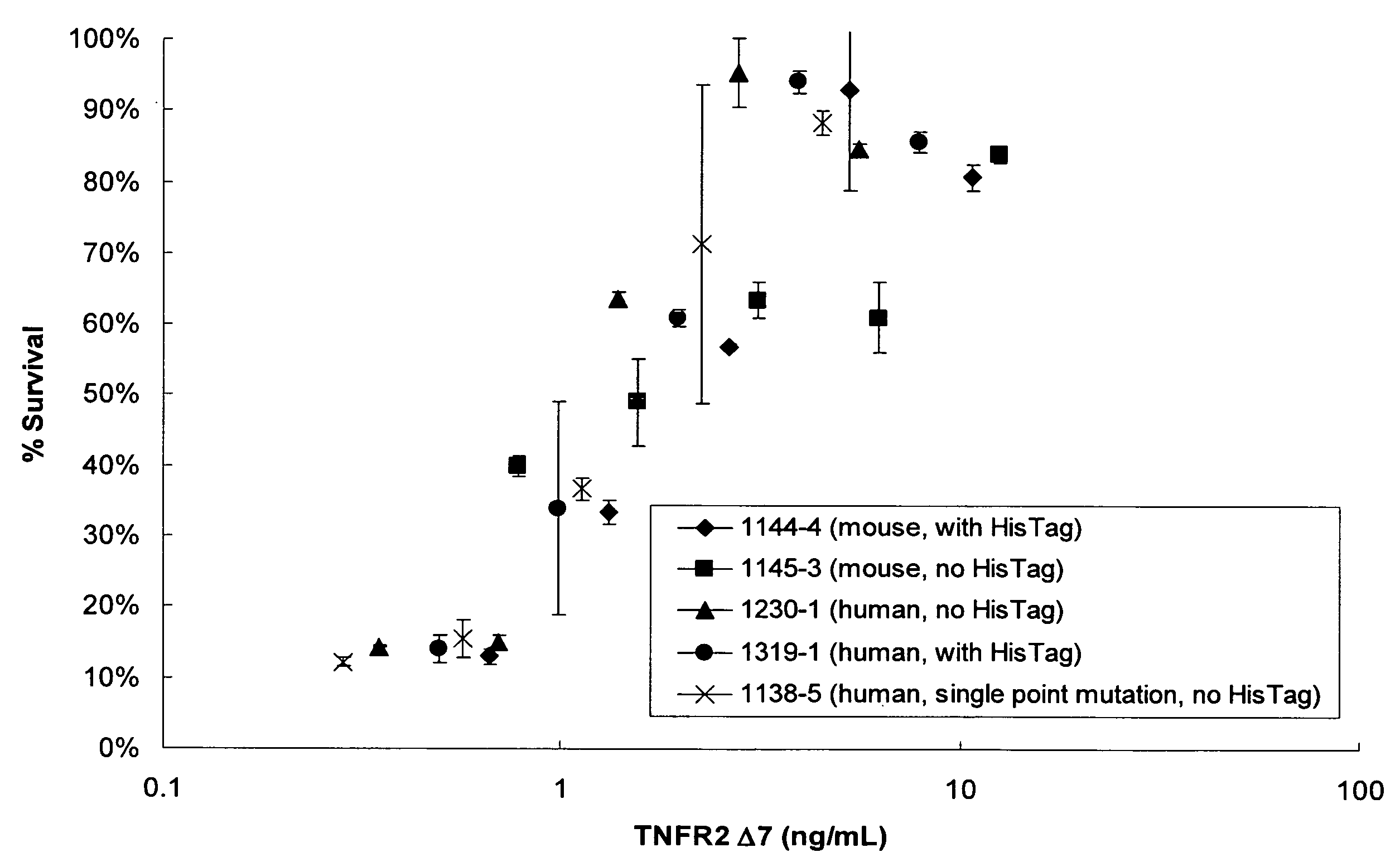
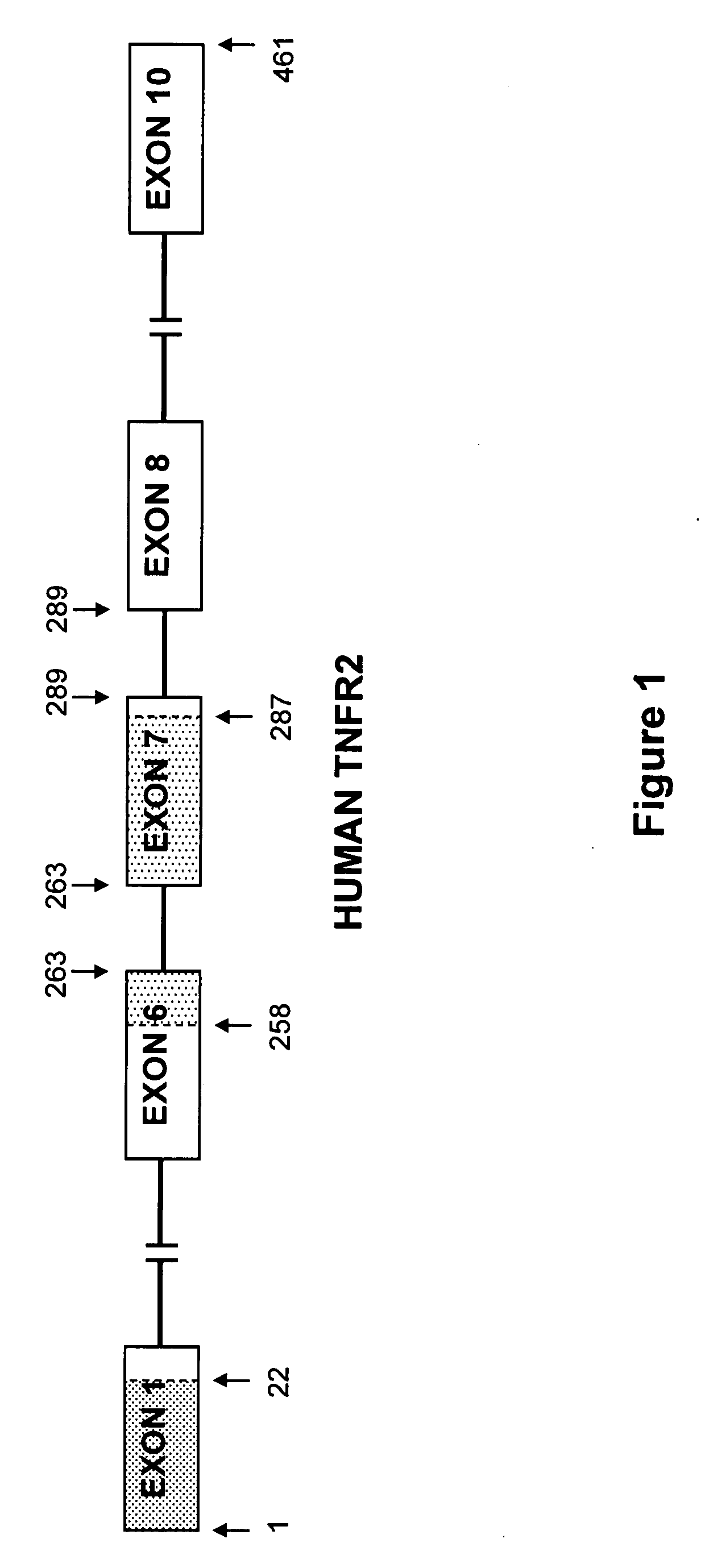
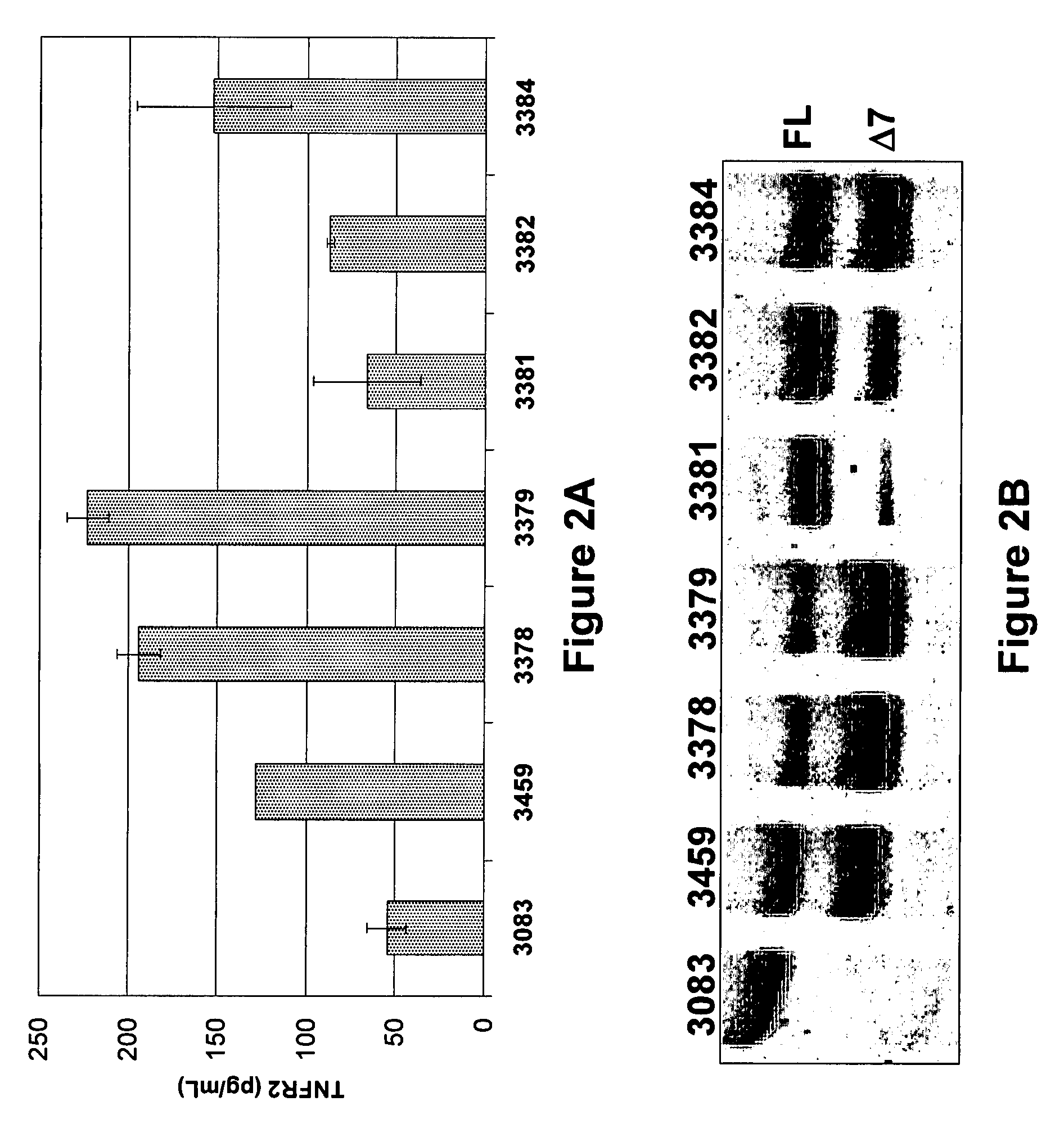
The present invention relates to tumor necrosis factor (TNF) antagonists and corresponding nucleic acids derived from tumor necrosis factor receptors (TNFRs) and their use in the treatment of inflammatory diseases. These proteins are soluble secreted decoy receptors that bind to TNF and prevent TNF from signaling to cells. In particular, the proteins are mammalian TNFRs that lack exon 7 and which can bind TNF and can act as a TNF antagonist.
Owner:NORTH CAROLINA AT CHAPEL HILL THE UNIV OF +2
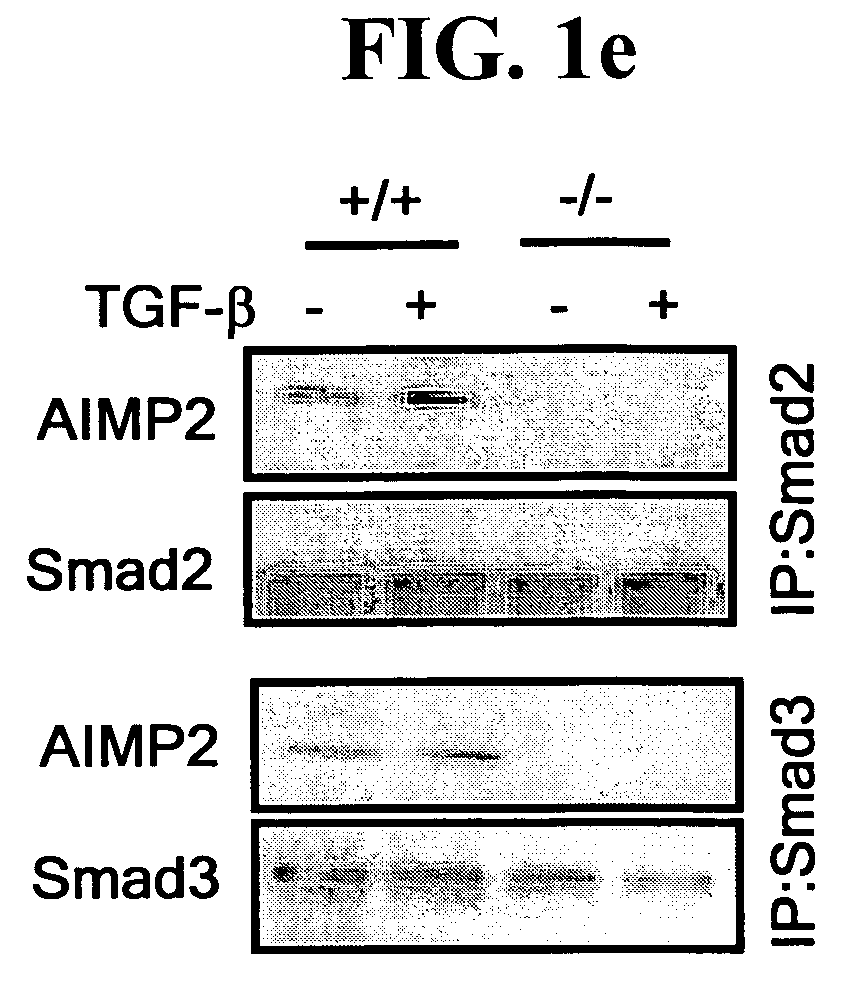
Antibody Specific to the AIMP2-DX2
InactiveUS20110117572A1Effective treatmentCompound screeningApoptosis detectionCancer cellCancers diagnosis
Owner:MEDICINAL BIOCONVERGENCE RES CENT
KIAA1217-RET fusion gene
ActiveCN105255927AActivate kinase domain activityHigh expressionHybrid peptidesDNA/RNA fragmentationRet geneNucleotide
The invention discloses a KIAA1217-RET fusion gene. The KIAA1217-RET fusion gene is formed by exons from number one to number eleven of a KIAA1217 gene and exons from number eight to number nineteen of an RET gene in a fusion mode, and the nucleotide sequence is shown in figure 1. Disclose of the fusion gene is favorable for further study of drive genetic modification not found in thyroid cancer, and the clinical pathological classification and treatment strategies of patients suffering from papillary thyroid carcinoma are optimized.
Owner:THE FIRST AFFILIATED HOSPITAL OF WENZHOU MEDICAL UNIV +3
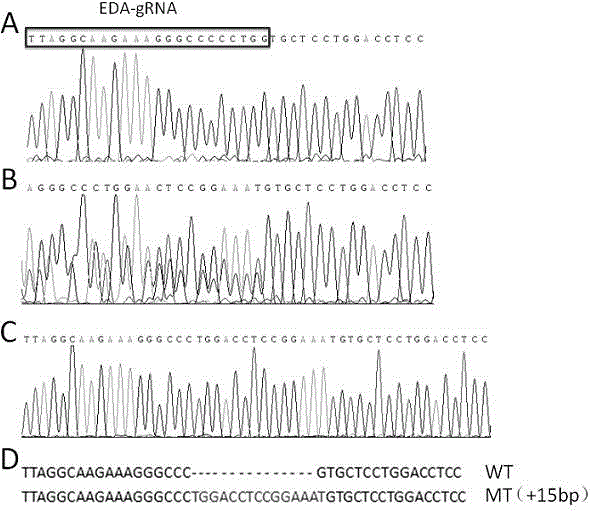
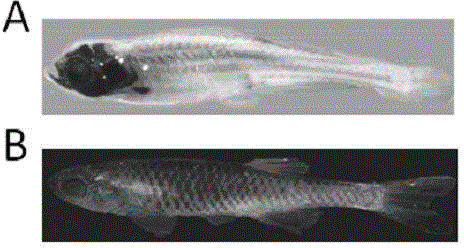
Crispr/Cas9-induced scale-missing zebra fish mode and establishment method
The invention relates to a Crispr / Cas9-induced scale-missing zebra fish mode. The mode is scale-missing zebra fish containing EDA gene exon 4-locus base insertion. Meanwhile, the invention also discloses an establishment method and the application of the Crispr / Cas9-induced scale-missing zebra fish mode. The established scale-missing zebra fish mode has a great application value in functional research of related genes of appendages of the skin, screening of medicines for treating ectoderm dysplasia such as human baldness, and the like.
Owner:CHINA ACAD OF SCI NORTHWEST HIGHLAND BIOLOGY INST
Method for cultivating low-cadmium-accumulation indica rice variety
ActiveCN106544357AAvoid safety hazardsPrecise and controllable mutation sitesNucleic acid vectorPlant peptidesTransgenesisMutant
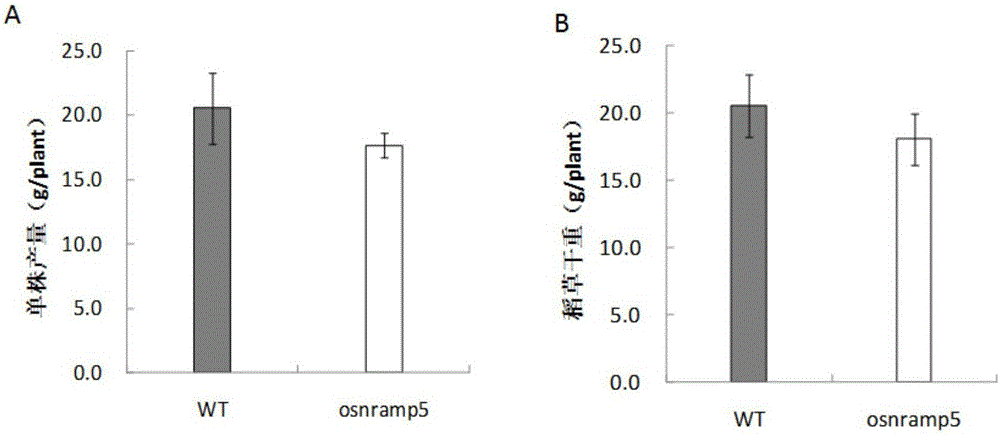
The invention discloses a method for cultivating a low-cadmium-accumulation indica rice variety. The method includes the steps of cloning part of the CDS sequence of OsNramp5 of an indica rice acceptor material; using a CRISPR / Cas9 system to select a target sequence according to an exon sequence; building pCRISPR / Cas9 recombinant vectors; leading the pCRISPR / Cas9 recombinant vectors into rice calluses to obtain transgene seedlings; screening transgene positive plants; obtaining mutant plants; and conducting seed propagation of the mutant plants, and separating afunction mutants containing no transgenic ingredients from plant progenies. By means of the method, the cadmium absorption major gene OsNramp5 of indica rice is directionally knocked out through the CRISPR / Cas9 technology, and the indica rice material containing no transgenic ingredients, having no significantly variant comprehensive agronomic characters and achieving a low cadmium content in rice is directionally cultivated. The method has the advantages that targeting is efficient, the breeding period is short, cost is low and practicality is high.
Owner:HUNAN HYBRID RICE RES CENT
Method for acquiring aromatic rice strain by targeting Badh2 gene via CRISPR/Cas9 gene editing technology
InactiveCN105505979AScalableEasy to operateVector-based foreign material introductionAngiosperms/flowering plantsAromatic riceGenomic DNA
The invention discloses a method for acquiring an aromatic rice strain by targeting a Badh2 gene via CRISPR / Cas9 gene editing technology. The method comprises the following steps: separately targeting sequences, recognizable by Cas9, of every exon and intron of an aromatic gene by using the CRISPR / Cas9 gene editing technology and then cutting a genomic DNA sequence to initiate DNA restoration so as to obtain an afunctional Badh2 gene; with the callus of Indica rice, japonica rice or glutinous rice as a receptor material of genetic transformation and an mature embryo, young ear, ovary or the like as explant, carrying out induction so as to obtain a diploid callus, introducing a targeting vector into cells of the callus by using an Agrobacterium mediated transformation method, screening and identifying positive plants and separating the plants from a T1 colony so as to obtain an aromatic rice strain; and with anther, pollen, unfertilized ovary or the like as explant, carrying out induction so as to obtain a haploid callus, introducing the targeting vector into cells of the callus by using the Agrobacterium mediated transformation method, screening a positive callus, reduplicating the positive callus by using colchicine, carrying out differentiation to form seedlings and identifying genetic transformation positive plants so as to eventually obtain the aromatic rice strain.
Owner:HUBEI UNIV
Method for targetedly integrating exogenous gene into target gene
InactiveCN104342457AOvercoming Random Integration ProblemsLess expensiveMicroinjection basedVector-based foreign material introductionMammalPosition effect
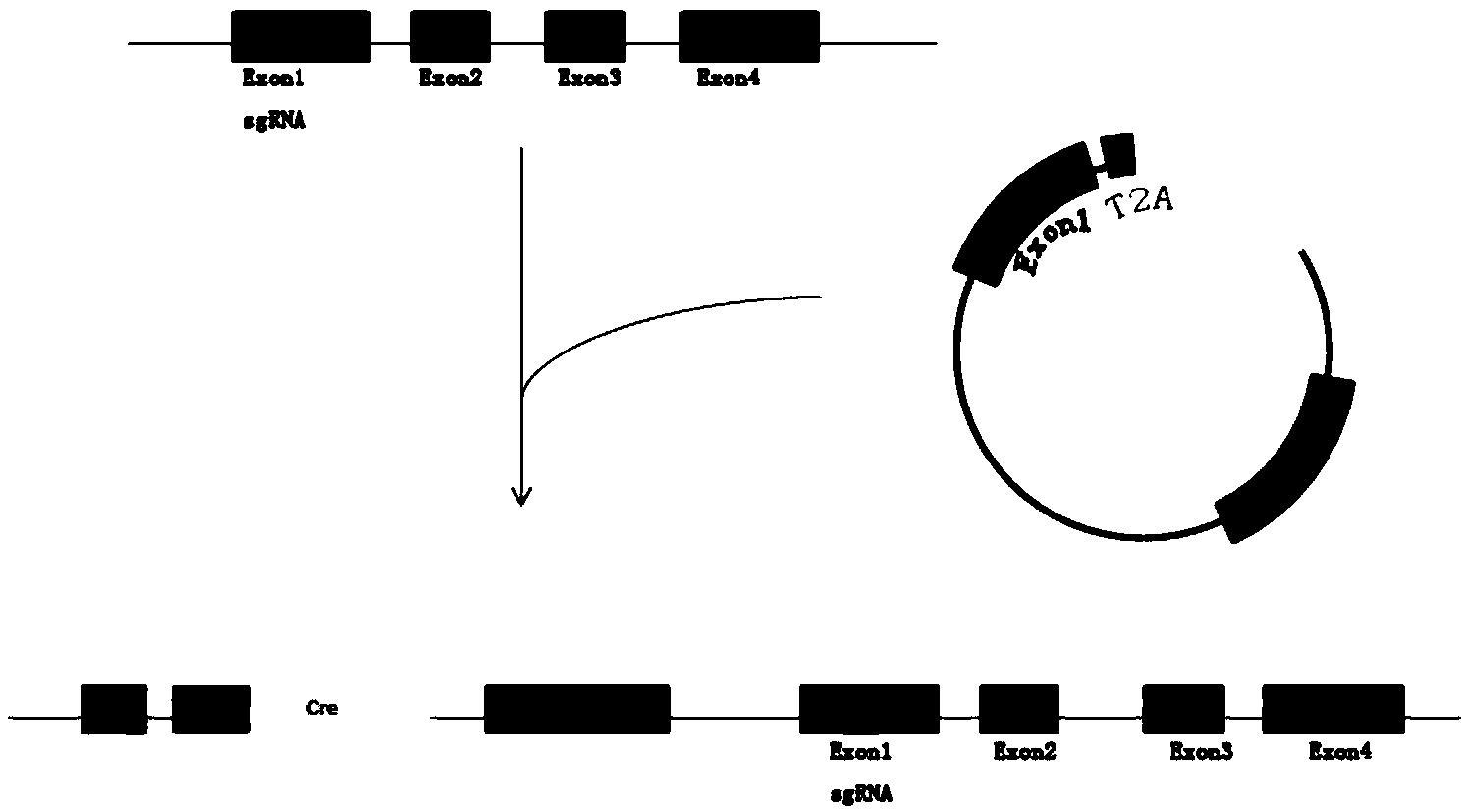
The invention discloses a method for targetedly integrating an exogenous gene into a target gene of a mammal, which is implemented by targetedly integrating an exogenous gene into a target gene by utilizing a CRISPR / Cas9 system, and especially targetedly integrating a Cre gene to a first exon of a mouse Enpp1 gene. The technique has the advantages of high integration efficiency (20%) and simple operation step, only needs to construct one exogenous gene vector, and can not be influenced by the position effect.
Owner:HANGZHOU NORMAL UNIVERSITY
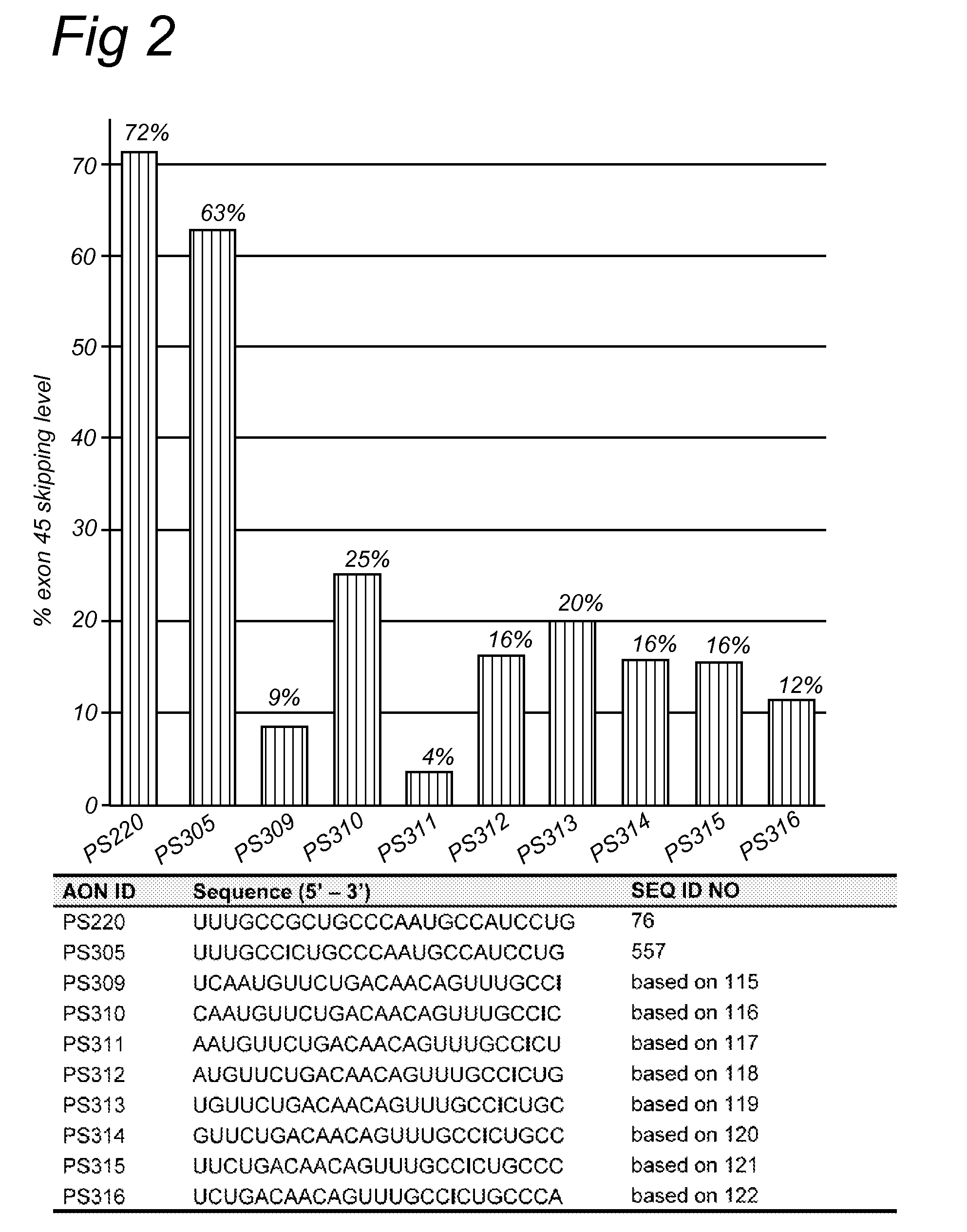
Oligonucleotide comprising an inosine for treating dmd
InactiveUS20120046342A1Avoid or decrease a potential multimerisation or aggregation of oligonucleotidesLow efficiencyOrganic active ingredientsSplicing alterationInosineNucleotide
The invention provides an oligonucleotide comprising an inosine, and / or a nucleotide containing a base able to form a wobble base pair or a functional equivalent thereof, wherein the oligonucleotide, or a functional equivalent thereof, comprises a sequence which is complementary to at least part of a dystrophin pre-m RNA exon or at least part of a non-exon region of a dystrophin pre-m RNA said part being a contiguous stretch comprising at least 8 nucleotides. The invention further provides the use of said oligonucleotide for preventing or treating DMD or BMD.
Owner:BIOMARIN TECH BV
Method for targeted knockout of specific gene of Suqin yellow chicken embryonic stem cell
InactiveCN104805118ASimple methodGood repeatabilityVector-based foreign material introductionForeign genetic material cellsDual promoterExon
The invention discloses a method for targeted knockout of a specific gene of a Suqin yellow chicken embryonic stem cell. The method comprises the following steps: firstly, checking out an exon sequence of a gene, cloning the gene, sequencing to obtain a complete exon sequence, designing a CRISPR / Cas9 knockout target site on the sequence as gRNA, and constructing a CRISPR / Cas9 dual-promoter knockout vector; performing SSA activity detection on the constructed Cas9 vector, transfecting a control group with an empty vector, detecting a luciferase signal, and obtaining a result that the more the luciferase activity is increased relative to the control group, the higher the gRNA shearing activity is shown; transfecting the CRISPR / Cas9 vector which is high in SSA activity with ESCs, using flow cytometry to screen a GFP-positive cell, extracting genome DNA, and designing a primer.
Owner:YANGZHOU UNIV
Mutagenesis methods
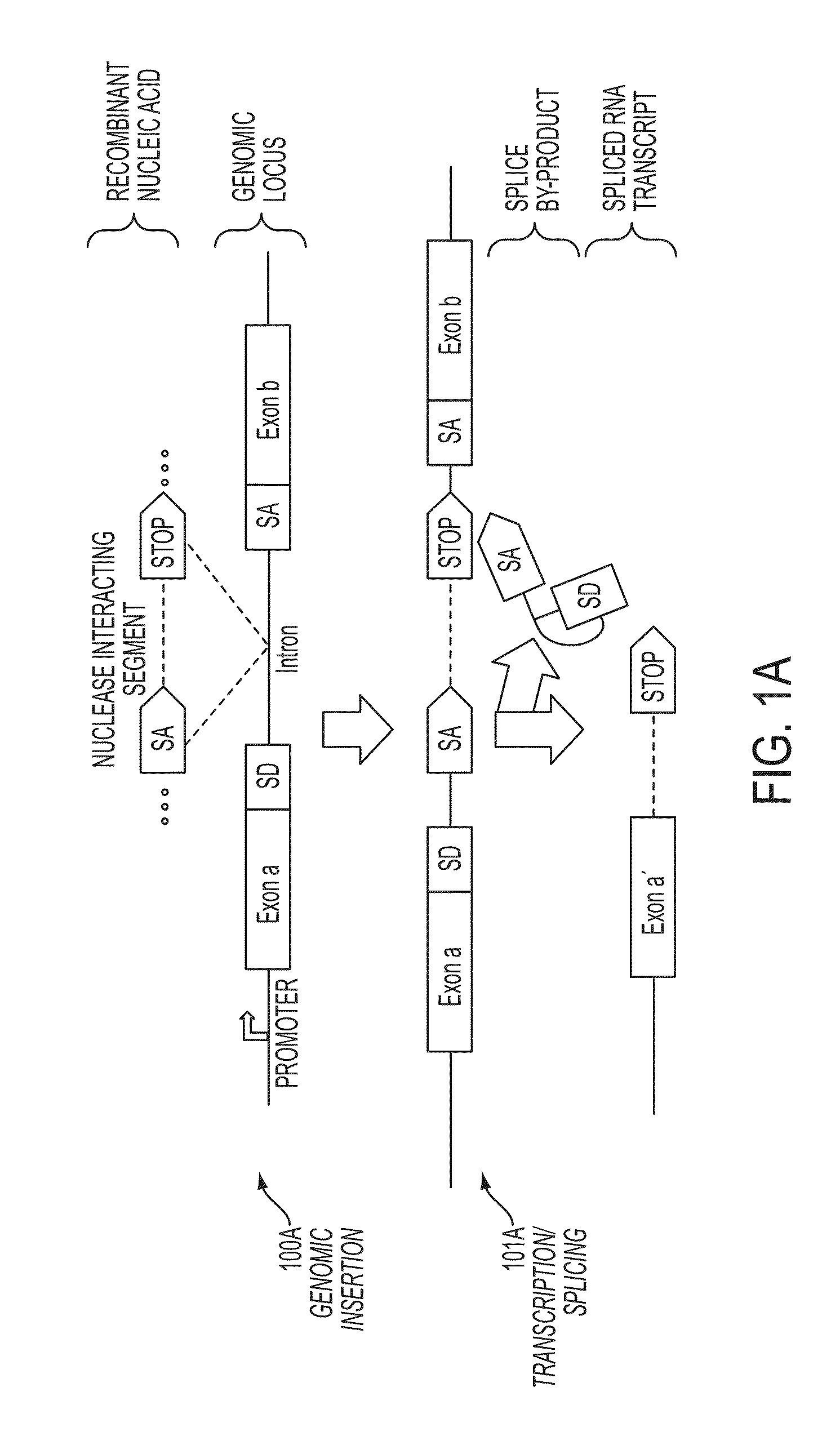
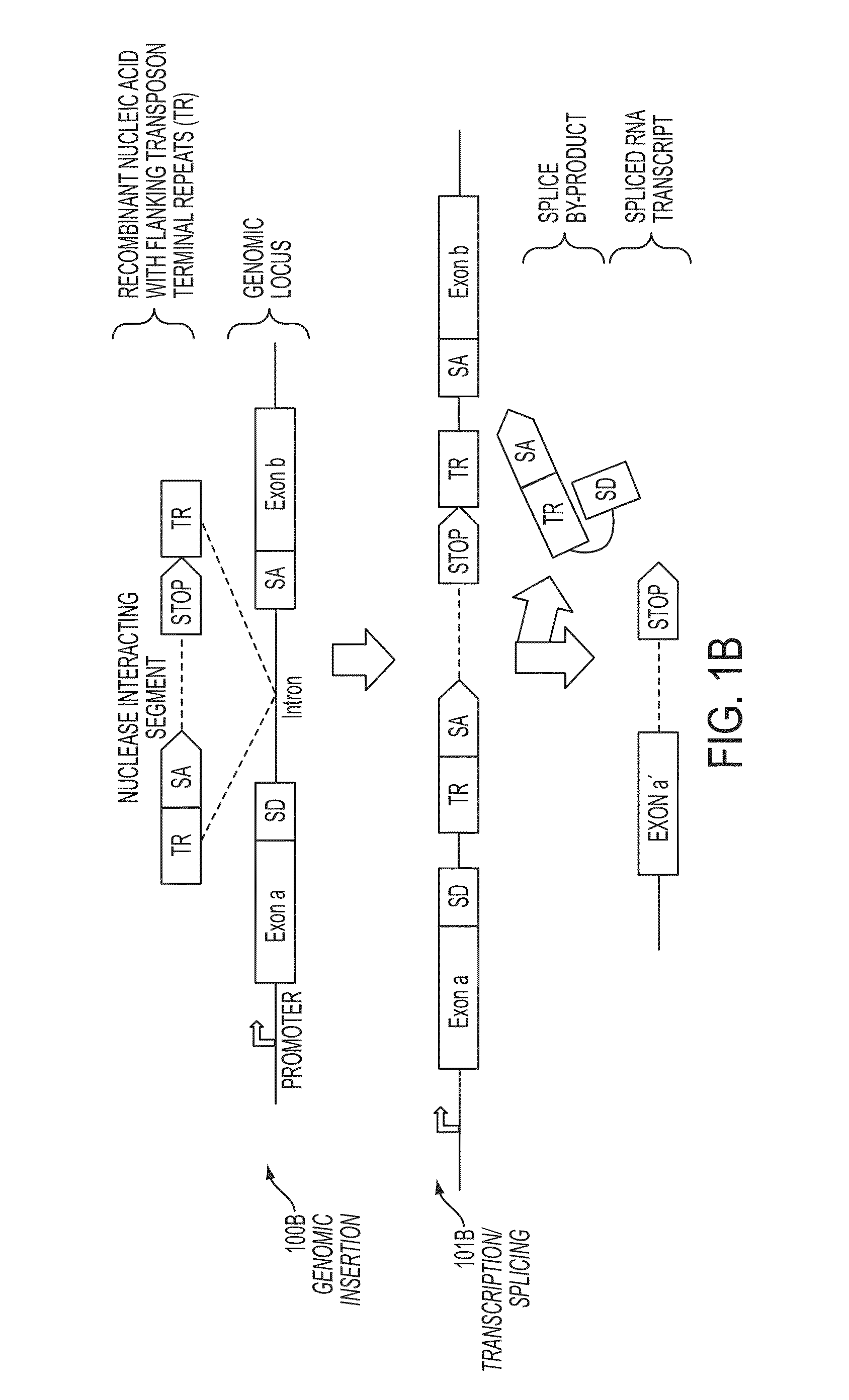
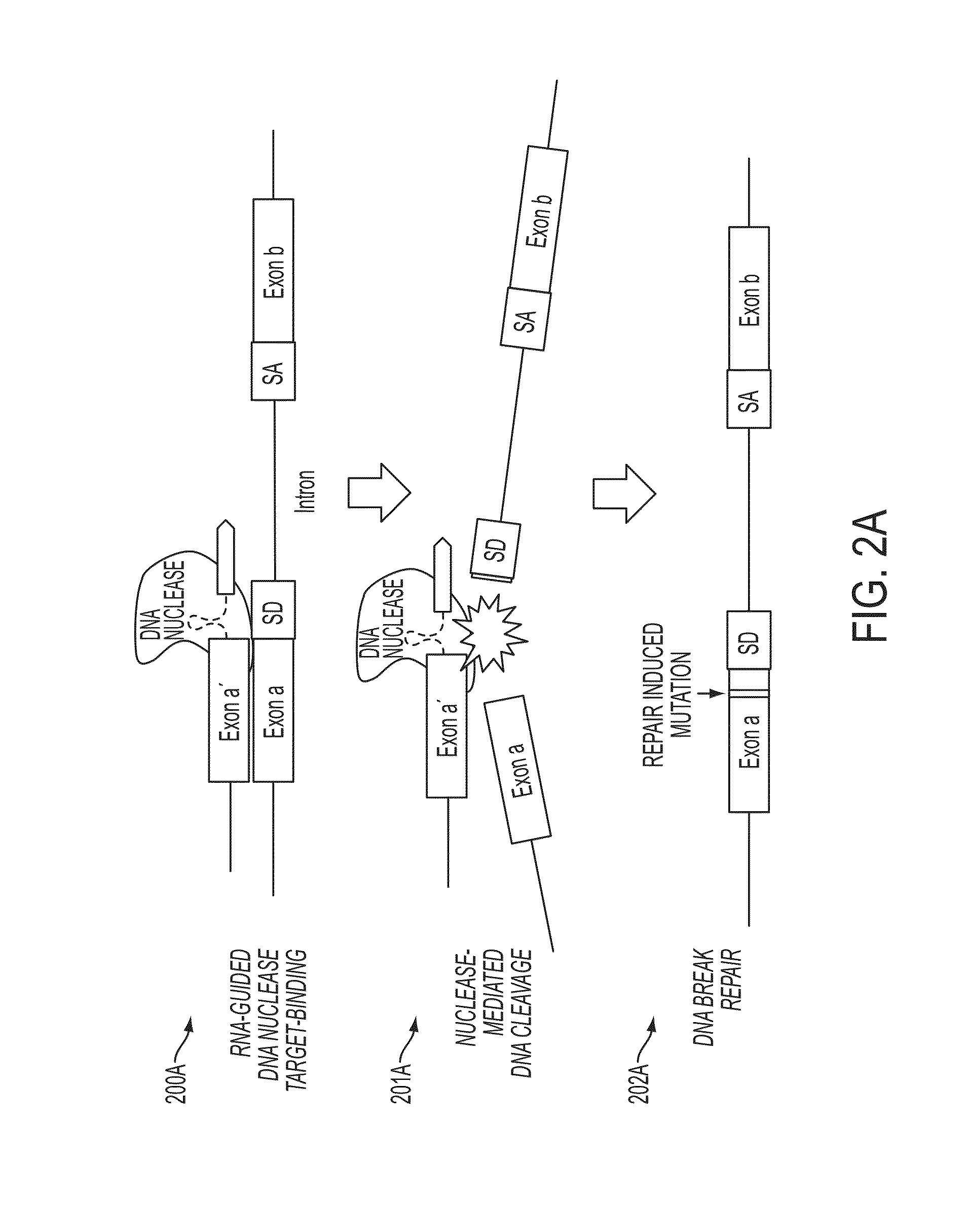
In some embodiments, aspects of the disclosure provide methods and compositions that are useful for modifying (e.g., mutating) one or more alleles of a genomic locus within a cell. In some embodiments, methods and compositions described herein involve producing a chimeric spliced RNA molecule that includes a transcribed exon spliced to a nuclease interacting RNA segment. In some embodiments, the chimeric spliced RNA guides a DNA modifying enzyme (e.g., a nuclease) to a genomic locus in a cell resulting in modification of the locus.
Owner:LAM THERAPEUTICS
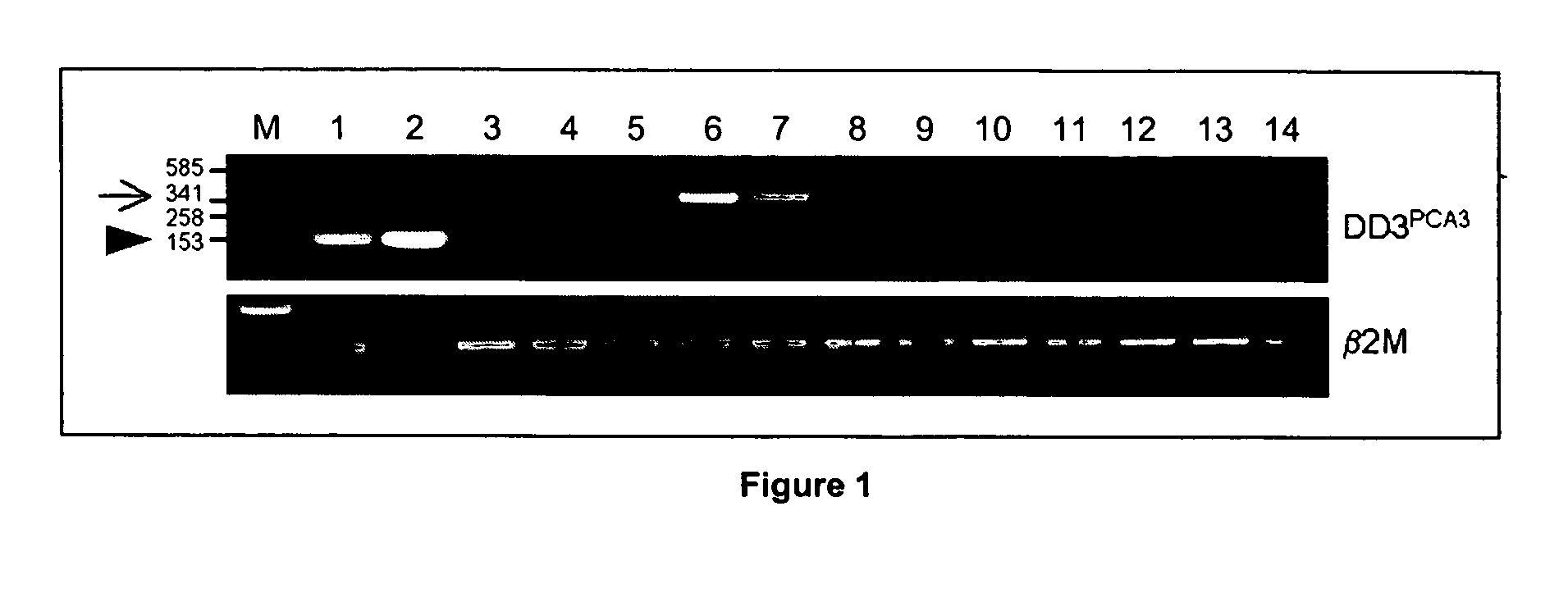
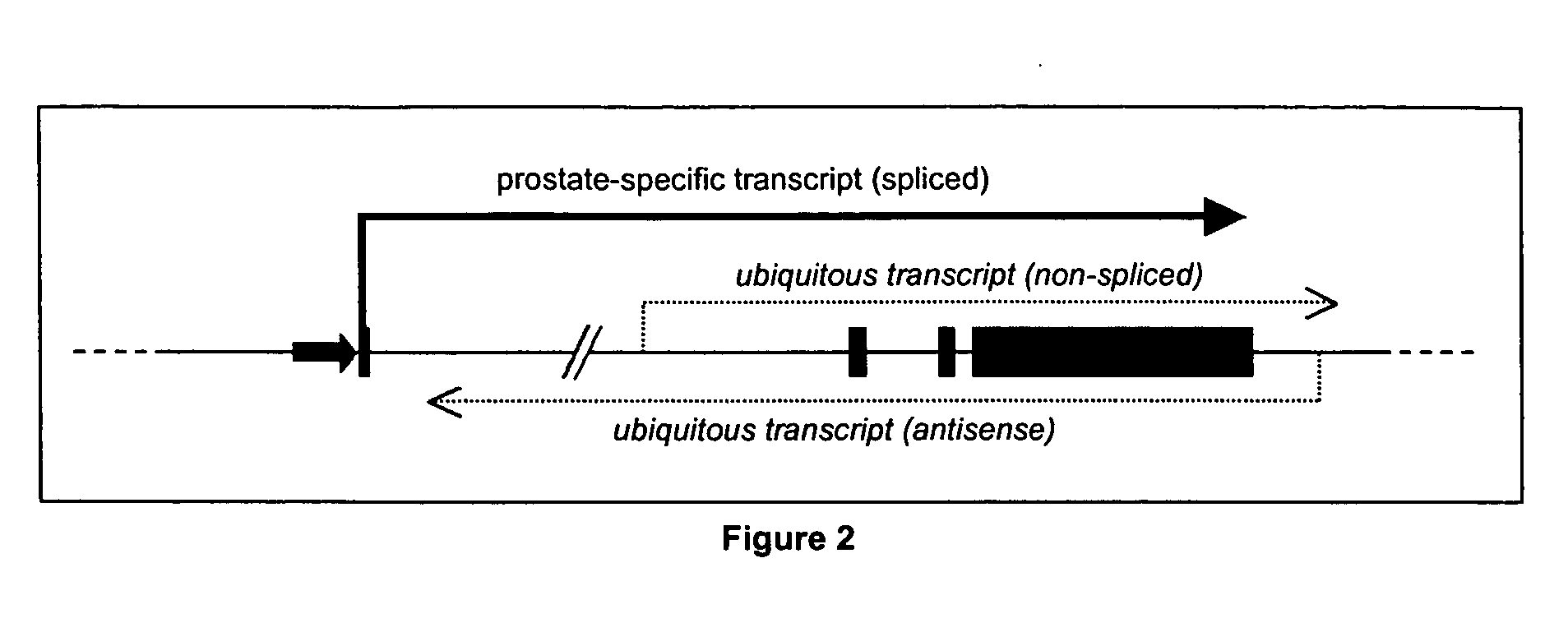
Specific method of prostate cancer detection based on PCA3 gene and kits therefor
InactiveUS20050164223A1Strong specificityHigh detection sensitivityMicrobiological testing/measurementNucleic Acid ProbesExon
The present invention relates, in general, to prostate cancer. More specifically, the present invention relates to a method to diagnose prostate cancer in a patient by detecting a PCA3 sequence, and more particularly a PCA3 RNA, the PCA3 sequence detected in a sample from the patient being specifically associated with prostate cancer. In a particular embodiment the method and kit enables an amplification of a PCA3 RNA through an exon-exon junction of a spliced PCA3 mRNA. The invention also related to methods and kits to detect such an amplified PCA3 RNA, using a probe which spans the amplified exon-exon junction. The invention also relates to kits containing nucleic acid primers and kits containing nucleic acid primers and nucleic acid probes to diagnose, assess, or prognose a human afflicted with prostate cancer. In particular the methods and kits are designed to detect a PCA3 RNA which lacks one intron or more, and in particular case is intron-less.
Owner:STICHTING KATHOLIEKE UNIV
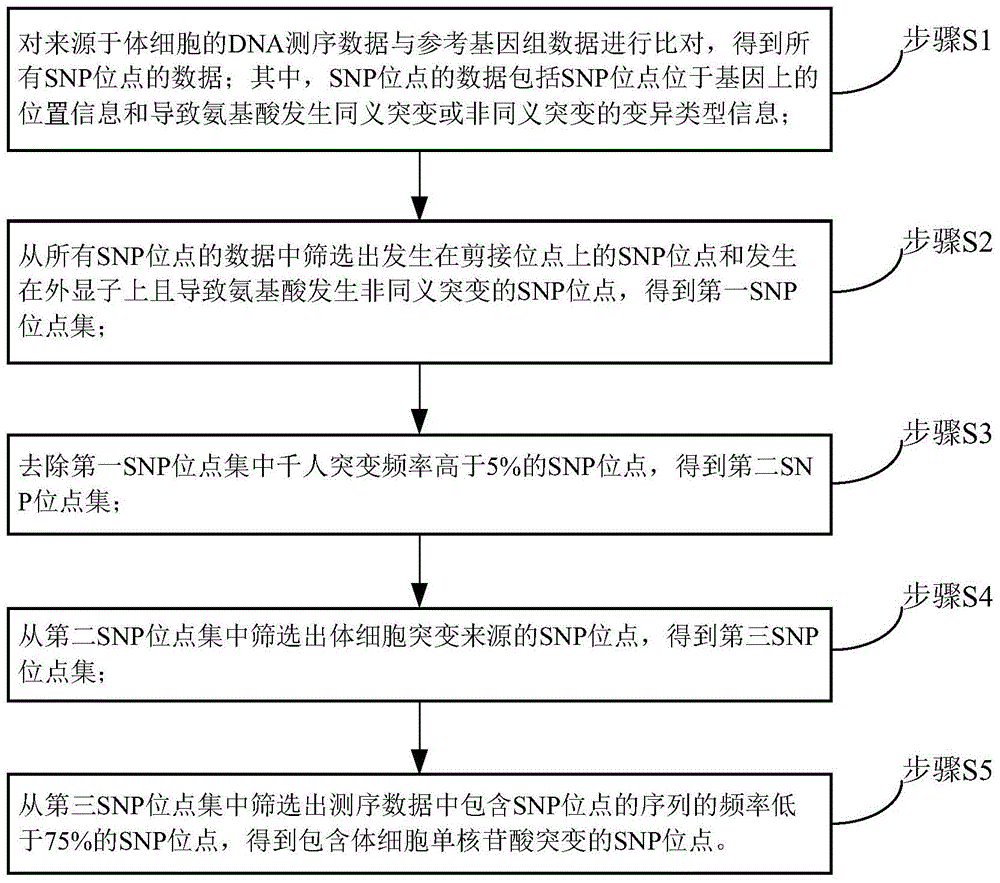
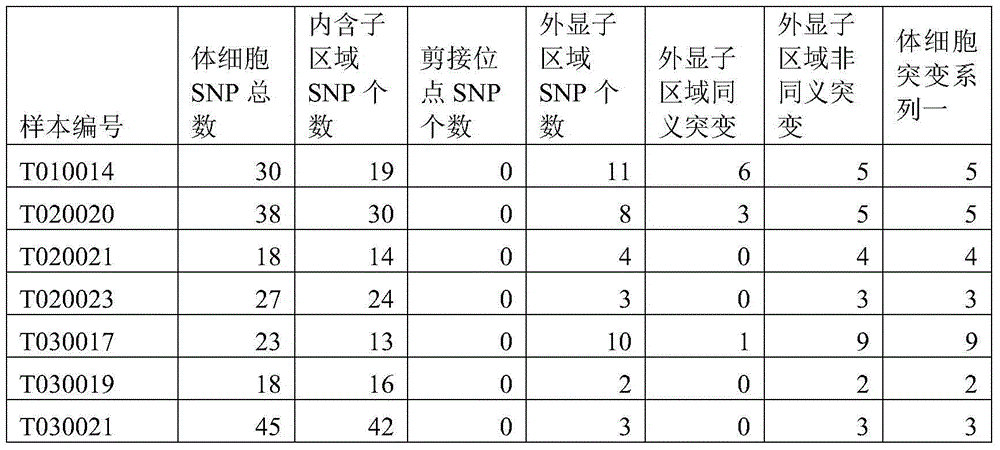
Method and device for detecting somatic cell SNP
ActiveCN104462869ARealize detectionSimple methodBioreactor/fermenter combinationsBiological substance pretreatmentsMutation frequencySimple sample
The invention discloses a method and device for detecting somatic cell SNP. The method comprises the steps of S1, comparing DNA sequencing data of somatic cells with reference genome data to obtain data of all SNP loci; S2, screening out SNP loci which occur on splicing loci and SNP loci which occur on exons and cause nonsynonymous mutation of amino acid to obtain a first SNP locus set; S3, removing SNP loci with the thousand-people mutation frequency higher than 5% in the first SNP locus set to obtain a second SNP locus set; S4, screening out SNP loci which are resources of somatic cell mutation from the second SNP locus set to obtain a third SNP locus set; S5, screening out SNP loci with the frequency of supporting sequences lower than 75% from the third SNP locus set to obtain SNP loci containing somatic cell SNP. By means of the method, somatic cell mutation can be detected through simple samples, and therefore the cost is lowered.
Owner:天津诺禾致源生物信息科技有限公司
Kit and application thereof in detection on hereditary bone disease genes
InactiveCN105586389ABioreactor/fermenter combinationsBiological substance pretreatmentsExonToxicology
The invention discloses a kit, an application thereof in detection on hereditary bone disease genes, and also a method and an apparatus for detection on the hereditary bone disease genes. The kit includes a probe which is fixed on a solid-phase substrate or is free in a solution. The probe can specially recognize exon regions of 722 or 363 genes, for example, the exon regions of the following nine genes: PHEX, ENPP1, FGF23, CLCN5, SLC34A3, DMP1, VDR, CYP2R1 and CYP27B1. The kit, and as well as the method and the apparatus for detection on the hereditary bone disease genes are used for one-time acquiring and / or detecting related genes of the hereditary bone disease and mutation status thereof.
Owner:天津华大基因科技有限公司 +1
Next generation sequencing-based detection panel and detection kit for pan-cancer targeting, chemotherapy and immune drugs and application thereof
ActiveCN109609647AReduce data volumeImprove uniformityMicrobiological testing/measurementGenes mutationCancer targeting
The invention discloses a next generation sequencing-based detection panel and detection kit for pan-cancer targeting, chemotherapy and immune drugs and application thereof. The detection panel includes gene mutations related to pan-cancer type, treatment and prognosis, tumor mutation load calculation related exon regions and microsatellite instability sites. According the technical solution of the present invention, the detection panel includes the gene mutations related to pan-cancer type, treatment and prognosis, the tumor mutation load calculation related exon regions and the microsatellite instability sites, and the gene information included is comprehensive; a variety of tumor mutations can be jointly detected; and the detection panel and detection kit can be used for the concomitantdiagnosis of targeted drugs, chemotherapeutics or immune drugs to obtain accurate results.
Owner:ZHENYUE BIOTECHNOLOGY JIANGSU CO LTD
Method for obtaining bmp2a gene knocked-out zebra fish through CRISPR/Cas9
The invention discloses a method for obtaining bmp2a gene knocked-out zebra fish through CRISPR / Cas9. A novel gRNA sequence is designed between a first exon and an intron of BMP2a, the gRNA sequence is GGAGCCCATCACTAGACTCTTGG, and enzyme digestion is HinfI. Compared with a traditional gene knockout technology, the CRISPR / Cas9 technology has the advantages of low toxicity, high accuracy, high efficiency, short success cycle and the like. Therefore, the BMP2a gene can be theoretically knocked out faster.
Owner:WUXI NO 2 PEOPLES HOSPITAL
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com