Oligonucleotide comprising an inosine for treating dmd
a technology of inosine and oligonucleotide, which is applied in the field of molecular biology and medicine, can solve the problems of calcium-activated proteases and fiber necrosis, progressive muscle wasting and weakness, and/or abnormal formation of dystrophins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Materials and Methods
[0155]AON design was based on (partly) overlapping open secondary structures of the target exon RNA as predicted by the m-fold program, on (partly) overlapping putative SR-protein binding sites as predicted by the ESE-finder software. AONs were synthesized by Prosensa Therapeutics B.V. (Leiden, Netherlands), and contain 2′-O-methyl RNA and full-length phosphorothioate (PS) backbones.
Tissue Culturing, Transfection and RT-PCR Analysis
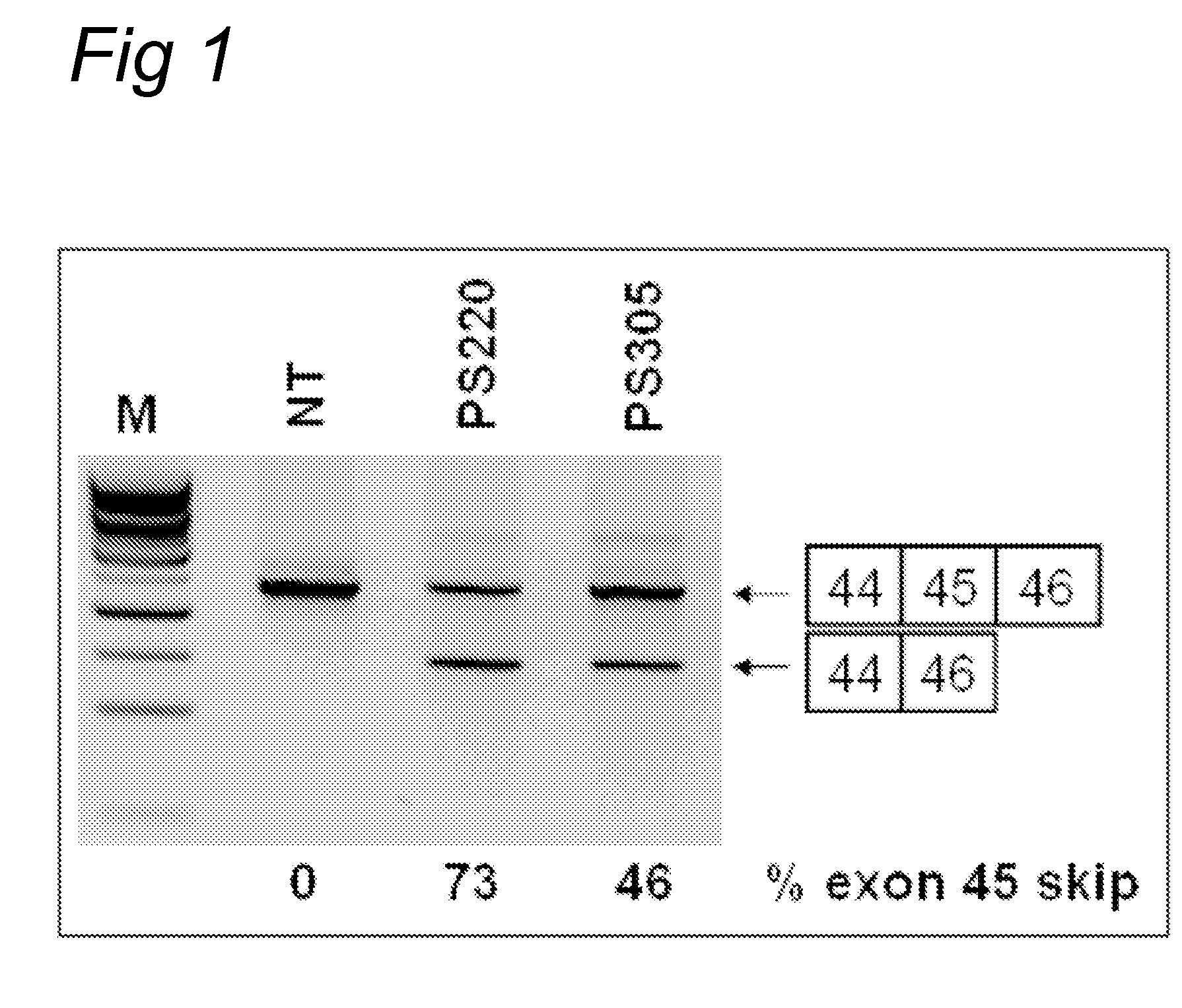
[0156]Myotube cultures derived from a healthy individual (“human control”) (examples 1, 3, and 4; exon 43, 50, 52 skipping) or a DMD patient carrying an exon 45 deletion (example 2; exon 46 skipping) were processed as described previously (Aartsma-Rus et al., Neuromuscul. Disord. 2002; 12: S71-77 and Hum Mol Genet. 2003; 12(8): 907-14). For the screening of AONs, myotube cultures were transfected with 200 nM for each AON (PS220 and PS305). Transfection reagent UNIFectylin (Prosensa Therapeutics BV, Netherlands) was used, with 2 μl UNI...
example 2
[0159]Materials and Methods
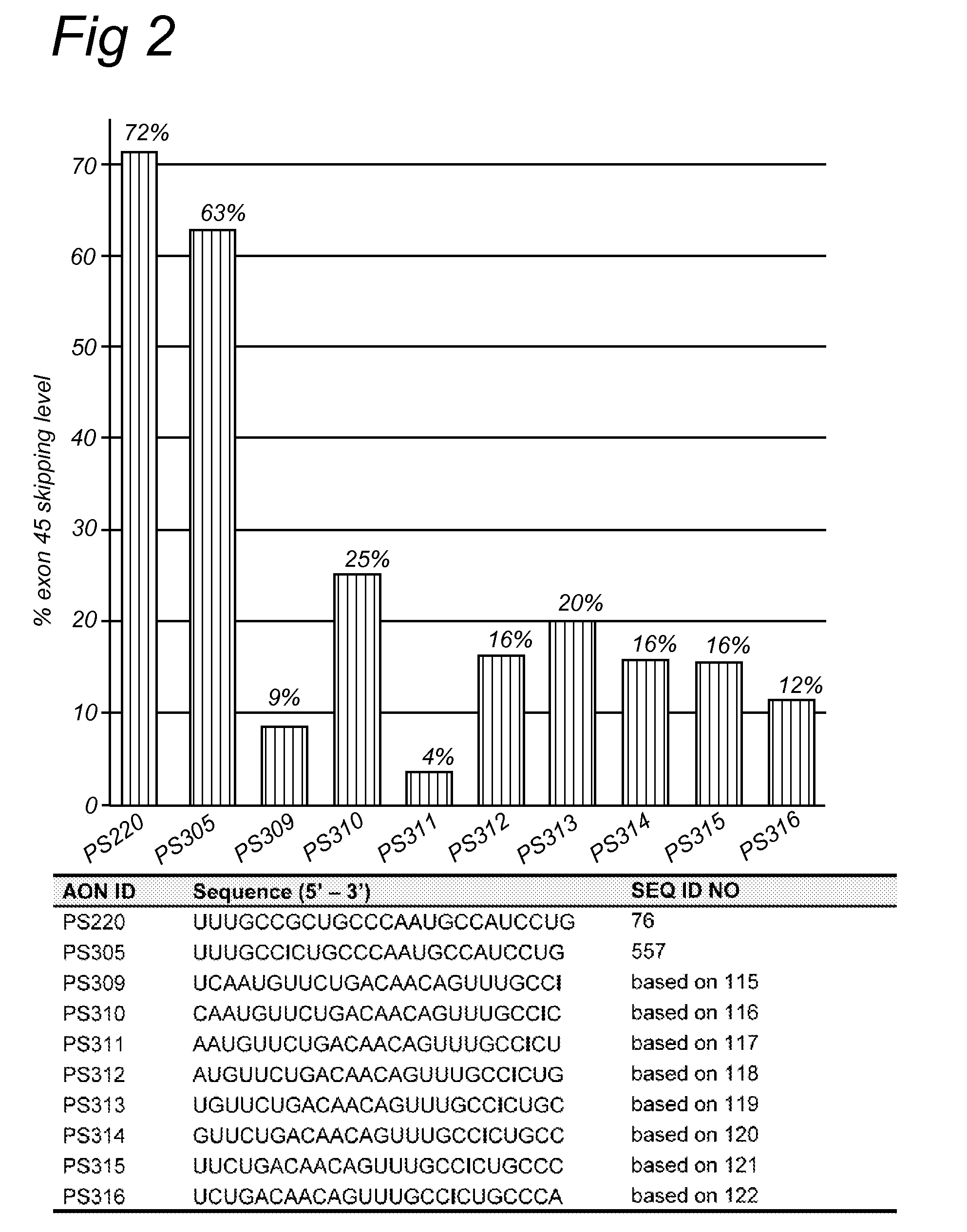
[0160]AON design was based on (partly) overlapping open secondary structures of the target exon 45 RNA as predicted by the m-fold program, on (partly) overlapping putative SR-protein binding sites as predicted by the ESE-finder software. AONs were synthesized by Prosensa Therapeutics B.V. (Leiden, Netherlands), and contain 2′-O-methyl RNA, full-length phosphorothioate (PS) backbones and one inosine for guanosine substitution.
Tissue Culturing, Transfection and RT-PCR Analysis
[0161]Myotube cultures derived from a healthy individual (“human control”) were processed as described previously (Aartsma-Rus et al., Neuromuscul. Disord. 2002; 12: S71-77 and Hum Mol Genet. 2003; 12(8): 907-14). For the screening of AONs, myotube cultures were transfected with 200 nM for each AON. Transfection reagent UNIFectylin (Prosensa Therapeutics BV, Netherlands) was used, with 2 μl UNIFectylin per μg AON. Exon skipping efficiencies were determined by nested RT-PCR analysis usin...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Dimensionless property | aaaaa | aaaaa |
| Dimensionless property | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com