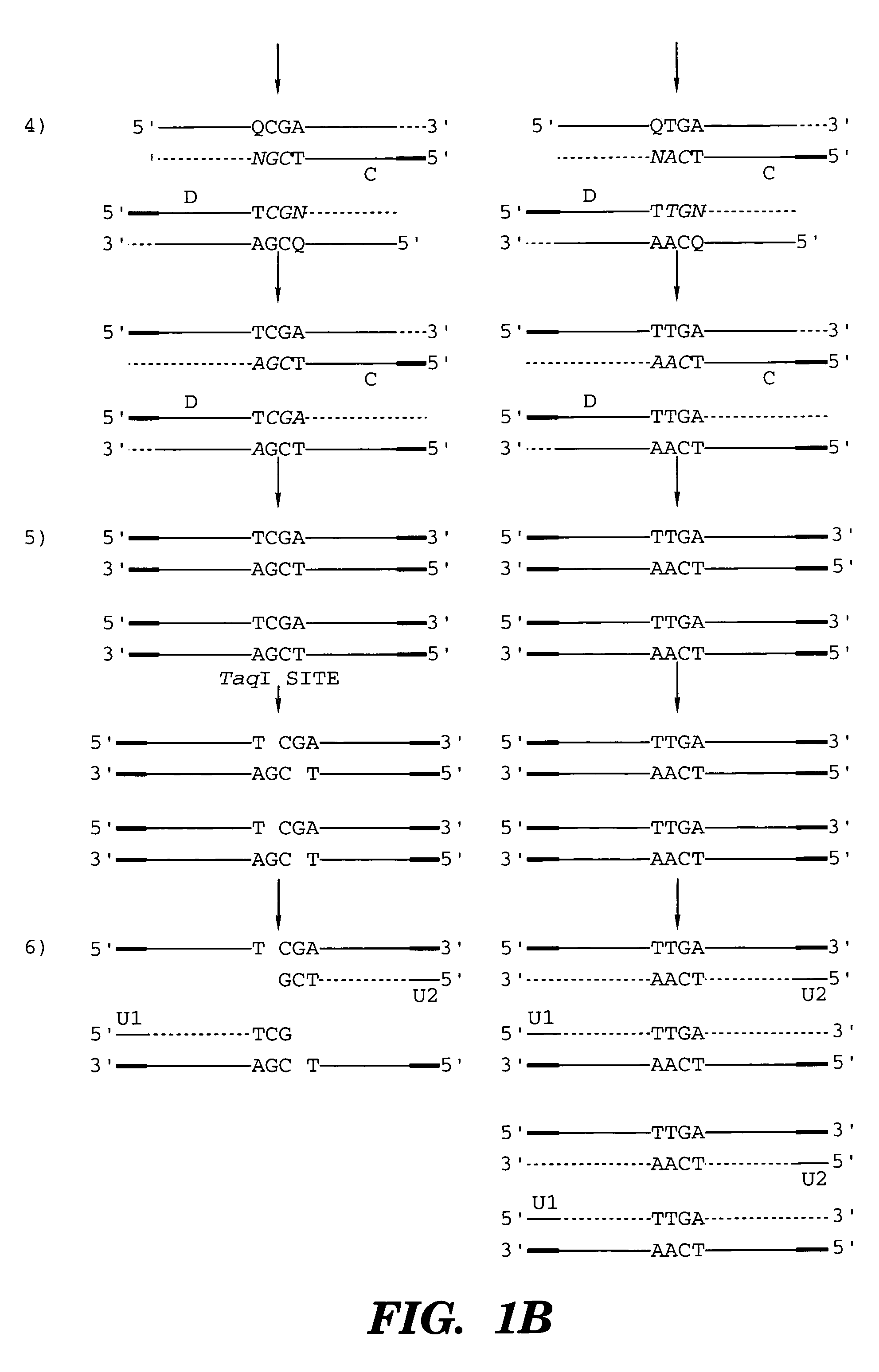
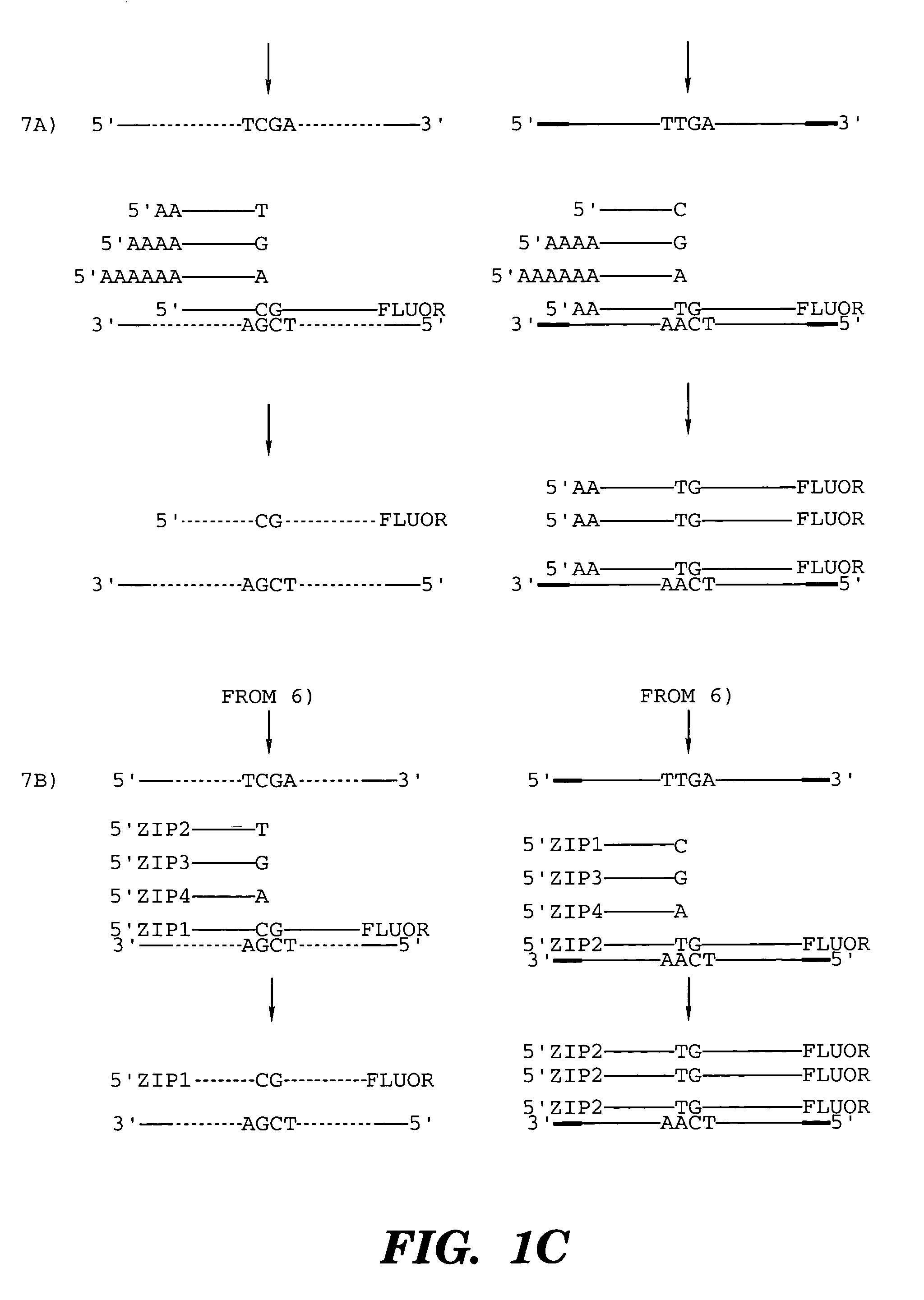
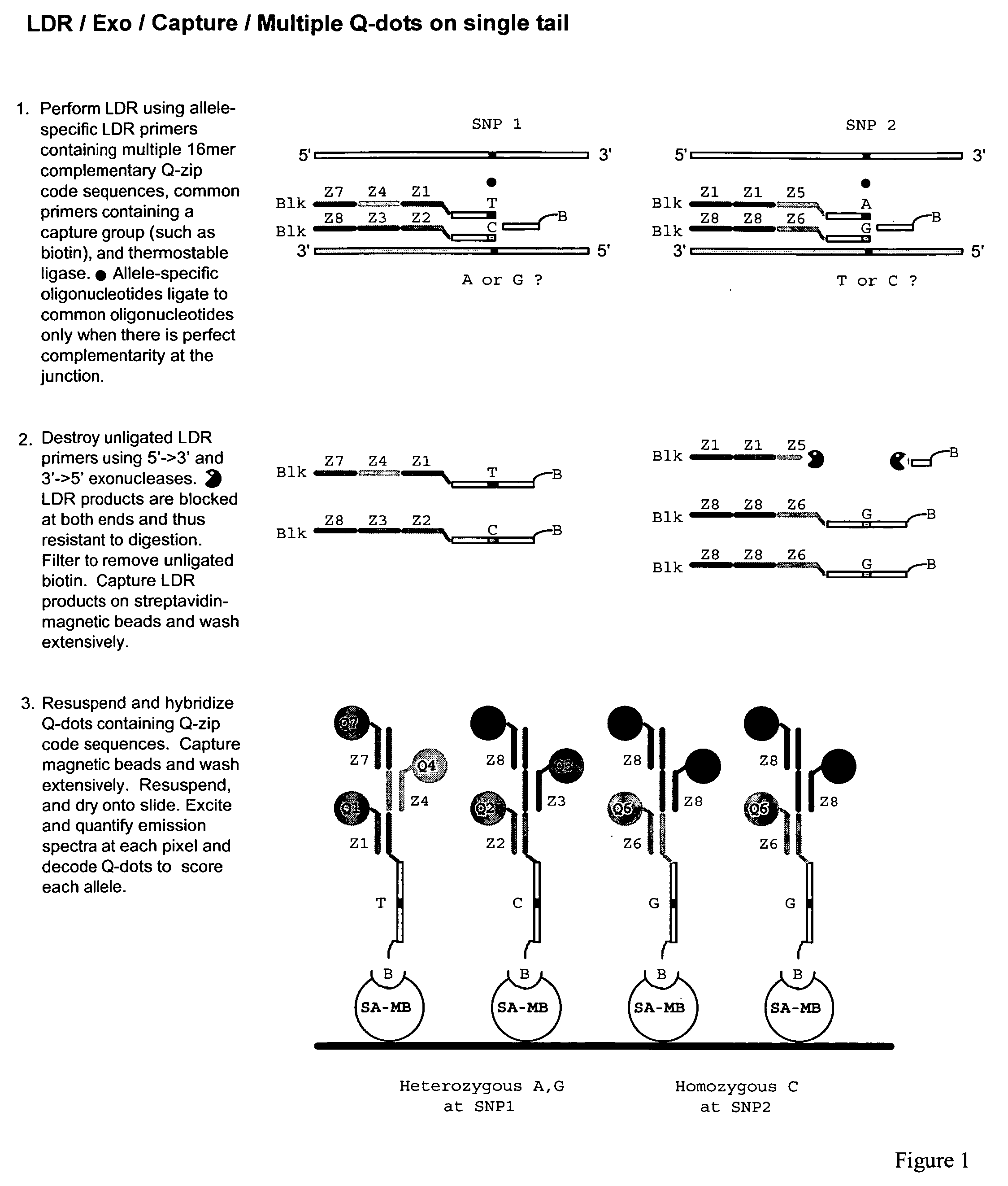
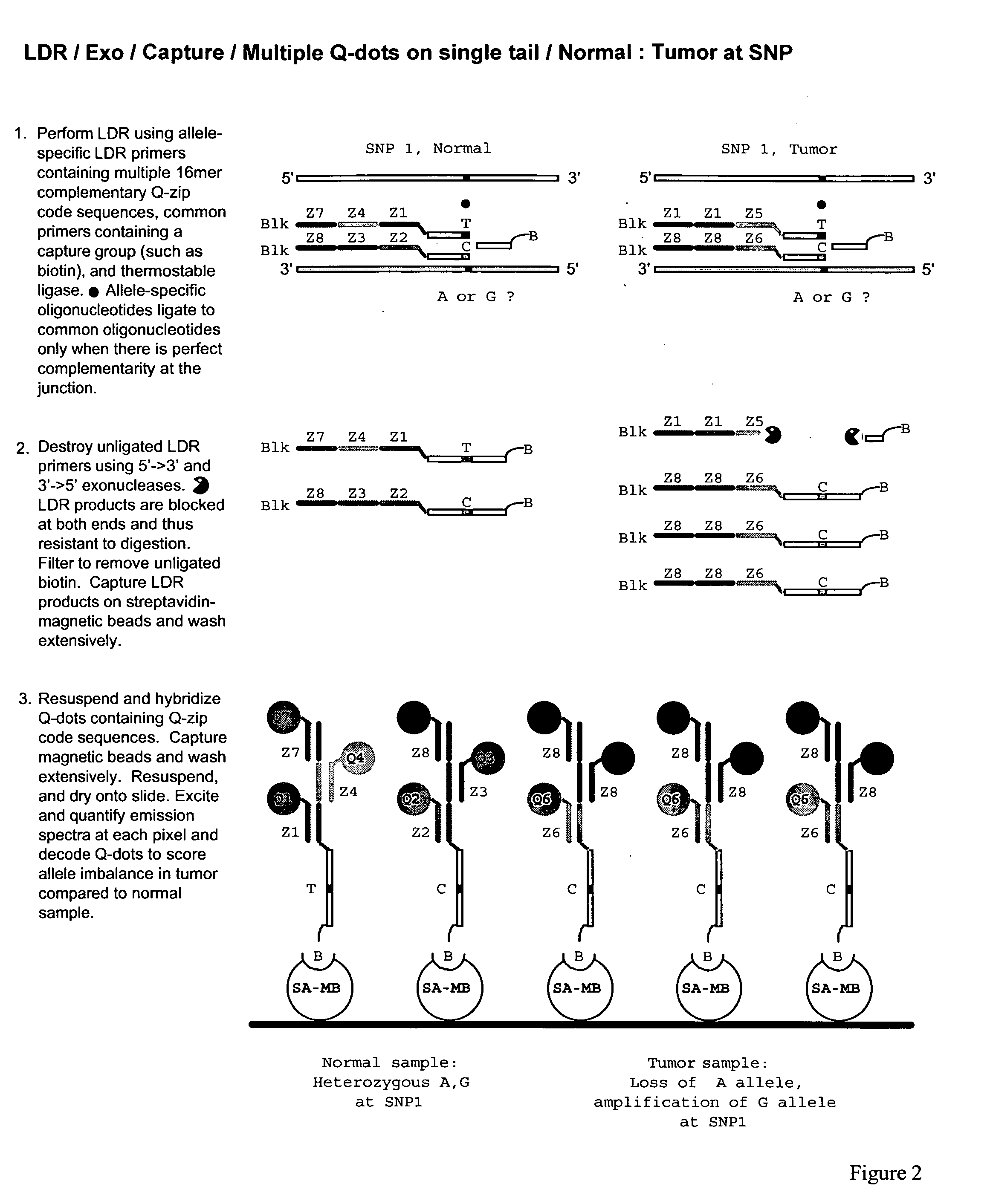
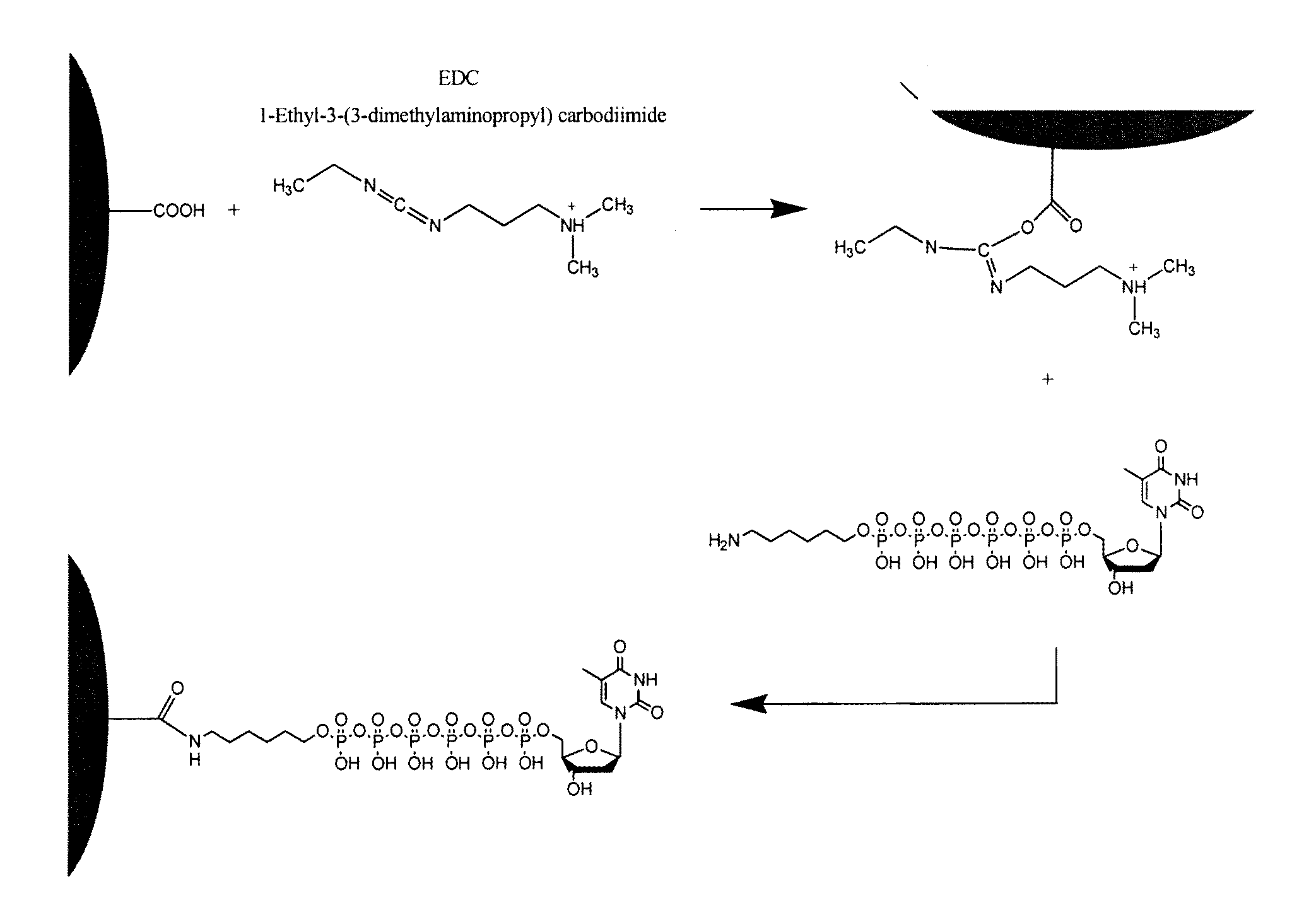
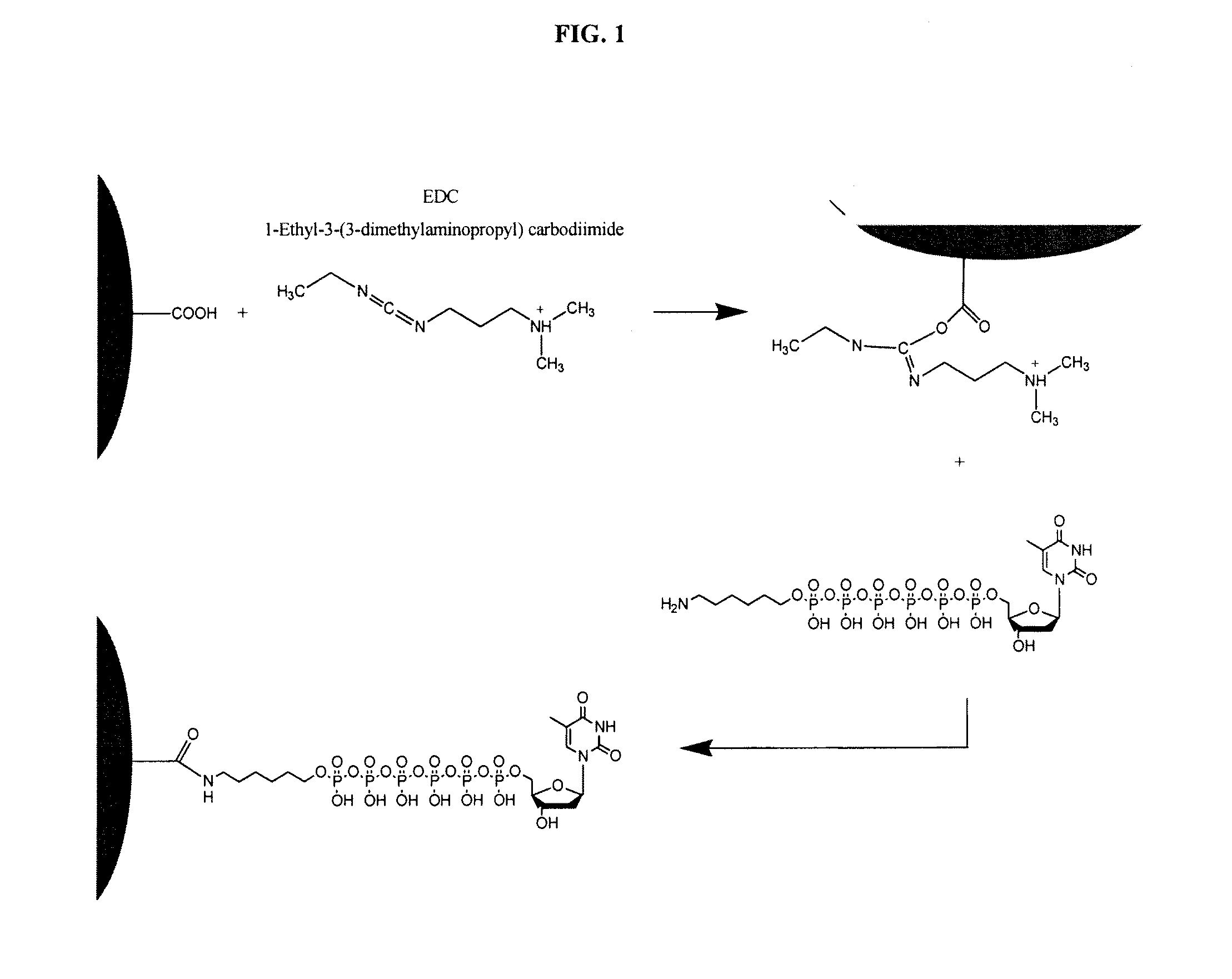
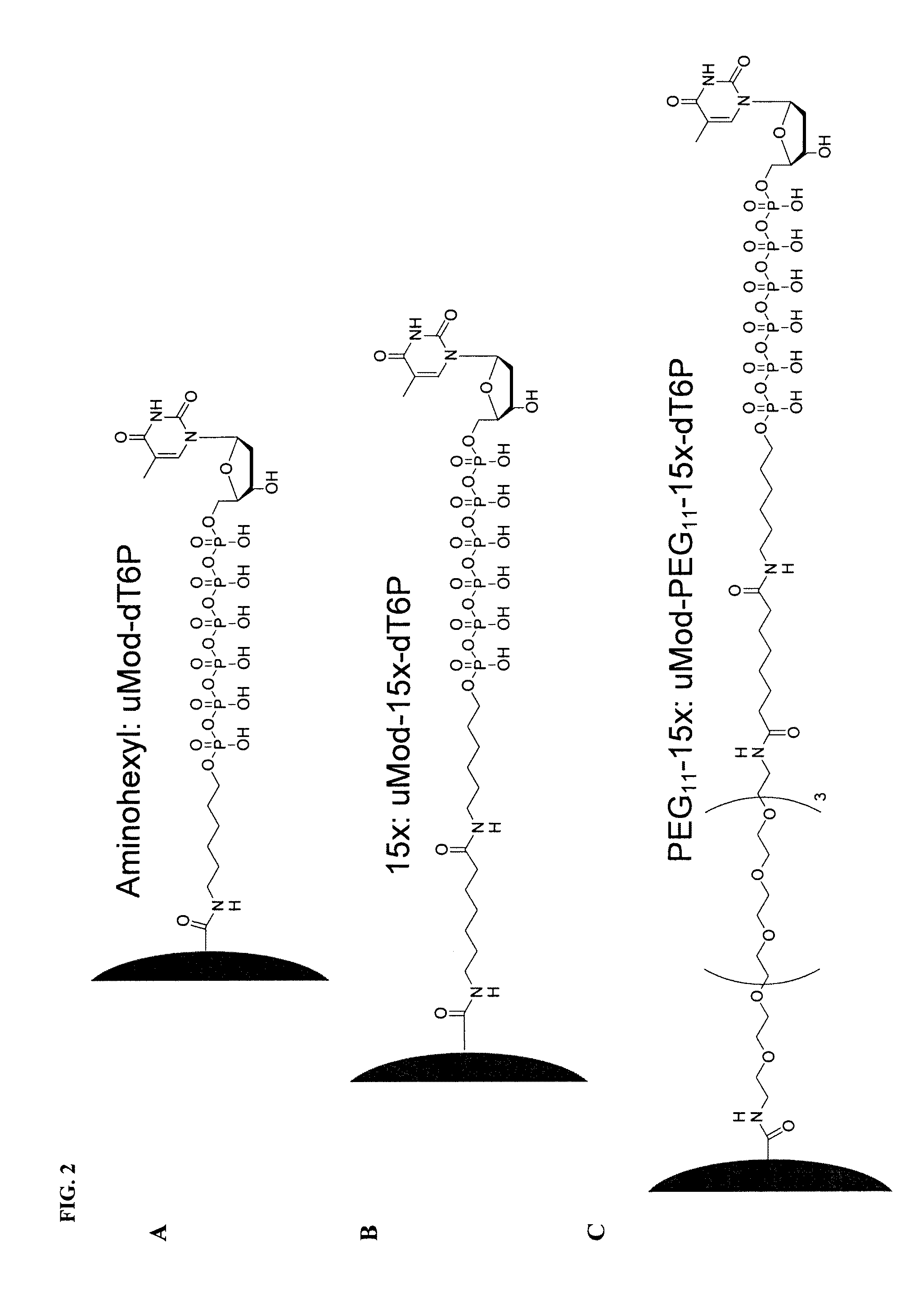
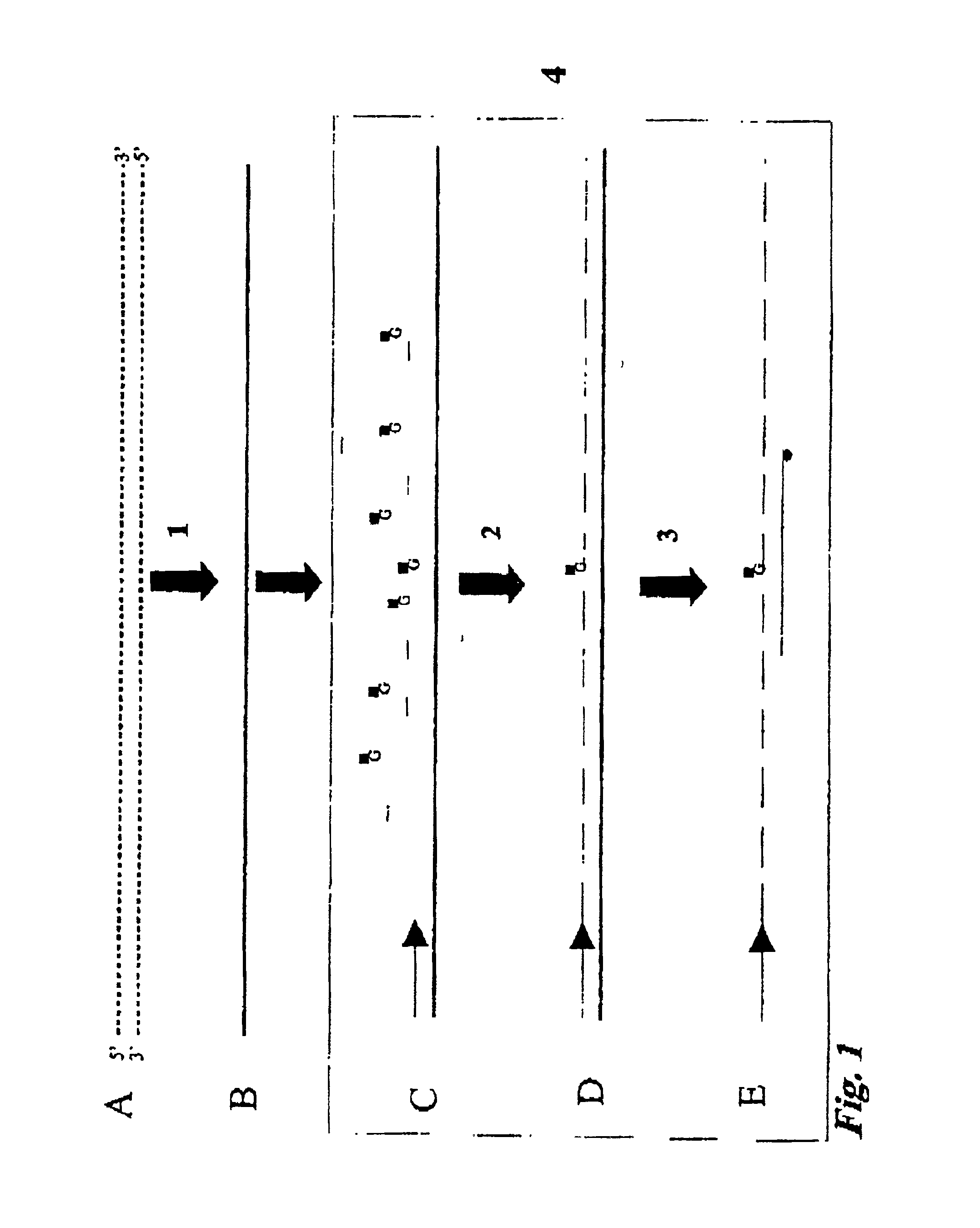
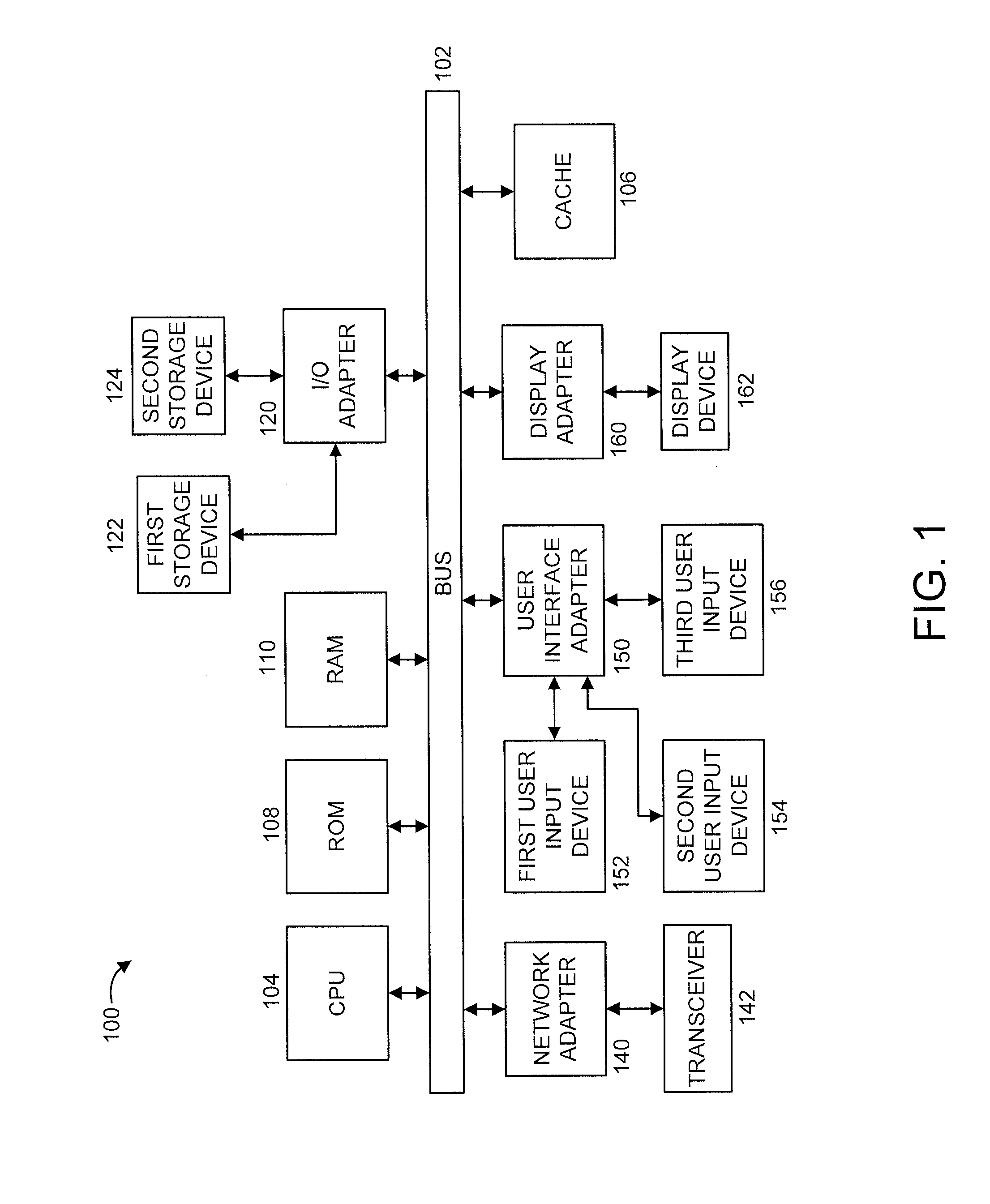
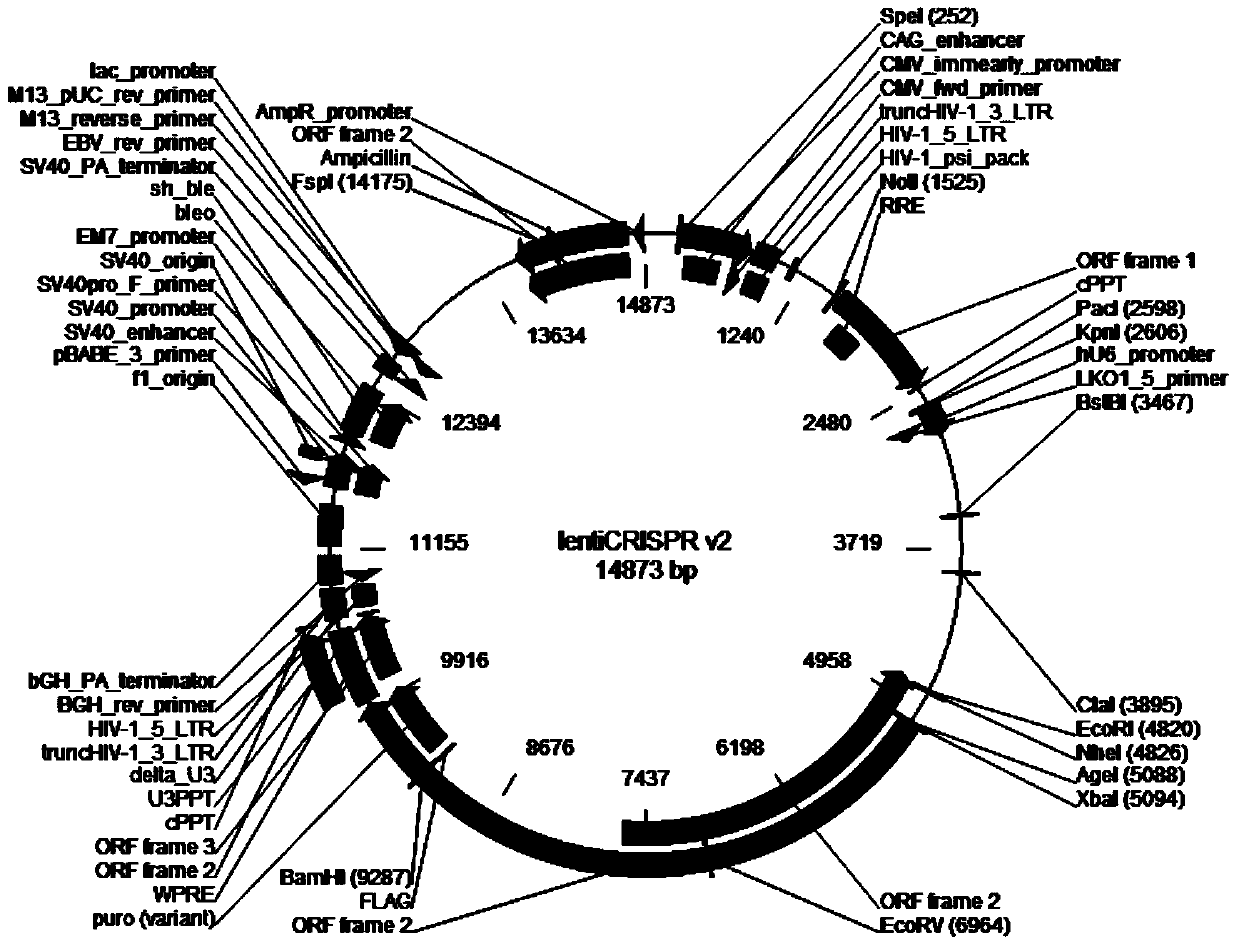
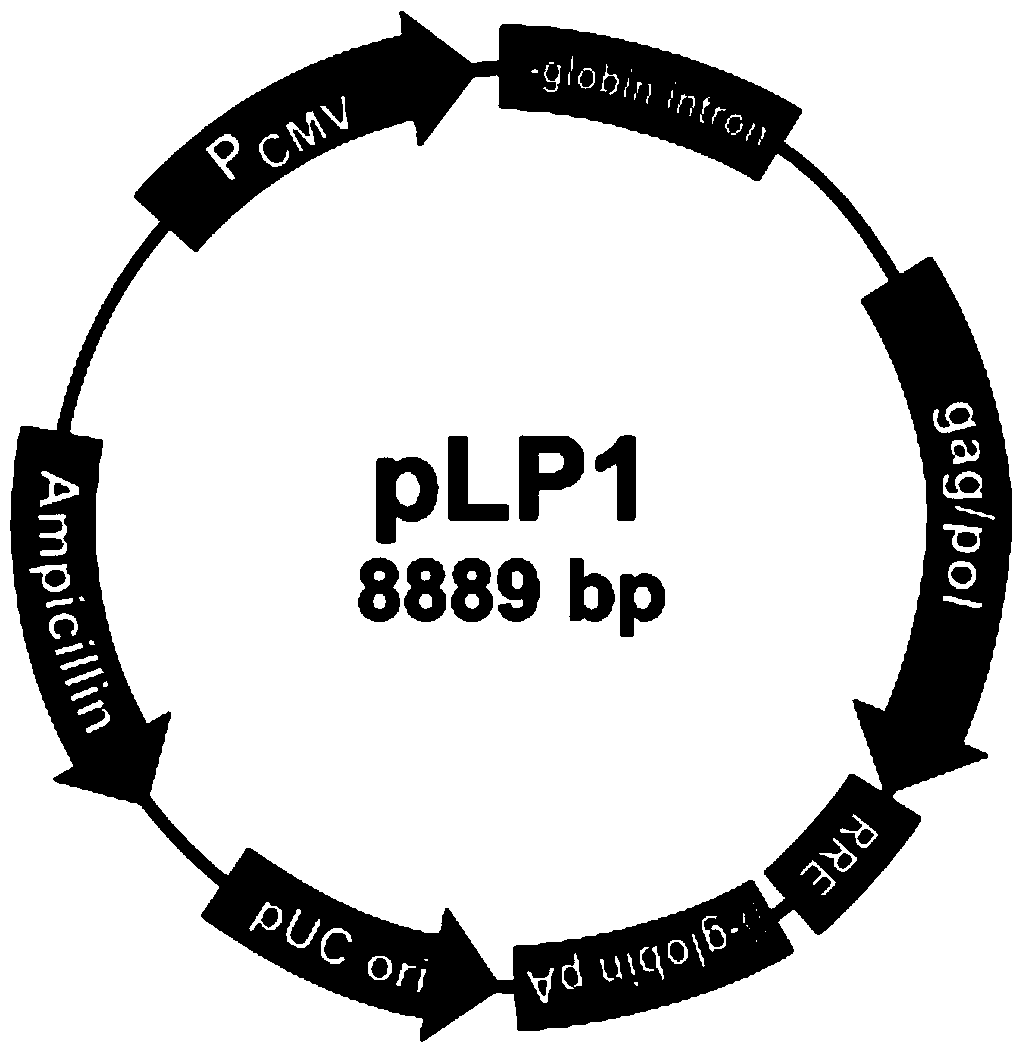
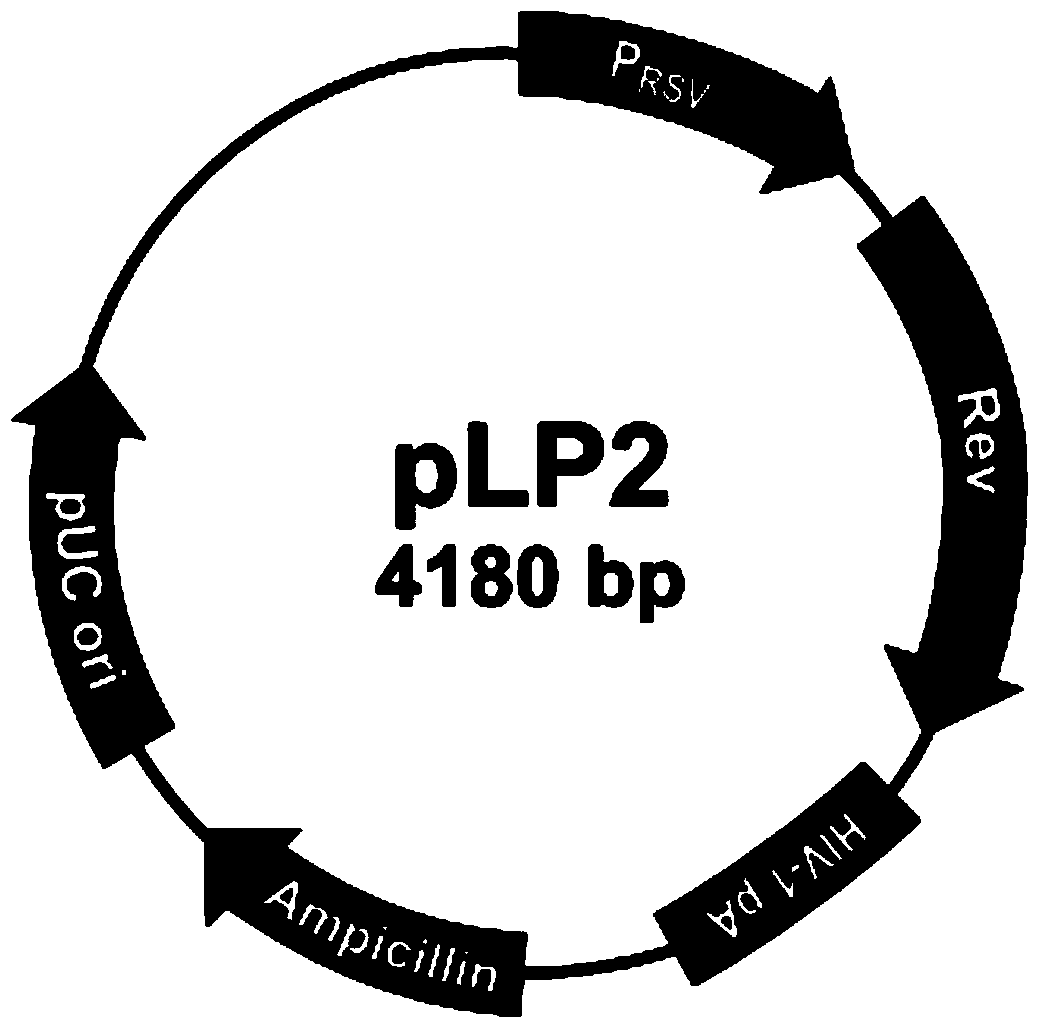
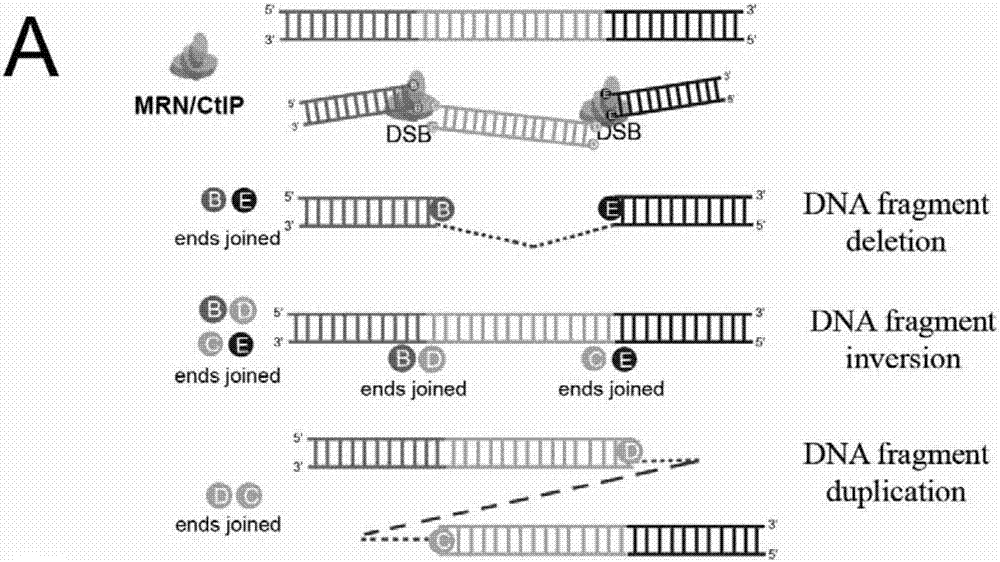
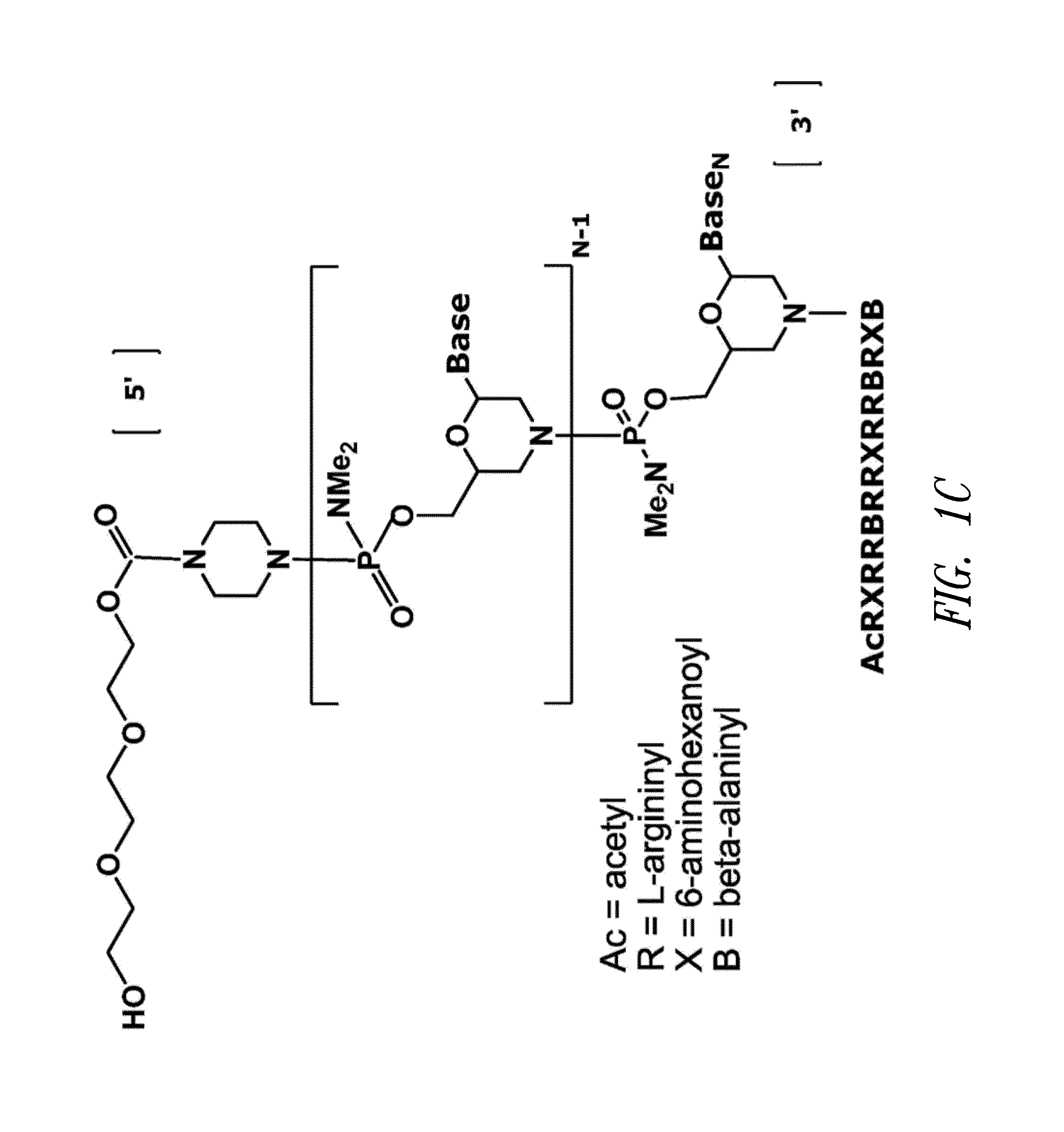
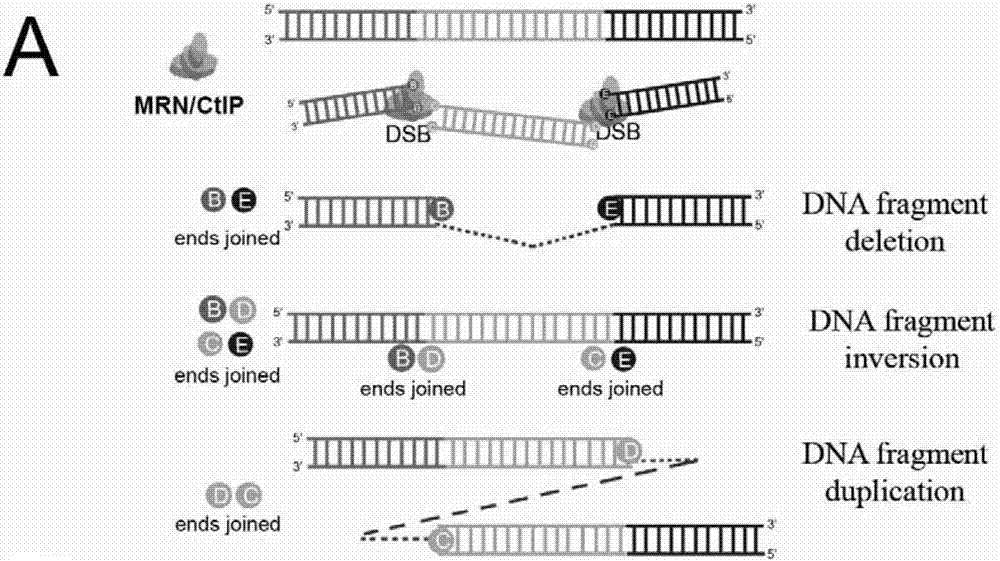
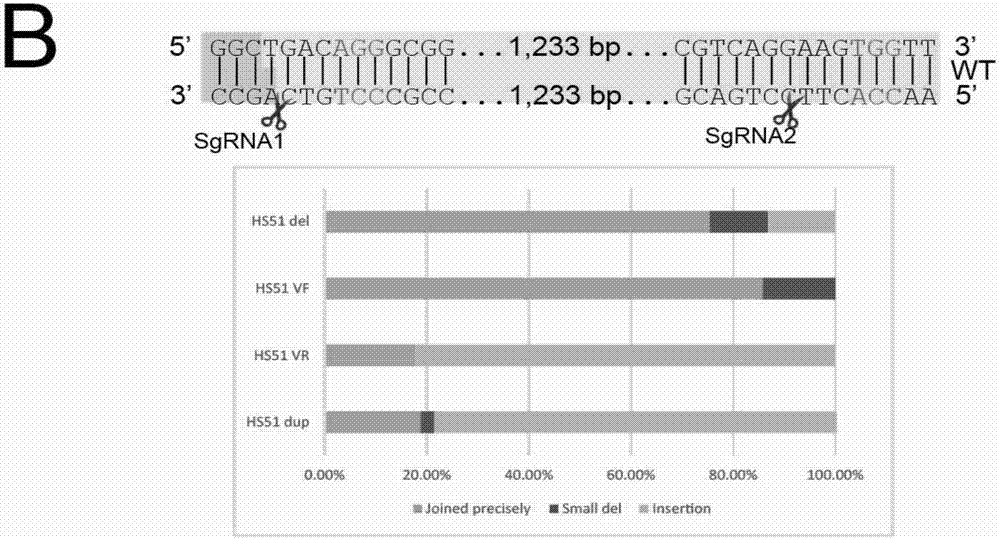
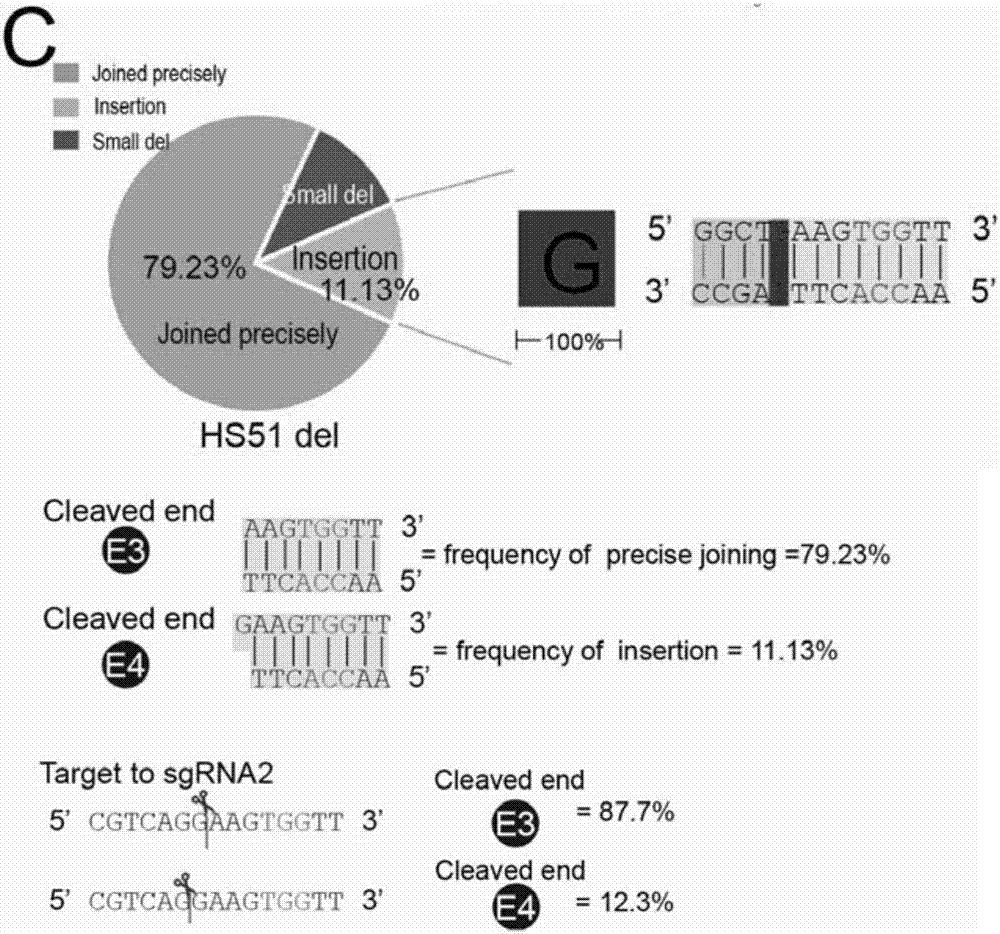
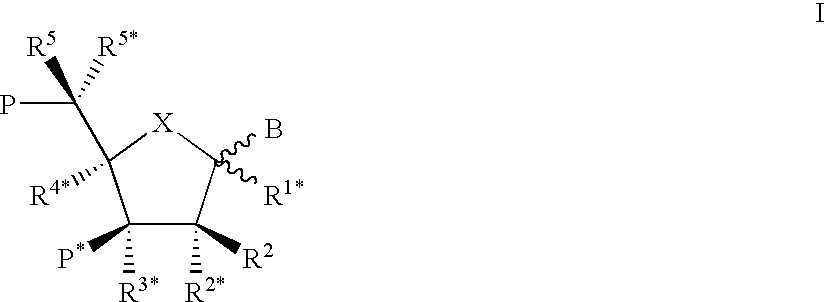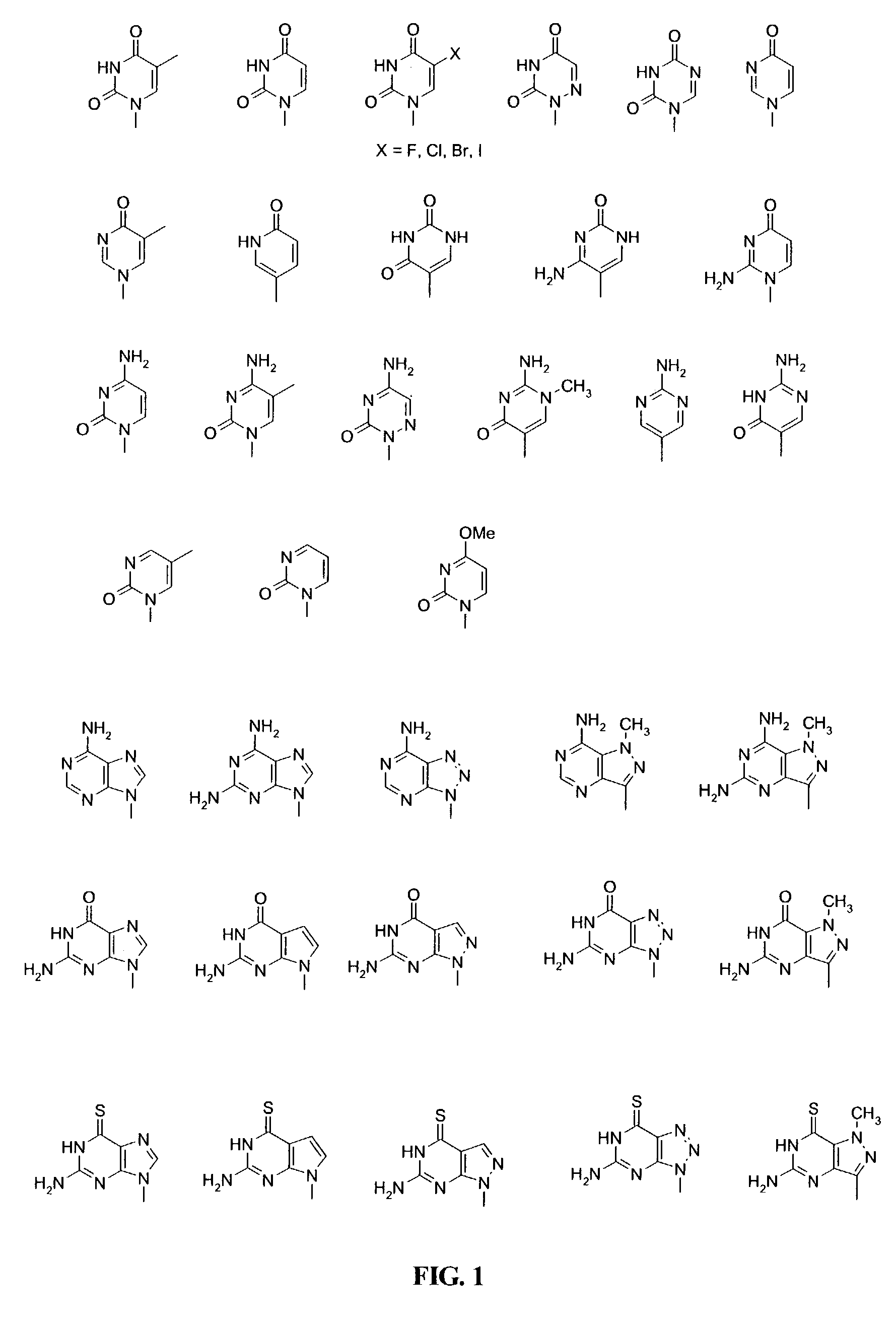Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
1369 results about "Base J" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
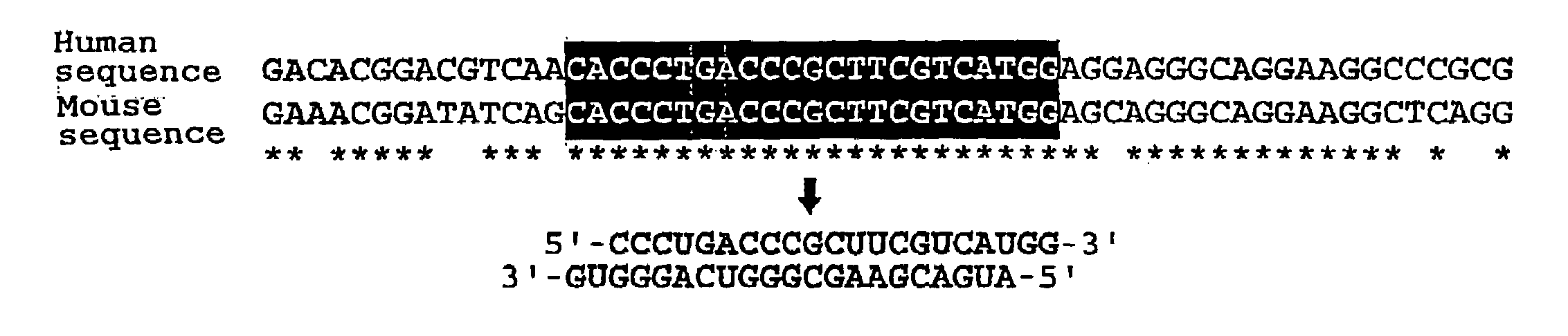
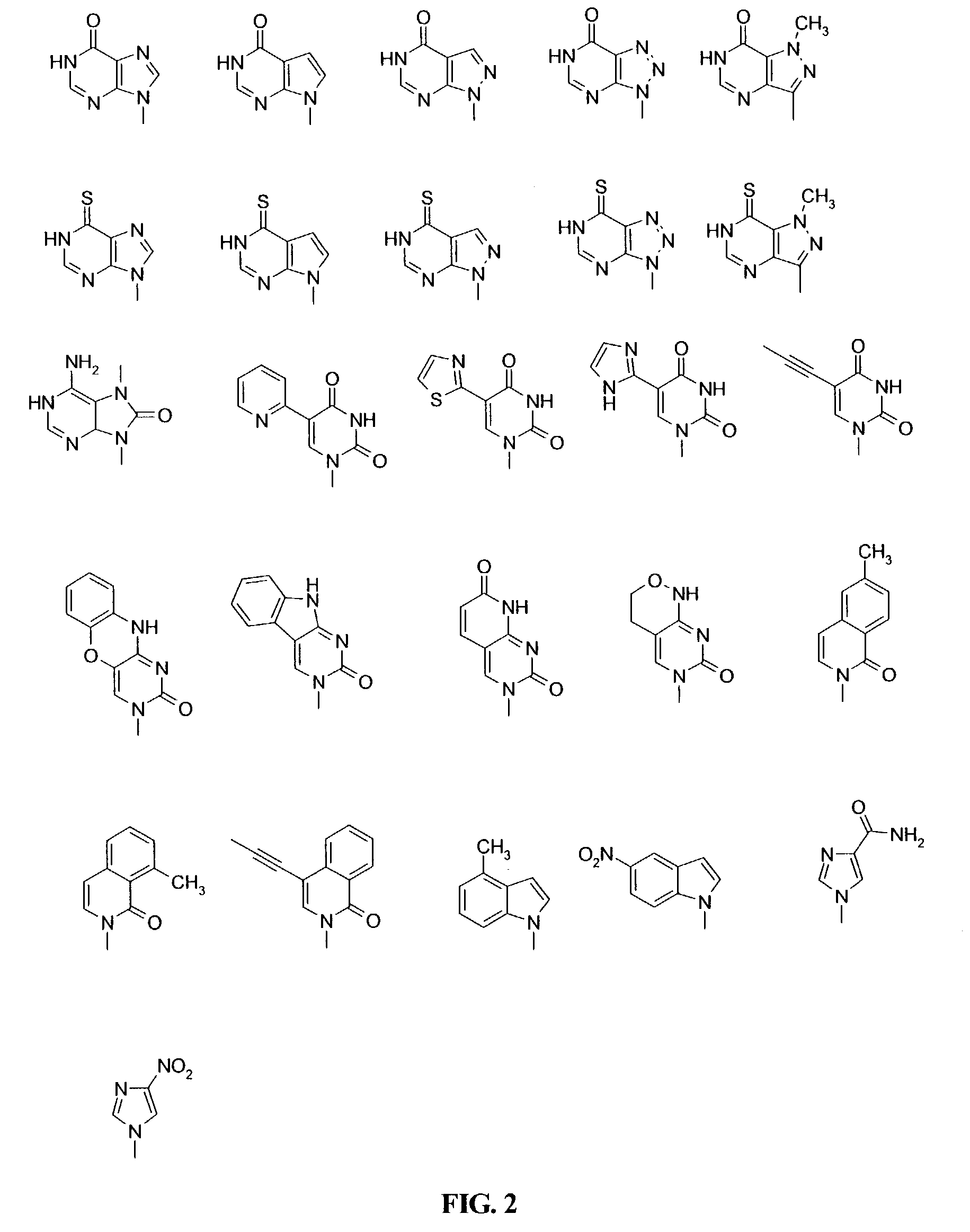
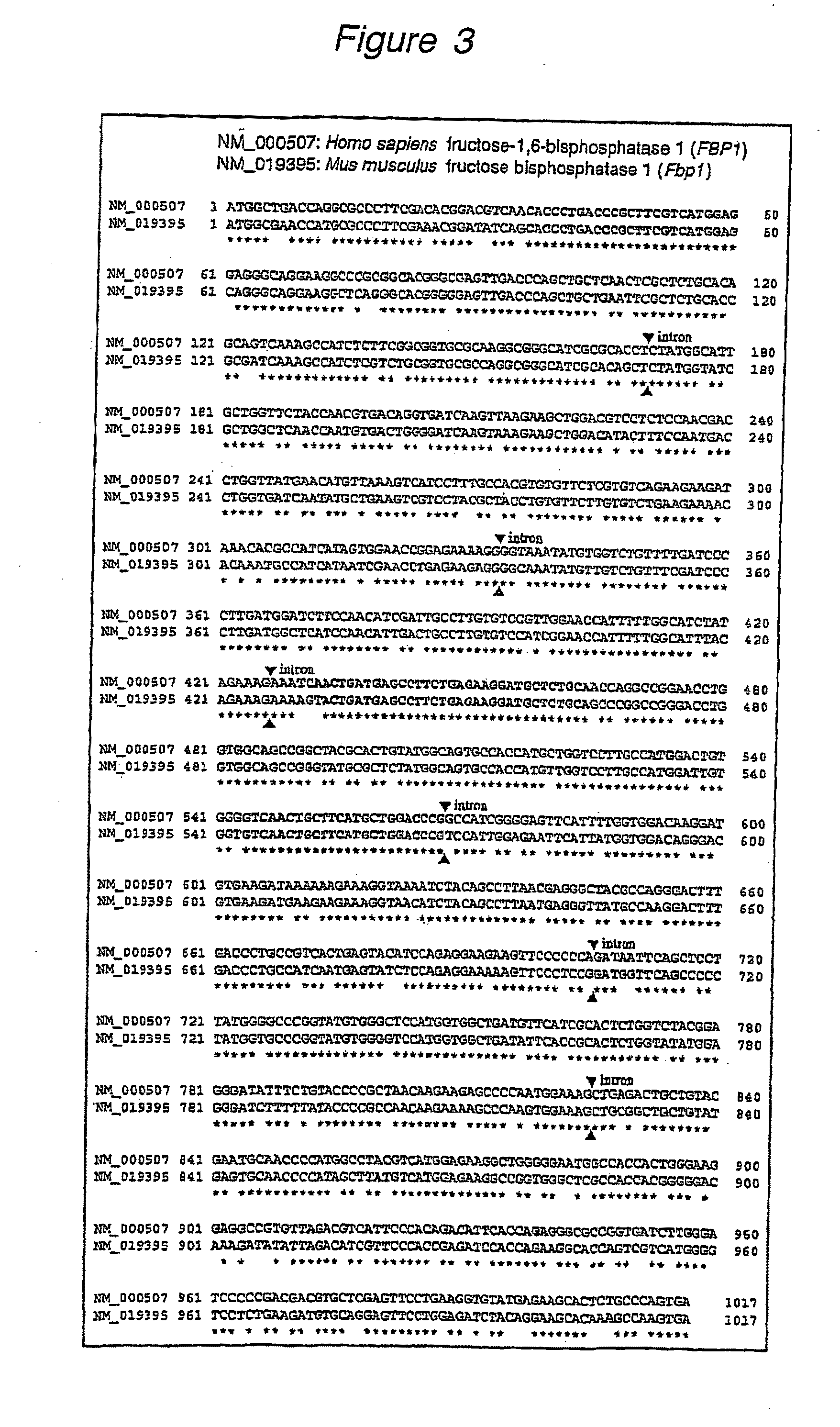
Β-D-Glucopyranosyloxymethyluracil or base J is a hypermodified nucleobase found in the DNA of kinetoplastids including the human pathogenic trypanosomes. It was discovered in 1993, in the trypanosome Trypanosoma brucei and was the first hypermodified nucleobase found in eukaryotic DNA; it has since been found in other kinetoplastids, including Leishmania. Within these organism Base J acts as a RNA polymerase II transcription terminator, with its removal in knockout cells being accompanied by a massive read-through at RNA polymerase II termination sites, which ultimately proves lethal to the cell.
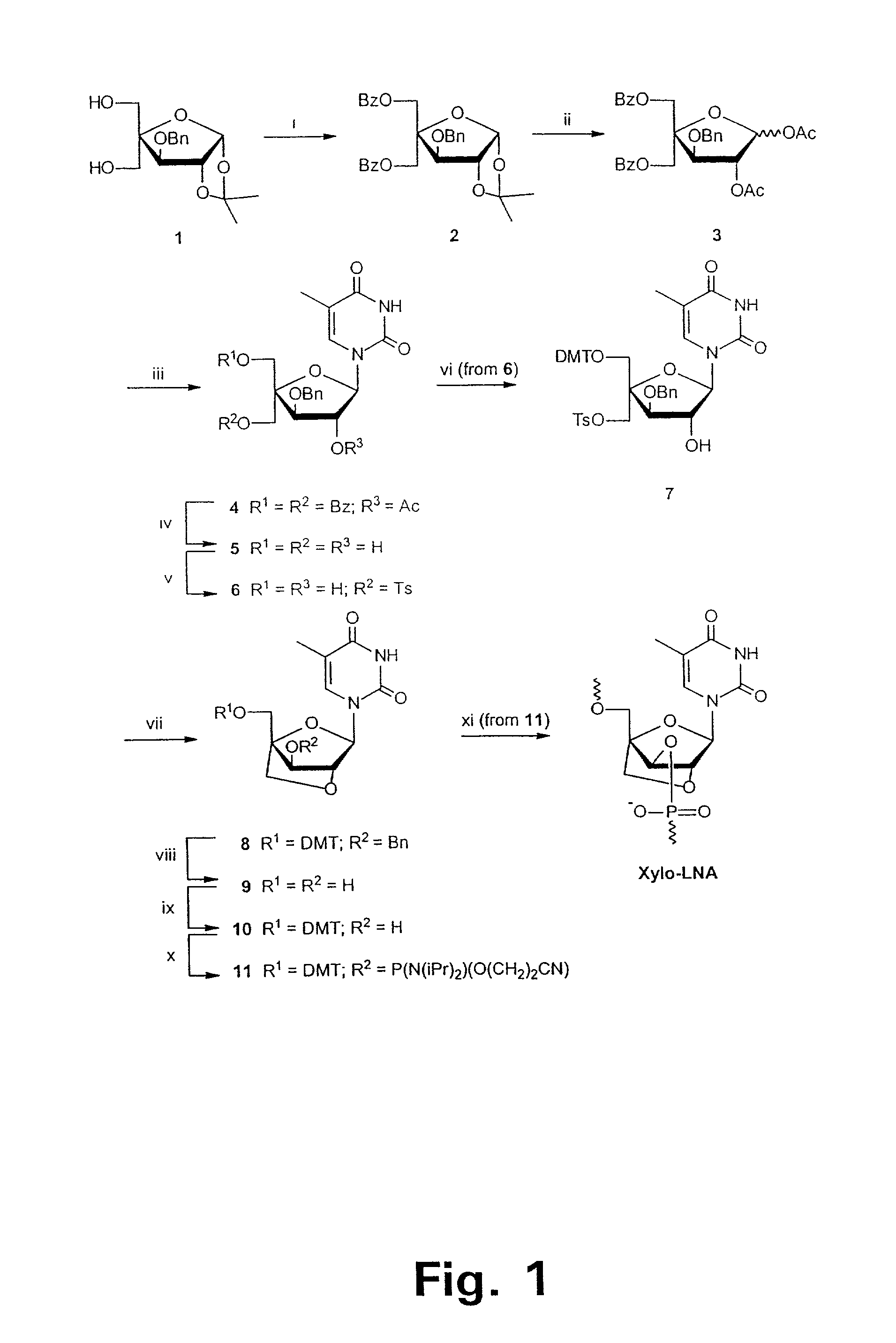
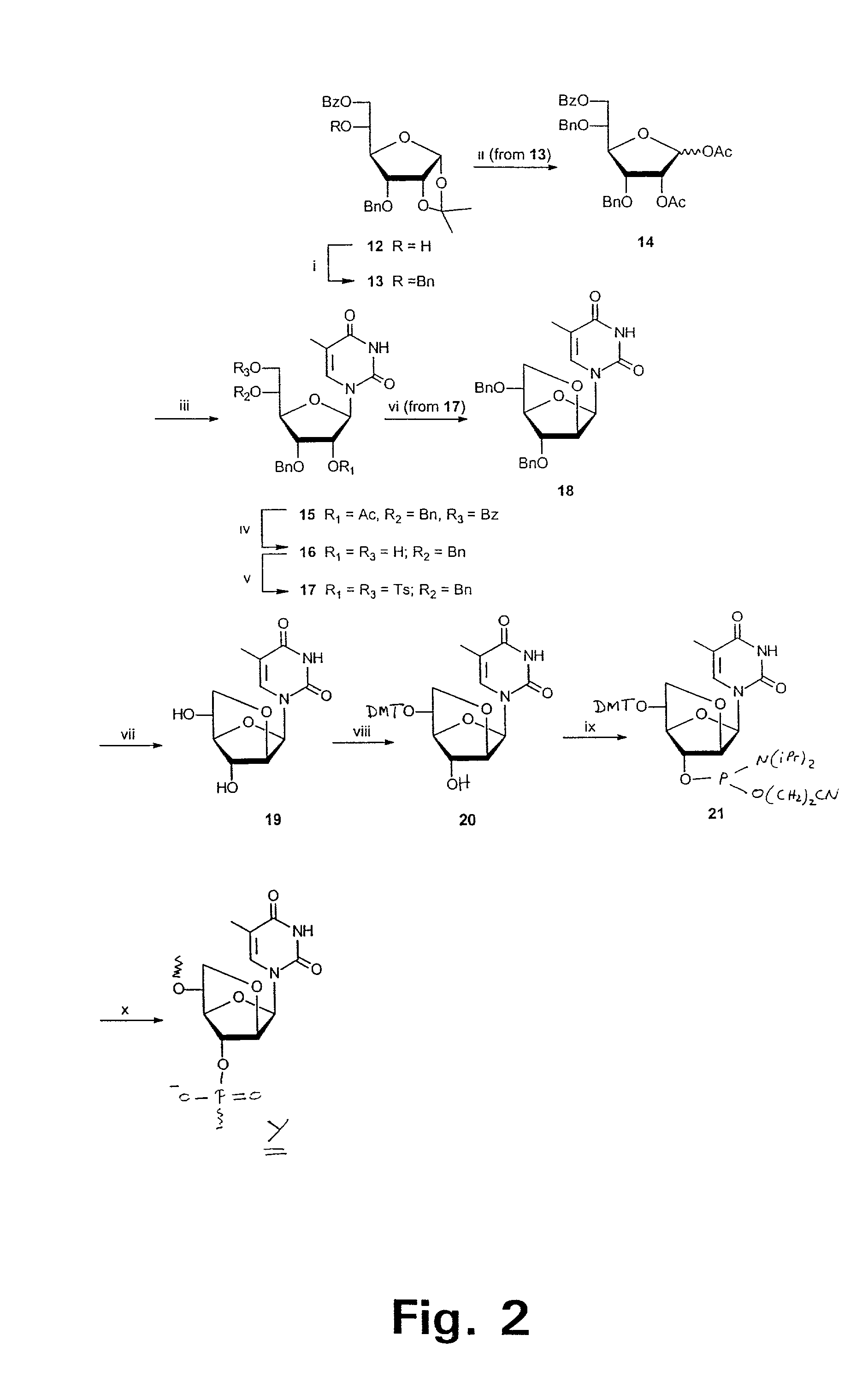
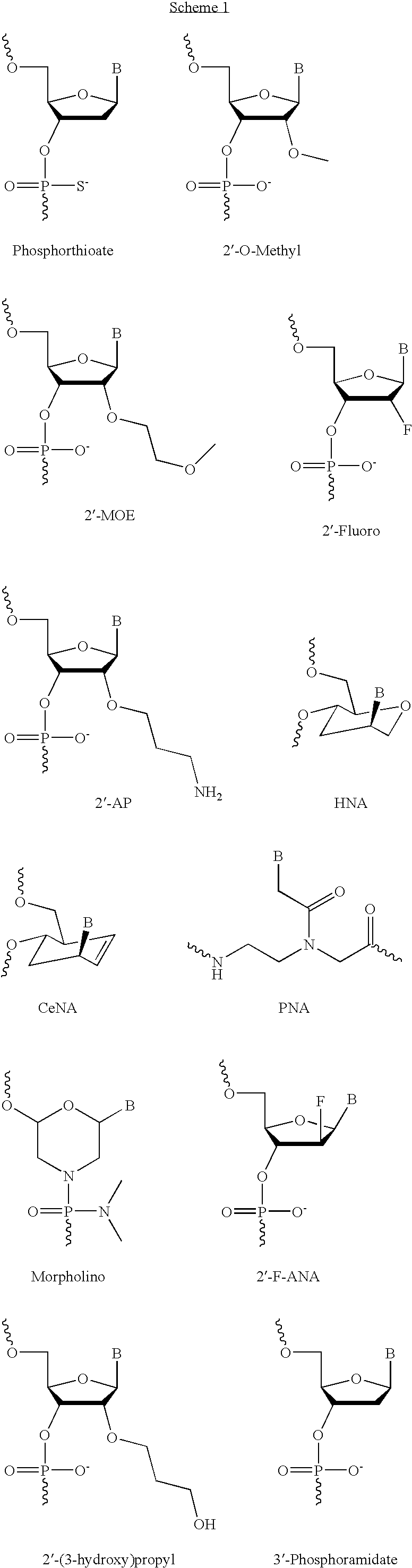
Xylo-LNA analogues
Based on the above and on the remarkable properties of the 2′-O,4′-C-methylene bridged LNA monomers it was decided to synthesise oligonucleotides comprising one or more 2′-O,4′-C-methylene-β-D-xylofuranosyl nucleotide monomer(s) as the first stereoisomer of LNA modified oligonucleotides. Modelling clearly indicated the xylo-LNA monomers to be locked in an N-type furanose conformation. Whereas the parent 2′-deoxy-β-D-xylofuranosyl nucleosides were shown to adopt mainly an N-type furanose conformation, the furanose ring of the 2′-deoxy-β-D-xylofuranosyl monomers present in xylo-DNA were shown by conformational analysis and computer modelling to prefer an S-type conformation thereby minimising steric repulsion between the nucleobase and the 3′-O-phopshate group (Seela, F.; Wömer, Rosemeyer, H. Helv. Chem. Acta 1994, 77, 883). As no report on the hybridisation properties and binding mode of xylo-configurated oligonucleotides in an RNA context was believed to exist, it was the aim to synthesise 2′-O,4′-C-methylene-β-D-xylofuranosyl nucleotide monomer and to study the thermal stability of oligonucleotides comprising this monomer. The results showed that fully modified or almost fully modified Xylo-LNA is useful for high-affinity targeting of complementary nucleic acids. When taking into consideration the inverted stereochemistry at C-3′ this is a surprising fact. It is likely that Xylo-LNA monomers, in a sequence context of Xylo-DNA monomers, should have an affinity-increasing effect.
Owner:QIAGEN GMBH
Polynucleotides for causing RNA interference and method for inhibiting gene expression using the same
InactiveUS20080113351A1High RNA interference effectLittle riskOrganic active ingredientsNervous disorderBase JNucleotide
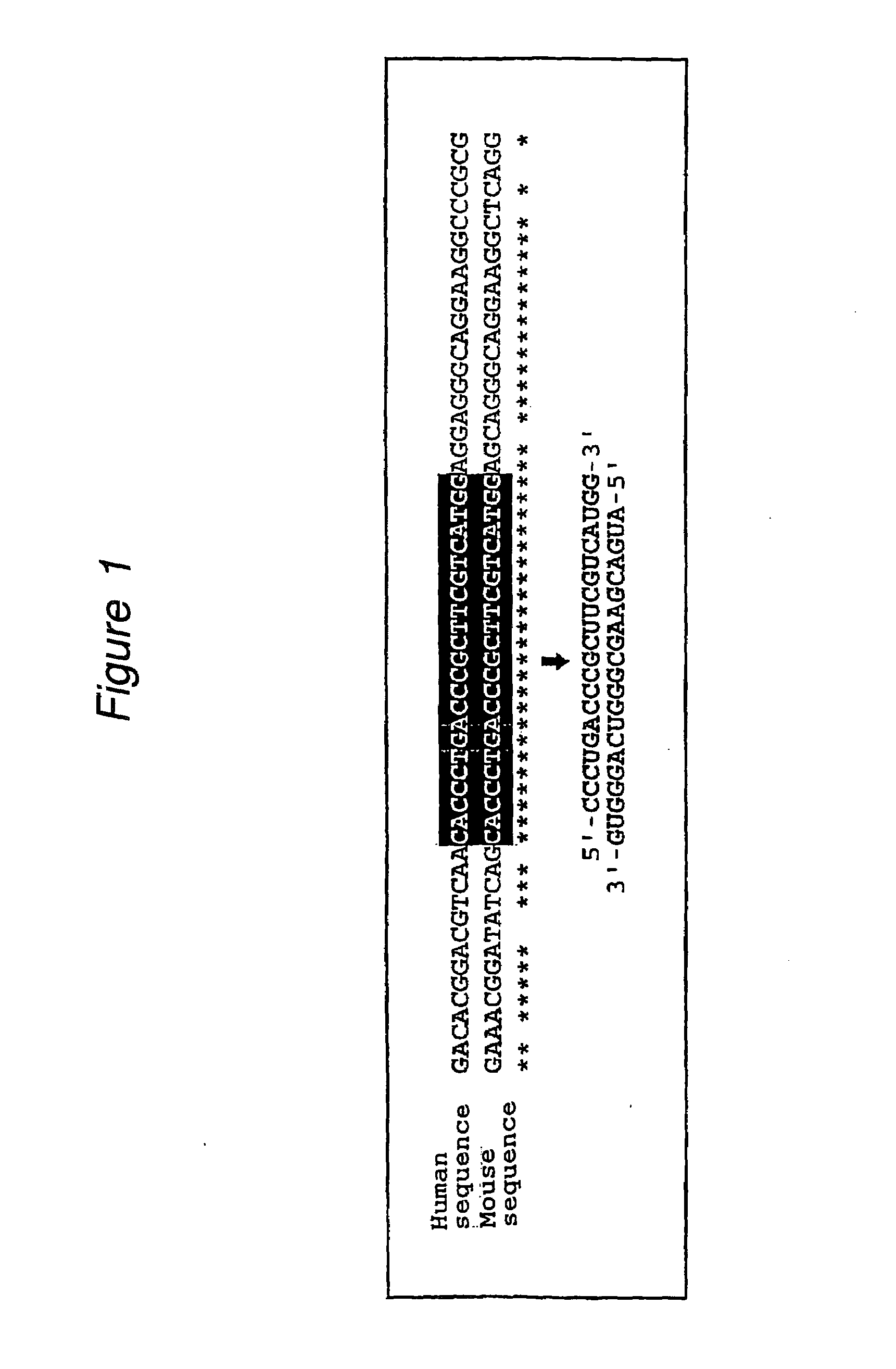
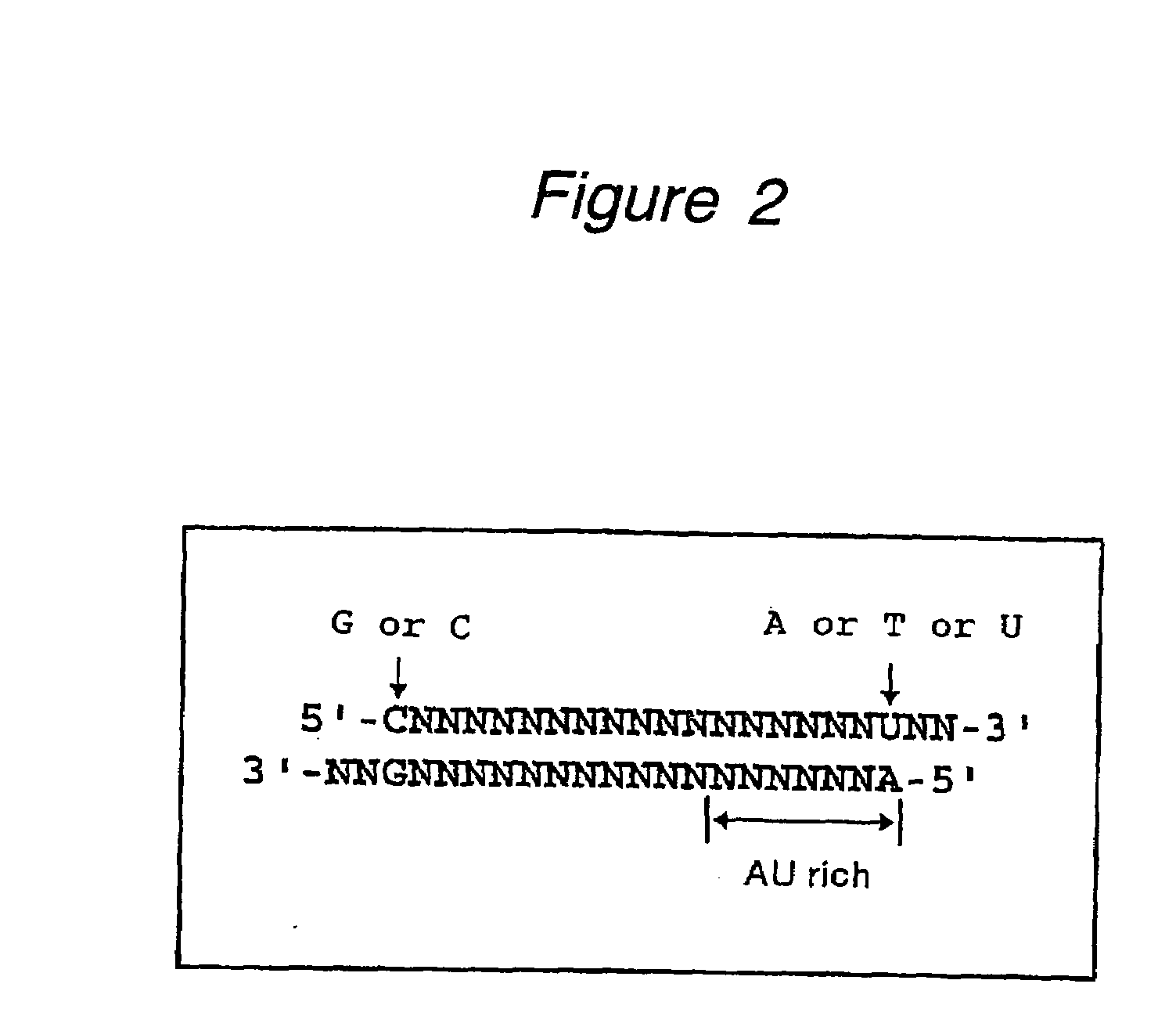
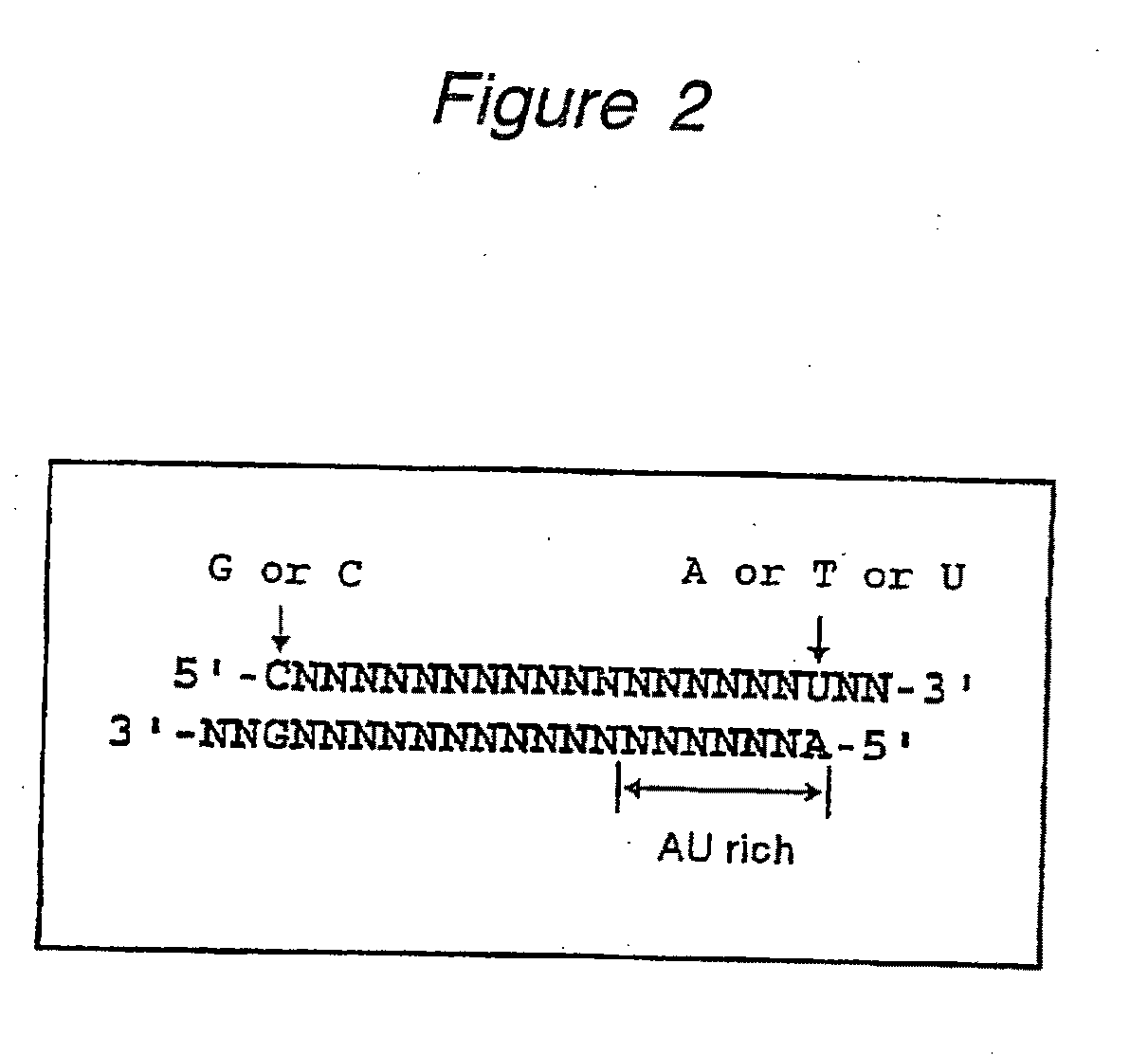
The present invention provides a polynucleotide that not only has a high RNA interference effect on its target gene, but also has a very small risk of causing RNA interference against a gene unrelated to the target gene. A sequence segment conforming to the following rules (a) to (d) is searched from the base sequences of a target gene for RNA interference and, based on the search results, a polynucleotide capable of causing RNAi is designed, synthesized, etc.:(a) The 3′ end base is adenine, thymine, or uracil,(b) The 5′ end base is guanine or cytosine,(c) A 7-base sequence from the 3′ end is rich in one or more types of bases selected from the group consisting of adenine, thymine, and uracil, and(d) The number of bases is within a range that allows RNA interference to occur without causing cytotoxicity.
Owner:ALPHAGEN
Method for producing complex DNA methylation fingerprints
InactiveUS6214556B1Accurately determineSugar derivativesMicrobiological testing/measurementFingerprintGenomic DNA
Method for characterizing, classifying and differentiating tissues and cell types, for predicting the behavior of tissues and groups of cells, and for identifying genes with changed expression. The method involves obtaining genomic DNA from a tissue sample, the genomic DNA subsequently being subjected to shearing, cleaved by means of a restriction endonuclease or not treated by either one of these methods. The base cytosine, but not 5-methylcytosine, from the thus-obtained genomic DNA is then converted into uracil by treatment with a bisulfite solution. Fractions of the thus-treated genomic DNA are then amplified using either very short or degenerated oligonucleotides or oligonuclcotides which are complementary to adaptor oligonucleotides that have been ligated to the ends of the cleaved DNA. The quantity of the remaining cytosines on the guanine-rich DNA strand and / or the quantity of guanines on the cytosine-rich DNA strand from the amplified fractions are then detected by hybridization or polymerase reaction, which quantities are such that the data generated thereby and automatically applied to a processing algorithm allow the drawing of conclusions concerning the phenotype of the sample material.
Owner:EPIGENOMICS AG
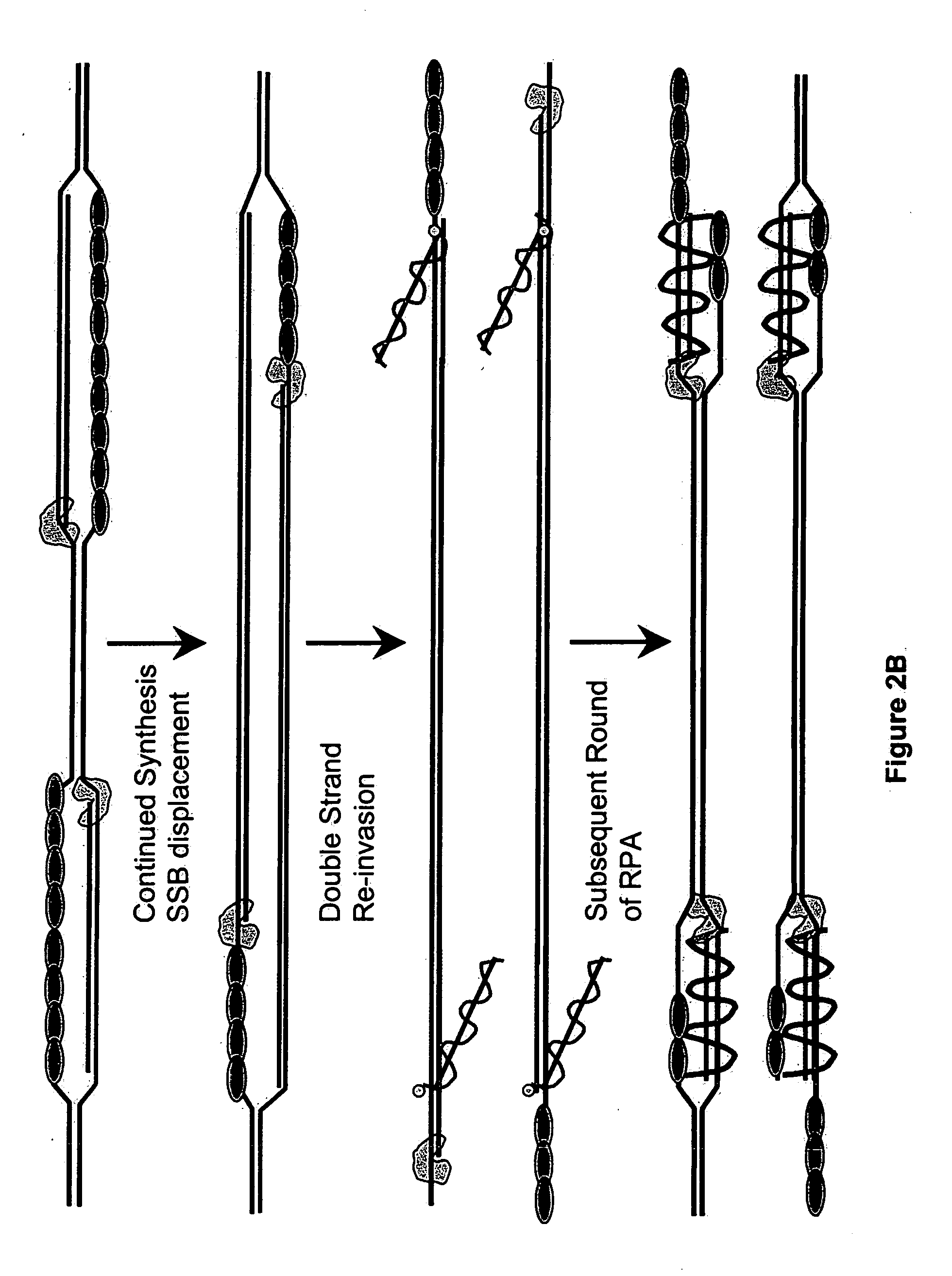
Recombinase polymerase amplification
ActiveUS20050112631A1HydrolasesMicrobiological testing/measurementRecombinase Polymerase AmplificationSingle strand
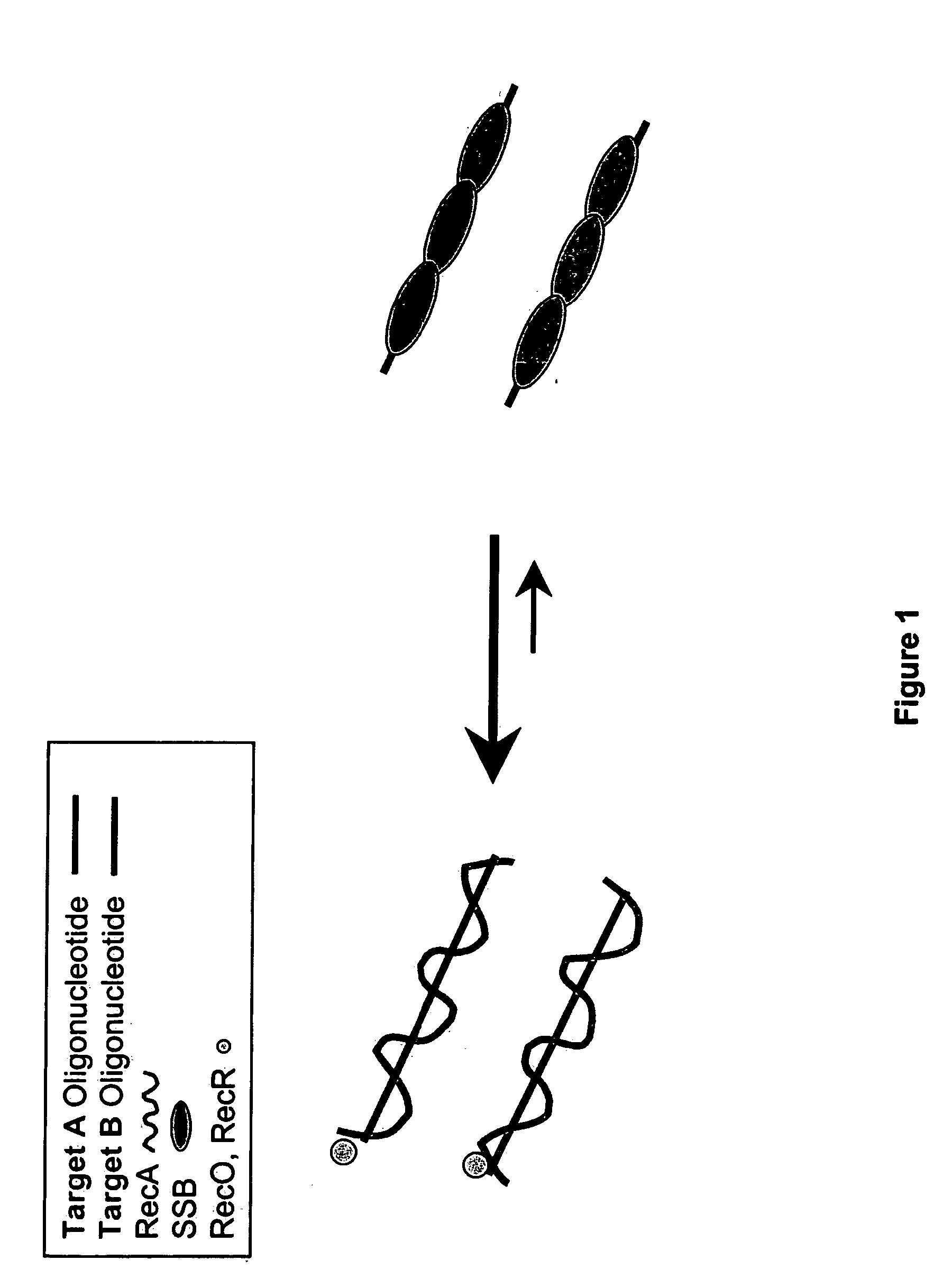
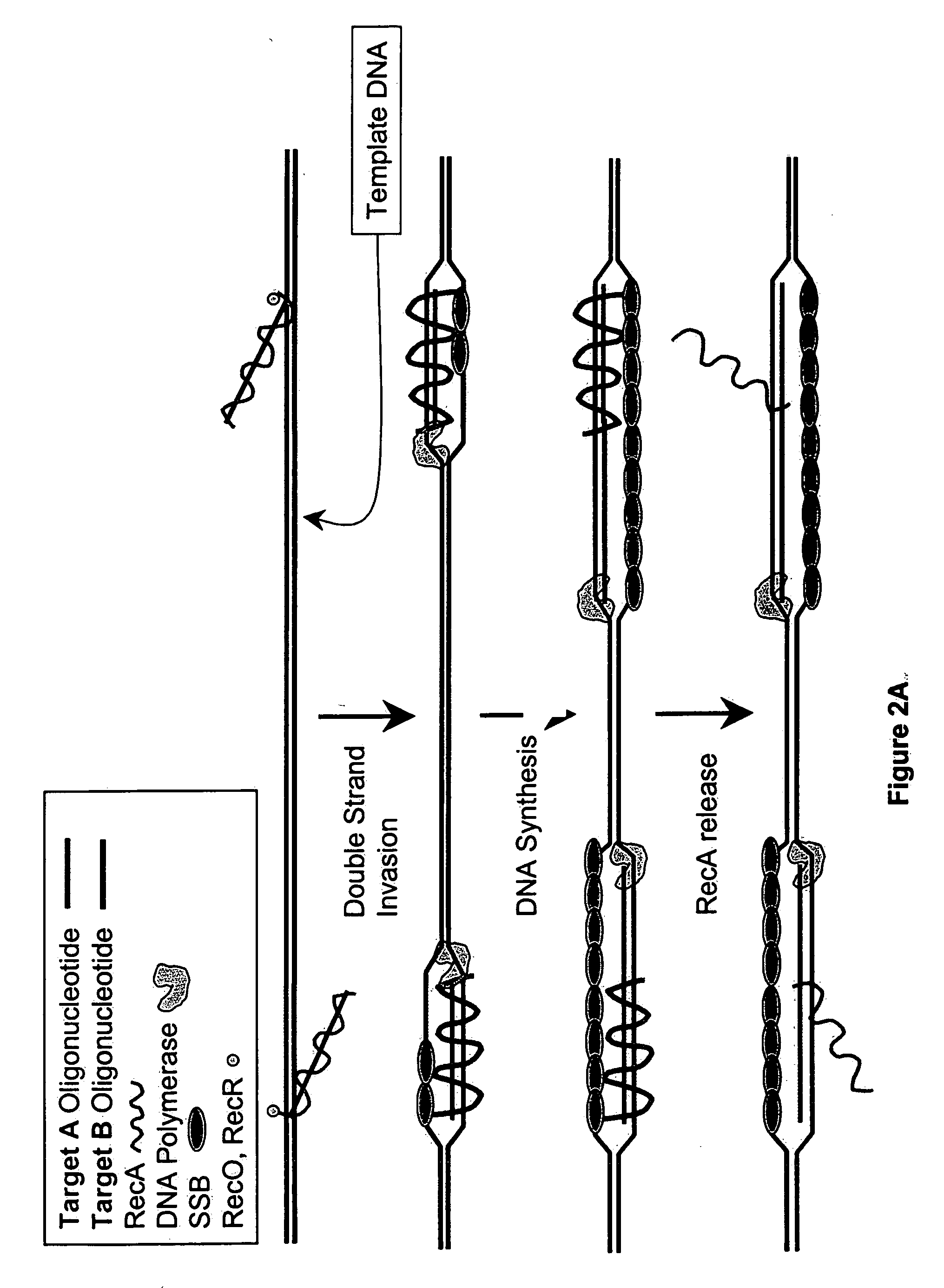
This disclosure describe three related novel methods for Recombinase-Polymerase Amplification (RPA) of a target DNA that exploit the properties of recombinase and related proteins, to invade double-stranded DNA with single stranded homologous DNA permitting sequence specific priming of DNA polymerase reactions. The disclosed methods have the advantage of not requiring thermocycling or thermophilic enzymes. Further, the improved processivity of the disclosed methods may allow amplification of DNA up to hundreds of megabases in length.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
PNA monomer and precursor
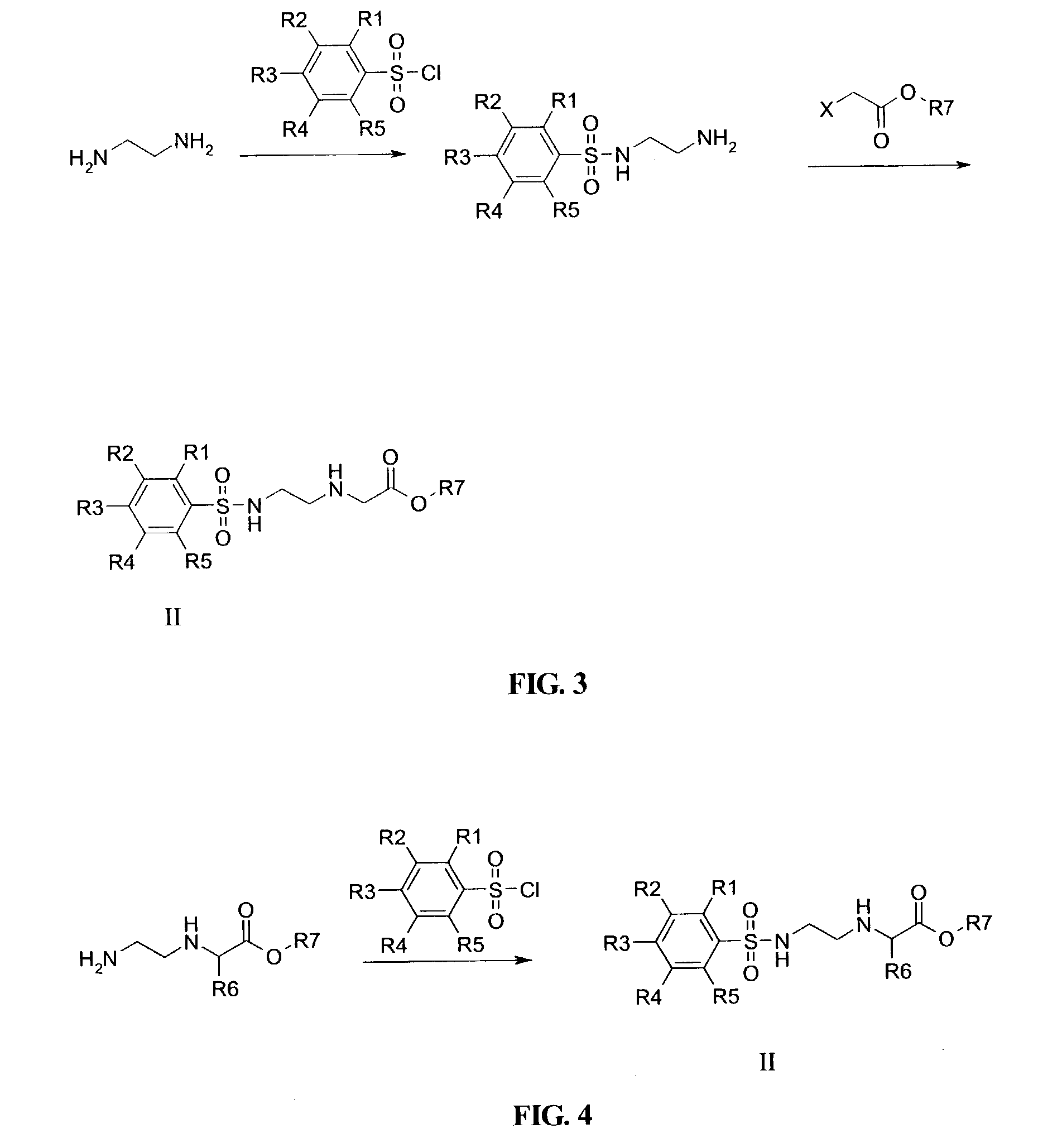
InactiveUS7022851B2Improve efficiencyImprove convenienceSugar derivativesPeptide/protein ingredientsBase JOligomer
This application relates to monomers of the general formula (I) for the preparation of PNA (peptide nucleic acid) oligomers and provides method for the synthesis of both predefined sequence PNA oligomers and random sequence PNA oligomers: whereinR1, R2, R3, R4, R5 is independently H, halogen, C1–C4 alkyl, nitro, nitrile, C1–C4 alkoxy, halogenated C1–C4 alkyl, or halogenated C1–C4 alkoxy, wherein at least one of R1, R3, and R5 is nitro;R6 is H or protected or unprotected side chain of natural or unnatural α-amino acid; andB is a natural or unnatural nucleobase, wherein when said nucleobase has an exocyclic amino function, said function is protected by protecting group which is labile to acids but stable to weak to medium bases.
Owner:PANAGENE INC
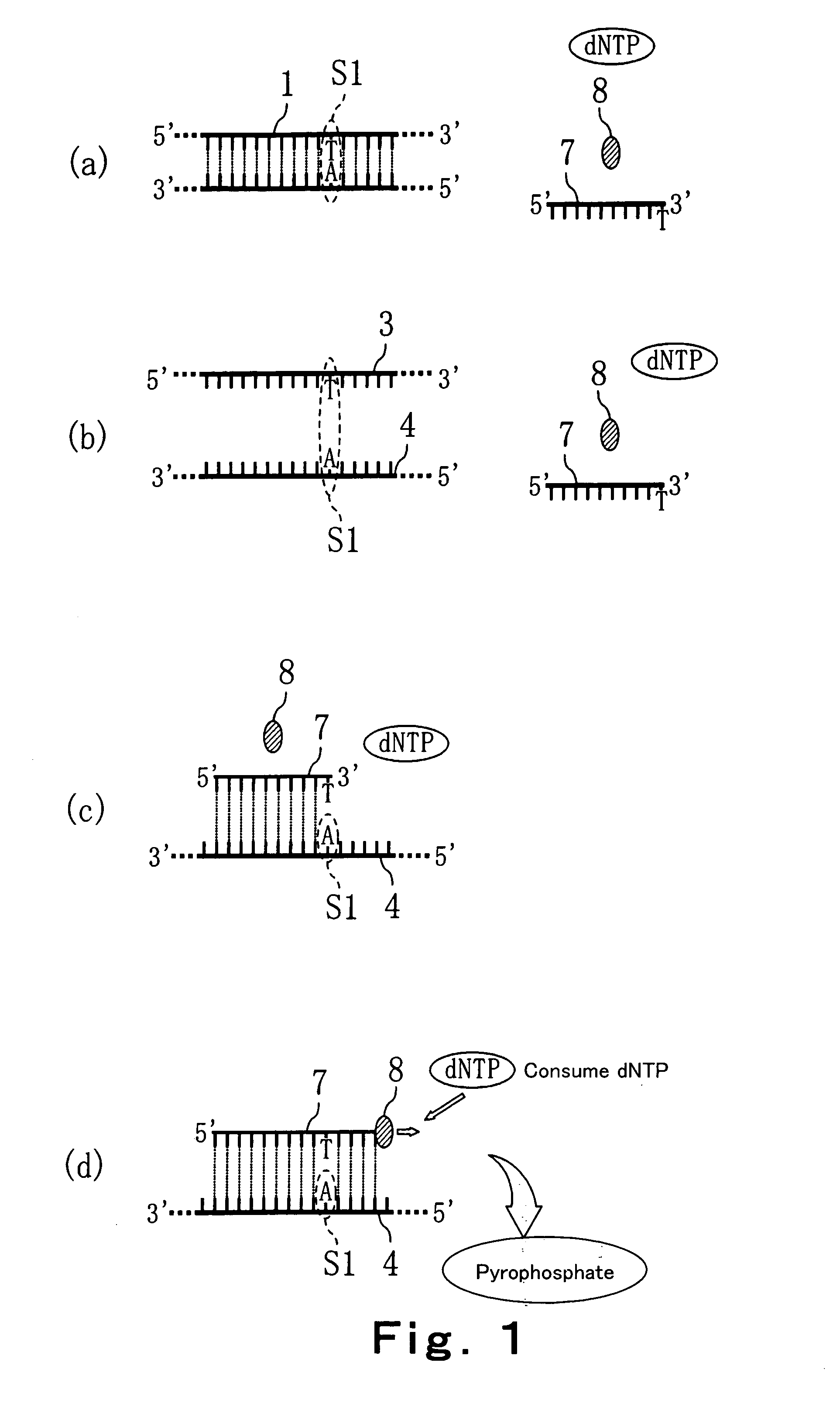
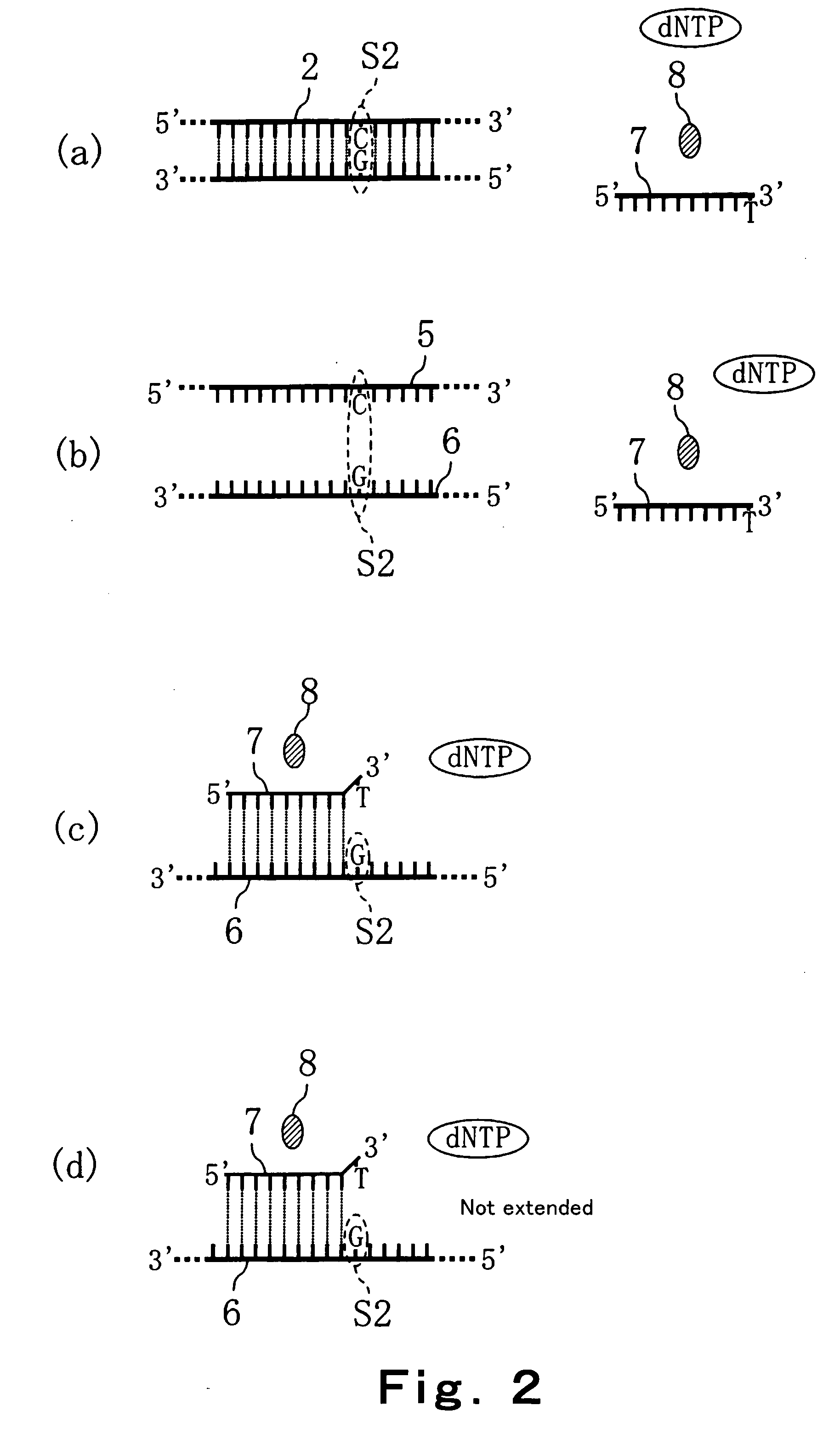
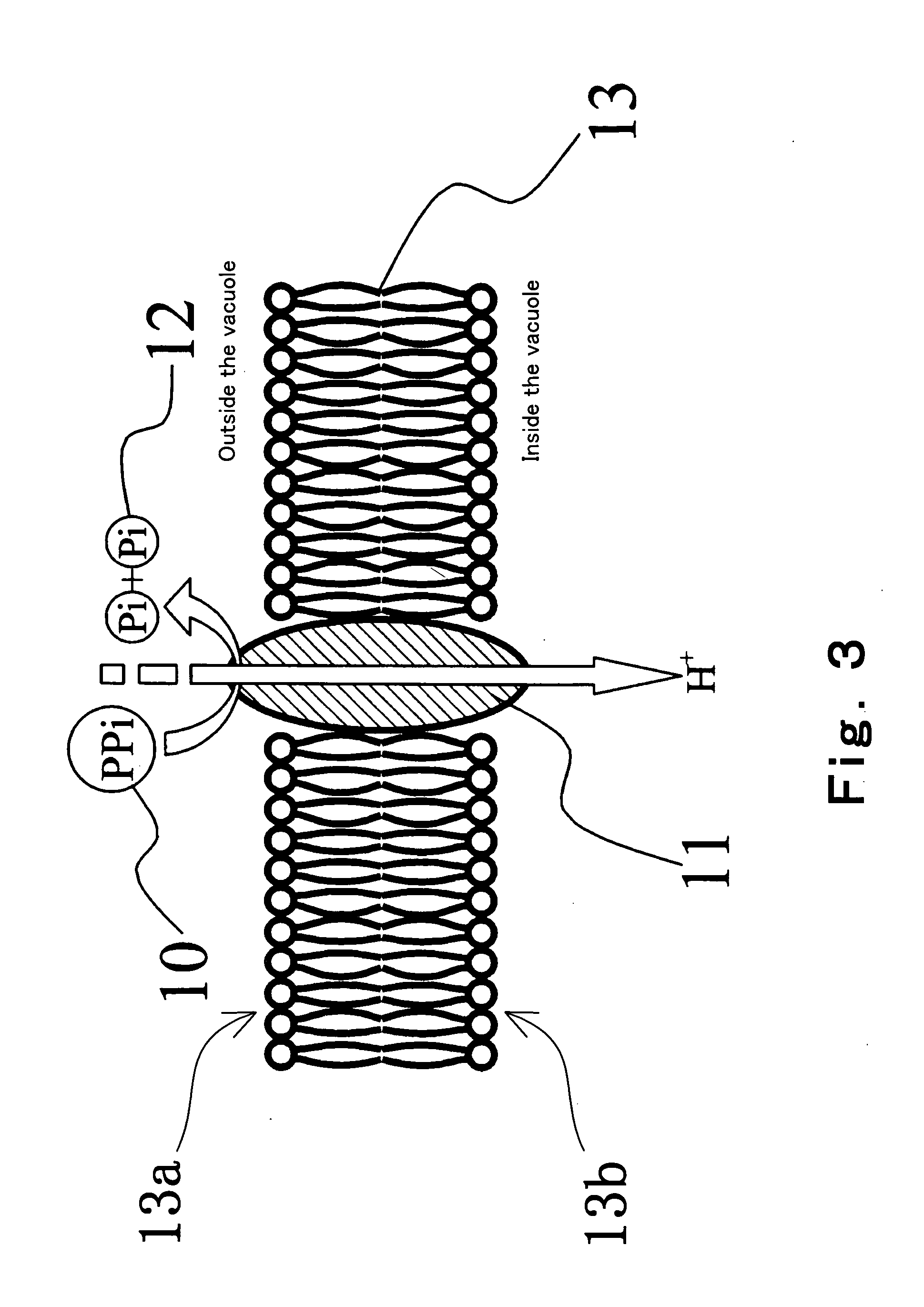
Method of detecting primer extension reaction, method of discriminating base type, device for discriminating base type, device for detecting pyrophosphate, method of detecting nucleic acid and tip for introducing sample solution
InactiveUS20050032075A1Decrease of H+ concentrationImprove concentrationMicrobiological testing/measurementMaterial analysisBase JNucleotide
Convenient techniques for discriminating the base type in a base sequence of a nucleic acid are provided. The technique includes the step (a) of preparing a sample solution containing a nucleic acid, a primer having a base sequence that includes a complementary binding region which complementarily binds to the nucleic acid, and a nucleotide; the step (b) of allowing the sample solution to stand under a condition to cause an extension reaction of the primer, and producing pyrophosphate when the extension reaction is caused; the step (c) of bringing the sample solution into contact with the front face of a H+ hardly permeable membrane having H+-pyrophosphatase, which penetrates from front to back of the membrane, of which active site that hydrolyzes pyrophosphate being exposed to the front face; the step (d) of measuring the H+ concentration of at least either one of the solution at the front face side of the H+ hardly permeable membrane or the solution at the back face side of the H+ hardly permeable membrane, in a state where the H+-pyrophosphatase is immersed in the solution; the step (e) of detecting the extension reaction on the basis of the result of measurement in the step (d) ; and the step (f) of discriminating the base type in the base sequence of the nucleic acid on the basis of the result of detection in the step (e).
Owner:PANASONIC CORP
Pyrophosphorolysis and incorporation of nucleotide method for nucleic acid detection
InactiveUS7090975B2Useful in detectionSugar derivativesMicrobiological testing/measurementDepolymerizationNucleic acid detection
Processes are disclosed using the depolymerization of a nucleic acid hybrid and incorporation of a suitable nucleotide to qualitatively and quantitatively analyze for the presence of predetermined nucleic acid target sequences. Applications of those processes include the detection of single nucleotide polymorphisms, identification of single base changes, genotyping, medical marker diagnostics, mirosequencing, and.
Owner:PROMEGA
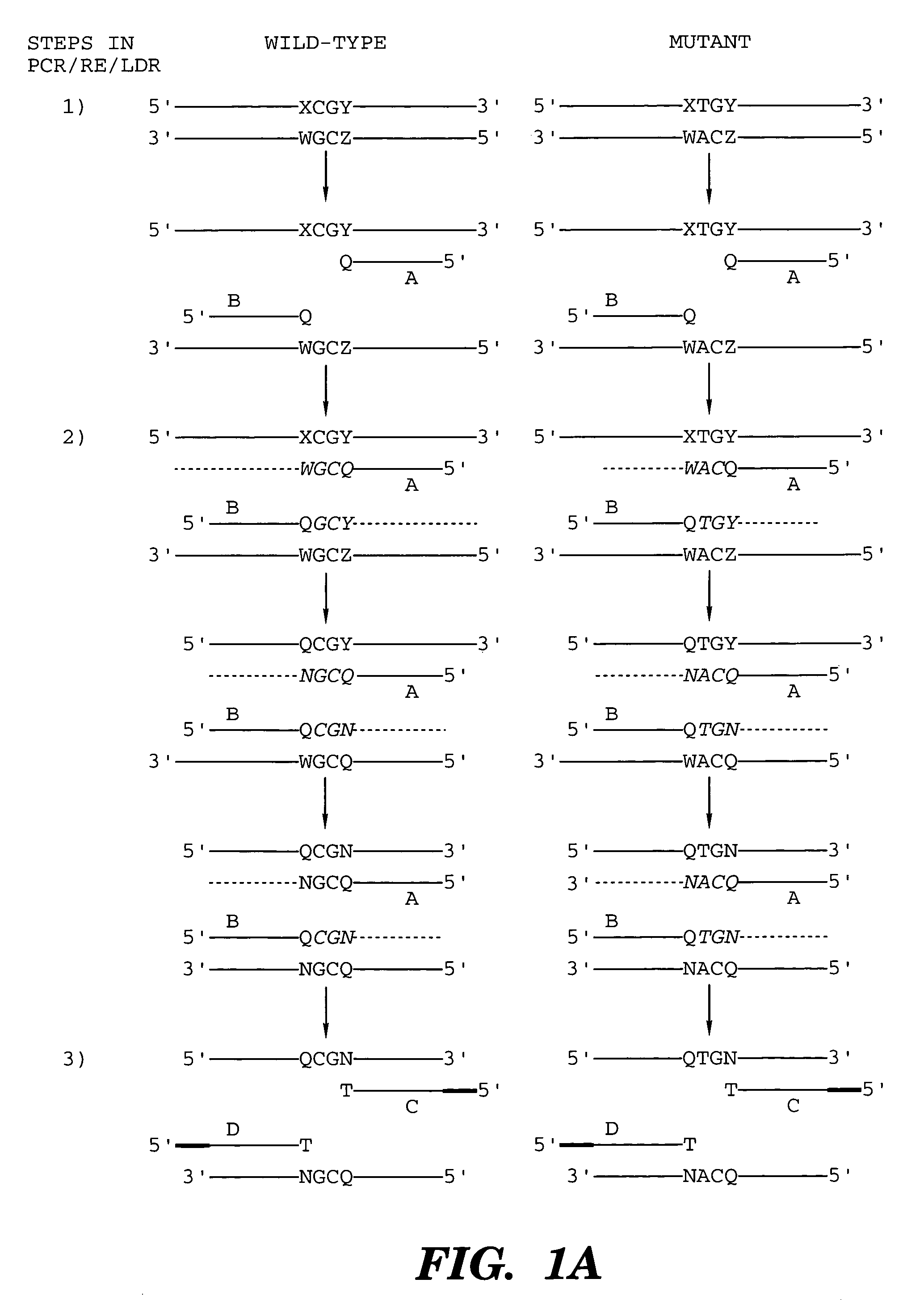
Coupled polymerase chain reaction-restriction-endonuclease digestion-ligase detection reaction process
InactiveUS7014994B1Sensitive highOptimizationSugar derivativesMicrobiological testing/measurementNucleotideWild type
The present invention provides a method for identifying one or more low abundance sequences differing by one or more single-base changes, insertions, or deletions, from a high abundance sequence in a plurality of target nucleotide sequences. The high abundance wild-type sequence is selectively removed using high fidelity polymerase chain reaction analog conversion, facilitated by optimal buffer conditions, to create a restriction endonuclease site in the high abundance wild-type gene, but not in the low abundance mutant gene. This allows for digestion of the high abundance DNA. Subsequently the low abundant mutant DNA is amplified and detected by the ligase detection reaction assay. The present invention also relates to a kit for carrying out this procedure.
Owner:LOUISIANA STATE UNIV +1
Methods for identifying target nucleic acid molecules
InactiveUS20050266417A1Rapid and accurate hybridizationReduce manufacturing costMicrobiological testing/measurementAgainst vector-borne diseasesSplice Site SNPTarget mrna
The present invention relates to methods for identifying target nucleic acid molecules differing by one or more single-base changes, insertions, deletions, or translocations; and identifying one or more target mRNA molecules differing by one or more splice site variations in a plurality of mRNA molecules. Also disclosed is a method of generating a linearly amplified representation of a whole genome. Other aspects of the present invention relate to labeled detection oligonucleotide probes and translational oligonucleotide probes as well as to methods of designing such probes.
Owner:CORNELL RES FOUNDATION INC
Oligomeric compounds for the modulation of Bcl-2
InactiveUS20050203042A1Comparable and enhanced biological effectInteresting biological propertiesBiocideOrganic active ingredientsDiseaseNucleobase
The present invention provides improved oligomeric compound, in particular oligonucleotide compounds, and methods for modulating the expression of the Bcl-2 gene in humans. In particular, this invention relates to oligomeric compounds of 10-30 nucleobases in length which comprise a target binding domain that is specifically hybridizable to a region ranging from base position No. 1459 (5′) to No. 1476 (3′) of the human Bcl-2 mRNA, said target binding domain having the formula: 5′-[(DNA / RNA)0-1-(LNA / LNA*)2-7-(DNA / RNA / LNA*)4-14-(LNA / LNA*)2-7-(DNA / RNA)0-1]-3 and said target binding domain comprising at least two LNA nucleotides or LNA analogue nucleotides linked by a phosphorothioate group (—O—P(O,S)—O—). In particular the oligo is predominantly or fully thiolated. The invention also provides the use of such oligomers or conjugates or chimera for the treatment of various diseases associated with the expression of the Bcl-2 gene, such as cancer.
Owner:SANTARIS PHARMA AS
Polynucleotides for causing RNA interference and method for inhibiting gene expression using the same
InactiveUS20110054005A1High RNA interference effectLittle riskOrganic active ingredientsNervous disorderBase JNucleotide
The present invention provides a polynucleotide that not only has a high RNA interference effect on its target gene, but also has a very small risk of causing RNA interference against a gene unrelated to the target gene. A sequence segment conforming to the following rules (a) to (d) is searched from the base sequences of a target gene for RNA interference and, based on the search results, a polynucleotide capable of causing RNAi is designed, synthesized, etc.:(a) The 3′ end base is adenine, thymine, or uracil,(b) The 5′ end base is guanine or cytosine,(c) A 7-base sequence from the 3′ end is rich in one or more types of bases selected from the group consisting of adenine, thymine, and uracil, and(d) The number of bases is within a range that allows RNA interference to occur without causing cytotoxicity.
Owner:BIO THINKTANK
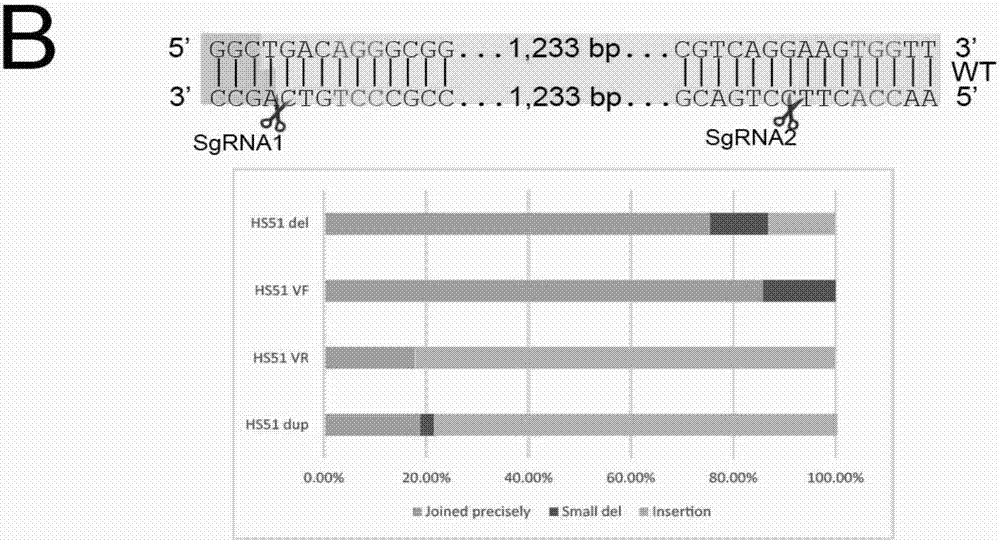
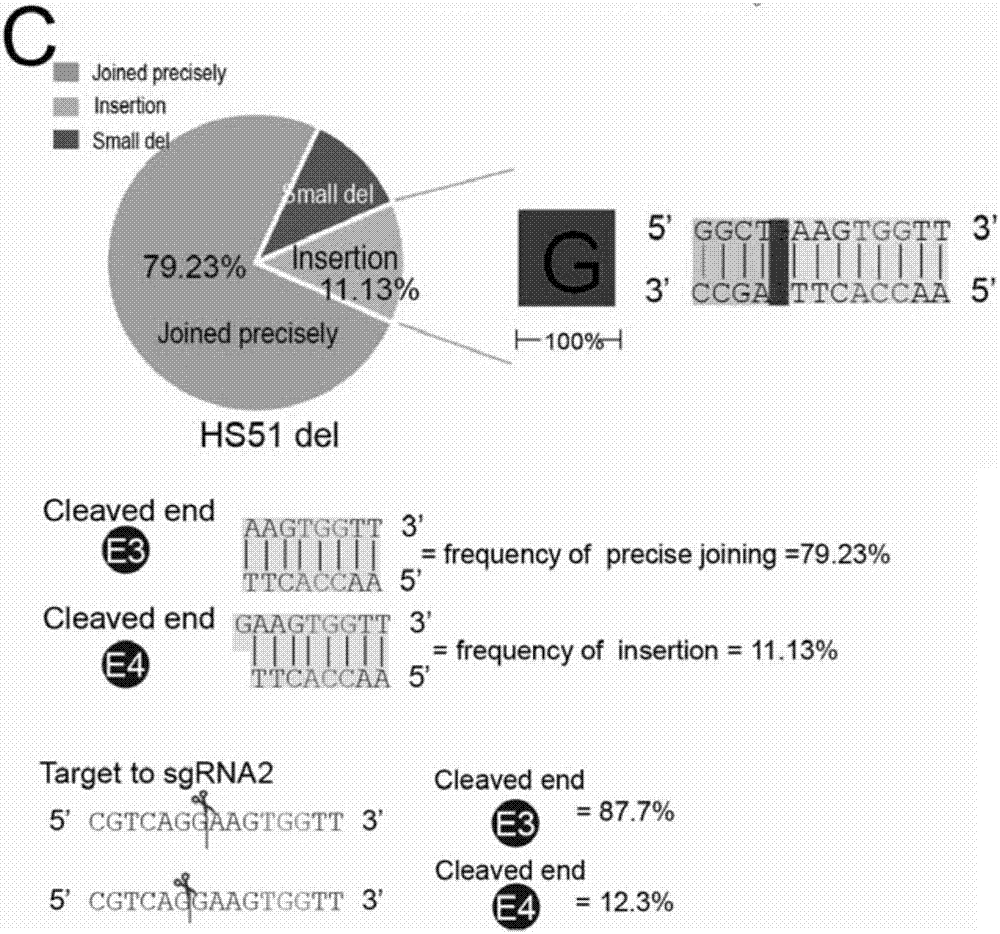
Method for knocking off animal myostatin gene by using CRISPR-Cas9 system
InactiveCN104531705AThe identification rules are simpleEasy to operateMicroinjection basedVector-based foreign material introductionBiotechnologyTranscription initiation site
The invention provides a method for knocking off an animal myostatin gene by using a CRISPR-Cas9 system. The method comprises the following steps: firstly, acquiring a DNA sequence aiming at an sgRNA recognition area of a second myostatin exon, wherein the base sequence of the DNA sequence is as shown in SEQ ID NO.1; secondly, establishing an sgRNA expression structure of the second myostatin exon, inserting a T7 starter before an sgRNA transcriptional start site, establishing an in-vitro transcription carrier of Cas9 protein, and regulating and controlling by using the T7 starter. Cas9 mRNA and sgRNA are obtained through the in-vitro transcription carrier of Cas9 and sgRNA, and the method can be used for knocking off the animal myostatin gene.
Owner:CHINA AGRI UNIV
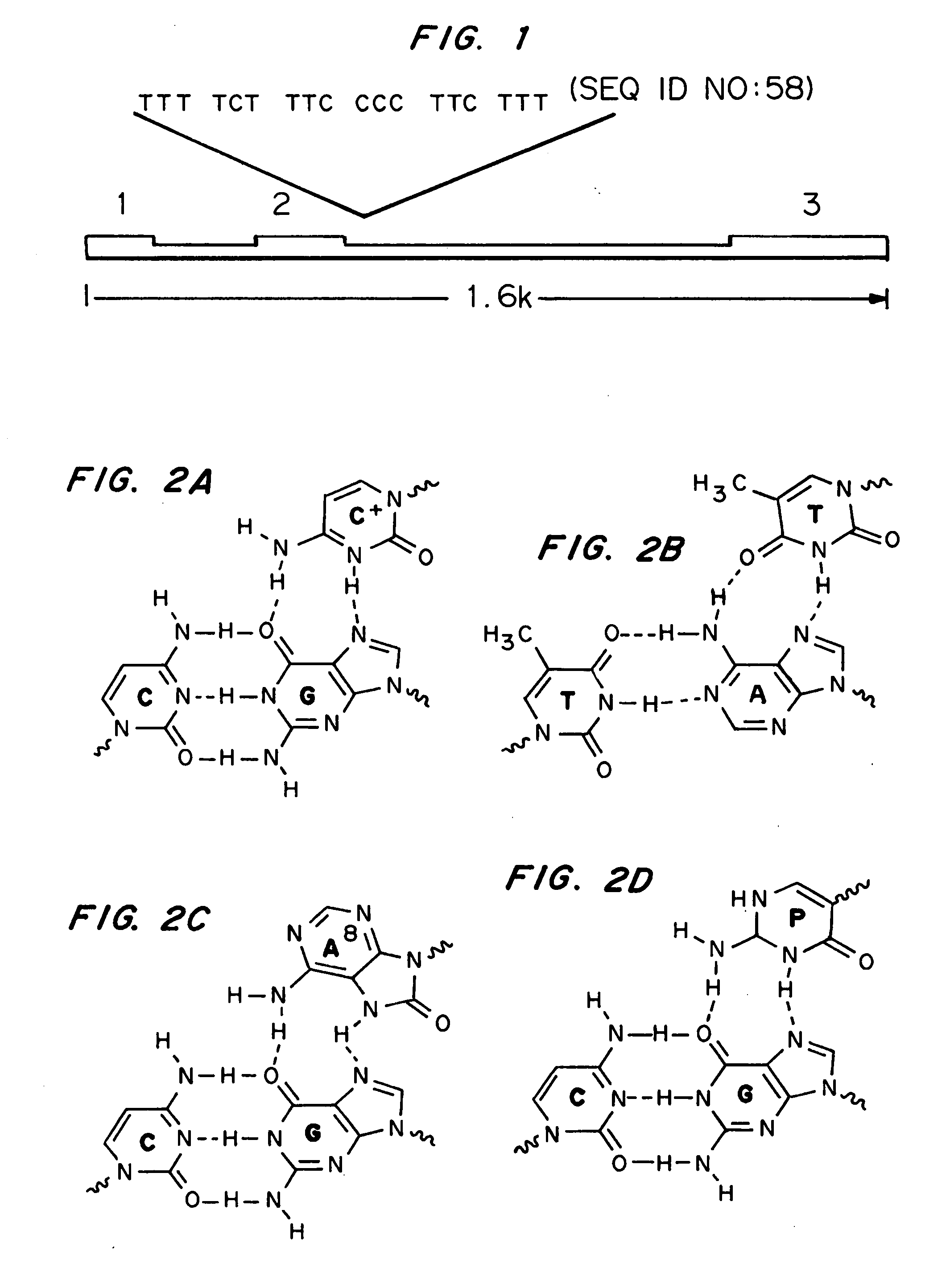
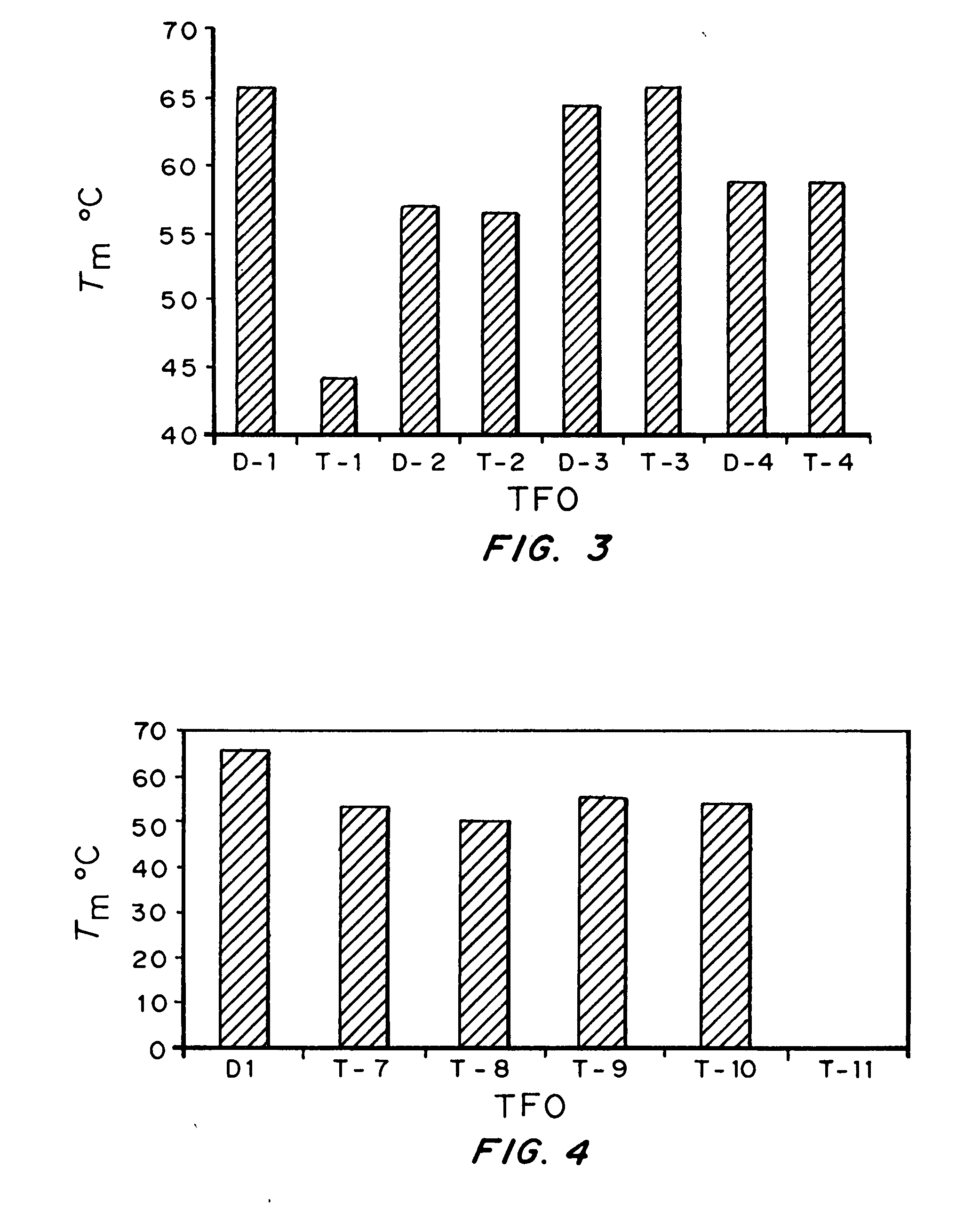
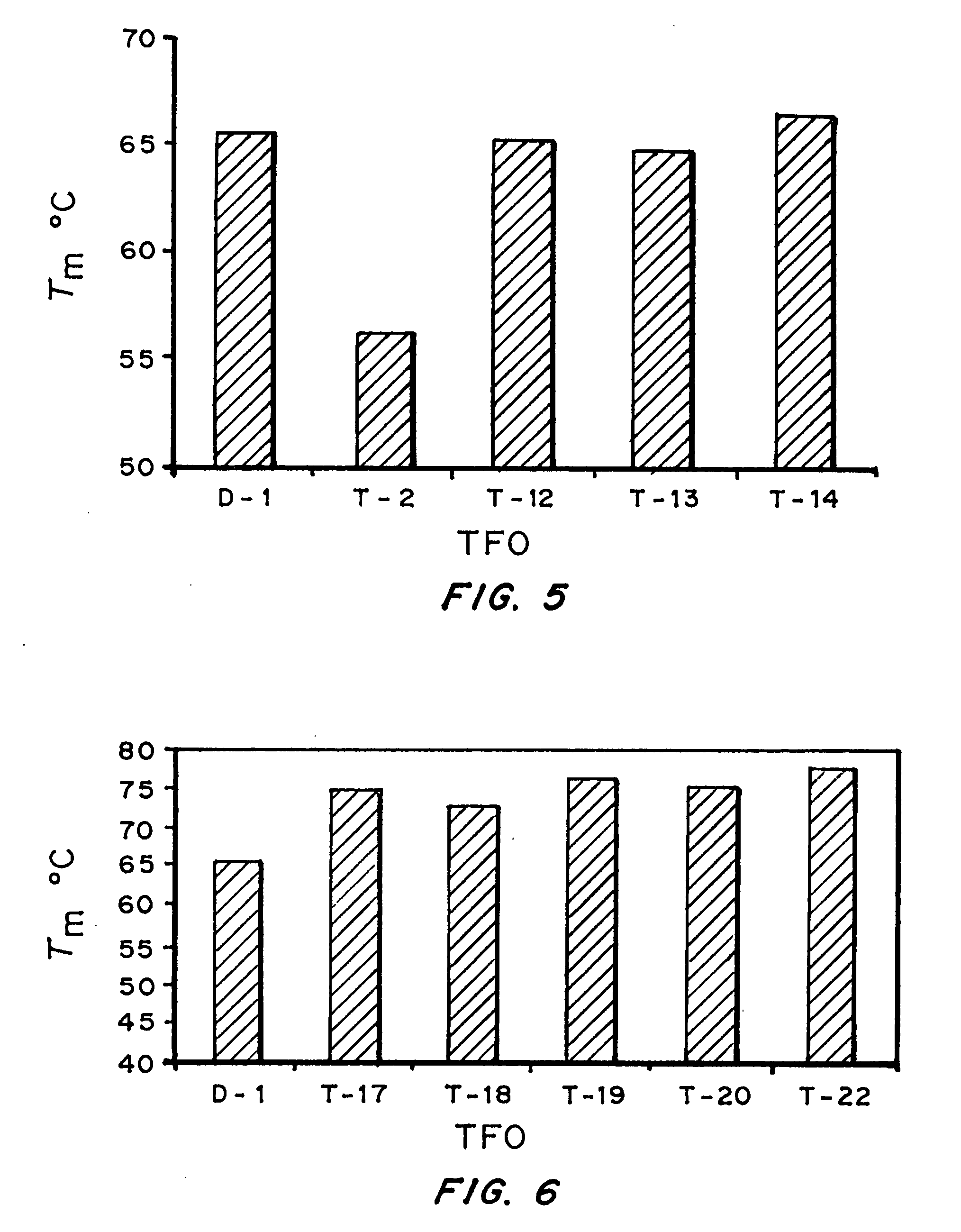
Modified triple-helix forming oligonucleotides for targeted mutagenesis
ActiveUS20070219122A1Enhance stabilityImprove stabilityPeptide/protein ingredientsGenetic material ingredientsMutagenic ProcessChemistry
High affinity, chemically modified triplex-forming oligonucleotides (TFOs) and methods for use thereof are disclosed. TFOs are defined as triplex-forming oligonucleotides which bind as third strands to duplex DNA in a sequence specific manner. Triplex-forming oligonucleotides may be comprised of any possible combination of nucleotides and modified nucleotides. Modified nucleotides may contain chemical modifications of the heterocyclic base, sugar moiety or phosphate moiety. A high affinity oligonucleotide (Kd≦2×10−8) which forms a triple strand with a specific DNA segment of a target gene DNA is generated. It is preferable that the Kd for the high affinity oligonucleotide is below 2×10−10. The nucleotide binds or hybridizes to a target sequence within a target gene or target region of a chromosome, forming a triplex region. The binding of the oligonucleotide to the target region stimulates mutations within or adjacent to the target region using cellular DNA synthesis, recombination, and repair mechanisms. The mutation generated activates, inactivates, or alters the activity and function of the target gene.
Owner:CONCORD +2
Phospholink nucleotides for sequencing applications
ActiveUS20100167299A1Bioreactor/fermenter combinationsBiological substance pretreatmentsNucleotideDNA sequencing
The present invention provides labeled phospholink nucleotides that can be used in place of naturally occurring nucleotide triphosphates or other analogs in template directed nucleic acid synthesis reactions and other nucleic acid reactions and various analyses based thereon, including DNA sequencing, single base identification, hybridization assays, and others.
Owner:PACIFIC BIOSCIENCES
Quantitative methylation detection in DNA samples
InactiveUS6960436B2Quick fixFast and cost-effective and accurateSugar derivativesMicrobiological testing/measurementPolymerase LGenomic DNA
Described is a method for methylation detection in a DNA sample. An isolated genomic DNA sample is treated in a manner capable of distinguishing methylated from unmethylated cytosine bases. The pretreated DNA is amplified using at least one oligonucleotide primer, a polymerase and a set of nucleotides of which at least one is labeled with a first type of label. A sequence-specific oligonucleotide probe, marked with a second type of label, hybridizes to the amplification product and a FRET reaction occurs if a labeled oligonucleotide is present in close proximity in the amplification product. The method determines the level of methylation of a sample by measuring the extent of fluorescence resonance energy transfer (FRET) between the donor and acceptor fluorophore.
Owner:EPIGENOMICS AG
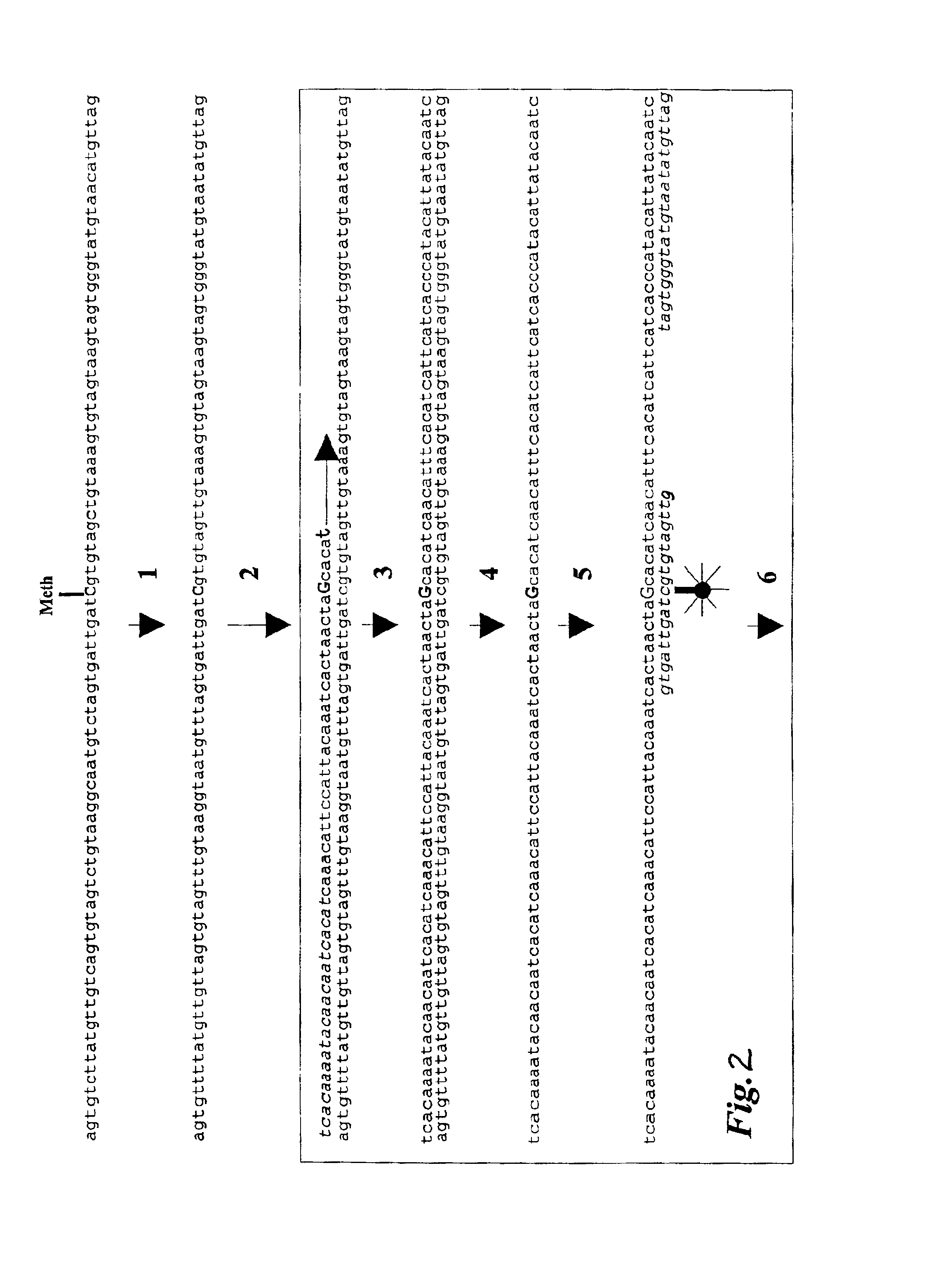
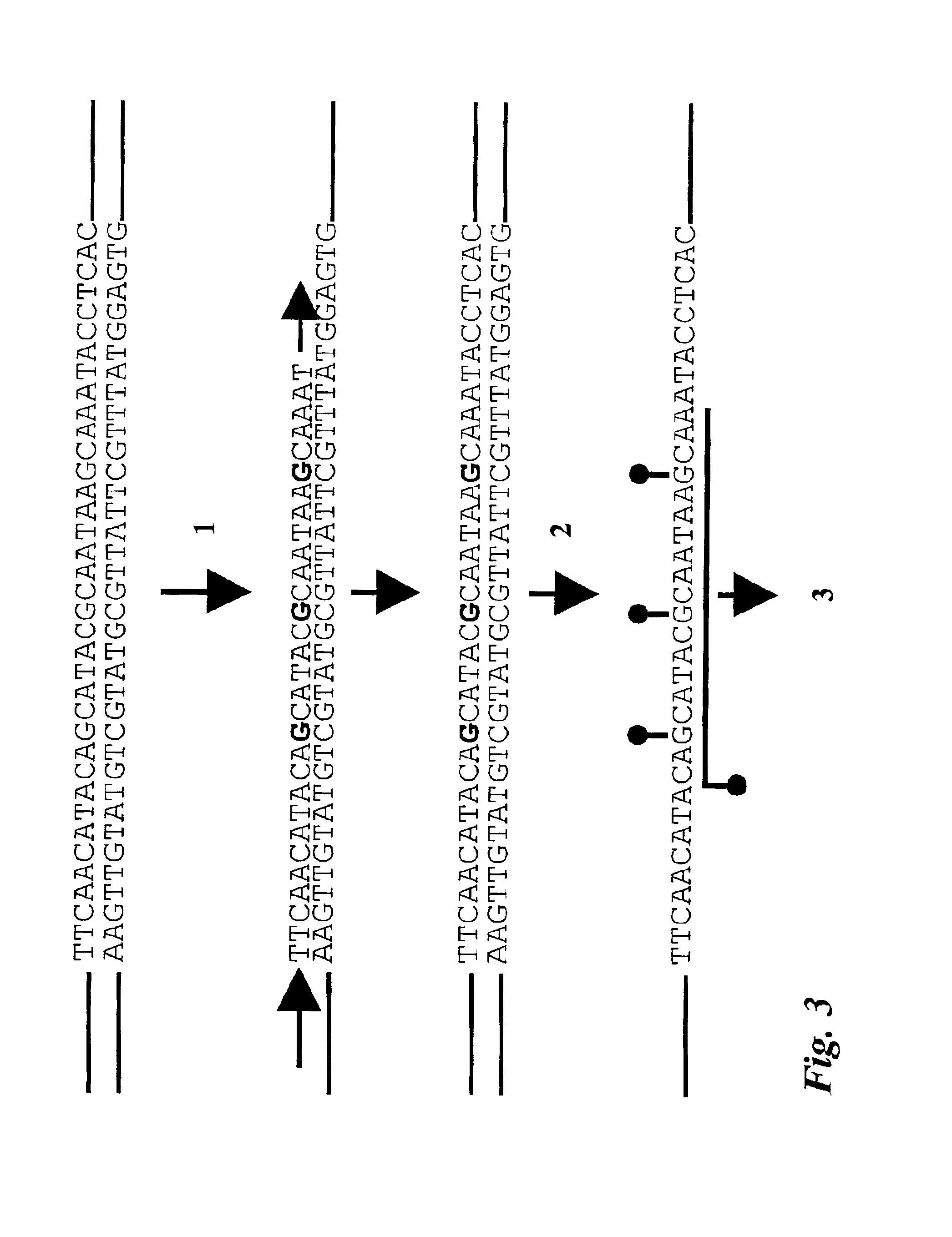
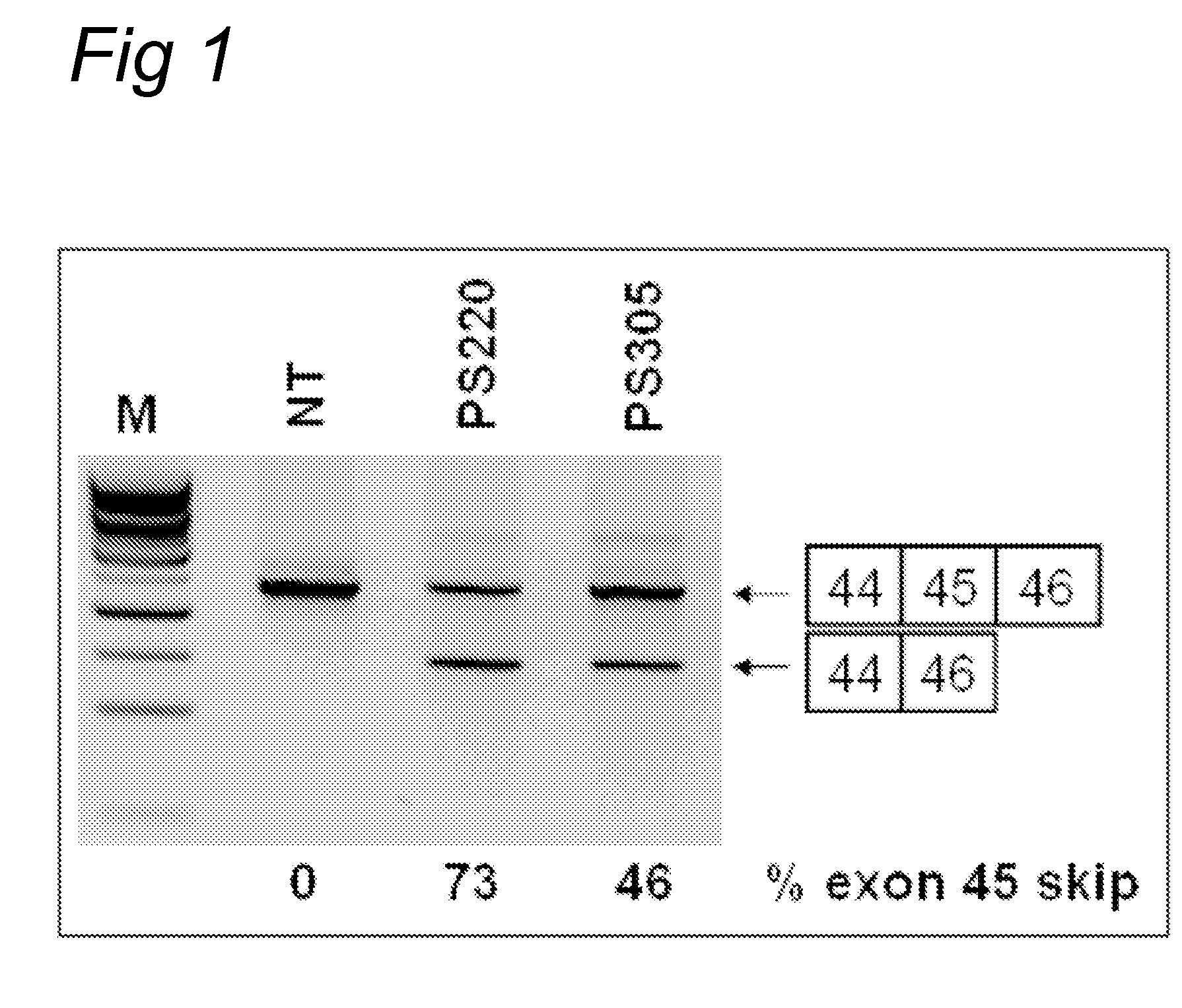
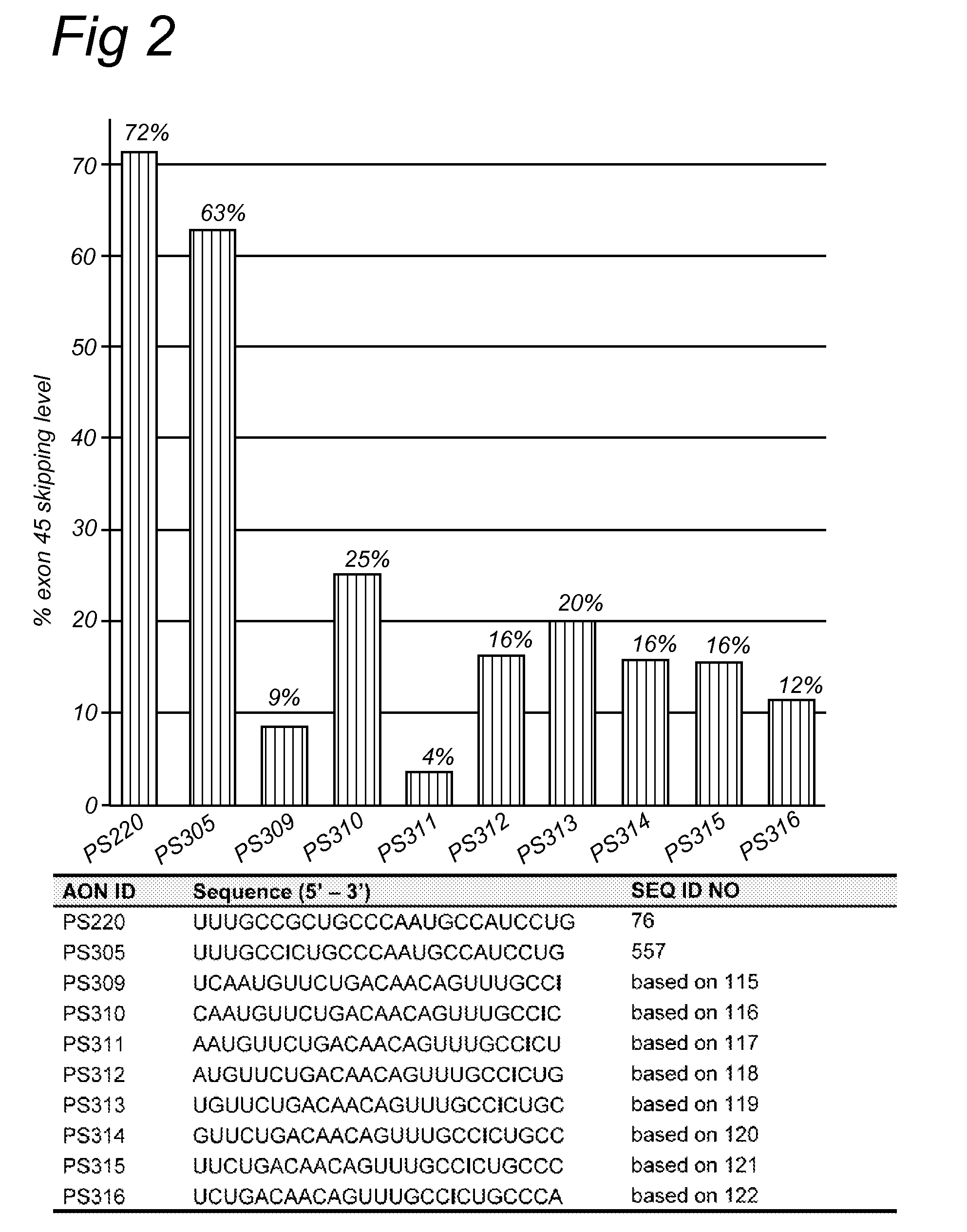
Oligonucleotide comprising an inosine for treating dmd
InactiveUS20120046342A1Avoid or decrease a potential multimerisation or aggregation of oligonucleotidesLow efficiencyOrganic active ingredientsSplicing alterationInosineNucleotide
The invention provides an oligonucleotide comprising an inosine, and / or a nucleotide containing a base able to form a wobble base pair or a functional equivalent thereof, wherein the oligonucleotide, or a functional equivalent thereof, comprises a sequence which is complementary to at least part of a dystrophin pre-m RNA exon or at least part of a non-exon region of a dystrophin pre-m RNA said part being a contiguous stretch comprising at least 8 nucleotides. The invention further provides the use of said oligonucleotide for preventing or treating DMD or BMD.
Owner:BIOMARIN TECH BV
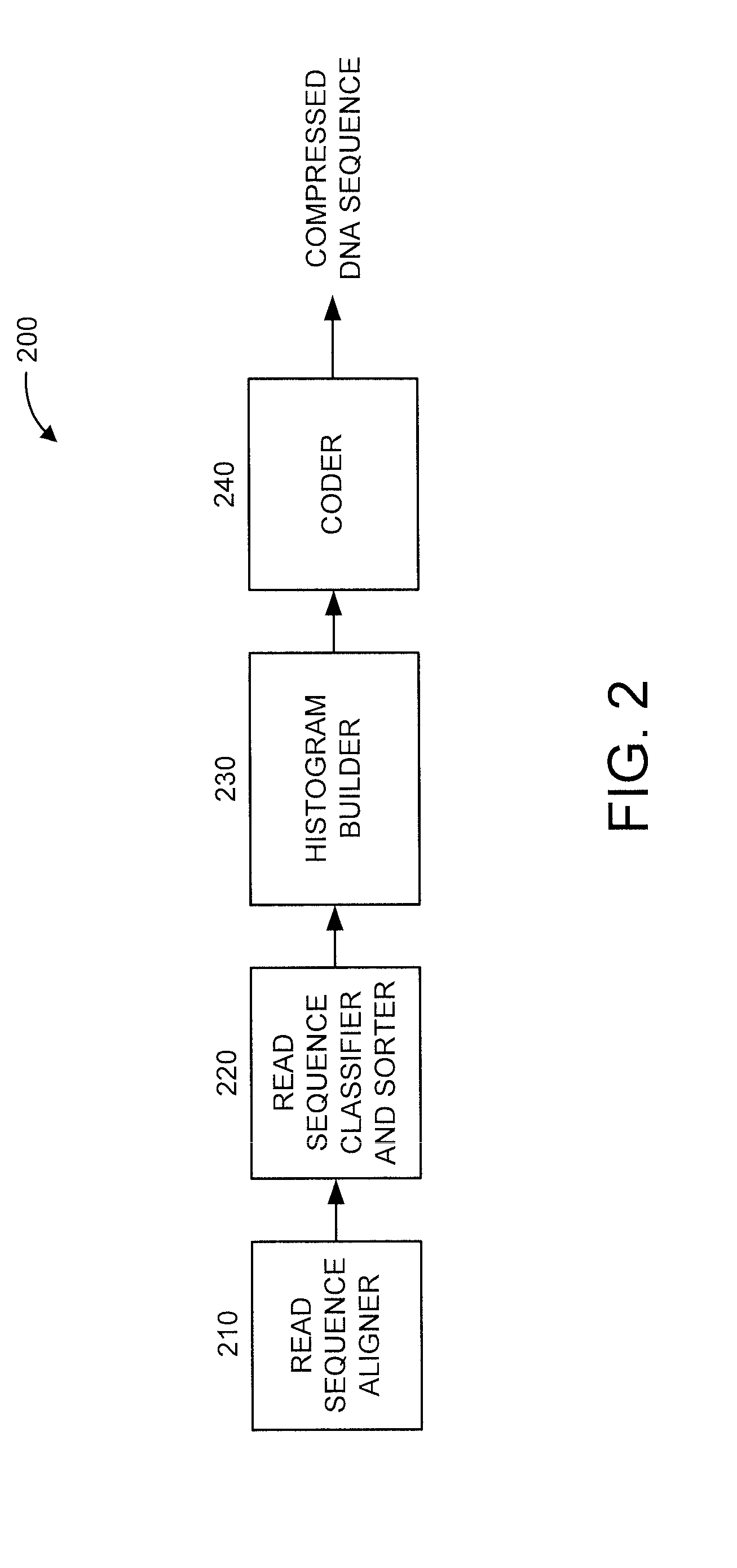
Lossless compression of DNA sequences
ActiveUS20150227686A1Nucleotide librariesMicrobiological testing/measurementLossless compressionHistogram
There is provided an apparatus and a processor-implemented method. The method includes aligning a reference genome with a plurality of DNA sequences. Each of the plurality of DNA sequences has a respective plurality of bases. The method further includes classifying and sorting the plurality of read sequences based on respective numbers of mismatched bases within the plurality of read sequences to obtain a plurality of re-arranged DNA sequences. The method also includes building a histogram based on respective positions of mismatched bases within the plurality of re-arranged DNA sequences. The method additionally includes coding at least some of the plurality of re-arranged DNA sequences based on the histogram.
Owner:IBM CORP
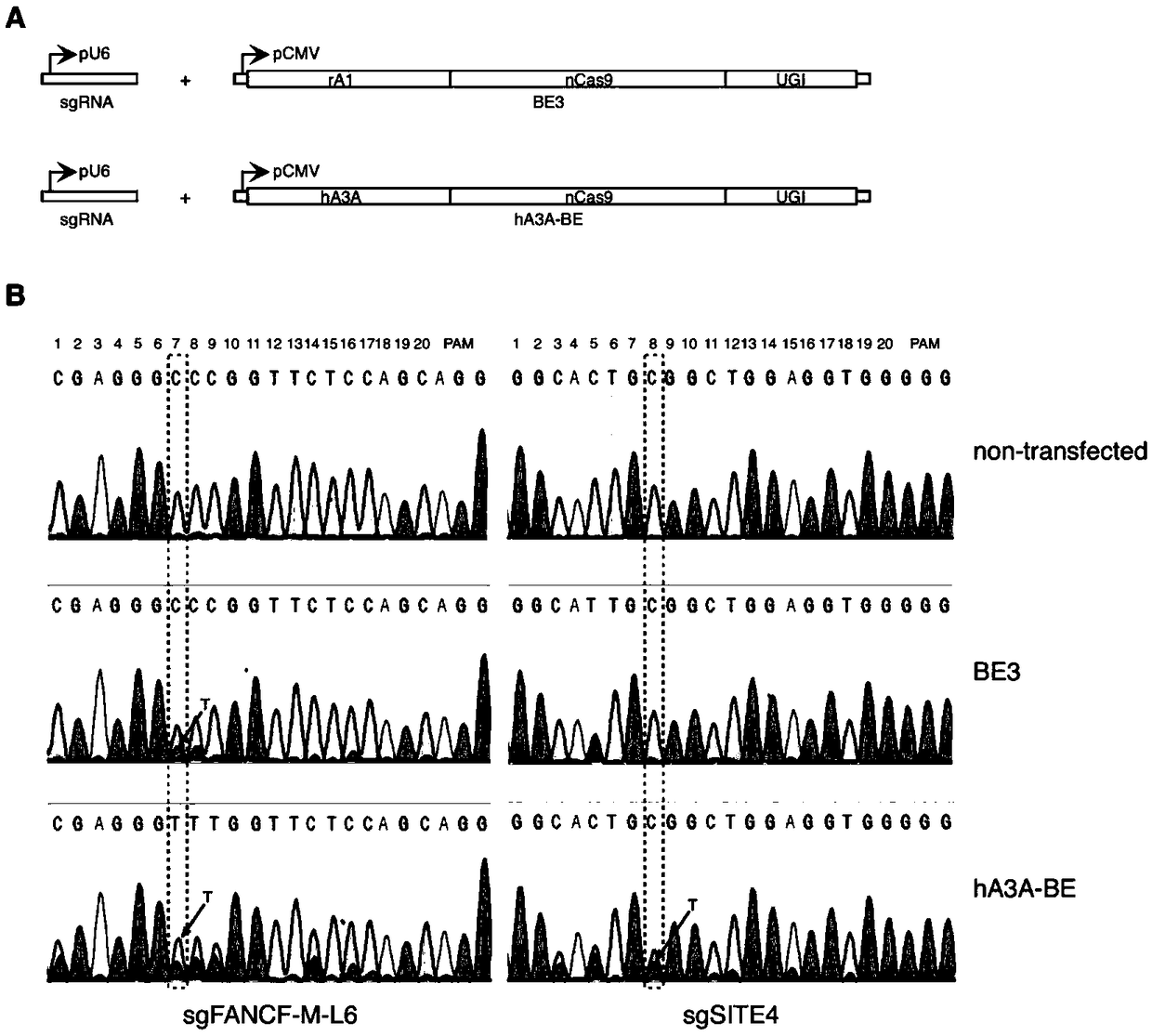
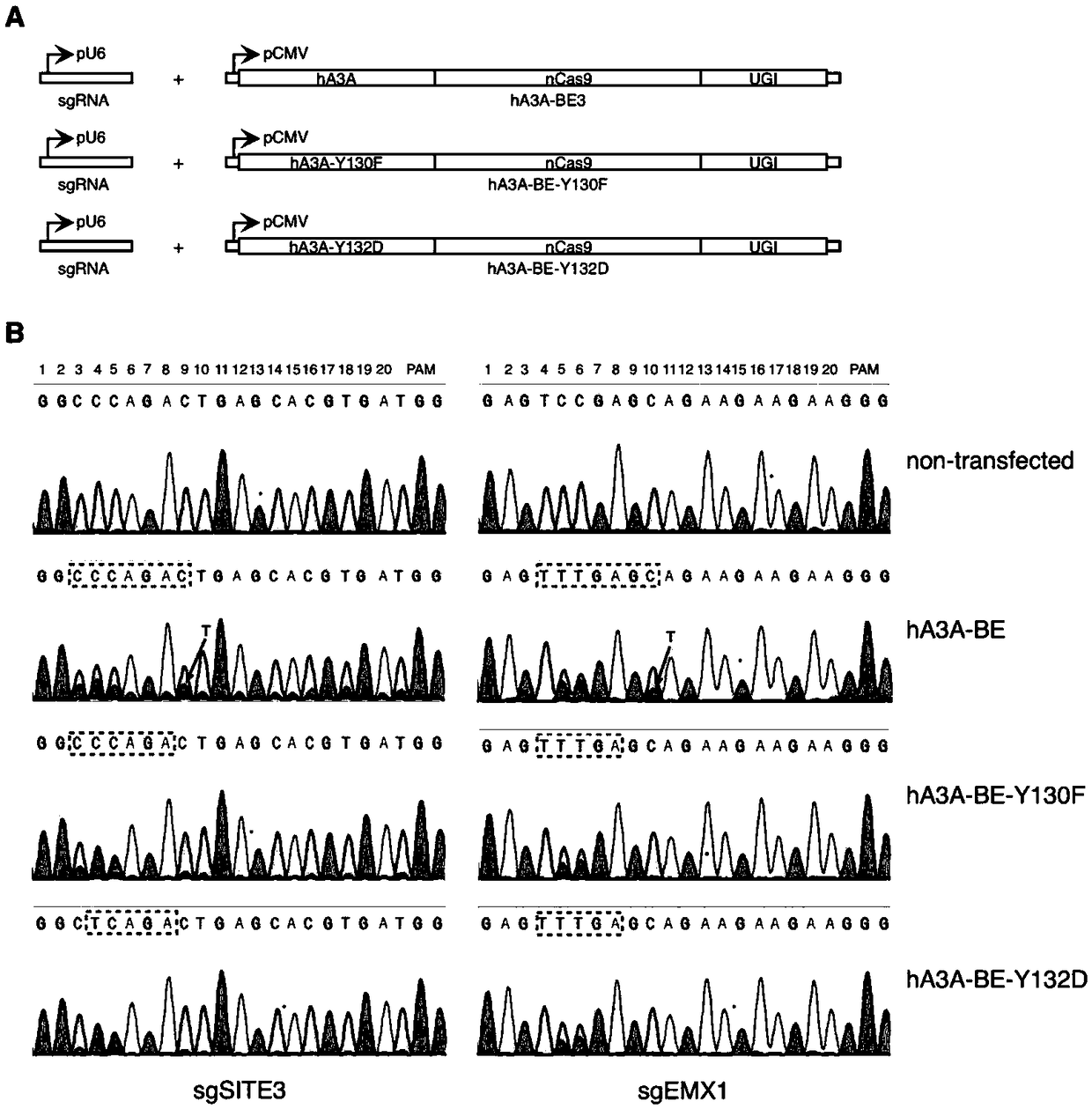
Gene base editor
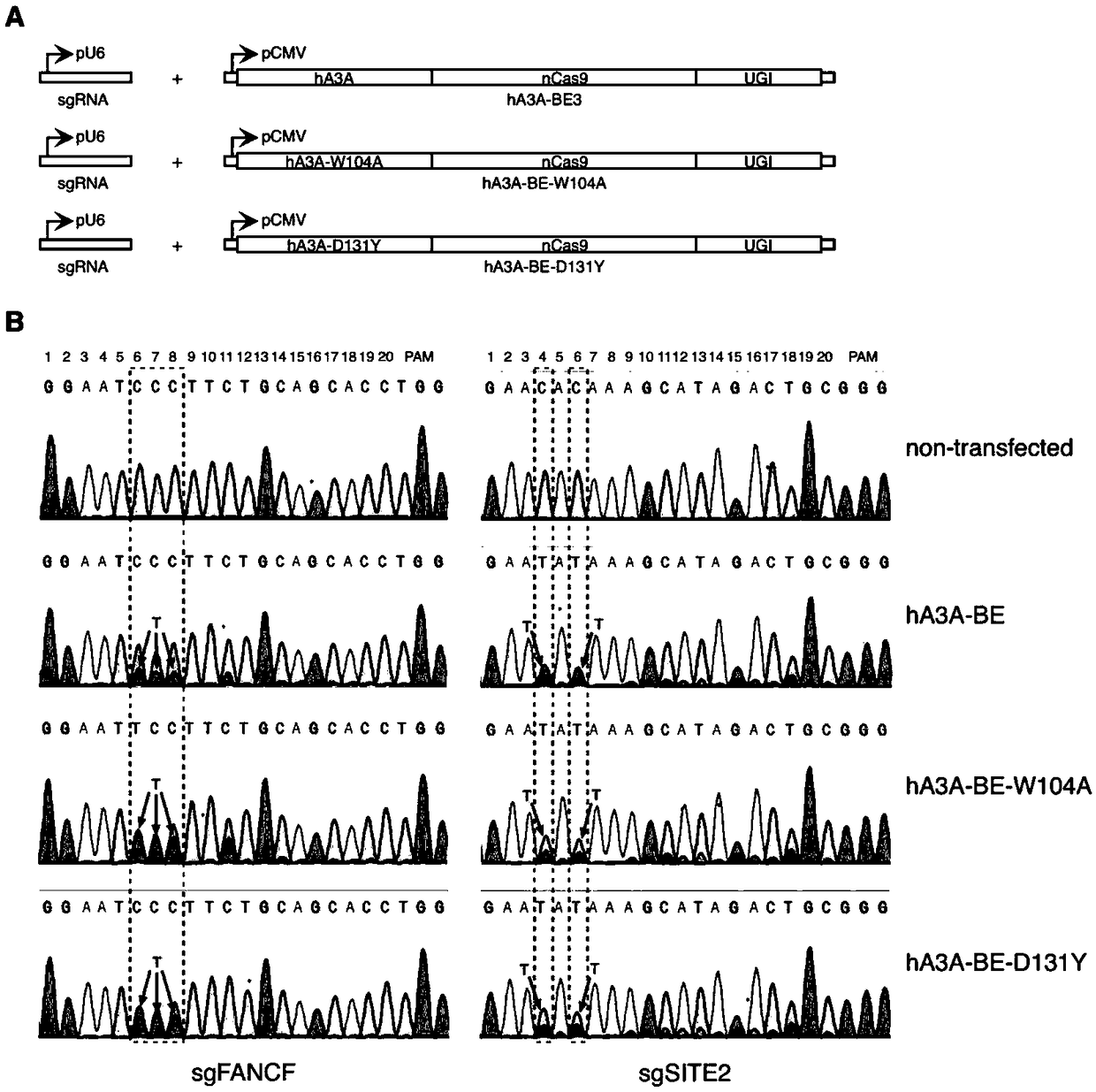
The invention provides a gene base editor, in particular relates to fusion protein. The fusion protein is characterized by comprising two fragments. The first fragment comprises APOBEC3A and the second fragment comprises CRISPR-related Cas protein. The base editor is capable of performing base edition in DNA and performing deamination on cytosine to be uracil, and the base editor still has higherediting efficiency even if cytosine is located at a GpC site or in a hypermethylated state.
Owner:SHANGHAI TECH UNIV
Methods and apparatus that increase sequencing-by-binding efficiency
ActiveUS9951385B1Improvement to accuracy and speed and costSatisfies needMicrobiological testing/measurementBiologyTernary complex
A method of nucleic acid detection including (a) contacting a primed template with a polymerase, nucleotide cognate of a first base type and nucleotide cognate of a second base type under ternary complex stabilizing conditions; (b) contacting the primed template nucleic acid with a polymerase, nucleotide cognate of the first base type and nucleotide cognate of a third base type under ternary complex stabilizing conditions; (c) examining products of (a) and (b) for signals produced by a ternary complex that includes the primed template nucleic acid, polymerase and next correct nucleotide, wherein signals for the product of (a) are ambiguous for the first and second base type, and signals for the product of (b) are ambiguous for the first and third base type; (d) disambiguating signals acquired in (c) to identify the next correct nucleotide.
Owner:PACIFIC BIOSCIENCES
Primer, detection method and detection reagent kit for detecting shigella
InactiveCN101153326AStrong specificityIncreased sensitivityMicrobiological testing/measurementDNA/RNA fragmentationFood borneLoop-mediated isothermal amplification
The invention relates to a technique for fast detecting food-borne pathogens based on a loop-mediated isothermal amplification, LAMP technique. A primer for detection of shigella can augment the specific base sequence of a target gene which is the ipaH-GenBank (accession no. M32063) of the shigella, and the primer is complementary to a part of or a complementary chain of the nucleic acid sequence on the 1095-1299bp loci on the target gene. The invention provides a primer unit having specificity to a specific gene fragment of the shigella, and through detecting whether or not the detecting specimen in a reagent box of the primer unit contains the specific gene fragment of the shigella, determines whether the shigella exists in the specimen or not.
Owner:ZHUHAI DISEASE PREVENTION & CONTROL CENT
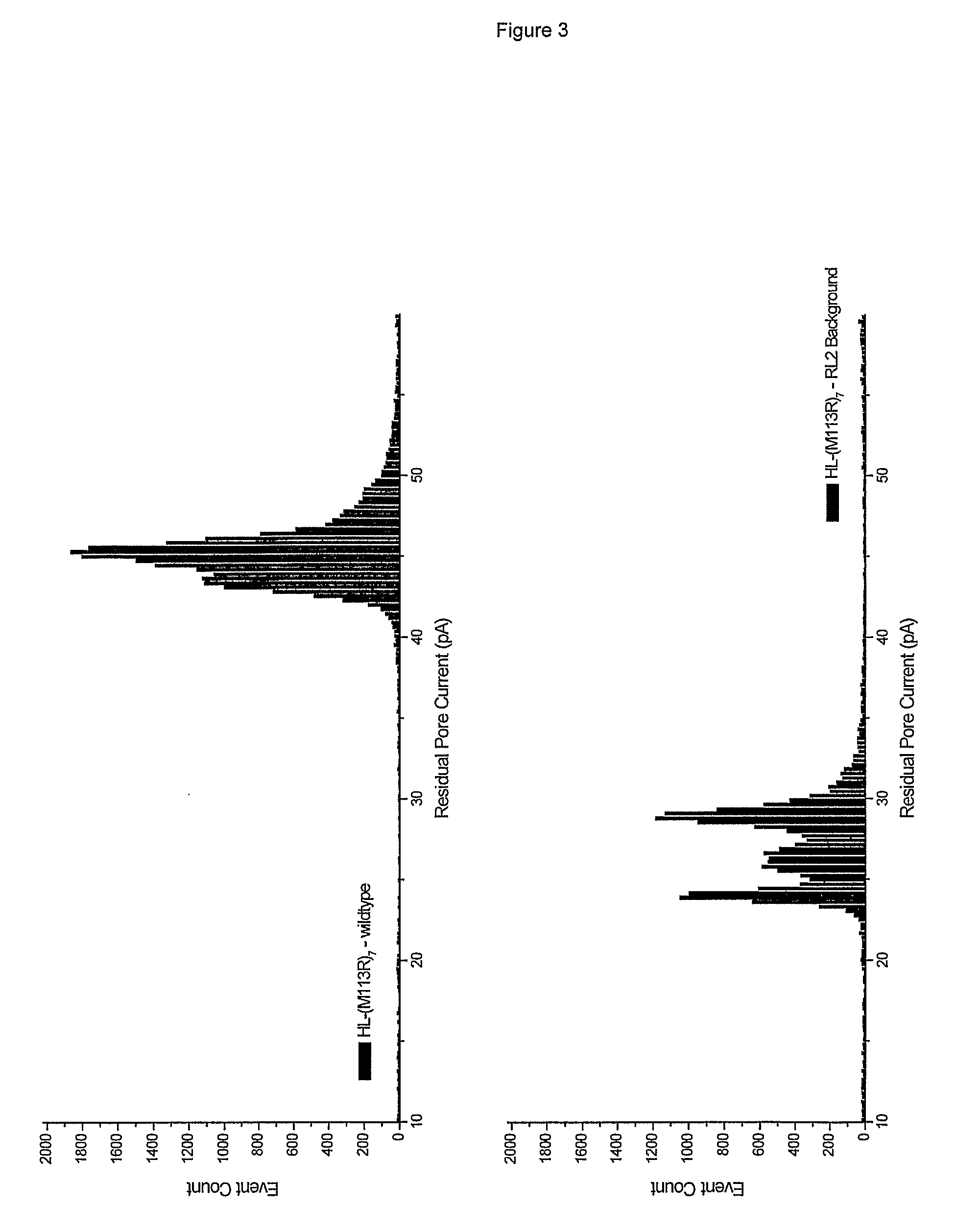
Base-detecting pore
ActiveUS9447152B2Enhanced interactionSugar derivativesMicrobiological testing/measurementBase JNucleotide
The invention relates to a mutant α-hemolysin (α-HL) pore which is useful for detecting one or more nucleotides by stochastic sensing. The pore is particularly useful for sequencing DNA or RNA. A molecular adaptor that allows detection of the nucleotide(s) is covalently attached to the pore. The pore is specifically modified to facilitate positioning of the adaptor and may be modified to facilitate covalent attachment.
Owner:OXFORD NANOPORE TECH LTD
Method for precisely editing genome DNA sequence
InactiveCN105567734ASolve the problem that the fixed-point insertion of genes cannot be realizedSolve the characteristicsNucleic acid vectorVector-based foreign material introductionGene targetsGene targeting
The invention provides a method for precisely editing a genome DNA sequence. A TALEN, CRISPR / Cas9 and other gene targeting technologies and homologous recombination technologies are utilized in a combined mode for precisely editing the genome DNA sequence. According to the method, the technical problem that in the prior art, a specific sequence cannot be deleted is solved, the specific base sequence can be seamlessly inserted or deleted in a fixed point mode, and seamless free modification of the genome DNA sequence of eukaryocyte is achieved for the first time.
Owner:WUHAN DANMIYOU BIOLOGICAL MEDICAL
Method for knocking out pig FGL2 genes with CRISPR-Cas9 specificity and sgRNA for specificity targeting FGL2 genes
ActiveCN105518139AKnockout fastKnockout precisionFermentationVector-based foreign material introductionExonBioinformatics
The invention discloses a method knocking out pig FGL2 genes with CRISPR-Cas9 specificity and sgRNA for specificity targeting FGL2 genes. The target sequence of the sgRNA of the specificity targeting FGL2 genes on the FGL2 genes meets a sequence arrangement rule of 5'-N(20) NGG- 3'; N(20)represents 20 continous basic groups; each N indicates A, T, C or G; the target sequence on the FGL2 is placed in exon encoding areas of the FGL2; and the target sequence on the FGL2 genes are unique. In the method knocking out pig FGL2genes with CRISPR-Cas9 specificity, FGL2genes of the pigs can be quickly, accurately and high-efficiently knocked out in a specific way; and problems of long term and high cost during gene knockout of the FGL2genes can be solved.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Cas9 nuclease G915F and application thereof
The invention belongs to the technical field of biologics, and particularly relates to a Cas9 nuclease G915F and application thereof. The Cas9 nuclease (Cas9-G915F) has activity of Cas9 nuclease and is applicable to a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) / Cas9 system, and the Cas9 nuclease (Cas9-G915F) is prepared by mutating glycine at a 915th site of wild Cas9 nuclease into phenylalanine. When the Cas9 nuclease (Cas9-G915F) is adopted to cut double chains of DNA (Deoxyribonucleic Acid), protruding fractured tail ends can be generated, an alkali group complemented with the protruding fractured tail ends can be added in a filling connection mode, and precise edition on specific positions of genome DNA fragments can be achieved.
Owner:SHANGHAI JIAO TONG UNIV
Antisense antiviral compound and method for treating influenza viral infection
ActiveUS8697858B2Improve throughputLimit deliveryAntibacterial agentsSugar derivativesInfluenzavirus CBase J
The present invention relates to antisense antiviral compounds and methods of their use and production in inhibition of growth of viruses of the Orthomyxoviridae family and in the treatment of a viral infection. The compounds are particularly useful in the treatment of influenza virus infection in a mammal. Exemplary antisense antiviral compounds are substantially uncharged, or partially positively charged, morpholino oligonucleotides having 1) a nuclease resistant backbone, 2) 12-40 nucleotide bases, and 3) a targeting sequence of at least 12 bases in length that hybridizes to a target region selected from the following: a) the 5′ or 3′ terminal 25 bases of the negative sense viral RNA segment of Influenzavirus A, Influenzavirus B and Influenzavirus C; b) the terminal 30 bases of the 5′ or 3′ terminus of the positive sense vcRNA; c) the 45 bases surrounding the AUG start codon of an influenza viral mRNA and; d) 50 bases surrounding the splice donor or acceptor sites of influenza mRNAs subject to alternative splicing.
Owner:SAREPTA THERAPEUTICS INC
Cas9 nuclease K918A and application thereof
The invention belongs to the technical field of biology and particularly relates to Cas9 nuclease and application. The Cas9 nuclease (Cas9-K918A) disclosed by the invention has the activity of the Cas9 nuclease, is suitable for a CRISPR / Cas9 system and is obtained by mutating 918th lysine of wild type Cas9 nuclease into alanine. The Cas9 nuclease (Cas9-K918A) is adopted for cutting DNA (Deoxyribonucleic Acid) double strands to generate a predominantly-broken terminal; a basic group complemented with the predominantly-broken terminal can be added in a connection filling-in mode, so that accurate edition of a specific position of a genomic DNA fragment can be realized.
Owner:SHANGHAI JIAO TONG UNIV
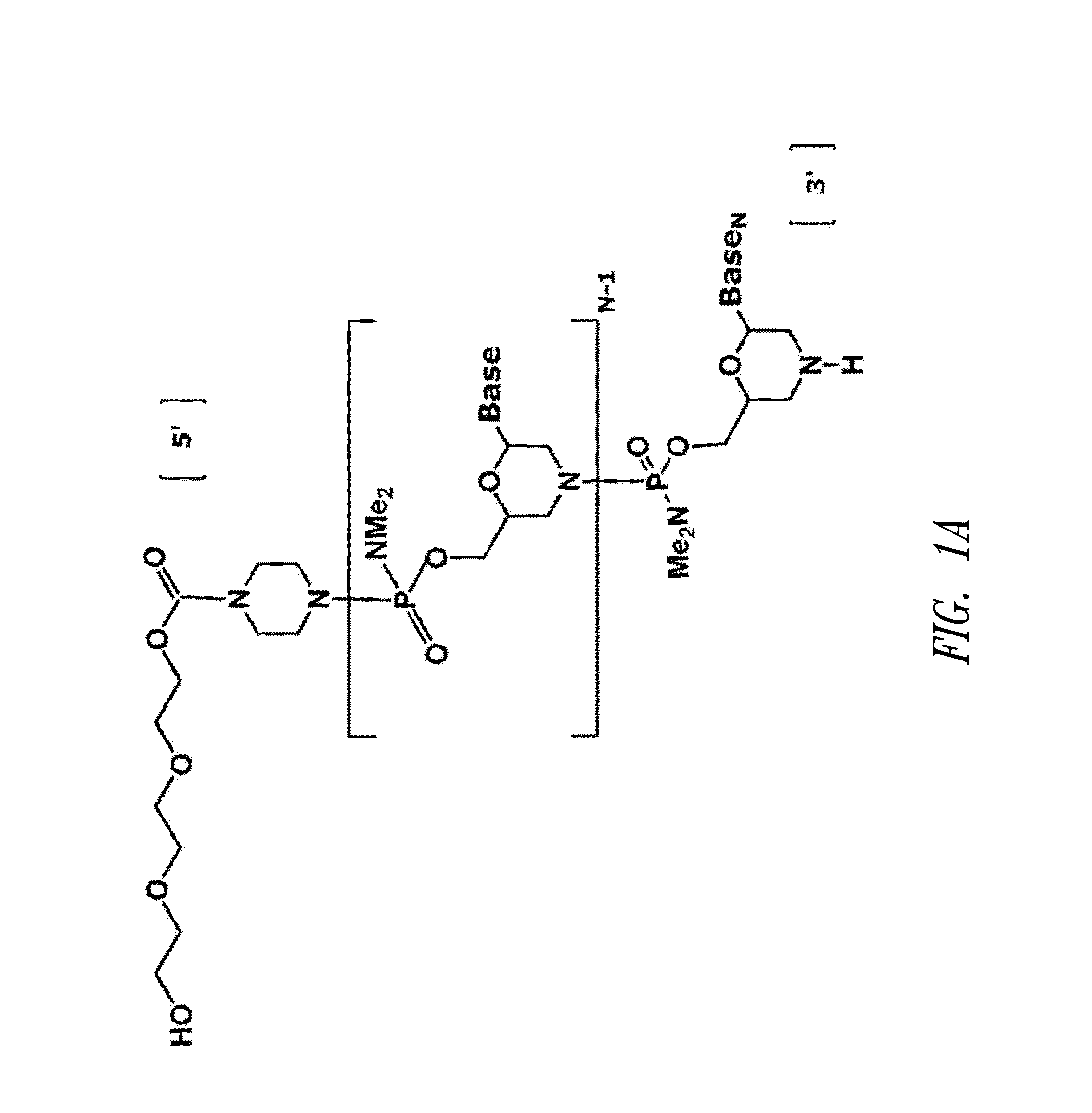
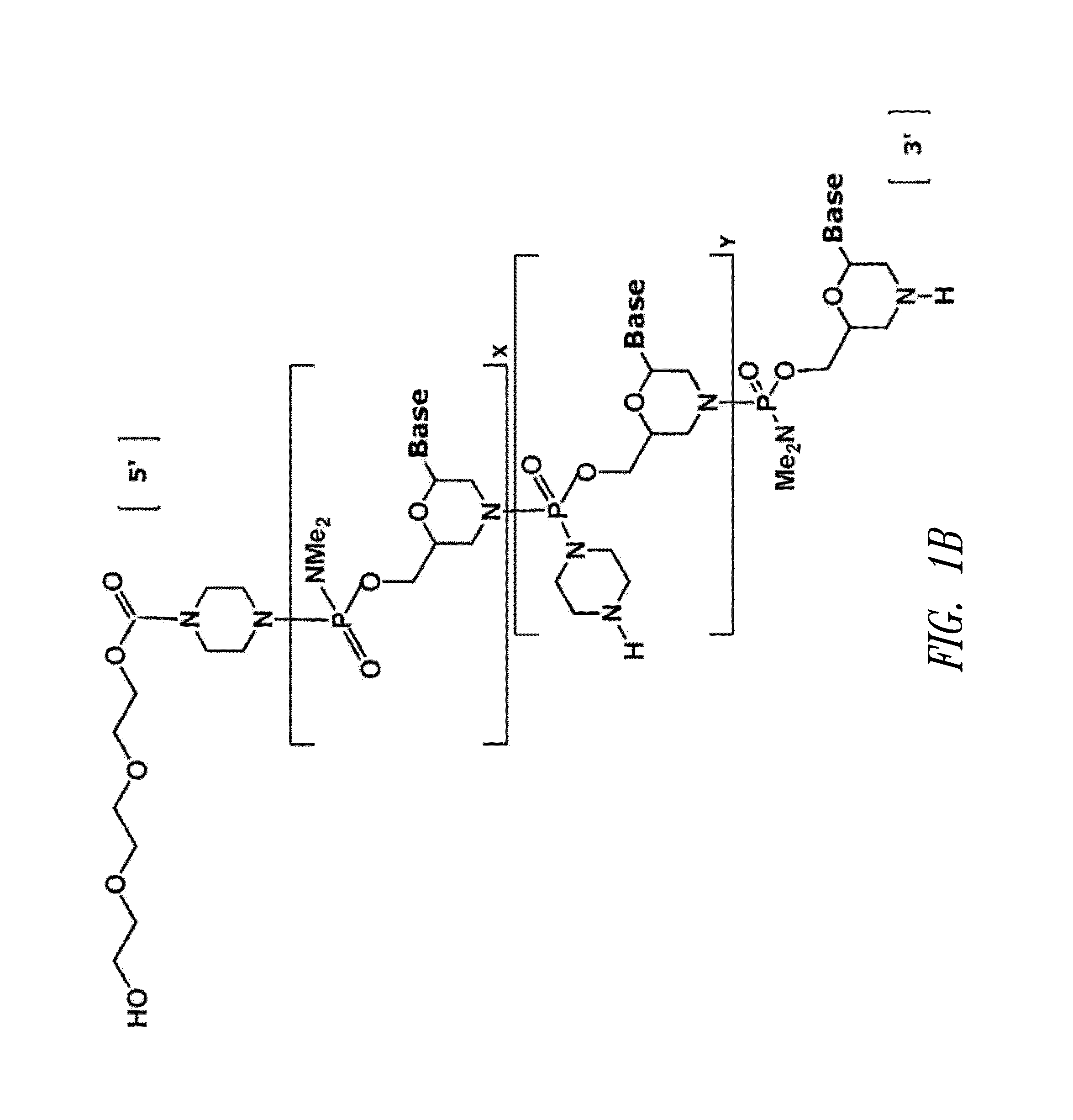
Phosphoramidite Compound And Method For Producing Oligo-Rna
ActiveUS20070282097A1Simple and high-yield methodSimple and high-yieldSugar derivativesOrganic compound preparationNitrogen atomPhosphoramidite
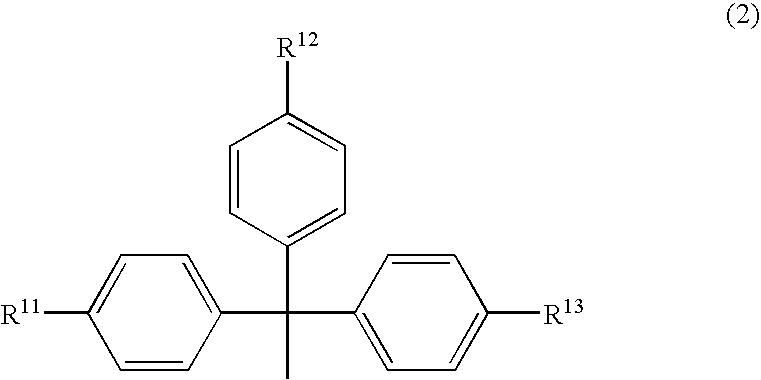
An object of the present invention is to provide a useful and novel phosphoramidite compound for the synthesis of oligo-RNA. A phosphoramidite compound represented by general formula (1), wherein: BX represents a nucleobase optionally having a protecting group; and R1 is a substituent represented by general formula (2), wherein: R11, R12 and R13 are the same or different and each represents hydrogen or alkoxy; R2a and R2b are the same or different and each represents alkyl, or R2a and R2b taken together with the adjacent nitrogen atom may form a 5- to 6-membered saturated amino cyclic group, the amino cyclic group optionally having an oxygen or sulfur atom as a ring-composing member in addition to the adjacent nitrogen atom; and WG1 and WG2 are the same or different and each represents an electron-withdrawing group.
Owner:NIPPON SHINYAKU CO LTD
Pseudonucleotide comprising an intercalator
InactiveUS20060014144A1Strong specificityHigh melting temperatureSugar derivativesMicrobiological testing/measurementNucleobaseNucleic acid analogue
The present invention relates to intercalator pseudonucleotides. Intercalator pseudonucleotides according to the invention are capable of being incorporated into the backbone of a nucleic acid or nucleic acid analogue and they comprise an intercalator comprising a flat conjugated system capable of co-stacking with nucleobases of DNA. The invention also relates to oligonucleotides or oligonucleotide analogues comprising at least one intercalator pseudo nucleotide. The invention furthermore relates to methods of synthesising intercalator pseudo nucleotides and methods of synthesising oligonucleotides or oligonucleotide analogues comprising at least one intercalator pseudonucleotide. In addtition, the invention describes methods of separating sequence specific DNA(s) from a mixture comprising nucleic acids, methods of detecting a sequence specific DNA (target DNA) in a mixture comprising nucleic acids and / or nucleic acid analogues and methods of detecting a sequence specific RNA in a mixture comprising nucleic acids and / or nucleic acid analogues. In particular said methods may involve the use of oligonucleotides comprising intercalator pseudo nucleotides. The invention furthermore relates to pairs of oligonucleotides or oligonucleotide analogues capable of hybridising to one another, wherein said pairs comprise at least one intercalator pseudonucleotide. Methods for inhibiting a DNAse and / or a RNAse and methods of modulating transcription of one or more specific genes are also described.
Owner:HUMAN GENETIC SIGNATURES PTY LTD
Enzymatic nucleic acid synthesis: compositions and methods
InactiveUS20070172860A1Pyrophosphorolysis can be preventedImprove fidelitySugar derivativesMicrobiological testing/measurementBase JAssay
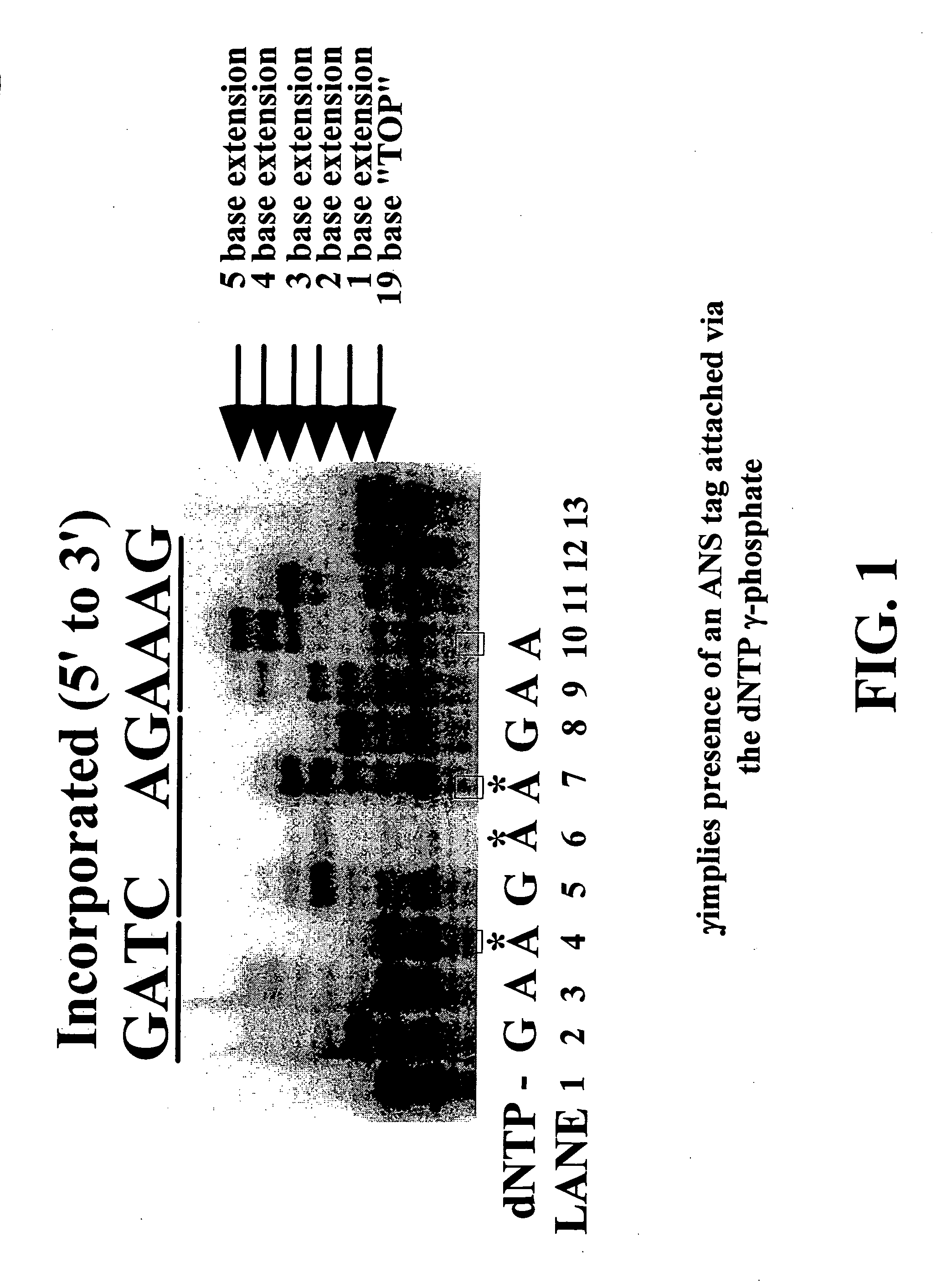
Nucleotide triphosphate probes containing a molecular and / or atomic tag on a a γ and / or β phosphate group and / or a base moiety having a detectable property are disclosed, and kits and method for using the tagged nucleotides in sequencing reactions and various assay. Also, phosphate and polyphosphate molecular fidelity altering agents are disclosed.
Owner:LIFE TECH CORP
CRISPR-Cas9 for targeting knockout of human intestinal cancer cell CNR1 gene, and specific sgRNA thereof
InactiveCN108588071AFacilitate the study of the mechanism of actionStable introduction of DNAFermentationIntestinal CancerLentivirus
The invention discloses a CRISPR-Cas9 for targeting knockout of a human intestinal cancer cell CNR1 gene, and specific sgRNA sequences thereof. Firstly, an sgRNA specifically targeting CNR1 gene second exon is obtained, and base sequences of the sgRNA are as shown in SEQ ID NO. 1; secondly, the sgRNA of the CNR1 gene is constructed to a lentiviral vector system, and the lentiviral vector system contains Cas9 protein; and finally, a CRISPR / Cas9 lentivirus containing the sgRNA is infected with human intestinal cancer HT-29 cells, and a cell strain with CNR1 protein expression level significantlybeing reduced is obtained. The operation steps are simple, good sgRNA targeting is achieved, and the cutting efficiency to the CNR1 gene is high; and in addition, the constructed CRISPR / Cas9 lentiviral system has the advantage of high knockout efficiency, and can specifically knock out the CNR1 gene, human intestinal cancer cells with the CNR1 gene being knocked out is obtained, and therefore a powerful tool for further study of the mechanism of action of CNR1 in intestinal cancer cells is provided.
Owner:OBIO TECH SHANGHAI CORP LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com