Improving sequence-specific antimicrobials by blocking DNA repair
a technology of endonuclease and antimicrobial agent, which is applied in the direction of antibacterial agents, viruses/bacteriophages, peptide/protein ingredients, etc., can solve the problems of insufficient use of nucleases and bacteria, and bacteria will be susceptible to antibiotics without dna repair, so as to improve the ability of endonuclease to kill bacteria.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Effect of Double Strand Breaks Introduced by Cas9 on Cell Death and Conditions for Survival to Such DNA Damage
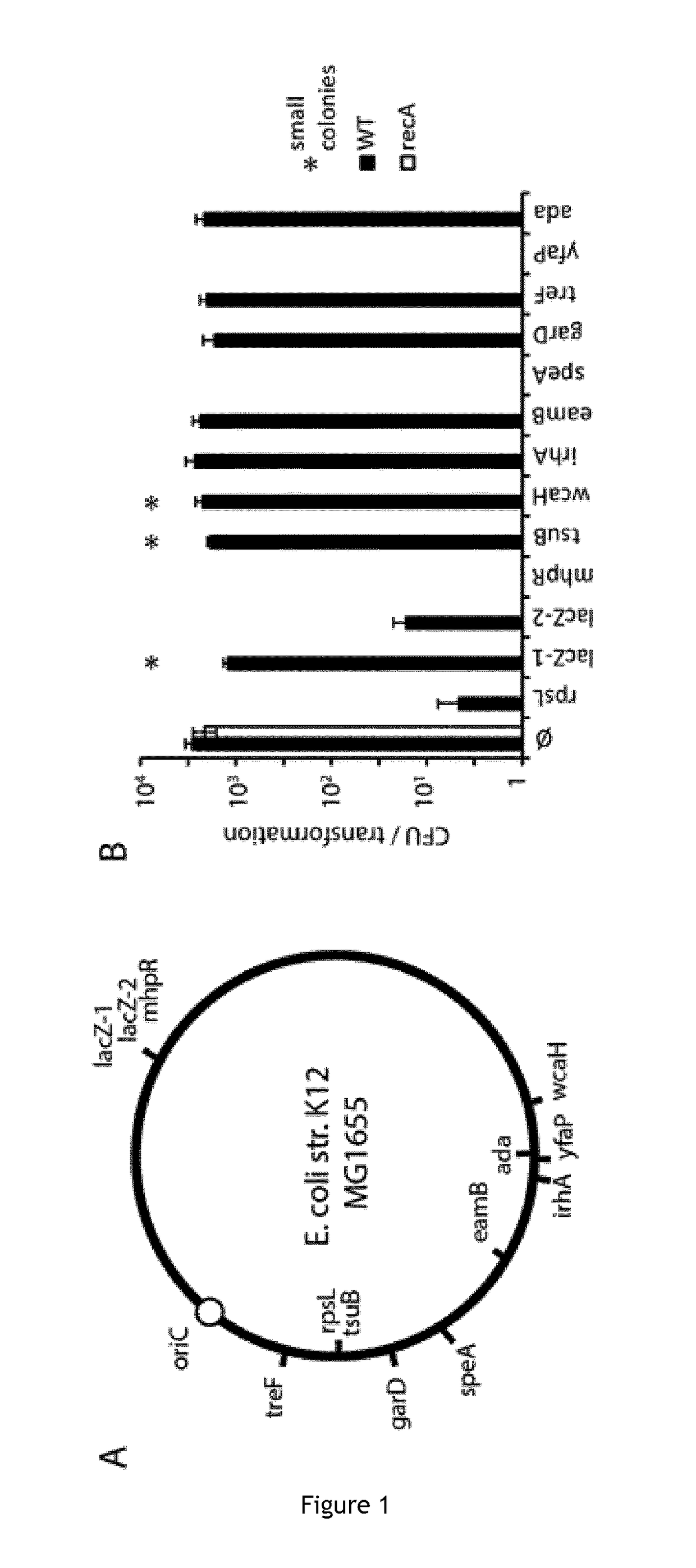
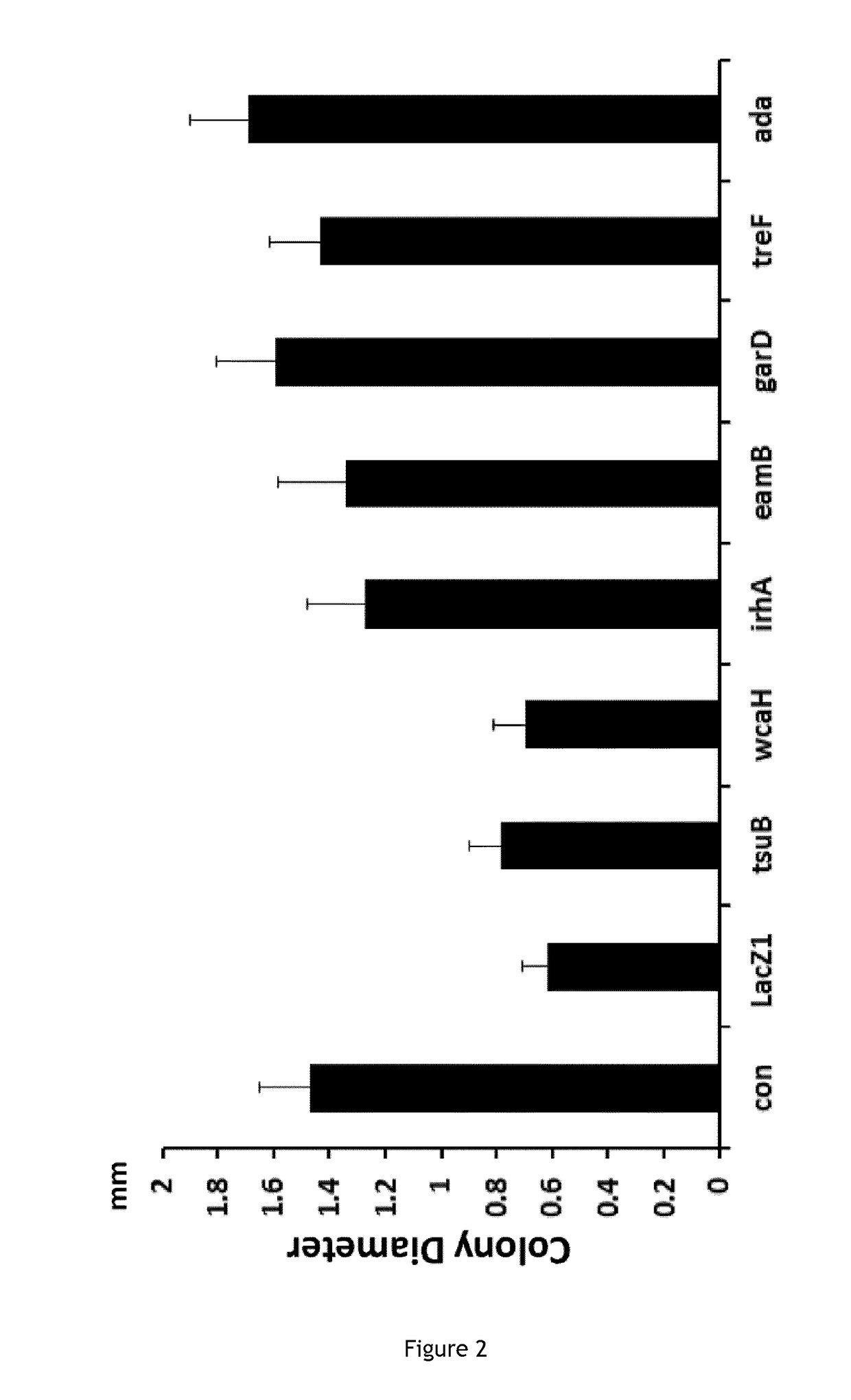
[0130]Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR associated (Cas) genes are the adaptive immune system of bacteria and archaea [1]. The RNA-guided Cas9 nuclease from Streptococcus pyogenes has emerged as a useful and versatile tool [2]. The ease with which it can be reprogrammed has in particular been driving its adoption for genome editing applications. Cas9 is guided by a small CRISPR RNA (crRNA) that is processed from the initial transcript of the CRISPR locus by Cas9 together with a trans-activating CRISPR RNA (tracrRNA) and the host RNAselII [3]. Both the tracrRNA and the processed crRNA remain bound to Cas9 and act as a complex to direct interference against target DNA molecules[4]. Alternatively, the crRNA and tracrRNA can be fused forming a chimeric single guide RNA (sgRNA) [4]. Cas9 scans DNA looking for a short sequence motif know...
example 2
Bacterial Strains and Media
[0134]E. coli strains were grown in Luria-Bertani (LB) broth (10 g Tryptone, 5 g Yeast Extract, 10 g NaCl, add ddH2O to 1000 ml, PH7.5, autoclaved). 1.5% LB Agar was used as solid medium. Different antibiotics (20 ug / ml chloramphenicol, 100 ug / ml carbenicillin, 50 ug / ml kanamycin) were used as needed. Plates containing IPTG (100 uM) and X-gal (40 ug / ml) were used for blue / white screening. Escherichia coli strain MG1656 (a ΔlacI-lacZ derivative of MG1655) was used as a cloning strain for plasmid pCas9::lacZ2 (see below). E. coli strains N4278 (MG1655 recB268::Tn10)29, MG1655 RecA::Tn10 and JJC443 (lexAind3 MalF::Tn10)30 are gifts from the Mazel lab.
example 3
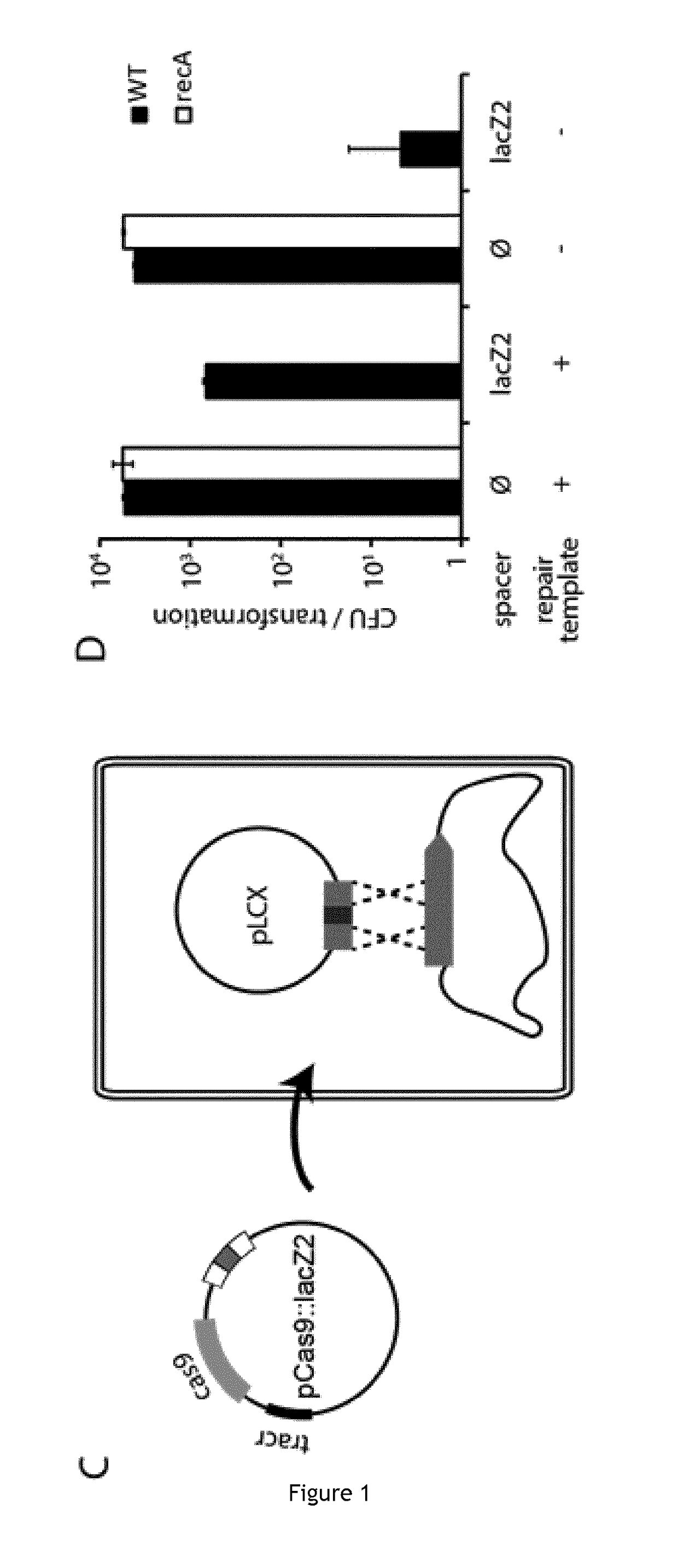
[0135]pCRRNA was assembled by amplification of pCRISPR using primer B299 / LC34 and of the tracrRNA fragment from pCas9 using primers LC35 / LC36, followed by Gibson assembly [31]. Novel spacers were cloned into pCRRNA or pCas9 plasmids as previously described [7]. The vector was digested with BsaI, followed by ligation of annealed oligonucleotides designed as follows: 5′-aaac+(target sequence)+g-3′ and 5′-aaaac+(reverse complement of the target sequence)-3′. A list of all spacers tested in this study is provided in (Table 2 in the present application was indicated with the number 4 in the text of the priority application corresponding to the table 2 of the priority application).
[0136]The pLCX plasmid was assembled from the pCRISPR backbone amplified using primers LC41 / LC42 and two lacZ fragments amplified from MG1655 genomic DNA using primers LC38 / LC39 and LC37 / LC40. The pZA31-sulA-GFP plasmid was assembled from pZA31-Luc [32] linearized with primers LC192 / LC193, the sul...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Composition | aaaaa | aaaaa |
| Electrical resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com