Whole-genome methylation non-bisulfite sequencing library, construction and applications thereof
A whole genome, sequencing library technology, applied in the field of bioinformatics, can solve the problems of low utilization rate of library sequencing data, difficult to achieve sequencing compatibility, base imbalance, etc., to achieve satisfactory coverage depth and solve base imbalance. , the effect of component reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
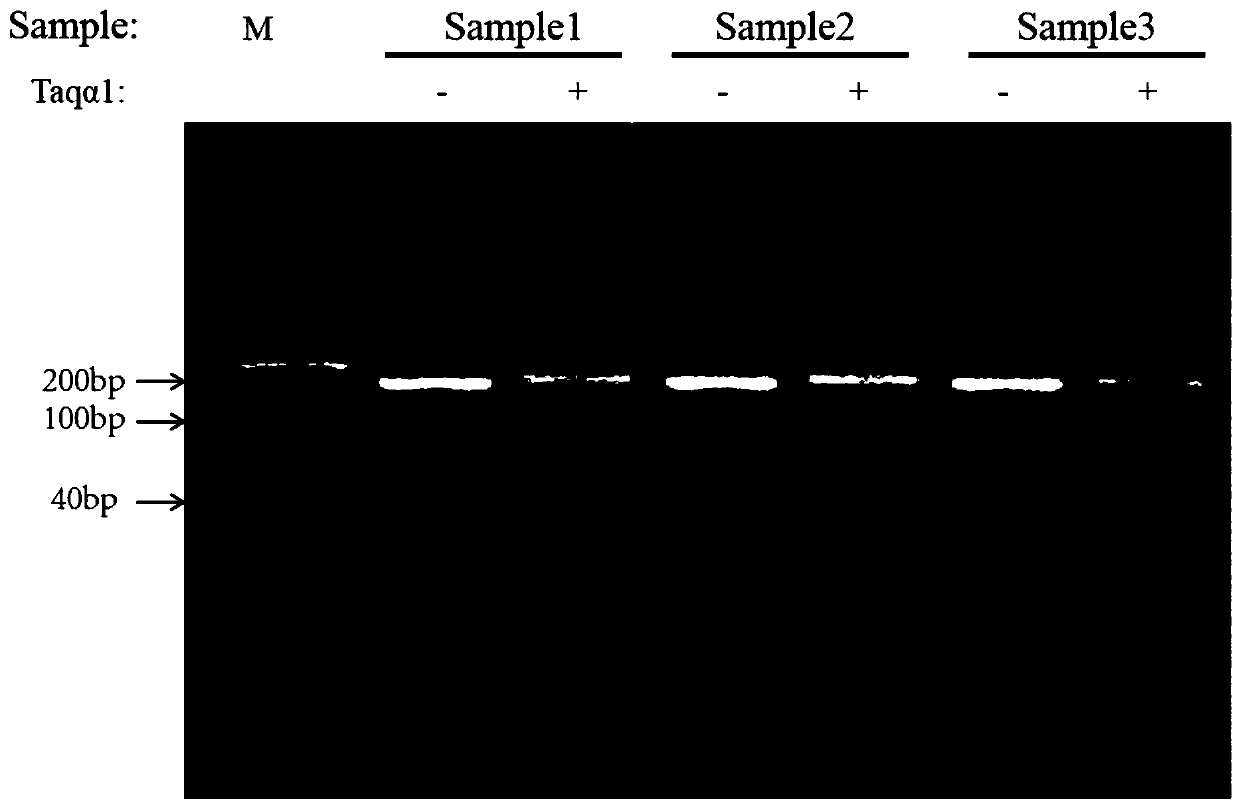
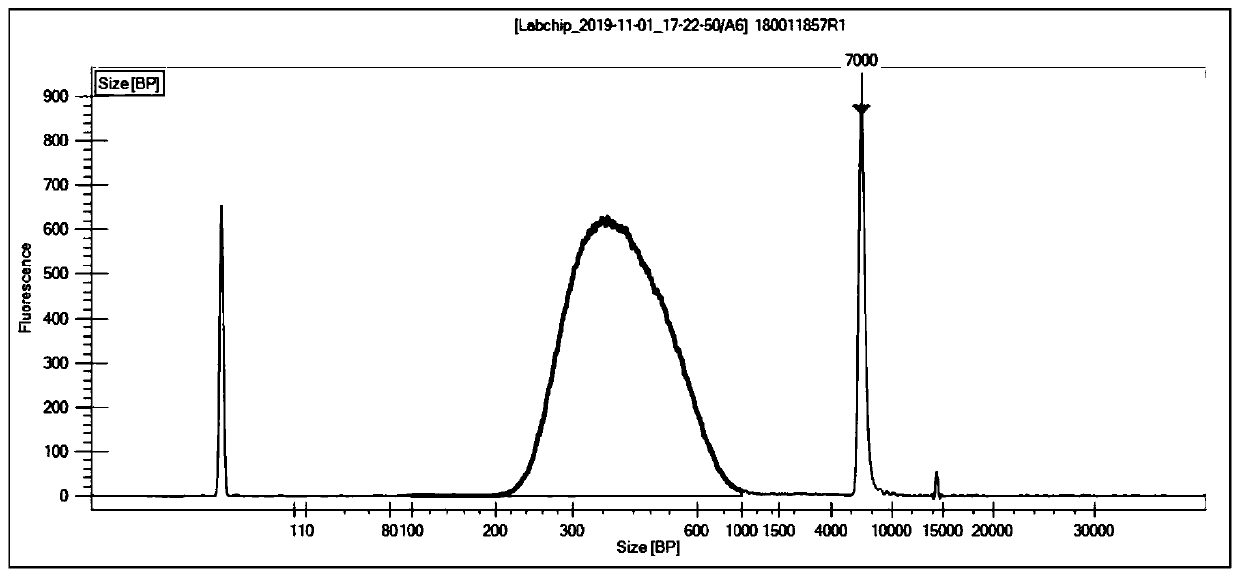
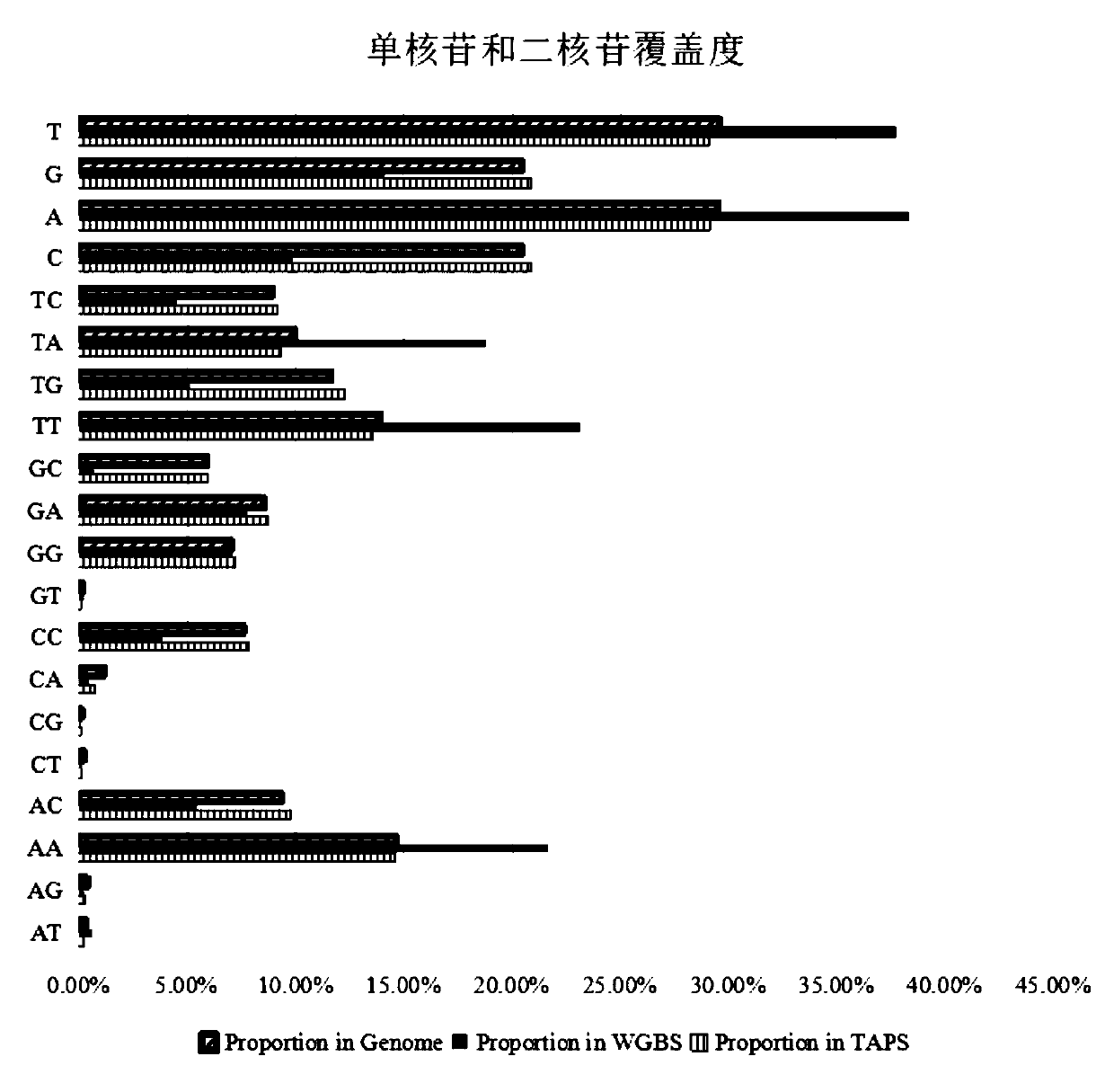
Image
Examples
Embodiment 1
[0043] Example 1 DNA methylation conversion kit
[0044] The DNA methylation conversion kit provided in this example includes an independently packaged TET enzyme oxidation buffer;
[0045] TET enzyme oxidation buffer (pH=8) is composed of the components in the following table 1:
[0046] Table 1. Composition of TET enzyme oxidation buffer
[0047]
[0048]
[0049] Further, the kit also includes Fe(NH 4 ) 2 (SO 4 ) 2 solution, the Fe(NH 4 ) 2 (SO 4 ) 2 Fe(NH 4 ) 2 (SO 4 ) 2 The concentration is 1.0mM.
[0050] Further, the kit also includes an independently packaged TET enzyme solution, and the concentration of the TET enzyme is 8 μM. The TET enzyme is Wisegene's TET1 enzyme.
[0051] Further, the kit also includes an independently packaged pyridine borane reducing agent;
[0052] The pyridine borane reducing agent consists of the individually packaged components in the following table 2:
[0053] Table 2, composition of pyridine borane reducing agent ...
Embodiment 2
[0055] Example 2 DNA methylation conversion kit
[0056] The DNA methylation conversion kit provided in this example includes an independently packaged TET enzyme oxidation buffer;
[0057] TET enzyme oxidation buffer (pH=8) is composed of the components in the following table 3:
[0058] Table 3, TET enzyme oxidation buffer composition
[0059] components concentration Tris–Cl 20mM NaCl 100mM 2-oxoglutarate 3.3mM ascorbic acid 6.67mM Adenosine triphosphate (ATP) 4mM
[0060] Further, the kit also includes Fe(NH 4 ) 2 (SO 4 ) 2 solution, the Fe(NH 4 ) 2 (SO 4 ) 2 Fe(NH 4 ) 2 (SO 4 ) 2 The concentration is 0.75mM.
[0061] Further, the kit also includes an independently packaged TET enzyme solution, and the concentration of the TET enzyme is 10.5 μM. The TET enzyme is NEB EM conversion module.
[0062] Further, the kit also includes an independently packaged pyridine borane reducing agent;
[0063] The pyridine...
Embodiment 3
[0066] Example 3 DNA methylation conversion kit
[0067] The DNA methylation conversion kit provided in this example includes an independently packaged TET enzyme oxidation buffer;
[0068] TET enzyme oxidation buffer (pH=8) is composed of the components in the following table 5:
[0069] Table 5. Composition of TET enzyme oxidation buffer
[0070] components concentration HEPES (4-Hydroxyethylpiperazineethanesulfonic acid) 167mM NaCl 333mM α-KG (α-ketoglutarate) 3.3mM ascorbic acid 6.67mM Adenosine triphosphate (ATP) 4mM
[0071] Further, the kit also includes Fe(NH 4 ) 2 (SO 4 ) 2 solution, the Fe(NH 4 ) 2 (SO 4 ) 2 Fe(NH 4 ) 2 (SO 4 ) 2 The concentration is 1.5mM.
[0072] Further, the kit also includes an independently packaged TET enzyme solution, and the concentration of the TET enzyme is 9 μM. The TET enzyme is NEB EM conversion module.
[0073] Further, the kit also includes an independently packaged py...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com