Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
50 results about "Chelex 100" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Chelex 100 is a chelating material from Bio-Rad used to purify other compounds via ion exchange. It is noteworthy for its ability to bind transition metal ions. It is a styrene-divinylbenzene co-polymer containing iminodiacetic acid groups.
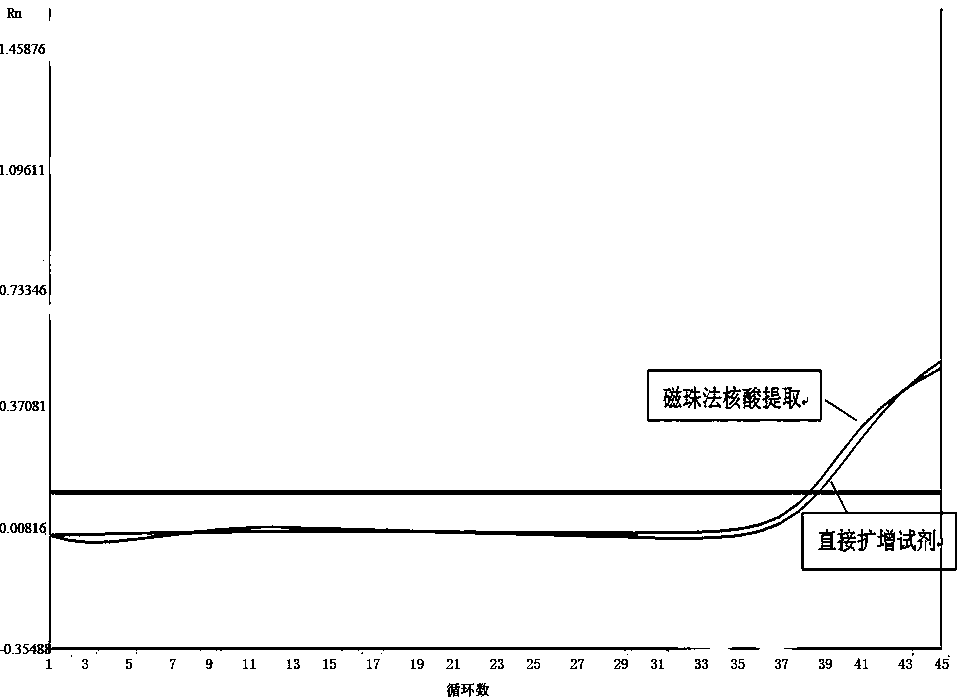
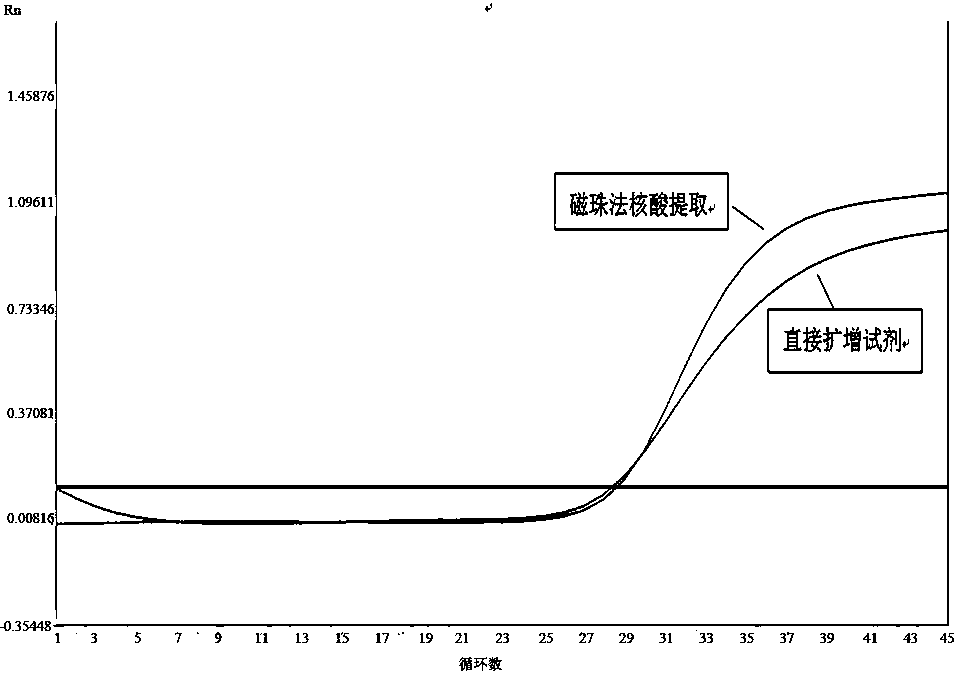
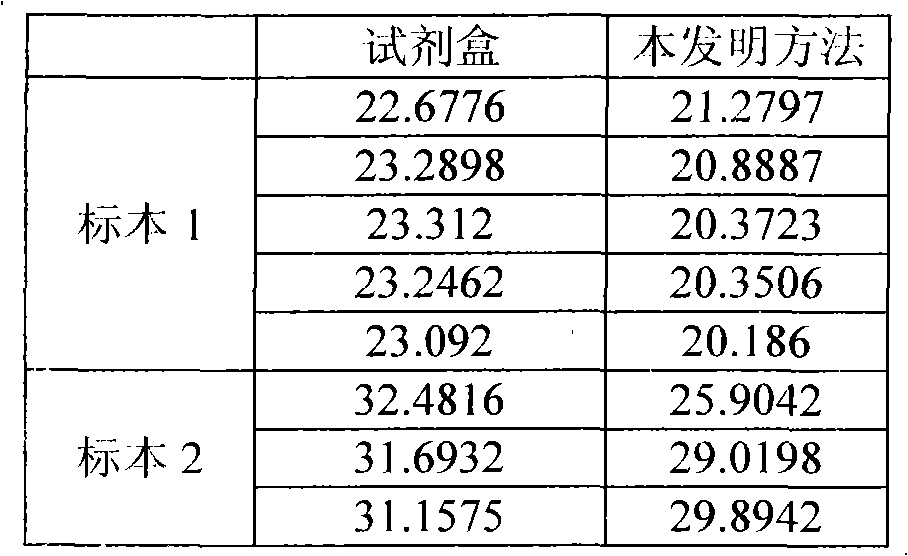
Method and kit for directly amplifying DNA (deoxyribonucleic acid) segment from biological material
The invention discloses a method and kit for directly amplifying a DNA (deoxyribonucleic acid) segment from a biological material. The kit disclosed by the invention comprises a lysis solution a, a lysis solution b, a lysis solution c and a neutralization solution, and can also comprise a 2*Taq PCR (polymerase chain reaction) mixed solution, wherein the lysis solution a comprises 50-70v% PEG (polyethylene glycol) 200 and 20-40mM KOH, and the pH value of the lysis solution a is 13.2-13.8; the lysis solution b comprises 5-10wt% chelex-100; the lysis solution c comprises 5-10wt% chelex-100 and 1-2wt% TritonX-100; and the neutralization solution comprises a 190-210 mmol / L Tris-HCl buffer of which the pH value is 6.8-7.5. The kit and method disclosed by the invention are applicable to many samples, such as bacteria, animal tissues, plant tissues, fungi, blood, saliva, hair and the like.
Owner:生工生物工程(上海)股份有限公司
Whole blood genomic DNA high-flux plate type extracting kit and extracting method
InactiveCN106636064AImprove throughputHigh-throughput Whole Blood DNA ExtractionDNA preparationMagnetic beadChelex 100
Owner:LUOYANG G N T BIOLOGICAL TECH CO LTD
Kit and method for quickly extracting DNA (deoxyribonucleic acid) from colla corii asini
ActiveCN104046616AIncrease concentrationHigh purityMicrobiological testing/measurementDNA preparationChelex 100A-DNA
The invention discloses a kit and a method for quickly extracting DNA (deoxyribonucleic acid) from colla corii asini. A combined technology of an SDS (sodium dodecyl sulfate)-protease K and Chelex-100 combined process DNA extraction technology and a Glassmilk DNA purification technology is applied to DNA extraction of the colla corii asini. The dosage of needed colla corii asini paste is controlled to be 0.3ml to 0.5ml, the weight of colla corii asini blocks can be controlled to be 0.2mg to 0.6mg, and extracted DNA is high in concentration, excellent in purity, stable and difficult to degrade; due to extracted DNA, sequence fragments of 100bp-500bp can be amplified, and subsequent molecular biology experiments of gene cloning, sequencing and the like can be carried out. According to the method, the operation is simple, the dosage is low, and the consumed time is short; the method is especially suitable for DNA extraction of various types of colla corii asini products with low nucleic acid content and high degradation degree caused by deep processing; a foundation is provided for realizing colla corii asini source identification by adopting a DNA molecular biology identification technology.
Owner:VEGETABLE RES INST OF SHANDONG ACADEMY OF AGRI SCI
Reagent and method for quick release of nucleic acid
The invention relates to a reagent and a method for the quick release of nucleic acid. The reagent for releasing the nucleic acid is prepared by dissolving 0.01 to 0.5 milliliter per liter (the concentration of the surface bioactive peptide in the sterile aqueous solution of KCl is 0.01 to 0.5 mM / L) of the surface bioactive peptide in 50 to 200 milliliter per liter of sterile aqueous solution of KCl, and adding 0.01 to 2 percent (the concentration of the SDS, the LLS or the Chelex-100 in sterile water is 0.01 to 2 percent) of the SDS, the LLS or the Chelex-100, and 0.05 to 1 percent (volume) of ethanol, wherein the percentages are calculated based on the volume of the sterile water. The method for releasing the nucleic acid comprises the following steps of: taking the reagent for releasing the nucleic acid, and performing equivalent dilution on the reagent by using the sterile water according to different experimental purposes; balancing the diluted reagent to the room temperature, and then mixing the reagent uniformly; sub-packaging the mixture into a sample tube to be tested; adding a sample to be treated in the sample tube; and absorbing and beating the sample repeatedly for 5 to 10 times by a pipettor. The reagent for releasing the nucleic acid is safe to use and has a low production cost; and the method for releasing the nucleic acid is simple and convenient to operate.
Owner:SANSURE BIOTECH INC
Sample conditioning fluid based on denaturation precipitation as well as application thereof and nucleic acid releasing method
ActiveCN103642919AGuaranteed stabilityImprove detection efficiencyMicrobiological testing/measurementChelex 100Potassium hydroxide
The invention belongs to the biotechnology field, and in particular relates to sample conditioning fluid based on denaturation precipitation as well as application thereof and a nucleic acid releasing method. The sample conditioning fluid comprises the components in percentage by mass: 1-15 percent of PEG400, 0.1-2 percent of surfactant, 1-20 percent of solid phase resin chelex-100, 3-20 percent of polymerase chain reaction (PCR) enhancer, 10-40 mmol / L of potassium hydroxide and 100-400 mmol / L of potassium chloride (KCL), wherein the solvent of the conditioning fluid is sterile water. The sample conditioning fluid is used for removing substances inhibiting PCR amplification based on denaturation precipitation, and the treated product can be subjected to direct amplification so as to guarantee the amplification stability; because of non-selectivity of the treated product based on the principle on the amplification, the fluid is suitable for taq enzymes, chemically-decorated and antibody-decorated hot start enzymes and the like and is particularly suitable for performing amplification on long sections and gene sections with complicated structure.
Owner:亚能生物技术(深圳)有限公司
Rapid nucleic acid extraction kit and applications thereof
The invention belongs to the technical field of biological nucleic acid detection, and particularly relates to a rapid nucleic acid extraction kit and applications thereof. The kit of the invention comprises a grinding rod, an extraction liquid, an extraction column, a micro ultra-filter and a centrifuge tube, wherein the extraction liquid is a DEPC aqueous solution containing chelex-100. The invention further relates to a use method for the kit, and applications of the kit in extractions of DNA or RNA from pathological tissue paraffin section samples, samples of blood stains and semen stains, tissue fluid samples, cerebrospinal fluid samples, blood samples, serum samples, plasma samples, urine samples, hydrops samples, and tissue cell samples. According to the present invention, content and quality of the nucleic acid extracted by the kit of the present invention can well meet requirements of follow-up reactions of PCR, RT-PCR and sequencing. With the present invention, limitation of complexity and time consuming of nucleic acid extraction in the prior art are overcome well, and characteristics of safety and environmental protection are provided.
Owner:上海源奇生物医药科技有限公司
Extraction-free direct amplification reagent for real-time fluorescent quantitation PCR and application thereof
ActiveCN109402239AEasy to operateShort processMicrobiological testing/measurementHuman DNA sequencingFluorescence
The invention provides an extraction-free direct amplification reagent for real-time fluorescent quantitation PCR and application thereof. The direct amplification reagent comprises the following components: sodium hydroxide (NaOH), Surfactin, dimethyl sulfoxide (DMSO), sucrose and chelex-100. When the reagent is used, there is no need to extract and purify nucleic acid from a sample, only the sample and the reagent need to be mixed according to the equal volume ratio of 1:1. The mixture can be directly used for fluorescent PCR detection. The extraction-free direct amplification reagent is applicable to nucleic acid extraction of human genome, bacteria and viruses in mouth swabs, nasopharyngeal swabs, serum, plasma, genital tract secretion and other samples. The reagent is suitable for both the fluorescence PCR of a probe method and the PCR of a dying method, is suitable for a variety of clinical applications and has the advantages of low cost, simple operation, accurate results and the like, and detection can be simple and quickly completed.
Owner:贝南生物科技(厦门)有限公司
Method for fast extracting AM epiphyte environment DNA in plant rhizosphere soil
The present invention relates to process of fast extracting AM fungal genome DNA in plant rhizosphere soil environment, and belongs to the field of molecular biology and applied microbiology. The process includes the first wet screening to eliminate great amount of PCR proliferation limiting factors from rhizosphere soil, collecting AM fungal related structures of rhizosphere soil for DNA extraction, mechanically breaking wall and adding CTAB to promote DNA release, and further purifying with Chelex-100 resin. The process is easy, simple and fast, and has great DNA extracting amount, capacity of being concentrated and purified and other advantages. The present invention has excellent application foreground.
Owner:YUNNAN UNIV
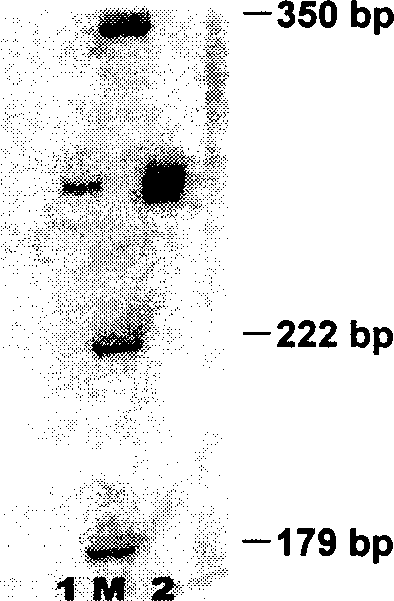
Spiroplasma pathogenic microorganism PCR fast checking technique
InactiveCN1733936AQuick checkIncreased sensitivityMicrobiological testing/measurementMaterial analysis by electric/magnetic meansPathogenic microorganismSpiroplasma
Disclosed is a PCR fast detection technique for Spiroplasma pathogen microorganisms comprising the following steps: (1) extracting template DNA in the detected sample with Chelex-100 method, (2) proceeding polymerase chain reaction with the 16S rDNA specific primer of the Spiroplasma, (3) subjecting the PCR product to electrophoresis detection, wherein the existence of Spiroplasma can be determined by the emergence of destination electrophoresis strip. The destination electrophoresis strip is at the position of 271bp, it can product certain diagnosis within 2-4 hours, and can carry out further semi-quantitative determination.
Owner:NANJING NORMAL UNIVERSITY
DGT probe and sediment/water interface micro-area detection method using same
InactiveCN104155215AAccurate Test ToolAccurate evaluation systemSurface/boundary effectDiffusionEutrophication
A DGT probe and sediment / water interface micro-area detection method using same. A three-layer fixed-glue DGT probe is provided in the invention. The probe includes sliver iodine-Chelex 100 fixed glue composed of sliver iodine glue and Chelex 100 fixed glue, wherein the sliver iodine glue and the Chelex 100 fixed glue are attached tightly to each other. The probe also includes hydrated ferric oxide fixed glue. Internal sides of both the sliver iodine-Chelex 100 fixed glue and the hydrated ferric oxide fixed glue are at least provided with substrates. External side of both the sliver iodine-Chelex 100 fixed glue and the hydrated ferric oxide fixed glue are at least provided with diffusion glue. The hydrated ferric oxide fixed glue is not arranged on the external side of the sliver iodine-Chelex 100 fixed glue. With the fixed glue DGT probe, a detection tool and an evaluation system which are more accurate in deep identification and determination of characteristics of lake phosphorus internal loading are provided and an important scientific basis for lake eutrophication control and water ecological restoration.
Owner:CHINESE RES ACAD OF ENVIRONMENTAL SCI
Method for rapidly and efficiently extracting DNA (Desoxvribose Nucleic Acid) from milk products such as liquid milk and milk powder
The invention belongs to the field of molecular biological technologies, particularly relates to the field of technologies of extracting DNA (Desoxvribose Nucleic Acid) from cattle and goat milk and milk powder, and discloses a method for rapidly and efficiently extracting DNA from cattle and goat liquid milk and milk powder. A Chelex-100 process and a Glassmilk (glass milk) DNA purification technology are perfectly combined and applied to extraction of trace DNA from cattle and goat liquid milk and milk powder for the first time. The collecting quantity of fresh liquid milk can be controlled to be 1-2 ml, the quantity of the milk powder can be controlled to be 1-10 mg, and the extracted DNA is high in concentration, extremely high in purity, and stable without being easily degraded. The extracted DNA can amplify more than 500-1000bp of a long-fragment gene sequence, and the obtained sequence is capable of developing subsequent molecular biology analysis. The method is simple in operation, low in consumption, short in time consumption, high in DNA purity, difficult in degradation, and convenient and practical in sampling; and the problem of difficulty in sampling blood and tissue can be prevented, no animal strsss reaction can be caused, and unnecessary loss caused by other sampling methods to the dairy enterprise and farmers is reduced.
Owner:黑龙江红星集团食品有限公司
Extraction method for microbial DNA in milk
The invention discloses an extraction method for microbial DNA in milk. The method comprises the following steps: adding 0.8 mL of a milk sample into 0.3 to 0.5 mL of an ES solution (EDTA with a pH value of 7.5 and concentration of 0.5 mol / L and SDS with concentration of 2%), carrying out uniform mixing and centrifugation and discarding supernatant; adding 0.8 to 1.2 mL of a PBS buffer solution, carrying out oscillation and shaking up, then carrying out centrifugation and discarding supernatant; adding 20 to 30 [mu]L of Staphylococcus lysozyme with a mass concentration of 0.02 mg / mL; adding 150 to 200 [mu]L of an aqueous Chelex-100 solution with a mass concentration of 10%, carrying out violent oscillation for 5 to 10 s and then carrying out heating in boiling water with a temperature of 100 DEG C for 8 to 12 min; and carrying out centrifugation and taking supernatant for a PCR reaction. The method provided by the invention employs a Chelex-100 process and can economically, rapidly, efficiently and directly extract bacterial DNA from the milk sample; and the bacterial DNA can be used for PCR detection of main pathogenic bacteria causing mastitis in cow.
Owner:CHINTEM TECH CONSULTING BEIJING CO LTD
Immobilized membrane for synchronously enriching phosphor and ferrum and preparation method thereof
InactiveCN104043409AImprove adsorption capacityShape stableOther chemical processesPreparing sample for investigationChelex 100Phosphoric acid
Provided is an immobilized membrane for synchronously enriching phosphor and ferrum. The immobilized membrane is a gel film which is prepared from a film-forming liquid in a gelling manner, wherein the film-forming liquid comprises zirconium hydroxide hydrate, Chelex-100 cation exchange resin and polyacrylamide. A preparation method of the immobilized membrane includes following steps: fully a mixing zirconium hydroxide hydrate powder, the Chelex-100 cation exchange resin and a polyacrylamide aqueous solution together to obtain a uniform membrane liquid, adding an appropriate amount of Tetramethylethylenediamine and ammonium persulfate, filling a glass mould with the membrane liquid, placing the glass mould horizontally at a low temperature of 2-4 DEG C for enabling zirconium hydroxide hydrate and Chelex to be freely precipitated, performing a heating process to 45+ / -5 DEG C and laying the glass mould up until the membrane liquid gels into a membrane. The immobilized membrane can synchronously enrich phosphor and ferrous ions with a high enriching capacity and can be used for a high resolution synchronous analysis of the phosphor and the ferrous ions in an environmental sample.
Owner:NANJING INST OF GEOGRAPHY & LIMNOLOGY
Method for high flux rapid extraction of vegetable single seed DNA
InactiveCN104845963AIncreased possibility of cross-contaminationNo harmDNA preparationWater bathsHigh flux
The invention discloses a method for high flux rapid extraction of vegetable single seed DNA, an early seedling single plant which is developed from a seed by breaking of hull, a to-be-tested sample single plant is ground into powder in a 1.5 mL pointed bottom centrifugal tube by a handheld electric disruptor, then100 mu L of 10 % Chelex-100 solution is added, the mixture is in water bath at 65 DEG C for 20 min, the mixture is quickly frozen for 10 min, and centrifuged for 3 min ate the speed of 13200 RPM / min, the supernatant is sucked to obtain sample DNA, and the sample DNA can be directly used for PCR amplification. According to the DNA extraction method, the whole course of sample DNA extraction can be finished in 35 minutes. The extracted DNA quality standards meet the requirements of the subsequent PCR amplification, the method is more suitable for the purpose of breeding industry variety breeding, high flux rapid extraction of DNA in the sample, and has the advantages of being efficient, fast, low in cost, simple in operation and the like.
Owner:TIANJIN INSTITUE OF QUALITY STANDARD & TESTING OF AGRICULTUAL PRODS
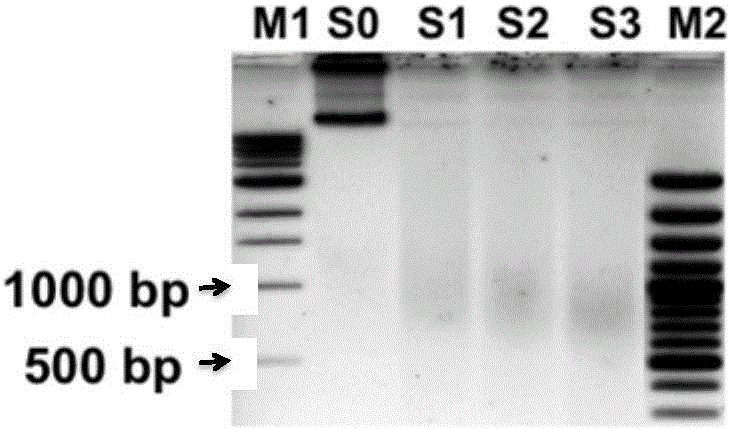
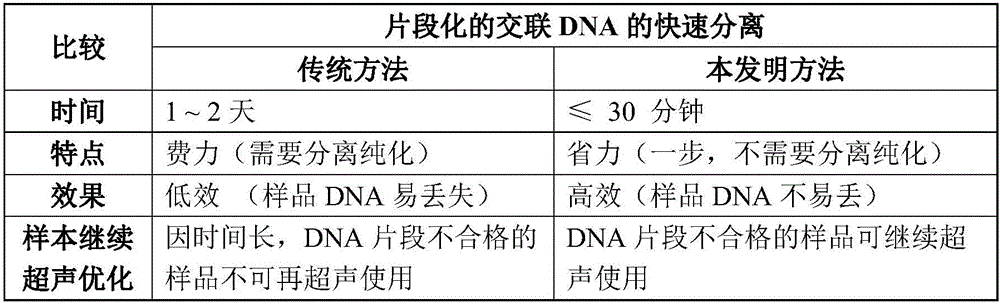
Rapid separation and detection kit and method for fragmented crosslinked DNA
InactiveCN105838783AIncrease success rateFast and effective quality control testingMicrobiological testing/measurementChelex 100A-DNA
The invention discloses a rapid separation and detection kit and method for fragmented crosslinked DNA. The kit includes a reagent for decrosslinking and separation extraction of fragmented DNA, and the reagent is Chelex 100. The kit can further include a protein degradation reagent, a DNA ladder marker, a sample loading buffer solution and / or an RNA degradation reagent. Chelex 100 can integrate multivalent metal ions, has high metal ion selectivity and binding force, can separate DNA and prevent DNA degradation. The rapid separation and detection method for fragmented crosslinked DNA can determine whether a to-be-detected sample needs further ultrasound or is used for a next step precipitation experiment according to the effect of rapid quality control ultrasound fragmentation, or can be used for ultrasound optimization of a plurality of to-be-detected samples so as to select optimum conditions. The kit and the method provided by the invention can be used for rapid detection of chromatin ultrasound fragmented sample quality in chromatin immunoprecipitation assay (ChIP) and other technologies, and can greatly improve the success rate of ChIP.
Owner:BEIJING JIAOTONG UNIV +1
Method for rapidly extracting plants sample DNA
InactiveCN101220390AIncrease cross contaminationReduce usageSugar derivativesMicrobiological testing/measurementSodium acetateCentrifugation
The invention discloses an improved method for extracting a plant sample DNA rapidly, which grinds the plant sample to be tested to powder in liquid nitrogen, then a certain amount of CTAB extraction buffer solution is added; the supernatant liquid is taken to be arranged in a centrifuge tube after being processed, a 3mol / L sodium acetate solution with one tenth of the volume of the supernatant liquid and isopropanol with 0.8 time of the volume of the supernatant liquid are added and the mixture is mixed evenly, the centrifugation is carried out for 5 to 10min and the supernatant liquid is discarded; 10 percent of Chelex-100 solution is added in the sediment for centrifugation, the taken supernatant liquid is the sample DNA which can be directly used for PCR amplification. The DNA extraction method which is adopted by the invention can complete the whole process of sample DNA extraction within 60 minutes. The quality standard of the extracted DNA meets the requirements of follow-up PCR amplification and is more applicable to the purpose of the rapid extraction of the sample DNA in production and scientific research processes.
Owner:CENT LAB TIANJIN ACADEMY OF AGRI SCI
Molecular detection technology of microsporidian on infected bumble bees
InactiveCN101603093AEasy extractionReduce lossesMicrobiological testing/measurementMicroorganism based processesEnzyme digestionChelex 100
The invention relates to molecular detection technology of microsporidian on infected bumble bees, which belongs to the technical field of animal inspection and quarantine and particularly relates to the technical field of molecular inspection for accurately and quickly detecting the microsporidian on imported and exported bumble bees. The molecular detection technology is characterized in that a Chelex-100 method is adopted to extract DNA; specific primers are used for carrying out PCR for one time; SSUrRNA genetic fragments of 540bp microsporidian are amplified; then, enzyme digestion of PCR products is carried out by restriction endonuclease Afa I; electrophoretic detection of enzyme digestion products are carried out by 2.0 percent agarose gel; and the type of the microsporidian parasitizing on the bumble bees can be detected directly through the length of fragments of the enzyme digestion products. The molecular detection technology can be applied to the inspection and quarantine of pathogen microsporidian on the imported and exported bumble bees.
Owner:BEE RES INST CHINESE ACAD OF AGRI SCI
Nucleic acid extraction method and extraction agent thereof
The invention relates to a nucleic acid extraction method. The method includes steps: well mixing a sample with an extraction agent proportionally, performing splitting decomposition for 3-5min at 85-95 DEG C, and uniformizing for 1-2 times during splitting decomposition; after particle settling, taking supernatant which is released nucleic acid. The extraction agent comprises Tris, EDTA, 3-[(3-cholesterol aminopropyl)dimethylamino]-1-propanesulfonic acid, NP-40, Tween-20, Chelex-100 and water, wherein the concentration of Tris is 5-10mM; the concentration of EDTA is 5-10mM; the volume concentration percentage of 3-[(3-cholesterol aminopropyl)dimethylamino]-1-propanesulfonic acid is 0.005%-0.02%; the concentration percentage of NP-40 is 0.01%-0.2%; the volume concentration percentage of Tween-20 is 0.01-0.2%; the volume content of Chelex-100 is 1-20%; PH of the extraction agent is 6-9.
Owner:BEIJING KINGHAWK PHARMA
Bacteria genome DNA extraction liquid, preparation and application thereof
ActiveCN101319213AEasy to makeEasy to storeFermentationPlant genotype modificationChelex 100DNA extraction
The invention discloses a bacteria genome DNA extracting solution in a blood and platelet product and a preparation method and an application method thereof. The bacteria genome DNA extracting solution consists of chelex-100, tween, NP-40, SDS, lysostaphin, a wall-breaking enzyme, a protease K and water. The extracting solution can be effectively used for extracting a bacteria genome DNA with high extraction efficiency. The preparation method and the application method are simple and convenient for operation.
Owner:SHANGHAI BLOOD CENT
Rapid nucleic acid extraction method for fluorescent quantitative PCR (polymerase chain reaction) detection
InactiveCN106987587AEasy to operateShorten the timeMicrobiological testing/measurementDNA preparationChelex 100Precipitation
The invention relates to a rapid nucleic acid extraction method for fluorescent quantitative PCR (polymerase chain reaction) detection. The method is applicable to extraction of DNA (deoxyribonucleic acid) of pathogenic microorganisms such as mycoplasmas, Gram negative bacteria and viruses, and the extracted product DNA can be used for fluorescent quantitative PCR detection. Samples of the pathogenic microorganisms such as the mycoplasmas, the Gram negative bacteria and the viruses are washed once by normal saline, solution containing 5% of Chelex-100, 0.5% of NP-40, 0.5% of Triton X-100 and 1mM of EDTA (ethylene diamine tetraacetic acid) is added in precipitation, the pH (potential of hydrogen) of the solution ranges from 11 to 12, the solution is re-suspended, heated for 10 minutes at the temperature of 80-90 DEG C and centrifuged for 5 minutes at the speed of 12000rpm, and supernatant is used for fluorescent quantitative PCR detection. Compared with an existing nucleic acid extraction method, the rapid nucleic acid extraction method has the advantages that operation is simple and rapid, cross infection among the samples is avoided, the DNA yield of the samples is increased, and fluorescent quantitative PCR amplification efficiency is improved.
Owner:江苏睿玻生物科技有限公司
Settling agent for improving extraction rate of free DNA (Deoxyribonucleic Acid) in blood plasma sample
The invention discloses a settling agent for improving the extraction rate of free DNA (Deoxyribonucleic Acid) in a blood plasma sample. The settling agent is prepared according to the following formula: 3.2 to 5.3 mg of human Cot-1 DNA and 1.4 to 2.2 mg of Chelex-100 cation exchange resin are added into each milliliter of the blood plasma sample. An experiment shows that the settling agent provided by the invention can be used for effectively removing interfering substances in blood plasma and remarkably improving the extraction yield of the free DNA, so that the detection rate of gene mutation of the free DNA of tumors can be improved (the extraction yield of the free DNA can reach 99 percent or more under the same extraction condition, so that the detection rate of the gene mutation ofthe free DNA of the tumors can be improved to 90 percent from 70 percent); and furthermore, the settling agent disclosed by the invention is convenient to use, and the extraction rate of the free DNAin the blood plasma sample can be remarkably improved only if the settling agent is added into the blood plasma sample and an existing extraction kit is used.
Owner:麦凯(浙江)医疗器械有限公司
Method for quickly and efficiently extracting deoxyribonucleic acid (DNA) of wheat stripe rust directly from infected wheat leaf blades
The invention discloses a method for quickly and efficiently extracting deoxyribonucleic acid (DNA) of wheat stripe rust directly from infected wheat leaf blades. The method is characterized in that 20mu l of prepared 20% chelex-100 suspension is drawn and injected into a 100mu l polymerase chain reaction (PCR) tube; 5mu l of chelex-100 supernate is drawn and dripped on the wheat leaf blades with wheat stripe rust uredispore and the spots of the leaf blades on which the chelex-100 supernate is dripped are smeared repeatedly to wash off the aecidiospore; the chelex-100 supernate containing the aecidiospore on the leaf blades is drawn back and added into the PCR tube containing 20mu l of 20% chelex-100 suspension; the mixture is drawn repeatedly and mixed evenly by a liquid-moving device; the mixture is put in boiling water bath for water bath for 2 minutes and then put in a vortex mixer to be oscillated and mixed evenly for 10 seconds; the mixture is subjected to water bath for 5 minutes and preserved at the temperature of minus 20 DEG C for later use; and the supernate can be used as templates of PCR. The invention provides an efficient and rapid method for extracting DNA of wheat stripe rust which is used as PCR amplification templates directly from infected wheat leaf blades.
Owner:YUNNAN AGRICULTURAL UNIVERSITY
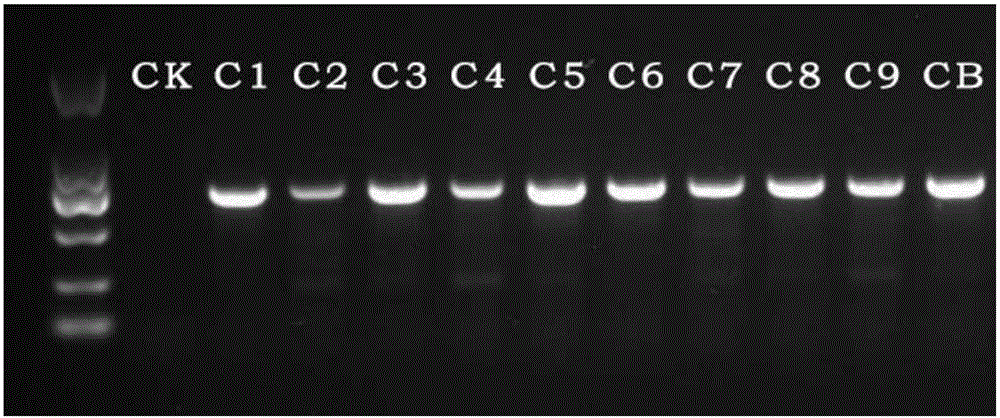
Method for quickly separating and detecting crosslinking DNA in paraffin-embedded tissue sample
InactiveCN105779438AHigh selectivityImprove bindingMicrobiological testing/measurementDNA preparationTissue sampleChelex 100
A method for quickly separating and detecting crosslinking DNA in a paraffin-embedded tissue sample comprises the following steps: adding ddH2O into the paraffin-embedded tissue sample, incubating at 60-80 DEG C, and removing supernate; adding a protein degrading agent, and incubating; adding a DNA de-crosslinking and separating and extracting agent, and vibrating, the DNA de-crosslinking and separating and extracting agent being Chelex 100; vibrating and incubating at a high temperature; centrifuging and reserving supernate; adding a sampling bearing buffer into the supernate; detecting electrophoretically. In the method, it is possible to estimate degradation degree and completeness of genome DNA of the tissue sample and determine the quality and uses of the sample through fragmented crosslinking DNA of the paraffin-embedded sample subjected to quick quality control; the method of the invention is a quick, low-rice and effective quality control detection method for paraffin-embedded sample DNA.
Owner:BEIJING JIAOTONG UNIV +1
Legionella DNA (deoxyribonucleic acid) extraction kit and method for extracting legionella DNA
InactiveCN103509785AReduce extraction costsAvoid cross contaminationDNA preparationEthylenediamineChelex 100
The invention relates to a method for extracting legionella DNA (deoxyribonucleic acid) from a clinic sample and an environmental water sample, and an extraction kit. The method comprises the following steps: denaturating protein in the samples by virtue of sodium azide and a Chelex-100 solution, after proteinase K is adequately digested, chelating non-nucleic acid organic matters by virtue of Chelex-100 solution, then centrifuging to remove Chelex particles, and separating other impurities from the DNA, thus dissolving the DNA of a supernatant in trihydroxymethyl aminomethane hydrochloride and ethylenediamine tetraacetic acid solution, and purifying and recovering the DNA. According to the method disclosed by the invention, a plurality of samples can be extracted simultaneously in a short time, the normal working time is less than 60 minutes, and DNA samples suitable for subsequent molecular biological analysis are obtained. The DNA samples obtained by the kit established by the method disclosed by the invention are high in purity; moreover, due to a single-tube operation, the pollution caused by transformation can be effectively avoided; the kit is low in cost and wide in use range.
Owner:SHANGHAI KINGMED DIAGNOSTICS INST
Method for detecting calcium retention capability of casein phosphopeptides products
InactiveCN104297184AThe determination method is simpleHigh sensitivityColor/spectral properties measurementsFood additivePotassium
The invention relates to a method for detecting calcium retention capability of casein phosphopeptides products, belonging to the field of food additives and detection methods of the food additives. The method for detecting calcium retention capability of casein phosphopeptides product comprises the following steps: removing calcium in CPP products by Chelex-100, adding deionized water and preparing the CPP products into a CPP solution with a certain concentration, stirring at the room temperature for 10 minutes, adding 5mL of a buffer solution prepared from monopotassium phosphate and potassium hydrogen phosphate with a pH value of 8.0 and uniformly stirring, then adding calcium chloride solution and uniformly mixing, standing the solution at 25 DEG C for 40 minutes until sediment is fully generated, subsequently centrifuging for 20 minutes at 2900rpm to remove the sediment in the solution, wherein the supernate contains calcium ions complexed with CPP; detecting the content of calcium in the supernate by virtue of an atomic absorption spectrometer and calculating the quantity of the calcium ions complexed with CPP with the certain concentration. The detection method has the characteristics of being simple and quick, high in sensitivity, high in reproducibility and accurate and reliable in result, therefore, the detection method can be applied to rapid and conventional analysis and detection of the calcium retention capability of casein phosphopeptides products.
Owner:西安莹朴生物科技股份有限公司
A method for rapidly extracting nucleic acid from biological samples
The present invention relates to a method for rapidly extracting nucleic acid from a biological sample. The method comprises: taking a biological sample, placing the sample in an aqueous solution of chelex-100 or a DEPC aqueous solution of chelex-100, and heating for 1-50 minutes at a temperature of 80-100 DEG C; placing an extraction column into a centrifuge tube, adding the resulting liquid to the extraction column, and centrifugating for 2-120 seconds at a rotation speed of 1000-20000 rotation / min; and taking the centrifugated liquid to carry out a PCR reaction or a RT-PCR reaction. According to the present invention, chelex-100 and a high temperature heating method are adopted to extract so as to eliminate a protease hydrolysis step, eliminate a paraffin section dewaxing step, remove an organic solvent xylene, and substantially shorten a time; and a retention membrane is placed in the extraction column so as to remove adverse effects on the PCR reaction or the RT-PCR reaction by the liquid paraffin and the chelex-100, and improve sensitivity of the PCR reaction or the RT-PCR reaction.
Owner:苏州云泰生物医药科技有限公司
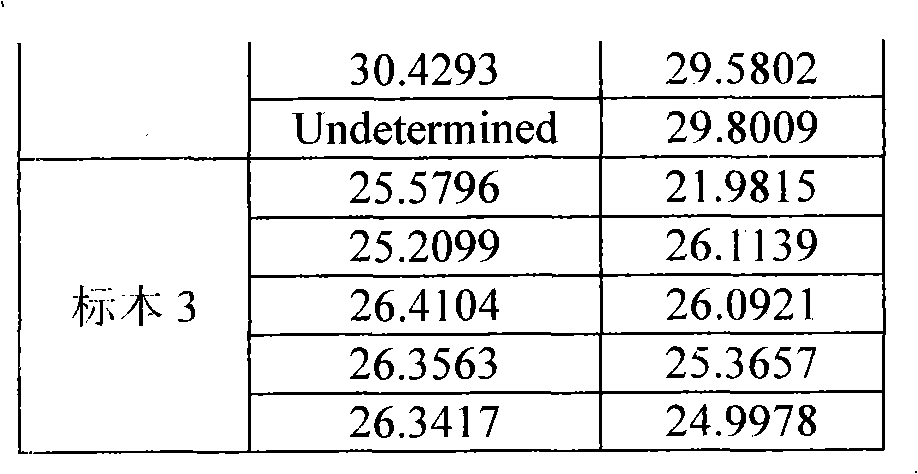
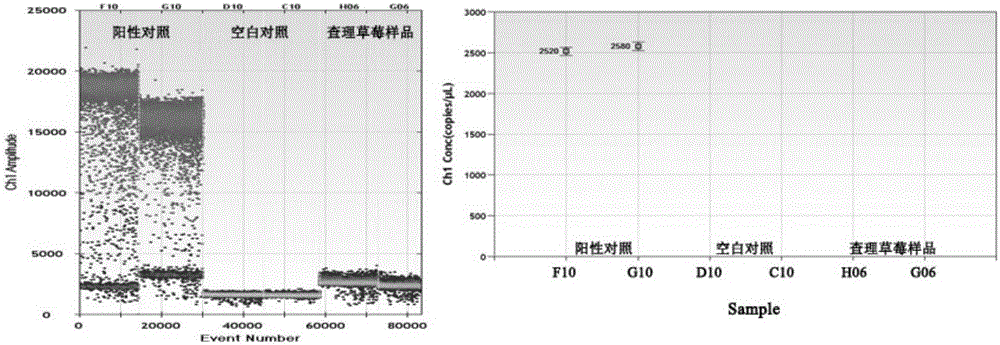
Method for rapidly and quantitatively screening salmonella in strawberry based on microdroplet digital PCR method
InactiveCN106244691AImprove detection efficiencyFast processMicrobiological testing/measurementMicroorganism based processesAgricultural scienceChelex 100
The invention discloses a method for rapidly and quantitatively screening salmonella in the strawberry based on a microdroplet digital PCR method. According to the method, salmonella invA virulence genes are adopted for synthesizing specific primers and probes, the bacterial DNA in the strawberry is extracted through a Chelex-100 method, and the salmonella is rapidly and quantitatively screened by adopting the microdroplet digital PCR technology. According to the established method, plasmids and a standard curve do not need to be constructed, so that the problem that the value determining accuracy of the sample is influenced by the quantity value deviation of the standard curve is avoided; in the whole detection process, the whole-process identification for the sample can be completed within 4h, therefore, the method has the advantages of being efficient, rapid, real-time, easy and convenient to operate, and a technical support is provided for rapidly and quantitatively screening the salmonella in the fresh fruits and vegetables, such as the strawberry.
Owner:TIANJIN INSTITUE OF QUALITY STANDARD & TESTING OF AGRICULTUAL PRODS
Extraction method of total genomic DNA of fish
The invention relates to an extraction method of total genomic DNA of fish. The extraction method comprises the following specific steps: A, taking the fish tissue containing DNA, washing, grinding, and placing into a centrifuging tube; B, adding with Chelex-100 solution, carrying out boiling water bath for 15-20 min, adding with proteinase K solution, and carrying out water bath at 50-60 DEG C for more then 3h; C, sucking liquid in the centrifuging tube, placing the liquid into another centrifuging tube, and carrying out centrifugal separation; D, taking the supernate, adding with RNase solution, carrying out water bath at 37 DEG C for more than 45 min; E, adding with Tris saturated phenol solution, uniformly mixing, carrying out centrifugal separation, and taking the supernate; F, adding with a suitable amount of chloroform and isoamylol mixed solution, uniformly mixing, carrying out centrifugal separation, and taking the upper solution; G, adding with alcohol, and placing for 1h or a longer time; H, carrying out centrifugal separation, and discharging the supernate; I, adding with alcohol, carrying out centrifugal separation, and discharging the supernate; J, removing the alcohol, adding with sterile water, and storing for later use. According to the extraction method, a reagent is convenient to fetch, the DNA extraction effect is good, the integrity of DNA is high, especially the extraction can be carried out by taking scale as the raw material, and limitation from the quantity of a sample is avoided.
Owner:NANCHANG UNIV
Method, kit and application for rapidly extracting genomic dna from hazelnut oil
ActiveCN106350510BAchieving amplified success rateFast and efficient extractionMicrobiological testing/measurementDNA preparationWater bathsDNA barcoding
The invention discloses a method for quickly extracting genome DNA from forest frog's oviduct, comprising the steps: (1) taking a forest frog's oviduct sample, adding Chelex-100 and protease K to the forest frog's oviduct sample, carrying out water bath heat insulation first, then terminating at high temperature, carrying out cell cracking, and centrifuging to obtain first supernatant, wherein the Chelex-100 is used for chelating non-nucleic acid organic matters in the forest frog's oviduct sample and inducing protein denaturation to facilitate the protease K to carry out enzymolysis, and the protease K is used for digesting protein in the forest frog's oviduct sample; (2) adding phenol / chloroform / isoamyl alcohol to the first supernatant for removing residual lipoid and protein; and (3) precipitating DNA by using isopropanol or absolute ethyl alcohol. The invention also provides a kit for quickly extracting genome DNA from the forest frog's oviduct, and application of a DNA bar code technology in identification of the forest frog's oviduct. The extraction method disclosed by the invention can realize quick and effective extraction of the genome DNA from dried forest frog's oviduct medicinal materials, and provides a basis for wider application of the DNA bar code technology.
Owner:哈尔滨市食品药品检验检测中心 +1
Method and kit for quickly extracting genome DNA from forest frog's oviduct, and application thereof
ActiveCN106350510AAchieving amplified success rateFast and efficient extractionMicrobiological testing/measurementDNA preparationWater bathsDNA barcoding
The invention discloses a method for quickly extracting genome DNA from forest frog's oviduct, comprising the steps: (1) taking a forest frog's oviduct sample, adding Chelex-100 and protease K to the forest frog's oviduct sample, carrying out water bath heat insulation first, then terminating at high temperature, carrying out cell cracking, and centrifuging to obtain first supernatant, wherein the Chelex-100 is used for chelating non-nucleic acid organic matters in the forest frog's oviduct sample and inducing protein denaturation to facilitate the protease K to carry out enzymolysis, and the protease K is used for digesting protein in the forest frog's oviduct sample; (2) adding phenol / chloroform / isoamyl alcohol to the first supernatant for removing residual lipoid and protein; and (3) precipitating DNA by using isopropanol or absolute ethyl alcohol. The invention also provides a kit for quickly extracting genome DNA from the forest frog's oviduct, and application of a DNA bar code technology in identification of the forest frog's oviduct. The extraction method disclosed by the invention can realize quick and effective extraction of the genome DNA from dried forest frog's oviduct medicinal materials, and provides a basis for wider application of the DNA bar code technology.
Owner:哈尔滨市食品药品检验检测中心 +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com