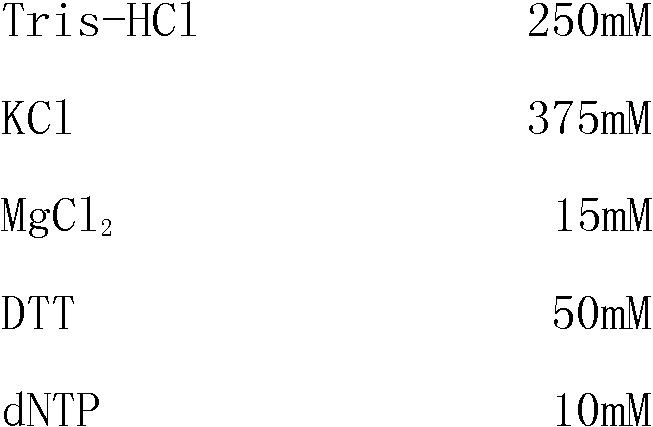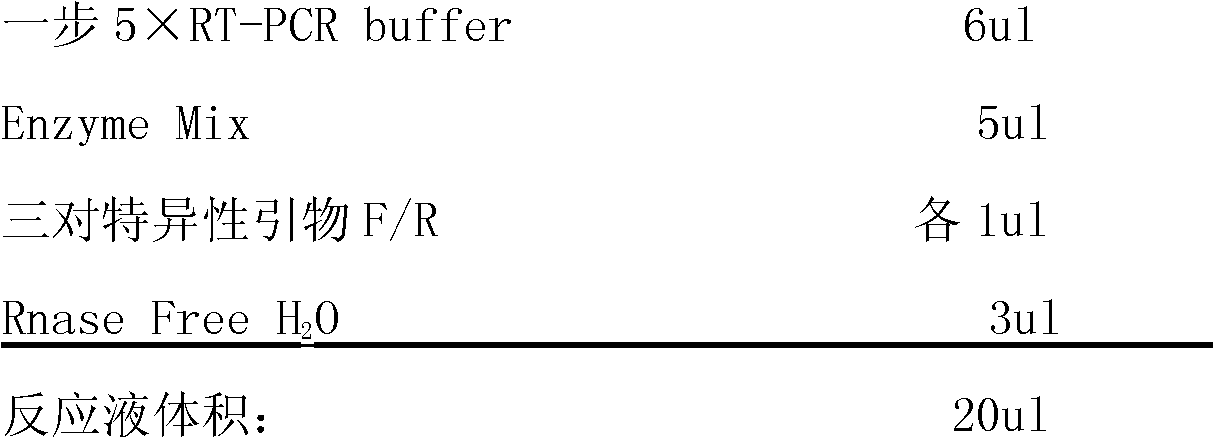Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
170 results about "Transcription (biology)" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Transcription is the first step of DNA based gene expression (gene is a short part of DNA that encodes for a protein), in which a particular segment of DNA is copied into RNA (especially mRNA) by the enzyme RNA polymerase. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript.
Plants having improved growth characteristics and methods for making the same
The present invention relates generally to the field of molecular biology and concerns a method for improving various plant growth characteristics by modulating expression in a plant of a nucleic acid encoding a GRP (Growth-Related Protein). The present invention also concerns plants having modulated expression of a nucleic acid encoding a GRP, which plants have improved growth characteristics relative to corresponding wild type plants or other control plants. The invention also provides constructs useful in the methods of the invention. The GRP may be one of the following: Seed Yield Regulator (SYR), FG-GAP1 CYP90B, CDC27, AT-hook transcription factors, DOF transcription factors and Cyclin Dependent Kinase Inhibitors (CKIs).
Owner:CROPDESIGN NV
Method for obtaining SSR (Simple Sequence Repeat) primer of lycium ruthenicum based on transcription sequencing
ActiveCN106754886AMicrobiological testing/measurementDNA preparationGenetic diversityMolecular breeding
The invention relates to a method for obtaining an SSR (Simple Sequence Repeat) primer of lycium ruthenicum based on transcription sequencing, and belongs to the technical field of molecular marking techniques of molecular genetics of biology. The method comprises the following steps of (1), carrying out transcriptome sequencing; (2), screening an EST-SSR (Expressed Sequence Tag-Simple Sequence Repeat) site; (3), carrying out primer design; (4), extracting DNA (Deoxyribonucleic Acid); (5), carrying out primer screening; (6), carrying out polymorphic primer screening by using 8-percent native polyacrylamide gel electrophoresis. The method for obtaining the SSR primer of the lycium ruthenicum based on the transcription sequencing, which is provided by the invention, can be used as a method for the correction of a linkage map of a genome between remote species and comparative mapping, and has higher utilization value in two aspects. The greatest advantages of the method are that the development is simple and quick and the expense is low. The general characteristic of the SSR of the lycium ruthenicum is specified by utilizing an SSR molecular marking technique; the SSR primer of the lycium ruthenicum is developed; the foundation is laid for carrying out the research on the genetic diversity, the linkage map construction and the affinity relationship of species resources of the lycium ruthenicum by utilizing an SSR molecular marker, so that the method is better applied to molecular breeding.
Owner:BEIJING FORESTRY UNIVERSITY
Detection kit and detection method for H1, H3 and H9 type avian influenza viruses
ActiveCN105296670AEasy to detectShorten detection timeMicrobiological testing/measurementForward primerRNA extraction
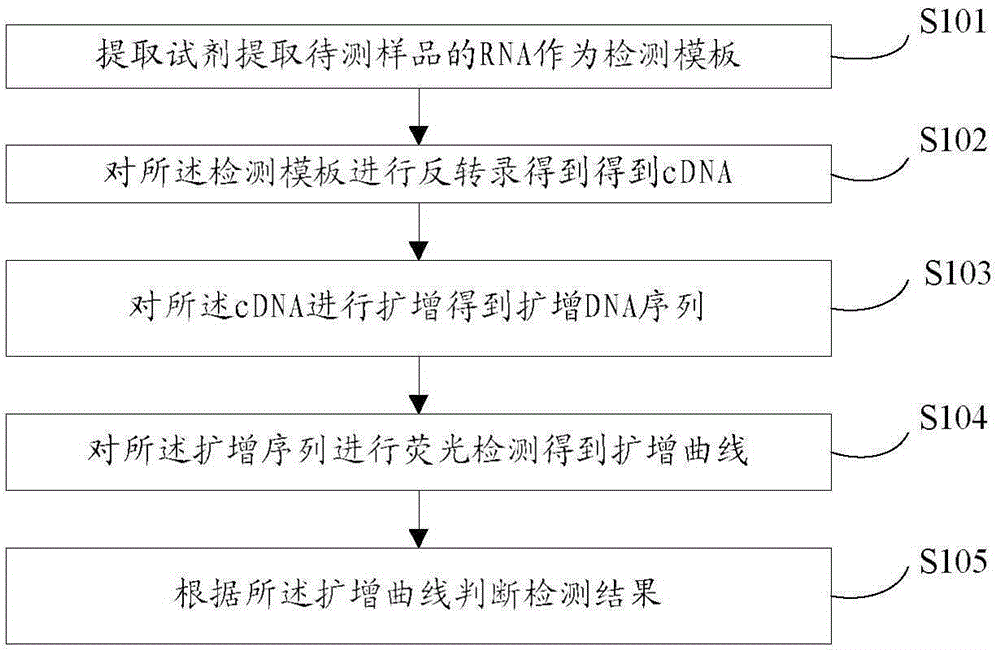
The invention discloses a detection kit and detection method for H1, H3 and H9 type avian influenza viruses and belongs to the technical field of biology. The kit comprises an RNA extraction kit, an RNA reverse transcription kit and an RPA detection kit. The RNA extraction kit is used for extracting RNA of a to-be-detected sample to be used as a detection template. The RNA reverse transcription kit is used for conducting reverse transcription on the RNA in the detection template to obtain cDNA. The RPA detection kit comprises primers and fluorescent dye. The primers are used for amplifying the cDNA to obtain an amplified DNA sequence. The primers comprise the forward primer and the reverse primer. The fluorescent dye is used for conducting fluorescence detection on the amplified DNA sequence. According to the detection kit and the detection method, the avian influenza viruses can be rapidly, easily and conveniently detected.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY SCI SHANXI ACAD OF AGRI SCI SAAS +1
Method for realizing efficient and fixed-point transgenosis by using TALEN (Transcription Activator-Like Effector Nuclease) technology
InactiveCN103266122ARealize point integrationEasy to operateVector-based foreign material introductionForeign genetic material cellsNucleaseTransgene
The invention relates to a method for realizing efficient and fixed-point transgenosis by using a TALEN (Transcription Activator-Like Effector Nuclease) technology, belonging to the field of molecular biology. The method comprises the following steps of: designing a TALEN target point with a specific sequence in a special position in an organism genome by using the TALEN technology, constructing a TALEN plasmid, and meanwhile, constructing an exogenous gene plasmid; then, co-transfecting the TALEN plasmid and an exogenous gene into recipient cells, and then incising in a to-be-integrated position of a recipient cell genome; and finally, automatically inserting an exogenous gene sequence to the exogenous gene at the incision of the recipient cell genome. The fixed-point integration of the exogenous gene on the genome is realized by using the method; and the method is simple in operation, high in controllability and strong in practicability.
Owner:YUNNAN AGRICULTURAL UNIVERSITY
Sarcoma fusion gene and/or mutation joint detection primer group and kit
ActiveCN112094915AAchieving Simultaneous DetectionHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationLigationTranscription (biology)
The invention relates to the technical field of molecular biology, and particularly discloses a sarcoma fusion gene joint detection primer group, kit and detection method. The kit provided by the invention comprises: 1) a special linker for DNA library construction; 2) a specific composite primer for detecting fusion genes and point mutation; and 3) a composite primer for library amplification. DNA and RNA of a to-be-detected sample are respectively extracted, reverse transcription is performed on RNA to synthesize cDNA, the cDNA is mixed with DNA, fragmentation, terminal repair and linker ligation are performed on the mixture, PCR amplification is performed on the product, a library is enriched, and high-throughput sequencing is performed. According to the fusion gene detection kit and detection method provided by the invention, various fusion genes or mutations related to sarcoma can be detected at one time, the detection efficiency is improved, the operation is convenient, the consumption time is short, and the sensitivity is relatively high and can reach 0.1%.
Owner:苏州科贝生物技术有限公司
Dual fluorescence quantification RT-PCR detection kit and application thereof
ActiveCN102230023AIncreased biosecurity riskPerfecting flu testingMicrobiological testing/measurementFluorescence/phosphorescenceSequence analysisReverse transcriptase
The invention provides a dual fluorescence quantification RT-PCR (reverse transcription-polymerase chain reaction) detection kit, comprising a deoxynucleoside triphosphate mixture, MgCl2, an RNA enzyme inhibitor, a Moloney murine leukemia virus reverse transcriptase, a DNA polymerase, a influenza virus standard and a reference substance. Based on sequence analysis of present pervasive A H1N1 influenza virus, the invention provides a multiple fluorescence quantification PCR molecular biology gene diagnosis method and a diagnostic kit which are rapid, specific, accurate and sensitive. In addition, one reaction tube can simultaneously detect and differentiate an influenza A virus or an influenza B virus in 2h, so as to improve influenza detection.
Owner:ZHEJIANG CENT FOR DISEASE CONTROL & PREVENTION
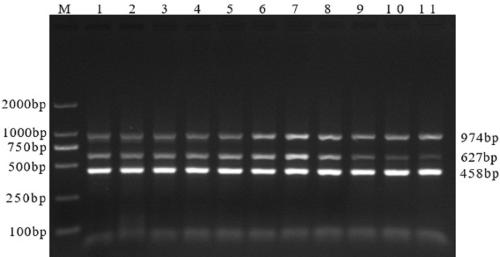
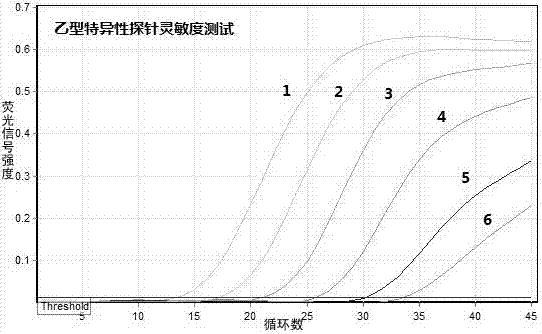
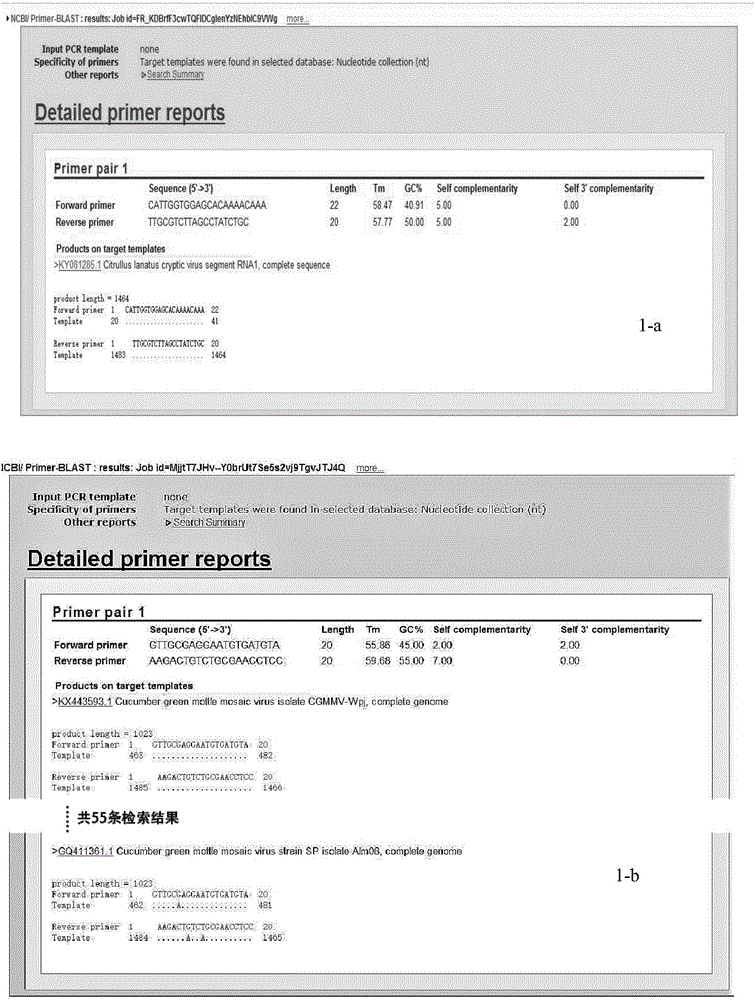
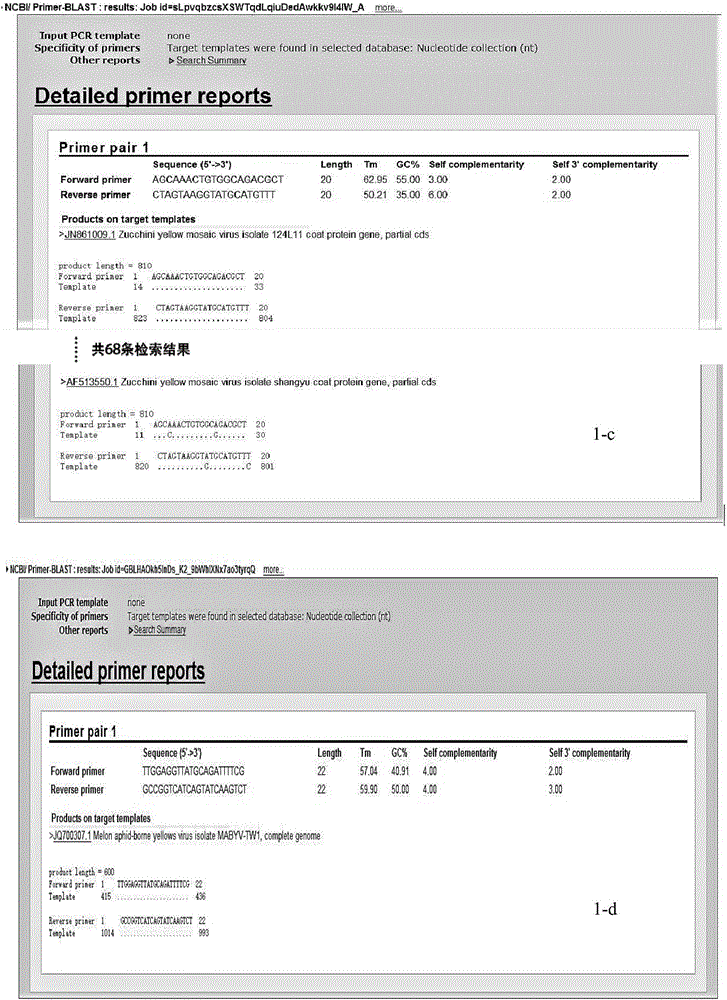
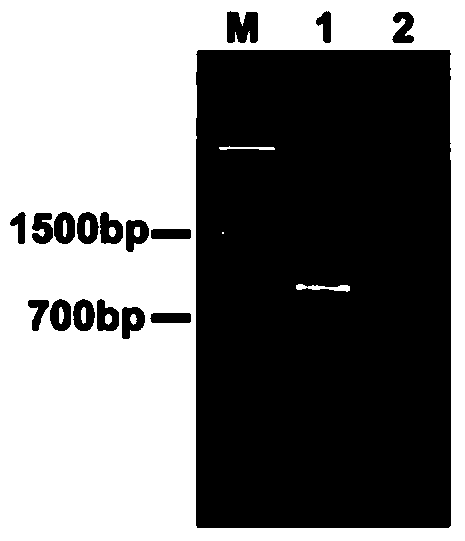
Multiple RT-PCR (Reverse Transcription-Polymerase Chain Reaction) method for synchronously detecting five watermelon viruses and application of method
InactiveCN106702029AWith characteristicsCompatibleMicrobiological testing/measurementMicroorganism based processesDisease epidemiologyPcr method
The invention belongs to the technical field of crop disease diagnosis and molecular biology and discloses a multiple RT-PCR (Reverse Transcription-Polymerase Chain Reaction) method for synchronously detecting five watermelon viruses. A PCR system contains five pairs of specific primers (SEQ ID NO1-10), and the five viruses are synchronously multiplied in one PCR system, so as to obtain specific bands 1464bp, 1023bp, 810bp, 600bp and 327bp and establish and optimize a multiple RT-PCR reaction system capable of synchronously detecting the viruses CiLCV, CGMMV, ZYMV, MABYV and WMV. A detection result indicates that the optimized multiple RT-PCR reaction system can detect a field sample quickly, efficiently and accurately. The method has a significant meaning for forcasting of occurrence and disease epidemiology of five common viruses such as CiLCV and the like on monitored watermelons.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
Preparation, detection and application of polyclonal antibody of Yam mild mosaic virus
ActiveCN103539856AEfficient detectionHighly sensitive and accurate detectionSerum immunoglobulinsMicrobiological testing/measurementSorbentSpecific immunity
The invention discloses preparation, detection and application of a polyclonal antibody of a Yam mild mosaic virus (YMMV). Based on a YMMV genome, the inventor purifies a nuclear inclusion protein / coat protein (NIb / CP) of the YMMV through prokaryotic expression and uses the nuclear inclusion protein / coat protein to prepare the polyclonal antibody for an immune rabbit. The polyclonal antibody can specifically identify the Nib / CP protein of YMMV through prokaryotic expression and the Nib / CP protein of the YMMV virus which infects common yam rhizome, and generates specific immunity response. On the basis, the inventor further builds a set of efficient, high sensitive and accurate IC-RT-PCR (Immunocapture-Reverse Transcription-Polymerase Chain Reaction, ELISA (Enzyme-Linked Immuno Sorbent Assay) and Western blot methods which can specifically detect the YMMV from a common yam rhizome plant which is infected by YMMV. By applying the invention, a material basis and a technical support can be provided for research on interaction of the YMMV and the host, quick detection of the virus and somatotype and molecular biology study, thereby laying a solid foundation for epidemiologically monitoring and preventing treating the virus.
Owner:GUANGXI UNIV
HPV nucleic acid detection kit based on enzyme-linked immunosorbent assay and application of HPV nucleic acid detection kit
InactiveCN104020291ALow costNo need for amplification and detection equipmentMicrobiological testing/measurementMaterial analysisViral loadBiology
The invention belongs to the technical fields of molecular biology, immunology and nucleic acid chemistry, and particularly relates to an HPV nucleic acid detection kit based on enzyme-linked immunosorbent assay and application of the HPV nucleic acid detection kit. The kit is based on an immunoassay method for detecting high-risk HPV16 type E6 and E7 RNA by using an Sdr monoclonal antibody; a detection process is realized as follows: amplification of a target sequence by using technologies such as inverse transcription and PCR is not needed, an unlabelled DNA probe is used for being hybridized with the HPV16 type E6 and E7 RNA to form a DNA:RNA heterozygote, and then the heterozygote is recognized by using the Sdr monoclonal antibody, and subsequent signal multiplication is performed. A detection method has the advantages of high speed, low cost, high sensitivity, and no need of complicated amplification and detection instruments, and is capable of detecting an initial viral load.
Owner:NANJING UNIV
Shark single-domain antibody targeting new coronavirus N protein as well as preparation method and application of shark single-domain antibody
ActiveCN113336844AHigh affinityImprove bindingImmunoglobulins against virusesAntiviralsTotal rnaSingle-domain antibody
The invention discloses a shark single-domain antibody targeting new coronavirus N protein as well as a preparation method and application thereof, and belongs to the technical field of biology. The amino acid sequence is as shown in SEQ ID NO: 1 or SEQ ID NO: 2, and the nucleotide sequence is as shown in SEQ ID NO: 3 or SEQ ID NO: 4; meanwhile, the invention also provides a preparation method of the shark single-domain antibody targeting the new coronavirus N protein, and the preparation method comprises the following steps: separating peripheral blood lymphocytes of immune stripe bamboo sharks, extracting total RNA, and carrying out reverse transcription to obtain cDNA; taking cDNA as a template, amplifying a vNAR fragment of mottled bamboo shark, and connecting with a vector to construct a phage library; screening positive clones for identifying the new coronavirus N protein from the phage library; and constructing an expression vector, and inducing to express the new coronavirus N protein single-domain antibody to finally obtain the shark single-domain antibody targeting the new coronavirus N protein.
Owner:MINJIANG UNIV
Plant expression vector and construction method thereof, and method for producing chicken alpha interferon by utilizing crowtoe as bioreactor
InactiveCN102121029AExtensive managementLong grass seasonFermentationHorticulture methodsBiotechnologyWestern blot
The invention relates to a plant expression vector and a construction method thereof, and a method for producing chicken alpha interferon by utilizing crowtoe as a bioreactor, belonging to the technical field of biology. The plane expression vector pSFRLH is shown as the figure 1 in the specification. In the invention, main stem sections, rachis stem sections and tender leaves of crowtoe are used as explants, and a constructed plane expression vector chicken alpha interferon gene ChIFN-alpha is subjected to genetic transformation and regeneration and then subjected to PCR (Polymerase Chain Reaction), RT-PCR (Reverse Transcription-Polymerase Chain Reaction), Southern blot, Western blot and ELISA (Enzyme-Linked Immuno Sorbent Assay). Proved by a detection result, the ChIFN-alpha gene is integrated into a crowtoe chromosome genome and correctly transcribed, translated and expressed, and recombinant protein can be neutralized by a ChIFN-alpha antibody. The invention lays the foundation for producing animal medical protein by utilizing crowtoe pasture grass as a bioreactor and provides a new thinking for the control of animal epidemic diseases.
Owner:GUIZHOU UNIV
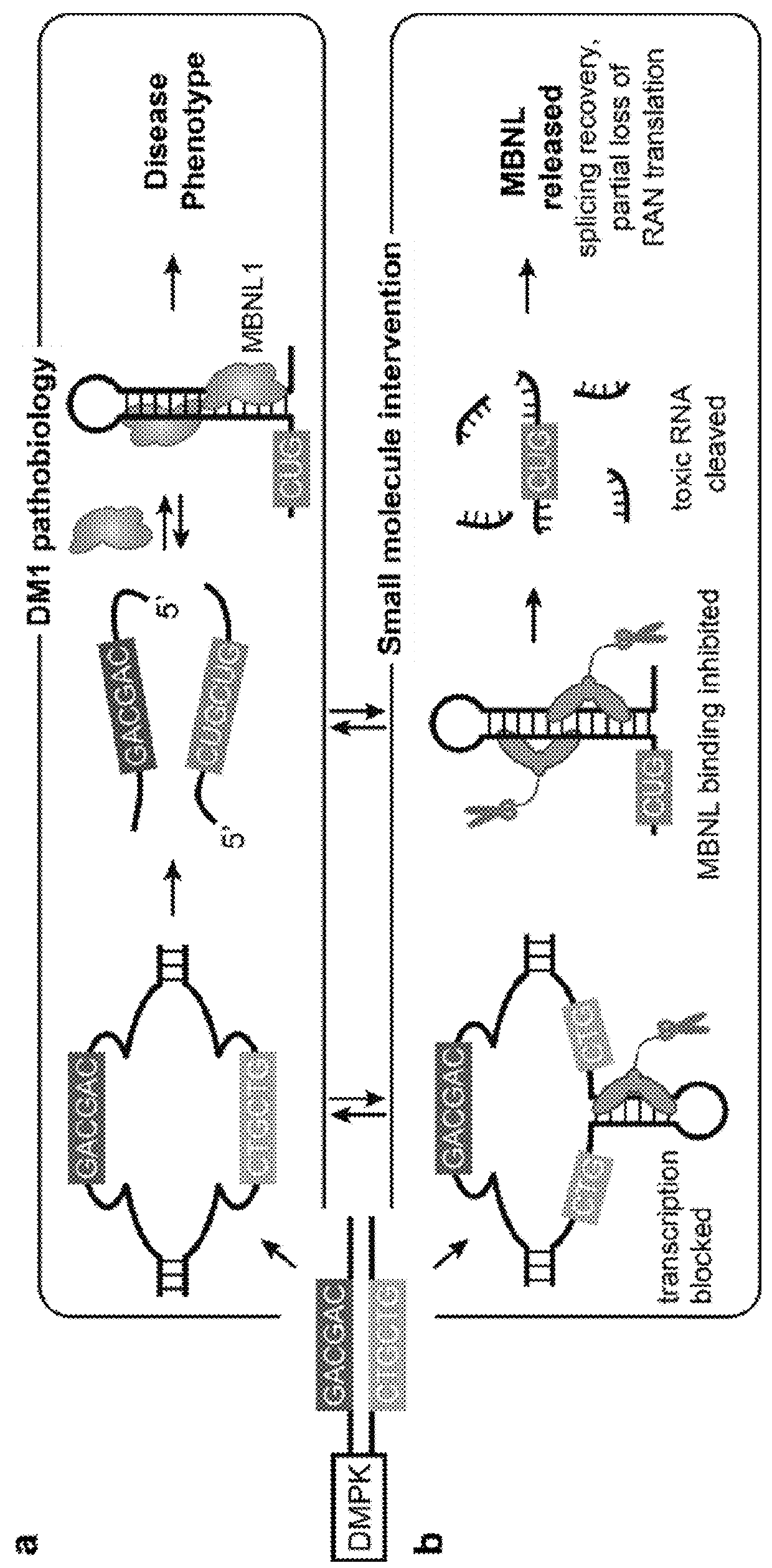
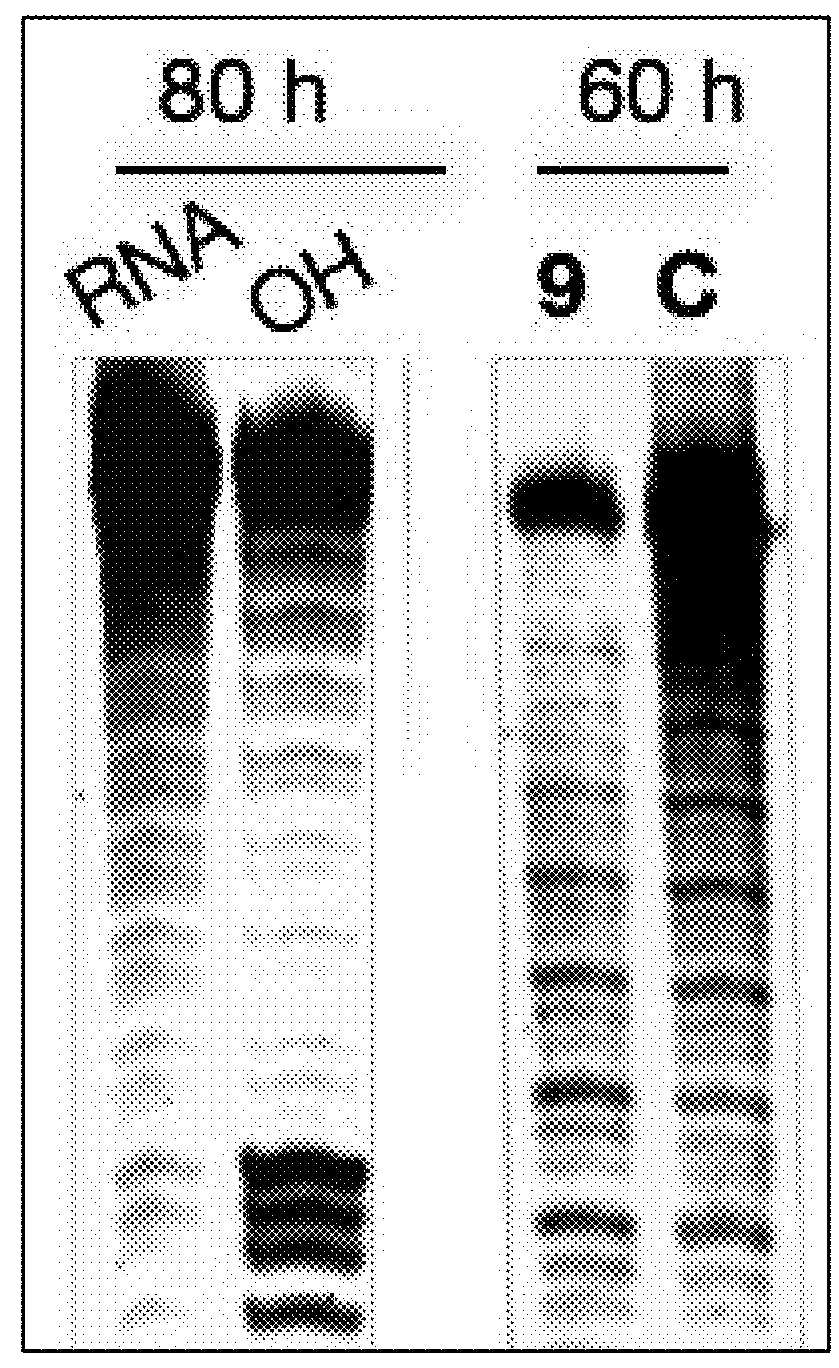
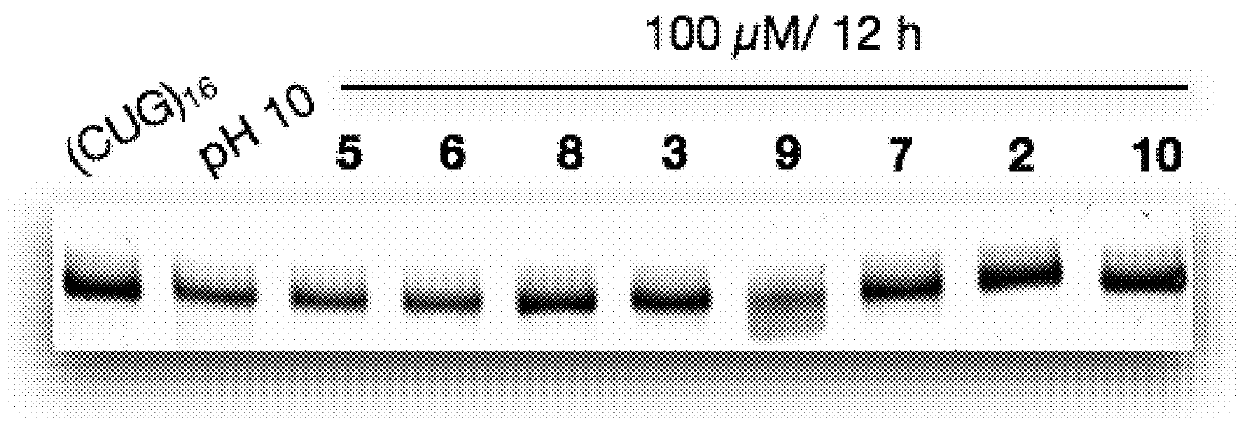
Compounds and methods for myotonic dystrophy therapy
The invention provides rationally designed multi-targeting therapeutic agents for myotonic dystrophy type 1 (DM1), an incurable neuromuscular disease that originates in an abnormal expansion of CTG repeats (CTGexp) in the DMPK gene. The rationally designed small molecules target the DM1 pathobiology in three distinct ways: (1) binding the expanded trinucleotide repeat, CTGexp, and inhibiting its transcription to the toxic CUGexp RNA, (2) binding the CUGexp RNA and releasing sequestered muscleblind-like protein (MBNL1), and (3) cleaving the toxic CUGexp in an RNase-like manner. Importantly, the compounds can reduce the levels of CUGexp in DM1 model cells and reverse two separate CUGexp-induced phenotypes of DM1.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS
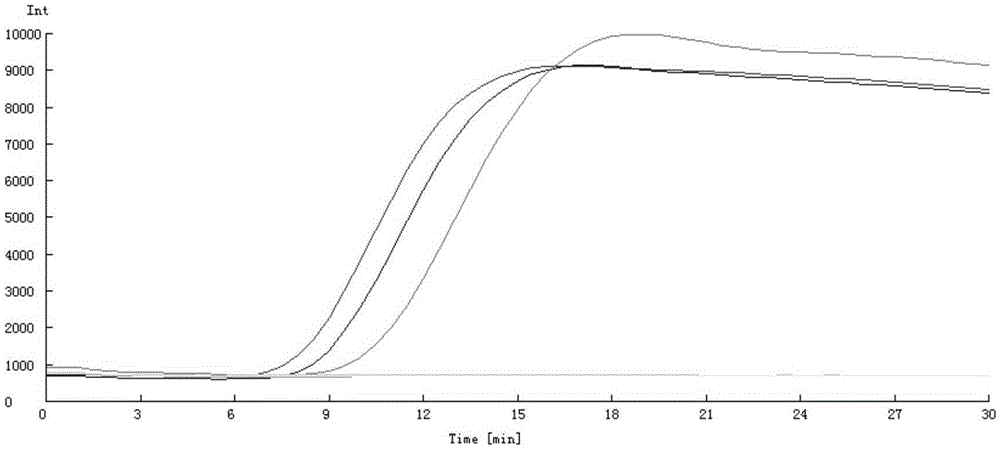
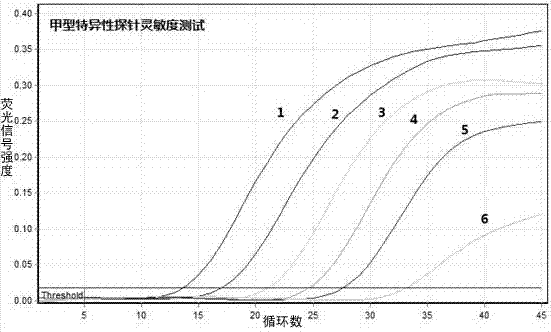
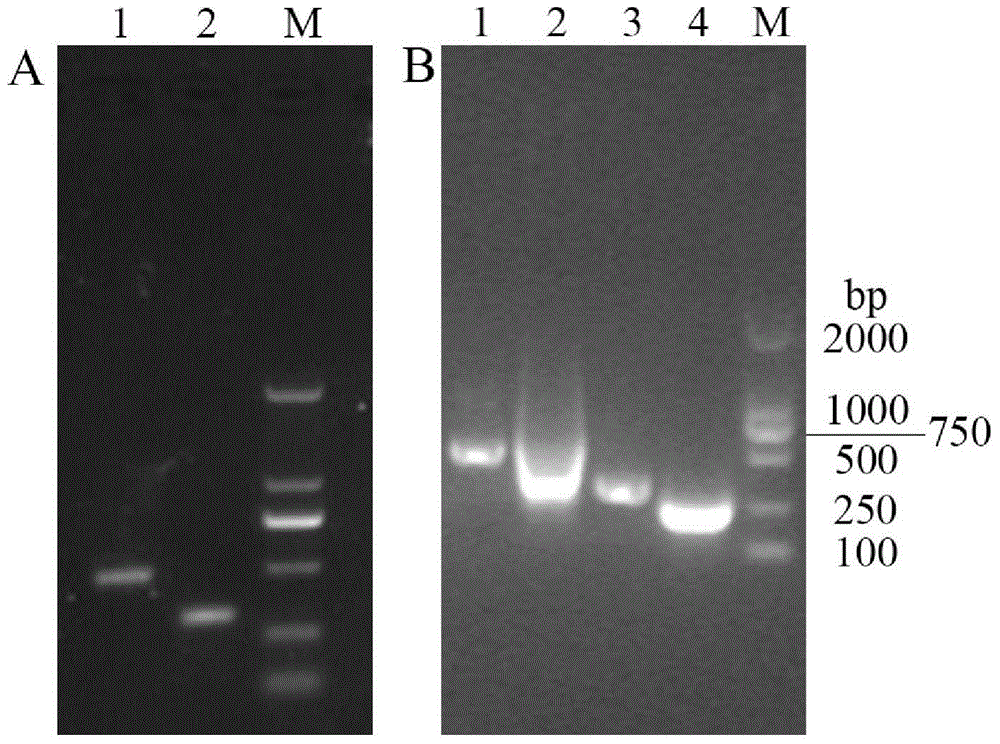
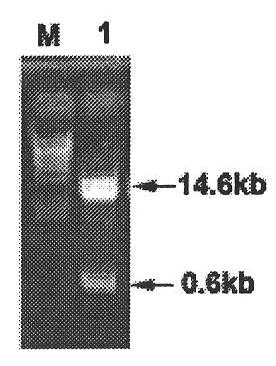
Method for constructing full-length infectious cDNA [complementary DNA (deoxyribonucleic acid)] of PEDV JS2008 (porcine epidemic diarrhea virus JS2008) strains and application of full-length infectious cDNA
ActiveCN107267532AImprove efficiencySsRNA viruses positive-senseVirus peptidesSilent mutationEnzyme digestion
The invention provides a method for constructing full-length infectious cDNA [complementary DNA (deoxyribonucleic acid)] of PEDV JS2008 (porcine epidemic diarrhea virus JS2008) strains and application of the full-length infectious cDNA, and belongs to the field of molecular biology and virology. The method for constructing the full-length infectious cDNA of the PEDV JS2008 strains includes steps of amplifying 6 fragments from reverse transcription products of total virus RNA (ribonucleic acid) of the PEDV JS2008 strains, inserting the fragments into vectors pSMART and carrying out silent mutation on enzyme digestion sites of incision enzymes PflmI at inappropriate locations; carrying out enzyme digestion on various recombinant vectors, recycling target cDNA fragments and connecting the target cDNA fragments with one another to obtain the full-length infectious cDNA of the PEDV JS2008 strains. The reverse transcription products are used as templates. The method for constructing the full-length infectious cDNA of the PEDV JS2008 strains and the application have the advantages that the method is ingenious, is high in efficiency and can be applied to rescuing porcine epidemic diarrhea viruses, and accordingly research on PEDV pathogenic mechanisms and novel vaccine can be carried out.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Complementary DNA (cDNA) sequence of tobacco NtFT1 genes and transient expression thereof for inducing tobacco early blossoming
The invention relates to a complementary DNA (cDNA) sequence of tobacco NtFT1 genes and transient expression thereof for inducing tobacco early blossoming, in particular to NtFT1 genes of tobacco FT (Flowering locus T) and application of the NtFT1 genes in inducing the tobacco early blossoming, and belongs to the field of genes in molecular biology. The invention discloses a full-length coded sequence of the NtFT1 genes derived from tobacco for controlling the plant blossom time and a protein sequence coded by using the full-length coded sequence. The NtFT1 genes are obtained by performing Blastn comparison and splice site analysis on the data searched from China tobacco genome sequencing planning data through homologous sequence searching. According to the cDNA sequence, a method for performing semi-quantitative polymerase chain reaction (PCR) or reverse transcription-polymerase chain reaction (RT-PCR) on NtFT1 gene specific primers is designed to clone the genes. According to the invention, a potato virus X (PVX) virus expression vector of the NtFT1 genes is also constructed, and the constructed virus expression vector is used for performing diafiltration and infestation on flue-cured tobacco varieties including Hongda and Yunnan tobaccos 87 and K326 through agrobacterium, and can induce the early blossoming of the varieties to prove that the genes have a function of inducing the early blossoming of cured tobaccos. When a PVX virus transient expression system of the NtFT1 genes is used for tobacco breeding, the growing time of the tobaccos can be shortened, and the fixed number of years for breeding a new tobacco variety is reduced.
Owner:YUNNAN ACAD OF TOBACCO AGRI SCI
RT(Reverse Transcription)-PCR (Polymerase Chain Reaction) kit for detecting parainfluenza virus in one step
InactiveCN102140550AShort detection time periodImprove detection efficiencyMicrobiological testing/measurementParainfluenza virusQualitative analysis
The invention discloses a RT (Reverse Transcription)-PCR (Polymerase Chain Reaction) kit for detecting parainfluenza virus in one step, and relates to a PCR kit in the technical field of biology. The kit comprises one-step 5xRT-PCRBuffer, a positive control substance, a negative control substance, four pairs of specific primers and EnzymeMix, wherein the sequences of the specific primers 1F and 1R designed for the parainfluenza virus A are respectively 5'-CCGGTAATTTCTCATACCTATG-3' and 5'-CCTTGGAGCGGAGTTGTTAAG-3'. The kit has the advantages of short detection time period, high detection efficiency, high specificity of the detected virus, high accuracy, simple operation, easy generalization and good repeatability of the experimental result; and the kit can carry out virus quantitative analysis, the detection sensitivity of the kit is higher than that of an immunology detection method.
Owner:WUHAN UNIV
Method for testing development process of silkworm embryos
InactiveCN103070141AAccurate measurementAvoid judgment errorsAnimal husbandryBiotechnologyTranscriptional expression
The invention discloses a method for testing the development process of silkworm embryos. The method specifically comprises the following steps of: (1), establishing a model expressing the incidence relation between the relative transcription expression quantity of the acetylcholine esterase type 1 (ace1) of a to-be-tested variety of the silkworm embryos and the development process of the embryos; (2), selecting at least one embryo from the to-be-tested silkworm embryos to use as a sample, testing the relative transcription expression quantity of the ace1 of the sample, and then contrasting with the model obtained in step (1) to determine the development process of the silkworm embryos according to the relative transcription expression quantity of the ace1 of the to-be-tested sample. The development stage of a whole graine can be accurately determined by the method through the test on the development process of a small quantity of silkworm embryos. Manual operation errors can be reduced, the test process is simplified, and a development process identification method is provided for molecular biology study of the silkworm embryos.
Owner:SUZHOU UNIV
Single-input multi-output gene loop, expression vector, host cell and application of host cell
InactiveCN110117614APromote proliferation and differentiationAvoid accidental injuryVectorsCytokines/lymphokines/interferonsEffector cellAntigen
The invention relates to the technical field of biology, and discloses a single-input multi-output gene loop. The gene loop selects two pairs of inhibitory transcription factors having good orthogonality and remarkable inhibition efficiency differences and DNAs combined with the inhibitory transcription factors for responding to the cell surface antigen density, and different functional proteins are driven to be expressed according to the intensity of input signals. Effect cells are improved through the single-input multi-output gene loop, and can be driven to differentially express differentfunctional proteins such as cell factors along with density changes of antigens of target cells, and a combination effect on the target cells is achieved. According to the single-input multi-output gene loop, the efficiency for cell treatment of a single antigen can be improved, and the single-input multi-output gene loop is considered in a method for cell treatment of multiple antigens.
Owner:FUDAN UNIV
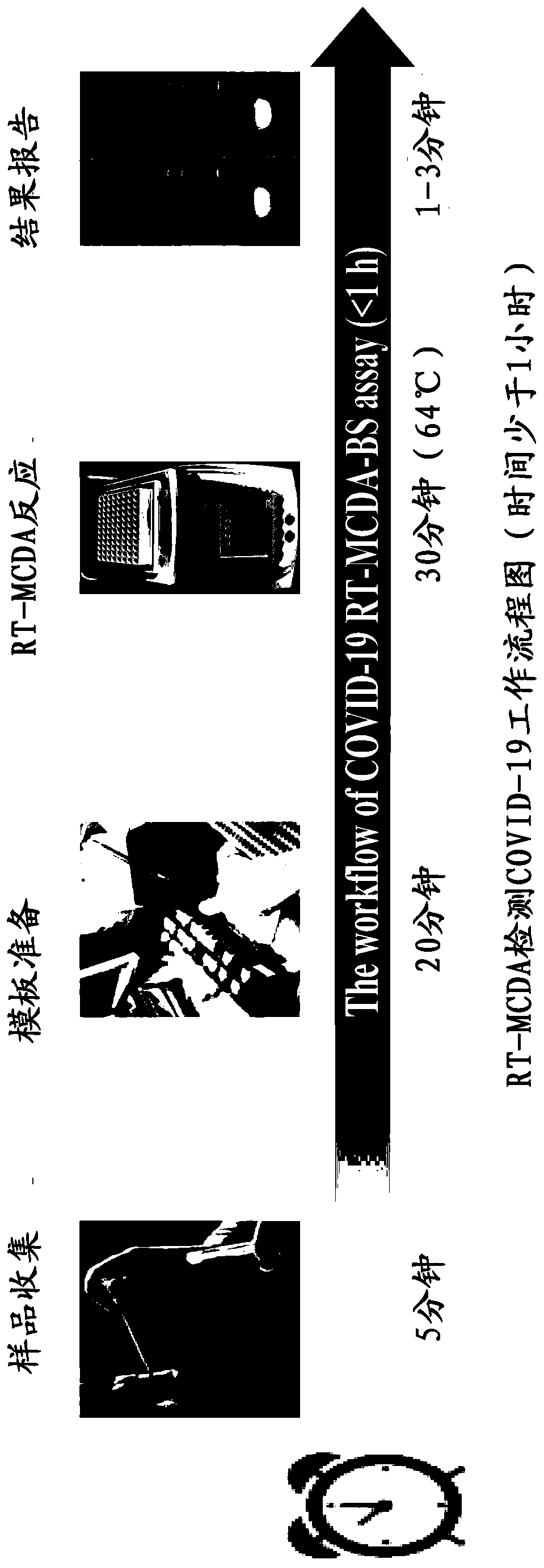
Method for detecting SARS-CoV-2 by combining reverse transcription multi-cross displacement amplification with nano-biology sensing
ActiveCN111518948AMicrobiological testing/measurementAgainst vector-borne diseasesOpen reading frameGenetic Crossing-Over
The invention discloses a method for detecting SARS-CoV-2 by combining reverse transcription multi-cross displacement amplification with nano-biology sensing. The method aims at an open reading frame1a / b(F1ab) fragment and two targets of an N gene of the SARS-CoV-2, a uniquely designed multi-cross displacement amplification primer is used, a digoxin half antigen is marked on the 5'-end of a F1absequence cross primer CP1 in the multi-cross displacement amplification, and a FITC (fluorescein) half antigen is marked on the 5'-end of an N gene cross primer CP1. The method aims at a situation that the Flab fragment of the 2019-novel coronavirus and a product amplified by the N gene can be visually detected through a macromolecule nano-biology sensor. The method is convenient, quick, sensitive and specific and is suitable for the specific detection of the 2019-novel coronavirus.
Owner:贵州省疾病预防控制中心
One-step RT-PCR (Reverse Transcription-Polymerase Chain Reaction) detection kit for influenza virus
InactiveCN102146485AShort measurement time periodImprove detection efficiencyMicrobiological testing/measurementVirus typeQualitative analysis
The invention discloses a one-step RT-PCR (Reverse Transcription-Polymerase Chain Reaction) detection kit for an influenza virus, relating to a PCR kit in the technical field of biology. The kit comprises one-step 5xRT-PCR Buffer, positive control, negative control, three pairs of specific primers and Enzyme Mix; a specific primer AF: 5'-ATTCTAACCGAGGTCGAAACGT-3' and a specific primer AR: 5'-GACAAAGCGTCTACGCTGCAGTC-3' are designed according to a gene M of an influenza virus type A; a specific primer is designed according to a gene NS of an influenza virus type B; and a specific primer is designed according to a gene HA of an influenza virus A type H1N1. The detection kit has a short detection time period, high detection efficiency, high virus detection specificity, high accuracy, higher detection sensitivity than an immunological detection method and high experiment result repeatability, can be applied to qualitative analysis of viruses, and is easy to operate and popularize.
Owner:WUHAN UNIV
Multiple RT-PCR (reverse transcription-polymerase chain reaction) detection primer group and kit for rapidly distinguishing PEDV (porcine epidemic diarrhea virus), PDCoV (porcine deltacoronavirus) and PReoV (porcine reovirus)
InactiveCN109439797AEasy to operateThe detection method is convenient and fastMicrobiological testing/measurementDNA/RNA fragmentationReverse transcriptaseBiology
The invention discloses a multiple RT-PCR (reverse transcription-polymerase chain reaction) detection primer group and kit for rapidly distinguishing PEDV (porcine epidemic diarrhea virus), PDCoV (porcine deltacoronavirus) and PReoV (porcine reovirus), and belongs to the technical field of animal virology and molecular biology. The kit comprises primer sequences as shown in SEQ ID 1-6, Prime STARHR buffer solution, a cDNA template and sterile water. The multiple RT-PCR detection kit of the PEDV, the PDCoV and the PReoV is amplified by multiple RT-PCR, three lesions are similar and serve as primary detection of RNA viruses, total detection time is controlled to be about 2 hours, the detection method is easy to operate, convenient and rapid, and rapid detection can be achieved.
Owner:GUANGXI VETERINARY RES INST
Preparation, detection and application of polyclonal antibody of yam virus X
ActiveCN103543264AEfficient detectionAccurate detectionMaterial analysisSpecific immunityWestern blot
The invention discloses preparation, detection and application of a polyclonal antibody of a virus of the potato virus X potyvirus for infecting yam virus X, (yam virus X, YVX). Based on the YVX genome, prokaryotic expression is utilized by an inventor to purify the CP (coat protein) and be used for immune a rabbit to prepare the polyclonal antibody; the polyclonal antibody can specifically recognize the CP of the YVX subjected to prokaryotic expression and the CP of the YVX virus infected by yam, and can generate a specificity immune reaction. Based on the preparation, an efficient, high-flexibility and accurate IC-RT-PCR (immunocapture-reverse transcription-polymerase chain reaction), ELISA (enzyme-linked immuno sorbent assay) and Western blot method is established by the inventor, and YVX can be detected specifically from a yam plant which is infected by the YVX. According to the invention, a material basis and a technical support can be provided for the mutual interaction research of the YVX and a host and the rapid detection, parting and molecular biology research of the virus, and a solid basis is developed for the prevalence monitoring, prevention and treatment of the virus.
Owner:GUANGXI UNIV
Circular RNA detection method and kit
ActiveCN111808932AEasy to operateHigh detection specificityMicrobiological testing/measurementTotal rnaGenetics
The invention relates to the field of molecular biology, and relates to a circular RNA detection method and kit. The circular RNA detection method comprises the following steps: S1, primer design; S2,reverse transcription reaction: taking 0.1-1 [mu]g of extracted total RNA, adding N6 Random Primer and RNase-Free H2O, performing mixing to obtain a reaction solution I, adding 5 x HT RT Buffer intothe reaction solution I, adding HT M-MLV Mix into the reaction solution I, and finally adding RNase-Free H2O to obtain cDNA; and S3, HS PCR amplification: performing qPCR detection by using the FSjodPrimers obtained in the step S1, 2 x SNP Taq Mix, 50 x ROX Mix *, the RNase-Free H2O and the cDNA obtained in the step S2.
Owner:GUANGZHOU GENESEED BIOTECH
Double RT-PCR (reverse transcription-polymerase chain reaction) detection method of CVB (chrysanthemum virus B) and CChMVd (chrysanthemum chlorotic mottle viroid)
InactiveCN102242225AEasy to operateHigh detection specificityMicrobiological testing/measurementMicroorganism based processesBiotechnologyDiseased plant
The invention discloses a double RT-PCR (reverse transcription-polymerase chain reaction) detection method of CVB (chrysanthemum virus B) and CChMVd (chrysanthemum chlorotic mottle viroid), belonging to the virus detection field of molecular biology. The method comprises the following steps of: (1) sampling from chrysanthemum plants, and extracting total RNA (ribonucleic acid); (2) respectively designing two pairs of specific primers for CVB and CChMVd: CVB-F and CVB-R; CChMVd-F and CChMVd-R; (3) carrying out reverse transcription by taking a random hexamer as a primer to obtain two kinds of cDNA (complementary deoxyribonucleic acid); and (4) amplifying the cDNA by utilizing the RT-PCR technology, and carrying out agarose gel electrophoresis on the amplified products to obtain a 665bp specific fragment for expressing infection of CVB and obtain a 206bp specific fragment for expressing infection of CChMVd. The detection method provided by the invention can be used for simultaneously detecting diseased plants which are compositely infected by CVB and CChMVd, and has the advantages of strong detection specificity, high sensitivity, simple process and cost saving.
Owner:NANJING AGRICULTURAL UNIVERSITY
Pathogen molecule detection method based on nanopore sequencing
PendingCN112646868AIncrease abundanceFast sequencingMicrobiological testing/measurementMicroorganism based processesComplementary deoxyribonucleic acidNasopharyngeal aspirate
The invention belongs to the technical field of biology, and discloses a pathogen molecule detection method based on nanopore sequencing. The pathogen molecule detection method based on nanopore sequencing comprises the following steps: obtaining a throat swab sample or a nasopharyngeal swab sample; carrying out nucleic acid extraction and purification on the obtained throat swab sample or nasopharyngeal swab sample; preparing cDNA (complementary deoxyribonucleic acid); amplifying and purifying the cDNA; preparing a nanopore library and carrying out sequencing; and carrying out bioinformatics analysis to obtain a pathogen molecule detection result. According to the invention, pathogenic molecule detection can be directly carried out on the basis of the clinical samples, the sequencing speed is high, and real-time monitoring can be carried out; and meanwhile, the sensitivity is high and the detection limit is low. According to the invention, with the addition of a universal joint and the one-step PCR amplification during the reverse transcription construction of the nanopore library, the unbiased amplification of all the sequences in the clinical samples can be achieved, so that the pathogenic molecule composition and the abundance in the samples are truly reflected, and more importantly, the detection sensitivity and the detection accuracy are substantially improved.
Owner:GANNAN MEDICAL UNIV
Method for fast detecting genome DNA residues in total RNA sample
ActiveCN108754010AImprove accuracyDetection is simple and fastMicrobiological testing/measurementDNA/RNA fragmentationInteinTotal rna
The invention provides a method for fast detecting genome DNA residues in a total RNA sample. The method comprises the steps that 1, according to genetic information of a to-be-detected biological material, a gene containing one or more intron sequence is selected as a target, and for the intron sequence, PCR primers are designed; 2, the total RNA of the to-be-detected biological material is extracted, the total RNA or cDNA obtained through reverse transcription of the total RNA is used as a template, and the primers in the step 1 are used for PCR amplification; 3, a PCR amplification productis analyzed. According to the method, only one pair of the primers are designed, so that whether or not the genome DNA residues exist in the total RNA and cDNA samples of the biological material can be detected simply, conveniently and fast; the method provides support for the accuracy of gene expression analysis and has important scientific significance for promoting fundamental research of the molecular biology.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Method for detecting gene with plant flavonoid biosynthesis positive regulation function
InactiveCN108893556AIncrease contentPromote accumulationMicrobiological testing/measurementBiotechnologyFlavonoid biosynthesis
The invention discloses a method for detecting a gene with a plant flavonoid biosynthesis positive regulation function. The method is characterized by including the steps: (1) screening out a candidate gene participating in plant flavonoid biosynthesis positive regulation from ARF genes; (2) verifying the plant flavonoid biosynthesis function of the candidate gene, and finally determining a targetgene. The method has the advantages that research methods of molecular biology, genetics and metabolite analysis are comprehensively utilized to determine a target gene and a key transcription factorregulated by an At5g62000 gene in biosynthesis ways of flavonol and proanthocyanidin (flavonoid metabolites), and a gene network of flavonoid biosynthesis regulated by auxin is expounded. The gene can be possibly used for culturing plant varieties with higher content of proanthocyanidin and flavonol.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
Ananas comosus sucrose phosphate synthase (Ac-SPS1) gene, coding protein and gene cloning method thereof
InactiveCN101899453AImprove qualityClear molecular mechanismEnzymesDNA preparationSucrose-phosphate synthasePolymerase chain reaction
The invention provides an ananas comosus sucrose phosphate synthase gene shown as SEQ ID NO:1 and coding protein thereof shown as SEQ ID NO:2, belonging to the technical field of biology. The invention also relates to a cloning method of the gene, and the method comprises the following steps of: extracting total RNA in pineapple fruits; detecting the total RNA in the ananas comosus fruits with agarose gel electrophoresis; carrying out reverse transcription to synthesize a first chain cDNA by using the total RNA as a template and Oligo dT as a primer; carrying out PCR (Polymerase Chain Reaction) amplification, and connecting a PCR product with a pGEM-T easy vector; converting; screening positive clones; and further verifying the screened positive clones by a PCR and then sequencing to obtain an Ac-SPS1 gene. The gene can provide a theoretical basis for researching the mechanism of sugar accumulation of the pineapple fruits from the molecular level.
Owner:SOUTH SUBTROPICAL CROPS RES INST CHINESE ACAD OF TROPICAL AGRI SCI
Primer combination and plasmid for simultaneously detecting 9 kinds of respiratory tract viruses and detection kit
InactiveCN112322790AQuick judgmentAccurate judgmentMicrobiological testing/measurementMicroorganism based processesRespiratory syncytial virus BReverse transcriptase
The invention discloses a primer combination and plasmid for simultaneously detecting 9 kinds of respiratory tract viruses and a detection kit and belongs to the technical field of biology. The primercombination comprises primer pairs for detecting an influenza a virus, an influenza b virus, an H7 influenza virus, an NL63 corona virus, a 229E corona virus, a type 2 parainfluenza virus, Bocavirus,a respiratory syncytial virus A, a respiratory syncytial virus B and a human internal-reference GAPDH gene. The detection kit comprises a 2*multiple reverse transcription PCR reaction solution, a reverse transcription PCR enzyme mixed solution, a primer mixed solution, non-RNAase water, a magnetic bead mixed solution, a reported buffer solution, a positive control and phycoerythrin-modified streptavidin. According to the primer combination and plasmid and the detection kit, by virtue of a liquid-phase chip technology, the manufactured detection kit can be used for carrying out qualitative analysis on a variety of respiratory tract viruses simultaneously, and the sensitivity and specificity of detection reach PCR levels; and types of respiratory tract virus infections can be rapidly and accurately judged, early etiologic diagnosis and treatment on clinical respiratory tract infections are facilitated, the shortening of a treatment cycle is facilitated, the medical expense is reduced, and good social benefits are generated.
Owner:GUANGZHOU MEDICAL UNIV
Whole genome cloning method of pelteobagrus fulvidraco bacillus-shaped virus
InactiveCN109735534AFacilitate subsequent purificationConvenient sequencingMicrobiological testing/measurementMicroorganism based processesPelteobagrusVirus
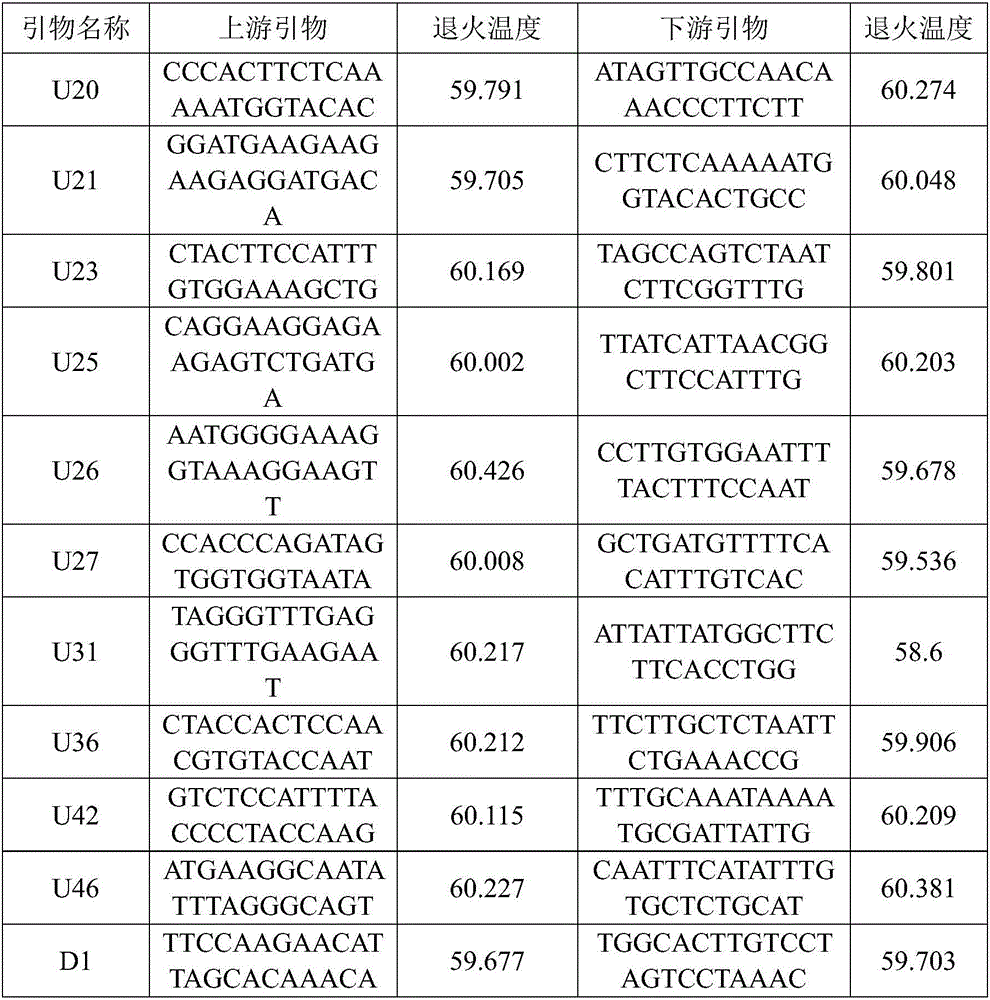
The invention relates to the technical field of biology and discloses a whole genome cloning method of pelteobagrus fulvidraco bacillus-shaped virus. The method comprises the following steps of: designing 14 specific primers for reverse transcription and 14 pairs of specific primers for PCR, wherein the primer sequences are shown as SEQ ID No.1-SEQ ID No.14; extracting pelteobagrus fulvidraco bacillus-shaped virus RNA; taking the extracted viral RNA as a template, respectively adding the extracted viral RNA into a reverse transcription reaction system and a PCR reaction system by using the primers, cloning the whole genome of the pelteobagrus fulvidraco bacillus-shaped virus by sections under reaction conditions, wherein 13 fragments of the amplified product are 2000bp in size, one fragment is 1000bp in size, a total of 14 fragments contain the entire genome sequence of the pelteobagrus fulvidraco bacillus-shaped virus. The invention provides a cloning method of the whole genome of thepelteobagrus fulvidraco bacillus-shaped virus. The method not only can be used for rapid cloning of the whole genome of the pelteobagrus fulvidraco bacillus-shaped virus, but also can be used for researching molecular biology pathogenic mechanism and molecular epidemiology investigation of the pelteobagrus fulvidraco bacillus-shaped virus, and is convenient to use and has good effect.
Owner:SHAOXING UNIVERSITY
Full-length cDNA nucleic acid linear amplification method and kit
ActiveCN103468670AAddresses deficiencies in active dynamicsHigh sensitivityDNA preparationFull length cdnaLinear amplification
The present invention relates to the field of molecular biology, and particularly discloses a full-length cDNA nucleic acid linear amplification method. The method comprises the following steps: separating and purifying mRNA, carrying out reverse transcription synthesis of first strand cDNA, carrying out first strand cDNA 3' terminal extension to obtain full-length first strand cDNA, enriching and purifying the full-length strand cDNA, synthesizing double-stranded cDNA, and the like, such that the full-length cDNA with both known terminal sequences is synthesized. The full-length cDNA nucleic acid linear amplification method has characteristics of high sensitivity and simpleness, and is suitable for efficient synthesis of the full-length cDNA.
Owner:SHANGHAI OFFO BIOPHARM CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com






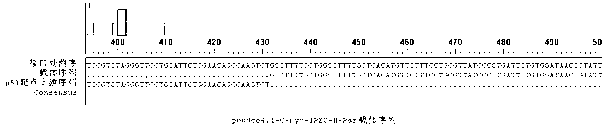
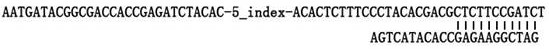












![Method for constructing full-length infectious cDNA [complementary DNA (deoxyribonucleic acid)] of PEDV JS2008 (porcine epidemic diarrhea virus JS2008) strains and application of full-length infectious cDNA Method for constructing full-length infectious cDNA [complementary DNA (deoxyribonucleic acid)] of PEDV JS2008 (porcine epidemic diarrhea virus JS2008) strains and application of full-length infectious cDNA](https://images-eureka-patsnap-com.libproxy1.nus.edu.sg/patent_img/57c6167a-9839-4561-aaaf-e44fa84810a4/HDA0001373833190000011.png)
![Method for constructing full-length infectious cDNA [complementary DNA (deoxyribonucleic acid)] of PEDV JS2008 (porcine epidemic diarrhea virus JS2008) strains and application of full-length infectious cDNA Method for constructing full-length infectious cDNA [complementary DNA (deoxyribonucleic acid)] of PEDV JS2008 (porcine epidemic diarrhea virus JS2008) strains and application of full-length infectious cDNA](https://images-eureka-patsnap-com.libproxy1.nus.edu.sg/patent_img/57c6167a-9839-4561-aaaf-e44fa84810a4/HDA0001373833190000012.png)
![Method for constructing full-length infectious cDNA [complementary DNA (deoxyribonucleic acid)] of PEDV JS2008 (porcine epidemic diarrhea virus JS2008) strains and application of full-length infectious cDNA Method for constructing full-length infectious cDNA [complementary DNA (deoxyribonucleic acid)] of PEDV JS2008 (porcine epidemic diarrhea virus JS2008) strains and application of full-length infectious cDNA](https://images-eureka-patsnap-com.libproxy1.nus.edu.sg/patent_img/57c6167a-9839-4561-aaaf-e44fa84810a4/HDA0001373833190000021.png)