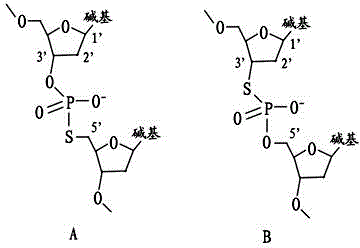
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
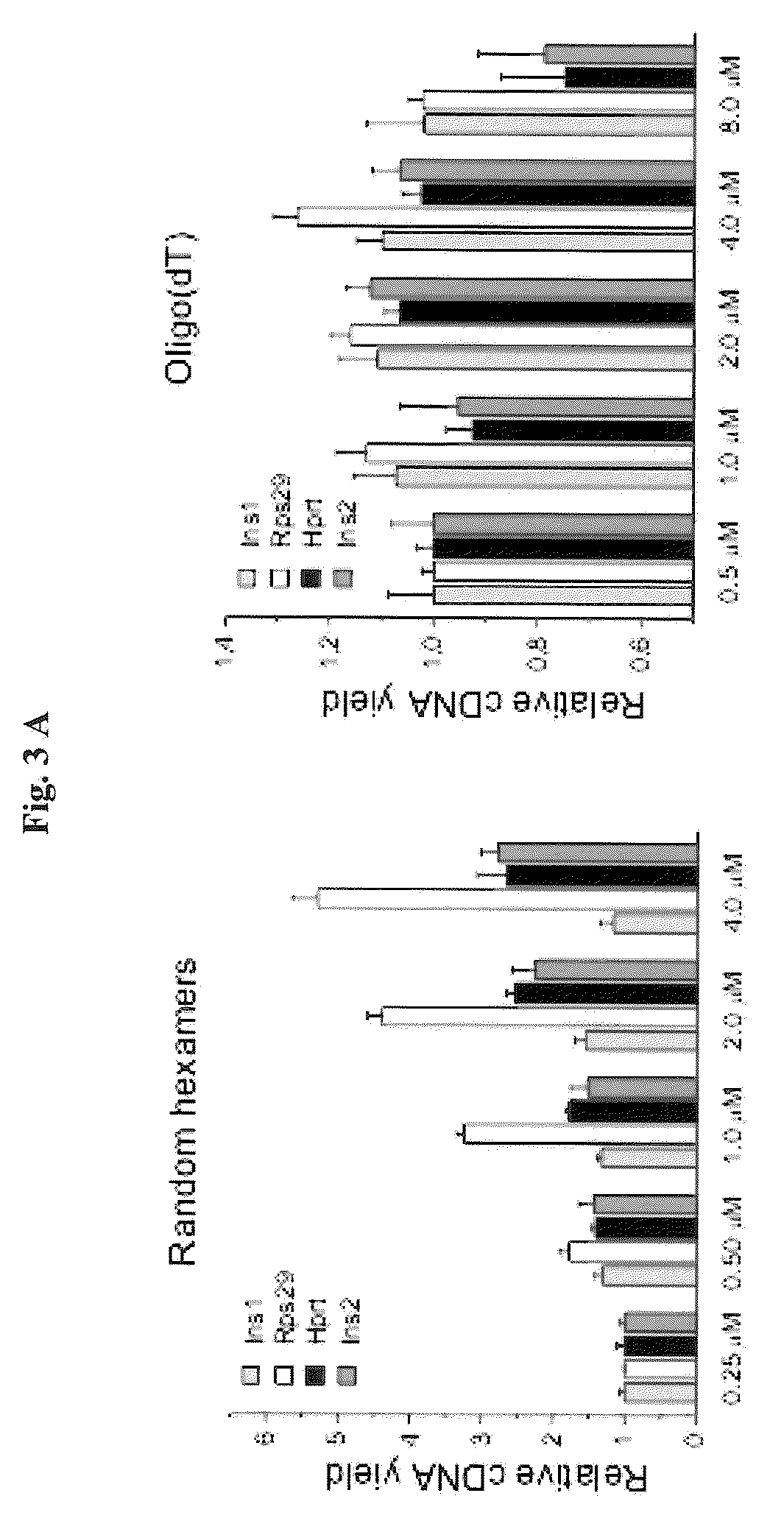
40 results about "Oligo dt" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
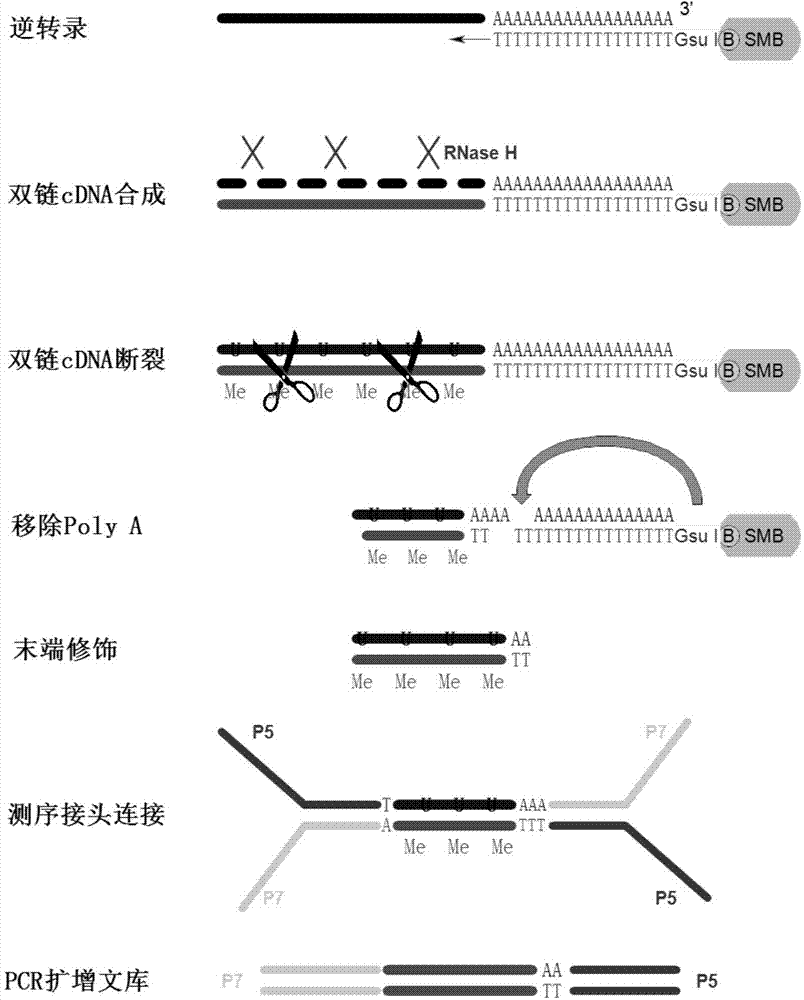
Oligo dT isolation is a very useful method for isolating sequenceswith a poly A tag. Normally people use this column on whole cellpreparations of mRNA but in this lab it is mainly used to isolatesynthetic RNA molecules that have a poly A portion. The celluloseresin has oligo-dT covalently attached to it which form specificWatson-Crick base...
Method for making full-length cDNA libraries
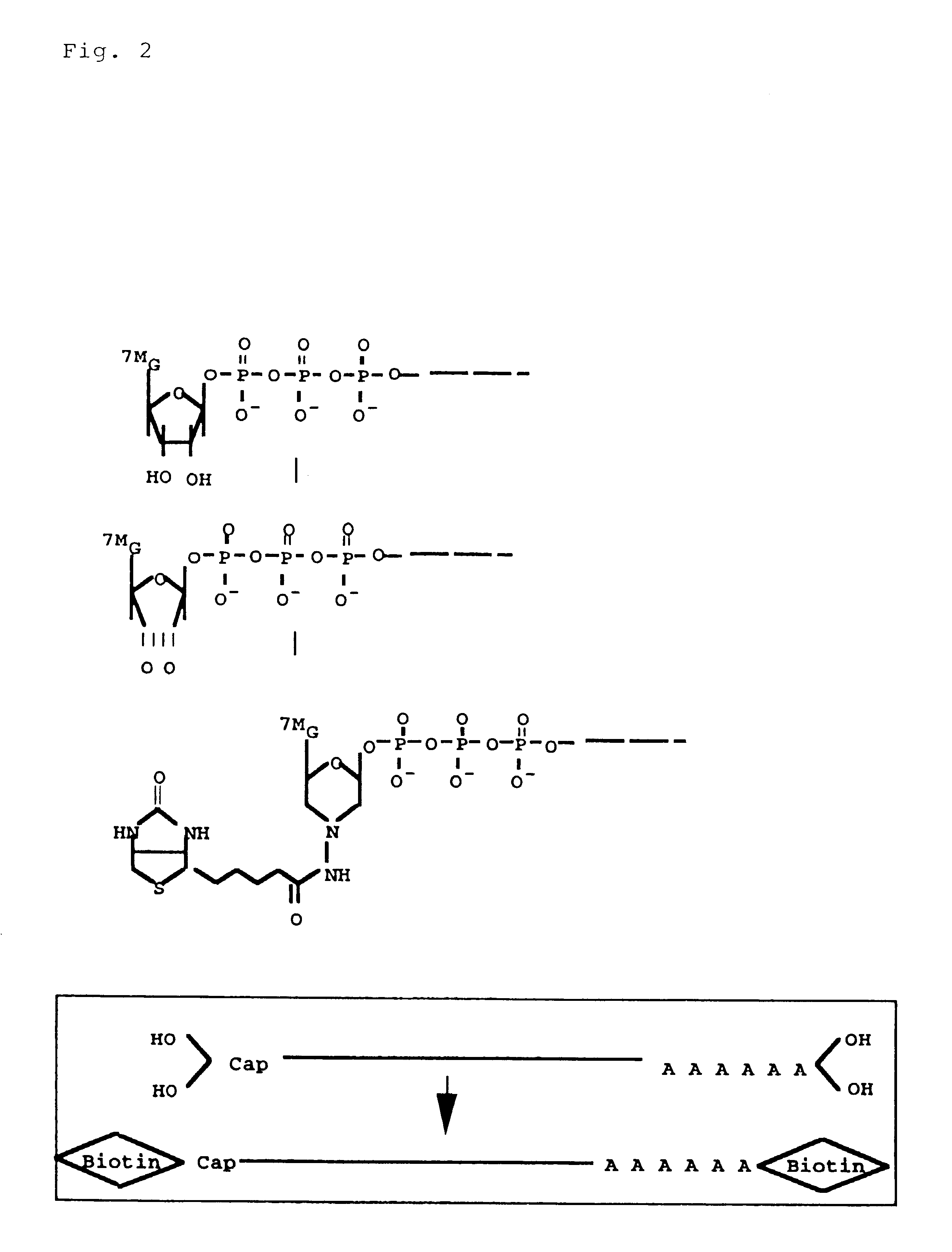
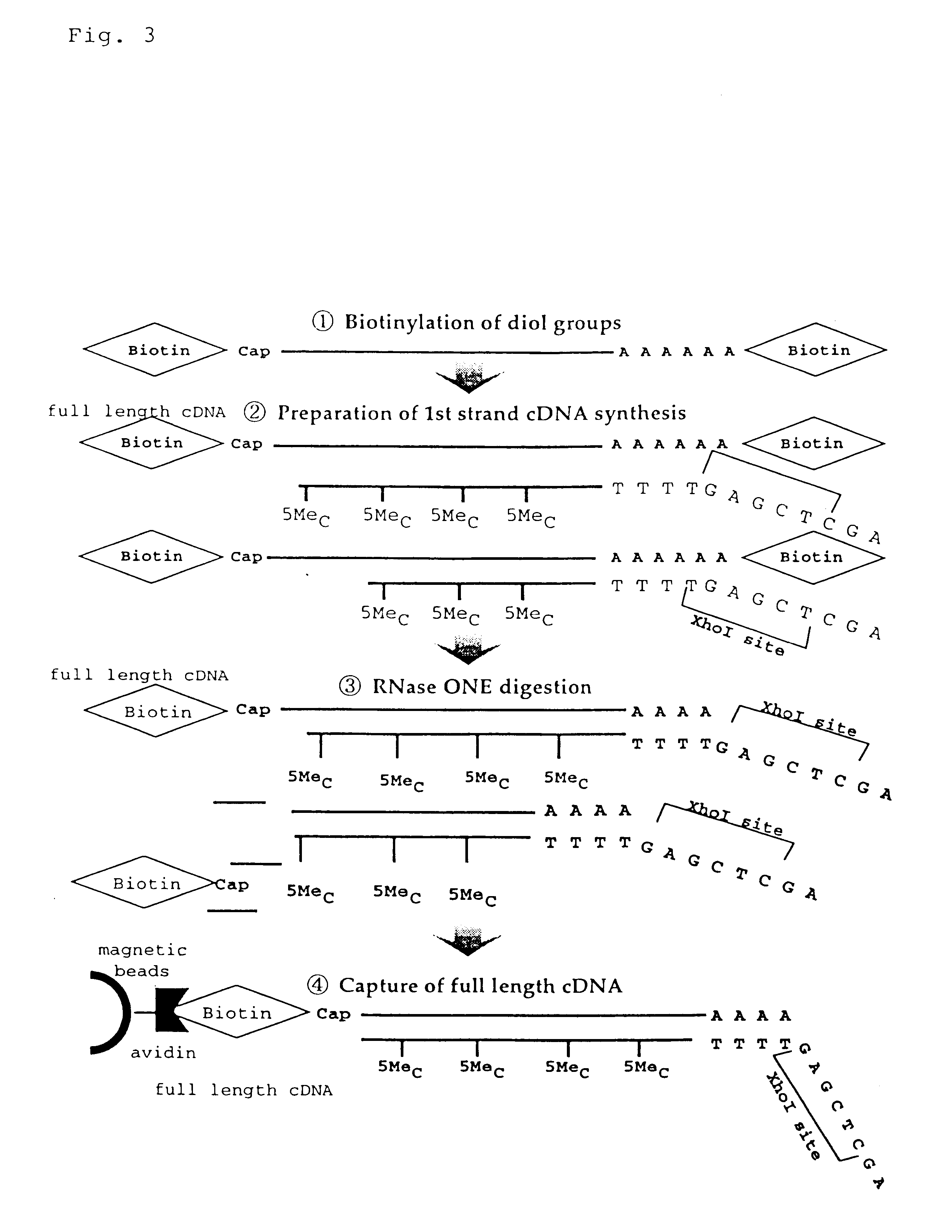
Disclosed is a method for making full-length CDNA libraries, which is for making libraries of cDNAs having a length corresponding to a full-length of mRNAs and comprises the following steps of; binding a tag molecule to a diol structure present in 5' Cap (7MeGpppN) sites of mRNAs, forming RNA-DNA hybrids by reverse transcription using primers such as oligo dT and the mRNAs connected with the tag molecule as templates, and separating RNA-DNA hybrids carrying a DNA corresponding to a full-length of mRNAs from the RNA-DNA hybrids formed above by using function of the tag molecule. To obtain mRNA connected with a tag molecule, the diol structure present in 5' Cap site of mRNA is subjected to a ring-open reaction by oxidation with sodium periodate to form a dialdehyde and the dialdehyde is reacted with a tag molecule having a hydrazine terminus. According to the present invention, there are provided a novel method capable of efficiently labeling 5' Cap site and a method for making full-length cDNA libraries utilizing the labeling method.
Owner:THE INST OF PHYSICAL & CHEM RES WAKO
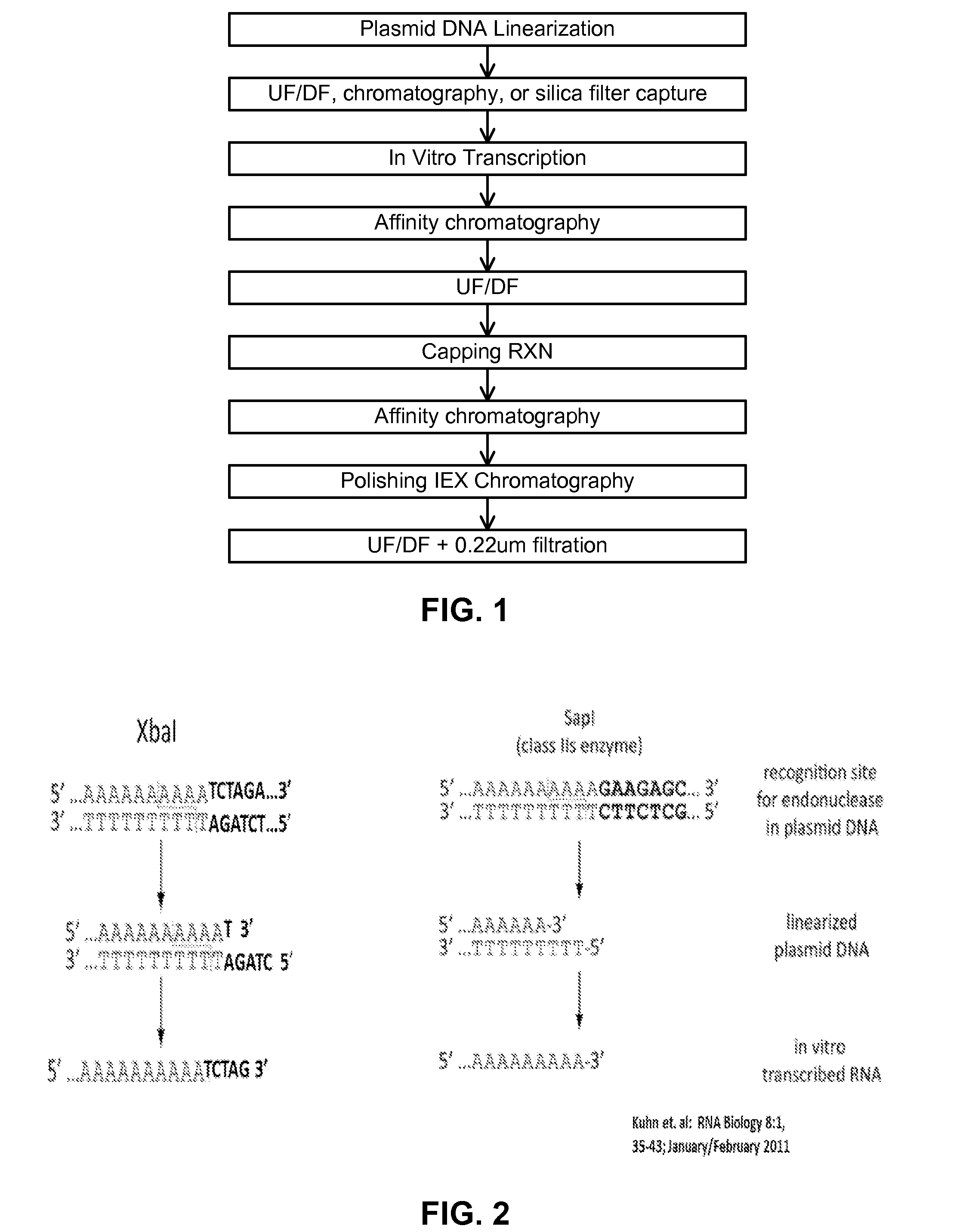
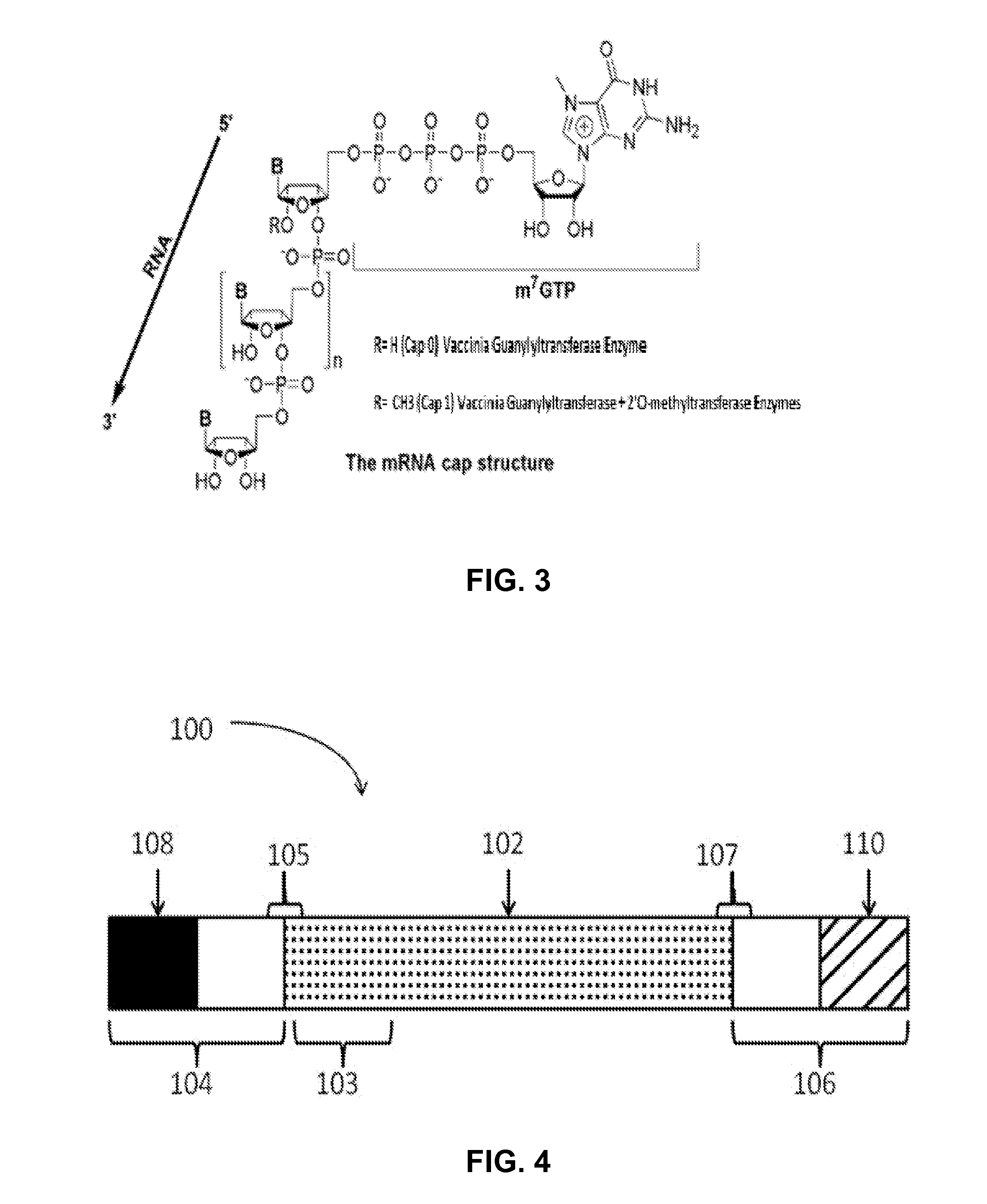
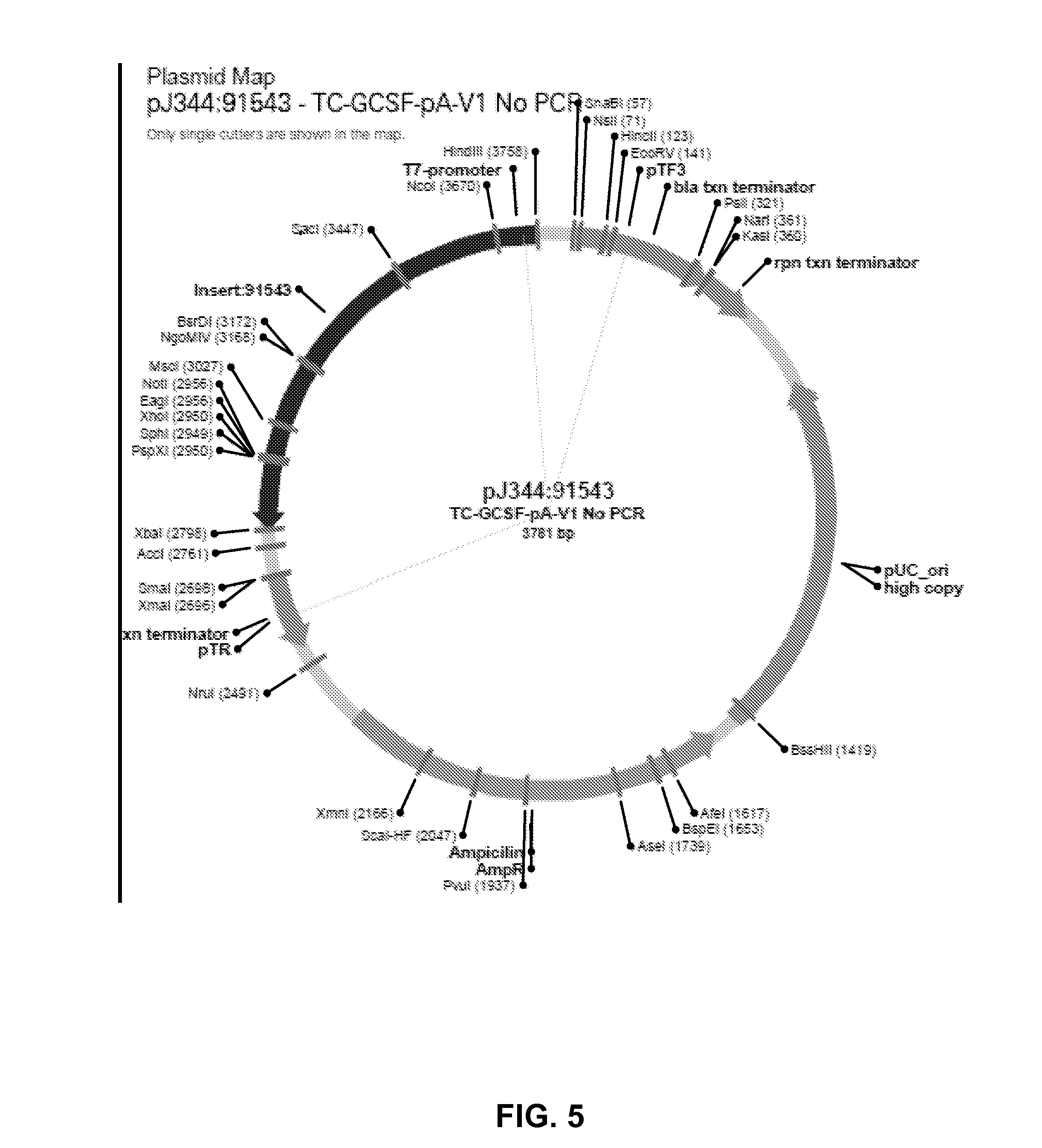
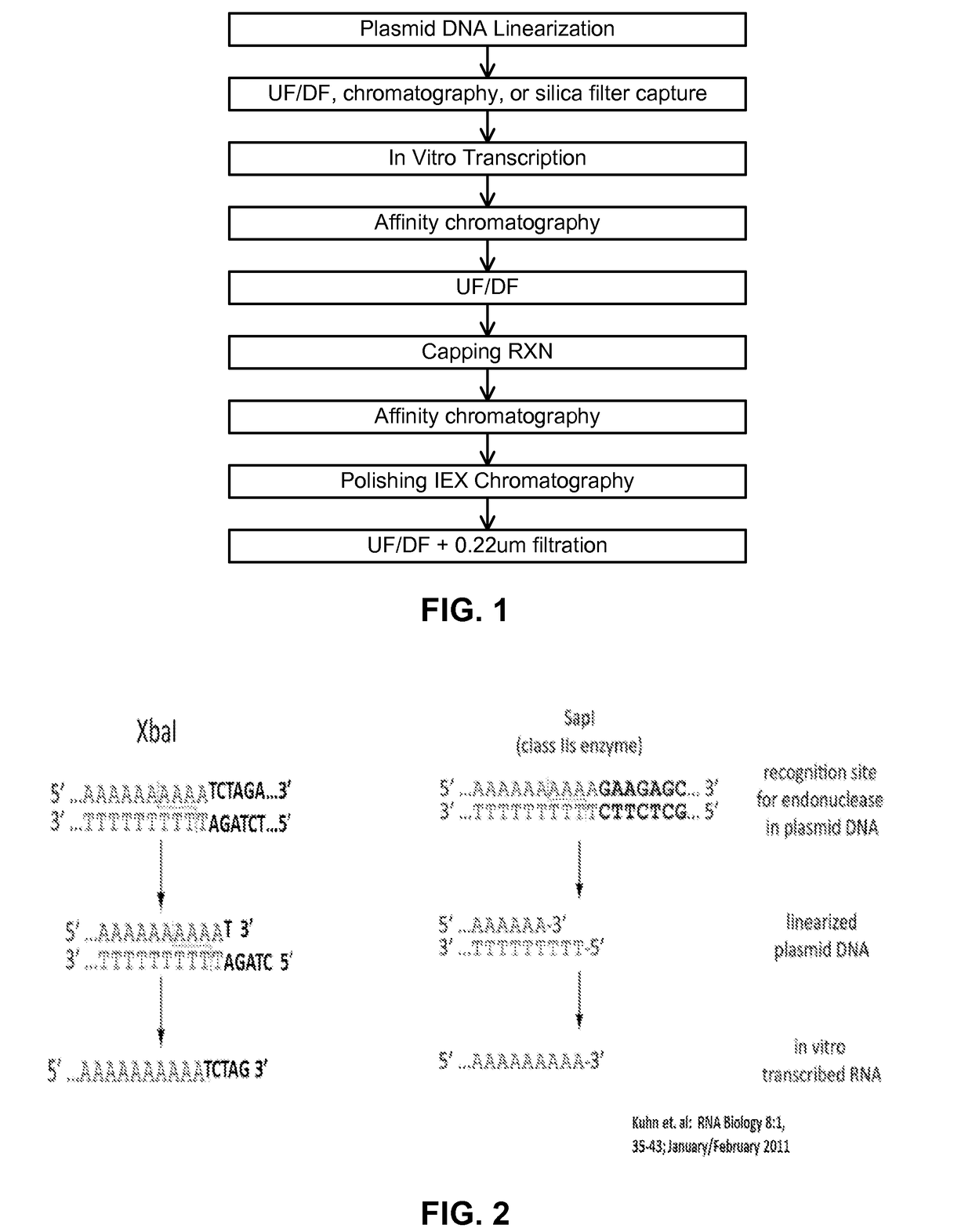
Manufacturing methods for production of RNA transcripts
ActiveUS20160024547A1High purityHigh potencyMicrobiological testing/measurementFermentationOligo dtIn vitro transcription
Owner:MODERNATX INC
Oligo dT primer and method for constructing cDNA library
ActiveCN102533752AImprove applicabilityIncrease success rateLibrary creationProtein nucleotide librariesCDNA librarySingle strand dna
The invention relates to the field of molecular biology and provides Oligo dT primer, wherein Oligo dT primer is a single-stranded DNA molecule containing continuous dT sequences and recognition sites for cutting. The invention further provides a method for constructing a cDNA library on the basis of the Oligo dT primer. The Oligo dT primer is excellent in applicability, can be applied to various different library schemes, and can further accurately locate the initiation site of mRNA molecule in poly A tail. The method for constructing the cDNA library can ensure that all mRNA molecules in a sample can show specificity in the constructed cDNA library.
Owner:盛司潼
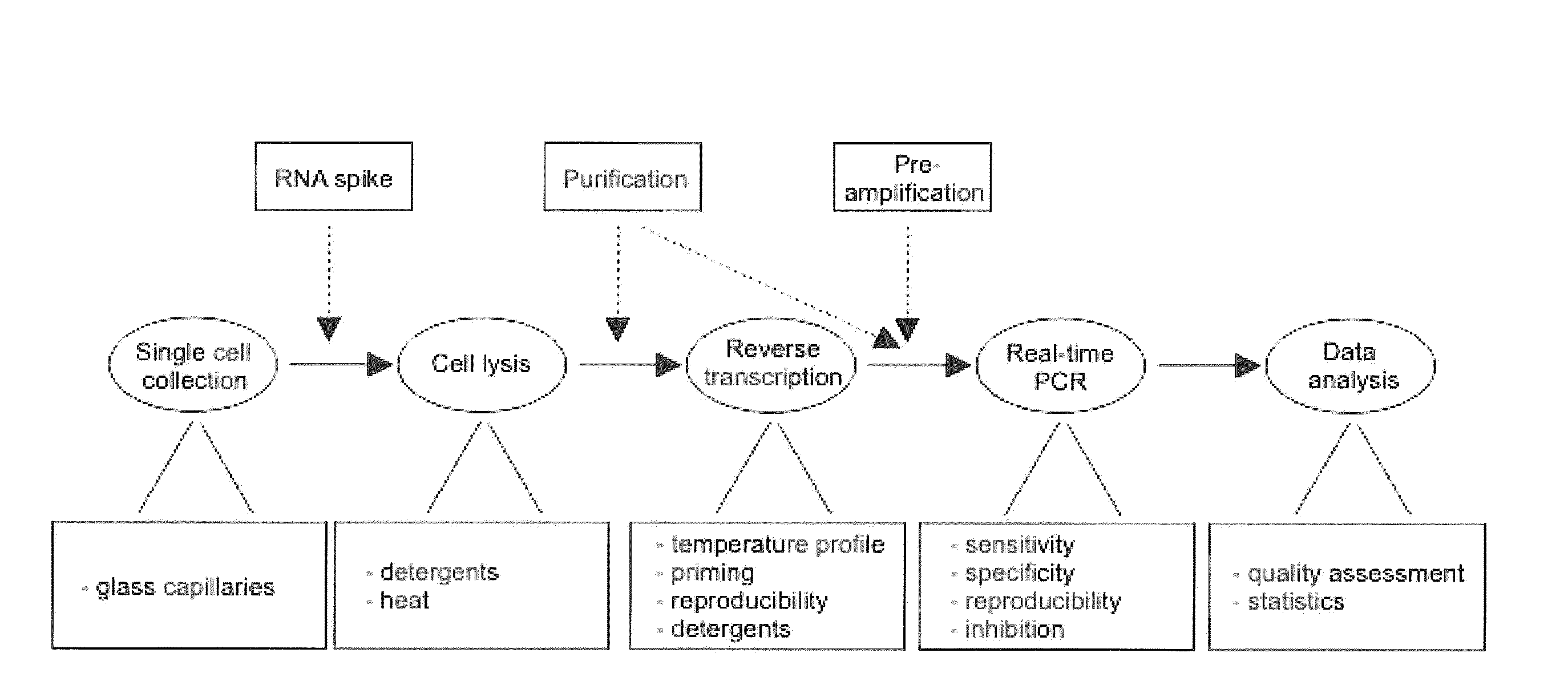
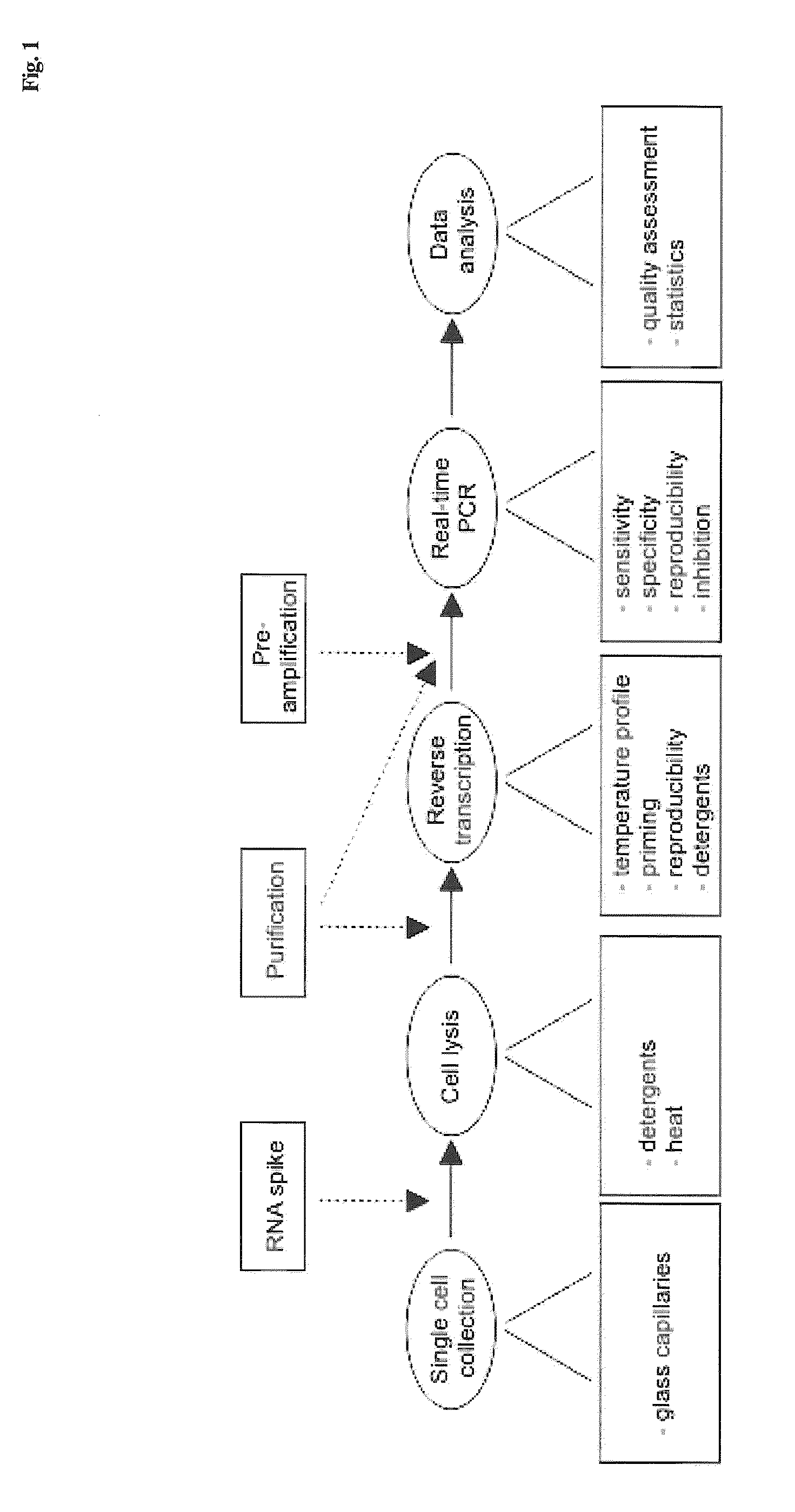
Lysis and reverse transcription for mRNA quantification
InactiveUS20110136180A1Prevent evaporationMicrobiological testing/measurementFermentationOligo dtBiology
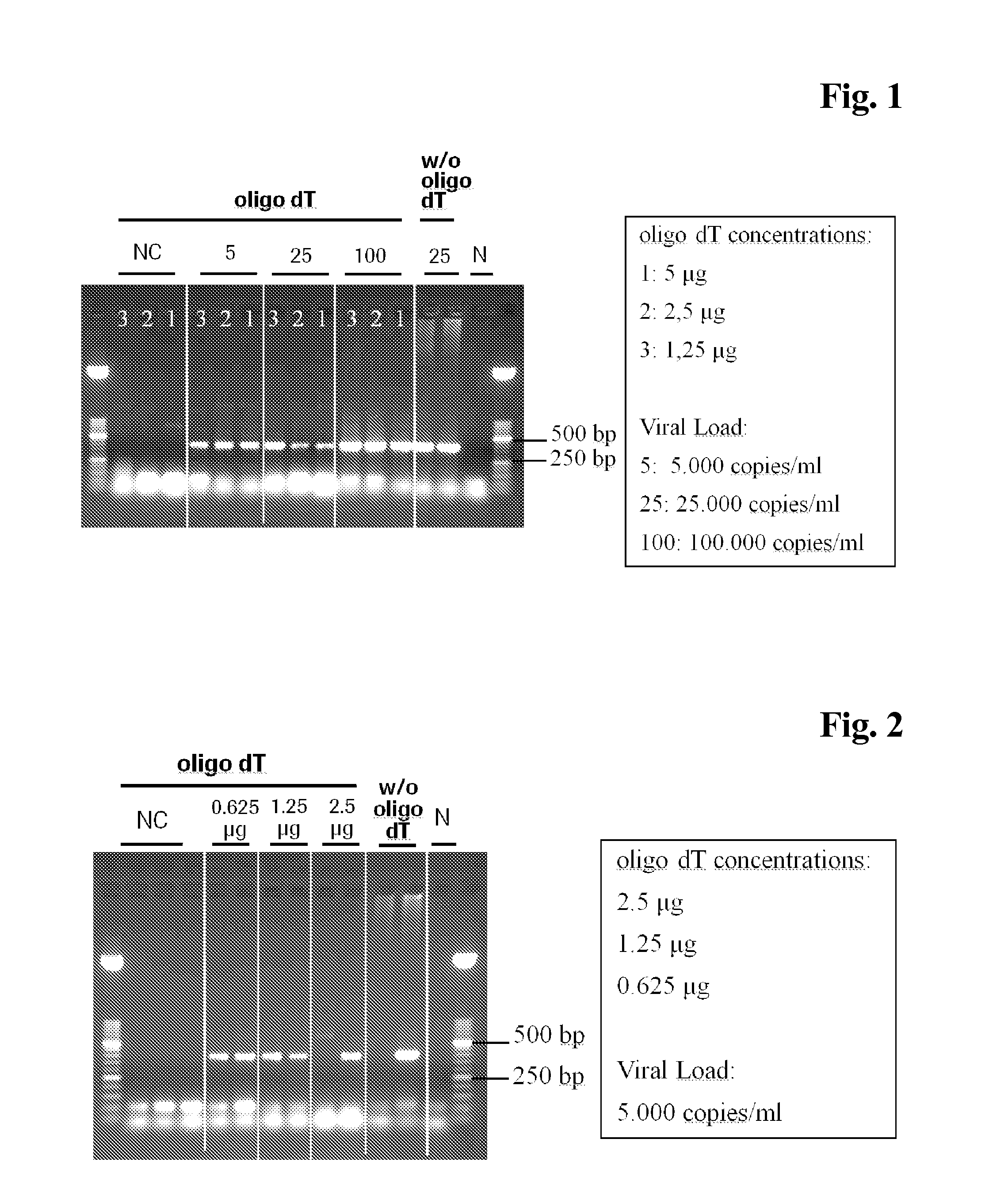
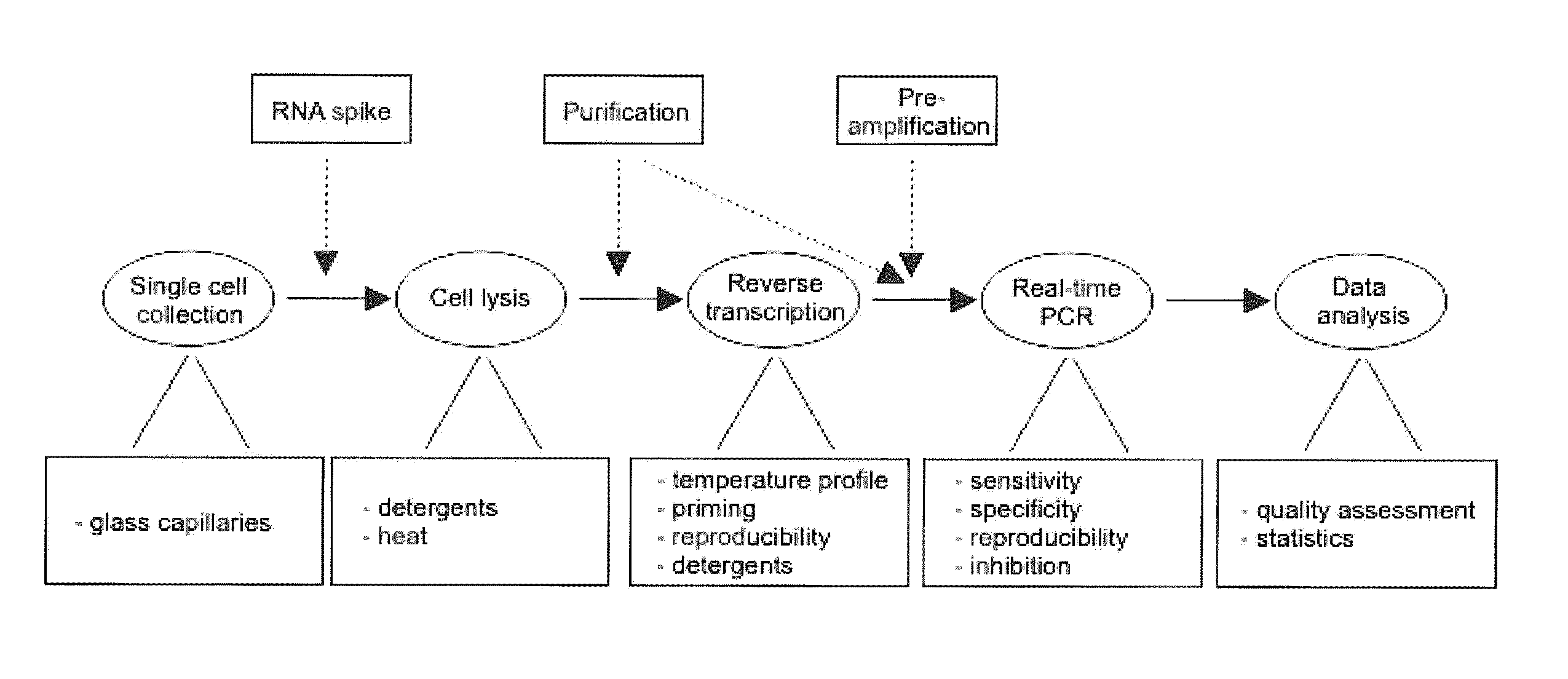
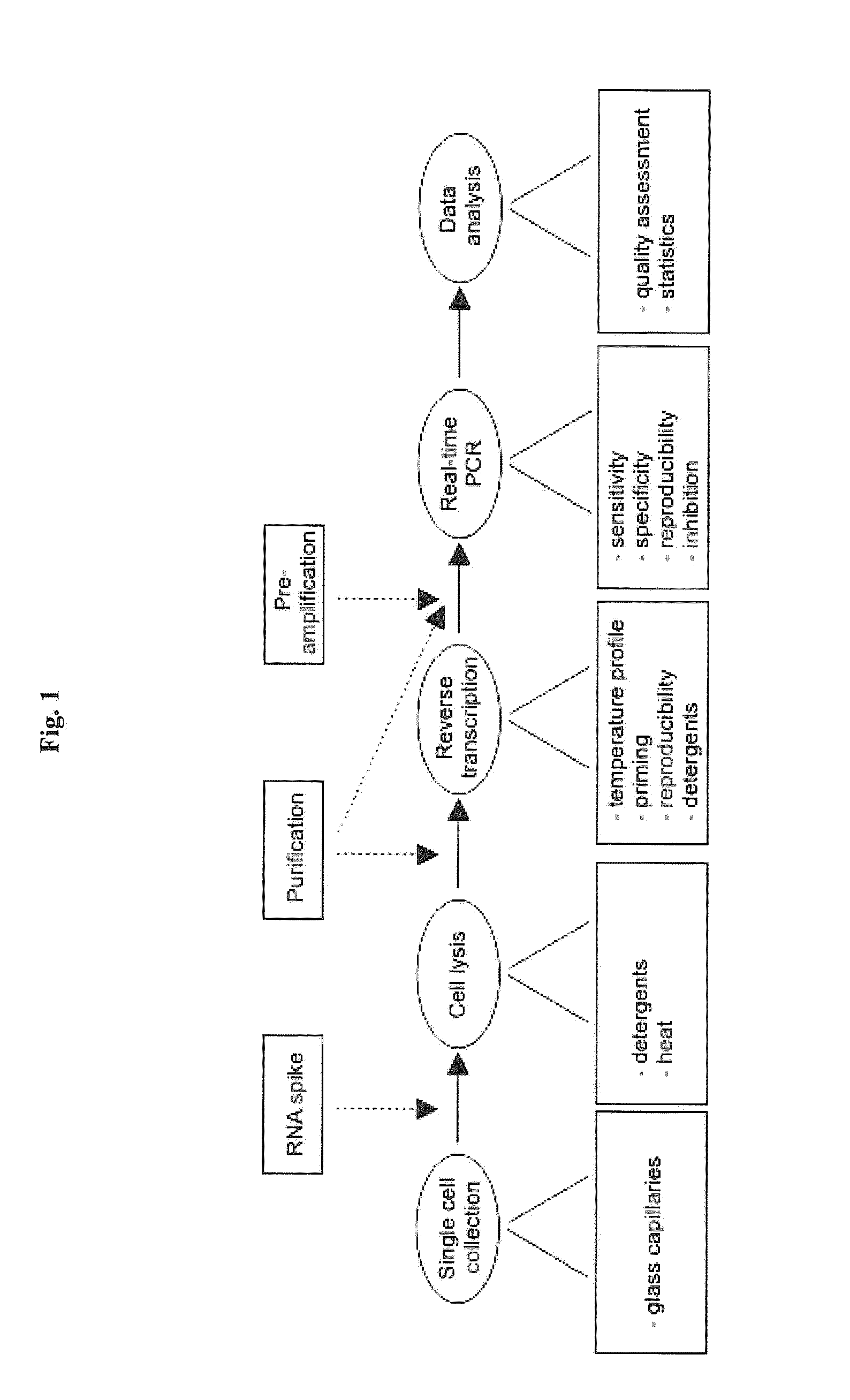
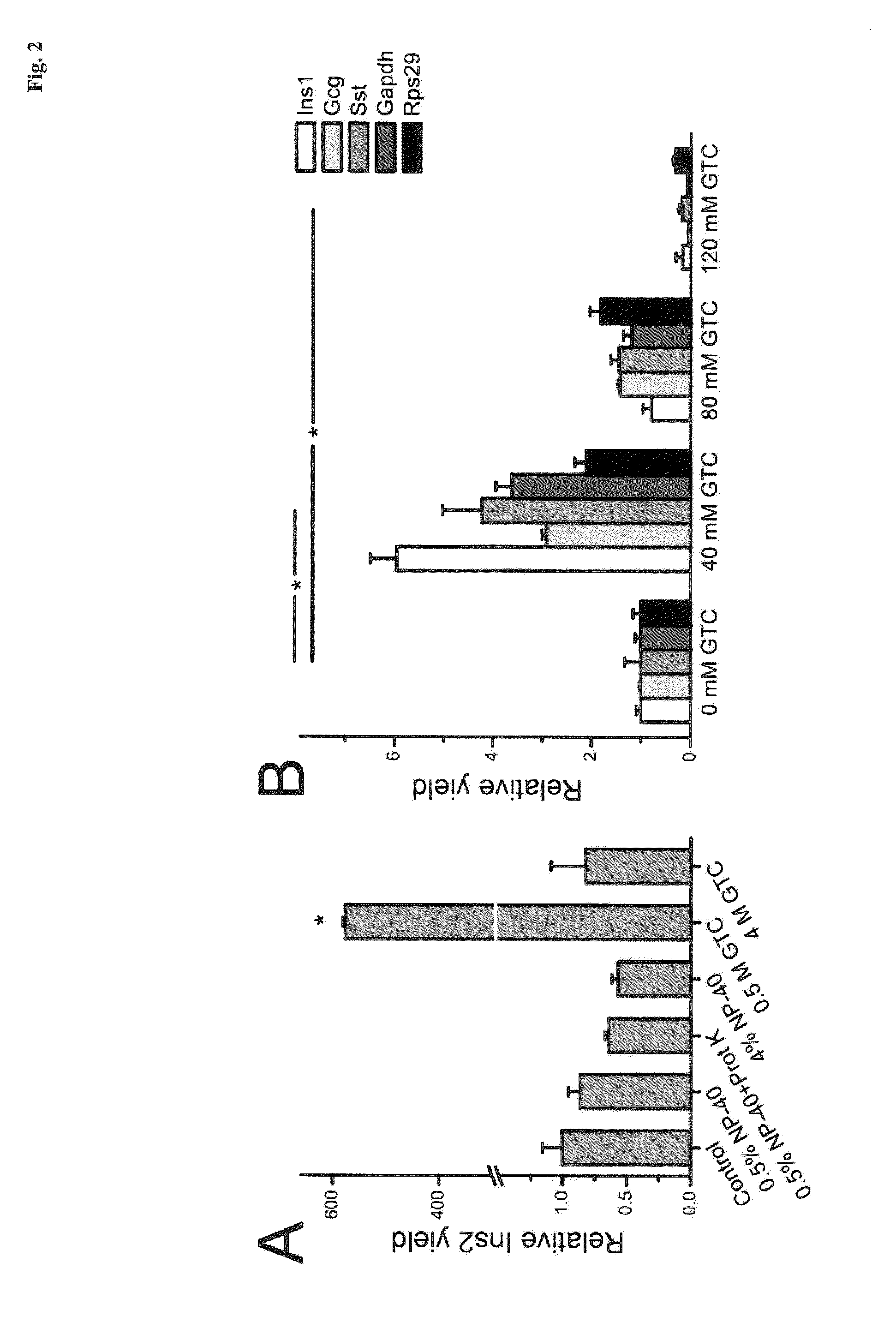
The present invention is directed to a method for performing an RT-PCR for amplifying a target RNA comprising the steps of a) lysis of a cellular sample which is supposed to contain the target RNA with a lysis buffer comprising between 0.2 M and 1 M Guanidine Thiocyanate, b) diluting the sample to an extend such that Guanidine Thiocyanate is present in a concentration of about 30 to 50 mM, c) reverse transcribing in the presence of a mixture of first strand cDNA synthesis primers, the mixture consisting of oligo dT primers and random primers, and d) subjecting the sample to multiple cycles of a thermocycling protocol and monitoring amplification of the first strand cDNA in real time, characterized in that steps a) to c) are done in the same vessel.
Owner:ROCHE MOLECULAR SYST INC
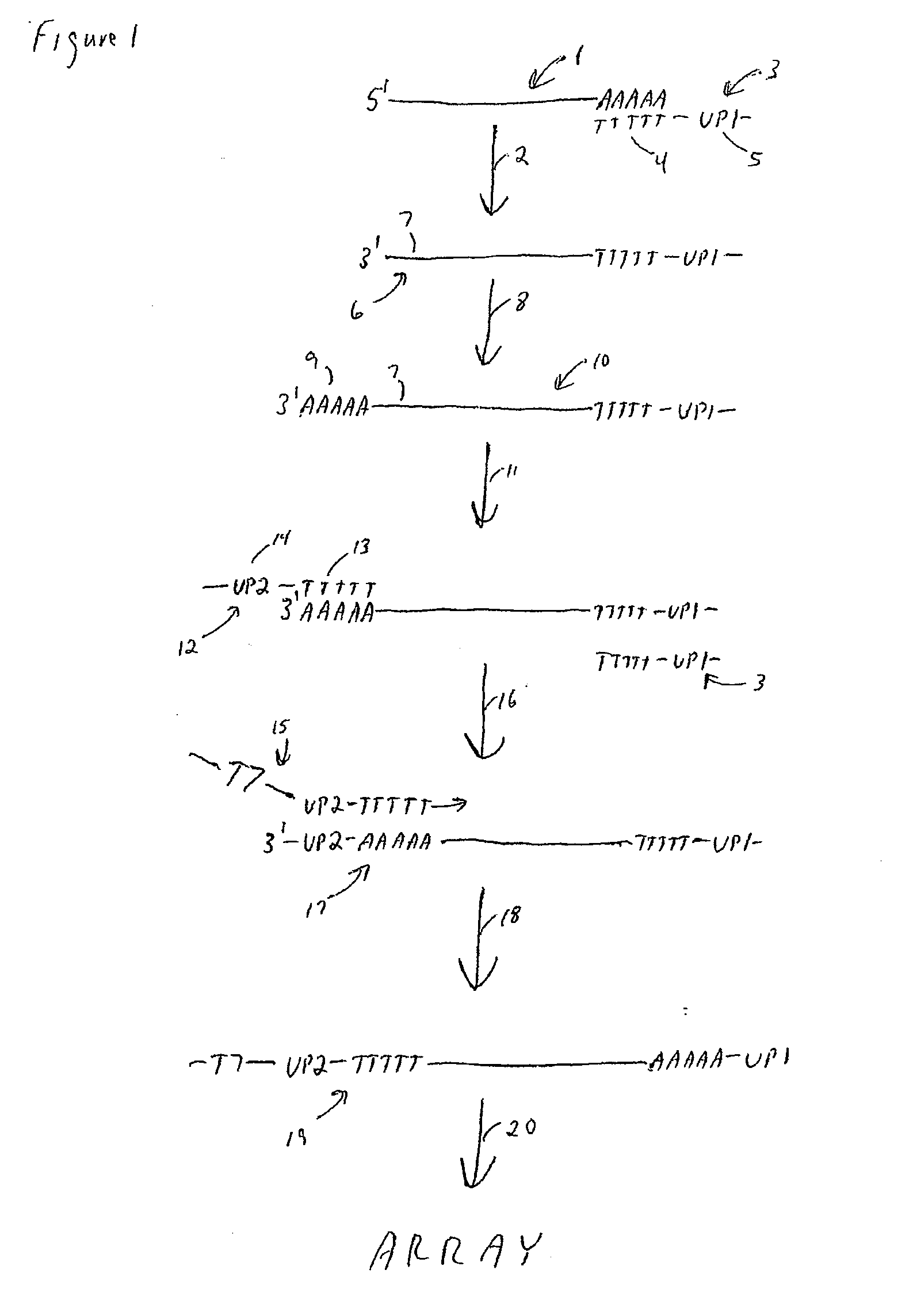
Method for amplifying monomorphic-tailed nucleic acids
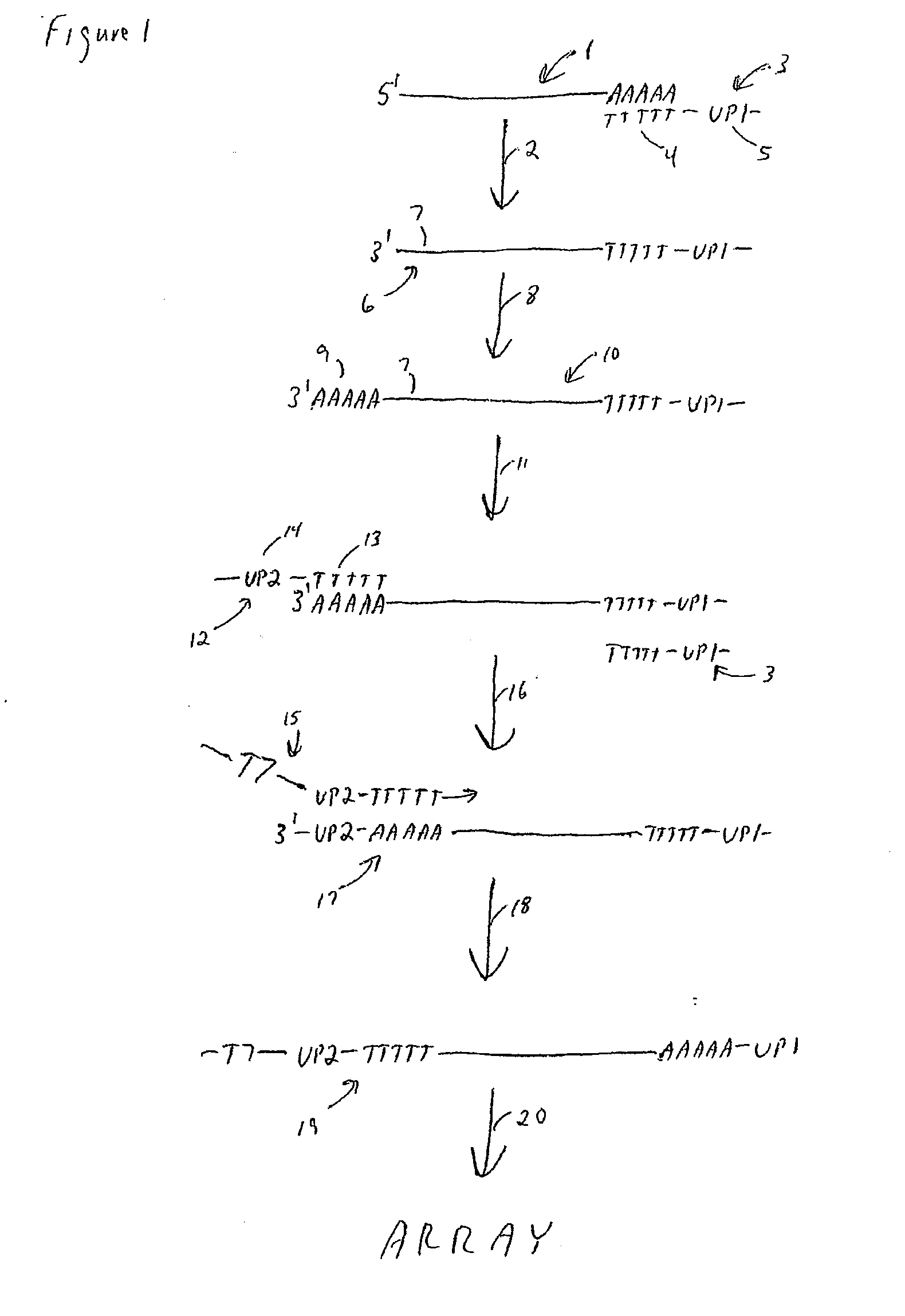
The present teachings provide methods for amplifying a plurality of target nucleic acids. In some embodiments, a first oligo-dT-universal primer comprising a 3′ oligo-dT portion and a first 5′ universal portion is used to reverse transcribe a plurality of 3′ poly-A tail-containing nucleic acids. A poly-A tail is added to the 3′ end of the first strand products to form a two-tailed reaction product. The two-tailed reaction product is amplified in a PCR, wherein the PCR comprises the first oligo-dT-universal primer, and a second oligo-dT-universal primer, wherein the second oligo-dT-universal primer comprises a 3′ oligo-dT portion and a second 5′ universal portion, wherein the second 5′ universal portion of the second oligo-dT-universal primer comprises a different sequence than the first 5′ universal portion of the first oligo-dT-universal primer. The present teachings also provide compositions and kits for amplifying target nucleic acids containing monomorphic tails.
Owner:APPL BIOSYSTEMS INC
Application of Oligo-DT Molecules To Avoid Generation of High Molecular PCR Products Induced By Poly-A Carrier
ActiveUS20150079600A1Inhibit irreversible bindingEnhances reversible bindingMicrobiological testing/measurementGel electrophoresisOligo dt
During RNA isolation carrier nucleic acids such as polyA-RNA are used in order to increase the yield of the isolated RNA. The carrier nucleic acids may lead to high molecular weight products which may interfere in subsequent steps, such as reverse transcription, PCR and gel electrophoresis. The present invention therefore refers to a method and a kit for reverse transcription and the use of a blocking nucleic acid molecule for blocking the carrier polyA-RNA.
Owner:ROCHE MOLECULAR SYST INC
Compositions and methods for entrapping protein on a surface
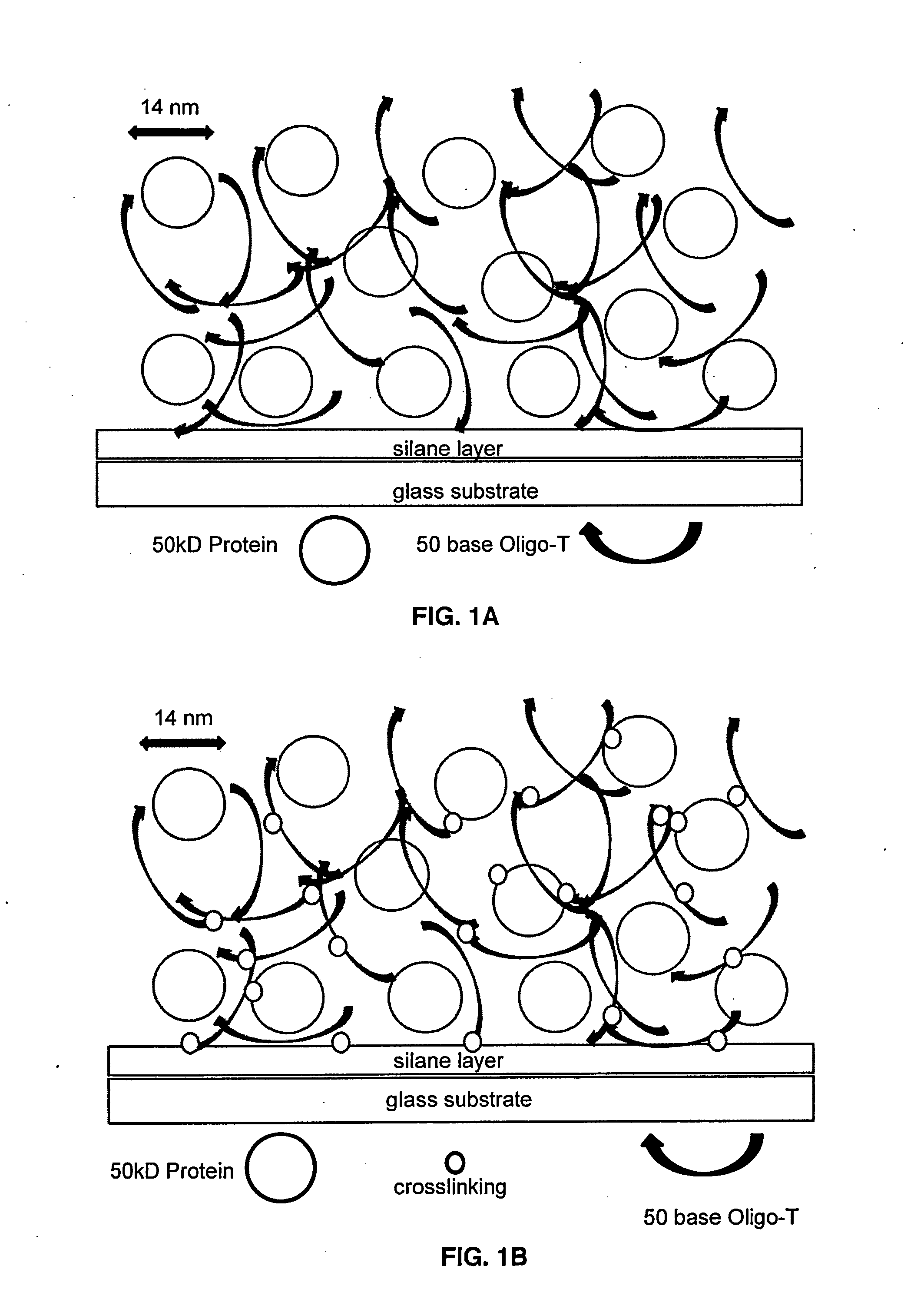
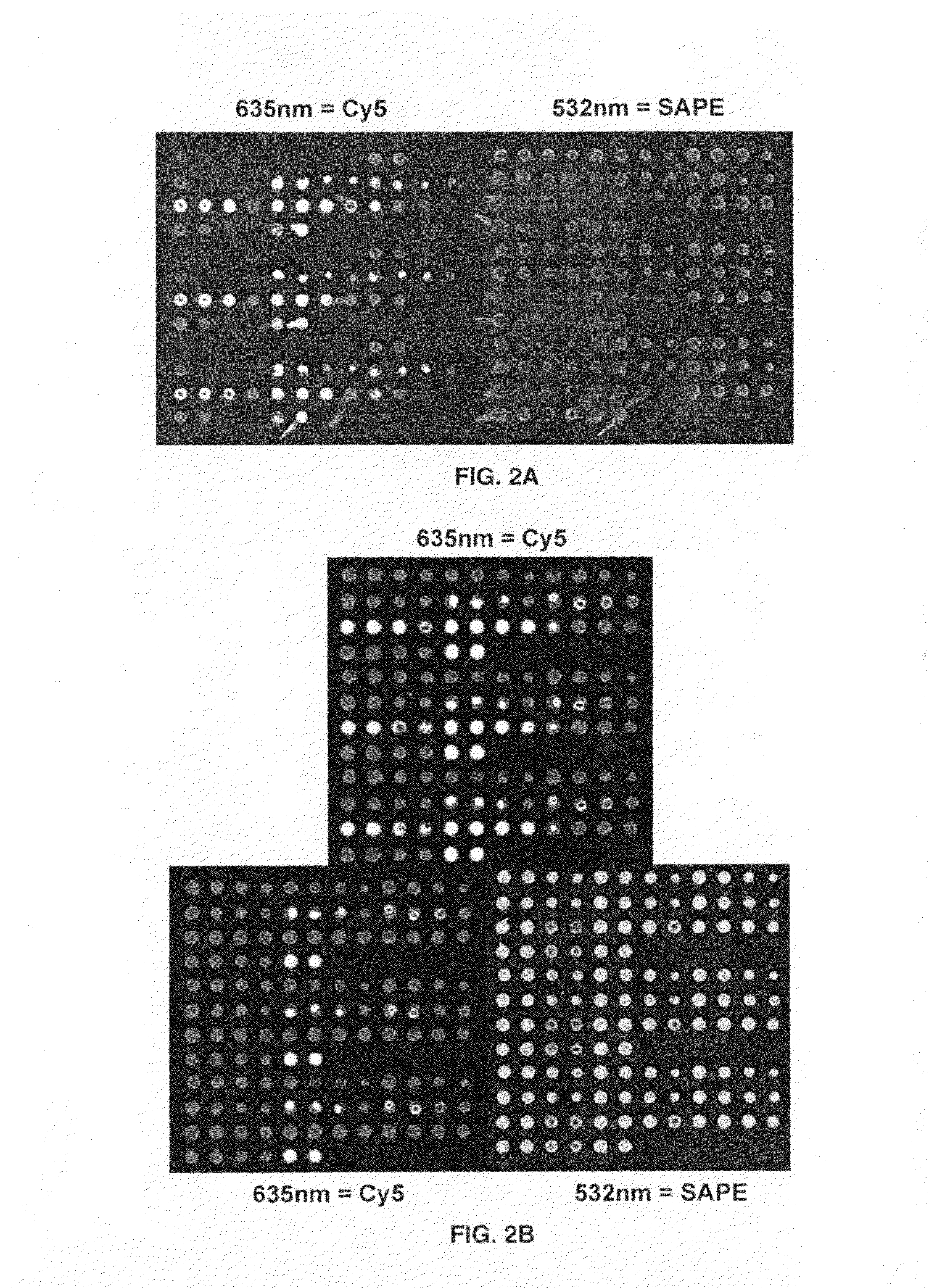
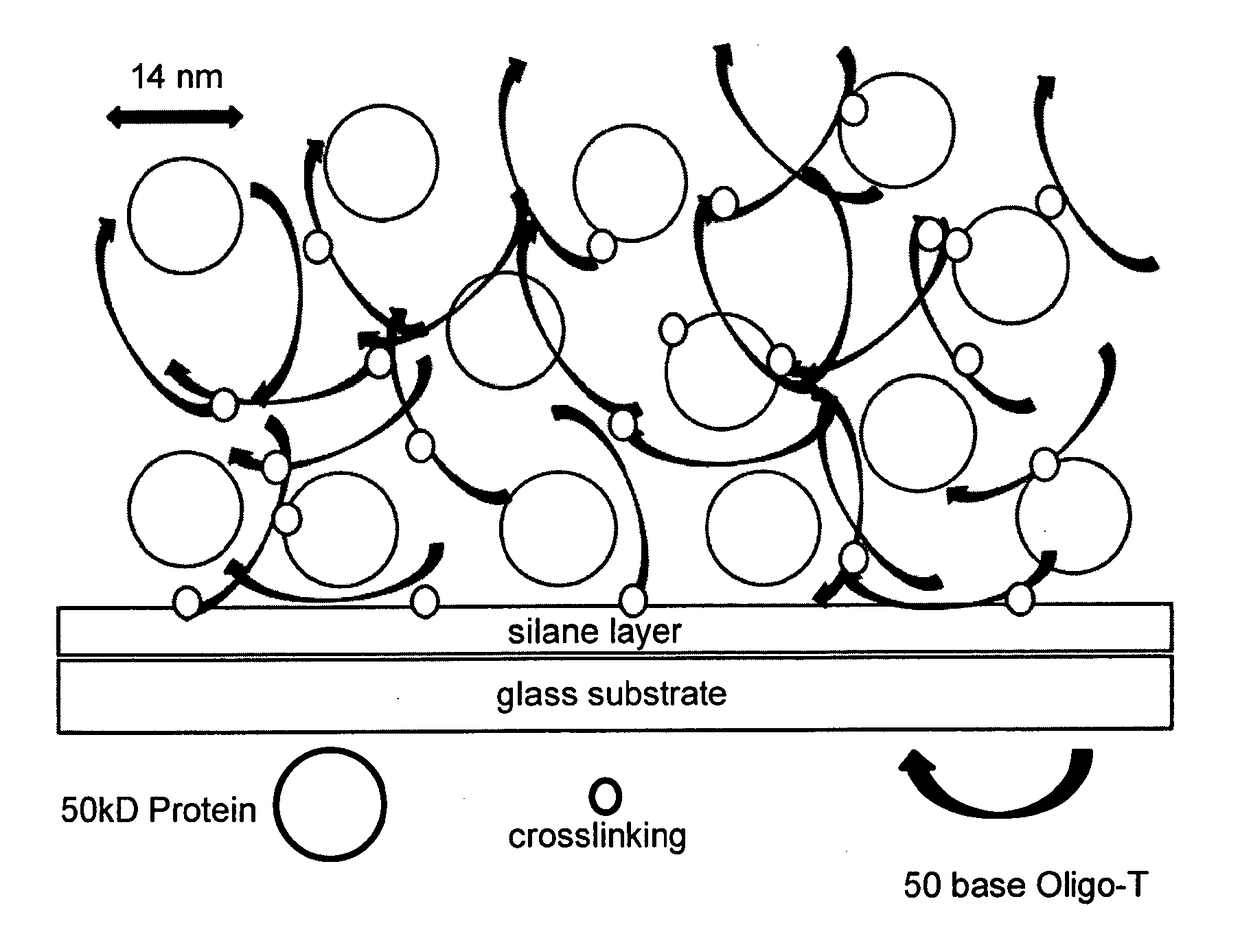
The present invention provides a formulation to link protein to a solid support that comprises one or more proteins, Oligo-dT and one or more non-volatile, water-soluble protein solvents, solutes or combination thereof in an aqueous solution. Further provided is a method of attaching a protein to a surface of a substrate. The formulations provided herein are contacted onto the substrate surface, printed thereon and air dried. The substrate surface is irradiated with UV light to induce thymidine photochemical crosslinking via the thymidine moieties of the Oligo-dT.
Owner:PURE TRANSPLANT SOLUTIONS L L C
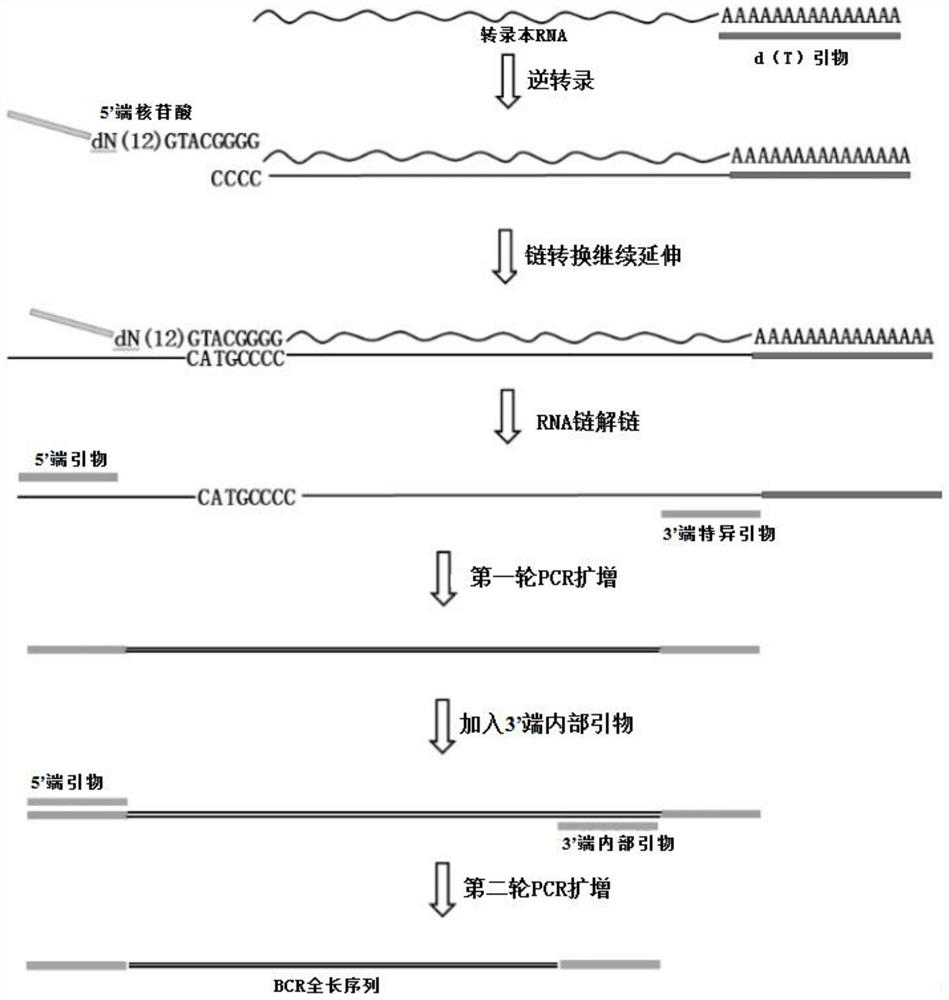
Three-generation library constructing and sequencing method for overall length amplification of BCR immunity group library
ActiveCN111662970ASimplify the experimental processQuantitatively accurateMicrobiological testing/measurementLibrary creationImmunityOligo dt
Owner:WUHAN FRASERGEN CO LTD
Single-cell mRNA quantification with real-time rt-pcr
The present invention is directed to a method for performing an RT-PCR for amplifying a target RNA comprising the steps of a) lysis of a cellular sample which is supposed to contain the target RNA with a lysis buffer comprising between 0.2 M and 1 M guanidine thiocyanate, b) diluting the sample to an extend such that guanidine thiocyanate is present in a concentration of about 30 to 50 mM, c) reverse transcribing in the presence of a mixture of first strand cDNA synthesis primers, the mixture consisting of oligo dT primers and random primers, and d) subjecting the sample to multiple cycles of a thermocycling protocol and monitoring amplification of the first strand cDNA in real time.
Owner:TATAA BIOCENT +1
Compositions and methods for entrapping protein on a surface
ActiveUS20170327599A1Sequential/parallel process reactionsCarrier-bound/immobilised peptidesWater solubleLink protein
The present invention provides a formulation to link protein to a solid support that comprises one or more proteins, Oligo-dT and one or more non-volatile, water-soluble protein solvents, solutes or combination thereof in an aqueous solution. Further provided is a method of attaching a protein to a surface of a substrate. The formulations provided herein are contacted onto the substrate surface, printed thereon and air dried. The substrate surface is irradiated with UV light to induce thymidine photochemical crosslinking via the thymidine moieties of the Oligo-dT.
Owner:PURE TRANSPLANT SOLUTIONS L L C
Method for detecting tumor microenvironment-associated antigen expression
InactiveCN106636392AStrong referenceMicrobiological testing/measurementDNA/RNA fragmentationAntigenFluorescence
The invention discloses a method for detecting tumor microenvironment-associated antigen expression. The method comprises the following steps of (1) extracting sample RNAs from solid tumor tissue samples; (2) checking the sample RNAs; (3) taking the extracted and qualified sample RNAs as templates, and carrying out inverse transcription of the RNAs by employing oligo dT as a primer to obtain tissue sample cDNAs; and (4) designing a qPCR detection primer, amplifying the tissue sample cDNAs through qPCR amplification and collecting fluorescence signals. The method has the beneficial effects that an inventor selects multiple tumor microenvironment-associated antigens, and correspondingly designs multiple antigen primers; and the expression conditions of the antigens in the tissue samples are specifically detected through current main gene expression analysis method and a fluorescent quantitative PCR method. Through the method, the microenvironment-associated antigen expression values of tumor patients are obtained and the expression values have high reference when a doctor provides the patients with targeted therapeutic schedules.
Owner:GUANGZHOU TAINUODI BIOTECH +2
Method for analyzing APA (Alternative Polyadenylation) based on 3T-seq (TT-mRNA Terminal Sequencing) within whole-genome range
InactiveCN103911449AAvoid damageHigh yieldMicrobiological testing/measurementBiotin-streptavidin complexPoly-A RNA
The invention relates to a method for detecting a PAS (Poly A Site) of an RNA (Ribose Nucleic Acid), and provides a method for analyzing APA (Alternative Polyadenylation) based on 3T-seq (TT-mRNA Terminal Sequencing) within a whole-genome range. The method for analyzing APA comprises the following steps: preparing magnetic beads containing oligo dT, i.e., mixing an oligo dT primer modified by 5' end biotin and the magnetic beads with streptavidin; mixing the magnetic beads with total RNA and screening RNA containing Poly A; performing reverse transcription, synthesizing the second strand of a double-stranded cDNA and breaking the double-stranded cDNA; removing the Poly A structure; after terminal modification, connecting a sequencing linker; and recovering libraries. According to the method for analyzing APA, RNA horizontal operation is reduced, a Poly A sequence is horizontally removed from the DNA, and the influence of the Poly structure on sequencing is eliminated; double-stranded DNA breaking enzyme is selected for treatment, and on the premise that the quality of the libraries is not affected, the DNA is broken horizontally; the experiment process is simplified, the experiment difficulty and cost are lowered and the method for analyzing APA is wide in application range.
Owner:SHANGHAI JIAO TONG UNIV
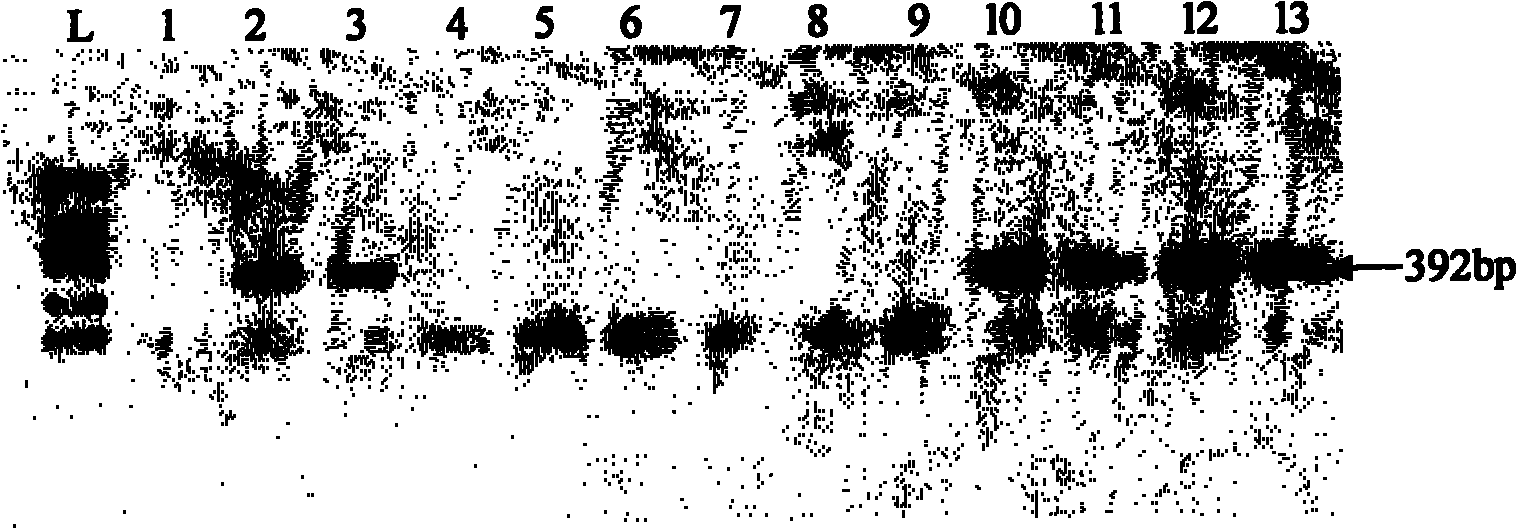
Broccoli Ogu cytoplasm quick identification kit and detection method thereof
InactiveCN102071250ALong identification periodShort identification periodMicrobiological testing/measurementDNA extractionTaq polymerase
The invention discloses a broccoli Ogu cytoplasm quick identification kit and a detection method thereof. The kit comprises a reagent A and a reagent B, wherein the reagent A contains a PCR (Polymerase Chain Reaction) buffer solution, Taq polymerase, dNTP and an Oligo-dT primer, and the reagent B is standard DL-2000DNA Ladder. The detection method comprises the following steps of: DNA extraction of a sample; (1) genome DNA extraction of the sample; (2) PCR amplification reaction; (3) agarose gel electrophoresis detection by which whether the sample is Ogu cytoplasm or not is determined according to the existence / absence of 392bp specific amplification bands. By using the broccoli Ogu cytoplasm quick identification kit, the steps from DNA extraction to the obtaining of detection results can be finished only in several hours, which is very convenient and quick; and by the broccoli Ogu cytoplasm quick identification kit, screening and identifying can be finished in seedling stage, thereby greatly shortening identification cycle. The broccoli Ogu cytoplasm quick identification kit has very important value in broccoli breeding practices.
Owner:WENZHOU VOCATIONAL COLLEGE OF SCI & TECH
Method for constructing circRNA high-throughput sequencing library of FFPE sample
InactiveCN108546700AImprove efficiencyAvoid demandMicrobiological testing/measurementLibrary creationMagnetic beadTotal rna
The invention discloses a method for constructing a circRNA high-throughput sequencing library of an FFPE (formalin-fixed and paraffin-embedded) sample. The method concretely comprises the following steps: (1) extracting total RNA from paraffin sections; (2) adding a PolyA end to the sample RNA; (3) capturing and separating linear RNA by Oligo dT magnetic beads; (4) further removing residual linear RNA; and (5) constructing the circRNA high-throughput sequencing library. The FFPE sample RNA has the characteristics of damages, low content and poor integrity, and the method for constructing thecircRNA high-throughput sequencing library of the FFPE sample has the advantages of simplicity in operation, short time, low cost, stableness in obtaining the library meeting high-throughput sequencing requirements, and avoiding of the problems of large sample size, high rRNA removal cost and low efficiency in the traditional library constructing methods.
Owner:广州密码子基因科技有限公司
Single cell MRNA quantification with real-time RT-PCR
Owner:TATAA BIOCENT +1
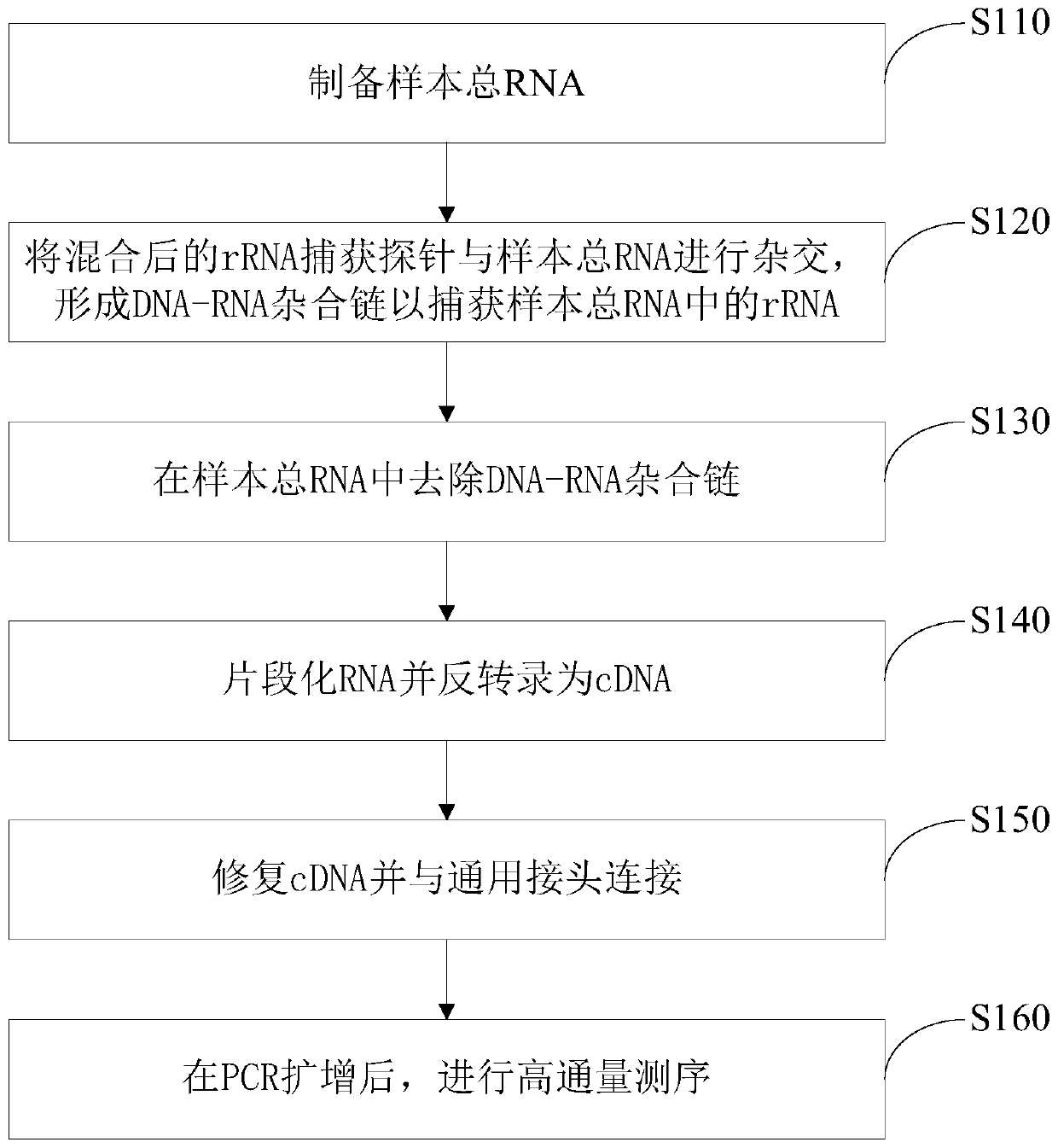
rRNA capture probe and application thereof
PendingCN110878298AEfficient removalSave the amount of sequencing dataMicrobiological testing/measurementDNA/RNA fragmentationMicrobiologyRibosomal RNA
The embodiment of the invention discloses an rRNA capture probe and an application method thereof. The rRNA capture probe is selected from one or more of the following three groups of probe sequences:a 5SrRNA probe sequence group including SEQ ID No. 1 and SEQ ID No. 2; a 16SrRNA probe sequence group including SEQ ID No. 3 to SEQ ID No. 24; and a 23SrRNA probe sequence group including SEQ ID No.25 to SEQ ID No. 76. Based on the capture probe, more than 90% of ribosomal RNA sequences in transcripts can be effectively removed; the technical problem that the prokaryotic mRNA does not contain polyA tail and cannot be enriched and separated by oligo dT is overcome; and the sequencing data volume and cost are greatly saved.
Owner:SHENZHEN E GENE TECH
Typing detection kit for three subtypes of BCR-ABL1 fusion gene
PendingCN111876486AHigh reverse transcription efficiencyReduce pollutionMicrobiological testing/measurementDNA/RNA fragmentationNucleotideNucleotide sequencing
The invention belongs to the field of genes, and particularly relates to a typing detection kit for three subtypes of a BCR-ABL1 fusion gene. Firstly, the invention discloses a primer probe composition for detecting three subtype genes of BCR-ABL1 at the same time, the three subtype genes of BCR-ABL1 are respectively P190, P210 and P230, and the detected nucleotide sequences are shown as SEQ ID NO: 1-9. A specific downstream primer of the ABL1 gene is adopted for reverse transcription, and the reverse transcription efficiency is higher than that of a random primer and oligo dT; three subtypesand internal control are qualitatively and quantitatively detected by multiple systems at the same time, so that pollution and waste possibly caused by sample adding for multiple times are reduced; amultiplex amplification system adopts the same downstream primer, so that the consistency of amplification efficiency is ensured as possible; and an internal control probe is placed in an amplicon, sothat the abnormal condition that the fusion ratio exceeds 100% is avoided.
Owner:PILOT GENE TECH HANGZHOU CO LTD
Chromatographic medium for capturing mRNA and preparation method thereof
PendingCN113797904AGood medium rigidityImprove surface hydrophilicityOther chemical processesSolid sorbent liquid separationBiochemistryOrganic chemistry
The invention discloses a chromatography medium for capturing mRNA and a preparation method thereof, the structure is SP-R-Oligo dT, SP is a basic medium, Oligo dT is an affinity ligand, and R is a spacer arm of the basic medium coupled Oligo dT. The method comprises the following steps: (1) activating a basic medium to obtain an activated medium with a large number of epoxy groups or aldehyde groups modified on the surface; (2) coupling the Oligo dT to an activation medium; and (3) carrying out sealing treatment on redundant aldehyde groups or epoxy groups to obtain the Oligo dT affinity medium. The medium is good in rigidity, good in surface hydrophilicity, high in loading capacity and suitable for downstream purification of mRNA prevention and treatment biological products.
Owner:SEPAX TECH
Oligo dT primer and method for constructing cDNA library
ActiveCN104388426AImprove applicabilityIncrease success rateLibrary creationProtein nucleotide librariesCDNA libraryUracil nucleotide
The invention relates to the field of molecular biology and provides an Oligo dT primer. The Oligo dT primer contains single-chain DNA molecules having a continuous dT sequence, wherein the DNA molecules contain recognition sites used for cutting; and the recognition sites used for cutting refer to uridine monophosphate, deoxyinosine or phosphorothioate bond. The invention also provides a method for constructing a cDNA library on the basis of the Oligo dT primer. The Oligo dT primer disclosed by the invention is high in applicability and can be applied to multiple different library construction schemes; the initiation sites at the polyA tails of mRNA molecules can be accurately positioned; and according to the method for constructing the cDNA library disclosed by the invention, all the mRNA molecules in samples can be specifically reflected in the constructed cDNA library.
Owner:盛司潼
Poly(A)-ClickSeq Click-Chemistry for Next Generation 3-End Sequencing Without RNA Enrichment or Fragmentation
ActiveUS20190256547A1Increase stringencyReduce generationSugar derivativesMicrobiological testing/measurementChemical ligationReverse transcriptase
The present invention includes a method and kit for cDNA synthesis of a 3′UTR / poly(A) tail junction of cellular RNA comprising: obtaining RNA comprising a 3′UTR / poly(A) junction and a poly(a) tail; combining the RNA with three terminating nucleotides of modified-deoxyGTP, modified-deoxyCTP and modified-deoxyATP, dNTPs, and adaptor sequence-oligo-dT; performing reverse transcription of the RNA with a reverse transcriptase primed with the adaptor sequence-oligo-dT to form terminated cDNA fragments that are stochastically terminated upstream of the 3′UTR / poly(A) junction, but not within the poly(A) tail; isolating the terminated cDNA fragments; chemically ligating a functionalized 5′ adaptor to the terminated cDNA; and amplifying the chemically-ligated cDNA into an amplification product, wherein the cDNA is enriched for sequences at the 3′UTR / poly(A) tail junction without fragmentation or enzymatic ligation.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Method for confirming 3.-terminus sequence of virus RNA (Ribonucleic Acid) molecule
InactiveCN102618531AOmit designOmit featureMicrobiological testing/measurementDNA preparationCompound aViral Reverse Transcription
A method for confirming a 3.-terminus sequence of a virus RNA (Ribonucleic Acid) molecule comprises the following steps of: extraction of virus RNA, tail transfer reaction of the virus RNA, purification of tailing virus RNA, reverse transcription reaction of the tailing virus RNA, and polymerase chain reaction amplification of a product of the virus reverse transcription reaction. According to the invention, after tailing is performed on the virus RNA by the tail transfer reaction, the 3.-terminus sequence of the virus RNA molecule can be accurately confirmed by the reverse transcription reaction, thus the method is convenient and quick. As an Oligo dT primer or a modified Oligo dT primer is utilized to perform the reverse transcription reaction, the process of designing and compounding a specific primer according to a specific virus RNA molecule is eliminated, so that the period of an experiment is simplified greatly, and the experiment cost is saved. Simultaneously, the method can be applied to confirming other 3.-terminus sequences of the RNA molecule the 3.-terminus of which does not have a PolyA tail structure.
Owner:SHAANXI NORMAL UNIV
Method for production of cDNA library having reduced content of cDNA clone derived from highly expressed gene
ActiveCN102124106AIncrease contentEfficiently obtainedNucleotide librariesMicrobiological testing/measurementCDNA libraryReverse transcriptase
Disclosed is a method for producing a cDNA library having a reduced content of a cDNA clone derived from a highly expressed gene with high efficiency. In a method for producing a cDNA library by using a double-stranded DNA primer containing an oligo-dT and also using mRNA as a template, the content of a cDNA clone derived from a highly expressed gene in the cDNA library can be reduced by allowing a probe to coexist, wherein the probe has such a property that the probe can bind to mRNA of a highly expressed target gene to inhibit the occurrence of a cDNA extension reaction with a reverse transcriptase.
Owner:HITACHI HIGH-TECH CORP +1
Sucrose synthetase gene Ac-Sy1 in pineapple fruits, encoded protein and gene cloning method thereof
The invention relates to a gene sequence of a sucrose synthetase gene Ac-Sy1 in pineapple fruits and a protein sequence encoded by the same, belonging to the technical of biology. The invention further relates to a cloning method of the gene, which comprises the following steps of: extracting the total RNA in the pineapple fruits by using a modified SDS method; detecting the total RNA in the pineapple fruits by using agarose gel electrophoresis; carrying out reverse transcription to synthesize a first chain cDNA by using the total RNA as a template and using Oligo dT as a primer for; using the cDNA as a template and carrying out PCR amplification by using the Ex-Taq of Takara; connecting a PCR product with a pGEM-T easy Vector, converting, screening the positive for cloning, further validating by PCR and sequencing to obtain the Ac-Sy1 gene. The gene can provide the theoretical evidence for scientifically adjusting and controlling the quality of the pineapple fruits.
Owner:SOUTH SUBTROPICAL CROPS RES INST CHINESE ACAD OF TROPICAL AGRI SCI
Genome-wide analysis of APA based on 3t-seq
InactiveCN103911449BAvoid damageHigh yieldMicrobiological testing/measurementPoly-A RNABiotin-streptavidin complex
The invention relates to a method for detecting a PAS (Poly A Site) of an RNA (Ribose Nucleic Acid), and provides a method for analyzing APA (Alternative Polyadenylation) based on 3T-seq (TT-mRNA Terminal Sequencing) within a whole-genome range. The method for analyzing APA comprises the following steps: preparing magnetic beads containing oligo dT, i.e., mixing an oligo dT primer modified by 5' end biotin and the magnetic beads with streptavidin; mixing the magnetic beads with total RNA and screening RNA containing Poly A; performing reverse transcription, synthesizing the second strand of a double-stranded cDNA and breaking the double-stranded cDNA; removing the Poly A structure; after terminal modification, connecting a sequencing linker; and recovering libraries. According to the method for analyzing APA, RNA horizontal operation is reduced, a Poly A sequence is horizontally removed from the DNA, and the influence of the Poly structure on sequencing is eliminated; double-stranded DNA breaking enzyme is selected for treatment, and on the premise that the quality of the libraries is not affected, the DNA is broken horizontally; the experiment process is simplified, the experiment difficulty and cost are lowered and the method for analyzing APA is wide in application range.
Owner:SHANGHAI JIAOTONG UNIV
Direct plant mRNA (messenger ribonucleic acid) extraction method
InactiveCN105802958ASimple processAccurate location extractionMicrobiological testing/measurementDNA preparationProcess complexityHead surface
The invention provides a direct plant mRNA (messenger ribonucleic acid) extraction method, belongs to the biomolecular level technology field, and particularly relates to a method for extracting plant mRNA directly.The direct plant mRNA extraction method is provided to solve the problems of process complexity and low efficiency of conventional plant mRNA extraction methods.A new technique of directly extracting solid-phase mRNA genes is provided, Oligo dT(15-25) is adsorbed on the head surface of an intradermal needle after a series of treatments including washing, incubating, culturing, embedding and drying of the intradermal needle, the treated intradermal needle is punched at a set position of a plant material to extract mRNA of the plant material.By the direct plant mRNA extraction method, plant mRNA extraction is realized, the mRNA can be extracted from the plant material in 2 minutes, and simplicity, convenience and rapidness of the operation process are achieved.
Owner:NORTHEAST FORESTRY UNIVERSITY
Method for amplifying monomorphic-tailed nucleic acids
The present teachings provide methods for amplifying a plurality of target nucleic acids. In some embodiments, a first oligo-dT-universal primer comprising a 3′ oligo-dT portion and a first 5′ universal portion is used to reverse transcribe a plurality of 3′ poly-A tail-containing nucleic acids. A poly-A tail is added to the 3′ end of the first strand products to form a two-tailed reaction product. The two-tailed reaction product is amplified in a PCR, wherein the PCR comprises the first oligo-dT-universal primer, and a second oligo-dT-universal primer, wherein the second oligo-dT-universal primer comprises a 3′ oligo-dT portion and a second 5′ universal portion, wherein the second 5′ universal portion of the second oligo-dT-universal primer comprises a different sequence than the first 5′ universal portion of the first oligo-dT-universal primer. The present teachings also provide compositions and kits for amplifying target nucleic acids containing monomorphic tails.
Owner:LIFE TECH CORP
Preparation method and application of microspheres for purifying mRNA (messenger ribonucleic acid)
PendingCN114307988ASeparableCapable of purifying mRNAIon-exchange process apparatusOther chemical processesEpoxyFluid phase
The invention discloses a preparation method and application of microspheres for purifying mRNA (messenger Ribonucleic Acid), and the preparation method of the microspheres for purifying mRNA comprises the following steps: S1) washing polystyrene microspheres with active groups on the surface by using a coupling buffer solution; s2) Oligo dT is added to react with the polystyrene microspheres, and after the reaction is finished, a coupling buffer solution is used for cleaning; and S3) adding a blocking reagent for reaction, and after the reaction is finished, cleaning with a coupling buffer solution to prepare the microspheres for purifying mRNA. The surface of the rigid polystyrene microsphere is modified with amino, carboxyl, epoxy group, active ester (NHS) or benzenesulfonyl, so that the rigid polystyrene microsphere has reaction activity and can quickly react with Oligo dT, and Oligo dT is fixed on the surface of the microsphere, so that the microsphere has mRNA separation and purification capabilities; and the microspheres have rigid structures, so that the microspheres can bear a certain pressure, and the Oligo dT microspheres are loaded into a chromatographic column, so that mRNA can be rapidly separated and purified on equipment such as liquid chromatography and the like.
Owner:SUZHOU KNOWLEDGE & BENEFIT TECH CO LTD
Multi-targeted priming for genome-wide gene expression assays
ActiveUS9127271B2Great quantity of reverse transcriptionMinimizing creationMicrobiological testing/measurementFermentationGenomeOligo dt
The present invention is a method of generating a library of expressed cDNA sequences using multi-targeted primers that are complementary to degenerate motifs present in a large proportion of corresponding mRNA sequences. Multi-targeting priming (MTP) for genome-wide gene expression assays provides selective targeting of multiple sequences and counter-selection against undesirable sequences. Priming with MTPs in addition to oligo-dT can result in higher sensitivity, a greater number of genes whose expression is well measured, a greater number of genes whose differences in gene expression are detected to be statistically, and a greater power to detect meager differences in expression.
Owner:TOWNSEND JEFFREY P
Preparation method and application of magnetic microspheres for purifying mRNA (messenger ribonucleic acid)
PendingCN114289003ASeparableCapable of purifying mRNAOther chemical processesAlkali metal oxides/hydroxidesEpoxyCarboxyl radical
The invention discloses a preparation method and application of magnetic microspheres for purifying mRNA (messenger ribonucleic acid). The preparation method of the magnetic microspheres for purifying mRNA comprises the following steps: S1) washing the magnetic microspheres with active groups on the surfaces by using a coupling buffer solution; s2) Oligo dT is added to react with the magnetic microspheres, and then a coupling buffer solution is used for cleaning; and S3) adding a blocking reagent for reaction, and after the reaction is finished, cleaning with a coupling buffer solution to prepare the magnetic microspheres for purifying mRNA. The surface of the magnetic microsphere is modified with amino, carboxyl, epoxy group, active ester (NHS) or benzenesulfonyl, so that the magnetic microsphere has reaction activity and can quickly react with Oligo dT, and Oligo dT is fixed on the surface of the magnetic microsphere, so that the magnetic microsphere has mRNA separation and purification capabilities, and because the magnetic microsphere has magnetism, in the presence of a magnetic field, the mRNA separation and purification capability of the magnetic microsphere is greatly improved. The magnetic microspheres can quickly move towards one side of a magnetic field, so that mRNA can be quickly separated and purified.
Owner:SUZHOU KNOWLEDGE & BENEFIT TECH CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com