Method for confirming 3.-terminus sequence of virus RNA (Ribonucleic Acid) molecule
A terminal sequence and virus technology, applied in the direction of DNA preparation, recombinant DNA technology, biochemical equipment and methods, to achieve the effect of saving experiment cost and simplifying experiment cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
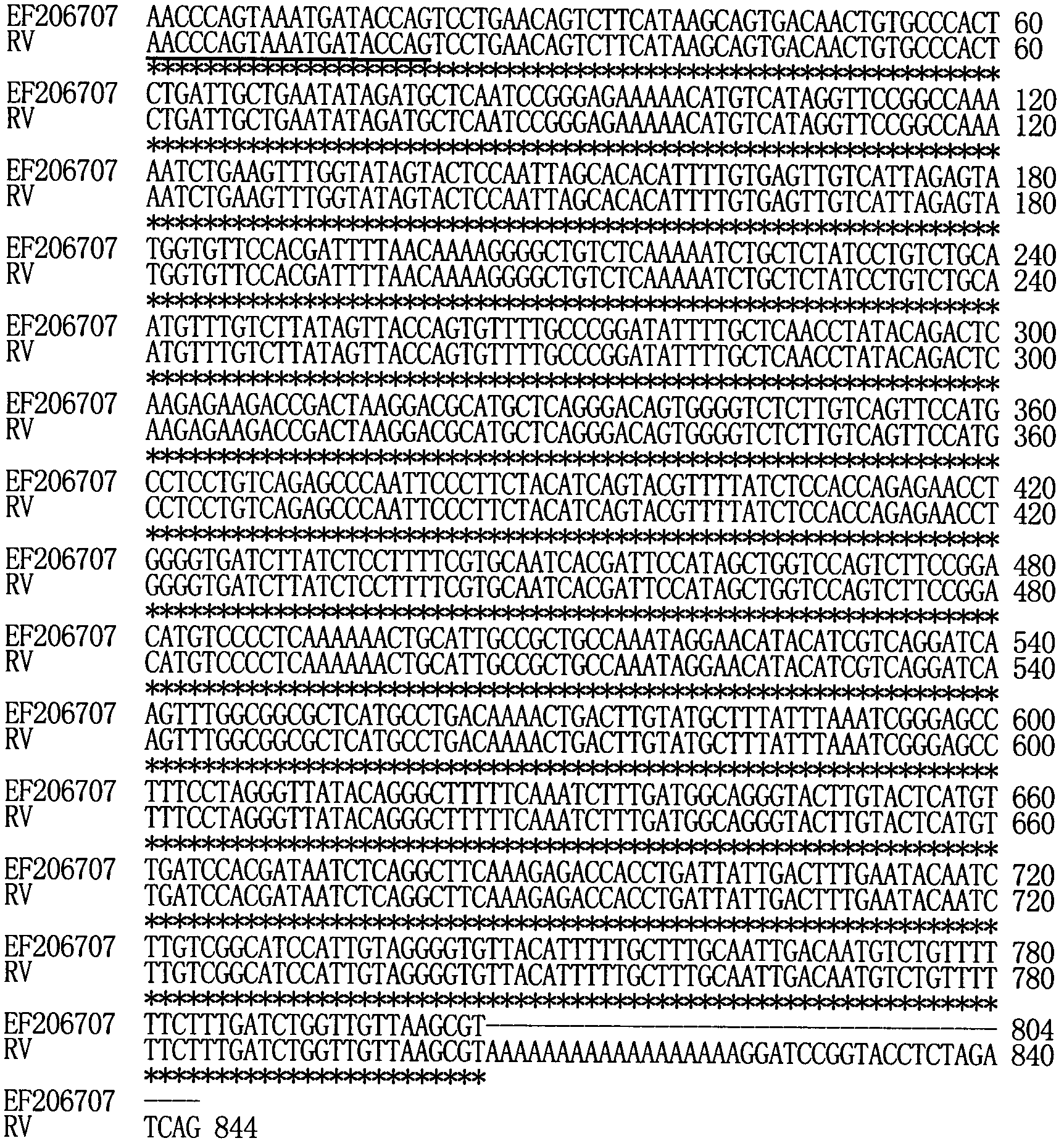
[0027] The method for obtaining the 3' end sequence of rabies virus (Rabies virus) genome is an example, and its steps are as follows:
[0028] 1. Extraction of rabies virus RNA
[0029] Rabies virus (ERA strain) was purchased from China Center for Veterinary Microorganism Culture Collection. After inoculating the hamster kidney cells BHK-21 with the virus, culture them in an incubator at 33-35°C for 5 days. The infected cell culture was collected, and the rabies virus RNA was extracted with the QIAamp Viral RNA Mini Kit kit according to the instructions. The QIAamp Viral RNA Mini Kit kit is a commercially available commodity sold by Qiagen. Rabies virus RNA was dissolved in 40 μL diethyl pyrocarbonate-treated water, 1 μL was taken to test the sample concentration, and the concentration of rabies virus RNA was quantified to 1 μg / μL.
[0030] 2. Terminal transfer reaction of rabies virus RNA
[0031] Add 7.5 μL of rabies virus RNA, 1 μL of deoxyadenosine triphosphate with a ...
Embodiment 2
[0042] Take the method for positioning the 3' end sequence of the influenza virus nucleoprotein (Nucleoprotein, NP) gene as an example, and its steps are as follows:
[0043] 1. Extraction of influenza virus RNA
[0044] Influenza virus (A / Hubei / 1190 / 2010 (H3N2)) was purchased from China Center for Type Culture Collection. After inoculating the chicken embryos with the virus, culture the chicken embryos in a 33-35°C incubator for 2-3 days. The allantoic fluid of chicken embryos was collected, and viral RNA was extracted with the QIAamp Viral RNA Mini Kit kit according to the instructions. The QIAamp Viral RNA Mini Kit kit is a commercially available commodity sold by Qiagen. Viral RNA was dissolved in 40 μL diethylpyrocarbonate-treated water, 1 μL was taken to test the sample concentration, and the concentration of influenza virus RNA was quantified to 1 μg / μL.
[0045] 2. Terminal transfer reaction of influenza virus RNA
[0046]Add 7.5 μg of influenza virus RNA, 1 μL of d...
Embodiment 3
[0057] The method for obtaining the 3' end sequence of the 28S rRNAα subunit of Schistosoma mansoni is an example, and the steps are as follows:
[0058] 1. Extraction of Schistosoma mansoni RNA
[0059] The RNA of Schistosoma mansoni was extracted by one-step method with Tizol reagent, and the quality of the obtained Schistosoma mansoni RNA was detected by formaldehyde denaturing gel electrophoresis. The Tizol reagent was a commercial product sold by Dalian Bao Biological Engineering Co., Ltd. Quantify the concentration of viral RNA to 1 µg / µL.
[0060] 2. Terminal transfer reaction of Schistosoma mansoni RNA
[0061] Add 7.5 μg of Schistosoma mansoni RNA, 1 μL of 10 mmol / L deoxyadenosine triphosphate, diethyl pyrocarbonate-treated water to 12.5 μL, and 10 mmol / L of deoxyadenosine triphosphate and diethyl pyrocarbonate in order to the centrifuge tube The volume ratio of ester-treated water and Schistosoma mansoni RNA is 1:4:7.5, mix well, incubate at 70°C for 15 minutes to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com