Circular RNA detection method and kit
A detection kit and detection method technology, applied in the field of molecular biology, can solve the problems of difficult design of SjodPrimers, false positives, and inability to accurately match cyclization sites.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
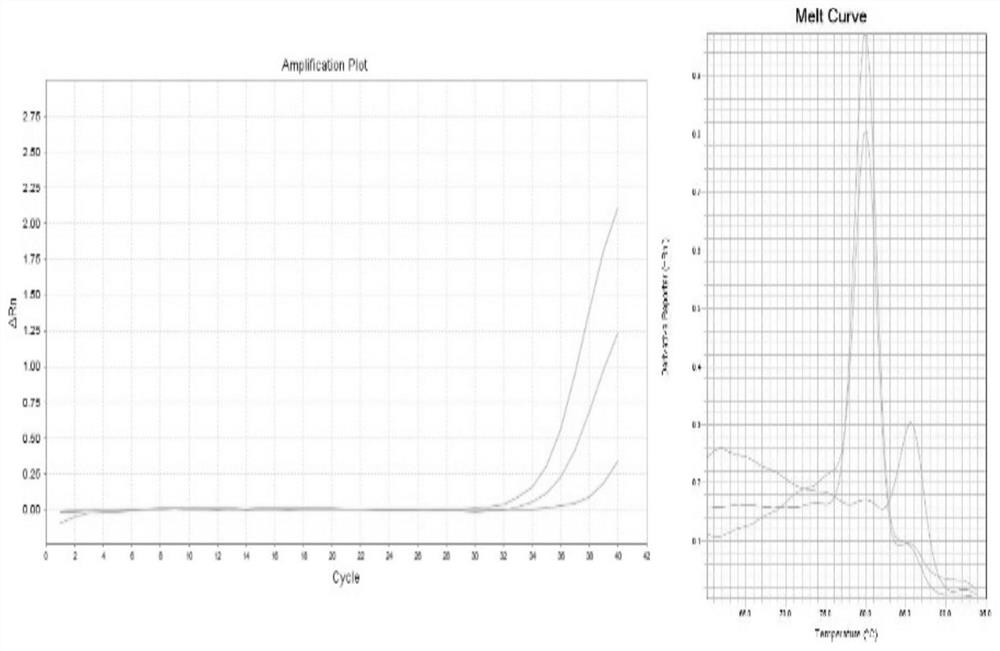
[0072] The circular RNA hsa_circ_0005836 is detected by qPCR, and the reverse transcription reaction is performed using the HT reverse transcription reagent of the present invention, and a conventional reverse transcription kit is used as a control.
[0073] 1. Primer design and synthesis
[0074] Obtain the circular RNA sequence and the Splice junction sequence from the circBase database, and input the 30bp sequence of the upstream 20bp and the downstream 10bp of the Splicejunction position into the primer design software, respectively design two pairs of primers, send the sequences to Shenggong Biosynthesis, and PAGE purification.
[0075] Divergent Primers:
[0076] 5836-DF:ATCATCAGGGCATCTATGTA
[0077] 5836-DR:ACTCATCCTTGAACCTTGCAG
[0078] Amplification size 122bp
[0079] FSjod Primers:
[0080] 5836-FF2:TTAGAACATGCATCTAAGGT
[0081] 5836-FR2:ATACTGTTTTCCTGCAGACAT
[0082] Amplification size 198bp
[0083] 2. RNA extraction and reverse transcription
Embodiment 2
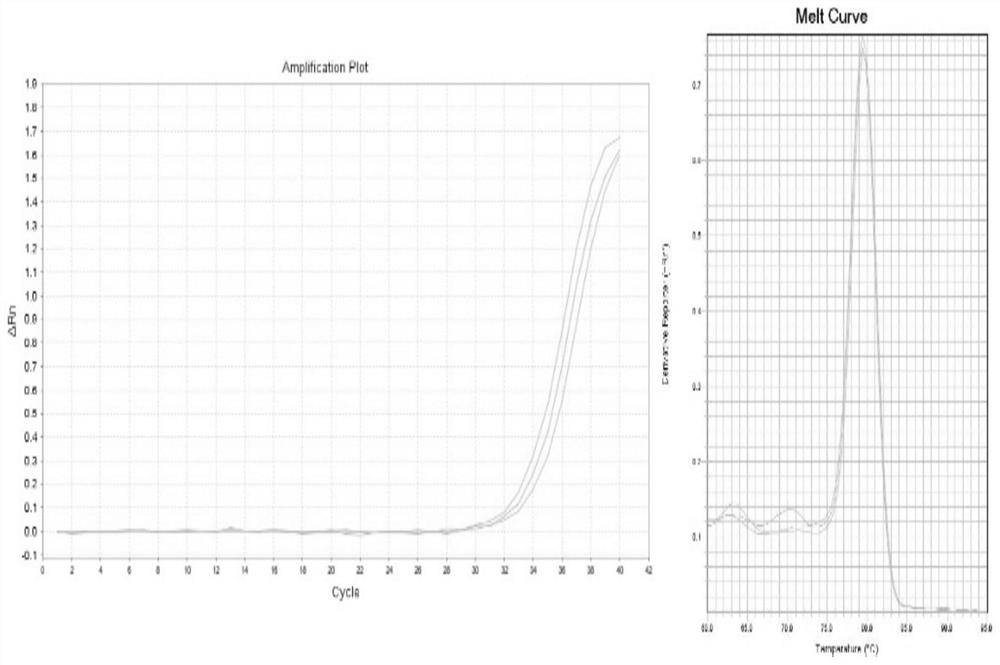
[0104] qPCR detects circular RNA hsa_circ_0007874, uses the HT reverse transcription reagent of the present invention to perform reverse transcription reaction, and uses a conventional reverse transcription kit as a control.
[0105] 1. Primer design and synthesis
[0106] Obtain the circular RNA sequence and the Splice junction sequence from the circBase database, and input the 30bp sequence of the upstream 20bp and the downstream 10bp of the Splicejunction position into the primer design software, respectively design two pairs of primers, send the sequences to Shenggong Biosynthesis, and PAGE purification.
[0107] Divergent Primers:
[0108] 7874-DF:GAGCTGTAGAAGATCTTATTC
[0109] 7874-DR:TAATGTACACCAGACTGGTC
[0110] Amplification size 193bp
[0111] FSjod Primers:
[0112] 7874-FF3:TCAGTGGGGTTGTTTTGGGTC
[0113] 7874-FR3: TGAGCTCTCAGACCCCACACA
[0114] Amplification size 184bp
[0115] 2. RNA extraction and reverse transcription
[0116] Same as Example 1.
[0117] 3. qPCR amplification
[0...
Embodiment 3
[0126] The circular RNA hsa_circ_0005836 is detected by qPCR, the HS PCR amplification reagent of the present invention is used for qPCR amplification, and the conventional qPCR amplification kit is used as a control.
[0127] 1. Primer design and synthesis
[0128] Obtain the circular RNA sequence and the Splice junction sequence from the circBase database, enter the 30bp sequence of the upstream 20bp and the downstream 10bp of the Splicejunction position into the primer design software, and design two sets of 1 base and 6 bases across the circularization site. For the FSjod primer, the sequence was sent to Shenggong Biosynthesis and purified by PAGE.
[0129] FSjod Primers:
[0130] 5836-FF1:TTTAGAACATGCATCTAAGG
[0131] 5836-FR1:ACTGTTTTCCTGCAGACATCT
[0132] Amplification size 197bp
[0133] 5836-FF6:GAACATGCATCTAAGGTTTAC
[0134] 5836-FR6: TGAAGTACATAGATGCCCTGA
[0135] Amplification size 150bp
[0136] 2. RNA extraction and reverse transcription
[0137] Same as Example 1.
[0138] 3. q...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com