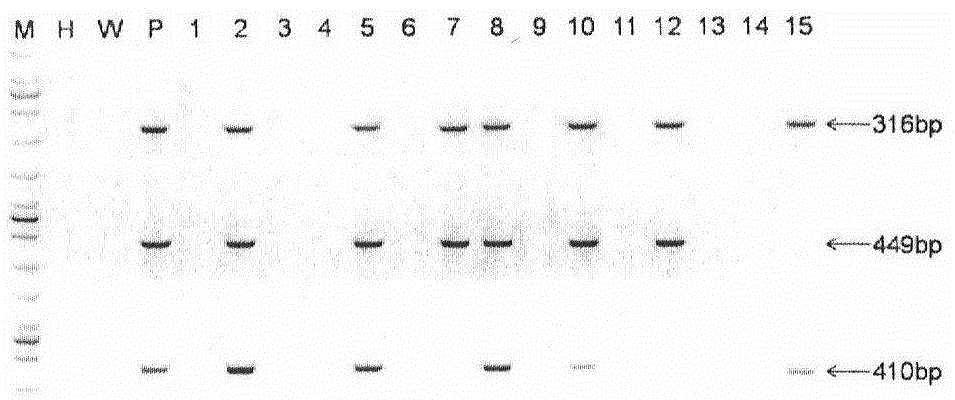
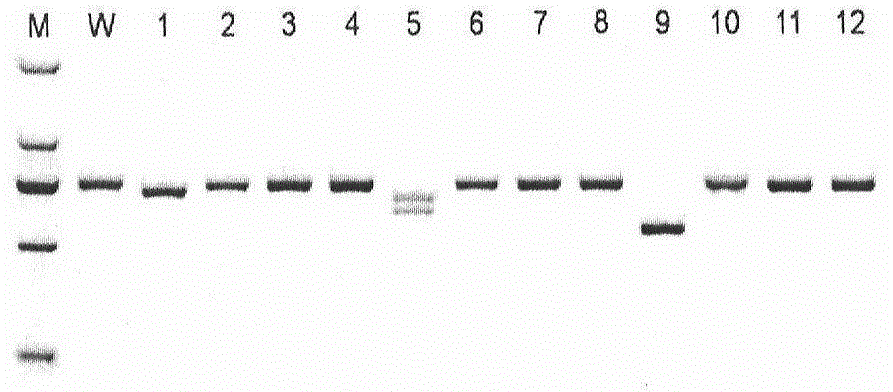
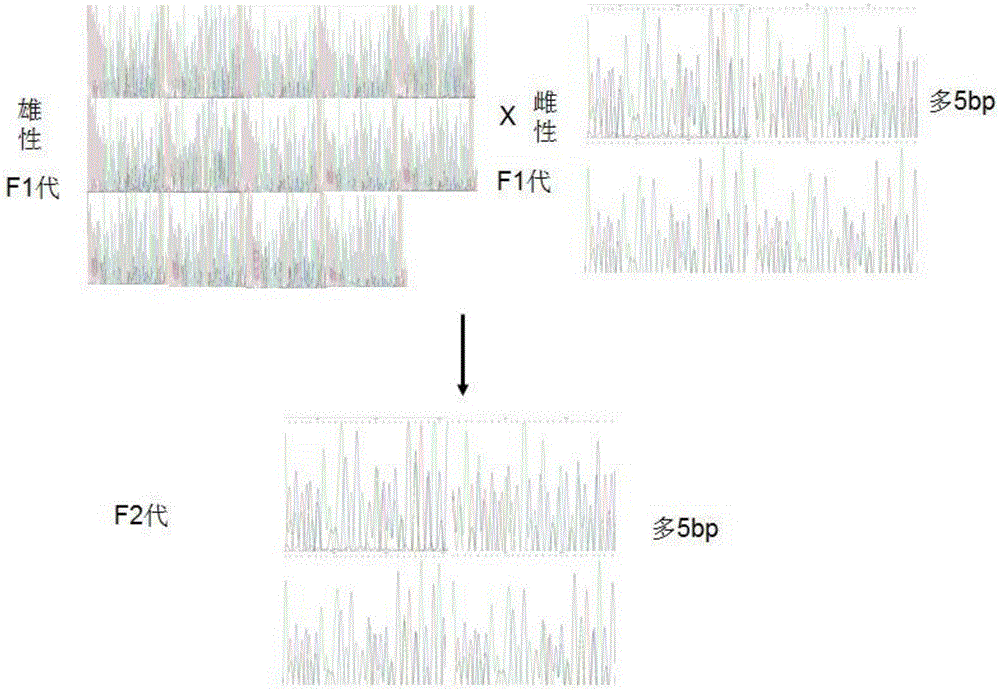
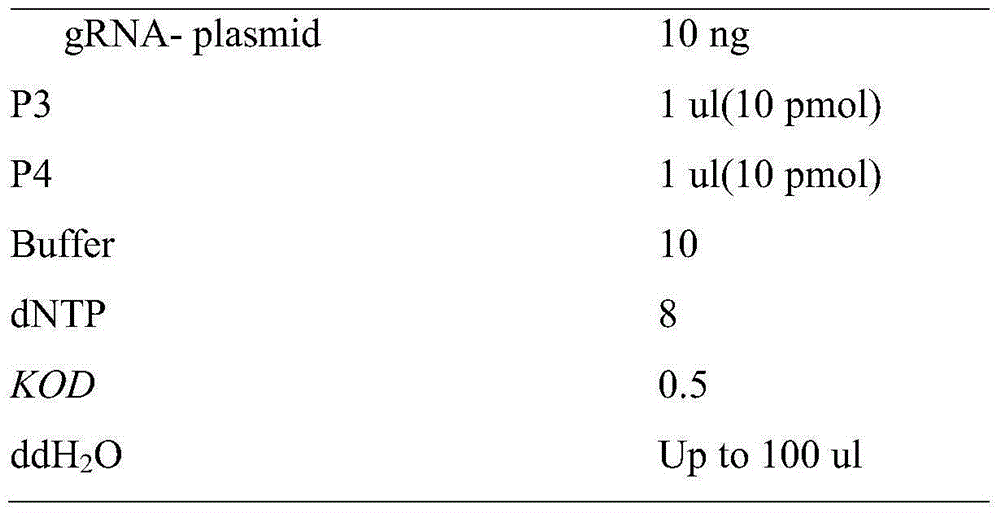
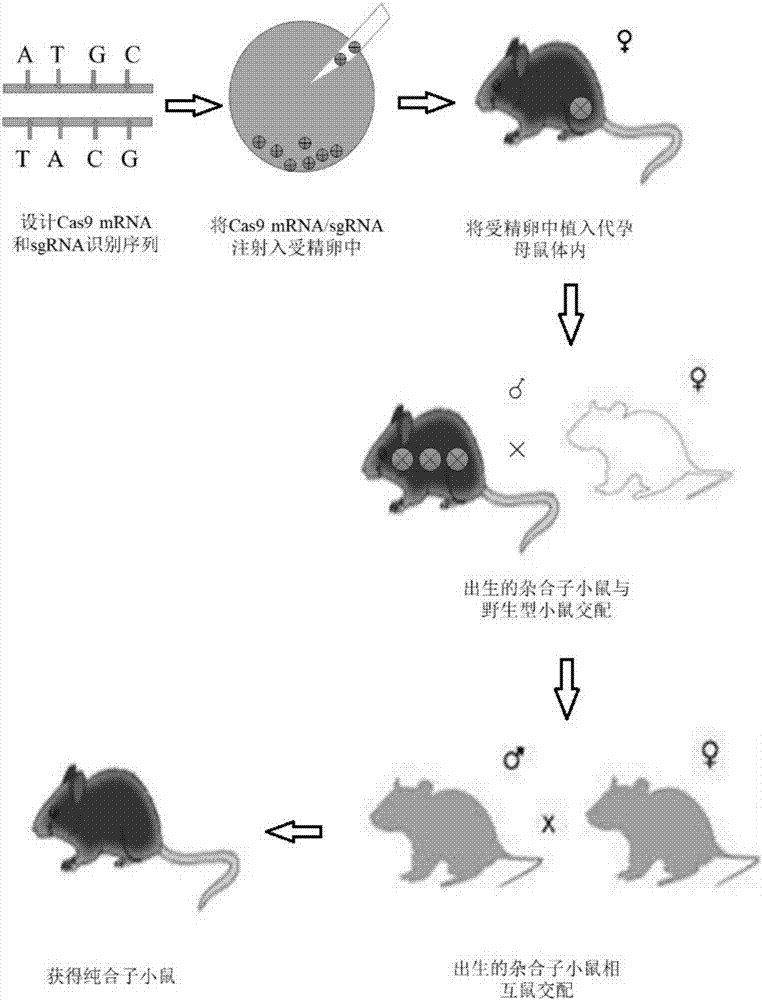
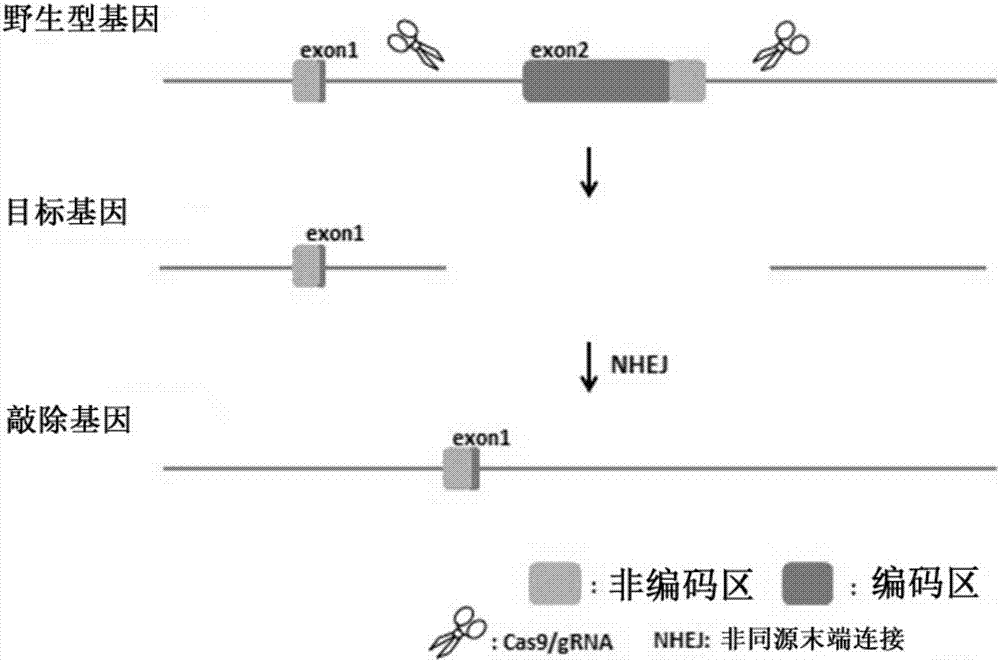
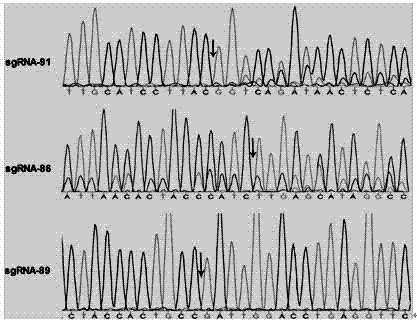
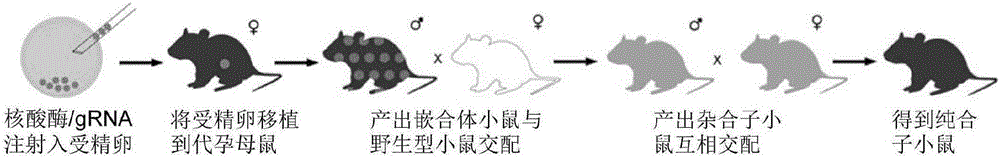
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
87 results about "Gene knockout technology" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Versus gene knockout. Knock-in technology is different from knockout technology in that knockout technology aims to either delete part of the DNA sequence or insert irrelevant DNA sequence information to disrupt the expression of a specific genetic locus.
Method for establishing humanized rat drug evaluation animal model
InactiveCN104593418AVector-based foreign material introductionAnimal husbandryLarge fragmentEngineered genetic
The invention provides a method for establishing a humanized rat drug evaluation animal model. According to the method, a multidrug resistance gene 1 (Abcb1)-knocked-out genetically engineered rat is obtained through a microinjection method by virtue of a CRISPR / Cas9 gene knockout technology and 153kb bacterial artificial chromosome (BAC) fragments containing a humanized Abcb1 promoter and cDNA is simultaneously inoculated into the rat genome through the microinjection method by virtue of a large fragment transgenic technology to obtain a transgenic rat capable of stably expressing human Abcb1 and the genetically engineered rat and the transgenic rat are hybridized to establish the humanized rat drug evaluation animal model. RT-PCR analysis shows that Abcb1 expression profiles of humanized Abcb1 rat are significantly different from those of the rat endogenous Abcb1. The method has the beneficial effects that the humanized rat capable of expressing human Abcb1 is obtained and the rat is used for expressing human Abcb1 genes and has closer expression profiles to those of human so that the model can be well used for the efficacy evaluation of newly developed drugs.
Owner:INST OF LAB ANIMAL SCI CHINESE ACAD OF MEDICAL SCI
Method for establishing obese rat animal model based on CRISPR (clustered regularly interspaced short palindromic repeat) gene knockout technology
InactiveCN103614415AImprove the level ofDeepen understanding of gene regulationVector-based foreign material introductionAnimal husbandryDiseaseLepr gene
The invention provides a method for establishing an obese rat animal model based on a CRISPR (clustered regularly interspaced short palindromic repeat) gene knockout technology. The method comprises the following steps: (1) establishing an Lep / Lepr gene knockout rat model; (2) carrying out the authentication and related analysis on the obese rat animal model; (3) evaluating the energy metabolism and the body fat rate of the obese rat animal model. According to the method, a CRISPR / Cas system is used for respectively or simultaneously knocking out Lep and LepR genes so as to obtain the rat model modified by a corresponding target gene, so that the understanding of gene regulation in the obesity morbidity process can be deepened, and the high-level animal model can be provided for the translational medicine and the new medicine research and development.
Owner:SUZHOU TONGSHAN BIO TECH
Preparation method of zebrafish with hepcidin gene knocked out by use of CRISPR / Cas9 technology
The present invention mainly relates to formation of zebrafish with hepcidin gene knocked out, by the use of CRISPR / Cas9 technology, a unique PAM region is designed, so that the hepcidin gene in the zebrafish is knocked out, and other genes are not accidentally injured. The first case of hepcidin knockout transgenic animal model zebrafish has great significance, the hepcidin is a major factor in the regulation of iron, once the hepcidin is knocked out, an animal model can be successfully molded into an iron overload animal model, the human factor intervention can be excluded, the hepcidin has great significance to iron expression researches, meanwhile compared with the traditional gene knockout technology, the CRISPR / Cas9 technology has low toxicity, high accuracy, high efficiency, short success cycle and other characteristics, and the hepcidin gene can be faster knocked out.
Owner:徐又佳
Construction method and application of Ifit3-eKO1 gene knockout mouse animal model
The invention relates to a method of constructing an Ifit3-eKO1 gene knockout mouse animal model and belongs to the biotechnical field. The method comprises the following steps: S1, determining specific target sites sgRNA1 and sgRNA2 of a to-be-knocked out gene of an Ifit3-eKO1 mouse, and performing in vitro transcription with Cas9 nuclease to mRNA; and S2, micro-injecting active sgRNA and Cas9RNA into an oosperm of the mouse to obtain the Ifit3-eKO1 gene knockout mouse. The method has the advantages that by using the CRISPR / Cas9 gene knockout technology, the Ifit3-eKO1 gene knockout mouse animal model is constructed for the first time, thereby providing a convenient, reliable and economical animal model for researching action of Ifit3 in tumorigenesis and development.
Owner:SHANGHAI TONGJI HOSPITAL
Method for establishing fragile X-syndrome non-human primate model on basis of CRISPR gene knockout technology
InactiveCN103642836APredictive effectReduce the risk of research and developmentVector-based foreign material introductionAnimal husbandryDiseaseFragile X chromosome
The invention discloses a method for establishing a fragile X-syndrome non-human primate model on the basis of a CRISPR gene knockout technology. The method comprises the following steps: (1) establishing a FMR1 gene knockout machin model; (2) carrying out identification and related functional analysis on the machin model; (3) carrying out tests on the nerve characteristics and learning and memorizing ability of the machin model. The method utilizes a CRISPR gene knockout technology to establish a fragile X-syndrome non-human primate model. The model fills the blank of non-human primate model, can effectively stimulate the pathological process of human diseases, can be used as an optimum animal model for researching human diseases, can effectively predict the effect of novel vaccine, novel drug or novel diagnostic reagent in clinical applications, and thus greatly reduces the risk of novel drug development.
Owner:SUZHOU TONGSHAN BIO TECH
Single guide ribonucleic acid (sgRNA) capable of effectively editing pig ROSA26 gene, and application of sgRNA
ActiveCN106916820AImprove securityEfficient and stable expressionAnimal husbandryDNA/RNA fragmentationBiotechnologyFibroblast
The invention discloses single guide ribonucleic acid (sgRNA) capable of effectively editing a pig ROSA26 gene, and application of the sgRNA. Based on the sgRNA capable of specifically identifying the pig ROSA26 gene in a pig genome, a cell line of enhanced green fluorescent protein (EGFP) site-specific integrated porcine fetal fibroblasts is successfully established by using a CRISPR / Cas9-mediated gene knock-in technique; the results show that the cell line can stably and efficiently express an EGFP gene; furthermore, in the preparation process of the cell line, exogenous promoter genes and positive and negative screening marker genes are not introduced into the cell line. Therefore, the sgRNA greatly improves the safety of transgenic pigs, and has important significance for eliminating the biological and food potential safety hazards of agricultural products of the transgenic pigs.
Owner:重庆吉渝科技有限公司
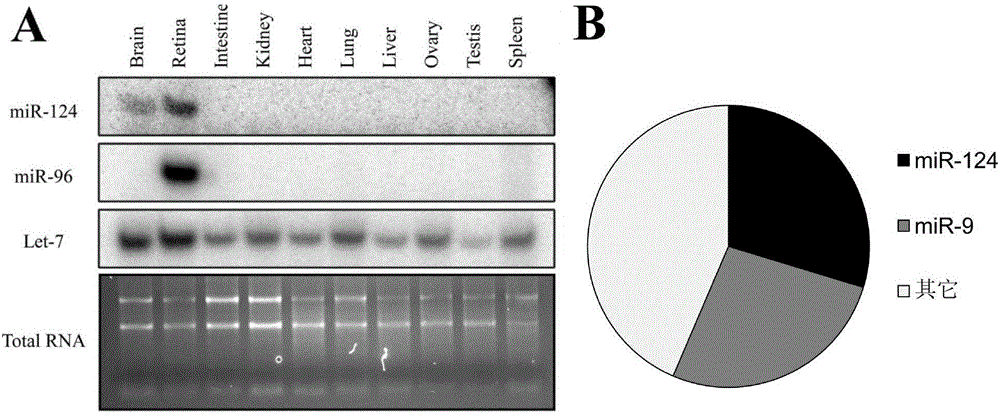
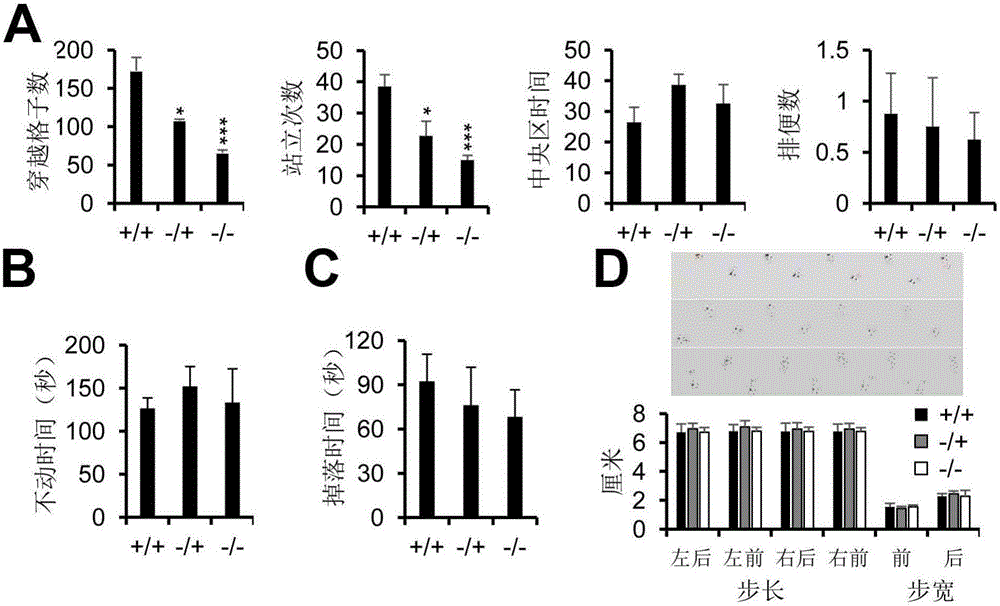
MiR-124 gene knockout murine animal models as well as construction method and application thereof
ActiveCN106172238AShort modeling timeSimple methodNucleic acid vectorVector-based foreign material introductionAmyloid beta proteinsKnockout mouse
The invention discloses miR-124 gene knockout murine animal models. The murine animal models refer to mice with miR-124-1, miR-124-2 and miR-124-3 genes knocked out. With the adoption of a Crispr-cas9 gene knockout technology, microRNA (micro ribonucleic acid) miR-124 genes with the highest expression quantity in the brains of the mice are knocked out. The obtained mice show obvious nervous system disease states such as reduction of locomotor activity, deterioration of learning and memorizing abilities, increase of soluble amyloid beta protein and the like. Therefore, the simple, reliable and economical animal models can be provided for research of nervous system disease pathology and screening of drugs for treating nervous system diseases.
Owner:CENT SOUTH UNIV
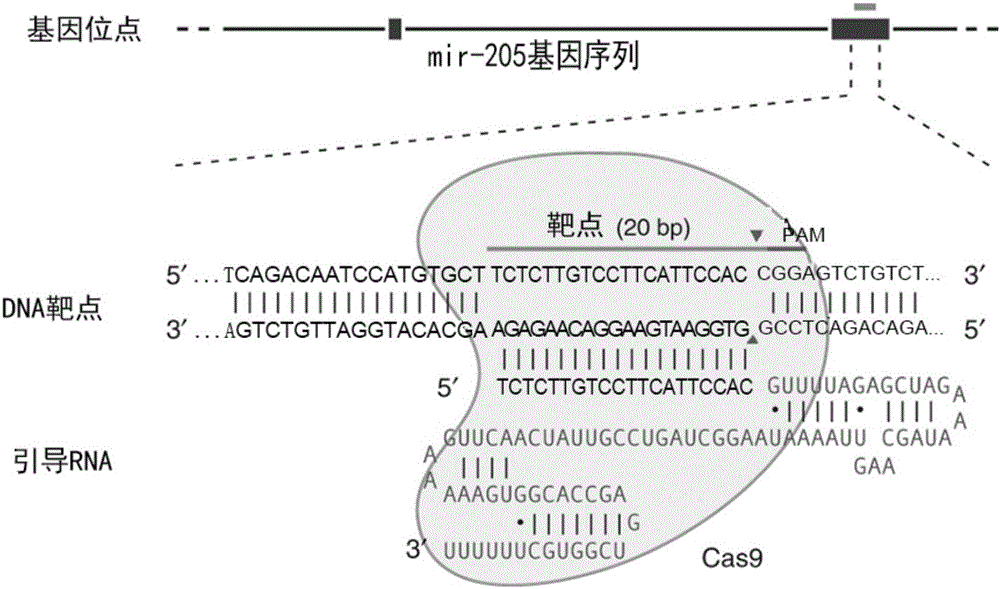
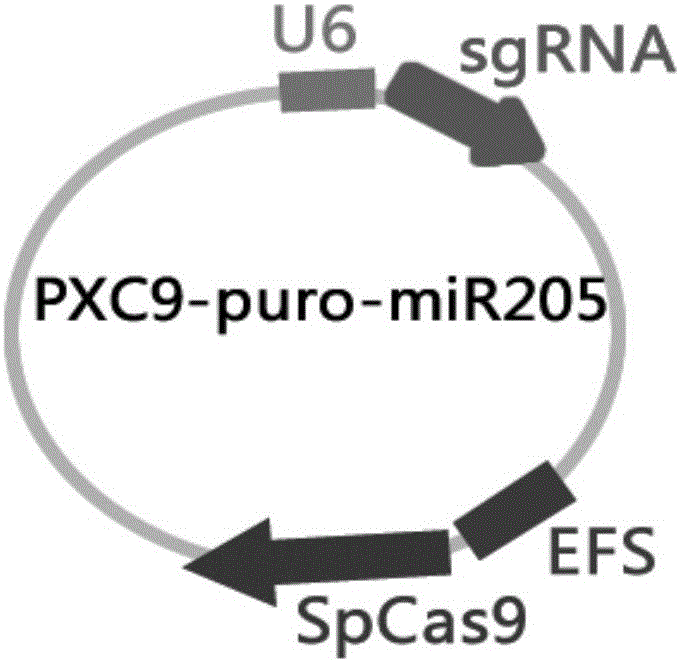
MiR-205 gene knockout kit based on CRISPR-Cas9 gene knockout technology
ActiveCN105112445AVector-based foreign material introductionDNA/RNA fragmentationEnzyme digestionFluorescence
The invention discloses a miR-205 gene knockout kit based on CRISPR-Cas9 gene knockout technology. According to the invention, an optimal CRISPR-Cas9 target sequence of a certain amount of miR-205 is obtained through target design software and an sgRNA single chain is synthesized in vitro; an insertion fragment is obtained through processing; then sgRNA is inoculated into a plasmid vector by using T4 ligase; and a miR-205 gene knockout cell strain is obtained through transfection of an LNCap cell strain, continuous drug screening and fluorescence detection. Heterogenous hybridization double strands are obtained by extracting DNAs of a cell, PCR amplification of miR-205, purification, denaturation of a PCR product and annealing; T7E1 enzyme digestion test is employed to determining shearing efficiency of a CRISPR-Cas9 system on miR-205; a verified optimized a miR-205 gene knockout CRISPR-Cas9 target sequence is obtained; and the kit is constructed on the basis of the target sequence and can be used for directional knockout of miR-205 genes. The kit has the characteristics of high gene knockout efficiency, fast speed, easiness and economic performance and has wide prospects in the aspects of construction of animal models and clinical research of medical science.
Owner:广州辉园苑医药科技有限公司
Method for obtaining bmp2a gene knocked-out zebra fish through CRISPR/Cas9
The invention discloses a method for obtaining bmp2a gene knocked-out zebra fish through CRISPR / Cas9. A novel gRNA sequence is designed between a first exon and an intron of BMP2a, the gRNA sequence is GGAGCCCATCACTAGACTCTTGG, and enzyme digestion is HinfI. Compared with a traditional gene knockout technology, the CRISPR / Cas9 technology has the advantages of low toxicity, high accuracy, high efficiency, short success cycle and the like. Therefore, the BMP2a gene can be theoretically knocked out faster.
Owner:WUXI NO 2 PEOPLES HOSPITAL
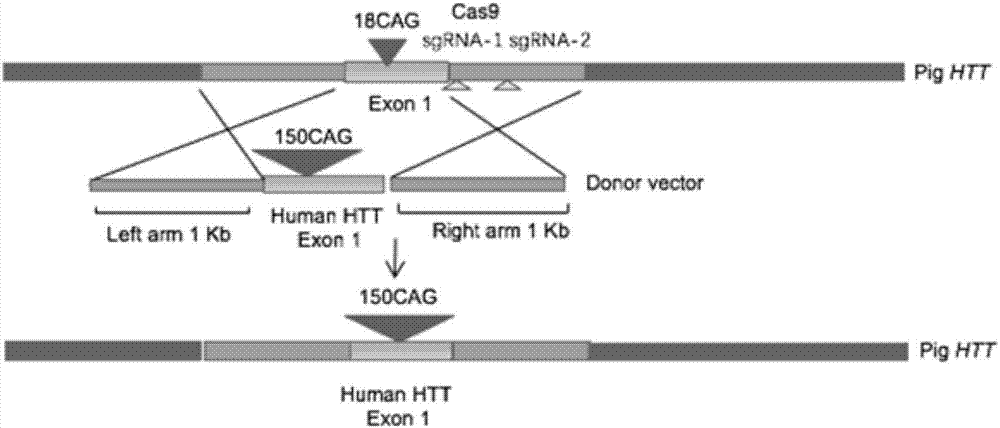
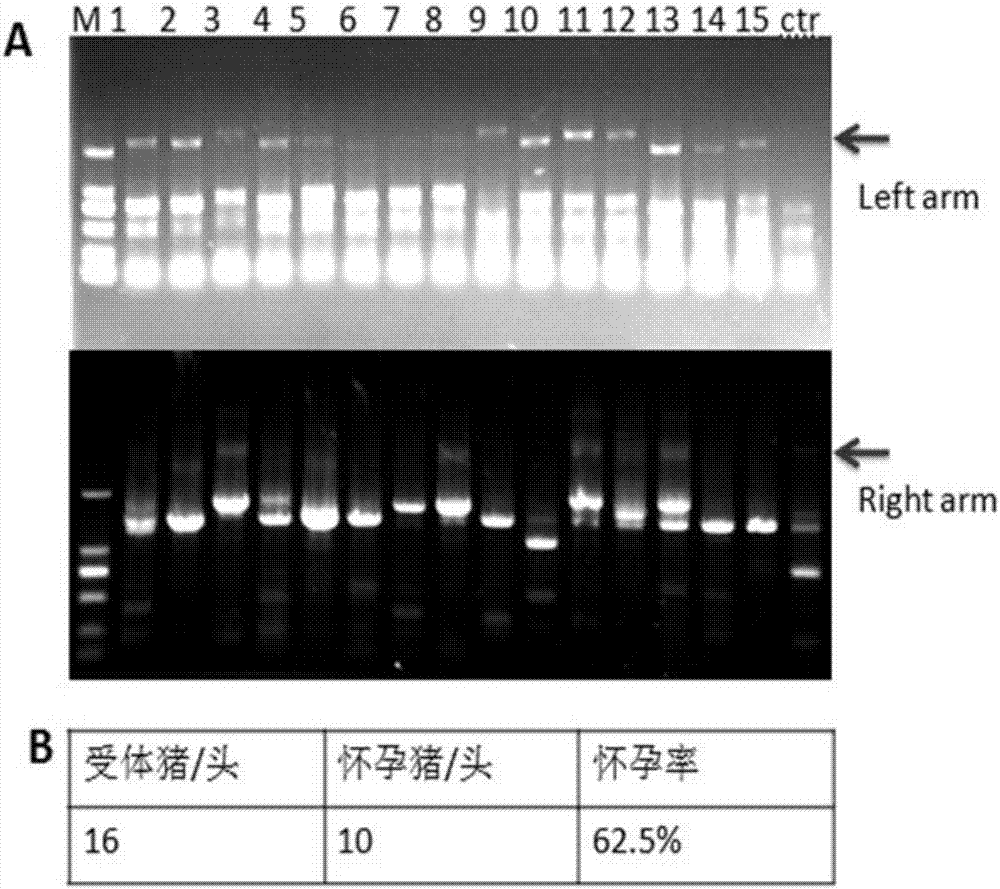
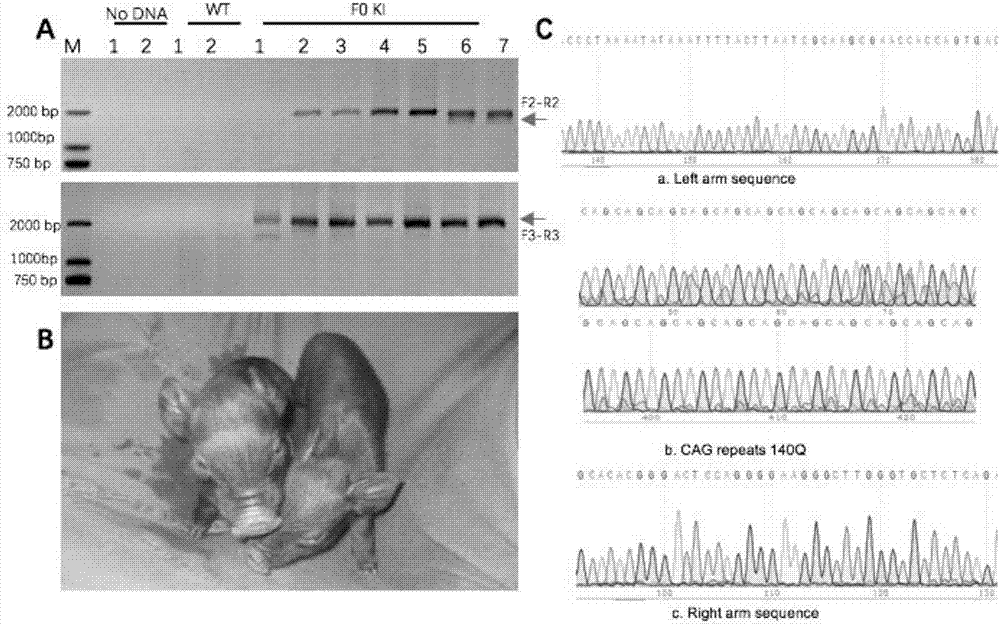
Recombinant vector for knock-in of human Huntington gene, construction method of recombinant vector and application of recombinant vector in construction of model pig
ActiveCN107988256AIncreased probability of knock-in positive clonesEfficient FeasibilityStable introduction of DNANucleic acid vectorHuman studiesExon
The invention discloses a recombinant vector for the knock-in of a human Huntington gene, a construction method of the recombinant vector and application of the recombinant vector in the constructionof a model pig. According to the recombinant vector, a human mutated Huntington exon gene is knocked in a fixed point manner for the first time, a virulence gene is knocked in by virtue of a CRISPR / Cas9 technique for the first time, and a donor vector is optimized by optimizing sfRNA, so that the probability that the gene is knocked into a positive cloned cell is increased; and by combining with apig cell nucleus transplantation technique, the probability that a directly obtained positive cell is knocked into the pig is increased, and a small human Huntington gene knock-in pig is obtained, thereby proving the efficient feasibility of the method for constructing gene modified pigs. The constructed Huntington gene knock-in model pig has behavioral characteristics such as respiratory disturbance and dyskinesia similar to human Huntington diseases, and stable heritable passage can be realized, so that a reliable model is provided for the research of the human Huntington diseases; and thenumber can be guaranteed so as to realize drug screening, gene treatment, stem cell treatment and the like, and the model can be a good human disease model.
Owner:JINAN UNIVERSITY +1
Construction method for Sqstm1 whole-genome knockout mouse animal model and application
The invention discloses a construction method for a Sqstm1 whole-genome knockout mouse animal model and application. The mouse animal model is a mouse of which the Sqstm1 gene is knocked out. On the basis of a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) / Cas9 gene knockout technology, the Sqstm1 gene knockout mouse model is constructed. The construction method comprises the following steps: S1: designing sgRNA (Ribonucleic Acid) and Cas9 RNA, carrying out in vitro transcription to obtain mRNA, carrying out microinjection on the sgRNA and the Cas9 RNA with activity into amouse oosperm, to obtain the Sqtm1 gene knockout mouse; S2: identifying the Sqtm1 gene knockout mouse animal model. On the basis of the CRISPR / Cas9 gene knockout technology, the Sqstm1 gene knockout mouse animal model is constructed for the first time, and a convenient, reliable and economic animal model is provided for researching a relationship between Sqstm1 and diseases including autophagy, tumors and the like.
Owner:SHANGHAI TONGJI HOSPITAL
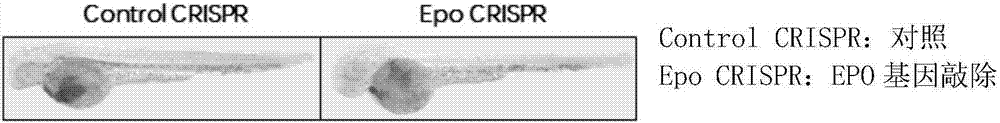
Method, primer and plasmid for constructing EPO gene knockout zebrafish animal model and preparation method of primer and plasmid
ActiveCN107217075ADeepen the understanding of the roleHigh gene knockout success rateHydrolasesStable introduction of DNAEscherichia coliExon
The invention provides a method, primer and plasmid for constructing an EPO gene knockout zebrafish animal model and a preparation method of the primer and the plasmid, and belongs to the field of biomedicine. The method includes the steps of 1) establishment of an EPO gene knockout zebrafish CRISPR oligomer sequence plasmid; and 2) establishment and culture of the EPO gene knockout zebrafish animal model on the basis of CRISPR gene knockout technology; wherein the step 1) comprises 1-1) design and synthesis of the primer aiming at an EPO exon 2 region target sequence and 1-2) synthesis of the Escherichia coli plasmid containing the EPO target sequence. The method for constructing the EPO gene knockout zebrafish animal model can be used to construct the EPO gene knockout zebrafish animal model, and facilitates further understanding and research of EPO.
Owner:THE FIRST AFFILIATED HOSPITAL OF MEDICAL COLLEGE OF XIAN JIAOTONG UNIV
Method for breeding high-producing strain of cellulase by gene knockout
ActiveCN103865949AReduce loadPromote productionFungiMicroorganism based processesHygromycin BAmylase
The invention discloses a method for breeding a high-producing strain of cellulase by gene knockout, and belongs to the field of enzyme engineering. The method for breeding the high-producing strain of cellulase by gene knockout comprises the step of knocking out an amylase gene of the cellulase producing strain. A high-producing strain of cellulase is aspergillus niger without the amylase gene. An amylase gene knockout plasmid is obtained by connecting the front half part of the recombinant segment amylase gene-hygromycin B-an expression unit of a resistant gene- the rear half part of the amylase gene to a pCAMBIA1300 plasmid. Then, the plasmid is converted into agrobacterium, and the amylase gene is knocked out by agrobacterium-mediated conversion of the aspergillus niger. By using the gene knockout technology, the amylase gene of the aspergillus niger is knocked out. After the amylase gene is knocked out, the cell load is reduced, thereby facilitating production of a lot of cellulase. The cellulase activity of the high-producing strain of cellulase obtained can reach 21U / g.
Owner:中农华威生物制药(湖北)有限公司
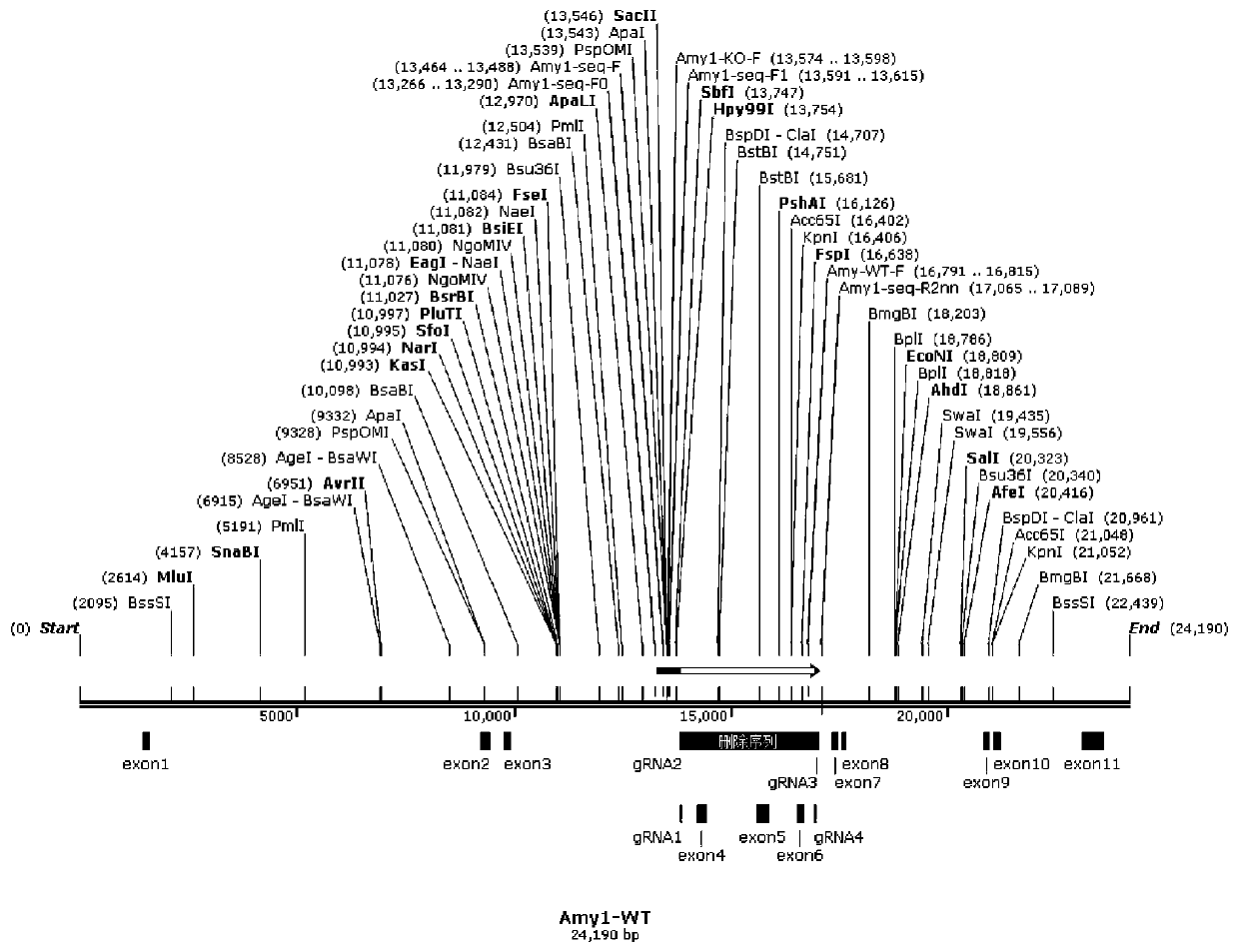
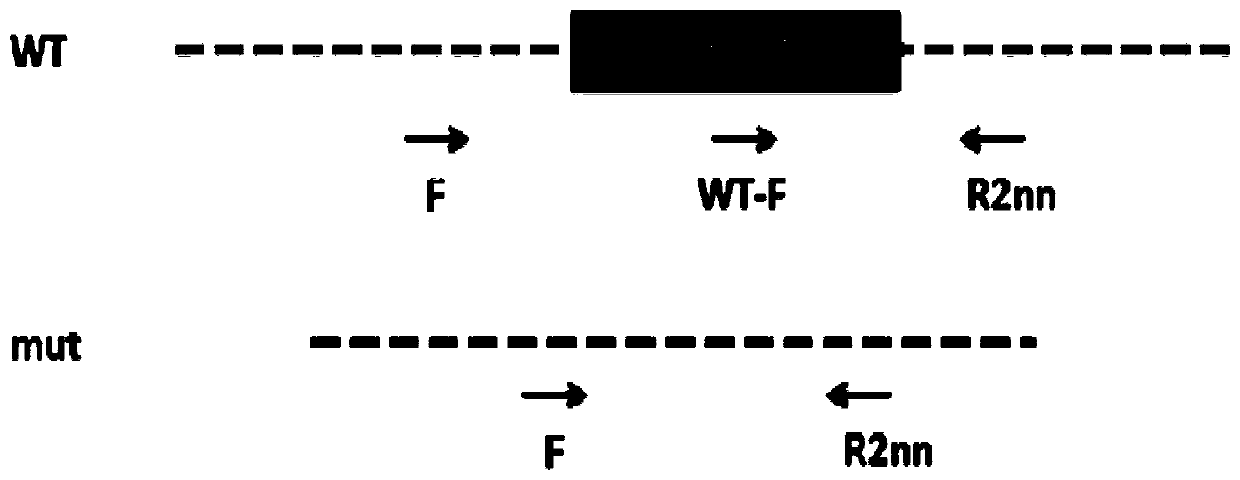
Construction method of Amy1 gene knockout mouse animal model and application
ActiveCN110885858AStable introduction of DNANucleic acid vectorBiotechnologyGene Knockout Techniques
The invention discloses a construction method of an Amy1 gene knockout mouse animal model and application. An Amy1 gene knockout mouse model is constructed based on a CRISPR / Cas9 gene knockout technology. The construction method comprises the following steps: step 1, designing sgRNA and Cas9 RNA, performing in-vitro transcription to obtain mRNA, and performing microinjection of the active sgRNA and Cas9 RNA on fertilized eggs of a mouse to obtain an Amy1 gene knockout mouse; and step 2, identifying the Amy1 gene knockout mouse animal model. The method has advantages: the CRISPR / Cas9 gene knockout technology is used and the mouse animal model with the Amy1 gene knocked out is established for the first time; and a convenient and reliable animal model is provided for researching the relationbetween the Amy1 gene and related diseases.
Owner:SHANGHAI JIAO TONG UNIV
Gene knock-in recombinant vector and preparation method thereof as well as method for preparing mouse model
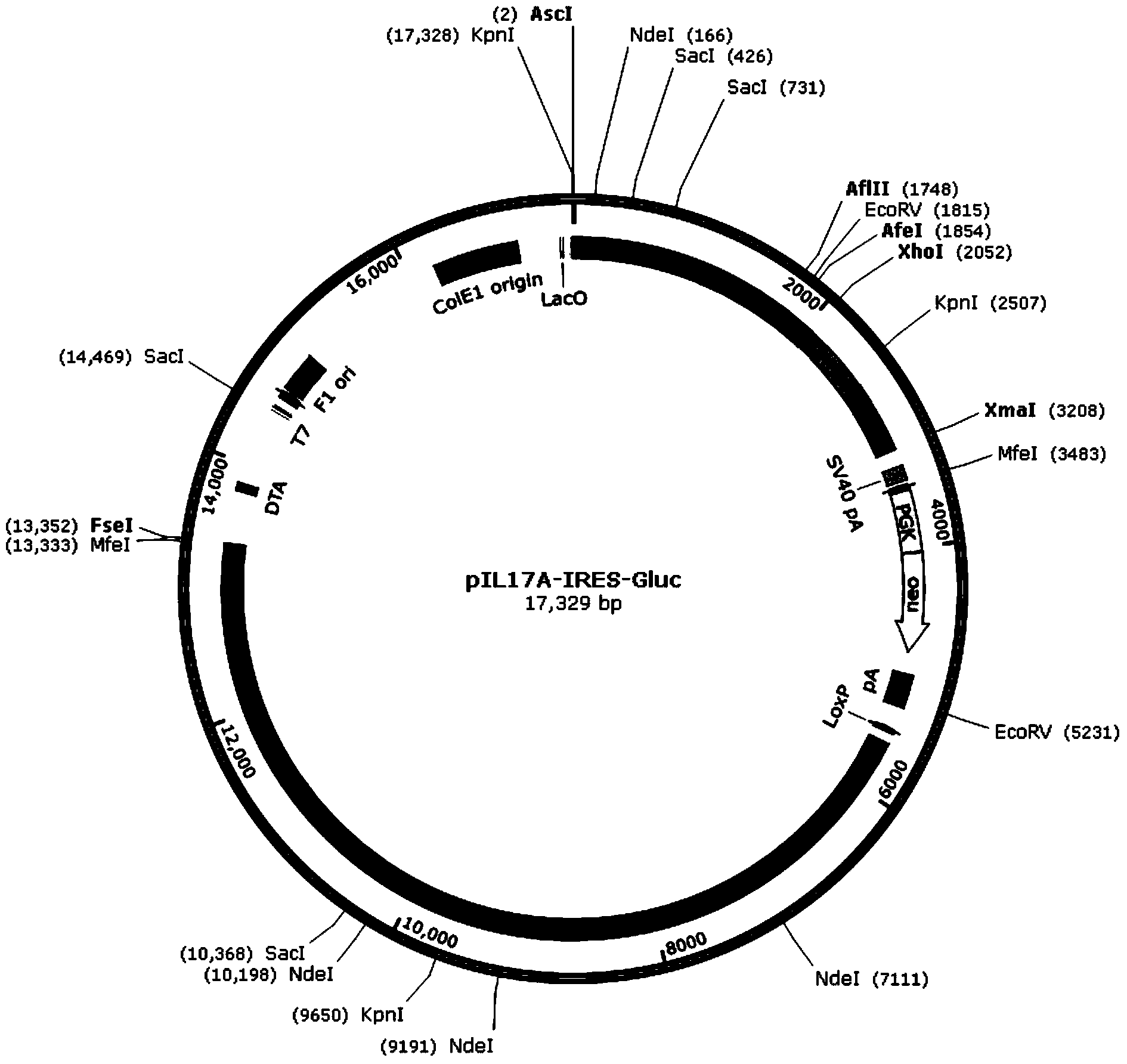
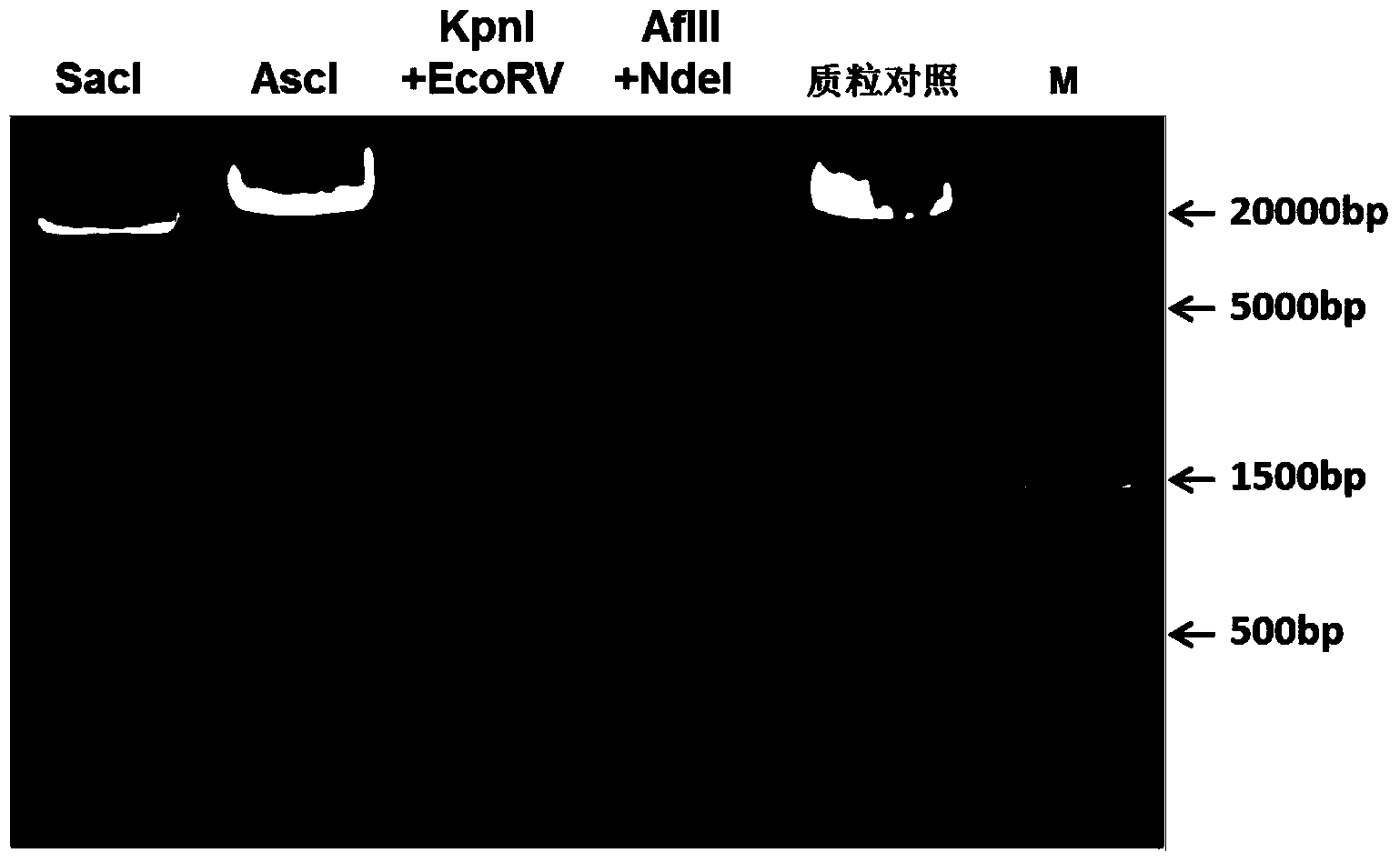
ActiveCN103642828AShorten the timeHigh detection sensitivityMicrobiological testing/measurementVector-based foreign material introductionWhite blood cellFluorescence microscope
The invention discloses a recombinant vector for gene knock-in. The recombinant vector comprises a DNA (deoxyribonucleic acid) fragment structure IL17A-IRES-luc, wherein IL17A is an encoding gene of interleukin 17A, IRES is internal ribosome entry site, and luc is an encoding gene of secreting luciferase. The invention also provides a method for preparing the gene knock-in recombinant vector and a mouse model prepared by using the recombinant vector, wherein the mouse model can be used for detecting the expression of the IL17A gene by detecting the expression of the secreting luciferase in blood or urine; a fluorescent microscope or other complicated equipment is not needed; the secreting luciferase emits light by virtue of an oxidizing reaction of substrate coelenterazine in the catalysis of luciferase, and laser light is not needed, so that the fluorescent background is far lower than GFP (green fluorescent protein) fluorescence, the detection sensitivity is higher, the background is cleaner, and the experiment results are more accurate.
Owner:BIOCYTOGEN PHARMACEUTICALS (BEIJING) CO LTD
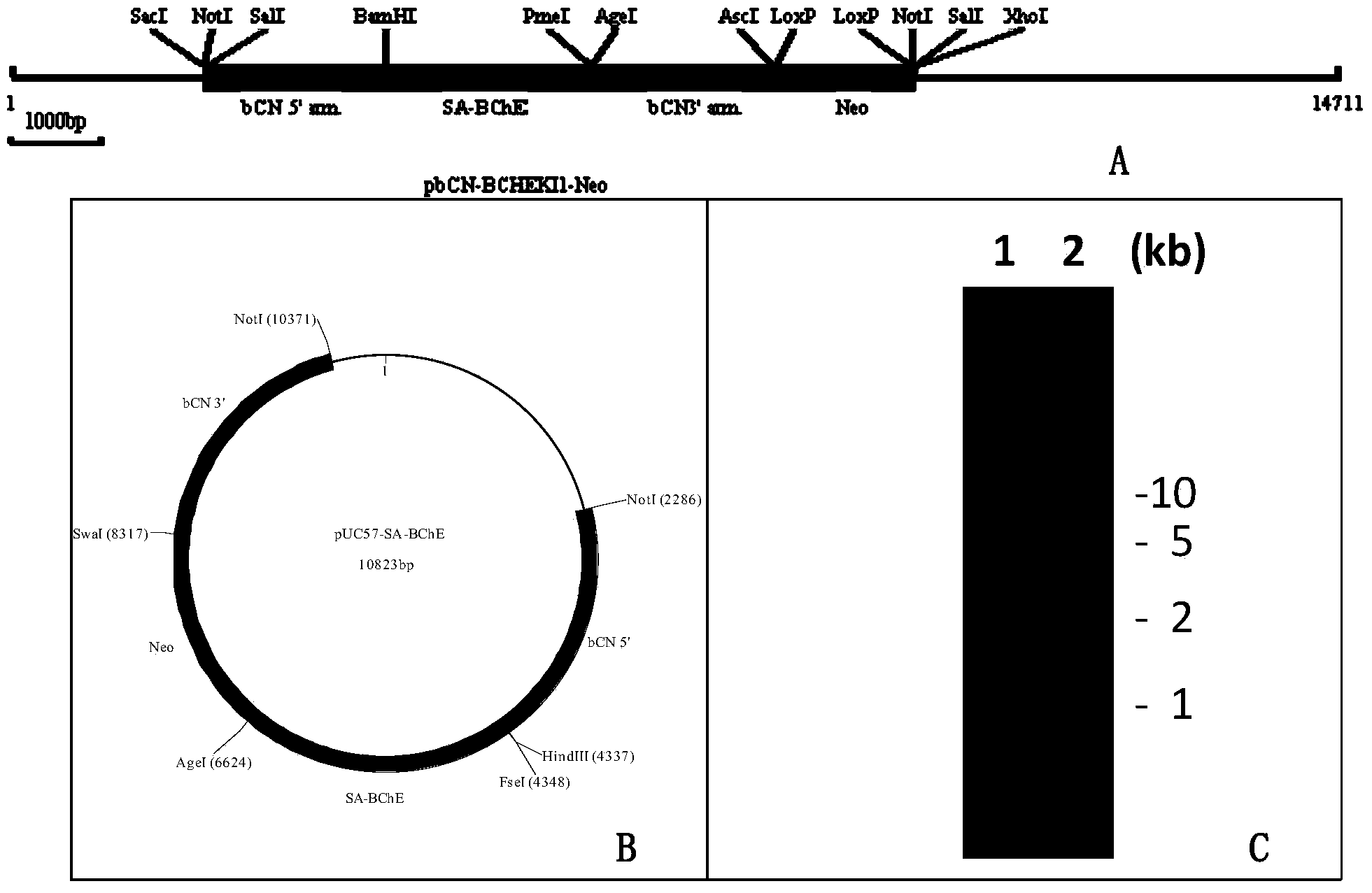
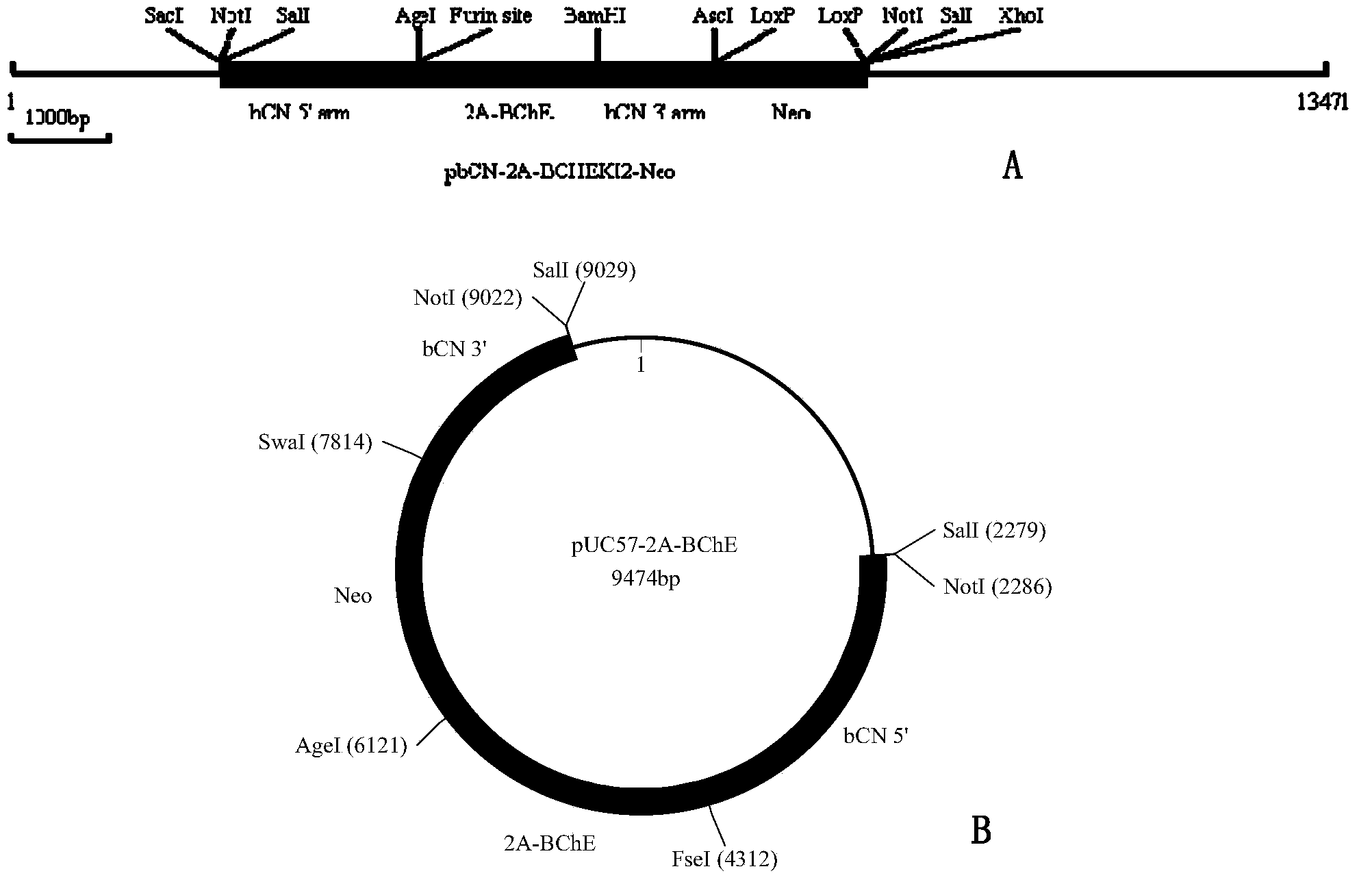
Method for producing recombinant human BChE (butyrylcholinesterase) from transgenic animals by using gene knock-in and nuclear transfer technologies
The invention discloses a method for producing recombinant human BChE (butyrylcholinesterase) from transgenic non-human mammals by using gene knock-in and nuclear transfer technologies, and particularly provides a donor construct used for specific expression of a mammary gland. The construct integrates DNA (deoxyribose nucleic acid) coding sequences of human BChE to a specific expression protein gene locus of the mammary gland in targeted manner and performs successful transfection on non-human mammal embryo fibroblasts, juvenile and adult somatic cells or embryonic stem cells, clone is performed by using a somatic cell nuclear transfer technology so as to obtain a new individual, and the carried human BChE gene enables the mammary gland of a non-human mammal to express stably and secrete holoenzyme or a monomer of the human BChE. Recombinant protein produced with the method can be used for preventing and treating nerve poison and organic phosphorus compound poisoning as well as apnea caused by cocaine poisoning and succinylcholine, and for detecting and removing residues of organic phosphorus compounds on vegetables, other crops, various article layers and the soil.
Owner:SHANGHAI JENOMED BIOTECH CO LTD
Attenuated Salmonella pullorum and application thereof
InactiveCN102154184ALow toxicityImproving immunogenicityAntibacterial agentsBacteriaHorizontal transmissionMicroorganism
The invention discloses an attenuated Salmonella pullorum and application thereof. By using gene knockout technology, the in vivo colonization gene and the main virulence gene of Salmonella pullorum are deleted, thus obtaining an dual-gene-deleted attenuated Salmonella pullorum SM091-DED strain; and the microbiological preservation number of the strain is CGMCC NO.4604. The virulence of the attenuated Salmonella pullorum disclosed by the invention is obviously reduced, and the in vivo colonization time is short after the attenuated Salmonella pullorum is inoculated into a host; the herd infection test shows that the attenuated Salmonella pullorum has no horizontal transmission capability; the attenuated Salmonella pullorum provides full cross protection for homotype and allotype high-virulence Salmonella pullorum; and after SPF (Specific Pathogen Free) chicks are immunized, the infection of the high-virulence strain can be effectively eliminated, and the herd infection can not be caused. Thus, the invention provides a safe, fine-immunogenicity and low-virulence Salmonellosis avium attenuated live vaccine for preventing Salmonellosis avium.
Owner:HARBIN VETERINARY RES INST CHINESE ACADEMY OF AGRI SCI
Method for regulating and controlling synthesis of EPS (extracellular polymeric substance) of clostridium acetobutylicum
The invention discloses a method for regulating and controlling synthesis of an extracellular polymeric substance of clostridium acetobutylicum. The method is characterized in that an intron sequence is interpolated into a gene Spo0A of the clostridium acetobutylicum by using a type II intron gene knockout technology. The invention provides a simple and efficient method for regulating and controlling EPS (Extracellular Polymeric Substance) synthesis; a recombinant strain obtained based on the method is relatively good in regulating and controlling effect on the EPS; in case of taking glucose as a carbon source, when cultivated in an anaerobic bottle with the volume of 100 ml by using a slide biomembrane cultivation method, a clostisdium acetobutylicum biomembrane is 5.81 mum in thickness, and the thickness of a strain biomembrane cultivated under the same condition is 24.6 mum.
Owner:NANJING UNIV OF TECH
Method for realizing genome editing and accurate and targeted knock-in of genes in fishes
ActiveCN108949830AImprove integration efficiencyLow costMicroinjection basedAnimal husbandryRandom mutationGenome editing
The invention provides a method for realizing genome editing and accurate and targeted knock-in of genes in fishes. A technology of producing DNA single-chain gap based on nicking enzyme is utilized,assisted with a set of homologous recombination repair factor RecOFAR, so as to realize high-efficient, accurate and targeted integration of genome editing and gene knock-in in the fishes. The problems that a current existing method is low in efficiency, and random mutation efficiency of target points is higher are overcome.
Owner:FUZHOU UNIV
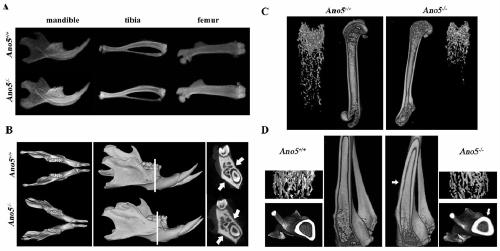
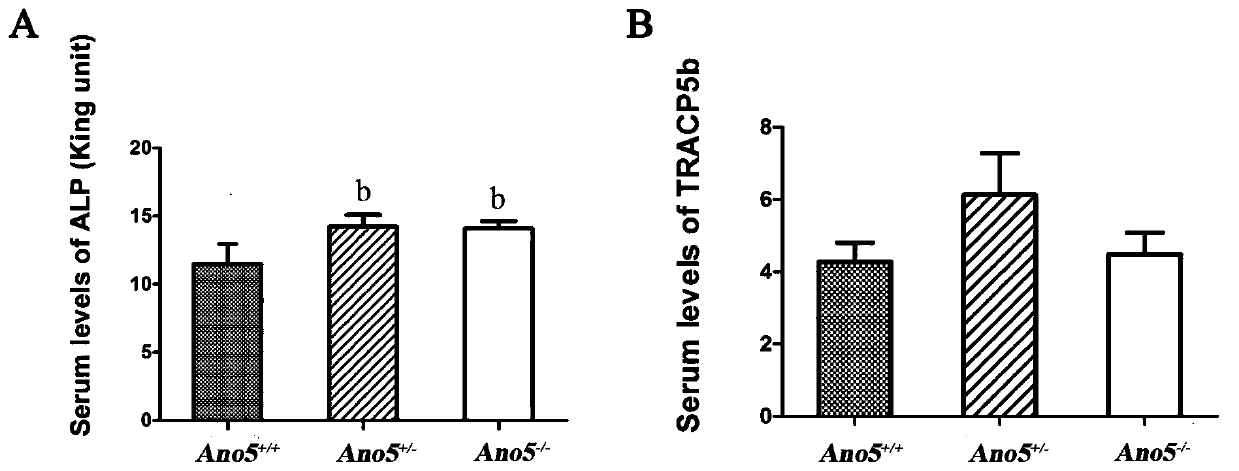
Construction method of Ano5 gene knockout mouse model
ActiveCN109694881AImprove stabilityStable introduction of DNAMicroorganism based processesEccentric hypertrophyKnockout animal
The invention provides a construction method of an Ano5 gene knockout mouse model. A CRISPR-Cas9 gene knockout technology is adopted to remove 11th and 12th exons of the Ano5 gene. After Ano5 gene knockout, mice all have clinical symptoms of human GDD patients such as lateral bending of shin bones, hyperplasia of cortical bone, hypertrophy of lower jawbone, increasing of alkaline phosphatase in blood, and the like. The established mouse model is used to simulate the human GDD disease, is stable, has stable heredity and similar symptoms of the human GDD disease, can be applied to research on GDD pathogenesis and GDD gene therapy, and is economic, simple and reliable.
Owner:BEIJING STOMATOLOGY HOSPITAL CAPITAL MEDICAL UNIV
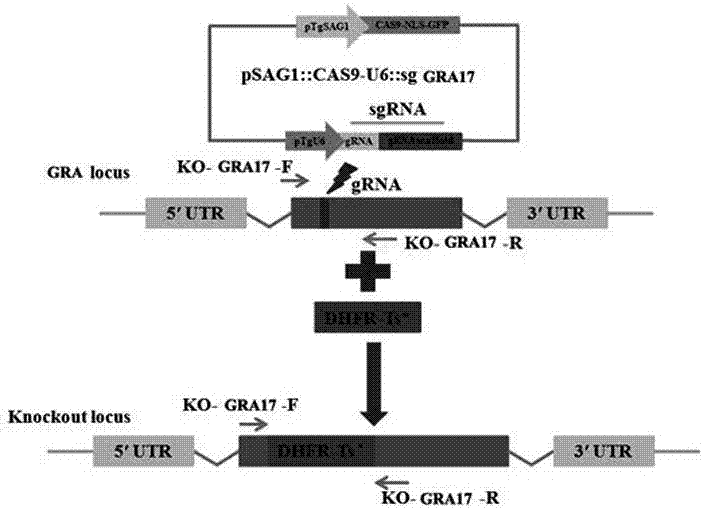
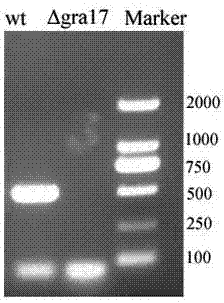
GRA17 gene deleted toxoplasma gondii attenuative mutant strain and preparation method thereof
The invention discloses a GRA17 gene deleted toxoplasma gondii attenuative mutant strain. The GRA17 gene deleted toxoplasma gondii attenuative mutant strain is LZdeltaGRA17, which is preserved in China Center for Type Culture Collection with a preservation number of CCTCC NO:V201656. The invention also provides a preparation method of the strain. According to the invention, the GRA17 gene deleted toxoplasma gondii attenuative mutant strain provided by the invention is established on a molecular biology basis, and is a gene deleted insect strain constructed based on the CRISPR / Cas9 gene knockout technology, the insect strain has attenuated toxicity, also loses the biological characteristic of lethality to mice, has enormous application value, and can be used as a candidate strain of attenuated live vaccines.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
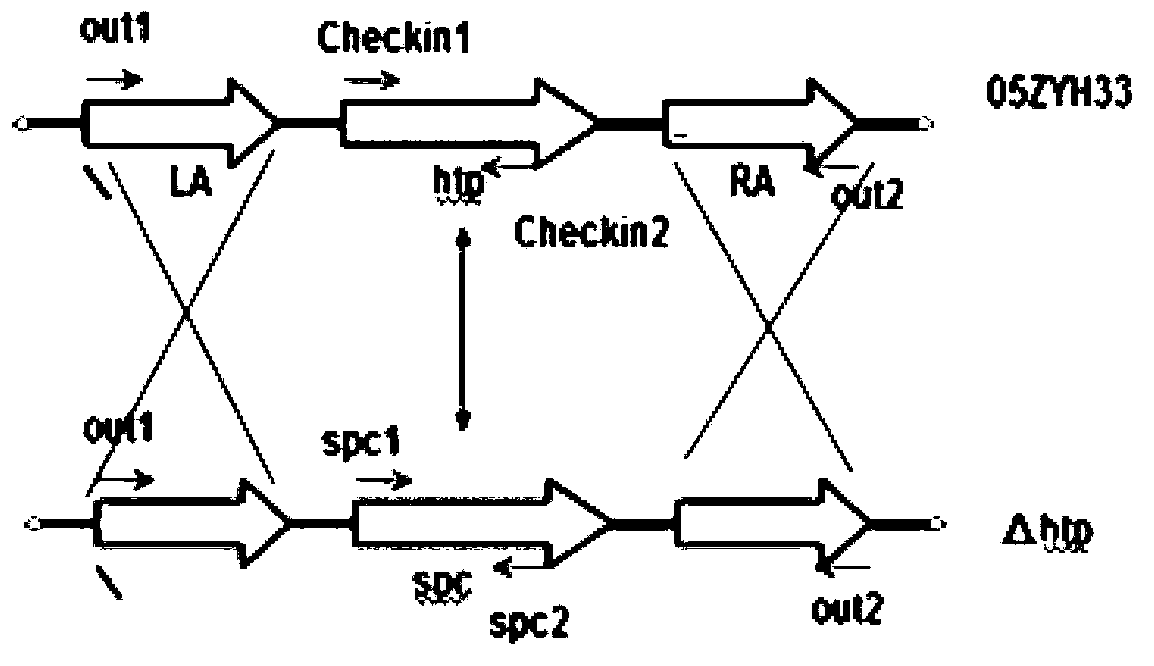
HtpsA-gene-knock-out mutant strain of Streptococcus suis serotype 2 and application thereof
InactiveCN103352015AConvenient lengthLow toxicityAntibacterial agentsBacterial antigen ingredientsBiotechnologyVirulent characteristics
The invention relates to an htpsA-gene-knock-out mutant strain of Streptococcus suis serotype 2 and application thereof, belonging to the field of genetic engineering. The mutant strain is 05ZYH33 delta htpsA with an accession number of CGMCC No. 7375. A preparation method for the mutant strain comprises the following steps: carrying out gene knockout on an htpsA gene by using a homologous recombination gene knockout technology, wherein the htpsA gene is located on chromosome 05ZYH33 of a wild S. suis 2 strain and codes histidine triad protein Htp; and determining that an obtained strain is a gene-knocked-out mutant strain through combined PCR product electrophoresis, RT-PCR and DNA sequencing identification and naming the obtained gene as 05ZYH33 delta htpsA. Pathogenicity of the mutant strain provided by the invention is researched through animal experiments, and results show that virulence of the mutant strain to a tested animal is substantially reduced; so the mutant strain can be used for developing an attenuated vaccine of Streptococcus suis serotype 2.
Owner:中国人民解放军南京军区军事医学研究所
Streptococcus suis serotype II htpsC gene knockout mutant strain and application thereof
InactiveCN103343102AReduce adhesionEnhanced phagocytosisAntibacterial agentsBacterial antigen ingredientsBiotechnologyStreptococcus suis serotype
The invention belongs the field of gene engineering, and relates to a streptococcus suis serotype II htpsC gene knockout mutant strain and an application thereof. The mutant strain 05ZYH33deltahtpsC has the preservation number of CGMCC (China General Microbiological Culture Collection Center) NO.7376. A preparation method of the mutant strain comprises the following step of: carrying out gene knockout on an htpsC gene of coded histidine tripolymer protein Htp of a chromosome of a S.suis II field strain 05ZYH33 by adopting a homologous recombinant gene knockout technology so as to obtain the strain, namely the gene knockout mutant strain through verification of combined PCR (polymerase chain reaction) product electrophoresis, RT (reverse transcription)-PCR and DNA (Deoxyribonucleic Acid) sequencing. By using the mutant strain to perform an animal experiment to research pathogenicity of the strain, results show that the toxicity of the mutant strain to the experiment animal is remarkably reduced, and the mutant strain can be applicable to developing a streptococcus suis serotype II attenuated vaccine.
Owner:中国人民解放军南京军区军事医学研究所
Pseudomonas plecoglossicida ExoU gene knockout mutant strain and application thereof
InactiveCN106047783AEffective prevention and controlReduced toxicityAntibacterial agentsBacteriaPseudocardium sachalinenseGenetic engineering
The invention belongs to the field of genetic engineering, and relates to a pseudomonas plecoglossicida ExoU gene knockout mutant strain and an application thereof. The mutant strain NB2011[delta]ExoU is preserved with preservation number of CGMCC No.12430; and a preparation method of the mutant strain comprises the following steps: conducting gene knocking-out on the ExoU gene of the pseudomonas plecoglossicida wild strain NB2011 by virtue of a homologous recombinant gene knockout technology, and conducting screening and identification by virtue of a PCR (polymerase chain reaction) technology, so that an obtained strain is determined as the gene knockout mutant strain. By researching pathogenicity of the mutant strain disclosed by the invention through animal experiments, results show that the toxicity of the mutant strain on experimental animals is significantly reduced; therefore, the mutant strain can be applicable to pseudomonas plecoglossicida hypo-toxic vaccines.
Owner:ZHEJIANG WANLI UNIV
Rice lesion mimic gene and encoded protein thereof
InactiveCN106243208AGood resistance to rice bacterial blightEasy to identifyPlant peptidesFermentationBiotechnologyNucleotide
The invention discloses a protein for controlling rice lesion mimic characters. The protein is formed by an amino acid sequence as shown by SEQ ID NO. 2 in a sequence table or a derivative sequence thereof. The invention further discloses a gene for encoding the protein. The gene is formed by a nucleotide sequence as show by SEQ ID NO. 1 in the sequence table. The lesion mimic gene provided by the invention has a good effect on resistance to rice bacterial blight, can be used for improving the bacterial blight resistance of nonglutinous rice, and provides a new way for breeding the nonglutinous rice capable of resisting bacterial blight; secondly, a new gene knockout technology is applied, a lesion mimic material under the background of japonica rice is acquired successfully, and a new method for utilizing the gene to culture japonica rice variety capable of resisting bacterial blight is provided. In addition, the gene provided by the invention is controlled by a single recessive gene, a transgene or filial generation can be simply identified and selected, and the convenience in application is realized.
Owner:SICHUAN AGRI UNIV
PROM1-KO mouse model construction method and application thereof
ActiveCN110257435AHigh knockout efficiencyNo distractionHydrolasesStable introduction of DNADiseaseKnockout animal
The invention relates to a PROM1-KO mouse model construction method and application thereof. A mouse model is a PROM1 gene knockout mouse. A PROM1 gene knockout mouse model is constructed based on CRISPR / Cas9 gene knockout technology. The construction method comprises the following steps: step one, designing sgRNA and Cas9mRNA, transcribing sgRNA and Cas9mRNA into mRNA in vitro, and microinjecting the active sgRNA and the active Cas9mRNA into mouse fertilized eggs to obtain PROM1-KO mice; and step two, identifying a PROM1-KO mouse animal model. The PROM1-KO mouse animal model is firstly constructed based on the CRISPR / Cas9 gene knockout technology, and a convenient, reliable and economical animal model is provided for researching the relationship between PROM1 and diseases such as hereditary retinopathy and tumors.
Owner:上海朗昇生物科技有限公司
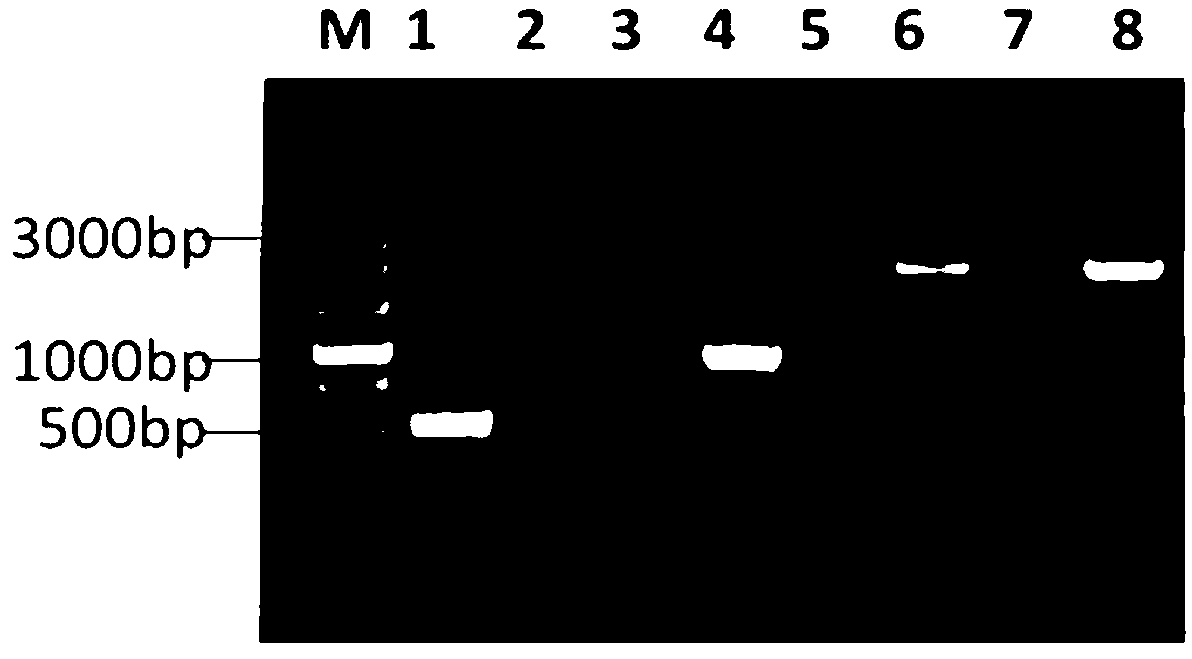
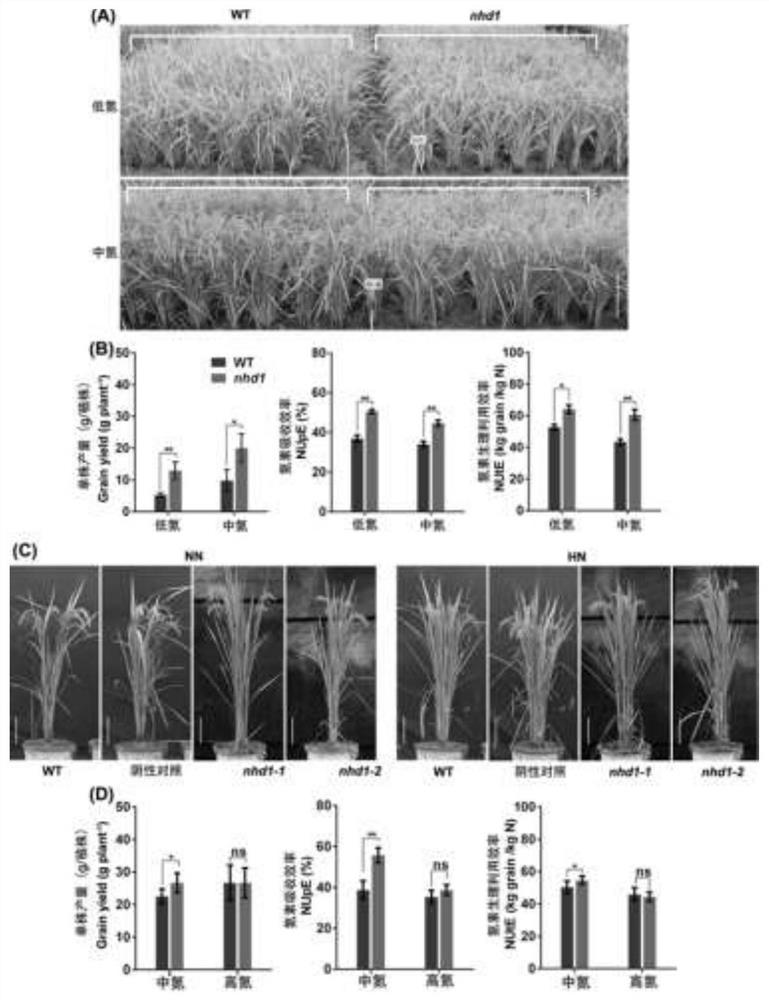
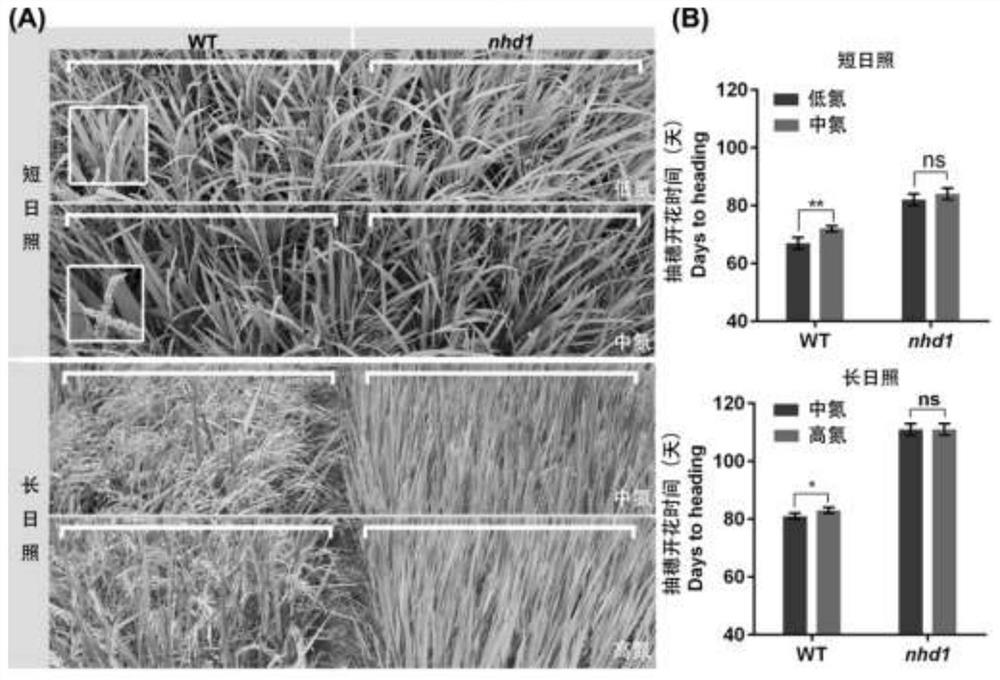
Application of photoperiod gene OsNhd1
ActiveCN113430208AProlong reproductive periodIncrease productionPlant peptidesFermentationBiotechnologyNitrogen assimilation
The invention discloses an application of a photoperiod gene OsNhd1. Rice (oryza sativa) photoperiod gene OsNhd1 is applied to regulation and control of the growth period, the yield and the nitrogen utilization efficiency of rice; and the rice photoperiod gene OsNhd1 is numbered as Os8g0157600 in an RAP-DB (Rice Annotation Project Database). According to the application, an OsNhd1 mutant plant is obtained through a gene knockout technology. Field tests and analysis prove that the mutant plant has the characteristics of prolonged growth period, increased yield and increased nitrogen utilization efficiency. The application proves that OsNhd1 not only directly regulates heading and flowering of rice, but also participates in nitrogen assimilation and transport processes of rice, which indicates that the gene plays an important role in coordinated regulation of growth period and nitrogen utilization efficiency, and establishes a theoretical basis for improvement of rice varieties in the future.
Owner:NANJING AGRICULTURAL UNIVERSITY
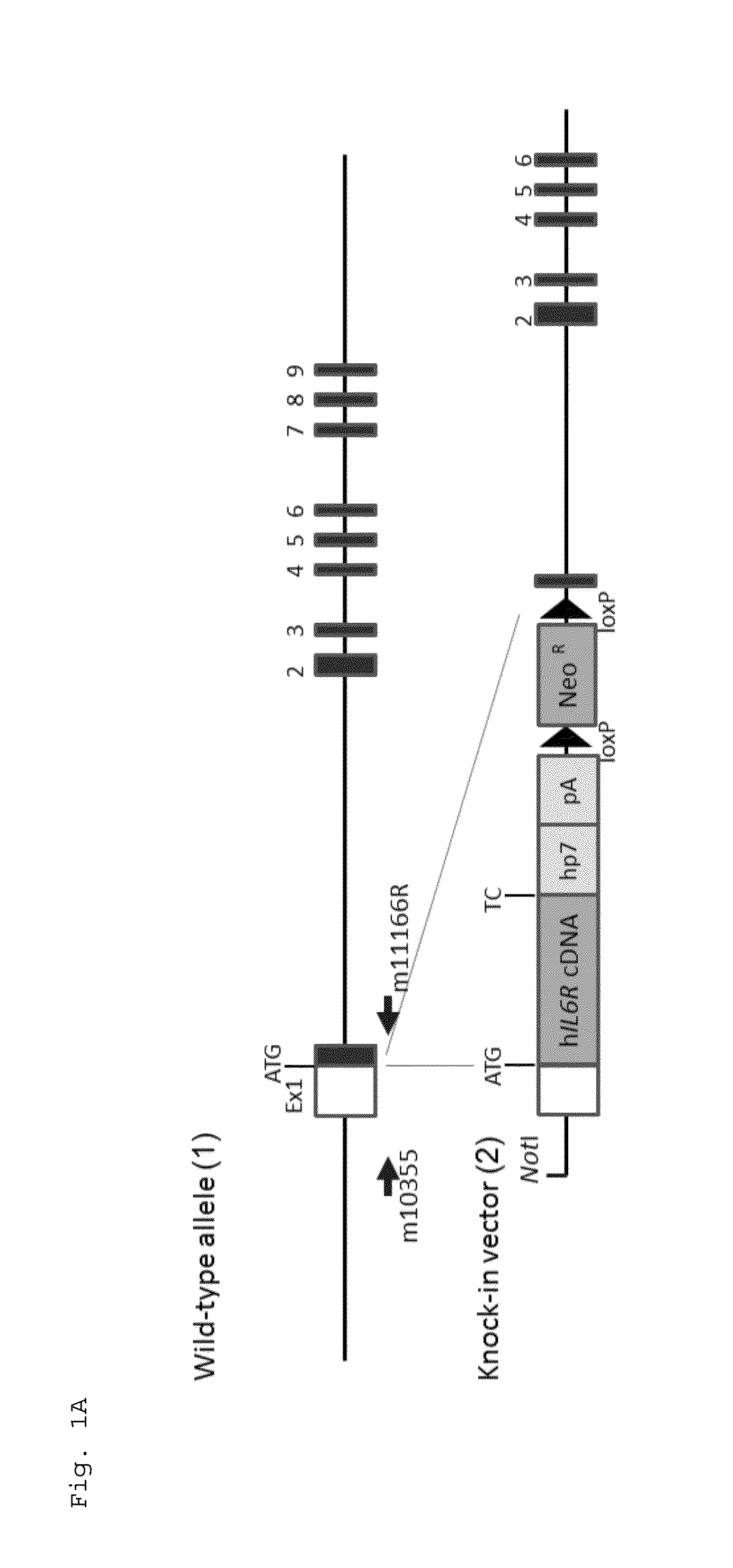
Gene knock-in non-human animal
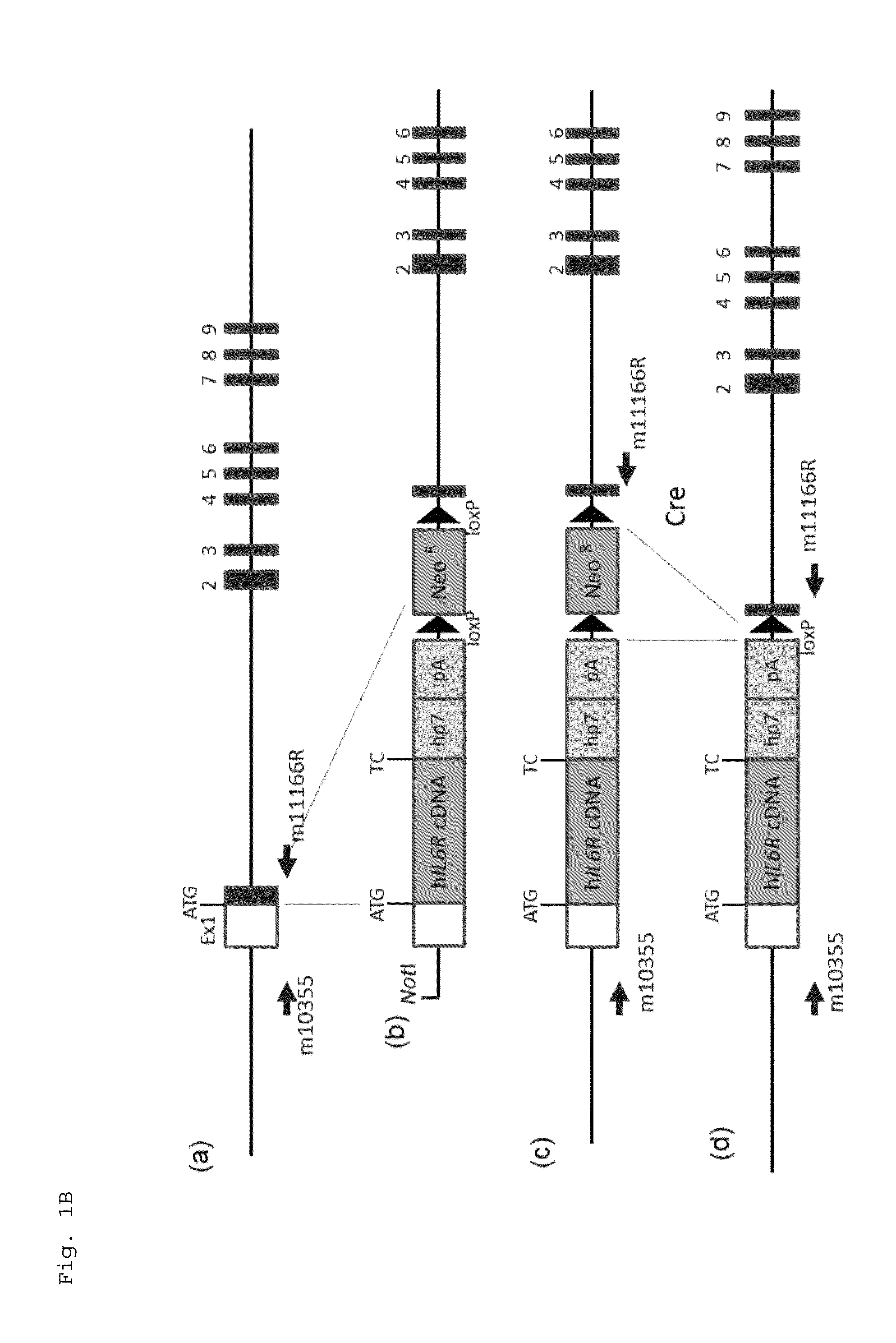
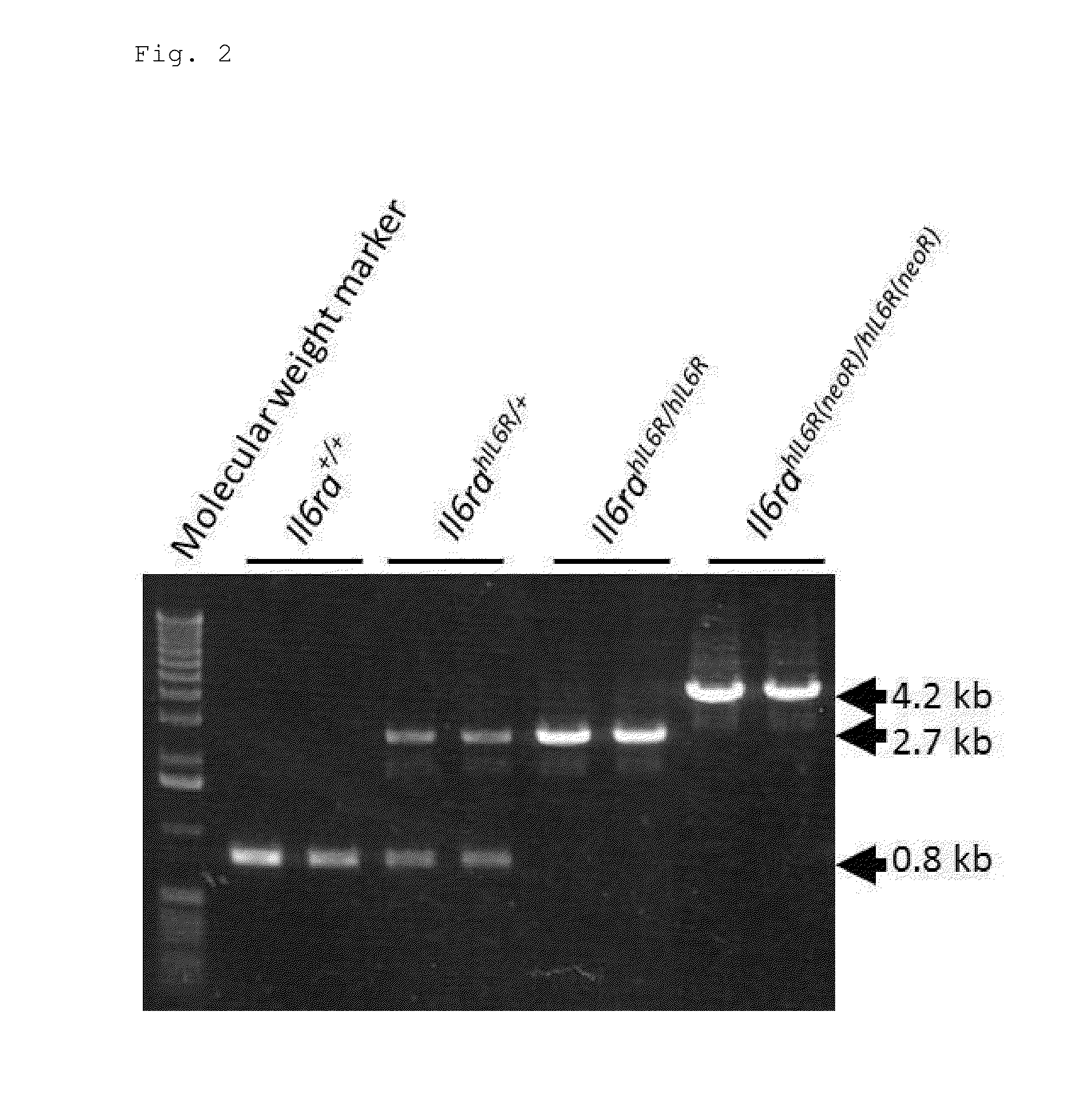
ActiveUS20150225481A1Suppressing expression of its endogenous geneAccurate assessmentCompounds screening/testingAntipyreticHuman animalA-DNA
The present invention provides a non-human animal in which a DNA comprising an hp7 sequence-encoding DNA and a poly A addition signal-encoding DNA added on the 3′ side of a DNA encoding an arbitrary foreign gene is inserted in the same reading frame as that of an arbitrary target gene present on the genome of the non-human animal.
Owner:CHUGAI PHARMA CO LTD
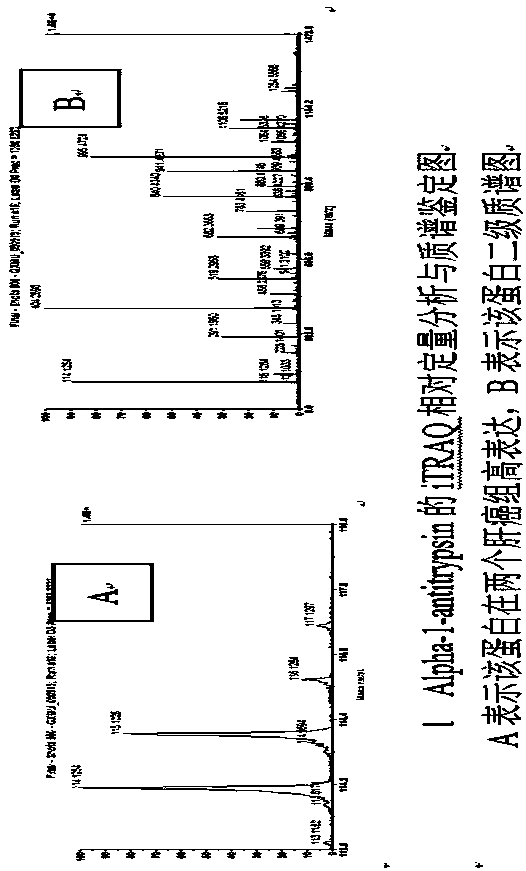
Method used for discovering and identifying liver cancer serum differential expression proteins and verifying marker proteins
InactiveCN107894507APropose feasibilityDisease diagnosisProtein markersCandidate Gene Association Study
The invention discloses a method used for discovering and identifying liver cancer serum differential expression proteins and verifying marker proteins. According to the method, iTRAQ labeling MALDI-TOF MS / MS technology is adopted for discovering and identifying liver cancer serum differential expression proteins and MRM verifying of marker proteins. According to the method, searching on effectivetumor markers possesses important meaning on canceration mechanism study, disease early stage diagnosis, and prognosis. Selected novel liver cancer protein markers are subjected to in vivo and in vitro experiments such as gene knockin and knockout, and nude mice transplanted cancer, influences of blocking or up-regulating of expression of candidate genes on tumor biological behavior are observed,and key proteins capable of influencing human liver cancer occurrence and development are positioned, the effects of the key proteins in a system signal network is analyzed based on bioinformatics analysis, and the feasibility, the liver cancer diagnosis value, and the clinical significance of the key proteins in clinical applications including liver cancer prevention, early stage diagnosis, andprognosis is evaluated based on MRM absolute quantitative verification.
Owner:南宁科城汇信息科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com