Method used for discovering and identifying liver cancer serum differential expression proteins and verifying marker proteins
A technique for differentially expressed proteins and differentially expressed proteins, which is applied in the field of discovery and identification of differentially expressed proteins in liver cancer serum and verification marker proteins, which can solve the problems that the precision and accuracy cannot meet the analysis requirements and affect the quantitative detection of low-abundance proteins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0021] Consists of the following steps:
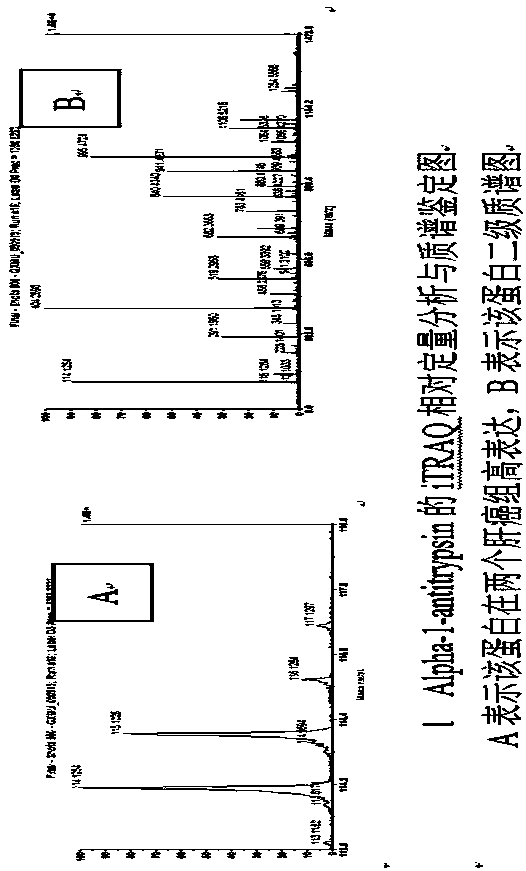
[0022] Using iTRAQ labeling combined with MALDI-TOF MS / MS technology to discover and identify differentially expressed proteins in liver cancer serum, MRM verification marker proteins: using iTRAQ technology to analyze the serum of 30 cases of liver cancer and 30 cases of normal controls, a total of 304 proteins were detected (Confidence >95%), 38 differential proteins were found (Pfigure 1 It shows that one of the proteins Alpha-1-antitrypsin is highly expressed in liver cancer; from the 38 differential proteins found from the above iTRAQ markers, MIDAS (MRM Initiated Detection And Sequencing, MIDAS) technology was used to complete the MRM qualitative verification of 14 differential proteins; The standard method completed the MRM quantitative verification of three proteins, which not only fully proved the existence of these 14 differential proteins in the mixed sample, but also obtained the quantitative information of the three protein...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com