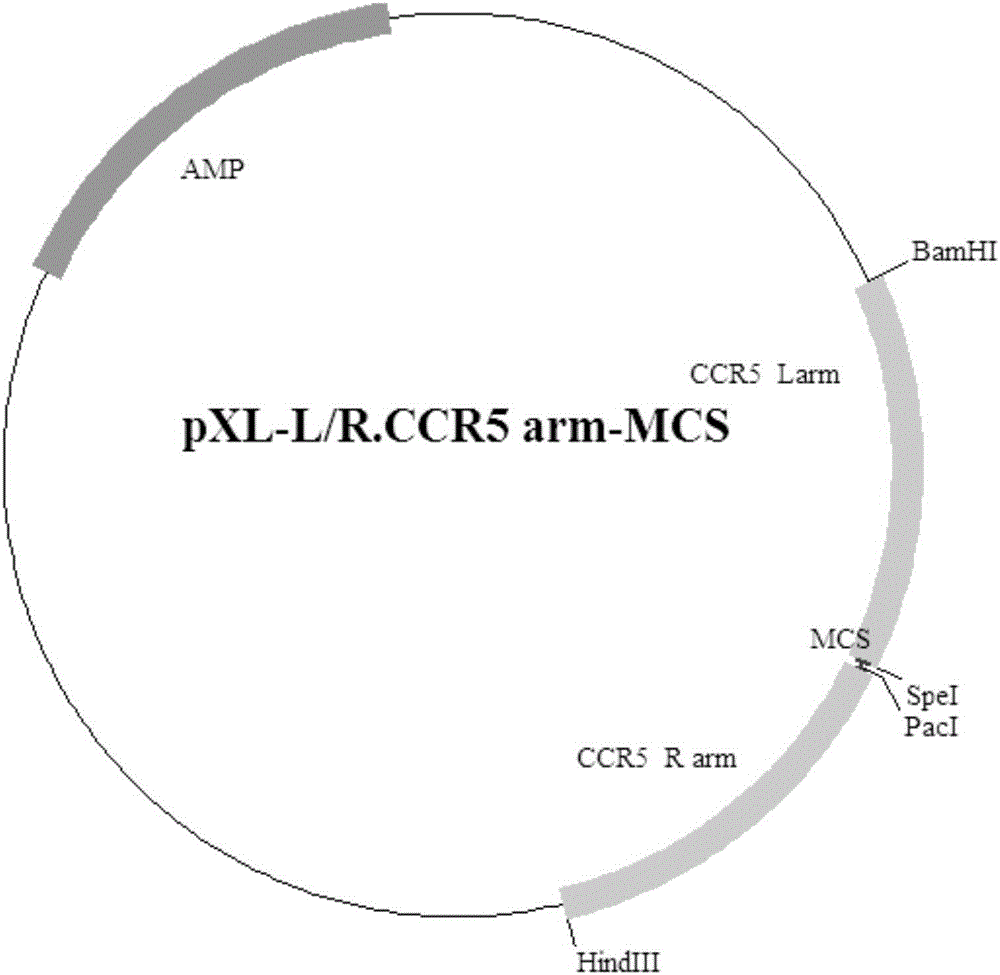
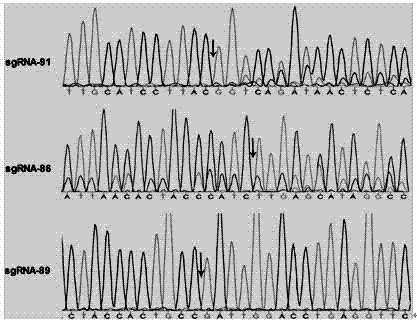
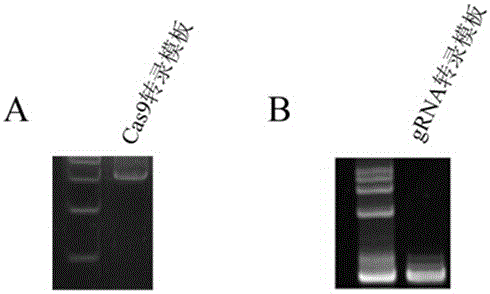
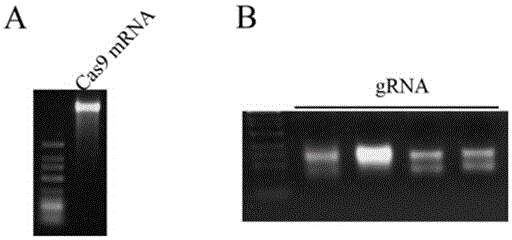
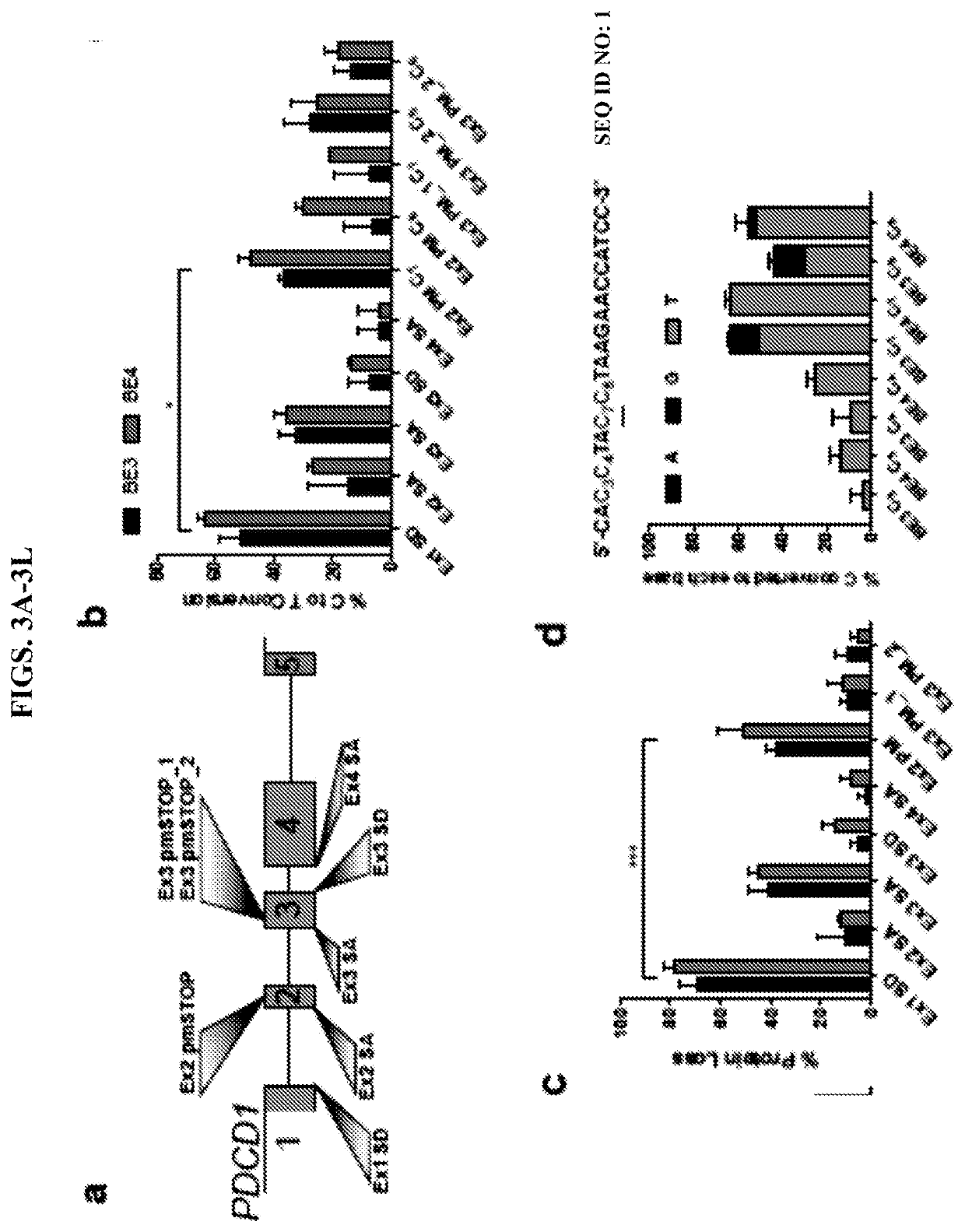
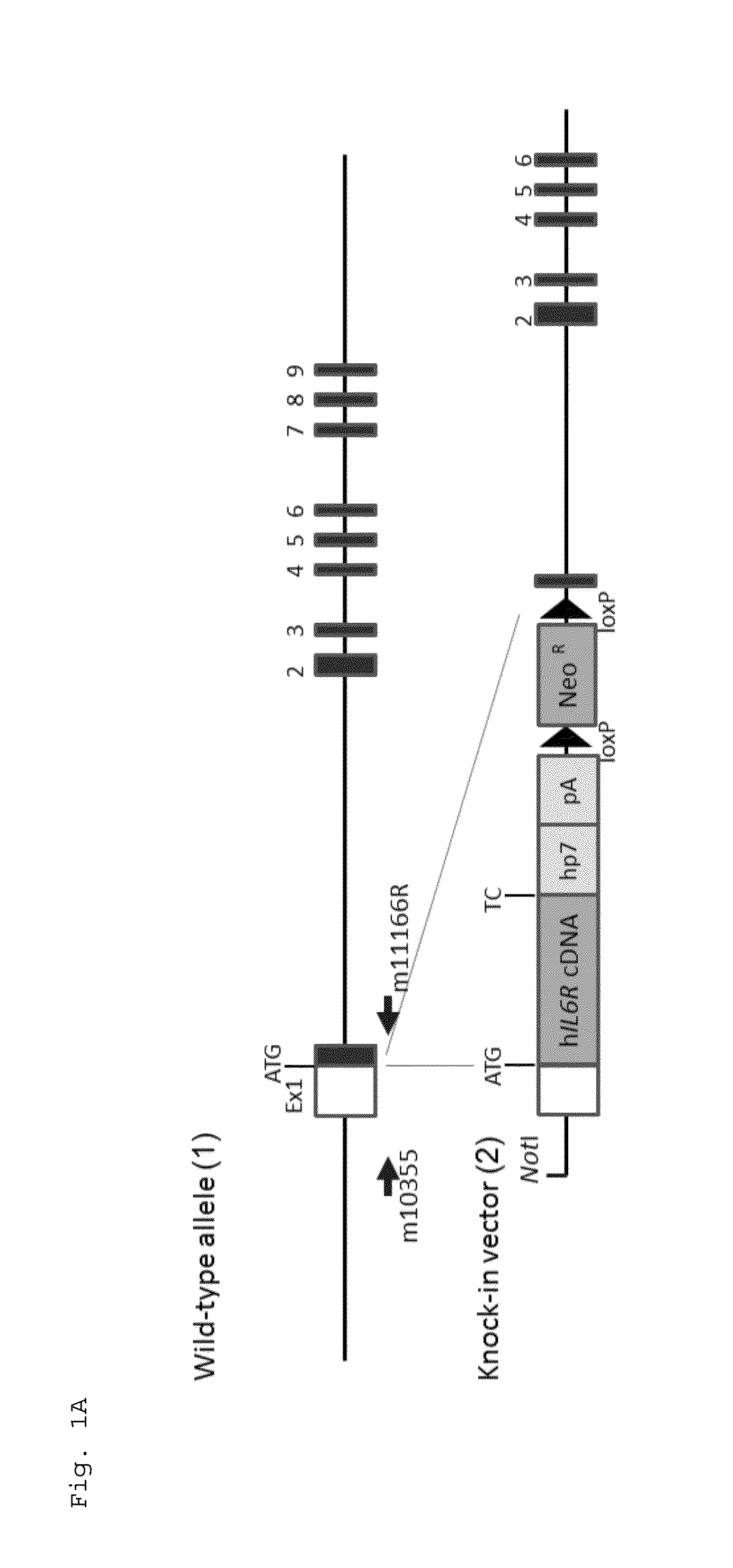
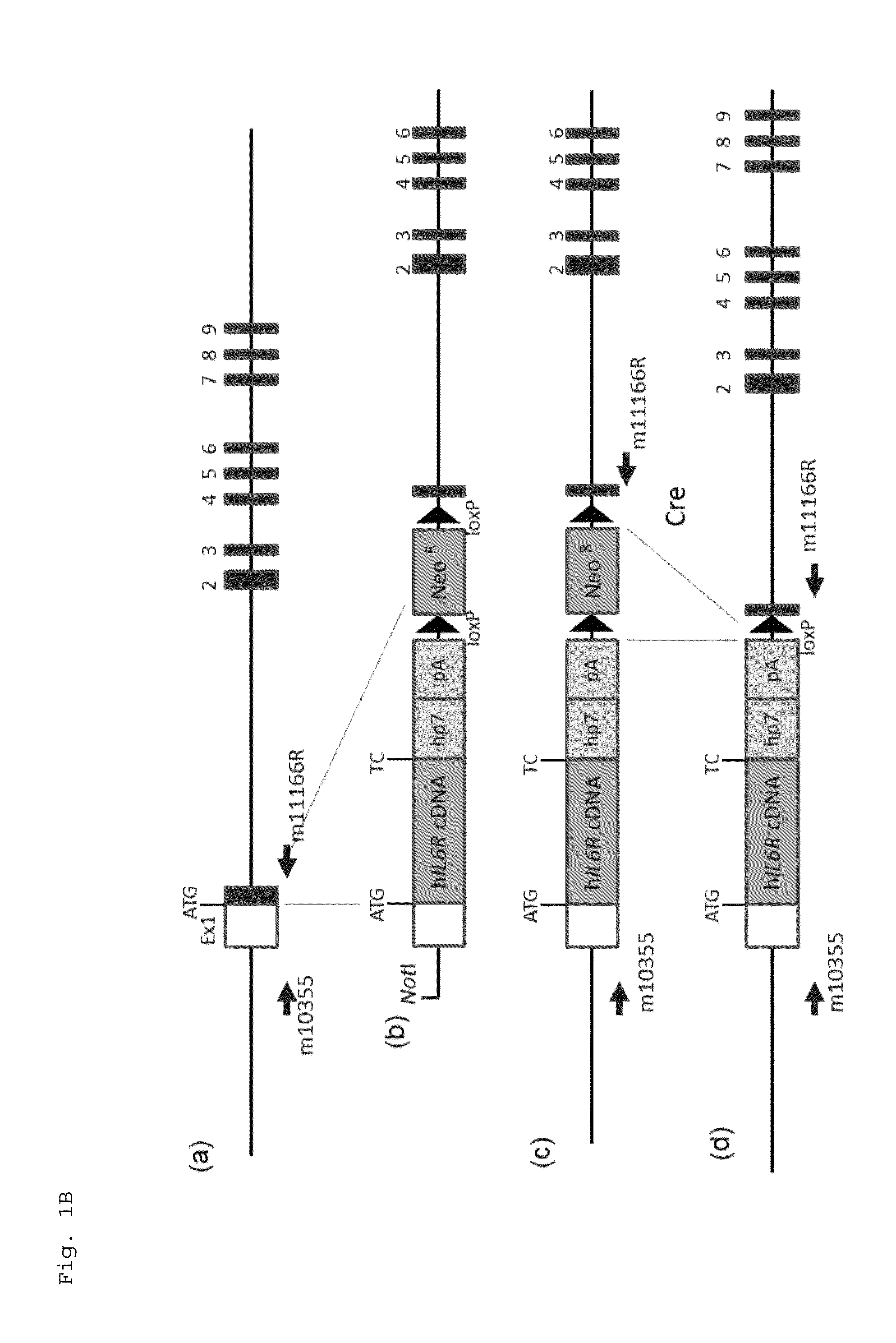
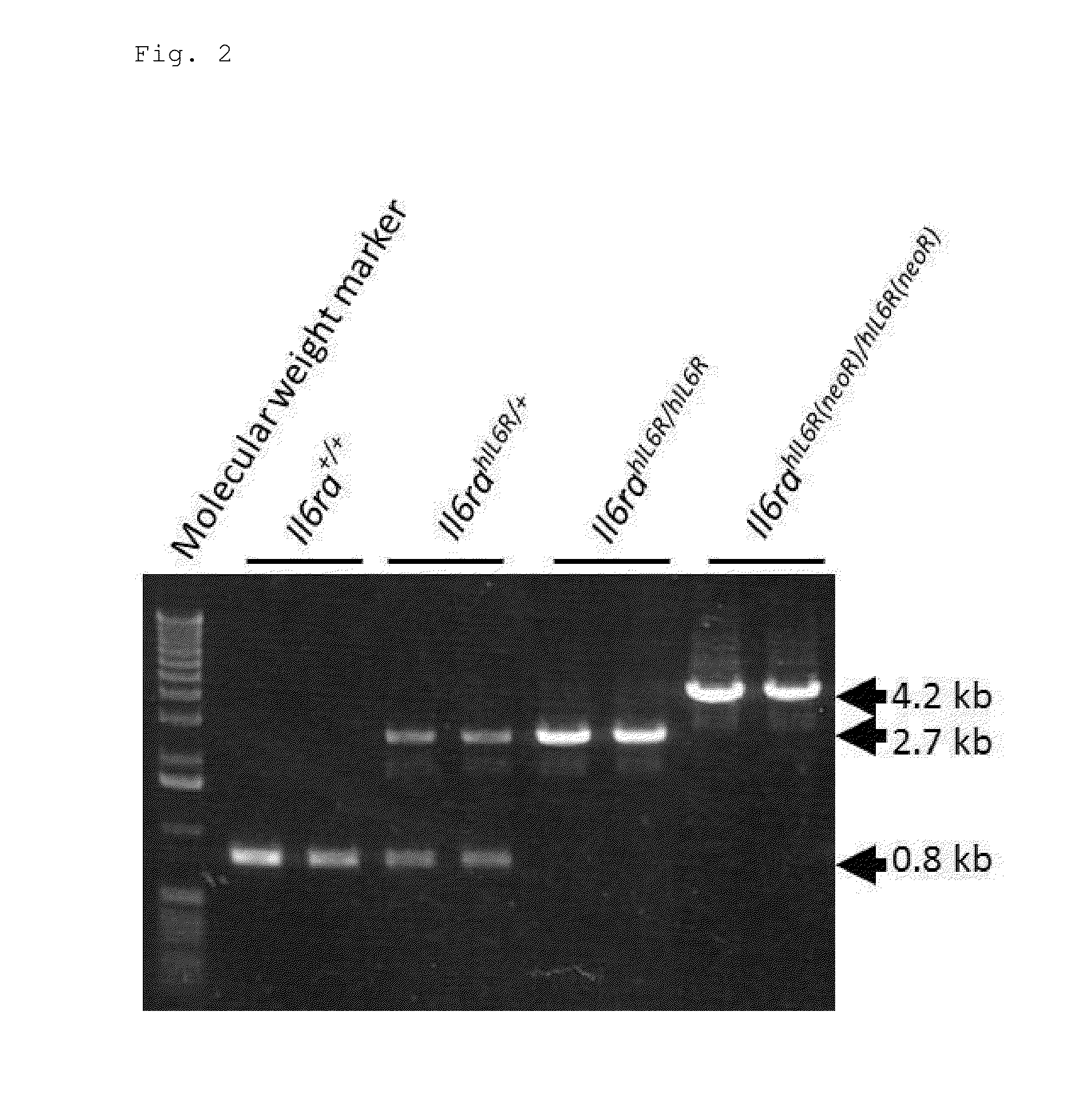
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
87 results about "Gene knockin" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In molecular cloning and biology, a knock-in (or gene knock-in) refers to a genetic engineering method that involves the one-for-one substitution of DNA sequence information in a genetic locus or the insertion of sequence information not found within the locus. Typically, this is done in mice since the technology for this process is more refined and there is a high degree of shared sequence complexity between mice and humans. The difference between knock-in technology and traditional transgenic techniques is that a knock-in involves a gene inserted into a specific locus, and is thus a "targeted" insertion.
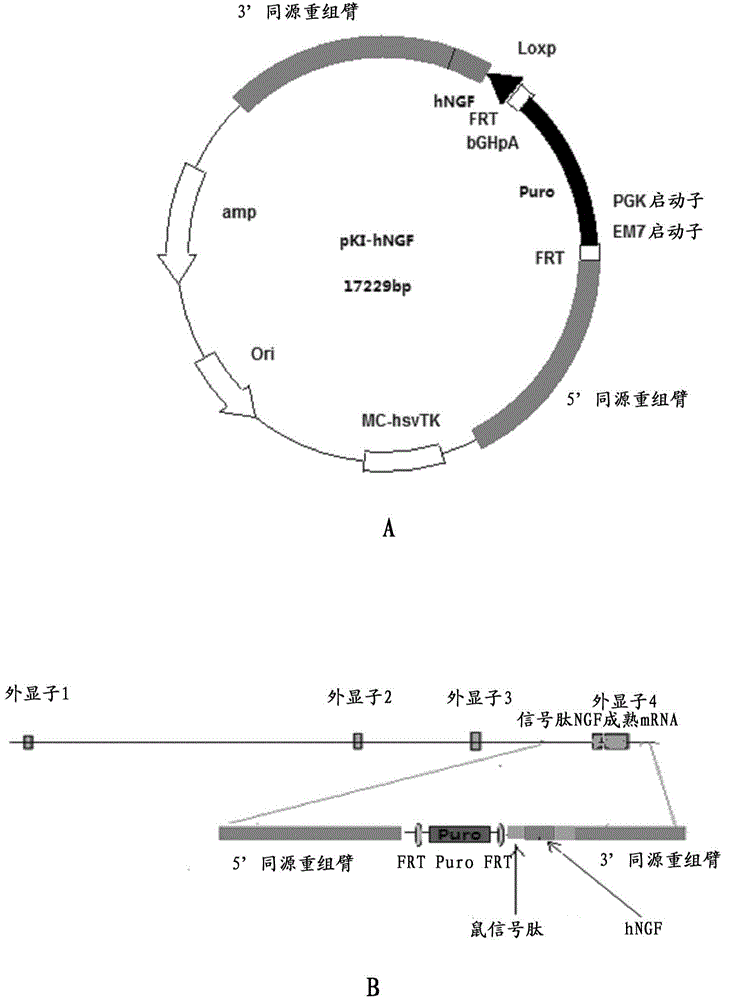
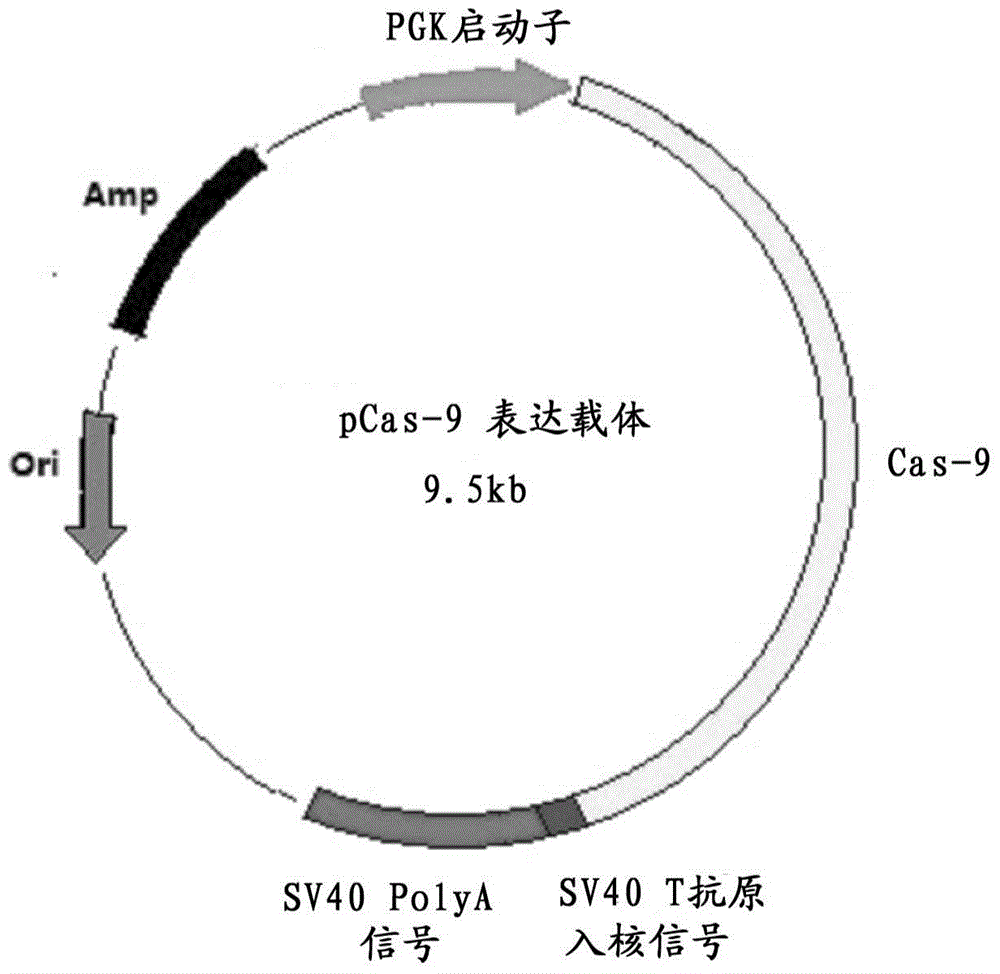
Preparation method for transgenic mice capable of producing human nerve growth factor
ActiveCN104561095AEasy to operateIncrease success rateAnimals/human peptidesVector-based foreign material introductionPlasmid dnaGenetically modified mouse
The invention discloses a method for producing transgenic mice through a homologous recombination technology. The method comprises the following steps: replacing mouse NGF genes on mouse chromosomes by human NGF genes through a Cas-9 / CRISPR gene knock-in technology, and obtaining the genes with homozygous human NGF genes through breeding and knocking the genes in mice, thus obtaining filial generation mice with salivary glands capable of secreting human NGF. Compared with the conventional targeting technology utilizing positive and negative double screening homologous recombination genes, the method disclosed by the invention is simple to operate, capable of being realized only by transfecting mouse embryonic stem cells by three plasmid DNAs or carrying out pronucleus injection on the zygotes of mice, and high in success rate which is up to 2-5% and remarkably higher than the mouse embryonic stem cell positive rate of 0.1% of the common gene targeting technology.
Owner:深圳市国创纳米抗体技术有限公司
Exogenous gene knocking-in and integrating system on basis of CRISPR/Cas9, method for establishing exogenous gene knocking-in and integrating system and application thereof
ActiveCN106191116ANucleic acid vectorVector-based foreign material introductionTarget geneIntegrated systems
The invention provides an exogenous gene knocking-in and integrating system on the basis of CRISPR / Cas9, a method for establishing the exogenous gene knocking-in and integrating system and application thereof. The exogenous gene knocking-in and integrating system comprises vectors with report / donor functions and Cas9 expression vectors. Each report / donor vector comprises two target gent homologous arms and an exogenous sequence fragment positioned between the two target gene homologous arms; homologous sequences, which are positioned on a target gene, of the two target gene homologous arms of each report / donor vector are respectively positioned on two sides of a target sequence of the target gene and are connected with the target sequence of the target gene; the exogenous sequence fragments comprise promoters, resistant genes, shorn peptide sequences, report genes and polyA tails which are sequentially arrayed, two SSA repair homologous sequences of each resistant gene are inserted into the resistant gene, and the target sequence of each target gene is inserted in a space between the two corresponding SSA repair homologous sequences. The exogenous gene knocking-in and integrating system, the method and the application have the advantages that exogenous genes can be integrated with endogenous gene sequences in an efficient site-directed and accurately targeted manner, and double-chromosome allelic gene double-knocking-in can be efficiently carried out.
Owner:成都中科奥格生物科技有限公司
Single guide ribonucleic acid (sgRNA) capable of effectively editing pig ROSA26 gene, and application of sgRNA
ActiveCN106916820AImprove securityEfficient and stable expressionAnimal husbandryDNA/RNA fragmentationBiotechnologyFibroblast
The invention discloses single guide ribonucleic acid (sgRNA) capable of effectively editing a pig ROSA26 gene, and application of the sgRNA. Based on the sgRNA capable of specifically identifying the pig ROSA26 gene in a pig genome, a cell line of enhanced green fluorescent protein (EGFP) site-specific integrated porcine fetal fibroblasts is successfully established by using a CRISPR / Cas9-mediated gene knock-in technique; the results show that the cell line can stably and efficiently express an EGFP gene; furthermore, in the preparation process of the cell line, exogenous promoter genes and positive and negative screening marker genes are not introduced into the cell line. Therefore, the sgRNA greatly improves the safety of transgenic pigs, and has important significance for eliminating the biological and food potential safety hazards of agricultural products of the transgenic pigs.
Owner:重庆吉渝科技有限公司
Preparation method of staphylococcus aureus CRISPR/Cas9 system and application of system in constructing mouse model
The invention discloses a preparation method of a staphylococcus aureus CRISPR / Cas9 system and an application of the system in constructing a genetically modified mouse model. The staphylococcus aureus CRISPR / Cas9 system is composed of two components, namely Cas9 mRNA and gRNA, wherein a preparation method of the Cas9 mRNA is achieved by adding a T7 promoter to the upstream region of original Cas9 coding DNA, and a preparation method of the gRNA is achieved by adding the T7 promoter to the upstream region of an original gRNA coding sequence. The staphylococcus aureus Cas9 mRNA and gRNA, which are injected to mouse fertilized embryos through micro-injection, can achieve gene editing and modification of various types, such as single-gene knockout, multi-gene knockout and / or gene knock-in and the like, on the mouse fertilized embryos; therefore, the CRISPR / Cas9 system has a good application prospect in the aspects of fertilized embryo gene editing and modification of such animals as mouse and the like as well as construction of animal models.
Owner:GUANGZHOU MAGIGEN BIOTECH
Development and applications of heat shock induced Cas9 enzyme transgene danio rerio
InactiveCN106434748AAchieve knockoutEnabling gene editing researchMicroinjection basedPeptidesWild typeTransgenic zebrafish
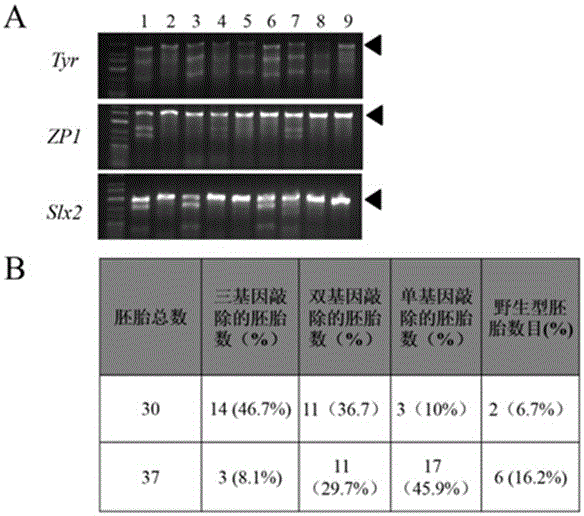
The present invention relates to the technical field of biology, particularly to development and applications of a heat shock induced Cas9 enzyme transgene danio rerio. The present invention firstly provides a Cas9 enzyme expression vector, which utilizes a heat shock induced promoter HSP70 to drive the expression of a downstream Cas9 gene. According to the present invention, the Cas9 enzyme expression vector and Tol2 mRNA are co-injected into wild type danio rerio single cell fertilized egg, and selection is performed to obtain the heat shock induced Cas9 enzyme transgene danio rerio, such that the gene editing research of the CRISPR-Cas9 system in the danio rerio is successfully achieved, and the know-out of the MC4R gene in the transgene danio rerio is firstly achieved; and the Cas9 enzyme expression vector is further suitable for heat shock induced gene knockout, gene knock-in, gene expression modification and other applications of other fishes.
Owner:CHONGQING INST OF GREEN & INTELLIGENT TECH CHINESE ACADEMY OF SCI
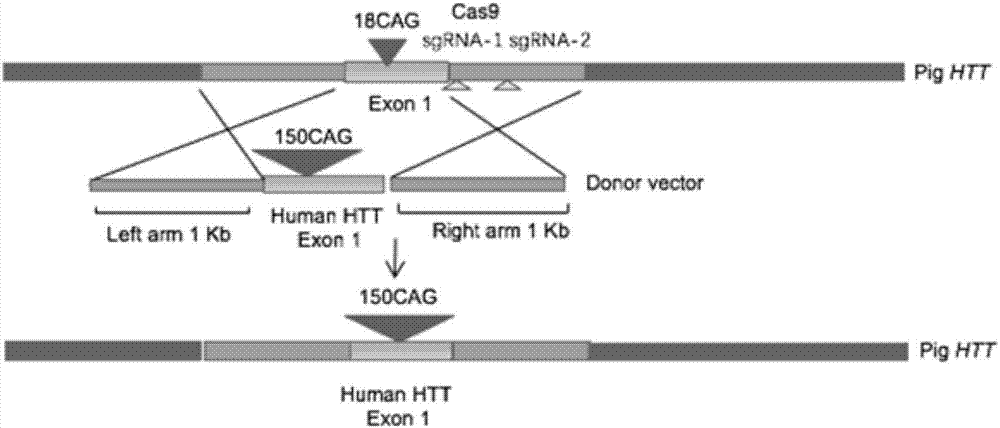
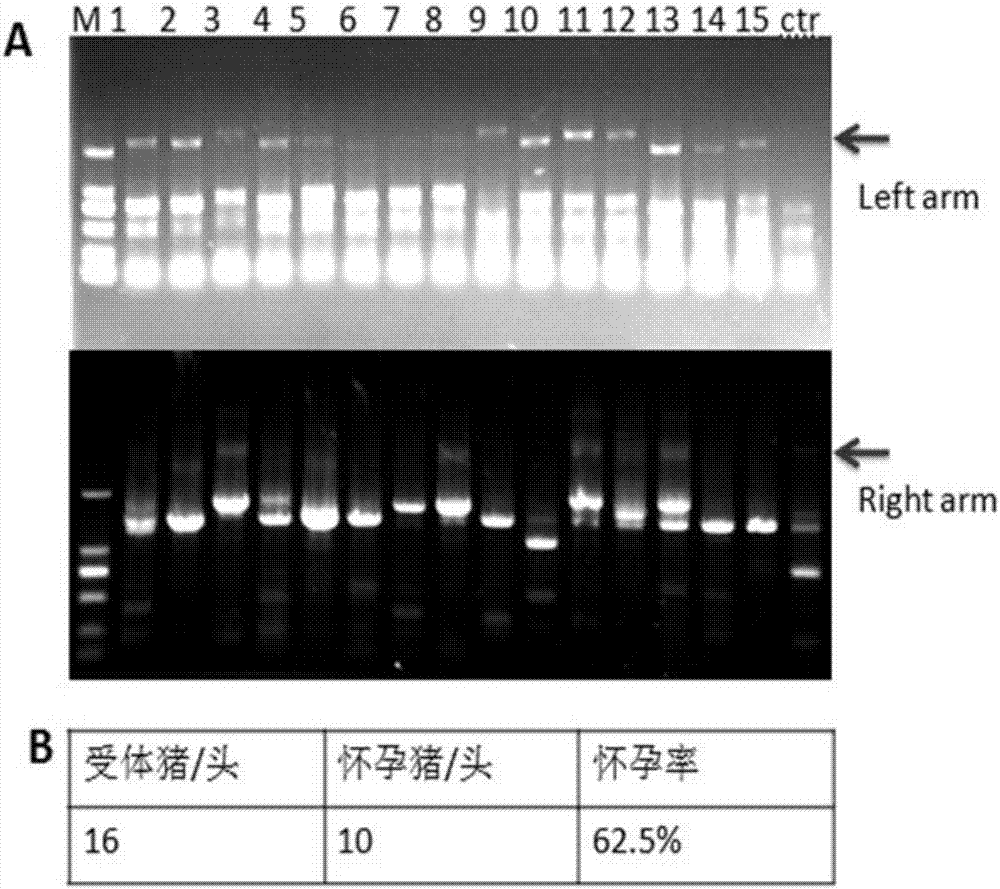
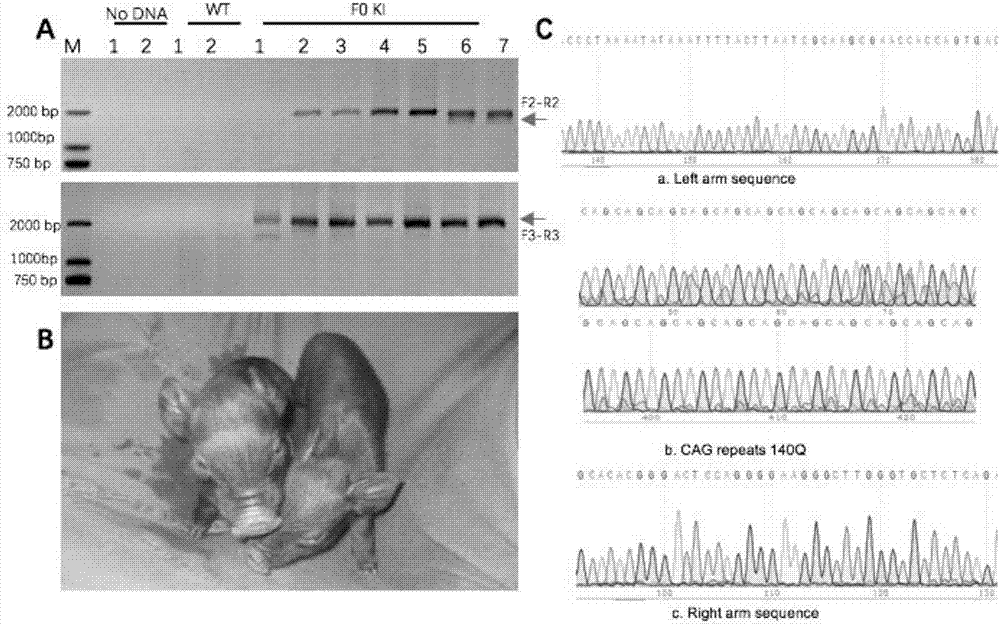
Recombinant vector for knock-in of human Huntington gene, construction method of recombinant vector and application of recombinant vector in construction of model pig
ActiveCN107988256AIncreased probability of knock-in positive clonesEfficient FeasibilityStable introduction of DNANucleic acid vectorHuman studiesExon
The invention discloses a recombinant vector for the knock-in of a human Huntington gene, a construction method of the recombinant vector and application of the recombinant vector in the constructionof a model pig. According to the recombinant vector, a human mutated Huntington exon gene is knocked in a fixed point manner for the first time, a virulence gene is knocked in by virtue of a CRISPR / Cas9 technique for the first time, and a donor vector is optimized by optimizing sfRNA, so that the probability that the gene is knocked into a positive cloned cell is increased; and by combining with apig cell nucleus transplantation technique, the probability that a directly obtained positive cell is knocked into the pig is increased, and a small human Huntington gene knock-in pig is obtained, thereby proving the efficient feasibility of the method for constructing gene modified pigs. The constructed Huntington gene knock-in model pig has behavioral characteristics such as respiratory disturbance and dyskinesia similar to human Huntington diseases, and stable heritable passage can be realized, so that a reliable model is provided for the research of the human Huntington diseases; and thenumber can be guaranteed so as to realize drug screening, gene treatment, stem cell treatment and the like, and the model can be a good human disease model.
Owner:JINAN UNIVERSITY +1
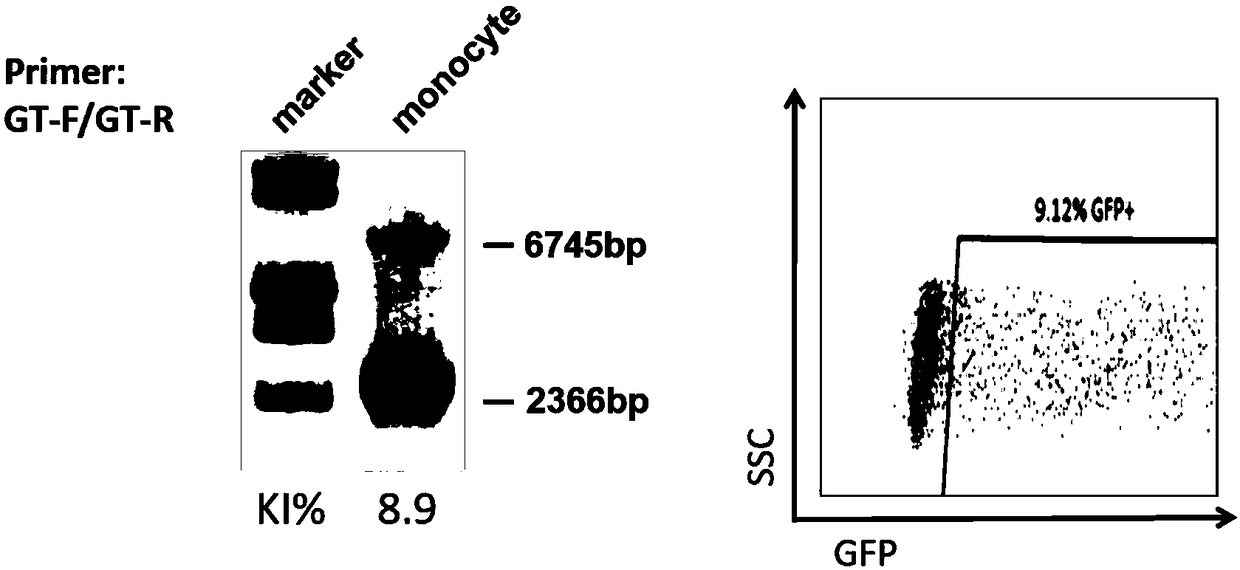
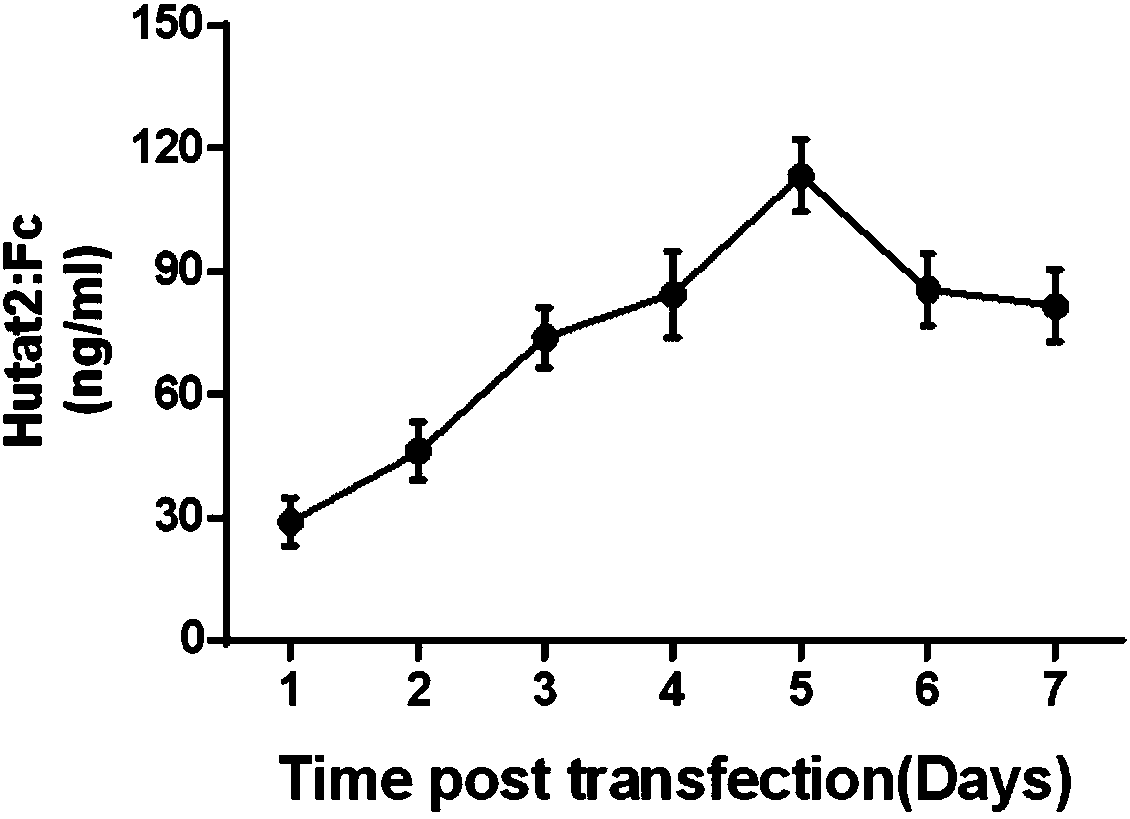
Experimental method for constructing Hutat2:Fc gene knock-in monocyte by using CRISPR/Cas9 technology
ActiveCN108559730ASimple methodImprove editing efficiencyGenetically modified cellsStable introduction of DNAExperimental methodsElectroporation
The invention relates to an experimental method for constructing a Hutat2:Fc gene knock-in monocyte by using a CRISPR / Cas9 technology. The experimental method is characterized by comprising the following steps of (1) CRISPR / Cas9 targeting plasmid construction; (2) donor plasmid construction; (3) primary generation monocyte sorting; (4) monocyte transfection through electroporation; and (5) knock-in efficiency identification. The experimental method for constructing the Hutat2:Fc gene knock-in monocyte by using the CRISPR / Cas9 technology has the advantages that the method is simple and implemented easily, subsequent cell screening is not needed, the higher gene editing efficiency can be obtained, the feasibility and application prospect of the experiment are greatly increased, and the application range is wide.
Owner:FOURTH MILITARY MEDICAL UNIVERSITY
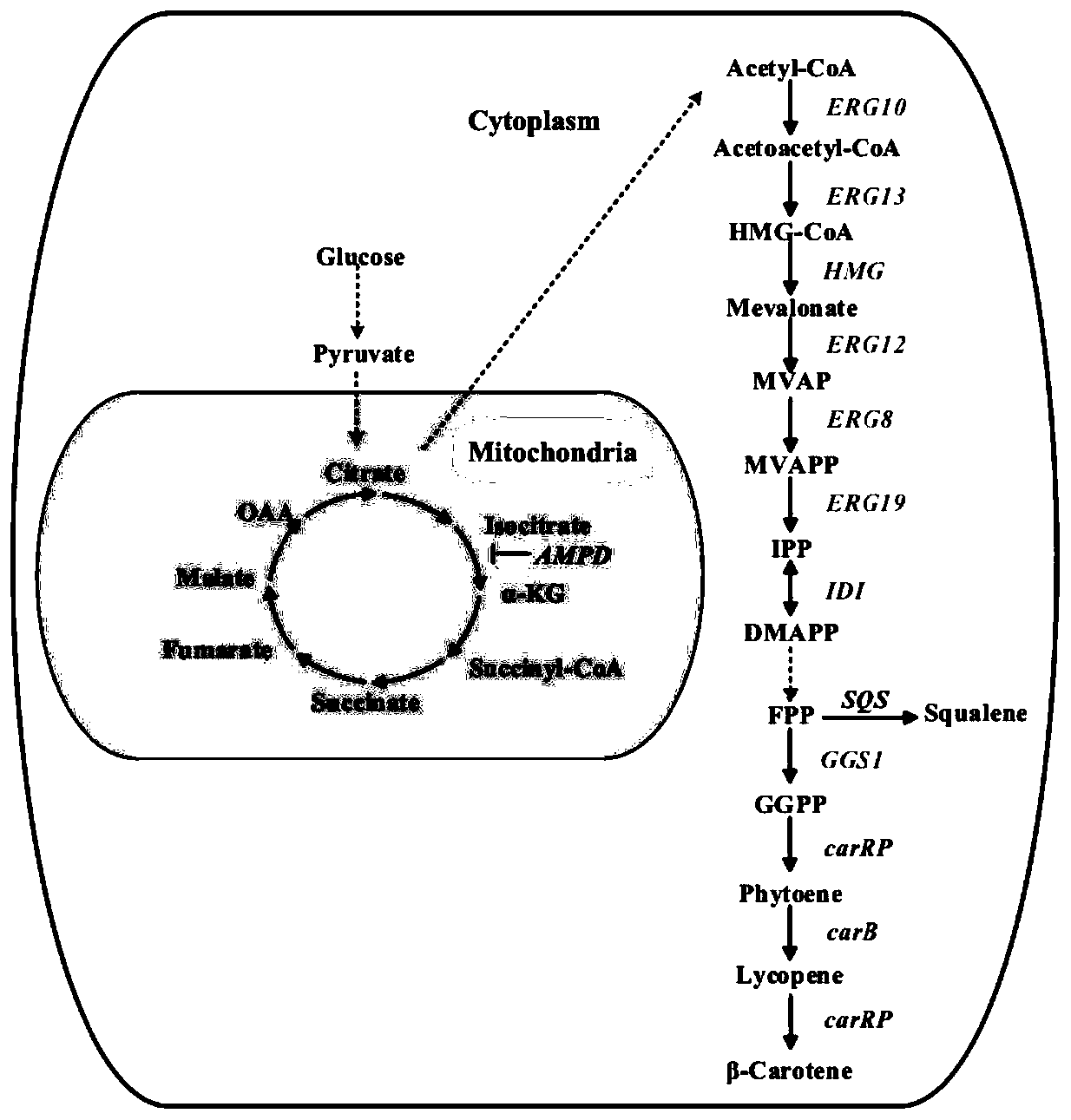
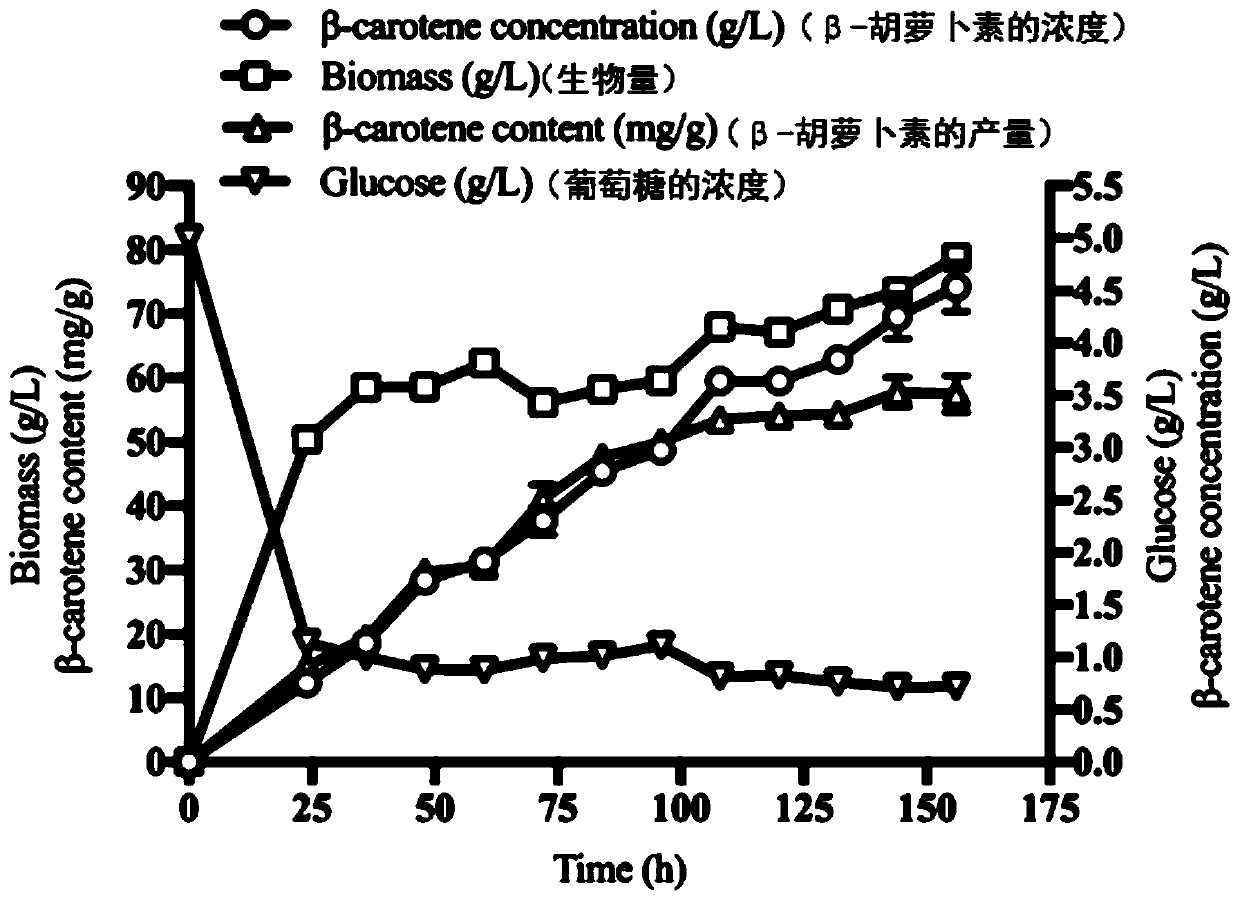
Yarrowia lipolytica genetic engineering bacterium for producing beta-carotene and application of yarrowia lipolytica genetic engineering bacterium
InactiveCN111321087AEasy to buildIncrease productionFungiStable introduction of DNAUracilMicrobiology
The invention provides a yarrowia lipolytica genetic engineering bacterium for producing beta-carotene. The yarrowia lipolytica genetic engineering bacterium for producing beta-carotene is prepared bythe following steps: knocking a carRP gene and a carB gene into chromosomes of uracil and leucine auxotrophic yarrowia lipolytica, then knocking a GGGS1 gene, a HMG gene, two copied carRP gene, one copied of carB gene, and an ERG13 gene into the chromosomes, and finally performing backfilling by two auxotrophic screening markers of uracil and leucine. Continuous feeding fermentation is performed,and the yield of beta-carotene can reach 4.5g / L. The yarrowia lipolytica genetic engineering bacteria constructed according to the invention has the advantages of simple and convenient operation andstable and reliable performance, and can be applied to large-scale commercial production, and the obtained beta-carotene can be safely used, so that a good prospect is achieved.
Owner:EAST CHINA UNIV OF SCI & TECH
Gene knock-in recombinant vector and preparation method thereof as well as method for preparing mouse model
ActiveCN103642828AShorten the timeHigh detection sensitivityMicrobiological testing/measurementVector-based foreign material introductionWhite blood cellFluorescence microscope
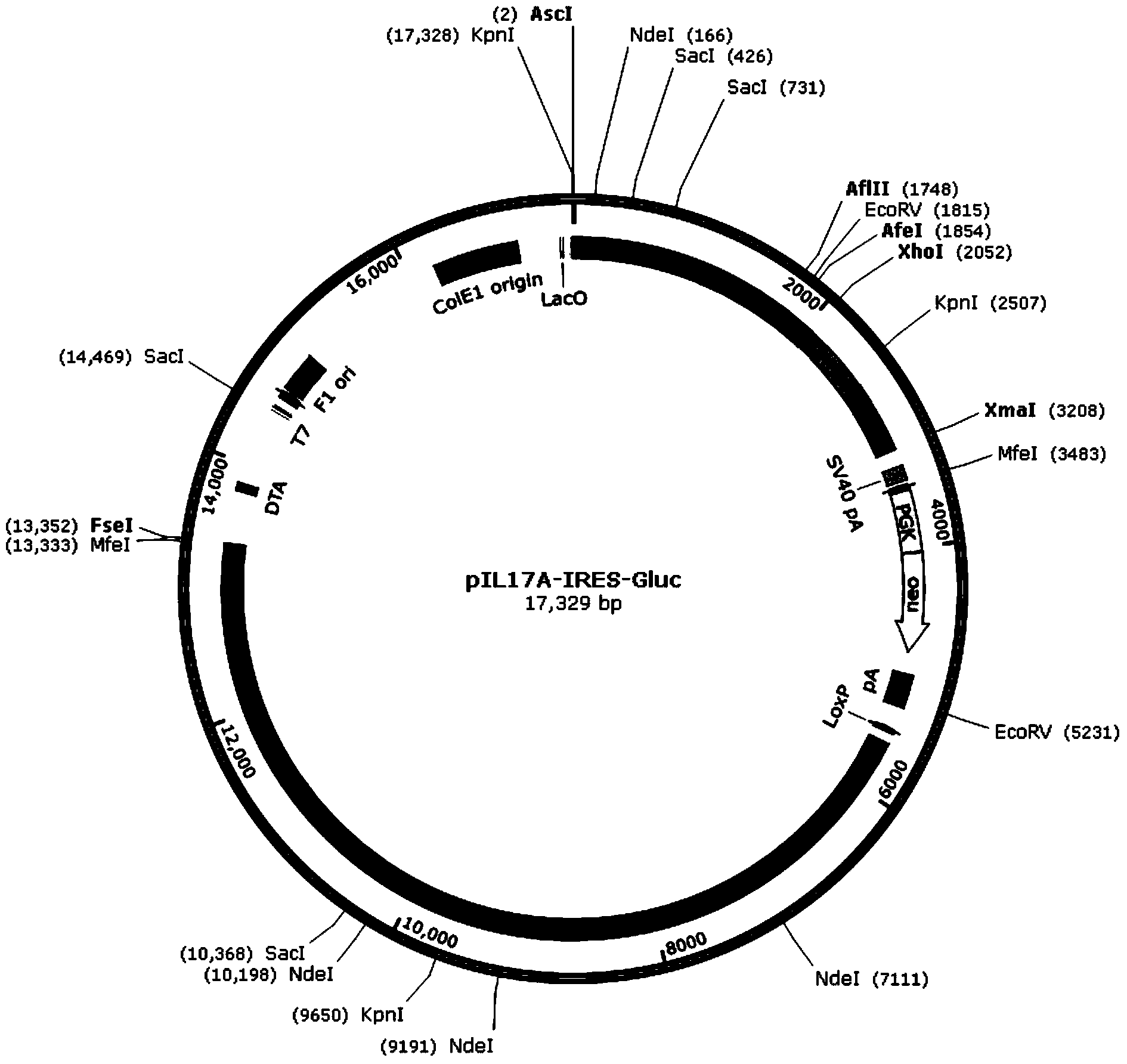
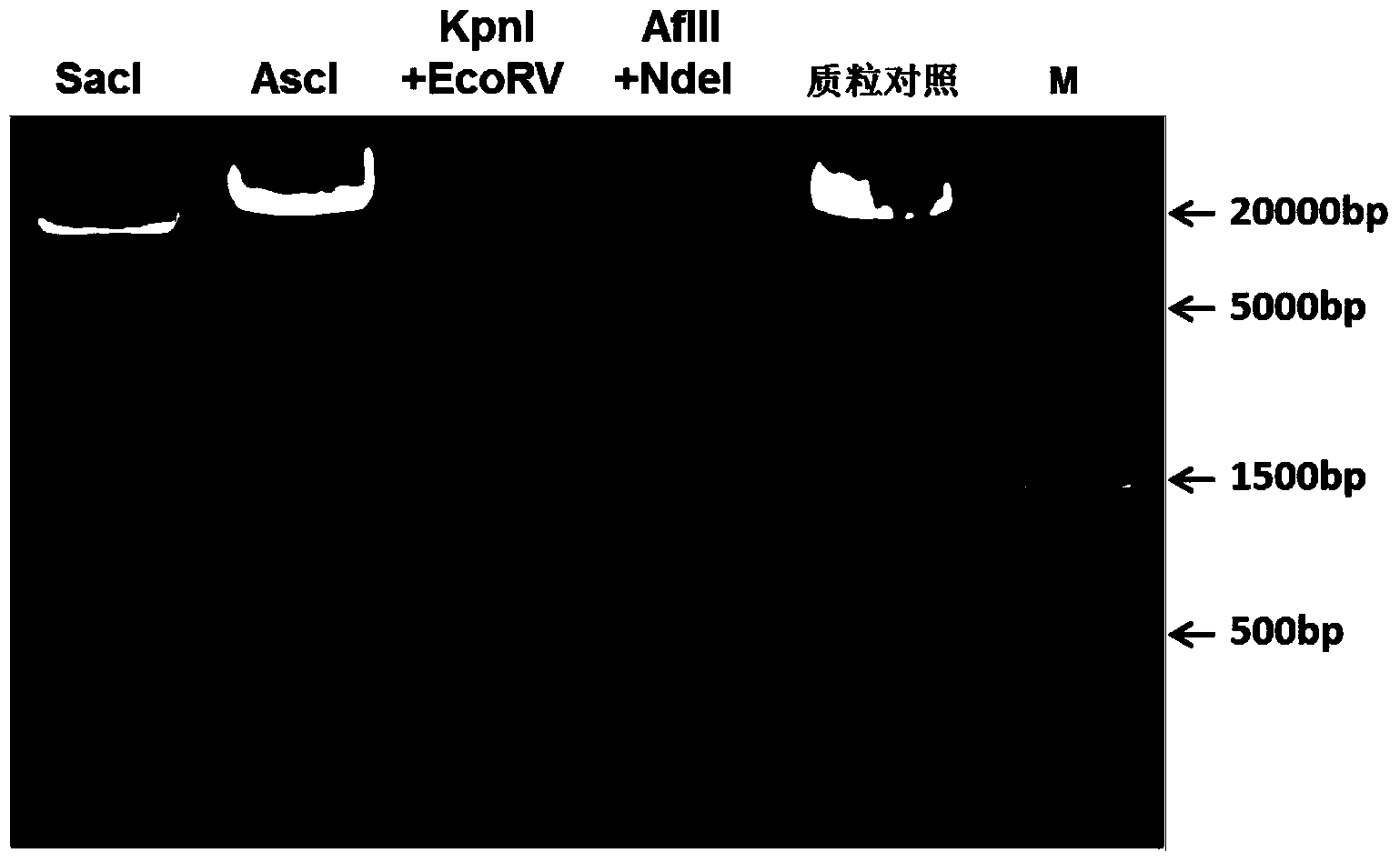
The invention discloses a recombinant vector for gene knock-in. The recombinant vector comprises a DNA (deoxyribonucleic acid) fragment structure IL17A-IRES-luc, wherein IL17A is an encoding gene of interleukin 17A, IRES is internal ribosome entry site, and luc is an encoding gene of secreting luciferase. The invention also provides a method for preparing the gene knock-in recombinant vector and a mouse model prepared by using the recombinant vector, wherein the mouse model can be used for detecting the expression of the IL17A gene by detecting the expression of the secreting luciferase in blood or urine; a fluorescent microscope or other complicated equipment is not needed; the secreting luciferase emits light by virtue of an oxidizing reaction of substrate coelenterazine in the catalysis of luciferase, and laser light is not needed, so that the fluorescent background is far lower than GFP (green fluorescent protein) fluorescence, the detection sensitivity is higher, the background is cleaner, and the experiment results are more accurate.
Owner:BIOCYTOGEN PHARMACEUTICALS (BEIJING) CO LTD
Construction method and use of enterovirus 71 type infection model based on hSCARB2 gene knock-in
ActiveCN106554972AIn-vivo testing preparationsVector-based foreign material introductionAntiviral drugPharmaceutical drug
The invention relates to a construction method of an enterovirus 71 type infection model based on hSCARB2 gene knock-in and use of the infection model, and especially provides an effective means for in vivo evaluation of EV71 vaccines and antiviral drugs.
Owner:NAT INST FOR FOOD & DRUG CONTROL
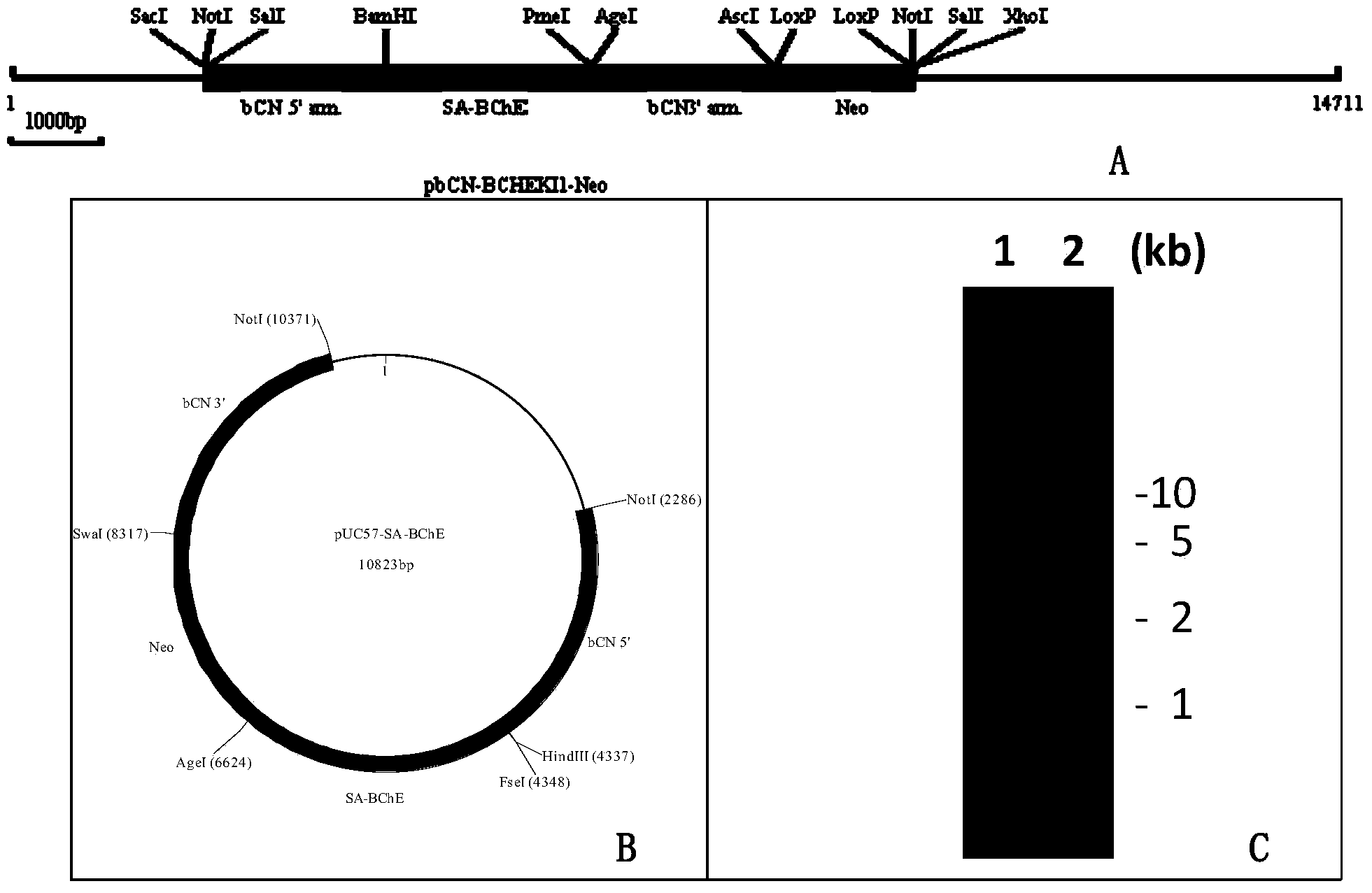
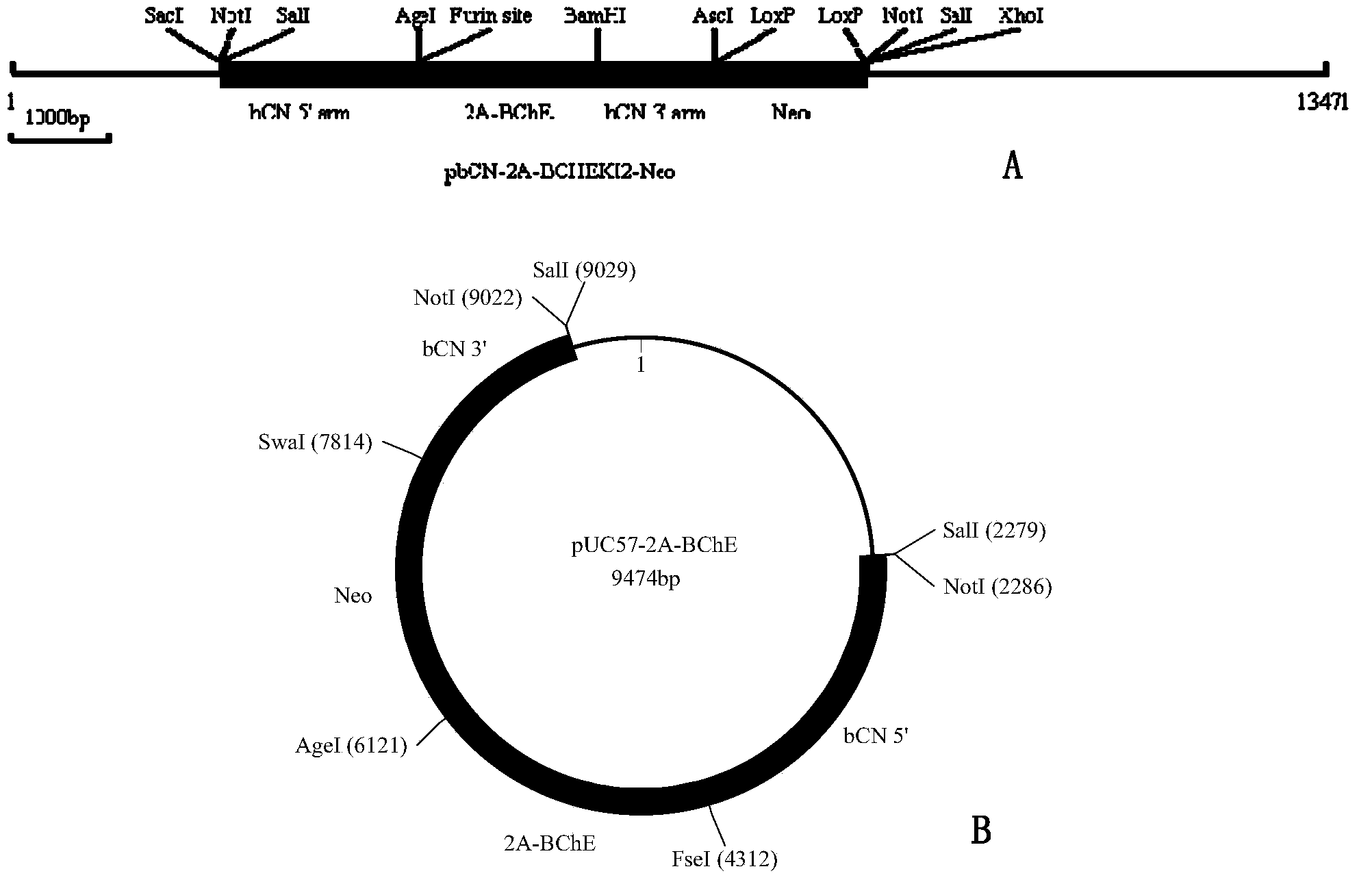
Method for producing recombinant human BChE (butyrylcholinesterase) from transgenic animals by using gene knock-in and nuclear transfer technologies
The invention discloses a method for producing recombinant human BChE (butyrylcholinesterase) from transgenic non-human mammals by using gene knock-in and nuclear transfer technologies, and particularly provides a donor construct used for specific expression of a mammary gland. The construct integrates DNA (deoxyribose nucleic acid) coding sequences of human BChE to a specific expression protein gene locus of the mammary gland in targeted manner and performs successful transfection on non-human mammal embryo fibroblasts, juvenile and adult somatic cells or embryonic stem cells, clone is performed by using a somatic cell nuclear transfer technology so as to obtain a new individual, and the carried human BChE gene enables the mammary gland of a non-human mammal to express stably and secrete holoenzyme or a monomer of the human BChE. Recombinant protein produced with the method can be used for preventing and treating nerve poison and organic phosphorus compound poisoning as well as apnea caused by cocaine poisoning and succinylcholine, and for detecting and removing residues of organic phosphorus compounds on vegetables, other crops, various article layers and the soil.
Owner:SHANGHAI JENOMED BIOTECH CO LTD
Method for realizing genome editing and accurate and targeted knock-in of genes in fishes
ActiveCN108949830AImprove integration efficiencyLow costMicroinjection basedAnimal husbandryRandom mutationGenome editing
The invention provides a method for realizing genome editing and accurate and targeted knock-in of genes in fishes. A technology of producing DNA single-chain gap based on nicking enzyme is utilized,assisted with a set of homologous recombination repair factor RecOFAR, so as to realize high-efficient, accurate and targeted integration of genome editing and gene knock-in in the fishes. The problems that a current existing method is low in efficiency, and random mutation efficiency of target points is higher are overcome.
Owner:FUZHOU UNIV
PirB gene-knock-in mouse animal model and construction method thereof
The invention discloses a PirB gene-knock-in mouse animal model, which comprises gRNA1 and gRNA2 for determining specific target sites to be knocked in by PirB gene, wherein the PirB gene is knocked into an intron 1 of ROSA26 gene of a C57BL / 6J mouse; the gene sequence of the gRNA1 is shown in SEQ ID NO.1; and the gene sequence of the gRNA2 is shown in SEQ ID NO.2. The invention further specifically discloses a construction method of the mouse animal model, the mouse animal model provides a good foundation for research on PirB gene function and in-vivo verification. Hybridization of PirB-knock-in animal model with different types of Cre mice can be used to study the functions of PirB in different organs or different cell types or different disease models.
Owner:XIAN MEDICAL UNIV
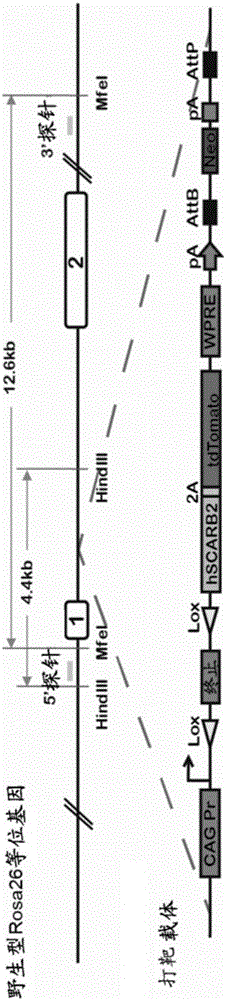
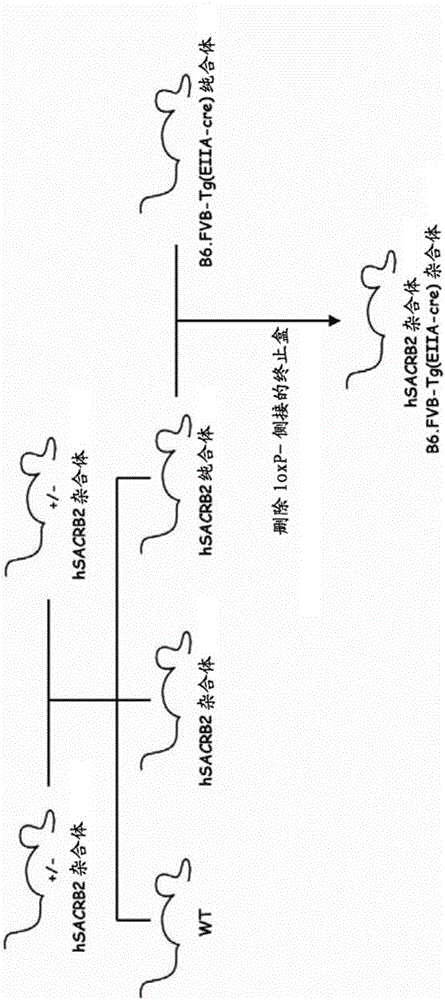
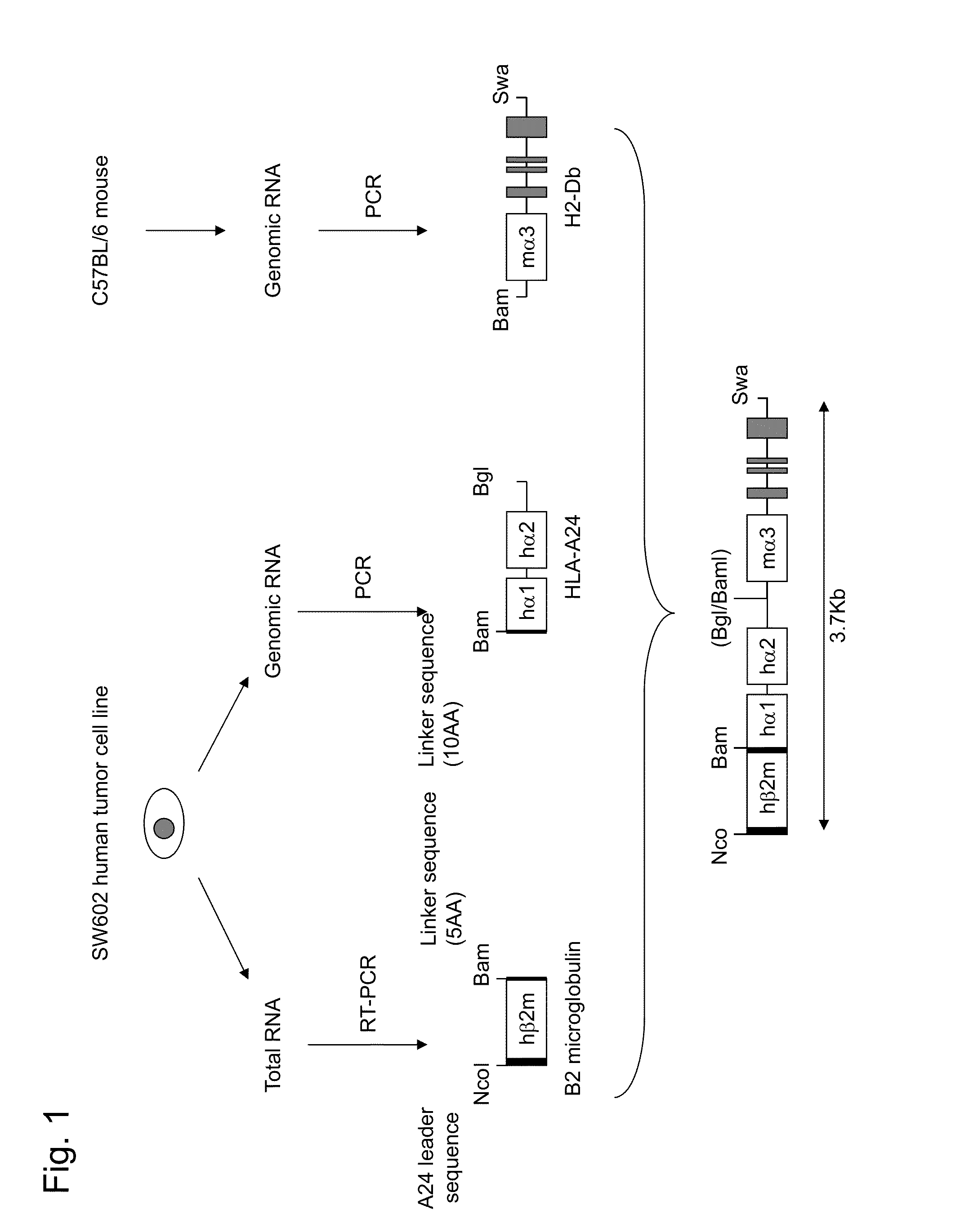
HLA class i-expressing non-human animal
ActiveUS20160227750A1Great advantagePolypeptide with localisation/targeting motifAntibody mimetics/scaffoldsBiotechnologyMHC class I
This invention provides an HLA class I-expressing non-human animal that can be prepared efficiently in a simple manner within a short period of time, in which the transgene copy number to be introduced thereinto and the integration site of transgene are regulated, and in which the expression level of the transgene and the site of expression thereof are regulated. The invention also provides a method for preparing such non-human animal. In such HLA class I gene knock-in non-human animal, an artificial chimeric gene encoding an artificial chimeric protein comprising a protein composed of β2 microglobulin, HLA class I α1 and α2 regions, and an MHC class I α3 region of a non-human animal ligated in such order from the N terminus is expressed under the control of a transcription regulatory region of β2 microglobulin gene of the non-human animal.
Owner:TAIHO PHARMA CO LTD
Gene knock-in composition and use method and application thereof
InactiveCN104946621AImprove the efficiency of homologous recombinationAvoid the disadvantages of instabilityRecombinant DNA-technologyForeign genetic material cellsGenome instabilityGenomic Stability
The invention provides a gene knock-in composition containing a single incision enzyme or polynucleotide coding the same, recombinase or polynucleotide coding the same, and an optional donor DNA containing an exogenous gene. The invention also provides a corresponding gene knock-in method and a method for preparing a transgenic non-human mammal model. The composition and the methods provided by the invention can be used for realizing efficient homologous recombination integration, and meanwhile various defects of insertion or deletion, genome instability and the like which are generated easily in the prior art can be avoided.
Owner:SHANGHAI INST OF BIOLOGICAL SCI CHINESE ACAD OF SCI +1
Insect targeted gene knock-in composition, use method and application thereof
ActiveCN105315352AImprove the efficiency of homologous recombinationDepsipeptidesGenetic engineeringNucleotideGenetically modified insect
The invention provides a composition used in insect targeted gene knock-in, which includes a single nickase or an encoding polynucleotide thereof; a recombinant factor or an encoding polynucleotide thereof; and optionally, a donor DNA of an exogenous gene. The invention also provides a corresponding gene knock-in method and a method for preparing transgenic insects. The composition and the method in the invention can achieve high-effective homologous recombination integration in insect cells and meanwhile can avoid the defects in gene knock-in technologies in the prior art. The composition also can stably and high-effectively achieve homologous recombination integration of a fragment being more than 10 kb in length in the insect cells, so that the composition can be used for transferring a genetic selection marker into the insect cells, preferably a visible genetic selection marker, which is convenient to determine strains of transgenic insects.
Owner:FUZHOU UNIV
Method for preparing general type CAR-T cells by using CRISPR/Cas9+AAV
InactiveCN109609551AImprove safety and controlReduce treatment time cost and capital costMammal material medical ingredientsBlood/immune system cellsCAR T-cell therapyAdeno-associated virus
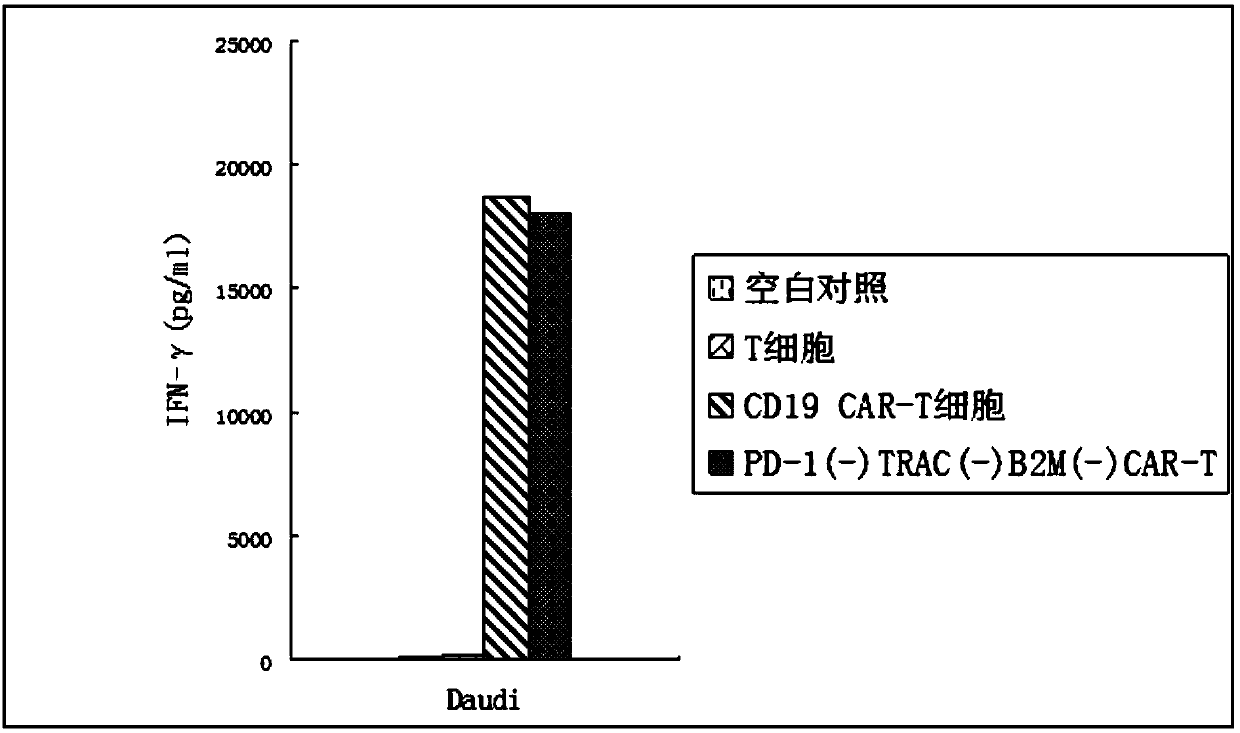
The invention relates to a method for preparing general type CAR-T cells by using CRISPR / Cas9+AAV, and belongs to the field of immunocytotherapy. The method utilizes CRISPR / Cas9+AAV to knock out an immunological checkpoint PD-1, a human leukocyte antigen (HLA) and TCR; multi-gene knockout is employed to prepare the general type CAR-T cells; and a CRISPR / Cas9 gene editing technology is used to perform precise gene knockout, and is used in combination with adeno-associated virus (AAV) to replace lentiviruses for precise gene knock-in and to avoid the potential risk of random integration of the lentiviruses. The method of the invention can realize multi-target gene knock-in and knock-out, construct the general type CAR-T cells for allogeneic treatment, and solve the problems existing in traditional CAR-T cell therapy such as risk of random insertion mutation of lentiviruses, high production cost and long preparation period for autologous CAR-T design and culture, and insufficient cell number and expansion ability.
Owner:广州白云山拜迪生物医药有限公司
High-efficient accurate targeted gene integration system and application thereof
ActiveCN110305908AIntegration of efficient genesAchieve targetedStable introduction of DNANucleic acid vectorTransposaseTransposon element
The invention discloses a high-efficient accurate targeted gene integration system and an application thereof. The high-efficient accurate targeted gene integration system comprises an expression exogenous fusion protein system, a guide RNA and a donor carrier, wherein the expression exogenous fusion protein system comprises a fusion protein formed by one of Cas9 or SB or Cas9 and SB, N123, N57 and SB100XE279D; the guide RNA is a long sequence of 20bp in a target genome region and two ends of the donor carrier; and two ends of the donor carrier guide RNA targeted sites, SB binding sites and middle target gene integration fragments are arranged at two ends of the donor carrier. Through the adoption of the high-efficient accurate targeted gene integration system disclosed by the invention, the targeting, the cutting and the integrating of gene can be simultaneously realized; and SB transposase is utilized to be combined with a transposon donor to form a tetramer, so that the donor carrier can be oriented to the target sites, and targeted integration of the gene is facilitated. According to the high-efficient accurate targeted gene integration system disclosed by the invention, in vivo linearization elements are utilized, so that in vitro linearization of the donor carrier can be realized, and the integration of the genes is facilitated; and the method for realizing gene typing isbased on the SB transposase and NHEJ in a gene repairing mechanism, so that the high-efficient accurate targeted gene integration system can realize gene integration efficiently.
Owner:SOUTHERN MEDICAL UNIVERSITY
Negative selection marker for escherichia coli gene knock-in
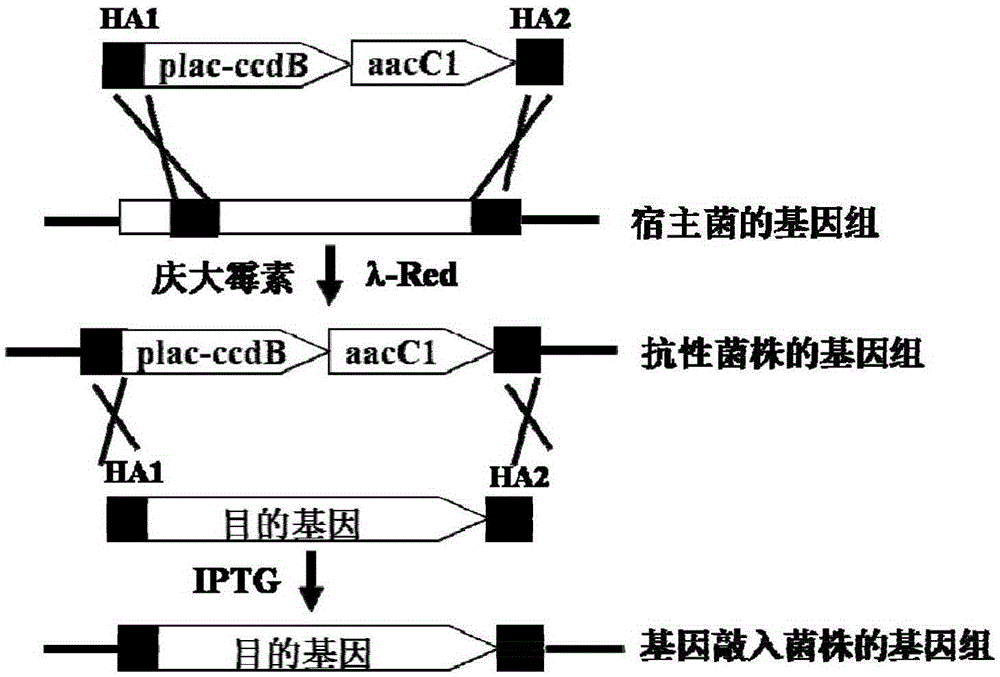
InactiveCN105238820AConvenient researchStable introduction of DNAMicroorganism based processesThiogalactosidesDNA fragmentation
The invention relates to a negative selection marker for escherichia coli gene knock-in, namely, the virulent gene ccdB driven by a plac promoter, and plac-ccdB and the gentamicin resistance gene aacC1 form a gene cassette for gene knock-in. In the first step, DNA fragments containing homologous arms plac-ccdB and aacC1 are obtained through amplification, and electro-transformed into escherichia coli for expression of recombinase. Under gentamicin screening, recombinase catalyzes homologous recombination and the DNA fragments are integrated into escherichia coli genome. In the second step, same homologous arms are utilized for amplification of foreign DNA fragments, ccdB is expressed in the bacterial strains obtained in the first step during the electrotransformation for expression of recombinase under the screening of isopropyl-beta-d-isopropylthiogalactoside, the toxicity serves as the feature of the negative selection marker, recombinase catalyzes homologous recombination among homologous arms, and foreign DNA fragments replace a plac-ccdB-aacC1 gene cassette to realize gene knock-in.
Owner:NANJING NORMAL UNIVERSITY
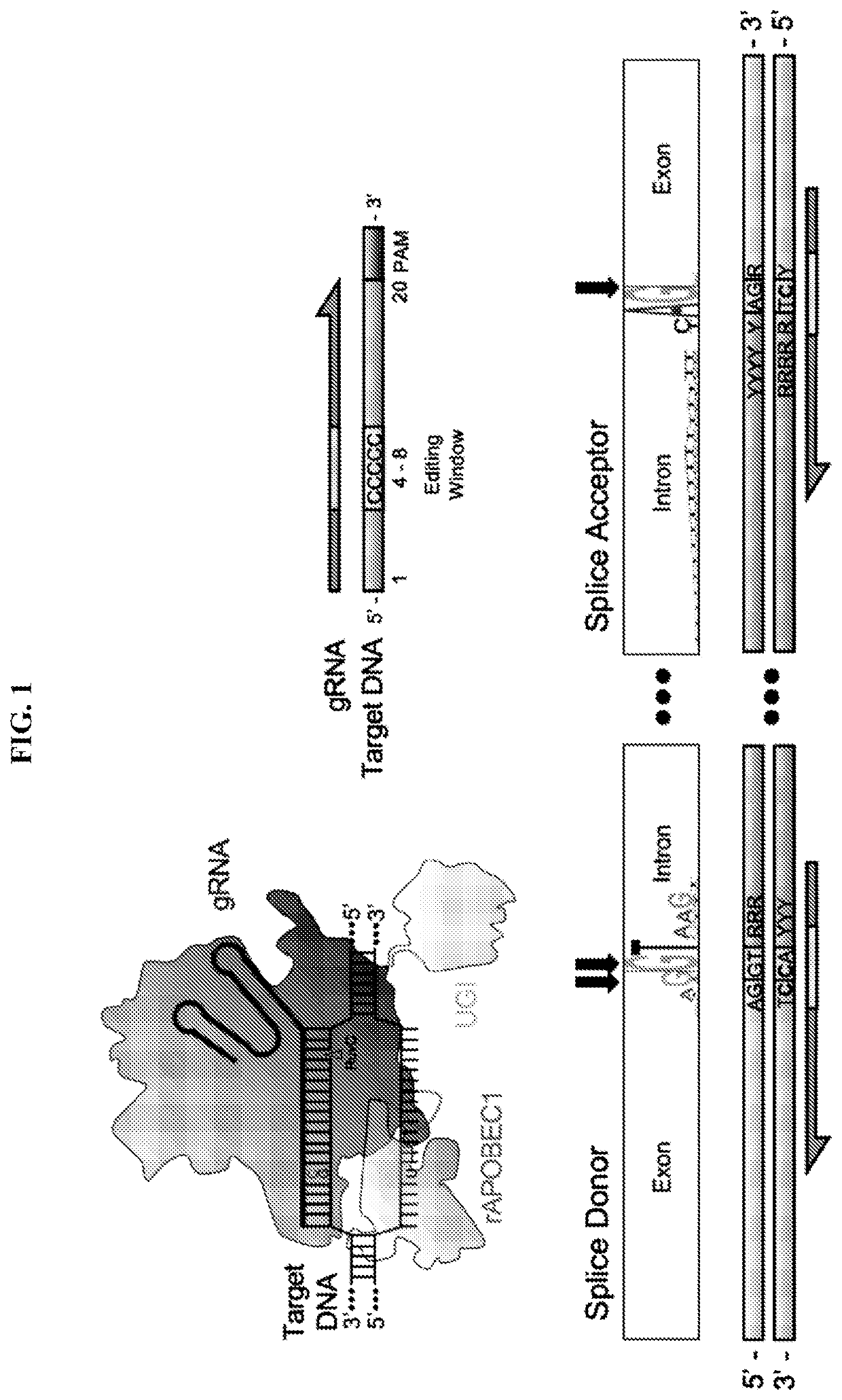
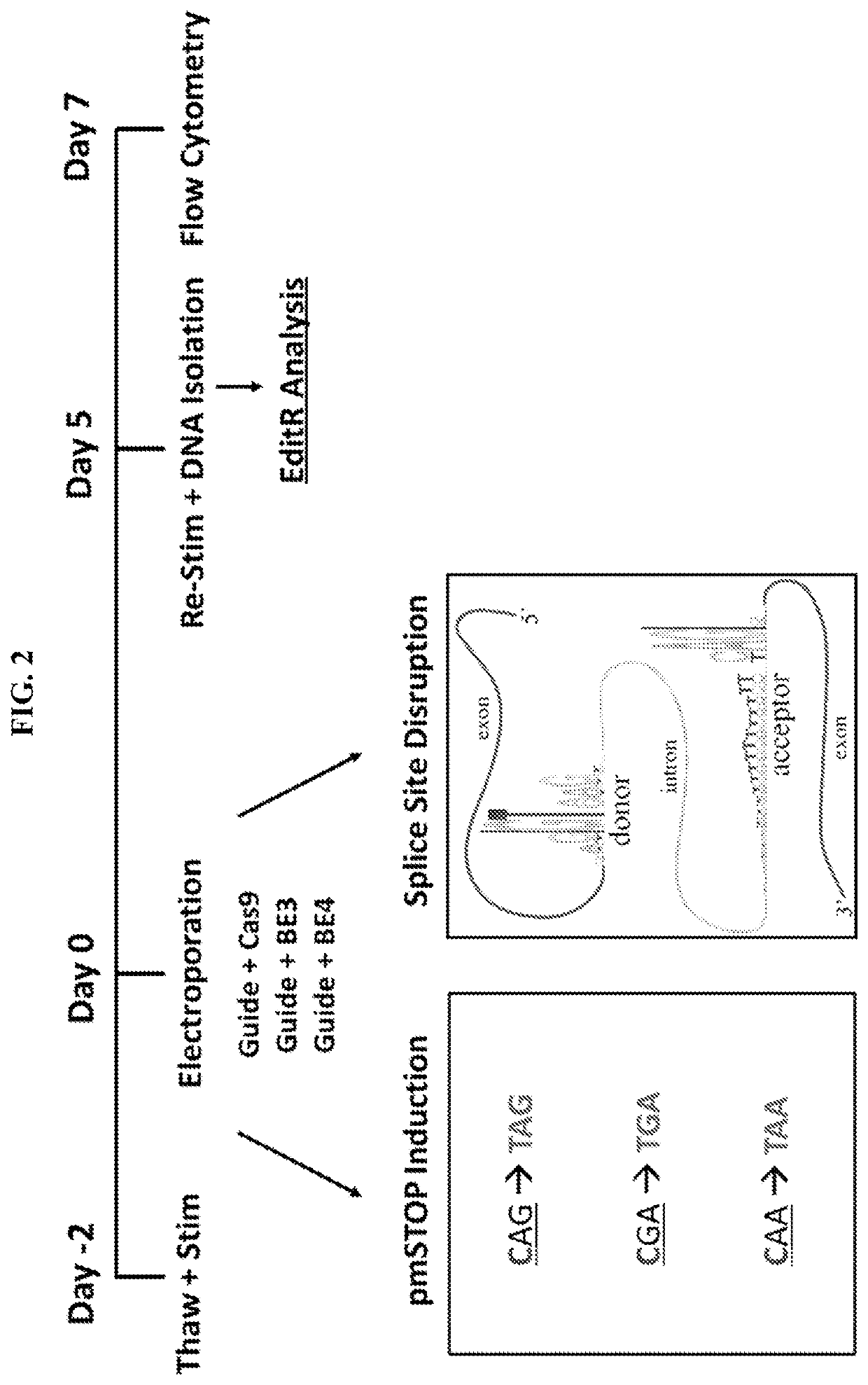
Lymphohematopoietic engineering using cas9 base editors
PendingUS20210040507A1Easily damagedGenetically modified cellsStable introduction of DNABase JEngineered genetic
Provided herein are methods and systems for targeted gene disruption (knock-out, missense mutation) and targeted gene knock-in in mammalian cells using base editors and guide RNAs (gRNAs) designed to target splice acceptor-splice donor sites. Also provided herein are universally acceptable genetically engineered cells comprising targeted disruptions in immunotherapy-related genes and comprising a CAR / TCR for therapeutic applications.
Owner:RGT UNIV OF MINNESOTA
Gene knock-in non-human animal
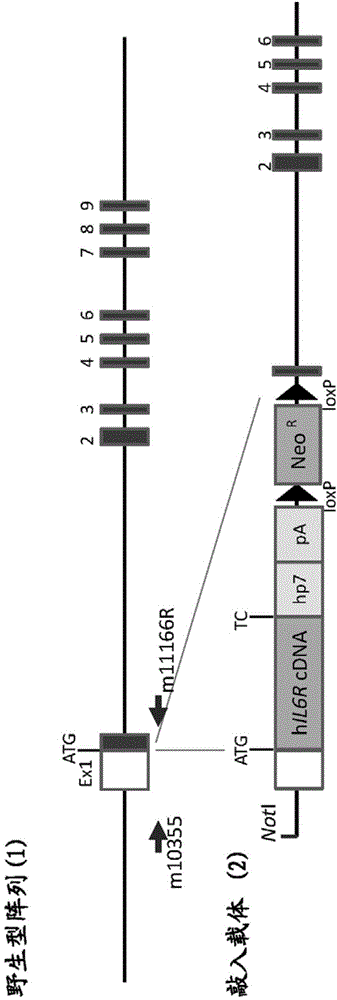
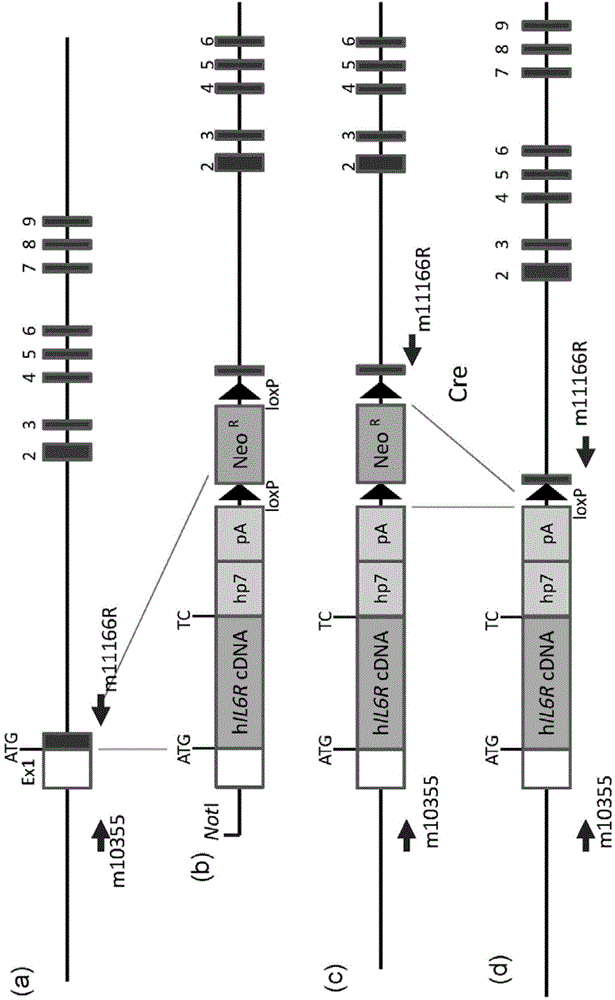
ActiveUS20150225481A1Suppressing expression of its endogenous geneAccurate assessmentCompounds screening/testingAntipyreticHuman animalA-DNA
The present invention provides a non-human animal in which a DNA comprising an hp7 sequence-encoding DNA and a poly A addition signal-encoding DNA added on the 3′ side of a DNA encoding an arbitrary foreign gene is inserted in the same reading frame as that of an arbitrary target gene present on the genome of the non-human animal.
Owner:CHUGAI PHARMA CO LTD
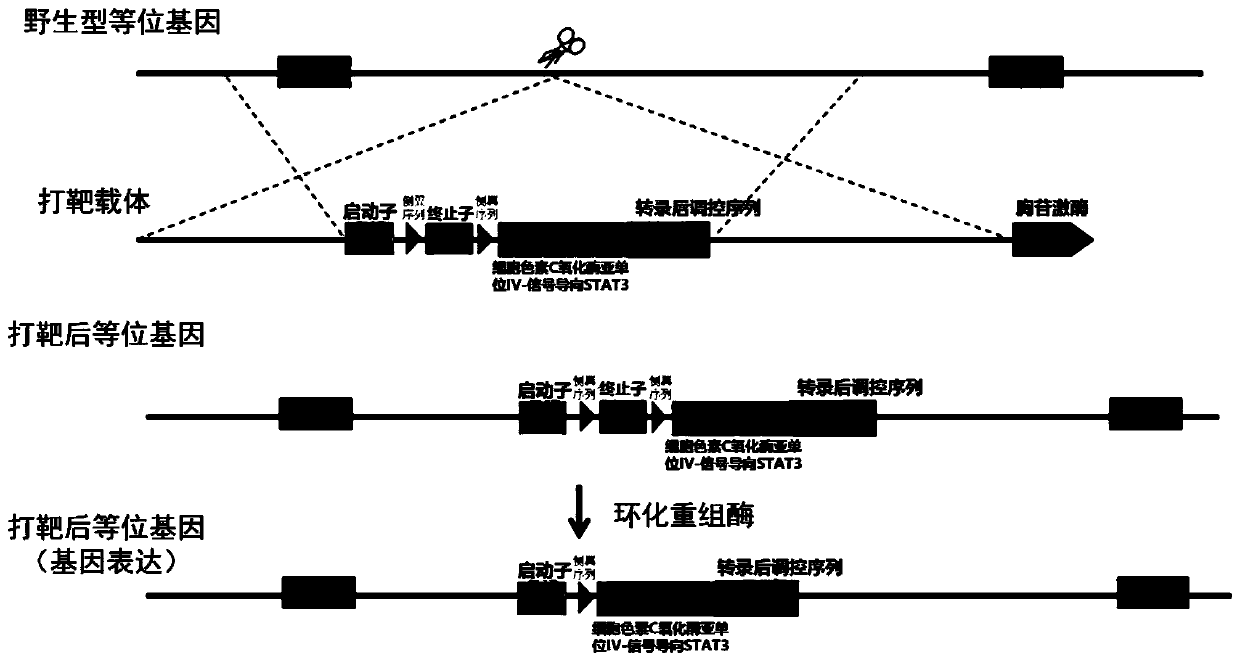
Construction method of STAT3 mitochondrial localization conditional gene knock-in mouse model
The invention relates to a construction method of an STAT3 mitochondrial localization conditional gene knock-in mouse model. The method comprises the following steps: firstly, constructing an STAT3 mitochondrial localization conditional gene knock-in recombinant targeting vector, transfecting the recombinant targeting vector to fertilized ova of a donor mouse, and then transplanting the donor mouse fertilized ova into the uterus of a fake pregnant mother mouse to produce a chimeric mouse; mating the chimeric mouse with a C57BL / 6J mouse to obtain an F1 generation; mating the F1 generation witha mouse of tissue-specific expression Cre recombinase to obtain a double-hybrid mouse; and are intercrossing the double-hybrid mouse and an STAT3<f / +> or STAT3<f / f> mouse, and inducing the obtained offspring by tamoxifen to obtain the STAT3 mitochondrial localization conditional gene knock-in mouse model. According to the mouse model disclosed by the invention, the STAT3 gene is expressed in specific tissues and cell types, and an ideal model is provided for researching the action mechanism of STAT3 in specific cell development and differentiation, body metabolism and tumor occurrence and development.
Owner:XUZHOU MEDICAL UNIV
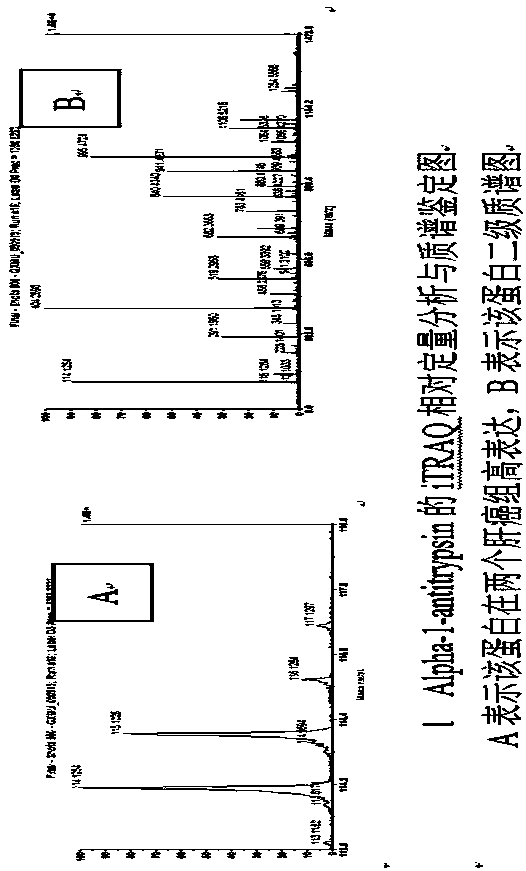
Method used for discovering and identifying liver cancer serum differential expression proteins and verifying marker proteins
InactiveCN107894507APropose feasibilityDisease diagnosisProtein markersCandidate Gene Association Study
The invention discloses a method used for discovering and identifying liver cancer serum differential expression proteins and verifying marker proteins. According to the method, iTRAQ labeling MALDI-TOF MS / MS technology is adopted for discovering and identifying liver cancer serum differential expression proteins and MRM verifying of marker proteins. According to the method, searching on effectivetumor markers possesses important meaning on canceration mechanism study, disease early stage diagnosis, and prognosis. Selected novel liver cancer protein markers are subjected to in vivo and in vitro experiments such as gene knockin and knockout, and nude mice transplanted cancer, influences of blocking or up-regulating of expression of candidate genes on tumor biological behavior are observed,and key proteins capable of influencing human liver cancer occurrence and development are positioned, the effects of the key proteins in a system signal network is analyzed based on bioinformatics analysis, and the feasibility, the liver cancer diagnosis value, and the clinical significance of the key proteins in clinical applications including liver cancer prevention, early stage diagnosis, andprognosis is evaluated based on MRM absolute quantitative verification.
Owner:南宁科城汇信息科技有限公司
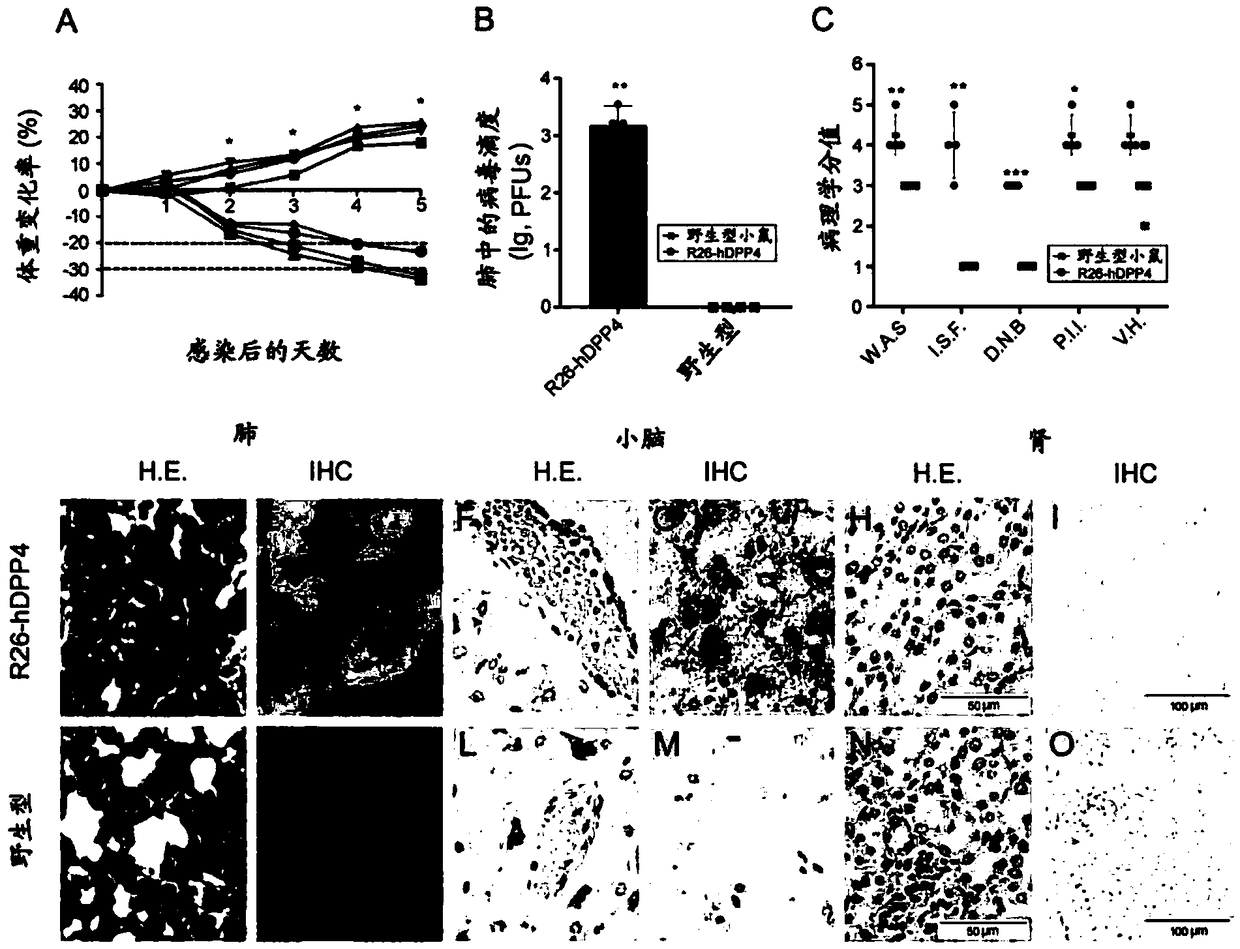
Human DPP4 gene knock-in mouse model and production method and use thereof
The present invention provides a human DPP4 gene knock-in mouse model and a production method and use thereof. Specifically, human DPP4 is accurately inserted into a safe harbor locus of a mouse genome without changing mouse DPP4 function to establish a novel homozygous mouse model in which a human DPP4 gene is knocked into the mouse safe harbor locus. The invention also provides the use of the mouse model in which the human DPP4 gene is knocked into the genome in infection of a mouse with a low dose MERS-CoV euvirus in a BSL-3 facility and evaluation of the in-vivo efficacy of anti-MERS-CoV antiviral agents and vaccines or infection of the mouse with pseudotype MERS-CoV virus in a biological laboratory lacking the BSL-3 facility and evaluation of the in-vivo efficacy of the anti-MERS-CoVantiviral agents and vaccines.
Owner:NAT INST FOR FOOD & DRUG CONTROL
Efficient and stable fixed-point integrated gene knock-in method based on pCAG-flox-neo vector
PendingCN114480457AEasy to replaceQuick buildPeptide-nucleic acidsGenetically modified cellsBiotechnologyEnzyme digestion
The invention provides an efficient and stable fixed-point integrated gene knock-in method based on a pCAG-flox-neo vector. The method comprises the following specific steps: (1) carrying out double enzyme digestion on a pCAG-STOP2 vector; the synthesized fragment is connected to a pCAG-STOP2 carrier; (2) carrying out enzyme digestion on the synthesized carrier again, and then connecting the carrier into polyA-loxP to obtain a pCAA-flox-neo carrier; (3) inserting the sgRNA sequence of the pig pH 11 site capable of being identified by the sgRNA and the PAM sequence into the T3 promoter, so as to obtain a final gene knock-in vector skeleton pCAG-floxP-neo-pH 11, wherein the gene knock-in vector skeleton pCAG-floxP-neo-pH 11 is obtained; and (4) carrying out PCR (Polymerase Chain Reaction) amplification on the required hKCNH2-T618I and hmuPKD2 fragments, and inserting the fragments into a pCAG-floxP-neo-pH11 vector skeleton in a one-step cloning manner. The invention establishes a method which is efficient, accurate, simple to operate and capable of knocking out the target gene at the same time, and has important scientific significance for researching the effect of the target gene in mammals or disease models.
Owner:ZHEJIANG UNIV
Escherichia coli for arsenic detection
ActiveCN104894042AReduce the background valueHigh background valueBacteriaMicrobiological testing/measurementEscherichia coliFluorescence
The invention relates to a microorganism method for detecting metallic-arsenic-like in water, in particular to an escherichia coli report bacterial strain culturing method improved through genetic engineering, and an escherichia coli culturing method for detecting arsenic in water. The detection bacterial strain is obtained by adopting a Red recombination system to knockout arsenic resistant arsB genes of wild type escherichia coli MC4100, and then adopting the gene knockin technology to substitute pars-arsR-gfpmut2 report genes to the positions of araB coding genes. The biological detection bacterial strain is named as E.coli WMC-011p. The lowest arsenic detection range meets the Integrated Wastewater Discharge Standard of GB8978-1996. The bacterial strain overcomes the shortcomings of high fluorescence background value, unstable detection signals, inaccurate results and the like of a biological detection bacterial strain based on a plasmid vector, and has the advantages of high specificity, high sensitivity, low cost and the like.
Owner:WENZHOU MEDICAL UNIV
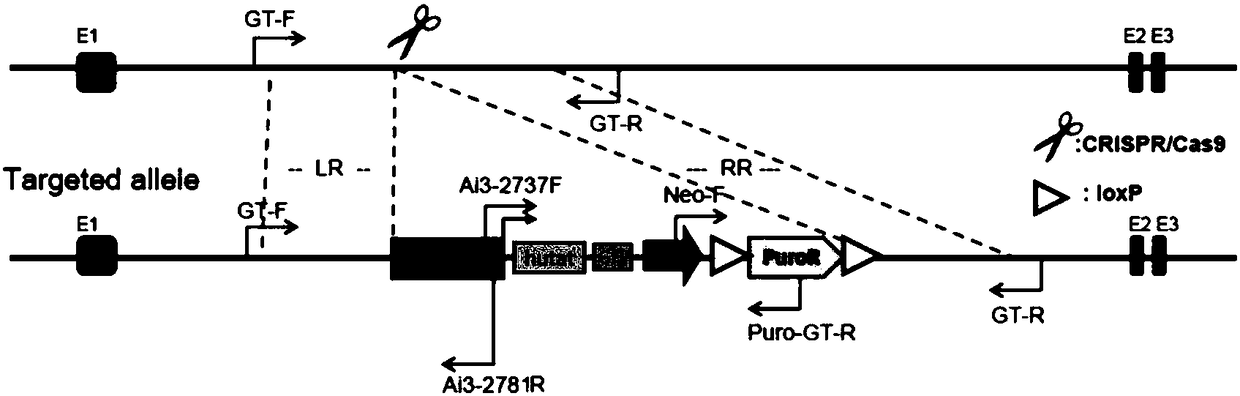
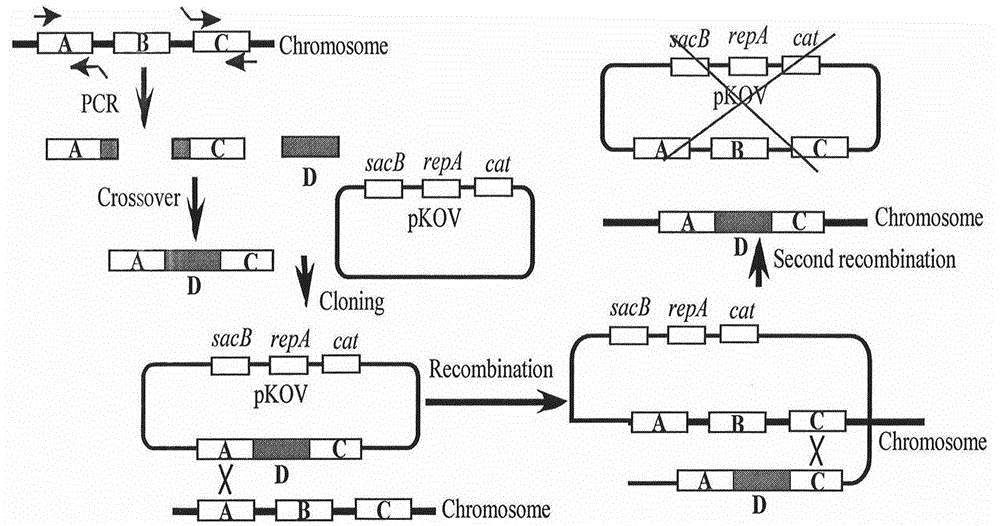
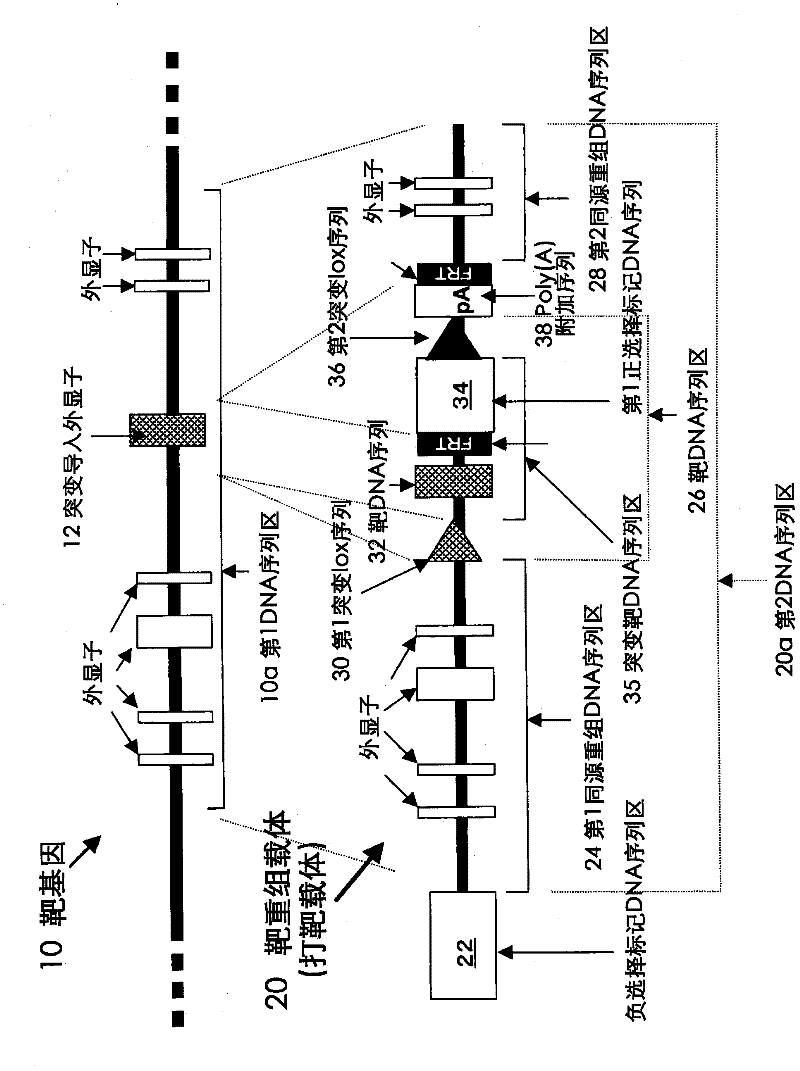
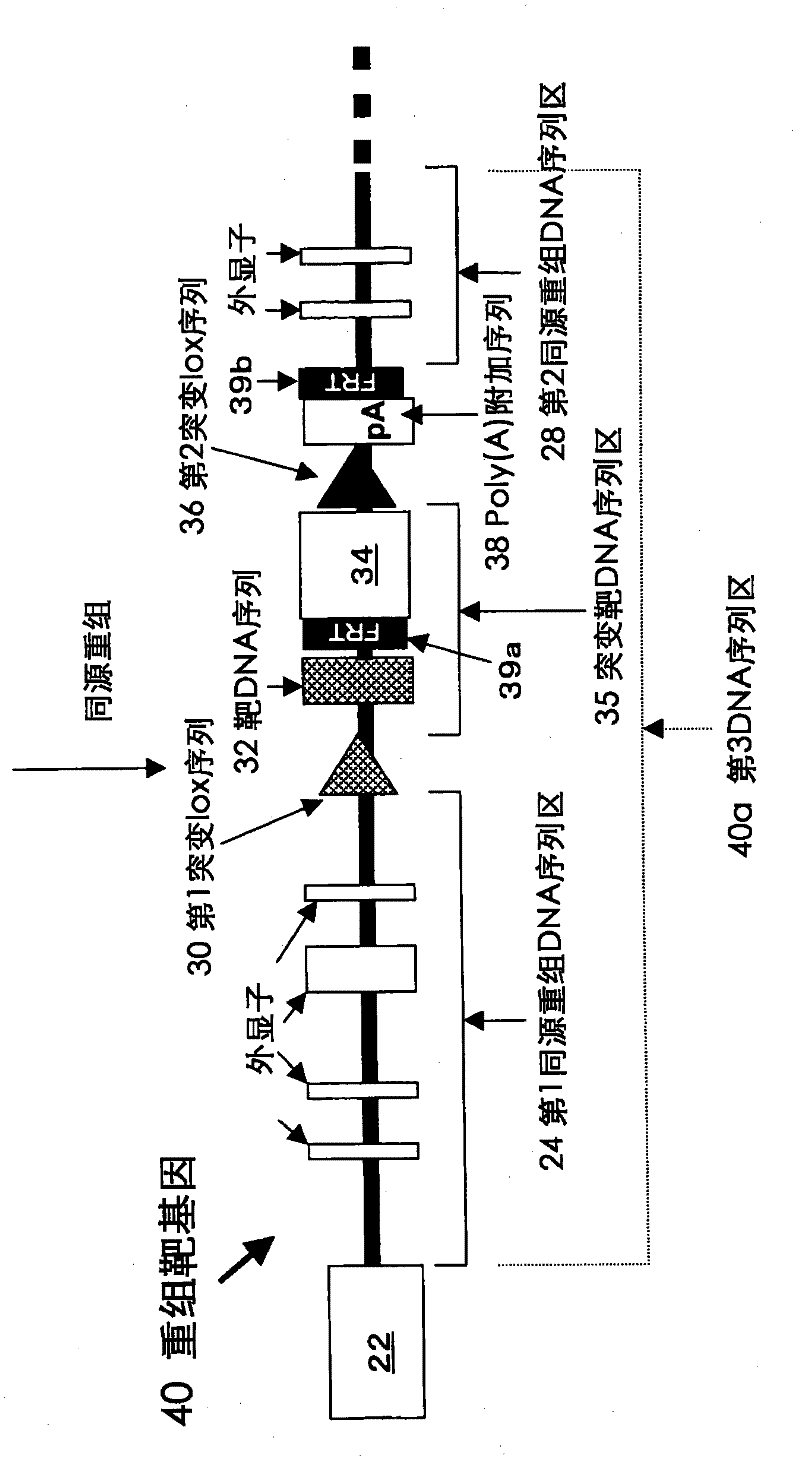
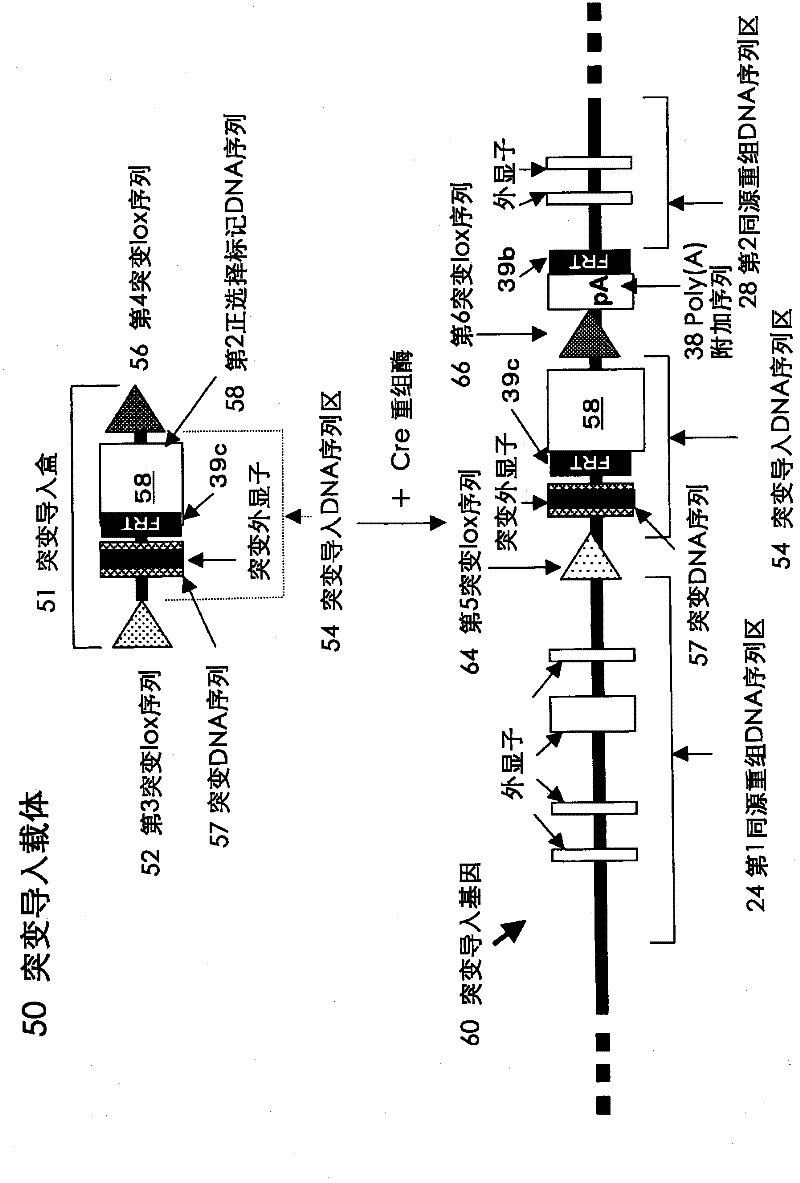
Method for introducing mutated gene, gene having mutation introduced therein, cassette for introducing mutation, vector for introducing mutation, and knock-in non-human mammal
InactiveCN102203249AQuick importCell receptors/surface-antigens/surface-determinantsNucleic acid vectorCre recombinaseExon
Disclosed is a method for introducing a mutation into a gene, which comprises the following steps: a homologous recombination step of carrying out the homologous recombination between a target gene into which the mutation is to be introduced and a target recombinant vector, thereby substituting an exon in the target gene into which the mutation is to be introduced by a target DNA sequence in the target recombinant vector; and a mutation introduction step of carrying out the specific recombination between the target DNA sequence in the resulting recombined target gene and a mutation introduction cassette of a mutation introduction vector carrying a mutated DNA sequence containing a mutated exon by the intervening action of Cre recombinase to substitute the target DNA sequence by the mutated DNA in the mutation introduction cassette, thereby producing a mutation-introduced gene into which the mutant DNA sequence has been introduced. The method enables the production of a knock-in non-human animal, such as a knock-in mouse, which carries the mutation-introduced gene.
Owner:FUKUOKA UNIV +1
Method for preparing gene knock-in cells
PendingUS20190169653A1High yieldShort timeHydrolasesStable introduction of DNAGenome editingAnimal use
The present inventors tried to prepare a knock-in animal using a genome editing system containing a nuclease in the form of protein and a DNA-targeting RNA and using a single-stranded DNA as a donor, and found that it is possible to obtain a cell in which the donor DNA has been knocked in with extremely high efficiency.
Owner:NAT UNIV CORP TOKYO MEDICAL & DENTAL UNIV
Gene knockin method and kit for gene knockin
InactiveUS20190225989A1Efficient preciseHydrolasesStable introduction of DNAGene knockinEndonuclease
A gene knockin method and a kit for gene knockin are provided. The method comprises (a) introducing a RNA-guided endonuclease that cleaves the chromosome at the insertion site into the cell; (b) introducing a guide RNA into the cell; and (c) introducing a donor plasmid into the cell, wherein the donor plasmid comprises the donor sequence flanked with a 5′ homology arm and a 3′ homology arm, a 5′ flanking sequence upstream of the 5′ homology arm, and a 3′ flanking sequence downstream of the 3′ homology arm, wherein the 5′ homology arm is homologous to a 5′ target sequence upstream of the insertion site on the genome and the 3′ homology arm is homologous to a 3′ target sequence downstream of the insertion site on the genome, wherein the guide RNA recognizes the insertion site, the 5′ flanking sequence, and the 3′ flanking sequence, wherein the RNA-guided endonuclease cleaves the donor plasmid at the 5′ flanking sequence and the 3′ flanking sequence, thereby producing a linear nucleic acid, wherein the donor sequence is inserted in to the genome at the insertion site through homology-directed repair.
Owner:INST OF HEMATOLOGY & BLOOD DISEASES HOSPITAL CAMS & PUMC
Gene knock-in non-human animal
The present invention provides a non-human animal in which a DNA comprising an hp7 sequence-encoding DNA and a poly A addition signal-encoding DNA added on the 3' side of a DNA encoding an arbitrary foreign gene is inserted in the same reading frame as that of an arbitrary target gene present on the genome of the non-human animal.
Owner:CHUGAI PHARMA CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com