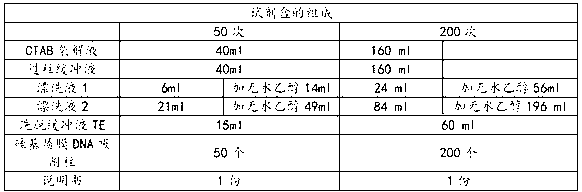
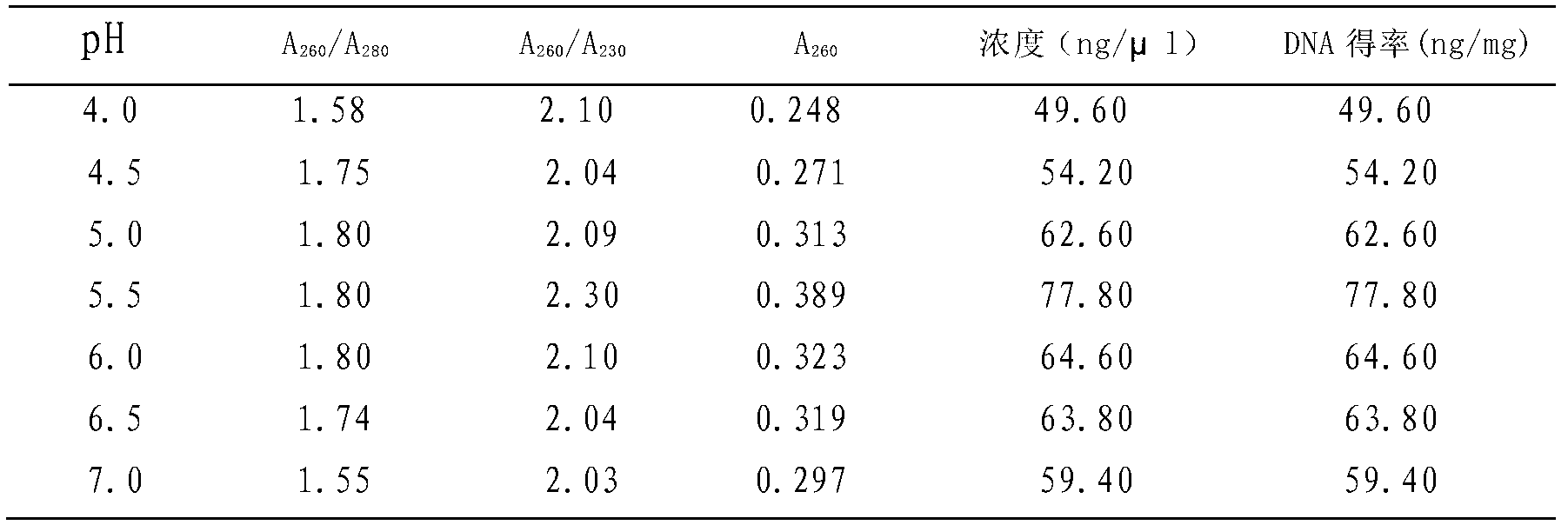
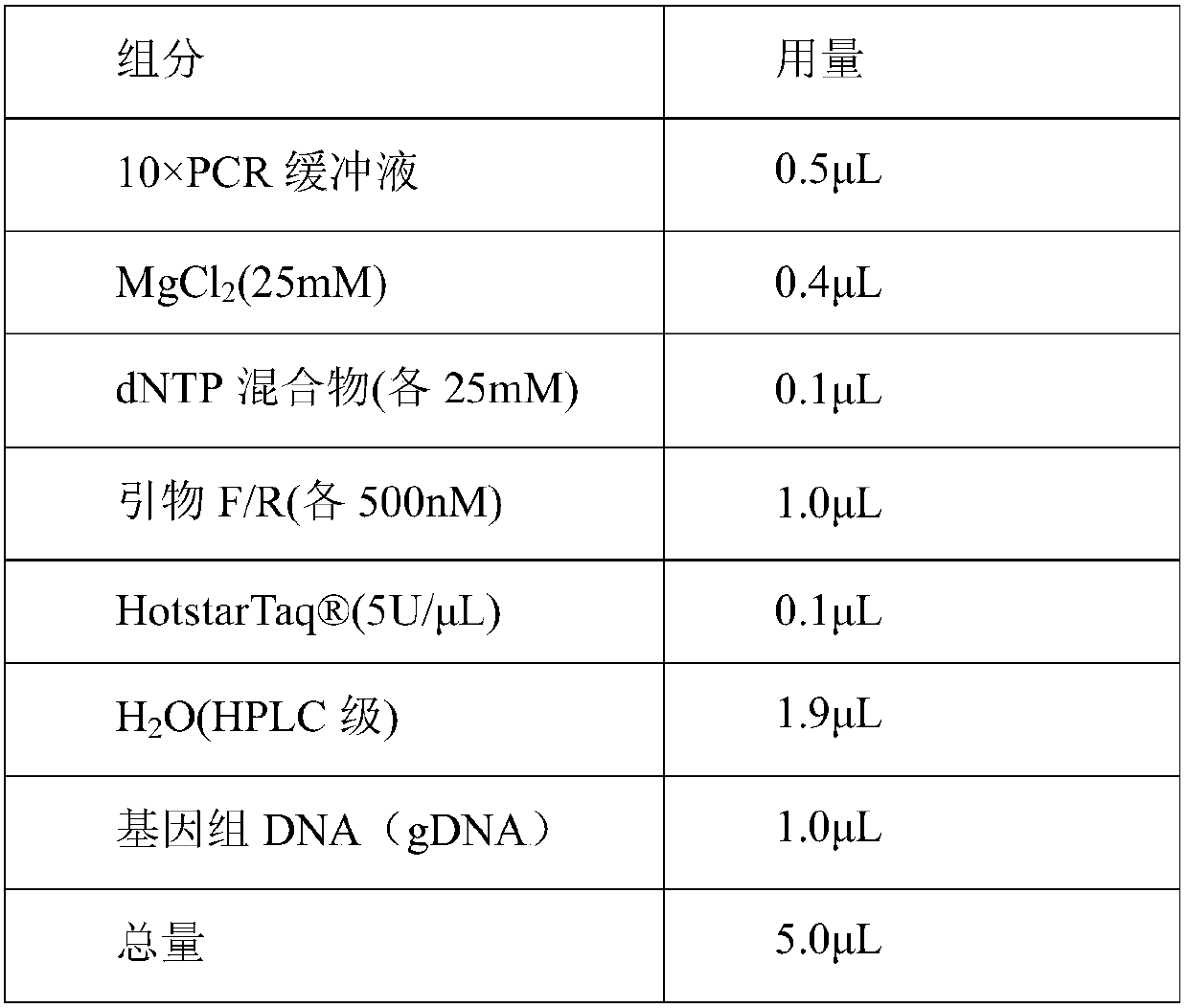
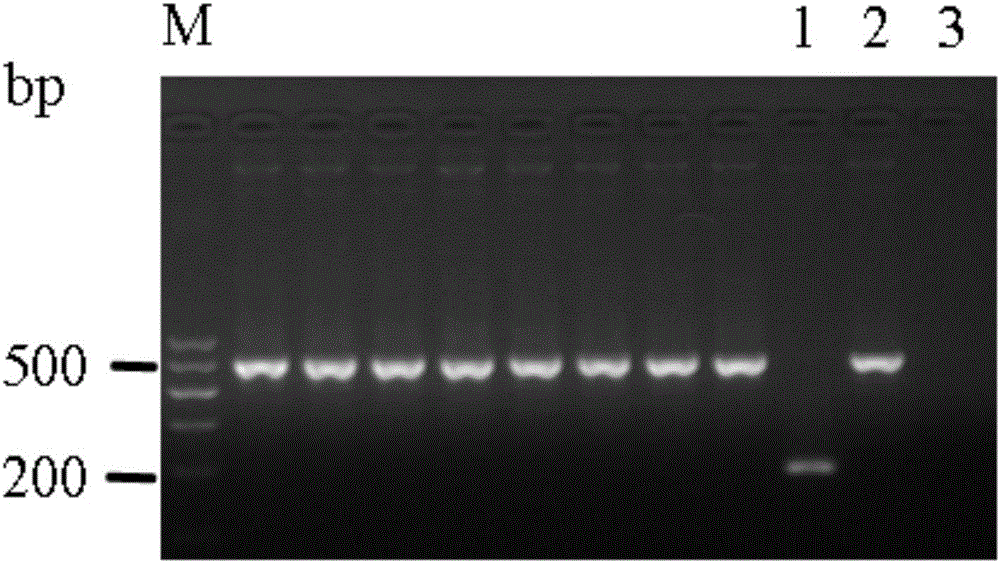
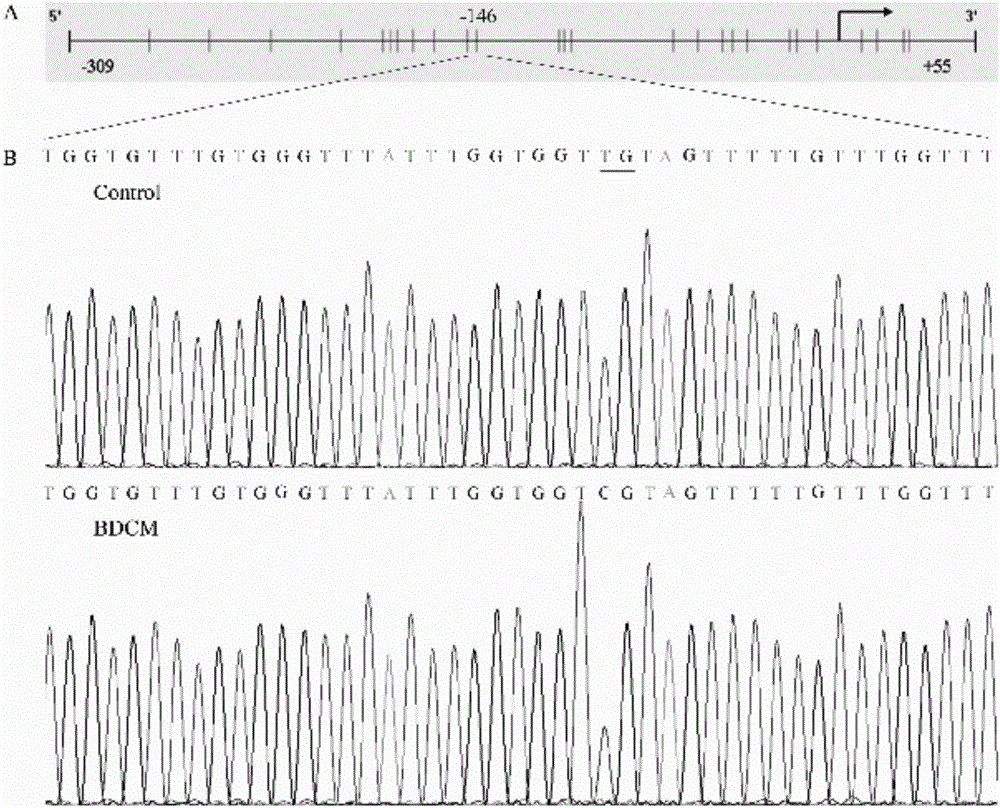
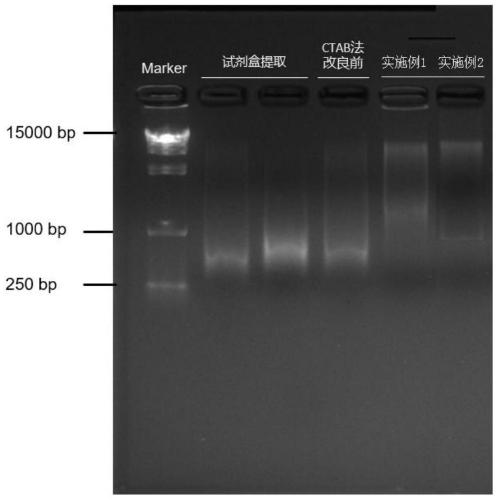
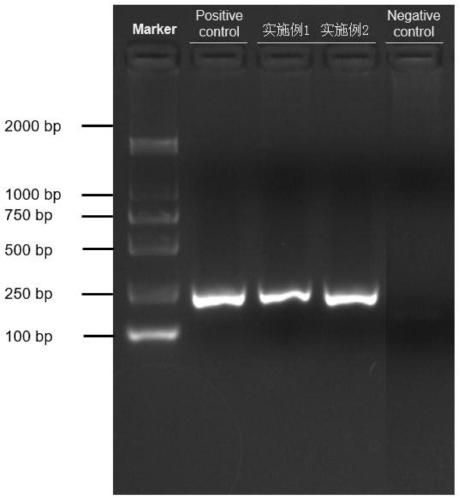
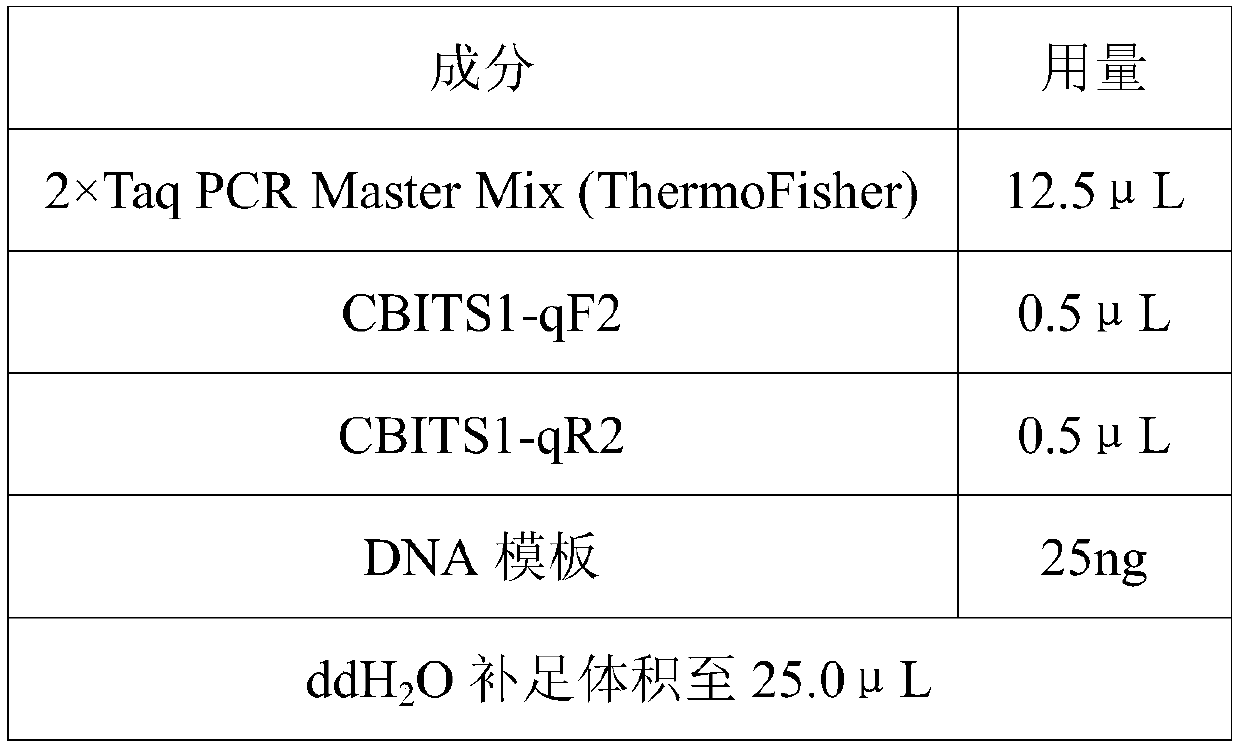
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
47 results about "Dna integrity" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA Integrity Number (DIN) is used for accurate and objective assessments of genomic DNA degradation. The broad size range allows analysis of genomic DNA samples from 200 to greater than 60,000 bp. Results may be obtained in less than 2 minutes per sample. Input sample QC allows saving of sequencing...
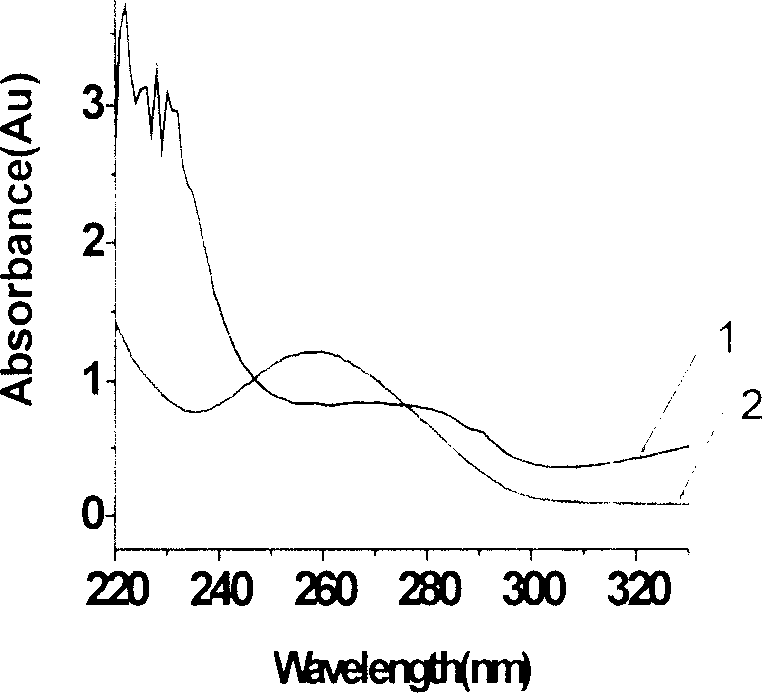
DNA integrity assay (DIA) for cancer diagnostics, using confocal fluorescence spectroscopy
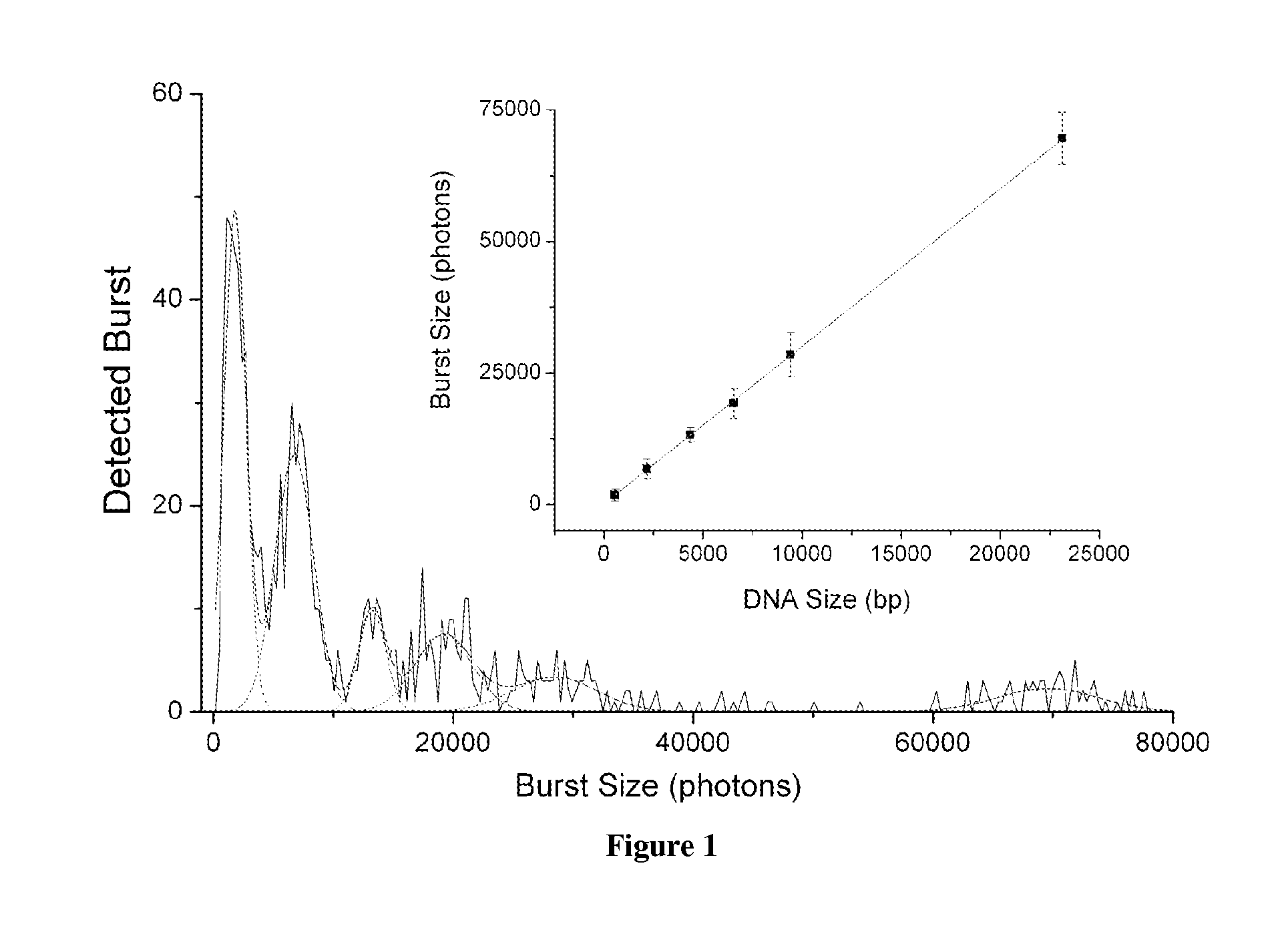
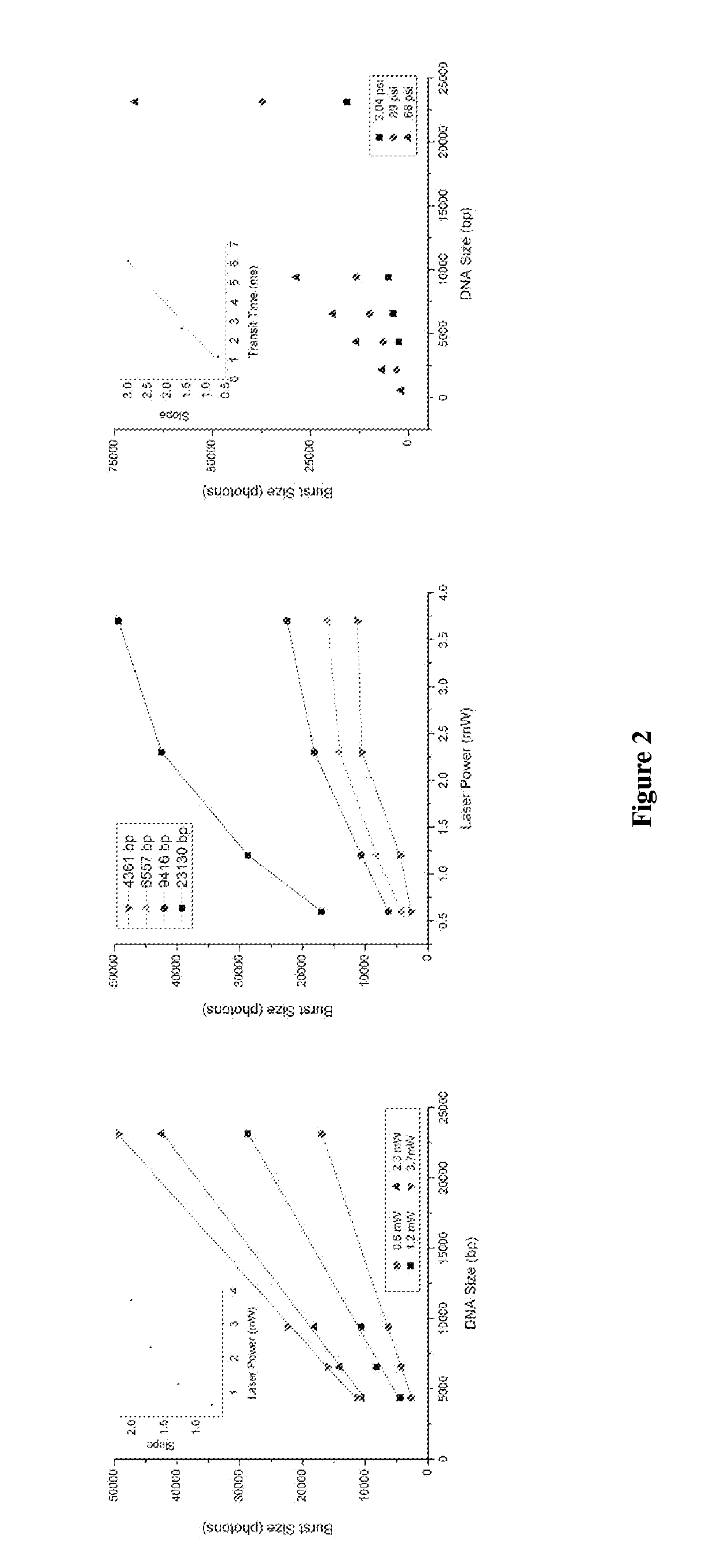
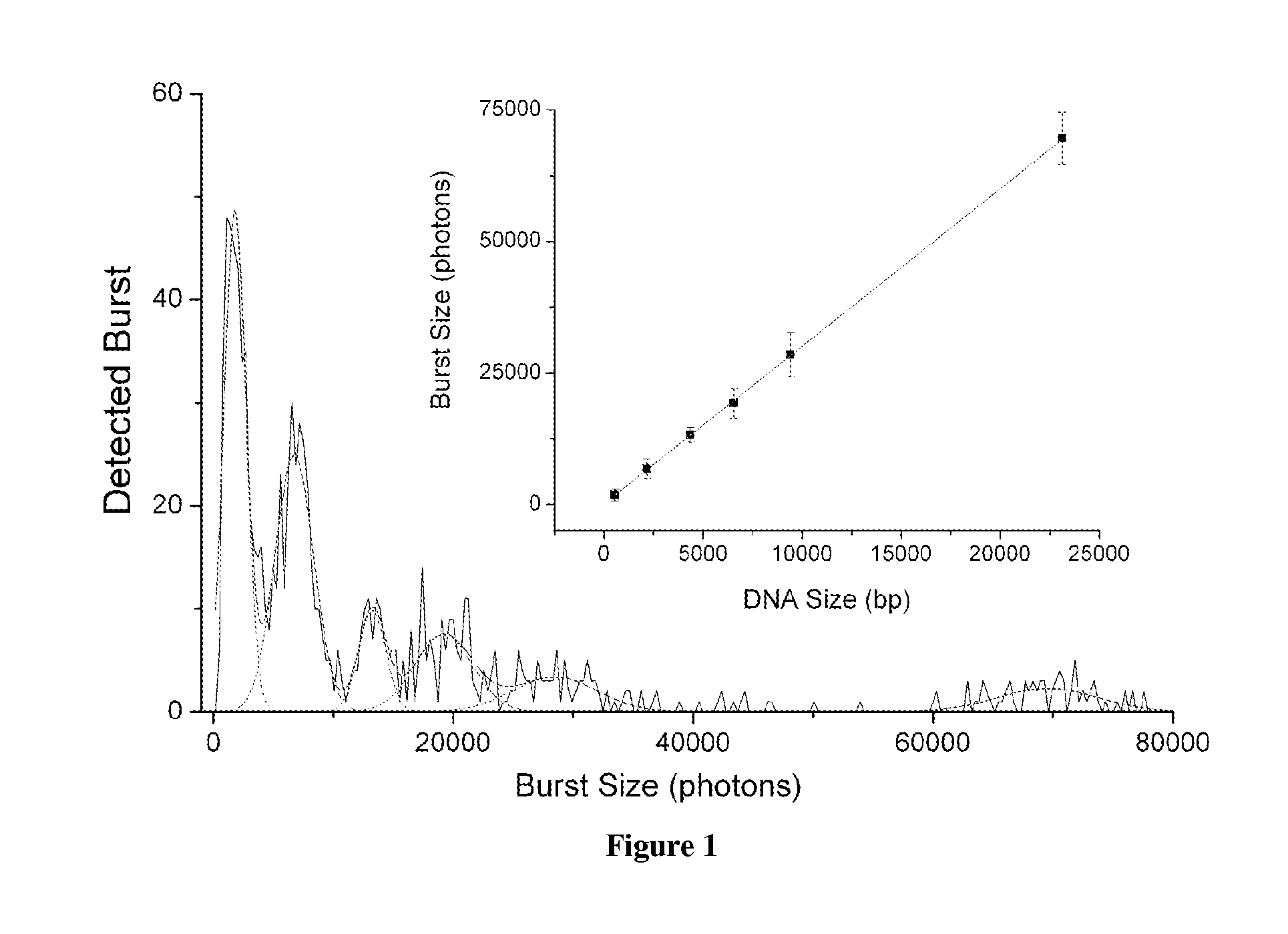
The present invention relates, e.g., to a method for determining the size distribution of DNA molecules in a sample comprising cell-free nucleic acid, comprising labeling the DNA with a fluorescent dye in a stoichiometric manner, subjecting the DNA to molecular spectroscopy (e.g., cylindrical illumination confocal spectroscopy), analyzing suitable fluorescent burst parameters of the labeled DNA, and conducting single molecule DNA integrity analysis of the labeled DNA molecules in the sample. In one embodiment of the invention, the method is used as a diagnostic method for detecting cancer.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
DNA integrity assay (DIA) for cancer diagnostics, using confocal fluorescence spectroscopy
ActiveUS20110171741A1Microbiological testing/measurementBiological testingFluorescence spectrometryMolecular spectroscopy
The present invention relates, e.g., to a method for determining the size distribution of DNA molecules in a sample comprising cell-free nucleic acid, comprising labeling the DNA with a fluorescent dye in a stoichiometric manner, subjecting the DNA to molecular spectroscopy (e.g., cylindrical illumination confocal spectroscopy), analyzing suitable fluorescent burst parameters of the labeled DNA, and conducting single molecule DNA integrity analysis of the labeled DNA molecules in the sample. In one embodiment of the invention, the method is used as a diagnostic method for detecting cancer.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Composite for preservation of human saliva and preparation method there of
InactiveCN102919218APollution suppressionAvoid pollutionDead animal preservationN-Acetyl-5-methoxytryptamineGenomic DNA
The invention relates to a composite for preservation of human saliva and a preparation method of the composite. The composite comprises the following components: Tris-HCl with pH being 6-8.5 (5-20mmol / L), EDTA (Ethylene Diamine Tetraacetic Acid) (0.5-2mmol / L), NaOAc (2.5-3.5mol / L), cane sugar (0.1-0.4mol / L), N-acetyl-5-methoxytryptamine (1-3mmol / L), propylparaben (1-4mg / mL), diazonium imidazolidinyl urea (1-3mg / mL), protease K(10mu g / mL) and a system with pH being 7.5-8.5. When used for preservation of human saliva, the composite is capable of inhibiting nuclease activity and preventing nuclease from being contaminated by external microbes or being oxidized by itself, thereby guaranteeing the integrality and the purity of genomic DNA (deoxyribonucleic acid). The composite is long in preservation period, wide in preservation temperature range and applicable to gene detection and related scientific researches.
Owner:湖北维达健基因技术有限公司
Kit for rapid extraction of plant seed DNA, and application thereof
The invention discloses a kit for rapid extraction of plant seed DNA, and an application thereof. The compositions of the kit comprise: a CTAB lysate, a silicon-based plasmalemma DNA adsorption column, an RNA enzyme, a buffer solution for passing through a column, a rinsing solution 1, a rinsing solution 2 and an eluting buffer solution; the compositions of the buffer solution for passing through the column include: 2.5-7.0 M of guanidine hydrochloride, 0.05 M of Tris-HCl, and 0.01 M of EDTA; and the pH value is 4.0-7.0. The invention further discloses the application of the kit for the rapid extraction of the high quality plant seed DNA. The kit can effectively improve yield and purity of the extraction of the plant seed DNA, has the extracted DNA with good integrity, high purity and high yield; while decreasing DNA extraction cost, the kit effectively improves extraction quality and extraction efficiency of the plant seed genomic DNA.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
Method for purifying DNA by using gold magnetism particles
ActiveCN101165182ALarge specific surface areaLarge fixed capacityDNA preparationMetal particleMicroparticle
The DNA purifying magnetic metal particle process includes the following steps: 1. preparing liquid sample containing DNA; 2. mixing magnetic metal particle, which is composite magnetic particle comprising a core of magnetic nanometer particle and a shell of Au, Ag or other noble metal, and has average size of 0.05-100 microns, and the liquid sample to form magnetic metal particle-DNA composition; 3. applying external magnetic field to separating DNA adsorbing magnetic metal particle from other components in the liquid sample; and 4. eluting DNA adsorbed onto the surface of the magnetic material. The process can separate and purify DNA effectively from various kinds of sample, and has the advantages of high purification rate, high DNA integrity, high purity and convenient operation.
Owner:XIAN GOLDMAG NANOBIOTECH
Method for quickly extracting fungal DNA for PCR amplification
InactiveCN101696411AReduce lossesImprove extraction efficiencyMicroorganism based processesDNA preparationBiotechnologyMolecular identification
The invention discloses a method for quickly extracting fungal DNA for PCR amplification, which comprises the steps of: (1) culturing fungi to obtain fungal mycelia; (2) placing the fungal myceliua into a centrifugal tube filled with a lysate; (3) alternately freezing and thawing the centrifugal tube; and (4) centrifuging the thawed fungal mycelia, taking a supernatant, adding 95 percent ethanol into the supernatant, mixing the mixture evenly, quickly freezing the mixture by liquid nitrogen, centrifuging the mixture, removing a supernatant, and air-drying a precipitate to obtain the fungal DNA. The method has simple operation, is time-saving and labor-saving, reduces the loss of the fungal DNA during the grinding, does not need phenol and chloroform for extraction, and avoids the contact of a toxic reagent. The DNA extracted by the method has good integrity, high purity and high yield, can be directly used for a PCR amplification reaction, and can efficiently amplify a target product for cloning and sequencing. The method improves the extraction efficiency of the fungal DNA, reduces the cost for the extraction of the fungal DNA, and can effectively improve the molecular identification and detection efficiency of the fungi.
Owner:INST OF FOREST ECOLOGY ENVIRONMENT & PROTECTION CHINESE ACAD OF FORESTRY
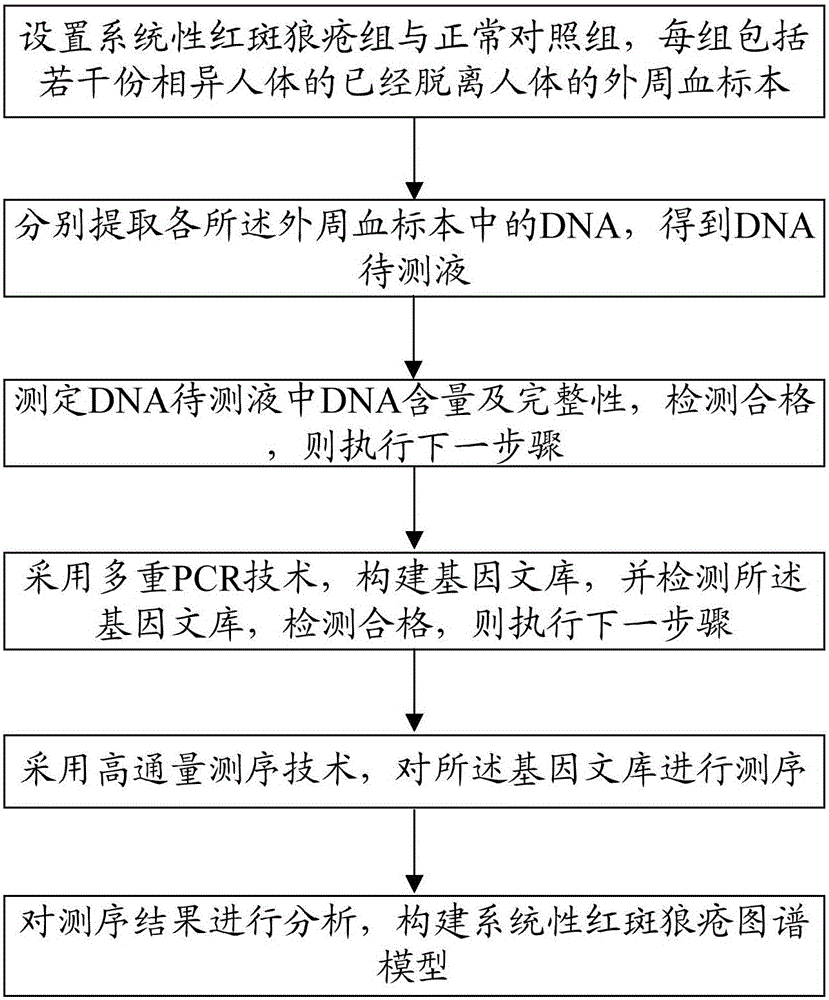
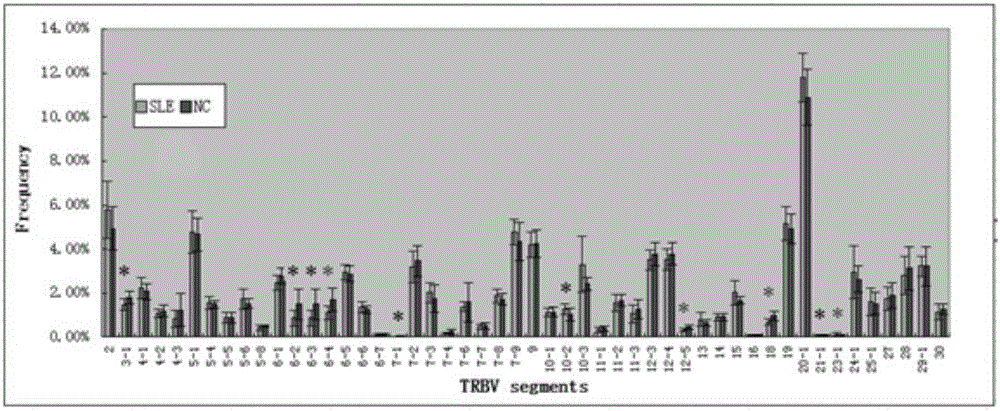
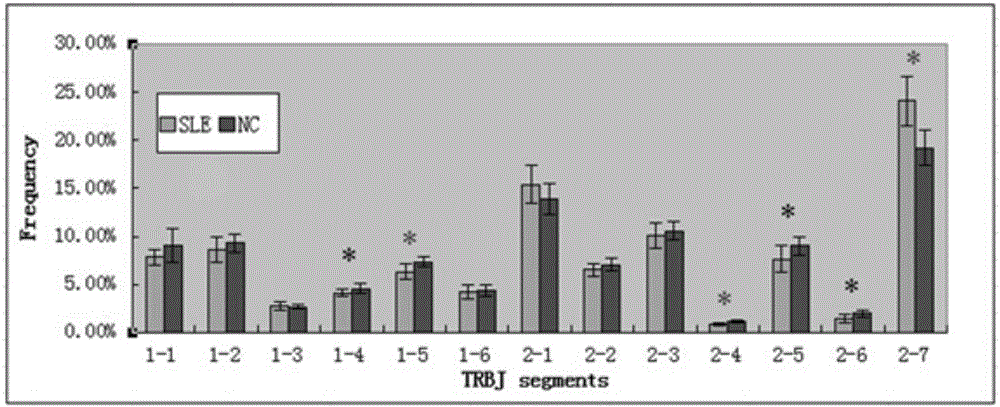
Construction method of systemic lupus erythematosus map model
ActiveCN105040111AReasonable designEfficient acquisitionMicrobiological testing/measurementLibrary creationHuman bodyPeripheral blood specimen
The invention relates to a construction method of a systemic lupus erythematosus map model. The construction method comprises: setting a systemic lupus erythematosus group and a control group, wherein each group comprises a plurality of peripheral blood samples isolated from human body and derived from different people; respectively extracting DNA from each peripheral blood sample to obtain DNA liquids to be detected; determining the DNA contents and the DNA integrity in the DNA liquids to be detected, detecting whether the sample is qualified, and performing the next step if the sample is qualified; constructing a gene library by using a multiplex PCR technology, detecting whether the gene library is qualified, and performing the next step if the gene library is qualified; carrying out sequencing on the gene library by using a high-throughput sequencing technology; and analyzing the sequencing results, and constructing the systemic lupus erythematosus map model. According to the construction method of the systemic lupus erythematosus map model of the present invention, the design is reasonable and feasible, and the SLE related information adopted as the middle results can be effectively obtained.
Owner:中国人民解放军联勤保障部队第九二四医院
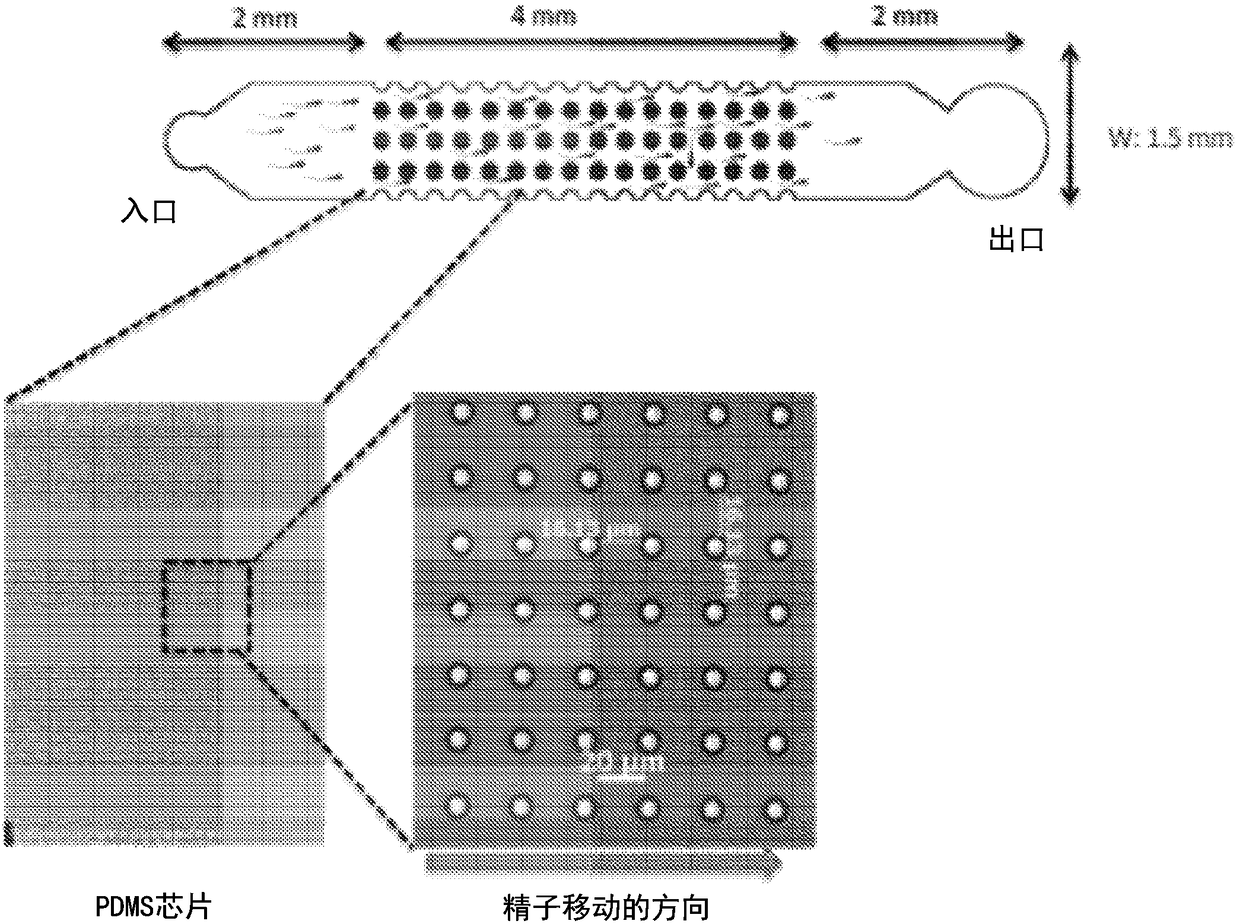
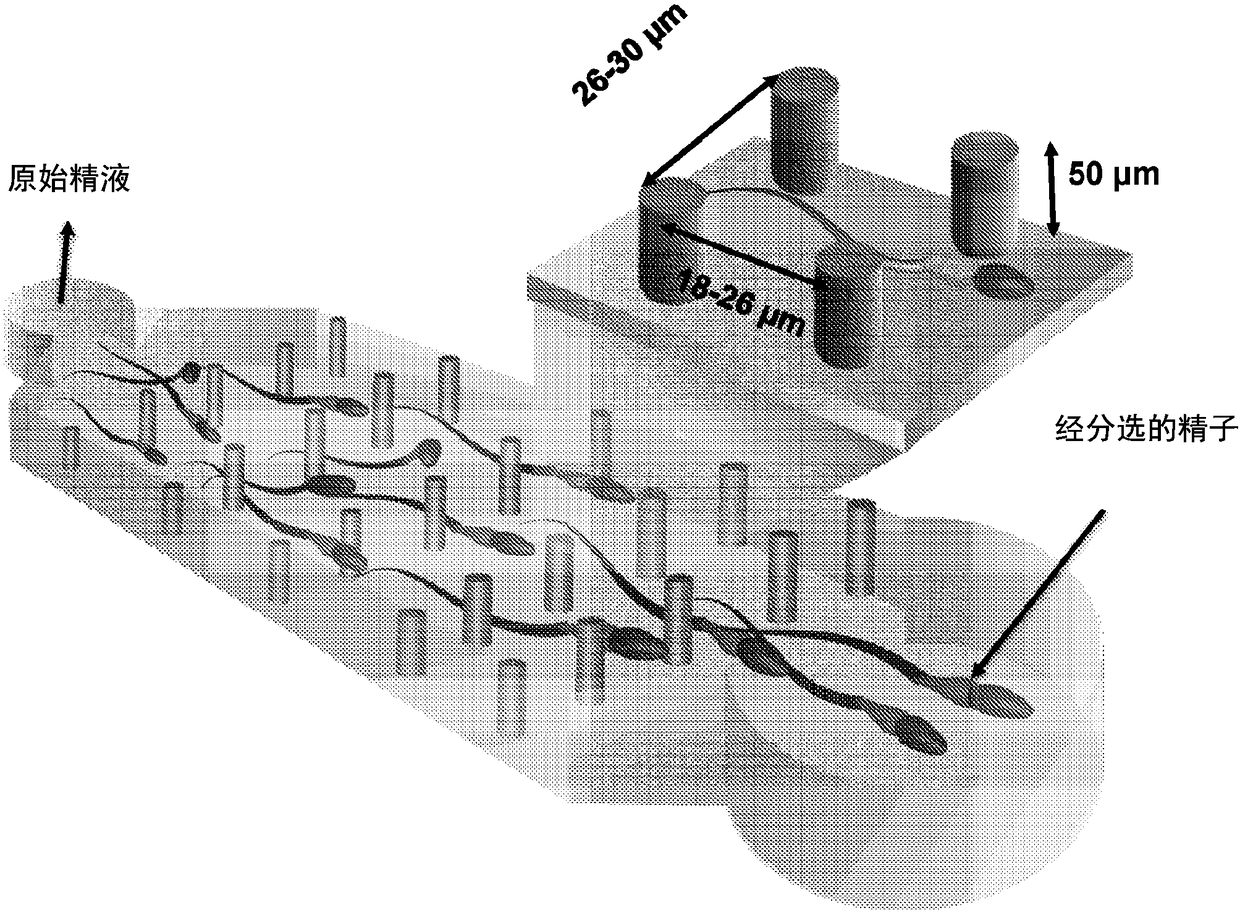
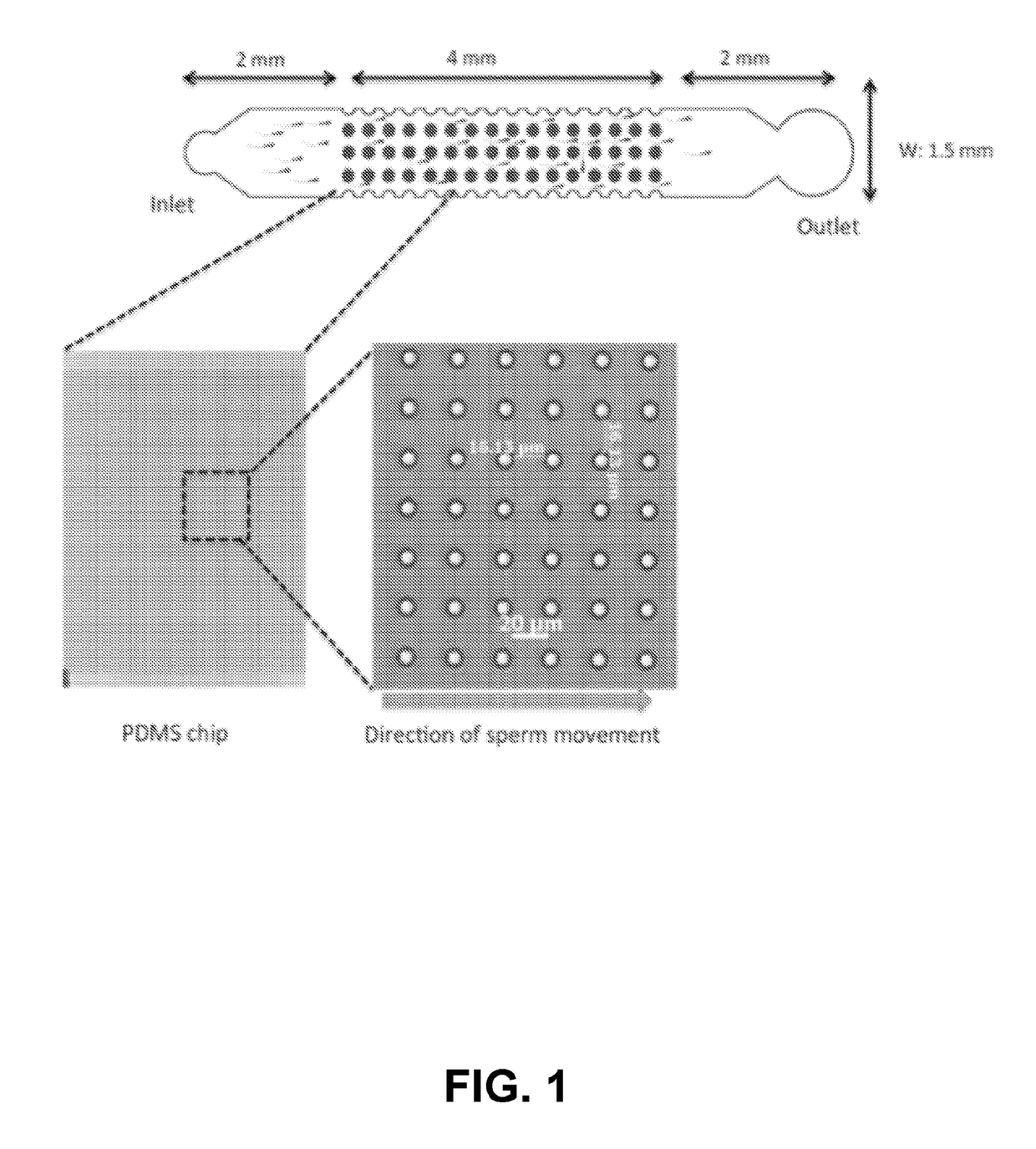
A micro-fluidic device for selective sorting of highly motile and morphologically normal sperm from unprocessed semen
A microfluidic chip is provided for self-sorting highly motile, morphologically normal sperm cell with high DNA integrity from a fresh semen sample. The sperm self-sorting microfluidic chip has one ormore inlet chambers, and sperm collection outlet chamber(s), and the middle of the channel features various micro-fabricated structures in different geometrical shapes and orientations, with varyingperiodicities and patterns, such as an array of micro-fabricated pillars that facilitate the transport of the active and healthy sperm into the outlet chamber.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV +1
Burkholderia gladioli molecular standard sample and preparation method thereof
InactiveCN105907753ASolve the shortage situationSolve the preparation technologyMicrobiological testing/measurementDNA preparationPenicillinGladiolus
The invention discloses a preparation method of a burkholderia gladioli molecular standard sample. The preparation method comprises the step of carrying out freeze drying on extracted genome nucleic acid of burkholderia gladioli, wherein the freeze drying conditions are as follows: a freeze dryer is started, and a cooling trap is started when temperature of a drying chamber is lowered to -35 DEG C; when the temperature of the cooling trap is lowered to -40 DEG C, a nucleic acid sample prefrozen for 2 hours at the temperature of -80 DEG C is put into the drying chamber and then vacuumized; when vacuum degree is reduced to 0.5Torr, freezing is finished; and the sample is dried at the temperature of 15 DEG C, the sample is taken out when the vacuum degree is reduced to 0.1Torr, a penicillin bottle is tightened to obtain a burkholderia gladioli genome nucleic acid standard sample, and the sample is kept in a dark place and stored at the temperature of 20 DEG C. The molecular standard sample disclosed by the invention has good physical and chemical properties and can be detected by adopting a specific primer through PCR process, and sensitivity reaches 10<-4>. Detection test shows that the standard sample has good DNA integrity, uniformity and stability, can be applied to import and export detection as a positive reference material, and accuracy of a detection result is improved.
Owner:INSPECTION & QUARANTINE TECH CENT SHANDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
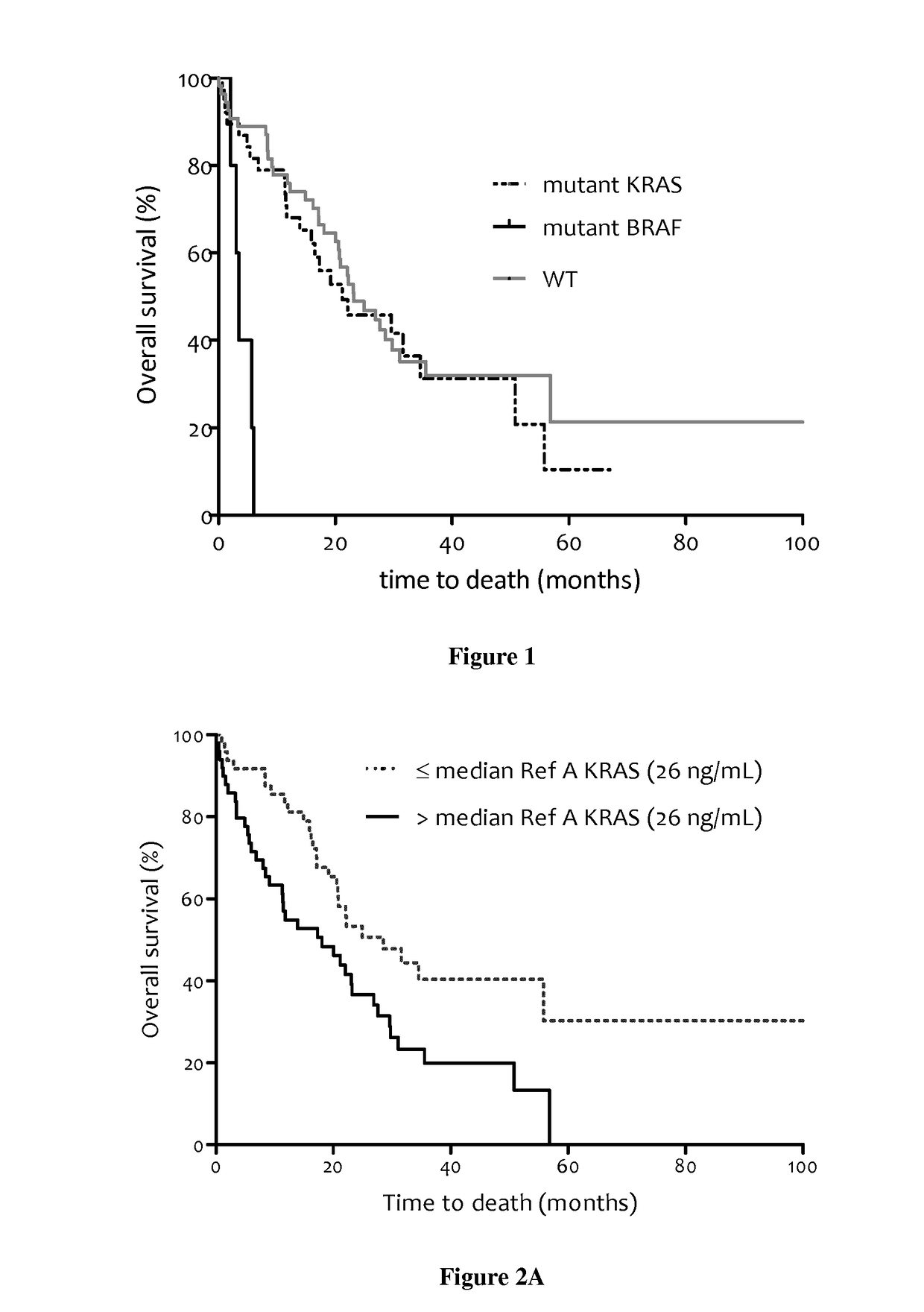
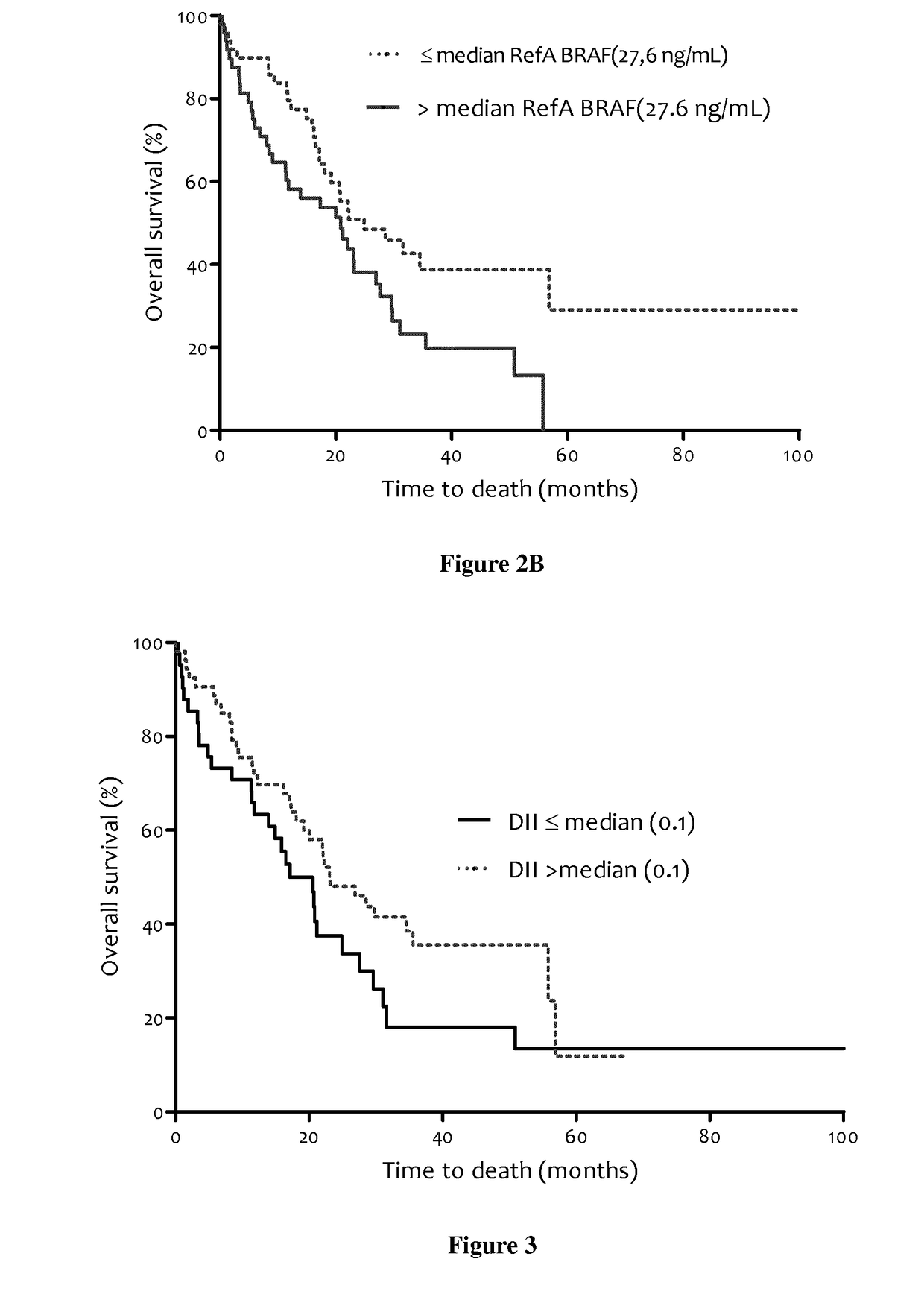
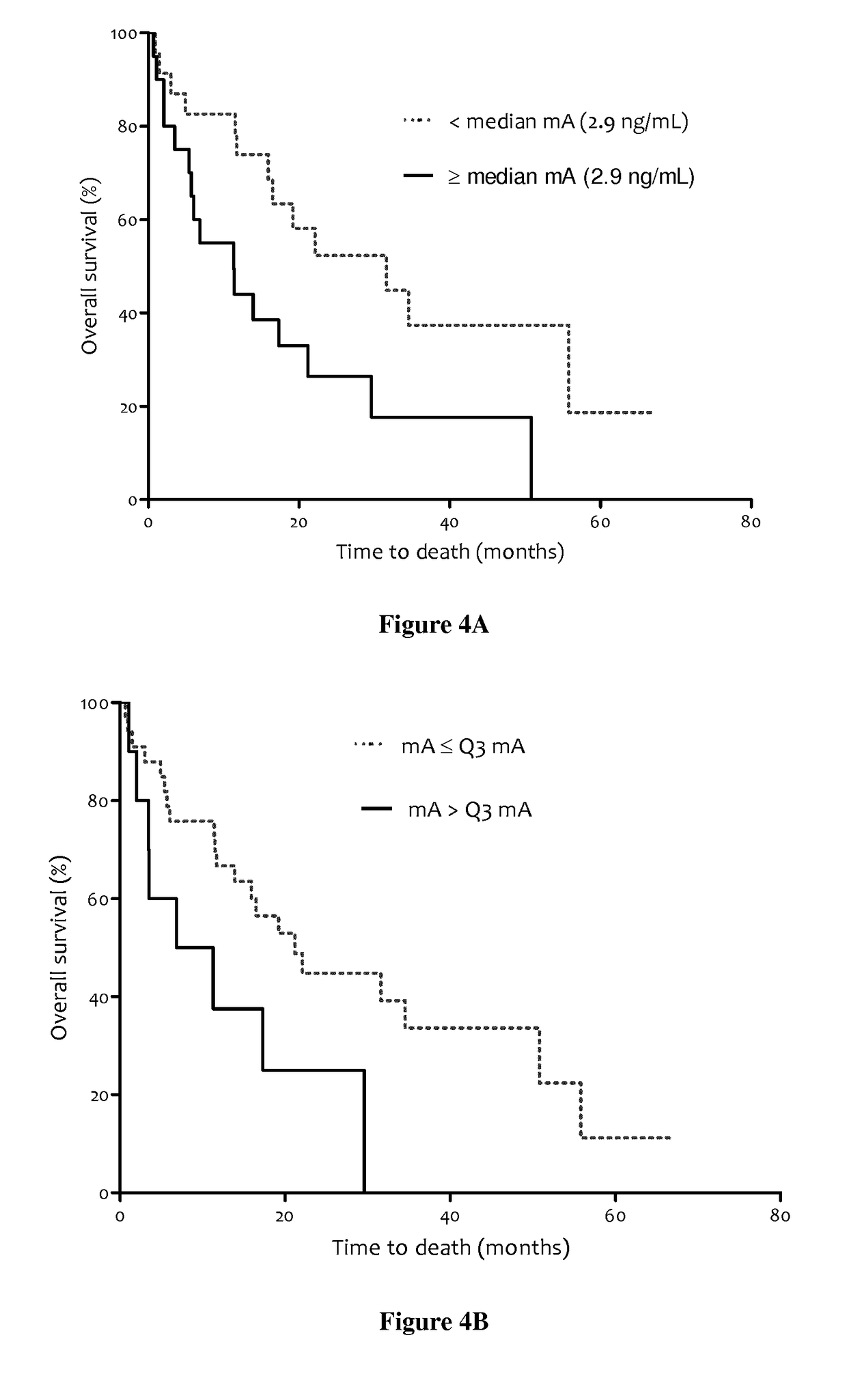
Methods for predicting the survival time of patients suffering from cancer
InactiveUS20170183742A1Short survival timeKeep for a long timeMicrobiological testing/measurementPatient survivalBiology
The present invention relates to methods for predicting the survival time of patients suffering from cancer. Said methods are based on the quantification and analysis of the cell free nucleic acids that are present in a sample from the patient and typically include the determination of the level of the mutant nucleic acid which contains a mutation of interest, the calculation of the mutation load for said mutation of interest, the calculation of the DNA integrity index or a combination thereof.
Owner:INST NAT DE LA SANTE & DE LA RECHERCHE MEDICALE (INSERM) +2
Micro-fluidic device for selective sorting of highly motile and morphologically normal sperm from unprocessed semen
A microfluidic chip is provided for self-sorting highly motile, morphologically normal sperm cell with high DNA integrity from a fresh semen sample. The sperm self-sorting microfluidic chip has one or more inlet chambers, and sperm collection outlet chamber(s), and the middle of the channel features various micro-fabricated structures in different geometrical shapes and orientations, with varying periodicities and patterns, such as an array of micro-fabricated pillars that facilitate the transport of the active and healthy sperm into the outlet chamber.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV +1
Oral swab preservation solution as well as preparation method and application thereof
An oral swab preservation solution comprises the following components: 5-50 mmol / L of Tris-HCl, 1-10 mmol / L of EDTA, 50 to 500 mmol / L of sodium chloride and 0.1 to 3% of SDS, wherein the pH range of the system ranges from 7 to 10. The oral swab preservation solution s clear in component, simple in preparation method, low in cost, good in stability and high in safety, free of toxic and side effects, easy to operate, convenient to store and transport, suitable for large-scale sampling and use, and wide in application prospect. The oral swab preservation solution is used for storing a human mouthswab sample, can be preserved at the temperature of 15-25 DEG C (room temperature.) for a long term, genomes in oral epithelial cells can be effectively protected from being damaged in a plurality ofweeks, the integrity of DNA (deoxyribonucleic acid) is good, and the oral swab sample is not polluted by microorganisms.
Owner:CHEERLAND BIOTECH CO LTD
Quick extraction method of plant genomic DNA (Deoxyribonucleic Acid)
ActiveCN107475248AHigh purityImprove integrityMicrobiological testing/measurementDNA preparationEnzyme digestionCell membrane
The invention discloses a quick extraction method of plant genomic DNA (Deoxyribonucleic Acid). The quick extraction method is characterized by comprising the following steps of (1), taking a plant tissue sample, adding liquid nitrogen, grinding the plant tissue sample into powder and collecting into a centrifugal tube, and the like. SDS (Sodium Dodecyl Sulfonate) in a Buffer A in the quick extraction method can be used for cracking a cell membrane to denature protein; a Buffer B can act with the SDS; impurities of the protein, a polysaccharide and the like can be effectively removed; moreover, PVP40 (Polyvinyl Pyrrolidone 40) in the Buffer A can be used for effectively removing substances of the polysaccharide and polyphenol in plant tissue; the purity of the DNA is improved; the completeness of the DNA is enabled to be better; the quick extraction method can be directly used for a PCR (Polymerase Chain Reaction) and various enzyme digestion reaction; meanwhile, the quick extraction method does not need to use organic solvents of phenol chloroform and the like in the entire extraction process; therefore, steps of ethanol precipitation and the like do not need to be carried out; the extraction method provided by the invention, compared with a conventional extraction method, can be used for more quickly, simply and conveniently extracting the DNA; the operation, on a single sample, of the quick extraction method can be completed within 1 hour and the extraction time is greatly shortened.
Owner:成都晟博源生物工程有限公司
Method for extracting edible mushroom DNA
The invention discloses a method for extracting edible mushroom DNA. The method comprises the following steps: (1) getting material: putting edible mushroom hyphae into a buffer solution; (2) heating: heating at the temperature of 90 to 110 DEG C; (3) centrifuging: centrifuging at the speed of 7000 to 9000 rpm for 5 to 10 min, and then extracting a supernatant liquor, wherein the supernatant liquor contains total DNA. The method for extracting the edible mushroom DNA is short in used time, simple and convenient to operate, low in cost and free from contamination to the environment, the prepared DNA is good in integrality and suitable for PCR reaction.
Owner:SHANGHAI ACAD OF AGRI SCI
Wu-he dipsacus asper ISSR-PCR molecule marking method
InactiveCN106434988AGood polymorphismReduce stepsMicrobiological testing/measurementDipsacus asperAgarose electrophoresis
The invention relates to a wu-he dipsacus asper ISSR-PCR molecule marking method. The method is characterized by comprising the following steps: 1, extracting the genome of the wu-he dipsacus asper, 2, conducting 1% agarose electrophoresis for 20 minutes, testing the integrity of the genome, at the same time using the DNA bar code ITS2 primer to conduct the PCR amplification of the extracted wu-he dipsacus asper DNA, verifying the purity of the extracted DNA, 3, based on the initial screened primer conducting the ISSR-PCR. Compared with the prior art, the reaction system created by the method has a simple optimization method. The amplified bands is both clear and stable with good polymorphism. The method compensates the shortcomings in the study of the genetic diversity of the wu-he dipsacus asper. The method can be used in the analysis of the genetic relationship and the verification of authenticity of the teasle, thus having good application value.
Owner:湖北省农业科学院中药材研究所
Amino acid buffer solution and plasma free DNA extraction kit
ActiveCN110129314AHigh recovery rateIncrease the adsorption areaMicrobiological testing/measurementDNA preparationShear injuryPolyethylene glycol
The invention discloses an amino acid buffer solution and a plasma free DNA extraction kit. The plasma free DNA extraction kit comprises the agents of a lysis solution, a proteinase K working liquid,an amino acid adsorption buffer solution, a washing solution, and a TE eluent; the amino acid adsorption buffer solution comprises 80-200 mM of amino acids, 0.1-0.3 M of a chaotropic salt, 5-15% of PEG (polyethylene glycol), 0.5-1.2 M of sodium chloride, 40-100 mM of Tris-HCl, and 20-50 mM of EDTA (ethylenediaminetetraacetic acid), and has pH of 2.1-2.4. the amino acids are combined with the chaotropic salt having a low concentration; pH of the buffer solution is controlled; therefore, extraction efficiency is improved. The use of PEG helps reduce shear injury of DNA and ensure DNA integrity.Therefore, extraction of free DNAs in plasma is free of inhibitor residue, has high quality, is simple to perform, and has low cost.
Owner:合肥欧创基因生物科技有限公司
Method for extracting microorganism total DNA (Deoxyribonucleic Acid) in pu'er tea piling fermentation process
The invention discloses a method for extracting microorganism total DNA (Deoxyribonucleic Acid) in a pu'er tea piling fermentation process at high quality. The method comprises the following steps of: separating microorganisms on the surface of pu'er tea by using ultrasonic wave and vortex oscillation; washing the microorganisms by using washing buffer solution; performing rough extraction on the microorganism total DNA by using DNA extract; and purifying the roughly extracted DNA. The method provided by the invention is low in cost and high in DNA extraction rate; the extracted DNA has high completeness and high purity and can meet the requirements of PCR (Polymerase Chain Reaction) without purification or dilution; the purified DNA can meet the requirements of molecular biology research; and meanwhile, the extracted DNA covers bacteria and fungi and the requirements of subsequent researches in microorganism diversity, functional genes, metagenomics and the like can be met.
Owner:KUNMING UNIV OF SCI & TECH
Extraction buffer solution and extraction method for extracting DNA from bones or teeth
InactiveCN108456676AImprove integrityHigh purityDNA preparationSodium Lauryl SarcosinateRoom temperature
The invention discloses a DNA extraction buffer solution which is formed by mixing a TES buffer solution, a sodium lauryl sarcosinate solution, a 1 mol / L DTT solution and a 10 mg / mL proteinase K solution in a volume ratio of (4-5):1:1:1. The invention also discloses a method for extracting DNA from bone or tooth samples in the presence of the DNA extraction buffer. The method comprises the following steps: cleaning impurities, pulverizing a sample at room temperature, adding the extraction buffer solution to cleave, adding an adsorption solution and adsorption beads for adsorption, rinsing adsorption beads, and adding an eluent for eluting. The invention also discloses a method of pretreating a sample during extraction of DNA from bones or teeth. The DNA extraction buffer solution and thecorresponding use method disclosed by the invention have the advantages thatthe efficiency of extracting DNA from bones and teeth is high, the extracted DNA is of good integrity and high purity, the operation is simple, rapid and safe, samples are free of crosscontamination, and the cost is low.
Owner:黄书琴
Composite for preservation of human saliva and preparation method thereof
InactiveCN102919218BGuaranteed stabilityIntegrity guaranteedDead animal preservationN-Acetyl-5-methoxytryptamineGenomic DNA
The invention relates to a composite for preservation of human saliva and a preparation method of the composite. The composite comprises the following components: Tris-HCl with pH being 6-8.5 (5-20mmol / L), EDTA (Ethylene Diamine Tetraacetic Acid) (0.5-2mmol / L), NaOAc (2.5-3.5mol / L), cane sugar (0.1-0.4mol / L), N-acetyl-5-methoxytryptamine (1-3mmol / L), propylparaben (1-4mg / mL), diazonium imidazolidinyl urea (1-3mg / mL), protease K(10mu g / mL) and a system with pH being 7.5-8.5. When used for preservation of human saliva, the composite is capable of inhibiting nuclease activity and preventing nuclease from being contaminated by external microbes or being oxidized by itself, thereby guaranteeing the integrality and the purity of genomic DNA (deoxyribonucleic acid). The composite is long in preservation period, wide in preservation temperature range and applicable to gene detection and related scientific researches.
Owner:湖北维达健基因技术有限公司
Method for purifying genomic deoxyribonucleic acid (DNA) of plant seeds by using goldmag particles
The invention provides a method for simply and quickly purifying genomic deoxyribonucleic acid (DNA) of various plant seeds by using a magnetic nano composite material. The method comprises the following steps of: performing sample lysis, namely adding 200 to 800 microliters of PVP (poly vinyl pyrrolidone) with mass-to-volume ratio of 2 to 5 percent, 2 to 5 percent of CTAB (cetyl trimethyl ammonium bromide) and beta-mercaptoethanol with volume fraction of 0.5 to 2 percent into a plant seed sample; blending, and performing full lysis for 5 to 30 min in a water bath; and adding 10 to 50 microliters of 10 mg / ml RNase solution, and thus obtaining sample lysis solution with the genomic DNA. The lysis solution is suitable for all plant seeds. The obtained genomic DNA does not contain an inhibitor which inhibits a downstream polymerase chain reaction (PCR); and DNA has high quality, high yield and high integrity.
Owner:XIAN GOLDMAG NANOBIOTECH
Molecular standard sample of rape stem canker pathogen and its preparation method
InactiveCN102277430AImprove integrityImprove uniformityMicrobiological testing/measurementMicroorganism based processesBiotechnologyRapeseed
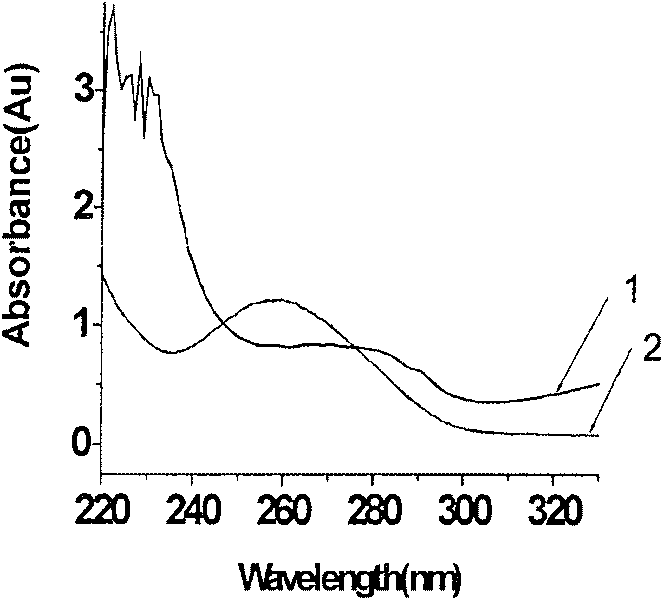
The invention belongs to the field of plant quarantine technology research. Molecular standard sample of canker canker of rapeseed. Rape canker sore molecular standard sample is prepared from high-purity DNA extracted from canker canker sore strain. The standard sample has the following physical and chemical properties: (1) shape: pure white powder; (2) each tube Content 5μg±0.5μg; (3) DNA purity: OD260 / 280=1.8~2; (4) Soluble in water or TE (Tris-EDTA) buffer. The rapeseed canker sore molecular standard sample of the present invention uses the rapeseed canker sore strain isolated by the laboratory as a raw material, and is cultured in large quantities under specific conditions to extract high-purity DNA, which is subpackaged and freeze-dried to obtain the rapeseed canker sore strain. Molecular standard product, with excellent physical and chemical properties, can be detected by PCR method with specific primers, and the sensitivity can reach 10-4 times. After testing, the standard product has good DNA integrity, uniformity and stability, and can be used in import and export testing as a positive reference substance to improve the accuracy of test results.
Owner:李鑫 +3
Reagent for extracting total DNA of lucid ganoderma and application of reagent
The invention relates provides a reagent for extracting total DNA of lucid ganoderma and the application of the reagent. The total DNA is rapidly and efficiently extracted from about 50 mg of myceliumand sporocarp sample by adopting the steps of crushing, cracking, DNA adsorption, rinsing, eluting and the like; for fresh dry samples, and total DNA suitable for ITS identification and other molecular biology experiments also can be extracted. Further experiment proves that the method also can be applied to edible-medicinal fungi such as cordyceps militaris, agaricus blazei murill and grifola frondosa. According to the method, the extraction time of single sample is shortened to 15 minutes; in addition, the extracted DNA has the advantages of good integrity and high purity, and can be directly used for downstream molecular biology experiments such as PCR, Real-Time PCR and molecular markers.
Owner:FUJIAN AGRI & FORESTRY UNIV
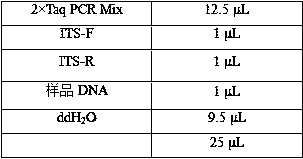
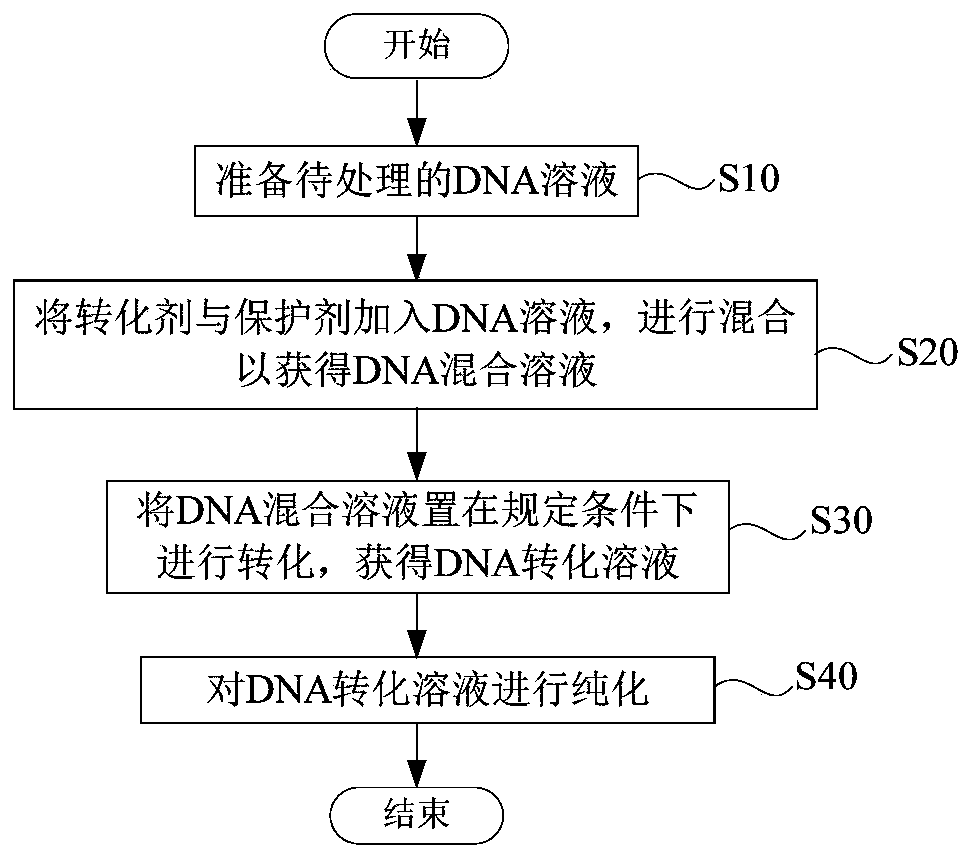
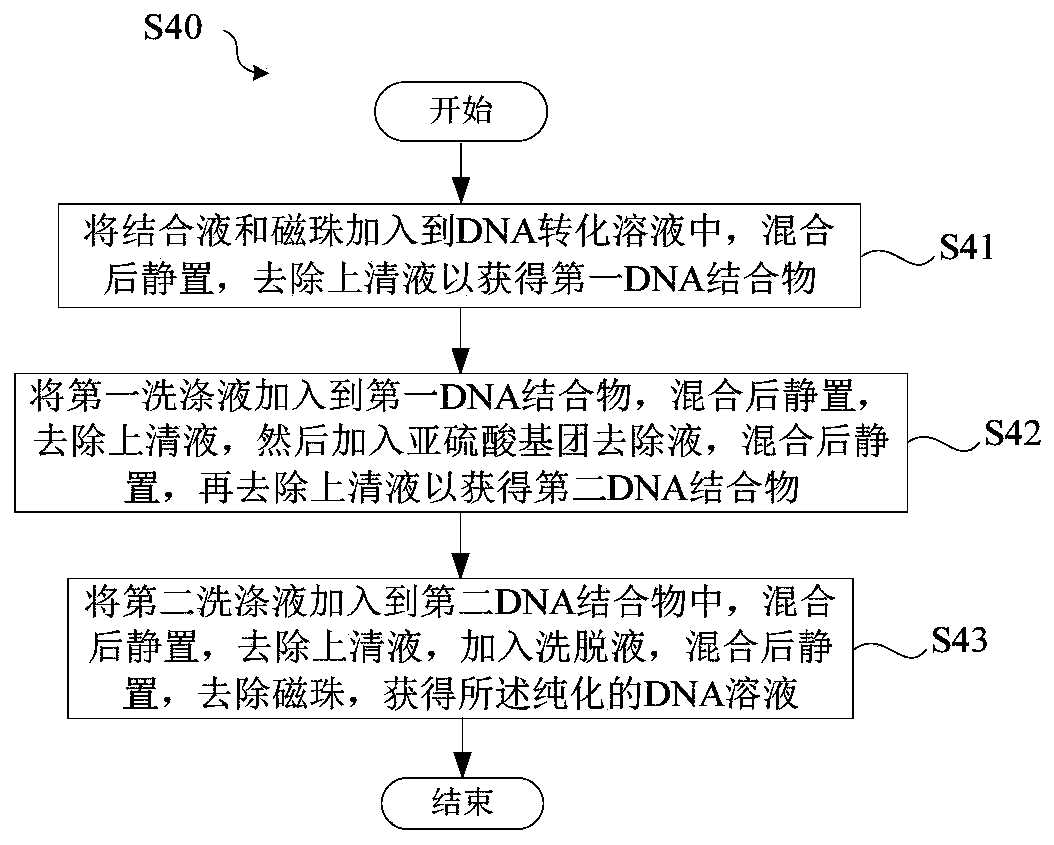
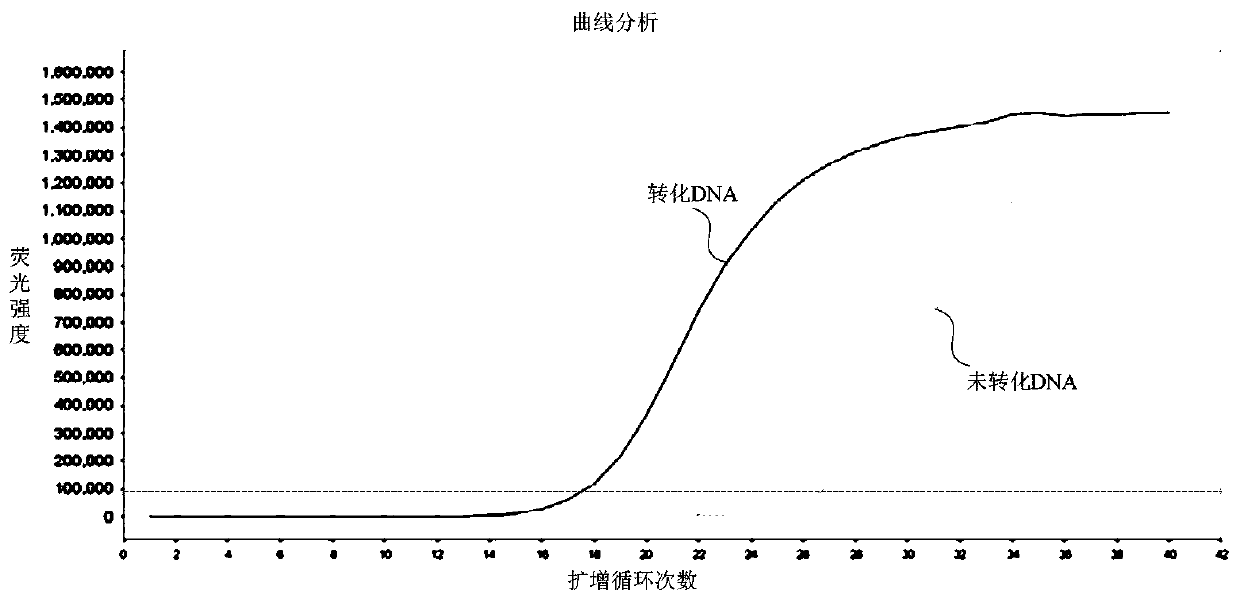
DNA sulfite conversion treatment method and kit
InactiveCN110079521AEfficient sulfite conversionRapid sulfite conversionMicrobiological testing/measurementDNA preparationDNA SolutionsSodium metabisulfite
The invention provides a DNA sulfite conversion treatment method. The method comprises the following steps of (a) preparing a DNA solution to be treated; (b) adding a conversion agent and a protectionagent into the DNA solution, and mixing to obtain a DNA mixed solution, wherein the conversion agent is a water solution of at least one of materials selected from sodium metabisulfite, sodium hydrogen sulfite, amine hydrogen sulfite, magnesium bisulfite and ammonium sulfite; the protection agent is a water solution of at least one of ingredients of hydroquinone and water-soluble vitamin E and mycose or agarose; (c) converting the DNA mixed solution under the specified condition to obtain a DNA conversion solution. According to the disclosure, the method with the advantages that the DNA sulfite conversion can be efficiently and fast realized, and the DNA integrity can be effectively ensured can be provided.
Owner:上海埃文生物科技有限公司
Biopsy culture solution and preparation method thereof
The invention belongs to the technical field of human assisted reproduction and relates to biopsy culture solution and a preparation method thereof. The biopsy culture solution is prepared by dissolving sucrose in concentration of 0.05-0.3mol / L into salt solution containing antibiotics and an indicator. By dissolution of sucrose in low molar ion into buffer salt solution without calcium and magnesium ions, embryo activity and plasma membrane integrality can be protected gently, embryo cell contraction is realized, adhesion to a biopsy needle is avoided, DNA integrality and activity can be improved after embryo biopsy, biopsy activity is protected, and accordingly biopsy sample detection effects are guaranteed, and the embryo implantation rate is increased. In addition, biopsy sample detection and assisted reproduction technology safety can be strongly guaranteed.
Owner:瑞柏生物(中国)股份有限公司
Reagent for extracting total DNAs of Grifola frondosa and application thereof
The invention provides a reagent for extracting total DNAs of Grifola frondosa and an application thereof. Total DNAs are fast and efficiently extracted from about 50mg of mycelium and sporocarp samples of Grifola frondosa through crushing and cleavage, DNA adsorption, rinsing and elution. The extraction time of a single sample is shortened to 15min, and the extracted DNAs have good integrity andhigh purity and can be directly used for downstream molecular biology experiments such as PCR, Real-Time PCR and molecular labeling. The toxic reagents such as chloroform, phenol and beta-mercaptoethanol are not used in the whole extraction process so that the researchers are protected from toxic reagents.
Owner:FUJIAN AGRI & FORESTRY UNIV
E-cadherin methylation detection method based on hydrosulphite sequencing method
InactiveCN105861718AReduce non-specific amplificationReduce the possibilityMicrobiological testing/measurementRenal cortexGel electrophoresis
The invention discloses an E-cadherin methylation detection method based on a hydrosulphite sequencing method. The E-cadherin methylation detection method includes the steps that firstly, rats which are 6-7 weeks old are selected, the kidneys of the killed rats are collected, connective tissue is sheared off, the renal cortex is taken and sheared into 50-100 mg tissue fragments, and the tissue fragments are quick-frozen with liquid nitrogen and stored in a -80 DEG C refrigerator; secondly, genome DNA is extracted with a genome DNA extraction reagent kit; thirdly, the concentration and purity of genome DNA are measured with a nucleic acid quantometer, the integrity of genome DNA is detected through 0.5%-1.0% sepharose gel electrophoresis, and DNA samples detected to be qualified are placed in a -20 DEG C--80 DEG C refrigerator for use; fourthly, genome DNA is modified with a methylation conversion reagent kit, and the concentration of modified genome DNA is determined with the nucleic acid quantometer; fifthly, PCR amplification is carried out under different experiment conditions; sixthly, samples used in follow-up steps are selected in the fifth step. By means of the E-cadherin methylation detection method, the success rate of E-cadherin methylation experiments is increased.
Owner:廖静
Traditional Chinese medicine bulbus fritillariae cirrhosae DNA extraction reagent and extraction method
The invention discloses a traditional Chinese medicine bulbus fritillariae cirrhosae DNA extraction reagent. The extraction reagent comprises a nuclear lysis solution, a CTAB buffer solution and a TEBuffer, wherein the nuclear lysis solution comprises Tris-HCl, EDTA, NaCl, PVP-40 and beta-mercaptoethanol, and is similar to the CTAB buffer solution in component. The DNA extraction reagent is suitable for extraction of traditional Chinese medicine bulbus fritillariae cirrhosae DNA, the removal rate of secondary metabolites is high, and the bulbus fritillariae cirrhosae DNA integrity is high. The invention further provides a traditional Chinese medicine bulbus fritillariae cirrhosae DNA extraction method. By adopting the extraction reagent, based on an existing CTAB method, improvement of anextraction reagent formula and optimization of extraction steps are carried out, so that high-purity and high-quality bulbus fritillariae cirrhosae total DNA can be obtained; and the extraction method is simple, easy to control and low in pollution.
Owner:康美华大基因技术有限公司 +1
Soil DNA extraction method for analyzing microbial community structure of polycyclic aromatic hydrocarbon polluted land
InactiveCN107254464ASimple extraction methodEasy to operateDNA preparationCommunity structureDna integrity
The invention discloses a soil DNA extraction method for analyzing the microbial community structure of a polycyclic aromatic hydrocarbon polluted land. The method comprises the following specific steps: 1, preparation of a reagent; 2, extraction of a general DNA. The extraction method is simple, convenient to operate, high in applicability and high in safety. For the characteristics of severe polycyclic aromatic hydrocarbon pollution, long-term pollution history, high humus content and the like in soil, firstly, beta-mercaptoethanol with an anti-oxidization effect at a volume ratio of 1.5 percent is added into a soil pretreatment buffer solution to reduce influence of impurities in the soil on DNA extraction; then a relatively mild method for crushing microbial cells through mechanical force of glass beads is adopted, so that the DNA is fully released; furthermore, no high shear force is caused on the DNA, so that the extracted genome DNA is relatively high in intactness.
Owner:BEIJING MUNICIPAL RES INST OF ENVIRONMENT PROTECTION
Method of RNA isolation from clinical samples
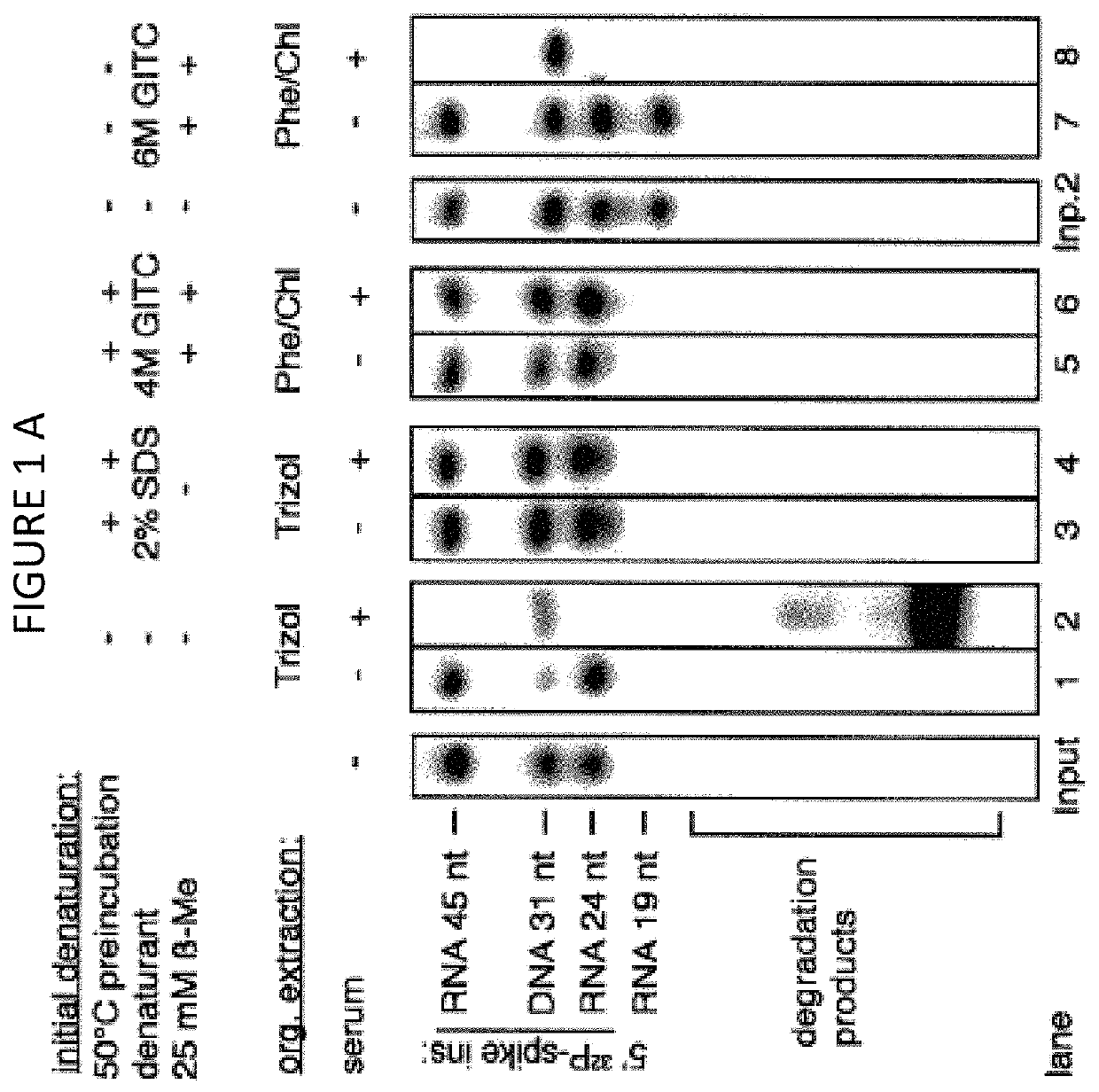
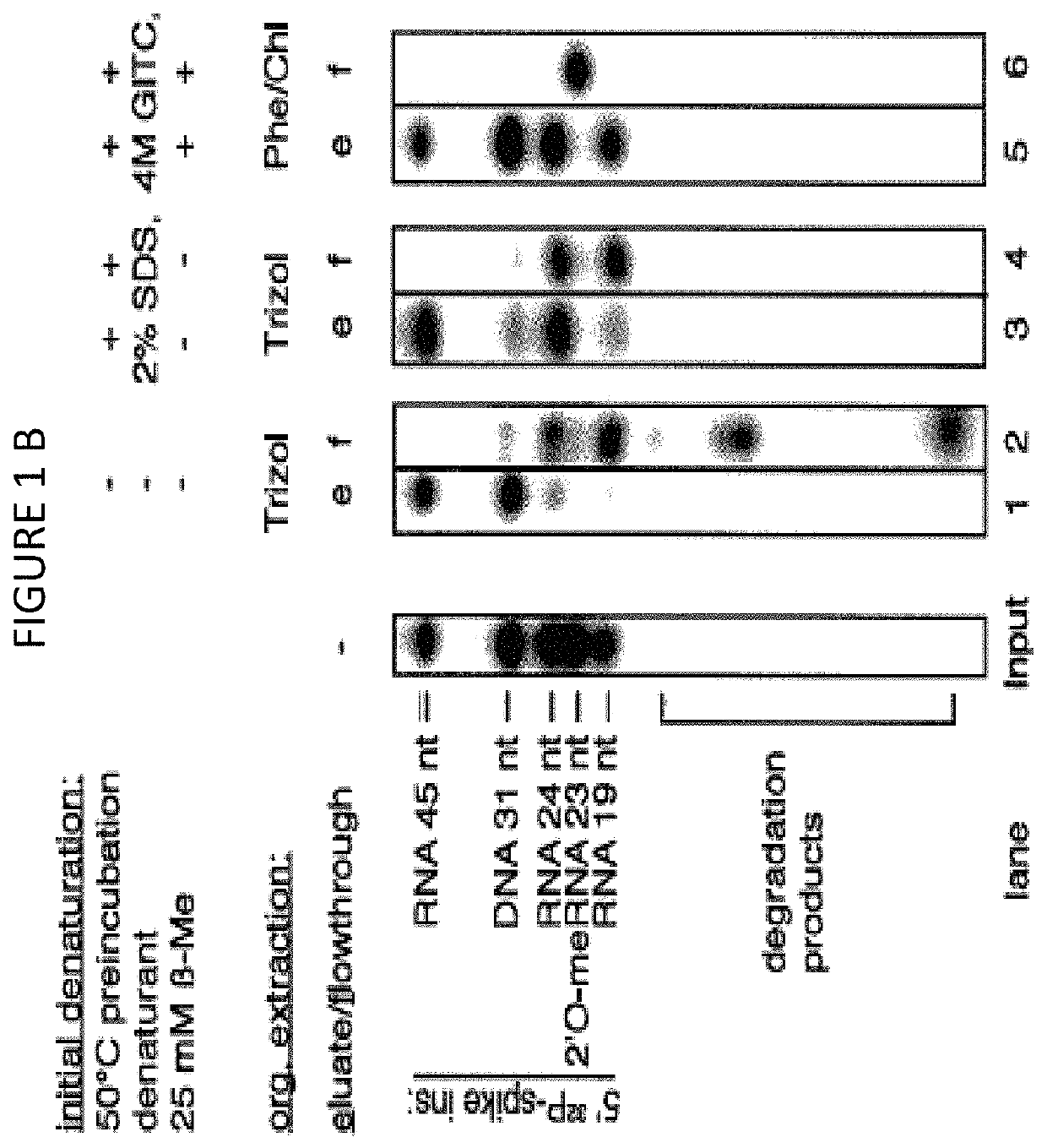
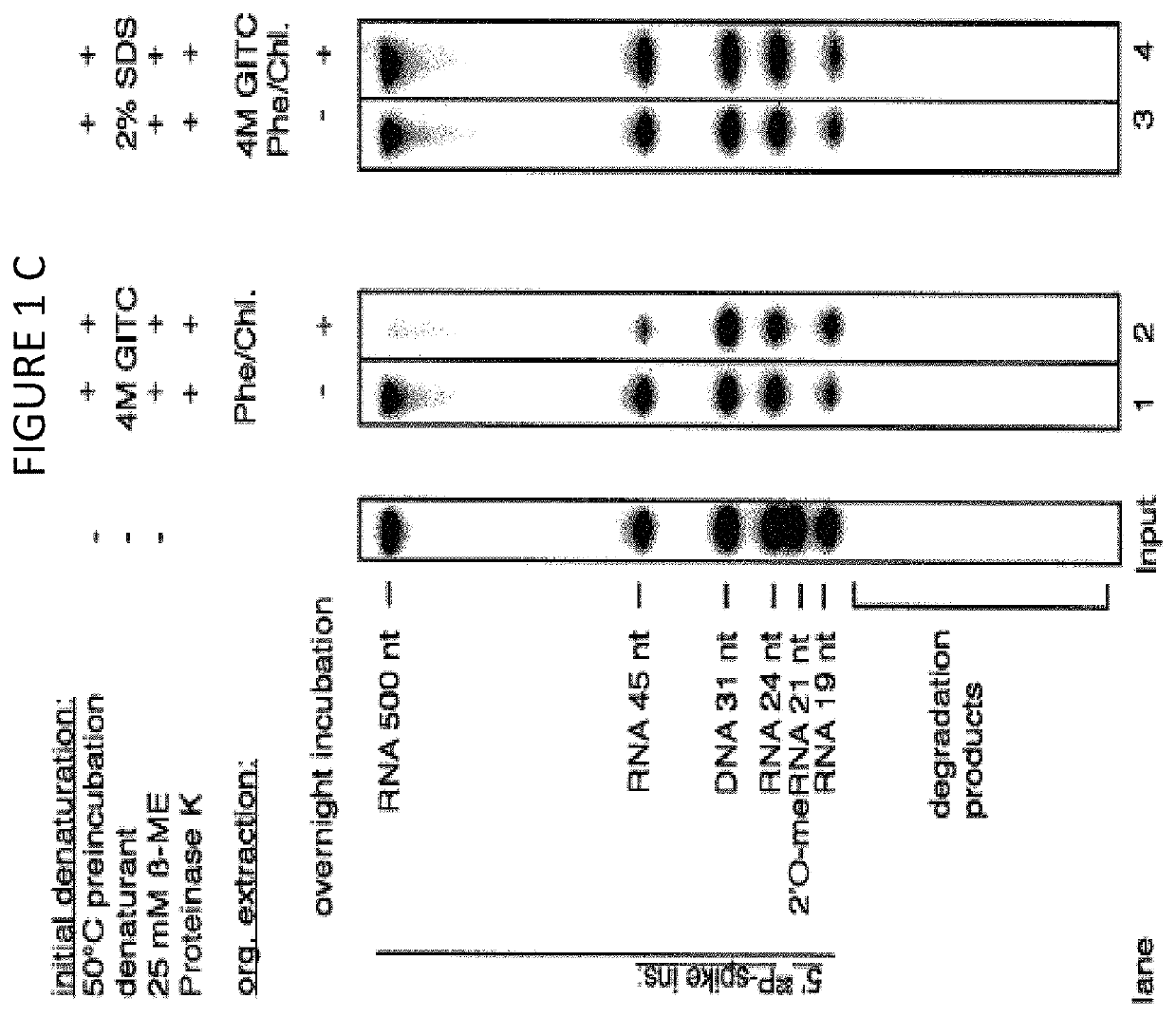
The disclosure provides methods for isolating nucleic acids from a biological fluid. In one aspect, the disclosure provides a method for isolating RNA. In another aspect, the disclosure provides a method for isolating DNA. In one aspect, the methods described herein utilize a protocol that combines a detergent-based initial denaturation, protease digestion, and organic extraction followed by column purification that maximizes RNA / DNA yield and preserves RNA / DNA integrity. In yet another aspect, the disclosure provides a kit for isolating RNA and / or DNA.
Owner:THE ROCKEFELLER UNIV
Method for purifying DNA by using gold magnetism particles
ActiveCN101165182BLarge specific surface areaLarge fixed capacityDNA preparationMetal particleParticle physics
Owner:XIAN GOLDMAG NANOBIOTECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com