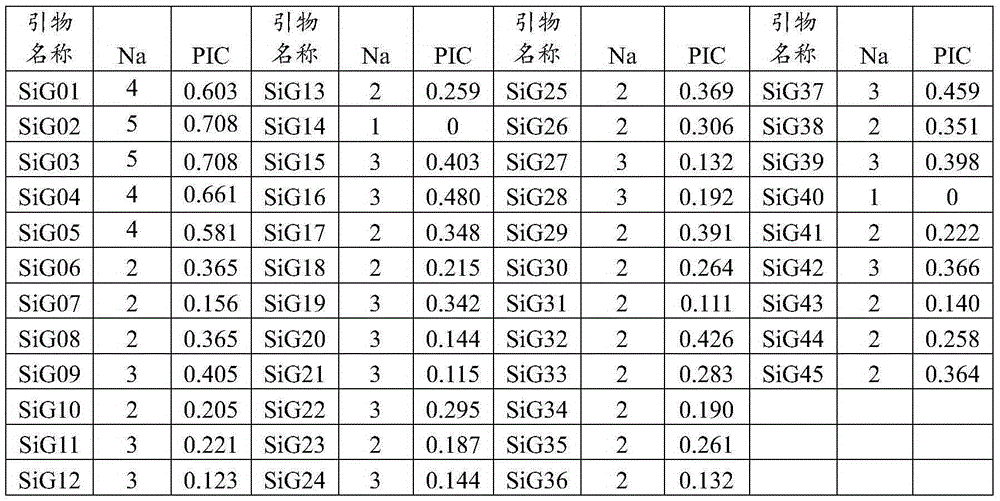
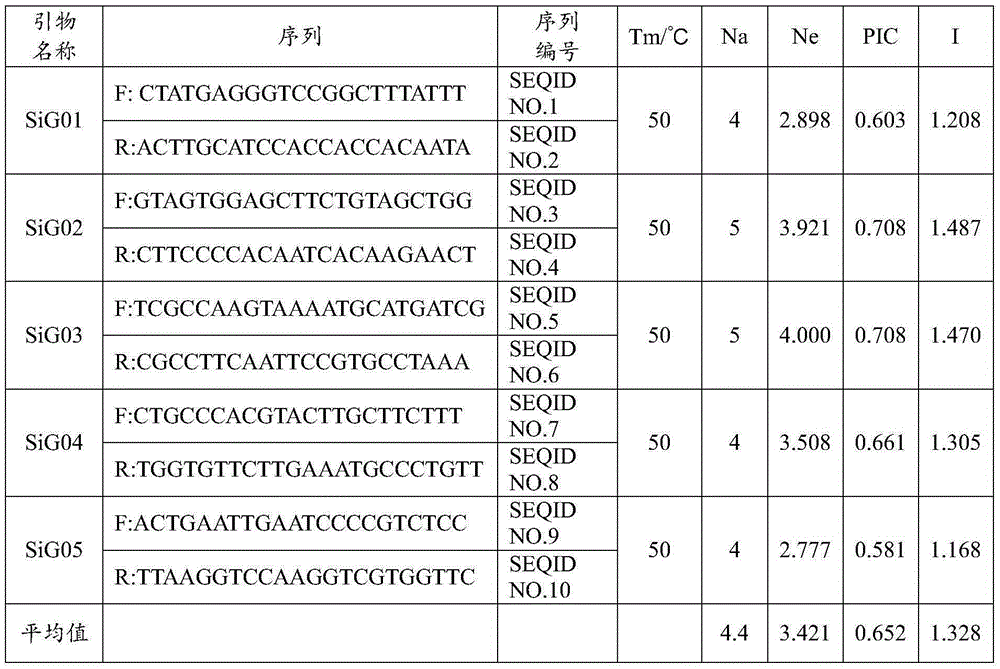
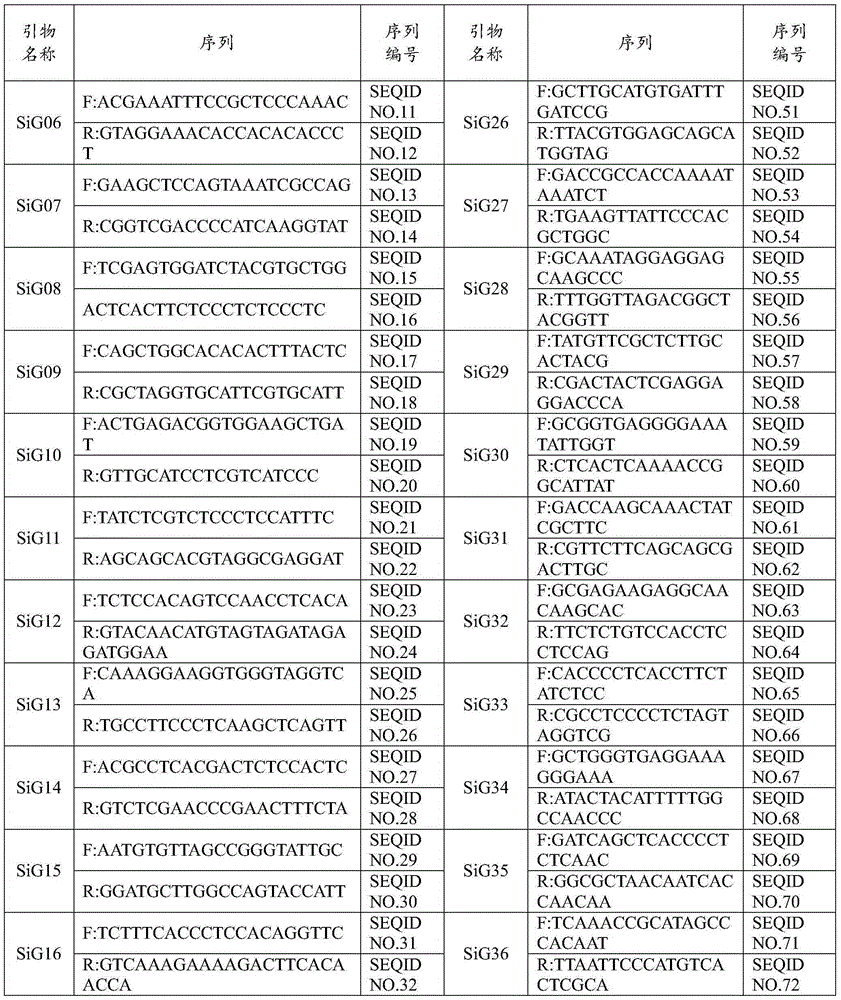
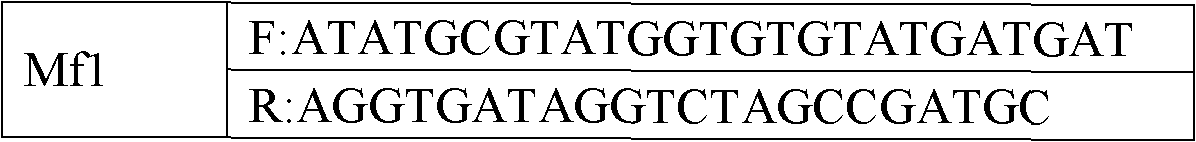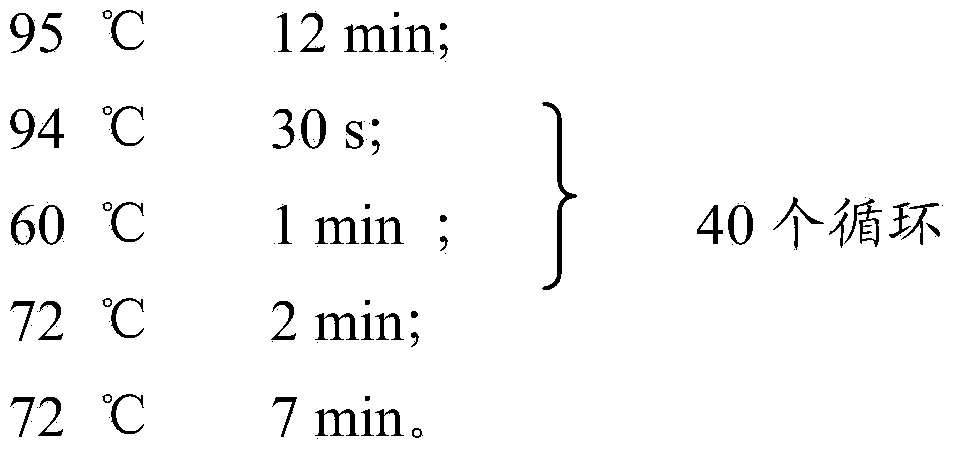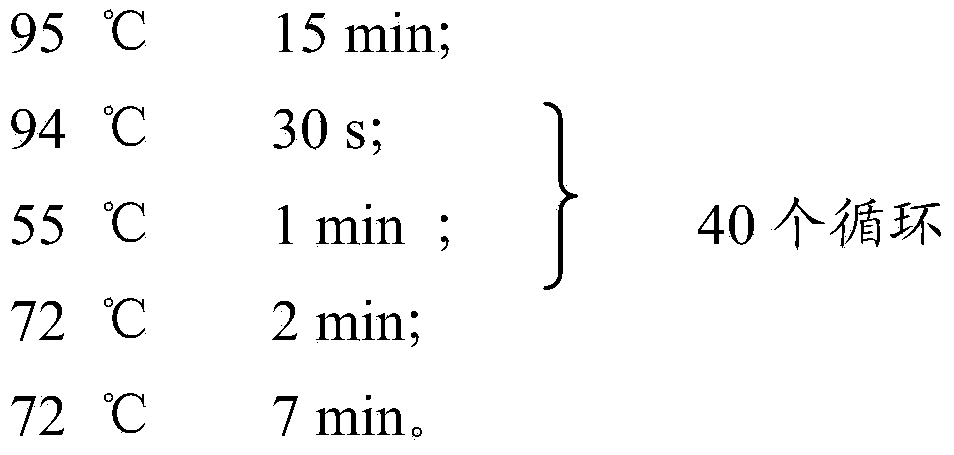Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
134results about How to "Good polymorphism" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Forensic medicine compound detection kit based on Y chromosome SNP (single nucleotide polymorphism) genetic marker
ActiveCN103131787AFast personal identificationGood polymorphismMicrobiological testing/measurementAlleleForensic science
The invention belongs to the field of forensic medicine genetics and in particular relates to a forensic medicine compound detection kit based on a Y chromosome SNP (single nucleotide polymorphism) genetic marker for individual recognition and genetic relationship identification by a legal medical expert. The forensic medicine compound detection kit provided by the invention is used for carrying out forensic medicine genetic relationship identification and individual recognition on human biology detection materials by utilizing a Y chromosome SNP genetic marker. According to the technical scheme for solving the technical problem, the forensic medicine compound detection kit based on the Y chromosome SNP genetic marker comprises a separated and packaged compound amplification primer mixture, a multiple single-basic-group extension reaction primer mixture, an allele typing standard mixture, a compound amplification reaction mixture and a single-basic-group extension reaction mixture. The kit provided by the invention can be applied to detection of common degradable materials in forensic medicine.
Owner:SICHUAN UNIV
Preparation method and use of simple sequence repeat (SSR) marker for screening miscanthus by magnetic bead enrichment process
InactiveCN102080130AEfficient enrichmentCo-dominance is goodMicrobiological testing/measurementMagnetic beadGenetic diversity
The invention discloses a preparation method and use of a simple sequence repeat (SSR) marker for screening miscanthus by a magnetic bead enrichment process, which comprises: 1) extracting genomic DNA of miscanthus floridulus which belongs to miscanthus; 2) incising the extracted total DNA by using restriction incision enzyme to obtain a genomic DNA; 3) intercrossing a probe and a target fragment; 4) enriching the hybrid DNA molecules having an SSR sequence by using magnetic beads; 5) amplifying and purifying the DNA fragment having the SSR sequence; 6) cloning and sequencing the DNA fragment; and 7) designing an SSR primer according to the flanking sequence of SSR, and designing a specific primer by using primer design software for amplifying the satellite fragment at the locus. The method is easy and convenient for operation; meanwhile, the SSR marker has the characteristics of high codominance, high polymorphism, multiple allele property and the like and can be used for analyzing the genetic diversity and genetic relationship of miscanthus and used in construction of genetic map of miscanthus, calibration of a target gene and drawing of a fingerprint.
Owner:湖北光芒能源植物有限公司
Forensic medicine compound detection kit based on Y chromosome SNP (single nucleotide polymorphism) genetic marker
ActiveCN103131787BFast personal identificationGood polymorphismMicrobiological testing/measurementAlleleForensic science
The invention belongs to the field of forensic medicine genetics and in particular relates to a forensic medicine compound detection kit based on a Y chromosome SNP (single nucleotide polymorphism) genetic marker for individual recognition and genetic relationship identification by a legal medical expert. The forensic medicine compound detection kit provided by the invention is used for carrying out forensic medicine genetic relationship identification and individual recognition on human biology detection materials by utilizing a Y chromosome SNP genetic marker. According to the technical scheme for solving the technical problem, the forensic medicine compound detection kit based on the Y chromosome SNP genetic marker comprises a separated and packaged compound amplification primer mixture, a multiple single-basic-group extension reaction primer mixture, an allele typing standard mixture, a compound amplification reaction mixture and a single-basic-group extension reaction mixture. The kit provided by the invention can be applied to detection of common degradable materials in forensic medicine.
Owner:SICHUAN UNIV
Screening and application of solanum melongene SSR (Simple Sequence Repeats) molecular marker core primers
ActiveCN110628931AHigh polymorphismGood polymorphismMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceSequence repeat
The invention discloses screening and application of solanum melongene SSR (Simple Sequence Repeats) molecular marker core primers. A solanum melongene SSR molecular marker core primer group comprises17 SSR molecular marker core primer pairs. The stable and reliable solanum melongene SSR primers with high polymorphism are screened by extracting DNA and SSR-PCR of solanum melongene core germplasmswith great property differences. The screened 17 SSR primer pairs are used for analyzing 106 parts of solanum melongene culture variety resources; through similarity calculation and clustering analysis, the result shows that the screened 17 pairs of primers can be used for accurately and efficiently identifying the solanum melongene variety; and the foundation is laid for the application of an SSR molecular marker technology to solanum melongene germplasm genetic relationship analysis and variety identification.
Owner:SHANGHAI ACAD OF AGRI SCI
Floral character associated molecular marker screening method of amenone form chrysanthemum and application of method
ActiveCN104313155AWide range of choicesHigh strengthMicrobiological testing/measurementPopulationTest material
The invention belongs to the biotechnical field and provides a floral character QTL (quantitative trait locus) molecular marker screening method of amenone form chrysanthemum. The method can be used for positioning and cloning excellent genes of floral characters of amenone form chrysanthemum and cultivating novel varieties of amenone form chrysanthemum. The method comprises the following steps: I, obtaining a test material and phenome data; II, constructing a linkage map of chrysanthemum; III, combining phenome data with a molecular genetic map for QTL analysis of floral characters of amenone form chrysanthemum; and IV, determining the floral character associated molecular marker of the amenone form chrysanthemum. By using 160 F1 segregation population which is obtained by taking an amenone form chrysanthemum variety QX-053 as a female parent and a non-menone form chrysanthemum variety Nannongjingyan as a male parent, a plurality of molecular markers which are remarkably associated with floral characters of amenone form chrysanthemum. The molecular markers which are associated with floral characters of amenone form chrysanthemum are obtained for fine-mapping and cloning excellent genes of floral characters of amenone form chrysanthemum so as to greatly improve the selection efficiency, so that the amenone form chrysanthemum cultivating process is accelerated.
Owner:NANJING AGRICULTURAL UNIVERSITY
EST-SSR core primer group for identifying variety of Chinese wolfberry and identification method and application thereof
ActiveCN106011228AGood polymorphismStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationVarieties of ChineseLycium chinense
The invention discloses an EST-SSR core primer group for identifying the variety of Chinese wolfberry and an identification method and an application thereof. Through screening of EST sequences of lycium ruthenicum and Ningxia wolfberry, a large number of SSR molecular markers are developed, and an SSR marker-meeting fluorescence detection technology system including 10 markers is established and is used for identification of the Chinese wolfberry variety and species. The 10 markers have the advantages of good polymorphism, stable amplification, clear amplification results and good repeatability, can be used for large-scale detection of authenticity of Chinese wolfberry seedlings and the purity of the seedlings, can provide a technical basis for standardization of seedling markets, and are conducive to development and use of the traditional Chinese medicine Chinese wolfberry.
Owner:SOUTH CHINA BOTANICAL GARDEN CHINESE ACADEMY OF SCI
Obtaining method and application of molecular markers related to chrysanthemum drought resistance
InactiveCN104498475AShort attenuation distancePrecise positioningMicrobiological testing/measurementDNA preparationResistant genotypeAgricultural science
The invention belongs to the biotechnology field, provides an obtaining method of molecular markers related to chrysanthemum drought resistance, and is used for positioning and cloning of a chrysanthemum drought resistance excellent gene and breeding of new chrysanthemum varieties. The obtaining method comprises the steps: a, identification of the drought resistance; b, analysis of a chrysanthemum variety group structure; c, analysis of chrysanthemum variety genetic relationship; and d, determination of the molecular markers closely related to the chrysanthemum drought resistance. The method simultaneously adopts three molecular marker techniques comprising SRAP, SCoT and EST-SSR, the principles are different with each other and each have pertinency, and obtained dyeing strips are good in polymorphism and high in repeatability, can cover the entire genome, have rich number of strips and are beneficial for obtaining the molecular markers closely related to the drought resistance. 8 molecular markers related to the chrysanthemum drought resistance are totally obtained, early selection, directional selection and accurate selection of excellent drought-resistant varieties are achieved, the work load can be reduced, and the selection efficiency of chrysanthemum drought resistant genotypes is greatly improved, so as to speed up the process of breeding.
Owner:NANJING AGRICULTURAL UNIVERSITY
Detection method for Apostichopus japonicas AjE101 micro-satellite DNA label
InactiveCN102140522AGood polymorphismImprove stabilityMicrobiological testing/measurementGerm plasmGenetic diversity
The invention belongs to the field of molecular biology DNA labeling technology and application, and in particular relates to Apostichopus japonicas express sequence tag EST micro-satellite label screening development and application. The invention provides Apostichopus japonicas AjE101 micro-satellite specific DNA primers, a polymerase chain reaction (PCR) reaction system by utilizing the primers, and a detection method for an Apostichopus japonicas AjE101 micro-satellite DNA label. The detection method comprises the following steps of: extracting genome of Litopenaeus vannamei Boone; designing the specific primers at two ends of an Apostichopus japonicas AjE101 micro-satellite DNA core sequence; and performing PCR amplification on genome DNA of different groups of the Litopenaeus vannamei Boone or individuals in the group by using the primers, analyzing products, and determining the genome of each individual so as to obtain an Apostichopus japonicas polymorphism genetic variation map. The invention is mainly applied to Apostichopus japonicas germ plasm resource and genetic diversity analysis, cular population genetics, construction of genetic maps and research of functional genes.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
SNP molecular marker relating to peach tree bleeding disease resistance
ActiveCN106636081AImprove accuracyOvercome areaMicrobiological testing/measurementDNA/RNA fragmentationResistant genesHigh humidity
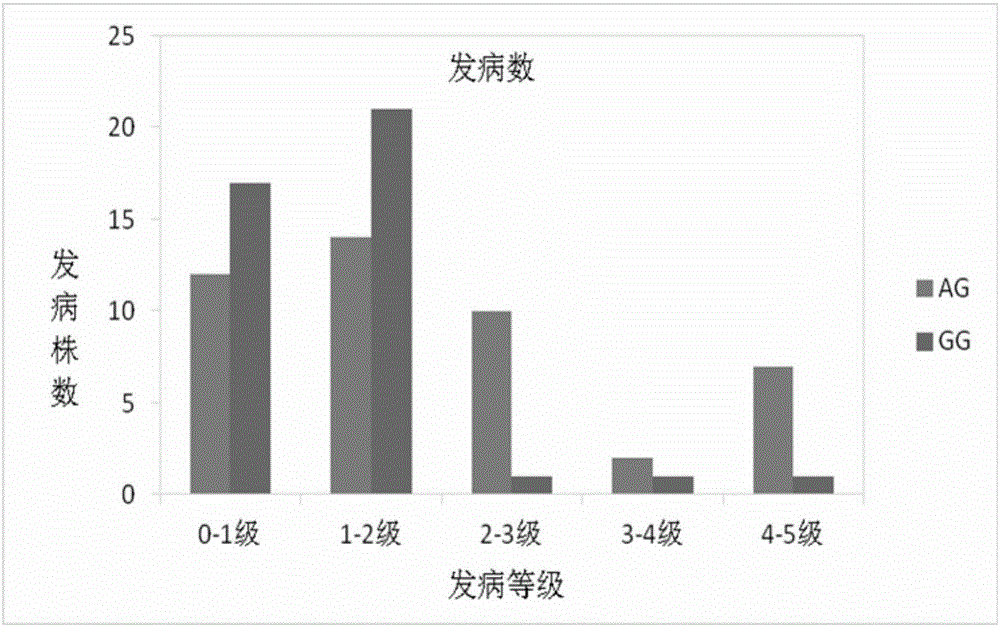
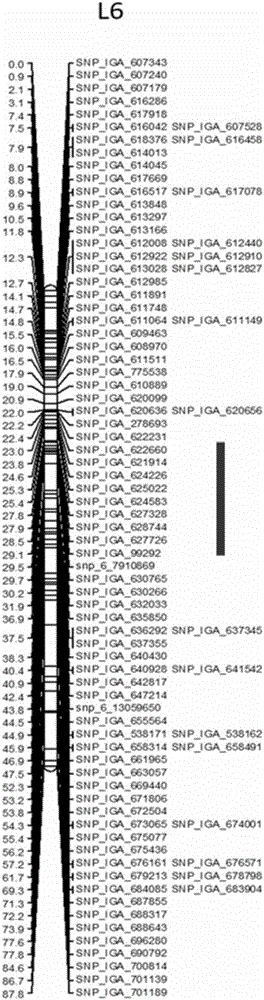
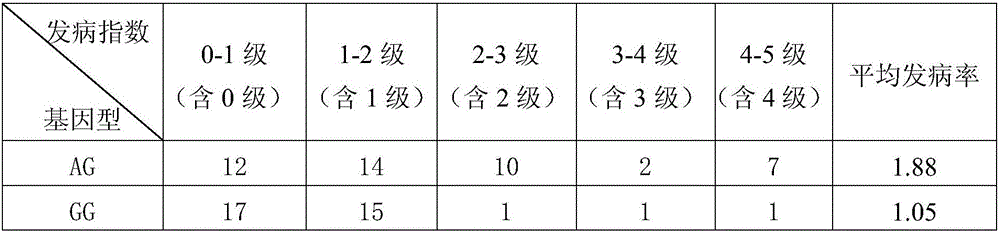
The invention discloses an SNP molecular marker relating to the peach tree bleeding disease resistance. The SNP molecular marker which has close linkage with the peach tree bleeding disease resistance related QTL is obtained by locating a peach tree bleeding disease resistance gene locus, and is located on the sixth chromosome of a peach; the locus is SNP_IGA_627726; the SNP locus is obviously related to the morbidity of the bleeding disease of different types of peaches and is high in polymorphism; a novel authentication method is provided for parent selection in the early stage and generation selection in the late hybridization stage in a bleeding disease resistance species selection process; the accuracy of bleeding disease resistance detection can be improved, the breeding period can be shortened, the breeding efficiency is improved, and implementation of seed selection of a new bleeding disease resistance species of the peach tree in a high temperature and high humidity district is accelerated.
Owner:SHANGHAI ACAD OF AGRI SCI
Chrysanthemum salt-tolerance associated molecular marker and obtaining method and application thereof
ActiveCN105112403ABroad genetic baseShort research cycleMicrobiological testing/measurementVector-based foreign material introductionSalt resistanceCut flowers
The invention discloses a chrysanthemum salt-tolerance associated molecular marker and an obtaining method and application thereof. The obtaining method comprises the steps that a, salt-tolerance identification is performed for two consecutive years; b, chrysanthemum variety population structure analysis is performed; c, chrysanthemum variety genetic relationship analysis is performed; d, the molecular marker closely associated with chrysanthemum salt-tolerance is determined. According to the obtaining method, genome-wide molecular marker scanning is performed on 159 cut-flower chrysanthemum varieties by utilizing the three molecular marker technologies of SRAP, SCoT and EST-SSR at the same time, the number of obtained dyed strips is large, the dyed strips are good in polymorphism and high in reproducibility, and the molecular marker which is closely associated with salt-tolerance can be obtained. Three marker loci detected through the obtaining method are remarkably associated with salt-tolerance of varieties and provide a certain reference for chrysanthemum salt-tolerance molecular marker-assisted selection.
Owner:NANJING AGRICULTURAL UNIVERSITY
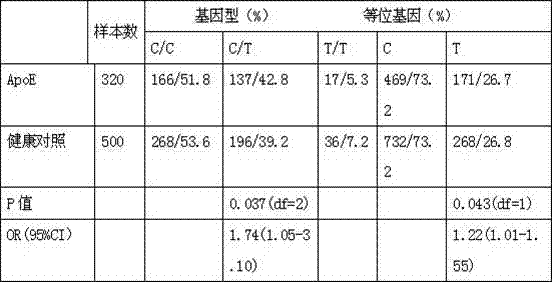
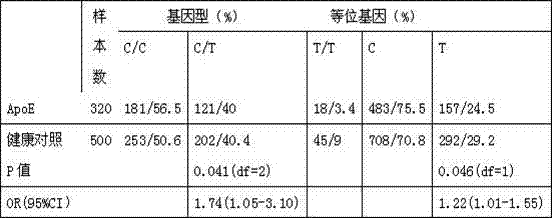
Kit for detecting APOE (Apoliprotein E) gene of Alzheimer disease
InactiveCN107447030AGood polymorphismImprove developmentMicrobiological testing/measurementDNA/RNA fragmentationDiagnosis methodsPolymerase L
The invention discloses a kit for detecting an APOE (Apoliprotein E) gene of Alzheimer disease, and belongs to the technical field of biological detection. The kit disclosed by the invention comprises a unit PCR detection reagent; the unit PCR detection reagent comprises a buffer solution, protease K, magnesium chloride stock solution, a primer, a probe and a polymerase; the primer is a primer pair of a reagent for detecting rs429358 and rs7412 gene segments of the APOE gene; and the kit for detecting the APOE gene of Alzheimer disease is enough to measure the polymorphism of the APOE gene by only a small amount of DNA samples based on a new mutational site related to the APOE of the APOE gene, and can be used in a gene diagnosis method for measuring the polymorphism of the APOE-related gene.
Owner:苏州康吉诊断试剂有限公司
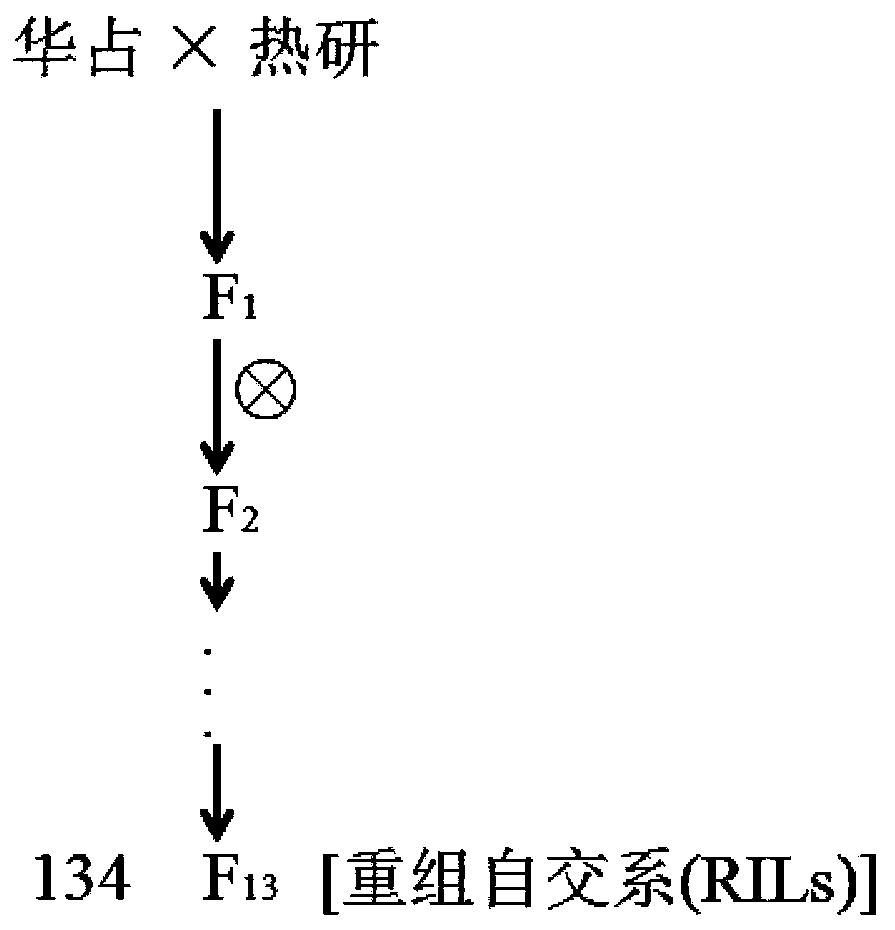
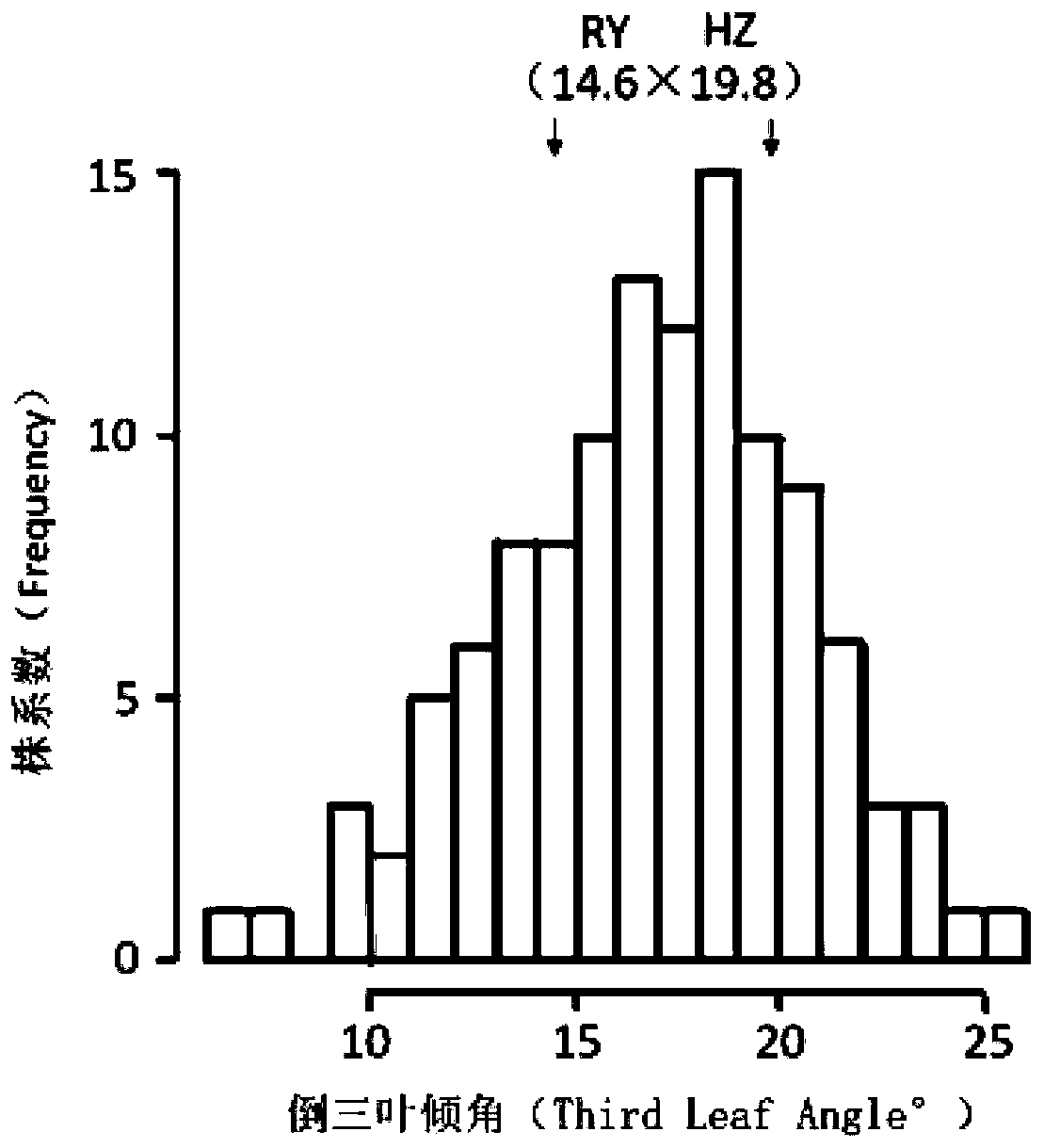
Molecular marker of major QTL, qTLA-9 for regulating angle of tilt of paddy rice leaves, and application thereof
InactiveCN109777886AHigh LOD valueHigh spectral densityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceIndel
The invention discloses a major QTL, qTLA-9, which is used for regulating the angle of tilt of paddy rice leaves, and locates between the mark Indel Tal-1 and on Indel Tal-2 on the ninth chromosome, and the genetic distance is 74.80 cm-98.33 cm, the physical distance is 17448080 bp-2293 8953 bp. 2293 8953 bp. The invention also provides a mark Indel of the major QTL, qTLA-9, which is used for regulating the angle of tilt of paddy rice leaves; the invention also provides the application of the major QTL: by developing the molecular marker closely linked to the major QTL, the test result that whether there are leaf inclination-related QTLs in rice varieties or strains can be attained, thereby speeding up the breeding process of excellent rice varieties.
Owner:ZHEJIANG NORMAL UNIVERSITY
Library building kit for high flux detection of STR genetic markers
ActiveCN106399496AFast Downstream Forensic ApplicationsLow costMicrobiological testing/measurementLibrary creationHigh fluxA-DNA
The invention provides a library building kit for high flux detection of STR genetic markers. The kit comprises multiple pairs of fusion primers for amplifying target STR loci, each pair of fusion primer respectively aims at different STR loci on a DNA template, and each pair of fusion primer comprises a sequencing joint sequence and a specific primer sequence which aims at the STR locus from a 5'end to a 3'end in order. The kit can be used for realizing construction of a STR locus amplicon sequencing library by one-time amplification, the operation process of library building is simplified, time and cost for building the library are reduced, and sequencing quality is not influenced.
Owner:承启医学(深圳)科技有限公司
SRAP molecule labeled primer for Megalobrama amblycephala family identification, method thereof, and application of primer
InactiveCN104293907AIncrease usageGood polymorphismMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceRepeatability
The invention discloses an SRAP molecule labeled primer for Megalobrama amblycephala family identification, a method thereof, and an application of the primer. The SRAP molecule labeled primer is selected from one of the following primer combinations: Me2 / Em18, Me3 / Em19, Me9 / Em13, Me9 / Em15, Me9 / Em16, Me11 / Em4, Me11 / Em15, Me11 / Em18, Me12 / Em13 and Me12 / Em16, and the sequences of all the primers are respectively shown in a sequence table. The disclosed SRAP molecular marker primer is used to identify the Megalobrama amblycephala family by carrying out SRAP molecule labeling of the Megalobrama amblycephala family, and a band obtained in the invention has the advantages of clearness, good polymorphism, high repeatability, simple operation and broad application prospect.
Owner:HUAZHONG AGRI UNIV
Wu-he dipsacus asper ISSR-PCR molecule marking method
InactiveCN106434988AGood polymorphismReduce stepsMicrobiological testing/measurementDipsacus asperAgarose electrophoresis
The invention relates to a wu-he dipsacus asper ISSR-PCR molecule marking method. The method is characterized by comprising the following steps: 1, extracting the genome of the wu-he dipsacus asper, 2, conducting 1% agarose electrophoresis for 20 minutes, testing the integrity of the genome, at the same time using the DNA bar code ITS2 primer to conduct the PCR amplification of the extracted wu-he dipsacus asper DNA, verifying the purity of the extracted DNA, 3, based on the initial screened primer conducting the ISSR-PCR. Compared with the prior art, the reaction system created by the method has a simple optimization method. The amplified bands is both clear and stable with good polymorphism. The method compensates the shortcomings in the study of the genetic diversity of the wu-he dipsacus asper. The method can be used in the analysis of the genetic relationship and the verification of authenticity of the teasle, thus having good application value.
Owner:湖北省农业科学院中药材研究所
Method for discriminating Chinese milk vetch variety by using SSR (Simple Sequence Repeat)fingerprint spectrum
InactiveCN101974516AAvoid interferenceIncrease diversityMicrobiological testing/measurementDNA preparationBiotechnologyMagnetic bead
The invention provides a method for discriminating Chinese milk vetch varieties by using an SSR (Simple Sequence Repeat)fingerprint spectrum. A specific SSR site and bilateral primer sequences thereof (SEQ.NO.1-18) in a Chinese milk vetch genome, and a fingerprint spectrum which is obtained by carrying out PCR (Polymerase Chain Reaction) amplification on screened primers can be used for discriminating Chinese milk vetch varieties. The discriminating method mainly comprises the following steps of: extracting Chinese milk vetch total DNA (Deoxyribonucleic Acid); screening the specific SSR site and designing primers from genome DNA by using magnetic-bead enrichment method; and finally obtaining 6 SSR site primer pairs which can form obvious banded difference by carrying out the PCR amplification on different varieties of different Chinese milk vetch. The constructed fingerprint spectrum can be effectively used for discriminating Chinese milk vetch varieties.
Owner:INST OF SOIL & FERTILIZER FUJIAN ACADEMY OF AGRI SCI
Genotype detection primer group and kit for mycobacterium tuberculosis
InactiveCN103834743AImprove resolutionStrong specificityMicrobiological testing/measurementMicroorganism based processesMycobacterium tuberculosis genotypeGenotype
The invention relates to the field of genotypes, and in particular relates to a genotype detection primer group and a kit for mycobacterium tuberculosis. The primer group comprises 22 primer pairs for detecting 22 VNTR loci in a mycobacterium tuberculosis genome. The primer group can be used for detecting the diversity of the 22 VNTR loci of the mycobacterium tuberculosis, so that the aim of genotyping the mycobacterium tuberculosis can be fulfilled. Experimental results show that high resolving power and high specificity are ensured when the primer group is used for mycobacterium tuberculosis genotype detection, each locus is high in polymorphism, and the genotyping index can reach 0.903.
Owner:中华人民共和国吉林出入境检验检疫局
Separation and identification method of cow mastitis pathogens
InactiveCN103642925AEfficient identificationIdentification is simple and economicalMicrobiological testing/measurementMicroorganism based processesSingle-strand conformation polymorphismSequence analysis
The invention relates to a separation and identification method of cow mastitis pathogens. Separation and identification on the Chinese Holstein cow mastitis pathogens are carried out by adopting a traditional bacterial culture method and a sequence analysis method and a double-primer polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) technology on the basis of pathogen 16S rRNA (ribosomal Ribonucleic Acid) genes. The identification result of the double-primer PCR-SSCP technology is basically the same as that of the sequence analysis method. The result of the double-primer PCR-SSCP technology is good in polymorphism, and is similar to the sequence analysis result. The separation and identification method is an efficient, simple and economic method for identifying the pathogens, and a theoretical foundation is provided for rapid identification of the Chinese Holstein cow mastitis pathogens. Thus, a theoretical basis is provided for diagnosis and treatment of cow mastitis.
Owner:ANHUI NORMAL UNIV
Method for rapidly detecting third exon single base mutation of myostatin gene
InactiveCN101724700AGood polymorphismPolymorphic shortcutMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMarker-assisted selection
The invention provides a method for rapidly detecting the third exon single nucleotide polymorphism (SNP) of a beef myostatin gene. In the method, aiming at the G-to-A mutation of a third exon of the myostatin gene, two groups of PCR primer pairs for respectively amplifying an upstream sequence and a downstream sequence of a G938A single base polymorphism site are designed, wherein the primer pairs are respectively provided with a mismatched primer for mutation site, and the 3' tail end of the primer is positioned on the mutation site. By utilizing the primers, different beef DNA samples can obtain different amplification results, thereby judging the polymorphism of the third exon of the myostatin gene. The method has low cost, and simple, rapid and accurate operation, is suitable for popularization and application, and is important to rapidly and accurately carry out marker-assisted selection of beef, accelerate the beef selecting and breeding process and research a rapid PCR detecting method of the field of molecular biology.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
Method for identifying foxtail millet variety by adopting SSR molecular marker technique and application
ActiveCN105385768AGood polymorphismHigh identification accuracyMicrobiological testing/measurementElectrophoresesNucleotide
The invention discloses a method for identifying a foxtail millet variety by adopting an SSR molecular marker technique and application, and belongs to the technical field of plant variety identification. The method comprises the steps that foxtail millet DNA is extracted, a PCR amplification reaction is performed through the foxtail millet DNA and 5 pairs of SSR core primers, and after electrophoresis detection is performed, the foxtail millet variety is identified according to an electrophoresis detection result; the nucleotide sequences of the SSR core primers are shown as SEQ ID NO.1-10. According to the method, the foxtail millet variety can be effectively distinguished, and an important way is supplied to further identification of a large number of the foxtail millet varieties.
Owner:HEILONGJIANG BAYI AGRICULTURAL UNIVERSITY
Set of SNP sites suitable for identifying variety and purity of hops and applications of set of SNP sites
InactiveCN107446991AGood polymorphismSimple and fast operationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceFingerprint database
The invention discloses a set of SNP sites suitable for identifying the variety and purity of hops. The set of SNP sites comprises five SNP sites for hops totally. Based on a whole genome sequencing result for 10 representative hop varieties at home and abroad, comparison and screening are carried out on the SNP sites of the whole genome, at present, the identification for the variety and purity of hops of 5 SNP sites with relatively good polymorphism is verified, the 5 SNP sites can identify the variety of the hops, meanwhile, a genotype result is stable, and after the follow-up verification work for the authenticity of the SNP sites is completed, an SNP site fingerprint database of the hops is complemented to be complete. With the SNP sites and the identification method provided by the invention, an identification system for the variety and the purity of hops, which realizes low cost, high efficiency and standardization is provided for the beer production industry.
Owner:CHINA NAT RES INST OF FOOD & FERMENTATION IND CO LTD
Amaranth EST-SSR marker primer and method for identifying amaranth varieties
InactiveCN108384880AGood repeatabilityClear bandMicrobiological testing/measurementDNA/RNA fragmentationSpectrum bandAgricultural science
The invention provides an amaranth EST-SSR marker primer and a method for identifying amaranth varieties. The nucleotide sequence (CATA)5F of the amaranth EST-SSR marker (CATA)5 is CACGTCCTTCATTCGGATCT, and the nucleotide sequence (CATA)5R of the amaranth EST-SSR marker (CATA)5 is AATCCCGGAGGTAGGATCAC. The amaranth EST-SSR marker (CATA)5 has good polymorphism among the amaranth varieties and is clear in spectrum band and stable and reliable. The amaranth EST-SSR marker (CATA)5 is also applicable to germplasm resource identification and genetic diversity analysis of more amaranth plants.
Owner:FUJIAN AGRI & FORESTRY UNIV
Scophthalmus maximus T170G single nucleotide polymorphic marking detection method
InactiveCN102586454AAccurate amplificationGood polymorphismMicrobiological testing/measurementNucleotideGenotype
The invention relates to a scophthalmus maximus T170G single nucleotide polymorphic marking detection method. The method comprises the following steps: extracting scophthalmus maximus genome DNA and diluting for later use; analyzing the sequences of EST, screening the sequence containing a candidate SNP locus, designing a specific primer at the two ends and designing a non-marking probe with the closed 3' end in front of and behind the locus (comprising the locus); performing asymmetric PCR amplification on the scophthalmus maximus group genome DNA by using the primer; hybridizing the amplification product with the non-marking probe with the closed 3' end; and placing the hybrid product on a LightScanner, and detecting and analyzing the melting curve to obtain the genetic polymorphism mapof the scophthalmus maximus. The method is applicable to detection technologies of scophthalmus maximus genetic marking, genealogy authentication, genetic linkage map construction and the like. According to the method which is convenient, quick and accurate, the scophthalmus maximus T170GSNP marked genetic variation map can be obtained quickly; and the genotype of each individual of the scophthalmus maximus can be detected intuitively.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Fluorescent SSR primer composition and application in constructing white wax new species molecular fingerprint spectrum thereof
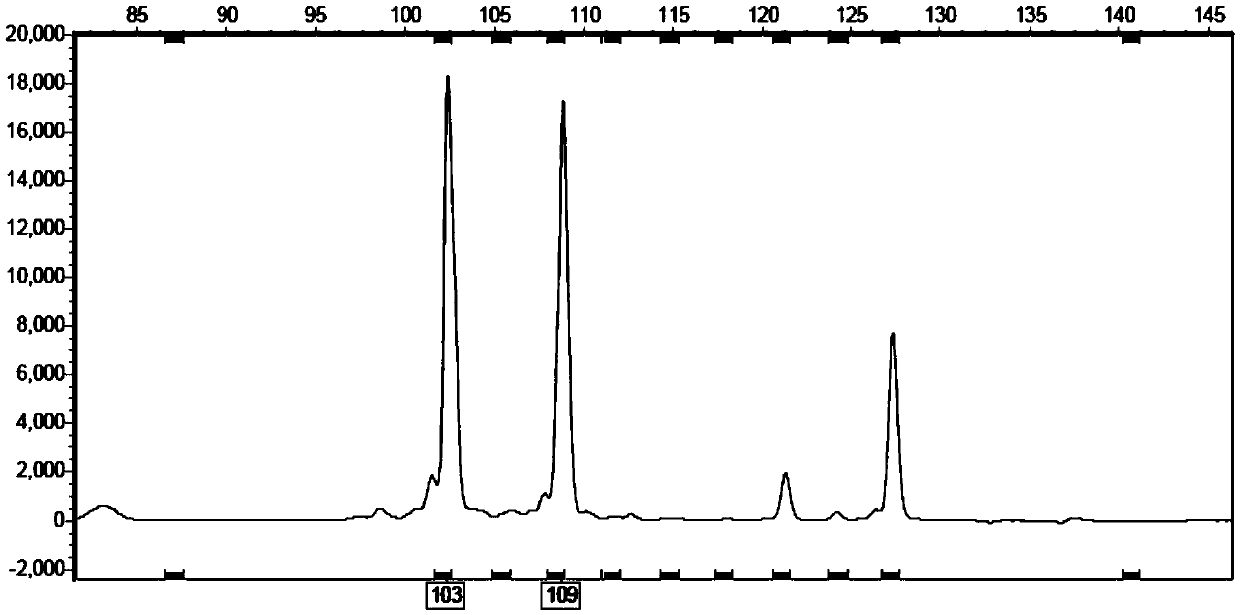
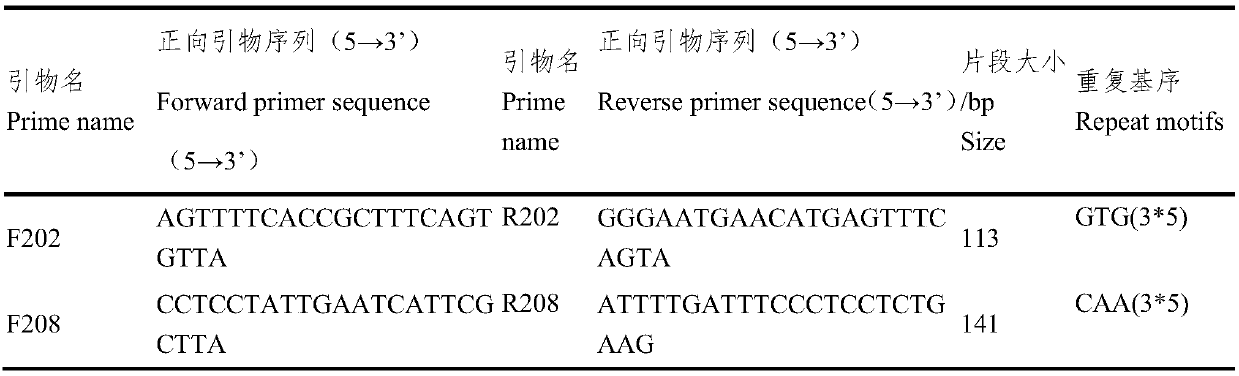
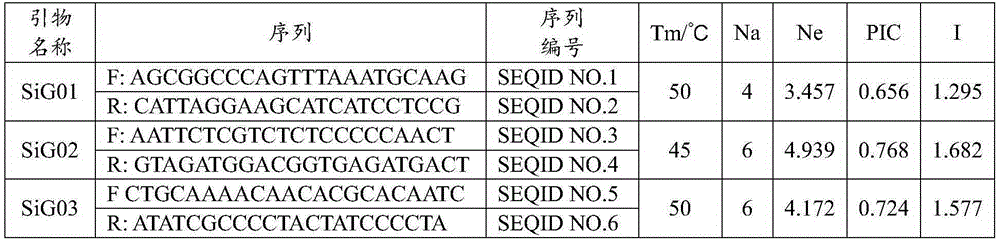
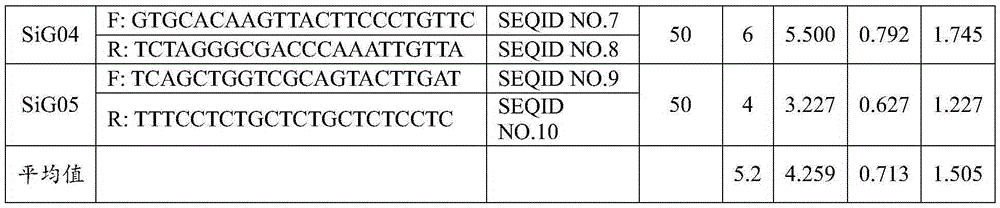
ActiveCN109652411ASpecific stabilityGood polymorphismMicrobiological testing/measurementDNA/RNA fragmentationWaxForward primer
The invention discloses a fluorescent SSR primer composition and application in constructing a white wax new species molecular fingerprint spectrum thereof. The fluorescent SSR primer composition is composed of SSR primer pairs of F202 / R202 and F208 / R208, FAM fluorescent marks are added to 5' ends of upstream primers of each pair of primers, and forward primers are composed of TP-M13 primers carrying fluorescent marks. According to the primer composition, the primer composition is obtained using a specific fingerprint spectrum constructing a white wax new species or white wax new species molecular ID card, and 20 white wax species can be completely distinguished to overcome the prior defects of morphological and physiological identification. The primer composition can provide scientific and effective reference bases for the further application researching of a DNA fingerprint spectrum on the white wax, the evaluation of white wax germ plasm resources, and the solving of species intellectual property right disputes of the white wax.
Owner:SHANDONG FOREST SCI RES INST +1
SSR molecular marker method for identifying foxtail millet variety and application
ActiveCN105385769AGood polymorphismGood identification can comeMicrobiological testing/measurementAgricultural scienceNucleotide
The invention discloses an SSR molecular marker method for identifying a foxtail millet variety and application, and belongs to the technical field of plant variety identification. The method comprises the steps that foxtail millet DNA is extracted, a PCR amplification reaction is performed through the foxtail millet DNA and 5 pairs of SSR core primers, and after electrophoresis detection is performed, the foxtail millet variety is identified according to an electrophoresis detection result; the nucleotide sequences of the SSR core primers are shown as SEQ ID NO.1-10. According to the method, the foxtail millet variety can be effectively distinguished, and an important way is supplied to further identification of a large number of the foxtail foxtail millet varieties.
Owner:HEILONGJIANG BAYI AGRICULTURAL UNIVERSITY
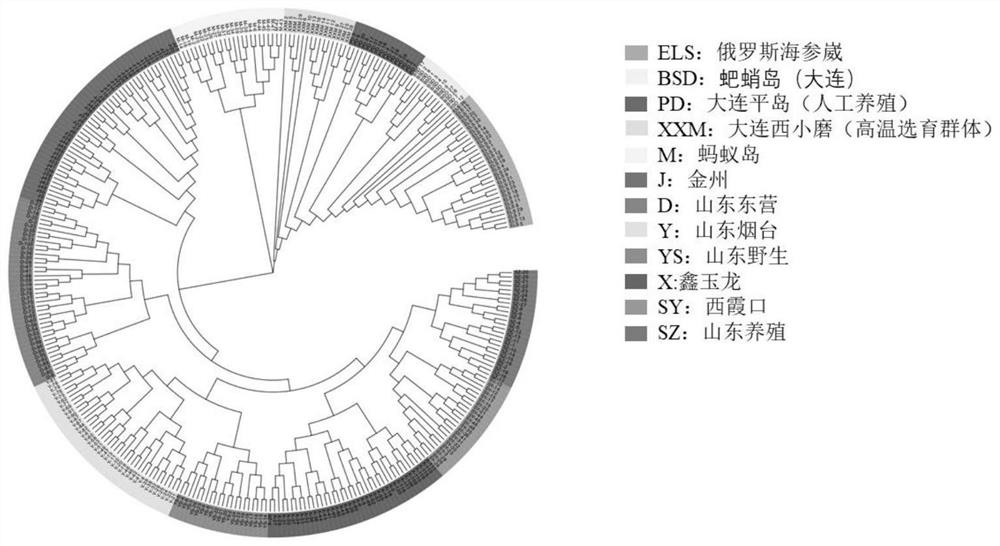
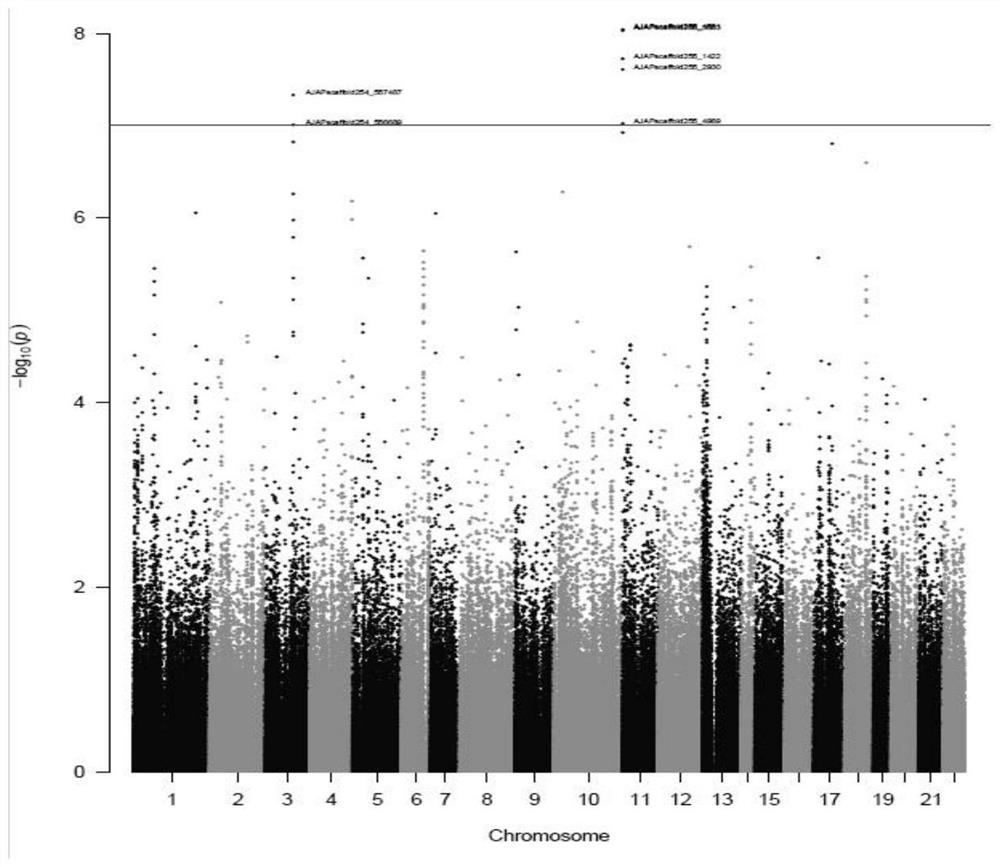
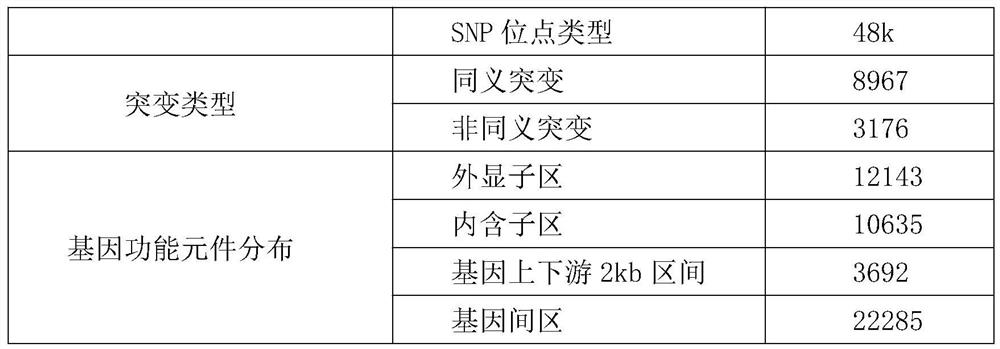
Whole-genome 50KSNP chip for apostichopus japonicus breeding and application
PendingCN113789391AGood targetingImprove consistencyMicrobiological testing/measurementProteomicsSample qualityBiotechnology
The invention discloses a whole-genome 50KSNP chip for apostichopus japonicus breeding and application. The method comprises the following steps: (1) development of the 50KSNP chip in a whole-genome range of apostichopus japonicus: constructing an apostichopus japonicus sample group, performing SNP typing in the whole-genome range, screening 50KSNP markers of the apostichopus japonicus, designing an HD-marker high-density chip and designing development probes to obtain a liquid-phase chip pool of 48K loci; (2) testing the accuracy and the typing effect of the chip, and ensuring the high accuracy and the good typing effect through DNA sample quality detection, HD-Marker chip detection and result analysis. The chip can be applied to genetic background analysis and character association analysis of the whole genome breeding chip of the apostichopus japonicus in different groups of the apostichopus japonicus.
Owner:OCEAN UNIV OF CHINA +1
Molecular marker and screening method of gift strain nile tilapia oreochromis niloticus streptococcus iniae infection resistant family
InactiveCN103642802AGuaranteed stabilityAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationScreening methodNile tilapia
The invention discloses a molecular marker and a screening method of gift strain nile tilapia oreochromis niloticus streptococcus iniae infection resistant family. The molecular marker is a sequence shown in SEQ ID NO.20; the screening method comprises five steps of streptococcus iniae infection, genome DNA extraction, primer screening, SRAP-PCR (Sequence-Related Amplified Polymorphism-Polymerase Chain Reaction) amplification and sequencing and analyzing, by the method, a parent family with stronger resistance to streptococcus iniae disease can be found more accurately and quickly, after enhanced cultivation, the parent family can be used as the parent for fingerlings propagation. The molecular marker screened out is prepared into a probe for detecting different parent families, the probe can sort out the streptococcus iniae disease resistant parent more simply and accurately, and selection of parent among different families is facilitated. Meanwhile, by using the bred disease-resistant parent as the propagation parent, quality of fingerlings can be improved further, and culture benefit of gift strain nile tilapia oreochromis niloticus can be increased.
Owner:FRESHWATER FISHERIES RES CENT OF CHINESE ACAD OF FISHERY SCI
12C<6+> ion beam radiation type breeding method for isatis tinctoria
ActiveCN103891597ASusceptible to mutationHigh mutation rateSeed and root treatmentPlant genotype modificationBiotechnologyWarm water
The invention relates to a 12C<6+> ion beam radiation type breeding method for an isatis tinctoria. The method comprises following steps of (1) selecting two kinds of parent materials according to a breeding objective, wherein one is a local germplasm with the relatively strong stress resistance, selected from local varieties, and the other is a breeding material selected from Anhui isatis tinctoria germplasm; (2) carrying out reciprocal cross on the two kinds of parent materials, then screening and reserving seeds for planting; (3) soaking the seeds in warm water, and then drying or naturally air drying, so as to obtain dried seeds; (4) soaking the dried full seeds, then air drying water on the surfaces of the seeds, irradiating a sample by a 12C<6+> ion beam, so as to obtain irradiated seeds; (5) planting the irradiated seeds in a field so as to obtain a first-generation group, carefully choosing 200 variation single plants according to the original parent; (6) continuously planting the variation single plants, harvesting, reserving seeds from second-generation single plants, continuously reserving seeds and breeding to obtain third-generations plants and fourth-generation plants; and (7) identifying and testing excellent mutation strains. The method has the high efficiency and realizes the purposes of synchronous improvement of multiple characters such as yield, quality and stress resistance within a short breeding period.
Owner:INST OF MODERN PHYSICS CHINESE ACADEMY OF SCI
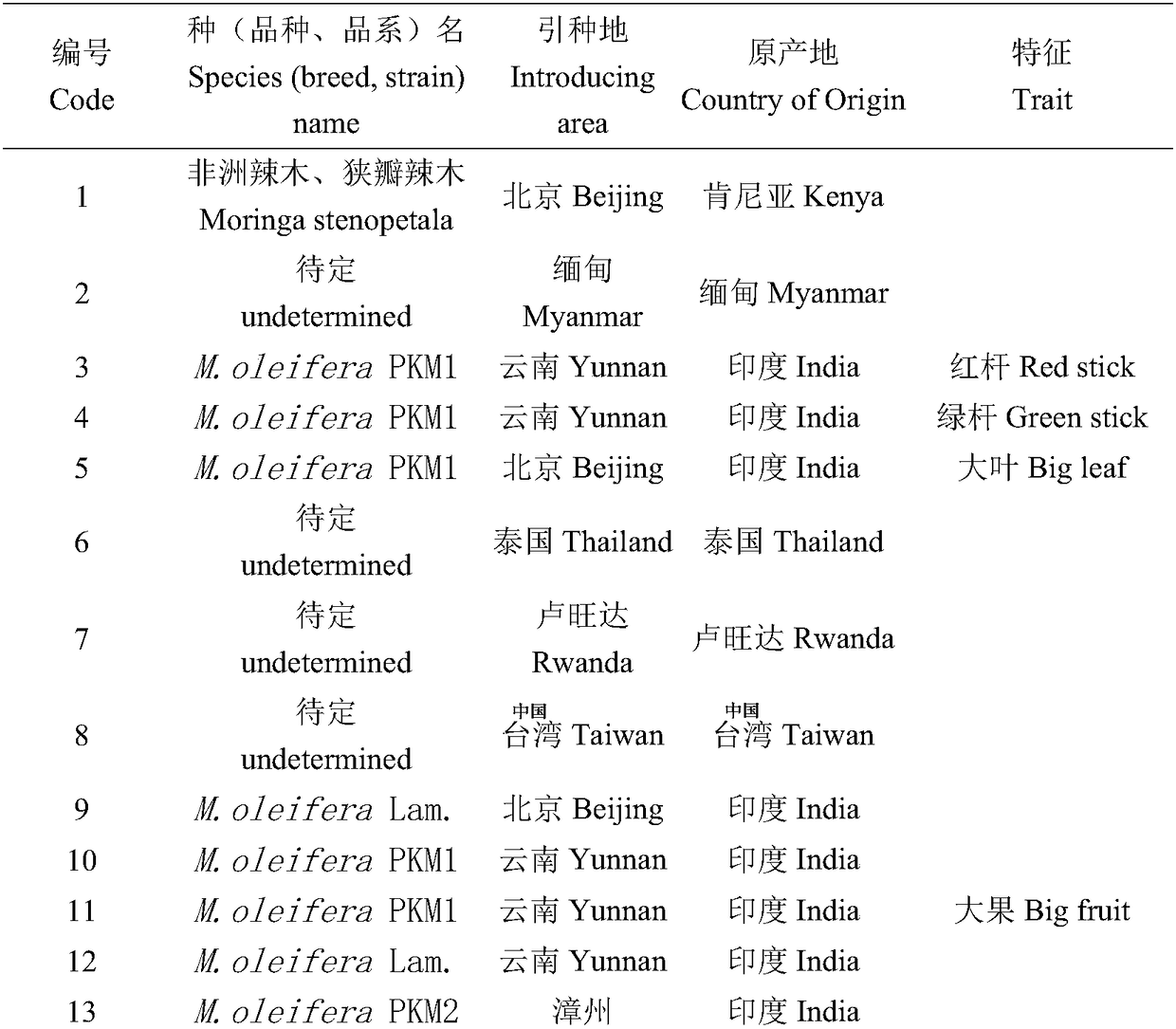
Reaction system and kit for analyzing moringa oleifera genetic relationship and application method thereof
ActiveCN108456719AGood polymorphismGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationGermplasmBiology
The invention provides a reaction system and kit for analyzing a moringa oleifera genetic relationship and application thereof. According to the technical scheme provided by the invention, 13 pairs ofprimers with good polymorphism and high definition are screened from 170 pairs of primers through an SRAP (Sequence Related Amplified Polymorphism) molecular marker technology and 18 parts of moringaoleifera DNAs (Deoxyribonucleic Acid) are marked; NTSYS software is used for finishing data analysis and UPGMA (Unweighted Pair-group Method with Arithmetic means) clustering; furthermore, the moringa oleifera genetic relationship and the genetic diversity are researched. A result shows that different moringa oleifera lines have abundant genetic diversity and an SRAP molecular marker is an effective means for analyzing themoringa oleifera germplasm genetic relationship; important technical supports are provided for moringa oleifera molecular assisted breeding and establishment of a genetic map database.
Owner:FUJIAN INST OF TROPICAL CROPS
Molecular marker of multieffect QTLs locus qTLS-4 for regulating and controlling size of rice leaf and application of molecular marker
ActiveCN112126703ASize predictionSpeed up breedingMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyA-DNA
The invention discloses a multieffect QTLs locus for regulating and controlling a size of a boot leaf of rice. The QTLs locus is located on a DNA fragment of a chromosome 4# of the rice, is called after qTLS-4, has a genetic distance of 87.1cM-97.4cM and a physical distance of 28747360bp-29507404bp and plays a role in regulating and controlling the size of the boot leaf of the rice. The inventionfurther discloses a molecular marker of the multieffect QTLs locus qTLS-4 for regulating and controlling the size of the boot leaf of the rice. The molecular marker comprises two pairs of molecular markers Indel Fls-1 and Indel Fls-2 which are closely chained. According to the molecular marker, whether a rice variety or line has the QTLs for regulating and controlling the size of the boot leaf ofthe rice or not is detected by using a molecular marker method, and thus, a breeding progress of excellent varieties of the rice is accelerated.
Owner:ZHEJIANG NORMAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com