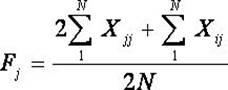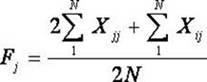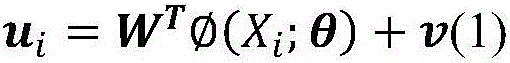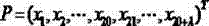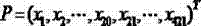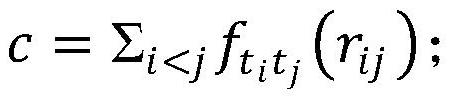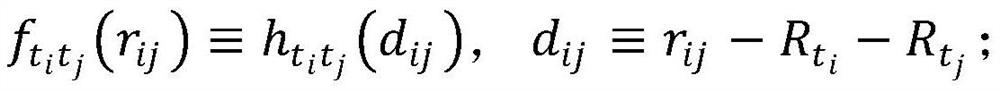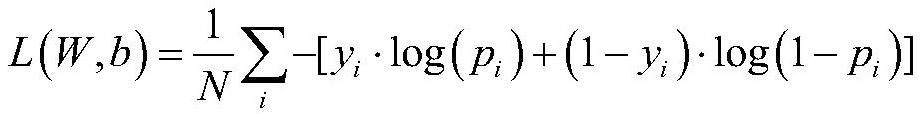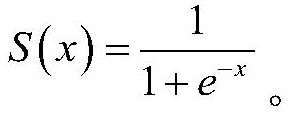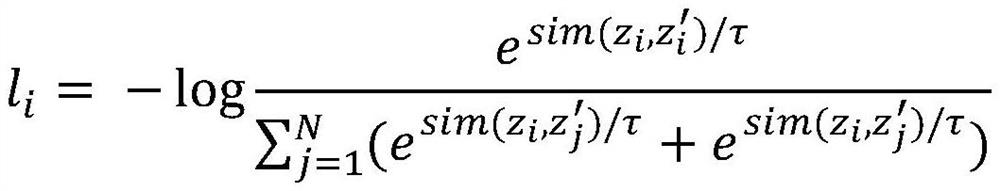Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
84 results about "Molecular Fingerprint" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Remote detection of substance delivery to cells
InactiveUS20050112065A1Highly preventive effectIncreasing effect on proton relaxivityDiagnostics using lightDispersion deliveryLipid formationElectrochemical gradient
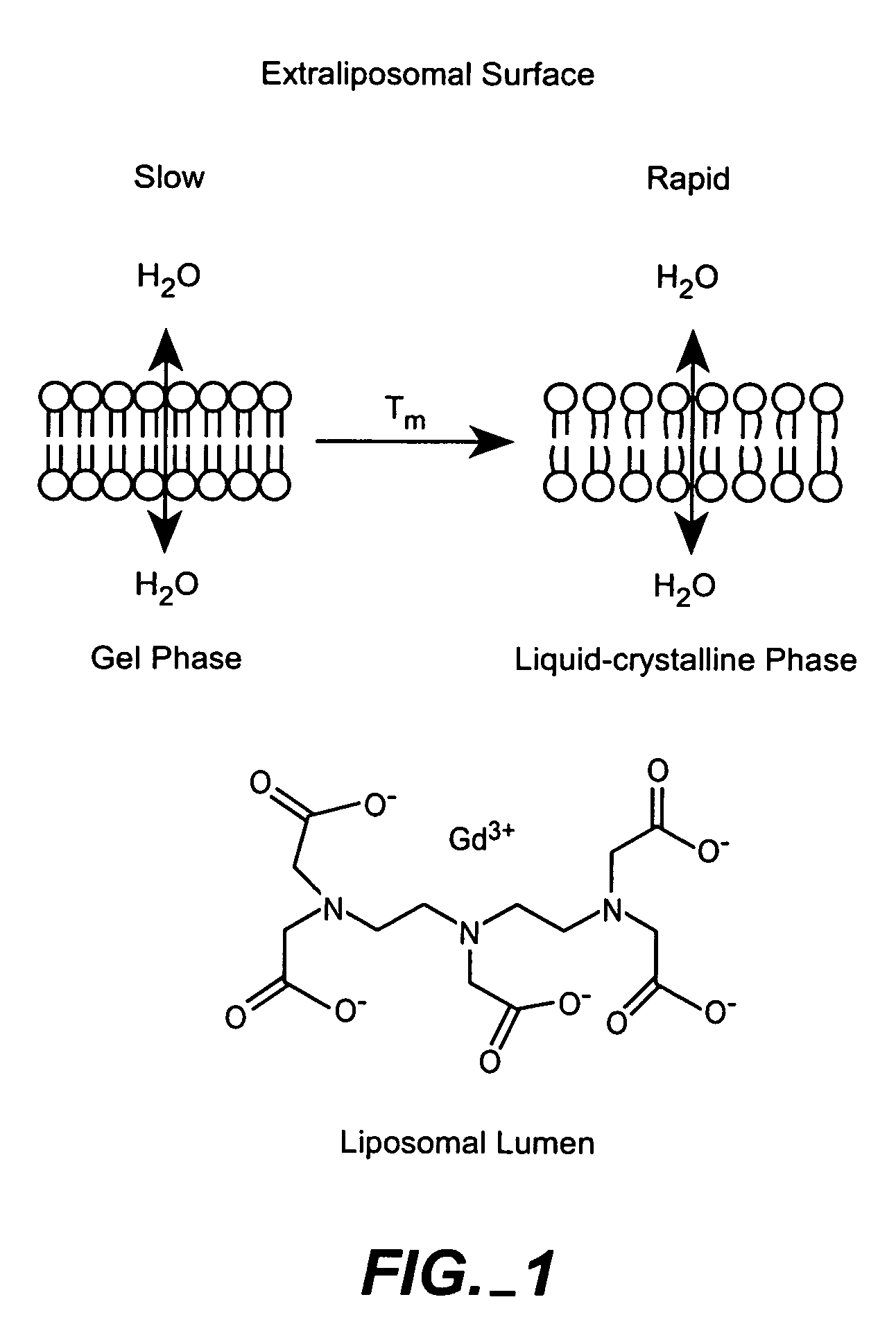
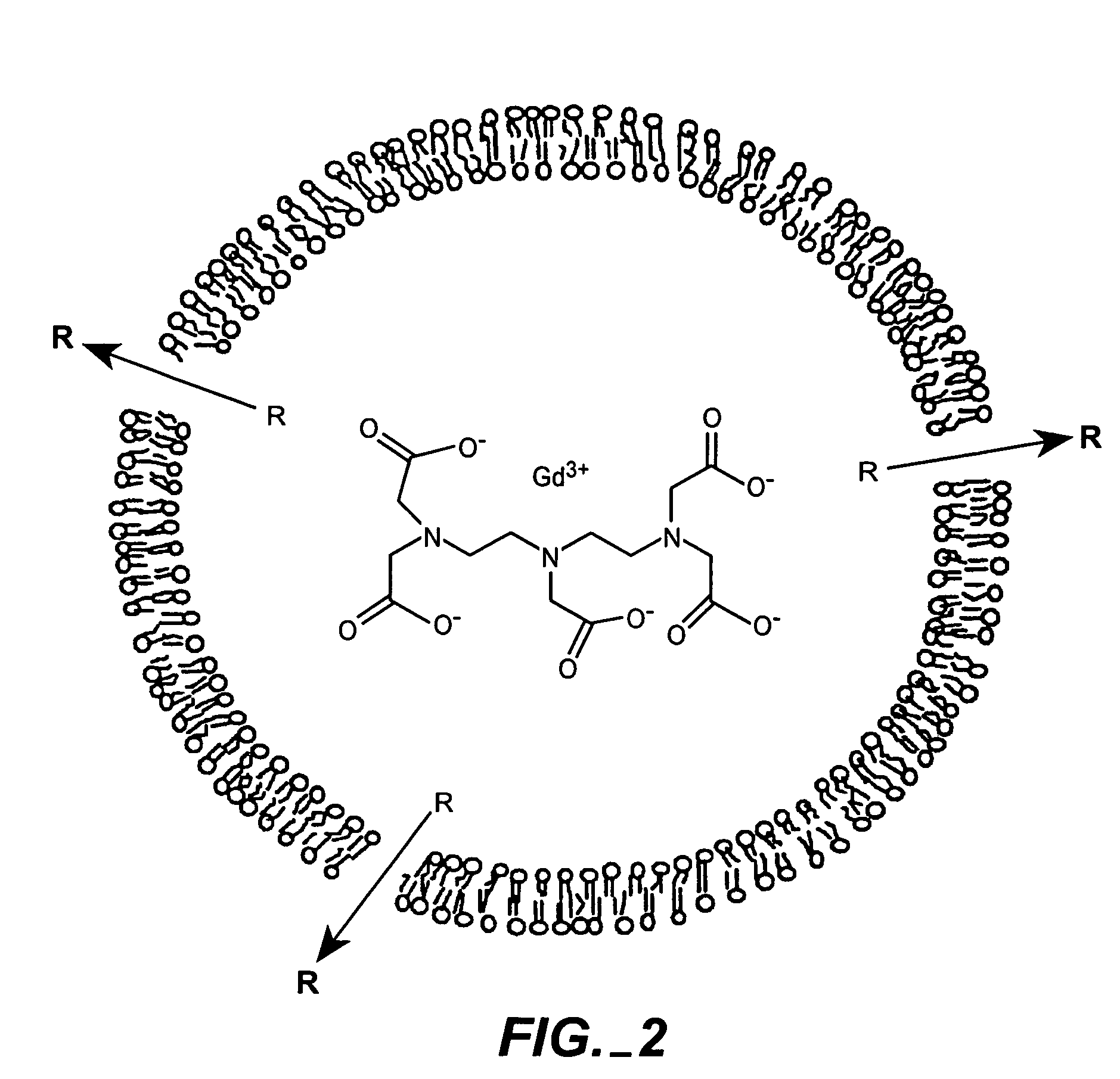
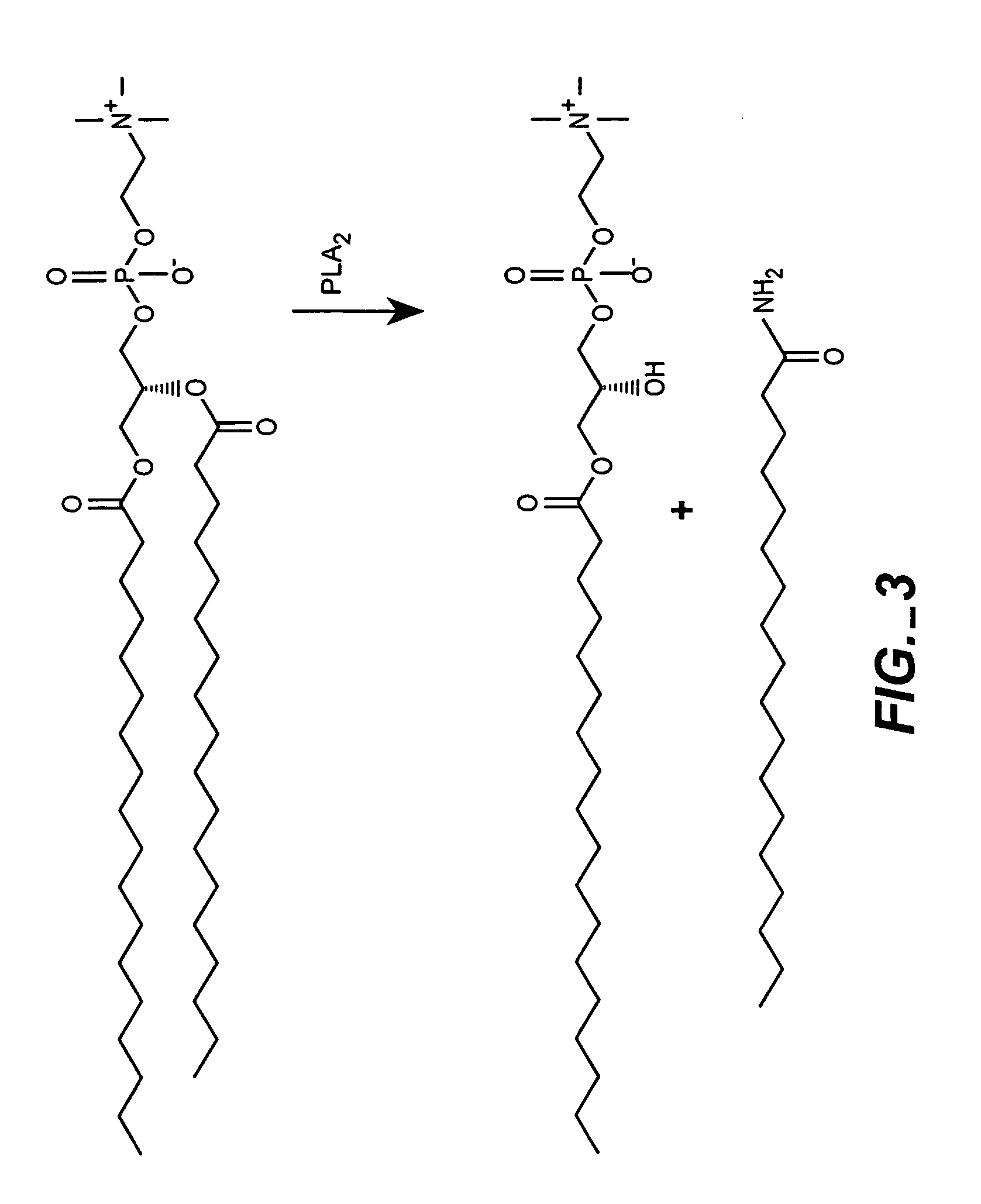
The present invention provides for the development of endocytosis-sensitive probes, and a remote method for measuring cellular endocytosis. These probes are based on the reduced water permeability of a nanoparticle or liposomal delivery system, and inherent degradability or disruption of barrier integrity upon endocytosis. The invention also provides for liposomes having combined therapeutic and diagnostic utilities by co-encapsulating ionically coupled diagnostic and therapeutic agents, in one embodiment, by a method using anionic chelators to prepare electrochemical gradients for loading of amphipathic therapeutic bases into liposomes already encapsulating an imaging agent. The invention provides for imaging of therapeutic liposomes by inserting a lipopolymer anchored, remotely sensing reporter molecules into liposomal lipid layer. The invention allows for an integrated delivery system capable of imaging molecular fingerprints in diseased tissues, treatment, and treatment monitoring.
Owner:SUTTER WEST BAY HOSPITALS
Small molecule drug virtual screening method based on deep migration learning and application thereof
ActiveCN110459274AImplement the buildMolecular designChemical machine learningDrug targetDrug biological activity
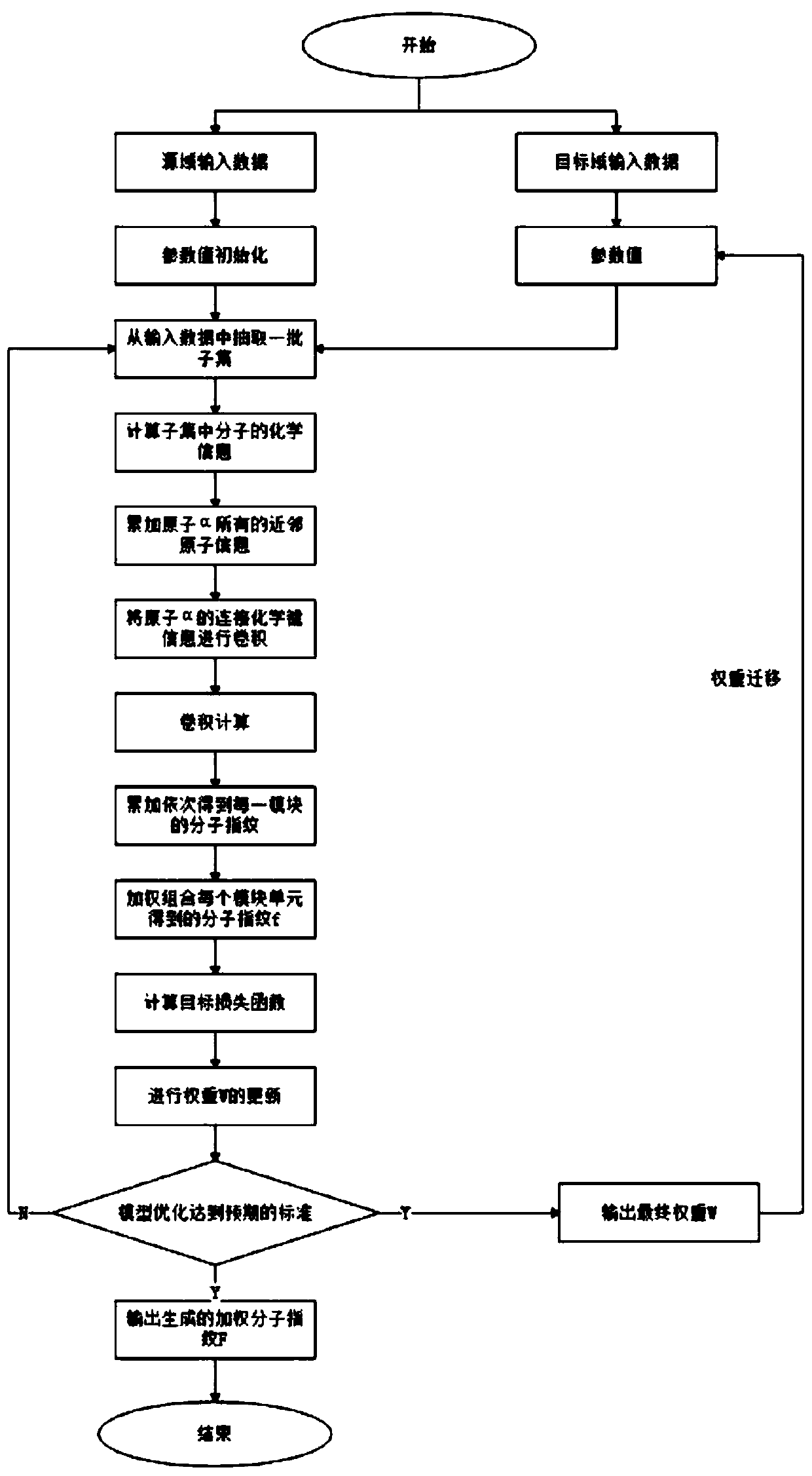
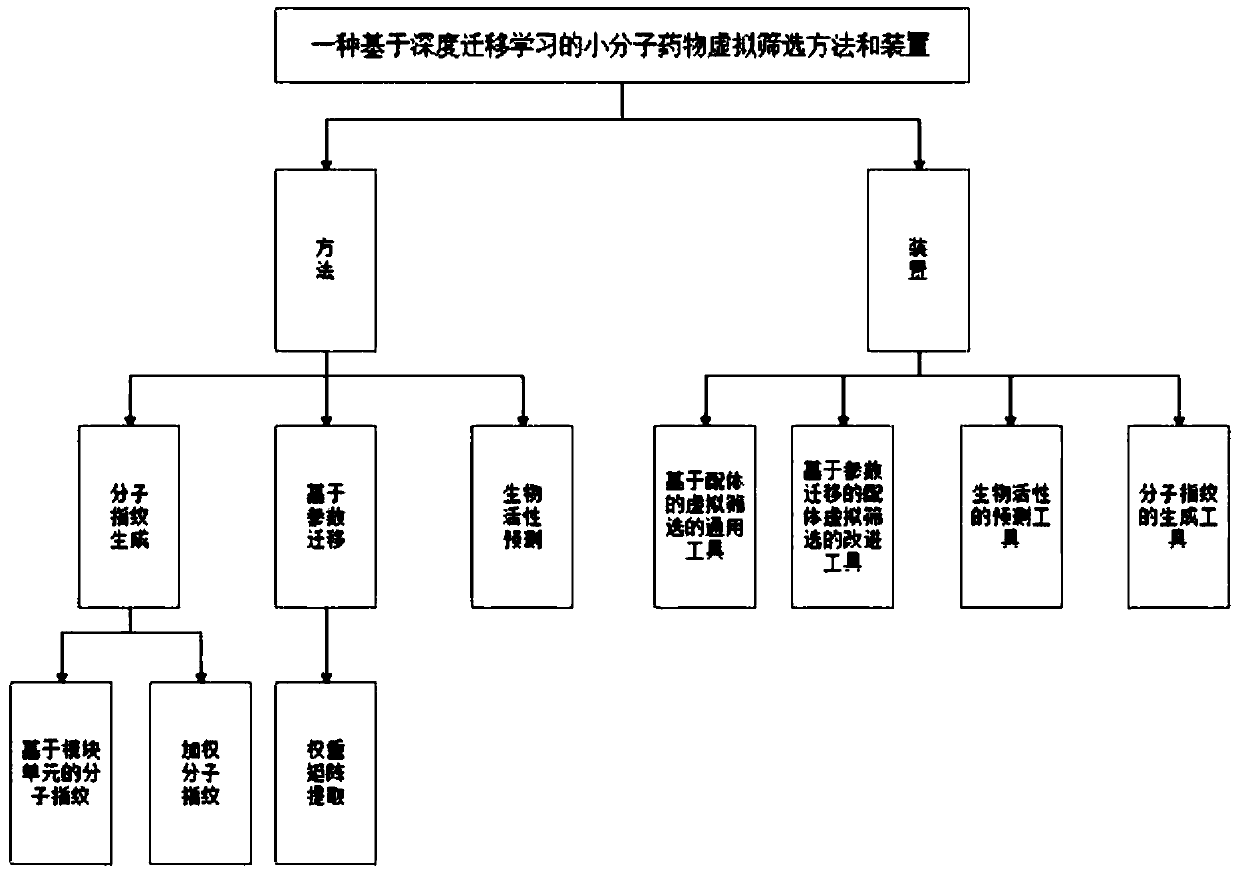
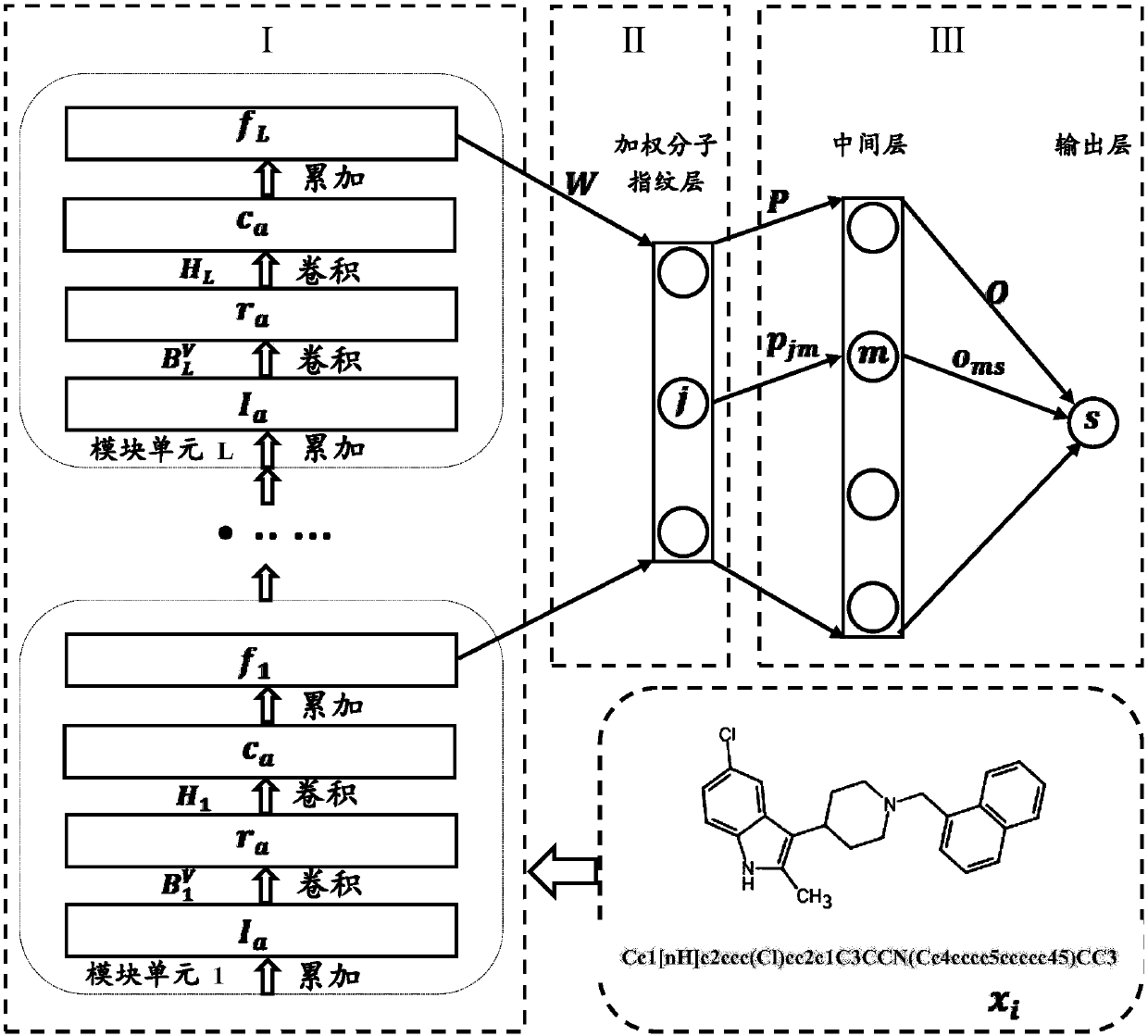
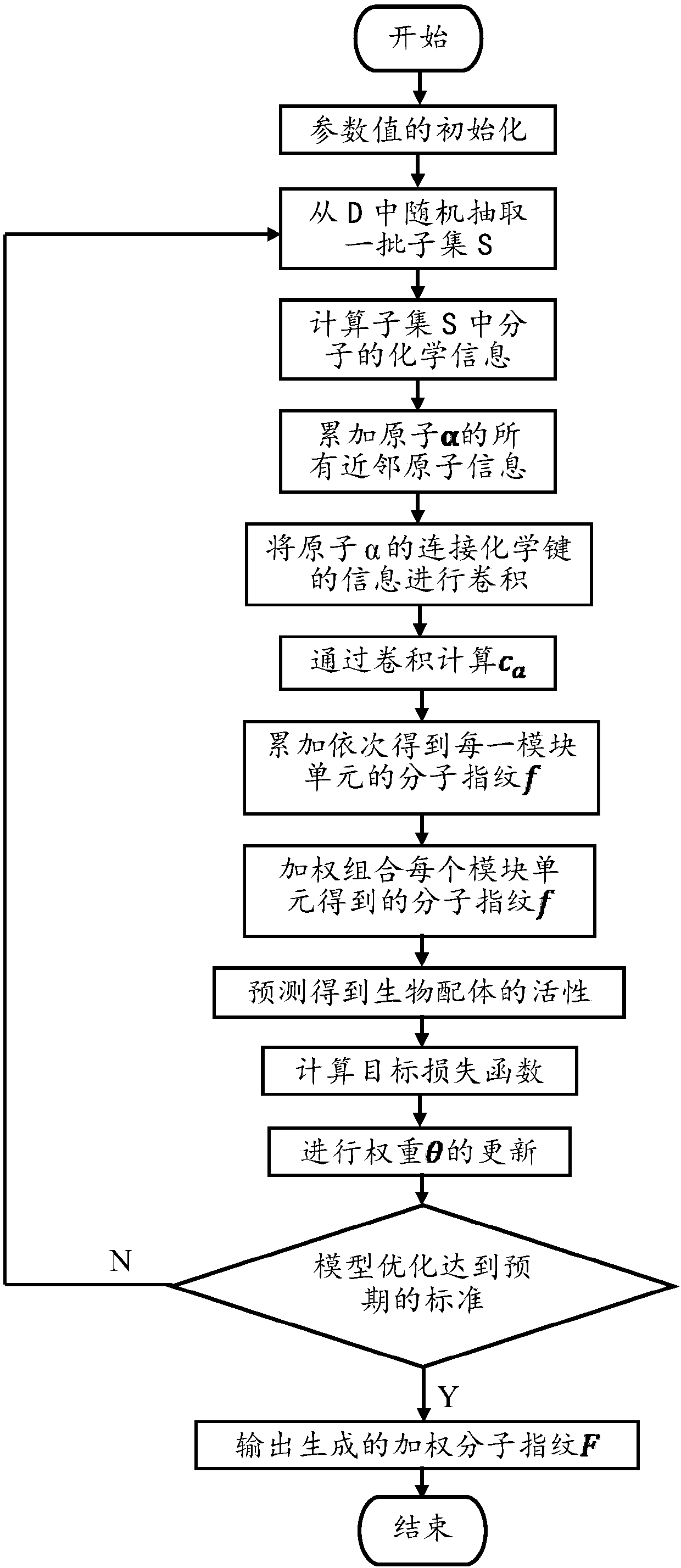
The invention discloses a small molecule drug virtual screening method based on deep migration learning and application thereof. A source domain is used as an input to be trained, converged and derived to obtain a weight matrix; a target domain is input into an improvement tool to serve as the initialization weight of the target domain; fine adjustment, training and convergence are conducted on the initialization weight and data in the target domain sequentially; a biological activity value of interaction of a lead compound and a drug target in the target domain is predicted, a target domain molecular fingerprint and a predicted value are obtained, and an evaluation index root mean square error and a correlation coefficient of a predicted result are output; the target domain is subjected to fine adjustment by repeating above steps, and the weight matrix of the source domain helps the target domain build a model. According to the small molecule drug virtual screening method and the application thereof, the effective virtual screening model can still be obtained under the condition that the information of a known active ligand sample is insufficient, and does not need to rely on a large number of data samples.
Owner:NANJING UNIV OF POSTS & TELECOMM
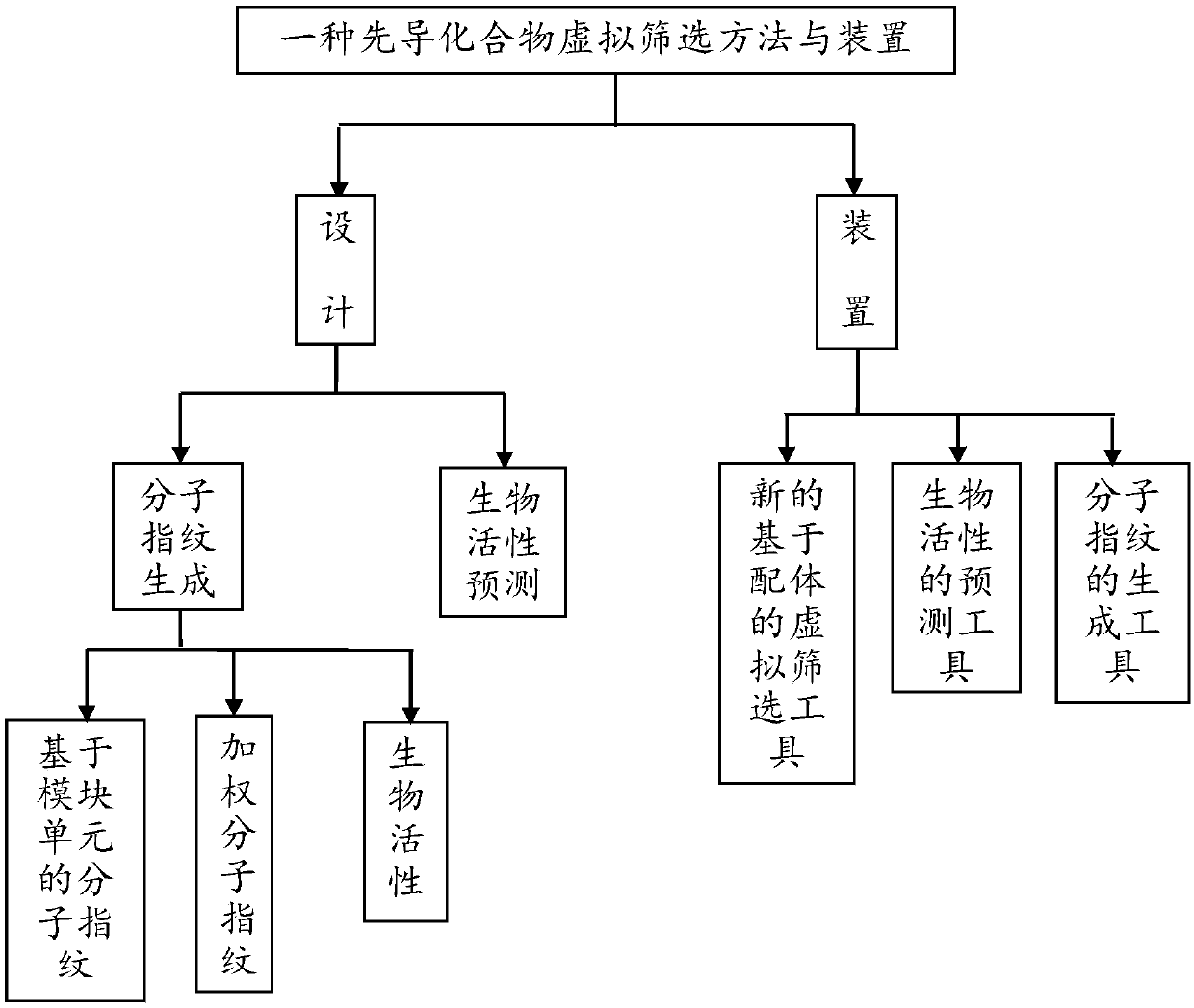
Lead compound virtual screening method and device
ActiveCN107862173AImprove performanceChemical property predictionChemical structure searchVirtual screeningMedicine
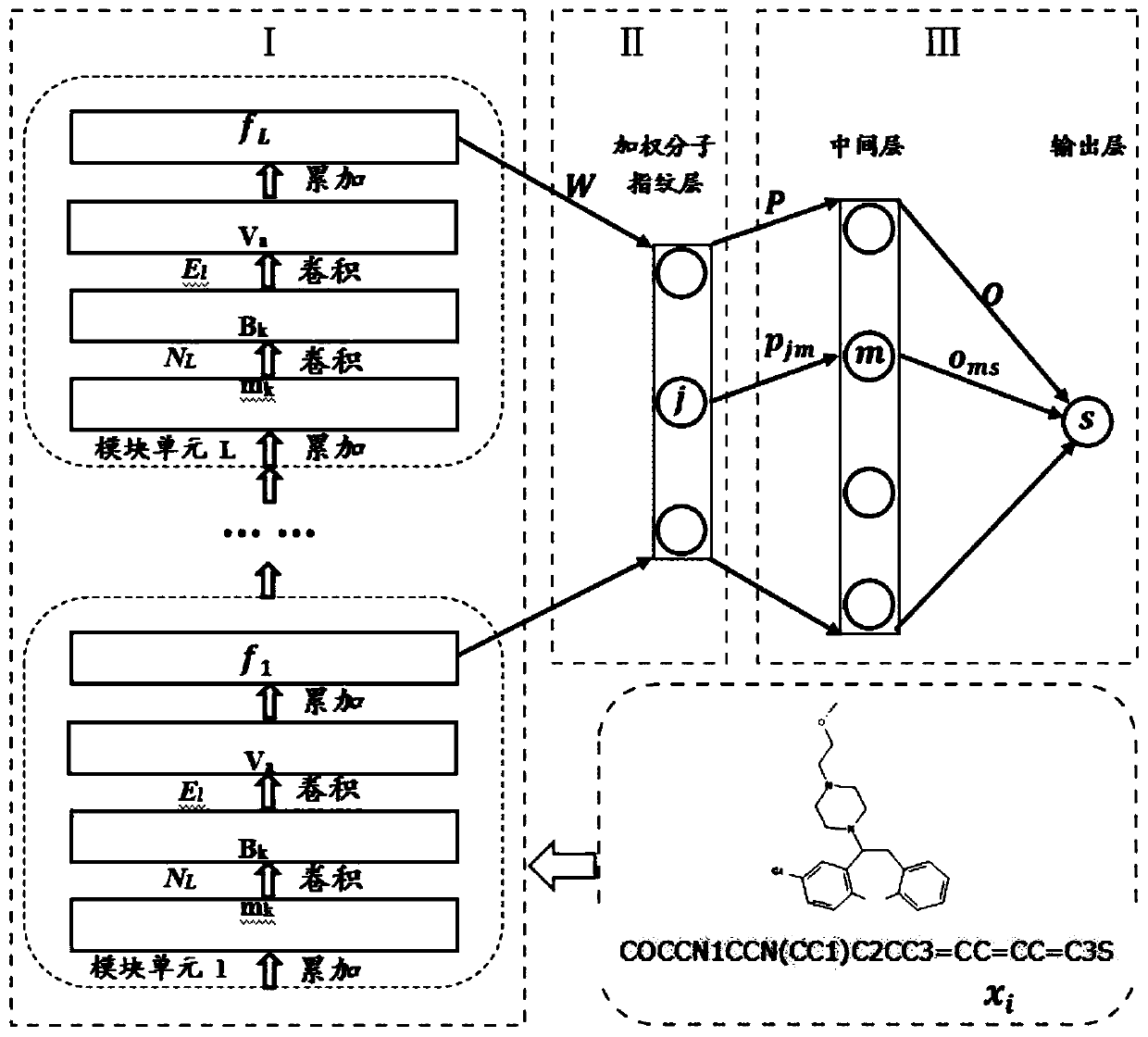
The invention discloses a lead compound virtual screening method and device. The method includes the steps of generation of molecular fingerprints of lead compounds on drug targets and bioactivity prediction of interaction between the lead compounds and the drug targets. The generation of the molecular fingerprints includes a molecular fingerprint part based on a module unit, a weighting molecularfingerprint part and a bioactivity part. During the bioactivity prediction, ligand molecular fingerprints and bioactivity values are utilized to serve as input of a random forest regression model, and a prediction model is constructed. Additionally, the device includes a universal tool for virtual screening on the basis of ligands, a prediction tool for the bioactivity generated when the lead compounds take effects on the drug targets, and a generation tool of the molecular fingerprints of the lead compounds on the drug targets. At present, molecular fingerprints which are excellent in performance and are used for the bioactivity prediction are often greater in length, and however, by adopting a designed deep learning algorithm, molecular fingerprints which are excellent in performance and smaller in length can be generated so that the best bioactivity prediction model of drug target ligands can be obtained.
Owner:NANJING UNIV OF POSTS & TELECOMM
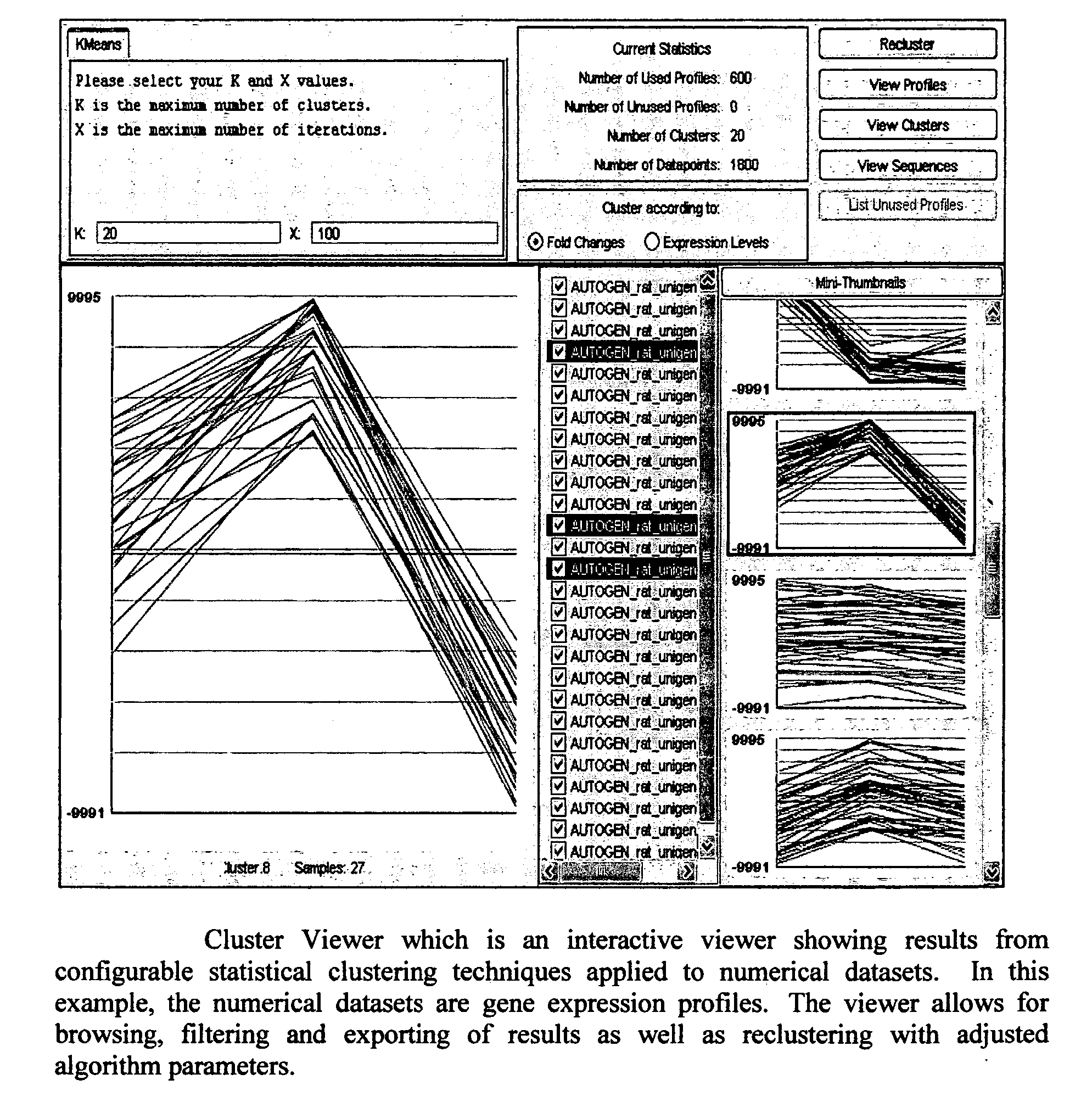
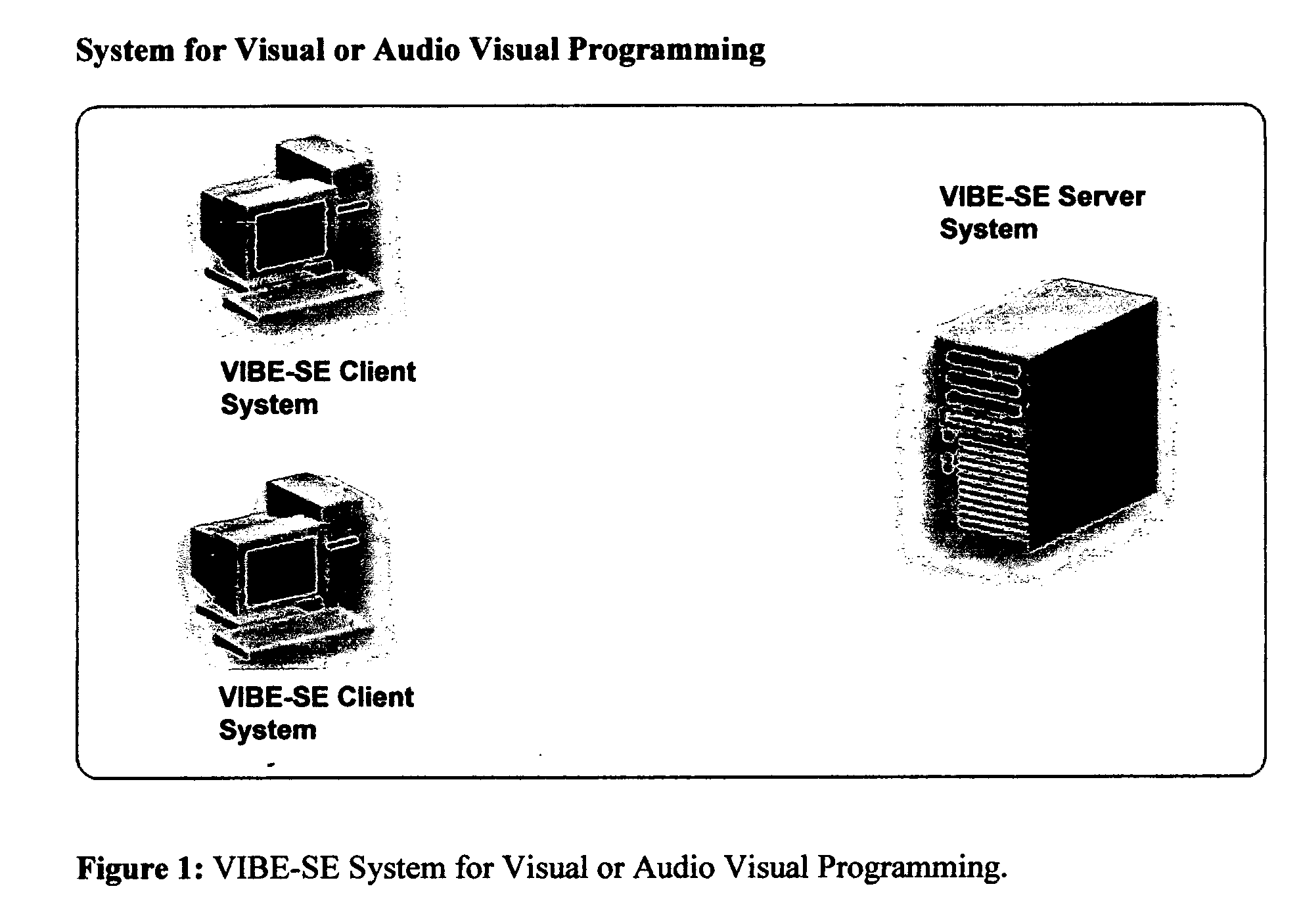
System and method using a visual or audio-visual programming environment to enable and optimize systems-level research in life sciences
InactiveUS20060190184A1Enabling and optimizing systems-level researchEnhances with user-level extensibilityData visualisationBiological testingAnalysis dataProteomic Profile
The current invention provides a visual or audio-visual programming environment for life science and bioinformatics. It is based on the VIBE platform, which is a flexible, extensible, and integrated workflow construction and management platform. The current invention enables researchers to consolidate molecular profiling data from complementary experimental techniques, intelligently reduce the volume of the data, construct disease-specific molecular fingerprints, construct relationship networks among functionally significant genomic, transcriptomic, metabonomic, and proteomic data, integrate information from existing biological databases into those networks, optimize the process through iterative feedback loops, and to generate and validate hypotheses based on the above process. The uses of this integrative, systems-based approach include, but are not limited to, the identification of potential biomarkers, characterization and classification of diseases and pathogens, and discovery of drug targets.
Owner:INCOGEN
Molecular marker method for identifying indica type rice and japonica rice by using rice grain
InactiveCN102031253AHigh speedSimple methodMicrobiological testing/measurementDNA preparationAgricultural scienceAgarose electrophoresis
The invention belongs to the technical field of biotype identification, in particular to a method for identifying indica type rice and japonica rice using rice grain by using a rice grain (rice) and inserting or deleting (InDel) a molecular marker. The method comprises the following steps of: extracting DNA from the rice grain, and comparing full-genome DNA sequences of indica type rice 93-11 andjaponica rice Nipponbare to obtain 40 pairs of specific InDel primers; and performing fragment amplification, electrophoretic separation and electrophoresis pattern analysis on the extracted DNA the in rice seed to identify the characteristics of the indica type rice and japonica rice of a rice sample. The method concretely comprises the following steps of: taking the DNA extracted from the rice grain as a template, and analyzing and counting molecular fingerprint patterns obtained on the basis of a polymerase chain reaction and agarose electrophoresis by using combination of the 40 pairs of specific InDel primers; and determining the characteristics of the indica type rice and japonica rice of the tested rice seed (sample) according to gene frequency (Fi or Fj) calculated by 93-11 genotype and japonica rice Nipponbare genotype molecular fingerprint on 40 InDel loci of the sample. The result can be obtained by only detecting the sample of one grain of seed, and the method is convenient and rapid, accurate in identification, and has good popularization and application prospect.
Owner:FUDAN UNIV
Construction method of HPLC fingerprint and DNA fingerprint of Saussurea Involucrata
InactiveCN101418348AQuality improvementTotal authenticityComponent separationMicrobiological testing/measurementHplc fingerprintHomoplantaginin
The invention relates to an HPLC fingerprint spectrum of Saussurea involucrate and a method for establishing a DNA fingerprint spectrum of the Saussurea involucrate. The invention is characterized in that the HPLC fingerprint spectrum comprises fingerprint spectrums of two extraction positions with different polarities of ethyl acetate and n-butyl alcohol, and the fingerprint spectrum of the extraction position of the ethyl acetate has 20 common peaks, wherein 7 peaks refer to scopoletin, rutoside, isoquercitrin, xylostein, quercetrin, homoplantaginin and hispidulin respectively; the fingerprint spectrum of the extraction position of the n-butyl alcohol has 11 common peaks, wherein 3 peaks refer to bergapten, scopoletin and rutoside; and the DNA fingerprint spectrum consists of an RAPD molecular fingerprint spectrum and an ISSR molecular fingerprint spectrum. The quality and the reality and falsity of the Saussurea involucrate can be universally and effectively detected in the aspects of chemistry and biology by combination of HPLC fingerprint and DNA fingerprint.
Owner:INST OF PROCESS ENG CHINESE ACAD OF SCI
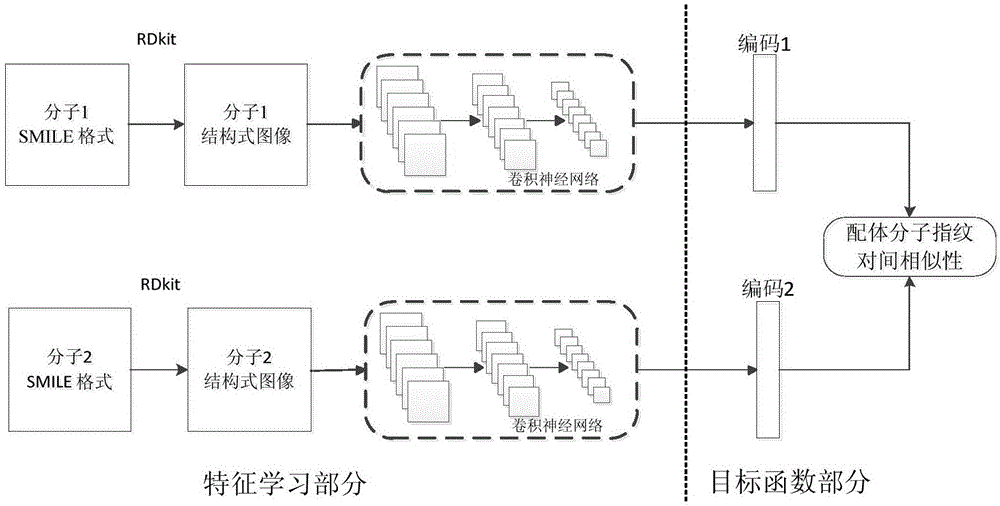
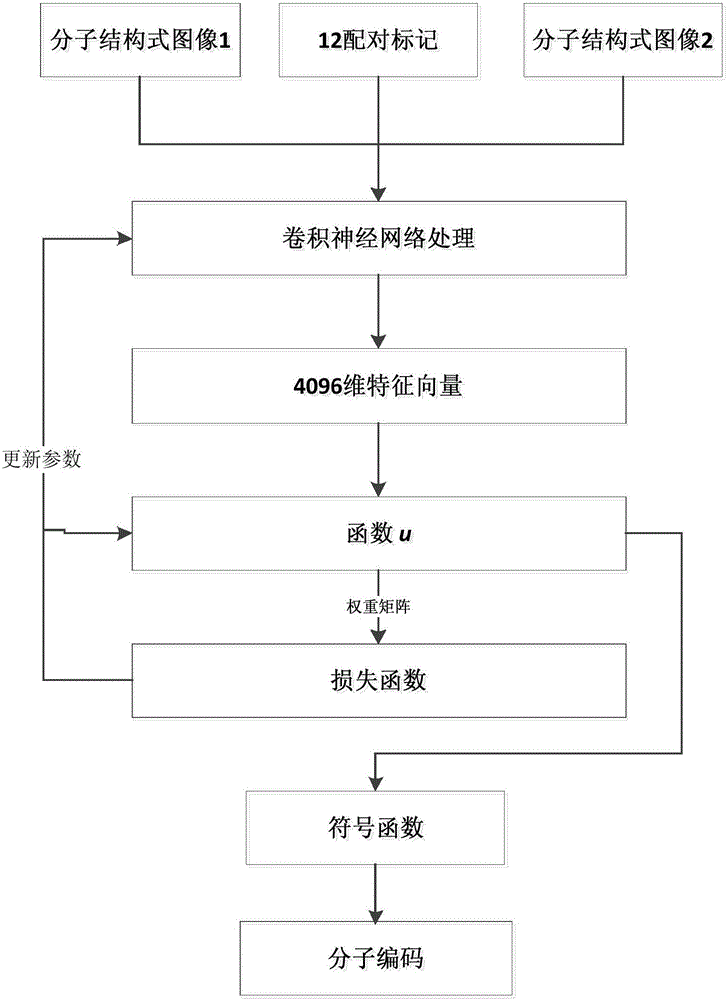
Ligand molecular fingerprint generation method based on deep Hash in drug screening
ActiveCN106777986AChemical property predictionMolecular entity identificationAlgorithmStructural formula
The invention discloses a ligand molecular fingerprint generation method based on deep Hash in drug screening. Firstly, a molecular structural formula image file is generated. Secondly, a pairwise label of a ligand molecular pair is defined, and a DPSH deep Hash learning model is trained. Finally, molecular fingerprint of new ligand molecules is predicted. A ligand molecular structural formula is converted into the image file, a target loss function is optimized by a deep Hash algorithm, and the molecular fingerprint is automatically generated. A first 'end-to-end' molecular fingerprint generation framework can be realized without manually extracting characteristics, and the method solves the problem that developers need to deeply understand field knowledge in an existing molecular fingerprint generation method. The universal molecular fingerprint generation framework is provided from a new perspective and serves as an important supplement for the existing molecular fingerprint generation method, and wider application of the molecular fingerprint to drug screening can be promoted.
Owner:NANJING UNIV OF POSTS & TELECOMM
Drug-target interaction prediction method based on graph convolution and word vector
ActiveCN110289050AImprove accuracyShorten the timeMolecular designChemical processes analysis/designAlgorithmProtein molecules
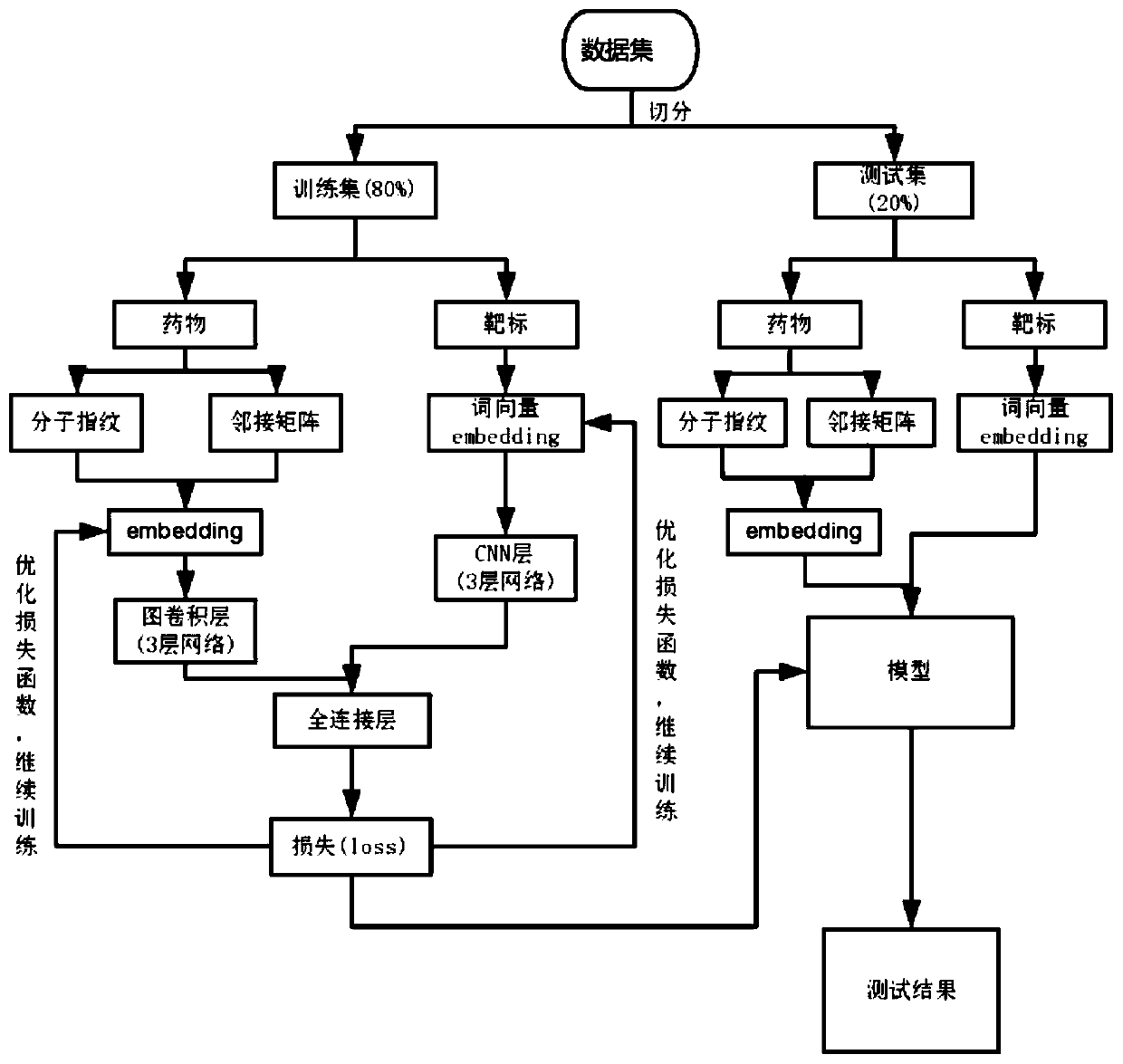
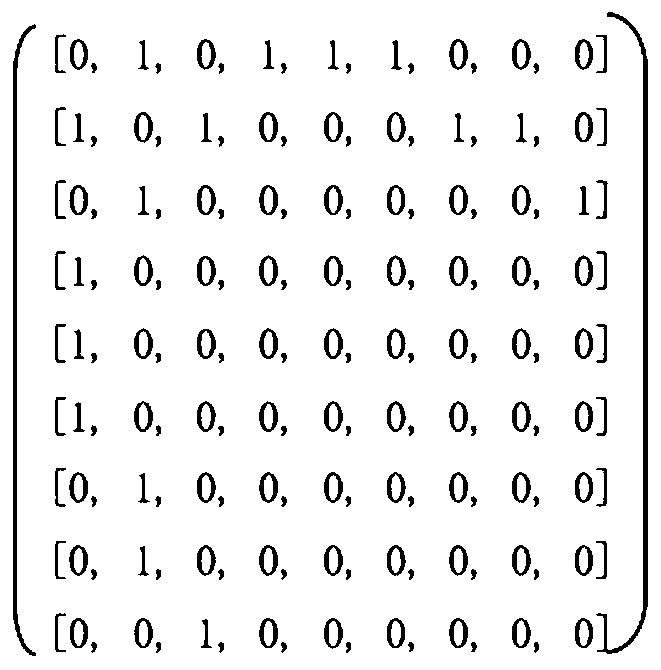
The invention provides a drug-target interaction prediction method based on graph convolution and word vectors. The method comprises the steps of: extracting molecular fingerprint features and adjacency matrix features from the medicine; training the features by utilizing graph convolution, cutting a protein molecular expression into groups, expressing the groups by utilizing a 100-dimensional vector, training word vector characteristics of the target by utilizing the CNN, and finally, combining the trained drug and the target together to carry out final result prediction. The method has the beneficial effects that more characteristics related to the medicine can be provided, so that higher accuracy is achieved; protein features are constructed by utilizing the word vectors, so that the feature construction time is greatly shortened; related information of a drug molecular diagram can be completely stored without losing characteristics; training time can be greatly shortened.
Owner:HUNAN UNIV
Virtual drug screening method and device, computing equipment and storage medium
ActiveCN111462833AFocus on physical and chemical propertiesFocus on dynamic featuresChemical property predictionMolecular designProtein targetAlgorithm
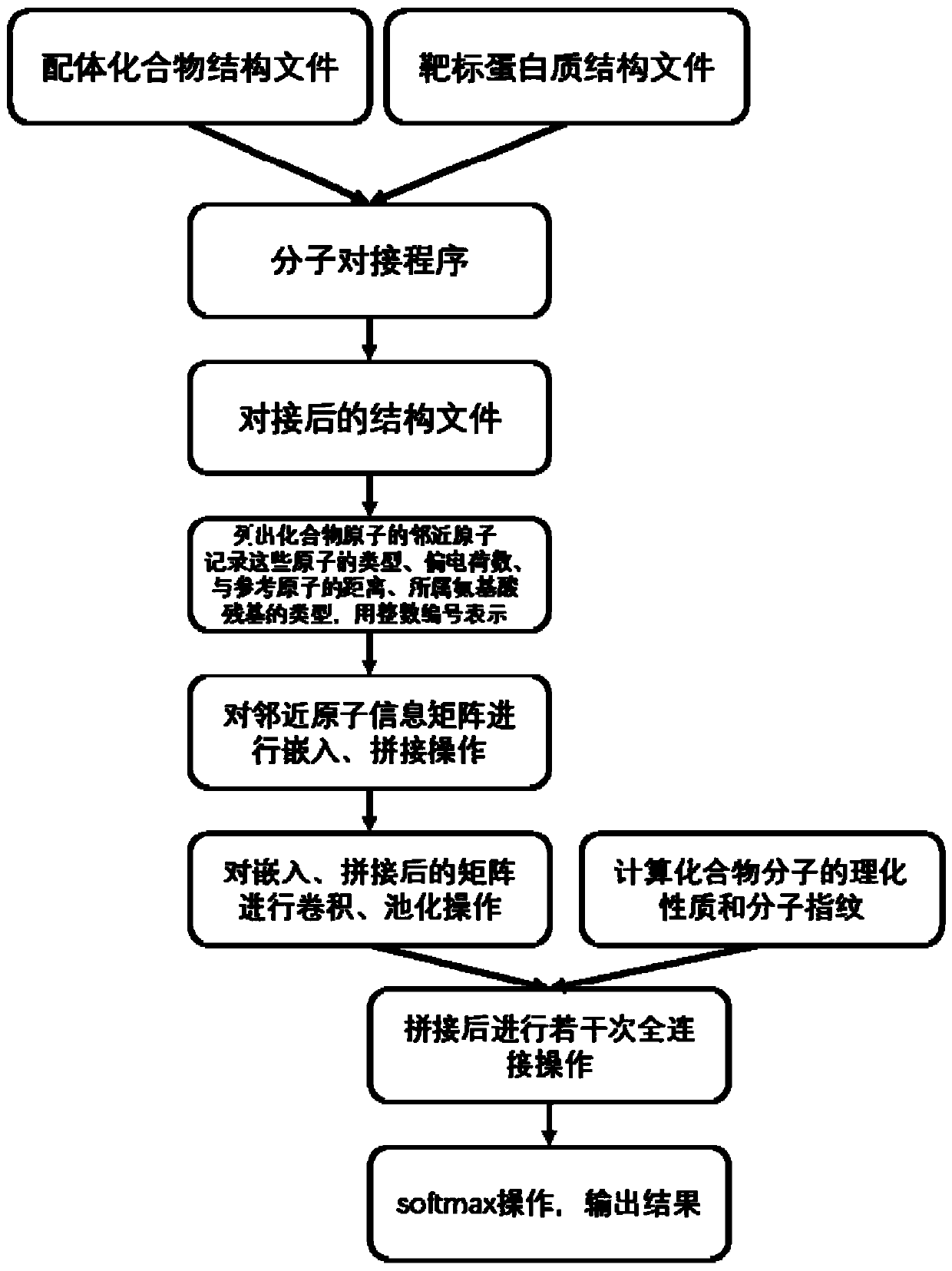
The invention discloses a virtual drug screening method and device, computing equipment and a storage medium. The method comprises the following steps: carrying out molecular docking on a ligand compound and a target protein; taking each atom contained in the docked compound molecules as a reference atom, determining a compound adjacent atom and a protein adjacent atom of each reference atom, recording corresponding predetermined structure information, and mapping the predetermined structure information into a structure information matrix group; performing embedding operation on the structureinformation matrix group by using a neural network, and obtaining a representation matrix of a compound-protein complex from the embedded structure information matrix group; carrying out convolution,bias and pooling on the representation matrix to obtain a structure vector; and splicing the structure vector with a physicochemical property vector representing the physicochemical property and the molecular fingerprint of the compound, and performing full-connection operation after neural network weighting and biasing to obtain a two-dimensional vector representing the inactivity and activity ofthe compound to the target protein so as to perform drug screening.
Owner:深圳智药信息科技有限公司
Compositions for cancer treatment and methods and uses for cancer treatment and prognosis
InactiveUS20200078401A1Strong cytotoxicityIncreased proliferationMicrobiological testing/measurementMammal material medical ingredientsPathway analysisTranscriptional analysis
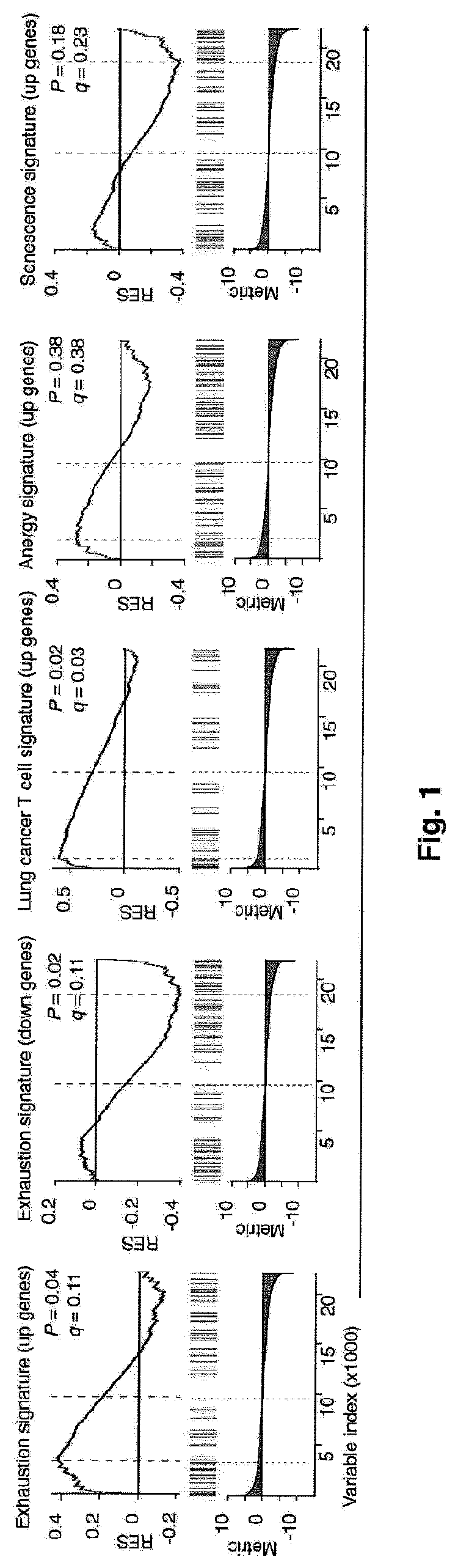
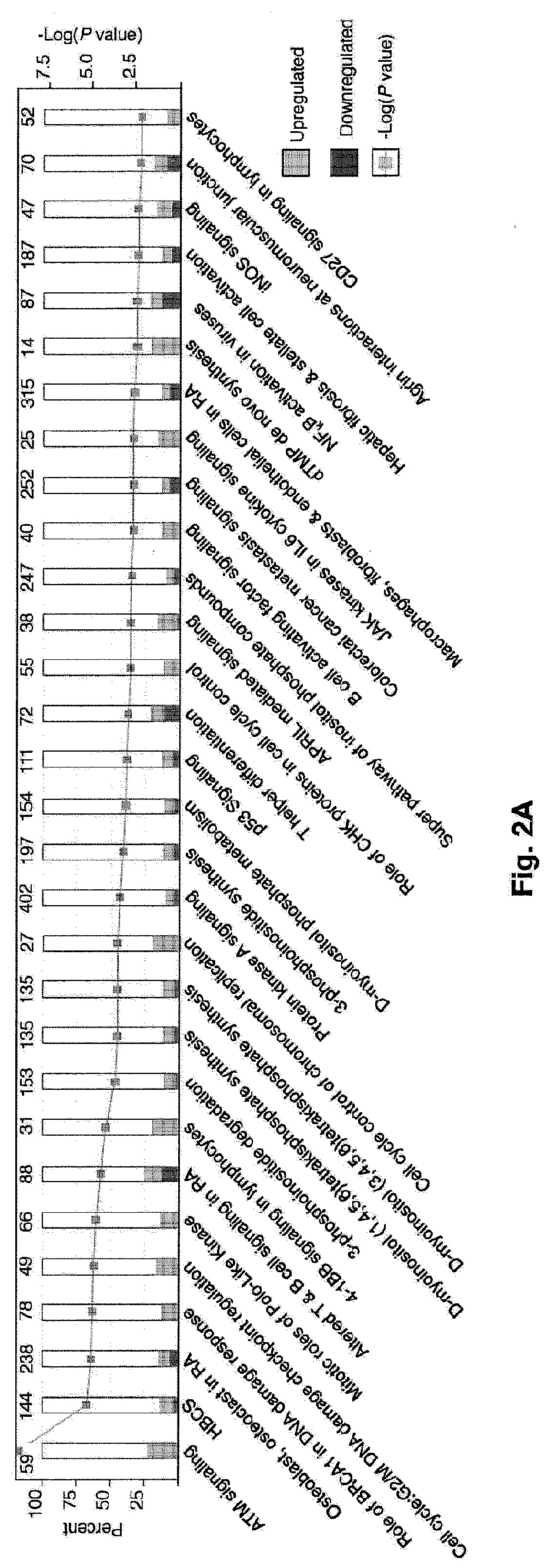
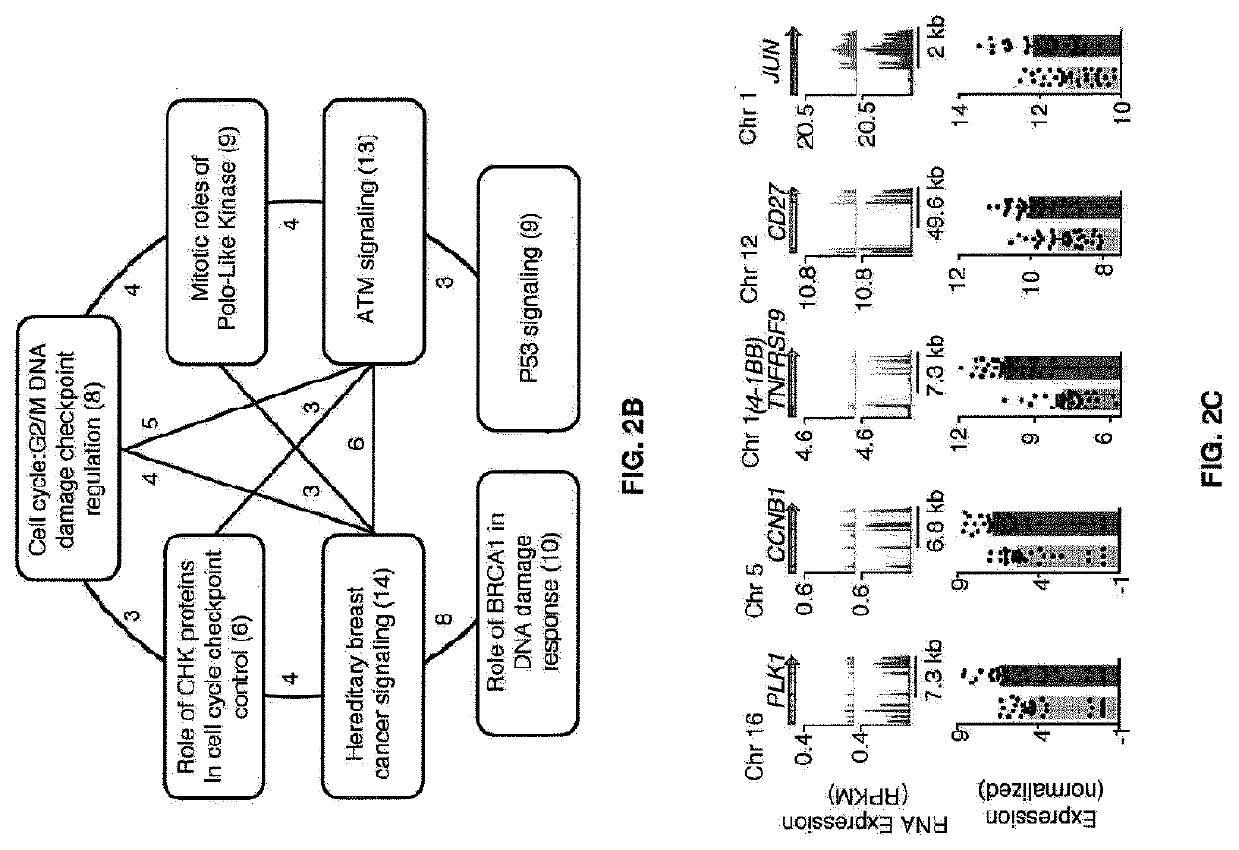
Global transcriptional profiling of CTLs in tumors and adjacent non-tumor tissue from treatment-naive patients with early stage lung cancer revealed molecular features associated with robustness of anti-tumor immune responses. Major differences in the transcriptional program of tumor-infiltrating CTLs were observed that are shared across tumor subtypes. Pathway analysis revealed enrichment of genes in cell cycle, T cell receptor (TCR) activation and co-stimulation pathways, indicating tumor-driven expansion of presumed tumor antigen-specific CTLs. Marked heterogeneity in the expression of molecules associated with TCR activation and immune checkpoints such as 4-1BB, PD1, TIM3, was also observed and their expression was positively correlated with the density of tumor-infiltrating CTLs. Transcripts linked to tissue-resident memory cells (TRM), such as CD 103, were enriched in tumors containing a high density of CTLs, and CTLs from CD 103high tumors displayed features of enhanced cytotoxicity, implying better anti-tumor activity. In an independent cohort of 689 lung cancer patients, patients with CD103high (TRM rich) tumors survived significantly longer. In summary, the molecular fingerprint of tumor-infiltrating CTLs at the site of primary tumor was defined and a number of novel targets identified that appear to be important in modulating the magnitude and specificity of anti-tumor immune responses in lung cancer.
Owner:LA JOLLA INST FOR ALLERGY & IMMUNOLOGY +1
Drug-target combined predicating method based on complexity and molecular fingerprints
InactiveCN102930179AOvercome the disadvantage of needing to know the three-dimensional structure of the proteinImprove prediction success rateSpecial data processing applicationsProtein targetAmbiguity
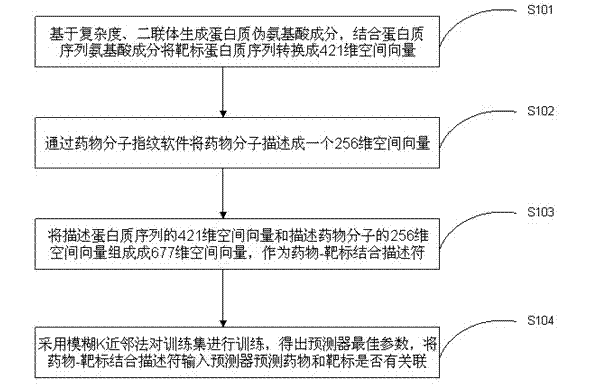
The invention discloses a drug-target combined predicating method based on complexity and molecular fingerprints. On the basis of the complexity and amino acid diad compositions, protein pseudo-amino acid compositions are generated, and protein sequence amino acid compositions are combined to convert target protein sequences into a 421 dimensional spatial vector; drug molecules are described into a 256 dimensional spatial vector through drug molecular fingerprint software; the 421 dimensional spatial vector which describes the protein sequences and the 256 dimensional spatial vector which describes the drug molecules are combined to form a 677 dimensional spatial vector serving as a drug-target combined descriptor; and an ambiguity K neighboring method is used for training a training set to obtain the best parameter of a predictor, and the drug-target combined descriptor is input into the predicator for predicating whether a drug is related to a target. According to the method, three dimensional structures of protein are not required to be measured, only one-dimensional sequences of the protein and the drug molecular fingerprints are required to predicate whether the drug and the protein can be combined, and the predicating success ratio is high.
Owner:JINGDEZHEN CERAMIC INSTITUTE
Local electromagnetic field enhancement device for Raman spectrum characterization as well as preparation method, application and utilization method thereof
InactiveCN108226133AEnhanced Raman scattering signalImprove adsorption capacityRaman scatteringScanning probe techniquesMolecular levelElectromagnetic field
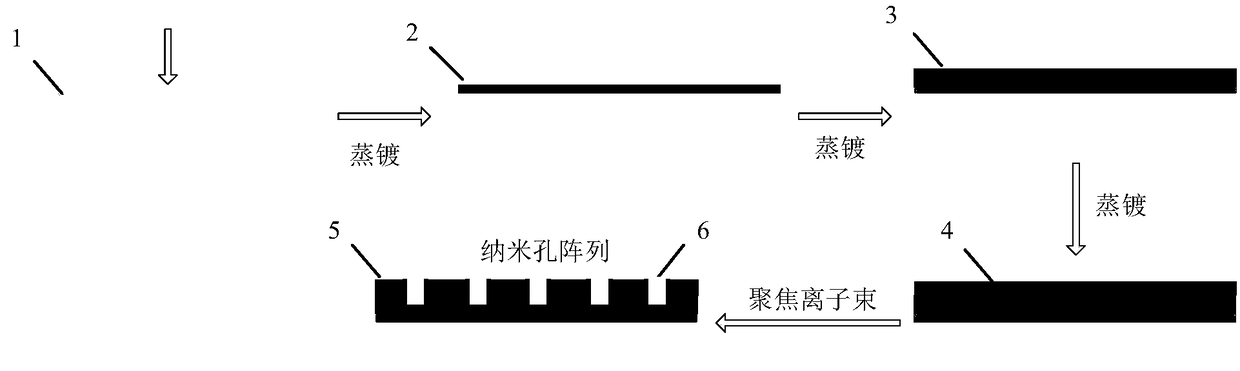
The invention relates to a local electromagnetic field enhancement device for Raman spectrum characterization as well as a preparation method, application and a utilization method thereof and belongsto the technical field of detection. The device comprises a composite metal nano-pore array substrate and an atomic force microscope probe, wherein the composite metal nano-pore array substrate is sequentially composed of a hard substrate, a chrome layer, a gold layer and a silver layer with the upper surface coated with a gold film from bottom to top; nano-pore arrays are arranged on the silver layer and the gold layer; the vertical depth of nano-pores is less than the sum of the thicknesses of the silver layer and the gold layer; pore walls of the nano-pores are coated with gold films; the surface of the atomic force microscope probe is coated with a silver film; the surface of a probe tip of the probe is provided with stripes. By adopting the device provided by the invention, single-molecule-grade molecular fingerprint spectrum characterization analysis can be realized, so that the disadvantage that configuration and conformation difference of a real single molecule on the surface of the substrate cannot be obtained by common Raman or surface enhanced scattering Raman is overcome; the local electromagnetic field enhancement device also can be applied to characterization of configuration and conformation of isomer molecules and discriminatory analysis of isomers in a single molecule level is realized.
Owner:CHONGQING INST OF GREEN & INTELLIGENT TECH CHINESE ACADEMY OF SCI
Online-internal-extraction electrospray-ionization mass-spectroscopy detection device for rapidly analyzing body fluid samples of human body
PendingCN106645374ALow costEasy to manufacturePreparing sample for investigationMaterial analysis by electric/magnetic meansCritical illnessSpectrograph
The invention relates to an online-internal-extraction electrospray-ionization mass-spectroscopy detection device for rapidly analyzing body fluid samples of a human body, and belongs to the technical field of chemical analysis. The detection device consists of an eluate introduction device, an in-situ sample enriching extractor, a sample introduction device, an electrospray ionization source and a mass spectrograph, wherein the eluate introduction device is used for introducing a flowing phase for sample eluting, the in-situ sample enriching extractor is used for carrying out sample loading and online extracting and enriching, the sample introduction device is used for introducing eluted samples into the electrospray ionization source for electrospraying, and the mass spectrograph is used for carrying out detection and analysis on online samples, so as to obtain mass-spectroscopy molecular fingerprint spectrogram information on the samples. According to the detection device, the detected human body fluid samples may be human body fluids such as blood, urine, sweat and saliva, do not need sample pretreatment, and are subjected to in-situ sample loading. The device provided by the invention is simple in structure and low-cost, the practicability is high, the detection time and economic cost are reduced greatly, and a technical support is provided for the rapid diagnosis and screening of critical illnesses.
Owner:JILIN UNIV
Remote detection of substance delivery to cells
InactiveUS20110104261A1Highly preventive effectImprove slackDiagnostics using lightDispersion deliveryImaging agentElectrochemical gradient
The present invention provides for the development of endocytosis-sensitive probes, and a remote method for measuring cellular endocytosis. These probes are based on the reduced water permeability of a nanoparticle or liposomal delivery system, and inherent degradability or disruption of barrier integrity upon endocytosis. The invention also provides for liposomes having combined therapeutic and diagnostic utilities by co-encapsulating ionically coupled diagnostic and therapeutic agents, in one embodiment, by a method using anionic chelators to prepare electrochemical gradients for loading of amphipathic therapeutic bases into liposomes already encapsulating an imaging agent. The invention provides for imaging of therapeutic liposomes by inserting a lipopolymer anchored, remotely sensing reporter molecules into liposomal lipid layer. The invention allows for an integrated delivery system capable of imaging molecular fingerprints in diseased tissues, treatment, and treatment monitoring.
Owner:SUTTER WEST BAY HOSPITALS +1
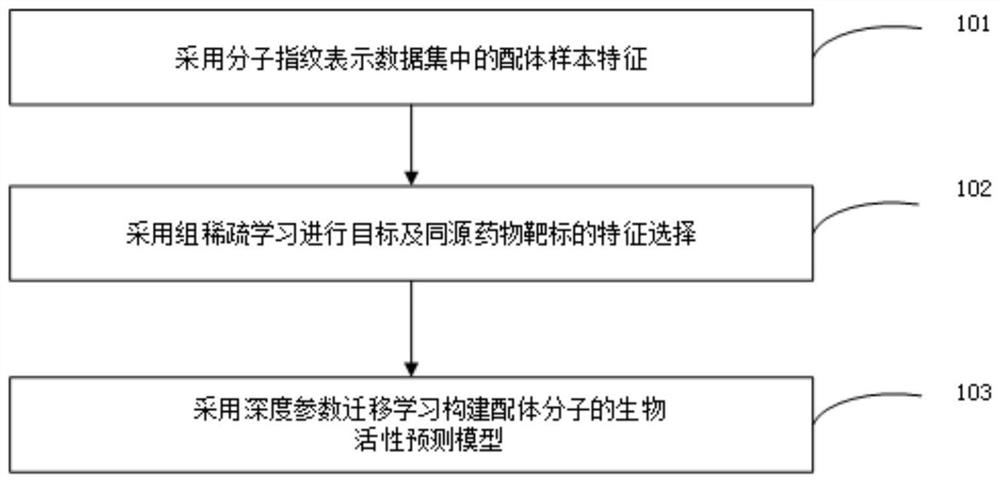
Small molecule drug virtual screening method and device based on deep parameter transfer learning
InactiveCN112086146ASolve the shortageGood forecastChemical property predictionMolecular designData setAlgorithm
The invention provides a small molecule drug virtual screening method and device based on deep parameter transfer learning. The method comprises the steps that molecular fingerprints represent ligandsample characteristics in a data set; feature selection is performed based on group sparse learning to obtain a key substructure; and the activity of the ligand small molecules is predicted based on deep parameter transfer learning. According to the invention, a good deep learning model is trained through similar drug targets rich in training samples, and the deep learning model of a target drug target is initialized by utilizing just learned model parameters according to the hypothesis that the similar drug targets are easy to fit to the similar deep learning model; and finally, the model isoptimized and updated by using limited training samples of the target drug target. The method based on deep parameter transfer learning can be used for trying to solve the problem of insufficient ligand samples in a drug virtual screening training data set, and has potential application value for virtual screening of new targets, understanding of interaction between ligands and the targets and optimization of ligand molecules.
Owner:NANJING UNIV OF POSTS & TELECOMM
Resonance raman spectroscopy analyzer instrument for biomolecules in tissues and cells
ActiveUS20170049327A1Increase vibration intensityDiagnostics using spectroscopySurgeryDiseaseResonance Raman spectroscopy
A method to detect vibrations associated with biomolecules in tissues and cellsuses Resonance Raman (RR) spectroscopy to measure specific biomolecules in tissue and cells signals. The changes of RR lines of key molecules present to the chemical conformations and change due to disease such as cancer and heart disease. Biomolecules are collagen, flavins, tryptophan, NADH, NAD, etc. The laser beams excite RR of vibration associated with absorption of the key native molecules in tissue (Tryptophan, NADH, Flavins, Collagen, carotenoids, porphyrins and others. The margin assessment and RR images in 2D and 3D regions are found by RR signals using position scanners. The intensity and the numbers of molecule fingerprints indicate the prescence of and the degree of the changes of chemical conformations.
Owner:ALFANO ROBERT R
Predication method of classification of drug in anatomical layer of ATC (Anatomical Therapeutic Chemical) system
InactiveCN103246824AImprove accuracyLow costSpecial data processing applicationsBiological systemDrug molecule
The invention provides a predication method of classification of a drug in an anatomical layer of an ATC (Anatomical Therapeutic Chemical) system. The theoretical basis of the invention is that compounds with the similar physic and chemical attributes have similar bioactivities; and ATC of the anatomical iatreusiology and chemical classification system of the official classification system of drugs are verified in the world health organization. The predication method comprises the steps of: mixing interaction information as well as similar information among molecules of the drug with fingerprint similar information of molecules of the drug; converting molecules of the drug into a digital sequence through a coding model; then establishing a 3883*42 dimension matrix based on a SVM (Support Vector Machine); normalizing the matrix; carrying out predictor studying by using the SVM; and further analyzing the classification of the sequence characteristics in the anatomical layer of the ATC system. The prediction method has the characteristics of high accuracy, relatively low cost, simple and convenient operation, and intuitional results, provides a new method for the research of new use of old drugs, and has a wide application space.
Owner:JINGDEZHEN CERAMIC INSTITUTE
Method for identifying cytoplasm type of cytoplasmic-genic male sterile rice by detecting mitochondrial DNA
InactiveCN102586435AEasy to identifyEfficient identificationMicrobiological testing/measurementBiotechnologyCytoplasm
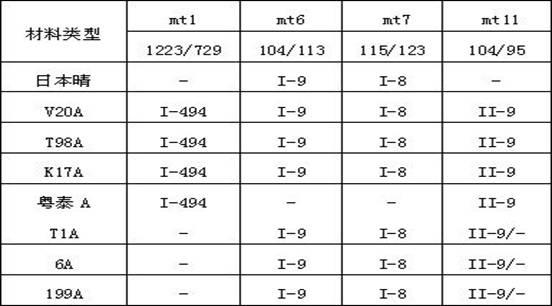
The invention discloses a method for identifying cytoplasm type of cytoplasmic-genic male sterile rice by detecting mitochondrial DNA. The method comprises the following steps of: extracting the mitochondrial DNA from rice leaves serving as materials; performing polymerase chain reaction (PCR) amplification by using mitochondrial primers mt1, mt6, mt7 and mt11, and cloning and sequencing amplification products; and comparing with molecular fingerprints of the mitochondrial DNA of the cytoplasmic-genic male sterile rice to easily and efficiently identify the cytoplasm type of the cytoplasmic-genic male sterile rice.
Owner:HUNAN NORMAL UNIVERSITY
Fluorescent SSR primer composition and application in constructing white wax new species molecular fingerprint spectrum thereof
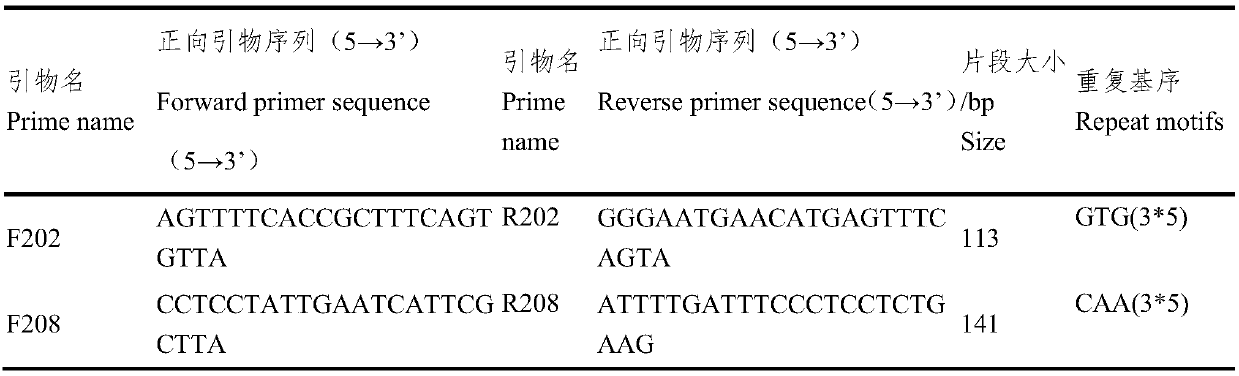
ActiveCN109652411ASpecific stabilityGood polymorphismMicrobiological testing/measurementDNA/RNA fragmentationWaxForward primer
The invention discloses a fluorescent SSR primer composition and application in constructing a white wax new species molecular fingerprint spectrum thereof. The fluorescent SSR primer composition is composed of SSR primer pairs of F202 / R202 and F208 / R208, FAM fluorescent marks are added to 5' ends of upstream primers of each pair of primers, and forward primers are composed of TP-M13 primers carrying fluorescent marks. According to the primer composition, the primer composition is obtained using a specific fingerprint spectrum constructing a white wax new species or white wax new species molecular ID card, and 20 white wax species can be completely distinguished to overcome the prior defects of morphological and physiological identification. The primer composition can provide scientific and effective reference bases for the further application researching of a DNA fingerprint spectrum on the white wax, the evaluation of white wax germ plasm resources, and the solving of species intellectual property right disputes of the white wax.
Owner:SHANDONG FOREST SCI RES INST +1
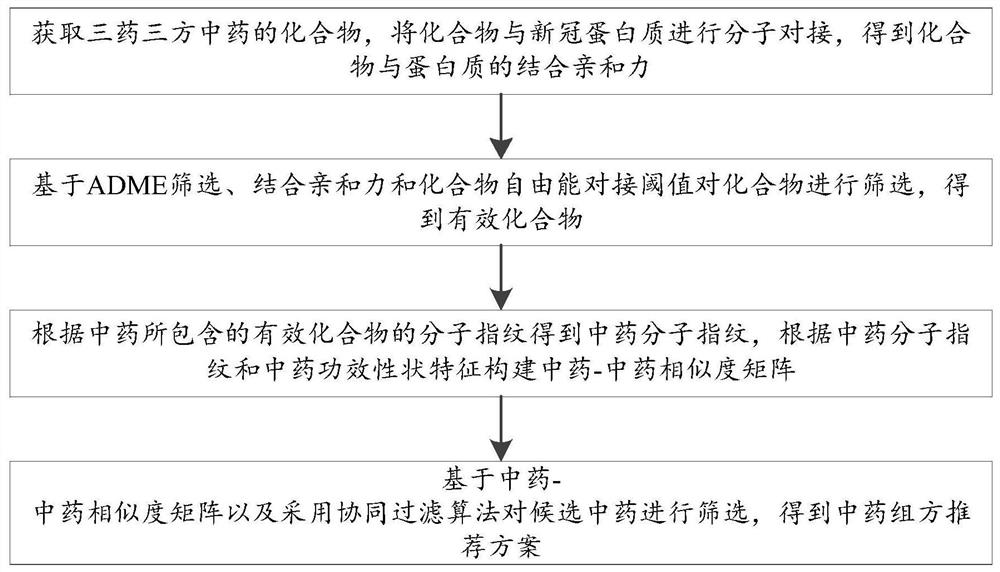
Novel coronavirus traditional Chinese medicine formula recommendation method and system based on collaborative filtering
ActiveCN112667922AImprove bindingLower free energyChemical property predictionDigital data information retrievalEfficacyTraditional medicine
The invention discloses a novel coronavirus traditional Chinese medicine formula recommendation method and system based on collaborative filtering, and the method comprises the steps: obtaining a compound of a three-medicine three-formula traditional Chinese medicine, carrying out the molecular docking of the compound and a new coronaprotein, and obtaining the binding affinity of the compound and the protein; based on ADME screening, screening the compound by combining affinity and a compound free energy docking threshold value to obtain an effective compound; obtaining traditional Chinese medicine molecular fingerprints according to the molecular fingerprints of the effective compounds contained in the traditional Chinese medicines, and constructing a traditional Chinese medicine similarity matrix according to the traditional Chinese medicine molecular fingerprints and traditional Chinese medicine efficacy trait characteristics; and screening the candidate traditional Chinese medicines based on the traditional Chinese medicine similarity matrix and a collaborative filtering algorithm to obtain a traditional Chinese medicine prescription recommendation scheme. A three-medicine three-prescription traditional Chinese medicine compound and new coronavirus protein are subjected to molecular docking, the combining capacity of traditional Chinese medicines and traditional Chinese medicine prescriptions and the new coronavirus protein is measured, meanwhile, traditional Chinese medicine prescriptions are recommended based on a collaborative filtering algorithm, and the effectiveness of the traditional Chinese medicine prescriptions on new coronavirus treatment is verified.
Owner:SHANDONG UNIV
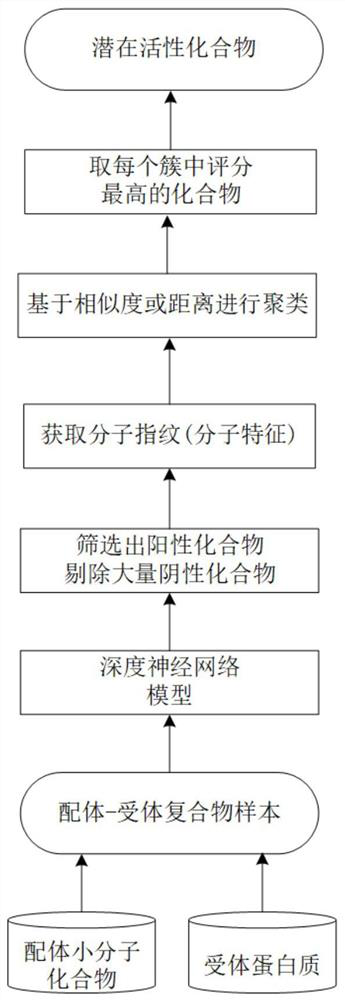
Ligand compound rapid pre-screening model based on deep learning
ActiveCN113393911AImprove pre-screening speedEfficient removalMolecular designCheminformatics data warehousingBinding siteReceptor
The invention provides a ligand compound rapid pre-screening model based on deep learning. The method includes the following steps: S1, constructing a data set, and encoding amino acid sequence information of binding sites of ligand compounds and receptor proteins; s2, combining the ligand and receptor structure vectors and inputting them into a deep neural network, outputting affinity scores, and training the model in a supervised learning mode; s3, sorting according to affinity scoring results, screening out positive compounds, and removing a large number of negative compounds; s4, obtaining molecular fingerprints (molecular characteristics) of positive compounds, and clustering based on a similarity or distance measurement method; and S5, taking the compound with the highest score in each cluster as a potential active compound. According to the method, rapid pre-screening of the ligand compound is achieved, redundancy can be removed through clustering analysis of the positive compound, the structural diversity of the ligand compound is guaranteed, and the prediction speed and accuracy are improved.
Owner:石家庄鲜虞数字生物科技有限公司 +3
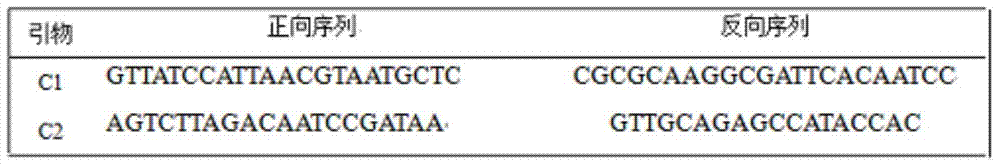
Method for detecting chloroplast DNAs to identify cytoplasmic male sterile line of hot pepper
InactiveCN104513856AEasy to identifyEfficient and accurate identificationMicrobiological testing/measurementBiotechnologyHot peppers
The invention discloses a method for detecting chloroplast DNAs to identify cytoplasmic male sterile line of hot pepper. The method comprises the following steps: taking the leaves of hot pepper as a material, extracting total DNAs in the leaves, carrying out PCR amplification by using chloroplast primers (C1 and C2), carrying out clone sequencing on amplification products, and then making a comparison with chloroplast DNA molecular fingerprints of the cytoplasmic male sterile line of hot pepper, namely identifying the cytoplasmic male sterile line of hot pepper simply, efficiently and accurately. The method for detecting chloroplast DNAs to identify the cytoplasmic male sterile line of hot pepper disclosed by the invention, the clone sequencing is carried out on the PCR amplification products of the chloroplast primers (C1 and C2) only when the cytoplasmic male sterile line of hot pepper is identified, and then a comparison is made, so that the cytoplasmic male sterile line of hot pepper can be identified simply, efficiently and accurately.
Owner:HUNAN UNIV OF ARTS & SCI +1
Mushroom bacterial standard detecting method
InactiveCN101134979AMicrobiological testing/measurementMaterial analysis by electric/magnetic meansDNA fragmentationMorphological trait
The present invention discloses standard Lentinus edodes seed detecting method, which includes 5 tested aspects including morphological characteristic, agronomic character, physiological character, quality character and molecular fingerprint of special DNA segment, and 36 test indexes, including 34 essential ones and 2 optional ones. Of the 34 essential indexes, there are 10 hypha character indexes and 24 cultivating character indexes.
Owner:上海市农业科学院食用菌研究所
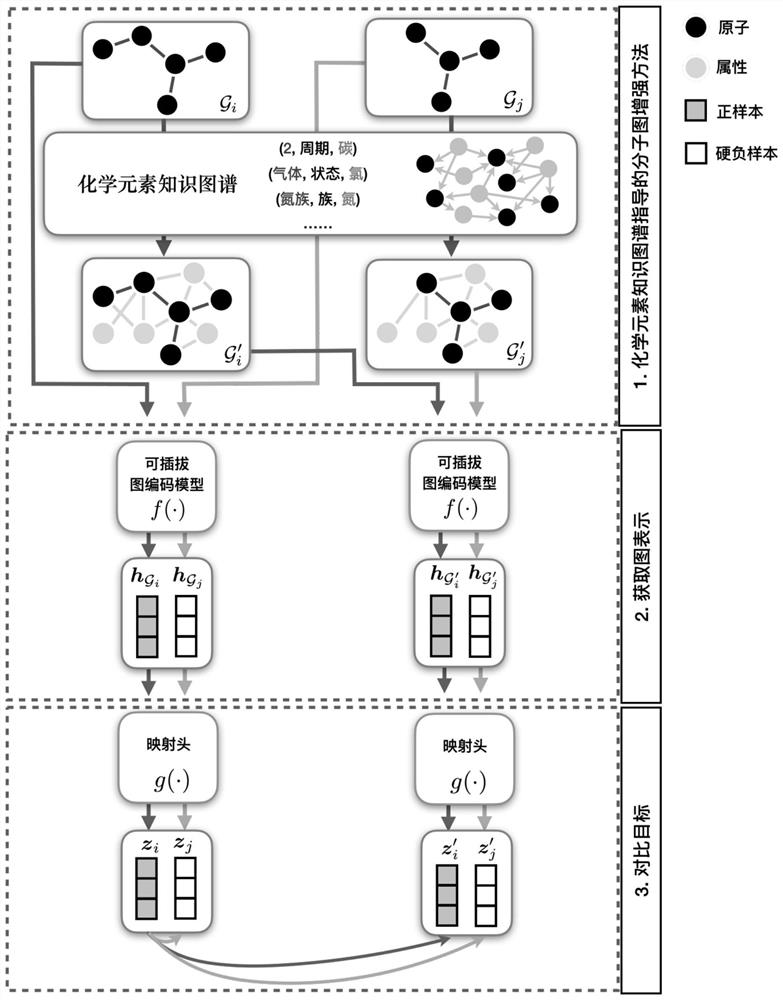
Molecular graph comparison learning method based on chemical element knowledge graph
PendingCN113990408APreserve basic structural informationEasy to learnChemical property predictionCharacter and pattern recognitionAlgorithmTheoretical computer science
The invention discloses a molecular graph comparison learning method based on a chemical element knowledge graph. The method comprises the following steps of: constructing a chemical element knowledge graph according to all chemical attributes of each chemical element in a periodic table of chemical elements; performing graph enhancement on the molecular graph by utilizing the chemical element knowledge graph to obtain a molecular enhancement graph; obtaining graph representations of the molecular graph and the molecular enhancement graph by using a pluggable representation model; adopting a hard negative sample mining technology to select other molecular graphs similar to the molecular graph in the molecular fingerprint space as negative samples; mapping the graph representation of positive sample pairs and the graph representation of negative sample pairs to the same space, constructing a contrast loss function by maximizing the consistency between the positive sample pairs and minimizing the consistency between the negative sample pairs, and performing optimization learning by using the contrast loss function; and forming a prediction model by using the parameter-determined pluggable representation model and a nonlinear classifier, and predicting the molecular properties of the molecular graph by using a parameter-finely-adjusted prediction model so as to improve the prediction accuracy of molecular properties.
Owner:ZHEJIANG UNIV
Genetic testing for predicting resistance of enterobacter species against antimicrobial agents
InactiveUS20180363030A1High unmet clinical needHigh-precision detectionMicrobiological testing/measurementDisease diagnosisBacteroidesEnterobacter species
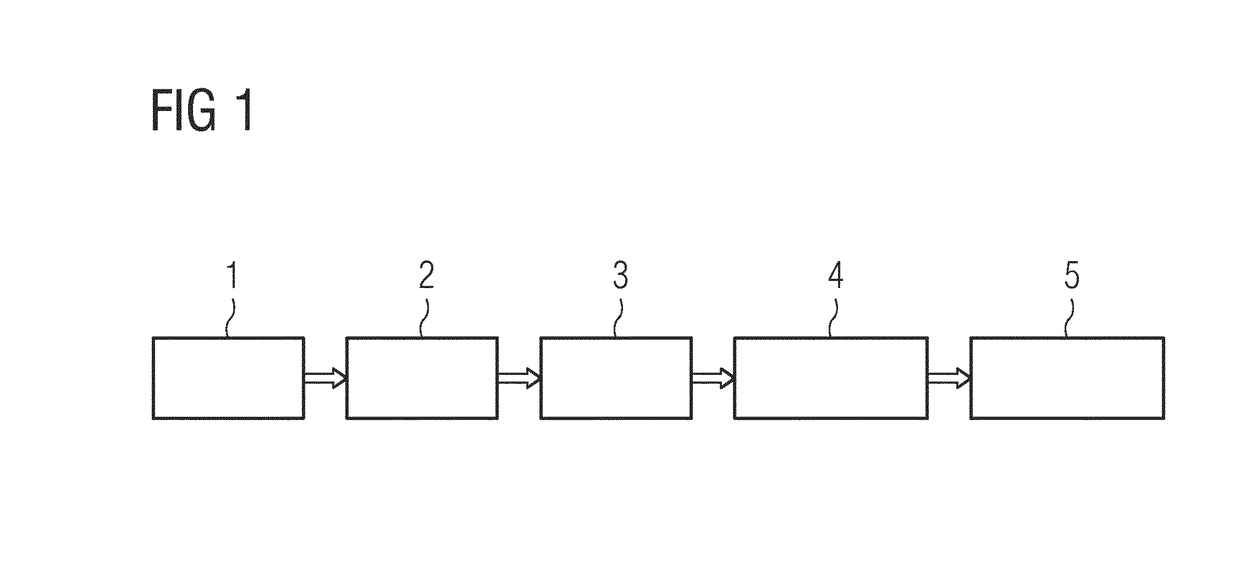
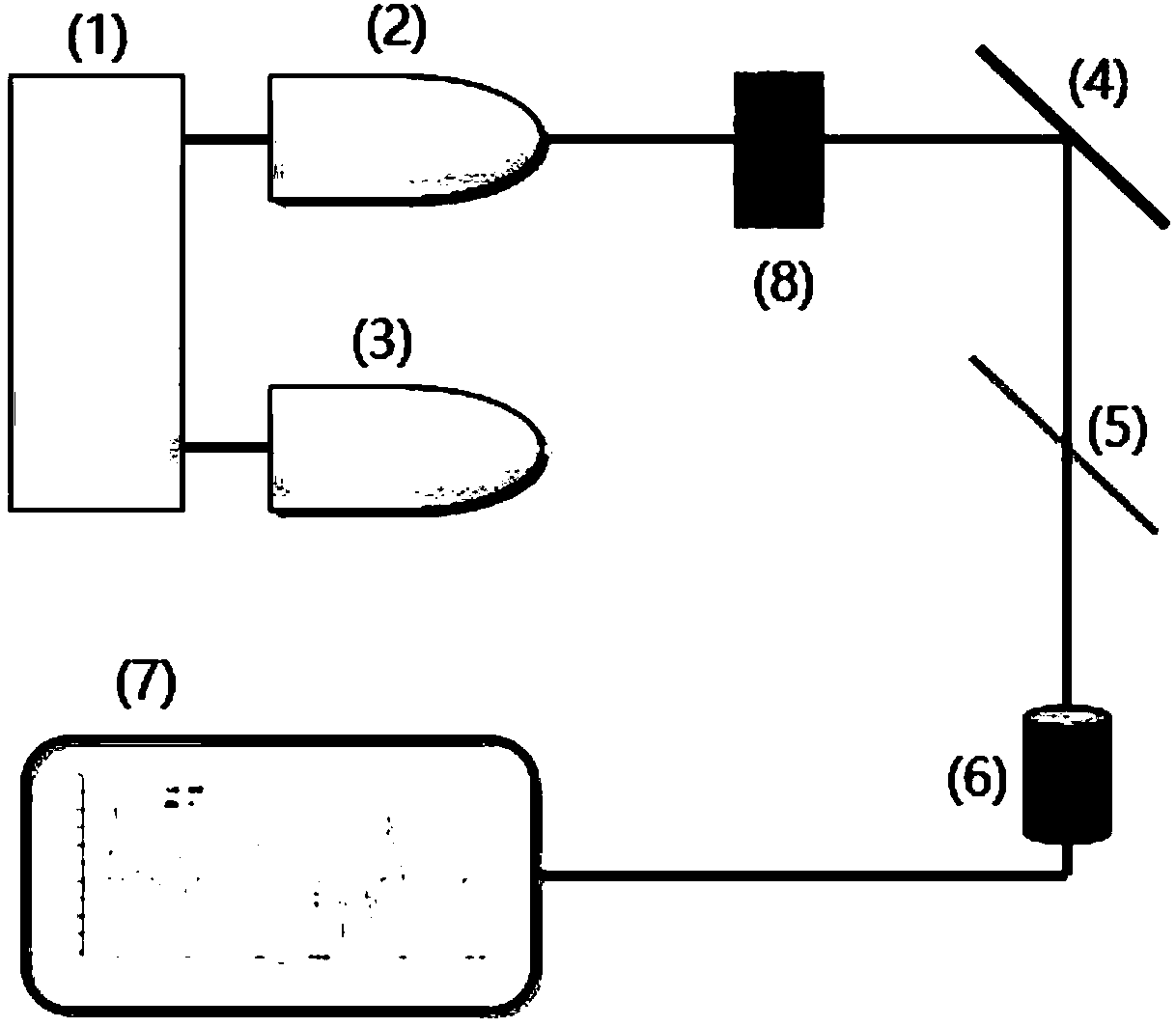
The invention relates to a method of determining an infection of a patient with Enterobacter species potentially resistant to antimicrobial drug treatment, a method of selecting a treatment of a patient suffering from an antibiotic resistant Enterobacter infection, and a method of determining an antibiotic resistance profile for bacterial microorganisms of Enterobacter species, as well as computer program products used in these methods. In an exemplary method, a sample (1), is used for molecular testing (2), and then a molecular fingerprint (3) is taken. The result is then compared to a reference library (4), and the result (5) is reported.
Owner:ARES GENETICS GMBH
Specificity molecule marker of thick wall cell verticil ZK7 strain and its application
InactiveCN1514021ARapid identificationQuick checkMicrobiological testing/measurementAgricultural scienceHybridization probe
A specific molecular marker of verticillium chlamydosporium ZK7 strain and its application are disclosed. The RAPD PCR amplification is used for molecular fingerprint analysis and comparison of said ZK7 strain, different reference strains and the fungus frequently encountered in soil to obtain the RAPD specific DNA fragments Vc 1200 and Vc 2000, which are then recovered, purified, cloned and sequenced. The specific primer SCAR-PCR is designed for PCR amplification. Said Vc 1200 and Vc2000 are marked by digoxin to obtain hybridization probes, which are used for the spot hydridization with the DNA extracted from the soil where the ZK7 strain is applied, so specifically detecting if there is ZK7 strain in soil and its quantity.
Owner:YUNNAN UNIV
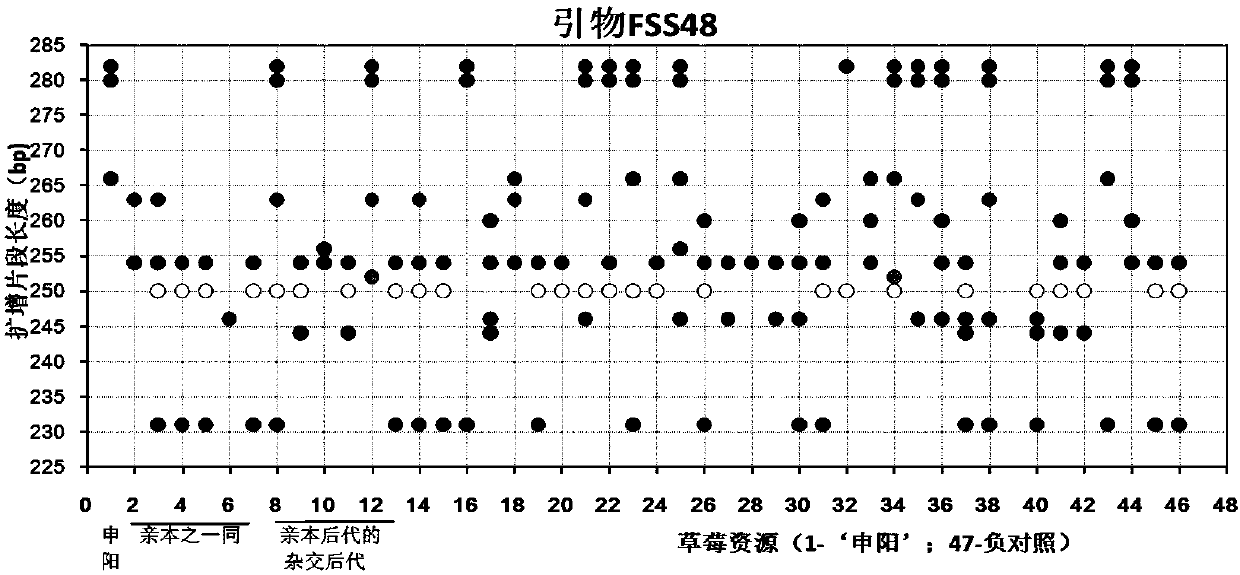
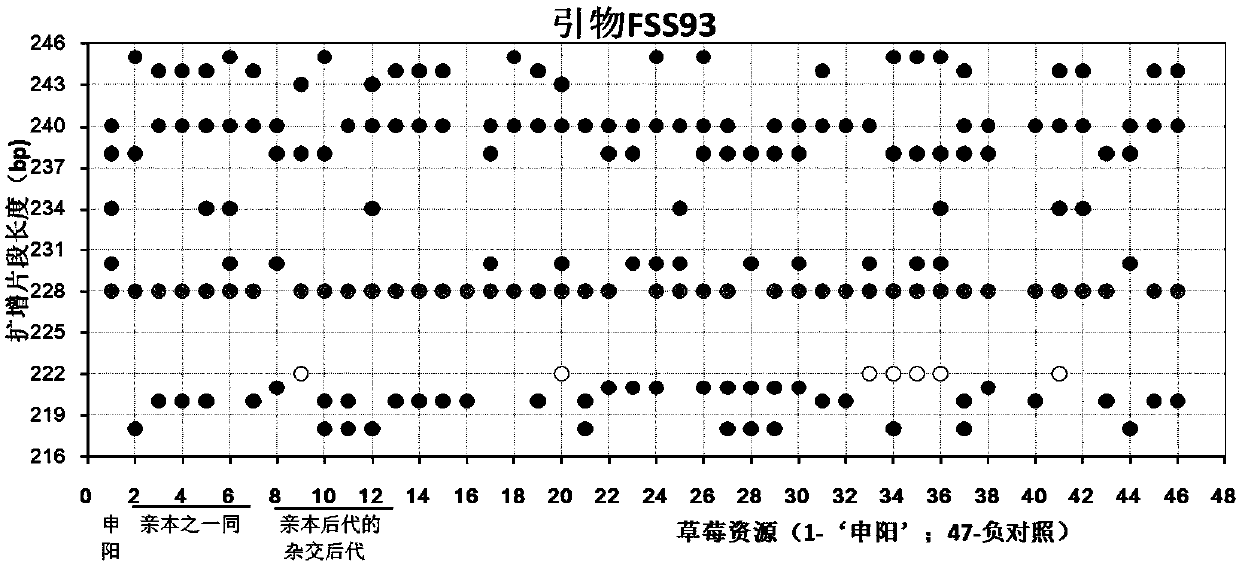
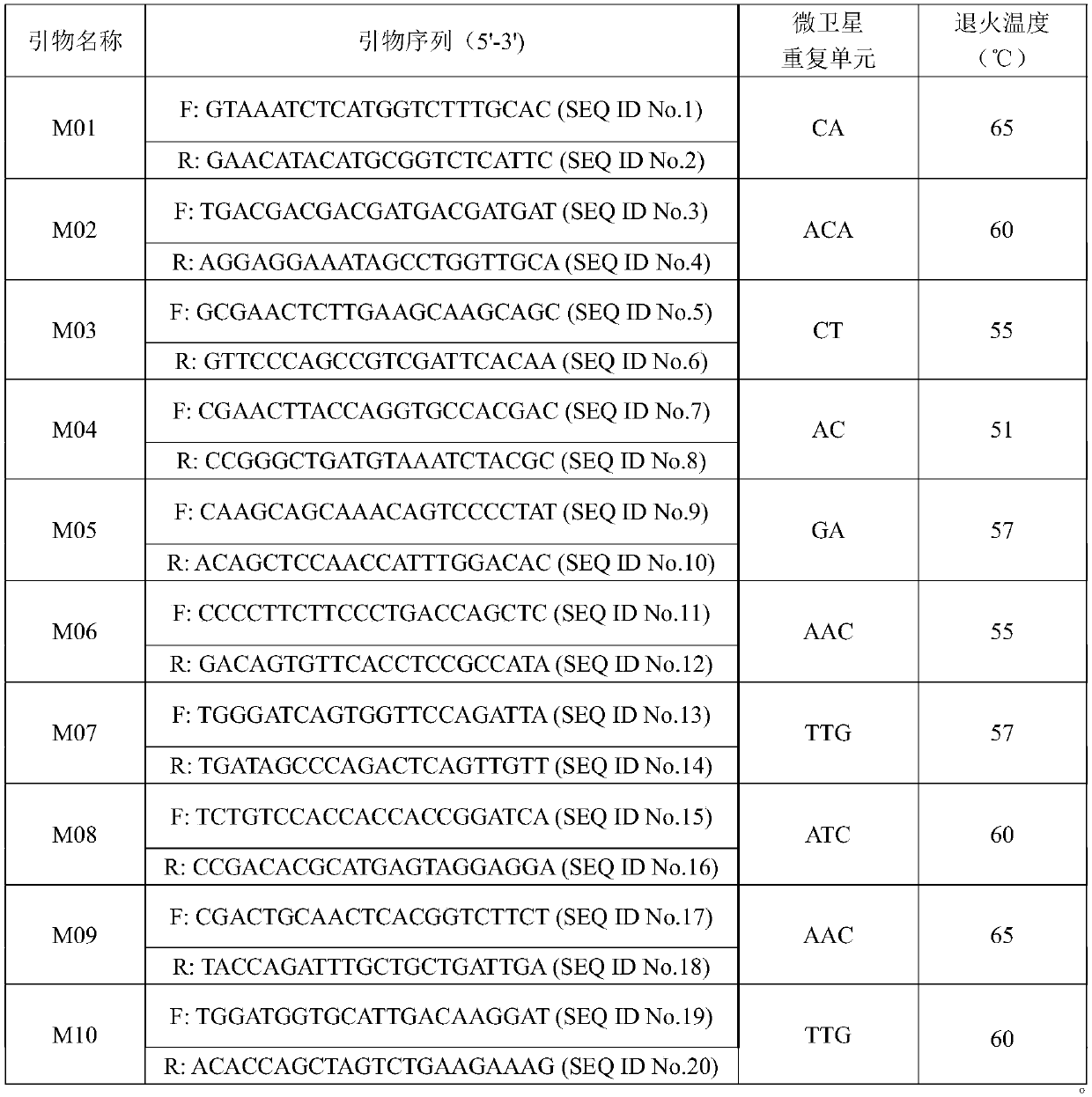
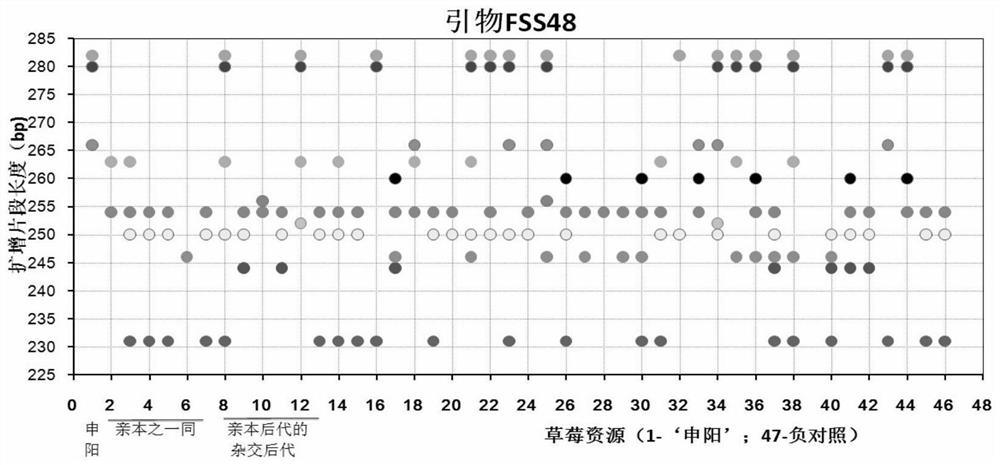
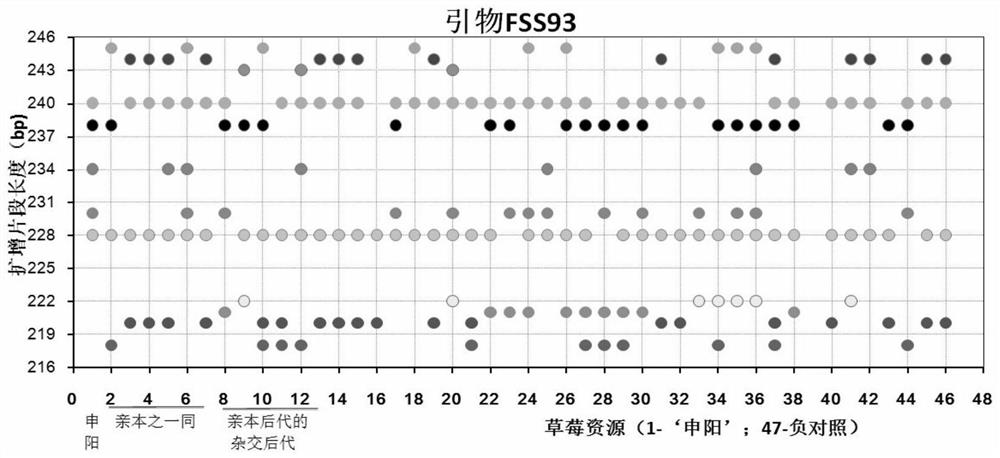
Construction method and application of SSR (Simple Sequence Repeat) molecular fingerprint map of strawberry hybrid
ActiveCN107663548AStrong dominantGood repeatabilityMicrobiological testing/measurementHorticulture methodsGrowth coefficientFragaria
The invention discloses a construction method and application of an SSR (Simple Sequence Repeat) molecular fingerprint map of strawberry hybrid. Eight pairs of SSR primer sequences are screened from 140 pairs of SSR primers which cover the whole genome of strawberries; the SSR molecular fingerprint map of the strawberry hybrid is constructed by utilizing the eight pairs of the SSR primer sequences; the purity of the seedlings of the strawberry hybrid Shenyang, a close relative sib line thereof, a non-near source variety, an excellent line and the like can be authenticated genetically; the SSRmolecular fingerprint map has the characteristics that the specificity is high, the polymorphism is stable, the band type combination is easy to identify and high in dominance, and can authenticate the strawberry hybrid accurately and quickly; moreover, culturing conditions are screened to control a somatic cell mutation risk which is possibly brought by tissue culture; the vitrification and the formation of calluses are stopped; the tissue culture proliferation generation number and the growth coefficient are inhibited; a micropropagation method of the strawberry hybrid Shenyang is determined; the Shenyang strawberry original seedlings with complete varietal purity can be micro-propagated quickly.
Owner:SHANGHAI ACAD OF AGRI SCI
Avena L. DNA (deoxyribonucleic acid) molecular fingerprint spectrum construction method and application
InactiveCN107557487AAvoid enteringEfficient identificationMicrobiological testing/measurementAgricultural scienceGenome
The invention discloses an avena L. DNA (deoxyribonucleic acid) molecular fingerprint spectrum construction method and application. According to the method, a set of specific SSR (simple sequence repeats) molecular marker primers (SEQ ID NO: 1-20) are designed aiming at avena L. genomes, and can be used for performing PCR (polymerase chain reaction) amplification on avena L. genome DNAs to generate amplification products with different lengths, and the lengths of the products are analyzed and detected to construct characteristic fingerprint spectrums of different avena L. varieties to implement variety identification. Compared with conventional morphological identification, the variety identification method provided by the invention has the advantages of low time consumption, no environmental influence, convenience, rapidness, accurate and reliable result and broad application prospect.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Construction method and application of ssr molecular fingerprint of "Shenyang" strawberry
ActiveCN107663548BStrong specificityStrong discriminationMicrobiological testing/measurementHorticulture methodsBiotechnologyFragaria
Owner:SHANGHAI ACAD OF AGRI SCI
Biological molecule detecting method based on optical comb coherent imaging analysis
ActiveCN107643263AQuick identificationAccurate identificationMaterial analysis by optical meansAction spectrumPhysics
The invention relates to a biological molecule detecting method based on optical comb coherent imaging analysis. According to the characteristics that molecules with different structures certainly have the same absorption spectrum, the wave coherent position of an absorption band and the strength of the absorption band reversely reflect features of the molecules on the structure, the biological molecule detecting method can be used for identifying a chemical group or the composition and structure of an unknown sample; and the absorption strength of the band corresponds to the content of the molecules, and therefore, the spectrum of an optical comb can carry out quantitative analysis and purity identification. Particularly by the measurement advantages of high reproducibility and high precision, the biological molecule detecting method can be used for spectrum calibration of millions of known biological molecules, and a spectral standard atlas database or a molecule fingerprint atlas database is established and is used as a 'fingerprint spectrum' of the biological molecules. The sample can be subjected to non-contact nondestructive detection. A pathogen is subjected to spectral analysis by a high-precision optical comb, the type and concentration of the pathogen can be recognized rapidly and accurately; and because the optical comb has a wide spectral range, the pathogen can bedetected at a time.
Owner:THE FIRST REHABILITATION HOSPITAL OF SHANGHAI +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com