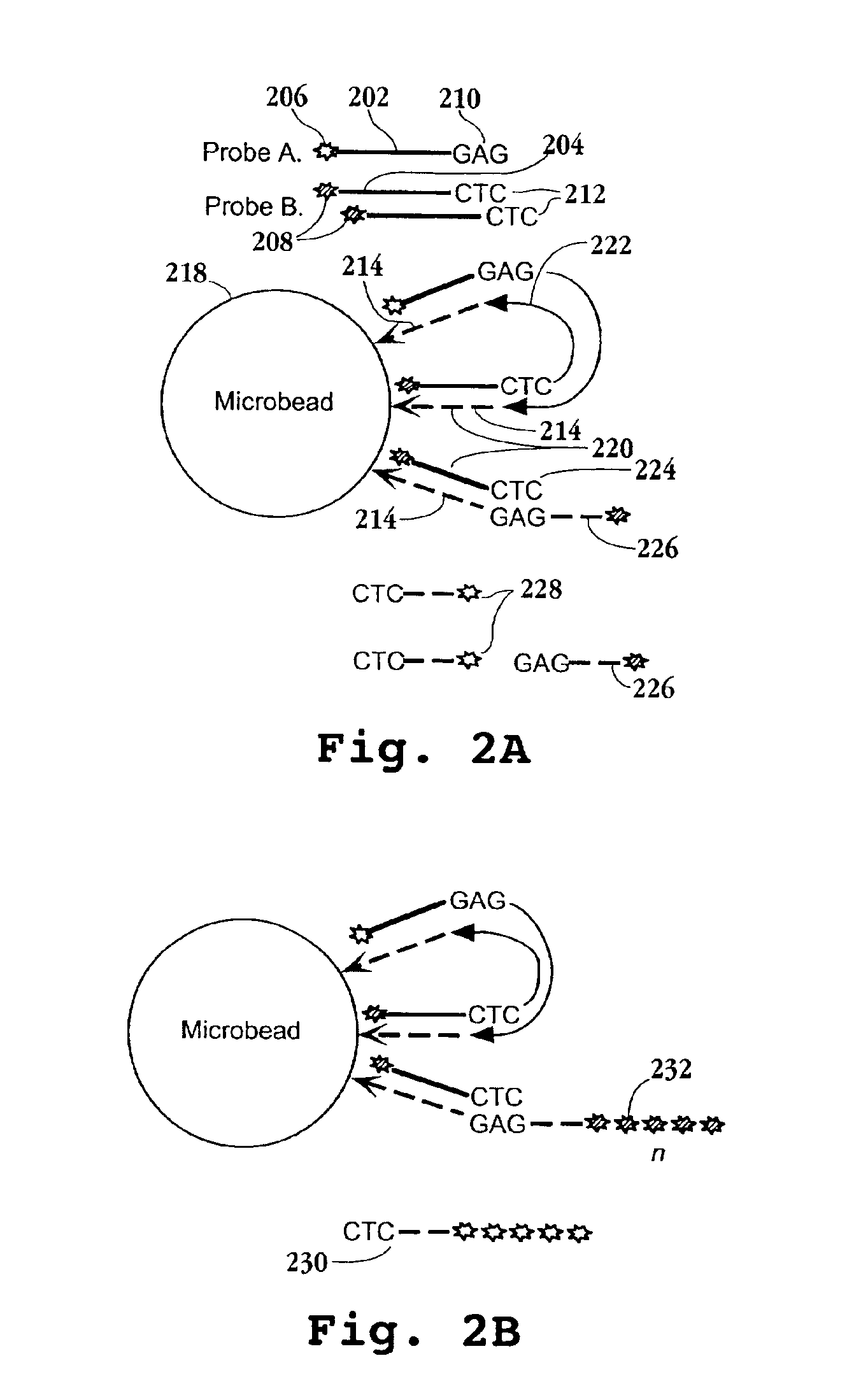
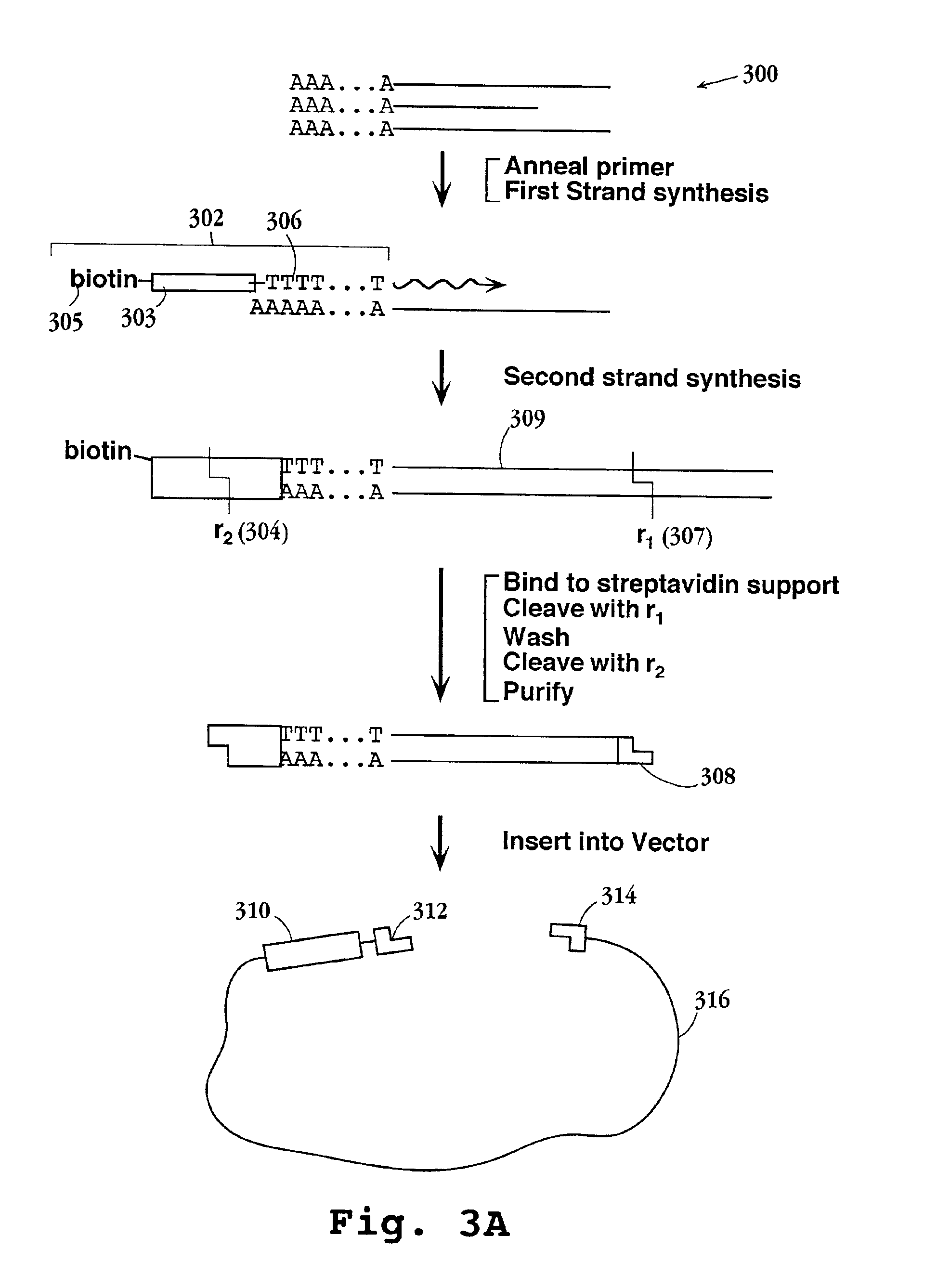
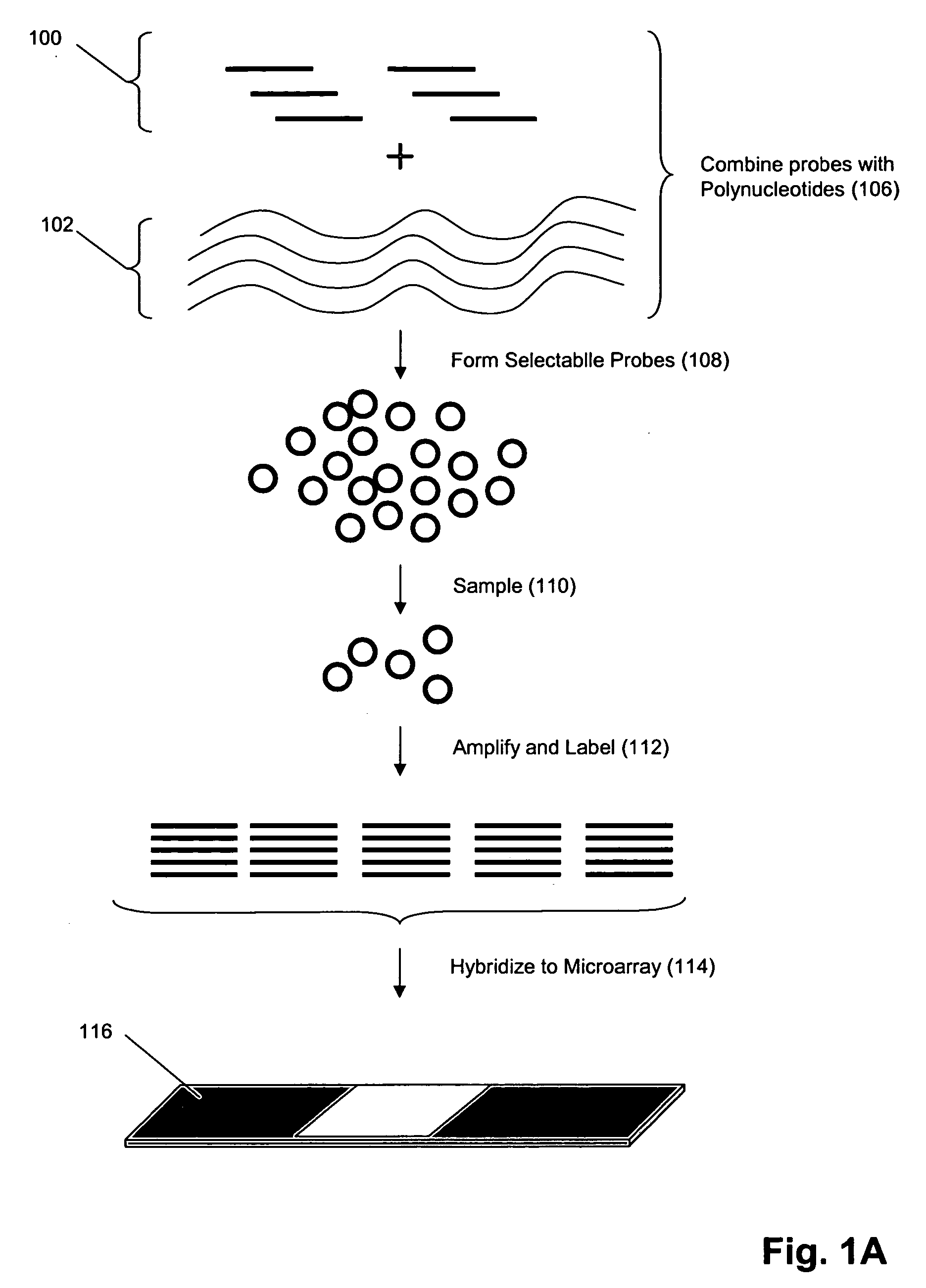
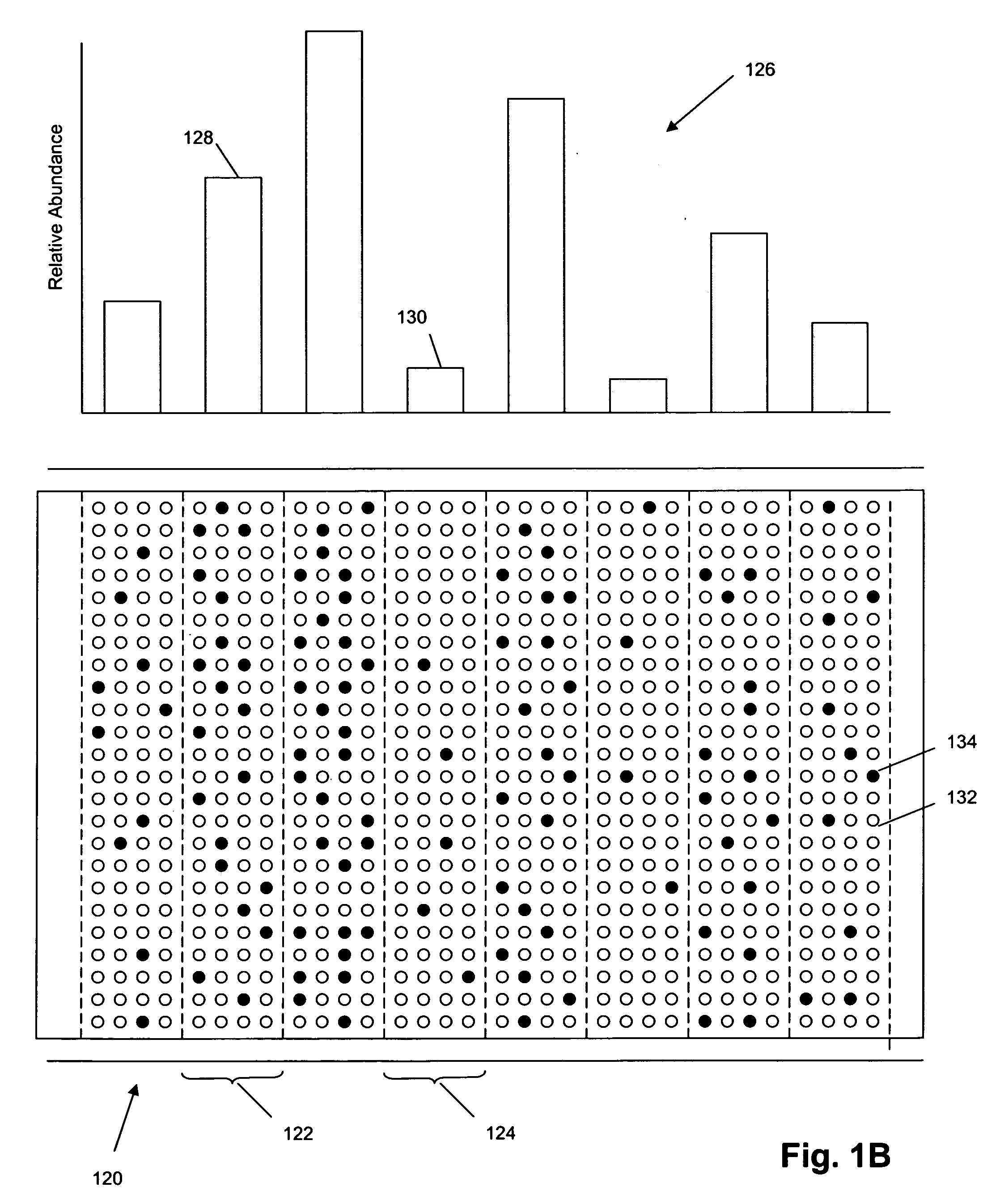
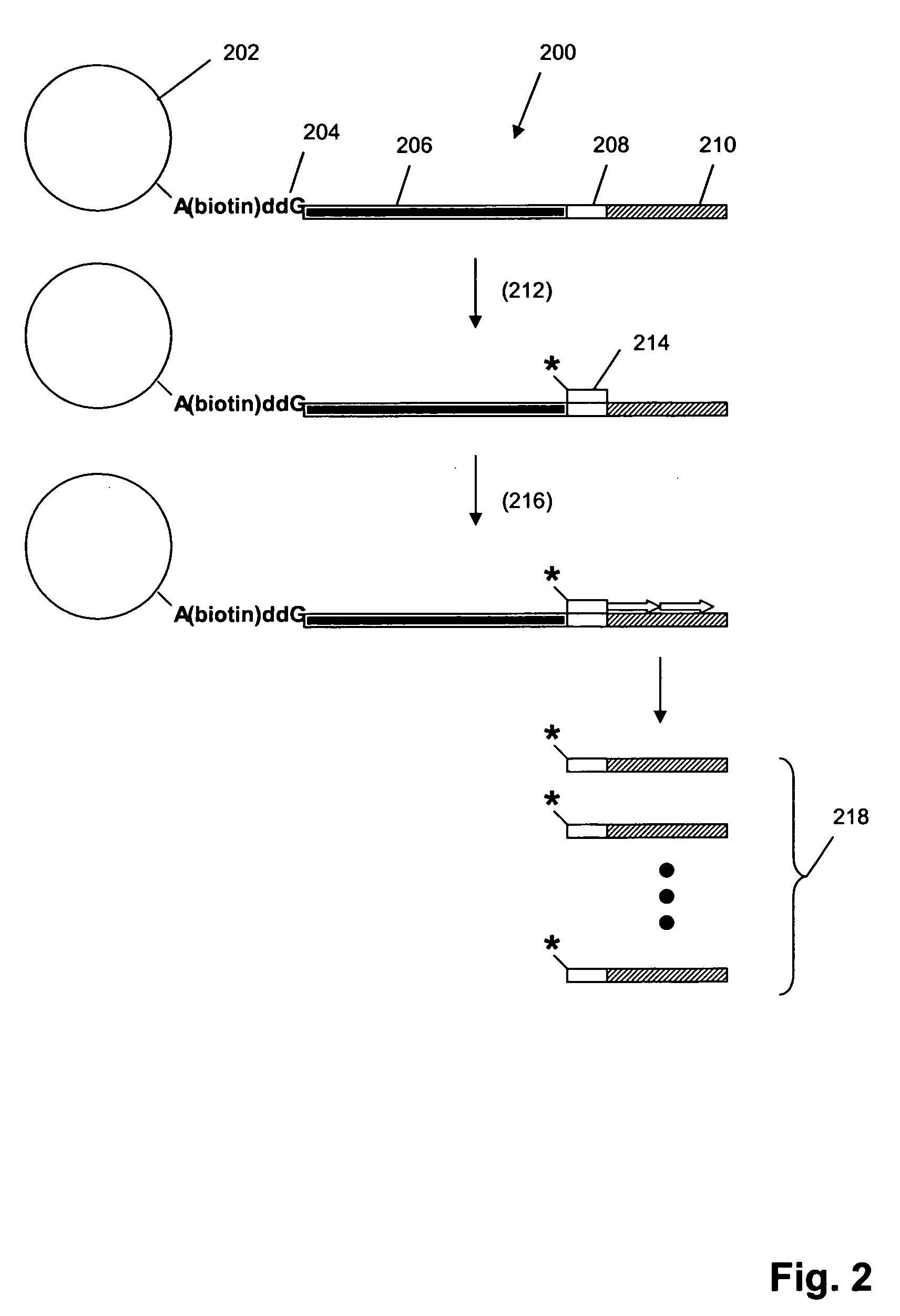
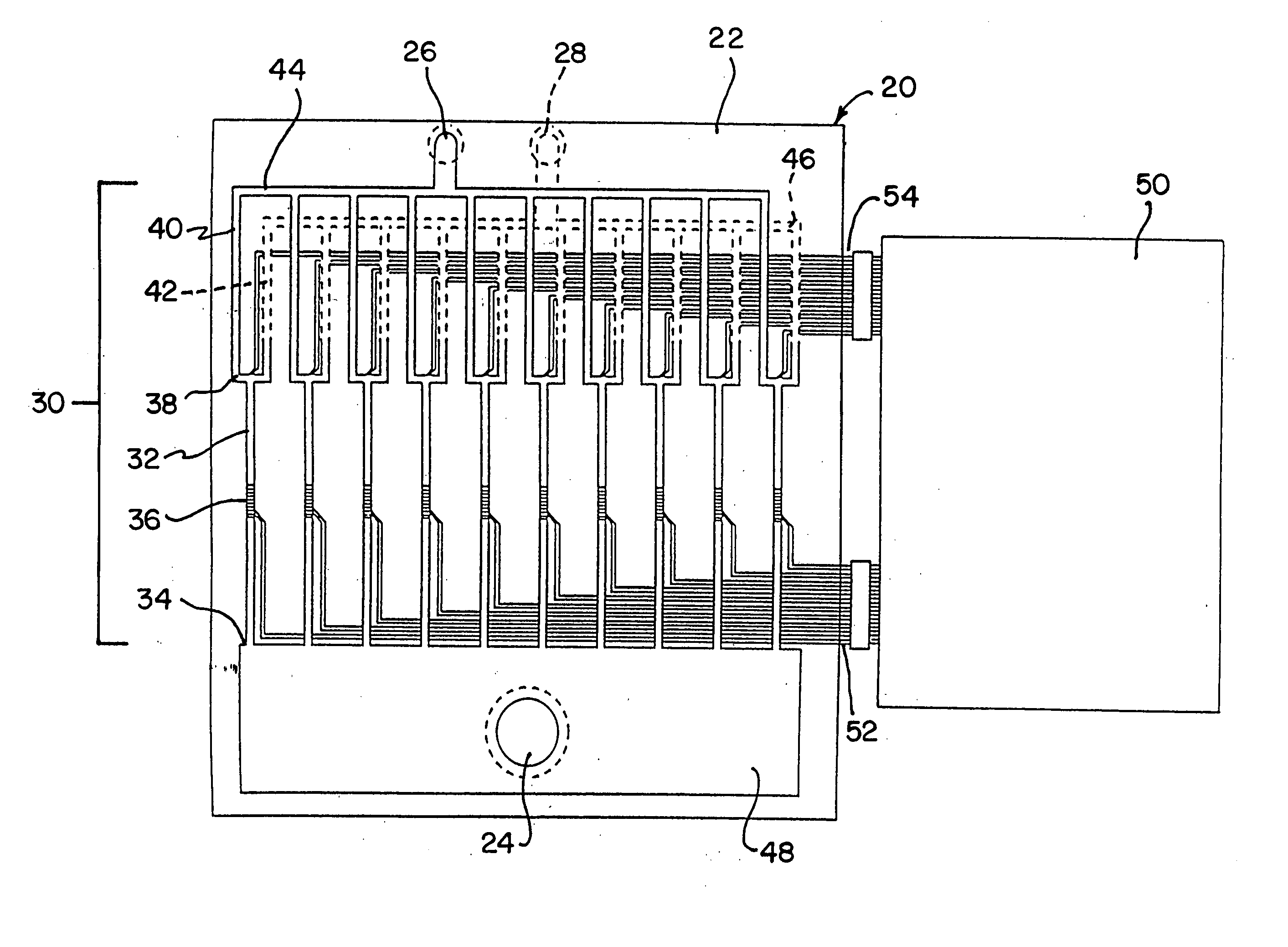
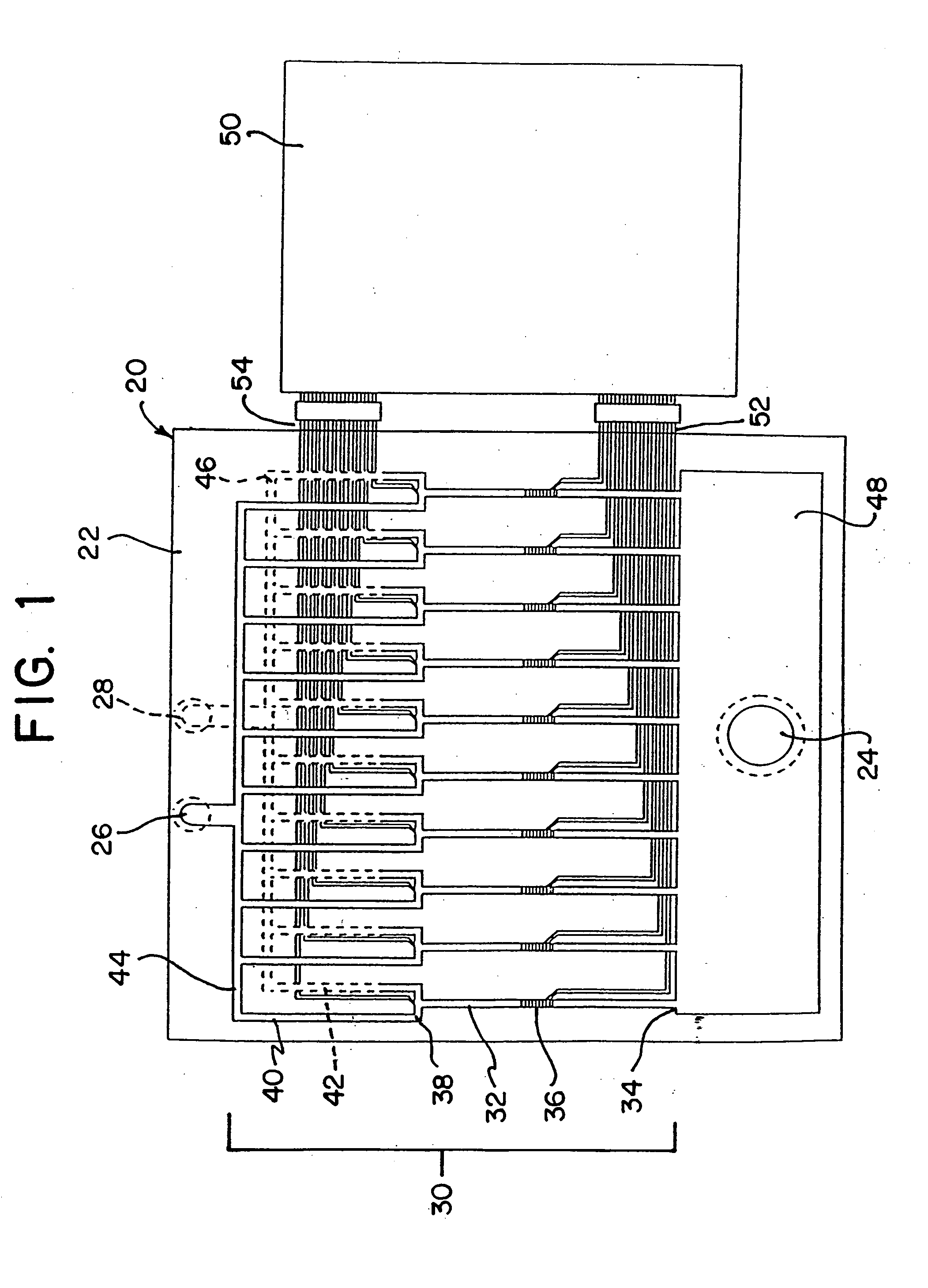
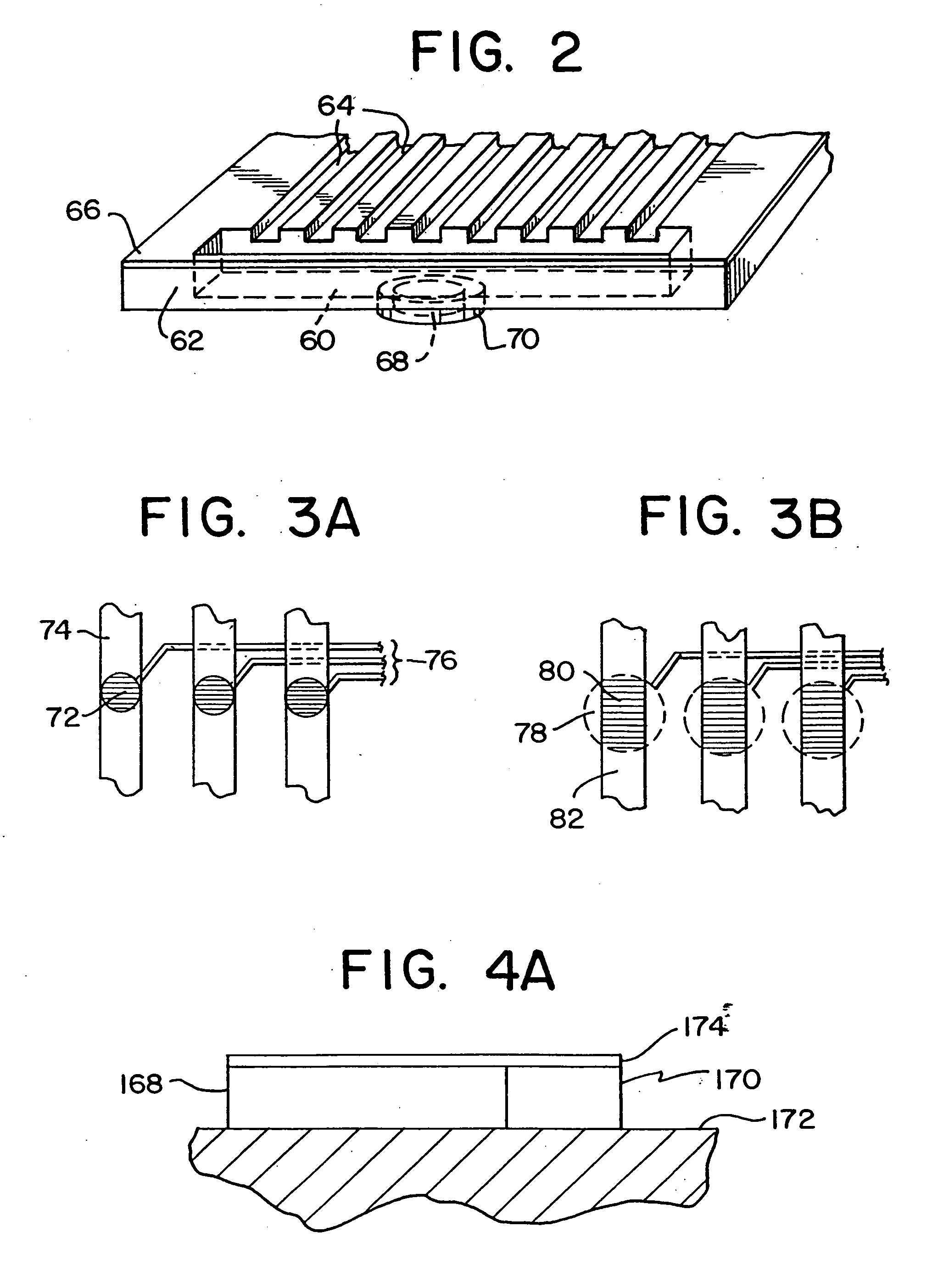
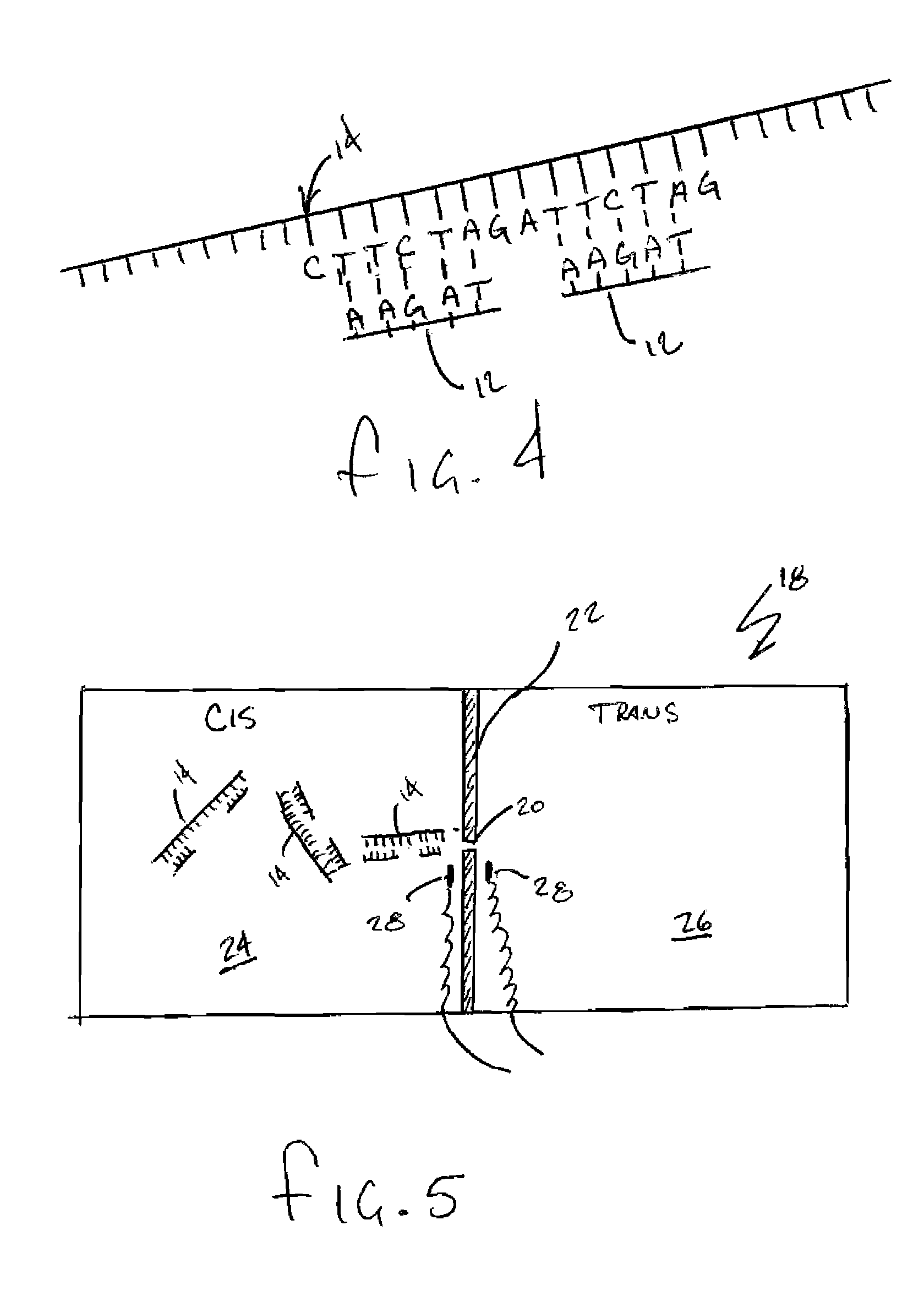
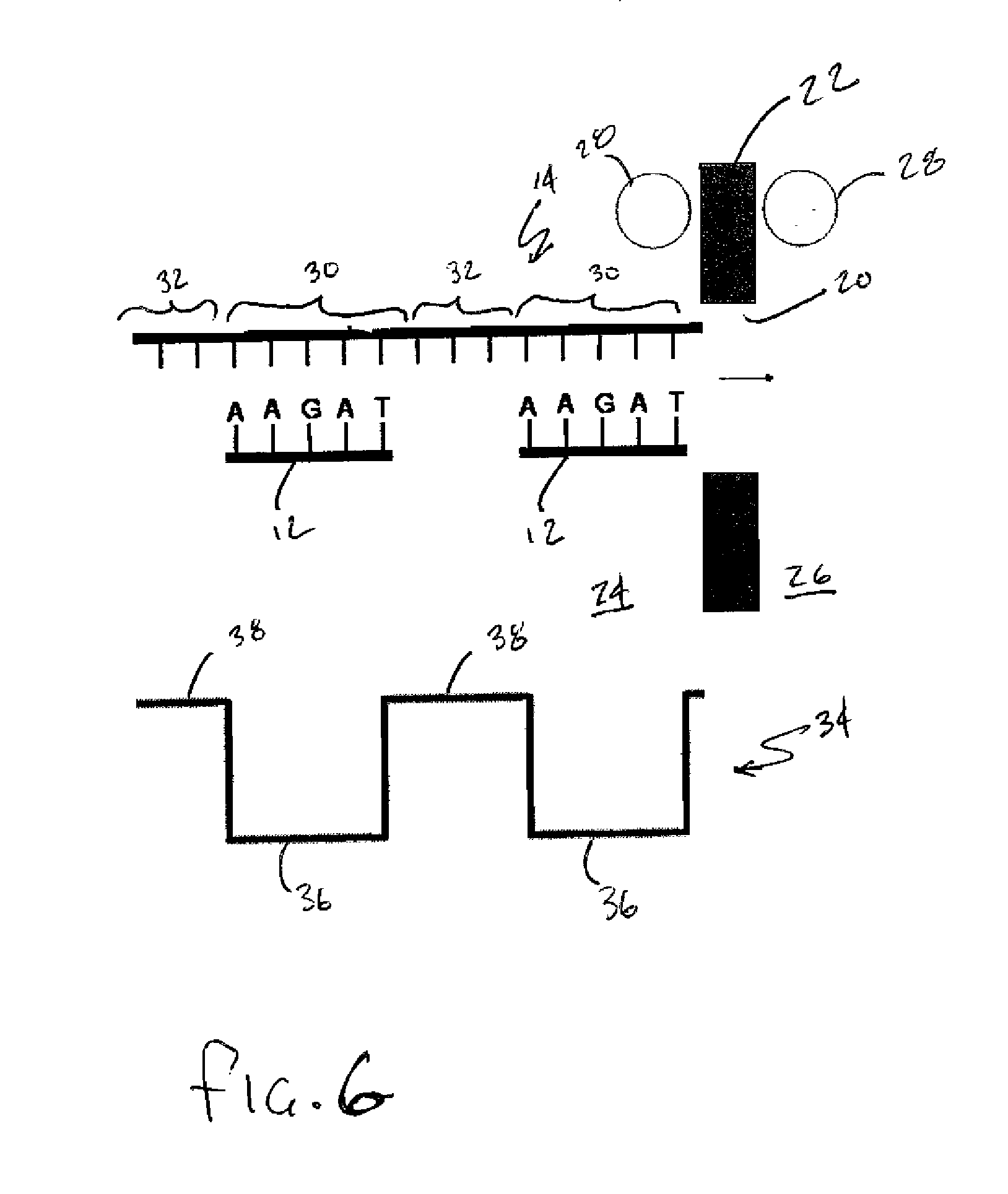
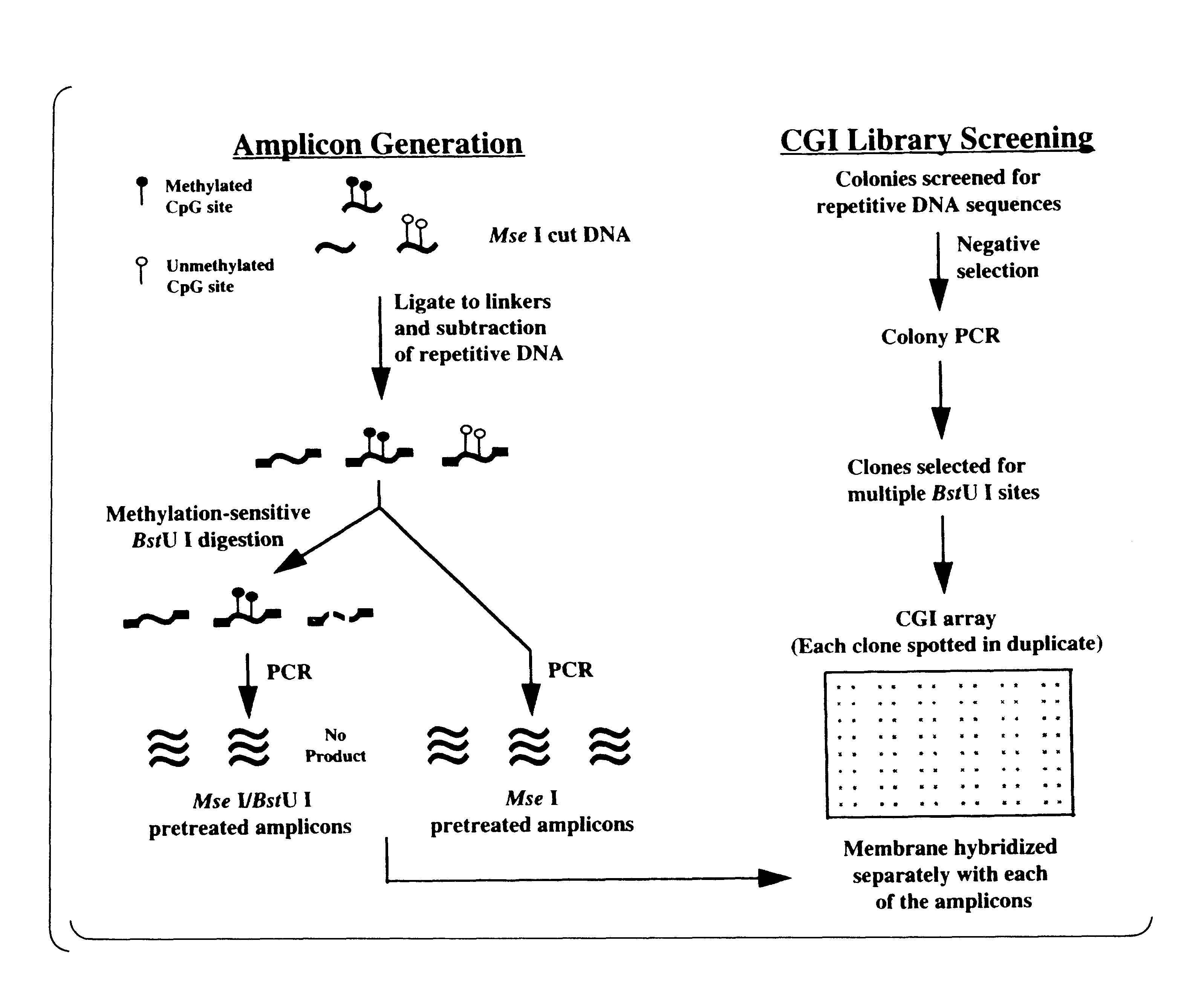
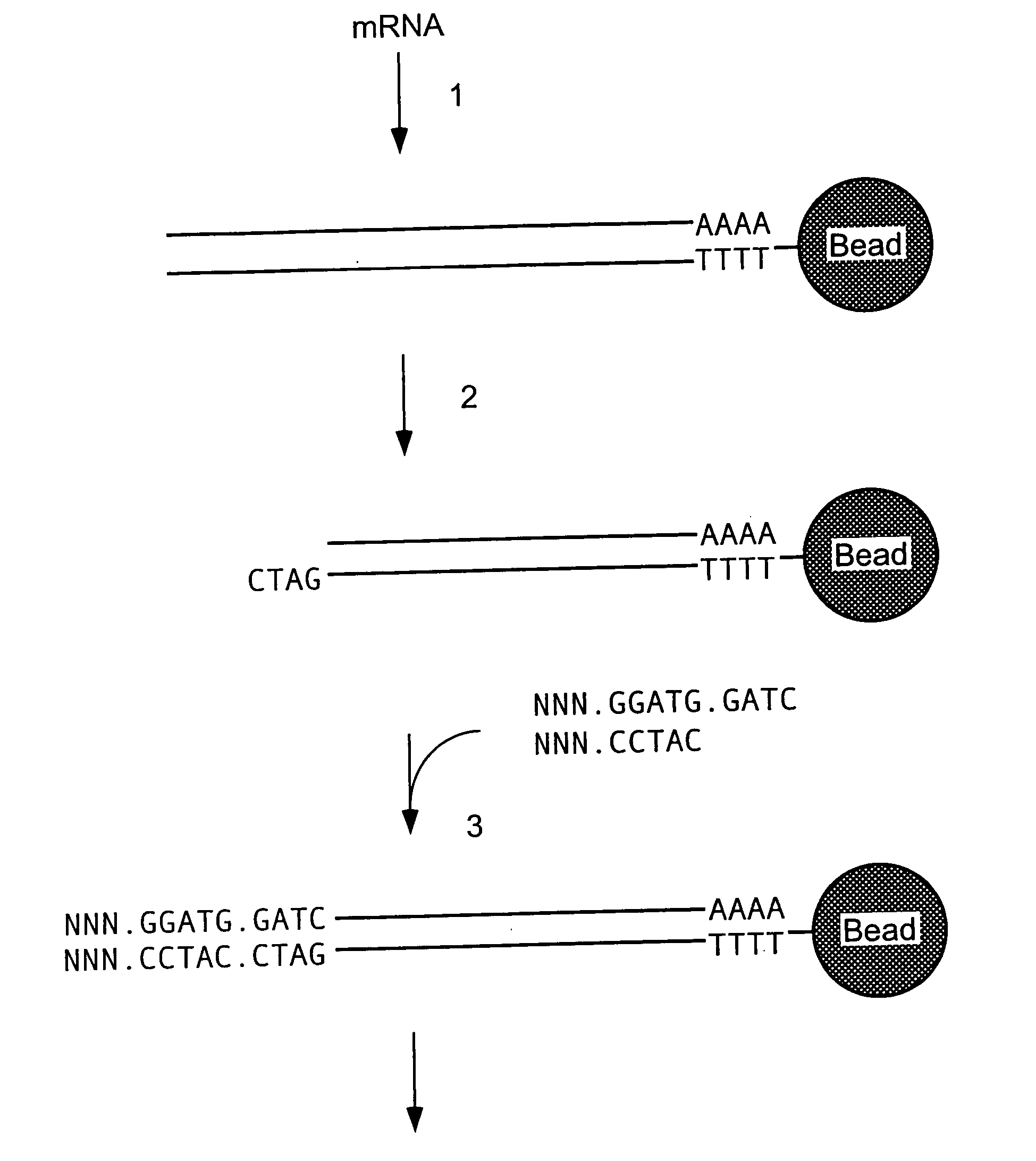
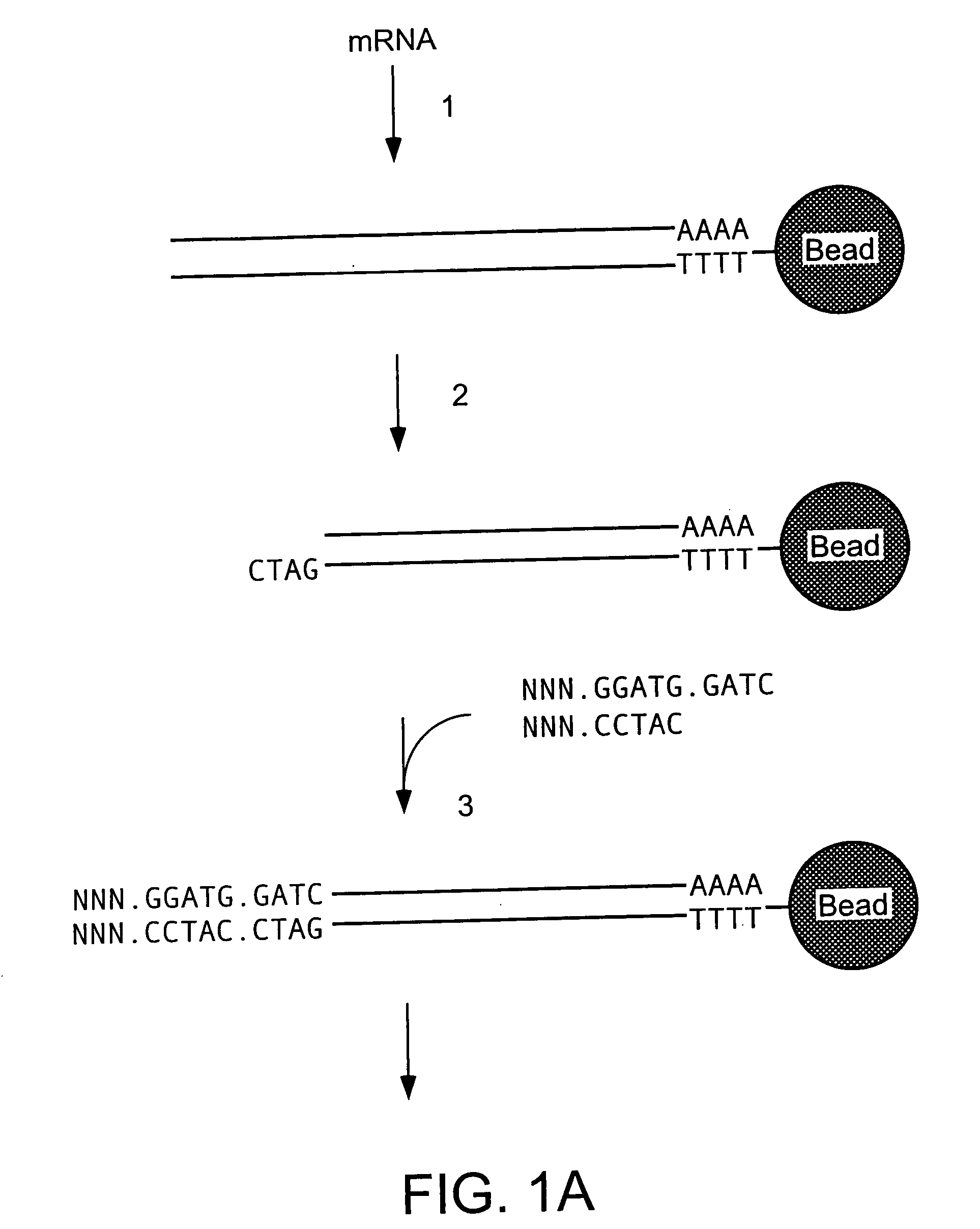
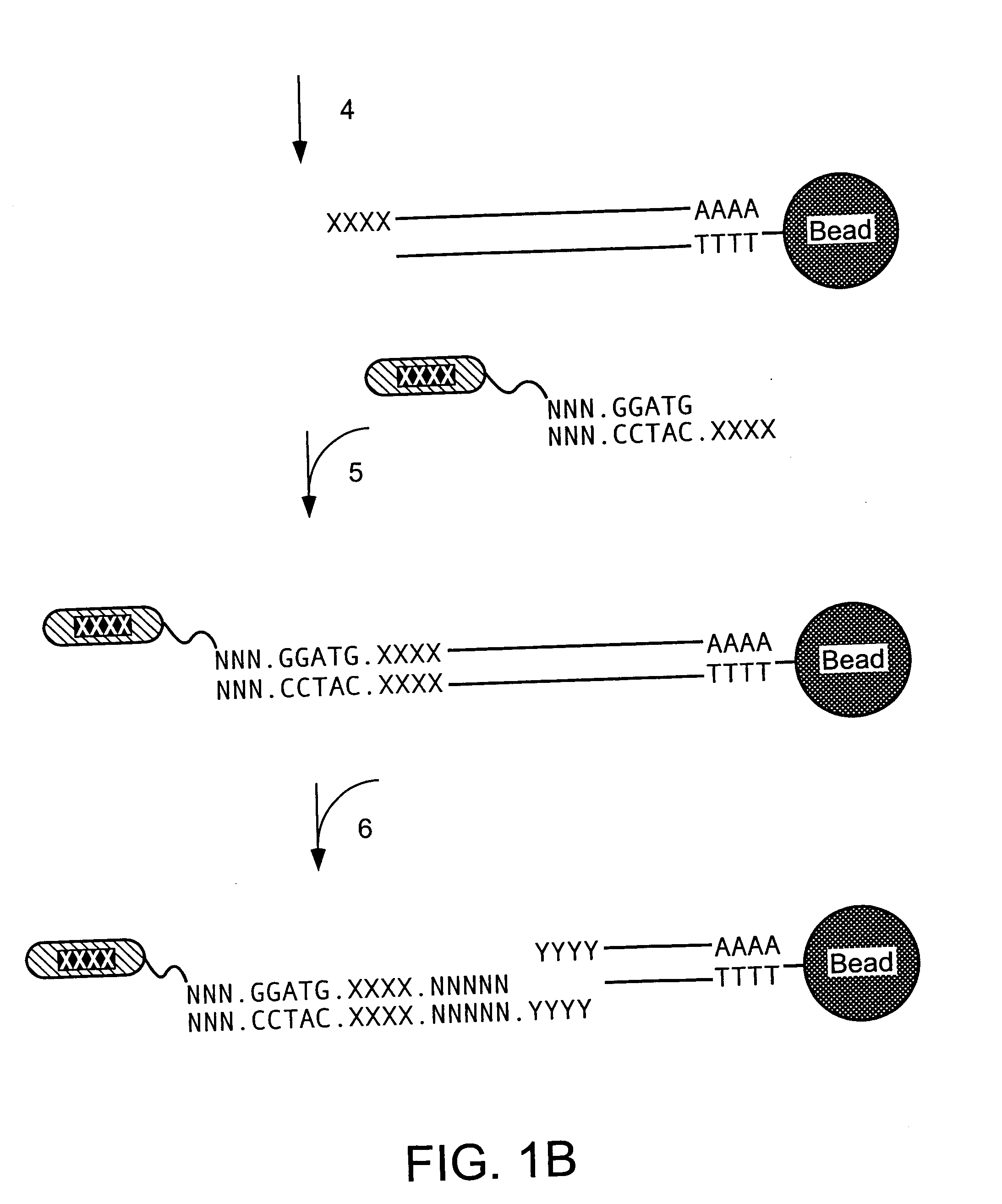
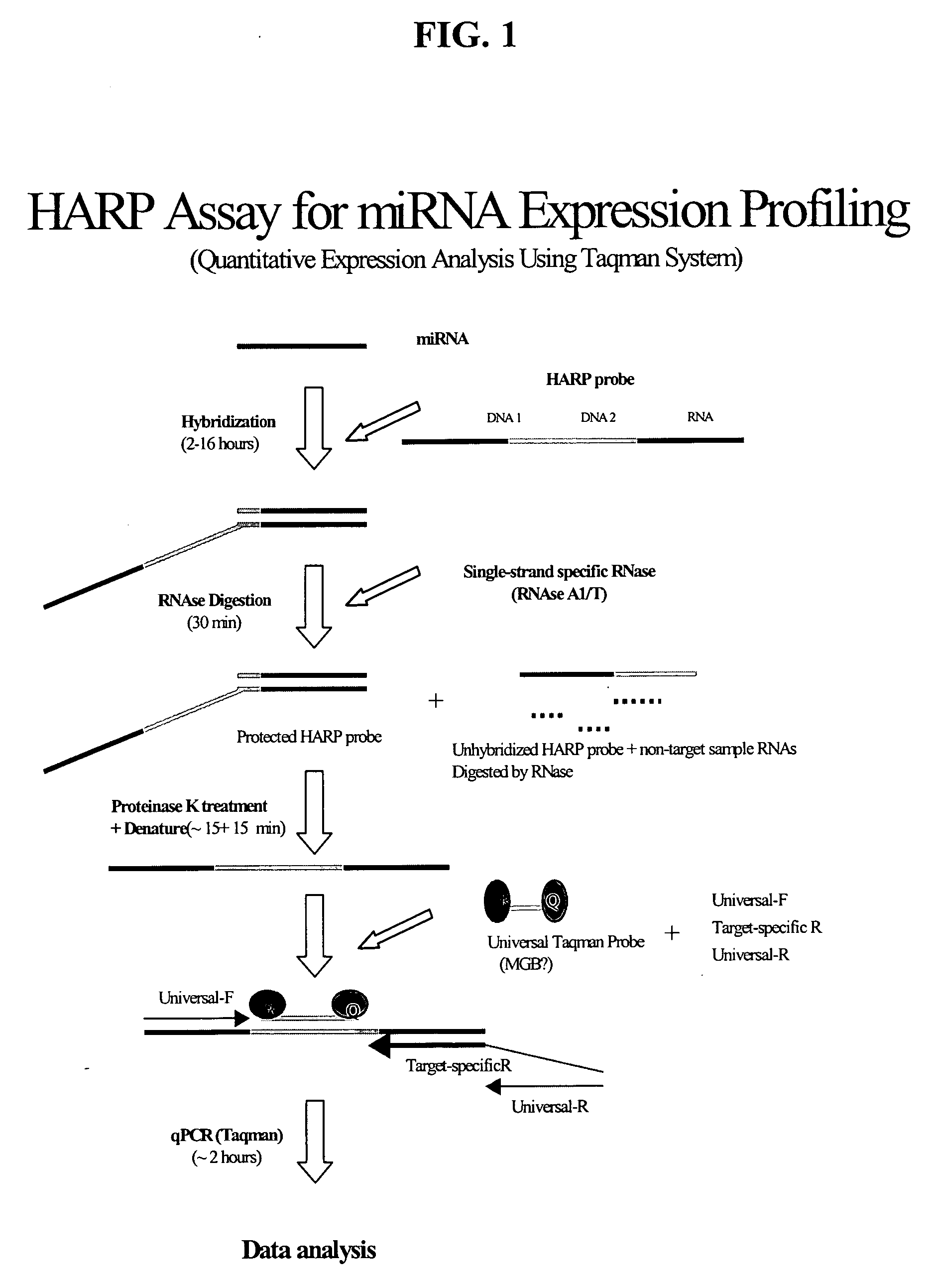
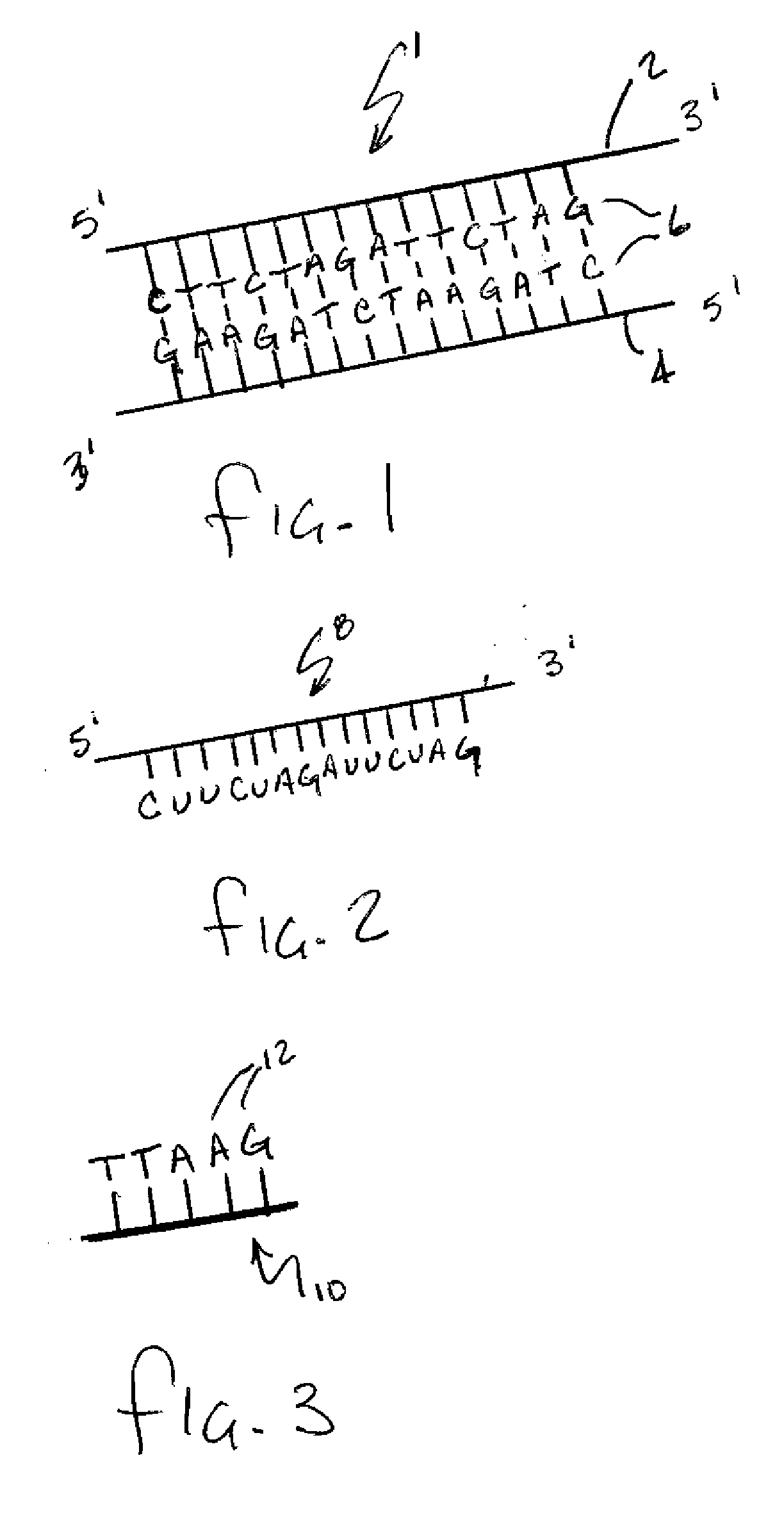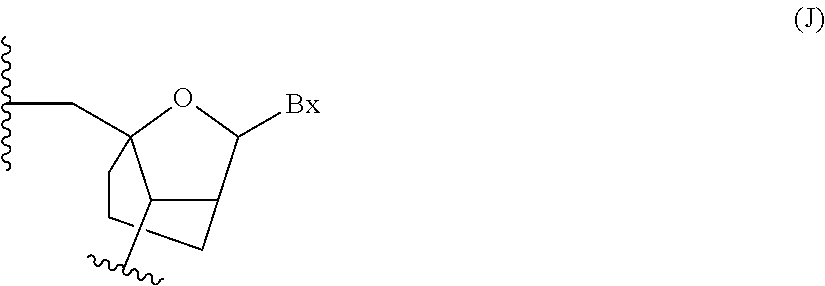Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
527 results about "Hybridization probe" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
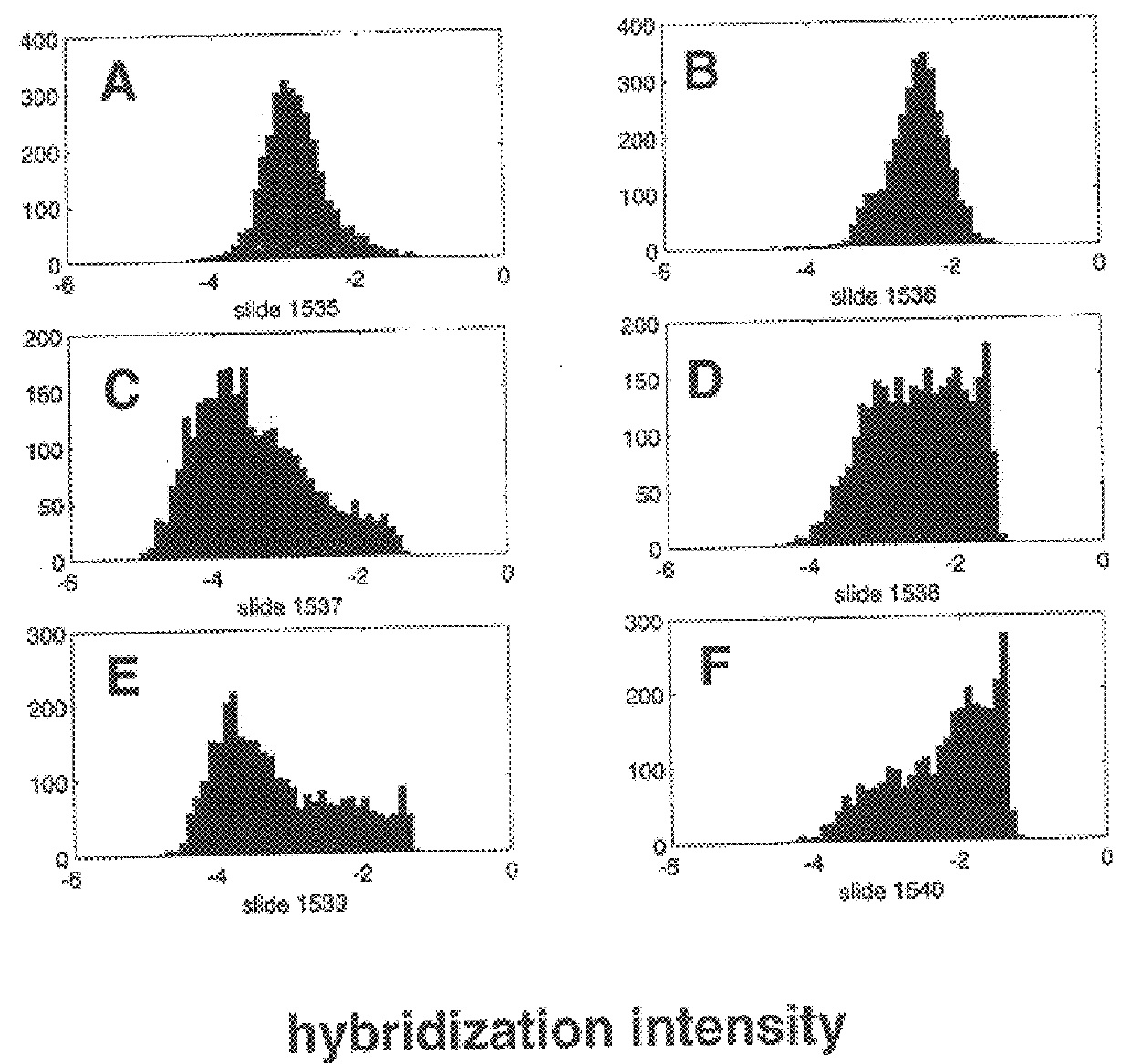
In molecular biology, a hybridization probe is a fragment of DNA or RNA of variable length (usually 100–10000 bases long) which can be radioactively or fluorescently labeled. It can then be used in DNA or RNA samples to detect the presence of nucleotide substances (the RNA target) that are complementary to the sequence in the probe. The probe thereby hybridizes to single-stranded nucleic acid (DNA or RNA) whose base sequence allows probe–target base pairing due to complementarity between the probe and target. The labeled probe is first denatured (by heating or under alkaline conditions such as exposure to sodium hydroxide) into single stranded DNA (ssDNA) and then hybridized to the target ssDNA (Southern blotting) or RNA (northern blotting) immobilized on a membrane or in situ. To detect hybridization of the probe to its target sequence, the probe is tagged (or "labeled") with a molecular marker of either radioactive or (more recently) fluorescent molecules; commonly used markers are ³²P (a radioactive isotope of phosphorus incorporated into the phosphodiester bond in the probe DNA) or digoxigenin, which is a non-radioactive, antibody-based marker. DNA sequences or RNA transcripts that have moderate to high sequence similarity to the probe are then detected by visualizing the hybridized probe via autoradiography or other imaging techniques. Normally, either X-ray pictures are taken of the filter, or the filter is placed under UV light. Detection of sequences with moderate or high similarity depends on how stringent the hybridization conditions were applied—high stringency, such as high hybridization temperature and low salt in hybridization buffers, permits only hybridization between nucleic acid sequences that are highly similar, whereas low stringency, such as lower temperature and high salt, allows hybridization when the sequences are less similar. Hybridization probes used in DNA microarrays refer to DNA covalently attached to an inert surface, such as coated glass slides or gene chips, to which a mobile cDNA target is hybridized.
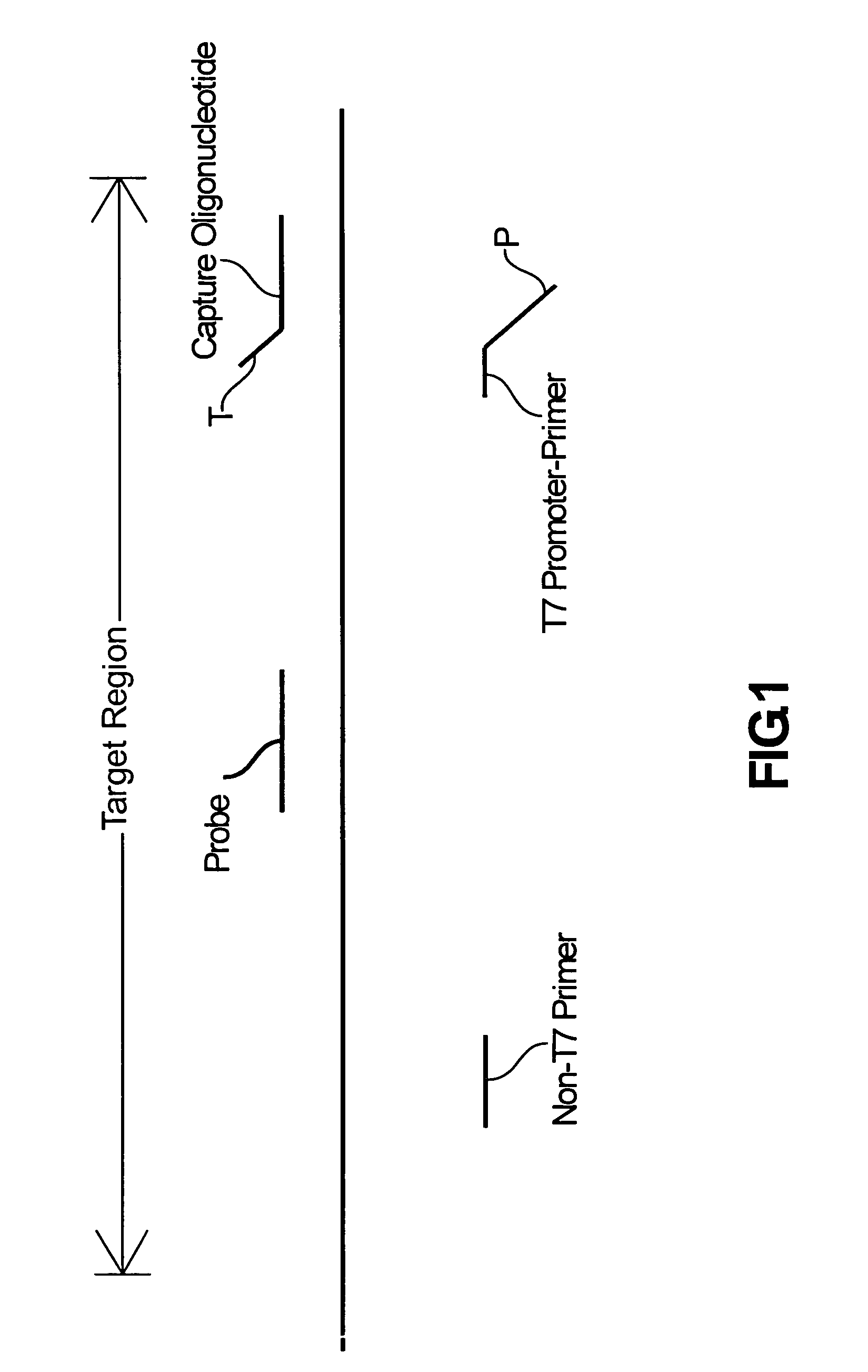
Integrated active flux microfluidic devices and methods
InactiveUS6767706B2Rapid and complete exposureQuick and accurate and inexpensive analysisBioreactor/fermenter combinationsFlow mixersAntigenHybridization probe
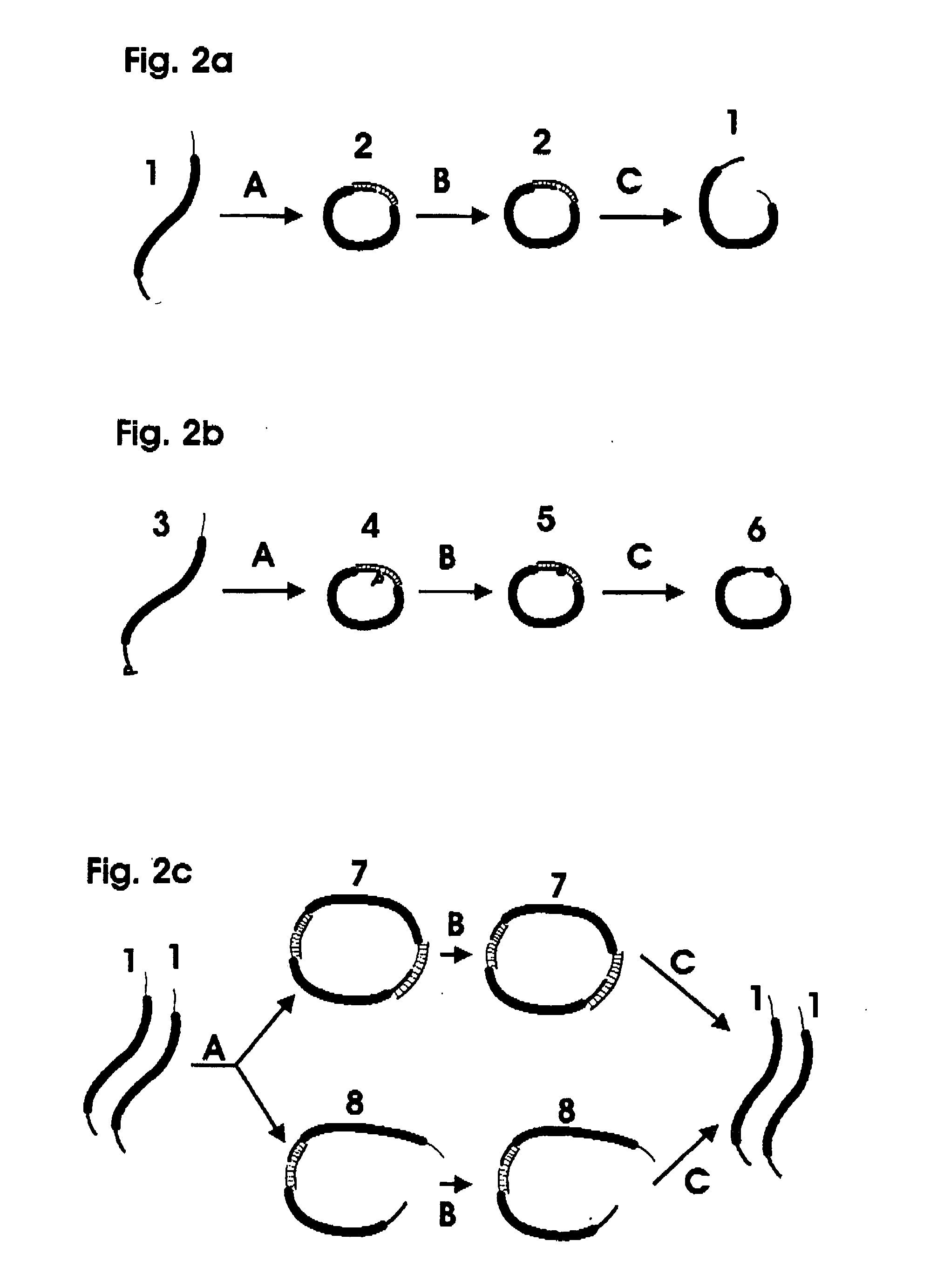
The invention relates to a microfabricated device for the rapid detection of DNA, proteins or other molecules associated with a particular disease. The devices and methods of the invention can be used for the simultaneous diagnosis of multiple diseases by detecting molecules (e.g. amounts of molecules), such as polynucleotides (e.g., DNA) or proteins (e.g., antibodies), by measuring the signal of a detectable reporter associated with hybridized polynucleotides or antigen / antibody complex. In the microfabricated device according to the invention, detection of the presence of molecules (i.e., polynucleotides, proteins, or antigen / antibody complexes) are correlated to a hybridization signal from an optically-detectable (e.g. fluorescent) reporter associated with the bound molecules. These hybridization signals can be detected by any suitable means, for example optical, and can be stored for example in a computer as a representation of the presence of a particular gene. Hybridization probes can be immobilized on a substrate that forms part of or is exposed to a channel or channels of the device that form a closed loop, for circulation of sample to actively contact complementary probes. Universal chips according to the invention can be fabricated not only with DNA but also with other molecules such as RNA, proteins, peptide nucleic acid (PNA) and polyamide molecules.
Owner:CALIFORNIA INST OF TECH
Method for determining relative abundance of nucleic acid sequences
InactiveUS6897023B2Sugar derivativesMicrobiological testing/measurementHybridization probeFluorescence
Disclosed are methods for identifying nucleic acid sequences which are of different abundances in different nucleic acid source populations, e.g. differentially expressed genes or genomic variations among individuals or populations of individuals. In one embodiment, probes derived from the source nucleic acid populations are derivatized with a terminal sample ID (SID) sequence characteristic of that population. Upon competitive hybridization of the probes to a reference or index nucleic acid library containing all the sequences in the populations being compared, the SID tags remain single stranded, and those from the different sources are then annealed to one another. Unhybridized (remainder) SID sequences are then quantified. By labeling such remainder SID sequences with a fluorescent dye, FACS sorting of beads containing the hybridized probes can be carried out. The signal ratio upon which such sorting is based is enhanced compared to competitive hybridization using labeled probes without SID sequences.
Owner:THE MOLECULAR SCI INST +1
Digital profiling of polynucleotide populations
InactiveUS20050250147A1Improved statistical confidenceOvercome deficienciesMicrobiological testing/measurementTransferasesAssayHybridization probe
The invention provides methods and compositions for hybridization-based assays that employ oligonucleotide tags, wherein probes specific for the same target polynucleotide are labeled with a plurality of different oligonucleotide tags. When probes are used in conjunction with a microarray, or like, readout platform, containing hybridization sites of tag complements, assay of a target polynucleotide results in a signal being generated from any of a plurality hybridization sites with predetermined addresses and the number of such sites generating a signal is proportional to the relative amount of the target polynucleotide in a population, test sample, or reaction volume, as the case may be. The invention provides methods and compositions for measuring amounts of selected target polynucleotides in a sample and for providing a digital readout of such amounts. Statistical confidence in measurements made by the present invention may be increased as much as desired by increasing the size of the sample of successfully hybridized and selected probes from which signals are generated.
Owner:PARALLELE BIOSCI
Integrated active flux microfluidic devices and methods
InactiveUS20040248167A1Increase speedImprove accuracyBioreactor/fermenter combinationsFlow mixersAntigenHybridization probe
The invention relates to a microfabricated device for the rapid detection of DNA, proteins or other molecules associated with a particular disease. The devices and methods of the invention can be used for the simultaneous diagnosis of multiple diseases by detecting molecules (e.g. amounts of molecules), such as polynucleotides (e.g., DNA) or proteins (e.g., antibodies), by measuring the signal of a detectable reporter associated with hybridized polynucleotides or antigen / antibody complex. In the microfabricated device according to the invention, detection of the presence of molecules (i.e., polynucleotides, proteins, or antigen / antibody complexes) are correlated to a hybridization signal from an optically-detectable (e.g. fluorescent) reporter associated with the bound molecules. These hybridization signals can be detected by any suitable means, for example optical, and can be stored for example in a computer as a representation of the presence of a particular gene. Hybridization probes can be immobilized on a substrate that forms part of or is exposed to a channel or channels of the device that form a closed loop, for circulation of sample to actively contact complementary probes. Universal chips according to the invention can be fabricated not only with DNA but also with other molecules such as RNA, proteins, peptide nucleic acid (PNA) and polyamide molecules.
Owner:CALIFORNIA INST OF TECH
Hybridization assisted nanopore sequencing
InactiveUS20070190542A1Eliminate needEasy to analyzeMicrobiological testing/measurementLaboratory glasswaresHybridization probeNucleic Acid Probes
A method of employing a nanopore structure in a manner that allows the detection of the positions (relative and / or absolute) of nucleic acid probes that are hybridized onto a single-stranded nucleic acid molecule. In accordance with the method the strand of interest is hybridized with a probe having a known sequence. The strand and hybridized probes are translocated through a nanopore. The fluctuations in current measured across the nanopore will vary as a function of time corresponding to the passing of a probe attachment point along the strand. These fluctuations in current are then used to determine the attachment positions of the probes along the strand of interest. This probe position data is then fed into a computer algorithm that returns the sequence of the strand of interest.
Owner:NABSYS 2 0 +1
Immunomodulatory oligonucleotides
Oligonucleotides containing unthylated CpG dinucleotides and therapeutic utilities based on their ability to stimulate an immune response in a subject are disclosed. Also disclosed are therapies for treating diseases associated with immune system activation that are initiated by unthylated CpG dinucleotides in a subject comprising administering to the subject oligonucleotides that do not contain unmethylated CpG sequences (i.e. methylated CpG sequences or no CpG sequence) to outcompete unmethylated CpG nucleic acids for binding. Further disclosed are methylated CpG containing dinucleotides for use antisense therapies or as in vivo hybridization probes, and immunoinhibitory oligonucleotides for use as antiviral therapeutics.
Owner:IOWA RES FOUND UNIV OF +3
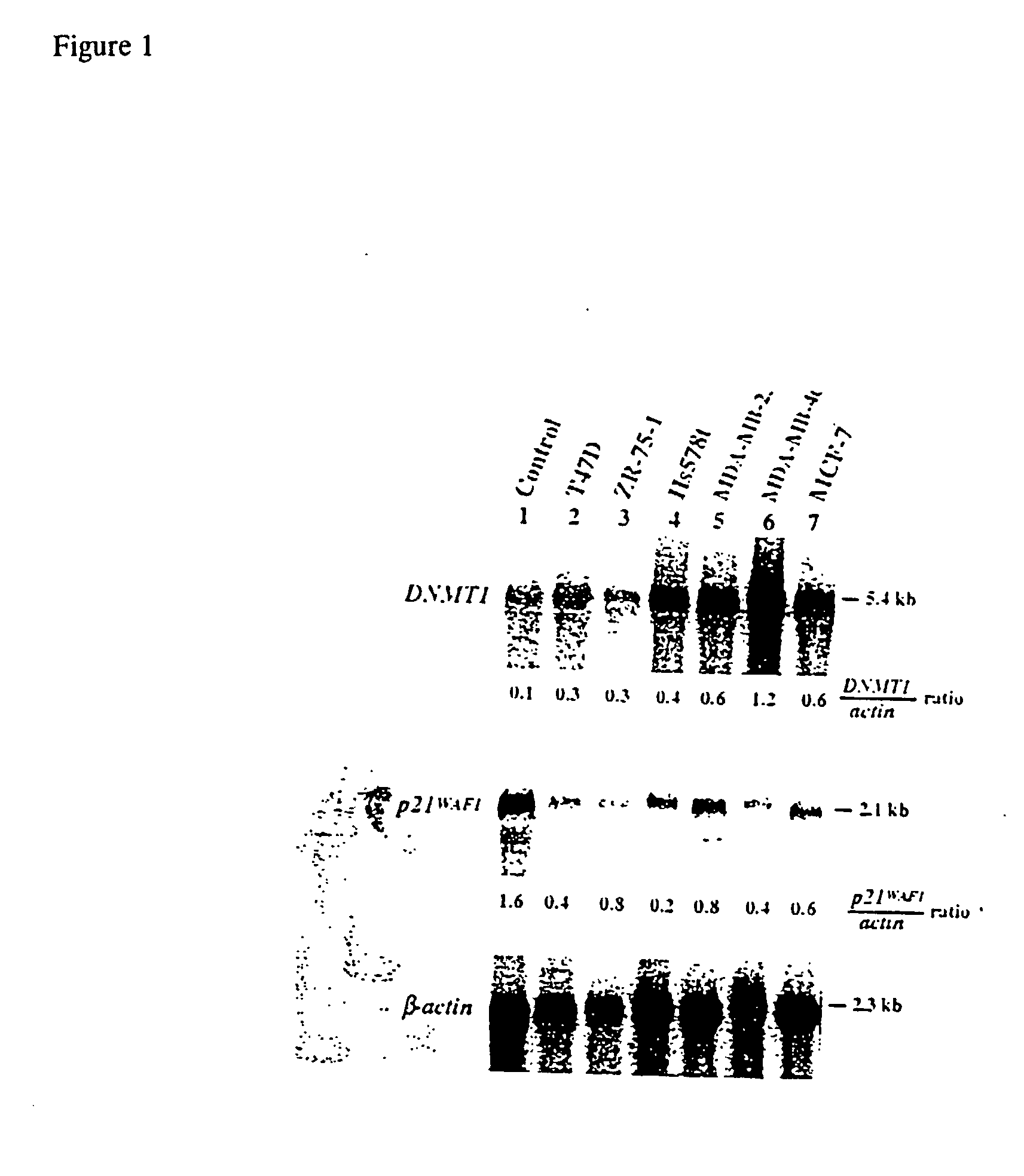
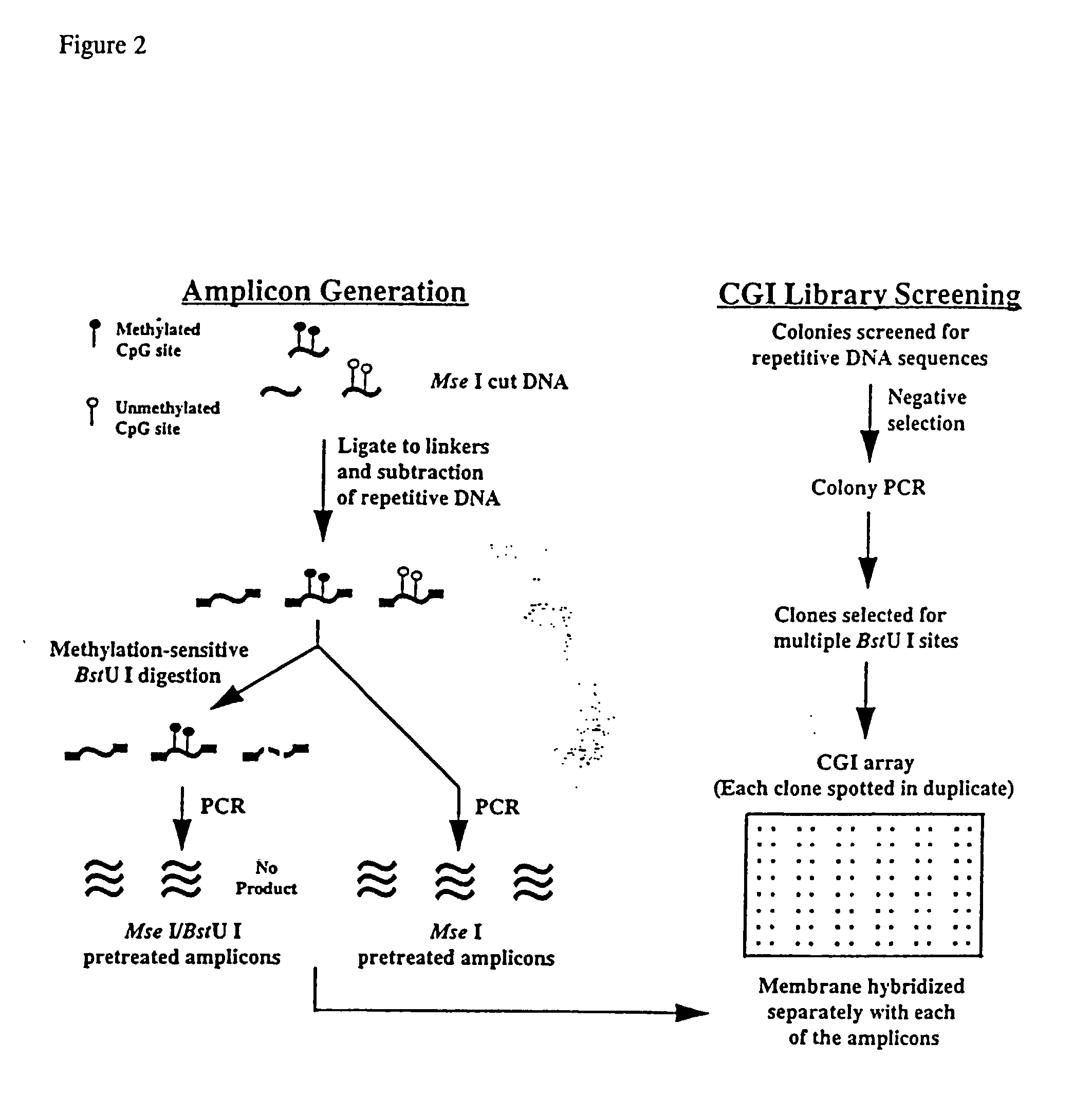
High-throughput methods for detecting DNA methylation
InactiveUS6605432B1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA methylationHybridization probe
The present invention provides a method of hybridization, differential methylation hybridization (DMH) for high throughput methylation analysis of multiple CpG island loci. DMH utilizes nucleic acid probes prepared from a cell sample to screen numerous CpG dinucleotide rich fragments affixed on a screening array. Positive hybridization signals indicate the presence of methylated sites. Methods of preparing the hybridization probes and screening array are also provided.
Owner:UNIVERSITY OF MISSOURI
Immunomodulatory oligonucleotides
InactiveUS20050004062A1Improve responseIncrease the number ofOrganic active ingredientsSugar derivativesHybridization probeIn vivo
Oligonucleotides containing unthylated CpG dinucleotides and therapeutic utilities based on their ability to stimulate an immune response in a subject are disclosed. Also disclosed are therapies for treating diseases associated with immune system activation that are initiated by unthylated CpG dinucleotides in a subject comprising administering to the subject oligonucleotides that do not contain unmethylated CpG sequences (i.e. methylated CpG sequences or no CpG sequence) to outcompete unmethylated CpG nucleic acids for binding. Further disclosed are methylated CpG containing dinucleotides for use antisense therapies or as in vivo hybridization probes, and immunoinhibitory oligonucleotides for use as antiviral therapeutics.
Owner:COLEY PHARM GRP INC +1
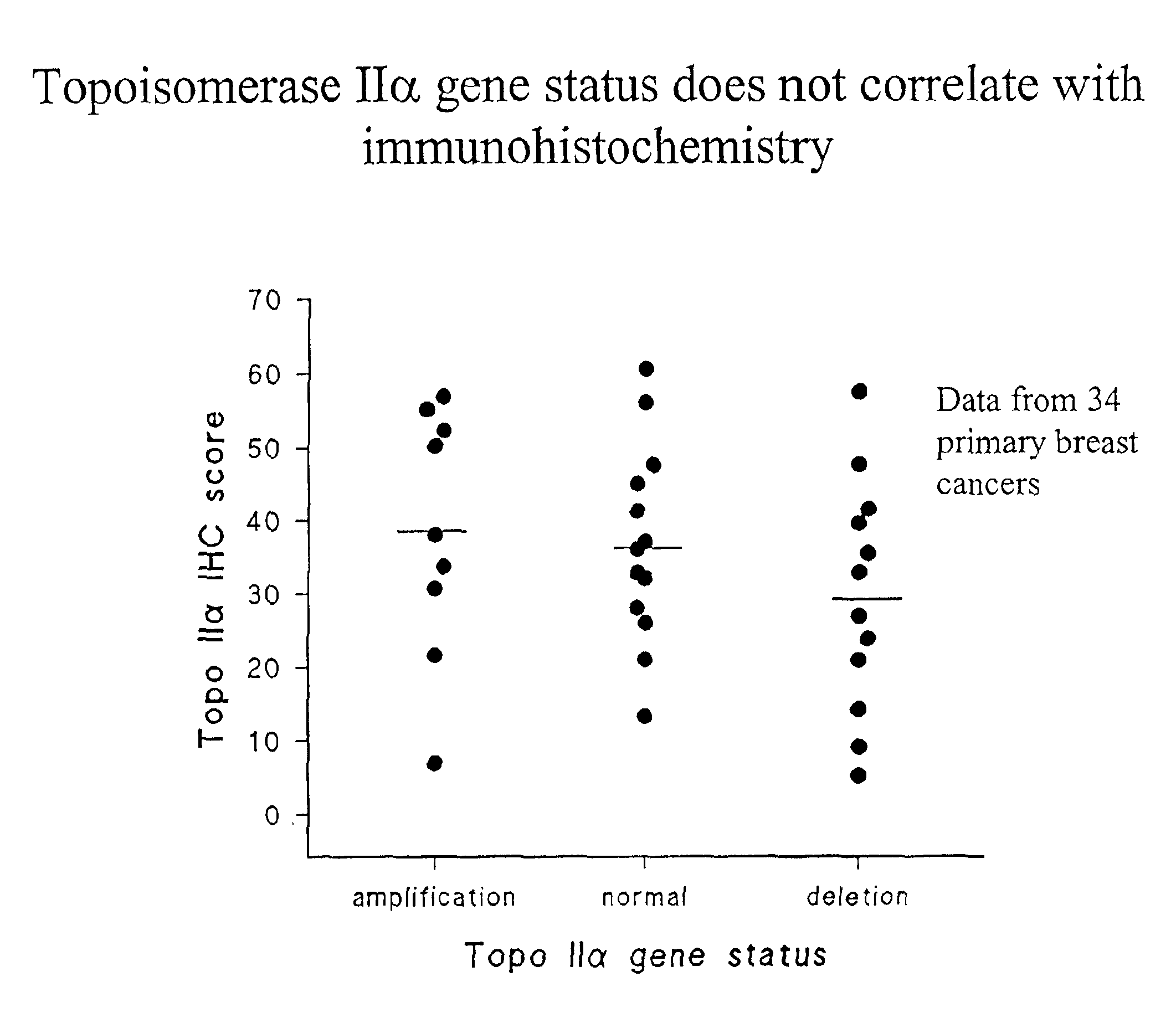
Identifying subjects suitable for topoisomerase II inhibitor treatment
InactiveUS6942970B2Suitable for treatmentSugar derivativesMicrobiological testing/measurementDNA underwindingHybridization probe
The present invention provides methods for diagnosing and treating cancer, and in particular methods for determining the susceptibility of subjects suspected of having breast cancer (or known to have breast cancer) to treatment with topoisomerase II inhibitors. The present invention also provides in situ hybridization probes for specifically detecting topoIIα gene sequences.
Owner:LIFE TECH CORP
Multipartite high-affinity nucleic acid probes
InactiveUS6451588B1Accurately determineBioreactor/fermenter combinationsBiological substance pretreatmentsHybridization probeNucleic Acid Probes
The invention provides a collection of probes useful for hybridizing to a target nucleic acid. The probes associate with each other, binding with high affinity to the target nucleic acid, to form three-way junctions and other complexes. At least one of the probes in each collection includes a nucleic acid analog. Methods using the probes in hybridization and as primers are also provided.
Owner:APPL BIOSYSTEMS INC
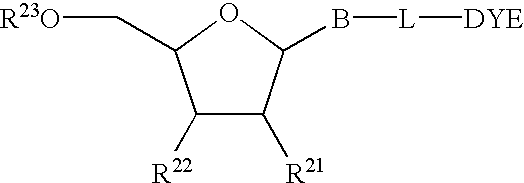
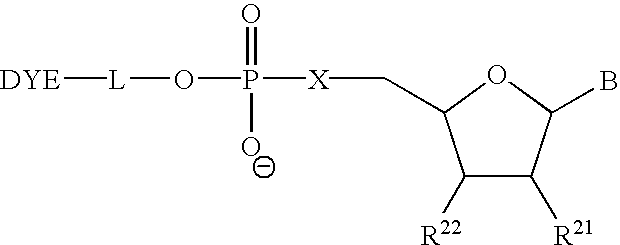
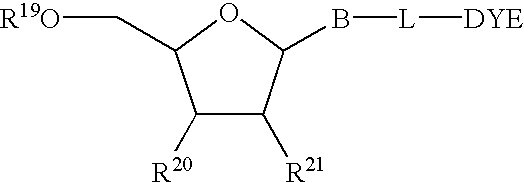
Mass label linked hybridisation probes
InactiveUS20050042625A1Increase the number ofEasy to separateBioreactor/fermenter combinationsBiological substance pretreatmentsHybridization probeMass spectrometry
An array of hybridization probes, each of which comprises a mass label linked to a known base sequence of predetermined length, wherein each mass label of the array, optionally together with the known base sequence, is relatable to that base sequence by mass spectrometry.
Owner:XZILLION
Methods and compositions for analyzing nucleic acids
InactiveUS20060078894A1Minimizes problemReduce capacityMicrobiological testing/measurementFermentationHybridization probeNucleic acid sequencing
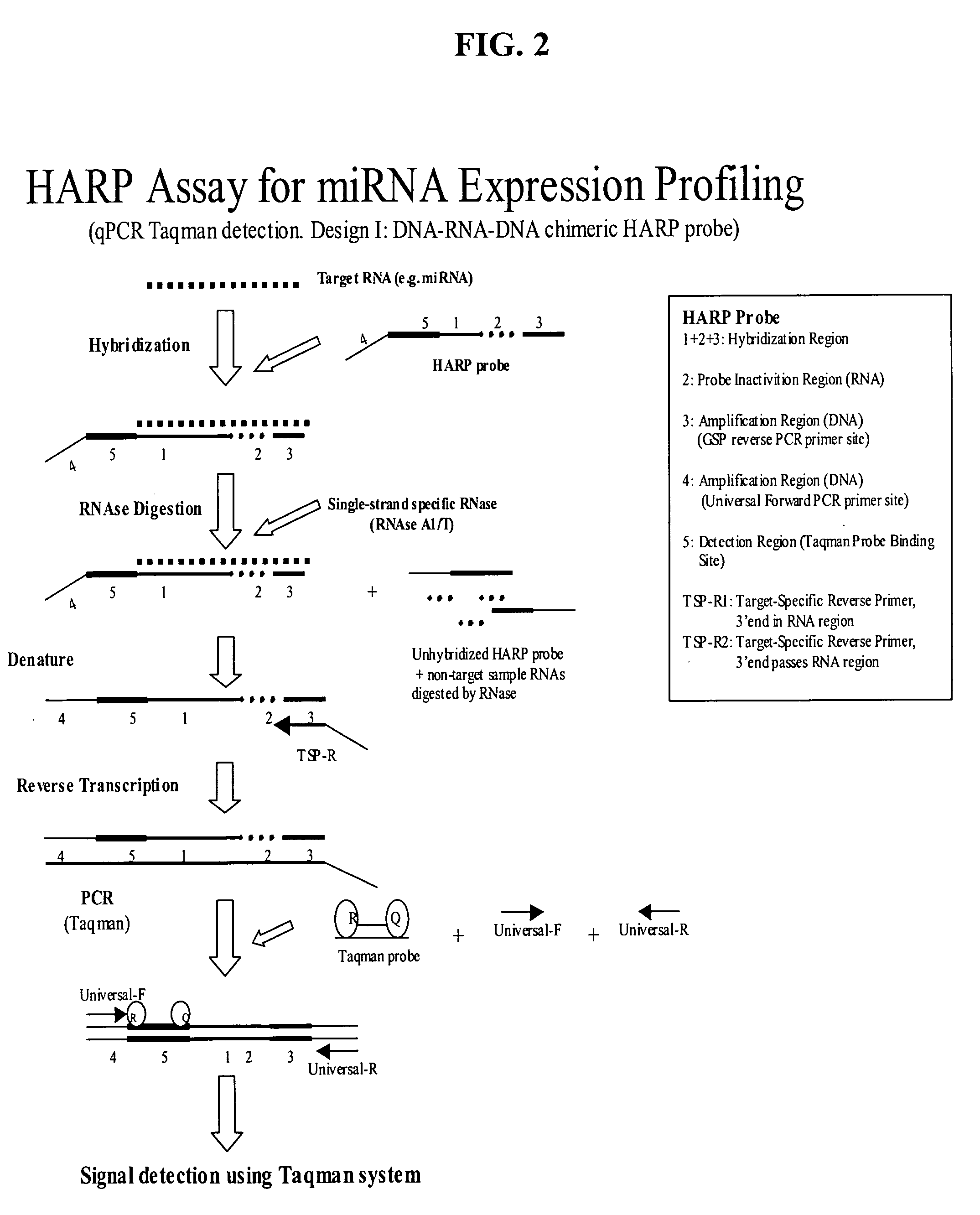
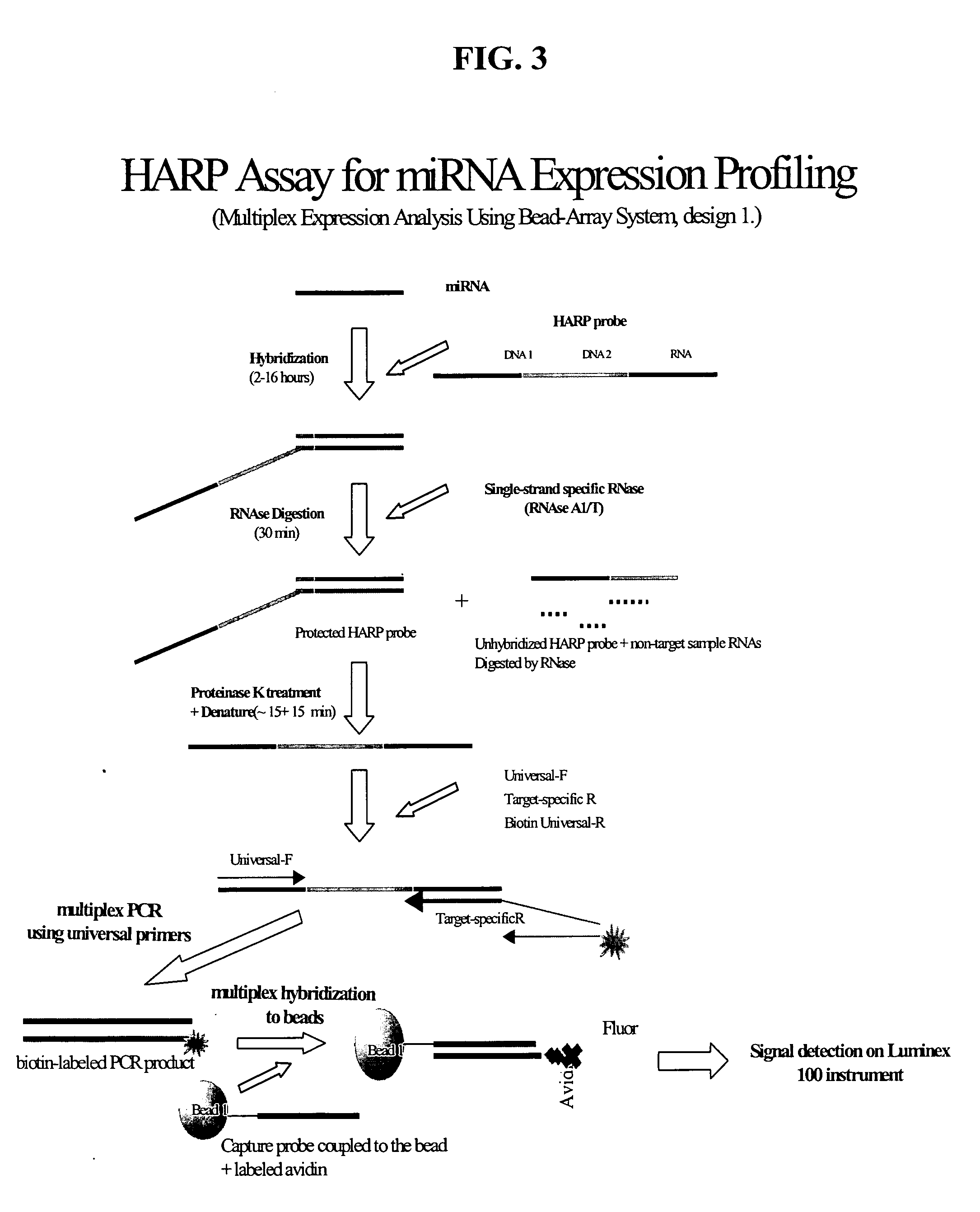
Methods and compositions for probe amplification to detect, identify, quantitate, and / or analyze a targeted nucleic acid sequence. After hybridization between a probe and the targeted nucleic acid, the probe is modified to distinguish hybridized probe from unhybridized probe. Thereafter, the probe is amplified. Moreover, in specific embodiments, the present invention involves a chimeric probe that is particularly effective when the targeted nucleic acid sequence is short and / or has a relatively low concentration, such as with an miRNA molecule.
Owner:ASURAGEN
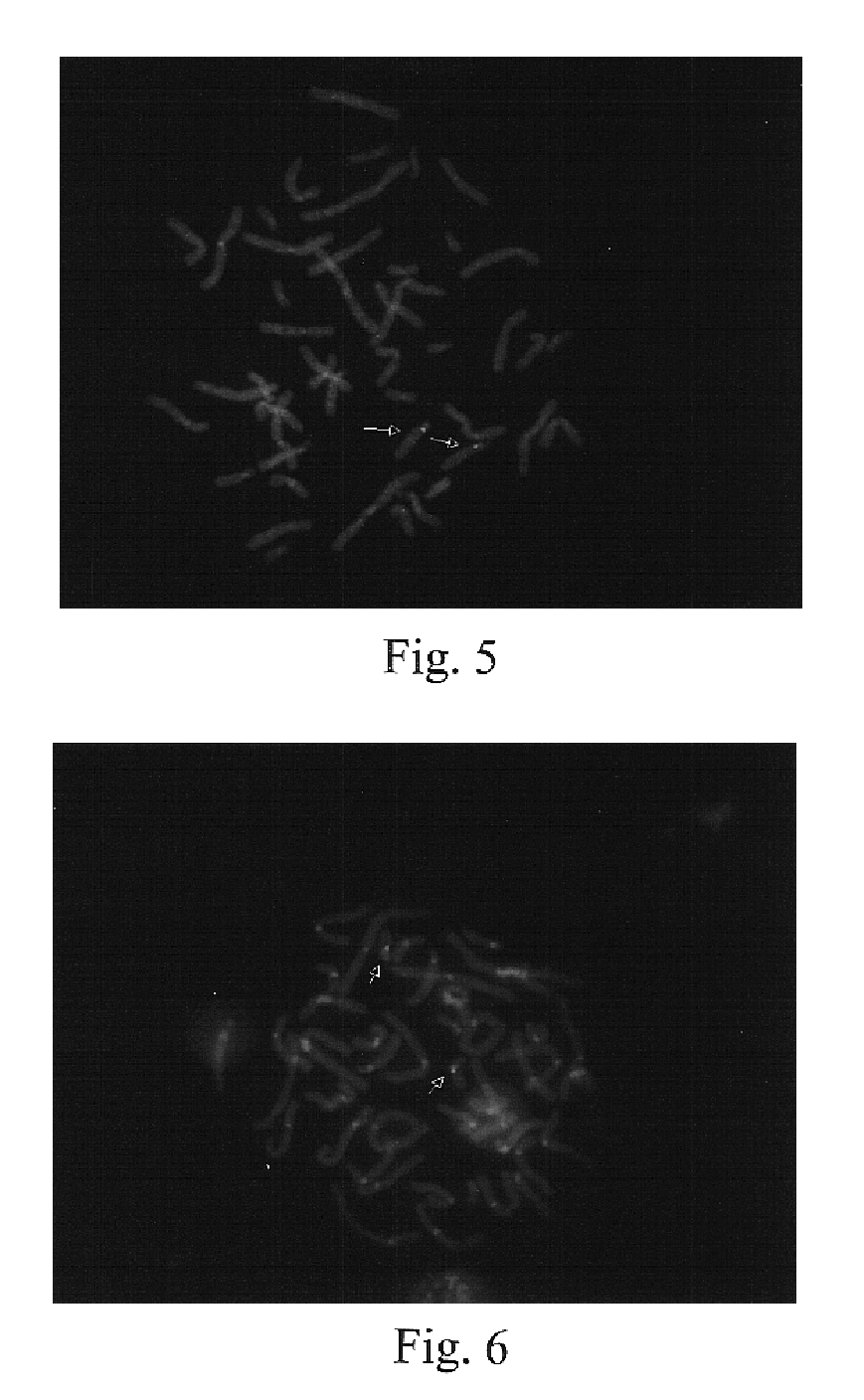
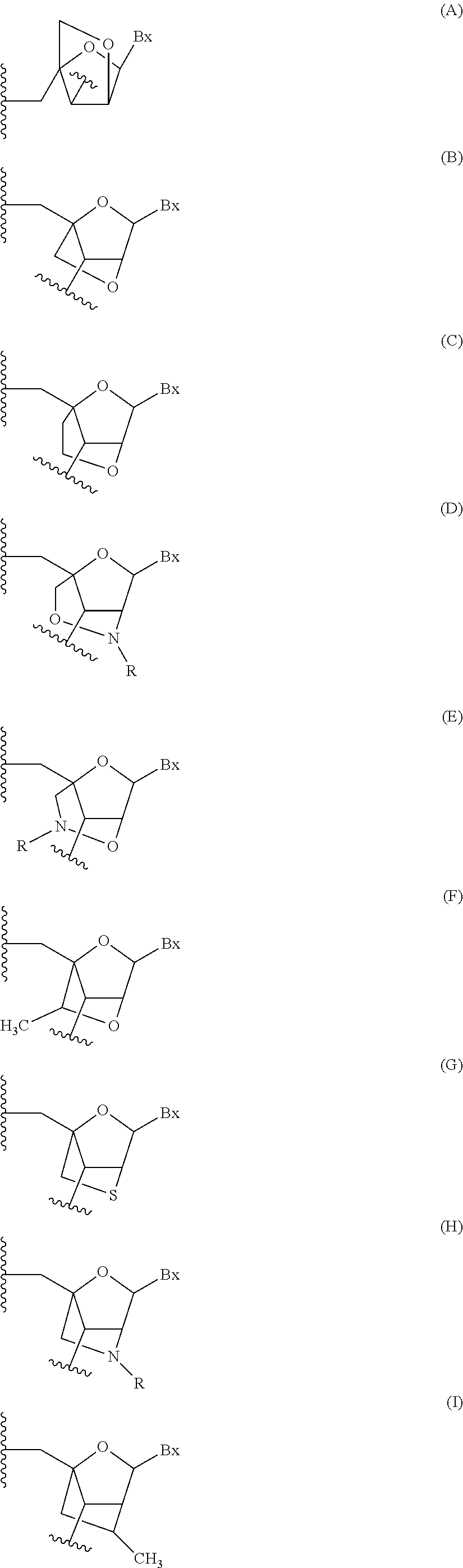
Chromosome structural abnormality localization with single copy probes
InactiveUS7014997B2Eliminates spurious hybridizationSugar derivativesMicrobiological testing/measurementGenomic SegmentHybridization probe
Nucleic acid (e.g., DNA) hybridization probes are described which comprise a labeled, single copy nucleic acid which hybridizes to a deduced single copy sequence interval in target nucleic acid of known sequence. The probes, which are essentially free of repetitive sequences, can be used in hybridization analyses without adding repetitive sequence-blocking nucleic acids. This allows rapid and accurate detection of chromosomal abnormalities. The probes are preferably designed by first determining the sequence of at least one single copy interval in a target nucleic acid sequence, and developing corresponding hybridization probes which hybridize to at least a part of the deduced single copy sequence. In practice, the sequences of the target and of known genomic repetitive sequence representatives are compared in order to deduce locations of the single copy sequence intervals. The single copy probes can be developed by any variety of methods, such as PCR amplification, restriction or exonuclease digestion of purified genomic fragments, or direct synthesis of DNA sequences. This is followed by labeling of the probes and hybridization to a target sequence.
Owner:CHILDRENS MERCY HOSPITAL
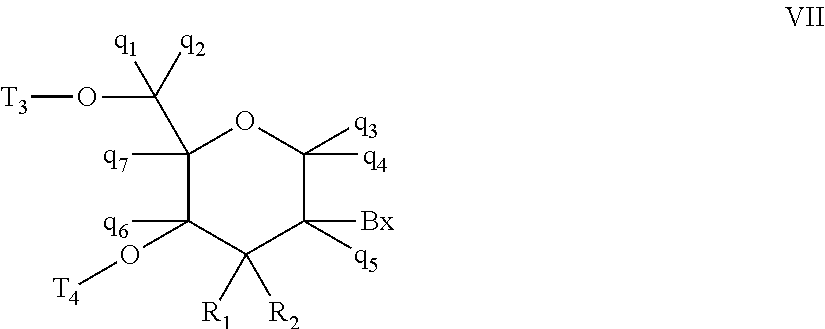
Oligomeric compounds comprising bicyclic nucleotides and uses thereof
The present invention provides oligomeric compounds. Certain such oligomeric compounds are useful for hybridizing to a complementary nucleic acid, including but not limited, to nucleic acids in a cell. In certain embodiments, hybridization results in modulation of the amount activity or expression of the target nucleic acid in a cell.
Owner:IONIS PHARMA INC
High-throughput methods for detecting DNA methylation
InactiveUS20030129602A1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA methylationHybridization probe
The present invention provides a method of hybridization, differential methylation hybridization (DMH) for high throughput methylation analysis of multiple CpG island loci. DMH utilizes nucleic acid probes prepared from a cell sample to screen numerous CpG dinucleotide rich fragments affixed on a screening array. Positive hybridization signals indicate the presence of methylated sites. Methods of preparing the hybridization probes and screening array are also provided.
Owner:UNIVERSITY OF MISSOURI
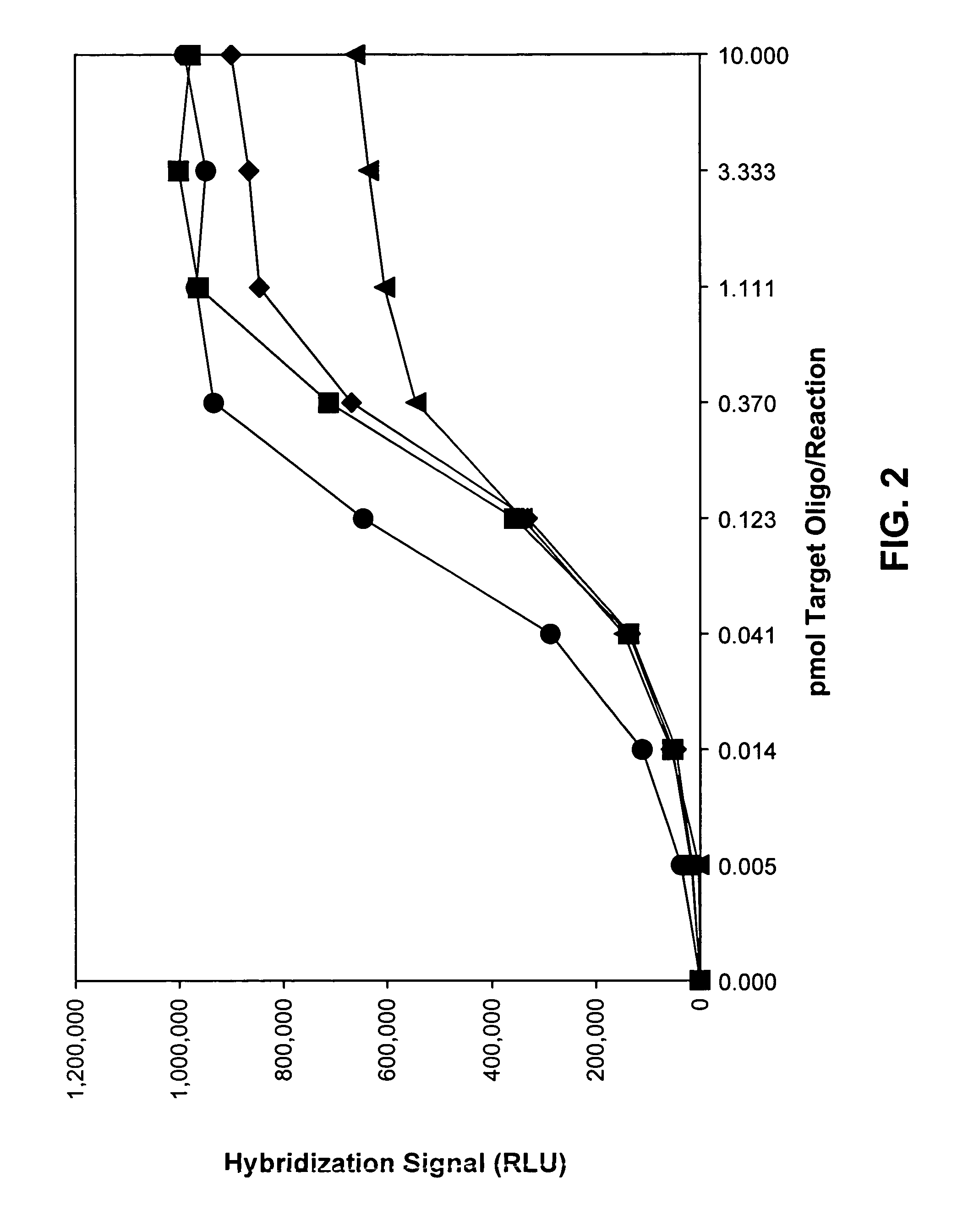
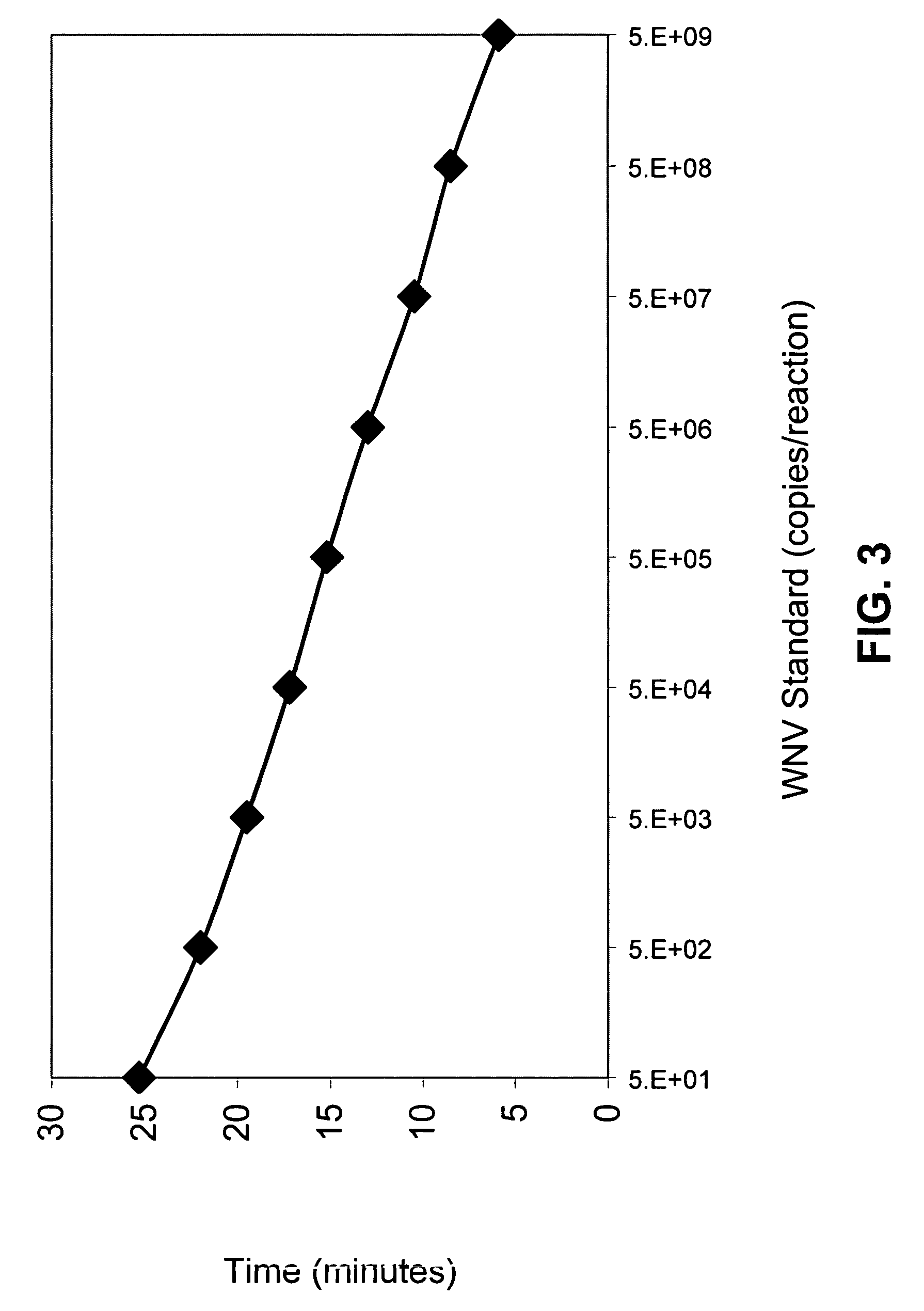
Compositions and methods for detecting West Nile virus
Compositions, methods and kits for detecting flavivirus nucleic acids. Particularly described are oligonucleotides that are useful as hybridization probes and amplification primers for detecting very low levels of West Nile virus nucleic acids.
Owner:GEN PROBE INC
Polymeric nucleic acid hybridization probes
InactiveUS20050112636A1Mismatch discriminationSugar derivativesMicrobiological testing/measurementHybridization probeNucleic Acid Probes
A novel polymeric nucleic acid probe improves detection sensitivity and specificity in a variety of hybridization platforms. The probe is made up of multiple short nucleic acid sequences (referred to as monomers) attached together to form a long polymeric probe for use in hybridization applications. For applications requiring immobilization of the probes to a surface, the polymeric probes are similar to long DNA probes in that they can be immobilized to a variety of surfaces without need for a chemical modification to the end of the probe. Because target nucleic acids hybridize to the relatively short monomers in the polymeric probe, the polymeric probes are more specific than long DNA probes. In addition, polymeric probes also improve the signal-to-background ratio by increasing the number of accessible monomer oligonucleotide probes immobilized per unit area on a surface.
Owner:ATOM SCI
Affinity-shifted probes for quantifying analyte polynucleotides
InactiveUS20080131892A1Good quantitative resultNegative effectMaterial analysis by observing effect on chemical indicatorMicrobiological testing/measurementAmpliconHybridization probe
Compositions, methods and devices for detecting and quantifying levels of an analyte polynucleotide in homogeneous assays using collections of soluble or immobilized hybridization probes. In certain preferred embodiments, the probes are immobilized in an array format. Polynucleotides may be quantified directly, or amplified in an in vitro nucleic acid amplification reaction prior to detection and quantitation. Amplification reactions may be performed in contact with the invented probes, and analyte amplicons quantified in real-time or end-point assays.
Owner:GEN PROBE INC
Automated hybridization/imaging device for fluorescent multiplex DNA sequencing
InactiveUS20020012910A1Easy to disassembleBioreactor/fermenter combinationsSludge treatmentHybridization probeSize fractionated
A method is disclosed for automated multiplex sequencing of DNA with an integrated automated imaging hybridization chamber system. This system comprises an hybridization chamber device for mounting a membrane containing size-fractionated multiplex sequencing reaction products, apparatus for fluid delivery to the chamber device, imaging apparatus for light delivery to the membrane and image recording of fluorescence emanating from the membrane while in the chamber device, and programmable controller apparatus for controlling operation of the system. The multiplex reaction products are hybridized with a probe, then an enzyme (such as alkaline phosphatase) is bound to a binding moiety on the probe, and a fluorogenic substrate (such as a benzothiazole derivative) is introduced into the chamber device by the fluid delivery apparatus. The enzyme converts the fluorogenic substrate into a fluorescent product which, when illuminated in the chamber device with a beam of light from the imaging apparatus, excites fluorescence of the fluorescent product to produce a pattern of hybridization. The pattern of hybridization is imaged by a CCD camera component of the imaging apparatus to obtain a series of digital signals. These signals are converted by the controller apparatus into a string of nucleotides corresponding to the nucleotide sequence an automated sequence reader. The method and apparatus are also applicable to other membrane-based applications such as colony and plaque hybridization and Southern, Northern, and Western blots.
Owner:WEISS ROBERT B +6
Monitoring of gene expression by detecting hybridization to nucleic acid arrays using anti-heteronucleic acid antibodies
InactiveUS6232068B1Nucleotide librariesMicrobiological testing/measurementHybridization probeHybridization Array
Owner:TULARIK INC +1
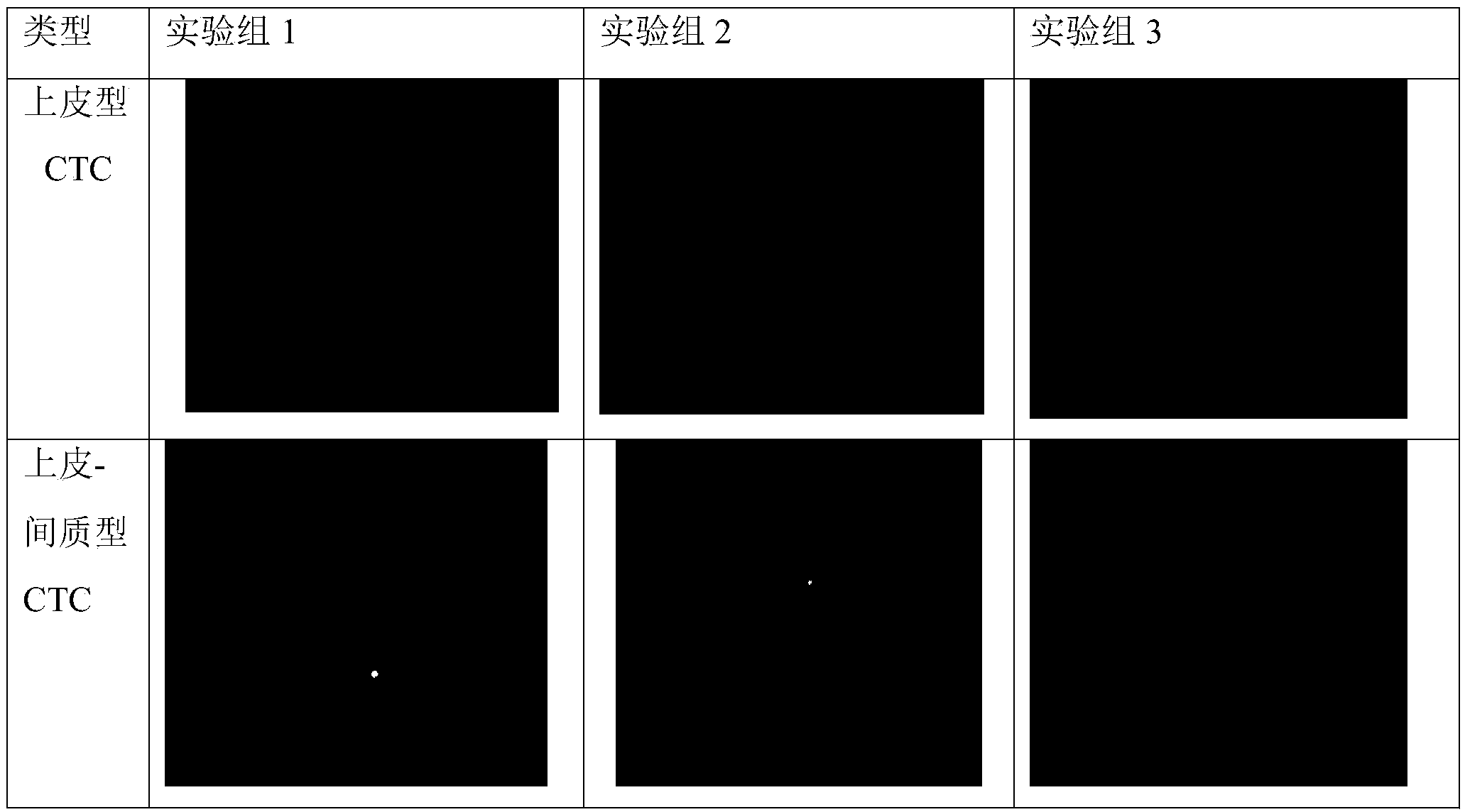
Circulating tumor cell identification kit and circulating tumor cell identification method
ActiveCN104031993AComprehensive detectionAvoid false negativesMicrobiological testing/measurementHybridization probeFluorescence
The invention discloses a circulating tumor cell identification kit and a circulating tumor cell identification method. The circulating tumor cell identification kit comprises a detection probe for detecting mRNA of an epithelial cell marking gene and / or mRNA of a mesenchymal cell marking gene, wherein the epithelial cell marking gene is selected from one or more of CDH2, VIMENTIN, FN1 and AKT2. Multiple RNA probes are adopted in the identification method, specific genes of a plurality of circulating tumor cells (CTCs) can be marked at the same time and are divided into genes I (epithelial genes), genes II (epithelial-mesenchymal genes) and genes III (mesenchymal genes), and false positive results caused by lack of part of the specific genes of the CTCs in a process that the CTCs enter peripheral blood for circulation are reduced; the method can be finished within eight hours, and a single copied mRNA hybridization probe is combined with a corresponding fluorescent probe by virtue of a signal amplification system, so that the detection sensitivity of in-situ RNA (Ribonucleic Acid) hybridization is remarkably improved.
Owner:SUREXAM BIO TECH
Immunomodulatory oligonucleotides
Oligonucleotides containing unthylated CpG dinucleotides and therapeutic utilities based on their ability to stimulate an immune response in a subject are disclosed. Also disclosed are therapies for treating diseases associated with immune system activation that are initiated by unthylated CpG dinucleotides in a subject comprising administering to the subject oligonucleotides that do not contain unmethylated CpG sequences (i.e. methylated CpG sequences or no CpG sequence) to outcompete unmethylated CpG nucleic acids for binding. Further disclosed are methylated CpG containing dinucleotides for use antisense therapies or as in vivo hybridization probes, and immunoinhibitory oligonucleotides for use as antiviral therapeutics.
Owner:UNIV OF IOWA RES FOUND +2
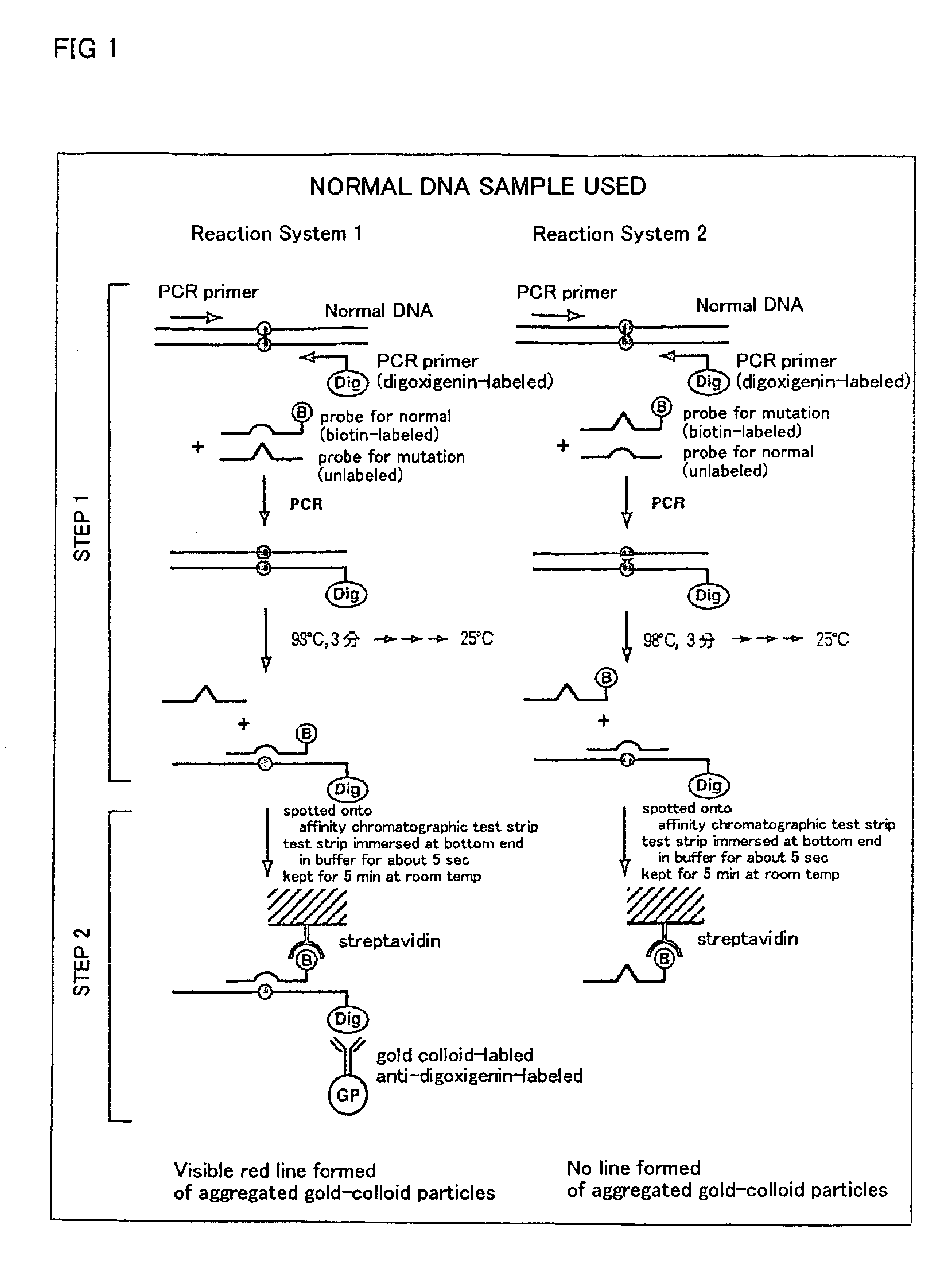
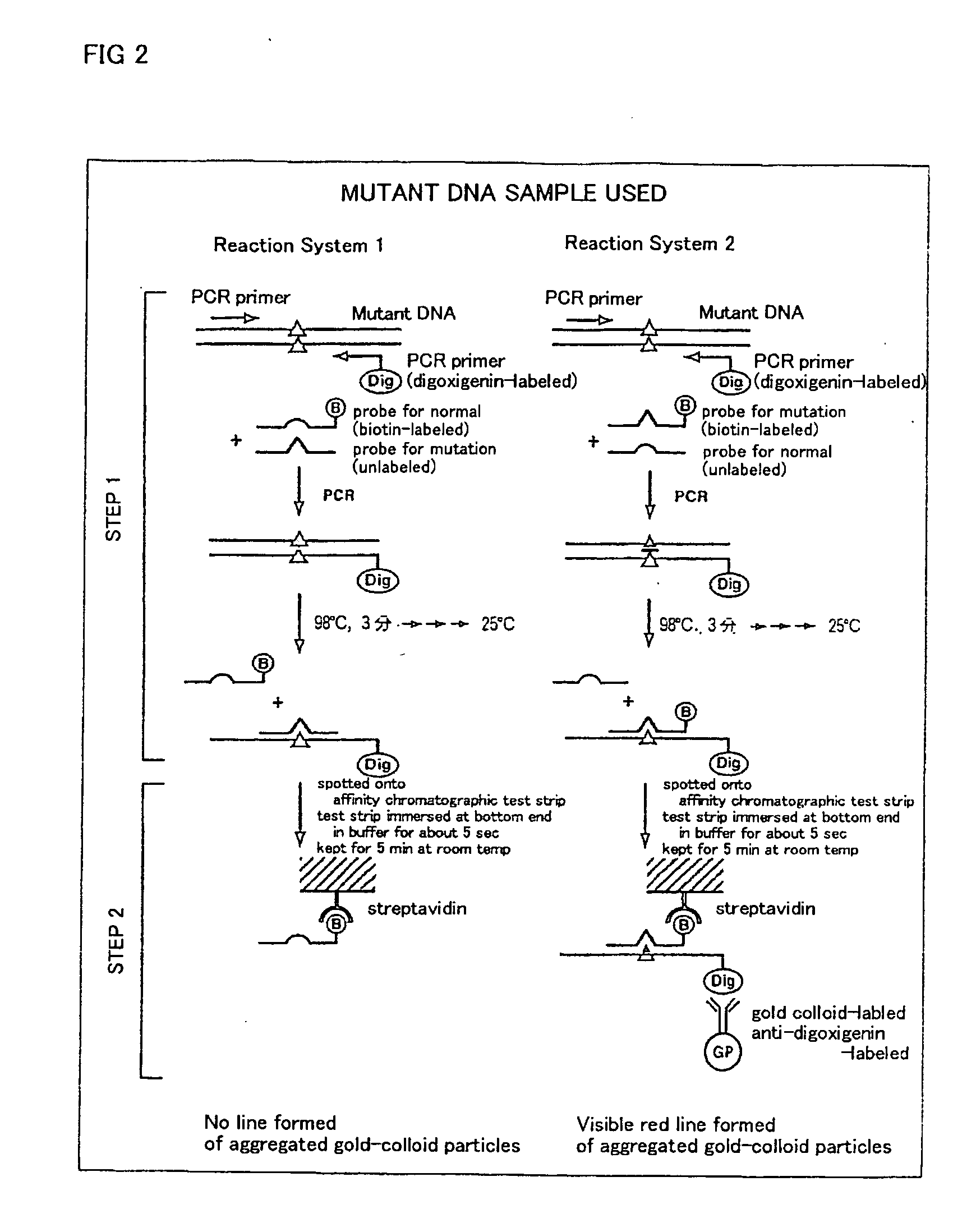
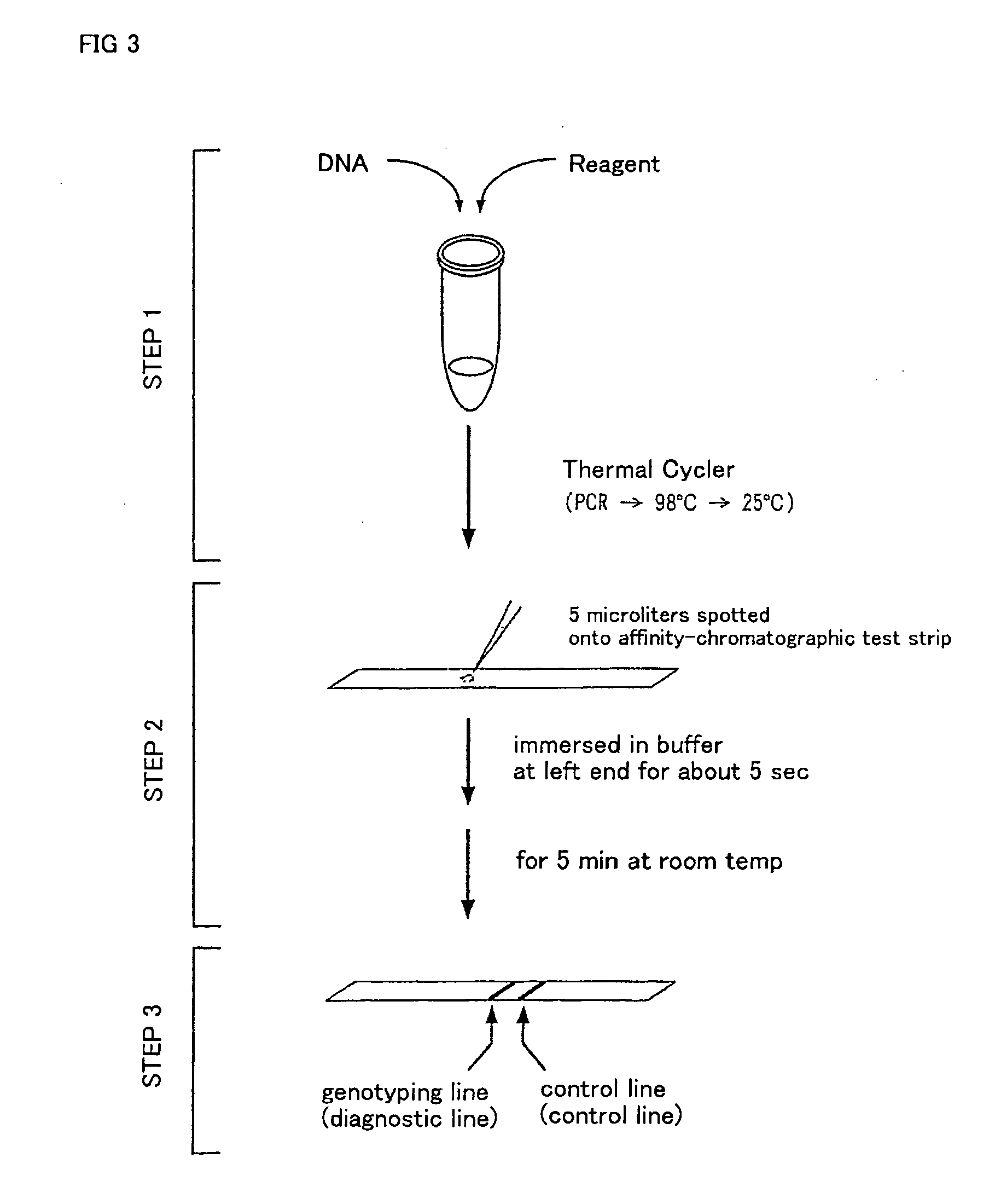
Method of detecting gene mutation
InactiveUS20060127907A1Simply and quickly detectingEasy to detectSugar derivativesMicrobiological testing/measurementHybridization probeDna amplification
DNA amplification and hybridization are successively carried out in a reaction system containing primers for the DNA amplification and hybridization probes, followed by detecting the hybrid in the reaction solution by affinity chromatography, wherein at least one of the primers to be used in the DNA amplification is labeled with a first labeling agent so that the amplified DNA will be labeled with the first labeling agent, a hybridization probe is labeled with a second labeling agent and contained in a reaction solution for effecting the DNA amplification, the base sequence of the hybridization probe is designed not to inhibit the DNA amplification, and a hybrid is detected by affinity chromatography with the use of the first and second labeling agents.
Owner:MATSUBARA YOICHI
System for multi color real time pcr
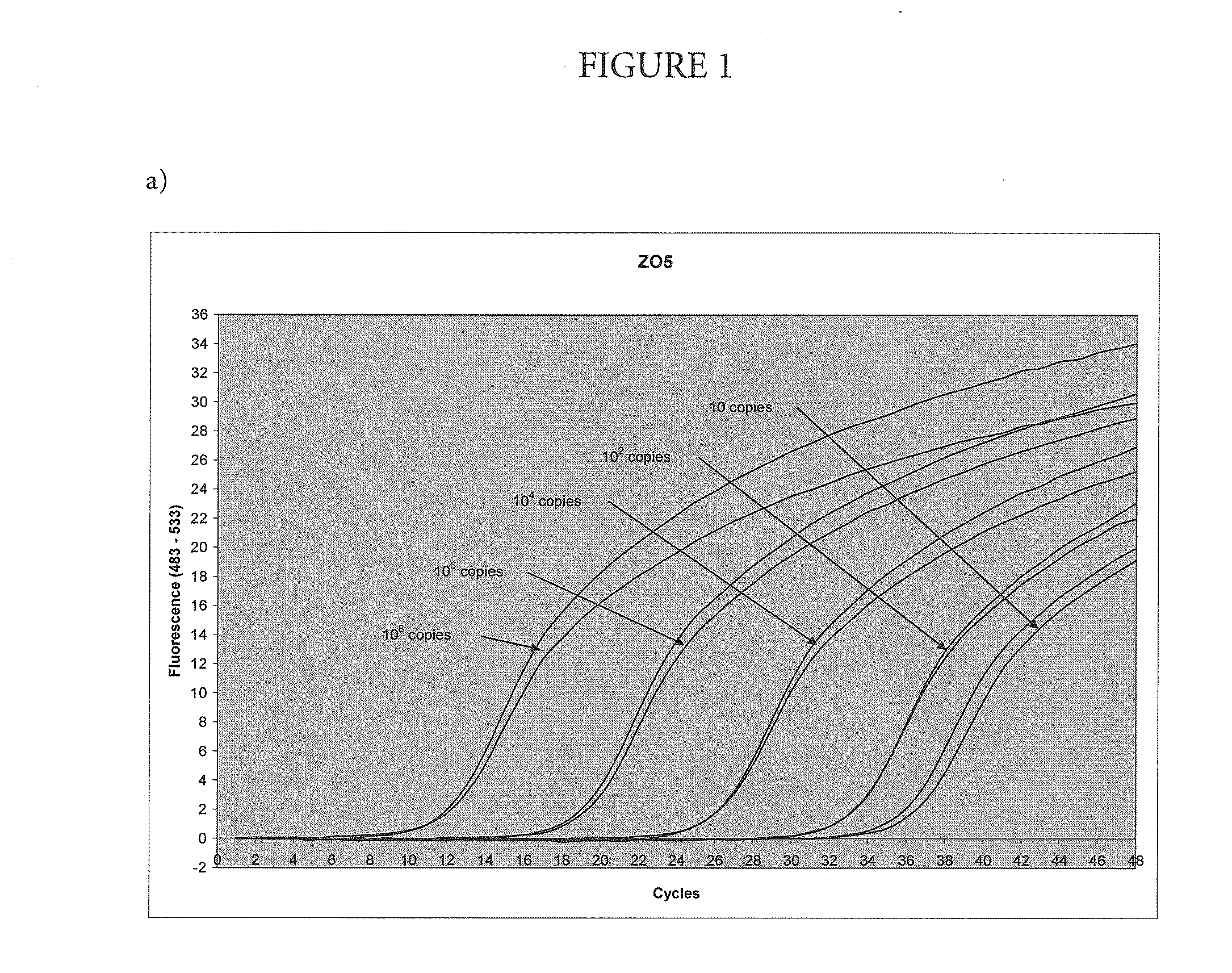
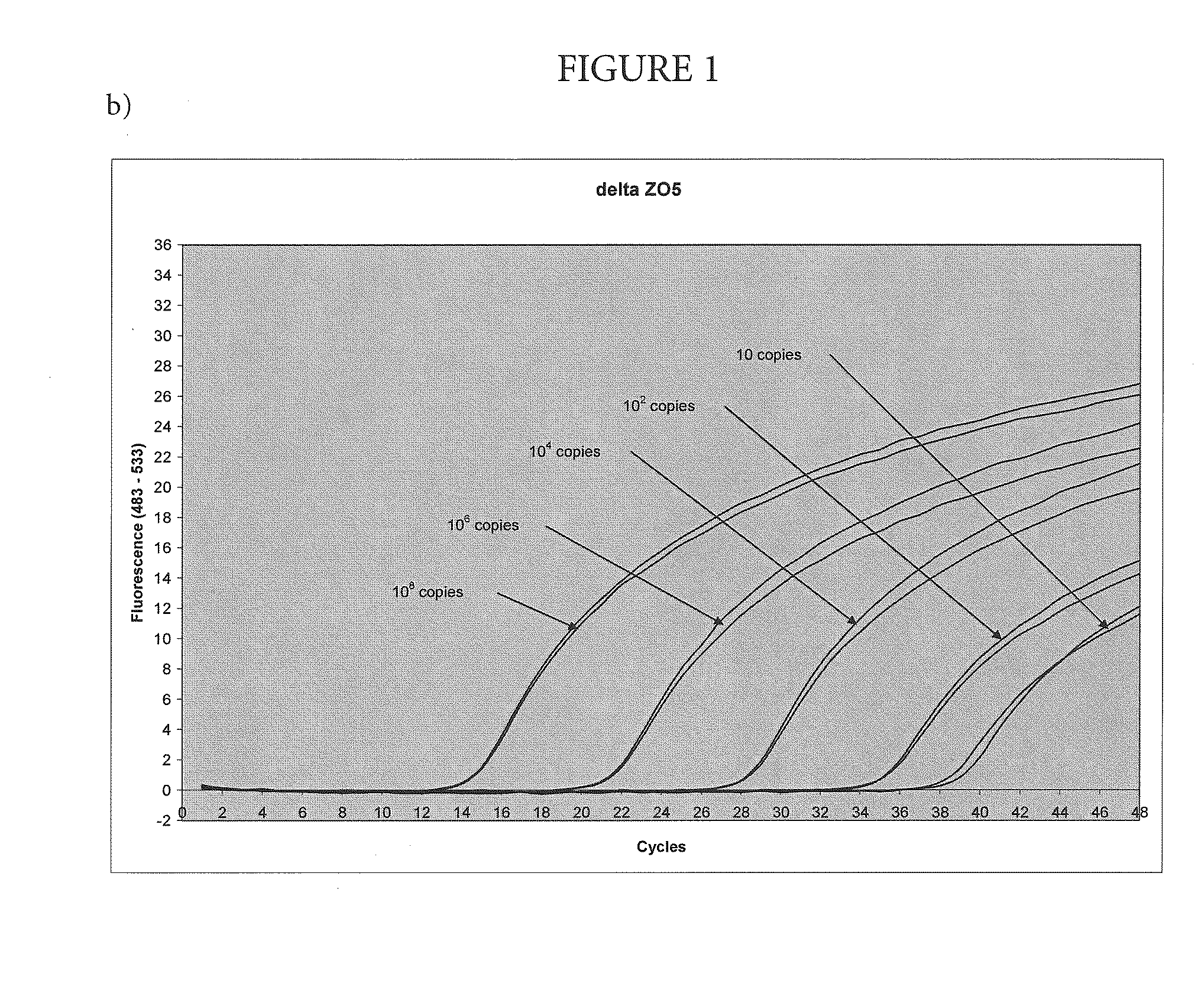
ActiveUS20060269922A1Enhanced signalLess-expensive setupBioreactor/fermenter combinationsBiological substance pretreatmentsReal-Time PCRsHybridization probe
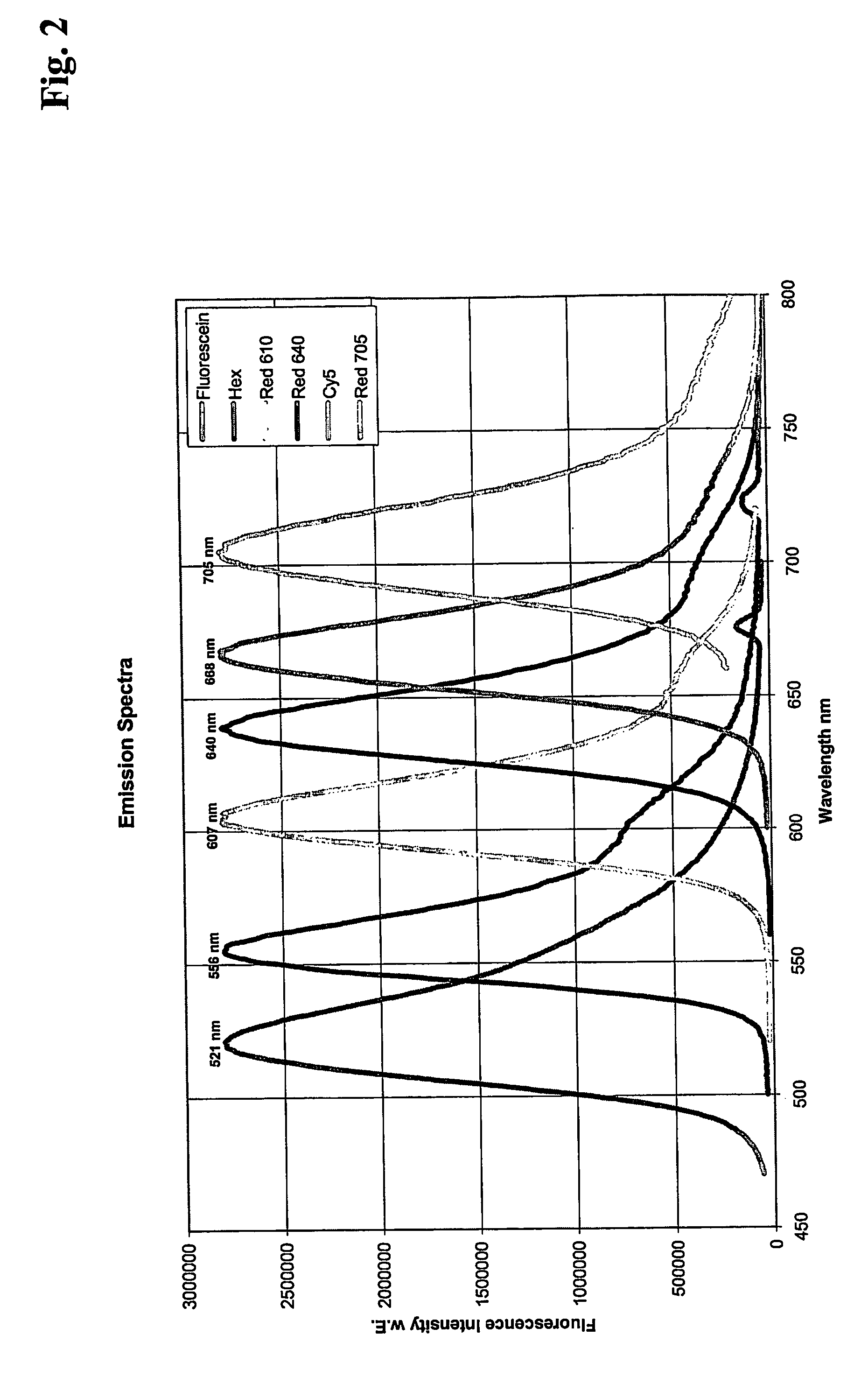
The invention is directed to a system for performing multi-color real time PCT, comprising a flexible real time PCT instrument and a specific composition or reaction mixture for performing multiplex PCR: In particular, the present invention is directed to a composition or reaction mixtuer which comprises at least 3, preferably 4-5 and most preferably exactly 4 pairs of FRET hybridization probes. Each pair of said hybridization probes consists of a FRET donor probe carrying a FRET donor moiety and a FRET acceptor probe carrying a FRET acceptor moiety having an emission maximum between 550 and 710 nm.
Owner:ROCHE MOLECULAR SYST INC
Method and probes for the detection of chromosome aberrations
InactiveUS7105294B2Lower melting temperatureRaise the ratioSugar derivativesMicrobiological testing/measurementIn situ hybridisationHybridization probe
A novel method for detecting chromosome aberrations is disclosed. More specifically, chromosome aberrations are detected by in situ hybridisation using at least two sets of hybridisation probes, at least one set comprising one or more peptide nucleic acid probes capable of hybridising to specific nucleic acid sequences related to a potential aberration in a chromosome, and at least one set comprising two or more peptide nucleic acid probes capable of hybridising to specific nucleic acid sequences related to another potential aberration in a chromosome. In particular, the method may be used for detecting chromosome aberrations in the form of breakpoints.
Owner:AGILENT TECH INC
Method for melting curve analysis of repetitive PCR products
InactiveUS6664064B1High sensitivitySugar derivativesMicrobiological testing/measurementHybridization probePolynucleotide
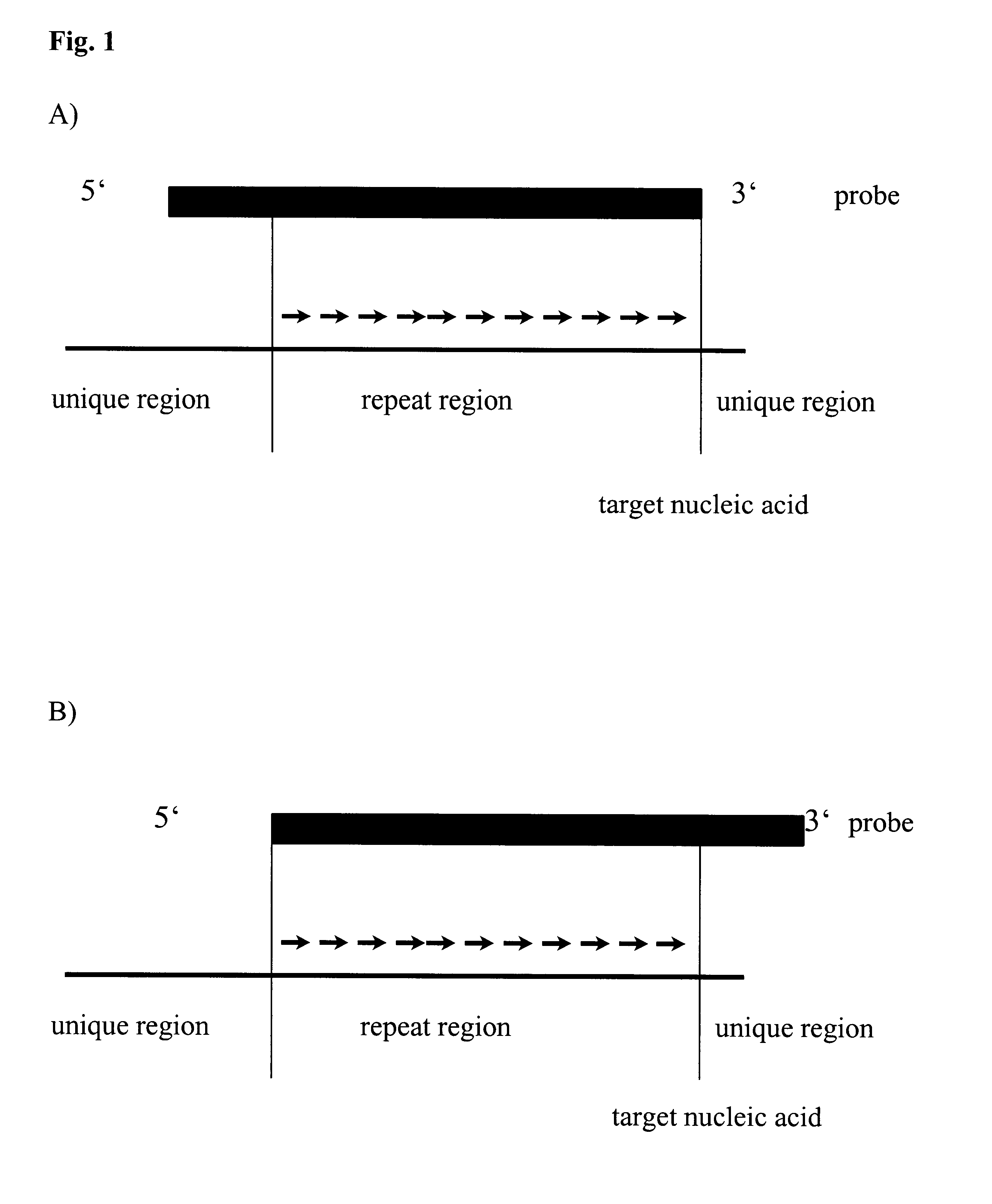
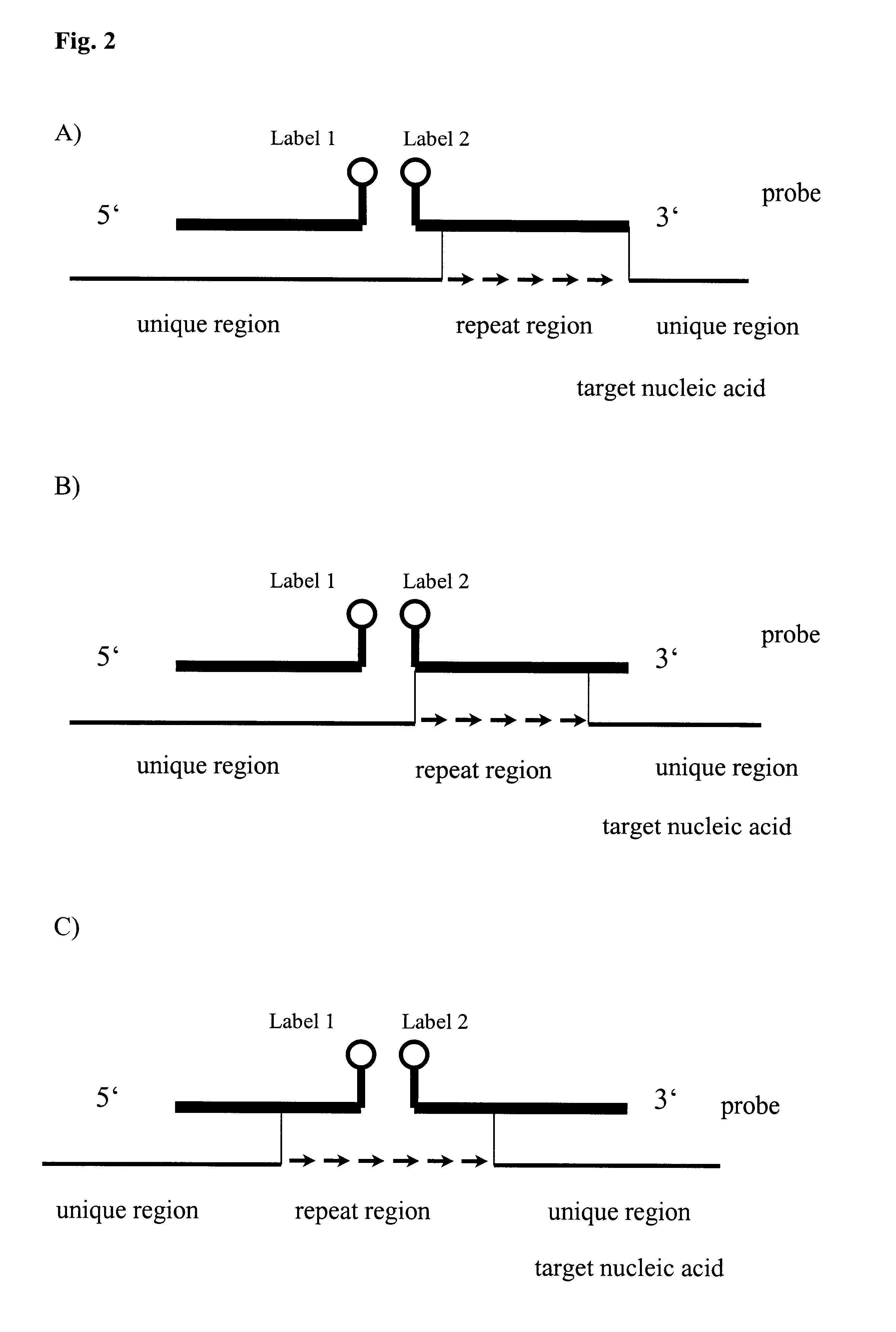
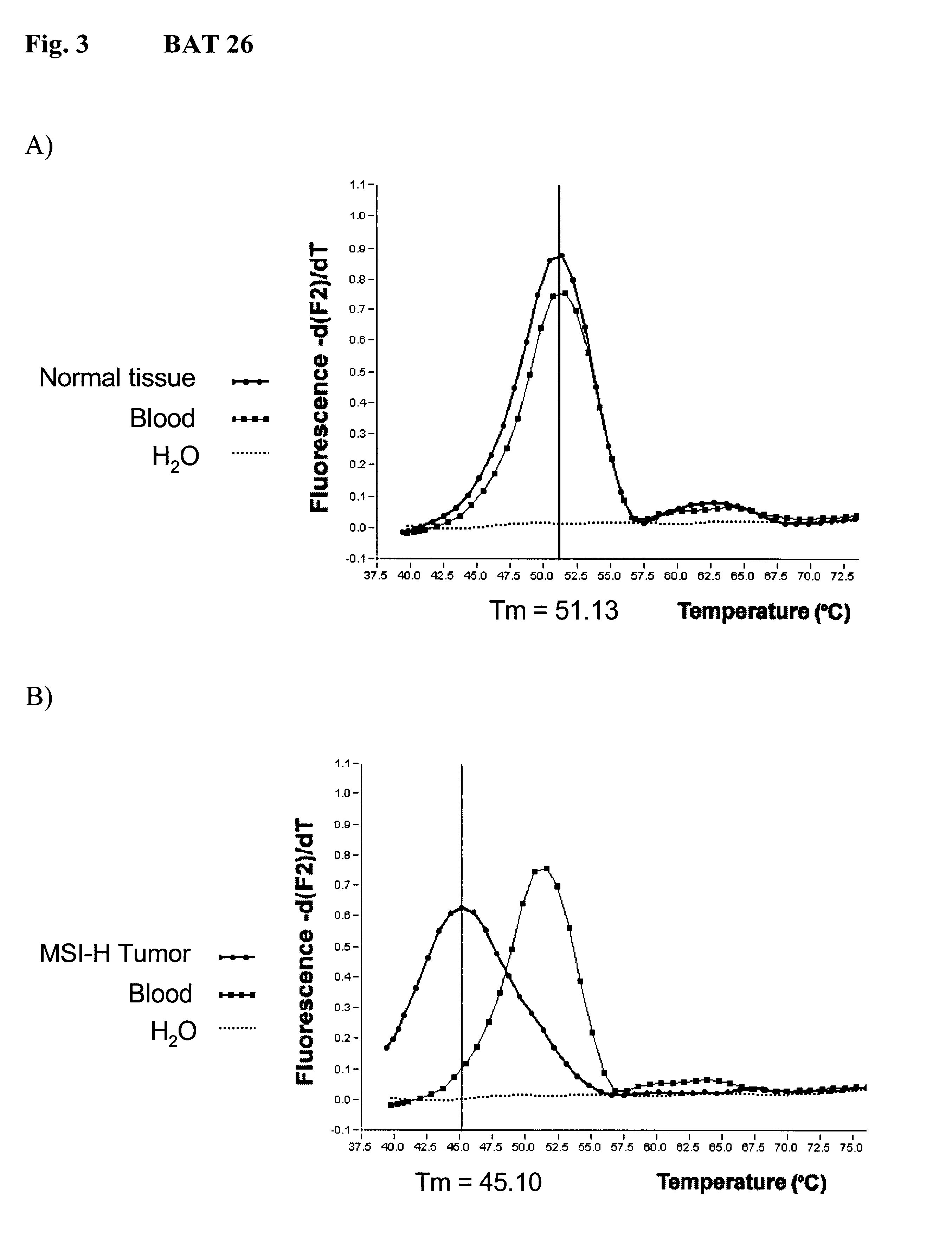
The invention relates to method, wherein the number of repeat sequences which are present in a sample is detemined by means of melting temperature analysis. More precisely, the invention relates to a method for analysis of a target nucleic acid consisting of repetitive and non repetitive sequences comprising (i) hybridization of at least one polynucleotide hybridization probe comprising a first segment which is complementary to a non repetitive region and a second segment which is compementary to an adjacent repetitive region, said second segment consisting of a defined number of repeats and (ii) determination of the melting point temperature of the hybrid which has been formed between the target nucleic acid and the at least one hybridization probe.
Owner:ROCHE DIAGNOSTICS OPERATIONS INC
Process for high throughput DNA methylation analysis
InactiveUS7112404B2Sugar derivativesMaterial analysis by observing effect on chemical indicatorDNA methylationHybridization probe
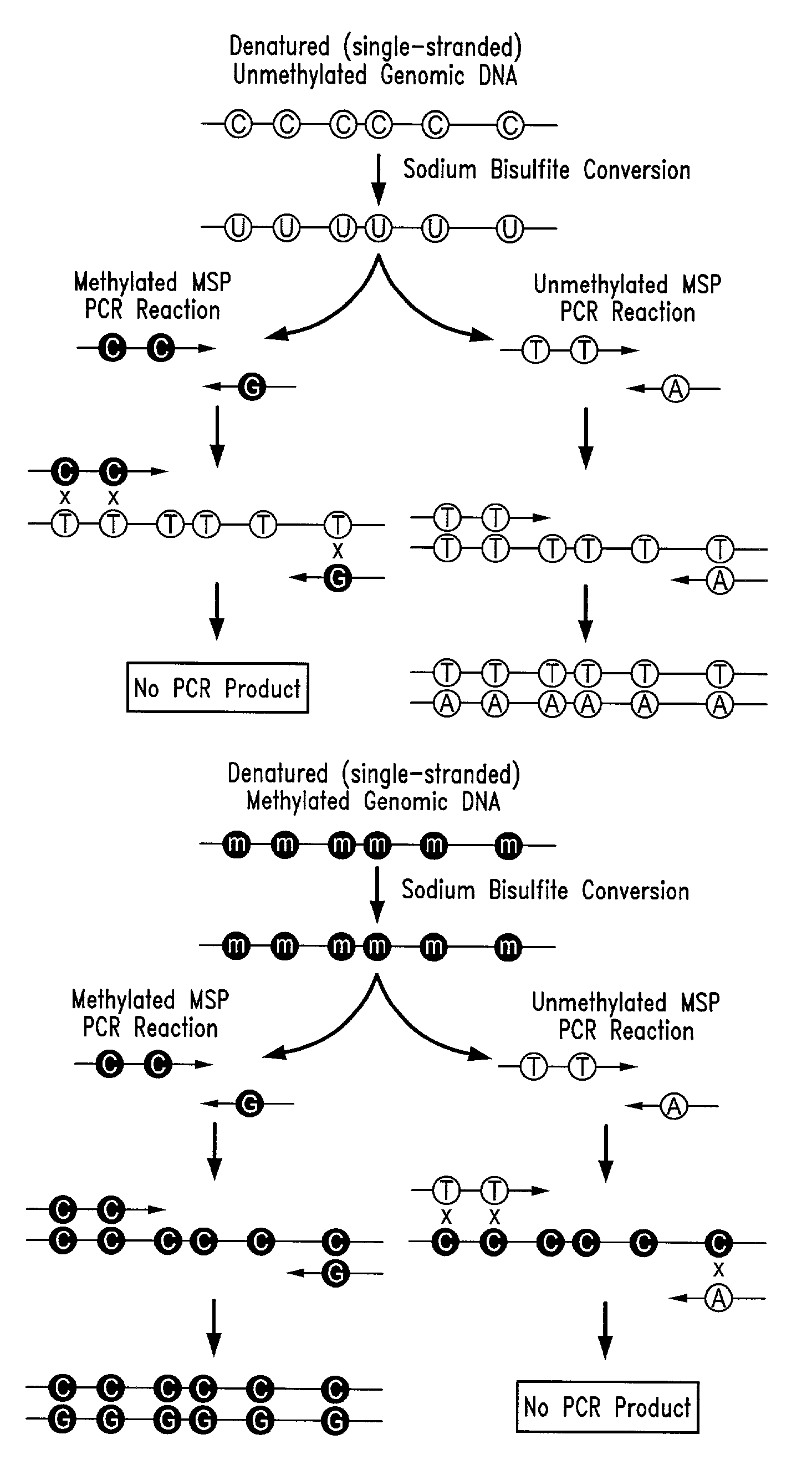
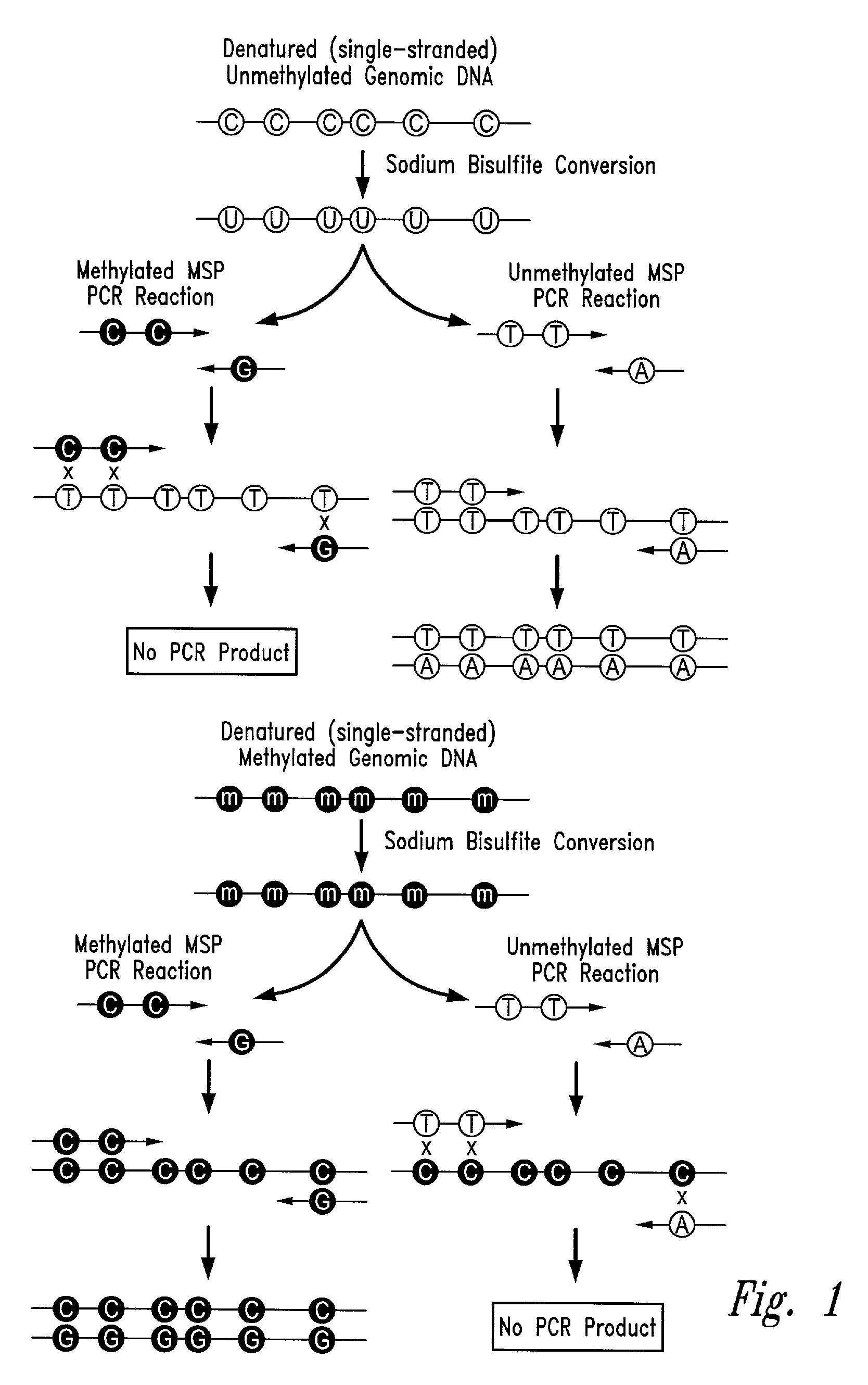
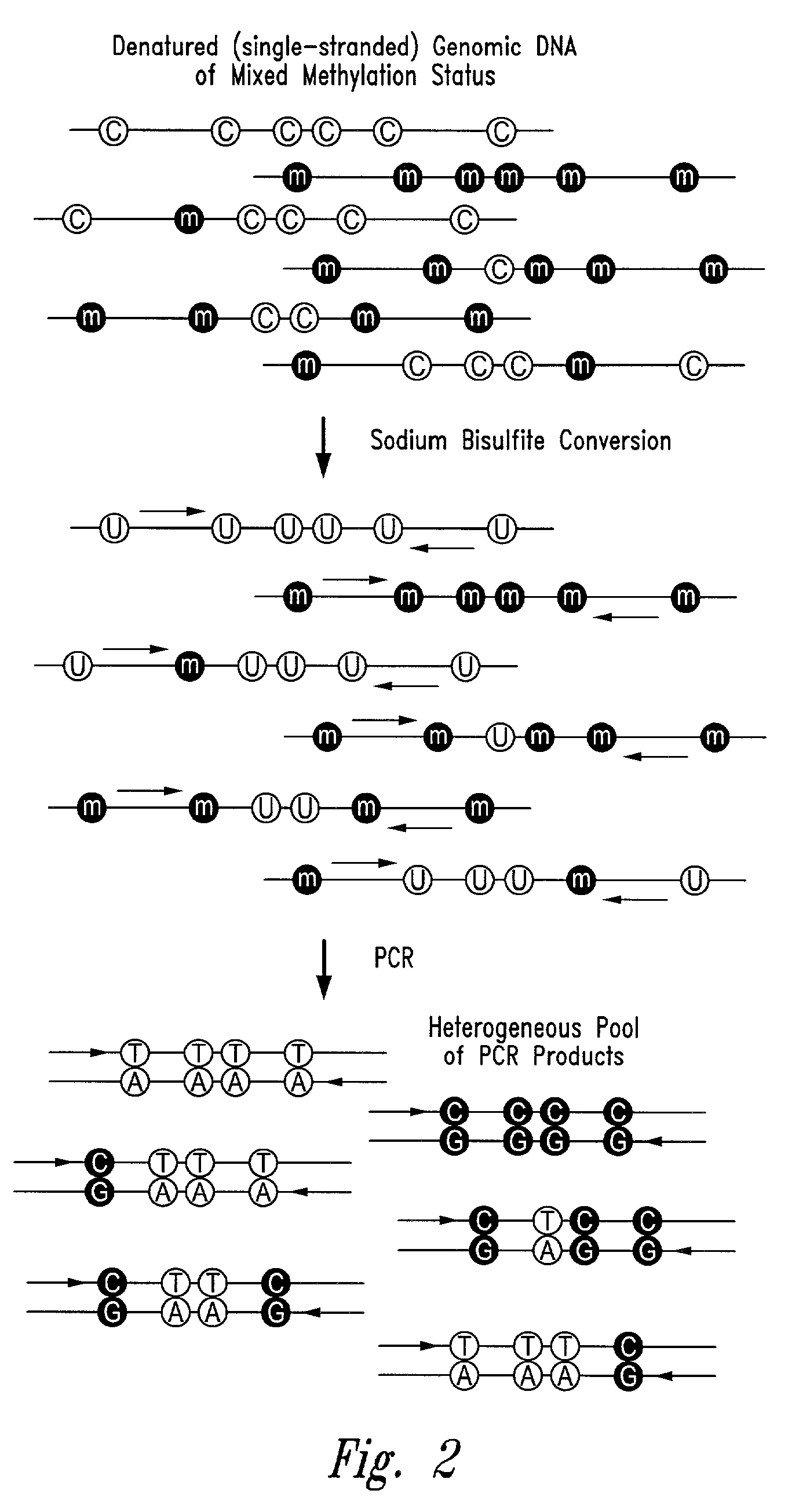
There is disclosed an improved high-throughput and quantitative process for determining methylation patterns in genomic DNA samples based on amplifying modified nucleic acid, and detecting methylated nucleic acid based on amplification-dependent displacement of specifically annealed hybridization probes. Specifically, the inventive process provides for treating genomic DNA samples with sodium bisulfite to create methylation-dependent sequence differences, followed by detection with fluorescence-based quantitative PCR techniques. The process is particularly well suited for the rapid analysis of a large number of nucleic acid samples, such as those from collections of tumor tissues.
Owner:UNIV OF SOUTHERN CALIFORNIA
Nucleic acid amplification oligonucleotides and probes to Lyme disease associated Borrelia
InactiveUS6074826AReduced thermal stabilityMaximize differenceSugar derivativesMicrobiological testing/measurementBorrelia gariniiHybridization probe
The present invention discloses hybridization assay probes, amplification primers, nucleic acid compositions and methods useful for detecting Borrelia nucleic acids. Hybridization assay probes and amplification primers that selectively detect Lyme disease-associated Borrelia and distinguish those Borrelia from Borrelia hermsii are disclosed. Other hybridization probes selectively detect Borrelia hermsii and not Lyme disease-associated Borrelia are also described.
Owner:GEN PROBE INC
Nuclease-Free Real-Time Detection of Nucleic Acids
The invention is a method for amplification and detection of nucleic acids using primers and at least one hybridization probe labeled with a first fluorescent moiety and a second moiety, capable of changing the fluorescence of said first fluorescent moiety. The method comprises the steps of effecting denaturation of said target, formation of hybrids between said primers and probe and said target and detecting the change in fluorescence of said first fluorescent moiety, upon formation of said hybrids. Reaction mixtures and kits for practicing the method of the present invention are also disclosed.
Owner:ROCHE MOLECULAR SYST INC
High-sensitivity real-time polymerase chain reaction for detection of nucleic acids
InactiveUS7252937B2Easy extractionImprove stabilitySugar derivativesMicrobiological testing/measurementClinical settingsHybridization probe
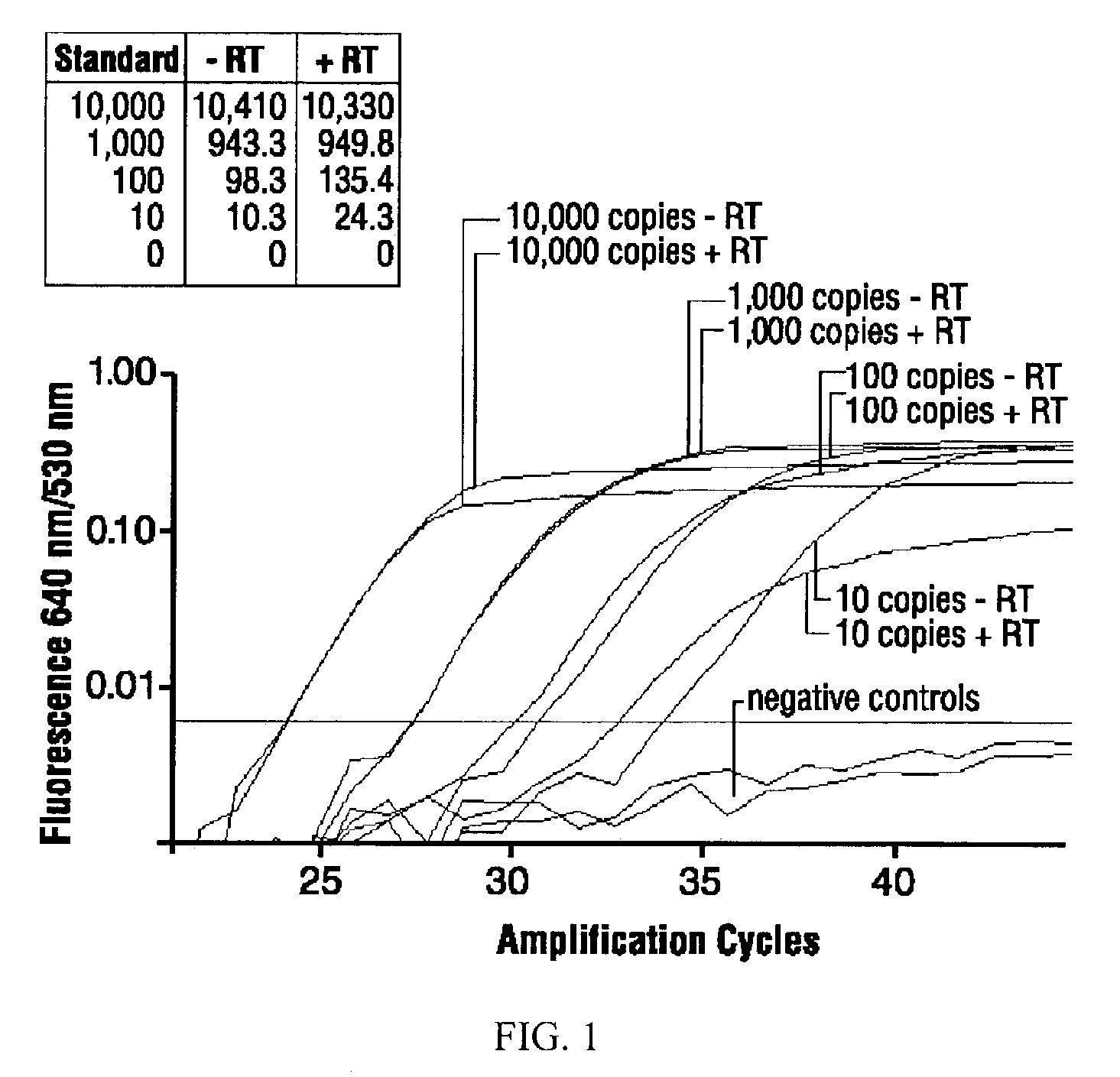
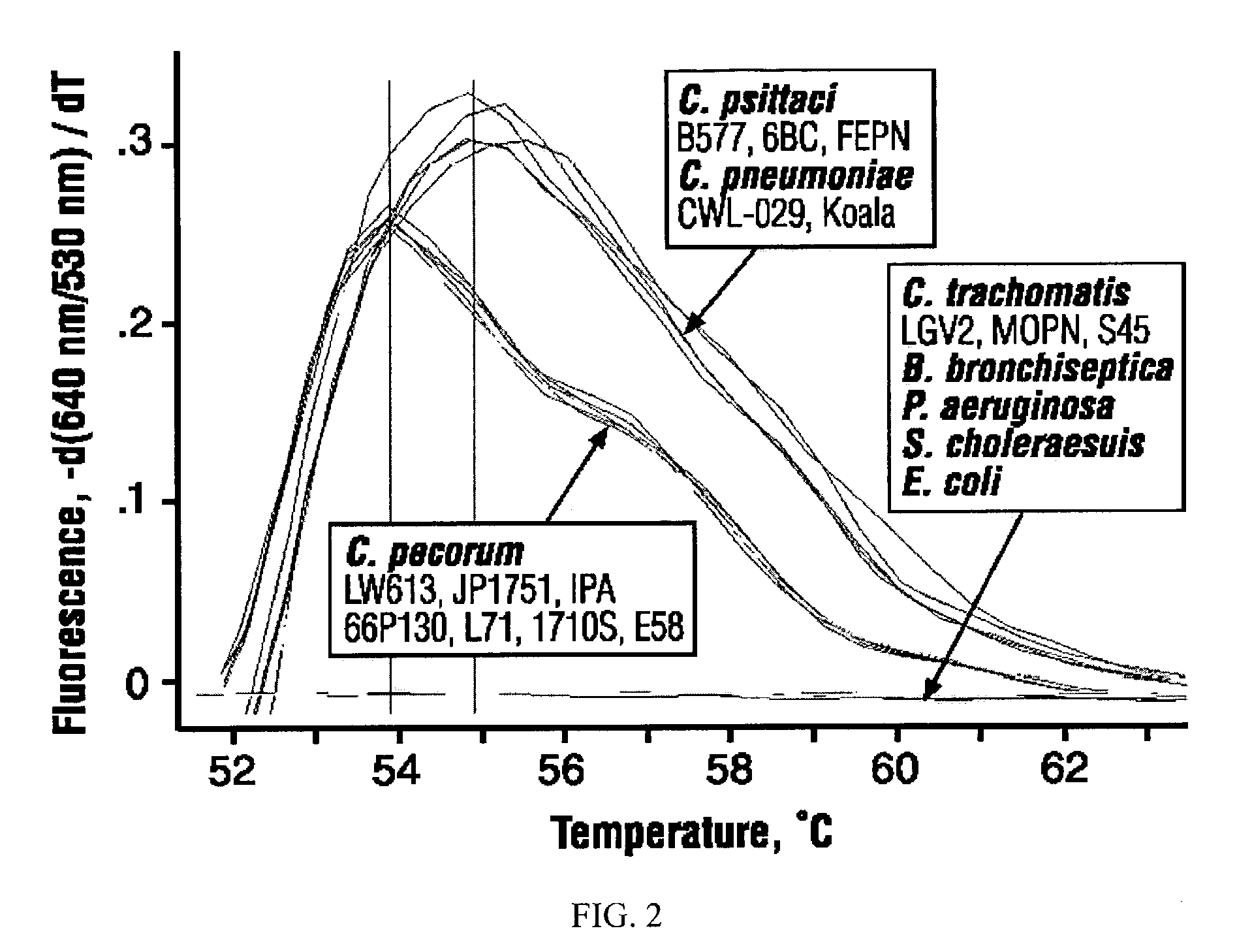
The invention features methods that are capable of detecting single target molecules in a sample input volume of, e.g., 5 μl, and of quantifying organismal, e.g., chiamydial, DNA. Desirably, these methods employ a single tube format coupled with fluorescent detection of amplicons. This approach facilitates the application of quantitative PCR (qPCR) to microbiological diagnosis in clinical settings. The invention also features primers and probes for the detection of Chlamydia. The use of specific hybridization probes with qPCR amplification provides the ability for identification of individual species or strains of microorganisms.
Owner:AUBURN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com