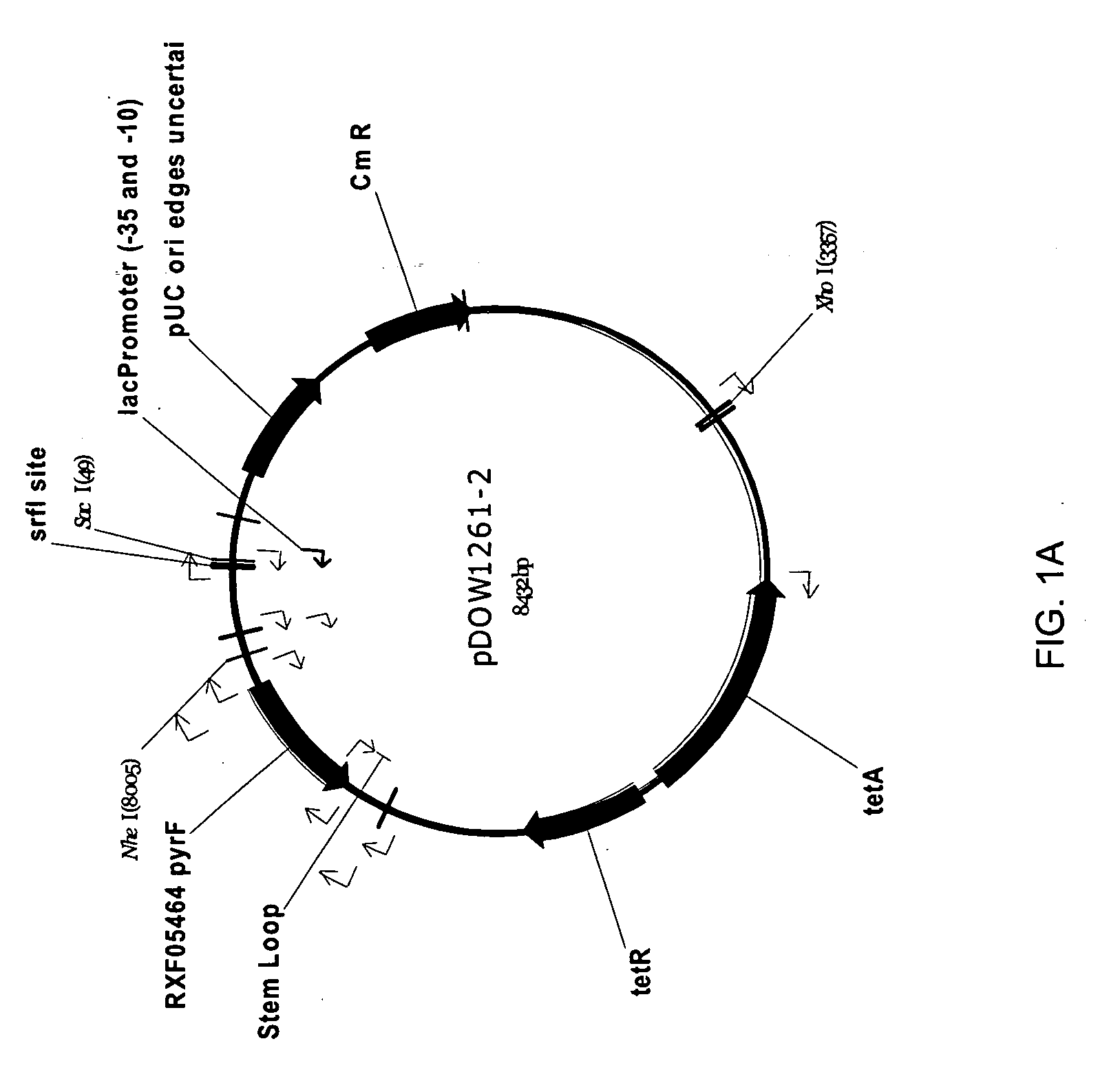
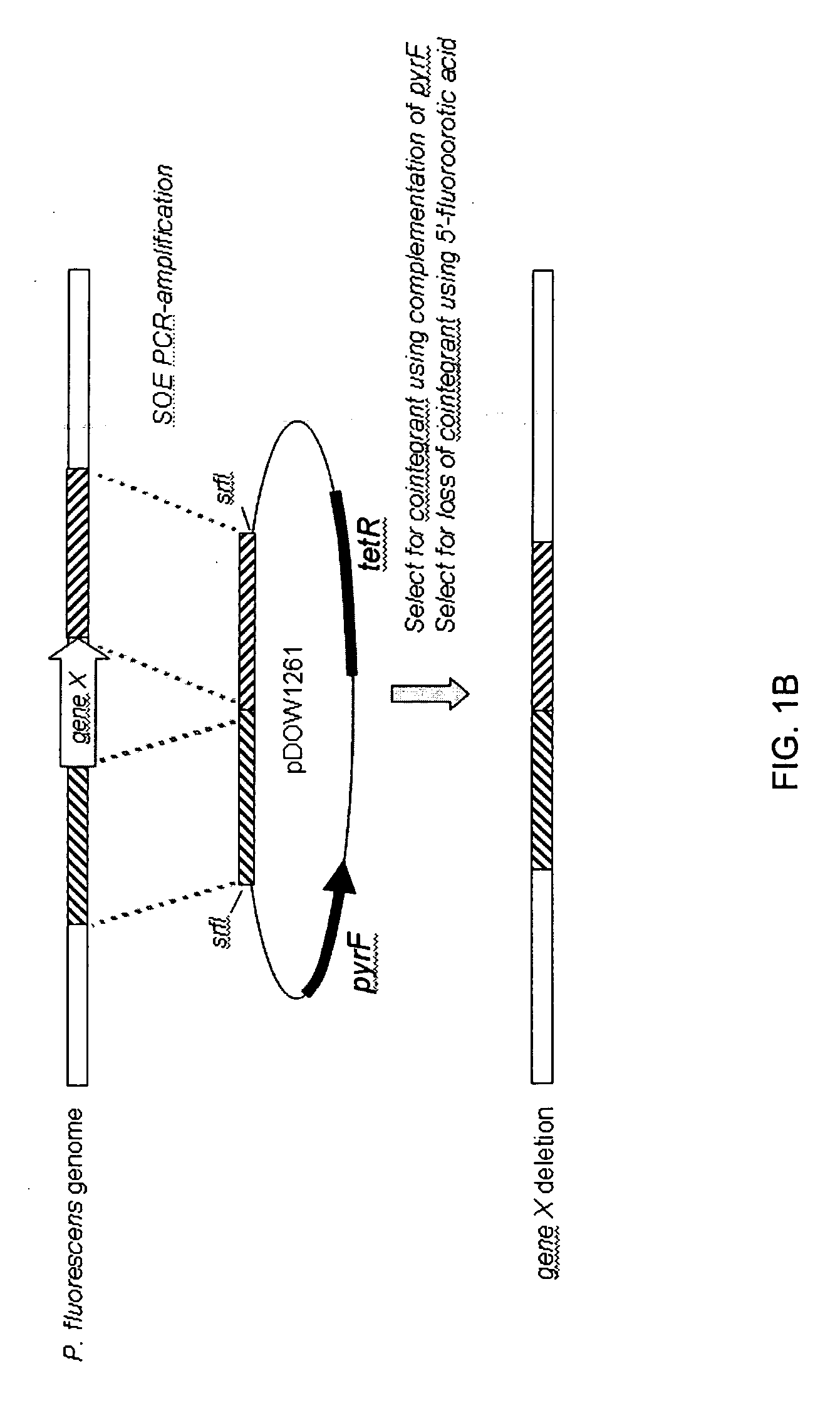
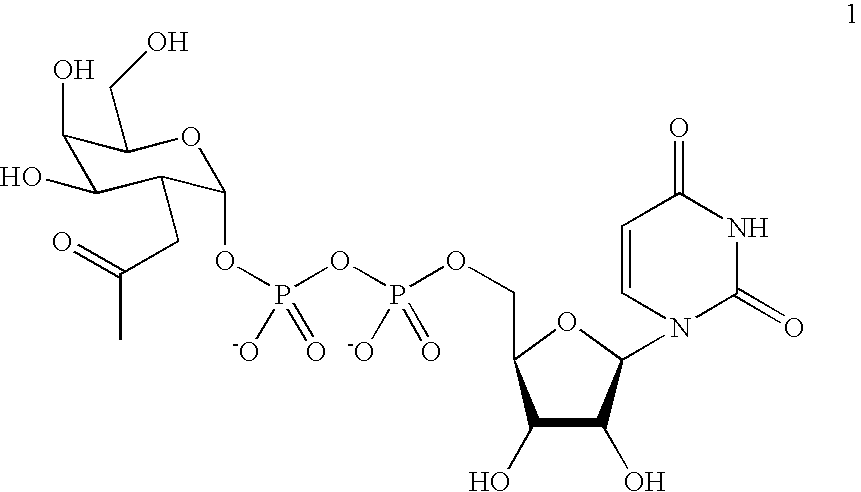
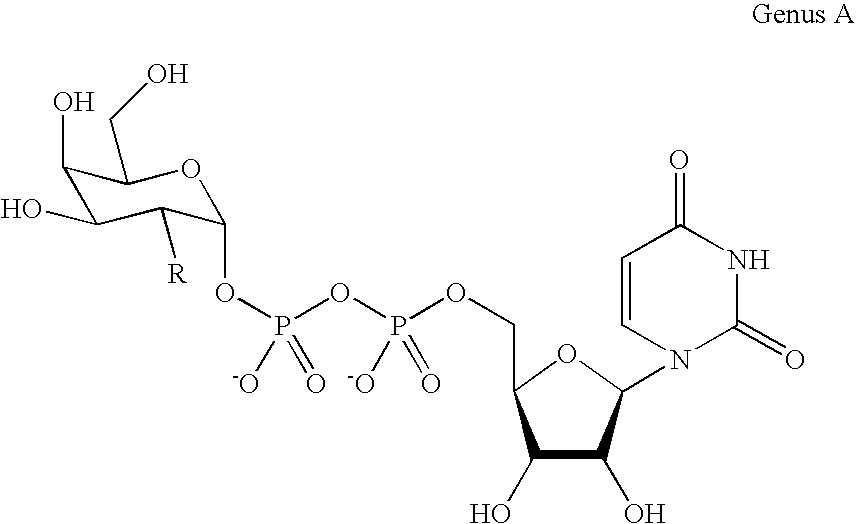
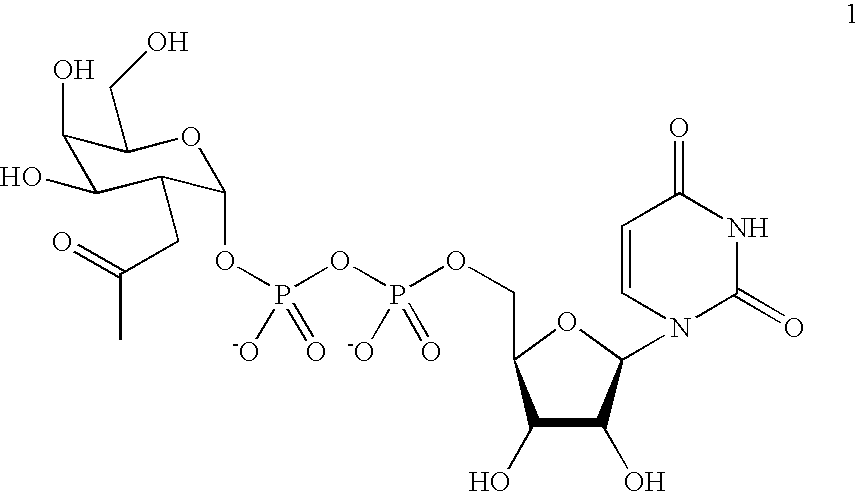
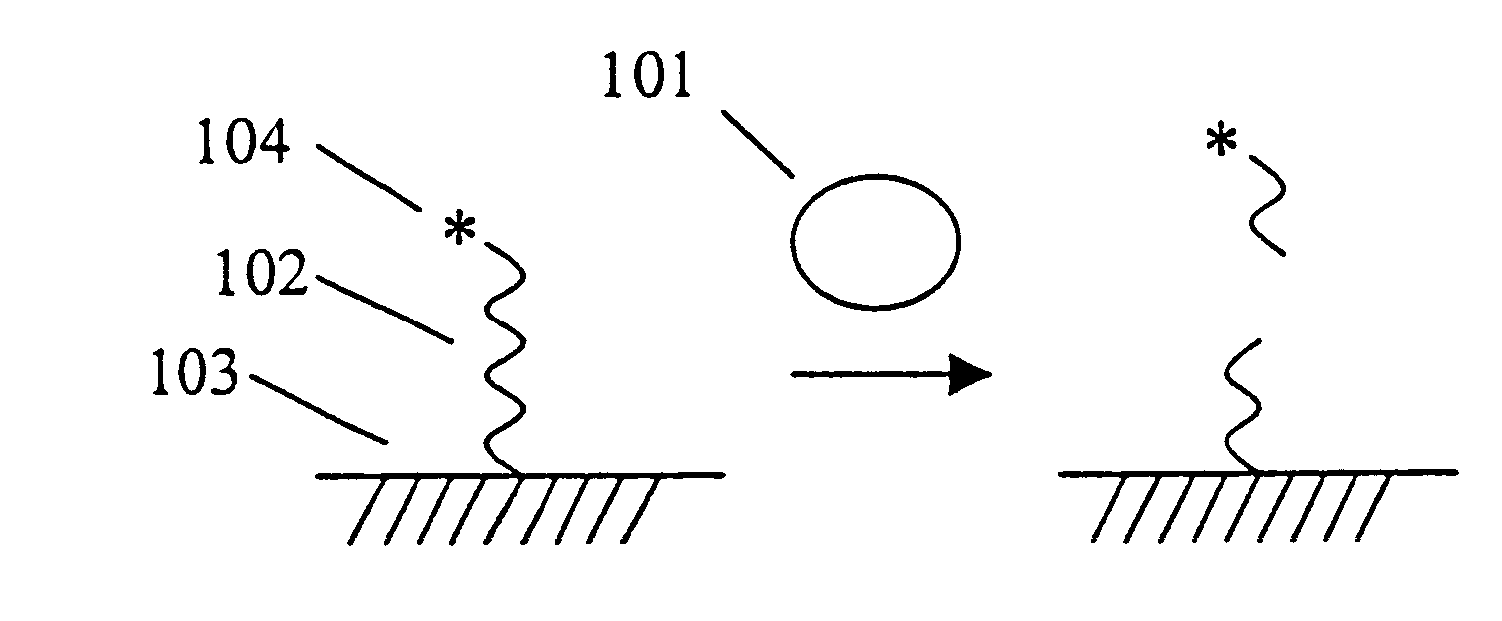
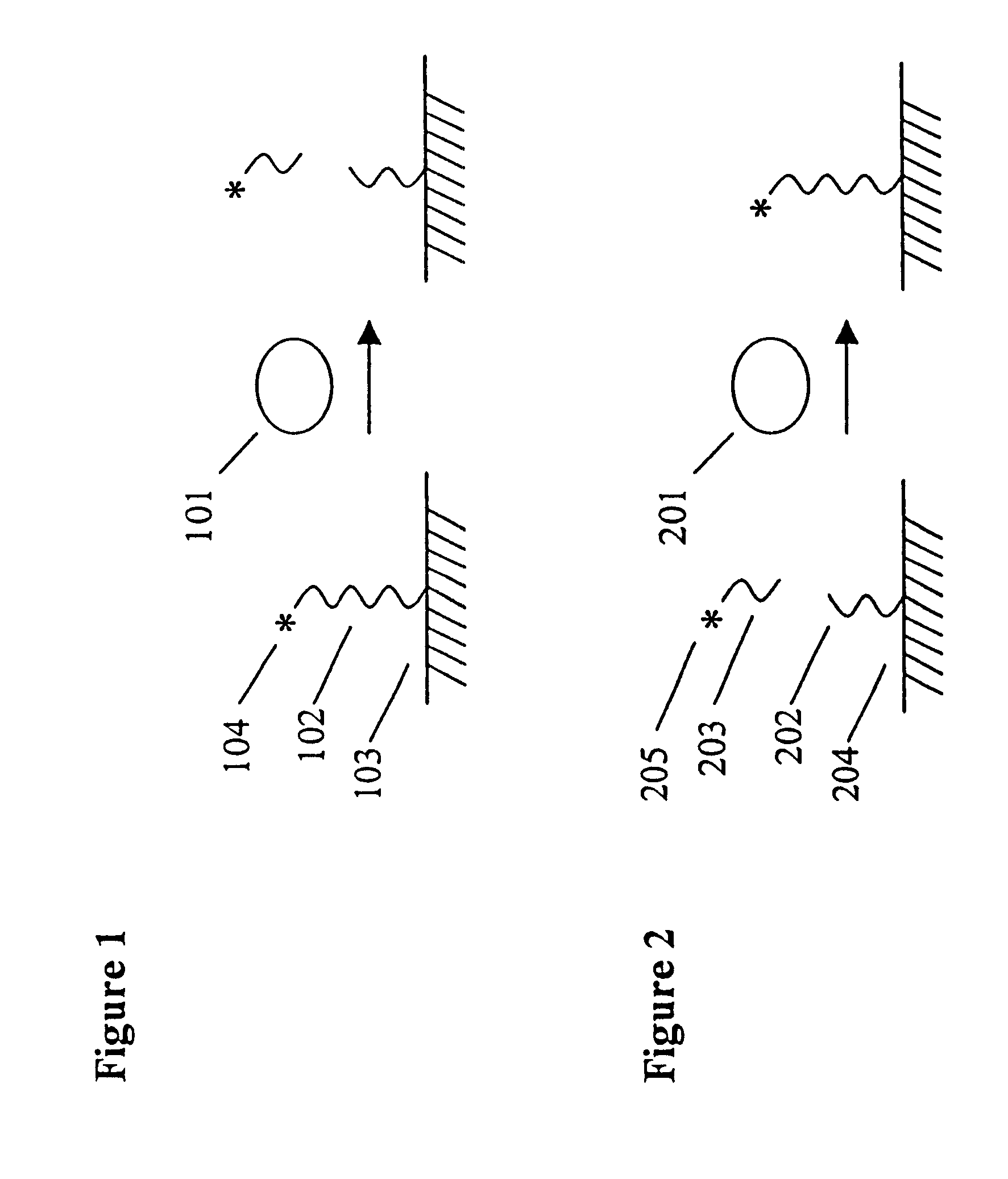
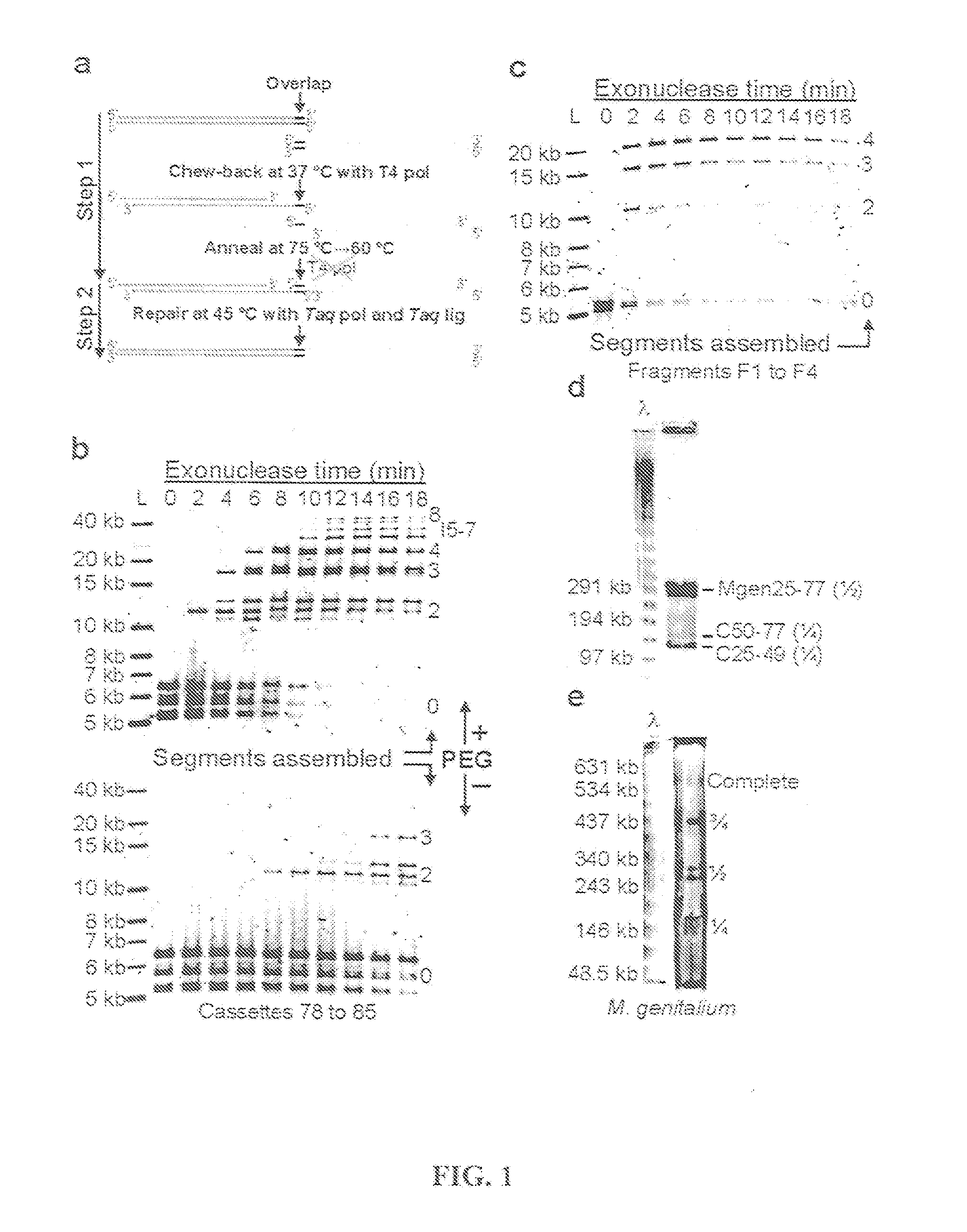Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
742 results about "Protein insertion" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
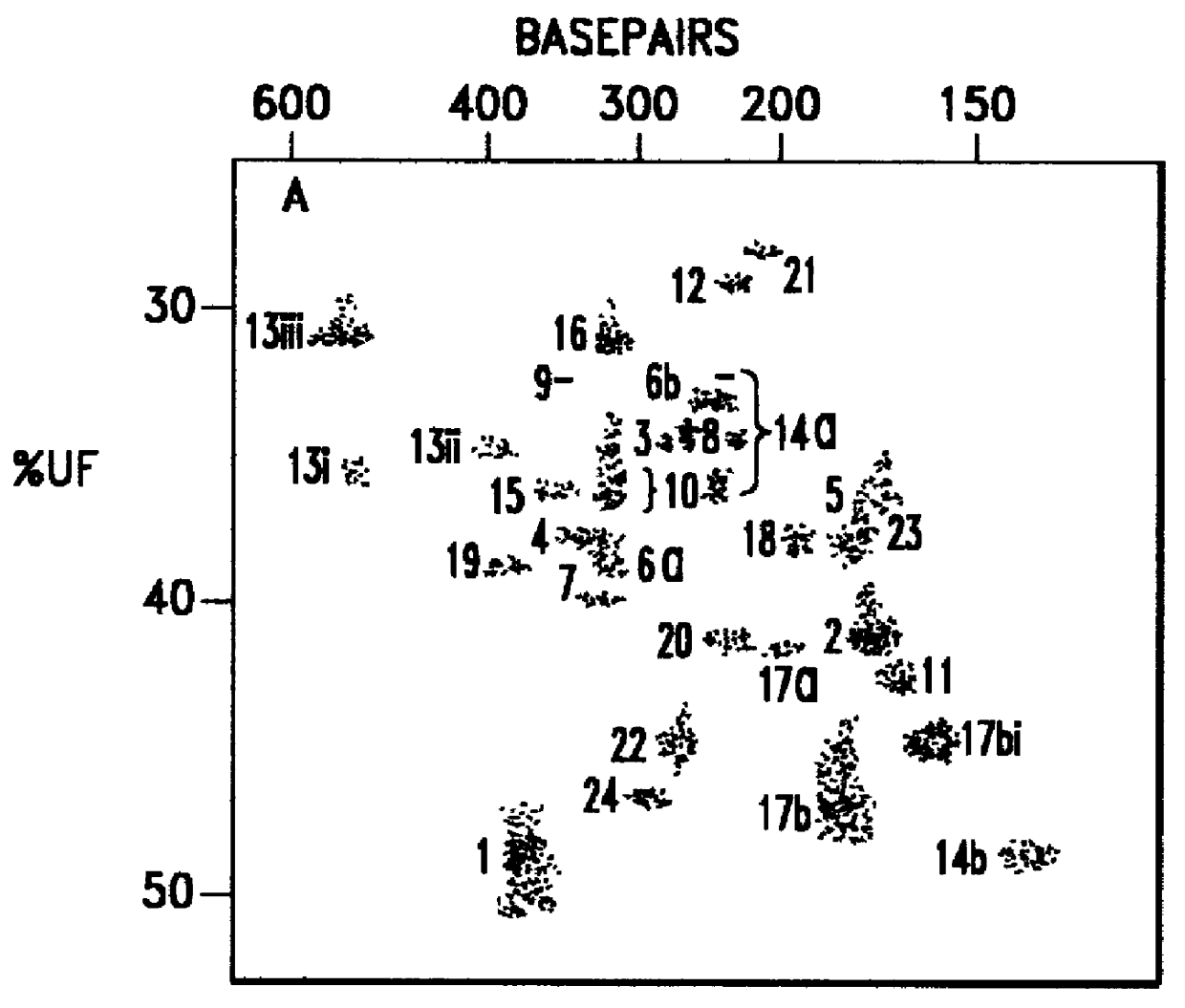
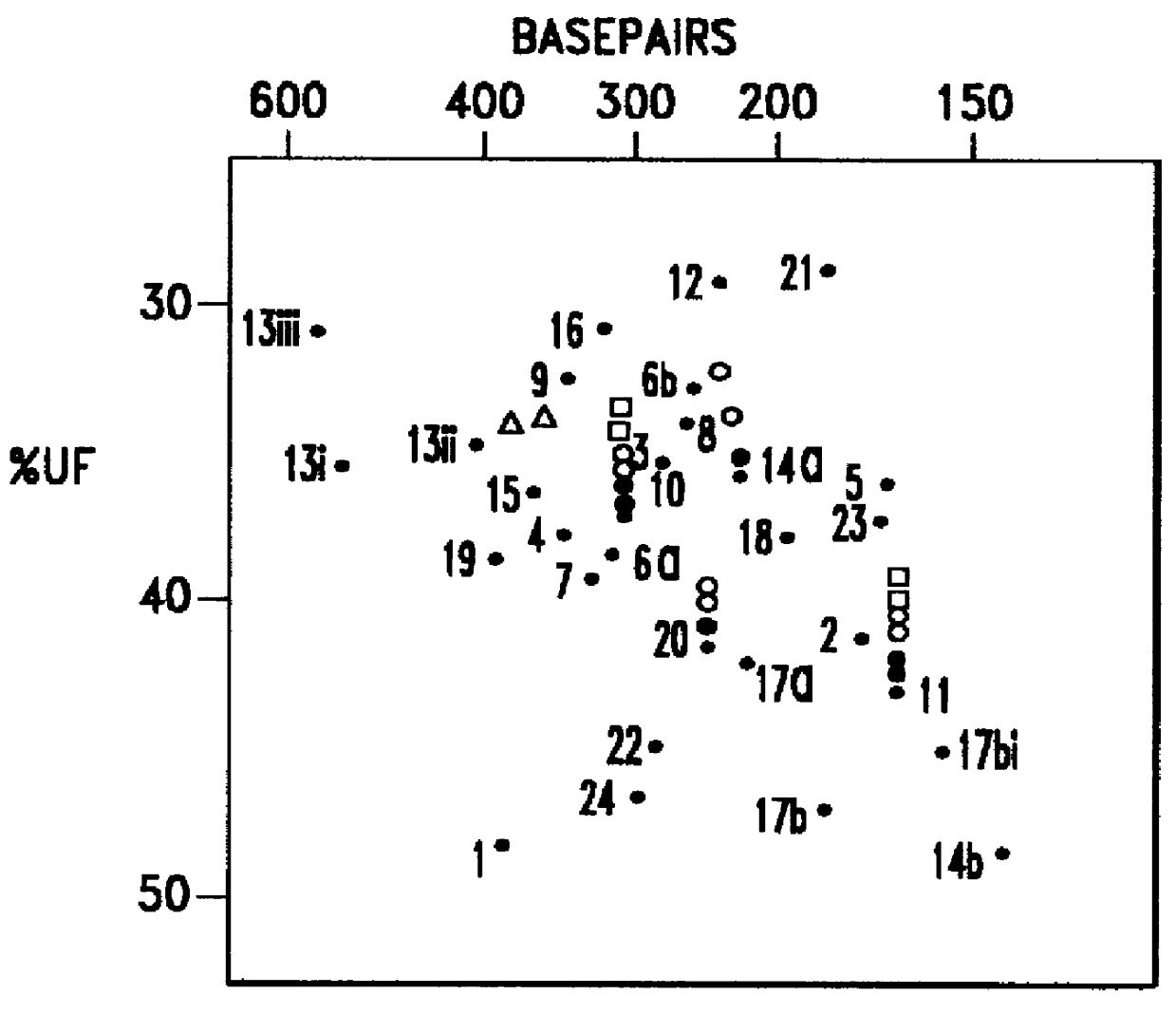
Thus, protein insertion into the membrane likely follows a two-stage process, with the ribosome driving the protein into the channel in the first stage and then it exiting laterally through the gate in the second. OmpLA – a challenge to the hypothesis. Membrane distortion induced by arginine of OmpLA.
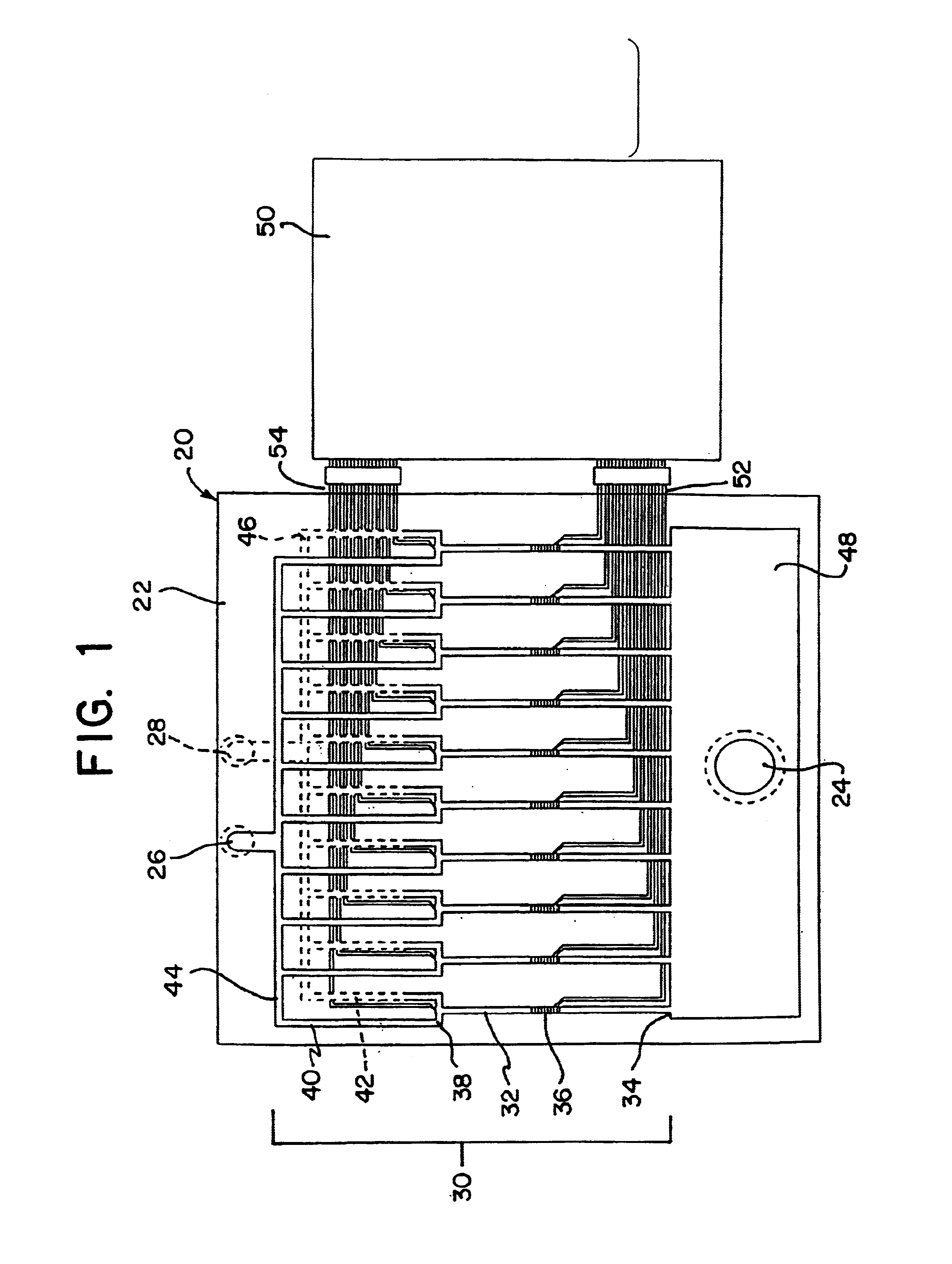
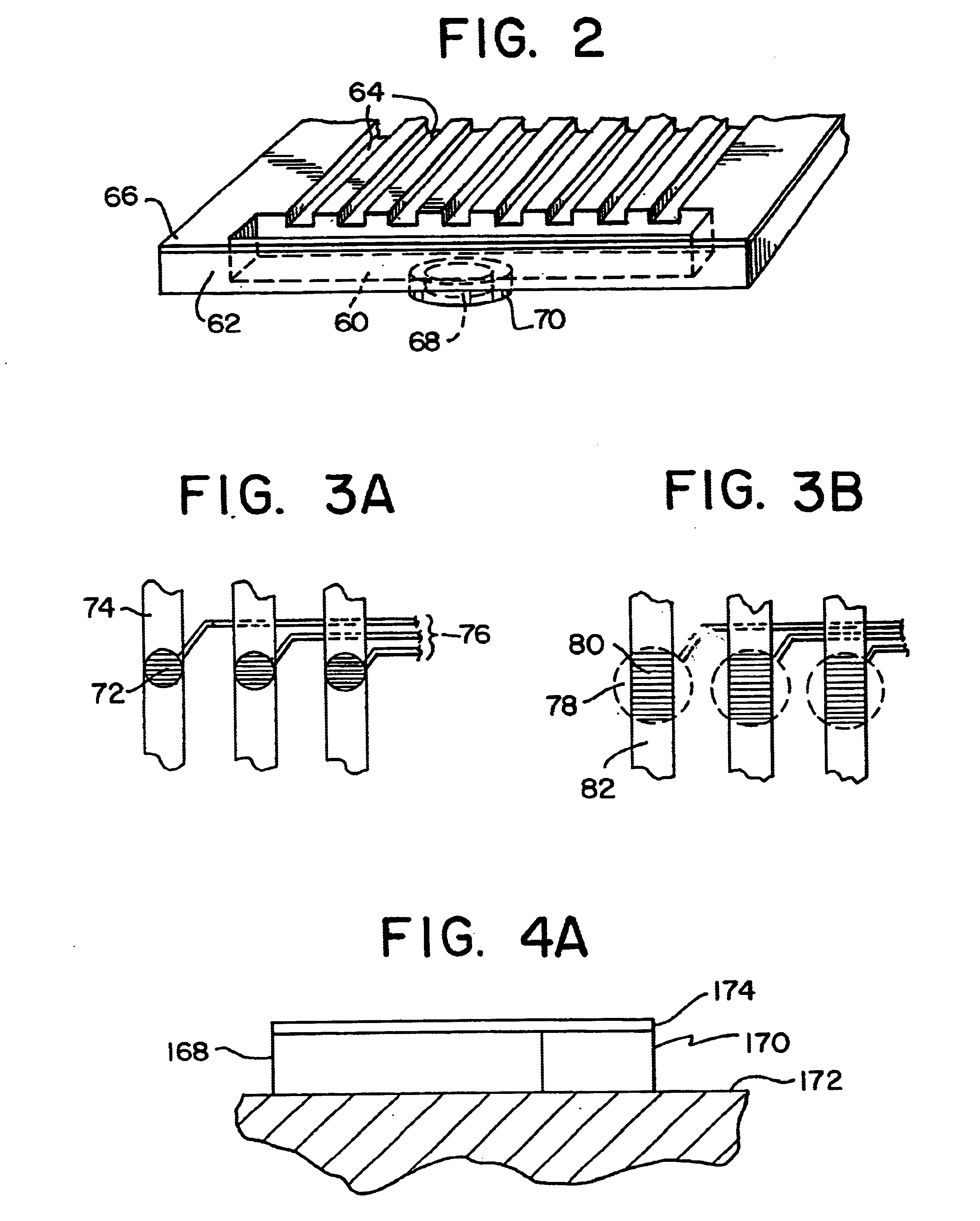
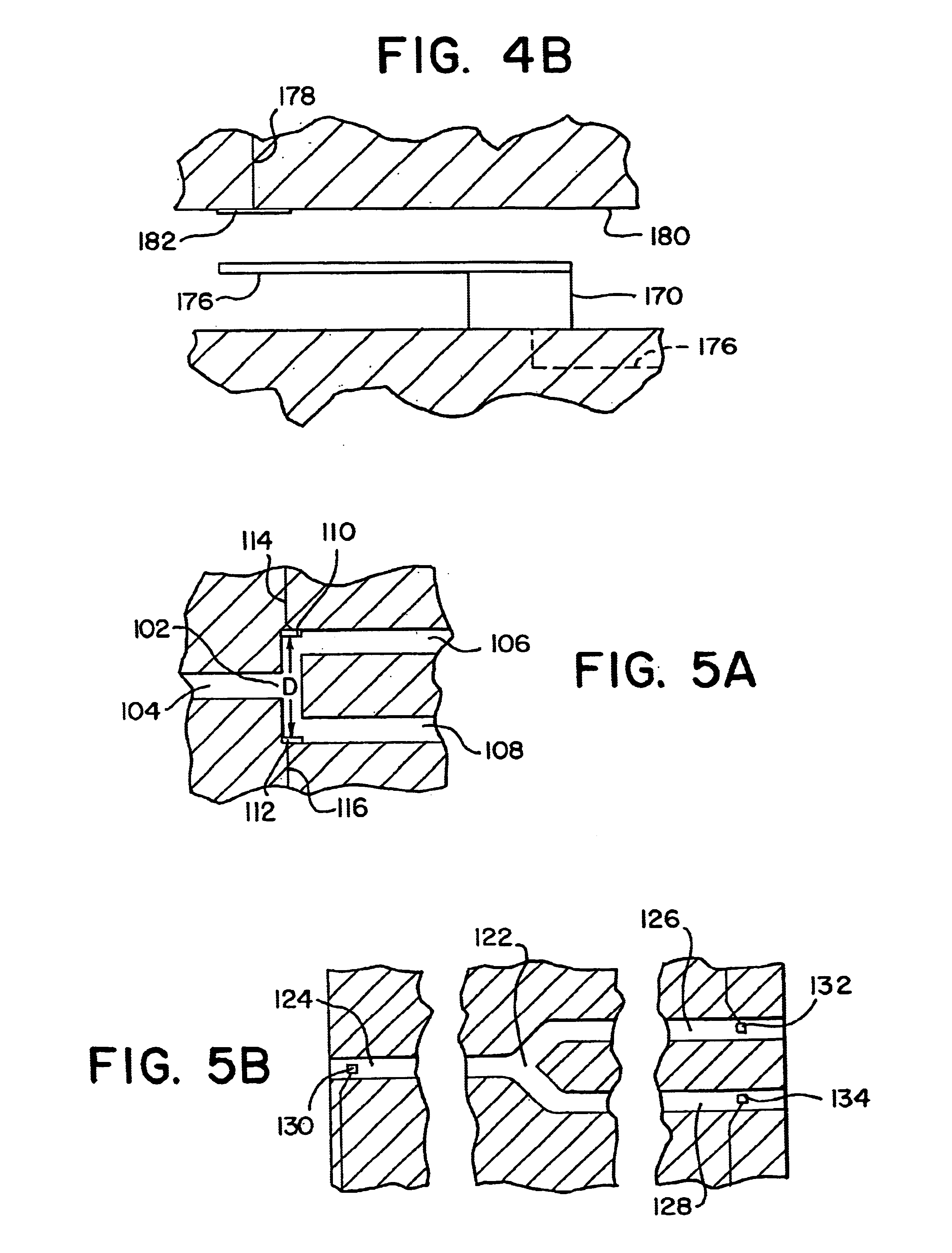
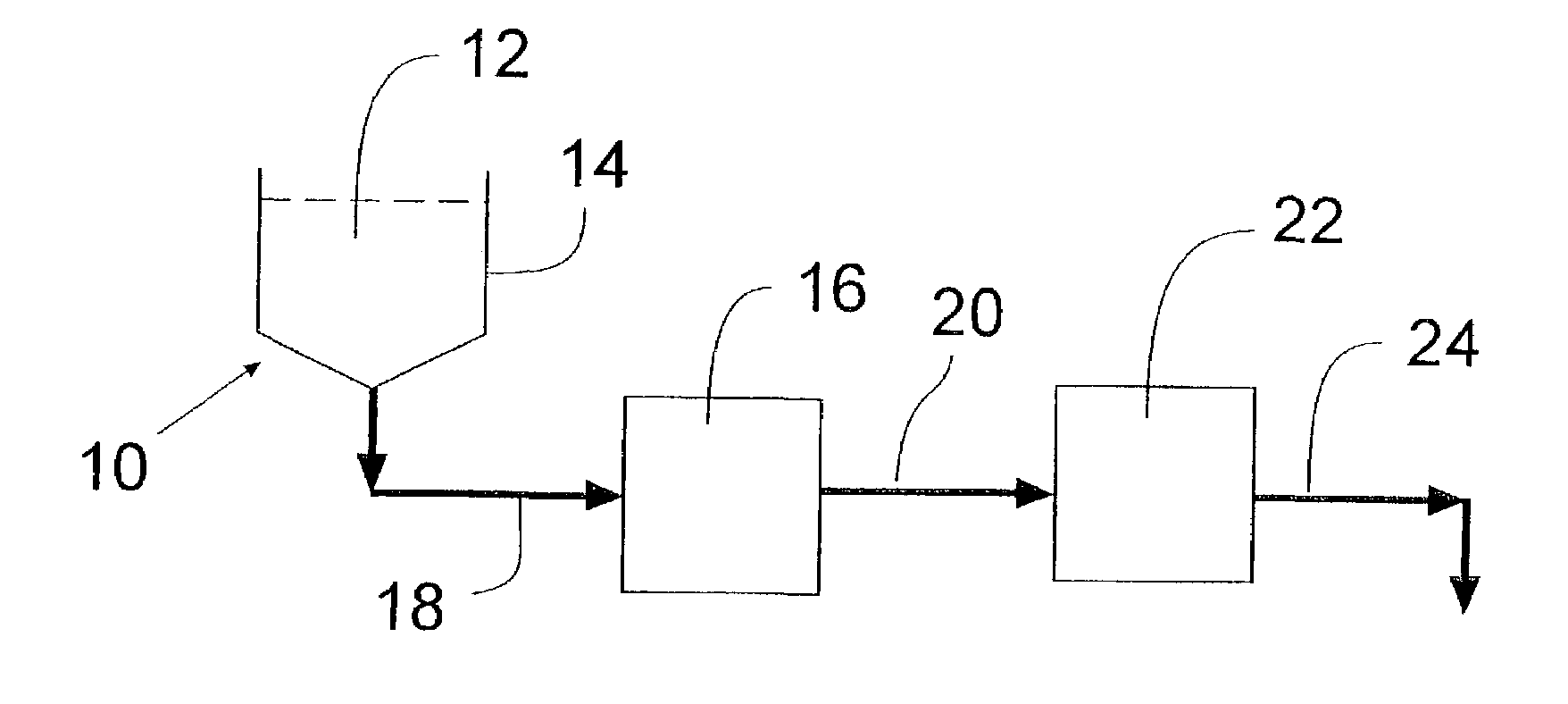
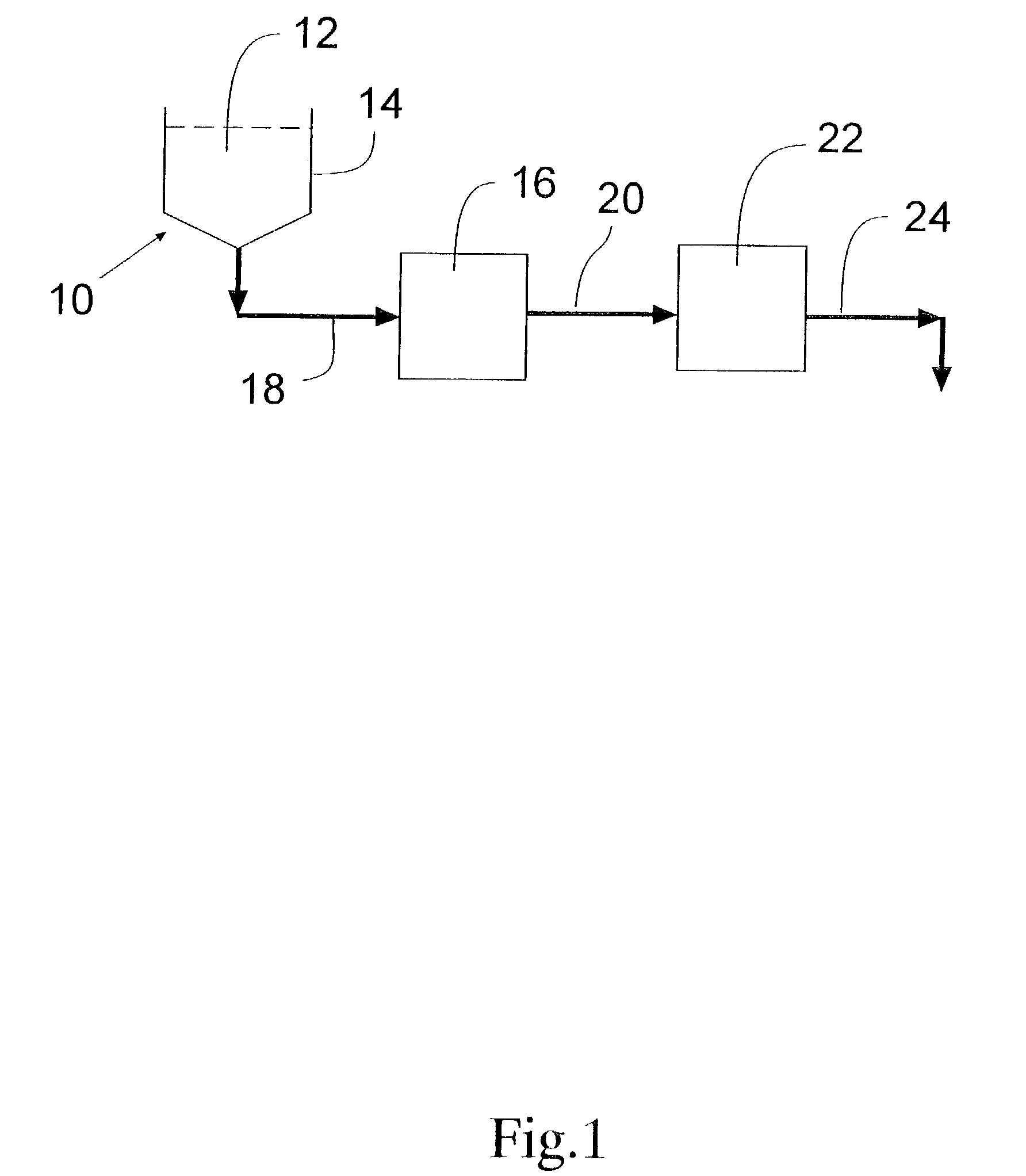
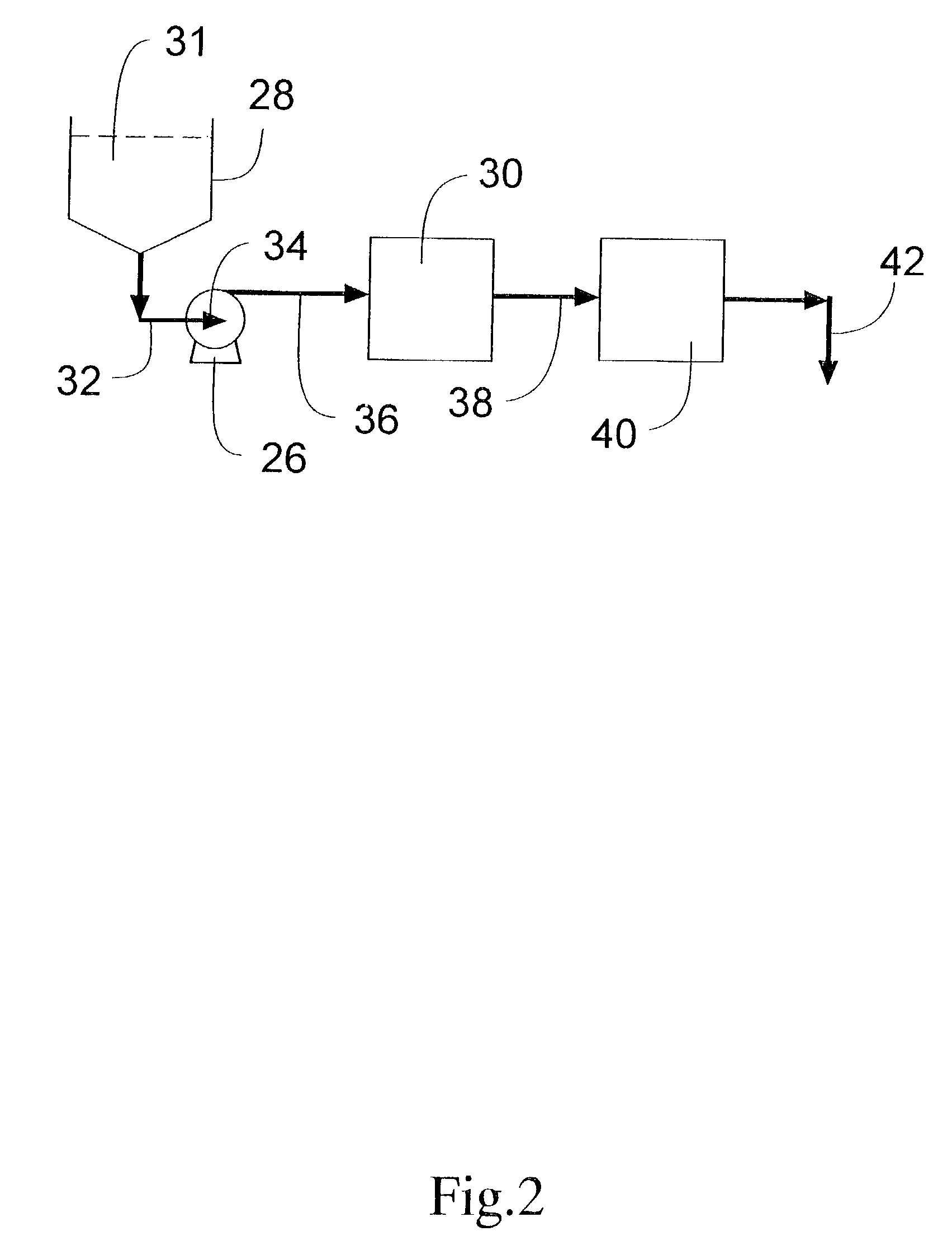
Integrated active flux microfluidic devices and methods
InactiveUS6767706B2Rapid and complete exposureQuick and accurate and inexpensive analysisBioreactor/fermenter combinationsFlow mixersAntigenHybridization probe
The invention relates to a microfabricated device for the rapid detection of DNA, proteins or other molecules associated with a particular disease. The devices and methods of the invention can be used for the simultaneous diagnosis of multiple diseases by detecting molecules (e.g. amounts of molecules), such as polynucleotides (e.g., DNA) or proteins (e.g., antibodies), by measuring the signal of a detectable reporter associated with hybridized polynucleotides or antigen / antibody complex. In the microfabricated device according to the invention, detection of the presence of molecules (i.e., polynucleotides, proteins, or antigen / antibody complexes) are correlated to a hybridization signal from an optically-detectable (e.g. fluorescent) reporter associated with the bound molecules. These hybridization signals can be detected by any suitable means, for example optical, and can be stored for example in a computer as a representation of the presence of a particular gene. Hybridization probes can be immobilized on a substrate that forms part of or is exposed to a channel or channels of the device that form a closed loop, for circulation of sample to actively contact complementary probes. Universal chips according to the invention can be fabricated not only with DNA but also with other molecules such as RNA, proteins, peptide nucleic acid (PNA) and polyamide molecules.
Owner:CALIFORNIA INST OF TECH
Immunoassay device with immuno-reference electrode
InactiveUS7723099B2Increase ionic strengthReduce distractionsBioreactor/fermenter combinationsBiological substance pretreatmentsProtein insertionNon specific
An electrochemical immunosensor system with reduced interference, comprising: a first immunosensor that generates an electrochemical signal based on the formation of a sandwich between an immobilized antibody, a target analyte and a labeled antibody, wherein a portion of the signal arises from non-specific binding of the labeled antibody in the region of the first immunosensor, anda second immunosensor that acts as an immuno-reference sensor and generates a signal that is the same as or predictably related to the degree of non-specific binding which occurs in the region of the first immunosensor, and has an immunocomplex between an immobilized antibody and an endogenous or exogenous protein that is in the sample and that is not the target analyte.
Owner:ABBOTT POINT CARE
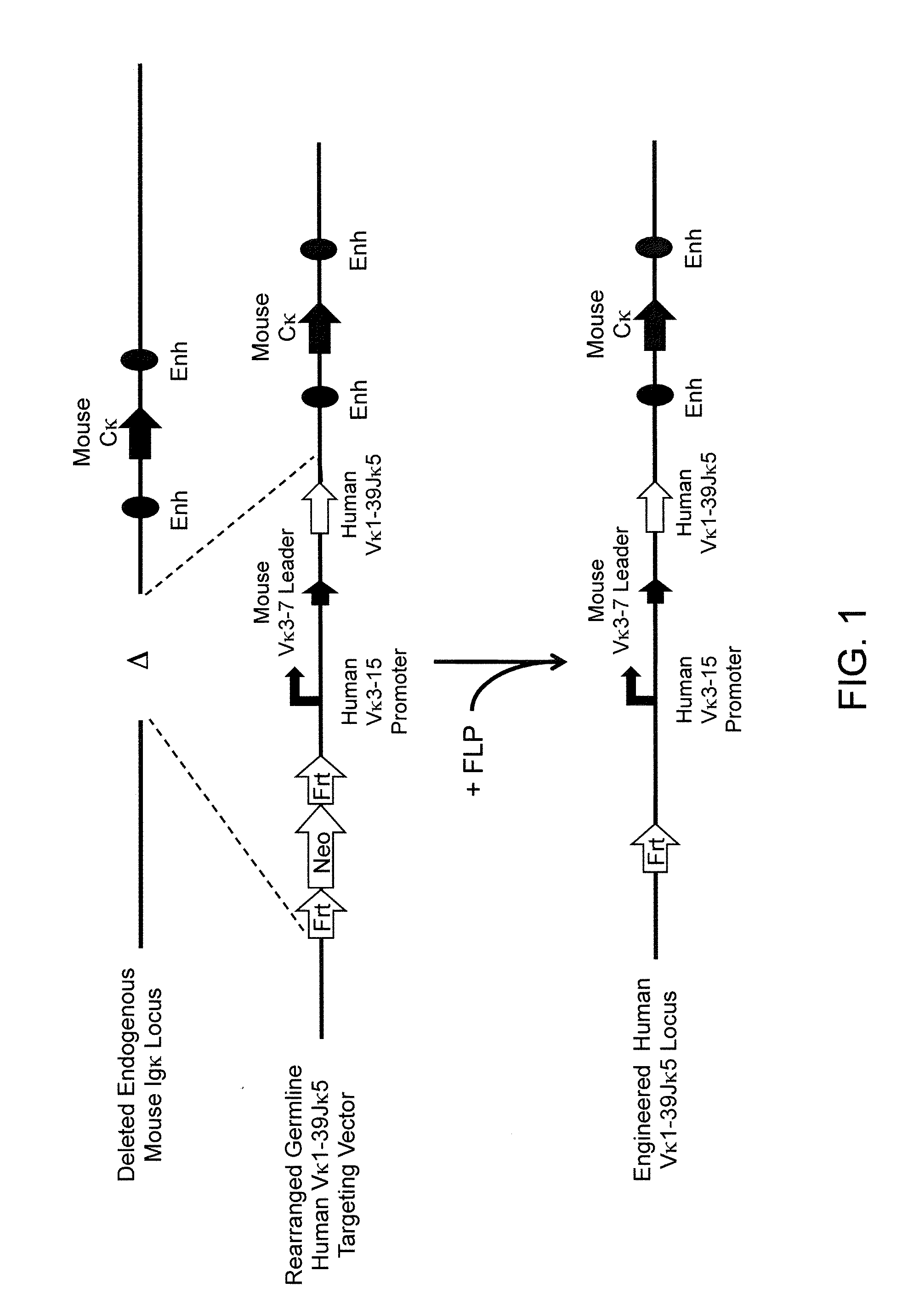
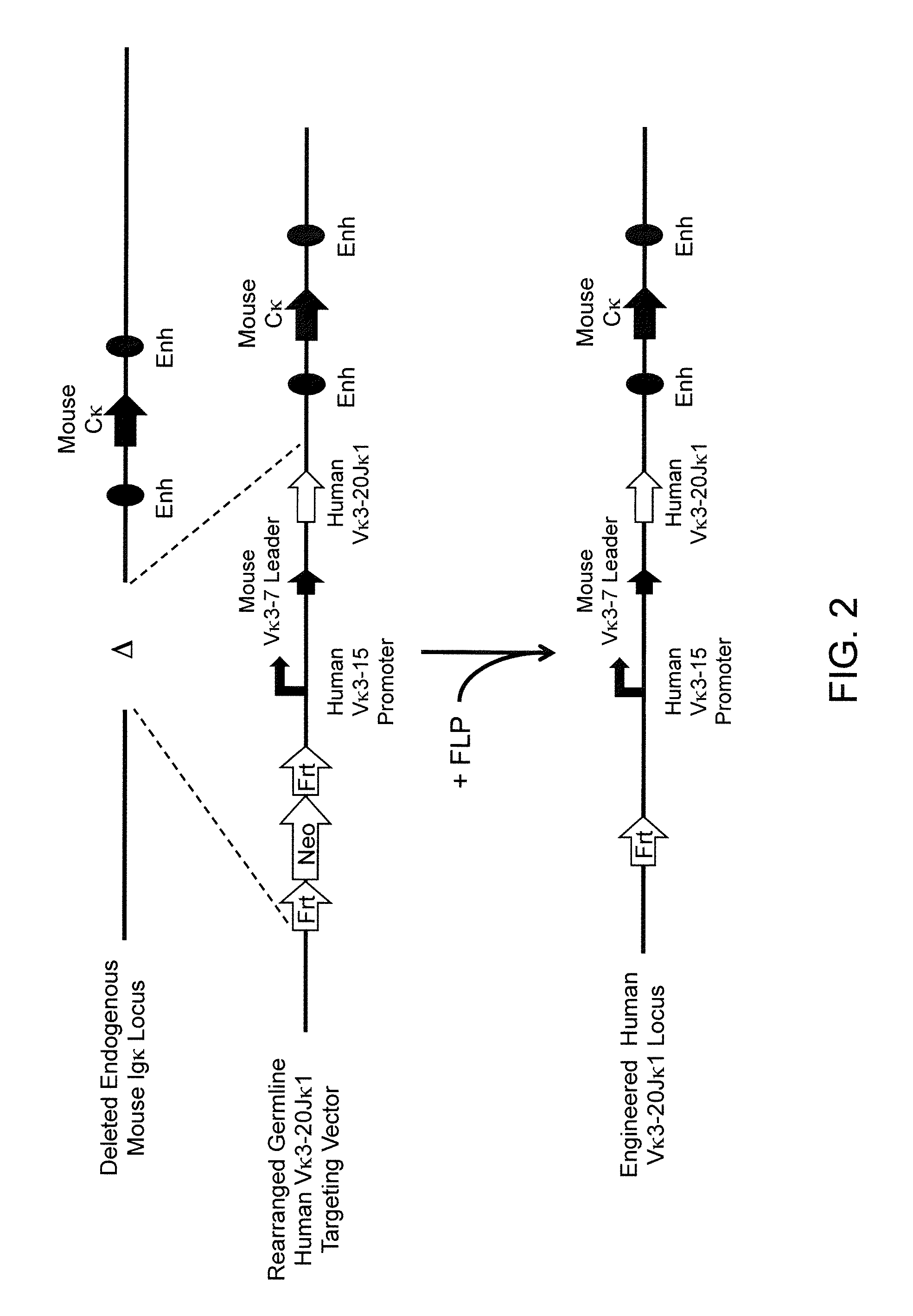
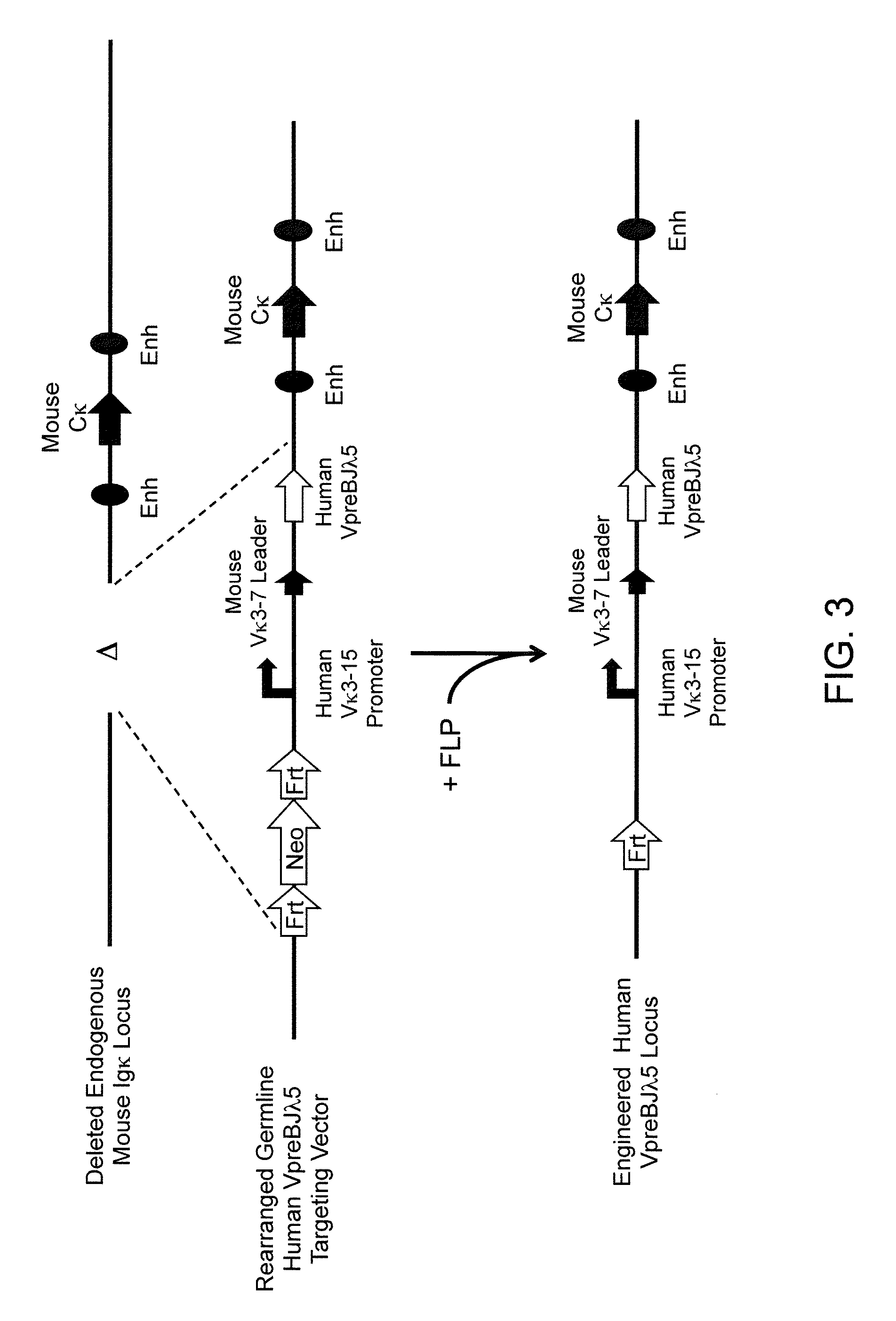
Methods For Making Fully Human Bispecific Antibodies Using A Common Light Chain
InactiveUS20130045492A1Reduce in quantitySimple methodAntibody mimetics/scaffoldsImmunoglobulins against cell receptors/antigens/surface-determinantsEpitopeProtein insertion
A genetically modified mouse is provided, wherein the mouse expresses an immunoglobulin light chain repertoire characterized by a limited number of light chain variable domains. Mice are provided that express just one or a few immunoglobulin light chain variable domains from a limited repertoire in their germline. Methods for making bispecific antibodies having universal light chains using mice as described herein, including human light chain variable regions, are provided. Methods for making human variable regions suitable for use in multispecific binding proteins, e.g., bispecific antibodies, and host cells are provided. Bispecific antibodies capable of binding first and second antigens are provided, wherein the first and second antigens are separate epitopes of a single protein or separate epitopes on two different proteins are provided.
Owner:REGENERON PHARM INC
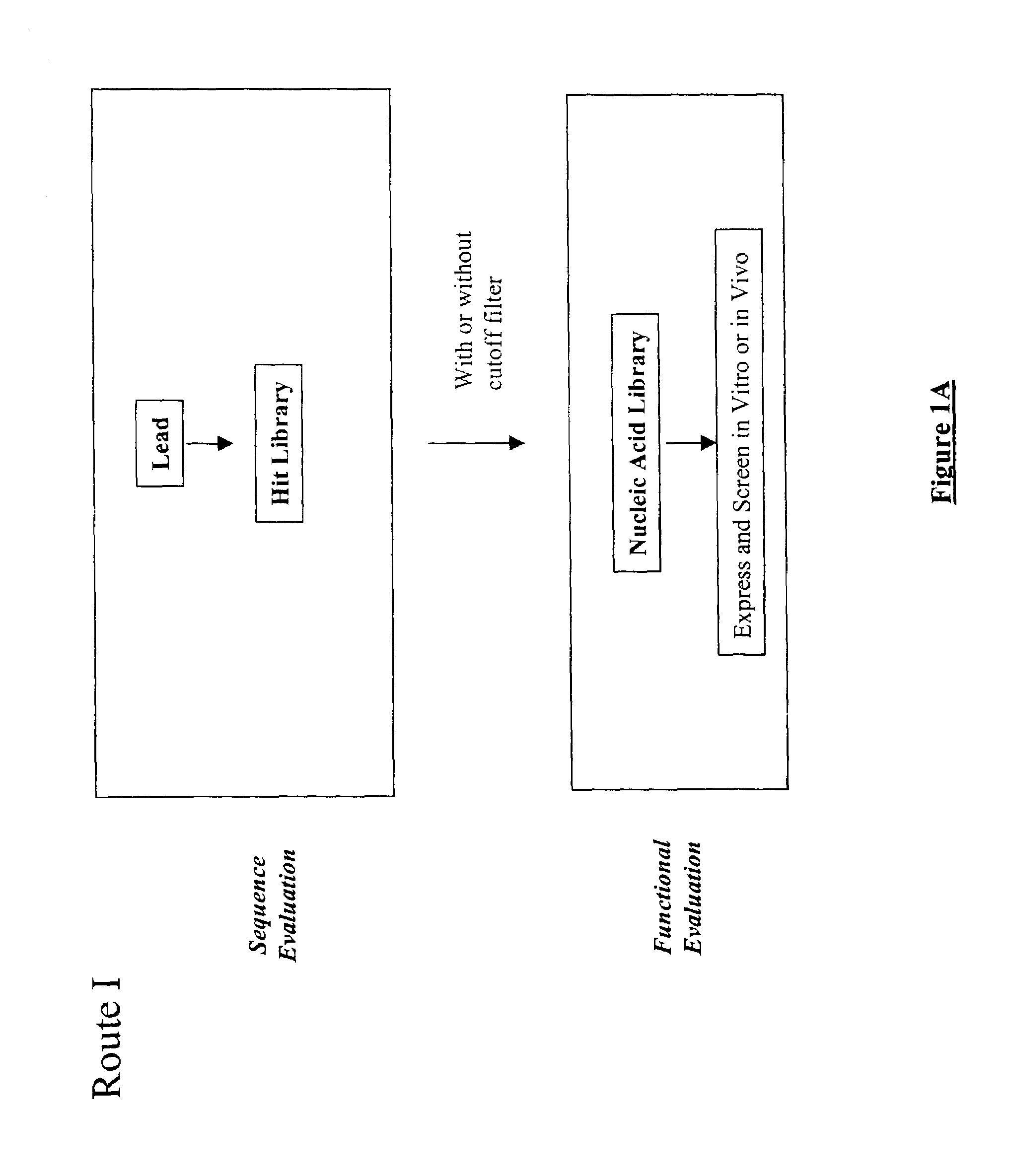
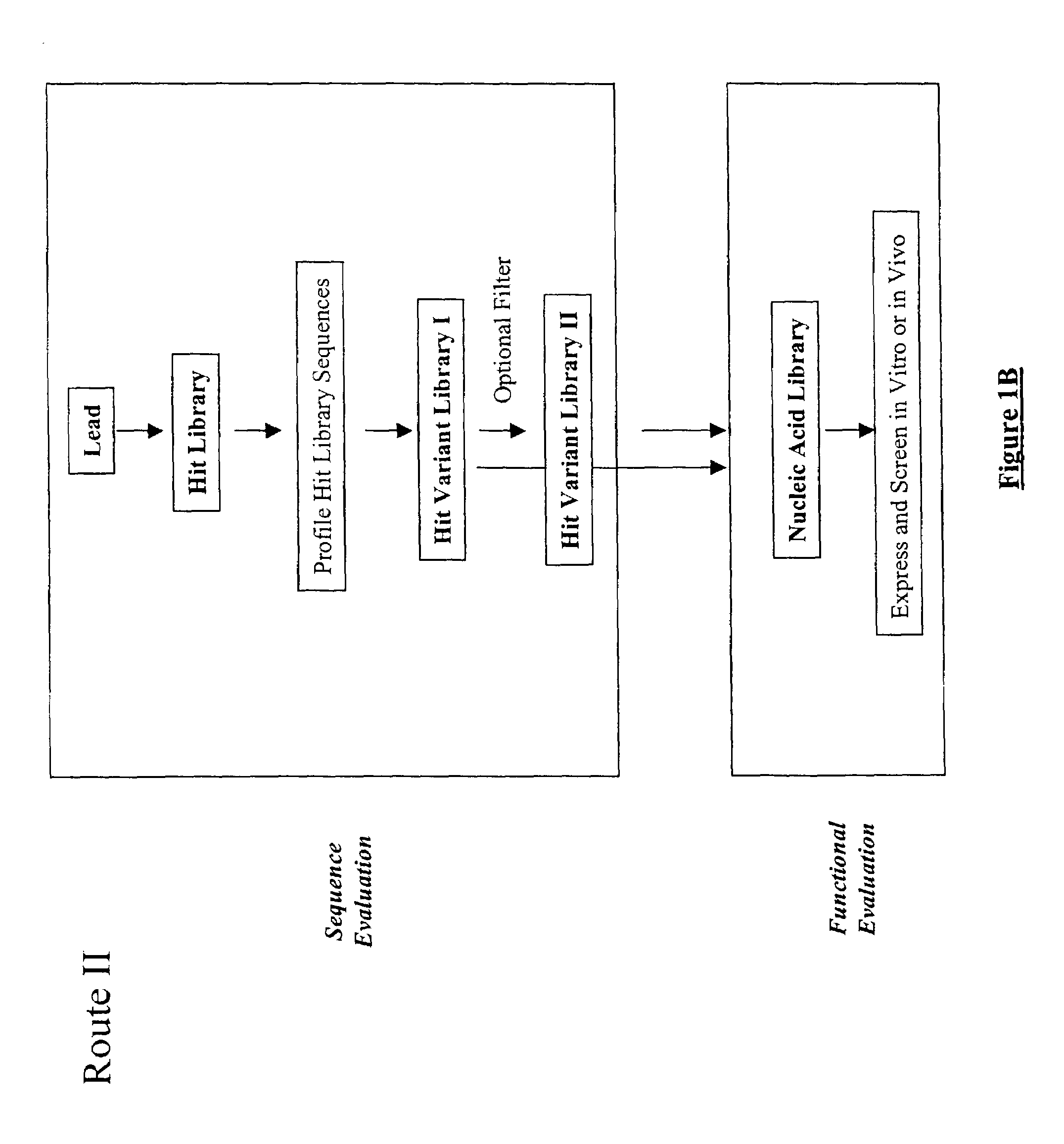
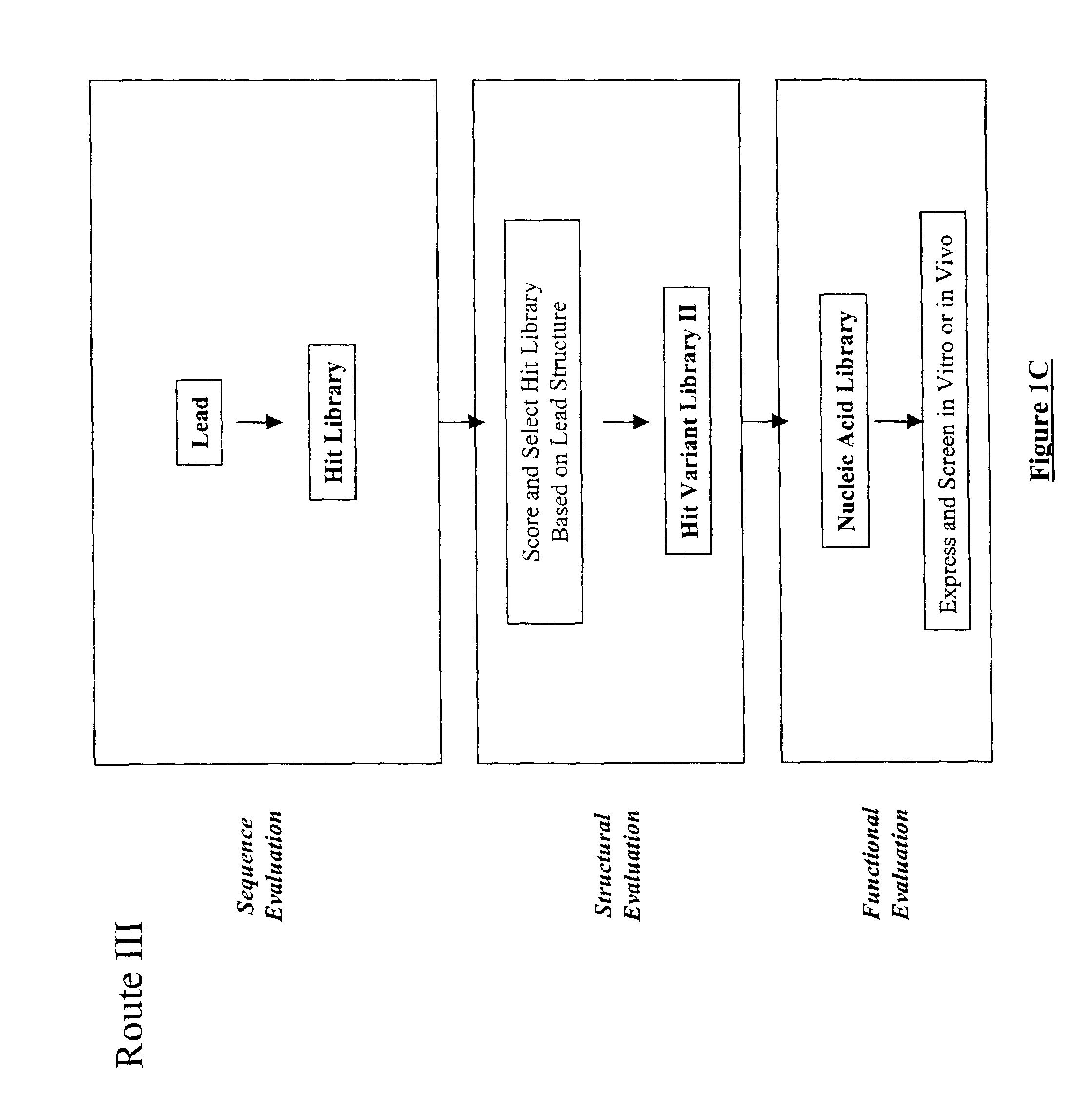
Structure-based selection and affinity maturation of antibody library
InactiveUS7117096B2High affinityImprove throughputPeptide librariesPeptide/protein ingredientsProtein insertionIn vivo
The present invention provides a structure-based methodology for efficiently generating and screening protein libraries for optimized proteins with desirable biological functions, such as antibodies with high binding affinity and low immunogenicity in humans. In one embodiment, a method is provided for constructing a library of antibody sequences based on a three dimensional structure of a lead antibody. The method comprises: providing an amino acid sequence of the variable region of the heavy chain (VH) or light chain (VL) of a lead antibody, the lead antibody having a known three dimensional structure which is defined as a lead structural template; identifying the amino acid sequences in the CDRs of the lead antibody; selecting one of the CDRs in the VH or VL region of the lead antibody; providing an amino acid sequence that comprises at least 3 consecutive amino acid residues in the selected CDR, the selected amino acid sequence being a lead sequence; comparing the lead sequence profile with a plurality of tester protein sequences; selecting from the plurality of tester protein sequences at least two peptide segments that have at least 10% sequence identity with lead sequence, the selected peptide segments forming a hit library; determining if a member of the hit library is structurally compatible with the lead structural template using a scoring function; and selecting the members of the hit library that score equal to or better than or equal to the lead sequence. The selected members of the hit library can be expressed in vitro or in vivo to produce a library of recombinant antibodies that can be screened for novel or improved function(s) over the lead antibody.
Owner:ABMAXIS
Electrochemical sensor using intercalative, redox-active moieties
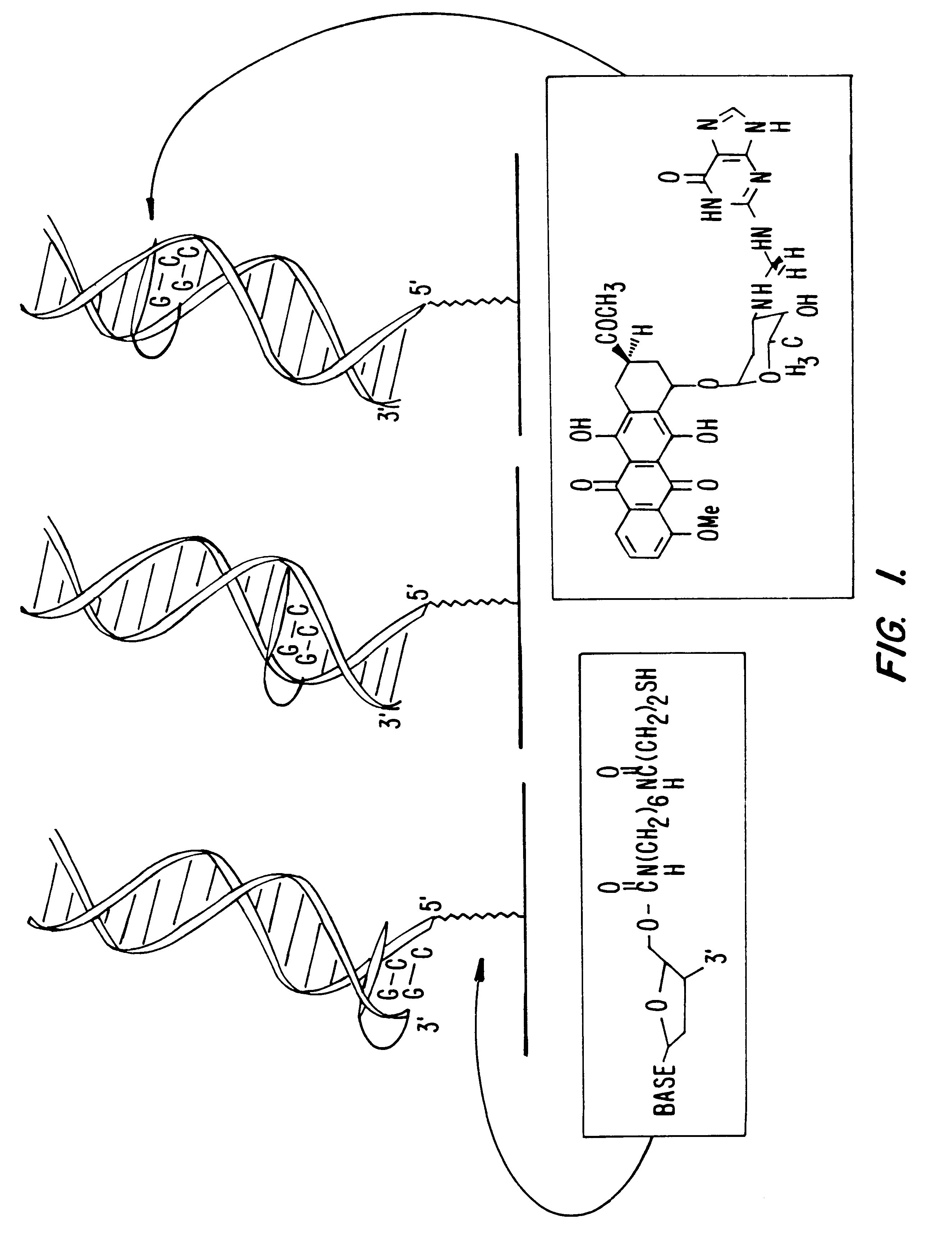
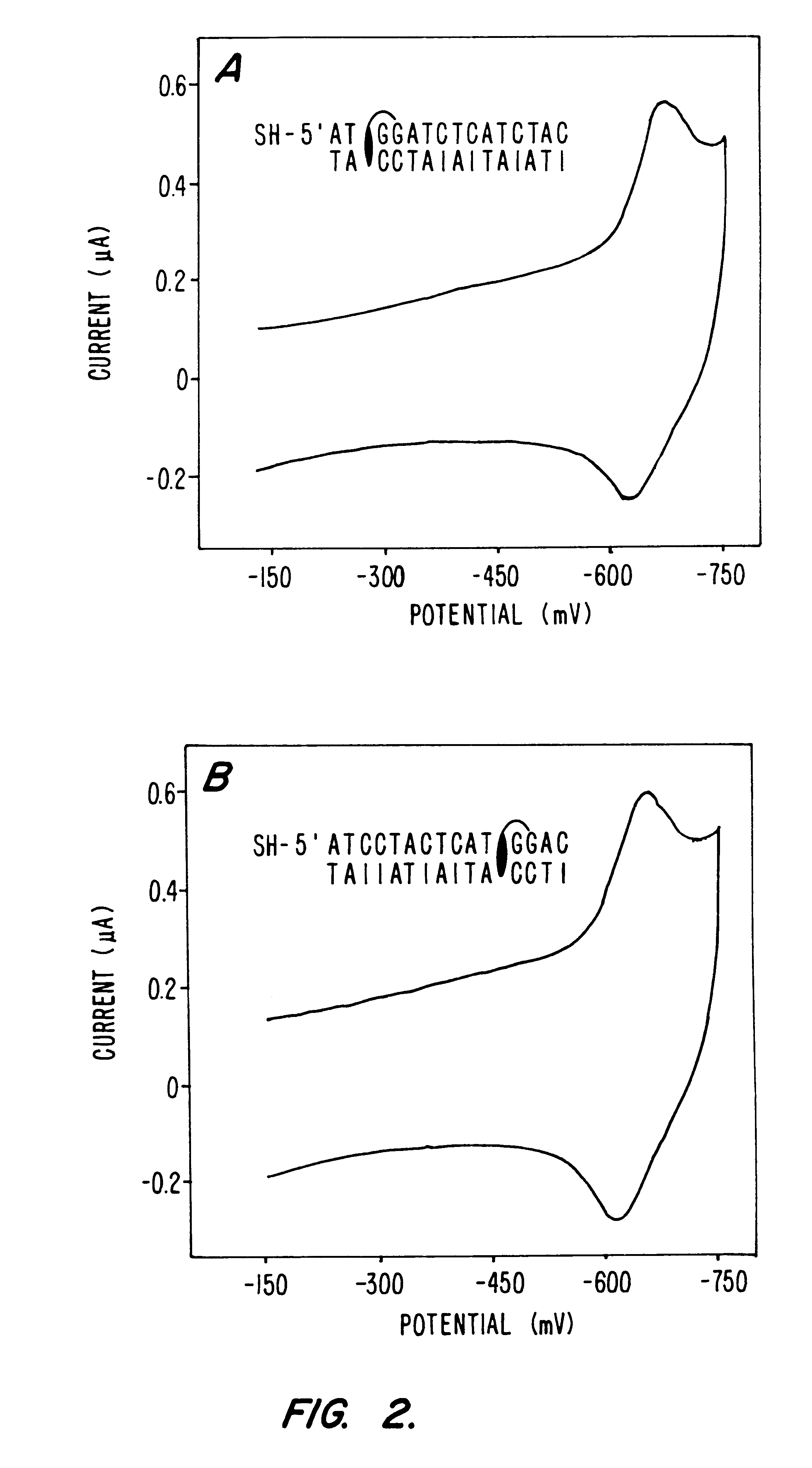
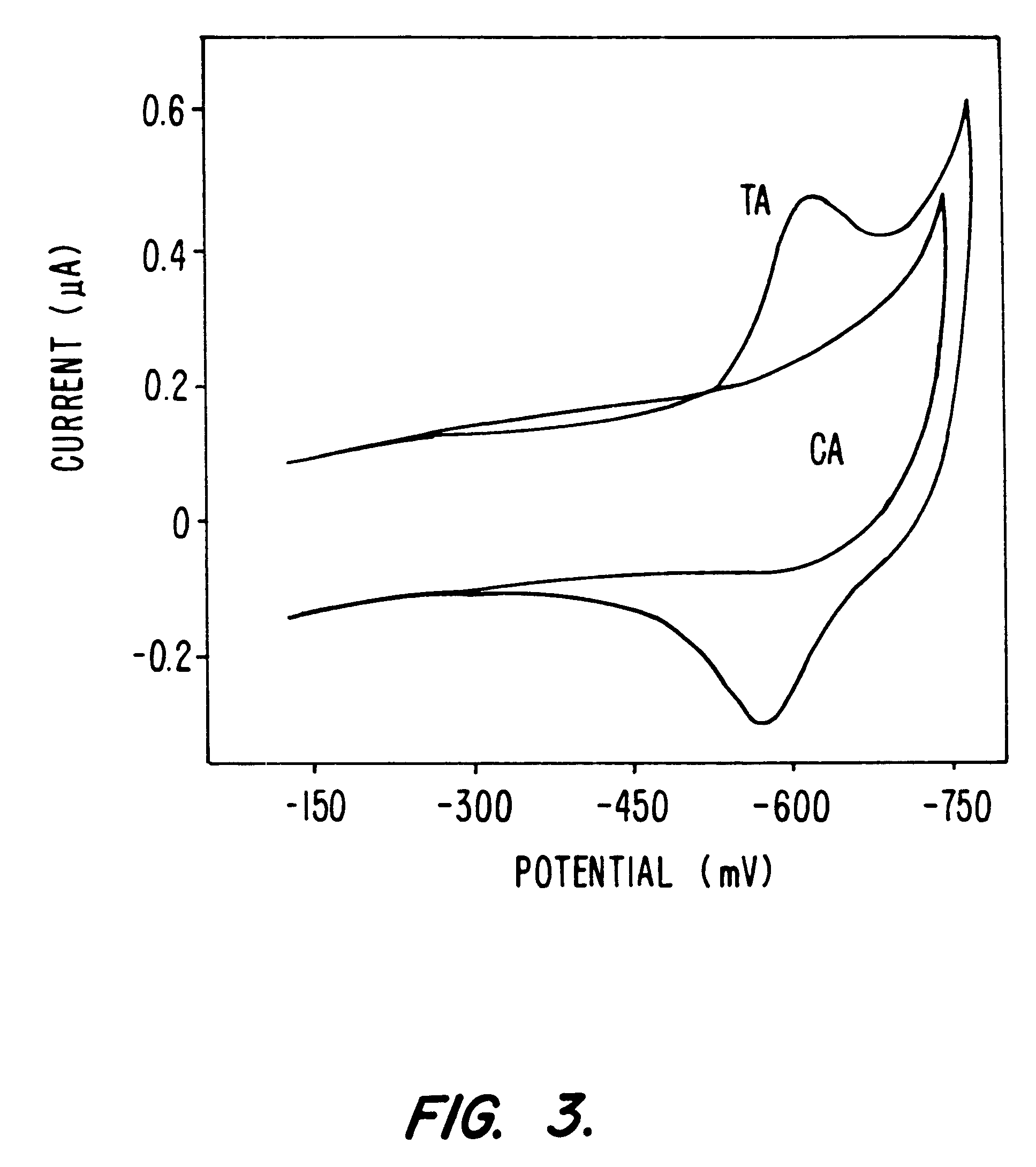
Compositions and methods for electrochemical detection and localization of genetic point mutations and other base-stacking perturbations within oligonucleotide duplexes adsorbed onto electrodes and their use in biosensing technologies are described. An intercalative, redox-active moiety (such as an intercalator or nucleic acid-binding protein) is adhered and / or crosslinked to immobilized DNA duplexes at different separations from an electrode and probed electrochemically in the presence or absence of a non-intercalative, redox-active moiety. Interruptions in DNA-mediated electron-transfer caused by base-stacking perturbations, such as mutations or binding of a protein to its recognition site are reflected in a difference in electrical current, charge and / or potential.
Owner:CALIFORNIA INST OF TECH
Method for rapidly screening microbial hosts to identify certain strains with improved yield and/or quality in the expression of heterologous proteins
ActiveUS20080269070A1High yieldQuality improvementBacteriaMicrobiological testing/measurementHeterologousADAMTS Proteins
The present invention provides an array for rapidly identifying a host cell population capable of producing heterologous protein with improved yield and / or quality. The array comprises one or more host cell populations that have been genetically modified to increase the expression of one or more target genes involved in protein production, decrease the expression of one or more target genes involved in protein degradation, or both. One or more of the strains in the array may express the heterologous protein of interest in a periplasm compartment, or may secrete the heterologous protein extracellularly through an outer cell wall. The strain arrays are useful for screening for improved expression of any protein of interest, including therapeutic proteins, hormones, a growth factors, extracellular receptors or ligands, proteases, kinases, blood proteins, chemokines, cytokines, antibodies and the like.
Owner:PFENEX
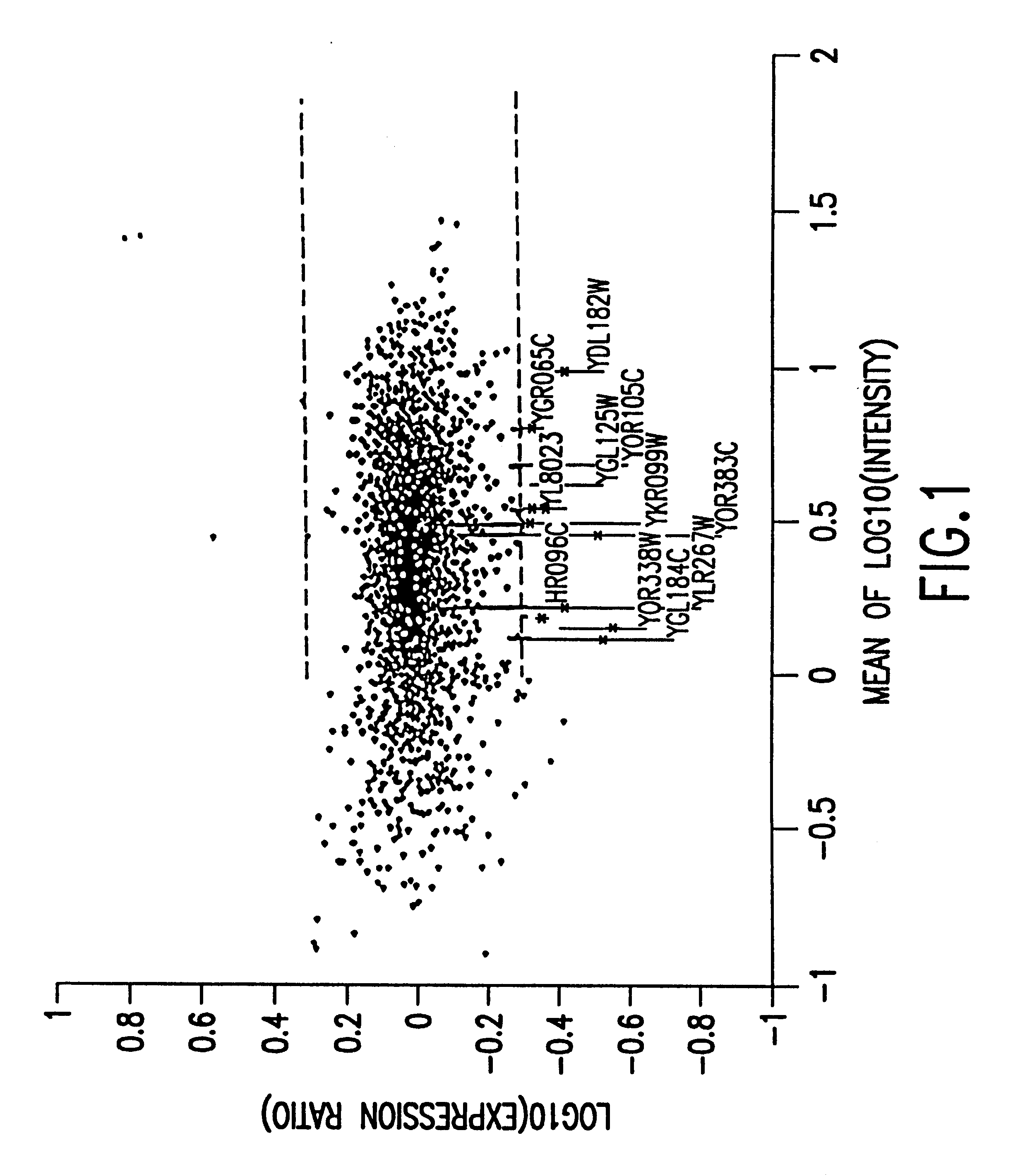
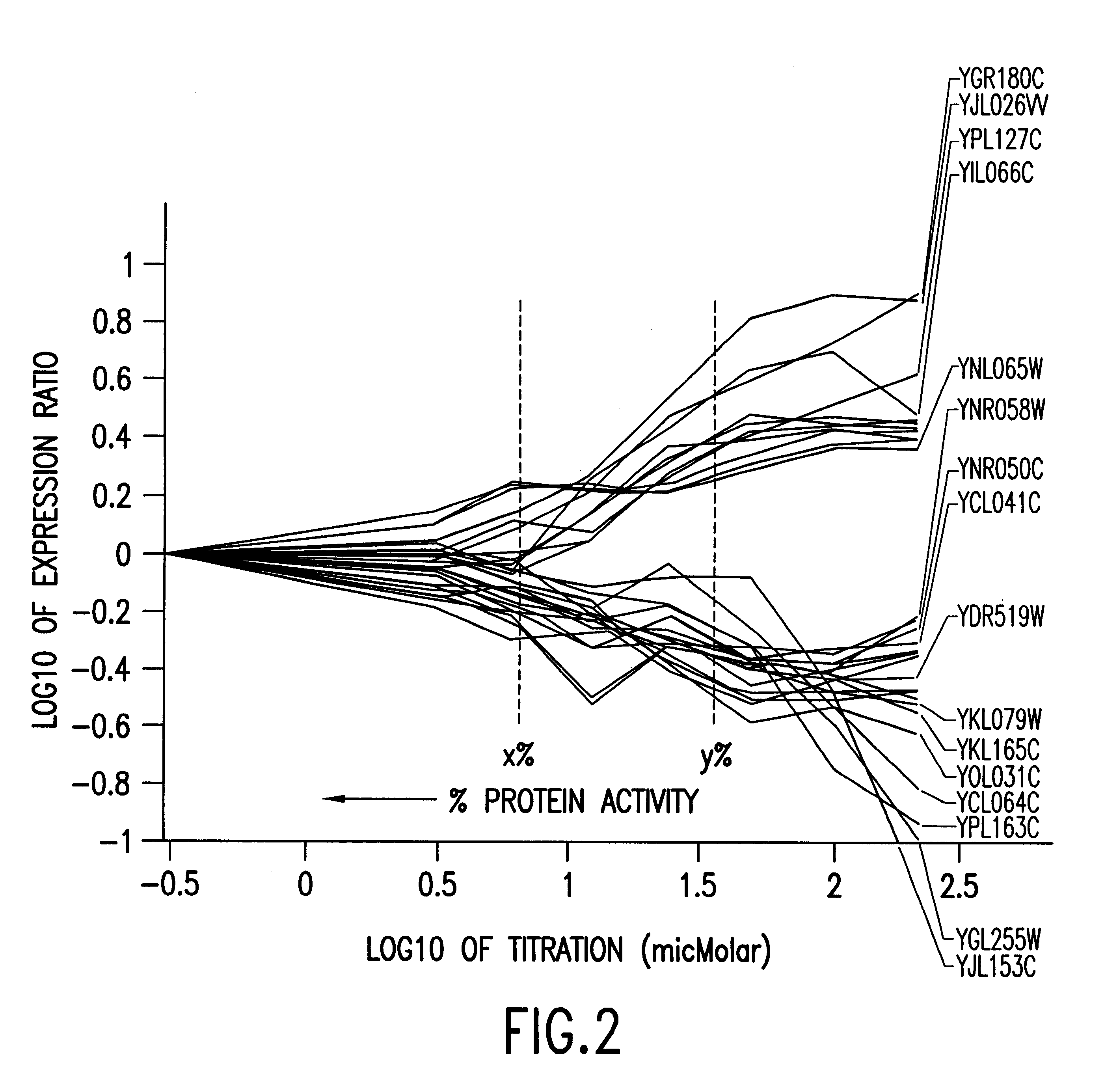
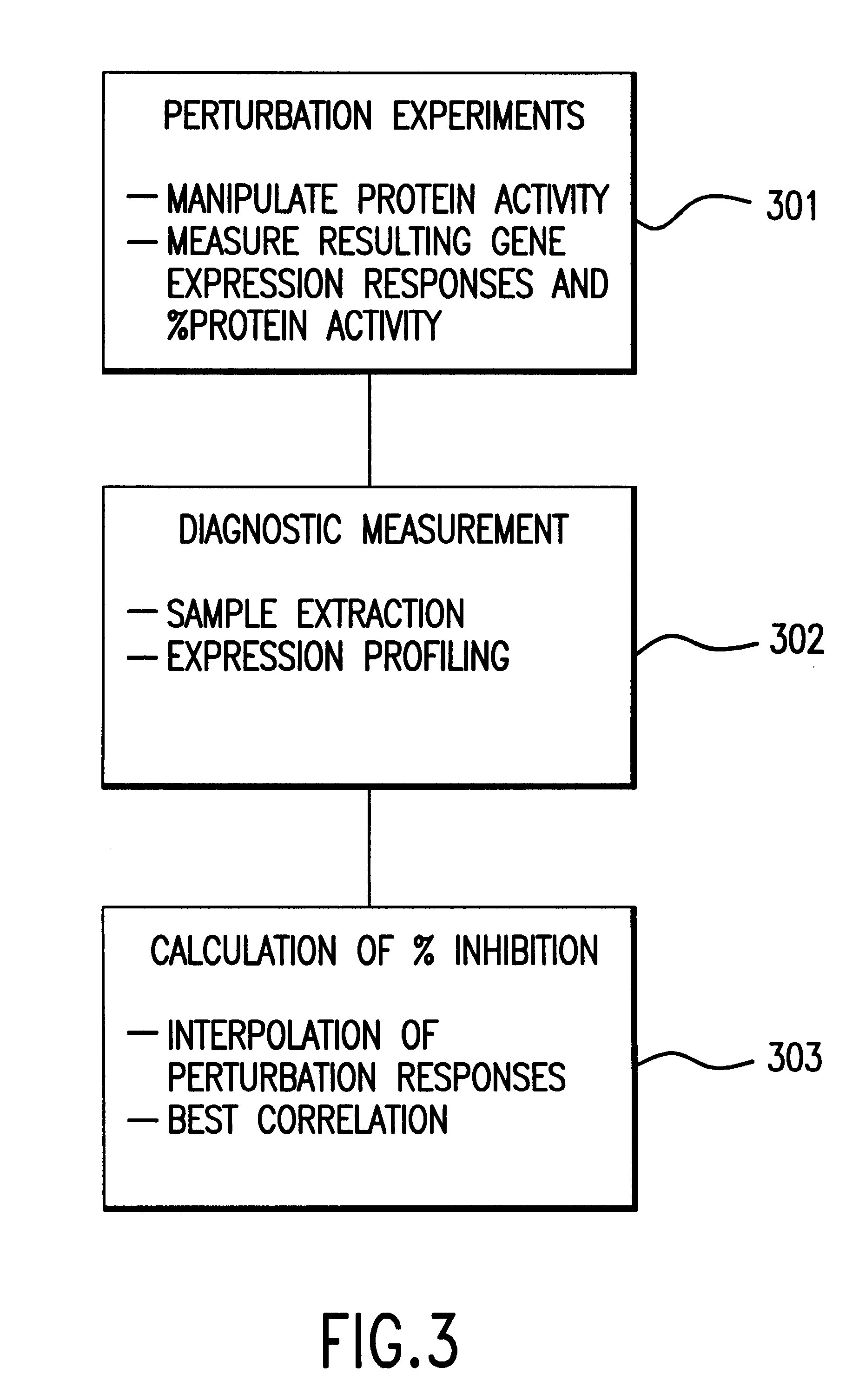
Methods of determining protein activity levels using gene expression profiles
InactiveUS6324479B1High similaritySugar derivativesPeptide/protein ingredientsProtein insertionDrug activity
The present invention provides methods for determining the level of protein activity in a cell by: (i) measuring abundances of cellular constituents in a cell in which the activity of a specific protein is to be determined so that a diagnostic profile is thus obtained; (ii) measuring abundances of cellular constituents that occur in a cell in response to perturbations in the activity of said protein to obtain response profiles and interpolating said response profiles to generate response curves; and (iii) determining a protein activity level at which the response profile extracted from the response curves best fits the measured diagnostic profile, according to some objective measure. In alternative embodiments, the present invention also provides methods for identifying individuals having genetic mutations or polymorphisms that disrupt protein activity, and methods for identifying drug activity in vivo by determining the activity levels of proteins which interact with said drugs.
Owner:MICROSOFT TECH LICENSING LLC
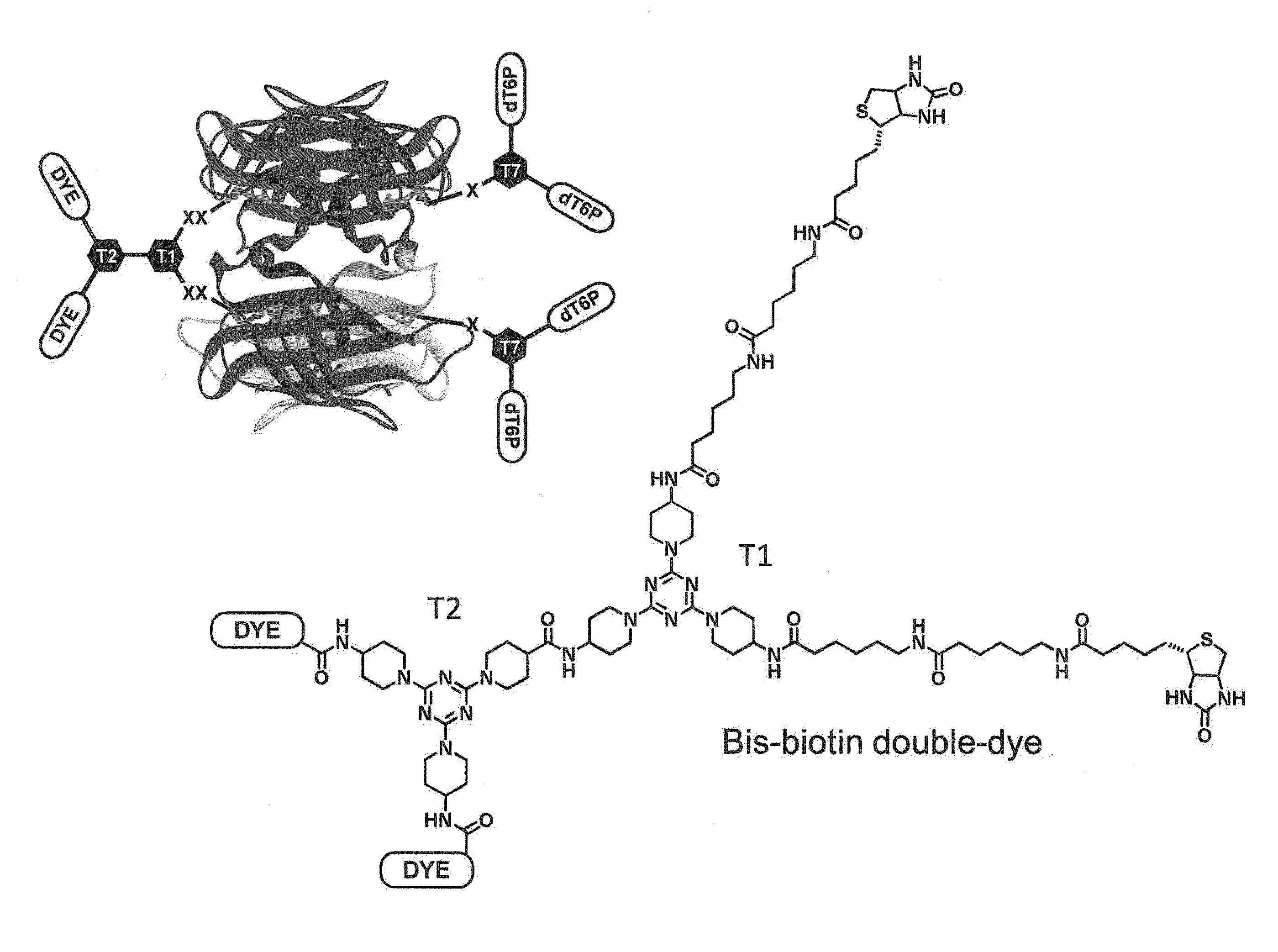
Polymerase enzyme substrates with protein shield
Compositions and methods are provided for nucleotide analogs comprising protein shields for improving enzyme photostability in single molecule real time sequencing. Nucleotide analogs of the invention have a protein shield between the dye moieties and nucleotide moieties of the analog. The protein prevents the direct interaction of the dye moiety with the enzyme carrying out nucleotide synthesis preventing photodamage to the enzyme. The nucleotide analogs of the invention can have multiple dyes and multiple nucleotide moieties.
Owner:PACIFIC BIOSCIENCES
Process for removing protein aggregates and virus from a protein solution
InactiveUS7118675B2Less-expensive to purchaseLess-expensive to operateComponent separationSolvent extractionProtein solutionFree protein
A process is provided for selectively removing protein aggregates from a protein solution in a normal flow (NFF) filtration process. Preferably, it relates to a process for selectively removing protein aggregates from a protein solution in a normal flow (NFF) filtration process and virus particles from a protein solution in a two-step filtration process. In a first step, a protein solution is filtered through one or more layers of adsorptive depth filters, charged or surface modified microporous membranes or a small bed of chromatography media in a normal flow filtration mode of operation, to produce a protein aggregate free stream. The aggregate free protein stream can then be filtered through one or more ultrafiltration membranes to retain virus particles at a retention level of at least 3 LRV and to allow passage therethrough of an aggregate free and virus free protein solution.
Owner:MILLIPORE CORP
Frozen tissue microarray technology for analysis RNA, DNA, and proteins
InactiveUS6893837B2Withdrawing sample devicesPreparing sample for investigationAnalysis dnaTissue Arrays
The invention disclosed herein improves upon existing tissue microarray technology by using frozen tissues embedded in tissue embedding compound as donor samples and arraying the specimens into a recipient block comprising tissue embedding compound. Tissue is not fixed prior to embedding, and sections from the array are evaluated without fixation or post-fixed according to the appropriate methodology used to analyze a specific gene at the DNA, RNA, and / or protein levels. Unlike paraffin tissue arrays which can be problematic for immunohistochemistry and for RNA in situ hybridization analyses, the disclosed methods allow optimal evaluation by each technique and uniform fixation across the array panel. The disclosed arrays work well for DNA, RNA, and protein analyses, and have significant qualitative and quantitative advantages over existing methods.
Owner:RGT UNIV OF CALIFORNIA
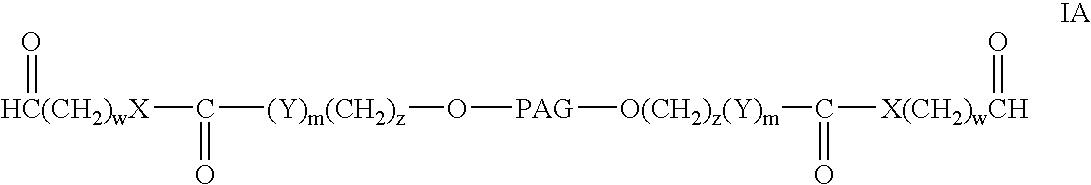
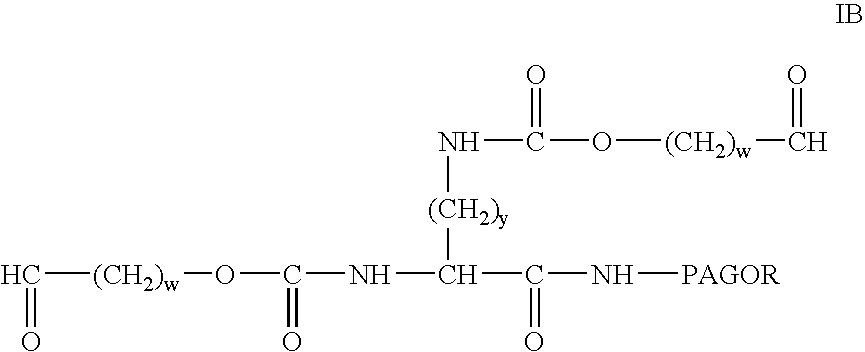
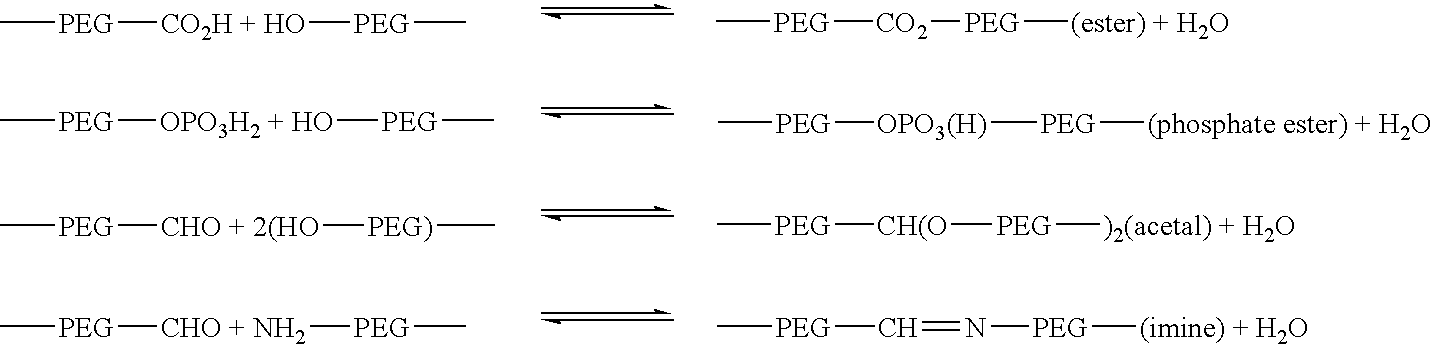
Bifunctional polyethylene glycol derivatives
InactiveUS20040115165A1Linkage stabilityDifficult to purifyOrganic chemistryPharmaceutical non-active ingredientsProtein insertionActive protein
The present invention provides novel heterobifunctional and monobifunctional polyethylene glycol derivatives for the pegylation of therapeutically active proteins. The heterobifunctional PEGs which bear two different functional groups as well as the monobifunctional PEGs which contain two similar functional groups, may be used for cross-linking purposes. The cross-linking may be intramolecular between two areas within the same molecule or intermolecular between two separate molecules. The pegylated protein conjugates that are produced, retain a substantial portion of their therapeutic activity and are less immunogenic than the protein from which the conjugate is derived. New syntheses for preparing such bifunctional derivatives are described.
Owner:SUN BIO INC
Defining biological states and related genes, proteins and patterns
InactiveUS20020169562A1Reduce the valueSugar derivativesMicrobiological testing/measurementDiseaseProtein insertion
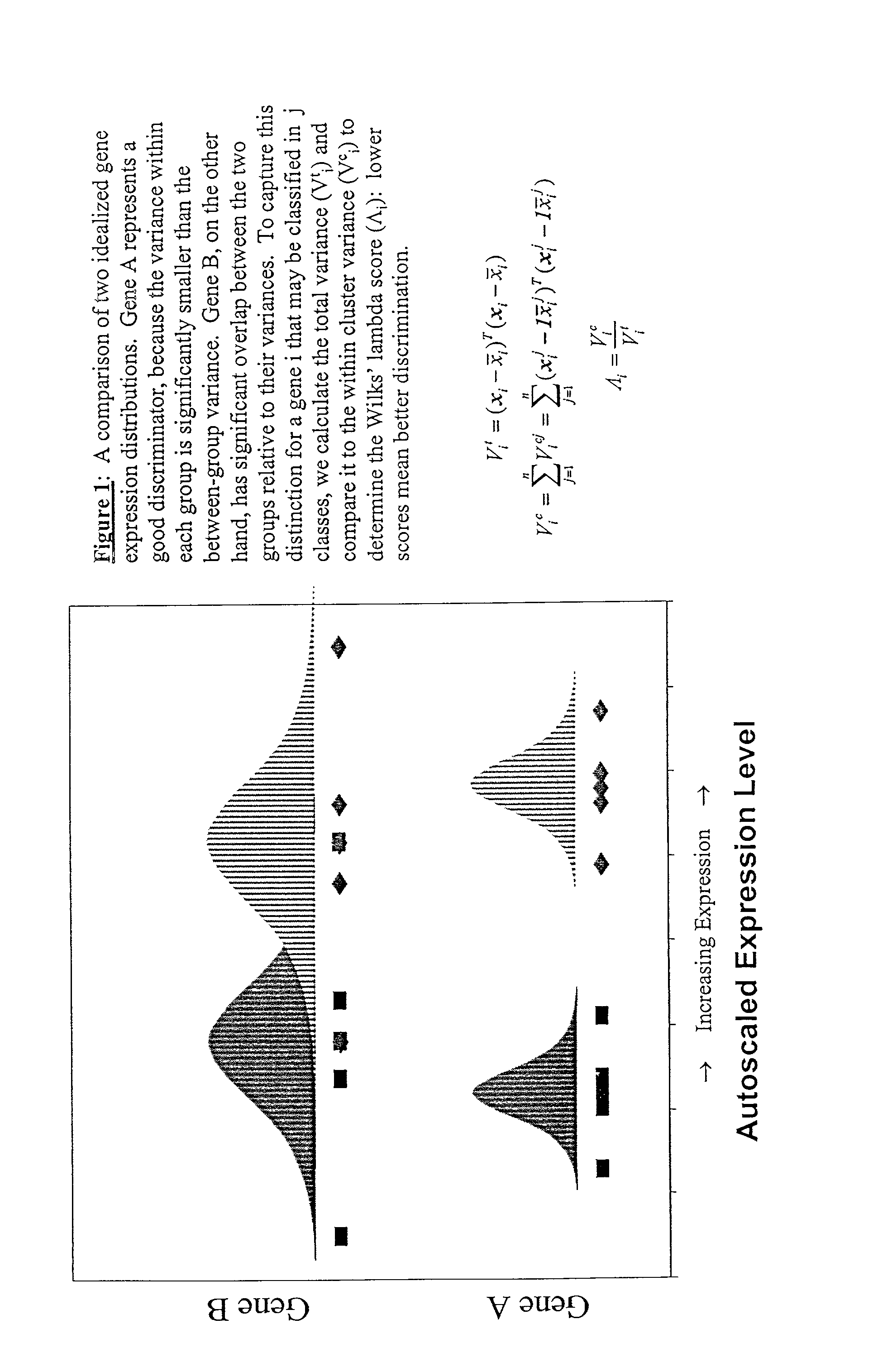
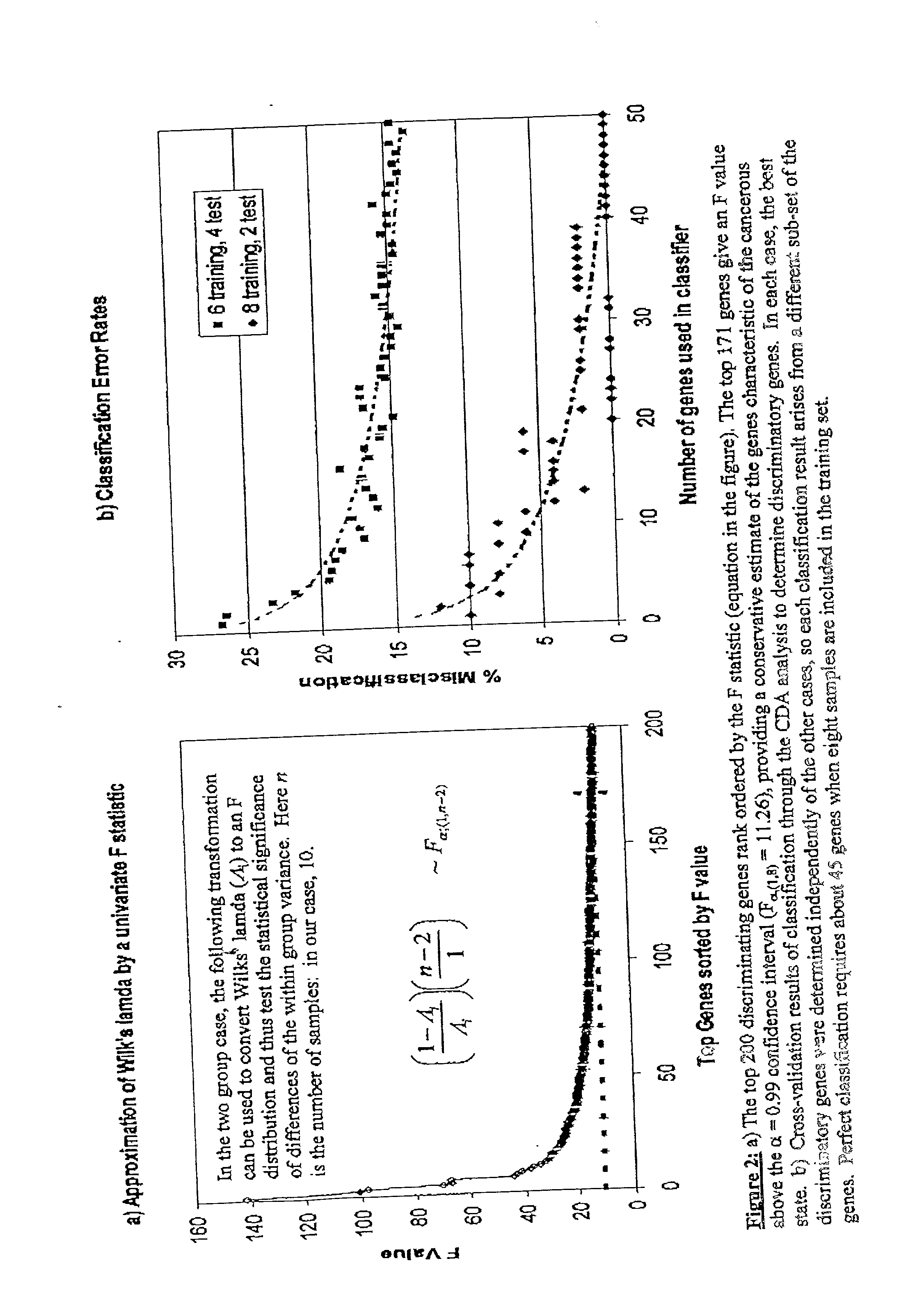
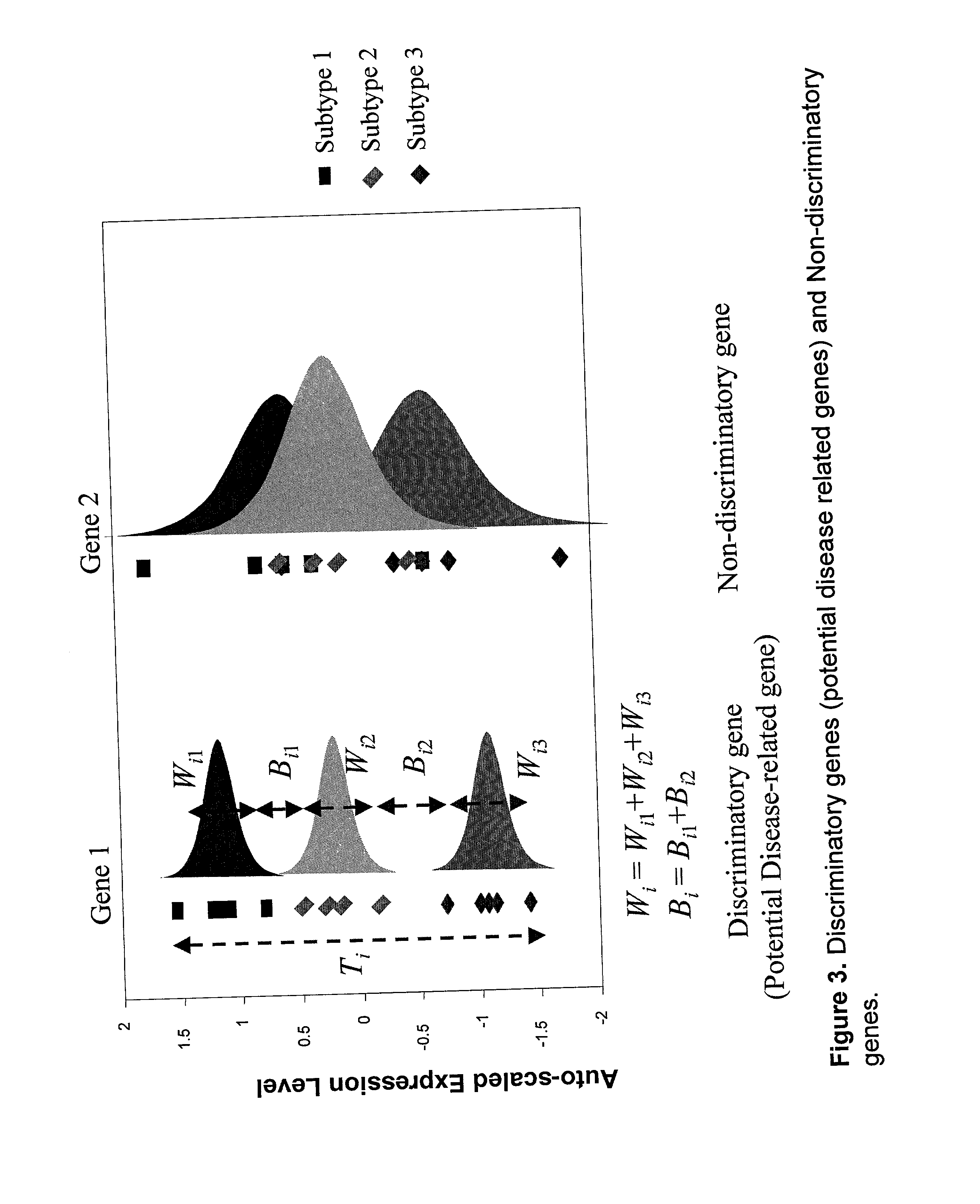
Disclosed are a variety of methods and computer systems for use in the analysis of gene and protein expression data. Also disclosed are methods for the definition of the cellular state of cells and tissues from multidimensional physiological data such as those obtained from gene expression measurements with DNA microarrays. A variety of classification methods can be applied to expression data to achieve this goal. Demonstrated is the application of several statistical tools including Wilks' lambda ratio of within-group to total variance, Fisher Discriminant Analysis, and the misclassification error rate to the identification of discriminating genes and the overall classification of expression data. Examples from several different cases demonstrate the ability of the method to produce well-separated groups in the projection space representing distinct physiological states. The method can be augmented and is useful in disease diagnosis, drug screening and bioprocessing applications.
Owner:MASSACHUSETTS INST OF TECH
Method and compositions for the detection of protein glycosylation
ActiveUS20050130235A1Sugar derivativesMicrobiological testing/measurementPost translationalProtein insertion
The invention provides methods and compositions for the rapid and sensitive detection of post-translationally modified proteins, and particularly of those with post-translational glycosylations. The methods can be used to detect O-GlcNAc posttranslational modifications on proteins on which such modifications were undetectable using other techniques. In one embodiment, the method exploits the ability of an engineered mutant of β-1,4-galactosyltransferase to selectively transfer an unnatural ketone functionality onto O-GlcNAc glycosylated proteins. Once transferred, the ketone moiety serves as a versatile handle for the attachment of biotin, thereby enabling detection of the modified protein. The approach permits the rapid visualization of proteins that are at the limits of detection using traditional methods. Further, the preferred embodiments can be used for detection of certain disease states, such as cancer, Alzheimer's disease, neurodegeneration, cardiovascular disease, and diabetes.
Owner:CALIFORNIA INST OF TECH
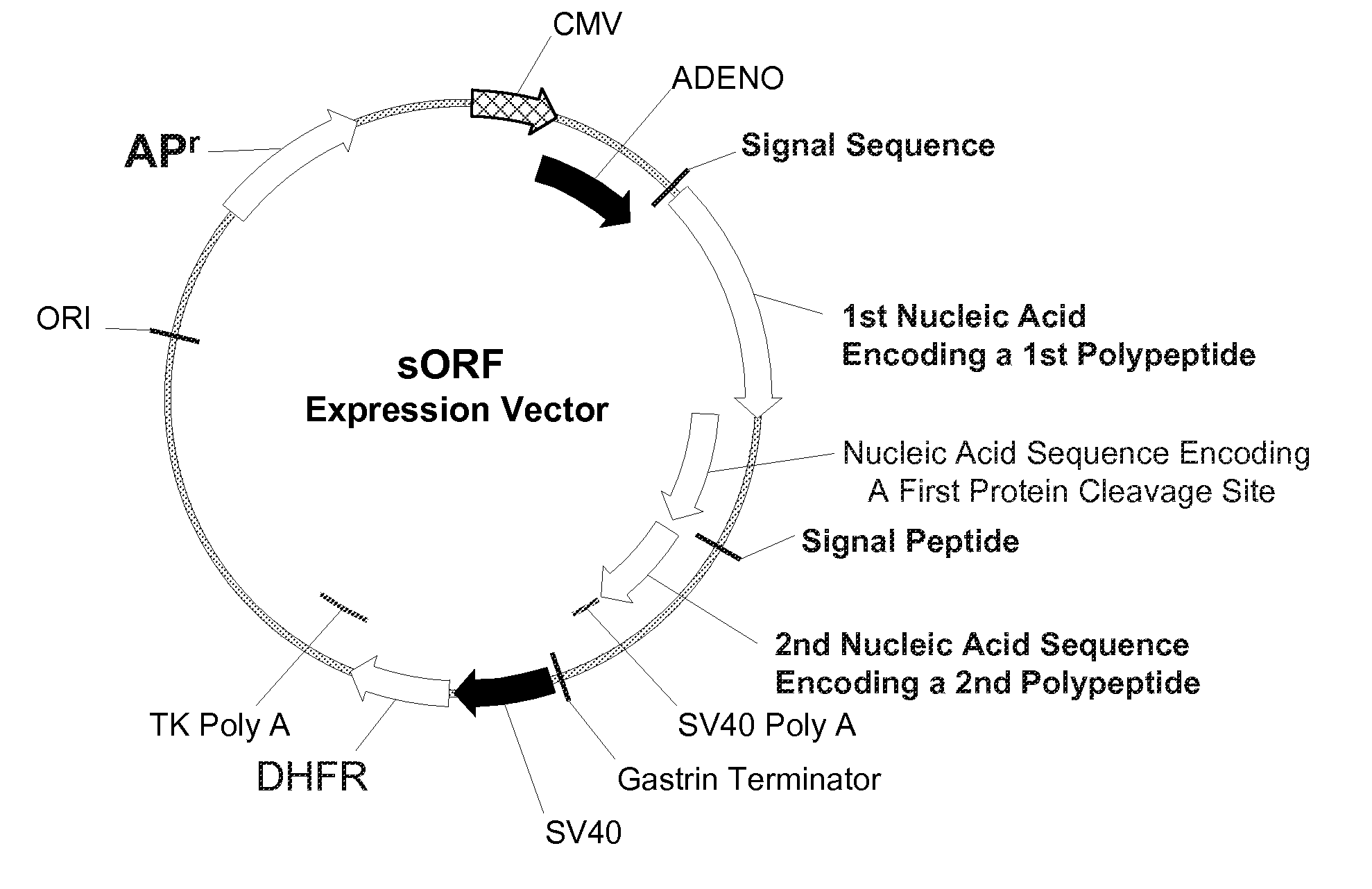
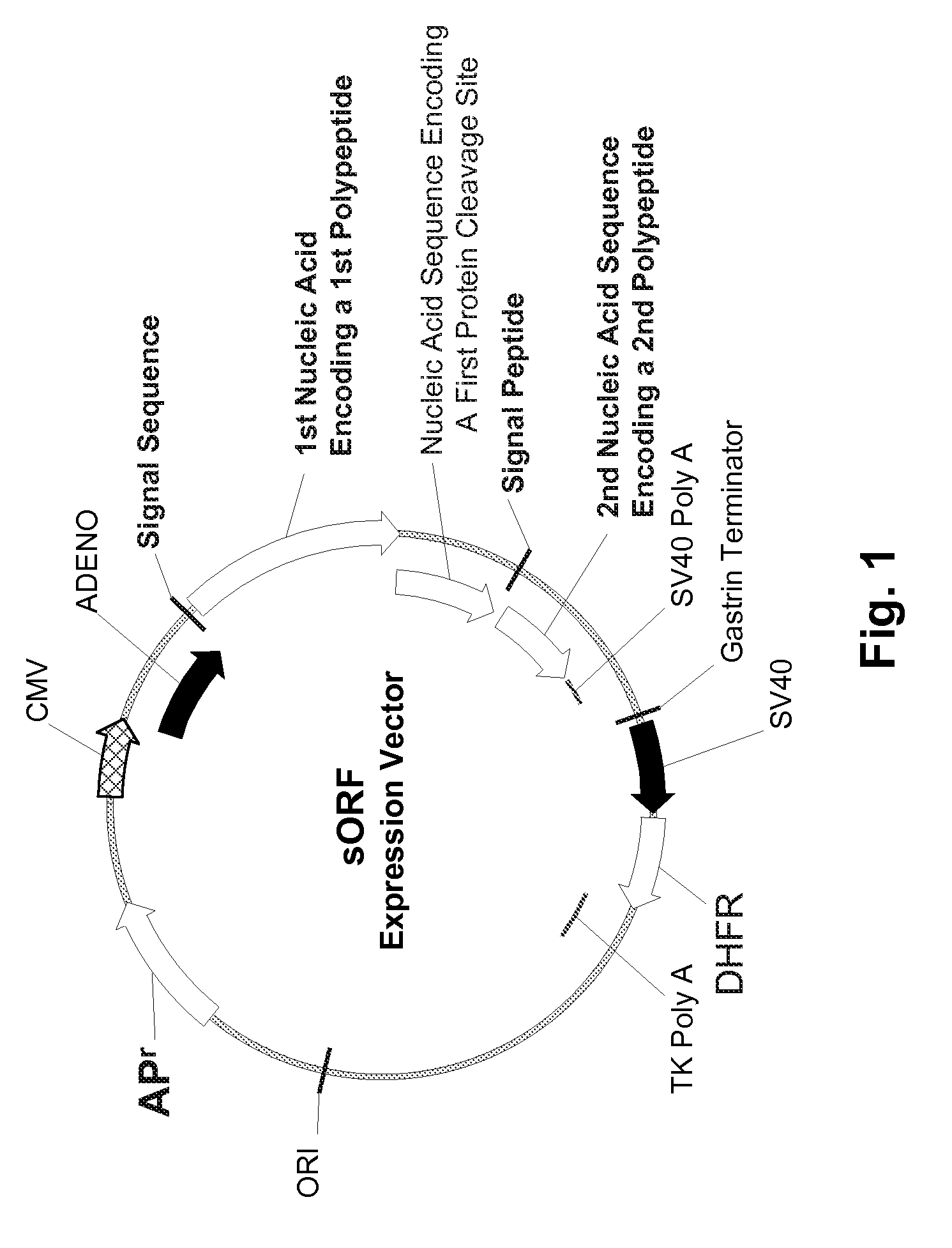
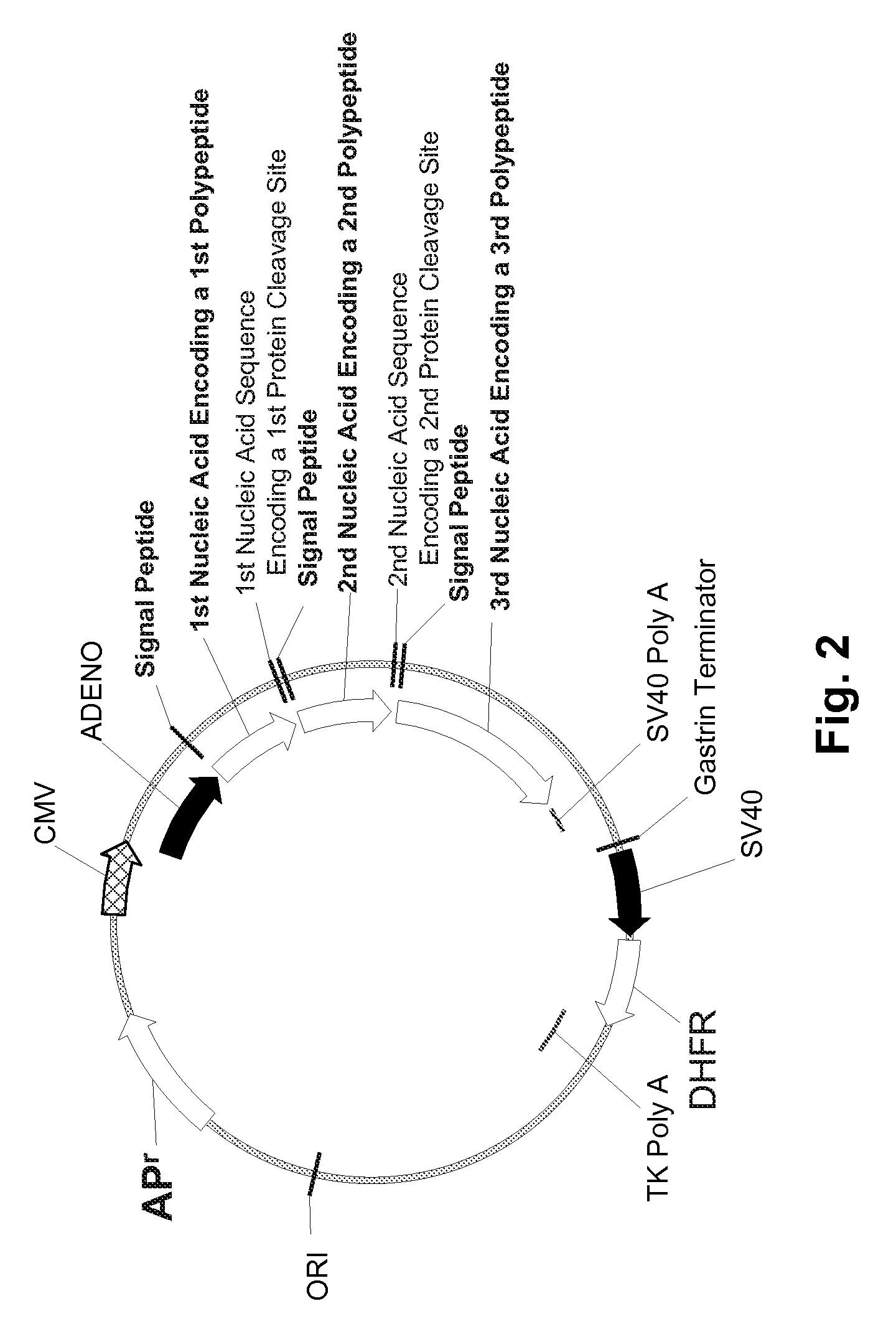
Multiple Gene Expression including sORF Constructs and Methods with Polyproteins, Pro-Proteins, and Proteolysis
InactiveUS20070065912A1Efficient expressionImprove economyFungiFusion with post-translational modification motifOpen reading frameADAMTS Proteins
Disclosed are useful constructs and methods for the expression of proteins using primary translation products that are processed within a recombinant host cell. Constructs comprising a single open reading frame (sORF) are described for protein expression including expression of multiple polypeptides. A primary translation product (a pro-protein or a polyprotein) contains polypeptides such as inteins or hedgehog family auto-processing domains, or variants thereof, inserted in frame between multiple protein subunits of interest. The primary product can also contain cleavage sequences such as other proteolytic cleavage or protease recognition sites, or signal peptides which contain recognition sequences for signal peptidases, separating at least two of the multiple protein subunits. The sequences of the inserted auto-processing polypeptides or cleavage sites can be manipulated to enhance the efficiency of expression of the separate multiple protein subunits. Also disclosed are independent aspects of conducting efficient expression, secretion, and / or multimeric assembly of proteins such as immunoglobulins. Where the polyprotein contains immunoglobulin heavy and light chain segments or fragments capable of antigen recognition, in an embodiment a selectable stoichiometric ratio is at least two copies of a light chain segment per heavy chain segment, with the result that the production of properly folded and assembled functional antibody is made. Modified signal peptides, including such from immunoglobulin light chains, are described.
Owner:ABBOTT LAB INC
Protein separation and display
InactiveUS20020098595A1Easy transferSuitable for automationSludge treatmentVolume/mass flow measurementProtein insertionImproved method
The present invention relates to multi-phase protein separation methods capable of resolving and characterizing large numbers of cellular proteins, including methods for efficiently facilitating the transfer of protein samples between separation phases. In particular, the present invention provides an automated system for the separation, identification, and characterization of protein samples. The present invention thus provides improved methods for the analysis of samples containing large numbers of proteins.
Owner:RGT UNIV OF MICHIGAN
Automatic protein and/or DNA analysis system and method
A fully automated protein and / or DNA gene fragment analyzing machine and method are disclosed in which, in a simple machine structure, a plurality of electrophoresis calls each containing such fragments, are robotically, under computer programming, inserted into an electrophoresis housing and subjected to voltage for producing electrophoretic migration in one dimension (horizontally), and then preferably after robotic 90 DEG rotating of the cells, are voltage migrated in an orthogonal direction to separate the fragments vertically, and then robotically presented to an optical image scanner for identifying the brightest fragments and classifying the same, with comparison with reference image fragment locations of normal or variant fragments, and for computer storing all relevant data.
Owner:ACAD OF APPL SCI INC
Assays for measuring nucleic acid binding proteins and enzyme activities
InactiveUS6312896B1Simple and accurate and reliable assayEfficient transferSugar derivativesMicrobiological testing/measurementProtein insertionNucleic acid binding protein
Owner:BIOVERIS CORP
Degradable heterobifunctional poly(ethylene glycol) acrylates
InactiveUS7166304B2Low release rateEasy to controlPowder deliveryPeptide preparation methodsProtein insertionEthylene glycol
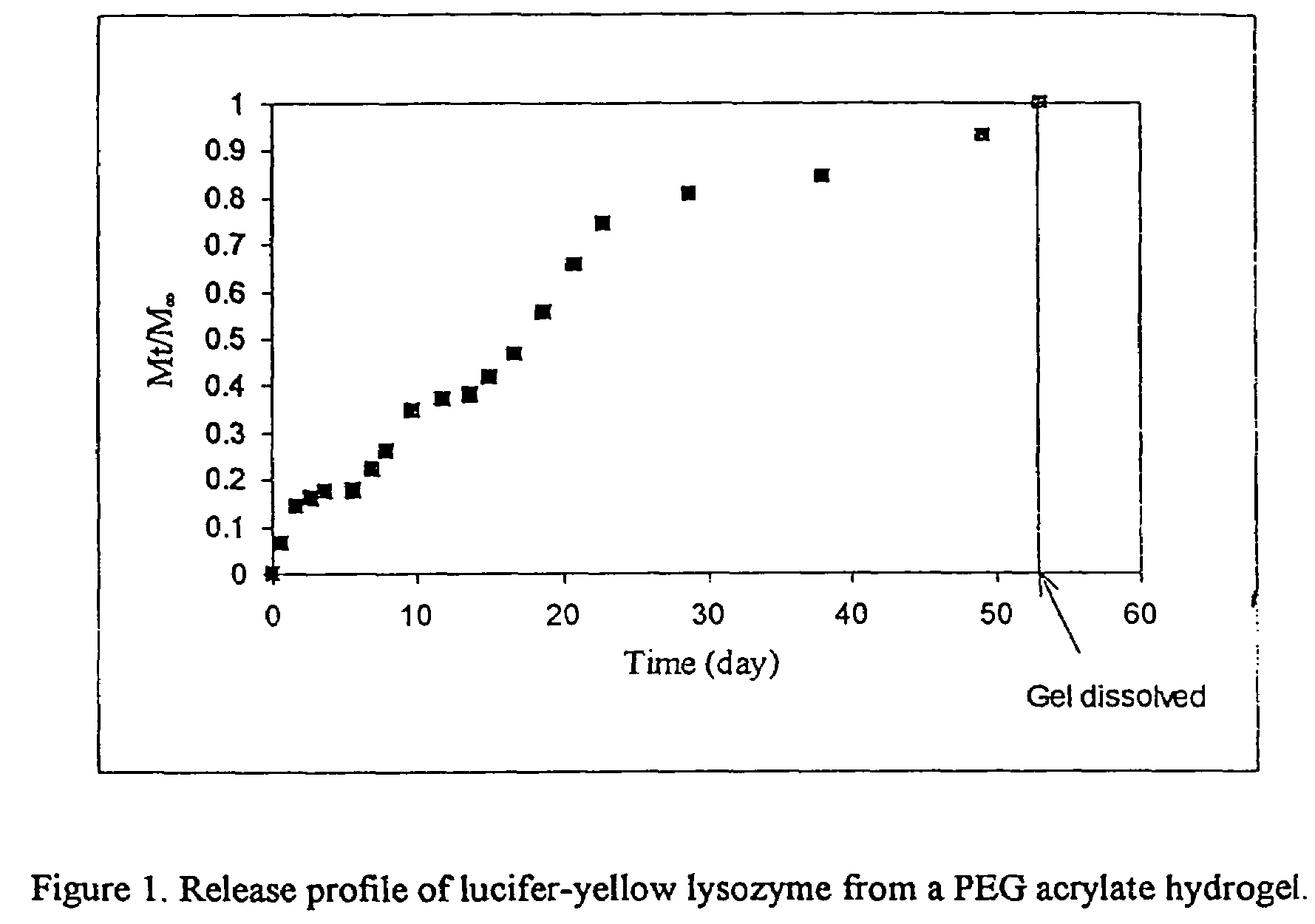
A heterobifunctional poly(ethylene glycol) is provided having a hydrolytically degradable linkage, a first terminus comprising an acrylate group, and a second terminus comprising a target such as a protein or pharmaceutical agent or a reactive moiety capable of coupling to a target. Hydrogels can be prepared. The hydrogels can be used as a carrier for a protein or a pharmaceutical agent that can be readily released in a controlled fashion.
Owner:NEKTAR THERAPEUTICS INC
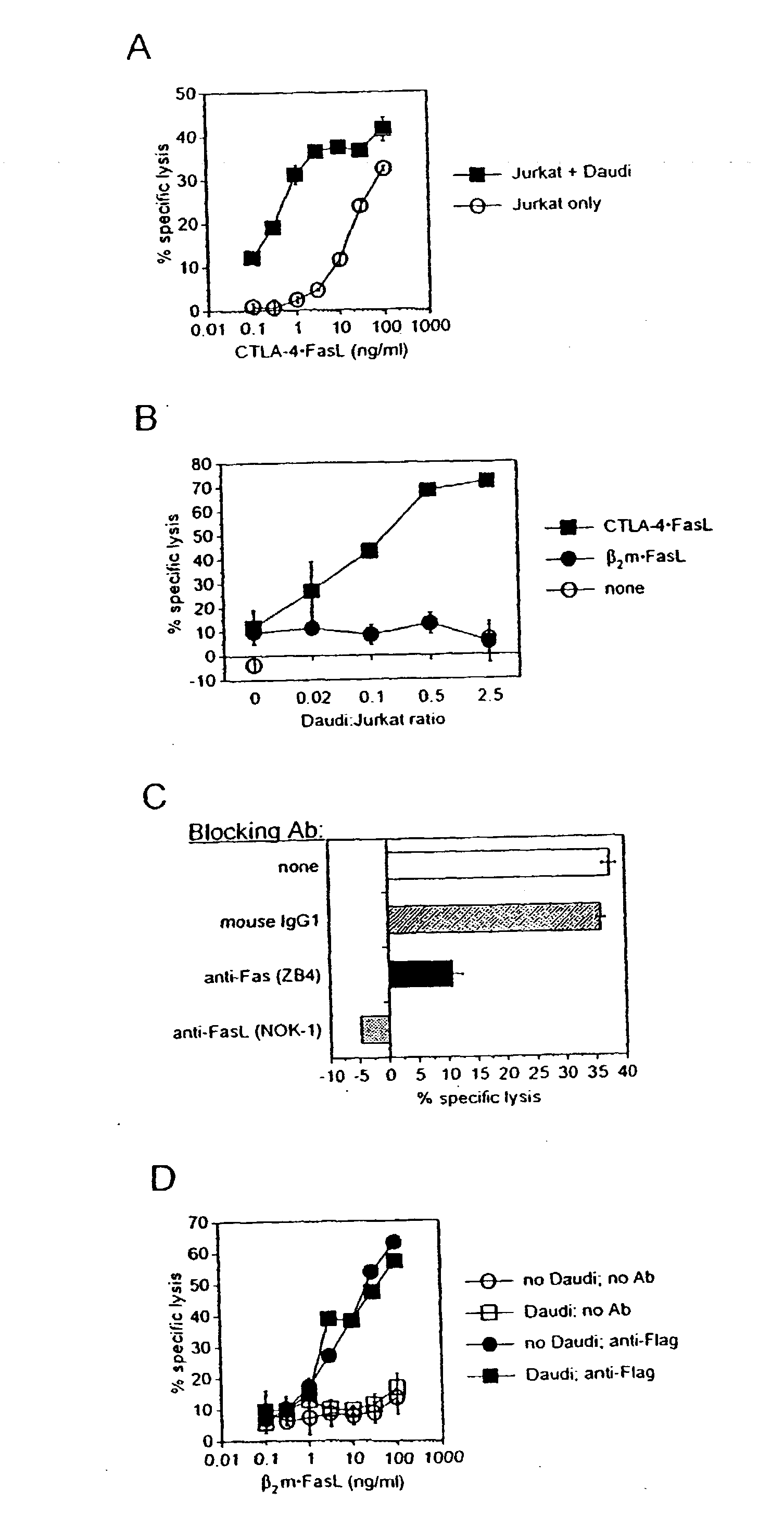
Novel chimeric proteins and methods for using the same
InactiveUS20030216546A1Compounds screening/testingPeptide/protein ingredientsDiseaseProtein insertion
Novel chimeric proteins are disclosed. The proteins comprise at least two portions. The first portion binds to a first cell and decreases the cell's ability to send a trans signal to a second cell; the second portion sends its own trans signal to the second cell. Methods for making and using these proteins in the treatment of cancer, viral infections, autoimmune and alloimmune diseases are also disclosed, as are pharmaceutical formulations comprising the novel chimeric proteins and genes. Either the proteins themselves or a genetic sequence encoding the protein can be administered. Other methods are also disclosed in which two molecular components result in decrement of a first trans signal from a first cell and the conferring of a second trans signal to a second cell.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
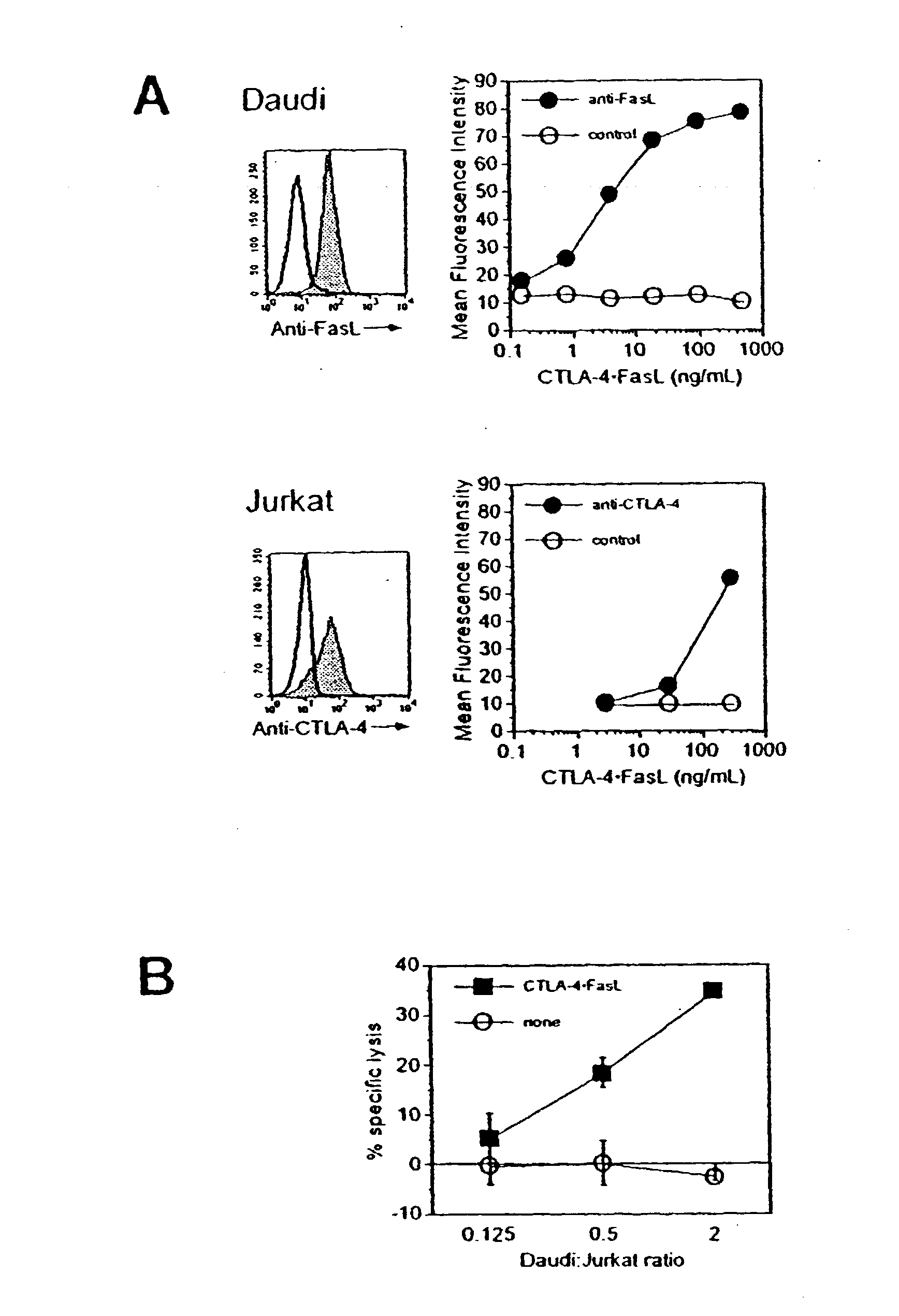
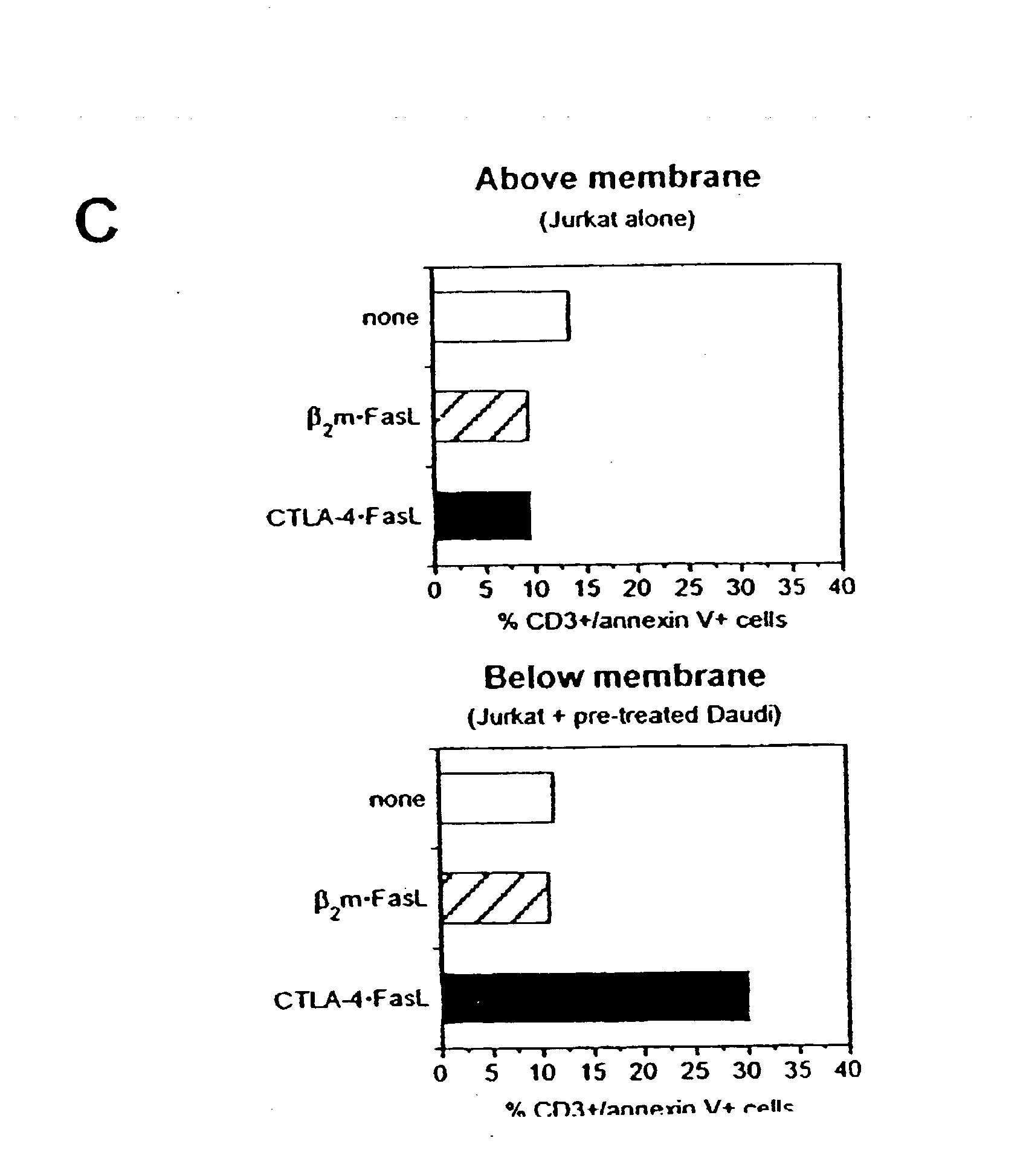
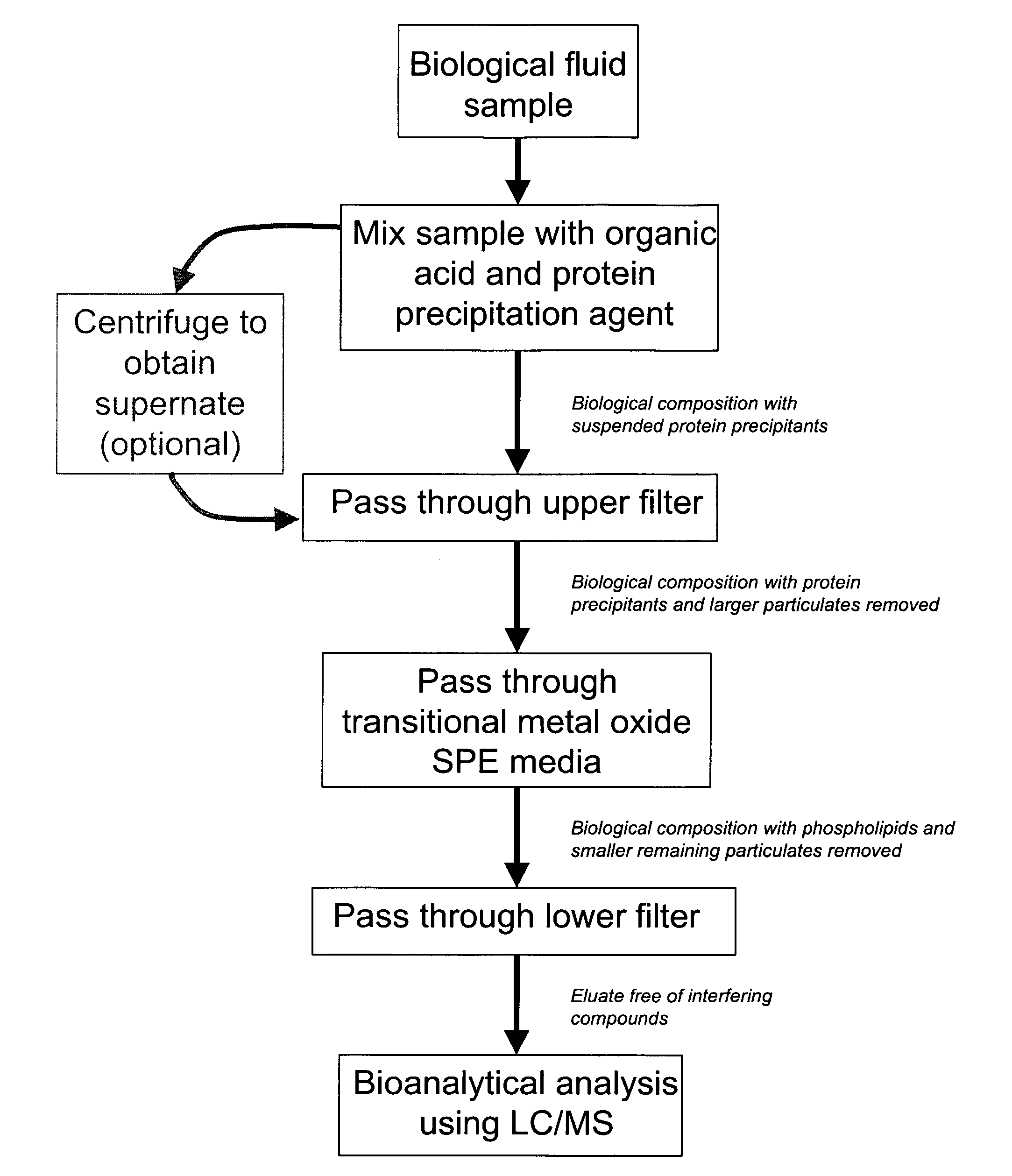
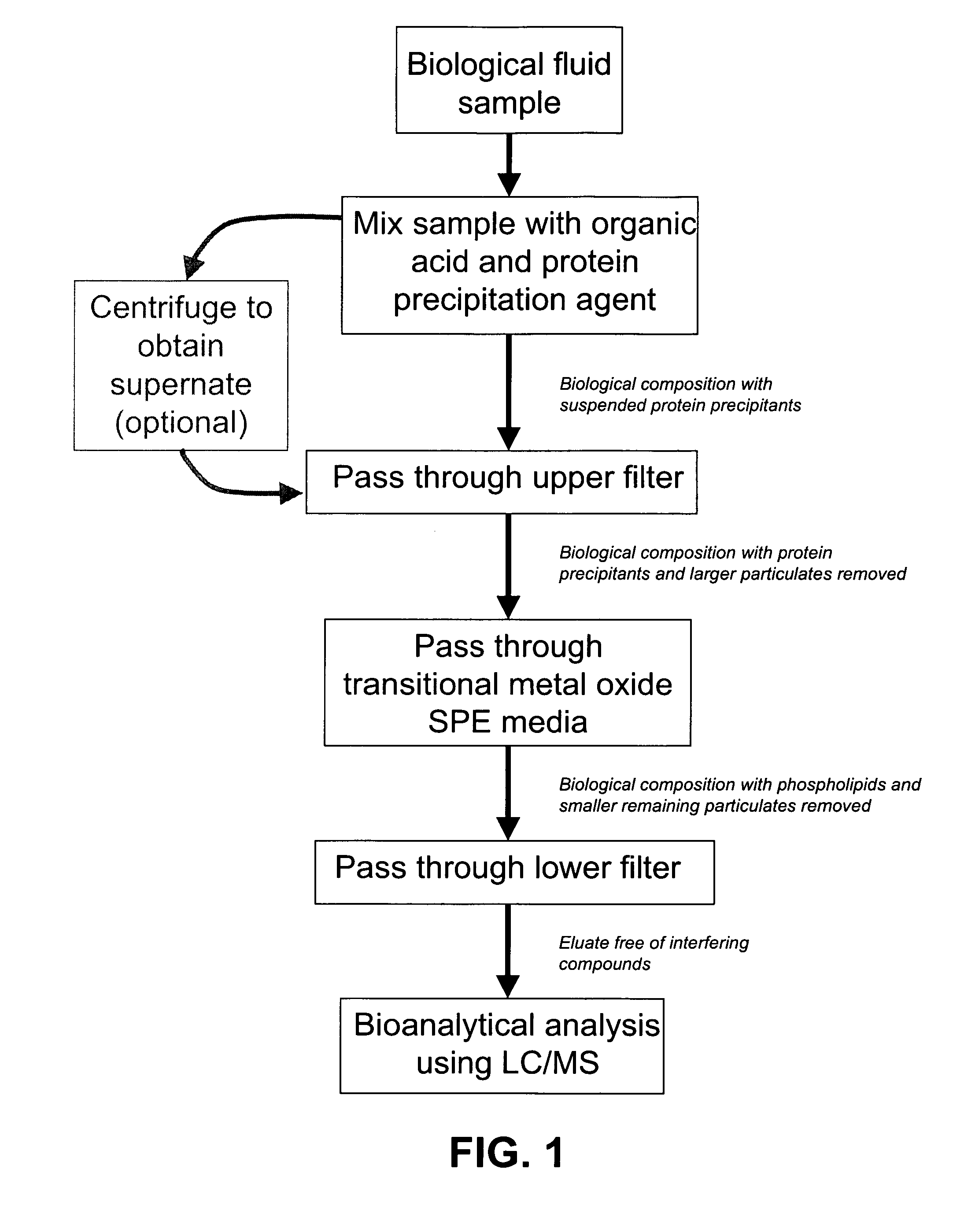
Compositions and methods for combining protein precipitation and solid phase extraction
ActiveUS20080213906A1Large specific surface areaEliminate later ion-suppressionComponent separationBiological testingProtein insertionPhosphate
A composition, method and device for the preparation of biological samples for subsequent LC-MS analysis using a combined and concurrent protein precipitation and solid phase extraction (SPE) process is described. Through an integrated combination of protein precipitation, filtration, and SPE using a novel zirconia-coated chromatographic media, interfering compounds, such as proteins and phosphate-containing compounds, are eliminated from the biological samples, affording a higher degree of analyte response during LC-MS analysis.
Owner:SIGMA ALDRICH CO LLC
Protein transistor device
ActiveUS20140048776A1Improve bindingGood reproducibilityNanoinformaticsSolid-state devicesProtein moleculesProtein insertion
The present invention discloses a protein transistor device, wherein an antibody molecule (antibody-antigen) is bonded to at least two gold nanoparticles in a high reproducible self-assembly way to form molecular junctions, and wherein the two gold nanoparticles are respectively joined to a drain and a source. The protein transistor device can be controlled to regulate current via applying a bias to the gate. The conformational change of the protein molecule will cause the variation of the charge transport characteristics of the protein transistor device. The protein transistor device can be further controlled by different optical fields via conjugating a quantum dot to the molecular junctions. Therefore, the present invention has diversified applications.
Owner:ALLBIO LIFE CO LTD
Gene recombination and hybrid protein development
InactiveUS20030032059A1Preserve stability and desired propertyComputationally tractableBiological testingSequence analysisBiopolymerProtein insertion
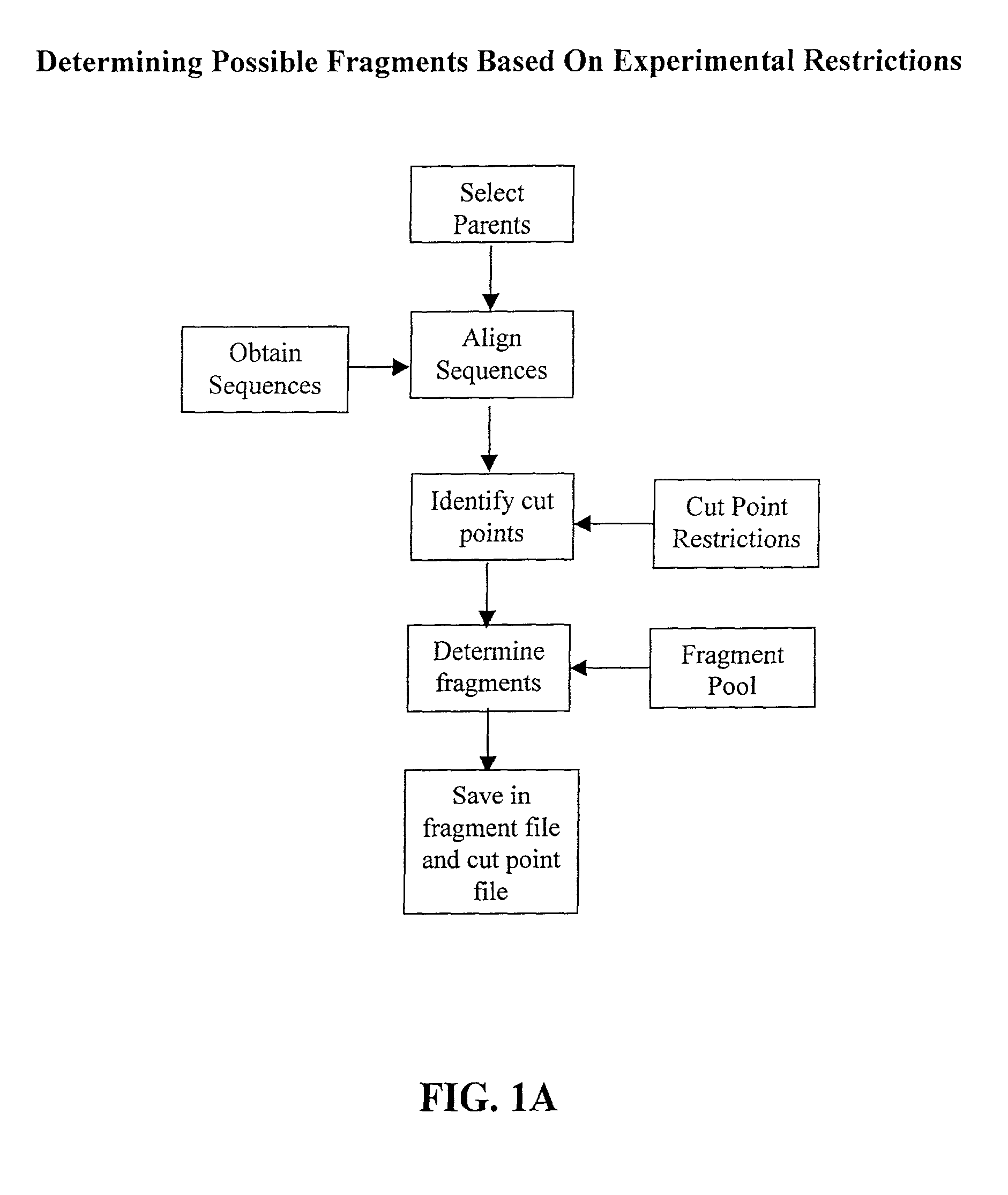
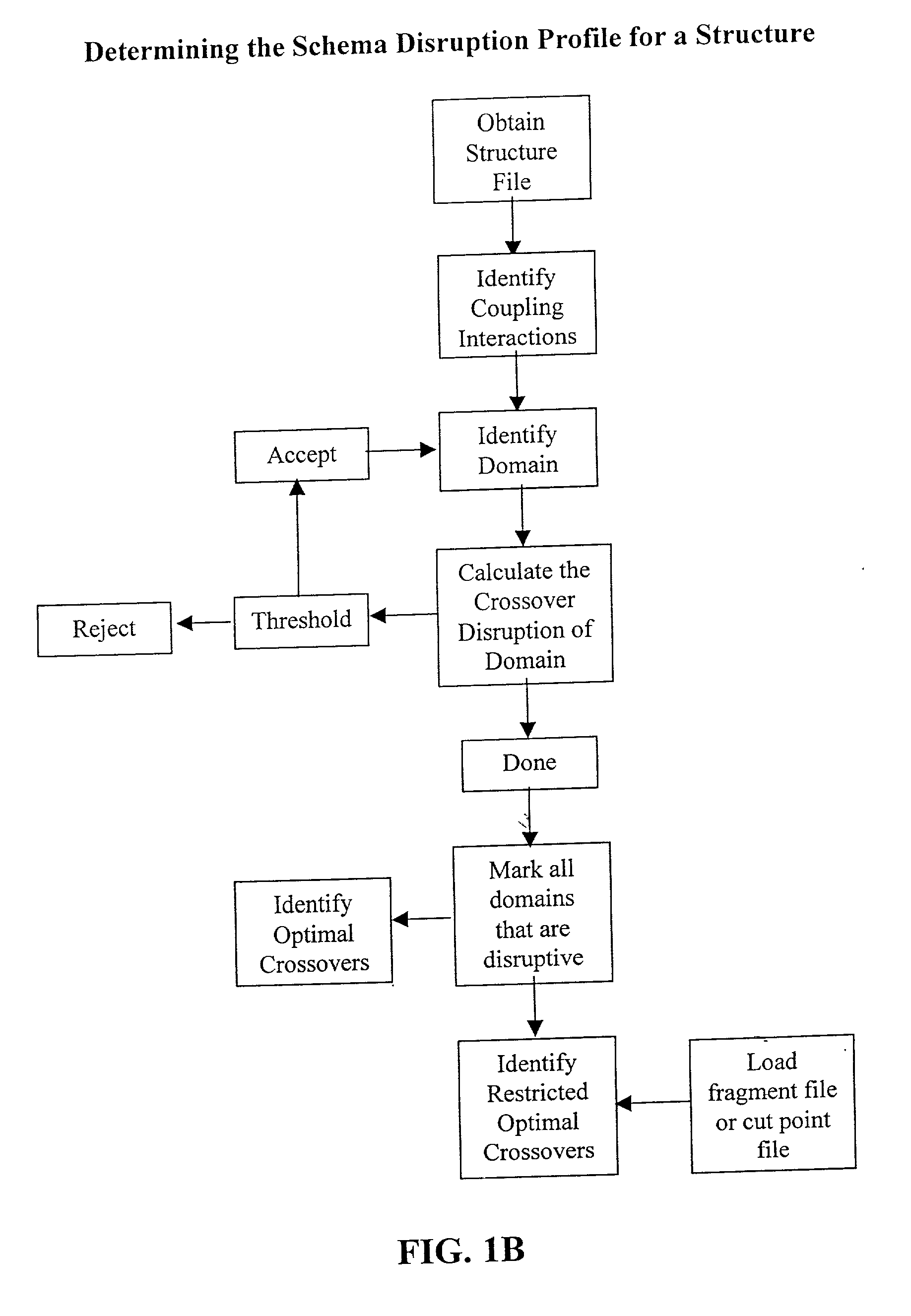
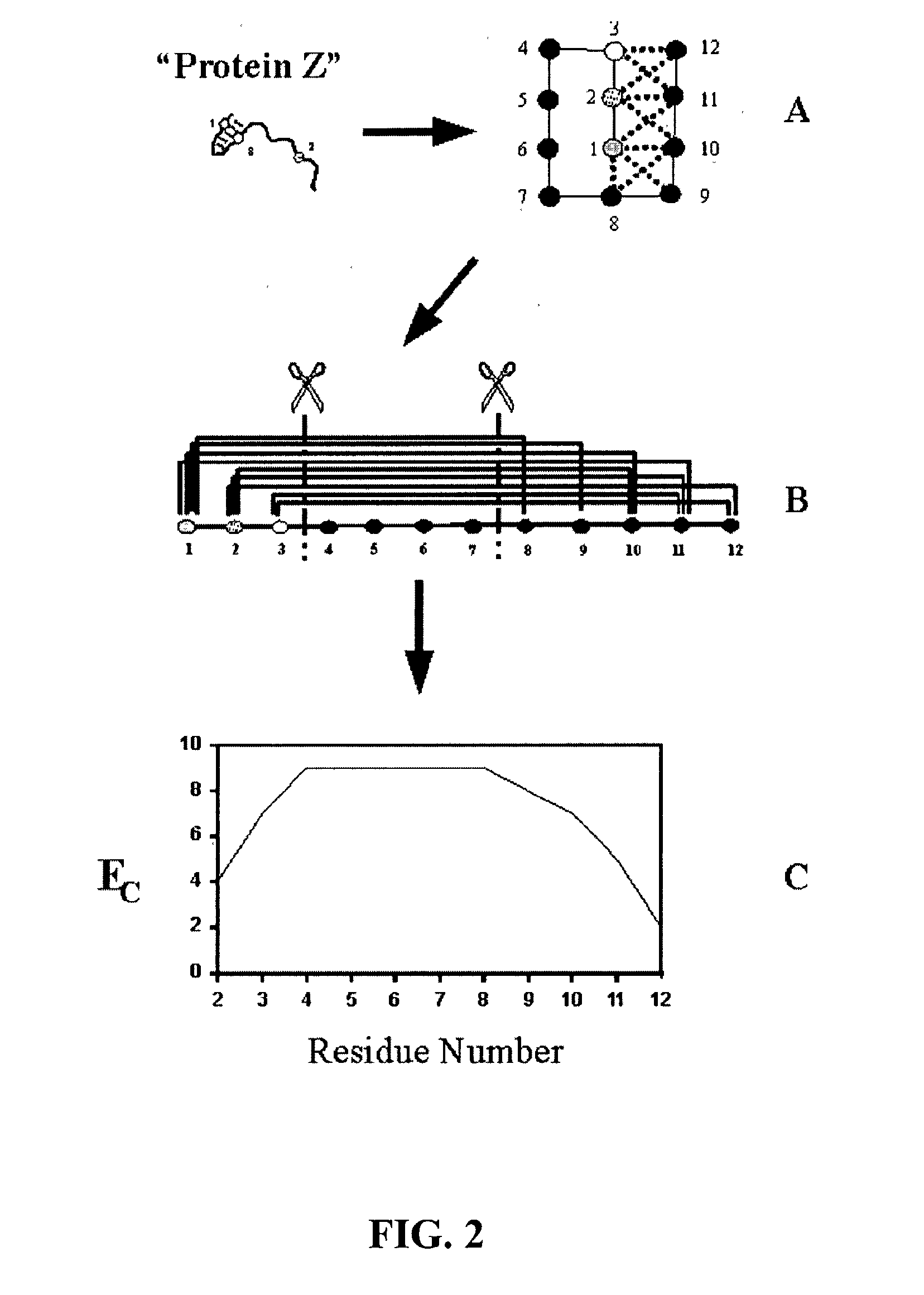
The invention relates to improved methods for directed evolution of polymers, including directed evolution of nucleic acids and proteins. Specifically, the methods of the invention include analytical methods for identifying "crossover locations" in a polymer. Crossovers at these locations are less likely to disrupt desirable properties of the protein, such as stability or functionality. The invention further provides improved methods for directed evolution wherein the polymer is selectively recombined at the identified "crossover locations". Crossover disruption profiles can be used to identify preferred crossover locations. Structural domains of a biopolymer can also be identified and analyzed, and domains can be organized into schema. Schema disruption profiles can be calculated, for example based on conformational energy or interatomic distances, and these can be used to identify preferred or candidate crossover locations. Computer systems for implementing analytical methods of the invention are also provided.
Owner:WANG ZHEN GANG +3
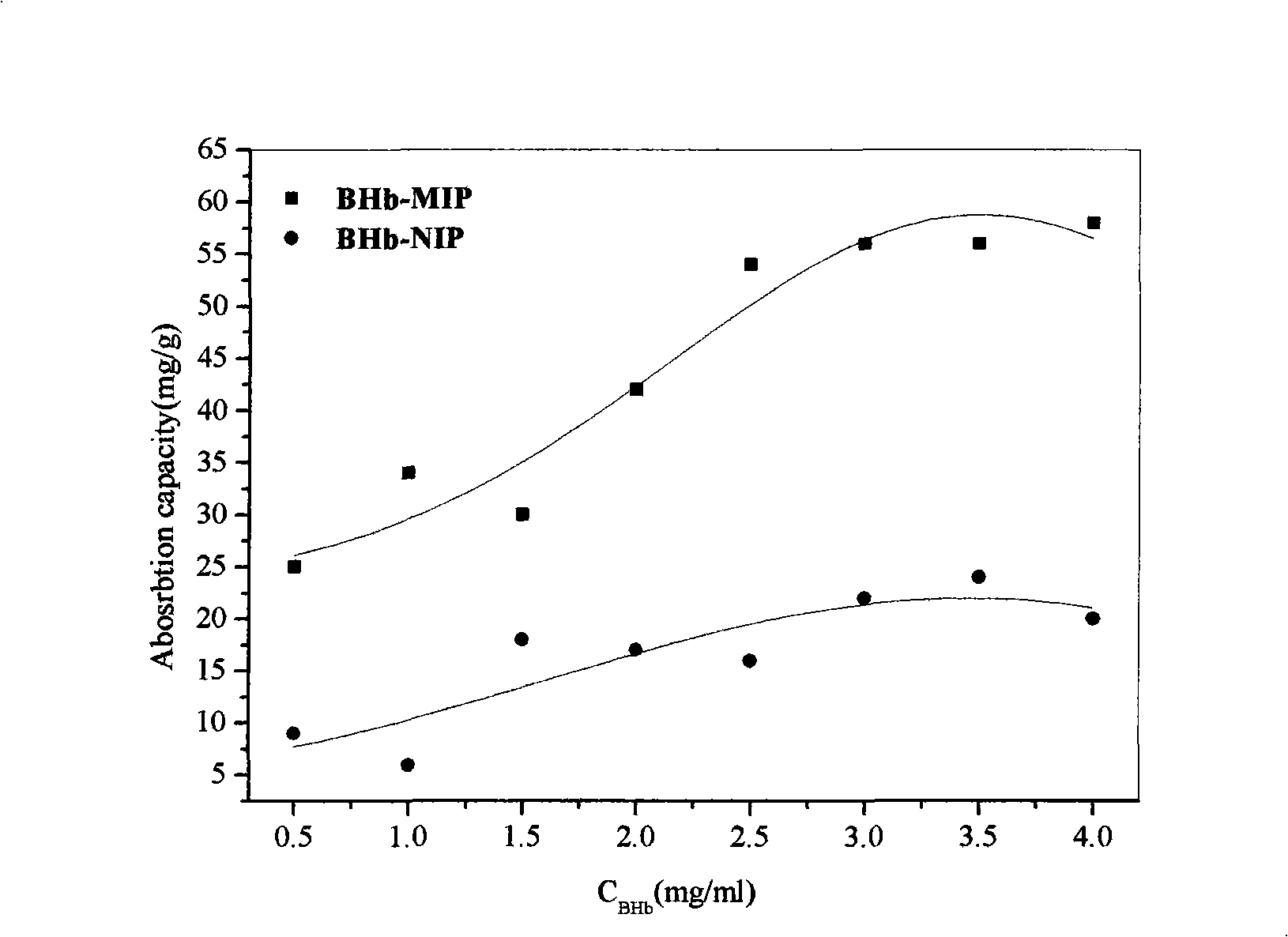
Method for preparing protein magnetic blotting nanospheres
InactiveCN101347721AAvoid complicated centrifugationSimplified centrifugation processOther chemical processesPolymerSerum protein
The invention relates to a preparation method of a protein magnetic imprinting nano sphere, in particular to a preparation method of a protein magnetic imprinting nano sphere which takes one of hemoglobin, muramidase or serum protein as template molecules; the preparation method includes the following steps: synthesizing magnetic nano particles which are Fe3O4 nano particles covered with silicon dioxide on the surfaces; carrying out silanization to the amino-groups on the surface of the magnetic sphere; using glutaric dialdehyde to connect protein; using silylation agent to fix the space structure of template protein; alkaline eluting the protein on the surface of the magnetic sphere to form imprinting sites. The hemoglobin, muramidase or serum protein magnetic nano imprinting polymer sphere prepared by the invention combines the precise and specific identification property of molecular imprinting and the quick separation property of the magnetic nano sphere under the action of an external magnetic field, integrates the advantages of molecular imprinting and the magnetic nano sphere, avoids complex centrifugation operation, plays vital roles in the separation of cells, protein and nucleic acid, the detection of biomolecules, tumor diagnosis and medicine targeting therapy and has promising application prospect in the field of isolation analysis.
Owner:NANKAI UNIV
Methods of modulating and identifying agents that modulate intracellular calcium
ActiveUS20090311720A1Increasing level of and expression of and activity of and molecular interactionReducing level of and expression of and activity of and molecular interactionSenses disorderNervous disorderProtein insertionCell biology
Methods are provided for identifying agents that modulate intracellular calcium. Also provided are methods of modulating calcium within cells and methods of identifying proteins involved in modulating intracellular calcium.
Owner:CALCIMEDICA
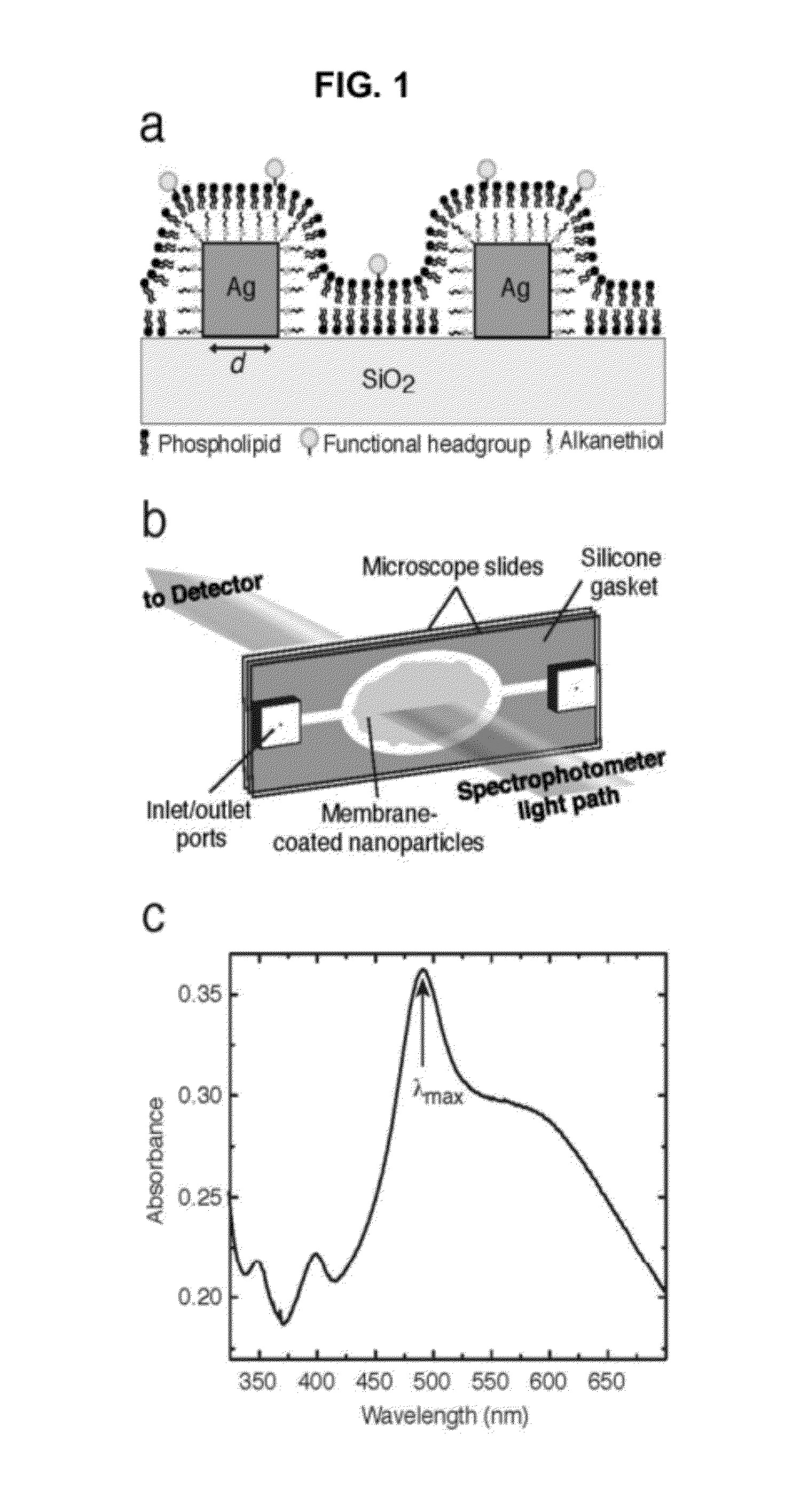
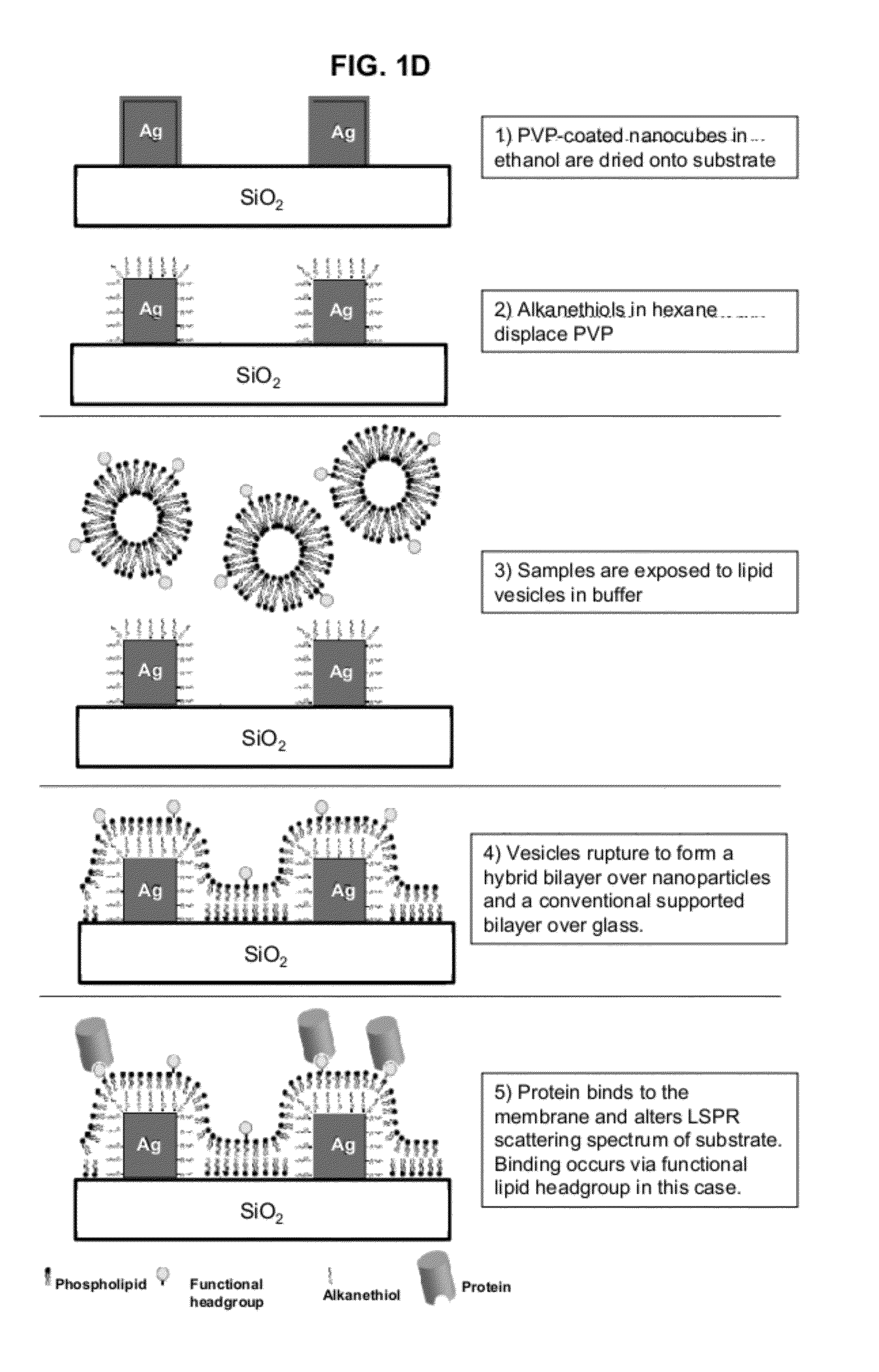
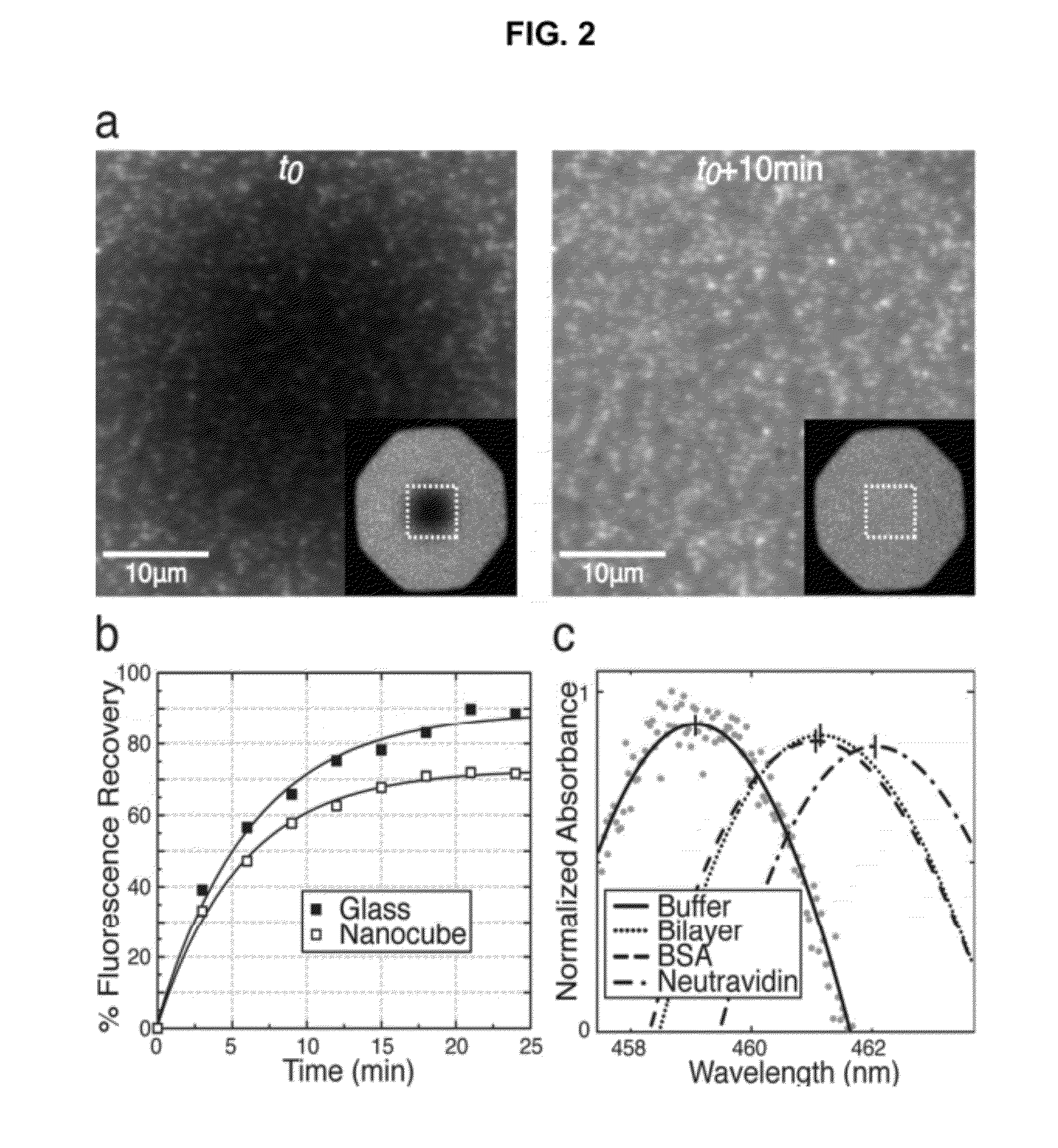
Plasmonic System for Detecting Binding of Biological Molecules
InactiveUS20120208174A1Simplicity of fabricationEasy to parallelizeMicrobiological testing/measurementScattering properties measurementsProtein insertionNanoparticle
Detection and characterization of molecular interactions on membrane surfaces is important to biological and pharmacological research. In one embodiment, silver nanocubes interfaced with glass-supported model membranes form a label-free sensor that measures protein binding to the membrane. The present device and technique utilizes plasmon resonance scattering of nanoparticles, which are chemically coupled to the membrane. In contrast to other plasmonic sensing techniques, this method features simple, solution-based device fabrication and readout. Static and dynamic protein / membrane binding are monitored and quantified.
Owner:RGT UNIV OF CALIFORNIA
Protein synthesis system for in-vitro protein synthesis, kit and preparation method for protein through in-vitro synthesis
ActiveCN108535489AIncrease productionLow costMicroorganism lysisBiological testingCell freeProtein insertion
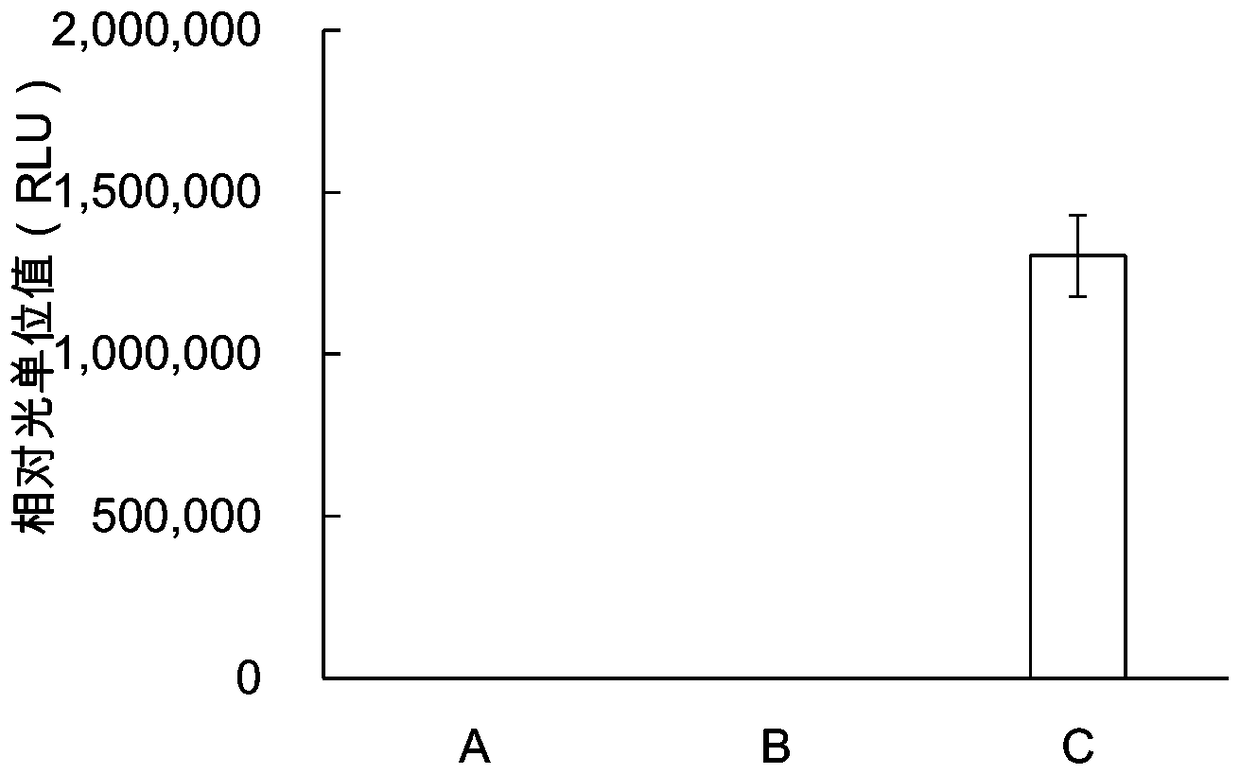
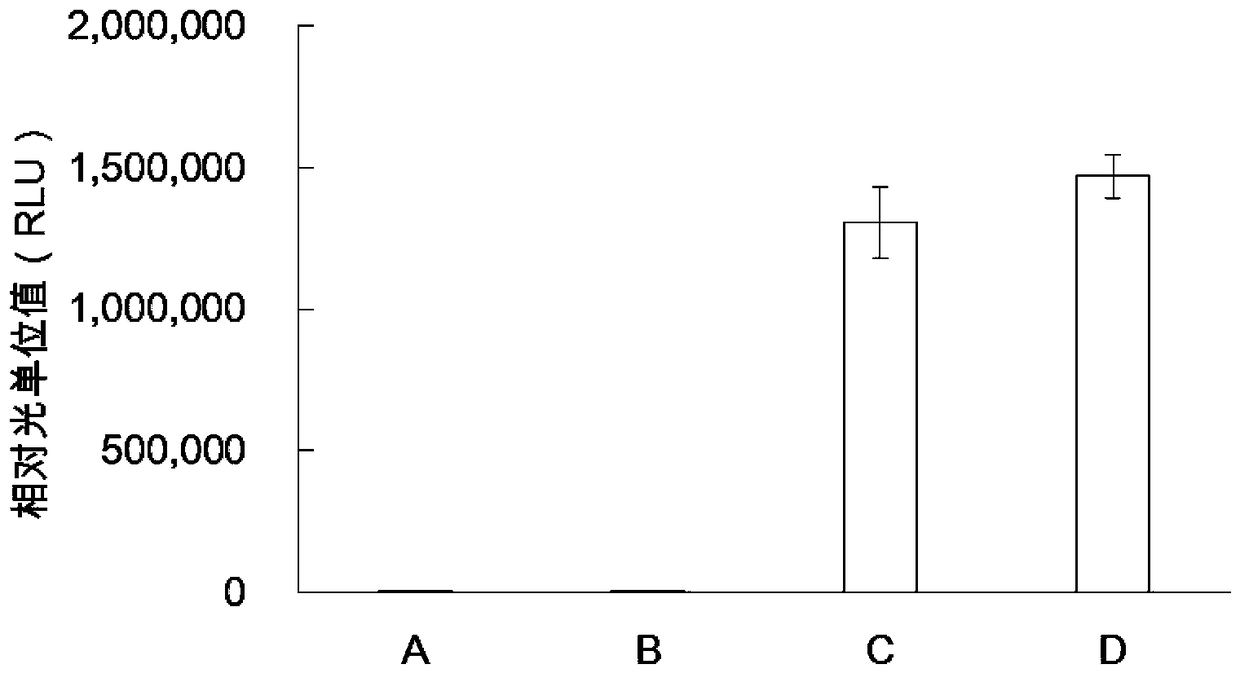
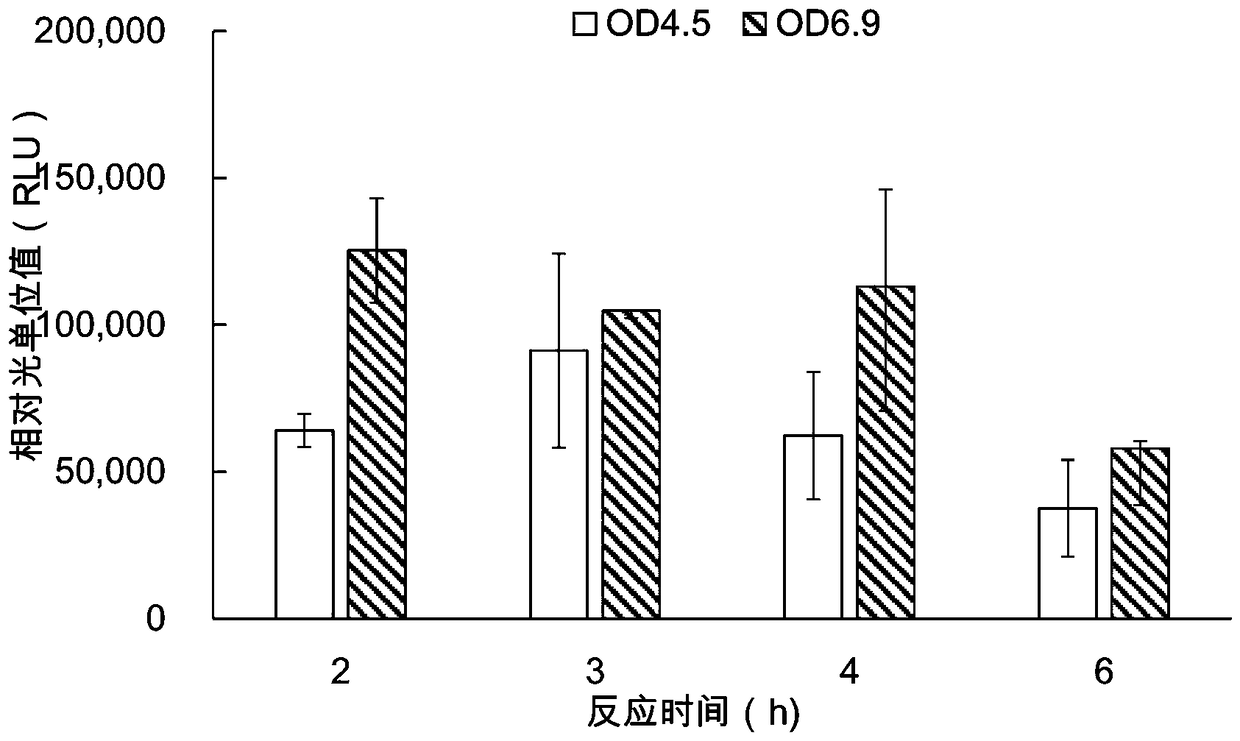
The invention provides a protein synthesis system for in-vitro protein synthesis, a kit and a preparation method for a protein through in-vitro synthesis. Specifically, the in-vitro cell-free expression system provided by the invention is capable of extremely efficiently synthesizing proteins and synthesizing complex proteins. Moreover, due to the in-vitro cell-free expression system disclosed bythe invention, the relative light unit value of the activity of the synthesized luciferase is higher than that of the conventional commercial system (such as a rabbit reticulocyte in-vitro expressionsystem) by at least one order of magnitude (more than or equal to 10 times or higher).
Owner:KANGMA SHANGHAI BIOTECH LTD
Method of identifying and locating immunobiologically-active linear peptides
InactiveUS6780598B1Without the need for time consuming and expensive testing regimesIncrease the number ofPeptide/protein ingredientsPeptide sourcesProtein insertionBiology
The present invention relates to identifying protein epitopes and more particularly to a novel method for identifying, determining the location, optimal length of amino acid residues and immunobiological potency of protein epitopes by fitting a hydrophilicity and / or hydrophobicity plot generated for the amino acid linear sequence of a polypeptide to a mathematically generated continuous curve thereby generating at least one set of potential epitopes which include ranked potential epitopes having a specific number of amino acid residues. The immunobiologically-active linear peptides are deemed the potential epitopes that exhibit the most alternating positioning about an equilibrium position when juxtaposed on the hydrophilicity and / or hydrophobicity plot and their optimal length corresponds to the specific number of amino acid residues in the set of ranked potential epitopes. The amino acid sequence of the protein epitopes of the present invention exhibit a hydrophobic-hydrophilic-hydrophobic motif
Owner:KOKOLUS WILLIAM J
Nucleic acid construct for endogenously expressing RNA polymerase in cells
The invention provides a nucleic acid construct for endogenously expressing RNA polymerase in cells, and particularly discovers a novel nucleic acid construct capable of greatly enhancing protein translation efficiency. The nucleic acid construct consists of a promoter with specific promotion intensity (such as RNR2, ADH1, GAPDH, TEF1, PGK1 and SED1) and coding sequences of proteins of RNAP (suchas T7RNAP, T3RNAP, T4RNAP and T5RNAP), if the nucleic acid construct of the invention is applied into a yeast extracorporeal protein synthesis system, the activity of synthesized luciferase is quite high relative to light unit value (RLU), and the effect can be the same as the effect of T7RNAP added exogenously. The effect can reach up to 6.6* 107.
Owner:KANGMA SHANGHAI BIOTECH LTD
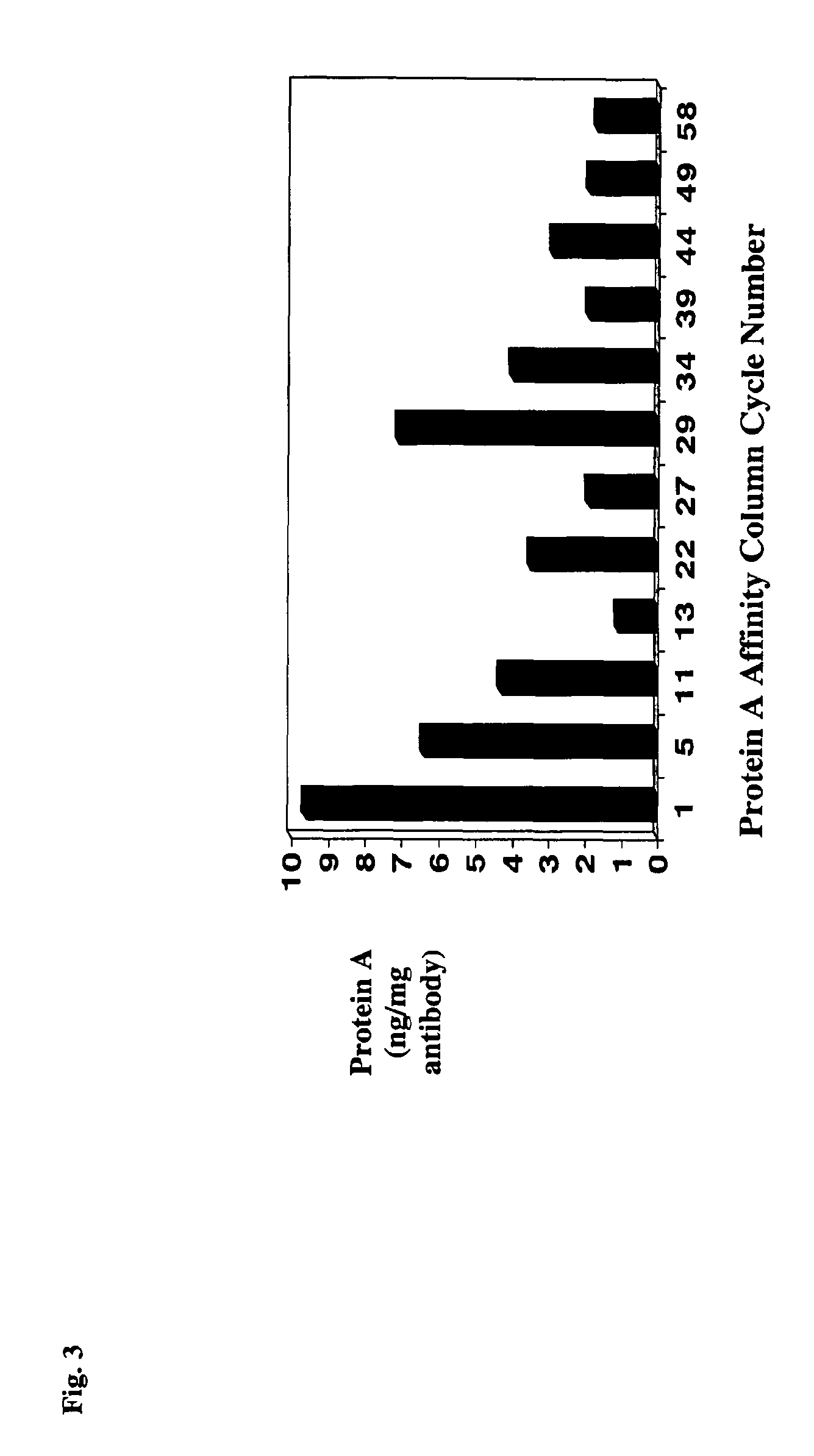
Antibody purification by protein a and ion exchange chromatography
InactiveUS7847071B2Serum immunoglobulinsSolid sorbent liquid separationIon chromatographyProtein insertion
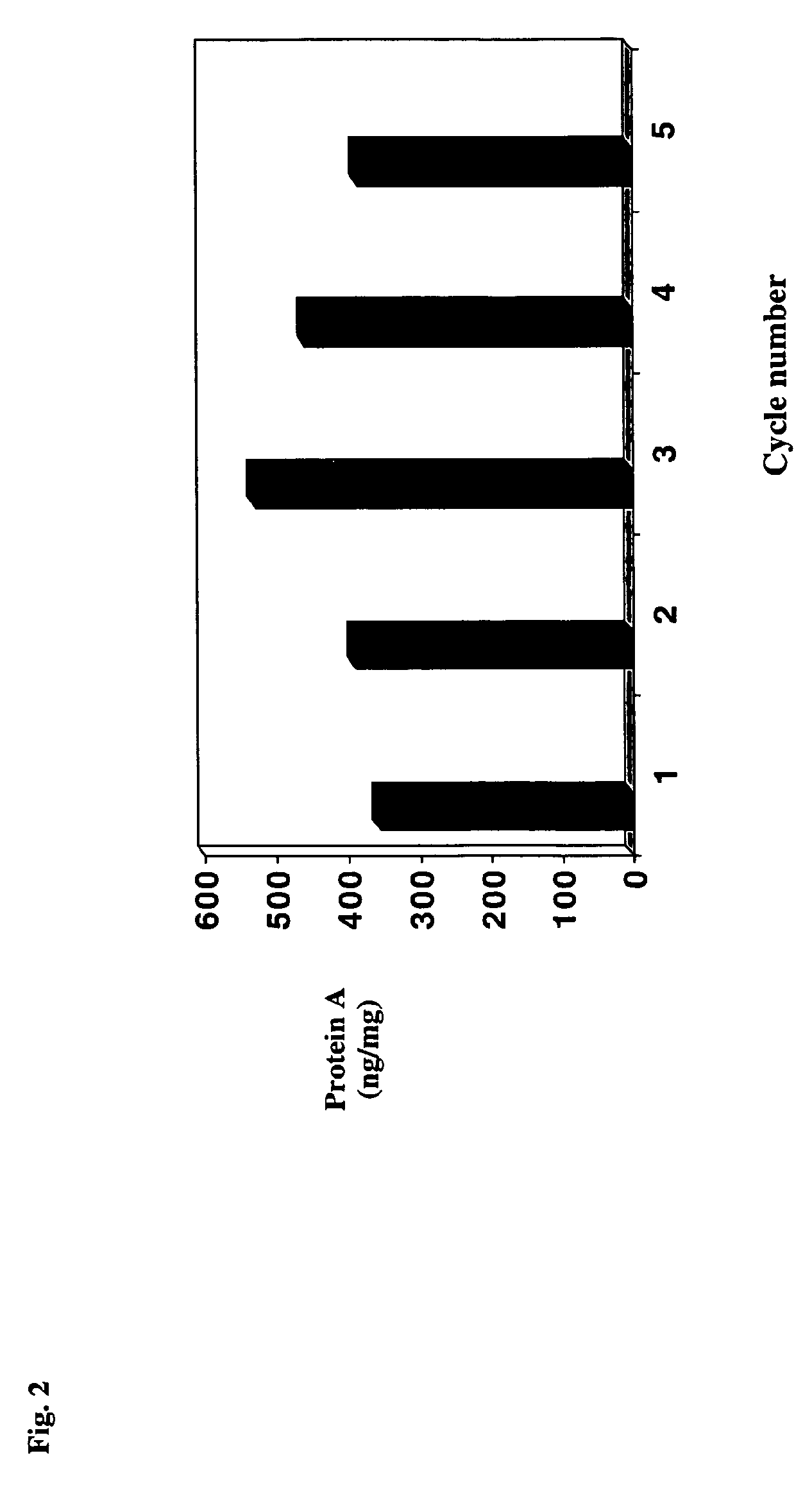
A novel method for selectively removing leaked protein A from antibody purified by means of protein A affinity chromatography is disclosed.
Owner:LONZA BIOLOGICS PLC
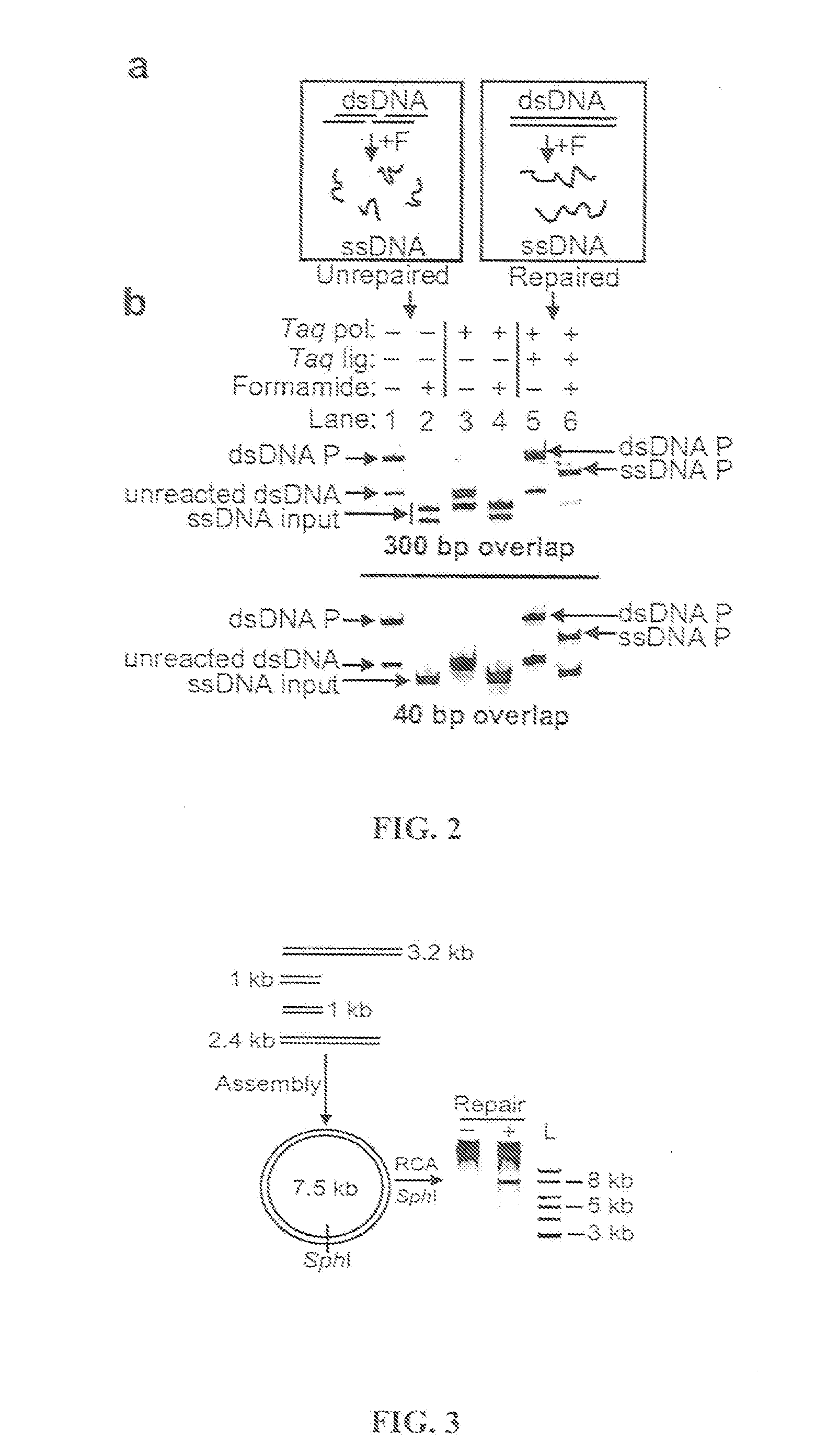
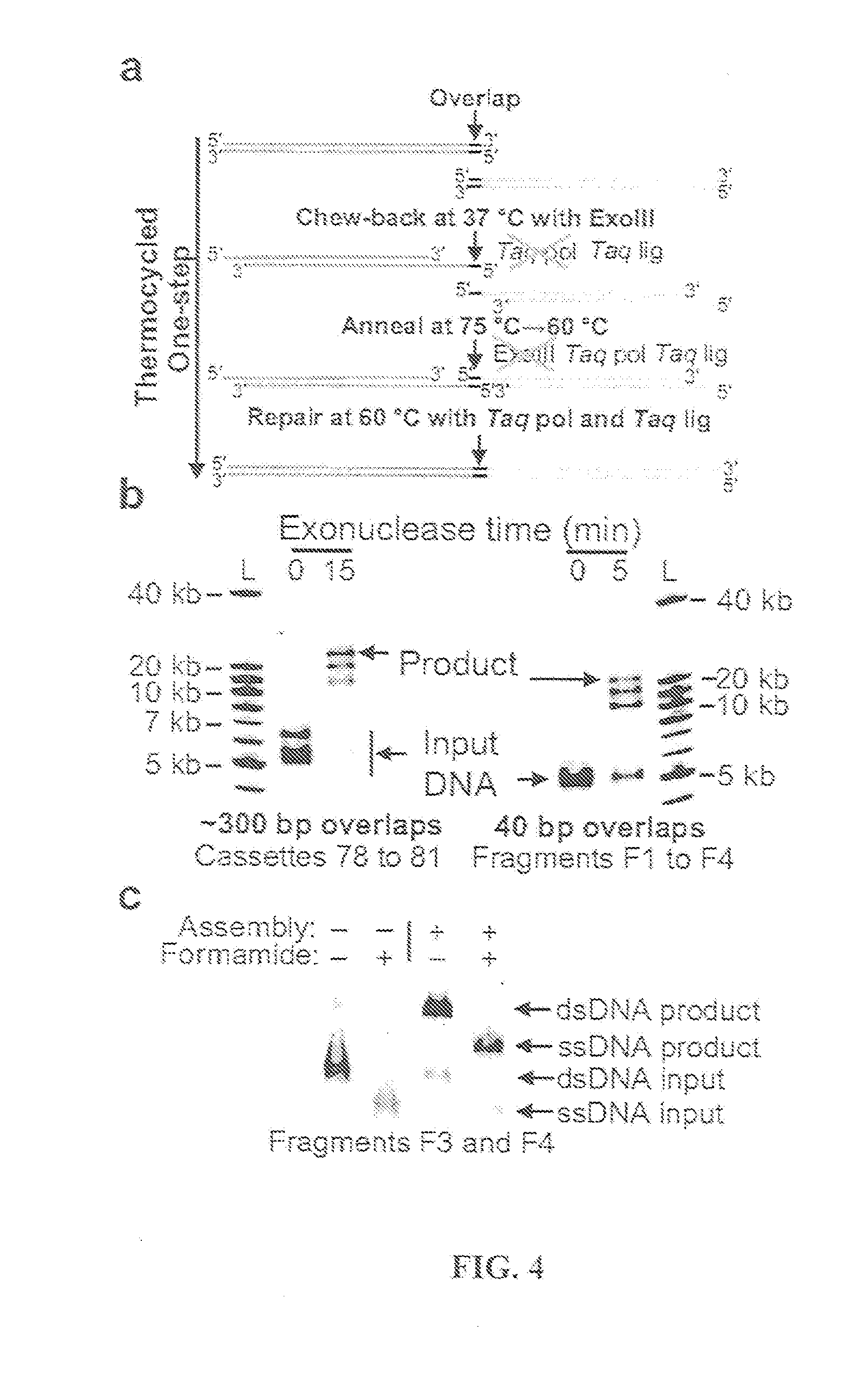
Methods for in vitro joining and combinatorial assembly of nucleic acid molecules
The present invention relates to methods of joining two or more double-stranded (ds) or single-stranded (ss) DNA molecules of interest in vitro, wherein the distal region of the first DNA molecule and the proximal region of the second DNA molecule of each pair share a region of sequence identity. The method allows the joining of a large number of DNA fragments, in a predetermined order and orientation, without the use of restriction enzymes. It can be used, e.g., to join synthetically produced sub-fragments of a gene or genome of interest. Kits for performing the method are also disclosed. The methods of joining DNA molecules may be used to generate combinatorial libraries useful to generate, for example, optimal protein expression through codon optimization, gene optimization, and pathway optimization.
Owner:SYNTHETIC GENOMICS INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com