Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
324 results about "Directed evolution" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
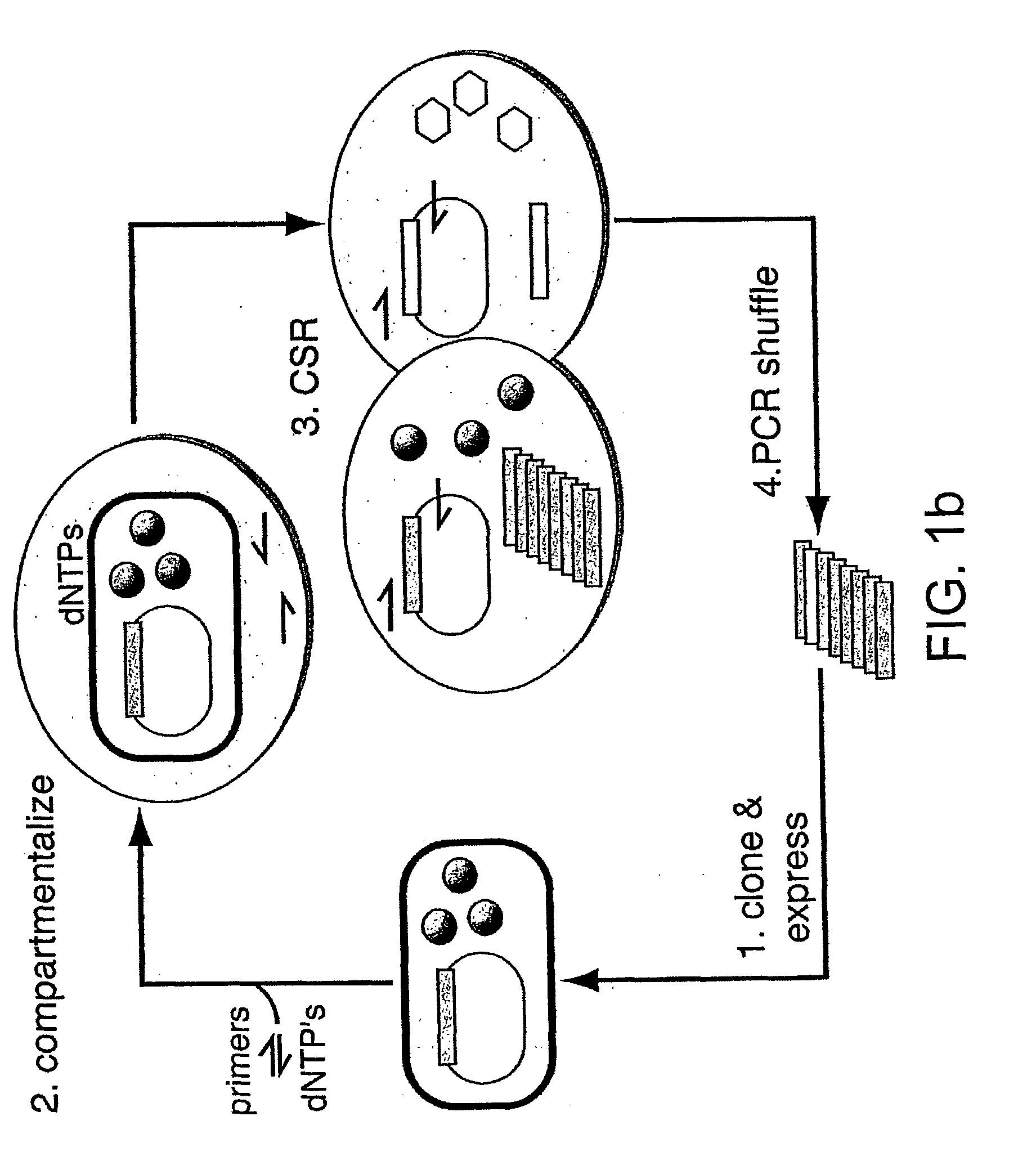
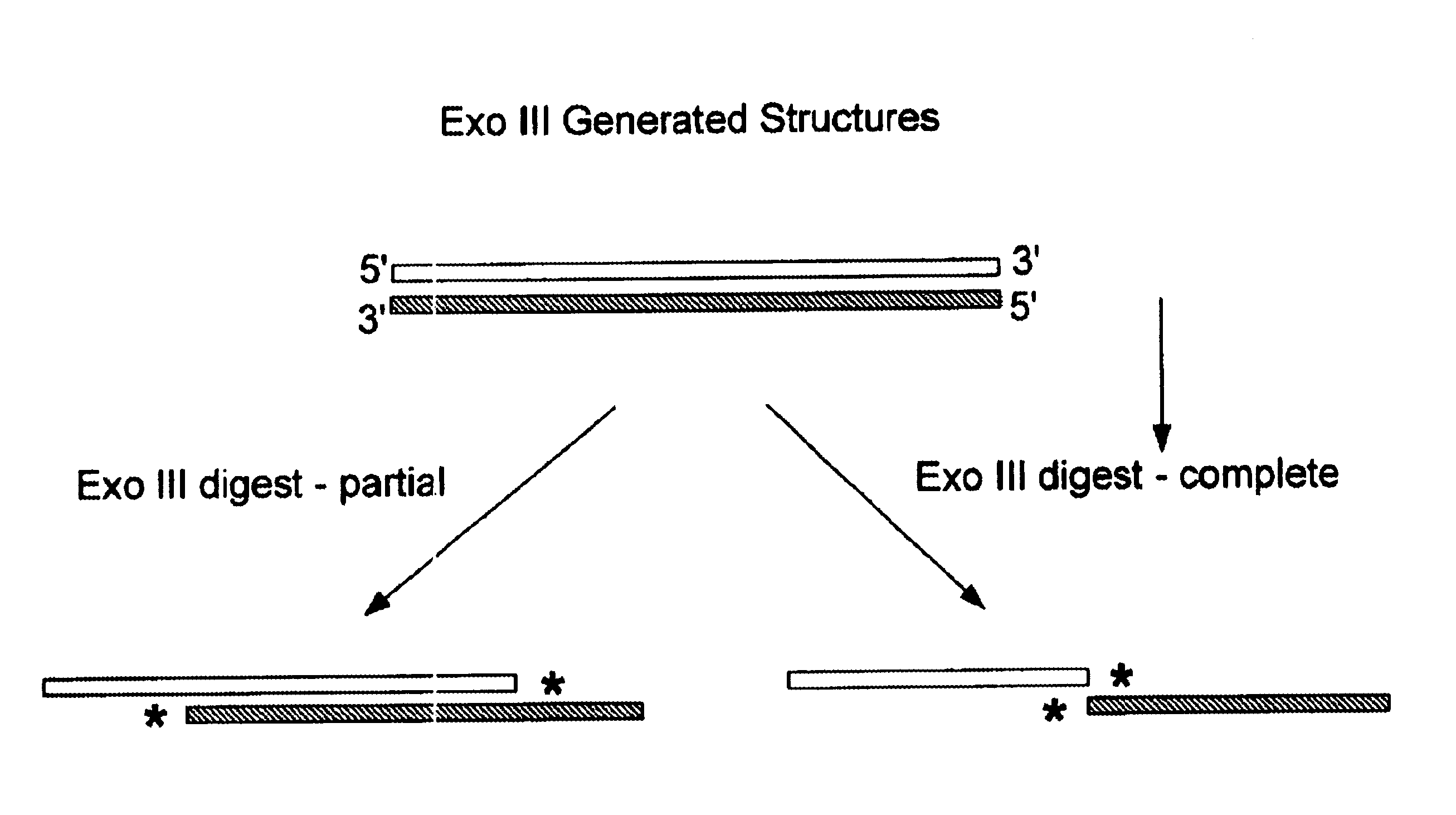
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis (creating a library of variants), selection (expressing those variants and isolating members with the desired function) and amplification (generating a template for the next round). It can be performed in vivo (in living organisms), or in vitro (in cells or free in solution). Directed evolution is used both for protein engineering as an alternative to rationally designing modified proteins, as well as studies of fundamental evolutionary principles in a controlled, laboratory environment.
Directed evolution of grg31 and grg36 epsp synthase enzymes
Compositions and methods for conferring herbicide resistance or tolerance to bacteria, plants, plant cells, tissues and seeds are provided. Compositions include polynucleotides encoding herbicide resistance or tolerance polypeptides, vectors comprising those polynucleotides, and host cells comprising the vectors. The nucleotide sequences of the invention can be used in DNA constructs or expression cassettes for transformation and expression in organisms, including microorganisms and plants. Compositions also include transformed bacteria, plants, plant cells, tissues, and seeds. In particular, isolated polynucleotides encoding glyphosate resistance or tolerance polypeptides are provided, particularly polypeptide variants of SEQ ID NO:2 and 4. Additionally, amino acid sequences corresponding to the polynucleotides are encompassed. In particular, the present invention provides for isolated polynucleotides containing nucleotide sequences encoding the amino acid sequence shown in SEQ ID NO:7-28, or the nucleotide sequence set forth in SEQ ID NO:29 or 30.
Owner:BASF AGRICULTURAL SOLUTIONS SEED LLC
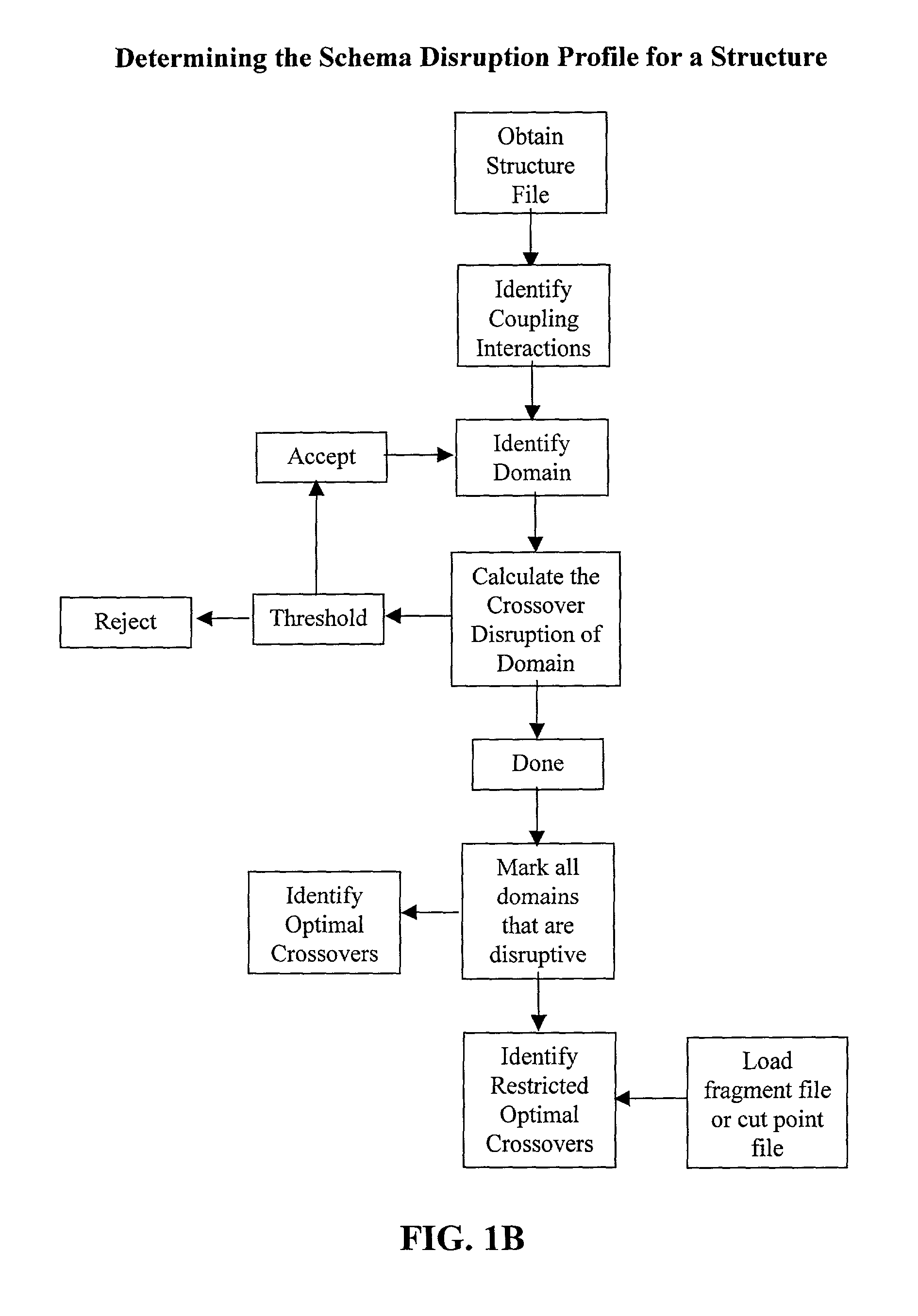
Optimization of crossover points for directed evolution
ActiveUS7620500B2Microbiological testing/measurementAnalogue computers for chemical processesNucleotideNucleotide sequencing
Owner:CODEXIS MAYFLOWER HLDG LLC
Directed evolution method
We describe a method of selecting an enzyme having replicase activity, the method comprising the steps of: (a) providing a pool of nucleic acids comprising members each encoding a replicase or a variant of the replicase; (b) subdividing the pool of nucleic acids into compartments, such that each compartment comprises a nucleic acid member of the pool together with the replicase or variant encoded by the nucleic acid member; (c) allowing nucleic acid replication to occur; and (d) detecting amplification of the nucleic acid member by the replicase. Methods for selecting agents capable of modulating replicase activity, and for selecting interacting polypeptides are also disclosed.
Owner:UK RES & INNOVATION LTD
Non-stochastic generation of genetic vaccines and enzymes
InactiveUS6713279B1Improve abilitiesPotent and direct effect on proliferation and Ig-synthesisAnimal cellsHydrolasesAntigenBiological property
This invention provides methods of obtaining novel polynucleotides and encoded polypeptides by use of non-stochastic methods of directed evolution (DirectEvolution(TM)). These methods include non-stochastic polynucleotide site-saturation mutagenesis (Gene Site Saturation Mutagenesis(TM)) and non-stochastic polynucleotide reassembly (GeneReassembly(TM)). Through use of the claimed methods, genetic vaccines, enzymes, and other desirable molecules can be evolved towards desirable properties. For example, vaccine vectors can be obtained that exhibit increased efficacy for use as genetic vaccines. Vectors obtained by using the methods can have, for example, enhanced antigen expression, increased uptake into a cell, increased stability in a cell, ability to tailor an immune response, and the like. This invention provides methods of obtaining novel enzymes that have optimized physical & / or biological properties. Furthermore, this invention provides methods of obtaining a variety of novel biologically active molecules, in the fields of antibiotics, pharmacotherapeutics, and transgenic traits.
Owner:BP CORP NORTH AMERICA INC
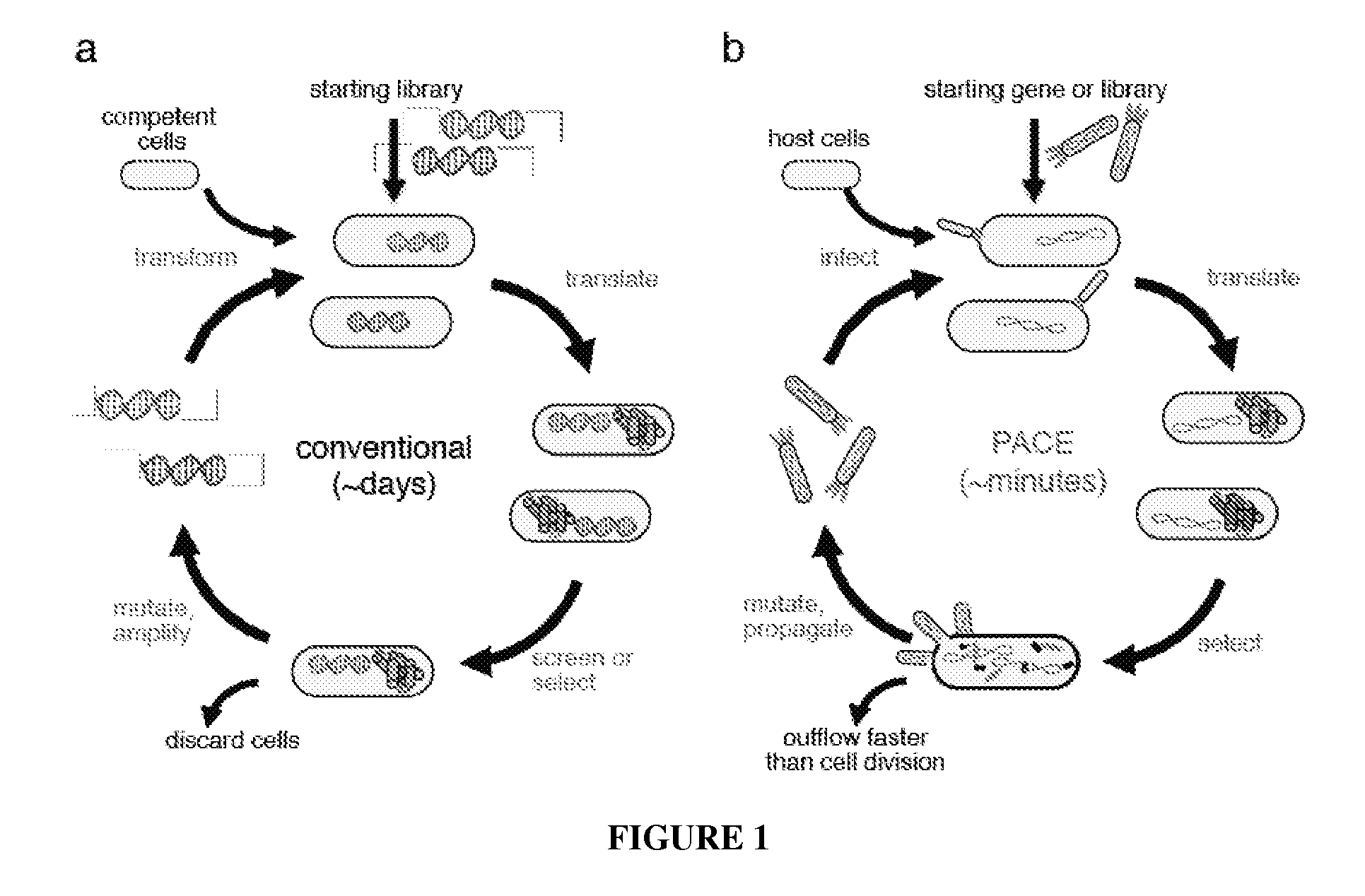
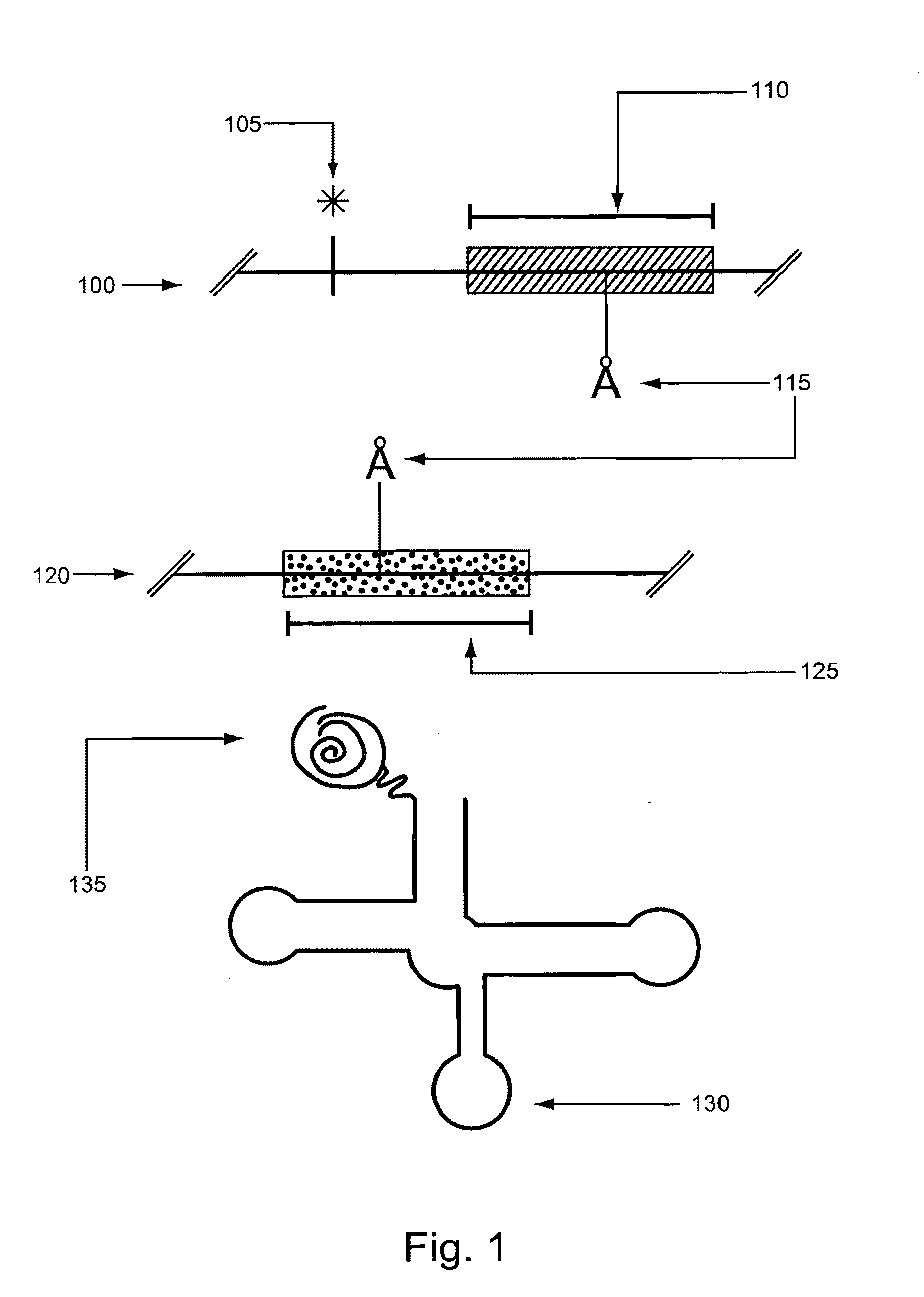
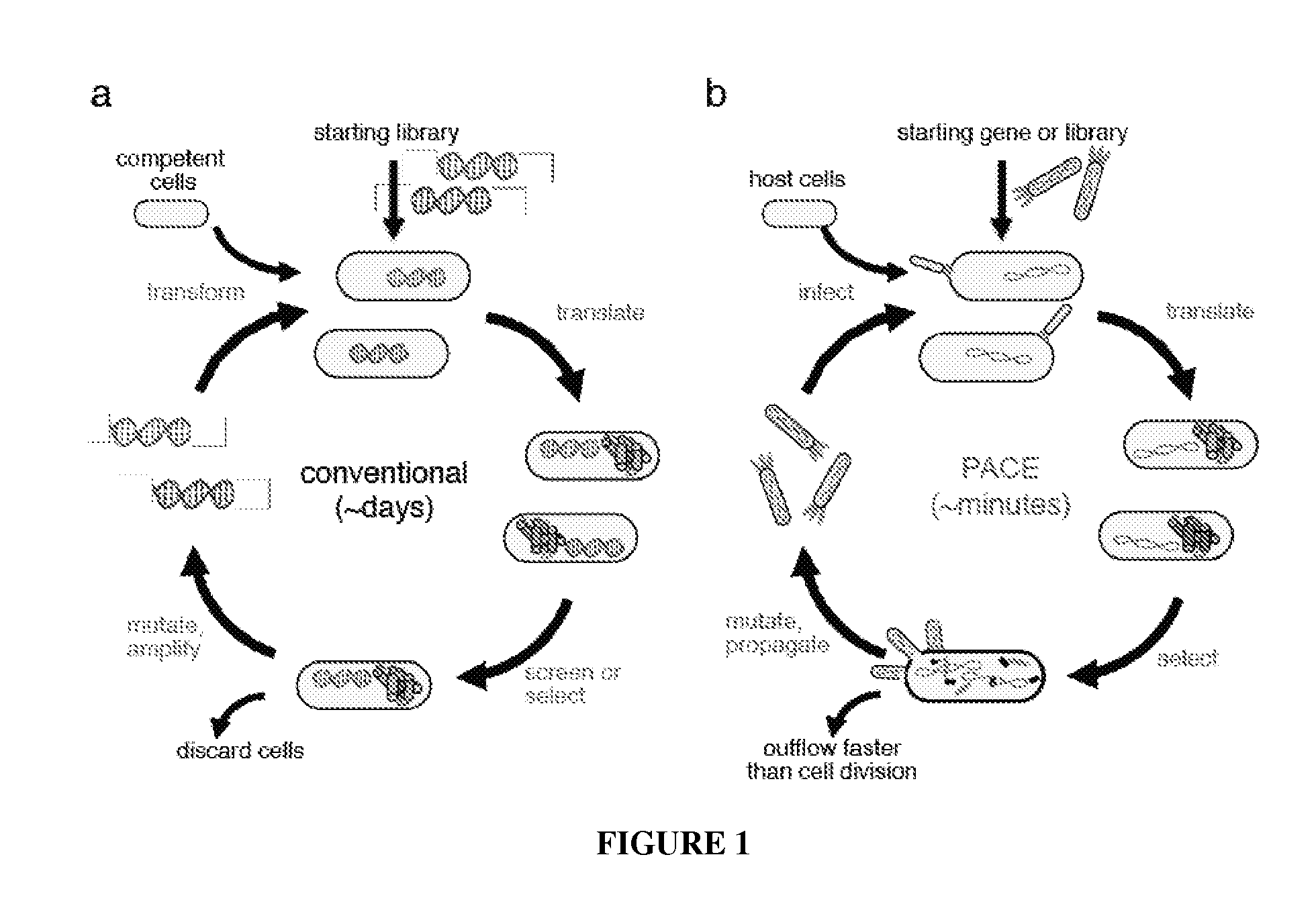
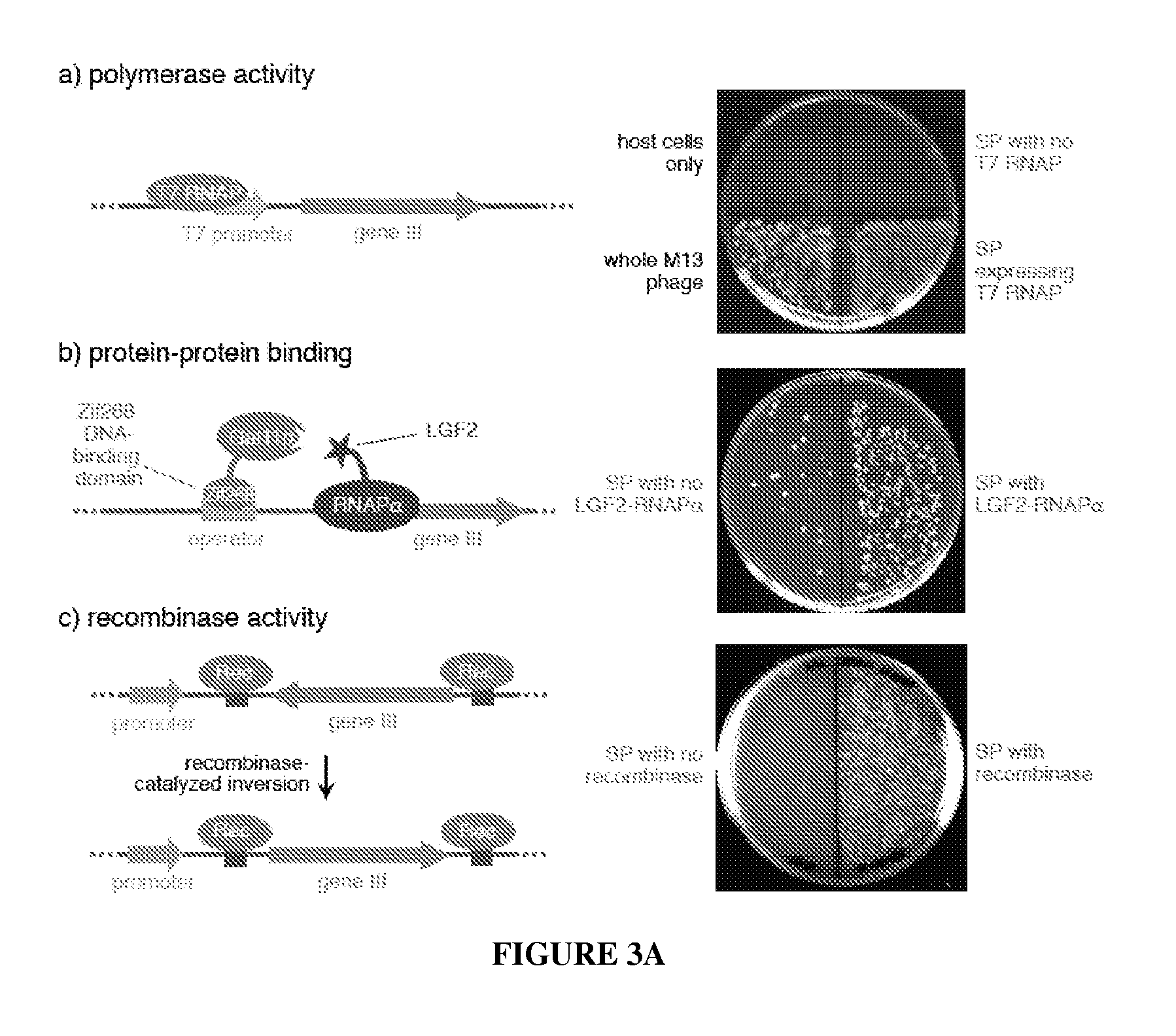
Continuous directed evolution of proteins and nucleic acids
ActiveUS9023594B2Microbiological testing/measurementMutant preparationCell biologyDirected evolution
The present invention discloses generalizable methods of evolving nucleic acids and proteins utilizing continuous directed evolution. The invention discloses methods of passing a nucleic acid from cell to cell in a desired function-dependent manner. The linkage of the desired function and passage of the nucleic acid from cell to cell allows for continuous selection and mutation of the nucleic acid.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Gene recombination and hybrid protein development
InactiveUS20020045175A1Preserve stabilityPreserve desired propertyMicrobiological testing/measurementBiological testingBiopolymerHybrid protein
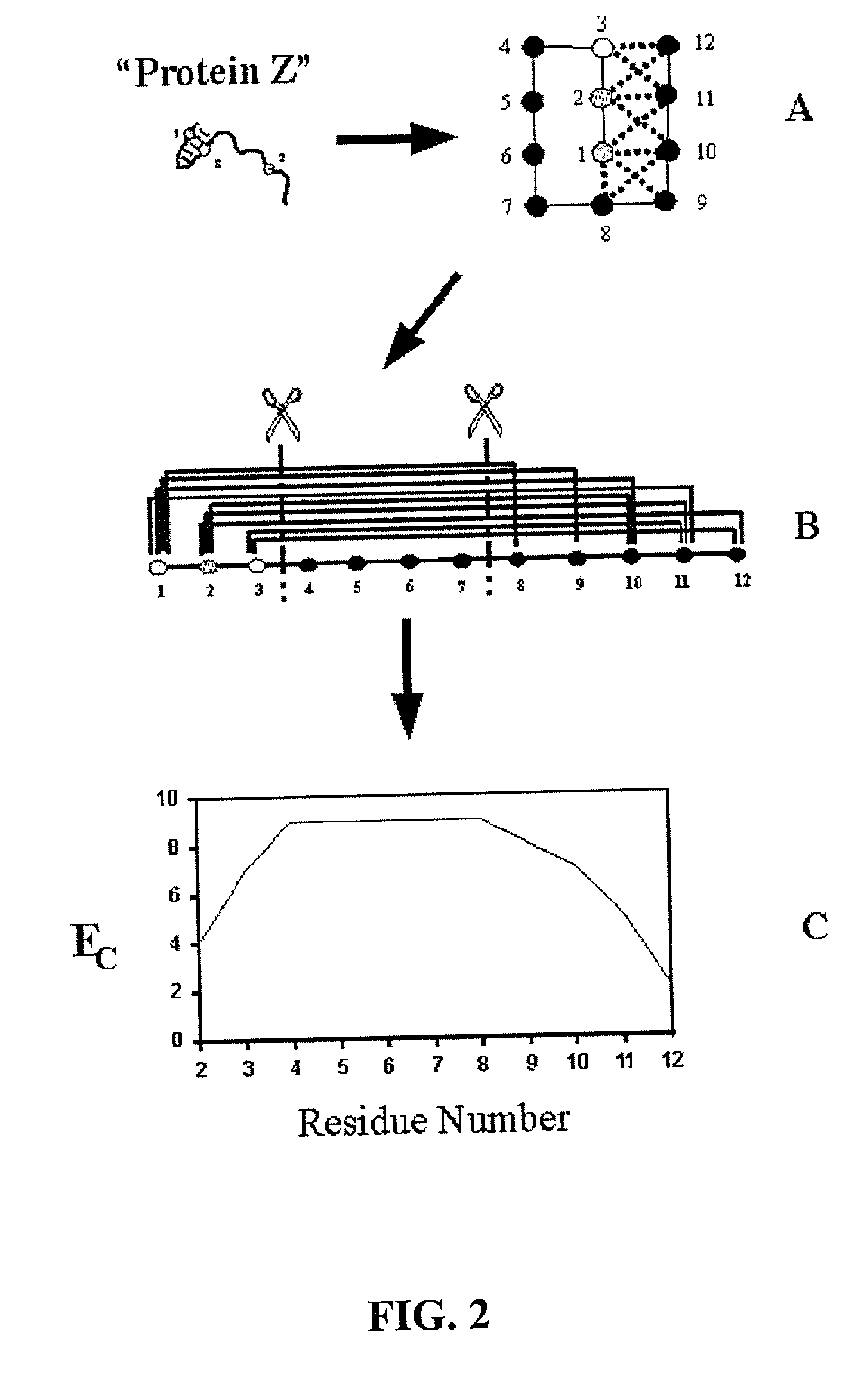
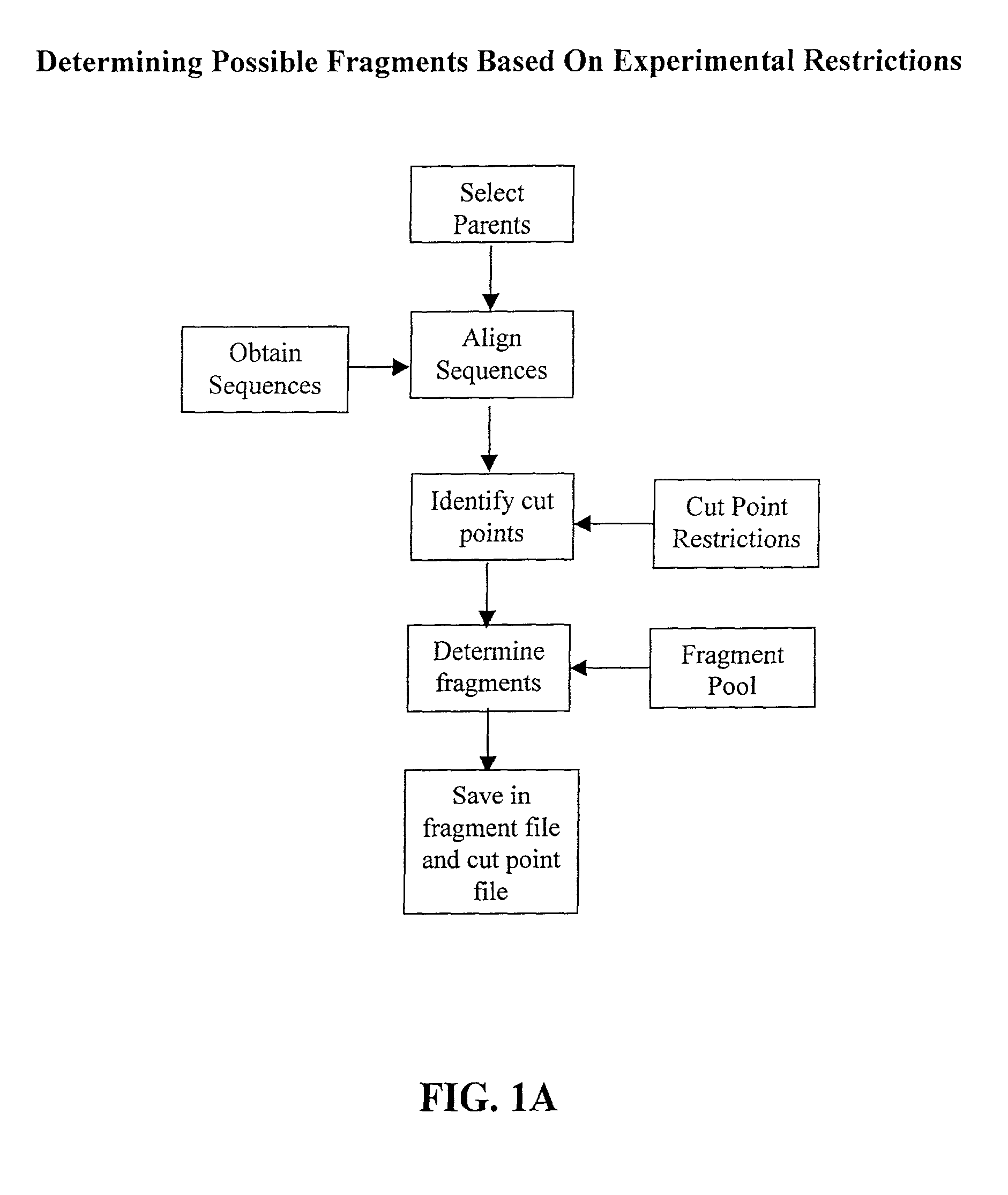
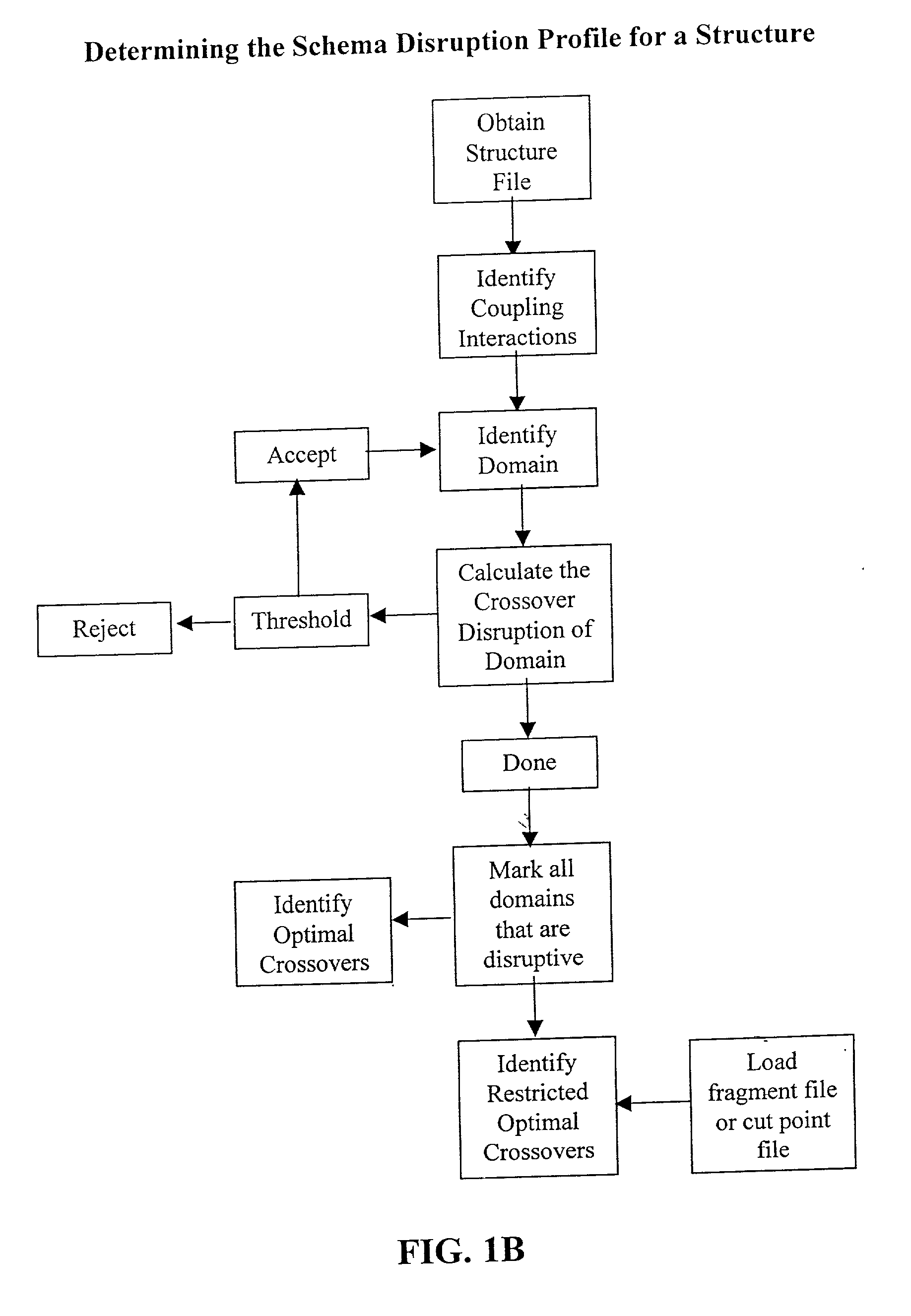
The invention relates to improved methods for directed evolution of polymers, including directed evolution of nucleic acids and proteins. Specifically, the methods of the invention include analytical methods for identifying "crossover locations" in a polymer. Crossovers at these locations are less likely to disrupt desirable properties of the protein, such as stability or functionality. The invention further provides improved methods for directed evolution wherein the polymer is selectively recombined at the identified "crossover locations". Crossover disruption profiles can be used to identify preferred crossover locations. Structural domains of a biopolymer can also be identified and analyzed, and domains can be organized into schema. Schema disruption profiles can be calculated, for example based on conformational energy or interatomic distances, and these can be used to identify preferred or candidate crossover locations. Computer systems for implementing analytical methods of the invention are also provided.
Owner:CALIFORNIA INST OF TECH
Screening methods
The invention relates to methods for the screening, identification and / or application of microorganisms of use in imparting beneficial properties to plants. In one embodiment, the method involves subjecting one or more plants to a growth medium in the presence of a set of microorganisms, selecting one or more plant, acquiring one or more microorganisms associated with the selected plant(s) and repeating the process one or more times. The method further involves the step of subjecting the one or more microorganisms to a conditioning and / or directed evolution process.
Owner:BIOCONSORTIA
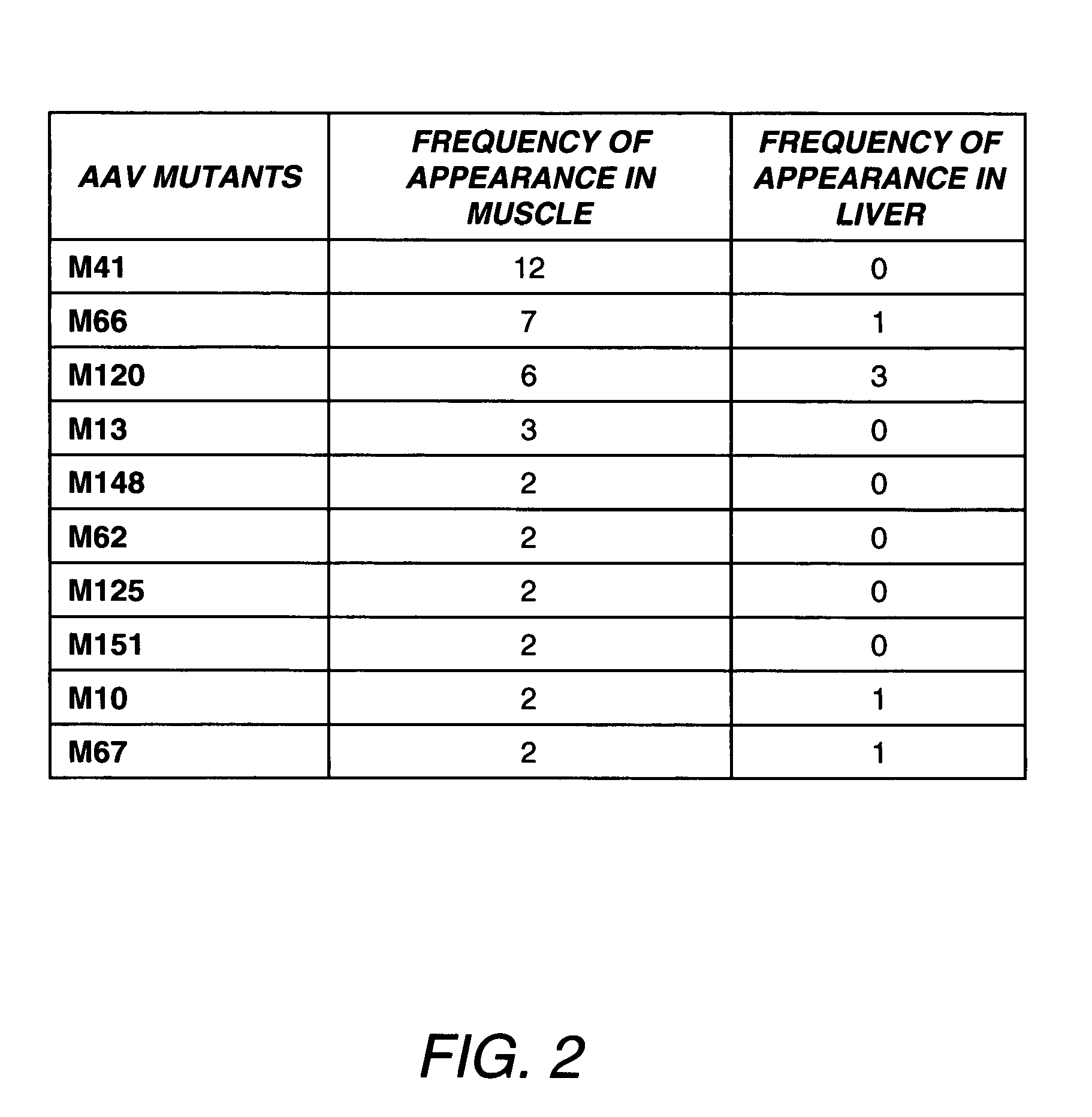
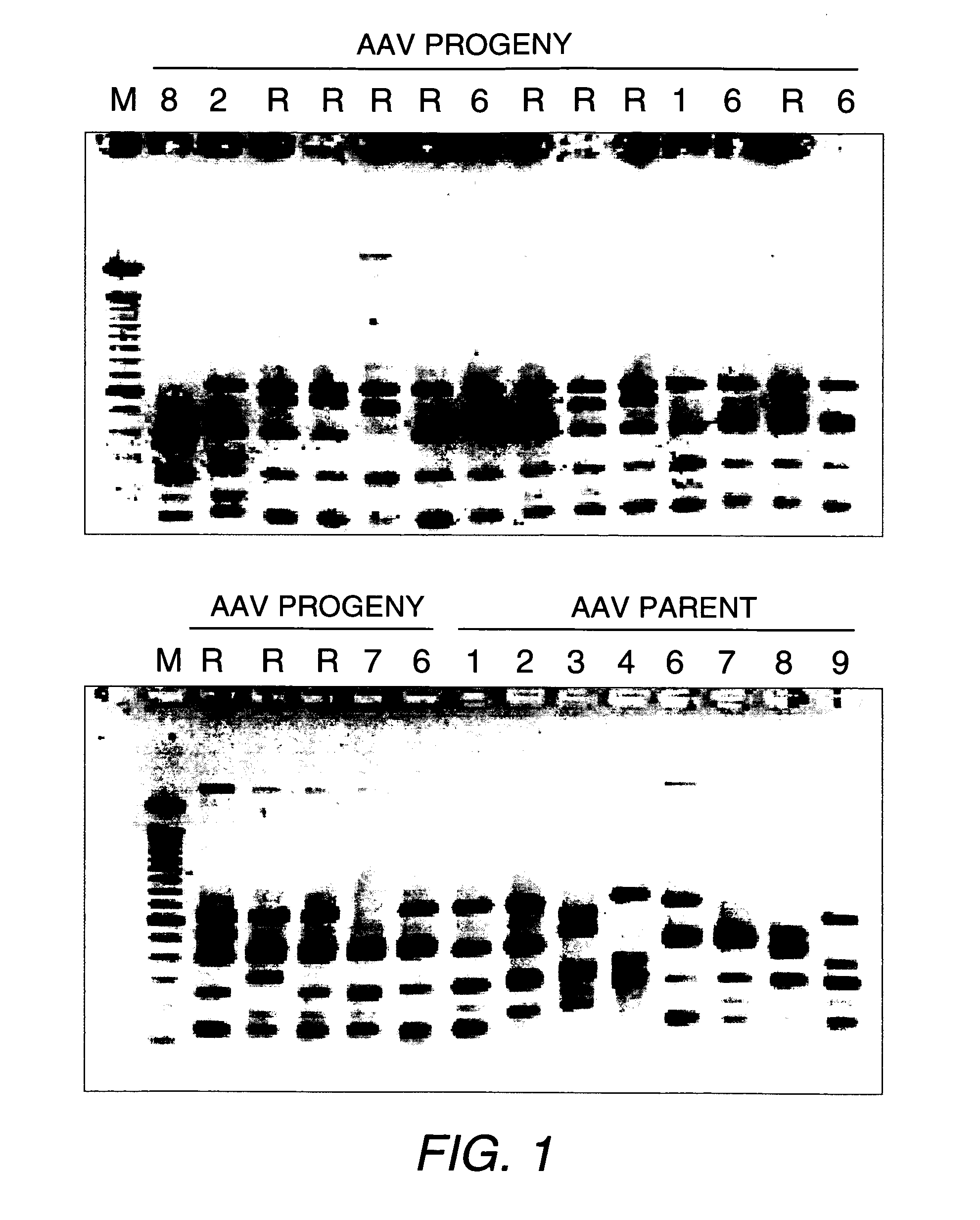
Directed evolution and in vivo panning of virus vectors
The present invention provides methods of achieving directed evolution of viruses by in vivo screening or “panning” to identify viruses comprising scrambled AAV capsids having characteristics of interest, e.g., tropism profile and / or neutralization profile (e.g., ability to evade neutralizing antibodies). The invention also provides scrambled AAV capsids and virus particles comprising the same.
Owner:THE UNIV OF NORTH CAROLINA AT CHAPEL HILL
Directed Evolution and In Vivo Panning of Virus Vectors
The present invention provides methods of achieving directed evolution of viruses by in vivo screening or “panning” to identify viruses comprising scrambled AAV capsids having characteristics of interest, e.g., tropism profile and / or neutralization profile (e.g., ability to evade neutralizing antibodies). The invention also provides scrambled AAV capsids and virus particles comprising the same.
Owner:THE UNIV OF NORTH CAROLINA AT CHAPEL HILL
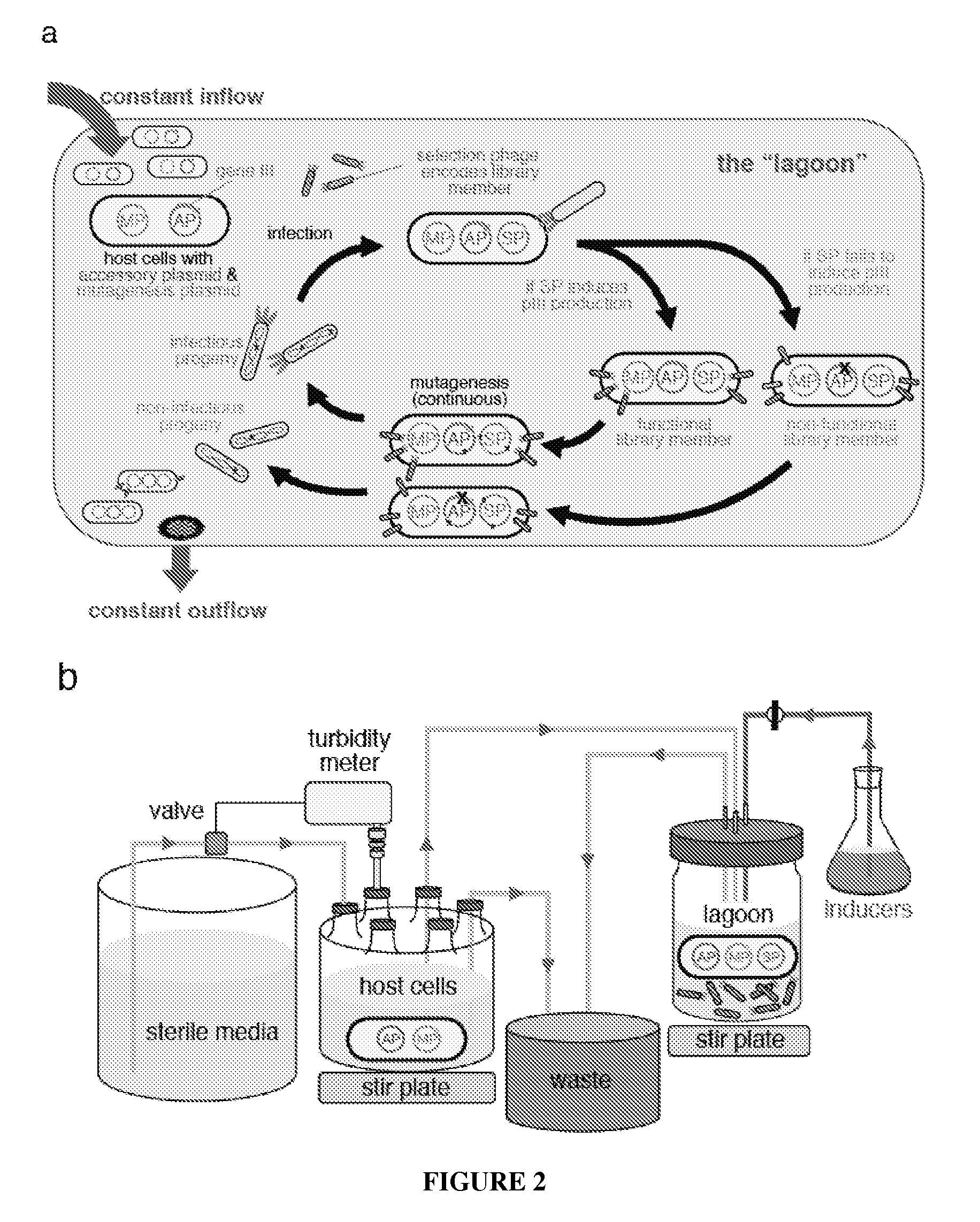
Continuous directed evolution
ActiveUS9394537B2Accelerating evolutionVirus peptidesDirected macromolecular evolutionViral vectorVirus
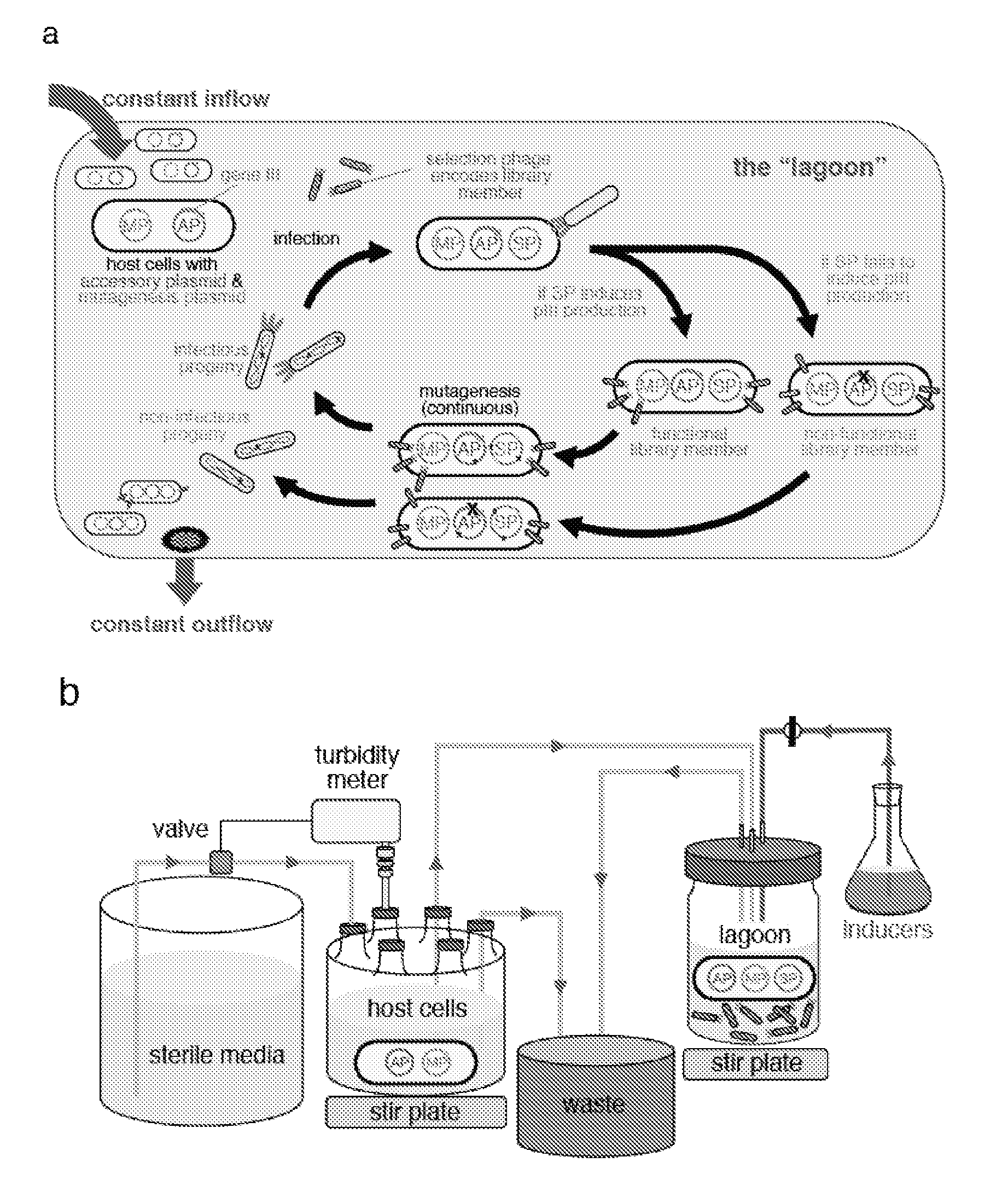
The invention provides systems, methods, reagents, apparatuses, vectors, and host cells for the continuous evolution of nucleic acids. For example, a lagoon is provided in which a population of viral vectors comprising a gene of interest replicates in a stream of host cells, wherein the viral vectors lack a gene encoding a protein required for the generation of infectious viral particles, and wherein that gene is expressed in the host cells under the control of a conditional promoter, the activity of which depends on a function of the gene of interest to be evolved. Some aspects of this invention provide evolved products obtained from continuous evolution procedures described herein. Kits containing materials for continuous evolution are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
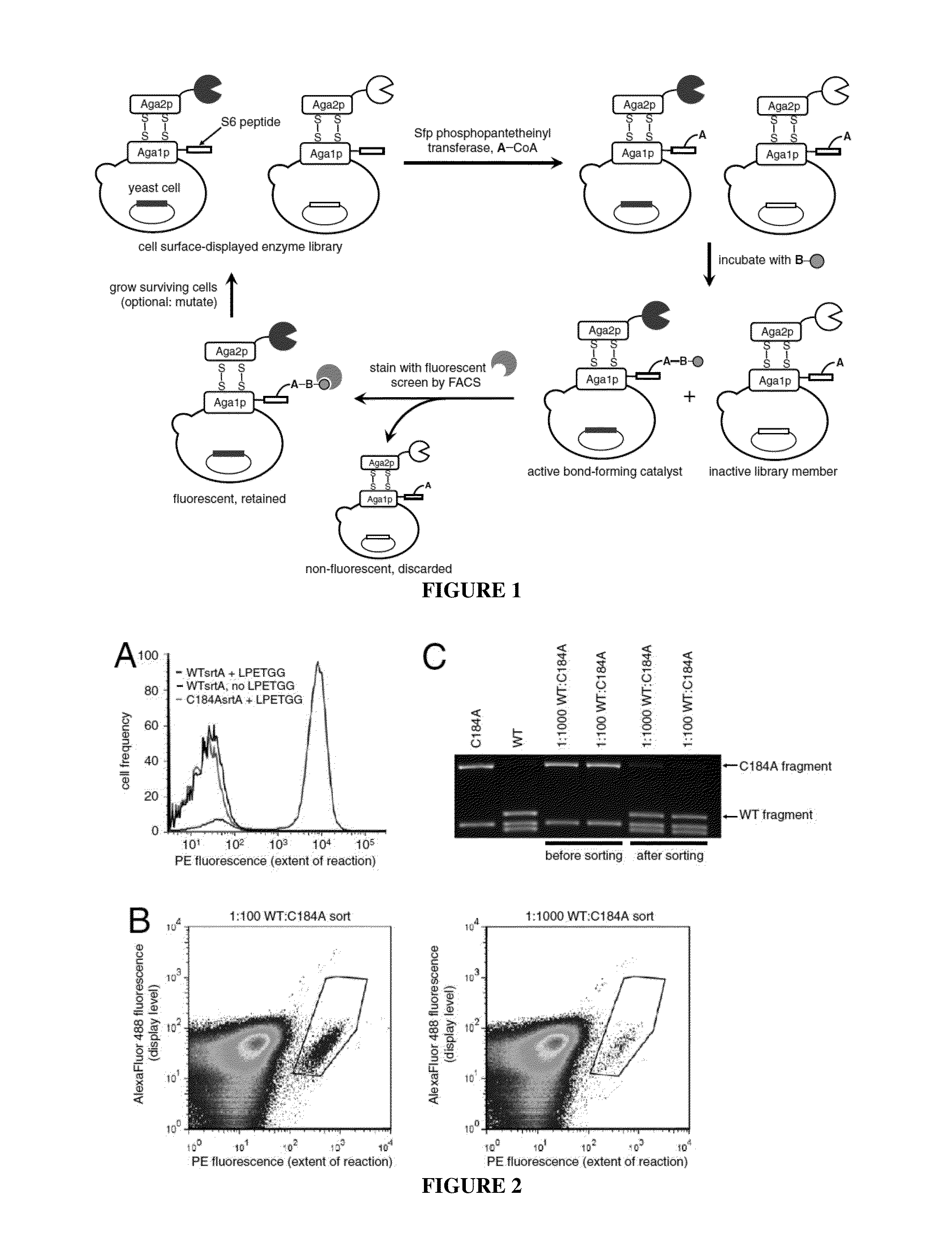
Evolution of bond-forming enzymes
Strategies, systems, methods, reagents, and kits for the directed evolution of bond-forming enzymes are provided herein. Evolution products, for example, evolved sortases exhibiting enhanced reaction kinetics and / or altered substrate preferences are also provided herein, as are methods for using such evolved bond-forming enzymes. Kits comprising materials, reagents, and cells for carrying out the directed evolution methods described herein are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Continuous directed evolution of proteins and nucleic acids
ActiveUS20110177495A1Microbiological testing/measurementMutant preparationDependent mannerDirected evolution
The present invention discloses generalizable methods of evolving nucleic acids and proteins utilizing continuous directed evolution. The invention discloses methods of passing a nucleic acid from cell to cell in a desired function-dependent manner. The linkage of the desired function and passage of the nucleic acid from cell to cell allows for continuous selection and mutation of the nucleic acid.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Detecting low-abundant analyte in microfluidic droplets
InactiveUS20150293102A1Count the number of beads more accurately and comfortablyMaximize useBiological material analysisBiomarker (petroleum)Molecular resolution
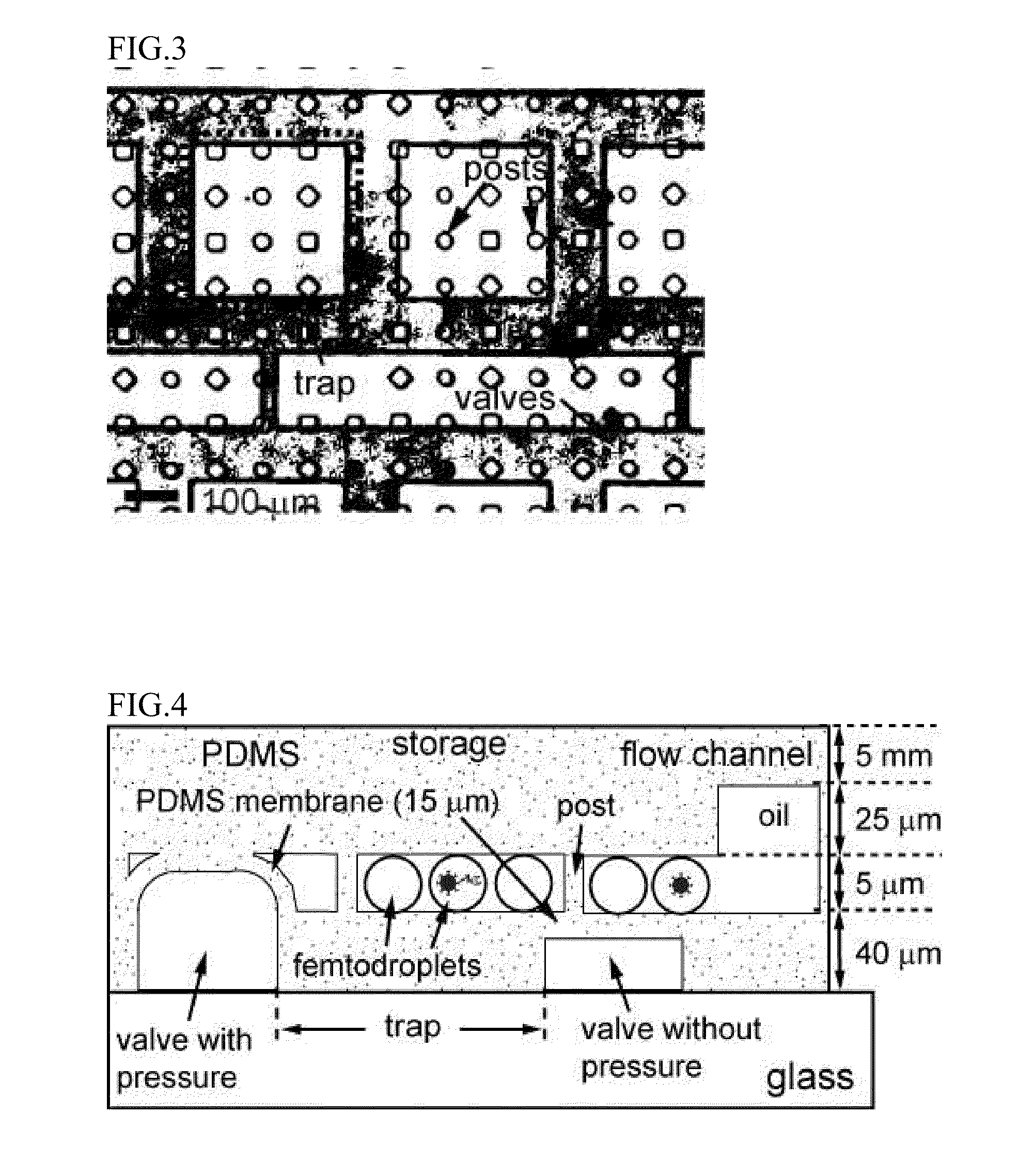
A method to produce aqueous droplets in oil and to manipulate the droplets for storage in the microfluidic device for certain amount of time to accumulate detectable amount of product produced by a single copy or plural copies of enzyme enclosed in the droplets, and to detect and measure the biomarkers in the antibody binding assay is disclosed. The method comprises: (1) generation of droplets in the microfluidic device, (2) storage of droplets in the microfluidic device, (3) measurement of activity of a single copy or plural copies of enzyme in the droplets, (4) individual molecule-counting immunoassay using the droplets.Applications can include the single molecule counting immunoassay, a platform for extremely high through digital PCR, a platform for directed evolution at individual molecule resolutions, nanoparticles synthesis, biodegradable polymer particle production and single molecule analysis.
Owner:SHIM JUNG UK
Computationally targeted evolutionary design
InactiveUS20010051855A1Readily and efficiently identifyComputationally tractableProteomicsBiological testingComputerized systemImproved method
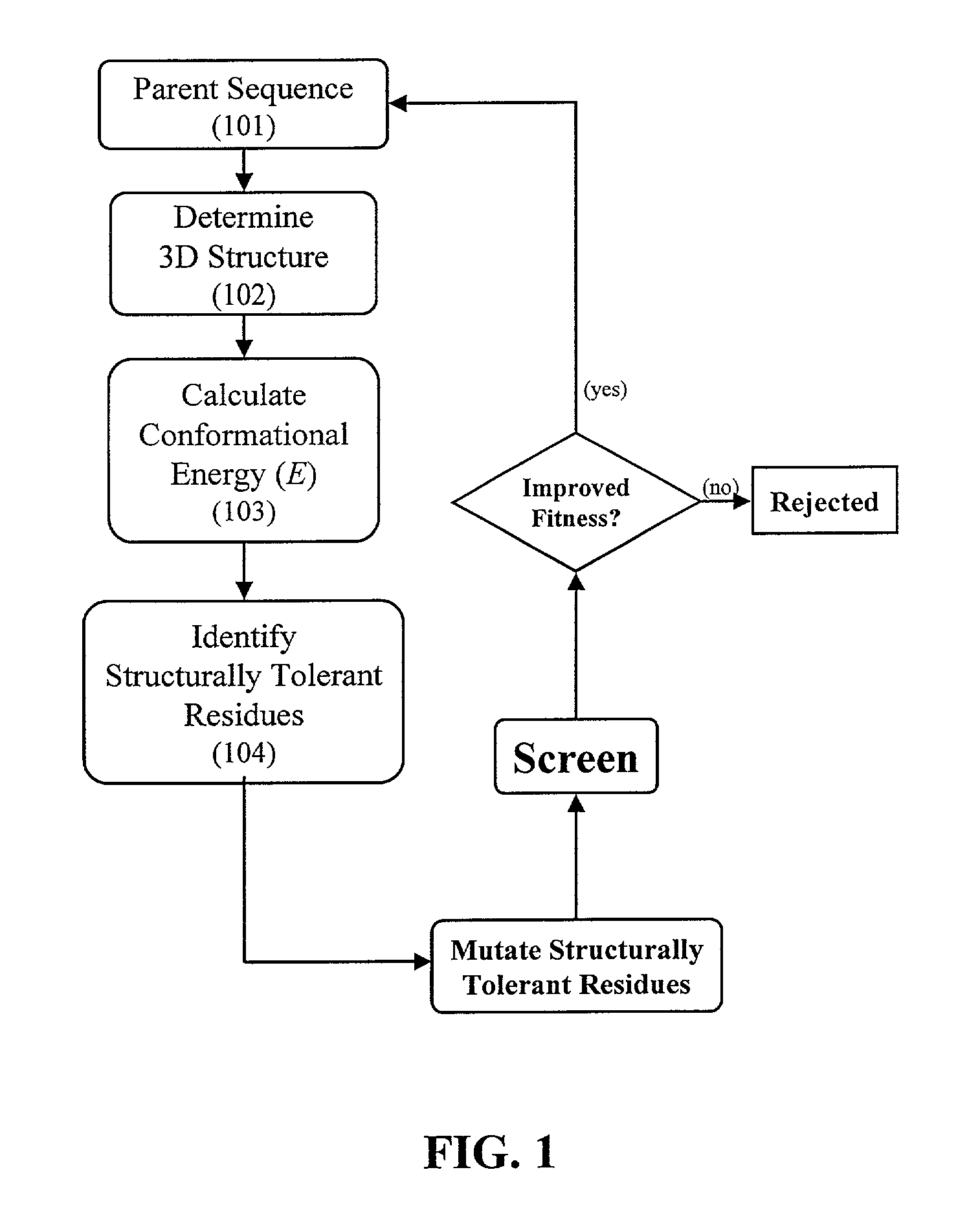
The invention relates to improved methods for directed evolution of polymers, including directed evolution of nucleic acids and proteins. Specifically, the methods of the invention include analytical methods for identifying "structurally tolerant" residues of a polymer. Mutations of these, structurally tolerant residues are less likely to adversely affect desirable properties of a polymer sequence. The invention further provides improved methods for directed evolution wherein the structurally tolerant residues of a polymer are selectively mutated. Computer systems for implementing analytical methods of the invention are also provided.
Owner:CALIFORNIA INST OF TECH
Gene recombination and hybrid protein development
InactiveUS20030032059A1Preserve stability and desired propertyComputationally tractableBiological testingSequence analysisBiopolymerProtein insertion
The invention relates to improved methods for directed evolution of polymers, including directed evolution of nucleic acids and proteins. Specifically, the methods of the invention include analytical methods for identifying "crossover locations" in a polymer. Crossovers at these locations are less likely to disrupt desirable properties of the protein, such as stability or functionality. The invention further provides improved methods for directed evolution wherein the polymer is selectively recombined at the identified "crossover locations". Crossover disruption profiles can be used to identify preferred crossover locations. Structural domains of a biopolymer can also be identified and analyzed, and domains can be organized into schema. Schema disruption profiles can be calculated, for example based on conformational energy or interatomic distances, and these can be used to identify preferred or candidate crossover locations. Computer systems for implementing analytical methods of the invention are also provided.
Owner:WANG ZHEN GANG +3
Directed evolution using proteins comprising unnatural amino acids
ActiveUS20090163368A1Enhanced and novel stericEnhanced and novel and chemicalPeptide librariesBacteriaAmino acidDirected evolution
The invention provides methods and compositions for screening polypeptide libraries that include variants comprising unnatural amino acids. In addition, the invention provides vector packaging systems and methods for packaging a nucleic acid in a vector. Compositions of vectors produced by the methods and systems are also provided
Owner:THE SCRIPPS RES INST
Continuous directed evolution
ActiveUS20130345064A1Infection efficiency can be decreasedAccelerating evolutionVirus peptidesDirected macromolecular evolutionViral vectorVirus
The invention provides systems, methods, reagents, apparatuses, vectors, and host cells for the continuous evolution of nucleic acids. For example, a lagoon is provided in which a population of viral vectors comprising a gene of interest replicates in a stream of host cells, wherein the viral vectors lack a gene encoding a protein required for the generation of infectious viral particles, and wherein that gene is expressed in the host cells under the control of a conditional promoter, the activity of which depends on a function of the gene of interest to be evolved. Some aspects of this invention provide evolved products obtained from continuous evolution procedures described herein. Kits containing materials for continuous evolution are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Saturation mutagenesis in directed evolution
InactiveUS20050100985A1Reduce complexityHigh activityHydrolasesMicrobiological testing/measurementNucleotideCombined use
Disclosed is a rapid and facilitated method of producing from a parentlal template polynucleotide, a set of mutagenized progeny polynculeotides whereby at each original codon position there is produced at least one substitute codon encoding each of the 20 naturally encoded amino acids. Accordingly, there is also provided a method of producing from a parental template polypeptide, a set of mutagenized progeny polypeptides wherein each of the 20 naturally encoded amino acids is represented at each original amino acid position. The method provided is termed site-saturation mutagenesis, or simply saturation mutagenesis, and can be used in combination with other mutagenization processes, such as, for example, a process wherein two or more related polynucleotides are introduced into a suitable host cell such that a hybrid polynucleotide is generated by recombination and reductive reassortment. Also provided are vector and expression vehicles incuding such polynucleotides, polypeptides expressed by the hybrid polynucleotides and a method for screening for hybrid polypeptides.
Owner:SHORT JAY
Engineering t cell receptors
InactiveUS20150191524A1Immunoglobulin superfamilySugar derivativesMajor histocompatibilityPeptide ligand
The use of model T cell receptors (TCRs) as scaffolds for in vitro engineering of novel specificities is provided. TCRs with de novo binding to a specific peptide-major histocompatibility complex (MHC) product can be isolated by: 1) mutagenizing a T cell receptor protein coding sequence to generate a variegated population of mutants (a library), 2) selection of the library of TCR mutants with the specific peptide-MHC, using a process of directed evolution and a “display” methodology (e.g., yeast, phage, mammalian cell) and the peptide-MHC ligand. The process can be repeated to identify TCR variants with improved affinity for the selecting peptide-MHC ligand.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS
Directed evolution of microorganisms
Owner:SCHELLENBERGER VOLKER +2
Evolution of bond-forming enzymes
ActiveUS20140057317A1High catalytic activityAltered substrate preferenceFermentationPeptidasesReagentDirected evolution
Strategies, systems, methods, reagents, and kits for the directed evolution of bond-forming enzymes are provided herein. Evolution products, for example, evolved sortases exhibiting enhanced reaction kinetics and / or altered substrate preferences are also provided herein, as are methods for using such evolved bond-forming enzymes. Kits comprising materials, reagents, and cells for carrying out the directed evolution methods described herein are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Accelerated directed evolution of microbial consortia for the development of desirable plant phenotypic traits
ActiveUS20150080261A1Efficient and fast and broadly applicableBiocideBacteriaPhenotypic traitPlant phenotyping
Owner:BIOCONSORTIA
Apparatus for forming and screening two-dimensional liquid droplet array, and use method thereof
The present invention discloses an apparatus for forming and screening a large-scale single-layer two-dimensional liquid droplet array, and a use method thereof. The apparatus comprises a micro-column array chip, a scraping plate for auxiliary spreading of a liquid droplet, a probe for controlling the liquid droplet, a three-dimensional translation table and other components, wherein a micro groove unit for trapping the single liquid droplet is formed by using the gap between micro-columns, and the liquid droplet is spread in the micro groove unit in a single-layer and single manner through gravity effect and capillary effect or through an auxiliary means so as to form the single-layer liquid droplet array. With the apparatus of the present invention, the accurate positioning and detection on the liquid droplet, the sucking on the target liquid droplet and the subsequent treatments can be performed. According to the present invention, the apparatus has characteristics of simple structure and easy operation; and the method has advantages of fast liquid droplet spreading, high flux, high liquid droplet trapping efficiency, liquid droplet position fixing and the like, and is suitable for single cell analysis and screening, single molecule analysis and screening, high-throughput gene screening, protein directed evolution, antibody screening, microbial research, drug screening, and other fields.
Owner:ZHEJIANG UNIV
Directed evolution of oxidase enzymes
InactiveUS7098010B1Improve propertiesHigh expressionSugar derivativesBacteriaLactose oxidaseNucleotide
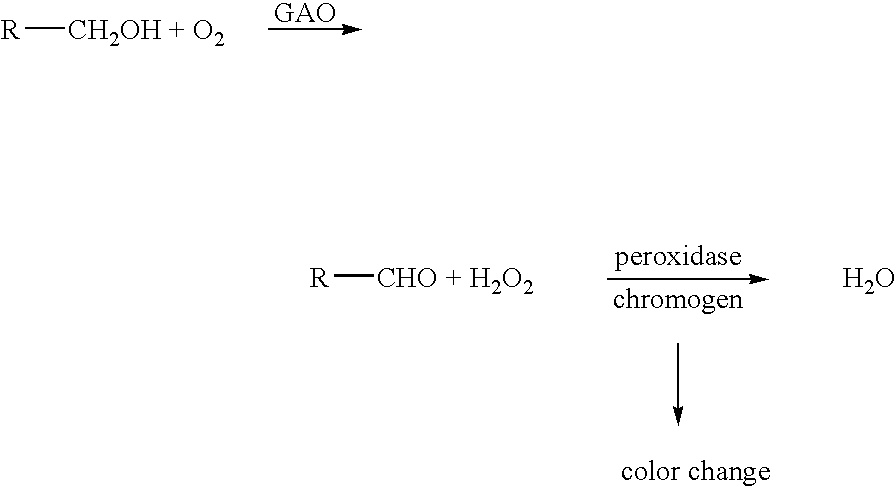
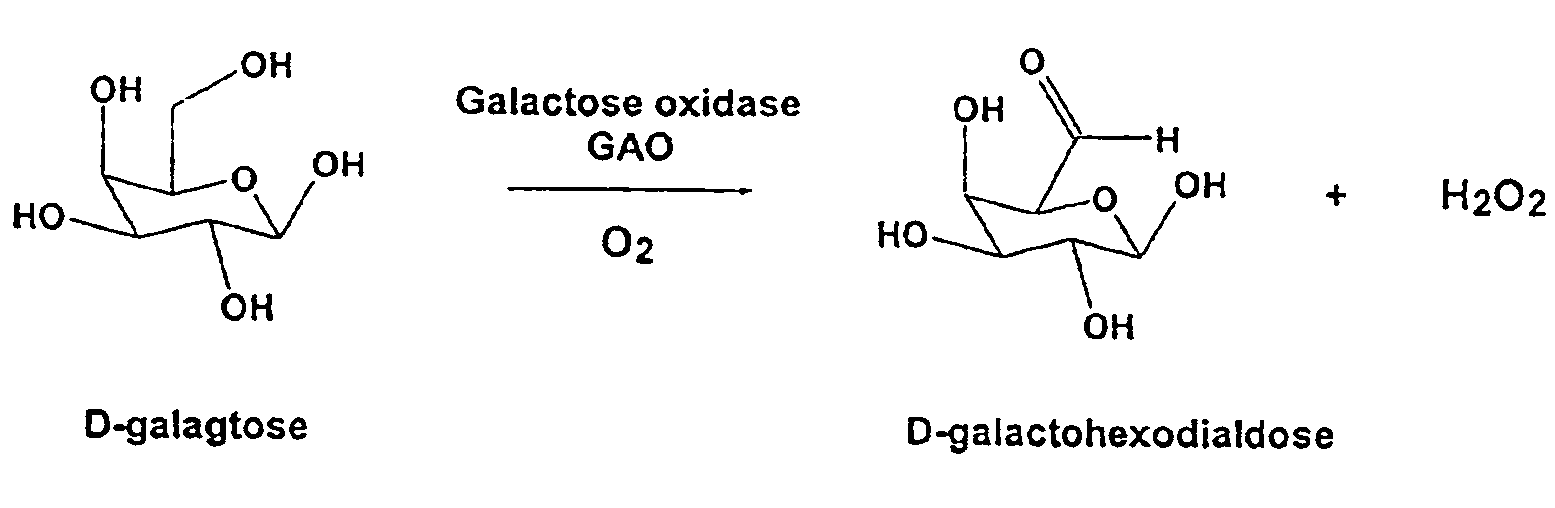
This invention relates to the expression of improved polynucleotide and polypeptide sequences encoding for eukaryotic enzymes, particularly oxidase enzymes. The enzymes are advantagoeusly produced in conventional or facile expression systems. Various methods for directed evolution of polynucleotide sequences can be used to obtain the improved sequences. The improved characteristics of the polypeptides or proteins generated in this manner include improved expression, enhanced activity toward one or more substrates, and increased thermal stability. In a particular embodiment, the invention relates to improved expression of the galactose oxidase gene and galactose oxidase enzymes. GAO mutants that are highly active and / or thermostable are disclosed.
Owner:CALIFORNIA INST OF TECH
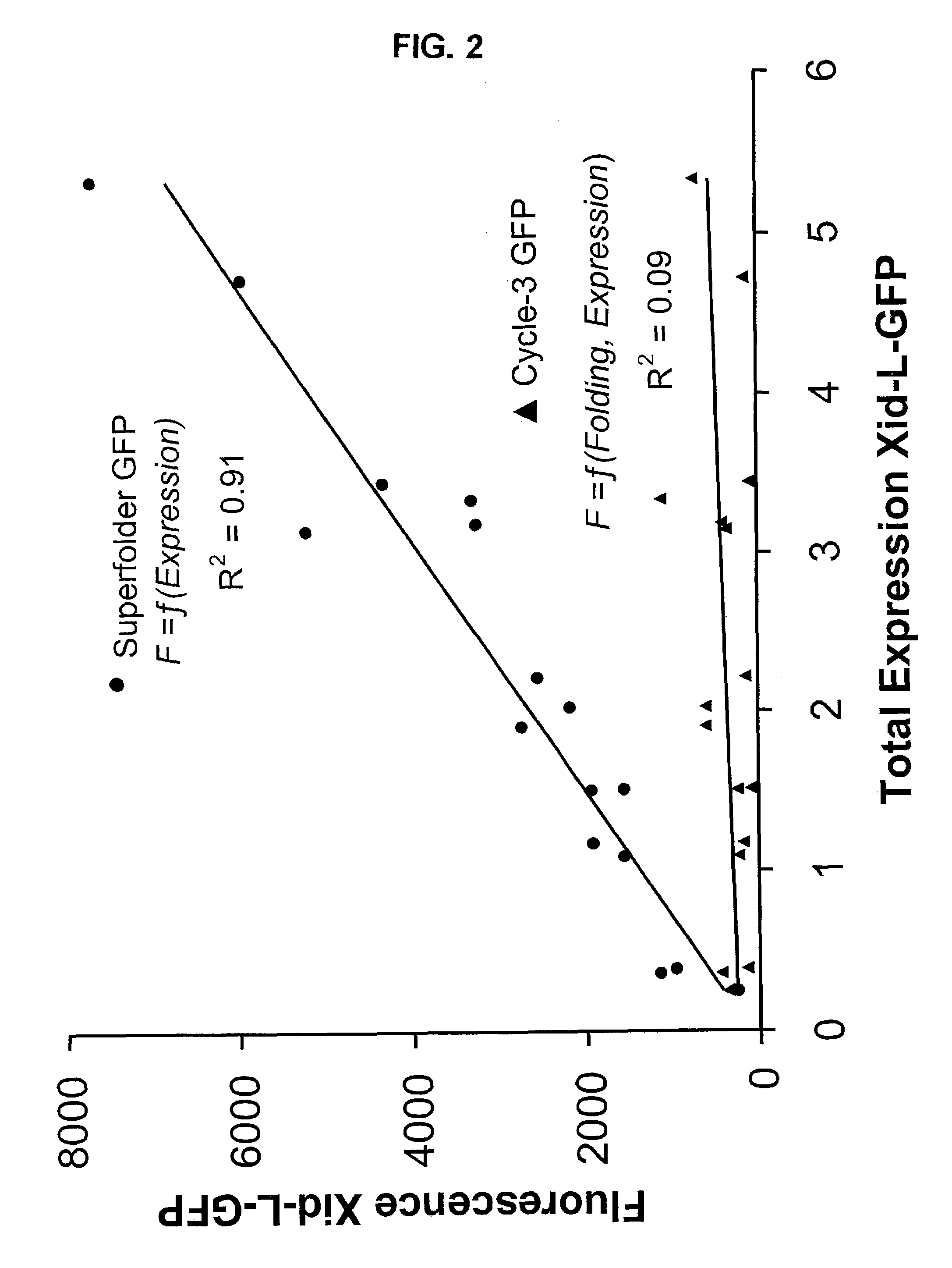
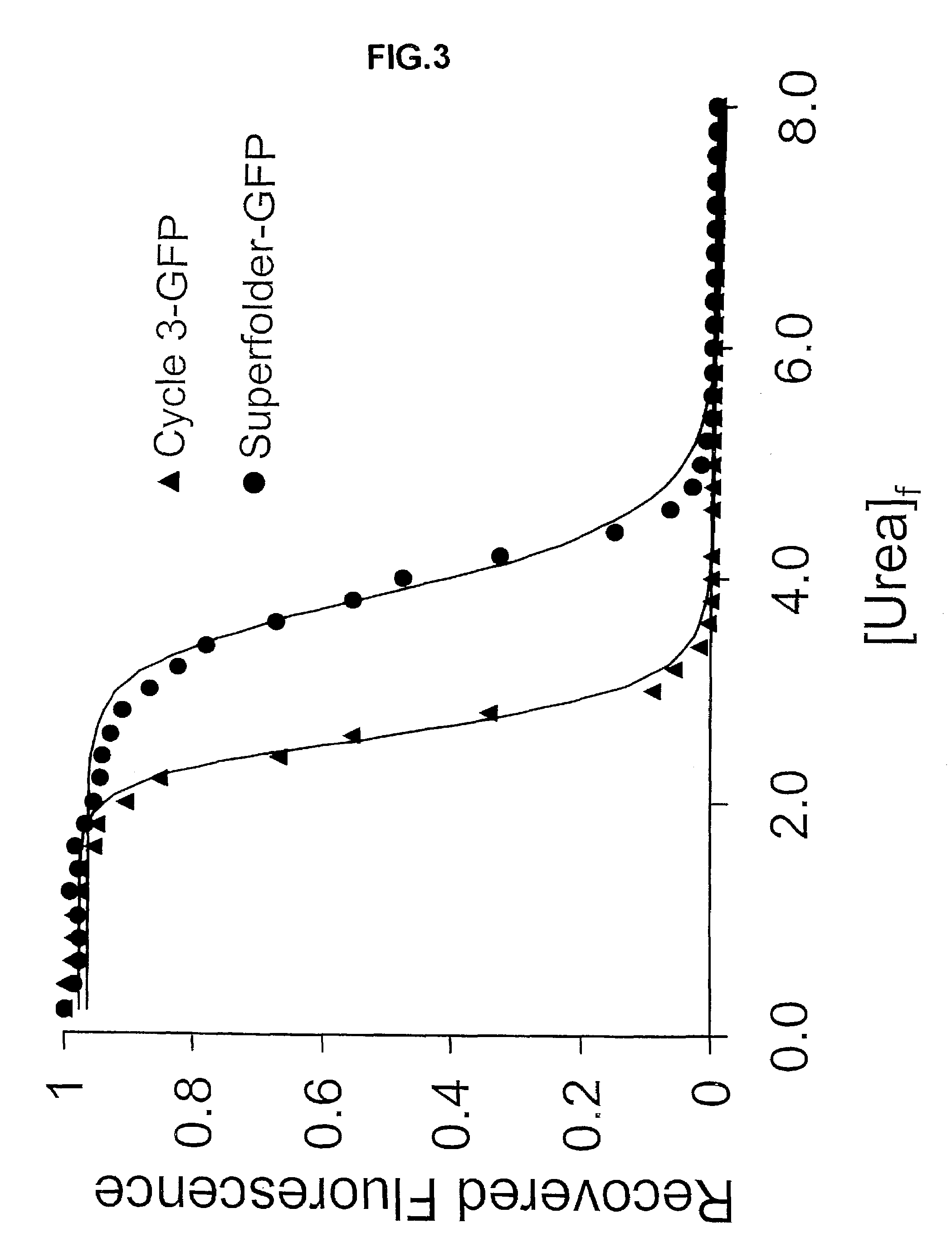
Directed evolution methods for improving polypeptide folding and solubility and superfolder fluorescent proteins generated thereby
InactiveUS7271241B2Improving foldingImproving solubility characteristicPeptide/protein ingredientsAntibody mimetics/scaffoldsSolubilityFluorescence
The current invention provides methods of improving folding of polypeptides using a poorly folding domain as a component of a fusion protein comprising the poorly folding domain and a polypeptide of interest to be improved. The invention also provides novel green fluorescent proteins (GFPs) and red fluorescent proteins that have enhanced folding properties.
Owner:TRIAD NAT SECURITY LLC
Computationally targeted evolutionary design
InactiveUS20050003389A1Readily and efficiently identifyComputationally tractableMicrobiological testing/measurementProteomicsImproved methodAnalysis method
The invention relates to improved methods for directed evolution of polymers, including directed evolution of nucleic acids and proteins. Specifically, the methods of the invention include analytical methods for identifying “structurally tolerant” residues of a polymer. Mutations of these, structurally tolerant residues are less likely to adversely affect desirable properties of a polymer sequence. The invention further provides improved methods for directed evolution wherein the structurally tolerant residues of a polymer are selectively mutated. Computer systems for implementing analytical methods of the invention are also provided.
Owner:WANG ZHEN GANG +3
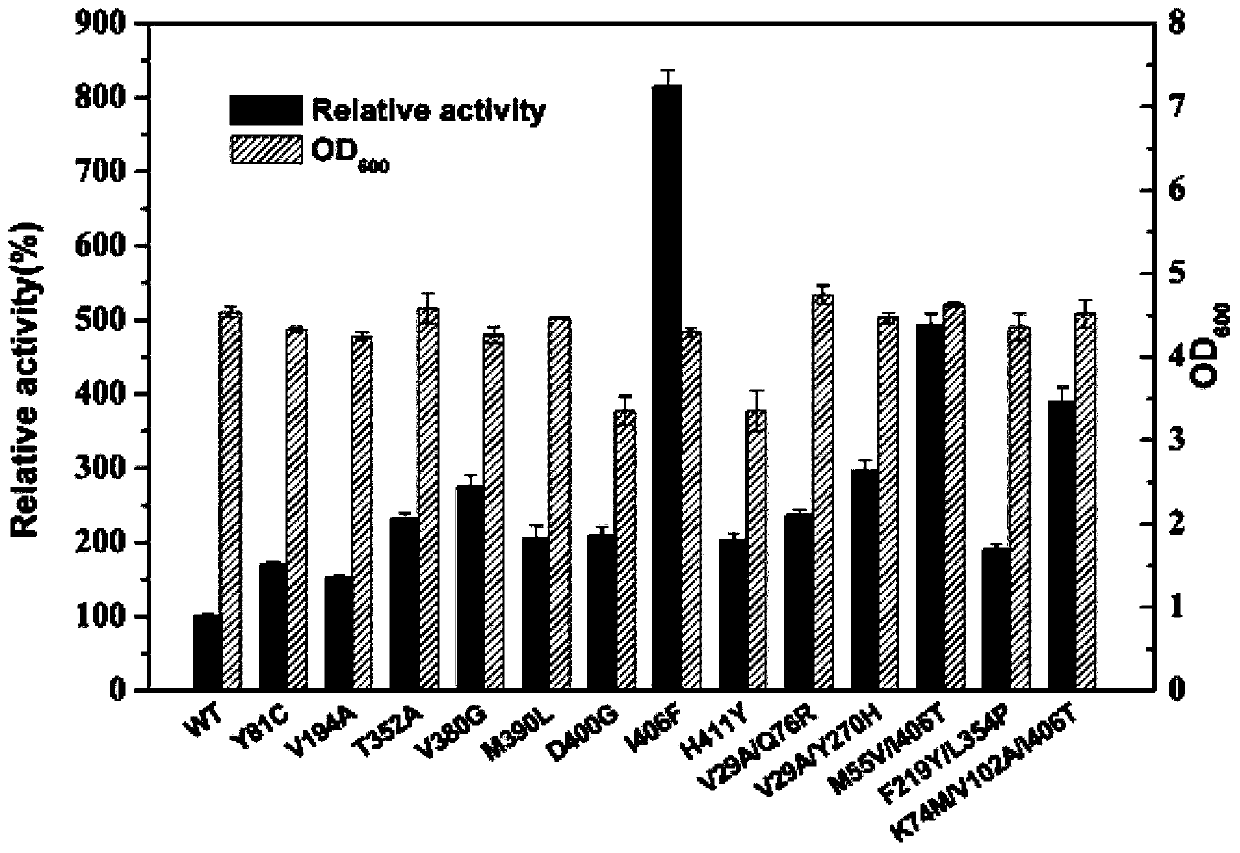
Glutamate dehydrogenase mutants and application thereof
ActiveCN110184246AHigh catalytic activitySolve the problem of low enzyme activityOxidoreductasesFermentationPseudomonas putidaThreonine
The present invention discloses glutamate dehydrogenase mutants and an application thereof. The glutamate dehydrogenase mutants are one of the following: mutating 402th lysine of an amino acid sequence shown in SEQ ID NO.1 to phenylalanine or aspartic acid; mutating 406th isoleucine to phenylalanine or threonine; conducting combined mutation of 121th threonine and 123th leucine; and conducting combined mutation of 379th alanine and 383th leucine. A molecular transformation method combining directed evolution with semi-rational design significantly improves catalytic activity of glutamate dehydrogenase derived from pseudomonas putida to 2-carbonyl-4-(hydroxymethylphosphono)butyric acid (PPO) and solves a problem of low glutamate dehydrogenase activity in preparation of L-glufosinate by a reductive amination method.
Owner:ZHEJIANG UNIV
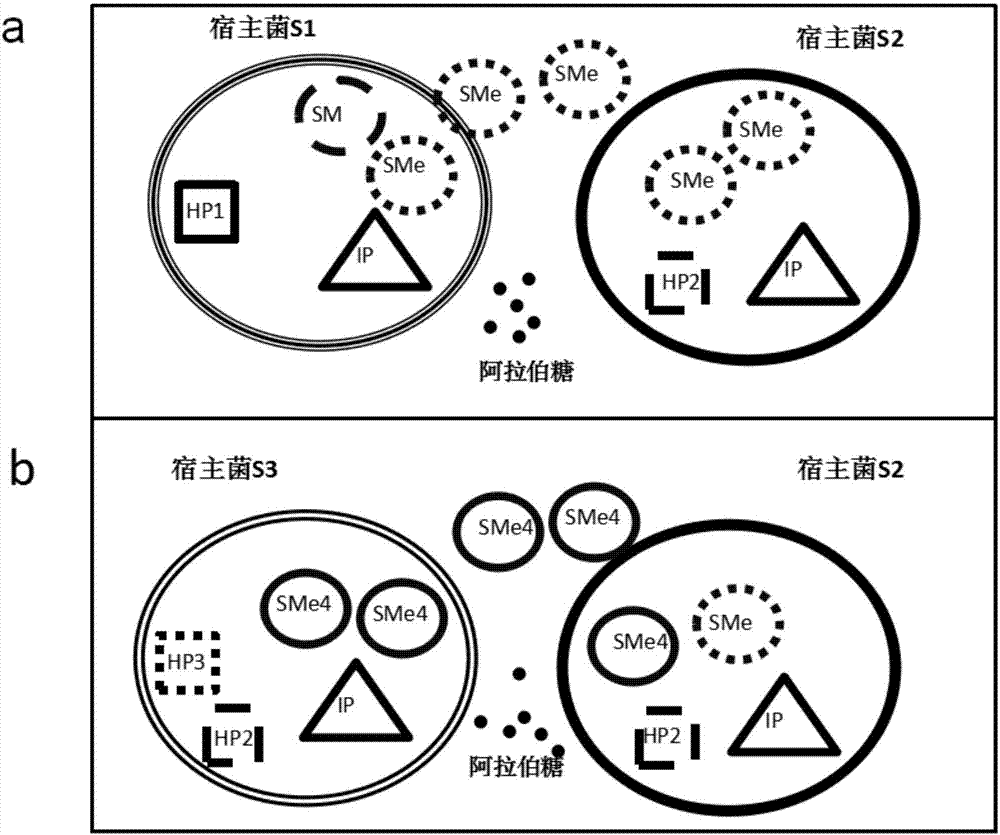
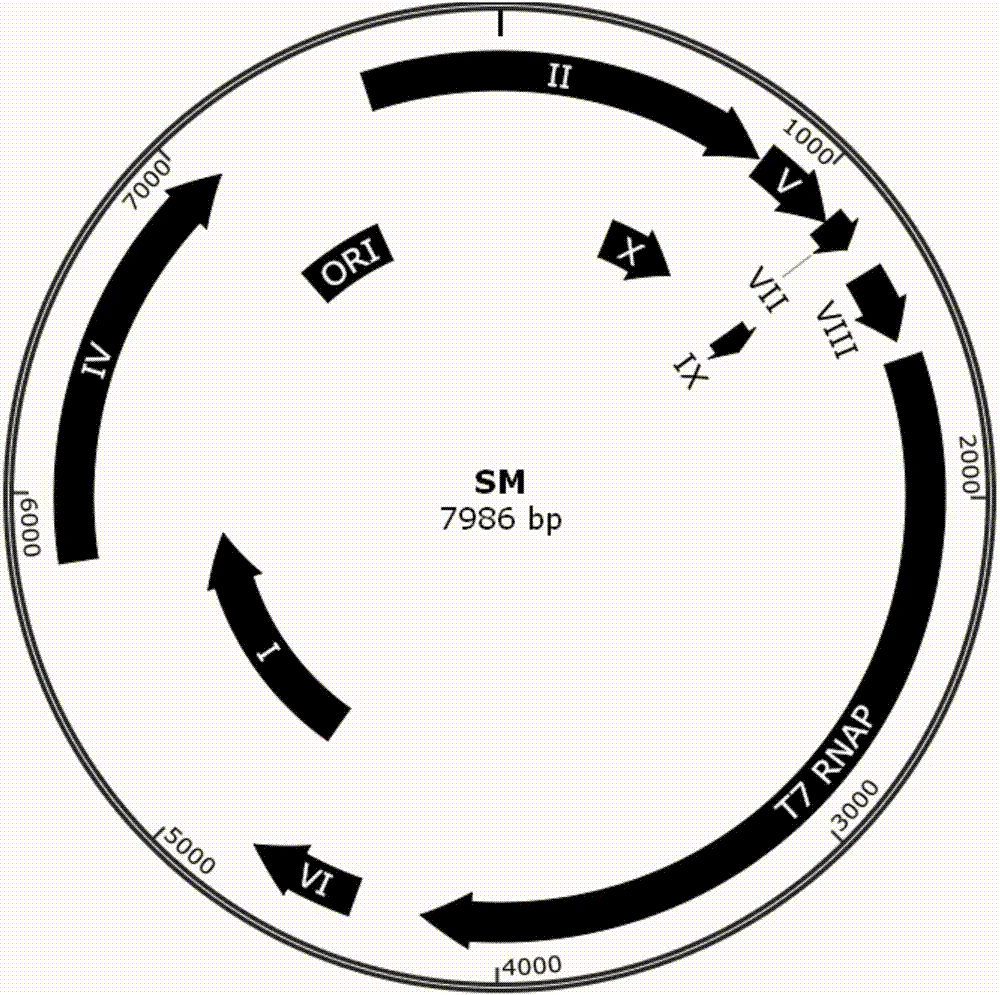
Phage-assisted multi-bacterial continuous directed evolution system and method
ActiveCN107418964AImprove evolutionary efficiencyBacteriaNucleic acid vectorGene ComponentBacteriophage
The invention discloses phage-assisted multi-bacterial continuous directed evolution system and method. The system includes: a phage SM which carries an evolution target gene, a plurality of assistant plasmids HP which support different SM proliferations before and after the evolution, and a plasmid IP which induces mutation. In the system, the different HPs are arranged in different host bacteria, so that proliferation and evolution efficiencies of the phage are effectively improved, and meanwhile, mutual interference between different gene components is avoided. The system and method have great practicability and can be applied to directed evolution of various genes.
Owner:SHENZHEN INST OF ADVANCED TECH CHINESE ACAD OF SCI
Method for preparing phosphatidylserine with docosahexaenoic acid at sn-2 bit
ActiveCN104004797AGood application effectIncreased specific enzyme activityFungiBacteriaDocosahexaenoic acidPhospholipase A2
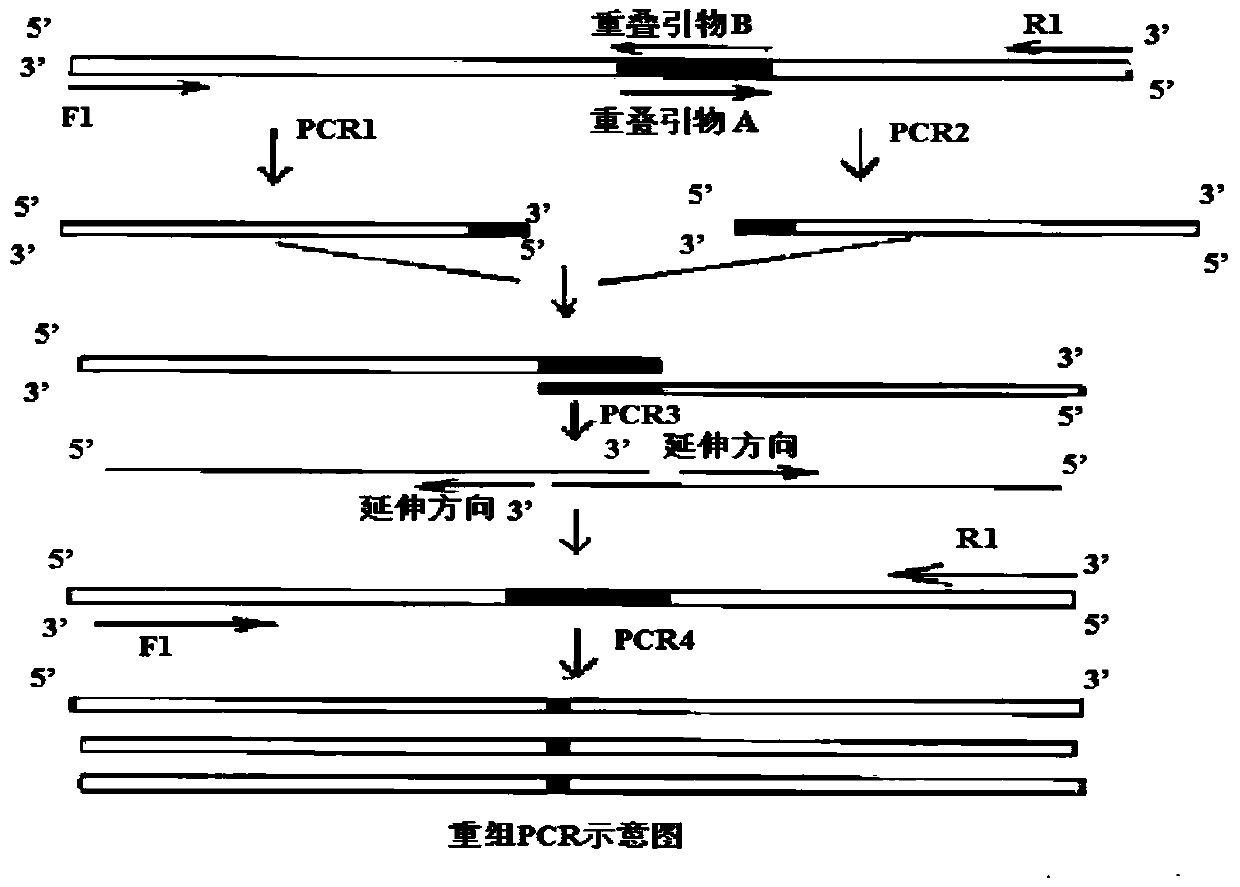
The invention relates to a method for preparing phosphatidylserine (2-DHA-PS) with docosahexaenoic acid at the sn-2 bit through high-activity phospholipase A2 and high-activity phospholipase D. Directed evolution is achieved through the overlapping PCR technology so that the high-activity phospholipase A2 and the high-activity phospholipase D can be obtained; the 2-DHA-PS is prepared through catalysis by means of the high-activity phospholipase A2 and the high-activity phospholipase D, phosphatidylserine is generated through phosphatidylcholine and serine under the catalysis of the high-activity phospholipase D first, and then the 2-DHA-PS is generated through the phosphatidylserine and the docosahexaenoic acid under the catalysis of the high-activity phospholipase A2. The relative content of the 2-DHA-PS in the product synthesized through the method is high, and the defects of an existing synthesizing method are effectively overcome.
Owner:TIANJIN UNIV OF SCI & TECH
Evolution of bt toxins
The disclosure provides amino acid sequence variants of Bacillus thuringiensis (Bt) toxins and methods of producing the same. Some aspects of this disclosure provide methods for generating Bt toxin variants by continuous directed evolution. Some aspects of this disclosure provide compositions and methods for pest control using the disclosed variant Bt toxins.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com