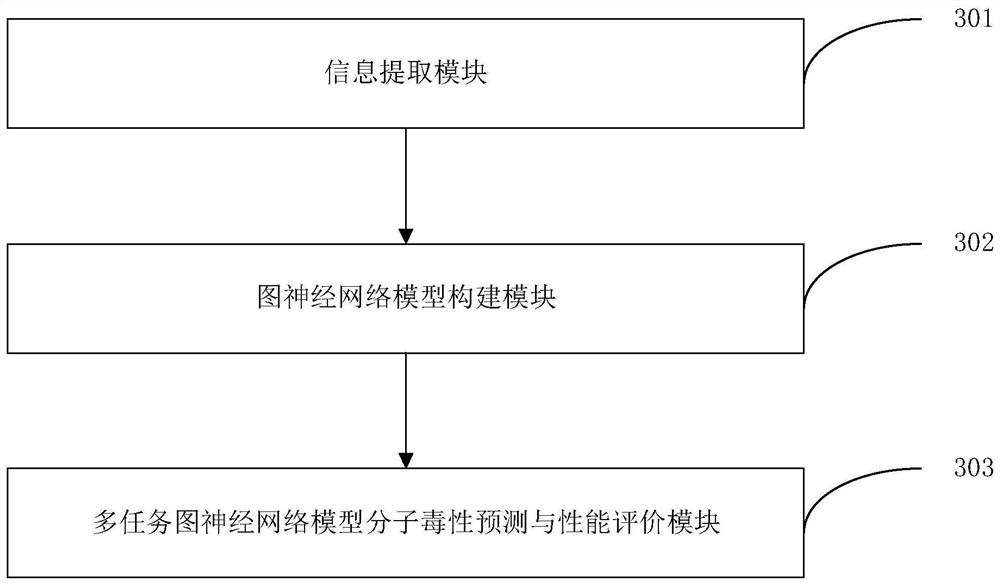
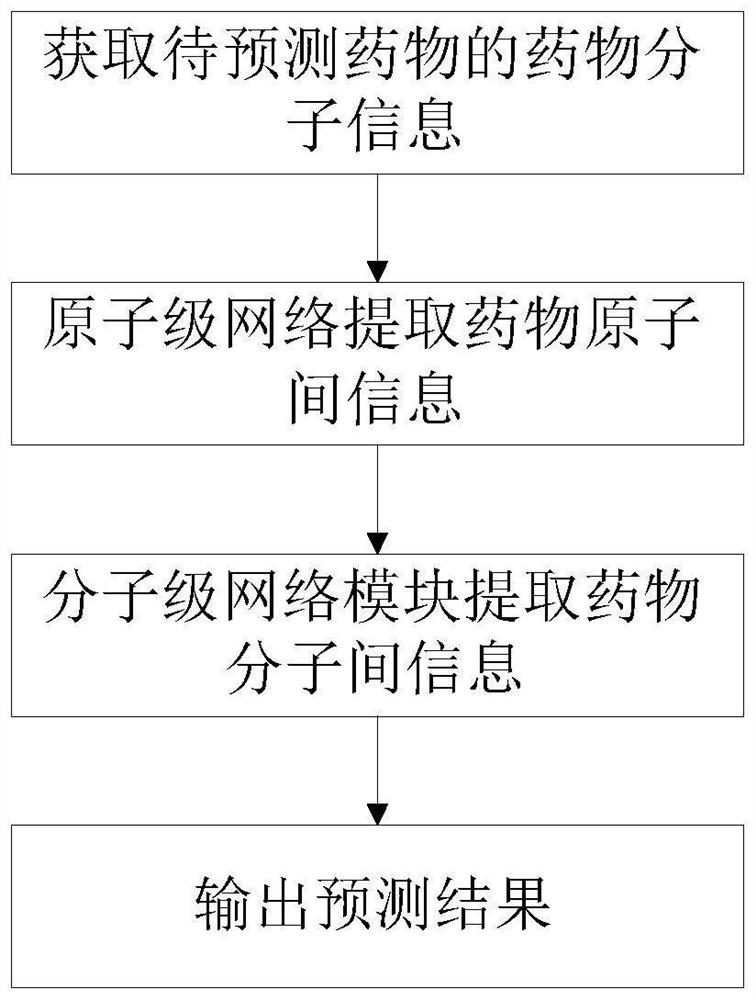
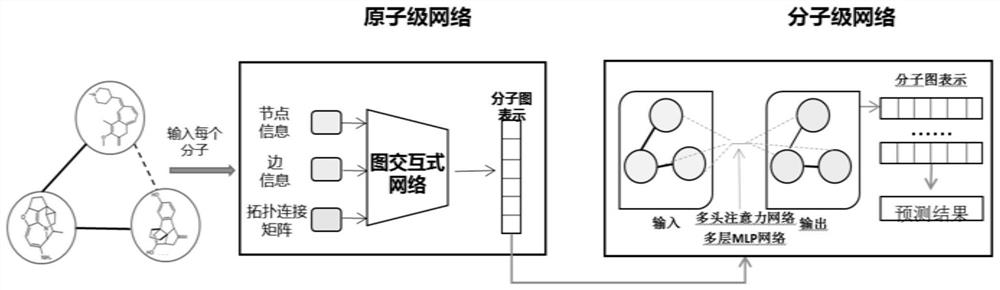
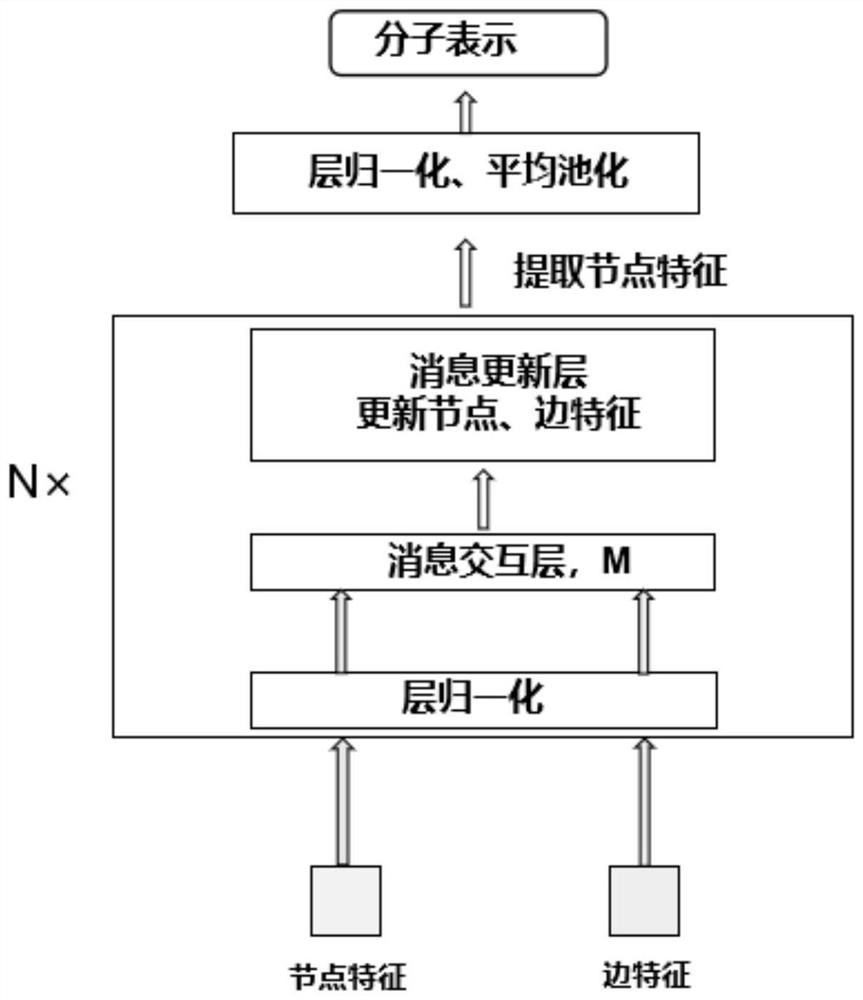
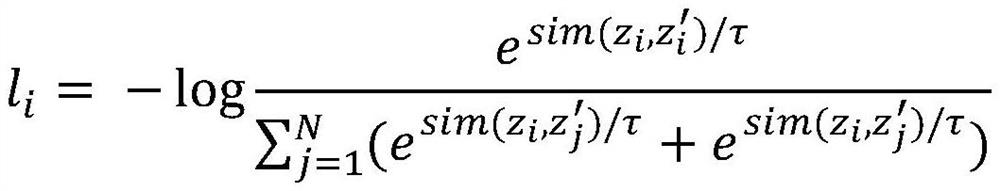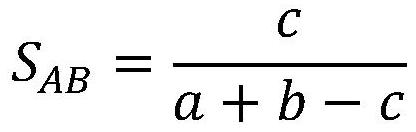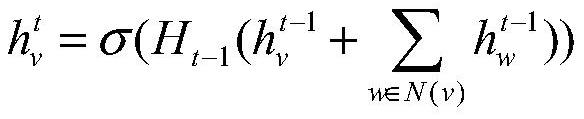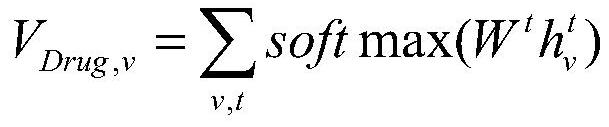Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
36 results about "Molecular graph" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
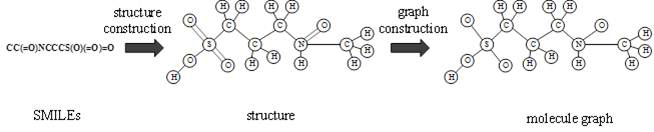
In chemical graph theory and in mathematical chemistry, a molecular graph or chemical graph is a representation of the structural formula of a chemical compound in terms of graph theory. A chemical graph is a labeled graph whose vertices correspond to the atoms of the compound and edges correspond to chemical bonds. Its vertices are labeled with the kinds of the corresponding atoms and edges are labeled with the types of bonds. For particular purposes any of the labelings may be ignored.
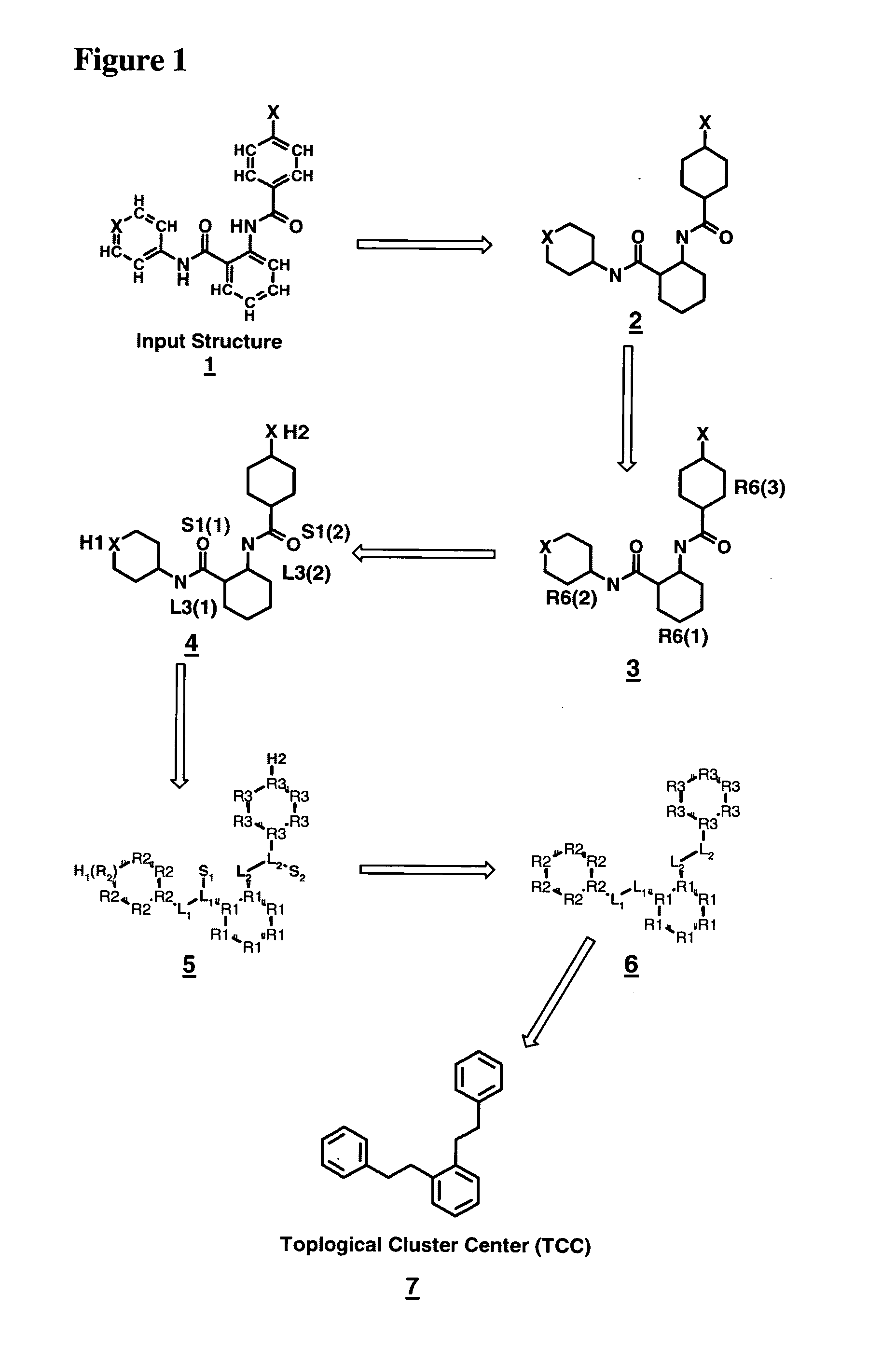
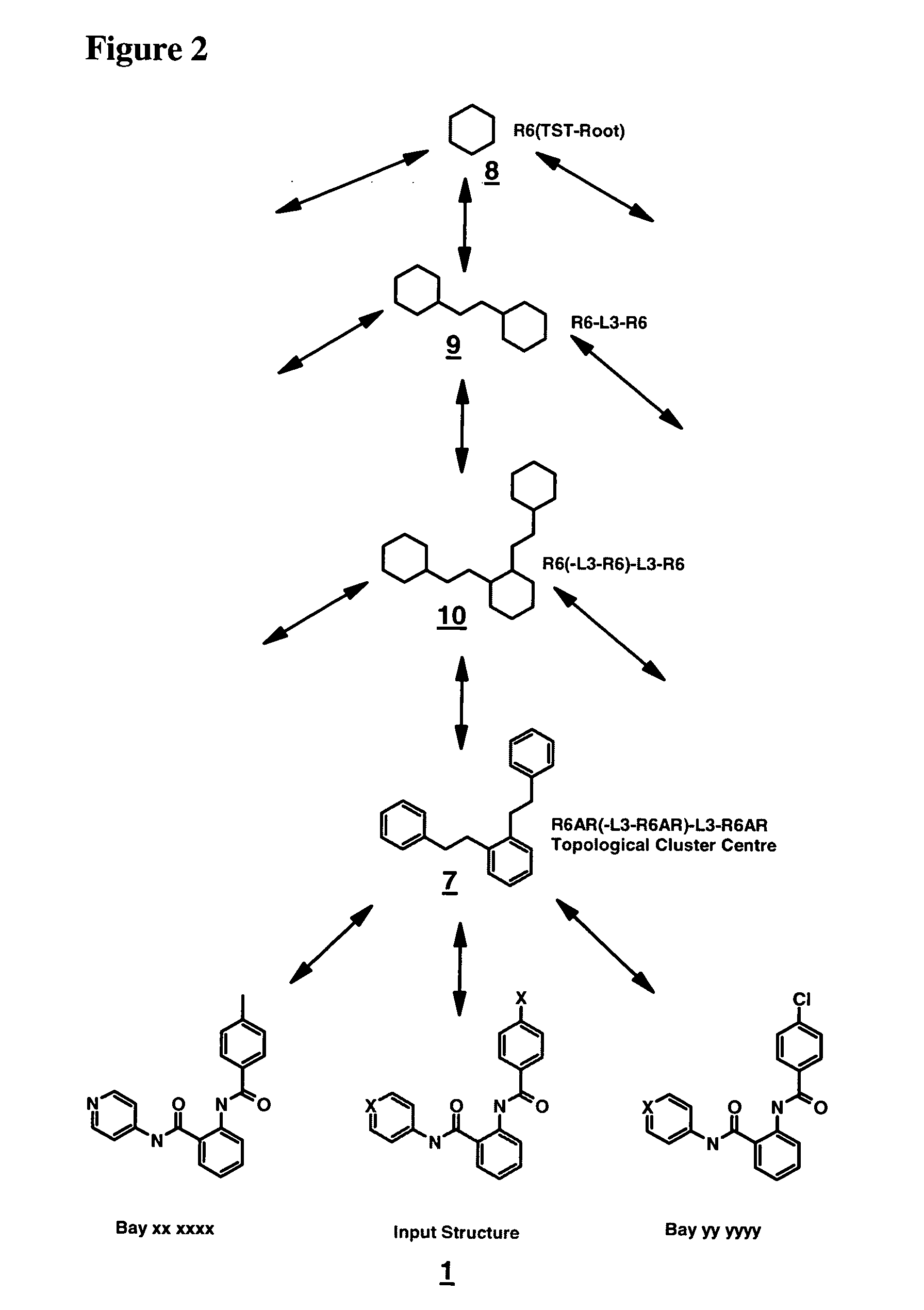
Method for generating a hierarchical topological tree of 2D or 3D-structural formulas of chemical compounds for property optimisation of chemical compounds
InactiveUS20070043511A1Easy to handleOrganic chemistry methodsBiological testingCADA compoundChemical compound
The invention concerns a new method for automatically and dynamically generating hierarchical topological trees of 2D- or 3D-structural formulas for structurally characterized chemical compounds, especially drug-like molecules, wherein the molecular graph of each 2D- or 3D-structure for a chemical compound is analyzed in terms of topological key features, the Largest Topological Substructure (LTS) and the proper Topological Cluster Centre (TCC) are created for each molecular graph, the ranking of the classes of topological key features and / or the ranking within each class of topological key features present in the TCC is used to generate a connected hierarchical Topological Sequence Path (TSP) of sentinel molecules from each molecular graph, and different molecular graphs and their Topological Sequence Paths (TSPs) share common vertices for common topological key features thus growing a Topological Structure Tree (TST), each chemical compound from the input stream is attached as a leaf node to the appropriate Largest Topological Substructure (LTS) node in the tree.
Owner:BAYER SCHERING PHARMA AG
Drug-target interaction prediction model method based on deep embedding learning of molecular graph and sequence
PendingCN113327644AImprove predictive performanceShorten screening timeNeural architecturesNeural learning methodsGraph neural networksDrug target
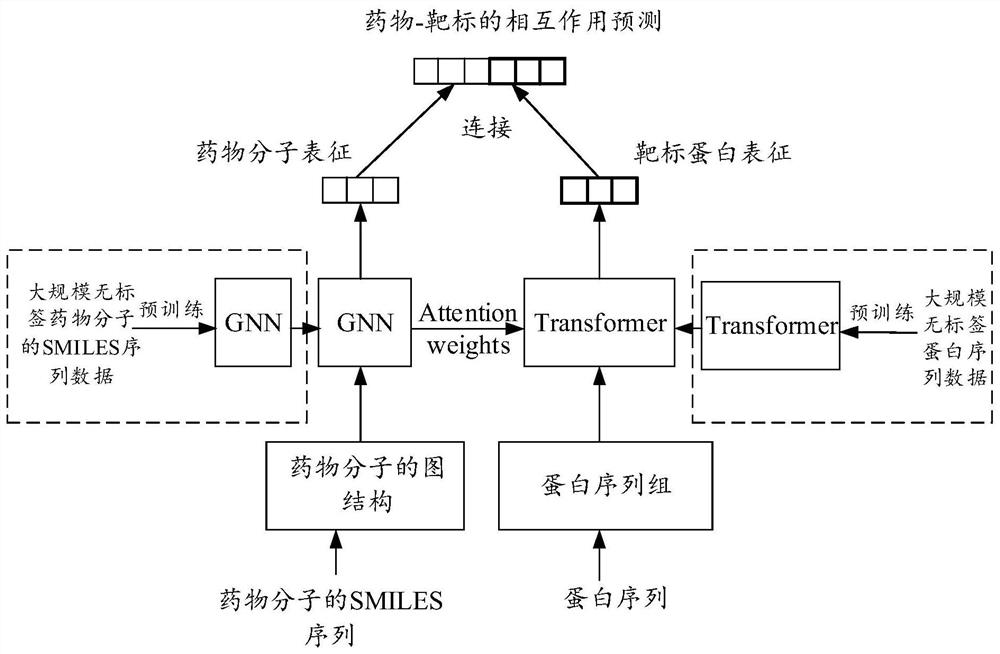
The invention provides a drug-target interaction prediction model method based on deep embedding learning of a molecular graph and a sequence, and the method comprises the steps: building a map neural network based on an attention mechanism and an attention-oriented bidirectional LSTM to predict the interaction, wherein in order to achieve the more effective training, a pre-training model BERT is utilized to extract embedded vector representation of each sub-sequence from a protein sequence, and meanwhile, a local breadth-first search algorithm is designed to extract sub-graph information of a drug molecular graph, so that a graph neural network learns higher feature information. On one hand, in the aspect of drug molecules, better spatial features can be learned based on the molecular graph; on the other hand, the protein sequence data size is large, larger protein space can be covered, and the generalization ability is improved.
Owner:SUN YAT SEN UNIV
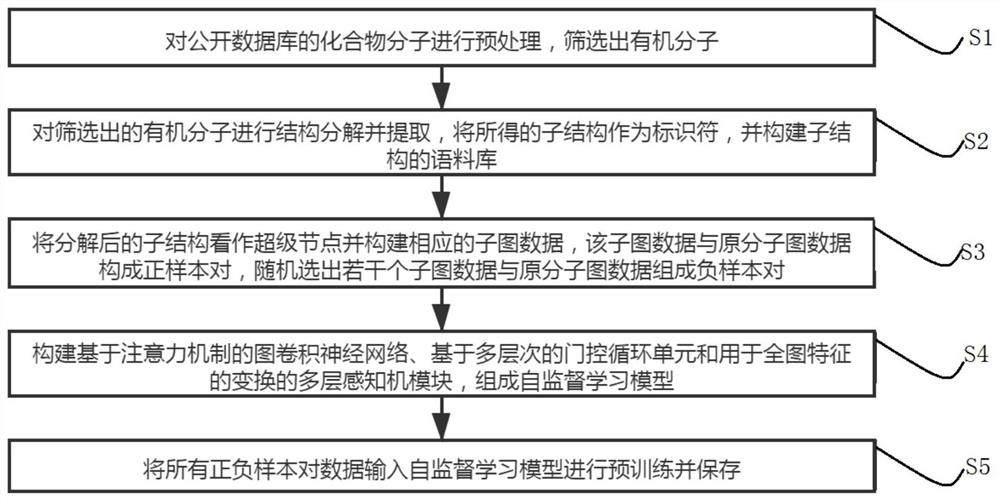
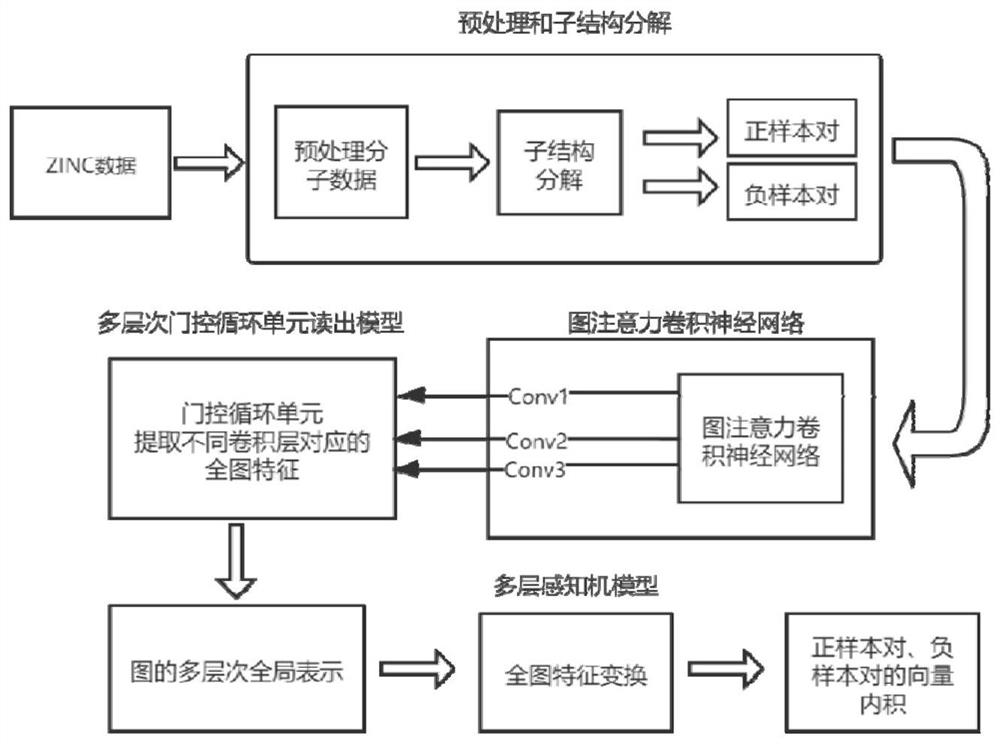
Self-supervised graph neural network pre-training method based on comparative learning
PendingCN114038517AImprove generalization abilitySolve the lack of generalization abilityMolecular entity identificationNeural architecturesGraph neural networksSupervised learning
The invention relates to a self-supervised graph neural network pre-training method based on comparative learning. The method comprises the steps of carrying out preprocessing on compound molecules of a public database, and screening out organic molecules; performing structural decomposition and extraction on the screened organic molecules, taking the obtained substructures as identifiers, and constructing a corpus of the substructures; taking the decomposed substructures as super nodes, and constructing corresponding subgraph data, wherein the subgraph data and the original molecular graph data form positive sample pairs, and a plurality of subgraph data are randomly selected to form negative sample pairs with the original molecular graph data; constructing a graph convolutional neural network based on an attention mechanism, and forming a self-supervised learning model based on a multi-level gating circulation unit and a multi-layer perceptron module; and inputting all positive and negative sample pair data into the self-supervised learning model for pre-training and storing, thereby facilitating fine adjustment of downstream tasks. The problem of insufficient generalization performance generated by deep learning model training in a scene lacking labeled drug molecules is solved.
Owner:JINAN UNIVERSITY
Drug and target interaction prediction method and device, equipment and storage medium
PendingCN113160894AImprove convenienceImprove efficiencyBiostatisticsCharacter and pattern recognitionFree proteinEngineering
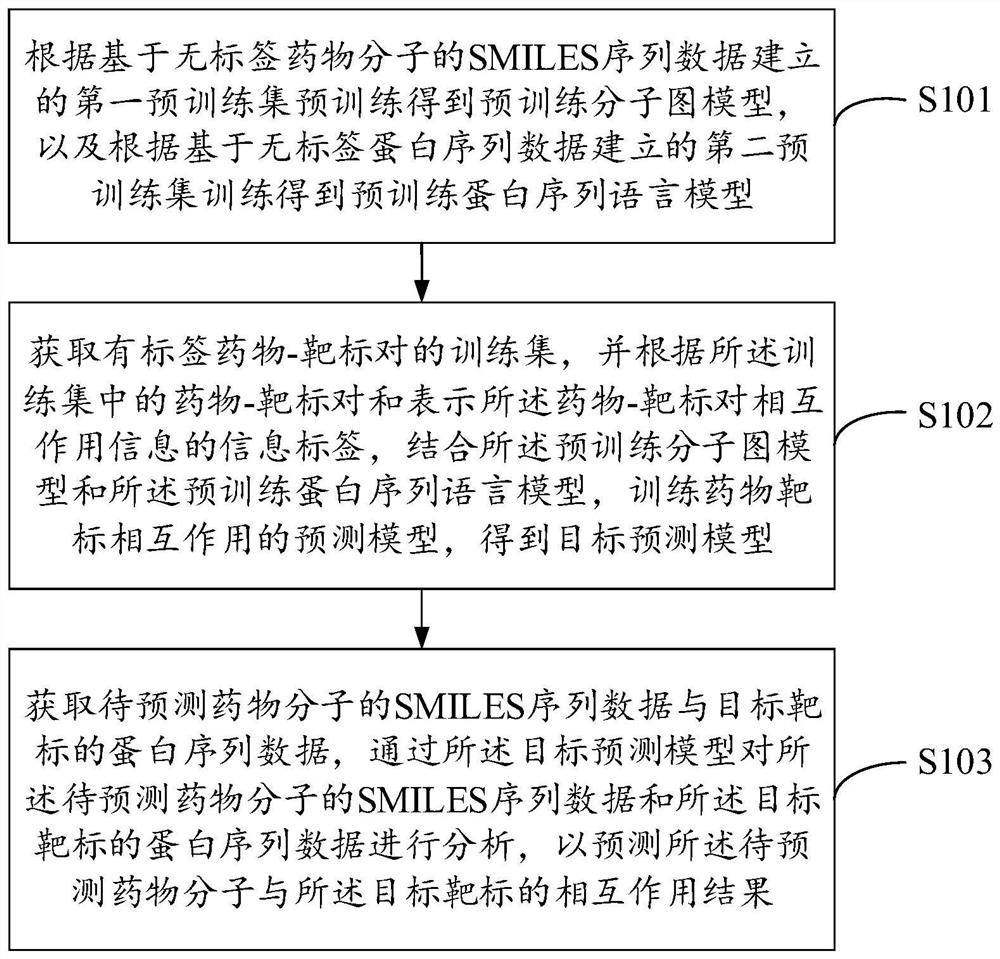
The invention belongs to the technical field of intelligent decision, and provides a drug and target interaction prediction method and device, equipment and a storage medium, and the method comprises the steps: carrying out the pre-training according to a first pre-training set established based on SMILES sequence data of label-free drug molecules, and obtaining a pre-training molecular graph model, training according to a second pre-training set established based on the label-free protein sequence data to obtain a pre-training protein sequence language model; acquiring a training set of labeled drug-target pairs, and training a prediction model of drug-target interaction according to the drug-target pairs in the training set and the information labels representing drug-target pair interaction information in combination with the pre-training molecular graph model and the pre-training protein sequence language model to obtain a target prediction model; and predicting the interaction between the to-be-predicted drug molecule and the target target through the target prediction model. The efficiency and the accuracy of drug and target interaction prediction can be improved.
Owner:PING AN TECH (SHENZHEN) CO LTD
Intelligent molecular design method based on auto-encoder and third-order graph convolution
PendingCN111428848AIncrease contributionBiomolecular computersNeural architecturesAlgorithmTheoretical computer science
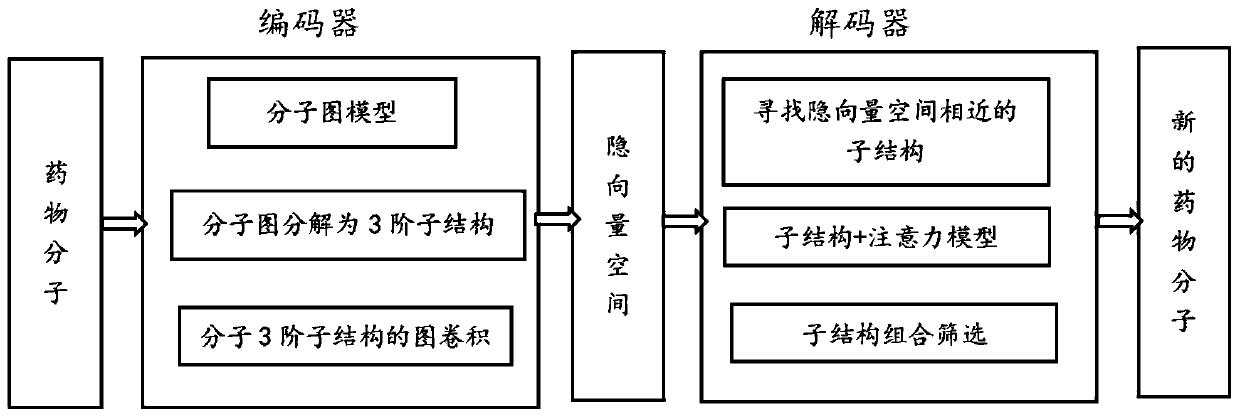
The invention provides an intelligent molecular design method based on an auto-encoder and third-order graph convolution. The method comprises the following steps: 1, enabling an encoder to express amedicine molecule in a molecular graph form, and enabling each molecular graph to be decomposed into a corresponding third-order substructure; 2, performing graph convolution on the third-order substructure by the encoder, and outputting an implicit vector of a molecule to acquire an implicit vector space; 3, training a decoder; and 4, enabling the decoder to find an implicit vector similar to themedicine molecule in the implicit vector space, and decoding the implicit vector to obtain a new medicine molecule. The method disclosed by the invention has the advantages that the three-order substructure of a molecule is subjected to spatial graph convolution by combining an auto-encoder and a graph convolution network method to find out an atom corresponding substructure, namely a functionalgroup, with high contribution degree to the molecule.
Owner:OCEAN UNIV OF CHINA +1
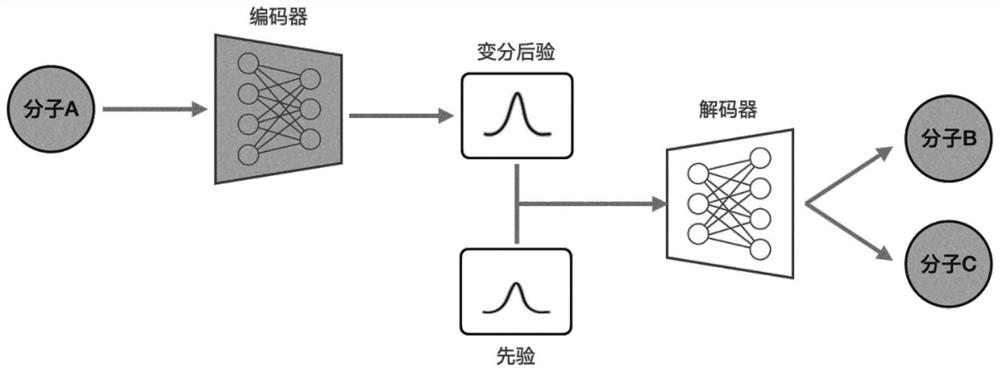
Molecular graph generation method based on variational auto-encoder and message passing neural network
PendingCN113327651AHigh unique rateImprove convenienceMolecular designNeural architecturesChemical databaseAlgorithm
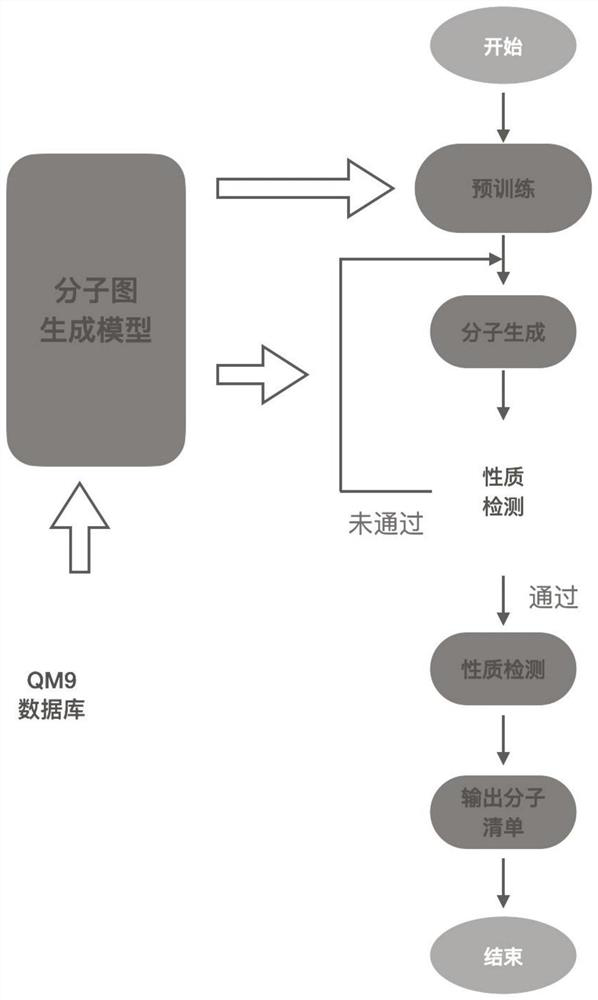
The invention discloses a molecular graph generation method based on a variational auto-encoder and a message passing neural network. The molecular graph generation method is used for molecular generation and molecular target characteristic optimization. According to the method, the message passing neural network is constructed into the encoder and the decoder of the variational auto-encoder, so that the running time and the occupied memory of the training process are further reduced; in addition, by constructing a potential space of the variational auto-encoder, molecular properties are allowed to be optimized; in a molecule generation experiment on a QM9 chemical database, the model can generate 100% effective compounds, and the novelty rate and the uniqueness rate are also very high; in a target optimization experiment on a QM9 chemical database, target characteristics can be further optimized.
Owner:SOUTHEAST UNIV
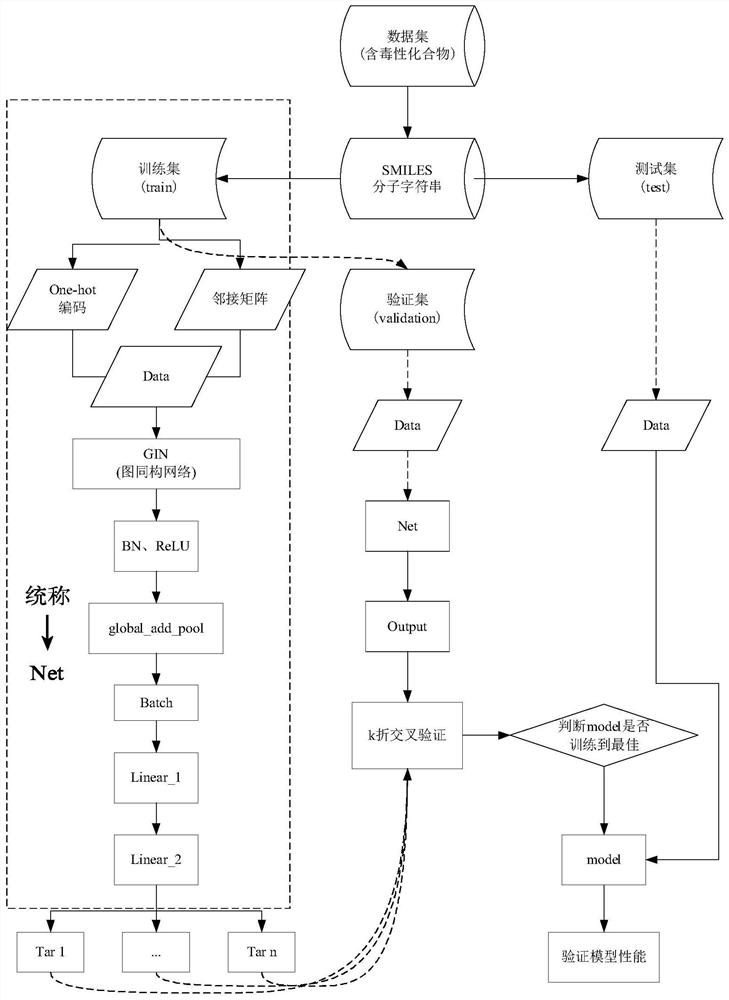
Method and apparatus for molecular toxicity prediction based on multi-task graph neural network
ActiveCN113257369ACombined associativityImprove performanceNeural architecturesChemical machine learningMulti-task learningGraph neural networks
The invention discloses a method and an apparatus for molecular toxicity prediction based on a multi-task graph neural network. The method comprises the following steps: S1, preparing a toxicity data set, and obtaining toxicity data represented by a chemical molecule standard expression; S2, generating an atomic node eigenvector by using the toxicity data which is obtained in the step S1 and represented by the chemical molecule standard expression; S3, generating a side information eigenvector by using the toxicity data which is obtained in the step S1 and represented by the chemical molecule standard expression; S4, on the basis of the atomic node eigenvector obtained in the step S2 and the side information eigenvector obtained in the step S3, constructing a molecular toxicity prediction model based on a multi-task graph neural network; S5, verifying performance of the model. According to the multi-task graph neural network designed for molecular toxicity datasets, an automatic learning molecular graph structure information model is constructed, and the performance of a toxicity prediction task can be improved by using a multi-task learning method in combination with the relevance between molecular toxicity tasks.
Owner:NANJING UNIV OF POSTS & TELECOMM
Deep learning drug interaction prediction method and device, medium and equipment
PendingCN114530258AImprove accuracyReduce adverse reactionsDrug referencesNeural architecturesDrug interactionBiochemistry
The invention provides a deep learning drug interaction prediction method and device, a medium and equipment. The method comprises the following steps: acquiring drug molecule information of two to-be-predicted drugs; the atomic-scale network encodes each piece of drug molecule information, captures interaction information between atoms and chemical bonds, and outputs an encoded drug molecule map to represent zatomj; the molecular level network uses a multi-head attention mechanism to extract the relationship between different drug molecules from each drug molecular map expression zatomj, and outputs a molecular map expression zmolj; and converting the output molecular maps zmol1 and zmol2 of the two drugs into a vector so as to obtain a drug interaction prediction result. According to the method, the problem that side information cannot be fully considered in a traditional framework can be solved, and relation information among different drug molecules can be captured, so that the accuracy of a prediction result is improved.
Owner:SOUTH CHINA UNIV OF TECH
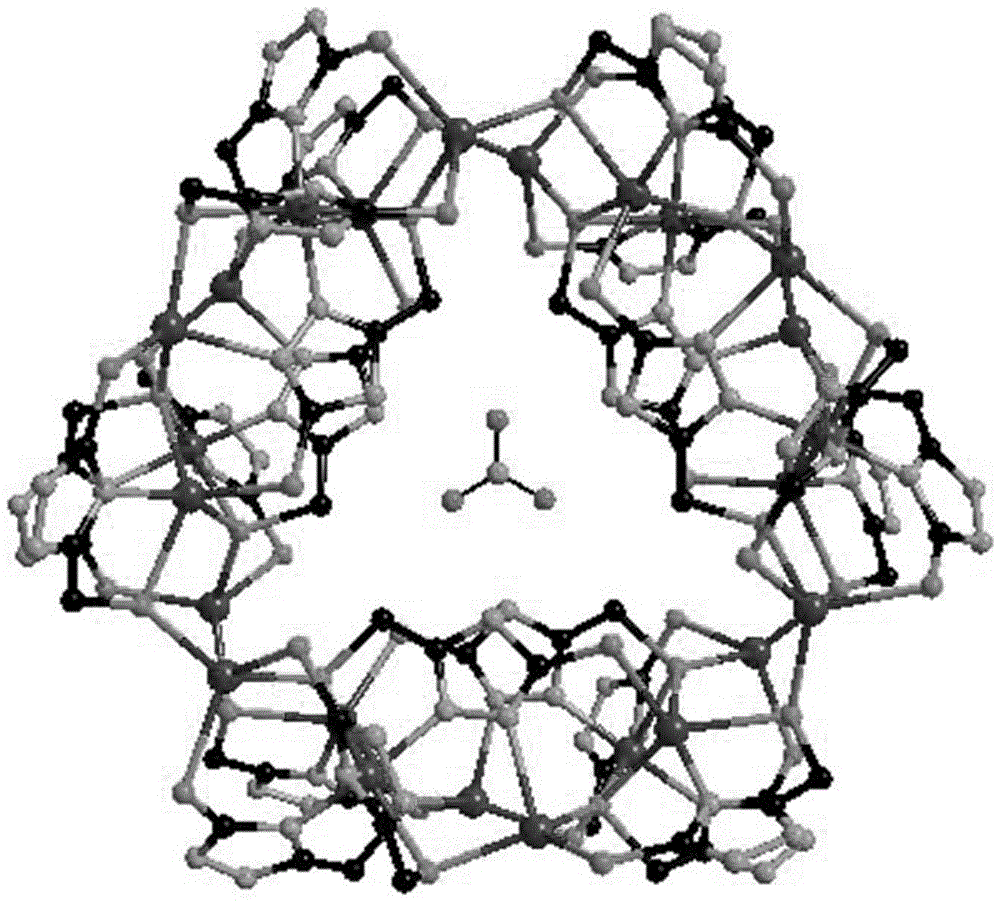
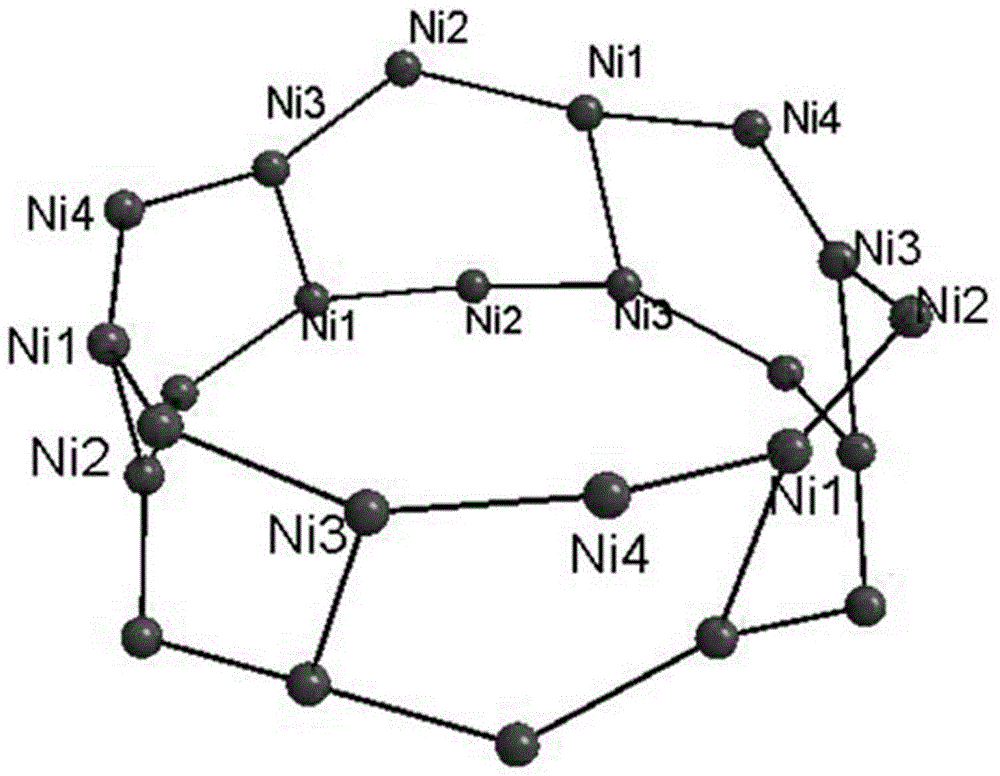
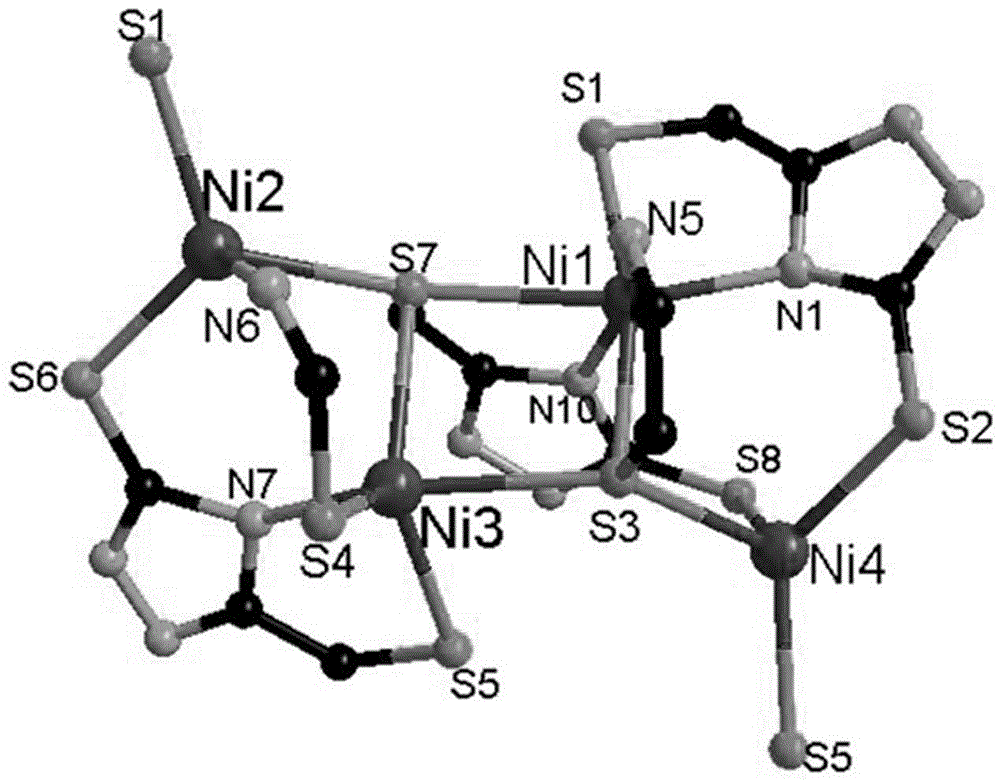
High nuclear nickel cluster based on sulphur-containing triazole ligand and preparation method thereof
The invention discloses a high nuclear nickel cluster based on a sulphur-containing triazole ligand and a preparation method thereof. The high nuclear nickel cluster can be known as a 3-thioketone-5-mercaptomethyl-1,2,4-triazole 24 nuclear nickel cluster from a molecular graph thereof. A method of adding second metal salt cadmium nitrate is adopted during preparation, so that the solubility of the insoluble sulphur-containing triazole cluster is improved. The nickel cluster has potential application values in the aspects of modern medicine, optics, magnetics and selective adsorption and separation of ions, and the preparation method has broad application prospects in preparation of insoluble cluster single crystals.
Owner:GUANGXI NORMAL UNIV
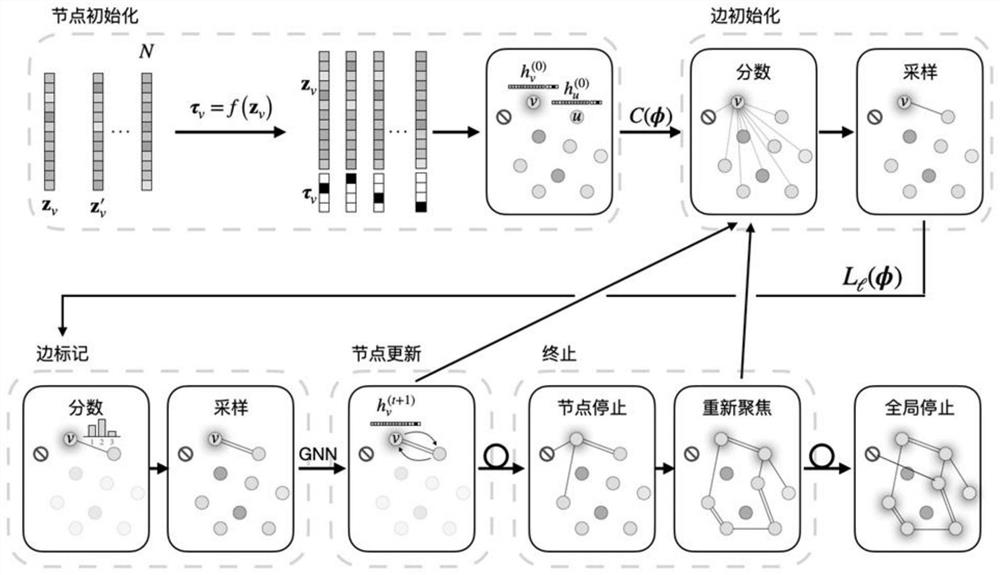
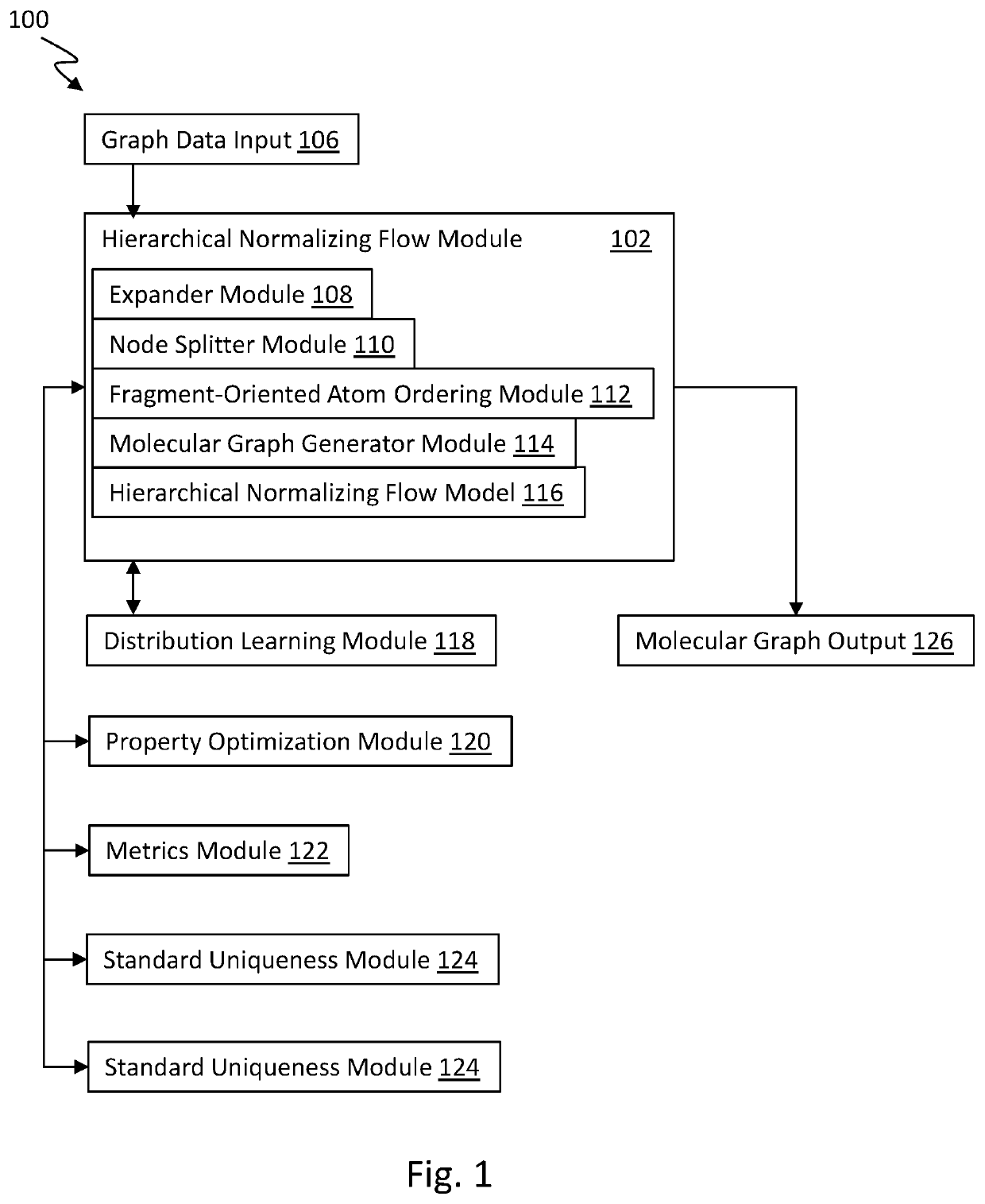
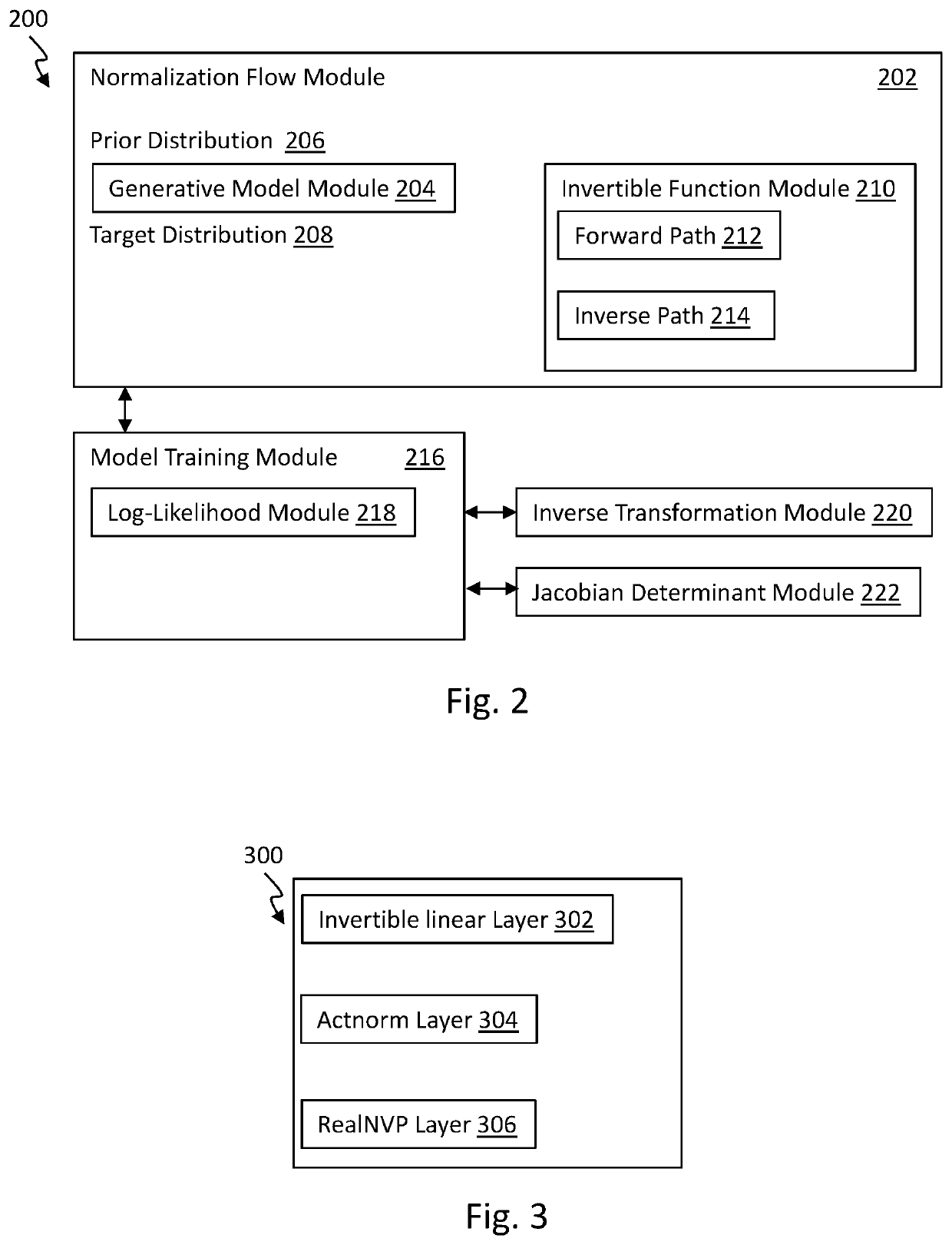
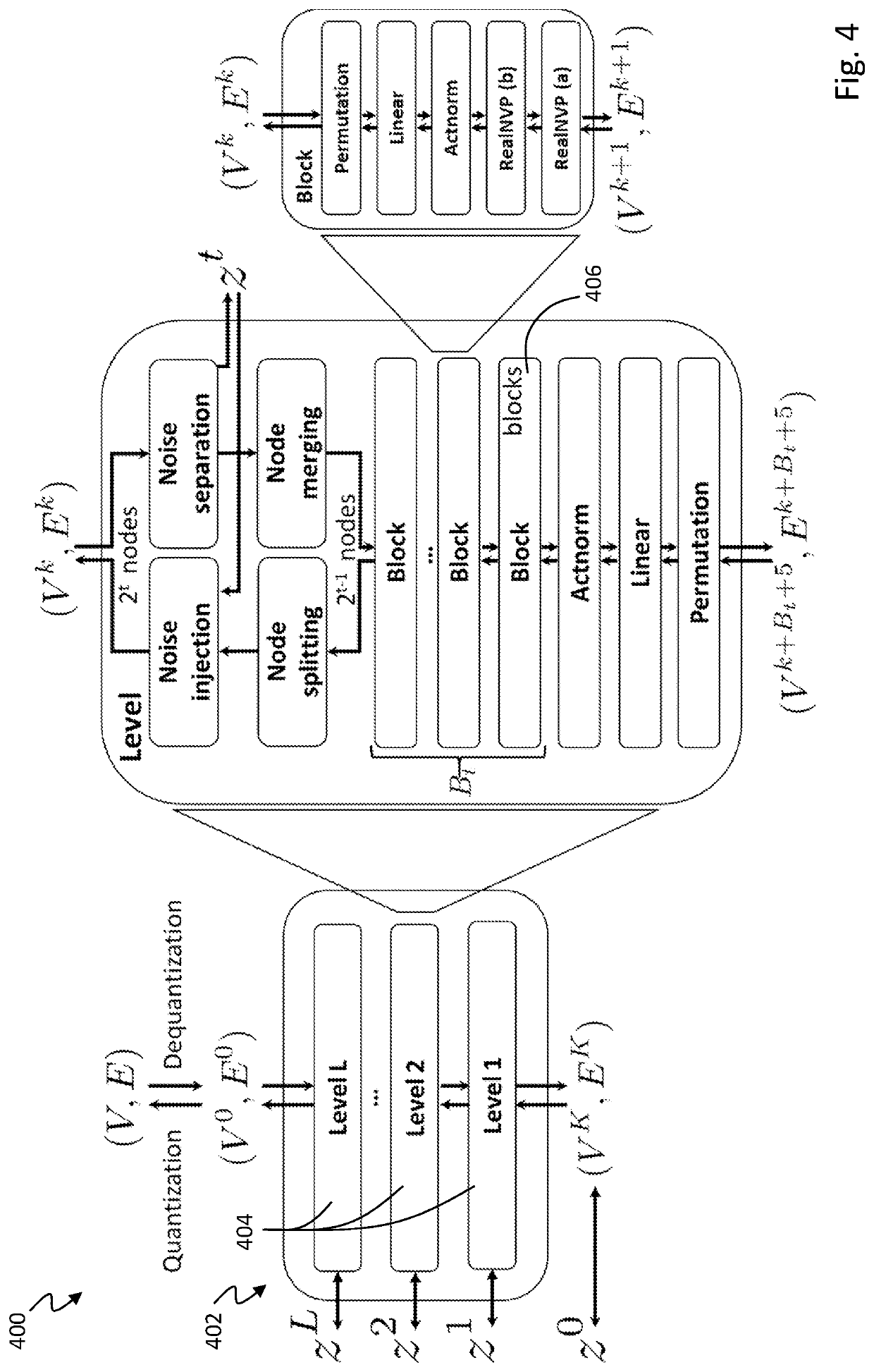
Graph normalizing flow for hierarchical molecular generation
PendingUS20210383898A1High trafficMolecular designDesign optimisation/simulationTheoretical computer scienceMolecular graph
A computing method for normalizing molecule graph data for hierarchical molecular generation can include: providing molecule graph data of a molecule having a node; recursively splitting the node into two nodes; iteratively recursively spilling other nodes into two nodes; generating generated molecular graph data of a generated molecule from node splitting; and providing a report with the generated molecular graph. A computing method can include: providing molecule graph data into a latent code generator having multiple levels with a forward and inverse; and generating latent codes by processing molecule graph data through multiple levels of operations, wherein each level of operations has a sequence of sublevels of operations in forward path and inverse path, wherein the sublevels of operations include node merging operation and node splitting operation; generating at least one molecular structure from latent codes; and outputting generate molecule graph data having the at least one molecular structure.
Owner:INSILICO MEDICINE IP LTD
Method for enhancing point-edge interaction of graph neural network
ActiveCN111860768AEnhanced interactionFocus on transferabilityCharacter and pattern recognitionMachine learningGraph structured dataAlgorithm
The invention provides a method for enhancing point-edge interaction of a graph neural network.; obtaining a directed molecular graph G and graph structure data thereof; according to the graph structure data, obtaining all node original features Xv in all and the graph structure data according to all and all created node original features Xv in all and the graph structure data according to all andall created node original features Xv in all and the graph structure data; the graph neural network is iterated to the Kth layer, a final node representation form h (v) of the directed molecular graph is obtained, k is larger than or equal to 1, and K is larger than k; the hidden representation from the adjacent node w of each arbitrary node v to the edge of the arbitrary node v is utilized, namely, the message vector of the arbitrary node v on the kth layer is created, so that the information of the edge is associated and transmitted with the node information, the embedding of the nodes andthe edge is updated in the neural network training process, and the transmissibility of the information between the nodes and the edge is concerned.
Owner:SUN YAT SEN UNIV
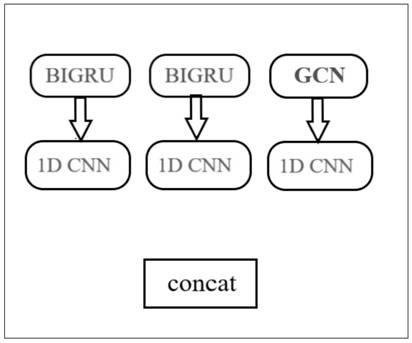
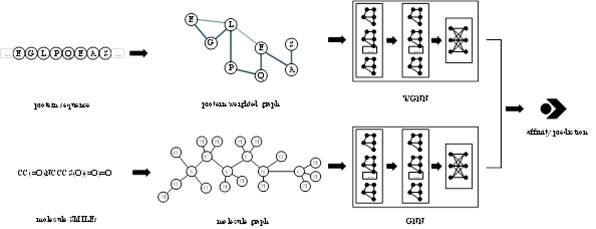
Novel deep learning model for predicting compound protein affinity, computer equipment and storage medium
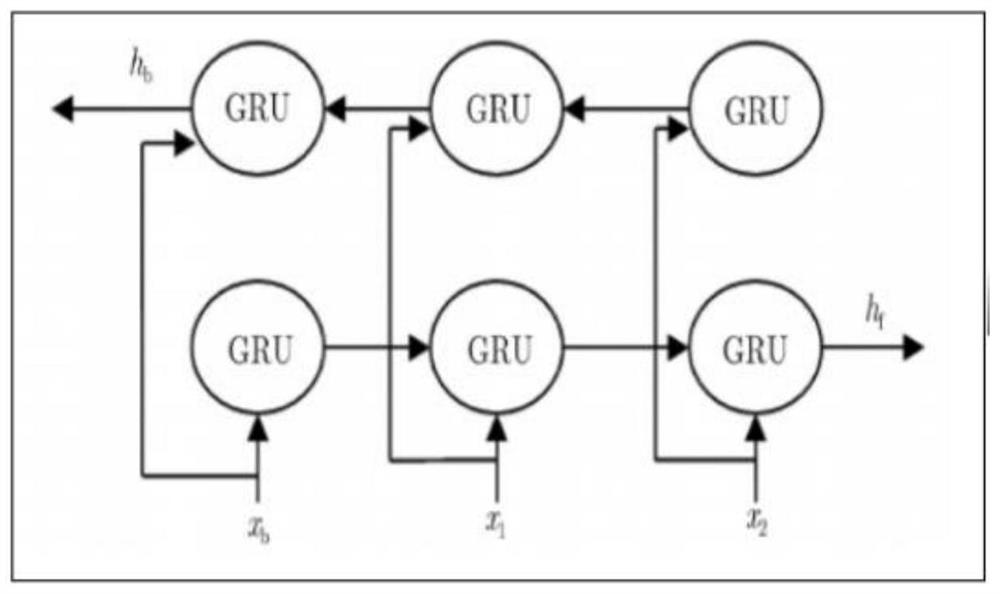
The invention discloses a novel deep learning model for predicting compound protein affinity. The novel depth model comprises a BiGRU (Bipolar Gated Recirculation Unit) model, a GCN (Graph Convolutional Neural Network) model and a CNN (Convolutional Neural Network) model, wherein the whole network architecture is BiGRU / BiGRU / GCN-CNN. The bidirectional gating cycle unit model comprises a sequence processing model composed of two gating cycle units (GRU), one input is forward input, the other input is reverse input, and the bidirectional gating cycle unit model is a bidirectional recurrent neural network with only an input gate and a forgetting gate. The input of the model is a compound one-dimensional SMILES sequence, a protein sequence and a compound two-dimensional molecular diagram, andthe three sequences are respectively input into a BiGRU / BiGRU / GCN model. BiGRU / BiGRU / GCN output represents a feature vector of the compound and a feature vector of the protein. The CNN model is composed of a convolution layer, a pooling layer and a full connection layer, and inputs of the model are a feature vector of a compound and a feature vector of a protein; The final output of the BiGRU / BiGRU / GCN-CNN model is a root mean square error value for predicting a compound protein affinity value.
Owner:CHINA UNIV OF PETROLEUM (EAST CHINA)
Small molecule drug hERG toxicity prediction method and device based on graph attention mechanism transfer learning
ActiveCN113192571AImprove performancePredictive attribute value contributes a lotChemical property predictionMolecular designData setAlgorithm
The invention discloses a graph attention mechanism transfer learning-based small molecule drug hERG toxicity prediction method and device; the method comprises the following steps: S1, data set preprocessing: enabling a to-be-detected drug-like compound to generate a fingerprint sequence through molecular fingerprint generation software; s2, acquiring atom and chemical bond characteristics through the fingerprint sequence generated in the step S1, and constructing a molecular graph and graph characteristics through the atom and chemical bond characteristics; s3, processing the molecular map obtained in the step S2 through a map attention mechanism, and generating a feature vector of each atom in the molecule; s4, generating a molecular feature vector through a graph attention mechanism and the feature of each atom. According to the method, a molecular graph structure is processed based on a graph attention mechanism, a substructure which contributes to a predicted attribute value is effectively obtained, and a source domain data set and a target domain data set are processed based on transfer learning; and thereby, the problem that the sample size is insufficient is effectively solved.
Owner:NANJING UNIV OF POSTS & TELECOMM
Drug small molecule property prediction method, device and equipment based on graph neural network
PendingCN113707236AGuaranteed property prediction accuracyFit closelyChemical property predictionMolecular entity identificationEngineeringGraph neural networks
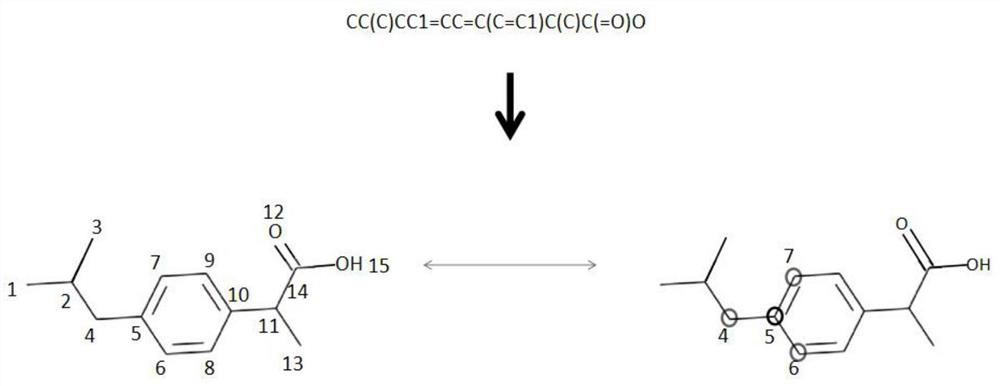
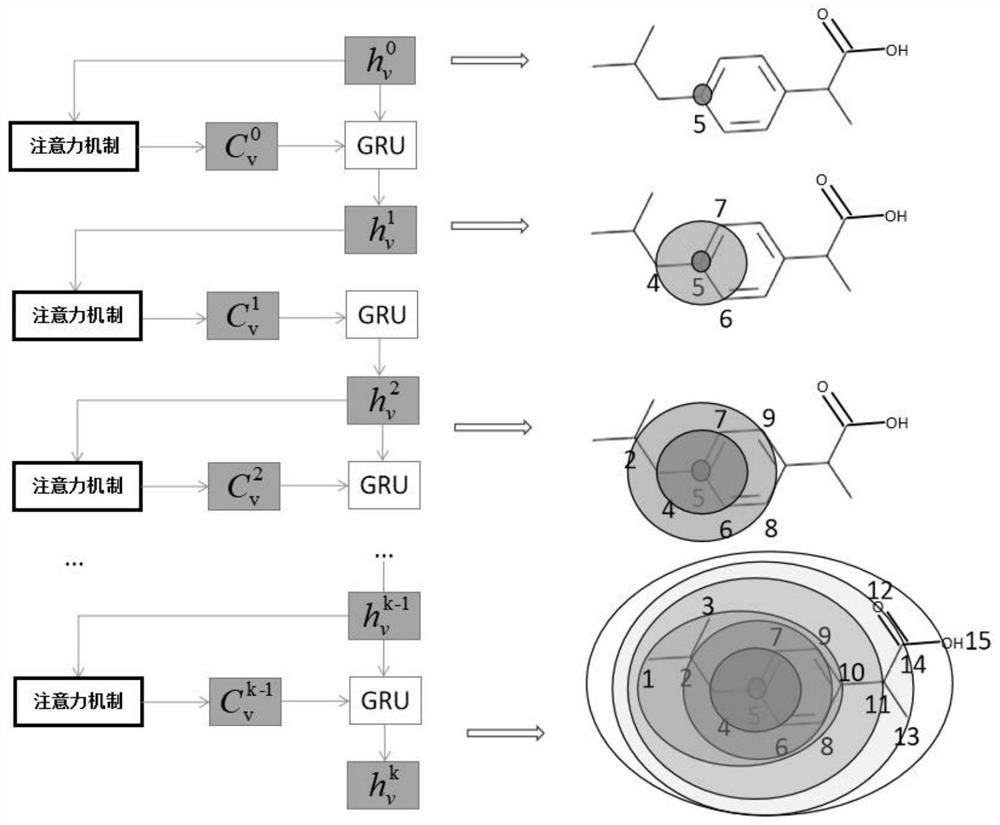
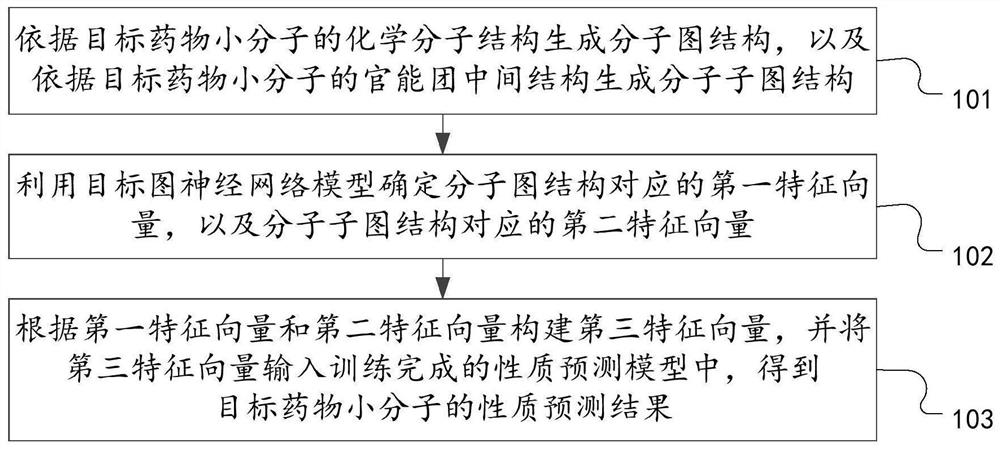
The invention discloses a drug small molecule property prediction method, device and equipment based on a graph neural network, relates to the technical field of artificial intelligence, and can solve the technical problems of low efficiency and low accuracy of drug small molecule property prediction at present. The method comprises the following steps: generating a molecular graph structure according to a chemical molecular structure of a target drug small molecule, and generating a molecular sub-graph structure according to a functional group intermediate structure of the target drug small molecule; determining a first feature vector corresponding to the molecular graph structure and a second feature vector corresponding to the molecular sub-graph structure by using a target graph neural network model; and constructing a third feature vector according to the first feature vector and the second feature vector, and inputting the third feature vector into a trained property prediction model to obtain a property prediction result of the target drug small molecule. The method is suitable for realizing intelligent prediction of drug micromolecule properties based on an artificial intelligence technology.
Owner:PING AN TECH (SHENZHEN) CO LTD
Article molecule generation method, device and equipment, and storage medium
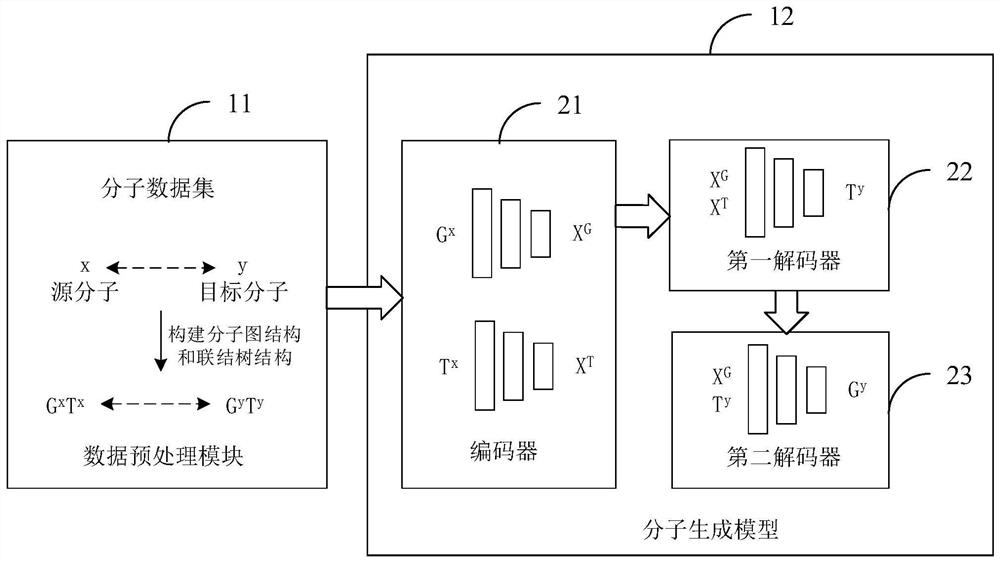
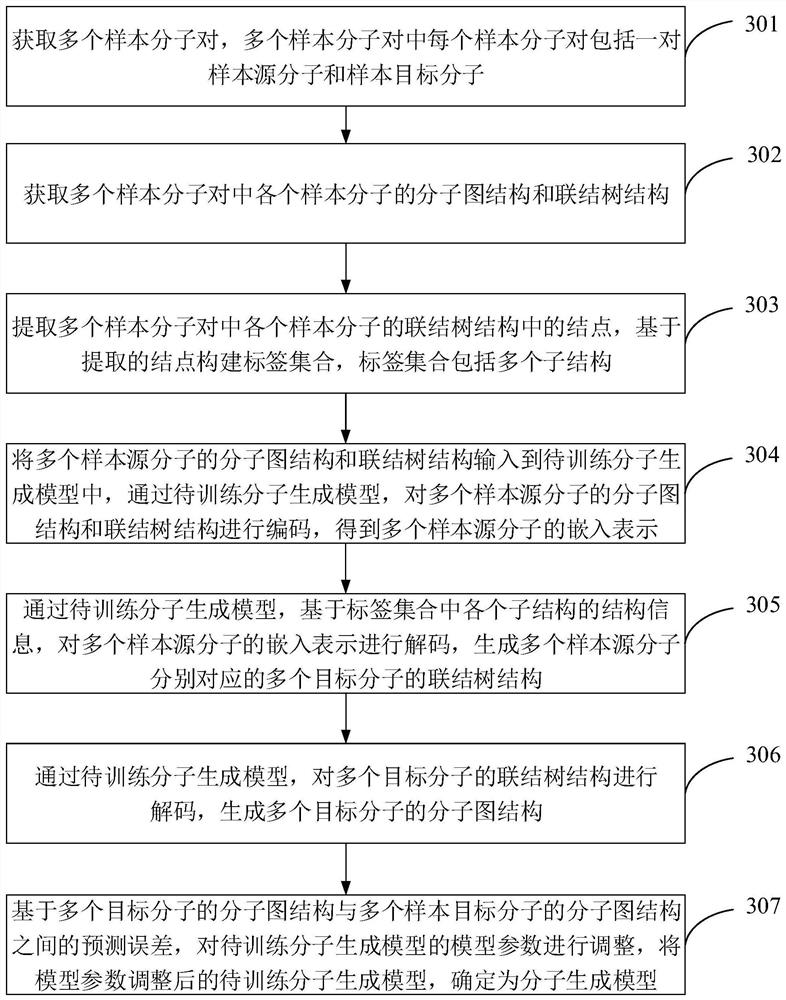
PendingCN112199884AMitigate the effects of imbalanceImprove rationalityDesign optimisation/simulationNeural architecturesTheoretical computer scienceMolecular graph
The invention is suitable for the technical field of computer aided article design, and provides an article molecule generation method, device and equipment and a storage medium. The method comprisesthe following steps: inputting a first molecular diagram structure and a first connection tree structure of a source molecule into a molecule generation model, and encoding the first molecular diagramstructure and the first connection tree structure through the molecule generation model to obtain embedded representation of the source molecule; decoding based on the structure information of each substructure in the label set and the embedded representation to generate a second connection tree structure of the target molecule; and decoding the second connection tree structure to obtain a secondmolecular diagram structure of the target molecule. The structure information of each substructure in the tag set and the embedded representation of the source molecule are combined for decoding, sothat the structure information of each substructure in the tag set can be well utilized to predict the connection tree structure of the target molecule, the reasonability of the prediction result is improved, and the influence of tag imbalance is relieved.
Owner:SHENZHEN INST OF ADVANCED TECH
Drug and target interaction prediction method and device, equipment and storage medium
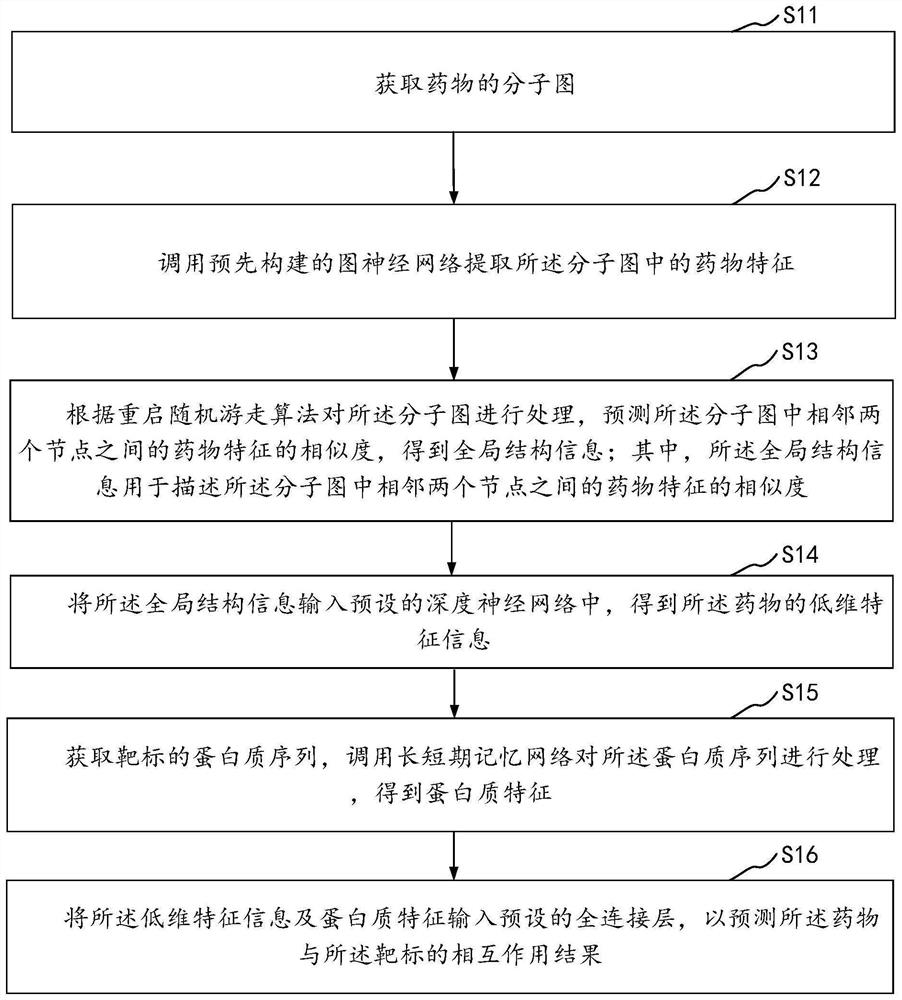
PendingCN114822683AImprove forecasting efficiencyImprove forecast accuracyMolecular designCharacter and pattern recognitionPredictive methodsEngineering
The invention relates to the technical field of neural networks of the artificial intelligence technology, and provides a drug and target interaction prediction method, device and equipment and a storage medium, and the method comprises the steps: calling a pre-constructed graph neural network to extract drug features in a molecular graph of a drug, processing the molecular map according to a restart random walk algorithm, predicting the similarity of drug characteristics between two adjacent nodes in the molecular map to obtain global structure information, inputting the global structure information into a preset deep neural network to obtain low-dimensional characteristic information of drugs, and obtaining a protein sequence of a target, and calling a long-short-term memory network to process the protein sequence to obtain protein features, and inputting the low-dimensional feature information and the protein features into a preset full-connection layer to predict an interaction result of the drug and the target so as to extract information contained in the drug and the protein sequence in a targeted manner. And the prediction efficiency of the interaction between the drug and the target is improved.
Owner:PING AN TECH (SHENZHEN) CO LTD
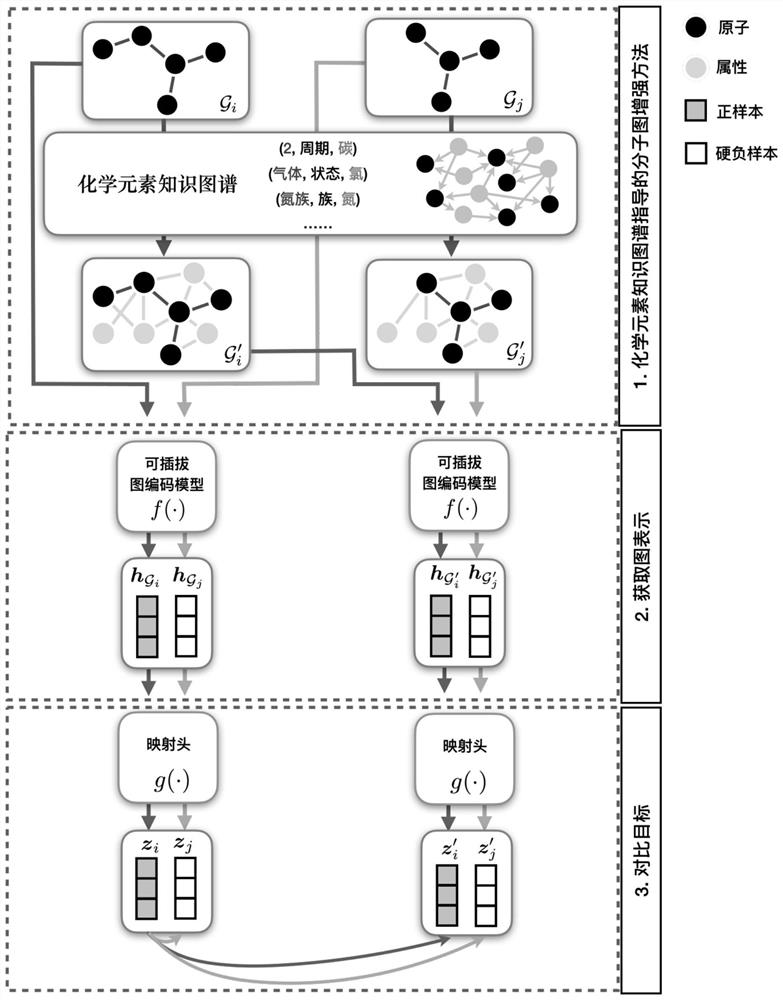
Molecular graph comparison learning method based on chemical element knowledge graph
PendingCN113990408APreserve basic structural informationEasy to learnChemical property predictionCharacter and pattern recognitionAlgorithmTheoretical computer science
The invention discloses a molecular graph comparison learning method based on a chemical element knowledge graph. The method comprises the following steps of: constructing a chemical element knowledge graph according to all chemical attributes of each chemical element in a periodic table of chemical elements; performing graph enhancement on the molecular graph by utilizing the chemical element knowledge graph to obtain a molecular enhancement graph; obtaining graph representations of the molecular graph and the molecular enhancement graph by using a pluggable representation model; adopting a hard negative sample mining technology to select other molecular graphs similar to the molecular graph in the molecular fingerprint space as negative samples; mapping the graph representation of positive sample pairs and the graph representation of negative sample pairs to the same space, constructing a contrast loss function by maximizing the consistency between the positive sample pairs and minimizing the consistency between the negative sample pairs, and performing optimization learning by using the contrast loss function; and forming a prediction model by using the parameter-determined pluggable representation model and a nonlinear classifier, and predicting the molecular properties of the molecular graph by using a parameter-finely-adjusted prediction model so as to improve the prediction accuracy of molecular properties.
Owner:ZHEJIANG UNIV
Intelligent prediction method for small molecule-protein binding affinity
PendingCN114333984ASolve the problem that it is difficult to quickly mine protein structure informationAccurate Sequence CharacterizationNeural architecturesNeural learning methodsAlgorithmTheoretical computer science
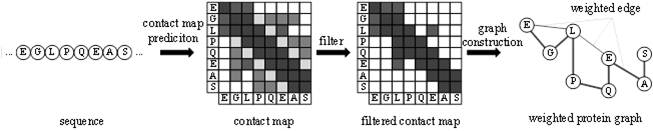
The invention provides a protein weight graph-based small molecule-protein binding affinity intelligent prediction method, which comprises the following steps of: firstly, constructing a small molecule graph based on SMILES, constructing a protein weight graph based on a sequence, and secondly, respectively extracting the characteristics of the small molecule graph and the protein weight graph by adopting two graph convolutional neural networks; and constructing a graph convolutional neural network to extract the features of the two, splicing the obtained feature vectors, and further predicting the affinity of the two. According to the protein weight graph constructed by the invention, more accurate representation of the sequence is realized, and the interaction among amino acid residues is more intuitively and effectively represented by a graph structure; the constructed protein weight graph does not need to be subjected to an extremely complex sequence alignment process, so that data processing is quicker, the method is suitable for a virtual screening process of a large molecular database, and more accurate small molecule-protein affinity prediction can be realized.
Owner:QINGDAO TECHNOLOGICAL UNIVERSITY
Directional molecule generation method based on graph neural network
PendingCN113140267AEnsure chemical validityMolecular designNeural architecturesAlgorithmGraph generation
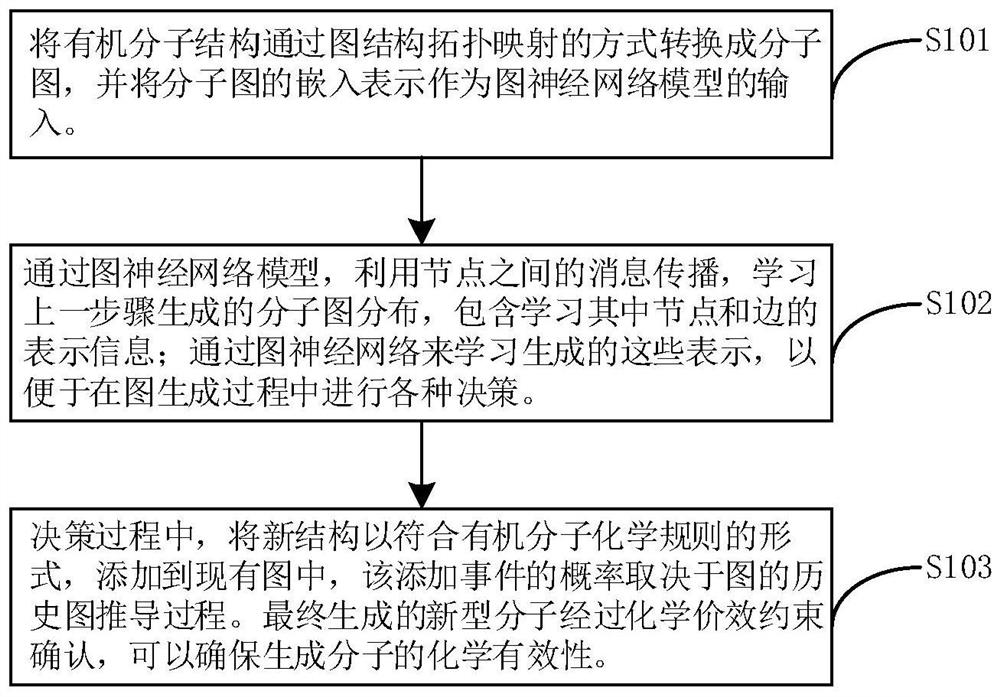
The invention relates to a directed molecule generation method based on a graph neural network, and relates to the technical field of material molecules. Comprising the following steps: converting an organic molecular structure graph into a molecular graph in a topological mapping mode, and taking embedded representation of the molecular graph as input of a graph neural network model; through a graph neural network model, learning the molecular graphs based on a message propagation process, including representations of nodes and edges in the molecular graphs; the generated representations are learned through a graph neural network so that various decisions can be made in the graph generation process; in the decision-making process, the new structure is added to the existing graph in a form conforming to the organic molecule chemical rule, and the probability of the addition event depends on the historical graph derivation process of the graph. The finally generated novel molecules are confirmed through chemical valence constraint, and the chemical effectiveness of the generated molecules can be ensured. According to the method, effective novel molecular structures with chemical properties similar to those of original molecules can be generated aiming at an organic molecule database.
Owner:BEIJING UNIV OF CHEM TECH
Data-mechanism driven material attribute prediction method of graph neural network
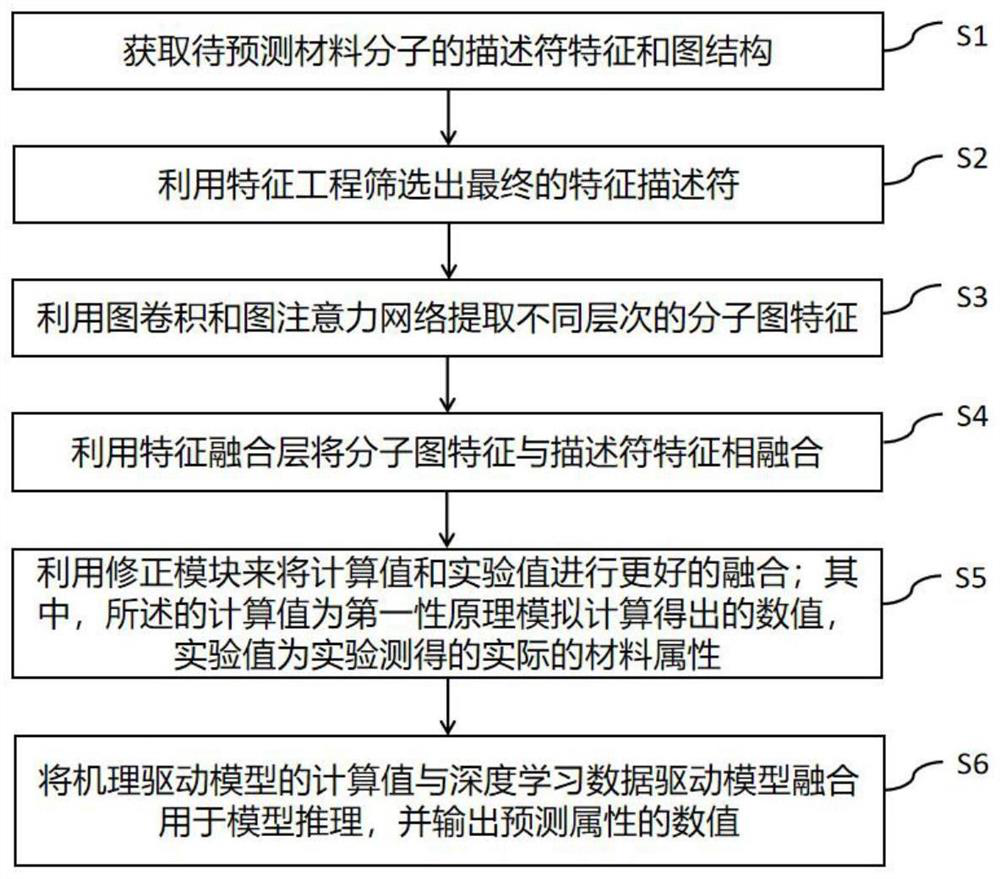
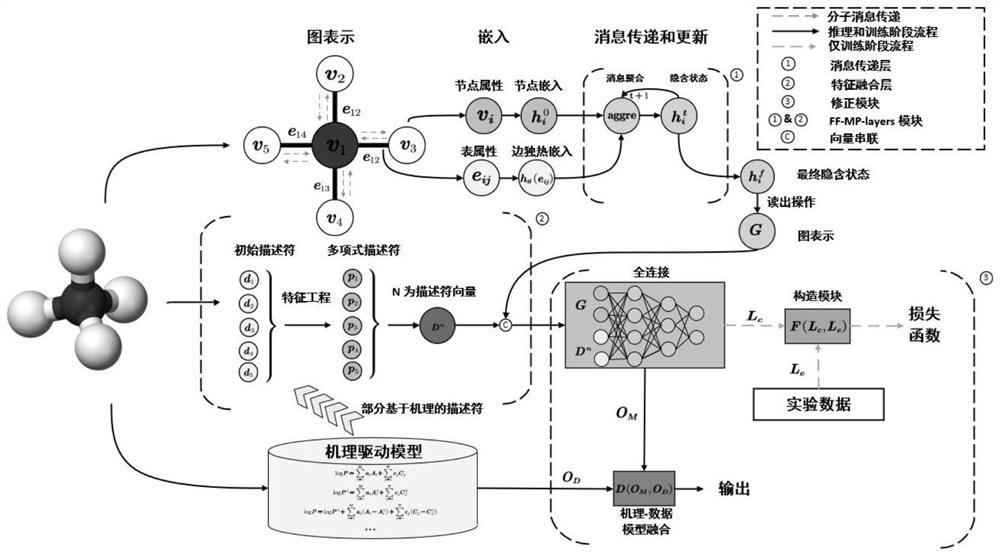
ActiveCN114818948AEasy to integrateImprove accuracyCharacter and pattern recognitionNeural architecturesPattern recognitionAlgorithm
The invention discloses a data-mechanism driven material attribute prediction method of a graph neural network. The method comprises the following steps: S1, obtaining descriptor features and a graph structure of a to-be-predicted material molecule; s2, a final feature descriptor is screened out through feature engineering; s3, using graph convolution and a graph attention network to extract different levels of molecular graph features; s4, fusing the molecular graph features with the descriptor features by using a feature fusion layer; s5, using a correction module to better fuse the calculated value and the experimental value; wherein the calculated value is a numerical value obtained through simulation calculation according to a first principle, and the experimental value is an actual material attribute measured through an experiment; and S6, fusing the calculated value of the mechanism-driven model and the deep learning data-driven model for model reasoning, and outputting a numerical value of a prediction attribute. According to the method, descriptor features of molecules and graph structure features are fused, and the problems that graph structure data information is incomplete and molecular attributes are ignored by the descriptor features are solved.
Owner:UNIV OF SCI & TECH BEIJING
System and method for molecular reconstruction from molecular probability distributions
A system and method comprising a transmoler that identifies common substructures of a given 3D conformer and predicts its structural information. First, based on contrastive learning, substructure embeddings are learned in an unsupervised manner. Secondly, a novel oriented 3D object regressor predicts the dimensions and directions of each substructure in a conformer as well as its fingerprint embedding which are used to create differentiable junction tree molecular graphs. Lastly, using the junction tree graphs, molecular representations such as DeepSMILES are generated which represent new and novel molecules. The system may also generate conformers directly from a pocket. A pocket may be input to the model and the model learns to generate structures which can fit that pocket by conditioning the generative system. Furthermore, structure-based contrastive embeddings generated for transmoler can be recycled in structure-based generative modelling.
Owner:RO5 INC
Drug molecule property prediction method, device and equipment based on comparative learning
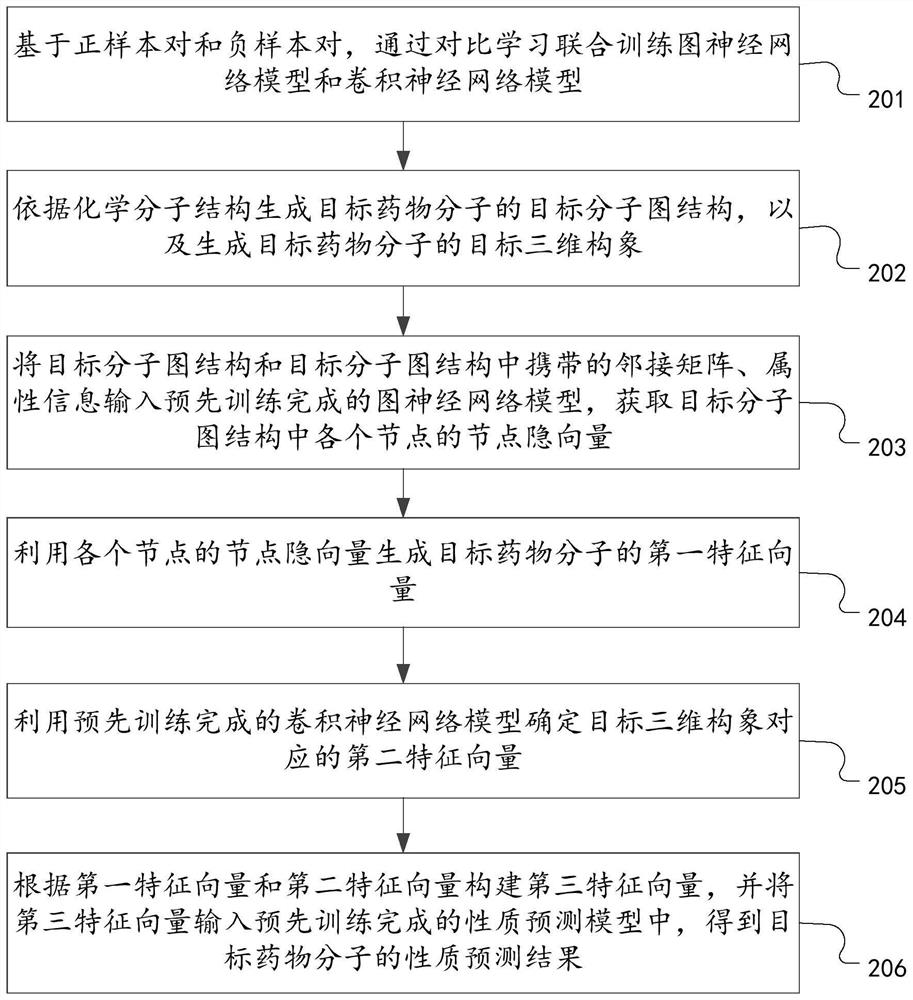
PendingCN114386694AComputationally efficientImprove generalization abilityMolecular designForecastingPredictive methodsPharmaceutical drug
The invention discloses a drug molecule property prediction method, device and equipment based on comparative learning, relates to the technical field of artificial intelligence, and can solve the technical problems of low efficiency and poor prediction performance of drug molecule property prediction at present. Comprising the following steps: generating a target molecular map structure of a target drug molecule according to a chemical molecular structure, and generating a target three-dimensional conformation of the target drug molecule; using the trained graph neural network model to determine a first feature vector corresponding to the target molecular graph structure; a second feature vector corresponding to the target three-dimensional conformation is determined through a trained convolutional neural network model, and the graph neural network model and the convolutional neural network model are obtained through comparative learning and joint training of a positive sample pair and a negative sample pair; and constructing a third feature vector according to the first feature vector and the second feature vector, and inputting the third feature vector into the trained property prediction model to obtain a property prediction result of the target drug molecule.
Owner:PING AN TECH (SHENZHEN) CO LTD
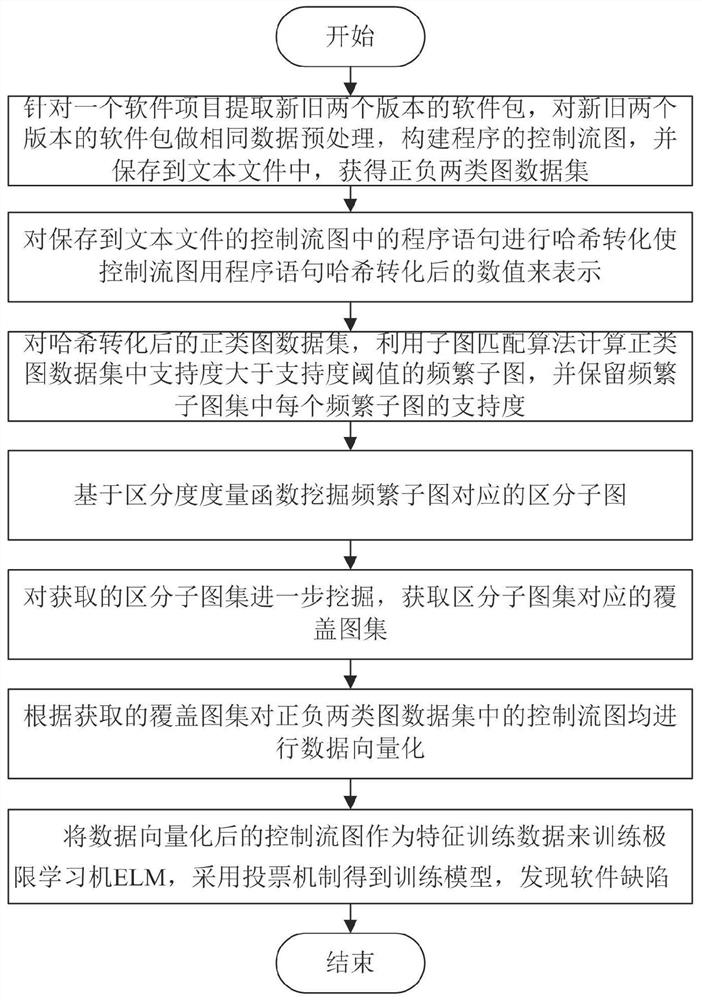
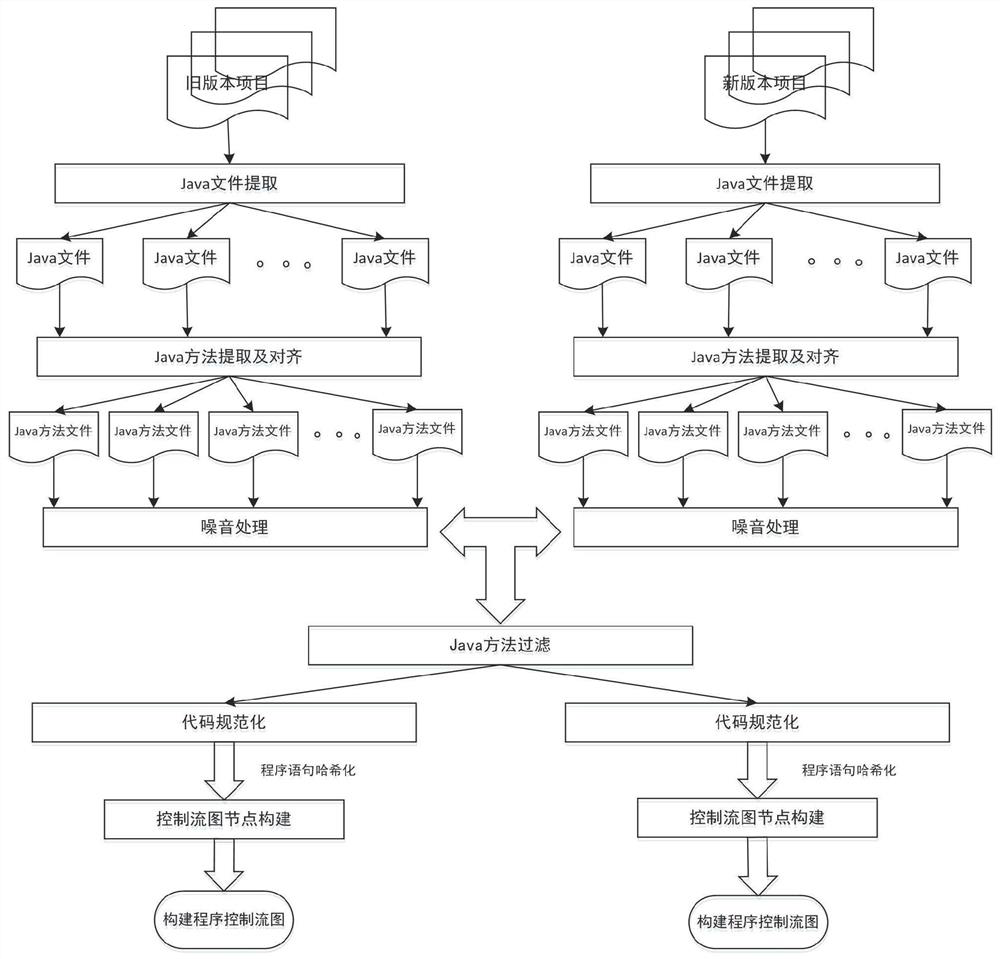
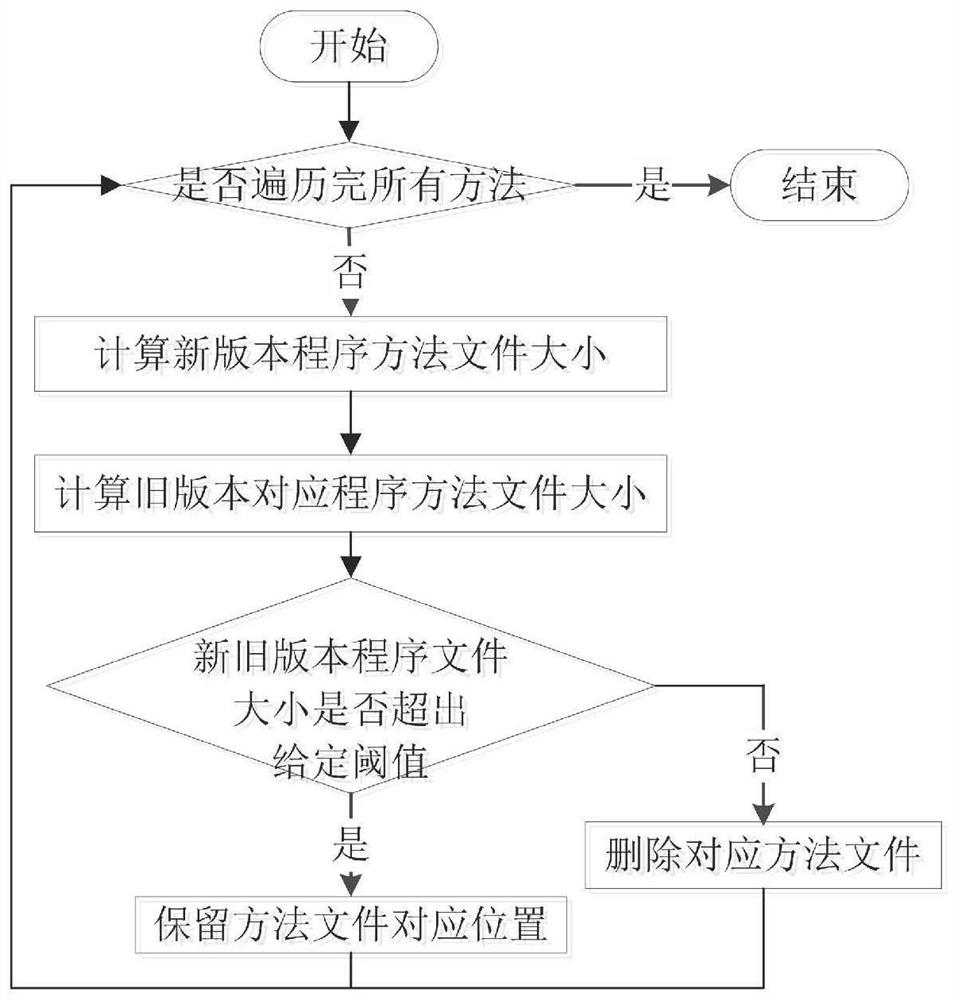
Software defect discovery method based on regional molecular graph mining
The invention provides a software defect discovery method based on regional molecular graph mining, and relates to the technical field of software engineering. The method comprises the following steps: firstly, extracting software packages of a new version and an old version from a software project, performing same data preprocessing on the software packages of the new version and the old version,constructing a control flow graph of a program, and storing the control flow graph into a text file to obtain a positive graph data set and a negative graph data set; performing hash conversion on the program statements stored in the control flow diagram of the text file, so that the control flow diagram is represented by numerical values obtained after Hash conversion of the program statements;carrying out overlay mining on the obtained hash-converted positive and negative graph data sets to obtain an overlay graph set; performing data vectorization on the control flow graphs in the positive and negative graph data sets according to the coverage graph set; and training an extreme learning machine by taking the control flow diagram subjected to data vectorization as feature training data, obtaining a training model by adopting a voting mechanism, and testing a to-be-tested program file through the tested training model to discover software defects.
Owner:NORTHEASTERN UNIV
Molecular feature determination method, related device and equipment
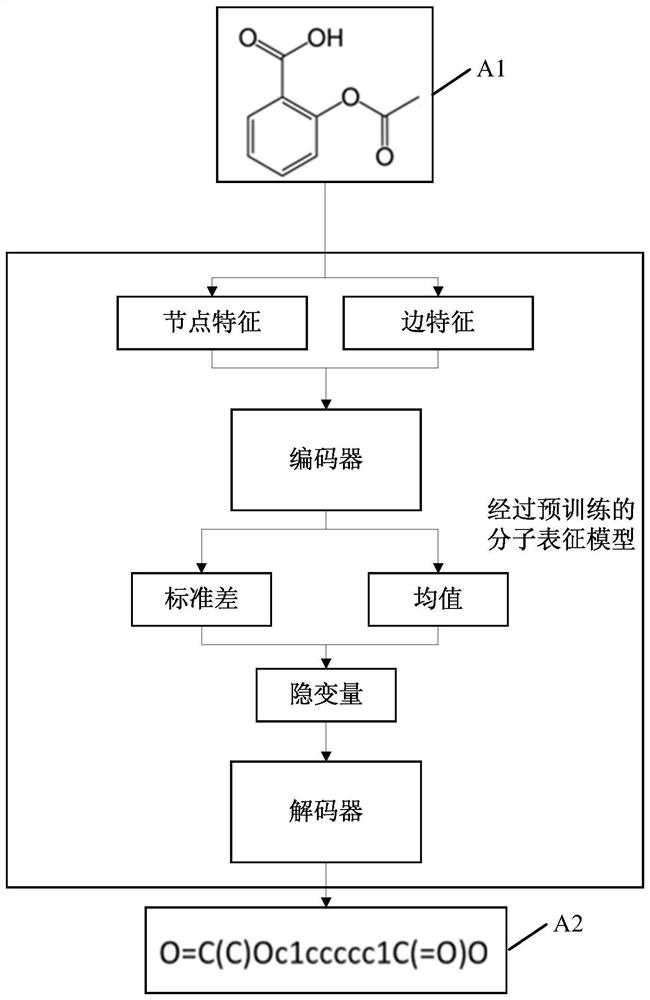
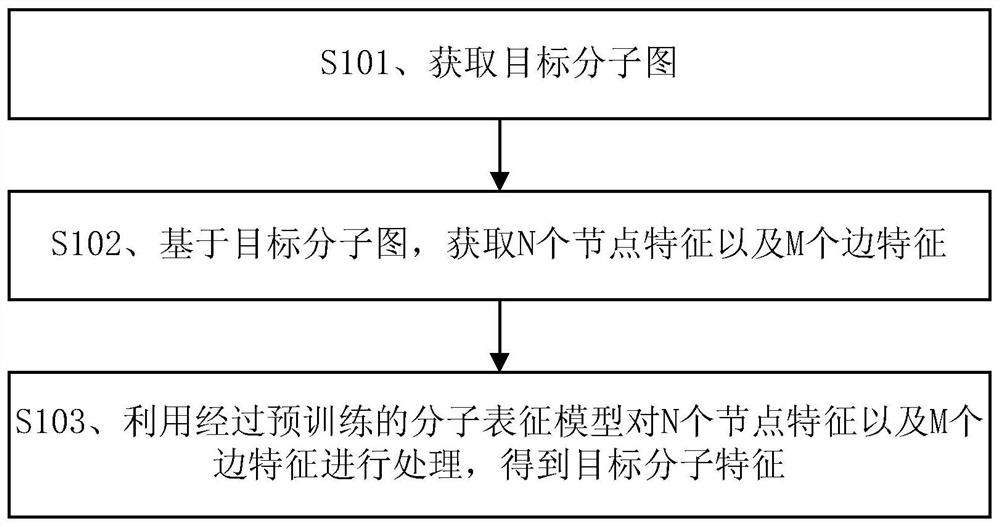
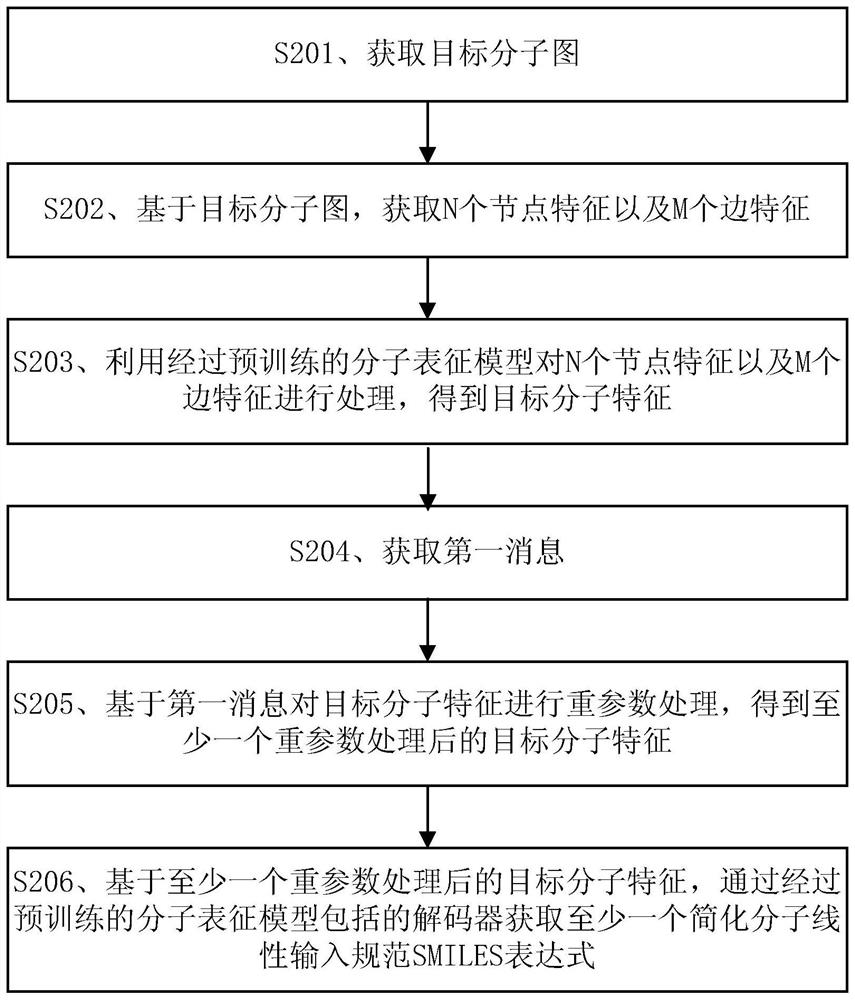
The embodiment of the invention discloses a molecular feature determination method, a related device and equipment, which are used for improving the accuracy of determined molecular features. In the method provided by the embodiment of the invention, a target molecular map is acquired, then N node features and M edge features are acquired based on the target molecular map, N and M are integers greater than 1, and the N node features and the M edge features are further processed by using a pre-trained molecular representation model to obtain target molecular features. The pre-trained molecular representation model comprises an encoder and a decoder, when the molecular representation model is pre-trained, the encoder is used for encoding features of a training molecular graph, the decoder is used for reconstructing an encoding result of the encoder into a simplified molecular linear input specification SMILES expression, and parameters of the encoder and the decoder are iteratively adjusted.
Owner:HUAWEI CLOUD COMPUTING TECH CO LTD
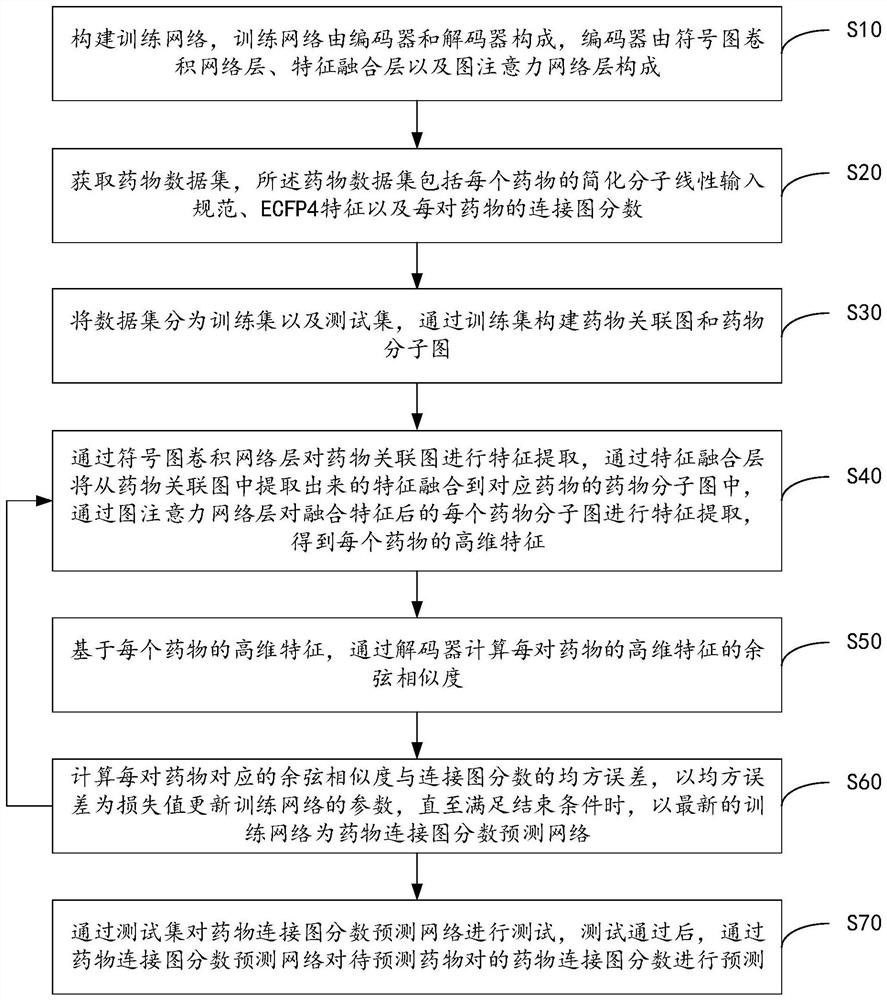
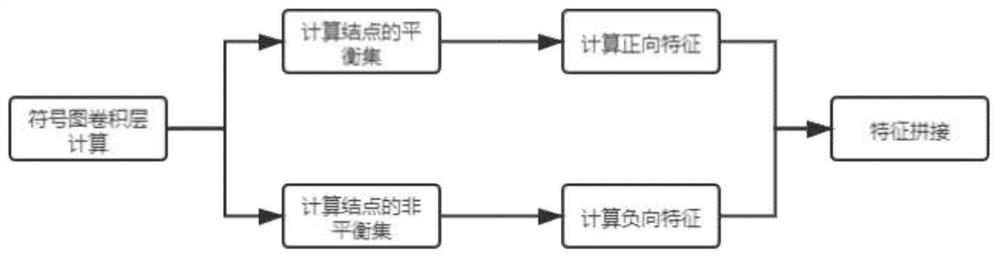
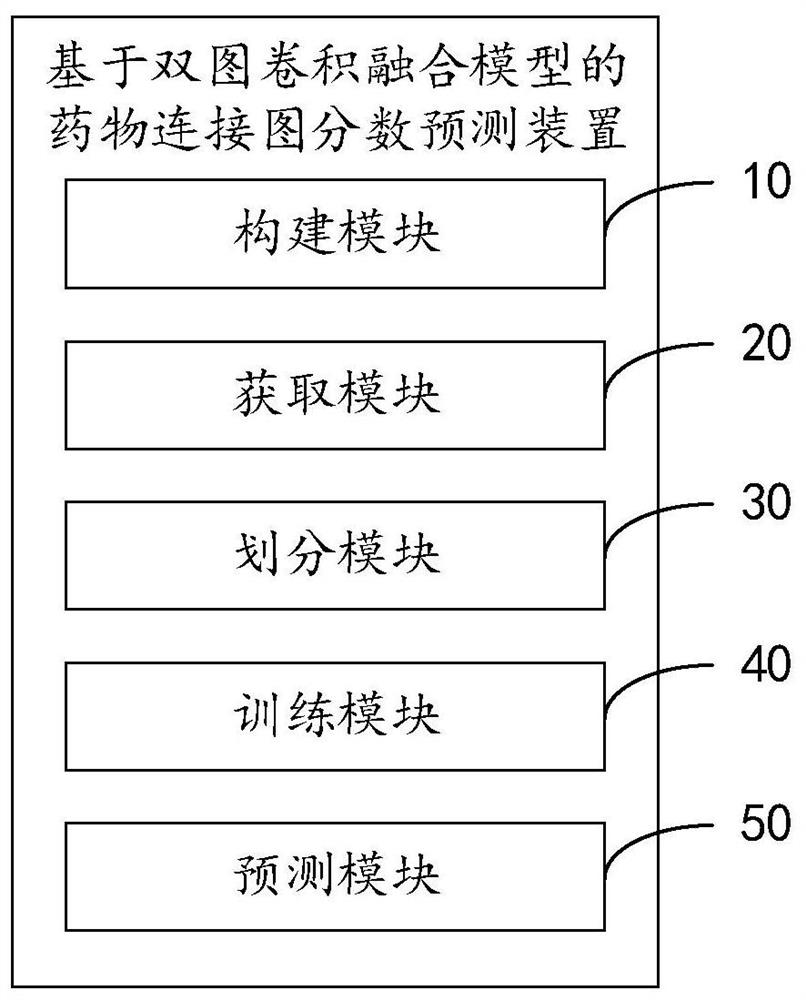
Drug connection graph score prediction method and device based on double-graph convolution fusion model
PendingCN113628696ARealize communication integrationAvoid time costMolecular designNeural architecturesCosine similarityAlgorithm
The invention provides a drug connection graph score prediction method and device based on a double-graph convolution fusion model. The method includes: after a drug association graph is trained by using a symbol graph convolutional network layer, fusing node features containing global information into a drug molecular graph of each drug through transformation of a full connection layer, then training the drug molecular graph fused with the global features by using a graph attention network layer, carrying out pooling operation, acquiring fusion features of a drug, realizing communication fusion of global information and local information, decoding the fusion features by calculating cosine similarity, comparing a predicted value with a true value, calculating an error, and performing back propagation and continuous iteration to obtain a drug connection diagram score prediction network for predicting drug connection diagram scores of the drug pairs. According to the method and device, the connection diagram score of the drug pair can be predicted quickly and accurately, candidate drugs can be screened, and time and capital costs can be reduced.
Owner:WUHAN UNIV
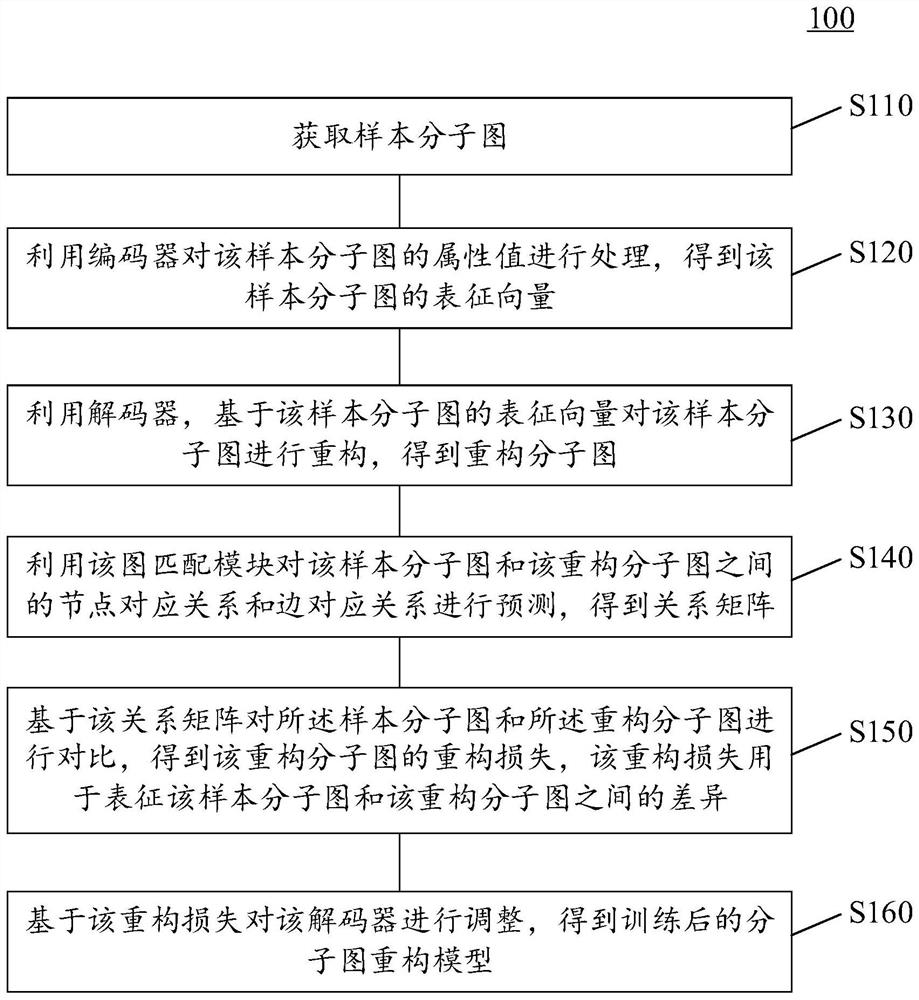
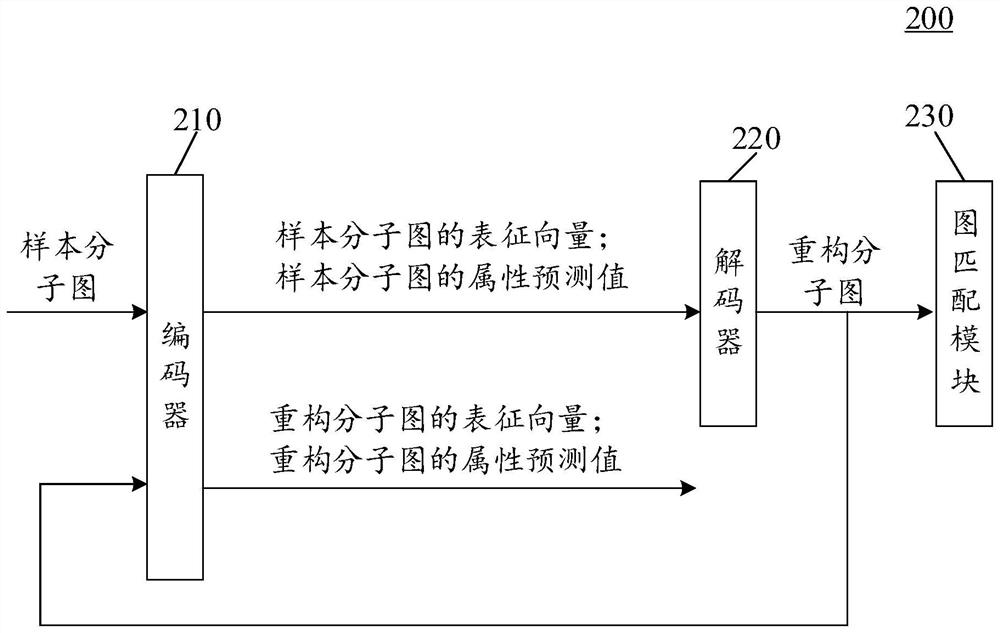
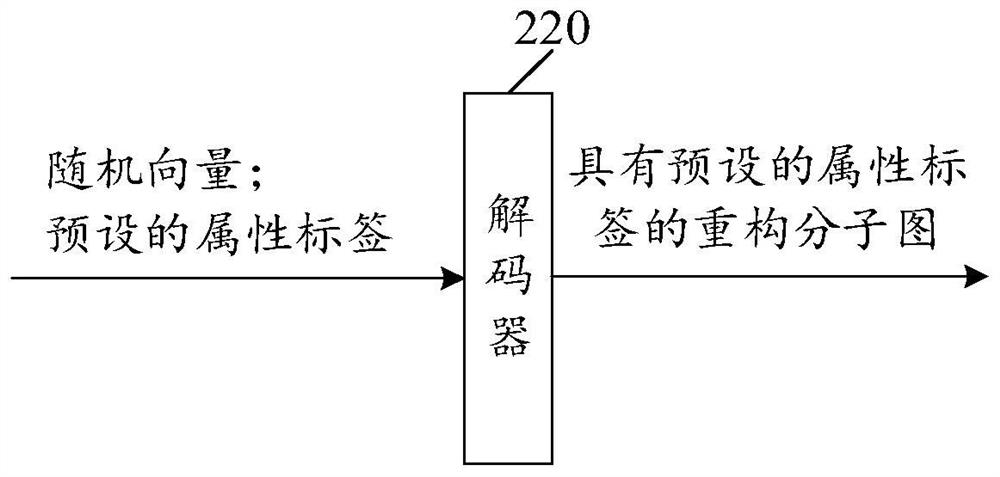
Training method and device of molecular graph reconstruction model and electronic equipment
PendingCN114334040AImprove guidanceReduce complexityMolecular designNeural architecturesEncoder decoderAlgorithm
The invention provides a molecular graph reconstruction model training method and device and electronic equipment, the method is suitable for the artificial intelligence medicine field, and the molecular graph reconstruction model comprises an encoder, a decoder and a graph matching module; in the training method provided by the invention, on one hand, a graph matching module is designed as a pre-training module, and a relation matrix output by the graph matching module is designed to be used for calculating the reconstruction loss between a sample molecular graph and a reconstruction molecular graph, on the other hand, a representation vector of the sample molecular graph output by an encoder is designed as an input vector of a decoder, and the reconstruction loss between the sample molecular graph and the reconstruction molecular graph is calculated. According to the molecular graph reconstruction model training method provided by the invention, the molecular graph reconstruction model can be suitable for a molecular discovery process, the matching complexity and resource consumption can be reduced, the matching accuracy is improved, and the guiding effect of a graph matching module on the molecular graph reconstruction model is further improved. In addition, the method provided by the invention can also improve the practicability and reconstruction effect of the molecular graph reconstruction model.
Owner:TENCENT TECH (SHENZHEN) CO LTD
A Molecular Structure Graph Retrieval Method Based on Evolutionary Computing Multi-View Fusion
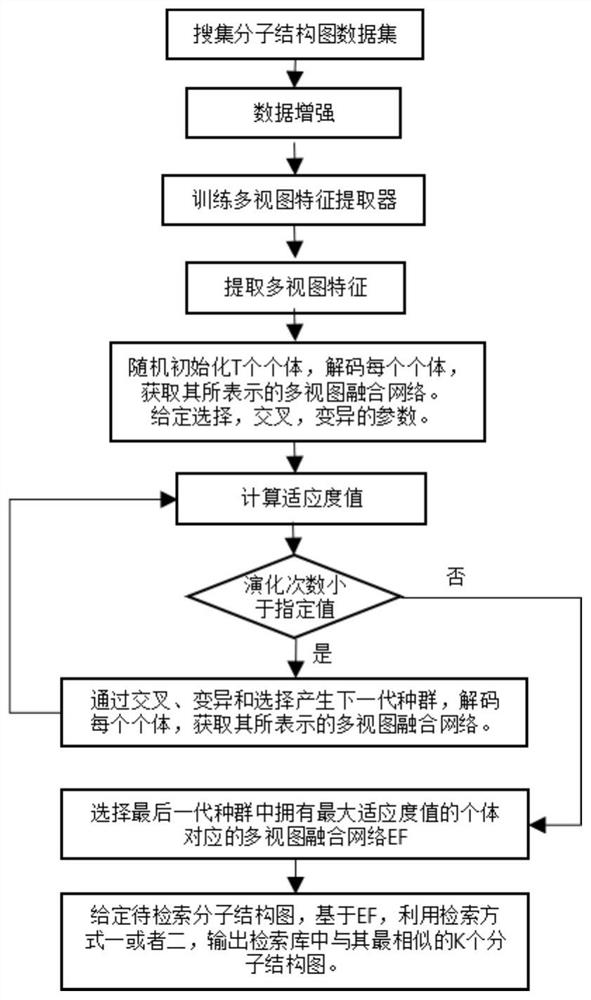
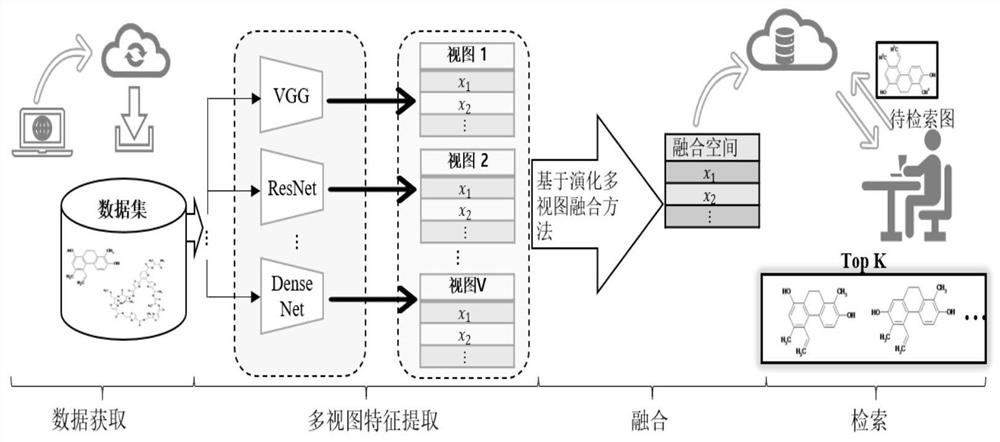
ActiveCN111897987BSearch impactEasy to useCharacter and pattern recognitionStill image data clustering/classificationData setAlgorithm
The invention relates to a method for retrieving molecular structure diagrams based on evolutionary calculation and multi-view fusion. Including: step 1, use operations such as rotation, translation, and scaling to enhance data; step 2, use the enhanced data to train multiple deep models as multi-view feature extractors; step 3, use the trained multi-view extractor to extract enhanced data The multi-view feature; step 4, search for a multi-view fusion model with excellent performance through the evolutionary algorithm to obtain the fusion feature of the classification model of the molecular structure diagram and the data set; step 5, directly use the classification model or use the fusion feature to calculate The ranking value of the similarity between the search graph and the molecular graph in the search database completes the search for the molecular structure graph to be retrieved. The invention is used to solve the problem of image-based chemical molecular structural formula retrieval without relying on molecular structural formula coding characters in the field of chemical informatics.
Owner:SHANXI UNIV
A Molecular Structure Prediction Method Based on Graph Convolutional Networks
ActiveCN113241130BSolve the problem of destroying molecular structure informationEasy to predictMolecular entity identificationMolecular designAlgorithmTheoretical computer science
The invention belongs to the technical field of machine learning, and in particular relates to a molecular structure prediction method based on a graph convolutional network. The present invention constructs a molecular graph and a molecular complete graph according to the SMILES of the input molecule, and correspondingly constructs a network model with two branches, one branch uses MLP for edge prediction, and the other branch includes graph convolutional network and MLP for The overall structural features of the branches are extracted. The invention uses graph convolution to extract molecular structure features, which can well extract the overall structural features of molecules, so as to better predict the structure; the double-branch model design is used to solve the problem that the complete graph destroys molecular structure information .
Owner:SOUTHWEST JIAOTONG UNIV
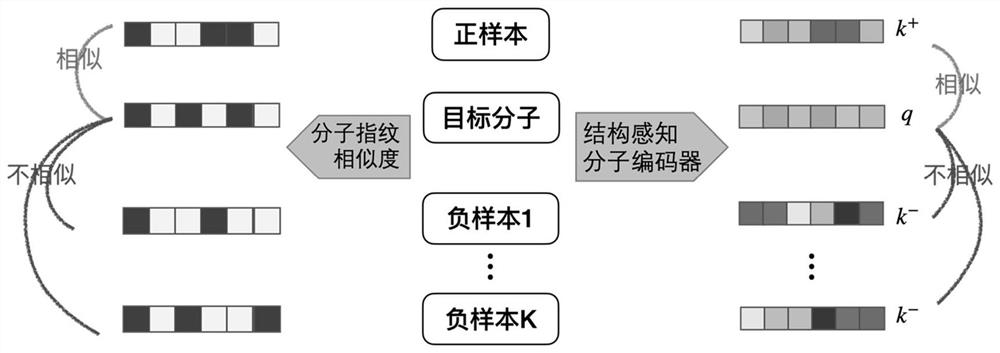
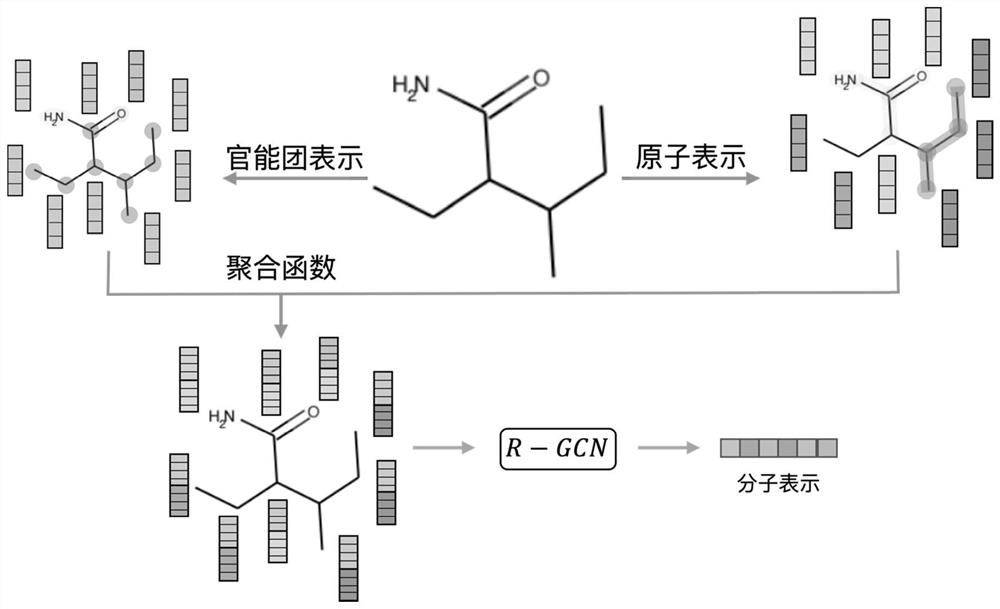
A Molecular Graph Representation Learning Method Based on Contrastive Learning
ActiveCN112669916BCapture comprehensiveEasy to obtainMolecular entity identificationChemical data visualisationAlgorithmLarge sample
The invention discloses a molecular graph representation learning method based on comparative learning, which includes: obtaining the molecular fingerprint representation of each molecule, and calculating the similarity between every two molecular fingerprints; Each atom matches the corresponding functional group; the molecular graph is modeled with the heterogeneous graph; the representation of each atom in the molecule and the representation of its functional group are encoded using the RGCN in the structure-aware molecular encoder, and the molecule is mapped to the feature through the aggregation function According to the fingerprint similarity between molecules, select positive and negative samples, and carry out comparative learning in the feature space; use the method of comparative learning to train on large sample molecular data sets, and obtain Structure-aware molecular encoders for downstream molecular property prediction tasks. The invention helps to capture richer molecular structure information and solve the problem of molecular attribute prediction.
Owner:ZHEJIANG UNIV
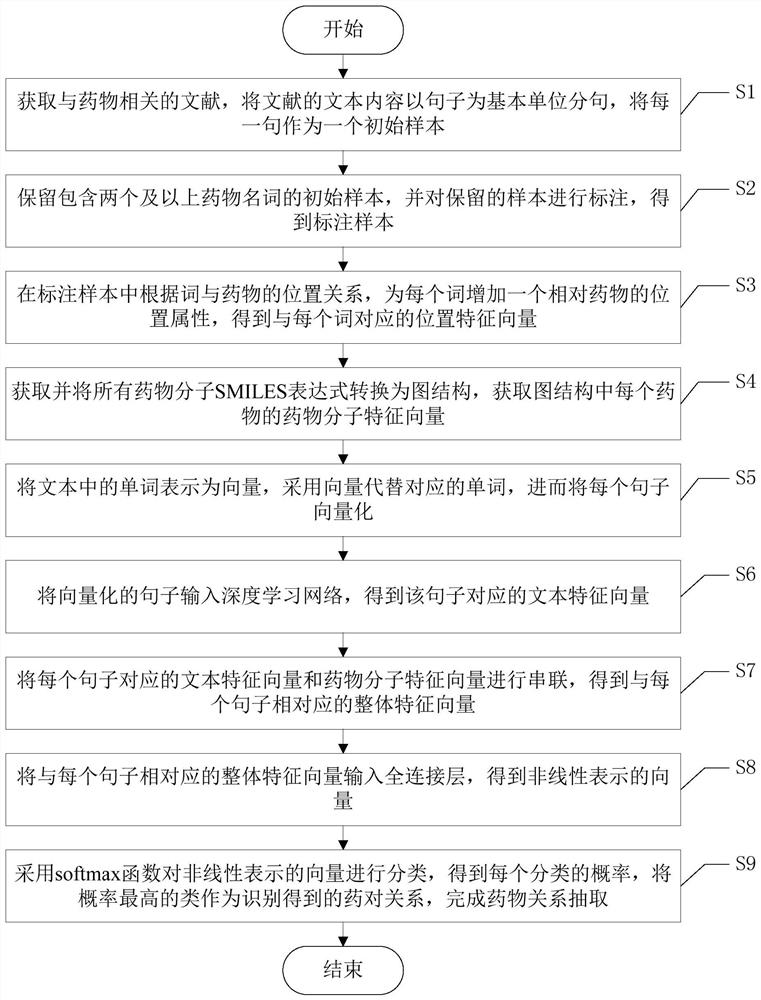
A drug relationship extraction method based on deep learning
ActiveCN111949792BImprove extraction accuracyReduced update lagMolecular designCharacter and pattern recognitionPharmaceutical drugRelationship extraction
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com