Self-supervised graph neural network pre-training method based on comparative learning
A neural network and pre-training technology, applied in the field of deep learning, can solve problems such as insufficient generalization performance and achieve good generalization effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
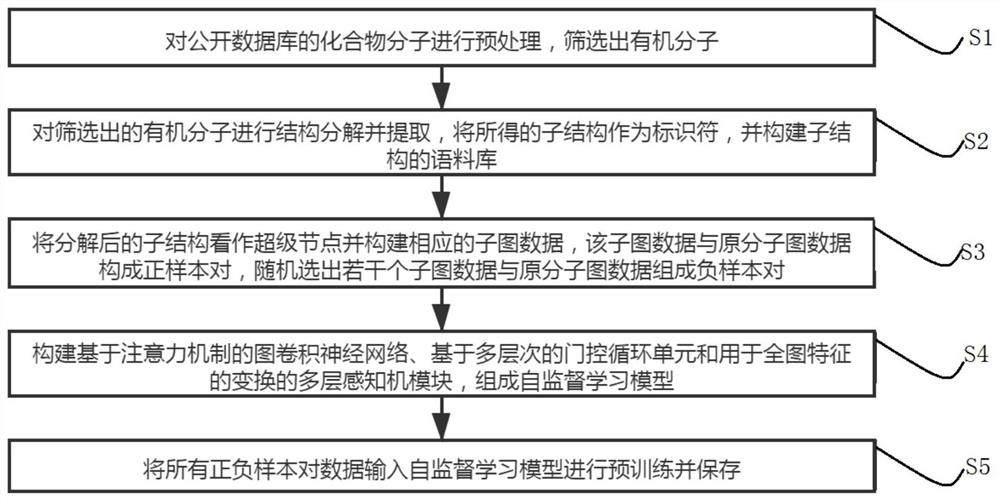
[0050] Such as figure 1 As shown, a self-supervised graph neural network pre-training method based on contrastive learning includes the following steps:
[0051] S1: Preprocessing the compound molecules in the public database to screen out organic molecules;
[0052] S2: Decompose and extract the structure of the screened organic molecules, use the obtained substructures as identifiers, and build a corpus of substructures;
[0053] S3: Treat the decomposed substructure as a super node and construct the corresponding subgraph data. The subgraph data and the original molecular graph data form a positive sample pair, and ten subgraph data are randomly selected to form a negative sample pair with the original molecular graph data. ;
[0054] S4: Construct a graph convolutional neural network based on an attention mechanism, a multi-level gated recurrent unit and a multi-layer perceptron module for the transformation of full-image features to form a self-supervised learning model...
Embodiment 2
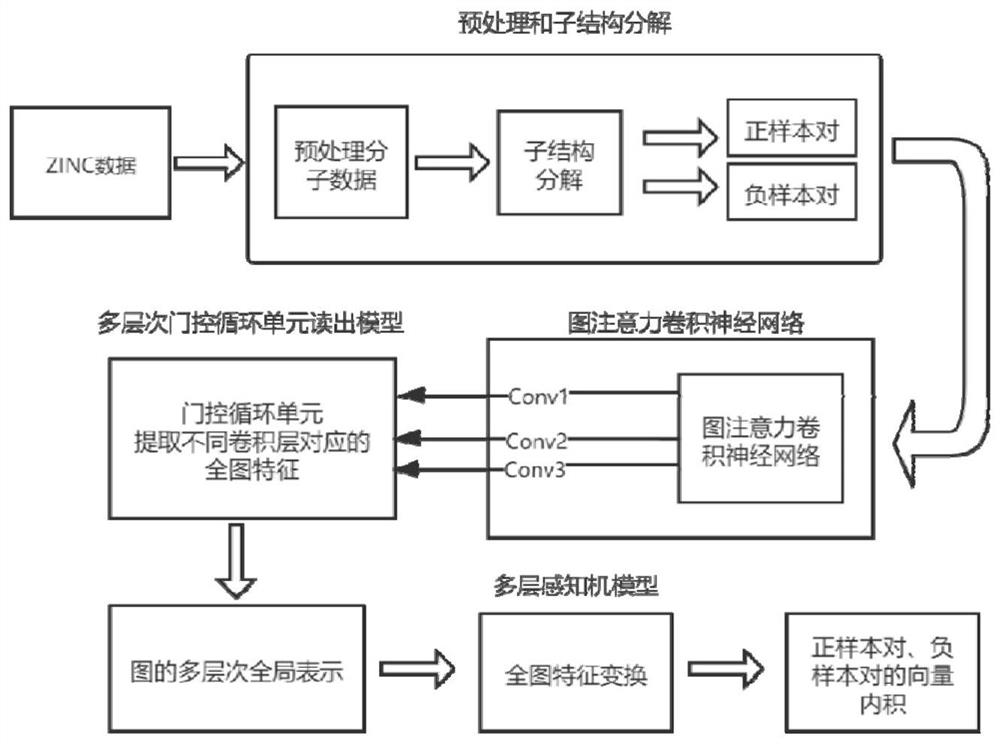
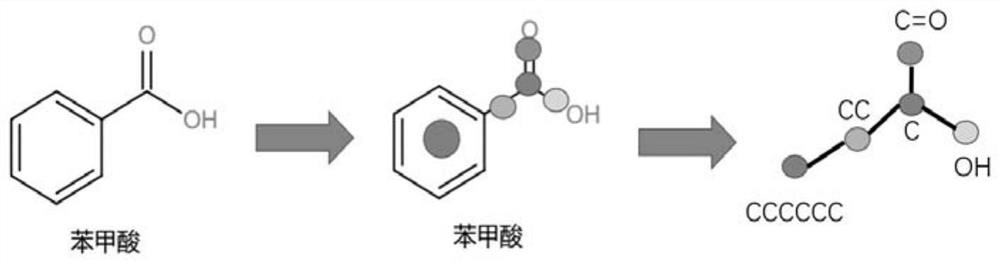
[0080] Such as image 3 As shown, the flow of molecules to obtain substructure subgraphs through substructure decomposition is as follows image 3 shown.
[0081] In this example, benzoic acid is used as an example to briefly illustrate the steps of substructure decomposition. Benzoic acid in the ZINC database is represented by the SMIELS string (C1=CC=CC=C1C(=O)O), which is converted to the molecular structure format by the Python open source chemical calculation toolkit RdKit. First, use Rdkit to obtain the atomic numbers corresponding to all the rings and functional groups in the molecule as the overall super node and store them in the hash table. Similarly, use all the keys in the molecule as the overall super node and store them in the hash table , and then record the connection relationship between super nodes, and agree that the nodes where more than three super nodes intersect are used as intermediate nodes and stored in the hash table, and the connected edges are ad...
Embodiment 3
[0085] Such as figure 2 As shown, in order to construct the positive and negative sample pairs, design the corresponding self-supervised training tasks, in the preprocessing and substructure decomposition steps, the substructure graph data and the original molecular graph data constitute positive sample pairs, and correspondingly randomly pass through the substructure graph data Collectively select ten sub-graph data and the original molecular graph data to form a negative sample pair, satisfying that the ratio of positive and negative samples is 1:10. Specifically, one original molecular graph data and its corresponding sub-structure graph data constitute a positive sample pair, randomly Select 10 substructure graph data and the original molecular graph data to form 10 negative sample pairs, which are used for the input of the subsequent graph attention convolutional neural network.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com