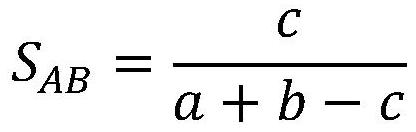A Molecular Graph Representation Learning Method Based on Contrastive Learning
A graph representation and molecular technology, applied in the field of graph representation learning, can solve problems such as molecular attribute prediction, and achieve the effect of enriching molecular structure information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0045] The present invention will be further described in detail below with reference to the accompanying drawings and embodiments. It should be noted that the following embodiments are intended to facilitate the understanding of the present invention, but do not limit it in any way.
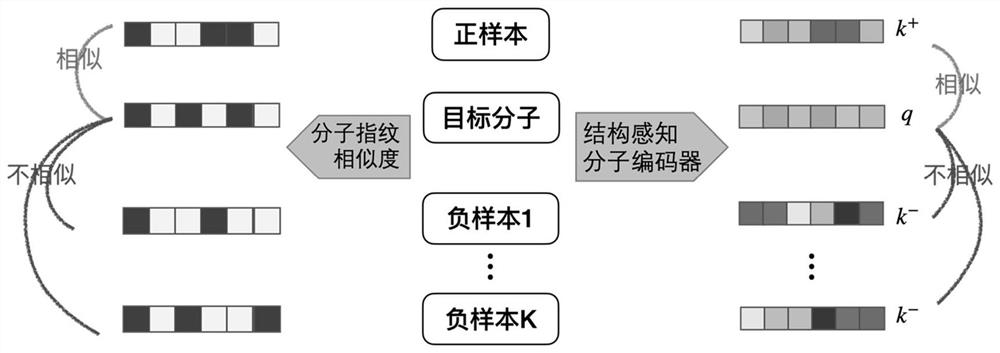
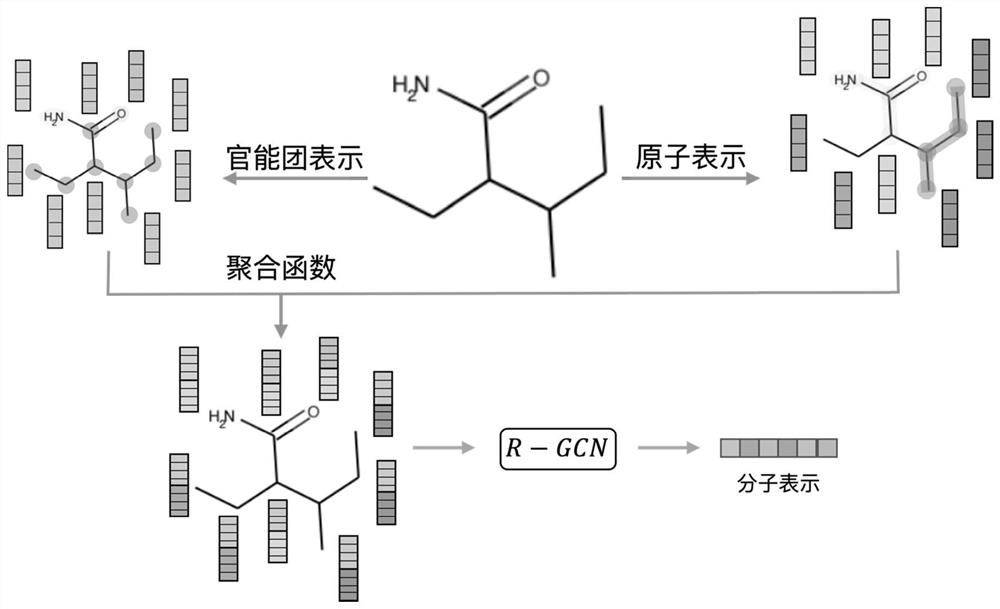
[0046] The molecular graph representation learning method based on comparative learning provided by the present invention can be used in application scenarios such as chemical molecular attribute prediction, virtual screening, etc., using the similarity of molecular fingerprints as a basis to select positive and negative samples, and comparing them with molecular data in the feature space, And directly encode the knowledge of functional groups in the chemical domain into the representation of molecules to obtain a discriminative molecular graph representation with chemical domain knowledge. The invention solves the problem of insufficient labeling data existing in supervised learning, and fully u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com