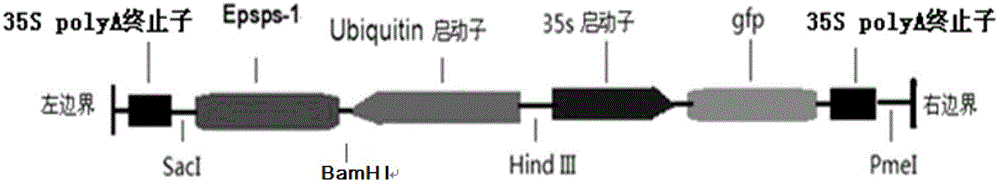
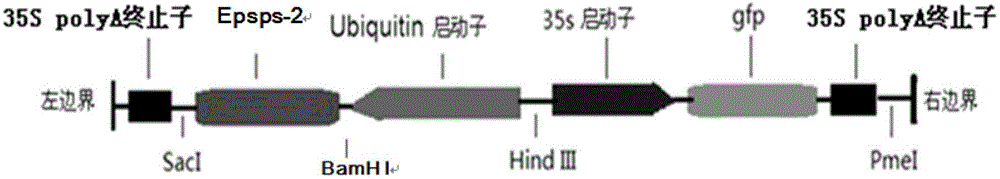
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
896 results about "Molecular identification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Molecular computational identification (MCID) is a technique in which molecules are used as means for identifying individual cells or nanodevices.
Near-infrared fluorescent molecular probe, synthesizing method and use thereof
InactiveCN101440282APrecise deliveryImplement diagnosticsMethine/polymethine dyesMicrobiological testing/measurementDiseaseMolecular identification
The invention relates to the field of specific molecular identification and diagnosis reagent and in particular discloses a near infrared fluorescent dye with structural formulas I and II; and the invention also discloses a near infrared molecular probe which is obtained through covalent bonding between the near infrared fluorescent dye with the structural formulas I and II and a ligand of specific molecules. The near infrared molecular probe can be used for early diagnosis of turmour diseases.
Owner:CHINA PHARM UNIV
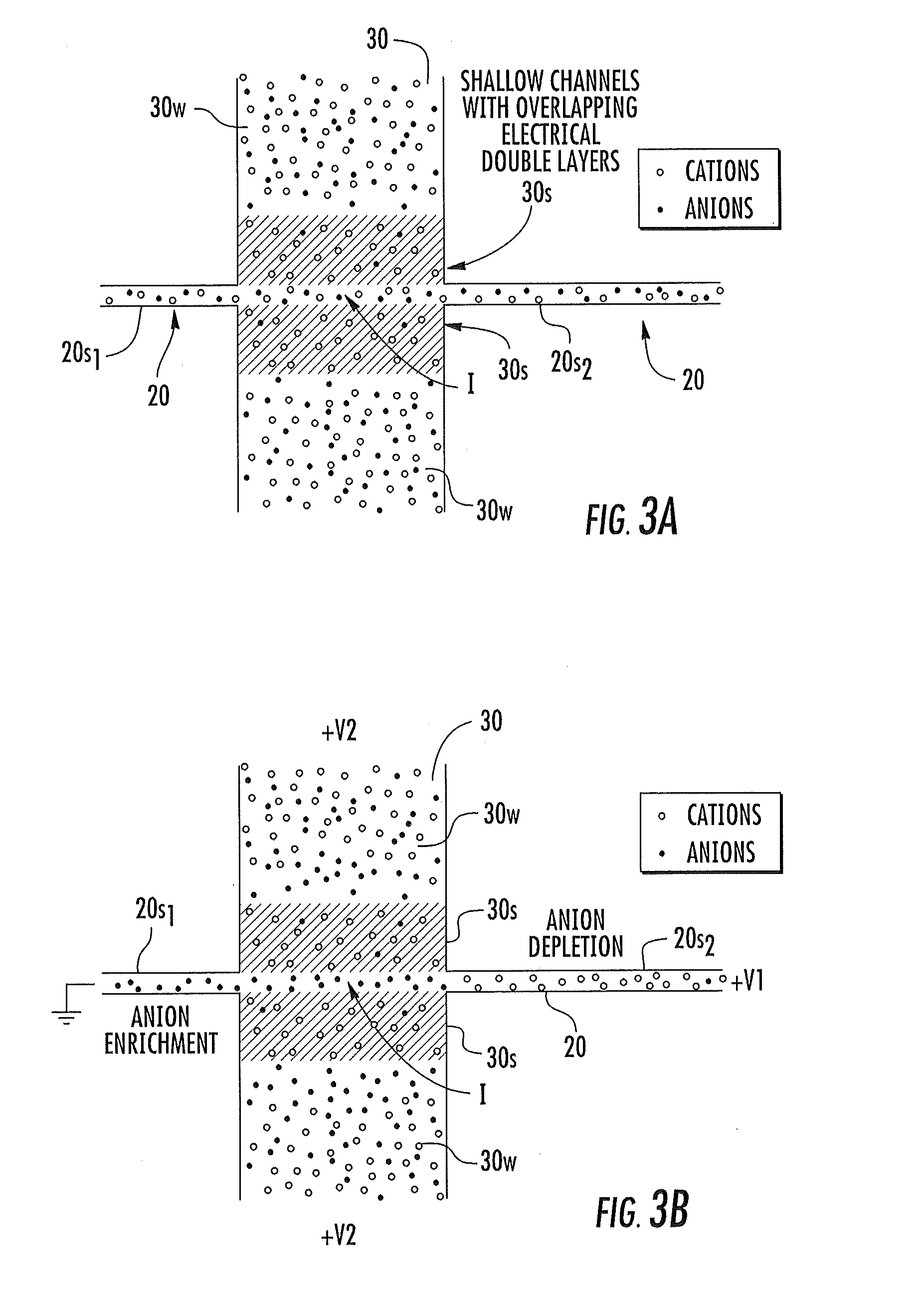
Nanofluidic devices with integrated components for the controlled capture, trapping, and transport of macromolecules and related methods of analysis
ActiveUS20140238856A1Easy to captureEasy to trapSludge treatmentVolume/mass flow measurementMolecular identificationTrapping
Devices for controlling the capture, trapping, and transport of macromolecules include at least one fluidic transport nanochannel that intersects and is in fluid communication with at least one transverse nanochannel with (shallow) regions and / or with integrated transverse electrodes that enable fine control of molecule transport dynamics and facilitates analyses of interest, e.g., molecular identification, length determination, localized (probe) mapping and the like.
Owner:THE UNIV OF NORTH CAROLINA AT CHAPEL HILL
Core SNP sites combination maizeSNP384 for building of maize DNA fingerprint database and molecular identification of varieties
ActiveCN104532359AImprove stabilityGood repeatabilityNucleotide librariesMicrobiological testing/measurementMolecular identificationAgricultural science
The invention discloses a core SNP sites combination maizeSNP384 for building of a maize DNA fingerprint database and molecular identification of varieties, and an application of the core SNP sites combination. The invention provides applications of 384 SNP sites in any one of the following conditions: (1) building of the maize DNA fingerprint database; (2) detecting of the authenticity of maize varieties; (3) genetic analysis of corn germplasm resources; and (4) molecular breeding of maize, wherein the physical positions of the 384 SNP sites are determined by comparison on the basis of a whole genome sequence of the maize variety B73; the version number of the whole genome sequence of the maize variety B73 is B73 RefGen V1; and the 384 SNP sites are MG001-MG384. An experiment proves that the 384 SNP sites can be applied to building of the maize variety DNA fingerprint database, identification of the variety authenticity, dividing of germplasm resource groups, and other related researches.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
A molecular identification method for Isaria cicadae
The invention relates to the field of biotechnology and discloses a molecular identification method for Isaria cicadae (CGMCCNO.3453). The method includes the steps of extracting genomic DNA of the strain, performing amplification of the DNA in the transcription spacers of the genome using primers ITS5 and ITS4, and recovering and purifying the obtained DNA fragments; performing bidirectional sequencing of the purified DNA fragments; aligning the obtained DNA sequences with SEQIDNO:3; and identifying the strain as Isaria cicadae (CGMCCNO.3453) if the ITS sequence of the strain has the same content and order of arrangement as SEQIDNO:3. The inventive method can rapidly, reliably and accurately identify Isaria cicadae (CGMCCNO.3453), and the identification method is not influenced by environmental factors.
Owner:ZHEJIANG BIOASIA PHARMA CO LTD
Tumor neoantigen detection method and device based on next-generation sequencing, and storage medium
ActiveCN108796055AQuality improvementMicrobiological testing/measurementSpecial data processing applicationsMolecular identificationClonality Analysis
Owner:深圳裕策生物科技有限公司
Molecular identification method of atlantic salmon
ActiveCN103122386ALower requirementEasy to operateMicrobiological testing/measurementMolecular identificationEnzyme digestion
The invention discloses a molecular identification method of an atlantic salmon. The molecular identification method comprises the following steps of 1, extraction of fish DNA (deoxyribonucleic acid); 2, PCR (polymerase chain reaction) augmentation; 3, enzyme digestion and electrophoresis detection; and 4, identification. The atlantic salmon can be accurately determined in four steps. The molecular identification method of the atlantic salmon has the characteristics of being simple to operate, rapid and accurate, low in price, low in requirement on an instrument and equipment, and the like.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Cultivating method of morchella importuna
The invention discloses a cultivating method of morchella importuna. The cultivating method of the morchella importuna comprises a bacterium culturing producing step, a cultivation material producing step, a hydration processing step, a managing and protecting step and the like. A maternal species (HKAS 62868) which can be identified as the morchella importuna (the number of the Genbank is JQ 618576) through a molecular identification can be used for cultivating cultispecieses. After the hydration processing is carried out on the cultispecieses, the cultispecieses are sowed on the face of the right land and then are earthed up. The cultivation materials can be produced and added in the field and the field can be normally managed for 80-120 days, so that sporocarps can be obtained. The cultivating method of the morchella importuna aims at providing an edible mushroom cultivating method which not only is beneficial to the sustainable development of ecology, but also helps to increase farmer income and a novel cultivating bacterial strain is provided for the industrialization development of the morchella importuna at the same time.
Owner:KUNMING INST OF BOTANY - CHINESE ACAD OF SCI
Preparation method of temperature sensitive adsorbent of halloysite magnetic composite material surface blotting
InactiveCN102580696AHigh mechanical strengthImprove adsorption capacityOther chemical processesComponent separationMolecular identificationHalloysite
The invention discloses a preparation method of a temperature sensitive adsorbent of halloysite magnetic composite material surface blotting, belonging to the technical field of environment functional material preparation. The preparation method comprises the following steps: preparing a ferroferric oxide / halloysite nanotube (Fe3O4 / HNTs) magnetic composite material through a simple and effective solvent thermosynthesis method; and then, performing ethylene modification on the magnetic composite material; and preparing the temperature sensitive adsorbent of halloysite nanotube magnetic composite material surface blotting through a radical polymerization process, and utilizing the adsorbent in selective identification and separation of 2,4,5-trichlorophenol in aqueous solution. The prepared temperature sensitive blotting adsorbent has excellent thermal and magnetic stability, sensitive magnetic and thermal induction effect, high adsorption capacity, excellent reversible adsorption / release function with temperature and obvious TCP molecular identification performance.
Owner:JIANGSU UNIV
DNA (deoxyribonucleic acid) barcoding based molecular identification method of leech
ActiveCN103898235AImprove accuracyHigh sensitivityMicrobiological testing/measurementMolecular identificationWhitmania pigra
The invention discloses a DNA (deoxyribonucleic acid) barcoding based molecular identification method and CO I sequence of a leech (whitmania pigra). Polymerase chain reaction (PCR) is carried out by using the method to amplify a CO I gene of the leech, a PCR product is sent to a sequencing company to be sequenced after being verified by agarose gel, the sequencing results are subjected to manual proofreading and sequence splicing and are compared with public sequences, and the tissue to be tested can be judged to be the source of whitmania pigra if the homology of the sequencing results and a gene sequence SEQ ID NO.1 is above 99%. The whitmania pigra variety is quickly and accurately identified by adopting the reliable DNA molecular identification technology, thus improving the accuracy and the reliability and ensuring the safety of the leech as a medicine.
Owner:MUDANJIANG YOUBO PHARMA CO LTD
Red yeast rice rich in Monacolin K and preparation method of red yeast rice
ActiveCN105112305AThe fermentation process is simpleFungiMicroorganism based processesBiotechnologyRed yeast rice
The invention discloses red yeast rice rich in Monacolin K and a preparation method of the red yeast rice. The preparation method comprises the steps of taking 40 monascus strains which are obtained in a separated mode as objects, and screening out a monascus strain, namely FG-8, wherein the monascus strain produces the Monacolin K in a high-yield mode and produces no citrinin, and the Monacolin K content of a fermentation product of the monascus strain is 2.46 mg / g and 2.29 times that of a type strain ACCC 30501. Through molecular identification, the strain is determined as red monascus. The red monascus, namely the FG-8, is treated as an original strain, 26 different varieties of rice raw materials are screened, when the FG-8 is founded, polished rice of a rice variety, namely 'California red rice', is treated as substrates for conducting fermentation, and the red yeast rice rich in the Monacolin K is obtained. The red yeast rice is simple in preparation method, the Monacolin K content measured under the optimum fermentation condition is 4.64 mg / g and approximately 8.30 times that of Furuta red yeast rice on the market, and no citrinin is detected.
Owner:FUJIAN AGRI & FORESTRY UNIV
Universal reader molecule for recognition tunneling
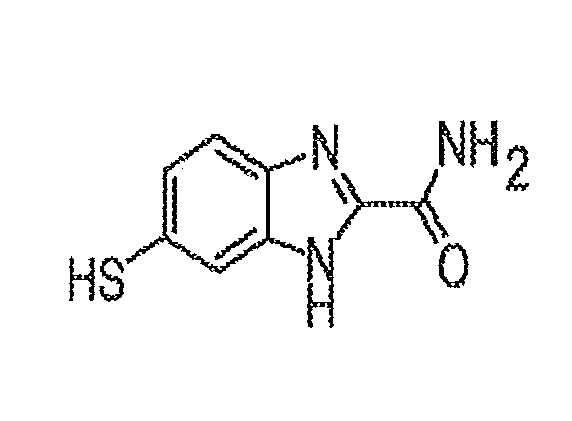
InactiveUS20160108002A1Enhanced signalOrganic chemistryMaterial analysisMolecular identificationBenzene
Some embodiments of the present disclosure are directed to a compound 5(6)-mercapto-1H-benzo[d]imidazole-2-carboxamide (“BIA”) which yields enhanced signals for recognition tunneling. Other embodiments are directed toward methods for producing such compounds as well as apparatuses and systems which utilize such compounds for recognition tunneling for molecule identification / sequencing (for example).
Owner:ARIZONA STATE UNIVERSITY
Molecular identification method for lasiohelea taiwana
ActiveCN103805609AAccurate identificationShorten identification timeMicrobiological testing/measurementFermentationBiotechnologyMolecular identification
The invention discloses a molecular identification method for lasiohelea taiwana. The method is characterized in that two different genes, namely a COI gene and an ITS2 gene are adopted to be combined, and the lasiohelea taiwana is subjected to species identification by comparison of the similarity of the gene sequences. By adopting the molecular identification method for lasiohelea taiwana provided by the invention, accurate and rapid identification of the lasiohelea taiwana is facilitated. A basis is provided for molecular identification of the lasiohelea taiwana. The new variety can play a certain role in transmission of plant pollen and environment harmless reaction.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
Method for extracting high-molecular-weight genome from animal feces
InactiveCN102586234AHigh molecular weightSimple methodDNA preparationSodium acetateMolecular identification
The invention relates to a method for extracting high-molecular-weight genome from animal feces. The method comprises the steps of sample treatment, bacterial cell lysis and extraction of DNA (Deoxyribonucleic Acid). Specifically, the method comprises the steps of: dissolving an animal feces sample with aseptic PBS (Phosphate Buffered Saline) buffer, carrying out low-speed centrifuging and filtering to remove food residues in the feces, collecting bacterial cells, performing wall breaking on the bacteria by adopting lysozyme, decolorizing peptidase, protease and SDS (Sodium Dodecyl Sulfate), extracting with a mixture of phenol, chloroform and isoamyl alcohol as well as a mixture of chloroform and isoamyl alcohol, precipitating with isopropanol and sodium acetate, washing and drying, dissolving, and digesting RNA (Ribonucleic Acid) with RNAse (Ribonuclease). According to the invention, the method is simple in operation; and the DNA molecular weight of the extracted genome achieves more than 40kb, so that the researches on microorganism molecule ecological diversity, molecular identification, target gene amplification, macro genome library construction and the like are met.
Owner:YUNNAN NORMAL UNIV
Preparation method of glyphosate resistant transgenic rice
InactiveCN104004781AReduce the number of copiesShort processPlant tissue cultureHorticulture methodsMolecular identificationGenetically modified rice
The invention provides a preparation method of glyphosate resistant transgenic rice. The method comprises the following steps: converting an expression vector containing glyphosate resistant gene into Agrobacterium tumefaciens, invading rice calluses by the Agrobacterium tumefaciens, transferring the calluses to a selective aluminum containing glyphosate for screening, choosing resistant calluses, differentiating, rooting, hardening-seedling, and transplanting to obtain glyphosate resistant transgenic rice. A molecular identification result shows that an exogenous gene is stably expressed. The obtained glyphosate resistant transgenic rice contains glyphosate herbicide resistant gene, contains no other screening marker genes, and can be used as a commercial herbicide resistant rice kind to reduce the reduce the later process and accelerate the breeding pace. The method has the advantages of simple operation, low copy number of transgenic plants, stable heredity properties, and good market application prospect.
Owner:CHINA NAT SEED GRP
Breeding design and identification method for short-haulm compact type cole suitable for mechanized harvest
InactiveCN101292626AQuickly and effectively design breeding technology systemReduce planting costsMicrobiological testing/measurementPlant genotype modificationStem lengthMolecular identification
The invention pertains to the field of rape breeding and discloses a method for seed selection and identification of a novel dwarf compact type rape adapting to high-density planting and mechanized harvesting. The rape of Variety 5148-2 with apreservation number of CCTCC-P200601 that is bred by Huazhong Agricultural University and yellow-seed rape of DH series YN90-1016 that is synthesized artificially in Canada are taken as seed-parents and crossbred to obtain F1. In inflorescence, the main anthotaxy of an F1 plant and the flower buds with a size ranging from 2.8mm to 3.8mm at the upper branches of the P1 plant are selected, and then chromosome doubling with colchicine and microspore culture are carried out successively to obtain the segregation populations of double haploid. Filed phenotypic characteristic identification are applied for years in various spots to the segregation populations; variant rape materials with five major traits, namely, stem length, branch trait, length of anthotaxy, quantity of silique and growth period differing from those of the common rape and dwarf, compact plant, smallish and acervate branches and main anthotaxy, concentrative florescence and compact pod bearing layers phenotypes are selected to obtain the novel dwarf compact type rape adapting to mechanized harvesting. The invention also discloses a method for molecular identification.
Owner:HUAZHONG AGRI UNIV
Method for identifying DNA bar code molecule of earthworm
InactiveCN103898234AIdentification is conducive to the realizationIdentification achievedMicrobiological testing/measurementMolecular identificationDNA barcoding
The invention discloses a method for identifying a DNA bar code molecule of an earthworm and a COI sequence of the DNA bar code molecule of the earthworm. The method comprises the steps of amplifying the gene of a sample COI through polymerase chain reaction (PCR), verifying the PCR product by agarose gel, then sending to a biological sequencing company for sequencing, according to the sequencing result, sequence splicing through manual correcting, comparing with a public sequence, then confirming the tissue to be tested as the source of pheretima aspergillum or pheretima vulgaris if the homology of the spliced sequence and the gene sequence SEQ ID NO.1-NO.4 is over 99%. By adopting the reliable DNA molecular identification technology, the method can rapidly and accurately identify the earthworm varieties specified by the 'pharmacopoeia of China' and non-medicinal material earthworm varieties without conducting morphological identification on the earthworm varieties, can further distinguish the pheretima aspergillum varieties from pheretima vulgaris varieties, improves the accuracy and reliability, and guarantees the safety of the earthworm as the medicinal material.
Owner:MUDANJIANG YOUBO PHARMA CO LTD
Molecular identification method for DNA bar code of lumbricus
InactiveCN106834467AIdentification is conducive to the realizationIdentification achievedMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationDNA barcoding
The invention discloses a molecular identification method for a DNA bar code of lumbricus and a CO I sequence thereof. The method comprises the following steps: amplifying a sample CO I gene through polymerase chain reaction (PCR); verifying the PCR product through agarose gel and then sending to a biological sequencing company for sequencing; manually correcting and splicing sequence for the sequencing result; comparing with a public sequence; and if the homology of the gene sequence SEQ ID NO.1-NO.4 is above 99%, confirming a to-be-detected tissue as a source of pheretima aspergillum or Shanghai lumbricus. According to the invention, a reliable DNA molecular identification technique is adopted; the morphological identification for lumbricus varieties is not required; the lumbricus varieties and non-drug lumbricus varieties specified in Chinese Pharmacopoeia can be quickly and accurately identified; besides, the pheretima aspergillum and the Shanghai lumbricus can be further distinguished; the accuracy and the reliability can be enhanced; the safety of the lumbricus used as a medicinal material can be guaranteed.
Owner:MUDANJIANG YOUBO PHARMA CO LTD
Fresh keeping liquid for effectively preventing sack expands of pickled pepper product and preparation method of fresh keeping liquid
InactiveCN106689350AAvoid swelling bagsAvoid Bulging Tank ProblemsFruit and vegetables preservationFood ingredientsMolecular identificationMicroorganism
The invention belongs to the technical field of food, in particular relates to the field of pickled vegetables in food, and particularly relates to fresh keeping liquid for effectively preventing sack expands of a pickled pepper product and a preparation method of the fresh keeping liquid. Microorganisms causing the gas production of the pickled pepper product are provided by various means for discriminating gas production germs such as traditional separation purification, molecular identification, duchenne small tube and the like to be pseudomonas, saccharomyces cerevisiae and kazachstania turicensis, an appropriate bacteriostatic agent can be selected on purpose, and the fresh keeping liquid for effectively preventing the sack expands of the pickled pepper products. Compared with the similar products (general pickled vegetable preservatives in the market), the fresh keeping liquid is more purposeful and better in effect, reduces the consumption of the preservatives and avoids the abuse risk of the preservatives; and moreover, the production method is simple, and the cost is low.
Owner:SICHUAN DONGPO CHINESE PAOCAI IND TECH RES INST
Microbial fermentation agent capable of being used for continuously fermenting pickles in multiple batches
ActiveCN106434432AAchieve standardizationRealize off-site productionBacteriaClimate change adaptationBiotechnologyMolecular identification
The invention belongs to the field of food, in particular belongs to the field of pickles in the food industry and in particular provides a microbial fermentation agent capable of being used for continuously fermenting pickles in multiple batches. Main microbes in Sichuan old saline water pickles are determined to be lactobacilluspentosus, lactobacillus brevis and kazachstaniae xigua through study by various advanced means, such as traditional separation and purification, molecular identification, high throughput measurement and quantitative PCR, and have a content ratio of 10 to 1 to 1. 1 / 10000-5 / 10000 of the microbes are added when the pickles are made. The microbial fermentation agent has the positive effects that compared with pickle fermentation agents (single strains) of the same kind, the microbial fermentation agent has the closer fermentation flavor to natural fermentation; and a stable fermentation system (equivalent to the pickle mother water) can be formed only by adding the fermentation agent when the pickles are made for the first time and can be used for continuously making at least 20 batches of pickles.
Owner:SICHUAN DONGPO CHINESE PAOCAI IND TECH RES INST
ISSR-SCAR (inter simple sequence repeat-sequence characterized amplified region) marker capable of identifying Wujiang brassica chinensis and identification method of marker
ActiveCN104894124AEasy to operateHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMolecular identification
The invention discloses an ISSR-SCAR (inter simple sequence repeat-sequence characterized amplified region) marker capable of identifying Wujiang brassica chinensis and an identification method of the marker. The method includes: performing total DNA (deoxyribose nucleic acid) extraction, PCR (polymerase chain reaction) amplification, primer selection and ISSR-SCAR detection on Wujiang brassica chinensis and other 10 brassica chinensis varieties, and finally determining an ISSR-SCAR molecular marker and a molecular identification method of the Wujiang brassica chinensis. A primer shown in the sequence such as SEQ ID NO:1 is used for amplification to obtain a specific fragment shown in the sequence such as SEQ ID NO:2. According to the sequence, a pair of primers in the sequences such as SEQ ID NO:3 and SEQ ID NO:4 is designed. The marker is simple to operate, stable and reliable, is the first molecular identification marker applied to the variety of Wujiang brassica chinensis, can be used for quickly identifying the variety and has significance of protecting germplasm resources of geographical indication products.
Owner:苏州市种子管理站
Molecular identification method for siniperca chuatsi and siniperca scherzeri and kit
InactiveCN101381768AImprove efficiencyImprove accuracyMicrobiological testing/measurementMolecular identificationAgricultural science
The invention discloses a method for molecular identification of Siniperca chuatsi basilewsky and Siniperca scherzeri Steindachner and a kit used by the same. The method for molecular identification comprises the following steps: DNA extraction, DNA fragment amplification, electrophoresis of PCR products and identification by a silver-staining method. The kit contains a pair of SSR primers and a standard map. The method for molecular identification can improve the efficiency and accuracy of identification of Siniperca chuatsi basilewsky and Siniperca scherzeri Steindachner, and is expected to solve the identification problem in the aquiculture of Siniperca chuatsi basilewsky and Siniperca scherzeri Steindachner and stimulate the sound development of Siniperca chuatsi culture industry.
Owner:CHANGSHA UNIVERSITY
Rice CYP704B2 gene mutant, as well as molecule identification method and applications thereof
ActiveCN104894144ABreeding benefitsMicrobiological testing/measurementFermentationGenetically modified riceAgricultural science
The invention provides a rice CYP704B2 gene mutant and applications thereof, belonging to the technical field of genetic engineering. A nonglutinous rice variety 93-11 is subjected to induced mutation by cobalt 60 radiation, so as to cause deletion of two G basic groups behind 1267 basic groups of a rice CYP704B2 gene; the rice CYP704B2 gene mutant is named as cyp704B2-2, and a nucleotide sequence thereof is shown as SEQ ID No.1 and further verifies that the mutant causes rice recessive genic male sterility; the rice CYP704B2 gene mutant can be used for preparing transgenic rice with recessive genic male sterility, and plays an important role in genetic improvement and breeding of rice germplasm resources. The invention also provides a molecular marker identification method of the mutant and applications of the method in breeding and seed production.
Owner:HAINAN BOLIAN RICE GENE TECH CO LTD
Molecular identification method of cytoplasm male sterile restoring line
InactiveCN104342434AClear resultThe resulting band is stableMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationMicrobiology
The invention belongs to the technical filed of biology, and relates to a molecular identification method of a cytoplasm male sterile restoring line, and concretely provides a CAPS-labeled primer used for identifying the cytoplasm male sterile restoring line, and a nucleotide sequence is shown as SEQ ID NO. 2 and SEQ ID NO.3. The method employs the CAPS label for a polymorphism detection of molecule level of a cytoplasm male sterile restoring line material, The method has the advantages of fast identification speed, high accuracy and simple operation, and is very suitable for indoor identification of purity of the cytoplasm male sterile restoring line material and molecular marker-assisted selection of a new restoring line material.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Molecular identification method based on transfer of GAI mRNA molecules between pear rootstock and scion
InactiveCN101942453AImprove accuracyFast wayMicrobiological testing/measurementPlant peptidesMolecular identificationPyrus betulaefolia
The invention discloses amino acid sequences of GAI proteins of Pyrus betulaefolia and Pyrus bretschneideri, and nucleotide sequences for coding the GAI proteins, wherein the sequences are disclosed as SEQ ID NO1-4 in the sequence table. The invention also discloses a molecular identification method based on transfer of GAI mRNA between pear rootstock and scion, which comprises the following steps: (1) micrografting seedlings in test tubes by using Pyrus bretschneideri as scion and using Pyrus betulaefolia as rootstock; (2) looking for the respective specific enzyme-digestion sites of the GAI genes of the Pyrus bretschneideri and the Pyrus betulaefolia, and respectively designing a primer at the upstream and downstream of the enzyme-digestion site; (3) and carrying out enzyme-digestion on the Pyrus betulaefolia, the Pyrus bretschneideri, and the RT-PCR product of the Pyrus betulaefolia and the Pyrus bretschneideri after grafting, and comparing the enzyme-digestion spectra before and after the grafting to identify the transfer conditions. The method is quick, sensitive, accurate, simple and convenient, and is applicable to other pear plants.
Owner:CHINA AGRI UNIV
PCR method for tracking source identification of origin components of eight kinds of animals
ActiveCN104946790AImprove featuresIncreased sensitivityMicrobiological testing/measurementMolecular identificationNucleotide
The invention belongs to the technical field of detection of related products of animal origin components including meat and source thereof, and particularly provides a pair of primers capable of detecting the origin components of eight kinds of animals of goat, sheep, buffalo, cattle, pig, deer, camel and yak at the same time in a PCR reaction, the molecular identification technology in the operation process, a kit comprising the primers, and an application method. The nucleotide sequences of the primers are shown in the SEQ ID 1 and the SEQ ID 3. The PCR method is based on the PCR technology, has the advantages of being simple in operation, strong in specificity, low in equipment requirement, and the like, importantly, can detect the origin components of the eight kinds of animals at the same time in once PCR, can be used for adulteration detection of the food meat and the meat products and the tracking source identification of the animal origin components in feed, is simple and quick, and improves the detection efficiency.
Owner:CHINA JILIANG UNIV +1
Molecular identification kit of meat product made of beef and application of molecular identification kit
ActiveCN103421890AStrong specificityLow costMicrobiological testing/measurementMolecular identificationAgricultural science
The invention belongs to the technical field of meat product molecular detection and particularly relates to a molecular identification method and kit of a meat product made of beef as well as application of the molecular identification kit. The molecular identification method comprises the following steps: (1) primer designing: designing a specific primer according to the sequence of the TNFRSF10A gene of a bos taurus, wherein the sequence of the specific primer is shown in SEQ ID NO: 2 and SEQ ID NO: 3. (2) creating a PCR amplified reaction method, that is, amplifying the genome DNA of the bos taurus through the specific primer so as to obtain amplified specific fragments; (3) analyzing the amplified PCR products, that is, analyzing the amplified PCR products through agarose gel electrophoresis; according to gel-imaging, observing whether target fragments appear or not, and judging whether the target fragments contain the components of the genome DNA of the bos taurus or not. The molecular identification method has the characteristics of simplicity, convenience, quickness, high specificity and relatively-low cost, and is applicable to grassroots units.
Owner:HUAZHONG AGRI UNIV
Rive CYP704B2 gene mutant, and molecular identification method and applications thereof
ActiveCN105002191ABreeding benefitsFermentationVector-based foreign material introductionMolecular identificationGenetically modified rice
The invention provides a rive CYP704B2 gene mutant, and applications thereof, and belongs to the technical field of gene engineering. According to the rive CYP704B2 gene mutant, indica rice 93-11 is subjected to cobalt 60 radiation-induced mutation so as to replace three G bases after the 794th base of rice CYP704B2 gene with one T base; the CYP704B2 gene mutant is named as cyp704B2-3; the nucleotide sequence of the CYP704B2 gene mutant is represented by SEQ ID No.1; it is further confirmed that rice recessive genic male sterility is induced by the CYP704B2 gene mutant; the CYP704B2 gene mutant can be used for culturing transgenic rice with recessive genic male sterility, and possesses great importance in rice genetic resource genetic improvement-breeding. The invention also provides a molecular marked identification method and applications of the rive CYP704B2 gene mutant in breeding.
Owner:HAINAN BOLIAN RICE GENE TECH CO LTD
DNA barcoding standard gene sequence of Rizhao Blepharipoda liberata and species identification method based thereon
InactiveCN103937802AFacilitate the realization of molecular identificationAchieve molecular identificationMicrobiological testing/measurementFermentationMolecular identificationForward primer
Relating to the technical field of species identification, the invention specifically relates to a DNA barcoding standard gene sequence of Rizhao Blepharipoda liberata and species identification method based on the gene sequence. The DNA barcoding standard gene is COI gene, which has a gene sequence shown as SEQIDNO.1. The identification method includes: conducting extraction separation of DNA on a to-be-tested tissue, and amplifying a target gene by polymerase chain reaction. A pair of primers used in the amplification reaction are shown as the following: a forward primer 5'-GGTCAACAAATCATAAAGATATTGG-3', and a reverse primer 5'-TAAACTTCAGGGTGACCAAAAAATCA-3'. The gene sequence obtained in the invention is in favor of realizing molecular identification of the Rizhao Blepharipoda liberata, and can effectively shorten the identification time of the Rizhao Blepharipoda liberata.
Owner:刘丰铭
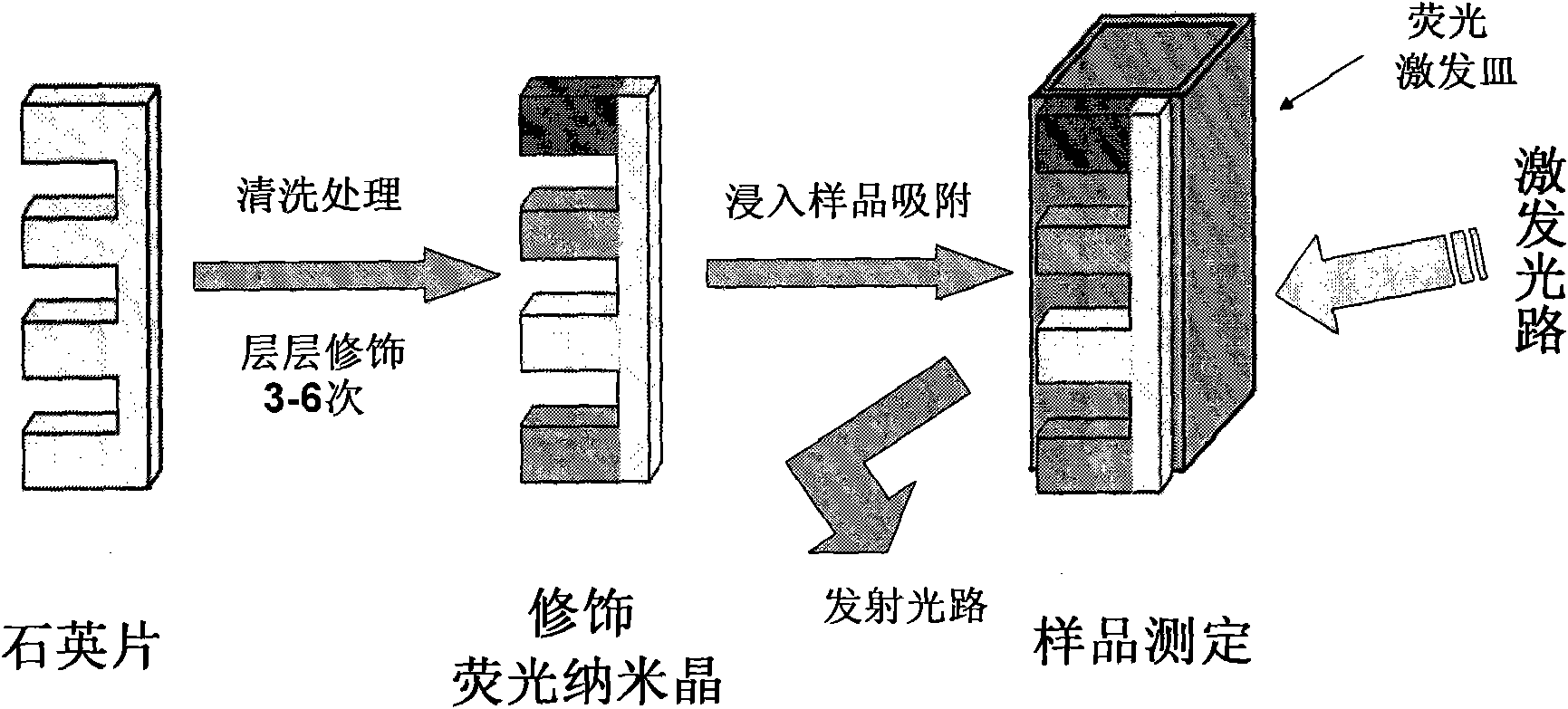
Research and application of molecular identification-based fluorescence nanocrystal quartz fluorescent sensor for high-selectivity multicomponent saccharide detection
InactiveCN102042975AIncreased sensitivityHigh selectivityFluorescence/phosphorescenceMolecular identificationFluorescence
The invention discloses a fluorescence nanocrystal quartz fluorescent sensor for molecular identification-based multicomponent simultaneous saccharide detection and a method for detecting saccharides by using the same. The invention provides a preparation method of a molecular identification-based fluorescence nanocrystal comb quartz plate, which comprises the following steps: selecting an identifier corresponding to the saccharides; preparing a fluorescence nanocrystal, and carrying out surface modification on the fluorescence nanocrystal according to the literature; and modifying the modified fluorescence nanocrystal onto surfaces of different probes of the comb quartz plate by using layer-by-layer accumulation surface modification technique. The invention also provides a method for multicomponent simultaneous saccharide detection, which comprises the following steps: immersing the modified quartz plate into a food sample solution which is simply pulpified, installing the quartz plate onto a sealed silica dish, and detecting the saccharides in the sample. The invention has the advantages of strong specificity, high sensitivity, short detection time and low cost. The saccharide fluorescence detection method has a quick and simple operation process, and the reaction and the result are automatically completed and recorded by instruments.
Owner:UNIV OF JINAN
Method for quickly extracting fungal DNA for PCR amplification
InactiveCN101696411AReduce lossesImprove extraction efficiencyMicroorganism based processesDNA preparationBiotechnologyMolecular identification
The invention discloses a method for quickly extracting fungal DNA for PCR amplification, which comprises the steps of: (1) culturing fungi to obtain fungal mycelia; (2) placing the fungal myceliua into a centrifugal tube filled with a lysate; (3) alternately freezing and thawing the centrifugal tube; and (4) centrifuging the thawed fungal mycelia, taking a supernatant, adding 95 percent ethanol into the supernatant, mixing the mixture evenly, quickly freezing the mixture by liquid nitrogen, centrifuging the mixture, removing a supernatant, and air-drying a precipitate to obtain the fungal DNA. The method has simple operation, is time-saving and labor-saving, reduces the loss of the fungal DNA during the grinding, does not need phenol and chloroform for extraction, and avoids the contact of a toxic reagent. The DNA extracted by the method has good integrity, high purity and high yield, can be directly used for a PCR amplification reaction, and can efficiently amplify a target product for cloning and sequencing. The method improves the extraction efficiency of the fungal DNA, reduces the cost for the extraction of the fungal DNA, and can effectively improve the molecular identification and detection efficiency of the fungi.
Owner:INST OF FOREST ECOLOGY ENVIRONMENT & PROTECTION CHINESE ACAD OF FORESTRY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com