Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
148 results about "Coi gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
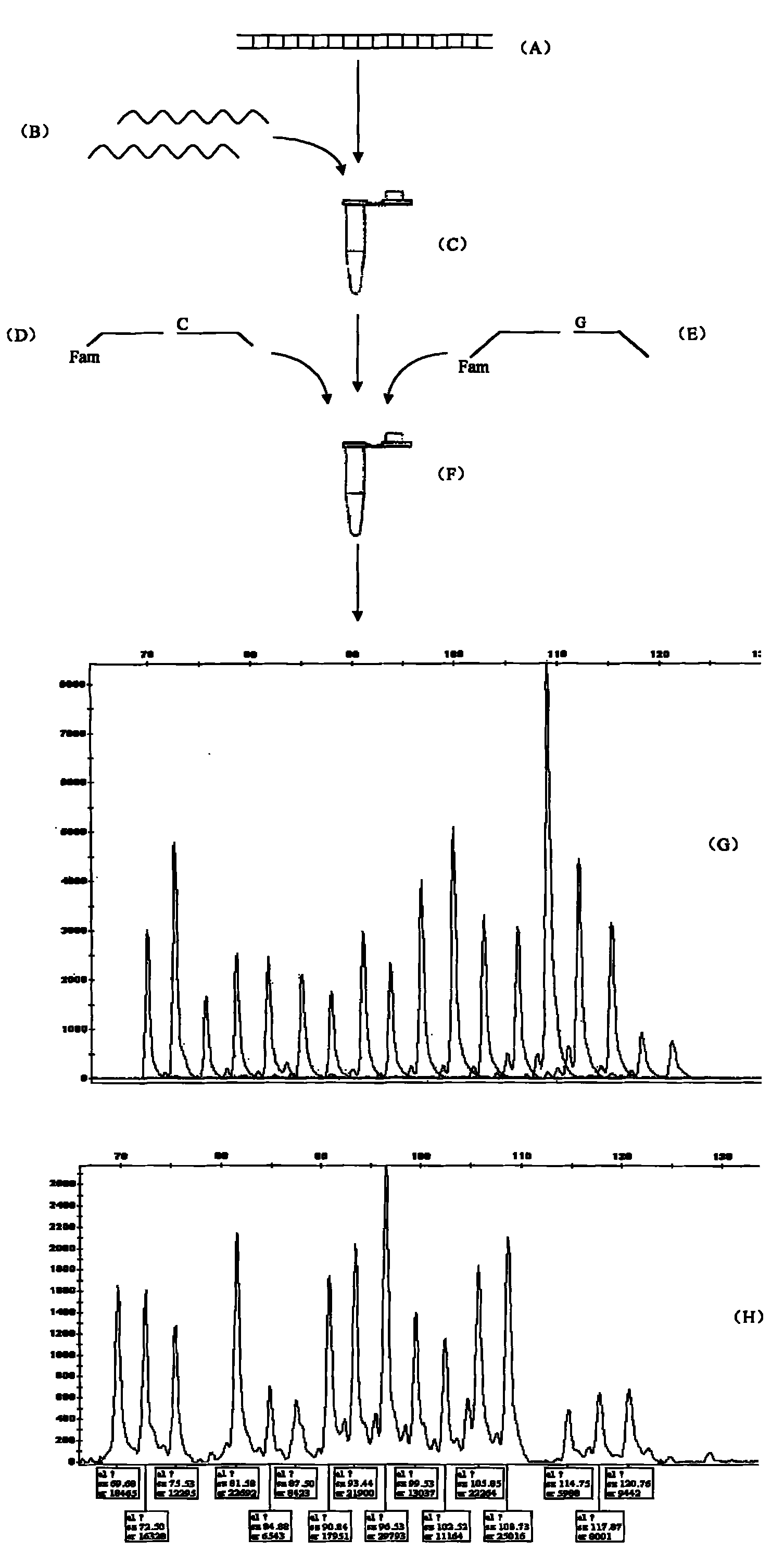
Cytochrome c Oxidase subunit I gene (COI), the gold standard in animal, insect DNA barcoding. Size of the Gene is 648-bp. This tool is used in the identification of species.
Verification method of sea horses
InactiveCN104017899AExclude phenotypic plasticityRule out genetic variabilityMicrobiological testing/measurementSystematicsOrganism
The invention relates to a verification method of sea horses, belonging to the field of molecular biology. The invention aims to provide a verification method of sea horses. The method is characterized by comprising the following steps: a. extracting sea horse total DNA (deoxyribonucleic acid); b. PCR (deoxyribonucleic acid) amplification: carrying out PCR amplification on the sea horse total DNA to obtain a COI gene sequence; c. carrying out PCR product sequencing; and d. carrying out sequence comparison to determine the species of the sea horse. The method is free from the restriction of morphological characteristics, can identify the species of the sea horse under the condition of being incapable of utilizing the traditional morphological classification process due to lack of morphological characteristics, has the advantages of high accuracy and the like, provides scientific references for sea horse systematics and sea horse resource researches in China, and also provides reliable references for biological diversity of sea horse resources and conservation of resources.
Owner:CHENGDU UNIV OF TRADITIONAL CHINESE MEDICINE
PCR special primer pair for identifying pig-cattle-sheep derived ingredients in livestock and poultry meat based on mitochondrion COI gene, PCR identification method and reagent kit
InactiveCN105755149AEfficient detectionAgainst adulterationMicrobiological testing/measurementDNA/RNA fragmentationAdditive ingredientBiology
The invention discloses a PCR special primer pair for identifying pig-cattle-sheep derived ingredients in livestock and poultry meat based on mitochondrion COI gene, an identification method and a reagent kit, and belongs to the technical field of food detection. According to difference sites of COI genes of 9 kinds of animals including pigs, cattle, sheep, rabbits, pigeons, quails, chickens, ducks and geese, specific primers of 3 kinds of animals including the pigs, the cattle and the sheep are designed and are shown in a sequence table SEQ ID NO. 1, a sequence table SEQ ID NO. 2, a sequence table SEQ ID NO. 3, a sequence table SEQ ID NO. 4, a sequence table SEQ ID NO. 5 and a sequence table SEQ ID NO. 6. The identification method comprises the following steps: extracting total DNA of species to be determined, and performing PCR amplification. The reagent kit comprises SEQ ID NO. (1-6). The primers of the PCR special primer pair disclosed by the invention perform specificity amplification only on objective fragments of respective species and cannot generate a reaction with other species; the method is high in sensitivity, simple, convenient and rapid, and the pig derived ingredients, the cattle derived ingredients and the sheep derived ingredients in the livestock and poultry meat can be effectively identified, so that the adulteration phenomena of beef and mutton are avoided. Besides, designed primers SEQ ID NO. (1-6) are matched with related reagents, so that the reagent kit can be made, and favorable industrialized production and application prospects can be obtained.
Owner:JIANGSU INST OF POULTRY SCI
Method for identifying DNA bar code molecule of earthworm
InactiveCN103898234AIdentification is conducive to the realizationIdentification achievedMicrobiological testing/measurementMolecular identificationDNA barcoding
The invention discloses a method for identifying a DNA bar code molecule of an earthworm and a COI sequence of the DNA bar code molecule of the earthworm. The method comprises the steps of amplifying the gene of a sample COI through polymerase chain reaction (PCR), verifying the PCR product by agarose gel, then sending to a biological sequencing company for sequencing, according to the sequencing result, sequence splicing through manual correcting, comparing with a public sequence, then confirming the tissue to be tested as the source of pheretima aspergillum or pheretima vulgaris if the homology of the spliced sequence and the gene sequence SEQ ID NO.1-NO.4 is over 99%. By adopting the reliable DNA molecular identification technology, the method can rapidly and accurately identify the earthworm varieties specified by the 'pharmacopoeia of China' and non-medicinal material earthworm varieties without conducting morphological identification on the earthworm varieties, can further distinguish the pheretima aspergillum varieties from pheretima vulgaris varieties, improves the accuracy and reliability, and guarantees the safety of the earthworm as the medicinal material.
Owner:MUDANJIANG YOUBO PHARMA CO LTD
Haplotype primer for identifying Q-shaped bemisia tabaci and identification method
InactiveCN102676680ASimple and Stable Molecular MarkersSolving the difficult problem of differentiating haplotype Q Bemisia tabaciMicrobiological testing/measurementDNA/RNA fragmentationGenomic DNADigestion
The invention relates to a haplotype primer for identifying Q-shaped bemisia tabaci and an identification method. The method comprises the following steps of: (1) extracting Q-shaped bemisia tabaci genomic DNA; (2) carrying out PCR (Polymerase Chain Reaction) amplification on mitochondrial COI (cytochrome oxidase subunit I) genes of the Q-shaped bemisia tabaci genomic DNA by taking the Q-shaped bemisia tabaci genomic DNA as a template; (3) digesting a PCR product prepared in step (2) by a restriction enzyme Hin1II (N1aIII) to obtain a digestion product; and (4) carrying out agarose gel electrophoresis analysis on the digestion product prepared in step (3). The restriction enzyme is a common restriction enzyme, a simple and stable digestion mark is provided for screening two types of haplotype Q-shaped bemisia tabaci, at the same time an identification technology for the two types of haplotype Q-shaped bemisia tabaci is developed and established, and a foundation is established for future population dynamic identification and biology and invasion mechanism researches of the two types of haplotype Q-shaped bemisia tabaci.
Owner:QINGDAO AGRI UNIV
Direct PCR (Polymerase Chain Reaction) detection method of Bombyx mori nuclear polyhedrosis pathogeny BmNPV
InactiveCN101886142AThe detection method is simpleEasy to scaleMicrobiological testing/measurementAgainst vector-borne diseasesDiseasePositive control
The invention discloses a direct PCR detection method of Bombyx mori nuclear polyhedrosis pathogeny BmNPV, comprising the following steps of: firstly, treating the blood of infected Bombyx mori by ethanol precipitation to remove a PCR reaction inhibitor; designing the PCR primer of a polyhedrin gene according to the genome analysis of the Bombyx mori nuclear polyhedrosis pathogeny BmNPV; carrying out PCR amplification on the polyhedrin gene, adding an anti-PCR reaction inhibitor BSA (Bull Serum Albumin) to a reaction system and simultaneously carrying out PCR reaction by the primer of a Bombyx mori chondriosome CO I gene to make positive control; respectively getting the blood of normal Bombyx mori and the inflected midguts of the Bombyx mori infected with white muscardine, green muscardine, a bacterial disease and a microsporidia disease and then carrying out PCR reaction specification analysis; and detecting the PCR reaction result by agar gel electrophoresis. The whole detection process has simple operation and can be completed only by 4 hours. The disease can be diagnosed at the early stage of BmNPV infection by adopting the direct PCR detection method to carry out periodical sampling detection so as to adopt a measure in time to prevent the disease from happening.
Owner:ANKANG UNIV
Method for screening veneridae mitochondria COI gene amplification primers
InactiveCN101979536AHigh variabilityLow amplification efficiencyDNA preparationDNA/RNA fragmentationPhylogenesisCoi gene
The invention discloses a method for screening veneridae mitochondria COI (cytochrome c oxidase subunit I) gene amplification primers. The method is characterized by comprising the following steps of: a, logging in National Center for Biotechnology Information, and downloading complete mitochondria sequences of veneridae shellfishes; b, comparing and analyzing the downloaded sequences by using BioEdit, and determining a conserved region of the sequences; c, designing a plurality of pairs of primers by using Primer Premier 5 in the conserved region and synthesizing the primers; d, extracting veneridae shellfish DNA, and performing PCR amplification on the synthesized primers under a corresponding reaction system; and 3, performing electrophoresis detection, and screening a group of cocktail primers with single stripe and high amplification efficiency, wherein the group of cocktail primers consists of a pair of degenerate primers and a pair of common primers. The veneridae mitochondria COI primers can effectively amplify destination fragments of different species of the veneridae, and have significance for COI sequence-based veneridae species classification, phylogenesis, systematicgeography research, veneridae germ plasm resource protection and fishery resource management.
Owner:OCEAN UNIV OF CHINA
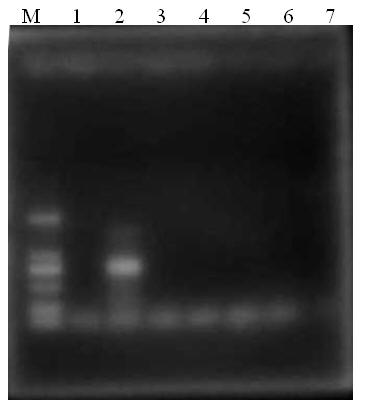
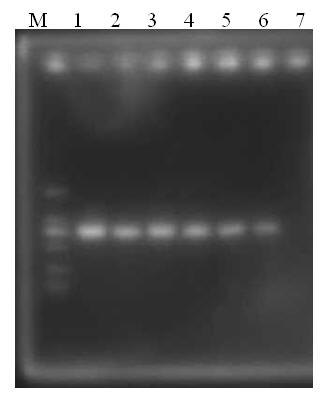
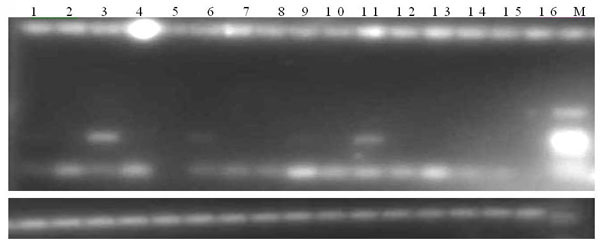
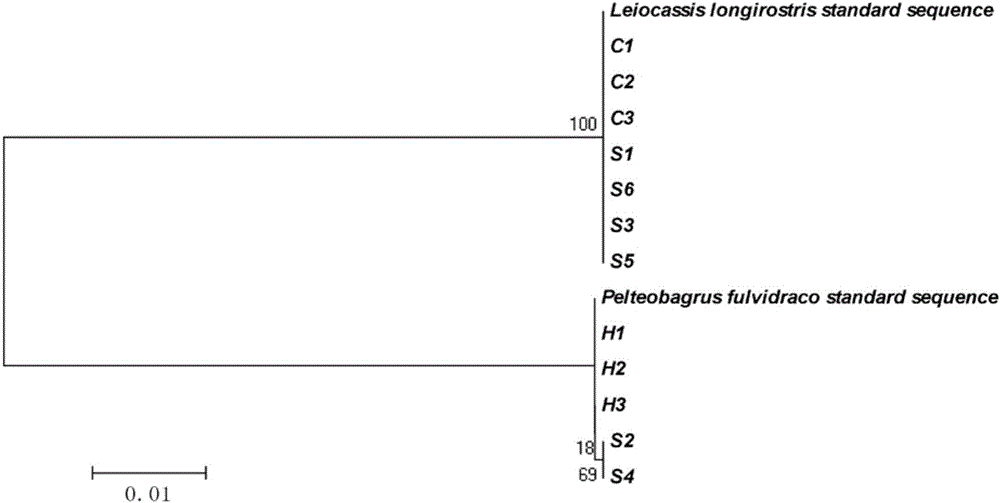
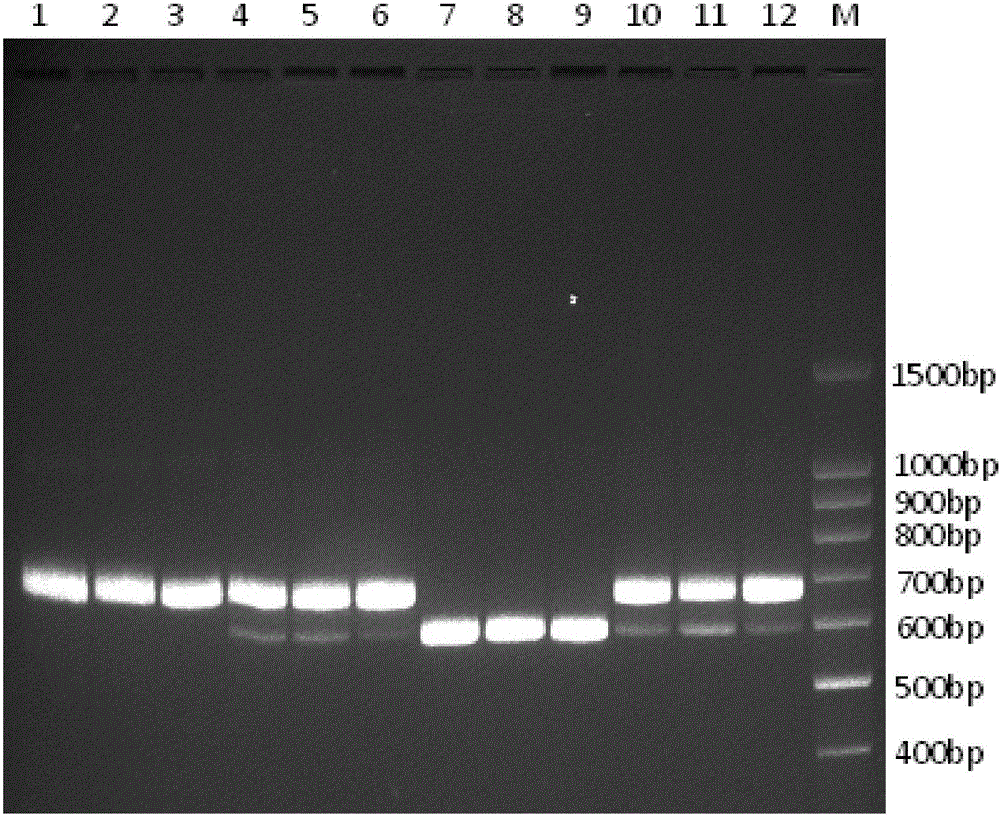
Primer group, kit and method for identifying tachysurus fulvidraco and leiocassis longirostris hybrid species
InactiveCN105695590AEfficient managementMonitor negative impactsMicrobiological testing/measurementDNA/RNA fragmentationAquaculture industryA-DNA
The invention provides a primer group for identifying a male parent and a female parent of tachysurus fulvidraco and leiocassis longirostris hybrid species. The primer group comprises a primer pair for amplifying COI genes and a primer pair for amplifying an ITS sequence, wherein the primer pair for amplifying COI genes consists of a primer COIF and a primer COIR; the primer pair for amplifying an ITS sequence consists of a primer ITSF and a primer ITSR. The invention also provides a kit which is for identifying the male parent and the female parent of tachysurus fulvidraco and leiocassis longirostris hybrid species and comprises the primer group. The kit also comprises any one or more of the following reagents: a buffer solution for PCR, dNTP, heat-resistant DNA polymerase, a cloning vector, a DNA ligase, and a buffer solution for ligation. The invention also provides a method for identifying the male parent and the female parent of tachysurus fulvidraco and leiocassis longirostris hybrid species. The primer group, the kit or the method can be used for accurately identifying the male parent and the female parent of the tachysurus fulvidraco and leiocassis longirostris hybrid species, thus effectively managing a breeding plan and monitoring negative influence thereof, and has great significance in sustainable development of aquaculture industry.
Owner:YANGTZE RIVER FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
DNA barcoding standard gene sequence of Rizhao Blepharipoda liberata and species identification method based thereon
InactiveCN103937802AFacilitate the realization of molecular identificationAchieve molecular identificationMicrobiological testing/measurementFermentationMolecular identificationForward primer
Relating to the technical field of species identification, the invention specifically relates to a DNA barcoding standard gene sequence of Rizhao Blepharipoda liberata and species identification method based on the gene sequence. The DNA barcoding standard gene is COI gene, which has a gene sequence shown as SEQIDNO.1. The identification method includes: conducting extraction separation of DNA on a to-be-tested tissue, and amplifying a target gene by polymerase chain reaction. A pair of primers used in the amplification reaction are shown as the following: a forward primer 5'-GGTCAACAAATCATAAAGATATTGG-3', and a reverse primer 5'-TAAACTTCAGGGTGACCAAAAAATCA-3'. The gene sequence obtained in the invention is in favor of realizing molecular identification of the Rizhao Blepharipoda liberata, and can effectively shorten the identification time of the Rizhao Blepharipoda liberata.
Owner:刘丰铭
High-throughput multi-LDR parting kit for detecting farmland ecological environment
InactiveCN101921828AGuaranteed to detectMaintain the ecological environmentMicrobiological testing/measurementEcological environmentPredation
The invention relates to a high-throughput multi-LDR parting kit for detecting farmland ecological environment, comprising mitochondria COI gene universal primers of 16 kinds of species in the farmland, specific probe sequences of the mitochondria COI gene universal primers of the 16 kinds of species in the farmland, and a reagent for PCR (polymerase chain reaction) proliferation and LDR (light dependent resistor) detection programs, wherein the mitochondria COI gene universal primers have the upstream of 5'-GGTCAACAAATCATAAAGATATTGG-3' and the downstream of 5'-taaacttcagggtgaccaaaaaatca-3'. In the invention, the kit effectively proliferates all target species sequences for the COI gene universal primers, and ensures that each target sequence and a low-copy template can be detected; the specific probe of each target species further ensures the specificity so as to totally distinguish each target sequence to realize multi-parting fixed quantity, thus the change of a farmland predation structure is known and the farmland ecological environment is maintained.
Owner:DONGHUA UNIV
Amplification primer of tridacnidae shellfish mitochondria COI (cytochrome oxidase I) gene
InactiveCN102816762AConducive to the study of genetic diversity of speciesLow amplification efficiencyDNA/RNA fragmentationGermplasmDesign software
The invention relates to an amplification primer of a tridacnidae shellfish mitochondria COI (cytochrome oxidase I) gene. The known 52 tridacnidae sequences in NCBI (National Center for Biotechnology Information) are utilized, two tridacnidae sequences obtained by amplifying a universal primer are jointly utilized, a CLUSTALW software is used for comparison analysis, a plurality of pairs of primers are designed by using a primer design software Primer Premier 5 in a determined conversation region, 43 tridacnidae individuals are repeatedly subjected to PCR (Polymerase Chain Reaction) experiments in a 10 mul system and are finally detected through agarose gel electrophoresis, and an efficient amplification primer of the tridacnidae shellfish mitochondria COI gene sequence is sieved. The efficient amplification primer is characterized in that CQF is 5'-GATATTGGAACCCTTTACTT-3'; and CQR is 5'-TTCAGGATTGTCCCAAGAATCA-3'. By using the tridacnidae shellfish mitochondria COI primer, the target fragments of different specifies of tridacnidae shellfishes can be efficiently amplified; and the tridacnidae shellfish mitochondria COI primer has important significance for identifying tridacnidae species and carrying out phylogenetic analysis by using a molecular method, and can provide reliable technical guidance for developing and protecting tridacnidae germplasm resources by using the molecular means.
Owner:OCEAN UNIV OF CHINA
Culicoides bellulus specific gene and molecular identification method thereof
InactiveCN105543249AGuaranteed accuracyEnsure reliabilityMicrobiological testing/measurementEnzymesMolecular identificationCulicoides
The invention discloses a culicoides bellulus specific gene and a molecular identification method thereof. The culicoides bellulus specific gene is characterized in that a molecular biology method is adopted to obtain the COI gene sequence of culicoides, and the species of the culicoides is identified by comparing gene sequence similarity. The ulicoides bellulus molecular identification method is beneficial for accurately and fast identifying culicoides bellulus.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
Primer and identifying method for identifying different genetic collateral series of aphelinid
InactiveCN103088136ASimple and Stable Molecular MarkersSolving the puzzle of differentiating genetic lineages of sunbeesMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestion3-deoxyribose
The invention relates to a primer and an identifying method for identifying different genetic collateral series of aphelinid. The identifying method comprises the following steps: (1), extracting aphelinid genome DNA (Deoxyribose Nucleic Acid); (2) carrying out PCR (Polymerase Chain Reaction) augmentation on a mitochondria COI (Cytochrome Oxidase Subunit) gene by adopting the aphelinid genome DNA as a template; (3), digesting the prepared PCR product by restriction enzyme Alu I to obtain enzyme-digested products; and (4), carrying out sepharose gel electrophoretic analysis on the prepared enzyme-digested products. The restriction enzyme is normal restriction enzyme, so that simple and stable enzyme digestion marks are provided for selecting the aphelinid with two genetic collateral series; and meanwhile, an identifying technology for the aphelinid with two genetic collateral series is established, so that the foundation is laid for population dynamics identification for the aphelinid with two genetic collateral series, the biology as well as the research of a mechanism of transmission.
Owner:QINGDAO AGRI UNIV
Screening method of Muricidae mitochondrion COI gene amplification primer
InactiveCN101942433AEfficient amplificationLow amplification efficiencyDNA preparationGerm plasmScreening method
The invention discloses a screening method of a Muricidae mitochondrion COI gene amplification primer, which is characterized by comprising the following steps: a. a COI general amplification primer is used for amplifying the COI sequence of parts of Muricidae species; b. the sequence of the COI general amplification product is compared with the COI sequence of the obtained parts of Muricidae species to find out all mutation bases of the Muricidae mitochondrion COI sequence on the COI general amplification primer sequence section so as to recombine different Muricidae COI amplification primers; and c. a pair of primers with best Muricidae species amplification effect is obtained by screening the newly-combined different COI amplification primers. The Muricidae mitochondrion COI gene amplification primer of the invention can effectively amplify the COI sequence of different specimens of the Muricidae, and has an important meaning for researching the species taxonomy, system development and systematic geography of Muricidae and protecting Muricidae germ plasm resource.
Owner:OCEAN UNIV OF CHINA
COI gene based PCR-RFLP discrimination method of seventeen economic sea cucumbers of Stichopodidae
ActiveCN103923972ALower requirementEasy to operateMicrobiological testing/measurementEnzyme digestionApplicability domain
The invention discloses a COI gene based PCR-RFLP discrimination method of seventeen economic sea cucumbers of Stichopodidae. The method comprises the following steps: 1, extracting DNA of a Stichopodidae sea cucumber to be detected as a detected sample, using the DNA as a template, and using primers COIe-F:5'-ATAATGATAGGAGGRTTTGG-3', COIe-R:5-GCTCGTGTRTCTACRTCCAT-3'PCR' to amplify a CO I fragment, wherein A is A / G; and 2, carrying out enzyme digestion on the CO I fragment by using a restrictive endonuclease group DdeI / SfcI or BstNI / Sau3AI, carrying out electrophoresis on the above obtained enzyme digestion product, estimating the size of each of the electrophoretic bands of samples to be detected, contrasting the number and the size of the electrophoretic bands in an electrophoretogram and the estimated value of the size with the reference band spectra of the seventeen sea cucumbers, and finding the standard spectral bands coupled with the number and the size of the electrophoretic bands, wherein the types represented by the standard spectral bands are the type of the detected sample. The method has the advantages of wide application range, simple operation, and accurate and fast detection, is especially suitable for the identification of a large batch of commodity sea cucumbers, so the method is a scientific method for identifying the sea cucumbers of Stichopodidae, and provides technologic supports for the standardization of the commodity market of the sea cucumbers and the protection of the sea cucumber resources.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
SS-COI (Sirex noctilio F. specific cytochrome oxidase I) primer pair and quick molecular detection method
InactiveCN104232634AReduce the need for technical expertiseThe identification method is fast and stableMicrobiological testing/measurementDNA/RNA fragmentationTremex fuscicornisSibling species
Owner:BEIJING FORESTRY UNIVERSITY
Method for identifying Frankliniella occidentalis species by utilizing PCR (Polymerase Chain Reaction)-RFLP (Restriction Fragment Length Polymorphism) technology
InactiveCN102392075AEasy and Stable Digestion TagSolve key technologiesMicrobiological testing/measurementTerminal restriction fragment length polymorphismElectrophoresis
The invention relates to a method for identifying Frankliniella occidentalis species by utilizing PCR (Polymerase Chain Reaction)-RFLP (Restriction Fragment Length Polymorphism) technology, comprising the following steps: (1) extracting Frankliniella occidentalis genome DNA; (2) conducting PCR amplification to the mitochondrion COI gene of the Frankliniella occidentalis genome DNA by taking the Frankliniella occidentalis genome DNA as a template; (3) conducting enzyme cutting to the PCR product prepared in step (2) by adopting restriction enzyme EarI to obtain an enzyme cutting product; and (4) conducting agar gel electrophoresis analysis on the enzyme cutting production prepared in step (3). The restriction enzyme is the common restriction enzyme, therefore, the method provides a simple, convenient and stable enzyme cutting marker for screening two types of Frankliniella occidentalis, and simultaneously explores and establishes an identification technology for the two types of Frankliniella occidentalis, and lays a foundation for identification on the population dynamics of the two types of Frankliniella occidentalis and research on biology and invasion mechanisms.
Owner:INST OF PLANT PROTECTION SHANDONG ACAD OF AGRI SCI
Specific gene and molecular identification method of culicoides cyancus
ActiveCN105483261AGuaranteed accuracyEnsure reliabilityMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationCulicoides
The invention discloses a specific gene and a molecular identification method of culicoides cyancus. The molecular identification method is characterized in that a COI gene sequence of the culicoides cyancus is acquired by adopting a molecular biology method, and species identification is performed on the culicoides cyancus through comparison of the gene sequence similarity. According to the molecular identification method of the culicoides cyancus, achievement of accurate and rapid identification on the culicoides cyancus is facilitated.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE
PCR primer for identifying codling moth cell line and identifying method of codling moth cell line
InactiveCN105255883ASimple Enzyme Digestion Identification MethodStable Enzyme Digestion Identification MethodMicrobiological testing/measurementDNA/RNA fragmentationForward primerAgricultural science
The invention provides a PCR primer for identifying a codling moth cell line and an identifying method of the codling moth cell line. A PCR primer pair for amplifying the mitochondria COI gene segments of the codling moth and other insect cell lines provided by the invention has a forward primer sequence as shown in SEQ ID NO:1 and has a reverse primer sequence as shown in SEQ ID NO: 2. The forward primer sequence after optimization is SEQ ID NO:3 and the reverse primer sequence is SEQ ID NO: 4. The invention also provides an identifying method for the codling moth cell line. According to the invention, the difference between the mitochondria COI gene segments of the codling moth and other insect cell lines is researched at the molecular level and the identifying method for the cell lines such as the codling moth, the dark holotricha parallela, the asparagus caterpillar, the cabbage looper is established.
Owner:QINGDAO AGRI UNIV
PCR amplification kit for detecting clonorchis sinensis metacercaria on basis of plastosome COI genes and amplification primer
InactiveCN105132414AStrong specificityHigh detection sensitivityMicrobiological testing/measurementDNA/RNA fragmentationClonorchis sinensisClonorchis sinensis Infections
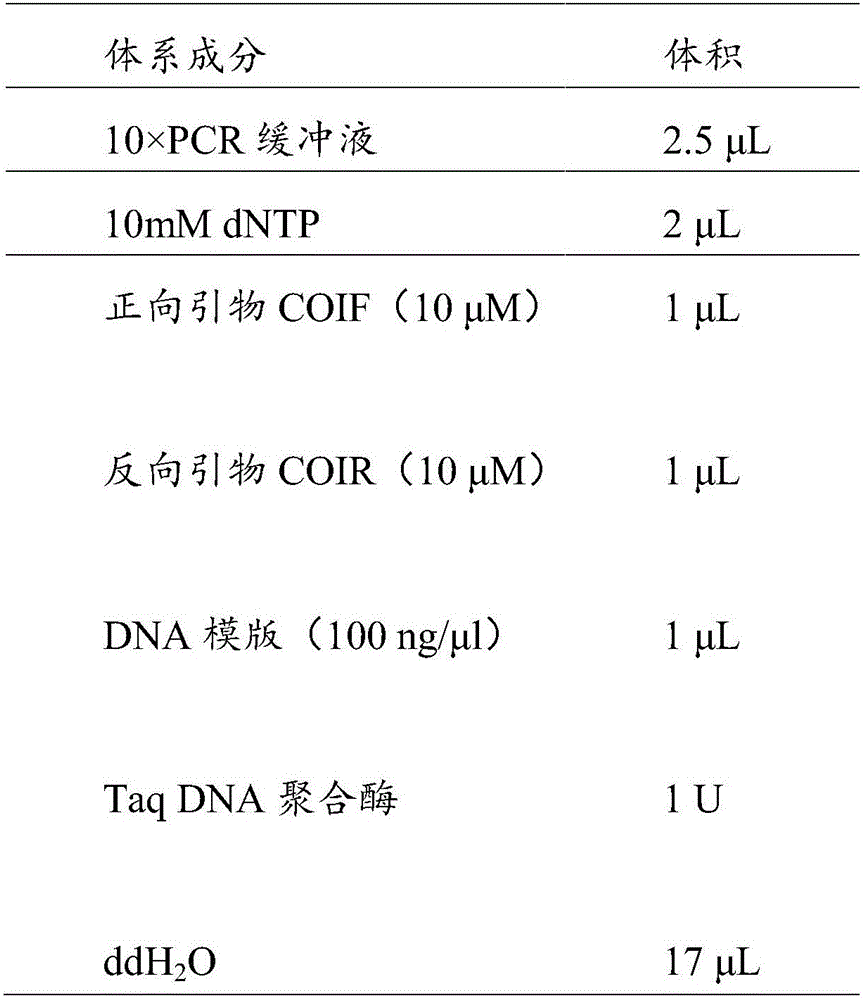
The invention discloses a PCR (Polymerase Chain Reaction) amplification kit for detecting clonorchis sinensis metacercaria on the basis of plastosome COI genes and an amplification primer. The PCR amplification kit comprises PCR amplification reaction liquid, wherein the PCR amplification reaction liquid includes 10mmol / L of Tris-HCl, 50mmol / L of KCL, 1.5mmol / L of MgCl<2>, 0.8mmol / L of dNTPs, 0.4[mu]mol / L of positive primers, 0.4[mu]mol / L of reverse primers, 20ng / [mu]L of DNA (Deoxyribonucleic Acid) templates and 0.075U / [mu]L of Taq enzyme. The PCR amplification kit has the advantages of high sensitivity, high specificity, high detection speed and high detection accuracy, and an effective technical measure is provided for diagnosis, treatment and prevention of clonorchis sinensis infection.
Owner:舟山出入境检验检疫局综合技术服务中心
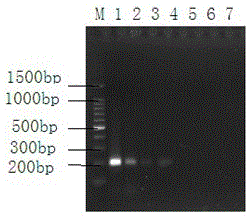
Method for detecting material sources of meat products on basis of short-sequence high-throughput sequencing
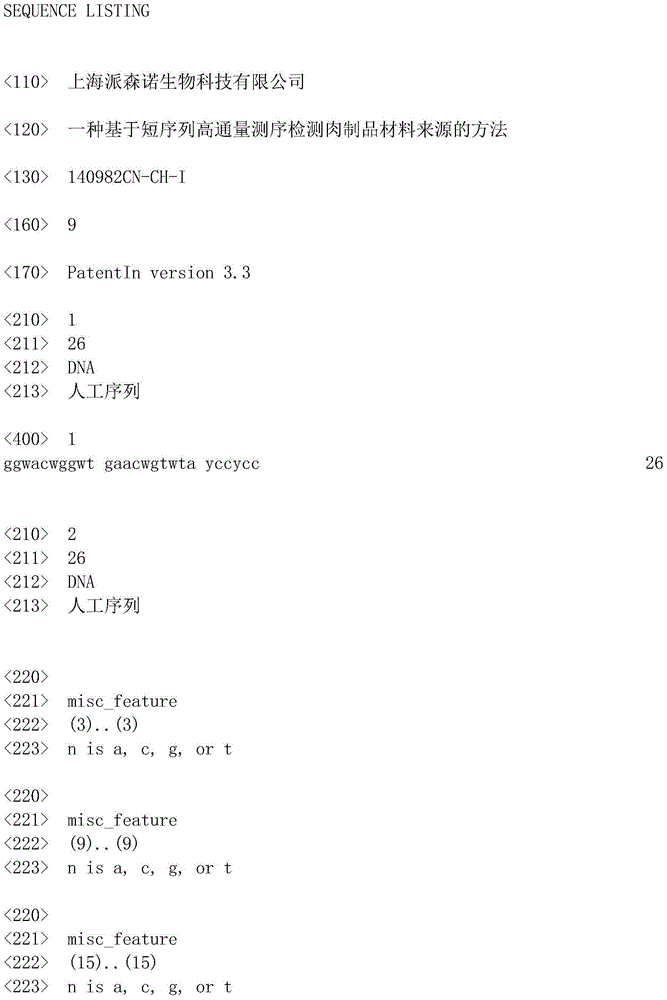
The invention belongs to the technical field of molecular biology and discloses a method for detecting material sources of meat products on the basis of short-sequence high-throughput sequencing. The method comprises the following steps that 1, genome DNA of the meat products to be detected is extracted; 2, universal primer sequences shown as SEQ ID NO:1 and SEQ ID NO:2 are designed and synthesized; 3, PCR amplification is carried out with the genome DNA of the meat products to be detected as a template to purify PCR products; 4, the PCR products are quantified, samples of the same quantity are mixed, a standard library is constructed, and then high-throughput sequencing is carried out; 5, COI sequence information of the meat products to be detected is obtained according to the sequencing result, so that the material sources of the meat products to be detected are obtained. By the utilization of the method, the material composition and the sources of the meat products can be identified, species can be well distinguished due to the difference between base sequences in the COI gene sequence, and the method is high in accuracy for identifying the material sources of the meat products.
Owner:SHANGHAI PASSION BIOTECHNOLOGY CO LTD
Method for identifying dried Pheretima aspergillum based on PCR-RFLP technology
InactiveCN109295170AEasy to operateMicrobiological testing/measurementEnzyme digestionPHERETIMA ASPERGILLUM
The invention discloses a method for identifying dried Pheretima aspergillum based on a PCR-RFLP technology. Universal primers of a COI gene are used to carry out PCR amplification to obtain mitochondrial cytochrome oxidase (COI) gene fragments of all kinds of earthworms, the sequences of the fragments are sequenced and compared, the complete sequences of the COI fragments of the different kinds of earthworms undergo restriction enzyme map analysis by DNAMAN software to finally screen a restriction enzyme RsaI, the restriction enzyme RsaI is used to carry out enzyme digestion on the above obtained PCR product, the enzyme digestion result of the PCR product is detected by 2% agarose gel, and the enzyme digestion maps are used to identify the dried Pheretima aspergillum and other varieties.The method is simple and reliable to operate, and provides a new technical way for the identification of dried Pheretima aspergillum products.
Owner:INST OF ZOOLOGY GUANGDONG ACAD OF SCI
Cockle shellfish mitochondria COI (cytochrome c oxidase I) gene amplification primer
ActiveCN102827839AConducive to the study of genetic diversityEfficient amplificationDNA/RNA fragmentationResource protectionGenetic diversity
The invention uses a universal primer for amplification to obtain 66 cardiidae mtCOI (cytochrome c oxidase I) sequences, uses a CLUSTALW software to compare and analyze, uses primer design software primer Premier5 to design 5 pairs of primers in a determined area, uses 216 cardiidae individuals to repeatedly conduct FCR reaction tests in a 10mu m system, finally is detected through agarose gel electrophoresis, and screens out ctockle shellfish mitochondria COI gene sequence efficient amplification primer. The cockle shellfish mitochondria COI gene amplification primer is characterized in that NGF5'-ACAAATCATAAAGATATTGG-3'NGR5'-TTCAGGGTGTCCGAAGAATCA-3' adopts the cockle shellfish mitochondria COI primer, objective sections of different species of cockle shellfish are effectively amplified, requirements of genetic diversity survey of cockles can be met, simultaneously reliable molecular technique instruments for germplasm resource protection and breeding industry development are provided.
Owner:OCEAN UNIV OF CHINA
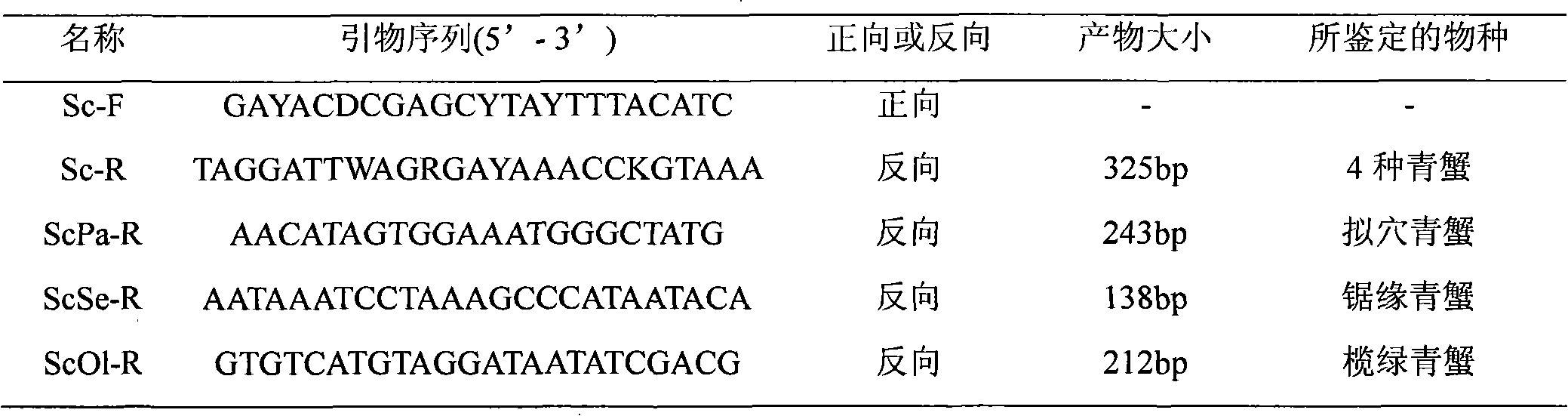
PCR method for rapid identification of four types of blue crabs of scylla
InactiveCN101792809ARapid identificationAccurate identificationMicrobiological testing/measurementGerm plasmRapid identification
The invention relates to a PCR method for the rapid identification of four types of blue crabs of Scylla, comprising the following steps: extracting the genome DNA of the four types of the blue crabs; (2) designing a species specific amplification primer according to the nucleotide sequence of the chondriosome (COI) gene of the four types of the blue crabs; (3) carrying out PCR amplification on the genome DNA of the four types of the blue crabs; and (4) utilizing 1.5% of agarose gel electrophoresis to detect PCR products, so as to rapidly identify the four types of the blue crabs according to the different sizes of the products. The method has simple and safe operation, clear amplification stripe and true and reliable results, and can rapidly identify the four types of the blue crabs, thus being widely used in the fileds of protection and management of blue crab germ plasm resources, inheritance breeding and the like.
Owner:EAST CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Sinilabeo rendahli DNA (deoxyribonucleic acid) bar code standard detection gene and application thereof
ActiveCN105483227AAccurate identificationIdentification is conducive to the realizationMicrobiological testing/measurementGenetic engineeringMolecular identificationNucleotide
The invention discloses a sinilabeo rendahli DNA (deoxyribonucleic acid) bar code standard detection gene and application thereof. The DNA bar code standard detection gene is of a COI gene of which a nucleotide sequence is SEQ ID NO.1. A sinilabeo rendahli DNA bar code species identification method disclosed by the invention adopts a reliable DNA molecular identification technology, and the sinilabeo rendahli is rapidly and accurately indentified. Compared with a traditional morphological identification method, the standard detection gene sequence obtained by the method is conducive to realizing the molecular identification of the sinilabeo rendahli, and can effectively shorten the identification time.
Owner:深圳华大海洋科技有限公司
Molecular biological variety identification method for Huangjiaji chicken
InactiveCN101974647AImprove accuracyShort identification timeMicrobiological testing/measurementNucleotideHaplotype
The invention discloses a molecular biological variety identification method for a Huangjiaji chicken, which comprises the following steps: accurately acquiring COI gene sequences of mitochondrion DNAs of a Huangjiaji chicken and a tested chicken based on gene cloning and DNA sequencing technologies, calculating variable sites, haplotypes and average nucleotide differences, nucleotide degrees of ramification and net genetic distances of the Huangjiaji chicken and the tested chicken by means of DnaSP4.10.7 software, constructing colony clustering charts and haplotype phylogenetic trees of the Huangjiaji chicken and the tested chicken, and determining whether the Huangjiaji chicken and the tested chicken are of the same variety according to the results of the constructed colony clustering charts and haplotype phylogenetic trees. By identifying whether the Huangjiaji chicken is true or false based on the precise software calculation and analysis results of the differences between varieties in genetic information, the invention has the characteristics of high accuracy, short identification time, low cost and the like.
Owner:MODERN NEW AGRI GROUP
Molecular marker for identifying coilia nasus and C. brachygnathus, and application of molecular marker
ActiveCN106282334AEasy to distinguishRapid Molecular Biology Identification TechnologyMicrobiological testing/measurementDNA/RNA fragmentationMolecular biology identification techniqueMolecular marker
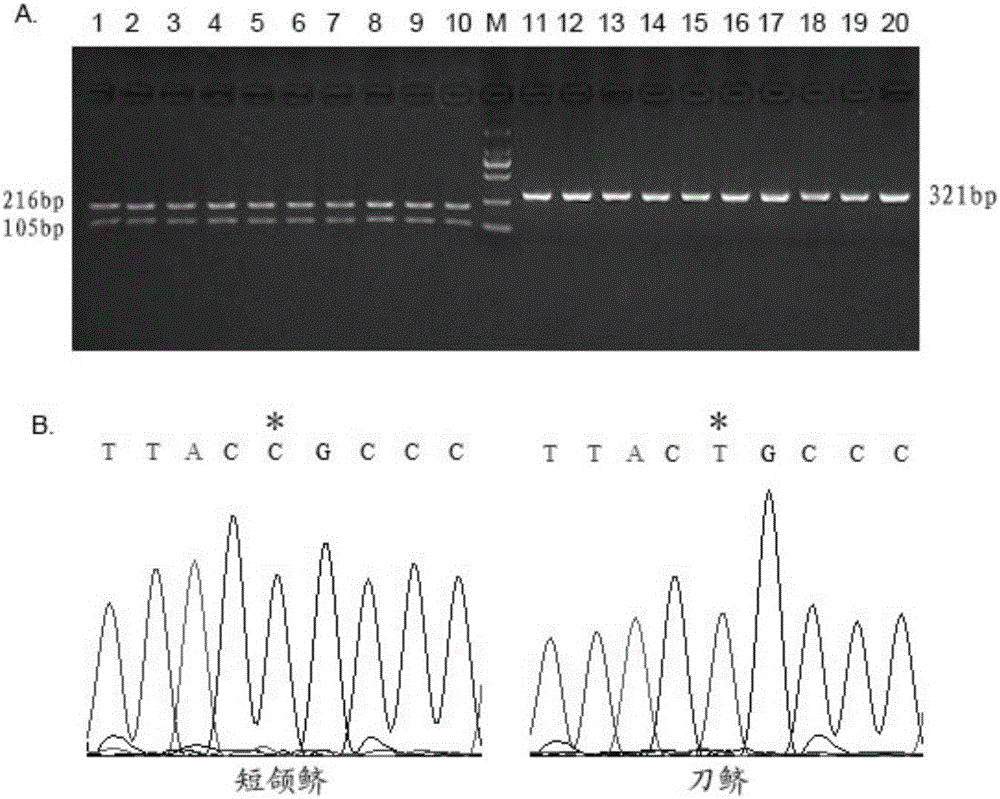
The invention discloses a molecular marker for identifying coilia nasus and C. brachygnathus, and application of the molecular marker. The molecular marker is positioned on a COI gene of a mitochondrion of the coilianasus or the C. brachygnathus, and is specifically positioned at the part of 236bp of the COI gene of the mitochondrion; base variation information shows that T is more than C; the variation site of the coilianasus is T, and the variation site of the C. brachygnathus is C. A method for identifying the coilianasus and the C. brachygnathus can be used to identify the coilianasus and the C. brachygnathus by detecting the molecular marker. The method overcomes the disadvantages of only using experiential observation to identify the coilianasus and the C. brachygnathus, and provides a rapid and reliable molecular biological identification technology for the accurate identification of the coilianasus and the C. brachygnathus.
Owner:FRESHWATER FISHERIES RES INSITUTE OF JIANGSUPROVINCE
Double PCR detection method for authenticity identification of cordyceps sinensis
ActiveCN107090502AStrong identification specificityImprove accuracyMicrobiological testing/measurementOphiocordyceps sinensisReaction system
The invention discloses a double PCR detection method for authenticity identification of cordyceps sinensis, and solves the problem that in the prior art, the identification of cordyceps sinensis is conducted only regarding fungi, and therefore the prior art is short of sufficient accuracy. According to the method, in the same PCR reaction system, ITS of cordyceps militaris and COI gene nucleic acid fragments which are hosts of cordyceps militaris are simultaneously detected, and an amplification primer group for identifying the COI gene nucleic acid fragments is COI-F: GGAAATCCHGGATCTTTAATT and COI-R: GATGCCCCMGARTGTGCAAT; an amplification primer group for identifying the ITS is GITS-F10': GTTGCCTCGGCGGGAC and CITS-R10'-2: CMTTTGCTTGCTTCTTGACTGAG. The double PCR detection method for the authenticity identification of cordyceps sinensis is convenient to operate, high in cordyceps sinensis identification specificity and accuracy, good in repeatability and suitable for popularization.
Owner:四川省农业科学院分析测试中心
Specific primers for amplifying plecoptera insect mitochondrion COI (cytochrome c oxidase I) gene
InactiveCN104946645AAccurate identificationImprove accuracyFermentationPlant genotype modificationMetapopulationGene
Owner:YANGZHOU UNIV
Method for screening tegillarca granosa mitochondria COI gene amplification primers
InactiveCN101942432AMeeting the needs of genetic diversity researchHigh amplification efficiencyDNA preparationBiotechnologyDesign software
The invention screens a pair of primers for amplification of a tegillarca granosa mitochondria COI sequence. The method comprises the following steps of: downloading the tegillarca granosa mitochondria COI sequence existing in the National Center of Biotechnology Information (NCBI); comparing and analyzing the tegillarca granosa mitochondria COI sequence by using BioEdit software; after determining a conserved domain, designing a plurality of pairs of primers by using primer design software Primer Premier 5; performing PCR reaction between the DNA of individual tegillarca granosa and the plurality of pairs of synthesized primers in a reaction system respectively; and finally, performing agarose gel electrophoresis detection to screen out a pair of primers COINF and COINR. Tests prove that the pair of primers has the advantages of single stripe and over 90-percent amplification efficiency, can meet the requirements of study on the genetic diversity of the tegillarca granosa, and plays an important role in the protection and utilization of tegillarca granosa resources, diversity estimation, genetic breeding and the like.
Owner:OCEAN UNIV OF CHINA
Molecular biological method for rapidly identifying Hoolock leuconedys and Nomascus leucogenys
InactiveCN106636319AAccurate identificationImprove case handling efficiencyMicrobiological testing/measurementMolecular identificationFeces
The invention discloses a molecular biological method for rapidly identifying Hoolock leuconedys and Nomascus leucogenys and belongs to the field of molecular biology. Genomic DNA is separated and extracted from to-be-detected objects and serves as a template; PCR is performed on the basis of a P1 primer pair and a P2 primer pair, and fragments A and B are obtained; a COI (cytochrome C oxidase I) gene complete sequence is obtained from the fragments A and B by splicing and cutting; species identification is performed according to the nucleotide sequence feature variation sites, disclosed for the first time, of the Hoolock leuconedys and the Nomascus leucogenys. Rapid and accurate molecular identification of the Hoolock leuconedys and the Nomascus leucogenys is realized for the first time, and various defects of the traditional morphological identification method are overcome. Identification objects include venous blood, feces, hair or urine samples of animals, and the method has the characteristics of being accurate, rapid and convenient, and has important function and significance for maintaining an ecosystem and protecting biodiversity.
Owner:SOUTHWEST FORESTRY UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com