Double PCR detection method for authenticity identification of cordyceps sinensis
A technology for authenticity identification of Cordyceps sinensis, applied in the biological field, achieves the effect of simple method, suitable for promotion, and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
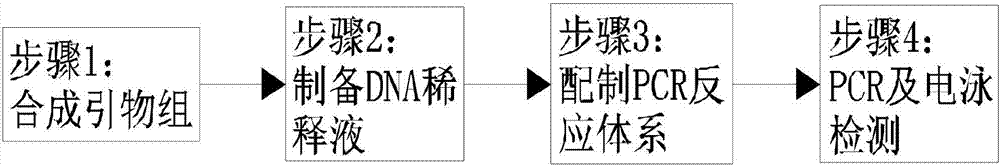
[0054] Cordyceps sinensis double PCR identification method, in the same PCR reaction system to simultaneously detect the ITS of Cordyceps fungus and the COI gene nucleic acid fragment of the Cordyceps fungus host, the steps are as follows:
[0055] Step 1: Synthesize an amplification primer set for identifying ITS, the amplification primer set is:
[0056] CITS-F10': GTTGCCTCGGCGGGAC
[0057] CITS-R10'-2: CMTTTGCTTGCTTCTTGACTGAG.
[0058] Synthesis is used to identify the amplification primer group of COI gene nucleic acid fragment, and this amplification primer group is:
[0059] COI-F: GGAAATCCHGGATCTTTAATT
[0060] COI-R: GATGCCCCMGARTGTGCAAT.
[0061] The concentration of both groups of primers was 10 μmol / l.
[0062] Step 2: Prepare DNA dilution;
[0063] DNA diluent of the sample to be tested: extract DNA from the sample to be tested and dilute to 50ng / μl DNA diluent;
[0064] Positive control substance DNA diluent: extract DNA from Cordyceps sinensis positive cont...
Embodiment 2
[0080] specificity experiment. The experimental method steps are as follows:
[0081] Step 1: same as embodiment 1
[0082] Step 2: Prepare DNA dilution;
[0083] DNA diluent of the sample to be tested: extract DNA from the sample to be tested, and dilute it to 50ng / μl DNA diluent; the samples to be tested are Cordyceps sinensis, bat moth larvae, Cordyceps mycelium, Liangshan Cordyceps, Yaxiang stick Cordyceps, Xinjiang Cordyceps, Cordyceps militaris (Cordyceps militaris), molded starch, corn, rice, soybeans, rapeseed, chicken, pigs.
[0084] Positive control substance DNA diluent and blank control are the same as in Example 1.
[0085] Step 3, step 4 are the same as embodiment 1.
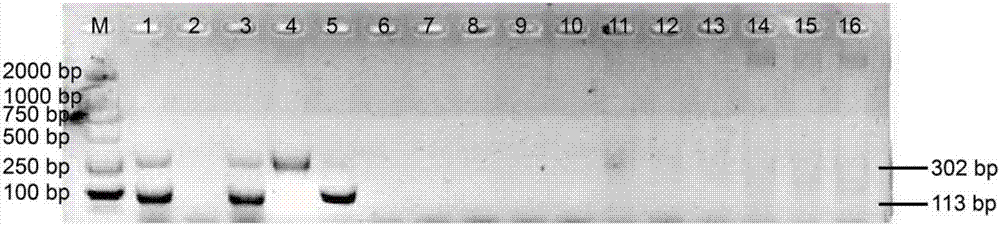
[0086] The results of the amplification are attached figure 2 shown. Only two target bands (lanes 1 and 3) were amplified in the Cordyceps positive control substance and samples of Cordyceps sinensis, and there were no target bands in the DNA of the blank control, counterfeit Cordyceps sinen...
Embodiment 3
[0088] Repeatable experiments.
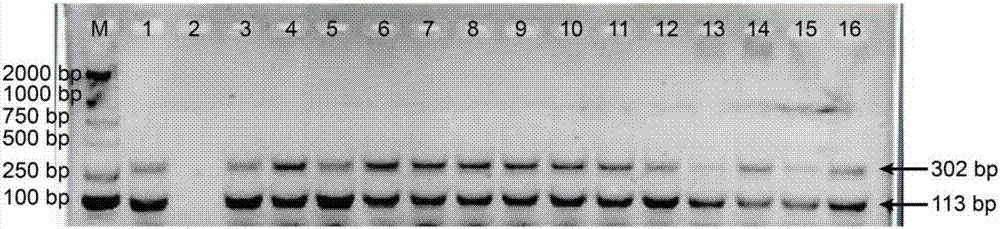
[0089] The method of Example 1 was used to detect the positive control sample of Cordyceps sinensis (the standard substance of Cordyceps sinensis purchased from China Institute for Food and Drug Control) and the samples of Cordyceps sinensis collected from Qinghai, Tibet, Sichuan and other places. The results are attached image 3 shown. All Cordyceps detection samples could amplify two (113bp and 302bp) target bands (lanes 1, 3-16), but no bands were amplified in the blank control (lane 2). And the result can be reproduced many times, which is repeatable. Therefore, the present invention is stable and reliable in distinguishing the authenticity of Cordyceps sinensis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com