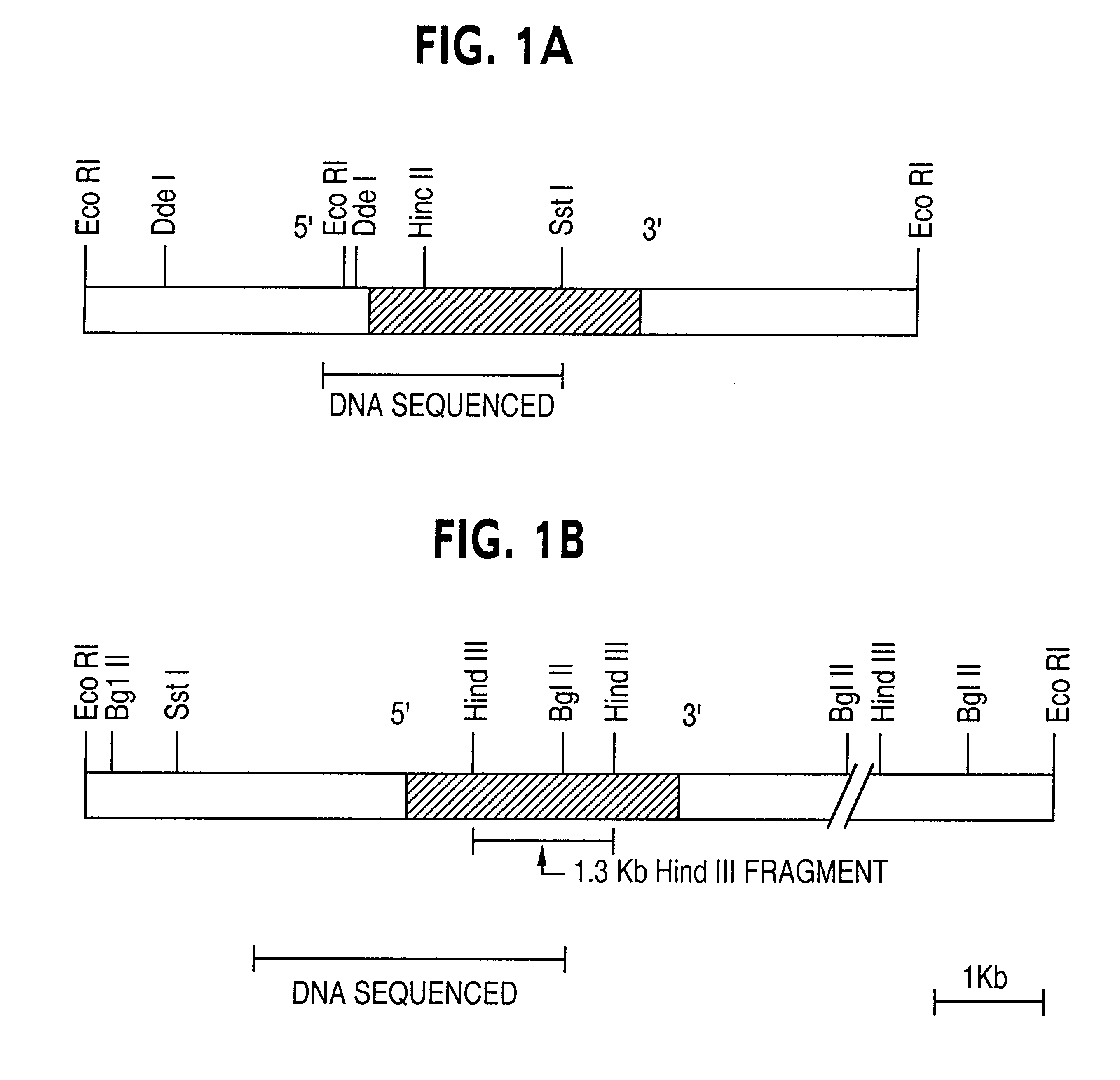
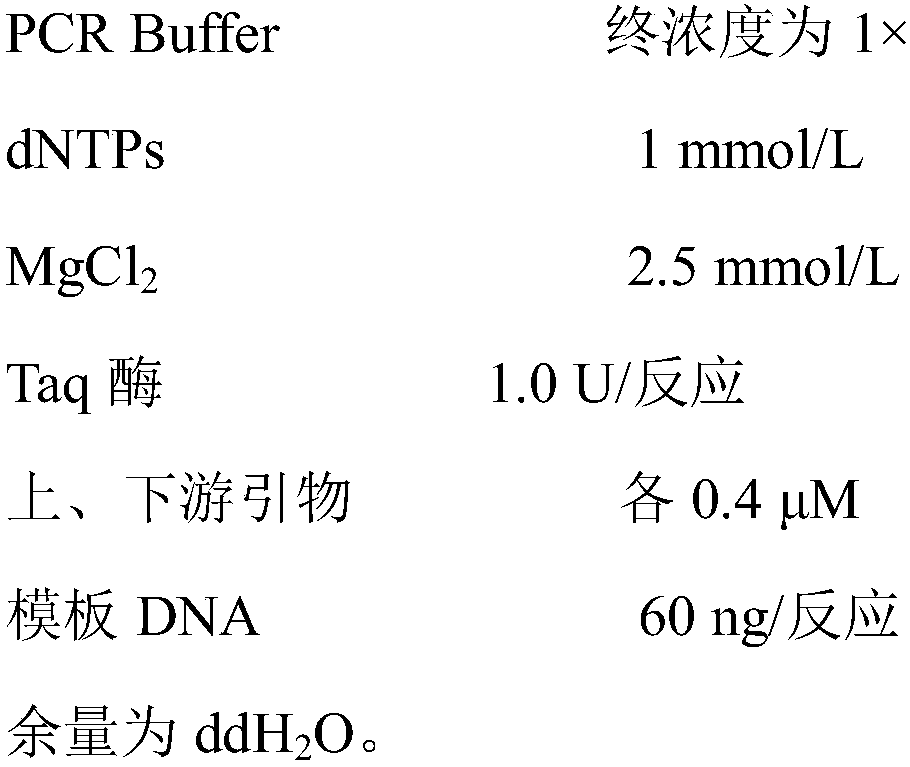
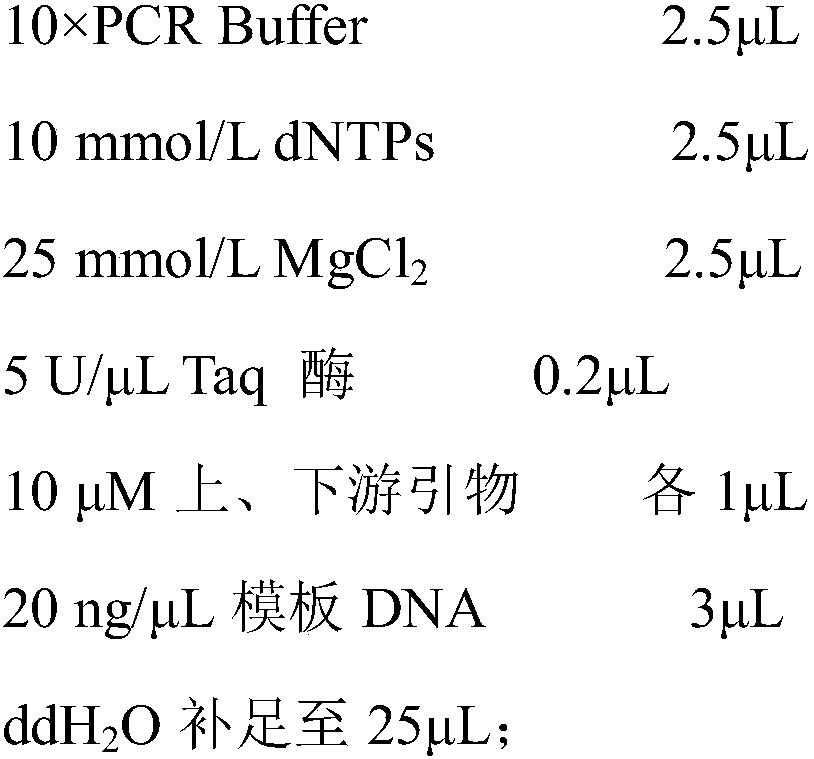
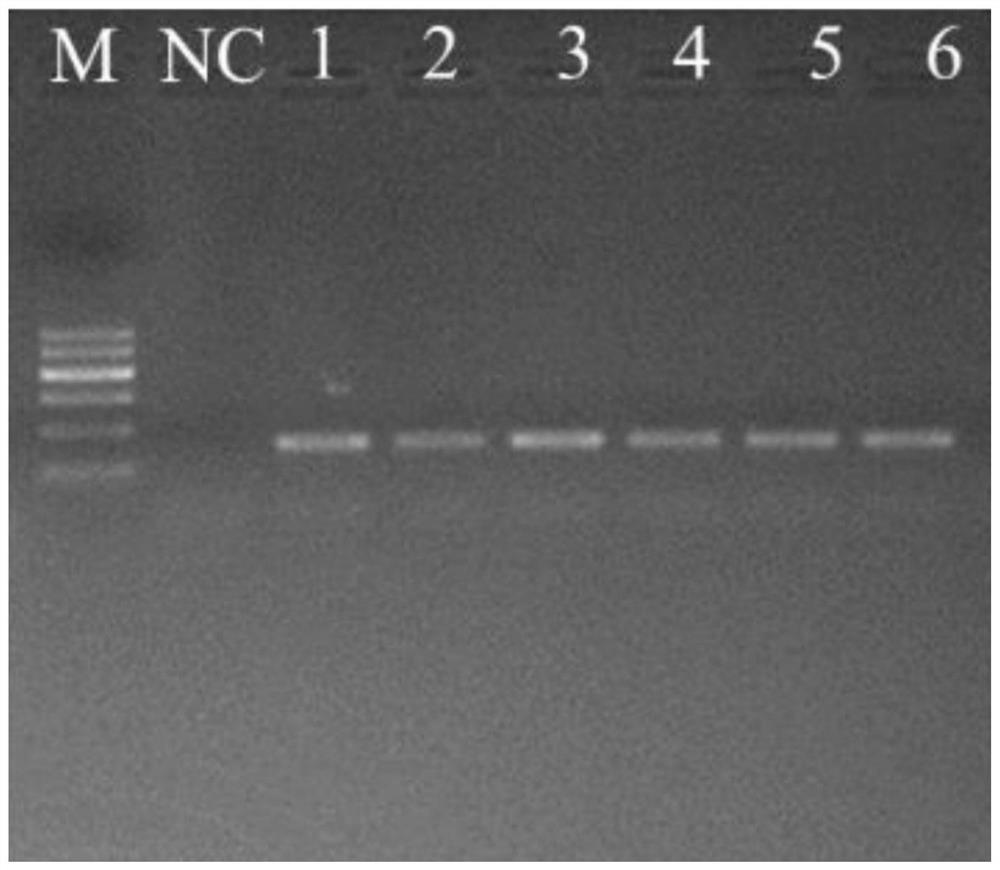
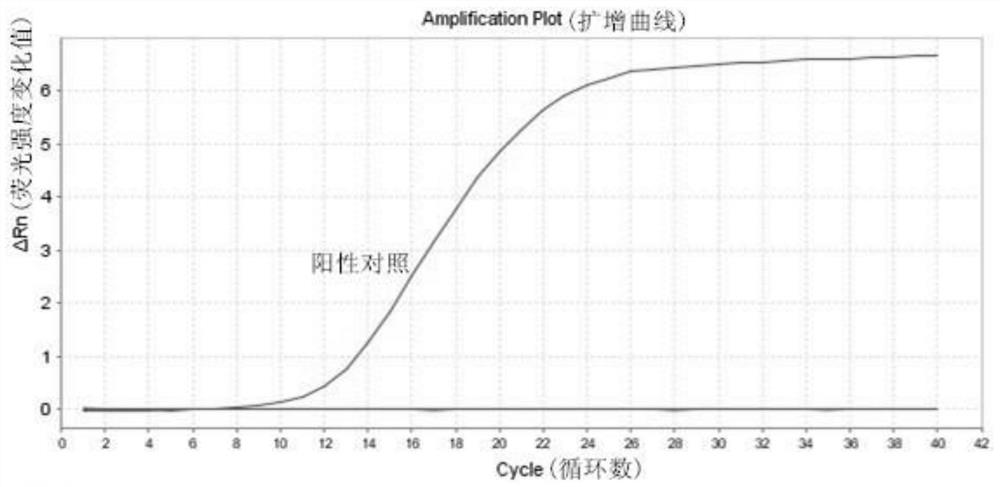
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
56 results about "Molecular Technique" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Antisense gene systems of pollination control for hybrid seed production
A process is described for producing hybrid seed using male-sterile plants created by employing molecular techniques to manipulate antisense DNA and other genes that are capable of controlling the production of fertile pollen in plants. Transformation techniques are used to introduce constructs containing antisense DNA and other genes into plants. Said plants are functionally male-sterile and are useful for the production of hybrid seed by the crossing of said male-sterile plants with pollen from male-fertile plants. Hybrid seed production is simplified and improved by this invention and can be extended to plant crop species for which commercially acceptable hybrid seed production methods are not currently available.
Owner:PIONEER HI BRED INT INC
Feature sequences, tag primer and identification method of carya illinoensis variety Nacono
ActiveCN107557369ALower requirementImprove reliabilityMicrobiological testing/measurementFermentationForward primerMolecular Technique
The invention relates to feature sequences and a molecular specificity tag primer of a pair of high-specificity feature sequences of carya illinoensis variety Nacono, a molecular specificity tag primer, and a method capable of being used for fast identifying the carya illinoensis variety Nacono. The molecular specificity tag primer has the sequence that a forward primer is 5'-GTCACTTCCCTTTCTCGGCA-3', and a reverse primer is 5'-CCCCGGGTGCAGGTATAAAA-3'. The molecular specificity tag primer provided by the invention can be used for performing fast early-stage identification on the carya illinoensis variety Nacono; the method is simple, fast and accurate, and belongs to an irreplaceable molecular technique for distinguishing the carya illinoensis variety through appearance features.
Owner:ZHEJIANG FORESTRY ACAD
Feature sequences, tag primer and identification method of carya illinoensis variety Van Deman
ActiveCN107557434ALower requirementImprove reliabilityMicrobiological testing/measurementDNA/RNA fragmentationForward primerGenetics
The invention relates to feature sequences and a molecular specificity tag primer of a pair of high-specificity feature sequences of carya illinoensis variety Van Deman, a molecular specificity tag primer, and a method capable of being used for fast identifying the carya illinoensis variety Van Deman. The molecular specificity tag primer has the sequence that a forward primer is 5'-ATGCAAGACGCTTGTGGAGA-3', and a reverse primer is 5'-CCGACCCGACCCGATAAAAT-3'. The molecular specificity tag primer provided by the invention can be used for performing fast early-stage identification on the carya illinoensis variety Van Deman; the method is simple, fast and accurate, and belongs to an irreplaceable molecular technique for distinguishing the carya illinoensis variety through appearance features.
Owner:ZHEJIANG FORESTRY ACAD
Method for identifying structure of yeast colony of Daqu starter or fermented grain of distilled spirit by using denaturing gradient electrophoresis
InactiveCN101570786AMicrobiological testing/measurementMaterial analysis by electric/magnetic meansBiotechnologyMolecular Technique
The invention relates a method for identifying a structure of a yeast colony of a Daqu starter or a fermented grain of distilled spirit by using denaturing gradient electrophoresis, which belongs to the field of microbial ecological techniques. The method comprises the following steps: extracting a genome DNA of the Daqu starter or fermented grain sample of the distilled spirit by using a bead milling method; by designing a general DGGE primer of yeasts in a common wine-making microbe, performing PCR amplification on a specific specificity DNA segment in a yeast ribosome; separating corresponding DNA segments of different varieties by DGGE electrophoresis; gel-cutting and recovering straps corresponding to preponderant microbes; performing the PRC application again, connecting a T carrierand selecting a positive clone; and after re-comparison by the DGGE electrophoresis, checking orders and identifying microbe information corresponding to gel-cutting straps. The method adopts a molecular technique without depending on cultivation, and has the characteristics of simplicity, convenience, quick speed, sensitivity and the like; besides, the method provides a detection method for identifying the structure of the yeast colony of the Daqu stater or the fermented grain of the distilled spirit, and further provides a reference frame for developing wine-brewing microbes oriented to distilled spirit flavors with different odor types.
Owner:JIANGNAN UNIV
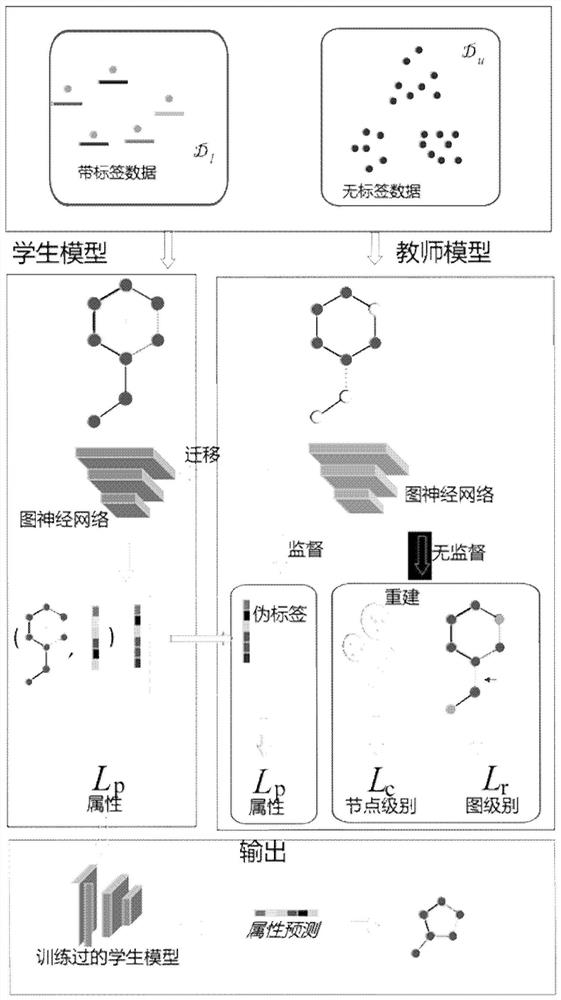
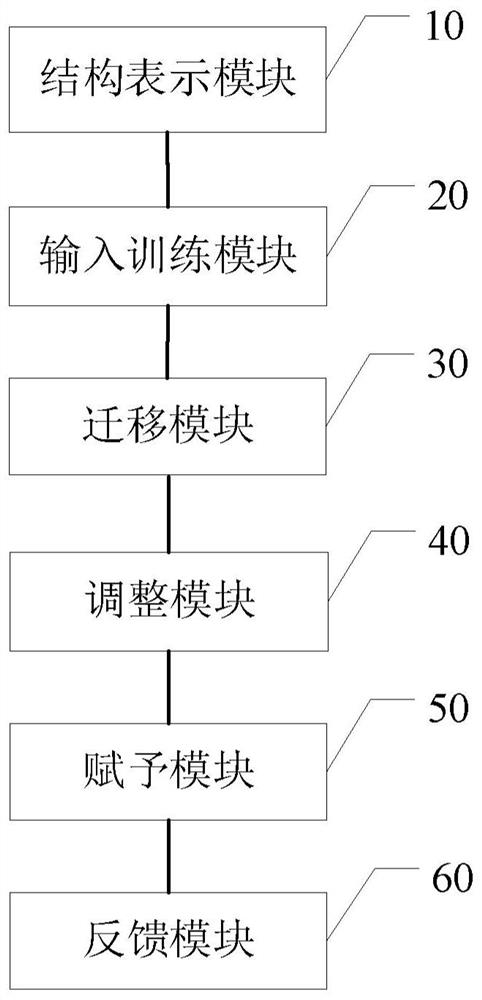
Molecular attribute determination method and device, electronic equipment and storage medium
ActiveCN111724867ASame prediction accuracyChemical property predictionChemical data visualisationAlgorithmTheoretical computer science
The invention discloses a molecular attribute determination method and device, electronic equipment and a storage medium, belonging to the technical field of molecules. The method comprise the following steps: S1, representing molecular attribute data of each labeled molecule and molecular structure data of each unlabeled molecule by using molecular topological structure diagrams; S2, inputting the molecular topological structure diagrams of all the labeled and unlabeled molecules into a pre-constructed teacher model, and training all the molecular topological structure diagrams by utilizing semi-supervised learning to obtain parameters of a teacher model; S3, migrating the parameters of the teacher model to a pre-constructed student model; S4, adjusting the student model by using labeledmolecules; S5, using the adjusted student model to assign labels to all the unlabeled molecules to obtain a labeled molecule set; S6, feeding back the labeled molecular set to the teacher model; and S7, repeatedly executing the steps S2 to S6 until the teacher model and the student model are both converged, and predicting molecular attributes by utilizing the converged student model. According tothe invention, the attributes of unknown molecules can be accurately predicted.
Owner:UNIV OF SCI & TECH OF CHINA
Method and a kit for determination of a microbial count
InactiveUS20050170346A1Rapid and reliable resultFast resultsMicrobiological testing/measurementAgainst vector-borne diseasesMicroorganismAssay
The invention relates to the field of estimating a microbial count in a sample using molecular techniques. The invention furthermore relates to a kit for performing such estimation and use of the estimation method for providing bacterial cell counts in milk and for diagnosing infectious conditions in animals and human beings. Furthermore, the invention relates to a method for quality control of food (especially milk and dairy products), feed and water samples using molecular techniques. Reproducible and reliable estimates of a microbial count can be obtained using a molecular hybridisation assay between a target nucleic acid sequence and a labelled probe. Proper calibration and quantitative recording of the signal produced by the label can estimate a microbial count. One important feature of the invention is the use of locked nucleic acid (LNA) monomers in the probes.
Owner:QUANTIBACT AS
Method for the identification by molecular techniques of genetic variants that encode no d antigen (d-) and altered c antigen (c+w)
InactiveUS20120172239A1Considerable efficiencySugar derivativesNucleotide librariesAntigenNucleotide
The invention relates to genotyping and blood cell antigen determination. In particular, the invention addresses discriminating the RHD*DIIIa-CE(4-7)-D or RHD*DIIIa-CE(4-7)-D)-like blood type variants, from RHD*DIIIa, RHD*DIVa-2 and other blood type variants. The invention provides methods for genotyping a subject, comprising:a) determining at least 4 markers in a sample that has been obtained from the subject, wherein the markers comprise:(i) the presence or absence of an RHCE*C allele;(ii) the presence or absence of an RHD / RHCE hybrid exon 3 (RHD / CE Hex03) allele;(iii) the absence of, or a single nucleotide polymorphism (SNP) variant within, any one of position 602 of exon 4, position 667 of exon 5, or position 819 of exon 6 of RHD; and(iv) the absence of, or SNP variant within, position 1048 of RHD exon 7. The invention also provides probes, primers and kits for use in such methods.
Owner:PROGENIKA BIOPHARMA SA
Cockle shellfish mitochondria COI (cytochrome c oxidase I) gene amplification primer
ActiveCN102827839AConducive to the study of genetic diversityEfficient amplificationDNA/RNA fragmentationResource protectionGenetic diversity
The invention uses a universal primer for amplification to obtain 66 cardiidae mtCOI (cytochrome c oxidase I) sequences, uses a CLUSTALW software to compare and analyze, uses primer design software primer Premier5 to design 5 pairs of primers in a determined area, uses 216 cardiidae individuals to repeatedly conduct FCR reaction tests in a 10mu m system, finally is detected through agarose gel electrophoresis, and screens out ctockle shellfish mitochondria COI gene sequence efficient amplification primer. The cockle shellfish mitochondria COI gene amplification primer is characterized in that NGF5'-ACAAATCATAAAGATATTGG-3'NGR5'-TTCAGGGTGTCCGAAGAATCA-3' adopts the cockle shellfish mitochondria COI primer, objective sections of different species of cockle shellfish are effectively amplified, requirements of genetic diversity survey of cockles can be met, simultaneously reliable molecular technique instruments for germplasm resource protection and breeding industry development are provided.
Owner:OCEAN UNIV OF CHINA
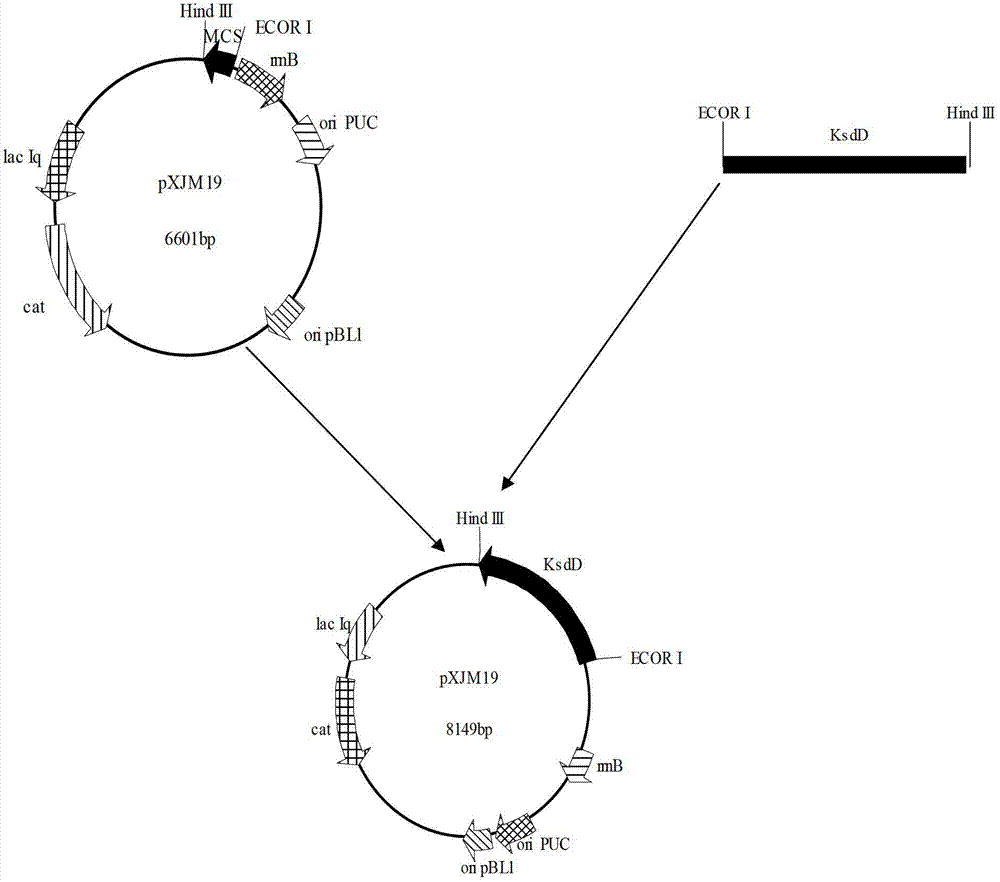
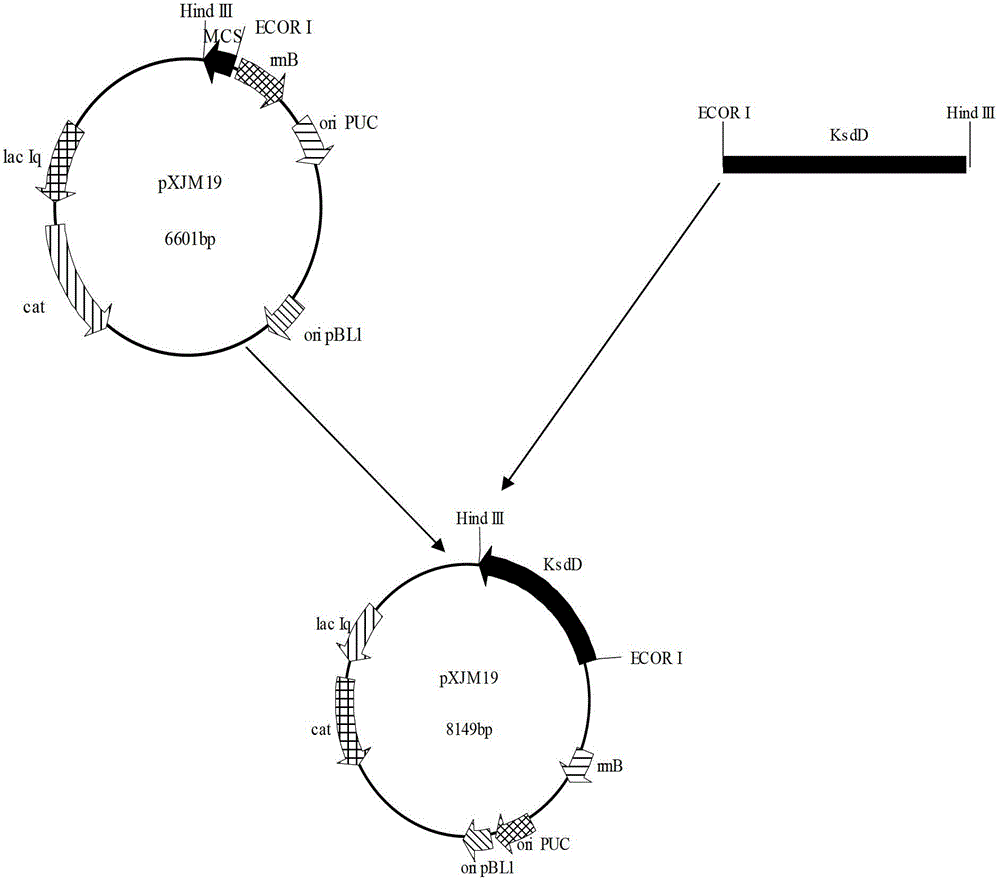
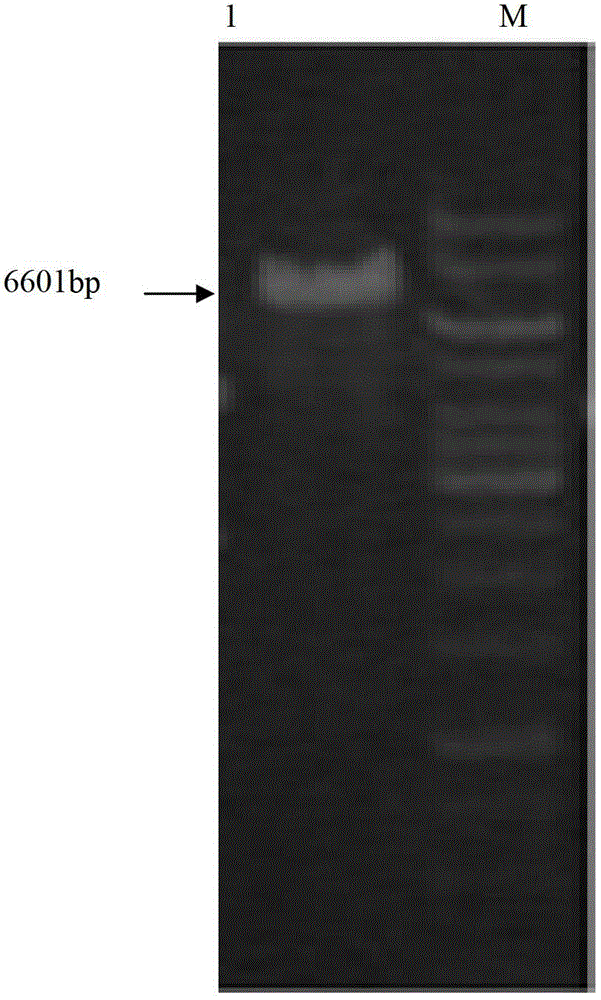
Constructing method and application of plasmid capable of being copied in arthrobacter simplex and expressing steroid compound A ring 1, 2 dehydrogenase genes
InactiveCN103205450AReduce manufacturing costHigh copy numberBacteriaMicroorganism based processesArthrobacter simplexMolecular Technique
The invention relates to application of plasmid capable of being copied in arthrobacter simplex and expressing steroid compound A ring 1, 2 dehydrogenase genes to constructing high-efficiency expression steroid compound A ring 1, 2 dehydrogenation arthrobacter simplex engineering bacteria. The plasmid is capable of being copied in arthrobacter simplex and efficiently expressing steroid compound A ring 1, 2 dehydrogenase genes and significantly practical, and basis is provided for further using plasmid pXJM19 to modify arthrobacter simplex through molecular techniques.
Owner:TIANJIN UNIV OF SCI & TECH
Nymphaea tetragona DNA fingerprint, primer to acquire same and construction method of Nymphaea tetragona DNA fingerprint
PendingCN110129472AHigh polymorphismGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationFingerprintMolecular mapping
The invention relates to Nymphaea tetragona DNA fingerprint, a primer to acquire the same and a construction method of the Nymphaea tetragona DNA fingerprint and belongs to the technical field of biology. The construction method of the Nymphaea tetragona DNA fingerprint comprises the specific steps of 1) extracting Nymphaea tetragona DNA; 2) performing ISSR-PCR (inter-simple sequence repeat-polymerase chain reaction) amplification reaction and electrophoresis detection, to be specific, amplifying Nymphaea tetragona DNA with an ISSR primer, and electrophoretically detecting the ISSR-PCR productwith 1.8% agarose gel, wherein the ISSR primer is from one or more of UBC840, UBC841, UBC811, UBC834, UBC835, UBC843, UBC844, UBC845, UBC873, and UBC880; 3) constructing DNA fingerprint. A stripe amplified with a Nymphaea tetragona ISSR-PCR system established herein has good repeatability and high polymorphism; the defects of domestic studies on Nymphaea tetragona germplasm resource genetic diversity and molecular mapping are overcome. The Nymphaea tetragona DNA fingerprint constructed via the method herein can act as basis to identify and classify corresponding primary germplasm resources ofNymphaea tetragona, and also guarantee the classification and identification of new germplasms and new varieties and the protection of intellectual properties at molecular technical level.
Owner:FLOWER RES INST GUANGXI ACADEMY OF AGRI SCI
Method for constructing yeast expression vector and preparing cellobiohydrolase by yeast expression vector and application thereof
The present invention discloses a method for constructing a yeast expression vector and preparing cellobiohydrolase by the yeast expression vector and an application thereof. A modern molecular technology successfully optimizes a gene sequence encoding the shii-take cellobiohydrolase, constructs the expression vector pPICZ alpha A-Cel7A of the cellobiohydrolase by expressed by yeasts in vitro, amdthe vector and pichia pastoris are utilized to prepare the cellobiohydrolase and thus provide technical supports for an application of the cellobiohydrolase in production. The recombinant pichia pastoris obtained by the method is high in enzyme yield, high in activity and excellent in effects.
Owner:JIANGXI AGRICULTURAL UNIVERSITY
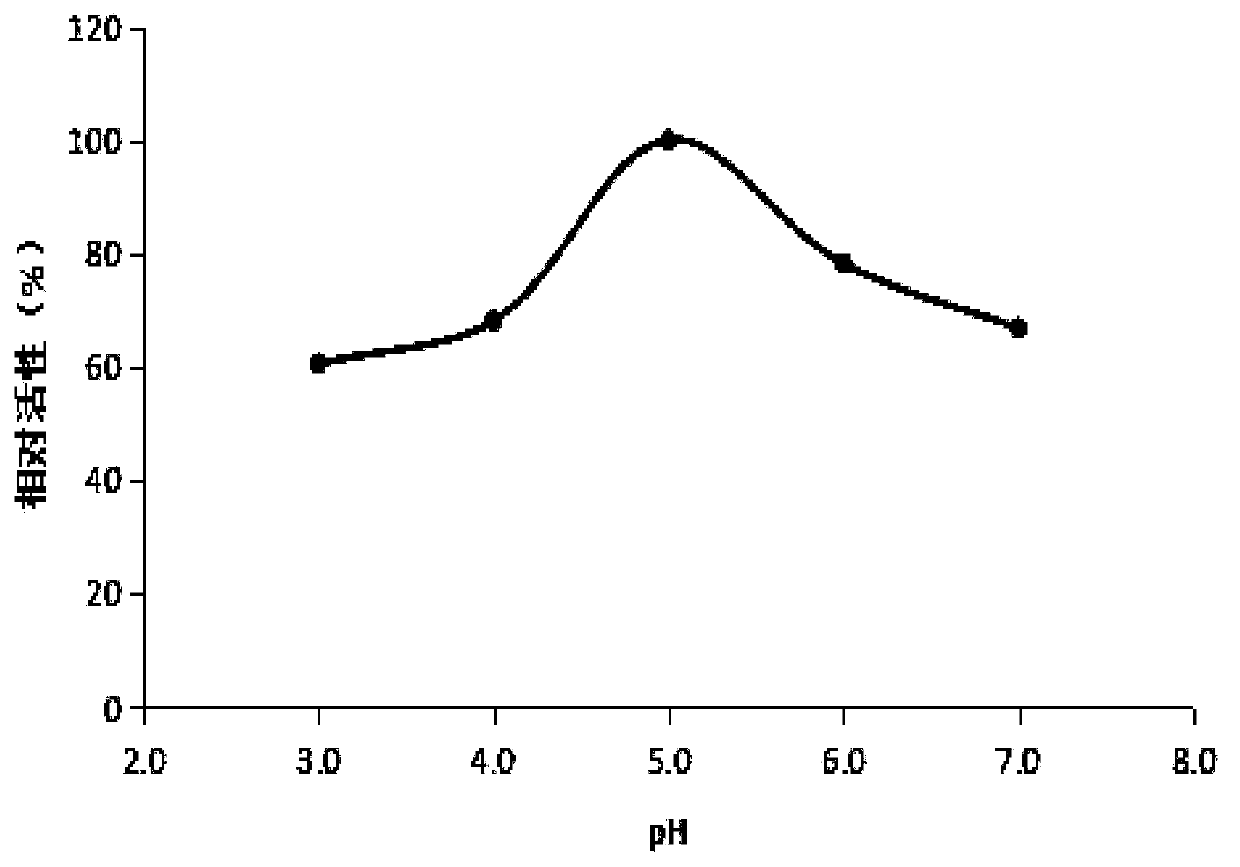
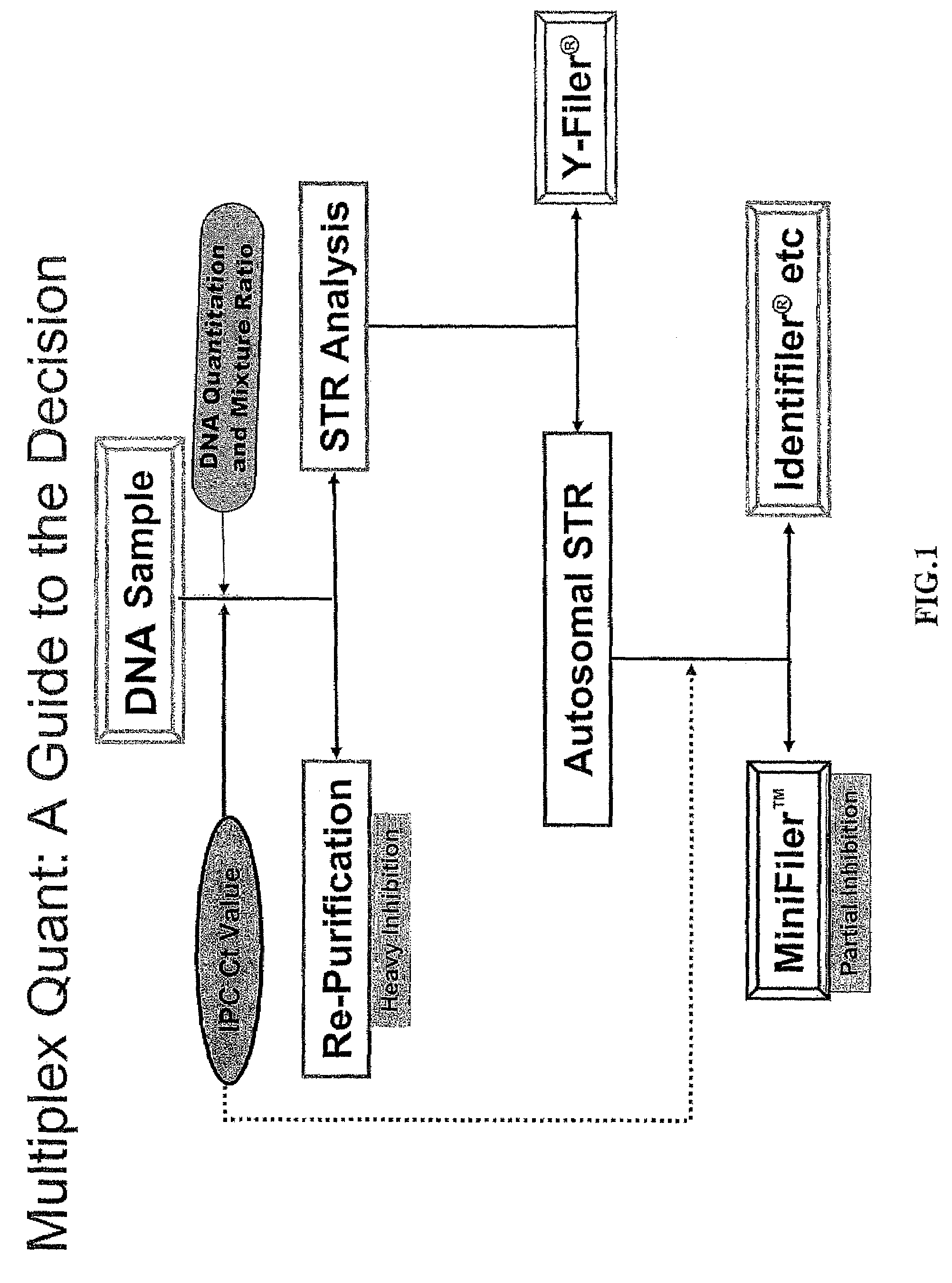
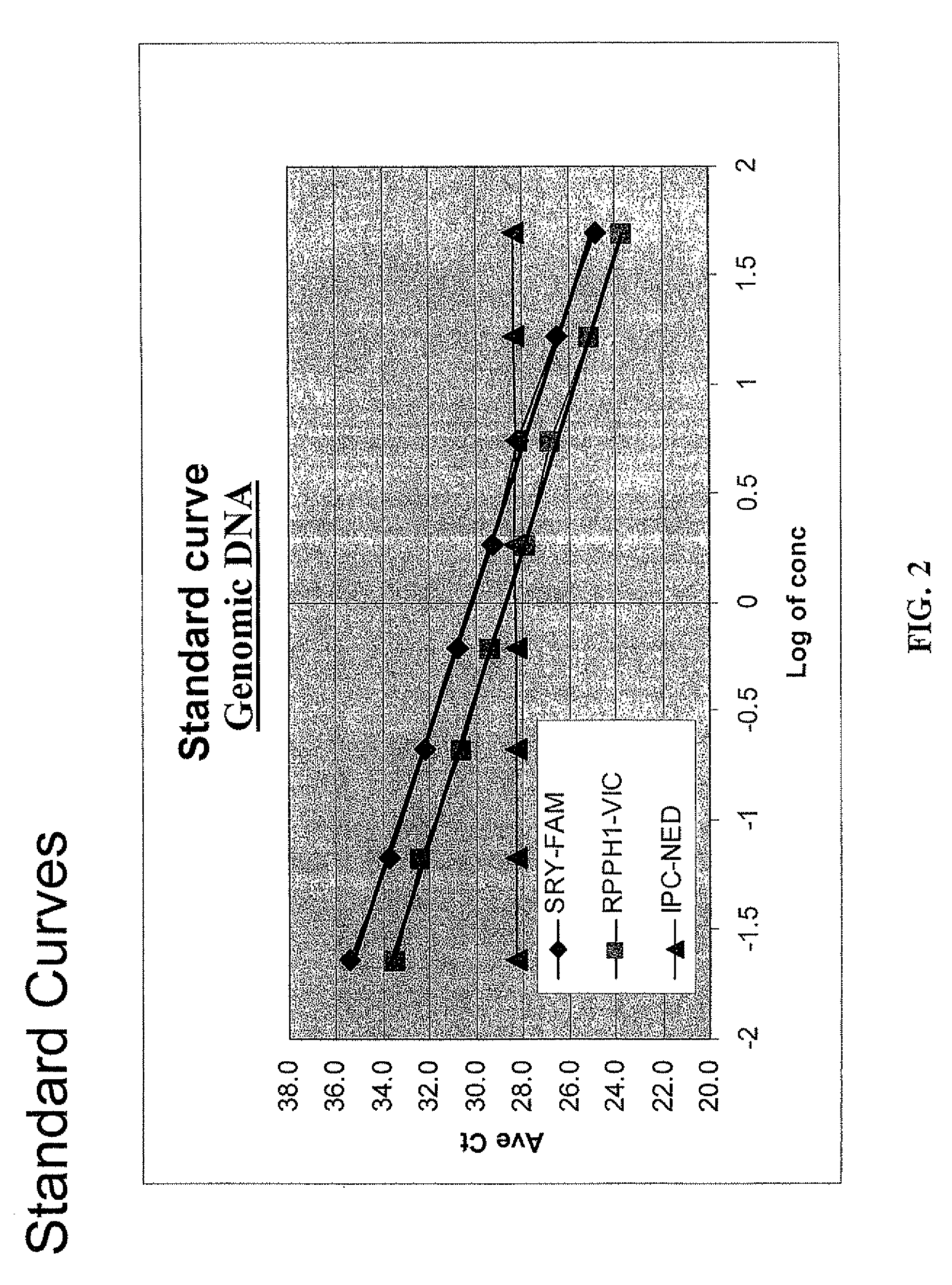
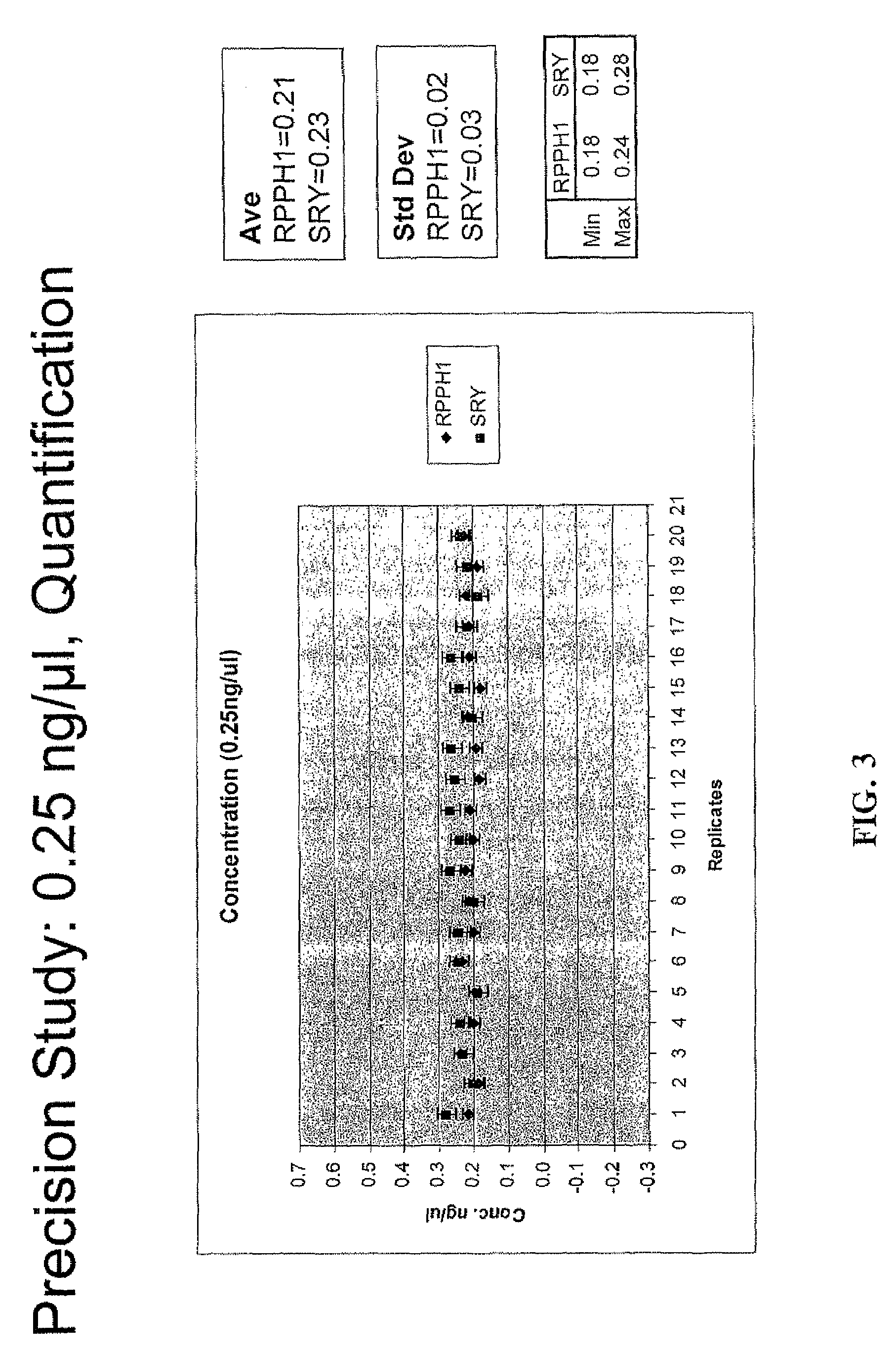
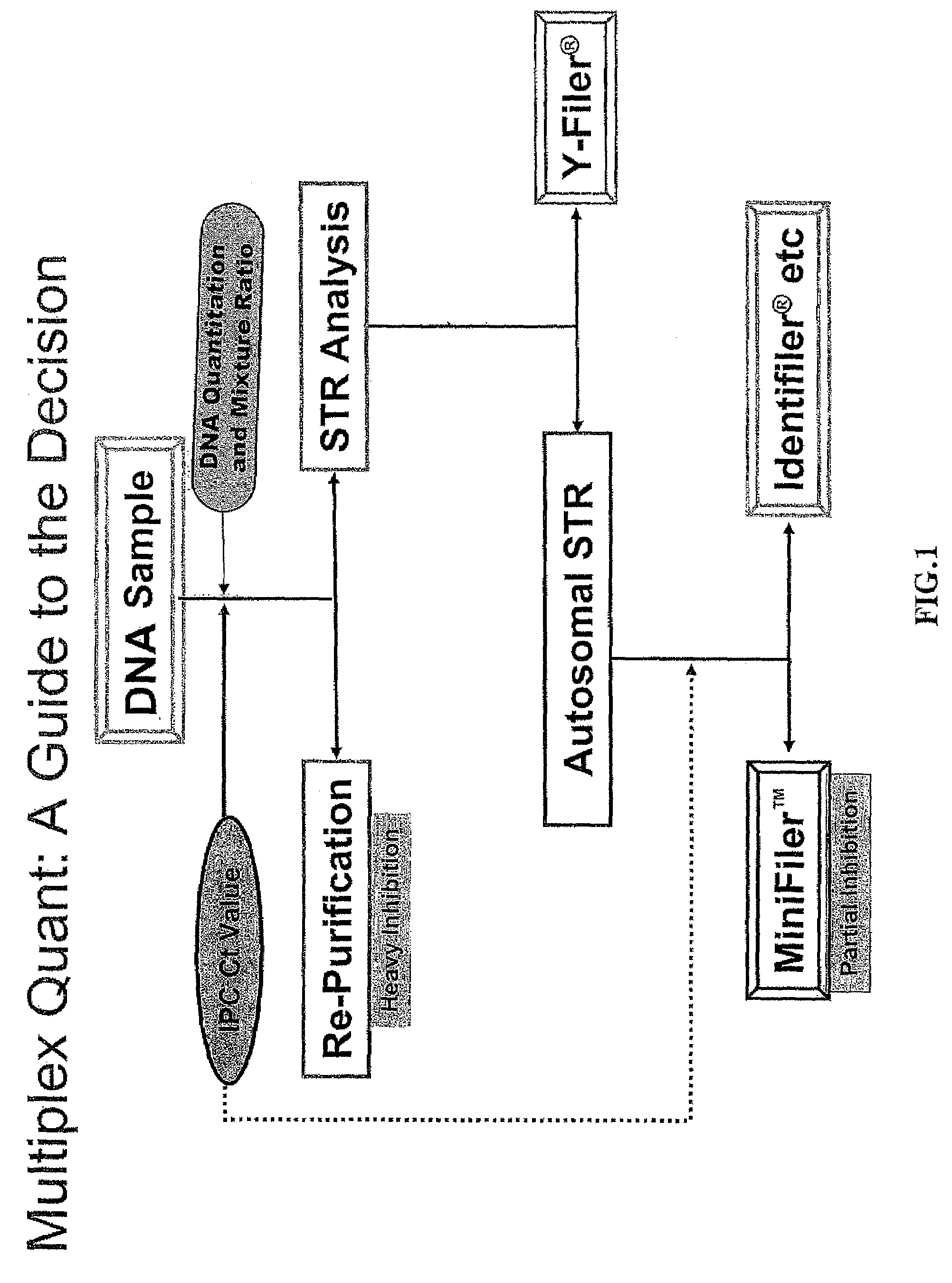
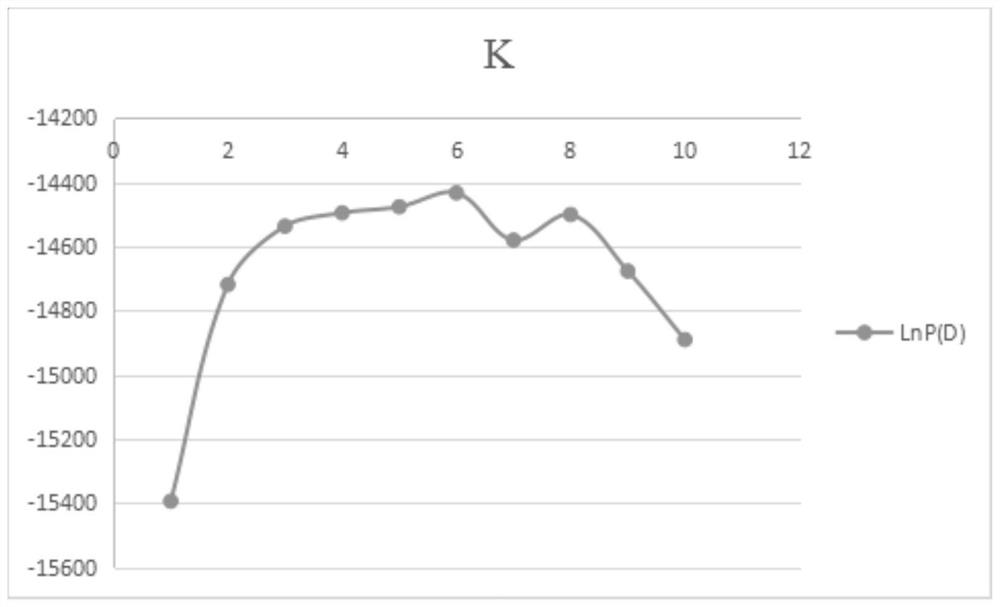
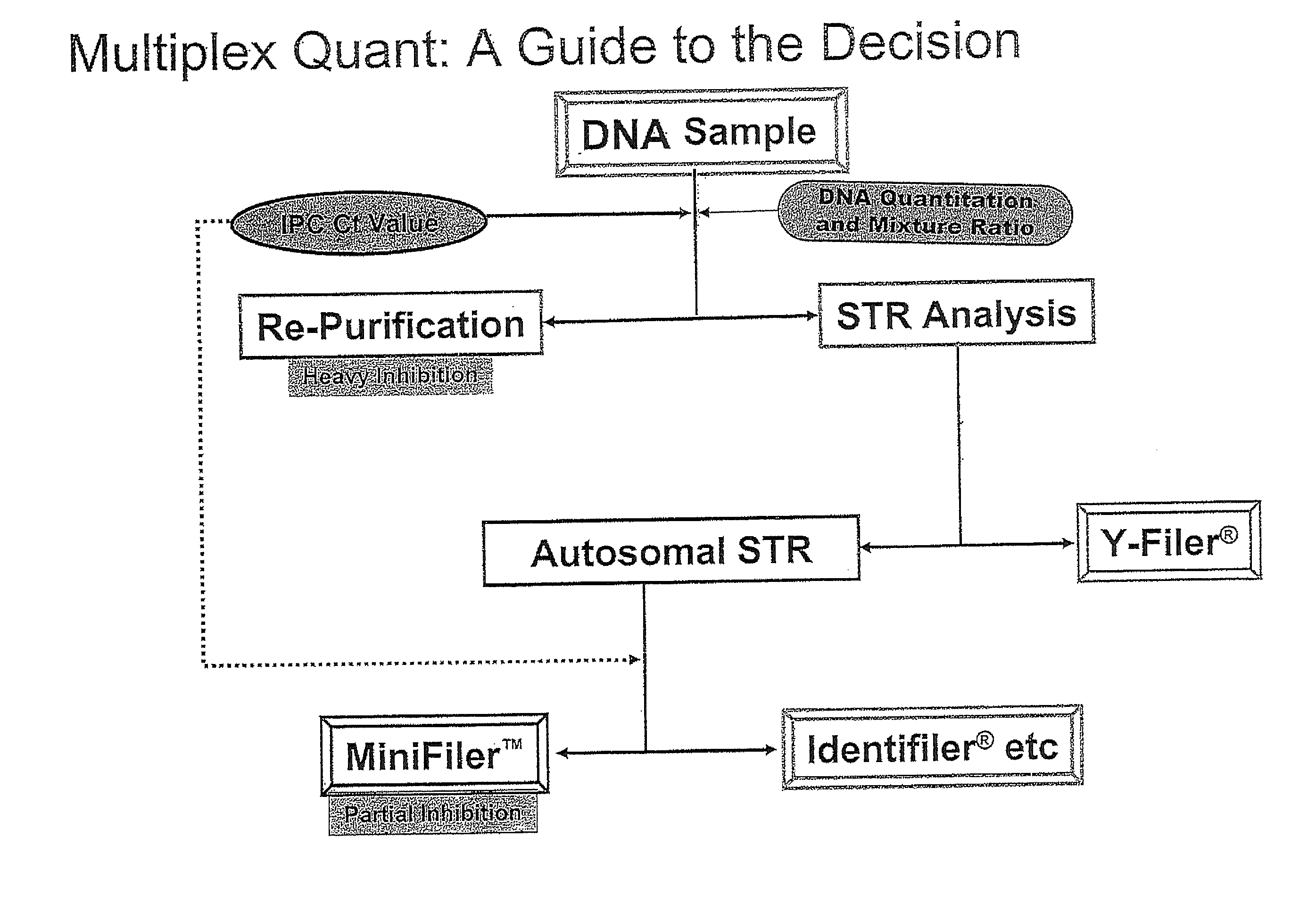
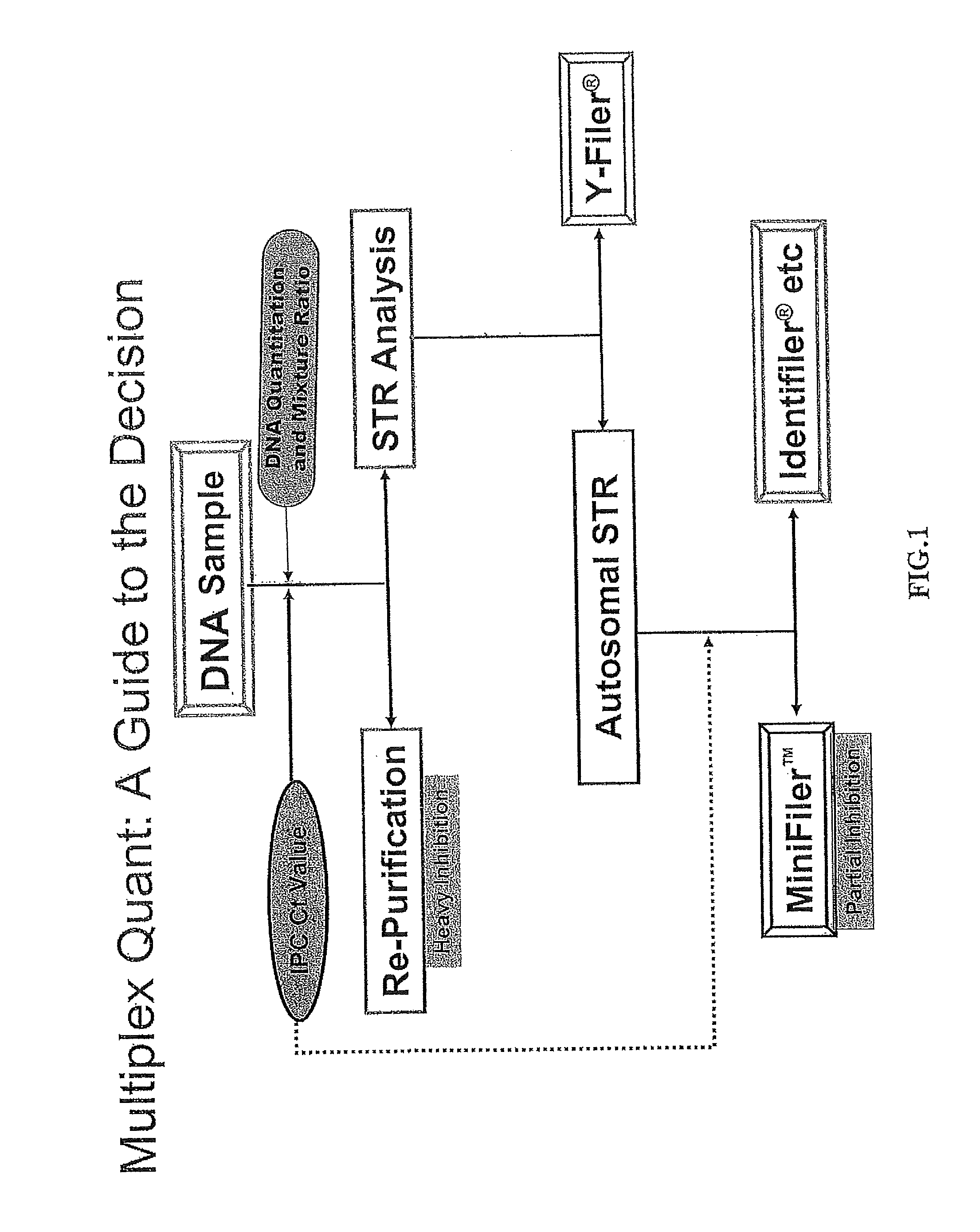
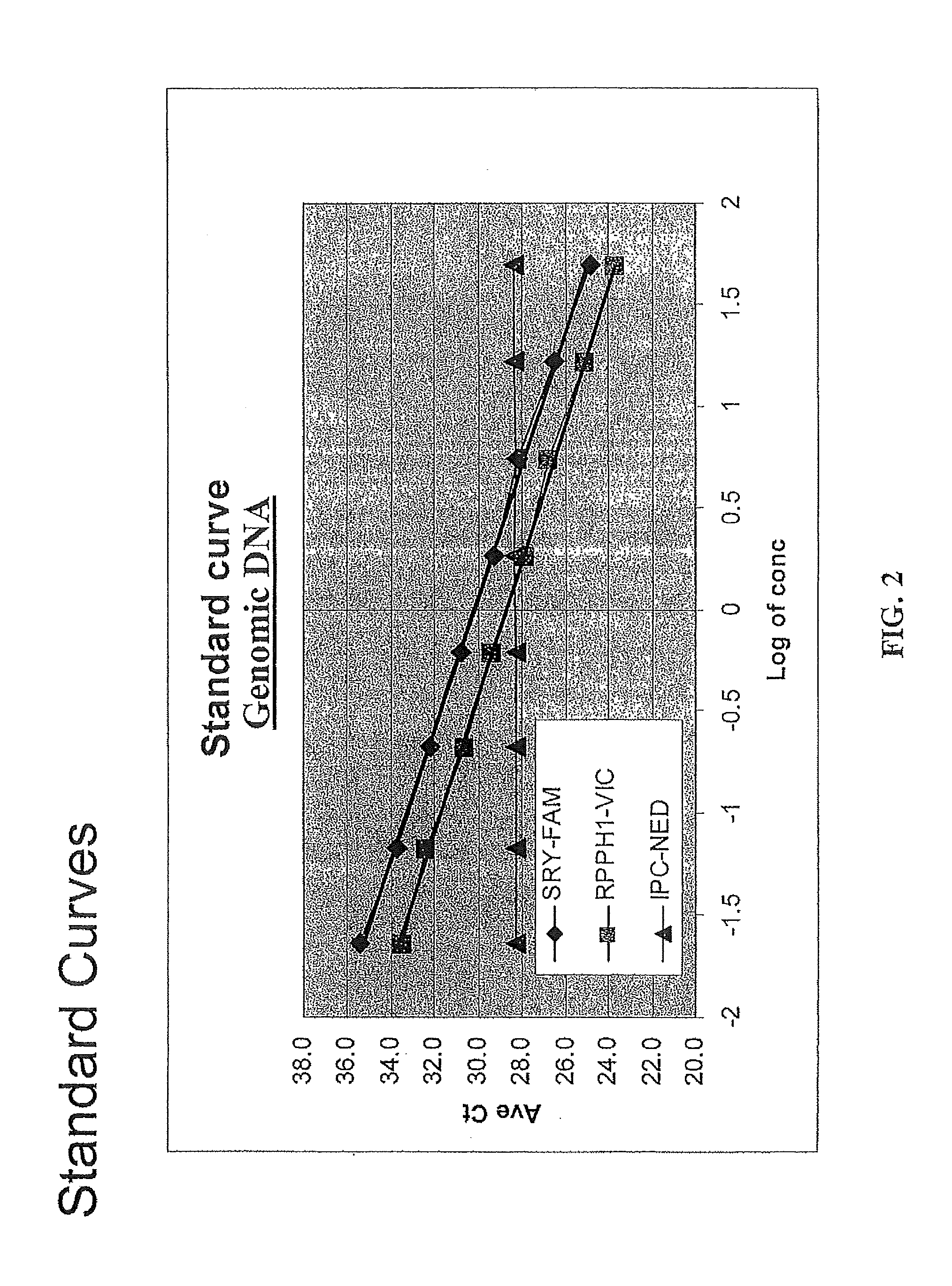
Multiplex compositions and methods for quantification of human nuclear DNA and human male DNA and detection of PCR inhibitors
ActiveUS8012691B2Sugar derivativesMicrobiological testing/measurementMolecular TechniqueQuantification methods
Owner:APPL BIOSYSTEMS INC
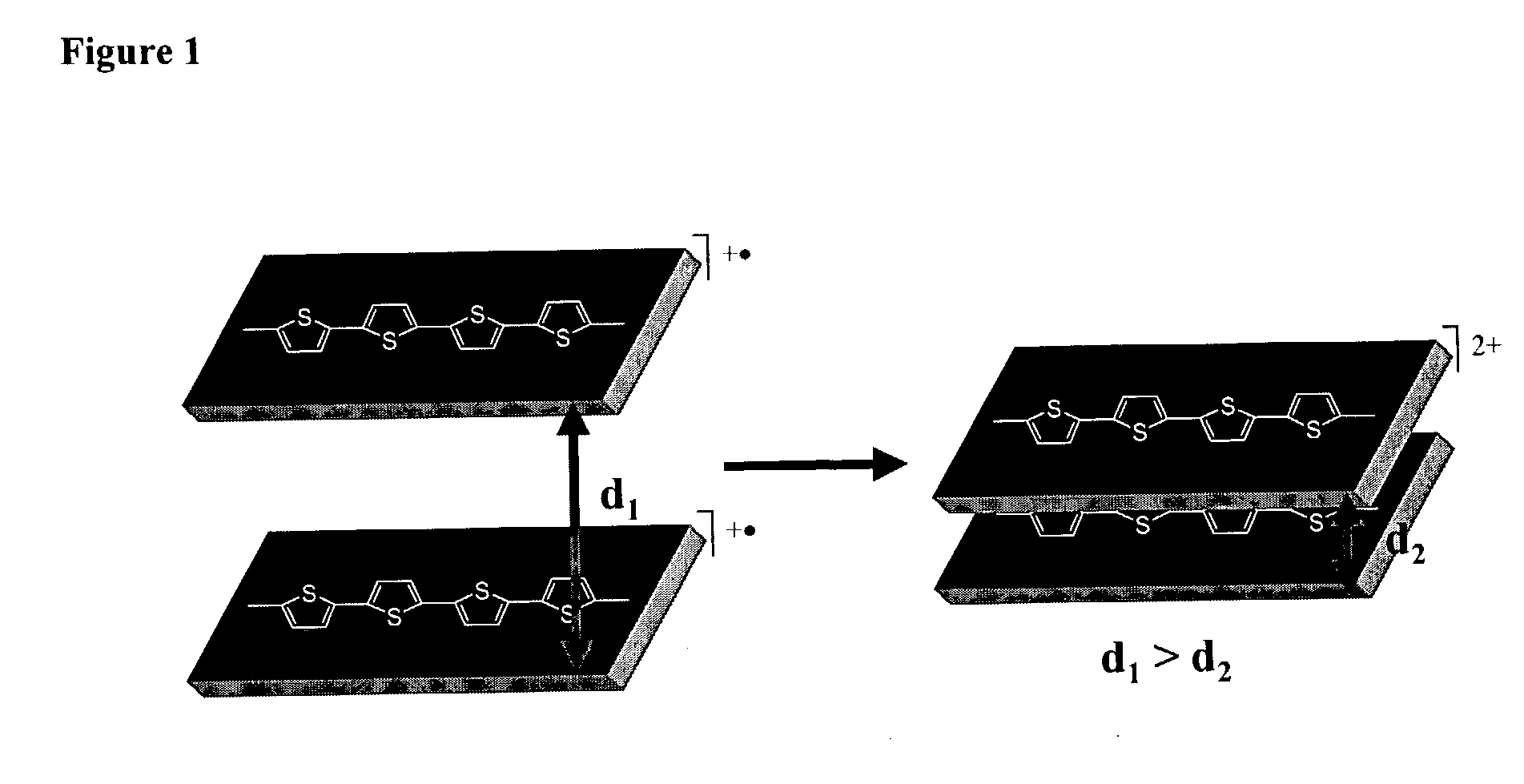
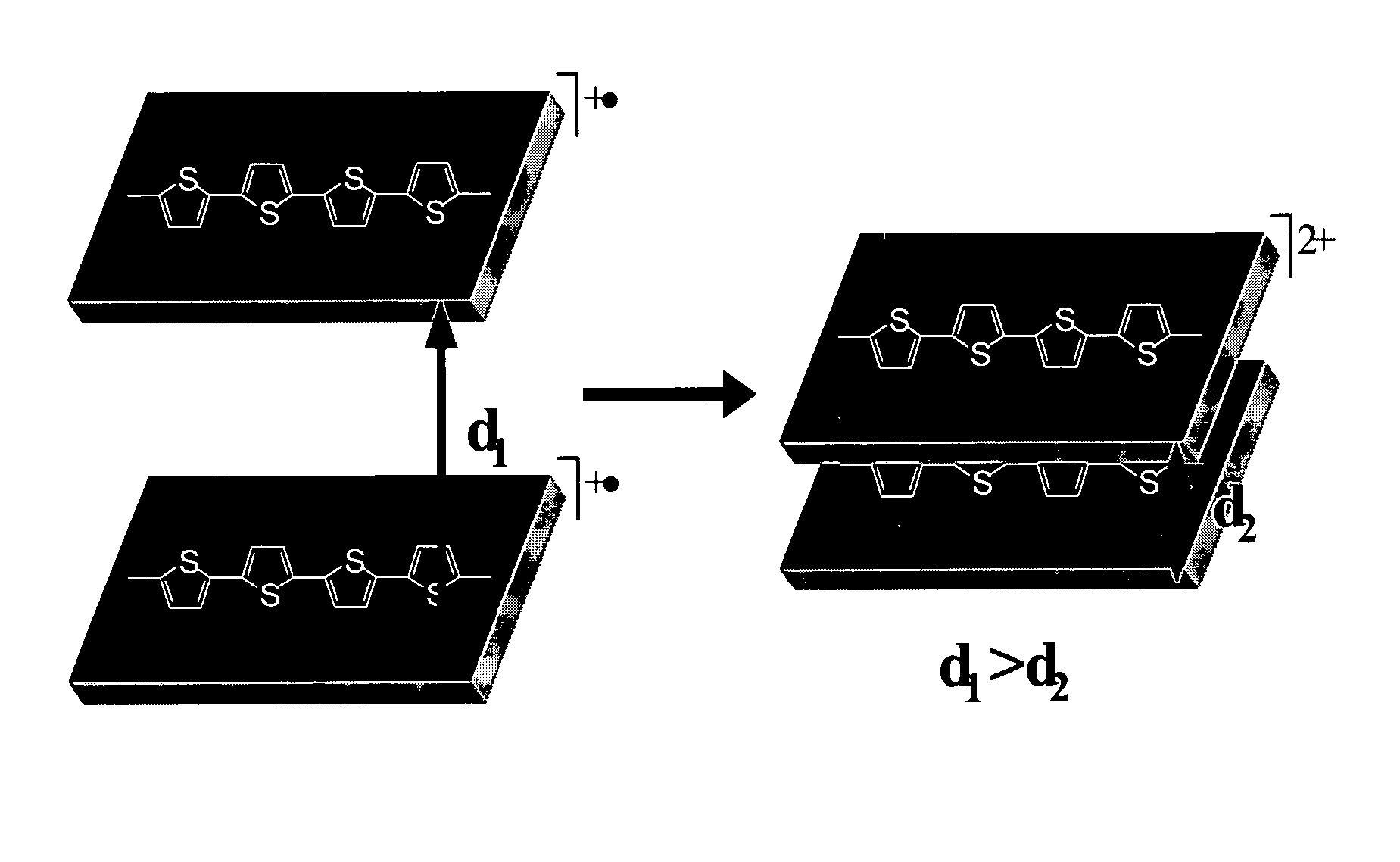
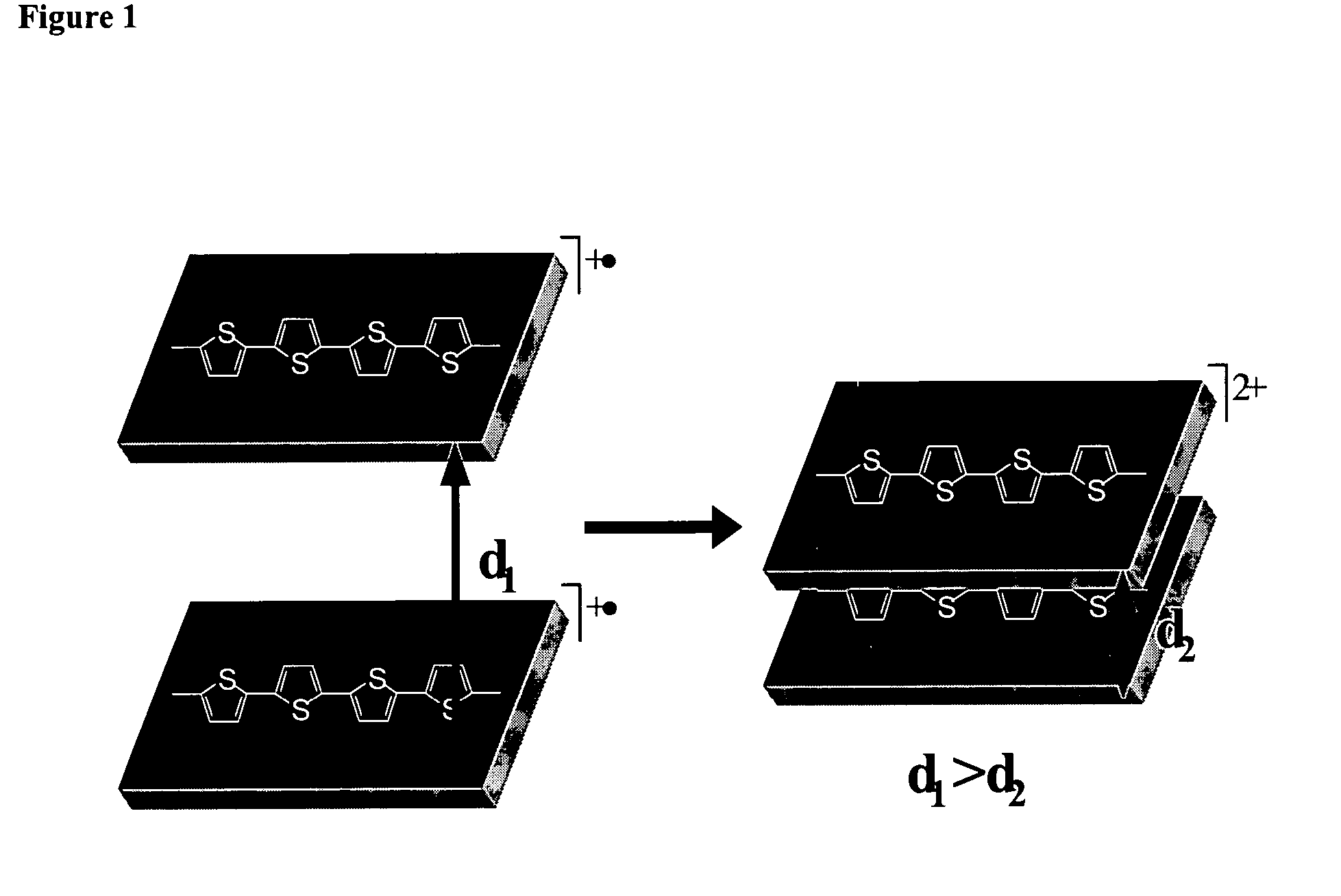
Molecular actuators, and methods of use thereof
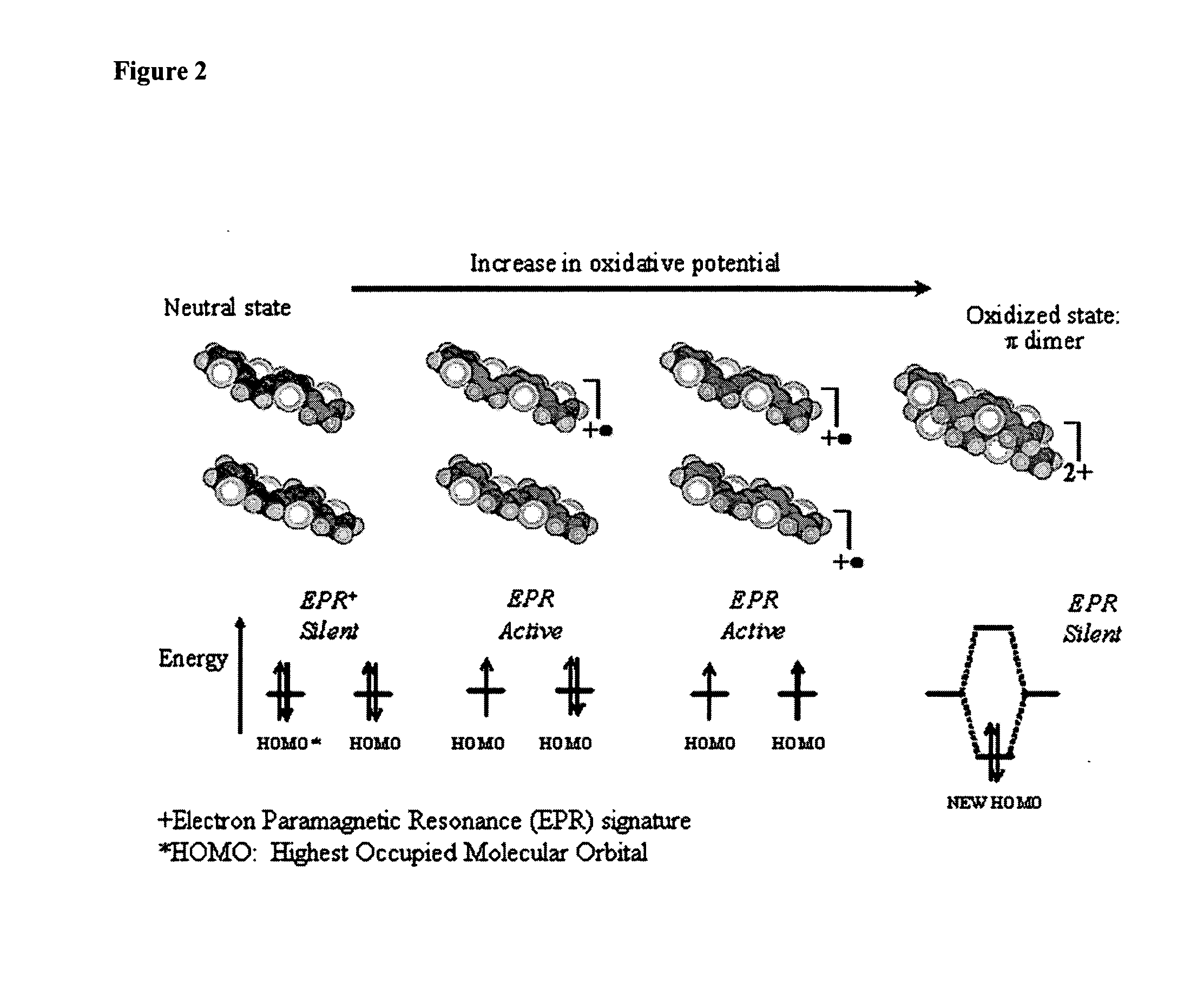
The synthesis of thiophene based conducting polymer molecular actuators, exhibiting electrically triggered molecular conformational transitions is reported. Actuation is believed to be the result of conformational rearrangement of the polymer backbone at the molecular level, not simply ion intercalation in the bulk polymer chain upon electrochemical activation. Molecular actuation results from π—π stacking of thiophene oligomers upon oxidation, producing a reversible molecular displacement that leads to surprising material properties, such as electrically controllable porosity and large strains. The existence of active molecular conformational changes is supported by in situ electrochemical data. Single molecule techniques have been used to characterize the molecular actuators.
Owner:MASSACHUSETTS INST OF TECH
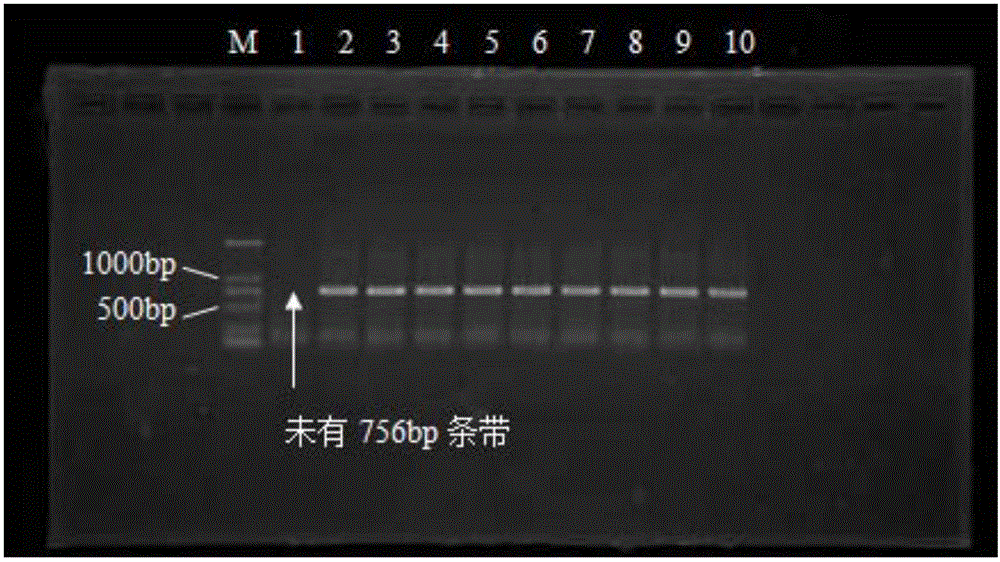
Specific labeled primer pair for rapidly identifying camellia oleifera improved variety Cenruan 2 and identifying method
ActiveCN106676175ARapid identificationEarly identificationMicrobiological testing/measurementDNA/RNA fragmentationCamellia oleiferaElectrophoresis
The invention discloses a specific labeled primer pair for rapidly identifying a camellia oleifera improved variety Cenruan 2. The specific labeled primer pair comprises an upstream primer and a downstream primer with the base sequences shown as SEQ. ID. No.1 and SEQ. ID. No.2 in a sequence table. Meanwhile, the invention further provides a corresponding identifying method. According to the method, the specific labeled primer pair is taken to perform PCR amplification on camellia oleifera improved variety series, if a DNA band with the length of 756 bp is not shown in an electrophoresis result, the to-be-detected camellia oleifera variety is Cenruan 2, otherwise, the variety is not Cenruan 2. Tests prove that the method has specificity, is only applicable to identification of camellia oleifera improved variety Cenruan series, can perform rapid early identification on the camellia oleifera improved variety Cenruan 2, has the characteristics of being simple, fast, accurate and the like, and is an irreplaceable molecular technique for distinguishing camellia oleifera improved variety on the basis of apparent characteristics.
Owner:GUANGXI FORESTRY RES INST
Molecular actuators, and methods of use thereof
The synthesis of thiophene based conducting polymer molecular actuators, exhibiting electrically triggered molecular conformational transitions is reported. Actuation is believed to be the result of conformational rearrangement of the polymer backbone at the molecular level, not simply ion intercalation in the bulk polymer chain upon electrochemical activation. Molecular actuation results from π-π stacking of thiophene oligomers upon oxidation, producing a reversible molecular displacement that leads to surprising material properties, such as electrically controllable porosity and large strains. The existence of active molecular conformational changes is supported by in situ electrochemical data. Single molecule techniques have been used to characterize the molecular actuators.
Owner:MASSACHUSETTS INST OF TECH +1
Primer probe combination for identifying mycobacterium abscessus and mycobacterium massiliense
InactiveCN108866216ARapid identificationStrong specificityMicrobiological testing/measurementMicroorganism based processesMycobacterium massilienseMycobacterium abscessus complex
The invention discloses a primer probe combination for identifying mycobacterium abscessus and mycobacterium massiliense. The primer probe combination provided by the invention consists of a single chain DNA shown as sequences 1 to 4 in a sequence table. The invention provides an identification method of mycobacterium abscessus and mycobacterium massiliense; the distinguishing on the mycobacteriumabscessus and the mycobacterium massiliense can be fast completed in the molecular level; the characteristics of simplicity, convenience, high speed, high specificity and high sensitivity are realized, so that the problem that the mycobacterium abscessus composite groups cannot be accurately and fast identified by the existing molecular technology is solved.
Owner:BEIJING CHEST HOSPITAL CAPITAL MEDICAL UNIV +1
Method for detecting environment DNA of giant salamander distribution
ActiveCN113088574AImprove featuresImprove efficiencyMicrobiological testing/measurementAgainst vector-borne diseasesElectrophoresesChinese giant salamander
The invention belongs to the technical field of molecules, and particularly relates to a method for detecting environment DNA of giant salamander distribution. According to the specific technical scheme, according to the method for detecting the environment DNA of giant salamander distribution, at least one pair of primers is used for conducting PCR amplification detection on a to-be-detected sample, if 158-224 bp positive bands appear in an electrophoresis result, it is considered that giant salamanders are distributed in the environment where the to-be-detected sample is sourced, the primer pair is AdCytbE1, and the sequences are SEQ ID NO: 1 and SEQ ID NO: 2. According to the method, environmental DNA of excrement, skin exfoliates, corpses and the like of Chinese giant salamanders in residual water is utilized, a molecular biological technology is combined, a primer with high specificity, high sensitivity and strong universality is developed, and a method capable of carrying out rapid and standardized detection aiming at the distribution of different species of giant salamanders is established.
Owner:CHENGDU INST OF BIOLOGY CHINESE ACAD OF S
Primer probe set for rapidly and efficiently detecting candida aurantiaca based on fluorescent PCR technology as well as kit and application thereof
PendingCN112063747AHigh detection sensitivityEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationForward primerPathogenic microorganism
The invention provides a primer probe set for rapidly and efficiently detecting candida aurantiaca based on fluorescent PCR technology as well as a kit and application thereof, and belongs to the technical field of pathogenic microorganism in-vitro molecules. The invention relates to a specific DNA fragment for detecting the candida aurantiaca, wherein the DNA fragment has a nucleotide sequence shown as SEQ ID NO: 1. The primer probe set for rapidly and efficiently detecting the candida aurantiaca based on the fluorescent PCR technology comprises a primer pair, and a probe; the primer pair consists of a forward primer with a nucleotide sequence as shown in SEQ ID NO: 2, and a reverse primer with a nucleotide sequence as shown in SEQ ID NO: 3; and the probe has a nucleotide sequence as shown in SEQ ID NO: 4. The primer probe set and the kit thereof are capable of rapidly and efficiently detecting the candida aurantiaca on basis of the fluorescent PCR technology; and thus, high detectionsensitivity, short detection time and simple operation can be realized.
Owner:杭州缔园生物技术有限公司
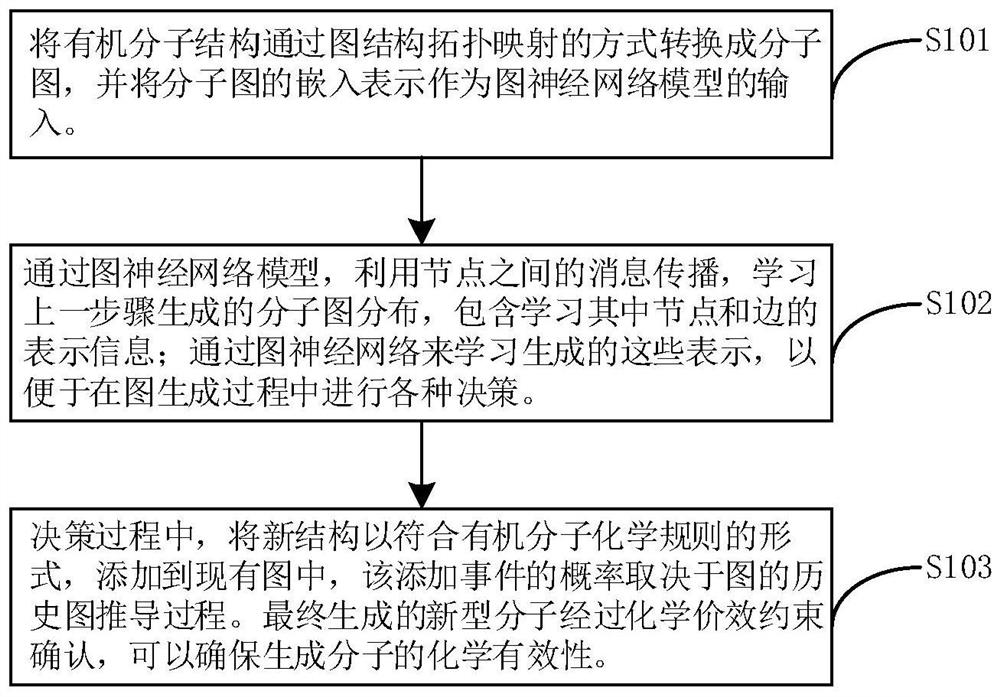
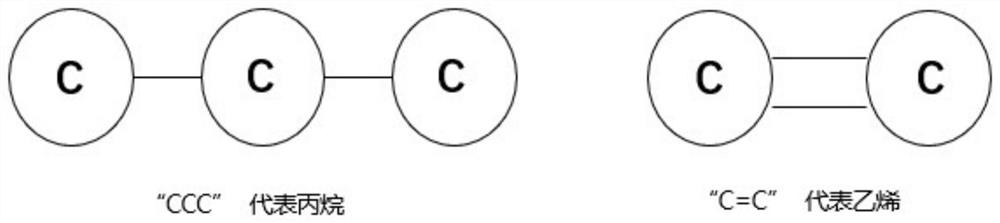
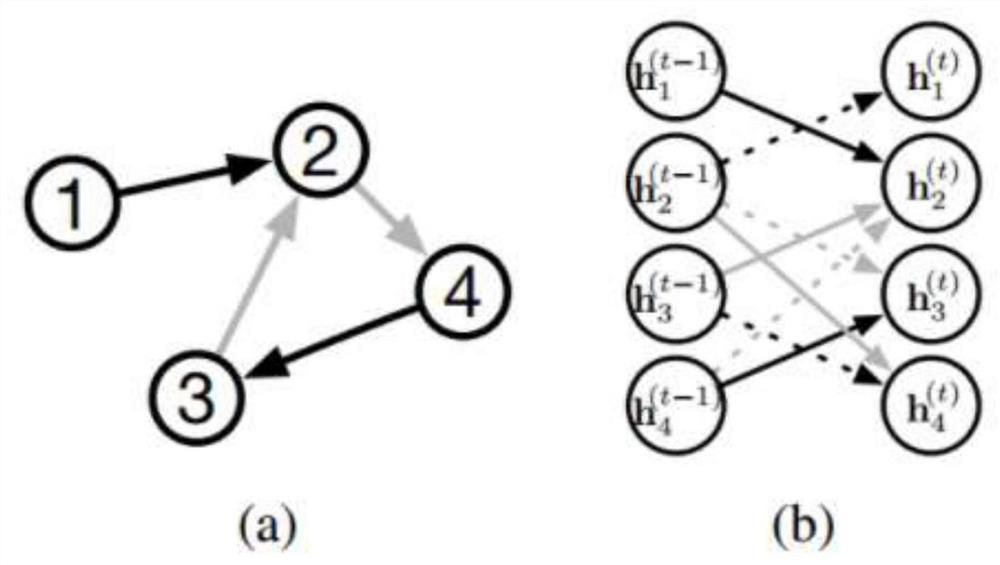
Directional molecule generation method based on graph neural network
PendingCN113140267AEnsure chemical validityMolecular designNeural architecturesAlgorithmGraph generation
The invention relates to a directed molecule generation method based on a graph neural network, and relates to the technical field of material molecules. Comprising the following steps: converting an organic molecular structure graph into a molecular graph in a topological mapping mode, and taking embedded representation of the molecular graph as input of a graph neural network model; through a graph neural network model, learning the molecular graphs based on a message propagation process, including representations of nodes and edges in the molecular graphs; the generated representations are learned through a graph neural network so that various decisions can be made in the graph generation process; in the decision-making process, the new structure is added to the existing graph in a form conforming to the organic molecule chemical rule, and the probability of the addition event depends on the historical graph derivation process of the graph. The finally generated novel molecules are confirmed through chemical valence constraint, and the chemical effectiveness of the generated molecules can be ensured. According to the method, effective novel molecular structures with chemical properties similar to those of original molecules can be generated aiming at an organic molecule database.
Owner:BEIJING UNIV OF CHEM TECH
Loach ploidy identification primer based on microsatellite polymorphism and application
ActiveCN107022630AImprove accuracySimple methodMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceSmall sample
The invention belongs to the field of molecular techniques for identification of fish polyploidy, and particularly discloses a loach ploidy identification primer based on microsatellite polymorphism and application. The developed primer used for difference and polymorphism of loaches of different ploidies is high in accuracy rate and stability, a method is simple and convenient, rapid identification for the ploidies of loaches can be completed only after regular PCR amplification and polyacrylamide gel electrophoresis are conducted, the cost is low, and the primer is suitable for identification of the ploidies of various loach samples including cryopreserved samples preserved for a long time, extremely small samples and the like; a technological support is provided for distinguishing of the ploidies of the loaches, utilization of loach genetic resources based on the ploidies, polyploid breeding, genetic research and the like.
Owner:HUAZHONG AGRI UNIV
Polymerase chain reaction (PCR) primer for cloning Actin gene of phalaris arundinacea linn and method
InactiveCN102586245AEasy accessDoes not appear to affect the hairpin structureMicrobiological testing/measurementFermentationNucleotideActin genes
The invention provides a pair of polymerase chain reaction (PCR) primers for cloning an Actin gene of phalaris arundinacea linn and a method, which belong to the field of molecular biology. Nucleotide sequences of the pair of PRC primers for cloning the Actin gene of the phalaris arundinacea linn are indicated as SEQ ID NO.1 and SEQ ID NO.2. The invention further provides the method for cloning the Actin gene of the phalaris arundinacea linn. The method includes extracting total ribonucleic acid (RNA) in a glass blade tissue of the phalaris arundinacea linn, and the RNA is inverse-transcripted into complementary deoxyribose nucleic acid (cDNA). The special primers are used for conducting a PCR to obtain a partial segment of the Actin gene with sequence length as 453bp. The gene segment can be used as an internal reference gene and provide convenience for research of other genes. The method has the advantages of being high in flexibility, strong in specificity, simple and fast and having good application value in the aspect of molecular technology breeding of the phalaris arundinacea linn.
Owner:CHINA AGRI UNIV
Multiplex compositions and methods for quantification of human nuclear DNA and human male DNA and detection of PCR inhibitors
ActiveUS20080268448A1Sugar derivativesMicrobiological testing/measurementMolecular TechniqueQuantification methods
The invention relates to a method for simultaneous quantification of human nuclear DNA and human male DNA in a biological sample while also detecting the presence of PCR inhibitors in a single reaction. The multiplex quantification method also provides a ratio of human nuclear and male DNA present in a biological sample. Such sample characterization is useful for achieving efficient and accurate results in downstream molecular techniques such as genotyping.
Owner:APPL BIOSYSTEMS INC
Spodoptera frugiperda polymorphic microsatellite molecular markers and primers thereof
InactiveCN111424099AContribute to molecular ecology researchGood polymorphismMicrobiological testing/measurementDNA/RNA fragmentationNucleotideGenetics
The invention discloses spodoptera frugiperda polymorphic microsatellite molecular markers and primers thereof. Nucleotide sequences of the microsatellite molecular markers are correspondingly shown as SEQ ID NO.1-12, and primer pairs of the microsatellite molecular marker are shown as SEQ ID NO.13-36. The invention further discloses a preparation method of the polymorphic microsatellite molecularmarker. According to the invention, polymorphism experimental verification is carried out on 33 sites by predicting four-base microsatellite sites with polymorphism in the genome of spodoptera frugiperda, and twelve four-base microsatellite markers with good polymorphism are obtained through screening for the first time, wherein the four-base microsatellite markers comprise four moderate polymorphism markers and eight high polymorphism markers, so that molecular technical support is provided for genetic research on invasion of spodoptera frugiperda, and molecular ecological research on the highly-multiphagous species, namely the grassland spodoptera litura, is facilitated.
Owner:SICHUAN UNIV
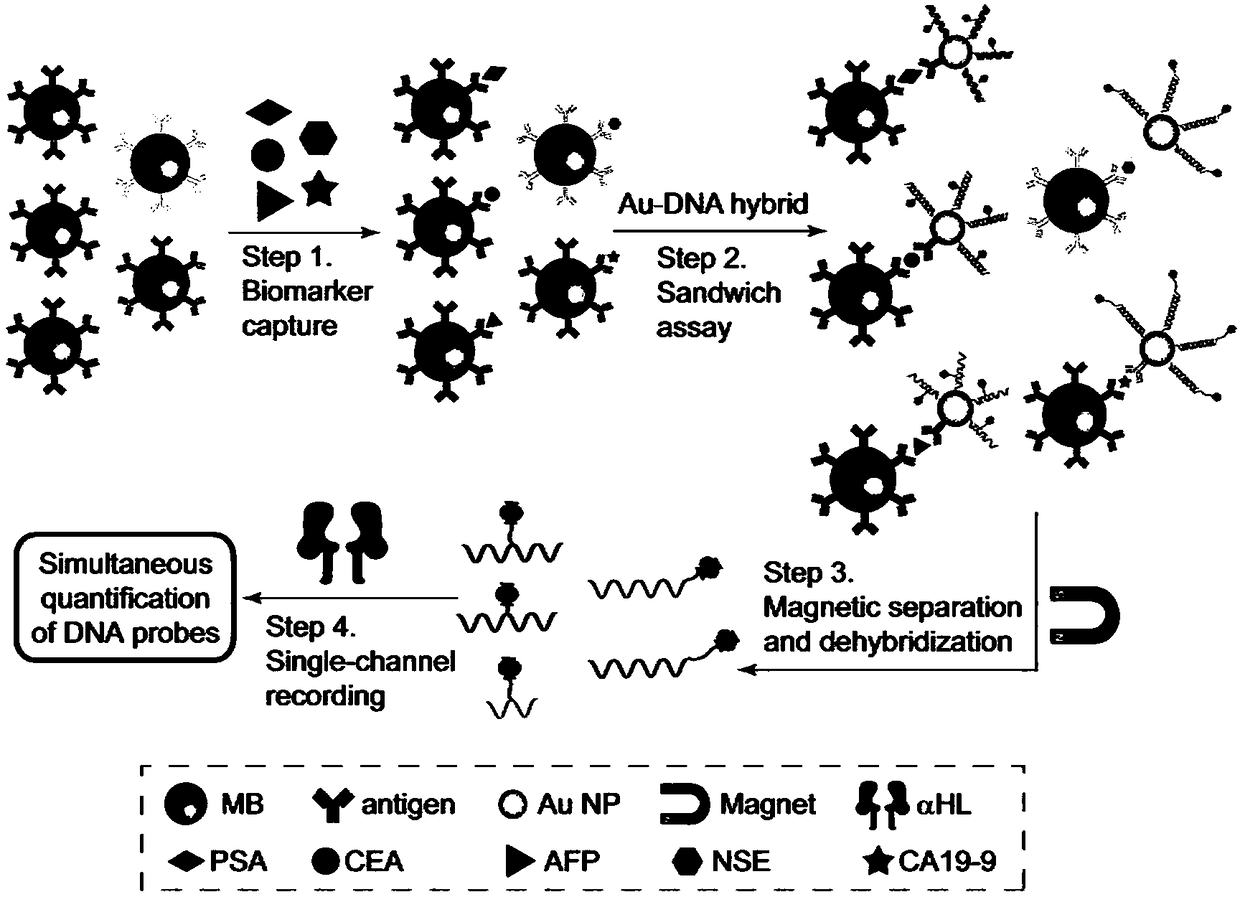
Cancer marker detection method combining DNA coding technology with nanopore technology
The invention discloses a cancer marker detection method combining DNA coding technology and nanopore technology. The cancer marker detection method comprises the following steps: firstly, synthesizing a DNA coding molecule, and modifying the group and the length of DNA by virtue of chemical modification changing the DNA base, so as to generate a characteristic current signal with higher discrimination when the DNA coding molecule passes through nanopores; in combination with immunoassay technology, adding cancer markers to form a sandwich structure with magnetic beads and nano gold; and thenreleasing the encoded DNA on the surface of nanogold, so as to detect the cancer markers separately and simultaneously. A high-throughput multi-component detection platform based on nanopore single molecule technology outputs corresponding characteristic current signals with different DNA coding molecules, so as to eliminate background interference as well as signal interference between the DNA coding molecules, and overcome the defects of fluorescent bleaching, short fluorescence lifetime, background interference, fluorescence interference between encoded microspheres and the like of fluorescence encoding of a traditional method.
Owner:INST OF CHEM CHINESE ACAD OF SCI +1
Antibody detection method for combining three-chain DNA molecular beacons with nanopore technology for combination
InactiveCN108467883AAccurately realize the designationRealize identificationMicrobiological testing/measurementDNA/RNA fragmentationPower flowInterference problem
The invention discloses an antibody detection method for combining a three-chain DNA molecular beacon with a nanopore technology for combination. The method comprises the steps that the three-chain DNA molecular beacon used for detecting an antibody is designed and synthesized, when the antibody is combined with the three-chain DNA molecular beacon, the molecular beacon structure is destroyed, anda probe is released; an antibody-antibody capture DAN molecular complex cannot penetrate through a nanopore due to too large volume, the probe passes through the nanopore and can generate a characteristic current signal, the signal specificity is extremely high, molecular recognition can be accurately achieved, and the background interference problem is avoided. In addition, the unique thermodynamics and kinetic properties of the molecular beacon are combined with the nanopore single molecular technique, and high sensitivity detection on the variable antibody is achieved. In addition, the method has the advantages that implementation is easy, reagent consumption is extremely low, and large-scale equipment is not needed.
Owner:INST OF CHEM CHINESE ACAD OF SCI +1
Quality control standard substance and quality control method for microorganism high-throughput DNA sequencing data
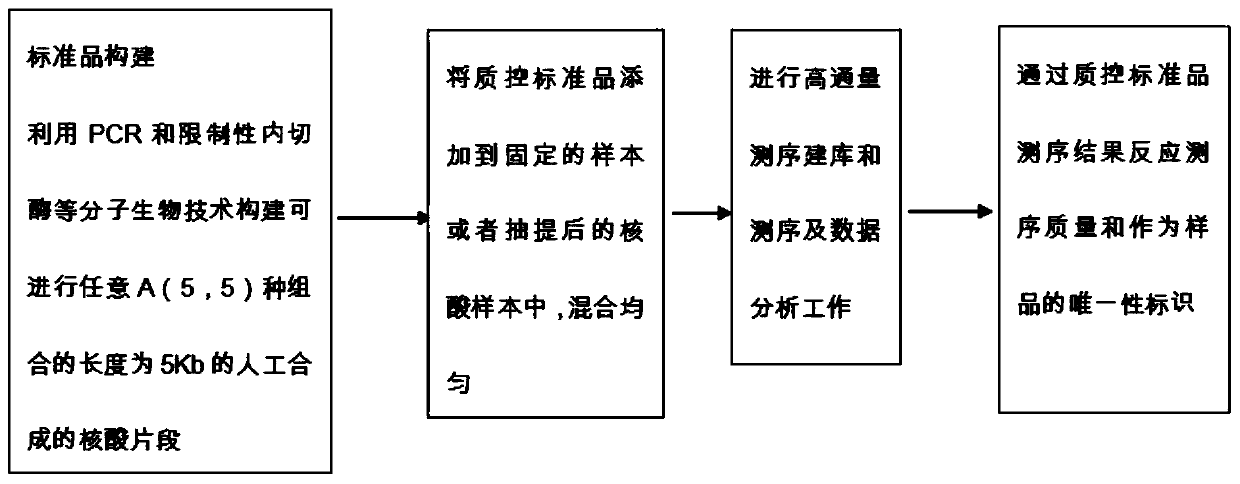
InactiveCN111500691AAvoid errors caused by confusionGuaranteed reliabilityMicrobiological testing/measurementDNA/RNA fragmentationMolecular TechniquePolynucleotide
The invention discloses a quality control standard substance and a quality control method for microorganism high-throughput DNA sequencing data. The quality control standard substance is an artificially synthesized polynucleotide sequence, the size of a nucleic acid molecule is 5Kb, the nucleic acid molecule is composed of five 1Kb fragments, the five 1Kb fragments can have A (5, 5) free combination modes, and different combination connection is carried out by adopting an overlap extension PCR molecular technology. According to the quality control method, the quality control standard substanceis utilized, and thus the quality control of factors such as laboratory management, personnel, detection experiments and bioinformatics analysis which influence the NGS detection result can be realized; meanwhile, errors caused by confusion of sample numbers in the sequencing process are prevented, and the test accuracy is improved; the quality control standard substance and a quality control method can be more effectively applied to various fields utilizing the high-throughput sequencing technology, the sequencing stability and accuracy can be effectively evaluated, and the sequencing data analysis reliability is ensured.
Owner:NAT INST FOR FOOD & DRUG CONTROL
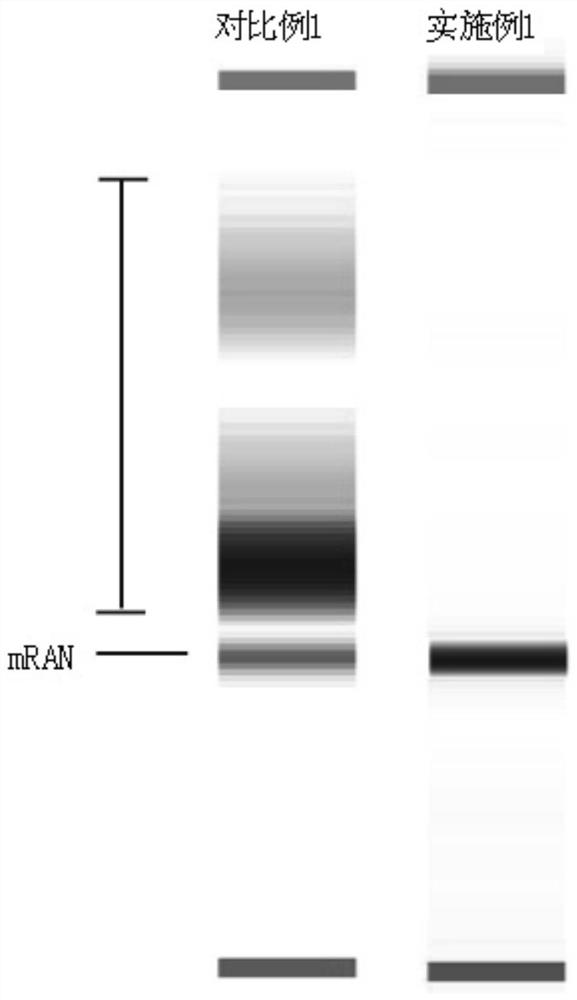
mRNA and its preparation method and application
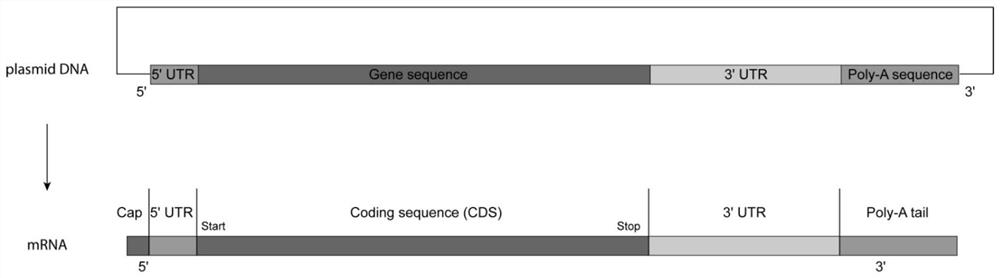
ActiveCN109628442BHigh purityEasy to purifyOrganic active ingredientsMicrobiological testing/measurementGeneticsA-DNA
The invention relates to the technical field of mRNA molecules, and specifically provides an mRNA and its preparation method and application. The preparation method of the mRNA comprises the following steps: providing a DNA transcription template; the DNA transcription template is composed of a sense strand and an antisense strand, wherein the sense strand comprises a promoter and a target DNA sequence connected to the promoter sequence, and the promoter The subsequence does not contain a stop codon, the target DNA sequence is identical to the target mRNA sequence, and the end of the target DNA sequence is connected with a polyadenylic acid sequence; under the action of RNA polymerase, the DNA transcription template is transcribed in vitro, Under the guidance of the promoter sequence, mRNA is transcribed. The preparation method of the present invention solves the problem of poor molecular homogeneity caused by the increase of by-product sequences during the Poly A tailing reaction of existing mRNA synthesis, and the preparation method also has the advantages of high preparation efficiency and easy purification of products.
Owner:GENEO MEDICINE CO LTD
Construction method and application of plasmid capable of replicating and expressing dehydrogenase gene at positions 1 and 2 of steroid compound a ring in Arthrobacter simplex
InactiveCN103205450BReduce manufacturing costHigh copy numberBacteriaMicroorganism based processesCompound aMolecular Technique
The invention relates to application of plasmid capable of being copied in arthrobacter simplex and expressing steroid compound A ring 1, 2 dehydrogenase genes to constructing high-efficiency expression steroid compound A ring 1, 2 dehydrogenation arthrobacter simplex engineering bacteria. The plasmid is capable of being copied in arthrobacter simplex and efficiently expressing steroid compound A ring 1, 2 dehydrogenase genes and significantly practical, and basis is provided for further using plasmid pXJM19 to modify arthrobacter simplex through molecular techniques.
Owner:TIANJIN UNIV OF SCI & TECH
Application of notoginseng ssr marker in determination of notoginseng saponin r1 content
ActiveCN109609675BEasy to carry outMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologySanchinosides
The present invention relates to the field of molecular technology, in particular to the use of notoginseng SSR markers in determining the content of notoginseng saponin R1, and notoginseng saponin R1 is associated with sites SSR35, SSR43, SSR89 and SSR2-6. The invention studies the correlation between the SSR marker and the notoginseng saponin R1, which is beneficial to the development of the notoginseng breeding with high content of the notoginseng saponin R1.
Owner:YUNNAN AGRICULTURAL UNIVERSITY
Multiplex compositions and methods for quantification of human nuclear DNA and human male DNA and detection of PCR inhibitors
ActiveUS20120015370A1Sugar derivativesMicrobiological testing/measurementMolecular TechniqueQuantification methods
The invention relates to a method for simultaneous quantification of human nuclear DNA and human male DNA in a biological sample while also detecting the presence of PCR inhibitors in a single reaction. The multiplex quantification method also provides a ratio of human nuclear and male DNA present in a biological sample. Such sample characterization is useful for achieving efficient and accurate results in downstream molecular techniques such as genotyping.
Owner:APPL BIOSYSTEMS INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com