Primer probe combination for identifying mycobacterium abscessus and mycobacterium massiliense
A primer probe, mycobacteria technology, applied in the direction of microorganism-based methods, microorganisms, recombinant DNA technology, etc., can solve the problem that the cure rate is only about 60%
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1, the design of primer probe combination
[0054] A large number of sequence analyzes and comparisons of Mycobacterium abscessus and Mycobacterium marseille were carried out, and the influencing factors of clarithromycin drug susceptibility of Mycobacterium abscessus and Mycobacterium marseille were analyzed to obtain a set of Primer-probe combinations for Bacillus and M. marseilles are as follows:
[0055] Primer F1 (sequence 1 of the sequence listing): 5'-GCGACGCCAGTGGGGCTGGT-3';
[0056] Primer R1 (sequence 2 of the sequence listing): 5'-CGCCGCCTGATCACCAGCAC-3';
[0057]Probe P1 (SEQ ID NO: 3 of the Sequence Listing): 5'-GCGGATCGTCGCCGAATCCGGT-3'
[0058] Probe P2 (Sequence 4 of the Sequence Listing) 5'-CGCCCTACCAAGTCACCAGCG-3'.
[0059] The 5' end of probe P1 was modified with FAM fluorescent group, and the 3' end was modified with BHQ1 quencher group.
[0060] The 5' end of probe P2 was modified with HEX fluorescent group, and the 3' end was modifie...
Embodiment 2
[0062] Embodiment 2, establishment of detection method
[0063] 1. Extract the total DNA of the sample to be tested.
[0064] 2. Using the total DNA obtained in step 1 as a template, perform fluorescent quantitative PCR detection.
[0065] Fluorescence quantitative PCR reaction system: primer F1, primer R1, probe P1, probe P2 each 1 μl, PCR buffer + dNTPs + Mg 2+ (PCR mix) 25μl, DNA template 4μl, RNase Free H 2 O 12 μl.
[0066] The concentration of each primer in the system is 500nM, and the concentration of each probe in the system is 250nM.
[0067] DNA was added to the reaction system in the form of DNA solution, and the concentration of DNA in the DNA solution was 10 ng / μl.
[0068] Fluorescent quantitative PCR reaction program: pre-denaturation at 95°C for 2 minutes, denaturation at 95°C for 5 seconds, annealing at 58°C for 20 seconds, extension at 72°C for 20 seconds, a total of 40 cycles.
[0069] At the same time, a positive control group and a negative control g...
Embodiment 3
[0079] Embodiment 3, standard bacterial strain detection
[0080] Strains to be tested: Mycobacterium abscessus (ATCC19977), Mycobacterium avium (ATCC25291), Mycobacterium intracellulare (ATCC13950), Mycobacterium kansasii (ATCC12478) and Mycobacterium marseille standard strain (CIP108297, purchased from Paris, France Stuart Institute).
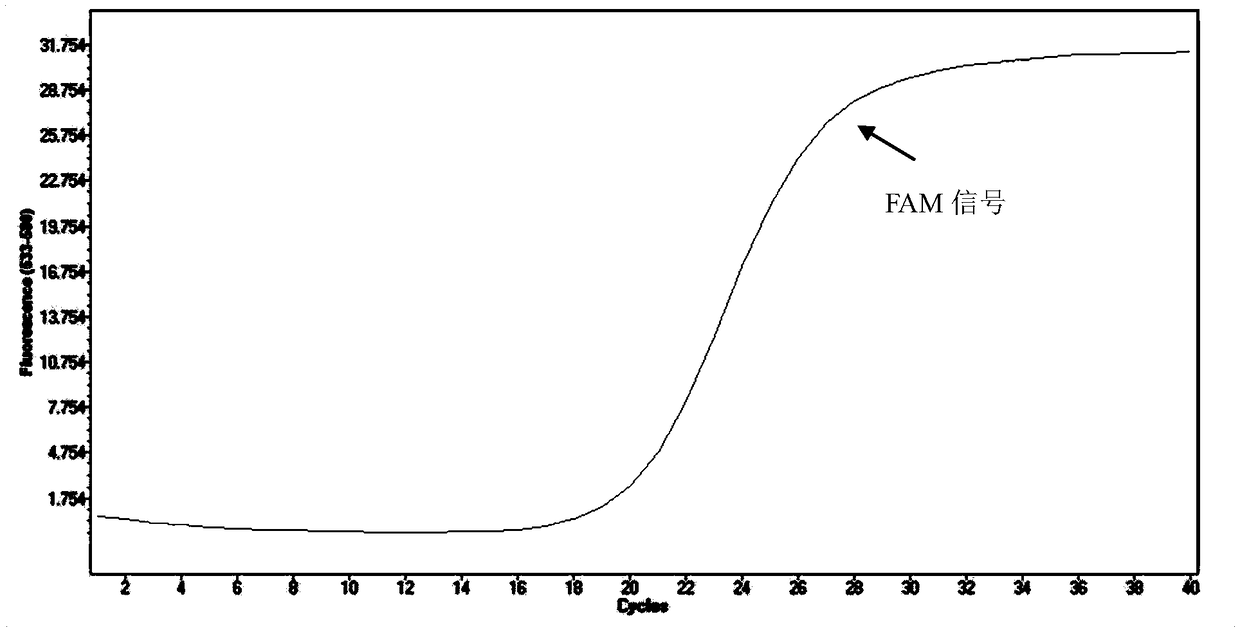
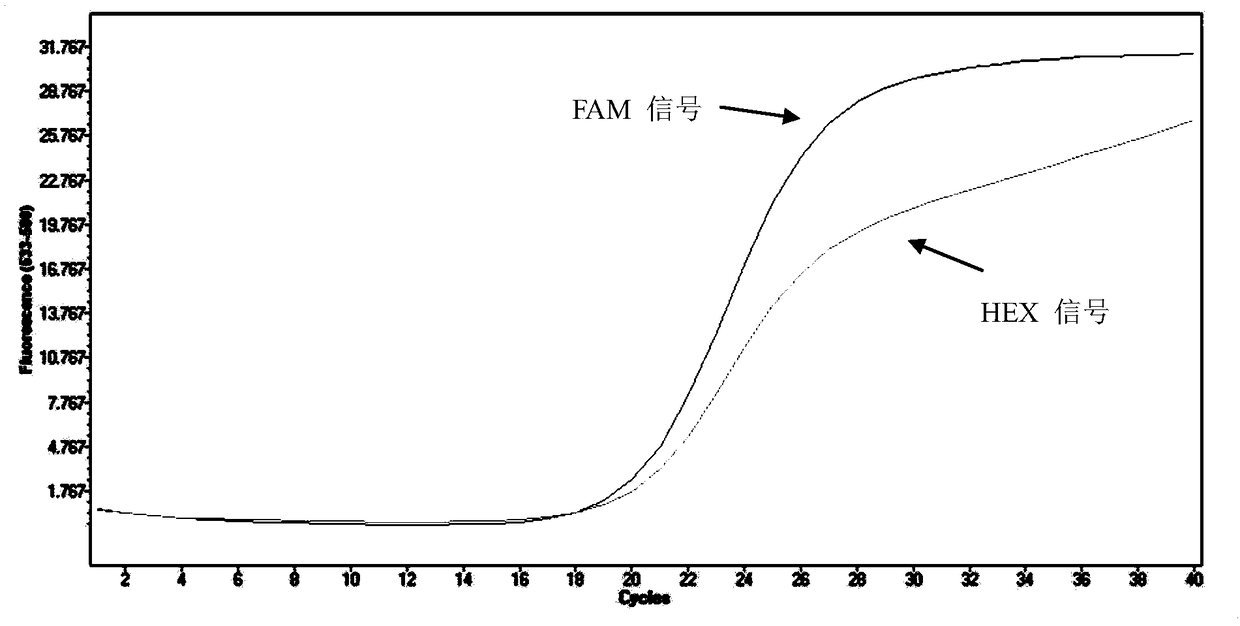
[0081] The method in Example 2 is adopted to detect, and the results show that when the sample to be tested is Mycobacterium avium, Mycobacterium intracellulare and Mycobacterium kansasii, the FAM signal is negative, and the HEX signal is negative simultaneously; when the sample to be tested is For Mycobacterium abscessus, the FAM signal is positive and the HEX signal is positive; when the sample to be tested is the standard strain of Mycobacterium marseille, the FAM signal is positive and the HEX signal is negative.
[0082] The above method has good specificity.
PUM
| Property | Measurement | Unit |
|---|---|---|
| separation | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com