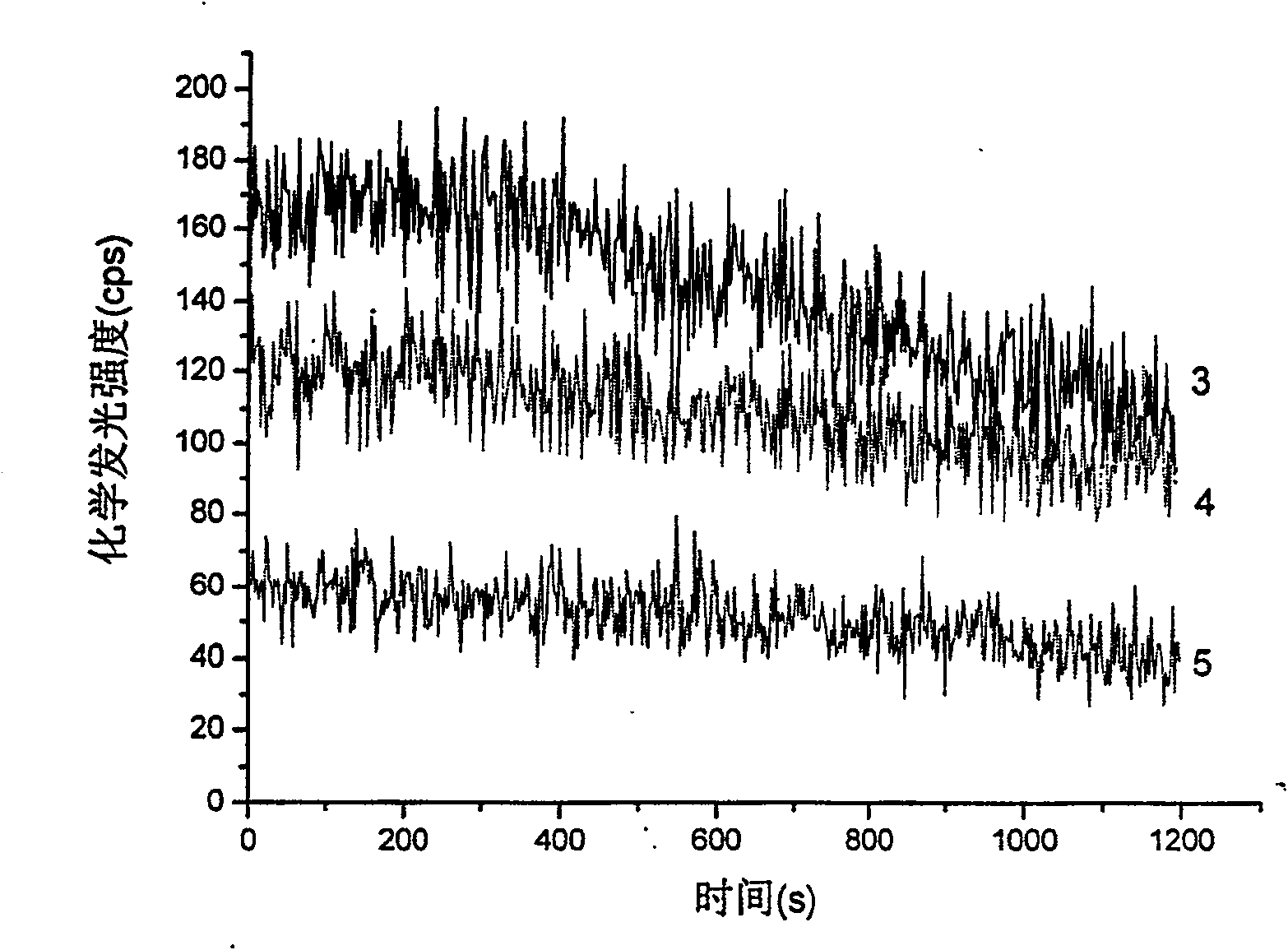
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
37results about How to "Short identification time" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Classification test method and classification test system for edible oil and swill-cooked dirty oil
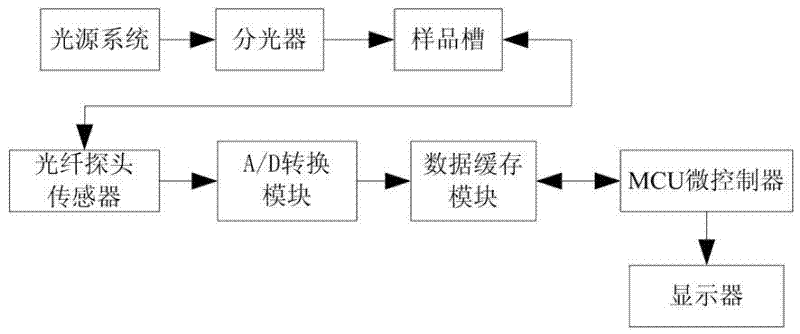
The invention discloses a classification test method and a classification test system for edible oil and swill-cooked dirty oil. The classification test method comprises the following steps that: Raman fingerprint databases for edible oil and swill-cooked dirty oil are first created, and the notable difference between the maximum Raman intensity values of the characteristic peaks of edible oil and swill-cooked dirty oil within a five-band wave number range and the threshold of the difference are then obtained by way of comparison; and in a practical test, the variety of a tested samples obtained by extracting the maximum Raman intensity value of the characteristic peak of the tested sample within the five-band wave number range and comparing the maximum Raman intensity value with the difference threshold. The classification test system comprises a light source system, an optical splitter, a sample box and a measurement control unit. The invention has the advantages of flexibility and high specificity, and can effectively test whether a tested sample is pure swill-cooked dirty oil, pure edible oil meeting the active national standard or edible oil mixed with swill-cooked dirty oil or not; and meanwhile, the operation is simple and convenient, the testing time is short, and swill-cooked dirty oil can be tested on the spot.
Owner:邹玉峰
Method for distinguishing green shell egg laying chicken and chicken green shell egg genotype as well as special fragment and primers
ActiveCN102876796AShort identification timeLow costMicrobiological testing/measurementDNA/RNA fragmentationMultiplex pcrsPolymerase chain reaction
The invention discloses a method for distinguishing green shell egg laying chicken and chicken green shell egg genotype as well as a special fragment and primers. The invention provides a method for identification or auxiliary identification of whether chicken to be detected is green shell egg chicken or not, in order to detect the genome DNA (deoxyribonucleic acid), if objective DNA molecules (sequence 4) are contained in the genome DNA, the chicken to be detected belongs to or is candidated as the green shell egg chicken, and when the objective DNA molecules are not contained in the genome DNA, the chicken to be detected does not belong to or is not candidated as the green shell egg chicken. Experiments prove that a 425bp short fragment is adopted, three primers are designed and synthesized according to the short fragment and are used for multiple PCR (polymerase chain reaction) amplification, whether the chicken to be detected lays green shell eggs or not can be distinguished through detecting whether amplification products contain the short fragment or not, and meanwhile, the chicken green shell egg genotype can be distinguished.
Owner:CHINA AGRI UNIV
Bacterium identification reagent kit as well as preparation method and uses thereof
InactiveCN101200755AHigh sensitivityImprove comparabilityMicrobiological testing/measurementBacteria identificationThiazole
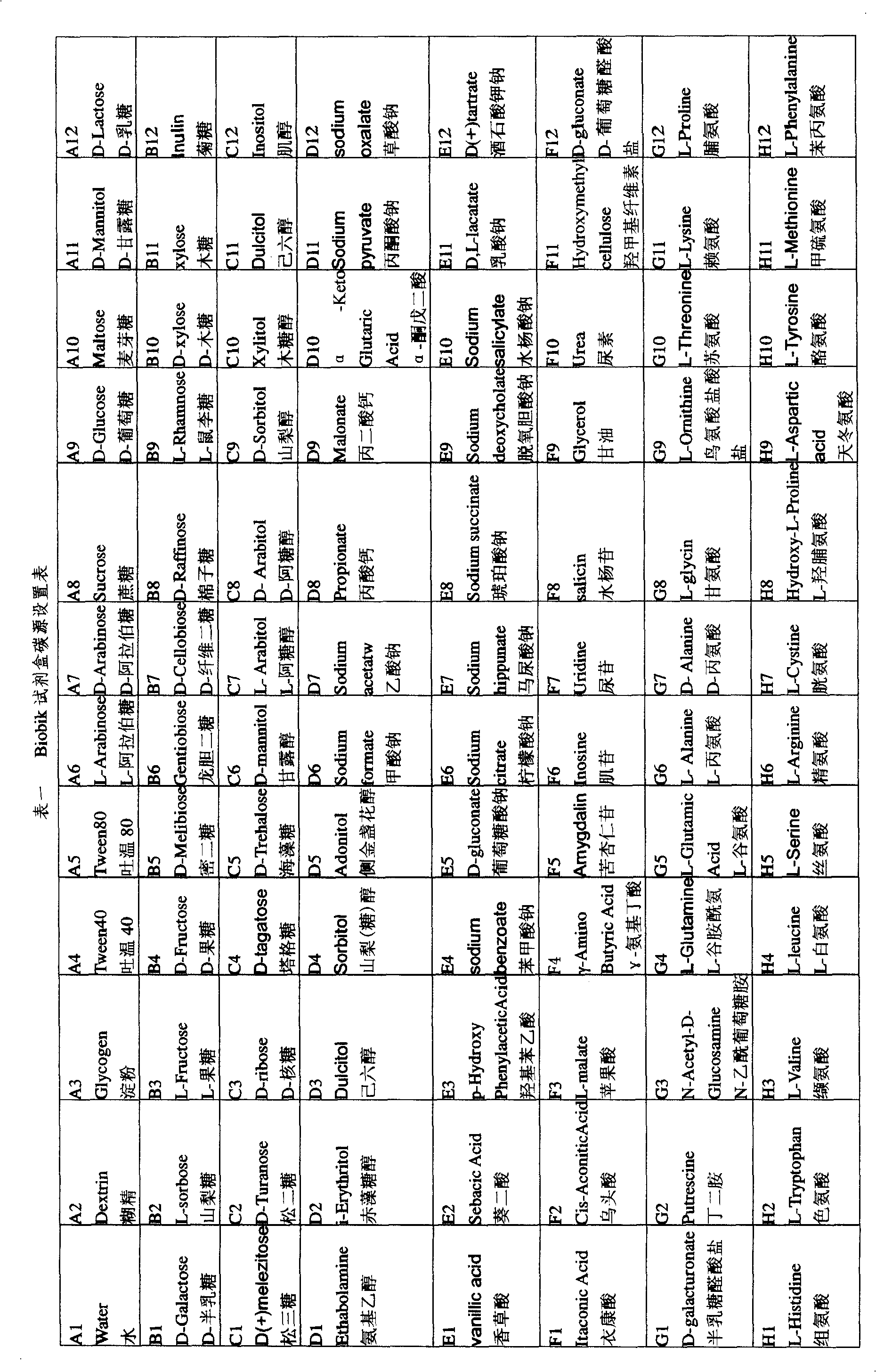
The invention discloses a bacteria identification kit, including indicator, culture medium and testing carbon sources. Thiazole blue is used as the indicator for indicating that whether bacteria can use the testing carbon sources. The invention is a kit which uses a 96-microtitor plate that is prearranged with the culture medium with 95 different carbon sources and is used for the rapid identification of carbon source utilization spectrum. One hole is not added with the carbon source for the comparison. The kit of the invention includes some carbon sources for detecting the specific physiological characteristics and common carbon sources. That using the thiazole blue as the indicator is convenient for naked eyes to observe that whether the bacteria grows and uses the carbon sources. And an eliasa can be used at the wavelength of 600nm to rapidly measure the growing condition of the bacteria. The kit of the invention has the advantages of low cost, short identification time, high sensitivity, convenient operation, accuracy, etc. the result can be made within 20-24 hours. And gram negative bacteria and positive bacteria can be identified.
Owner:SOUTH CHINA AGRI UNIV
Method for rapid identification of sweet potato stem rot resistance
InactiveCN103555813APracticalEasy to operateMicrobiological testing/measurementMicroorganism based processesDiseaseRapid identification
The invention relates to a method for rapid identification of sweet potato stem rot resistance. The method comprises the operation steps of: trimming a sweet potato stem or side branch into a stem branch with 3-4 extended leaves, and conducting pre-culture for 2-3 days under room temperature; when fibrous roots grow out of the stem branch, placing the stem branch horizontally, stabbing a small hole by a pipettor tip between two sections in the stem branch center without penetrating the stem branch, then placing the stem branch horizontally in a sealed plastic box paved with filter paper steeped by sterile water, and making the stem branch side stabbed with the small hole upward, absorbing 15-25 microliters of a 1*10<8>cfu / mL stem rot pathogenic bacterial suspension, and injecting the suspension into the small hole for inoculated culture; and after 20-24h of culture, observing the disease condition and conducting disease grade identification on the sweet potato stem branch. The method provided by the invention has the advantages of simple and fast operation, strong practicability, strong operability, and high identification result repeatability and reliability, and has potential social and economic significance for screening excellent germplasm resources resisting sweet potato stem rot.
Owner:CROP RES INST GUANGDONG ACAD OF AGRI SCI
Application of ITS (internal transcribed spacer) sequence as DNA barcode in identifying Jiaxing Zuili (Prunus salicina lindl.) and identifying method
ActiveCN105671185AImprove detection accuracyGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationPrunus salicina
The invention discloses application of ITS (internal transcribed spacer) sequence as a DNA barcode in identifying Jiaxing Zuili (Prunus salicina lindl.) and an identifying method and belongs to the technical field of molecular identification. The identifying method comprises: amplifying ITS sequence of a sample to be tested by using PCR (polymerase chain reaction) and carrying out sequencing; results show that if both basic group 165bp and basic group 195bp of the ITS sequence are 'T', it is determined that the sample to be tested is Jiaxing Zuili. The invention also discloses a method for identifying early maturing and late maturing varieties of Jiaxing Zuili; if the basic group sequence of the ITS sequence is shown as in SEQ ID No. 1, it is determined that the sample to be tested is an early maturing variety of Jiaxing Zuili; if the basic group sequence of the ITS sequence is shown as in SEQ ID No. 2, it is determined that the sample to be tested is a late maturing variety of Jiaxing Zuili. The ITS sequence is used herein as the DNA barcode to identify Jiaxing Zuili, detection accuracy is high, detection repeatability is high, and identifying time is short.
Owner:ZHEJIANG JIAXING AGRI SCI ACADEMY INST
Watermelon hybridization breeding and hybrid identification method
InactiveCN105613256AShort identification timeAvoid economic lossPlant genotype modificationHybrid speciesFirst generation
The invention discloses a watermelon hybridization breeding and hybrid identification method. The method comprises the following steps that 1, watermelon leaf yellow spot inheritable characters are introduced into a male parent of a hybridized combination, and the male parent is made to express the leaf yellow spot inheritable characters; 2, the male parent with the leaf yellow spot inheritable characters is hybridized with a female parent with no leaf yellow spot inheritable characters to obtain the first generation of hybrid, and the new combination or variety of watermelon is obtained; 3, further seeding and emergence observation are further performed on seeds of the first generation of hybrid, single plants with no yellow spots on leaves are not hybridization plants, single plants with yellow spots on leaves are hybridization plants, and therefore the purity of hybridization seeds can be identified. The obtained hybridization seeds have the good yield and quality, non-hybridization seedlings can be accurately recognized and removed in a nursery stage, economic losses brought to production due to insufficient seed purity are avoided, hybridization identification can be performed, the requirement for rapid and visual watermelon hybridization identification is met, and the good market prospect is achieved.
Owner:ZHENGZHOU FRUIT RES INST CHINESE ACADEMY OF AGRI SCI
Quick tubercle myco-bacillus culture medium
InactiveCN1858238ARapid cultivationShort identification timeMicrobiological testing/measurementBiotechnologyMalachite green
The present invention relates to a kind of quick tubercle mycobacillus culture medium. The quick tubercle mycobacillus culture medium has tryptone, sodium gluconate, pyritinol hydrochloride, water solution of malachite green, glycerin, agar, calf blood and other components. The quick tubercle mycobacillus culture medium has short bacterial culture time and short spawn identification time, and may be used in quick culturing and identification of tubercle mycobacillus and non-tubercle mycobacillus, the drug sensitivity test and the long term preservation of tubercle mycobacillus and non-tubercle mycobacillus. The culture medium has simple preparation process, easy observation of culture result and wide application range.
Owner:李洪敏
Fingerprint spectrum of butterfly-leaf platycladus orientalis SSR marker and construction method and application thereof
ActiveCN112029894AShort identification timeHigh identification accuracyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention discloses a fingerprint spectrum of a butterfly-leaf platycladus orientalis SSR marker and a construction method and application thereof. The invention provides a set of primers, which are composed of a primer pair 1 to a primer pair 8; and the nucleotide sequences of the primer pair 1 to the primer pair 8 are respectively sequence 1 to sequence 16 in a sequence table. The SSR markersuitable for butterfly-leaf platycladus orientalis is developed based on platycladus orientalis transcriptome sequencing, and the construction method and application of the fingerprint spectrum of the butterfly-leaf platycladus orientalis SSR marker are established. Compared with morphological detection, the technology has the advantages that the identification result is not affected by climate,environment, human factors and tree age, identification and differentiation can be effectively carried out in the seedling stage, the identification time is short, the identification accuracy is high,the detection efficiency is high, the stability is good, and the technology has important significance and good application effect on genetic specificity identification of butterfly-leaf platycladusorientalis and other platycladus orientalis germplasm.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Method for poducing transgene seeds of hybrid paddy rice in multiple resistances
InactiveCN1625939AStatistical purityShort identification timeFermentationGenetic engineeringBiotechnologyResistant genes
A process for preparing the seeds of multi-resistance transgenic rice includes transferring the anti-herbicide gene and the anti-borer gene to the restoring line of three-line or two-line crossed rice, transferring the plant hopper-resistant gene to the maintenance line of three-line crossed rice, hybridizing between sterile line and maintenance line, and hybridizing between transgenic restoring line and transgenic sterile line.
Owner:天津市水稻研究所 +2
Method for identifying glutenin macro-polymer content in wheat
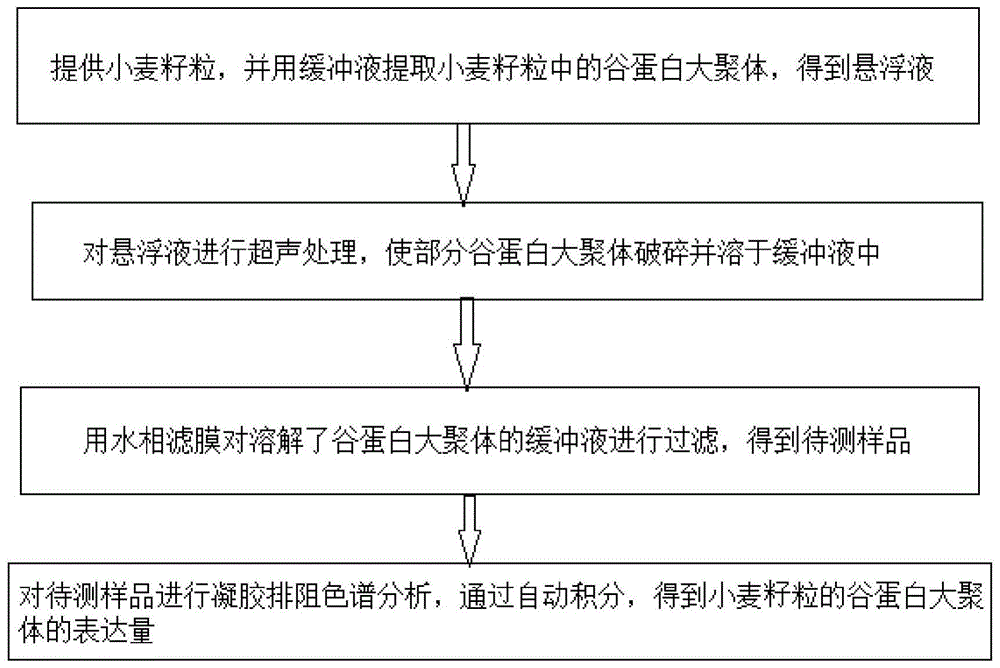
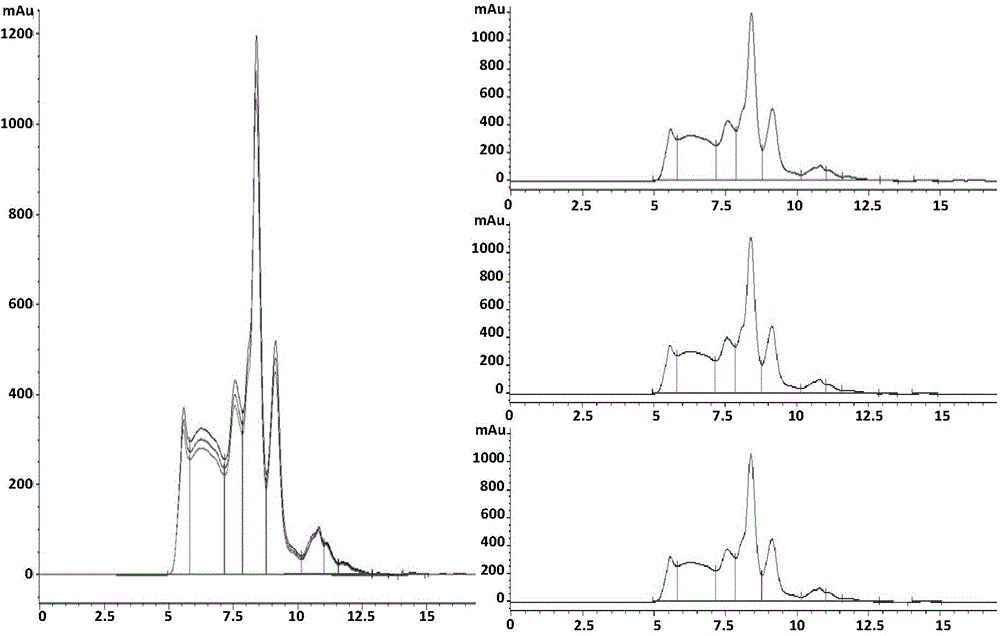
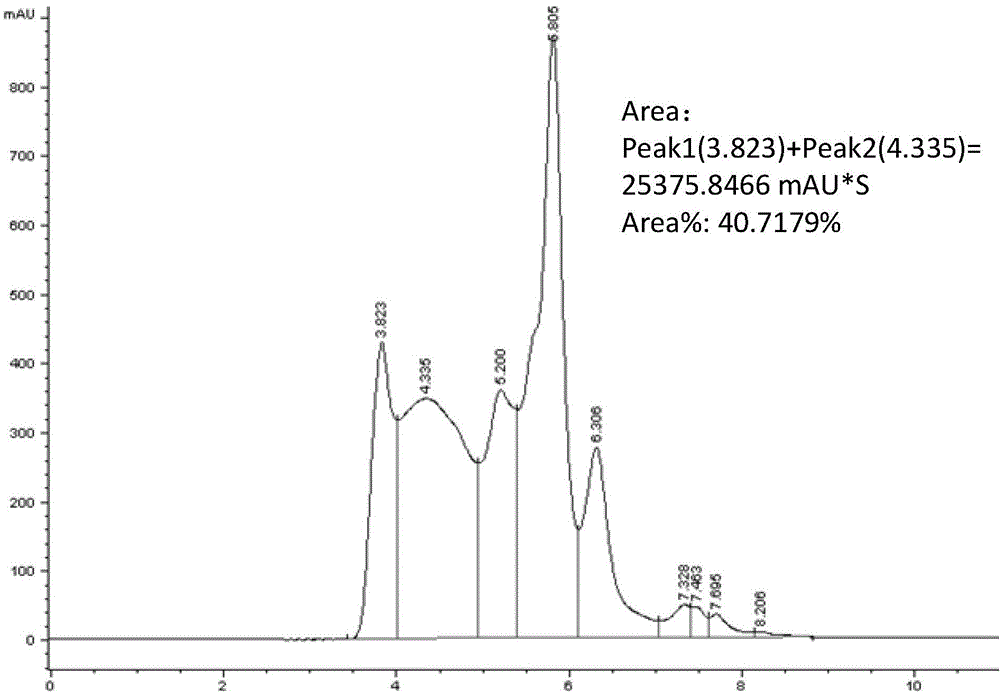
InactiveCN104458959AShort identification timeImprove accuracyComponent separationGluteninWheat glutelin
The invention discloses a method for identifying a glutenin macro-polymer in wheat. The method comprises the following steps: S1: providing wheat grains and extracting a glutenin macro-polymer in the wheat grains by virtue of a buffer solution to obtain a solution; S2: carrying out ultrasonic treatment on the suspension so that partial glutenin macro-polymer is broken and is dissolved in the buffer solution; S3: filtering the buffer solution with the dissolved glutenin macro-polymer by virtue of an aqueous filter membrane so as to obtain a to-be-detected sample; and S4: carrying out size exclusion chromatography analysis on the to-be-detected sample, and implementing automatic integration to obtain the expression quantity of the glutenin macro-polymer in the wheat grains. According to the embodiment disclosed by the invention, the method can be used for rapidly and accurately identifying the content of the glutenin macro-polymer in the wheat grains in the different stages of grain development, and can be used for effectively screening the varieties of the glutenin macro-polymer according to different expression quantities in mature grains.
Owner:CAPITAL NORMAL UNIVERSITY
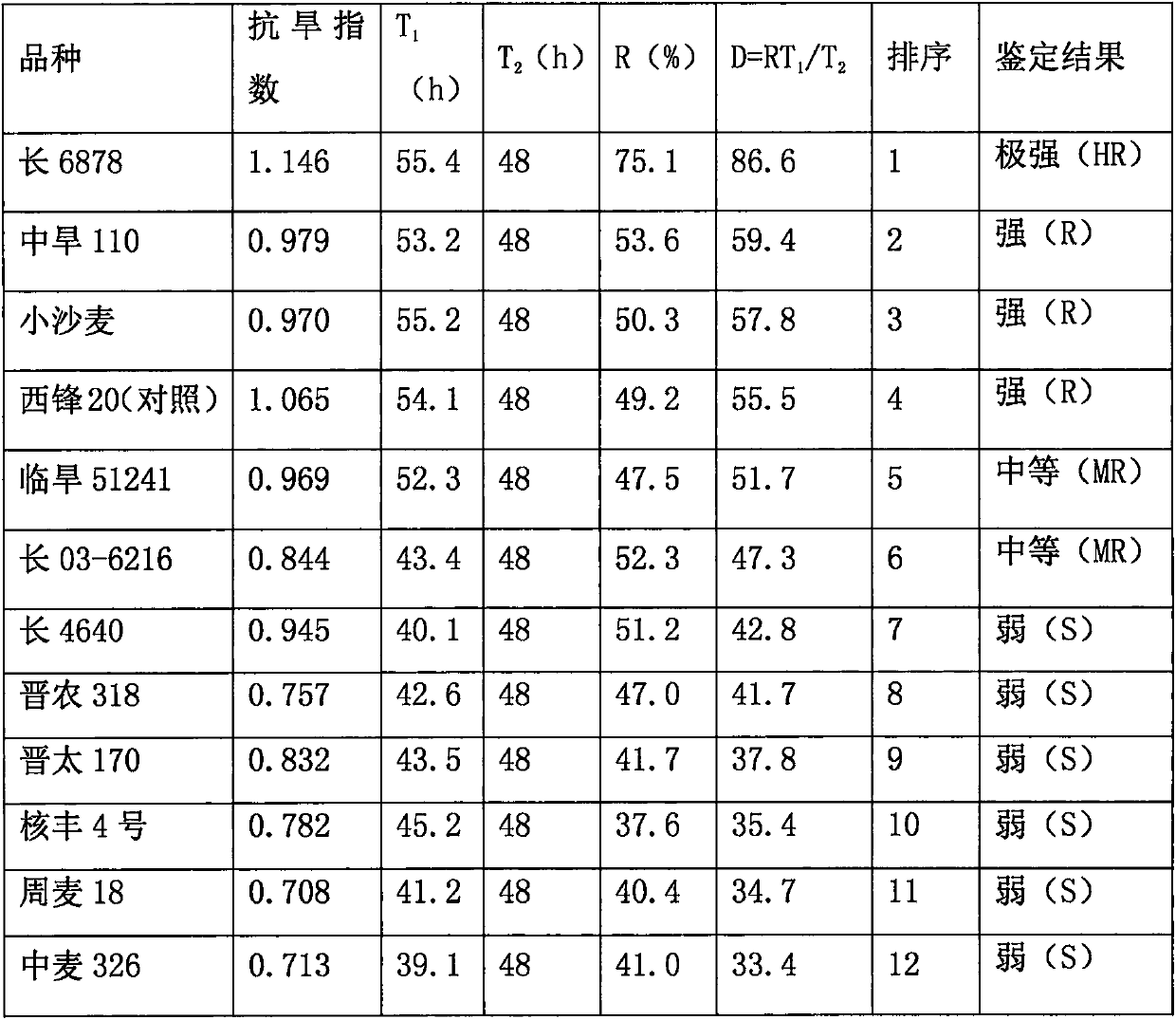
Method for quickly identifying drought resistance of wheat by using green recovering rate of leaves
InactiveCN107873435AThe results are consistentQuick filterGrowth substratesCulture mediaAgricultural scienceRapidity
The invention discloses a method for quickly identifying the drought resistance of wheat by using the green recovering rate of leaves. Drought stress treatment is performed in a three-leaf period of wheat, the time from dryness to green recovering of the leaves and the ratio of the leaves which recover green are detected after rehydration, and the drought resistance of the wheat can be identifiedquickly through comparison and rating; the result of drought resistance identification is in good accordance with the result of a method for identifying drought resistance indexes. The method has theadvantages of rapidity, high efficiency, low cost, easy operation and the like, field water management of farmers can be guided during production, and through the cooperation with other drought resistance identifying methods in scientific researches, excellent drought-resistant genetic resources can be selected quickly in a batch of materials.
Owner:COASTAL AGRI RES INST HEBEI ACAD OF AGRI & FORESTRY SCI
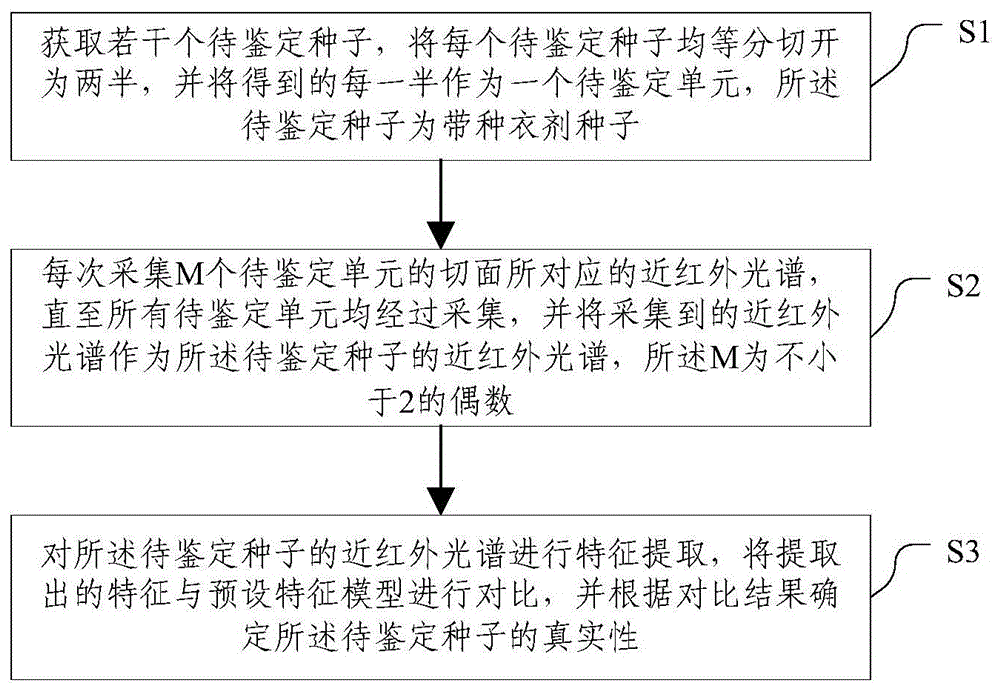
Method and system for rapidly identifying authenticity of seeds with seed coatings
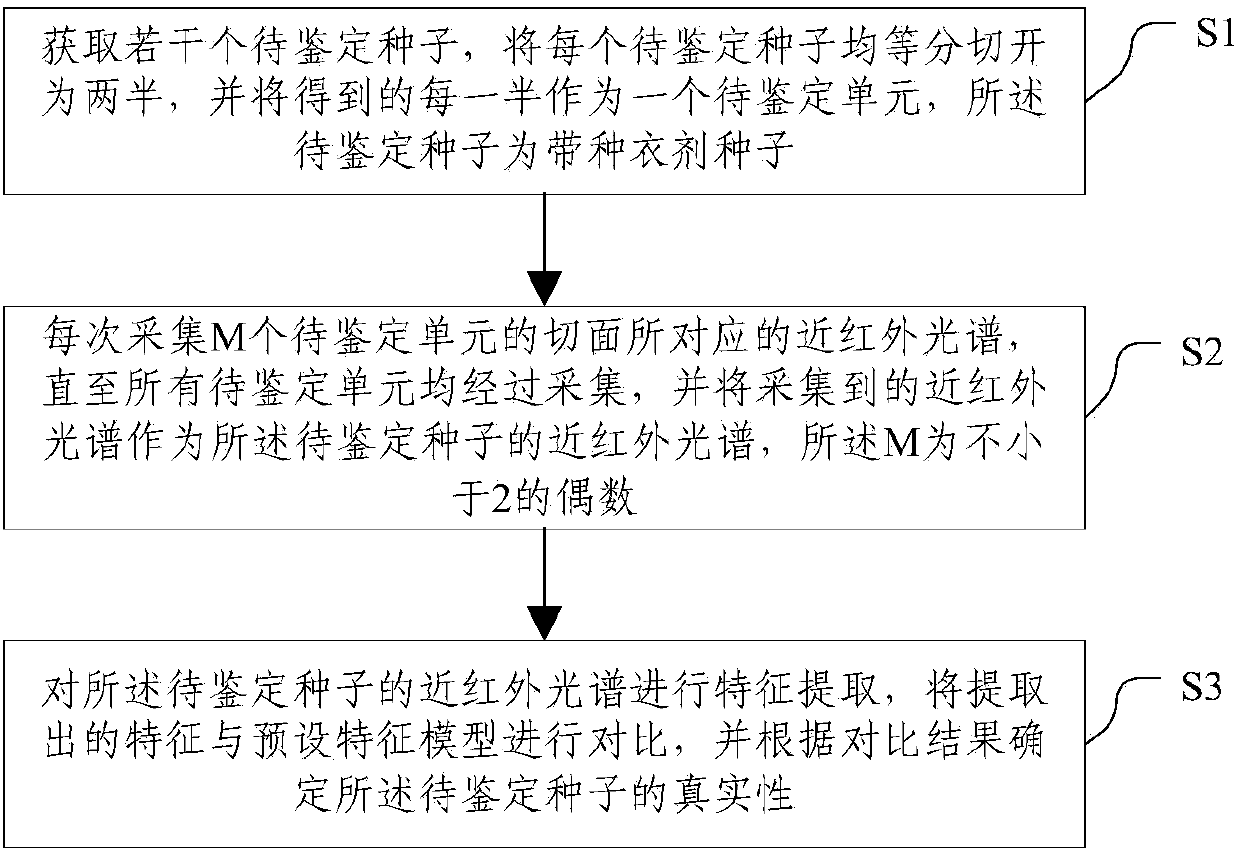
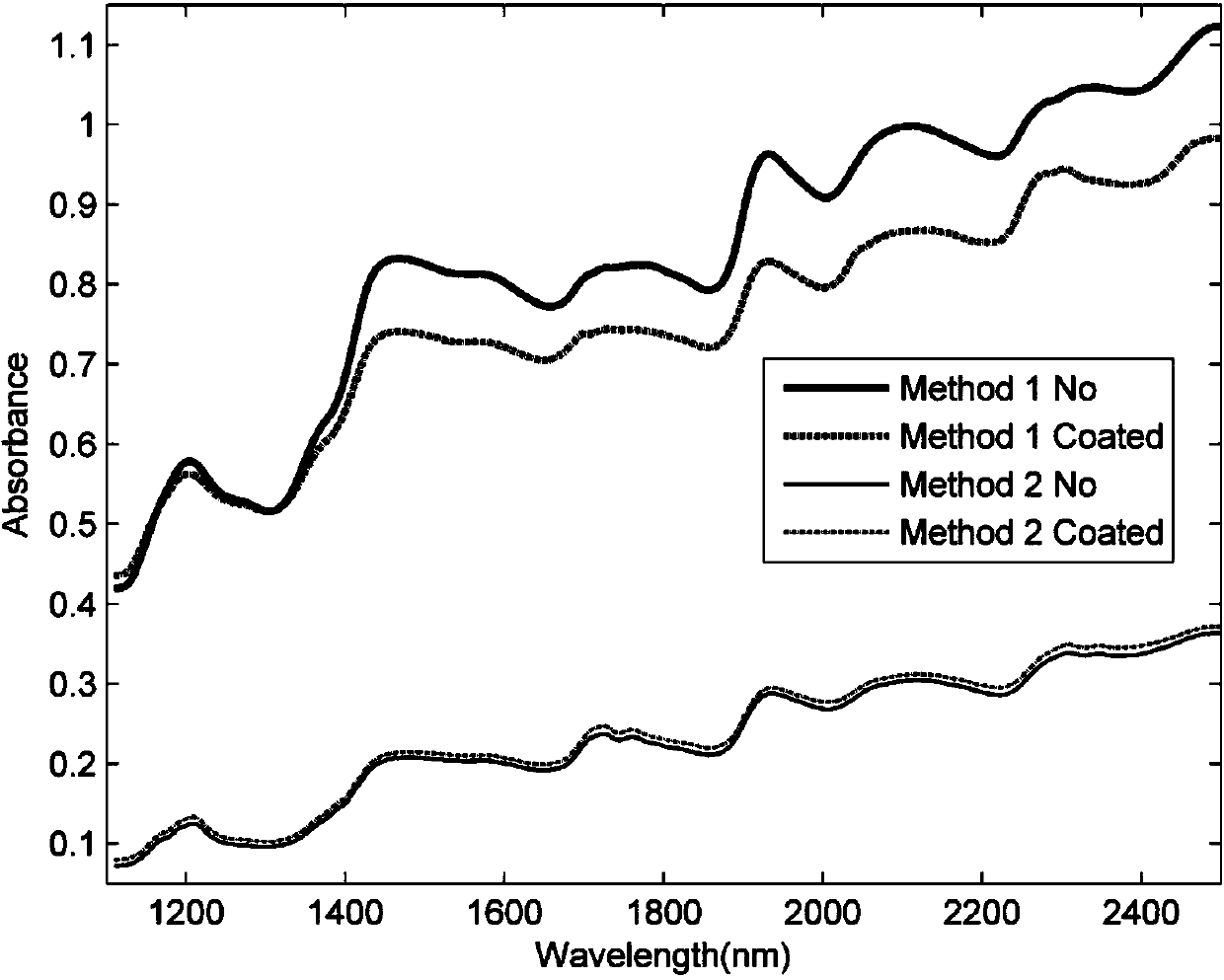
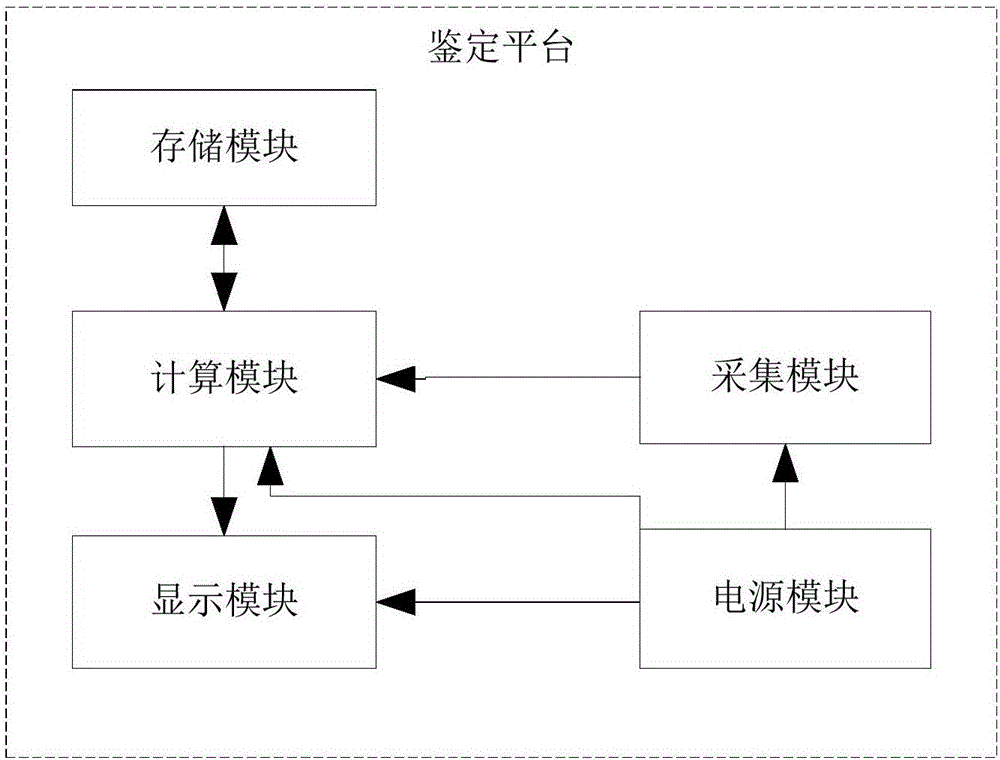
InactiveCN104198428AAchieving authenticity verificationShort identification timeMaterial analysis by optical meansFeature extractionBiology
The invention discloses a method and a system for rapidly identifying the authenticity of seeds with seed coatings, and relates to the technical field of seed authenticity identification. The method comprises the steps of S1, acquiring a plurality of seeds to be identified, and cutting each seed to be identified into two equal halves, wherein each half is taken as a unit to be indentified; S2, acquiring near-infrared spectrums corresponding to the cut surfaces of the M units to be indentified every time, wherein the acquired near-infrared spectrums are taken as near-infrared spectrums of the seeds to be identified; and S3, carrying out feature extraction on the near-infrared spectrums of the seeds to be identified, comparing extracted features with a preset characteristic model, and determining the authenticity of the seeds to be identified according to the comparison results. According to the method and the system, the seeds to be identified are treated, the near-infrared spectrums of the cut surfaces of the seeds to be identified are acquired, and the seeds to be identified can be identified according to the near-infrared spectrums, so that the authenticity of the seeds can be identified by the near-infrared spectrums, and the identification time is short.
Owner:CHINA AGRI UNIV
Method and platform for intelligently identifying redwood species
InactiveCN107179297ACause damageRapid identificationScattering properties measurementsReflection spectroscopyDiffuse reflection
The invention discloses a method and a platform for intelligently identifying redwood species, and aims to solve the technical problem that the redwood species are rapidly identified without human intervention and loss. The method includes the steps: acquiring spectrum data of redwood samples: acquiring a surface diffuse reflection spectrum of each redwood sample in advance, performing mark on surface diffuse reflection spectrums corresponding to the redwood samples, and storing the spectrum data of the redwood samples; building redwood spectrum classification models; performing classification on the spectrum data of the redwood samples to obtain and store redwood spectrum model data corresponding to each redwood species; identifying the redwood species in real time: acquiring surface diffuse reflection spectrums of redwood samples to be detected to obtain spectrum data of the redwood samples to be detected, and comparing spectrum data of the redwood samples to be detected and the redwood spectrum model data to obtain species results of the redwood samples to be detected.
Owner:北京贝叶科技有限公司
Rapid screening method for identifying cold resistance of tomatoes and application
PendingCN109618896ASuitable for growth and developmentFast identificationGrowth substratesCulture mediaCold treatmentVideo monitoring
The invention belongs to the technical field of tomato cold resistance identification, and discloses a rapid screening method for identifying cold resistance of tomatoes and an application; the rapidscreening system comprises a video monitoring module, a temperature detection module, a main control module, a culture module, a sterilization module, a watering module, a cold treatment module, a conductivity measurement module and a display module. According to the invention, a plurality of mixtures are adopted to prepare the culture medium, so that the fertilizer is better suitable for the growth and development of tomatoes; meanwhile, by utilizing the relative conductivity of isolated leaves, the damage degree of the low temperature to the cell membranes can be reflected, and by combiningthe recovery growth experiment after cold injury, the direct tolerance capability of the cells to the low temperature injury is identified, and the growth recovery capability of the material after thelow temperature is released is identified.
Owner:WENZHOU VOCATIONAL COLLEGE OF SCI & TECH
Microbial identification system and method based on Logistic four-parameter model and MASCA algorithm
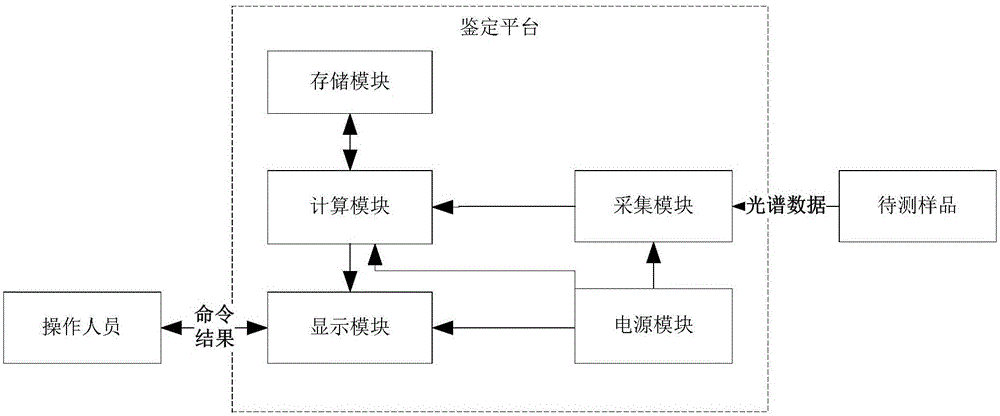
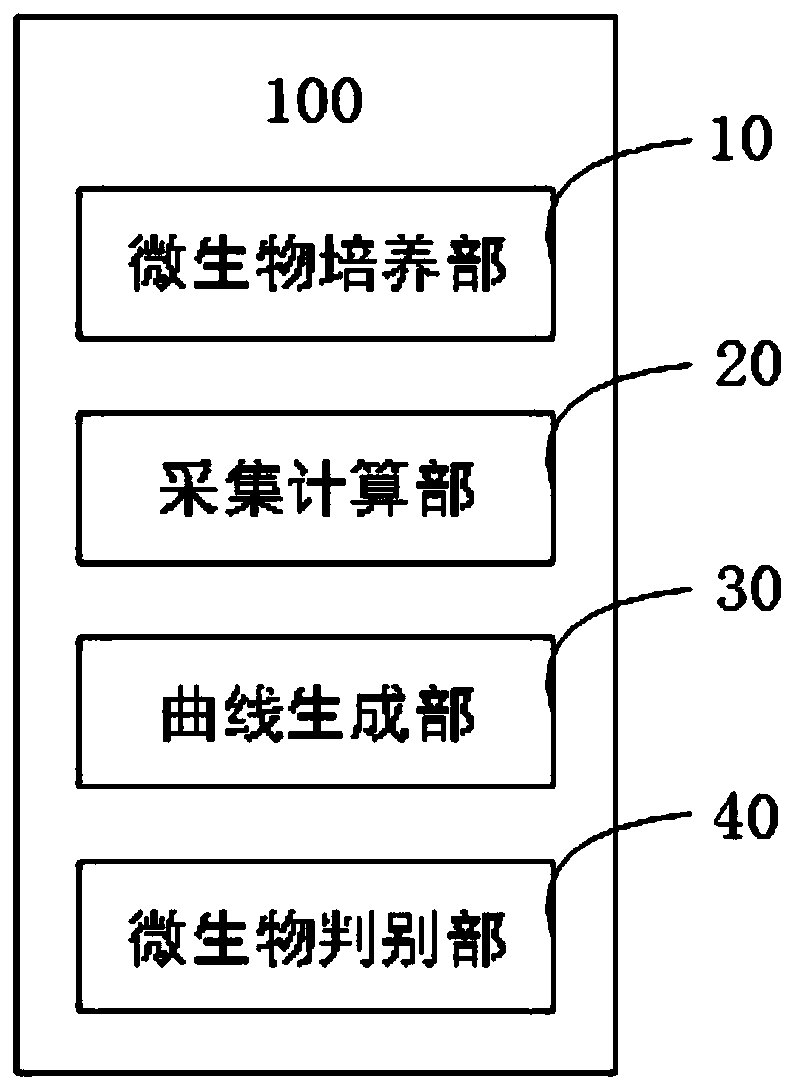
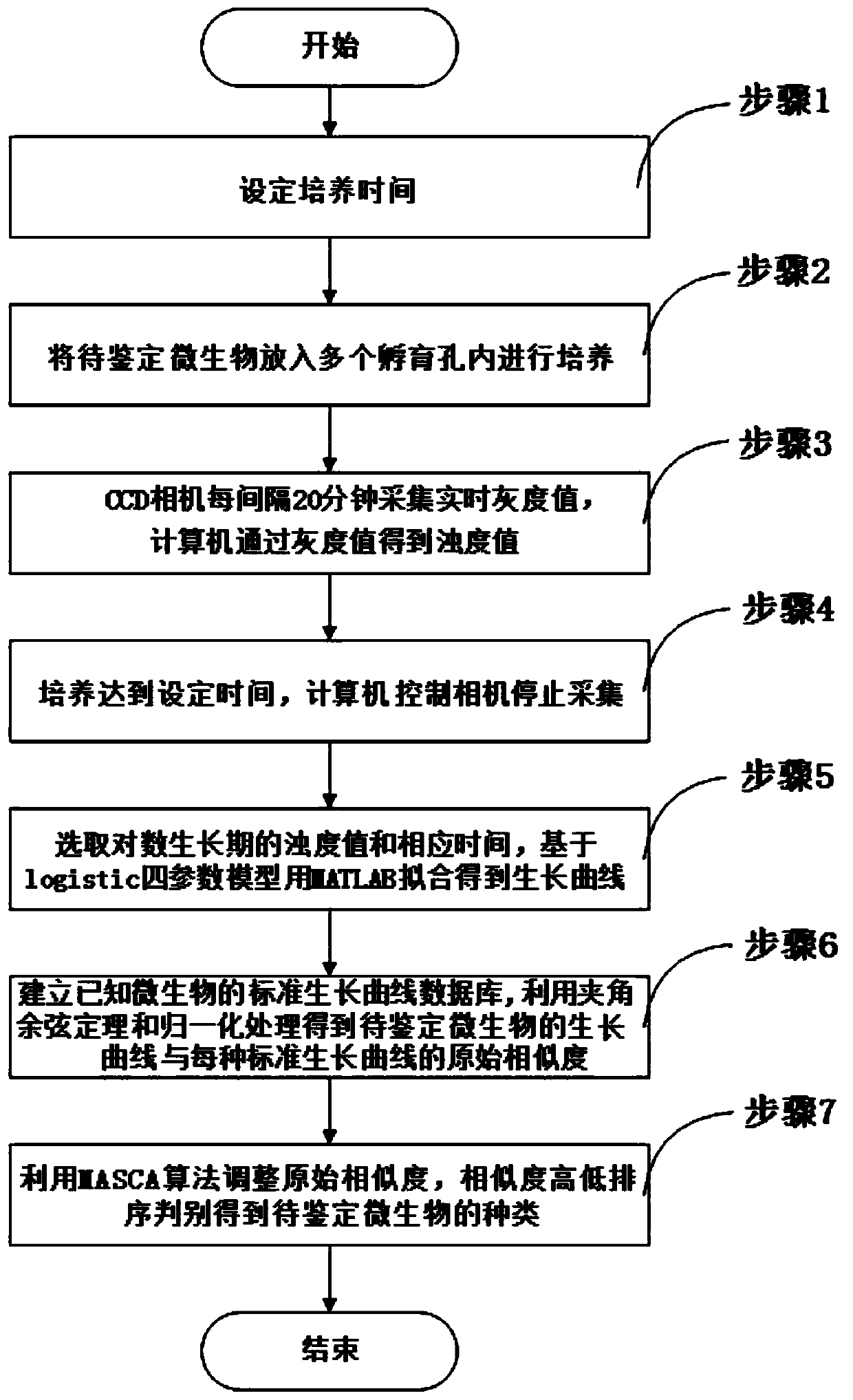
InactiveCN110218626AAvoid acquisition timeAccurate growth curveBioreactor/fermenter combinationsBiological substance pretreatmentsLaboratory cultureCcd camera
The invention provides a microbial identification system and method based on a Logistic four-parameter model and a MASCA algorithm. The system comprises a microbial culture part, an acquisition calculation part, a curve generating part and a microbial judgement part. The microbial culture part is provided with multiple incubation holes and used for culturing to-be-identified microorganisms, wherein the incubation holes are formed in an incubation plate, and a base liquid is placed in each incubation hole; the acquisition calculation part is provided with a CCD camera and a computer, wherein the CCD camera is used for acquiring an image of a bacterial liquid of the to-be-identified microorganisms in each incubation hole and obtaining the gray value of the image, and the computer is connected with the CCD camera and used for converting the gray values into microorganism bacterial liquid turbidity values on the basis of a predetermined rule; the curve generating part is used for generating a growth curve according to the turbidity values of the microorganism bacterial liquids during logarithmic growth period and the to-be-identified microorganisms in each incubation hole during corresponding growth time; the microbial judgement part is used for matching the growth curve with a standard growth curve of known microorganisms in a standard growth curve database and correspondingly obtaining the species of the to-be-identified microorganisms. According to the method, the microorganism identification system is adopted for identification, and finally the species of the to-be-identified microorganisms is obtained.
Owner:UNIV OF SHANGHAI FOR SCI & TECH
DNA (Deoxyribonucleic Acid) bar code for identifying orobanche coerulescens, and method and application for identifying orobanche coerulescens
ActiveCN106520968AImprove detection accuracyGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationOrobanche coerulescensMolecular identification
The invention relates to the technical field of molecular identification, in particular to a DNA (Deoxyribonucleic Acid) bar code for identifying orobanche coerulescens, and a method and application for identifying the orobanche coerulescens. The method for identifying the orobanche coerulescens comprises the following steps of utilizing PCR (Polymerase Chain Reaction) for amplifying an ITS2 sequence of a sample to be tested and sequencing, wherein a PCR primer amplification sequence of an ITS2 gene is SEQ ID NO.2 and SEQ ID NO.3; and if a 16bp basic group is T, a 28bp basic group is C, a 30bp basic group is T, a 32bp basic group is T, a 35bp basic group is C, and a 187bp basic group is C, determining the sample to be tested is the orobanche coerulescens. Compared with a traditional morphological and micro-morphological identification method and a DNA fingerprint marker, the method provided by the invention utilizes the ITS2 sequence for identifying the orobanche coerulescens, and an identification result can be directly obtained through carrying out PCR amplification and sequencing on the ITS2 sequence, so that the testing accuracy is high, the repeatability is high, and the identification time is short.
Owner:INSPECTION & QUARANTINE TECH CENT OF FUJIAN ENTRY EXIT INSPECTION & QUARANTINE BUREAU
DNA (deoxyribonucleic acid) bar code and application of bar code to identification of muscadine
ActiveCN107779521AVersatileImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationAgricultural science
The invention discloses a DNA (deoxyribonucleic acid) bar code. The nucleotide sequence of the DNA bar code is as shown in SEQ ID NO.1. According to the DNA bar code, an ITS sequence of a muscadine isfirstly disclosed, available molecular markers are provided for species identification of the muscadine, and the DNA bar code further comprises parts of conserved sequences and has universality whenbeing used for species identification of grapes. The invention discloses a method and a primer for preparing the nucleotide sequence of the DNA bar code and can acquire sequences of an ITS area of themuscadine. The invention further discloses a molecular identification method of the muscadine and a detection primer and a kit for identifying the muscadine. PCR (polymerase chain reaction) amplification is performed by the aid of the detection primer or the kit to obtain sequences of ITS areas of the grapes, an amplification sequence and the sequence of the DNA bar code of the muscadine are compared or applied to construction of phylogenetic trees of the grapes, so that grape varieties are accurately identified, and identifying processes are simple to operate, high in efficiency and good inrepeatability.
Owner:NANJING XIAOZHUANG UNIV
Optical disc device
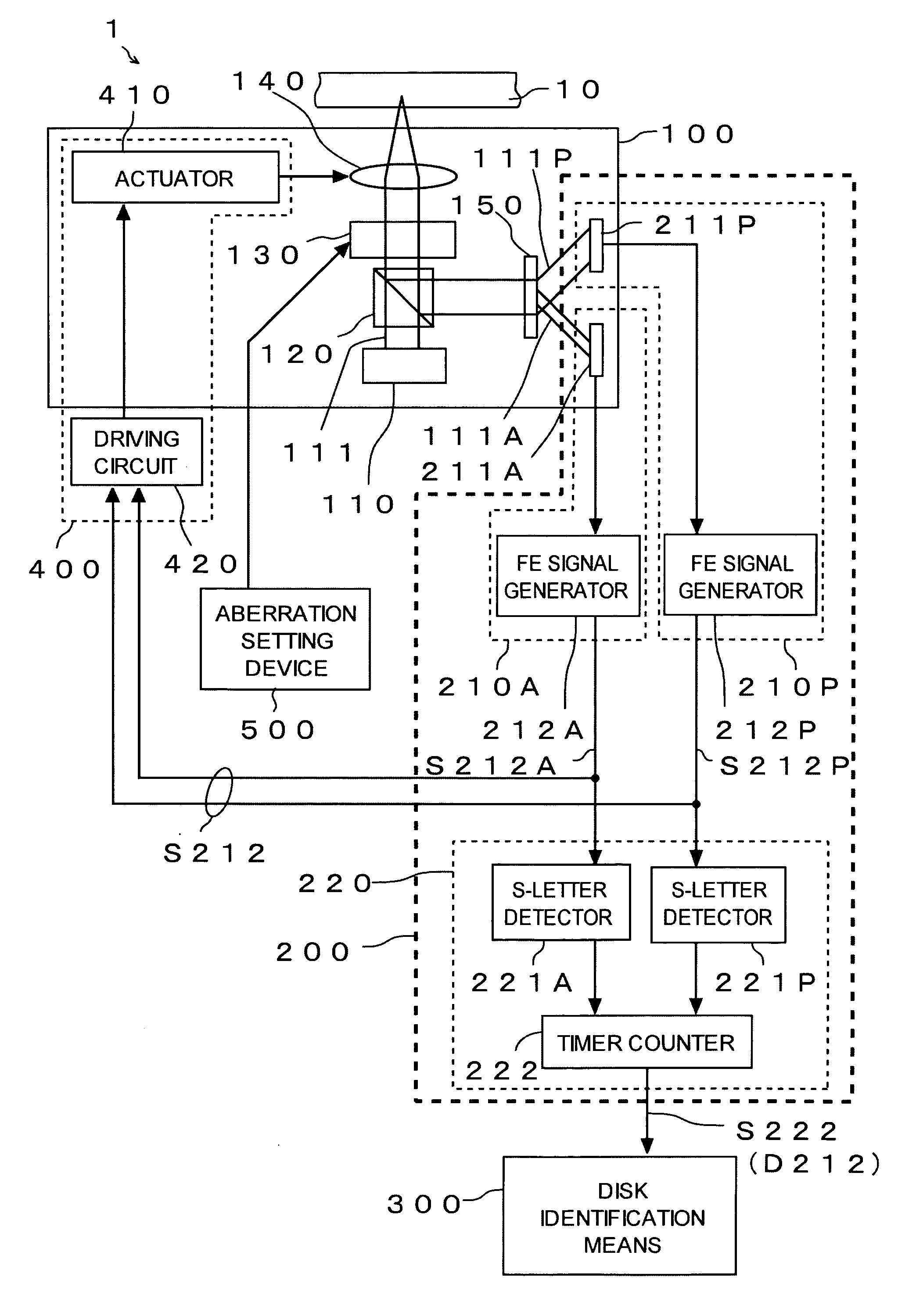
InactiveUS20060176786A1Easy to operateSmall processing loadRecord information storageOptical beam guiding meansOptoelectronicsTime difference
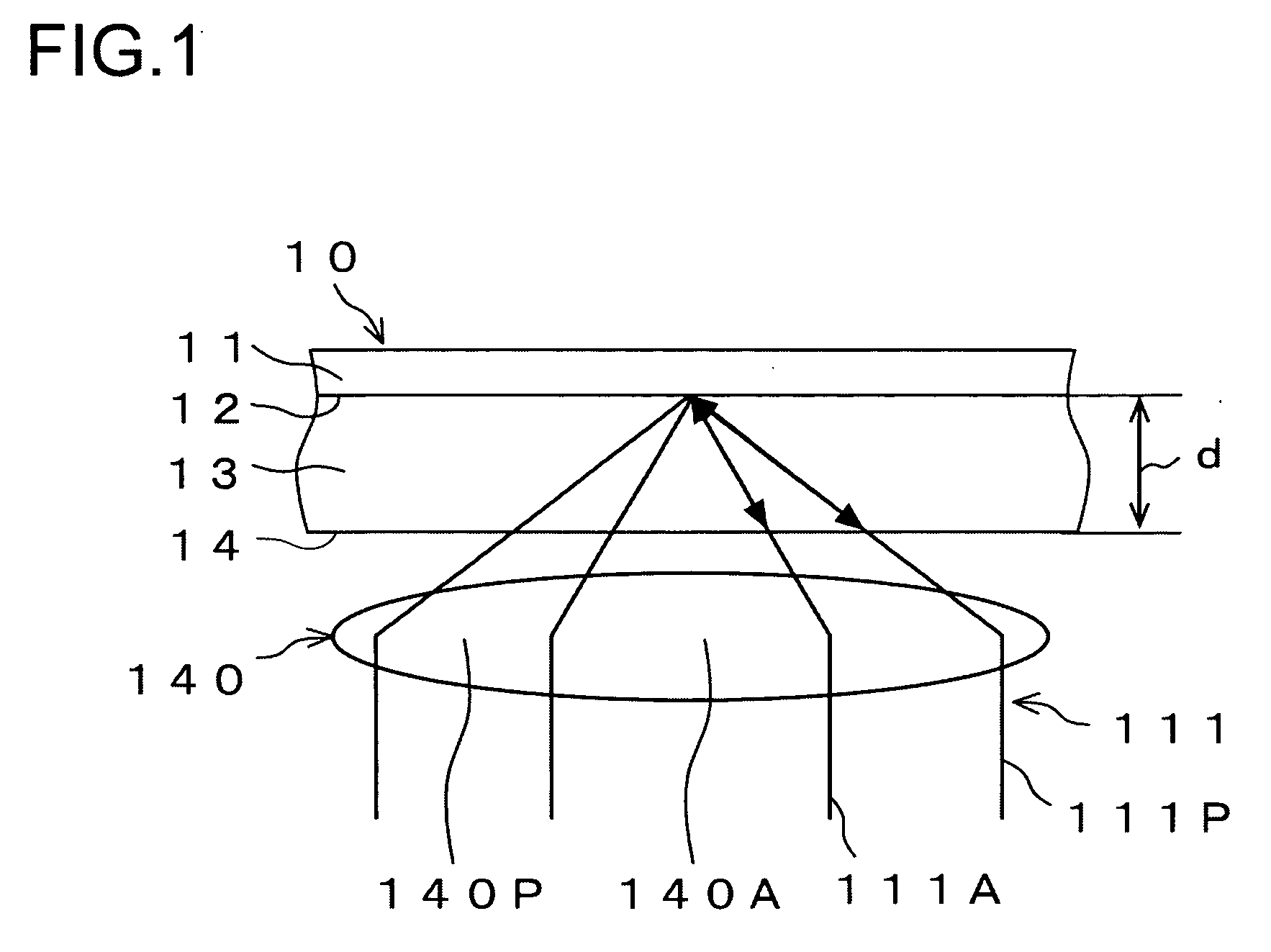
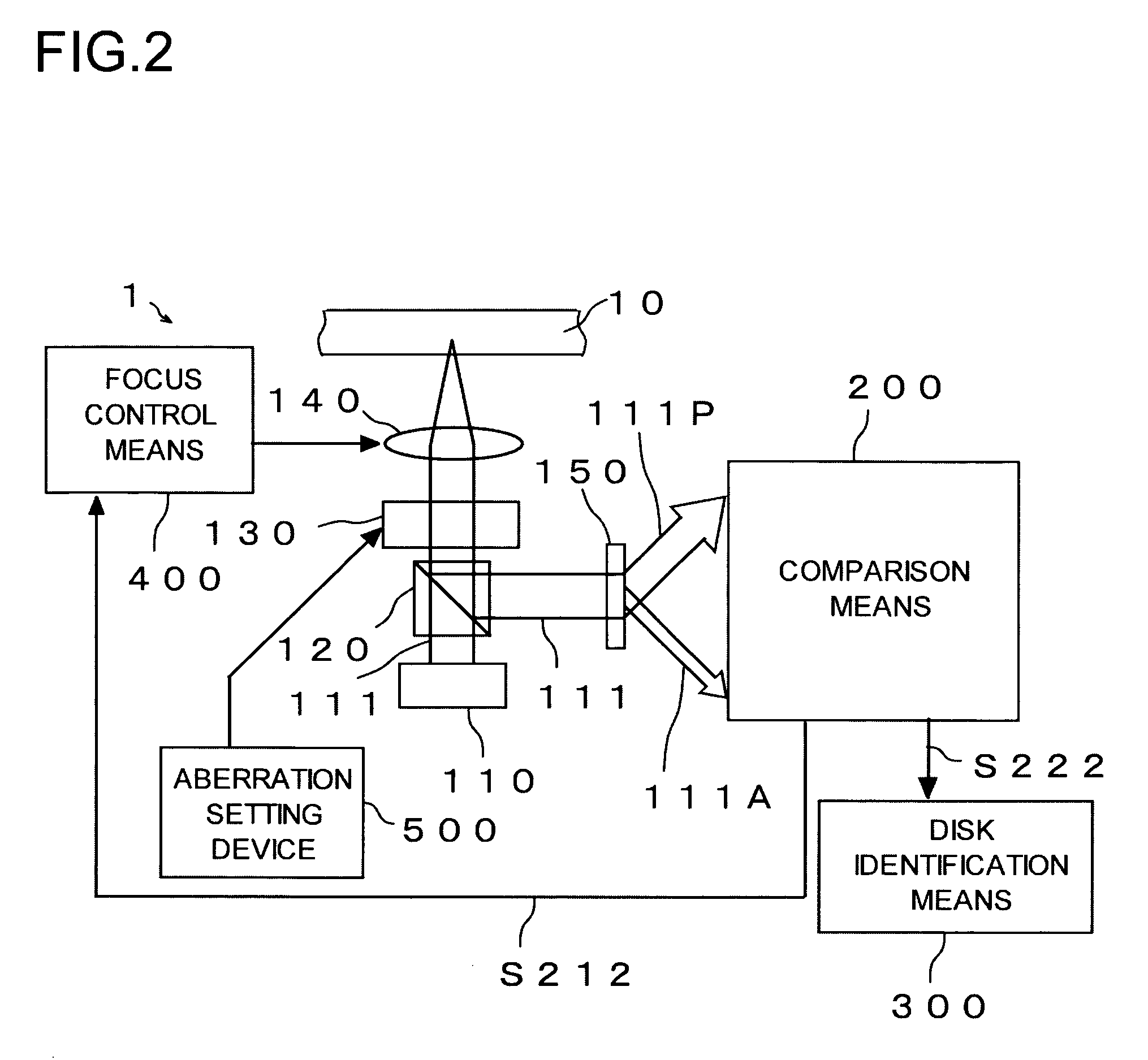
Light (111) emitted from a light source (110) is transmitted through an objective lens (140), reflected from an optical disc (10), transmitted again through the objective lens (140), and thereafter divided into central light (111A) transmitted through a central part of the objective lens (140) and peripheral light (111P) transmitted through a peripheral part of the objective lens (140) by a hologram (150). A central light FE signal (S212A) is generated from the central light (111A) and a peripheral light FE signal (S212P) is generated from the peripheral light (111P). In a focus search operation, an S-letter detector (221A) detects a time point at a zero level of the central light FE signal (S212A), and an S-letter detector (221P) detects a time point at a zero level of the peripheral light FE signal (S212P). A time difference is measured by a timer counter (222) in terms of elapsed time. Disc identification means (300) identifies a type of the optical disc (10) based on the time difference.
Owner:FUNAI ELECTRIC CO LTD
A dna barcode and its use in identifying muscadine grapes
ActiveCN107779521BVersatileImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMolecular identification
The invention discloses a DNA barcode, the nucleotide sequence of which is shown in SEQ ID NO.1. The DNA barcode disclosed the ITS sequence of Muscadine grape for the first time, which provided an effective molecular marker for the species identification of Muscadine grape. In addition, the DNA barcode also included some conserved sequences, which is universal when applied to the species identification of grapevine. The invention discloses a method for preparing the nucleotide sequence of the DNA barcode and primers, which can be used to obtain the sequence of the ITS region of muscadine grapes. The invention also discloses a molecular identification method of muscadine grapes, detection primers and a kit for identifying muscadine grapes, using the detection primers or kits to carry out PCR amplification to obtain the sequence of the grape ITS2 region, and then combining the amplified sequence with the muscadine The DNA barcode sequences of grapes are compared or applied to construct a phylogenetic tree of grapes to achieve accurate identification of grape varieties, and the identification process is simple, efficient and repeatable.
Owner:NANJING XIAOZHUANG UNIV
Molecular biological variety identification method for Huangjiaji chicken
InactiveCN101974647BImprove accuracyShort identification timeMicrobiological testing/measurementHaplotypeGenetic distance
The invention discloses a molecular biological variety identification method for a Huangjiaji chicken, which comprises the following steps: accurately acquiring COI gene sequences of mitochondrion DNAs of a Huangjiaji chicken and a tested chicken based on gene cloning and DNA sequencing technologies, calculating variable sites, haplotypes and average nucleotide differences, nucleotide degrees of ramification and net genetic distances of the Huangjiaji chicken and the tested chicken by means of DnaSP4.10.7 software, constructing colony clustering charts and haplotype phylogenetic trees of the Huangjiaji chicken and the tested chicken, and determining whether the Huangjiaji chicken and the tested chicken are of the same variety according to the results of the constructed colony clustering charts and haplotype phylogenetic trees. By identifying whether the Huangjiaji chicken is true or false based on the precise software calculation and analysis results of the differences between varieties in genetic information, the invention has the characteristics of high accuracy, short identification time, low cost and the like.
Owner:MODERN NEW AGRI GROUP
Method and system for rapid identification of authenticity of seeds with seed coating agent
InactiveCN104198428BAchieving authenticity verificationShort identification timeMaterial analysis by optical meansFeature extractionRapid identification
The invention discloses a method and system for quickly identifying the authenticity of seeds with a seed coating agent, and relates to the technical field of seed authenticity identification. The method includes: S1: obtaining several seeds to be identified, and dividing each seed to be identified equally Divide it into two halves, and use each half as a unit to be identified; S2: Collect the near-infrared spectra corresponding to the sections of M units to be identified each time, and use the collected near-infrared spectra as the seeds to be identified S3: performing feature extraction on the near-infrared spectrum of the seeds to be identified, comparing the extracted features with a preset feature model, and determining the authenticity of the seeds to be identified according to the comparison results. The present invention collects the near-infrared spectrum of the cut surface of the seed to be identified by processing the seed to be identified, and realizes the identification of the seed to be identified according to the near-infrared spectrum, which not only realizes the authenticity identification of the seed by using the near-infrared spectrum, but also shortens the identification time .
Owner:CHINA AGRI UNIV
Pepper anthracnose resistance living body identification method and application
PendingCN114606292AReliable true resistance levelControllable environmental conditionsMicrobiological testing/measurementBiological material analysisSporelingDisease
The invention provides a pepper anthracnose resistance living body identification method and application. The method specifically comprises the steps of cultivating pepper seedlings, preparing pepper colletotrichum spore suspension, inoculating pepper colletotrichum living bodies, managing after inoculation, investigating illness conditions, evaluating variety resistance and the like. The method comprises the following steps: inoculating pepper seedlings by spraying colletotrichum spore suspension when the pepper seedlings grow to have 4-6 true leaves, investigating disease indexes of each plant according to an established pepper anthracnose seedling stage disease grading standard in the fifth to seventh days after inoculation, and evaluating the anti-infection characteristics of different pepper varieties. The method provided by the invention is a pepper anthracnose living body inoculation identification method, the operation steps are simple and efficient, the identification period is short, the result is reliable and stable, the repeatability is high, the resistance level of an identification material can be accurately reflected, and a technical support is provided for pepper anthracnose resistance resource screening and disease-resistant variety breeding and evaluation.
Owner:天津市农业科学院
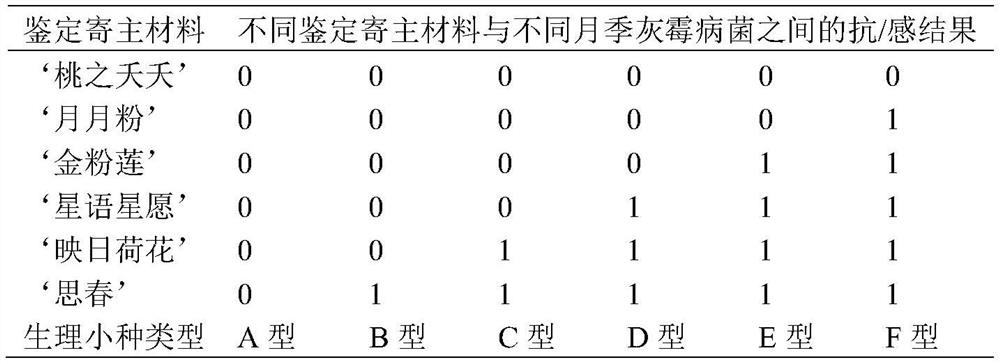
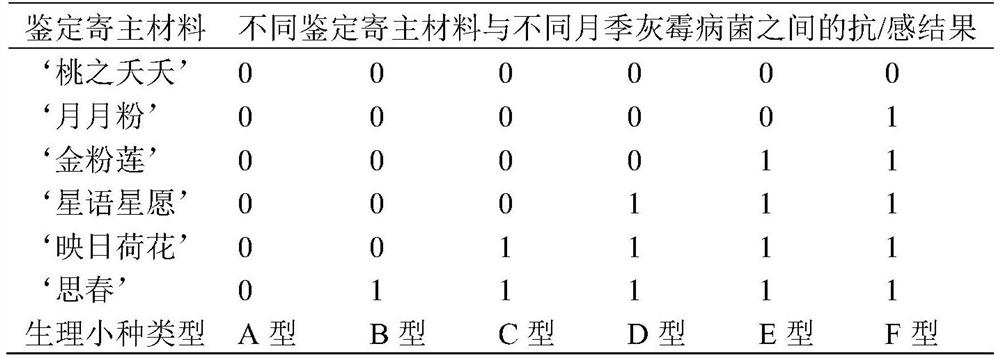
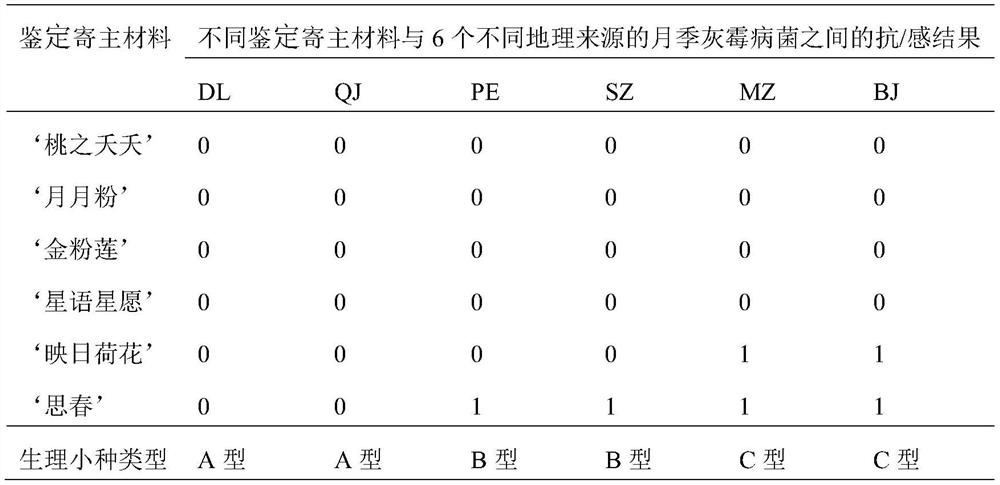
Identification method of Chinese rose botrytis cinerea physiological race
InactiveCN112501240AAccurate and efficient identificationLow costMicrobiological testing/measurementSporelingBotryotinia polyblastis
The invention discloses an identification method of a Chinese rose botrytis cinerea physiological race. The method comprises the steps of: carrying out separation, purification and propagation expansion on botrytis cinerea; preparing Chinese rose botrytis cinerea spore suspension; adopting 6 parts of identification host materials; carrying out in-vitro petal identification; and grading petal disease conditions. According to the method, only by six parts of identification host materials, the Chinese rose botrytis cinerea physiological race can be efficiently and accurately identified in variousregions of China, the cost is saved, and labor and time are saved.
Owner:FLOWER RES INST OF YUNNAN ACAD OF AGRI SCI
The application of its sequence as a dna barcode in the identification of Jiaxing Plum and its identification method
ActiveCN105671185BImprove detection accuracyGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationPrunus salicinaMolecular identification
The invention discloses application of ITS (internal transcribed spacer) sequence as a DNA barcode in identifying Jiaxing Zuili (Prunus salicina lindl.) and an identifying method and belongs to the technical field of molecular identification. The identifying method comprises: amplifying ITS sequence of a sample to be tested by using PCR (polymerase chain reaction) and carrying out sequencing; results show that if both basic group 165bp and basic group 195bp of the ITS sequence are 'T', it is determined that the sample to be tested is Jiaxing Zuili. The invention also discloses a method for identifying early maturing and late maturing varieties of Jiaxing Zuili; if the basic group sequence of the ITS sequence is shown as in SEQ ID No. 1, it is determined that the sample to be tested is an early maturing variety of Jiaxing Zuili; if the basic group sequence of the ITS sequence is shown as in SEQ ID No. 2, it is determined that the sample to be tested is a late maturing variety of Jiaxing Zuili. The ITS sequence is used herein as the DNA barcode to identify Jiaxing Zuili, detection accuracy is high, detection repeatability is high, and identifying time is short.
Owner:ZHEJIANG JIAXING AGRI SCI ACADEMY INST
Method for rapidly identifying tomato bacterial wilt seedling stage resistance through injection inoculation method
PendingCN113125643AThe method is simple and fastImprove identification efficiencyTesting plants/treesBiotechnologyRalstonia solanacearum
The invention belongs to the technical field of agricultural breeding, and provides a method for rapidly identifying tomato bacterial wilt seedling stage resistance by an injection inoculation method, wherein the method comprises the steps of tomato material seedling preparation, ralstonia solanacearum inoculation liquid preparation, injection inoculation and disease resistance observation and identification. The injection inoculation comprises the steps: when a tomato seedling grows to have 5-6 leaves, sucking ralstonia solanacearum inoculation bacterial liquid by using an injector, aligning a needle head of an injector to the middle of a first stem of the seedling, puncturing the stem along the inclined lower direction of the stem, returning the needle head to a position close to a puncturing point in the stem, and injecting the bacterial liquid until the bacterial liquid overflows from the stem. According to the method, the bacterial wilt resistance of a breeding population material can be rapidly identified in the seedling stage, the tomato bacterial wilt resistance breeding efficiency is improved, and a simple, convenient and rapid detection method can be provided for research on tomato bacterial wilt resistance germplasm resource screening, a disease-resistant molecular mechanism and the like.
Owner:NINGBO ACAD OF AGRI SCI
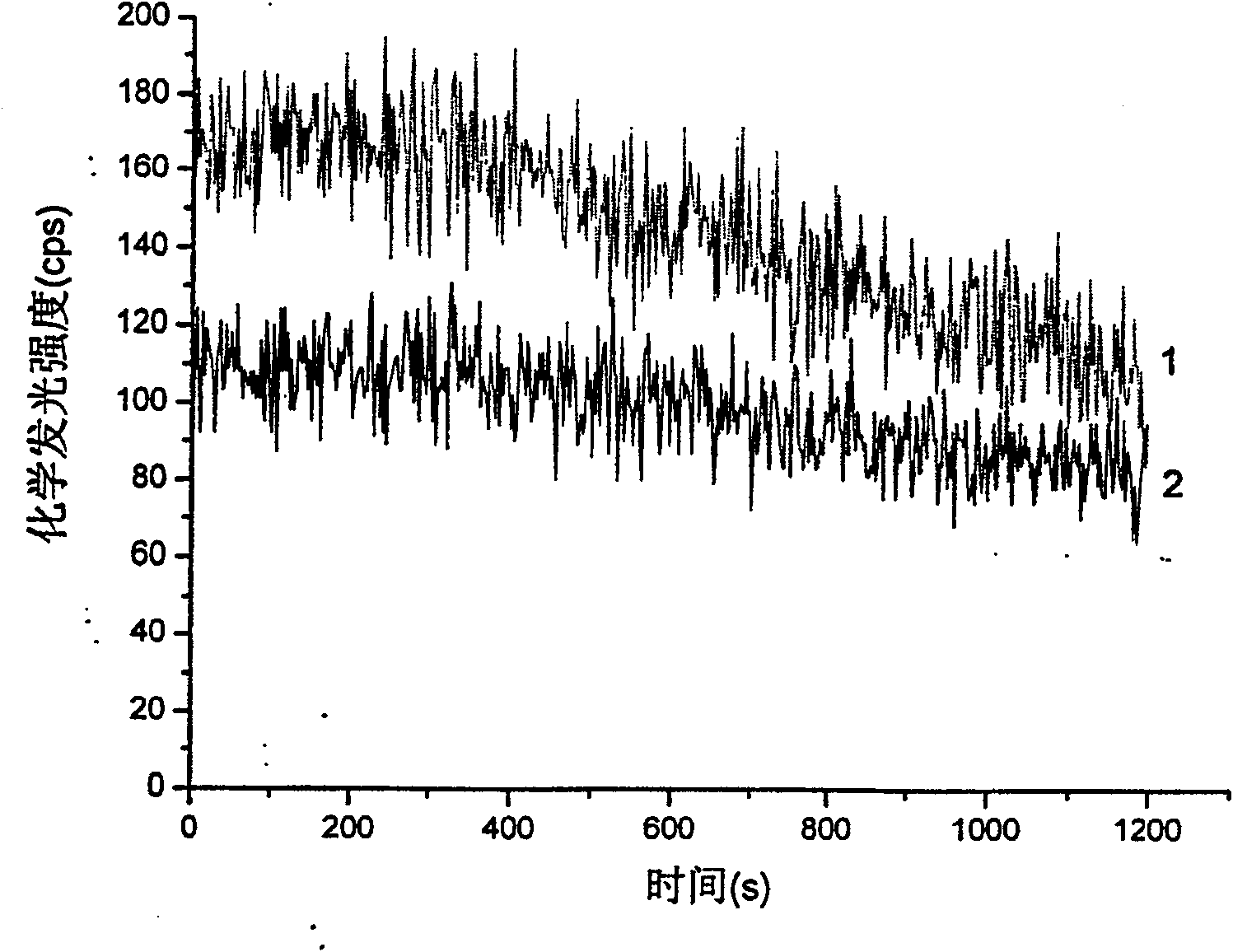
True or false and new or old paddy rice seed identifying method
InactiveCN100573110CEasy to measureRapid determinationChemiluminescene/bioluminescencePreparing sample for investigationUltimate tensile strengthBiology
Owner:SOUTH CHINA NORMAL UNIVERSITY
Bacterium identification reagent kit as well as preparation method and uses thereof
InactiveCN101200755BHigh sensitivityRapid and direct determination of growth statusMicrobiological testing/measurementBiotechnologyBacteria identification
Owner:SOUTH CHINA AGRI UNIV
A Method for Quickly Identifying Drought Resistance of Wheat Using Leaf Regreening Ratio
InactiveCN107873435BHigh simulationAvoid errorsGrowth substratesCulture mediaGermplasmGenetic resources
The invention discloses a method for quickly identifying the drought resistance of wheat by using the green recovering rate of leaves. Drought stress treatment is performed in a three-leaf period of wheat, the time from dryness to green recovering of the leaves and the ratio of the leaves which recover green are detected after rehydration, and the drought resistance of the wheat can be identifiedquickly through comparison and rating; the result of drought resistance identification is in good accordance with the result of a method for identifying drought resistance indexes. The method has theadvantages of rapidity, high efficiency, low cost, easy operation and the like, field water management of farmers can be guided during production, and through the cooperation with other drought resistance identifying methods in scientific researches, excellent drought-resistant genetic resources can be selected quickly in a batch of materials.
Owner:COASTAL AGRI RES INST HEBEI ACAD OF AGRI & FORESTRY SCI
Method for fast identifying type of gene alternative splicing and application thereof
InactiveCN105821121AReduce experimental stepsEasy interpretation of resultsMicrobiological testing/measurementTissue sampleRapid identification
The invention provides a method for fast identifying the type of gene alternative splicing. The method comprises the steps of preprocessing, sample detection and data analysis. Compared with the prior art, the method has the advantages of being wide in application range, small in number of experiment steps, short in time consumed in detection, simple in result interpretation, large in detection flux, high in accuracy, small in error and low in detecting cost. Meanwhile, according to the detecting method, inverse transcription, amplification and qualititation of a gene segment are completed in a closed tube at one step, and the method is very simple, convenient to use, suitable for clinical sample studies and especially suitable for analyzing the alternative splicing types of clinical peripheral blood and tissue sample genes. Clinical experiments verify that the method is fast, efficient and practical, has high theory and application value on the aspect of studying gene alternative splicing, and therefore application prospects are very wide.
Owner:HUNAN UNIV OF SCI & TECH
A rapid judgment method of "fried charcoal storage" prepared from orientalis leaves
ActiveCN107907623BEfficient separationShort identification timeComponent separationAdditive ingredientCarbon storage
The invention relates to a rapid judgment method for 'carbonizing' of processing of cacumen biotae. A method for simultaneously identifying quercitrin and quercetin by TLC is used as a measuring basisof 'carbonizing' of the cacumen biotae, a method of secondary extending on the same thin layer is adopted, two ingredients including the quercitrin and the quercetin are identified simultaneously, and thus, it shows that requirements of 'carbonizing' of the cacumen biotae are met. By the method, the two ingredients including the quercitrin and the quercetin which show 'carbonizing' of the cacumenbiotae can be simultaneously identified on the same thin layer plate, and thus, judgment basis of 'carbonizing' of the cacumen biotae can be obtained. The identifying time is short, the method is accurate, stable and feasible, effective separation of the quercitrin from the quercetin in cacumen platycladi carbonisatum can be realized, and after separation, spots are clear.
Owner:SHANDONG ACAD OF CHINESE MEDICINE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com