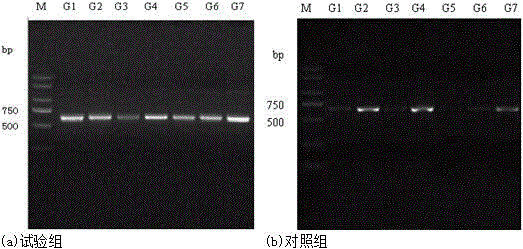
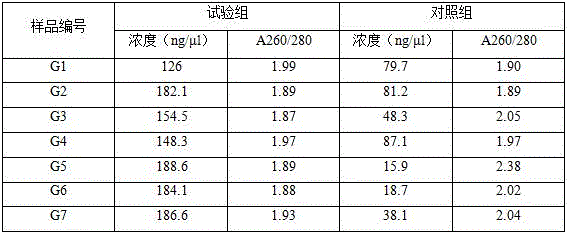
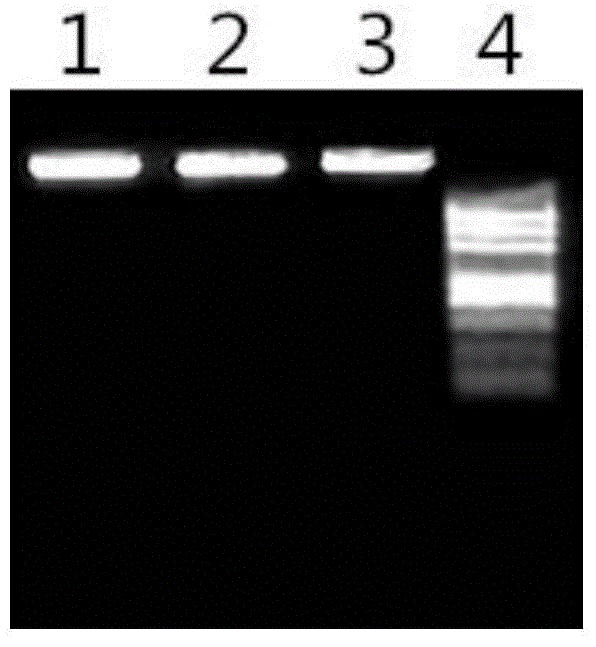
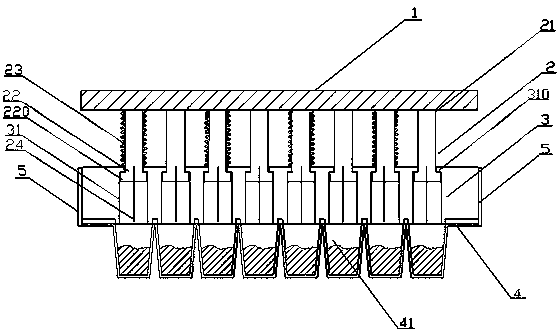
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
83 results about "Dna adsorption" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA adsorption takes place mainly via the phosphate backbone, although the bases might also have moderate contributions. Peptide nucleic acids (PNAs) with an amide backbone cannot be adsorbed. DNA adsorption is strongly affected by inorganic anions, where phosphate and citrate can strongly inhibit DNA adsorption.
Multifunctional integrated microfluidic nucleic acid analysis chip and preparation and analysis method thereof
ActiveCN105349401AQuick buildEasy to operateBioreactor/fermenter combinationsBiological substance pretreatmentsInjection mouldingMultiple layer
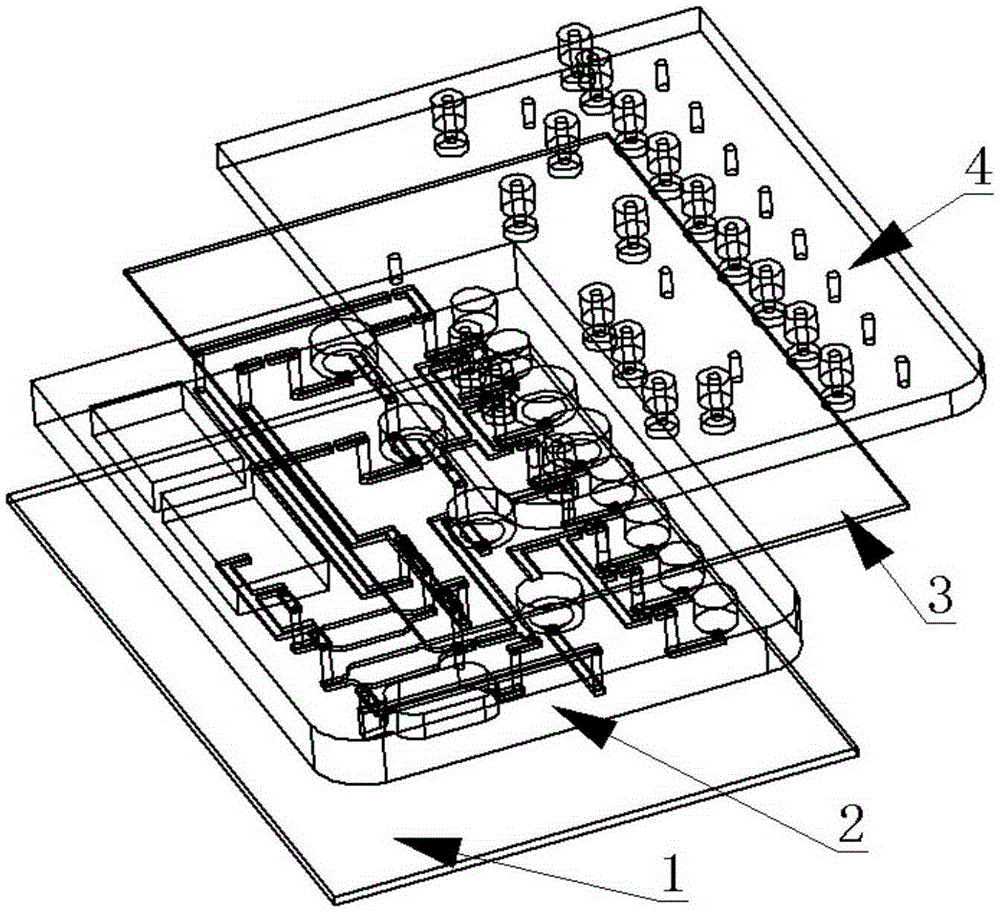
The invention provides a multifunctional integrated microfluidic nucleic acid analysis chip. The chip is prepared by preparing a combination mould according to 3D printing or micromachining technology, generating a microfluidic substrate according to an injection moulding method, and completing bonding of multiple layers including a chip substrate, a liquid circuit layer, an elastic film, a gas circuit layer and the like by means of plasma cleaning to finally obtain the complete chip. The chip integrates various structures including reagent cavities, mixing cavities, a splitting cavity, a DNA (deoxyribonucleic acid) extraction cavity, amplification reaction cavities, a microvalve, a microchannel and the like and has functions of sample loading, reagent mixing, heating for splitting, DNA adsorption, cleaning, eluting, amplification reaction and the like. The multifunctional integrated microfluidic nucleic acid analysis chip has the advantages that totally-closed automatic operation of 'sample input-result output' is realized, complicated manual operations are avoided, and detection efficiency is improved.
Owner:合肥中科易康达生物医学有限公司
Preparation method for surface plasmon resonance DNA sensor based on graphene oxide
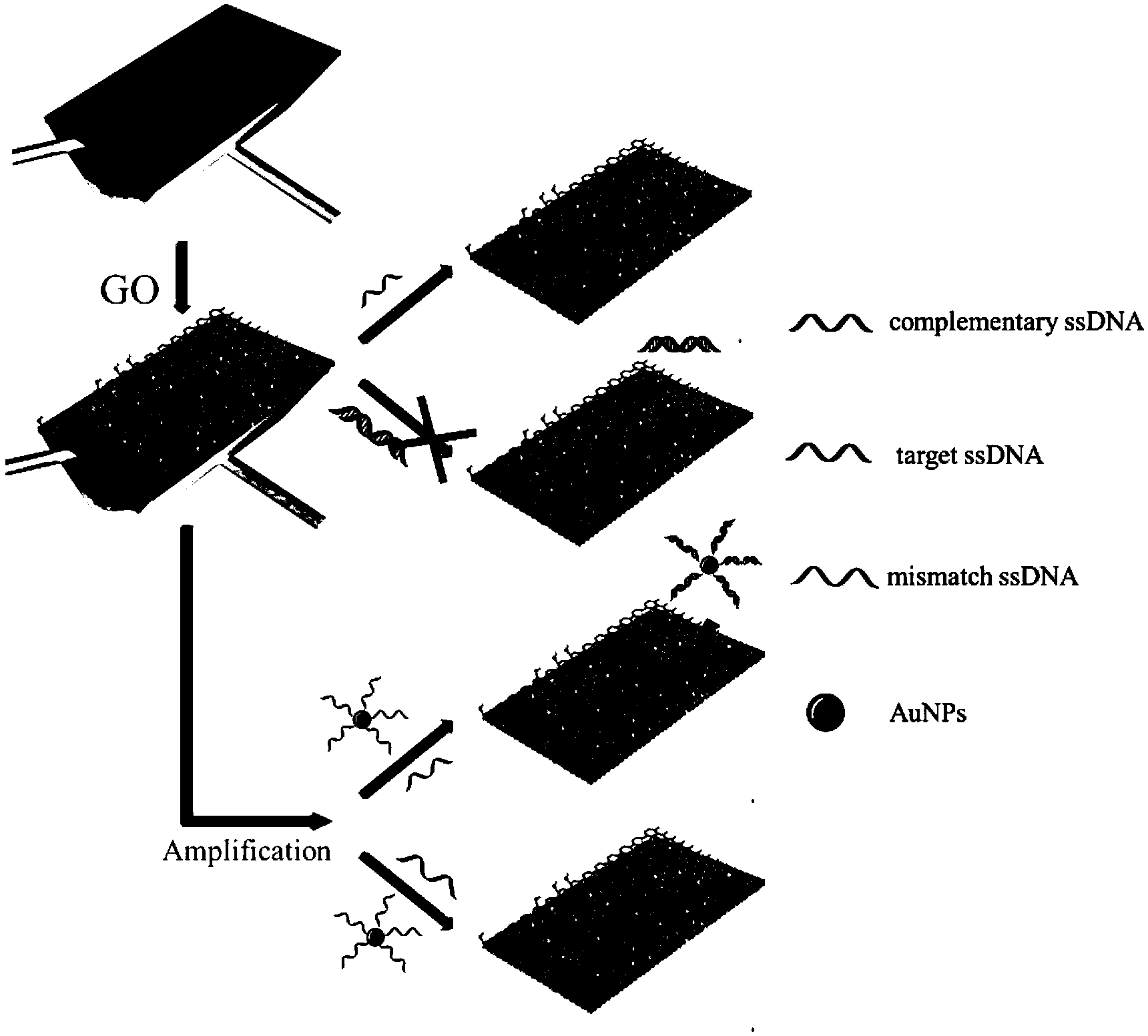
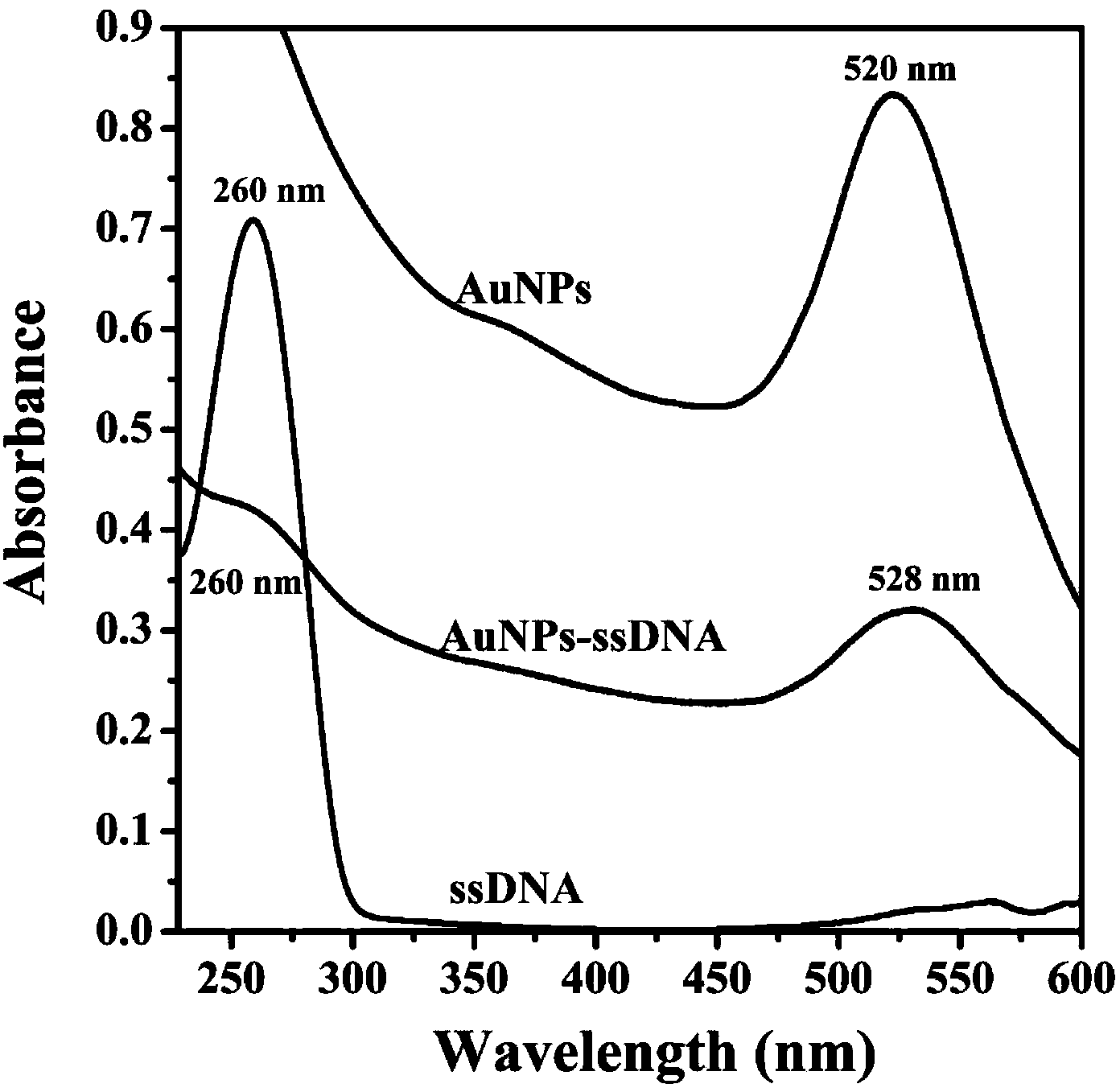
InactiveCN103411933ASensitive detectionSolving the difficult problem of lowering the detection limitAnalysis by material excitationResonance angleDna adsorption
The invention discloses a preparation method for a surface plasmon resonance (SPR) DNA sensor based on graphene oxide, and belongs to the technical field of nanometer material biology. The technical problem to be solved in the preparation method is that the distinctive effect of graphene oxide-DNA is utilized, the surface plasmon resonance technology and the signal amplification mechanism of the AuNPs are applied, based on the competitive inhibition method, the phenomenon that the single-stranded DNA with different concentrations is absorbed on the surface of a sensing chip to cause SPR spectral change, and the single-stranded DNA is detected through the linear change of the resonance angle. Through the adoption of the preparation method, the SPR technology is adopted, the GO (Graphene Oxide) is utilized to assemble the surface of the chip, the single-stranded DNA is sensitively detected through adopting the competitive inhibition method and the AuNPs signal amplification effect, the concentration of the single-stranded DNA is quantitatively detected through analyzing the change of the SPR peak, and the detection limit is ultra-low. The preparation method has the advantages as follows: the instrument and equipment are low in cost, the cost is low, the operation is simple, the efficiency is high, the precision is high, and the detection limit is ultra-low.
Owner:JILIN UNIV
Kit and extraction method for quickly extracting microbial genome DNA from animal fecal microorganisms
InactiveCN104531680AReduce extraction timeSimplify the experimental stepsDNA preparationDecompositionAnimal feces
The invention discloses a kit and an extraction method for quickly extracting microbial genome DNA from animal fecal microorganisms. The extraction method for extracting microbial genome DNA from animal fecal microorganisms comprises the following five steps: pre-treatment of feces; splitting decomposition of microbial cells in feces; dissociation of DNA and abstraction of impure proteins; DNA adsorption by a DNA adsorption column and removal of impurities; and elution of DNA. The kit disclosed by the invention is capable of absorbing the genome DNA by a silicone mold adsorption column and removing impurities such as proteins by using special deproteinization liquid and wash liquid. Compared with conventional extraction methods for extracting microbial genome DNA from animal fecal microorganisms, the microbial genome DNA from animal fecal microorganisms can be extracted within 4 hours, so that not only is a lot of extraction time shortened, but also the experimental steps are simplified. Therefore, the microbial genome DNA from animal fecal microorganisms can be effectively, quickly and economically extracted.
Owner:FUJIAN NORMAL UNIV
Kit for rapid extraction of plant seed DNA, and application thereof
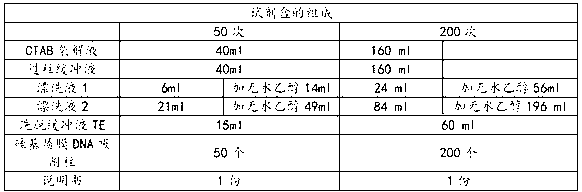
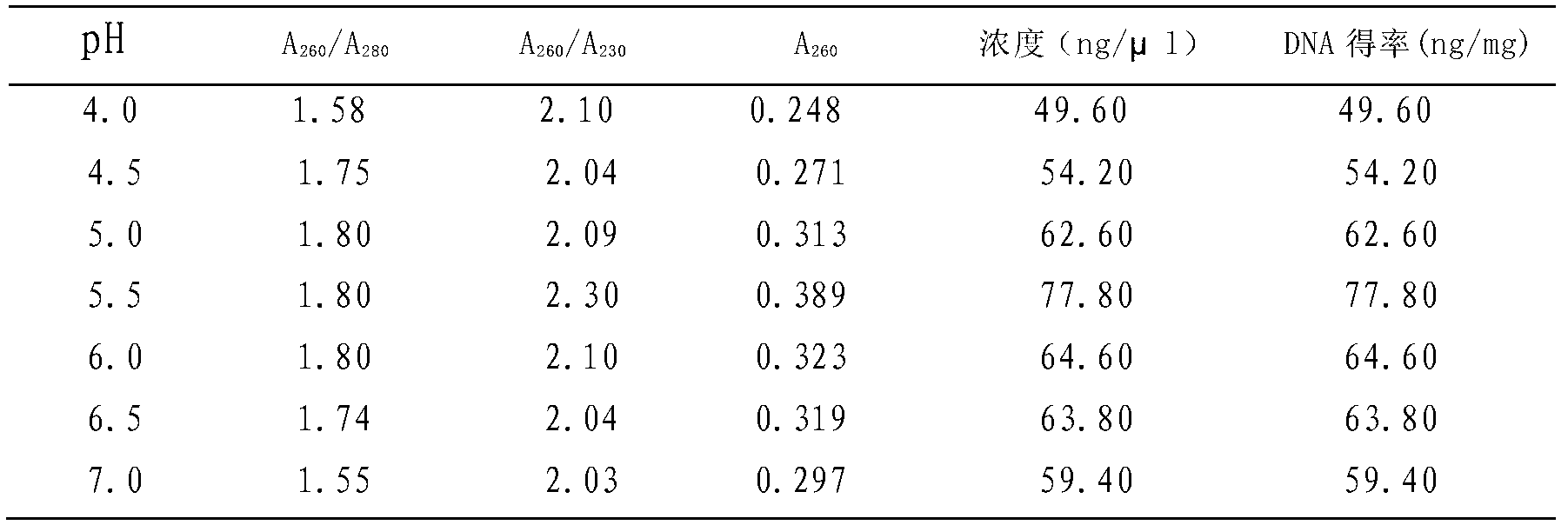
The invention discloses a kit for rapid extraction of plant seed DNA, and an application thereof. The compositions of the kit comprise: a CTAB lysate, a silicon-based plasmalemma DNA adsorption column, an RNA enzyme, a buffer solution for passing through a column, a rinsing solution 1, a rinsing solution 2 and an eluting buffer solution; the compositions of the buffer solution for passing through the column include: 2.5-7.0 M of guanidine hydrochloride, 0.05 M of Tris-HCl, and 0.01 M of EDTA; and the pH value is 4.0-7.0. The invention further discloses the application of the kit for the rapid extraction of the high quality plant seed DNA. The kit can effectively improve yield and purity of the extraction of the plant seed DNA, has the extracted DNA with good integrity, high purity and high yield; while decreasing DNA extraction cost, the kit effectively improves extraction quality and extraction efficiency of the plant seed genomic DNA.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
Souvenir containing DNA and production thereof
InactiveCN1817263AQuick extractionHigh puritySpecial ornamental structuresBraceletsDna adsorptionChemistry
A souvenir containing DNA is composed of a full-sealed container and the resin bead or glass bead in which the DNA is adsorbed. Its preparing process includes such steps as extracting DNA from a human body, adsorbing the DNA by resin bead or glass bead, loading the resin bead or glass bead in said container, sealing and vacuumizing.
Owner:哈尔滨基太生物芯片开发有限责任公司
Extraction method of rosewood heartwood genome DNA (deoxyribonucleic acid)
The invention discloses an extraction method of rosewood heartwood genome DNA (deoxyribonucleic acid). The extraction method comprises the following steps: grinding the rosewood heartwood into powder in liquid nitrogen; adding CTAB (Cetyltrimethyl Ammonium Bromide) splitting liquid to the obtained powder for splitting; adding cellulase to a split product for continuous splitting, centrifuging the obtained split product, taking supernatant, adding a mixed solution of phenol, chloroform and isoamyl alcohol, centrifuging and taking supernatant; transferring the obtained supernatant to a DNA adsorption column, centrifuging, discarding lower liquid, washing the adsorption column by using ethyl alcohol, centrifuging and discarding lower liquid; adding a conventional reagent for dissolving the DNA to the DNA adsorption column, and centrifuging, thus obtaining the rosewood heartwood genome DNA. The extraction method of the rosewood heartwood genome DNA is used for solving the defect of low quality of the genome DNA extracted from the rosewood heartwood by using a conventional genome DNA extraction method, extracting the genome DNA from the rosewood heartwood and preparing the high-quality genome DNA; the obtained genome DNA has extremely low pollution and can be used for PCR (Polymerase Chain Reaction) amplification identification and the like.
Owner:SHANGHAI INST OF ORGANIC CHEM CHINESE ACAD OF SCI +1
DNA extracting method needing no tube transferring
InactiveCN106434639ASimplify the extraction processAvoid lostMicrobiological testing/measurementDNA preparationElectrophoresisDna adsorption
The invention belongs to the technical field of nucleic acid extraction and purification and particularly relates to a DNA extracting method needing no tube transferring. The DNA extracting method includes the steps of firstly, preparing reagents, to be more specific, preparing lysate, washing liquid and eluent, and preparing a combined tube; secondly, extracting DNA, to be more specific, placing a sample into the inner-layer splitting tube of the combined tube, adding the lysate and protease k, then performing splitting, centrifuging, allowing liquid in the inner-layer splitting tube to be combined by a nucleic acid absorption film when the liquid penetrates the nucleic acid absorption film in a middle-layer DNA absorption tube, and separating to obtain purified DNA; thirdly, performing PCR amplification and electrophoresis. The DNA extracting method has the advantages that the tube transferring is not needed, the DNA extracting steps are simplified, and detecting time is shortened; DNA loss and cross contamination risks possibly caused by the tube transferring are avoided.
Owner:ANHUI SENPENG BIOTECHNOLOGY CO LTD
Kit for extracting microbial genome DNA from soil and its method
InactiveCN1936005AReduce lossesReduce physical health damageFermentationPlant genotype modificationImpurityMicroorganism
The invention relates to an agent box to distill microorganism gene group DNA from earth and the distilling method. The agent box includes agent A(immersion fluid), agent B(pretreatment fluid), agent C(cracking fluid), and agent D that is made up of RNA enzyme A, agent E Al2(SO4)3, agent F hexadecyl trimethyl amine, and agent G(DNA cleaning fluid). The distilling method includes the following steps: dipping the sample, taking pretreatment to the sample, cracking the microorganism, removing humus, taking DNA coupling, removing protein impurity, adhering DNA, washing DNA, and taking elution to DNA. The advantages of the invention are rapid, little DNA loss, little harmful to researchers, high yield, and low cost, etc.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Bacteriophage genome DNA (deoxyribonucleic acid) extraction kit and method
ActiveCN105567678AMeet the needs of molecular experiments such as sequencingEliminate Interference ProblemsDNA preparationProtein structureDNA Contamination
The invention provides a bacteriophage genome DNA (deoxyribonucleic acid) extraction kit and method. The kit comprises a reagent A (chloroform), a reagent B (DNase I), a reagent C (RNase A), a reagent D (precipitation buffer solution), a reagent E (pyrolysis buffer solution), a reagent F (primary pyrolysis main solution), a reagent G (secondary pyrolysis solution), a reagent H (impurity removal solution), a reagent I (DNA combination buffer solution), a reagent J (rinsing buffer solution) and a reagent K (DNA eluate). The extraction method comprises the following steps: removal of bacterium cells and fragments, degradation of bacterium nucleic acid, bacteriophage particle precipitation, bacteriophage capsid protein structure damage and hydrolysis, impurity removal, DNA liquid separation, DNA adsorption, cleaning of adsorbed DNA and elution of adsorbed DNA. The kit and method can be widely used for extracting the bacteriophage genome DNA, and has the advantages of high extraction speed, high extracted DNA yield and high purity, and the extracted DNA can not be polluted by the host bacterium genome DNA.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Kit and method for quickly extracting DNA of plant by use of DNA adsorption column
The invention relates to a kit and a method for quickly extracting a DNA of a plant by use of a DNA adsorption column, and belongs to the technical field of biology. The kit comprises such components as a splitting liquor, a leaching liquor, a precipitator, a deproteinizing liquor, a rinsing liquor, an eluent and the DNA adsorption column. The method comprises pretreatment of a plant tissue sample, dissociation of DNA and removal of impurity proteins, adsorption of DNA by use of the DNA adsorption column and removal of impurities, and DNA elution. The kit can be used for quickly extracting the genome DNA of the plant, and the obtained genome DNA has relatively high purity.
Owner:FUJIAN NORMAL UNIV
Method for quickly extracting and preserving silica gel membrane type DNA
InactiveCN101649317AAvoid Experimental Operational ImpactLong-term storage at room temperatureSugar derivativesDNA preparationDna adsorptionSuppressor
The invention discloses a method for quickly extracting and preserving a silica gel membrane type DNA. By utilizing the characteristic of a silica gel membrane that the silica gel membrane adsorbs DNAunder the conditions of high salt and low pH and releases the DNA under the conditions of low salt and high pH, the invention does not need a process of extracting the DNA in advance after the silicagel membrane is processed by a novel solution containing a cell cracking agent and a nuclease suppressor, cells are directly lysed, the DNA is adsorbed on the silica gel membrane, the silica gel membrane adsorbing the DNA can be preserved for a long time because of the existence of the nuclease suppressor and the degradation of the DNA is avoided. In the extracting process, the pure DNA is adsorbed on the membrane by the elution function of eluent I after salt and cell residues on the membrane are rinsed. By the elution function of eluent II, the DNA combined on the membrane is eluted into the solution. The whole DNA extracting and preserving process has quick operation and low price and is simple and convenient.
Owner:林桂兰
Method and reagent for improving rate of extraction of DNAs of castoff cells in case trace sample
ActiveCN102229927AFacilitates efficient extractionFully lysedDNA preparationMagnetic beadPolyethylene glycol
Owner:深圳柏悦基因科技有限公司
Ribonucleic acid (RNA) extraction kit without deoxyribonucleic acid (DNA) residues and RNA extraction method
ActiveCN102703437AImproved DNA removal effectSolve the problem of DNA residueDNA preparationRNA extractionDna adsorption
The invention discloses a ribonucleic acid (RNA) extraction kit without deoxyribonucleic acid (DNA) residues and an RNA extraction method. The RNA extraction kit without the DNA residues comprises RNA eluent, DNA eluent, at least one DNA adsorption column which is matched with the RNA eluent and at least one RNA adsorption column which is matched with the DNA eluent. The RNA extraction method at least comprises the following two steps of: (A), eluting RNA on the DNA adsorption column by using the RNA eluent; and (B), eluting DNA on the RNA adsorption column by using the DNA eluent. The RNA extraction method is simple and quick; and the extracted RNA is high in purity, DNA residues are a few and cannot be seen by naked eyes under a common ultraviolet lamp.
Owner:BEIJING AIDLAB BIOTECH
DNA extraction kit for soil urine and method
InactiveCN107058296AIncrease productionReduce lossesMicrobiological testing/measurementDNA preparationDecompositionDna adsorption
The invention relates to a DNA extraction kit for soil urine and a method. The method comprises the following steps: 1) DNA extraction: a urine-containing soil sample is subjected to splitting decomposition with a soil lysate and proteinase K, soil and a liquid supernatant are separated centrifugally, a binding solution is added to the liquid supernatant, a silicon-based DNA adsorbing material is added after the materials are mixed, the silicon-based DNA adsorbing material and a liquid are separated, the silicon-based DNA adsorbing material is rinsed, and DNA is eluted; 2) PCR amplification and electrophoresis: DNA purified in the step 1) is subjected to PCR amplification, and DNA is detected in electrophoresis apparatus. With adoption of the kit and the method, DNA adsorption by silicon dioxide sourced from sample soil is eliminated, so that unnecessary DNA loss is reduced, and high-quality and high-yield DNA from soil urine can be obtained.
Owner:ANHUI SENPENG BIOTECHNOLOGY CO LTD
Process for extracting DNA from biological samples
InactiveCN1634965ASimple processLow costSugar derivativesMicrobiological testing/measurementDna adsorptionDNA extraction
This invention relates to a biological sample DNA extraction method, which comprises the following steps: a, leading the micro magnetic leads with carboxyl mark into the micro channel of the chip and absorbing and fixing it inside the channel to form the micro flow chip of the sample; b, mixing the sample and DNA absorbing liquid and then leading into the chip and mixing it with the micro magnetic leads, wherein the DNA in sample is absorbed on the leads with buffer liquid flowing in for washing and then collecting the washing liquid to get the high purified DNA.
Owner:SHANGHAI INST OF MICROSYSTEM & INFORMATION TECH CHINESE ACAD OF SCI
Kit for rapidly extracting genomes of animal excrement by virtue of CTAB method
InactiveCN104975004AQuick extractionEfficient extractionDNA preparationMicrobial GenomesMicroorganism
The invention provides a kit for rapidly extracting genomes of animal excrement by virtue of a CTAB method. The kit is utilized for carrying out pretreating the excrement, splitting microbial cells in the excrement, dissociating DNA, abstracting impurity protein, adsorbing DNA by virtue of a DNA adsorption column, removing impurities and eluting DNA; after cells are disrupted, protein impurities are removed by virtue of a CTAB solution, meanwhile, genome DNA is adsorbed by virtue of a silicone mold adsorption column, and finally, the residual protein impurities are eluted by virtue of special deproteinization liquid and rinsing liquid. A conventional animal excrement microbial genome extraction method needs to be finished within 14 hours or longer, and the genomes are easily degraded due to the long-playing operation. Compared with the conventional animal excrement microbial genome extraction method, the kit has the advantages that the animal excrement genomes can be extracted within only 4 hours, so that plenty of extraction time is saved, meanwhile, experimental steps are simplified, and the microbial genomes of the animal excrement can be relatively effectively, rapidly and economically extracted.
Owner:FUJIAN NORMAL UNIV
Kit for rapidly extracting sludge microbial genome DNA and extracting method
The invention discloses a kit for rapidly extracting sludge microbial genome DNA and an extracting method. The sludge microbial genome DNA is extracted by the following steps: breaking sludge microbial cells, removing impurity proteins, adsorbing DNA by using a DNA adsorption column, and removing impurities and eluting the DNA. The conventional method for extracting the sludge microbial genome DNA often uses a method for precipitating the genome by using anhydrous ethanol, the operating time is above 12 hours, and the genome DNA is easily degraded. The kit mainly uses a silicone mould adsorption column for absorbing the genome DNA, and then uses a specific protein-removing solution and a specific bleaching solution for removing the proteins and other impurities; the kit can extract the sludge microbial genome DNA within 4 hours, so that the kit is more convenient and quicker.
Owner:FUJIAN NORMAL UNIV
An extracting method of an alveolar lavage fluid metagenome
ActiveCN106244580AEliminate distractionsHigh precisionDNA preparationAlveolar lavage fluidDna adsorption
A safe, simple, convenient, low-pollution and good-repeatability extracting method of alveolar lavage fluid metagenome DNAs is provided. The method includes steps of liquefying a sample, crushing cells, isolating target DNAs, removing impurity proteins and salt ions, performing DNA adsorption and performing DNA elution. The method can increase the yield and purity of the metagenome DNAs and can meet PCR, DNA library construction, gene sequencing, and other molecular biological experiments.
Owner:厦门基源医疗科技有限公司
Kit for extracting fungal genome DNA
InactiveCN104975006AQuick extractionSuitable for commercial productionDNA preparationFungal genomeFungal gene
The invention relates to a kit for extracting fungal genome DNA and belongs to the technical field of biology. The kit for extracting the fungal genome DNA comprises the specific steps of crushing fungal cells, dissociating DNA and removing impure proteins, adsorbing DNA by using a DNA adsorbing column, removing impurities and eluting DNA, finally, so as to obtain a liquid which is the fungal genome DNA which is subjected to electrophoresis detection in 1% sepharose gel. The conventional steps for extracting the fungal genome DNA are tedious, consume a long time, and can extract the fungal genome DNA in about 18 hours generally. The method provided by the invention, compared with the conventional fungal genome extraction method, only needs about 7 hours to extract the fungal genome DNA, so that a lot of extraction time is saved, and the fungal genome DNA can be extracted more effectively, quickly and economically. Meanwhile, the kit is suitable for commercial production and has a certain economic value.
Owner:FUJIAN NORMAL UNIV
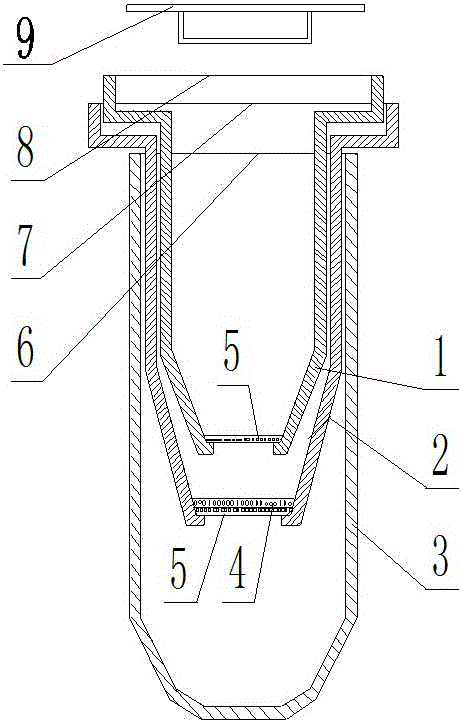
Batch and fast DNA extraction method and extraction device
PendingCN108384780AAvoid mutual contaminationFast extractionBioreactor/fermenter combinationsBiological substance pretreatmentsDna adsorptionDNA extraction
The invention provides a batch and fast DNA extraction method and extraction device, and relates to the technical field of a biological device. DNA extraction liquid is added into a deep hole plate; an extraction device is pressed for several seconds; filter paper falls into the extraction liquid; after 3 to 5 seconds, DNA is adsorbed onto the filter paper. The device comprises a press plate, a filter paper groove plate, a protection pipe and elastic posts uniformly distributed on the press plate at equal intervals; the quantity and the space of the elastic posts are identical to the quantityand spacing of holes on the deep hole plate; the head parts of the elastic posts are fixed at one side of the press plate; the filter paper is clamped in the lower end longitudinal center shaft position of the clamping part at the other end; the clamping part is inserted into a downward extending protection pipe of the filter paper groove plate; a spring is sleeved on the outer periphery of the head of the elastic post, so that a certain distance is formed between the filter paper groove plate and the press plate; a slide resistance block for preventing the clamping part from sliding out extends in the axis direction from the top end of the protection pipe; anti-slip bulges corresponding to the slide resistance blocks are arranged on the outer periphery of the clamping part. The method andthe device have the beneficial effects that the fast and batch DNA adsorption is realized; the adhesion between adjacent pieces of filter paper can be prevented; the sample crossed pollution is prevented; the operation is simple; the application range is wide.
Owner:TOBACCO RES INST CHIN AGRI SCI ACAD
Preparation of graphene-based composite material and application thereof in chemiluminiscence detecting DNA (Deoxyribonucleic Acid) content
InactiveCN105771890AImprove adsorption capacityImprove efficiencyOther chemical processesChemiluminescene/bioluminescenceDna adsorptionSingle strand dna
The invention discloses preparation of a graphene-based composite material and application thereof in chemiluminiscence detecting the DNA (Deoxyribonucleic Acid) content. The main technical characteristic is characterized in that according to a method, magnetism is modified on the surface of graphene oxide, and a magnetic graphite base composite material with a conjugate adsorption capacity on a single-stranded DNA molecule is synthetized, the preparation process is simple, the conditions are easy to control, and the production cost is low. The invention provides a new method for detecting DNA. The synthetized Fe3O4@SiO2 / GO composite material can realize quick adsorption on the single-stranded DNA, is strong in adsorption capacity, and high in efficiency; the time for the single-stranded DNA adsorption to achieve the balance is only 10 to 15min, and the maximal adsorption capacity is the saturated adsorption amount achieving the magnitude order 10<-7> to 10 <-6>mol / g. The method for detecting DNA is low in detection line, can achieve 10<-9>mol / L, and has the advantages of high sensitivity, high selectivity, good reproducibility, no pollution on the environment and the like.
Owner:UNIV OF JINAN
Total DNA extraction method for Eremochloa ophiuroides (Munro.) Hack.
InactiveCN103436525AImprove adsorption capacityQuality improvementDNA preparationPolyphenolFine powder
The invention discloses a total DNA extraction method for Eremochloa ophiuroides (Munro.) Hack. and relates to the technology of extracting total DNA from plants in the field of molecular biology. The total DNA extraction method mainly comprises the following steps: adding a cell nucleus extraction buffer solution into Eremochloa ophiuroides (Munro.) Hack. leaves which are ground into fine powder, and achieving DNA adsorption and elution and DNA purification and separation through silicon beads. According to the invention, the silicon beads are used for effectively adsorbing DNA, removing secondary metabolism products, such as polysaccharose, polyphenols, pigment and protein, and extracting to obtain high-quality Eremochloa ophiuroides (Munro.) Hack. total DNA. The total DNA extraction method can extract high-quality Eremochloa ophiuroides (Munro.) Hack. total DNA from Eremochloa ophiuroides (Munro.) Hack. tissues preserved at low temperature and fresh Eremochloa ophiuroides (Munro.) Hack. tissues, and is suitable for extracting high-quality total DNA from Eremochloa ophiuroides (Munro.) Hack.
Owner:GUANGXI FORESTRY RES INST
DNA specimen normal temperature preservation preparation method and packaging equipment
InactiveCN106672441AAvoid destructionPrevent overflowPackaging under vacuum/special atmosphereShock-sensitive articlesDna adsorptionNitrogen
The invention relates to a DNA preservation method and equipment, in particular to a DNA specimen normal temperature preservation preparation method and packaging equipment. The DNA specimen normal temperature preservation preparation method comprises the following steps: enabling DNA to be adsorbed on an adsorption film, taking out the adsorption film, placing the adsorption film in a 3ML disinfection clean specimen bottle, then placing the specimen bottle on a DNA normal temperature preservation automatic packaging machine, and placing the DNA normal temperature preservation automatic packaging machine in a vacuum drying box chamber; carrying out first vacuumizing on the vacuum drying box chamber to 0.005MPa, raising the temperature to 10-35 DEG C, and drying for 30mins; filling nitrogen into the vacuum drying box chamber until the pressure is 0.1MPa; carrying out second vacuumizing to 0.005MPa; filling the nitrogen the vacuum drying box chamber secondly until the pressure is 0.1MPa; carrying out third vacuumizing to 0.005MPa; automatically plugging a rubber plug into a bottle mouth of the specimen bottle by the DNA normal temperature preservation automatic packaging machine; and taking out the DNA normal temperature preservation automatic packaging machine and the specimen bottle, adding a metal cover cap at the external of the specimen bottle, and permanently packaging the external of the metal cover cap and the neck of the specimen bottle together with resin. According to the method, harmful gas in the packaging environment is greatly lowered, the DNA is prevented from being damaged by high temperature, and automatic packaging is finished in the vacuum environment.
Owner:浙江丽生生物科技有限公司
Method for extracting genomic DNA (deoxyribonucleic acid) from whole blood on basis of improved immunomagnetic bead method
The invention relates to a method for extracting genomic DNA (deoxyribonucleic acid) from whole blood on basis of an improved immunomagnetic bead method.The method includes steps: erythrocyte splitting, leukocyte splitting, DNA adsorption, DNA purification and DNA elution.DNA concentration and purity are measured by a DNA quantometer, sequencing of PCR (polymerase chain reaction) amplification products is realized by a sequencer, and HLA (human leukocyte antigen) classification result analysis is performed through relevant analysis software.The method has the advantages of high DNA concentration uniformity, high purity, substantial increase of HLA classification result success rate, and high acceptability of a signal-to-noise ratio, thereby being widely applicable to extraction of the genomic DNA from whole blood samples.
Owner:武汉血液中心
Real-time fluorescent quantitative PCR (polymerase chain reaction) detection kit for shrimp white spot syndrome virus
InactiveCN102952902AHigh detection sensitivityOptimizationMicrobiological testing/measurementMicroorganism based processesAbsorption columnWhite spot syndrome
The invention discloses a real-time fluorescent quantitative PCR (polymerase chain reaction) detection kit for shrimp white spot syndrome virus. The kit comprises the following reagents of a reagent A, a reagent B, a reagent C, a reagent D, a reagent E, a reagent F, a reagent G, sterilized double distilled water, and a DNA (deoxyribonucleic acid) absorption column, wherein the reagent A is prepared by adding water into hexadecyl trimethyl ammonium bromide, NaCl (sodium chloride), ethylene diamine tetraacetate, and hydroxymethyl aminomethane-hydrochloric acid, the reagent B adopts protease K, a solvent of the reagent B is the sterilized double distilled water, the reagent C is an ethanol water solution, the reagent D is prepared by mixing a PCR reaction buffer containing MgCl2 (magnesium chloride), Taq (thermos aquaticus) DNA polymerase and the sterilized double distilled water, the reagent E is prepared by mixing a primer P1, a primer P2 and deoxyribonucleotide, the reagent F is a fluorescent probe premixed solution, and the reagent G adopts a shrimp white spot syndrome virus positive plasmid. The kit has the advantages that the detection sensitivity is high, the kit is used for the early infection and tracking monitoring of the cultivation shrimp white spot syndrome virus, and the kit is also used for the related basic scientific research of virus multiplication.
Owner:天津市水生动物疫病预防控制中心
Kit for extracting gram-positive bacterium genome
InactiveCN105154430AFewer extraction stepsReduce extraction timeDNA preparationBiotechnologyDna adsorption
The invention provides a kit for extracting a gram-positive bacterium genome, and belongs to the field of biotechnology. The kit comprises the following specific steps: breaking gram-positive bacterium cells, dissociating DNA and removing impurity protein, adsorbing the DNA by virtue of a DNA adsorption column and removing impurities, and eluting the DNA. Different from a conventional kit for extracting a gram-positive bacterium genome which is relatively complex in extraction step and relatively long in extraction time consumed which generally requires about 12h, the kit for extracting the gram-positive bacterium genome, compared with a conventional kit in method for extracting gram-positive bacterium genome DNA, is capable of extracting the gram-positive bacterium genome DNA for only about 2h, so as to save a lot of extraction time; and meanwhile, the kit can simplify experimental steps, and can be used for extracting the gram-positive bacterium genome DNA more effectively, rapidly and economically.
Owner:FUJIAN NORMAL UNIV
Kit and method for extracting pectin-rich polysaccharide plant DNA
The invention relates to a kit and a method for extracting pectin-rich polysaccharide plant DNA. The kit comprises a desaccharifying buffer solution, a lysate, leaching liquor, a column buffer solution, a protein-free solution, a rinsing solution, an eluant and a DNA adsorption column. An extraction method comprises the following steps: removing polysaccharide from a plant, extracting DNA, subsequently extracting and removing polysaccharide again by filtering with a column, thereby acquiring the extracted DNA with high quality and no impurity. The kit and the method provided by the invention are suitable for the extraction of plant genome DNA of mass secondary metabolites rich in polysaccharide, polyphenol, and the like. The kit has the characteristics of a low cost, simple, convenient andquick operation, high quality, high efficiency, high applicability, and the like.
Owner:四川省植物工程研究院
Kit and method for extracting collybia radicata genome DNA (Deoxyribonucleic Acid)
The invention discloses a kit and method for extracting collybia radicata genome DNA (Deoxyribonucleic Acid). The kit is prepared from the following components: an equilibrium liquid including 1 percent of Na2SiO3.9H2O and 1 percent of NaOH, lysate including 5.0M of guanidine hydrochloride, 25 percent of isopropanol and 1 percent of PVP K30, a cleaning solution including 20 mM of NaCl, 20 mM of Tris-HCl and 85 percent of ethanol with pH of 7.5, eluent including 10 mM of Tris-HCl with the pH of 8.5 and a DNA adsorption column. The reagent for extracting the collybia radicata genome DNA does notcontain toxic substances such as phenol and chloroform, so that higher safety is realized; the extraction method is simple and efficient and the extraction time is shortened to 15 minutes; an extracted product has high purity and good integrity, and can be directly used for molecular biology and genetic research.
Owner:FUJIAN AGRI & FORESTRY UNIV
Reagent for extracting total DNA of bamboo shoots and application
The invention discloses a reagent for extracting total DNA of bamboo shoots and an application. The method comprises the steps of column balancing, lysing crackinga sample, absorbing the DNA, cleaning, and eluting the DNA. Compared with the traditional DNA extraction method such as a the CTAB method, the a SDS method, the an alkali lysising method and a conventional kit, the method provided byof the invention has the advantages of less consumption of samples, high purity of the DAN samples, good completeness, simple operation and the like, the extraction time can be reduced from 1 to 2 hours to 15 minutes, in the whole extraction process, no toxic reagent such as chloroform, phenol, and beta-mercaptoethanol and the like is used, and the research personnel is ensured enabled not to be harmed by the toxic reagent. An experiment result shows that the reagent is suitable for extracting the total DNA of various fungi such as the bamboo shoots.
Owner:FUJIAN AGRI & FORESTRY UNIV
Method for reclaiming DNA from agarose gel
InactiveCN101274952AOvercoming pollutionOvercome the disadvantage of easy cross-contaminationSugar derivativesSugar derivatives preparationElectrophoresisDna adsorption
The invention relates to a method for recycling DNA from agarose gel. The method for recycling DNA samples from agarose gel after electrophoresis comprises the following steps in sequence: first, stripped absorbent material of wedge type is prepared, then inserted into gel, sucks out the DNA samples, and then the method is finished through centrifugal collection, DNA absorption, DNA washing, DNA elution, etc; the invention overcomes the defects existing in the prior art of complicated operation of cutting gel for recycling and easy cross contamination when operating a plurality of samples, provides a method for recycling liner or annular DNA from agarose gel after electrophoresis, can recycle DNA segments for purification from agarose gel within 15min, has high pureness and can meet the subsequent operation of molecular cloning, enzyme analysis, marking, PCR, etc.; the method adopts easy equipment, has convenient operation, high recycling rate and low cost and can put reagent in kits.
Owner:GUIYANG MEDICAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com