Process for extracting DNA from biological samples
A biological sample and chip technology, which is applied in biochemical equipment and methods, microbial determination/inspection, sugar derivatives, etc. The effect of less reagent consumption, enhanced safety, and simple chip fabrication
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
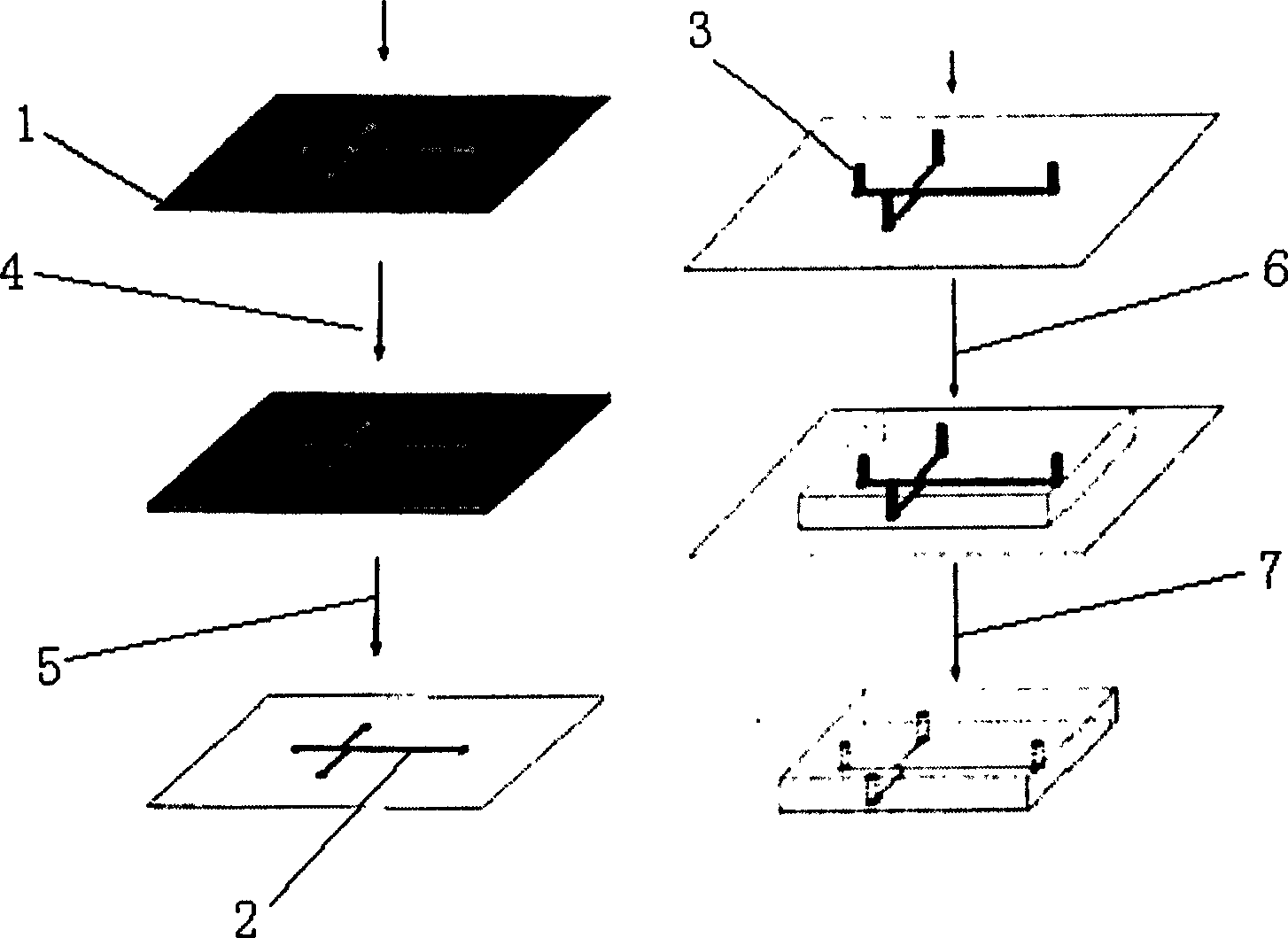
[0030] see figure 1 , the preparation method of polydimethylsiloxane chip, comprises the steps:
[0031] (1) Preparation of the mould: transfer the designed chip structure to a mask 1 that adopts a silicon wafer or a glass wafer as a base material and is coated with a photoresist, then photoetches 4, develops 5 by a conventional method, and obtains A mold provided with protruding runners 2;
[0032] Said photoresist is a conventional material used in photolithography, such as SU8 negative photoresist; (SU-8, 2025, Microchem).
[0033](2) Preparation of the chip: place the small glass column 3 on the top of the protruding runner 2 of the mold, then cast the mixture of polydimethylsiloxane and curing agent on the mold 6, solidify, and demould 7, that is Obtain a polydimethylsiloxane sheet with micro-channels, bond and package with another polydimethylsiloxane sheet or glass sheet, and finally form a chip made of polydimethylsiloxane ,Such as figure 2 , the channel formed by...
Embodiment 1
[0035] The DNA in the Escherichia coli culture solution is extracted, and the steps are as follows:
[0036] 1) Introduce 5ul micro-magnetic bead dilution into the chip, place a magnet under the channel to absorb the magnetic beads, pressurize the liquid after three minutes of adsorption to discharge the liquid from the chip, and fix the magnetic beads in the chip through the adsorption of the magnet, such as image 3 ;
[0037] 2) Escherichia coli Ecoli DH5α was inoculated and cultured overnight on a shaking table.
[0038] 3) Take away the magnet, add 20 μL of the bacterial solution to 50 μL of adsorption buffer (4% Triton X-100, 0.5M NaCl, 20% PEG 8000), slowly introduce into the chip, mix and then introduce into the chip, and place it at 37°C 15 minutes, then place a magnet under the channel to absorb the magnetic beads, pressurize at the same time to discharge the liquid at a flow rate of 2 μL / min, collect the recovered liquid, and obtain the sample 1
[0039] 4) A magn...
Embodiment 2
[0046] The DNA in the PCR product is extracted, and the steps are as follows:
[0047] 1) Introduce 5ul micro-magnetic bead dilution into the chip, place a magnet under the channel to absorb the magnetic beads, pressurize the liquid after three minutes of adsorption to discharge the liquid from the chip, and fix the magnetic beads in the chip through the adsorption of the magnet, such as image 3 ;
[0048] 2) Take away the magnet, add 10 μL of the PCR product to 20 μL of adsorption buffer and slowly introduce it into the microchannel of the chip, mix it, and place it at 37 ° C for 10 minutes, then place a magnet under the microchannel to absorb the magnetic beads, and pressurize at the same time The liquid is discharged so that the flow rate is 2 μL / min;
[0049] 3) A magnet is still placed under the chip, and 20ul of 70% ethanol is introduced into the microchannel for flow cleaning;
[0050] 4) Place the chip in a 37°C incubator for 2 minutes to dry the magnetic beads;
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com