Extraction method of rosewood heartwood genome DNA (deoxyribonucleic acid)
An extraction method and genome technology are applied in the extraction field of mahogany heartwood genomic DNA, which can solve the problems of extracting high-quality genomic DNA, and achieve the effect of less pollution and high quality.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Extracting Genomic DNA from the Heartwood of Dalbergia cochinchinensis
[0030] Reagent preparation and use requirements for the extraction process are as follows:
[0031] The formula of CTAB lysate is: 2g CTAB, 5g PVP, 8.18g NaCl, 0.74g Na 2 EDTA.2H 2 O, 2 mL of β-mercaptoethanol, 10 mL of 1 mol / L Tris-HCl (pH 8.0), add water to make up to 100 mL.
[0032] Grinding Bowl: Washed and then autoclaved.
[0033] Mix phenol: chloroform: isoamyl alcohol in a volume ratio of 25:24:1 to prepare 10 ml of a solution.
[0034] The extraction method of rosewood Dalbergia cochinchinensis genomic DNA comprises the following steps:
[0035] (1) Weigh 300mg of rosewood Dalbergia cochinchinensis heartwood chips, put them into the weighed chips after being ground and cooled, and grind them into powder in liquid nitrogen.
[0036] (2) Add 700 μl of 65°C preheated CTAB lysate to the powder, place it at 65°C for 5 hours, and invert the EP tube every 30 minutes during this pe...
Embodiment 2
[0042] Example 2 The quality verification of Dalbergia codica chinensis genomic DNA
[0043] 1. The reagent preparation and process of the verification process are as follows:
[0044] 1% agarose: Weigh 0.1g of agarose and dissolve in 10ml of 1×TAE.
[0045] The formula of 50×TAE buffer solution is: Tris242g, Na 2 EDTA.2H 2 O37.2g, add 800ml of deionized water, stir and dissolve, then add 57.1ml of acetic acid.
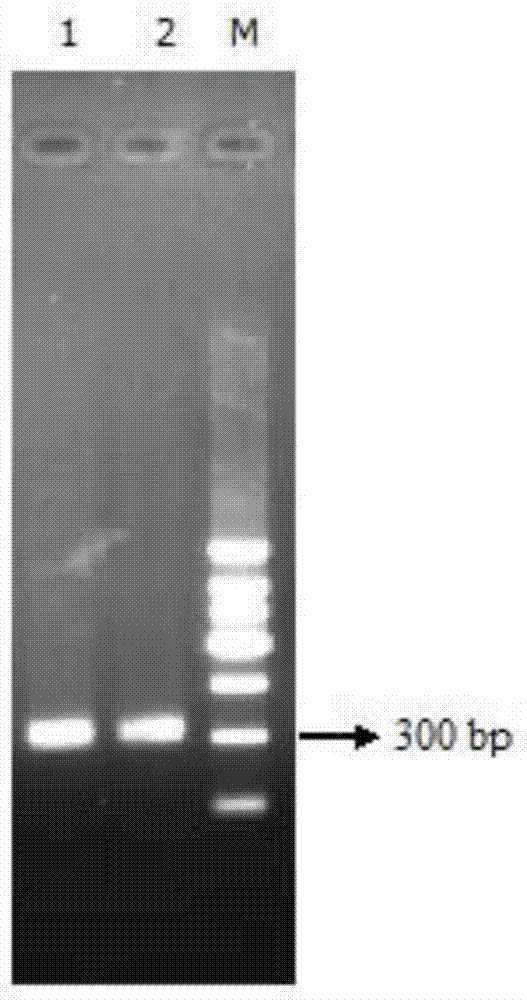
[0046] According to the reported Dalbergia cochinchinensis ITS2 sequence, design PCR identification upstream amplification primer and downstream amplification primer, the sequence of amplification primer is respectively as shown in SEQ ID NO:1 and SEQ ID NO:2 in the sequence list, PCR amplification The size of the amplification product was 364bp.
[0047] Synthetic PCR identification primers: PCR identification upstream amplification primer sequence (as shown in SEQ ID NO: 1 in the sequence listing) is: 5'-CTTGCATCGATGAAGAACGTAG-3';
[0048] The downstream amplif...
Embodiment 3
[0058] (1) Weigh 200mg of rosewood Dalbergia cochinchinensis heartwood chips, put them into the weighed chips after being ground and cooled, and grind them into powder in liquid nitrogen.
[0059] (2) Add 700 μl of 65°C preheated CTAB lysate to the powder, place it at 65°C for 3 hours, and invert the EP tube every 30 minutes during this period.
[0060] (3) Add cellulase at a final concentration of 30 U / g, bathe in 50°C water for 1 hour, and invert the EP tube continuously during this period.
[0061] (4) After cooling at room temperature, centrifuge at 10,000 rpm for 5 minutes, carefully absorb the supernatant and measure the volume of the supernatant, add the same volume of phenol: chloroform: isoamyl alcohol mixed solution = 25:24:1, mix well and then centrifuge at 12,000 rpm for 5 minutes.
[0062] (5) Transfer all supernatants to DNA adsorption columns in batches, centrifuge at 12,000 rpm for 1 min and discard the supernatant.
[0063] (6) Wash off the protein on the ads...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com