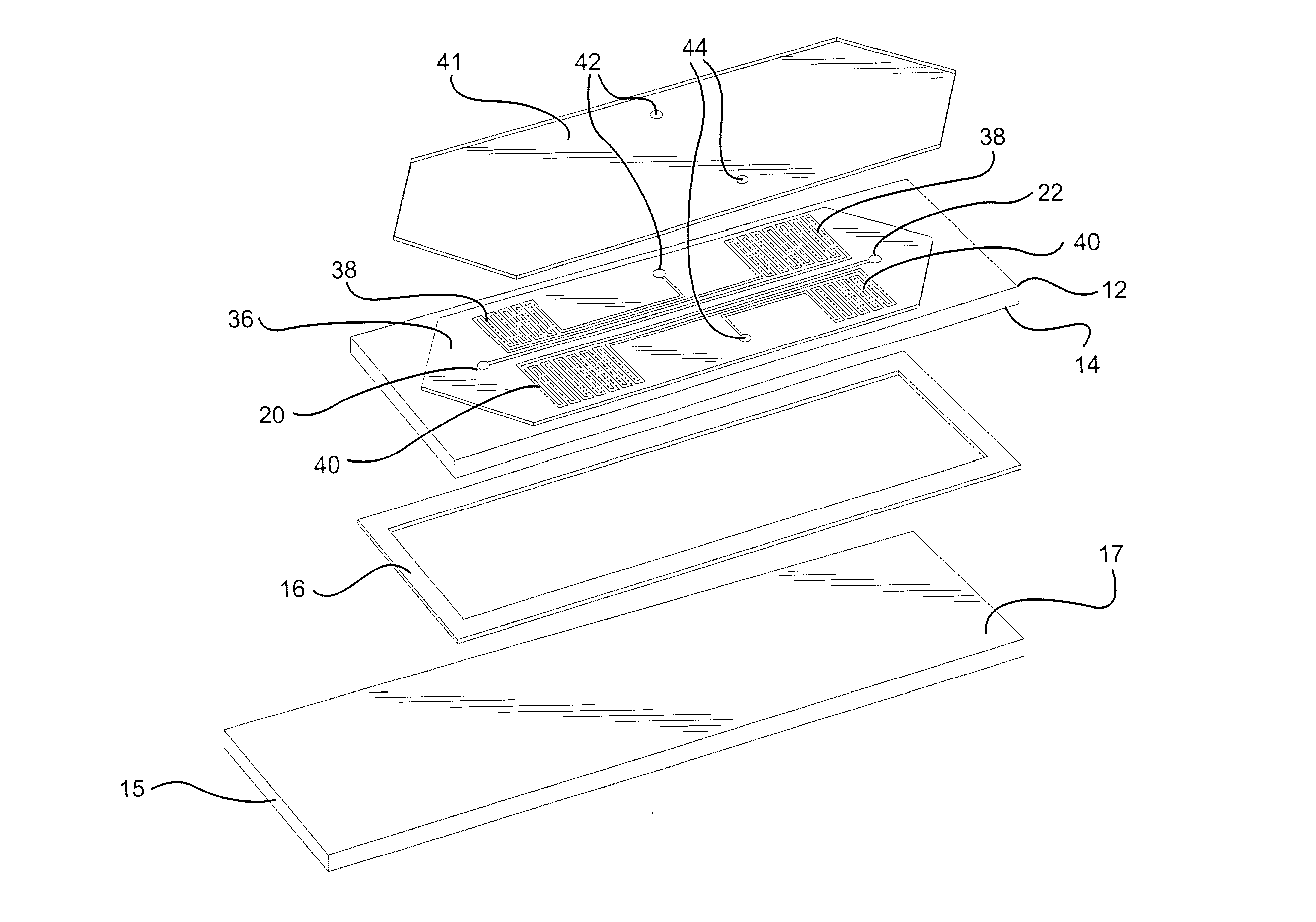
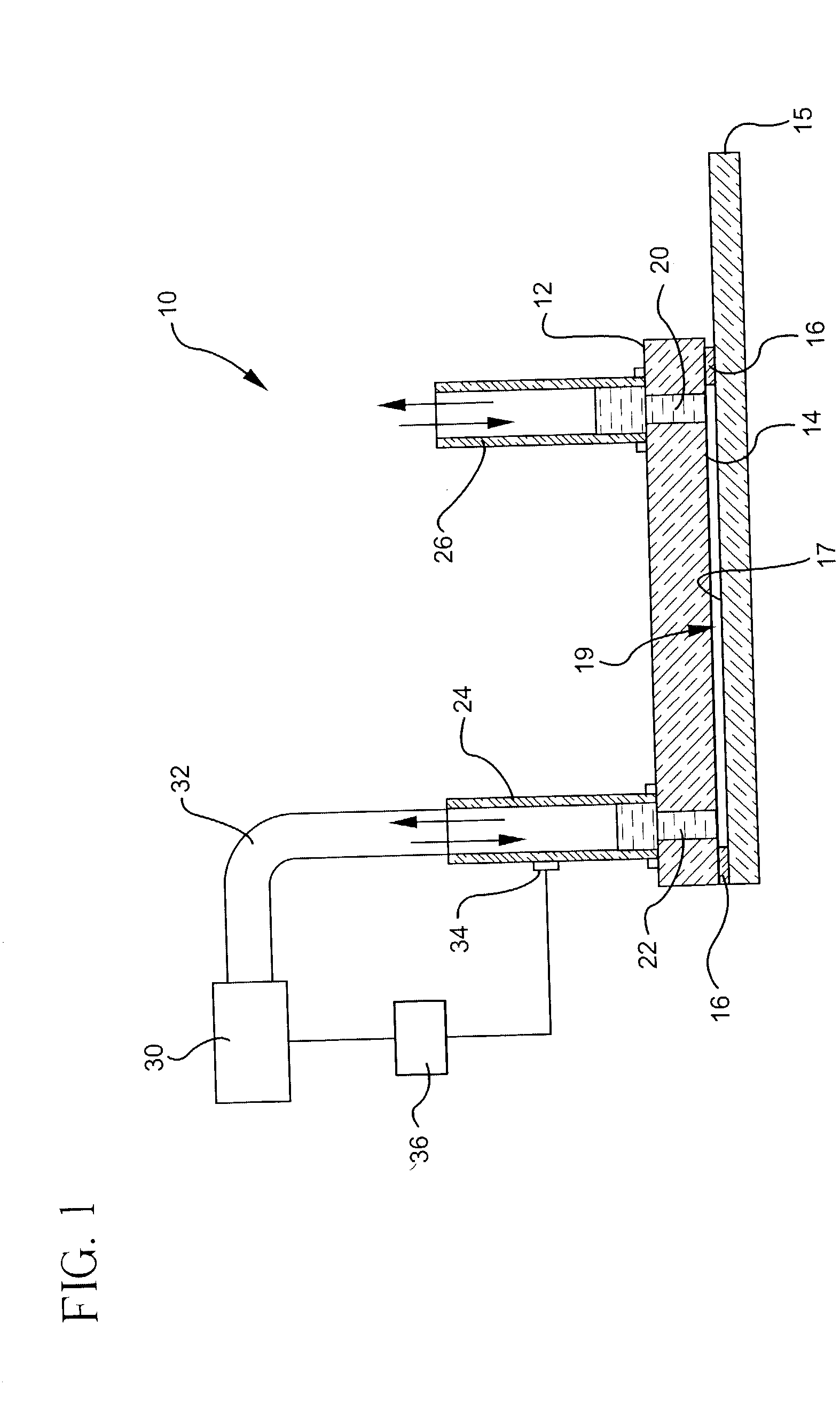
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
92 results about "Microarray hybridization" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
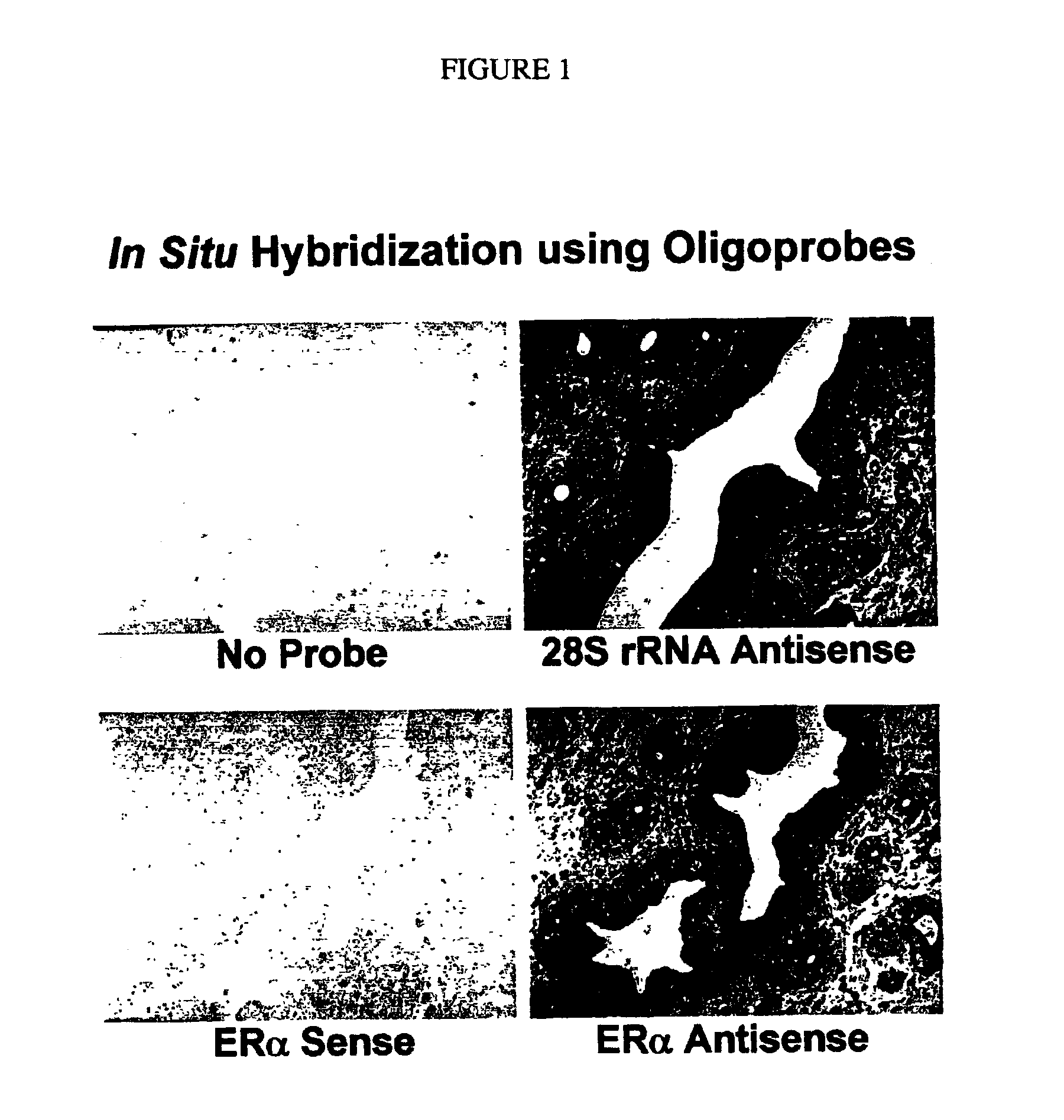
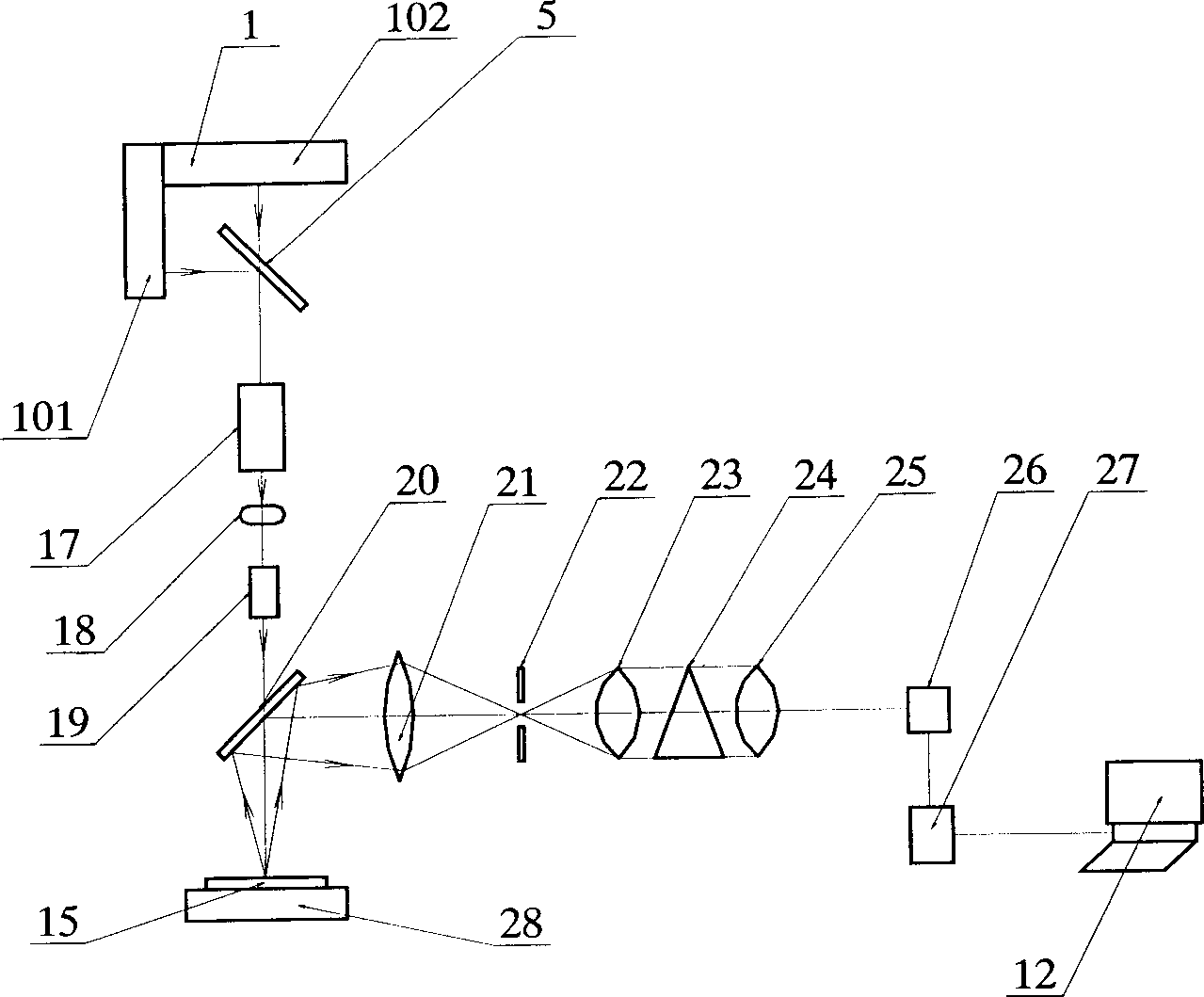
Microarray Hybridization. Introduction. The use of microarrays allows the identification of organisms in an ecological sample by hybridizing DNA extracted from the sample to a set of known DNA specimens arrayed on a solid support, usually a microscope slide (reviewed in Call et al. 2003).
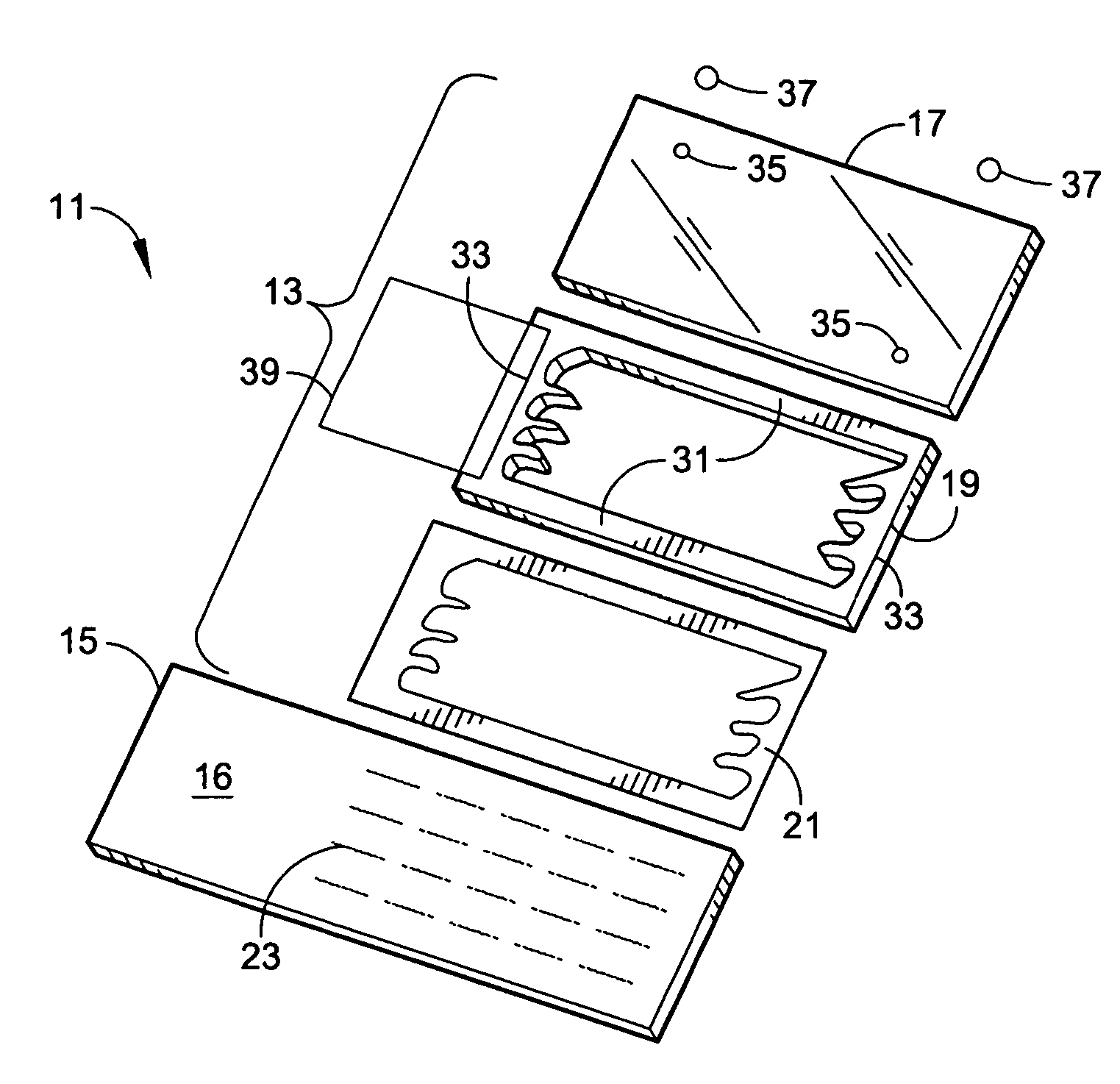
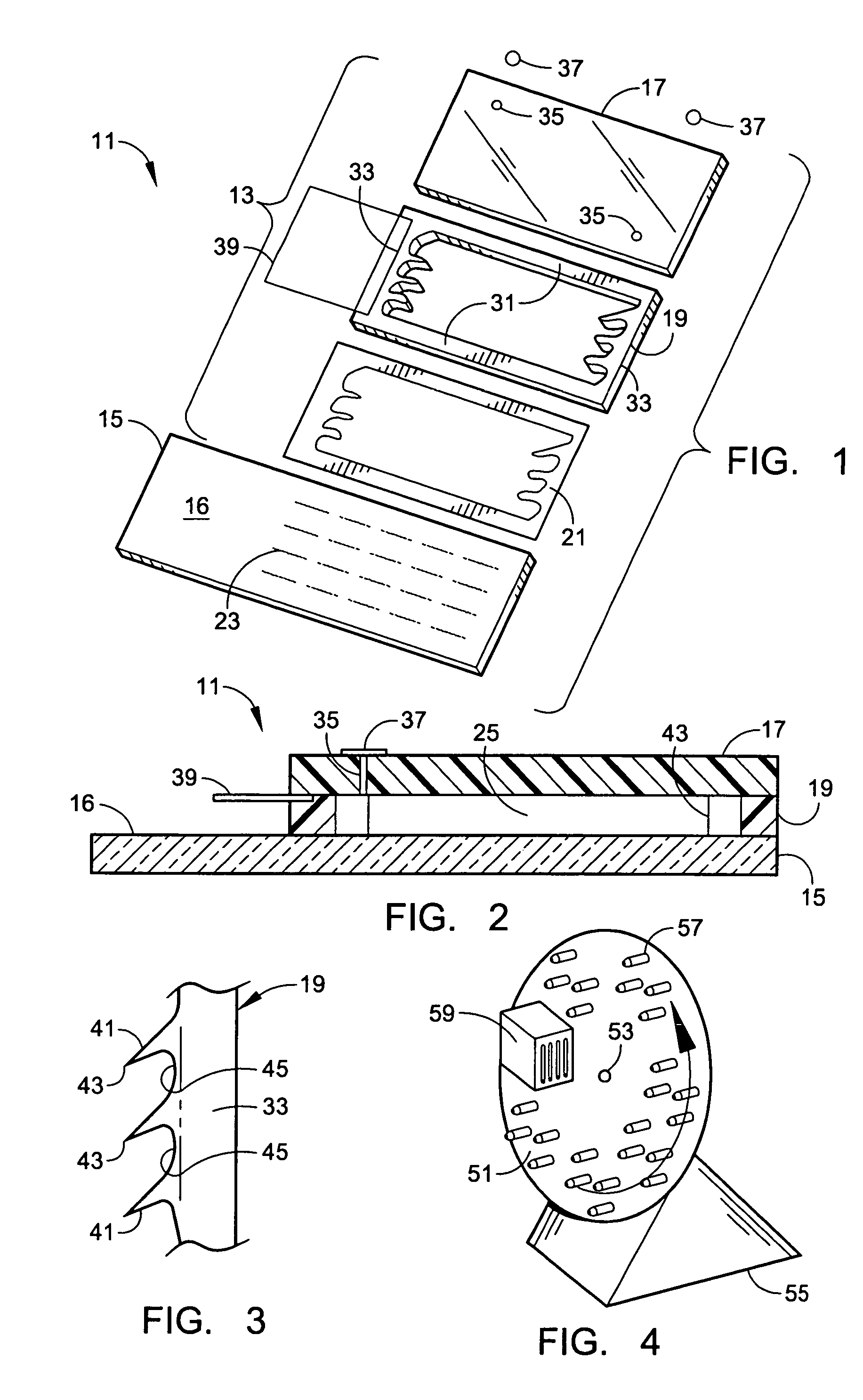
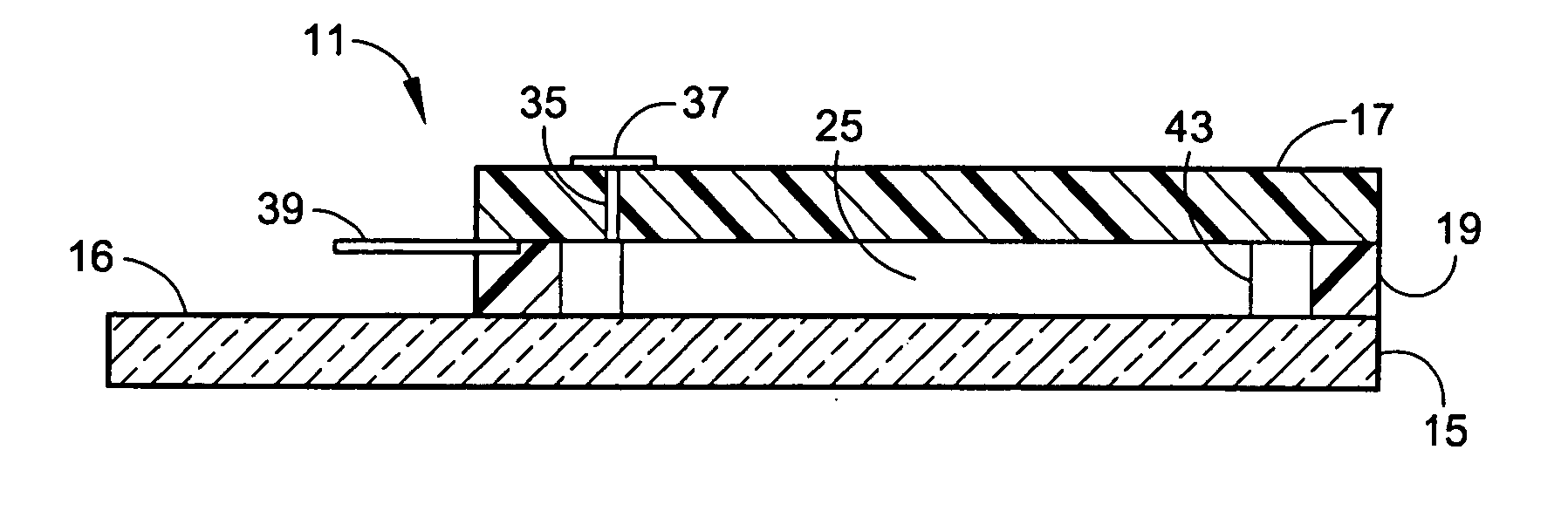
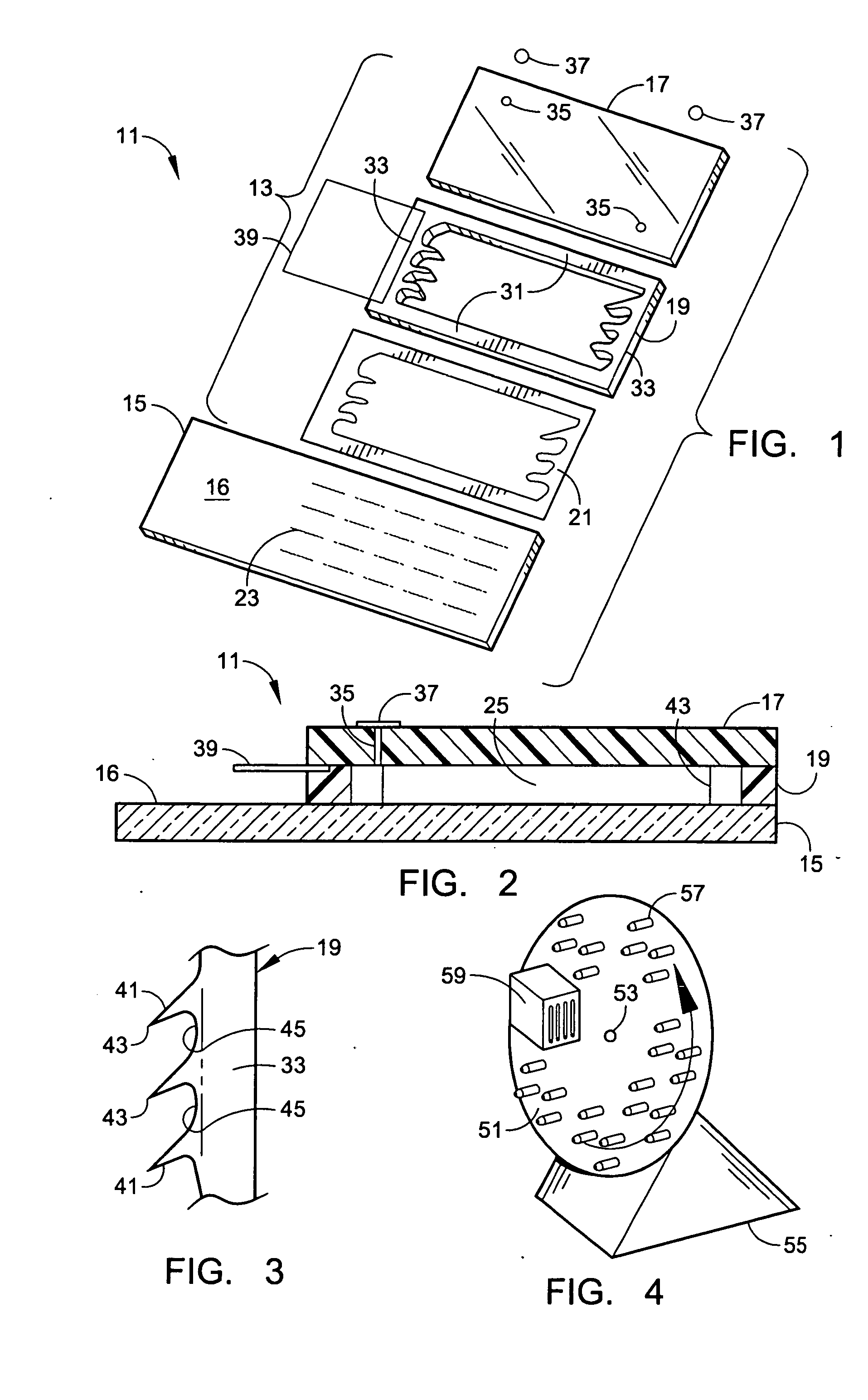
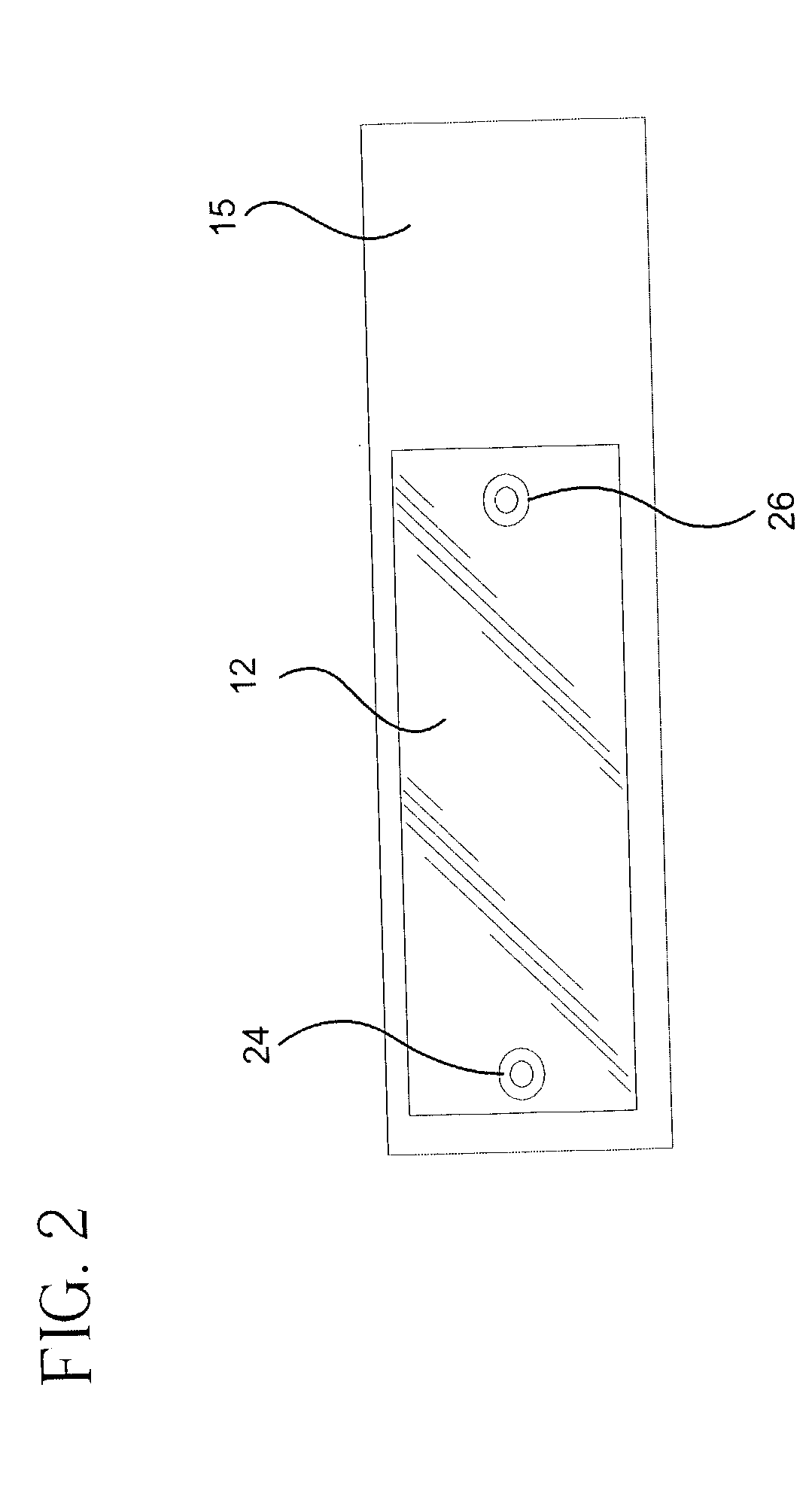
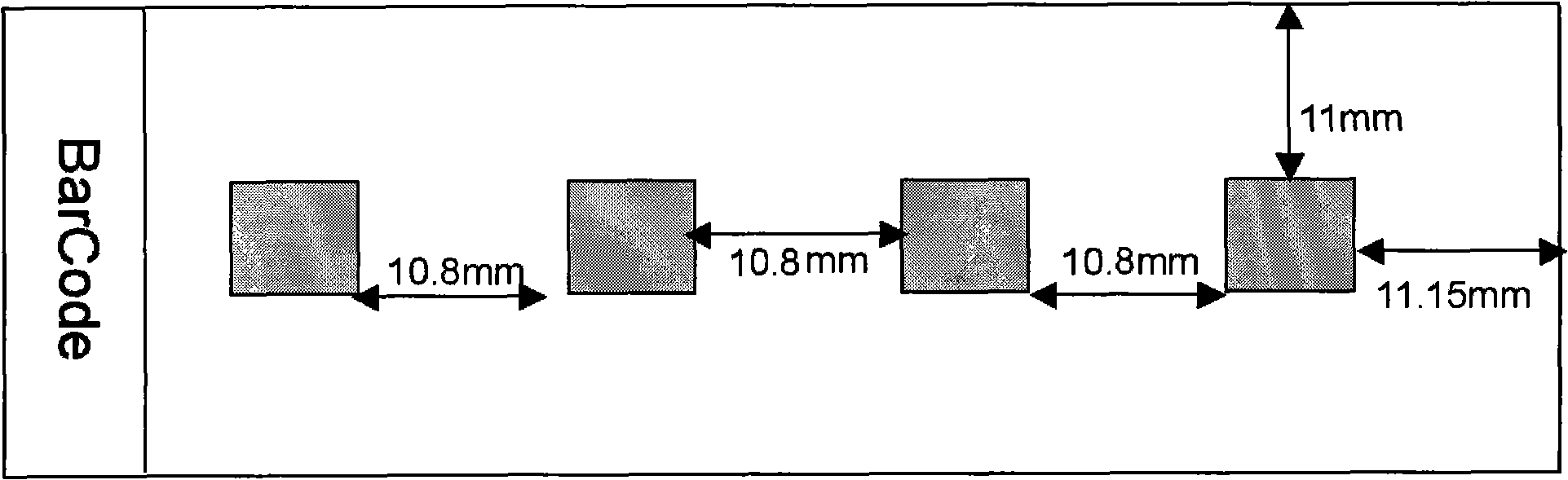
Microarray hybridization device having bubble-fracturing elements
InactiveUS7238521B2Degree of improvementImprove uniformityBioreactor/fermenter combinationsRotating receptacle mixersMicrobubblesHybridization reaction
A novel hybridization device that improves the efficiency and consistency of microarray hybridization reactions by achieving a greater degree of internal mixing of target solution. The device provides a gasket-and-cover-type chamber wherein solution mixing is achieved by the creation of a multitude of microbubbles. One or more of the inner walls that define the chamber contain bubble-rupturing elements that extend into the chamber and terminate in sharp edges. They are typically located on opposite sides of a rectangular chamber and are pointed in a direction opposing bubble movement. Their interference with larger bubbles causes their breakup into microbubbles which travel separate and distinct paths as a result of external agitation and thereby provide improved solution mixing that results in a uniform distribution of target molecules to the probe molecules bound to the substrate. The sensitivity and consistency of the hybridization reaction is significantly increased.
Owner:BIOCEPT INC
Microarray hybridization device
InactiveUS20050112589A1Increase the degree of mixingImprove uniformityRotating receptacle mixersBioreactor/fermenter combinationsMicrobubblesHybridization reaction
A novel hybridization device that improves the efficiency and consistency of microarray hybridization reactions by achieving a greater degree of internal mixing of target solution. The device provides a gasket-and-cover-type chamber wherein solution mixing is achieved by the creation of a multitude of microbubbles. One or more of the inner walls that define the chamber contain bubble-rupturing elements that extend into the chamber and terminate in sharp edges. They are typically located on opposite sides of a rectangular chamber and are pointed in a direction opposing bubble movement. Their interference with larger bubbles causes their breakup into microbubbles which travel separate and distinct paths as a result of external agitation and thereby provide improved solution mixing that results in a uniform distribution of target molecules to the probe molecules bound to the substrate. The sensitivity and consistency of the hybridization reaction is significantly increased.
Owner:BIOCEPT INC
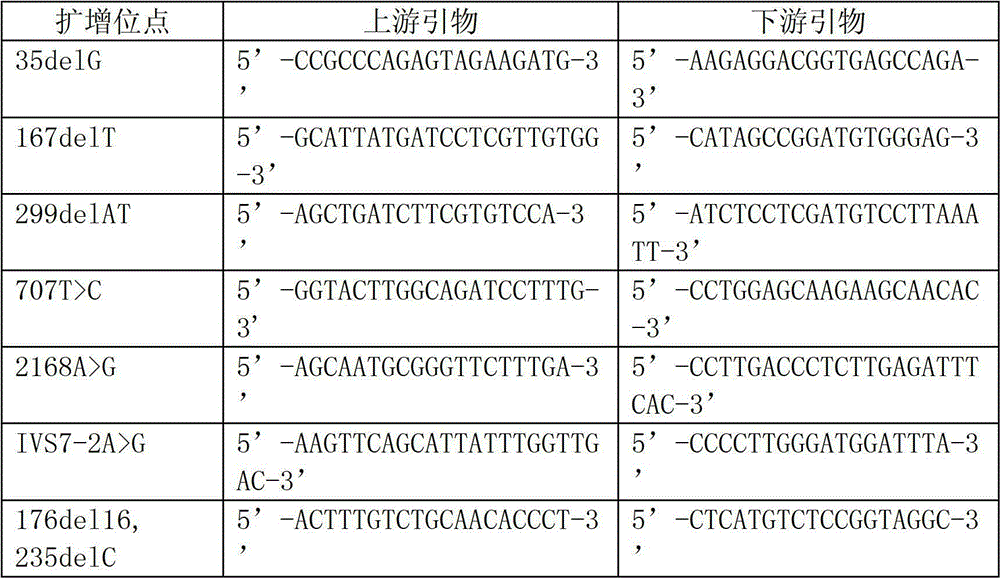
Upland cotton SNP marker and application thereof
InactiveCN105349537AImprove detection efficiencyImprove work efficiencyNucleotide librariesMicrobiological testing/measurementAgricultural scienceHomologous sequence
The invention discloses an upland cotton SNP marker and application thereof, and belongs to the field of upland cotton SNP marker development. The upland cotton SNP marker is obtained by developing a single-copy SNP marker of an upland cotton genome via chip hybridization and homologous sequence comparison, and picking out SNP markers in a linkage disequilibrium recession distance via linkage disequilibrium analysis. The SNP marker provided by the invention can be applied to analysis of genetic diversity and group structure, or to germplasm identification of upland cotton, is single-copy, and has no remarkable linkage disequilibrium among markers, so that the efficiency of detection, adopting the SNP marker, during analysis of genetic diversity, group structure or fingerprint identification can be greatly improved, and the work efficiency is higher.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Preparing method for tr-gene products detecting oligonucleotides chip and use thereof
InactiveCN1584049AGuaranteed specificityTo achieve the purpose of mutual verificationMicrobiological testing/measurementHeterologousOligonucleotide chip
A method for preparing oligonucleotide chip for transgene product detection and use are disclosed. It includes: designing specific oligonucleotide probe and related primer by heterogeneric inserting gene, species internal standard gene, specific boundary sequence in transgene product gene set, fixing probe on glass slide to form transgene product detecting chip, amplifying DNA of plant to be tested by related primmer, marking by fluorescence, and hybridizing with chip. It can be use to acquire variable information.
Owner:国家质量监督检验检疫总局动植物检疫实验所 +2
Microvolume biochemical reaction chamber
InactiveUS20040101870A1High sensitivityLower the volumeBioreactor/fermenter combinationsBiological substance pretreatmentsReagentChamber method
Methods and apparatus for performing biomolecular reactions using microvolumes of reagents are disclosed. The apparatus and methods include a chamber having a height less than 50 microns and means for mixing the extremely small volume of fluid in the chamber. The decreased volumes combined with mixing greatly improved microarray hybridization signal strength.
Owner:CORNING INC
Nucleic acid sequencing process based on micro array chip
InactiveCN1932033AMature technologyEasy to implementMicrobiological testing/measurementGenomic sequencingNucleotide
The nucleic acid sequencing process based on nucleic acid micro array chip is for sequencing genome and detecting gene transcription and expression spectrum of some specific species, and detecting various genome DNA modifying spectrum. Different DNA segments to be sequenced are fixed onto one solid chip to form nucleic acid micro array, and each of the DNA segments is made to contain the same common sequence as the complementary sequence of the sequencing primer. After crossing the common sequencing primer and the chip, some mixed nucleotide solution with certain ratio of fluorescent marker ddNTP and dNTP is injected to the chip to terminate on-chip base reaction with a micro flow controller by means of Sanger end terminating process. Once the termination is finished, one fluorescent signal is obtained through scanning to obtain the base extending information in different sample points on the chip. All the sequencing maps are finally superposed to obtain the complete nucleic acid segment information.
Owner:SOUTHEAST UNIV
Reagents and methods for automated hybridization
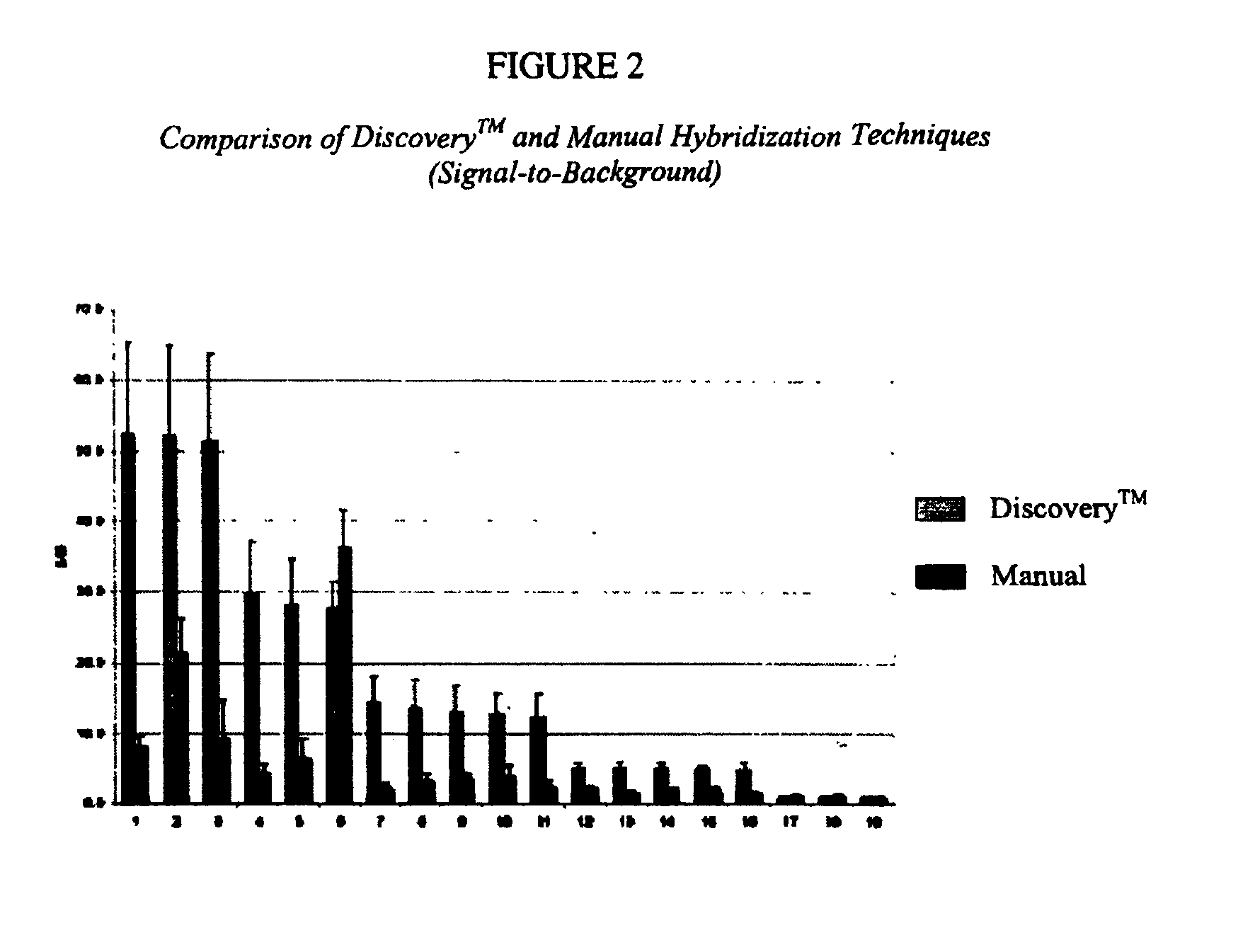
The invention provides novel reagents, reagent kits, and methods for automated hybridization. More particularly, the invention provides reagents, reagent kits, and methods for automated in situ hybridization and automated hybridization on a microarray. The use of automated instruments for in situ hybridization and microarray hybridization dramatically reduces the amount of labor and time involved and also facilitates standardization of protocols and consistency between results.
Owner:VENTANA MEDICAL SYST INC +1
Gene chip of aquatic product cultivation pathogenic bacterium
InactiveCN101691608AReduce volumeRapid Test InterpretationNucleotide librariesMicrobiological testing/measurementBacteroidesPositive control
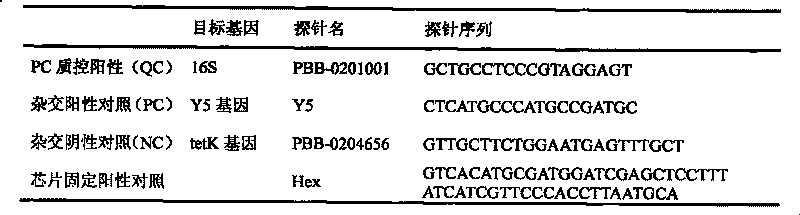
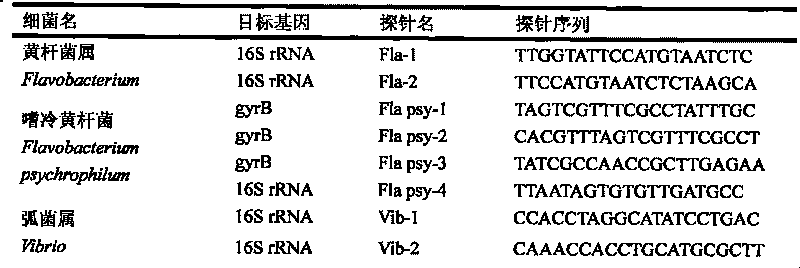
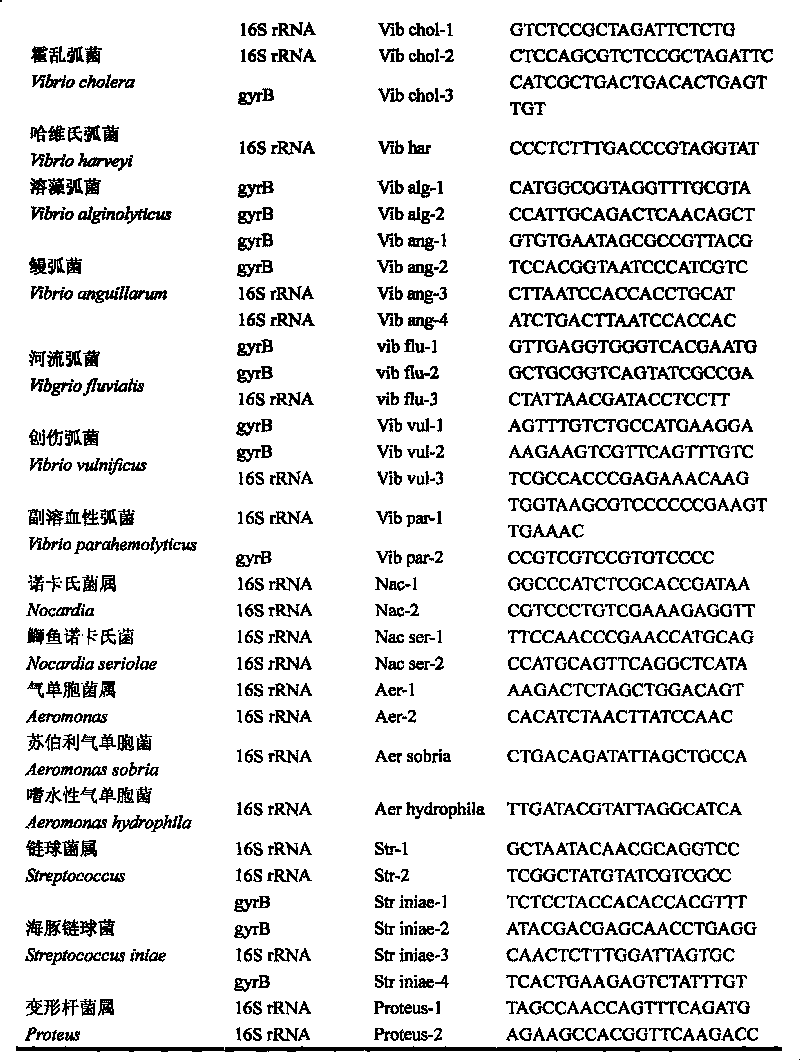
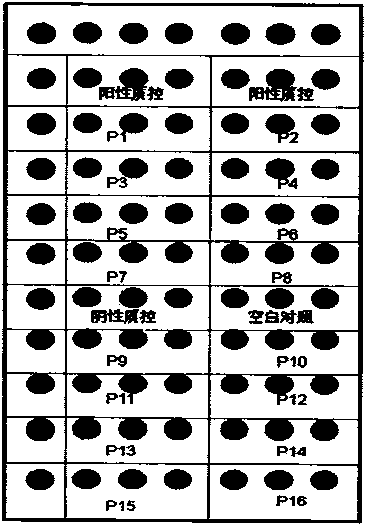
The invention discloses a gene chip of aquatic product cultivation pathogenic bacterium, comprising a solid phase carrier which is modified chemically, a detection probe and a quality control probe are distributed on the solid phase carrier in a dot matrix way; the detection probe comprises specificity 16S rDNA sequences and / or gyrB gene sequences of vibrio, comma bacillus, vibrio harveyi, vibrio alginolyicus, vibrio anguillarum, vibrio parahemolyticus, nocardia, nacardia seriolea, aeromonas, hydrophilic aeromonas, streptococcus and dolphin streptococcus, which are to be detected, the quality control probe includes PCR positive, chip fixed positive control, chip hybridizing negative control, chip hybridizing positive control and chip hybridizing blank control; the gene chip has the advantages of small volume and high flux, can detect known and unknown germs of the vibrio, the nocardia, the aeromonas and the streptococcus, and can detect specific germs with multiple kinds, and the simpleness and rapidness and specificity of the germs can be detected, and automatic detection can be carried out after detection software is additionally arranged.
Owner:NINGBO UNIV +2
Portable nucleic acid analysis system and high-performance microfluidic electroactive polymer actuators
PendingUS20170058324A1Easy to buildHeating or cooling apparatusMicrobiological testing/measurementElastomerActive polymer
Devices, systems and methods for the parallel detection of a set of distinct nucleic acid sequences use multiple sequence amplification and simultaneous hybridization readout. An automated nucleic acid analysis system comprises in microfluidic connection sample lysis, purification, PCR and detection modules configured to detect in parallel distinct nucleic acid sequences via multiple sequence amplification and simultaneous microarray hybridization readout. High performance microfluidic electroactive polymer (μEAP) actuators comprising a dead-end fluid chamber in which the floor of the chamber is an electrode covered with an EAP layer of dielectric elastomer are configured for particle sorting.
Owner:SRI INTERNATIONAL
Method for building sequencing library by hybridization
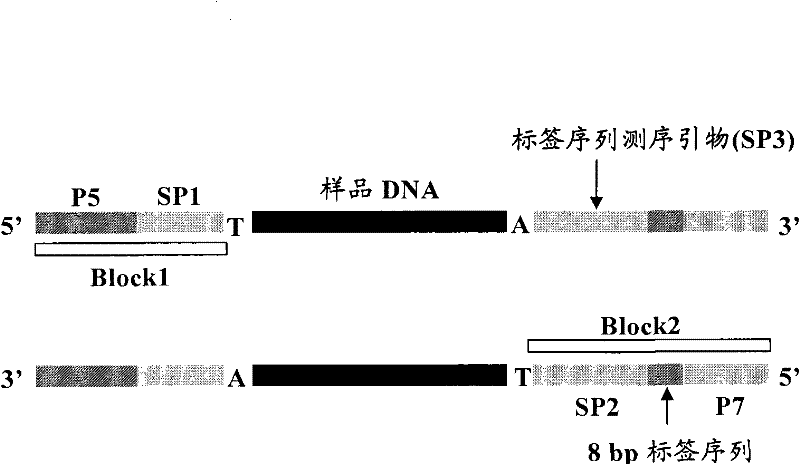
ActiveCN102409047AImprove introduction efficiencyAvoid wastingMicrobiological testing/measurementLibrary creationPcr methodBioinformatics
The invention provides indexes for DNA sequencing, especially for a sequence acquisition technology, and a method for introducing the indexes by adopting a PCR (polymerase chain reaction) method. The invention also provides adapter block sequences for blocking adapters and application of the adapter block sequences in a NimbleGen chip hybridization system, an Agilent liquid hybridization system and a NimbleGen EZ liquid hybridization system. The invention further provides a method for building a library and / or sequencing by using the indexes and / or adapter block sequence, and the library built by the method.
Owner:BGI TECH SOLUTIONS
Application of miRNA (Micro-Ribonucleic Acid) expression profile in predicting sensibilities of lung cancer patients to radiotherapy
InactiveCN101824464AReduce overtreatmentLittle side effectsMicrobiological testing/measurementAbnormal tissue growthTotal rna
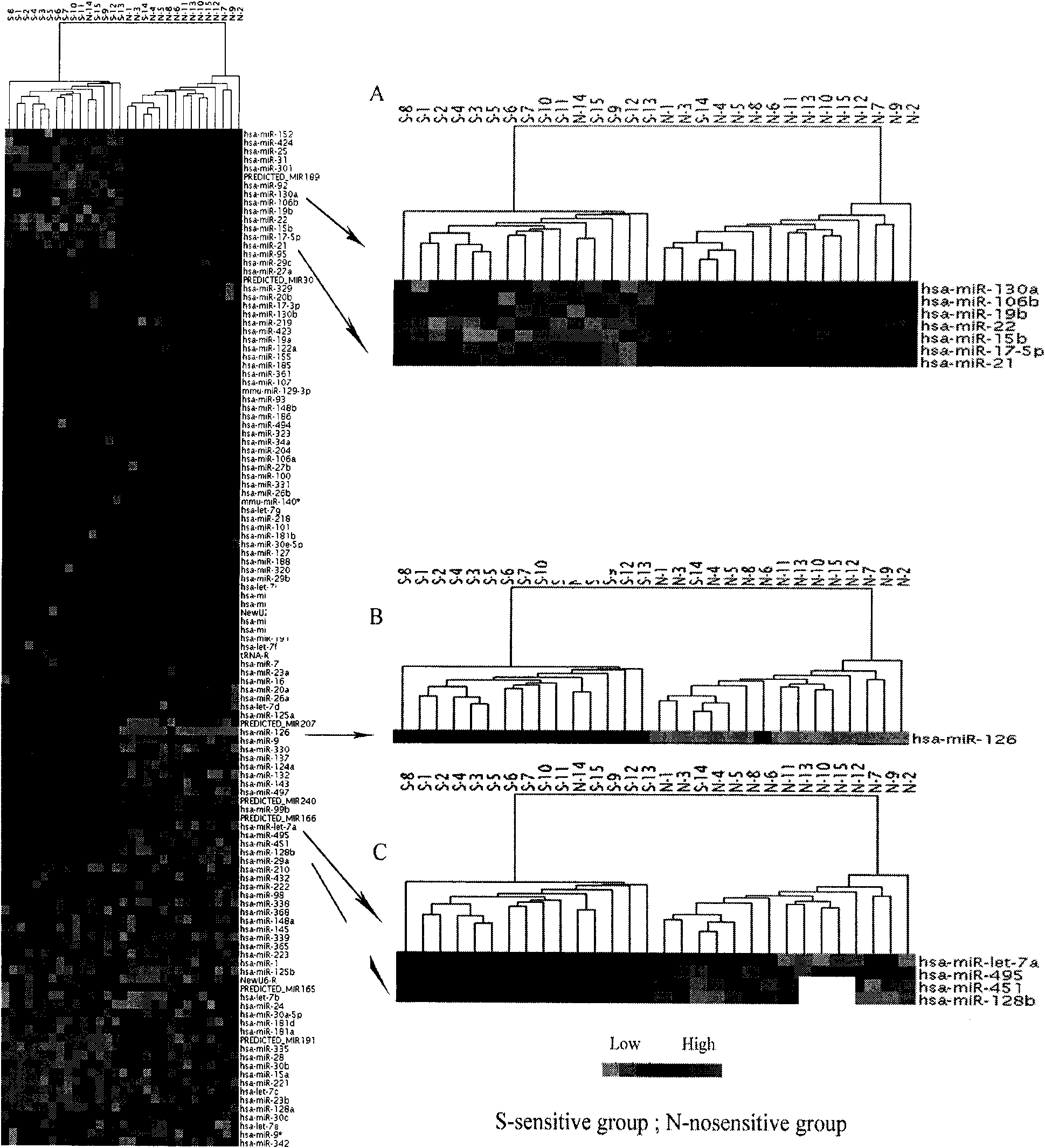
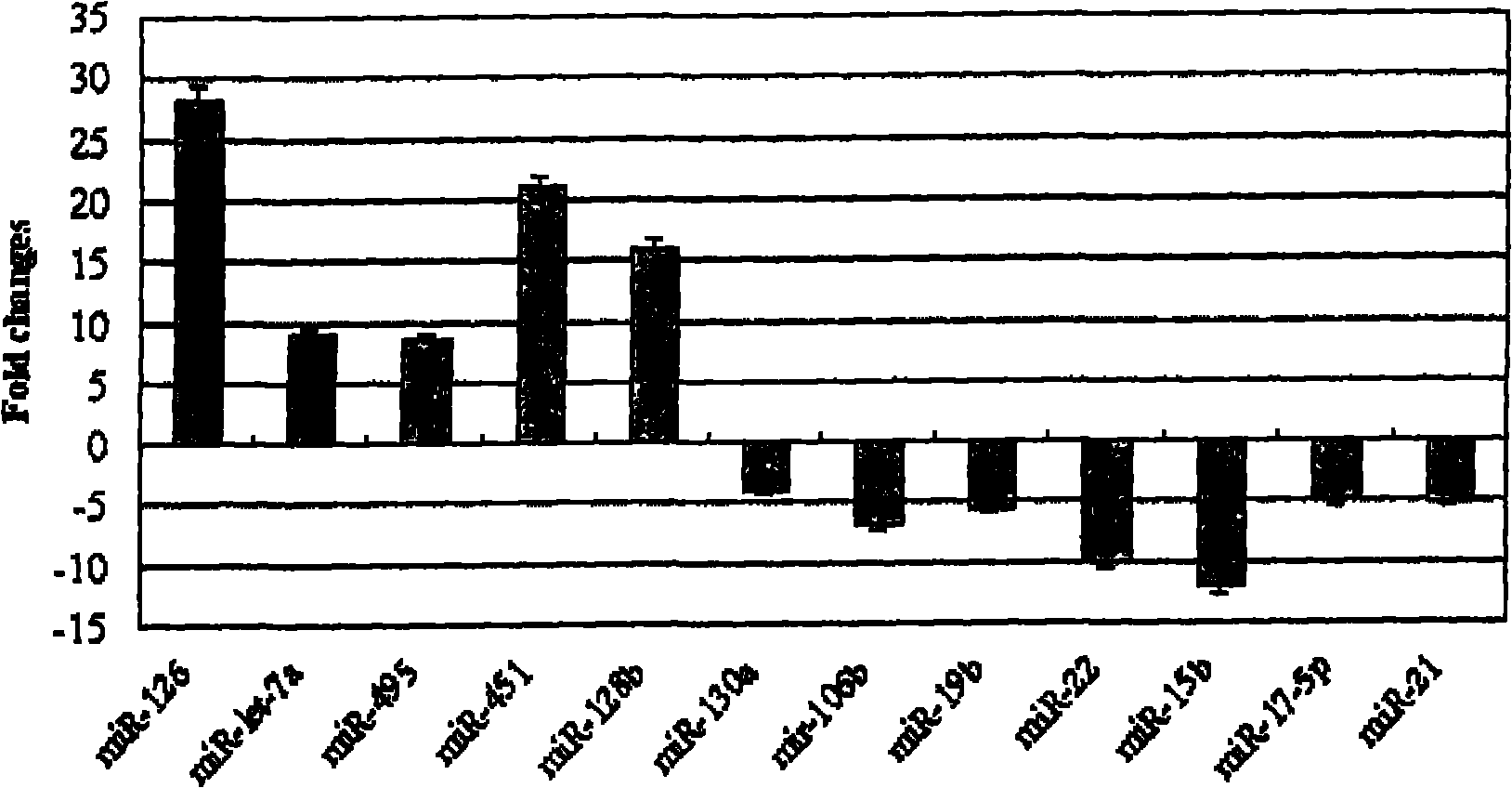
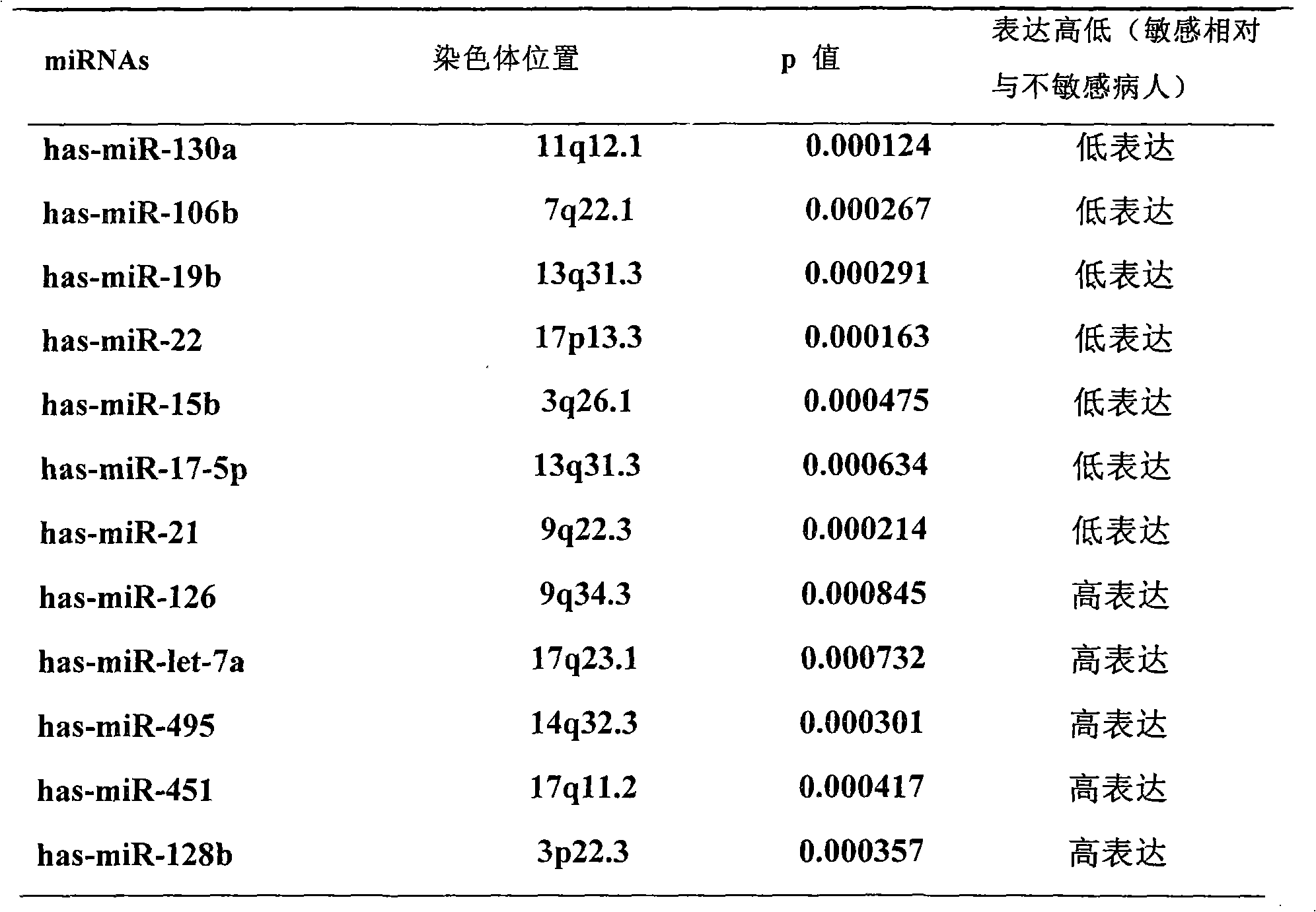
The invention discloses application of a miRNA (Micro-Ribonucleic Acid) expression profile in predicting the sensibilities of lung cancer patients to the radiotherapy. A preparation method for the invention comprises the following steps of: selecting lung cancer patients who are clinically unusually sensitive and highly tolerant to the radiotherapy, drawing 5ml of peripheral blood, and extracting total RNA by using a TRIZOL kit (Invitrogen); then hybridizing by using a MicroRNA expression profile chip; carrying out data processing, and analyzing the difference of MicroRNA expression profiles of radiotherapy sensitive and non-sensitive patients by using a cluster analysis method; and finally, checking a result by using a Real-time PCR (Polymerase Chain Reaction) method. The inventor finds that an expression profile consisting of 12 miRNA can regulate and control the expression of genes relevant to tumor radiosensitivity and be used for predicting the sensitivity of the lung cancer patients to the radiotherapy and providing an experimental foundation of an early period for the individualized radiotherapy of lung cancer.
Owner:INST OF RADIATION MEDICINE CHINESE ACADEMY OF MEDICAL SCI
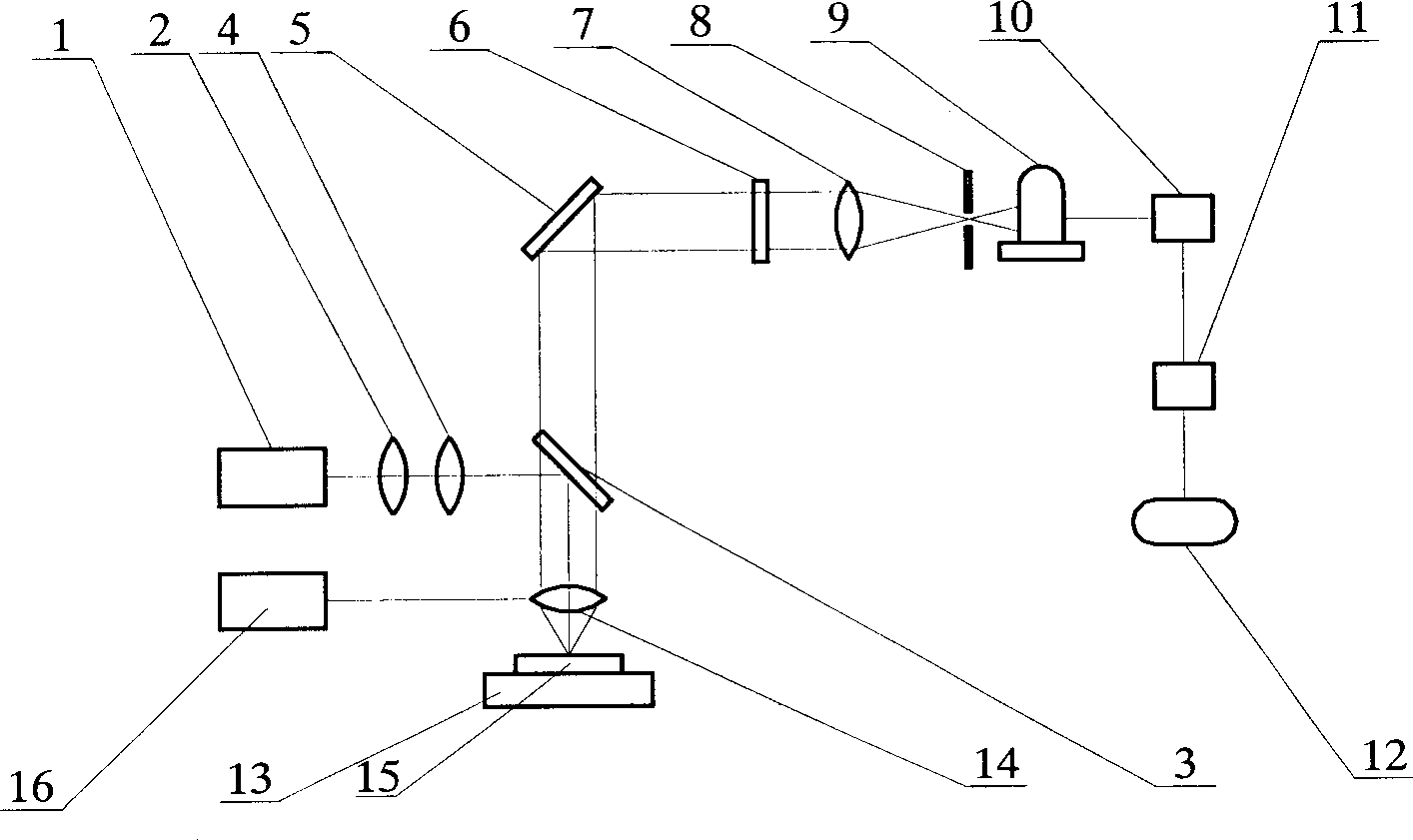
Multispectral imaging gene chip scanning instrument
InactiveCN1375691ASimplified Scan DirectionShorten detection timeEmission spectroscopyRadiation pyrometryFluorescenceImaging lens
The gene chip scanister mainly is used for detecting fluorescently-labeled post-hybridization gene biological chip, including the following steps: a laser beam is saped by linear shaper and passed through a reflector with slit to form a linear laser beam on the tested chip; the optical signal fed back by tested chip is passed through the reflector with slit and reflected, then passed through a photographic optical system and image in the slit diaphragm position; when it is excited by using laser beam with multiple wavelength, said signal is further passed through first imaging lens, dispersing element and second imaging lens and reflected on the detector of electric charge coupler with face array or line array. As compared with existent technology, it not only can simplify scanning direction, save detection time, but also can raise the resolution and accuracy of the detection.
Owner:SHANGHAI INST OF OPTICS & FINE MECHANICS CHINESE ACAD OF SCI
Medicine and bacterium resistant detection chip, method for preparation and application thereof
InactiveCN1635150AEasy to detectQuick checkMicrobiological testing/measurementBacteria identificationInfective disorder
The invention relates to a detection chip for familiar resistant organisms and its preparation and application methods belonging to microorganism detection field. The invention employs DAN chip method including fixing the synthesized oligonucleotide probes for clinical detection of familiar resistant organisms on the slide surface to form a dot-plot, hybridizing the pending sample DNA with the chip to obtain a great deal of gene sequence information relative to the bacteria identification and drug tolerance, identifying the kinds of the familiar pathogenicbacterias existing in the clinical sample, thus to implement the identification of the clinical familiar pathogenicbacterias and detection of the main drug resistant spectrum. The invention is simple, convenient, quick, sensitive and specific, and can obviously shorten the disease diagnosis time.
Owner:AFFILIATED HUSN HOSPITAL OF FUDAN UNIV +1
Method for preparing detection chip and method for detecting pathogen by using the same
InactiveCN101240347ANot easy to storeEnhanced signalMicrobiological testing/measurementMultiplex pcrsBiology
The invention discloses a preparation of a detection chip, comprising the steps of: 1) designing oligonucleotide probe of bacteria; 2) preparing the chip. The invention further discloses a method for detecting pathogene in multiple food samples with the detection chip, comprising the following steps: 1) designing a primer; 2) extracting DNA from samples to be detected, and getting extracting solution; 3) multi-PCR amplifying of target gene; 4) hybridization detecting of chip; and 5) detecting and analyzing of hybridization result. The detection step of the invention is simple, easy to operate, and produces high correct rate result.
Owner:ZHEJIANG UNIV
Gene chip and reagent box for detecting food-borne virus
InactiveCN101328505AGuaranteed reproducibilityGuaranteed accuracyMicrobiological testing/measurementAgainst vector-borne diseasesFood borneRotavirus RNA
The invention relates to a gene chip for detecting food-home virus and a kit, belonging to the inspection field. The surface of a solid phase carrier is fixed with a plurality of detection probes and quality control contrast probes, wherein, the detection probes consist of the probes for detecting hepatitis A virus, human astrovirus, norwalk virus G1, norwalk virus G2, rotavirus, polio virus 1, polio virus 2 and polio virus 3; and the quality control contrast probes consist of a spotting positive quality control probe, a chip hybridization positive quality control probe and a chip negative quality control probe. The gene chip and the kit have the advantages of: (1) high throughout: five common viruses are integrated and detected simultaneously, and the practicability is strong; (2) rapidness: the detection time is only 4 hours; (3) specificity: the false positive caused by the cross reaction is avoided; (4) flexibility: the detection flexibility of the chip is 10<8> virus particles per gram of the tissue sample, which is higher than that of RT-PCR; and (5) good repetitiveness and stable results. The gene chip and the kit can be widely applied to the food safety inspection of the inspections system.
Owner:PEOPLES REPUBLIC OF CHINA BEIJING ENTRY EXIT INSPECTION & QUARANTINE BUREAU +1
Method for noninvasive prenatal disgnosis of congenital deafness genetic disease
InactiveCN103436609AEasy to diagnoseRelieve painNucleotide librariesMicrobiological testing/measurementPrenatal diagnosisPhysiology
The invention belongs to the technical field of gene diagnosis, and provides a method for noninvasive prenatal disgnosis of a congenital deafness genetic disease through the technology taking gene chips as a platform. Each microarray gene chip comprises a chip base and probes fixed on the chip base, and the probes have the nucleotide sequences which are as shown in the table 1-2. The diagnosis method comprises the steps as follows: preparation of to-be-tested DNA, multiple PCR amplification, chip hybridization, data processing and image analysis, chip scanning and result obtaining. At the same time, the invention comprises a kit for the noninvasive prenatal disgnosis of the congenital deafness genetic disease, and the kit comprises the microarray gene chips; the method for noninvasive prenatal disgnosis of the congenital deafness genetic disease is simple and convenient; detection has the characteristics of high throughput, good specificity and high sensitivity, the situation that whether each locus belongs to the wild type or the mutant type can be quickly screened, and the diagnosis of the congenital deafness genetic disease can be improved to the gene level; since the noninvasive prenatal disgnosis technology is adopted, the cost is reduced, the time is saved, and the pain of patients is further reduced.
Owner:康盈创新生物技术(北京)有限公司
Gene chip detection method of transgenic maize
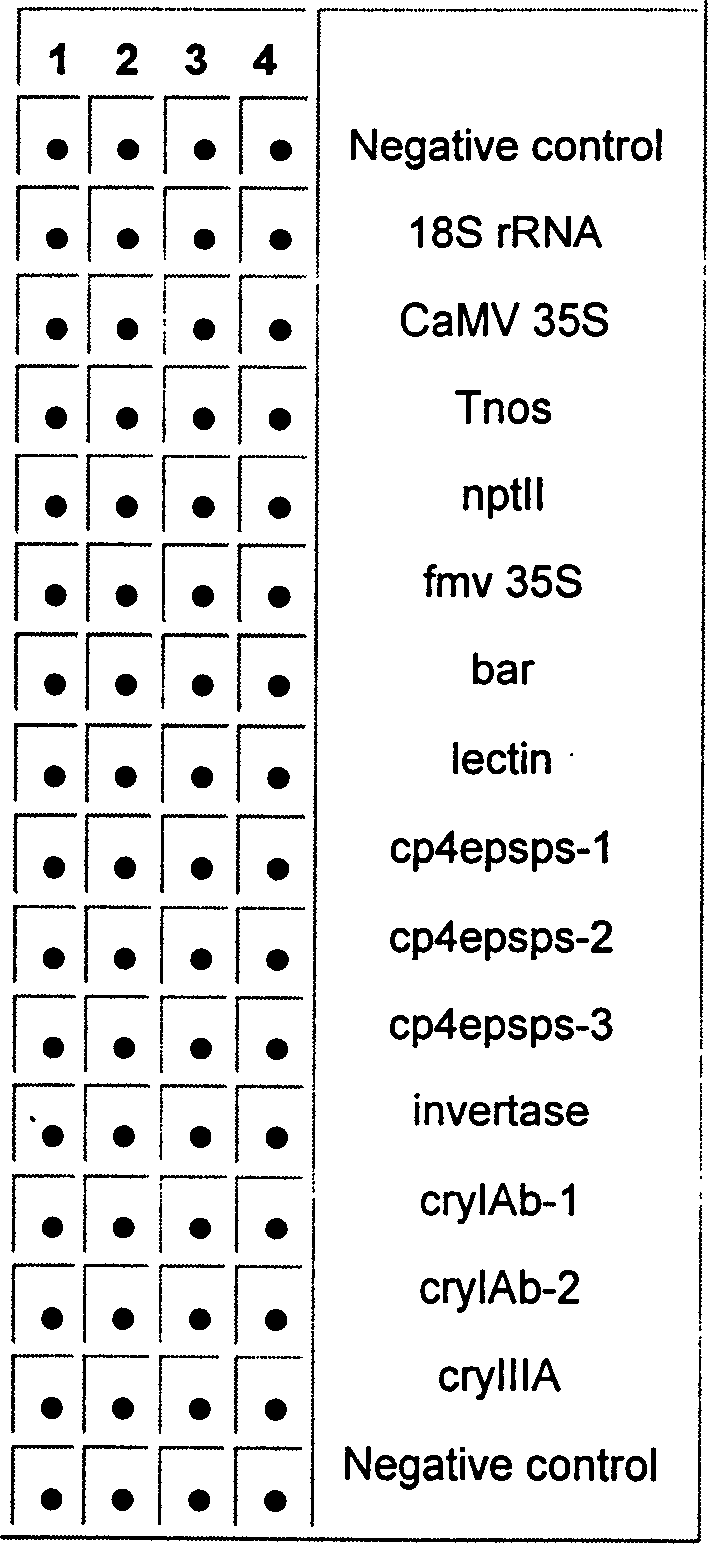
The invention discloses a gene chip detection method of transgenic maize. The main steps comprise primer and probe determination, chip preparation, PCR (polymerase chain reaction) amplification, PCR product marking, chip hybridization, chip scanning and data processing. By adopting the detection method, sequences of seven main exogenous DNAs (deoxyribonucleic acid) commonly used in transgenic maize, such as promoters, terminators, insect-resistant genes, glyphosate-resistant genes and the like, can be detected and analyzed through one step, so that the detection method of transgenic maize has the advantages of high throughput, miniaturization, automation, rapidity, high efficiency and high sensitivity, and can be widely applied to related fields such as import-export inspection and quarantine, and the like.
Owner:威海华康新材料科技有限公司
Gene chip for non-invasive prenatal diagnosis of high-risk hereditary hearing loss and preparation method
InactiveCN102719538AEasy to operateHigh detection specificityNucleotide librariesMicrobiological testing/measurementPrenatal diagnosisNon invasive
The invention relates to a gene chip for non-invasive prenatal diagnosis of high-risk hereditary hearing loss and a preparation method in the technical field of gene diagnosis. The gene chip comprises a substrate and probes fixed on the substrate, wherein the probes are nucleotide sequences shown as SEQ ID NO.1-24. Usage of the gene chip includes the steps of sample preparation, multiplex PCR (polymerase chain reaction) amplification, ligase detection reaction, chip hybridization, chip scanning and result obtaining. Beside, the invention further relates to a reagent kit comprising the gene chip. The gene chip for diagnosis is simple in operation steps, only requires the multiplex PCR reaction, the ligase detection reaction and hybrid scanning once, is high in detection specificity and good in stability so as to be capable of correctly distinguishing homozygotes from heterozygotes of each locus and high in repeatability of repeated experiments, is short in detection time and low in cost as the substrate can be universally used for other chips by means of universal chip technology.
Owner:SHANGHAI JIAO TONG UNIV +1
Chip-based 5-hydroxymethylated cytosine detection method with high flux and high sensitivity
InactiveCN103409514AEnable high-throughput detectionGuaranteed accuracyMicrobiological testing/measurementModified dnaCytosine
The invention discloses a chip-based 5-hydroxymethylated cytosine detection method with high flux and high sensitivity. The method comprises steps of fixing rolling circle amplification primers to a chip to form a micro array; treating genome DNA to be detected by using beta-glucosyltransferase; subjecting all 5hmC in the genome DNA to galactosylated modification; treating the modified DNA by using sodium sulfite; placing open-loop probes used for detecting different C sites into the same reaction tube to carry out hybridization connection; subjecting products after connection and the chip to hybridization; subjecting specific primers of the fixed chip to rolling circle amplification by using the connected probe as the template; subjecting different fluorescence detection probes and the chip to hybridization; and determining whether hydroxymethylation happens to the different C sites of the genome DNA and the hydroxymethylation frequency according to fluorescence types and intensity of different matrix dots on the chip. The method has strong operationality, strong specificity, high sensitivity, accurate detection results, high detection efficiency, low detection cost and wide application range.
Owner:XUZHOU MEDICAL COLLEGE
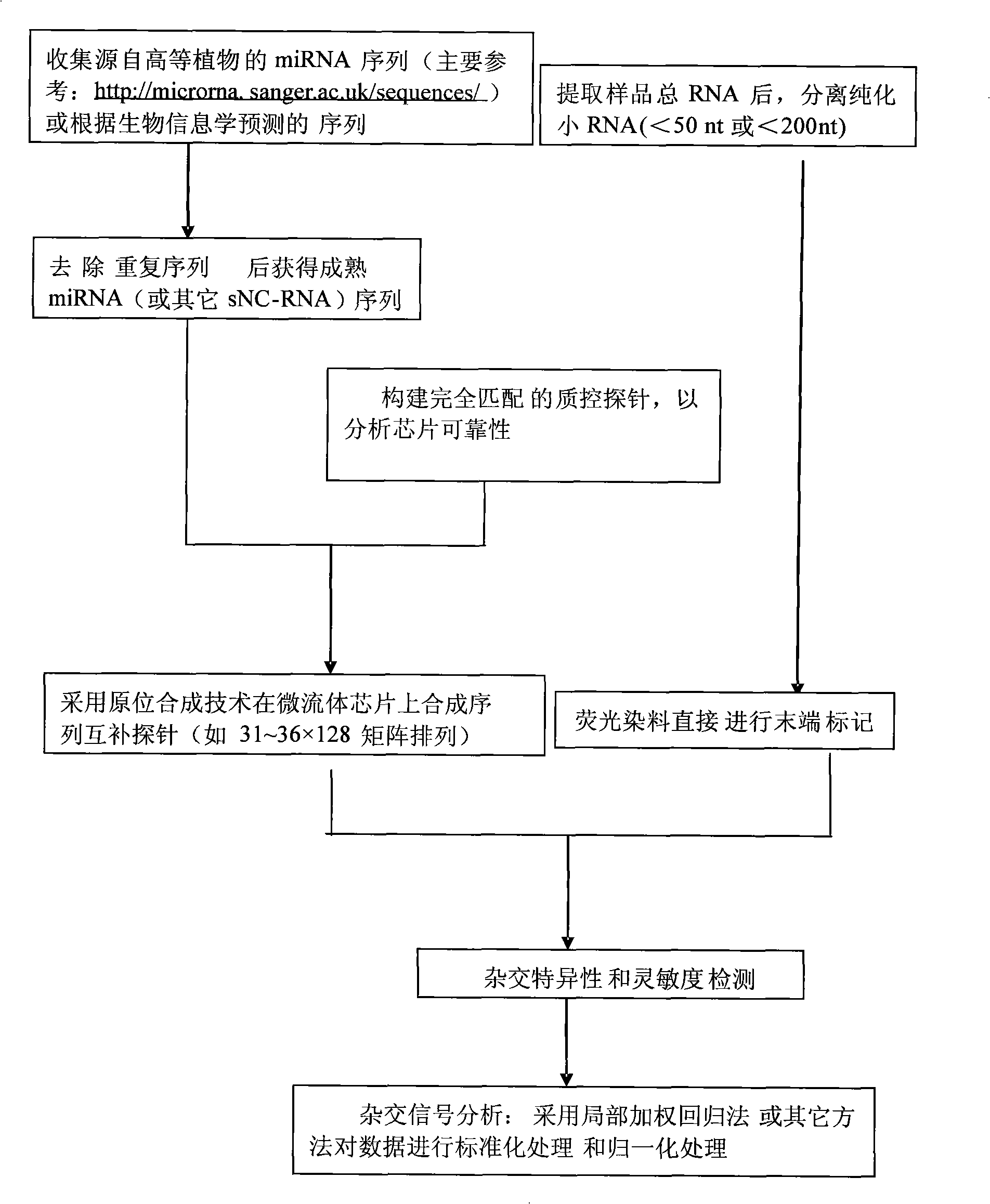
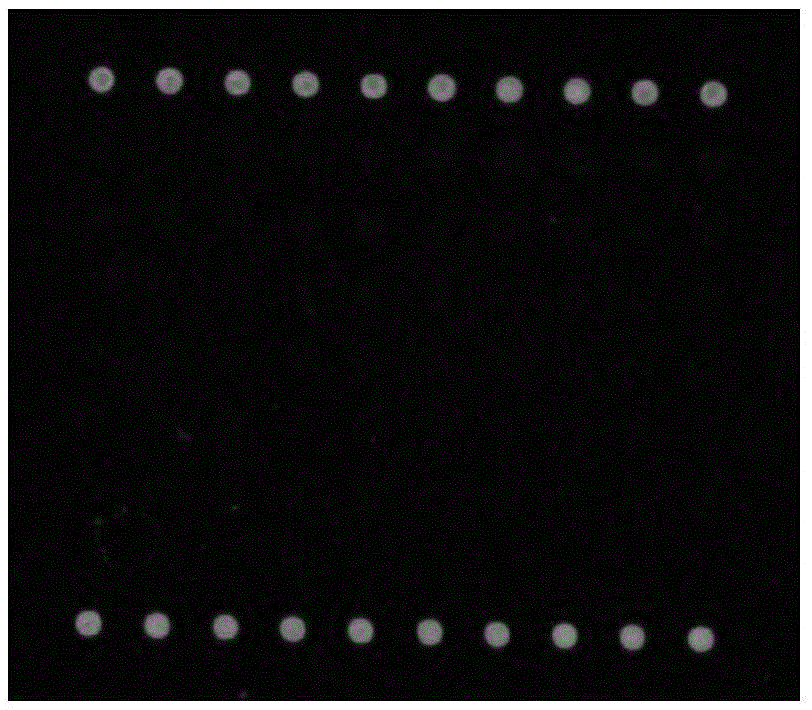
Detection process for synthesizing microfluid chip by in-situ method
InactiveCN101285100AHigh sensitivityImprove throughputMicrobiological testing/measurementOligonucleotide chipSense strand
The invention discloses a detection method synthesizing microfluidic chips in situ, which comprises the following steps that: 1) a sequence of non-coding small RNAs reported on the prior model plant is utilized, sequence information of miRNA is preferably selected, and complementary oligonucleotide hybridization probes are designed according to the sequence of a sense strand or an antisense strand of miRNA; 2) an oligonucleotide chip is prepared by an in-site synthetic method; 3) small RNAs smaller than a 50 oligonucleotide group are extracted from a plant to be detected, then are hybridized with the probes in the chip after making end labeling, and the result of chip hybridization is obtained to perform qualitative and quantitative analysis. The detection method can be used for the validation of miRNA (and other sNC-RNAs) obtained through the forecast by a bioinformatics method, can also be used for the detection and identification of miRNA (and other sNC-RNAs) in new plant species, and can also be used for the determination of miRNA (and other sNC-RNAs) variation relation under different treatment conditions (eg. under the condition of virus infection).
Owner:ZHEJIANG SCI-TECH UNIV
Micro-array chip detection method for 10 on-melon diseases and used chip probe
InactiveCN105297141APromote the development of import and export tradeEffective control of intrusionNucleotide librariesMicrobiological testing/measurementDiseaseFluorescence
The invention discloses a micro-array chip for 10 on-melon diseases, and also discloses a micro-array chip detection method for the 10 on-melon diseases by using the chip. The following steps are executed in sequence for each to-be-tested sample: 1) extracting the DNA of the to-be-tested sample; 2) performing a PCR reaction on a primer fluorescent marker; 3) mixing volume-equal PCR amplification products corresponding to at least two germs obtained by step 2); 4) chip-hybridizing and washing: denaturating one PCR amplification product corresponding to each germ obtained by step 2) or a mixture obtained by step 3) at 95 DEG C for 5min, and then ice-bathing for 5min to obtain after-pretreatment PCR amplification products; then chip-hybridizing and washing the after-pretreatment PCR amplification products, wherein the chip-hybridizing is performed at 45 DEG C for 1h; 5) scanning the chip to acquire a detection result.
Owner:新疆出入境检验检疫局检验检疫技术中心
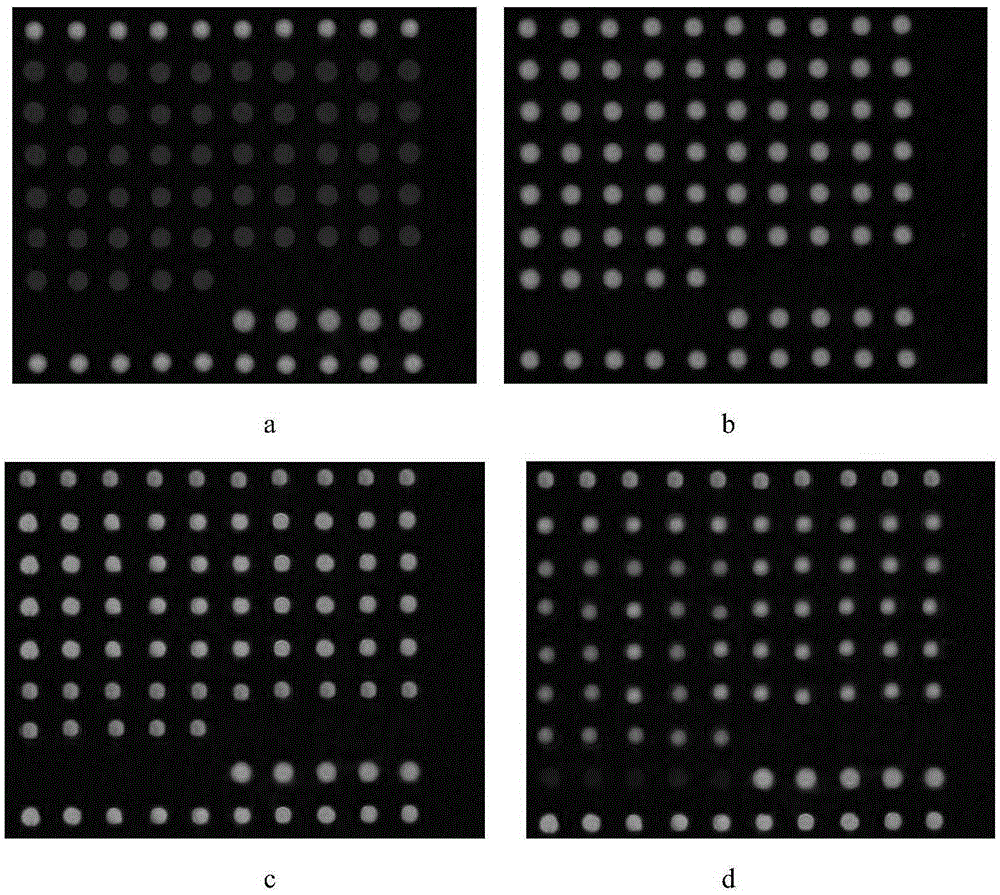
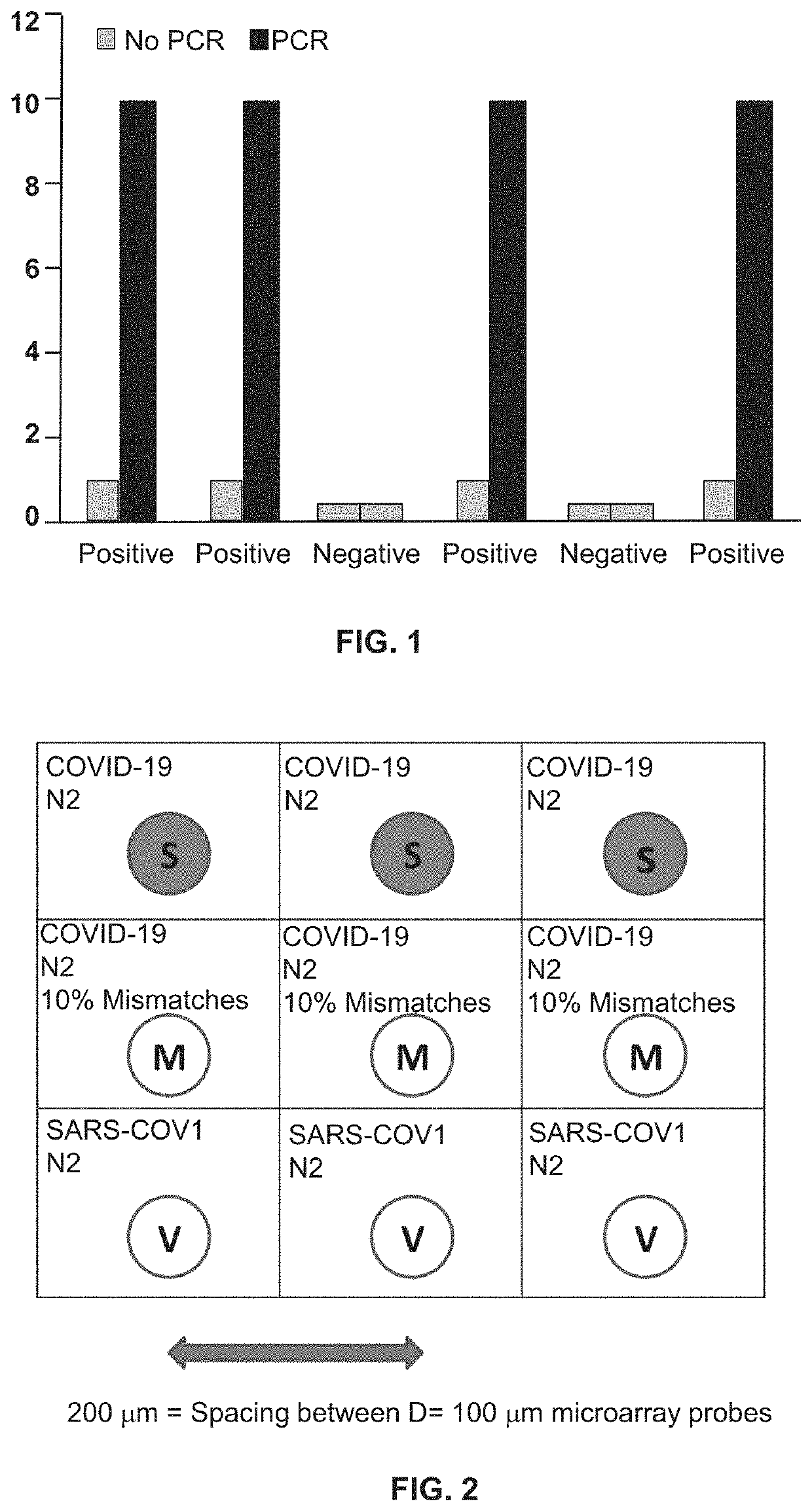
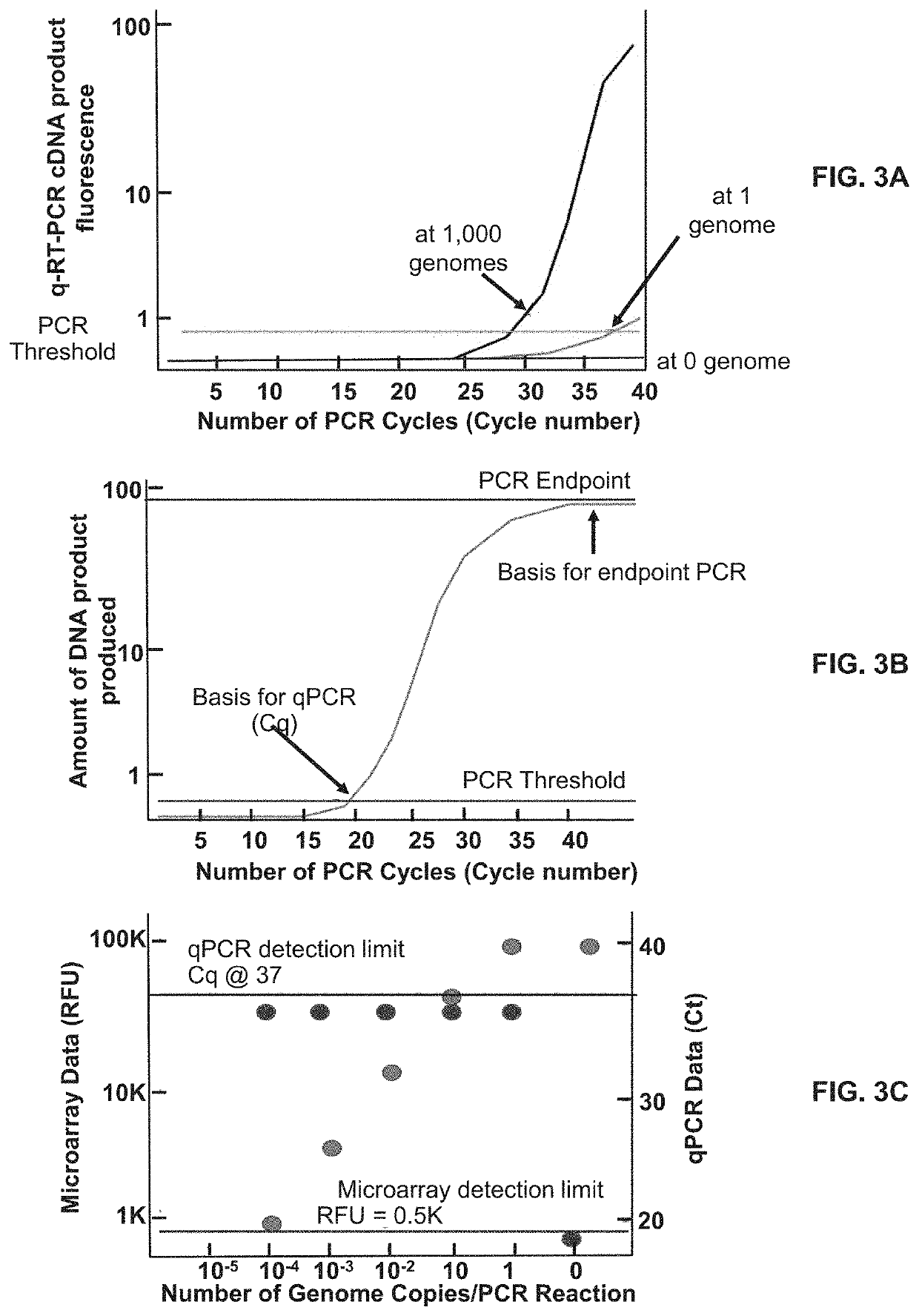
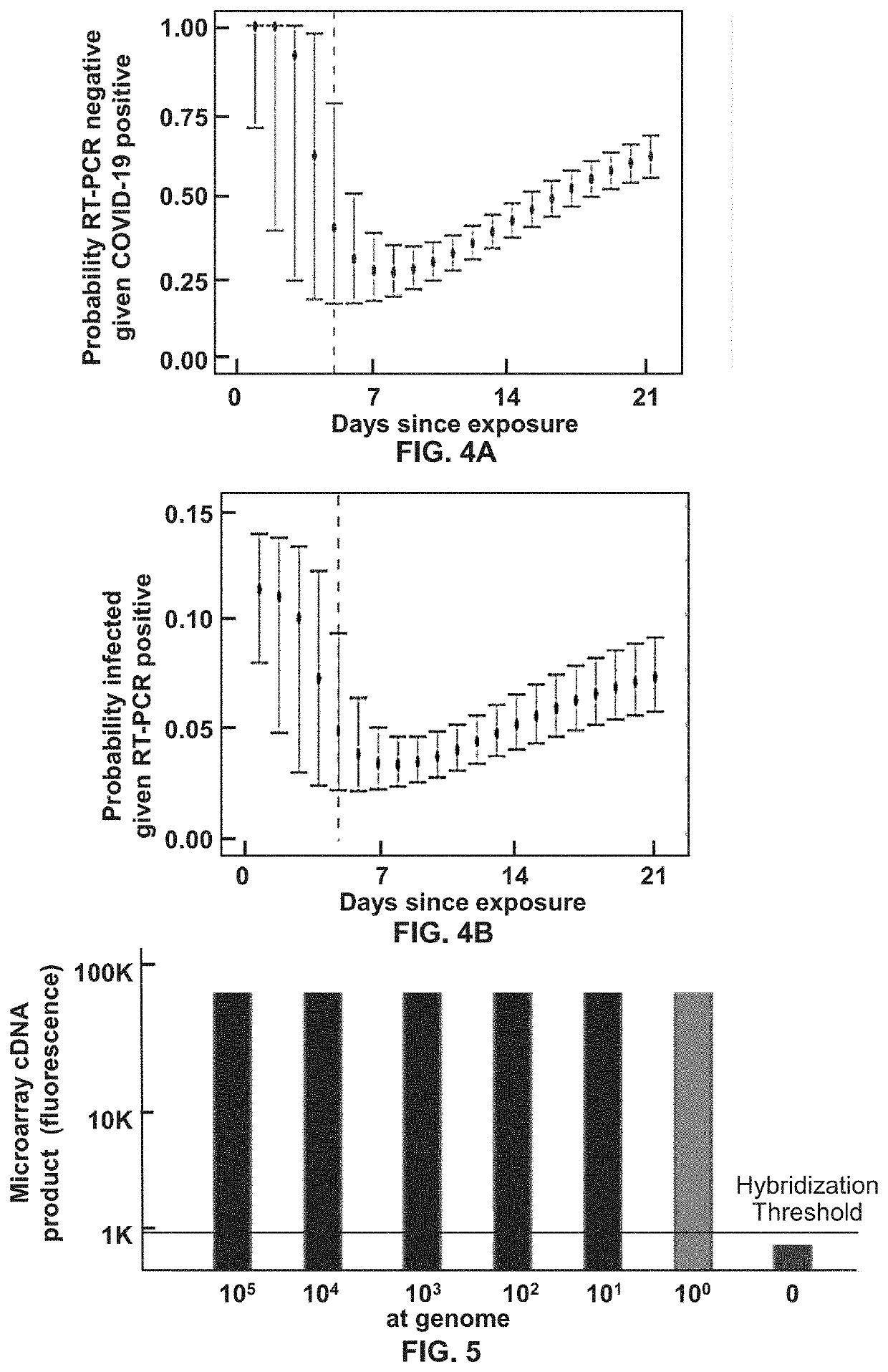
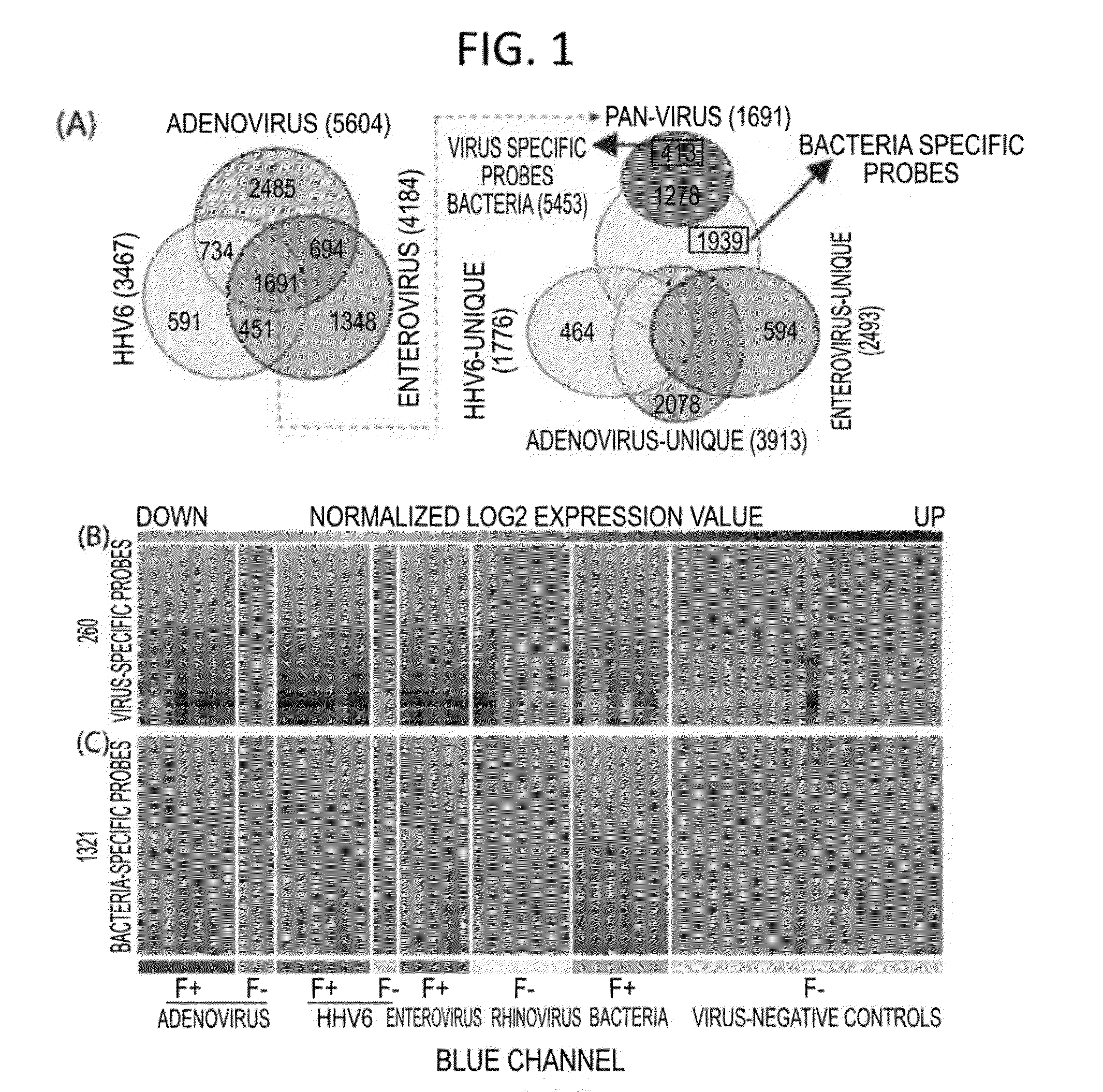
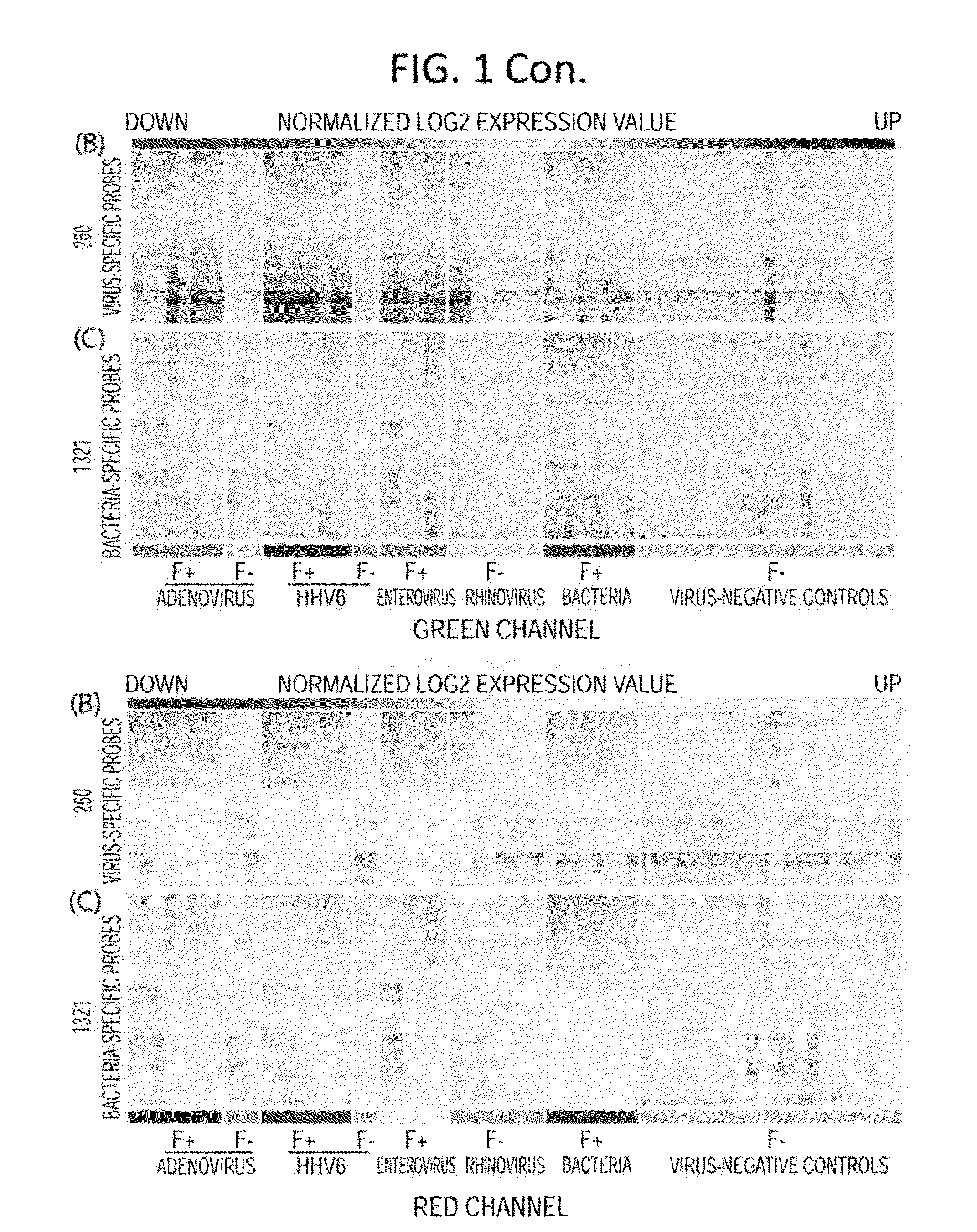
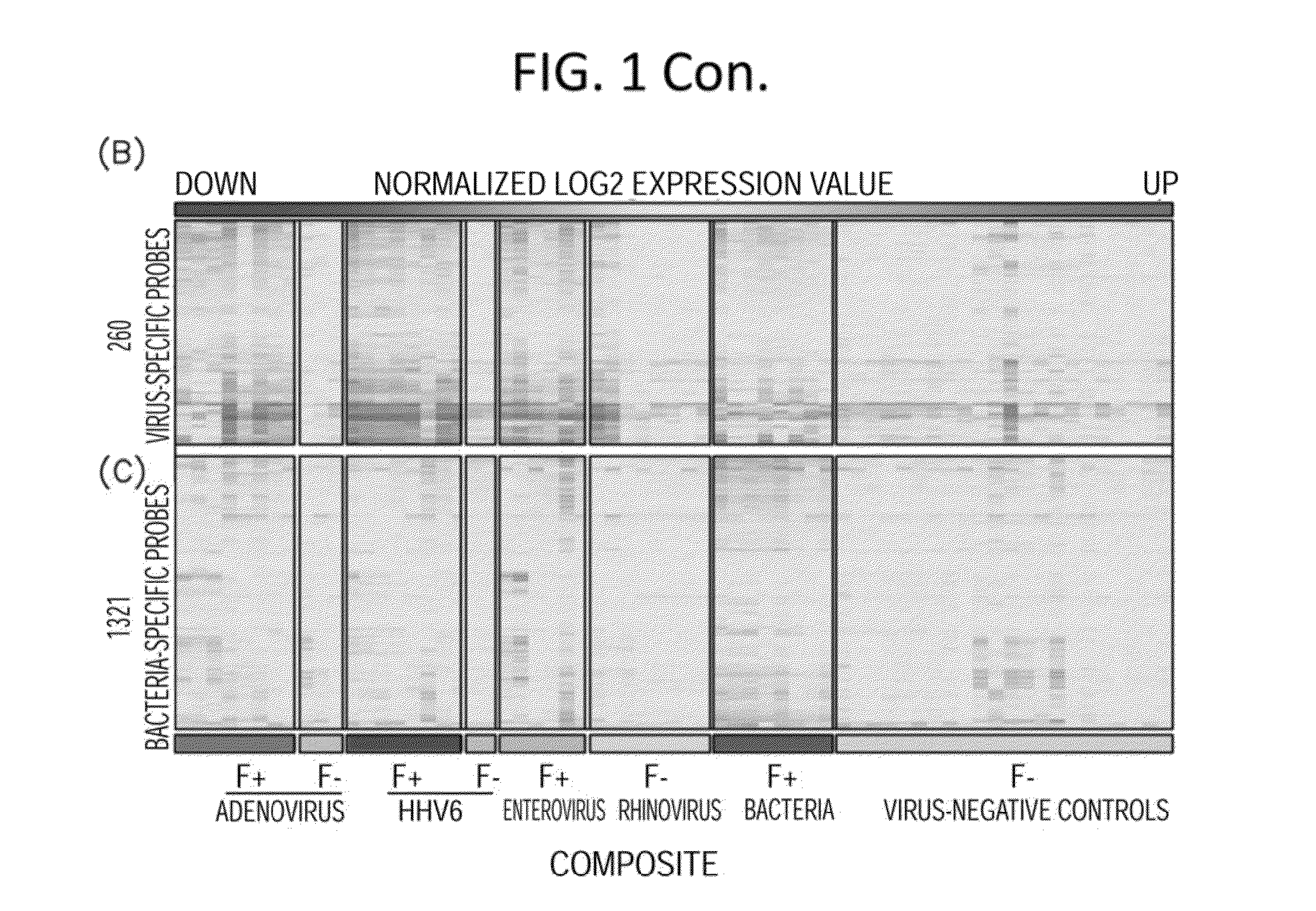
Methods for Detecting Low Levels of Covid-19 Virus
PendingUS20220364157A1Low thresholdMicrobiological testing/measurementDiseaseRespiratory tract disease
Provided herein is a method for detecting the presence of a COVID-19 virus in a human sample or an environmental sample having one or more viruses and bacterial pathogens. Samples are processed to obtain total nucleic acids. A combined reverse transcription and asymmetric PCR amplification reaction is performed to obtain fluorescent labeled COVID-19 virus specific amplicons. The amplicons are detected by microarray hybridization near the lowest limit of detection. Also provided is a method for detecting concurrently with COVID-19 virus, the presence of respiratory disease-causing pathogens including viruses, bacteria and fungus in a single assay using the above method.
Owner:PATHOGENDX INC
Transgenic product low-density gene chip detecting method
InactiveCN1580782AGuaranteed concentrationLow mismatch rateBiological testingExogenous DNALow density
The low-density gene chip detection method of transgenic product relates to a detection method for transgenic product by uisng low-density gene chip. Said method includes the following steps: selecting primer and probe; processng chip carrier, preparing chip; dissolving probein ultrapure water; adding sample application liquid; fixing the probeon the treated chip carrier, sampling; multigene PCR amplification; labeling MPCR product; chip hybridization experiment of MPCR product; scanning analysis and data processing. As compared with traditional detection method said invention has the characteristics of quick seep, high efficiency and high automation level, and can simultaneously make detection and analysis of common 9 exogenous DNA sequences of promoter, terminator, screening marker gene, weedicide-resisting gene and insect-resisting gene, etc. in transgenic products.
Owner:XIAMEN UNIV +1
Portable nucleic acid analysis system and high-performance microfluidic electroactive polymer actuators
ActiveUS20170030859A1Easy to buildHeating or cooling apparatusMicrobiological testing/measurementActive polymerLysis
Devices, systems and methods for the parallel detection of a set of distinct nucleic acid sequences use multiple sequence amplification and simultaneous hybridization readout. An automated nucleic acid analysis system comprises in microfiuidic connection sample lysis, purification, PCR and detection modules configured to detect in parallel distinct nucleic acid sequences via multiple sequence amplification and simultaneous microarray hybridization readout. High performance microfluidic electroactive polymer (μEAP) actuators comprising a dead-end fluid chamber in which a surface of the chamber is an electrode covered with an EAP layer of dielectric elastomer are configured for particle sorting.
Owner:SRI INTERNATIONAL
Determination of one section vagina bacillus Gardner DNA sequence (3) specificity and its application
InactiveCN1493698AFast wayAccurate methodSugar derivativesMicrobiological testing/measurementOrganismBiomedical technology
The specificity and PCR amplification primer of a viginal Gardener's bacillus DNA sequence (3) fragment are determined by searching and analyzing the data of gene database. Said sequence with 196 bp in length is obtained by PCR amplification and clone of said viginal Gardener's bacillus infected specimen. It can be used to diagnose the viginal Gardener's bacillus with high speed and correctness and low cost.
Owner:昆明寰基生物芯片开发有限公司
A method for detecting structural variation in target regions of the genome
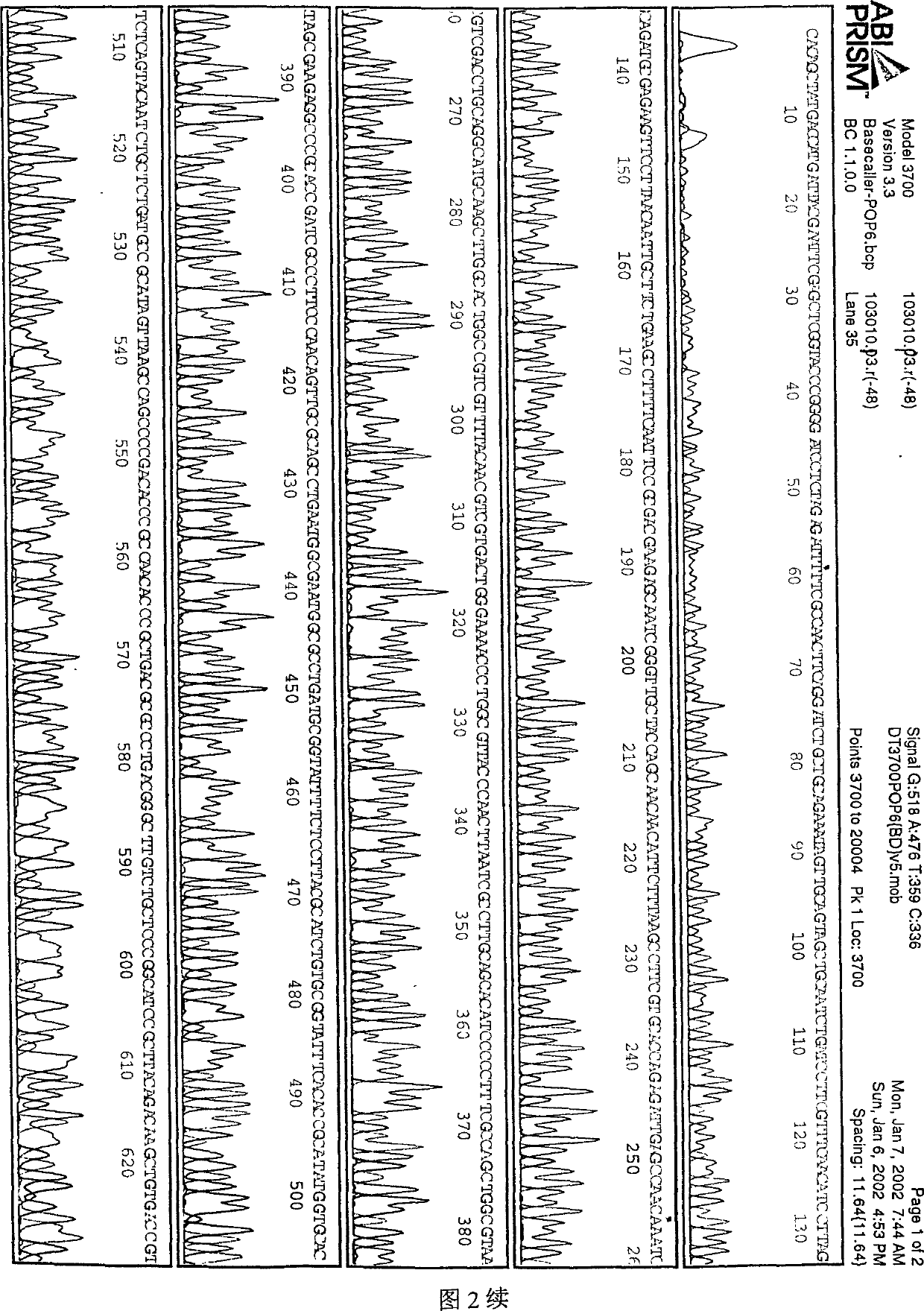
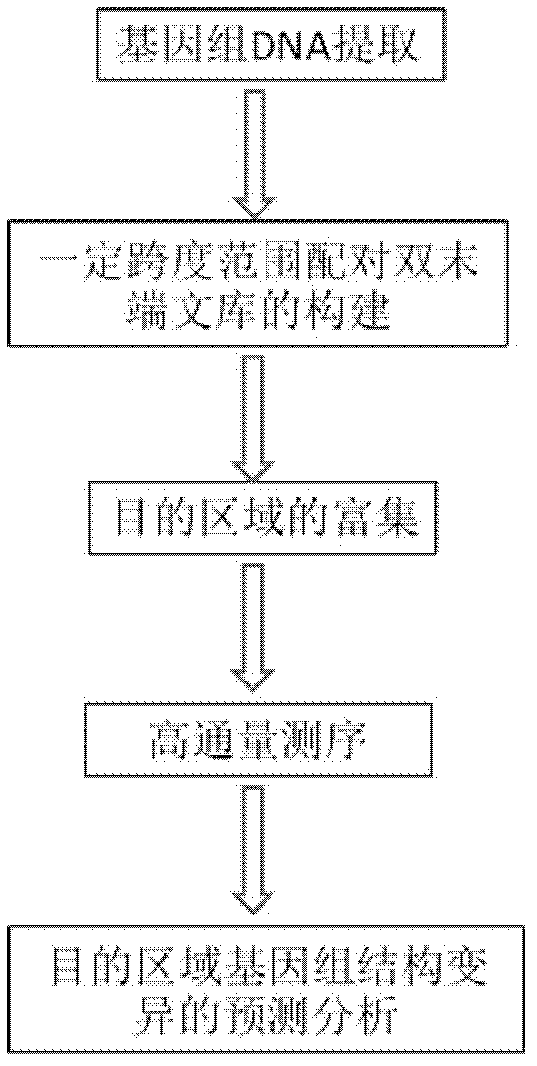
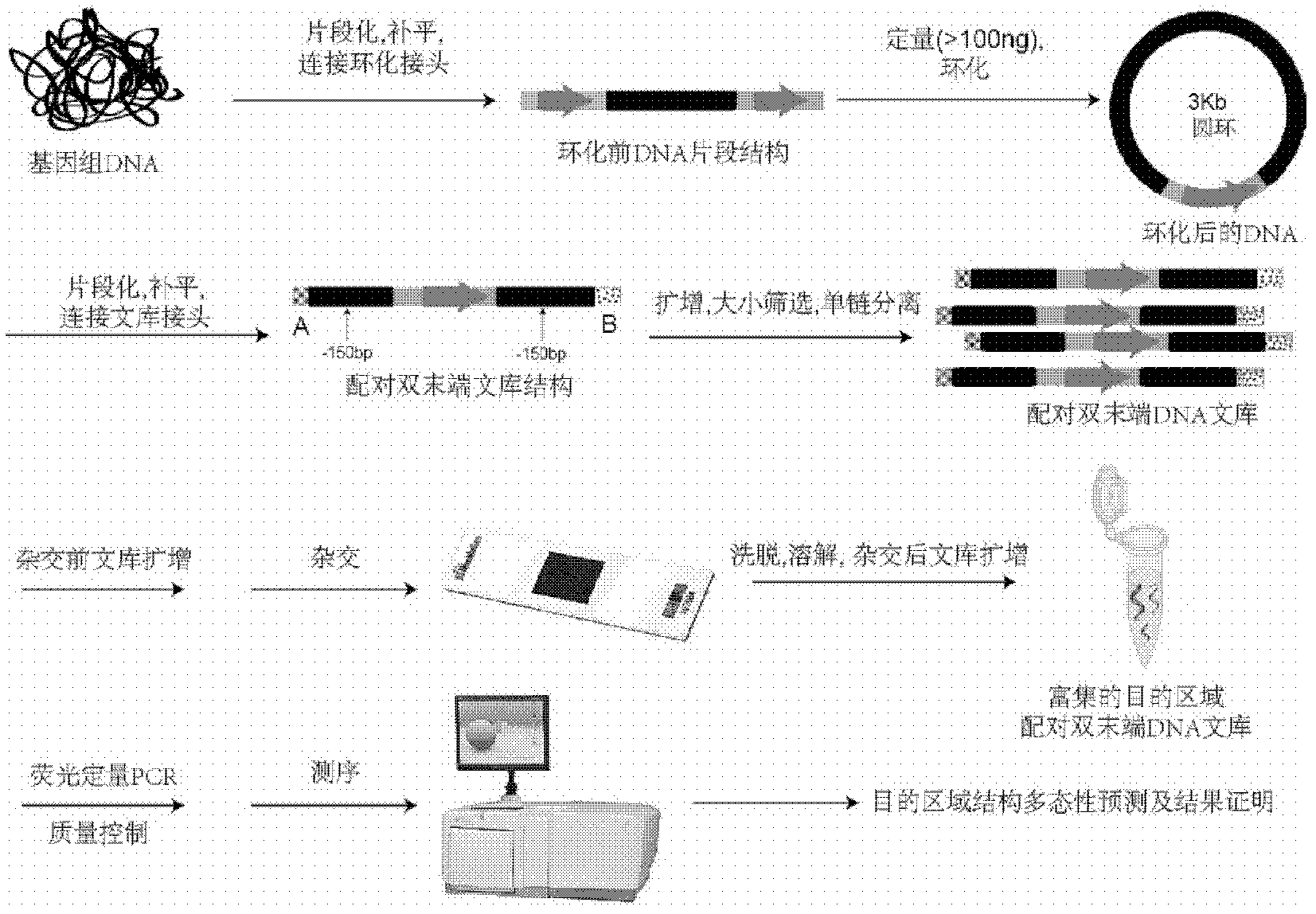
InactiveCN102286632AHigh sensitivityHigh resolutionMicrobiological testing/measurementGeneComputational biology
The invention provides a method for detecting the structural variation of the genome target region. The present invention skillfully combines double-end library construction technology, chip hybridization technology and high-throughput sequencing technology together, so that the structural variation of any interested target region can be detected with high sensitivity and high resolution.
Owner:SUN YAT SEN UNIV
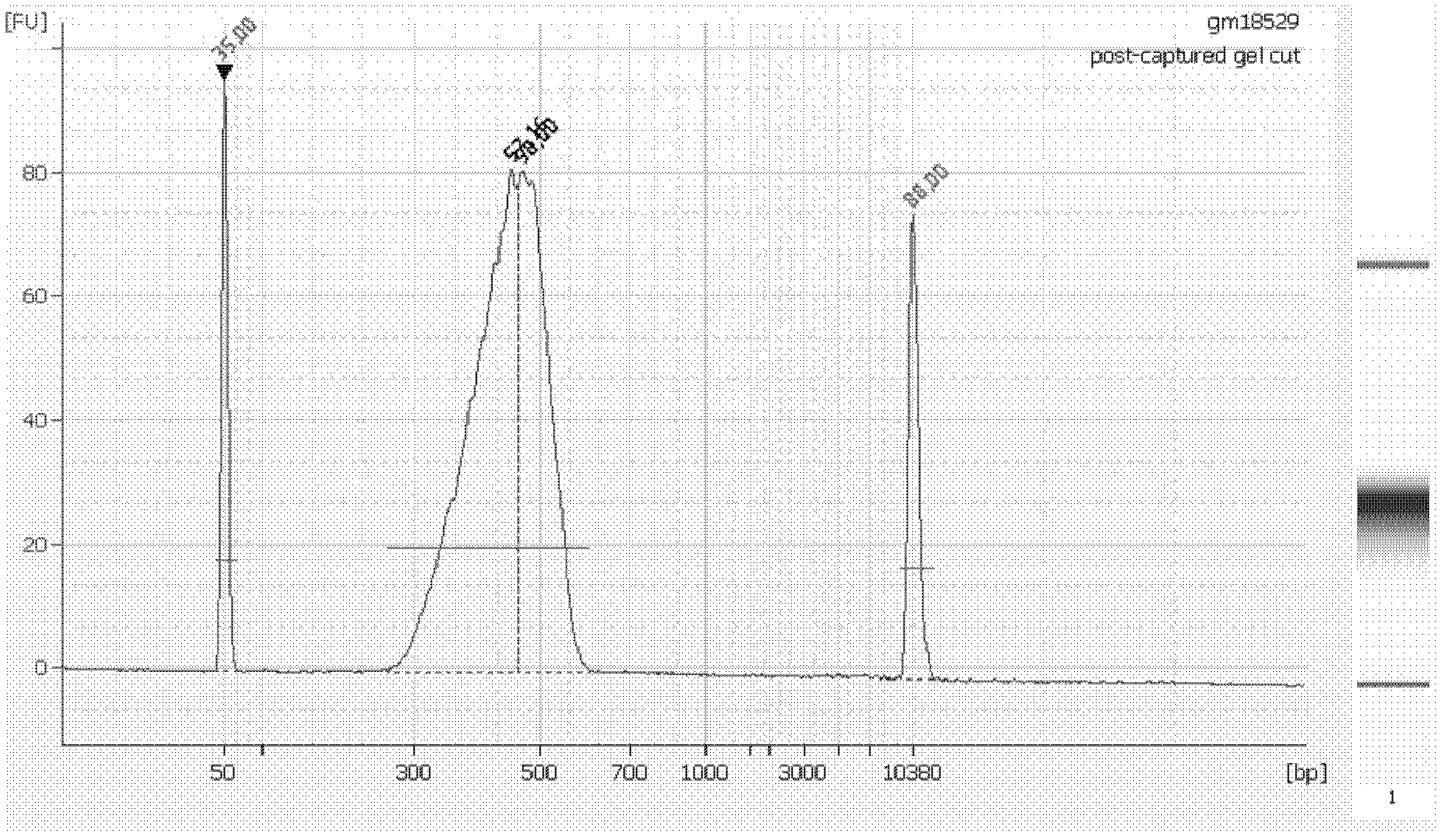
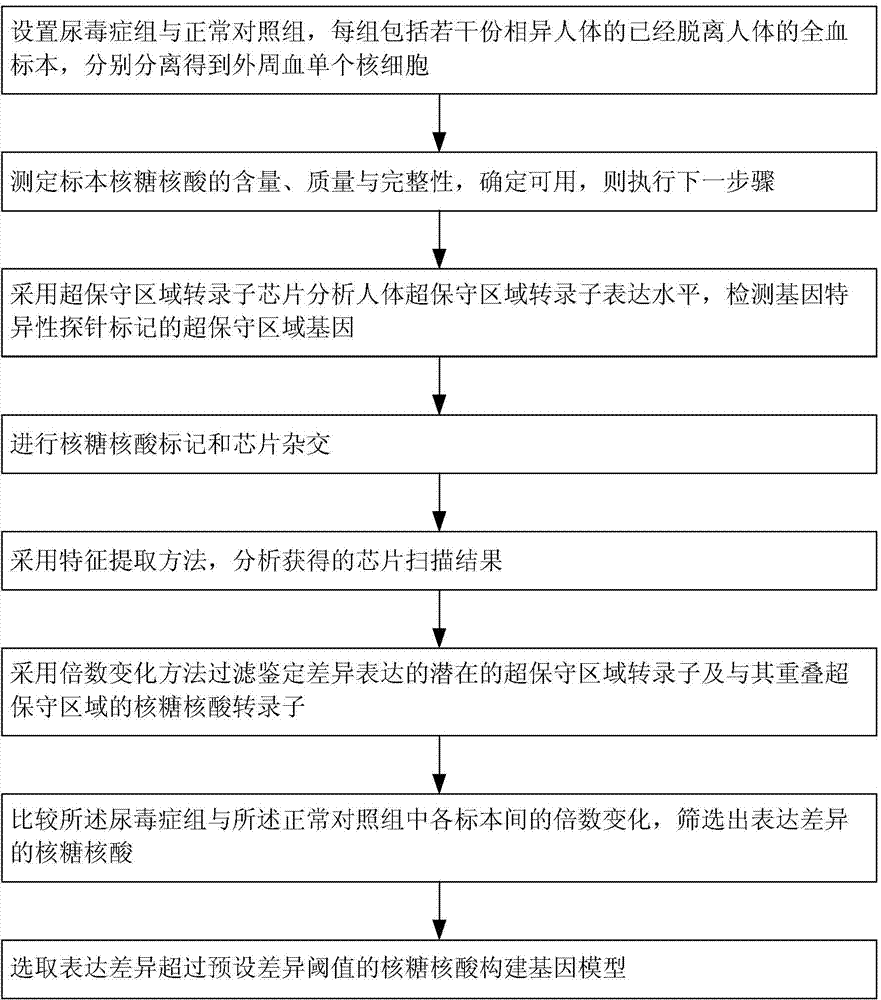
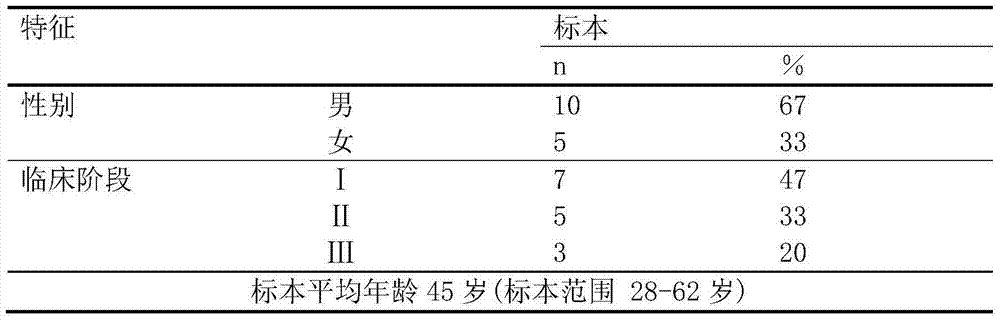
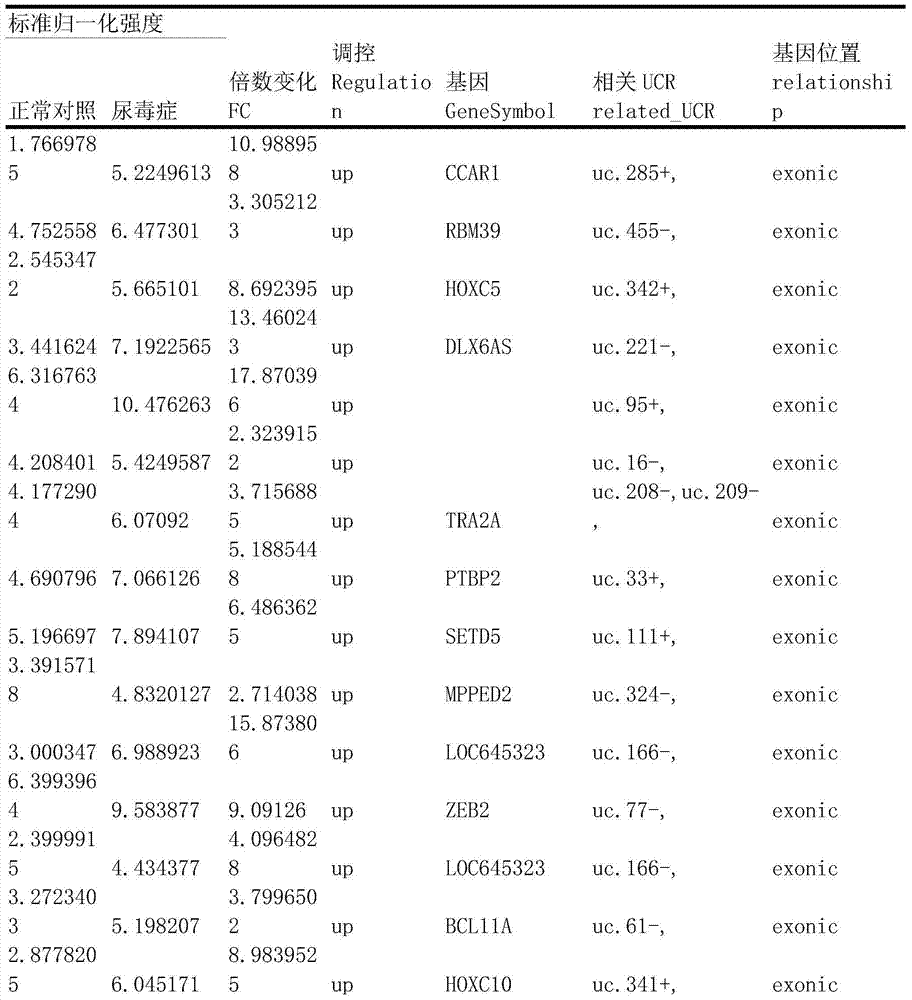
Method for constructing transcription expression gene model of ultra-conserved region of uremic peripheral blood mononuclear cell, and application of model
ActiveCN103937879AMicrobiological testing/measurementBlood/immune system cellsHuman bodyPeripheral blood mononuclear cell
The invention relates to a method for constructing a transcription expression gene model of an ultra-conserved region of a uremic peripheral blood mononuclear cell, and an application of the model. The construction method comprises the following steps: arranging a uremia group and a normal control group, wherein each of the above groups comprises a plurality of whole blood specimens separate from different human bodies, and respectively separating to obtain the uremic peripheral blood mononuclear cell; determining the ribonucleic acid content, the mass and the integrity of the specimens; analyzing the transcripton expression level of the human body ultra-conserved region by adopting an ultra-conserved region transcripton chip, and detecting a gene specificity probe marked ultra-conserved region gene; carrying out ribonucleic acid marking and chip hybridization; analyzing the obtained chip scanning result; filtering a latent ultra-conserved region transcripton and a ribonucleic acid transcripton of the ultra-conserved region overlapping with the ultra-conserved region transcripton for dientifyign differential expression; comparing the multiple change among the specimen in each of the uremia group and the normal control group, and screening expression difference ribonucleic acid; and selecting ribonucleic acid with the expression difference exceeding a preset difference threshold to construct the gene model.
Owner:中国人民解放军联勤保障部队第九二四医院
Computer software to assist in identifying SNPS with microarrays
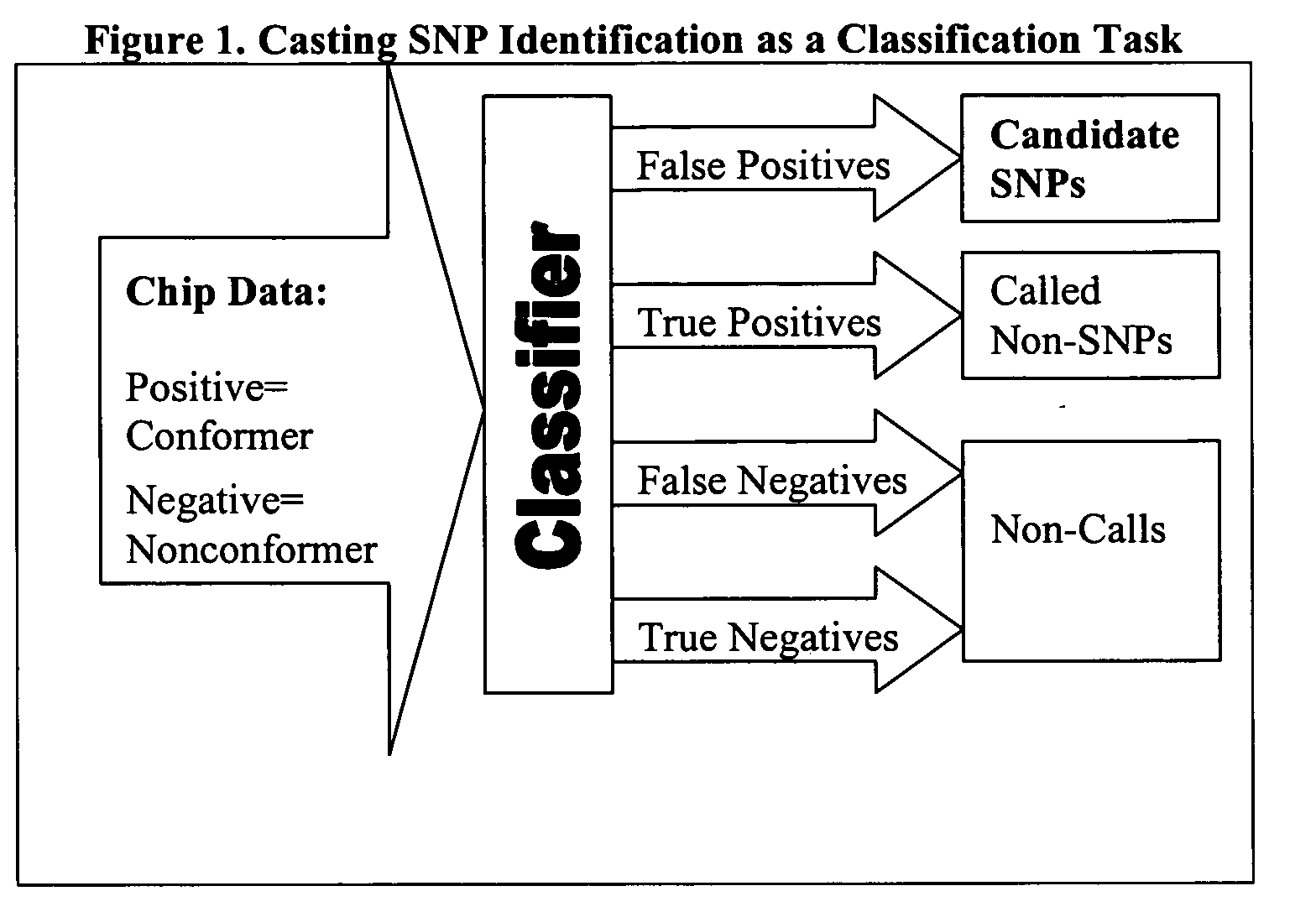
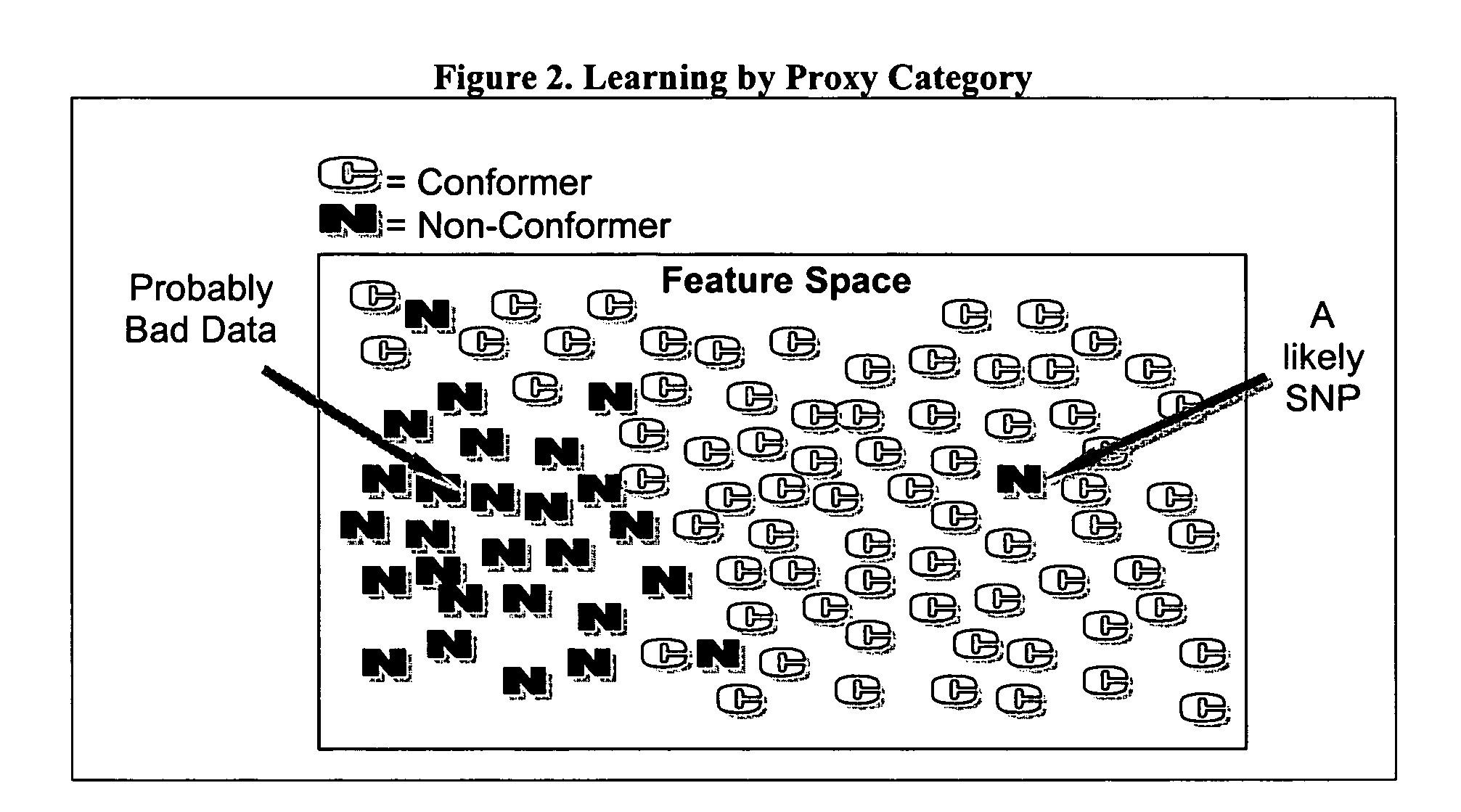
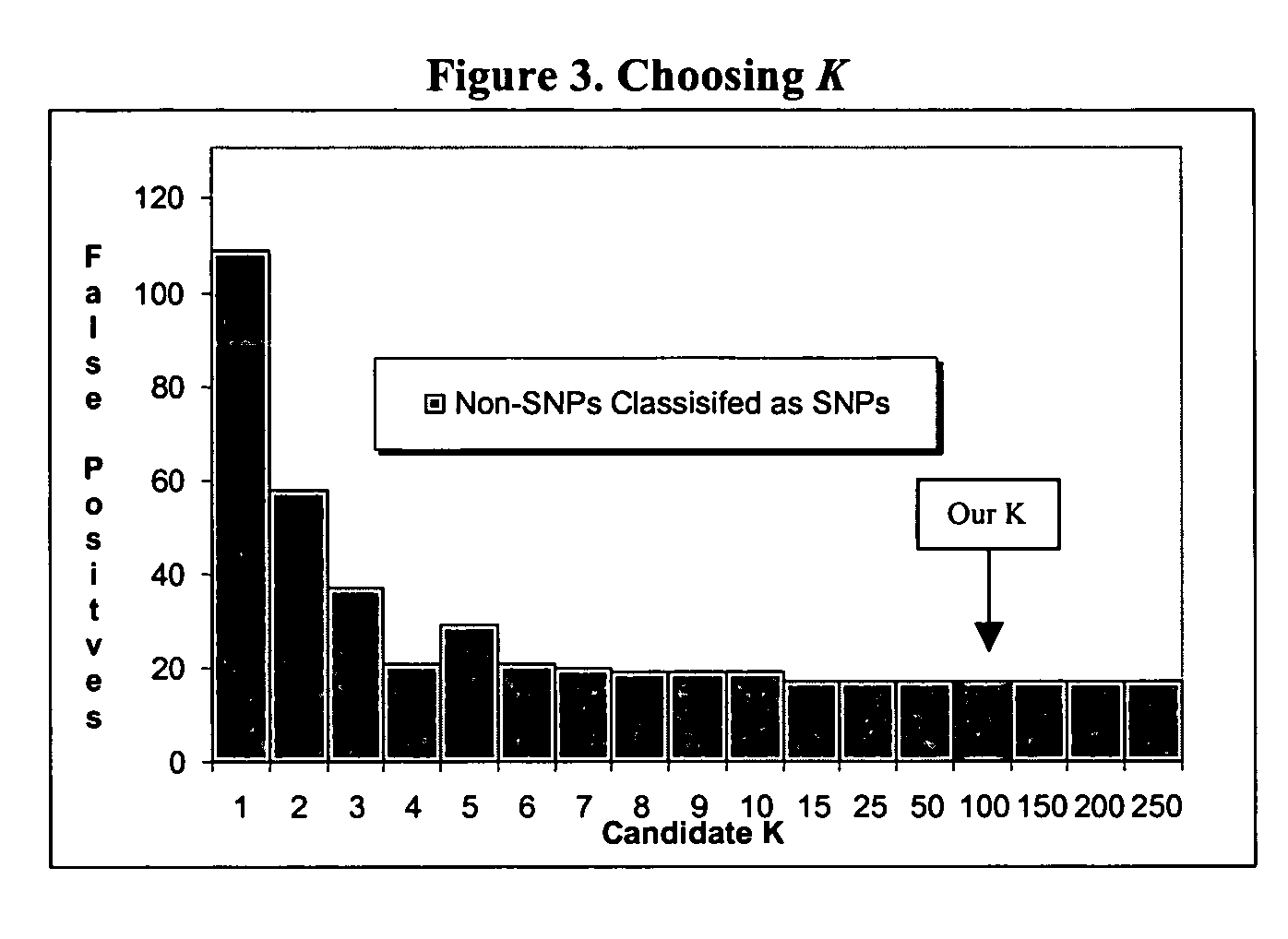
InactiveUS20060014164A1Improve predictabilityNanotechMicrobiological testing/measurementSensing dataComputer software
The present invention is a method to assist in the identification of single nucleotide polymorphisms (SNP) from microarray hybridization data. Data from hybridization protocols run on microarrays often have variations in the data resulting from variations in hybridization conditions and efficiencies and variations in optical intensities. An algorithm is described to screen the results to identify those data points most likely to be real SNPs as opposed to variations in the hybridization or sensing data.
Owner:ROCHE NIMBLEGEN
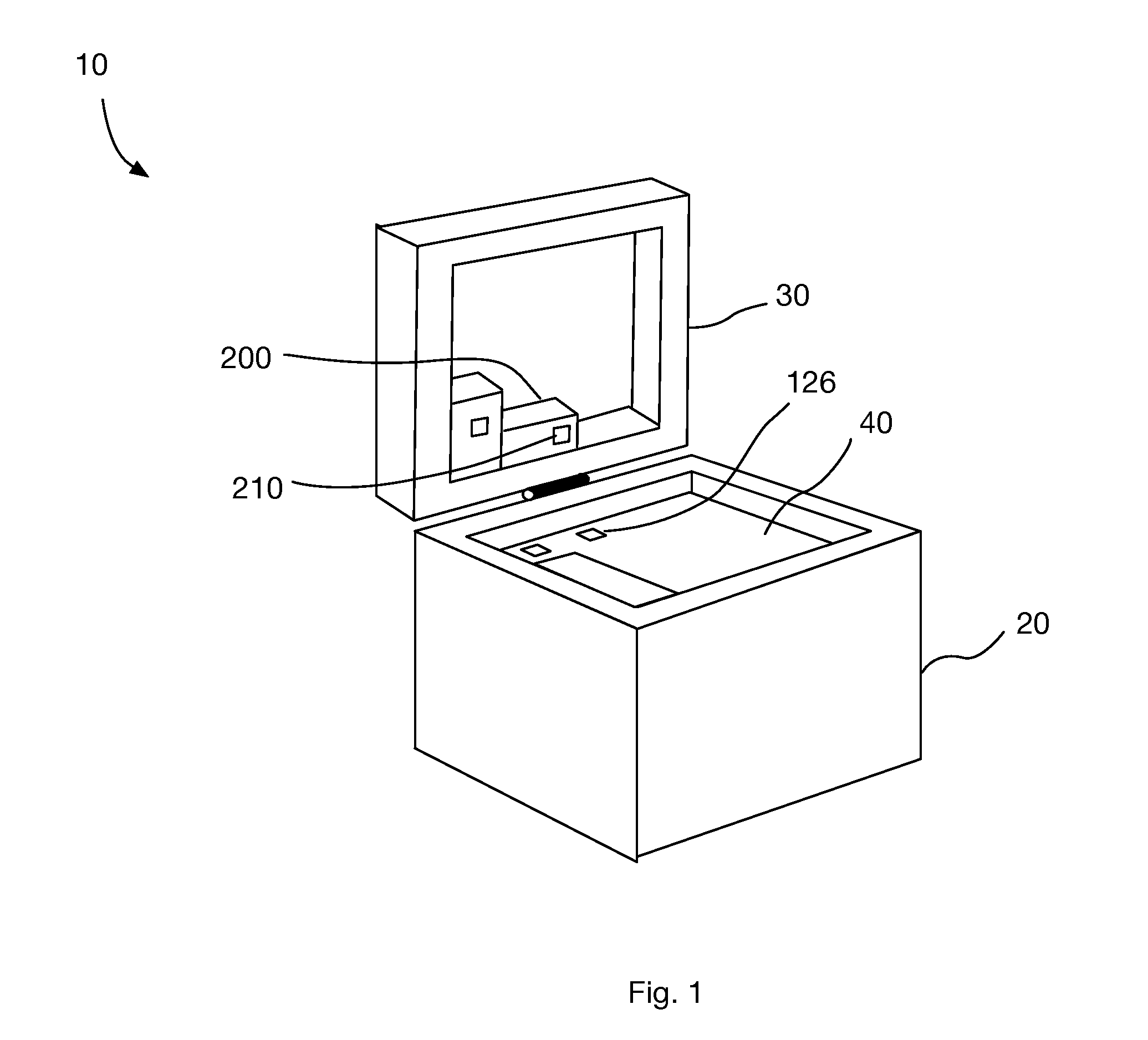
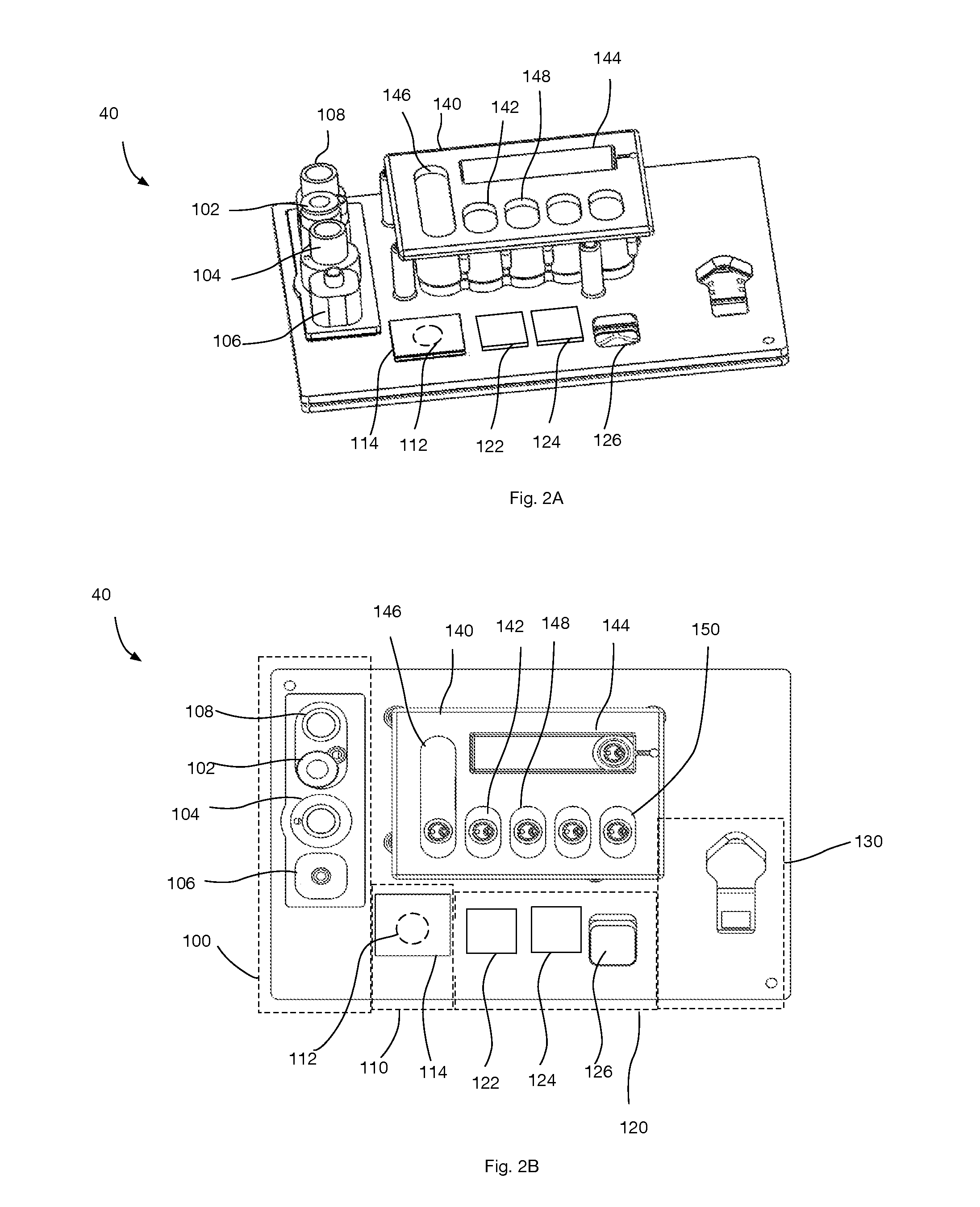
Microarray hybridization elution instrument
ActiveCN105176790AWell mixedAvoid pollutionBioreactor/fermenter combinationsBiological substance pretreatmentsBiologyMicroarray hybridization
A biochip microarray hybridization elution instrument is composed of a hybridization instrument, an elution instrument, accessories hybridization boxes, washing liquid bottles and a waste liquid bottle and is used for completion of microarray hybridization elution. The hybridization instrument can be used for setting the temperature and the time, six hybridization boxes are placed at the same time, and left and right oscillation can be performed after a power supply is switched on; the elution instrument adopts pipeline heating and can automatically set the elution temperature, the eluant amount, the number of elution times and the elution time. Needles are arranged on a needle seat directly facing to the upper side of an object stage, a group is composed of two needles, the needles totally comprise 20 groups, each 10 groups form a unit, and the units can be independently controlled. The two needles are both hollow, one is used for adding an eluant in the washing liquid bottles, and the other one is used for taking out waste liquid in the waste liquid bottle. The upper side of each reaction chamber of an upper cover of each hybridization box is provided with a sample adding hole in accordance with the position of the needle on the elution instrument. The full set of device is easy to use, after hybridization, elution is completed under the condition without disassembly of the hybridization boxes and chips, cross contamination is not generated, and the device is a good microarray hybridization elution device.
Owner:KUNMING HUANJI BIOLOGICAL CHIP IND
Diagnostic methods for infectious disease using endogenous gene expression
Disclosed are methods of diagnosis of a pathogen-associated disease. These methods comprise: providing a biological sample from a human subject; determining presence, absence and / or quantity of a bacterial pathogen, a viral pathogen, or a combination thereof, by a pathogen culture, a serum antibody detection test, a pathogen antigen detection test, a pathogen DNA and / or RNA detection test, or a combination thereof; determining in the sample, expression levels of at least one endogenous gene in which aberrant expression levels are associated with infection with a pathogen, by a microarray hybridization assay, RNA-seq assay, polymerase chain reaction assay, a LAMP assay, a ligase chain reaction assay, a Southern, Northern, or Western blot assay, an ELISA or a combination thereof. The subject can be diagnosed with the disease if the subject comprises the pathogen and an aberrant level of expression of an endogenous gene.
Owner:WASHINGTON UNIV IN SAINT LOUIS
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com