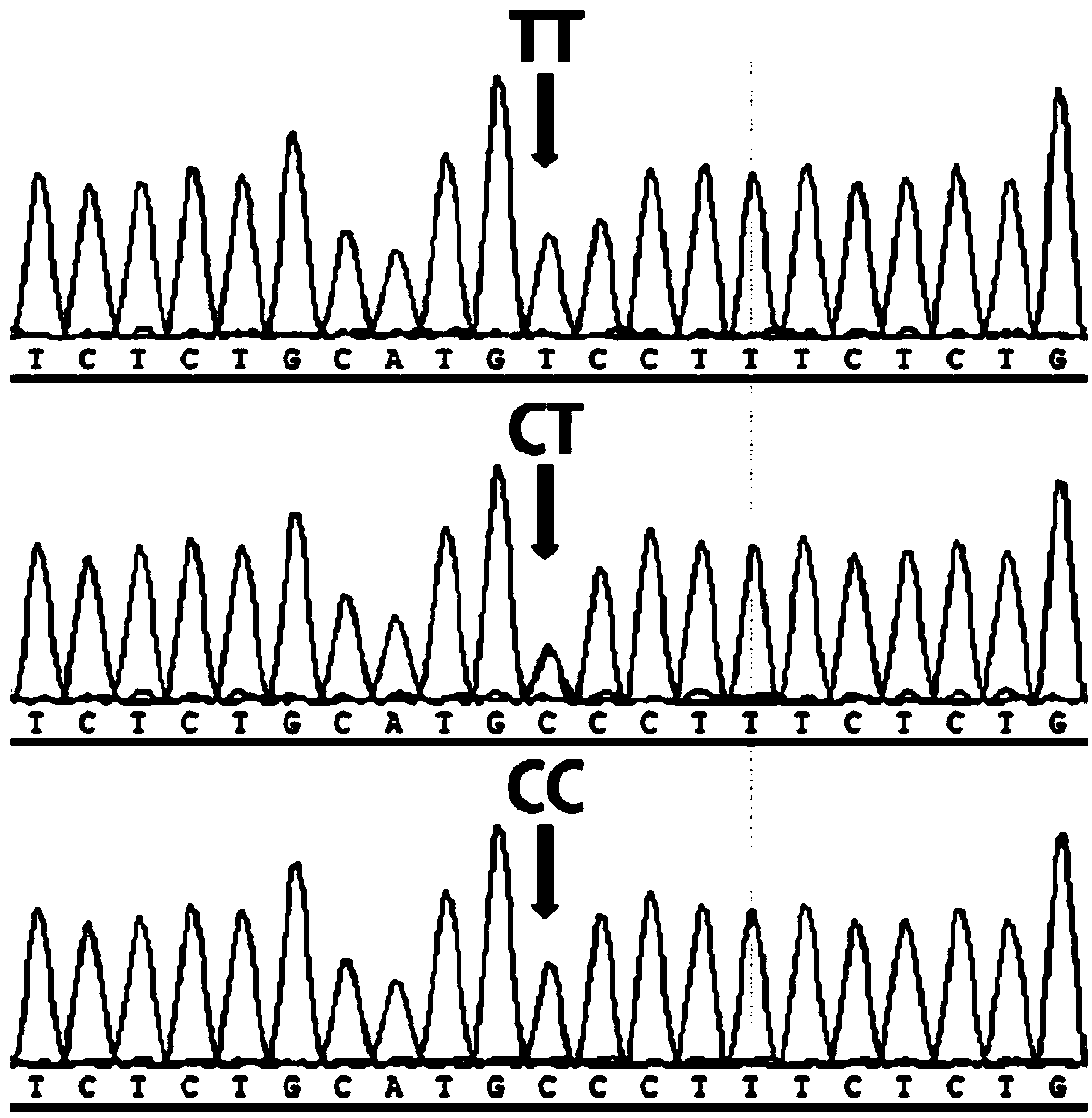
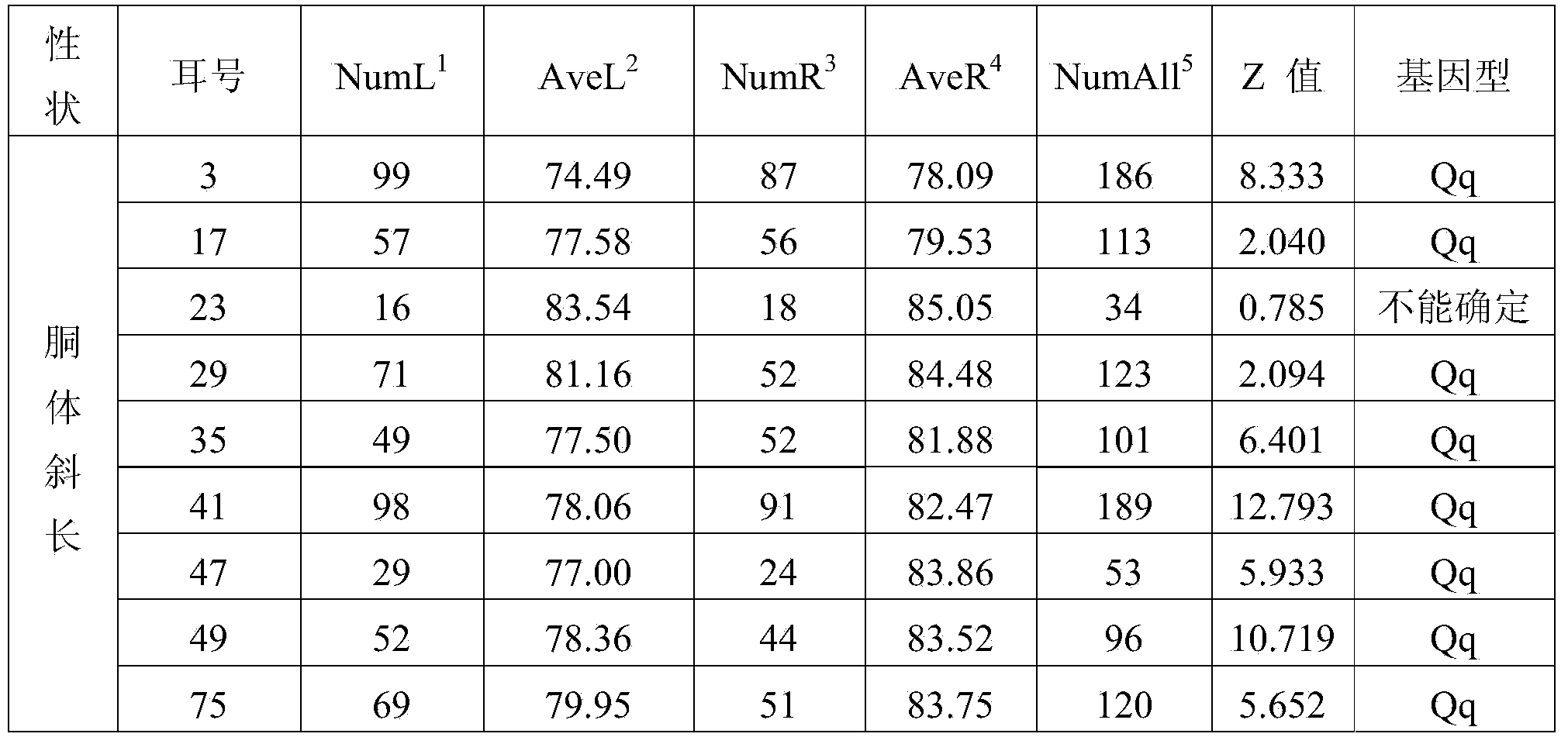
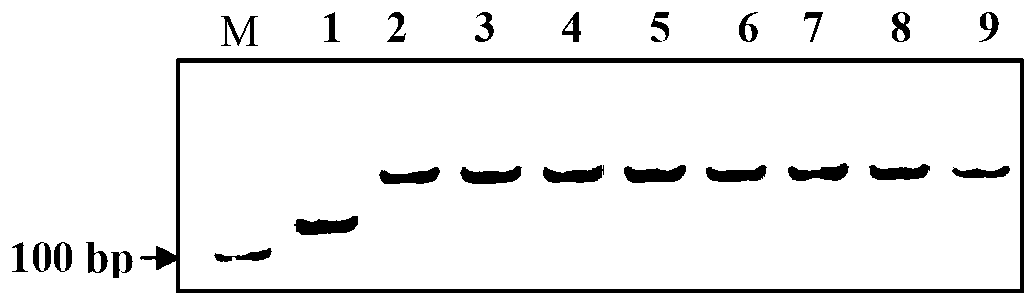
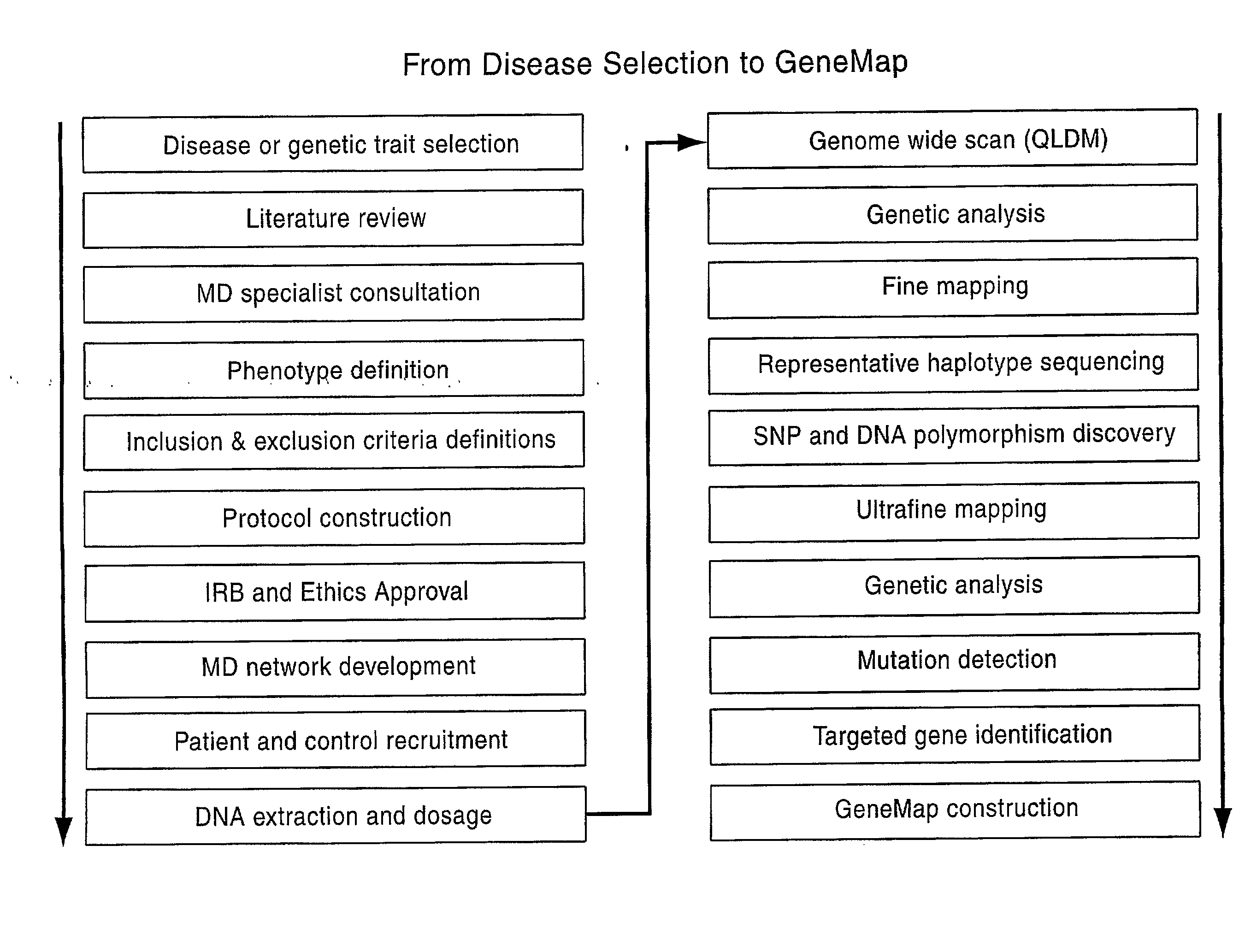
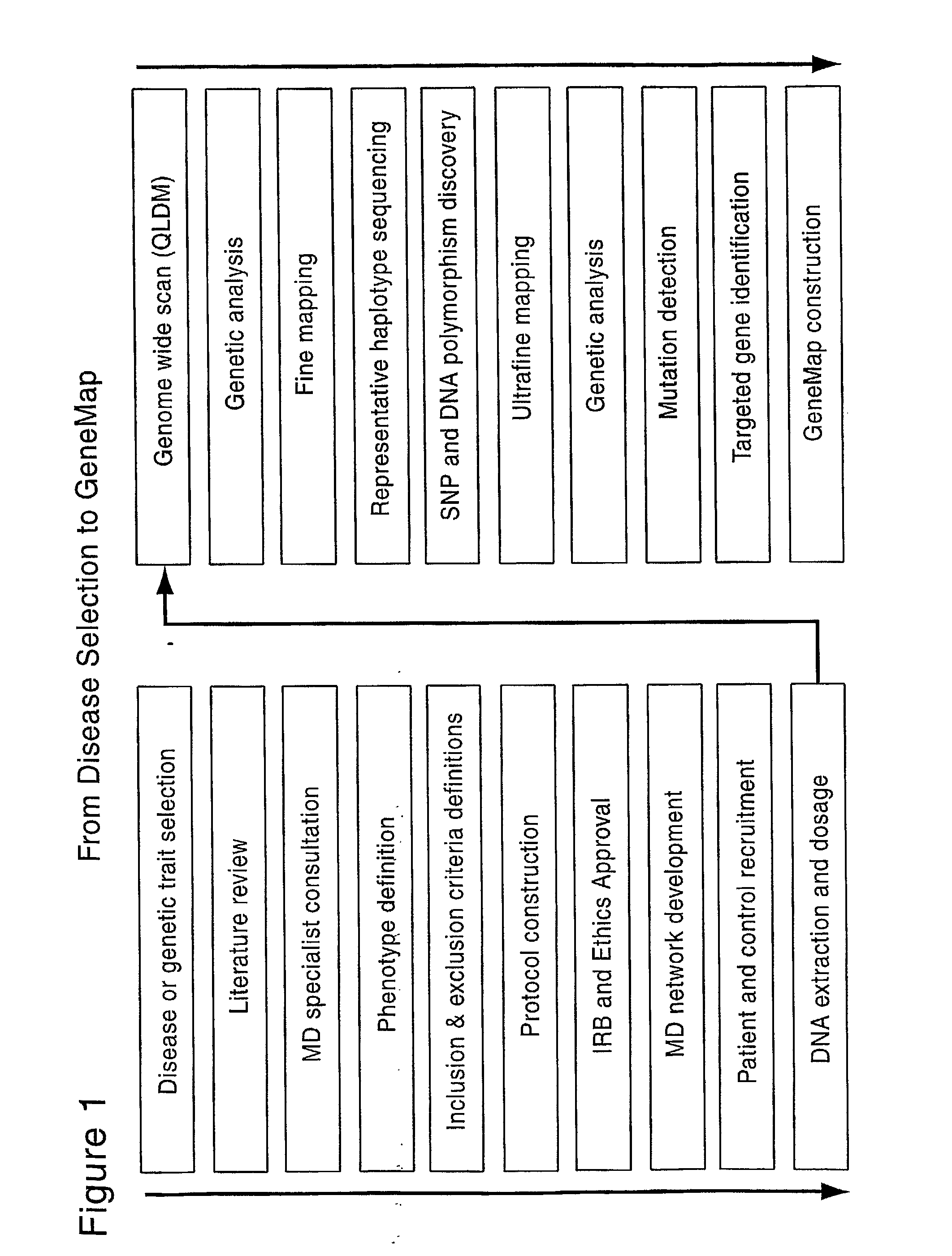
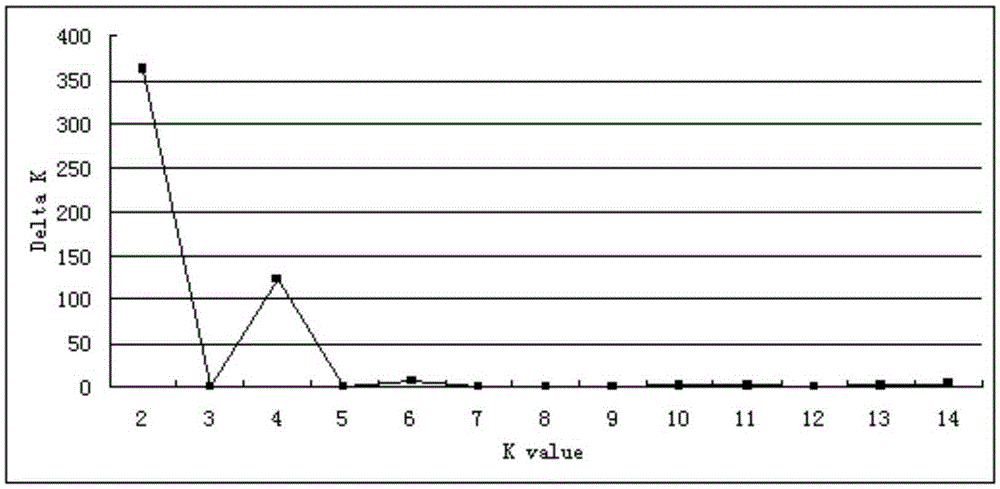
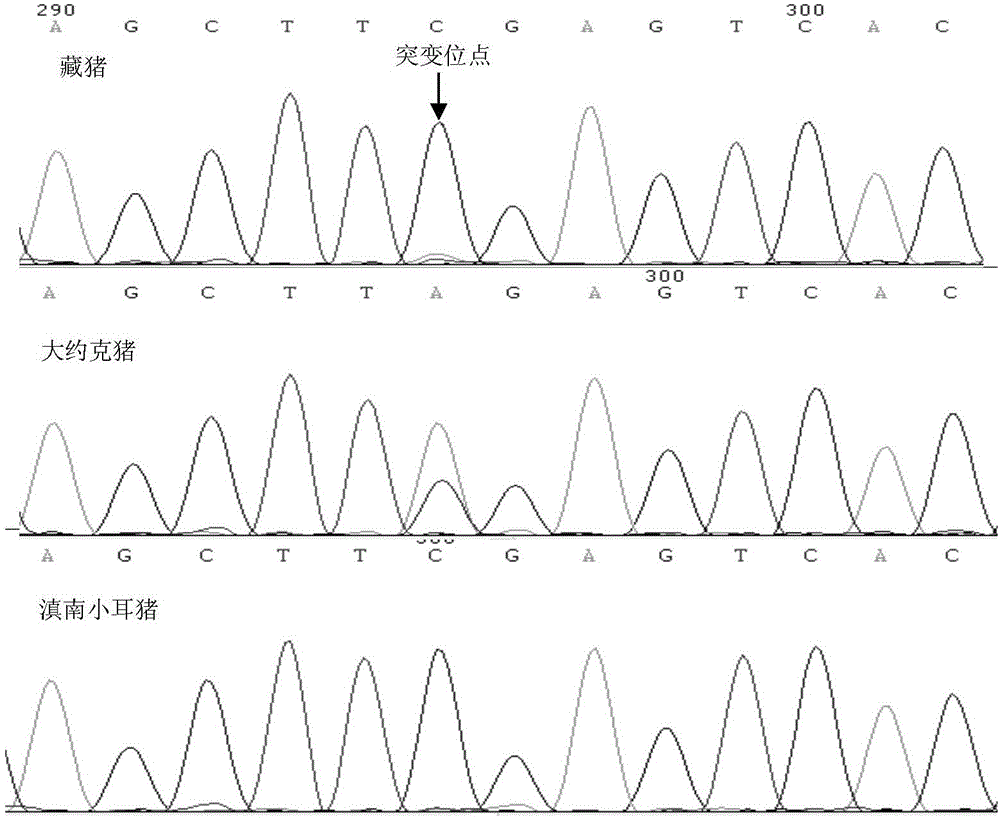
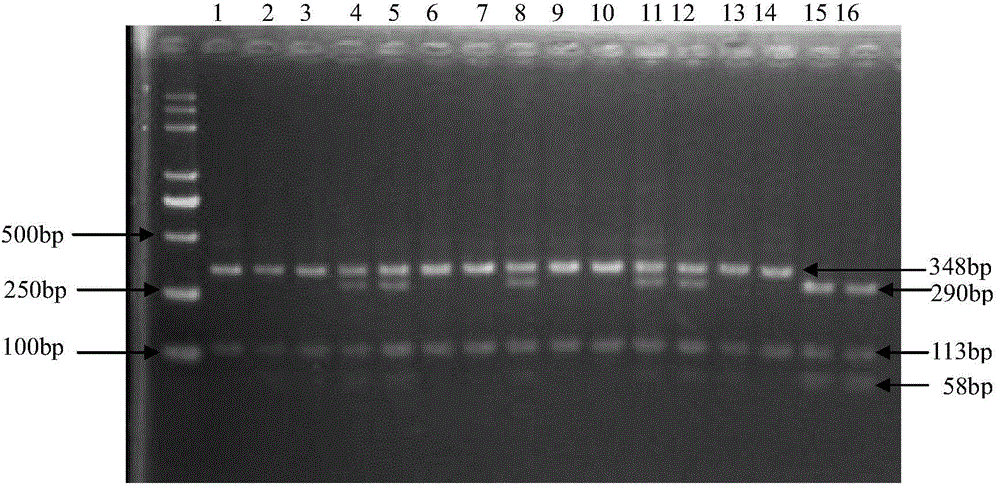
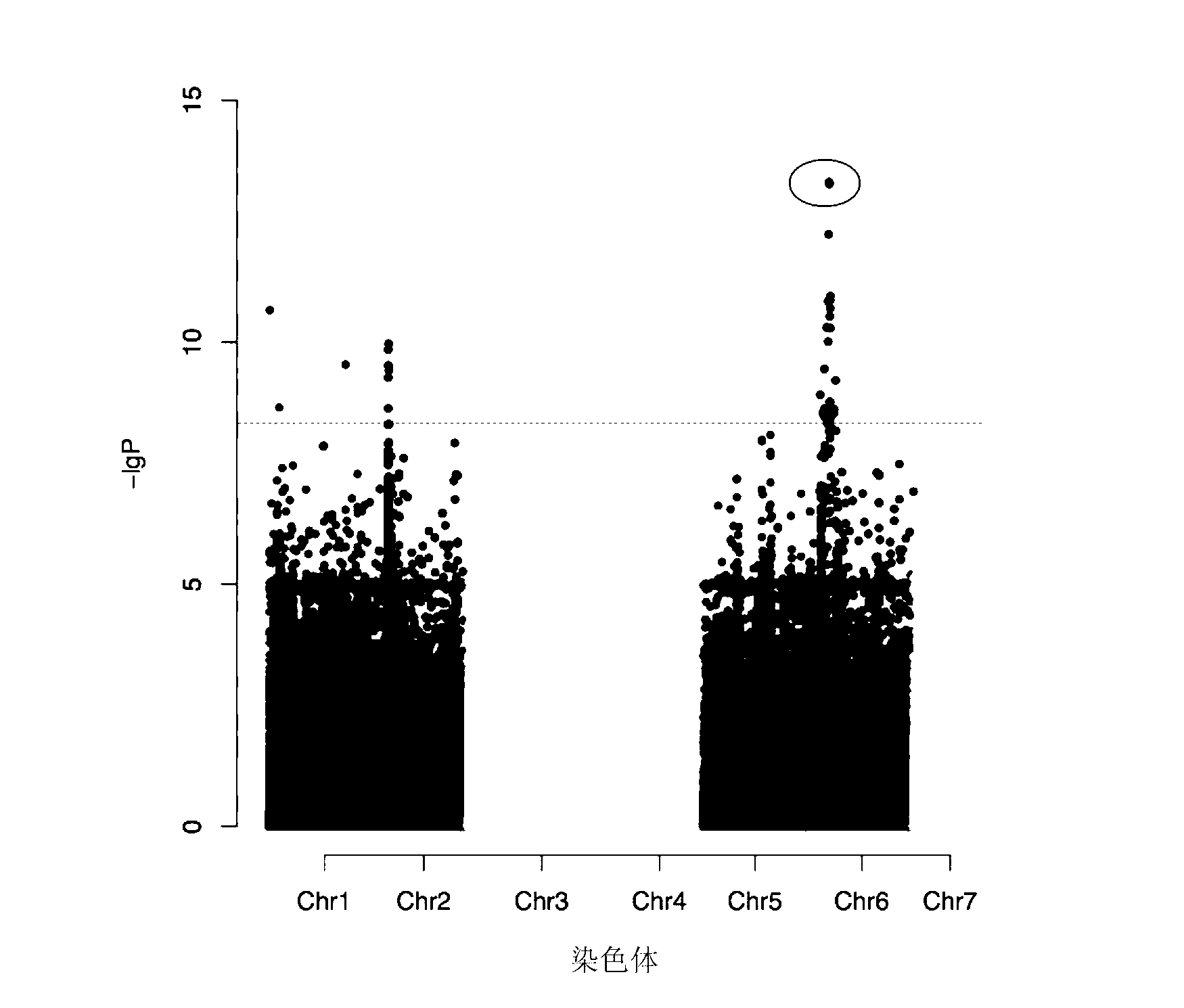
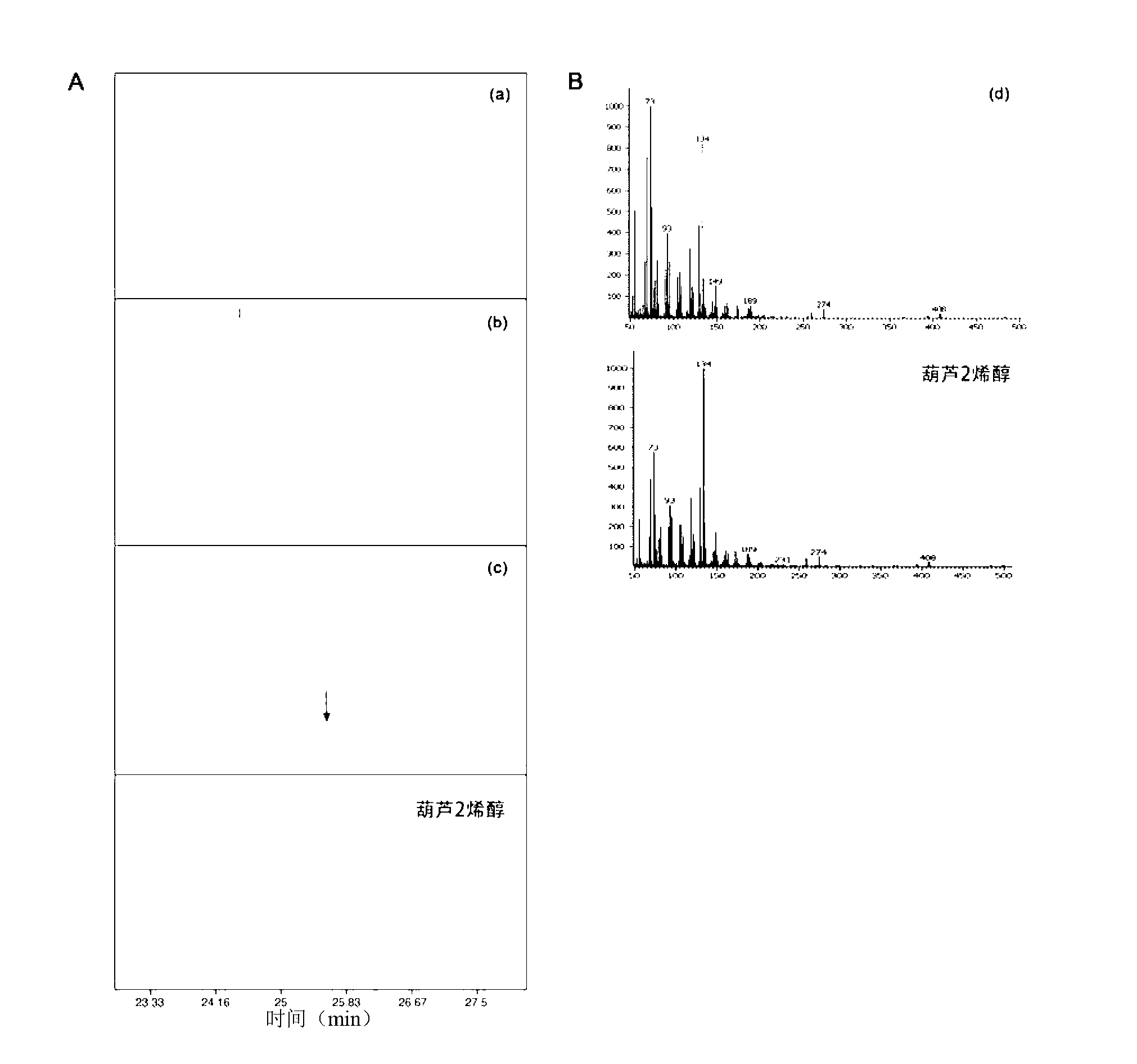
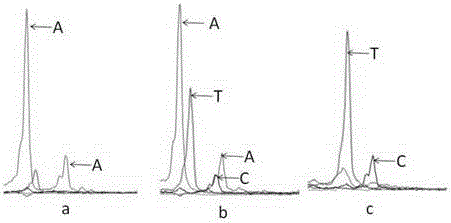
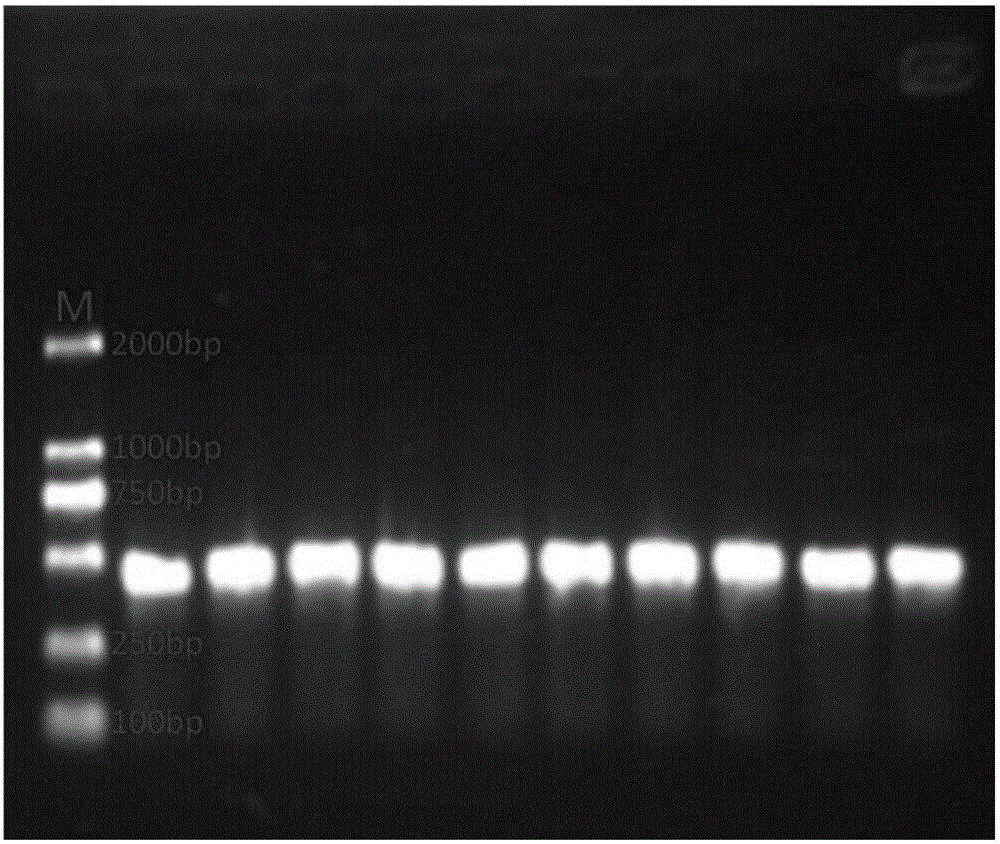
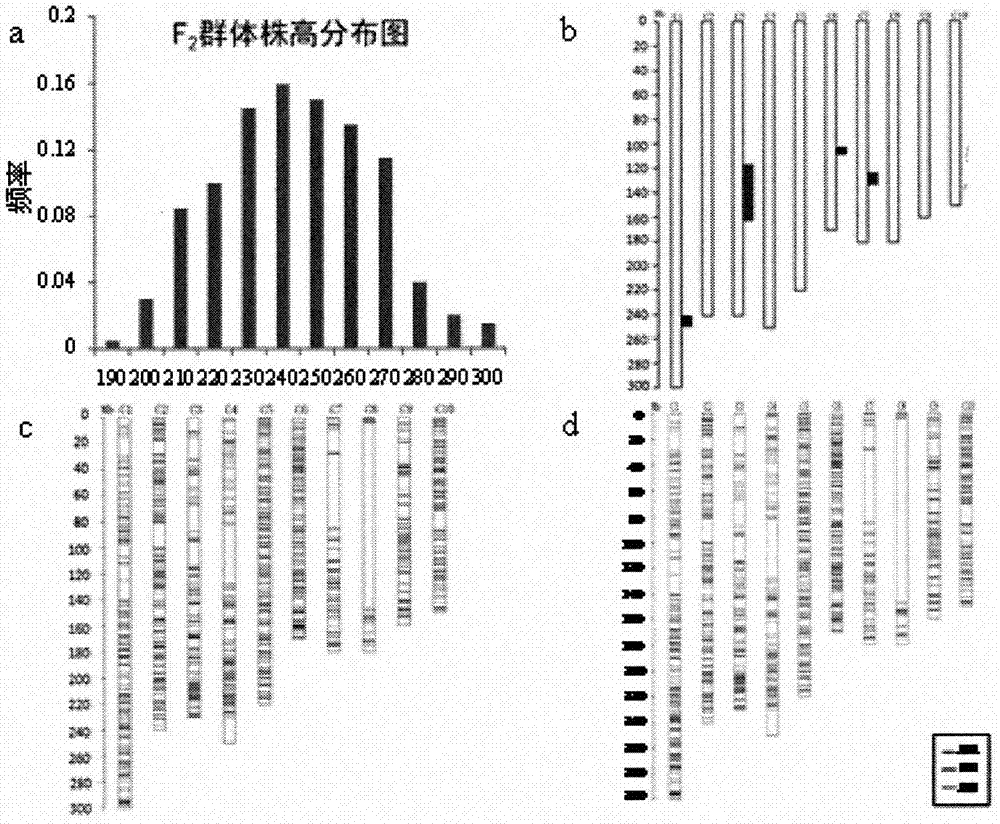
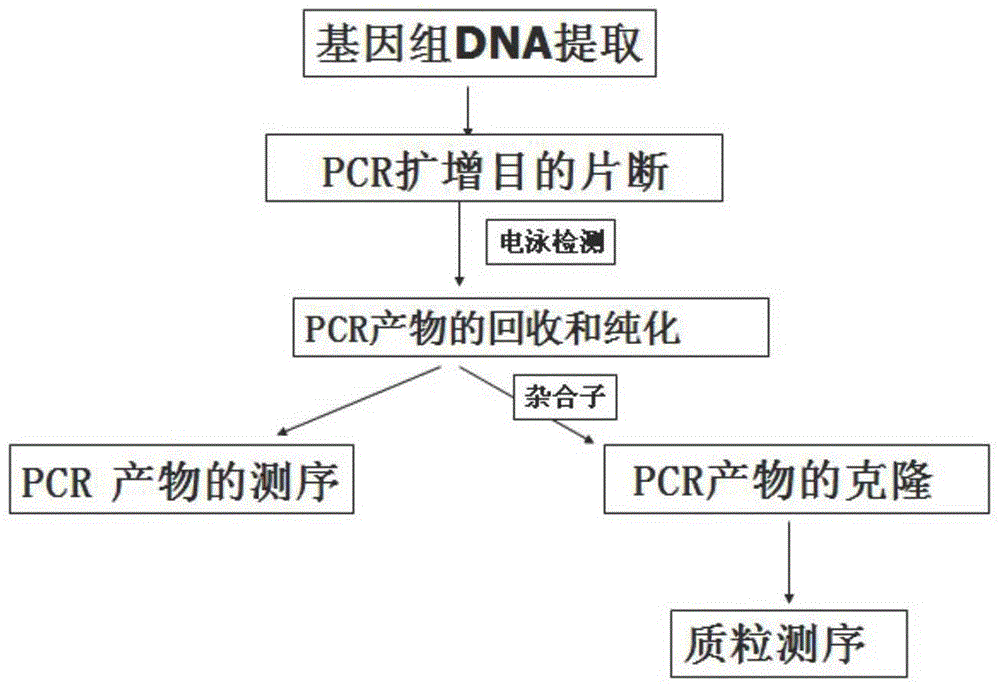
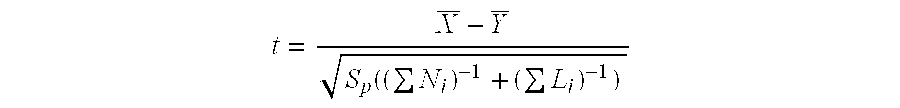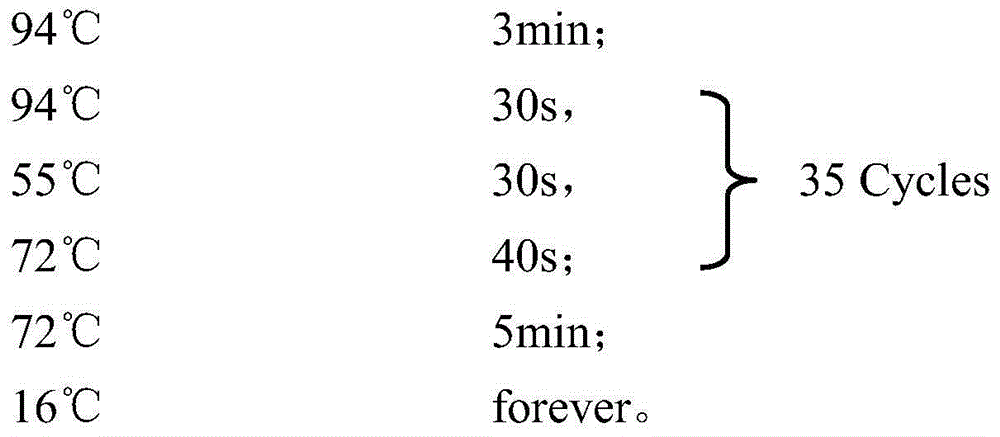Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
757 results about "Snp markers" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Rice whole genome SNP chip and application thereof
ActiveCN102747138AImprove throughputGood repeatabilityNucleotide librariesMicrobiological testing/measurementManufacturing technologyGermplasm
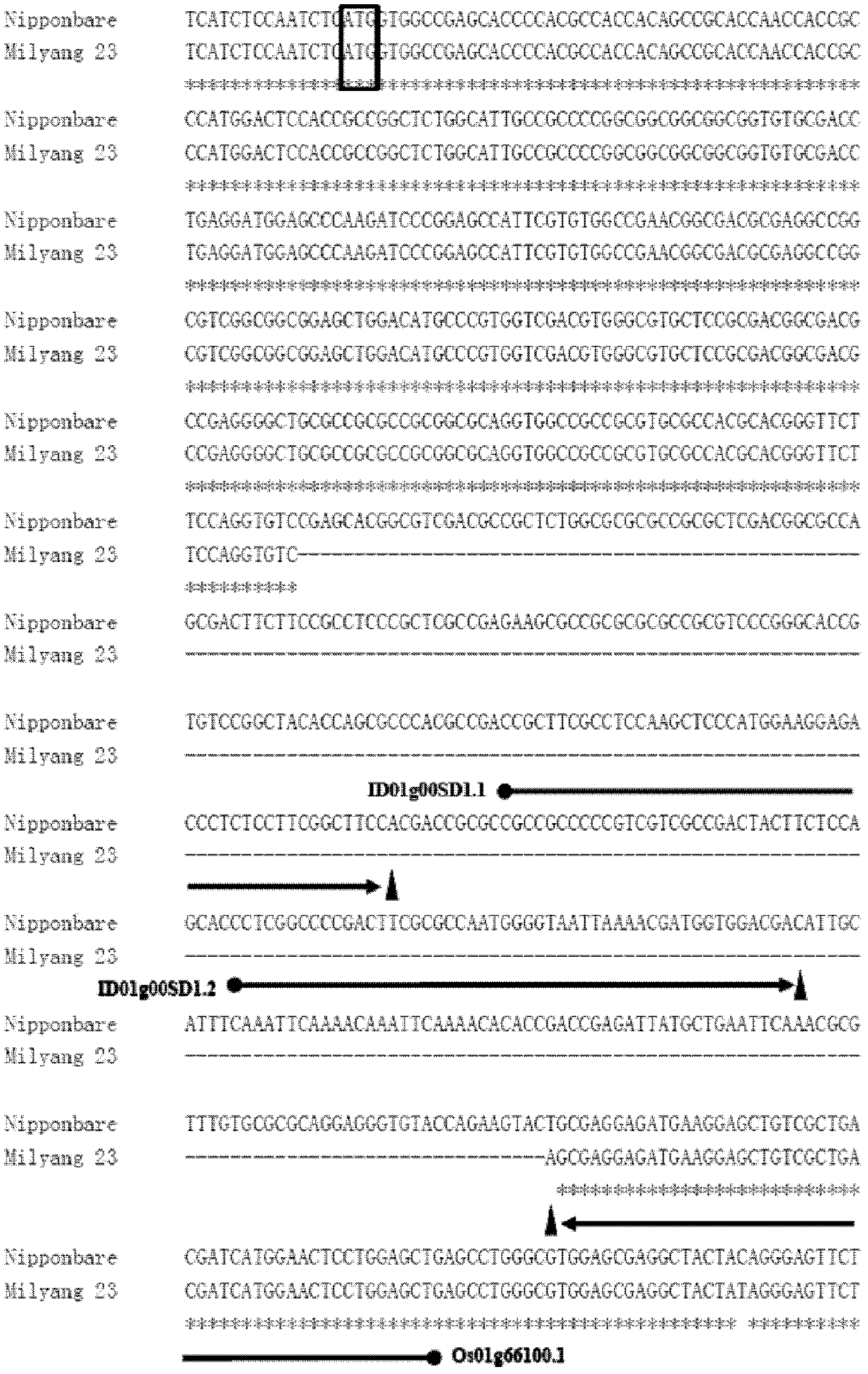
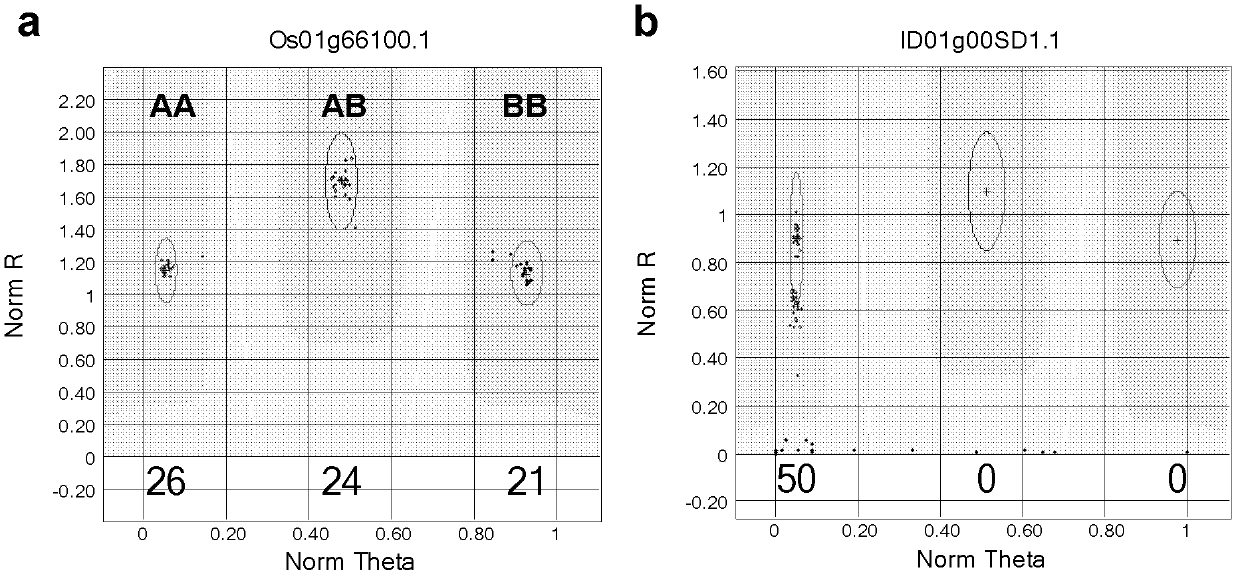
The present invention discloses a rice whole genome SNP chip and an application thereof. A method for preparing the chip comprises: (1) obtaining a first class of probes on a chip, wherein sequencing is performed to obtain a parental genome sequence, resequencing data of other rice varieties in a public database are combined, a Nipponbare genome is adopted as a reference sequence, a MAQ software is adopted to match and analyze all the sequencing data, and finally a SNP marker is screened; (2) obtaining a second class of probes on the chip, wherein a rice function gene is obtained from the public database, sequence difference reflecting gene function is searched, and a SNP / INDEL probe is designed according to the sequence difference; (3) adopting an infinium chip manufacturing technology to produce a SNP chip; and (4) testing accuracy and application efficiency of the chip. The chip of the present invention can be applicable for rice germplasm resource molecule marker fingerprint analysis, seed authenticity detection, filial generation genotyping, and other related researches.
Owner:先正达集团股份有限公司
Major SNP (single nucleotide polymorphism) marker influencing growth traits of pigs and application thereof in genetic improvement of productivity of breeding pigs
The invention provides a major SNP (single nucleotide polymorphism) marker influencing the growth traits of pigs. The SNP marker is located at a nucleotide sequence of an HMGA1 gene of a pig chromosome 7, a locus of the SNP marker is the nucleotide mutation of C857-G857 with an SEQ ID NO:1 sequence labeling position of 857, corresponding to a 34983991st nucleotide locus C> on chromosome 7 in a reference sequence in an international pig genome version 10.2, G mutation, or one of seven other loci completely linked with the locus. The invention also provides the application of the SNP marker in the genetic improvement of growth traits of breeding pigs, and provides the application of an SNP molecular marker with a linkage disequilibrium degree (r2) with the SNP marker of greater than 0.8 in the genetic improvement of growth traits of breeding pigs. The growth traits include one or more of the length and height of a living body of a pig, carcass length, carcass weight, daily gain and head weight. According to the invention, the breeding process of breeding pigs can be accelerated, the productivity of breeding pigs can be effectively improved, and remarkable economic benefits can be obtained.
Owner:JIANGXI AGRICULTURAL UNIVERSITY
Rice blast resistance gene Pi9 gene specificity molecular marker Pi9SNP as well as preparation and application thereof
ActiveCN103305510AStrong specificityImprove throughputMicrobiological testing/measurementDNA preparationAgricultural scienceNucleotide
The invention discloses a rice blast resistance gene Pi9 gene specificity molecular marker Pi9SNP as well as preparation and application thereof. The molecular marker Pi9SNP is prepared by amplifying from total DNA (Deoxyribonucleic Acid) of a rice blast resistant variety with rice blast resistance gene Pi9 by using a primer pair F1 and R1 so as to obtain a nucleotide segment I, and carrying out digestion on the nucleotide segment I by using restriction enzyme Hind III, so as to obtain a nucleotide segment II which is in specificity banding pattern with the obtained rice blast resistance gene Pi9. The molecular marker is the first one which is developed for marking Pi9 gene specificity SNP (Signal Nucleotide Polymorphism) for an internal sequence of the Pig gene, and has the following advantages that the specificity is high, the cost is low and the flux is high in practical application, and the molecular marker is widely applied to colonies of different genetic backgrounds. By utilizing the molecular marker, the efficiency in auxiliary molecular marker selective breeding, gene pyramiding breeding and transgenosis breeding is improved.
Owner:PLANT PROTECTION RES INST OF GUANGDONG ACADEMY OF AGRI SCI
Genemap of the human genes associated with longevity
InactiveUS20090305900A1Electrolysis componentsVolume/mass flow measurementGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of SNP markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's longevity, their protection against age-related diseases and / or their response to a particular drug or drugs.
Owner:BELOUCHI ABDELMAJID +12
Upland cotton SNP marker and application thereof
InactiveCN105349537AImprove detection efficiencyImprove work efficiencyNucleotide librariesMicrobiological testing/measurementAgricultural scienceHomologous sequence
The invention discloses an upland cotton SNP marker and application thereof, and belongs to the field of upland cotton SNP marker development. The upland cotton SNP marker is obtained by developing a single-copy SNP marker of an upland cotton genome via chip hybridization and homologous sequence comparison, and picking out SNP markers in a linkage disequilibrium recession distance via linkage disequilibrium analysis. The SNP marker provided by the invention can be applied to analysis of genetic diversity and group structure, or to germplasm identification of upland cotton, is single-copy, and has no remarkable linkage disequilibrium among markers, so that the efficiency of detection, adopting the SNP marker, during analysis of genetic diversity, group structure or fingerprint identification can be greatly improved, and the work efficiency is higher.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Gene-specific molecular marker Pi2SNP of rice blast-resistant gene Pi2 as well as preparation method and application thereof
ActiveCN103320437AStrong specificityHigh sequence identityMicrobiological testing/measurementDNA preparationAgricultural scienceEnzyme digestion
The invention discloses a gene-specific molecular marker Pi2SNP of a rice blast-resistant gene Pi2 as well as a preparation method and application thereof. The molecular marker Pi2SNP is a nucleotide fragment II in a specific banding pattern with the rice blast-resistant gene Pi2 obtained by amplifying F1 and R1 from the total DNA (Deoxyribose Nucleic Acid) of the rice blast-resistant variety carrying rice blast-resistant gene Pi2 by use of a primer to obtain a nucleotide fragment I and then performing enzyme digestion of the nucleotide fragment I by use of restriction endonuclease Pst I or Hinf I. The molecular marker is the first Pi2 gene-specific SNP marker developed for the sequence in the Pi2 gene, and has the advantages of high specificity and low cost and high flux in practical application, and can be widely applied to the colonies with different inheritance backgrounds. By adopting the molecular marker, the utilization efficiency of the gene in molecular marker-assisted selective breeding, gene pyramiding breeding and transgenic breeding can be improved.
Owner:PLANT PROTECTION RES INST OF GUANGDONG ACADEMY OF AGRI SCI
Novel genes and markers associated to type 2 diabetes mellitus
InactiveUS20070059722A1Aggressive managementReduce caloric intakeCompound screeningApoptosis detectionGene productHaplotype
Genes, SNP markers and haplotypes of susceptibility or predisposition to T2D and subdiagnosis of T2D are disclosed. Methods for diagnosis, prediction of clinical course and efficacy of treatments for T2D using polymorphisms in the T2D risk genes are also disclosed. The genes, gene products and agents of the invention are also useful for monitoring the effectiveness of prevention and treatment of T2D. Kits are also provided for the diagnosis, selecting treatment and assessing prognosis of T2D.
Owner:DSM IP ASSETS BV
SNP (single-nucleotide polymorphism) markers and primer pairs for mouse inbred line identification, and application thereof
InactiveCN104975105ALow costQuality improvementMicrobiological testing/measurementDNA/RNA fragmentationAgricultural sciencePrimary screening
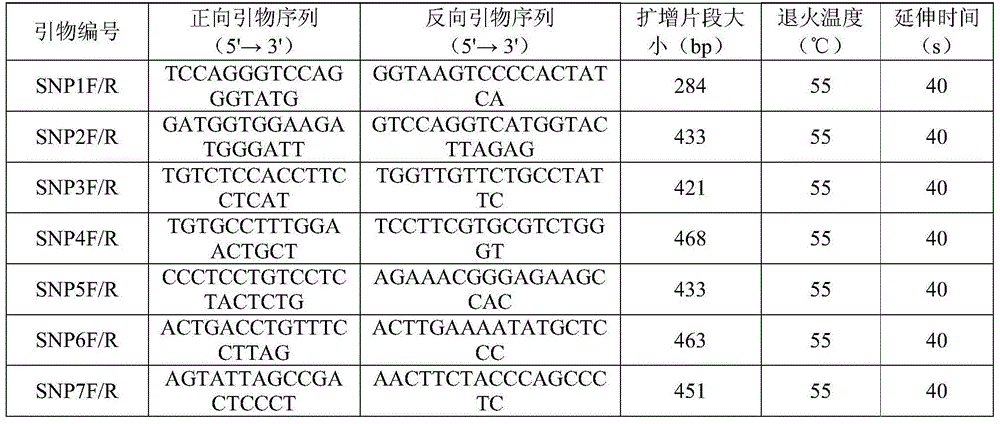
The invention discloses SNP (single-nucleotide polymorphism) markers and primer pairs for mouse inbred line identification, and application thereof. 7 screened SNP sites, including SNP1, SNP2, SNP3, SNP4, SNP5, SNP6 and SNP7, are utilized to design 7 primer pairs of which the sequences are disclosed as SEQ ID NO:1-SEQ ID NO:14; and each primer pair can amplify genome sequences disclosed as SEQ ID NO:15-SEQ ID NO:21 of the mouse inbred line under the same conditions. The sequences amplified by each primer pair contain one specific SNP marker. The application of the SNP marker and primer pairs can achieve the goals of quickly performing primary screening and lowering the detection cost, thereby promoting the inbred line mouse genetic quality monitoring standardization progress.
Owner:SOUTH CHINA UNIV OF TECH +1
SNP marker associated with pig growth rate, and applications thereof
ActiveCN104404133ASpeed up the breeding processThe detection method is accurateMicrobiological testing/measurementDNA/RNA fragmentationAnimal scienceSnp markers
The present invention provides a SNP marker associated with pig growth rate, and application thereof, wherein the SNP marker is located on the 201 bp position of the pig LXR[alpha] gene exon 5, and the growth rate of the pig with the base A at the position is significantly higher than the growth rate of the pig with the base C at the position. According to the present invention, the polymerase chain reaction (PCR) and the sequencing technology are adopted to screen the SNP marker associated with pig growth rate, the restriction fragment length polymorphism (RFLP) method is adopted to detect the genotype of the pig gene to be detected, and the breeding of the predominant variety of the pig with the rapid growth is performed according to the genotype so as to accelerate the pig breeding process.
Owner:CHINA AGRI UNIV
SNP (Single Nucleotide Polymorphism) related to chick carcass trait and application thereof
InactiveCN102250889AFast and accurate auxiliary screeningShorten the timeMicrobiological testing/measurementAnimal husbandrySnp markersMolecular marker
The invention relates to the field of molecular biology detection, in particular to SNP (Single Nucleotide Polymorphism) related to chick carcass trait and application thereof. The invention provides 10 SNP markers of a chick SREBF2 gene and a method for detecting the carcass trait of a chick colony by applying the SNP markers. The invention further provides a primer for detecting the SNP markersand a kit comprising the same. The SNP markers provided by the invention can be applied to molecular marker assistant breeding of chicken, can assist in screening quickly and accurately, and have theadvantages of early screening, saving in time, low cost and high accuracy.
Owner:SICHUAN AGRI UNIV
SNP molecular marker associated with chromosome 6 and fiber strength of upland cotton
ActiveCN106868131AShorten the breeding cycleImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMolecular breeding
The invention belongs to the technical field of molecular breeding of cotton and discloses an SNP molecular marker associated with fiber strength of upland cotton as well as detection and application thereof. The SNP molecular marker is obtained by taking stable RIL group of cotton as the material through a genome resequencing method. When the SNP markers are used for carrying out molecular marker-assisted breeding selection, the breeding period can be greatly shortened; the breeding efficiency of the cotton can be improved; the cotton fiber strength can be improved.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
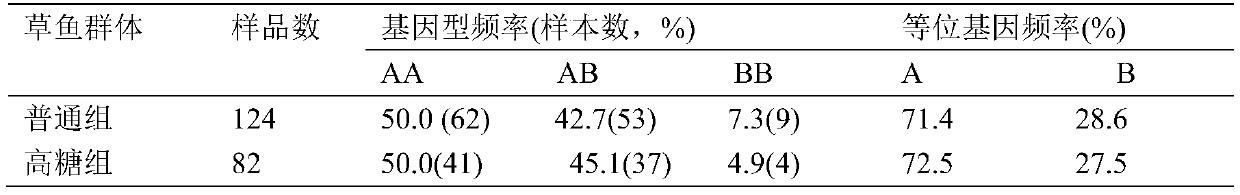
SNP marker related to sugar tolerance of grass carps and application of SNP marker
ActiveCN105505927AEasy identificationEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
The invention provides an SNP marker related to the sugar tolerance of grass carps and application of the SNP marker. The SNP marker comprises two SNP loci, namely H3 and H4, wherein H3 is located at position 1817 from the 5' end of the nucleotide sequence shown as SEQ ID NO:1 in the specification, and base at the position 1817 is T or C; H4 is located at position 1818 from the 5' end of the nucleotide sequence shown as SEQ ID NO:1 in the specification, and base at the position 1818 is G or T. The SNP marker provided by the invention is closely related to the sugar tolerance of the grass carps, and can be effectively used for identification or selective breeding of high-sugar-tolerance grass carp species.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
SNP (single-nucleotide polymorphism) marker set for identifying pig breeds and application thereof
InactiveCN105603089AMicrobiological testing/measurementDNA/RNA fragmentationPrincipal component analysisCorrelation analysis
The invention discloses an SNP (single-nucleotide polymorphism) marker set for identifying pig breeds. The application method mainly comprises the following steps: by using 200 purebred Large White and Landrace individuals as study objects, carrying out genome SNP 60K chip scanning typing on all the samples of the two breeds; after carrying out quality control on all the data according to the requirements, screening out SNP marker sites capable of identifying Landrace and Large White, and carrying out genome correlation analysis to screen out 16 SNP sites for identifying Landrace and Large White; and carrying out verification on another 500 individuals by principal component analysis and cluster analysis, wherein the accuracy of the 16 SNP sites reaches 99%, which indicates that the 16 SNP sites can be used for identifying Large White and Landrace.
Owner:ZHANGZHOU AONONG MODERN AGRI DEV CO LTD
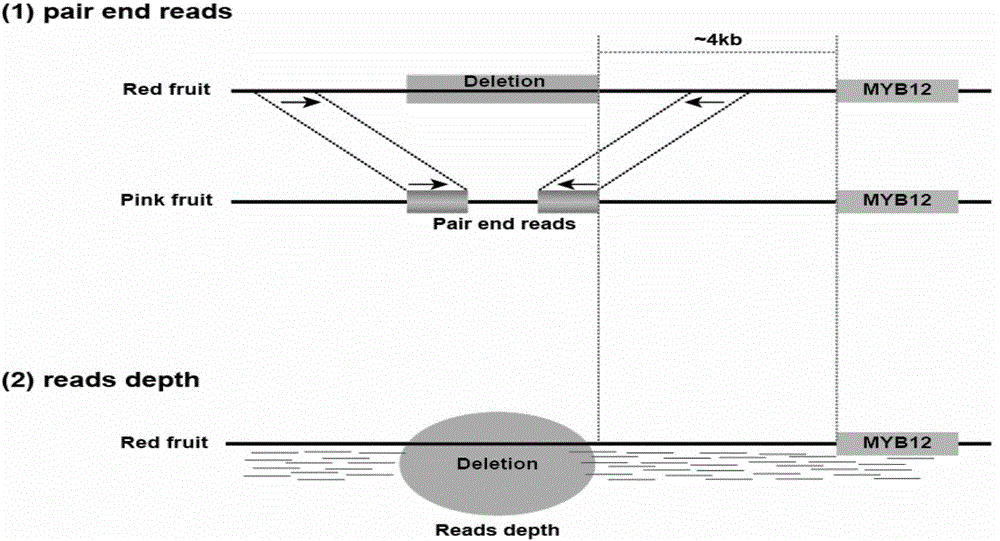
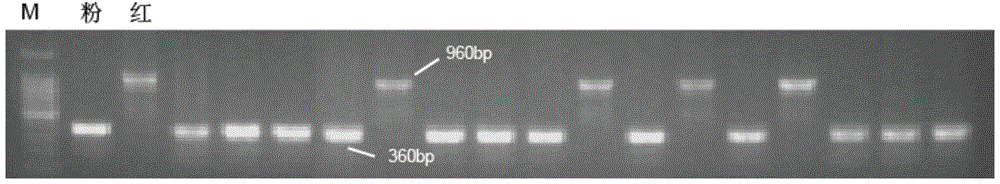
Molecular marker related to color property of tomato fruit and application thereof
ActiveCN104087576ASpeed up the breeding processReliable resultsMicrobiological testing/measurementDNA/RNA fragmentationStart codonAgricultural science
The invention provides a molecular marker related to the color property of tomato fruit and application thereof. The molecular marker includes 2 SNP markers and 2 InDel markers. SNP-p is located at site 8616bp of the upstream of initiator codon of a tomato SlMYB 12 gene, wherein bases which are C represent red tomatoes and bases which are A represent pink tomatoes. SNP-s is located at site 307bp of the downstream of the initiator codon of the tomato SlMYB 12 gene, wherein bases which are T represent pink tomatoes and bases which are C represent red tomatoes. InDel-p is a sequence located at about site 4kp of the upstream of the initiator codon of the tomato SlMYB 12 gene and having a size of 603 bp, wherein pink tomatoes are deleted in the sequence. InDel-i is located at site 248bp of the downstream of the initiator codon of the tomato SlMYB 12 gene, wherein A is located behind the site and is a pink tomato. The molecular marker can be applied to molecular marker-assisted breeding of tomatoes and can substantially accelerate the breeding process of tomatoes.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
SNP (Single Nucleotide Polymorphism) marker related to bitter character of cucumber and application of SNP marker
ActiveCN103013966ASpeed up the breeding processEasy to identifyFungiBacteriaPhenotypic traitBitter taste
The invention provides an SNP (Single Nucleotide Polymorphism) marker related to the bitter character of cucumber and application of the SNP marker. The SNP marker is located at the NO.1178bp of the gene sequence for coding the Csa6G088690 protein of cucumber; cucumber with basic groups G at the NO.1178bp is bitter; and cucumber with basic groups A at the NO.1178bp is not bitter. According to the invention, a genome wide association study (GWAS) method is adopted to screen out the SNP marker related to the bitter taste character of cucumber, a derived cleaved amplified polymorphic sequences (dCAPS) method is adopted to detect the gene types of cucumber to be detected, and bitter or non-bitter cucumber variety breeding is carried out according to the gene types, so as to speed up the breeding process of non-bitter cucumber. The SNP marker is more accurate and reliable in result than the traditional method of subjectively judging whether cucumber is bitter or not, is simpler and more convenient than HPLC (High Performance Liquid Chromatography) for identifying bitter substances, and can be used for large-scale screening of breeding materials. Besides, the invention discovers the committed step for the Csa6G088690 protein of cucumber to control the synthesis pathway of bitter of cucumber for the first time.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
SNP markers related to grass carp growth speed and application thereof
ActiveCN106381331AEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationStart codonNucleotide
Through amplification of a part of fragments of an NPY gene, and through an SNaPshot SNP typing method, provided is SNP markers related to the grass carp growth speed, wherein S1 is located at a 639th locus from an initiation codon (ATG, with an ATG first nucleotide as the first) in an intron 1 sequence of the NPY gene, and a base at the position is T or A; S2 is located at an 892nd locus from the initiation codon in the intron 1 sequence of the NPY gene, and a base at the position is C or A. The two SNP loci can be used for identification or breeding of grass carp with faster growth speed, and lay the foundation for screening and establishment of grass carp fast-growth new varieties.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
Method for extensively screening scallop SNP
InactiveCN101845489AThe principle is safe and reliableReliable Large-Scale Screening RunsMicrobiological testing/measurementMolecular geneticsPhenol
The invention belongs to a method for extensively screening scallop SNP in the technical field of the scallop DNA molecular genetic marker, which comprises the following steps that: solution D and phenol-chloroform are firstly used for extracting a plurality of Chlamys farreri RNA, and a ultraviolet specrophotometer is used for measuring the concentration of the mixture of solution D and the phenol-chloroform which are uniformly mixed; a scallop full-length cDNA sample is re-established and comprises the synthesis of a cDNA first chain and the synthesis and augmentation of a cDNA second chain; then homogenization of full-length cDNA is performed, and ultrasonic breaking is performed; then sequence measuring joints are connected, and terminal repair, joint connection and sample augmentation are included; finally a biological software is used for analyze the data to obtain an SNP marker; then an SNP marker to be screened is selected to design a primer; and DNA of individual scallop is extracted to be equivalently mixed to be used as a template, conventional colony sequence measuring is undertaken after the PCR augmentation, and the position point with the occurring frequency of single base difference being 10 percent or more is defined as an SNP marker. The method has safe and reliable principle, simple and controllable flow procedures, reliable running of large-scale screening, excellent effect and strong practicability.
Owner:OCEAN UNIV OF CHINA
Chinese population linkage analysis single nucleotide polymorphism (SNP) marker sets and use method and application thereof
InactiveCN102121046AEnsure reliabilityEnsure comprehensivenessMicrobiological testing/measurementDNA/RNA fragmentationGenetic linkage disequilibriumInternational HapMap Project
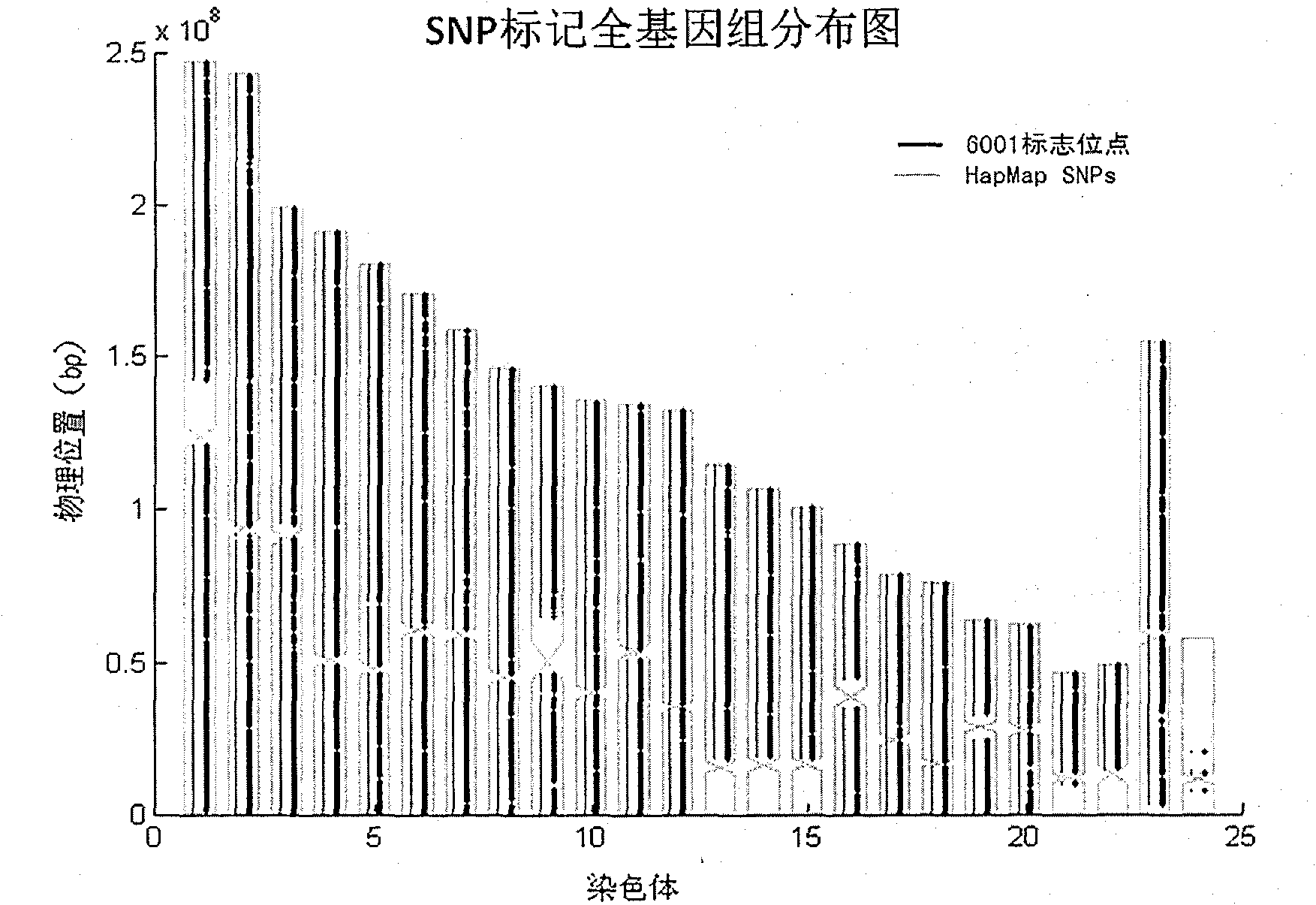
The invention relates to Chinese population linkage analysis single nucleotide polymorphism (SNP) marker sets and a use method and application thereof. On the basis of hundreds of millions of Chinese Han population data results in the mass data of the International HapMap Project, medium-density and high-density SNP marker sets for linkage analysis are constructed and optimized, according to the statistical comparisons of multiple parameters such as the linkage disequilibrium, the polymorphism level, the typing success rate, the distribution position and density of genomes and the functional characteristic, and the multi-level selections and experimental verifications. The two marker sets separately contain 3000 and 6001 loci, wherein the 6001 loci contain the 3000 loci. The SNP sets aim at the Han genetic background in design, have high polymorphism in Chinese and can realize the aim of efficiently marking the Chinese family sample genomes. The selection of polymorphic loci is based on the neutral evolution principle, and all the loci are in a non-gene function region, thus the influence of evolution on the gene function can be avoided. Meanwhile, the characteristics that the marking loci have high typing detectability and can uniformly cover the whole genomes can ensure that the whole genomes can be screened completely and new pathogenic genes can be located and found. The two sets of SNP markers are used to customize probes or chips and perform whole-genome genotyping to family samples; and the typing data are used for linkage analysis, and the haplotyping and fine locating of the linkage candidate region are also adopted, thus the use method has more accurate locating result than the traditional method while the cost is lower and the speed is higher. The distribution and coverage of the 6001 SNP marker set in human chromosomes are shown in the appended drawings.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION +1
SNP (single nucleotide polymorphism) marker for evaluating growth performance of ctenopharyngodonidella, primer and evaluation method
ActiveCN106755527AEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationGenetic exchangeIntein
The invention provides an SNP (single nucleotide polymorphism) marker (comprising SNP1 and SNP4) for evaluating growth performance of ctenopharyngodonidella and a primer pair for amplifying a gene segment where an SNP locus is positioned by virtue of methods of molecular genetics and molecular biology. SNP1 is positioned at the 940th locus in a promoter of a complete sequence of a MyoD (myogenic determining factor) gene of the ctenopharyngodonidella, and a base T is inserted or deficient at the position; SNP4 is positioned in a 107th locus in a first intron of the complete sequence of the MyoD gene of the ctenopharyngodonidella, and a base at the position is A or T. The growth performance of the ctenopharyngodonidella is determined by detecting haplotypes of the two SNP loci. The adopted haplotypes are mutated according to bases generated in the MyoD gene, so that genetic exchange and further phenotype verification are avoided. By virtue of the haplotypes of the SNP marker, rapidly growing ctenopharyngodonidella can be simply and rapidly identified, and rapidly growing ctenopharyngodonidella of a new line can also be guided to be bred.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
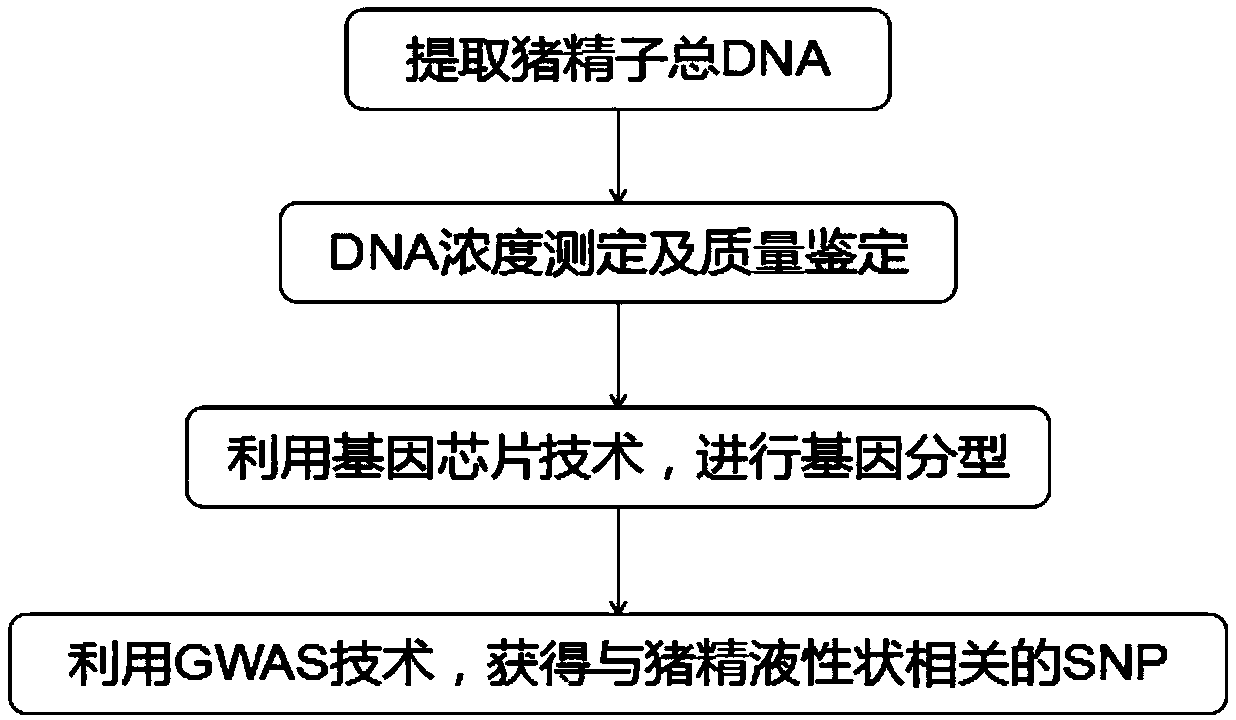
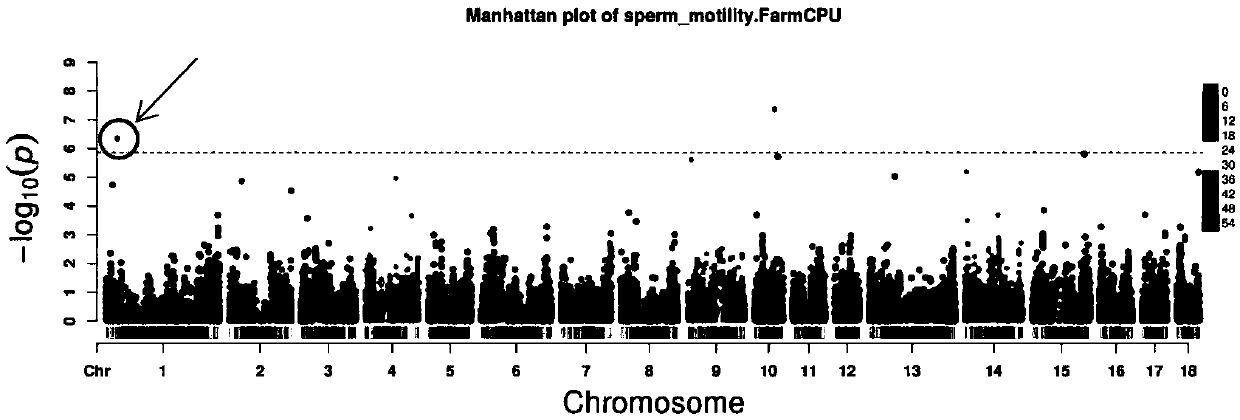
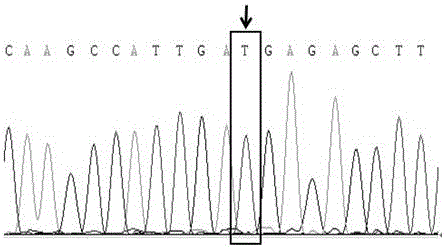
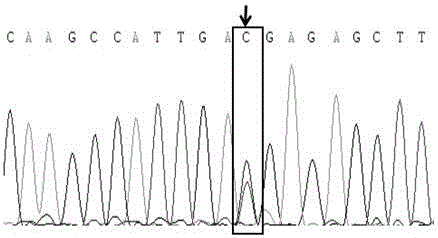
Molecular marker relative to sperm viability and total sperm number of boar and application
ActiveCN109576380AHigh sensitivityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationScreening methodBOAR
The invention belongs to the technical field of screening of molecular markers of boars, and particularly relates to a molecular marker relative to sperm viability and a total sperm number of a boar and application. The molecular marker is cloned from a gene segment with a login number of ALGA 0001958. Through a gene chip technology, a gene is typed, screening is conducted to obtain the molecularmarker relative to characters of the sperm viability and the total sperm number of the boar, a marked nucleotide sequence is as shown in SEQ ID NO:1, A / G allele mutation exists in a base position of asite 101 of the sequence, and when nucleotide of the site 101 on the nucleotide sequence as shown in the SEQ ID NO:1 is A, the fact that the boar has higher sperm viability and a larger total sperm number is judged. The invention discloses a screening method of a molecular marker relative to a boar seminal fluid character and correlation analysis thereof. New SNP marker resources are provided formarker assisted selection of the characters of the sperm viability and the total sperm number of the boars.
Owner:HUAZHONG AGRI UNIV
SNP marker related with low dissolved oxygen survivability of litopenaeus vannamei, screening and applications thereof
InactiveCN105936937ARaise the genetic levelBreeding needs Accurate early selection of vannamei breeding materialMicrobiological testing/measurementDNA/RNA fragmentationSurvivabilityAquatic animal
The invention relates to an SNP marker related with low dissolved oxygen survivability of litopenaeus vannamei, screening and applications thereof, and belongs to the technical field of molecular marker assisted selection of aquatic animals. An SNP molecular marker is cloned from SOD gene. The SNP marker is that the R in the 118-th base from the 5' terminal of the nucleotide sequence represented by SEQ ID No.1 is C or T. The SNP marker is prominently related with the character of tolerating low dissolved oxygen of litopenaeus vannamei, and thus can be applied to the molecular marker assisted selection so as to screen litopenaeus vannamei with strong performance of tolerating low dissolved oxygen. The provided SNP marker can be used to carry out early stage screening according to the actual needs, thus the efficiency and accuracy of breeding are effectively improved, the genetic level of litopenaeus vannamei is improved, species with strong stress resistance can be accurately and efficiently bred, the SNP marker is practical and the application range is wide.
Owner:GUANGDONG OCEAN UNIVERSITY
SNP (single nucleotide polymorphism) marker combination and identification method of small Meishan pig and raw meat product
ActiveCN107699624AEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyRaw meat
The invention provides an SNP (single nucleotide polymorphism) marker combination and identification method of a small Meishan pig and a raw meat product. The method comprises the following steps: extracting a genome DNA of raw pork or a meat product, performing AGE (agarose gel electrophoresis) and Sanger sequencing after performing PCR (polymerase chain reaction) amplification on the genome DNA,and identifying the small Meishan pig and the meat product thereof according to an SNP genotype of a characteristic site of a sequencing result, wherein in the step of Sanger sequencing, peculiar mutation occurs at identified sites: SNP6, SNP12, SNP16, SNP71 and SNP79. According to the method, the problem that an identification method of the small Meishan pig and the meat product thereof does notexist in the prior art is solved.
Owner:SHANGHAI JIAO TONG UNIV
SNP (Single Nucleotide Polymorphism) marker, kit and method for rapidly identifying panax traditional Chinese medicines including panax stipuleanatus and panax quinquefolius
ActiveCN106591459AStrong specificityHigh amplification efficiencyMicrobiological testing/measurementDNA/RNA fragmentationPanax stipuleanatusMicrobiology
Owner:INST OF MEDICINAL PLANT DEV CHINESE ACADEMY OF MEDICAL SCI
Corn whole genome SNP chip and application thereof
ActiveCN108004344AEffective SNP siteMicrobiological testing/measurementDNA/RNA fragmentationLarge-Scale SequencingGermplasm
The invention relates to the field of molecular biology and genome breeding, and particularly discloses a corn whole genome SNP chip and application thereof. SNP loci covering a genetic expression area and SNP loci which are uniformly distributed in the whole genome are collected, after sifting, a high-throughput Infinium SNP chip of Illumina is prepared, and the chip comprises 500 probes of SNP markers shown in a detection list 1. By means of the selected SNP loci based on a large-scale sequencing result, specific signals which have reasonable frequency distribution, a low miss rate and low false positives are obtained, wherein 2 / 3 SNP loci are distributed in all the corn warm zone and tropical zone materials, and can be effectively used for selecting and utilizing the tropical zone materials; 426 SNP loci are functional markers, and are obviously relevant to kernel substance accumulation gene expression; an Infinium platform of Illumina has the advantages of being high in SNP typingdetection rate, high in repetitive rate, simple in detection and high in accuracy. The corn whole genome SNP chip can be used for corn germplasm resource molecular marker fingerprint analysis, corn linkage map construction, QTL locating, corn breeding material background selection and the like.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Method for acquiring chicken whole genome high-density SNP marker sites
ActiveCN105238859ALow costGood repeatabilityMicrobiological testing/measurementGenomic sequencingGenetic engineering
The invention belongs to the technical field of genetic engineering, and provides a method for acquiring chicken whole genome high-density SNP marker sites. The method comprises the following steps: (1) predicting distribution of restriction fragments acquired from double-restriction chicken genomes of EcoRI and MseI; (2) designing a universal joint, a barcode joint and a PCR amplification primer according to the distribution characteristics of the restriction fragments of EcoRI and MseI; (3) constructing a simplified genome sequencing library; (4) sequencing by virtue of the library constructed in the step (3) through a computer; and (5) according to a sequencing result, acquiring SNP marker sites. The method disclosed by the invention provides a universal strategy for the construction of a whole genome high-density SNP map by virtue of double-restriction GBS for different varieties of chickens, so that the cost on acquiring every SNP marker site is reduced by an order of magnitude compared with a conventional chip technology; and the method is stable in technology and high in repeatability.
Owner:CHINA AGRI UNIV
Molecular marker capable of influencing intramuscular fat content of Duroc boar and application
ActiveCN108315433AImprove meat qualityIncrease profitMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyIntramuscular fat
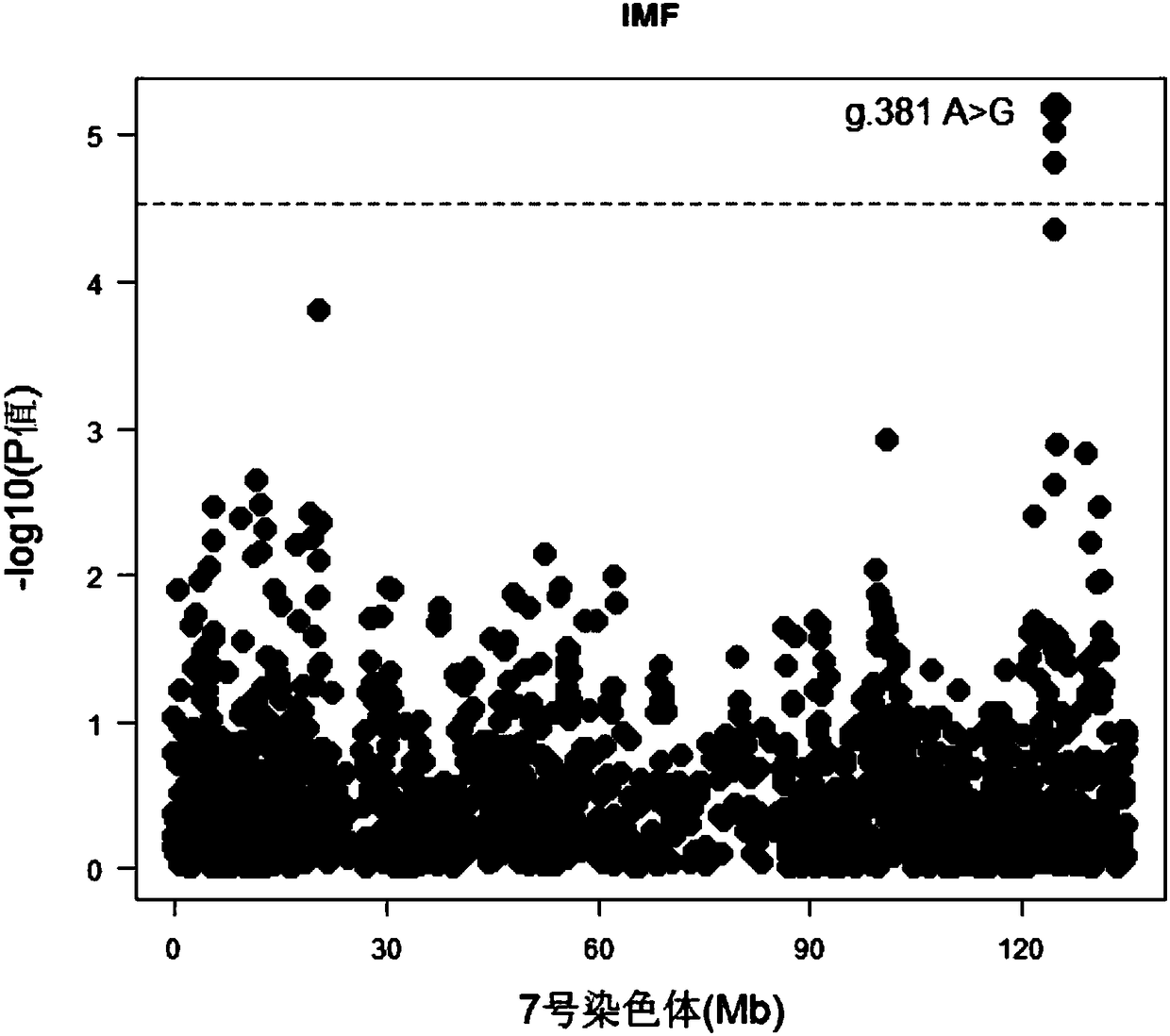
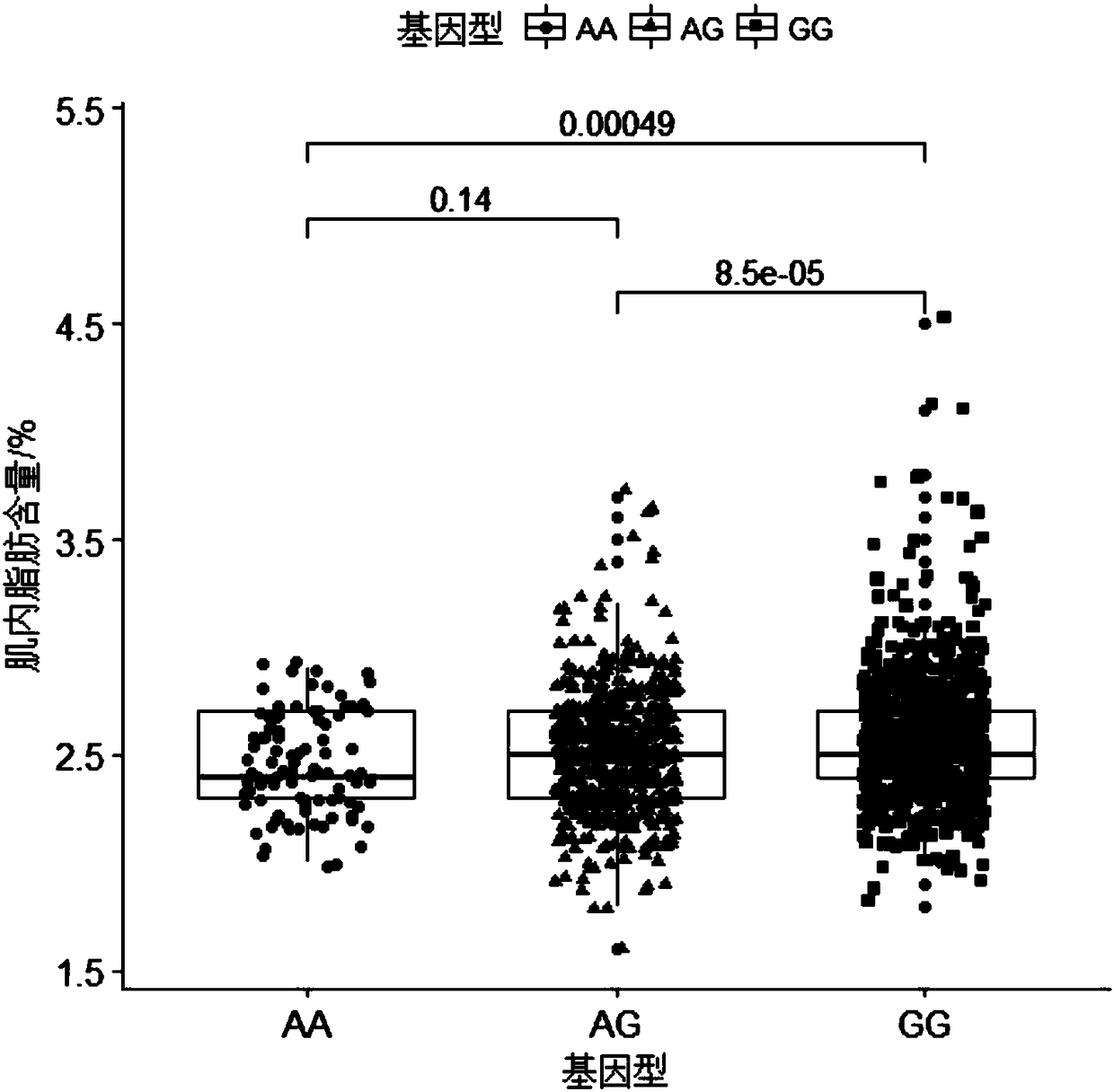
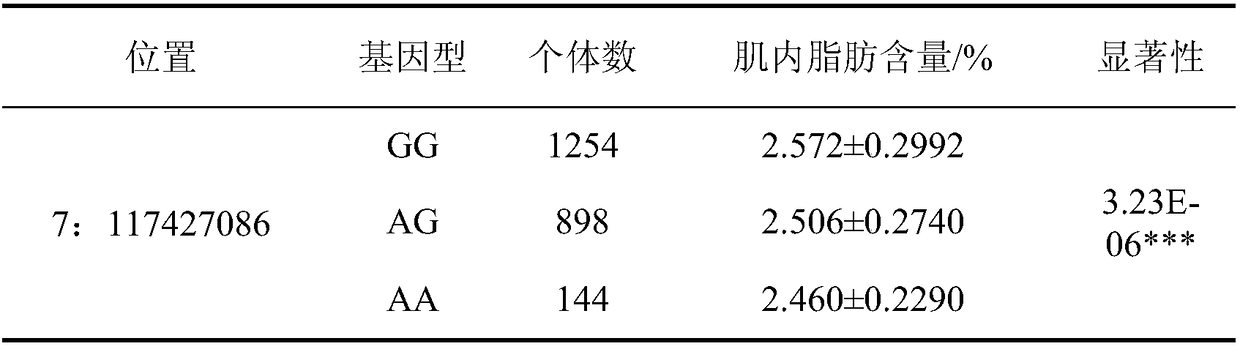
The invention belongs to the fields of molecular biological technology and molecular marker technology and relates to an SNP molecular marker related to intramuscular fat content of a Duroc boar. Thesite of the SNP marker is the 117427086th nucleotide site on the chromosome 7 of the International Pig Reference Genome 11.1 version, and the basic group of the site is G or A. By preferably selectingthe dominant allele of the SNP, the frequency of the dominant allele can be increased generation by generation, the intramuscular fat content of the boar can be increased, and the genetic improvementprogress of the boar can be accelerated, thereby effectively improving the economic benefit of boar breeding.
Owner:SOUTH CHINA AGRI UNIV
SNP site relevant to Canadian Duroc backfat thickness and applications of SNP site
InactiveCN106520943AEasy to breedMicrobiological testing/measurementDNA/RNA fragmentationGenotypeSnp markers
The invention discloses an SNP site relevant to the Canadian Duroc backfat thickness and applications of the SNP site. The SNP is located in the CACNA1E gene on the swine No.9 chromosome, the SNP site of the CACNA1E gene is ALGA0055091 (G / A), the individual with the genotype AA of the site has the backfat thinner than that of the individual with the genotype GG of the site, on the basis, ALGA0055091 can be taken as an SNP site, through the selection for the genotype, the selection for the Canadian Duroc backfat thickness can be accelerated, and meanwhile, the genetic progress for improving the meat factor of the Canadian Duroc is accelerated.
Owner:ZHANGZHOU AONONG MODERN AGRI DEV CO LTD +6
Novel genes and markers in type 2 diabetes and obesity
ActiveUS20070292412A1Reducing and minimizing debilitating effectDiagnosis can be and efficient and safeOrganic active ingredientsNervous disorderDiseaseNovel gene
Genes, SNP markers and haplotypes of susceptibility or predisposition to T2D and subdiagnosis of T2D and related medical conditions are disclosed. Methods for diagnosis, prediction of clinical course and efficacy of treatments for T2D, obesity and related phenotypes using polymorphisms in the risk genes are also disclosed. The genes, gene products and agents of the invention are also useful for monitoring the effectiveness of prevention and treatment of T2D and related traits. Kits are also provided for the diagnosis, selecting treatment and assessing prognosis of T2D. Novel methods for prevention and treatment of metabolic diseases such as T2D based on the disclosed T2D genes, polypeptides and related pathways are also disclosed.
Owner:DSM IP ASSETS BV
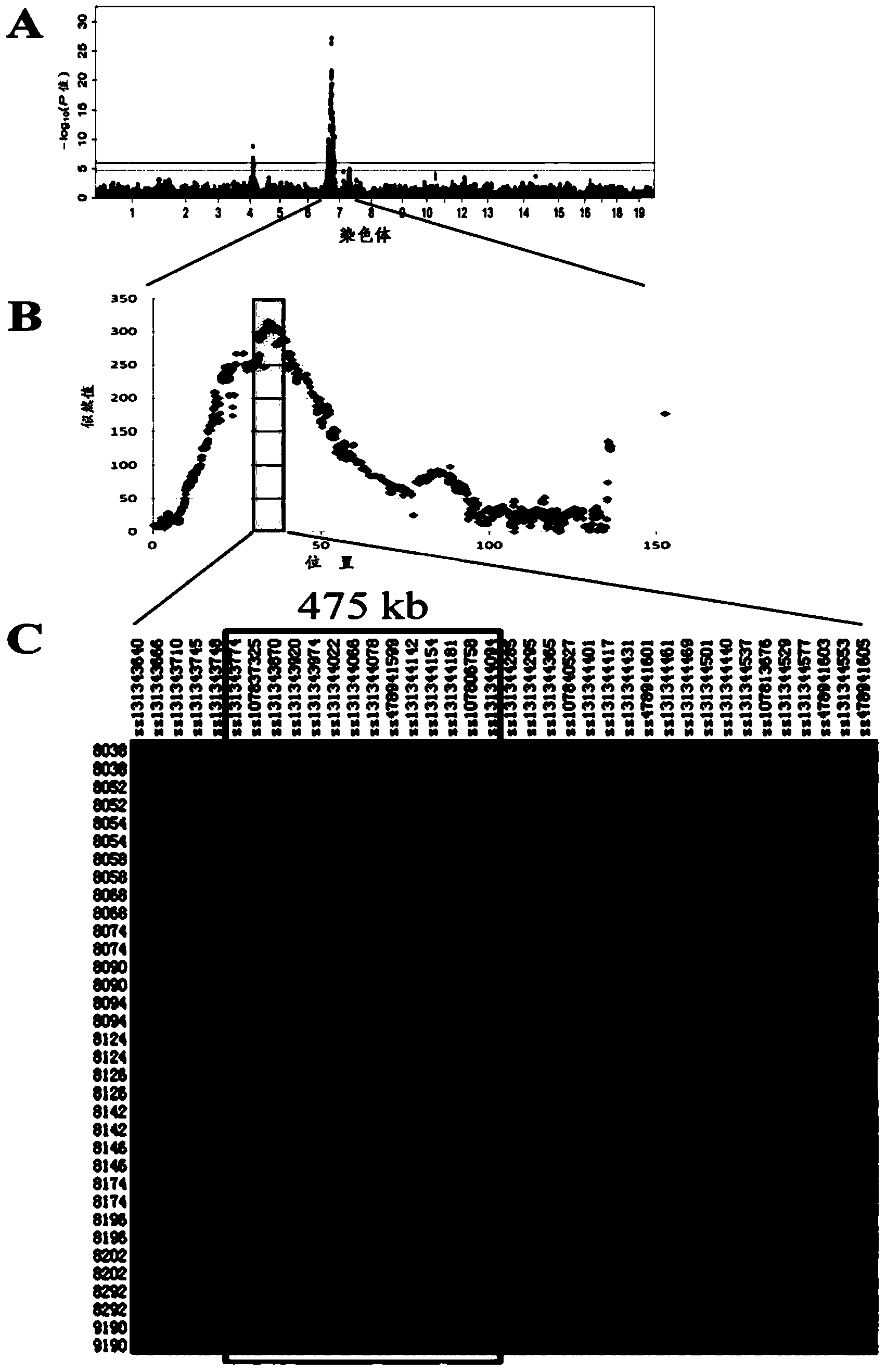
SNP (simple nucleotide polymorphism) modular marker for rice low cadmium accumulation gene OsHMA3 and application thereof
ActiveCN106480228AAccurate measurementOvercome expensiveMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCadmium Cation
The invention relates to an SNP (simple nucleotide polymorphism) modular marker for rice low cadmium accumulation gene OsHMA3; a detection site of the marker is at base 7431781 of chromosome 7 of rice. The invention also provides a primer set and method for detecting the SNP marker; the primer set has a sequence shown as in SEQ ID NO. 2-4. The SNP molecular marker and the primer set and method have the advantages that the SNP molecular marker is suitable for predicting the cadmium content of rice kernels before rice not bearing fruits, and suitable for accurate screening, such that breeding of low-cadmium-content varieties of rice is significantly promoted; the detection method is accurate and reliable and is simple to perform; the detection of SNP sites for rice OsHMA3 gene provides scientific basis for the breeding or improvement of low-cadmium-content kernel varieties of rice.
Owner:HUAZHI RICE BIO TECH CO LTD
SNP (single nucleotide polymorphism) marker and method for identifying MC1R (melanocortin receptor 1) genes of pig breeds and colors of pig breeds as well as application
PendingCN105648071AThe screening method is accurateEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationUnknown SourceScreening method
The invention discloses an SNP (single nucleotide polymorphism) marker for identifying MC1R (melanocortin receptor 1) genes of pig breeds and colors of the pig breeds and further discloses an SNP marker primer for identifying the MC1R genes of the pig breeds and the colors of the pig breeds as well as an SNP marker method for identifying the MC1R genes of the pig breeds and the colors of the pig breeds. The molecular genetic marker is not limited by ages, sexes and the like of pigs, can be applied to early breeding of pure breeds of local black hogs and can further be used for performing pig breed and color identification and genetic relationship deduction on pork samples from unknown sources. A screening method is accurate, operation is simple, the cost is low, and the efficiency is high.
Owner:KUNMING UNIV +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com