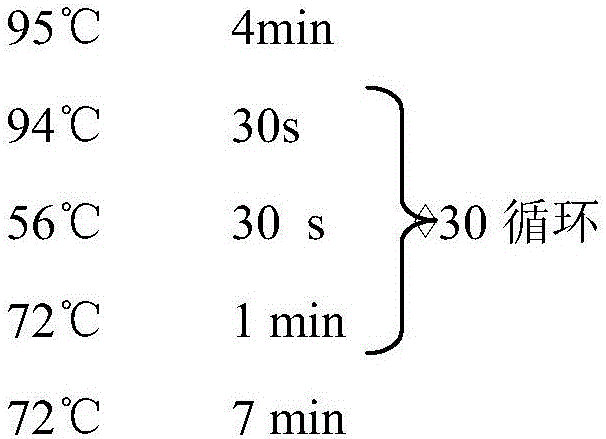SNP (single nucleotide polymorphism) marker for evaluating growth performance of ctenopharyngodonidella, primer and evaluation method
A growth performance, grass carp technology, applied in the field of grass carp myogenic determinant sequence-related SNP marker screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] 1. Obtaining the complete sequence of grass carp MyodD gene
[0049] According to the full-length cDNA sequence of the grass carp MyoD gene (GenBank Accession No. JQ793893.1), the Primer Premier 5.0 software was used to design specific primers to amplify the gene intron and promoter.
[0050] Cloning of MyoD gene intron:
[0051] Design specific primers iMyOD-F: 5′-CACATAAAGATGGAGTTGTCGGA-3′, iMyOD-R: 5′-ATGCTTCTTGATTCAAATTGTCC-3′, respectively use the extracted grass carp genomic DNA and muscle cDNA as templates for PCR amplification, and the reaction system is 50 μL: PCR SuperMix High Fidelity (Invitrogen, catalog number: 12532-016) 45 μL, template 2 μL, upstream and downstream primers 1 μL, sterilized deionized water 1 μL. PCR amplification parameters: 94°C for 2min; 94°C for 30s, 62°C for 30s, 68°C for 90min, a total of 35 cycles; extension at 72°C for 10min to terminate. The amplified product was detected by electrophoresis in 1.0% agarose gel, and the target fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com