SNP (Single Nucleotide Polymorphism) marker, kit and method for rapidly identifying panax traditional Chinese medicines including panax stipuleanatus and panax quinquefolius
A kit and ginseng technology are applied in the field of SNP labeling and kits to achieve the effects of rapid identification, high specificity and high amplification efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1: identification method of Pingbian Radix Notoginseng raw material
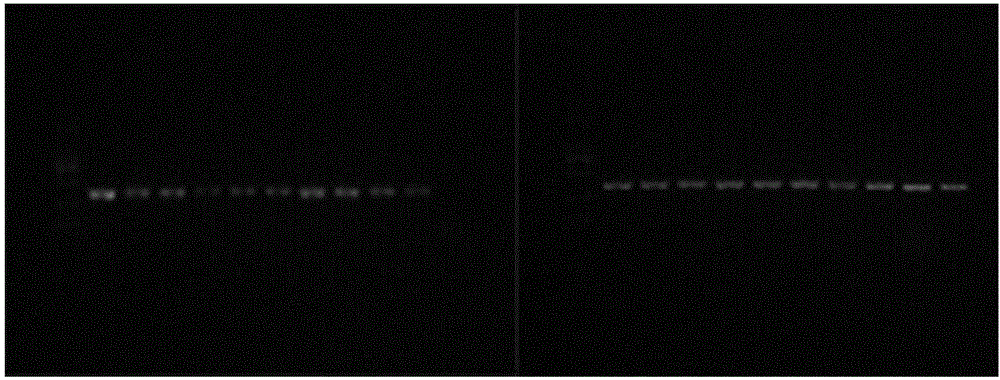
[0033] The samples of Panax genus were collected from different producing areas, and the specific medicinal materials and collection locations represented by each sample are shown in Table 1. 25mg samples were taken and ground on MM400 ball mill (Retsch, Germany), and the total DNA was extracted with the plant genomic DNA kit of Tiangen Biochemical Technology (Beijing) Co., Ltd.
[0034] Table 1: Panax samples
[0035] serial number Sample serial number herbs sampling location 1 RS1-6 ginseng Fusong County, Jilin Province 2 BB1-6 Bamboo ginseng Hubei Enshi 3 R1-3 Bamboo ginseng Japan 4 ZZ1-6 Ginseng Gansu 5 XZ1-6 Ginseng Nyingchi, Tibet 6 JZ1-5 Ginger notoginseng Myanmar 7 XY1-2 Panax ginseng Mount Emei, Sichuan 8 PB1-13 Panax notoginseng Yunnan
[0036] PCR amplification
[0037] The primer...
Embodiment 2
[0050] Embodiment 2: identification method of Pingbian Sanqi decoction pieces
[0051] Compared with Example 1, the only difference is that the Pingbian notoginseng decoction pieces are used as samples, DNA is extracted and identified, and the above two SNP sites can also be specifically detected. During the sequence comparison, it was found that the 88th base from the 5' end of the sequence of SEQ ID NO.1 of Pingbian Sanqi Decoction Pieces is A, and the 117th base is C.
Embodiment 3
[0052] Embodiment 3: Identification of Pingbian Panax notoginseng powder
[0053] Compared with Example 1, the only difference is that the Pingbian notoginseng powder is used as a sample, DNA is extracted and identified, and the above two SNP sites can also be specifically detected as a result. During the sequence comparison, it was found that the 88th base from the 5' end of the sequence of SEQ ID NO.1 of Pingbian Sanqi Decoction Pieces is A, and the 117th base is C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com