Corn whole genome SNP chip and application thereof
A whole genome, chip technology, applied in the fields of molecular biology and genome breeding, can solve the problems of limited application scope, high cost and difficult to meet the needs of large-scale breeding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1 Application of corn SNP chips in molecular marker fingerprint analysis of corn germplasm resources
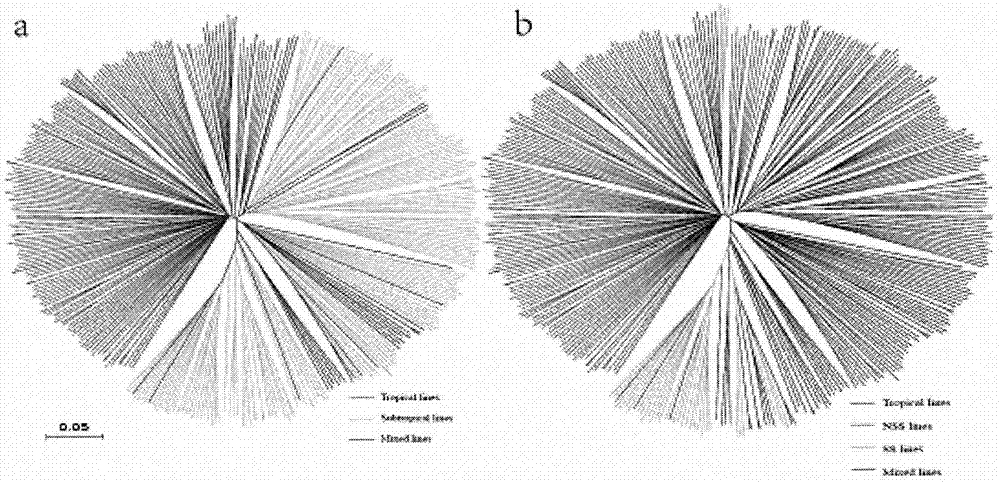
[0093] Using the corn SNP chip, the present invention conducts molecular marker fingerprint analysis on 368 corn inbred lines from a wide range of sources. Microarray detected 368 maize inbred lines. By calculating the deletion rate and MAF of each SNP marker, the sites with a deletion rate greater than 5% and a MAF less than 20% were deleted, leaving 3987 high-quality sites, accounting for 66.5% of the total number of sites ( 3987 / 6000). According to the genotyping results, the 368 maize inbred lines were clustered and analyzed, and it was found that all 150 tropical materials were clustered into one group, and the remaining 126 temperate materials were clustered into one group ( figure 2 a). The temperate materials were further clustered, and it was found that all 32 materials were clustered into the SS (Stiff Stalk) series, and the remaining 94 materials ...
Embodiment 2
[0095] Example 2 Application of Maize SNP Chip in Linkage Map Construction and QTL Mapping
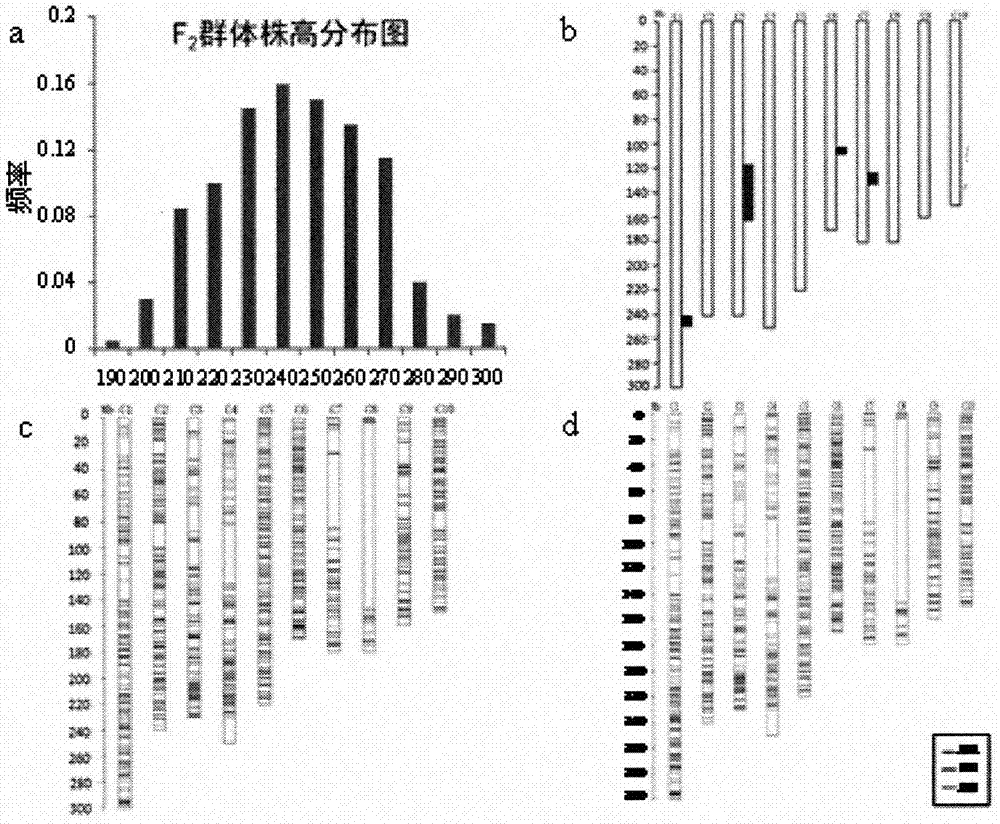
[0096] In this study, the inbred lines Huangzaosi (HZS) and 1462 were used as parents to construct the F2 population, and the F2 population and its parents were genotyped using the maize SNP chip. After analysis, there are 1248 homozygous SNPs that are different and homozygous between the parents Huangzaosi (HZS) and 1462, 220 SNPs in the F2 population, and 1028 SNPs that were finally used for linkage map construction and QTL mapping. Accounting for 19.8% of effective loci (1028 / 5179). The total length of the constructed linkage map is 1596.94cM, including a total of 655 bins (bins are recombination units), and the average length of each bin is 2.4cM. Due to severe partial segregation on the 8th chromosome, most of the segment of the 8th chromosome is blank. The results show that the SNP typing of the population can be effectively carried out by the corn SNP chip.
[0097] The two p...
Embodiment 3
[0100] Example 3 Application of Corn SNP Chip in Mixed Pond Location
[0101] The object of this study is a maize mutant barren stalk with degenerated male and female spikes. The mutant shows that both male and female spikes are degenerated and do not differentiate into spikelets or florets. The mutant material was derived from a natural mutation of an inbred line, and the phenotype in the self-bred offspring showed a 3:1 segregation. The mutant material was used to construct a backcross population with maize inbred lines B73, Mo17, and Zheng58 to obtain a BC3F1 population. Mutants and wild-types in the population were pooled for every 10 individual plants, repeated 5 times, and the whole genome DNA was purified and quantified for microarray genotyping. According to the analysis of "Principle of Consistency between Phenotype and Genotyping", in theory, the marker closely linked to the target gene in the mutant should be a homozygous mutation site, while the corresponding site ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com